Best Matches in Database for Each Motif (Highest to Lowest)
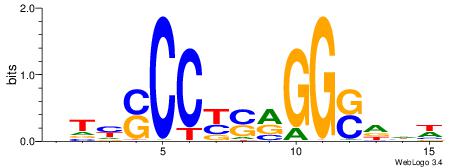
| Dataset #: 1 | Motif ID: 2 | Motif name: Motif 2 |
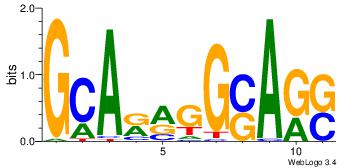
| Original motif | Reverse complement motif |
| Consensus sequence: GCARRGGSAGS | Consensus sequence: SCTSCCKMTGC |
 |
 |
Best Matches for Motif ID 2 (Highest to Lowest)
| Motif ID: | UP00153 |
| Motif name: | Pitx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.0454693 |
Alignment:
HHDRTTAATCCCKHBVH
------SCTSCCKMTGC
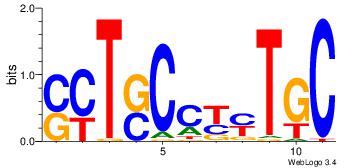
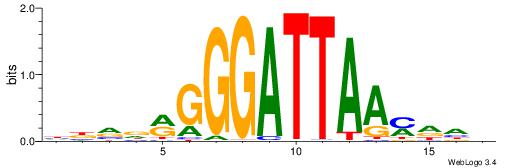
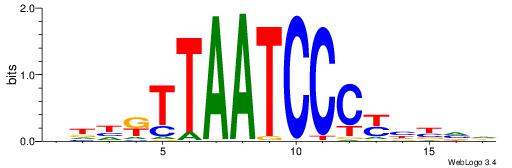
| Original motif | Reverse complement motif |
| Consensus sequence: HBVDRGGGATTAAMDHH | Consensus sequence: HHDRTTAATCCCKHBVH |
 |
 |
| Motif ID: | UP00085 |
| Motif name: | Sfpi1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0458219 |
Alignment:
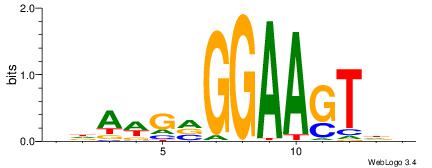
BDAWGVGGAAGTDH
GCARRGGSAGS---
| Original motif | Reverse complement motif |
| Consensus sequence: BDAWGVGGAAGTDH | Consensus sequence: HDACTTCCBCWTDV |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 11 |
| Similarity score: | 0.0463674 |
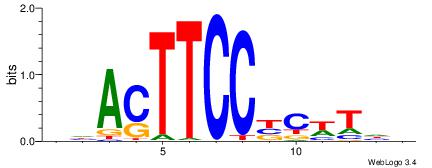
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
-------SCTSCCKMTGC----
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0465857 |
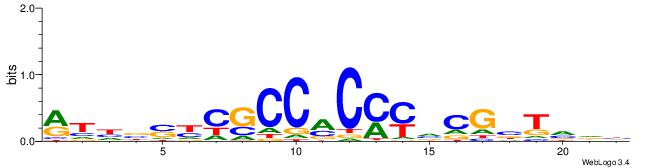
Alignment:
BADHBDHCGCCCMCGCAHHDBBV
-----SCTSCCKMTGC-------
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
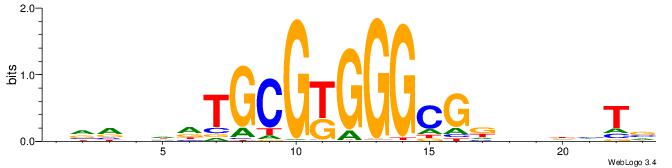 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0473355 |
Alignment:
BBYVVCAGCTGCBVHHD
-SCTSCCKMTGC-----
| Original motif | Reverse complement motif |
| Consensus sequence: BBYVVCAGCTGCBVHHD | Consensus sequence: HHDVVGCAGCTGVBKVB |
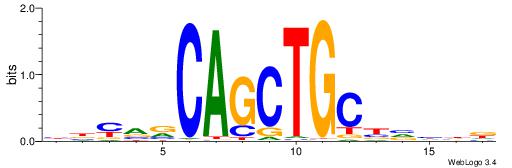 |
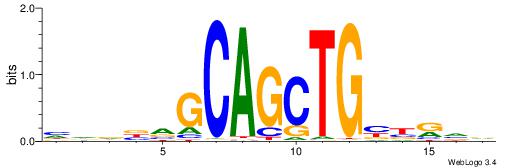 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 11 |
| Number of overlap: | 11 |
| Similarity score: | 0.0487126 |
Alignment:
VHYTCCDWCCGCGTCMMTHWHV
-SCTSCCKMTGC----------
| Original motif | Reverse complement motif |
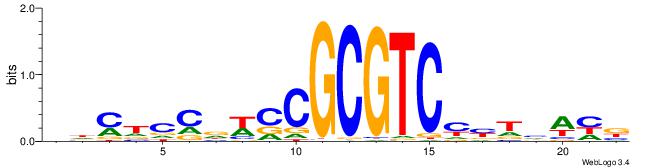
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
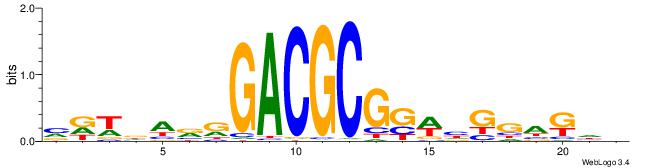 |
 |
| Motif ID: | UP00010 |
| Motif name: | Tcfap2b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0497623 |
Alignment:
HHKGCCYSRGGCWAD
----GCARRGGSAGS
| Original motif | Reverse complement motif |
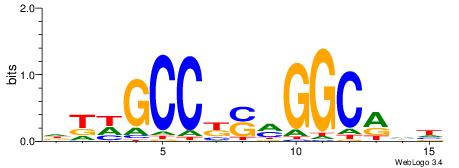
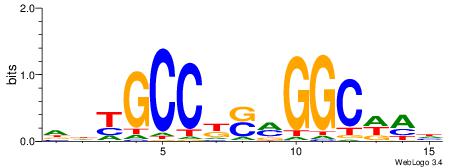
| Consensus sequence: DTWGCCKSMGGCRHH | Consensus sequence: HHKGCCYSRGGCWAD |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0507547 |
Alignment:
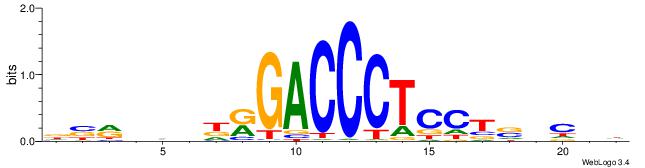
DVMHHDHKGACCCTCCTSVCBH
-----------SCTSCCKMTGC
| Original motif | Reverse complement motif |
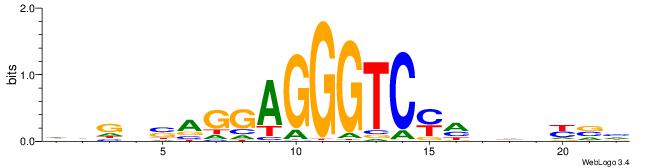
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Motif ID: | UP00055 |
| Motif name: | Hbp1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0508648 |
Alignment:
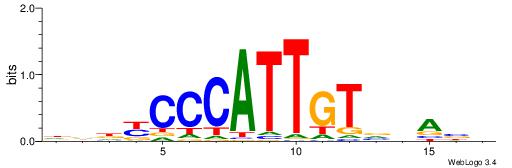
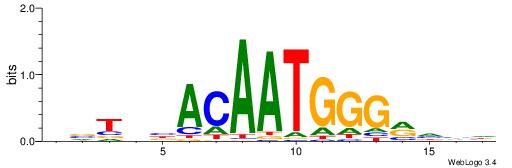
HBTHVACAATGGGMDHD
-----GCARRGGSAGS-
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCCATTGTVHABH | Consensus sequence: HBTHVACAATGGGMDHD |
 |
 |
| Motif ID: | UP00005 |
| Motif name: | Tcfap2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0512938 |
Alignment:
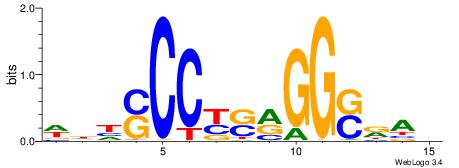
WDHSCCYSRGGSRAD
----GCARRGGSAGS
| Original motif | Reverse complement motif |
| Consensus sequence: WDHSCCYSRGGSRAD | Consensus sequence: DTMSCCKSMGGSHDW |
 |
 |
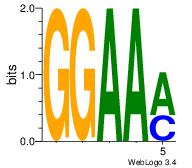
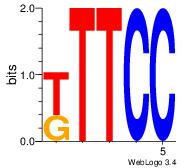
| Dataset #: 1 | Motif ID: 4 | Motif name: Motif 4 |
| Original motif | Reverse complement motif |
| Consensus sequence: GGAAM | Consensus sequence: YTTCC |
 |
 |
Best Matches for Motif ID 4 (Highest to Lowest)
| Motif ID: | UP00424 |
| Motif name: | Gm4881 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0 |
Alignment:
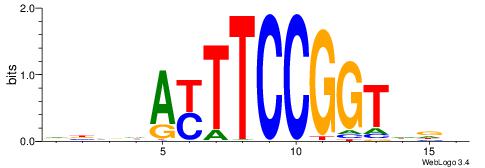
VBBHAYTTCCGGTDDD
-----YTTCC------
| Original motif | Reverse complement motif |
| Consensus sequence: VBBHAYTTCCGGTDDD | Consensus sequence: DHDACCGGAAMTHBVB |
 |
 |
| Motif ID: | UP00416 |
| Motif name: | Fli1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 5 |
| Similarity score: | 0.00163794 |
Alignment:
VBBVAYTTCCGGTDDH
-----YTTCC------
| Original motif | Reverse complement motif |
| Consensus sequence: VBBVAYTTCCGGTDDH | Consensus sequence: DHDACCGGAAKTBBVV |
 |
 |
| Motif ID: | UP00422 |
| Motif name: | Etv3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.00234412 |
Alignment:
VBBHAYTTCCGGTHDH
-----YTTCC------
| Original motif | Reverse complement motif |
| Consensus sequence: VBBHAYTTCCGGTHDH | Consensus sequence: DHHACCGGAAKTHBBB |
 |
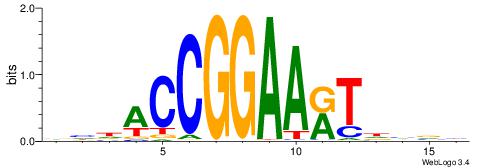 |
| Motif ID: | UP00411 |
| Motif name: | Erg |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.00589761 |
Alignment:
VBBVAYTTCCGGTDDB
-----YTTCC------
| Original motif | Reverse complement motif |
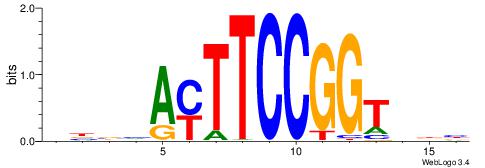
| Consensus sequence: VBBVAYTTCCGGTDDB | Consensus sequence: BDDACCGGAAKTVBVB |
 |
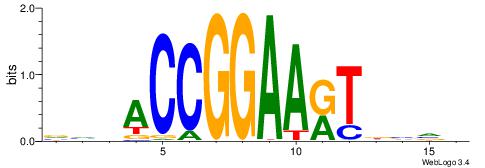 |
| Motif ID: | UP00418 |
| Motif name: | Etv6 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 5 |
| Similarity score: | 0.00691345 |
Alignment:
DVHBTACTTCCGGHTHB
------YTTCC------
| Original motif | Reverse complement motif |
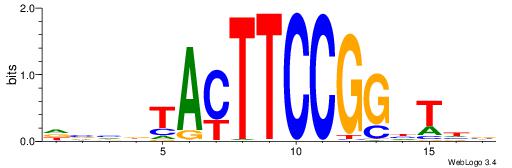
| Consensus sequence: DVHBTACTTCCGGHTHB | Consensus sequence: BHAHCCGGAAGTABDVD |
 |
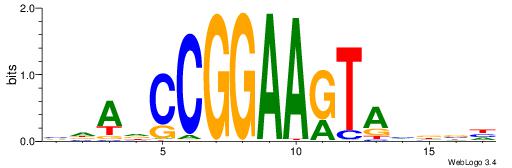 |
| Motif ID: | UP00415 |
| Motif name: | Elk4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 5 |
| Similarity score: | 0.00722176 |
Alignment:
VBBHRYTTCCGGTDDH
-----YTTCC------
| Original motif | Reverse complement motif |
| Consensus sequence: VBBHRYTTCCGGTDDH | Consensus sequence: DHDACCGGAAKKHBVB |
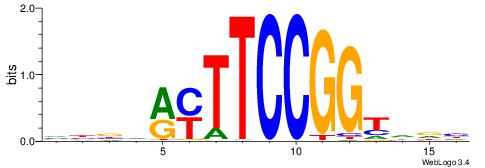 |
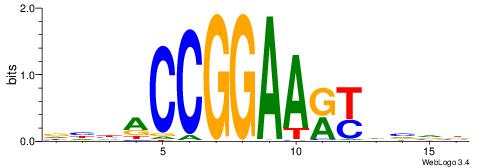 |
| Motif ID: | UP00413 |
| Motif name: | Elf4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 5 |
| Similarity score: | 0.0153114 |
Alignment:
DBBHACTTCCGGKWTH
-----YTTCC------
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGKWTH | Consensus sequence: HAWYCCGGAAGTHBBD |
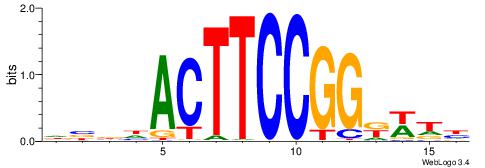 |
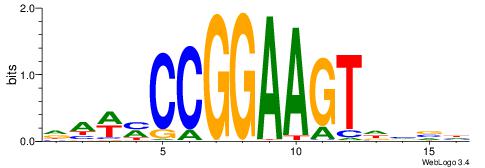 |
| Motif ID: | UP00404 |
| Motif name: | Elf2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 5 |
| Similarity score: | 0.0153458 |
Alignment:
DBBHACTTCCGGDWDB
-----YTTCC------
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGDWDB | Consensus sequence: BDWDCCGGAAGTHBBD |
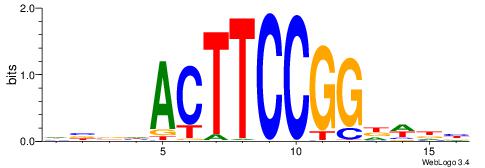 |
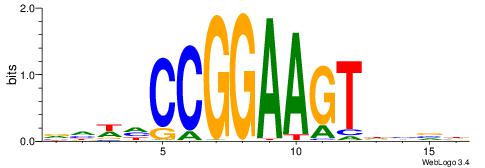 |
| Motif ID: | UP00417 |
| Motif name: | Etv4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.0219684 |
Alignment:
VBBHAYWTCCGGTDDB
-----YTTCC------
| Original motif | Reverse complement motif |
| Consensus sequence: VBBHAYWTCCGGTDDB | Consensus sequence: BHHACCGGAWKTHBVV |
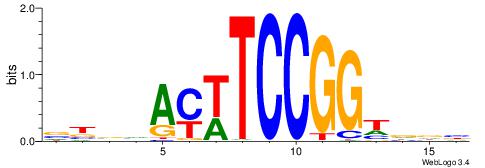 |
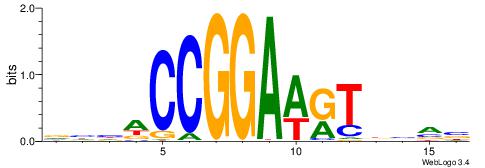 |
| Motif ID: | UP00419 |
| Motif name: | Spic |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 5 |
| Similarity score: | 0.0224017 |
Alignment:
DWACTTCCRCTTTB
---YTTCC------
| Original motif | Reverse complement motif |
| Consensus sequence: VAAAGMGGAAGTWD | Consensus sequence: DWACTTCCRCTTTB |
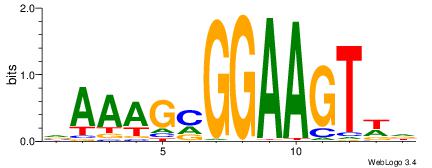 |
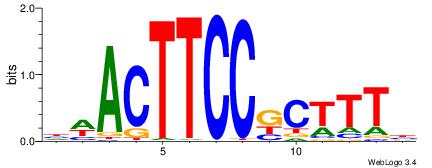 |
| Dataset #: 2 | Motif ID: 7 | Motif name: Motif 7 |
| Original motif | Reverse complement motif |
| Consensus sequence: HCAGRRVACASAG | Consensus sequence: CTSTGTBKMCTGH |
 |
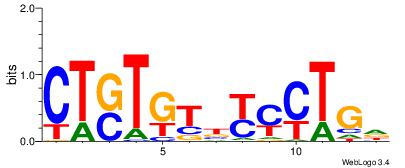 |
Best Matches for Motif ID 7 (Highest to Lowest)
| Motif ID: | UP00028 |
| Motif name: | Tcfap2e_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0445121 |
Alignment:
HDCDBRAAAAAADD
-HCAGRRVACASAG
| Original motif | Reverse complement motif |
| Consensus sequence: HDCDBRAAAAAADD | Consensus sequence: DDTTTTTTMBDGDH |
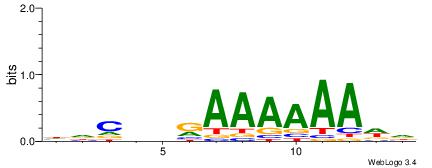 |
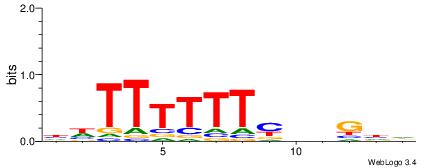 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 10 |
| Number of overlap: | 13 |
| Similarity score: | 0.0448793 |
Alignment:
GTVBDAATTGTVTGHGDDHKVB
---------CTSTGTBKMCTGH
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
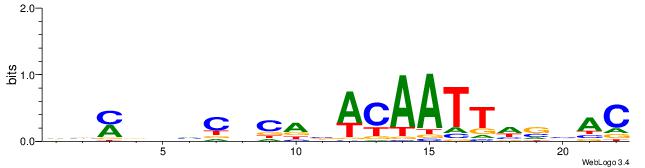 |
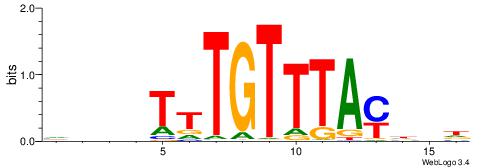
| Motif ID: | UP00041 |
| Motif name: | Foxj1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0454552 |
Alignment:
DDDHTTTGTTTAMHDH
---CTSTGTBKMCTGH
| Original motif | Reverse complement motif |
| Consensus sequence: HDHRTAAACAAAHDDD | Consensus sequence: DDDHTTTGTTTAMHDH |
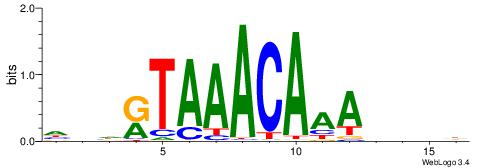 |
 |
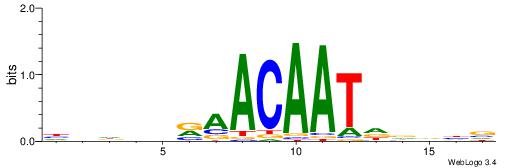
| Motif ID: | UP00016 |
| Motif name: | Sry_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0481412 |
Alignment:
HHDHDRAACAATDDVHV
HCAGRRVACASAG----
| Original motif | Reverse complement motif |
| Consensus sequence: HHDHDRAACAATDDVHV | Consensus sequence: VHVHDATTGTTMHDDDH |
 |
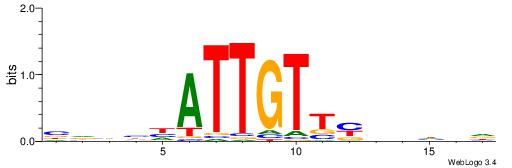 |
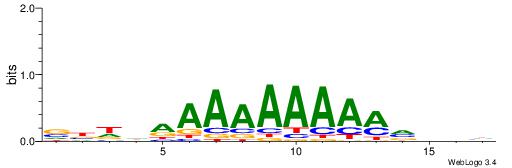
| Motif ID: | UP00077 |
| Motif name: | Srf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0545707 |
Alignment:
VBDDAAAAAAAAMVBHH
-HCAGRRVACASAG---
| Original motif | Reverse complement motif |
| Consensus sequence: VBDDAAAAAAAAMVBHH | Consensus sequence: HHVBYTTTTTTTTDDVV |
 |
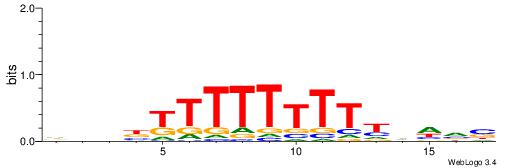 |
| Motif ID: | UP00058 |
| Motif name: | Tcf3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0553073 |
Alignment:
DVBSBDAHAHAHAHH
-HCAGRRVACASAG-
| Original motif | Reverse complement motif |
| Consensus sequence: DVBSBDAHAHAHAHH | Consensus sequence: HHTHTHTHTDBSBVD |
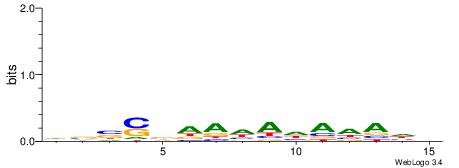 |
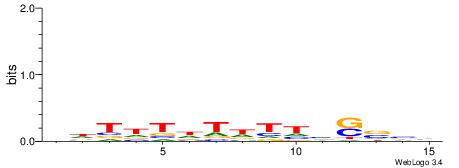 |
| Motif ID: | UP00091 |
| Motif name: | Sox5_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0562585 |
Alignment:
DHDCATMATTDASDD
--HCAGRRVACASAG
| Original motif | Reverse complement motif |
| Consensus sequence: DHDCATMATTDASDD | Consensus sequence: DHSTDAATYATGDHD |
 |
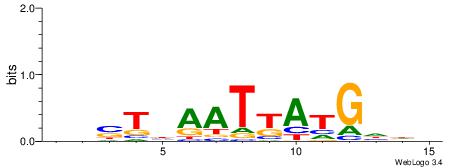 |
| Motif ID: | UP00012 |
| Motif name: | Bbx_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 13 |
| Similarity score: | 0.0564984 |
Alignment:
VHVHBTGTTAACVHWVH
--CTSTGTBKMCTGH--
| Original motif | Reverse complement motif |
| Consensus sequence: HVWHBGTTAACABHBDV | Consensus sequence: VHVHBTGTTAACVHWVH |
 |
 |
| Motif ID: | UP00073 |
| Motif name: | Foxa2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 13 |
| Similarity score: | 0.0571073 |
Alignment:
VVDAATAACAAAVVB
HCAGRRVACASAG--
| Original motif | Reverse complement motif |
| Consensus sequence: VVDAATAACAAAVVB | Consensus sequence: BVVTTTGTTATTDBB |
 |
 |
| Motif ID: | UP00054 |
| Motif name: | Tcf7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 13 |
| Similarity score: | 0.0574997 |
Alignment:
HHDTTTVTAATAYBD
CTSTGTBKMCTGH--
| Original motif | Reverse complement motif |
| Consensus sequence: HBKTATTABAAAHHH | Consensus sequence: HHDTTTVTAATAYBD |
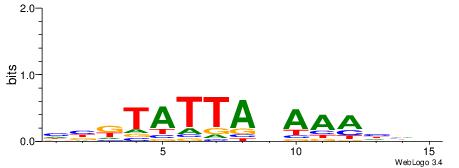 |
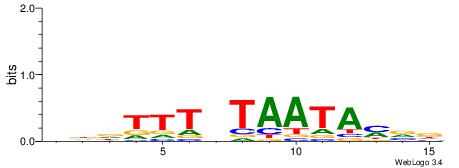 |
| Dataset #: 2 | Motif ID: 41 | Motif name: Motif 41 |
| Original motif | Reverse complement motif |
| Consensus sequence: ACTTTGG | Consensus sequence: CCAAAGT |
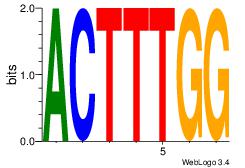 |
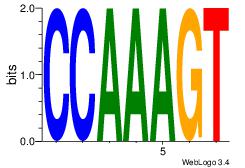 |
Best Matches for Motif ID 41 (Highest to Lowest)
| Motif ID: | UP00066 |
| Motif name: | Hnf4a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 7 |
| Similarity score: | 0 |
Alignment:
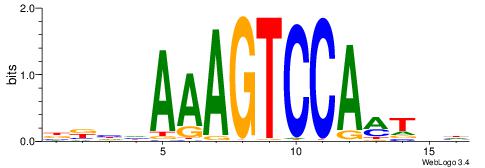
BBHHAAAGTCCAMTHH
--CCAAAGT-------
| Original motif | Reverse complement motif |
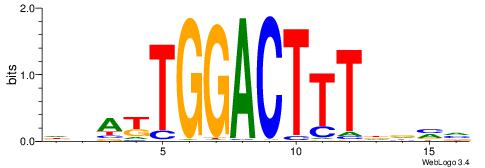
| Consensus sequence: BBHHAAAGTCCAMTHH | Consensus sequence: HHAYTGGACTTTHDBV |
 |
 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 7 |
| Similarity score: | 0.0128381 |
Alignment:
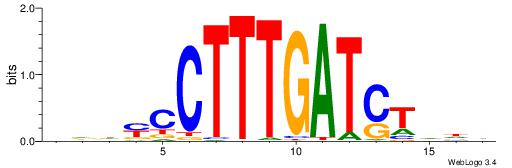
VDDAGATCAAAGGKDDD
------CCAAAGT----
| Original motif | Reverse complement motif |
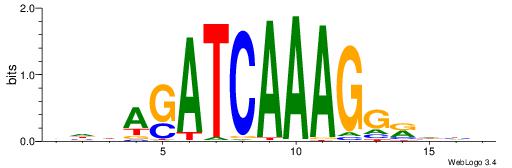
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 7 |
| Similarity score: | 0.0140846 |
Alignment:
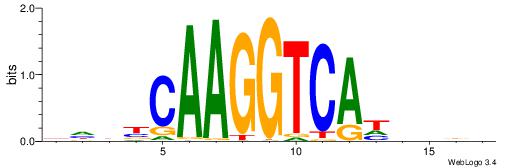
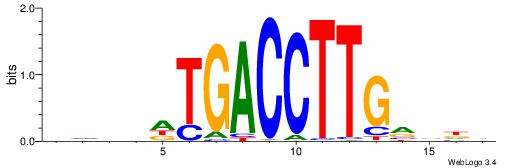
DHDBCAAGGTCAHVBDH
---CCAAAGT-------
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBCAAGGTCAHVBDH | Consensus sequence: HHBVHTGACCTTGVDHD |
 |
 |
| Motif ID: | UP00083 |
| Motif name: | Tcf7l2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0.0141156 |
Alignment:
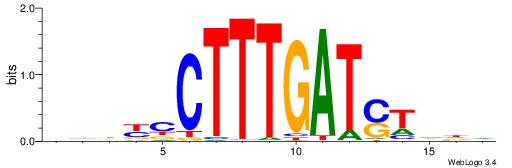
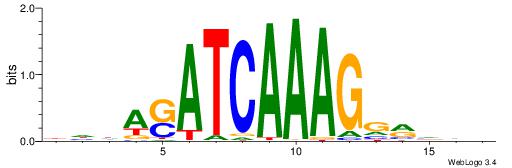
BDDASATCAAAGGMDDD
------CCAAAGT----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATSTDDV | Consensus sequence: BDDASATCAAAGGMDDD |
 |
 |
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0.0159798 |
Alignment:
HHBBMCTTTGTTHDDBH
----ACTTTGG------
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
 |
 |
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0.0181605 |
Alignment:
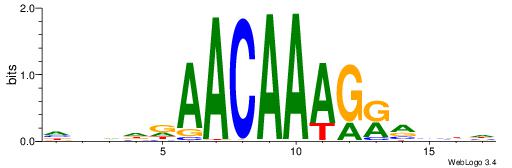
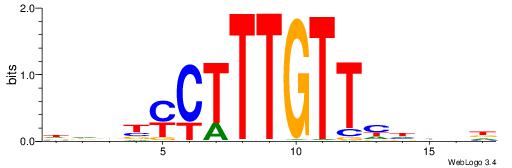
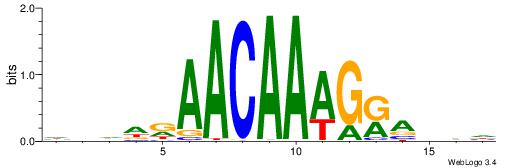
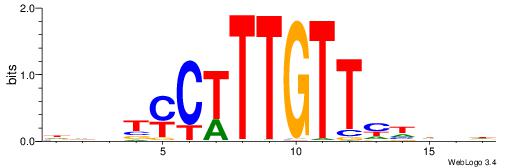
HHBTMCTTTGTTMDDVH
----ACTTTGG------
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
 |
 |
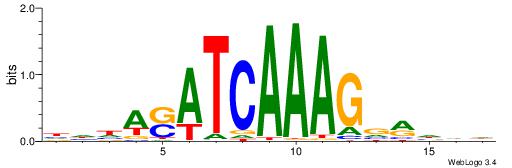
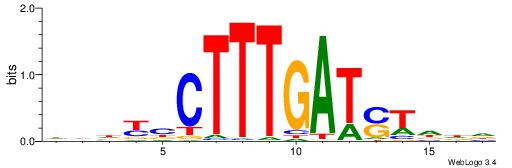
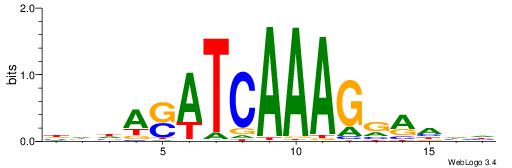
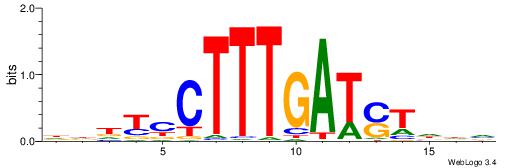
| Motif ID: | UP00058 |
| Motif name: | Tcf3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0.0200761 |
Alignment:
HHHKVCTTTGATSTHHH
----ACTTTGG------
| Original motif | Reverse complement motif |
| Consensus sequence: HHHASATCAAAGVRHHH | Consensus sequence: HHHKVCTTTGATSTHHH |
 |
 |
| Motif ID: | UP00054 |
| Motif name: | Tcf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0.0230245 |
Alignment:
HHHTCCTTTGATSTHHV
----ACTTTGG------
| Original motif | Reverse complement motif |
| Consensus sequence: BHHASATCAAAGGAHHH | Consensus sequence: HHHTCCTTTGATSTHHV |
 |
 |
| Motif ID: | UP00413 |
| Motif name: | Elf4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.0283328 |
Alignment:
HAWYCCGGAAGTHBBD
-----CCAAAGT----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGKWTH | Consensus sequence: HAWYCCGGAAGTHBBD |
 |
 |
| Motif ID: | UP00418 |
| Motif name: | Etv6 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.0287289 |
Alignment:
BHAHCCGGAAGTABDVD
-----CCAAAGT-----
| Original motif | Reverse complement motif |
| Consensus sequence: DVHBTACTTCCGGHTHB | Consensus sequence: BHAHCCGGAAGTABDVD |
 |
 |
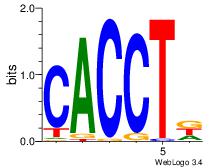
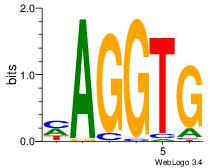
| Dataset #: 3 | Motif ID: 55 | Motif name: ZEB1 |
| Original motif | Reverse complement motif |
| Consensus sequence: CACCTD | Consensus sequence: HAGGTG |
 |
 |
Best Matches for Motif ID 55 (Highest to Lowest)
| Motif ID: | UP00068 |
| Motif name: | Eomes_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
VVBVCKACACCTHVBV
-------CACCTD---
| Original motif | Reverse complement motif |
| Consensus sequence: VBVDAGGTGTYGVBBV | Consensus sequence: VVBVCKACACCTHVBV |
 |
 |
| Motif ID: | UP00199 |
| Motif name: | Six4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.00433068 |
Alignment:
BBVHVGGTGTCATHDDT
---HAGGTG--------
| Original motif | Reverse complement motif |
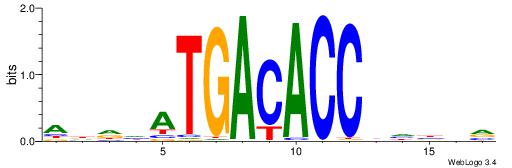
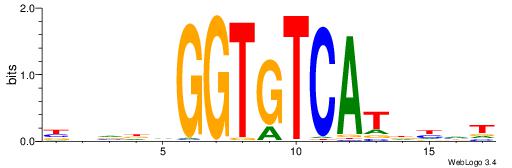
| Consensus sequence: ADDHATGACACCBHBBV | Consensus sequence: BBVHVGGTGTCATHDDT |
 |
 |
| Motif ID: | UP00068 |
| Motif name: | Eomes_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.00524081 |
Alignment:
HHHTTTCACACCTDHBV
--------CACCTD---
| Original motif | Reverse complement motif |
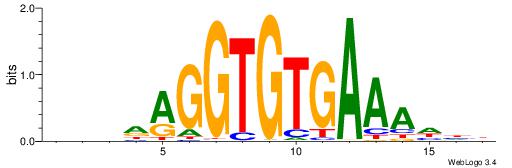
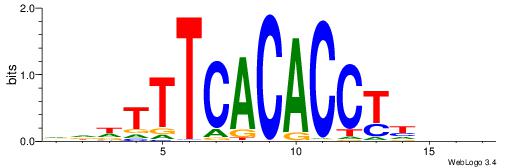
| Consensus sequence: BVHDAGGTGTGAAAHHH | Consensus sequence: HHHTTTCACACCTDHBV |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.00911301 |
Alignment:
VBHHVCAGGTGCDVDHD
-----HAGGTG------
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
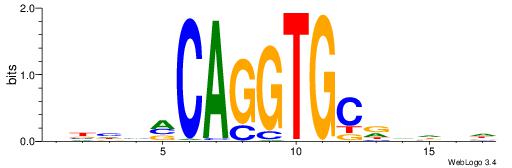 |
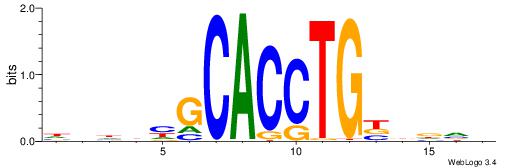 |
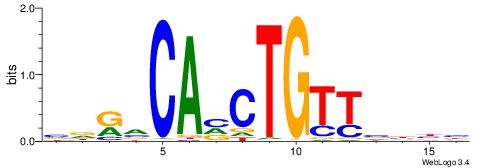
| Motif ID: | UP00047 |
| Motif name: | Zbtb7b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.0233556 |
Alignment:
BDDRVGACCACCHBDVB
--------CACCTD---
| Original motif | Reverse complement motif |
| Consensus sequence: BDDRVGACCACCHBDVB | Consensus sequence: BBDVHGGTGGTCBKDDB |
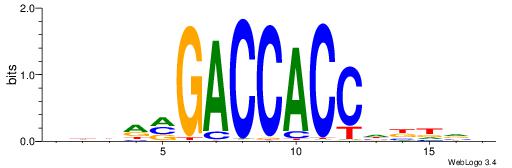 |
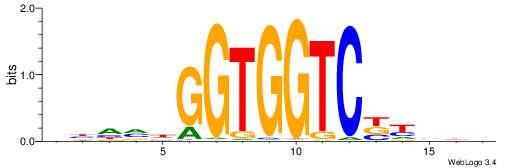 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 11 |
| Number of overlap: | 6 |
| Similarity score: | 0.0233928 |
Alignment:
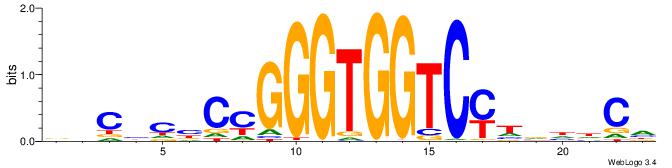
BHCBCBCCGGGTGGTCYHVHDCH
-------HAGGTG----------
| Original motif | Reverse complement motif |
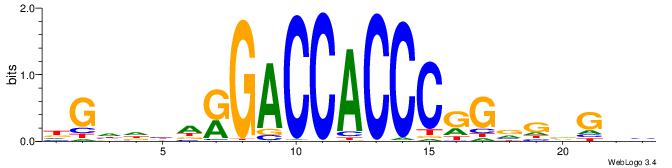
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
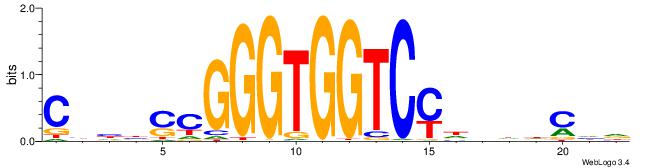
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.0254338 |
Alignment:
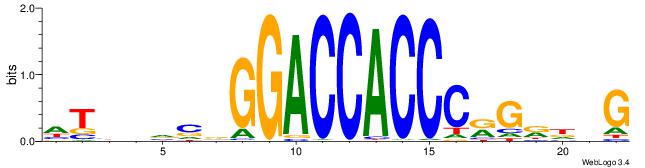
HTBVVVDGGACCACCCRGRDBG
-----------CACCTD-----
| Original motif | Reverse complement motif |
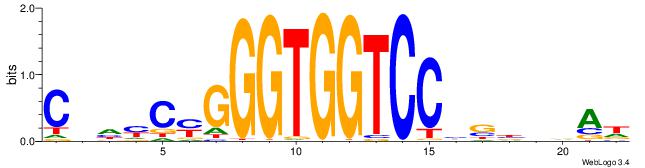
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
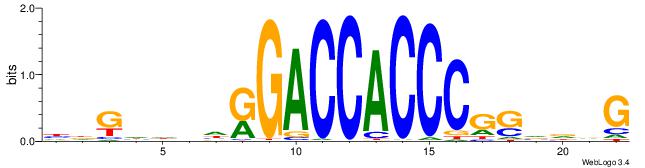
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.0290143 |
Alignment:
HDGHHBHRGACCACCCGSVDVG
-----------CACCTD-----
| Original motif | Reverse complement motif |
| Consensus sequence: HDGHHBHRGACCACCCGSVDVG | Consensus sequence: CVHBSCGGGTGGTCMHVHHCDH |
 |
 |
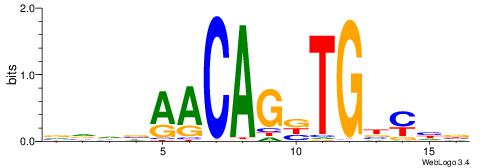
| Motif ID: | UP00036 |
| Motif name: | Myf6_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0309673 |
Alignment:
BDKVCABCTGTTBDBV
----CACCTD------
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBAACAGBTGBYHB | Consensus sequence: BDKVCABCTGTTBDBV |
 |
 |
| Motif ID: | UP00249 |
| Motif name: | Nkx2-5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.031502 |
Alignment:
HDADCCACTTRAAWTT
-----CACCTD-----
| Original motif | Reverse complement motif |
| Consensus sequence: HDADCCACTTRAAWTT | Consensus sequence: AAWTTMAAGTGGHTDH |
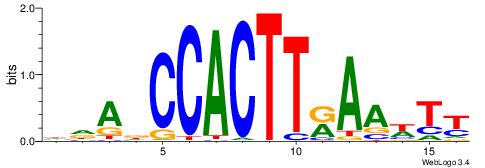 |
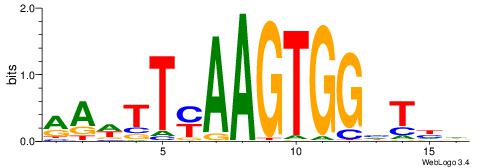 |
| Dataset #: 3 | Motif ID: 61 | Motif name: NR4A2 |
| Original motif | Reverse complement motif |
| Consensus sequence: AAGGTCAC | Consensus sequence: GTGACCTT |
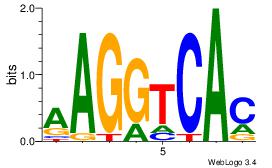 |
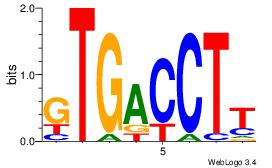 |
Best Matches for Motif ID 61 (Highest to Lowest)
| Motif ID: | UP00048 |
| Motif name: | Rara |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HHVGTGACCTTTDVDD
---GTGACCTT-----
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCACVHD | Consensus sequence: HHVGTGACCTTTDVDD |
 |
 |
| Motif ID: | UP00009 |
| Motif name: | Nr2f2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0074527 |
Alignment:
BHVDTGACCTTTDVDD
---GTGACCTT-----
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCAHVHB | Consensus sequence: BHVDTGACCTTTDVDD |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.01542 |
Alignment:
DHDBCAAGGTCAHVBDH
-----AAGGTCAC----
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBCAAGGTCAHVBDH | Consensus sequence: HHBVHTGACCTTGVDHD |
 |
 |
| Motif ID: | UP00009 |
| Motif name: | Nr2f2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0395644 |
Alignment:
VBVBCSGGGTCAVBBH
-----AAGGTCAC---
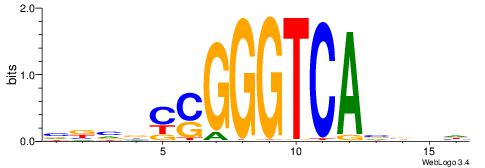
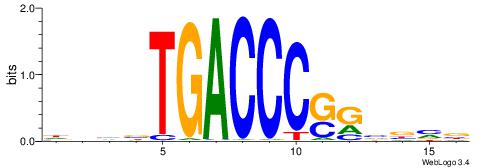
| Original motif | Reverse complement motif |
| Consensus sequence: VBVBCSGGGTCAVBBH | Consensus sequence: HVBVTGACCCSGBVBV |
 |
 |
| Motif ID: | UP00082 |
| Motif name: | Zfp187_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.0407874 |
Alignment:
DDGMCCTBGTCCHYHV
GTGACCTT--------
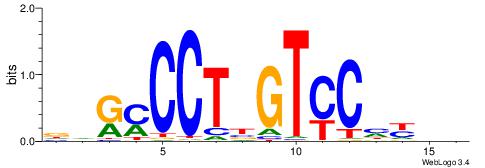
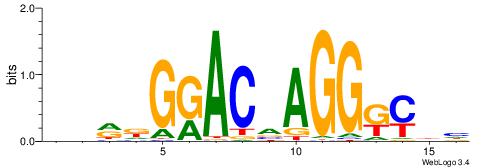
| Original motif | Reverse complement motif |
| Consensus sequence: DDGMCCTBGTCCHYHV | Consensus sequence: VHMDGGACVAGGRCDH |
 |
 |
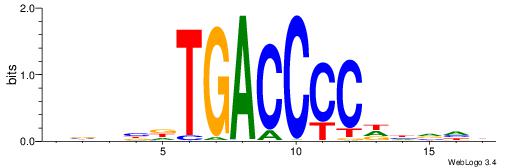
| Motif ID: | UP00053 |
| Motif name: | Rxra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0422933 |
Alignment:
DBHBDTGACCCCDHVHB
----GTGACCTT-----
| Original motif | Reverse complement motif |
| Consensus sequence: DBHBDTGACCCCDHVHB | Consensus sequence: VHBHDGGGGTCAHBHBD |
 |
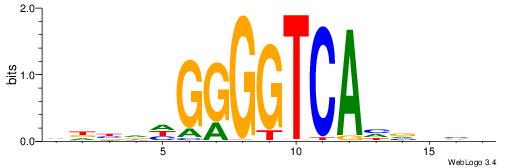 |
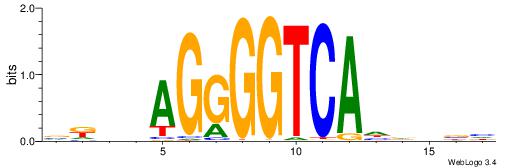
| Motif ID: | UP00079 |
| Motif name: | Esrra_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0447444 |
Alignment:
VKBBAGGGGTCAHDBBH
-----AAGGTCAC----
| Original motif | Reverse complement motif |
| Consensus sequence: VKBBAGGGGTCAHDBBH | Consensus sequence: DBBHHTGACCCCTBBYV |
 |
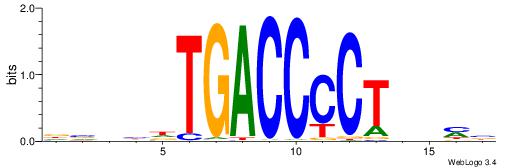 |
| Motif ID: | UP00066 |
| Motif name: | Hnf4a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0481418 |
Alignment:
BBHHAAAGTCCAMTHH
-----AAGGTCAC---
| Original motif | Reverse complement motif |
| Consensus sequence: BBHHAAAGTCCAMTHH | Consensus sequence: HHAYTGGACTTTHDBV |
 |
 |
| Motif ID: | UP00197 |
| Motif name: | Hoxc9 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0497679 |
Alignment:
VDHHTTAATGACCBHV
-------GTGACCTT-
| Original motif | Reverse complement motif |
| Consensus sequence: VDVGGTCATTAAHHDB | Consensus sequence: VDHHTTAATGACCBHV |
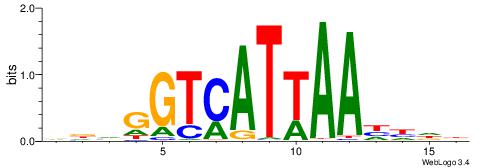 |
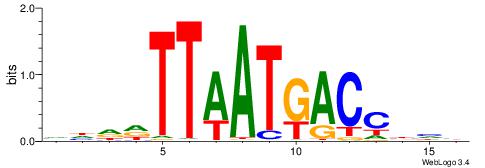 |
| Motif ID: | UP00035 |
| Motif name: | Hic1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0500062 |
Alignment:
DBDDRTGCCCAWDHDV
----GTGACCTT----
| Original motif | Reverse complement motif |
| Consensus sequence: DBDDRTGCCCAWDHDV | Consensus sequence: VHHDWTGGGCAMDHBH |
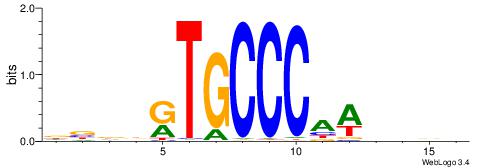 |
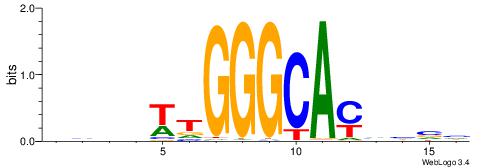 |
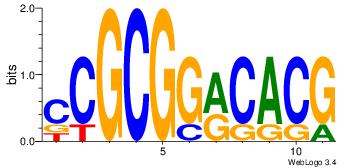
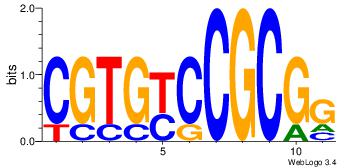
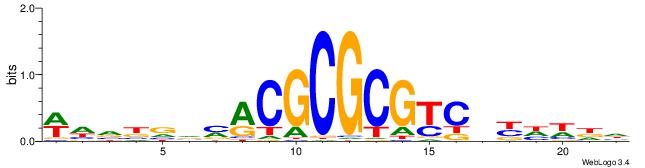
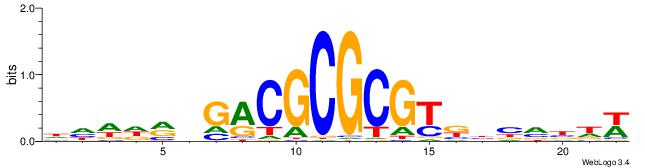
| Dataset #: 4 | Motif ID: 79 | Motif name: cCGCGGrCACG |
| Original motif | Reverse complement motif |
| Consensus sequence: CCGCGGRCACG | Consensus sequence: CGTGKCCGCGG |
 |
 |
Best Matches for Motif ID 79 (Highest to Lowest)
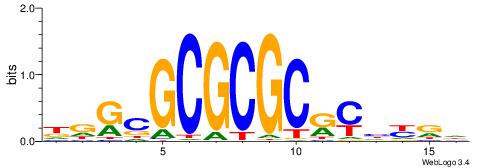
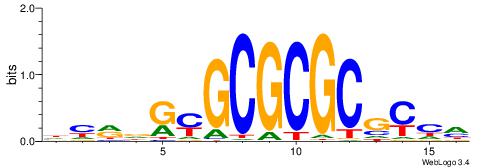
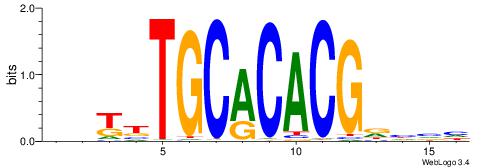
| Motif ID: | UP00065 |
| Motif name: | Zfp161_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0220206 |
Alignment:
DDGCGCGCGCRCHYRD
----CCGCGGRCACG-
| Original motif | Reverse complement motif |
| Consensus sequence: DDGCGCGCGCRCHYRD | Consensus sequence: DMMDGMGCGCGCGCHD |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 11 |
| Similarity score: | 0.024575 |
Alignment:
VHYTCCDWCCGCGTCMMTHWHV
---CGTGKCCGCGG--------
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00042 |
| Motif name: | Gm397_second |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0279366 |
Alignment:
DVMVGCACACACKCVH
--CCGCGGRCACG---
| Original motif | Reverse complement motif |
| Consensus sequence: DVMVGCACACACKCVH | Consensus sequence: HBGYGTGTGTGCVRVD |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0285381 |
Alignment:
VHTGCGGGGGCGGH
-CCGCGGRCACG--
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0335378 |
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
--------CCGCGGRCACG----
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0340428 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
-------CCGCGGRCACG-----
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00065 |
| Motif name: | Zfp161_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0345855 |
Alignment:
GYCGCGCARBGCVB
-CCGCGGRCACG--
| Original motif | Reverse complement motif |
| Consensus sequence: GYCGCGCARBGCVB | Consensus sequence: VVGCVMTGCGCGKC |
 |
 |
| Motif ID: | UP00097 |
| Motif name: | Mtf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0350434 |
Alignment:
DBDHCGTGTGCAAMDH
---CCGCGGRCACG--
| Original motif | Reverse complement motif |
| Consensus sequence: DBDHCGTGTGCAAMDH | Consensus sequence: HDYTTGCACACGDHBH |
 |
 |
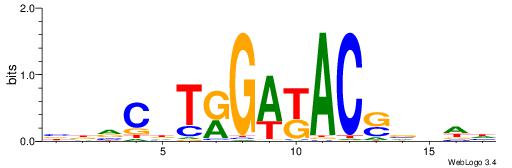
| Motif ID: | UP00076 |
| Motif name: | Rfxdc2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0359928 |
Alignment:
HDDVSGTRTCMADGBVB
----CGTGKCCGCGG--
| Original motif | Reverse complement motif |
| Consensus sequence: BBVCDTRGAKACSVDDH | Consensus sequence: HDDVSGTRTCMADGBVB |
 |
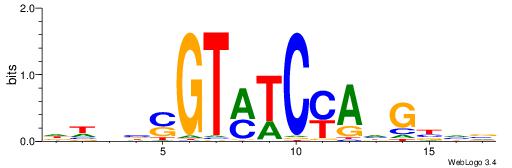 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0361963 |
Alignment:
HDWHMHGACGCGCGTRBVVBHW
---------CCGCGGRCACG--
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
 |
 |
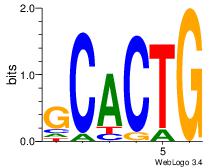
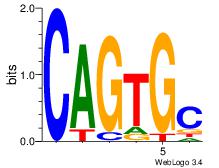
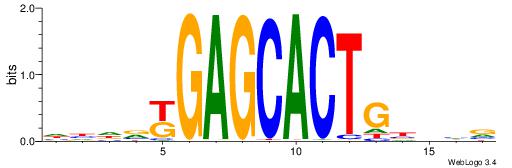
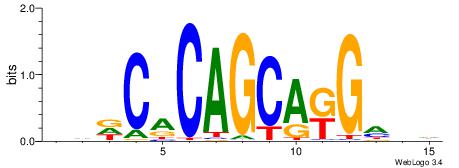
| Dataset #: 5 | Motif ID: 84 | Motif name: TFW3 |
| Original motif | Reverse complement motif |
| Consensus sequence: GCACTG | Consensus sequence: CAGTGC |
 |
 |
Best Matches for Motif ID 84 (Highest to Lowest)
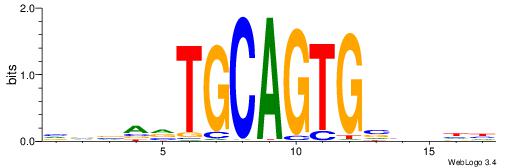
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
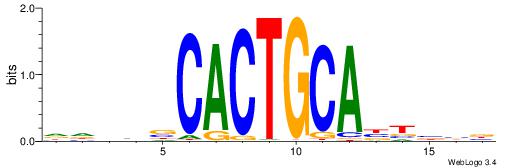
Alignment:
BBBAVTGCAGTGBBVDD
-------CAGTGC----
| Original motif | Reverse complement motif |
| Consensus sequence: DDBBBCACTGCABTBBB | Consensus sequence: BBBAVTGCAGTGBBVDD |
 |
 |
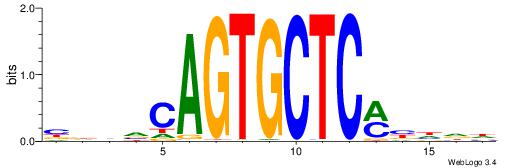
| Motif ID: | UP00095 |
| Motif name: | Zfp691_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.020963 |
Alignment:
BDHVCAGTGCTCMBHDD
----CAGTGC-------
| Original motif | Reverse complement motif |
| Consensus sequence: BDHVCAGTGCTCMBHDD | Consensus sequence: DDHBYGAGCACTGBHHB |
 |
 |
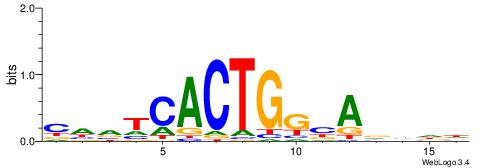
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.0248871 |
Alignment:
DDHHTBCCAGTGABHG
-------CAGTGC---
| Original motif | Reverse complement motif |
| Consensus sequence: CHVTCACTGGBADHDD | Consensus sequence: DDHHTBCCAGTGABHG |
 |
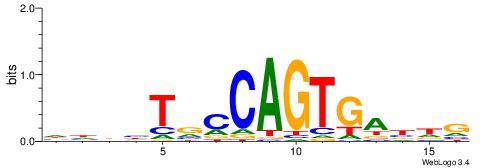 |
| Motif ID: | UP00065 |
| Motif name: | Zfp161_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0303167 |
Alignment:
GYCGCGCARBGCVB
------CAGTGC--
| Original motif | Reverse complement motif |
| Consensus sequence: GYCGCGCARBGCVB | Consensus sequence: VVGCVMTGCGCGKC |
 |
 |
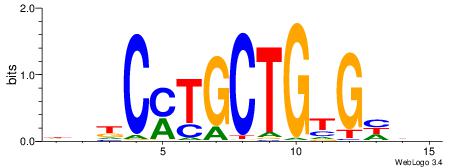
| Motif ID: | UP00006 |
| Motif name: | Zic3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.0420592 |
Alignment:
DDDCACAGCAKGHVD
--GCACTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: DDDCACAGCAKGHVD | Consensus sequence: DVHCYTGCTGTGHDH |
 |
 |
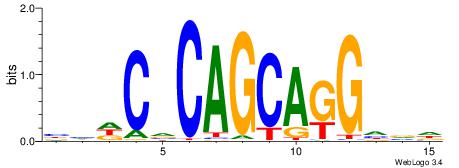
| Motif ID: | UP00102 |
| Motif name: | Zic1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 6 |
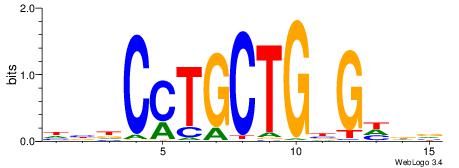
| Similarity score: | 0.0499082 |
Alignment:
BHDCVCAGCAGGHVD
--GCACTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: BHDCVCAGCAGGHVD | Consensus sequence: DVHCCTGCTGBGDDB |
 |
 |
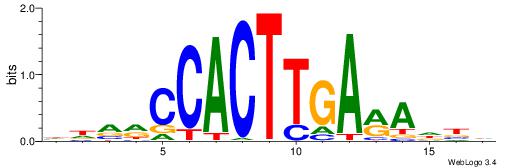
| Motif ID: | UP00231 |
| Motif name: | Nkx2-2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.0507672 |
Alignment:
DVDTKTCAAGTGGKBDH
-------CAGTGC----
| Original motif | Reverse complement motif |
| Consensus sequence: HDVRCCACTTGARADBD | Consensus sequence: DVDTKTCAAGTGGKBDH |
 |
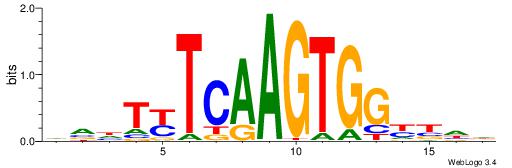 |
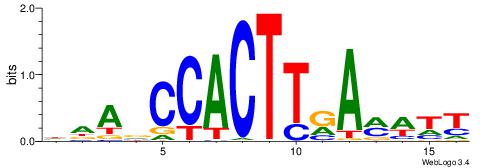
| Motif ID: | UP00107 |
| Motif name: | Nkx2-4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0519322 |
Alignment:
AAWYTMAAGTGGVTDH
------CAGTGC----
| Original motif | Reverse complement motif |
| Consensus sequence: HDAVCCACTTRAMWTT | Consensus sequence: AAWYTMAAGTGGVTDH |
 |
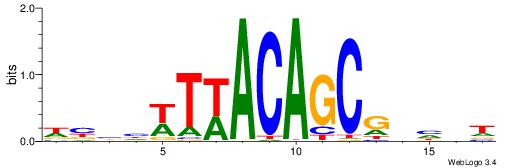
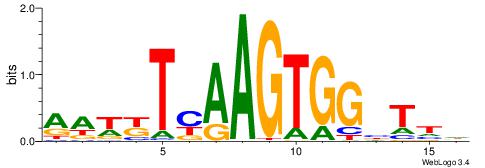 |
| Motif ID: | UP00193 |
| Motif name: | Rhox11_2205.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.0530479 |
Alignment:
WVVHWTWACAGCGHHVT
--------CAGTGC---
| Original motif | Reverse complement motif |
| Consensus sequence: AVDHCGCTGTWAWDVVW | Consensus sequence: WVVHWTWACAGCGHHVT |
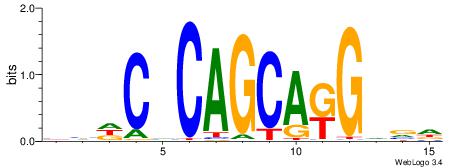
 |
 |
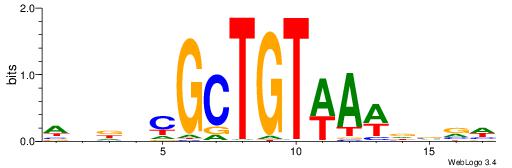
| Motif ID: | UP00057 |
| Motif name: | Zic2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.0539075 |
Alignment:
HHDCVCAGCAKGVVD
--GCACTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: HHDCVCAGCAKGVVD | Consensus sequence: DVBCYTGCTGBGDDD |
 |
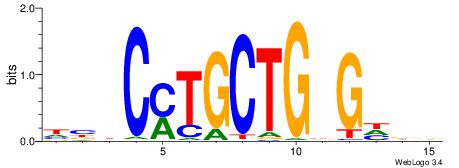 |
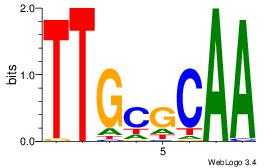
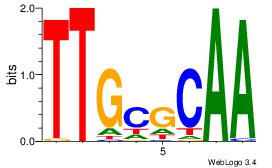
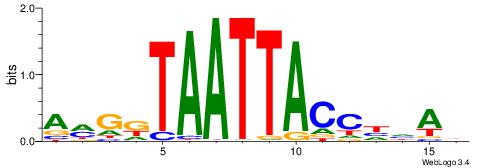
| Dataset #: 5 | Motif ID: 85 | Motif name: TFW1 |
| Original motif | Reverse complement motif |
| Consensus sequence: TTGCGCAA | Consensus sequence: TTGCGCAA |
 |
 |
Best Matches for Motif ID 85 (Highest to Lowest)
| Motif ID: | UP00045 |
| Motif name: | Mafb_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.00957029 |
Alignment:
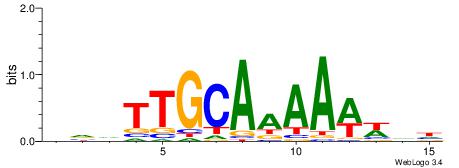
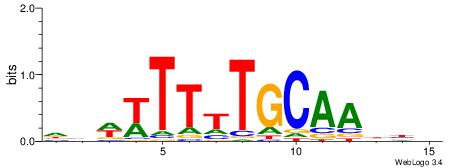
HDWWTTTTGCAADBD
----TTGCGCAA---
| Original motif | Reverse complement motif |
| Consensus sequence: HVDTTGCAAAAWWDH | Consensus sequence: HDWWTTTTGCAADBD |
 |
 |
| Motif ID: | UP00097 |
| Motif name: | Mtf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0184619 |
Alignment:
DBDHCGTGTGCAAMDH
-----TTGCGCAA---
| Original motif | Reverse complement motif |
| Consensus sequence: DBDHCGTGTGCAAMDH | Consensus sequence: HDYTTGCACACGDHBH |
 |
 |
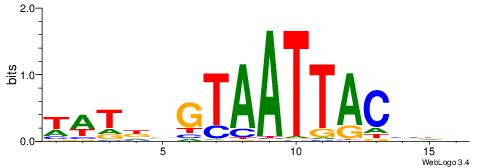
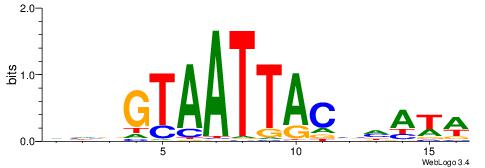
| Motif ID: | UP00259 |
| Motif name: | Hoxb6 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0226004 |
Alignment:
WWTDBGTAATTACVDB
-TTGCGCAA-------
| Original motif | Reverse complement motif |
| Consensus sequence: WWTDBGTAATTACVDB | Consensus sequence: VDVGTAATTACBDAWW |
 |
 |
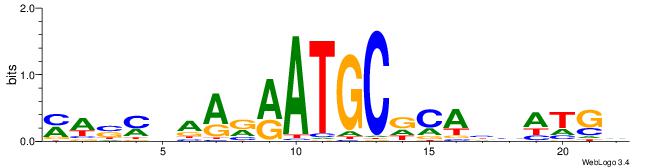
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0239956 |
Alignment:
DCATDBTGHGCATKMTTBRSWR
-----TTGCGCAA---------
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
 |
 |
| Motif ID: | UP00035 |
| Motif name: | Hic1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0261465 |
Alignment:
VHHDWTGGGCAMDHBH
----TTGCGCAA----
| Original motif | Reverse complement motif |
| Consensus sequence: DBDDRTGCCCAWDHDV | Consensus sequence: VHHDWTGGGCAMDHBH |
 |
 |
| Motif ID: | UP00004 |
| Motif name: | Sox14_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0272314 |
Alignment:
BKBASACAATRGHBB
-TTGCGCAA------
| Original motif | Reverse complement motif |
| Consensus sequence: BKBASACAATRGHBB | Consensus sequence: BBDCMATTGTSTBRB |
 |
 |
| Motif ID: | UP00055 |
| Motif name: | Hbp1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.0278778 |
Alignment:
HBTHVACAATGGGMDHD
-TTGCGCAA--------
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCCATTGTVHABH | Consensus sequence: HBTHVACAATGGGMDHD |
 |
 |
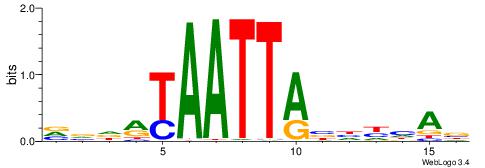
| Motif ID: | UP00138 |
| Motif name: | Bsx |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0281625 |
Alignment:
VVRDYAATTRBYBVAD
-------TTGCGCAA-
| Original motif | Reverse complement motif |
| Consensus sequence: VVRDYAATTRBYBVAD | Consensus sequence: HTVVKBKAATTMHMBV |
 |
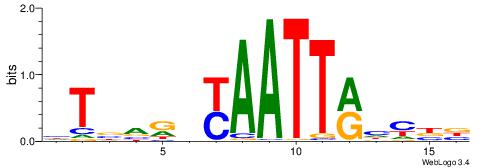 |
| Motif ID: | UP00126 |
| Motif name: | Dlx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.030109 |
Alignment:
VVDRTAATTABHHHAB
-------TTGCGCAA-
| Original motif | Reverse complement motif |
| Consensus sequence: VVDRTAATTABHHHAB | Consensus sequence: BTDHHBTAATTAKDVV |
 |
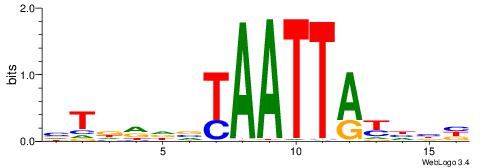 |
| Motif ID: | UP00182 |
| Motif name: | Hoxa6 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0308523 |
Alignment:
AMGKTAATTACCHHAD
-------TTGCGCAA-
| Original motif | Reverse complement motif |
| Consensus sequence: AMGKTAATTACCHHAD | Consensus sequence: DTHHGGTAATTAYCYT |
 |
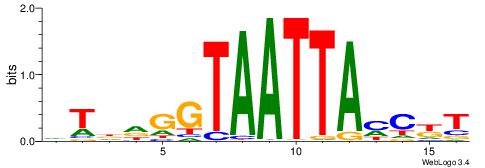 |
| Dataset #: 5 | Motif ID: 87 | Motif name: TFF1 |
| Original motif | Reverse complement motif |
| Consensus sequence: AGGACAAAGAVV | Consensus sequence: VVTCTTTGTCCT |
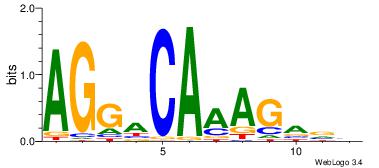 |
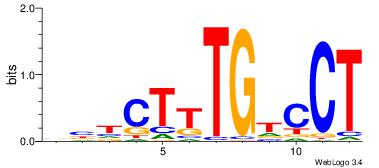 |
Best Matches for Motif ID 87 (Highest to Lowest)
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0258536 |
Alignment:
HBDDDAACAAAGRVBHH
---AGGACAAAGAVV--
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
 |
 |
| Motif ID: | UP00072 |
| Motif name: | IRC900814_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0258585 |
Alignment:
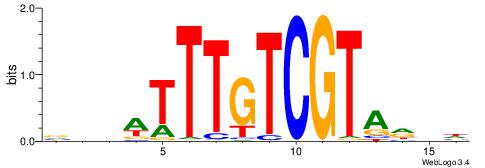
DDHTACGACAAAWDDB
----AGGACAAAGAVV
| Original motif | Reverse complement motif |
| Consensus sequence: DDHTACGACAAAWDDB | Consensus sequence: BHDWTTTGTCGTAHDD |
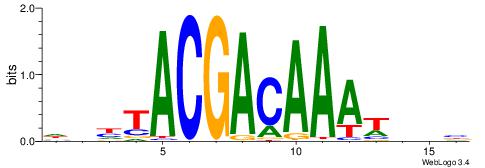 |
 |
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0284165 |
Alignment:
HBDDRAACAAAGRABHH
---AGGACAAAGAVV--
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
 |
 |
| Motif ID: | UP00101 |
| Motif name: | Sox12_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0317868 |
Alignment:
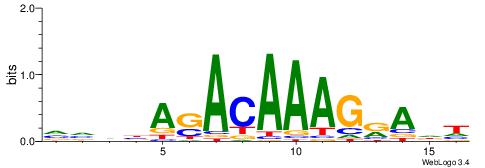
VVDBASACAAAGRADH
---AGGACAAAGAVV-
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBASACAAAGRADH | Consensus sequence: HDTMCTTTGTSTVDBB |
 |
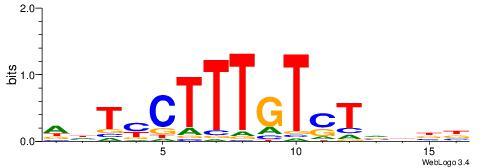 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0318041 |
Alignment:
VDDAGATCAAAGGKDDD
---AGGACAAAGAVV--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
 |
 |
| Motif ID: | UP00058 |
| Motif name: | Tcf3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0329392 |
Alignment:
HHHASATCAAAGVRHHH
---AGGACAAAGAVV--
| Original motif | Reverse complement motif |
| Consensus sequence: HHHASATCAAAGVRHHH | Consensus sequence: HHHKVCTTTGATSTHHH |
 |
 |
| Motif ID: | UP00054 |
| Motif name: | Tcf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0344802 |
Alignment:
BHHASATCAAAGGAHHH
---AGGACAAAGAVV--
| Original motif | Reverse complement motif |
| Consensus sequence: BHHASATCAAAGGAHHH | Consensus sequence: HHHTCCTTTGATSTHHV |
 |
 |
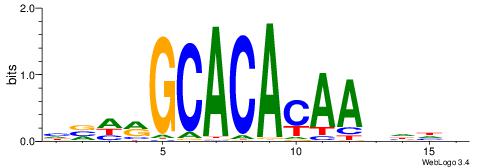
| Motif ID: | UP00026 |
| Motif name: | Zscan4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0344812 |
Alignment:
BHHHTTGTGTGCKHVD
-VVTCTTTGTCCT---
| Original motif | Reverse complement motif |
| Consensus sequence: HVHRGCACACAAHHHV | Consensus sequence: BHHHTTGTGTGCKHVD |
 |
 |
| Motif ID: | UP00083 |
| Motif name: | Tcf7l2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0344876 |
Alignment:
DDDYCCTTTGATSTDDV
--VVTCTTTGTCCT---
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATSTDDV | Consensus sequence: BDDASATCAAAGGMDDD |
 |
 |
| Motif ID: | UP00020 |
| Motif name: | Atf1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0352035 |
Alignment:
VDATGACGADYADH
--AGGACAAAGAVV
| Original motif | Reverse complement motif |
| Consensus sequence: VDATGACGADYADH | Consensus sequence: DDTMDTCGTCATDV |
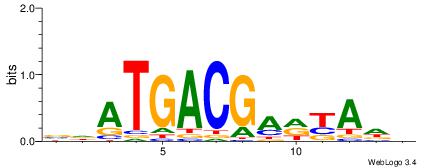 |
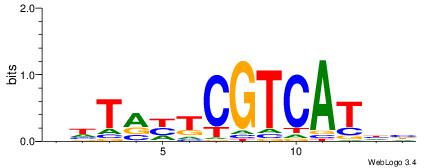 |