Best Matches in Database for Each Motif (Highest to Lowest)
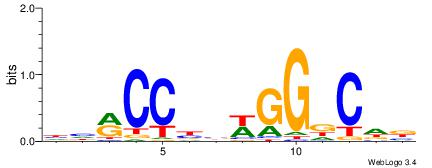
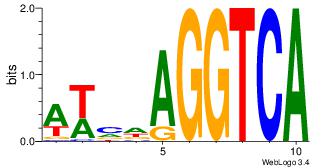
| Dataset #: 1 | Motif ID: 1 | Motif name: HNF4A |
| Original motif | Reverse complement motif |
| Consensus sequence: RGGBCAAAGKYCA | Consensus sequence: TGMYCTTTGBCCK |
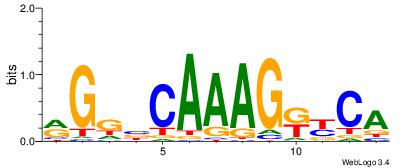 |
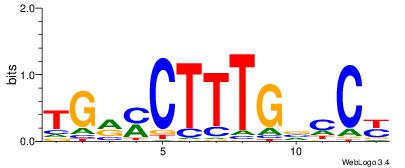 |
Best Matches for Motif ID 1 (Highest to Lowest)
| Motif ID: | UP00058 |
| Motif name: | Tcf3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0357776 |
Alignment:
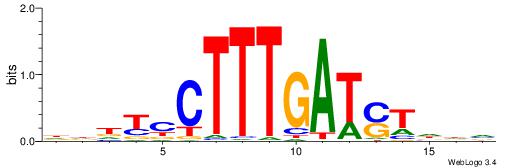
HHHASATCAAAGVRHHH
---RGGBCAAAGKYCA-
| Original motif | Reverse complement motif |
| Consensus sequence: HHHASATCAAAGVRHHH | Consensus sequence: HHHKVCTTTGATSTHHH |
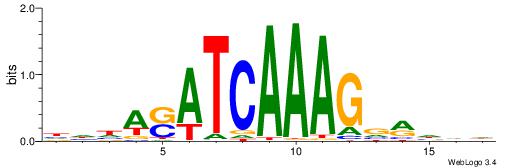 |
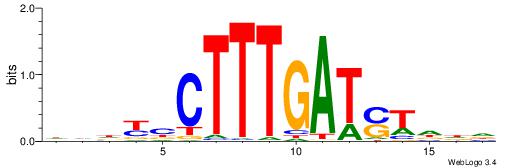 |
| Motif ID: | UP00054 |
| Motif name: | Tcf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0357792 |
Alignment:
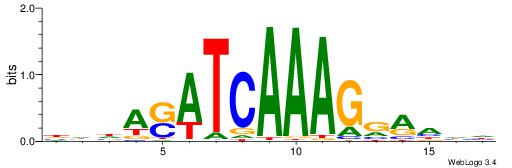
BHHASATCAAAGGAHHH
---RGGBCAAAGKYCA-
| Original motif | Reverse complement motif |
| Consensus sequence: BHHASATCAAAGGAHHH | Consensus sequence: HHHTCCTTTGATSTHHV |
 |
 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 13 |
| Similarity score: | 0.0359011 |
Alignment:
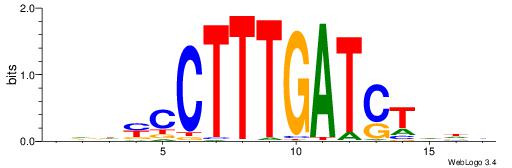
VDDAGATCAAAGGKDDD
---RGGBCAAAGKYCA-
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
 |
 |
| Motif ID: | UP00083 |
| Motif name: | Tcf7l2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 13 |
| Similarity score: | 0.0360786 |
Alignment:
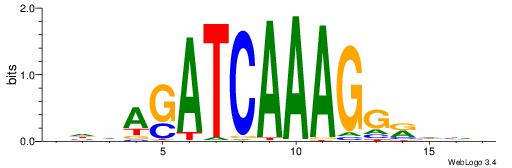
BDDASATCAAAGGMDDD
---RGGBCAAAGKYCA-
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATSTDDV | Consensus sequence: BDDASATCAAAGGMDDD |
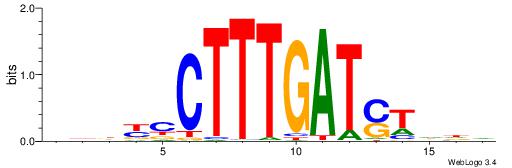 |
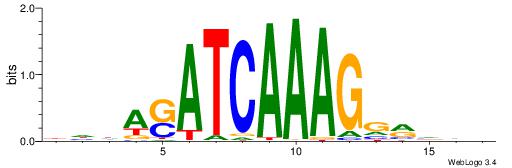 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 13 |
| Similarity score: | 0.0405764 |
Alignment:
HVDDVCAGATGKYHBBV
-RGGBCAAAGKYCA---
| Original motif | Reverse complement motif |
| Consensus sequence: HVDDVCAGATGKYHBBV | Consensus sequence: VBBDMYCATCTGVHHBH |
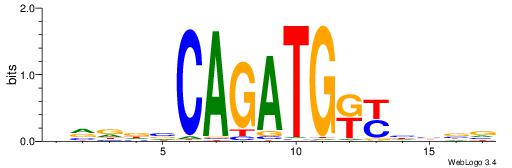 |
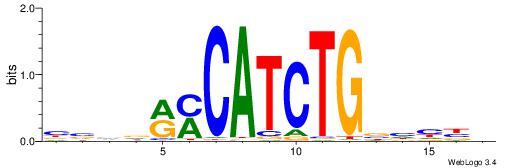 |
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0409915 |
Alignment:
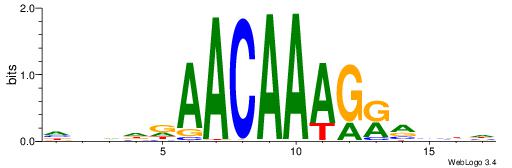
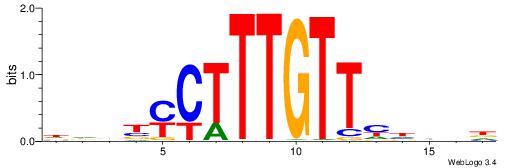
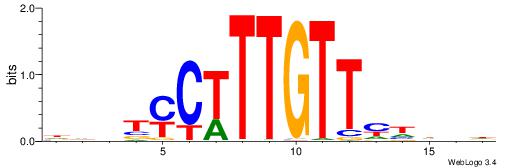
HHBBMCTTTGTTHDDBH
-TGMYCTTTGBCCK---
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
 |
 |
| Motif ID: | UP00101 |
| Motif name: | Sox12_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0413218 |
Alignment:
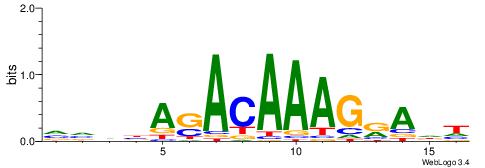
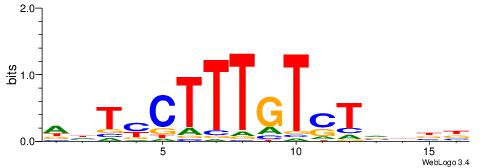
HDTMCTTTGTSTVDBB
TGMYCTTTGBCCK---
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBASACAAAGRADH | Consensus sequence: HDTMCTTTGTSTVDBB |
 |
 |
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 13 |
| Similarity score: | 0.0421105 |
Alignment:
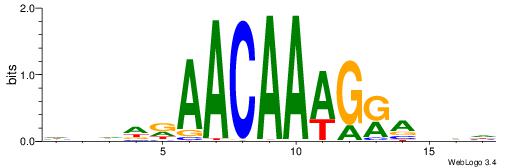
HHBTMCTTTGTTMDDVH
-TGMYCTTTGBCCK---
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
 |
 |
| Motif ID: | UP00086 |
| Motif name: | Irf3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0442004 |
Alignment:
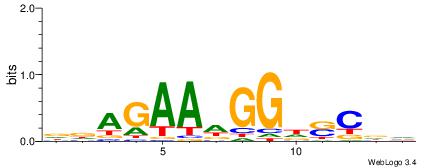
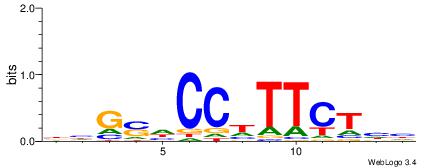
DDAGAADGGDSCDV
RGGBCAAAGKYCA-
| Original motif | Reverse complement motif |
| Consensus sequence: DDAGAADGGDSCDV | Consensus sequence: BHGSDCCDTTCTHH |
 |
 |
| Motif ID: | UP00082 |
| Motif name: | Zfp187_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 13 |
| Similarity score: | 0.0446884 |
Alignment:
VHMDGGACVAGGRCDH
---RGGBCAAAGKYCA
| Original motif | Reverse complement motif |
| Consensus sequence: DDGMCCTBGTCCHYHV | Consensus sequence: VHMDGGACVAGGRCDH |
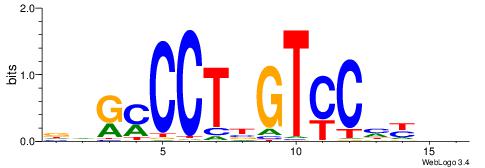 |
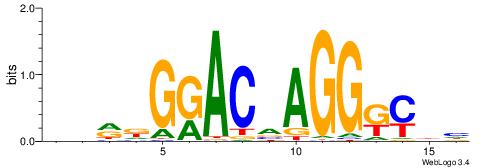 |
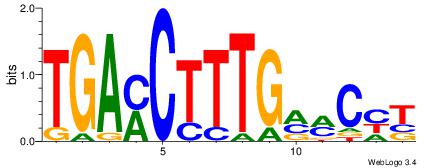
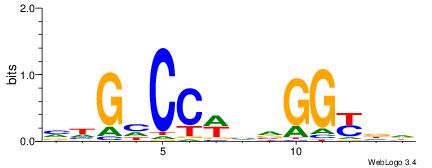
| Dataset #: 1 | Motif ID: 2 | Motif name: NR2F1 |
| Original motif | Reverse complement motif |
| Consensus sequence: TGAMCTTTGMMCYT | Consensus sequence: AKGYYCAAAGRTCA |
 |
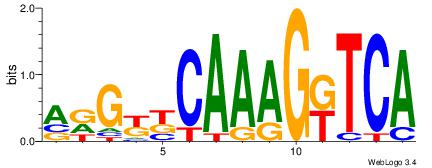 |
Best Matches for Motif ID 2 (Highest to Lowest)
| Motif ID: | UP00083 |
| Motif name: | Tcf7l2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0452364 |
Alignment:
DDDYCCTTTGATSTDDV
-TGAMCTTTGMMCYT--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATSTDDV | Consensus sequence: BDDASATCAAAGGMDDD |
 |
 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0453449 |
Alignment:
VDDAGATCAAAGGKDDD
--AKGYYCAAAGRTCA-
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 14 |
| Similarity score: | 0.0461358 |
Alignment:
DVMHHDHKGACCCTCCTSVCBH
-------TGAMCTTTGMMCYT-
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
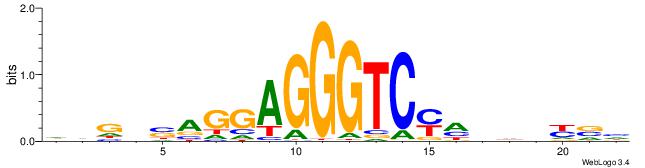 |
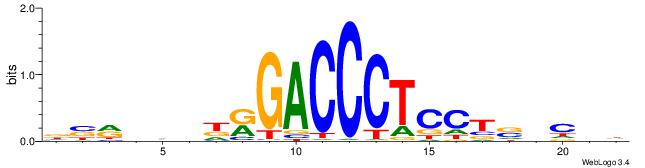 |
| Motif ID: | UP00054 |
| Motif name: | Tcf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0461768 |
Alignment:
BHHASATCAAAGGAHHH
--AKGYYCAAAGRTCA-
| Original motif | Reverse complement motif |
| Consensus sequence: BHHASATCAAAGGAHHH | Consensus sequence: HHHTCCTTTGATSTHHV |
 |
 |
| Motif ID: | UP00058 |
| Motif name: | Tcf3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0473628 |
Alignment:
HHHASATCAAAGVRHHH
--AKGYYCAAAGRTCA-
| Original motif | Reverse complement motif |
| Consensus sequence: HHHASATCAAAGVRHHH | Consensus sequence: HHHKVCTTTGATSTHHH |
 |
 |
| Motif ID: | UP00082 |
| Motif name: | Zfp187_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0480188 |
Alignment:
DDGMCCTBGTCCHYHV
TGAMCTTTGMMCYT--
| Original motif | Reverse complement motif |
| Consensus sequence: DDGMCCTBGTCCHYHV | Consensus sequence: VHMDGGACVAGGRCDH |
 |
 |
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0495447 |
Alignment:
HHBBMCTTTGTTHDDBH
-TGAMCTTTGMMCYT--
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
 |
 |
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0499033 |
Alignment:
HHBTMCTTTGTTMDDVH
-TGAMCTTTGMMCYT--
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
 |
 |
| Motif ID: | UP00023 |
| Motif name: | Sox30_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0511534 |
Alignment:
DHDBAACAATDRHHHH
--TGAMCTTTGMMCYT
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBAACAATDRHHHH | Consensus sequence: HHHHMHATTGTTBDHD |
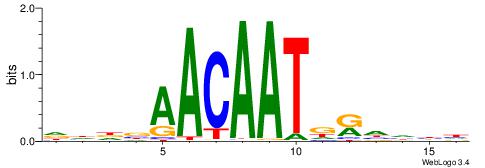 |
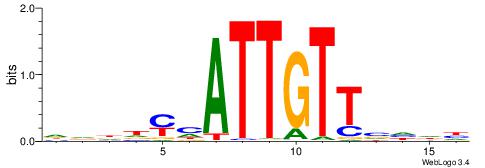 |
| Motif ID: | UP00005 |
| Motif name: | Tcfap2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0516973 |
Alignment:
BDGHCCWBVGGKDH
AKGYYCAAAGRTCA
| Original motif | Reverse complement motif |
| Consensus sequence: HHRCCBBWGGDCDB | Consensus sequence: BDGHCCWBVGGKDH |
 |
 |
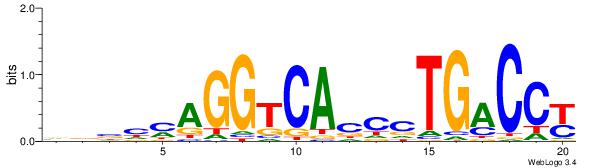
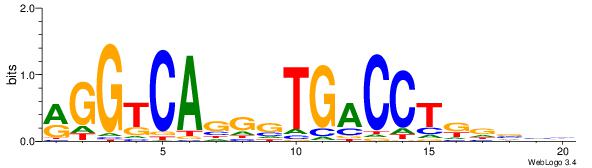
| Dataset #: 1 | Motif ID: 3 | Motif name: ESR1 |
| Original motif | Reverse complement motif |
| Consensus sequence: VDBHMAGGTCACCCTGACCY | Consensus sequence: MGGTCAGGGTGACCTRDBHV |
 |
 |
Best Matches for Motif ID 3 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 20 |
| Similarity score: | 0.0658405 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
VDBHMAGGTCACCCTGACCY---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
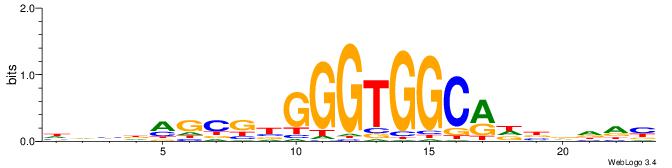 |
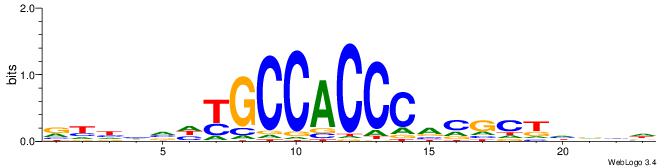 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 20 |
| Similarity score: | 0.0689748 |
Alignment:
BDBBVBVGTAWCCAAGVBHBAGB
VDBHMAGGTCACCCTGACCY---
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
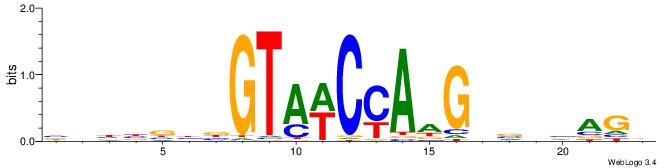 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 20 |
| Similarity score: | 0.0701247 |
Alignment:
CBDMCMGGGTGGTCCHVBVBAH
MGGTCAGGGTGACCTRDBHV--
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
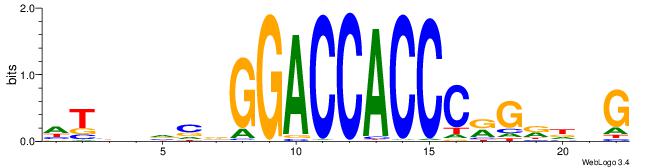 |
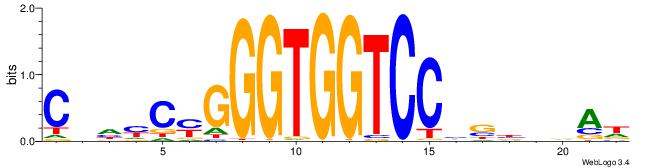 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 20 |
| Similarity score: | 0.0711267 |
Alignment:
HVGVSAGGAGGGTCYHHHHYVH
MGGTCAGGGTGACCTRDBHV--
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 20 |
| Similarity score: | 0.0742427 |
Alignment:
CVHBSCGGGTGGTCMHVHHCDH
-VDBHMAGGTCACCCTGACCY-
| Original motif | Reverse complement motif |
| Consensus sequence: HDGHHBHRGACCACCCGSVDVG | Consensus sequence: CVHBSCGGGTGGTCMHVHHCDH |
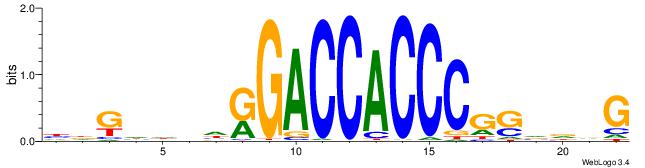 |
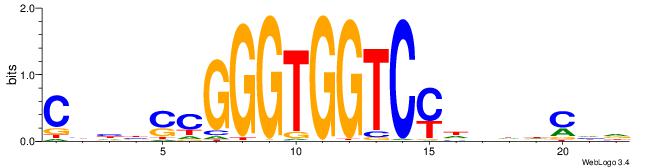 |
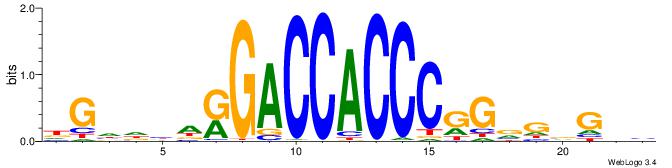
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 20 |
| Similarity score: | 0.0747053 |
Alignment:
ABBBBVVRGACCACCCACRDBBM
--VDBHMAGGTCACCCTGACCY-
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
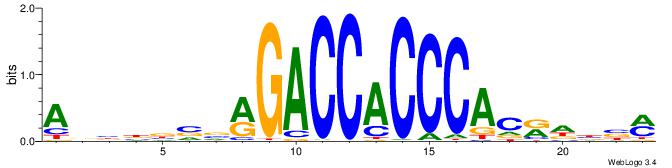 |
 |
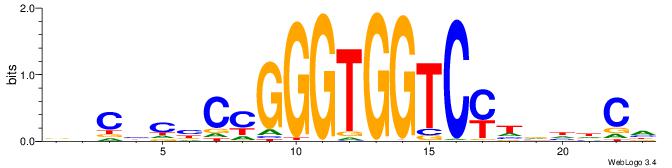
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 20 |
| Similarity score: | 0.0749165 |
Alignment:
BHCBCBCCGGGTGGTCYHVHDCH
---VDBHMAGGTCACCCTGACCY
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
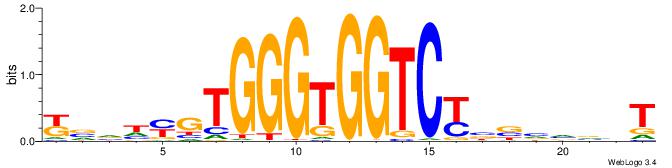
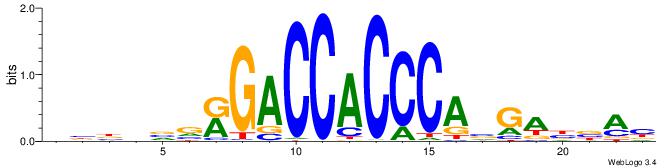
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 20 |
| Similarity score: | 0.0759307 |
Alignment:
BBBDVVRGACCACCCAVGABBAB
-VDBHMAGGTCACCCTGACCY--
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 20 |
| Similarity score: | 0.0762689 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
-MGGTCAGGGTGACCTRDBHV--
| Original motif | Reverse complement motif |
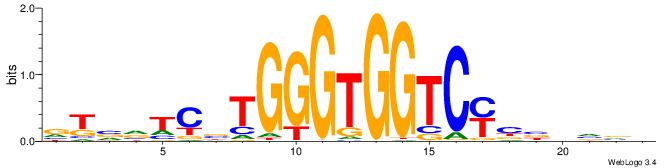
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
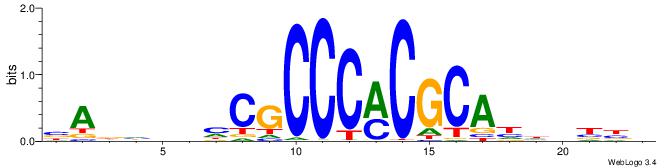 |
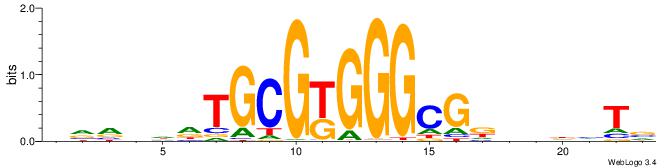 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 20 |
| Similarity score: | 0.0780113 |
Alignment:
GTVBDAATTGTVTGHGDDHKVB
VDBHMAGGTCACCCTGACCY--
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
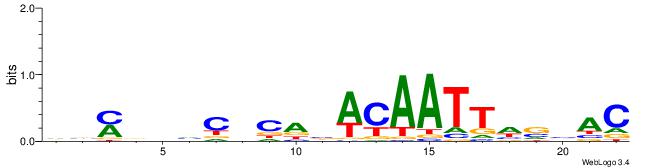 |
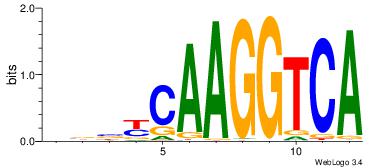
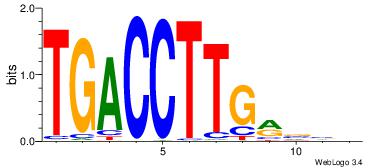
| Dataset #: 1 | Motif ID: 4 | Motif name: Esrrb |
| Original motif | Reverse complement motif |
| Consensus sequence: VBBYCAAGGTCA | Consensus sequence: TGACCTTGMBBB |
 |
 |
Best Matches for Motif ID 4 (Highest to Lowest)
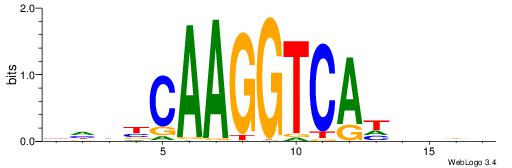
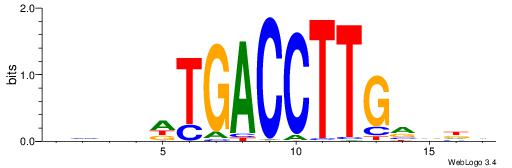
| Motif ID: | UP00079 |
| Motif name: | Esrra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 12 |
| Similarity score: | 0 |
Alignment:
HHBVHTGACCTTGVDHD
-----TGACCTTGMBBB
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBCAAGGTCAHVBDH | Consensus sequence: HHBVHTGACCTTGVDHD |
 |
 |
| Motif ID: | UP00009 |
| Motif name: | Nr2f2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0205471 |
Alignment:
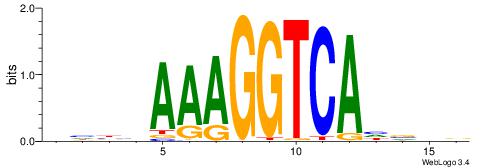
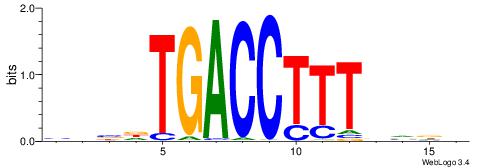
DHBHAAAGGTCAHVHB
VBBYCAAGGTCA----
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCAHVHB | Consensus sequence: BHVDTGACCTTTDVDD |
 |
 |
| Motif ID: | UP00048 |
| Motif name: | Rara |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0214372 |
Alignment:
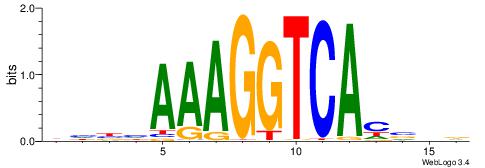
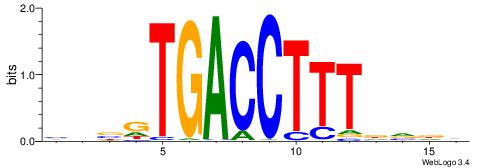
DHBHAAAGGTCACVHD
VBBYCAAGGTCA----
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCACVHD | Consensus sequence: HHVGTGACCTTTDVDD |
 |
 |
| Motif ID: | UP00009 |
| Motif name: | Nr2f2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 0.0430759 |
Alignment:
HVBVTGACCCSGBVBV
----TGACCTTGMBBB
| Original motif | Reverse complement motif |
| Consensus sequence: VBVBCSGGGTCAVBBH | Consensus sequence: HVBVTGACCCSGBVBV |
 |
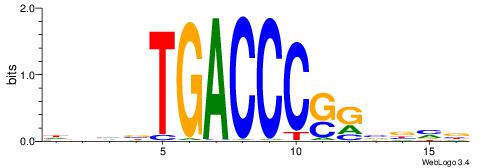 |
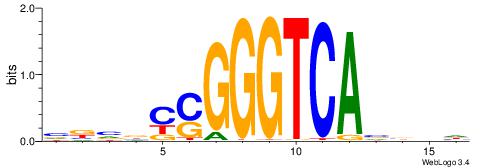
| Motif ID: | UP00066 |
| Motif name: | Hnf4a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0451776 |
Alignment:
BDBVMGGGGTCAABHDV
VBBYCAAGGTCA-----
| Original motif | Reverse complement motif |
| Consensus sequence: BDBVMGGGGTCAABHDV | Consensus sequence: BHHVTTGACCCCYVVDB |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 12 |
| Similarity score: | 0.0482496 |
Alignment:
VKBBAGGGGTCAHDBBH
VBBYCAAGGTCA-----
| Original motif | Reverse complement motif |
| Consensus sequence: VKBBAGGGGTCAHDBBH | Consensus sequence: DBBHHTGACCCCTBBYV |
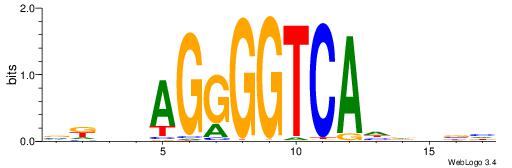 |
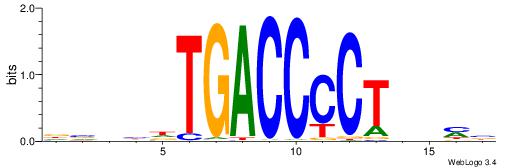 |
| Motif ID: | UP00053 |
| Motif name: | Rxra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0499025 |
Alignment:
DBHBDTGACCCCDHVHB
-----TGACCTTGMBBB
| Original motif | Reverse complement motif |
| Consensus sequence: DBHBDTGACCCCDHVHB | Consensus sequence: VHBHDGGGGTCAHBHBD |
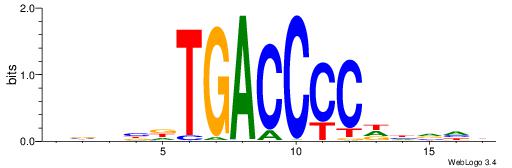 |
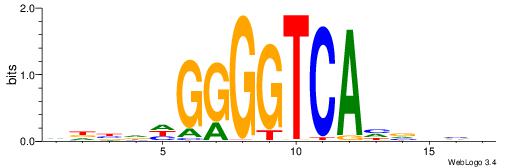 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 12 |
| Similarity score: | 0.0525354 |
Alignment:
DVMHHDHKGACCCTCCTSVCBH
-------TGACCTTGMBBB---
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
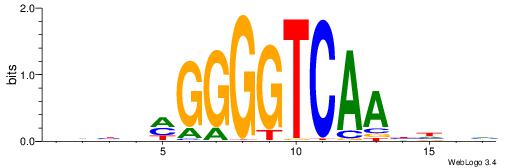
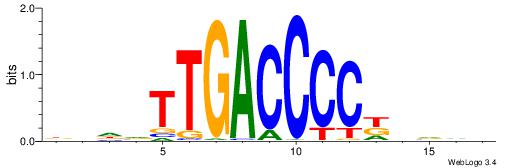
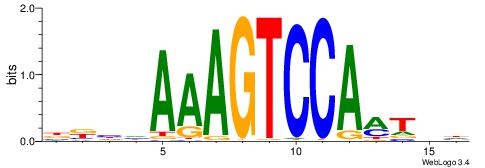
| Motif ID: | UP00066 |
| Motif name: | Hnf4a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0580553 |
Alignment:
HHAYTGGACTTTHDBV
----TGACCTTGMBBB
| Original motif | Reverse complement motif |
| Consensus sequence: BBHHAAAGTCCAMTHH | Consensus sequence: HHAYTGGACTTTHDBV |
 |
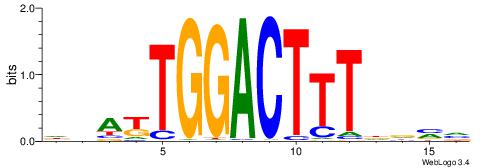 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0609738 |
Alignment:
BDBBVBVGTAWCCAAGVBHBAGB
--------TGACCTTGMBBB---
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
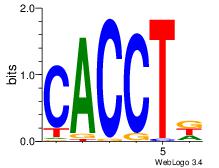
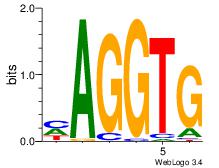
| Dataset #: 1 | Motif ID: 5 | Motif name: ZEB1 |
| Original motif | Reverse complement motif |
| Consensus sequence: CACCTD | Consensus sequence: HAGGTG |
 |
 |
Best Matches for Motif ID 5 (Highest to Lowest)
| Motif ID: | UP00068 |
| Motif name: | Eomes_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
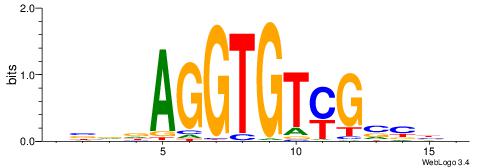
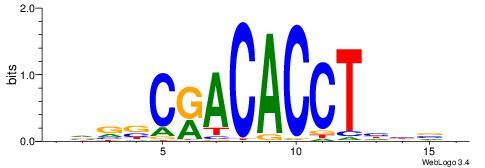
VVBVCKACACCTHVBV
-------CACCTD---
| Original motif | Reverse complement motif |
| Consensus sequence: VBVDAGGTGTYGVBBV | Consensus sequence: VVBVCKACACCTHVBV |
 |
 |
| Motif ID: | UP00199 |
| Motif name: | Six4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.00433068 |
Alignment:
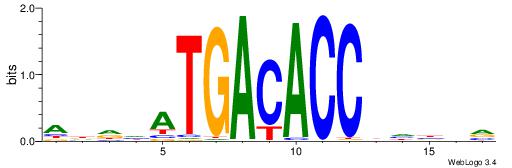
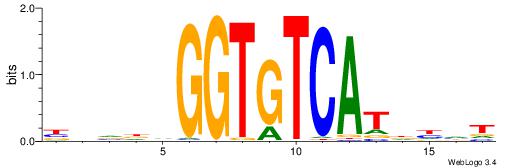
BBVHVGGTGTCATHDDT
---HAGGTG--------
| Original motif | Reverse complement motif |
| Consensus sequence: ADDHATGACACCBHBBV | Consensus sequence: BBVHVGGTGTCATHDDT |
 |
 |
| Motif ID: | UP00068 |
| Motif name: | Eomes_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 6 |
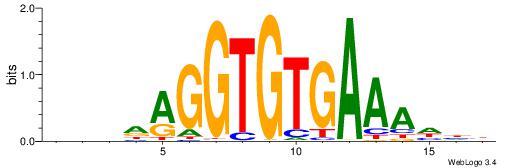
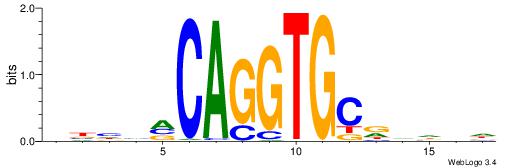
| Similarity score: | 0.00524081 |
Alignment:
HHHTTTCACACCTDHBV
--------CACCTD---
| Original motif | Reverse complement motif |
| Consensus sequence: BVHDAGGTGTGAAAHHH | Consensus sequence: HHHTTTCACACCTDHBV |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 6 |
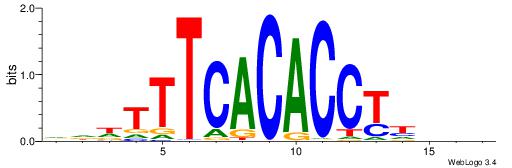
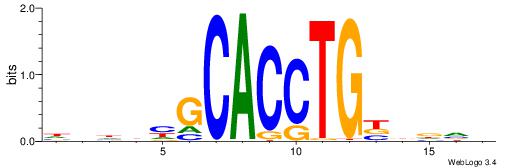
| Similarity score: | 0.00911301 |
Alignment:
VBHHVCAGGTGCDVDHD
-----HAGGTG------
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Motif ID: | UP00047 |
| Motif name: | Zbtb7b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.0233556 |
Alignment:
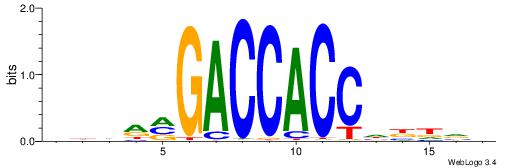
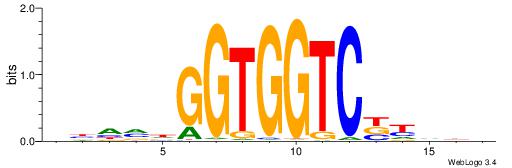
BDDRVGACCACCHBDVB
--------CACCTD---
| Original motif | Reverse complement motif |
| Consensus sequence: BDDRVGACCACCHBDVB | Consensus sequence: BBDVHGGTGGTCBKDDB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 11 |
| Number of overlap: | 6 |
| Similarity score: | 0.0233928 |
Alignment:
BHCBCBCCGGGTGGTCYHVHDCH
-------HAGGTG----------
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 12 |
| Number of overlap: | 6 |
| Similarity score: | 0.0254338 |
Alignment:
HTBVVVDGGACCACCCRGRDBG
-----------CACCTD-----
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 12 |
| Number of overlap: | 6 |
| Similarity score: | 0.0290143 |
Alignment:
HDGHHBHRGACCACCCGSVDVG
-----------CACCTD-----
| Original motif | Reverse complement motif |
| Consensus sequence: HDGHHBHRGACCACCCGSVDVG | Consensus sequence: CVHBSCGGGTGGTCMHVHHCDH |
 |
 |
| Motif ID: | UP00036 |
| Motif name: | Myf6_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0309673 |
Alignment:
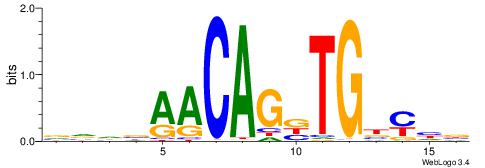
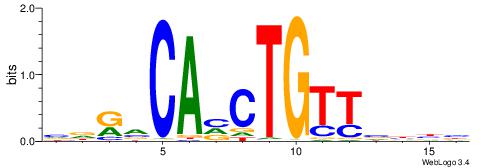
VVDBAACAGBTGBYHB
------HAGGTG----
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBAACAGBTGBYHB | Consensus sequence: BDKVCABCTGTTBDBV |
 |
 |
| Motif ID: | UP00249 |
| Motif name: | Nkx2-5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.031502 |
Alignment:
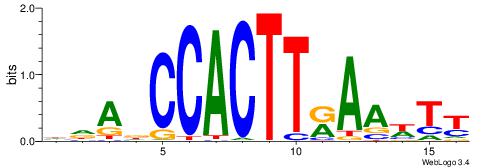
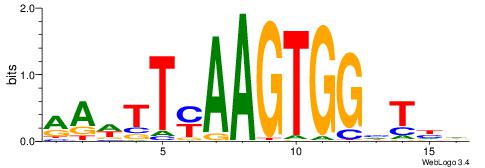
HDADCCACTTRAAWTT
-----CACCTD-----
| Original motif | Reverse complement motif |
| Consensus sequence: HDADCCACTTRAAWTT | Consensus sequence: AAWTTMAAGTGGHTDH |
 |
 |
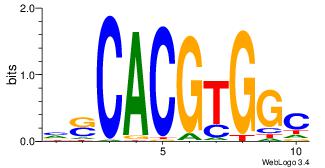
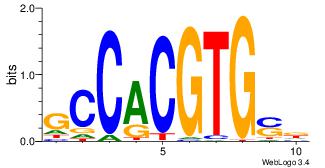
| Dataset #: 1 | Motif ID: 6 | Motif name: Mycn |
| Original motif | Reverse complement motif |
| Consensus sequence: HSCACGTGGC | Consensus sequence: GCCACGTGSD |
 |
 |
Best Matches for Motif ID 6 (Highest to Lowest)
| Motif ID: | UP00060 |
| Motif name: | Max_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
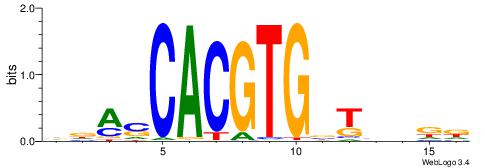
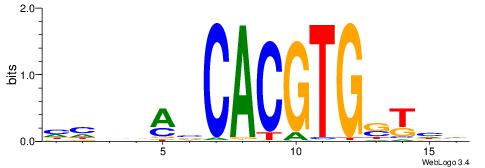
Alignment:
DDASCACGTGBTBVDD
--HSCACGTGGC----
| Original motif | Reverse complement motif |
| Consensus sequence: DDASCACGTGBTBVDD | Consensus sequence: HHVBABCACGTGSTHD |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.00783793 |
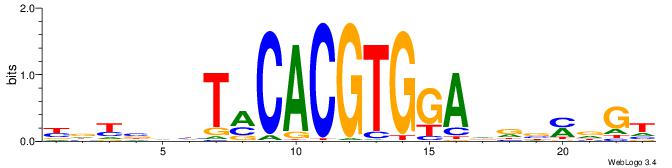
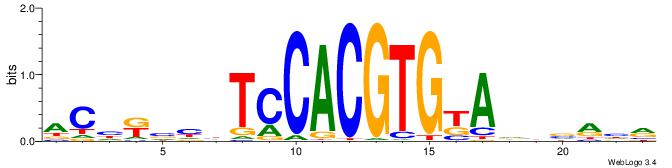
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
-------GCCACGTGSD------
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
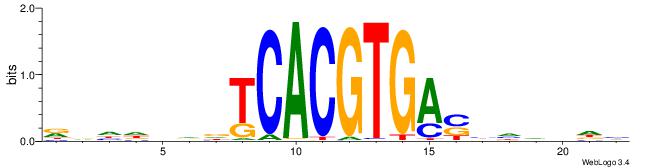
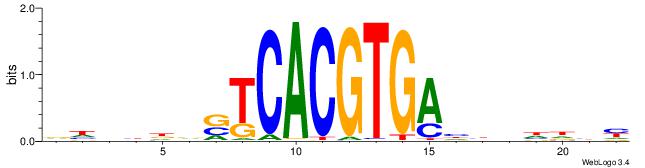
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0132858 |
Alignment:
DDHBHBSTCACGTGAHBBDHHM
------GCCACGTGSD------
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0227933 |
Alignment:
VBHHVCAGGTGCDVDHD
---GCCACGTGSD----
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
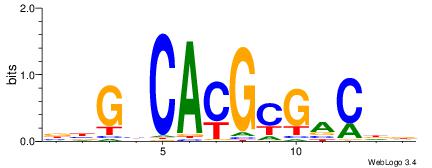
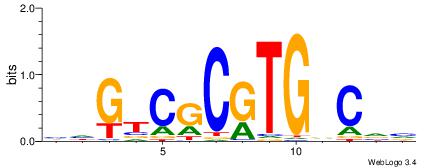
| Motif ID: | UP00060 |
| Motif name: | Max_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0257011 |
Alignment:
BBGHCACGCGACDD
--HSCACGTGGC--
| Original motif | Reverse complement motif |
| Consensus sequence: BBGHCACGCGACDD | Consensus sequence: HDGTCGCGTGDCVB |
 |
 |
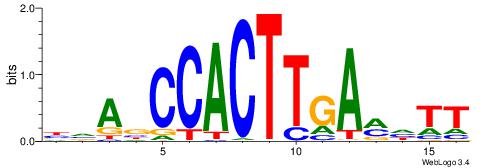
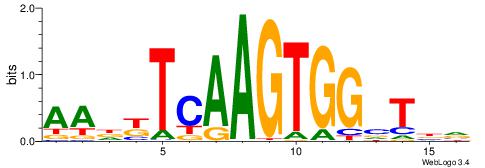
| Motif ID: | UP00165 |
| Motif name: | Titf1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0344833 |
Alignment:
BVABCCACTTGAMHTT
---GCCACGTGSD---
| Original motif | Reverse complement motif |
| Consensus sequence: BVABCCACTTGAMHTT | Consensus sequence: AAHYTCAAGTGGBTBV |
 |
 |
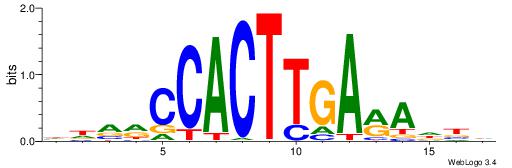
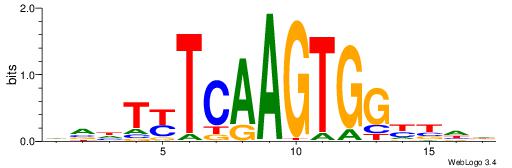
| Motif ID: | UP00231 |
| Motif name: | Nkx2-2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.035228 |
Alignment:
HDVRCCACTTGARADBD
---GCCACGTGSD----
| Original motif | Reverse complement motif |
| Consensus sequence: HDVRCCACTTGARADBD | Consensus sequence: DVDTKTCAAGTGGKBDH |
 |
 |
| Motif ID: | UP00084 |
| Motif name: | Gmeb1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0358581 |
Alignment:
BVVDVRYACGTRYVBHB
----GCCACGTGSD---
| Original motif | Reverse complement motif |
| Consensus sequence: BHBBKKACGTMMVDBVB | Consensus sequence: BVVDVRYACGTRYVBHB |
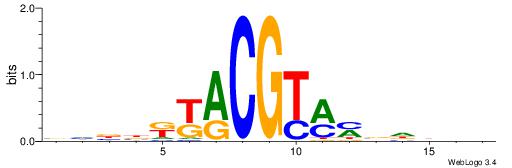 |
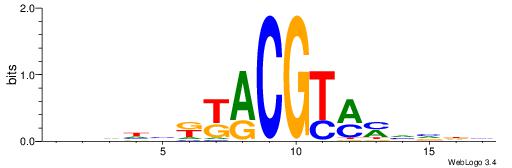 |
| Motif ID: | UP00084 |
| Motif name: | Gmeb1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.03725 |
Alignment:
HHMACKACGTMGBVCD
---HSCACGTGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: DGVBCRACGTYGTYHH | Consensus sequence: HHMACKACGTMGBVCD |
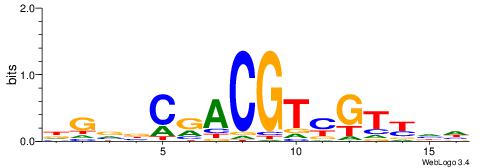 |
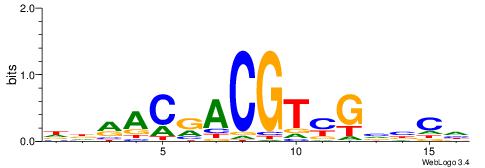 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0383187 |
Alignment:
WHVBVVMACGCGCGTCDYHWDH
------HSCACGTGGC------
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
 |
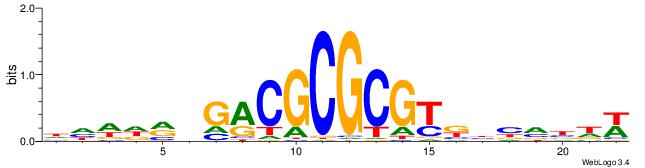 |
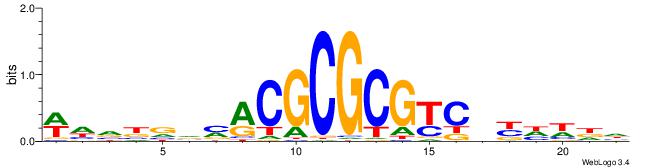
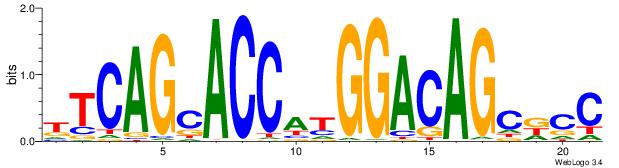
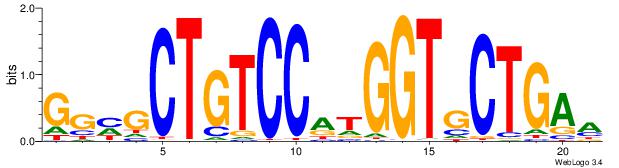
| Dataset #: 1 | Motif ID: 7 | Motif name: REST |
| Original motif | Reverse complement motif |
| Consensus sequence: TTCAGCACCATGGACAGCKCC | Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
 |
 |
Best Matches for Motif ID 7 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 21 |
| Similarity score: | 0.0699178 |
Alignment:
BBBDVVRGACCACCCAVGABBAB
--TTCAGCACCATGGACAGCKCC
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 21 |
| Similarity score: | 0.0734685 |
Alignment:
VMYDHDGMCCHCCKBGVVAAVH
GGYGCTGTCCATGGTGCTGAA-
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
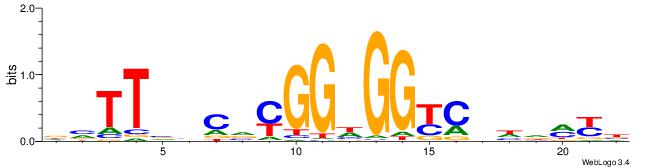 |
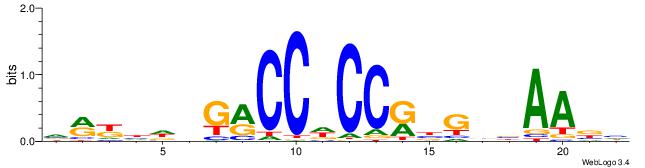 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 21 |
| Similarity score: | 0.0740652 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
-TTCAGCACCATGGACAGCKCC-
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
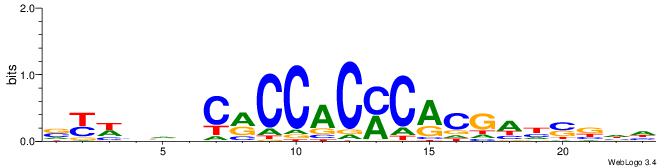 |
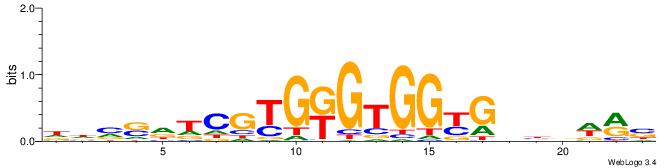 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 21 |
| Similarity score: | 0.0750152 |
Alignment:
MHHHWHYTMCCHCCCACVAABVH
--TTCAGCACCATGGACAGCKCC
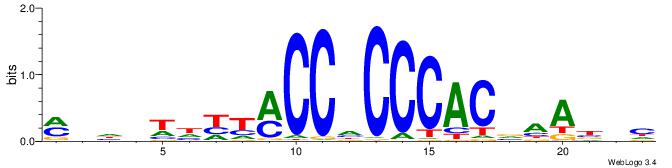
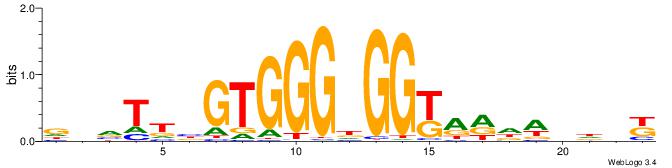
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 21 |
| Similarity score: | 0.0751449 |
Alignment:
BHCBCBCCGGGTGGTCYHVHDCH
GGYGCTGTCCATGGTGCTGAA--
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 21 |
| Similarity score: | 0.075832 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
-TTCAGCACCATGGACAGCKCC-
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 21 |
| Similarity score: | 0.0773446 |
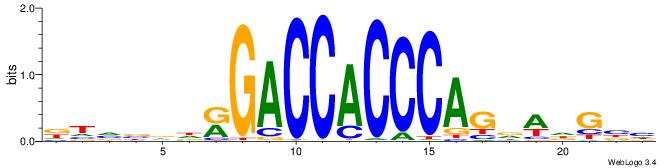
Alignment:
DTVBDDRGACCACCCAGDWDGBH
--TTCAGCACCATGGACAGCKCC
| Original motif | Reverse complement motif |
| Consensus sequence: DTVBDDRGACCACCCAGDWDGBH | Consensus sequence: DBCDWHCTGGGTGGTCMDHBBAH |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 21 |
| Similarity score: | 0.0775367 |
Alignment:
YDYBDHTMCACGTGGADDBMDGT
--TTCAGCACCATGGACAGCKCC
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 21 |
| Similarity score: | 0.077977 |
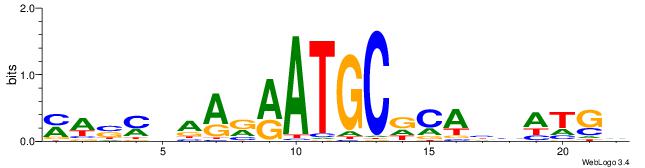
Alignment:
DCATDBTGHGCATKMTTBRSWR
-GGYGCTGTCCATGGTGCTGAA
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
 |
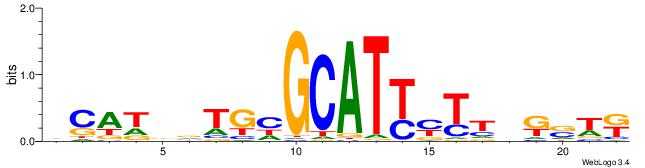 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 21 |
| Similarity score: | 0.0785969 |
Alignment:
BADHBDHCGCCCMCGCAHHDBBV
--GGYGCTGTCCATGGTGCTGAA
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
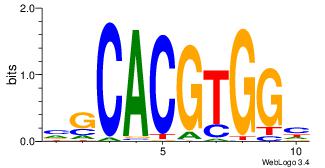
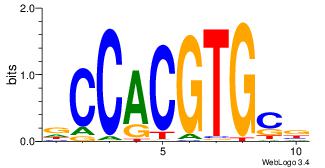
| Dataset #: 1 | Motif ID: 8 | Motif name: Myc |
| Original motif | Reverse complement motif |
| Consensus sequence: VGCACGTGGH | Consensus sequence: DCCACGTGCV |
 |
 |
Best Matches for Motif ID 8 (Highest to Lowest)
| Motif ID: | UP00060 |
| Motif name: | Max_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
DDASCACGTGBTBVDD
--VGCACGTGGH----
| Original motif | Reverse complement motif |
| Consensus sequence: DDASCACGTGBTBVDD | Consensus sequence: HHVBABCACGTGSTHD |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.00631885 |
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
-------DCCACGTGCV------
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.00821668 |
Alignment:
DDHBHBSTCACGTGAHBBDHHM
------DCCACGTGCV------
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0140005 |
Alignment:
DHDBHGCACCTGBDDVB
----VGCACGTGGH---
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0308912 |
Alignment:
BBGHCACGCGACDD
--VGCACGTGGH--
| Original motif | Reverse complement motif |
| Consensus sequence: BBGHCACGCGACDD | Consensus sequence: HDGTCGCGTGDCVB |
 |
 |
| Motif ID: | UP00165 |
| Motif name: | Titf1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0311723 |
Alignment:
BVABCCACTTGAMHTT
---DCCACGTGCV---
| Original motif | Reverse complement motif |
| Consensus sequence: BVABCCACTTGAMHTT | Consensus sequence: AAHYTCAAGTGGBTBV |
 |
 |
| Motif ID: | UP00249 |
| Motif name: | Nkx2-5 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.031308 |
Alignment:
HDADCCACTTRAAWTT
---DCCACGTGCV---
| Original motif | Reverse complement motif |
| Consensus sequence: HDADCCACTTRAAWTT | Consensus sequence: AAWTTMAAGTGGHTDH |
 |
 |
| Motif ID: | UP00231 |
| Motif name: | Nkx2-2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0334845 |
Alignment:
HDVRCCACTTGARADBD
---DCCACGTGCV----
| Original motif | Reverse complement motif |
| Consensus sequence: HDVRCCACTTGARADBD | Consensus sequence: DVDTKTCAAGTGGKBDH |
 |
 |
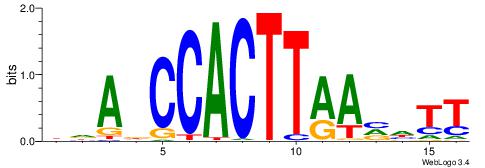
| Motif ID: | UP00147 |
| Motif name: | Nkx2-6 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0345548 |
Alignment:
DDADCCACTTAAVHTT
---DCCACGTGCV---
| Original motif | Reverse complement motif |
| Consensus sequence: DDADCCACTTAAVHTT | Consensus sequence: AAHVTTAAGTGGHTDD |
 |
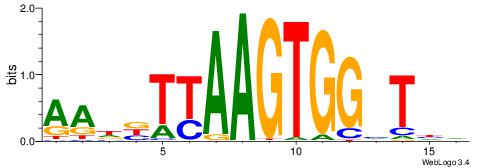 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0363818 |
Alignment:
VDWDARRGACGCGGWHGGAKHV
-----VGCACGTGGH-------
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
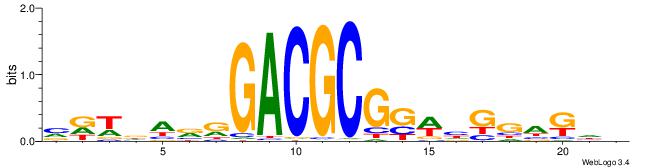 |
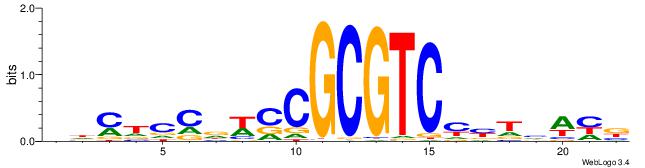 |
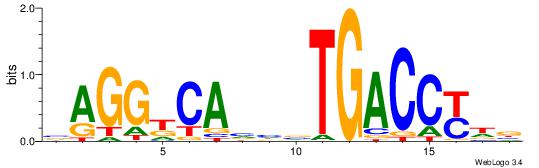
| Dataset #: 1 | Motif ID: 9 | Motif name: ESR2 |
| Original motif | Reverse complement motif |
| Consensus sequence: VHRGGTCABDBTGMCCTB | Consensus sequence: BAGGYCABHBTGACCKHV |
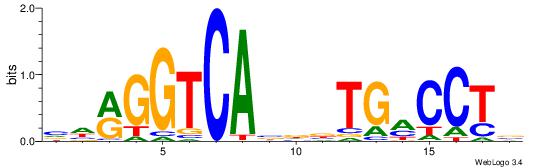 |
 |
Best Matches for Motif ID 9 (Highest to Lowest)
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0443894 |
Alignment:
GTVBDAATTGTVTGHGDDHKVB
---VHRGGTCABDBTGMCCTB-
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.049219 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
---VHRGGTCABDBTGMCCTB--
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0509182 |
Alignment:
DDHBHBSTCACGTGAHBBDHHM
--VHRGGTCABDBTGMCCTB--
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.0527075 |
Alignment:
DVTTVVCVYGGHGGYCHHDMYB
-BAGGYCABHBTGACCKHV---
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
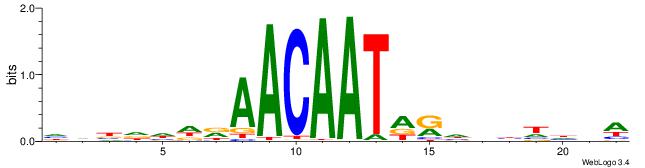
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.0541044 |
Alignment:
WBHHHHHMDATTGTTHDHDVHH
-BAGGYCABHBTGACCKHV---
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
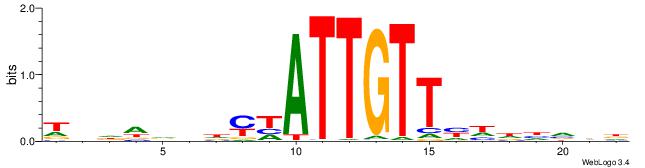 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0541108 |
Alignment:
VCTBHBVCTTGGWTACVVVVVDB
--BAGGYCABHBTGACCKHV---
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.0543733 |
Alignment:
VDWDARRGACGCGGWHGGAKHV
---VHRGGTCABDBTGMCCTB-
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.0548087 |
Alignment:
BHCBCBCCGGGTGGTCYHVHDCH
-BAGGYCABHBTGACCKHV----
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.055453 |
Alignment:
DCATDBTGHGCATKMTTBRSWR
----VHRGGTCABDBTGMCCTB
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
 |
 |
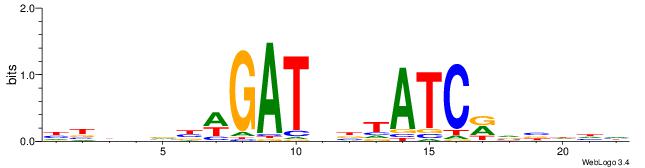
| Motif ID: | UP00032 |
| Motif name: | Gata3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0566426 |
Alignment:
HBHDVBWGATHBTATCRVBHHH
-VHRGGTCABDBTGMCCTB---
| Original motif | Reverse complement motif |
| Consensus sequence: HBHDVBWGATHBTATCRVBHHH | Consensus sequence: HHHBBMGATAVHATCWVVDHVH |
 |
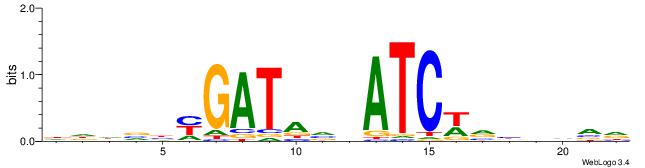 |
| Dataset #: 1 | Motif ID: 10 | Motif name: NR1H2RXRA |
| Original motif | Reverse complement motif |
| Consensus sequence: AAAGGTCAAAGGTCAAC | Consensus sequence: GTTGACCTTTGACCTTT |
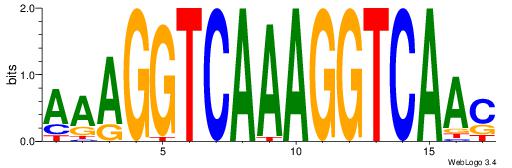 |
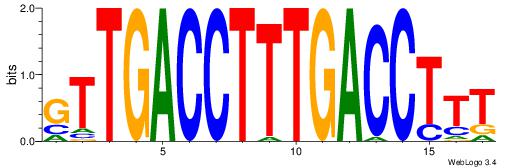 |
Best Matches for Motif ID 10 (Highest to Lowest)
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0538452 |
Alignment:
DVMHHDHKGACCCTCCTSVCBH
-----GTTGACCTTTGACCTTT
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0545098 |
Alignment:
BDHDDADAGATAAGADHBDHVD
-----AAAGGTCAAAGGTCAAC
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
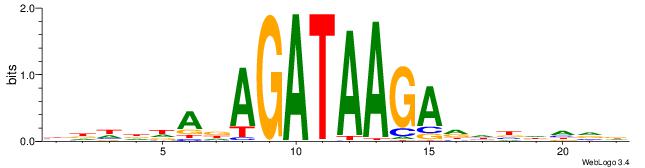 |
 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.521114 |
Alignment:
VDDAGATCAAAGGKDDD-
-AAAGGTCAAAGGTCAAC
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
 |
 |
| Motif ID: | UP00083 |
| Motif name: | Tcf7l2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.527825 |
Alignment:
BDDASATCAAAGGMDDD-
-GTTGACCTTTGACCTTT
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATSTDDV | Consensus sequence: BDDASATCAAAGGMDDD |
 |
 |
| Motif ID: | UP00054 |
| Motif name: | Tcf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.531154 |
Alignment:
BHHASATCAAAGGAHHH-
-AAAGGTCAAAGGTCAAC
| Original motif | Reverse complement motif |
| Consensus sequence: BHHASATCAAAGGAHHH | Consensus sequence: HHHTCCTTTGATSTHHV |
 |
 |
| Motif ID: | UP00058 |
| Motif name: | Tcf3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.531876 |
Alignment:
-HHHASATCAAAGVRHHH
AAAGGTCAAAGGTCAAC-
| Original motif | Reverse complement motif |
| Consensus sequence: HHHASATCAAAGVRHHH | Consensus sequence: HHHKVCTTTGATSTHHH |
 |
 |
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.544403 |
Alignment:
HBDDRAACAAAGRABHH-
-AAAGGTCAAAGGTCAAC
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
 |
 |
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.544565 |
Alignment:
-HHBBMCTTTGTTHDDBH
GTTGACCTTTGACCTTT-
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
 |
 |
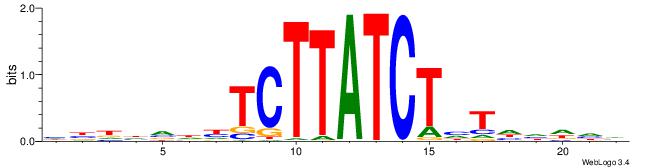
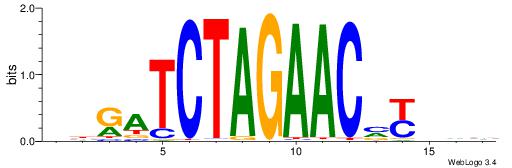
| Motif ID: | UP00019 |
| Motif name: | Zbtb12_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.550511 |
Alignment:
-BBGATCTAGAACBKHDB
GTTGACCTTTGACCTTT-
| Original motif | Reverse complement motif |
| Consensus sequence: BDHRBGTTCTAGATCVB | Consensus sequence: BBGATCTAGAACBKHDB |
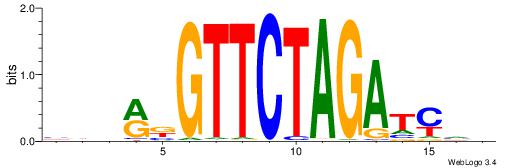 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.552064 |
Alignment:
-HVDDVCAGATGKYHBBV
AAAGGTCAAAGGTCAAC-
| Original motif | Reverse complement motif |
| Consensus sequence: HVDDVCAGATGKYHBBV | Consensus sequence: VBBDMYCATCTGVHHBH |
 |
 |
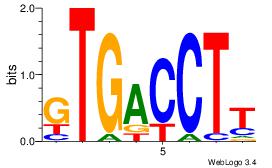
| Dataset #: 1 | Motif ID: 11 | Motif name: NR4A2 |
| Original motif | Reverse complement motif |
| Consensus sequence: AAGGTCAC | Consensus sequence: GTGACCTT |
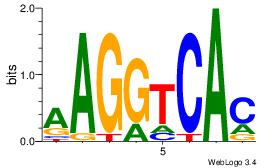 |
 |
Best Matches for Motif ID 11 (Highest to Lowest)
| Motif ID: | UP00048 |
| Motif name: | Rara |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HHVGTGACCTTTDVDD
---GTGACCTT-----
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCACVHD | Consensus sequence: HHVGTGACCTTTDVDD |
 |
 |
| Motif ID: | UP00009 |
| Motif name: | Nr2f2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0074527 |
Alignment:
BHVDTGACCTTTDVDD
---GTGACCTT-----
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCAHVHB | Consensus sequence: BHVDTGACCTTTDVDD |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.01542 |
Alignment:
DHDBCAAGGTCAHVBDH
-----AAGGTCAC----
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBCAAGGTCAHVBDH | Consensus sequence: HHBVHTGACCTTGVDHD |
 |
 |
| Motif ID: | UP00009 |
| Motif name: | Nr2f2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0395644 |
Alignment:
VBVBCSGGGTCAVBBH
-----AAGGTCAC---
| Original motif | Reverse complement motif |
| Consensus sequence: VBVBCSGGGTCAVBBH | Consensus sequence: HVBVTGACCCSGBVBV |
 |
 |
| Motif ID: | UP00082 |
| Motif name: | Zfp187_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.0407874 |
Alignment:
DDGMCCTBGTCCHYHV
GTGACCTT--------
| Original motif | Reverse complement motif |
| Consensus sequence: DDGMCCTBGTCCHYHV | Consensus sequence: VHMDGGACVAGGRCDH |
 |
 |
| Motif ID: | UP00053 |
| Motif name: | Rxra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0422933 |
Alignment:
DBHBDTGACCCCDHVHB
----GTGACCTT-----
| Original motif | Reverse complement motif |
| Consensus sequence: DBHBDTGACCCCDHVHB | Consensus sequence: VHBHDGGGGTCAHBHBD |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0447444 |
Alignment:
VKBBAGGGGTCAHDBBH
-----AAGGTCAC----
| Original motif | Reverse complement motif |
| Consensus sequence: VKBBAGGGGTCAHDBBH | Consensus sequence: DBBHHTGACCCCTBBYV |
 |
 |
| Motif ID: | UP00066 |
| Motif name: | Hnf4a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0481418 |
Alignment:
BBHHAAAGTCCAMTHH
-----AAGGTCAC---
| Original motif | Reverse complement motif |
| Consensus sequence: BBHHAAAGTCCAMTHH | Consensus sequence: HHAYTGGACTTTHDBV |
 |
 |
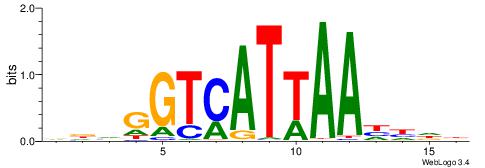
| Motif ID: | UP00197 |
| Motif name: | Hoxc9 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0497679 |
Alignment:
VDHHTTAATGACCBHV
-------GTGACCTT-
| Original motif | Reverse complement motif |
| Consensus sequence: VDVGGTCATTAAHHDB | Consensus sequence: VDHHTTAATGACCBHV |
 |
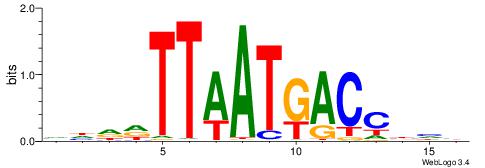 |
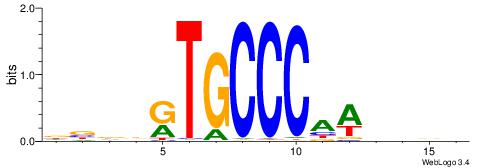
| Motif ID: | UP00035 |
| Motif name: | Hic1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0500062 |
Alignment:
DBDDRTGCCCAWDHDV
----GTGACCTT----
| Original motif | Reverse complement motif |
| Consensus sequence: DBDDRTGCCCAWDHDV | Consensus sequence: VHHDWTGGGCAMDHBH |
 |
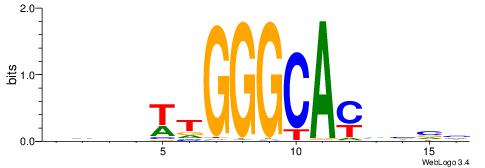 |
| Dataset #: 1 | Motif ID: 12 | Motif name: RORA_1 |
| Original motif | Reverse complement motif |
| Consensus sequence: AWVDAGGTCA | Consensus sequence: TGACCTDVWT |
 |
 |
Best Matches for Motif ID 12 (Highest to Lowest)
| Motif ID: | UP00009 |
| Motif name: | Nr2f2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0154749 |
Alignment:
DHBHAAAGGTCAHVHB
--AWVDAGGTCA----
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCAHVHB | Consensus sequence: BHVDTGACCTTTDVDD |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0176661 |
Alignment:
HHBVHTGACCTTGVDHD
-----TGACCTDVWT--
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBCAAGGTCAHVBDH | Consensus sequence: HHBVHTGACCTTGVDHD |
 |
 |
| Motif ID: | UP00048 |
| Motif name: | Rara |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0189599 |
Alignment:
DHBHAAAGGTCACVHD
--AWVDAGGTCA----
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCACVHD | Consensus sequence: HHVGTGACCTTTDVDD |
 |
 |
| Motif ID: | UP00009 |
| Motif name: | Nr2f2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0331999 |
Alignment:
VBVBCSGGGTCAVBBH
--AWVDAGGTCA----
| Original motif | Reverse complement motif |
| Consensus sequence: VBVBCSGGGTCAVBBH | Consensus sequence: HVBVTGACCCSGBVBV |
 |
 |
| Motif ID: | UP00066 |
| Motif name: | Hnf4a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0341771 |
Alignment:
BDBVMGGGGTCAABHDV
--AWVDAGGTCA-----
| Original motif | Reverse complement motif |
| Consensus sequence: BDBVMGGGGTCAABHDV | Consensus sequence: BHHVTTGACCCCYVVDB |
 |
 |
| Motif ID: | UP00053 |
| Motif name: | Rxra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0363566 |
Alignment:
DBHBDTGACCCCDHVHB
-----TGACCTDVWT--
| Original motif | Reverse complement motif |
| Consensus sequence: DBHBDTGACCCCDHVHB | Consensus sequence: VHBHDGGGGTCAHBHBD |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0391284 |
Alignment:
VKBBAGGGGTCAHDBBH
--AWVDAGGTCA-----
| Original motif | Reverse complement motif |
| Consensus sequence: VKBBAGGGGTCAHDBBH | Consensus sequence: DBBHHTGACCCCTBBYV |
 |
 |
| Motif ID: | UP00035 |
| Motif name: | Hic1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0444205 |
Alignment:
DBDDRTGCCCAWDHDV
-----TGACCTDVWT-
| Original motif | Reverse complement motif |
| Consensus sequence: DBDDRTGCCCAWDHDV | Consensus sequence: VHHDWTGGGCAMDHBH |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0461434 |
Alignment:
DDHBHBSTCACGTGAHBBDHHM
AWVDAGGTCA------------
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0489571 |
Alignment:
BHCBCBCCGGGTGGTCYHVHDCH
-----------TGACCTDVWT--
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
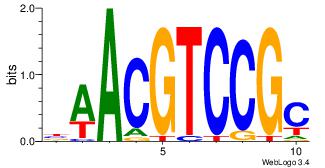
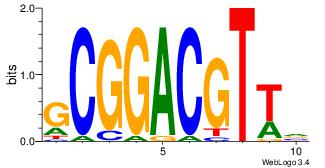
| Dataset #: 1 | Motif ID: 13 | Motif name: MIZF |
| Original motif | Reverse complement motif |
| Consensus sequence: BAACGTCCGC | Consensus sequence: GCGGACGTTV |
 |
 |
Best Matches for Motif ID 13 (Highest to Lowest)
| Motif ID: | UP00419 |
| Motif name: | Spic |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
VAAAGMGGAAGTWD
BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: VAAAGMGGAAGTWD | Consensus sequence: DWACTTCCRCTTTB |
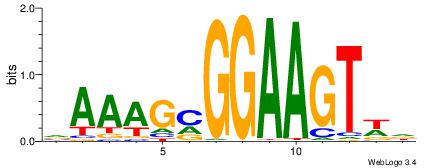 |
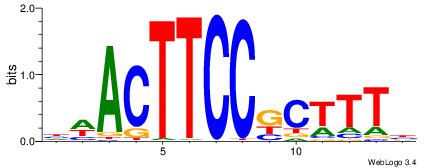 |
| Motif ID: | UP00409 |
| Motif name: | Elf5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.00379999 |
Alignment:
DWACTTCCRSDTWV
BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: BWAHSMGGAAGTWD | Consensus sequence: DWACTTCCRSDTWV |
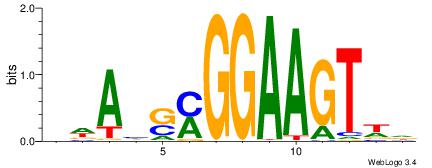 |
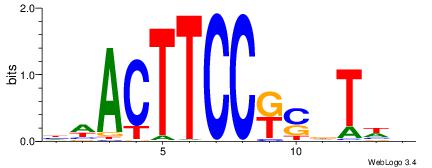 |
| Motif ID: | UP00413 |
| Motif name: | Elf4 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0124101 |
Alignment:
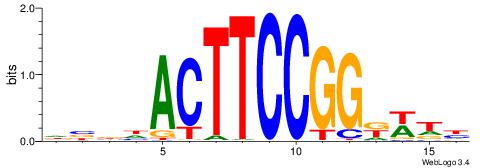
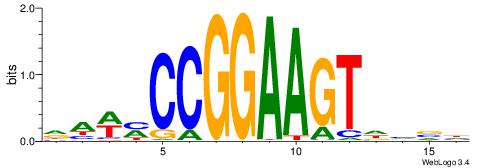
DBBHACTTCCGGKWTH
--BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGKWTH | Consensus sequence: HAWYCCGGAAGTHBBD |
 |
 |
| Motif ID: | UP00404 |
| Motif name: | Elf2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0128348 |
Alignment:
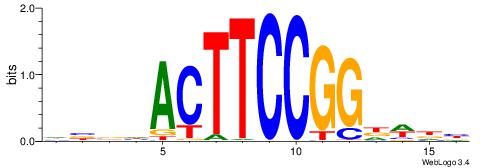
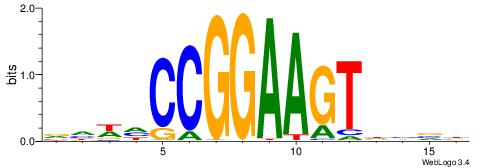
DBBHACTTCCGGDWDB
--BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGDWDB | Consensus sequence: BDWDCCGGAAGTHBBD |
 |
 |
| Motif ID: | UP00085 |
| Motif name: | Sfpi1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0141516 |
Alignment:
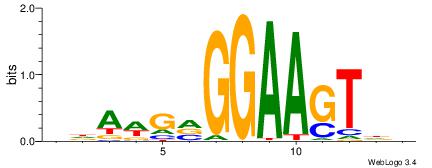
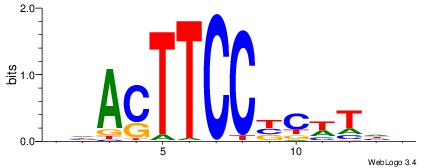
BDAWGVGGAAGTDH
BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: BDAWGVGGAAGTDH | Consensus sequence: HDACTTCCBCWTDV |
 |
 |
| Motif ID: | UP00423 |
| Motif name: | Gm5454 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0144665 |
Alignment:
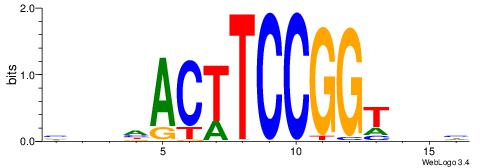
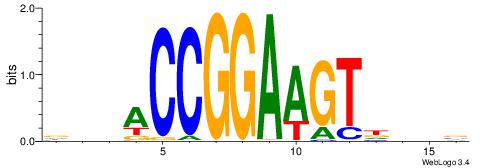
DVHACCGGAAGTHDDV
----GCGGACGTTV--
| Original motif | Reverse complement motif |
| Consensus sequence: VDHHACTTCCGGTHVH | Consensus sequence: DVHACCGGAAGTHDDV |
 |
 |
| Motif ID: | UP00412 |
| Motif name: | Etv5 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0155825 |
Alignment:
DHHACCGGAWGTTDDV
----GCGGACGTTV--
| Original motif | Reverse complement motif |
| Consensus sequence: VDHAACWTCCGGTHHH | Consensus sequence: DHHACCGGAWGTTDDV |
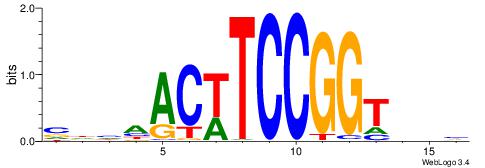 |
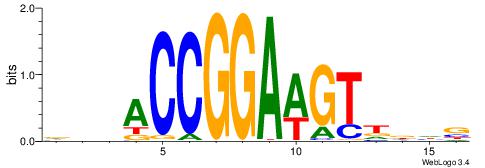 |
| Motif ID: | UP00015 |
| Motif name: | Ehf_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0162018 |
Alignment:
VBDABCCGGAAGTDD
-----GCGGACGTTV
| Original motif | Reverse complement motif |
| Consensus sequence: VBDABCCGGAAGTDD | Consensus sequence: DDACTTCCGGBTHBB |
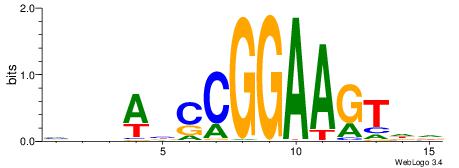 |
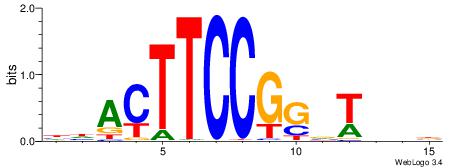 |
| Motif ID: | UP00418 |
| Motif name: | Etv6 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0171234 |
Alignment:
DVHBTACTTCCGGHTHB
---BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: DVHBTACTTCCGGHTHB | Consensus sequence: BHAHCCGGAAGTABDVD |
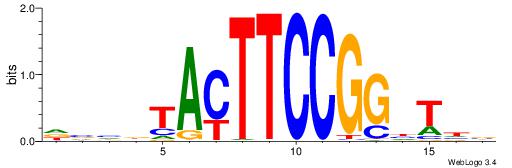 |
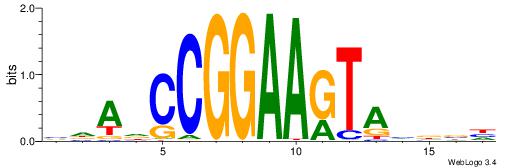 |
| Motif ID: | UP00421 |
| Motif name: | Etv1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0184147 |
Alignment:
MVHBACCGGAAGTVDVH
-----GCGGACGTTV--
| Original motif | Reverse complement motif |
| Consensus sequence: MVHBACCGGAAGTVDVH | Consensus sequence: HBDBACTTCCGGTBHVY |
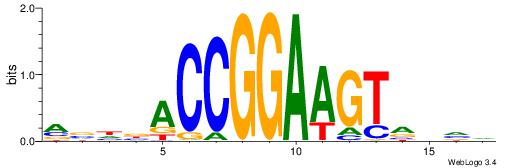 |
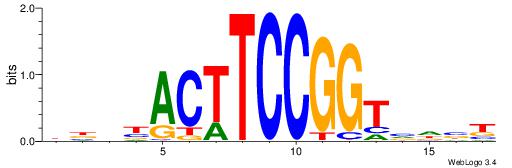 |
| Dataset #: 1 | Motif ID: 14 | Motif name: PPARGRXRA |
| Original motif | Reverse complement motif |
| Consensus sequence: BTRGGDCARAGGKCA | Consensus sequence: TGRCCTKTGHCCKAB |
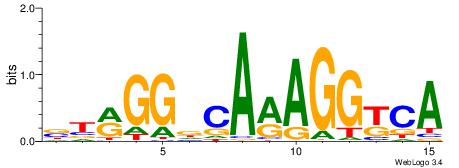 |
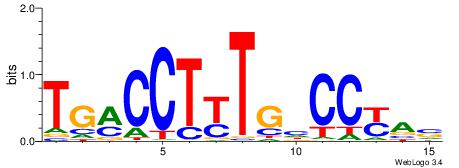 |
Best Matches for Motif ID 14 (Highest to Lowest)
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0304954 |
Alignment:
VDDAGATCAAAGGKDDD
-BTRGGDCARAGGKCA-
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
 |
 |
| Motif ID: | UP00058 |
| Motif name: | Tcf3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0319475 |
Alignment:
HHHASATCAAAGVRHHH
-BTRGGDCARAGGKCA-
| Original motif | Reverse complement motif |
| Consensus sequence: HHHASATCAAAGVRHHH | Consensus sequence: HHHKVCTTTGATSTHHH |
 |
 |
| Motif ID: | UP00054 |
| Motif name: | Tcf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.032779 |
Alignment:
BHHASATCAAAGGAHHH
-BTRGGDCARAGGKCA-
| Original motif | Reverse complement motif |
| Consensus sequence: BHHASATCAAAGGAHHH | Consensus sequence: HHHTCCTTTGATSTHHV |
 |
 |
| Motif ID: | UP00082 |
| Motif name: | Zfp187_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0334185 |
Alignment:
VHMDGGACVAGGRCDH
BTRGGDCARAGGKCA-
| Original motif | Reverse complement motif |
| Consensus sequence: DDGMCCTBGTCCHYHV | Consensus sequence: VHMDGGACVAGGRCDH |
 |
 |
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0352539 |
Alignment:
HBDDDAACAAAGRVBHH
-BTRGGDCARAGGKCA-
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
 |
 |
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0358782 |
Alignment:
HBDDRAACAAAGRABHH
-BTRGGDCARAGGKCA-
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
 |
 |
| Motif ID: | UP00101 |
| Motif name: | Sox12_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0374729 |
Alignment:
HDTMCTTTGTSTVDBB
TGRCCTKTGHCCKAB-
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBASACAAAGRADH | Consensus sequence: HDTMCTTTGTSTVDBB |
 |
 |
| Motif ID: | UP00083 |
| Motif name: | Tcf7l2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0379757 |
Alignment:
BDDASATCAAAGGMDDD
-BTRGGDCARAGGKCA-
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATSTDDV | Consensus sequence: BDDASATCAAAGGMDDD |
 |
 |
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0382733 |
Alignment:
DHHDGGGCGRGGKHBH
BTRGGDCARAGGKCA-
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
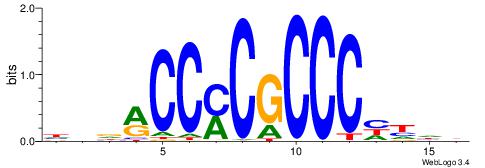 |
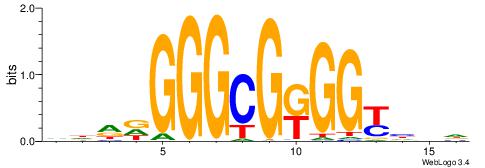 |
| Motif ID: | UP00055 |
| Motif name: | Hbp1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 15 |
| Similarity score: | 0.0397168 |
Alignment:
HBTHVACAATGGGMDHD
BTRGGDCARAGGKCA--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCCATTGTVHABH | Consensus sequence: HBTHVACAATGGGMDHD |
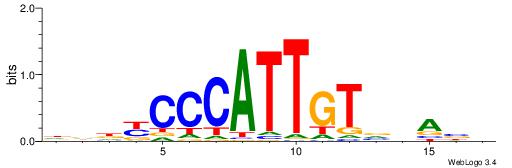 |
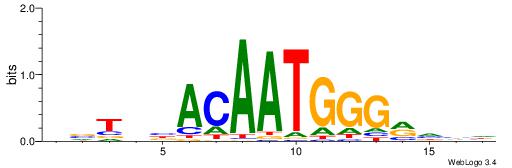 |
| Dataset #: 1 | Motif ID: 15 | Motif name: Tcfcp2l1 |
| Original motif | Reverse complement motif |
| Consensus sequence: CCAGYYHVADCCRG | Consensus sequence: CKGGDTBDMMCTGG |
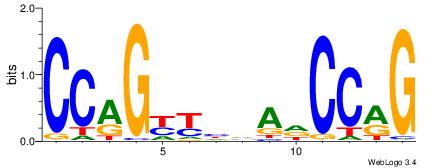 |
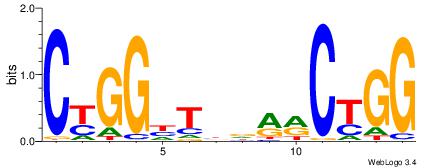 |
Best Matches for Motif ID 15 (Highest to Lowest)
| Motif ID: | UP00032 |
| Motif name: | Gata3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 14 |
| Similarity score: | 0.0527249 |
Alignment:
HBHDVBWGATHBTATCRVBHHH
----CCAGYYHVADCCRG----
| Original motif | Reverse complement motif |
| Consensus sequence: HBHDVBWGATHBTATCRVBHHH | Consensus sequence: HHHBBMGATAVHATCWVVDHVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0551691 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
--------CCAGYYHVADCCRG
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00005 |
| Motif name: | Tcfap2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0560983 |
Alignment:
HHRCCBBWGGDCDB
CCAGYYHVADCCRG
| Original motif | Reverse complement motif |
| Consensus sequence: HHRCCBBWGGDCDB | Consensus sequence: BDGHCCWBVGGKDH |
 |
 |
| Motif ID: | UP00013 |
| Motif name: | Gabpa_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0566467 |
Alignment:
CYDBYWTCCSMHBVVH
CCAGYYHVADCCRG--
| Original motif | Reverse complement motif |
| Consensus sequence: CYDBYWTCCSMHBVVH | Consensus sequence: DBVVDRSGGAWKVHKG |
 |
 |
| Motif ID: | UP00075 |
| Motif name: | Sox15_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0577636 |
Alignment:
BBVAATGDMATTHVA
-CCAGYYHVADCCRG
| Original motif | Reverse complement motif |
| Consensus sequence: BBVAATGDMATTHVA | Consensus sequence: TVDAATYDCATTVVV |
 |
 |
| Motif ID: | UP00096 |
| Motif name: | Sox13_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0587893 |
Alignment:
DDDHTGGGTGGGHAHHD
---CKGGDTBDMMCTGG
| Original motif | Reverse complement motif |
| Consensus sequence: DDDHTGGGTGGGHAHHD | Consensus sequence: DHHTHCCCACCCAHDDH |
 |
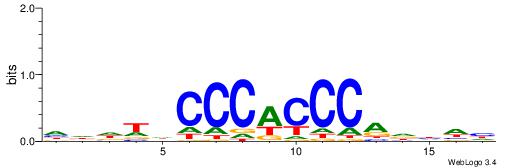 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0590238 |
Alignment:
VVYDDDCHCAVACAATTDBVAC
--CCAGYYHVADCCRG------
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 14 |
| Similarity score: | 0.0596904 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
--------CCAGYYHVADCCRG-
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0597695 |
Alignment:
BBBDVVRGACCACCCAVGABBAB
---------CCAGYYHVADCCRG
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0598357 |
Alignment:
HVGVSAGGAGGGTCYHHHHYVH
-------CKGGDTBDMMCTGG-
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Dataset #: 1 | Motif ID: 16 | Motif name: CTCF |
| Original motif | Reverse complement motif |
| Consensus sequence: YDRCCASYAGRKGGCRSYV | Consensus sequence: BMSMGCCYMCTKSTGGMHM |
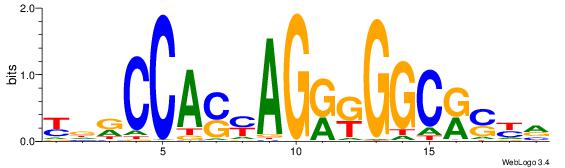 |
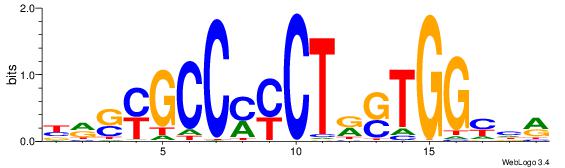 |
Best Matches for Motif ID 16 (Highest to Lowest)
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 19 |
| Similarity score: | 0.0409369 |
Alignment:
BADHBDHCGCCCMCGCAHHDBBV
----BMSMGCCYMCTKSTGGMHM
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 19 |
| Similarity score: | 0.0409815 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
-YDRCCASYAGRKGGCRSYV---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 19 |
| Similarity score: | 0.0431951 |
Alignment:
VMYDHDGMCCHCCKBGVVAAVH
---BMSMGCCYMCTKSTGGMHM
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 19 |
| Similarity score: | 0.0469794 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
---BMSMGCCYMCTKSTGGMHM
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 19 |
| Similarity score: | 0.0482204 |
Alignment:
BHCBCBCCGGGTGGTCYHVHDCH
YDRCCASYAGRKGGCRSYV----
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 19 |
| Similarity score: | 0.0491604 |
Alignment:
HHBSHTCGTGRGTGGKGDBVWMV
-YDRCCASYAGRKGGCRSYV---
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 19 |
| Similarity score: | 0.0511535 |
Alignment:
BBBDVVRGACCACCCAVGABBAB
---BMSMGCCYMCTKSTGGMHM-
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 19 |
| Similarity score: | 0.0520741 |
Alignment:
HVGVSAGGAGGGTCYHHHHYVH
YDRCCASYAGRKGGCRSYV---
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 19 |
| Similarity score: | 0.0532334 |
Alignment:
MHHHWHYTMCCHCCCACVAABVH
YDRCCASYAGRKGGCRSYV----
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 19 |
| Similarity score: | 0.0535675 |
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
--BMSMGCCYMCTKSTGGMHM--
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Dataset #: 2 | Motif ID: 17 | Motif name: TgTGgTGTCACAGTGCTCC |
| Original motif | Reverse complement motif |
| Consensus sequence: TGTGGTGTCACAGTGCTCC | Consensus sequence: GGAGCACTGTGACACCACA |
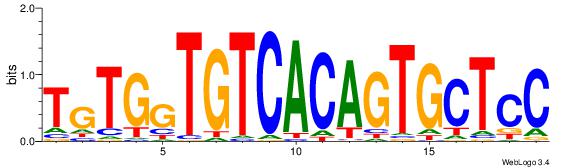 |
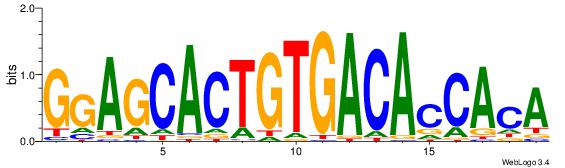 |
Best Matches for Motif ID 17 (Highest to Lowest)
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 19 |
| Similarity score: | 0.043204 |
Alignment:
BDBBVBVGTAWCCAAGVBHBAGB
---TGTGGTGTCACAGTGCTCC-
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 18 |
| Similarity score: | 0.553914 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB-
-----TGTGGTGTCACAGTGCTCC
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00192 |
| Motif name: | Six1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 1.04528 |
Alignment:
--AVHDHTGATACCCMDHH
GGAGCACTGTGACACCACA
| Original motif | Reverse complement motif |
| Consensus sequence: DHDRGGGTATCAHDHBT | Consensus sequence: AVHDHTGATACCCMDHH |
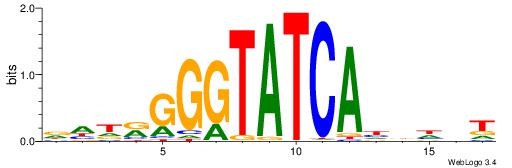 |
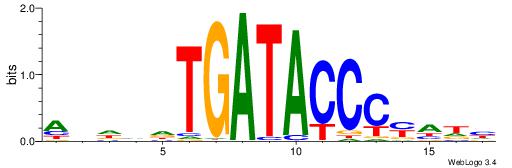 |
| Motif ID: | UP00193 |
| Motif name: | Rhox11_2205.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 1.0459 |
Alignment:
--WVVHWTWACAGCGHHVT
TGTGGTGTCACAGTGCTCC
| Original motif | Reverse complement motif |
| Consensus sequence: AVDHCGCTGTWAWDVVW | Consensus sequence: WVVHWTWACAGCGHHVT |
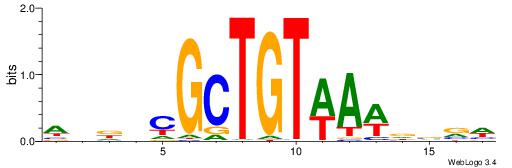 |
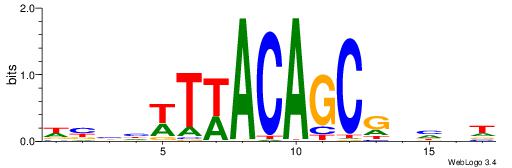 |
| Motif ID: | UP00193 |
| Motif name: | Rhox11_1765.2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 1.0462 |
Alignment:
AVDVCGCTGTWAWDVVW--
--TGTGGTGTCACAGTGCTCC
| Original motif | Reverse complement motif |
| Consensus sequence: AVDVCGCTGTWAWDVVW | Consensus sequence: WVVHWTWACAGCGBHBT |
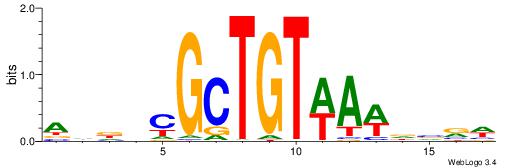 |
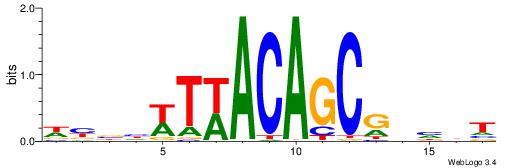 |
| Motif ID: | UP00068 |
| Motif name: | Eomes_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 1.04631 |
Alignment:
--HHHTTTCACACCTDHBV
GGAGCACTGTGACACCACA--
| Original motif | Reverse complement motif |
| Consensus sequence: BVHDAGGTGTGAAAHHH | Consensus sequence: HHHTTTCACACCTDHBV |
 |
 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 1.04807 |
Alignment:
--VDDAGATCAAAGGKDDD
GGAGCACTGTGACACCACA-
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 1.04831 |
Alignment:
YBVDMGTGGGTGGTCKVVBVBBT--
--GGAGCACTGTGACACCACA----
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Motif ID: | UP00053 |
| Motif name: | Rxra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 1.04867 |
Alignment:
--DBHBDTGACCCCDHVHB
TGTGGTGTCACAGTGCTCC--
| Original motif | Reverse complement motif |
| Consensus sequence: DBHBDTGACCCCDHVHB | Consensus sequence: VHBHDGGGGTCAHBHBD |
 |
 |
| Motif ID: | UP00388 |
| Motif name: | Six6_2267.4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 1.0491 |
Alignment:
--AHWDHTGATACCCKATD
GGAGCACTGTGACACCACA
| Original motif | Reverse complement motif |
| Consensus sequence: DATRGGGTATCAHDWHT | Consensus sequence: AHWDHTGATACCCKATD |
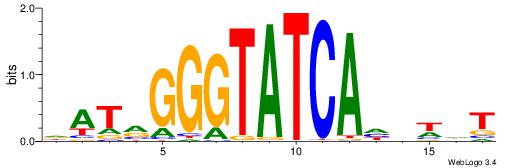 |
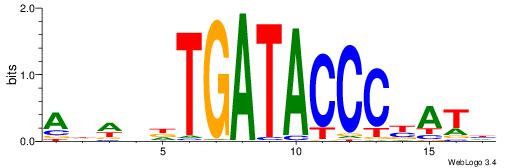 |
| Dataset #: 2 | Motif ID: 18 | Motif name: sscCCCGCGcs |
| Original motif | Reverse complement motif |
| Consensus sequence: BSCCCCGCGBS | Consensus sequence: SBCGCGGGGSB |
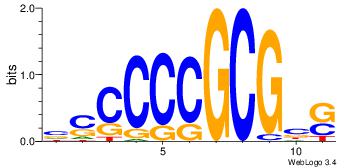 |
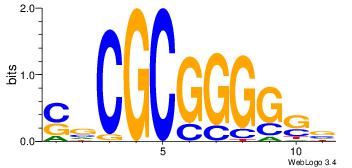 |
Best Matches for Motif ID 18 (Highest to Lowest)
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.00934222 |
Alignment:
BHBHDTGGCGGGGBDHD
----SBCGCGGGGSB--
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
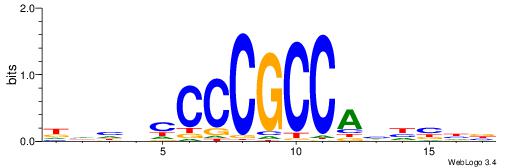 |
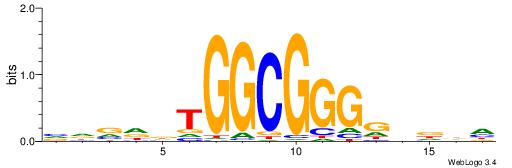 |
| Motif ID: | UP00006 |
| Motif name: | Zic3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0105793 |
Alignment:
BMCCCCCGGGGGGGB
-BSCCCCGCGBS---
| Original motif | Reverse complement motif |
| Consensus sequence: BMCCCCCGGGGGGGB | Consensus sequence: VCCCCCCCGGGGGRB |
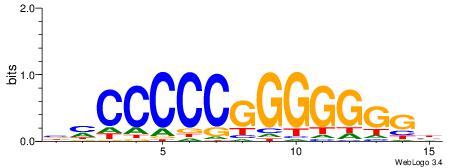 |
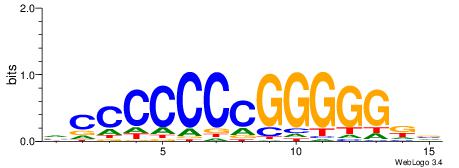 |
| Motif ID: | UP00057 |
| Motif name: | Zic2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0112489 |
Alignment:
VCCCCCCCGGGGGRB
---SBCGCGGGGSB-
| Original motif | Reverse complement motif |
| Consensus sequence: BMCCCCCGGGGGGGB | Consensus sequence: VCCCCCCCGGGGGRB |
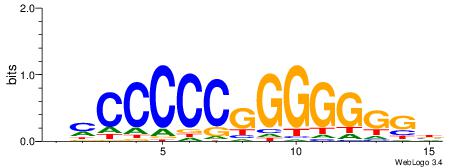 |
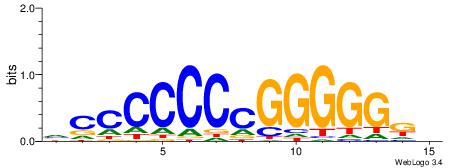 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.014467 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
------SBCGCGGGGSB------
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0150561 |
Alignment:
HDWHMHGACGCGCGTRBVVBHW
--------SBCGCGGGGSB---
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
 |
 |
| Motif ID: | UP00065 |
| Motif name: | Zfp161_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0169049 |
Alignment:
DMMDGMGCGCGCGCHD
--BSCCCCGCGBS---
| Original motif | Reverse complement motif |
| Consensus sequence: DDGCGCGCGCRCHYRD | Consensus sequence: DMMDGMGCGCGCGCHD |
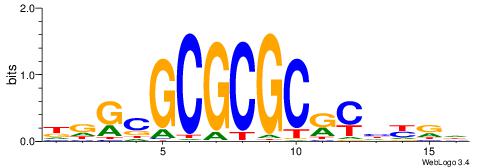 |
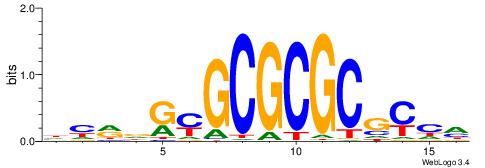 |
| Motif ID: | UP00102 |
| Motif name: | Zic1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0186112 |
Alignment:
BHCCCCCGGGGGGG
-BSCCCCGCGBS--
| Original motif | Reverse complement motif |
| Consensus sequence: BHCCCCCGGGGGGG | Consensus sequence: CCCCCCCGGGGGHB |
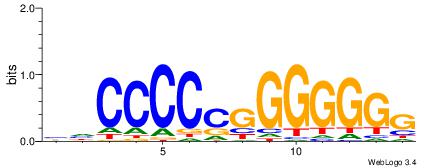 |
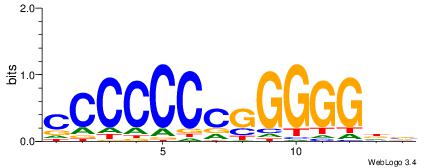 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 11 |
| Similarity score: | 0.0192401 |
Alignment:
VHYTCCDWCCGCGTCMMTHWHV
----BSCCCCGCGBS-------
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0204596 |
Alignment:
BHHDYGGGGGGGGBVD
--SBCGCGGGGSB---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
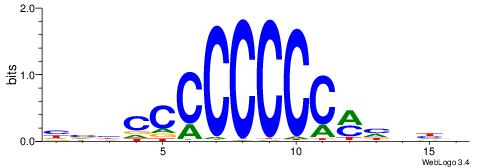 |
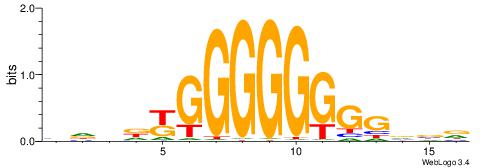 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0232114 |
Alignment:
DDRMHBACGCGYGCGTDGBBDB
-------SBCGCGGGGSB----
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
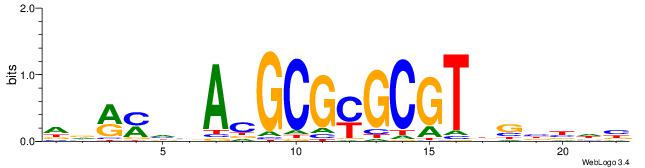 |
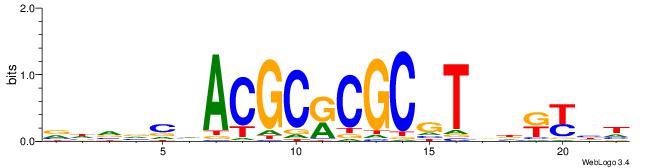 |
| Dataset #: 2 | Motif ID: 19 | Motif name: atactttggc |
| Original motif | Reverse complement motif |
| Consensus sequence: ATACTTTGGC | Consensus sequence: GCCAAAGTAT |
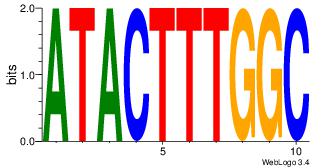 |
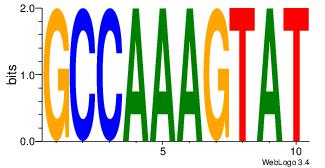 |
Best Matches for Motif ID 19 (Highest to Lowest)
| Motif ID: | UP00156 |
| Motif name: | Msx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0221581 |
Alignment:
ADHKCTAATTKGTCDBV
----ATACTTTGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: VVDGACYAATTAGYDHT | Consensus sequence: ADHKCTAATTKGTCDBV |
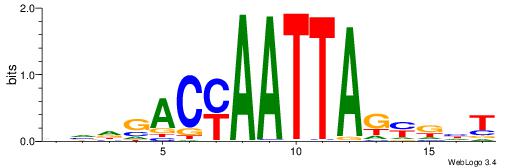 |
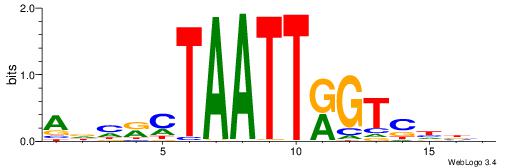 |
| Motif ID: | UP00101 |
| Motif name: | Sox12_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.027519 |
Alignment:
HDTMCTTTGTSTVDBB
-ATACTTTGGC-----
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBASACAAAGRADH | Consensus sequence: HDTMCTTTGTSTVDBB |
 |
 |
| Motif ID: | UP00066 |
| Motif name: | Hnf4a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0285786 |
Alignment:
BBHHAAAGTCCAMTHH
-GCCAAAGTAT-----
| Original motif | Reverse complement motif |
| Consensus sequence: BBHHAAAGTCCAMTHH | Consensus sequence: HHAYTGGACTTTHDBV |
 |
 |
| Motif ID: | UP00418 |
| Motif name: | Etv6 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.030488 |
Alignment:
BHAHCCGGAAGTABDVD
----GCCAAAGTAT---
| Original motif | Reverse complement motif |
| Consensus sequence: DVHBTACTTCCGGHTHB | Consensus sequence: BHAHCCGGAAGTABDVD |
 |
 |
| Motif ID: | UP00083 |
| Motif name: | Tcf7l2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0314685 |
Alignment:
BDDASATCAAAGGMDDD
-----GCCAAAGTAT--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATSTDDV | Consensus sequence: BDDASATCAAAGGMDDD |
 |
 |
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0317293 |
Alignment:
HHBTMCTTTGTTMDDVH
--ATACTTTGGC-----
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
 |
 |
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.031914 |
Alignment:
HHBBMCTTTGTTHDDBH
--ATACTTTGGC-----
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
 |
 |
| Motif ID: | UP00035 |
| Motif name: | Hic1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0323012 |
Alignment:
DBBATGCCAACCHVHH
-----GCCAAAGTAT-
| Original motif | Reverse complement motif |
| Consensus sequence: DBBATGCCAACCHVHH | Consensus sequence: DDBHGGTTGGCATVBD |
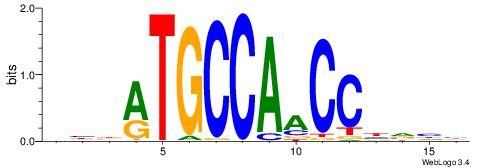 |
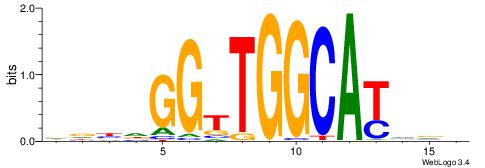 |
| Motif ID: | UP00146 |
| Motif name: | Pou6f1_3733.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.032571 |
Alignment:
VHHVATAATGAGSTDDH
----ATACTTTGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: VHHVATAATGAGSTDDH | Consensus sequence: DHDASCTCATTATVHHB |
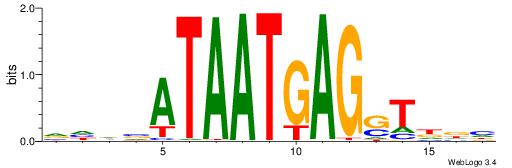 |
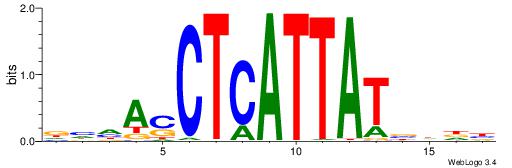 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0330221 |
Alignment:
DDDYCCTTTGATCTDDV
--ATACTTTGGC-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
 |
 |
| Dataset #: 2 | Motif ID: 20 | Motif name: CagTGCTCCACTGTGgT |
| Original motif | Reverse complement motif |
| Consensus sequence: CAGTGCTCCACTGTGBT | Consensus sequence: ABCACAGTGGAGCACTG |
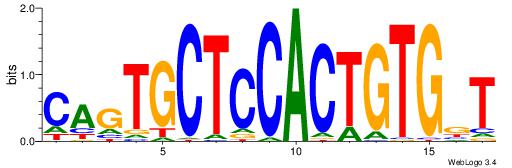 |
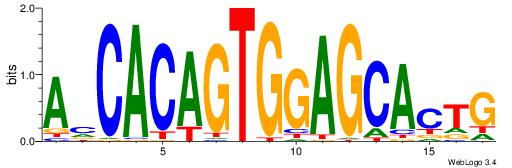 |
Best Matches for Motif ID 20 (Highest to Lowest)
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 17 |
| Similarity score: | 0.0419306 |
Alignment:
CBDMCMGGGTGGTCCHVBVBAH
--ABCACAGTGGAGCACTG---
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0501904 |
Alignment:
WBHHHHHMDATTGTTHDHDVHH
-----ABCACAGTGGAGCACTG
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 0.0504778 |
Alignment:
HGDHVHKGACCACCCGGBGBGDB
--CAGTGCTCCACTGTGBT----
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0513094 |
Alignment:
DDDTWCCCCCCGGDRVH
CAGTGCTCCACTGTGBT
| Original motif | Reverse complement motif |
| Consensus sequence: DDDTWCCCCCCGGDRVH | Consensus sequence: HVKDCCGGGGGGWADDD |
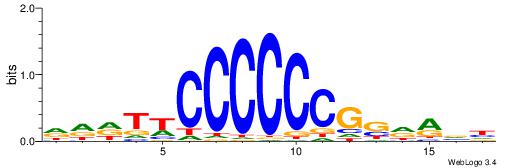 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 0.0516506 |
Alignment:
VMYDHDGMCCHCCKBGVVAAVH
-CAGTGCTCCACTGTGBT----
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 0.0523515 |
Alignment:
VHVRCAYGATGCATCDTVVADTH
--ABCACAGTGGAGCACTG----
| Original motif | Reverse complement motif |
| Consensus sequence: HADTBVADGATGCATCMTGMVHB | Consensus sequence: VHVRCAYGATGCATCDTVVADTH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 0.053668 |
Alignment:
CVHBSCGGGTGGTCMHVHHCDH
--ABCACAGTGGAGCACTG---
| Original motif | Reverse complement motif |
| Consensus sequence: HDGHHBHRGACCACCCGSVDVG | Consensus sequence: CVHBSCGGGTGGTCMHVHHCDH |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 17 |
| Similarity score: | 0.0539016 |
Alignment:
YDYBDHTMCACGTGGADDBMDGT
-----ABCACAGTGGAGCACTG-
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00042 |
| Motif name: | Gm397_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0558205 |
Alignment:
HHRTATGTGCACATHHD
ABCACAGTGGAGCACTG
| Original motif | Reverse complement motif |
| Consensus sequence: HHDATGTGCACATAMDH | Consensus sequence: HHRTATGTGCACATHHD |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 17 |
| Similarity score: | 0.0558674 |
Alignment:
BDVBCDACGCKCGCGTBHRKHD
----CAGTGCTCCACTGTGBT-
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
 |
 |
| Dataset #: 2 | Motif ID: 21 | Motif name: AmTGTGACACCACAGT |
| Original motif | Reverse complement motif |
| Consensus sequence: AMTGTGACACCACAGT | Consensus sequence: ACTGTGGTGTCACART |
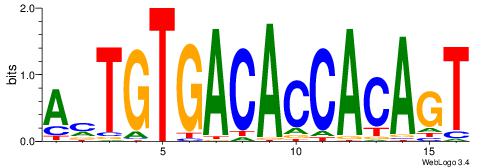 |
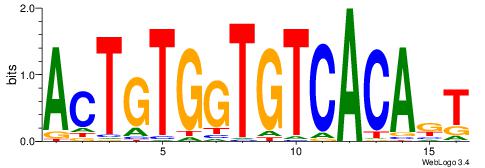 |
Best Matches for Motif ID 21 (Highest to Lowest)
| Motif ID: | UP00020 |
| Motif name: | Atf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0273934 |
Alignment:
DBDRTGACGTCAYHRD
AMTGTGACACCACAGT
| Original motif | Reverse complement motif |
| Consensus sequence: DBDRTGACGTCAYHRD | Consensus sequence: DMDMTGACGTCAKHBD |
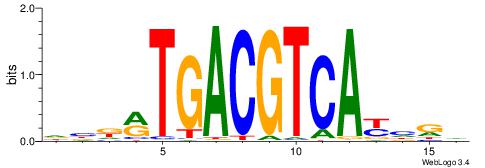 |
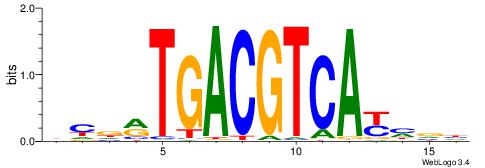 |
| Motif ID: | UP00159 |
| Motif name: | Six2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0303245 |
Alignment:
AHWDGTGATACCCMRTD
-AMTGTGACACCACAGT
| Original motif | Reverse complement motif |
| Consensus sequence: DAKRGGGTATCACDWHT | Consensus sequence: AHWDGTGATACCCMRTD |
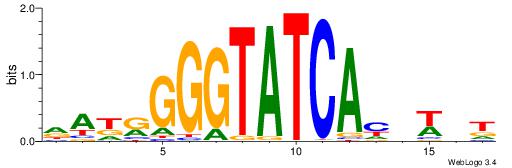 |
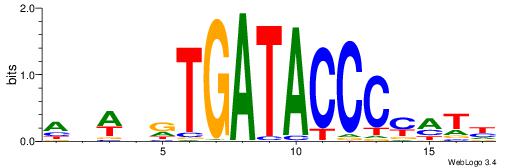 |
| Motif ID: | UP00103 |
| Motif name: | Jundm2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0344928 |
Alignment:
BBGATGACGTCAYCVH
AMTGTGACACCACAGT
| Original motif | Reverse complement motif |
| Consensus sequence: BBGATGACGTCAYCVH | Consensus sequence: HVGMTGACGTCATCBB |
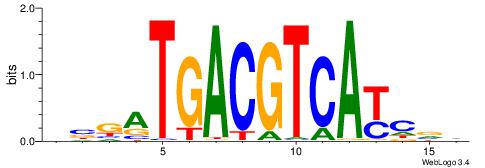 |
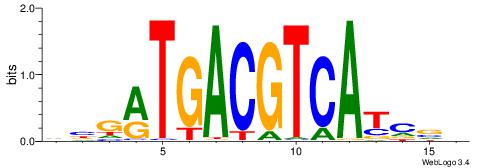 |
| Motif ID: | UP00122 |
| Motif name: | Tgif1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0348371 |
Alignment:
DBVVAGCTGTCAWWVTH
ACTGTGGTGTCACART-
| Original motif | Reverse complement motif |
| Consensus sequence: DABWWTGACAGCTVVBD | Consensus sequence: DBVVAGCTGTCAWWVTH |
 |
 |
| Motif ID: | UP00195 |
| Motif name: | Six3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0349083 |
Alignment:
DATRGGGTATCAHHWHT
ACTGTGGTGTCACART-
| Original motif | Reverse complement motif |
| Consensus sequence: DATRGGGTATCAHHWHT | Consensus sequence: AHWHDTGATACCCKATH |
 |
 |
| Motif ID: | UP00388 |
| Motif name: | Six6_2267.4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0354092 |
Alignment:
DATRGGGTATCAHDWHT
ACTGTGGTGTCACART-
| Original motif | Reverse complement motif |
| Consensus sequence: DATRGGGTATCAHDWHT | Consensus sequence: AHWDHTGATACCCKATD |
 |
 |
| Motif ID: | UP00192 |
| Motif name: | Six1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0354313 |
Alignment:
DHDRGGGTATCAHDHBT
ACTGTGGTGTCACART-
| Original motif | Reverse complement motif |
| Consensus sequence: DHDRGGGTATCAHDHBT | Consensus sequence: AVHDHTGATACCCMDHH |
 |
 |
| Motif ID: | UP00199 |
| Motif name: | Six4 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0355151 |
Alignment:
ADDHATGACACCBHBBV
-AMTGTGACACCACAGT
| Original motif | Reverse complement motif |
| Consensus sequence: ADDHATGACACCBHBBV | Consensus sequence: BBVHVGGTGTCATHDDT |
 |
 |
| Motif ID: | UP00053 |
| Motif name: | Rxra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0362492 |
Alignment:
DBHBDTGACCCCDHVHB
-AMTGTGACACCACAGT
| Original motif | Reverse complement motif |
| Consensus sequence: DBHBDTGACCCCDHVHB | Consensus sequence: VHBHDGGGGTCAHBHBD |
 |
 |
| Motif ID: | UP00388 |
| Motif name: | Six6_2267.5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0374881 |
Alignment:
DDDRGGGTATCAWHWHT
-AMTGTGACACCACAGT
| Original motif | Reverse complement motif |
| Consensus sequence: DDDRGGGTATCAWHWHT | Consensus sequence: AHWHWTGATACCCKDDD |
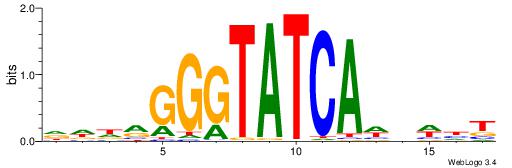 |
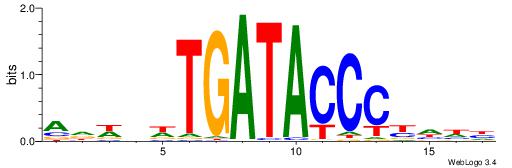 |
| Dataset #: 2 | Motif ID: 22 | Motif name: CACTGTGrYrtCACAGTGswsCAcT |
| Original motif | Reverse complement motif |
| Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT | Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
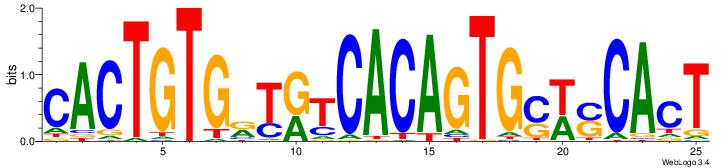 |
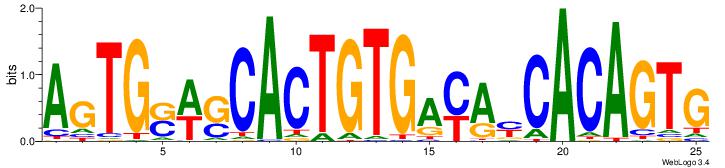 |
Best Matches for Motif ID 22 (Highest to Lowest)
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 21 |
| Similarity score: | 0.0558366 |
Alignment:
----HHBDHDDAACAATDRHHHHHBW
CACTGTGRTRTCACAGTGSWSCACT-
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 20 |
| Similarity score: | 0.55477 |
Alignment:
-----VMYDHDGMCCHCCKBGVVAAVH
AGTGSWSCACTGTGAMAMCACAGTG-----
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 19 |
| Similarity score: | 1.05722 |
Alignment:
------DTVBDDRGACCACCCAGDWDGBH
AGTGSWSCACTGTGAMAMCACAGTG----
| Original motif | Reverse complement motif |
| Consensus sequence: DTVBDDRGACCACCCAGDWDGBH | Consensus sequence: DBCDWHCTGGGTGGTCMDHBBAH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 19 |
| Similarity score: | 1.05946 |
Alignment:
HGDHVHKGACCACCCGGBGBGDB------
------AGTGSWSCACTGTGAMAMCACAGTG
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 18 |
| Similarity score: | 1.5496 |
Alignment:
HTBVVVDGGACCACCCRGRDBG-------
-------CACTGTGRTRTCACAGTGSWSCACT
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 18 |
| Similarity score: | 1.55149 |
Alignment:
HADTBVADGATGCATCMTGMVHB-------
-------CACTGTGRTRTCACAGTGSWSCACT
| Original motif | Reverse complement motif |
| Consensus sequence: HADTBVADGATGCATCMTGMVHB | Consensus sequence: VHVRCAYGATGCATCDTVVADTH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 18 |
| Similarity score: | 1.55607 |
Alignment:
-------CVHBSCGGGTGGTCMHVHHCDH
AGTGSWSCACTGTGAMAMCACAGTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: HDGHHBHRGACCACCCGSVDVG | Consensus sequence: CVHBSCGGGTGGTCMHVHHCDH |
 |
 |
| Motif ID: | UP00228 |
| Motif name: | Bapx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 2.04247 |
Alignment:
DTBVTTAAGTGGTTHHV--------
-AGTGSWSCACTGTGAMAMCACAGTG
| Original motif | Reverse complement motif |
| Consensus sequence: VHHAACCACTTAAVVAH | Consensus sequence: DTBVTTAAGTGGTTHHV |
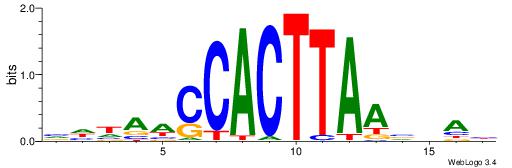 |
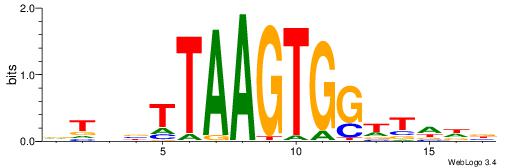 |
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 2.04316 |
Alignment:
--------DDBBBCACTGCABTBBB
CACTGTGRTRTCACAGTGSWSCACT--
| Original motif | Reverse complement motif |
| Consensus sequence: DDBBBCACTGCABTBBB | Consensus sequence: BBBAVTGCAGTGBBVDD |
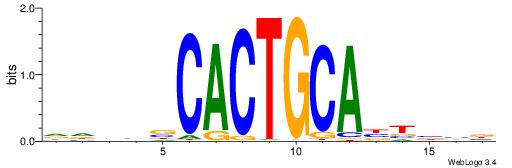 |
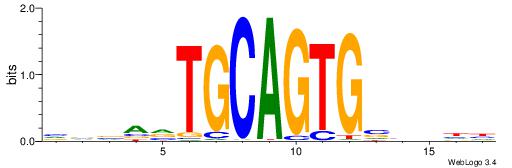 |
| Motif ID: | UP00193 |
| Motif name: | Rhox11_1765.2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 17 |
| Similarity score: | 2.04346 |
Alignment:
WVVHWTWACAGCGBHBT--------
---AGTGSWSCACTGTGAMAMCACAGTG
| Original motif | Reverse complement motif |
| Consensus sequence: AVDVCGCTGTWAWDVVW | Consensus sequence: WVVHWTWACAGCGBHBT |
 |
 |
| Dataset #: 2 | Motif ID: 23 | Motif name: TGGAGCACTGTGACAcCACAGTGg |
| Original motif | Reverse complement motif |
| Consensus sequence: TGGAGCACTGTGACAVCACAGTGG | Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
 |
 |
Best Matches for Motif ID 23 (Highest to Lowest)
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 21 |
| Similarity score: | 0.0391954 |
Alignment:
BDBBVBVGTAWCCAAGVBHBAGB---
---CCACTGTGVTGTCACAGTGCTCCA
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 21 |
| Similarity score: | 0.0400271 |
Alignment:
YBVDMGTGGGTGGTCKVVBVBBT---
--CCACTGTGVTGTCACAGTGCTCCA
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 20 |
| Similarity score: | 0.535486 |
Alignment:
----DBCDWHCTGGGTGGTCMDHBBAH
CCACTGTGVTGTCACAGTGCTCCA----
| Original motif | Reverse complement motif |
| Consensus sequence: DTVBDDRGACCACCCAGDWDGBH | Consensus sequence: DBCDWHCTGGGTGGTCMDHBBAH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 20 |
| Similarity score: | 0.536069 |
Alignment:
----BTBVTCVTGGGTGGTCMVVDVBB
CCACTGTGVTGTCACAGTGCTCCA----
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 20 |
| Similarity score: | 0.539704 |
Alignment:
----HGDHVHKGACCACCCGGBGBGDB
TGGAGCACTGTGACAVCACAGTGG---
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 18 |
| Similarity score: | 1.53819 |
Alignment:
------DVTTVVCVYGGHGGYCHHDMYB
TGGAGCACTGTGACAVCACAGTGG------
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00122 |
| Motif name: | Tgif1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 2.02534 |
Alignment:
-------DABWWTGACAGCTVVBD
TGGAGCACTGTGACAVCACAGTGG--
| Original motif | Reverse complement motif |
| Consensus sequence: DABWWTGACAGCTVVBD | Consensus sequence: DBVVAGCTGTCAWWVTH |
 |
 |
| Motif ID: | UP00159 |
| Motif name: | Six2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 2.02571 |
Alignment:
AHWDGTGATACCCMRTD-------
--CCACTGTGVTGTCACAGTGCTCCA
| Original motif | Reverse complement motif |
| Consensus sequence: DAKRGGGTATCACDWHT | Consensus sequence: AHWDGTGATACCCMRTD |
 |
 |
| Motif ID: | UP00192 |
| Motif name: | Six1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 2.02892 |
Alignment:
AVHDHTGATACCCMDHH-------
--CCACTGTGVTGTCACAGTGCTCCA
| Original motif | Reverse complement motif |
| Consensus sequence: DHDRGGGTATCAHDHBT | Consensus sequence: AVHDHTGATACCCMDHH |
 |
 |
| Motif ID: | UP00076 |
| Motif name: | Rfxdc2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 17 |
| Similarity score: | 2.0292 |
Alignment:
HDDVSGTRTCMADGBVB-------
---CCACTGTGVTGTCACAGTGCTCCA
| Original motif | Reverse complement motif |
| Consensus sequence: BBVCDTRGAKACSVDDH | Consensus sequence: HDDVSGTRTCMADGBVB |
 |
 |
| Dataset #: 2 | Motif ID: 24 | Motif name: ssCCCCGCSssk |
| Original motif | Reverse complement motif |
| Consensus sequence: SBCCCCGCCSBB | Consensus sequence: BBSGGCGGGGBS |
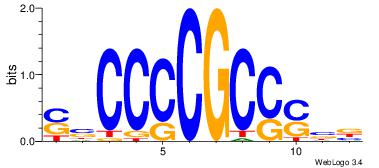 |
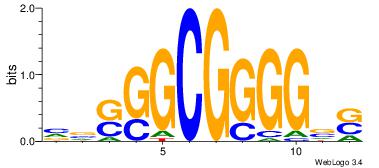 |
Best Matches for Motif ID 24 (Highest to Lowest)
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0114847 |
Alignment:
DHHBCCCCGCCAHHBHB
--SBCCCCGCCSBB---
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
 |
 |
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0165262 |
Alignment:
HBDRCCMCGCCCHHHD
--SBCCCCGCCSBB--
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0276137 |
Alignment:
HVBCCCCCCCCMHHHB
--SBCCCCGCCSBB--
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00003 |
| Motif name: | E2F3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0328578 |
Alignment:
WHSGCGCGCCHTDHB
-SBCCCCGCCSBB--
| Original motif | Reverse complement motif |
| Consensus sequence: VHDADGGCGCGCSHW | Consensus sequence: WHSGCGCGCCHTDHB |
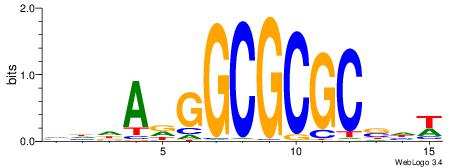 |
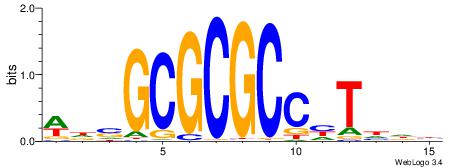 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 0.0336734 |
Alignment:
BHHBHCCGCCCCDHHHH
-SBCCCCGCCSBB----
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
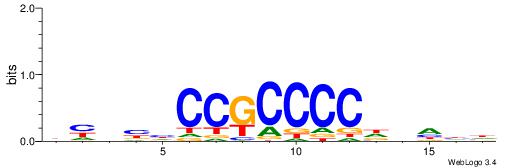 |
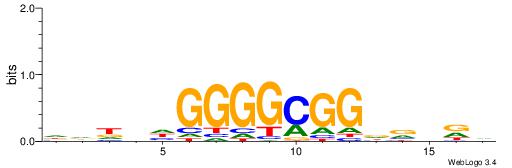 |
| Motif ID: | UP00001 |
| Motif name: | E2F2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0345737 |
Alignment:
HHVGCGCGCCKTDHB
-SBCCCCGCCSBB--
| Original motif | Reverse complement motif |
| Consensus sequence: VHDARGGCGCGCVHH | Consensus sequence: HHVGCGCGCCKTDHB |
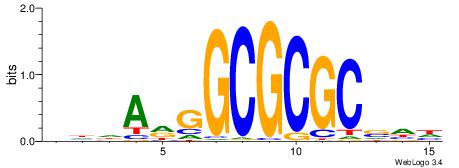 |
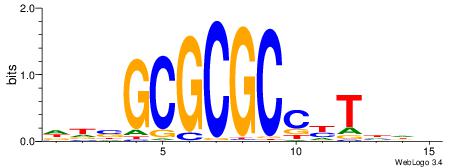 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0347468 |
Alignment:
HVDVGGGHGGGGDBHB
--BBSGGCGGGGBS--
| Original motif | Reverse complement motif |
| Consensus sequence: BHVDCCCCDCCCBDBH | Consensus sequence: HVDVGGGHGGGGDBHB |
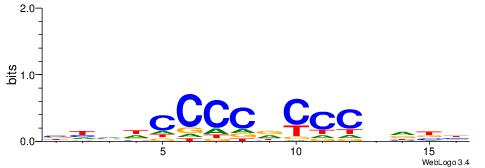 |
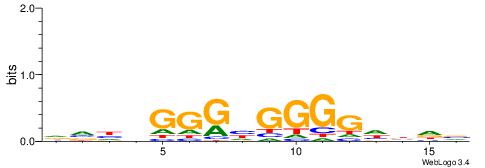 |
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0358124 |
Alignment:
BHHHWGGGGCGGBBVH
--BBSGGCGGGGBS--
| Original motif | Reverse complement motif |
| Consensus sequence: HBBBCCGCCCCWHHHV | Consensus sequence: BHHHWGGGGCGGBBVH |
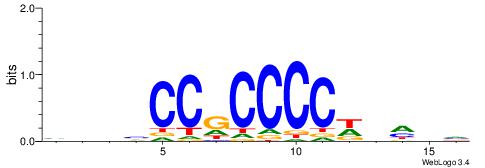 |
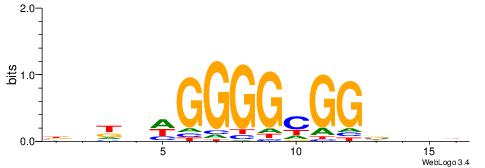 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 12 |
| Similarity score: | 0.0365354 |
Alignment:
BADHBDHCGCCCMCGCAHHDBBV
--------SBCCCCGCCSBB---
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 12 |
| Similarity score: | 0.0389727 |
Alignment:
HDWHMHGACGCGCGTRBVVBHW
-----BBSGGCGGGGBS-----
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
 |
 |
| Dataset #: 2 | Motif ID: 25 | Motif name: wwCCAmAGTCmt |
| Original motif | Reverse complement motif |
| Consensus sequence: DDCCAMAGTCHB | Consensus sequence: VDGACTRTGGDD |
 |
 |
Best Matches for Motif ID 25 (Highest to Lowest)
| Motif ID: | UP00079 |
| Motif name: | Esrra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 0.00927341 |
Alignment:
DHDBCAAGGTCAHVBDH
-DDCCAMAGTCHB----
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBCAAGGTCAHVBDH | Consensus sequence: HHBVHTGACCTTGVDHD |
 |
 |
| Motif ID: | UP00066 |
| Motif name: | Hnf4a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0157554 |
Alignment:
BBHHAAAGTCCAMTHH
DDCCAMAGTCHB----
| Original motif | Reverse complement motif |
| Consensus sequence: BBHHAAAGTCCAMTHH | Consensus sequence: HHAYTGGACTTTHDBV |
 |
 |
| Motif ID: | UP00023 |
| Motif name: | Sox30_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0218941 |
Alignment:
DHDBAACAATDRHHHH
--VDGACTRTGGDD--
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBAACAATDRHHHH | Consensus sequence: HHHHMHATTGTTBDHD |
 |
 |
| Motif ID: | UP00028 |
| Motif name: | Tcfap2e_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.023236 |
Alignment:
DTHGCCTSAGGCHHH
--DDCCAMAGTCHB-
| Original motif | Reverse complement motif |
| Consensus sequence: HHHGCCTSAGGCDAD | Consensus sequence: DTHGCCTSAGGCHHH |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 12 |
| Similarity score: | 0.0235871 |
Alignment:
HHBDHDDAACAATDRHHHHHBW
-----VDGACTRTGGDD-----
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00095 |
| Motif name: | Zfp691_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0240897 |
Alignment:
VHBDDAGGAGTCWBBTD
--DDCCAMAGTCHB---
| Original motif | Reverse complement motif |
| Consensus sequence: DABBWGACTCCTHDVHV | Consensus sequence: VHBDDAGGAGTCWBBTD |
 |
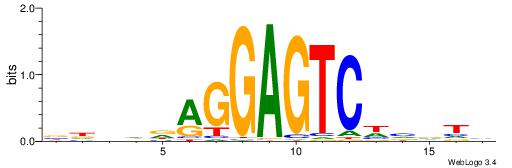 |
| Motif ID: | UP00004 |
| Motif name: | Sox14_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.025783 |
Alignment:
BKBASACAATRGHBB
--VDGACTRTGGDD-
| Original motif | Reverse complement motif |
| Consensus sequence: BKBASACAATRGHBB | Consensus sequence: BBDCMATTGTSTBRB |
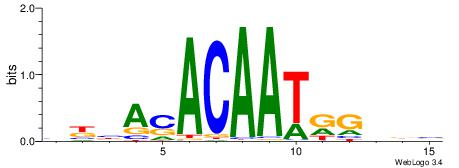 |
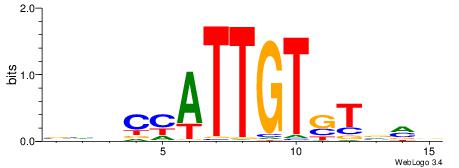 |
| Motif ID: | UP00081 |
| Motif name: | Mybl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.026472 |
Alignment:
HDDDHAACCGTTAHHHD
---VDGACTRTGGDD--
| Original motif | Reverse complement motif |
| Consensus sequence: HDDDHAACCGTTAHHHD | Consensus sequence: DHHHTAACGGTTHDHDH |
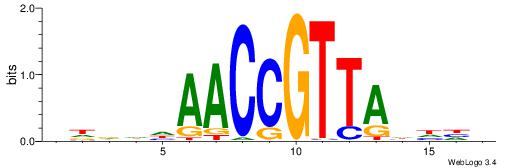 |
 |
| Motif ID: | UP00052 |
| Motif name: | Osr2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.027027 |
Alignment:
HHHHGCTACKGTHVHH
---DDCCAMAGTCHB-
| Original motif | Reverse complement motif |
| Consensus sequence: HHVHACRGTAGCHHHD | Consensus sequence: HHHHGCTACKGTHVHH |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0270662 |
Alignment:
HVHBVTGTCTGGDDHDD
--VDGACTRTGGDD---
| Original motif | Reverse complement motif |
| Consensus sequence: HDHDDCCAGACABBHVH | Consensus sequence: HVHBVTGTCTGGDDHDD |
 |
 |
| Dataset #: 2 | Motif ID: 26 | Motif name: CAcCACAGTGGAGCAct |
| Original motif | Reverse complement motif |
| Consensus sequence: CAHCACAGTGGAGCACT | Consensus sequence: AGTGCTCCACTGTGDTG |
 |
 |
Best Matches for Motif ID 26 (Highest to Lowest)
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 0.0381337 |
Alignment:
CBDMCMGGGTGGTCCHVBVBAH
-CAHCACAGTGGAGCACT----
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0393208 |
Alignment:
BBBAVTGCAGTGBBVDD
AGTGCTCCACTGTGDTG
| Original motif | Reverse complement motif |
| Consensus sequence: DDBBBCACTGCABTBBB | Consensus sequence: BBBAVTGCAGTGBBVDD |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 0.0441166 |
Alignment:
HHBDHDDAACAATDRHHHHHBW
----CAHCACAGTGGAGCACT-
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 17 |
| Similarity score: | 0.0477617 |
Alignment:
HGDHVHKGACCACCCGGBGBGDB
---AGTGCTCCACTGTGDTG---
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00231 |
| Motif name: | Nkx2-2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0479423 |
Alignment:
HDVRCCACTTGARADBD
CAHCACAGTGGAGCACT
| Original motif | Reverse complement motif |
| Consensus sequence: HDVRCCACTTGARADBD | Consensus sequence: DVDTKTCAAGTGGKBDH |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 17 |
| Similarity score: | 0.0484681 |
Alignment:
VHVRCAYGATGCATCDTVVADTH
-CAHCACAGTGGAGCACT-----
| Original motif | Reverse complement motif |
| Consensus sequence: HADTBVADGATGCATCMTGMVHB | Consensus sequence: VHVRCAYGATGCATCDTVVADTH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0487437 |
Alignment:
HHBSHTCGTGRGTGGKGDBVWMV
AGTGCTCCACTGTGDTG------
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.048891 |
Alignment:
DHDBHGCACCTGBDDVB
AGTGCTCCACTGTGDTG
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 17 |
| Similarity score: | 0.0491352 |
Alignment:
CVHBSCGGGTGGTCMHVHHCDH
-CAHCACAGTGGAGCACT----
| Original motif | Reverse complement motif |
| Consensus sequence: HDGHHBHRGACCACCCGSVDVG | Consensus sequence: CVHBSCGGGTGGTCMHVHHCDH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 17 |
| Similarity score: | 0.0515016 |
Alignment:
DVTTVVCVYGGHGGYCHHDMYB
---CAHCACAGTGGAGCACT--
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Dataset #: 2 | Motif ID: 27 | Motif name: AgTGCTCCACTGTGgTGTCACAgT |
| Original motif | Reverse complement motif |
| Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT | Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
 |
 |
Best Matches for Motif ID 27 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 23 |
| Similarity score: | 0.0509731 |
Alignment:
BBBDVVRGACCACCCAVGABBAB-
ACTGTGACACCACAGTGGAGCACT
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 21 |
| Similarity score: | 1.0488 |
Alignment:
DTVBDDRGACCACCCAGDWDGBH---
---AGTGCTCCACTGTGGTGTCACAGT
| Original motif | Reverse complement motif |
| Consensus sequence: DTVBDDRGACCACCCAGDWDGBH | Consensus sequence: DBCDWHCTGGGTGGTCMDHBBAH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 21 |
| Similarity score: | 1.05087 |
Alignment:
---HGDHVHKGACCACCCGGBGBGDB
ACTGTGACACCACAGTGGAGCACT---
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 18 |
| Similarity score: | 2.54748 |
Alignment:
CBDMCMGGGTGGTCCHVBVBAH------
------ACTGTGACACCACAGTGGAGCACT
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Motif ID: | UP00045 |
| Motif name: | Mafb_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 3.0367 |
Alignment:
-------HAAWDTGCTGACDWARH
AGTGCTCCACTGTGGTGTCACAGT
| Original motif | Reverse complement motif |
| Consensus sequence: HAAWDTGCTGACDWARH | Consensus sequence: DMTWDGTCAGCADWTTH |
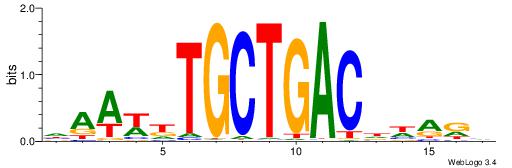 |
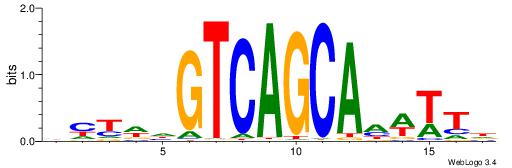 |
| Motif ID: | UP00055 |
| Motif name: | Hbp1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 3.04028 |
Alignment:
DDDYCCCATTGTVHABH-------
-AGTGCTCCACTGTGGTGTCACAGT
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCCATTGTVHABH | Consensus sequence: HBTHVACAATGGGMDHD |
 |
 |
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 17 |
| Similarity score: | 3.04347 |
Alignment:
-------DDBBBCACTGCABTBBB
AGTGCTCCACTGTGGTGTCACAGT-------
| Original motif | Reverse complement motif |
| Consensus sequence: DDBBBCACTGCABTBBB | Consensus sequence: BBBAVTGCAGTGBBVDD |
 |
 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 17 |
| Similarity score: | 3.04662 |
Alignment:
VDDAGATCAAAGGKDDD-------
---ACTGTGACACCACAGTGGAGCACT
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
 |
 |
| Motif ID: | UP00070 |
| Motif name: | Gcm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 3.04663 |
Alignment:
DDBVHATAGGGGRBRMB-------
-AGTGCTCCACTGTGGTGTCACAGT
| Original motif | Reverse complement motif |
| Consensus sequence: DDBVHATAGGGGRBRMB | Consensus sequence: BYMBKCCCCTATDVBHD |
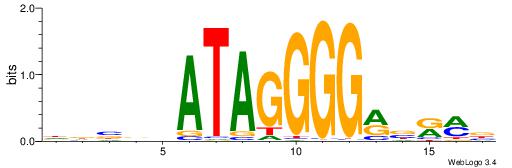 |
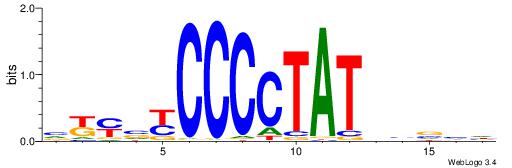 |
| Motif ID: | UP00148 |
| Motif name: | Hdx |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 17 |
| Similarity score: | 3.04907 |
Alignment:
-------BDBDHTGATTTCDHYDD
ACTGTGACACCACAGTGGAGCACT-------
| Original motif | Reverse complement motif |
| Consensus sequence: DDKDHGAAATCAHHBHV | Consensus sequence: BDBDHTGATTTCDHYDD |
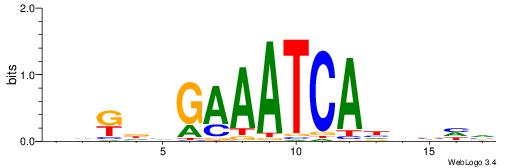 |
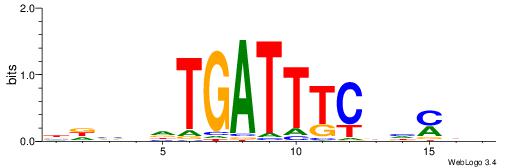 |
| Dataset #: 2 | Motif ID: 28 | Motif name: ssGCkTGCssk |
| Original motif | Reverse complement motif |
| Consensus sequence: SSGCKTGCSSK | Consensus sequence: YSSGCAYGCSS |
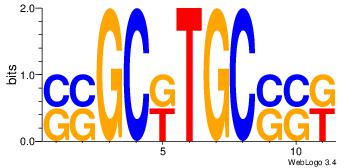 |
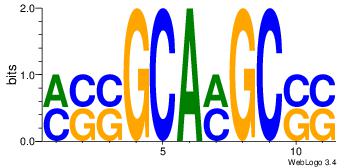 |
Best Matches for Motif ID 28 (Highest to Lowest)
| Motif ID: | UP00003 |
| Motif name: | E2F3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.00463328 |
Alignment:
VHDADGGCGCGCSHW
----SSGCKTGCSSK
| Original motif | Reverse complement motif |
| Consensus sequence: VHDADGGCGCGCSHW | Consensus sequence: WHSGCGCGCCHTDHB |
 |
 |
| Motif ID: | UP00094 |
| Motif name: | Zfp128_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.00482124 |
Alignment:
BHHDKGGCGTACCCBHD
----SSGCKTGCSSK--
| Original motif | Reverse complement motif |
| Consensus sequence: BHHDKGGCGTACCCBHD | Consensus sequence: DHVGGGTACGCCRDHDV |
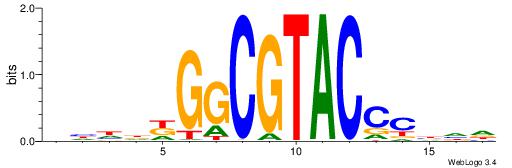 |
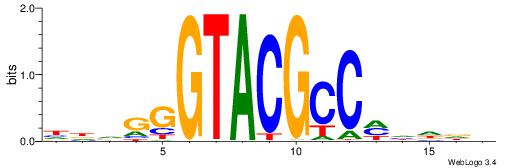 |
| Motif ID: | UP00035 |
| Motif name: | Hic1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.00562779 |
Alignment:
VHHDWTGGGCAMDHBH
-----YSSGCAYGCSS
| Original motif | Reverse complement motif |
| Consensus sequence: DBDDRTGCCCAWDHDV | Consensus sequence: VHHDWTGGGCAMDHBH |
 |
 |
| Motif ID: | UP00065 |
| Motif name: | Zfp161_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.00595984 |
Alignment:
DDGCGCGCGCRCHYRD
--SSGCKTGCSSK---
| Original motif | Reverse complement motif |
| Consensus sequence: DDGCGCGCGCRCHYRD | Consensus sequence: DMMDGMGCGCGCGCHD |
 |
 |
| Motif ID: | UP00001 |
| Motif name: | E2F2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.00821725 |
Alignment:
HHVGCGCGCCKTDHB
-SSGCKTGCSSK---
| Original motif | Reverse complement motif |
| Consensus sequence: VHDARGGCGCGCVHH | Consensus sequence: HHVGCGCGCCKTDHB |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0109259 |
Alignment:
BBGHCACGCGACDD
YSSGCAYGCSS---
| Original motif | Reverse complement motif |
| Consensus sequence: BBGHCACGCGACDD | Consensus sequence: HDGTCGCGTGDCVB |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0150199 |
Alignment:
VDWDARRGACGCGGWHGGAKHV
---YSSGCAYGCSS--------
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0160778 |
Alignment:
HHDGBCACGCCTWDB
-YSSGCAYGCSS---
| Original motif | Reverse complement motif |
| Consensus sequence: BDWAGGCGTGBCHHD | Consensus sequence: HHDGBCACGCCTWDB |
 |
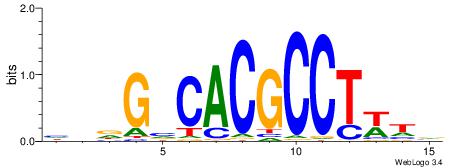 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0171602 |
Alignment:
HDWHMHGACGCGCGTRBVVBHW
---------SSGCKTGCSSK--
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0172713 |
Alignment:
DCATDBTGHGCATKMTTBRSWR
------YSSGCAYGCSS-----
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
 |
 |
| Dataset #: 2 | Motif ID: 29 | Motif name: cCGCGGrCACG |
| Original motif | Reverse complement motif |
| Consensus sequence: CCGCGGRCACG | Consensus sequence: CGTGKCCGCGG |
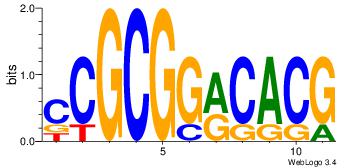 |
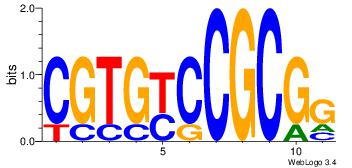 |
Best Matches for Motif ID 29 (Highest to Lowest)
| Motif ID: | UP00065 |
| Motif name: | Zfp161_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0220206 |
Alignment:
DDGCGCGCGCRCHYRD
----CCGCGGRCACG-
| Original motif | Reverse complement motif |
| Consensus sequence: DDGCGCGCGCRCHYRD | Consensus sequence: DMMDGMGCGCGCGCHD |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 11 |
| Similarity score: | 0.024575 |
Alignment:
VHYTCCDWCCGCGTCMMTHWHV
---CGTGKCCGCGG--------
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00042 |
| Motif name: | Gm397_second |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0279366 |
Alignment:
DVMVGCACACACKCVH
--CCGCGGRCACG---
| Original motif | Reverse complement motif |
| Consensus sequence: DVMVGCACACACKCVH | Consensus sequence: HBGYGTGTGTGCVRVD |
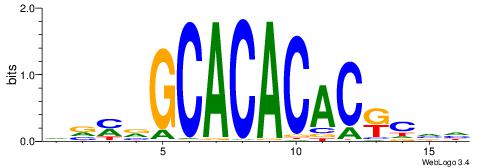 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0285381 |
Alignment:
VHTGCGGGGGCGGH
-CCGCGGRCACG--
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
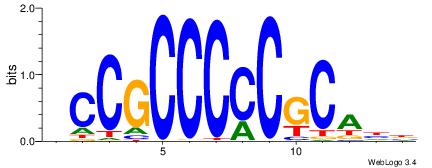 |
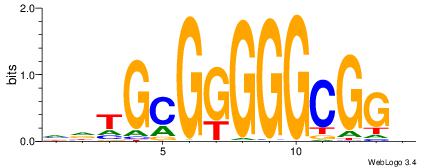 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 11 |
| Similarity score: | 0.0335378 |
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
--------CCGCGGRCACG----
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0340428 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
-------CCGCGGRCACG-----
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00065 |
| Motif name: | Zfp161_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0345855 |
Alignment:
GYCGCGCARBGCVB
-CCGCGGRCACG--
| Original motif | Reverse complement motif |
| Consensus sequence: GYCGCGCARBGCVB | Consensus sequence: VVGCVMTGCGCGKC |
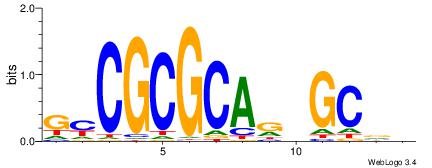 |
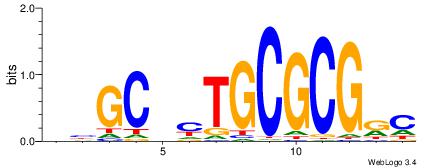 |
| Motif ID: | UP00097 |
| Motif name: | Mtf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0350434 |
Alignment:
DBDHCGTGTGCAAMDH
---CCGCGGRCACG--
| Original motif | Reverse complement motif |
| Consensus sequence: DBDHCGTGTGCAAMDH | Consensus sequence: HDYTTGCACACGDHBH |
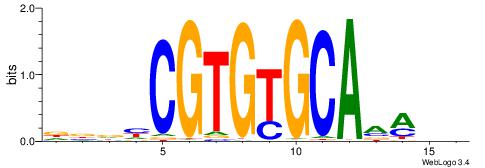 |
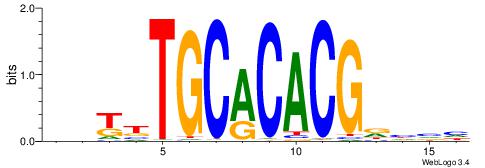 |
| Motif ID: | UP00076 |
| Motif name: | Rfxdc2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0359928 |
Alignment:
HDDVSGTRTCMADGBVB
----CGTGKCCGCGG--
| Original motif | Reverse complement motif |
| Consensus sequence: BBVCDTRGAKACSVDDH | Consensus sequence: HDDVSGTRTCMADGBVB |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0361963 |
Alignment:
HDWHMHGACGCGCGTRBVVBHW
---------CCGCGGRCACG--
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
 |
 |
| Dataset #: 2 | Motif ID: 30 | Motif name: taaacgatgcc |
| Original motif | Reverse complement motif |
| Consensus sequence: TAAACGATGCC | Consensus sequence: GGCATCGTTTA |
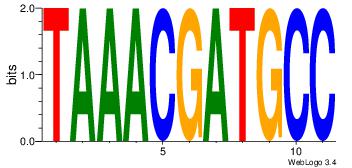 |
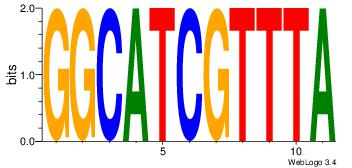 |
Best Matches for Motif ID 30 (Highest to Lowest)
| Motif ID: | UP00114 |
| Motif name: | Homez |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0207645 |
Alignment:
HDTVAAAACGATRWWHT
----TAAACGATGCC--
| Original motif | Reverse complement motif |
| Consensus sequence: AHWWMATCGTTTTBADD | Consensus sequence: HDTVAAAACGATRWWHT |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0294316 |
Alignment:
VVVWTTGTTTAMATTWD
GGCATCGTTTA------
| Original motif | Reverse complement motif |
| Consensus sequence: DWAATRTAAACAAWVVB | Consensus sequence: VVVWTTGTTTAMATTWD |
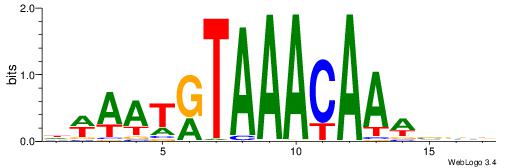 |
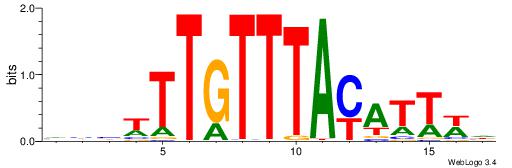 |
| Motif ID: | UP00039 |
| Motif name: | Foxj3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.030091 |
Alignment:
HDHADGTAAACAAAVVH
------TAAACGATGCC
| Original motif | Reverse complement motif |
| Consensus sequence: HDHADGTAAACAAAVVH | Consensus sequence: DVVTTTGTTTACDTHDH |
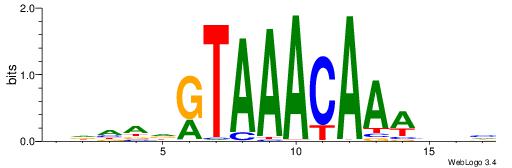 |
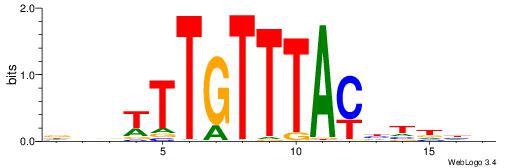 |
| Motif ID: | UP00025 |
| Motif name: | Foxk1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.0306614 |
Alignment:
BBVTTTGTTTACATTDD
GGCATCGTTTA------
| Original motif | Reverse complement motif |
| Consensus sequence: DDAATGTAAACAAAVVB | Consensus sequence: BBVTTTGTTTACATTDD |
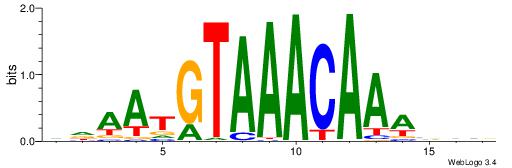 |
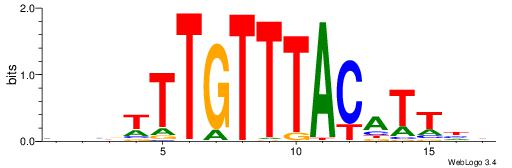 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 10 |
| Number of overlap: | 11 |
| Similarity score: | 0.0342477 |
Alignment:
DBBVDDDRTRGCGTCBBYTHGA
---------GGCATCGTTTA--
| Original motif | Reverse complement motif |
| Consensus sequence: DBBVDDDRTRGCGTCBBYTHGA | Consensus sequence: TCDAMVBGACGCMAKDDDVBBD |
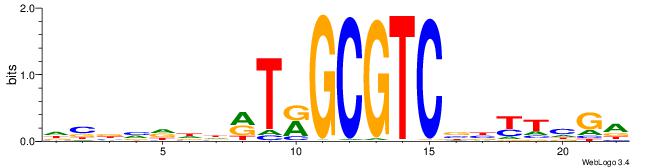 |
 |
| Motif ID: | UP00040 |
| Motif name: | Irf5_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0345017 |
Alignment:
BDKGTTTCGKTWDDD
--GGCATCGTTTA--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDWAYCGAAACYDV | Consensus sequence: BDKGTTTCGKTWDDD |
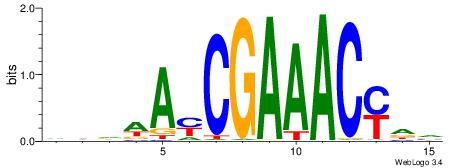 |
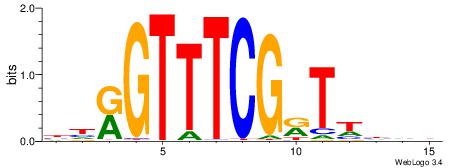 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0351616 |
Alignment:
VHVRCAYGATGCATCDTVVADTH
--TAAACGATGCC----------
| Original motif | Reverse complement motif |
| Consensus sequence: HADTBVADGATGCATCMTGMVHB | Consensus sequence: VHVRCAYGATGCATCDTVVADTH |
 |
 |
| Motif ID: | UP00232 |
| Motif name: | Dobox4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0371486 |
Alignment:
DAWKGGGTATCTAWHWH
-----GGCATCGTTTA-
| Original motif | Reverse complement motif |
| Consensus sequence: HWHWTAGATACCCYWTD | Consensus sequence: DAWKGGGTATCTAWHWH |
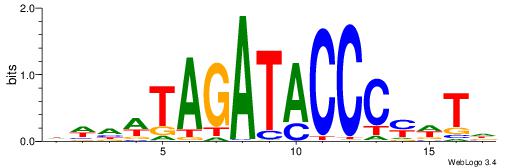 |
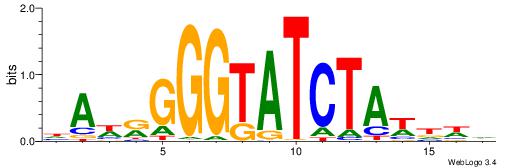 |
| Motif ID: | UP00073 |
| Motif name: | Foxa2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.0384798 |
Alignment:
BDVTTTKTTTACWTWHH
GGCATCGTTTA------
| Original motif | Reverse complement motif |
| Consensus sequence: HHWAWGTAAAYAAAVDB | Consensus sequence: BDVTTTKTTTACWTWHH |
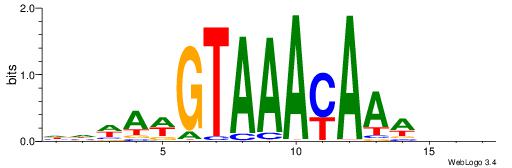 |
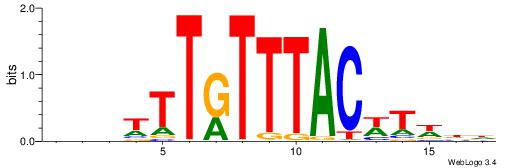 |
| Motif ID: | UP00011 |
| Motif name: | Irf6_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0388532 |
Alignment:
DHDBTKGTTTCGHTHDD
-----GGCATCGTTTA-
| Original motif | Reverse complement motif |
| Consensus sequence: HDDAHCGAAACYAVDDD | Consensus sequence: DHDBTKGTTTCGHTHDD |
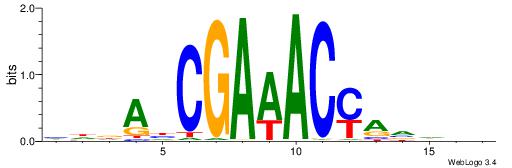 |
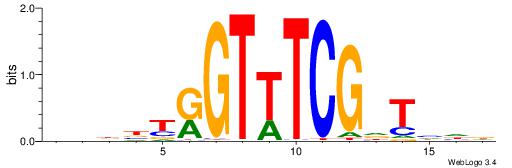 |
| Dataset #: 2 | Motif ID: 31 | Motif name: ctatacggacg |
| Original motif | Reverse complement motif |
| Consensus sequence: CTATACGGACG | Consensus sequence: CGTCCGTATAG |
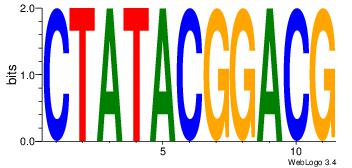 |
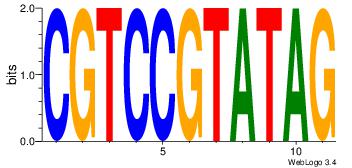 |
Best Matches for Motif ID 31 (Highest to Lowest)
| Motif ID: | UP00038 |
| Motif name: | Spdef_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.027939 |
Alignment:
VVHHATCCGGATGKHV
--CTATACGGACG---
| Original motif | Reverse complement motif |
| Consensus sequence: VHRCATCCGGATHHBB | Consensus sequence: VVHHATCCGGATGKHV |
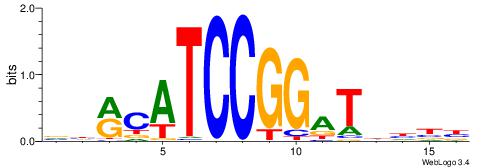 |
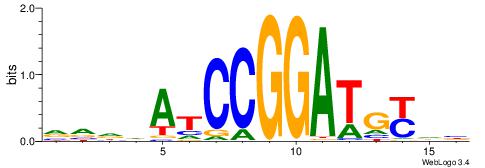 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 12 |
| Number of overlap: | 11 |
| Similarity score: | 0.0308121 |
Alignment:
VDWDARRGACGCGGWHGGAKHV
CTATACGGACG-----------
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00413 |
| Motif name: | Elf4 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0328673 |
Alignment:
HAWYCCGGAAGTHBBD
CTATACGGACG-----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGKWTH | Consensus sequence: HAWYCCGGAAGTHBBD |
 |
 |
| Motif ID: | UP00418 |
| Motif name: | Etv6 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0334005 |
Alignment:
BHAHCCGGAAGTABDVD
------CGTCCGTATAG
| Original motif | Reverse complement motif |
| Consensus sequence: DVHBTACTTCCGGHTHB | Consensus sequence: BHAHCCGGAAGTABDVD |
 |
 |
| Motif ID: | UP00409 |
| Motif name: | Elf5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0336391 |
Alignment:
BWAHSMGGAAGTWD
CTATACGGACG---
| Original motif | Reverse complement motif |
| Consensus sequence: BWAHSMGGAAGTWD | Consensus sequence: DWACTTCCRSDTWV |
 |
 |
| Motif ID: | UP00097 |
| Motif name: | Mtf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0346533 |
Alignment:
DBDHCGTGTGCAAMDH
----CGTCCGTATAG-
| Original motif | Reverse complement motif |
| Consensus sequence: DBDHCGTGTGCAAMDH | Consensus sequence: HDYTTGCACACGDHBH |
 |
 |
| Motif ID: | UP00135 |
| Motif name: | Hoxc12 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0360998 |
Alignment:
DDVHWTTTACGACCKHH
----CTATACGGACG--
| Original motif | Reverse complement motif |
| Consensus sequence: HHRGGTCGTAAAWHBDH | Consensus sequence: DDVHWTTTACGACCKHH |
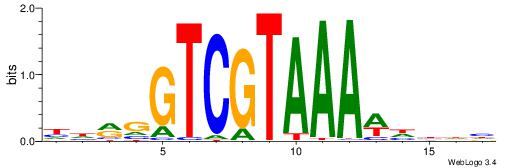 |
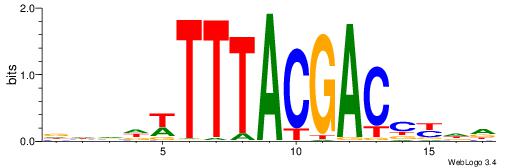 |
| Motif ID: | UP00084 |
| Motif name: | Gmeb1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0365466 |
Alignment:
BVVDVRYACGTRYVBHB
----CGTCCGTATAG--
| Original motif | Reverse complement motif |
| Consensus sequence: BHBBKKACGTMMVDBVB | Consensus sequence: BVVDVRYACGTRYVBHB |
 |
 |
| Motif ID: | UP00015 |
| Motif name: | Ehf_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0367131 |
Alignment:
VBDABCCGGAAGTDD
-CTATACGGACG---
| Original motif | Reverse complement motif |
| Consensus sequence: VBDABCCGGAAGTDD | Consensus sequence: DDACTTCCGGBTHBB |
 |
 |
| Motif ID: | UP00421 |
| Motif name: | Etv1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0373 |
Alignment:
HBDBACTTCCGGTBHVY
-----CGTCCGTATAG-
| Original motif | Reverse complement motif |
| Consensus sequence: MVHBACCGGAAGTVDVH | Consensus sequence: HBDBACTTCCGGTBHVY |
 |
 |
| Dataset #: 2 | Motif ID: 32 | Motif name: gCGCGCsCgsG |
| Original motif | Reverse complement motif |
| Consensus sequence: GCGCGCSCGCG | Consensus sequence: CGCGSGCGCGC |
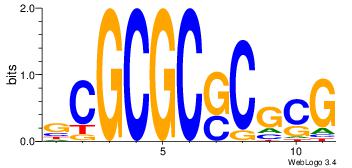 |
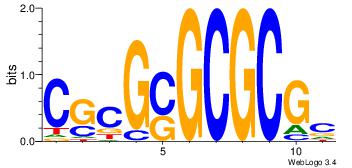 |
Best Matches for Motif ID 32 (Highest to Lowest)
| Motif ID: | UP00065 |
| Motif name: | Zfp161_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0 |
Alignment:
DMMDGMGCGCGCGCHD
----GCGCGCSCGCG-
| Original motif | Reverse complement motif |
| Consensus sequence: DDGCGCGCGCRCHYRD | Consensus sequence: DMMDGMGCGCGCGCHD |
 |
 |
| Motif ID: | UP00003 |
| Motif name: | E2F3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0102009 |
Alignment:
WHSGCGCGCCHTDHB
CGCGSGCGCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: VHDADGGCGCGCSHW | Consensus sequence: WHSGCGCGCCHTDHB |
 |
 |
| Motif ID: | UP00001 |
| Motif name: | E2F2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0124455 |
Alignment:
HHVGCGCGCCKTDHB
CGCGSGCGCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: VHDARGGCGCGCVHH | Consensus sequence: HHVGCGCGCCKTDHB |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 11 |
| Similarity score: | 0.0192204 |
Alignment:
WHVBVVMACGCGCGTCDYHWDH
----CGCGSGCGCGC-------
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 11 |
| Similarity score: | 0.0231982 |
Alignment:
DDRMHBACGCGYGCGTDGBBDB
-------CGCGSGCGCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0289369 |
Alignment:
HCCGCCCCCGCAHB
-GCGCGCSCGCG--
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.0297254 |
Alignment:
BADHBDHCGCCCMCGCAHHDBBV
------GCGCGCSCGCG------
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00088 |
| Motif name: | Plagl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0316913 |
Alignment:
BHDGGGGSSCCCCBVV
-CGCGSGCGCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: BHDGGGGSSCCCCBVV | Consensus sequence: VBVGGGGSSCCCCHHV |
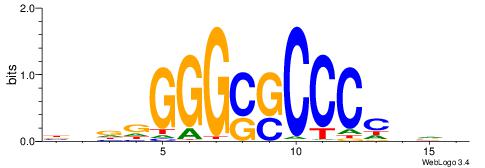 |
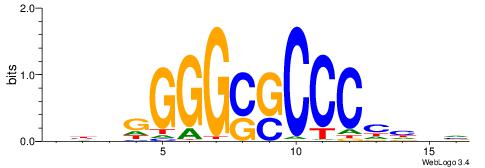 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0323627 |
Alignment:
BVVHAGGGGGCGRDHHB
----CGCGSGCGCGC--
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
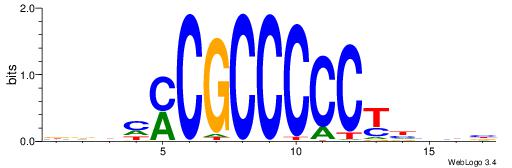 |
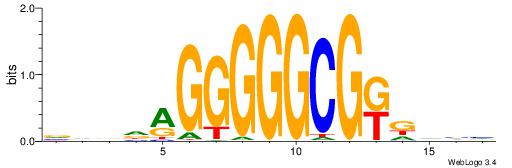 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.039112 |
Alignment:
VHYTCCDWCCGCGTCMMTHWHV
-----CGCGSGCGCGC------
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Dataset #: 2 | Motif ID: 33 | Motif name: srACyCCGAyr |
| Original motif | Reverse complement motif |
| Consensus sequence: SRACYCCGAYR | Consensus sequence: KKTCGGKGTKS |
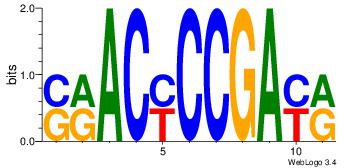 |
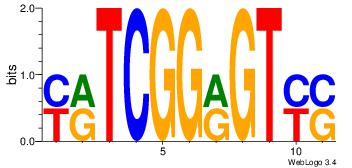 |
Best Matches for Motif ID 33 (Highest to Lowest)
| Motif ID: | UP00024 |
| Motif name: | Glis2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0129914 |
Alignment:
HKBBGGGGGGTCVHHH
--KKTCGGKGTKS---
| Original motif | Reverse complement motif |
| Consensus sequence: HHHVGACCCCCCVBRD | Consensus sequence: HKBBGGGGGGTCVHHH |
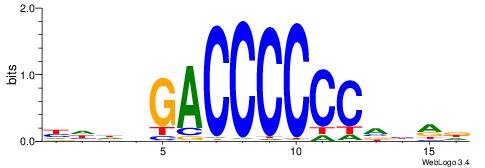 |
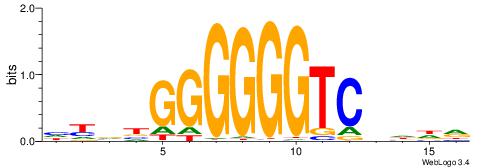 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.013032 |
Alignment:
VDWDARRGACGCGGWHGGAKHV
------SRACYCCGAYR-----
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00070 |
| Motif name: | Gcm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0132459 |
Alignment:
BYMBKCCCCTATDVBHD
--SRACYCCGAYR----
| Original motif | Reverse complement motif |
| Consensus sequence: DDBVHATAGGGGRBRMB | Consensus sequence: BYMBKCCCCTATDVBHD |
 |
 |
| Motif ID: | UP00070 |
| Motif name: | Gcm1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0135374 |
Alignment:
HVVHACCCGCATVDHD
--SRACYCCGAYR---
| Original motif | Reverse complement motif |
| Consensus sequence: HVVHACCCGCATVDHD | Consensus sequence: DHDVATGCGGGTHVVH |
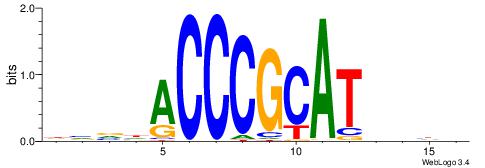 |
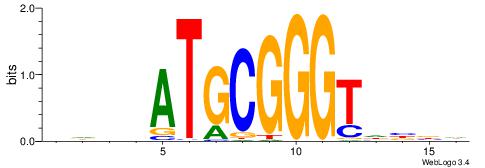 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0136908 |
Alignment:
DKKWDACCCCCAHTDDV
---SRACYCCGAYR---
| Original motif | Reverse complement motif |
| Consensus sequence: DKKWDACCCCCAHTDDV | Consensus sequence: VDDAHTGGGGGTHWYYD |
 |
 |
| Motif ID: | UP00047 |
| Motif name: | Zbtb7b_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0146089 |
Alignment:
VHWTTHGGGGGGVDD
--KKTCGGKGTKS--
| Original motif | Reverse complement motif |
| Consensus sequence: DDVCCCCCCHAAWHB | Consensus sequence: VHWTTHGGGGGGVDD |
 |
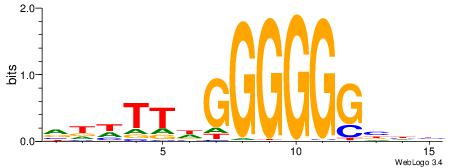 |
| Motif ID: | UP00040 |
| Motif name: | Irf5_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0148195 |
Alignment:
BDKGTTTCGKTWDDD
----KKTCGGKGTKS
| Original motif | Reverse complement motif |
| Consensus sequence: DDDWAYCGAAACYDV | Consensus sequence: BDKGTTTCGKTWDDD |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 11 |
| Similarity score: | 0.014859 |
Alignment:
ABBBBVVRGACCACCCACRDBBM
--------SRACYCCGAYR----
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0153296 |
Alignment:
GKRGGGGGGGGGGBD
KKTCGGKGTKS----
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
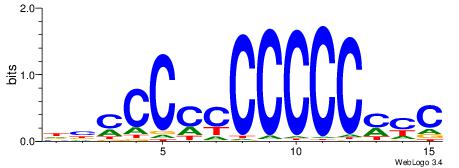 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 10 |
| Number of overlap: | 11 |
| Similarity score: | 0.0155548 |
Alignment:
DBCDWHCTGGGTGGTCMDHBBAH
---KKTCGGKGTKS---------
| Original motif | Reverse complement motif |
| Consensus sequence: DTVBDDRGACCACCCAGDWDGBH | Consensus sequence: DBCDWHCTGGGTGGTCMDHBBAH |
 |
 |