Best Matches in Database for Each Motif (Highest to Lowest)
| Dataset #: 1 | Motif ID: 1 | Motif name: C001 |
| Original motif | Reverse complement motif |
| Consensus sequence: WWWAAAAAWWAAAAAWAAAAAWWAAAAHWW | Consensus sequence: WWHTTTTWWTTTTTWTTTTTWWTTTTTWWW |
 |
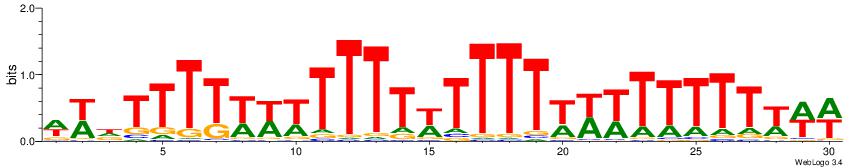 |
Best Matches for Motif ID 1 (Highest to Lowest)
| Motif ID: | UP00528 |
| Motif name: | Foxm1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 23 |
| Similarity score: | 0.0846473 |
Alignment:
-------HADTBVADGATGCATCMTGMVHB
WWWAAAAAWWAAAAAWAAAAAWWAAAAHWW--
| Original motif | Reverse complement motif |
| Consensus sequence: HADTBVADGATGCATCMTGMVHB | Consensus sequence: VHVRCAYGATGCATCDTVVADTH |
 |
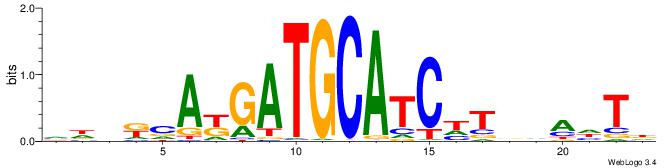 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 22 |
| Similarity score: | 0.561689 |
Alignment:
--------HHBDHDDAACAATDRHHHHHBW
WWWAAAAAWWAAAAAWAAAAAWWAAAAHWW-
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
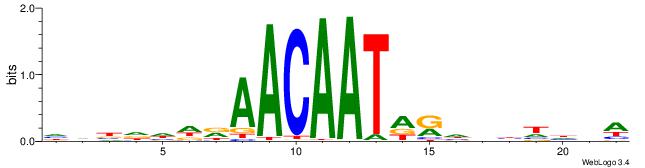 |
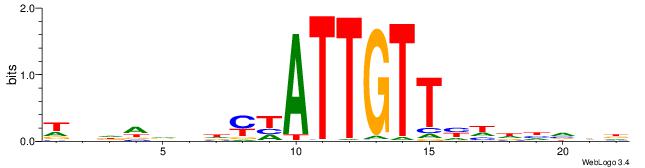 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 22 |
| Similarity score: | 0.566383 |
Alignment:
--------HBHDVHDTCTTATCTHTDDHDV
WWHTTTTWWTTTTTWTTTTTWWTTTTTWWW-----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
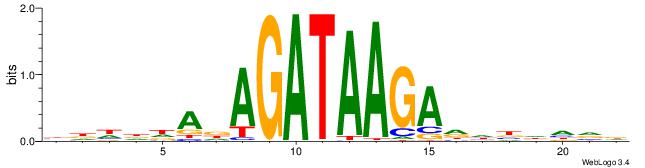 |
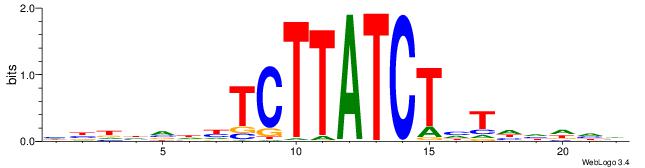 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 22 |
| Similarity score: | 0.575767 |
Alignment:
HBHDVBWGATHBTATCRVBHHH--------
------WWWAAAAAWWAAAAAWAAAAAWWAAAAHWW
| Original motif | Reverse complement motif |
| Consensus sequence: HBHDVBWGATHBTATCRVBHHH | Consensus sequence: HHHBBMGATAVHATCWVVDHVH |
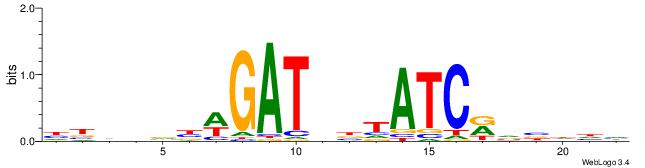 |
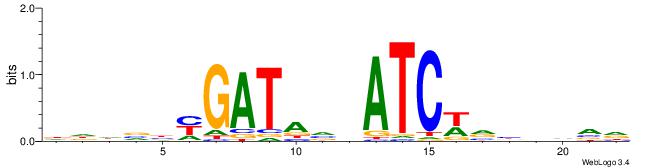 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 22 |
| Similarity score: | 0.578275 |
Alignment:
--------VVYDDDCHCAVACAATTDBVAC
WWHTTTTWWTTTTTWTTTTTWWTTTTTWWW-----
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
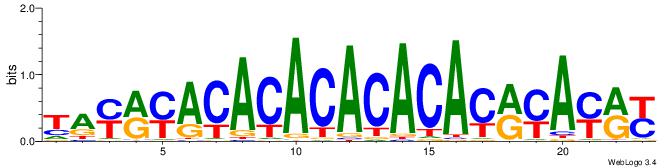
| Dataset #: 1 | Motif ID: 2 | Motif name: C002 |
| Original motif | Reverse complement motif |
| Consensus sequence: TAYRYACACACACACACRYAYRY | Consensus sequence: MKKTKKGTGTGTGTGTGTKKKTA |
 |
 |
Best Matches for Motif ID 2 (Highest to Lowest)
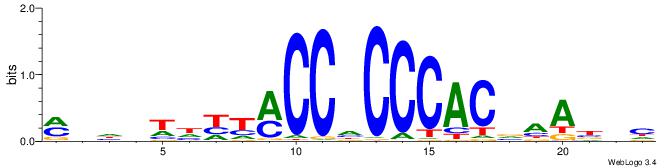
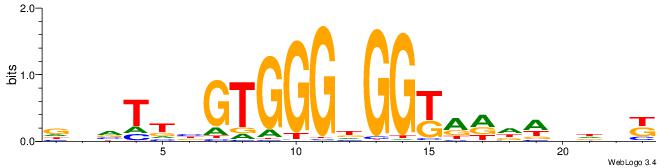
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 23 |
| Similarity score: | 0.0589959 |
Alignment:
MHHHWHYTMCCHCCCACVAABVH
TAYRYACACACACACACRYAYRY
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
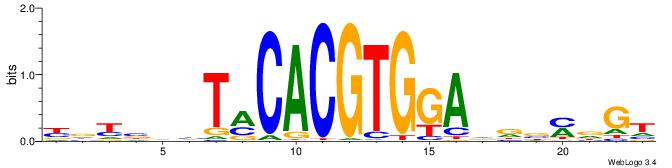
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 23 |
| Similarity score: | 0.0678722 |
Alignment:
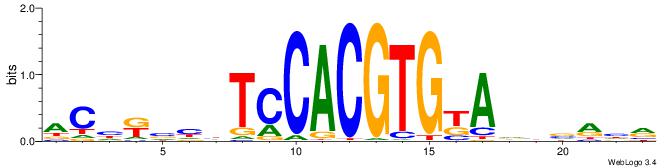
YDYBDHTMCACGTGGADDBMDGT
TAYRYACACACACACACRYAYRY
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 23 |
| Similarity score: | 0.0739216 |
Alignment:
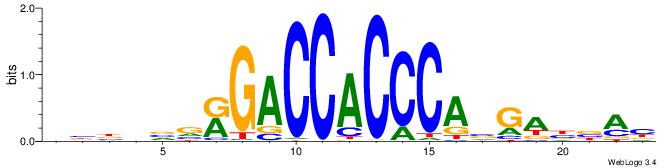
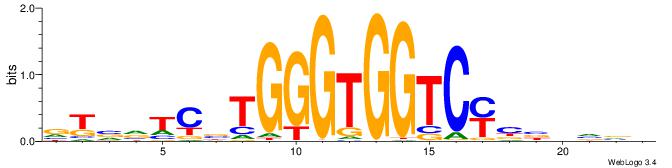
BBBDVVRGACCACCCAVGABBAB
TAYRYACACACACACACRYAYRY
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 23 |
| Similarity score: | 0.077586 |
Alignment:
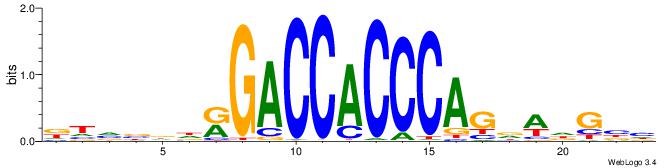
DTVBDDRGACCACCCAGDWDGBH
TAYRYACACACACACACRYAYRY
| Original motif | Reverse complement motif |
| Consensus sequence: DTVBDDRGACCACCCAGDWDGBH | Consensus sequence: DBCDWHCTGGGTGGTCMDHBBAH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 22 |
| Similarity score: | 0.55938 |
Alignment:
-HVBDAGCGBGGGTGGCRDBDDDB
MKKTKKGTGTGTGTGTGTKKKTA-
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
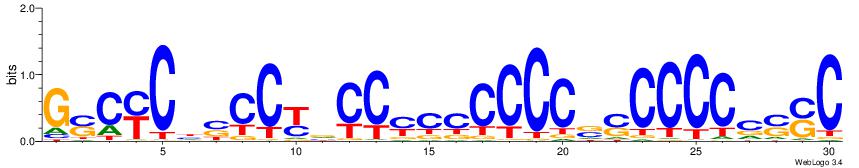
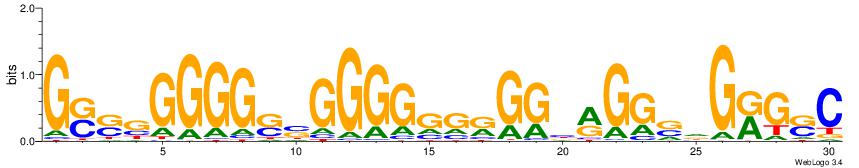
| Dataset #: 1 | Motif ID: 3 | Motif name: C003 |
| Original motif | Reverse complement motif |
| Consensus sequence: GSCYCHSCCYVCCYCCCCCCSSCCCCCSSC | Consensus sequence: GSSGGGGGSSGGGGGGKGGVMGGSHGKGSC |
 |
 |
Best Matches for Motif ID 3 (Highest to Lowest)
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 23 |
| Similarity score: | 0.0504417 |
Alignment:
-------VYWVVHCRCCACMCACGAHSBHH
GSCYCHSCCYVCCYCCCCCCSSCCCCCSSC-
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 23 |
| Similarity score: | 0.0508168 |
Alignment:
-------MHHHWHYTMCCHCCCACVAABVH
GSCYCHSCCYVCCYCCCCCCSSCCCCCSSC--
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 23 |
| Similarity score: | 0.0518257 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH-------
-GSSGGGGGSSGGGGGGKGGVMGGSHGKGSC
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 23 |
| Similarity score: | 0.05306 |
Alignment:
-------BADHBDHCGCCCMCGCAHHDBBV
GSCYCHSCCYVCCYCCCCCCSSCCCCCSSC
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 23 |
| Similarity score: | 0.0591633 |
Alignment:
-------BTBVTCVTGGGTGGTCMVVDVBB
GSSGGGGGSSGGGGGGKGGVMGGSHGKGSC-----
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
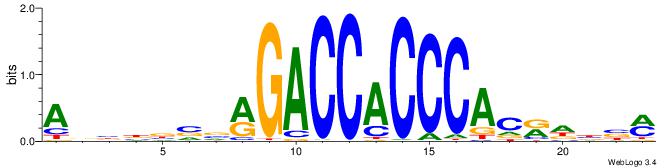
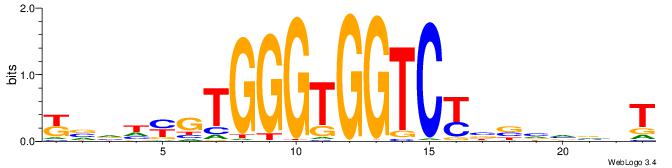
| Dataset #: 1 | Motif ID: 4 | Motif name: C004 |
| Original motif | Reverse complement motif |
| Consensus sequence: SSSSSCSSSSSCCSSSSCCSCSSSSSS | Consensus sequence: SSSSSSGSGGSSSSGGSSSSSGSSSSS |
 |
 |
Best Matches for Motif ID 4 (Highest to Lowest)
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 23 |
| Similarity score: | 0.0378869 |
Alignment:
HHBSHTCGTGRGTGGKGDBVWMV----
-SSSSSSGSGGSSSSGGSSSSSGSSSSS
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 23 |
| Similarity score: | 0.042097 |
Alignment:
----HVBDAGCGBGGGTGGCRDBDDDB
SSSSSCSSSSSCCSSSSCCSCSSSSSS----
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 23 |
| Similarity score: | 0.042858 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB----
--SSSSSSGSGGSSSSGGSSSSSGSSSSS
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 23 |
| Similarity score: | 0.0467324 |
Alignment:
BTBVTCVTGGGTGGTCMVVDVBB----
--SSSSSSGSGGSSSSGGSSSSSGSSSSS
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 23 |
| Similarity score: | 0.0486423 |
Alignment:
----ABBBBVVRGACCACCCACRDBBM
SSSSSCSSSSSCCSSSSCCSCSSSSSS---
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
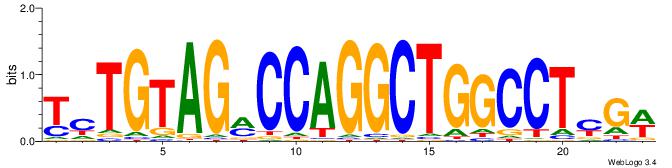
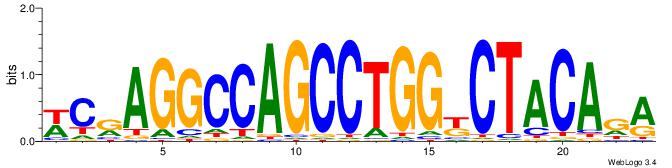
| Dataset #: 1 | Motif ID: 5 | Motif name: C005 |
| Original motif | Reverse complement motif |
| Consensus sequence: TCTGTAGMCCAGGCTGGCCTYGW | Consensus sequence: WCKAGGCCAGCCTGGYCTACAGA |
 |
 |
Best Matches for Motif ID 5 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 23 |
| Similarity score: | 0.0620662 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
TCTGTAGMCCAGGCTGGCCTYGW
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 22 |
| Similarity score: | 0.553936 |
Alignment:
-DTVBDDRGACCACCCAGDWDGBH
TCTGTAGMCCAGGCTGGCCTYGW-
| Original motif | Reverse complement motif |
| Consensus sequence: DTVBDDRGACCACCCAGDWDGBH | Consensus sequence: DBCDWHCTGGGTGGTCMDHBBAH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 22 |
| Similarity score: | 0.556669 |
Alignment:
-BBBDVVRGACCACCCAVGABBAB
TCTGTAGMCCAGGCTGGCCTYGW-
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
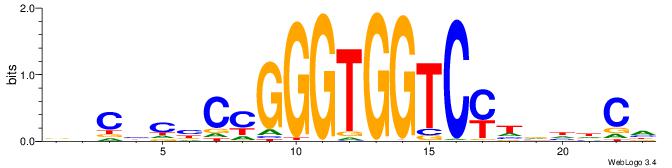
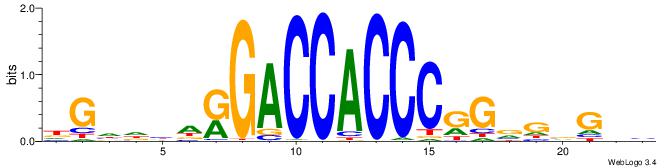
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 22 |
| Similarity score: | 0.55849 |
Alignment:
HGDHVHKGACCACCCGGBGBGDB-
-WCKAGGCCAGCCTGGYCTACAGA
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
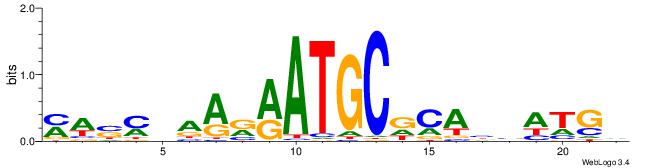
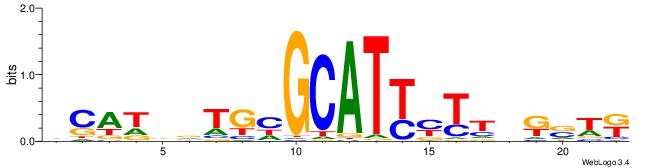
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 22 |
| Similarity score: | 0.56544 |
Alignment:
DCATDBTGHGCATKMTTBRSWR-
WCKAGGCCAGCCTGGYCTACAGA
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
 |
 |
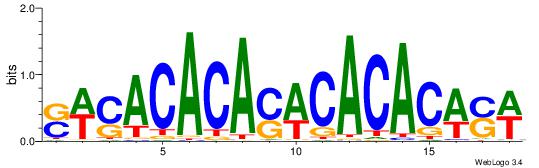
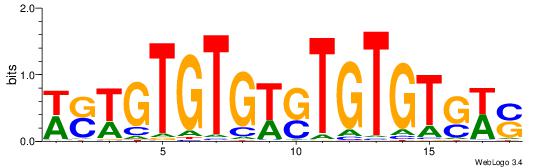
| Dataset #: 1 | Motif ID: 6 | Motif name: C006 |
| Original motif | Reverse complement motif |
| Consensus sequence: SWCACACASWCACACWSW | Consensus sequence: WSWGTGTGWSTGTGTGWS |
 |
 |
Best Matches for Motif ID 6 (Highest to Lowest)
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0361661 |
Alignment:
VVYDDDCHCAVACAATTDBVAC
--SWCACACASWCACACWSW--
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0406554 |
Alignment:
BADHBDHCGCCCMCGCAHHDBBV
---SWCACACASWCACACWSW--
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.040868 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
-SWCACACASWCACACWSW----
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0421858 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
WSWGTGTGWSTGTGTGWS-----
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0427432 |
Alignment:
VCTBHBVCTTGGWTACVVVVVDB
---SWCACACASWCACACWSW--
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
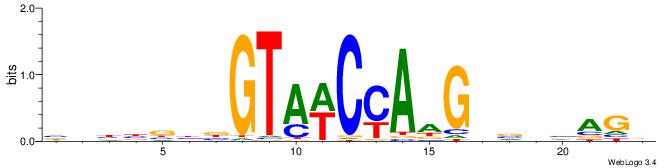 |
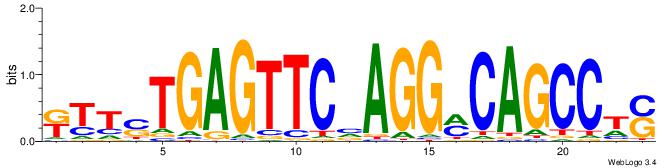
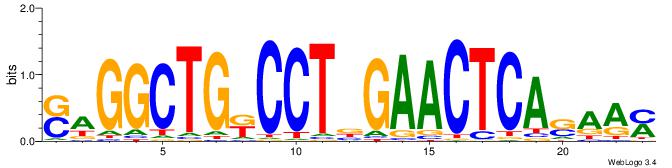
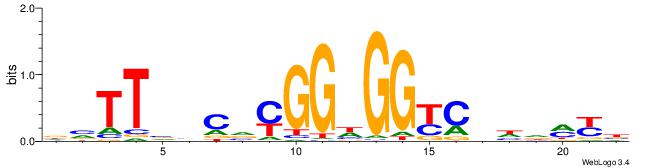
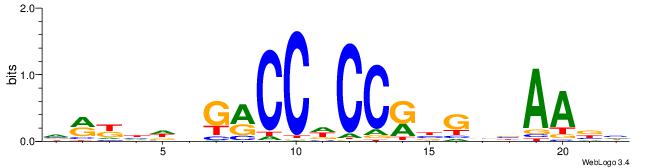
| Dataset #: 1 | Motif ID: 7 | Motif name: C007 |
| Original motif | Reverse complement motif |
| Consensus sequence: KTTSTGAGTTCVAGGMCAGCCTS | Consensus sequence: SAGGCTGYCCTVGAACTCASAAY |
 |
 |
Best Matches for Motif ID 7 (Highest to Lowest)
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 22 |
| Similarity score: | 0.0642913 |
Alignment:
VVYDDDCHCAVACAATTDBVAC-
-SAGGCTGYCCTVGAACTCASAAY
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 22 |
| Similarity score: | 0.0664801 |
Alignment:
-VCTBHBVCTTGGWTACVVVVVDB
KTTSTGAGTTCVAGGMCAGCCTS-
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
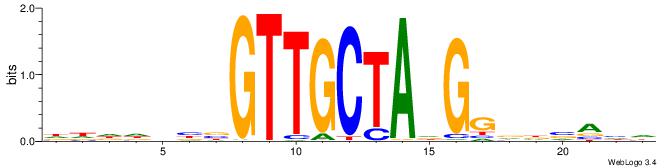
| Motif ID: | UP00098 |
| Motif name: | Rfx3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 22 |
| Similarity score: | 0.0672967 |
Alignment:
-HDTBHHCCHTAGCAACVVDHHHH
SAGGCTGYCCTVGAACTCASAAY-
| Original motif | Reverse complement motif |
| Consensus sequence: HDTBHHCCHTAGCAACVVDHHHH | Consensus sequence: HHHHDVVGTTGCTAHGGDHBAHH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 22 |
| Similarity score: | 0.0707277 |
Alignment:
-DVVTTVGTGGGHGGYAMHWHHHY
KTTSTGAGTTCVAGGMCAGCCTS-
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 21 |
| Similarity score: | 0.562337 |
Alignment:
VMYDHDGMCCHCCKBGVVAAVH--
--SAGGCTGYCCTVGAACTCASAAY
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
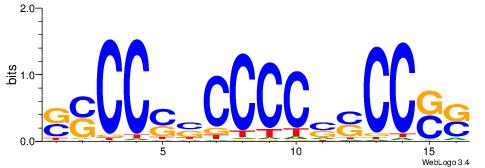
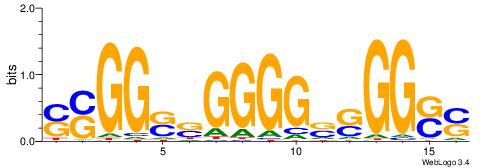
| Dataset #: 1 | Motif ID: 8 | Motif name: C008 |
| Original motif | Reverse complement motif |
| Consensus sequence: SSCCSSCCCCSSCCSS | Consensus sequence: SSGGSSGGGGSSGGSS |
 |
 |
Best Matches for Motif ID 8 (Highest to Lowest)
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0353177 |
Alignment:
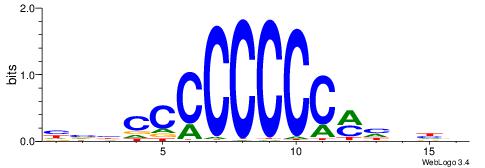
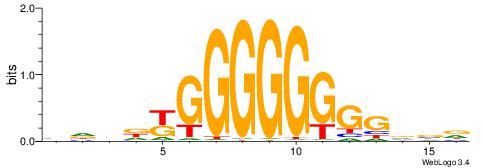
HVBCCCCCCCCMHHHB
SSCCSSCCCCSSCCSS
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 16 |
| Similarity score: | 0.0353208 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
--SSCCSSCCCCSSCCSS----
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 16 |
| Similarity score: | 0.0416492 |
Alignment:
HHBSHTCGTGRGTGGKGDBVWMV
-------SSGGSSGGGGSSGGSS
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 16 |
| Similarity score: | 0.0427346 |
Alignment:
DVVTTVGTGGGHGGYAMHWHHHY
SSGGSSGGGGSSGGSS-------
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0428588 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
-SSGGSSGGGGSSGGSS------
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Dataset #: 1 | Motif ID: 9 | Motif name: C009 |
| Original motif | Reverse complement motif |
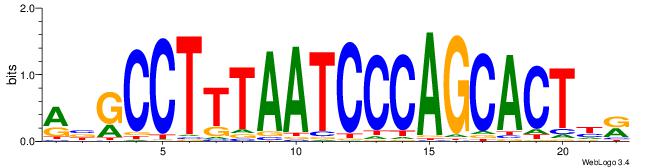
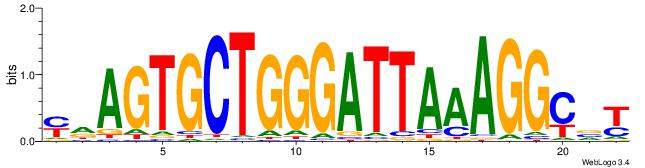
| Consensus sequence: ABGCCTTTAATCCCAGCACTHR | Consensus sequence: MHAGTGCTGGGATTAAAGGCBT |
 |
 |
Best Matches for Motif ID 9 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 22 |
| Similarity score: | 0.0664875 |
Alignment:
DVVTTVGTGGGHGGYAMHWHHHY
MHAGTGCTGGGATTAAAGGCBT-
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 21 |
| Similarity score: | 0.56321 |
Alignment:
HBHDVBWGATHBTATCRVBHHH-
-ABGCCTTTAATCCCAGCACTHR
| Original motif | Reverse complement motif |
| Consensus sequence: HBHDVBWGATHBTATCRVBHHH | Consensus sequence: HHHBBMGATAVHATCWVVDHVH |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 21 |
| Similarity score: | 0.565627 |
Alignment:
-BADHBDHCGCCCMCGCAHHDBBV
ABGCCTTTAATCCCAGCACTHR--
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 21 |
| Similarity score: | 0.56702 |
Alignment:
HVGVSAGGAGGGTCYHHHHYVH-
-ABGCCTTTAATCCCAGCACTHR
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
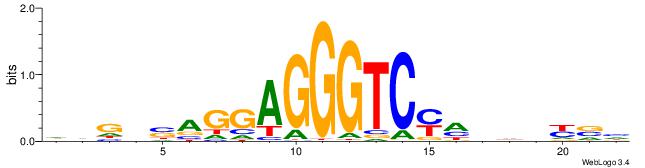 |
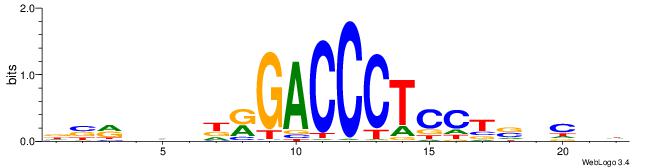 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 20 |
| Similarity score: | 1.06431 |
Alignment:
--BDBBVBVGTAWCCAAGVBHBAGB
MHAGTGCTGGGATTAAAGGCBT---
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Dataset #: 1 | Motif ID: 10 | Motif name: C010 |
| Original motif | Reverse complement motif |
| Consensus sequence: WWWAAAAWWAAAAAAAAAAWW | Consensus sequence: WWTTTTTTTTTTWWTTTTWWW |
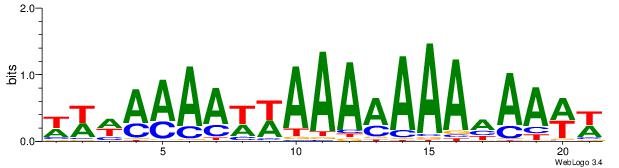 |
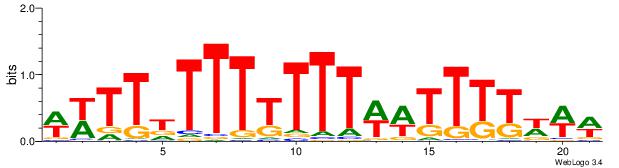 |
Best Matches for Motif ID 10 (Highest to Lowest)
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 21 |
| Similarity score: | 0.0528064 |
Alignment:
HBHDVHDTCTTATCTHTDDHDV
WWTTTTTTTTTTWWTTTTWWW-
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 21 |
| Similarity score: | 0.0586273 |
Alignment:
HHBDHDDAACAATDRHHHHHBW
-WWWAAAAWWAAAAAAAAAAWW
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 21 |
| Similarity score: | 0.0606713 |
Alignment:
HHHBBMGATAVHATCWVVDHVH
-WWWAAAAWWAAAAAAAAAAWW
| Original motif | Reverse complement motif |
| Consensus sequence: HBHDVBWGATHBTATCRVBHHH | Consensus sequence: HHHBBMGATAVHATCWVVDHVH |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 21 |
| Similarity score: | 0.0617422 |
Alignment:
VVYDDDCHCAVACAATTDBVAC
WWWAAAAWWAAAAAAAAAAWW-
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 21 |
| Similarity score: | 0.065491 |
Alignment:
HDTBHHCCHTAGCAACVVDHHHH
WWWAAAAWWAAAAAAAAAAWW--
| Original motif | Reverse complement motif |
| Consensus sequence: HDTBHHCCHTAGCAACVVDHHHH | Consensus sequence: HHHHDVVGTTGCTAHGGDHBAHH |
 |
 |
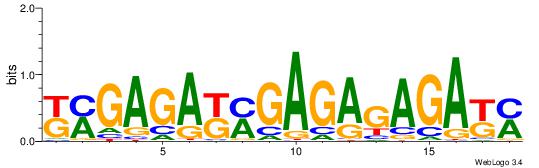
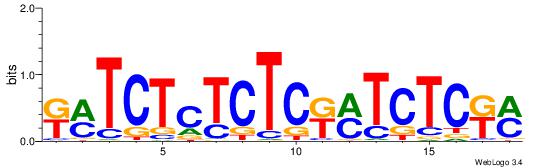
| Dataset #: 1 | Motif ID: 11 | Motif name: C011 |
| Original motif | Reverse complement motif |
| Consensus sequence: KMGAGAKMGAGAGAGAKM | Consensus sequence: RRTCTCTCTCRRTCTCRR |
 |
 |
Best Matches for Motif ID 11 (Highest to Lowest)
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.0404807 |
Alignment:
VVYDDDCHCAVACAATTDBVAC
---RRTCTCTCTCRRTCTCRR-
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.0471227 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
----RRTCTCTCTCRRTCTCRR-
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0474739 |
Alignment:
BDBBVBVGTAWCCAAGVBHBAGB
RRTCTCTCTCRRTCTCRR-----
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0475854 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
--KMGAGAKMGAGAGAGAKM--
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0500023 |
Alignment:
HHBSHTCGTGRGTGGKGDBVWMV
---KMGAGAKMGAGAGAGAKM--
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
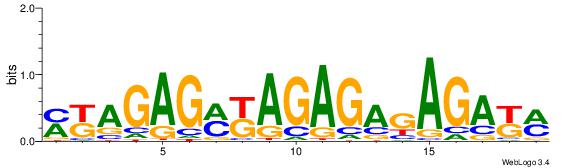
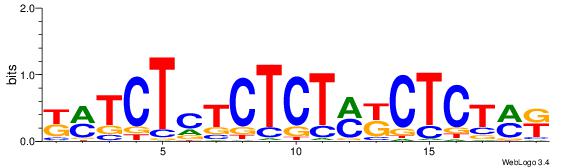
| Dataset #: 1 | Motif ID: 12 | Motif name: C012 |
| Original motif | Reverse complement motif |
| Consensus sequence: MKAGAGMKAGAGAGAGAKM | Consensus sequence: YRTCTCTCTCTRYCTCTRR |
 |
 |
Best Matches for Motif ID 12 (Highest to Lowest)
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 19 |
| Similarity score: | 0.0419739 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
-MKAGAGMKAGAGAGAGAKM--
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 19 |
| Similarity score: | 0.0446412 |
Alignment:
HBHDVHDTCTTATCTHTDDHDV
YRTCTCTCTCTRYCTCTRR---
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 19 |
| Similarity score: | 0.0455704 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
--MKAGAGMKAGAGAGAGAKM--
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 19 |
| Similarity score: | 0.0502954 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
--MKAGAGMKAGAGAGAGAKM--
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 19 |
| Similarity score: | 0.0510365 |
Alignment:
HHBSHTCGTGRGTGGKGDBVWMV
--MKAGAGMKAGAGAGAGAKM--
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Dataset #: 1 | Motif ID: 13 | Motif name: C013 |
| Original motif | Reverse complement motif |
| Consensus sequence: WVCTGGAACTCACTHTGTAGACCAGGM | Consensus sequence: YCCTGGTCTACADAGTGAGTTCCAGVW |
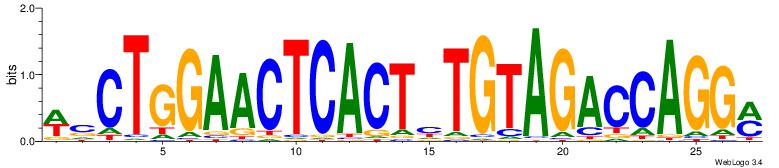 |
 |
Best Matches for Motif ID 13 (Highest to Lowest)
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 22 |
| Similarity score: | 0.0456555 |
Alignment:
-----MWSMBAARRATGCDCABHATGD
WVCTGGAACTCACTHTGTAGACCAGGM-----
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 22 |
| Similarity score: | 0.0503111 |
Alignment:
VVYDDDCHCAVACAATTDBVAC-----
-----YCCTGGTCTACADAGTGAGTTCCAGVW
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 22 |
| Similarity score: | 0.0518427 |
Alignment:
-----WHVBVVMACGCGCGTCDYHWDH
WVCTGGAACTCACTHTGTAGACCAGGM-----
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
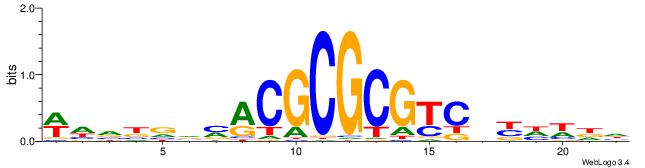 |
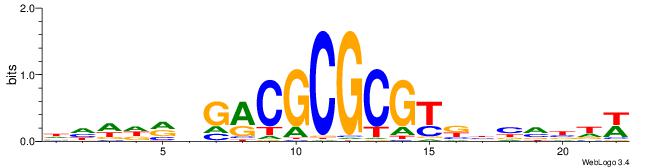 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 21 |
| Similarity score: | 0.546981 |
Alignment:
------BHCBCBCCGGGTGGTCYHVHDCH
WVCTGGAACTCACTHTGTAGACCAGGM------
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 21 |
| Similarity score: | 0.549369 |
Alignment:
------BTBVTCVTGGGTGGTCMVVDVBB
YCCTGGTCTACADAGTGAGTTCCAGVW--
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
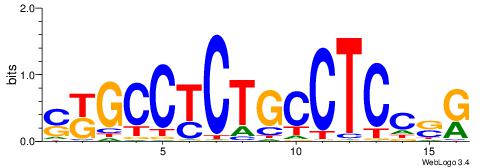
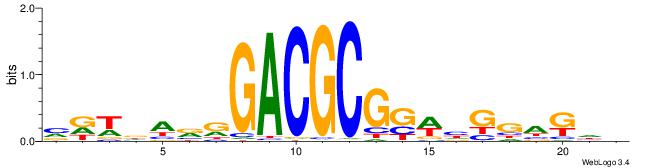
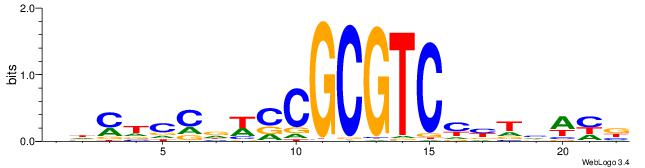
| Dataset #: 1 | Motif ID: 14 | Motif name: C014 |
| Original motif | Reverse complement motif |
| Consensus sequence: YSGGAGGCAGAGGCMS | Consensus sequence: SYGCCTCTGCCTCCSK |
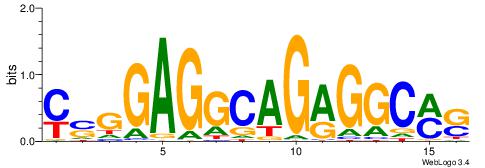 |
 |
Best Matches for Motif ID 14 (Highest to Lowest)
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 16 |
| Similarity score: | 0.0331356 |
Alignment:
VHYTCCDWCCGCGTCMMTHWHV
--SYGCCTCTGCCTCCSK----
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
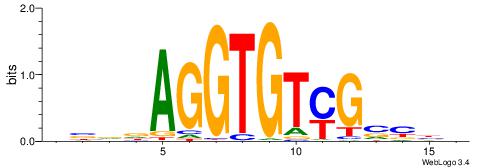
| Motif ID: | UP00068 |
| Motif name: | Eomes_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.034812 |
Alignment:
VVBVCKACACCTHVBV
SYGCCTCTGCCTCCSK
| Original motif | Reverse complement motif |
| Consensus sequence: VBVDAGGTGTYGVBBV | Consensus sequence: VVBVCKACACCTHVBV |
 |
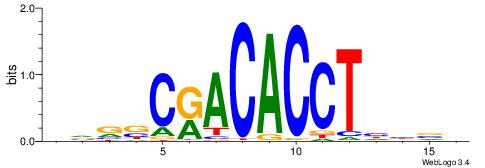 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 16 |
| Similarity score: | 0.034835 |
Alignment:
HVGVSAGGAGGGTCYHHHHYVH
YSGGAGGCAGAGGCMS------
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
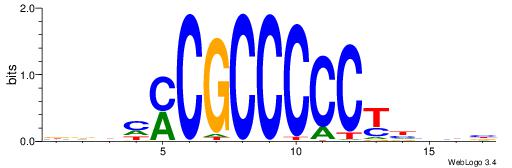
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.034915 |
Alignment:
BDHHMCGCCCCCTHVBB
-SYGCCTCTGCCTCCSK
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
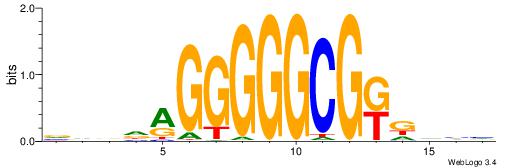 |
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0350751 |
Alignment:
BBBAVTGCAGTGBBVDD
-SYGCCTCTGCCTCCSK
| Original motif | Reverse complement motif |
| Consensus sequence: DDBBBCACTGCABTBBB | Consensus sequence: BBBAVTGCAGTGBBVDD |
 |
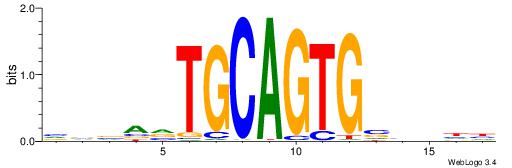 |
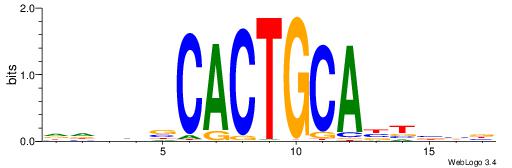
| Dataset #: 1 | Motif ID: 15 | Motif name: C015 |
| Original motif | Reverse complement motif |
| Consensus sequence: DVAGCCWKGGCTACAVAGCK | Consensus sequence: RGCTVTGTAGCCYWGGCTVD |
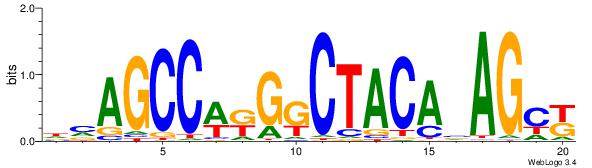 |
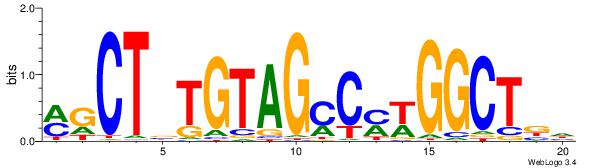 |
Best Matches for Motif ID 15 (Highest to Lowest)
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 20 |
| Similarity score: | 0.0486537 |
Alignment:
VCTBHBVCTTGGWTACVVVVVDB
--DVAGCCWKGGCTACAVAGCK-
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 20 |
| Similarity score: | 0.0536153 |
Alignment:
HDTBHHCCHTAGCAACVVDHHHH
--DVAGCCWKGGCTACAVAGCK-
| Original motif | Reverse complement motif |
| Consensus sequence: HDTBHHCCHTAGCAACVVDHHHH | Consensus sequence: HHHHDVVGTTGCTAHGGDHBAHH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 20 |
| Similarity score: | 0.0624325 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
-RGCTVTGTAGCCYWGGCTVD--
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
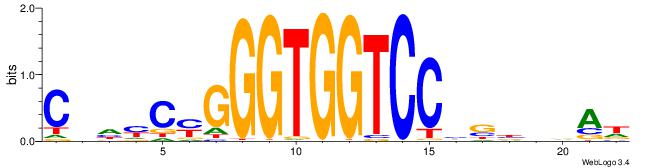
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 20 |
| Similarity score: | 0.0635845 |
Alignment:
CBDMCMGGGTGGTCCHVBVBAH
--RGCTVTGTAGCCYWGGCTVD
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
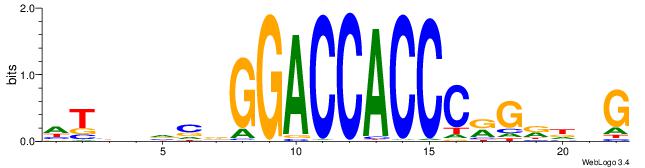 |
 |
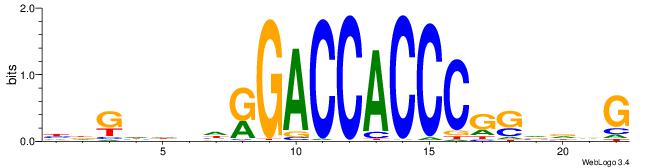
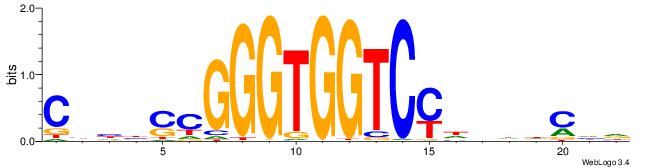
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 20 |
| Similarity score: | 0.0640763 |
Alignment:
CVHBSCGGGTGGTCMHVHHCDH
--RGCTVTGTAGCCYWGGCTVD
| Original motif | Reverse complement motif |
| Consensus sequence: HDGHHBHRGACCACCCGSVDVG | Consensus sequence: CVHBSCGGGTGGTCMHVHHCDH |
 |
 |
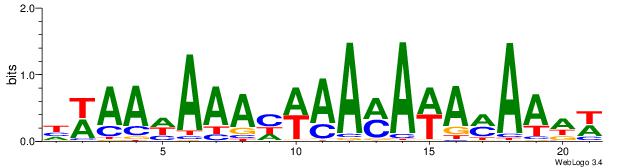
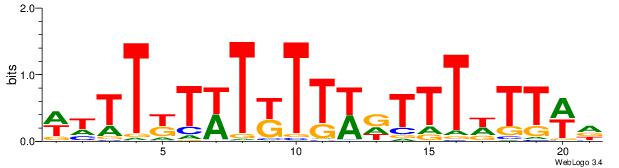
| Dataset #: 1 | Motif ID: 16 | Motif name: C016 |
| Original motif | Reverse complement motif |
| Consensus sequence: HWAAWAAAHWAAMAWAMAAWW | Consensus sequence: WWTTYTWTYTTWDTTTWTTWH |
 |
 |
Best Matches for Motif ID 16 (Highest to Lowest)
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 21 |
| Similarity score: | 0.0445695 |
Alignment:
HHBDHDDAACAATDRHHHHHBW
-HWAAWAAAHWAAMAWAMAAWW
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 21 |
| Similarity score: | 0.0472803 |
Alignment:
HBHDVHDTCTTATCTHTDDHDV
WWTTYTWTYTTWDTTTWTTWH-
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 21 |
| Similarity score: | 0.0518988 |
Alignment:
HBHDVBWGATHBTATCRVBHHH
-HWAAWAAAHWAAMAWAMAAWW
| Original motif | Reverse complement motif |
| Consensus sequence: HBHDVBWGATHBTATCRVBHHH | Consensus sequence: HHHBBMGATAVHATCWVVDHVH |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 21 |
| Similarity score: | 0.0523549 |
Alignment:
VVYDDDCHCAVACAATTDBVAC
HWAAWAAAHWAAMAWAMAAWW-
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 21 |
| Similarity score: | 0.0599775 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
-HWAAWAAAHWAAMAWAMAAWW-
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
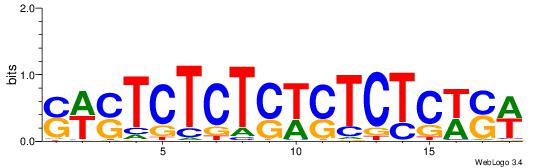
| Dataset #: 1 | Motif ID: 17 | Motif name: C017 |
| Original motif | Reverse complement motif |
| Consensus sequence: WSWGAGASWSAGAGASWS | Consensus sequence: SWSTCTCTSWSTCTCWSW |
 |
 |
Best Matches for Motif ID 17 (Highest to Lowest)
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0370092 |
Alignment:
VDWDARRGACGCGGWHGGAKHV
--WSWGAGASWSAGAGASWS--
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0399153 |
Alignment:
VVYDDDCHCAVACAATTDBVAC
--SWSTCTCTSWSTCTCWSW--
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 18 |
| Similarity score: | 0.0422644 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
WSWGAGASWSAGAGASWS-----
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.0425861 |
Alignment:
HHBSHTCGTGRGTGGKGDBVWMV
----WSWGAGASWSAGAGASWS-
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0439386 |
Alignment:
BDBBVBVGTAWCCAAGVBHBAGB
--WSWGAGASWSAGAGASWS---
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
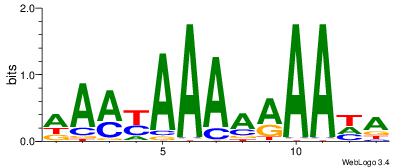
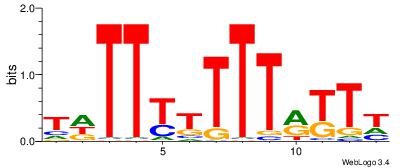
| Dataset #: 1 | Motif ID: 18 | Motif name: C018 |
| Original motif | Reverse complement motif |
| Consensus sequence: AAMYAAAAAAAHA | Consensus sequence: THTTTTTTTMYTT |
 |
 |
Best Matches for Motif ID 18 (Highest to Lowest)
| Motif ID: | UP00028 |
| Motif name: | Tcfap2e_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0145388 |
Alignment:
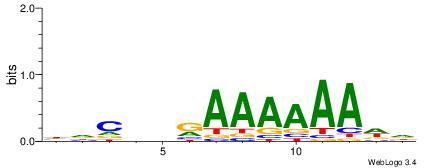
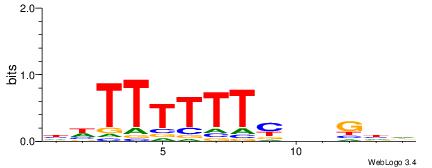
HDCDBRAAAAAADD
-AAMYAAAAAAAHA
| Original motif | Reverse complement motif |
| Consensus sequence: HDCDBRAAAAAADD | Consensus sequence: DDTTTTTTMBDGDH |
 |
 |
| Motif ID: | UP00077 |
| Motif name: | Srf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0166921 |
Alignment:
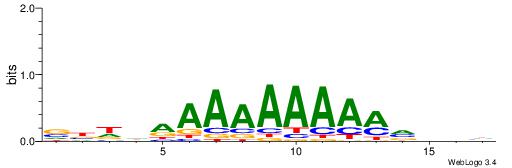
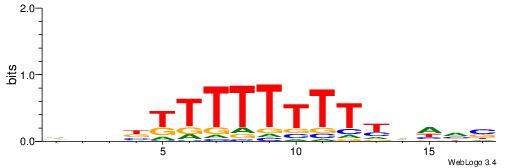
VBDDAAAAAAAAMVBHH
----AAMYAAAAAAAHA
| Original motif | Reverse complement motif |
| Consensus sequence: VBDDAAAAAAAAMVBHH | Consensus sequence: HHVBYTTTTTTTTDDVV |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 13 |
| Similarity score: | 0.0192633 |
Alignment:
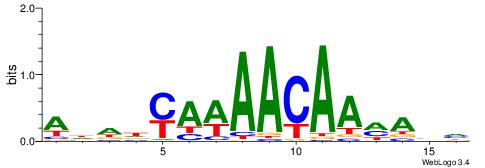
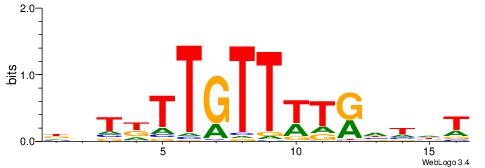
ADDDYAWAACAAMAHH
-AAMYAAAAAAAHA--
| Original motif | Reverse complement motif |
| Consensus sequence: ADDDYAWAACAAMAHH | Consensus sequence: HDTYTTGTTWTKDDDT |
 |
 |
| Motif ID: | UP00134 |
| Motif name: | Hoxb13 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0204511 |
Alignment:
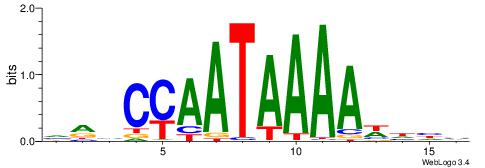
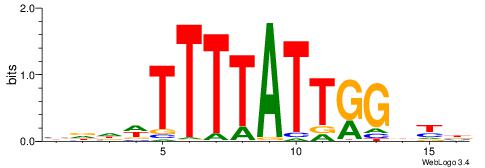
VRHCCAATAAAAWHHV
-AAMYAAAAAAAHA--
| Original motif | Reverse complement motif |
| Consensus sequence: VRHCCAATAAAAWHHV | Consensus sequence: VDHWTTTTATTGGDKB |
 |
 |
| Motif ID: | UP00041 |
| Motif name: | Foxj1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0217367 |
Alignment:
HDHRTAAACAAAHDDD
-AAMYAAAAAAAHA--
| Original motif | Reverse complement motif |
| Consensus sequence: HDHRTAAACAAAHDDD | Consensus sequence: DDDHTTTGTTTAMHDH |
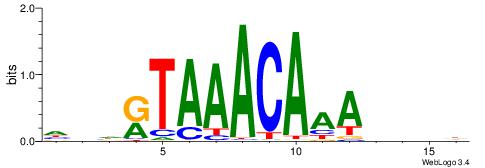 |
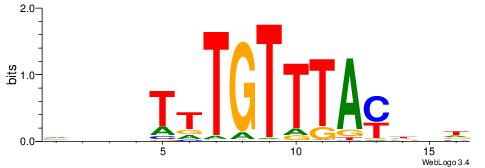 |
| Dataset #: 1 | Motif ID: 19 | Motif name: C019 |
| Original motif | Reverse complement motif |
| Consensus sequence: MWYCCCAGCACTTGGGAGGCAG | Consensus sequence: CTGCCTCCCAAGTGCTGGGMWR |
 |
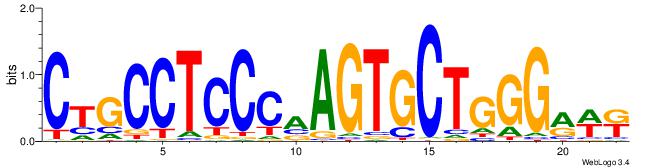 |
Best Matches for Motif ID 19 (Highest to Lowest)
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 22 |
| Similarity score: | 0.041956 |
Alignment:
YDYBDHTMCACGTGGADDBMDGT
MWYCCCAGCACTTGGGAGGCAG-
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 22 |
| Similarity score: | 0.0441925 |
Alignment:
HHBSHTCGTGRGTGGKGDBVWMV
CTGCCTCCCAAGTGCTGGGMWR-
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 22 |
| Similarity score: | 0.0552063 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
MWYCCCAGCACTTGGGAGGCAG
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 22 |
| Similarity score: | 0.0596824 |
Alignment:
HTBVVVDGGACCACCCRGRDBG
MWYCCCAGCACTTGGGAGGCAG
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 22 |
| Similarity score: | 0.0599404 |
Alignment:
DDHBHBSTCACGTGAHBBDHHM
CTGCCTCCCAAGTGCTGGGMWR
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
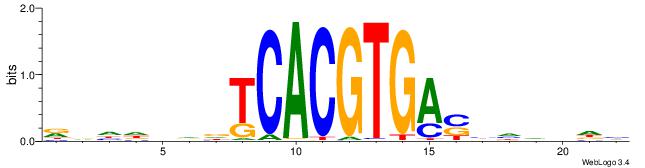 |
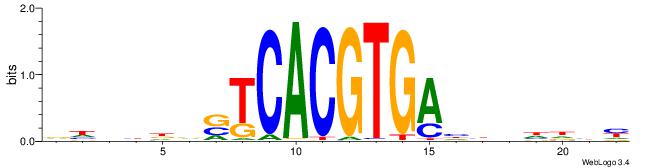 |
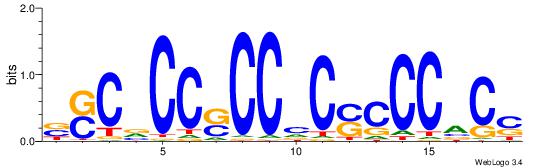
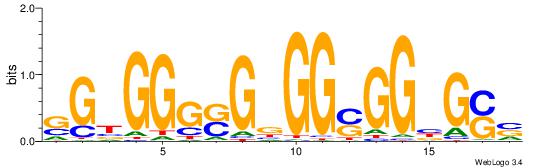
| Dataset #: 1 | Motif ID: 20 | Motif name: C020 |
| Original motif | Reverse complement motif |
| Consensus sequence: BSCVCCSCCVCSCCCACS | Consensus sequence: SGTGGGSGVGGSGGVGSB |
 |
 |
Best Matches for Motif ID 20 (Highest to Lowest)
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0350167 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
SGTGGGSGVGGSGGVGSB----
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.0412841 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
-SGTGGGSGVGGSGGVGSB----
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0415056 |
Alignment:
BADHBDHCGCCCMCGCAHHDBBV
--BSCVCCSCCVCSCCCACS---
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.0438926 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
-BSCVCCSCCVCSCCCACS----
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 18 |
| Similarity score: | 0.0475246 |
Alignment:
MHHHWHYTMCCHCCCACVAABVH
BSCVCCSCCVCSCCCACS-----
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
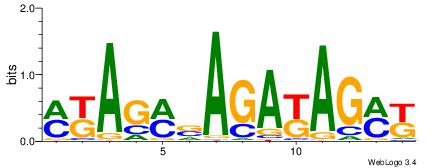
| Dataset #: 1 | Motif ID: 21 | Motif name: C021 |
| Original motif | Reverse complement motif |
| Consensus sequence: MKAGMVAGAKAGMK | Consensus sequence: RYCTRTCTVYCTRY |
 |
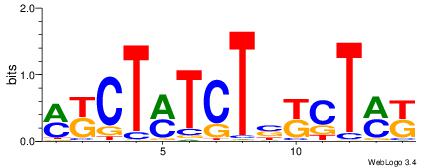 |
Best Matches for Motif ID 21 (Highest to Lowest)
| Motif ID: | UP00101 |
| Motif name: | Sox12_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0269394 |
Alignment:
HDTMCTTTGTSTVDBB
--RYCTRTCTVYCTRY
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBASACAAAGRADH | Consensus sequence: HDTMCTTTGTSTVDBB |
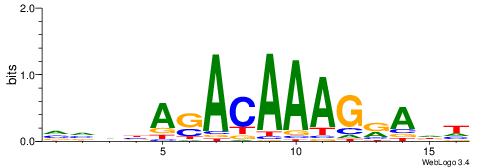 |
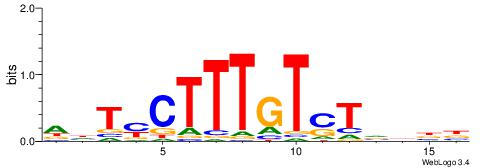 |
| Motif ID: | >UP00011 |
| Motif name: | Irf6_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0331495 |
Alignment:
VBBACTCYCGGKHDV
-MKAGMVAGAKAGMK
| Original motif | Reverse complement motif |
| Consensus sequence: VBBACTCYCGGKHDV | Consensus sequence: VDDRCCGMGAGTBBB |
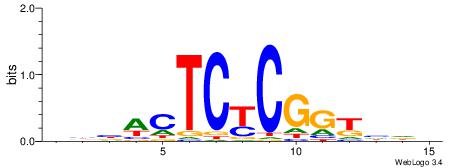 |
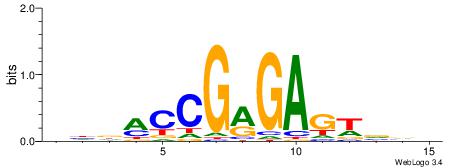 |
| Motif ID: | UP00040 |
| Motif name: | Irf5_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0341976 |
Alignment:
BHDAHCGAGARTHBV
-MKAGMVAGAKAGMK
| Original motif | Reverse complement motif |
| Consensus sequence: BHDAHCGAGARTHBV | Consensus sequence: VBHAKTCTCGHTHHV |
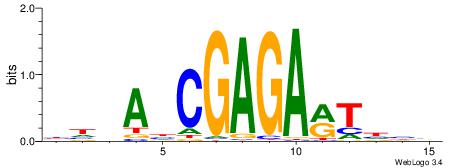 |
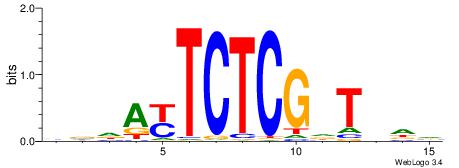 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0359613 |
Alignment:
HVDDVCAGATGKYHBBV
MKAGMVAGAKAGMK---
| Original motif | Reverse complement motif |
| Consensus sequence: HVDDVCAGATGKYHBBV | Consensus sequence: VBBDMYCATCTGVHHBH |
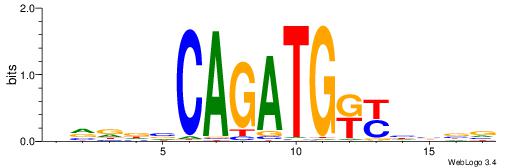 |
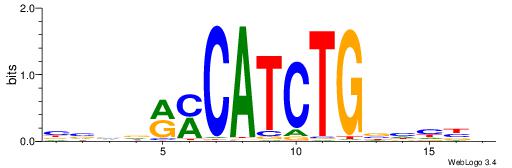 |
| Motif ID: | UP00042 |
| Motif name: | Gm397_second |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0368158 |
Alignment:
HBGYGTGTGTGCVRVD
--RYCTRTCTVYCTRY
| Original motif | Reverse complement motif |
| Consensus sequence: DVMVGCACACACKCVH | Consensus sequence: HBGYGTGTGTGCVRVD |
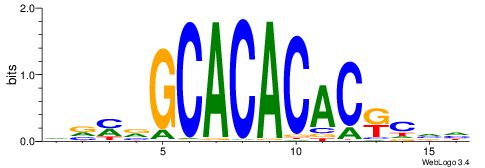 |
 |
| Dataset #: 1 | Motif ID: 22 | Motif name: C022 |
| Original motif | Reverse complement motif |
| Consensus sequence: BGCCYCBGSCYCCCSVS | Consensus sequence: SVSGGGKGSCBGMGGCV |
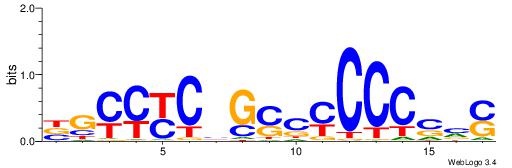 |
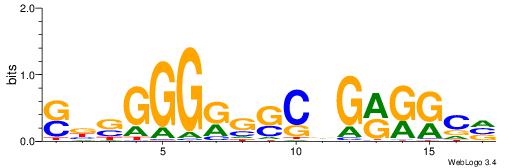 |
Best Matches for Motif ID 22 (Highest to Lowest)
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 17 |
| Similarity score: | 0.0352062 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
BGCCYCBGSCYCCCSVS-----
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0384374 |
Alignment:
BHHBHCCGCCCCDHHHH
BGCCYCBGSCYCCCSVS
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
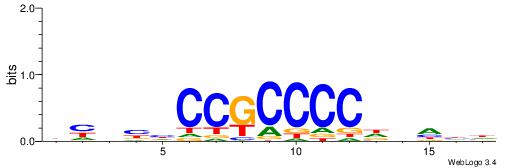 |
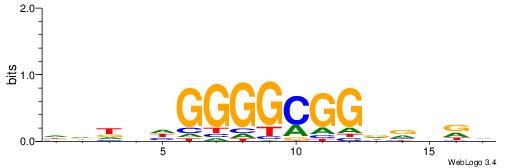 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 17 |
| Similarity score: | 0.0394824 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
------SVSGGGKGSCBGMGGCV
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0470502 |
Alignment:
BDHHMCGCCCCCTHVBB
BGCCYCBGSCYCCCSVS
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 17 |
| Similarity score: | 0.0477827 |
Alignment:
HHBSHTCGTGRGTGGKGDBVWMV
------SVSGGGKGSCBGMGGCV
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Dataset #: 1 | Motif ID: 23 | Motif name: C023 |
| Original motif | Reverse complement motif |
| Consensus sequence: SSCCSCBHCSCCSS | Consensus sequence: SSGGSGDBGSGGSS |
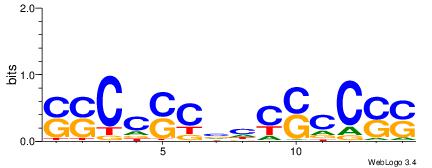 |
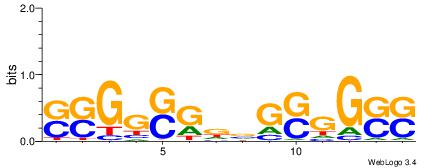 |
Best Matches for Motif ID 23 (Highest to Lowest)
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0233667 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
---SSCCSCBHCSCCSS-----
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0308308 |
Alignment:
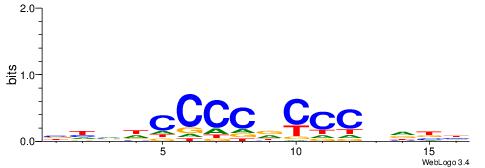
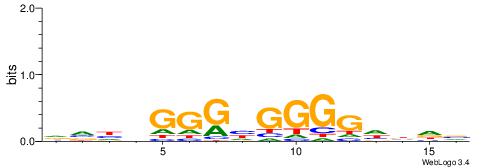
HVDVGGGHGGGGDBHB
SSGGSGDBGSGGSS--
| Original motif | Reverse complement motif |
| Consensus sequence: BHVDCCCCDCCCBDBH | Consensus sequence: HVDVGGGHGGGGDBHB |
 |
 |
| Motif ID: | UP00013 |
| Motif name: | Gabpa_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
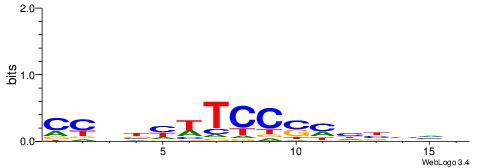
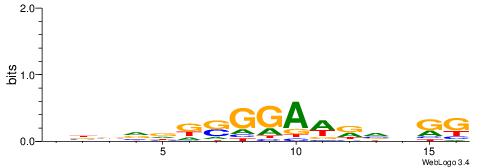
| Similarity score: | 0.0321962 |
Alignment:
DBVVDRSGGAWKVHKG
--SSGGSGDBGSGGSS
| Original motif | Reverse complement motif |
| Consensus sequence: CYDBYWTCCSMHBVVH | Consensus sequence: DBVVDRSGGAWKVHKG |
 |
 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0322741 |
Alignment:
GKRGGGGGGGGGGBD
-SSGGSGDBGSGGSS
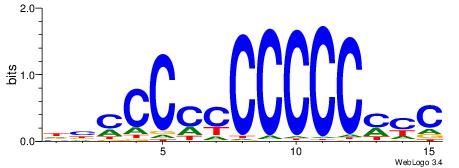
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0324541 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
-------SSCCSCBHCSCCSS--
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Dataset #: 1 | Motif ID: 24 | Motif name: C024 |
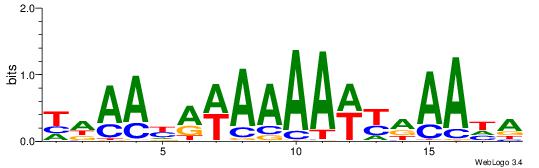
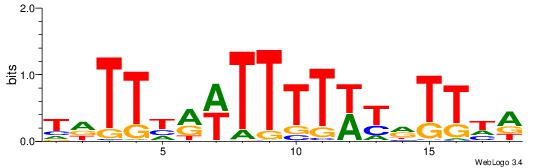
| Original motif | Reverse complement motif |
| Consensus sequence: TDAAYRWAAAAWYDAAHA | Consensus sequence: THTTDMWTTTTWKMTTDA |
 |
 |
Best Matches for Motif ID 24 (Highest to Lowest)
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0410905 |
Alignment:
WBHHHHHMDATTGTTHDHDVHH
--THTTDMWTTTTWKMTTDA--
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.0441479 |
Alignment:
VVYDDDCHCAVACAATTDBVAC
----TDAAYRWAAAAWYDAAHA
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0508186 |
Alignment:
HBHDVHDTCTTATCTHTDDHDV
THTTDMWTTTTWKMTTDA----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0546986 |
Alignment:
HHHBBMGATAVHATCWVVDHVH
--TDAAYRWAAAAWYDAAHA--
| Original motif | Reverse complement motif |
| Consensus sequence: HBHDVBWGATHBTATCRVBHHH | Consensus sequence: HHHBBMGATAVHATCWVVDHVH |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0553 |
Alignment:
MWSMBAARRATGCDCABHATGD
TDAAYRWAAAAWYDAAHA----
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
 |
 |
| Dataset #: 1 | Motif ID: 25 | Motif name: C025 |
| Original motif | Reverse complement motif |
| Consensus sequence: KMGAKAGAGMGAKM | Consensus sequence: RRTCRCTCTRTCRR |
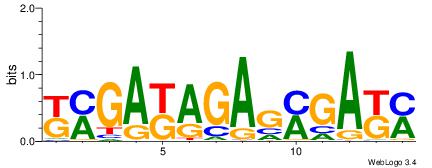 |
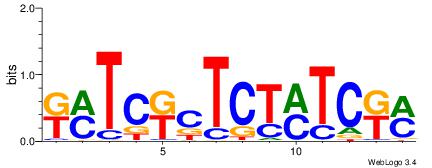 |
Best Matches for Motif ID 25 (Highest to Lowest)
| Motif ID: | UP00080 |
| Motif name: | Gata5_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.036512 |
Alignment:
VVBHRAGATATCWDHBB
RRTCRCTCTRTCRR---
| Original motif | Reverse complement motif |
| Consensus sequence: VVBHRAGATATCWDHBB | Consensus sequence: VVHHWGATATCTMHBBV |
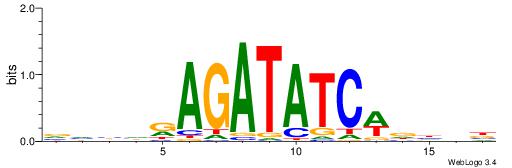 |
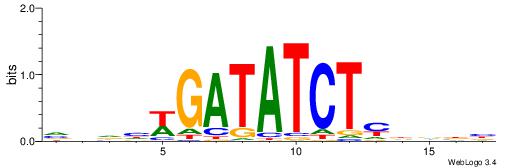 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0377627 |
Alignment:
VVYDDDCHCAVACAATTDBVAC
-----RRTCRCTCTRTCRR---
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 10 |
| Number of overlap: | 14 |
| Similarity score: | 0.0384238 |
Alignment:
VCTBHBVCTTGGWTACVVVVVDB
---------KMGAKAGAGMGAKM
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Motif ID: | UP00100 |
| Motif name: | Gata6_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0394703 |
Alignment:
HKCBSCGATATCGVBBC
RRTCRCTCTRTCRR---
| Original motif | Reverse complement motif |
| Consensus sequence: GBBVCGATATCGSVGYD | Consensus sequence: HKCBSCGATATCGVBBC |
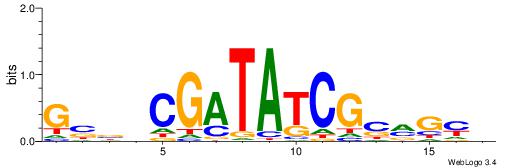 |
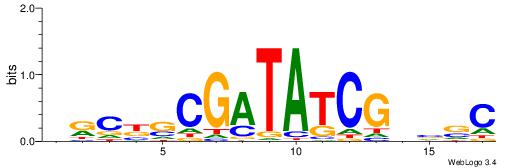 |
| Motif ID: | UP00040 |
| Motif name: | Irf5_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0405383 |
Alignment:
BHDAHCGAGARTHBV
KMGAKAGAGMGAKM-
| Original motif | Reverse complement motif |
| Consensus sequence: BHDAHCGAGARTHBV | Consensus sequence: VBHAKTCTCGHTHHV |
 |
 |
| Dataset #: 1 | Motif ID: 26 | Motif name: C026 |
| Original motif | Reverse complement motif |
| Consensus sequence: KMGAGAKMKMGAGAKM | Consensus sequence: RRTCTCRRRRTCTCRR |
 |
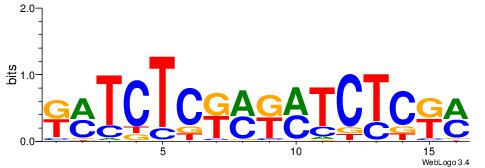 |
Best Matches for Motif ID 26 (Highest to Lowest)
| Motif ID: | UP00095 |
| Motif name: | Zfp691_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0454643 |
Alignment:
VHBDDAGGAGTCWBBTD
RRTCTCRRRRTCTCRR-
| Original motif | Reverse complement motif |
| Consensus sequence: DABBWGACTCCTHDVHV | Consensus sequence: VHBDDAGGAGTCWBBTD |
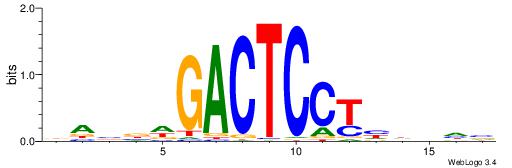 |
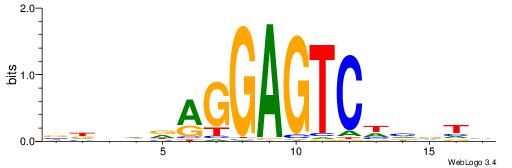 |
| Motif ID: | UP00037 |
| Motif name: | Zfp105_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0459821 |
Alignment:
HDMAVTTATTGARVCMH
KMGAGAKMKMGAGAKM-
| Original motif | Reverse complement motif |
| Consensus sequence: DYGVKTCAATAABTYDD | Consensus sequence: HDMAVTTATTGARVCMH |
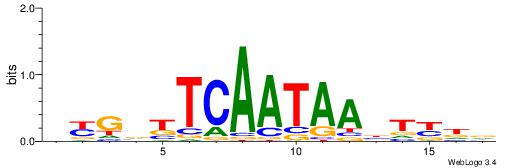 |
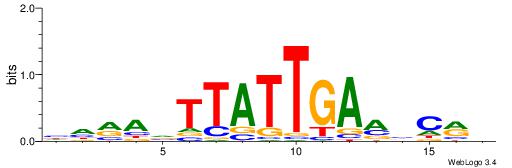 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 16 |
| Similarity score: | 0.0470026 |
Alignment:
BDBBVBVGTAWCCAAGVBHBAGB
------RRTCTCRRRRTCTCRR-
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Motif ID: | UP00100 |
| Motif name: | Gata6_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0472484 |
Alignment:
HKCBSCGATATCGVBBC
RRTCTCRRRRTCTCRR-
| Original motif | Reverse complement motif |
| Consensus sequence: GBBVCGATATCGSVGYD | Consensus sequence: HKCBSCGATATCGVBBC |
 |
 |
| Motif ID: | UP00080 |
| Motif name: | Gata5_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0489582 |
Alignment:
VVHHWGATATCTMHBBV
-KMGAGAKMKMGAGAKM
| Original motif | Reverse complement motif |
| Consensus sequence: VVBHRAGATATCWDHBB | Consensus sequence: VVHHWGATATCTMHBBV |
 |
 |
| Dataset #: 1 | Motif ID: 27 | Motif name: C027 |
| Original motif | Reverse complement motif |
| Consensus sequence: MRCAGRGAAACCCTGTCTYDW | Consensus sequence: WHKAGACAGGGTTTCMCTGKY |
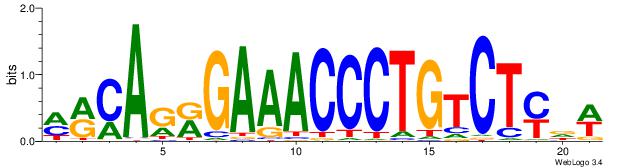 |
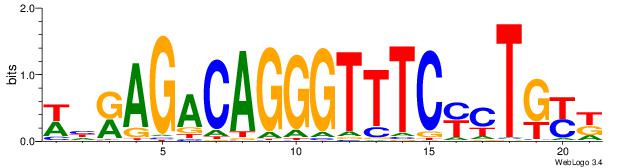 |
Best Matches for Motif ID 27 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 21 |
| Similarity score: | 0.0379369 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
-WHKAGACAGGGTTTCMCTGKY-
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 21 |
| Similarity score: | 0.0428251 |
Alignment:
HGDHVHKGACCACCCGGBGBGDB
--MRCAGRGAAACCCTGTCTYDW
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 21 |
| Similarity score: | 0.0432221 |
Alignment:
VCTBHBVCTTGGWTACVVVVVDB
-WHKAGACAGGGTTTCMCTGKY-
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 21 |
| Similarity score: | 0.0454185 |
Alignment:
DVMHHDHKGACCCTCCTSVCBH
MRCAGRGAAACCCTGTCTYDW-
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 21 |
| Similarity score: | 0.0491194 |
Alignment:
HBHDVBWGATHBTATCRVBHHH
-WHKAGACAGGGTTTCMCTGKY
| Original motif | Reverse complement motif |
| Consensus sequence: HBHDVBWGATHBTATCRVBHHH | Consensus sequence: HHHBBMGATAVHATCWVVDHVH |
 |
 |
| Dataset #: 1 | Motif ID: 28 | Motif name: C028 |
| Original motif | Reverse complement motif |
| Consensus sequence: BVGASSCASAGVSABV | Consensus sequence: VBTSVCTSTGSSTCVB |
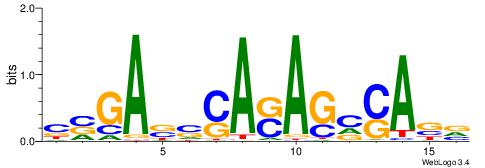 |
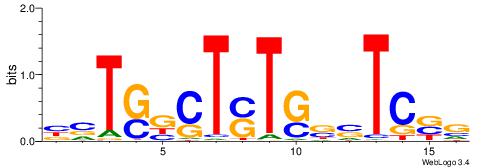 |
Best Matches for Motif ID 28 (Highest to Lowest)
| Motif ID: | UP00000 |
| Motif name: | Smad3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0351716 |
Alignment:
HDHDDCCAGACABBHVH
BVGASSCASAGVSABV-
| Original motif | Reverse complement motif |
| Consensus sequence: HDHDDCCAGACABBHVH | Consensus sequence: HVHBVTGTCTGGDDHDD |
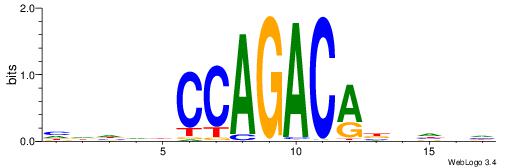 |
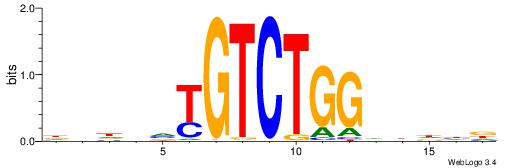 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 16 |
| Similarity score: | 0.0393573 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
---BVGASSCASAGVSABV---
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00013 |
| Motif name: | Gabpa_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0397911 |
Alignment:
CYDBYWTCCSMHBVVH
VBTSVCTSTGSSTCVB
| Original motif | Reverse complement motif |
| Consensus sequence: CYDBYWTCCSMHBVVH | Consensus sequence: DBVVDRSGGAWKVHKG |
 |
 |
| Motif ID: | UP00011 |
| Motif name: | Irf6_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0399886 |
Alignment:
DHDBTKGTTTCGHTHDD
-VBTSVCTSTGSSTCVB
| Original motif | Reverse complement motif |
| Consensus sequence: HDDAHCGAAACYAVDDD | Consensus sequence: DHDBTKGTTTCGHTHDD |
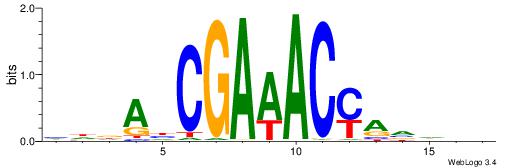 |
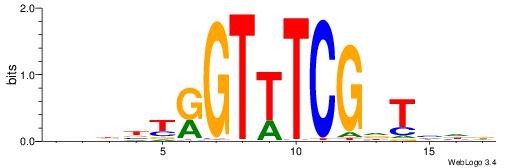 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 16 |
| Similarity score: | 0.0408182 |
Alignment:
GTVBDAATTGTVTGHGDDHKVB
--VBTSVCTSTGSSTCVB----
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Dataset #: 1 | Motif ID: 29 | Motif name: C029 |
| Original motif | Reverse complement motif |
| Consensus sequence: AAWWWAAAAWWWAA | Consensus sequence: TTWWWTTTTWWWTT |
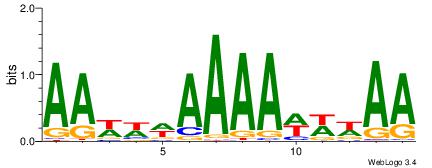 |
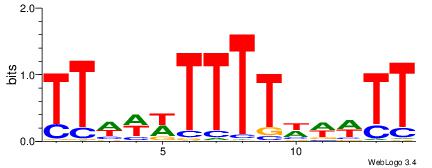 |
Best Matches for Motif ID 29 (Highest to Lowest)
| Motif ID: | UP00121 |
| Motif name: | Hoxd10 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0387466 |
Alignment:
HDDDATTTTATTKVRDB
TTWWWTTTTWWWTT---
| Original motif | Reverse complement motif |
| Consensus sequence: VDKVYAATAAAATDDDH | Consensus sequence: HDDDATTTTATTKVRDB |
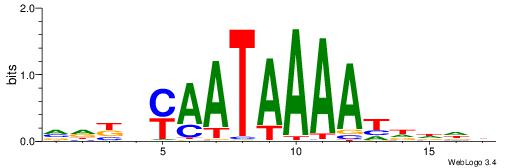 |
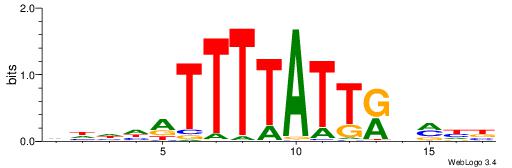 |
| Motif ID: | UP00090 |
| Motif name: | Elf3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0414949 |
Alignment:
DDHBTTTTTTTTHDAVH
TTWWWTTTTWWWTT---
| Original motif | Reverse complement motif |
| Consensus sequence: DBTHHAAAAAAAAVHDH | Consensus sequence: DDHBTTTTTTTTHDAVH |
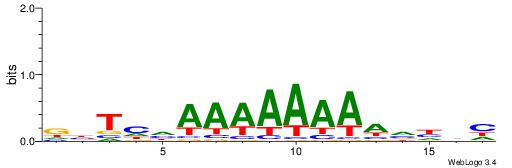 |
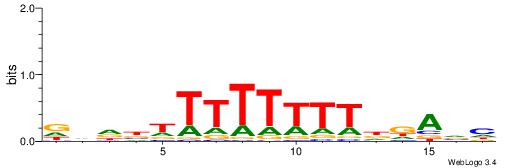 |
| Motif ID: | UP00077 |
| Motif name: | Srf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0415039 |
Alignment:
VBDDAAAAAAAAMVBHH
---AAWWWAAAAWWWAA
| Original motif | Reverse complement motif |
| Consensus sequence: VBDDAAAAAAAAMVBHH | Consensus sequence: HHVBYTTTTTTTTDDVV |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0434847 |
Alignment:
ADDDYAWAACAAMAHH
AAWWWAAAAWWWAA--
| Original motif | Reverse complement motif |
| Consensus sequence: ADDDYAWAACAAMAHH | Consensus sequence: HDTYTTGTTWTKDDDT |
 |
 |
| Motif ID: | UP00255 |
| Motif name: | Dbx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.045306 |
Alignment:
HARTHAATTAWTAVDHD
-AAWWWAAAAWWWAA--
| Original motif | Reverse complement motif |
| Consensus sequence: HARTHAATTAWTAVDHD | Consensus sequence: DHDBTAWTAATTHAKTH |
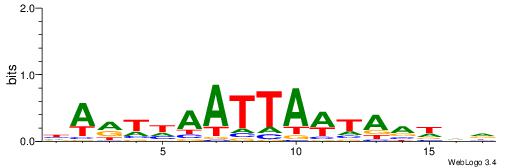 |
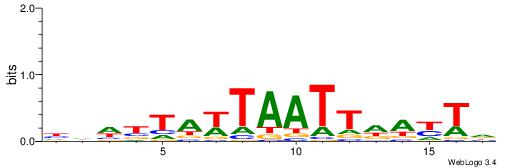 |
| Dataset #: 1 | Motif ID: 30 | Motif name: C030 |
| Original motif | Reverse complement motif |
| Consensus sequence: WYAAAAAMAAAAAAAAWAM | Consensus sequence: YTWTTTTTTTTYTTTTTMW |
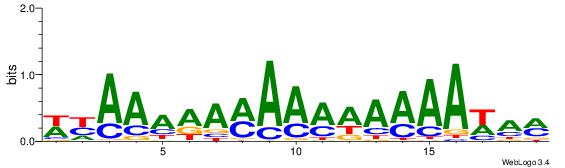 |
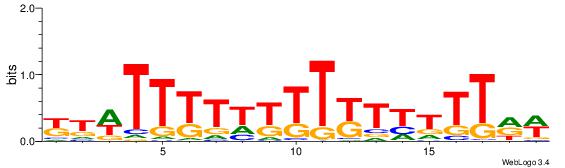 |
Best Matches for Motif ID 30 (Highest to Lowest)
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 19 |
| Similarity score: | 0.0369294 |
Alignment:
HHBDHDDAACAATDRHHHHHBW
--WYAAAAAMAAAAAAAAWAM-
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 19 |
| Similarity score: | 0.0399523 |
Alignment:
GTVBDAATTGTVTGHGDDHKVB
---YTWTTTTTTTTYTTTTTMW
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 19 |
| Similarity score: | 0.0431733 |
Alignment:
HBHDVHDTCTTATCTHTDDHDV
--YTWTTTTTTTTYTTTTTMW-
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 19 |
| Similarity score: | 0.0503657 |
Alignment:
HHHBBMGATAVHATCWVVDHVH
---WYAAAAAMAAAAAAAAWAM
| Original motif | Reverse complement motif |
| Consensus sequence: HBHDVBWGATHBTATCRVBHHH | Consensus sequence: HHHBBMGATAVHATCWVVDHVH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 19 |
| Similarity score: | 0.0507131 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
--WYAAAAAMAAAAAAAAWAM--
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Dataset #: 1 | Motif ID: 31 | Motif name: C031 |
| Original motif | Reverse complement motif |
| Consensus sequence: WHAAAMRWWYAAAMAWW | Consensus sequence: WWTYTTTMWWKYTTTHW |
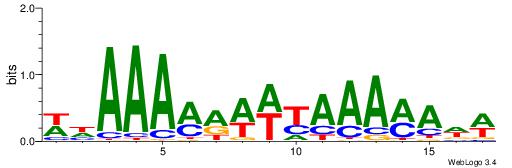 |
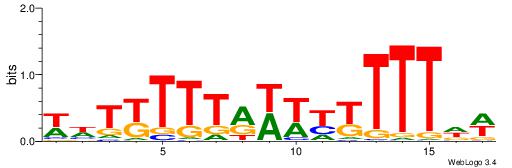 |
Best Matches for Motif ID 31 (Highest to Lowest)
| Motif ID: | UP00077 |
| Motif name: | Srf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0458825 |
Alignment:
VBDDAAAAAAAAMVBHH
WHAAAMRWWYAAAMAWW
| Original motif | Reverse complement motif |
| Consensus sequence: VBDDAAAAAAAAMVBHH | Consensus sequence: HHVBYTTTTTTTTDDVV |
 |
 |
| Motif ID: | UP00090 |
| Motif name: | Elf3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0488194 |
Alignment:
DDHBTTTTTTTTHDAVH
WWTYTTTMWWKYTTTHW
| Original motif | Reverse complement motif |
| Consensus sequence: DBTHHAAAAAAAAVHDH | Consensus sequence: DDHBTTTTTTTTHDAVH |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 17 |
| Similarity score: | 0.0501404 |
Alignment:
WBHHHHHMDATTGTTHDHDVHH
WWTYTTTMWWKYTTTHW-----
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00255 |
| Motif name: | Dbx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0539477 |
Alignment:
DHDBTAWTAATTHAKTH
WWTYTTTMWWKYTTTHW
| Original motif | Reverse complement motif |
| Consensus sequence: HARTHAATTAWTAVDHD | Consensus sequence: DHDBTAWTAATTHAKTH |
 |
 |
| Motif ID: | UP00078 |
| Motif name: | Arid3a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0545289 |
Alignment:
DHHYTTTAATTAAHBBV
WWTYTTTMWWKYTTTHW
| Original motif | Reverse complement motif |
| Consensus sequence: VBBHTTAATTAAAMHHH | Consensus sequence: DHHYTTTAATTAAHBBV |
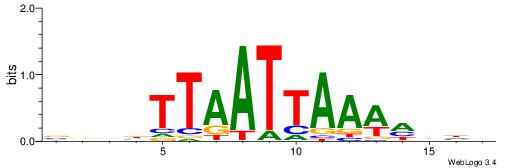 |
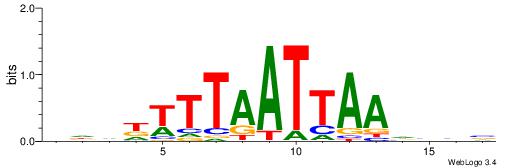 |
| Dataset #: 1 | Motif ID: 32 | Motif name: C032 |
| Original motif | Reverse complement motif |
| Consensus sequence: HWAAWYDAAAMWRWAMWR | Consensus sequence: KWYTWKWYTTTDMWTTWH |
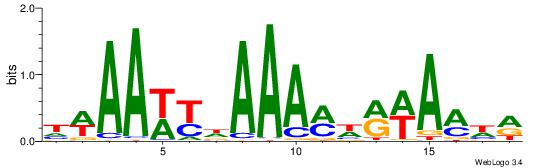 |
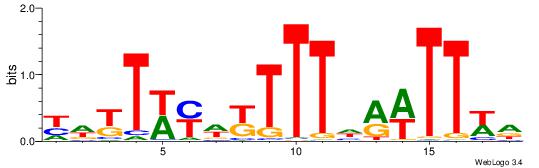 |
Best Matches for Motif ID 32 (Highest to Lowest)
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0461423 |
Alignment:
WBHHHHHMDATTGTTHDHDVHH
---KWYTWKWYTTTDMWTTWH-
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.050828 |
Alignment:
GTVBDAATTGTVTGHGDDHKVB
---HWAAWYDAAAMWRWAMWR-
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.0516978 |
Alignment:
HBHDVHDTCTTATCTHTDDHDV
-KWYTWKWYTTTDMWTTWH---
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0517018 |
Alignment:
HBHDVBWGATHBTATCRVBHHH
----HWAAWYDAAAMWRWAMWR
| Original motif | Reverse complement motif |
| Consensus sequence: HBHDVBWGATHBTATCRVBHHH | Consensus sequence: HHHBBMGATAVHATCWVVDHVH |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0667428 |
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
--HWAAWYDAAAMWRWAMWR---
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Dataset #: 1 | Motif ID: 33 | Motif name: C033 |
| Original motif | Reverse complement motif |
| Consensus sequence: WWAAWYAAAMRWAAAW | Consensus sequence: WTTTWKYTTTMWTTWW |
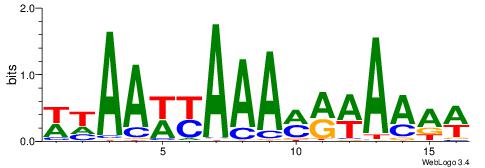 |
 |
Best Matches for Motif ID 33 (Highest to Lowest)
| Motif ID: | UP00061 |
| Motif name: | Foxl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0226221 |
Alignment:
VVVWTTGTTTAMATTWD
-WTTTWKYTTTMWTTWW
| Original motif | Reverse complement motif |
| Consensus sequence: DWAATRTAAACAAWVVB | Consensus sequence: VVVWTTGTTTAMATTWD |
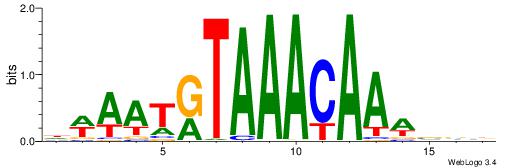 |
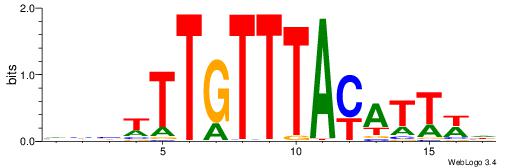 |
| Motif ID: | UP00073 |
| Motif name: | Foxa2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0262386 |
Alignment:
BDVTTTKTTTACWTWHH
-WTTTWKYTTTMWTTWW
| Original motif | Reverse complement motif |
| Consensus sequence: HHWAWGTAAAYAAAVDB | Consensus sequence: BDVTTTKTTTACWTWHH |
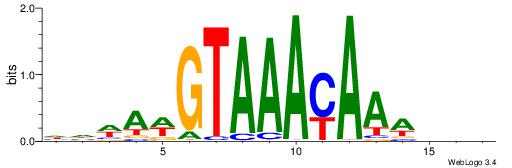 |
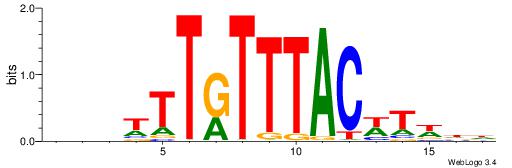 |
| Motif ID: | UP00025 |
| Motif name: | Foxk1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0287316 |
Alignment:
BBVTTTGTTTACATTDD
-WTTTWKYTTTMWTTWW
| Original motif | Reverse complement motif |
| Consensus sequence: DDAATGTAAACAAAVVB | Consensus sequence: BBVTTTGTTTACATTDD |
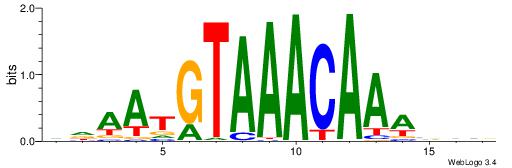 |
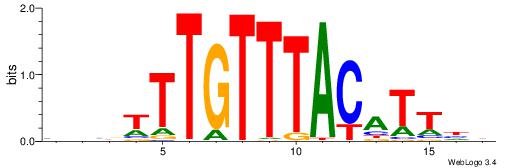 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0296838 |
Alignment:
ADDDYAWAACAAMAHH
WWAAWYAAAMRWAAAW
| Original motif | Reverse complement motif |
| Consensus sequence: ADDDYAWAACAAMAHH | Consensus sequence: HDTYTTGTTWTKDDDT |
 |
 |
| Motif ID: | UP00077 |
| Motif name: | Srf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0300495 |
Alignment:
VBDDAAAAAAAAMVBHH
-WWAAWYAAAMRWAAAW
| Original motif | Reverse complement motif |
| Consensus sequence: VBDDAAAAAAAAMVBHH | Consensus sequence: HHVBYTTTTTTTTDDVV |
 |
 |
| Dataset #: 1 | Motif ID: 34 | Motif name: C034 |
| Original motif | Reverse complement motif |
| Consensus sequence: SCACTGSGGAGSMAGAGS | Consensus sequence: SCTCTRSCTCCSCAGTGS |
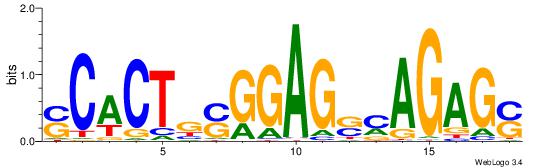 |
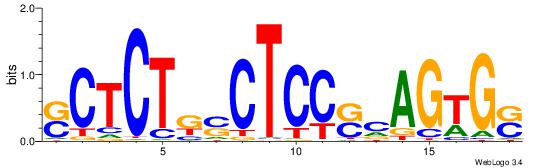 |
Best Matches for Motif ID 34 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0434619 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
---SCACTGSGGAGSMAGAGS--
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0468771 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
--SCTCTRSCTCCSCAGTGS--
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0486672 |
Alignment:
BADHBDHCGCCCMCGCAHHDBBV
---SCTCTRSCTCCSCAGTGS--
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0500626 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
--SCTCTRSCTCCSCAGTGS---
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0522233 |
Alignment:
VCTBHBVCTTGGWTACVVVVVDB
-----SCTCTRSCTCCSCAGTGS
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Dataset #: 1 | Motif ID: 35 | Motif name: C035 |
| Original motif | Reverse complement motif |
| Consensus sequence: WWACCYKGTCTMRAAAWW | Consensus sequence: WWTTTMYAGACRKGGTWW |
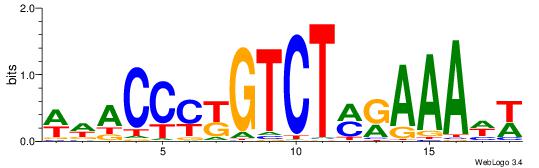 |
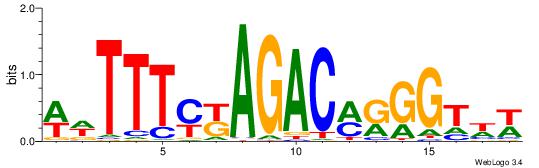 |
Best Matches for Motif ID 35 (Highest to Lowest)
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.0424694 |
Alignment:
HBHDVHDTCTTATCTHTDDHDV
----WWACCYKGTCTMRAAAWW
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0529352 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
---WWTTTMYAGACRKGGTWW--
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 18 |
| Similarity score: | 0.0530184 |
Alignment:
YDYBDHTMCACGTGGADDBMDGT
WWTTTMYAGACRKGGTWW-----
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0572016 |
Alignment:
VCTBHBVCTTGGWTACVVVVVDB
WWACCYKGTCTMRAAAWW-----
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.0574877 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
----WWTTTMYAGACRKGGTWW-
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Dataset #: 1 | Motif ID: 36 | Motif name: C036 |
| Original motif | Reverse complement motif |
| Consensus sequence: MKAGMKMGAGAKMKAGMK | Consensus sequence: RRCTRRRTCTCRRRCTRY |
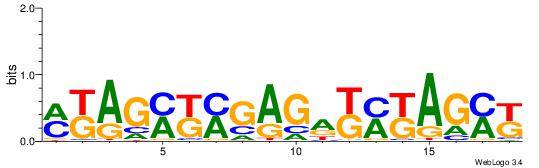 |
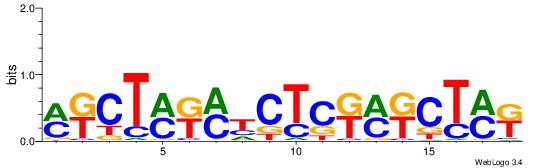 |
Best Matches for Motif ID 36 (Highest to Lowest)
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0426031 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
----RRCTRRRTCTCRRRCTRY
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.0454827 |
Alignment:
GTVBDAATTGTVTGHGDDHKVB
----RRCTRRRTCTCRRRCTRY
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0473662 |
Alignment:
WHVBVVMACGCGCGTCDYHWDH
RRCTRRRTCTCRRRCTRY----
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0480971 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
--MKAGMKMGAGAKMKAGMK---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0493902 |
Alignment:
VDWDARRGACGCGGWHGGAKHV
-RRCTRRRTCTCRRRCTRY---
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Dataset #: 1 | Motif ID: 37 | Motif name: C037 |
| Original motif | Reverse complement motif |
| Consensus sequence: SVCCGBCMCCSYCMCCVB | Consensus sequence: BVGGYGKSGGYGBCGGVS |
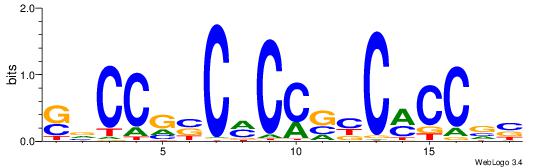 |
 |
Best Matches for Motif ID 37 (Highest to Lowest)
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.0265916 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
-BVGGYGKSGGYGBCGGVS---
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0363116 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
--BVGGYGKSGGYGBCGGVS---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0376504 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
---SVCCGBCMCCSYCMCCVB--
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.0409627 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
----BVGGYGKSGGYGBCGGVS-
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0454859 |
Alignment:
VMYDHDGMCCHCCKBGVVAAVH
----BVGGYGKSGGYGBCGGVS
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Dataset #: 1 | Motif ID: 38 | Motif name: C038 |
| Original motif | Reverse complement motif |
| Consensus sequence: RRRATCCDSCTGCCTYT | Consensus sequence: AKAGGCAGSDGGATKKK |
 |
 |
Best Matches for Motif ID 38 (Highest to Lowest)
| Motif ID: | UP00528 |
| Motif name: | Foxm1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0332889 |
Alignment:
VHVRCAYGATGCATCDTVVADTH
------AKAGGCAGSDGGATKKK
| Original motif | Reverse complement motif |
| Consensus sequence: HADTBVADGATGCATCMTGMVHB | Consensus sequence: VHVRCAYGATGCATCDTVVADTH |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0415981 |
Alignment:
HVKDCCGGGGGGWADDD
AKAGGCAGSDGGATKKK
| Original motif | Reverse complement motif |
| Consensus sequence: DDDTWCCCCCCGGDRVH | Consensus sequence: HVKDCCGGGGGGWADDD |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0467833 |
Alignment:
HVDDVCAGATGKYHBBV
AKAGGCAGSDGGATKKK
| Original motif | Reverse complement motif |
| Consensus sequence: HVDDVCAGATGKYHBBV | Consensus sequence: VBBDMYCATCTGVHHBH |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 17 |
| Similarity score: | 0.0476081 |
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
---RRRATCCDSCTGCCTYT---
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 0.0482666 |
Alignment:
DDRMHBACGCGYGCGTDGBBDB
-RRRATCCDSCTGCCTYT----
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
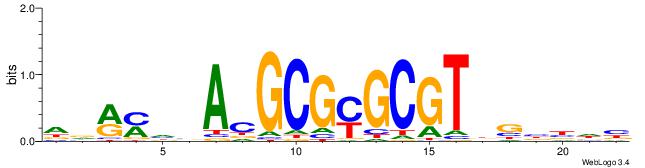 |
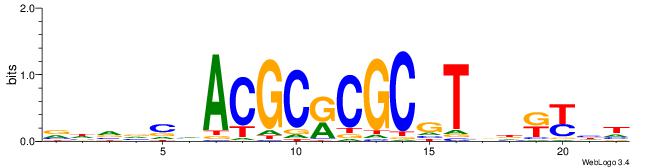 |
| Dataset #: 1 | Motif ID: 39 | Motif name: C039 |
| Original motif | Reverse complement motif |
| Consensus sequence: SMAGRSCAGSSYGGBS | Consensus sequence: SBCCKSSCTGSMCTRS |
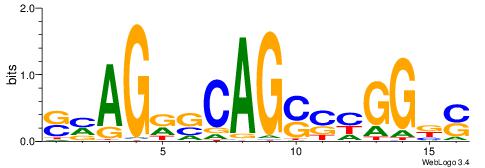 |
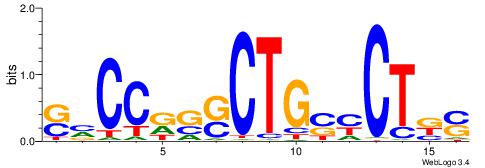 |
Best Matches for Motif ID 39 (Highest to Lowest)
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 16 |
| Similarity score: | 0.0381161 |
Alignment:
HTBVVVDGGACCACCCRGRDBG
-----SMAGRSCAGSSYGGBS-
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 16 |
| Similarity score: | 0.0406922 |
Alignment:
BHCBCBCCGGGTGGTCYHVHDCH
---SBCCKSSCTGSMCTRS----
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0416812 |
Alignment:
HVHBVTGTCTGGDDHDD
-SBCCKSSCTGSMCTRS
| Original motif | Reverse complement motif |
| Consensus sequence: HDHDDCCAGACABBHVH | Consensus sequence: HVHBVTGTCTGGDDHDD |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0417968 |
Alignment:
HDGHHBHRGACCACCCGSVDVG
-----SMAGRSCAGSSYGGBS-
| Original motif | Reverse complement motif |
| Consensus sequence: HDGHHBHRGACCACCCGSVDVG | Consensus sequence: CVHBSCGGGTGGTCMHVHHCDH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 16 |
| Similarity score: | 0.0424976 |
Alignment:
YBVDMGTGGGTGGTCKVVBVBBT
--SBCCKSSCTGSMCTRS-----
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Dataset #: 1 | Motif ID: 40 | Motif name: C040 |
| Original motif | Reverse complement motif |
| Consensus sequence: RSTGASYTCMARSYCAGY | Consensus sequence: KCTGMSKTYGAMSTCASM |
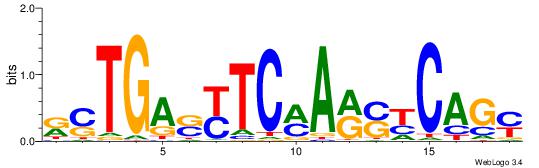 |
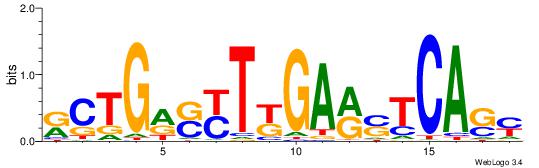 |
Best Matches for Motif ID 40 (Highest to Lowest)
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.0426851 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
RSTGASYTCMARSYCAGY----
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0443113 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
-----KCTGMSKTYGAMSTCASM
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.0450319 |
Alignment:
DTVBDDRGACCACCCAGDWDGBH
----RSTGASYTCMARSYCAGY-
| Original motif | Reverse complement motif |
| Consensus sequence: DTVBDDRGACCACCCAGDWDGBH | Consensus sequence: DBCDWHCTGGGTGGTCMDHBBAH |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 18 |
| Similarity score: | 0.0455665 |
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
KCTGMSKTYGAMSTCASM-----
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0457782 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
---RSTGASYTCMARSYCAGY--
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Dataset #: 1 | Motif ID: 41 | Motif name: C041 |
| Original motif | Reverse complement motif |
| Consensus sequence: CSGAGGCMKAGGCASS | Consensus sequence: SSTGCCTYRGCCTCSG |
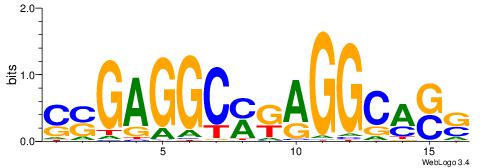 |
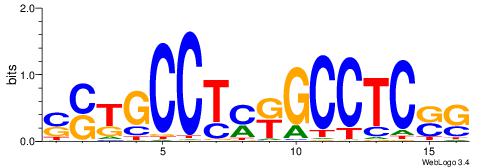 |
Best Matches for Motif ID 41 (Highest to Lowest)
| Motif ID: | UP00098 |
| Motif name: | Rfx3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 16 |
| Similarity score: | 0.0297471 |
Alignment:
HDTBHHCCHTAGCAACVVDHHHH
--SSTGCCTYRGCCTCSG-----
| Original motif | Reverse complement motif |
| Consensus sequence: HDTBHHCCHTAGCAACVVDHHHH | Consensus sequence: HHHHDVVGTTGCTAHGGDHBAHH |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0308327 |
Alignment:
VHYTCCDWCCGCGTCMMTHWHV
-SSTGCCTYRGCCTCSG-----
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 16 |
| Similarity score: | 0.0317547 |
Alignment:
DBBVDDDRTRGCGTCBBYTHGA
-SSTGCCTYRGCCTCSG-----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBVDDDRTRGCGTCBBYTHGA | Consensus sequence: TCDAMVBGACGCMAKDDDVBBD |
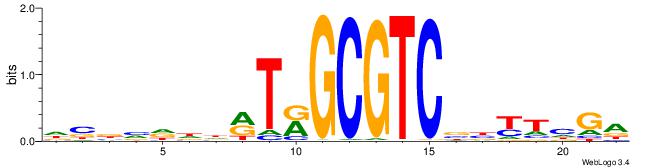 |
 |
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0317742 |
Alignment:
DHHDGGGCGRGGKHBH
CSGAGGCMKAGGCASS
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
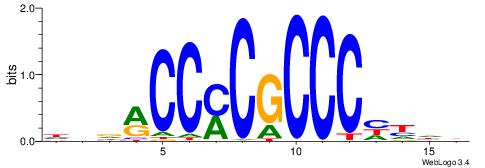 |
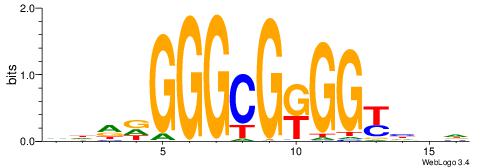 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0321641 |
Alignment:
BHVDCCCCDCCCBDBH
CSGAGGCMKAGGCASS
| Original motif | Reverse complement motif |
| Consensus sequence: BHVDCCCCDCCCBDBH | Consensus sequence: HVDVGGGHGGGGDBHB |
 |
 |
| Dataset #: 1 | Motif ID: 42 | Motif name: C042 |
| Original motif | Reverse complement motif |
| Consensus sequence: WGTCCKRGAACYMACTCW | Consensus sequence: WGAGTRMGTTCKRGGACW |
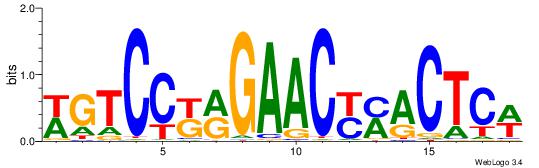 |
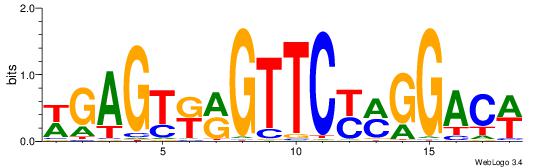 |
Best Matches for Motif ID 42 (Highest to Lowest)
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.0266184 |
Alignment:
YBVDMGTGGGTGGTCKVVBVBBT
----WGAGTRMGTTCKRGGACW-
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.0330025 |
Alignment:
HDTBHHCCHTAGCAACVVDHHHH
----WGTCCKRGAACYMACTCW-
| Original motif | Reverse complement motif |
| Consensus sequence: HDTBHHCCHTAGCAACVVDHHHH | Consensus sequence: HHHHDVVGTTGCTAHGGDHBAHH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.03417 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
-WGTCCKRGAACYMACTCW----
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.0347873 |
Alignment:
HHBSHTCGTGRGTGGKGDBVWMV
----WGAGTRMGTTCKRGGACW-
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.035399 |
Alignment:
VCTBHBVCTTGGWTACVVVVVDB
----WGTCCKRGAACYMACTCW-
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Dataset #: 1 | Motif ID: 43 | Motif name: C043 |
| Original motif | Reverse complement motif |
| Consensus sequence: WWAAWAAAAAARAATW | Consensus sequence: WATTMTTTTTTWTTWW |
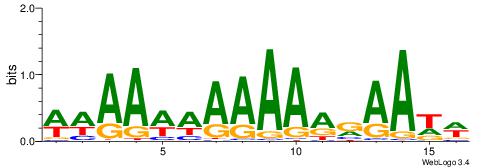 |
 |
Best Matches for Motif ID 43 (Highest to Lowest)
| Motif ID: | UP00077 |
| Motif name: | Srf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0232401 |
Alignment:
VBDDAAAAAAAAMVBHH
-WWAAWAAAAAARAATW
| Original motif | Reverse complement motif |
| Consensus sequence: VBDDAAAAAAAAMVBHH | Consensus sequence: HHVBYTTTTTTTTDDVV |
 |
 |
| Motif ID: | UP00054 |
| Motif name: | Tcf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0288687 |
Alignment:
BHHASATCAAAGGAHHH
-WWAAWAAAAAARAATW
| Original motif | Reverse complement motif |
| Consensus sequence: BHHASATCAAAGGAHHH | Consensus sequence: HHHTCCTTTGATSTHHV |
 |
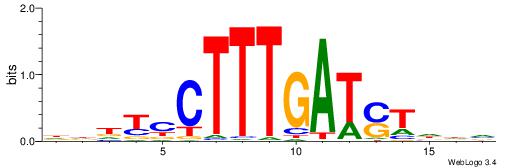 |
| Motif ID: | UP00255 |
| Motif name: | Dbx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0296812 |
Alignment:
DHDBTAWTAATTHAKTH
-WATTMTTTTTTWTTWW
| Original motif | Reverse complement motif |
| Consensus sequence: HARTHAATTAWTAVDHD | Consensus sequence: DHDBTAWTAATTHAKTH |
 |
 |
| Motif ID: | UP00051 |
| Motif name: | Sox8_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0320831 |
Alignment:
DDATBWATTGTTHHDDD
-WATTMTTTTTTWTTWW
| Original motif | Reverse complement motif |
| Consensus sequence: DDATBWATTGTTHHDDD | Consensus sequence: DDDHDAACAATWBATDD |
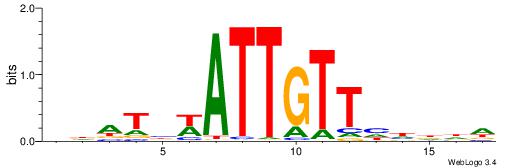 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0327018 |
Alignment:
HDTYTTGTTWTKDDDT
WATTMTTTTTTWTTWW
| Original motif | Reverse complement motif |
| Consensus sequence: ADDDYAWAACAAMAHH | Consensus sequence: HDTYTTGTTWTKDDDT |
 |
 |
| Dataset #: 1 | Motif ID: 44 | Motif name: C044 |
| Original motif | Reverse complement motif |
| Consensus sequence: BVAGAGGCAGRVSGATB | Consensus sequence: BATCSVMCTGCCTCTVB |
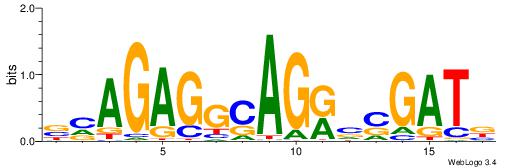 |
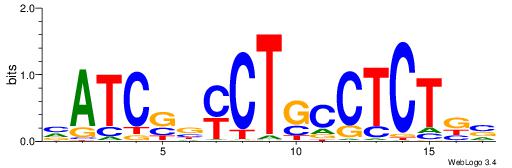 |
Best Matches for Motif ID 44 (Highest to Lowest)
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 0.0438724 |
Alignment:
VDWDARRGACGCGGWHGGAKHV
----BVAGAGGCAGRVSGATB-
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 0.046578 |
Alignment:
VMYDHDGMCCHCCKBGVVAAVH
-BATCSVMCTGCCTCTVB----
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.046833 |
Alignment:
DDBBBCACTGCABTBBB
BATCSVMCTGCCTCTVB
| Original motif | Reverse complement motif |
| Consensus sequence: DDBBBCACTGCABTBBB | Consensus sequence: BBBAVTGCAGTGBBVDD |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 17 |
| Similarity score: | 0.049551 |
Alignment:
TCDAMVBGACGCMAKDDDVBBD
----BVAGAGGCAGRVSGATB-
| Original motif | Reverse complement motif |
| Consensus sequence: DBBVDDDRTRGCGTCBBYTHGA | Consensus sequence: TCDAMVBGACGCMAKDDDVBBD |
 |
 |
| Motif ID: | UP00095 |
| Motif name: | Zfp691_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0496109 |
Alignment:
DABBWGACTCCTHDVHV
BATCSVMCTGCCTCTVB
| Original motif | Reverse complement motif |
| Consensus sequence: DABBWGACTCCTHDVHV | Consensus sequence: VHBDDAGGAGTCWBBTD |
 |
 |
| Dataset #: 1 | Motif ID: 45 | Motif name: C045 |
| Original motif | Reverse complement motif |
| Consensus sequence: RYAGMCMKGKCTRY | Consensus sequence: MKAGYCYRGRCTKK |
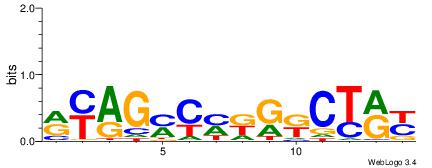 |
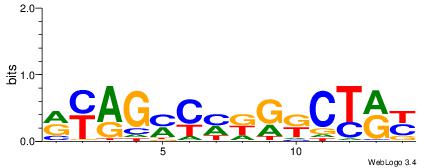 |
Best Matches for Motif ID 45 (Highest to Lowest)
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0303562 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
-RYAGMCMKGKCTRY-------
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00086 |
| Motif name: | Irf3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0355506 |
Alignment:
DDAGAADGGDSCDV
RYAGMCMKGKCTRY
| Original motif | Reverse complement motif |
| Consensus sequence: DDAGAADGGDSCDV | Consensus sequence: BHGSDCCDTTCTHH |
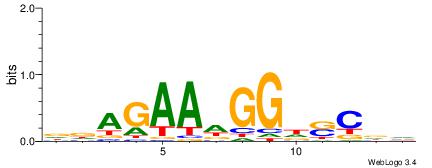 |
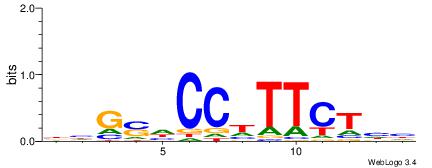 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0391606 |
Alignment:
HVDVGGGHGGGGDBHB
-MKAGYCYRGRCTKK-
| Original motif | Reverse complement motif |
| Consensus sequence: BHVDCCCCDCCCBDBH | Consensus sequence: HVDVGGGHGGGGDBHB |
 |
 |
| Motif ID: | UP00084 |
| Motif name: | Gmeb1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0399089 |
Alignment:
BVVDVRYACGTRYVBHB
--MKAGYCYRGRCTKK-
| Original motif | Reverse complement motif |
| Consensus sequence: BHBBKKACGTMMVDBVB | Consensus sequence: BVVDVRYACGTRYVBHB |
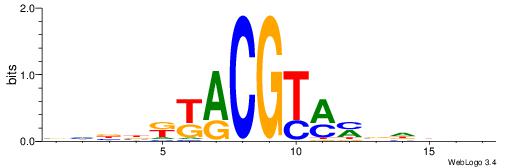 |
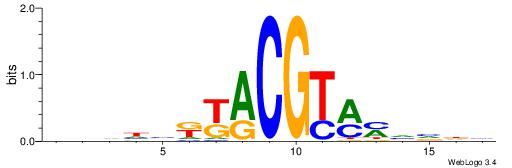 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 14 |
| Similarity score: | 0.0406925 |
Alignment:
GTVBDAATTGTVTGHGDDHKVB
------MKAGYCYRGRCTKK--
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Dataset #: 1 | Motif ID: 46 | Motif name: C046 |
| Original motif | Reverse complement motif |
| Consensus sequence: KSTGKCCTRGAACWSM | Consensus sequence: YSWGTTCKAGGYCASY |
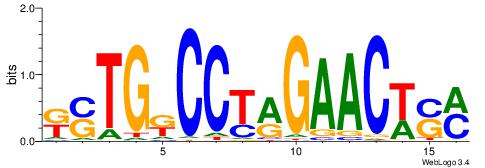 |
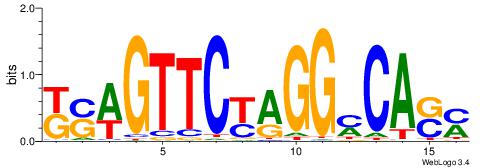 |
Best Matches for Motif ID 46 (Highest to Lowest)
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.033819 |
Alignment:
DCTTTCKAGGAATHHV
KSTGKCCTRGAACWSM
| Original motif | Reverse complement motif |
| Consensus sequence: DCTTTCKAGGAATHHV | Consensus sequence: VHHATTCCTYGAAAGD |
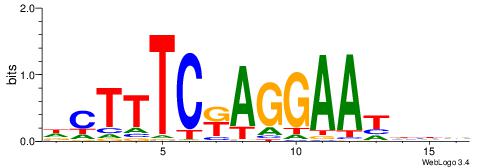 |
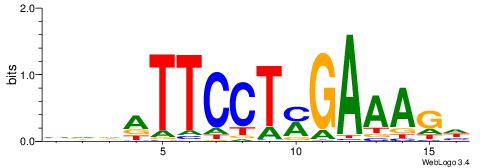 |
| Motif ID: | UP00414 |
| Motif name: | Ets1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0383006 |
Alignment:
BDDDHACCGGAARTVDD
KSTGKCCTRGAACWSM-
| Original motif | Reverse complement motif |
| Consensus sequence: BDDDHACCGGAARTVDD | Consensus sequence: DDBAMTTCCGGTDHDHB |
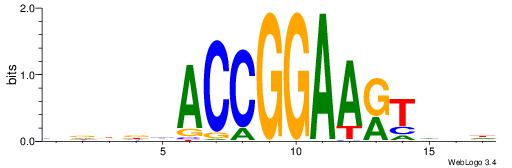 |
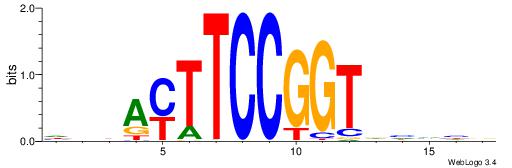 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 16 |
| Similarity score: | 0.0428174 |
Alignment:
DVVTTVGTGGGHGGYAMHWHHHY
---YSWGTTCKAGGYCASY----
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 16 |
| Similarity score: | 0.0436721 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
--YSWGTTCKAGGYCASY-----
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00213 |
| Motif name: | Hoxa9 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0441221 |
Alignment:
WDTAWTTTWATGKCCGD
-YSWGTTCKAGGYCASY
| Original motif | Reverse complement motif |
| Consensus sequence: DCGGYCATWAAAWTADW | Consensus sequence: WDTAWTTTWATGKCCGD |
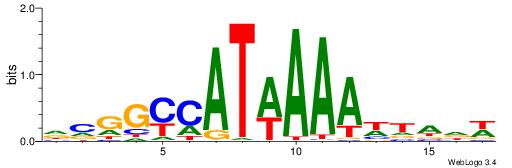 |
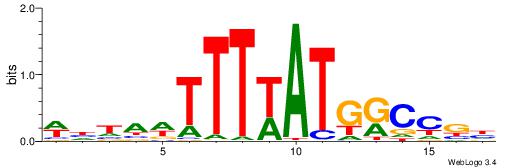 |
| Dataset #: 1 | Motif ID: 47 | Motif name: C047 |
| Original motif | Reverse complement motif |
| Consensus sequence: TSGAAMTCACTMKGTASA | Consensus sequence: TSTACRYAGTGAYTTCSA |
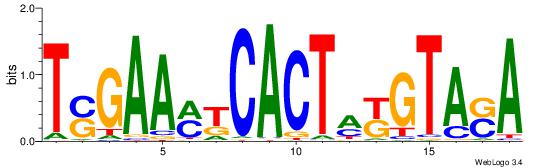 |
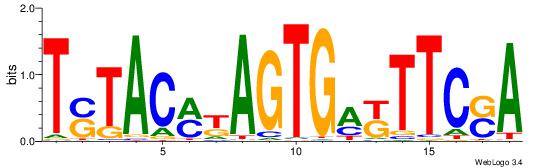 |
Best Matches for Motif ID 47 (Highest to Lowest)
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0369958 |
Alignment:
DCATDBTGHGCATKMTTBRSWR
---TSGAAMTCACTMKGTASA-
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.045149 |
Alignment:
TCDAMVBGACGCMAKDDDVBBD
----TSTACRYAGTGAYTTCSA
| Original motif | Reverse complement motif |
| Consensus sequence: DBBVDDDRTRGCGTCBBYTHGA | Consensus sequence: TCDAMVBGACGCMAKDDDVBBD |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0467709 |
Alignment:
VVYDDDCHCAVACAATTDBVAC
--TSTACRYAGTGAYTTCSA--
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.047063 |
Alignment:
DVTTVVCVYGGHGGYCHHDMYB
--TSTACRYAGTGAYTTCSA--
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 18 |
| Similarity score: | 0.0479788 |
Alignment:
BDBBVBVGTAWCCAAGVBHBAGB
-----TSGAAMTCACTMKGTASA
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Dataset #: 1 | Motif ID: 48 | Motif name: C048 |
| Original motif | Reverse complement motif |
| Consensus sequence: SGAGGCWSWGGCAGSC | Consensus sequence: GSCTGCCWSWGCCTCS |
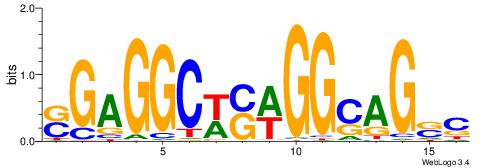 |
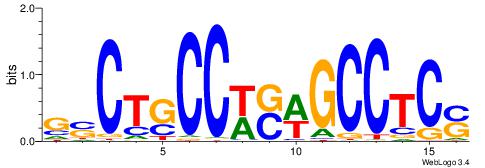 |
Best Matches for Motif ID 48 (Highest to Lowest)
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 16 |
| Similarity score: | 0.0237024 |
Alignment:
VDWDARRGACGCGGWHGGAKHV
------SGAGGCWSWGGCAGSC
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0327172 |
Alignment:
DVMHHDHKGACCCTCCTSVCBH
GSCTGCCWSWGCCTCS------
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 16 |
| Similarity score: | 0.034796 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
---SGAGGCWSWGGCAGSC----
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 16 |
| Similarity score: | 0.036242 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
---GSCTGCCWSWGCCTCS---
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0393403 |
Alignment:
HDTBHHCCHTAGCAACVVDHHHH
-GSCTGCCWSWGCCTCS------
| Original motif | Reverse complement motif |
| Consensus sequence: HDTBHHCCHTAGCAACVVDHHHH | Consensus sequence: HHHHDVVGTTGCTAHGGDHBAHH |
 |
 |
| Dataset #: 1 | Motif ID: 49 | Motif name: C049 |
| Original motif | Reverse complement motif |
| Consensus sequence: WAWWAAAATAAWT | Consensus sequence: AWTTATTTTWWTW |
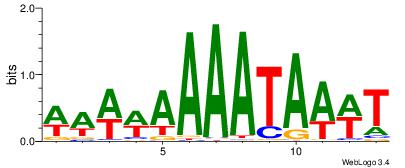 |
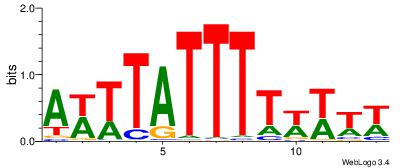 |
Best Matches for Motif ID 49 (Highest to Lowest)
| Motif ID: | UP00061 |
| Motif name: | Foxl1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0235016 |
Alignment:
ADDDYAWAACAAMAHH
-WAWWAAAATAAWT--
| Original motif | Reverse complement motif |
| Consensus sequence: ADDDYAWAACAAMAHH | Consensus sequence: HDTYTTGTTWTKDDDT |
 |
 |
| Motif ID: | UP00073 |
| Motif name: | Foxa2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 13 |
| Similarity score: | 0.0251228 |
Alignment:
HHWAWGTAAAYAAAVDB
--WAWWAAAATAAWT--
| Original motif | Reverse complement motif |
| Consensus sequence: HHWAWGTAAAYAAAVDB | Consensus sequence: BDVTTTKTTTACWTWHH |
 |
 |
| Motif ID: | UP00121 |
| Motif name: | Hoxd10 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 13 |
| Similarity score: | 0.0269927 |
Alignment:
HDDDATTTTATTKVRDB
AWTTATTTTWWTW----
| Original motif | Reverse complement motif |
| Consensus sequence: VDKVYAATAAAATDDDH | Consensus sequence: HDDDATTTTATTKVRDB |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 13 |
| Similarity score: | 0.0270626 |
Alignment:
DWAATRTAAACAAWVVB
--WAWWAAAATAAWT--
| Original motif | Reverse complement motif |
| Consensus sequence: DWAATRTAAACAAWVVB | Consensus sequence: VVVWTTGTTTAMATTWD |
 |
 |
| Motif ID: | UP00037 |
| Motif name: | Zfp105_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0290157 |
Alignment:
HHHTTRTTDTTTKDH
-AWTTATTTTWWTW-
| Original motif | Reverse complement motif |
| Consensus sequence: HDYAAAHAAMAADHD | Consensus sequence: HHHTTRTTDTTTKDH |
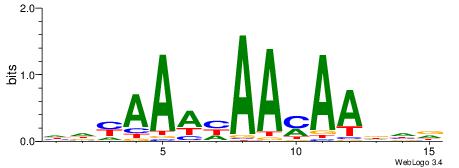 |
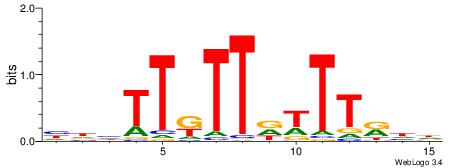 |
| Dataset #: 1 | Motif ID: 50 | Motif name: C050 |
| Original motif | Reverse complement motif |
| Consensus sequence: CBGCCKCYSCCTCYMG | Consensus sequence: CYKGAGGSMGRGGCVG |
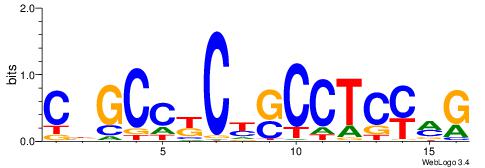 |
 |
Best Matches for Motif ID 50 (Highest to Lowest)
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 16 |
| Similarity score: | 0.0274857 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
-----CBGCCKCYSCCTCYMG-
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 16 |
| Similarity score: | 0.0294264 |
Alignment:
VHYTCCDWCCGCGTCMMTHWHV
--CBGCCKCYSCCTCYMG----
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0295783 |
Alignment:
BHHDYGGGGGGGGBVD
CYKGAGGSMGRGGCVG
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0315534 |
Alignment:
HVDVGGGHGGGGDBHB
CYKGAGGSMGRGGCVG
| Original motif | Reverse complement motif |
| Consensus sequence: BHVDCCCCDCCCBDBH | Consensus sequence: HVDVGGGHGGGGDBHB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 16 |
| Similarity score: | 0.0322386 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
-----CBGCCKCYSCCTCYMG--
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Dataset #: 1 | Motif ID: 51 | Motif name: C051 |
| Original motif | Reverse complement motif |
| Consensus sequence: SGCCCCCHCCCS | Consensus sequence: SGGGHGGGGGCS |
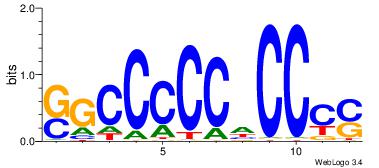 |
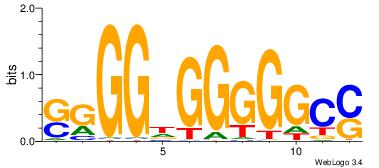 |
Best Matches for Motif ID 51 (Highest to Lowest)
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.00935805 |
Alignment:
DBCCCCCCCCCCMYC
-SGCCCCCHCCCS--
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0122535 |
Alignment:
DHHDGGGCGRGGKHBH
---SGGGHGGGGGCS-
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0145224 |
Alignment:
HVBCCCCCCCCMHHHB
-SGCCCCCHCCCS---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0145616 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
---SGCCCCCHCCCS-------
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 12 |
| Similarity score: | 0.0226961 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
---SGCCCCCHCCCS--------
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Dataset #: 1 | Motif ID: 52 | Motif name: C052 |
| Original motif | Reverse complement motif |
| Consensus sequence: GAGTBVSAGGCMAGVC | Consensus sequence: GVCTYGCCTSVVACTC |
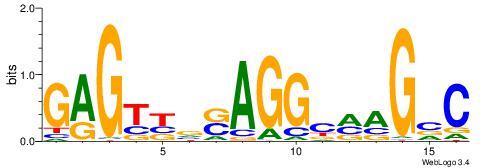 |
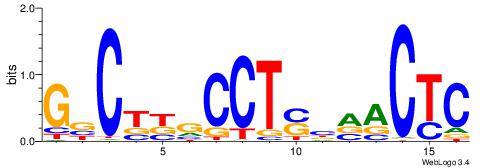 |
Best Matches for Motif ID 52 (Highest to Lowest)
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 16 |
| Similarity score: | 0.0423398 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
----GAGTBVSAGGCMAGVC--
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 16 |
| Similarity score: | 0.047746 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
--GVCTYGCCTSVVACTC-----
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00064 |
| Motif name: | Sox18_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0490125 |
Alignment:
DBVBTRAATTYADDHV
GAGTBVSAGGCMAGVC
| Original motif | Reverse complement motif |
| Consensus sequence: DBVBTRAATTYADDHV | Consensus sequence: VDHDTKAATTMABBBH |
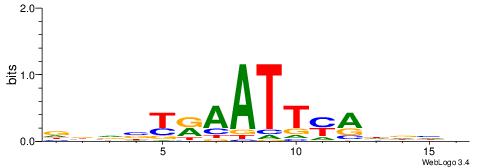 |
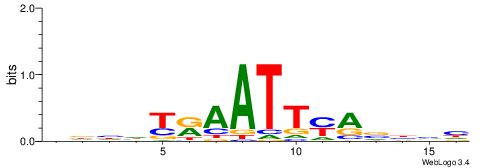 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 16 |
| Similarity score: | 0.0499529 |
Alignment:
BADHBDHCGCCCMCGCAHHDBBV
GVCTYGCCTSVVACTC-------
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0535021 |
Alignment:
MHHHWHYTMCCHCCCACVAABVH
GAGTBVSAGGCMAGVC-------
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Dataset #: 1 | Motif ID: 53 | Motif name: C053 |
| Original motif | Reverse complement motif |
| Consensus sequence: RMACRGAGAAAYCCTGKY | Consensus sequence: MRCAGGMTTTCTCKGTYK |
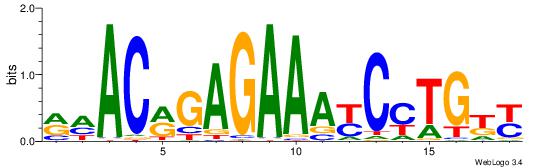 |
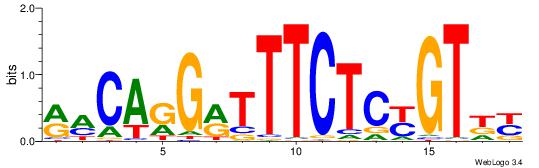 |
Best Matches for Motif ID 53 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0388232 |
Alignment:
HGDHVHKGACCACCCGGBGBGDB
--MRCAGGMTTTCTCKGTYK---
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.0410442 |
Alignment:
HDTBHHCCHTAGCAACVVDHHHH
----RMACRGAGAAAYCCTGKY-
| Original motif | Reverse complement motif |
| Consensus sequence: HDTBHHCCHTAGCAACVVDHHHH | Consensus sequence: HHHHDVVGTTGCTAHGGDHBAHH |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0413339 |
Alignment:
DDRMHBACGCGYGCGTDGBBDB
---MRCAGGMTTTCTCKGTYK-
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0440408 |
Alignment:
VCTBHBVCTTGGWTACVVVVVDB
RMACRGAGAAAYCCTGKY-----
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0442508 |
Alignment:
CBDMCMGGGTGGTCCHVBVBAH
-RMACRGAGAAAYCCTGKY---
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Dataset #: 1 | Motif ID: 54 | Motif name: C054 |
| Original motif | Reverse complement motif |
| Consensus sequence: SSCCDCCCCCCCSS | Consensus sequence: SSGGGGGGGHGGSS |
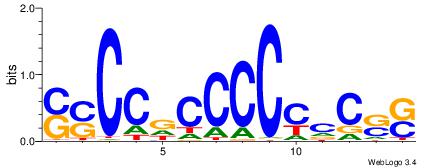 |
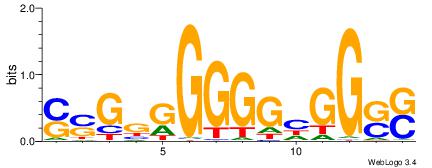 |
Best Matches for Motif ID 54 (Highest to Lowest)
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0162118 |
Alignment:
HBBBCCGCCCCWHHHV
--SSCCDCCCCCCCSS
| Original motif | Reverse complement motif |
| Consensus sequence: HBBBCCGCCCCWHHHV | Consensus sequence: BHHHWGGGGCGGBBVH |
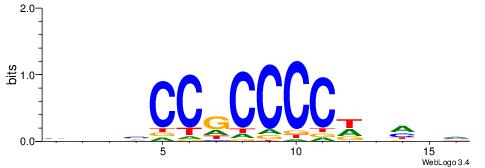 |
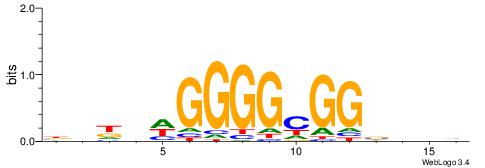 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0185646 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
---SSCCDCCCCCCCSS-----
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0201409 |
Alignment:
DBCCCCCCCCCCMYC
-SSCCDCCCCCCCSS
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0221949 |
Alignment:
HHHHDGGGGCGGDBHDV
SSGGGGGGGHGGSS---
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0265213 |
Alignment:
HVBCCCCCCCCMHHHB
-SSCCDCCCCCCCSS-
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Dataset #: 1 | Motif ID: 55 | Motif name: C055 |
| Original motif | Reverse complement motif |
| Consensus sequence: MKAGAGMKMKAGAGMK | Consensus sequence: RYCTCTRRRRCTCTRR |
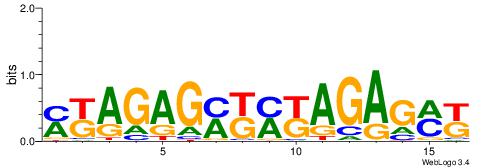 |
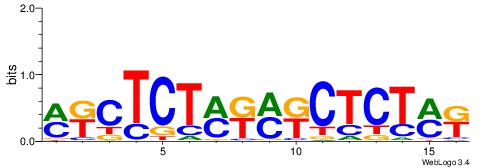 |
Best Matches for Motif ID 55 (Highest to Lowest)
| Motif ID: | UP00101 |
| Motif name: | Sox12_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.039204 |
Alignment:
VVDBASACAAAGRADH
MKAGAGMKMKAGAGMK
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBASACAAAGRADH | Consensus sequence: HDTMCTTTGTSTVDBB |
 |
 |
| Motif ID: | UP00080 |
| Motif name: | Gata5_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0398738 |
Alignment:
VVBHRAGATATCWDHBB
-MKAGAGMKMKAGAGMK
| Original motif | Reverse complement motif |
| Consensus sequence: VVBHRAGATATCWDHBB | Consensus sequence: VVHHWGATATCTMHBBV |
 |
 |
| Motif ID: | UP00019 |
| Motif name: | Zbtb12_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0427919 |
Alignment:
BBGATCTAGAACBKHDB
-RYCTCTRRRRCTCTRR
| Original motif | Reverse complement motif |
| Consensus sequence: BDHRBGTTCTAGATCVB | Consensus sequence: BBGATCTAGAACBKHDB |
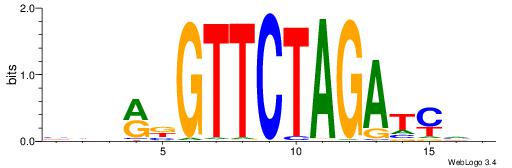 |
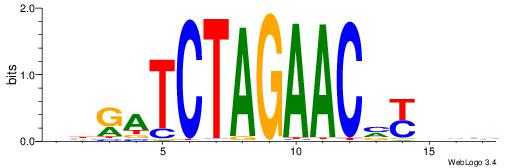 |
| Motif ID: | UP00075 |
| Motif name: | Sox15_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0433178 |
Alignment:
HDDDDAACAATWRRBHD
-MKAGAGMKMKAGAGMK
| Original motif | Reverse complement motif |
| Consensus sequence: HDDDDAACAATWRRBHD | Consensus sequence: DHVKMWATTGTTHDHDH |
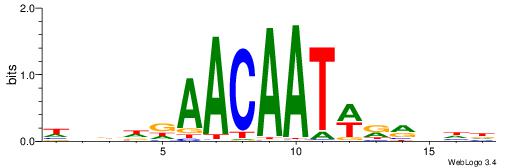 |
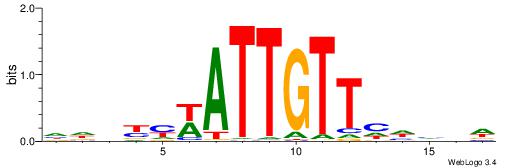 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 16 |
| Similarity score: | 0.0454282 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
-------RYCTCTRRRRCTCTRR
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Dataset #: 1 | Motif ID: 56 | Motif name: C056 |
| Original motif | Reverse complement motif |
| Consensus sequence: AWMAAAYWRAMAAWH | Consensus sequence: HWTTYTKWMTTTYWT |
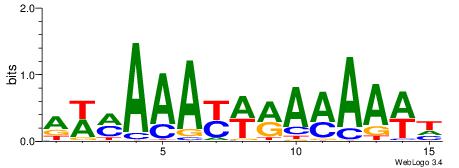 |
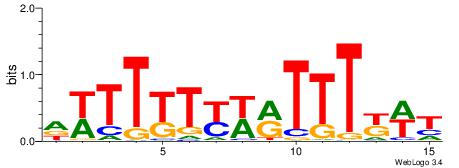 |
Best Matches for Motif ID 56 (Highest to Lowest)
| Motif ID: | UP00077 |
| Motif name: | Srf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0213582 |
Alignment:
VBDDAAAAAAAAMVBHH
-AWMAAAYWRAMAAWH-
| Original motif | Reverse complement motif |
| Consensus sequence: VBDDAAAAAAAAMVBHH | Consensus sequence: HHVBYTTTTTTTTDDVV |
 |
 |
| Motif ID: | UP00037 |
| Motif name: | Zfp105_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0251847 |
Alignment:
HDYAAAHAAMAADHD
AWMAAAYWRAMAAWH
| Original motif | Reverse complement motif |
| Consensus sequence: HDYAAAHAAMAADHD | Consensus sequence: HHHTTRTTDTTTKDH |
 |
 |
| Motif ID: | UP00090 |
| Motif name: | Elf3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0274993 |
Alignment:
DBTHHAAAAAAAAVHDH
-AWMAAAYWRAMAAWH-
| Original motif | Reverse complement motif |
| Consensus sequence: DBTHHAAAAAAAAVHDH | Consensus sequence: DDHBTTTTTTTTHDAVH |
 |
 |
| Motif ID: | UP00255 |
| Motif name: | Dbx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.027785 |
Alignment:
HARTHAATTAWTAVDHD
-AWMAAAYWRAMAAWH-
| Original motif | Reverse complement motif |
| Consensus sequence: HARTHAATTAWTAVDHD | Consensus sequence: DHDBTAWTAATTHAKTH |
 |
 |
| Motif ID: | UP00121 |
| Motif name: | Hoxd10 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.02926 |
Alignment:
HDDDATTTTATTKVRDB
-HWTTYTKWMTTTYWT-
| Original motif | Reverse complement motif |
| Consensus sequence: VDKVYAATAAAATDDDH | Consensus sequence: HDDDATTTTATTKVRDB |
 |
 |
| Dataset #: 1 | Motif ID: 57 | Motif name: C057 |
| Original motif | Reverse complement motif |
| Consensus sequence: WRMARYAAMARYAAMAWH | Consensus sequence: HWTRTTMKTYTTMKTYKW |
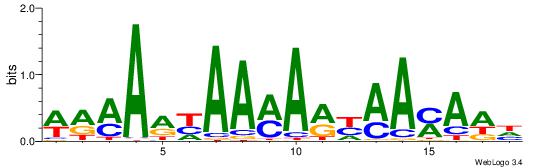 |
 |
Best Matches for Motif ID 57 (Highest to Lowest)
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.0381479 |
Alignment:
HHBDHDDAACAATDRHHHHHBW
-WRMARYAAMARYAAMAWH---
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.052401 |
Alignment:
VVYDDDCHCAVACAATTDBVAC
----WRMARYAAMARYAAMAWH
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0540821 |
Alignment:
DCATDBTGHGCATKMTTBRSWR
--HWTRTTMKTYTTMKTYKW--
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0542401 |
Alignment:
HBHDVHDTCTTATCTHTDDHDV
--HWTRTTMKTYTTMKTYKW--
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0552305 |
Alignment:
HHHBBMGATAVHATCWVVDHVH
---WRMARYAAMARYAAMAWH-
| Original motif | Reverse complement motif |
| Consensus sequence: HBHDVBWGATHBTATCRVBHHH | Consensus sequence: HHHBBMGATAVHATCWVVDHVH |
 |
 |
| Dataset #: 1 | Motif ID: 58 | Motif name: C058 |
| Original motif | Reverse complement motif |
| Consensus sequence: CYCCBCSGVCYCCBC | Consensus sequence: GVGGMGVCSGBGGMG |
 |
 |
Best Matches for Motif ID 58 (Highest to Lowest)
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 15 |
| Similarity score: | 0.0246506 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
-GVGGMGVCSGBGGMG------
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00036 |
| Motif name: | Myf6_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0266507 |
Alignment:
GGDGBGVCTGTKVVB
GVGGMGVCSGBGGMG
| Original motif | Reverse complement motif |
| Consensus sequence: VVVRACAGVCBCDCC | Consensus sequence: GGDGBGVCTGTKVVB |
 |
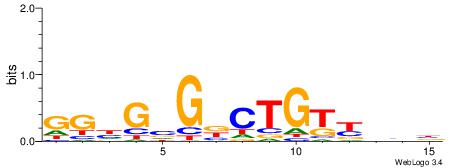 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0296277 |
Alignment:
BHHBHCCGCCCCDHHHH
CYCCBCSGVCYCCBC--
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0309574 |
Alignment:
BDHHMCGCCCCCTHVBB
--CYCCBCSGVCYCCBC
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0331968 |
Alignment:
BHVDCCCCDCCCBDBH
CYCCBCSGVCYCCBC-
| Original motif | Reverse complement motif |
| Consensus sequence: BHVDCCCCDCCCBDBH | Consensus sequence: HVDVGGGHGGGGDBHB |
 |
 |
| Dataset #: 1 | Motif ID: 59 | Motif name: C059 |
| Original motif | Reverse complement motif |
| Consensus sequence: SYCCDCCCCCRG | Consensus sequence: CMGGGGGDGGKS |
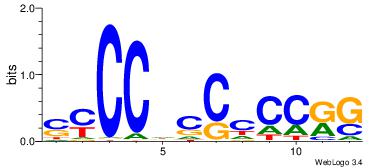 |
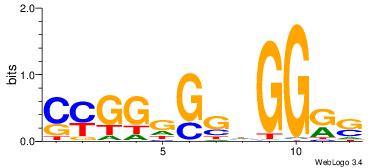 |
Best Matches for Motif ID 59 (Highest to Lowest)
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 0.0197332 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
------SYCCDCCCCCRG----
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0224264 |
Alignment:
HHHHDGGGGCGGDBHDV
--CMGGGGGDGGKS---
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
 |
 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 0.0231684 |
Alignment:
HVDVGGGHGGGGDBHB
CMGGGGGDGGKS----
| Original motif | Reverse complement motif |
| Consensus sequence: BHVDCCCCDCCCBDBH | Consensus sequence: HVDVGGGHGGGGDBHB |
 |
 |
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.023387 |
Alignment:
BHHHWGGGGCGGBBVH
--CMGGGGGDGGKS--
| Original motif | Reverse complement motif |
| Consensus sequence: HBBBCCGCCCCWHHHV | Consensus sequence: BHHHWGGGGCGGBBVH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 12 |
| Similarity score: | 0.0246213 |
Alignment:
HHBSHTCGTGRGTGGKGDBVWMV
-----CMGGGGGDGGKS------
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Dataset #: 1 | Motif ID: 60 | Motif name: C060 |
| Original motif | Reverse complement motif |
| Consensus sequence: SMGGCAKMGGCAKS | Consensus sequence: SYTGCCYYTGCCYS |
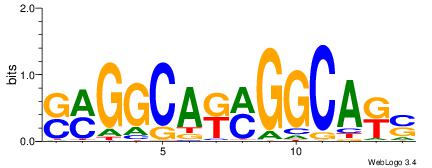 |
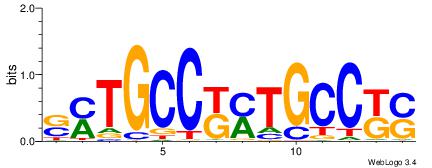 |
Best Matches for Motif ID 60 (Highest to Lowest)
| Motif ID: | UP00010 |
| Motif name: | Tcfap2b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0309254 |
Alignment:
HHKGCCYSRGGCWAD
SYTGCCYYTGCCYS-
| Original motif | Reverse complement motif |
| Consensus sequence: DTWGCCKSMGGCRHH | Consensus sequence: HHKGCCYSRGGCWAD |
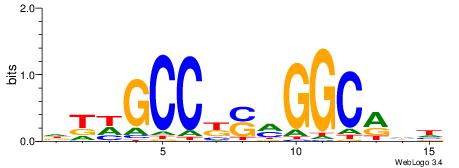 |
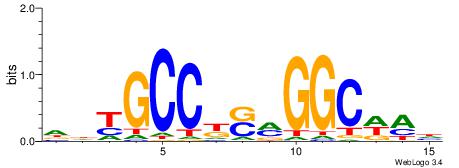 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0349665 |
Alignment:
HHBVHTGACCTTGVDHD
---SYTGCCYYTGCCYS
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBCAAGGTCAHVBDH | Consensus sequence: HHBVHTGACCTTGVDHD |
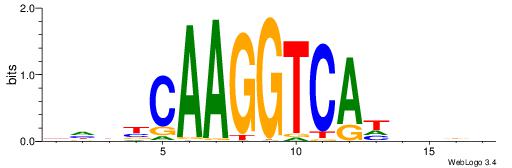 |
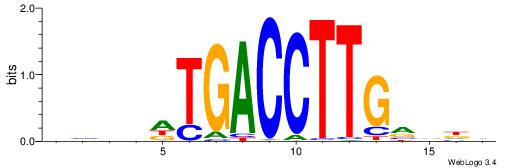 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 14 |
| Similarity score: | 0.0406726 |
Alignment:
DVMHHDHKGACCCTCCTSVCBH
-----SYTGCCYYTGCCYS---
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Motif ID: | UP00028 |
| Motif name: | Tcfap2e_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0419455 |
Alignment:
DTHGCCTSAGGCHHH
SYTGCCYYTGCCYS-
| Original motif | Reverse complement motif |
| Consensus sequence: HHHGCCTSAGGCDAD | Consensus sequence: DTHGCCTSAGGCHHH |
 |
 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0436588 |
Alignment:
BHVDCCCCDCCCBDBH
SYTGCCYYTGCCYS--
| Original motif | Reverse complement motif |
| Consensus sequence: BHVDCCCCDCCCBDBH | Consensus sequence: HVDVGGGHGGGGDBHB |
 |
 |
| Dataset #: 1 | Motif ID: 61 | Motif name: C061 |
| Original motif | Reverse complement motif |
| Consensus sequence: MRGMMBAGGCHGGCCK | Consensus sequence: RGGCCDGCCTBRRCMY |
 |
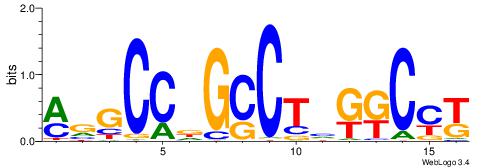 |
Best Matches for Motif ID 61 (Highest to Lowest)
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0354983 |
Alignment:
DDRMHBACGCGYGCGTDGBBDB
MRGMMBAGGCHGGCCK------
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 16 |
| Similarity score: | 0.0371599 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
-----RGGCCDGCCTBRRCMY--
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0397188 |
Alignment:
BBBDVVRGACCACCCAVGABBAB
------RGGCCDGCCTBRRCMY-
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 16 |
| Similarity score: | 0.0410046 |
Alignment:
ABBBBVVRGACCACCCACRDBBM
-------RGGCCDGCCTBRRCMY
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0416624 |
Alignment:
VDWDARRGACGCGGWHGGAKHV
-----MRGMMBAGGCHGGCCK-
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Dataset #: 1 | Motif ID: 62 | Motif name: C062 |
| Original motif | Reverse complement motif |
| Consensus sequence: WWAMAWRMAMAYWAAHWW | Consensus sequence: WWDTTWMTRTRKWTYTWW |
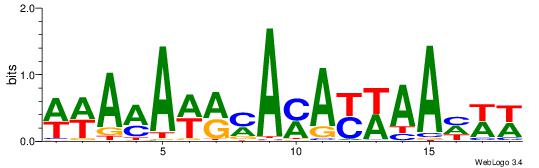 |
 |
Best Matches for Motif ID 62 (Highest to Lowest)
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0496099 |
Alignment:
WBHHHHHMDATTGTTHDHDVHH
-WWDTTWMTRTRKWTYTWW---
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0618489 |
Alignment:
HBHDVHDTCTTATCTHTDDHDV
---WWDTTWMTRTRKWTYTWW-
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0618699 |
Alignment:
MWSMBAARRATGCDCABHATGD
-WWAMAWRMAMAYWAAHWW---
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0626028 |
Alignment:
VVYDDDCHCAVACAATTDBVAC
---WWAMAWRMAMAYWAAHWW-
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.065917 |
Alignment:
HHHBBMGATAVHATCWVVDHVH
---WWAMAWRMAMAYWAAHWW-
| Original motif | Reverse complement motif |
| Consensus sequence: HBHDVBWGATHBTATCRVBHHH | Consensus sequence: HHHBBMGATAVHATCWVVDHVH |
 |
 |
| Dataset #: 1 | Motif ID: 63 | Motif name: C063 |
| Original motif | Reverse complement motif |
| Consensus sequence: MAAKMWAAAAWKHAA | Consensus sequence: TTHRWTTTTWYRTTY |
 |
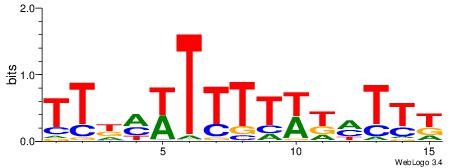 |
Best Matches for Motif ID 63 (Highest to Lowest)
| Motif ID: | UP00061 |
| Motif name: | Foxl1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0344456 |
Alignment:
HDTYTTGTTWTKDDDT
TTHRWTTTTWYRTTY-
| Original motif | Reverse complement motif |
| Consensus sequence: ADDDYAWAACAAMAHH | Consensus sequence: HDTYTTGTTWTKDDDT |
 |
 |
| Motif ID: | UP00039 |
| Motif name: | Foxj3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0345147 |
Alignment:
DVHWDTGTTWTGDTBDH
TTHRWTTTTWYRTTY--
| Original motif | Reverse complement motif |
| Consensus sequence: HDBAHCAWAACADWDVD | Consensus sequence: DVHWDTGTTWTGDTBDH |
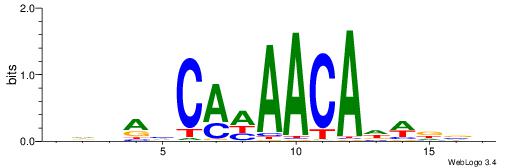 |
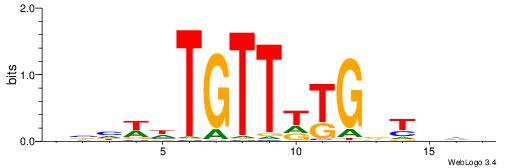 |
| Motif ID: | UP00090 |
| Motif name: | Elf3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0355538 |
Alignment:
DDHBTTTTTTTTHDAVH
--TTHRWTTTTWYRTTY
| Original motif | Reverse complement motif |
| Consensus sequence: DBTHHAAAAAAAAVHDH | Consensus sequence: DDHBTTTTTTTTHDAVH |
 |
 |
| Motif ID: | UP00255 |
| Motif name: | Dbx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0356888 |
Alignment:
DHDBTAWTAATTHAKTH
--TTHRWTTTTWYRTTY
| Original motif | Reverse complement motif |
| Consensus sequence: HARTHAATTAWTAVDHD | Consensus sequence: DHDBTAWTAATTHAKTH |
 |
 |
| Motif ID: | UP00213 |
| Motif name: | Hoxa9 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0367298 |
Alignment:
WDTAWTTTWATGKCCGD
TTHRWTTTTWYRTTY--
| Original motif | Reverse complement motif |
| Consensus sequence: DCGGYCATWAAAWTADW | Consensus sequence: WDTAWTTTWATGKCCGD |
 |
 |
| Dataset #: 1 | Motif ID: 64 | Motif name: C064 |
| Original motif | Reverse complement motif |
| Consensus sequence: DAAAKAAAAMAAK | Consensus sequence: RTTYTTTTRTTTD |
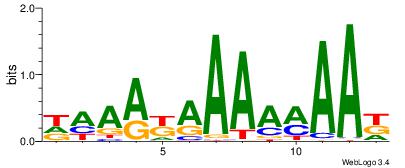 |
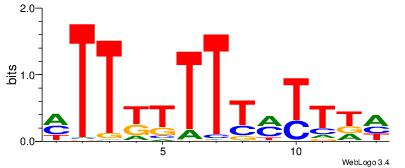 |
Best Matches for Motif ID 64 (Highest to Lowest)
| Motif ID: | UP00077 |
| Motif name: | Srf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 13 |
| Similarity score: | 0.0175156 |
Alignment:
HHVBYTTTTTTTTDDVV
--RTTYTTTTRTTTD--
| Original motif | Reverse complement motif |
| Consensus sequence: VBDDAAAAAAAAMVBHH | Consensus sequence: HHVBYTTTTTTTTDDVV |
 |
 |
| Motif ID: | UP00028 |
| Motif name: | Tcfap2e_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0210094 |
Alignment:
HDCDBRAAAAAADD
DAAAKAAAAMAAK-
| Original motif | Reverse complement motif |
| Consensus sequence: HDCDBRAAAAAADD | Consensus sequence: DDTTTTTTMBDGDH |
 |
 |
| Motif ID: | UP00025 |
| Motif name: | Foxk1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.030589 |
Alignment:
HDDTGTTKTTKTHHD
--RTTYTTTTRTTTD
| Original motif | Reverse complement motif |
| Consensus sequence: HHHAYAAYAACAHHH | Consensus sequence: HDDTGTTKTTKTHHD |
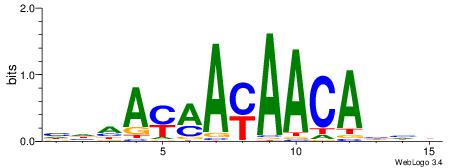 |
 |
| Motif ID: | UP00090 |
| Motif name: | Elf3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 13 |
| Similarity score: | 0.0322312 |
Alignment:
DDHBTTTTTTTTHDAVH
-RTTYTTTTRTTTD---
| Original motif | Reverse complement motif |
| Consensus sequence: DBTHHAAAAAAAAVHDH | Consensus sequence: DDHBTTTTTTTTHDAVH |
 |
 |
| Motif ID: | UP00180 |
| Motif name: | Hoxd13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0326524 |
Alignment:
HDHHTTTTATTKKHDD
---RTTYTTTTRTTTD
| Original motif | Reverse complement motif |
| Consensus sequence: HDHYYAATAAAAHHHH | Consensus sequence: HDHHTTTTATTKKHDD |
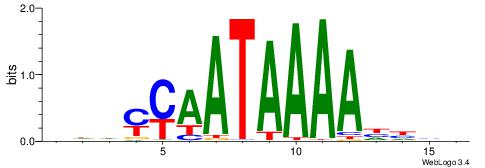 |
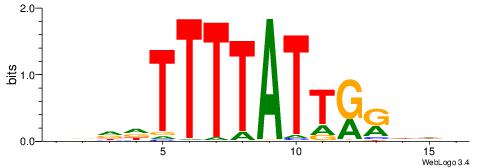 |
| Dataset #: 1 | Motif ID: 65 | Motif name: C065 |
| Original motif | Reverse complement motif |
| Consensus sequence: MSTGAGWTCMASK | Consensus sequence: YSTRGAWCTCASR |
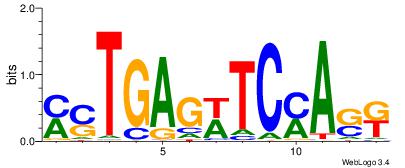 |
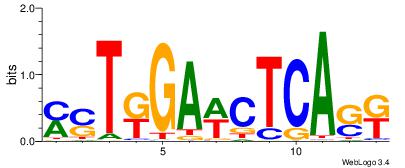 |
Best Matches for Motif ID 65 (Highest to Lowest)
| Motif ID: | UP00080 |
| Motif name: | Gata5_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 13 |
| Similarity score: | 0.0144176 |
Alignment:
VVHHWGATATCTMHBBV
--MSTGAGWTCMASK--
| Original motif | Reverse complement motif |
| Consensus sequence: VVBHRAGATATCWDHBB | Consensus sequence: VVHHWGATATCTMHBBV |
 |
 |
| Motif ID: | UP00148 |
| Motif name: | Hdx |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 13 |
| Similarity score: | 0.0160219 |
Alignment:
DDKDHGAAATCAHHBHV
-YSTRGAWCTCASR---
| Original motif | Reverse complement motif |
| Consensus sequence: DDKDHGAAATCAHHBHV | Consensus sequence: BDBDHTGATTTCDHYDD |
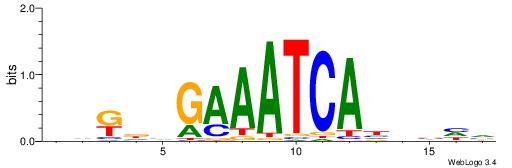 |
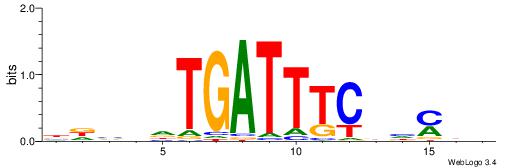 |
| Motif ID: | UP00066 |
| Motif name: | Hnf4a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 13 |
| Similarity score: | 0.0188193 |
Alignment:
BDBVMGGGGTCAABHDV
--MSTGAGWTCMASK--
| Original motif | Reverse complement motif |
| Consensus sequence: BDBVMGGGGTCAABHDV | Consensus sequence: BHHVTTGACCCCYVVDB |
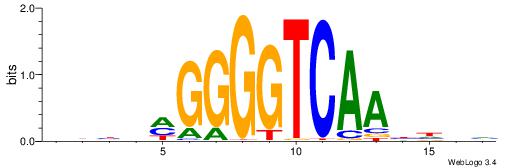 |
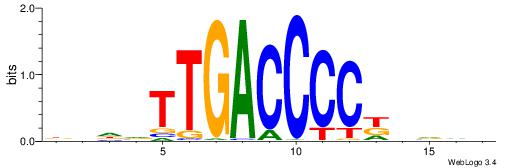 |
| Motif ID: | UP00020 |
| Motif name: | Atf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 13 |
| Similarity score: | 0.0224584 |
Alignment:
DMDMTGACGTCAKHBD
--MSTGAGWTCMASK-
| Original motif | Reverse complement motif |
| Consensus sequence: DBDRTGACGTCAYHRD | Consensus sequence: DMDMTGACGTCAKHBD |
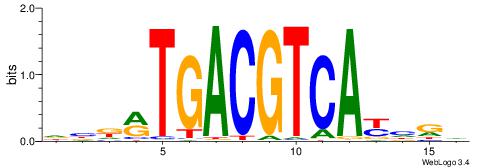 |
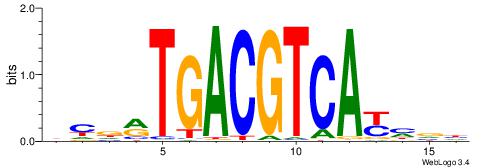 |
| Motif ID: | UP00054 |
| Motif name: | Tcf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 13 |
| Similarity score: | 0.0232284 |
Alignment:
BHHASATCAAAGGAHHH
YSTRGAWCTCASR----
| Original motif | Reverse complement motif |
| Consensus sequence: BHHASATCAAAGGAHHH | Consensus sequence: HHHTCCTTTGATSTHHV |
 |
 |
| Dataset #: 1 | Motif ID: 66 | Motif name: C066 |
| Original motif | Reverse complement motif |
| Consensus sequence: MAMTCCCMSKGCCTCTK | Consensus sequence: YAGAGGCRSYGGGARTR |
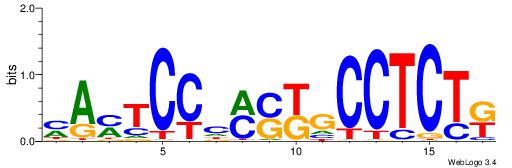 |
 |
Best Matches for Motif ID 66 (Highest to Lowest)
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 17 |
| Similarity score: | 0.03808 |
Alignment:
VDWDARRGACGCGGWHGGAKHV
-----YAGAGGCRSYGGGARTR
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 17 |
| Similarity score: | 0.0499224 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
---YAGAGGCRSYGGGARTR---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0510562 |
Alignment:
HHHHDGGGGCGGDBHDV
YAGAGGCRSYGGGARTR
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 17 |
| Similarity score: | 0.0517099 |
Alignment:
HHBSHTCGTGRGTGGKGDBVWMV
---YAGAGGCRSYGGGARTR---
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 0.0522208 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
--YAGAGGCRSYGGGARTR---
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Dataset #: 1 | Motif ID: 67 | Motif name: C067 |
| Original motif | Reverse complement motif |
| Consensus sequence: WDAAAAAARKAAAD | Consensus sequence: DTTTRKTTTTTTDW |
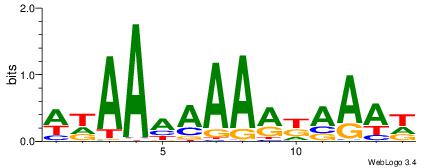 |
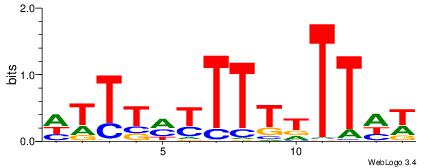 |
Best Matches for Motif ID 67 (Highest to Lowest)
| Motif ID: | UP00077 |
| Motif name: | Srf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0232781 |
Alignment:
VBDDAAAAAAAAMVBHH
---WDAAAAAARKAAAD
| Original motif | Reverse complement motif |
| Consensus sequence: VBDDAAAAAAAAMVBHH | Consensus sequence: HHVBYTTTTTTTTDDVV |
 |
 |
| Motif ID: | UP00037 |
| Motif name: | Zfp105_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0263721 |
Alignment:
HDYAAAHAAMAADHD
-WDAAAAAARKAAAD
| Original motif | Reverse complement motif |
| Consensus sequence: HDYAAAHAAMAADHD | Consensus sequence: HHHTTRTTDTTTKDH |
 |
 |
| Motif ID: | UP00014 |
| Motif name: | Sox17_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0301541 |
Alignment:
DDWTBAATTGTTDDB
-DTTTRKTTTTTTDW
| Original motif | Reverse complement motif |
| Consensus sequence: VDDAACAATTVAWHD | Consensus sequence: DDWTBAATTGTTDDB |
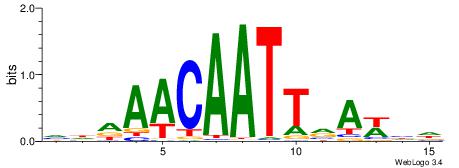 |
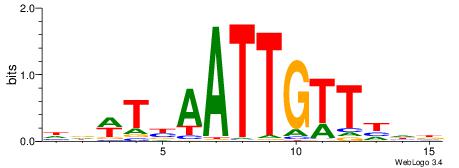 |
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0307495 |
Alignment:
HBDDDAACAAAGRVBHH
---WDAAAAAARKAAAD
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
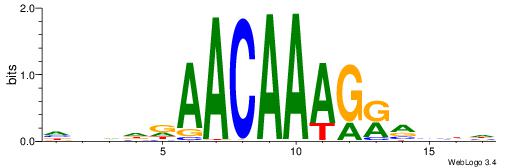 |
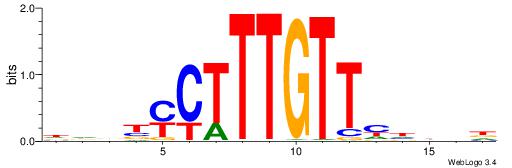 |
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0312958 |
Alignment:
HBDDRAACAAAGRABHH
---WDAAAAAARKAAAD
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
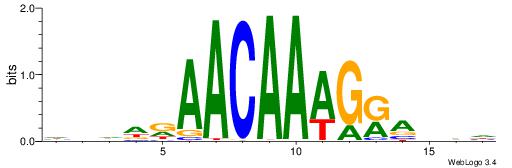 |
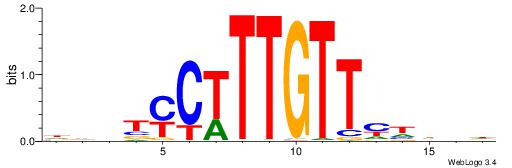 |
| Dataset #: 1 | Motif ID: 68 | Motif name: C068 |
| Original motif | Reverse complement motif |
| Consensus sequence: BGCYKCTGCMTCCM | Consensus sequence: YGGAYGCAGYKGCV |
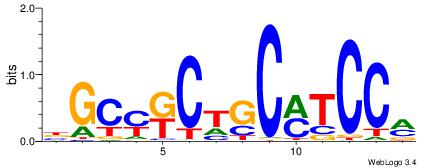 |
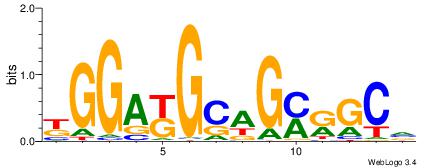 |
Best Matches for Motif ID 68 (Highest to Lowest)
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0242069 |
Alignment:
VHYTCCDWCCGCGTCMMTHWHV
---BGCYKCTGCMTCCM-----
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.02581 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
---BGCYKCTGCMTCCM-----
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0294733 |
Alignment:
BBBAVTGCAGTGBBVDD
-YGGAYGCAGYKGCV--
| Original motif | Reverse complement motif |
| Consensus sequence: DDBBBCACTGCABTBBB | Consensus sequence: BBBAVTGCAGTGBBVDD |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 10 |
| Number of overlap: | 14 |
| Similarity score: | 0.0297174 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
BGCYKCTGCMTCCM---------
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0300511 |
Alignment:
BHHBHCCGCCCCDHHHH
BGCYKCTGCMTCCM---
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
 |
 |
| Dataset #: 1 | Motif ID: 69 | Motif name: C069 |
| Original motif | Reverse complement motif |
| Consensus sequence: AHAAMAAYWDYAAMAAHD | Consensus sequence: DHTTYTTMDWMTTYTTHT |
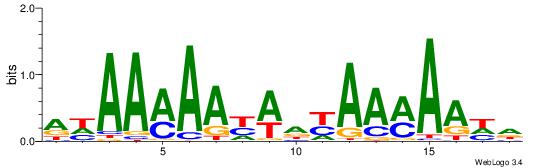 |
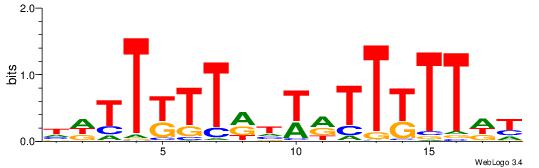 |
Best Matches for Motif ID 69 (Highest to Lowest)
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0547241 |
Alignment:
HHBDHDDAACAATDRHHHHHBW
--AHAAMAAYWDYAAMAAHD--
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0556229 |
Alignment:
HBHDVHDTCTTATCTHTDDHDV
-DHTTYTTMDWMTTYTTHT---
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0564427 |
Alignment:
VVYDDDCHCAVACAATTDBVAC
AHAAMAAYWDYAAMAAHD----
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.058939 |
Alignment:
HHHBBMGATAVHATCWVVDHVH
-AHAAMAAYWDYAAMAAHD---
| Original motif | Reverse complement motif |
| Consensus sequence: HBHDVBWGATHBTATCRVBHHH | Consensus sequence: HHHBBMGATAVHATCWVVDHVH |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.064465 |
Alignment:
DCATDBTGHGCATKMTTBRSWR
-DHTTYTTMDWMTTYTTHT---
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
 |
 |
| Dataset #: 1 | Motif ID: 70 | Motif name: C070 |
| Original motif | Reverse complement motif |
| Consensus sequence: GVGABAGAVAGASVK | Consensus sequence: RBSTCTVTCTBTCVC |
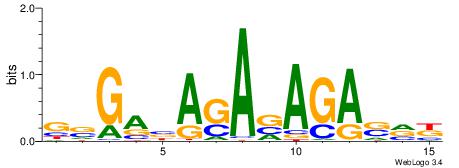 |
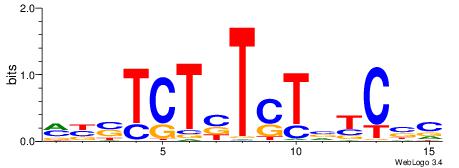 |
Best Matches for Motif ID 70 (Highest to Lowest)
| Motif ID: | >UP00011 |
| Motif name: | Irf6_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0371635 |
Alignment:
VBBACTCYCGGKHDV
RBSTCTVTCTBTCVC
| Original motif | Reverse complement motif |
| Consensus sequence: VBBACTCYCGGKHDV | Consensus sequence: VDDRCCGMGAGTBBB |
 |
 |
| Motif ID: | UP00101 |
| Motif name: | Sox12_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0394179 |
Alignment:
VVDBASACAAAGRADH
-GVGABAGAVAGASVK
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBASACAAAGRADH | Consensus sequence: HDTMCTTTGTSTVDBB |
 |
 |
| Motif ID: | UP00018 |
| Motif name: | Irf4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0452113 |
Alignment:
VVBAYTCTCGGWHDB
RBSTCTVTCTBTCVC
| Original motif | Reverse complement motif |
| Consensus sequence: VVBAYTCTCGGWHDB | Consensus sequence: BHHWCCGAGAMTVVB |
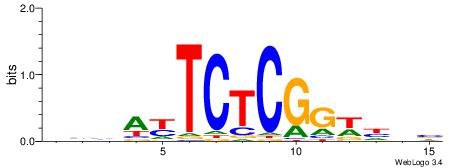 |
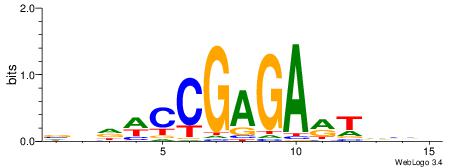 |
| Motif ID: | UP00077 |
| Motif name: | Srf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0480451 |
Alignment:
VBDDAAAAAAAAMVBHH
GVGABAGAVAGASVK--
| Original motif | Reverse complement motif |
| Consensus sequence: VBDDAAAAAAAAMVBHH | Consensus sequence: HHVBYTTTTTTTTDDVV |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 15 |
| Similarity score: | 0.0486114 |
Alignment:
HVHBVTGTCTGGDDHDD
--RBSTCTVTCTBTCVC
| Original motif | Reverse complement motif |
| Consensus sequence: HDHDDCCAGACABBHVH | Consensus sequence: HVHBVTGTCTGGDDHDD |
 |
 |
| Dataset #: 1 | Motif ID: 71 | Motif name: C071 |
| Original motif | Reverse complement motif |
| Consensus sequence: SVCCACSVSCACCBS | Consensus sequence: SBGGTGSVSGTGGVS |
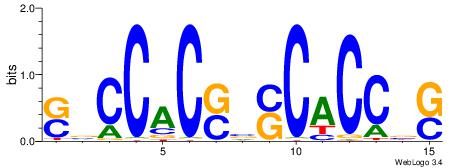 |
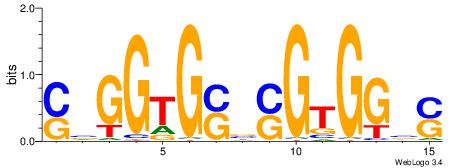 |
Best Matches for Motif ID 71 (Highest to Lowest)
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0205519 |
Alignment:
HVBCCCCCCCCMHHHB
-SVCCACSVSCACCBS
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 15 |
| Similarity score: | 0.020698 |
Alignment:
BDHHMCGCCCCCTHVBB
SVCCACSVSCACCBS--
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0211186 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
-SBGGTGSVSGTGGVS------
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 15 |
| Similarity score: | 0.022472 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
------SVCCACSVSCACCBS--
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 15 |
| Similarity score: | 0.0250115 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
------SVCCACSVSCACCBS--
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Dataset #: 1 | Motif ID: 72 | Motif name: C072 |
| Original motif | Reverse complement motif |
| Consensus sequence: SWAGCCVDGGCW | Consensus sequence: WGCCHVGGCTWS |
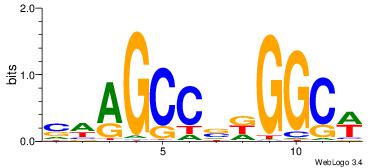 |
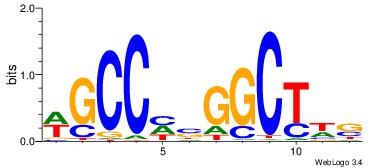 |
Best Matches for Motif ID 72 (Highest to Lowest)
| Motif ID: | UP00010 |
| Motif name: | Tcfap2b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.00558864 |
Alignment:
HHKGCCYSRGGCWAD
-SWAGCCVDGGCW--
| Original motif | Reverse complement motif |
| Consensus sequence: DTWGCCKSMGGCRHH | Consensus sequence: HHKGCCYSRGGCWAD |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.00743548 |
Alignment:
BDWAGGCGTGBCHHD
-SWAGCCVDGGCW--
| Original motif | Reverse complement motif |
| Consensus sequence: BDWAGGCGTGBCHHD | Consensus sequence: HHDGBCACGCCTWDB |
 |
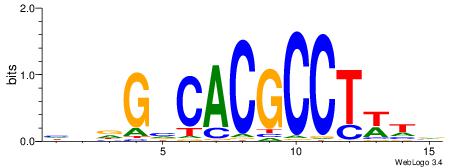 |
| Motif ID: | UP00028 |
| Motif name: | Tcfap2e_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0141542 |
Alignment:
HHHGCCTSAGGCDAD
-SWAGCCVDGGCW--
| Original motif | Reverse complement motif |
| Consensus sequence: HHHGCCTSAGGCDAD | Consensus sequence: DTHGCCTSAGGCHHH |
 |
 |
| Motif ID: | UP00087 |
| Motif name: | Tcfap2c_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0187577 |
Alignment:
WHHSCCYSRGGSDAD
--WGCCHVGGCTWS-
| Original motif | Reverse complement motif |
| Consensus sequence: WHHSCCYSRGGSDAD | Consensus sequence: DTHSCCKSMGGSHHW |
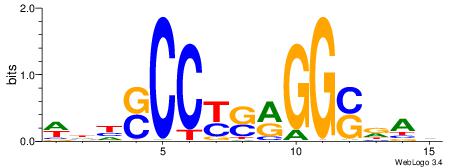 |
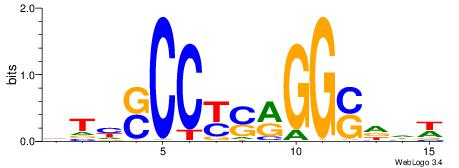 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0187696 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
-SWAGCCVDGGCW---------
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Dataset #: 1 | Motif ID: 73 | Motif name: C073 |
| Original motif | Reverse complement motif |
| Consensus sequence: YMWGSMYAGCCRKSKCTR | Consensus sequence: KAGYSYKGGCTKRSCWYK |
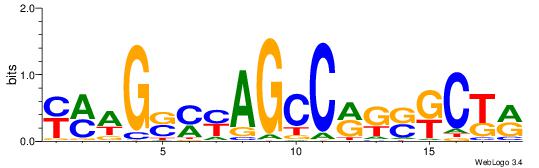 |
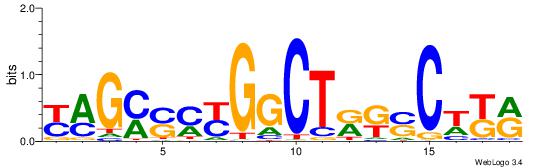 |
Best Matches for Motif ID 73 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.0271167 |
Alignment:
HGDHVHKGACCACCCGGBGBGDB
----YMWGSMYAGCCRKSKCTR-
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 18 |
| Similarity score: | 0.0280669 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
-----KAGYSYKGGCTKRSCWYK
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0284377 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
---YMWGSMYAGCCRKSKCTR--
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0313979 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
--KAGYSYKGGCTKRSCWYK--
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.032852 |
Alignment:
BBBDVVRGACCACCCAVGABBAB
----YMWGSMYAGCCRKSKCTR-
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Dataset #: 1 | Motif ID: 74 | Motif name: C074 |
| Original motif | Reverse complement motif |
| Consensus sequence: CCAGGCWGGCSKS | Consensus sequence: SRSGCCWGCCTGG |
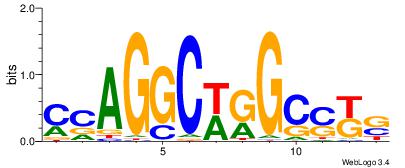 |
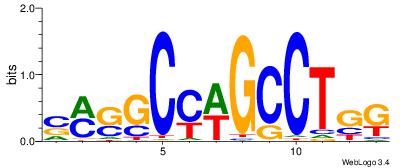 |
Best Matches for Motif ID 74 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 13 |
| Similarity score: | 0.0257723 |
Alignment:
BBBDVVRGACCACCCAVGABBAB
-----SRSGCCWGCCTGG-----
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 13 |
| Similarity score: | 0.0270649 |
Alignment:
DDRMHBACGCGYGCGTDGBBDB
-----SRSGCCWGCCTGG----
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0283276 |
Alignment:
BHBHDTGGCGGGGBDHD
---CCAGGCWGGCSKS-
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
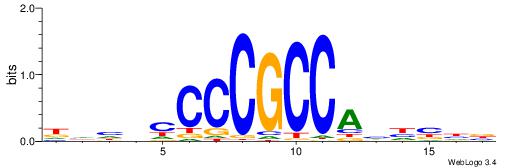 |
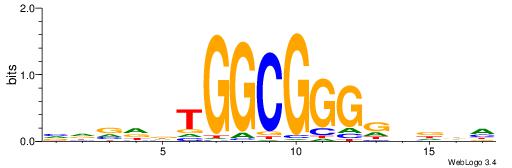 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0313235 |
Alignment:
HHDGBCACGCCTWDB
-SRSGCCWGCCTGG-
| Original motif | Reverse complement motif |
| Consensus sequence: BDWAGGCGTGBCHHD | Consensus sequence: HHDGBCACGCCTWDB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 13 |
| Similarity score: | 0.0327573 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
------CCAGGCWGGCSKS----
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Dataset #: 1 | Motif ID: 75 | Motif name: C075 |
| Original motif | Reverse complement motif |
| Consensus sequence: MGGCAGMSGCAGSCK | Consensus sequence: YGSCTGCSYCTGCCY |
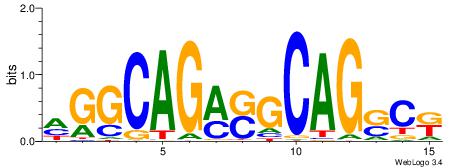 |
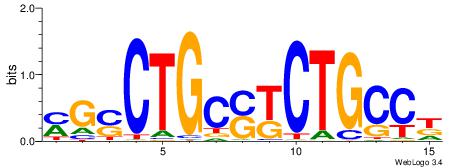 |
Best Matches for Motif ID 75 (Highest to Lowest)
| Motif ID: | UP00099 |
| Motif name: | Ascl2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 15 |
| Similarity score: | 0.0343574 |
Alignment:
BBYVVCAGCTGCBVHHD
--MGGCAGMSGCAGSCK
| Original motif | Reverse complement motif |
| Consensus sequence: BBYVVCAGCTGCBVHHD | Consensus sequence: HHDVVGCAGCTGVBKVB |
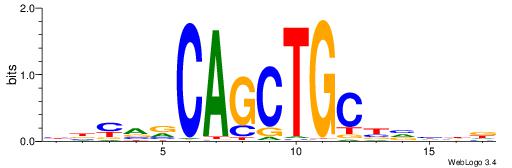 |
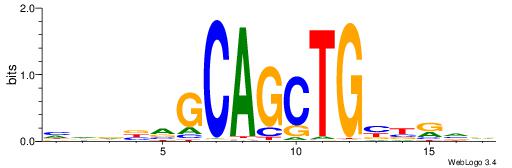 |
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0352972 |
Alignment:
HBBBCCGCCCCWHHHV
-YGSCTGCSYCTGCCY
| Original motif | Reverse complement motif |
| Consensus sequence: HBBBCCGCCCCWHHHV | Consensus sequence: BHHHWGGGGCGGBBVH |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0363915 |
Alignment:
HHBVHTGACCTTGVDHD
-YGSCTGCSYCTGCCY-
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBCAAGGTCAHVBDH | Consensus sequence: HHBVHTGACCTTGVDHD |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 15 |
| Similarity score: | 0.036959 |
Alignment:
BHHBHCCGCCCCDHHHH
--YGSCTGCSYCTGCCY
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0381017 |
Alignment:
DDRMHBACGCGYGCGTDGBBDB
MGGCAGMSGCAGSCK-------
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
 |
 |
| Dataset #: 1 | Motif ID: 76 | Motif name: C076 |
| Original motif | Reverse complement motif |
| Consensus sequence: ASCTRCCTCYBCSB | Consensus sequence: VSGBKGAGGMAGST |
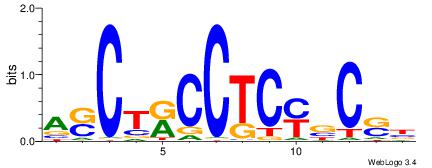 |
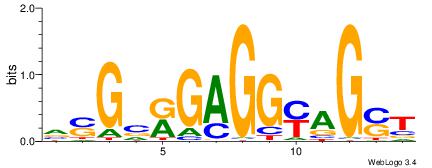 |
Best Matches for Motif ID 76 (Highest to Lowest)
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0266758 |
Alignment:
BHHHWGGGGCGGBBVH
VSGBKGAGGMAGST--
| Original motif | Reverse complement motif |
| Consensus sequence: HBBBCCGCCCCWHHHV | Consensus sequence: BHHHWGGGGCGGBBVH |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0272671 |
Alignment:
HHHHDGGGGCGGDBHDV
VSGBKGAGGMAGST---
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
 |
 |
| Motif ID: | UP00013 |
| Motif name: | Gabpa_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0302132 |
Alignment:
DBVVDRSGGAWKVHKG
VSGBKGAGGMAGST--
| Original motif | Reverse complement motif |
| Consensus sequence: CYDBYWTCCSMHBVVH | Consensus sequence: DBVVDRSGGAWKVHKG |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0328017 |
Alignment:
BHHDYGGGGGGGGBVD
-VSGBKGAGGMAGST-
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0330521 |
Alignment:
HVKDCCGGGGGGWADDD
---VSGBKGAGGMAGST
| Original motif | Reverse complement motif |
| Consensus sequence: DDDTWCCCCCCGGDRVH | Consensus sequence: HVKDCCGGGGGGWADDD |
 |
 |
| Dataset #: 1 | Motif ID: 77 | Motif name: C077 |
| Original motif | Reverse complement motif |
| Consensus sequence: SBCCCCGGSSCCCCCB | Consensus sequence: BGGGGGSSCCGGGGBS |
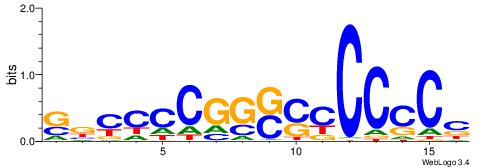 |
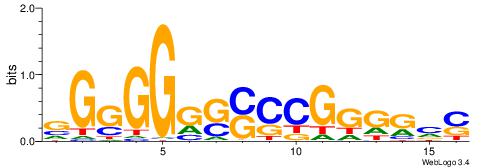 |
Best Matches for Motif ID 77 (Highest to Lowest)
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0480753 |
Alignment:
BDHHMCGCCCCCTHVBB
SBCCCCGGSSCCCCCB-
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0484396 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
-------BGGGGGSSCCGGGGBS
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0490479 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
SBCCCCGGSSCCCCCB------
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0535184 |
Alignment:
HHHHDGGGGCGGDBHDV
-BGGGGGSSCCGGGGBS
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0543092 |
Alignment:
VDWDARRGACGCGGWHGGAKHV
SBCCCCGGSSCCCCCB------
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Dataset #: 1 | Motif ID: 78 | Motif name: C078 |
| Original motif | Reverse complement motif |
| Consensus sequence: YRGAYAGAGRGAYR | Consensus sequence: KMTCKCTCTMTCKM |
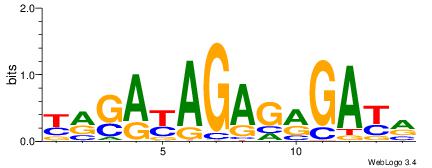 |
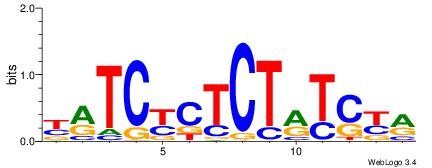 |
Best Matches for Motif ID 78 (Highest to Lowest)
| Motif ID: | >UP00011 |
| Motif name: | Irf6_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0289623 |
Alignment:
VDDRCCGMGAGTBBB
-KMTCKCTCTMTCKM
| Original motif | Reverse complement motif |
| Consensus sequence: VBBACTCYCGGKHDV | Consensus sequence: VDDRCCGMGAGTBBB |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0334841 |
Alignment:
BHHDTCCCACTCBDBV
--KMTCKCTCTMTCKM
| Original motif | Reverse complement motif |
| Consensus sequence: BBHBGAGTGGGAHHDB | Consensus sequence: BHHDTCCCACTCBDBV |
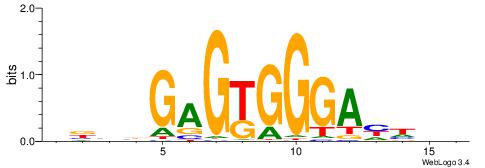 |
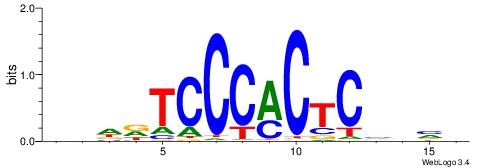 |
| Motif ID: | UP00040 |
| Motif name: | Irf5_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0339287 |
Alignment:
BHDAHCGAGARTHBV
YRGAYAGAGRGAYR-
| Original motif | Reverse complement motif |
| Consensus sequence: BHDAHCGAGARTHBV | Consensus sequence: VBHAKTCTCGHTHHV |
 |
 |
| Motif ID: | UP00078 |
| Motif name: | Arid3a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0352486 |
Alignment:
HHVVRTATCRDVDHD
-YRGAYAGAGRGAYR
| Original motif | Reverse complement motif |
| Consensus sequence: HHVVRTATCRDVDHD | Consensus sequence: DHDBDKGATAMVVDH |
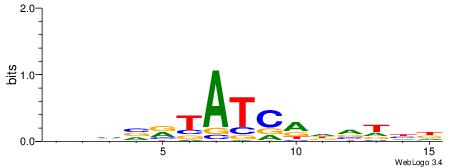 |
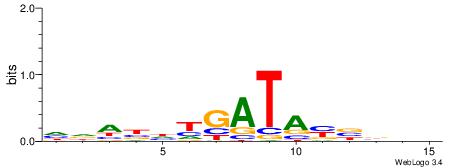 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 14 |
| Similarity score: | 0.0392148 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
-------YRGAYAGAGRGAYR-
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Dataset #: 1 | Motif ID: 79 | Motif name: C079 |
| Original motif | Reverse complement motif |
| Consensus sequence: ADAAAKWAAAAWAKARW | Consensus sequence: WKTRTWTTTTWRTTTDT |
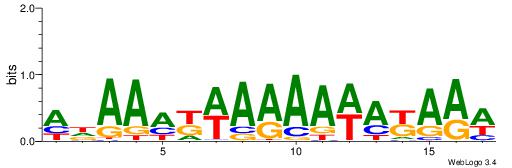 |
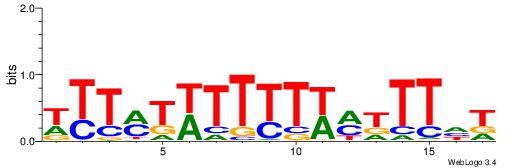 |
Best Matches for Motif ID 79 (Highest to Lowest)
| Motif ID: | UP00077 |
| Motif name: | Srf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0265367 |
Alignment:
VBDDAAAAAAAAMVBHH
ADAAAKWAAAAWAKARW
| Original motif | Reverse complement motif |
| Consensus sequence: VBDDAAAAAAAAMVBHH | Consensus sequence: HHVBYTTTTTTTTDDVV |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 0.0319437 |
Alignment:
WBHHHHHMDATTGTTHDHDVHH
----WKTRTWTTTTWRTTTDT-
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00090 |
| Motif name: | Elf3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0359246 |
Alignment:
DBTHHAAAAAAAAVHDH
ADAAAKWAAAAWAKARW
| Original motif | Reverse complement motif |
| Consensus sequence: DBTHHAAAAAAAAVHDH | Consensus sequence: DDHBTTTTTTTTHDAVH |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.036697 |
Alignment:
BDHDDADAGATAAGADHBDHVD
-----ADAAAKWAAAAWAKARW
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Motif ID: | UP00025 |
| Motif name: | Foxk1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0381106 |
Alignment:
DDAATGTAAACAAAVVB
ADAAAKWAAAAWAKARW
| Original motif | Reverse complement motif |
| Consensus sequence: DDAATGTAAACAAAVVB | Consensus sequence: BBVTTTGTTTACATTDD |
 |
 |
| Dataset #: 1 | Motif ID: 80 | Motif name: C080 |
| Original motif | Reverse complement motif |
| Consensus sequence: WWAAVAAMMHAAWW | Consensus sequence: WWTTHRYTTBTTWW |
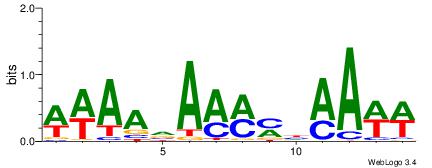 |
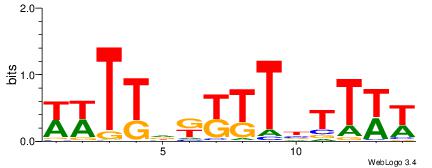 |
Best Matches for Motif ID 80 (Highest to Lowest)
| Motif ID: | UP00077 |
| Motif name: | Srf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0245388 |
Alignment:
VBDDAAAAAAAAMVBHH
---WWAAVAAMMHAAWW
| Original motif | Reverse complement motif |
| Consensus sequence: VBDDAAAAAAAAMVBHH | Consensus sequence: HHVBYTTTTTTTTDDVV |
 |
 |
| Motif ID: | UP00170 |
| Motif name: | Isl2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0341915 |
Alignment:
HWDTTAATYKMTTHHR
-WWTTHRYTTBTTWW-
| Original motif | Reverse complement motif |
| Consensus sequence: MHHAAYYMATTAADWH | Consensus sequence: HWDTTAATYKMTTHHR |
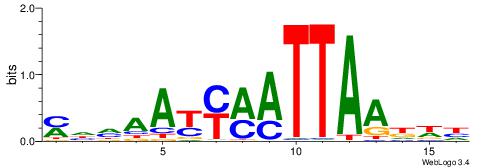 |
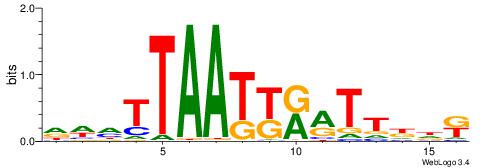 |
| Motif ID: | UP00090 |
| Motif name: | Elf3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0349632 |
Alignment:
DBTHHAAAAAAAAVHDH
---WWAAVAAMMHAAWW
| Original motif | Reverse complement motif |
| Consensus sequence: DBTHHAAAAAAAAVHDH | Consensus sequence: DDHBTTTTTTTTHDAVH |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0362632 |
Alignment:
ADDDYAWAACAAMAHH
WWAAVAAMMHAAWW--
| Original motif | Reverse complement motif |
| Consensus sequence: ADDDYAWAACAAMAHH | Consensus sequence: HDTYTTGTTWTKDDDT |
 |
 |
| Motif ID: | UP00037 |
| Motif name: | Zfp105_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0380401 |
Alignment:
HDYAAAHAAMAADHD
WWAAVAAMMHAAWW-
| Original motif | Reverse complement motif |
| Consensus sequence: HDYAAAHAAMAADHD | Consensus sequence: HHHTTRTTDTTTKDH |
 |
 |
| Dataset #: 1 | Motif ID: 81 | Motif name: C081 |
| Original motif | Reverse complement motif |
| Consensus sequence: BCCSHCCCSCMG | Consensus sequence: CYGSGGGHSGGB |
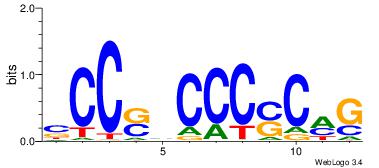 |
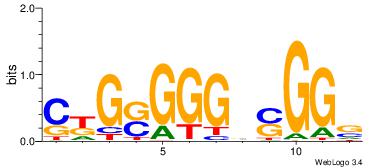 |
Best Matches for Motif ID 81 (Highest to Lowest)
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0133435 |
Alignment:
DBCCCCCCCCCCMYC
--BCCSHCCCSCMG-
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0173124 |
Alignment:
HBBBCCGCCCCWHHHV
---BCCSHCCCSCMG-
| Original motif | Reverse complement motif |
| Consensus sequence: HBBBCCGCCCCWHHHV | Consensus sequence: BHHHWGGGGCGGBBVH |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0193295 |
Alignment:
BHHBHCCGCCCCDHHHH
----BCCSHCCCSCMG-
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0214278 |
Alignment:
HVBCCCCCCCCMHHHB
-BCCSHCCCSCMG---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 12 |
| Similarity score: | 0.0230163 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
----CYGSGGGHSGGB------
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Dataset #: 1 | Motif ID: 82 | Motif name: C082 |
| Original motif | Reverse complement motif |
| Consensus sequence: WKAAARAAARAAW | Consensus sequence: WTTKTTTMTTTYW |
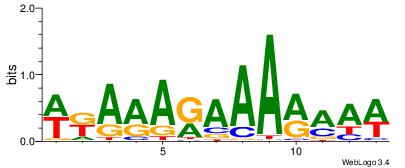 |
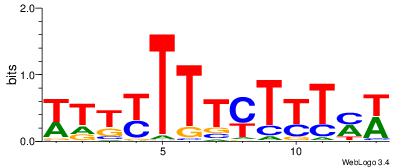 |
Best Matches for Motif ID 82 (Highest to Lowest)
| Motif ID: | UP00077 |
| Motif name: | Srf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 13 |
| Similarity score: | 0.0105695 |
Alignment:
VBDDAAAAAAAAMVBHH
--WKAAARAAARAAW--
| Original motif | Reverse complement motif |
| Consensus sequence: VBDDAAAAAAAAMVBHH | Consensus sequence: HHVBYTTTTTTTTDDVV |
 |
 |
| Motif ID: | UP00028 |
| Motif name: | Tcfap2e_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0217842 |
Alignment:
DDTTTTTTMBDGDH
-WTTKTTTMTTTYW
| Original motif | Reverse complement motif |
| Consensus sequence: HDCDBRAAAAAADD | Consensus sequence: DDTTTTTTMBDGDH |
 |
 |
| Motif ID: | UP00037 |
| Motif name: | Zfp105_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0234988 |
Alignment:
HHHTTRTTDTTTKDH
--WTTKTTTMTTTYW
| Original motif | Reverse complement motif |
| Consensus sequence: HDYAAAHAAMAADHD | Consensus sequence: HHHTTRTTDTTTKDH |
 |
 |
| Motif ID: | UP00090 |
| Motif name: | Elf3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 13 |
| Similarity score: | 0.0265681 |
Alignment:
DDHBTTTTTTTTHDAVH
-WTTKTTTMTTTYW---
| Original motif | Reverse complement motif |
| Consensus sequence: DBTHHAAAAAAAAVHDH | Consensus sequence: DDHBTTTTTTTTHDAVH |
 |
 |
| Motif ID: | UP00097 |
| Motif name: | Mtf1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0267248 |
Alignment:
VDTHTTTHHTATDH
WTTKTTTMTTTYW-
| Original motif | Reverse complement motif |
| Consensus sequence: HDATAHDAAAHADV | Consensus sequence: VDTHTTTHHTATDH |
 |
 |
| Dataset #: 1 | Motif ID: 83 | Motif name: C083 |
| Original motif | Reverse complement motif |
| Consensus sequence: CKBCCTSYRSCTCMMG | Consensus sequence: CYRGAGSMKSAGGBRG |
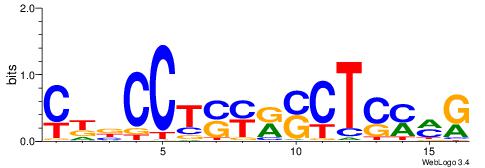 |
 |
Best Matches for Motif ID 83 (Highest to Lowest)
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 16 |
| Similarity score: | 0.0269608 |
Alignment:
VHYTCCDWCCGCGTCMMTHWHV
--CKBCCTSYRSCTCMMG----
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0336468 |
Alignment:
HBBBCCGCCCCWHHHV
CKBCCTSYRSCTCMMG
| Original motif | Reverse complement motif |
| Consensus sequence: HBBBCCGCCCCWHHHV | Consensus sequence: BHHHWGGGGCGGBBVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 16 |
| Similarity score: | 0.0351892 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
-CKBCCTSYRSCTCMMG-----
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.035551 |
Alignment:
HVDVGGGHGGGGDBHB
CYRGAGSMKSAGGBRG
| Original motif | Reverse complement motif |
| Consensus sequence: BHVDCCCCDCCCBDBH | Consensus sequence: HVDVGGGHGGGGDBHB |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.035682 |
Alignment:
BHHBHCCGCCCCDHHHH
-CKBCCTSYRSCTCMMG
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
 |
 |
| Dataset #: 1 | Motif ID: 84 | Motif name: C084 |
| Original motif | Reverse complement motif |
| Consensus sequence: SCTGCMKCTGCVB | Consensus sequence: VVGCAGYRGCAGS |
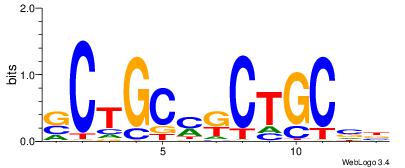 |
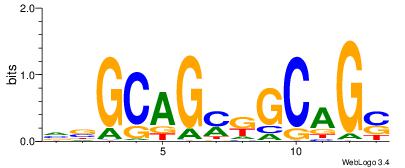 |
Best Matches for Motif ID 84 (Highest to Lowest)
| Motif ID: | UP00099 |
| Motif name: | Ascl2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 13 |
| Similarity score: | 0.0229768 |
Alignment:
HHDVVGCAGCTGVBKVB
---VVGCAGYRGCAGS-
| Original motif | Reverse complement motif |
| Consensus sequence: BBYVVCAGCTGCBVHHD | Consensus sequence: HHDVVGCAGCTGVBKVB |
 |
 |
| Motif ID: | UP00036 |
| Motif name: | Myf6_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0256725 |
Alignment:
VVVRACAGVCBCDCC
SCTGCMKCTGCVB--
| Original motif | Reverse complement motif |
| Consensus sequence: VVVRACAGVCBCDCC | Consensus sequence: GGDGBGVCTGTKVVB |
 |
 |
| Motif ID: | UP00013 |
| Motif name: | Gabpa_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0310274 |
Alignment:
CYDBYWTCCSMHBVVH
SCTGCMKCTGCVB---
| Original motif | Reverse complement motif |
| Consensus sequence: CYDBYWTCCSMHBVVH | Consensus sequence: DBVVDRSGGAWKVHKG |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 13 |
| Similarity score: | 0.0325376 |
Alignment:
BHHBHCCGCCCCDHHHH
----SCTGCMKCTGCVB
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
 |
 |
| Motif ID: | UP00102 |
| Motif name: | Zic1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0348713 |
Alignment:
BHDCVCAGCAGGHVD
--VVGCAGYRGCAGS
| Original motif | Reverse complement motif |
| Consensus sequence: BHDCVCAGCAGGHVD | Consensus sequence: DVHCCTGCTGBGDDB |
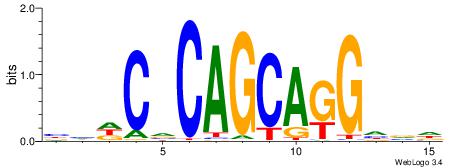 |
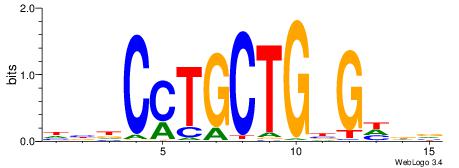 |
| Dataset #: 1 | Motif ID: 85 | Motif name: C085 |
| Original motif | Reverse complement motif |
| Consensus sequence: VSGRSBHAGARDVSGRS | Consensus sequence: SKCSVHMTCTDBSKCSV |
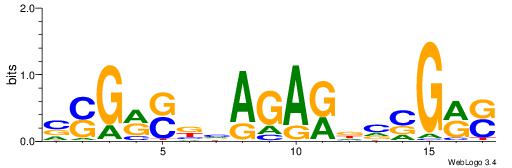 |
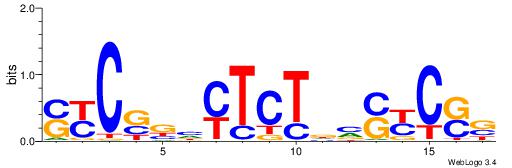 |
Best Matches for Motif ID 85 (Highest to Lowest)
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 17 |
| Similarity score: | 0.0345982 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
----SKCSVHMTCTDBSKCSV-
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0368478 |
Alignment:
BBYVVCAGCTGCBVHHD
SKCSVHMTCTDBSKCSV
| Original motif | Reverse complement motif |
| Consensus sequence: BBYVVCAGCTGCBVHHD | Consensus sequence: HHDVVGCAGCTGVBKVB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0387638 |
Alignment:
HHBSHTCGTGRGTGGKGDBVWMV
------VSGRSBHAGARDVSGRS
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 0.0394694 |
Alignment:
GTVBDAATTGTVTGHGDDHKVB
---SKCSVHMTCTDBSKCSV--
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00016 |
| Motif name: | Sry_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0403194 |
Alignment:
HHDHDRAACAATDDVHV
VSGRSBHAGARDVSGRS
| Original motif | Reverse complement motif |
| Consensus sequence: HHDHDRAACAATDDVHV | Consensus sequence: VHVHDATTGTTMHDDDH |
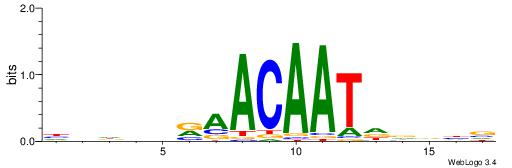 |
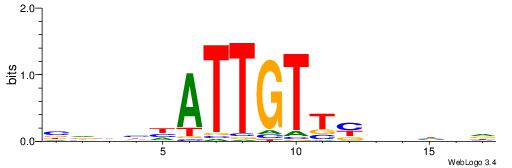 |
| Dataset #: 1 | Motif ID: 86 | Motif name: C086 |
| Original motif | Reverse complement motif |
| Consensus sequence: CSGGAGKCWGMGGSASG | Consensus sequence: CSTSCCYCWGRCTCCSG |
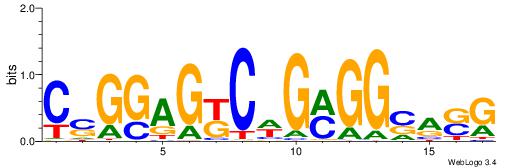 |
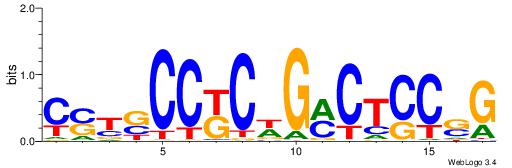 |
Best Matches for Motif ID 86 (Highest to Lowest)
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 17 |
| Similarity score: | 0.0432994 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
----CSTSCCYCWGRCTCCSG--
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 17 |
| Similarity score: | 0.0456174 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
-CSGGAGKCWGMGGSASG----
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 17 |
| Similarity score: | 0.0460804 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
----CSTSCCYCWGRCTCCSG--
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 0.052654 |
Alignment:
VDWDARRGACGCGGWHGGAKHV
----CSGGAGKCWGMGGSASG-
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 0.0530373 |
Alignment:
BDBBVBVGTAWCCAAGVBHBAGB
----CSGGAGKCWGMGGSASG--
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Dataset #: 1 | Motif ID: 87 | Motif name: C087 |
| Original motif | Reverse complement motif |
| Consensus sequence: WRGACCRSSYTGGCYW | Consensus sequence: WKGCCAMSSKGGTCKW |
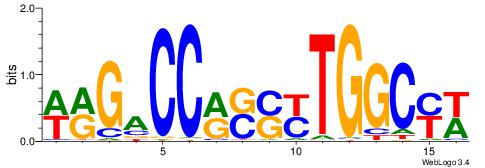 |
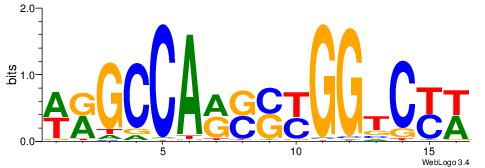 |
Best Matches for Motif ID 87 (Highest to Lowest)
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 16 |
| Similarity score: | 0.0240508 |
Alignment:
DVTTVVCVYGGHGGYCHHDMYB
--WKGCCAMSSKGGTCKW----
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 16 |
| Similarity score: | 0.0273083 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
-----WKGCCAMSSKGGTCKW--
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0299939 |
Alignment:
HVGVSAGGAGGGTCYHHHHYVH
------WRGACCRSSYTGGCYW
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 16 |
| Similarity score: | 0.0316295 |
Alignment:
BBBDVVRGACCACCCAVGABBAB
-----WRGACCRSSYTGGCYW--
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 16 |
| Similarity score: | 0.031888 |
Alignment:
HGDHVHKGACCACCCGGBGBGDB
-----WRGACCRSSYTGGCYW--
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Dataset #: 1 | Motif ID: 88 | Motif name: C088 |
| Original motif | Reverse complement motif |
| Consensus sequence: SKCACTCWGTAGAMS | Consensus sequence: SYTCTACWGAGTGYS |
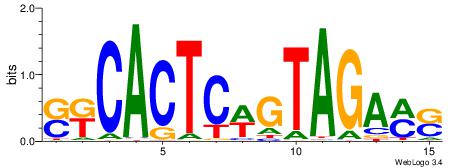 |
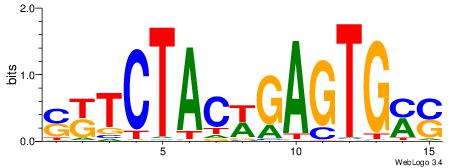 |
Best Matches for Motif ID 88 (Highest to Lowest)
| Motif ID: | UP00201 |
| Motif name: | Emx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.028571 |
Alignment:
WVVVCTAATTAGYBGHB
-SYTCTACWGAGTGYS-
| Original motif | Reverse complement motif |
| Consensus sequence: WVVVCTAATTAGYBGHB | Consensus sequence: BHCBMCTAATTAGBVVW |
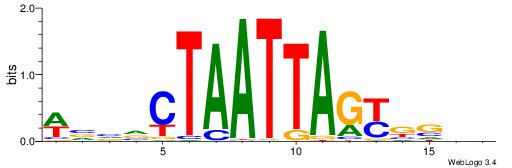 |
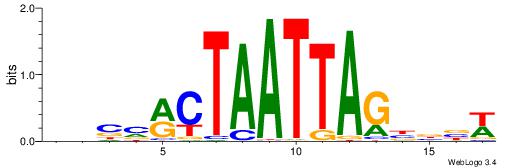 |
| Motif ID: | UP00123 |
| Motif name: | Hlxb9 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0306813 |
Alignment:
BBASTAATTAGTGVHV
SYTCTACWGAGTGYS-
| Original motif | Reverse complement motif |
| Consensus sequence: BBASTAATTAGTGVHV | Consensus sequence: VDVCACTAATTASTVB |
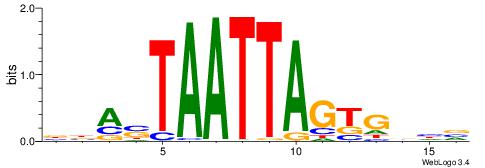 |
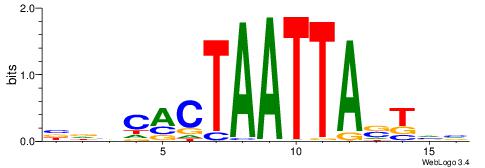 |
| Motif ID: | UP00162 |
| Motif name: | Evx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0319373 |
Alignment:
WVHVCTAATTAGYBVDV
-SYTCTACWGAGTGYS-
| Original motif | Reverse complement motif |
| Consensus sequence: WVHVCTAATTAGYBVDV | Consensus sequence: VDVBMCTAATTAGBHVW |
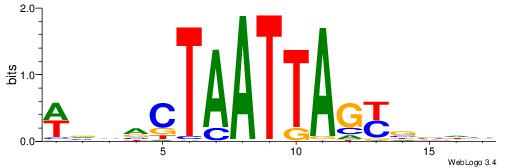 |
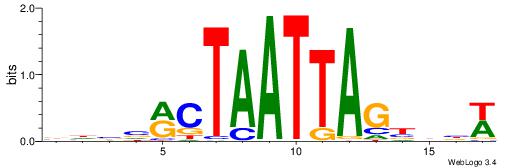 |
| Motif ID: | UP00132 |
| Motif name: | Evx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0320303 |
Alignment:
WRBGCTAATTAGCRGBB
-SYTCTACWGAGTGYS-
| Original motif | Reverse complement motif |
| Consensus sequence: BVCMGCTAATTAGCBKW | Consensus sequence: WRBGCTAATTAGCRGBB |
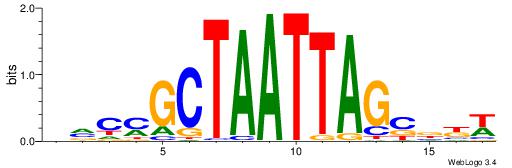 |
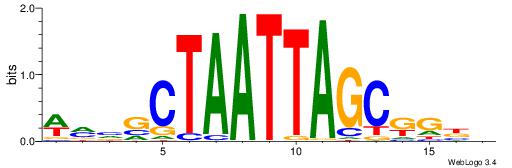 |
| Motif ID: | UP00116 |
| Motif name: | Rhox6 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0322217 |
Alignment:
WVHVYTAATTARTBSDH
-SYTCTACWGAGTGYS-
| Original motif | Reverse complement motif |
| Consensus sequence: WVHVYTAATTARTBSDH | Consensus sequence: DDSBAKTAATTAMVDVW |
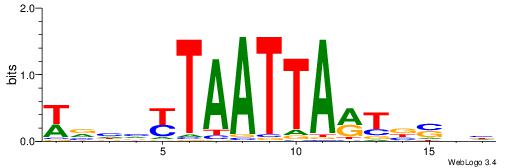 |
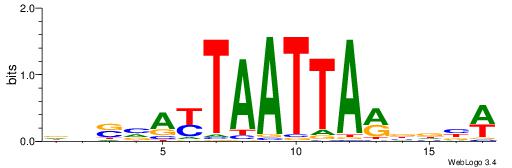 |
| Dataset #: 1 | Motif ID: 89 | Motif name: C089 |
| Original motif | Reverse complement motif |
| Consensus sequence: YKCACTMYRDAGRCMR | Consensus sequence: MRGKCTDMKYAGTGRK |
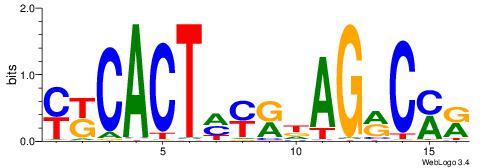 |
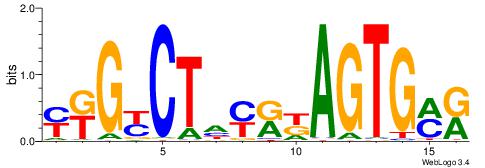 |
Best Matches for Motif ID 89 (Highest to Lowest)
| Motif ID: | UP00132 |
| Motif name: | Evx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0412748 |
Alignment:
WRBGCTAATTAGCRGBB
MRGKCTDMKYAGTGRK-
| Original motif | Reverse complement motif |
| Consensus sequence: BVCMGCTAATTAGCBKW | Consensus sequence: WRBGCTAATTAGCRGBB |
 |
 |
| Motif ID: | UP00131 |
| Motif name: | Gbx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0424502 |
Alignment:
DVDVKCTAATTAGMVVD
-MRGKCTDMKYAGTGRK
| Original motif | Reverse complement motif |
| Consensus sequence: DVVRCTAATTAGYVDBD | Consensus sequence: DVDVKCTAATTAGMVVD |
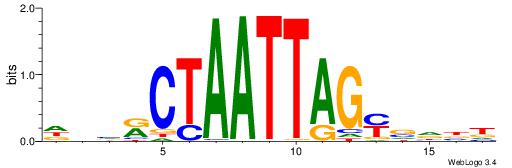 |
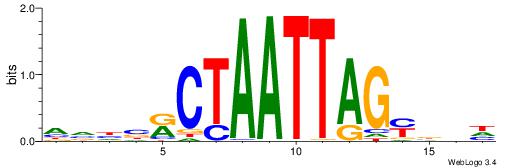 |
| Motif ID: | UP00201 |
| Motif name: | Emx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0439615 |
Alignment:
WVVVCTAATTAGYBGHB
MRGKCTDMKYAGTGRK-
| Original motif | Reverse complement motif |
| Consensus sequence: WVVVCTAATTAGYBGHB | Consensus sequence: BHCBMCTAATTAGBVVW |
 |
 |
| Motif ID: | UP00184 |
| Motif name: | Lhx8 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0454659 |
Alignment:
CAHHGCTAATTAGVVVH
MRGKCTDMKYAGTGRK-
| Original motif | Reverse complement motif |
| Consensus sequence: HVVVCTAATTAGCDDTG | Consensus sequence: CAHHGCTAATTAGVVVH |
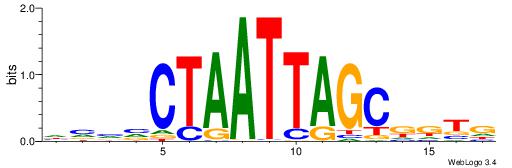 |
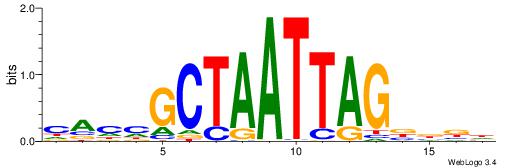 |
| Motif ID: | UP00084 |
| Motif name: | Gmeb1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0461544 |
Alignment:
DGVBCRACGTYGTYHH
YKCACTMYRDAGRCMR
| Original motif | Reverse complement motif |
| Consensus sequence: DGVBCRACGTYGTYHH | Consensus sequence: HHMACKACGTMGBVCD |
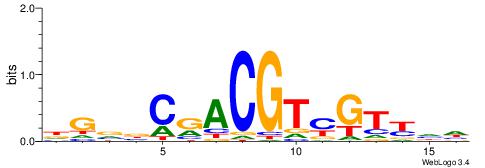 |
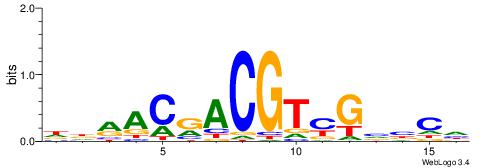 |
| Dataset #: 1 | Motif ID: 90 | Motif name: C090 |
| Original motif | Reverse complement motif |
| Consensus sequence: WAAWWAAADWWWAW | Consensus sequence: WTWWWDTTTWWTTW |
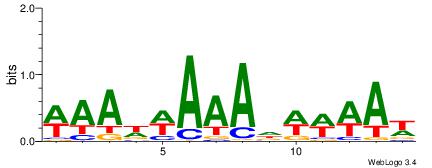 |
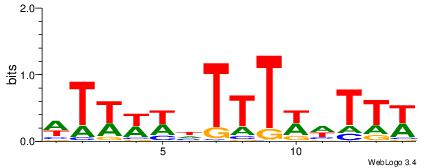 |
Best Matches for Motif ID 90 (Highest to Lowest)
| Motif ID: | UP00090 |
| Motif name: | Elf3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0393475 |
Alignment:
DDHBTTTTTTTTHDAVH
WTWWWDTTTWWTTW---
| Original motif | Reverse complement motif |
| Consensus sequence: DBTHHAAAAAAAAVHDH | Consensus sequence: DDHBTTTTTTTTHDAVH |
 |
 |
| Motif ID: | UP00255 |
| Motif name: | Dbx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0396489 |
Alignment:
HARTHAATTAWTAVDHD
WAAWWAAADWWWAW---
| Original motif | Reverse complement motif |
| Consensus sequence: HARTHAATTAWTAVDHD | Consensus sequence: DHDBTAWTAATTHAKTH |
 |
 |
| Motif ID: | UP00077 |
| Motif name: | Srf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0396638 |
Alignment:
HHVBYTTTTTTTTDDVV
WTWWWDTTTWWTTW---
| Original motif | Reverse complement motif |
| Consensus sequence: VBDDAAAAAAAAMVBHH | Consensus sequence: HHVBYTTTTTTTTDDVV |
 |
 |
| Motif ID: | UP00097 |
| Motif name: | Mtf1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0432915 |
Alignment:
VDTHTTTHHTATDH
WTWWWDTTTWWTTW
| Original motif | Reverse complement motif |
| Consensus sequence: HDATAHDAAAHADV | Consensus sequence: VDTHTTTHHTATDH |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0464108 |
Alignment:
HDTYTTGTTWTKDDDT
-WTWWWDTTTWWTTW-
| Original motif | Reverse complement motif |
| Consensus sequence: ADDDYAWAACAAMAHH | Consensus sequence: HDTYTTGTTWTKDDDT |
 |
 |
| Dataset #: 1 | Motif ID: 91 | Motif name: C091 |
| Original motif | Reverse complement motif |
| Consensus sequence: KMACTCAMKYTGTAKM | Consensus sequence: YRTACAMRYTGAGTYY |
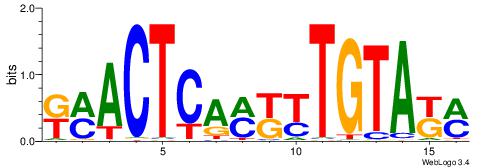 |
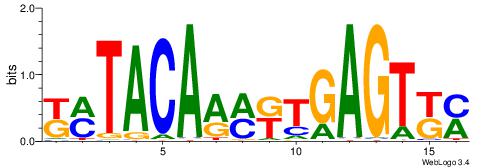 |
Best Matches for Motif ID 91 (Highest to Lowest)
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 16 |
| Similarity score: | 0.0271799 |
Alignment:
YDYBDHTMCACGTGGADDBMDGT
----YRTACAMRYTGAGTYY---
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0333934 |
Alignment:
VMYDHDGMCCHCCKBGVVAAVH
YRTACAMRYTGAGTYY------
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0347276 |
Alignment:
HADTBVADGATGCATCMTGMVHB
KMACTCAMKYTGTAKM-------
| Original motif | Reverse complement motif |
| Consensus sequence: HADTBVADGATGCATCMTGMVHB | Consensus sequence: VHVRCAYGATGCATCDTVVADTH |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 16 |
| Similarity score: | 0.0370054 |
Alignment:
VVYDDDCHCAVACAATTDBVAC
--YRTACAMRYTGAGTYY----
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00165 |
| Motif name: | Titf1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0386898 |
Alignment:
BVABCCACTTGAMHTT
KMACTCAMKYTGTAKM
| Original motif | Reverse complement motif |
| Consensus sequence: BVABCCACTTGAMHTT | Consensus sequence: AAHYTCAAGTGGBTBV |
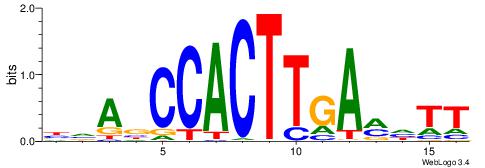 |
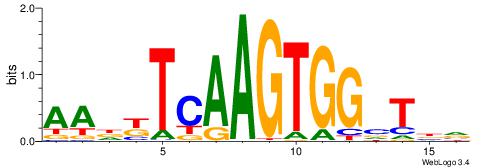 |
| Dataset #: 1 | Motif ID: 92 | Motif name: C092 |
| Original motif | Reverse complement motif |
| Consensus sequence: RYCAGRSTGGCSYCGARY | Consensus sequence: KKTCGMSGCCASKCTGMM |
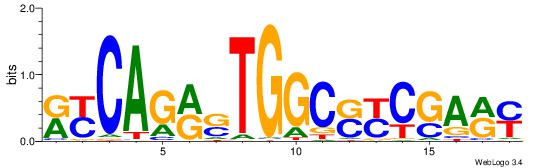 |
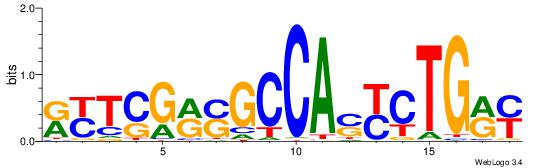 |
Best Matches for Motif ID 92 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0336539 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
-----RYCAGRSTGGCSYCGARY
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0406327 |
Alignment:
CBDMCMGGGTGGTCCHVBVBAH
--RYCAGRSTGGCSYCGARY--
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.0436535 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
----RYCAGRSTGGCSYCGARY
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.0459987 |
Alignment:
BHCBCBCCGGGTGGTCYHVHDCH
----RYCAGRSTGGCSYCGARY-
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.0476871 |
Alignment:
BTBVTCVTGGGTGGTCMVVDVBB
----RYCAGRSTGGCSYCGARY-
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Dataset #: 1 | Motif ID: 93 | Motif name: C093 |
| Original motif | Reverse complement motif |
| Consensus sequence: AKCCASMBSCCKSTGCMT | Consensus sequence: ARGCASYGGSVYSTGGYT |
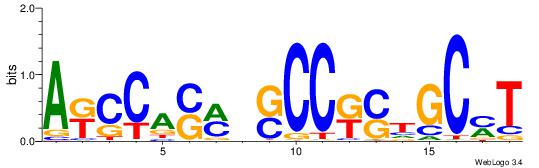 |
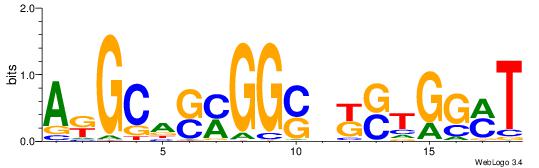 |
Best Matches for Motif ID 93 (Highest to Lowest)
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0389257 |
Alignment:
BADHBDHCGCCCMCGCAHHDBBV
AKCCASMBSCCKSTGCMT-----
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0409936 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
---AKCCASMBSCCKSTGCMT--
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0442874 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
--AKCCASMBSCCKSTGCMT--
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0446823 |
Alignment:
DDRMHBACGCGYGCGTDGBBDB
ARGCASYGGSVYSTGGYT----
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.0463423 |
Alignment:
WHVBVVMACGCGCGTCDYHWDH
---ARGCASYGGSVYSTGGYT-
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
 |
 |
| Dataset #: 1 | Motif ID: 94 | Motif name: C094 |
| Original motif | Reverse complement motif |
| Consensus sequence: SSHCCCSGCCCCSG | Consensus sequence: CSGGGGCSGGGDSS |
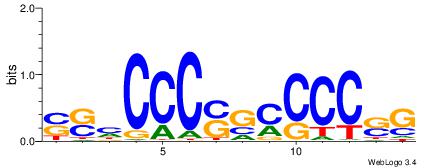 |
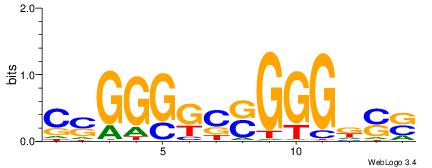 |
Best Matches for Motif ID 94 (Highest to Lowest)
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0170835 |
Alignment:
BHHBHCCGCCCCDHHHH
SSHCCCSGCCCCSG---
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0203409 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
---SSHCCCSGCCCCSG-----
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0229345 |
Alignment:
HVDVGGGHGGGGDBHB
-CSGGGGCSGGGDSS-
| Original motif | Reverse complement motif |
| Consensus sequence: BHVDCCCCDCCCBDBH | Consensus sequence: HVDVGGGHGGGGDBHB |
 |
 |
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0239192 |
Alignment:
DHHDGGGCGRGGKHBH
-CSGGGGCSGGGDSS-
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0245603 |
Alignment:
HVBCCCCCCCCMHHHB
-SSHCCCSGCCCCSG-
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Dataset #: 1 | Motif ID: 95 | Motif name: C095 |
| Original motif | Reverse complement motif |
| Consensus sequence: KCCAKMSTGGTSKMCAGM | Consensus sequence: RCTGYYSACCASYYTGGY |
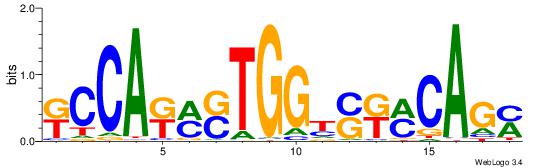 |
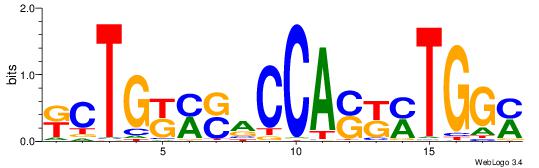 |
Best Matches for Motif ID 95 (Highest to Lowest)
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0278633 |
Alignment:
VMYDHDGMCCHCCKBGVVAAVH
RCTGYYSACCASYYTGGY----
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0281669 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
RCTGYYSACCASYYTGGY----
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.033095 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
----RCTGYYSACCASYYTGGY-
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 18 |
| Similarity score: | 0.036897 |
Alignment:
HHBSHTCGTGRGTGGKGDBVWMV
-----KCCAKMSTGGTSKMCAGM
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.0379359 |
Alignment:
HGDHVHKGACCACCCGGBGBGDB
-RCTGYYSACCASYYTGGY----
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Dataset #: 1 | Motif ID: 96 | Motif name: C096 |
| Original motif | Reverse complement motif |
| Consensus sequence: MRMCMYGSCYRGCCYK | Consensus sequence: RKGGCMMGSCKRGRKY |
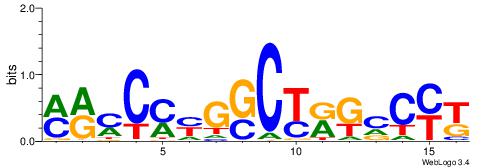 |
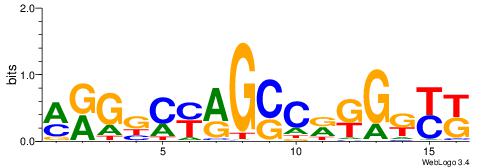 |
Best Matches for Motif ID 96 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 16 |
| Similarity score: | 0.0346607 |
Alignment:
HGDHVHKGACCACCCGGBGBGDB
-----RKGGCMMGSCKRGRKY--
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0356309 |
Alignment:
BHHBHCCGCCCCDHHHH
-MRMCMYGSCYRGCCYK
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 16 |
| Similarity score: | 0.0379898 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
--MRMCMYGSCYRGCCYK----
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0407626 |
Alignment:
DHHBCCCCGCCAHHBHB
-MRMCMYGSCYRGCCYK
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0416066 |
Alignment:
DDDTWCCCCCCGGDRVH
-RKGGCMMGSCKRGRKY
| Original motif | Reverse complement motif |
| Consensus sequence: DDDTWCCCCCCGGDRVH | Consensus sequence: HVKDCCGGGGGGWADDD |
 |
 |
| Dataset #: 1 | Motif ID: 97 | Motif name: C097 |
| Original motif | Reverse complement motif |
| Consensus sequence: WBAASWWAAAAWWSAAVW | Consensus sequence: WVTTSWWTTTTWWSTTBW |
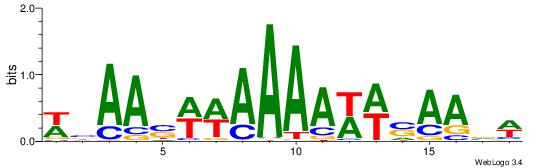 |
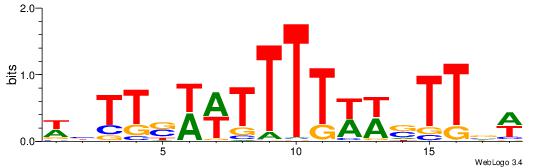 |
Best Matches for Motif ID 97 (Highest to Lowest)
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.032986 |
Alignment:
WBHHHHHMDATTGTTHDHDVHH
---WVTTSWWTTTTWWSTTBW-
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.0395253 |
Alignment:
HBHDVHDTCTTATCTHTDDHDV
WVTTSWWTTTTWWSTTBW----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0483356 |
Alignment:
HHHBBMGATAVHATCWVVDHVH
----WBAASWWAAAAWWSAAVW
| Original motif | Reverse complement motif |
| Consensus sequence: HBHDVBWGATHBTATCRVBHHH | Consensus sequence: HHHBBMGATAVHATCWVVDHVH |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.0488925 |
Alignment:
VVYDDDCHCAVACAATTDBVAC
----WBAASWWAAAAWWSAAVW
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.0549213 |
Alignment:
MWSMBAARRATGCDCABHATGD
-WBAASWWAAAAWWSAAVW---
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
 |
 |
| Dataset #: 1 | Motif ID: 98 | Motif name: C098 |
| Original motif | Reverse complement motif |
| Consensus sequence: AAAAWWAMAAWWAAWH | Consensus sequence: HWTTWWTTYTWWTTTT |
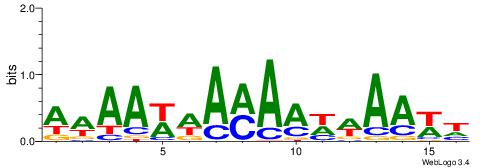 |
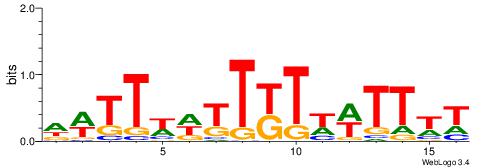 |
Best Matches for Motif ID 98 (Highest to Lowest)
| Motif ID: | UP00077 |
| Motif name: | Srf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0386561 |
Alignment:
VBDDAAAAAAAAMVBHH
-AAAAWWAMAAWWAAWH
| Original motif | Reverse complement motif |
| Consensus sequence: VBDDAAAAAAAAMVBHH | Consensus sequence: HHVBYTTTTTTTTDDVV |
 |
 |
| Motif ID: | UP00051 |
| Motif name: | Sox8_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0410043 |
Alignment:
DDDHDAACAATWBATDD
AAAAWWAMAAWWAAWH-
| Original motif | Reverse complement motif |
| Consensus sequence: DDATBWATTGTTHHDDD | Consensus sequence: DDDHDAACAATWBATDD |
 |
 |
| Motif ID: | UP00090 |
| Motif name: | Elf3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0419783 |
Alignment:
DBTHHAAAAAAAAVHDH
AAAAWWAMAAWWAAWH-
| Original motif | Reverse complement motif |
| Consensus sequence: DBTHHAAAAAAAAVHDH | Consensus sequence: DDHBTTTTTTTTHDAVH |
 |
 |
| Motif ID: | UP00255 |
| Motif name: | Dbx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0419915 |
Alignment:
HARTHAATTAWTAVDHD
AAAAWWAMAAWWAAWH-
| Original motif | Reverse complement motif |
| Consensus sequence: HARTHAATTAWTAVDHD | Consensus sequence: DHDBTAWTAATTHAKTH |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 16 |
| Similarity score: | 0.0430251 |
Alignment:
HHBDHDDAACAATDRHHHHHBW
--AAAAWWAMAAWWAAWH----
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Dataset #: 1 | Motif ID: 99 | Motif name: C099 |
| Original motif | Reverse complement motif |
| Consensus sequence: MYCCCMGCACTCKGGARK | Consensus sequence: RKTCCRGAGTGCYGGGKR |
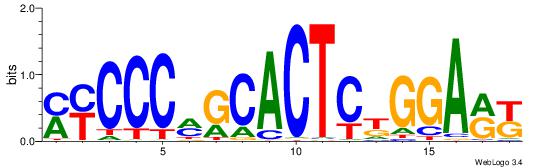 |
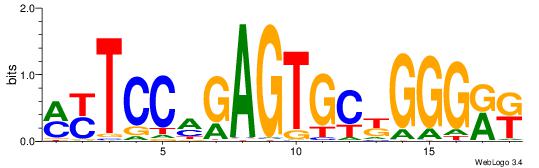 |
Best Matches for Motif ID 99 (Highest to Lowest)
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0318658 |
Alignment:
HHBSHTCGTGRGTGGKGDBVWMV
---RKTCCRGAGTGCYGGGKR--
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0366211 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
----MYCCCMGCACTCKGGARK
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0404912 |
Alignment:
BDVBCDACGCKCGCGTBHRKHD
--MYCCCMGCACTCKGGARK--
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0438598 |
Alignment:
MHHHWHYTMCCHCCCACVAABVH
---MYCCCMGCACTCKGGARK--
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0443498 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
---RKTCCRGAGTGCYGGGKR--
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Dataset #: 1 | Motif ID: 100 | Motif name: C100 |
| Original motif | Reverse complement motif |
| Consensus sequence: WDWCSCTGTSTCHW | Consensus sequence: WHGASACAGSGWDW |
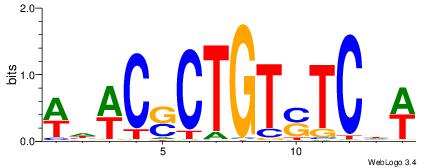 |
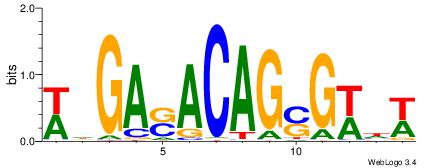 |
Best Matches for Motif ID 100 (Highest to Lowest)
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0211347 |
Alignment:
CHVTCACTGGBADHDD
-WDWCSCTGTSTCHW-
| Original motif | Reverse complement motif |
| Consensus sequence: CHVTCACTGGBADHDD | Consensus sequence: DDHHTBCCAGTGABHG |
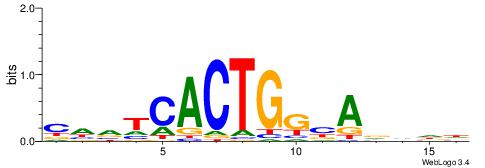 |
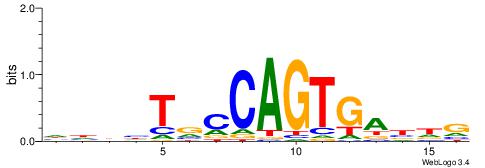 |
| Motif ID: | UP00101 |
| Motif name: | Sox12_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0255389 |
Alignment:
HDTMCTTTGTSTVDBB
-WDWCSCTGTSTCHW-
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBASACAAAGRADH | Consensus sequence: HDTMCTTTGTSTVDBB |
 |
 |
| Motif ID: | UP00210 |
| Motif name: | Mrg2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0263585 |
Alignment:
DBHTTGACAGSTVDTD
-WHGASACAGSGWDW-
| Original motif | Reverse complement motif |
| Consensus sequence: DADBASCTGTCAAHVH | Consensus sequence: DBHTTGACAGSTVDTD |
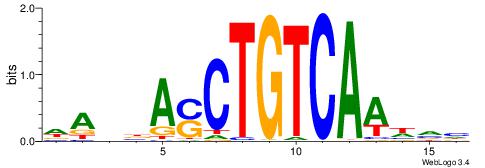 |
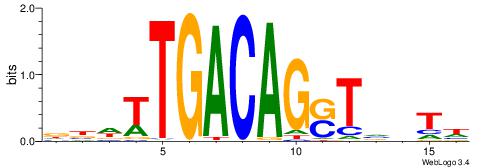 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 10 |
| Number of overlap: | 14 |
| Similarity score: | 0.0316143 |
Alignment:
VCTBHBVCTTGGWTACVVVVVDB
---------WHGASACAGSGWDW
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Motif ID: | UP00186 |
| Motif name: | Meis1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0316571 |
Alignment:
DBWTTGACAGSTBVTW
-WHGASACAGSGWDW-
| Original motif | Reverse complement motif |
| Consensus sequence: WAVBASCTGTCAAWVH | Consensus sequence: DBWTTGACAGSTBVTW |
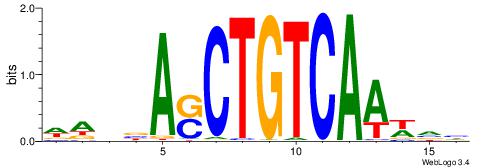 |
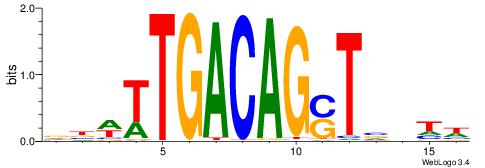 |
| Dataset #: 1 | Motif ID: 101 | Motif name: C101 |
| Original motif | Reverse complement motif |
| Consensus sequence: YTSCCTSKSVSTCCCAR | Consensus sequence: MTGGGASVSYSAGGSAK |
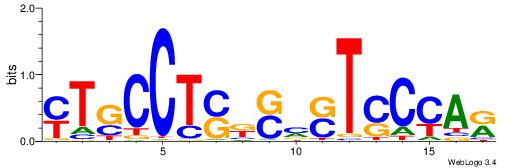 |
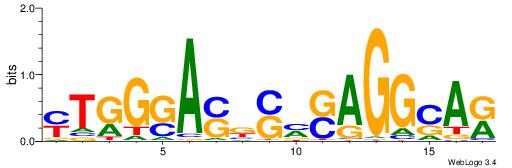 |
Best Matches for Motif ID 101 (Highest to Lowest)
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 0.0379768 |
Alignment:
VDWDARRGACGCGGWHGGAKHV
---MTGGGASVSYSAGGSAK--
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 17 |
| Similarity score: | 0.043481 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
-MTGGGASVSYSAGGSAK-----
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0446789 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
-----YTSCCTSKSVSTCCCAR
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 17 |
| Similarity score: | 0.0447452 |
Alignment:
DVTTVVCVYGGHGGYCHHDMYB
--YTSCCTSKSVSTCCCAR---
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 0.0464938 |
Alignment:
BADHBDHCGCCCMCGCAHHDBBV
-MTGGGASVSYSAGGSAK-----
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Dataset #: 1 | Motif ID: 102 | Motif name: C102 |
| Original motif | Reverse complement motif |
| Consensus sequence: SCMCCSSSVCCMSS | Consensus sequence: SSRGGVSSSGGYGS |
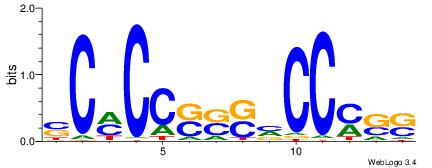 |
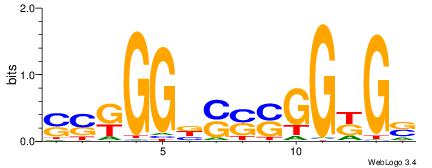 |
Best Matches for Motif ID 102 (Highest to Lowest)
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0109986 |
Alignment:
DBCCCCCCCCCCMYC
-SCMCCSSSVCCMSS
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 14 |
| Similarity score: | 0.0204348 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
--SCMCCSSSVCCMSS------
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0214073 |
Alignment:
VMYDHDGMCCHCCKBGVVAAVH
--------SCMCCSSSVCCMSS
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0236835 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
--------SCMCCSSSVCCMSS-
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.024744 |
Alignment:
HVBCCCCCCCCMHHHB
SCMCCSSSVCCMSS--
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Dataset #: 1 | Motif ID: 103 | Motif name: C103 |
| Original motif | Reverse complement motif |
| Consensus sequence: RRAGRAACCYTGYY | Consensus sequence: MMCAMGGTTKCTKM |
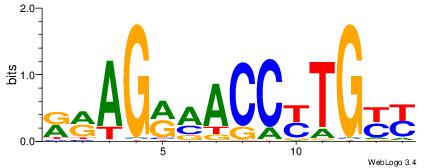 |
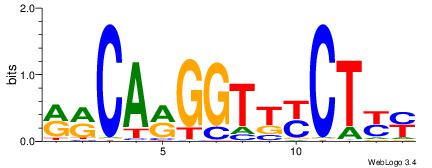 |
Best Matches for Motif ID 103 (Highest to Lowest)
| Motif ID: | UP00098 |
| Motif name: | Rfx3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 14 |
| Similarity score: | 0.0387119 |
Alignment:
HHHHDVVGTTGCTAHGGDHBAHH
-MMCAMGGTTKCTKM--------
| Original motif | Reverse complement motif |
| Consensus sequence: HDTBHHCCHTAGCAACVVDHHHH | Consensus sequence: HHHHDVVGTTGCTAHGGDHBAHH |
 |
 |
| Motif ID: | UP00036 |
| Motif name: | Myf6_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0405304 |
Alignment:
VVDBAACAGBTGBYHB
RRAGRAACCYTGYY--
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBAACAGBTGBYHB | Consensus sequence: BDKVCABCTGTTBDBV |
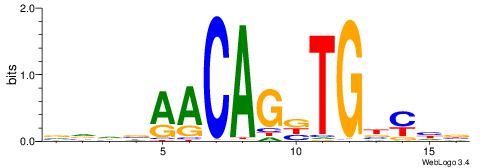 |
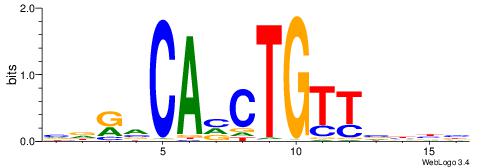 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 14 |
| Similarity score: | 0.0407907 |
Alignment:
BDBBVBVGTAWCCAAGVBHBAGB
----RRAGRAACCYTGYY-----
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Motif ID: | UP00056 |
| Motif name: | Rfx4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0414662 |
Alignment:
HVVGTAWCKADGBHH
-MMCAMGGTTKCTKM
| Original motif | Reverse complement motif |
| Consensus sequence: HHBCHTRGWTACVVH | Consensus sequence: HVVGTAWCKADGBHH |
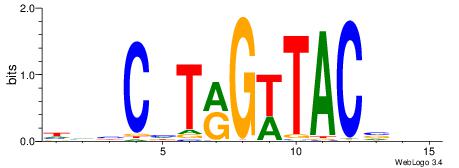 |
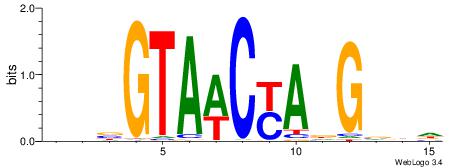 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 14 |
| Similarity score: | 0.0421016 |
Alignment:
DDRMHBACGCGYGCGTDGBBDB
---MMCAMGGTTKCTKM-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
 |
 |
| Dataset #: 1 | Motif ID: 104 | Motif name: C104 |
| Original motif | Reverse complement motif |
| Consensus sequence: SSCCYCCCCDCCSS | Consensus sequence: SSGGHGGGGKGGSS |
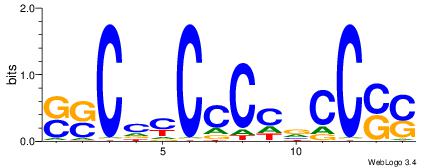 |
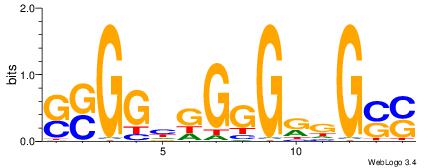 |
Best Matches for Motif ID 104 (Highest to Lowest)
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 14 |
| Similarity score: | 0.0212662 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
----SSCCYCCCCDCCSS----
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0241093 |
Alignment:
BHHDYGGGGGGGGBVD
SSGGHGGGGKGGSS--
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0261421 |
Alignment:
DBCCCCCCCCCCMYC
SSCCYCCCCDCCSS-
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0264558 |
Alignment:
HBBBCCGCCCCWHHHV
--SSCCYCCCCDCCSS
| Original motif | Reverse complement motif |
| Consensus sequence: HBBBCCGCCCCWHHHV | Consensus sequence: BHHHWGGGGCGGBBVH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 14 |
| Similarity score: | 0.0275752 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
-------SSCCYCCCCDCCSS--
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Dataset #: 1 | Motif ID: 105 | Motif name: C105 |
| Original motif | Reverse complement motif |
| Consensus sequence: TGCCKMBSCCKMCCVA | Consensus sequence: TVGGRYGGSBRRGGCA |
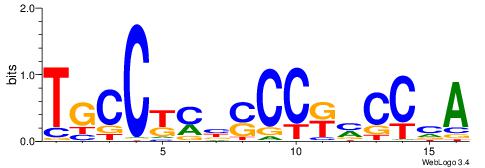 |
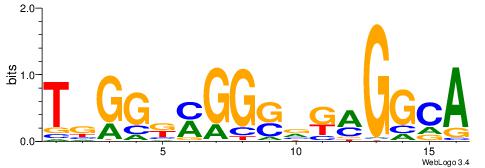 |
Best Matches for Motif ID 105 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0398676 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
-TVGGRYGGSBRRGGCA------
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 16 |
| Similarity score: | 0.0413056 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
--TVGGRYGGSBRRGGCA----
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0445913 |
Alignment:
HHBSHTCGTGRGTGGKGDBVWMV
------TVGGRYGGSBRRGGCA-
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0479431 |
Alignment:
BADHBDHCGCCCMCGCAHHDBBV
-TGCCKMBSCCKMCCVA------
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0484323 |
Alignment:
BHBHDTGGCGGGGBDHD
TVGGRYGGSBRRGGCA-
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
 |
 |
| Dataset #: 1 | Motif ID: 106 | Motif name: C106 |
| Original motif | Reverse complement motif |
| Consensus sequence: MSGCAGWGGCASK | Consensus sequence: RSTGCCWCTGCSY |
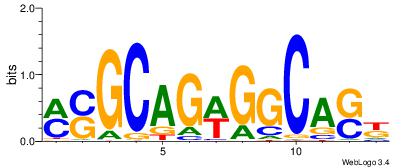 |
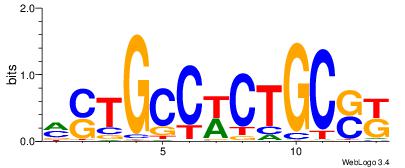 |
Best Matches for Motif ID 106 (Highest to Lowest)
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0221156 |
Alignment:
BBBAVTGCAGTGBBVDD
RSTGCCWCTGCSY----
| Original motif | Reverse complement motif |
| Consensus sequence: DDBBBCACTGCABTBBB | Consensus sequence: BBBAVTGCAGTGBBVDD |
 |
 |
| Motif ID: | UP00010 |
| Motif name: | Tcfap2b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 13 |
| Similarity score: | 0.026668 |
Alignment:
HHKGCCYSRGGCWAD
RSTGCCWCTGCSY--
| Original motif | Reverse complement motif |
| Consensus sequence: DTWGCCKSMGGCRHH | Consensus sequence: HHKGCCYSRGGCWAD |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0273606 |
Alignment:
VDWDARRGACGCGGWHGGAKHV
--------MSGCAGWGGCASK-
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 13 |
| Similarity score: | 0.0282689 |
Alignment:
DDHHTBCCAGTGABHG
--RSTGCCWCTGCSY-
| Original motif | Reverse complement motif |
| Consensus sequence: CHVTCACTGGBADHDD | Consensus sequence: DDHHTBCCAGTGABHG |
 |
 |
| Motif ID: | UP00036 |
| Motif name: | Myf6_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 13 |
| Similarity score: | 0.0289018 |
Alignment:
GGDGBGVCTGTKVVB
RSTGCCWCTGCSY--
| Original motif | Reverse complement motif |
| Consensus sequence: VVVRACAGVCBCDCC | Consensus sequence: GGDGBGVCTGTKVVB |
 |
 |
| Dataset #: 1 | Motif ID: 107 | Motif name: C107 |
| Original motif | Reverse complement motif |
| Consensus sequence: CHBBCCTYBAABCCCDG | Consensus sequence: CDGGGBTTVMAGGVBHG |
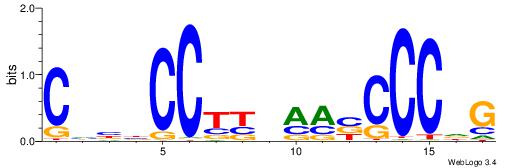 |
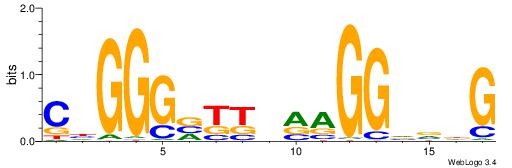 |
Best Matches for Motif ID 107 (Highest to Lowest)
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 17 |
| Similarity score: | 0.0580885 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
-CDGGGBTTVMAGGVBHG----
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0584165 |
Alignment:
MHHHWHYTMCCHCCCACVAABVH
CHBBCCTYBAABCCCDG------
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 17 |
| Similarity score: | 0.0604761 |
Alignment:
VCTBHBVCTTGGWTACVVVVVDB
--CHBBCCTYBAABCCCDG----
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 0.0618639 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
----CHBBCCTYBAABCCCDG--
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 17 |
| Similarity score: | 0.0669075 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
----CHBBCCTYBAABCCCDG--
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Dataset #: 1 | Motif ID: 108 | Motif name: C108 |
| Original motif | Reverse complement motif |
| Consensus sequence: RBACWGASAWASVY | Consensus sequence: MVSTWTSTCWGTBK |
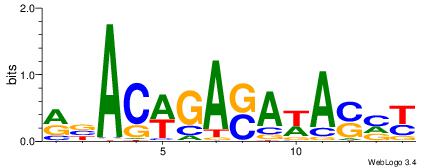 |
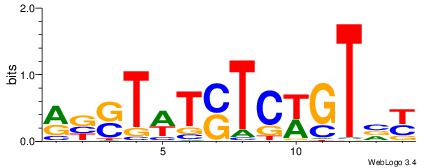 |
Best Matches for Motif ID 108 (Highest to Lowest)
| Motif ID: | UP00101 |
| Motif name: | Sox12_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0263171 |
Alignment:
HDTMCTTTGTSTVDBB
--MVSTWTSTCWGTBK
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBASACAAAGRADH | Consensus sequence: HDTMCTTTGTSTVDBB |
 |
 |
| Motif ID: | UP00018 |
| Motif name: | Irf4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0315601 |
Alignment:
VVBAYTCTCGGWHDB
MVSTWTSTCWGTBK-
| Original motif | Reverse complement motif |
| Consensus sequence: VVBAYTCTCGGWHDB | Consensus sequence: BHHWCCGAGAMTVVB |
 |
 |
| Motif ID: | >UP00011 |
| Motif name: | Irf6_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0320561 |
Alignment:
VDDRCCGMGAGTBBB
-RBACWGASAWASVY
| Original motif | Reverse complement motif |
| Consensus sequence: VBBACTCYCGGKHDV | Consensus sequence: VDDRCCGMGAGTBBB |
 |
 |
| Motif ID: | UP00073 |
| Motif name: | Foxa2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0348659 |
Alignment:
VVDAATAACAAAVVB
-RBACWGASAWASVY
| Original motif | Reverse complement motif |
| Consensus sequence: VVDAATAACAAAVVB | Consensus sequence: BVVTTTGTTATTDBB |
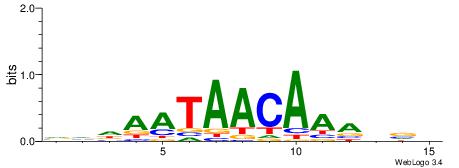 |
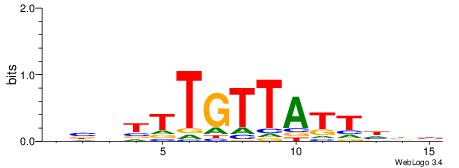 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0352792 |
Alignment:
HVHBVTGTCTGGDDHDD
MVSTWTSTCWGTBK---
| Original motif | Reverse complement motif |
| Consensus sequence: HDHDDCCAGACABBHVH | Consensus sequence: HVHBVTGTCTGGDDHDD |
 |
 |
| Dataset #: 1 | Motif ID: 109 | Motif name: C109 |
| Original motif | Reverse complement motif |
| Consensus sequence: RMCWSGCTGSCCKY | Consensus sequence: MRGGSCAGCSWGRK |
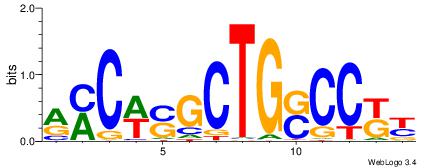 |
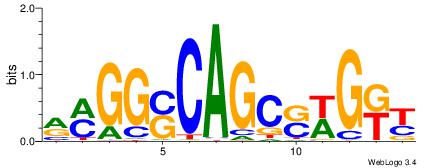 |
Best Matches for Motif ID 109 (Highest to Lowest)
| Motif ID: | UP00006 |
| Motif name: | Zic3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0232278 |
Alignment:
DDDCACAGCAKGHVD
MRGGSCAGCSWGRK-
| Original motif | Reverse complement motif |
| Consensus sequence: DDDCACAGCAKGHVD | Consensus sequence: DVHCYTGCTGTGHDH |
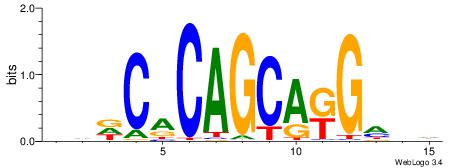 |
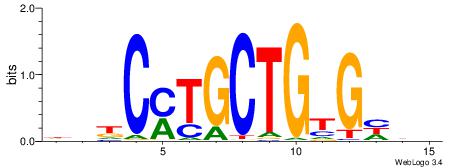 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 14 |
| Similarity score: | 0.0232672 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
----MRGGSCAGCSWGRK-----
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0233983 |
Alignment:
VBBDMYCATCTGVHHBH
---RMCWSGCTGSCCKY
| Original motif | Reverse complement motif |
| Consensus sequence: HVDDVCAGATGKYHBBV | Consensus sequence: VBBDMYCATCTGVHHBH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0243539 |
Alignment:
HTBVVVDGGACCACCCRGRDBG
------MRGGSCAGCSWGRK--
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Motif ID: | UP00057 |
| Motif name: | Zic2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.024667 |
Alignment:
HHDCVCAGCAKGVVD
MRGGSCAGCSWGRK-
| Original motif | Reverse complement motif |
| Consensus sequence: HHDCVCAGCAKGVVD | Consensus sequence: DVBCYTGCTGBGDDD |
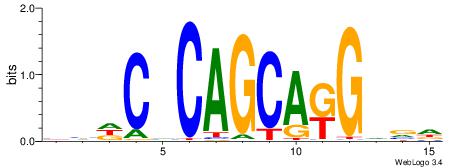 |
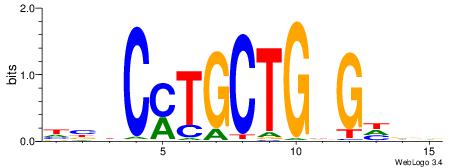 |
| Dataset #: 1 | Motif ID: 110 | Motif name: C110 |
| Original motif | Reverse complement motif |
| Consensus sequence: YYCTKTCTCMAARR | Consensus sequence: KKTTRGAGAYAGKM |
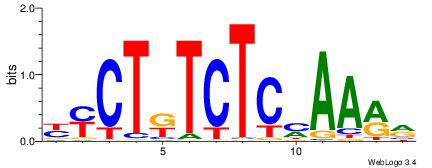 |
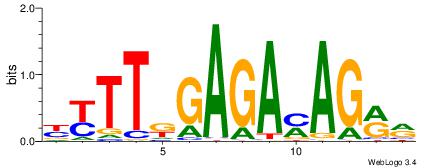 |
Best Matches for Motif ID 110 (Highest to Lowest)
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.030626 |
Alignment:
BDHDDADAGATAAGADHBDHVD
-KKTTRGAGAYAGKM-------
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0328671 |
Alignment:
VHHATTCCTYGAAAGD
-YYCTKTCTCMAARR-
| Original motif | Reverse complement motif |
| Consensus sequence: DCTTTCKAGGAATHHV | Consensus sequence: VHHATTCCTYGAAAGD |
 |
 |
| Motif ID: | UP00076 |
| Motif name: | Rfxdc2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0362358 |
Alignment:
BBVCDTRGAKACSVDDH
--KKTTRGAGAYAGKM-
| Original motif | Reverse complement motif |
| Consensus sequence: BBVCDTRGAKACSVDDH | Consensus sequence: HDDVSGTRTCMADGBVB |
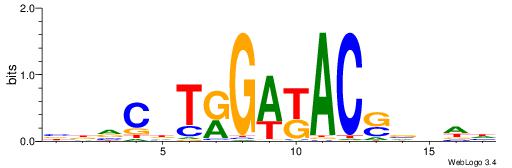 |
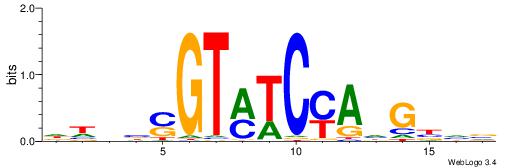 |
| Motif ID: | UP00101 |
| Motif name: | Sox12_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0396775 |
Alignment:
HDTMCTTTGTSTVDBB
--YYCTKTCTCMAARR
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBASACAAAGRADH | Consensus sequence: HDTMCTTTGTSTVDBB |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0396815 |
Alignment:
HBHDVBWGATHBTATCRVBHHH
KKTTRGAGAYAGKM--------
| Original motif | Reverse complement motif |
| Consensus sequence: HBHDVBWGATHBTATCRVBHHH | Consensus sequence: HHHBBMGATAVHATCWVVDHVH |
 |
 |
| Dataset #: 1 | Motif ID: 111 | Motif name: C111 |
| Original motif | Reverse complement motif |
| Consensus sequence: TBATWYCTGARWTCSM | Consensus sequence: YSGAWKTCAGMWATBA |
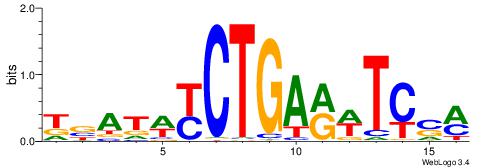 |
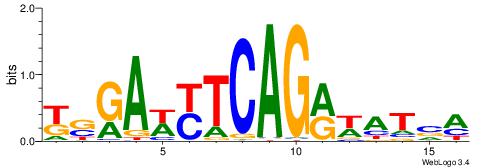 |
Best Matches for Motif ID 111 (Highest to Lowest)
| Motif ID: | UP00045 |
| Motif name: | Mafb_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0382144 |
Alignment:
DMTWDGTCAGCADWTTH
YSGAWKTCAGMWATBA-
| Original motif | Reverse complement motif |
| Consensus sequence: HAAWDTGCTGACDWARH | Consensus sequence: DMTWDGTCAGCADWTTH |
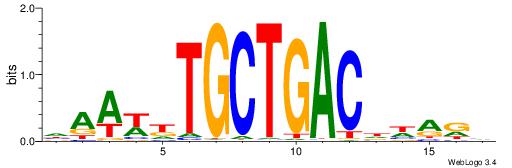 |
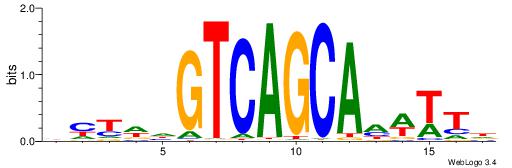 |
| Motif ID: | UP00410 |
| Motif name: | Elk1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0436521 |
Alignment:
VVHBACCGGAAGTDBHD
TBATWYCTGARWTCSM-
| Original motif | Reverse complement motif |
| Consensus sequence: VVHBACCGGAAGTDBHD | Consensus sequence: DHBDACTTCCGGTVHVB |
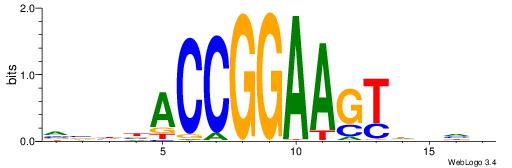 |
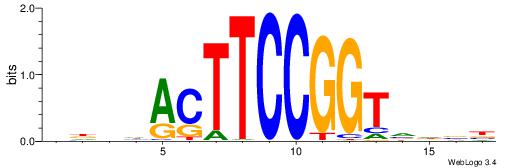 |
| Motif ID: | UP00013 |
| Motif name: | Gabpa_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0439267 |
Alignment:
HVHHACCGGAAGTDHHV
TBATWYCTGARWTCSM-
| Original motif | Reverse complement motif |
| Consensus sequence: HVHHACCGGAAGTDHHV | Consensus sequence: BHHHACTTCCGGTHHBD |
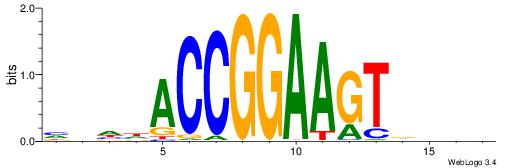 |
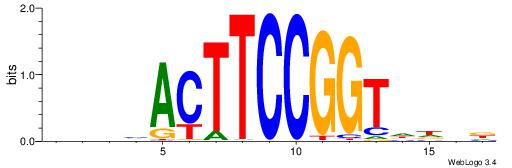 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 16 |
| Similarity score: | 0.0485736 |
Alignment:
BDHDDADAGATAAGADHBDHVD
----YSGAWKTCAGMWATBA--
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Motif ID: | UP00193 |
| Motif name: | Rhox11_1765.2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0486964 |
Alignment:
WVVHWTWACAGCGBHBT
-YSGAWKTCAGMWATBA
| Original motif | Reverse complement motif |
| Consensus sequence: AVDVCGCTGTWAWDVVW | Consensus sequence: WVVHWTWACAGCGBHBT |
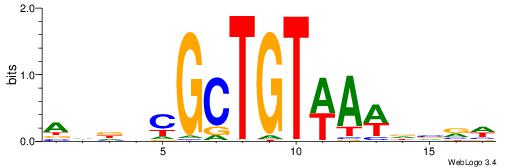 |
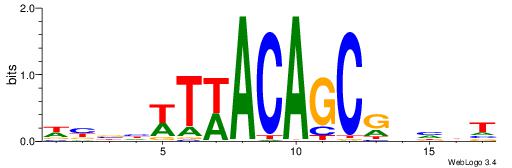 |
| Dataset #: 1 | Motif ID: 112 | Motif name: C112 |
| Original motif | Reverse complement motif |
| Consensus sequence: CCBCYSSSGCCCCSS | Consensus sequence: SSGGGGCSSSKGBGG |
 |
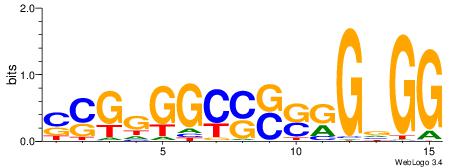 |
Best Matches for Motif ID 112 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 15 |
| Similarity score: | 0.0368115 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
SSGGGGCSSSKGBGG--------
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 15 |
| Similarity score: | 0.038423 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
--CCBCYSSSGCCCCSS-----
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0430481 |
Alignment:
HHHHDGGGGCGGDBHDV
--SSGGGGCSSSKGBGG
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
 |
 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0450061 |
Alignment:
BHVDCCCCDCCCBDBH
CCBCYSSSGCCCCSS-
| Original motif | Reverse complement motif |
| Consensus sequence: BHVDCCCCDCCCBDBH | Consensus sequence: HVDVGGGHGGGGDBHB |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0460082 |
Alignment:
HVBCCCCCCCCMHHHB
CCBCYSSSGCCCCSS-
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Dataset #: 1 | Motif ID: 113 | Motif name: C113 |
| Original motif | Reverse complement motif |
| Consensus sequence: CHCCSCSVSCCCC | Consensus sequence: GGGGSVSGSGGDG |
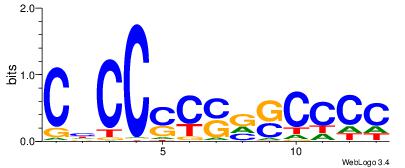 |
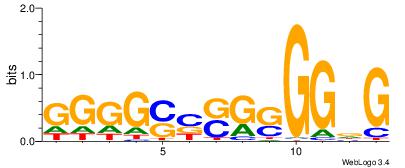 |
Best Matches for Motif ID 113 (Highest to Lowest)
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0317901 |
Alignment:
DBCCCCCCCCCCMYC
-CHCCSCSVSCCCC-
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0384426 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
-GGGGSVSGSGGDG---------
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0386858 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
GGGGSVSGSGGDG---------
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 11 |
| Number of overlap: | 13 |
| Similarity score: | 0.0387928 |
Alignment:
DVVTTVGTGGGHGGYAMHWHHHY
GGGGSVSGSGGDG----------
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0424015 |
Alignment:
HVBCCCCCCCCMHHHB
---CHCCSCSVSCCCC
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Dataset #: 1 | Motif ID: 114 | Motif name: C114 |
| Original motif | Reverse complement motif |
| Consensus sequence: SRARCCMKGTCTYS | Consensus sequence: SMAGACRRGGMTKS |
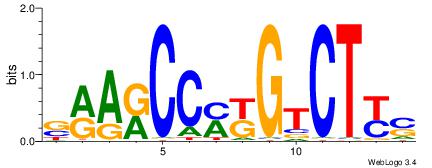 |
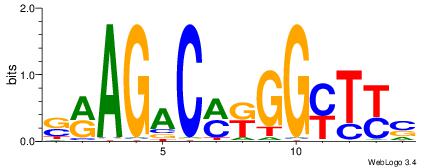 |
Best Matches for Motif ID 114 (Highest to Lowest)
| Motif ID: | UP00082 |
| Motif name: | Zfp187_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0219685 |
Alignment:
DDGMCCTBGTCCHYHV
SRARCCMKGTCTYS--
| Original motif | Reverse complement motif |
| Consensus sequence: DDGMCCTBGTCCHYHV | Consensus sequence: VHMDGGACVAGGRCDH |
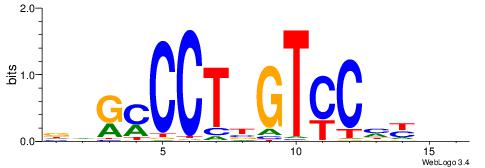 |
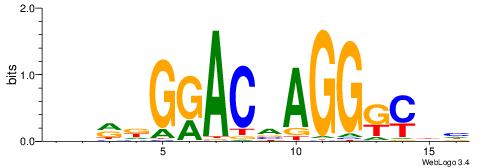 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0320328 |
Alignment:
BDWAGGCGTGBCHHD
-SMAGACRRGGMTKS
| Original motif | Reverse complement motif |
| Consensus sequence: BDWAGGCGTGBCHHD | Consensus sequence: HHDGBCACGCCTWDB |
 |
 |
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0320453 |
Alignment:
HBDRCCMCGCCCHHHD
SRARCCMKGTCTYS--
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0338574 |
Alignment:
BBHBGAGTGGGAHHDB
-SMAGACRRGGMTKS-
| Original motif | Reverse complement motif |
| Consensus sequence: BBHBGAGTGGGAHHDB | Consensus sequence: BHHDTCCCACTCBDBV |
 |
 |
| Motif ID: | UP00009 |
| Motif name: | Nr2f2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0350465 |
Alignment:
VBVBCSGGGTCAVBBH
SRARCCMKGTCTYS--
| Original motif | Reverse complement motif |
| Consensus sequence: VBVBCSGGGTCAVBBH | Consensus sequence: HVBVTGACCCSGBVBV |
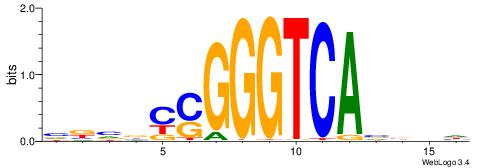 |
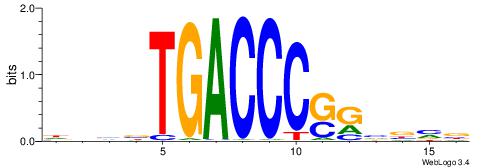 |
| Dataset #: 1 | Motif ID: 115 | Motif name: C115 |
| Original motif | Reverse complement motif |
| Consensus sequence: MDRGKGSTGGGHK | Consensus sequence: RHCCCASCRCKDY |
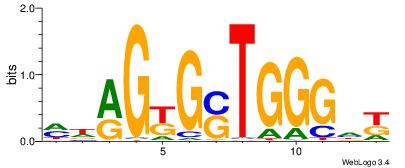 |
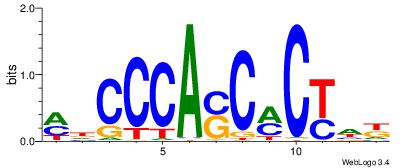 |
Best Matches for Motif ID 115 (Highest to Lowest)
| Motif ID: | UP00099 |
| Motif name: | Ascl2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 13 |
| Similarity score: | 0.0107043 |
Alignment:
HVDVGGGHGGGGDBHB
-MDRGKGSTGGGHK--
| Original motif | Reverse complement motif |
| Consensus sequence: BHVDCCCCDCCCBDBH | Consensus sequence: HVDVGGGHGGGGDBHB |
 |
 |
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0119418 |
Alignment:
BHHHWGGGGCGGBBVH
--MDRGKGSTGGGHK-
| Original motif | Reverse complement motif |
| Consensus sequence: HBBBCCGCCCCWHHHV | Consensus sequence: BHHHWGGGGCGGBBVH |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0142708 |
Alignment:
DHHBCCCCGCCAHHBHB
---RHCCCASCRCKDY-
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 13 |
| Similarity score: | 0.0145702 |
Alignment:
HHHHDGGGGCGGDBHDV
--MDRGKGSTGGGHK--
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0200599 |
Alignment:
BDWAGGCGTGBCHHD
-MDRGKGSTGGGHK-
| Original motif | Reverse complement motif |
| Consensus sequence: BDWAGGCGTGBCHHD | Consensus sequence: HHDGBCACGCCTWDB |
 |
 |
| Dataset #: 1 | Motif ID: 116 | Motif name: C116 |
| Original motif | Reverse complement motif |
| Consensus sequence: WWMAWACACWMAWW | Consensus sequence: WWTRWGTGTWTRWW |
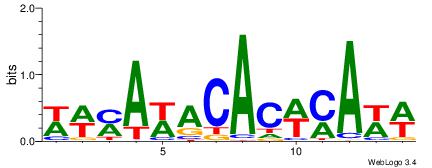 |
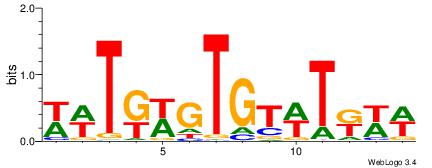 |
Best Matches for Motif ID 116 (Highest to Lowest)
| Motif ID: | UP00055 |
| Motif name: | Hbp1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0242601 |
Alignment:
HHDWTSAATGAATDDB
WWMAWACACWMAWW--
| Original motif | Reverse complement motif |
| Consensus sequence: HHDWTSAATGAATDDB | Consensus sequence: VDHATTCATTSAWDDH |
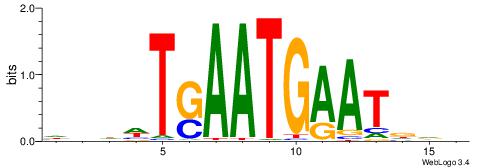 |
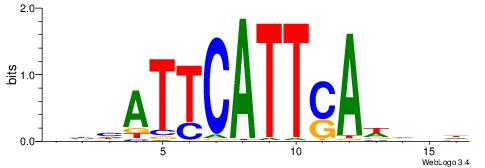 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.02629 |
Alignment:
DWAATRTAAACAAWVVB
WWMAWACACWMAWW---
| Original motif | Reverse complement motif |
| Consensus sequence: DWAATRTAAACAAWVVB | Consensus sequence: VVVWTTGTTTAMATTWD |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 14 |
| Similarity score: | 0.0267723 |
Alignment:
GTVBDAATTGTVTGHGDDHKVB
------WWTRWGTGTWTRWW--
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00228 |
| Motif name: | Bapx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0270117 |
Alignment:
DTBVTTAAGTGGTTHHV
---WWTRWGTGTWTRWW
| Original motif | Reverse complement motif |
| Consensus sequence: VHHAACCACTTAAVVAH | Consensus sequence: DTBVTTAAGTGGTTHHV |
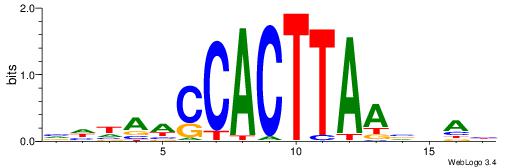 |
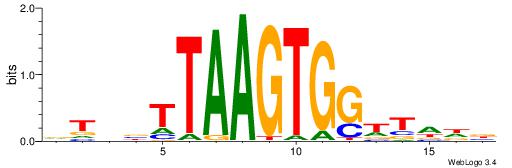 |
| Motif ID: | UP00037 |
| Motif name: | Zfp105_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0271642 |
Alignment:
HDYAAAHAAMAADHD
WWMAWACACWMAWW-
| Original motif | Reverse complement motif |
| Consensus sequence: HDYAAAHAAMAADHD | Consensus sequence: HHHTTRTTDTTTKDH |
 |
 |
| Dataset #: 1 | Motif ID: 117 | Motif name: C117 |
| Original motif | Reverse complement motif |
| Consensus sequence: CMCWRSCTGYCCKG | Consensus sequence: CYGGKCAGSKWGYG |
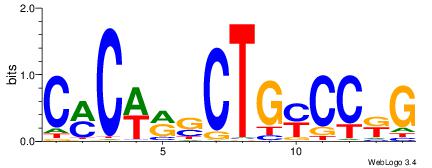 |
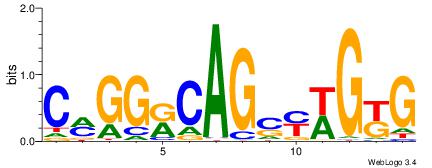 |
Best Matches for Motif ID 117 (Highest to Lowest)
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.023372 |
Alignment:
DDHHTBCCAGTGABHG
--CYGGKCAGSKWGYG
| Original motif | Reverse complement motif |
| Consensus sequence: CHVTCACTGGBADHDD | Consensus sequence: DDHHTBCCAGTGABHG |
 |
 |
| Motif ID: | UP00092 |
| Motif name: | Myb_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0238724 |
Alignment:
DBVKGGCAGTTGGTHB
-CYGGKCAGSKWGYG-
| Original motif | Reverse complement motif |
| Consensus sequence: BDACCAACTGCCRBBH | Consensus sequence: DBVKGGCAGTTGGTHB |
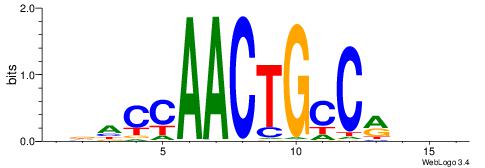 |
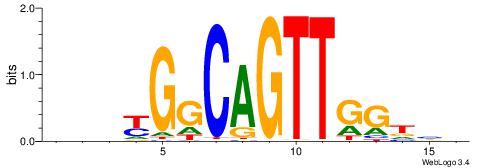 |
| Motif ID: | UP00057 |
| Motif name: | Zic2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0250247 |
Alignment:
HHDCVCAGCAKGVVD
CYGGKCAGSKWGYG-
| Original motif | Reverse complement motif |
| Consensus sequence: HHDCVCAGCAKGVVD | Consensus sequence: DVBCYTGCTGBGDDD |
 |
 |
| Motif ID: | UP00081 |
| Motif name: | Mybl1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0262709 |
Alignment:
BHCGGCAGTTGGBBB
CYGGKCAGSKWGYG-
| Original motif | Reverse complement motif |
| Consensus sequence: BBVCCAACTGCCGHB | Consensus sequence: BHCGGCAGTTGGBBB |
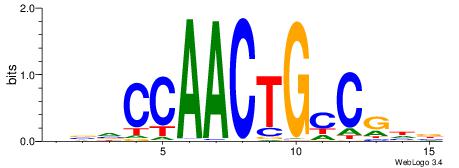 |
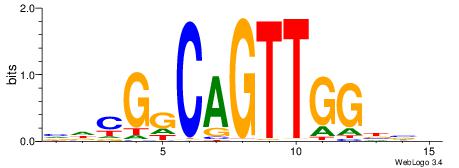 |
| Motif ID: | UP00006 |
| Motif name: | Zic3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0274658 |
Alignment:
DVHCYTGCTGTGHDH
-CMCWRSCTGYCCKG
| Original motif | Reverse complement motif |
| Consensus sequence: DDDCACAGCAKGHVD | Consensus sequence: DVHCYTGCTGTGHDH |
 |
 |
| Dataset #: 1 | Motif ID: 118 | Motif name: C118 |
| Original motif | Reverse complement motif |
| Consensus sequence: GMCTGCSWSTGCCKC | Consensus sequence: GRGGCASWSGCAGYC |
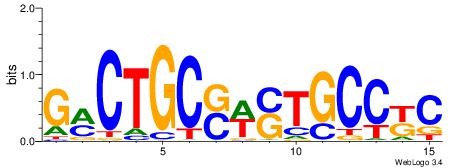 |
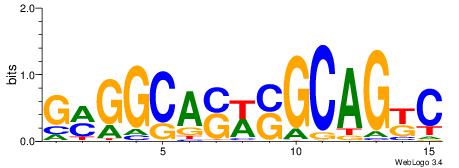 |
Best Matches for Motif ID 118 (Highest to Lowest)
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 15 |
| Similarity score: | 0.0439545 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
---GMCTGCSWSTGCCKC-----
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 15 |
| Similarity score: | 0.0446432 |
Alignment:
VHYTCCDWCCGCGTCMMTHWHV
GMCTGCSWSTGCCKC-------
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0451734 |
Alignment:
CHVTCACTGGBADHDD
-GMCTGCSWSTGCCKC
| Original motif | Reverse complement motif |
| Consensus sequence: CHVTCACTGGBADHDD | Consensus sequence: DDHHTBCCAGTGABHG |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 15 |
| Similarity score: | 0.0458009 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
---GMCTGCSWSTGCCKC-----
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00414 |
| Motif name: | Ets1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 15 |
| Similarity score: | 0.0501418 |
Alignment:
BDDDHACCGGAARTVDD
GRGGCASWSGCAGYC--
| Original motif | Reverse complement motif |
| Consensus sequence: BDDDHACCGGAARTVDD | Consensus sequence: DDBAMTTCCGGTDHDHB |
 |
 |
| Dataset #: 1 | Motif ID: 119 | Motif name: C119 |
| Original motif | Reverse complement motif |
| Consensus sequence: SWRTYCCAGRACWS | Consensus sequence: SWGTMCTGGKAKWS |
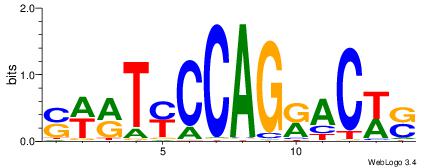 |
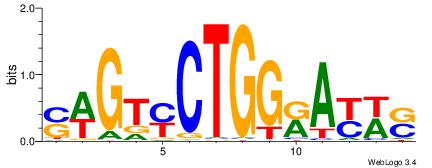 |
Best Matches for Motif ID 119 (Highest to Lowest)
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0288751 |
Alignment:
CHVTCACTGGBADHDD
-SWGTMCTGGKAKWS-
| Original motif | Reverse complement motif |
| Consensus sequence: CHVTCACTGGBADHDD | Consensus sequence: DDHHTBCCAGTGABHG |
 |
 |
| Motif ID: | UP00040 |
| Motif name: | Irf5_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0294274 |
Alignment:
BHDAHCGAGARTHBV
SWRTYCCAGRACWS-
| Original motif | Reverse complement motif |
| Consensus sequence: BHDAHCGAGARTHBV | Consensus sequence: VBHAKTCTCGHTHHV |
 |
 |
| Motif ID: | UP00019 |
| Motif name: | Zbtb12_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.031753 |
Alignment:
BDHRBGTTCTAGATCVB
---SWRTYCCAGRACWS
| Original motif | Reverse complement motif |
| Consensus sequence: BDHRBGTTCTAGATCVB | Consensus sequence: BBGATCTAGAACBKHDB |
 |
 |
| Motif ID: | UP00018 |
| Motif name: | Irf4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0323112 |
Alignment:
BHHWCCGAGAMTVVB
SWRTYCCAGRACWS-
| Original motif | Reverse complement motif |
| Consensus sequence: VVBAYTCTCGGWHDB | Consensus sequence: BHHWCCGAGAMTVVB |
 |
 |
| Motif ID: | UP00203 |
| Motif name: | Pknox1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0328626 |
Alignment:
VVWTTGACAGSTSBKD
-SWRTYCCAGRACWS-
| Original motif | Reverse complement motif |
| Consensus sequence: DRVSASCTGTCAAWVV | Consensus sequence: VVWTTGACAGSTSBKD |
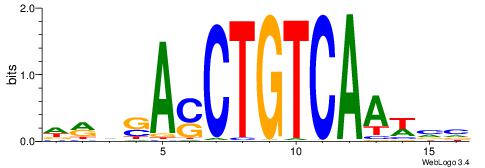 |
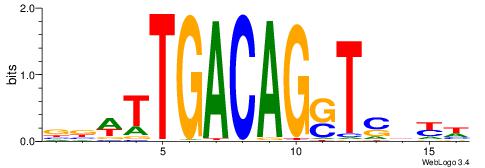 |
| Dataset #: 1 | Motif ID: 120 | Motif name: C120 |
| Original motif | Reverse complement motif |
| Consensus sequence: TGARACMSKGTYTCA | Consensus sequence: TGAMACRSYGTKTCA |
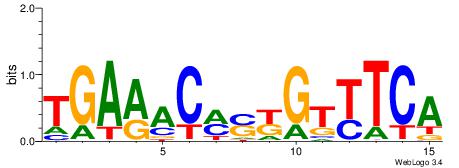 |
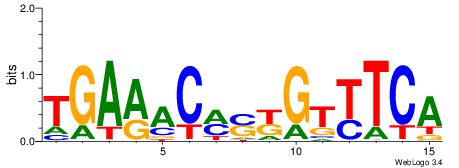 |
Best Matches for Motif ID 120 (Highest to Lowest)
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 15 |
| Similarity score: | 0.0456935 |
Alignment:
BDBBVBVGTAWCCAAGVBHBAGB
------TGARACMSKGTYTCA--
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Motif ID: | UP00036 |
| Motif name: | Myf6_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.049534 |
Alignment:
GGDGBGVCTGTKVVB
TGAMACRSYGTKTCA
| Original motif | Reverse complement motif |
| Consensus sequence: VVVRACAGVCBCDCC | Consensus sequence: GGDGBGVCTGTKVVB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 15 |
| Similarity score: | 0.0503421 |
Alignment:
VMYDHDGMCCHCCKBGVVAAVH
----TGAMACRSYGTKTCA---
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 15 |
| Similarity score: | 0.0525165 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
------TGARACMSKGTYTCA-
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00059 |
| Motif name: | Arid5a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0539819 |
Alignment:
BDYDVAATACGADDBVV
--TGARACMSKGTYTCA
| Original motif | Reverse complement motif |
| Consensus sequence: BDYDVAATACGADDBVV | Consensus sequence: BBVDDTCGTATTVDMHB |
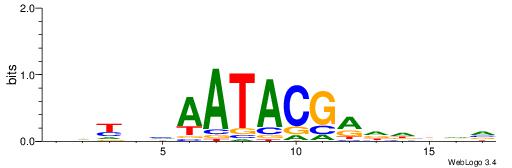 |
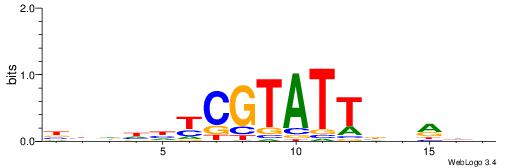 |
| Dataset #: 1 | Motif ID: 121 | Motif name: C121 |
| Original motif | Reverse complement motif |
| Consensus sequence: MRCCCTRGCYSTCCYK | Consensus sequence: RKGGASMGCKAGGGMY |
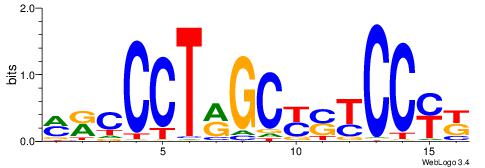 |
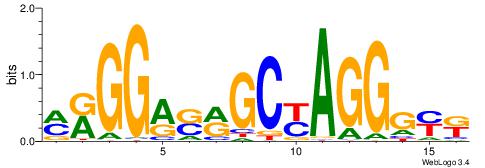 |
Best Matches for Motif ID 121 (Highest to Lowest)
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 16 |
| Similarity score: | 0.0432795 |
Alignment:
WHVBVVMACGCGCGTCDYHWDH
--RKGGASMGCKAGGGMY----
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0435346 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
------RKGGASMGCKAGGGMY
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.043859 |
Alignment:
CBDMCMGGGTGGTCCHVBVBAH
-MRCCCTRGCYSTCCYK-----
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 16 |
| Similarity score: | 0.0450464 |
Alignment:
BHCBCBCCGGGTGGTCYHVHDCH
---MRCCCTRGCYSTCCYK----
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0451554 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
-------RKGGASMGCKAGGGMY
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Dataset #: 1 | Motif ID: 122 | Motif name: C122 |
| Original motif | Reverse complement motif |
| Consensus sequence: YAGCCCWGSCWSBCCTR | Consensus sequence: MAGGVSWGSCWGGGCTK |
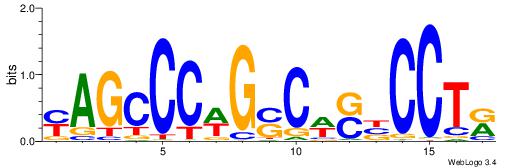 |
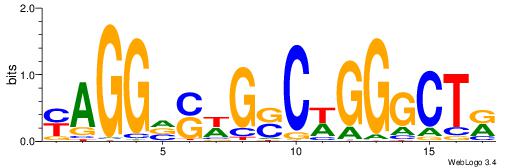 |
Best Matches for Motif ID 122 (Highest to Lowest)
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 17 |
| Similarity score: | 0.0374003 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
MAGGVSWGSCWGGGCTK------
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 0.0431179 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
----YAGCCCWGSCWSBCCTR--
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 17 |
| Similarity score: | 0.0445547 |
Alignment:
YBVDMGTGGGTGGTCKVVBVBBT
MAGGVSWGSCWGGGCTK------
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 17 |
| Similarity score: | 0.0459573 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
-----MAGGVSWGSCWGGGCTK
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 0.0473282 |
Alignment:
BHCBCBCCGGGTGGTCYHVHDCH
--YAGCCCWGSCWSBCCTR----
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Dataset #: 1 | Motif ID: 123 | Motif name: C123 |
| Original motif | Reverse complement motif |
| Consensus sequence: GAAMTCWGAAATC | Consensus sequence: GATTTCWGARTTC |
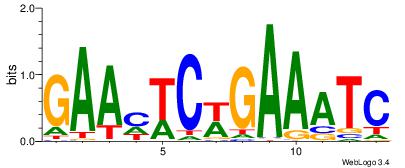 |
 |
Best Matches for Motif ID 123 (Highest to Lowest)
| Motif ID: | UP00015 |
| Motif name: | Ehf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 13 |
| Similarity score: | 0.0397393 |
Alignment:
WDVDABTTCCKAHSDW
---GATTTCWGARTTC
| Original motif | Reverse complement motif |
| Consensus sequence: WDVDABTTCCKAHSDW | Consensus sequence: WDSHTYGGAAVTDVDW |
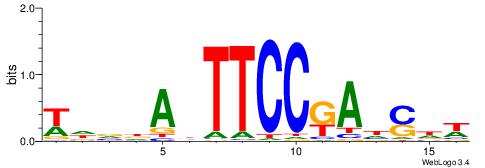 |
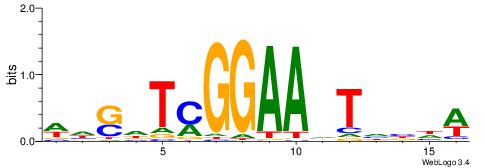 |
| Motif ID: | UP00409 |
| Motif name: | Elf5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0411525 |
Alignment:
BWAHSMGGAAGTWD
GAAMTCWGAAATC-
| Original motif | Reverse complement motif |
| Consensus sequence: BWAHSMGGAAGTWD | Consensus sequence: DWACTTCCRSDTWV |
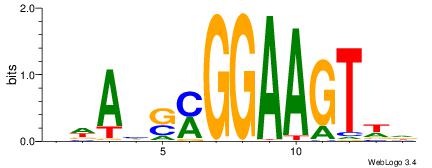 |
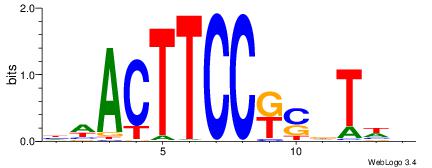 |
| Motif ID: | UP00015 |
| Motif name: | Ehf_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0417179 |
Alignment:
VBDABCCGGAAGTDD
-GAAMTCWGAAATC-
| Original motif | Reverse complement motif |
| Consensus sequence: VBDABCCGGAAGTDD | Consensus sequence: DDACTTCCGGBTHBB |
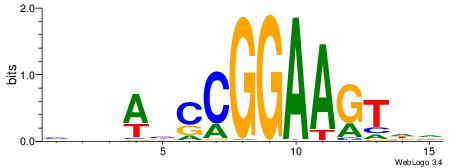 |
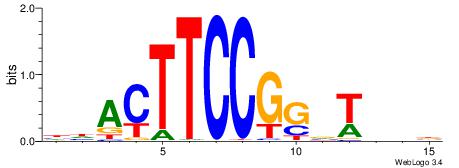 |
| Motif ID: | UP00413 |
| Motif name: | Elf4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 13 |
| Similarity score: | 0.0418022 |
Alignment:
DBBHACTTCCGGKWTH
---GATTTCWGARTTC
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGKWTH | Consensus sequence: HAWYCCGGAAGTHBBD |
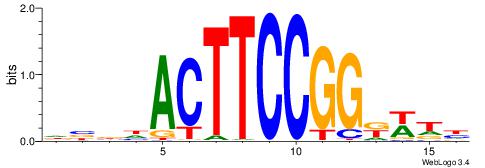 |
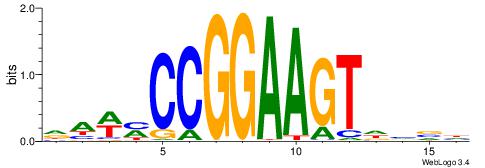 |
| Motif ID: | UP00418 |
| Motif name: | Etv6 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 13 |
| Similarity score: | 0.0418622 |
Alignment:
BHAHCCGGAAGTABDVD
GAAMTCWGAAATC----
| Original motif | Reverse complement motif |
| Consensus sequence: DVHBTACTTCCGGHTHB | Consensus sequence: BHAHCCGGAAGTABDVD |
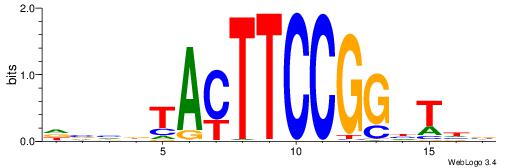 |
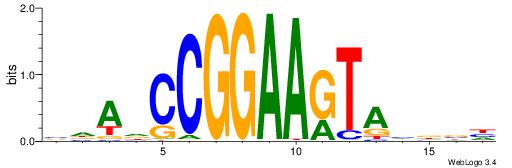 |
| Dataset #: 1 | Motif ID: 124 | Motif name: C124 |
| Original motif | Reverse complement motif |
| Consensus sequence: WSACSRSGCTGSYSTCSW | Consensus sequence: WSGASKSCAGCSMSGTSW |
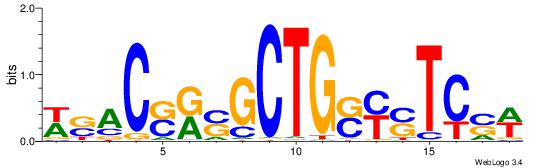 |
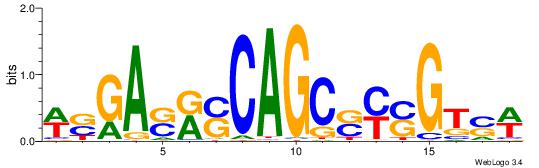 |
Best Matches for Motif ID 124 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0329213 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
--WSGASKSCAGCSMSGTSW---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0329451 |
Alignment:
MWSMBAARRATGCDCABHATGD
-WSACSRSGCTGSYSTCSW---
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0351741 |
Alignment:
BDVBCDACGCKCGCGTBHRKHD
-WSACSRSGCTGSYSTCSW---
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0362523 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
--WSGASKSCAGCSMSGTSW--
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0378518 |
Alignment:
WHVBVVMACGCGCGTCDYHWDH
--WSACSRSGCTGSYSTCSW--
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
 |
 |
| Dataset #: 1 | Motif ID: 125 | Motif name: C125 |
| Original motif | Reverse complement motif |
| Consensus sequence: SKASWYASAGRWASMS | Consensus sequence: SYSTWKCTSTMWSTRS |
 |
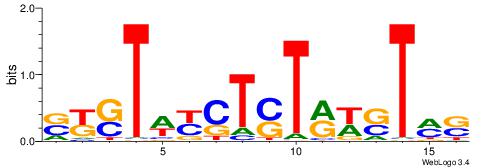 |
Best Matches for Motif ID 125 (Highest to Lowest)
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 16 |
| Similarity score: | 0.0304657 |
Alignment:
GTVBDAATTGTVTGHGDDHKVB
-SYSTWKCTSTMWSTRS-----
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00122 |
| Motif name: | Tgif1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.033763 |
Alignment:
DBVVAGCTGTCAWWVTH
SYSTWKCTSTMWSTRS-
| Original motif | Reverse complement motif |
| Consensus sequence: DABWWTGACAGCTVVBD | Consensus sequence: DBVVAGCTGTCAWWVTH |
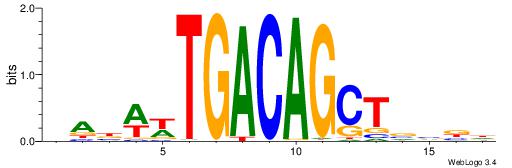 |
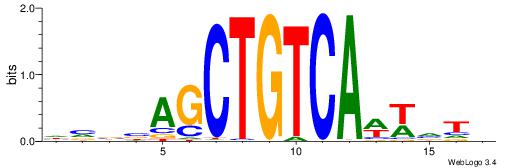 |
| Motif ID: | UP00014 |
| Motif name: | Sox17_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0342041 |
Alignment:
VHBHHCATTCATRHVDD
SKASWYASAGRWASMS-
| Original motif | Reverse complement motif |
| Consensus sequence: VHBHHCATTCATRHVDD | Consensus sequence: DDBDKATGAATGHDBHV |
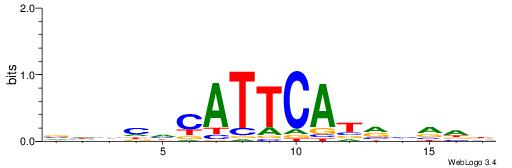 |
 |
| Motif ID: | UP00077 |
| Motif name: | Srf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0346406 |
Alignment:
VBDDAAAAAAAAMVBHH
SKASWYASAGRWASMS-
| Original motif | Reverse complement motif |
| Consensus sequence: VBDDAAAAAAAAMVBHH | Consensus sequence: HHVBYTTTTTTTTDDVV |
 |
 |
| Motif ID: | UP00084 |
| Motif name: | Gmeb1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0359108 |
Alignment:
BHBBKKACGTMMVDBVB
-SKASWYASAGRWASMS
| Original motif | Reverse complement motif |
| Consensus sequence: BHBBKKACGTMMVDBVB | Consensus sequence: BVVDVRYACGTRYVBHB |
 |
 |
| Dataset #: 1 | Motif ID: 126 | Motif name: C126 |
| Original motif | Reverse complement motif |
| Consensus sequence: SVWGRCSMKSCTGGYS | Consensus sequence: SMCCAGSYYSGMCWBS |
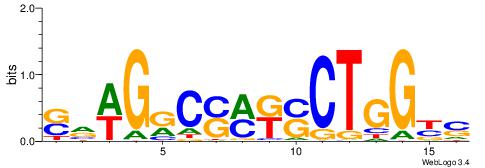 |
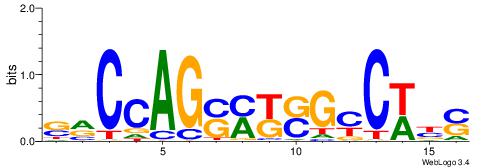 |
Best Matches for Motif ID 126 (Highest to Lowest)
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0246313 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
SVWGRCSMKSCTGGYS------
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0257483 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
------SMCCAGSYYSGMCWBS-
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 16 |
| Similarity score: | 0.0279168 |
Alignment:
VMYDHDGMCCHCCKBGVVAAVH
--SVWGRCSMKSCTGGYS----
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 16 |
| Similarity score: | 0.0307173 |
Alignment:
DVMHHDHKGACCCTCCTSVCBH
-----SVWGRCSMKSCTGGYS-
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 16 |
| Similarity score: | 0.0329173 |
Alignment:
HGDHVHKGACCACCCGGBGBGDB
----SVWGRCSMKSCTGGYS---
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Dataset #: 1 | Motif ID: 127 | Motif name: C127 |
| Original motif | Reverse complement motif |
| Consensus sequence: CKCCYYTHATCCCMG | Consensus sequence: CYGGGATHAMMGGRG |
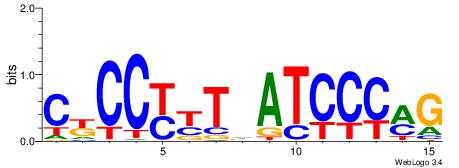 |
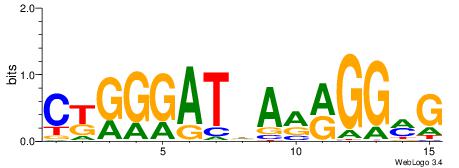 |
Best Matches for Motif ID 127 (Highest to Lowest)
| Motif ID: | UP00219 |
| Motif name: | Cutl1_3494.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0320609 |
Alignment:
DBWSDTKATCAAVCGGT
--CYGGGATHAMMGGRG
| Original motif | Reverse complement motif |
| Consensus sequence: ACCGVTTGATYAHSWBD | Consensus sequence: DBWSDTKATCAAVCGGT |
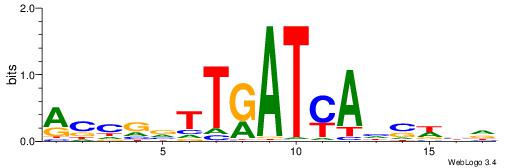 |
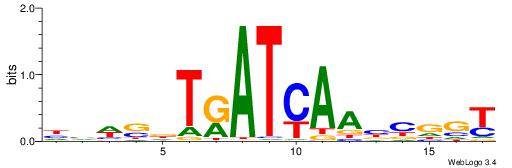 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0338391 |
Alignment:
VDDAGATCAAAGGKDDD
CYGGGATHAMMGGRG--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
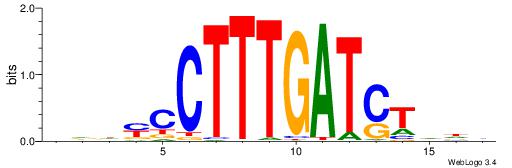 |
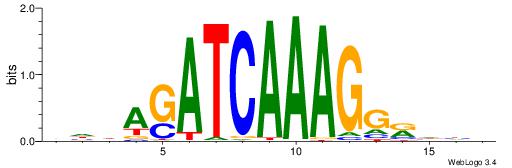 |
| Motif ID: | UP00014 |
| Motif name: | Sox17_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 15 |
| Similarity score: | 0.0345074 |
Alignment:
DDBDKATGAATGHDBHV
CYGGGATHAMMGGRG--
| Original motif | Reverse complement motif |
| Consensus sequence: VHBHHCATTCATRHVDD | Consensus sequence: DDBDKATGAATGHDBHV |
 |
 |
| Motif ID: | UP00058 |
| Motif name: | Tcf3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0345461 |
Alignment:
HHHKVCTTTGATSTHHH
CYGGGATHAMMGGRG--
| Original motif | Reverse complement motif |
| Consensus sequence: HHHASATCAAAGVRHHH | Consensus sequence: HHHKVCTTTGATSTHHH |
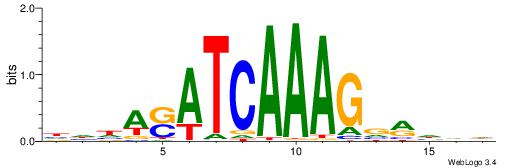 |
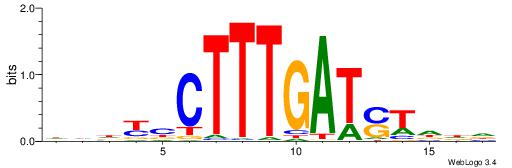 |
| Motif ID: | UP00083 |
| Motif name: | Tcf7l2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 15 |
| Similarity score: | 0.0348977 |
Alignment:
BDDASATCAAAGGMDDD
CYGGGATHAMMGGRG--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATSTDDV | Consensus sequence: BDDASATCAAAGGMDDD |
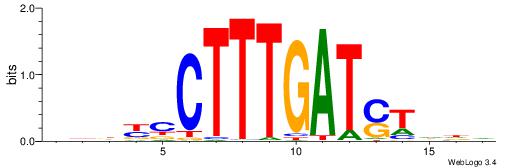 |
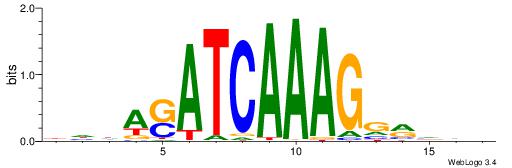 |
| Dataset #: 1 | Motif ID: 128 | Motif name: C128 |
| Original motif | Reverse complement motif |
| Consensus sequence: SYCTSRSTGTCSYSGARS | Consensus sequence: SKTCSMSGACASKSAGMS |
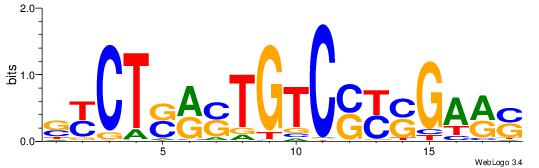 |
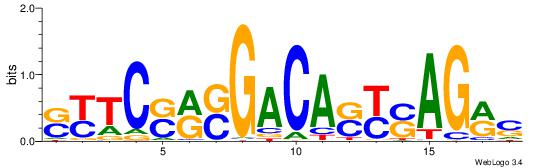 |
Best Matches for Motif ID 128 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0345039 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
-----SYCTSRSTGTCSYSGARS
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0355993 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
----SYCTSRSTGTCSYSGARS
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 18 |
| Similarity score: | 0.040905 |
Alignment:
VCTBHBVCTTGGWTACVVVVVDB
-----SYCTSRSTGTCSYSGARS
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 18 |
| Similarity score: | 0.0416039 |
Alignment:
HHBSHTCGTGRGTGGKGDBVWMV
-----SYCTSRSTGTCSYSGARS
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0436679 |
Alignment:
VDWDARRGACGCGGWHGGAKHV
SKTCSMSGACASKSAGMS----
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Dataset #: 1 | Motif ID: 129 | Motif name: C129 |
| Original motif | Reverse complement motif |
| Consensus sequence: KKGCTAYRYRGAGAMM | Consensus sequence: RRTCTCKKMMTAGCRY |
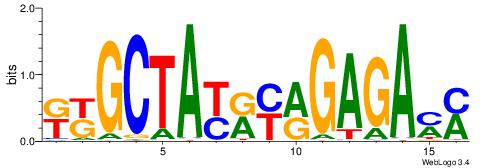 |
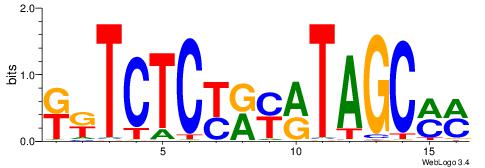 |
Best Matches for Motif ID 129 (Highest to Lowest)
| Motif ID: | UP00131 |
| Motif name: | Gbx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0195892 |
Alignment:
DVDVKCTAATTAGMVVD
RRTCTCKKMMTAGCRY-
| Original motif | Reverse complement motif |
| Consensus sequence: DVVRCTAATTAGYVDBD | Consensus sequence: DVDVKCTAATTAGMVVD |
 |
 |
| Motif ID: | UP00136 |
| Motif name: | Prrx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0328703 |
Alignment:
DDDHKCTAATTAGCHBT
RRTCTCKKMMTAGCRY-
| Original motif | Reverse complement motif |
| Consensus sequence: AVHGCTAATTAGYDDDD | Consensus sequence: DDDHKCTAATTAGCHBT |
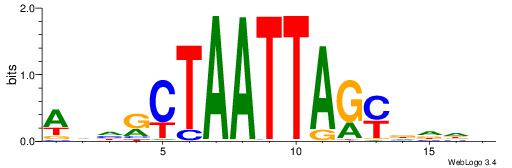 |
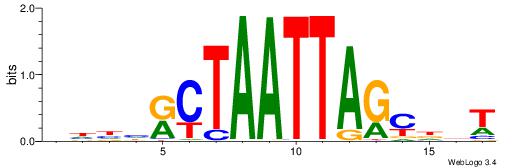 |
| Motif ID: | UP00132 |
| Motif name: | Evx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0356184 |
Alignment:
BVCMGCTAATTAGCBKW
RRTCTCKKMMTAGCRY-
| Original motif | Reverse complement motif |
| Consensus sequence: BVCMGCTAATTAGCBKW | Consensus sequence: WRBGCTAATTAGCRGBB |
 |
 |
| Motif ID: | UP00211 |
| Motif name: | Pou3f3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0360591 |
Alignment:
HADDTATGCATAATDDA
-KKGCTAYRYRGAGAMM
| Original motif | Reverse complement motif |
| Consensus sequence: HADDTATGCATAATDDA | Consensus sequence: TDDATTATGCATADDTH |
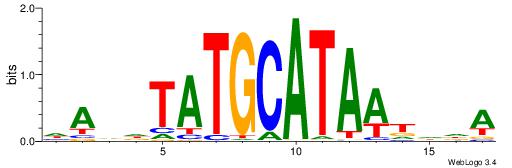 |
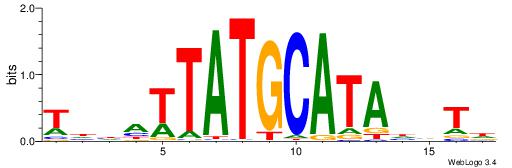 |
| Motif ID: | UP00178 |
| Motif name: | Og2x |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0365623 |
Alignment:
BHDHBMKAATTMKVVCG
RRTCTCKKMMTAGCRY-
| Original motif | Reverse complement motif |
| Consensus sequence: CGVVYYAATTRRBHDHB | Consensus sequence: BHDHBMKAATTMKVVCG |
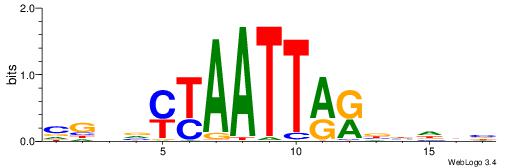 |
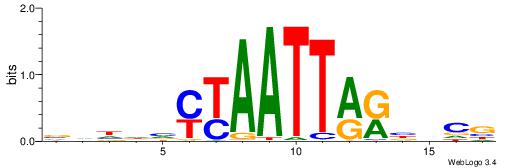 |
| Dataset #: 1 | Motif ID: 130 | Motif name: C130 |
| Original motif | Reverse complement motif |
| Consensus sequence: TKCCWGGVCWGCMA | Consensus sequence: TRGCWGBCCWGGYA |
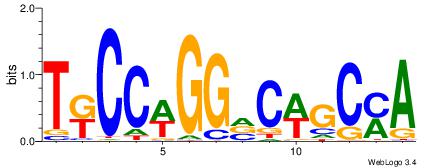 |
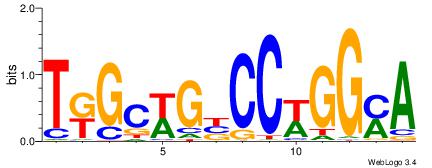 |
Best Matches for Motif ID 130 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 14 |
| Similarity score: | 0.0417596 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
-TKCCWGGVCWGCMA--------
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0438533 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
------TKCCWGGVCWGCMA---
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0449212 |
Alignment:
HHHHDVVGTTGCTAHGGDHBAHH
TRGCWGBCCWGGYA---------
| Original motif | Reverse complement motif |
| Consensus sequence: HDTBHHCCHTAGCAACVVDHHHH | Consensus sequence: HHHHDVVGTTGCTAHGGDHBAHH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0463369 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
-------TKCCWGGVCWGCMA-
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 14 |
| Similarity score: | 0.0472891 |
Alignment:
YBVDMGTGGGTGGTCKVVBVBBT
------TRGCWGBCCWGGYA---
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Dataset #: 1 | Motif ID: 131 | Motif name: C131 |
| Original motif | Reverse complement motif |
| Consensus sequence: WWAADAARVHAAWW | Consensus sequence: WWTTHBKTTDTTWW |
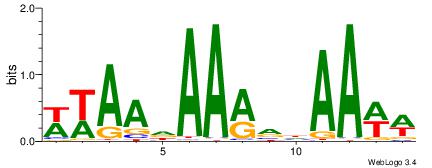 |
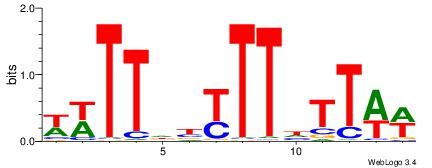 |
Best Matches for Motif ID 131 (Highest to Lowest)
| Motif ID: | UP00077 |
| Motif name: | Srf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0256716 |
Alignment:
VBDDAAAAAAAAMVBHH
--WWAADAARVHAAWW-
| Original motif | Reverse complement motif |
| Consensus sequence: VBDDAAAAAAAAMVBHH | Consensus sequence: HHVBYTTTTTTTTDDVV |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0279274 |
Alignment:
HDTYTTGTTWTKDDDT
WWTTHBKTTDTTWW--
| Original motif | Reverse complement motif |
| Consensus sequence: ADDDYAWAACAAMAHH | Consensus sequence: HDTYTTGTTWTKDDDT |
 |
 |
| Motif ID: | UP00028 |
| Motif name: | Tcfap2e_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0294273 |
Alignment:
HDCDBRAAAAAADD
WWAADAARVHAAWW
| Original motif | Reverse complement motif |
| Consensus sequence: HDCDBRAAAAAADD | Consensus sequence: DDTTTTTTMBDGDH |
 |
 |
| Motif ID: | UP00014 |
| Motif name: | Sox17_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0331143 |
Alignment:
DDWTBAATTGTTDDB
WWTTHBKTTDTTWW-
| Original motif | Reverse complement motif |
| Consensus sequence: VDDAACAATTVAWHD | Consensus sequence: DDWTBAATTGTTDDB |
 |
 |
| Motif ID: | UP00037 |
| Motif name: | Zfp105_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0331856 |
Alignment:
HHHTTRTTDTTTKDH
-WWTTHBKTTDTTWW
| Original motif | Reverse complement motif |
| Consensus sequence: HDYAAAHAAMAADHD | Consensus sequence: HHHTTRTTDTTTKDH |
 |
 |
| Dataset #: 1 | Motif ID: 132 | Motif name: C132 |
| Original motif | Reverse complement motif |
| Consensus sequence: WCAGRGWGAGAWCYAGGW | Consensus sequence: WCCTKGWTCTCWCMCTGW |
 |
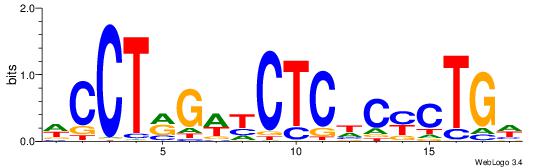 |
Best Matches for Motif ID 132 (Highest to Lowest)
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0464813 |
Alignment:
VVYDDDCHCAVACAATTDBVAC
WCCTKGWTCTCWCMCTGW----
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.0491336 |
Alignment:
HVGVSAGGAGGGTCYHHHHYVH
-WCAGRGWGAGAWCYAGGW---
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0511816 |
Alignment:
DCATDBTGHGCATKMTTBRSWR
WCCTKGWTCTCWCMCTGW----
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0524801 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
--WCAGRGWGAGAWCYAGGW---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.0525963 |
Alignment:
DVTTVVCVYGGHGGYCHHDMYB
---WCAGRGWGAGAWCYAGGW-
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Dataset #: 1 | Motif ID: 133 | Motif name: C133 |
| Original motif | Reverse complement motif |
| Consensus sequence: AWMAWHDACACHRWMAW | Consensus sequence: WTYWKHGTGTDHWTRWT |
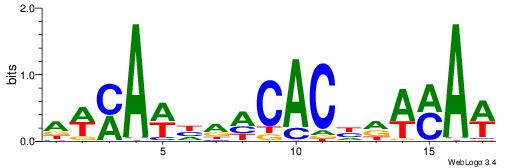 |
 |
Best Matches for Motif ID 133 (Highest to Lowest)
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 0.039607 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
-AWMAWHDACACHRWMAW-----
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 17 |
| Similarity score: | 0.0412538 |
Alignment:
WBHHHHHMDATTGTTHDHDVHH
----WTYWKHGTGTDHWTRWT-
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00039 |
| Motif name: | Foxj3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0441279 |
Alignment:
DVHWDTGTTWTGDTBDH
WTYWKHGTGTDHWTRWT
| Original motif | Reverse complement motif |
| Consensus sequence: HDBAHCAWAACADWDVD | Consensus sequence: DVHWDTGTTWTGDTBDH |
 |
 |
| Motif ID: | UP00119 |
| Motif name: | Nkx2-9 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0448373 |
Alignment:
TTTDAAGTACTTVAADH
WTYWKHGTGTDHWTRWT
| Original motif | Reverse complement motif |
| Consensus sequence: TTTDAAGTACTTVAADH | Consensus sequence: HDTTBAAGTACTTDAAA |
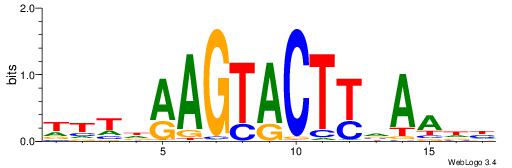 |
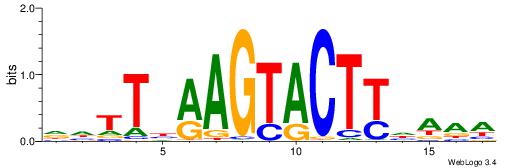 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 0.0454758 |
Alignment:
GTVBDAATTGTVTGHGDDHKVB
-WTYWKHGTGTDHWTRWT----
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Dataset #: 1 | Motif ID: 134 | Motif name: C134 |
| Original motif | Reverse complement motif |
| Consensus sequence: CCBCCCCSCCC | Consensus sequence: GGGSGGGGBGG |
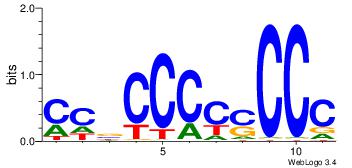 |
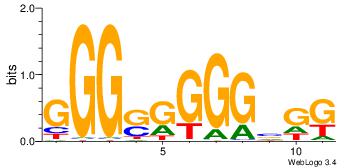 |
Best Matches for Motif ID 134 (Highest to Lowest)
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.00418064 |
Alignment:
GKRGGGGGGGGGGBD
----GGGSGGGGBGG
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0057444 |
Alignment:
BHHDYGGGGGGGGBVD
-----GGGSGGGGBGG
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0142081 |
Alignment:
DHHBCCCCGCCAHHBHB
-CCBCCCCSCCC-----
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
 |
 |
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0188242 |
Alignment:
HBBBCCGCCCCWHHHV
CCBCCCCSCCC-----
| Original motif | Reverse complement motif |
| Consensus sequence: HBBBCCGCCCCWHHHV | Consensus sequence: BHHHWGGGGCGGBBVH |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0189121 |
Alignment:
HHHHDGGGGCGGDBHDV
------GGGSGGGGBGG
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
 |
 |
| Dataset #: 1 | Motif ID: 135 | Motif name: C135 |
| Original motif | Reverse complement motif |
| Consensus sequence: KKCCKAGMACBMASMM | Consensus sequence: RYSTYVGTYCTYGGRY |
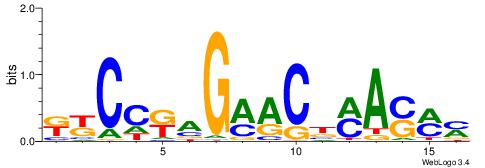 |
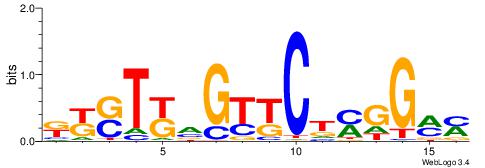 |
Best Matches for Motif ID 135 (Highest to Lowest)
| Motif ID: | UP00390 |
| Motif name: | Tcf1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0428575 |
Alignment:
VDDTTAGTTAACBVRSD
RYSTYVGTYCTYGGRY-
| Original motif | Reverse complement motif |
| Consensus sequence: HSKBVGTTAACTAADDB | Consensus sequence: VDDTTAGTTAACBVRSD |
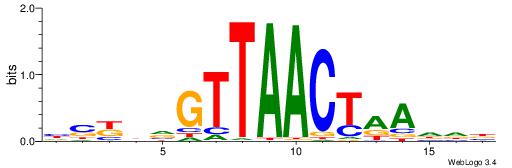 |
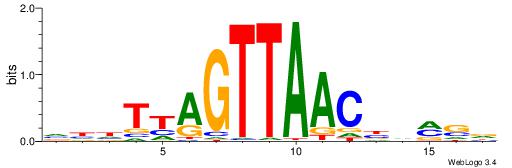 |
| Motif ID: | UP00389 |
| Motif name: | Nkx3-1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0437525 |
Alignment:
BDTTTAAGTACTTDDDD
-RYSTYVGTYCTYGGRY
| Original motif | Reverse complement motif |
| Consensus sequence: DDHDAAGTACTTAAADB | Consensus sequence: BDTTTAAGTACTTDDDD |
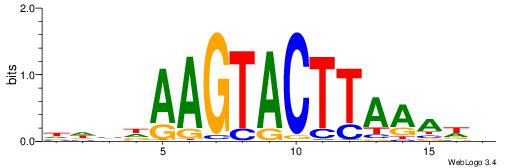 |
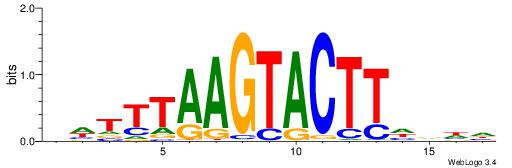 |
| Motif ID: | UP00100 |
| Motif name: | Gata6_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0456644 |
Alignment:
GBBVCGATATCGSVGYD
-KKCCKAGMACBMASMM
| Original motif | Reverse complement motif |
| Consensus sequence: GBBVCGATATCGSVGYD | Consensus sequence: HKCBSCGATATCGVBBC |
 |
 |
| Motif ID: | UP00090 |
| Motif name: | Elf3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0458473 |
Alignment:
DDHBTTTTTTTTHDAVH
-RYSTYVGTYCTYGGRY
| Original motif | Reverse complement motif |
| Consensus sequence: DBTHHAAAAAAAAVHDH | Consensus sequence: DDHBTTTTTTTTHDAVH |
 |
 |
| Motif ID: | UP00053 |
| Motif name: | Rxra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0463964 |
Alignment:
DBHBDTGACCCCDHVHB
KKCCKAGMACBMASMM-
| Original motif | Reverse complement motif |
| Consensus sequence: DBHBDTGACCCCDHVHB | Consensus sequence: VHBHDGGGGTCAHBHBD |
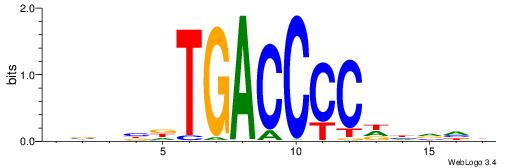 |
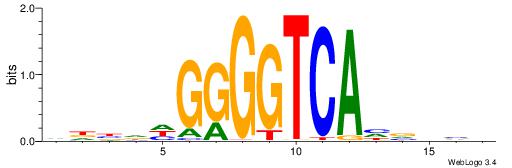 |
| Dataset #: 1 | Motif ID: 136 | Motif name: C136 |
| Original motif | Reverse complement motif |
| Consensus sequence: GDGGVKSMHGGGRMT | Consensus sequence: AYKCCCDRSYVCCHC |
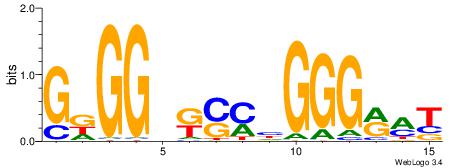 |
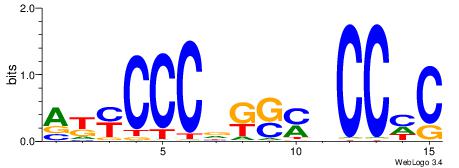 |
Best Matches for Motif ID 136 (Highest to Lowest)
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 15 |
| Similarity score: | 0.0296869 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
AYKCCCDRSYVCCHC-------
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00036 |
| Motif name: | Myf6_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0329478 |
Alignment:
VVVRACAGVCBCDCC
GDGGVKSMHGGGRMT
| Original motif | Reverse complement motif |
| Consensus sequence: VVVRACAGVCBCDCC | Consensus sequence: GGDGBGVCTGTKVVB |
 |
 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0360132 |
Alignment:
GKRGGGGGGGGGGBD
GDGGVKSMHGGGRMT
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Motif ID: | UP00082 |
| Motif name: | Zfp187_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0383817 |
Alignment:
DDGMCCTBGTCCHYHV
AYKCCCDRSYVCCHC-
| Original motif | Reverse complement motif |
| Consensus sequence: DDGMCCTBGTCCHYHV | Consensus sequence: VHMDGGACVAGGRCDH |
 |
 |
| Motif ID: | UP00096 |
| Motif name: | Sox13_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0391249 |
Alignment:
DDDHTGGGTGGGHAHHD
GDGGVKSMHGGGRMT--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDHTGGGTGGGHAHHD | Consensus sequence: DHHTHCCCACCCAHDDH |
 |
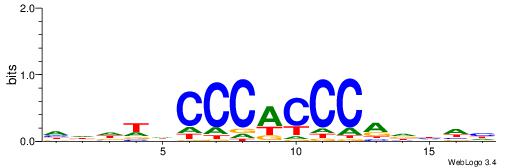 |
| Dataset #: 1 | Motif ID: 137 | Motif name: C137 |
| Original motif | Reverse complement motif |
| Consensus sequence: RAAYTCAGRAAY | Consensus sequence: MTTKCTGAKTTK |
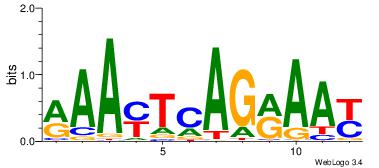 |
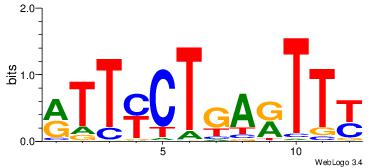 |
Best Matches for Motif ID 137 (Highest to Lowest)
| Motif ID: | UP00085 |
| Motif name: | Sfpi1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.00902566 |
Alignment:
HHAWTTHVGGAACH
-RAAYTCAGRAAY-
| Original motif | Reverse complement motif |
| Consensus sequence: HHAWTTHVGGAACH | Consensus sequence: DGTTCCVDAAWTHD |
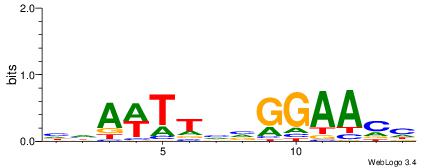 |
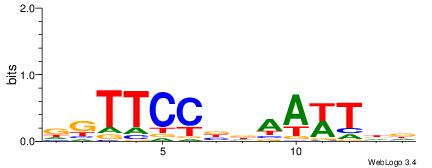 |
| Motif ID: | UP00101 |
| Motif name: | Sox12_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 0.0179124 |
Alignment:
VVDBASACAAAGRADH
----RAAYTCAGRAAY
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBASACAAAGRADH | Consensus sequence: HDTMCTTTGTSTVDBB |
 |
 |
| Motif ID: | UP00185 |
| Motif name: | Pbx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 12 |
| Similarity score: | 0.0179708 |
Alignment:
BVDWHWTTGATGRKTKH
-----MTTKCTGAKTTK
| Original motif | Reverse complement motif |
| Consensus sequence: HYAYMCATCAAWHWDVV | Consensus sequence: BVDWHWTTGATGRKTKH |
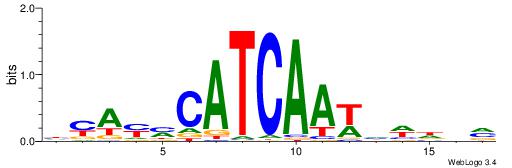 |
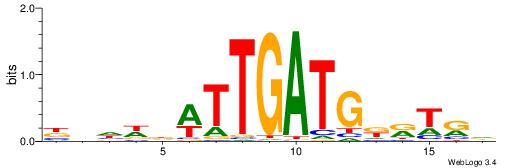 |
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 0.0209132 |
Alignment:
HBDDDAACAAAGRVBHH
----RAAYTCAGRAAY-
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
 |
 |
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 0.0212068 |
Alignment:
HBDDRAACAAAGRABHH
----RAAYTCAGRAAY-
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
 |
 |
| Dataset #: 1 | Motif ID: 138 | Motif name: C138 |
| Original motif | Reverse complement motif |
| Consensus sequence: AAGAAARSAAG | Consensus sequence: CTTSKTTTCTT |
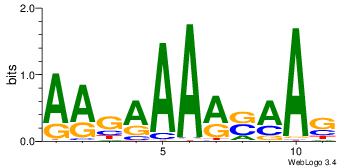 |
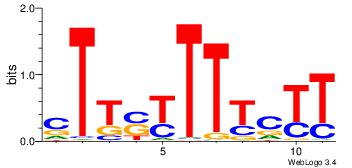 |
Best Matches for Motif ID 138 (Highest to Lowest)
| Motif ID: | UP00077 |
| Motif name: | Srf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0334943 |
Alignment:
HHVBYTTTTTTTTDDVV
--CTTSKTTTCTT----
| Original motif | Reverse complement motif |
| Consensus sequence: VBDDAAAAAAAAMVBHH | Consensus sequence: HHVBYTTTTTTTTDDVV |
 |
 |
| Motif ID: | UP00037 |
| Motif name: | Zfp105_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0441276 |
Alignment:
HHHTTRTTDTTTKDH
--CTTSKTTTCTT--
| Original motif | Reverse complement motif |
| Consensus sequence: HDYAAAHAAMAADHD | Consensus sequence: HHHTTRTTDTTTKDH |
 |
 |
| Motif ID: | UP00028 |
| Motif name: | Tcfap2e_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0449117 |
Alignment:
HDCDBRAAAAAADD
--AAGAAARSAAG-
| Original motif | Reverse complement motif |
| Consensus sequence: HDCDBRAAAAAADD | Consensus sequence: DDTTTTTTMBDGDH |
 |
 |
| Motif ID: | UP00171 |
| Motif name: | Msx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0452028 |
Alignment:
HHVTTAATTKGTTHTV
--CTTSKTTTCTT---
| Original motif | Reverse complement motif |
| Consensus sequence: VAHAACYAATTAABHH | Consensus sequence: HHVTTAATTKGTTHTV |
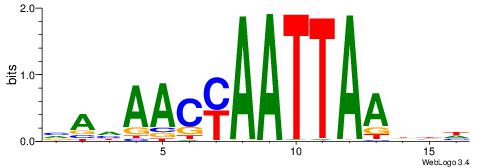 |
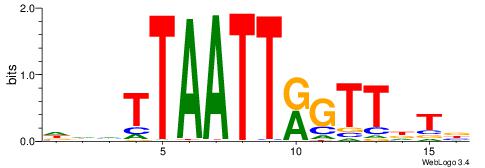 |
| Motif ID: | UP00025 |
| Motif name: | Foxk1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0456684 |
Alignment:
HDDTGTTKTTKTHHD
-CTTSKTTTCTT---
| Original motif | Reverse complement motif |
| Consensus sequence: HHHAYAAYAACAHHH | Consensus sequence: HDDTGTTKTTKTHHD |
 |
 |
| Dataset #: 1 | Motif ID: 139 | Motif name: C139 |
| Original motif | Reverse complement motif |
| Consensus sequence: WWMAAABWWWVAAAAWW | Consensus sequence: WWTTTTVWWWBTTTYWW |
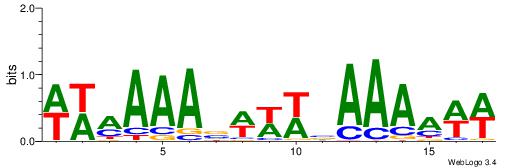 |
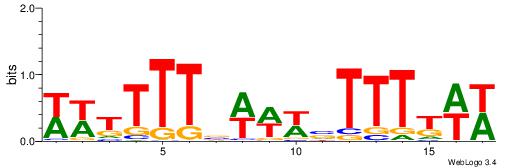 |
Best Matches for Motif ID 139 (Highest to Lowest)
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 0.0481917 |
Alignment:
HHBDHDDAACAATDRHHHHHBW
---WWMAAABWWWVAAAAWW--
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00077 |
| Motif name: | Srf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0496844 |
Alignment:
VBDDAAAAAAAAMVBHH
WWMAAABWWWVAAAAWW
| Original motif | Reverse complement motif |
| Consensus sequence: VBDDAAAAAAAAMVBHH | Consensus sequence: HHVBYTTTTTTTTDDVV |
 |
 |
| Motif ID: | UP00255 |
| Motif name: | Dbx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0509582 |
Alignment:
DHDBTAWTAATTHAKTH
WWTTTTVWWWBTTTYWW
| Original motif | Reverse complement motif |
| Consensus sequence: HARTHAATTAWTAVDHD | Consensus sequence: DHDBTAWTAATTHAKTH |
 |
 |
| Motif ID: | UP00090 |
| Motif name: | Elf3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0532816 |
Alignment:
DBTHHAAAAAAAAVHDH
WWMAAABWWWVAAAAWW
| Original motif | Reverse complement motif |
| Consensus sequence: DBTHHAAAAAAAAVHDH | Consensus sequence: DDHBTTTTTTTTHDAVH |
 |
 |
| Motif ID: | UP00037 |
| Motif name: | Zfp105_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.05356 |
Alignment:
HDMAVTTATTGARVCMH
WWMAAABWWWVAAAAWW
| Original motif | Reverse complement motif |
| Consensus sequence: DYGVKTCAATAABTYDD | Consensus sequence: HDMAVTTATTGARVCMH |
 |
 |
| Dataset #: 1 | Motif ID: 140 | Motif name: C140 |
| Original motif | Reverse complement motif |
| Consensus sequence: BSCCBCVSSSBCCYHVG | Consensus sequence: CVDMGGBSSSVGBGGSB |
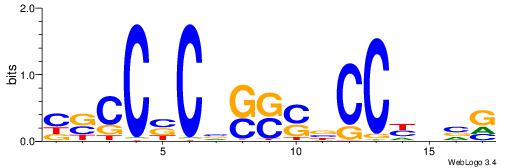 |
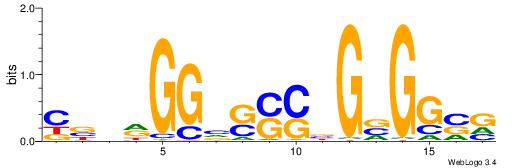 |
Best Matches for Motif ID 140 (Highest to Lowest)
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0510096 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
BSCCBCVSSSBCCYHVG-----
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 17 |
| Similarity score: | 0.0520625 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
--CVDMGGBSSSVGBGGSB----
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0523971 |
Alignment:
BHHBHCCGCCCCDHHHH
BSCCBCVSSSBCCYHVG
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 17 |
| Similarity score: | 0.0544357 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
------BSCCBCVSSSBCCYHVG
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 0.0585258 |
Alignment:
HHBSHTCGTGRGTGGKGDBVWMV
--CVDMGGBSSSVGBGGSB----
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Dataset #: 1 | Motif ID: 141 | Motif name: C141 |
| Original motif | Reverse complement motif |
| Consensus sequence: YMATCCYRSYRCTTGKR | Consensus sequence: KYCAAGKMSKMGGATRK |
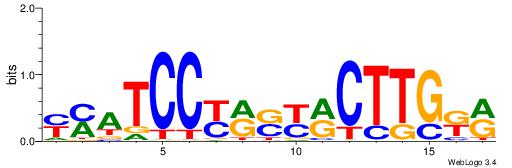 |
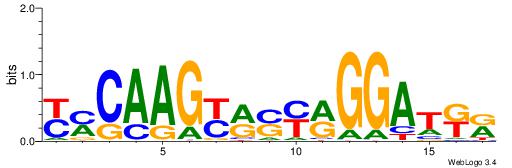 |
Best Matches for Motif ID 141 (Highest to Lowest)
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 0.048695 |
Alignment:
VDWDARRGACGCGGWHGGAKHV
-KYCAAGKMSKMGGATRK----
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 0.0533376 |
Alignment:
BDVBCDACGCKCGCGTBHRKHD
---KYCAAGKMSKMGGATRK--
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0540672 |
Alignment:
BHHBHCCGCCCCDHHHH
YMATCCYRSYRCTTGKR
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0558883 |
Alignment:
HDWHMHGACGCGCGTRBVVBHW
KYCAAGKMSKMGGATRK-----
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0562075 |
Alignment:
BVVHAGGGGGCGRDHHB
KYCAAGKMSKMGGATRK
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Dataset #: 1 | Motif ID: 142 | Motif name: C142 |
| Original motif | Reverse complement motif |
| Consensus sequence: WHAAAAAWTAAAAWW | Consensus sequence: WWTTTTAWTTTTTHW |
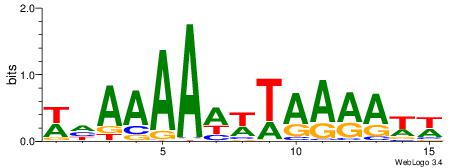 |
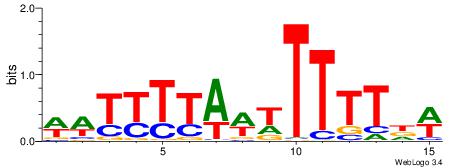 |
Best Matches for Motif ID 142 (Highest to Lowest)
| Motif ID: | UP00077 |
| Motif name: | Srf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0209943 |
Alignment:
HHVBYTTTTTTTTDDVV
WWTTTTAWTTTTTHW--
| Original motif | Reverse complement motif |
| Consensus sequence: VBDDAAAAAAAAMVBHH | Consensus sequence: HHVBYTTTTTTTTDDVV |
 |
 |
| Motif ID: | UP00255 |
| Motif name: | Dbx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 15 |
| Similarity score: | 0.0296488 |
Alignment:
HARTHAATTAWTAVDHD
WHAAAAAWTAAAAWW--
| Original motif | Reverse complement motif |
| Consensus sequence: HARTHAATTAWTAVDHD | Consensus sequence: DHDBTAWTAATTHAKTH |
 |
 |
| Motif ID: | UP00044 |
| Motif name: | Mafk_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0298134 |
Alignment:
MBHTGCADTTTTTHV
WWTTTTAWTTTTTHW
| Original motif | Reverse complement motif |
| Consensus sequence: VHAAAAADTGCAHBR | Consensus sequence: MBHTGCADTTTTTHV |
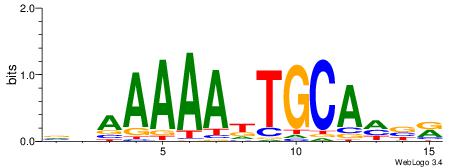 |
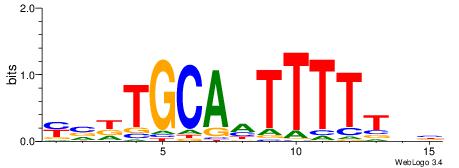 |
| Motif ID: | UP00090 |
| Motif name: | Elf3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.02982 |
Alignment:
DDHBTTTTTTTTHDAVH
-WWTTTTAWTTTTTHW-
| Original motif | Reverse complement motif |
| Consensus sequence: DBTHHAAAAAAAAVHDH | Consensus sequence: DDHBTTTTTTTTHDAVH |
 |
 |
| Motif ID: | UP00037 |
| Motif name: | Zfp105_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0336901 |
Alignment:
HHHTTRTTDTTTKDH
WWTTTTAWTTTTTHW
| Original motif | Reverse complement motif |
| Consensus sequence: HDYAAAHAAMAADHD | Consensus sequence: HHHTTRTTDTTTKDH |
 |
 |
| Dataset #: 1 | Motif ID: 143 | Motif name: C143 |
| Original motif | Reverse complement motif |
| Consensus sequence: MKTGCTKSSMTTAAMK | Consensus sequence: RRTTAAYSSRAGCARY |
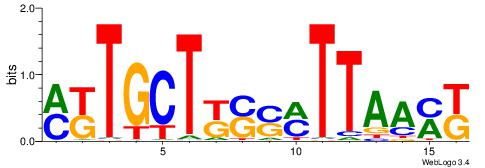 |
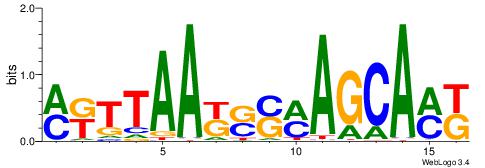 |
Best Matches for Motif ID 143 (Highest to Lowest)
| Motif ID: | UP00032 |
| Motif name: | Gata3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 16 |
| Similarity score: | 0.043815 |
Alignment:
HBHDVBWGATHBTATCRVBHHH
---RRTTAAYSSRAGCARY---
| Original motif | Reverse complement motif |
| Consensus sequence: HBHDVBWGATHBTATCRVBHHH | Consensus sequence: HHHBBMGATAVHATCWVVDHVH |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 16 |
| Similarity score: | 0.0460848 |
Alignment:
HDTBHHCCHTAGCAACVVDHHHH
RRTTAAYSSRAGCARY-------
| Original motif | Reverse complement motif |
| Consensus sequence: HDTBHHCCHTAGCAACVVDHHHH | Consensus sequence: HHHHDVVGTTGCTAHGGDHBAHH |
 |
 |
| Motif ID: | UP00190 |
| Motif name: | Nkx2-3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0485356 |
Alignment:
HHTTAAGTACTTAAMK
MKTGCTKSSMTTAAMK
| Original motif | Reverse complement motif |
| Consensus sequence: YYTTAAGTACTTAAHD | Consensus sequence: HHTTAAGTACTTAAMK |
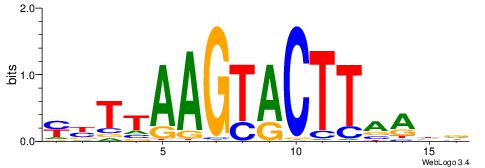 |
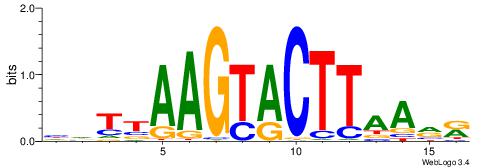 |
| Motif ID: | UP00421 |
| Motif name: | Etv1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0513932 |
Alignment:
MVHBACCGGAAGTVDVH
RRTTAAYSSRAGCARY-
| Original motif | Reverse complement motif |
| Consensus sequence: MVHBACCGGAAGTVDVH | Consensus sequence: HBDBACTTCCGGTBHVY |
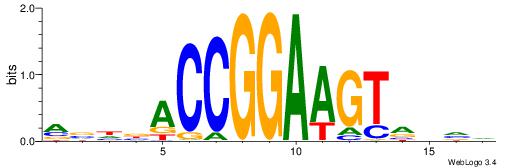 |
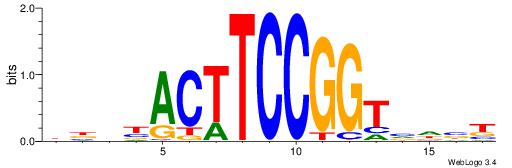 |
| Motif ID: | UP00410 |
| Motif name: | Elk1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.051952 |
Alignment:
VVHBACCGGAAGTDBHD
RRTTAAYSSRAGCARY-
| Original motif | Reverse complement motif |
| Consensus sequence: VVHBACCGGAAGTDBHD | Consensus sequence: DHBDACTTCCGGTVHVB |
 |
 |
| Dataset #: 1 | Motif ID: 144 | Motif name: C144 |
| Original motif | Reverse complement motif |
| Consensus sequence: GACCAGRSYRGYCTC | Consensus sequence: GAGKCKKSKCTGGTC |
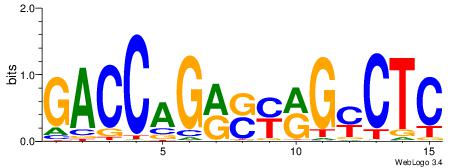 |
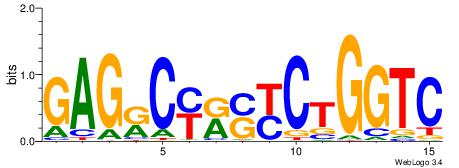 |
Best Matches for Motif ID 144 (Highest to Lowest)
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 15 |
| Similarity score: | 0.0365285 |
Alignment:
YBVDMGTGGGTGGTCKVVBVBBT
GAGKCKKSKCTGGTC--------
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 15 |
| Similarity score: | 0.0367839 |
Alignment:
BTBVTCVTGGGTGGTCMVVDVBB
-GAGKCKKSKCTGGTC-------
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0372292 |
Alignment:
DBCDWHCTGGGTGGTCMDHBBAH
-GAGKCKKSKCTGGTC-------
| Original motif | Reverse complement motif |
| Consensus sequence: DTVBDDRGACCACCCAGDWDGBH | Consensus sequence: DBCDWHCTGGGTGGTCMDHBBAH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 15 |
| Similarity score: | 0.037305 |
Alignment:
CBDMCMGGGTGGTCCHVBVBAH
-GACCAGRSYRGYCTC------
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0395722 |
Alignment:
DVVTTVGTGGGHGGYAMHWHHHY
-GAGKCKKSKCTGGTC-------
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Dataset #: 1 | Motif ID: 145 | Motif name: C145 |
| Original motif | Reverse complement motif |
| Consensus sequence: WWCADCAMCHAMWW | Consensus sequence: WWRTHGYTGDTGWW |
 |
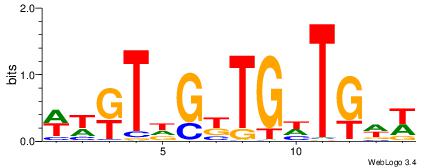 |
Best Matches for Motif ID 145 (Highest to Lowest)
| Motif ID: | UP00037 |
| Motif name: | Zfp105_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0335902 |
Alignment:
HDYAAAHAAMAADHD
-WWCADCAMCHAMWW
| Original motif | Reverse complement motif |
| Consensus sequence: HDYAAAHAAMAADHD | Consensus sequence: HHHTTRTTDTTTKDH |
 |
 |
| Motif ID: | UP00069 |
| Motif name: | Sox1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0379211 |
Alignment:
HBDTAATTGTTABBV
WWRTHGYTGDTGWW-
| Original motif | Reverse complement motif |
| Consensus sequence: HBDTAATTGTTABBV | Consensus sequence: VBVTAACAATTADVD |
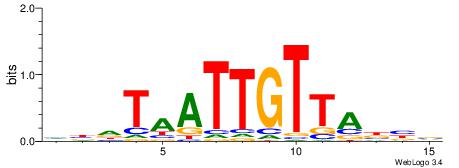 |
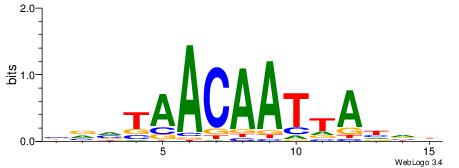 |
| Motif ID: | UP00185 |
| Motif name: | Pbx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0393885 |
Alignment:
HYAYMCATCAAWHWDVV
---WWCADCAMCHAMWW
| Original motif | Reverse complement motif |
| Consensus sequence: HYAYMCATCAAWHWDVV | Consensus sequence: BVDWHWTTGATGRKTKH |
 |
 |
| Motif ID: | UP00091 |
| Motif name: | Sox5_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0396706 |
Alignment:
DHDCATMATTDASDD
-WWCADCAMCHAMWW
| Original motif | Reverse complement motif |
| Consensus sequence: DHDCATMATTDASDD | Consensus sequence: DHSTDAATYATGDHD |
 |
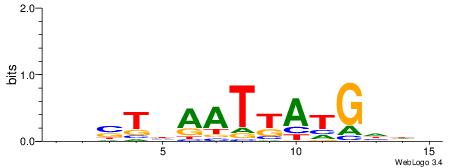 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0406825 |
Alignment:
VDVVATCAATCAHBHD
-WWCADCAMCHAMWW-
| Original motif | Reverse complement motif |
| Consensus sequence: VDVVATCAATCAHBHD | Consensus sequence: DHVDTGATTGATVBDV |
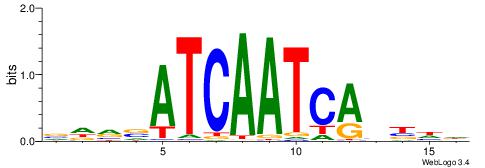 |
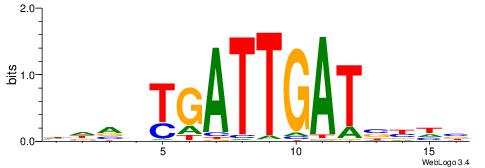 |
| Dataset #: 1 | Motif ID: 146 | Motif name: C146 |
| Original motif | Reverse complement motif |
| Consensus sequence: YRGCSAGGGSTAYR | Consensus sequence: KMTASCCCTSGCKK |
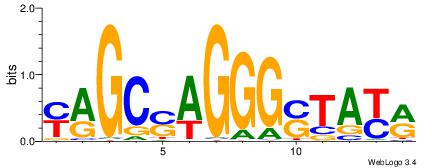 |
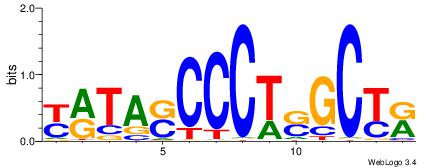 |
Best Matches for Motif ID 146 (Highest to Lowest)
| Motif ID: | UP00022 |
| Motif name: | Zfp740_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0330877 |
Alignment:
HVKDCCGGGGGGWADDD
--YRGCSAGGGSTAYR-
| Original motif | Reverse complement motif |
| Consensus sequence: DDDTWCCCCCCGGDRVH | Consensus sequence: HVKDCCGGGGGGWADDD |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 14 |
| Similarity score: | 0.0345026 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
-----YRGCSAGGGSTAYR----
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0369871 |
Alignment:
DHHDGGGCGRGGKHBH
--YRGCSAGGGSTAYR
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0374088 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
--YRGCSAGGGSTAYR------
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00096 |
| Motif name: | Sox13_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0400162 |
Alignment:
DDDHTGGGTGGGHAHHD
---YRGCSAGGGSTAYR
| Original motif | Reverse complement motif |
| Consensus sequence: DDDHTGGGTGGGHAHHD | Consensus sequence: DHHTHCCCACCCAHDDH |
 |
 |
| Dataset #: 1 | Motif ID: 147 | Motif name: C147 |
| Original motif | Reverse complement motif |
| Consensus sequence: CYWGGKCTACMCAGRG | Consensus sequence: CKCTGYGTAGYCCWKG |
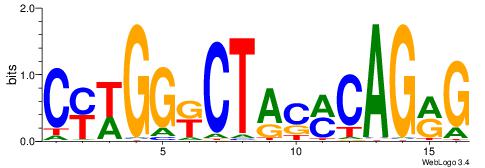 |
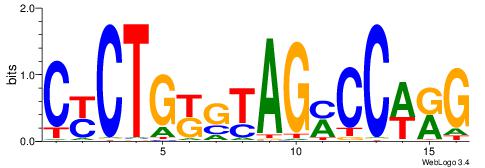 |
Best Matches for Motif ID 147 (Highest to Lowest)
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 16 |
| Similarity score: | 0.0270313 |
Alignment:
HTBVVVDGGACCACCCRGRDBG
----CYWGGKCTACMCAGRG--
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 16 |
| Similarity score: | 0.029263 |
Alignment:
DTVBDDRGACCACCCAGDWDGBH
---CYWGGKCTACMCAGRG----
| Original motif | Reverse complement motif |
| Consensus sequence: DTVBDDRGACCACCCAGDWDGBH | Consensus sequence: DBCDWHCTGGGTGGTCMDHBBAH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 16 |
| Similarity score: | 0.0296975 |
Alignment:
HGDHVHKGACCACCCGGBGBGDB
---CYWGGKCTACMCAGRG----
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 16 |
| Similarity score: | 0.0322159 |
Alignment:
HDGHHBHRGACCACCCGSVDVG
----CYWGGKCTACMCAGRG--
| Original motif | Reverse complement motif |
| Consensus sequence: HDGHHBHRGACCACCCGSVDVG | Consensus sequence: CVHBSCGGGTGGTCMHVHHCDH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 16 |
| Similarity score: | 0.0386994 |
Alignment:
BBBDVVRGACCACCCAVGABBAB
---CYWGGKCTACMCAGRG----
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Dataset #: 1 | Motif ID: 148 | Motif name: C148 |
| Original motif | Reverse complement motif |
| Consensus sequence: SCMSBGACACHGSCAS | Consensus sequence: STGSCDGTGTCBSYGS |
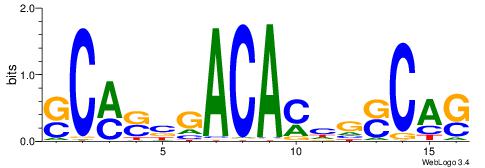 |
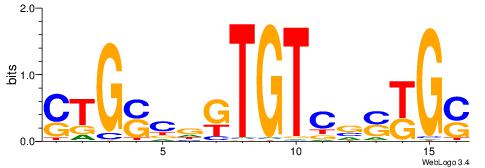 |
Best Matches for Motif ID 148 (Highest to Lowest)
| Motif ID: | UP00068 |
| Motif name: | Eomes_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0245227 |
Alignment:
VBVDAGGTGTYGVBBV
STGSCDGTGTCBSYGS
| Original motif | Reverse complement motif |
| Consensus sequence: VBVDAGGTGTYGVBBV | Consensus sequence: VVBVCKACACCTHVBV |
 |
 |
| Motif ID: | UP00100 |
| Motif name: | Gata6_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0325938 |
Alignment:
HKCBSCGATATCGVBBC
-STGSCDGTGTCBSYGS
| Original motif | Reverse complement motif |
| Consensus sequence: GBBVCGATATCGSVGYD | Consensus sequence: HKCBSCGATATCGVBBC |
 |
 |
| Motif ID: | UP00042 |
| Motif name: | Gm397_second |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0369917 |
Alignment:
DVMVGCACACACKCVH
SCMSBGACACHGSCAS
| Original motif | Reverse complement motif |
| Consensus sequence: DVMVGCACACACKCVH | Consensus sequence: HBGYGTGTGTGCVRVD |
 |
 |
| Motif ID: | UP00016 |
| Motif name: | Sry_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0372098 |
Alignment:
HHDHDRAACAATDDVHV
-SCMSBGACACHGSCAS
| Original motif | Reverse complement motif |
| Consensus sequence: HHDHDRAACAATDDVHV | Consensus sequence: VHVHDATTGTTMHDDDH |
 |
 |
| Motif ID: | UP00101 |
| Motif name: | Sox12_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.037346 |
Alignment:
VVDBASACAAAGRADH
SCMSBGACACHGSCAS
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBASACAAAGRADH | Consensus sequence: HDTMCTTTGTSTVDBB |
 |
 |
| Dataset #: 1 | Motif ID: 149 | Motif name: C149 |
| Original motif | Reverse complement motif |
| Consensus sequence: TKAACTCAGRMR | Consensus sequence: KYKCTGAGTTYA |
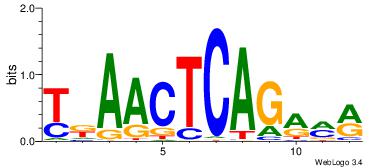 |
 |
Best Matches for Motif ID 149 (Highest to Lowest)
| Motif ID: | UP00148 |
| Motif name: | Hdx |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 0.0197225 |
Alignment:
BDBDHTGATTTCDHYDD
-KYKCTGAGTTYA----
| Original motif | Reverse complement motif |
| Consensus sequence: DDKDHGAAATCAHHBHV | Consensus sequence: BDBDHTGATTTCDHYDD |
 |
 |
| Motif ID: | UP00020 |
| Motif name: | Atf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0267132 |
Alignment:
DMDMTGACGTCAKHBD
KYKCTGAGTTYA----
| Original motif | Reverse complement motif |
| Consensus sequence: DBDRTGACGTCAYHRD | Consensus sequence: DMDMTGACGTCAKHBD |
 |
 |
| Motif ID: | UP00014 |
| Motif name: | Sox17_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0270855 |
Alignment:
DDBDKATGAATGHDBHV
--KYKCTGAGTTYA---
| Original motif | Reverse complement motif |
| Consensus sequence: VHBHHCATTCATRHVDD | Consensus sequence: DDBDKATGAATGHDBHV |
 |
 |
| Motif ID: | UP00064 |
| Motif name: | Sox18_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0271199 |
Alignment:
DBVBTRAATTYADDHV
KYKCTGAGTTYA----
| Original motif | Reverse complement motif |
| Consensus sequence: DBVBTRAATTYADDHV | Consensus sequence: VDHDTKAATTMABBBH |
 |
 |
| Motif ID: | UP00045 |
| Motif name: | Mafb_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 0.0301995 |
Alignment:
DMTWDGTCAGCADWTTH
-TKAACTCAGRMR----
| Original motif | Reverse complement motif |
| Consensus sequence: HAAWDTGCTGACDWARH | Consensus sequence: DMTWDGTCAGCADWTTH |
 |
 |
| Dataset #: 1 | Motif ID: 150 | Motif name: C150 |
| Original motif | Reverse complement motif |
| Consensus sequence: YYCMRSSACASYCARR | Consensus sequence: KKTGKSTGTSSKRGMM |
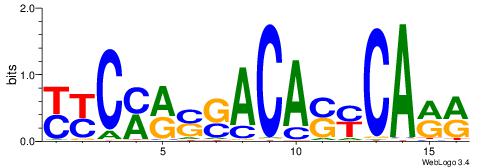 |
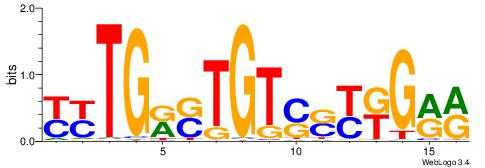 |
Best Matches for Motif ID 150 (Highest to Lowest)
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 16 |
| Similarity score: | 0.022327 |
Alignment:
ABBBBVVRGACCACCCACRDBBM
---YYCMRSSACASYCARR----
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 16 |
| Similarity score: | 0.0227877 |
Alignment:
BBBDVVRGACCACCCAVGABBAB
--YYCMRSSACASYCARR-----
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0235838 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
-YYCMRSSACASYCARR------
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 16 |
| Similarity score: | 0.0250268 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
-YYCMRSSACASYCARR------
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 16 |
| Similarity score: | 0.0265224 |
Alignment:
DTVBDDRGACCACCCAGDWDGBH
--YYCMRSSACASYCARR-----
| Original motif | Reverse complement motif |
| Consensus sequence: DTVBDDRGACCACCCAGDWDGBH | Consensus sequence: DBCDWHCTGGGTGGTCMDHBBAH |
 |
 |
| Dataset #: 1 | Motif ID: 151 | Motif name: C151 |
| Original motif | Reverse complement motif |
| Consensus sequence: SWGGCWGKCCWGGVWS | Consensus sequence: SWBCCWGGYCWGCCWS |
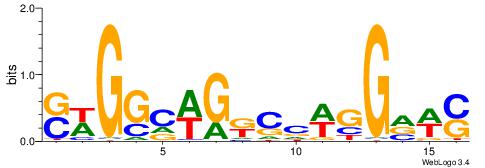 |
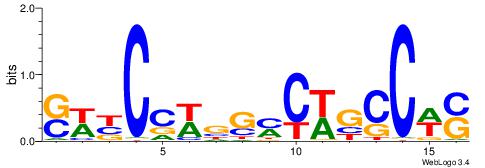 |
Best Matches for Motif ID 151 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 16 |
| Similarity score: | 0.0274084 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
-------SWGGCWGKCCWGGVWS
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0288558 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
SWBCCWGGYCWGCCWS------
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 16 |
| Similarity score: | 0.0368282 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
SWBCCWGGYCWGCCWS-------
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 16 |
| Similarity score: | 0.0424675 |
Alignment:
BBBDVVRGACCACCCAVGABBAB
-SWBCCWGGYCWGCCWS------
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 16 |
| Similarity score: | 0.04298 |
Alignment:
ABBBBVVRGACCACCCACRDBBM
--SWBCCWGGYCWGCCWS-----
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Dataset #: 1 | Motif ID: 152 | Motif name: C152 |
| Original motif | Reverse complement motif |
| Consensus sequence: KWRRKWRARAWMRAWM | Consensus sequence: YWTKYWTKTKWYKKWR |
 |
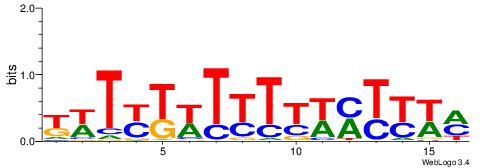 |
Best Matches for Motif ID 152 (Highest to Lowest)
| Motif ID: | UP00077 |
| Motif name: | Srf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0218448 |
Alignment:
HHVBYTTTTTTTTDDVV
YWTKYWTKTKWYKKWR-
| Original motif | Reverse complement motif |
| Consensus sequence: VBDDAAAAAAAAMVBHH | Consensus sequence: HHVBYTTTTTTTTDDVV |
 |
 |
| Motif ID: | UP00090 |
| Motif name: | Elf3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.038475 |
Alignment:
DDHBTTTTTTTTHDAVH
-YWTKYWTKTKWYKKWR
| Original motif | Reverse complement motif |
| Consensus sequence: DBTHHAAAAAAAAVHDH | Consensus sequence: DDHBTTTTTTTTHDAVH |
 |
 |
| Motif ID: | UP00051 |
| Motif name: | Sox8_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0392698 |
Alignment:
DDATBWATTGTTHHDDD
-YWTKYWTKTKWYKKWR
| Original motif | Reverse complement motif |
| Consensus sequence: DDATBWATTGTTHHDDD | Consensus sequence: DDDHDAACAATWBATDD |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0401556 |
Alignment:
HDTYTTGTTWTKDDDT
YWTKYWTKTKWYKKWR
| Original motif | Reverse complement motif |
| Consensus sequence: ADDDYAWAACAAMAHH | Consensus sequence: HDTYTTGTTWTKDDDT |
 |
 |
| Motif ID: | UP00255 |
| Motif name: | Dbx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0410382 |
Alignment:
HARTHAATTAWTAVDHD
KWRRKWRARAWMRAWM-
| Original motif | Reverse complement motif |
| Consensus sequence: HARTHAATTAWTAVDHD | Consensus sequence: DHDBTAWTAATTHAKTH |
 |
 |
| Dataset #: 1 | Motif ID: 153 | Motif name: C153 |
| Original motif | Reverse complement motif |
| Consensus sequence: YYAGRVTGGCBYTGR | Consensus sequence: MCAMBGCCAVMCTKM |
 |
 |
Best Matches for Motif ID 153 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 15 |
| Similarity score: | 0.0398267 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
------YYAGRVTGGCBYTGR--
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 15 |
| Similarity score: | 0.0410774 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
--MCAMBGCCAVMCTKM------
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00035 |
| Motif name: | Hic1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0449339 |
Alignment:
DDBHGGTTGGCATVBD
MCAMBGCCAVMCTKM-
| Original motif | Reverse complement motif |
| Consensus sequence: DBBATGCCAACCHVHH | Consensus sequence: DDBHGGTTGGCATVBD |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0460502 |
Alignment:
BBHBGAGTGGGAHHDB
-YYAGRVTGGCBYTGR
| Original motif | Reverse complement motif |
| Consensus sequence: BBHBGAGTGGGAHHDB | Consensus sequence: BHHDTCCCACTCBDBV |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0461303 |
Alignment:
VMYDHDGMCCHCCKBGVVAAVH
-MCAMBGCCAVMCTKM------
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |