Top 10 Significant Motifs - Global Matching (Highest to Lowest)
| Dataset #: 3 | Motif ID: 69 | Motif name: NKX3-1 |
| Original motif Consensus sequence: ATACTTA | Reverse complement motif Consensus sequence: TAAGTAT |
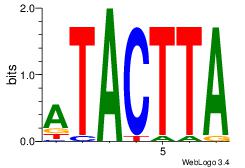 |
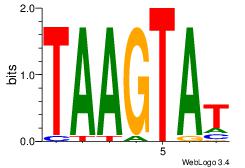 |
Best Matches for Top Significant Motif ID 69 (Highest to Lowest)
| Dataset #: | 4 |
| Motif ID: | 90 |
| Motif name: | ymTACATAyw |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 7 |
| Similarity score: | 0 |
Alignment:
DBTATGTAHH
--TAAGTAT-
| Original motif Consensus sequence: HHTACATABD | Reverse complement motif Consensus sequence: DBTATGTAHH |
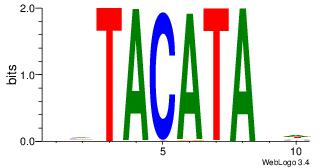 |
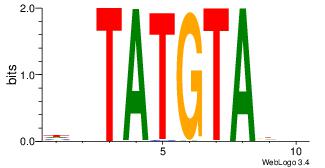 |
| Dataset #: | 1 |
| Motif ID: | 10 |
| Motif name: | Motif 10 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 7 |
| Similarity score: | 0.00274846 |
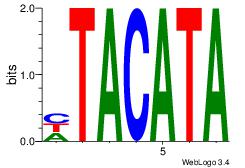
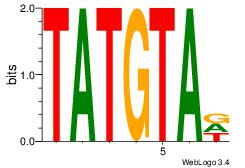
Alignment:
TATGTAD
TAAGTAT
| Original motif Consensus sequence: HTACATA | Reverse complement motif Consensus sequence: TATGTAD |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 34 |
| Motif name: | Motif 34 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 7 |
| Similarity score: | 0.00883486 |
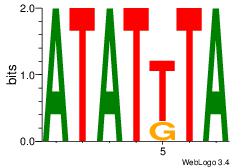
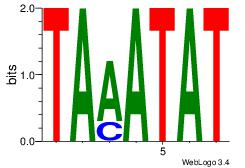
Alignment:
ATATTTA
ATACTTA
| Original motif Consensus sequence: ATATTTA | Reverse complement motif Consensus sequence: TAAATAT |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 106 |
| Motif name: | wtAATTAAtw |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 7 |
| Similarity score: | 0.0100679 |
Alignment:
HATTAATTAD
---TAAGTAT
| Original motif Consensus sequence: DTAATTAATH | Reverse complement motif Consensus sequence: HATTAATTAD |
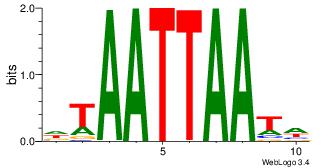 |
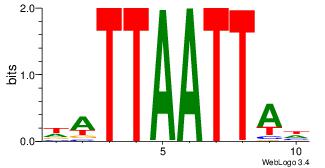 |
| Dataset #: | 4 |
| Motif ID: | 100 |
| Motif name: | wwTAATAAww |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 7 |
| Similarity score: | 0.0316418 |
Alignment:
DDTTATTAHD
--TAAGTAT-
| Original motif Consensus sequence: DHTAATAADD | Reverse complement motif Consensus sequence: DDTTATTAHD |
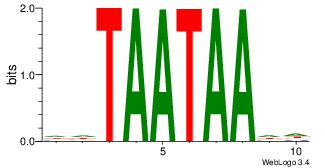 |
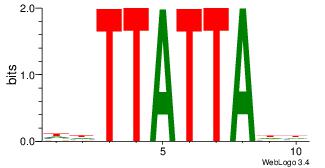 |
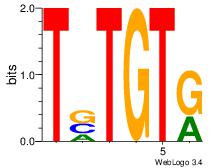
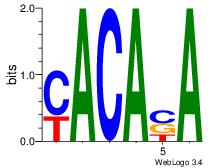
| Dataset #: 2 | Motif ID: 53 | Motif name: Motif 53 |
| Original motif Consensus sequence: TSTGTR | Reverse complement motif Consensus sequence: MACASA |
 |
 |
Best Matches for Top Significant Motif ID 53 (Highest to Lowest)
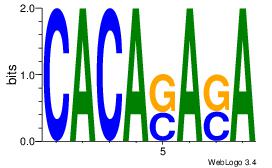
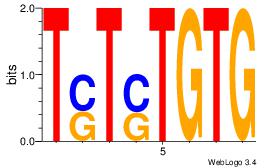
| Dataset #: | 1 |
| Motif ID: | 7 |
| Motif name: | Motif 7 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.00228262 |
Alignment:
CACASASA
--MACASA
| Original motif Consensus sequence: CACASASA | Reverse complement motif Consensus sequence: TSTSTGTG |
 |
 |
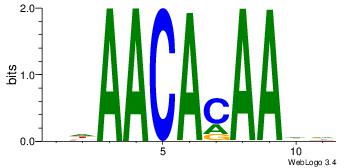
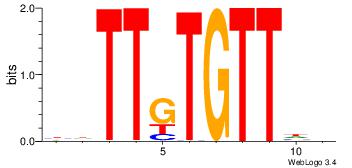
| Dataset #: | 4 |
| Motif ID: | 114 |
| Motif name: | awAACAcAAwa |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.0122629 |
Alignment:
DDAACACAADV
--MACASA---
| Original motif Consensus sequence: DDAACACAADV | Reverse complement motif Consensus sequence: BDTTGTGTTDD |
 |
 |
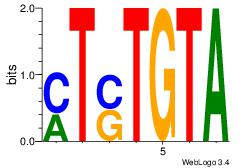
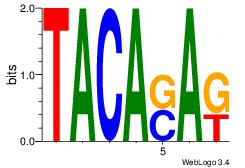
| Dataset #: | 1 |
| Motif ID: | 23 |
| Motif name: | Motif 23 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.0137023 |
Alignment:
MTSTGTA
-TSTGTR
| Original motif Consensus sequence: MTSTGTA | Reverse complement motif Consensus sequence: TACASAR |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 2 |
| Motif name: | Motif 2 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.0252334 |
Alignment:
GTGTGTGT
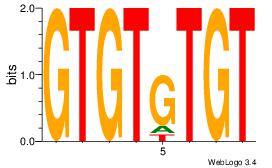
-TSTGTR-
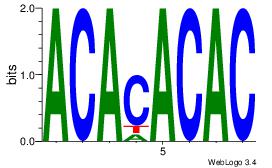
| Original motif Consensus sequence: ACACACAC | Reverse complement motif Consensus sequence: GTGTGTGT |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 12 |
| Motif name: | Motif 12 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.0262273 |
Alignment:
MTGTGTGC
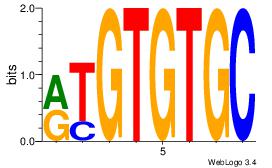
-TSTGTR-
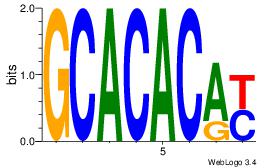
| Original motif Consensus sequence: GCACACAY | Reverse complement motif Consensus sequence: MTGTGTGC |
 |
 |
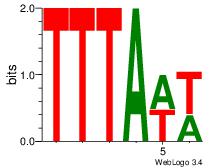
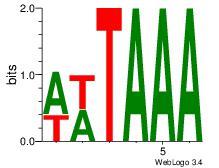
| Dataset #: 1 | Motif ID: 25 | Motif name: Motif 25 |
| Original motif Consensus sequence: TTTAWW | Reverse complement motif Consensus sequence: WWTAAA |
 |
 |
Best Matches for Top Significant Motif ID 25 (Highest to Lowest)
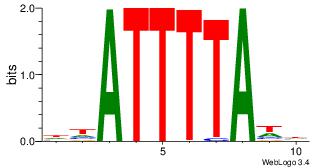
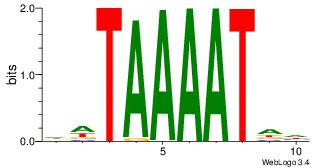
| Dataset #: | 4 |
| Motif ID: | 91 |
| Motif name: | wtATTTTAww |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0116421 |
Alignment:
HWTAAAATHD
WWTAAA----
| Original motif Consensus sequence: DHATTTTAWH | Reverse complement motif Consensus sequence: HWTAAAATHD |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 108 |
| Motif name: | wwATkTTTAww |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0118254 |
Alignment:
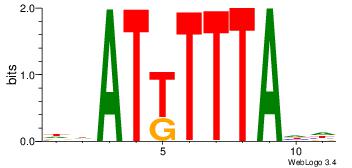
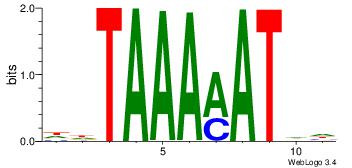
DHATKTTTAHH
-----TTTAWW
| Original motif Consensus sequence: DHATKTTTAHH | Reverse complement motif Consensus sequence: HHTAAARATHD |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 70 |
| Motif name: | Foxq1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0158082 |
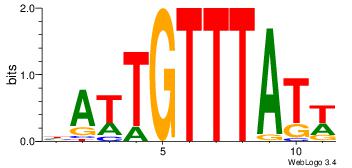
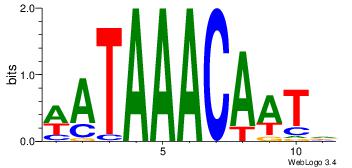
Alignment:
HATTGTTTATW
-----TTTAWW
| Original motif Consensus sequence: HATTGTTTATW | Reverse complement motif Consensus sequence: WATAAACAATH |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 55 |
| Motif name: | Motif 55 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0307675 |
Alignment:
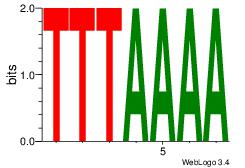
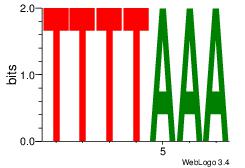
TTTTAAA
-TTTAWW
| Original motif Consensus sequence: TTTAAAA | Reverse complement motif Consensus sequence: TTTTAAA |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 67 |
| Motif name: | FOXI1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.0347747 |
Alignment:
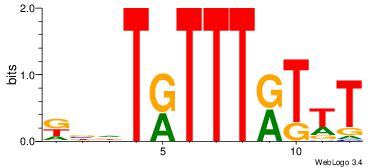
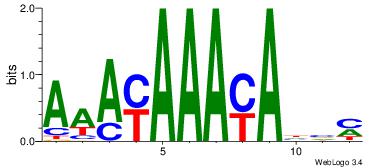
KBDTRTTTRTTT
-----TTTAWW-
| Original motif Consensus sequence: KBDTRTTTRTTT | Reverse complement motif Consensus sequence: AAAMAAAMADBY |
 |
 |
| Dataset #: 1 | Motif ID: 24 | Motif name: Motif 24 |
| Original motif Consensus sequence: CACGB | Reverse complement motif Consensus sequence: BCGTG |
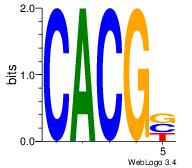 |
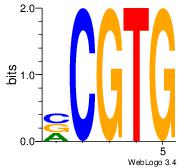 |
Best Matches for Top Significant Motif ID 24 (Highest to Lowest)
| Dataset #: | 2 |
| Motif ID: | 56 |
| Motif name: | Motif 56 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 5 |
| Similarity score: | 0 |
Alignment:
TCCGTG
-BCGTG
| Original motif Consensus sequence: CACGGA | Reverse complement motif Consensus sequence: TCCGTG |
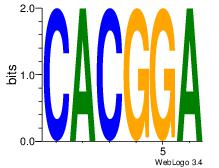 |
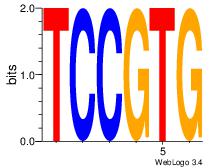 |
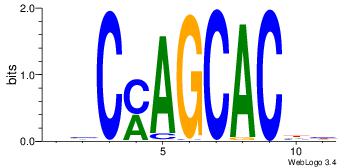
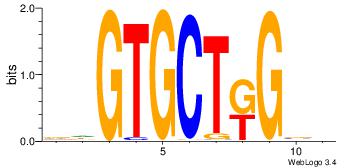
| Dataset #: | 4 |
| Motif ID: | 115 |
| Motif name: | ysCmAGCACwy |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 5 |
| Similarity score: | 0.0186397 |
Alignment:
DHGTGCTRGVH
BCGTG------
| Original motif Consensus sequence: HVCMAGCACHH | Reverse complement motif Consensus sequence: DHGTGCTRGVH |
 |
 |
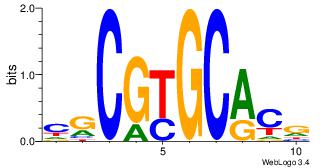
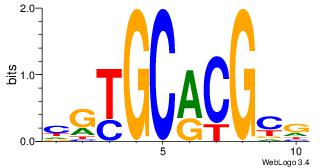
| Dataset #: | 4 |
| Motif ID: | 107 |
| Motif name: | crCGyGCRcg |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 5 |
| Similarity score: | 0.019278 |
Alignment:
CGTGCACGCG
----CACGB-
| Original motif Consensus sequence: CGCGTGCACG | Reverse complement motif Consensus sequence: CGTGCACGCG |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 58 |
| Motif name: | Motif 58 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 5 |
| Similarity score: | 0.0339655 |
Alignment:
CACACACACACACA
CACGB---------
| Original motif Consensus sequence: CACACACACACACA | Reverse complement motif Consensus sequence: TGTGTGTGTGTGTG |
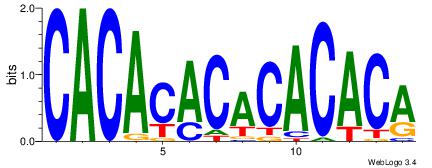 |
 |
| Dataset #: | 4 |
| Motif ID: | 93 |
| Motif name: | cmCARCACTwr |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.0385683 |
Alignment:
VHAGTGCTGHB
-BCGTG-----
| Original motif Consensus sequence: BHCAGCACTHV | Reverse complement motif Consensus sequence: VHAGTGCTGHB |
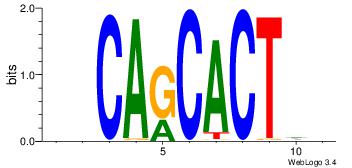 |
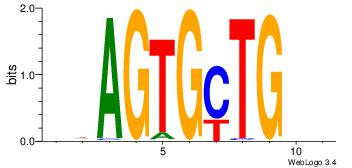 |
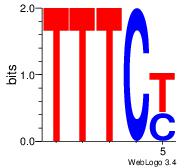
| Dataset #: 2 | Motif ID: 54 | Motif name: Motif 54 |
| Original motif Consensus sequence: RGAAA | Reverse complement motif Consensus sequence: TTTCK |
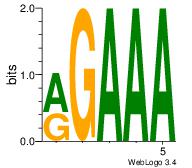 |
 |
Best Matches for Top Significant Motif ID 54 (Highest to Lowest)
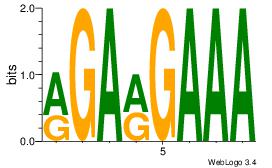
| Dataset #: | 1 |
| Motif ID: | 26 |
| Motif name: | Motif 26 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 5 |
| Similarity score: | 0 |
Alignment:
RGARGAAA
---RGAAA
| Original motif Consensus sequence: RGARGAAA | Reverse complement motif Consensus sequence: TTTCKTCK |
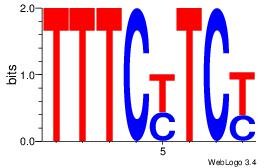
 |
 |
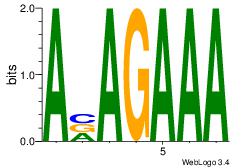
| Dataset #: | 1 |
| Motif ID: | 28 |
| Motif name: | Motif 28 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 5 |
| Similarity score: | 0.0199405 |
Alignment:
TTTCTVT
TTTCK--
| Original motif Consensus sequence: AVAGAAA | Reverse complement motif Consensus sequence: TTTCTVT |
 |
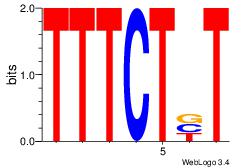 |
| Dataset #: | 1 |
| Motif ID: | 33 |
| Motif name: | Motif 33 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 5 |
| Similarity score: | 0.0284722 |
Alignment:
CMTTTCC
TTTCK--
| Original motif Consensus sequence: CMTTTCC | Reverse complement motif Consensus sequence: GGAAARG |
 |
 |
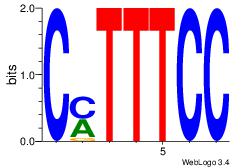
| Dataset #: | 1 |
| Motif ID: | 46 |
| Motif name: | Motif 46 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 5 |
| Similarity score: | 0.0284722 |
Alignment:
GGAAASA
RGAAA--
| Original motif Consensus sequence: GGAAASA | Reverse complement motif Consensus sequence: TSTTTCC |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 44 |
| Motif name: | Motif 44 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 5 |
| Similarity score: | 0.0339244 |
Alignment:
CATTTCT
--TTTCK
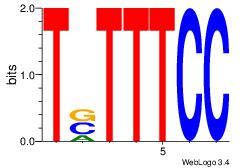
| Original motif Consensus sequence: AGAAATG | Reverse complement motif Consensus sequence: CATTTCT |
 |
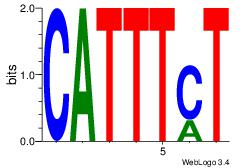 |
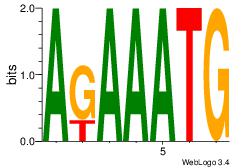
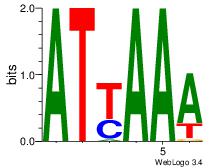
| Dataset #: 3 | Motif ID: 64 | Motif name: ARID3A |
| Original motif Consensus sequence: ATYAAA | Reverse complement motif Consensus sequence: TTTMAT |
 |
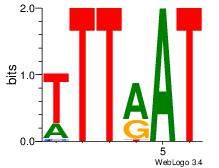 |
Best Matches for Top Significant Motif ID 64 (Highest to Lowest)
| Dataset #: | 4 |
| Motif ID: | 106 |
| Motif name: | wtAATTAAtw |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.0114914 |
Alignment:
DTAATTAATH
---ATYAAA-
| Original motif Consensus sequence: DTAATTAATH | Reverse complement motif Consensus sequence: HATTAATTAD |
 |
 |
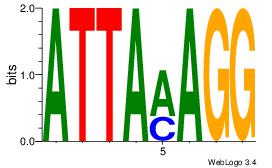
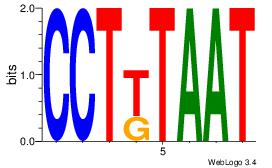
| Dataset #: | 1 |
| Motif ID: | 20 |
| Motif name: | Motif 20 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.015396 |
Alignment:
ATTAMAGG
ATYAAA--
| Original motif Consensus sequence: ATTAMAGG | Reverse complement motif Consensus sequence: CCTYTAAT |
 |
 |
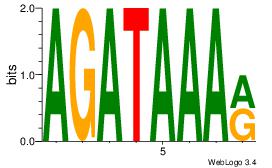
| Dataset #: | 1 |
| Motif ID: | 49 |
| Motif name: | Motif 49 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0340365 |
Alignment:
AGATAAAR
--ATYAAA
| Original motif Consensus sequence: AGATAAAR | Reverse complement motif Consensus sequence: KTTTATCT |
 |
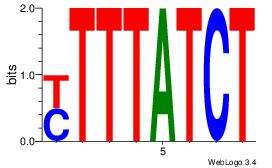 |
| Dataset #: | 1 |
| Motif ID: | 25 |
| Motif name: | Motif 25 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0368439 |
Alignment:
WWTAAA
ATYAAA
| Original motif Consensus sequence: TTTAWW | Reverse complement motif Consensus sequence: WWTAAA |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 100 |
| Motif name: | wwTAATAAww |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0394923 |
Alignment:
DHTAATAADD
TTTMAT----
| Original motif Consensus sequence: DHTAATAADD | Reverse complement motif Consensus sequence: DDTTATTAHD |
 |
 |
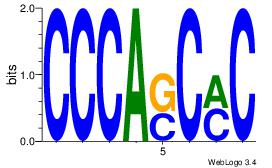
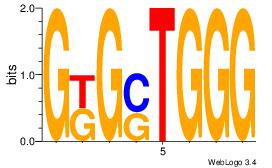
| Dataset #: 1 | Motif ID: 6 | Motif name: Motif 6 |
| Original motif Consensus sequence: CCCASCMC | Reverse complement motif Consensus sequence: GYGSTGGG |
 |
 |
Best Matches for Top Significant Motif ID 6 (Highest to Lowest)
| Dataset #: | 4 |
| Motif ID: | 115 |
| Motif name: | ysCmAGCACwy |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0212781 |
Alignment:
DHGTGCTRGVH
--GYGSTGGG-
| Original motif Consensus sequence: HVCMAGCACHH | Reverse complement motif Consensus sequence: DHGTGCTRGVH |
 |
 |
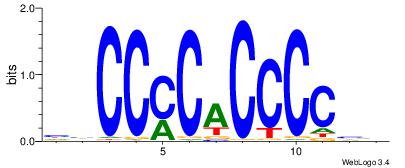
| Dataset #: | 4 |
| Motif ID: | 111 |
| Motif name: | ccCCmCaCCCCcc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0266863 |
Alignment:
HBCCCCACCCCBH
---CCCASCMC--
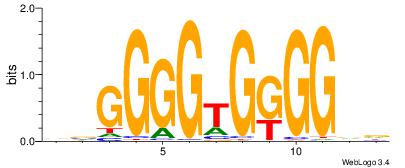
| Original motif Consensus sequence: HBCCCCACCCCBH | Reverse complement motif Consensus sequence: DBGGGGTGGGGBD |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 93 |
| Motif name: | cmCARCACTwr |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.030511 |
Alignment:
BHCAGCACTHV
CCCASCMC---
| Original motif Consensus sequence: BHCAGCACTHV | Reverse complement motif Consensus sequence: VHAGTGCTGHB |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 119 |
| Motif name: | cbCCAGCTCmyk |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0322039 |
Alignment:
BBCCAGCTCMBD
-CCCASCMC---
| Original motif Consensus sequence: BBCCAGCTCMBD | Reverse complement motif Consensus sequence: HBYGAGCTGGBB |
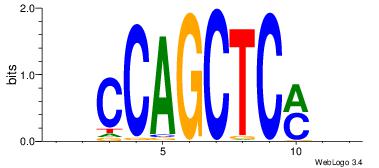 |
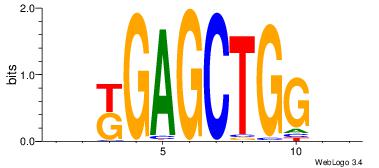 |
| Dataset #: | 4 |
| Motif ID: | 101 |
| Motif name: | csCACCCCgg |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.0415267 |
Alignment:
HVCACCCCBB
CCCASCMC--
| Original motif Consensus sequence: HVCACCCCBB | Reverse complement motif Consensus sequence: BBGGGGTGVD |
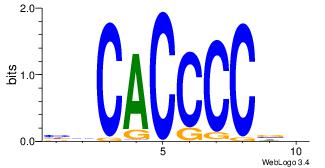 |
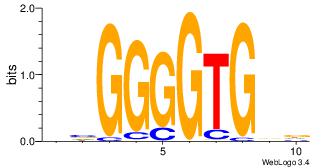 |
| Dataset #: 4 | Motif ID: 94 | Motif name: yrTCWCAAyr |
| Original motif Consensus sequence: HVTCACAAYD | Reverse complement motif Consensus sequence: HKTTGTGABH |
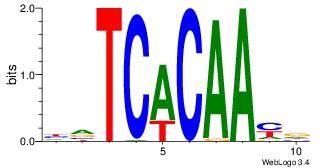 |
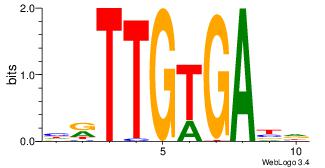 |
Best Matches for Top Significant Motif ID 94 (Highest to Lowest)
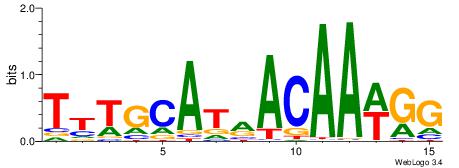
| Dataset #: | 3 |
| Motif ID: | 76 |
| Motif name: | Sox2 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.00762497 |
Alignment:
CCWTTGTYATGCAAA
----HKTTGTGABH-
| Original motif Consensus sequence: CCWTTGTYATGCAAA | Reverse complement motif Consensus sequence: TTTGCATMACAAWGG |
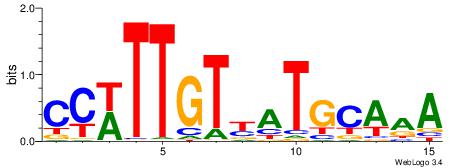 |
 |
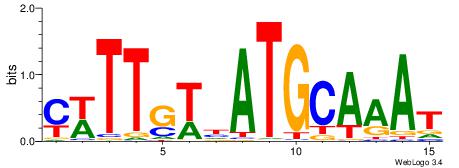
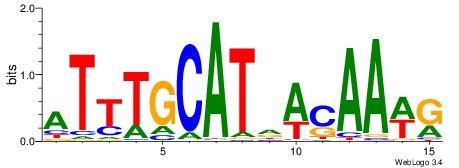
| Dataset #: | 3 |
| Motif ID: | 80 |
| Motif name: | Pou5f1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0127574 |
Alignment:
CWTTGTHATGCAAAT
-----HKTTGTGABH
| Original motif Consensus sequence: CWTTGTHATGCAAAT | Reverse complement motif Consensus sequence: ATTTGCATHACAAWG |
 |
 |
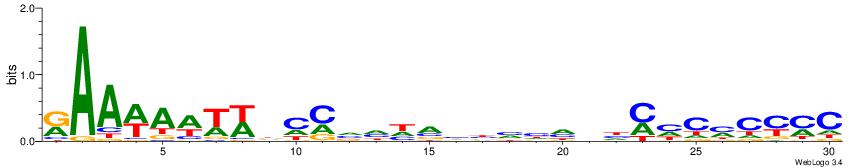
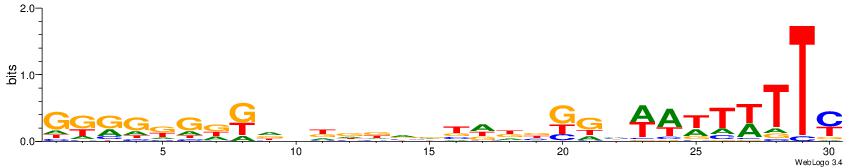
| Dataset #: | 3 |
| Motif ID: | 81 |
| Motif name: | Pax4 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0426407 |
Alignment:
RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC
-------------------HVTCACAAYD-
| Original motif Consensus sequence: RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC | Reverse complement motif Consensus sequence: GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM |
 |
 |
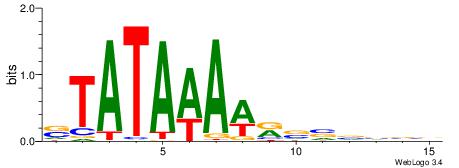
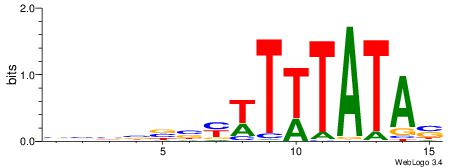
| Dataset #: | 3 |
| Motif ID: | 62 |
| Motif name: | TBP |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0429648 |
Alignment:
STATAAAWRVVVVBV
-HVTCACAAYD----
| Original motif Consensus sequence: STATAAAWRVVVVBV | Reverse complement motif Consensus sequence: VBVVVVMWTTTATAS |
 |
 |
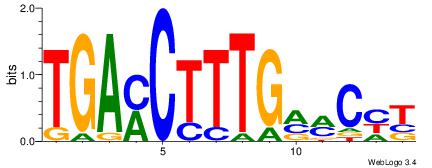
| Dataset #: | 3 |
| Motif ID: | 83 |
| Motif name: | NR2F1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0463536 |
Alignment:
TGAMCTTTGMMCYT
HKTTGTGABH----
| Original motif Consensus sequence: TGAMCTTTGMMCYT | Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
 |
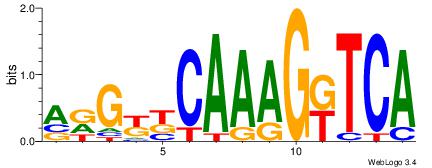 |
| Dataset #: 2 | Motif ID: 55 | Motif name: Motif 55 |
| Original motif Consensus sequence: TTTAAAA | Reverse complement motif Consensus sequence: TTTTAAA |
 |
 |
Best Matches for Top Significant Motif ID 55 (Highest to Lowest)
| Dataset #: | 4 |
| Motif ID: | 120 |
| Motif name: | wtTTTwAAAaw |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 7 |
| Similarity score: | 0.00286145 |
Alignment:
HHTTTWAAADD
--TTTAAAA--
| Original motif Consensus sequence: HHTTTWAAADD | Reverse complement motif Consensus sequence: DDTTTWAAAHH |
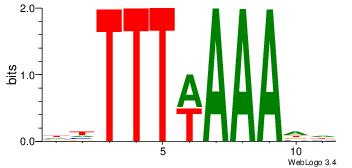 |
 |
| Dataset #: | 1 |
| Motif ID: | 9 |
| Motif name: | Motif 9 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 7 |
| Similarity score: | 0.0142562 |
Alignment:
TTTWWAAA
-TTTAAAA
| Original motif Consensus sequence: TTTWWAAA | Reverse complement motif Consensus sequence: TTTWWAAA |
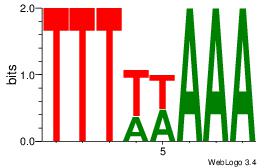 |
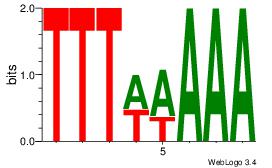 |
| Dataset #: | 4 |
| Motif ID: | 91 |
| Motif name: | wtATTTTAww |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 7 |
| Similarity score: | 0.036821 |
Alignment:
HWTAAAATHD
TTTAAAA---
| Original motif Consensus sequence: DHATTTTAWH | Reverse complement motif Consensus sequence: HWTAAAATHD |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 108 |
| Motif name: | wwATkTTTAww |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 7 |
| Similarity score: | 0.0428268 |
Alignment:
DHATKTTTAHH
----TTTTAAA
| Original motif Consensus sequence: DHATKTTTAHH | Reverse complement motif Consensus sequence: HHTAAARATHD |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 62 |
| Motif name: | TBP |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 7 |
| Similarity score: | 0.058454 |
Alignment:
VBVVVVMWTTTATAS
-------TTTTAAA-
| Original motif Consensus sequence: STATAAAWRVVVVBV | Reverse complement motif Consensus sequence: VBVVVVMWTTTATAS |
 |
 |
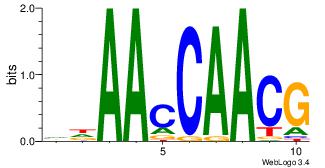
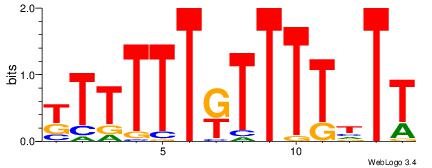
| Dataset #: 4 | Motif ID: 96 | Motif name: mdAAcCAACG |
| Original motif Consensus sequence: VDAACCAACG | Reverse complement motif Consensus sequence: CGTTGGTTDB |
 |
 |
Best Matches for Top Significant Motif ID 96 (Highest to Lowest)
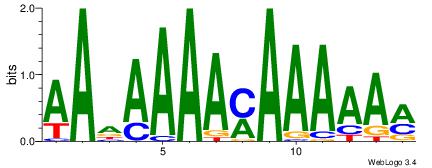
| Dataset #: | 2 |
| Motif ID: | 60 |
| Motif name: | Motif 60 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0244161 |
Alignment:
YTTTTTRTTTTDTT
--CGTTGGTTDB--
| Original motif Consensus sequence: AADAAAAMAAAAAM | Reverse complement motif Consensus sequence: YTTTTTRTTTTDTT |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 70 |
| Motif name: | Foxq1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0266383 |
Alignment:
HATTGTTTATW
-CGTTGGTTDB
| Original motif Consensus sequence: HATTGTTTATW | Reverse complement motif Consensus sequence: WATAAACAATH |
 |
 |
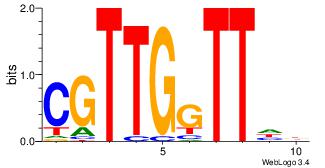
| Dataset #: | 3 |
| Motif ID: | 67 |
| Motif name: | FOXI1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.036655 |
Alignment:
AAAMAAAMADBY
VDAACCAACG--
| Original motif Consensus sequence: KBDTRTTTRTTT | Reverse complement motif Consensus sequence: AAAMAAAMADBY |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 83 |
| Motif name: | NR2F1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0376121 |
Alignment:
TGAMCTTTGMMCYT
CGTTGGTTDB----
| Original motif Consensus sequence: TGAMCTTTGMMCYT | Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 76 |
| Motif name: | Sox2 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0382008 |
Alignment:
TTTGCATMACAAWGG
VDAACCAACG-----
| Original motif Consensus sequence: CCWTTGTYATGCAAA | Reverse complement motif Consensus sequence: TTTGCATMACAAWGG |
 |
 |