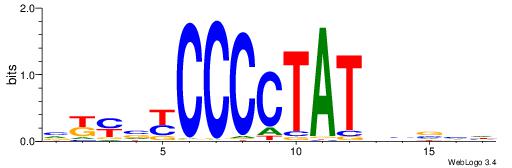
Best Matches in Database for Each Motif (Highest to Lowest)
| Dataset #: 1 | Motif ID: 1 | Motif name: Motif 1 |
| Original motif | Reverse complement motif |
| Consensus sequence: AGRKGGCR | Consensus sequence: KGCCYKCT |
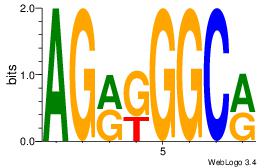 |
 |
Best Matches for Motif ID 1 (Highest to Lowest)
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
BVVHAGGGGGCGRDHHB
----AGRKGGCR-----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0152094 |
Alignment:
VKBBAGGGGTCAHDBBH
----AGRKGGCR-----
| Original motif | Reverse complement motif |
| Consensus sequence: VKBBAGGGGTCAHDBBH | Consensus sequence: DBBHHTGACCCCTBBYV |
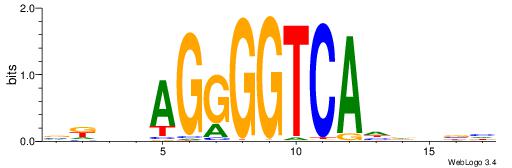 |
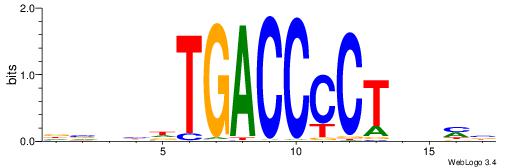 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0160089 |
Alignment:
VHTGCGGGGGCGGH
----AGRKGGCR--
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
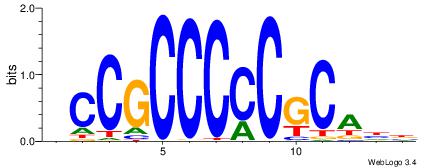 |
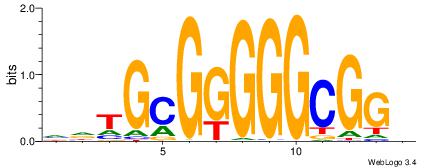 |
| Motif ID: | UP00010 |
| Motif name: | Tcfap2b_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0244919 |
Alignment:
HTGCCMTVKGGCMD
-KGCCYKCT-----
| Original motif | Reverse complement motif |
| Consensus sequence: DYGCCYBARGGCAH | Consensus sequence: HTGCCMTVKGGCMD |
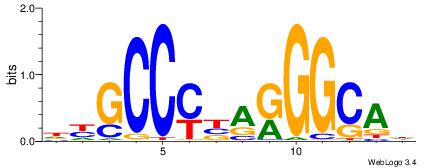 |
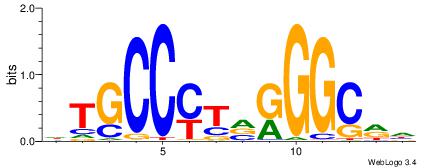 |
| Motif ID: | UP00070 |
| Motif name: | Gcm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0267457 |
Alignment:
DDBVHATAGGGGRBRMB
-------AGRKGGCR--
| Original motif | Reverse complement motif |
| Consensus sequence: DDBVHATAGGGGRBRMB | Consensus sequence: BYMBKCCCCTATDVBHD |
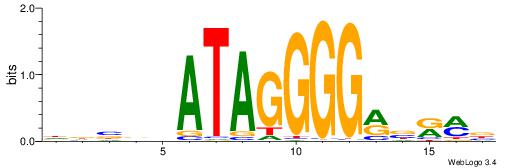 |
 |
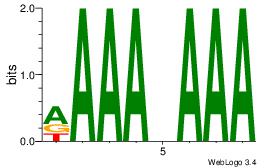
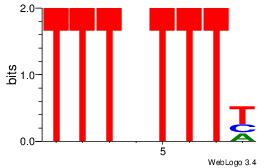
| Dataset #: 1 | Motif ID: 2 | Motif name: Motif 2 |
| Original motif | Reverse complement motif |
| Consensus sequence: AAAAHAAA | Consensus sequence: TTTDTTTT |
 |
 |
Best Matches for Motif ID 2 (Highest to Lowest)
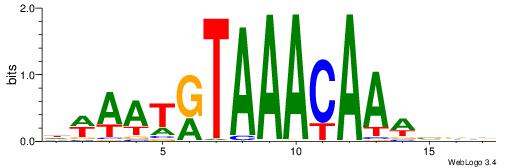
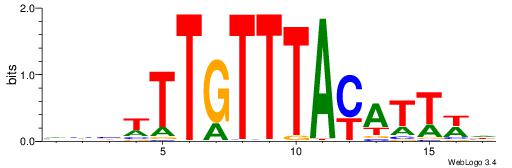
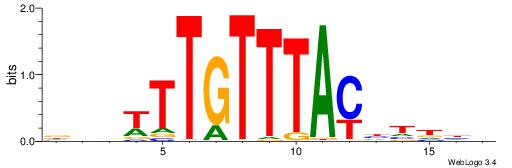
| Motif ID: | UP00061 |
| Motif name: | Foxl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
VVVWTTGTTTAMATTWD
---TTTDTTTT------
| Original motif | Reverse complement motif |
| Consensus sequence: DWAATRTAAACAAWVVB | Consensus sequence: VVVWTTGTTTAMATTWD |
 |
 |
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.000495784 |
Alignment:
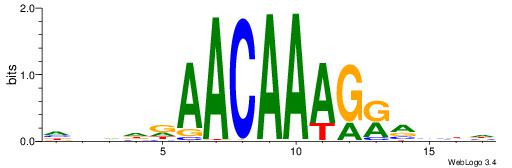
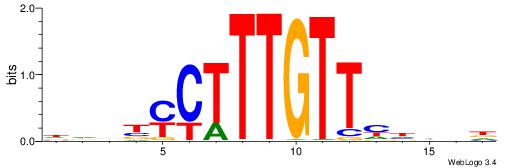
HBDDDAACAAAGRVBHH
---AAAAHAAA------
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
 |
 |
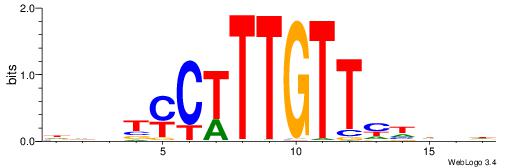
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.000916442 |
Alignment:
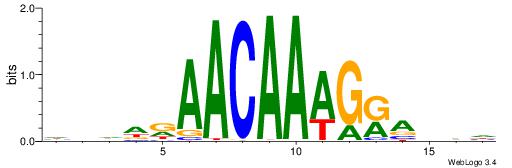
HBDDRAACAAAGRABHH
---AAAAHAAA------
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
 |
 |
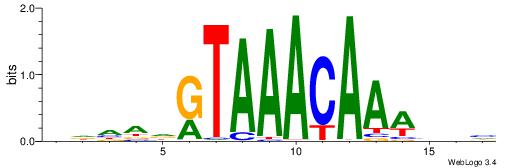
| Motif ID: | UP00039 |
| Motif name: | Foxj3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.00246818 |
Alignment:
HDHADGTAAACAAAVVH
------AAAAHAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: HDHADGTAAACAAAVVH | Consensus sequence: DVVTTTGTTTACDTHDH |
 |
 |
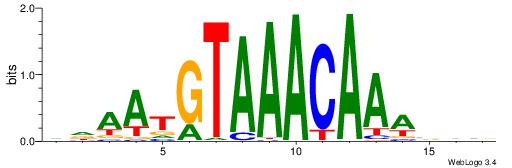
| Motif ID: | UP00025 |
| Motif name: | Foxk1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.00320862 |
Alignment:
DDAATGTAAACAAAVVB
------AAAAHAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: DDAATGTAAACAAAVVB | Consensus sequence: BBVTTTGTTTACATTDD |
 |
 |
| Dataset #: 1 | Motif ID: 3 | Motif name: Motif 3 |
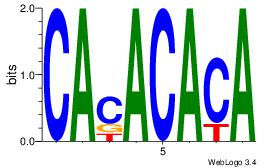
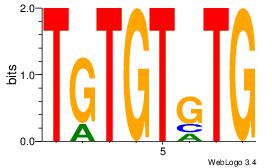
| Original motif | Reverse complement motif |
| Consensus sequence: CACACACA | Consensus sequence: TGTGTGTG |
 |
 |
Best Matches for Motif ID 3 (Highest to Lowest)
| Motif ID: | UP00042 |
| Motif name: | Gm397_second |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
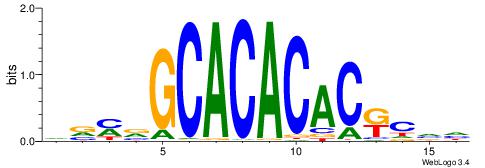
Alignment:
HBGYGTGTGTGCVRVD
---TGTGTGTG-----
| Original motif | Reverse complement motif |
| Consensus sequence: DVMVGCACACACKCVH | Consensus sequence: HBGYGTGTGTGCVRVD |
 |
 |
| Motif ID: | UP00026 |
| Motif name: | Zscan4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0271943 |
Alignment:
BHHHTTGTGTGCKHVD
-----TGTGTGTG---
| Original motif | Reverse complement motif |
| Consensus sequence: HVHRGCACACAAHHHV | Consensus sequence: BHHHTTGTGTGCKHVD |
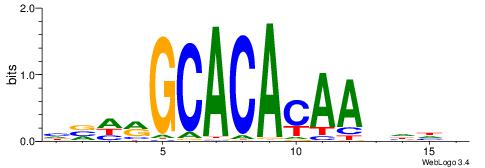 |
 |
| Motif ID: | UP00026 |
| Motif name: | Zscan4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.0336839 |
Alignment:
DHVDTGTGCACAYAHDH
--------CACACACA-
| Original motif | Reverse complement motif |
| Consensus sequence: DHVDTGTGCACAYAHDH | Consensus sequence: HDHTMTGTGCACADVHD |
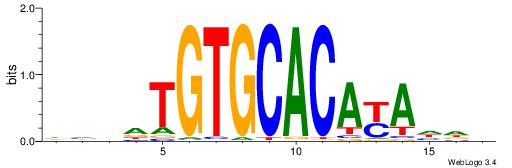 |
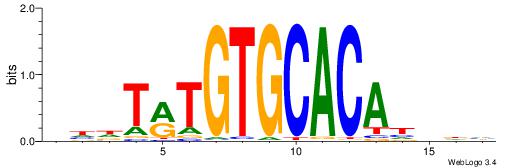 |
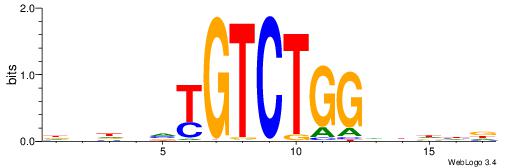
| Motif ID: | UP00000 |
| Motif name: | Smad3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.0355485 |
Alignment:
HVHBVTGTCTGGDDHDD
---TGTGTGTG------
| Original motif | Reverse complement motif |
| Consensus sequence: HDHDDCCAGACABBHVH | Consensus sequence: HVHBVTGTCTGGDDHDD |
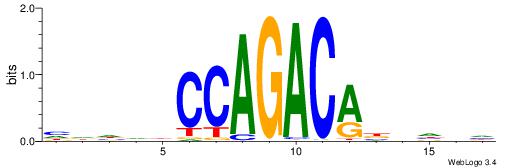 |
 |
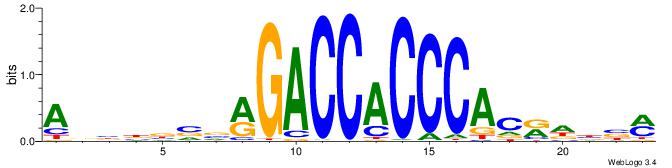
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 12 |
| Number of overlap: | 8 |
| Similarity score: | 0.0404116 |
Alignment:
ABBBBVVRGACCACCCACRDBBM
-----------CACACACA----
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
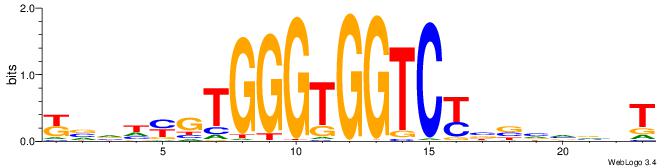 |
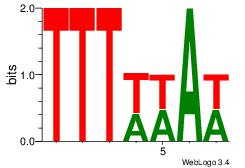
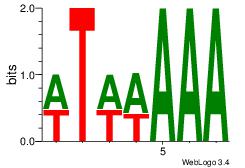
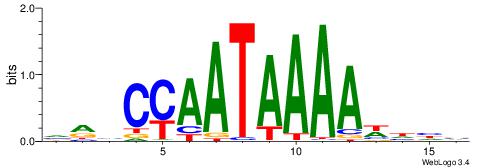
| Dataset #: 1 | Motif ID: 4 | Motif name: Motif 4 |
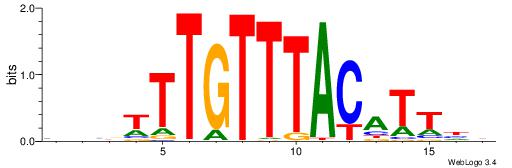
| Original motif | Reverse complement motif |
| Consensus sequence: TTTWWAW | Consensus sequence: WTWWAAA |
 |
 |
Best Matches for Motif ID 4 (Highest to Lowest)
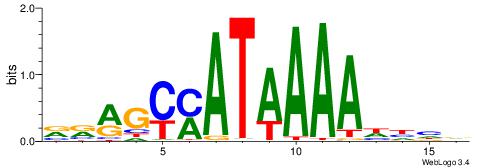
| Motif ID: | UP00207 |
| Motif name: | Hoxb9 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 7 |
| Similarity score: | 0 |
Alignment:
HDHHTTTTATRGCKMV
---TTTWWAW------
| Original motif | Reverse complement motif |
| Consensus sequence: VRRGCMATAAAAHHHD | Consensus sequence: HDHHTTTTATRGCKMV |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 7 |
| Similarity score: | 0.000691918 |
Alignment:
VVVWTTGTTTAMATTWD
-------TTTWWAW---
| Original motif | Reverse complement motif |
| Consensus sequence: DWAATRTAAACAAWVVB | Consensus sequence: VVVWTTGTTTAMATTWD |
 |
 |
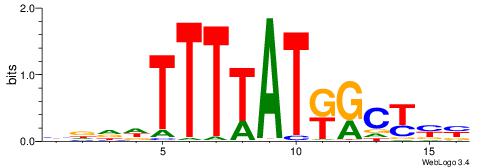
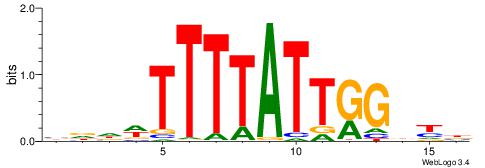
| Motif ID: | UP00134 |
| Motif name: | Hoxb13 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 7 |
| Similarity score: | 0.00184871 |
Alignment:
VDHWTTTTATTGGDKB
---TTTWWAW------
| Original motif | Reverse complement motif |
| Consensus sequence: VRHCCAATAAAAWHHV | Consensus sequence: VDHWTTTTATTGGDKB |
 |
 |
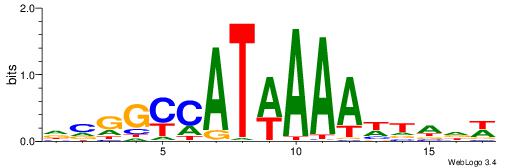
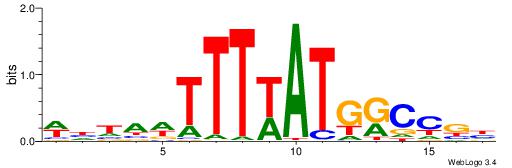
| Motif ID: | UP00213 |
| Motif name: | Hoxa9 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 7 |
| Similarity score: | 0.00275206 |
Alignment:
WDTAWTTTWATGKCCGD
----TTTWWAW------
| Original motif | Reverse complement motif |
| Consensus sequence: DCGGYCATWAAAWTADW | Consensus sequence: WDTAWTTTWATGKCCGD |
 |
 |
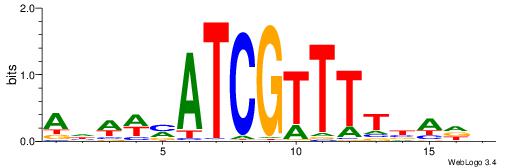
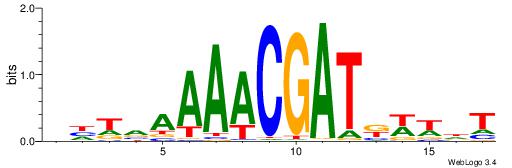
| Motif ID: | UP00114 |
| Motif name: | Homez |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 7 |
| Similarity score: | 0.00499979 |
Alignment:
AHWWMATCGTTTTBADD
---------TTTWWAW-
| Original motif | Reverse complement motif |
| Consensus sequence: AHWWMATCGTTTTBADD | Consensus sequence: HDTVAAAACGATRWWHT |
 |
 |
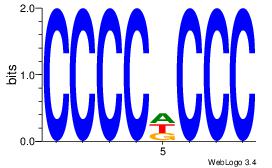
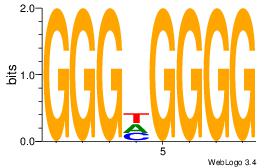
| Dataset #: 1 | Motif ID: 5 | Motif name: Motif 5 |
| Original motif | Reverse complement motif |
| Consensus sequence: CCCCDCCC | Consensus sequence: GGGDGGGG |
 |
 |
Best Matches for Motif ID 5 (Highest to Lowest)
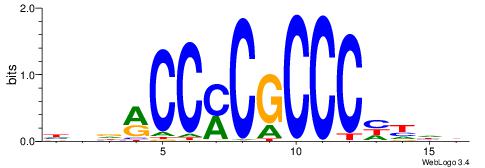
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
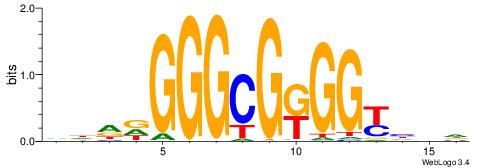
HBDRCCMCGCCCHHHD
----CCCCDCCC----
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0165797 |
Alignment:
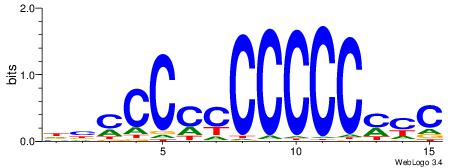
GKRGGGGGGGGGGBD
-----GGGDGGGG--
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.026633 |
Alignment:
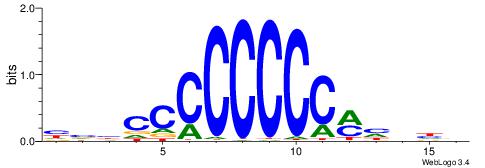
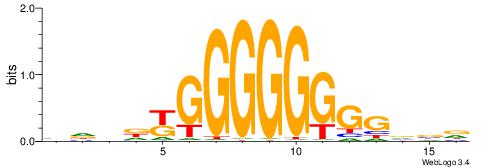
BHHDYGGGGGGGGBVD
-----GGGDGGGG---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
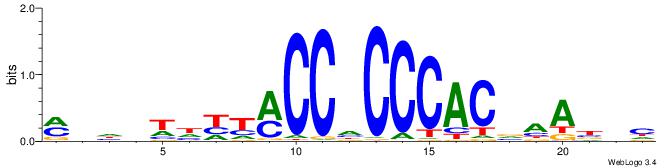
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0273225 |
Alignment:
MHHHWHYTMCCHCCCACVAABVH
-------CCCCDCCC--------
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
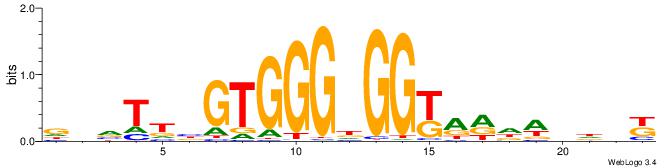 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0364166 |
Alignment:
VHTGCGGGGGCGGH
-----GGGDGGGG-
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
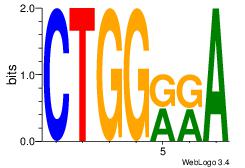
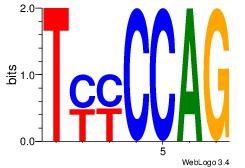
| Dataset #: 1 | Motif ID: 6 | Motif name: Motif 6 |
| Original motif | Reverse complement motif |
| Consensus sequence: CTGGRRA | Consensus sequence: TMMCCAG |
 |
 |
Best Matches for Motif ID 6 (Highest to Lowest)
| Motif ID: | UP00000 |
| Motif name: | Smad3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 7 |
| Similarity score: | 0.00121838 |
Alignment:
HVHBVTGTCTGGDDHDD
--------CTGGRRA--
| Original motif | Reverse complement motif |
| Consensus sequence: HDHDDCCAGACABBHVH | Consensus sequence: HVHBVTGTCTGGDDHDD |
 |
 |
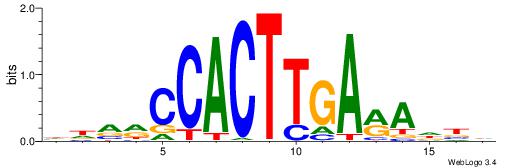
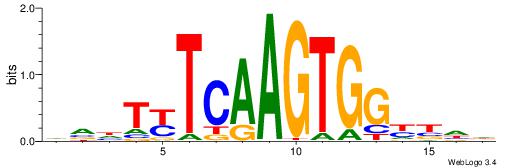
| Motif ID: | UP00231 |
| Motif name: | Nkx2-2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 7 |
| Similarity score: | 0.00386806 |
Alignment:
HDVRCCACTTGARADBD
-------CTGGRRA---
| Original motif | Reverse complement motif |
| Consensus sequence: HDVRCCACTTGARADBD | Consensus sequence: DVDTKTCAAGTGGKBDH |
 |
 |
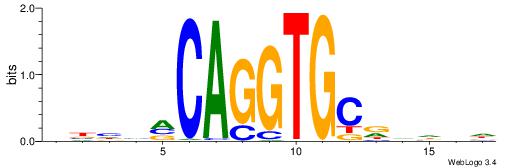
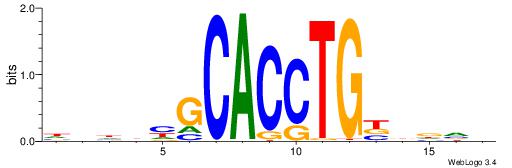
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 7 |
| Similarity score: | 0.00420562 |
Alignment:
VBHHVCAGGTGCDVDHD
-TMMCCAG---------
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Motif ID: | UP00161 |
| Motif name: | Hmbox1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.00428647 |
Alignment:
BDDBTTAACTAGWDKHV
-----TMMCCAG-----
| Original motif | Reverse complement motif |
| Consensus sequence: VHRDWCTAGTTAABDDB | Consensus sequence: BDDBTTAACTAGWDKHV |
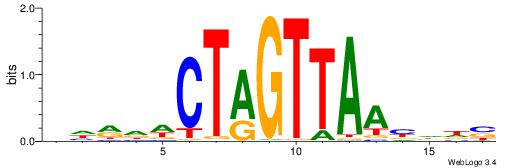 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 7 |
| Similarity score: | 0.010788 |
Alignment:
BDBBVBVGTAWCCAAGVBHBAGB
--------TMMCCAG--------
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
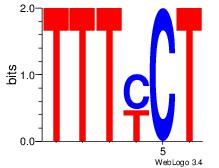
| Dataset #: 1 | Motif ID: 7 | Motif name: Motif 7 |
| Original motif | Reverse complement motif |
| Consensus sequence: AGRAAA | Consensus sequence: TTTMCT |
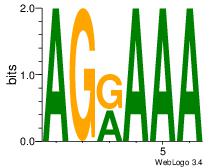 |
 |
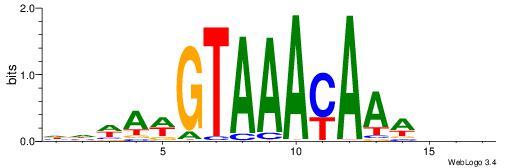
Best Matches for Motif ID 7 (Highest to Lowest)
| Motif ID: | UP00073 |
| Motif name: | Foxa2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
BDVTTTKTTTACWTWHH
-------TTTMCT----
| Original motif | Reverse complement motif |
| Consensus sequence: HHWAWGTAAAYAAAVDB | Consensus sequence: BDVTTTKTTTACWTWHH |
 |
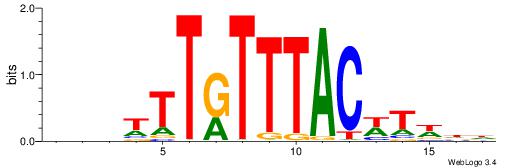 |
| Motif ID: | UP00039 |
| Motif name: | Foxj3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.0125029 |
Alignment:
DVVTTTGTTTACDTHDH
-------TTTMCT----
| Original motif | Reverse complement motif |
| Consensus sequence: HDHADGTAAACAAAVVH | Consensus sequence: DVVTTTGTTTACDTHDH |
 |
 |
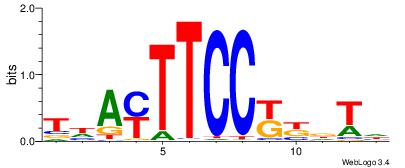
| Motif ID: | UP00090 |
| Motif name: | Elf3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.0161149 |
Alignment:
DAHMMGGAARTDA
----AGRAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: DAHMMGGAARTDA | Consensus sequence: TDAMTTCCYYDTD |
 |
 |
| Motif ID: | UP00025 |
| Motif name: | Foxk1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0165207 |
Alignment:
BBVTTTGTTTACATTDD
-------TTTMCT----
| Original motif | Reverse complement motif |
| Consensus sequence: DDAATGTAAACAAAVVB | Consensus sequence: BBVTTTGTTTACATTDD |
 |
 |
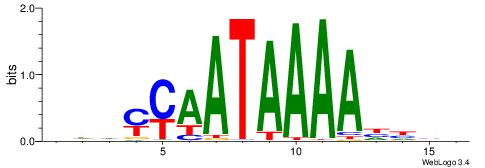
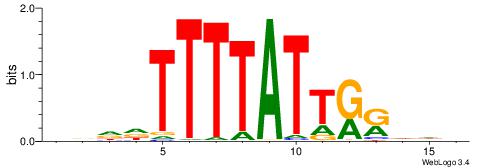
| Motif ID: | UP00180 |
| Motif name: | Hoxd13 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0170044 |
Alignment:
HDHYYAATAAAAHHHH
------AGRAAA----
| Original motif | Reverse complement motif |
| Consensus sequence: HDHYYAATAAAAHHHH | Consensus sequence: HDHHTTTTATTKKHDD |
 |
 |
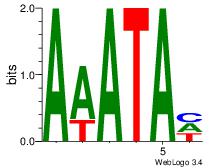
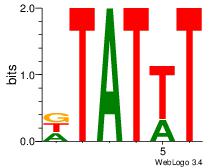
| Dataset #: 1 | Motif ID: 8 | Motif name: Motif 8 |
| Original motif | Reverse complement motif |
| Consensus sequence: AAATAH | Consensus sequence: DTATTT |
 |
 |
Best Matches for Motif ID 8 (Highest to Lowest)
| Motif ID: | UP00073 |
| Motif name: | Foxa2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
HHWAWGTAAAYAAAVDB
-------AAATAH----
| Original motif | Reverse complement motif |
| Consensus sequence: HHWAWGTAAAYAAAVDB | Consensus sequence: BDVTTTKTTTACWTWHH |
 |
 |
| Motif ID: | UP00166 |
| Motif name: | Barhl1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.00310877 |
Alignment:
DDVKTAATTGGTTDTH
---DTATTT-------
| Original motif | Reverse complement motif |
| Consensus sequence: HAHAACCAATTARBDH | Consensus sequence: DDVKTAATTGGTTDTH |
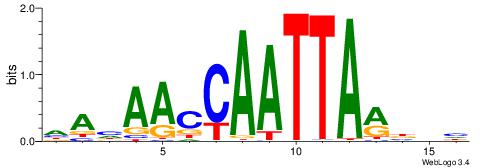 |
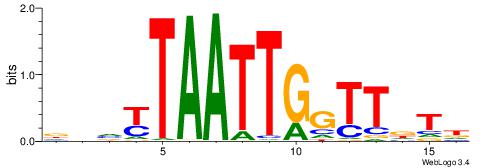 |
| Motif ID: | UP00242 |
| Motif name: | Hoxc8 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.00420475 |
Alignment:
BHDDDGTAATTAHHDT
-------AAATAH---
| Original motif | Reverse complement motif |
| Consensus sequence: BHDDDGTAATTAHHDT | Consensus sequence: AHDHTAATTACHHHHV |
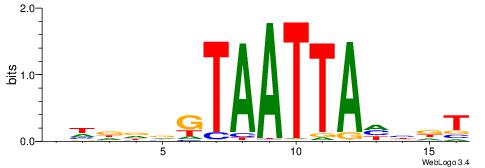 |
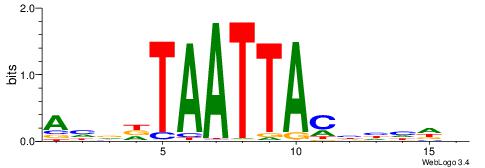 |
| Motif ID: | UP00179 |
| Motif name: | Pou2f3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.00508605 |
Alignment:
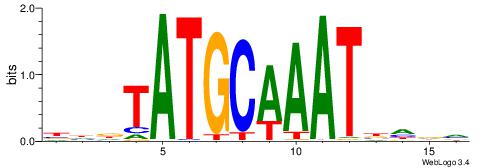
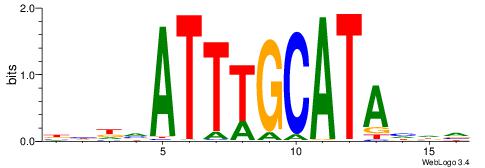
BHDTATGCAAATBHDV
--------AAATAH--
| Original motif | Reverse complement motif |
| Consensus sequence: BHDTATGCAAATBHDV | Consensus sequence: BHHVATTTGCATAHHV |
 |
 |
| Motif ID: | UP00202 |
| Motif name: | Dlx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.00517242 |
Alignment:
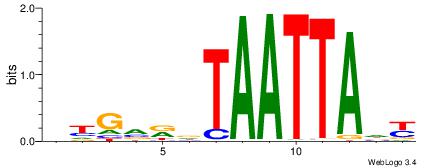
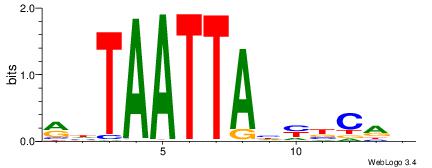
MDTAATTAHMBCHB
---AAATAH-----
| Original motif | Reverse complement motif |
| Consensus sequence: BHGVRDTAATTADY | Consensus sequence: MDTAATTAHMBCHB |
 |
 |
| Dataset #: 1 | Motif ID: 9 | Motif name: Motif 9 |
| Original motif | Reverse complement motif |
| Consensus sequence: GGTGGCWS | Consensus sequence: SWGCCACC |
 |
 |
Best Matches for Motif ID 9 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 11 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
----------GGTGGCWS-----
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0128982 |
Alignment:
HTBVVVDGGACCACCCRGRDBG
-------SWGCCACC-------
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
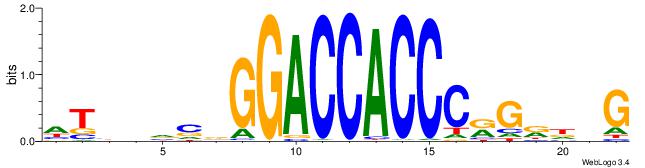 |
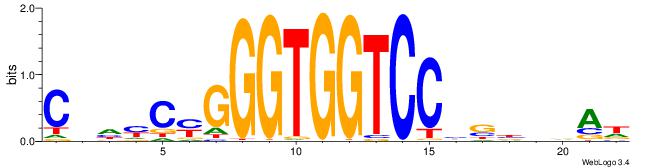 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 10 |
| Number of overlap: | 8 |
| Similarity score: | 0.0130995 |
Alignment:
HGDHVHKGACCACCCGGBGBGDB
------SWGCCACC---------
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
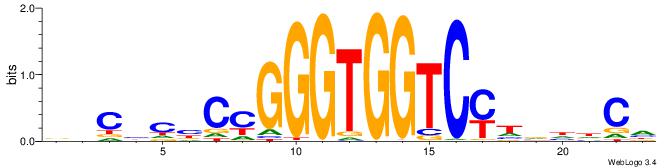 |
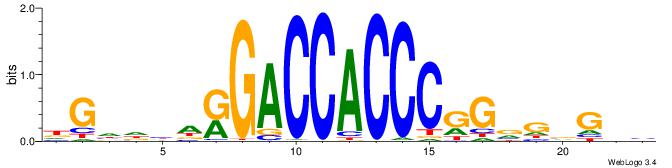 |
| Motif ID: | UP00035 |
| Motif name: | Hic1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0142671 |
Alignment:
DBBATGCCAACCHVHH
---SWGCCACC-----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBATGCCAACCHVHH | Consensus sequence: DDBHGGTTGGCATVBD |
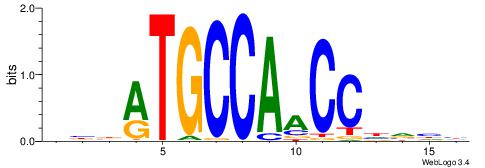 |
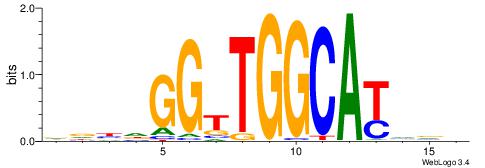 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0142765 |
Alignment:
HDGHHBHRGACCACCCGSVDVG
-------SWGCCACC-------
| Original motif | Reverse complement motif |
| Consensus sequence: HDGHHBHRGACCACCCGSVDVG | Consensus sequence: CVHBSCGGGTGGTCMHVHHCDH |
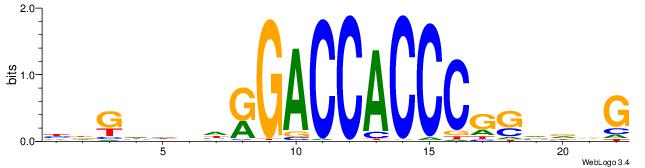 |
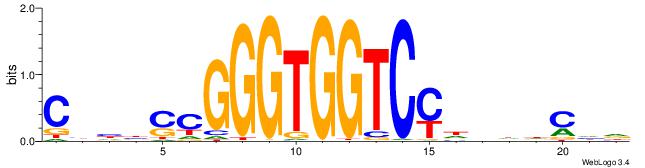 |
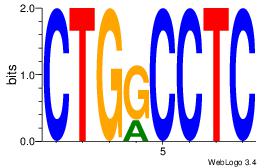
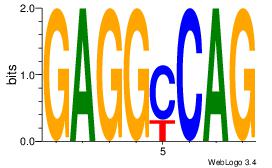
| Dataset #: 1 | Motif ID: 10 | Motif name: Motif 10 |
| Original motif | Reverse complement motif |
| Consensus sequence: CTGGCCTC | Consensus sequence: GAGGCCAG |
 |
 |
Best Matches for Motif ID 10 (Highest to Lowest)
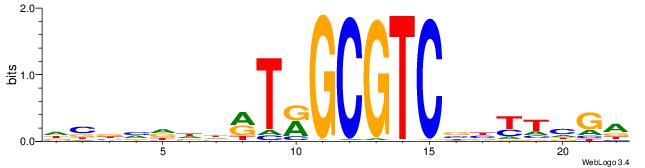
| Motif ID: | UP00527 |
| Motif name: | Foxn4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
TCDAMVBGACGCMAKDDDVBBD
-------GAGGCCAG-------
| Original motif | Reverse complement motif |
| Consensus sequence: DBBVDDDRTRGCGTCBBYTHGA | Consensus sequence: TCDAMVBGACGCMAKDDDVBBD |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.000107403 |
Alignment:
DBBHHTGACCCCTBBYV
----CTGGCCTC-----
| Original motif | Reverse complement motif |
| Consensus sequence: VKBBAGGGGTCAHDBBH | Consensus sequence: DBBHHTGACCCCTBBYV |
 |
 |
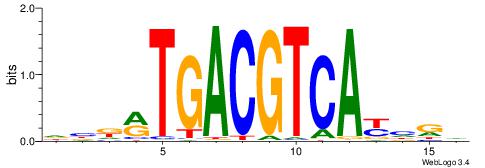
| Motif ID: | UP00020 |
| Motif name: | Atf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
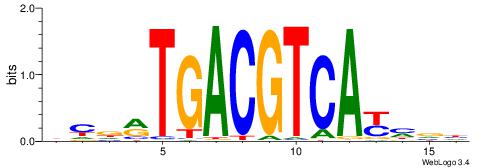
| Number of overlap: | 8 |
| Similarity score: | 0.00300031 |
Alignment:
DMDMTGACGTCAKHBD
---CTGGCCTC-----
| Original motif | Reverse complement motif |
| Consensus sequence: DBDRTGACGTCAYHRD | Consensus sequence: DMDMTGACGTCAKHBD |
 |
 |
| Motif ID: | UP00053 |
| Motif name: | Rxra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
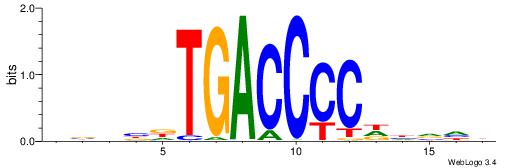
| Number of overlap: | 8 |
| Similarity score: | 0.00367302 |
Alignment:
DBHBDTGACCCCDHVHB
----CTGGCCTC-----
| Original motif | Reverse complement motif |
| Consensus sequence: DBHBDTGACCCCDHVHB | Consensus sequence: VHBHDGGGGTCAHBHBD |
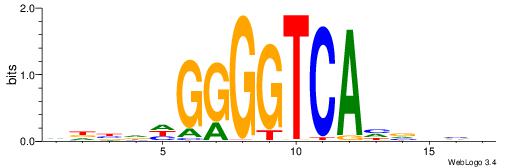
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
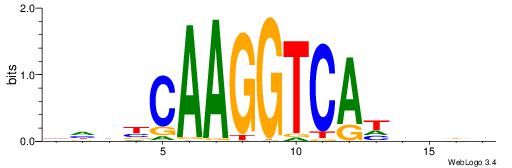
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.00526703 |
Alignment:
HHBVHTGACCTTGVDHD
----CTGGCCTC-----
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBCAAGGTCAHVBDH | Consensus sequence: HHBVHTGACCTTGVDHD |
 |
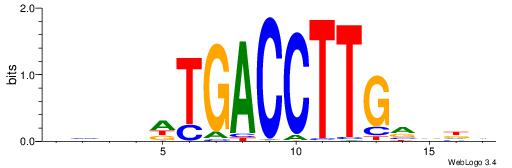 |
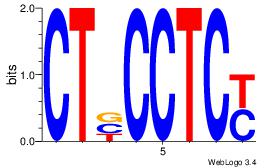
| Dataset #: 1 | Motif ID: 11 | Motif name: Motif 11 |
| Original motif | Reverse complement motif |
| Consensus sequence: CTBCCTCY | Consensus sequence: MGAGGBAG |
 |
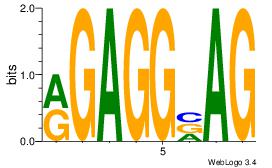 |
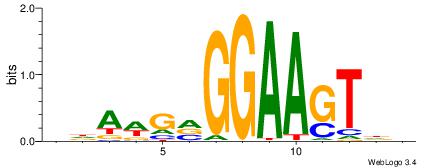
Best Matches for Motif ID 11 (Highest to Lowest)
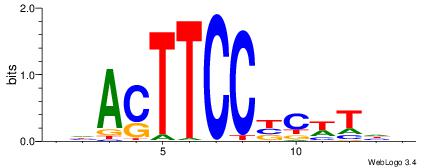
| Motif ID: | UP00085 |
| Motif name: | Sfpi1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HDACTTCCBCWTDV
---CTBCCTCY---
| Original motif | Reverse complement motif |
| Consensus sequence: BDAWGVGGAAGTDH | Consensus sequence: HDACTTCCBCWTDV |
 |
 |
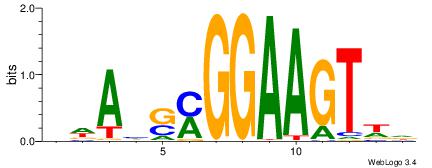
| Motif ID: | UP00409 |
| Motif name: | Elf5 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.000275689 |
Alignment:
BWAHSMGGAAGTWD
---MGAGGBAG---
| Original motif | Reverse complement motif |
| Consensus sequence: BWAHSMGGAAGTWD | Consensus sequence: DWACTTCCRSDTWV |
 |
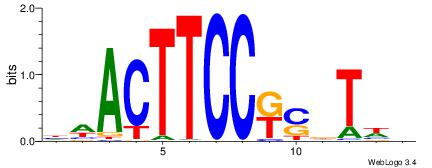 |
| Motif ID: | UP00419 |
| Motif name: | Spic |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.000518708 |
Alignment:
DWACTTCCRCTTTB
---CTBCCTCY---
| Original motif | Reverse complement motif |
| Consensus sequence: VAAAGMGGAAGTWD | Consensus sequence: DWACTTCCRCTTTB |
 |
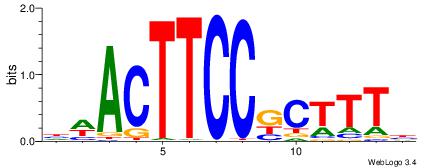 |
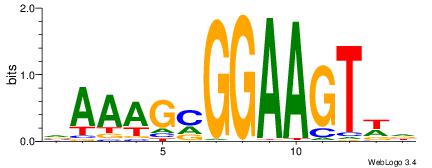
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.00485464 |
Alignment:
BVVHAGGGGGCGRDHHB
----MGAGGBAG-----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.00614662 |
Alignment:
DHHDGGGCGRGGKHBH
-------MGAGGBAG-
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
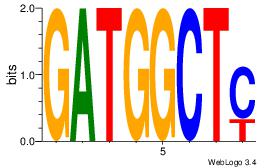
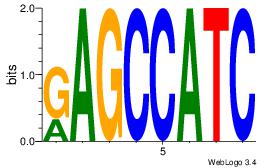
| Dataset #: 1 | Motif ID: 12 | Motif name: Motif 12 |
| Original motif | Reverse complement motif |
| Consensus sequence: GATGGCTC | Consensus sequence: GAGCCATC |
 |
 |
Best Matches for Motif ID 12 (Highest to Lowest)
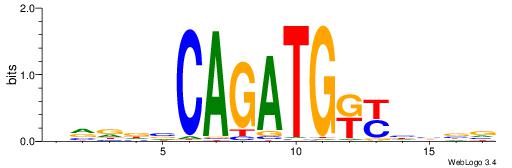
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
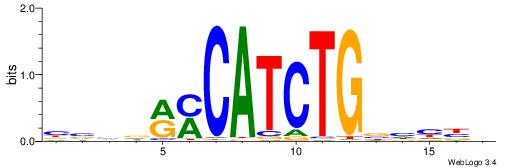
HVDDVCAGATGKYHBBV
-------GATGGCTC--
| Original motif | Reverse complement motif |
| Consensus sequence: HVDDVCAGATGKYHBBV | Consensus sequence: VBBDMYCATCTGVHHBH |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.00155237 |
Alignment:
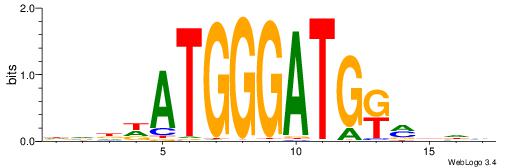
DVDDATGGGATGKMDDV
--------GATGGCTC-
| Original motif | Reverse complement motif |
| Consensus sequence: DVDDATGGGATGKMDDV | Consensus sequence: BDDYYCATCCCATDDBD |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.00582365 |
Alignment:
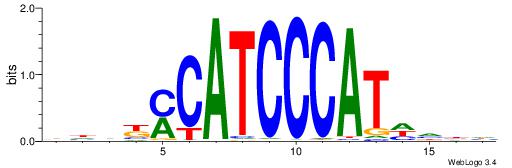
VHVRCAYGATGCATCDTVVADTH
-------GAGCCATC--------
| Original motif | Reverse complement motif |
| Consensus sequence: HADTBVADGATGCATCMTGMVHB | Consensus sequence: VHVRCAYGATGCATCDTVVADTH |
 |
 |
| Motif ID: | UP00103 |
| Motif name: | Jundm2_secondary |
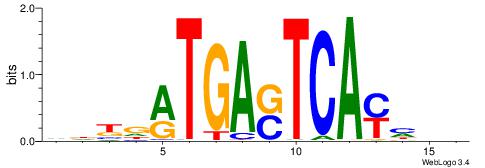
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.00695523 |
Alignment:
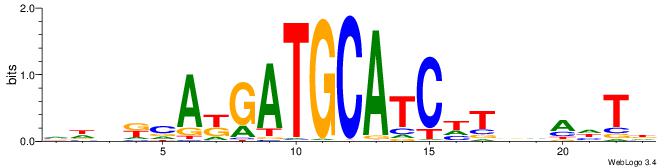
HDDKTGASTCAKHRDB
-----GAGCCATC---
| Original motif | Reverse complement motif |
| Consensus sequence: VDKDRTGASTCAYHDH | Consensus sequence: HDDKTGASTCAKHRDB |
 |
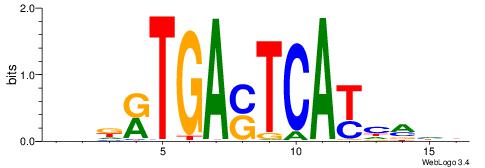 |
| Motif ID: | UP00035 |
| Motif name: | Hic1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.00724333 |
Alignment:
DBBATGCCAACCHVHH
---GAGCCATC-----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBATGCCAACCHVHH | Consensus sequence: DDBHGGTTGGCATVBD |
 |
 |
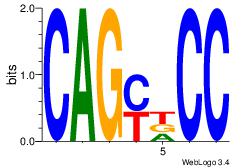
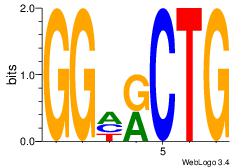
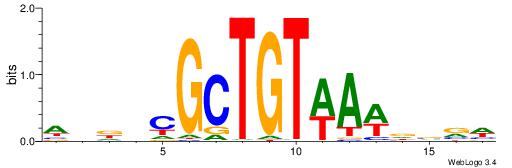
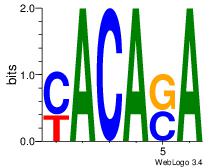
| Dataset #: 1 | Motif ID: 13 | Motif name: Motif 13 |
| Original motif | Reverse complement motif |
| Consensus sequence: CAGYDCC | Consensus sequence: GGDKCTG |
 |
 |
Best Matches for Motif ID 13 (Highest to Lowest)
| Motif ID: | UP00205 |
| Motif name: | Pknox2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 7 |
| Similarity score: | 0.0202873 |
Alignment:
HAVSRSCTGTCAATHB
--GGDKCTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: HAVSRSCTGTCAATHB | Consensus sequence: VHATTGACAGSKSVTH |
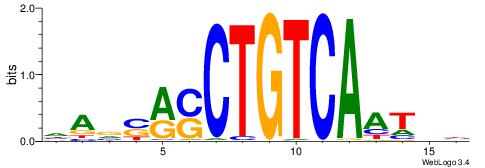 |
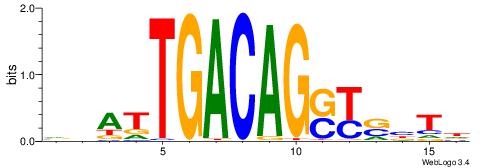 |
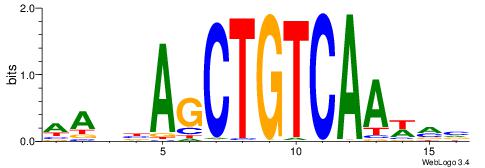
| Motif ID: | UP00122 |
| Motif name: | Tgif1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 7 |
| Similarity score: | 0.022836 |
Alignment:
DABWWTGACAGCTVVBD
--------CAGYDCC--
| Original motif | Reverse complement motif |
| Consensus sequence: DABWWTGACAGCTVVBD | Consensus sequence: DBVVAGCTGTCAWWVTH |
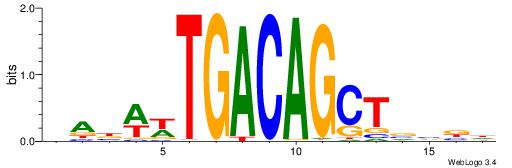 |
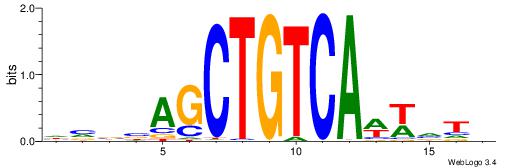 |
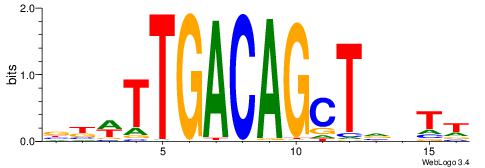
| Motif ID: | UP00258 |
| Motif name: | Tgif2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 7 |
| Similarity score: | 0.0283472 |
Alignment:
VBWTTGACAGCTVBTT
-------CAGYDCC--
| Original motif | Reverse complement motif |
| Consensus sequence: AABBAGCTGTCAAWVV | Consensus sequence: VBWTTGACAGCTVBTT |
 |
 |
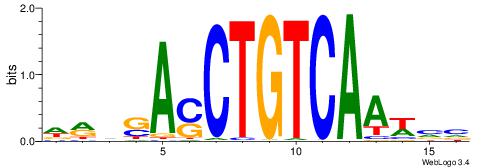
| Motif ID: | UP00203 |
| Motif name: | Pknox1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 7 |
| Similarity score: | 0.0289437 |
Alignment:
VVWTTGACAGSTSBKD
-------CAGYDCC--
| Original motif | Reverse complement motif |
| Consensus sequence: DRVSASCTGTCAAWVV | Consensus sequence: VVWTTGACAGSTSBKD |
 |
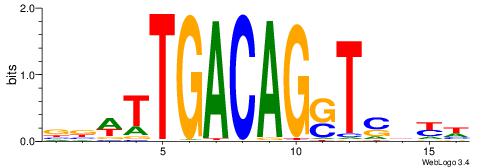 |
| Motif ID: | UP00193 |
| Motif name: | Rhox11_2205.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 7 |
| Similarity score: | 0.0291958 |
Alignment:
WVVHWTWACAGCGHHVT
--------CAGYDCC--
| Original motif | Reverse complement motif |
| Consensus sequence: AVDHCGCTGTWAWDVVW | Consensus sequence: WVVHWTWACAGCGHHVT |
 |
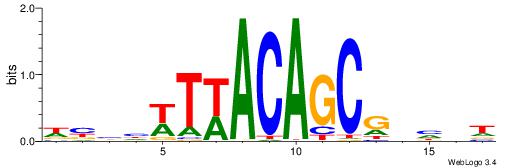 |
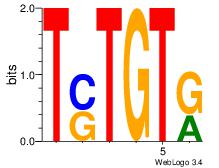
| Dataset #: 1 | Motif ID: 14 | Motif name: Motif 14 |
| Original motif | Reverse complement motif |
| Consensus sequence: TSTGTR | Consensus sequence: MACASA |
 |
 |
Best Matches for Motif ID 14 (Highest to Lowest)
| Motif ID: | UP00026 |
| Motif name: | Zscan4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
HVHRGCACACAAHHHV
-----MACASA-----
| Original motif | Reverse complement motif |
| Consensus sequence: HVHRGCACACAAHHHV | Consensus sequence: BHHHTTGTGTGCKHVD |
 |
 |
| Motif ID: | UP00042 |
| Motif name: | Gm397_second |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.00200057 |
Alignment:
DVMVGCACACACKCVH
-----MACASA-----
| Original motif | Reverse complement motif |
| Consensus sequence: DVMVGCACACACKCVH | Consensus sequence: HBGYGTGTGTGCVRVD |
 |
 |
| Motif ID: | UP00026 |
| Motif name: | Zscan4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.0148405 |
Alignment:
DHVDTGTGCACAYAHDH
--------MACASA---
| Original motif | Reverse complement motif |
| Consensus sequence: DHVDTGTGCACAYAHDH | Consensus sequence: HDHTMTGTGCACADVHD |
 |
 |
| Motif ID: | UP00042 |
| Motif name: | Gm397_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.0236149 |
Alignment:
HHRTATGTGCACATHHD
---TSTGTR--------
| Original motif | Reverse complement motif |
| Consensus sequence: HHDATGTGCACATAMDH | Consensus sequence: HHRTATGTGCACATHHD |
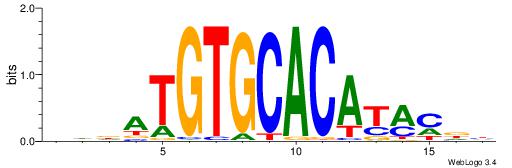 |
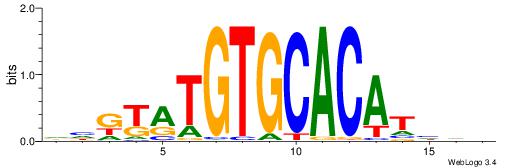 |
| Motif ID: | UP00041 |
| Motif name: | Foxj1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0266195 |
Alignment:
HDHRTAAACAAAHDDD
------MACASA----
| Original motif | Reverse complement motif |
| Consensus sequence: HDHRTAAACAAAHDDD | Consensus sequence: DDDHTTTGTTTAMHDH |
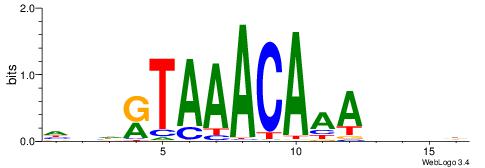 |
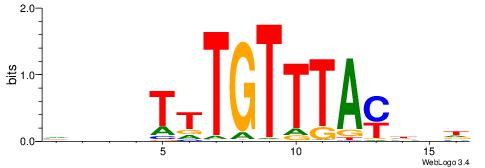 |
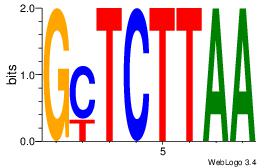
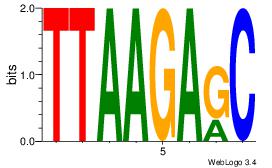
| Dataset #: 1 | Motif ID: 15 | Motif name: Motif 15 |
| Original motif | Reverse complement motif |
| Consensus sequence: GCTCTTAA | Consensus sequence: TTAAGAGC |
 |
 |
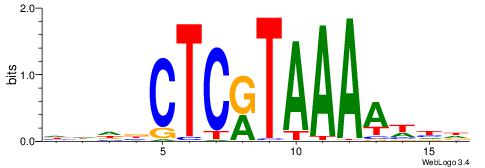
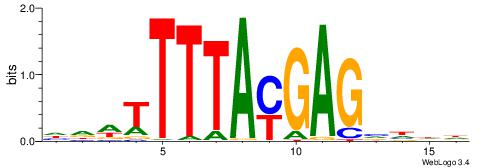
Best Matches for Motif ID 15 (Highest to Lowest)
| Motif ID: | UP00173 |
| Motif name: | Hoxc13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
DHWTTTTAMGAGBDHH
-----TTAAGAGC---
| Original motif | Reverse complement motif |
| Consensus sequence: HHDBCTCRTAAAAWHD | Consensus sequence: DHWTTTTAMGAGBDHH |
 |
 |
| Motif ID: | UP00183 |
| Motif name: | Hoxa13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.00510293 |
Alignment:
DVWTTTTACGAGBHDH
-----TTAAGAGC---
| Original motif | Reverse complement motif |
| Consensus sequence: HDHBCTCGTAAAAWBD | Consensus sequence: DVWTTTTACGAGBHDH |
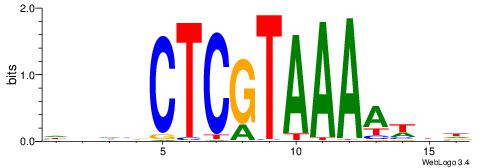 |
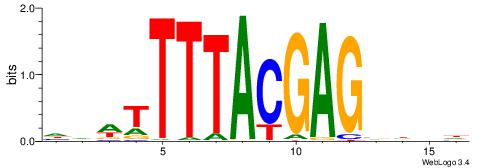 |
| Motif ID: | UP00228 |
| Motif name: | Bapx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0182341 |
Alignment:
DTBVTTAAGTGGTTHHV
----TTAAGAGC-----
| Original motif | Reverse complement motif |
| Consensus sequence: VHHAACCACTTAAVVAH | Consensus sequence: DTBVTTAAGTGGTTHHV |
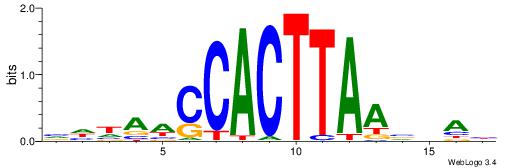 |
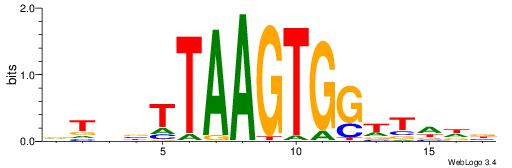 |
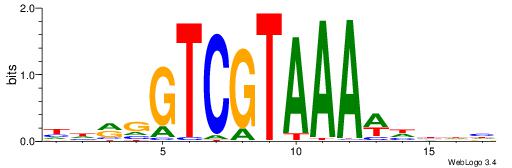
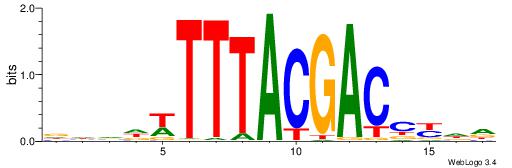
| Motif ID: | UP00135 |
| Motif name: | Hoxc12 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0206445 |
Alignment:
DDVHWTTTACGACCKHH
------TTAAGAGC---
| Original motif | Reverse complement motif |
| Consensus sequence: HHRGGTCGTAAAWHBDH | Consensus sequence: DDVHWTTTACGACCKHH |
 |
 |
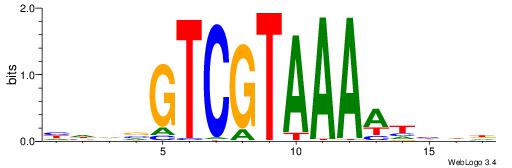
| Motif ID: | UP00177 |
| Motif name: | Hoxd12 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.023305 |
Alignment:
DHDHTTTTACGACVDHD
------TTAAGAGC---
| Original motif | Reverse complement motif |
| Consensus sequence: HHDVGTCGTAAAAHHHD | Consensus sequence: DHDHTTTTACGACVDHD |
 |
 |
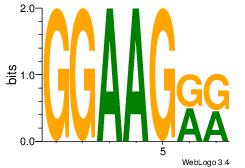
| Dataset #: 1 | Motif ID: 16 | Motif name: Motif 16 |
| Original motif | Reverse complement motif |
| Consensus sequence: GGAAGRR | Consensus sequence: MMCTTCC |
 |
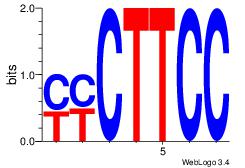 |
Best Matches for Motif ID 16 (Highest to Lowest)
| Motif ID: | UP00409 |
| Motif name: | Elf5 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 7 |
| Similarity score: | 0 |
Alignment:
DWACTTCCRSDTWV
-MMCTTCC------
| Original motif | Reverse complement motif |
| Consensus sequence: BWAHSMGGAAGTWD | Consensus sequence: DWACTTCCRSDTWV |
 |
 |
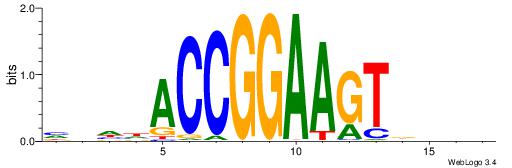
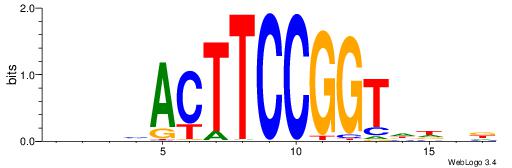
| Motif ID: | UP00013 |
| Motif name: | Gabpa_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 7 |
| Similarity score: | 0.00097547 |
Alignment:
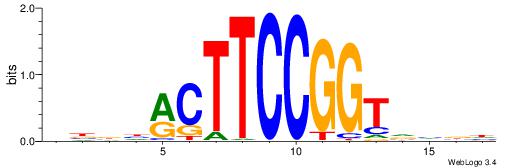
BHHHACTTCCGGTHHBD
---MMCTTCC-------
| Original motif | Reverse complement motif |
| Consensus sequence: HVHHACCGGAAGTDHHV | Consensus sequence: BHHHACTTCCGGTHHBD |
 |
 |
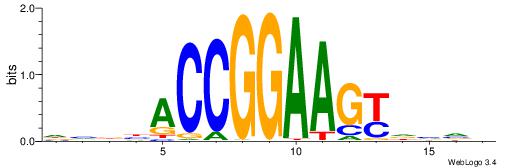
| Motif ID: | UP00420 |
| Motif name: | Elk3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 7 |
| Similarity score: | 0.00131169 |
Alignment:
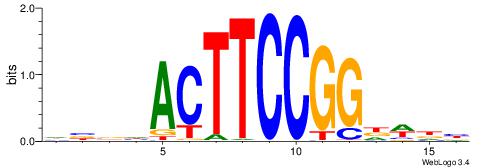
VBVDMCTTCCGGTVBVD
---MMCTTCC-------
| Original motif | Reverse complement motif |
| Consensus sequence: DVBBACCGGAAGYDVVV | Consensus sequence: VBVDMCTTCCGGTVBVD |
 |
 |
| Motif ID: | UP00419 |
| Motif name: | Spic |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 7 |
| Similarity score: | 0.00204701 |
Alignment:
DWACTTCCRCTTTB
-MMCTTCC------
| Original motif | Reverse complement motif |
| Consensus sequence: VAAAGMGGAAGTWD | Consensus sequence: DWACTTCCRCTTTB |
 |
 |
| Motif ID: | UP00404 |
| Motif name: | Elf2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 7 |
| Similarity score: | 0.00230178 |
Alignment:
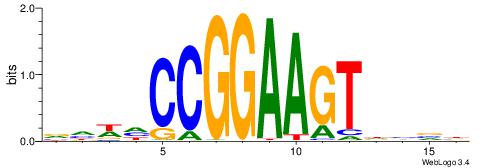
DBBHACTTCCGGDWDB
---MMCTTCC------
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGDWDB | Consensus sequence: BDWDCCGGAAGTHBBD |
 |
 |
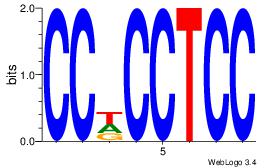
| Dataset #: 1 | Motif ID: 17 | Motif name: Motif 17 |
| Original motif | Reverse complement motif |
| Consensus sequence: CCDCCTCC | Consensus sequence: GGAGGDGG |
 |
 |
Best Matches for Motif ID 17 (Highest to Lowest)
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.00331161 |
Alignment:
GKRGGGGGGGGGGBD
---GGAGGDGG----
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.00990566 |
Alignment:
HCCGCCCCCGCAHB
-CCDCCTCC-----
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0127556 |
Alignment:
BDHHMCGCCCCCTHVBB
----CCDCCTCC-----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0192546 |
Alignment:
BHHDYGGGGGGGGBVD
-----GGAGGDGG---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 10 |
| Number of overlap: | 8 |
| Similarity score: | 0.0193256 |
Alignment:
MHHHWHYTMCCHCCCACVAABVH
---------CCDCCTCC------
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
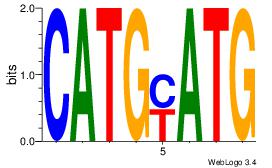
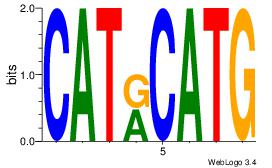
| Dataset #: 1 | Motif ID: 18 | Motif name: Motif 18 |
| Original motif | Reverse complement motif |
| Consensus sequence: CATGYATG | Consensus sequence: CATKCATG |
 |
 |
Best Matches for Motif ID 18 (Highest to Lowest)
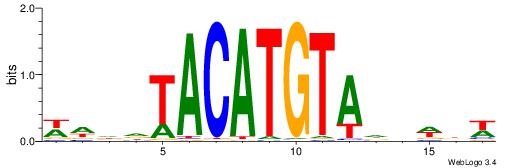
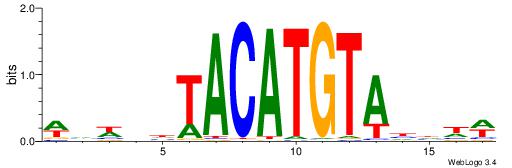
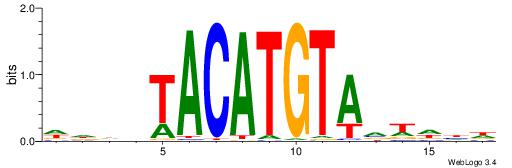
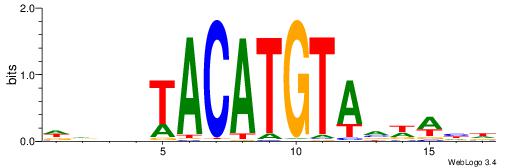
| Motif ID: | UP00236 |
| Motif name: | Irx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0094584 |
Alignment:
WVWDDTACATGTADDDW
-------CATGYATG--
| Original motif | Reverse complement motif |
| Consensus sequence: WDDDTACATGTADDWBW | Consensus sequence: WVWDDTACATGTADDDW |
 |
 |
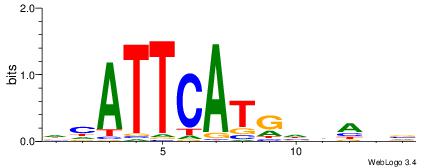
| Motif ID: | UP00051 |
| Motif name: | Sox8_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0108518 |
Alignment:
DHATTCATGHVABV
-CATKCATG-----
| Original motif | Reverse complement motif |
| Consensus sequence: DHATTCATGHVABV | Consensus sequence: VBTVHCATGAATDD |
 |
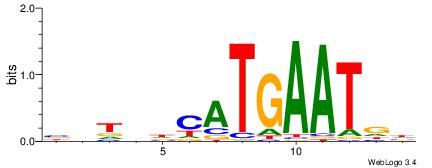 |
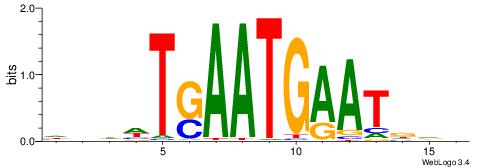
| Motif ID: | UP00055 |
| Motif name: | Hbp1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.0133688 |
Alignment:
VDHATTCATTSAWDDH
--CATKCATG------
| Original motif | Reverse complement motif |
| Consensus sequence: HHDWTSAATGAATDDB | Consensus sequence: VDHATTCATTSAWDDH |
 |
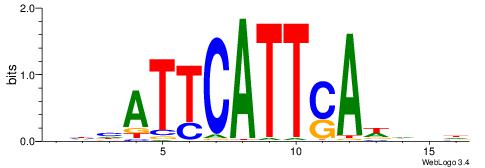 |
| Motif ID: | UP00223 |
| Motif name: | Irx3_2226.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0143625 |
Alignment:
DHDWBTACATGTAHDDD
-------CATGYATG--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDHTACATGTAVWDHD | Consensus sequence: DHDWBTACATGTAHDDD |
 |
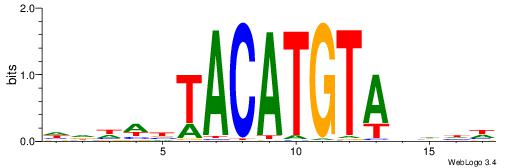 |
| Motif ID: | UP00223 |
| Motif name: | Irx3_0920.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0163294 |
Alignment:
DDWWBTACATGTADDDD
-------CATGYATG--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDDTACATGTAVWWHD | Consensus sequence: DDWWBTACATGTADDDD |
 |
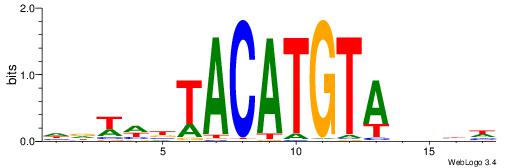 |
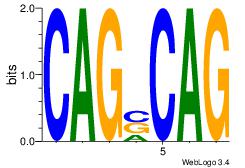
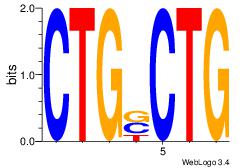
| Dataset #: 1 | Motif ID: 19 | Motif name: Motif 19 |
| Original motif | Reverse complement motif |
| Consensus sequence: CAGSCAG | Consensus sequence: CTGSCTG |
 |
 |
Best Matches for Motif ID 19 (Highest to Lowest)
| Motif ID: | UP00000 |
| Motif name: | Smad3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0 |
Alignment:
HDHDDCCAGACABBHVH
------CAGSCAG----
| Original motif | Reverse complement motif |
| Consensus sequence: HDHDDCCAGACABBHVH | Consensus sequence: HVHBVTGTCTGGDDHDD |
 |
 |
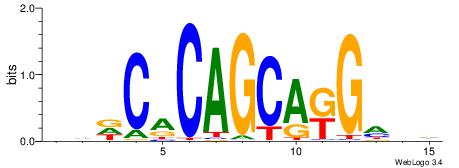
| Motif ID: | UP00006 |
| Motif name: | Zic3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 7 |
| Similarity score: | 0.0064943 |
Alignment:
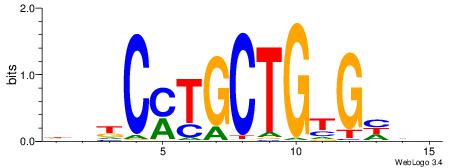
DVHCYTGCTGTGHDH
---CTGSCTG-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDCACAGCAKGHVD | Consensus sequence: DVHCYTGCTGTGHDH |
 |
 |
| Motif ID: | UP00057 |
| Motif name: | Zic2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.00786726 |
Alignment:
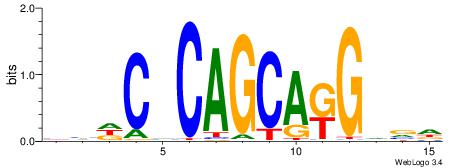
DVBCYTGCTGBGDDD
---CTGSCTG-----
| Original motif | Reverse complement motif |
| Consensus sequence: HHDCVCAGCAKGVVD | Consensus sequence: DVBCYTGCTGBGDDD |
 |
 |
| Motif ID: | UP00092 |
| Motif name: | Myb_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 7 |
| Similarity score: | 0.0123596 |
Alignment:
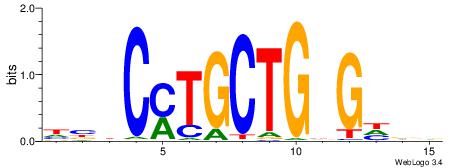
DBVKGGCAGTTGGTHB
--CAGSCAG-------
| Original motif | Reverse complement motif |
| Consensus sequence: BDACCAACTGCCRBBH | Consensus sequence: DBVKGGCAGTTGGTHB |
 |
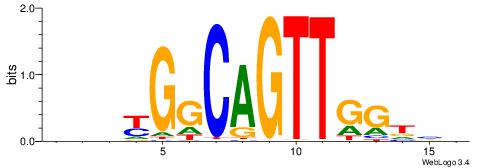 |
| Motif ID: | UP00102 |
| Motif name: | Zic1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 7 |
| Similarity score: | 0.0127545 |
Alignment:
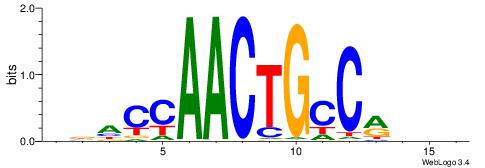
DVHCCTGCTGBGDDB
---CTGSCTG-----
| Original motif | Reverse complement motif |
| Consensus sequence: BHDCVCAGCAGGHVD | Consensus sequence: DVHCCTGCTGBGDDB |
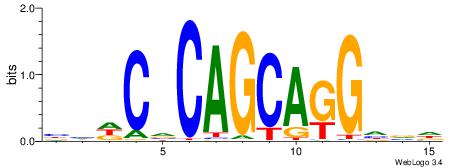 |
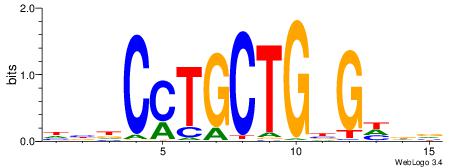 |
| Dataset #: 1 | Motif ID: 20 | Motif name: Motif 20 |
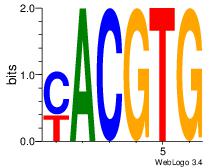
| Original motif | Reverse complement motif |
| Consensus sequence: CACGTR | Consensus sequence: MACGTG |
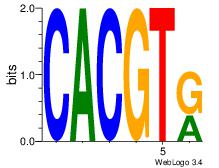 |
 |
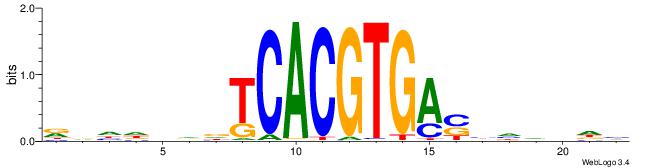
Best Matches for Motif ID 20 (Highest to Lowest)
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
DDHBHBSTCACGTGAHBBDHHM
--------CACGTR--------
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
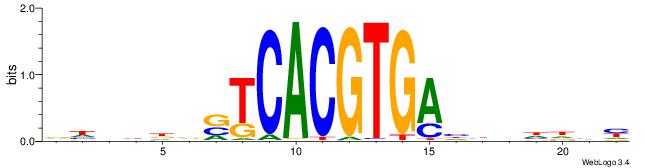 |
| Motif ID: | UP00060 |
| Motif name: | Max_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.00504882 |
Alignment:
HHVBABCACGTGSTHD
------CACGTR----
| Original motif | Reverse complement motif |
| Consensus sequence: DDASCACGTGBTBVDD | Consensus sequence: HHVBABCACGTGSTHD |
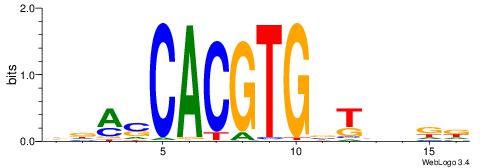 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.0050835 |
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
---------CACGTR--------
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0336216 |
Alignment:
DHDBHGCACCTGBDDVB
------CACGTR-----
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Motif ID: | UP00249 |
| Motif name: | Nkx2-5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.0339468 |
Alignment:
HDADCCACTTRAAWTT
-----CACGTR-----
| Original motif | Reverse complement motif |
| Consensus sequence: HDADCCACTTRAAWTT | Consensus sequence: AAWTTMAAGTGGHTDH |
 |
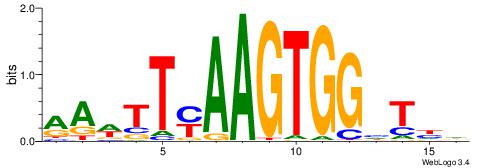 |
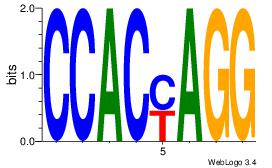
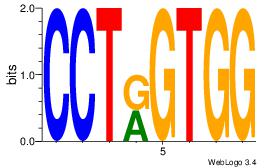
| Dataset #: 1 | Motif ID: 21 | Motif name: Motif 21 |
| Original motif | Reverse complement motif |
| Consensus sequence: CCACYAGG | Consensus sequence: CCTKGTGG |
 |
 |
Best Matches for Motif ID 21 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HGDHVHKGACCACCCGGBGBGDB
---------CCACYAGG------
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 11 |
| Number of overlap: | 8 |
| Similarity score: | 0.00212209 |
Alignment:
CBDMCMGGGTGGTCCHVBVBAH
----CCTKGTGG----------
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 11 |
| Number of overlap: | 8 |
| Similarity score: | 0.00654113 |
Alignment:
CVHBSCGGGTGGTCMHVHHCDH
----CCTKGTGG----------
| Original motif | Reverse complement motif |
| Consensus sequence: HDGHHBHRGACCACCCGSVDVG | Consensus sequence: CVHBSCGGGTGGTCMHVHHCDH |
 |
 |
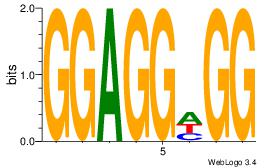
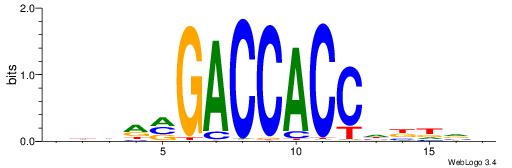
| Motif ID: | UP00047 |
| Motif name: | Zbtb7b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.00890824 |
Alignment:
BDDRVGACCACCHBDVB
-------CCACYAGG--
| Original motif | Reverse complement motif |
| Consensus sequence: BDDRVGACCACCHBDVB | Consensus sequence: BBDVHGGTGGTCBKDDB |
 |
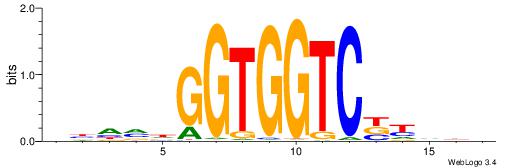 |
| Motif ID: | UP00102 |
| Motif name: | Zic1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0167011 |
Alignment:
DVHCCTGCTGBGDDB
---CCTKGTGG----
| Original motif | Reverse complement motif |
| Consensus sequence: BHDCVCAGCAGGHVD | Consensus sequence: DVHCCTGCTGBGDDB |
 |
 |
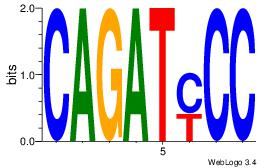
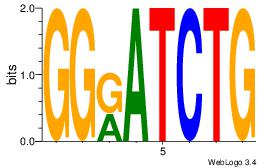
| Dataset #: 1 | Motif ID: 22 | Motif name: Motif 22 |
| Original motif | Reverse complement motif |
| Consensus sequence: CAGATYCC | Consensus sequence: GGKATCTG |
 |
 |
Best Matches for Motif ID 22 (Highest to Lowest)
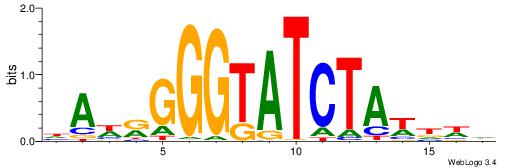
| Motif ID: | UP00232 |
| Motif name: | Dobox4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
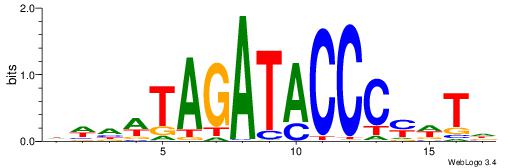
Alignment:
DAWKGGGTATCTAWHWH
-----GGKATCTG----
| Original motif | Reverse complement motif |
| Consensus sequence: HWHWTAGATACCCYWTD | Consensus sequence: DAWKGGGTATCTAWHWH |
 |
 |
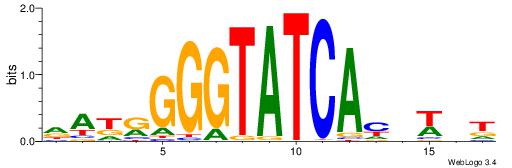
| Motif ID: | UP00159 |
| Motif name: | Six2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0241128 |
Alignment:
DAKRGGGTATCACDWHT
-----GGKATCTG----
| Original motif | Reverse complement motif |
| Consensus sequence: DAKRGGGTATCACDWHT | Consensus sequence: AHWDGTGATACCCMRTD |
 |
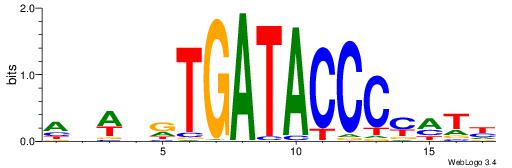 |
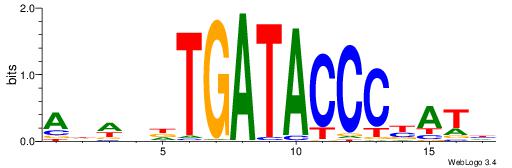
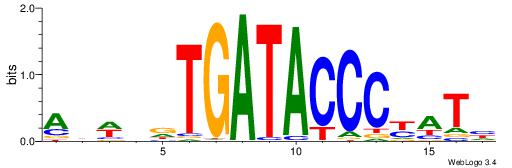
| Motif ID: | UP00388 |
| Motif name: | Six6_2267.4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0250604 |
Alignment:
DATRGGGTATCAHDWHT
-----GGKATCTG----
| Original motif | Reverse complement motif |
| Consensus sequence: DATRGGGTATCAHDWHT | Consensus sequence: AHWDHTGATACCCKATD |
 |
 |
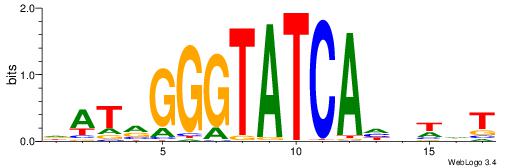
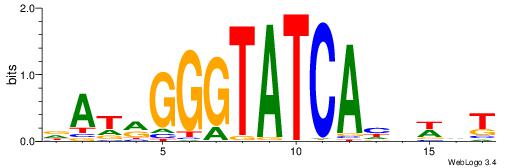
| Motif ID: | UP00195 |
| Motif name: | Six3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0259075 |
Alignment:
DATRGGGTATCAHHWHT
-----GGKATCTG----
| Original motif | Reverse complement motif |
| Consensus sequence: DATRGGGTATCAHHWHT | Consensus sequence: AHWHDTGATACCCKATH |
 |
 |
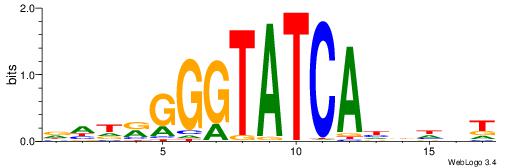
| Motif ID: | UP00192 |
| Motif name: | Six1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0264035 |
Alignment:
DHDRGGGTATCAHDHBT
-----GGKATCTG----
| Original motif | Reverse complement motif |
| Consensus sequence: DHDRGGGTATCAHDHBT | Consensus sequence: AVHDHTGATACCCMDHH |
 |
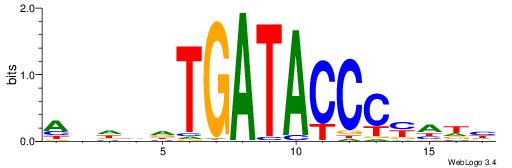 |
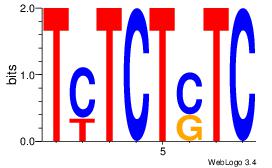
| Dataset #: 1 | Motif ID: 23 | Motif name: Motif 23 |
| Original motif | Reverse complement motif |
| Consensus sequence: GASAGAGA | Consensus sequence: TCTCTSTC |
 |
 |
Best Matches for Motif ID 23 (Highest to Lowest)
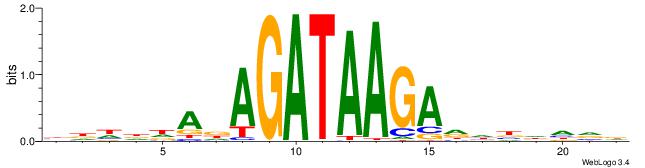
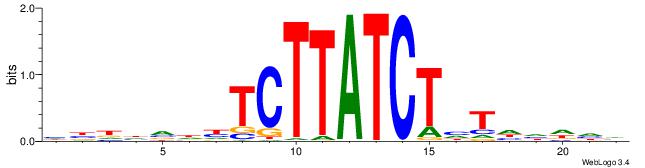
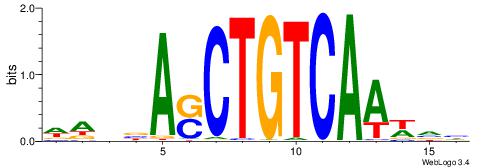
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HBHDVHDTCTTATCTHTDDHDV
----------TCTCTSTC----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
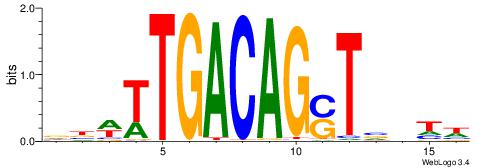
| Motif ID: | UP00040 |
| Motif name: | Irf5_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.00356941 |
Alignment:
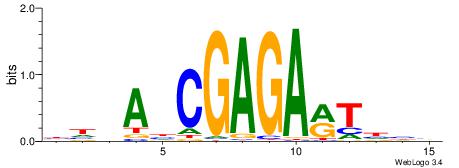
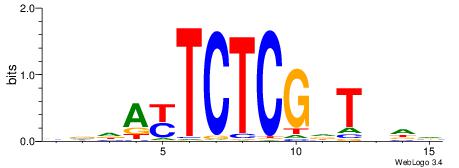
BHDAHCGAGARTHBV
--GASAGAGA-----
| Original motif | Reverse complement motif |
| Consensus sequence: BHDAHCGAGARTHBV | Consensus sequence: VBHAKTCTCGHTHHV |
 |
 |
| Motif ID: | UP00258 |
| Motif name: | Tgif2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0059307 |
Alignment:
AABBAGCTGTCAAWVV
---TCTCTSTC-----
| Original motif | Reverse complement motif |
| Consensus sequence: AABBAGCTGTCAAWVV | Consensus sequence: VBWTTGACAGCTVBTT |
 |
 |
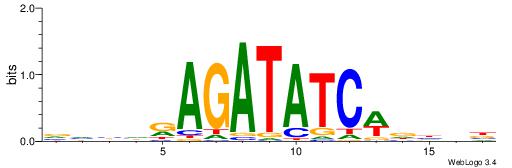
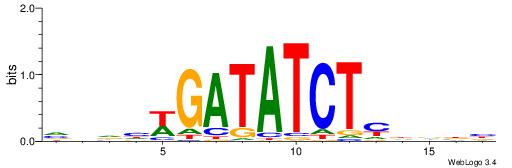
| Motif ID: | UP00080 |
| Motif name: | Gata5_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.00994332 |
Alignment:
VVHHWGATATCTMHBBV
-------TCTCTSTC--
| Original motif | Reverse complement motif |
| Consensus sequence: VVBHRAGATATCWDHBB | Consensus sequence: VVHHWGATATCTMHBBV |
 |
 |
| Motif ID: | UP00186 |
| Motif name: | Meis1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0114008 |
Alignment:
WAVBASCTGTCAAWVH
---TCTCTSTC-----
| Original motif | Reverse complement motif |
| Consensus sequence: WAVBASCTGTCAAWVH | Consensus sequence: DBWTTGACAGSTBVTW |
 |
 |
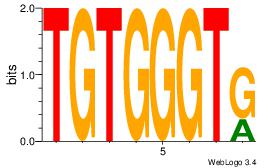
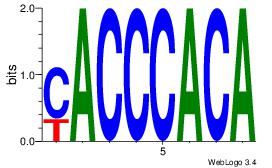
| Dataset #: 1 | Motif ID: 24 | Motif name: Motif 24 |
| Original motif | Reverse complement motif |
| Consensus sequence: TGTGGGTG | Consensus sequence: CACCCACA |
 |
 |
Best Matches for Motif ID 24 (Highest to Lowest)
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 12 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
ABBBBVVRGACCACCCACRDBBM
-----------CACCCACA----
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
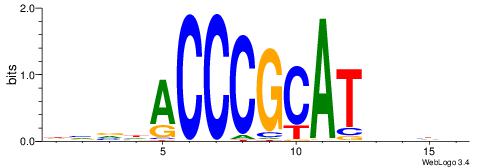
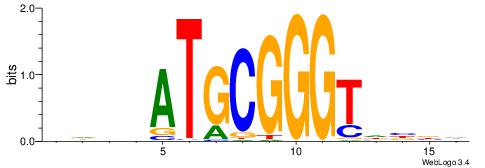
| Motif ID: | UP00070 |
| Motif name: | Gcm1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.00199912 |
Alignment:
DHDVATGCGGGTHVVH
-----TGTGGGTG---
| Original motif | Reverse complement motif |
| Consensus sequence: HVVHACCCGCATVDHD | Consensus sequence: DHDVATGCGGGTHVVH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 11 |
| Number of overlap: | 8 |
| Similarity score: | 0.00682225 |
Alignment:
MHHHWHYTMCCHCCCACVAABVH
----------CACCCACA-----
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
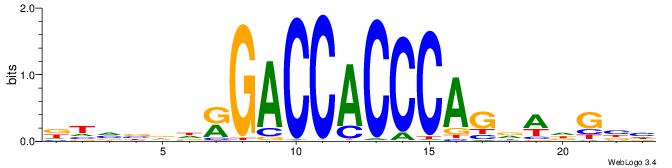
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 11 |
| Number of overlap: | 8 |
| Similarity score: | 0.0105432 |
Alignment:
DTVBDDRGACCACCCAGDWDGBH
----------CACCCACA-----
| Original motif | Reverse complement motif |
| Consensus sequence: DTVBDDRGACCACCCAGDWDGBH | Consensus sequence: DBCDWHCTGGGTGGTCMDHBBAH |
 |
 |
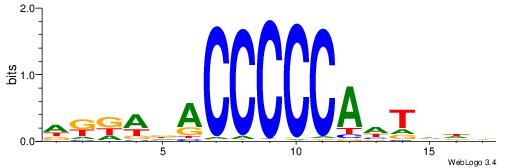
| Motif ID: | UP00021 |
| Motif name: | Zfp281_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.011455 |
Alignment:
DKKWDACCCCCAHTDDV
----CACCCACA-----
| Original motif | Reverse complement motif |
| Consensus sequence: DKKWDACCCCCAHTDDV | Consensus sequence: VDDAHTGGGGGTHWYYD |
 |
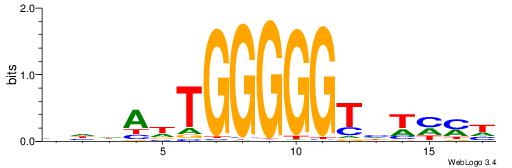 |
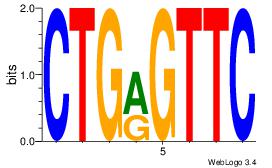
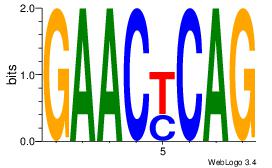
| Dataset #: 1 | Motif ID: 25 | Motif name: Motif 25 |
| Original motif | Reverse complement motif |
| Consensus sequence: CTGRGTTC | Consensus sequence: GAACKCAG |
 |
 |
Best Matches for Motif ID 25 (Highest to Lowest)
| Motif ID: | UP00019 |
| Motif name: | Zbtb12_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.015191 |
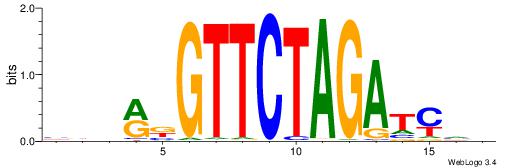
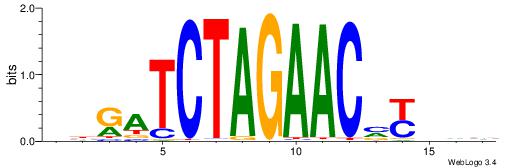
Alignment:
BDHRBGTTCTAGATCVB
-CTGRGTTC--------
| Original motif | Reverse complement motif |
| Consensus sequence: BDHRBGTTCTAGATCVB | Consensus sequence: BBGATCTAGAACBKHDB |
 |
 |
| Motif ID: | UP00006 |
| Motif name: | Zic3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0235944 |
Alignment:
DDDCACAGCAKGHVD
GAACKCAG-------
| Original motif | Reverse complement motif |
| Consensus sequence: DDDCACAGCAKGHVD | Consensus sequence: DVHCYTGCTGTGHDH |
 |
 |
| Motif ID: | UP00102 |
| Motif name: | Zic1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0252114 |
Alignment:
BHDCVCAGCAGGHVD
GAACKCAG-------
| Original motif | Reverse complement motif |
| Consensus sequence: BHDCVCAGCAGGHVD | Consensus sequence: DVHCCTGCTGBGDDB |
 |
 |
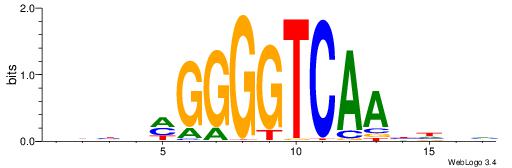
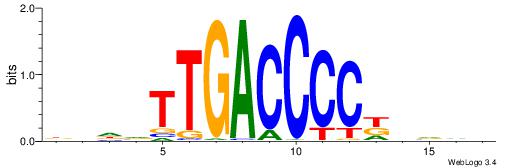
| Motif ID: | UP00066 |
| Motif name: | Hnf4a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0255154 |
Alignment:
BDBVMGGGGTCAABHDV
---CTGRGTTC------
| Original motif | Reverse complement motif |
| Consensus sequence: BDBVMGGGGTCAABHDV | Consensus sequence: BHHVTTGACCCCYVVDB |
 |
 |
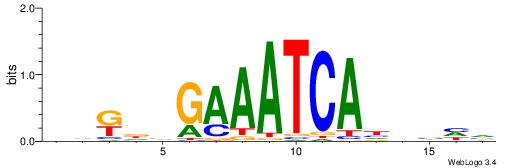
| Motif ID: | UP00148 |
| Motif name: | Hdx |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0260414 |
Alignment:
BDBDHTGATTTCDHYDD
----CTGRGTTC-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDKDHGAAATCAHHBHV | Consensus sequence: BDBDHTGATTTCDHYDD |
 |
 |
| Dataset #: 1 | Motif ID: 26 | Motif name: Motif 26 |
| Original motif | Reverse complement motif |
| Consensus sequence: CAGAGGAY | Consensus sequence: KTCCTCTG |
 |
 |
Best Matches for Motif ID 26 (Highest to Lowest)
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HVDDVCAGATGKYHBBV
-----CAGAGGAY----
| Original motif | Reverse complement motif |
| Consensus sequence: HVDDVCAGATGKYHBBV | Consensus sequence: VBBDMYCATCTGVHHBH |
 |
 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.00361871 |
Alignment:
DDDYCCTTTGATCTDDV
--KTCCTCTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
 |
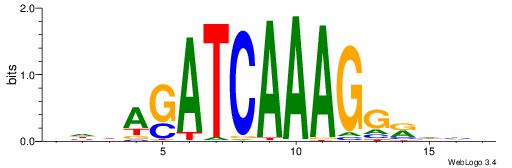 |
| Motif ID: | UP00083 |
| Motif name: | Tcf7l2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.00571944 |
Alignment:
DDDYCCTTTGATSTDDV
--KTCCTCTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATSTDDV | Consensus sequence: BDDASATCAAAGGMDDD |
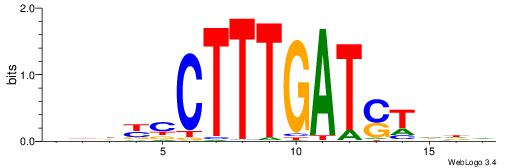 |
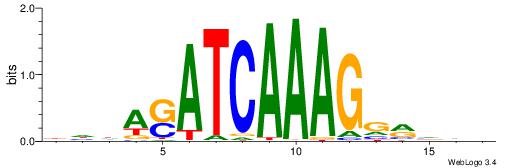 |
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.00988532 |
Alignment:
HHBTMCTTTGTTMDDVH
--KTCCTCTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
 |
 |
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0118578 |
Alignment:
HHBBMCTTTGTTHDDBH
--KTCCTCTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
 |
 |
| Dataset #: 1 | Motif ID: 27 | Motif name: Motif 27 |
| Original motif | Reverse complement motif |
| Consensus sequence: CSGCCGCC | Consensus sequence: GGCGGCSG |
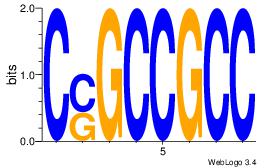 |
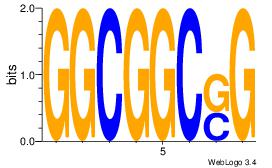 |
Best Matches for Motif ID 27 (Highest to Lowest)
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.012469 |
Alignment:
HCCGCCCCCGCAHB
-CSGCCGCC-----
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.012605 |
Alignment:
BVVHAGGGGGCGRDHHB
-----GGCGGCSG----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.024557 |
Alignment:
TCDAMVBGACGCMAKDDDVBBD
-----CSGCCGCC---------
| Original motif | Reverse complement motif |
| Consensus sequence: DBBVDDDRTRGCGTCBBYTHGA | Consensus sequence: TCDAMVBGACGCMAKDDDVBBD |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.0251911 |
Alignment:
DHHBCCCCGCCAHHBHB
---CSGCCGCC------
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
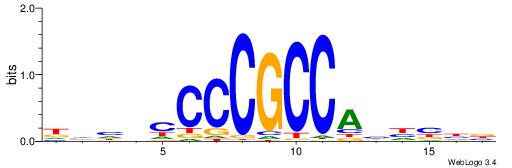 |
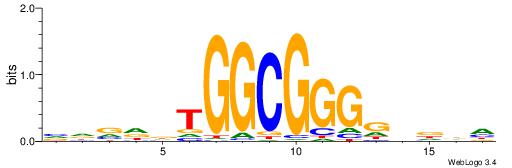 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.026934 |
Alignment:
DBCCCCCCCCCCMYC
----CSGCCGCC---
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Dataset #: 1 | Motif ID: 28 | Motif name: Motif 28 |
| Original motif | Reverse complement motif |
| Consensus sequence: CTTCCTG | Consensus sequence: CAGGAAG |
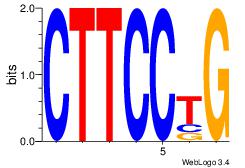 |
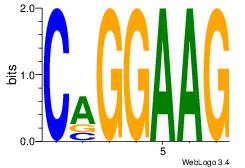 |
Best Matches for Motif ID 28 (Highest to Lowest)
| Motif ID: | UP00413 |
| Motif name: | Elf4 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0 |
Alignment:
DBBHACTTCCGGKWTH
-----CTTCCTG----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGKWTH | Consensus sequence: HAWYCCGGAAGTHBBD |
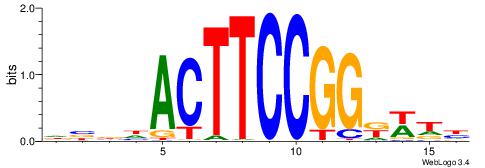 |
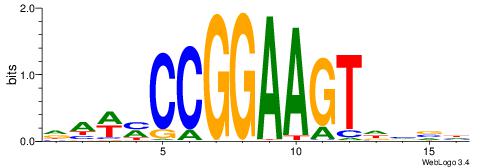 |
| Motif ID: | UP00404 |
| Motif name: | Elf2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.00352958 |
Alignment:
DBBHACTTCCGGDWDB
-----CTTCCTG----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGDWDB | Consensus sequence: BDWDCCGGAAGTHBBD |
 |
 |
| Motif ID: | UP00013 |
| Motif name: | Gabpa_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.00394394 |
Alignment:
HVHHACCGGAAGTDHHV
-----CAGGAAG-----
| Original motif | Reverse complement motif |
| Consensus sequence: HVHHACCGGAAGTDHHV | Consensus sequence: BHHHACTTCCGGTHHBD |
 |
 |
| Motif ID: | UP00420 |
| Motif name: | Elk3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.00419952 |
Alignment:
DVBBACCGGAAGYDVVV
-----CAGGAAG-----
| Original motif | Reverse complement motif |
| Consensus sequence: DVBBACCGGAAGYDVVV | Consensus sequence: VBVDMCTTCCGGTVBVD |
 |
 |
| Motif ID: | UP00410 |
| Motif name: | Elk1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.006051 |
Alignment:
VVHBACCGGAAGTDBHD
-----CAGGAAG-----
| Original motif | Reverse complement motif |
| Consensus sequence: VVHBACCGGAAGTDBHD | Consensus sequence: DHBDACTTCCGGTVHVB |
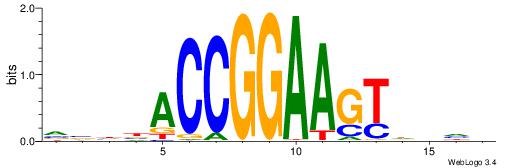 |
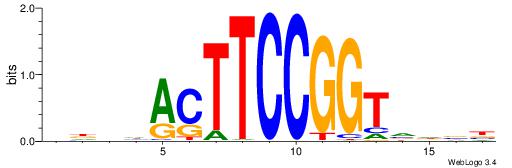 |
| Dataset #: 1 | Motif ID: 29 | Motif name: Motif 29 |
| Original motif | Reverse complement motif |
| Consensus sequence: CATTTCY | Consensus sequence: MGAAATG |
 |
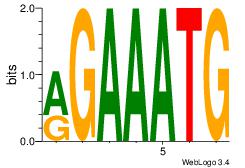 |
Best Matches for Motif ID 29 (Highest to Lowest)
| Motif ID: | UP00411 |
| Motif name: | Erg |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 7 |
| Similarity score: | 0 |
Alignment:
VBBVAYTTCCGGTDDB
---CATTTCY------
| Original motif | Reverse complement motif |
| Consensus sequence: VBBVAYTTCCGGTDDB | Consensus sequence: BDDACCGGAAKTVBVB |
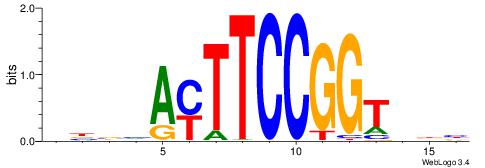 |
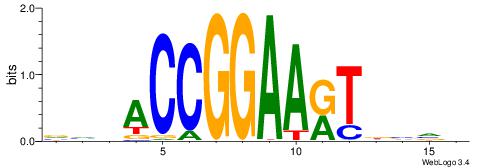 |
| Motif ID: | UP00424 |
| Motif name: | Gm4881 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 7 |
| Similarity score: | 3.61876e-06 |
Alignment:
VBBHAYTTCCGGTDDD
---CATTTCY------
| Original motif | Reverse complement motif |
| Consensus sequence: VBBHAYTTCCGGTDDD | Consensus sequence: DHDACCGGAAMTHBVB |
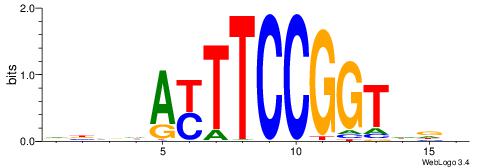 |
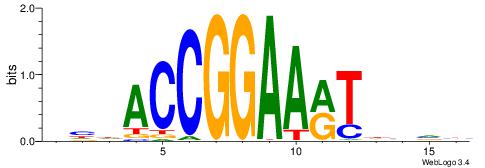 |
| Motif ID: | UP00148 |
| Motif name: | Hdx |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0.000285971 |
Alignment:
DDKDHGAAATCAHHBHV
----MGAAATG------
| Original motif | Reverse complement motif |
| Consensus sequence: DDKDHGAAATCAHHBHV | Consensus sequence: BDBDHTGATTTCDHYDD |
 |
 |
| Motif ID: | UP00418 |
| Motif name: | Etv6 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0.00137994 |
Alignment:
DVHBTACTTCCGGHTHB
----CATTTCY------
| Original motif | Reverse complement motif |
| Consensus sequence: DVHBTACTTCCGGHTHB | Consensus sequence: BHAHCCGGAAGTABDVD |
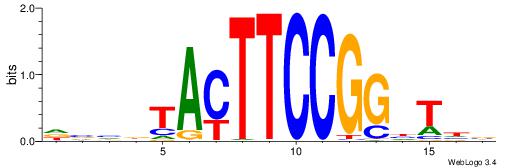 |
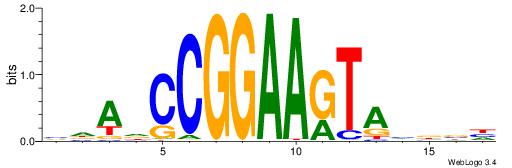 |
| Motif ID: | UP00422 |
| Motif name: | Etv3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 7 |
| Similarity score: | 0.00221043 |
Alignment:
VBBHAYTTCCGGTHDH
---CATTTCY------
| Original motif | Reverse complement motif |
| Consensus sequence: VBBHAYTTCCGGTHDH | Consensus sequence: DHHACCGGAAKTHBBB |
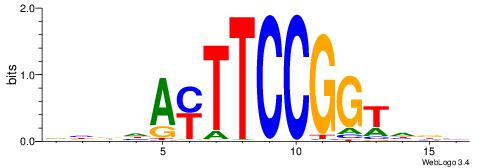 |
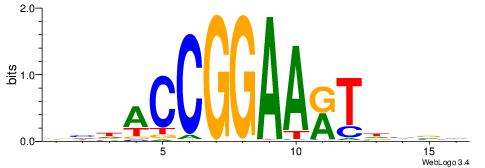 |
| Dataset #: 1 | Motif ID: 30 | Motif name: Motif 30 |
| Original motif | Reverse complement motif |
| Consensus sequence: CWGCAGC | Consensus sequence: GCTGCWG |
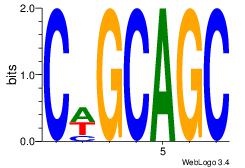 |
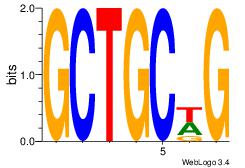 |
Best Matches for Motif ID 30 (Highest to Lowest)
| Motif ID: | UP00052 |
| Motif name: | Osr2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0 |
Alignment:
HHHHGCTACKGTHVHH
----GCTGCWG-----
| Original motif | Reverse complement motif |
| Consensus sequence: HHVHACRGTAGCHHHD | Consensus sequence: HHHHGCTACKGTHVHH |
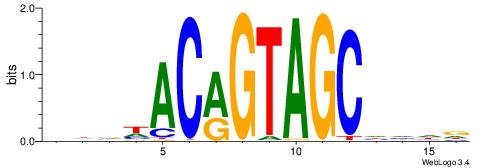 |
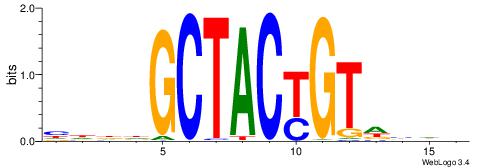 |
| Motif ID: | UP00027 |
| Motif name: | Osr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.00152865 |
Alignment:
DHBHGCTACKGTHDHH
----GCTGCWG-----
| Original motif | Reverse complement motif |
| Consensus sequence: HHDHACRGTAGCHVHD | Consensus sequence: DHBHGCTACKGTHDHH |
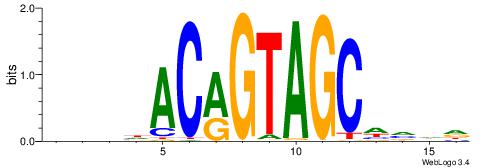 |
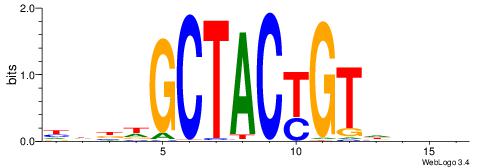 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 7 |
| Similarity score: | 0.00352181 |
Alignment:
HHDVVGCAGCTGVBKVB
---CWGCAGC-------
| Original motif | Reverse complement motif |
| Consensus sequence: BBYVVCAGCTGCBVHHD | Consensus sequence: HHDVVGCAGCTGVBKVB |
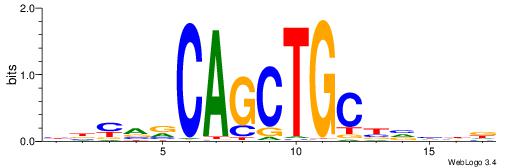 |
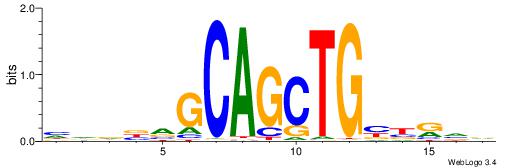 |
| Motif ID: | UP00102 |
| Motif name: | Zic1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 7 |
| Similarity score: | 0.0201867 |
Alignment:
BHDCVCAGCAGGHVD
-----CWGCAGC---
| Original motif | Reverse complement motif |
| Consensus sequence: BHDCVCAGCAGGHVD | Consensus sequence: DVHCCTGCTGBGDDB |
 |
 |
| Motif ID: | UP00057 |
| Motif name: | Zic2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.0241363 |
Alignment:
DVBCYTGCTGBGDDD
---GCTGCWG-----
| Original motif | Reverse complement motif |
| Consensus sequence: HHDCVCAGCAKGVVD | Consensus sequence: DVBCYTGCTGBGDDD |
 |
 |
| Dataset #: 1 | Motif ID: 31 | Motif name: Motif 31 |
| Original motif | Reverse complement motif |
| Consensus sequence: CATGYACA | Consensus sequence: TGTKCATG |
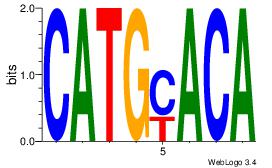 |
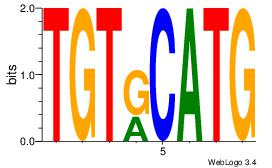 |
Best Matches for Motif ID 31 (Highest to Lowest)
| Motif ID: | UP00150 |
| Motif name: | Irx6 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.00505399 |
Alignment:
HAHDTACATGTAVDWHT
------CATGYACA---
| Original motif | Reverse complement motif |
| Consensus sequence: HAHDTACATGTAVDWHT | Consensus sequence: AHWDBTACATGTADHTH |
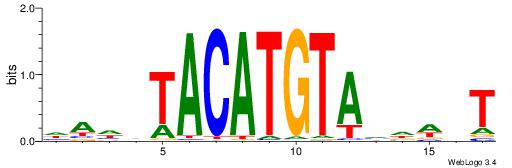 |
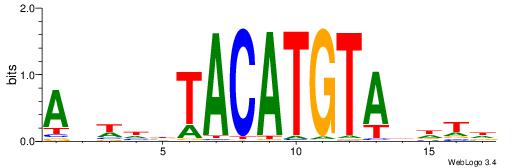 |
| Motif ID: | UP00097 |
| Motif name: | Mtf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.00507655 |
Alignment:
HDYTTGCACACGDHBH
--CATGYACA------
| Original motif | Reverse complement motif |
| Consensus sequence: DBDHCGTGTGCAAMDH | Consensus sequence: HDYTTGCACACGDHBH |
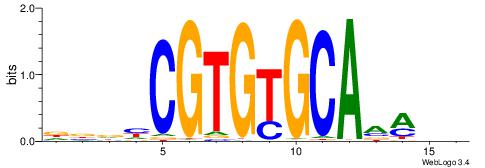 |
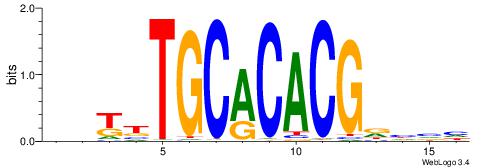 |
| Motif ID: | UP00223 |
| Motif name: | Irx3_2226.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.00836655 |
Alignment:
DHDWBTACATGTAHDDD
-------CATGYACA--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDHTACATGTAVWDHD | Consensus sequence: DHDWBTACATGTAHDDD |
 |
 |
| Motif ID: | UP00179 |
| Motif name: | Pou2f3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.00847056 |
Alignment:
BHDTATGCAAATBHDV
---CATGYACA-----
| Original motif | Reverse complement motif |
| Consensus sequence: BHDTATGCAAATBHDV | Consensus sequence: BHHVATTTGCATAHHV |
 |
 |
| Motif ID: | UP00191 |
| Motif name: | Pou2f2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.00973489 |
Alignment:
BHDTATGCAAATDHDV
---CATGYACA-----
| Original motif | Reverse complement motif |
| Consensus sequence: BHDTATGCAAATDHDV | Consensus sequence: BHHDATTTGCATAHHV |
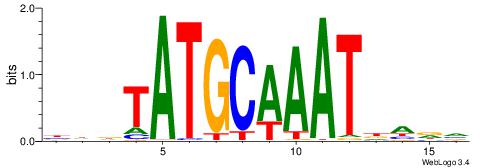 |
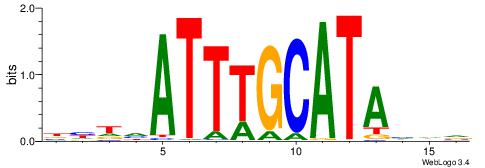 |
| Dataset #: 1 | Motif ID: 32 | Motif name: Motif 32 |
| Original motif | Reverse complement motif |
| Consensus sequence: KAATAAA | Consensus sequence: TTTATTY |
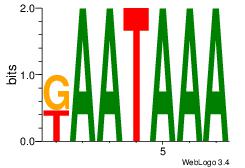 |
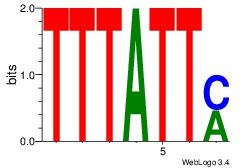 |
Best Matches for Motif ID 32 (Highest to Lowest)
| Motif ID: | UP00180 |
| Motif name: | Hoxd13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0 |
Alignment:
HDHHTTTTATTKKHDD
-----TTTATTY----
| Original motif | Reverse complement motif |
| Consensus sequence: HDHYYAATAAAAHHHH | Consensus sequence: HDHHTTTTATTKKHDD |
 |
 |
| Motif ID: | UP00133 |
| Motif name: | Cdx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.000531578 |
Alignment:
HRHTTTTATYMCCVBD
----TTTATTY-----
| Original motif | Reverse complement motif |
| Consensus sequence: DVVGGYMATAAAAHKH | Consensus sequence: HRHTTTTATYMCCVBD |
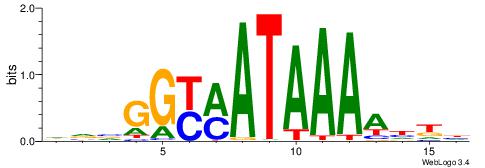 |
 |
| Motif ID: | UP00134 |
| Motif name: | Hoxb13 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.00373425 |
Alignment:
VRHCCAATAAAAWHHV
----KAATAAA-----
| Original motif | Reverse complement motif |
| Consensus sequence: VRHCCAATAAAAWHHV | Consensus sequence: VDHWTTTTATTGGDKB |
 |
 |
| Motif ID: | UP00121 |
| Motif name: | Hoxd10 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0.00617561 |
Alignment:
VDKVYAATAAAATDDDH
----KAATAAA------
| Original motif | Reverse complement motif |
| Consensus sequence: VDKVYAATAAAATDDDH | Consensus sequence: HDDDATTTTATTKVRDB |
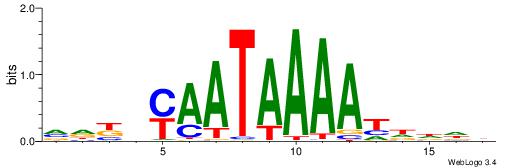 |
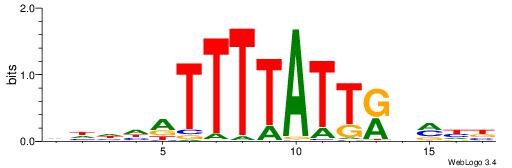 |
| Motif ID: | UP00217 |
| Motif name: | Hoxa10 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.00873461 |
Alignment:
DHBGYAATAAAATDHW
----KAATAAA-----
| Original motif | Reverse complement motif |
| Consensus sequence: DHBGYAATAAAATDHW | Consensus sequence: WDDATTTTATTMCBHD |
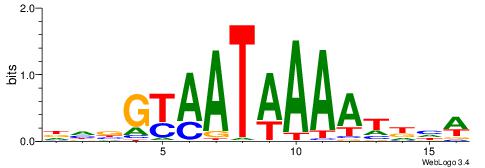 |
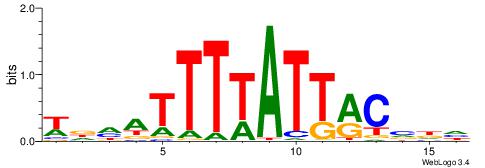 |
| Dataset #: 1 | Motif ID: 33 | Motif name: Motif 33 |
| Original motif | Reverse complement motif |
| Consensus sequence: CTGGCTWC | Consensus sequence: GWAGCCAG |
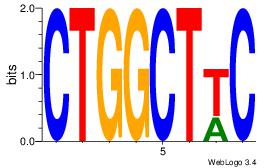 |
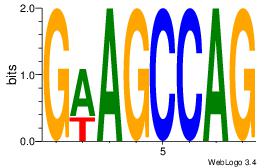 |
Best Matches for Motif ID 33 (Highest to Lowest)
| Motif ID: | UP00527 |
| Motif name: | Foxn4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
TCDAMVBGACGCMAKDDDVBBD
-------GWAGCCAG-------
| Original motif | Reverse complement motif |
| Consensus sequence: DBBVDDDRTRGCGTCBBYTHGA | Consensus sequence: TCDAMVBGACGCMAKDDDVBBD |
 |
 |
| Motif ID: | UP00045 |
| Motif name: | Mafb_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.00527751 |
Alignment:
DMTWDGTCAGCADWTTH
--GWAGCCAG-------
| Original motif | Reverse complement motif |
| Consensus sequence: HAAWDTGCTGACDWARH | Consensus sequence: DMTWDGTCAGCADWTTH |
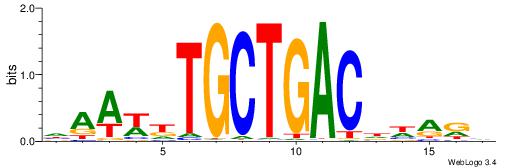 |
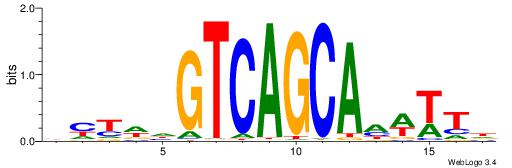 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0073813 |
Alignment:
HDHDDCCAGACABBHVH
-GWAGCCAG--------
| Original motif | Reverse complement motif |
| Consensus sequence: HDHDDCCAGACABBHVH | Consensus sequence: HVHBVTGTCTGGDDHDD |
 |
 |
| Motif ID: | UP00052 |
| Motif name: | Osr2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.00888762 |
Alignment:
HHVHACRGTAGCHHHD
-------GWAGCCAG-
| Original motif | Reverse complement motif |
| Consensus sequence: HHVHACRGTAGCHHHD | Consensus sequence: HHHHGCTACKGTHVHH |
 |
 |
| Motif ID: | UP00027 |
| Motif name: | Osr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.00951832 |
Alignment:
HHDHACRGTAGCHVHD
-------GWAGCCAG-
| Original motif | Reverse complement motif |
| Consensus sequence: HHDHACRGTAGCHVHD | Consensus sequence: DHBHGCTACKGTHDHH |
 |
 |
| Dataset #: 1 | Motif ID: 34 | Motif name: Motif 34 |
| Original motif | Reverse complement motif |
| Consensus sequence: TGKCCACA | Consensus sequence: TGTGGYCA |
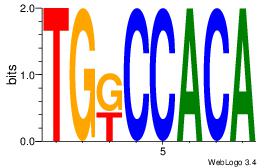 |
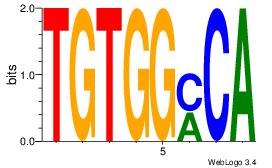 |
Best Matches for Motif ID 34 (Highest to Lowest)
| Motif ID: | UP00026 |
| Motif name: | Zscan4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HDHTMTGTGCACADVHD
-----TGKCCACA----
| Original motif | Reverse complement motif |
| Consensus sequence: DHVDTGTGCACAYAHDH | Consensus sequence: HDHTMTGTGCACADVHD |
 |
 |
| Motif ID: | UP00042 |
| Motif name: | Gm397_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.00132554 |
Alignment:
HHRTATGTGCACATHHD
-----TGKCCACA----
| Original motif | Reverse complement motif |
| Consensus sequence: HHDATGTGCACATAMDH | Consensus sequence: HHRTATGTGCACATHHD |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.0066252 |
Alignment:
DBCDWHCTGGGTGGTCMDHBBAH
---------TGTGGYCA------
| Original motif | Reverse complement motif |
| Consensus sequence: DTVBDDRGACCACCCAGDWDGBH | Consensus sequence: DBCDWHCTGGGTGGTCMDHBBAH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 10 |
| Number of overlap: | 8 |
| Similarity score: | 0.00683926 |
Alignment:
BTBVTCVTGGGTGGTCMVVDVBB
---------TGTGGYCA------
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
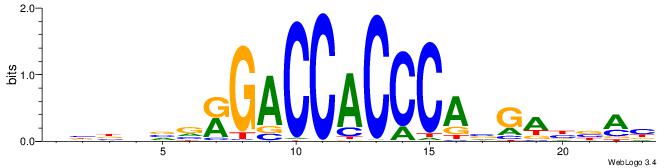 |
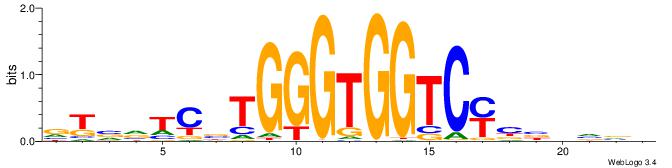 |
| Motif ID: | UP00053 |
| Motif name: | Rxra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.00716059 |
Alignment:
VHBHDGGGGTCAHBHBD
----TGTGGYCA-----
| Original motif | Reverse complement motif |
| Consensus sequence: DBHBDTGACCCCDHVHB | Consensus sequence: VHBHDGGGGTCAHBHBD |
 |
 |
| Dataset #: 1 | Motif ID: 35 | Motif name: Motif 35 |
| Original motif | Reverse complement motif |
| Consensus sequence: ACAACCAY | Consensus sequence: KTGGTTGT |
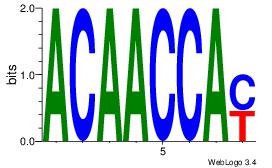 |
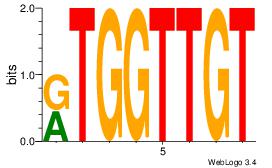 |
Best Matches for Motif ID 35 (Highest to Lowest)
| Motif ID: | UP00047 |
| Motif name: | Zbtb7b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
BDDRVGACCACCHBDVB
------ACAACCAY---
| Original motif | Reverse complement motif |
| Consensus sequence: BDDRVGACCACCHBDVB | Consensus sequence: BBDVHGGTGGTCBKDDB |
 |
 |
| Motif ID: | UP00227 |
| Motif name: | Duxl |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.00363031 |
Alignment:
VHVVGTTGATTGGGTMK
-----KTGGTTGT----
| Original motif | Reverse complement motif |
| Consensus sequence: YRACCCAATCAACVVHV | Consensus sequence: VHVVGTTGATTGGGTMK |
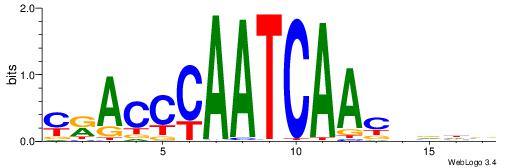 |
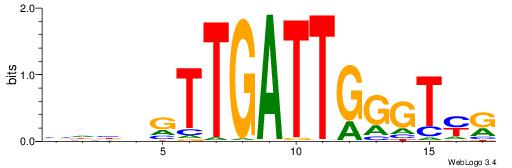 |
| Motif ID: | UP00166 |
| Motif name: | Barhl1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.00780833 |
Alignment:
DDVKTAATTGGTTDTH
-------KTGGTTGT-
| Original motif | Reverse complement motif |
| Consensus sequence: HAHAACCAATTARBDH | Consensus sequence: DDVKTAATTGGTTDTH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 10 |
| Number of overlap: | 8 |
| Similarity score: | 0.0118241 |
Alignment:
YBVDMGTGGGTGGTCKVVBVBBT
------KTGGTTGT---------
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0118666 |
Alignment:
CBDMCMGGGTGGTCCHVBVBAH
-----KTGGTTGT---------
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Dataset #: 1 | Motif ID: 36 | Motif name: Motif 36 |
| Original motif | Reverse complement motif |
| Consensus sequence: TCCTGGRA | Consensus sequence: TMCCAGGA |
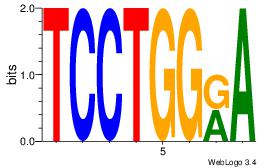 |
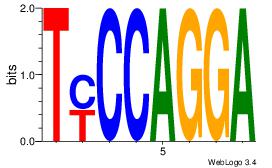 |
Best Matches for Motif ID 36 (Highest to Lowest)
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
VHHATTCCTYGAAAGD
-----TCCTGGRA---
| Original motif | Reverse complement motif |
| Consensus sequence: DCTTTCKAGGAATHHV | Consensus sequence: VHHATTCCTYGAAAGD |
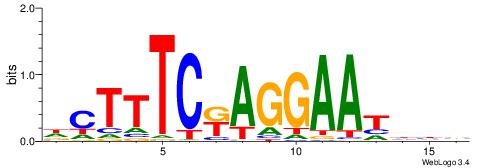 |
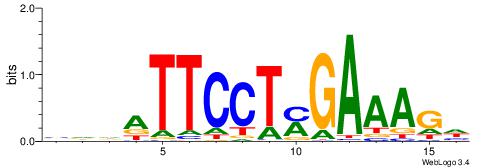 |
| Motif ID: | UP00038 |
| Motif name: | Spdef_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.00573581 |
Alignment:
VDWYTAGGATGTHVDH
-TMCCAGGA-------
| Original motif | Reverse complement motif |
| Consensus sequence: DDBHACATCCTAKWDV | Consensus sequence: VDWYTAGGATGTHVDH |
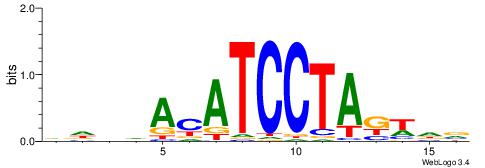 |
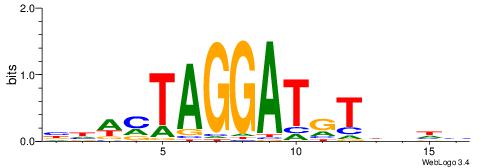 |
| Motif ID: | UP00413 |
| Motif name: | Elf4 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.00675491 |
Alignment:
HAWYCCGGAAGTHBBD
--TCCTGGRA------
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGKWTH | Consensus sequence: HAWYCCGGAAGTHBBD |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0109341 |
Alignment:
BDDYYCATCCCATDDBD
-------TMCCAGGA--
| Original motif | Reverse complement motif |
| Consensus sequence: DVDDATGGGATGKMDDV | Consensus sequence: BDDYYCATCCCATDDBD |
 |
 |
| Motif ID: | UP00410 |
| Motif name: | Elk1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.012227 |
Alignment:
DHBDACTTCCGGTVHVB
-------TCCTGGRA--
| Original motif | Reverse complement motif |
| Consensus sequence: VVHBACCGGAAGTDBHD | Consensus sequence: DHBDACTTCCGGTVHVB |
 |
 |
| Dataset #: 1 | Motif ID: 37 | Motif name: Motif 37 |
| Original motif | Reverse complement motif |
| Consensus sequence: CTGAGCYA | Consensus sequence: TKGCTCAG |
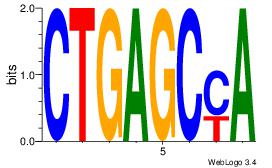 |
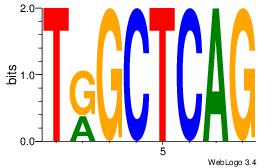 |
Best Matches for Motif ID 37 (Highest to Lowest)
| Motif ID: | UP00098 |
| Motif name: | Rfx3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HHHHDVVGTTGCTAHGGDHBAHH
--------TKGCTCAG-------
| Original motif | Reverse complement motif |
| Consensus sequence: HDTBHHCCHTAGCAACVVDHHHH | Consensus sequence: HHHHDVVGTTGCTAHGGDHBAHH |
 |
 |
| Motif ID: | UP00103 |
| Motif name: | Jundm2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.00321157 |
Alignment:
HDDKTGASTCAKHRDB
----TKGCTCAG----
| Original motif | Reverse complement motif |
| Consensus sequence: VDKDRTGASTCAYHDH | Consensus sequence: HDDKTGASTCAKHRDB |
 |
 |
| Motif ID: | UP00076 |
| Motif name: | Rfxdc2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.00414765 |
Alignment:
DVSGTTGCTAHGVBB
----TKGCTCAG---
| Original motif | Reverse complement motif |
| Consensus sequence: BBVCHTAGCAACSVD | Consensus sequence: DVSGTTGCTAHGVBB |
 |
 |
| Motif ID: | UP00056 |
| Motif name: | Rfx4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.00415736 |
Alignment:
DVSGTTGCTAHGGHV
----TKGCTCAG---
| Original motif | Reverse complement motif |
| Consensus sequence: BHCCHTAGCAACSVD | Consensus sequence: DVSGTTGCTAHGGHV |
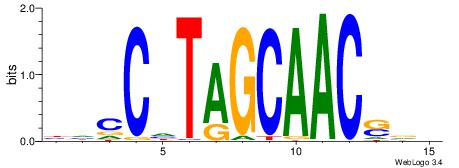 |
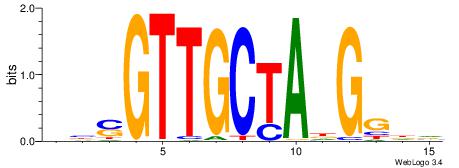 |
| Motif ID: | UP00045 |
| Motif name: | Mafb_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0106486 |
Alignment:
DMTWDGTCAGCADWTTH
--TKGCTCAG-------
| Original motif | Reverse complement motif |
| Consensus sequence: HAAWDTGCTGACDWARH | Consensus sequence: DMTWDGTCAGCADWTTH |
 |
 |
| Dataset #: 1 | Motif ID: 38 | Motif name: Motif 38 |
| Original motif | Reverse complement motif |
| Consensus sequence: GAGTTACA | Consensus sequence: TGTAACTC |
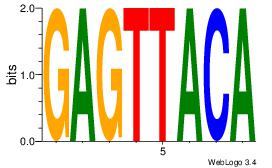 |
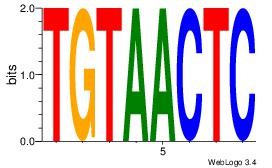 |
Best Matches for Motif ID 38 (Highest to Lowest)
| Motif ID: | UP00056 |
| Motif name: | Rfx4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HHBCHTRGWTACVVH
-----GAGTTACA--
| Original motif | Reverse complement motif |
| Consensus sequence: HHBCHTRGWTACVVH | Consensus sequence: HVVGTAWCKADGBHH |
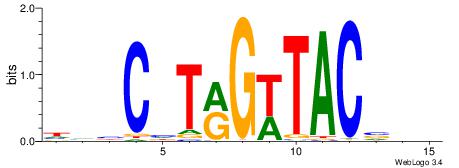 |
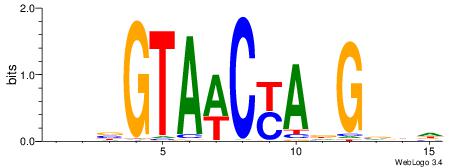 |
| Motif ID: | UP00103 |
| Motif name: | Jundm2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.00296274 |
Alignment:
VDKDRTGASTCAYHDH
------GAGTTACA--
| Original motif | Reverse complement motif |
| Consensus sequence: VDKDRTGASTCAYHDH | Consensus sequence: HDDKTGASTCAKHRDB |
 |
 |
| Motif ID: | UP00193 |
| Motif name: | Rhox11_1765.2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.00944938 |
Alignment:
AVDVCGCTGTWAWDVVW
-------TGTAACTC--
| Original motif | Reverse complement motif |
| Consensus sequence: AVDVCGCTGTWAWDVVW | Consensus sequence: WVVHWTWACAGCGBHBT |
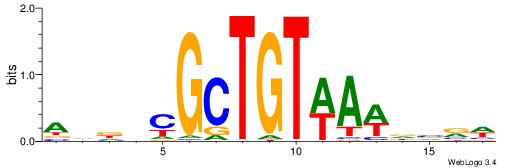 |
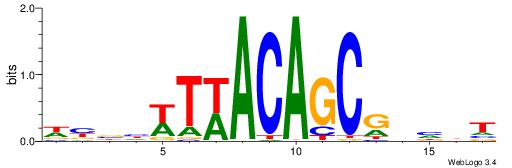 |
| Motif ID: | UP00205 |
| Motif name: | Pknox2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.00964683 |
Alignment:
HAVSRSCTGTCAATHB
-------TGTAACTC-
| Original motif | Reverse complement motif |
| Consensus sequence: HAVSRSCTGTCAATHB | Consensus sequence: VHATTGACAGSKSVTH |
 |
 |
| Motif ID: | UP00236 |
| Motif name: | Irx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.00974779 |
Alignment:
WVWDDTACATGTADDDW
-GAGTTACA--------
| Original motif | Reverse complement motif |
| Consensus sequence: WDDDTACATGTADDWBW | Consensus sequence: WVWDDTACATGTADDDW |
 |
 |
| Dataset #: 1 | Motif ID: 39 | Motif name: Motif 39 |
| Original motif | Reverse complement motif |
| Consensus sequence: BAGAAA | Consensus sequence: TTTCTB |
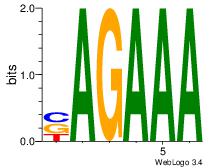 |
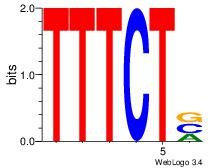 |
Best Matches for Motif ID 39 (Highest to Lowest)
| Motif ID: | UP00040 |
| Motif name: | Irf5_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
BDKGTTTCGKTWDDD
----TTTCTB-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDWAYCGAAACYDV | Consensus sequence: BDKGTTTCGKTWDDD |
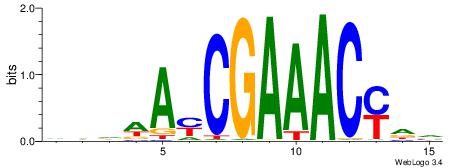 |
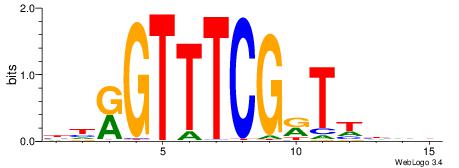 |
| Motif ID: | UP00018 |
| Motif name: | Irf4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.00208221 |
Alignment:
VDDAYCGAAACYDVA
----BAGAAA-----
| Original motif | Reverse complement motif |
| Consensus sequence: VDDAYCGAAACYDVA | Consensus sequence: TBDKGTTTCGMTDHV |
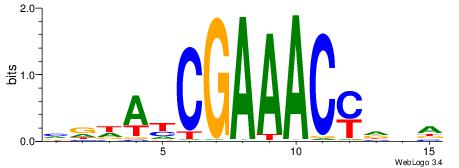 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 11 |
| Number of overlap: | 6 |
| Similarity score: | 0.00450619 |
Alignment:
BDHDDADAGATAAGADHBDHVD
------BAGAAA----------
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Motif ID: | UP00074 |
| Motif name: | Isgf3g_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.00517682 |
Alignment:
TDAGTTTCGWTTKDV
----TTTCTB-----
| Original motif | Reverse complement motif |
| Consensus sequence: VDRAAWCGAAACTDA | Consensus sequence: TDAGTTTCGWTTKDV |
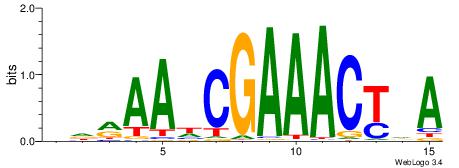 |
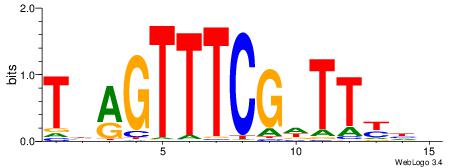 |
| Motif ID: | UP00011 |
| Motif name: | Irf6_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.00721031 |
Alignment:
HDDAHCGAAACYAVDDD
----BAGAAA-------
| Original motif | Reverse complement motif |
| Consensus sequence: HDDAHCGAAACYAVDDD | Consensus sequence: DHDBTKGTTTCGHTHDD |
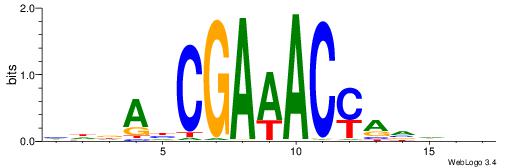 |
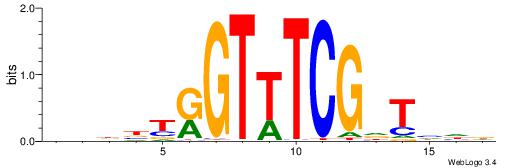 |
| Dataset #: 1 | Motif ID: 40 | Motif name: Motif 40 |
| Original motif | Reverse complement motif |
| Consensus sequence: ARTCCCAG | Consensus sequence: CTGGGAMT |
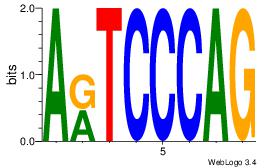 |
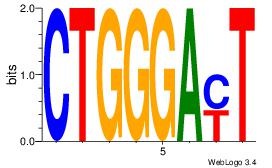 |
Best Matches for Motif ID 40 (Highest to Lowest)
| Motif ID: | UP00216 |
| Motif name: | Obox1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
DHDGTTAATCCCCKTDD
------ARTCCCAG---
| Original motif | Reverse complement motif |
| Consensus sequence: DDARGGGGATTAACDHH | Consensus sequence: DHDGTTAATCCCCKTDD |
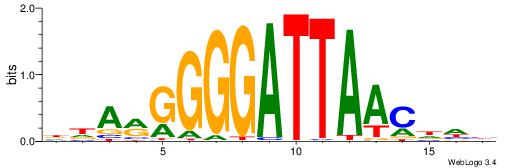 |
 |
| Motif ID: | UP00229 |
| Motif name: | Otx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.00997246 |
Alignment:
VHHWATTAATCCCMDHH
-------ARTCCCAG--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDRGGGATTAATWHHB | Consensus sequence: VHHWATTAATCCCMDHH |
 |
 |
| Motif ID: | UP00160 |
| Motif name: | Obox3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.0101137 |
Alignment:
HHWGTTAATCCCCCBBH
------ARTCCCAG---
| Original motif | Reverse complement motif |
| Consensus sequence: HBVGGGGGATTAACWHH | Consensus sequence: HHWGTTAATCCCCCBBH |
 |
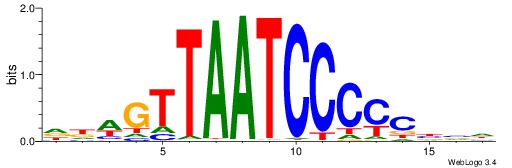 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0101427 |
Alignment:
BDDYYCATCCCATDDBD
-----ARTCCCAG----
| Original motif | Reverse complement motif |
| Consensus sequence: DVDDATGGGATGKMDDV | Consensus sequence: BDDYYCATCCCATDDBD |
 |
 |
| Motif ID: | UP00208 |
| Motif name: | Obox5_2284.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0112114 |
Alignment:
DHHAHTTAATCCCKVDH
-------ARTCCCAG--
| Original motif | Reverse complement motif |
| Consensus sequence: HDVRGGGATTAAHTHHH | Consensus sequence: DHHAHTTAATCCCKVDH |
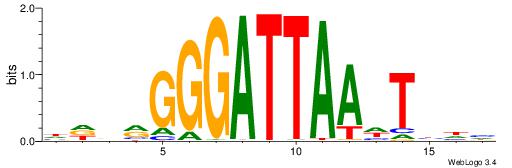 |
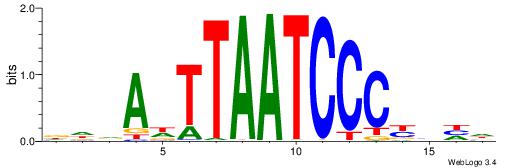 |
| Dataset #: 1 | Motif ID: 41 | Motif name: Motif 41 |
| Original motif | Reverse complement motif |
| Consensus sequence: CCTGCTGK | Consensus sequence: YCAGCAGG |
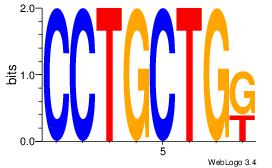 |
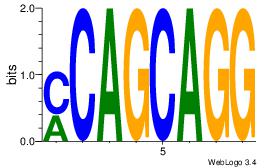 |
Best Matches for Motif ID 41 (Highest to Lowest)
| Motif ID: | UP00102 |
| Motif name: | Zic1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
DVHCCTGCTGBGDDB
---CCTGCTGK----
| Original motif | Reverse complement motif |
| Consensus sequence: BHDCVCAGCAGGHVD | Consensus sequence: DVHCCTGCTGBGDDB |
 |
 |
| Motif ID: | UP00057 |
| Motif name: | Zic2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0043517 |
Alignment:
DVBCYTGCTGBGDDD
---CCTGCTGK----
| Original motif | Reverse complement motif |
| Consensus sequence: HHDCVCAGCAKGVVD | Consensus sequence: DVBCYTGCTGBGDDD |
 |
 |
| Motif ID: | UP00006 |
| Motif name: | Zic3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.00761363 |
Alignment:
DVHCYTGCTGTGHDH
---CCTGCTGK----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDCACAGCAKGHVD | Consensus sequence: DVHCYTGCTGTGHDH |
 |
 |
| Motif ID: | UP00045 |
| Motif name: | Mafb_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.0402767 |
Alignment:
DMTWDGTCAGCADWTTH
------YCAGCAGG---
| Original motif | Reverse complement motif |
| Consensus sequence: HAAWDTGCTGACDWARH | Consensus sequence: DMTWDGTCAGCADWTTH |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0490068 |
Alignment:
VBHHVCAGGTGCDVDHD
-YCAGCAGG--------
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Dataset #: 1 | Motif ID: 42 | Motif name: Motif 42 |
| Original motif | Reverse complement motif |
| Consensus sequence: CGCGCSG | Consensus sequence: CSGCGCG |
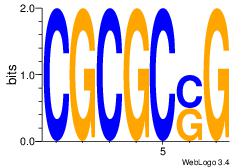 |
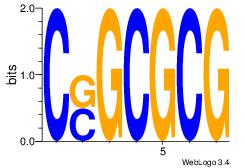 |
Best Matches for Motif ID 42 (Highest to Lowest)
| Motif ID: | UP00003 |
| Motif name: | E2F3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 7 |
| Similarity score: | 0 |
Alignment:
WHSGCGCGCCHTDHB
-CSGCGCG-------
| Original motif | Reverse complement motif |
| Consensus sequence: VHDADGGCGCGCSHW | Consensus sequence: WHSGCGCGCCHTDHB |
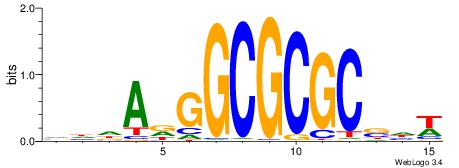 |
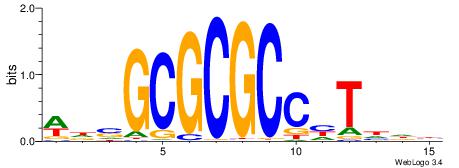 |
| Motif ID: | UP00001 |
| Motif name: | E2F2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0.000129734 |
Alignment:
HHVGCGCGCCKTDHB
----CGCGCSG----
| Original motif | Reverse complement motif |
| Consensus sequence: VHDARGGCGCGCVHH | Consensus sequence: HHVGCGCGCCKTDHB |
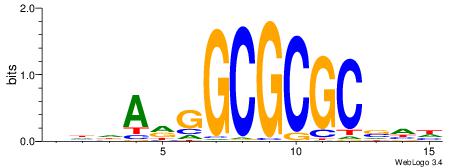 |
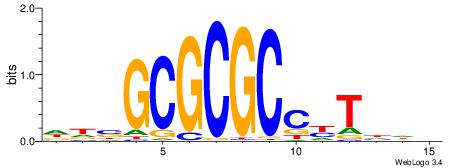 |
| Motif ID: | UP00065 |
| Motif name: | Zfp161_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 7 |
| Similarity score: | 0.0145336 |
Alignment:
DMMDGMGCGCGCGCHD
-------CGCGCSG--
| Original motif | Reverse complement motif |
| Consensus sequence: DDGCGCGCGCRCHYRD | Consensus sequence: DMMDGMGCGCGCGCHD |
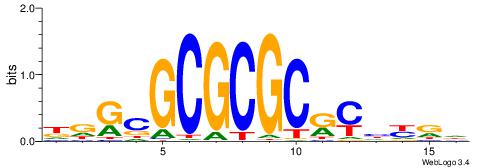 |
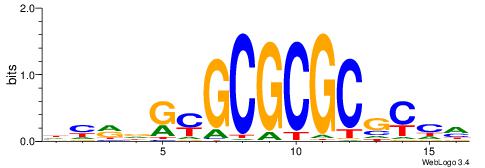 |
| Motif ID: | UP00065 |
| Motif name: | Zfp161_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 7 |
| Similarity score: | 0.016879 |
Alignment:
VVGCVMTGCGCGKC
-----CSGCGCG--
| Original motif | Reverse complement motif |
| Consensus sequence: GYCGCGCARBGCVB | Consensus sequence: VVGCVMTGCGCGKC |
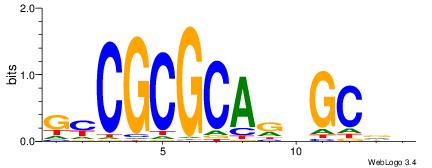 |
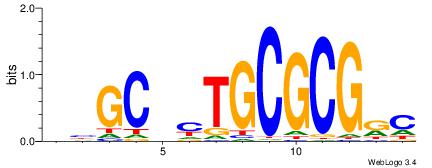 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 7 |
| Similarity score: | 0.0219936 |
Alignment:
HDWHMHGACGCGCGTRBVVBHW
-------CSGCGCG--------
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
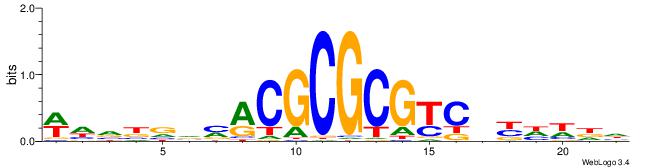 |
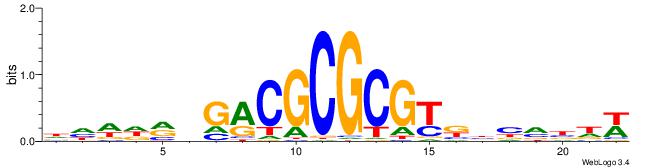 |
| Dataset #: 1 | Motif ID: 43 | Motif name: Motif 43 |
| Original motif | Reverse complement motif |
| Consensus sequence: ACATYTA | Consensus sequence: TAMATGT |
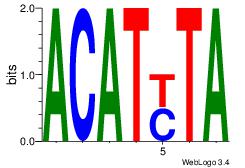 |
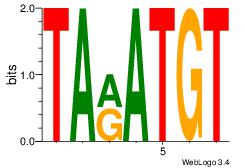 |
Best Matches for Motif ID 43 (Highest to Lowest)
| Motif ID: | UP00236 |
| Motif name: | Irx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0 |
Alignment:
WVWDDTACATGTADDDW
-----TAMATGT-----
| Original motif | Reverse complement motif |
| Consensus sequence: WDDDTACATGTADDWBW | Consensus sequence: WVWDDTACATGTADDDW |
 |
 |
| Motif ID: | UP00150 |
| Motif name: | Irx6 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 3.6652e-05 |
Alignment:
AHWDBTACATGTADHTH
-----TAMATGT-----
| Original motif | Reverse complement motif |
| Consensus sequence: HAHDTACATGTAVDWHT | Consensus sequence: AHWDBTACATGTADHTH |
 |
 |
| Motif ID: | UP00223 |
| Motif name: | Irx3_2226.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.000501187 |
Alignment:
DHDWBTACATGTAHDDD
-----TAMATGT-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDHTACATGTAVWDHD | Consensus sequence: DHDWBTACATGTAHDDD |
 |
 |
| Motif ID: | UP00223 |
| Motif name: | Irx3_0920.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.00389424 |
Alignment:
DDWWBTACATGTADDDD
-----TAMATGT-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDDTACATGTAVWWHD | Consensus sequence: DDWWBTACATGTADDDD |
 |
 |
| Motif ID: | UP00250 |
| Motif name: | Irx5 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.0071841 |
Alignment:
WVTDHWACATGTWHDDD
-----TAMATGT-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDHWACATGTWHDABW | Consensus sequence: WVTDHWACATGTWHDDD |
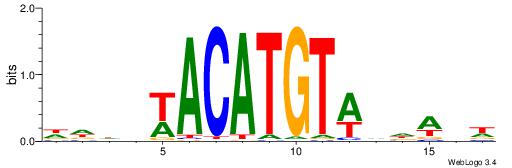 |
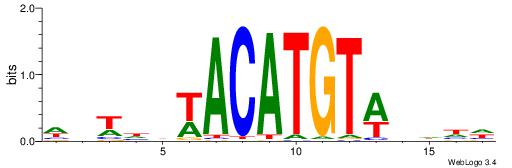 |
| Dataset #: 1 | Motif ID: 44 | Motif name: Motif 44 |
| Original motif | Reverse complement motif |
| Consensus sequence: GDAAACA | Consensus sequence: TGTTTDC |
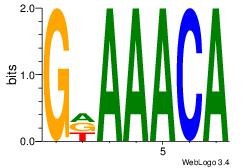 |
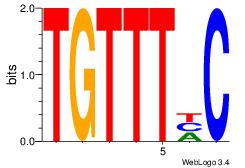 |
Best Matches for Motif ID 44 (Highest to Lowest)
| Motif ID: | UP00025 |
| Motif name: | Foxk1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0 |
Alignment:
BBVTTTGTTTACATTDD
-----TGTTTDC-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDAATGTAAACAAAVVB | Consensus sequence: BBVTTTGTTTACATTDD |
 |
 |
| Motif ID: | UP00073 |
| Motif name: | Foxa2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.00709733 |
Alignment:
BDVTTTKTTTACWTWHH
-----TGTTTDC-----
| Original motif | Reverse complement motif |
| Consensus sequence: HHWAWGTAAAYAAAVDB | Consensus sequence: BDVTTTKTTTACWTWHH |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.00723594 |
Alignment:
DWAATRTAAACAAWVVB
-----GDAAACA-----
| Original motif | Reverse complement motif |
| Consensus sequence: DWAATRTAAACAAWVVB | Consensus sequence: VVVWTTGTTTAMATTWD |
 |
 |
| Motif ID: | UP00039 |
| Motif name: | Foxj3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.0079365 |
Alignment:
HDHADGTAAACAAAVVH
-----GDAAACA-----
| Original motif | Reverse complement motif |
| Consensus sequence: HDHADGTAAACAAAVVH | Consensus sequence: DVVTTTGTTTACDTHDH |
 |
 |
| Motif ID: | UP00041 |
| Motif name: | Foxj1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 7 |
| Similarity score: | 0.0162324 |
Alignment:
DDDHTTTGTTTAMHDH
------TGTTTDC---
| Original motif | Reverse complement motif |
| Consensus sequence: HDHRTAAACAAAHDDD | Consensus sequence: DDDHTTTGTTTAMHDH |
 |
 |
| Dataset #: 1 | Motif ID: 45 | Motif name: Motif 45 |
| Original motif | Reverse complement motif |
| Consensus sequence: GSAGAGA | Consensus sequence: TCTCTSC |
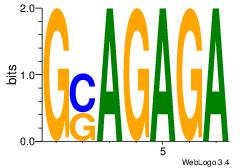 |
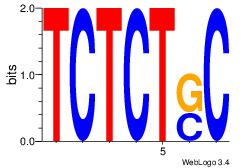 |
Best Matches for Motif ID 45 (Highest to Lowest)
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0 |
Alignment:
DDBBBCACTGCABTBBB
----TCTCTSC------
| Original motif | Reverse complement motif |
| Consensus sequence: DDBBBCACTGCABTBBB | Consensus sequence: BBBAVTGCAGTGBBVDD |
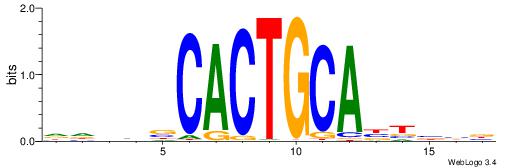 |
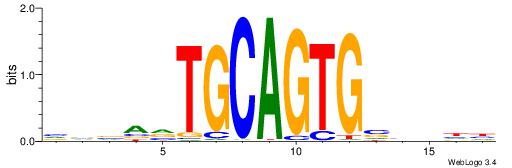 |
| Motif ID: | UP00018 |
| Motif name: | Irf4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.00147657 |
Alignment:
BHHWCCGAGAMTVVB
---GSAGAGA-----
| Original motif | Reverse complement motif |
| Consensus sequence: VVBAYTCTCGGWHDB | Consensus sequence: BHHWCCGAGAMTVVB |
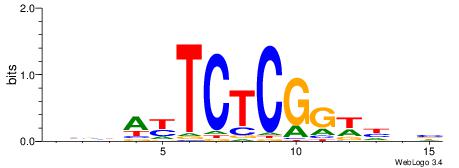 |
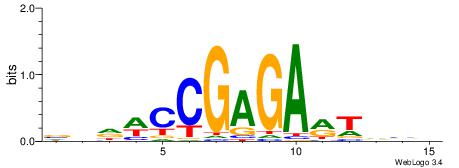 |
| Motif ID: | UP00040 |
| Motif name: | Irf5_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.00225471 |
Alignment:
BHDAHCGAGARTHBV
---GSAGAGA-----
| Original motif | Reverse complement motif |
| Consensus sequence: BHDAHCGAGARTHBV | Consensus sequence: VBHAKTCTCGHTHHV |
 |
 |
| Motif ID: | UP00026 |
| Motif name: | Zscan4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 7 |
| Similarity score: | 0.00336112 |
Alignment:
HDHTMTGTGCACADVHD
---TCTCTSC-------
| Original motif | Reverse complement motif |
| Consensus sequence: DHVDTGTGCACAYAHDH | Consensus sequence: HDHTMTGTGCACADVHD |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.00351001 |
Alignment:
BDHDDADAGATAAGADHBDHVD
-----GSAGAGA----------
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Dataset #: 2 | Motif ID: 46 | Motif name: Motif 46 |
| Original motif | Reverse complement motif |
| Consensus sequence: AGRKGGCR | Consensus sequence: KGCCYKCT |
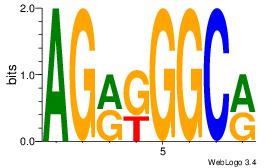 |
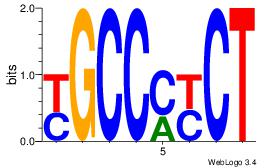 |
Best Matches for Motif ID 46 (Highest to Lowest)
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
BVVHAGGGGGCGRDHHB
----AGRKGGCR-----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0154192 |
Alignment:
VKBBAGGGGTCAHDBBH
----AGRKGGCR-----
| Original motif | Reverse complement motif |
| Consensus sequence: VKBBAGGGGTCAHDBBH | Consensus sequence: DBBHHTGACCCCTBBYV |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0160089 |
Alignment:
VHTGCGGGGGCGGH
----AGRKGGCR--
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
| Motif ID: | UP00010 |
| Motif name: | Tcfap2b_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0241611 |
Alignment:
HTGCCMTVKGGCMD
-KGCCYKCT-----
| Original motif | Reverse complement motif |
| Consensus sequence: DYGCCYBARGGCAH | Consensus sequence: HTGCCMTVKGGCMD |
 |
 |
| Motif ID: | UP00070 |
| Motif name: | Gcm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0267457 |
Alignment:
DDBVHATAGGGGRBRMB
-------AGRKGGCR--
| Original motif | Reverse complement motif |
| Consensus sequence: DDBVHATAGGGGRBRMB | Consensus sequence: BYMBKCCCCTATDVBHD |
 |
 |
| Dataset #: 2 | Motif ID: 47 | Motif name: Motif 47 |
| Original motif | Reverse complement motif |
| Consensus sequence: CAGYDCC | Consensus sequence: GGDKCTG |
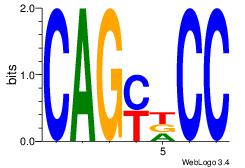 |
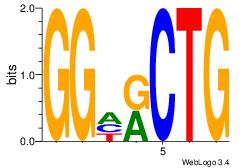 |
Best Matches for Motif ID 47 (Highest to Lowest)
| Motif ID: | UP00205 |
| Motif name: | Pknox2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 7 |
| Similarity score: | 0.0213831 |
Alignment:
HAVSRSCTGTCAATHB
--GGDKCTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: HAVSRSCTGTCAATHB | Consensus sequence: VHATTGACAGSKSVTH |
 |
 |
| Motif ID: | UP00122 |
| Motif name: | Tgif1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 7 |
| Similarity score: | 0.0245571 |
Alignment:
DABWWTGACAGCTVVBD
--------CAGYDCC--
| Original motif | Reverse complement motif |
| Consensus sequence: DABWWTGACAGCTVVBD | Consensus sequence: DBVVAGCTGTCAWWVTH |
 |
 |
| Motif ID: | UP00203 |
| Motif name: | Pknox1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 7 |
| Similarity score: | 0.0300471 |
Alignment:
VVWTTGACAGSTSBKD
-------CAGYDCC--
| Original motif | Reverse complement motif |
| Consensus sequence: DRVSASCTGTCAAWVV | Consensus sequence: VVWTTGACAGSTSBKD |
 |
 |
| Motif ID: | UP00258 |
| Motif name: | Tgif2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 7 |
| Similarity score: | 0.0300683 |
Alignment:
VBWTTGACAGCTVBTT
-------CAGYDCC--
| Original motif | Reverse complement motif |
| Consensus sequence: AABBAGCTGTCAAWVV | Consensus sequence: VBWTTGACAGCTVBTT |
 |
 |
| Motif ID: | UP00193 |
| Motif name: | Rhox11_2205.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 7 |
| Similarity score: | 0.0305033 |
Alignment:
WVVHWTWACAGCGHHVT
--------CAGYDCC--
| Original motif | Reverse complement motif |
| Consensus sequence: AVDHCGCTGTWAWDVVW | Consensus sequence: WVVHWTWACAGCGHHVT |
 |
 |
| Dataset #: 2 | Motif ID: 48 | Motif name: Motif 48 |
| Original motif | Reverse complement motif |
| Consensus sequence: CCACYAGR | Consensus sequence: MCTKGTGG |
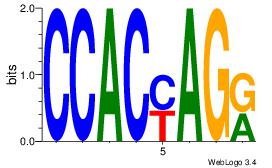 |
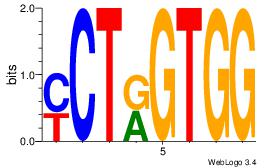 |
Best Matches for Motif ID 48 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HGDHVHKGACCACCCGGBGBGDB
---------CCACYAGR------
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 11 |
| Number of overlap: | 8 |
| Similarity score: | 0.000875703 |
Alignment:
CBDMCMGGGTGGTCCHVBVBAH
----MCTKGTGG----------
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Motif ID: | UP00047 |
| Motif name: | Zbtb7b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.00261558 |
Alignment:
BDDRVGACCACCHBDVB
-------CCACYAGR--
| Original motif | Reverse complement motif |
| Consensus sequence: BDDRVGACCACCHBDVB | Consensus sequence: BBDVHGGTGGTCBKDDB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.00296024 |
Alignment:
CVHBSCGGGTGGTCMHVHHCDH
----MCTKGTGG----------
| Original motif | Reverse complement motif |
| Consensus sequence: HDGHHBHRGACCACCCGSVDVG | Consensus sequence: CVHBSCGGGTGGTCMHVHHCDH |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0161251 |
Alignment:
VBHHVCAGGTGCDVDHD
----MCTKGTGG-----
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Dataset #: 2 | Motif ID: 49 | Motif name: Motif 49 |
| Original motif | Reverse complement motif |
| Consensus sequence: CTGGCCTC | Consensus sequence: GAGGCCAG |
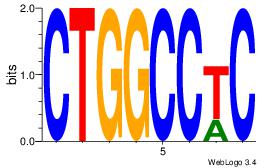 |
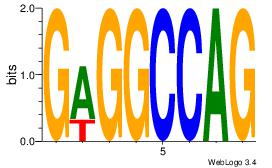 |
Best Matches for Motif ID 49 (Highest to Lowest)
| Motif ID: | UP00527 |
| Motif name: | Foxn4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
TCDAMVBGACGCMAKDDDVBBD
-------GAGGCCAG-------
| Original motif | Reverse complement motif |
| Consensus sequence: DBBVDDDRTRGCGTCBBYTHGA | Consensus sequence: TCDAMVBGACGCMAKDDDVBBD |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.00199504 |
Alignment:
DBBHHTGACCCCTBBYV
----CTGGCCTC-----
| Original motif | Reverse complement motif |
| Consensus sequence: VKBBAGGGGTCAHDBBH | Consensus sequence: DBBHHTGACCCCTBBYV |
 |
 |
| Motif ID: | UP00035 |
| Motif name: | Hic1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0108368 |
Alignment:
DDBHGGTTGGCATVBD
------CTGGCCTC--
| Original motif | Reverse complement motif |
| Consensus sequence: DBBATGCCAACCHVHH | Consensus sequence: DDBHGGTTGGCATVBD |
 |
 |
| Motif ID: | UP00045 |
| Motif name: | Mafb_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0117596 |
Alignment:
DMTWDGTCAGCADWTTH
--GAGGCCAG-------
| Original motif | Reverse complement motif |
| Consensus sequence: HAAWDTGCTGACDWARH | Consensus sequence: DMTWDGTCAGCADWTTH |
 |
 |
| Motif ID: | UP00053 |
| Motif name: | Rxra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0118075 |
Alignment:
VHBHDGGGGTCAHBHBD
-----GAGGCCAG----
| Original motif | Reverse complement motif |
| Consensus sequence: DBHBDTGACCCCDHVHB | Consensus sequence: VHBHDGGGGTCAHBHBD |
 |
 |
| Dataset #: 2 | Motif ID: 50 | Motif name: Motif 50 |
| Original motif | Reverse complement motif |
| Consensus sequence: AGRAA | Consensus sequence: TTMCT |
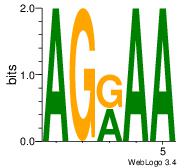 |
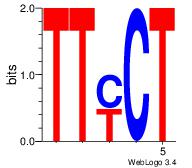 |
Best Matches for Motif ID 50 (Highest to Lowest)
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 5 |
| Similarity score: | 0 |
Alignment:
DCTTTCKAGGAATHHV
-------AGRAA----
| Original motif | Reverse complement motif |
| Consensus sequence: DCTTTCKAGGAATHHV | Consensus sequence: VHHATTCCTYGAAAGD |
 |
 |
| Motif ID: | UP00409 |
| Motif name: | Elf5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 5 |
| Similarity score: | 0.00150766 |
Alignment:
BWAHSMGGAAGTWD
-----AGRAA----
| Original motif | Reverse complement motif |
| Consensus sequence: BWAHSMGGAAGTWD | Consensus sequence: DWACTTCCRSDTWV |
 |
 |
| Motif ID: | UP00090 |
| Motif name: | Elf3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 5 |
| Similarity score: | 0.00378425 |
Alignment:
TDAMTTCCYYDTD
----TTMCT----
| Original motif | Reverse complement motif |
| Consensus sequence: DAHMMGGAARTDA | Consensus sequence: TDAMTTCCYYDTD |
 |
 |
| Motif ID: | UP00181 |
| Motif name: | Barx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 10 |
| Number of overlap: | 5 |
| Similarity score: | 0.00386973 |
Alignment:
WTHHMSKAATTMVKHY
---------TTMCT--
| Original motif | Reverse complement motif |
| Consensus sequence: MHRVYAATTRSYDHAW | Consensus sequence: WTHHMSKAATTMVKHY |
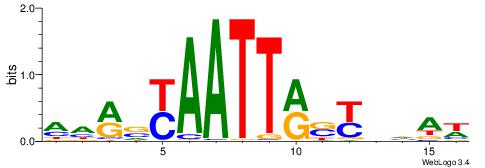 |
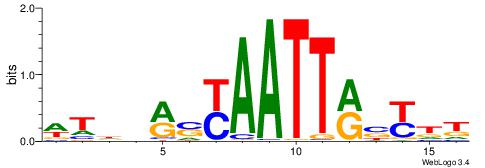 |
| Motif ID: | UP00085 |
| Motif name: | Sfpi1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 5 |
| Similarity score: | 0.00623266 |
Alignment:
BDAWGVGGAAGTDH
-----AGRAA----
| Original motif | Reverse complement motif |
| Consensus sequence: BDAWGVGGAAGTDH | Consensus sequence: HDACTTCCBCWTDV |
 |
 |
| Dataset #: 2 | Motif ID: 51 | Motif name: Motif 51 |
| Original motif | Reverse complement motif |
| Consensus sequence: GAGTTACA | Consensus sequence: TGTAACTC |
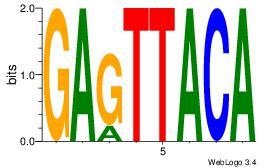 |
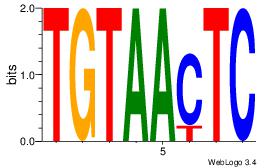 |
Best Matches for Motif ID 51 (Highest to Lowest)
| Motif ID: | UP00103 |
| Motif name: | Jundm2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
VDKDRTGASTCAYHDH
------GAGTTACA--
| Original motif | Reverse complement motif |
| Consensus sequence: VDKDRTGASTCAYHDH | Consensus sequence: HDDKTGASTCAKHRDB |
 |
 |
| Motif ID: | UP00193 |
| Motif name: | Rhox11_1765.2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.00122205 |
Alignment:
AVDVCGCTGTWAWDVVW
-------TGTAACTC--
| Original motif | Reverse complement motif |
| Consensus sequence: AVDVCGCTGTWAWDVVW | Consensus sequence: WVVHWTWACAGCGBHBT |
 |
 |
| Motif ID: | UP00205 |
| Motif name: | Pknox2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0014195 |
Alignment:
HAVSRSCTGTCAATHB
-------TGTAACTC-
| Original motif | Reverse complement motif |
| Consensus sequence: HAVSRSCTGTCAATHB | Consensus sequence: VHATTGACAGSKSVTH |
 |
 |
| Motif ID: | UP00236 |
| Motif name: | Irx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.00152046 |
Alignment:
WVWDDTACATGTADDDW
-GAGTTACA--------
| Original motif | Reverse complement motif |
| Consensus sequence: WDDDTACATGTADDWBW | Consensus sequence: WVWDDTACATGTADDDW |
 |
 |
| Motif ID: | UP00193 |
| Motif name: | Rhox11_2205.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.00217941 |
Alignment:
AVDHCGCTGTWAWDVVW
-------TGTAACTC--
| Original motif | Reverse complement motif |
| Consensus sequence: AVDHCGCTGTWAWDVVW | Consensus sequence: WVVHWTWACAGCGHHVT |
 |
 |
| Dataset #: 2 | Motif ID: 52 | Motif name: Motif 52 |
| Original motif | Reverse complement motif |
| Consensus sequence: AAATAAAW | Consensus sequence: WTTTATTT |
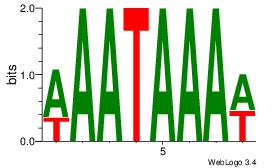 |
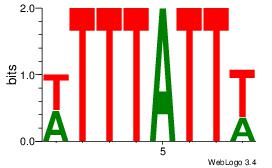 |
Best Matches for Motif ID 52 (Highest to Lowest)
| Motif ID: | UP00133 |
| Motif name: | Cdx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
DVVGGYMATAAAAHKH
-----AAATAAAW---
| Original motif | Reverse complement motif |
| Consensus sequence: DVVGGYMATAAAAHKH | Consensus sequence: HRHTTTTATYMCCVBD |
 |
 |
| Motif ID: | UP00180 |
| Motif name: | Hoxd13 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.00105399 |
Alignment:
HDHYYAATAAAAHHHH
----AAATAAAW----
| Original motif | Reverse complement motif |
| Consensus sequence: HDHYYAATAAAAHHHH | Consensus sequence: HDHHTTTTATTKKHDD |
 |
 |
| Motif ID: | UP00134 |
| Motif name: | Hoxb13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.00468315 |
Alignment:
VDHWTTTTATTGGDKB
----WTTTATTT----
| Original motif | Reverse complement motif |
| Consensus sequence: VRHCCAATAAAAWHHV | Consensus sequence: VDHWTTTTATTGGDKB |
 |
 |
| Motif ID: | UP00217 |
| Motif name: | Hoxa10 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.00632827 |
Alignment:
DHBGYAATAAAATDHW
----AAATAAAW----
| Original motif | Reverse complement motif |
| Consensus sequence: DHBGYAATAAAATDHW | Consensus sequence: WDDATTTTATTMCBHD |
 |
 |
| Motif ID: | UP00121 |
| Motif name: | Hoxd10 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.00650679 |
Alignment:
HDDDATTTTATTKVRDB
-----WTTTATTT----
| Original motif | Reverse complement motif |
| Consensus sequence: VDKVYAATAAAATDDDH | Consensus sequence: HDDDATTTTATTKVRDB |
 |
 |
| Dataset #: 2 | Motif ID: 53 | Motif name: Motif 53 |
| Original motif | Reverse complement motif |
| Consensus sequence: CAGATCCC | Consensus sequence: GGGATCTG |
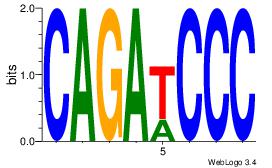 |
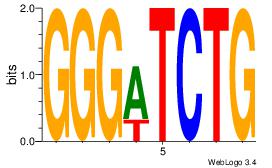 |
Best Matches for Motif ID 53 (Highest to Lowest)
| Motif ID: | UP00232 |
| Motif name: | Dobox4 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HWHWTAGATACCCYWTD
----CAGATCCC-----
| Original motif | Reverse complement motif |
| Consensus sequence: HWHWTAGATACCCYWTD | Consensus sequence: DAWKGGGTATCTAWHWH |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.0114177 |
Alignment:
DVDDATGGGATGKMDDV
------GGGATCTG---
| Original motif | Reverse complement motif |
| Consensus sequence: DVDDATGGGATGKMDDV | Consensus sequence: BDDYYCATCCCATDDBD |
 |
 |
| Motif ID: | UP00019 |
| Motif name: | Zbtb12_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0161888 |
Alignment:
BDHRBGTTCTAGATCVB
---GGGATCTG------
| Original motif | Reverse complement motif |
| Consensus sequence: BDHRBGTTCTAGATCVB | Consensus sequence: BBGATCTAGAACBKHDB |
 |
 |
| Motif ID: | UP00265 |
| Motif name: | Pitx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0173992 |
Alignment:
VGGGGGATTAGCDDBC
---GGGATCTG-----
| Original motif | Reverse complement motif |
| Consensus sequence: VGGGGGATTAGCDDBC | Consensus sequence: GBHDGCTAATCCCCCB |
 |
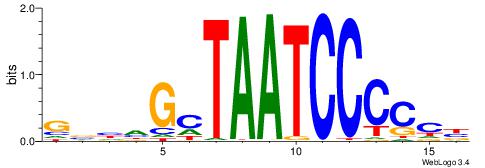 |
| Motif ID: | UP00024 |
| Motif name: | Glis2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0190616 |
Alignment:
HHHVGACCCCCCVBRD
--CAGATCCC------
| Original motif | Reverse complement motif |
| Consensus sequence: HHHVGACCCCCCVBRD | Consensus sequence: HKBBGGGGGGTCVHHH |
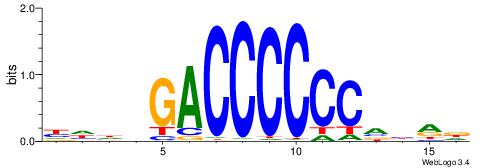 |
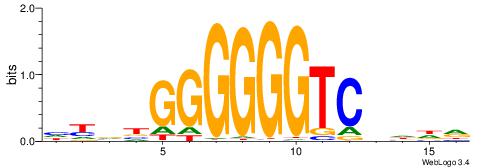 |
| Dataset #: 2 | Motif ID: 54 | Motif name: Motif 54 |
| Original motif | Reverse complement motif |
| Consensus sequence: ATTYY | Consensus sequence: MKAAT |
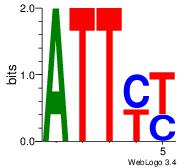 |
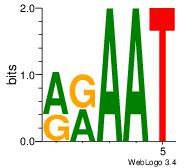 |
Best Matches for Motif ID 54 (Highest to Lowest)
| Motif ID: | UP00253 |
| Motif name: | Rax |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 5 |
| Similarity score: | 0 |
Alignment:
DBSBKMTAATTAKBGSW
-----MKAAT-------
| Original motif | Reverse complement motif |
| Consensus sequence: WSCVYTAATTARYBSVH | Consensus sequence: DBSBKMTAATTAKBGSW |
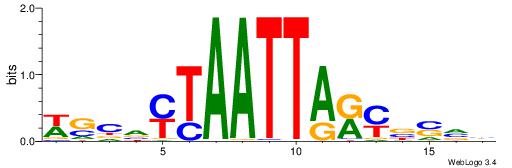 |
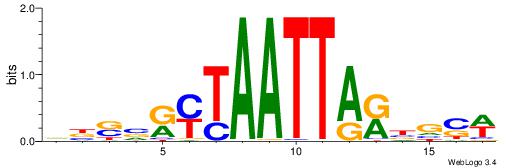 |
| Motif ID: | UP00163 |
| Motif name: | En2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.00258272 |
Alignment:
HHVBAMTAATTAKBDCW
-----MKAAT-------
| Original motif | Reverse complement motif |
| Consensus sequence: WGHVYTAATTARTBVHH | Consensus sequence: HHVBAMTAATTAKBDCW |
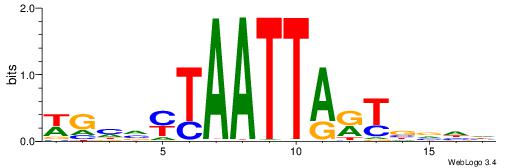 |
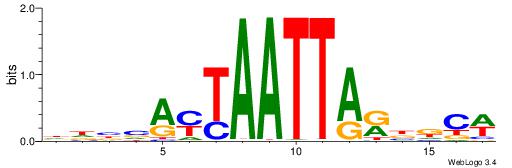 |
| Motif ID: | UP00178 |
| Motif name: | Og2x |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.00347324 |
Alignment:
BHDHBMKAATTMKVVCG
-----MKAAT-------
| Original motif | Reverse complement motif |
| Consensus sequence: CGVVYYAATTRRBHDHB | Consensus sequence: BHDHBMKAATTMKVVCG |
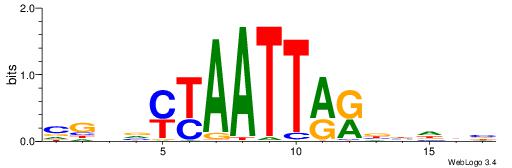 |
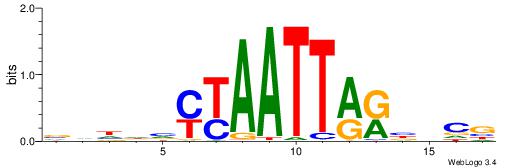 |
| Motif ID: | UP00108 |
| Motif name: | Alx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 5 |
| Similarity score: | 0.00511465 |
Alignment:
VHVAKMTAATTAKTHBW
-----MKAAT-------
| Original motif | Reverse complement motif |
| Consensus sequence: WVHAYTAATTARYTVHV | Consensus sequence: VHVAKMTAATTAKTHBW |
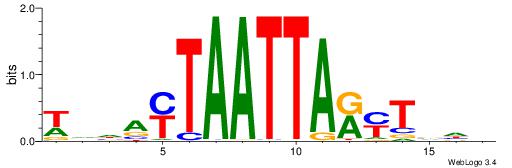 |
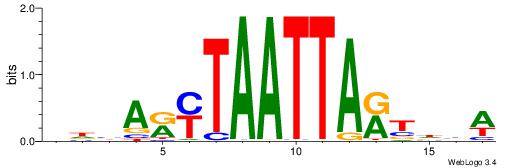 |
| Motif ID: | UP00139 |
| Motif name: | Nkx1-2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 5 |
| Similarity score: | 0.0054602 |
Alignment:
WGHAMTAATTAKKVBDH
----MKAAT--------
| Original motif | Reverse complement motif |
| Consensus sequence: DDBVRYTAATTARTDCW | Consensus sequence: WGHAMTAATTAKKVBDH |
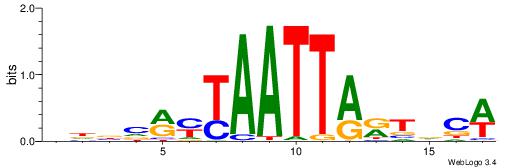 |
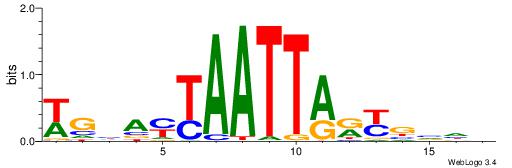 |
| Dataset #: 2 | Motif ID: 55 | Motif name: Motif 55 |
| Original motif | Reverse complement motif |
| Consensus sequence: GCTCACAA | Consensus sequence: TTGTGAGC |
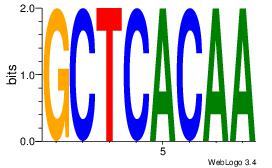 |
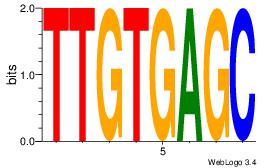 |
Best Matches for Motif ID 55 (Highest to Lowest)
| Motif ID: | UP00026 |
| Motif name: | Zscan4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HVHRGCACACAAHHHV
----GCTCACAA----
| Original motif | Reverse complement motif |
| Consensus sequence: HVHRGCACACAAHHHV | Consensus sequence: BHHHTTGTGTGCKHVD |
 |
 |
| Motif ID: | UP00042 |
| Motif name: | Gm397_second |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0097578 |
Alignment:
DVMVGCACACACKCVH
----GCTCACAA----
| Original motif | Reverse complement motif |
| Consensus sequence: DVMVGCACACACKCVH | Consensus sequence: HBGYGTGTGTGCVRVD |
 |
 |
| Motif ID: | UP00095 |
| Motif name: | Zfp691_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.0133529 |
Alignment:
DDHBYGAGCACTGBHHB
-TTGTGAGC--------
| Original motif | Reverse complement motif |
| Consensus sequence: BDHVCAGTGCTCMBHDD | Consensus sequence: DDHBYGAGCACTGBHHB |
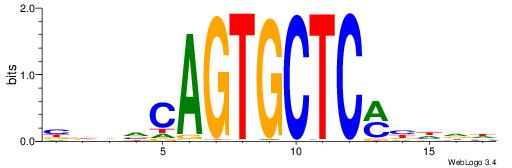 |
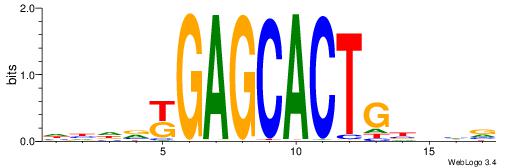 |
| Motif ID: | UP00258 |
| Motif name: | Tgif2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0172769 |
Alignment:
VBWTTGACAGCTVBTT
---TTGTGAGC-----
| Original motif | Reverse complement motif |
| Consensus sequence: AABBAGCTGTCAAWVV | Consensus sequence: VBWTTGACAGCTVBTT |
 |
 |
| Motif ID: | UP00045 |
| Motif name: | Mafb_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.0197606 |
Alignment:
DMTWDGTCAGCADWTTH
---TTGTGAGC------
| Original motif | Reverse complement motif |
| Consensus sequence: HAAWDTGCTGACDWARH | Consensus sequence: DMTWDGTCAGCADWTTH |
 |
 |
| Dataset #: 2 | Motif ID: 56 | Motif name: Motif 56 |
| Original motif | Reverse complement motif |
| Consensus sequence: CCACATGG | Consensus sequence: CCATGTGG |
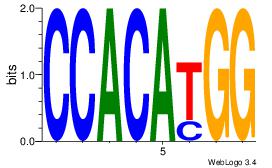 |
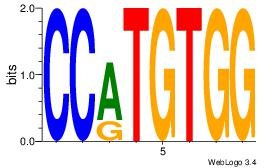 |
Best Matches for Motif ID 56 (Highest to Lowest)
| Motif ID: | UP00010 |
| Motif name: | Tcfap2b_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HTGCCMTVKGGCMD
---CCATGTGG---
| Original motif | Reverse complement motif |
| Consensus sequence: DYGCCYBARGGCAH | Consensus sequence: HTGCCMTVKGGCMD |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.000569341 |
Alignment:
YDYBDHTMCACGTGGADDBMDGT
-------CCACATGG--------
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0020225 |
Alignment:
DDASCACGTGBTBVDD
---CCACATGG-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDASCACGTGBTBVDD | Consensus sequence: HHVBABCACGTGSTHD |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.00356885 |
Alignment:
HVDDVCAGATGKYHBBV
----CCACATGG-----
| Original motif | Reverse complement motif |
| Consensus sequence: HVDDVCAGATGKYHBBV | Consensus sequence: VBBDMYCATCTGVHHBH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 11 |
| Number of overlap: | 8 |
| Similarity score: | 0.00437787 |
Alignment:
HTBVVVDGGACCACCCRGRDBG
----------CCACATGG----
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Dataset #: 2 | Motif ID: 57 | Motif name: Motif 57 |
| Original motif | Reverse complement motif |
| Consensus sequence: ACACACAY | Consensus sequence: KTGTGTGT |
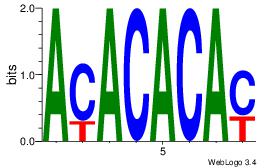 |
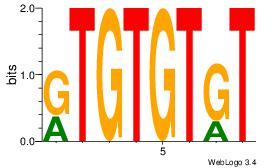 |
Best Matches for Motif ID 57 (Highest to Lowest)
| Motif ID: | UP00042 |
| Motif name: | Gm397_second |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HBGYGTGTGTGCVRVD
--KTGTGTGT------
| Original motif | Reverse complement motif |
| Consensus sequence: DVMVGCACACACKCVH | Consensus sequence: HBGYGTGTGTGCVRVD |
 |
 |
| Motif ID: | UP00026 |
| Motif name: | Zscan4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0175328 |
Alignment:
BHHHTTGTGTGCKHVD
------KTGTGTGT--
| Original motif | Reverse complement motif |
| Consensus sequence: HVHRGCACACAAHHHV | Consensus sequence: BHHHTTGTGTGCKHVD |
 |
 |
| Motif ID: | UP00041 |
| Motif name: | Foxj1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0353784 |
Alignment:
HDHRTAAACAAAHDDD
-----ACACACAY---
| Original motif | Reverse complement motif |
| Consensus sequence: HDHRTAAACAAAHDDD | Consensus sequence: DDDHTTTGTTTAMHDH |
 |
 |
| Motif ID: | UP00026 |
| Motif name: | Zscan4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0359654 |
Alignment:
DHVDTGTGCACAYAHDH
-------ACACACAY--
| Original motif | Reverse complement motif |
| Consensus sequence: DHVDTGTGCACAYAHDH | Consensus sequence: HDHTMTGTGCACADVHD |
 |
 |
| Motif ID: | UP00039 |
| Motif name: | Foxj3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0382549 |
Alignment:
DVVTTTGTTTACDTHDH
--KTGTGTGT-------
| Original motif | Reverse complement motif |
| Consensus sequence: HDHADGTAAACAAAVVH | Consensus sequence: DVVTTTGTTTACDTHDH |
 |
 |
| Dataset #: 2 | Motif ID: 58 | Motif name: Motif 58 |
| Original motif | Reverse complement motif |
| Consensus sequence: TACATGCA | Consensus sequence: TGCATGTA |
 |
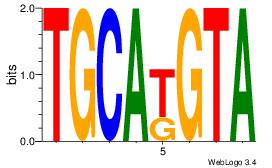 |
Best Matches for Motif ID 58 (Highest to Lowest)
| Motif ID: | UP00150 |
| Motif name: | Irx6 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
AHWDBTACATGTADHTH
-----TACATGCA----
| Original motif | Reverse complement motif |
| Consensus sequence: HAHDTACATGTAVDWHT | Consensus sequence: AHWDBTACATGTADHTH |
 |
 |
| Motif ID: | UP00236 |
| Motif name: | Irx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.000109499 |
Alignment:
WVWDDTACATGTADDDW
-----TACATGCA----
| Original motif | Reverse complement motif |
| Consensus sequence: WDDDTACATGTADDWBW | Consensus sequence: WVWDDTACATGTADDDW |
 |
 |
| Motif ID: | UP00223 |
| Motif name: | Irx3_2226.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.000755361 |
Alignment:
DHDWBTACATGTAHDDD
-----TACATGCA----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDHTACATGTAVWDHD | Consensus sequence: DHDWBTACATGTAHDDD |
 |
 |
| Motif ID: | UP00223 |
| Motif name: | Irx3_0920.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.00254146 |
Alignment:
DDDDTACATGTAVWWHD
----TGCATGTA-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDDTACATGTAVWWHD | Consensus sequence: DDWWBTACATGTADDDD |
 |
 |
| Motif ID: | UP00250 |
| Motif name: | Irx5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0106901 |
Alignment:
WVTDHWACATGTWHDDD
-----TACATGCA----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDHWACATGTWHDABW | Consensus sequence: WVTDHWACATGTWHDDD |
 |
 |
| Dataset #: 2 | Motif ID: 59 | Motif name: Motif 59 |
| Original motif | Reverse complement motif |
| Consensus sequence: CCCCRCCC | Consensus sequence: GGGKGGGG |
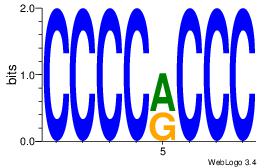 |
 |
Best Matches for Motif ID 59 (Highest to Lowest)
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HBDRCCMCGCCCHHHD
----CCCCRCCC----
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0189858 |
Alignment:
GKRGGGGGGGGGGBD
-----GGGKGGGG--
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0254924 |
Alignment:
BHHDYGGGGGGGGBVD
-----GGGKGGGG---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0331118 |
Alignment:
MHHHWHYTMCCHCCCACVAABVH
-------CCCCRCCC--------
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0351247 |
Alignment:
VHTGCGGGGGCGGH
-----GGGKGGGG-
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
| Dataset #: 2 | Motif ID: 60 | Motif name: Motif 60 |
| Original motif | Reverse complement motif |
| Consensus sequence: CTGGAR | Consensus sequence: MTCCAG |
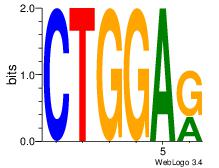 |
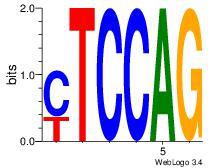 |
Best Matches for Motif ID 60 (Highest to Lowest)
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
BBBAVTGCAGTGBBVDD
----MTCCAG-------
| Original motif | Reverse complement motif |
| Consensus sequence: DDBBBCACTGCABTBBB | Consensus sequence: BBBAVTGCAGTGBBVDD |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.00563465 |
Alignment:
HDHDDCCAGACABBHVH
---MTCCAG--------
| Original motif | Reverse complement motif |
| Consensus sequence: HDHDDCCAGACABBHVH | Consensus sequence: HVHBVTGTCTGGDDHDD |
 |
 |
| Motif ID: | UP00416 |
| Motif name: | Fli1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.00856965 |
Alignment:
DHDACCGGAAKTBBVV
----CTGGAR------
| Original motif | Reverse complement motif |
| Consensus sequence: VBBVAYTTCCGGTDDH | Consensus sequence: DHDACCGGAAKTBBVV |
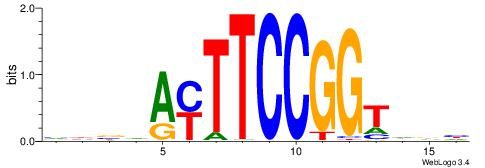 |
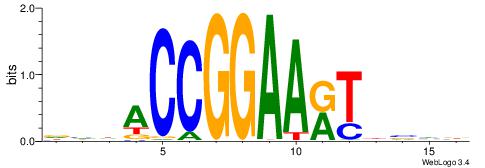 |
| Motif ID: | UP00411 |
| Motif name: | Erg |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.00868366 |
Alignment:
BDDACCGGAAKTVBVB
----CTGGAR------
| Original motif | Reverse complement motif |
| Consensus sequence: VBBVAYTTCCGGTDDB | Consensus sequence: BDDACCGGAAKTVBVB |
 |
 |
| Motif ID: | UP00231 |
| Motif name: | Nkx2-2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.0091158 |
Alignment:
HDVRCCACTTGARADBD
-------CTGGAR----
| Original motif | Reverse complement motif |
| Consensus sequence: HDVRCCACTTGARADBD | Consensus sequence: DVDTKTCAAGTGGKBDH |
 |
 |
| Dataset #: 2 | Motif ID: 61 | Motif name: Motif 61 |
| Original motif | Reverse complement motif |
| Consensus sequence: AGATGGY | Consensus sequence: KCCATCT |
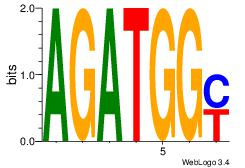 |
 |
Best Matches for Motif ID 61 (Highest to Lowest)
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 7 |
| Similarity score: | 0 |
Alignment:
HVDDVCAGATGKYHBBV
------AGATGGY----
| Original motif | Reverse complement motif |
| Consensus sequence: HVDDVCAGATGKYHBBV | Consensus sequence: VBBDMYCATCTGVHHBH |
 |
 |
| Motif ID: | UP00036 |
| Motif name: | Myf6_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 7 |
| Similarity score: | 0.0380154 |
Alignment:
BDKVCABCTGTTBDBV
--KCCATCT-------
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBAACAGBTGBYHB | Consensus sequence: BDKVCABCTGTTBDBV |
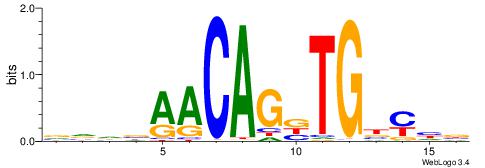 |
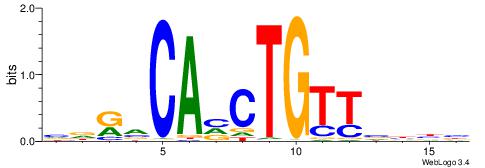 |
| Motif ID: | UP00081 |
| Motif name: | Mybl1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 7 |
| Similarity score: | 0.0385991 |
Alignment:
BHCGGCAGTTGGBBB
------AGATGGY--
| Original motif | Reverse complement motif |
| Consensus sequence: BBVCCAACTGCCGHB | Consensus sequence: BHCGGCAGTTGGBBB |
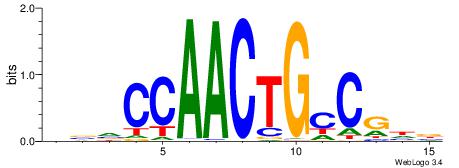 |
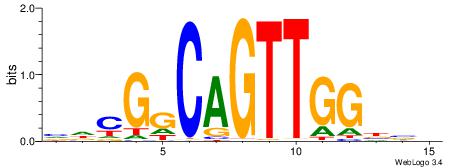 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 7 |
| Similarity score: | 0.0422018 |
Alignment:
BDDYYCATCCCATDDBD
---KCCATCT-------
| Original motif | Reverse complement motif |
| Consensus sequence: DVDDATGGGATGKMDDV | Consensus sequence: BDDYYCATCCCATDDBD |
 |
 |
| Motif ID: | UP00092 |
| Motif name: | Myb_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 7 |
| Similarity score: | 0.0463999 |
Alignment:
DBVKGGCAGTTGGTHB
-------AGATGGY--
| Original motif | Reverse complement motif |
| Consensus sequence: BDACCAACTGCCRBBH | Consensus sequence: DBVKGGCAGTTGGTHB |
 |
 |
| Dataset #: 2 | Motif ID: 62 | Motif name: Motif 62 |
| Original motif | Reverse complement motif |
| Consensus sequence: CGCCVCC | Consensus sequence: GGVGGCG |
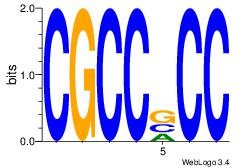 |
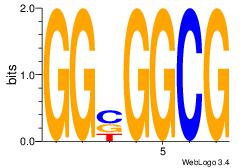 |
Best Matches for Motif ID 62 (Highest to Lowest)
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0 |
Alignment:
BVVHAGGGGGCGRDHHB
-----GGVGGCG-----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 7 |
| Similarity score: | 0.0105671 |
Alignment:
HCCGCCCCCGCAHB
--CGCCVCC-----
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.0302678 |
Alignment:
DHHDGGGCGRGGKHBH
-----GGVGGCG----
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 10 |
| Number of overlap: | 7 |
| Similarity score: | 0.0347726 |
Alignment:
BADHBDHCGCCCMCGCAHHDBBV
-------CGCCVCC---------
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
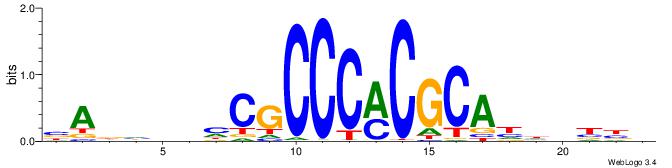 |
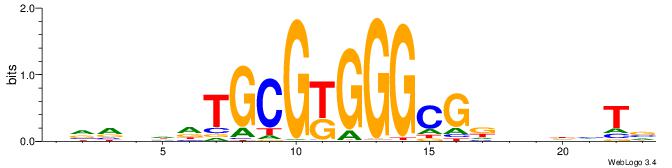 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 7 |
| Similarity score: | 0.0384581 |
Alignment:
DHHBCCCCGCCAHHBHB
-------CGCCVCC---
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
 |
 |
| Dataset #: 2 | Motif ID: 63 | Motif name: Motif 63 |
| Original motif | Reverse complement motif |
| Consensus sequence: AGAGGGWG | Consensus sequence: CWCCCTCT |
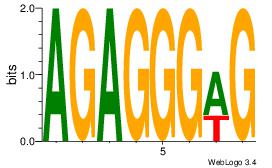 |
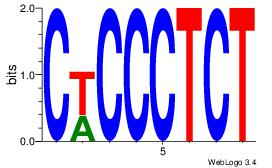 |
Best Matches for Motif ID 63 (Highest to Lowest)
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
BVVHAGGGGGCGRDHHB
----AGAGGGWG-----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 11 |
| Number of overlap: | 8 |
| Similarity score: | 0.00709886 |
Alignment:
DVVTTVGTGGGHGGYAMHWHHHY
-----AGAGGGWG----------
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 12 |
| Number of overlap: | 8 |
| Similarity score: | 0.0107485 |
Alignment:
YBVDMGTGGGTGGTCKVVBVBBT
----AGAGGGWG-----------
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0126141 |
Alignment:
VHTGCGGGGGCGGH
----AGAGGGWG--
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0180504 |
Alignment:
GKRGGGGGGGGGGBD
--AGAGGGWG-----
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Dataset #: 2 | Motif ID: 64 | Motif name: Motif 64 |
| Original motif | Reverse complement motif |
| Consensus sequence: CATATRCA | Consensus sequence: TGMATATG |
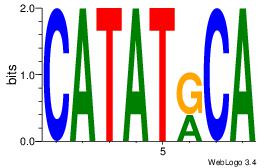 |
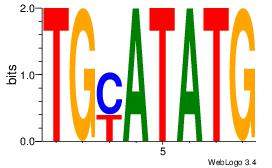 |
Best Matches for Motif ID 64 (Highest to Lowest)
| Motif ID: | UP00211 |
| Motif name: | Pou3f3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HADDTATGCATAATDDA
------TGMATATG---
| Original motif | Reverse complement motif |
| Consensus sequence: HADDTATGCATAATDDA | Consensus sequence: TDDATTATGCATADDTH |
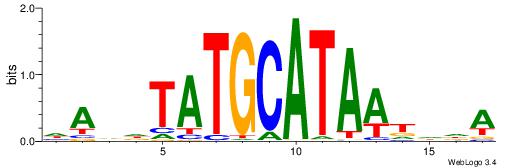 |
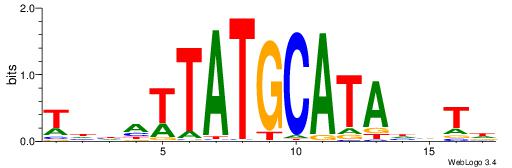 |
| Motif ID: | UP00179 |
| Motif name: | Pou2f3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.00980027 |
Alignment:
BHDTATGCAAATBHDV
-----TGMATATG---
| Original motif | Reverse complement motif |
| Consensus sequence: BHDTATGCAAATBHDV | Consensus sequence: BHHVATTTGCATAHHV |
 |
 |
| Motif ID: | UP00191 |
| Motif name: | Pou2f2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.011527 |
Alignment:
BHDTATGCAAATDHDV
-----TGMATATG---
| Original motif | Reverse complement motif |
| Consensus sequence: BHDTATGCAAATDHDV | Consensus sequence: BHHDATTTGCATAHHV |
 |
 |
| Motif ID: | UP00097 |
| Motif name: | Mtf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0243027 |
Alignment:
HDYTTGCACACGDHBH
----TGMATATG----
| Original motif | Reverse complement motif |
| Consensus sequence: DBDHCGTGTGCAAMDH | Consensus sequence: HDYTTGCACACGDHBH |
 |
 |
| Motif ID: | UP00042 |
| Motif name: | Gm397_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0250687 |
Alignment:
HHRTATGTGCACATHHD
---CATATRCA------
| Original motif | Reverse complement motif |
| Consensus sequence: HHDATGTGCACATAMDH | Consensus sequence: HHRTATGTGCACATHHD |
 |
 |
| Dataset #: 2 | Motif ID: 65 | Motif name: Motif 65 |
| Original motif | Reverse complement motif |
| Consensus sequence: ARAACA | Consensus sequence: TGTTMT |
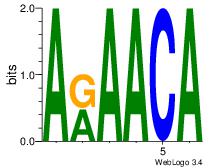 |
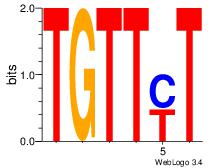 |
Best Matches for Motif ID 65 (Highest to Lowest)
| Motif ID: | UP00101 |
| Motif name: | Sox12_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
DWTHAVAACAATHA
----ARAACA----
| Original motif | Reverse complement motif |
| Consensus sequence: THATTGTTVTHAWH | Consensus sequence: DWTHAVAACAATHA |
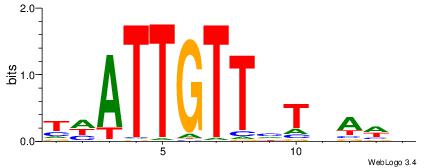 |
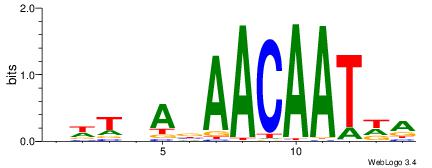 |
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.000395587 |
Alignment:
HBDDRAACAAAGRABHH
---ARAACA--------
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
 |
 |
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.00357565 |
Alignment:
HHBBMCTTTGTTHDDBH
--------TGTTMT---
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.00753019 |
Alignment:
HHBDHDDAACAATDRHHHHHBW
-----ARAACA-----------
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
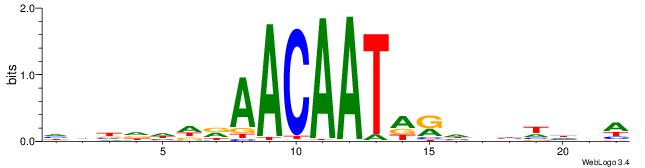 |
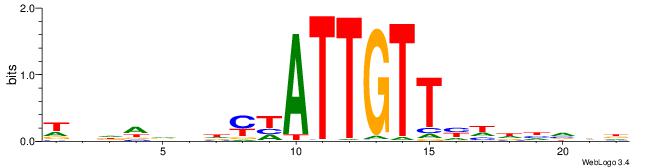 |
| Motif ID: | UP00039 |
| Motif name: | Foxj3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0083324 |
Alignment:
DVHWDTGTTWTGDTBDH
-----TGTTMT------
| Original motif | Reverse complement motif |
| Consensus sequence: HDBAHCAWAACADWDVD | Consensus sequence: DVHWDTGTTWTGDTBDH |
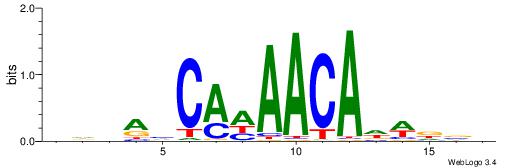 |
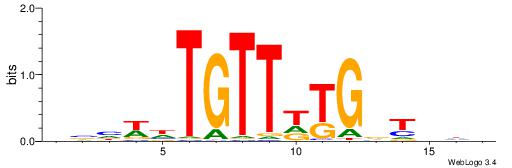 |
| Dataset #: 2 | Motif ID: 66 | Motif name: Motif 66 |
| Original motif | Reverse complement motif |
| Consensus sequence: TATAAATR | Consensus sequence: KATTTATA |
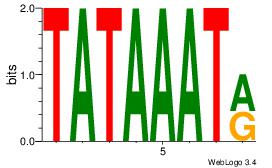 |
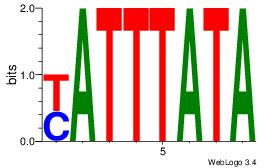 |
Best Matches for Motif ID 66 (Highest to Lowest)
| Motif ID: | UP00118 |
| Motif name: | Pou4f3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HVTTATTAATKADKHC
---TATAAATR-----
| Original motif | Reverse complement motif |
| Consensus sequence: HVTTATTAATKADKHC | Consensus sequence: GHYHTYATTAATAAVH |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.00672344 |
Alignment:
DWAATRTAAACAAWVVB
----TATAAATR-----
| Original motif | Reverse complement motif |
| Consensus sequence: DWAATRTAAACAAWVVB | Consensus sequence: VVVWTTGTTTAMATTWD |
 |
 |
| Motif ID: | UP00154 |
| Motif name: | Dlx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.00785197 |
Alignment:
BHVVDATAATTACMVHH
----TATAAATR-----
| Original motif | Reverse complement motif |
| Consensus sequence: BHVVDATAATTACMVHH | Consensus sequence: DHVRGTAATTATHVVDV |
 |
 |
| Motif ID: | UP00016 |
| Motif name: | Sry_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.00840021 |
Alignment:
VDDDATTATAATHDHV
------TATAAATR--
| Original motif | Reverse complement motif |
| Consensus sequence: BHDHATTATAATDDDV | Consensus sequence: VDDDATTATAATHDHV |
 |
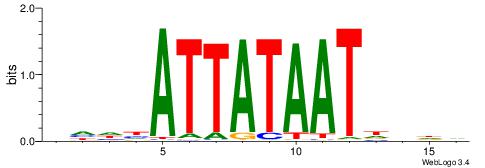 |
| Motif ID: | UP00110 |
| Motif name: | Dlx4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.011539 |
Alignment:
GHVRGTAATTATDVVGV
-----KATTTATA----
| Original motif | Reverse complement motif |
| Consensus sequence: BCVVDATAATTACMVHC | Consensus sequence: GHVRGTAATTATDVVGV |
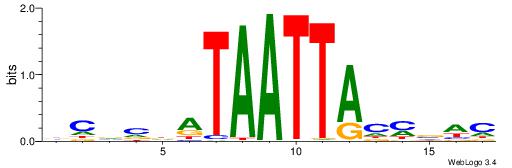 |
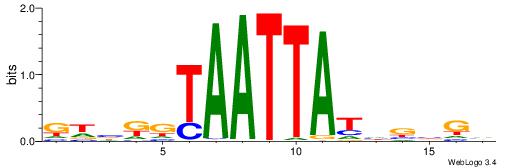 |
| Dataset #: 2 | Motif ID: 67 | Motif name: Motif 67 |
| Original motif | Reverse complement motif |
| Consensus sequence: SYGCCCYCTDSTGG | Consensus sequence: CCASHAGKGGGCKS |
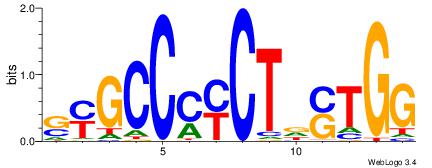 |
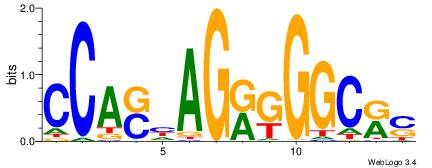 |
Best Matches for Motif ID 67 (Highest to Lowest)
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.036534 |
Alignment:
BHHDYGGGGGGGGBVD
CCASHAGKGGGCKS--
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 14 |
| Similarity score: | 0.0371579 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
-----SYGCCCYCTDSTGG---
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0410702 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
---CCASHAGKGGGCKS------
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0411664 |
Alignment:
DDDTWCCCCCCGGDRVH
--SYGCCCYCTDSTGG-
| Original motif | Reverse complement motif |
| Consensus sequence: DDDTWCCCCCCGGDRVH | Consensus sequence: HVKDCCGGGGGGWADDD |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0419746 |
Alignment:
BBHBGAGTGGGAHHDB
CCASHAGKGGGCKS--
| Original motif | Reverse complement motif |
| Consensus sequence: BBHBGAGTGGGAHHDB | Consensus sequence: BHHDTCCCACTCBDBV |
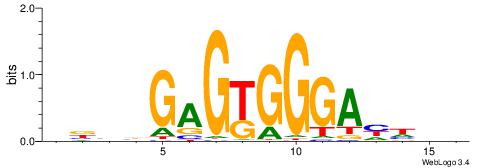 |
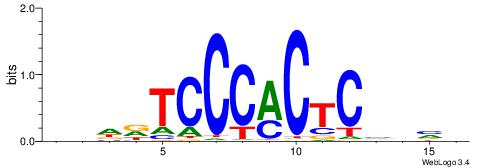 |
| Dataset #: 2 | Motif ID: 68 | Motif name: Motif 68 |
| Original motif | Reverse complement motif |
| Consensus sequence: WAAWAAAWAWWWAA | Consensus sequence: TTWWWTWTTTWTTW |
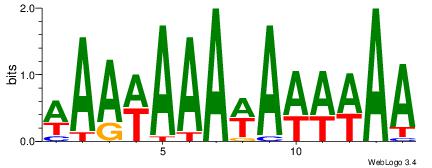 |
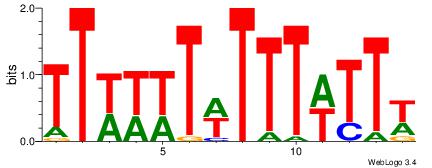 |
Best Matches for Motif ID 68 (Highest to Lowest)
| Motif ID: | UP00073 |
| Motif name: | Foxa2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0252297 |
Alignment:
HHWAWGTAAAYAAAVDB
---WAAWAAAWAWWWAA
| Original motif | Reverse complement motif |
| Consensus sequence: HHWAWGTAAAYAAAVDB | Consensus sequence: BDVTTTKTTTACWTWHH |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0252358 |
Alignment:
HDTYTTGTTWTKDDDT
TTWWWTWTTTWTTW--
| Original motif | Reverse complement motif |
| Consensus sequence: ADDDYAWAACAAMAHH | Consensus sequence: HDTYTTGTTWTKDDDT |
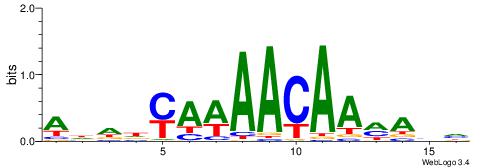 |
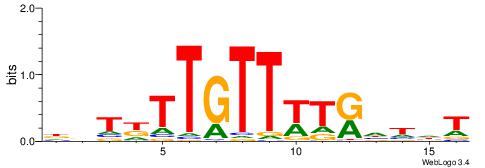 |
| Motif ID: | UP00039 |
| Motif name: | Foxj3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0267996 |
Alignment:
HDHADGTAAACAAAVVH
---WAAWAAAWAWWWAA
| Original motif | Reverse complement motif |
| Consensus sequence: HDHADGTAAACAAAVVH | Consensus sequence: DVVTTTGTTTACDTHDH |
 |
 |
| Motif ID: | UP00244 |
| Motif name: | Tlx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0275942 |
Alignment:
WWDTTATTAATTHATTV
--TTWWWTWTTTWTTW-
| Original motif | Reverse complement motif |
| Consensus sequence: BAATHAATTAATAAHWW | Consensus sequence: WWDTTATTAATTHATTV |
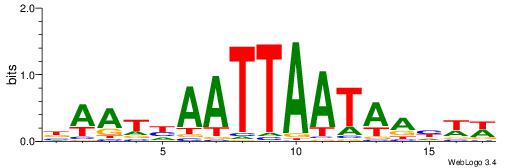 |
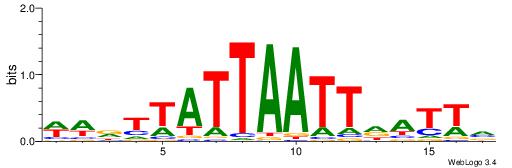 |
| Motif ID: | UP00041 |
| Motif name: | Foxj1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0288501 |
Alignment:
HDHRTAAACAAAHDDD
-WAAWAAAWAWWWAA-
| Original motif | Reverse complement motif |
| Consensus sequence: HDHRTAAACAAAHDDD | Consensus sequence: DDDHTTTGTTTAMHDH |
 |
 |
| Dataset #: 2 | Motif ID: 69 | Motif name: Motif 69 |
| Original motif | Reverse complement motif |
| Consensus sequence: CCTGGARCTGGAGT | Consensus sequence: ACTCCAGMTCCAGG |
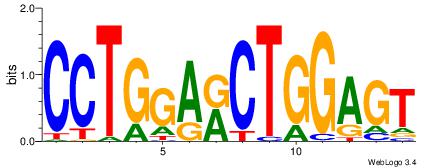 |
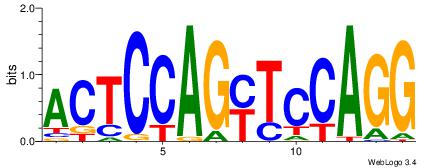 |
Best Matches for Motif ID 69 (Highest to Lowest)
| Motif ID: | UP00007 |
| Motif name: | Egr1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0303891 |
Alignment:
BHHDTCCCACTCBDBV
--ACTCCAGMTCCAGG
| Original motif | Reverse complement motif |
| Consensus sequence: BBHBGAGTGGGAHHDB | Consensus sequence: BHHDTCCCACTCBDBV |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.031849 |
Alignment:
DHDBCAAGGTCAHVBDH
-ACTCCAGMTCCAGG--
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBCAAGGTCAHVBDH | Consensus sequence: HHBVHTGACCTTGVDHD |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0328055 |
Alignment:
TCDAMVBGACGCMAKDDDVBBD
--------ACTCCAGMTCCAGG
| Original motif | Reverse complement motif |
| Consensus sequence: DBBVDDDRTRGCGTCBBYTHGA | Consensus sequence: TCDAMVBGACGCMAKDDDVBBD |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0343241 |
Alignment:
HVHBVTGTCTGGDDHDD
-CCTGGARCTGGAGT--
| Original motif | Reverse complement motif |
| Consensus sequence: HDHDDCCAGACABBHVH | Consensus sequence: HVHBVTGTCTGGDDHDD |
 |
 |
| Motif ID: | UP00076 |
| Motif name: | Rfxdc2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0347793 |
Alignment:
HDDVSGTRTCMADGBVB
ACTCCAGMTCCAGG---
| Original motif | Reverse complement motif |
| Consensus sequence: BBVCDTRGAKACSVDDH | Consensus sequence: HDDVSGTRTCMADGBVB |
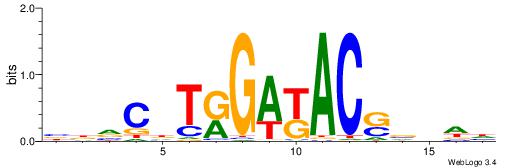 |
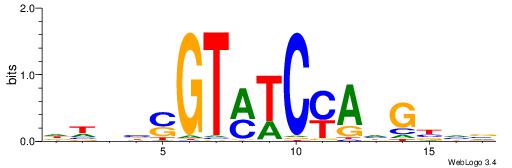 |
| Dataset #: 3 | Motif ID: 70 | Motif name: Ar |
| Original motif | Reverse complement motif |
| Consensus sequence: HWDAGHACRHHVTGTHCCHVMV | Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
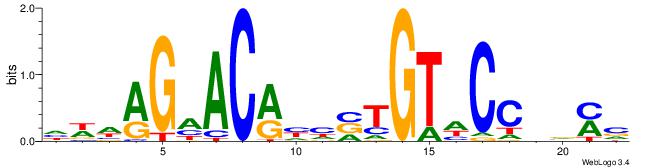 |
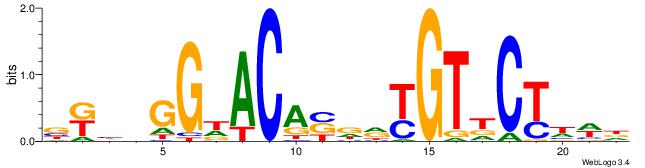 |
Best Matches for Motif ID 70 (Highest to Lowest)
| Motif ID: | UP00032 |
| Motif name: | Gata3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 22 |
| Similarity score: | 0.0676991 |
Alignment:
HBHDVBWGATHBTATCRVBHHH
HWDAGHACRHHVTGTHCCHVMV
| Original motif | Reverse complement motif |
| Consensus sequence: HBHDVBWGATHBTATCRVBHHH | Consensus sequence: HHHBBMGATAVHATCWVVDHVH |
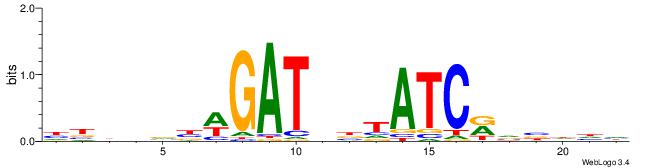 |
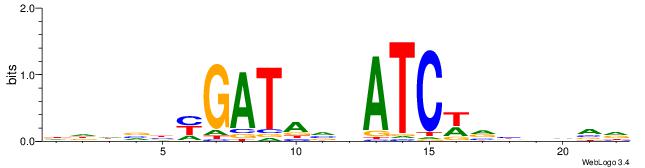 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 22 |
| Similarity score: | 0.0692146 |
Alignment:
VVYDDDCHCAVACAATTDBVAC
VRVDGGHACAVDDKGTHCTDWH
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
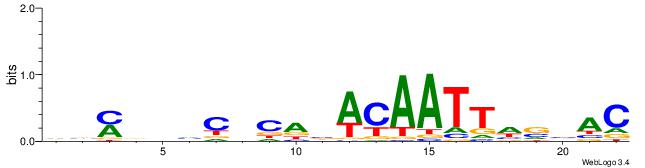 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 22 |
| Similarity score: | 0.0695612 |
Alignment:
DDHBHBSTCACGTGAHBBDHHM
VRVDGGHACAVDDKGTHCTDWH
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 22 |
| Similarity score: | 0.0771346 |
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
-HWDAGHACRHHVTGTHCCHVMV
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 22 |
| Similarity score: | 0.0772045 |
Alignment:
HDWHMHGACGCGCGTRBVVBHW
VRVDGGHACAVDDKGTHCTDWH
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
 |
 |
| Dataset #: 3 | Motif ID: 71 | Motif name: Arnt |
| Original motif | Reverse complement motif |
| Consensus sequence: CACGTG | Consensus sequence: CACGTG |
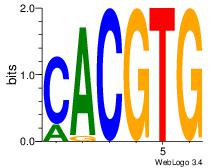 |
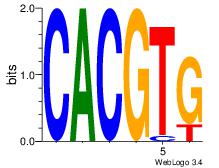 |
Best Matches for Motif ID 71 (Highest to Lowest)
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
RDHDBVDTCACGTGASBHVHDH
--------CACGTG--------
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.0026425 |
Alignment:
YDYBDHTMCACGTGGADDBMDGT
--------CACGTG---------
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0044422 |
Alignment:
HHVBABCACGTGSTHD
------CACGTG----
| Original motif | Reverse complement motif |
| Consensus sequence: DDASCACGTGBTBVDD | Consensus sequence: HHVBABCACGTGSTHD |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0342153 |
Alignment:
DHDBHGCACCTGBDDVB
------CACGTG-----
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Motif ID: | UP00097 |
| Motif name: | Mtf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.0471928 |
Alignment:
HDYTTGCACACGDHBH
--------CACGTG--
| Original motif | Reverse complement motif |
| Consensus sequence: DBDHCGTGTGCAAMDH | Consensus sequence: HDYTTGCACACGDHBH |
 |
 |
| Dataset #: 3 | Motif ID: 72 | Motif name: ArntAhr |
| Original motif | Reverse complement motif |
| Consensus sequence: YGCGTG | Consensus sequence: CACGCM |
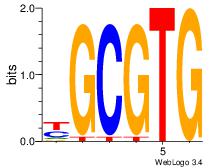 |
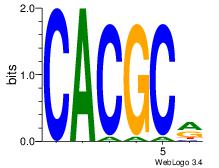 |
Best Matches for Motif ID 72 (Highest to Lowest)
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
------YGCGTG-----------
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.00183587 |
Alignment:
HHDGBCACGCCTWDB
-----CACGCM----
| Original motif | Reverse complement motif |
| Consensus sequence: BDWAGGCGTGBCHHD | Consensus sequence: HHDGBCACGCCTWDB |
 |
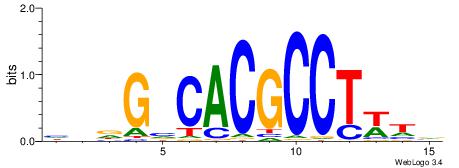 |
| Motif ID: | UP00060 |
| Motif name: | Max_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.00348048 |
Alignment:
HDGTCGCGTGDCVB
----YGCGTG----
| Original motif | Reverse complement motif |
| Consensus sequence: BBGHCACGCGACDD | Consensus sequence: HDGTCGCGTGDCVB |
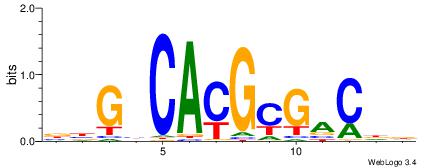 |
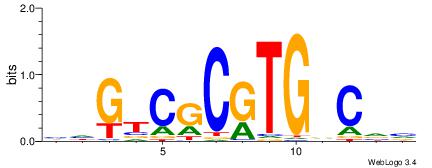 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.00526158 |
Alignment:
TCDAMVBGACGCMAKDDDVBBD
-------CACGCM---------
| Original motif | Reverse complement motif |
| Consensus sequence: DBBVDDDRTRGCGTCBBYTHGA | Consensus sequence: TCDAMVBGACGCMAKDDDVBBD |
 |
 |
| Motif ID: | UP00042 |
| Motif name: | Gm397_second |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.00558171 |
Alignment:
HBGYGTGTGTGCVRVD
---YGCGTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: DVMVGCACACACKCVH | Consensus sequence: HBGYGTGTGTGCVRVD |
 |
 |
| Dataset #: 3 | Motif ID: 73 | Motif name: CREB1 |
| Original motif | Reverse complement motif |
| Consensus sequence: TGACGTCA | Consensus sequence: TGACGTCA |
 |
 |
Best Matches for Motif ID 73 (Highest to Lowest)
| Motif ID: | UP00103 |
| Motif name: | Jundm2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
BBGATGACGTCAYCVH
----TGACGTCA----
| Original motif | Reverse complement motif |
| Consensus sequence: BBGATGACGTCAYCVH | Consensus sequence: HVGMTGACGTCATCBB |
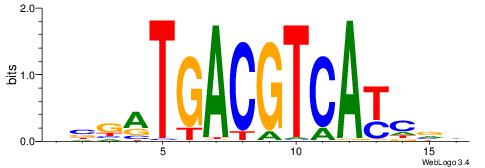 |
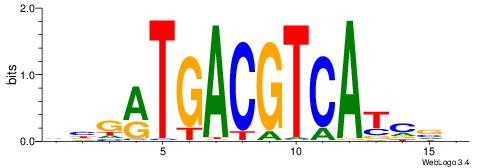 |
| Motif ID: | UP00020 |
| Motif name: | Atf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 2.53244e-05 |
Alignment:
DBDRTGACGTCAYHRD
----TGACGTCA----
| Original motif | Reverse complement motif |
| Consensus sequence: DBDRTGACGTCAYHRD | Consensus sequence: DMDMTGACGTCAKHBD |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.029362 |
Alignment:
TCDAMVBGACGCMAKDDDVBBD
------TGACGTCA--------
| Original motif | Reverse complement motif |
| Consensus sequence: DBBVDDDRTRGCGTCBBYTHGA | Consensus sequence: TCDAMVBGACGCMAKDDDVBBD |
 |
 |
| Motif ID: | UP00020 |
| Motif name: | Atf1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0350871 |
Alignment:
DDTMDTCGTCATDV
---TGACGTCA---
| Original motif | Reverse complement motif |
| Consensus sequence: VDATGACGADYADH | Consensus sequence: DDTMDTCGTCATDV |
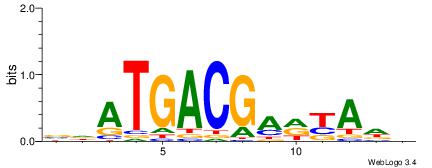 |
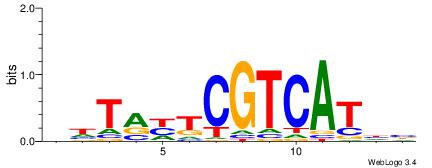 |
| Motif ID: | UP00084 |
| Motif name: | Gmeb1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0378098 |
Alignment:
BHBBKKACGTMMVDBVB
----TGACGTCA-----
| Original motif | Reverse complement motif |
| Consensus sequence: BHBBKKACGTMMVDBVB | Consensus sequence: BVVDVRYACGTRYVBHB |
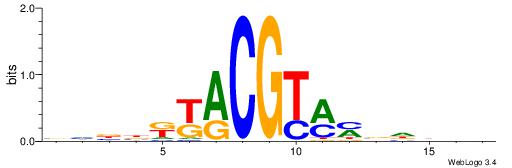 |
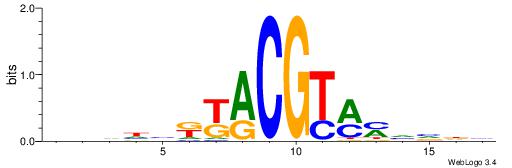 |
| Dataset #: 3 | Motif ID: 74 | Motif name: CTCF |
| Original motif | Reverse complement motif |
| Consensus sequence: YDRCCASYAGRKGGCRSYV | Consensus sequence: BMSMGCCYMCTKSTGGMHM |
 |
 |
Best Matches for Motif ID 74 (Highest to Lowest)
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 19 |
| Similarity score: | 0.0409369 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
----BMSMGCCYMCTKSTGGMHM
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 19 |
| Similarity score: | 0.0409815 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
-YDRCCASYAGRKGGCRSYV---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 19 |
| Similarity score: | 0.0431951 |
Alignment:
VMYDHDGMCCHCCKBGVVAAVH
---BMSMGCCYMCTKSTGGMHM
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
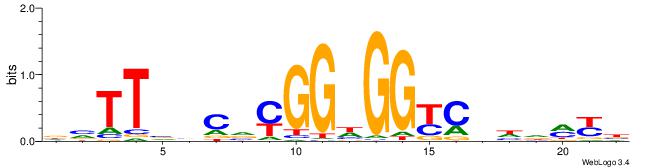 |
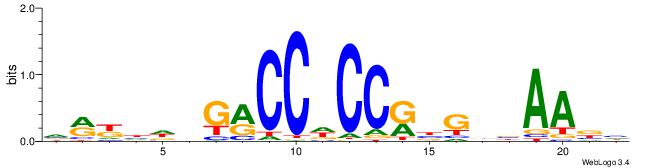 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 19 |
| Similarity score: | 0.0469794 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
---BMSMGCCYMCTKSTGGMHM
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 19 |
| Similarity score: | 0.0482204 |
Alignment:
BHCBCBCCGGGTGGTCYHVHDCH
YDRCCASYAGRKGGCRSYV----
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Dataset #: 3 | Motif ID: 75 | Motif name: Ddit3Cebpa |
| Original motif | Reverse complement motif |
| Consensus sequence: VDRTGCAATMCC | Consensus sequence: GGRATTGCAKHB |
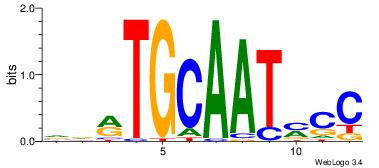 |
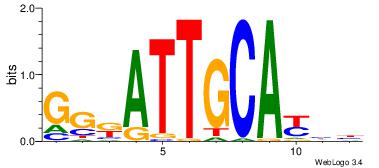 |
Best Matches for Motif ID 75 (Highest to Lowest)
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.026968 |
Alignment:
BBBAVTGCAGTGBBVDD
--VDRTGCAATMCC---
| Original motif | Reverse complement motif |
| Consensus sequence: DDBBBCACTGCABTBBB | Consensus sequence: BBBAVTGCAGTGBBVDD |
 |
 |
| Motif ID: | UP00035 |
| Motif name: | Hic1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0308236 |
Alignment:
DDBHGGTTGGCATVBD
---GGRATTGCAKHB-
| Original motif | Reverse complement motif |
| Consensus sequence: DBBATGCCAACCHVHH | Consensus sequence: DDBHGGTTGGCATVBD |
 |
 |
| Motif ID: | UP00083 |
| Motif name: | Tcf7l2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0323758 |
Alignment:
VDVVATCAATCRBBHD
-VDRTGCAATMCC---
| Original motif | Reverse complement motif |
| Consensus sequence: VDVVATCAATCRBBHD | Consensus sequence: DHVBKGATTGATVBDV |
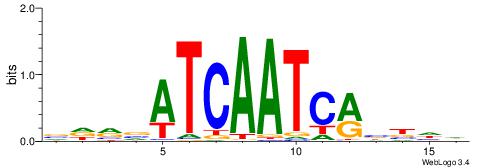 |
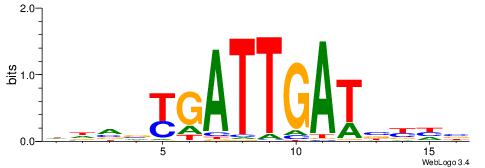 |
| Motif ID: | UP00016 |
| Motif name: | Sry_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0357538 |
Alignment:
HHDHDRAACAATDDVHV
---VDRTGCAATMCC--
| Original motif | Reverse complement motif |
| Consensus sequence: HHDHDRAACAATDDVHV | Consensus sequence: VHVHDATTGTTMHDDDH |
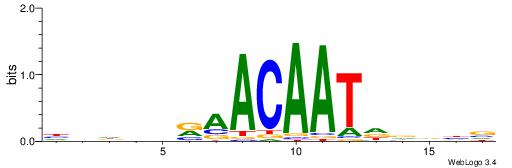 |
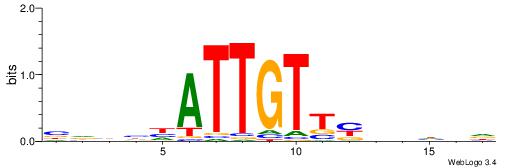 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0387064 |
Alignment:
VDVVATCAATCAHBHD
-VDRTGCAATMCC---
| Original motif | Reverse complement motif |
| Consensus sequence: VDVVATCAATCAHBHD | Consensus sequence: DHVDTGATTGATVBDV |
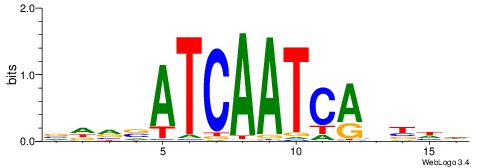 |
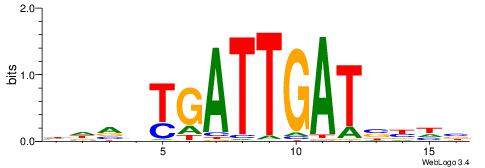 |
| Dataset #: 3 | Motif ID: 76 | Motif name: E2F1 |
| Original motif | Reverse complement motif |
| Consensus sequence: TTTSGCGC | Consensus sequence: GCGCSAAA |
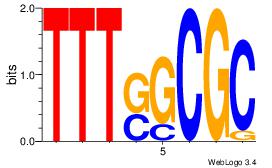 |
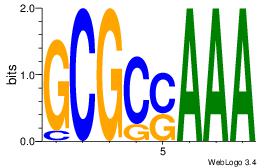 |
Best Matches for Motif ID 76 (Highest to Lowest)
| Motif ID: | UP00001 |
| Motif name: | E2F2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HVBWYGGCGCCAADVBB
------GCGCSAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBWYGGCGCCAADVBB | Consensus sequence: BBBDTTGGCGCCKWVVD |
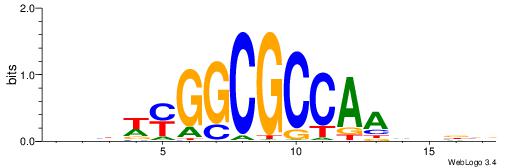 |
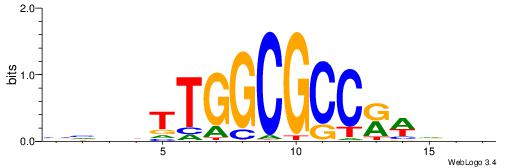 |
| Motif ID: | UP00003 |
| Motif name: | E2F3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.00182028 |
Alignment:
HBHWYGGCGCCAMDVBB
------GCGCSAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: HBHWYGGCGCCAMDVBB | Consensus sequence: BBBDYTGGCGCCKWDBD |
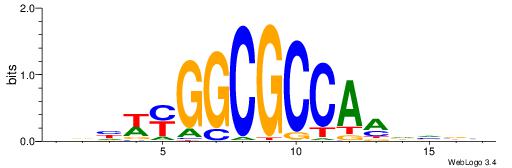 |
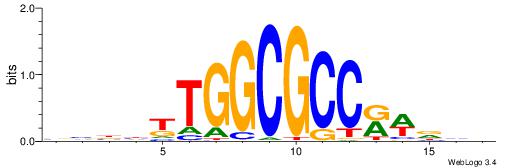 |
| Motif ID: | UP00072 |
| Motif name: | IRC900814_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0237639 |
Alignment:
BHDWTTTGTCGTAHDD
----TTTSGCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: DDHTACGACAAAWDDB | Consensus sequence: BHDWTTTGTCGTAHDD |
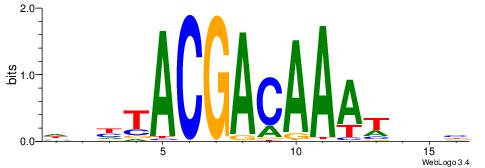 |
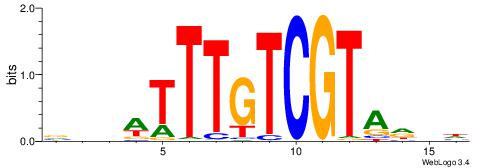 |
| Motif ID: | UP00003 |
| Motif name: | E2F3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0275636 |
Alignment:
WHSGCGCGCCHTDHB
-----GCGCSAAA--
| Original motif | Reverse complement motif |
| Consensus sequence: VHDADGGCGCGCSHW | Consensus sequence: WHSGCGCGCCHTDHB |
 |
 |
| Motif ID: | UP00065 |
| Motif name: | Zfp161_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.028109 |
Alignment:
DMMDGMGCGCGCGCHD
--------GCGCSAAA
| Original motif | Reverse complement motif |
| Consensus sequence: DDGCGCGCGCRCHYRD | Consensus sequence: DMMDGMGCGCGCGCHD |
 |
 |
| Dataset #: 3 | Motif ID: 77 | Motif name: EBF1 |
| Original motif | Reverse complement motif |
| Consensus sequence: MCCCMAGGGA | Consensus sequence: TCCCTYGGGY |
 |
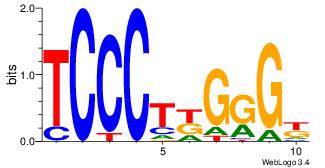 |
Best Matches for Motif ID 77 (Highest to Lowest)
| Motif ID: | UP00057 |
| Motif name: | Zic2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0191678 |
Alignment:
VCCCCCCCGGGGGRB
---MCCCMAGGGA--
| Original motif | Reverse complement motif |
| Consensus sequence: BMCCCCCGGGGGGGB | Consensus sequence: VCCCCCCCGGGGGRB |
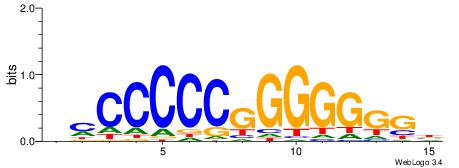 |
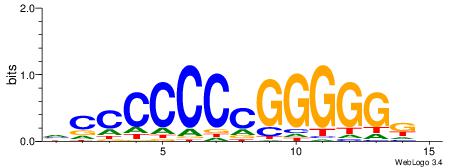 |
| Motif ID: | UP00102 |
| Motif name: | Zic1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0198894 |
Alignment:
CCCCCCCGGGGGHB
--MCCCMAGGGA--
| Original motif | Reverse complement motif |
| Consensus sequence: BHCCCCCGGGGGGG | Consensus sequence: CCCCCCCGGGGGHB |
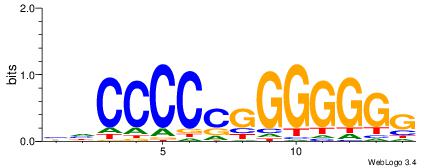 |
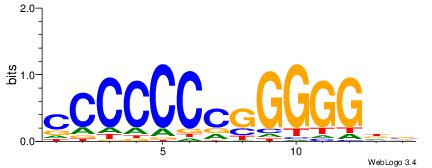 |
| Motif ID: | UP00006 |
| Motif name: | Zic3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0208394 |
Alignment:
VCCCCCCCGGGGGRB
---MCCCMAGGGA--
| Original motif | Reverse complement motif |
| Consensus sequence: BMCCCCCGGGGGGGB | Consensus sequence: VCCCCCCCGGGGGRB |
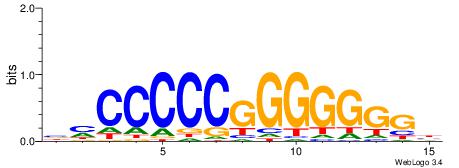 |
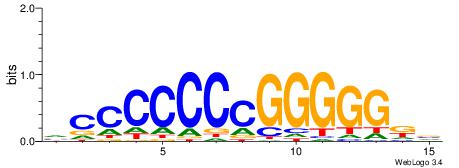 |
| Motif ID: | UP00010 |
| Motif name: | Tcfap2b_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0215181 |
Alignment:
HTGCCMTVKGGCMD
--TCCCTYGGGY--
| Original motif | Reverse complement motif |
| Consensus sequence: DYGCCYBARGGCAH | Consensus sequence: HTGCCMTVKGGCMD |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 13 |
| Number of overlap: | 10 |
| Similarity score: | 0.0271543 |
Alignment:
HTBVVVDGGACCACCCRGRDBG
MCCCMAGGGA------------
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Dataset #: 3 | Motif ID: 78 | Motif name: Egr1 |
| Original motif | Reverse complement motif |
| Consensus sequence: TTTSGCGC | Consensus sequence: GCGCSAAA |
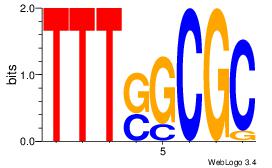 |
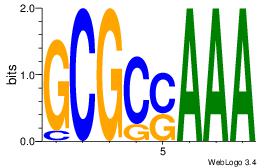 |
Best Matches for Motif ID 78 (Highest to Lowest)
| Motif ID: | UP00001 |
| Motif name: | E2F2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HVBWYGGCGCCAADVBB
------GCGCSAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBWYGGCGCCAADVBB | Consensus sequence: BBBDTTGGCGCCKWVVD |
 |
 |
| Motif ID: | UP00003 |
| Motif name: | E2F3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.00182028 |
Alignment:
HBHWYGGCGCCAMDVBB
------GCGCSAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: HBHWYGGCGCCAMDVBB | Consensus sequence: BBBDYTGGCGCCKWDBD |
 |
 |
| Motif ID: | UP00072 |
| Motif name: | IRC900814_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0237639 |
Alignment:
DDHTACGACAAAWDDB
----GCGCSAAA----
| Original motif | Reverse complement motif |
| Consensus sequence: DDHTACGACAAAWDDB | Consensus sequence: BHDWTTTGTCGTAHDD |
 |
 |
| Motif ID: | UP00003 |
| Motif name: | E2F3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0275636 |
Alignment:
WHSGCGCGCCHTDHB
-----GCGCSAAA--
| Original motif | Reverse complement motif |
| Consensus sequence: VHDADGGCGCGCSHW | Consensus sequence: WHSGCGCGCCHTDHB |
 |
 |
| Motif ID: | UP00065 |
| Motif name: | Zfp161_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.028109 |
Alignment:
DMMDGMGCGCGCGCHD
--------GCGCSAAA
| Original motif | Reverse complement motif |
| Consensus sequence: DDGCGCGCGCRCHYRD | Consensus sequence: DMMDGMGCGCGCGCHD |
 |
 |
| Dataset #: 3 | Motif ID: 79 | Motif name: ELK1 |
| Original motif | Reverse complement motif |
| Consensus sequence: VDDCCGGAAR | Consensus sequence: MTTCCGGHBV |
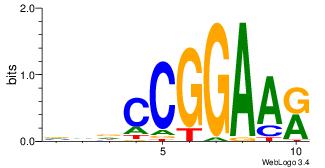 |
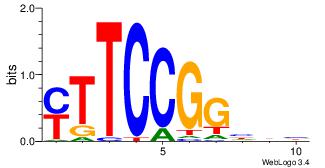 |
Best Matches for Motif ID 79 (Highest to Lowest)
| Motif ID: | UP00414 |
| Motif name: | Ets1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.00796762 |
Alignment:
BDDDHACCGGAARTVDD
---VDDCCGGAAR----
| Original motif | Reverse complement motif |
| Consensus sequence: BDDDHACCGGAARTVDD | Consensus sequence: DDBAMTTCCGGTDHDHB |
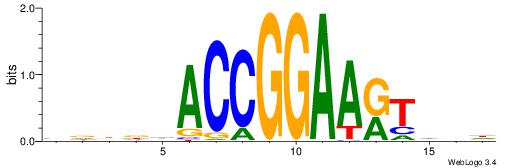 |
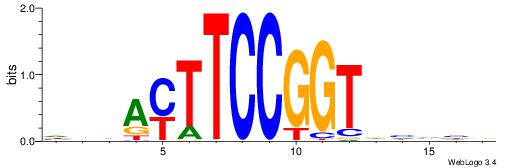 |
| Motif ID: | UP00015 |
| Motif name: | Ehf_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0101295 |
Alignment:
VBDABCCGGAAGTDD
--VDDCCGGAAR---
| Original motif | Reverse complement motif |
| Consensus sequence: VBDABCCGGAAGTDD | Consensus sequence: DDACTTCCGGBTHBB |
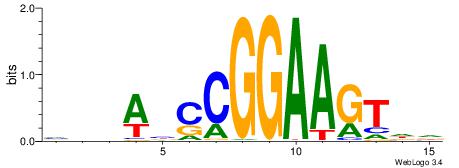 |
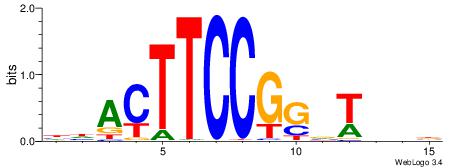 |
| Motif ID: | UP00411 |
| Motif name: | Erg |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0105752 |
Alignment:
BDDACCGGAAKTVBVB
-VDDCCGGAAR-----
| Original motif | Reverse complement motif |
| Consensus sequence: VBBVAYTTCCGGTDDB | Consensus sequence: BDDACCGGAAKTVBVB |
 |
 |
| Motif ID: | UP00404 |
| Motif name: | Elf2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.010693 |
Alignment:
BDWDCCGGAAGTHBBD
-VDDCCGGAAR-----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGDWDB | Consensus sequence: BDWDCCGGAAGTHBBD |
 |
 |
| Motif ID: | UP00423 |
| Motif name: | Gm5454 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.011366 |
Alignment:
DVHACCGGAAGTHDDV
-VDDCCGGAAR-----
| Original motif | Reverse complement motif |
| Consensus sequence: VDHHACTTCCGGTHVH | Consensus sequence: DVHACCGGAAGTHDDV |
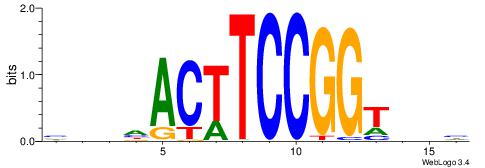 |
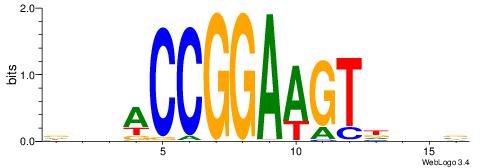 |
| Dataset #: 3 | Motif ID: 80 | Motif name: ELK4 |
| Original motif | Reverse complement motif |
| Consensus sequence: ACCGGAAGT | Consensus sequence: ACTTCCGGT |
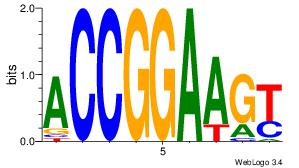 |
 |
Best Matches for Motif ID 80 (Highest to Lowest)
| Motif ID: | UP00013 |
| Motif name: | Gabpa_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 9 |
| Similarity score: | 0 |
Alignment:
HVHHACCGGAAGTDHHV
----ACCGGAAGT----
| Original motif | Reverse complement motif |
| Consensus sequence: HVHHACCGGAAGTDHHV | Consensus sequence: BHHHACTTCCGGTHHBD |
 |
 |
| Motif ID: | UP00414 |
| Motif name: | Ets1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 9 |
| Similarity score: | 0.0025276 |
Alignment:
BDDDHACCGGAARTVDD
-----ACCGGAAGT---
| Original motif | Reverse complement motif |
| Consensus sequence: BDDDHACCGGAARTVDD | Consensus sequence: DDBAMTTCCGGTDHDHB |
 |
 |
| Motif ID: | UP00421 |
| Motif name: | Etv1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 9 |
| Similarity score: | 0.00356917 |
Alignment:
MVHBACCGGAAGTVDVH
----ACCGGAAGT----
| Original motif | Reverse complement motif |
| Consensus sequence: MVHBACCGGAAGTVDVH | Consensus sequence: HBDBACTTCCGGTBHVY |
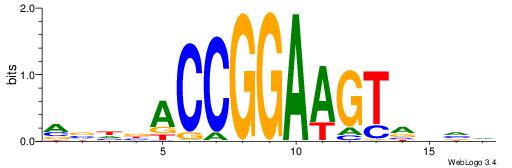 |
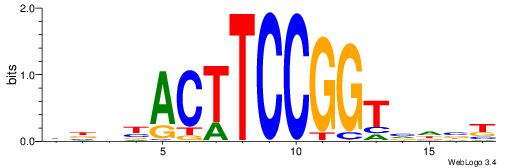 |
| Motif ID: | UP00410 |
| Motif name: | Elk1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 9 |
| Similarity score: | 0.00371937 |
Alignment:
VVHBACCGGAAGTDBHD
----ACCGGAAGT----
| Original motif | Reverse complement motif |
| Consensus sequence: VVHBACCGGAAGTDBHD | Consensus sequence: DHBDACTTCCGGTVHVB |
 |
 |
| Motif ID: | UP00420 |
| Motif name: | Elk3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 9 |
| Similarity score: | 0.0045671 |
Alignment:
DVBBACCGGAAGYDVVV
----ACCGGAAGT----
| Original motif | Reverse complement motif |
| Consensus sequence: DVBBACCGGAAGYDVVV | Consensus sequence: VBVDMCTTCCGGTVBVD |
 |
 |
| Dataset #: 3 | Motif ID: 81 | Motif name: ESR1 |
| Original motif | Reverse complement motif |
| Consensus sequence: VDBHMAGGTCACCCTGACCY | Consensus sequence: MGGTCAGGGTGACCTRDBHV |
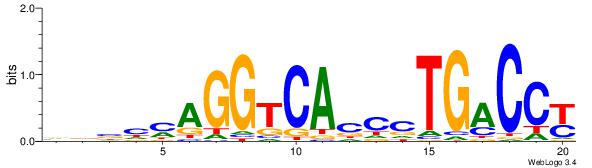 |
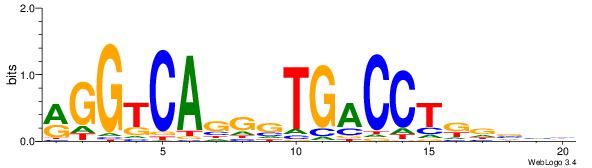 |
Best Matches for Motif ID 81 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 20 |
| Similarity score: | 0.0658405 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
VDBHMAGGTCACCCTGACCY---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 20 |
| Similarity score: | 0.0689748 |
Alignment:
BDBBVBVGTAWCCAAGVBHBAGB
VDBHMAGGTCACCCTGACCY---
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 20 |
| Similarity score: | 0.0701247 |
Alignment:
CBDMCMGGGTGGTCCHVBVBAH
MGGTCAGGGTGACCTRDBHV--
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 20 |
| Similarity score: | 0.0711267 |
Alignment:
HVGVSAGGAGGGTCYHHHHYVH
MGGTCAGGGTGACCTRDBHV--
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
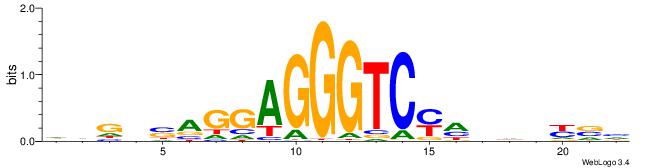 |
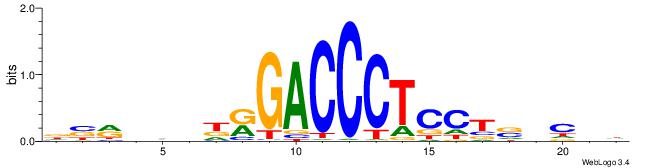 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 20 |
| Similarity score: | 0.0742427 |
Alignment:
CVHBSCGGGTGGTCMHVHHCDH
-VDBHMAGGTCACCCTGACCY-
| Original motif | Reverse complement motif |
| Consensus sequence: HDGHHBHRGACCACCCGSVDVG | Consensus sequence: CVHBSCGGGTGGTCMHVHHCDH |
 |
 |
| Dataset #: 3 | Motif ID: 82 | Motif name: ESR2 |
| Original motif | Reverse complement motif |
| Consensus sequence: VHRGGTCABDBTGMCCTB | Consensus sequence: BAGGYCABHBTGACCKHV |
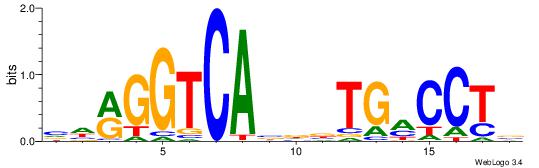 |
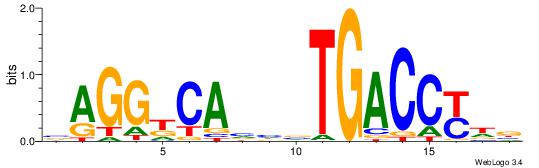 |
Best Matches for Motif ID 82 (Highest to Lowest)
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0443894 |
Alignment:
GTVBDAATTGTVTGHGDDHKVB
---VHRGGTCABDBTGMCCTB-
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.049219 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
---VHRGGTCABDBTGMCCTB--
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0509182 |
Alignment:
DDHBHBSTCACGTGAHBBDHHM
--VHRGGTCABDBTGMCCTB--
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.0527075 |
Alignment:
DVTTVVCVYGGHGGYCHHDMYB
-BAGGYCABHBTGACCKHV---
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.0541044 |
Alignment:
WBHHHHHMDATTGTTHDHDVHH
-BAGGYCABHBTGACCKHV---
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Dataset #: 3 | Motif ID: 83 | Motif name: Esrrb |
| Original motif | Reverse complement motif |
| Consensus sequence: VBBYCAAGGTCA | Consensus sequence: TGACCTTGMBBB |
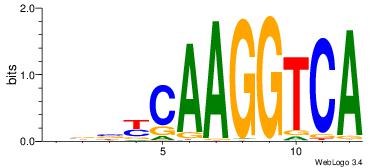 |
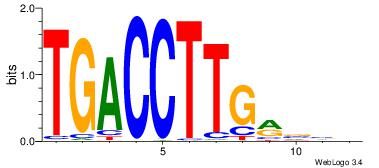 |
Best Matches for Motif ID 83 (Highest to Lowest)
| Motif ID: | UP00079 |
| Motif name: | Esrra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 12 |
| Similarity score: | 0 |
Alignment:
HHBVHTGACCTTGVDHD
-----TGACCTTGMBBB
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBCAAGGTCAHVBDH | Consensus sequence: HHBVHTGACCTTGVDHD |
 |
 |
| Motif ID: | UP00009 |
| Motif name: | Nr2f2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0205471 |
Alignment:
DHBHAAAGGTCAHVHB
VBBYCAAGGTCA----
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCAHVHB | Consensus sequence: BHVDTGACCTTTDVDD |
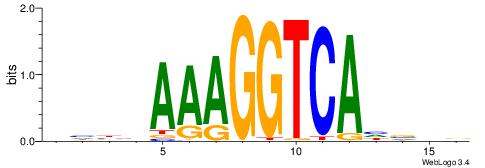 |
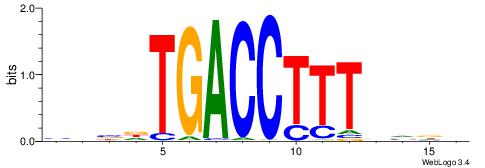 |
| Motif ID: | UP00048 |
| Motif name: | Rara |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0214372 |
Alignment:
DHBHAAAGGTCACVHD
VBBYCAAGGTCA----
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCACVHD | Consensus sequence: HHVGTGACCTTTDVDD |
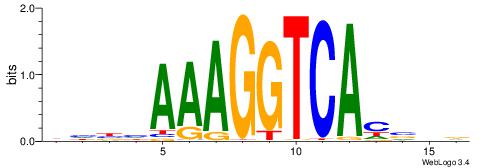 |
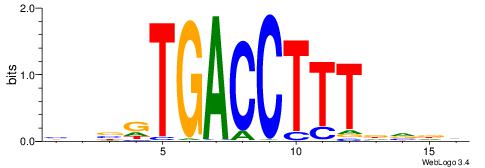 |
| Motif ID: | UP00009 |
| Motif name: | Nr2f2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 0.0430759 |
Alignment:
HVBVTGACCCSGBVBV
----TGACCTTGMBBB
| Original motif | Reverse complement motif |
| Consensus sequence: VBVBCSGGGTCAVBBH | Consensus sequence: HVBVTGACCCSGBVBV |
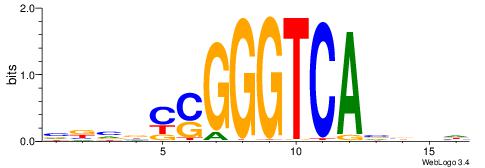 |
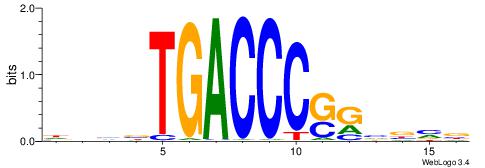 |
| Motif ID: | UP00066 |
| Motif name: | Hnf4a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0451776 |
Alignment:
BDBVMGGGGTCAABHDV
VBBYCAAGGTCA-----
| Original motif | Reverse complement motif |
| Consensus sequence: BDBVMGGGGTCAABHDV | Consensus sequence: BHHVTTGACCCCYVVDB |
 |
 |
| Dataset #: 3 | Motif ID: 84 | Motif name: GABPA |
| Original motif | Reverse complement motif |
| Consensus sequence: CCGGAAGTGVV | Consensus sequence: VVCACTTCCGG |
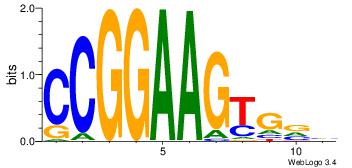 |
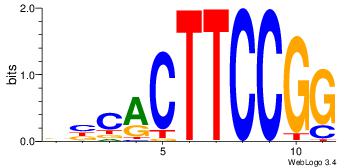 |
Best Matches for Motif ID 84 (Highest to Lowest)
| Motif ID: | UP00013 |
| Motif name: | Gabpa_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0 |
Alignment:
BHHHACTTCCGGTHHBD
-VVCACTTCCGG-----
| Original motif | Reverse complement motif |
| Consensus sequence: HVHHACCGGAAGTDHHV | Consensus sequence: BHHHACTTCCGGTHHBD |
 |
 |
| Motif ID: | UP00410 |
| Motif name: | Elk1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.000136484 |
Alignment:
VVHBACCGGAAGTDBHD
-----CCGGAAGTGVV-
| Original motif | Reverse complement motif |
| Consensus sequence: VVHBACCGGAAGTDBHD | Consensus sequence: DHBDACTTCCGGTVHVB |
 |
 |
| Motif ID: | UP00420 |
| Motif name: | Elk3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.000219706 |
Alignment:
DVBBACCGGAAGYDVVV
-----CCGGAAGTGVV-
| Original motif | Reverse complement motif |
| Consensus sequence: DVBBACCGGAAGYDVVV | Consensus sequence: VBVDMCTTCCGGTVBVD |
 |
 |
| Motif ID: | UP00421 |
| Motif name: | Etv1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.000932023 |
Alignment:
MVHBACCGGAAGTVDVH
-----CCGGAAGTGVV-
| Original motif | Reverse complement motif |
| Consensus sequence: MVHBACCGGAAGTVDVH | Consensus sequence: HBDBACTTCCGGTBHVY |
 |
 |
| Motif ID: | UP00404 |
| Motif name: | Elf2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.00527483 |
Alignment:
DBBHACTTCCGGDWDB
-VVCACTTCCGG----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGDWDB | Consensus sequence: BDWDCCGGAAGTHBBD |
 |
 |
| Dataset #: 3 | Motif ID: 85 | Motif name: Hand1Tcfe2a |
| Original motif | Reverse complement motif |
| Consensus sequence: BRTCTGGMWT | Consensus sequence: AWRCCAGAMB |
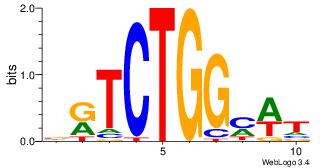 |
 |
Best Matches for Motif ID 85 (Highest to Lowest)
| Motif ID: | UP00000 |
| Motif name: | Smad3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
HVHBVTGTCTGGDDHDD
-----BRTCTGGMWT--
| Original motif | Reverse complement motif |
| Consensus sequence: HDHDDCCAGACABBHVH | Consensus sequence: HVHBVTGTCTGGDDHDD |
 |
 |
| Motif ID: | UP00404 |
| Motif name: | Elf2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0172295 |
Alignment:
BDWDCCGGAAGTHBBD
-AWRCCAGAMB-----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGDWDB | Consensus sequence: BDWDCCGGAAGTHBBD |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0178635 |
Alignment:
VBBDMYCATCTGVHHBH
------BRTCTGGMWT-
| Original motif | Reverse complement motif |
| Consensus sequence: HVDDVCAGATGKYHBBV | Consensus sequence: VBBDMYCATCTGVHHBH |
 |
 |
| Motif ID: | UP00040 |
| Motif name: | Irf5_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.022871 |
Alignment:
VBHAKTCTCGHTHHV
---BRTCTGGMWT--
| Original motif | Reverse complement motif |
| Consensus sequence: BHDAHCGAGARTHBV | Consensus sequence: VBHAKTCTCGHTHHV |
 |
 |
| Motif ID: | UP00038 |
| Motif name: | Spdef_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0241958 |
Alignment:
VHRCATCCGGATHHBB
---BRTCTGGMWT---
| Original motif | Reverse complement motif |
| Consensus sequence: VHRCATCCGGATHHBB | Consensus sequence: VVHHATCCGGATGKHV |
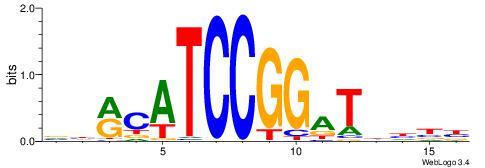 |
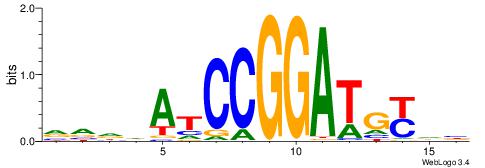 |
| Dataset #: 3 | Motif ID: 86 | Motif name: HIF1AARNT |
| Original motif | Reverse complement motif |
| Consensus sequence: VBACGTGV | Consensus sequence: VCACGTBV |
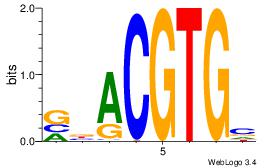 |
 |
Best Matches for Motif ID 86 (Highest to Lowest)
| Motif ID: | UP00097 |
| Motif name: | Mtf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HDYTTGCACACGDHBH
-------VCACGTBV-
| Original motif | Reverse complement motif |
| Consensus sequence: DBDHCGTGTGCAAMDH | Consensus sequence: HDYTTGCACACGDHBH |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.000574345 |
Alignment:
HHVBABCACGTGSTHD
-----VCACGTBV---
| Original motif | Reverse complement motif |
| Consensus sequence: DDASCACGTGBTBVDD | Consensus sequence: HHVBABCACGTGSTHD |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.00642455 |
Alignment:
YDYBDHTMCACGTGGADDBMDGT
-------VBACGTGV--------
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00084 |
| Motif name: | Gmeb1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0131171 |
Alignment:
BVVDVRYACGTRYVBHB
-----VBACGTGV----
| Original motif | Reverse complement motif |
| Consensus sequence: BHBBKKACGTMMVDBVB | Consensus sequence: BVVDVRYACGTRYVBHB |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.014006 |
Alignment:
RDHDBVDTCACGTGASBHVHDH
-------VCACGTBV-------
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Dataset #: 3 | Motif ID: 87 | Motif name: HNF4A |
| Original motif | Reverse complement motif |
| Consensus sequence: RGGBCAAAGKYCA | Consensus sequence: TGMYCTTTGBCCK |
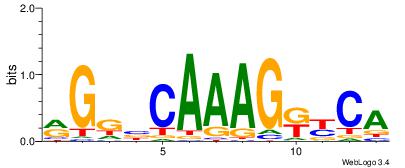 |
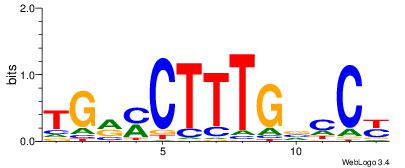 |
Best Matches for Motif ID 87 (Highest to Lowest)
| Motif ID: | UP00058 |
| Motif name: | Tcf3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0357776 |
Alignment:
HHHASATCAAAGVRHHH
---RGGBCAAAGKYCA-
| Original motif | Reverse complement motif |
| Consensus sequence: HHHASATCAAAGVRHHH | Consensus sequence: HHHKVCTTTGATSTHHH |
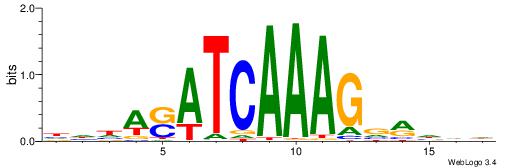 |
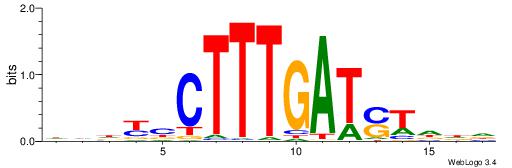 |
| Motif ID: | UP00054 |
| Motif name: | Tcf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0357792 |
Alignment:
BHHASATCAAAGGAHHH
---RGGBCAAAGKYCA-
| Original motif | Reverse complement motif |
| Consensus sequence: BHHASATCAAAGGAHHH | Consensus sequence: HHHTCCTTTGATSTHHV |
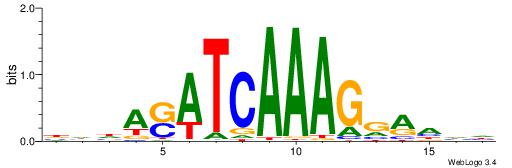 |
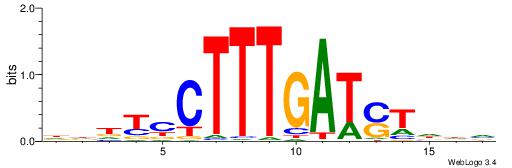 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 13 |
| Similarity score: | 0.0359011 |
Alignment:
VDDAGATCAAAGGKDDD
---RGGBCAAAGKYCA-
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
 |
 |
| Motif ID: | UP00083 |
| Motif name: | Tcf7l2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 13 |
| Similarity score: | 0.0360786 |
Alignment:
BDDASATCAAAGGMDDD
---RGGBCAAAGKYCA-
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATSTDDV | Consensus sequence: BDDASATCAAAGGMDDD |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 13 |
| Similarity score: | 0.0405764 |
Alignment:
HVDDVCAGATGKYHBBV
-RGGBCAAAGKYCA---
| Original motif | Reverse complement motif |
| Consensus sequence: HVDDVCAGATGKYHBBV | Consensus sequence: VBBDMYCATCTGVHHBH |
 |
 |
| Dataset #: 3 | Motif ID: 88 | Motif name: INSM1 |
| Original motif | Reverse complement motif |
| Consensus sequence: TGYCAGGGGGCR | Consensus sequence: MGCCCCCTGMCA |
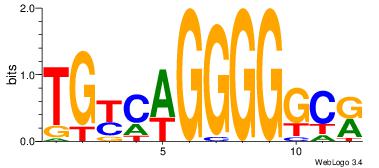 |
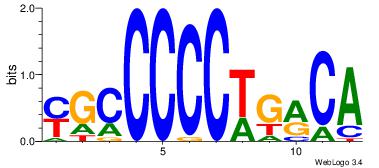 |
Best Matches for Motif ID 88 (Highest to Lowest)
| Motif ID: | UP00022 |
| Motif name: | Zfp740_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.00085101 |
Alignment:
HVKDCCGGGGGGWADDD
--TGYCAGGGGGCR---
| Original motif | Reverse complement motif |
| Consensus sequence: DDDTWCCCCCCGGDRVH | Consensus sequence: HVKDCCGGGGGGWADDD |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 12 |
| Similarity score: | 0.00406431 |
Alignment:
BVVHAGGGGGCGRDHHB
TGYCAGGGGGCR-----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00047 |
| Motif name: | Zbtb7b_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.00486866 |
Alignment:
VHWTTHGGGGGGVDD
--TGYCAGGGGGCR-
| Original motif | Reverse complement motif |
| Consensus sequence: DDVCCCCCCHAAWHB | Consensus sequence: VHWTTHGGGGGGVDD |
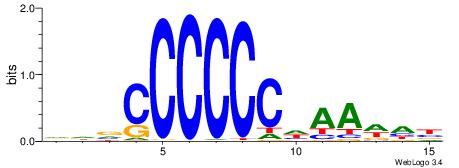 |
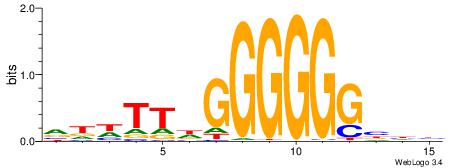 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.00862553 |
Alignment:
VKBBAGGGGTCAHDBBH
TGYCAGGGGGCR-----
| Original motif | Reverse complement motif |
| Consensus sequence: VKBBAGGGGTCAHDBBH | Consensus sequence: DBBHHTGACCCCTBBYV |
 |
 |
| Motif ID: | UP00024 |
| Motif name: | Glis2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.00936306 |
Alignment:
HKBBGGGGGGTCVHHH
-TGYCAGGGGGCR---
| Original motif | Reverse complement motif |
| Consensus sequence: HHHVGACCCCCCVBRD | Consensus sequence: HKBBGGGGGGTCVHHH |
 |
 |
| Dataset #: 3 | Motif ID: 89 | Motif name: Klf4 |
| Original motif | Reverse complement motif |
| Consensus sequence: DGGGYGKGGC | Consensus sequence: GCCYCMCCCD |
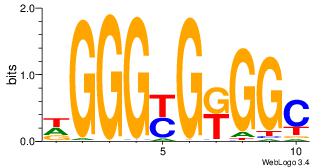 |
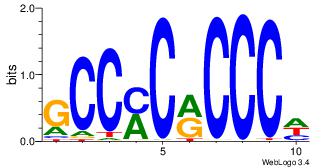 |
Best Matches for Motif ID 89 (Highest to Lowest)
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
DHHDGGGCGRGGKHBH
---DGGGYGKGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0153308 |
Alignment:
DBCCCCCCCCCCMYC
---GCCYCMCCCD--
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0186791 |
Alignment:
BDWAGGCGTGBCHHD
--DGGGYGKGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: BDWAGGCGTGBCHHD | Consensus sequence: HHDGBCACGCCTWDB |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0218399 |
Alignment:
HVBCCCCCCCCMHHHB
--GCCYCMCCCD----
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0249586 |
Alignment:
HCCGCCCCCGCAHB
GCCYCMCCCD----
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
| Dataset #: 3 | Motif ID: 90 | Motif name: Mafb |
| Original motif | Reverse complement motif |
| Consensus sequence: GCTGACDB | Consensus sequence: BHGTCAGC |
 |
 |
Best Matches for Motif ID 90 (Highest to Lowest)
| Motif ID: | UP00044 |
| Motif name: | Mafk_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.00415984 |
Alignment:
DWAAAWTGCTGACTH
-------GCTGACDB
| Original motif | Reverse complement motif |
| Consensus sequence: DWAAAWTGCTGACTH | Consensus sequence: HAGTCAGCAWTTTWD |
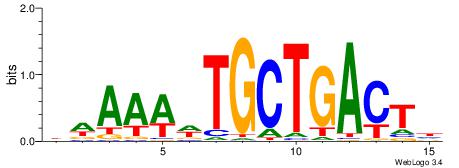 |
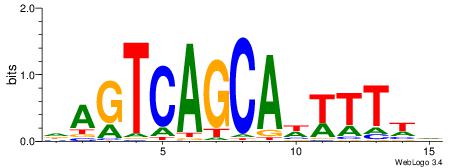 |
| Motif ID: | UP00045 |
| Motif name: | Mafb_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.012743 |
Alignment:
HAAWDTGCTGACDWARH
------GCTGACDB---
| Original motif | Reverse complement motif |
| Consensus sequence: HAAWDTGCTGACDWARH | Consensus sequence: DMTWDGTCAGCADWTTH |
 |
 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.0222535 |
Alignment:
HHDVVGCAGCTGVBKVB
--------GCTGACDB-
| Original motif | Reverse complement motif |
| Consensus sequence: BBYVVCAGCTGCBVHHD | Consensus sequence: HHDVVGCAGCTGVBKVB |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0269322 |
Alignment:
VHYTCCDWCCGCGTCMMTHWHV
----BHGTCAGC----------
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
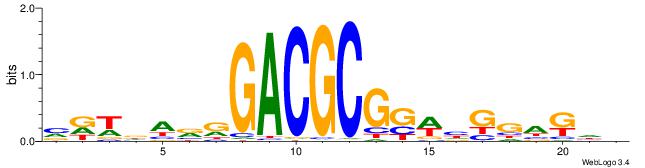 |
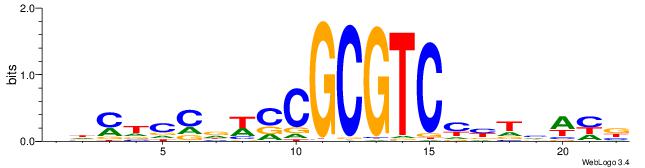 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0319883 |
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
----BHGTCAGC-----------
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Dataset #: 3 | Motif ID: 91 | Motif name: MAX |
| Original motif | Reverse complement motif |
| Consensus sequence: DAHCACGTGD | Consensus sequence: BCACGTGDTD |
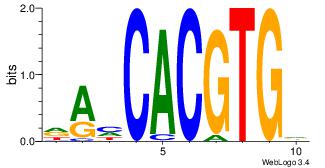 |
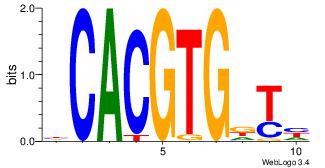 |
Best Matches for Motif ID 91 (Highest to Lowest)
| Motif ID: | UP00060 |
| Motif name: | Max_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
HHVBABCACGTGSTHD
---DAHCACGTGD---
| Original motif | Reverse complement motif |
| Consensus sequence: DDASCACGTGBTBVDD | Consensus sequence: HHVBABCACGTGSTHD |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 10 |
| Similarity score: | 0.018544 |
Alignment:
DDHBHBSTCACGTGAHBBDHHM
-------BCACGTGDTD-----
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 10 |
| Similarity score: | 0.0267839 |
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
------DAHCACGTGD-------
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0321448 |
Alignment:
DHDBHGCACCTGBDDVB
-----BCACGTGDTD--
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Motif ID: | UP00036 |
| Motif name: | Myf6_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.042685 |
Alignment:
VVDBAACAGBTGBYHB
---DAHCACGTGD---
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBAACAGBTGBYHB | Consensus sequence: BDKVCABCTGTTBDBV |
 |
 |
| Dataset #: 3 | Motif ID: 92 | Motif name: MIZF |
| Original motif | Reverse complement motif |
| Consensus sequence: BAACGTCCGC | Consensus sequence: GCGGACGTTV |
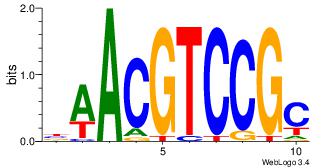 |
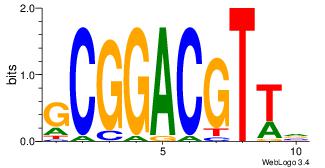 |
Best Matches for Motif ID 92 (Highest to Lowest)
| Motif ID: | UP00419 |
| Motif name: | Spic |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
VAAAGMGGAAGTWD
BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: VAAAGMGGAAGTWD | Consensus sequence: DWACTTCCRCTTTB |
 |
 |
| Motif ID: | UP00409 |
| Motif name: | Elf5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.00379999 |
Alignment:
DWACTTCCRSDTWV
BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: BWAHSMGGAAGTWD | Consensus sequence: DWACTTCCRSDTWV |
 |
 |
| Motif ID: | UP00413 |
| Motif name: | Elf4 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0124101 |
Alignment:
DBBHACTTCCGGKWTH
--BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGKWTH | Consensus sequence: HAWYCCGGAAGTHBBD |
 |
 |
| Motif ID: | UP00404 |
| Motif name: | Elf2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0128348 |
Alignment:
DBBHACTTCCGGDWDB
--BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGDWDB | Consensus sequence: BDWDCCGGAAGTHBBD |
 |
 |
| Motif ID: | UP00085 |
| Motif name: | Sfpi1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0141516 |
Alignment:
BDAWGVGGAAGTDH
BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: BDAWGVGGAAGTDH | Consensus sequence: HDACTTCCBCWTDV |
 |
 |
| Dataset #: 3 | Motif ID: 93 | Motif name: Myb |
| Original motif | Reverse complement motif |
| Consensus sequence: GRCVGTTG | Consensus sequence: CAACVGMC |
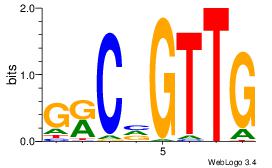 |
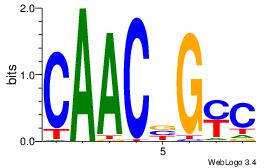 |
Best Matches for Motif ID 93 (Highest to Lowest)
| Motif ID: | UP00081 |
| Motif name: | Mybl1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
BHCGGCAGTTGGBBB
---GRCVGTTG----
| Original motif | Reverse complement motif |
| Consensus sequence: BBVCCAACTGCCGHB | Consensus sequence: BHCGGCAGTTGGBBB |
 |
 |
| Motif ID: | UP00092 |
| Motif name: | Myb_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.00802457 |
Alignment:
DBVKGGCAGTTGGTHB
----GRCVGTTG----
| Original motif | Reverse complement motif |
| Consensus sequence: BDACCAACTGCCRBBH | Consensus sequence: DBVKGGCAGTTGGTHB |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0217124 |
Alignment:
HDTBHHCCHTAGCAACVVDHHHH
------------CAACVGMC---
| Original motif | Reverse complement motif |
| Consensus sequence: HDTBHHCCHTAGCAACVVDHHHH | Consensus sequence: HHHHDVVGTTGCTAHGGDHBAHH |
 |
 |
| Motif ID: | UP00227 |
| Motif name: | Duxl |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.035989 |
Alignment:
YRACCCAATCAACVVHV
GRCVGTTG---------
| Original motif | Reverse complement motif |
| Consensus sequence: YRACCCAATCAACVVHV | Consensus sequence: VHVVGTTGATTGGGTMK |
 |
 |
| Motif ID: | UP00081 |
| Motif name: | Mybl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0421486 |
Alignment:
HDDDHAACCGTTAHHHD
-----GRCVGTTG----
| Original motif | Reverse complement motif |
| Consensus sequence: HDDDHAACCGTTAHHHD | Consensus sequence: DHHHTAACGGTTHDHDH |
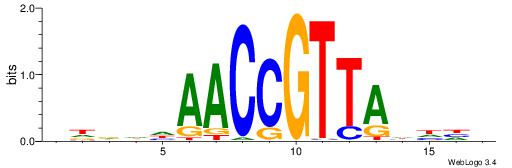 |
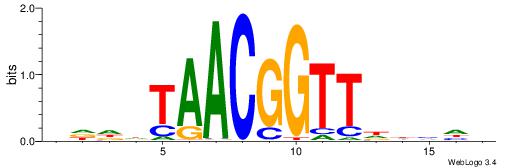 |
| Dataset #: 3 | Motif ID: 94 | Motif name: Myc |
| Original motif | Reverse complement motif |
| Consensus sequence: VGCACGTGGH | Consensus sequence: DCCACGTGCV |
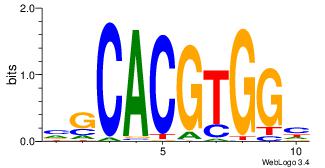 |
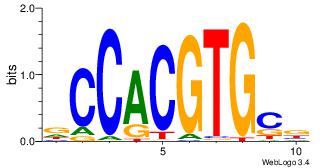 |
Best Matches for Motif ID 94 (Highest to Lowest)
| Motif ID: | UP00060 |
| Motif name: | Max_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
DDASCACGTGBTBVDD
--VGCACGTGGH----
| Original motif | Reverse complement motif |
| Consensus sequence: DDASCACGTGBTBVDD | Consensus sequence: HHVBABCACGTGSTHD |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 10 |
| Similarity score: | 0.00631885 |
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
-------DCCACGTGCV------
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.00821668 |
Alignment:
DDHBHBSTCACGTGAHBBDHHM
------DCCACGTGCV------
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0140005 |
Alignment:
DHDBHGCACCTGBDDVB
----VGCACGTGGH---
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0308912 |
Alignment:
BBGHCACGCGACDD
--VGCACGTGGH--
| Original motif | Reverse complement motif |
| Consensus sequence: BBGHCACGCGACDD | Consensus sequence: HDGTCGCGTGDCVB |
 |
 |
| Dataset #: 3 | Motif ID: 95 | Motif name: MYCMAX |
| Original motif | Reverse complement motif |
| Consensus sequence: RASCACGTGGT | Consensus sequence: ACCACGTGSTM |
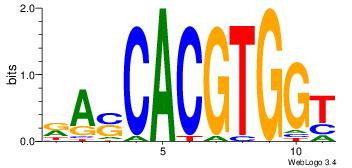 |
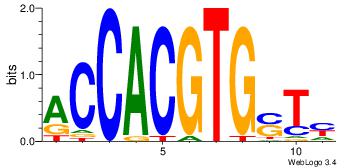 |
Best Matches for Motif ID 95 (Highest to Lowest)
| Motif ID: | UP00060 |
| Motif name: | Max_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0 |
Alignment:
HHVBABCACGTGSTHD
---RASCACGTGGT--
| Original motif | Reverse complement motif |
| Consensus sequence: DDASCACGTGBTBVDD | Consensus sequence: HHVBABCACGTGSTHD |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 11 |
| Similarity score: | 0.025477 |
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
-------ACCACGTGSTM-----
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.033078 |
Alignment:
RDHDBVDTCACGTGASBHVHDH
-----RASCACGTGGT------
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0375583 |
Alignment:
VBHHVCAGGTGCDVDHD
---ACCACGTGSTM---
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0452419 |
Alignment:
HVDDVCAGATGKYHBBV
--RASCACGTGGT----
| Original motif | Reverse complement motif |
| Consensus sequence: HVDDVCAGATGKYHBBV | Consensus sequence: VBBDMYCATCTGVHHBH |
 |
 |
| Dataset #: 3 | Motif ID: 96 | Motif name: Mycn |
| Original motif | Reverse complement motif |
| Consensus sequence: HSCACGTGGC | Consensus sequence: GCCACGTGSD |
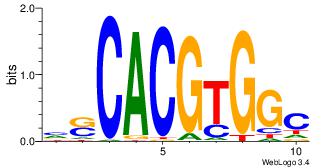 |
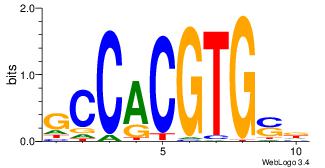 |
Best Matches for Motif ID 96 (Highest to Lowest)
| Motif ID: | UP00060 |
| Motif name: | Max_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
DDASCACGTGBTBVDD
--HSCACGTGGC----
| Original motif | Reverse complement motif |
| Consensus sequence: DDASCACGTGBTBVDD | Consensus sequence: HHVBABCACGTGSTHD |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.00783793 |
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
-------GCCACGTGSD------
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0132858 |
Alignment:
DDHBHBSTCACGTGAHBBDHHM
------GCCACGTGSD------
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0227933 |
Alignment:
VBHHVCAGGTGCDVDHD
---GCCACGTGSD----
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0257011 |
Alignment:
BBGHCACGCGACDD
--HSCACGTGGC--
| Original motif | Reverse complement motif |
| Consensus sequence: BBGHCACGCGACDD | Consensus sequence: HDGTCGCGTGDCVB |
 |
 |
| Dataset #: 3 | Motif ID: 97 | Motif name: Myf |
| Original motif | Reverse complement motif |
| Consensus sequence: MRGCARCWGSWG | Consensus sequence: CWSCWGMTGCKR |
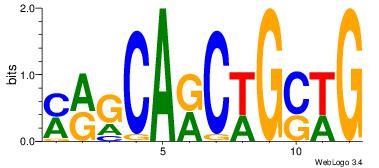 |
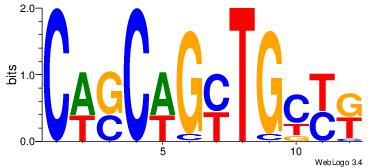 |
Best Matches for Motif ID 97 (Highest to Lowest)
| Motif ID: | UP00099 |
| Motif name: | Ascl2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0 |
Alignment:
BBYVVCAGCTGCBVHHD
--CWSCWGMTGCKR---
| Original motif | Reverse complement motif |
| Consensus sequence: BBYVVCAGCTGCBVHHD | Consensus sequence: HHDVVGCAGCTGVBKVB |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0150333 |
Alignment:
VBHHVCAGGTGCDVDHD
--CWSCWGMTGCKR---
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Motif ID: | UP00036 |
| Motif name: | Myf6_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 0.0199226 |
Alignment:
VVDBAACAGBTGBYHB
MRGCARCWGSWG----
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBAACAGBTGBYHB | Consensus sequence: BDKVCABCTGTTBDBV |
 |
 |
| Motif ID: | UP00081 |
| Motif name: | Mybl1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0255212 |
Alignment:
BBVCCAACTGCCGHB
-MRGCARCWGSWG--
| Original motif | Reverse complement motif |
| Consensus sequence: BBVCCAACTGCCGHB | Consensus sequence: BHCGGCAGTTGGBBB |
 |
 |
| Motif ID: | UP00057 |
| Motif name: | Zic2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0289533 |
Alignment:
DVBCYTGCTGBGDDD
-CWSCWGMTGCKR--
| Original motif | Reverse complement motif |
| Consensus sequence: HHDCVCAGCAKGVVD | Consensus sequence: DVBCYTGCTGBGDDD |
 |
 |
| Dataset #: 3 | Motif ID: 98 | Motif name: MZF1_1-4 |
| Original motif | Reverse complement motif |
| Consensus sequence: BGGGGA | Consensus sequence: TCCCCV |
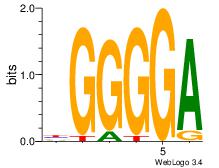 |
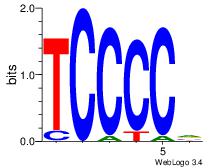 |
Best Matches for Motif ID 98 (Highest to Lowest)
| Motif ID: | UP00176 |
| Motif name: | Crx |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
YBWDGGGGATTARCCD
---BGGGGA-------
| Original motif | Reverse complement motif |
| Consensus sequence: YBWDGGGGATTARCCD | Consensus sequence: DGGMTAATCCCCDWBK |
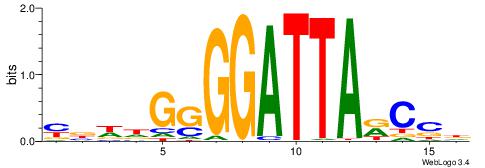 |
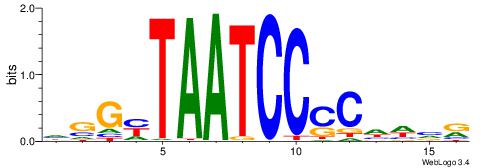 |
| Motif ID: | UP00265 |
| Motif name: | Pitx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.00193291 |
Alignment:
GBHDGCTAATCCCCCB
---------TCCCCV-
| Original motif | Reverse complement motif |
| Consensus sequence: VGGGGGATTAGCDDBC | Consensus sequence: GBHDGCTAATCCCCCB |
 |
 |
| Motif ID: | UP00239 |
| Motif name: | Obox2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.00284735 |
Alignment:
HTARTTAATCCCCCKBD
--------TCCCCV---
| Original motif | Reverse complement motif |
| Consensus sequence: DBRGGGGGATTAAMTAH | Consensus sequence: HTARTTAATCCCCCKBD |
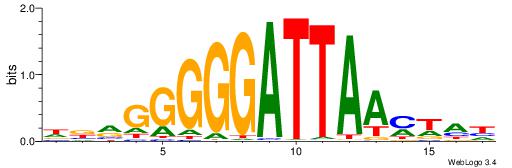 |
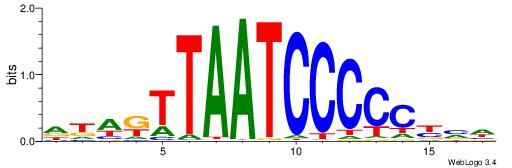 |
| Motif ID: | UP00216 |
| Motif name: | Obox1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.00461976 |
Alignment:
DHDGTTAATCCCCKTDD
--------TCCCCV---
| Original motif | Reverse complement motif |
| Consensus sequence: DDARGGGGATTAACDHH | Consensus sequence: DHDGTTAATCCCCKTDD |
 |
 |
| Motif ID: | UP00160 |
| Motif name: | Obox3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.00641818 |
Alignment:
HHWGTTAATCCCCCBBH
--------TCCCCV---
| Original motif | Reverse complement motif |
| Consensus sequence: HBVGGGGGATTAACWHH | Consensus sequence: HHWGTTAATCCCCCBBH |
 |
 |
| Dataset #: 3 | Motif ID: 99 | Motif name: MZF1_5-13 |
| Original motif | Reverse complement motif |
| Consensus sequence: BKAGGGGDAD | Consensus sequence: BTHCCCCTYB |
 |
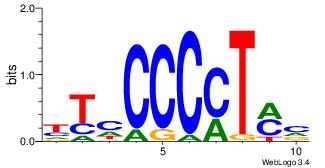 |
Best Matches for Motif ID 99 (Highest to Lowest)
| Motif ID: | UP00022 |
| Motif name: | Zfp740_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0090535 |
Alignment:
DDDTWCCCCCCGGDRVH
---BTHCCCCTYB----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDTWCCCCCCGGDRVH | Consensus sequence: HVKDCCGGGGGGWADDD |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.00910178 |
Alignment:
BBHBGAGTGGGAHHDB
---BKAGGGGDAD---
| Original motif | Reverse complement motif |
| Consensus sequence: BBHBGAGTGGGAHHDB | Consensus sequence: BHHDTCCCACTCBDBV |
 |
 |
| Motif ID: | UP00070 |
| Motif name: | Gcm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0101177 |
Alignment:
DDBVHATAGGGGRBRMB
-----BKAGGGGDAD--
| Original motif | Reverse complement motif |
| Consensus sequence: DDBVHATAGGGGRBRMB | Consensus sequence: BYMBKCCCCTATDVBHD |
 |
 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0103225 |
Alignment:
GKRGGGGGGGGGGBD
BKAGGGGDAD-----
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0126235 |
Alignment:
HVDVGGGHGGGGDBHB
-----BKAGGGGDAD-
| Original motif | Reverse complement motif |
| Consensus sequence: BHVDCCCCDCCCBDBH | Consensus sequence: HVDVGGGHGGGGDBHB |
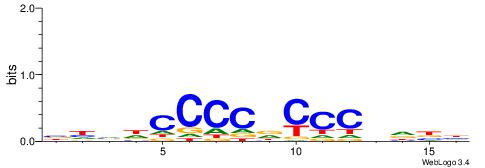 |
 |
| Dataset #: 3 | Motif ID: 100 | Motif name: NFE2L2 |
| Original motif | Reverse complement motif |
| Consensus sequence: RTGACWHAGCA | Consensus sequence: TGCTDWGTCAK |
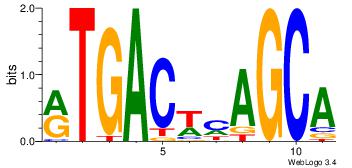 |
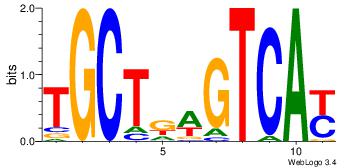 |
Best Matches for Motif ID 100 (Highest to Lowest)
| Motif ID: | UP00103 |
| Motif name: | Jundm2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0252843 |
Alignment:
HDDKTGASTCAKHRDB
---RTGACWHAGCA--
| Original motif | Reverse complement motif |
| Consensus sequence: VDKDRTGASTCAYHDH | Consensus sequence: HDDKTGASTCAKHRDB |
 |
 |
| Motif ID: | UP00020 |
| Motif name: | Atf1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0449032 |
Alignment:
VDATGACGADYADH
--RTGACWHAGCA-
| Original motif | Reverse complement motif |
| Consensus sequence: VDATGACGADYADH | Consensus sequence: DDTMDTCGTCATDV |
 |
 |
| Motif ID: | UP00009 |
| Motif name: | Nr2f2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0453065 |
Alignment:
HVBVTGACCCSGBVBV
---RTGACWHAGCA--
| Original motif | Reverse complement motif |
| Consensus sequence: VBVBCSGGGTCAVBBH | Consensus sequence: HVBVTGACCCSGBVBV |
 |
 |
| Motif ID: | UP00045 |
| Motif name: | Mafb_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.04674 |
Alignment:
HAAWDTGCTGACDWARH
-----TGCTDWGTCAK-
| Original motif | Reverse complement motif |
| Consensus sequence: HAAWDTGCTGACDWARH | Consensus sequence: DMTWDGTCAGCADWTTH |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0473193 |
Alignment:
DDHBHBSTCACGTGAHBBDHHM
-----------RTGACWHAGCA
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Dataset #: 3 | Motif ID: 101 | Motif name: NFIC |
| Original motif | Reverse complement motif |
| Consensus sequence: TTGGCD | Consensus sequence: DGCCAA |
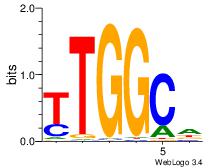 |
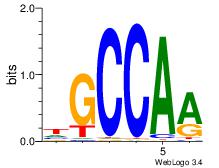 |
Best Matches for Motif ID 101 (Highest to Lowest)
| Motif ID: | UP00035 |
| Motif name: | Hic1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
DDBHGGTTGGCATVBD
------TTGGCD----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBATGCCAACCHVHH | Consensus sequence: DDBHGGTTGGCATVBD |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0174701 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
------DGCCAA-----------
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00003 |
| Motif name: | E2F3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0231918 |
Alignment:
BBBDYTGGCGCCKWDBD
----TTGGCD-------
| Original motif | Reverse complement motif |
| Consensus sequence: HBHWYGGCGCCAMDVBB | Consensus sequence: BBBDYTGGCGCCKWDBD |
 |
 |
| Motif ID: | UP00001 |
| Motif name: | E2F2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0258343 |
Alignment:
BBBDTTGGCGCCKWVVD
----TTGGCD-------
| Original motif | Reverse complement motif |
| Consensus sequence: HVBWYGGCGCCAADVBB | Consensus sequence: BBBDTTGGCGCCKWVVD |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.0266372 |
Alignment:
HDGTCGCGTGDCVB
-------TTGGCD-
| Original motif | Reverse complement motif |
| Consensus sequence: BBGHCACGCGACDD | Consensus sequence: HDGTCGCGTGDCVB |
 |
 |
| Dataset #: 3 | Motif ID: 102 | Motif name: NF-kappaB |
| Original motif | Reverse complement motif |
| Consensus sequence: GGGRMTTYCC | Consensus sequence: GGMAAYKCCC |
 |
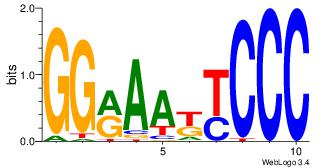 |
Best Matches for Motif ID 102 (Highest to Lowest)
| Motif ID: | UP00021 |
| Motif name: | Zfp281_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0379362 |
Alignment:
DKKWDACCCCCAHTDDV
-------GGGRMTTYCC
| Original motif | Reverse complement motif |
| Consensus sequence: DKKWDACCCCCAHTDDV | Consensus sequence: VDDAHTGGGGGTHWYYD |
 |
 |
| Motif ID: | UP00176 |
| Motif name: | Crx |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0387084 |
Alignment:
DGGMTAATCCCCDWBK
-GGMAAYKCCC-----
| Original motif | Reverse complement motif |
| Consensus sequence: YBWDGGGGATTARCCD | Consensus sequence: DGGMTAATCCCCDWBK |
 |
 |
| Motif ID: | UP00265 |
| Motif name: | Pitx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0428696 |
Alignment:
GBHDGCTAATCCCCCB
---GGMAAYKCCC---
| Original motif | Reverse complement motif |
| Consensus sequence: VGGGGGATTAGCDDBC | Consensus sequence: GBHDGCTAATCCCCCB |
 |
 |
| Motif ID: | UP00422 |
| Motif name: | Etv3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0433644 |
Alignment:
DHHACCGGAAKTHBBB
-----GGGRMTTYCC-
| Original motif | Reverse complement motif |
| Consensus sequence: VBBHAYTTCCGGTHDH | Consensus sequence: DHHACCGGAAKTHBBB |
 |
 |
| Motif ID: | UP00035 |
| Motif name: | Hic1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0443841 |
Alignment:
DBDDRTGCCCAWDHDV
GGMAAYKCCC------
| Original motif | Reverse complement motif |
| Consensus sequence: DBDDRTGCCCAWDHDV | Consensus sequence: VHHDWTGGGCAMDHBH |
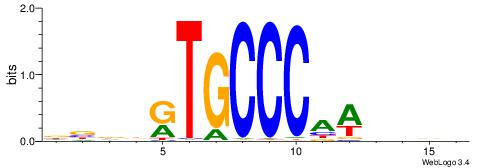 |
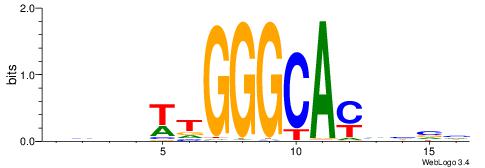 |
| Dataset #: 3 | Motif ID: 103 | Motif name: NFKB1 |
| Original motif | Reverse complement motif |
| Consensus sequence: GGGGRTTCCCC | Consensus sequence: GGGGAAKCCCC |
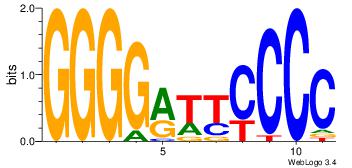 |
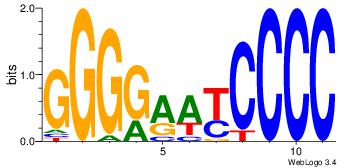 |
Best Matches for Motif ID 103 (Highest to Lowest)
| Motif ID: | UP00021 |
| Motif name: | Zfp281_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0371794 |
Alignment:
VDDAHTGGGGGTHWYYD
------GGGGRTTCCCC
| Original motif | Reverse complement motif |
| Consensus sequence: DKKWDACCCCCAHTDDV | Consensus sequence: VDDAHTGGGGGTHWYYD |
 |
 |
| Motif ID: | UP00088 |
| Motif name: | Plagl1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0407901 |
Alignment:
DBHGGGGGGTACCHHHD
----GGGGRTTCCCC--
| Original motif | Reverse complement motif |
| Consensus sequence: DBHGGGGGGTACCHHHD | Consensus sequence: DHDDGGTACCCCCCHBH |
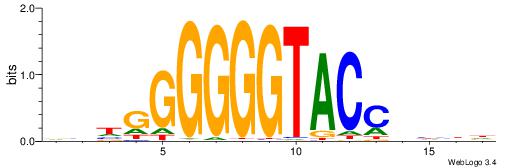 |
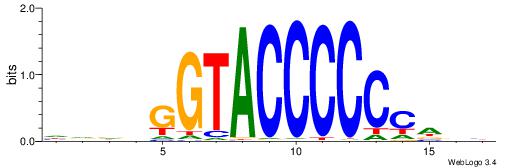 |
| Motif ID: | UP00088 |
| Motif name: | Plagl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0459696 |
Alignment:
BHDGGGGSSCCCCBVV
--GGGGRTTCCCC---
| Original motif | Reverse complement motif |
| Consensus sequence: BHDGGGGSSCCCCBVV | Consensus sequence: VBVGGGGSSCCCCHHV |
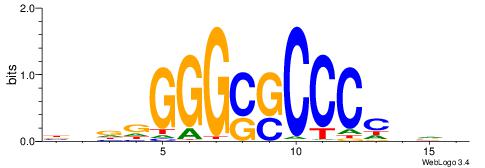 |
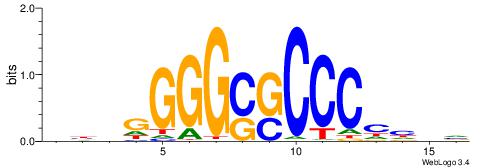 |
| Motif ID: | UP00176 |
| Motif name: | Crx |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0464973 |
Alignment:
DGGMTAATCCCCDWBK
-GGGGAAKCCCC----
| Original motif | Reverse complement motif |
| Consensus sequence: YBWDGGGGATTARCCD | Consensus sequence: DGGMTAATCCCCDWBK |
 |
 |
| Motif ID: | UP00265 |
| Motif name: | Pitx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.048533 |
Alignment:
GBHDGCTAATCCCCCB
---GGGGAAKCCCC--
| Original motif | Reverse complement motif |
| Consensus sequence: VGGGGGATTAGCDDBC | Consensus sequence: GBHDGCTAATCCCCCB |
 |
 |
| Dataset #: 3 | Motif ID: 104 | Motif name: NFYA |
| Original motif | Reverse complement motif |
| Consensus sequence: VBBRRCCAATSRGVDB | Consensus sequence: BHVCKSATTGGMKBVV |
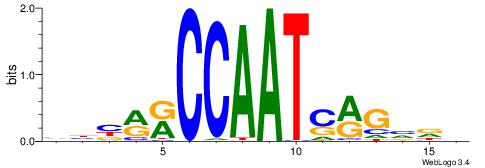 |
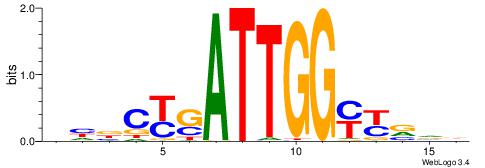 |
Best Matches for Motif ID 104 (Highest to Lowest)
| Motif ID: | UP00083 |
| Motif name: | Tcf7l2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0368059 |
Alignment:
DHVBKGATTGATVBDV
BHVCKSATTGGMKBVV
| Original motif | Reverse complement motif |
| Consensus sequence: VDVVATCAATCRBBHD | Consensus sequence: DHVBKGATTGATVBDV |
 |
 |
| Motif ID: | UP00220 |
| Motif name: | Nkx1-1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0372416 |
Alignment:
WGHRCYAATTRGYGSDV
-BHVCKSATTGGMKBVV
| Original motif | Reverse complement motif |
| Consensus sequence: WGHRCYAATTRGYGSDV | Consensus sequence: BHSCMCKAATTMGMDCW |
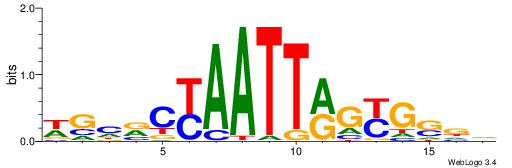 |
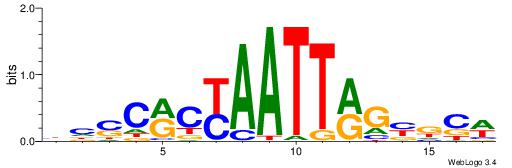 |
| Motif ID: | UP00156 |
| Motif name: | Msx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0372531 |
Alignment:
VVDGACYAATTAGYDHT
VBBRRCCAATSRGVDB-
| Original motif | Reverse complement motif |
| Consensus sequence: VVDGACYAATTAGYDHT | Consensus sequence: ADHKCTAATTKGTCDBV |
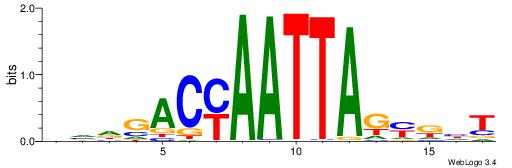 |
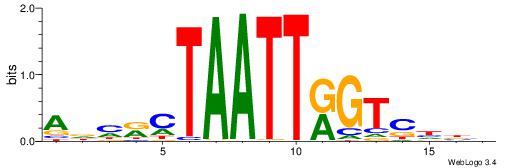 |
| Motif ID: | UP00055 |
| Motif name: | Hbp1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0374245 |
Alignment:
HBTHVACAATGGGMDHD
VBBRRCCAATSRGVDB-
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCCATTGTVHABH | Consensus sequence: HBTHVACAATGGGMDHD |
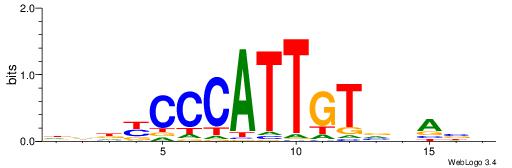 |
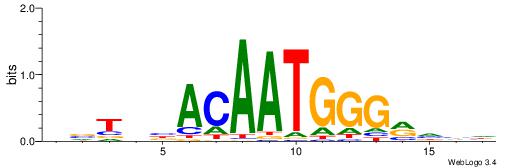 |
| Motif ID: | UP00162 |
| Motif name: | Evx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0384724 |
Alignment:
WVHVCTAATTAGYBVDV
-BHVCKSATTGGMKBVV
| Original motif | Reverse complement motif |
| Consensus sequence: WVHVCTAATTAGYBVDV | Consensus sequence: VDVBMCTAATTAGBHVW |
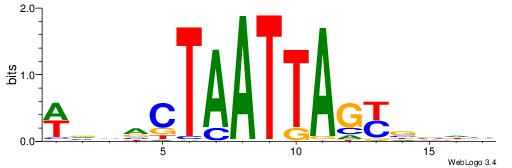 |
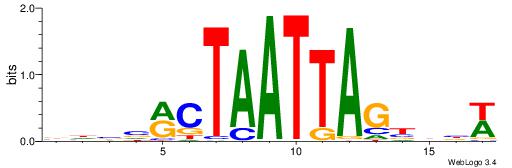 |
| Dataset #: 3 | Motif ID: 105 | Motif name: NHLH1 |
| Original motif | Reverse complement motif |
| Consensus sequence: VCGCAGCTGCGB | Consensus sequence: VCGCAGCTGCGV |
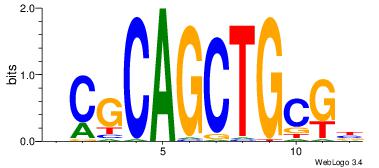 |
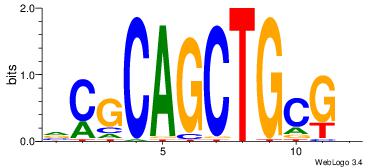 |
Best Matches for Motif ID 105 (Highest to Lowest)
| Motif ID: | UP00099 |
| Motif name: | Ascl2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0 |
Alignment:
HHDVVGCAGCTGVBKVB
---VCGCAGCTGCGB--
| Original motif | Reverse complement motif |
| Consensus sequence: BBYVVCAGCTGCBVHHD | Consensus sequence: HHDVVGCAGCTGVBKVB |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0128197 |
Alignment:
VBHHVCAGGTGCDVDHD
--VCGCAGCTGCGV---
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0308854 |
Alignment:
VBBDMYCATCTGVHHBH
---VCGCAGCTGCGB--
| Original motif | Reverse complement motif |
| Consensus sequence: HVDDVCAGATGKYHBBV | Consensus sequence: VBBDMYCATCTGVHHBH |
 |
 |
| Motif ID: | UP00036 |
| Motif name: | Myf6_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0327062 |
Alignment:
VVDBAACAGBTGBYHB
---VCGCAGCTGCGB-
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBAACAGBTGBYHB | Consensus sequence: BDKVCABCTGTTBDBV |
 |
 |
| Motif ID: | UP00102 |
| Motif name: | Zic1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0363264 |
Alignment:
DVHCCTGCTGBGDDB
-VCGCAGCTGCGV--
| Original motif | Reverse complement motif |
| Consensus sequence: BHDCVCAGCAGGHVD | Consensus sequence: DVHCCTGCTGBGDDB |
 |
 |
| Dataset #: 3 | Motif ID: 106 | Motif name: Nkx3-2 |
| Original motif | Reverse complement motif |
| Consensus sequence: BTRAGTGVV | Consensus sequence: BVCACTKAH |
 |
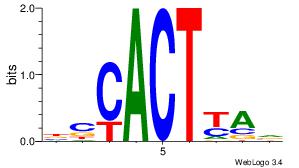 |
Best Matches for Motif ID 106 (Highest to Lowest)
| Motif ID: | UP00147 |
| Motif name: | Nkx2-6 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 9 |
| Similarity score: | 0 |
Alignment:
AAHVTTAAGTGGHTDD
----BTRAGTGVV---
| Original motif | Reverse complement motif |
| Consensus sequence: DDADCCACTTAAVHTT | Consensus sequence: AAHVTTAAGTGGHTDD |
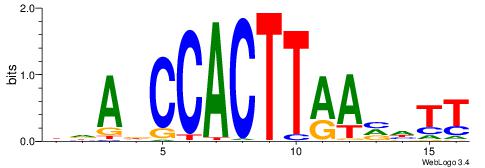 |
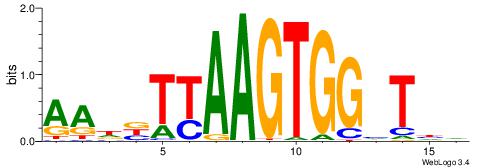 |
| Motif ID: | UP00228 |
| Motif name: | Bapx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 9 |
| Similarity score: | 0.00188665 |
Alignment:
VHHAACCACTTAAVVAH
----BVCACTKAH----
| Original motif | Reverse complement motif |
| Consensus sequence: VHHAACCACTTAAVVAH | Consensus sequence: DTBVTTAAGTGGTTHHV |
 |
 |
| Motif ID: | UP00249 |
| Motif name: | Nkx2-5 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 9 |
| Similarity score: | 0.00571144 |
Alignment:
HDADCCACTTRAAWTT
---BVCACTKAH----
| Original motif | Reverse complement motif |
| Consensus sequence: HDADCCACTTRAAWTT | Consensus sequence: AAWTTMAAGTGGHTDH |
 |
 |
| Motif ID: | UP00107 |
| Motif name: | Nkx2-4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 9 |
| Similarity score: | 0.00990848 |
Alignment:
HDAVCCACTTRAMWTT
---BVCACTKAH----
| Original motif | Reverse complement motif |
| Consensus sequence: HDAVCCACTTRAMWTT | Consensus sequence: AAWYTMAAGTGGVTDH |
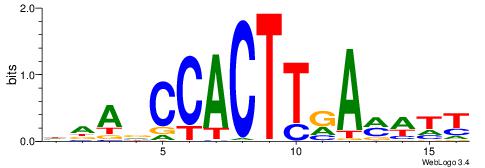 |
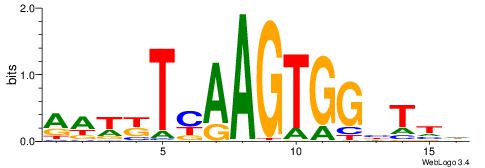 |
| Motif ID: | UP00165 |
| Motif name: | Titf1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 9 |
| Similarity score: | 0.0125254 |
Alignment:
BVABCCACTTGAMHTT
---BVCACTKAH----
| Original motif | Reverse complement motif |
| Consensus sequence: BVABCCACTTGAMHTT | Consensus sequence: AAHYTCAAGTGGBTBV |
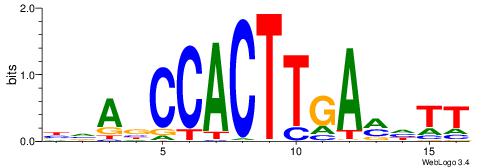 |
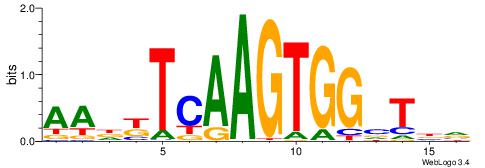 |
| Dataset #: 3 | Motif ID: 107 | Motif name: NR2F1 |
| Original motif | Reverse complement motif |
| Consensus sequence: TGAMCTTTGMMCYT | Consensus sequence: AKGYYCAAAGRTCA |
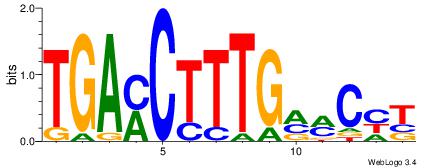 |
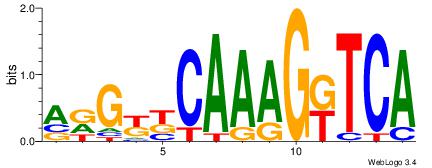 |
Best Matches for Motif ID 107 (Highest to Lowest)
| Motif ID: | UP00083 |
| Motif name: | Tcf7l2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0452364 |
Alignment:
DDDYCCTTTGATSTDDV
-TGAMCTTTGMMCYT--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATSTDDV | Consensus sequence: BDDASATCAAAGGMDDD |
 |
 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0453449 |
Alignment:
VDDAGATCAAAGGKDDD
--AKGYYCAAAGRTCA-
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0461358 |
Alignment:
DVMHHDHKGACCCTCCTSVCBH
-------TGAMCTTTGMMCYT-
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Motif ID: | UP00054 |
| Motif name: | Tcf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0461768 |
Alignment:
BHHASATCAAAGGAHHH
--AKGYYCAAAGRTCA-
| Original motif | Reverse complement motif |
| Consensus sequence: BHHASATCAAAGGAHHH | Consensus sequence: HHHTCCTTTGATSTHHV |
 |
 |
| Motif ID: | UP00058 |
| Motif name: | Tcf3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0473628 |
Alignment:
HHHASATCAAAGVRHHH
--AKGYYCAAAGRTCA-
| Original motif | Reverse complement motif |
| Consensus sequence: HHHASATCAAAGVRHHH | Consensus sequence: HHHKVCTTTGATSTHHH |
 |
 |
| Dataset #: 3 | Motif ID: 108 | Motif name: NR4A2 |
| Original motif | Reverse complement motif |
| Consensus sequence: AAGGTCAC | Consensus sequence: GTGACCTT |
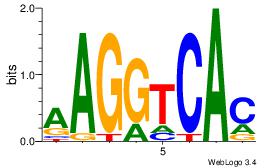 |
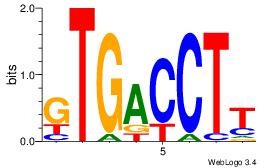 |
Best Matches for Motif ID 108 (Highest to Lowest)
| Motif ID: | UP00048 |
| Motif name: | Rara |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HHVGTGACCTTTDVDD
---GTGACCTT-----
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCACVHD | Consensus sequence: HHVGTGACCTTTDVDD |
 |
 |
| Motif ID: | UP00009 |
| Motif name: | Nr2f2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0074527 |
Alignment:
DHBHAAAGGTCAHVHB
-----AAGGTCAC---
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCAHVHB | Consensus sequence: BHVDTGACCTTTDVDD |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.01542 |
Alignment:
DHDBCAAGGTCAHVBDH
-----AAGGTCAC----
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBCAAGGTCAHVBDH | Consensus sequence: HHBVHTGACCTTGVDHD |
 |
 |
| Motif ID: | UP00009 |
| Motif name: | Nr2f2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0395644 |
Alignment:
VBVBCSGGGTCAVBBH
-----AAGGTCAC---
| Original motif | Reverse complement motif |
| Consensus sequence: VBVBCSGGGTCAVBBH | Consensus sequence: HVBVTGACCCSGBVBV |
 |
 |
| Motif ID: | UP00082 |
| Motif name: | Zfp187_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.0407874 |
Alignment:
DDGMCCTBGTCCHYHV
GTGACCTT--------
| Original motif | Reverse complement motif |
| Consensus sequence: DDGMCCTBGTCCHYHV | Consensus sequence: VHMDGGACVAGGRCDH |
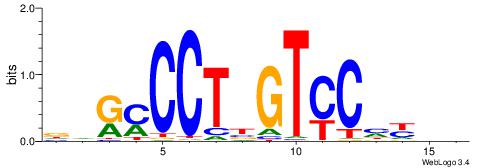 |
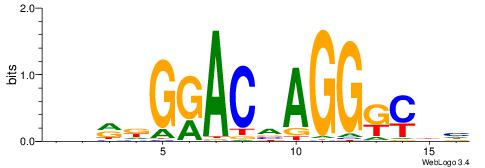 |
| Dataset #: 3 | Motif ID: 109 | Motif name: Pax5 |
| Original motif | Reverse complement motif |
| Consensus sequence: DGVBCABTGDWGCGKRRCSR | Consensus sequence: MSGKKRCGCWDCABTGBBCD |
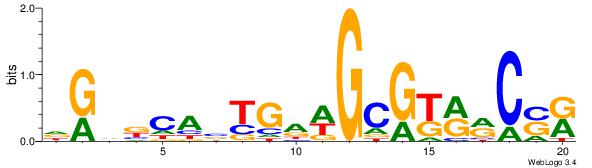 |
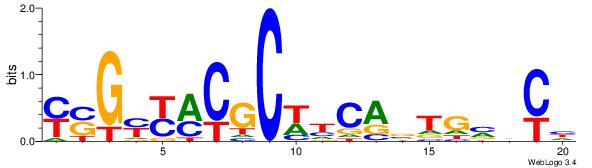 |
Best Matches for Motif ID 109 (Highest to Lowest)
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 20 |
| Similarity score: | 0.0499537 |
Alignment:
DDRMHBACGCGYGCGTDGBBDB
-DGVBCABTGDWGCGKRRCSR-
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
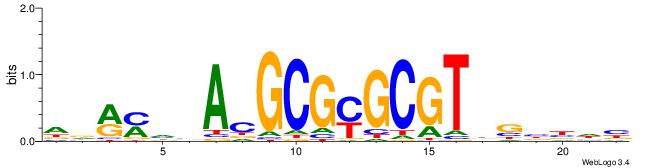 |
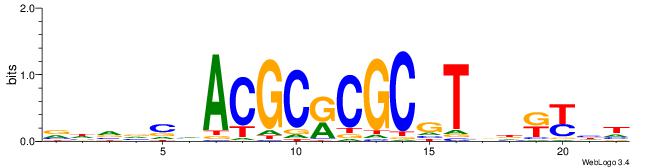 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 20 |
| Similarity score: | 0.0520196 |
Alignment:
WHVBVVMACGCGCGTCDYHWDH
DGVBCABTGDWGCGKRRCSR--
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 20 |
| Similarity score: | 0.0542764 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
-DGVBCABTGDWGCGKRRCSR-
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 20 |
| Similarity score: | 0.0560636 |
Alignment:
DCATDBTGHGCATKMTTBRSWR
--MSGKKRCGCWDCABTGBBCD
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
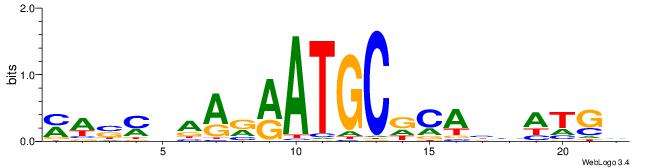 |
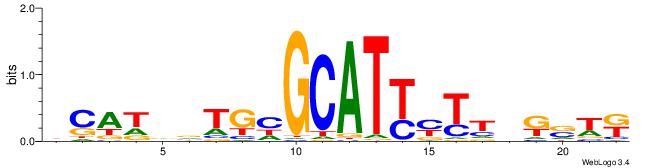 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 20 |
| Similarity score: | 0.0595671 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
DGVBCABTGDWGCGKRRCSR---
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
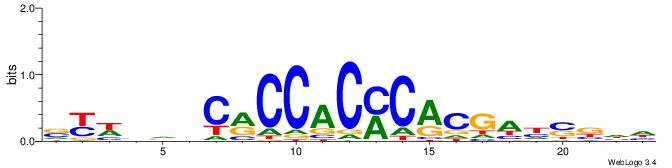 |
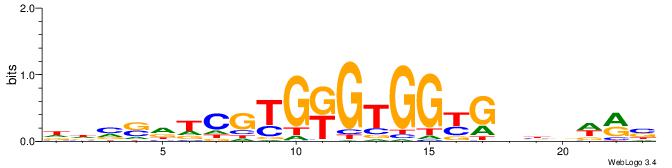 |
| Dataset #: 3 | Motif ID: 110 | Motif name: PLAG1 |
| Original motif | Reverse complement motif |
| Consensus sequence: GGGGCCCAAGGGGG | Consensus sequence: CCCCCTTGGGCCCC |
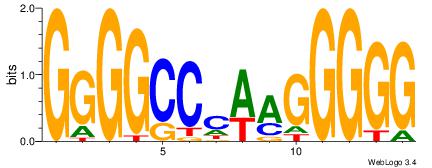 |
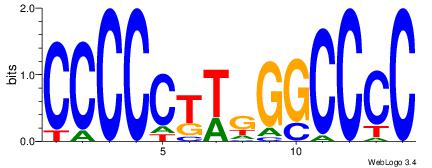 |
Best Matches for Motif ID 110 (Highest to Lowest)
| Motif ID: | UP00102 |
| Motif name: | Zic1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0499525 |
Alignment:
CCCCCCCGGGGGHB
CCCCCTTGGGCCCC
| Original motif | Reverse complement motif |
| Consensus sequence: BHCCCCCGGGGGGG | Consensus sequence: CCCCCCCGGGGGHB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0540613 |
Alignment:
BBBDVVRGACCACCCAVGABBAB
---------CCCCCTTGGGCCCC
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00057 |
| Motif name: | Zic2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0548411 |
Alignment:
BMCCCCCGGGGGGGB
-CCCCCTTGGGCCCC
| Original motif | Reverse complement motif |
| Consensus sequence: BMCCCCCGGGGGGGB | Consensus sequence: VCCCCCCCGGGGGRB |
 |
 |
| Motif ID: | UP00005 |
| Motif name: | Tcfap2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0556785 |
Alignment:
HHRCCBBWGGDCDB
GGGGCCCAAGGGGG
| Original motif | Reverse complement motif |
| Consensus sequence: HHRCCBBWGGDCDB | Consensus sequence: BDGHCCWBVGGKDH |
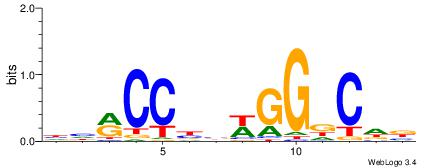 |
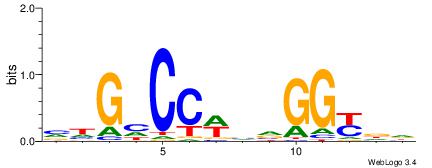 |
| Motif ID: | UP00006 |
| Motif name: | Zic3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.056976 |
Alignment:
BMCCCCCGGGGGGGB
-CCCCCTTGGGCCCC
| Original motif | Reverse complement motif |
| Consensus sequence: BMCCCCCGGGGGGGB | Consensus sequence: VCCCCCCCGGGGGRB |
 |
 |
| Dataset #: 3 | Motif ID: 111 | Motif name: PPARG |
| Original motif | Reverse complement motif |
| Consensus sequence: STAGGTCACBGTGACCYABT | Consensus sequence: ABTMGGTCACBGTGACCTAS |
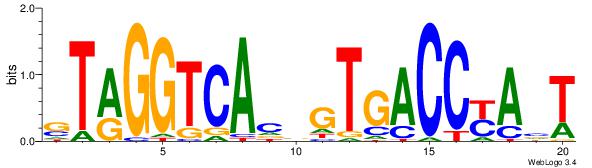 |
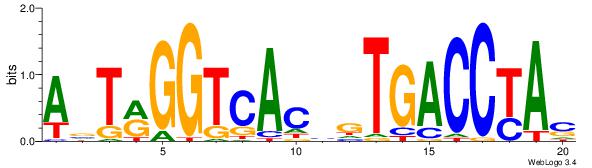 |
Best Matches for Motif ID 111 (Highest to Lowest)
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 20 |
| Similarity score: | 0.0610439 |
Alignment:
BDBBVBVGTAWCCAAGVBHBAGB
--ABTMGGTCACBGTGACCTAS-
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 20 |
| Similarity score: | 0.0641524 |
Alignment:
GTVBDAATTGTVTGHGDDHKVB
--ABTMGGTCACBGTGACCTAS
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 20 |
| Similarity score: | 0.0680441 |
Alignment:
VDWDARRGACGCGGWHGGAKHV
--ABTMGGTCACBGTGACCTAS
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 20 |
| Similarity score: | 0.0685319 |
Alignment:
HHHHDVVGTTGCTAHGGDHBAHH
--ABTMGGTCACBGTGACCTAS-
| Original motif | Reverse complement motif |
| Consensus sequence: HDTBHHCCHTAGCAACVVDHHHH | Consensus sequence: HHHHDVVGTTGCTAHGGDHBAHH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 20 |
| Similarity score: | 0.0686444 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
---STAGGTCACBGTGACCYABT
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Dataset #: 3 | Motif ID: 112 | Motif name: RELA |
| Original motif | Reverse complement motif |
| Consensus sequence: GGGRATTTCC | Consensus sequence: GGAAATKCCC |
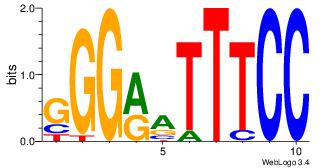 |
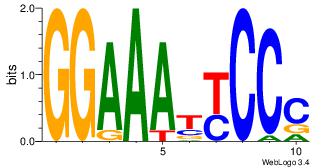 |
Best Matches for Motif ID 112 (Highest to Lowest)
| Motif ID: | UP00424 |
| Motif name: | Gm4881 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0152117 |
Alignment:
VBBHAYTTCCGGTDDD
GGGRATTTCC------
| Original motif | Reverse complement motif |
| Consensus sequence: VBBHAYTTCCGGTDDD | Consensus sequence: DHDACCGGAAMTHBVB |
 |
 |
| Motif ID: | UP00416 |
| Motif name: | Fli1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0156652 |
Alignment:
VBBVAYTTCCGGTDDH
GGGRATTTCC------
| Original motif | Reverse complement motif |
| Consensus sequence: VBBVAYTTCCGGTDDH | Consensus sequence: DHDACCGGAAKTBBVV |
 |
 |
| Motif ID: | UP00422 |
| Motif name: | Etv3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0163451 |
Alignment:
VBBHAYTTCCGGTHDH
GGGRATTTCC------
| Original motif | Reverse complement motif |
| Consensus sequence: VBBHAYTTCCGGTHDH | Consensus sequence: DHHACCGGAAKTHBBB |
 |
 |
| Motif ID: | UP00415 |
| Motif name: | Elk4 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.018032 |
Alignment:
VBBHRYTTCCGGTDDH
GGGRATTTCC------
| Original motif | Reverse complement motif |
| Consensus sequence: VBBHRYTTCCGGTDDH | Consensus sequence: DHDACCGGAAKKHBVB |
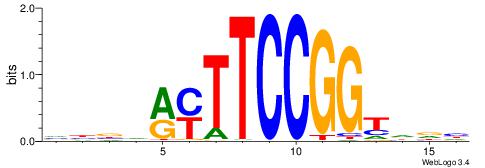 |
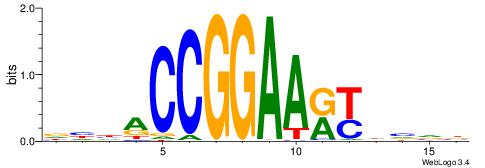 |
| Motif ID: | UP00411 |
| Motif name: | Erg |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0206291 |
Alignment:
VBBVAYTTCCGGTDDB
GGGRATTTCC------
| Original motif | Reverse complement motif |
| Consensus sequence: VBBVAYTTCCGGTDDB | Consensus sequence: BDDACCGGAAKTVBVB |
 |
 |
| Dataset #: 3 | Motif ID: 113 | Motif name: REST |
| Original motif | Reverse complement motif |
| Consensus sequence: TTCAGCACCATGGACAGCKCC | Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
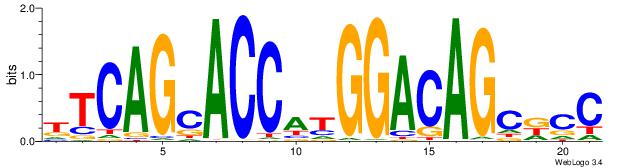 |
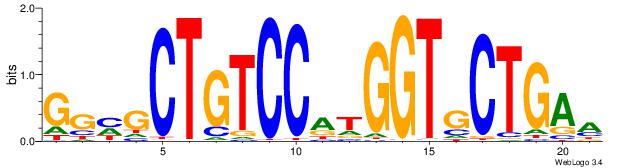 |
Best Matches for Motif ID 113 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 21 |
| Similarity score: | 0.0699178 |
Alignment:
BBBDVVRGACCACCCAVGABBAB
--TTCAGCACCATGGACAGCKCC
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 21 |
| Similarity score: | 0.0734685 |
Alignment:
VMYDHDGMCCHCCKBGVVAAVH
GGYGCTGTCCATGGTGCTGAA-
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 21 |
| Similarity score: | 0.0740652 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
-TTCAGCACCATGGACAGCKCC-
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 21 |
| Similarity score: | 0.0750152 |
Alignment:
MHHHWHYTMCCHCCCACVAABVH
--TTCAGCACCATGGACAGCKCC
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 21 |
| Similarity score: | 0.0751449 |
Alignment:
BHCBCBCCGGGTGGTCYHVHDCH
GGYGCTGTCCATGGTGCTGAA--
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Dataset #: 3 | Motif ID: 114 | Motif name: RUNX1 |
| Original motif | Reverse complement motif |
| Consensus sequence: BBYTGTGGTTT | Consensus sequence: AAACCACAKVB |
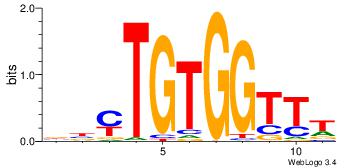 |
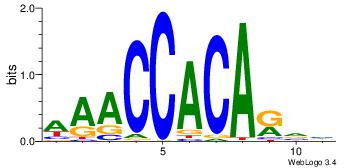 |
Best Matches for Motif ID 114 (Highest to Lowest)
| Motif ID: | UP00070 |
| Motif name: | Gcm1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0126586 |
Alignment:
HVVHACCCGCATVDHD
---AAACCACAKVB--
| Original motif | Reverse complement motif |
| Consensus sequence: HVVHACCCGCATVDHD | Consensus sequence: DHDVATGCGGGTHVVH |
 |
 |
| Motif ID: | UP00073 |
| Motif name: | Foxa2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0139444 |
Alignment:
BVVTTTGTTATTDBB
--BBYTGTGGTTT--
| Original motif | Reverse complement motif |
| Consensus sequence: VVDAATAACAAAVVB | Consensus sequence: BVVTTTGTTATTDBB |
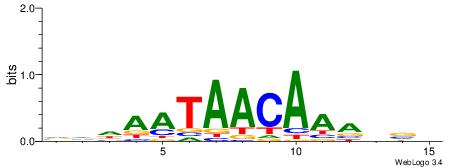 |
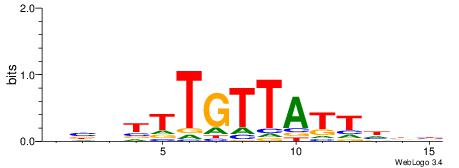 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.0141619 |
Alignment:
DVTTVVCVYGGHGGYCHHDMYB
------BBYTGTGGTTT-----
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00047 |
| Motif name: | Zbtb7b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.015194 |
Alignment:
BBDVHGGTGGTCBKDDB
--BBYTGTGGTTT----
| Original motif | Reverse complement motif |
| Consensus sequence: BDDRVGACCACCHBDVB | Consensus sequence: BBDVHGGTGGTCBKDDB |
 |
 |
| Motif ID: | UP00025 |
| Motif name: | Foxk1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0158291 |
Alignment:
HDDTGTTKTTKTHHD
----AAACCACAKVB
| Original motif | Reverse complement motif |
| Consensus sequence: HHHAYAAYAACAHHH | Consensus sequence: HDDTGTTKTTKTHHD |
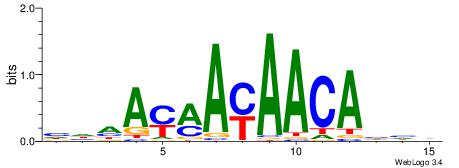 |
 |
| Dataset #: 3 | Motif ID: 115 | Motif name: RXRRAR_DR5 |
| Original motif | Reverse complement motif |
| Consensus sequence: RGKTCABVVRGAGGTCA | Consensus sequence: TGACCTCKVVBTGAYCK |
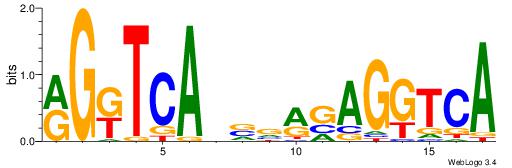 |
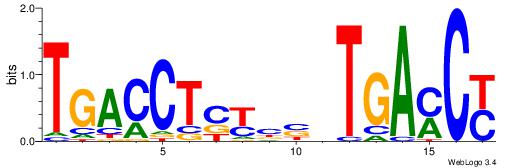 |
Best Matches for Motif ID 115 (Highest to Lowest)
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0585087 |
Alignment:
DVTTVVCVYGGHGGYCHHDMYB
RGKTCABVVRGAGGTCA-----
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0709955 |
Alignment:
GTVBDAATTGTVTGHGDDHKVB
-----RGKTCABVVRGAGGTCA
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.075055 |
Alignment:
DVVTTVGTGGGHGGYAMHWHHHY
RGKTCABVVRGAGGTCA------
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 17 |
| Similarity score: | 0.0758824 |
Alignment:
BBBDVVRGACCACCCAVGABBAB
------RGKTCABVVRGAGGTCA
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 17 |
| Similarity score: | 0.0759953 |
Alignment:
HBHDVBWGATHBTATCRVBHHH
TGACCTCKVVBTGAYCK-----
| Original motif | Reverse complement motif |
| Consensus sequence: HBHDVBWGATHBTATCRVBHHH | Consensus sequence: HHHBBMGATAVHATCWVVDHVH |
 |
 |
| Dataset #: 3 | Motif ID: 116 | Motif name: SP1 |
| Original motif | Reverse complement motif |
| Consensus sequence: CCCCKCCCCC | Consensus sequence: GGGGGYGGGG |
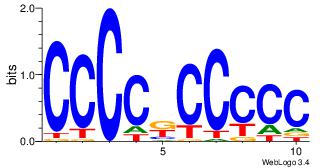 |
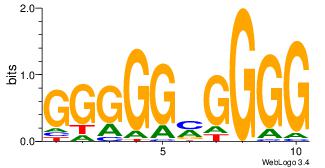 |
Best Matches for Motif ID 116 (Highest to Lowest)
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0231018 |
Alignment:
DBCCCCCCCCCCMYC
--CCCCKCCCCC---
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0303207 |
Alignment:
HBDRCCMCGCCCHHHD
----CCCCKCCCCC--
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0323007 |
Alignment:
HHHHDGGGGCGGDBHDV
----GGGGGYGGGG---
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
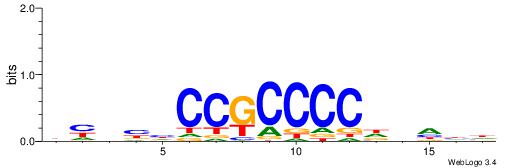 |
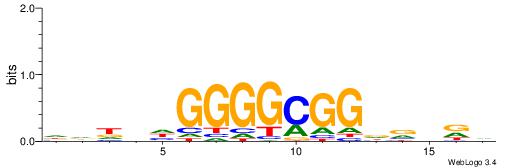 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0340973 |
Alignment:
HVDVGGGHGGGGDBHB
--GGGGGYGGGG----
| Original motif | Reverse complement motif |
| Consensus sequence: BHVDCCCCDCCCBDBH | Consensus sequence: HVDVGGGHGGGGDBHB |
 |
 |
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0351051 |
Alignment:
BHHHWGGGGCGGBBVH
----GGGGGYGGGG--
| Original motif | Reverse complement motif |
| Consensus sequence: HBBBCCGCCCCWHHHV | Consensus sequence: BHHHWGGGGCGGBBVH |
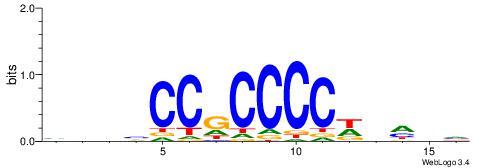 |
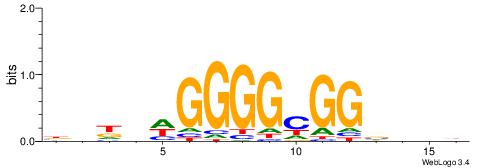 |
| Dataset #: 3 | Motif ID: 117 | Motif name: Spz1 |
| Original motif | Reverse complement motif |
| Consensus sequence: AGGGTAWCAGC | Consensus sequence: GCTGWTACCCT |
 |
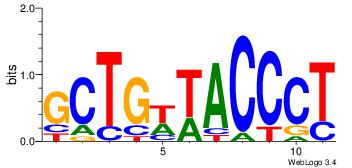 |
Best Matches for Motif ID 117 (Highest to Lowest)
| Motif ID: | UP00388 |
| Motif name: | Six6_2267.4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.00457598 |
Alignment:
AHWDHTGATACCCKATD
---GCTGWTACCCT---
| Original motif | Reverse complement motif |
| Consensus sequence: DATRGGGTATCAHDWHT | Consensus sequence: AHWDHTGATACCCKATD |
 |
 |
| Motif ID: | UP00195 |
| Motif name: | Six3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.00651969 |
Alignment:
DATRGGGTATCAHHWHT
---AGGGTAWCAGC---
| Original motif | Reverse complement motif |
| Consensus sequence: DATRGGGTATCAHHWHT | Consensus sequence: AHWHDTGATACCCKATH |
 |
 |
| Motif ID: | UP00388 |
| Motif name: | Six6_2267.5 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0083787 |
Alignment:
AHWHWTGATACCCKDDD
---GCTGWTACCCT---
| Original motif | Reverse complement motif |
| Consensus sequence: DDDRGGGTATCAWHWHT | Consensus sequence: AHWHWTGATACCCKDDD |
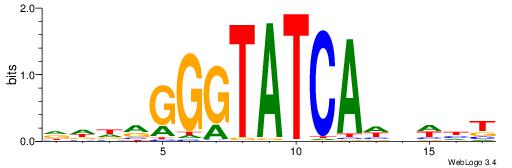 |
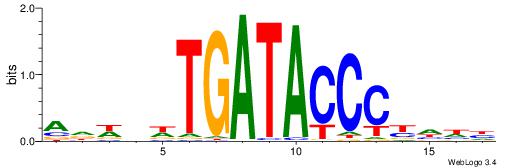 |
| Motif ID: | UP00192 |
| Motif name: | Six1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0113267 |
Alignment:
DHDRGGGTATCAHDHBT
---AGGGTAWCAGC---
| Original motif | Reverse complement motif |
| Consensus sequence: DHDRGGGTATCAHDHBT | Consensus sequence: AVHDHTGATACCCMDHH |
 |
 |
| Motif ID: | UP00159 |
| Motif name: | Six2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0117598 |
Alignment:
DAKRGGGTATCACDWHT
---AGGGTAWCAGC---
| Original motif | Reverse complement motif |
| Consensus sequence: DAKRGGGTATCACDWHT | Consensus sequence: AHWDGTGATACCCMRTD |
 |
 |
| Dataset #: 3 | Motif ID: 118 | Motif name: SRF |
| Original motif | Reverse complement motif |
| Consensus sequence: GCCCATATATGG | Consensus sequence: CCATATATGGGC |
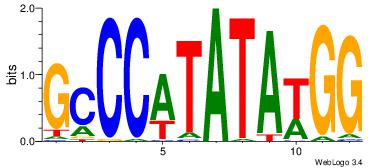 |
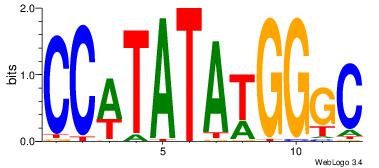 |
Best Matches for Motif ID 118 (Highest to Lowest)
| Motif ID: | UP00077 |
| Motif name: | Srf_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0 |
Alignment:
THCCWTATAWGGHH
--CCATATATGGGC
| Original motif | Reverse complement motif |
| Consensus sequence: HHCCWTATAWGGHA | Consensus sequence: THCCWTATAWGGHH |
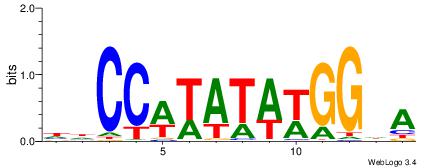 |
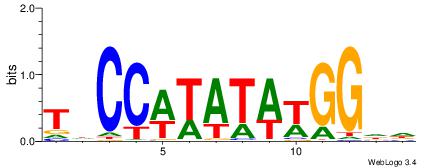 |
| Motif ID: | UP00070 |
| Motif name: | Gcm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0318541 |
Alignment:
BYMBKCCCCTATDVBHD
----GCCCATATATGG-
| Original motif | Reverse complement motif |
| Consensus sequence: DDBVHATAGGGGRBRMB | Consensus sequence: BYMBKCCCCTATDVBHD |
 |
 |
| Motif ID: | UP00080 |
| Motif name: | Gata5_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0503431 |
Alignment:
HDDDCTGATAAGRVDDD
---CCATATATGGGC--
| Original motif | Reverse complement motif |
| Consensus sequence: HDDDCTGATAAGRVDDD | Consensus sequence: DDHBKCTTATCAGDDDH |
 |
 |
| Motif ID: | UP00133 |
| Motif name: | Cdx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0528496 |
Alignment:
DVVGGYMATAAAAHKH
---GCCCATATATGG-
| Original motif | Reverse complement motif |
| Consensus sequence: DVVGGYMATAAAAHKH | Consensus sequence: HRHTTTTATYMCCVBD |
 |
 |
| Motif ID: | UP00213 |
| Motif name: | Hoxa9 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0533104 |
Alignment:
WDTAWTTTWATGKCCGD
---CCATATATGGGC--
| Original motif | Reverse complement motif |
| Consensus sequence: DCGGYCATWAAAWTADW | Consensus sequence: WDTAWTTTWATGKCCGD |
 |
 |
| Dataset #: 3 | Motif ID: 119 | Motif name: Stat3 |
| Original motif | Reverse complement motif |
| Consensus sequence: TTCCAGGAAG | Consensus sequence: CTTCCTGGAA |
 |
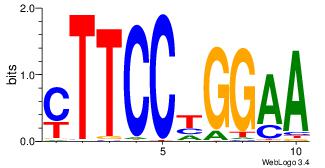 |
Best Matches for Motif ID 119 (Highest to Lowest)
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
VHHATTCCTYGAAAGD
---CTTCCTGGAA---
| Original motif | Reverse complement motif |
| Consensus sequence: DCTTTCKAGGAATHHV | Consensus sequence: VHHATTCCTYGAAAGD |
 |
 |
| Motif ID: | UP00413 |
| Motif name: | Elf4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.00702689 |
Alignment:
DBBHACTTCCGGKWTH
-----CTTCCTGGAA-
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGKWTH | Consensus sequence: HAWYCCGGAAGTHBBD |
 |
 |
| Motif ID: | UP00410 |
| Motif name: | Elk1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.00785398 |
Alignment:
DHBDACTTCCGGTVHVB
-----CTTCCTGGAA--
| Original motif | Reverse complement motif |
| Consensus sequence: VVHBACCGGAAGTDBHD | Consensus sequence: DHBDACTTCCGGTVHVB |
 |
 |
| Motif ID: | UP00013 |
| Motif name: | Gabpa_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.00988664 |
Alignment:
BHHHACTTCCGGTHHBD
-----CTTCCTGGAA--
| Original motif | Reverse complement motif |
| Consensus sequence: HVHHACCGGAAGTDHHV | Consensus sequence: BHHHACTTCCGGTHHBD |
 |
 |
| Motif ID: | UP00404 |
| Motif name: | Elf2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0105941 |
Alignment:
DBBHACTTCCGGDWDB
-----CTTCCTGGAA-
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGDWDB | Consensus sequence: BDWDCCGGAAGTHBBD |
 |
 |
| Dataset #: 3 | Motif ID: 120 | Motif name: T |
| Original motif | Reverse complement motif |
| Consensus sequence: CTAGGTGTGAA | Consensus sequence: TTCACACCTAG |
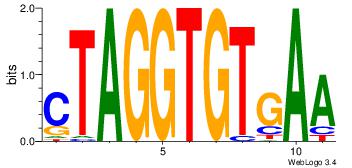 |
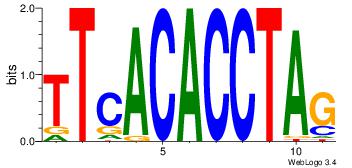 |
Best Matches for Motif ID 120 (Highest to Lowest)
| Motif ID: | UP00068 |
| Motif name: | Eomes_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0 |
Alignment:
BVHDAGGTGTGAAAHHH
--CTAGGTGTGAA----
| Original motif | Reverse complement motif |
| Consensus sequence: BVHDAGGTGTGAAAHHH | Consensus sequence: HHHTTTCACACCTDHBV |
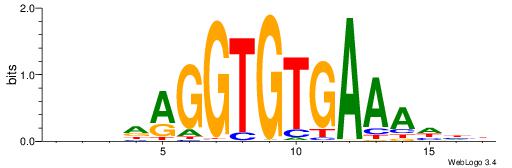 |
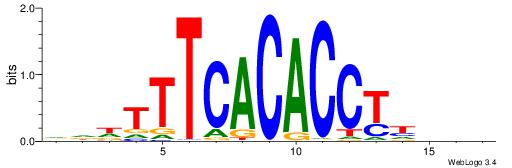 |
| Motif ID: | UP00258 |
| Motif name: | Tgif2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0253823 |
Alignment:
VBWTTGACAGCTVBTT
---TTCACACCTAG--
| Original motif | Reverse complement motif |
| Consensus sequence: AABBAGCTGTCAAWVV | Consensus sequence: VBWTTGACAGCTVBTT |
 |
 |
| Motif ID: | UP00199 |
| Motif name: | Six4 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0278802 |
Alignment:
BBVHVGGTGTCATHDDT
--CTAGGTGTGAA----
| Original motif | Reverse complement motif |
| Consensus sequence: ADDHATGACACCBHBBV | Consensus sequence: BBVHVGGTGTCATHDDT |
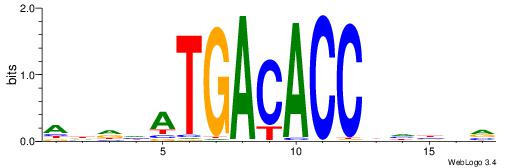 |
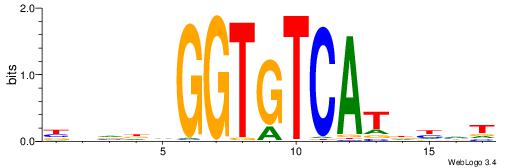 |
| Motif ID: | UP00186 |
| Motif name: | Meis1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0349057 |
Alignment:
WAVBASCTGTCAAWVH
--CTAGGTGTGAA---
| Original motif | Reverse complement motif |
| Consensus sequence: WAVBASCTGTCAAWVH | Consensus sequence: DBWTTGACAGSTBVTW |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0381312 |
Alignment:
VBHHVCAGGTGCDVDHD
----CTAGGTGTGAA--
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Dataset #: 3 | Motif ID: 121 | Motif name: TAL1TCF3 |
| Original motif | Reverse complement motif |
| Consensus sequence: HVAMCATCTGKT | Consensus sequence: ARCAGATGRTVD |
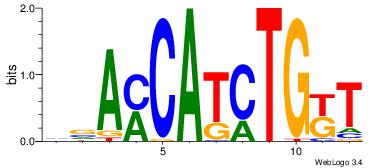 |
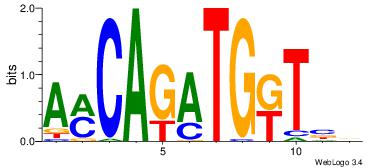 |
Best Matches for Motif ID 121 (Highest to Lowest)
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0117104 |
Alignment:
VBBDMYCATCTGVHHBH
--HVAMCATCTGKT---
| Original motif | Reverse complement motif |
| Consensus sequence: HVDDVCAGATGKYHBBV | Consensus sequence: VBBDMYCATCTGVHHBH |
 |
 |
| Motif ID: | UP00036 |
| Motif name: | Myf6_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0121214 |
Alignment:
VVDBAACAGBTGBYHB
HVAMCATCTGKT----
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBAACAGBTGBYHB | Consensus sequence: BDKVCABCTGTTBDBV |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.024819 |
Alignment:
HHVBABCACGTGSTHD
----ARCAGATGRTVD
| Original motif | Reverse complement motif |
| Consensus sequence: DDASCACGTGBTBVDD | Consensus sequence: HHVBABCACGTGSTHD |
 |
 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.025362 |
Alignment:
HHDVVGCAGCTGVBKVB
----ARCAGATGRTVD-
| Original motif | Reverse complement motif |
| Consensus sequence: BBYVVCAGCTGCBVHHD | Consensus sequence: HHDVVGCAGCTGVBKVB |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0322545 |
Alignment:
VBHHVCAGGTGCDVDHD
---ARCAGATGRTVD--
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Dataset #: 3 | Motif ID: 122 | Motif name: Tcfcp2l1 |
| Original motif | Reverse complement motif |
| Consensus sequence: CCAGYYHVADCCRG | Consensus sequence: CKGGDTBDMMCTGG |
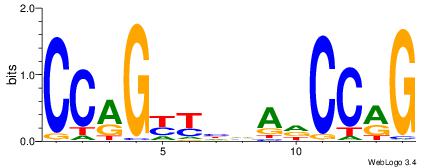 |
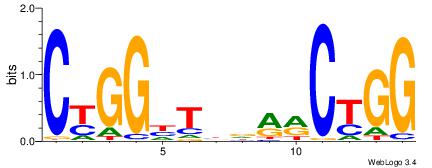 |
Best Matches for Motif ID 122 (Highest to Lowest)
| Motif ID: | UP00032 |
| Motif name: | Gata3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 14 |
| Similarity score: | 0.0527249 |
Alignment:
HBHDVBWGATHBTATCRVBHHH
----CCAGYYHVADCCRG----
| Original motif | Reverse complement motif |
| Consensus sequence: HBHDVBWGATHBTATCRVBHHH | Consensus sequence: HHHBBMGATAVHATCWVVDHVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0551691 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
--------CCAGYYHVADCCRG
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00005 |
| Motif name: | Tcfap2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0560983 |
Alignment:
HHRCCBBWGGDCDB
CCAGYYHVADCCRG
| Original motif | Reverse complement motif |
| Consensus sequence: HHRCCBBWGGDCDB | Consensus sequence: BDGHCCWBVGGKDH |
 |
 |
| Motif ID: | UP00013 |
| Motif name: | Gabpa_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0566467 |
Alignment:
CYDBYWTCCSMHBVVH
CCAGYYHVADCCRG--
| Original motif | Reverse complement motif |
| Consensus sequence: CYDBYWTCCSMHBVVH | Consensus sequence: DBVVDRSGGAWKVHKG |
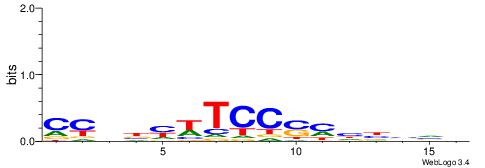 |
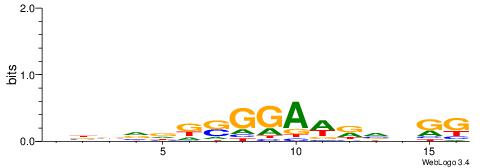 |
| Motif ID: | UP00075 |
| Motif name: | Sox15_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0577636 |
Alignment:
BBVAATGDMATTHVA
-CCAGYYHVADCCRG
| Original motif | Reverse complement motif |
| Consensus sequence: BBVAATGDMATTHVA | Consensus sequence: TVDAATYDCATTVVV |
 |
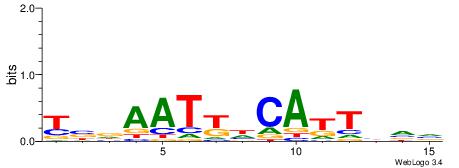 |
| Dataset #: 3 | Motif ID: 123 | Motif name: TFAP2A |
| Original motif | Reverse complement motif |
| Consensus sequence: GCCBBVRGS | Consensus sequence: SCMVBBGGC |
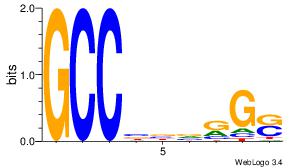 |
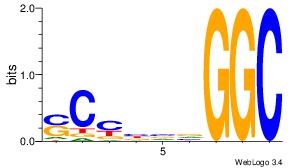 |
Best Matches for Motif ID 123 (Highest to Lowest)
| Motif ID: | UP00010 |
| Motif name: | Tcfap2b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 9 |
| Similarity score: | 0.0115546 |
Alignment:
HHKGCCYSRGGCWAD
---GCCBBVRGS---
| Original motif | Reverse complement motif |
| Consensus sequence: DTWGCCKSMGGCRHH | Consensus sequence: HHKGCCYSRGGCWAD |
 |
 |
| Motif ID: | UP00010 |
| Motif name: | Tcfap2b_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 9 |
| Similarity score: | 0.0150447 |
Alignment:
DYGCCYBARGGCAH
--GCCBBVRGS---
| Original motif | Reverse complement motif |
| Consensus sequence: DYGCCYBARGGCAH | Consensus sequence: HTGCCMTVKGGCMD |
 |
 |
| Motif ID: | UP00005 |
| Motif name: | Tcfap2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 9 |
| Similarity score: | 0.0151527 |
Alignment:
WDHSCCYSRGGSRAD
---GCCBBVRGS---
| Original motif | Reverse complement motif |
| Consensus sequence: WDHSCCYSRGGSRAD | Consensus sequence: DTMSCCKSMGGSHDW |
 |
 |
| Motif ID: | UP00087 |
| Motif name: | Tcfap2c_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 9 |
| Similarity score: | 0.016909 |
Alignment:
WHHSCCYSRGGSDAD
---GCCBBVRGS---
| Original motif | Reverse complement motif |
| Consensus sequence: WHHSCCYSRGGSDAD | Consensus sequence: DTHSCCKSMGGSHHW |
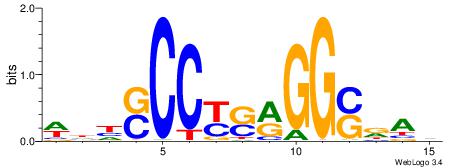 |
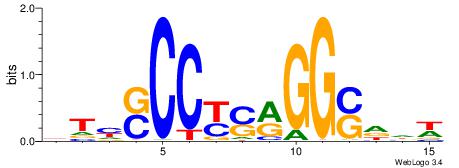 |
| Motif ID: | UP00087 |
| Motif name: | Tcfap2c_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 9 |
| Similarity score: | 0.0170259 |
Alignment:
BHGCCCDAGGGCWD
--GCCBBVRGS---
| Original motif | Reverse complement motif |
| Consensus sequence: BHGCCCDAGGGCWD | Consensus sequence: HWGCCCTDGGGCDB |
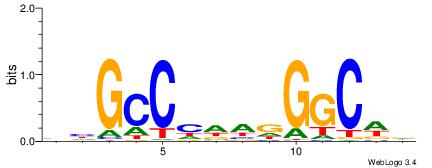 |
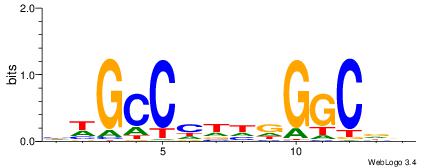 |
| Dataset #: 3 | Motif ID: 124 | Motif name: TLX1NFIC |
| Original motif | Reverse complement motif |
| Consensus sequence: TGGCASBDHGCCAA | Consensus sequence: TTGGCHDBSTGCCA |
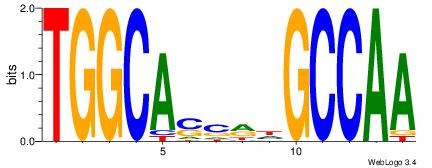 |
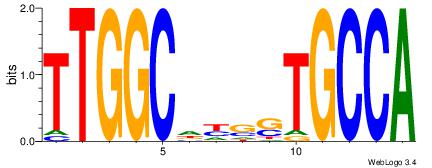 |
Best Matches for Motif ID 124 (Highest to Lowest)
| Motif ID: | UP00010 |
| Motif name: | Tcfap2b_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0679595 |
Alignment:
HTGCCMTVKGGCMD
TTGGCHDBSTGCCA
| Original motif | Reverse complement motif |
| Consensus sequence: DYGCCYBARGGCAH | Consensus sequence: HTGCCMTVKGGCMD |
 |
 |
| Motif ID: | UP00010 |
| Motif name: | Tcfap2b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0689546 |
Alignment:
HHKGCCYSRGGCWAD
-TGGCASBDHGCCAA
| Original motif | Reverse complement motif |
| Consensus sequence: DTWGCCKSMGGCRHH | Consensus sequence: HHKGCCYSRGGCWAD |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0749445 |
Alignment:
HDTBHHCCHTAGCAACVVDHHHH
---------TGGCASBDHGCCAA
| Original motif | Reverse complement motif |
| Consensus sequence: HDTBHHCCHTAGCAACVVDHHHH | Consensus sequence: HHHHDVVGTTGCTAHGGDHBAHH |
 |
 |
| Motif ID: | UP00087 |
| Motif name: | Tcfap2c_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0753915 |
Alignment:
HWGCCCTDGGGCDB
TTGGCHDBSTGCCA
| Original motif | Reverse complement motif |
| Consensus sequence: BHGCCCDAGGGCWD | Consensus sequence: HWGCCCTDGGGCDB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0779719 |
Alignment:
DVTTVVCVYGGHGGYCHHDMYB
--TTGGCHDBSTGCCA------
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Dataset #: 3 | Motif ID: 125 | Motif name: TP53 |
| Original motif | Reverse complement motif |
| Consensus sequence: MSGGACATGYCCGGGCATGT | Consensus sequence: ACATGCCCGGKCATGTCCSR |
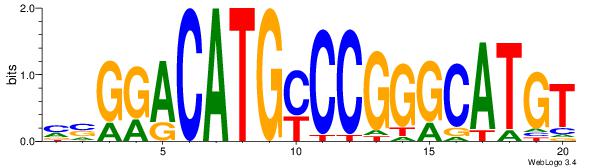 |
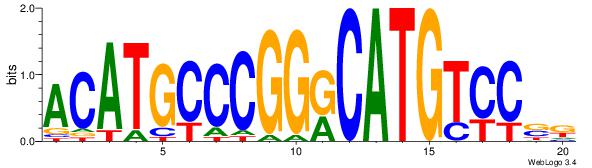 |
Best Matches for Motif ID 125 (Highest to Lowest)
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 20 |
| Similarity score: | 0.0503863 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
---ACATGCCCGGKCATGTCCSR
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 20 |
| Similarity score: | 0.0537648 |
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
MSGGACATGYCCGGGCATGT---
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 20 |
| Similarity score: | 0.0545224 |
Alignment:
DDRMHBACGCGYGCGTDGBBDB
MSGGACATGYCCGGGCATGT--
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 19 |
| Similarity score: | 0.554849 |
Alignment:
HGDHVHKGACCACCCGGBGBGDB-
-ACATGCCCGGKCATGTCCSR---
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 19 |
| Similarity score: | 0.5552 |
Alignment:
-BTBVTCVTGGGTGGTCMVVDVBB
---ACATGCCCGGKCATGTCCSR-
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Dataset #: 3 | Motif ID: 126 | Motif name: USF1 |
| Original motif | Reverse complement motif |
| Consensus sequence: CACGTGR | Consensus sequence: MCACGTG |
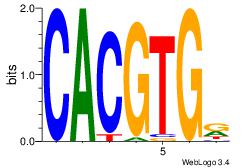 |
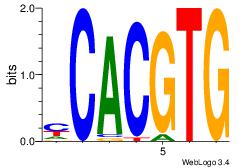 |
Best Matches for Motif ID 126 (Highest to Lowest)
| Motif ID: | UP00060 |
| Motif name: | Max_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0 |
Alignment:
DDASCACGTGBTBVDD
----CACGTGR-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDASCACGTGBTBVDD | Consensus sequence: HHVBABCACGTGSTHD |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 7 |
| Similarity score: | 0.00626993 |
Alignment:
YDYBDHTMCACGTGGADDBMDGT
--------CACGTGR--------
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 7 |
| Similarity score: | 0.00965663 |
Alignment:
DDHBHBSTCACGTGAHBBDHHM
--------CACGTGR-------
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0.0311072 |
Alignment:
VBHHVCAGGTGCDVDHD
----MCACGTG------
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0.0449596 |
Alignment:
BBGHCACGCGACDD
---MCACGTG----
| Original motif | Reverse complement motif |
| Consensus sequence: BBGHCACGCGACDD | Consensus sequence: HDGTCGCGTGDCVB |
 |
 |
| Dataset #: 3 | Motif ID: 127 | Motif name: YY1 |
| Original motif | Reverse complement motif |
| Consensus sequence: RCCATB | Consensus sequence: BATGGM |
 |
 |
Best Matches for Motif ID 127 (Highest to Lowest)
| Motif ID: | UP00033 |
| Motif name: | Zfp410_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
DVDDATGGGATGKMDDV
--------BATGGM---
| Original motif | Reverse complement motif |
| Consensus sequence: DVDDATGGGATGKMDDV | Consensus sequence: BDDYYCATCCCATDDBD |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.00378844 |
Alignment:
HVDDVCAGATGKYHBBV
-------BATGGM----
| Original motif | Reverse complement motif |
| Consensus sequence: HVDDVCAGATGKYHBBV | Consensus sequence: VBBDMYCATCTGVHHBH |
 |
 |
| Motif ID: | UP00207 |
| Motif name: | Hoxb9 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.0208704 |
Alignment:
VRRGCMATAAAAHHHD
---RCCATB-------
| Original motif | Reverse complement motif |
| Consensus sequence: VRRGCMATAAAAHHHD | Consensus sequence: HDHHTTTTATRGCKMV |
 |
 |
| Motif ID: | UP00077 |
| Motif name: | Srf_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.023319 |
Alignment:
HHCCWTATAWGGHA
-------BATGGM-
| Original motif | Reverse complement motif |
| Consensus sequence: HHCCWTATAWGGHA | Consensus sequence: THCCWTATAWGGHH |
 |
 |
| Motif ID: | UP00213 |
| Motif name: | Hoxa9 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.0242984 |
Alignment:
DCGGYCATWAAAWTADW
---RCCATB--------
| Original motif | Reverse complement motif |
| Consensus sequence: DCGGYCATWAAAWTADW | Consensus sequence: WDTAWTTTWATGKCCGD |
 |
 |
| Dataset #: 3 | Motif ID: 128 | Motif name: ZEB1 |
| Original motif | Reverse complement motif |
| Consensus sequence: CACCTD | Consensus sequence: HAGGTG |
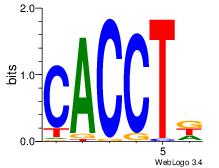 |
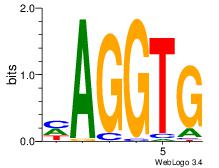 |
Best Matches for Motif ID 128 (Highest to Lowest)
| Motif ID: | UP00068 |
| Motif name: | Eomes_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
VVBVCKACACCTHVBV
-------CACCTD---
| Original motif | Reverse complement motif |
| Consensus sequence: VBVDAGGTGTYGVBBV | Consensus sequence: VVBVCKACACCTHVBV |
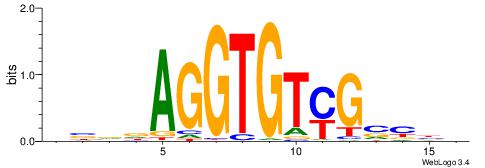 |
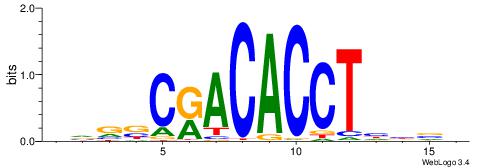 |
| Motif ID: | UP00199 |
| Motif name: | Six4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.00433068 |
Alignment:
BBVHVGGTGTCATHDDT
---HAGGTG--------
| Original motif | Reverse complement motif |
| Consensus sequence: ADDHATGACACCBHBBV | Consensus sequence: BBVHVGGTGTCATHDDT |
 |
 |
| Motif ID: | UP00068 |
| Motif name: | Eomes_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.00524081 |
Alignment:
HHHTTTCACACCTDHBV
--------CACCTD---
| Original motif | Reverse complement motif |
| Consensus sequence: BVHDAGGTGTGAAAHHH | Consensus sequence: HHHTTTCACACCTDHBV |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.00911301 |
Alignment:
VBHHVCAGGTGCDVDHD
-----HAGGTG------
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Motif ID: | UP00047 |
| Motif name: | Zbtb7b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.0233556 |
Alignment:
BDDRVGACCACCHBDVB
--------CACCTD---
| Original motif | Reverse complement motif |
| Consensus sequence: BDDRVGACCACCHBDVB | Consensus sequence: BBDVHGGTGGTCBKDDB |
 |
 |
| Dataset #: 3 | Motif ID: 129 | Motif name: Zfp423 |
| Original motif | Reverse complement motif |
| Consensus sequence: GSMMCCYARGGKKKC | Consensus sequence: GYRYCCMTKGGYRSC |
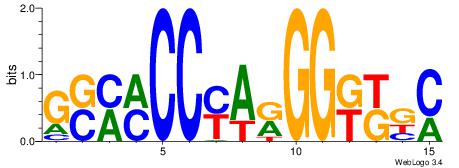 |
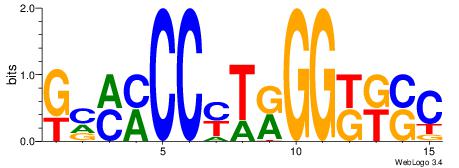 |
Best Matches for Motif ID 129 (Highest to Lowest)
| Motif ID: | UP00057 |
| Motif name: | Zic2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0178077 |
Alignment:
BMCCCCCGGGGGGGB
GSMMCCYARGGKKKC
| Original motif | Reverse complement motif |
| Consensus sequence: BMCCCCCGGGGGGGB | Consensus sequence: VCCCCCCCGGGGGRB |
 |
 |
| Motif ID: | UP00006 |
| Motif name: | Zic3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0213948 |
Alignment:
BMCCCCCGGGGGGGB
GSMMCCYARGGKKKC
| Original motif | Reverse complement motif |
| Consensus sequence: BMCCCCCGGGGGGGB | Consensus sequence: VCCCCCCCGGGGGRB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0255241 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
-------GSMMCCYARGGKKKC-
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00087 |
| Motif name: | Tcfap2c_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0261399 |
Alignment:
WHHSCCYSRGGSDAD
GSMMCCYARGGKKKC
| Original motif | Reverse complement motif |
| Consensus sequence: WHHSCCYSRGGSDAD | Consensus sequence: DTHSCCKSMGGSHHW |
 |
 |
| Motif ID: | UP00005 |
| Motif name: | Tcfap2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0266201 |
Alignment:
WDHSCCYSRGGSRAD
GSMMCCYARGGKKKC
| Original motif | Reverse complement motif |
| Consensus sequence: WDHSCCYSRGGSRAD | Consensus sequence: DTMSCCKSMGGSHDW |
 |
 |
| Dataset #: 3 | Motif ID: 130 | Motif name: Zfx |
| Original motif | Reverse complement motif |
| Consensus sequence: BBVGCCBVGGCCTV | Consensus sequence: VAGGCCBBGGCVBB |
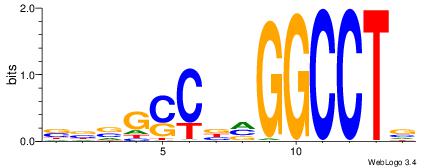 |
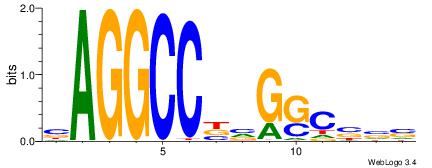 |
Best Matches for Motif ID 130 (Highest to Lowest)
| Motif ID: | UP00005 |
| Motif name: | Tcfap2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0596136 |
Alignment:
HHRCCBBWGGDCDB
BBVGCCBVGGCCTV
| Original motif | Reverse complement motif |
| Consensus sequence: HHRCCBBWGGDCDB | Consensus sequence: BDGHCCWBVGGKDH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0603516 |
Alignment:
ABBBBVVRGACCACCCACRDBBM
BBVGCCBVGGCCTV---------
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0604694 |
Alignment:
BDHHMCGCCCCCTHVBB
---VAGGCCBBGGCVBB
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0606067 |
Alignment:
HBDRCCMCGCCCHHHD
BBVGCCBVGGCCTV--
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 14 |
| Similarity score: | 0.0606878 |
Alignment:
HVGVSAGGAGGGTCYHHHHYVH
--------VAGGCCBBGGCVBB
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Dataset #: 3 | Motif ID: 131 | Motif name: znf143 |
| Original motif | Reverse complement motif |
| Consensus sequence: BAHYTCCCAKMATGCMWYGC | Consensus sequence: GCMWRGCATYRTGGGAMHTB |
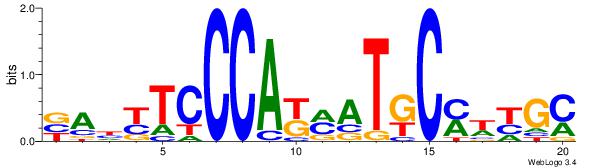 |
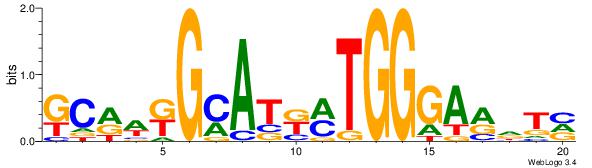 |
Best Matches for Motif ID 131 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 20 |
| Similarity score: | 0.0673743 |
Alignment:
MHHHWHYTMCCHCCCACVAABVH
---BAHYTCCCAKMATGCMWYGC
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 20 |
| Similarity score: | 0.0687663 |
Alignment:
DVTTVVCVYGGHGGYCHHDMYB
GCMWRGCATYRTGGGAMHTB--
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 20 |
| Similarity score: | 0.0697414 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
-GCMWRGCATYRTGGGAMHTB--
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 20 |
| Similarity score: | 0.0698272 |
Alignment:
DBCDWHCTGGGTGGTCMDHBBAH
GCMWRGCATYRTGGGAMHTB---
| Original motif | Reverse complement motif |
| Consensus sequence: DTVBDDRGACCACCCAGDWDGBH | Consensus sequence: DBCDWHCTGGGTGGTCMDHBBAH |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 20 |
| Similarity score: | 0.072957 |
Alignment:
HHBDHDDAACAATDRHHHHHBW
-GCMWRGCATYRTGGGAMHTB-
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Dataset #: 3 | Motif ID: 132 | Motif name: ZNF354C |
| Original motif | Reverse complement motif |
| Consensus sequence: MTCCAC | Consensus sequence: GTGGAY |
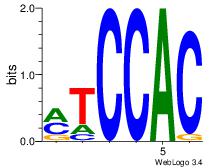 |
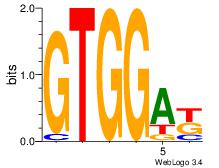 |
Best Matches for Motif ID 132 (Highest to Lowest)
| Motif ID: | UP00249 |
| Motif name: | Nkx2-5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
HDADCCACTTRAAWTT
--MTCCAC--------
| Original motif | Reverse complement motif |
| Consensus sequence: HDADCCACTTRAAWTT | Consensus sequence: AAWTTMAAGTGGHTDH |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 12 |
| Number of overlap: | 6 |
| Similarity score: | 0.00247916 |
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
------MTCCAC-----------
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00147 |
| Motif name: | Nkx2-6 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.00337038 |
Alignment:
DDADCCACTTAAVHTT
--MTCCAC--------
| Original motif | Reverse complement motif |
| Consensus sequence: DDADCCACTTAAVHTT | Consensus sequence: AAHVTTAAGTGGHTDD |
 |
 |
| Motif ID: | UP00228 |
| Motif name: | Bapx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.00628534 |
Alignment:
VHHAACCACTTAAVVAH
---MTCCAC--------
| Original motif | Reverse complement motif |
| Consensus sequence: VHHAACCACTTAAVVAH | Consensus sequence: DTBVTTAAGTGGTTHHV |
 |
 |
| Motif ID: | UP00231 |
| Motif name: | Nkx2-2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 10 |
| Number of overlap: | 6 |
| Similarity score: | 0.0113442 |
Alignment:
HDVRCCACTTGARADBD
--MTCCAC---------
| Original motif | Reverse complement motif |
| Consensus sequence: HDVRCCACTTGARADBD | Consensus sequence: DVDTKTCAAGTGGKBDH |
 |
 |
| Dataset #: 4 | Motif ID: 133 | Motif name: shAGrGGGCAgy |
| Original motif | Reverse complement motif |
| Consensus sequence: SHAGRGGGCABH | Consensus sequence: DBTGCCCKCTDS |
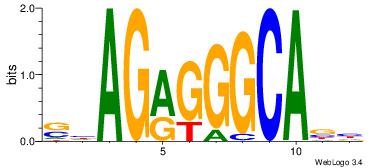 |
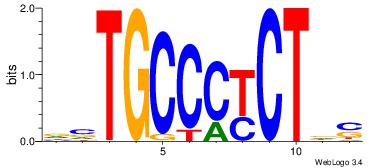 |
Best Matches for Motif ID 133 (Highest to Lowest)
| Motif ID: | UP00079 |
| Motif name: | Esrra_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0 |
Alignment:
VKBBAGGGGTCAHDBBH
--SHAGRGGGCABH---
| Original motif | Reverse complement motif |
| Consensus sequence: VKBBAGGGGTCAHDBBH | Consensus sequence: DBBHHTGACCCCTBBYV |
 |
 |
| Motif ID: | UP00009 |
| Motif name: | Nr2f2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0084293 |
Alignment:
DHBHAAAGGTCAHVHB
--SHAGRGGGCABH--
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCAHVHB | Consensus sequence: BHVDTGACCTTTDVDD |
 |
 |
| Motif ID: | UP00048 |
| Motif name: | Rara |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.00844318 |
Alignment:
DHBHAAAGGTCACVHD
--SHAGRGGGCABH--
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCACVHD | Consensus sequence: HHVGTGACCTTTDVDD |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.00952396 |
Alignment:
BVVHAGGGGGCGRDHHB
--SHAGRGGGCABH---
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00066 |
| Motif name: | Hnf4a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.011648 |
Alignment:
BDBVMGGGGTCAABHDV
--SHAGRGGGCABH---
| Original motif | Reverse complement motif |
| Consensus sequence: BDBVMGGGGTCAABHDV | Consensus sequence: BHHVTTGACCCCYVVDB |
 |
 |
| Dataset #: 4 | Motif ID: 134 | Motif name: ssCGwGCGss |
| Original motif | Reverse complement motif |
| Consensus sequence: BSCGWGCGBV | Consensus sequence: VBCGCWCGSB |
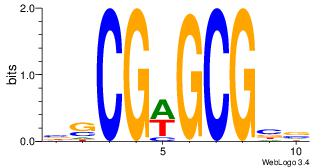 |
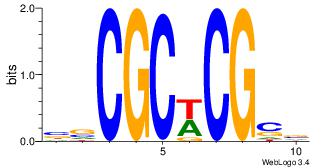 |
Best Matches for Motif ID 134 (Highest to Lowest)
| Motif ID: | UP00065 |
| Motif name: | Zfp161_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
DDGCGCGCGCRCHYRD
---BSCGWGCGBV---
| Original motif | Reverse complement motif |
| Consensus sequence: DDGCGCGCGCRCHYRD | Consensus sequence: DMMDGMGCGCGCGCHD |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 10 |
| Similarity score: | 0.00413791 |
Alignment:
DDRMHBACGCGYGCGTDGBBDB
-------BSCGWGCGBV-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.013065 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
------BSCGWGCGBV-------
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0147607 |
Alignment:
HDWHMHGACGCGCGTRBVVBHW
------BSCGWGCGBV------
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
 |
 |
| Motif ID: | UP00001 |
| Motif name: | E2F2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0160468 |
Alignment:
VHDARGGCGCGCVHH
---BSCGWGCGBV--
| Original motif | Reverse complement motif |
| Consensus sequence: VHDARGGCGCGCVHH | Consensus sequence: HHVGCGCGCCKTDHB |
 |
 |
| Dataset #: 4 | Motif ID: 135 | Motif name: ssCGGCCGss |
| Original motif | Reverse complement motif |
| Consensus sequence: BSCGGCCGSV | Consensus sequence: VSCGGCCGSB |
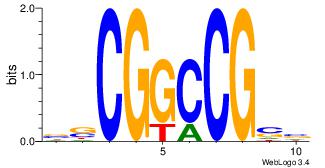 |
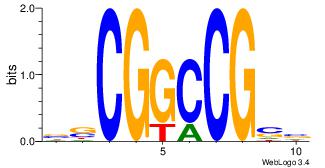 |
Best Matches for Motif ID 135 (Highest to Lowest)
| Motif ID: | UP00099 |
| Motif name: | Ascl2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.013915 |
Alignment:
HHDVVGCAGCTGVBKVB
----VSCGGCCGSB---
| Original motif | Reverse complement motif |
| Consensus sequence: BBYVVCAGCTGCBVHHD | Consensus sequence: HHDVVGCAGCTGVBKVB |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0179182 |
Alignment:
BHBHDTGGCGGGGBDHD
---BSCGGCCGSV----
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
 |
 |
| Motif ID: | UP00036 |
| Motif name: | Myf6_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0197811 |
Alignment:
VVVRACAGVCBCDCC
---BSCGGCCGSV--
| Original motif | Reverse complement motif |
| Consensus sequence: VVVRACAGVCBCDCC | Consensus sequence: GGDGBGVCTGTKVVB |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0198393 |
Alignment:
BHHBHCCGCCCCDHHHH
---BSCGGCCGSV----
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
 |
 |
| Motif ID: | UP00094 |
| Motif name: | Zfp128_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0205643 |
Alignment:
DHVGGGTACGCCRDHDV
--BSCGGCCGSV-----
| Original motif | Reverse complement motif |
| Consensus sequence: BHHDKGGCGTACCCBHD | Consensus sequence: DHVGGGTACGCCRDHDV |
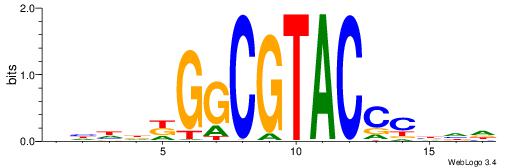 |
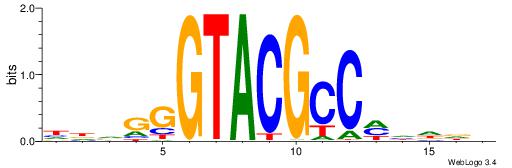 |
| Dataset #: 4 | Motif ID: 136 | Motif name: dwCAGAAGwh |
| Original motif | Reverse complement motif |
| Consensus sequence: DHCAGAAGDH | Consensus sequence: HDCTTCTGHD |
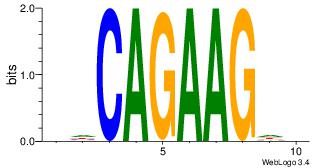 |
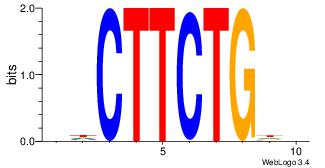 |
Best Matches for Motif ID 136 (Highest to Lowest)
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
HVDDVCAGATGKYHBBV
---DHCAGAAGDH----
| Original motif | Reverse complement motif |
| Consensus sequence: HVDDVCAGATGKYHBBV | Consensus sequence: VBBDMYCATCTGVHHBH |
 |
 |
| Motif ID: | UP00404 |
| Motif name: | Elf2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.00342014 |
Alignment:
BDWDCCGGAAGTHBBD
---DHCAGAAGDH---
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGDWDB | Consensus sequence: BDWDCCGGAAGTHBBD |
 |
 |
| Motif ID: | UP00052 |
| Motif name: | Osr2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.00452111 |
Alignment:
HHHHGCTACKGTHVHH
---HDCTTCTGHD---
| Original motif | Reverse complement motif |
| Consensus sequence: HHVHACRGTAGCHHHD | Consensus sequence: HHHHGCTACKGTHVHH |
 |
 |
| Motif ID: | UP00027 |
| Motif name: | Osr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.00483126 |
Alignment:
DHBHGCTACKGTHDHH
---HDCTTCTGHD---
| Original motif | Reverse complement motif |
| Consensus sequence: HHDHACRGTAGCHVHD | Consensus sequence: DHBHGCTACKGTHDHH |
 |
 |
| Motif ID: | UP00015 |
| Motif name: | Ehf_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.00574168 |
Alignment:
DDACTTCCGGBTHBB
-HDCTTCTGHD----
| Original motif | Reverse complement motif |
| Consensus sequence: VBDABCCGGAAGTDD | Consensus sequence: DDACTTCCGGBTHBB |
 |
 |
| Dataset #: 4 | Motif ID: 137 | Motif name: rgCGCCmyCTgs |
| Original motif | Reverse complement motif |
| Consensus sequence: VGCGCCCYCTDS | Consensus sequence: SHAGKGGGCGCB |
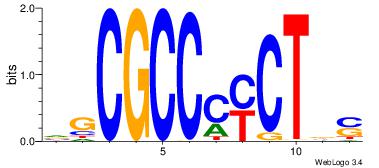 |
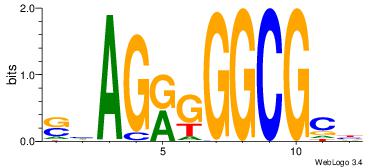 |
Best Matches for Motif ID 137 (Highest to Lowest)
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0 |
Alignment:
BVVHAGGGGGCGRDHHB
--SHAGKGGGCGCB---
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00068 |
| Motif name: | Eomes_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0223356 |
Alignment:
VVBVCKACACCTHVBV
--VGCGCCCYCTDS--
| Original motif | Reverse complement motif |
| Consensus sequence: VBVDAGGTGTYGVBBV | Consensus sequence: VVBVCKACACCTHVBV |
 |
 |
| Motif ID: | UP00003 |
| Motif name: | E2F3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0255543 |
Alignment:
WHSGCGCGCCHTDHB
--VGCGCCCYCTDS-
| Original motif | Reverse complement motif |
| Consensus sequence: VHDADGGCGCGCSHW | Consensus sequence: WHSGCGCGCCHTDHB |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.027032 |
Alignment:
VKBBAGGGGTCAHDBBH
--SHAGKGGGCGCB---
| Original motif | Reverse complement motif |
| Consensus sequence: VKBBAGGGGTCAHDBBH | Consensus sequence: DBBHHTGACCCCTBBYV |
 |
 |
| Motif ID: | UP00001 |
| Motif name: | E2F2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0274822 |
Alignment:
VHDARGGCGCGCVHH
-SHAGKGGGCGCB--
| Original motif | Reverse complement motif |
| Consensus sequence: VHDARGGCGCGCVHH | Consensus sequence: HHVGCGCGCCKTDHB |
 |
 |
| Dataset #: 4 | Motif ID: 138 | Motif name: grCCACyAGAkG |
| Original motif | Reverse complement motif |
| Consensus sequence: DDCCACYAGAKG | Consensus sequence: CYTCTMGTGGHH |
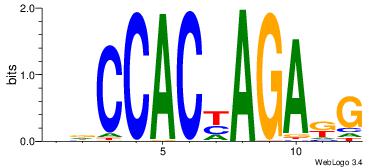 |
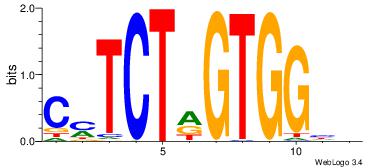 |
Best Matches for Motif ID 138 (Highest to Lowest)
| Motif ID: | UP00165 |
| Motif name: | Titf1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0 |
Alignment:
AAHYTCAAGTGGBTBV
--CYTCTMGTGGHH--
| Original motif | Reverse complement motif |
| Consensus sequence: BVABCCACTTGAMHTT | Consensus sequence: AAHYTCAAGTGGBTBV |
 |
 |
| Motif ID: | UP00231 |
| Motif name: | Nkx2-2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.00409843 |
Alignment:
HDVRCCACTTGARADBD
--DDCCACYAGAKG---
| Original motif | Reverse complement motif |
| Consensus sequence: HDVRCCACTTGARADBD | Consensus sequence: DVDTKTCAAGTGGKBDH |
 |
 |
| Motif ID: | UP00107 |
| Motif name: | Nkx2-4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.00589166 |
Alignment:
AAWYTMAAGTGGVTDH
--CYTCTMGTGGHH--
| Original motif | Reverse complement motif |
| Consensus sequence: HDAVCCACTTRAMWTT | Consensus sequence: AAWYTMAAGTGGVTDH |
 |
 |
| Motif ID: | UP00249 |
| Motif name: | Nkx2-5 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.00660985 |
Alignment:
AAWTTMAAGTGGHTDH
--CYTCTMGTGGHH--
| Original motif | Reverse complement motif |
| Consensus sequence: HDADCCACTTRAAWTT | Consensus sequence: AAWTTMAAGTGGHTDH |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 12 |
| Similarity score: | 0.00783877 |
Alignment:
DDHBHBSTCACGTGAHBBDHHM
-----CYTCTMGTGGHH-----
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Dataset #: 4 | Motif ID: 139 | Motif name: mkCTyTTCsg |
| Original motif | Reverse complement motif |
| Consensus sequence: HBCTTTTCBD | Consensus sequence: HBGAAAAGBD |
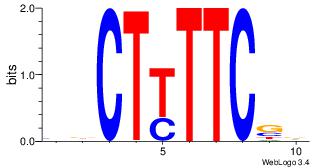 |
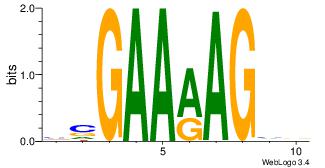 |
Best Matches for Motif ID 139 (Highest to Lowest)
| Motif ID: | UP00100 |
| Motif name: | Gata6_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
DHDVDWGATAAGADDHV
----HBGAAAAGBD---
| Original motif | Reverse complement motif |
| Consensus sequence: DHDVDWGATAAGADDHV | Consensus sequence: VHDDTCTTATCWHBDHD |
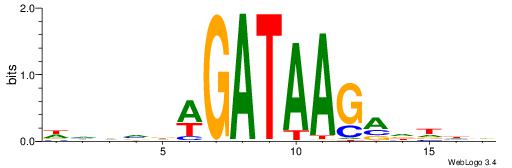 |
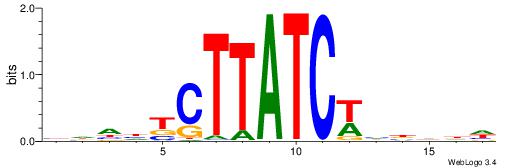 |
| Motif ID: | UP00080 |
| Motif name: | Gata5_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.000223307 |
Alignment:
DDHBKCTTATCAGDDDH
---HBCTTTTCBD----
| Original motif | Reverse complement motif |
| Consensus sequence: HDDDCTGATAAGRVDDD | Consensus sequence: DDHBKCTTATCAGDDDH |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.00555961 |
Alignment:
BDHDDADAGATAAGADHBDHVD
------HBGAAAAGBD------
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Motif ID: | UP00018 |
| Motif name: | Irf4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0104391 |
Alignment:
TBDKGTTTCGMTDHV
-HBCTTTTCBD----
| Original motif | Reverse complement motif |
| Consensus sequence: VDDAYCGAAACYDVA | Consensus sequence: TBDKGTTTCGMTDHV |
 |
 |
| Motif ID: | UP00422 |
| Motif name: | Etv3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.012889 |
Alignment:
VBBHAYTTCCGGTHDH
-HBCTTTTCBD-----
| Original motif | Reverse complement motif |
| Consensus sequence: VBBHAYTTCCGGTHDH | Consensus sequence: DHHACCGGAAKTHBBB |
 |
 |
| Dataset #: 4 | Motif ID: 140 | Motif name: vkCKCTkCGk |
| Original motif | Reverse complement motif |
| Consensus sequence: VDCTCTKCGB | Consensus sequence: BCGYAGAGDV |
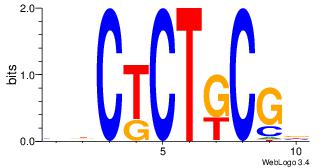 |
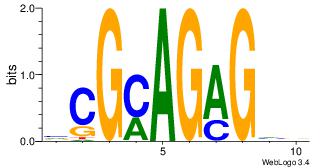 |
Best Matches for Motif ID 140 (Highest to Lowest)
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
DDBBBCACTGCABTBBB
---VDCTCTKCGB----
| Original motif | Reverse complement motif |
| Consensus sequence: DDBBBCACTGCABTBBB | Consensus sequence: BBBAVTGCAGTGBBVDD |
 |
 |
| Motif ID: | UP00065 |
| Motif name: | Zfp161_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.00813165 |
Alignment:
VVGCVMTGCGCGKC
-VDCTCTKCGB---
| Original motif | Reverse complement motif |
| Consensus sequence: GYCGCGCARBGCVB | Consensus sequence: VVGCVMTGCGCGKC |
 |
 |
| Motif ID: | UP00057 |
| Motif name: | Zic2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0158498 |
Alignment:
HHDCVCAGCAKGVVD
-VDCTCTKCGB----
| Original motif | Reverse complement motif |
| Consensus sequence: HHDCVCAGCAKGVVD | Consensus sequence: DVBCYTGCTGBGDDD |
 |
 |
| Motif ID: | UP00102 |
| Motif name: | Zic1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0176179 |
Alignment:
BHDCVCAGCAGGHVD
-VDCTCTKCGB----
| Original motif | Reverse complement motif |
| Consensus sequence: BHDCVCAGCAGGHVD | Consensus sequence: DVHCCTGCTGBGDDB |
 |
 |
| Motif ID: | UP00006 |
| Motif name: | Zic3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0202331 |
Alignment:
DDDCACAGCAKGHVD
-VDCTCTKCGB----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDCACAGCAKGHVD | Consensus sequence: DVHCYTGCTGTGHDH |
 |
 |
| Dataset #: 4 | Motif ID: 141 | Motif name: raCAAAACam |
| Original motif | Reverse complement motif |
| Consensus sequence: DACAAAACAH | Consensus sequence: HTGTTTTGTD |
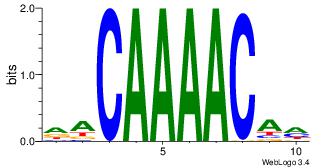 |
 |
Best Matches for Motif ID 141 (Highest to Lowest)
| Motif ID: | UP00039 |
| Motif name: | Foxj3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
DVHWDTGTTWTGDTBDH
----HTGTTTTGTD---
| Original motif | Reverse complement motif |
| Consensus sequence: HDBAHCAWAACADWDVD | Consensus sequence: DVHWDTGTTWTGDTBDH |
 |
 |
| Motif ID: | UP00040 |
| Motif name: | Irf5_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.00529846 |
Alignment:
BDKGTTTCGKTWDDD
-HTGTTTTGTD----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDWAYCGAAACYDV | Consensus sequence: BDKGTTTCGKTWDDD |
 |
 |
| Motif ID: | UP00018 |
| Motif name: | Irf4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.010321 |
Alignment:
TBDKGTTTCGMTDHV
--HTGTTTTGTD---
| Original motif | Reverse complement motif |
| Consensus sequence: VDDAYCGAAACYDVA | Consensus sequence: TBDKGTTTCGMTDHV |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0116411 |
Alignment:
ADDDYAWAACAAMAHH
--DACAAAACAH----
| Original motif | Reverse complement motif |
| Consensus sequence: ADDDYAWAACAAMAHH | Consensus sequence: HDTYTTGTTWTKDDDT |
 |
 |
| Motif ID: | UP00011 |
| Motif name: | Irf6_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0133918 |
Alignment:
DHDBTKGTTTCGHTHDD
----HTGTTTTGTD---
| Original motif | Reverse complement motif |
| Consensus sequence: HDDAHCGAAACYAVDDD | Consensus sequence: DHDBTKGTTTCGHTHDD |
 |
 |
| Dataset #: 4 | Motif ID: 142 | Motif name: ctCTTAACyw |
| Original motif | Reverse complement motif |
| Consensus sequence: HHCTTAACHD | Consensus sequence: DDGTTAAGHD |
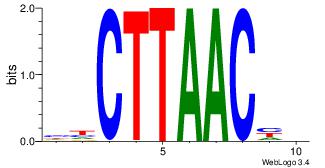 |
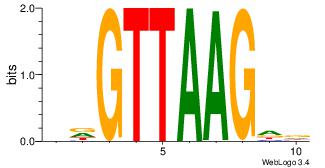 |
Best Matches for Motif ID 142 (Highest to Lowest)
| Motif ID: | UP00100 |
| Motif name: | Gata6_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
DHDVDWGATAAGADDHV
----DDGTTAAGHD---
| Original motif | Reverse complement motif |
| Consensus sequence: DHDVDWGATAAGADDHV | Consensus sequence: VHDDTCTTATCWHBDHD |
 |
 |
| Motif ID: | UP00390 |
| Motif name: | Tcf1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.00372807 |
Alignment:
HSKBVGTTAACTAADDB
---HHCTTAACHD----
| Original motif | Reverse complement motif |
| Consensus sequence: HSKBVGTTAACTAADDB | Consensus sequence: VDDTTAGTTAACBVRSD |
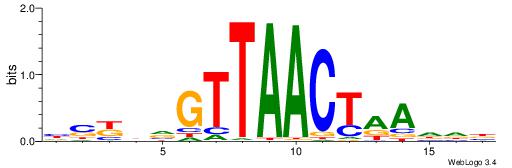 |
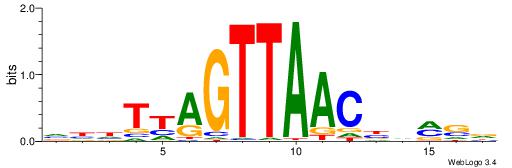 |
| Motif ID: | UP00222 |
| Motif name: | Tcf2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.00564182 |
Alignment:
DBBBGTTAACTAGBHBT
--HHCTTAACHD-----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBBGTTAACTAGBHBT | Consensus sequence: ABDBCTAGTTAACVBBD |
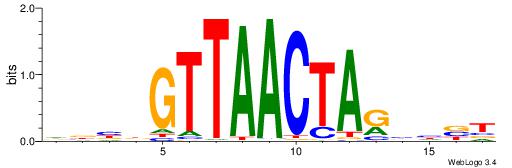 |
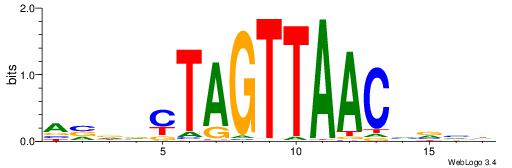 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.00635188 |
Alignment:
BDHDDADAGATAAGADHBDHVD
------DDGTTAAGHD------
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Motif ID: | UP00012 |
| Motif name: | Bbx_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.00788123 |
Alignment:
HVWHBGTTAACABHBDV
---DDGTTAAGHD----
| Original motif | Reverse complement motif |
| Consensus sequence: HVWHBGTTAACABHBDV | Consensus sequence: VHVHBTGTTAACVHWVH |
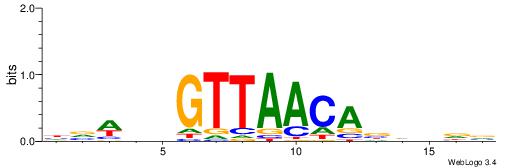 |
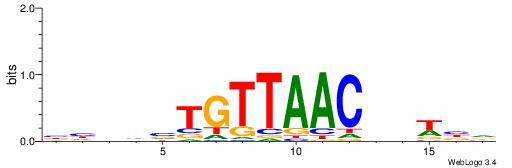 |
| Dataset #: 4 | Motif ID: 143 | Motif name: AgmAGAGGGCrscAGak |
| Original motif | Reverse complement motif |
| Consensus sequence: AGMAGAGGGCASCAGAK | Consensus sequence: RTCTGSTGCCCTCTYCT |
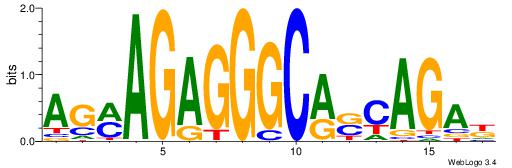 |
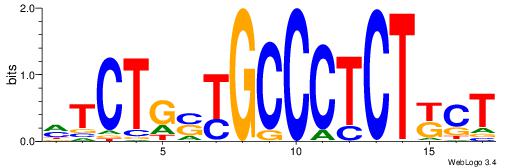 |
Best Matches for Motif ID 143 (Highest to Lowest)
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 0.0294896 |
Alignment:
BADHBDHCGCCCMCGCAHHDBBV
-RTCTGSTGCCCTCTYCT-----
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 0.0339644 |
Alignment:
HVGVSAGGAGGGTCYHHHHYVH
---AGMAGAGGGCASCAGAK--
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 17 |
| Similarity score: | 0.0353444 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
------AGMAGAGGGCASCAGAK
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 0.0356077 |
Alignment:
TCDAMVBGACGCMAKDDDVBBD
---AGMAGAGGGCASCAGAK--
| Original motif | Reverse complement motif |
| Consensus sequence: DBBVDDDRTRGCGTCBBYTHGA | Consensus sequence: TCDAMVBGACGCMAKDDDVBBD |
 |
 |
| Motif ID: | UP00088 |
| Motif name: | Plagl1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0356806 |
Alignment:
DBHGGGGGGTACCHHHD
AGMAGAGGGCASCAGAK
| Original motif | Reverse complement motif |
| Consensus sequence: DBHGGGGGGTACCHHHD | Consensus sequence: DHDDGGTACCCCCCHBH |
 |
 |
| Dataset #: 4 | Motif ID: 144 | Motif name: ctCTrsyGCCmCCTast |
| Original motif | Reverse complement motif |
| Consensus sequence: HDCTGSYGCCMCCTAST | Consensus sequence: ASTAGGYGGCMSCAGDD |
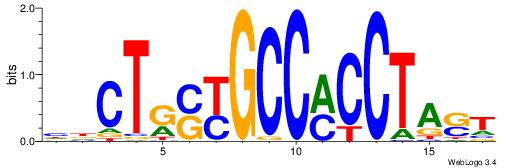 |
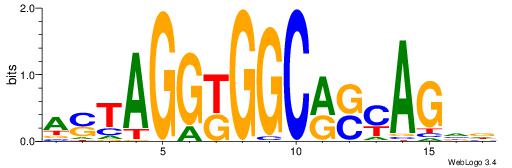 |
Best Matches for Motif ID 144 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 17 |
| Similarity score: | 0.0268744 |
Alignment:
BTBVTCVTGGGTGGTCMVVDVBB
-----ASTAGGYGGCMSCAGDD-
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 17 |
| Similarity score: | 0.0280132 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
HDCTGSYGCCMCCTAST------
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 17 |
| Similarity score: | 0.0281897 |
Alignment:
DVVTTVGTGGGHGGYAMHWHHHY
-----ASTAGGYGGCMSCAGDD-
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 0.0307583 |
Alignment:
YBVDMGTGGGTGGTCKVVBVBBT
----ASTAGGYGGCMSCAGDD--
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 17 |
| Similarity score: | 0.033913 |
Alignment:
DBCDWHCTGGGTGGTCMDHBBAH
-----ASTAGGYGGCMSCAGDD-
| Original motif | Reverse complement motif |
| Consensus sequence: DTVBDDRGACCACCCAGDWDGBH | Consensus sequence: DBCDWHCTGGGTGGTCMDHBBAH |
 |
 |
| Dataset #: 4 | Motif ID: 145 | Motif name: grCCACyAGAkG |
| Original motif | Reverse complement motif |
| Consensus sequence: DDCCACYAGAKG | Consensus sequence: CYTCTKGTGGHH |
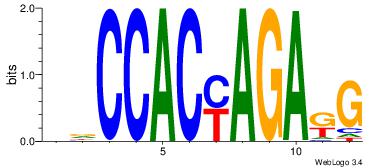 |
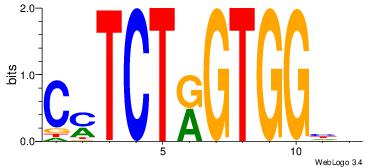 |
Best Matches for Motif ID 145 (Highest to Lowest)
| Motif ID: | UP00231 |
| Motif name: | Nkx2-2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0 |
Alignment:
HDVRCCACTTGARADBD
--DDCCACYAGAKG---
| Original motif | Reverse complement motif |
| Consensus sequence: HDVRCCACTTGARADBD | Consensus sequence: DVDTKTCAAGTGGKBDH |
 |
 |
| Motif ID: | UP00165 |
| Motif name: | Titf1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.00595864 |
Alignment:
AAHYTCAAGTGGBTBV
--CYTCTKGTGGHH--
| Original motif | Reverse complement motif |
| Consensus sequence: BVABCCACTTGAMHTT | Consensus sequence: AAHYTCAAGTGGBTBV |
 |
 |
| Motif ID: | UP00249 |
| Motif name: | Nkx2-5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.00794079 |
Alignment:
HDADCCACTTRAAWTT
--DDCCACYAGAKG--
| Original motif | Reverse complement motif |
| Consensus sequence: HDADCCACTTRAAWTT | Consensus sequence: AAWTTMAAGTGGHTDH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 0.0107822 |
Alignment:
HGDHVHKGACCACCCGGBGBGDB
-------DDCCACYAGAKG----
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 12 |
| Similarity score: | 0.0113877 |
Alignment:
DDHBHBSTCACGTGAHBBDHHM
-----CYTCTKGTGGHH-----
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Dataset #: 4 | Motif ID: 146 | Motif name: myrGYGCCmCCTast |
| Original motif | Reverse complement motif |
| Consensus sequence: VBAGCGCCMCCTAST | Consensus sequence: ASTAGGYGGCGCTBB |
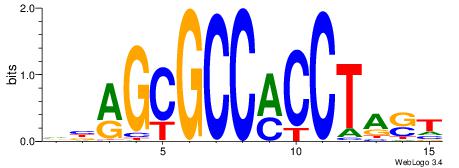 |
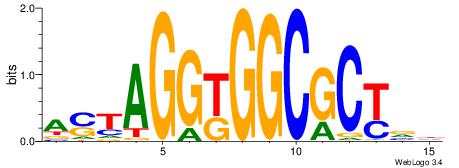 |
Best Matches for Motif ID 146 (Highest to Lowest)
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.015769 |
Alignment:
BDHHMCGCCCCCTHVBB
-VBAGCGCCMCCTAST-
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00068 |
| Motif name: | Eomes_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0179196 |
Alignment:
VBVDAGGTGTYGVBBV
-ASTAGGYGGCGCTBB
| Original motif | Reverse complement motif |
| Consensus sequence: VBVDAGGTGTYGVBBV | Consensus sequence: VVBVCKACACCTHVBV |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 15 |
| Similarity score: | 0.020063 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
--VBAGCGCCMCCTAST------
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 15 |
| Similarity score: | 0.0236839 |
Alignment:
HTBVVVDGGACCACCCRGRDBG
----VBAGCGCCMCCTAST---
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Motif ID: | UP00003 |
| Motif name: | E2F3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0240418 |
Alignment:
WHSGCGCGCCHTDHB
VBAGCGCCMCCTAST
| Original motif | Reverse complement motif |
| Consensus sequence: VHDADGGCGCGCSHW | Consensus sequence: WHSGCGCGCCHTDHB |
 |
 |
| Dataset #: 4 | Motif ID: 147 | Motif name: asCAGrkGGCrsy |
| Original motif | Reverse complement motif |
| Consensus sequence: ASCAGRGGGCRSB | Consensus sequence: BSKGCCCMCTGST |
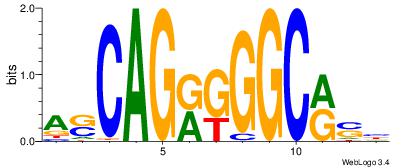 |
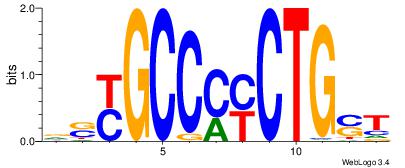 |
Best Matches for Motif ID 147 (Highest to Lowest)
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 13 |
| Similarity score: | 4.36149e-05 |
Alignment:
BVVHAGGGGGCGRDHHB
-ASCAGRGGGCRSB---
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 13 |
| Similarity score: | 0.00629602 |
Alignment:
HVDDVCAGATGKYHBBV
---ASCAGRGGGCRSB-
| Original motif | Reverse complement motif |
| Consensus sequence: HVDDVCAGATGKYHBBV | Consensus sequence: VBBDMYCATCTGVHHBH |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0114104 |
Alignment:
DBBHHTGACCCCTBBYV
---BSKGCCCMCTGST-
| Original motif | Reverse complement motif |
| Consensus sequence: VKBBAGGGGTCAHDBBH | Consensus sequence: DBBHHTGACCCCTBBYV |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0149411 |
Alignment:
VHTGCGGGGGCGGH
-ASCAGRGGGCRSB
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0154138 |
Alignment:
BBYVVCAGCTGCBVHHD
---ASCAGRGGGCRSB-
| Original motif | Reverse complement motif |
| Consensus sequence: BBYVVCAGCTGCBVHHD | Consensus sequence: HHDVVGCAGCTGVBKVB |
 |
 |
| Dataset #: 4 | Motif ID: 148 | Motif name: wwTwAAAAww |
| Original motif | Reverse complement motif |
| Consensus sequence: DDTWAAAAHH | Consensus sequence: HHTTTTWADD |
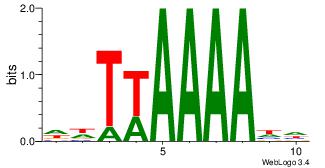 |
 |
Best Matches for Motif ID 148 (Highest to Lowest)
| Motif ID: | UP00180 |
| Motif name: | Hoxd13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0170004 |
Alignment:
HDHHTTTTATTKKHDD
--HHTTTTWADD----
| Original motif | Reverse complement motif |
| Consensus sequence: HDHYYAATAAAAHHHH | Consensus sequence: HDHHTTTTATTKKHDD |
 |
 |
| Motif ID: | UP00134 |
| Motif name: | Hoxb13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0201634 |
Alignment:
VDHWTTTTATTGGDKB
-HHTTTTWADD-----
| Original motif | Reverse complement motif |
| Consensus sequence: VRHCCAATAAAAWHHV | Consensus sequence: VDHWTTTTATTGGDKB |
 |
 |
| Motif ID: | UP00121 |
| Motif name: | Hoxd10 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0216837 |
Alignment:
HDDDATTTTATTKVRDB
---HHTTTTWADD----
| Original motif | Reverse complement motif |
| Consensus sequence: VDKVYAATAAAATDDDH | Consensus sequence: HDDDATTTTATTKVRDB |
 |
 |
| Motif ID: | UP00213 |
| Motif name: | Hoxa9 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0217118 |
Alignment:
WDTAWTTTWATGKCCGD
--HHTTTTWADD-----
| Original motif | Reverse complement motif |
| Consensus sequence: DCGGYCATWAAAWTADW | Consensus sequence: WDTAWTTTWATGKCCGD |
 |
 |
| Motif ID: | UP00078 |
| Motif name: | Arid3a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0227023 |
Alignment:
VBBHTTAATTAAAMHHH
------DDTWAAAAHH-
| Original motif | Reverse complement motif |
| Consensus sequence: VBBHTTAATTAAAMHHH | Consensus sequence: DHHYTTTAATTAAHBBV |
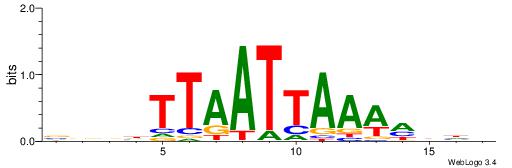 |
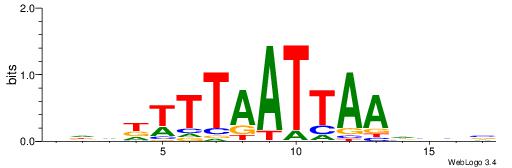 |
| Dataset #: 4 | Motif ID: 149 | Motif name: asmAGRGGGCrCTGsmkc |
| Original motif | Reverse complement motif |
| Consensus sequence: ASMAGAGGGCRCTGSABH | Consensus sequence: DBTSCAGMGCCCTCTRST |
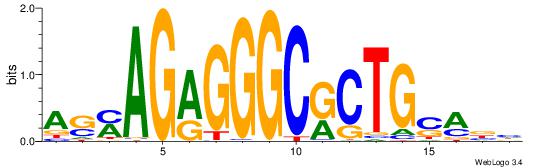 |
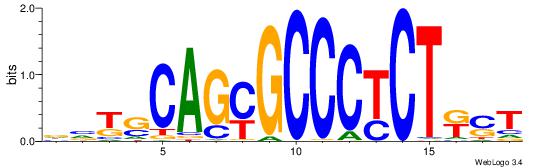 |
Best Matches for Motif ID 149 (Highest to Lowest)
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.0559948 |
Alignment:
MWSMBAARRATGCDCABHATGD
---ASMAGAGGGCRCTGSABH-
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0570957 |
Alignment:
DDRMHBACGCGYGCGTDGBBDB
----ASMAGAGGGCRCTGSABH
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.0582215 |
Alignment:
HVGVSAGGAGGGTCYHHHHYVH
---ASMAGAGGGCRCTGSABH-
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.0613486 |
Alignment:
YBVDMGTGGGTGGTCKVVBVBBT
-ASMAGAGGGCRCTGSABH----
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0613771 |
Alignment:
MHHHWHYTMCCHCCCACVAABVH
---DBTSCAGMGCCCTCTRST--
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Dataset #: 4 | Motif ID: 150 | Motif name: waATwAAAATAww |
| Original motif | Reverse complement motif |
| Consensus sequence: DHATWAAAATAHD | Consensus sequence: DHTATTTTWATHD |
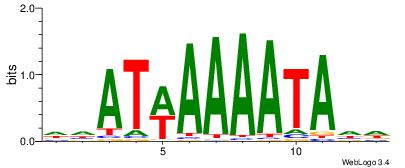 |
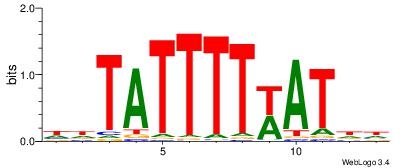 |
Best Matches for Motif ID 150 (Highest to Lowest)
| Motif ID: | UP00071 |
| Motif name: | Sox21_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.00363797 |
Alignment:
HHHHATTATAATWDDD
--DHATWAAAATAHD-
| Original motif | Reverse complement motif |
| Consensus sequence: HHHHATTATAATWDDD | Consensus sequence: HDDWATTATAATHHHH |
 |
 |
| Motif ID: | UP00016 |
| Motif name: | Sry_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 13 |
| Similarity score: | 0.0060438 |
Alignment:
VDDDATTATAATHDHV
-DHTATTTTWATHD--
| Original motif | Reverse complement motif |
| Consensus sequence: BHDHATTATAATDDDV | Consensus sequence: VDDDATTATAATHDHV |
 |
 |
| Motif ID: | UP00004 |
| Motif name: | Sox14_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.008022 |
Alignment:
DDDWATTATAATTHDH
-DHTATTTTWATHD--
| Original motif | Reverse complement motif |
| Consensus sequence: DHHAATTATAATWDDH | Consensus sequence: DDDWATTATAATTHDH |
 |
 |
| Motif ID: | UP00213 |
| Motif name: | Hoxa9 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.00895732 |
Alignment:
DCGGYCATWAAAWTADW
----DHATWAAAATAHD
| Original motif | Reverse complement motif |
| Consensus sequence: DCGGYCATWAAAWTADW | Consensus sequence: WDTAWTTTWATGKCCGD |
 |
 |
| Motif ID: | UP00121 |
| Motif name: | Hoxd10 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0122884 |
Alignment:
HDDDATTTTATTKVRDB
DHTATTTTWATHD----
| Original motif | Reverse complement motif |
| Consensus sequence: VDKVYAATAAAATDDDH | Consensus sequence: HDDDATTTTATTKVRDB |
 |
 |
| Dataset #: 4 | Motif ID: 151 | Motif name: agrCCAGmAGrg |
| Original motif | Reverse complement motif |
| Consensus sequence: HVGCCAGMAGRG | Consensus sequence: CKCTRCTGGCVH |
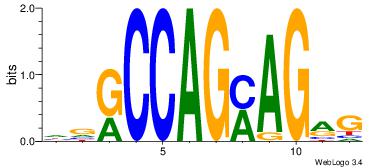 |
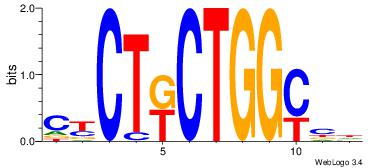 |
Best Matches for Motif ID 151 (Highest to Lowest)
| Motif ID: | UP00052 |
| Motif name: | Osr2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.00527025 |
Alignment:
HHHHGCTACKGTHVHH
---CKCTRCTGGCVH-
| Original motif | Reverse complement motif |
| Consensus sequence: HHVHACRGTAGCHHHD | Consensus sequence: HHHHGCTACKGTHVHH |
 |
 |
| Motif ID: | UP00013 |
| Motif name: | Gabpa_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.00879859 |
Alignment:
HVHHACCGGAAGTDHHV
--HVGCCAGMAGRG---
| Original motif | Reverse complement motif |
| Consensus sequence: HVHHACCGGAAGTDHHV | Consensus sequence: BHHHACTTCCGGTHHBD |
 |
 |
| Motif ID: | UP00027 |
| Motif name: | Osr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0106437 |
Alignment:
DHBHGCTACKGTHDHH
---CKCTRCTGGCVH-
| Original motif | Reverse complement motif |
| Consensus sequence: HHDHACRGTAGCHVHD | Consensus sequence: DHBHGCTACKGTHDHH |
 |
 |
| Motif ID: | UP00045 |
| Motif name: | Mafb_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0125184 |
Alignment:
DMTWDGTCAGCADWTTH
---HVGCCAGMAGRG--
| Original motif | Reverse complement motif |
| Consensus sequence: HAAWDTGCTGACDWARH | Consensus sequence: DMTWDGTCAGCADWTTH |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0127651 |
Alignment:
VBBDMYCATCTGVHHBH
----CKCTRCTGGCVH-
| Original motif | Reverse complement motif |
| Consensus sequence: HVDDVCAGATGKYHBBV | Consensus sequence: VBBDMYCATCTGVHHBH |
 |
 |
| Dataset #: 4 | Motif ID: 152 | Motif name: yrCATGCAyr |
| Original motif | Reverse complement motif |
| Consensus sequence: BRCATGCABD | Consensus sequence: HVTGCATGKV |
 |
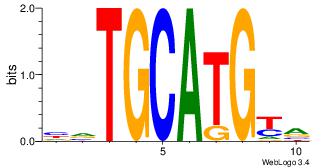 |
Best Matches for Motif ID 152 (Highest to Lowest)
| Motif ID: | UP00236 |
| Motif name: | Irx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
WVWDDTACATGTADDDW
-----BRCATGCABD--
| Original motif | Reverse complement motif |
| Consensus sequence: WDDDTACATGTADDWBW | Consensus sequence: WVWDDTACATGTADDDW |
 |
 |
| Motif ID: | UP00223 |
| Motif name: | Irx3_2226.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.000793267 |
Alignment:
DHDWBTACATGTAHDDD
-----BRCATGCABD--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDHTACATGTAVWDHD | Consensus sequence: DHDWBTACATGTAHDDD |
 |
 |
| Motif ID: | UP00223 |
| Motif name: | Irx3_0920.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.00151885 |
Alignment:
DDWWBTACATGTADDDD
-----BRCATGCABD--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDDTACATGTAVWWHD | Consensus sequence: DDWWBTACATGTADDDD |
 |
 |
| Motif ID: | UP00150 |
| Motif name: | Irx6 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.00440523 |
Alignment:
HAHDTACATGTAVDWHT
--HVTGCATGKV-----
| Original motif | Reverse complement motif |
| Consensus sequence: HAHDTACATGTAVDWHT | Consensus sequence: AHWDBTACATGTADHTH |
 |
 |
| Motif ID: | UP00250 |
| Motif name: | Irx5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.00456694 |
Alignment:
WVTDHWACATGTWHDDD
-----BRCATGCABD--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDHWACATGTWHDABW | Consensus sequence: WVTDHWACATGTWHDDD |
 |
 |
| Dataset #: 4 | Motif ID: 153 | Motif name: scAGrkGGCGcy |
| Original motif | Reverse complement motif |
| Consensus sequence: SHAGRGGGCGCB | Consensus sequence: VGCGCCCMCTDS |
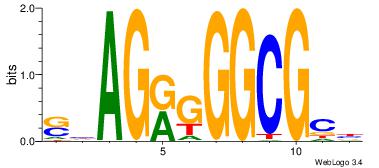 |
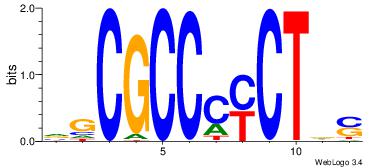 |
Best Matches for Motif ID 153 (Highest to Lowest)
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0 |
Alignment:
BDHHMCGCCCCCTHVBB
---VGCGCCCMCTDS--
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0220034 |
Alignment:
VHTGCGGGGGCGGH
VGCGCCCMCTDS--
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
| Motif ID: | UP00068 |
| Motif name: | Eomes_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0243258 |
Alignment:
VBVDAGGTGTYGVBBV
--SHAGRGGGCGCB--
| Original motif | Reverse complement motif |
| Consensus sequence: VBVDAGGTGTYGVBBV | Consensus sequence: VVBVCKACACCTHVBV |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0256904 |
Alignment:
DBBHHTGACCCCTBBYV
---VGCGCCCMCTDS--
| Original motif | Reverse complement motif |
| Consensus sequence: VKBBAGGGGTCAHDBBH | Consensus sequence: DBBHHTGACCCCTBBYV |
 |
 |
| Motif ID: | UP00003 |
| Motif name: | E2F3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0276084 |
Alignment:
WHSGCGCGCCHTDHB
--VGCGCCCMCTDS-
| Original motif | Reverse complement motif |
| Consensus sequence: VHDADGGCGCGCSHW | Consensus sequence: WHSGCGCGCCHTDHB |
 |
 |
| Dataset #: 4 | Motif ID: 154 | Motif name: csCsCCTCCcc |
| Original motif | Reverse complement motif |
| Consensus sequence: VBCCCCTCCHB | Consensus sequence: BDGGAGGGGBV |
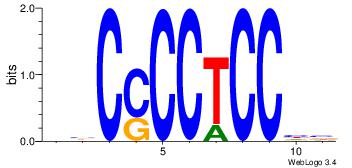 |
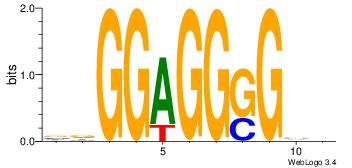 |
Best Matches for Motif ID 154 (Highest to Lowest)
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0 |
Alignment:
BHBHDTGGCGGGGBDHD
----BDGGAGGGGBV--
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.000861882 |
Alignment:
HVBCCCCCCCCMHHHB
--VBCCCCTCCHB---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.00225908 |
Alignment:
DHHDGGGCGRGGKHBH
---BDGGAGGGGBV--
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.00928712 |
Alignment:
BDHHMCGCCCCCTHVBB
---VBCCCCTCCHB---
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00047 |
| Motif name: | Zbtb7b_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0113601 |
Alignment:
VHWTTHGGGGGGVDD
----BDGGAGGGGBV
| Original motif | Reverse complement motif |
| Consensus sequence: DDVCCCCCCHAAWHB | Consensus sequence: VHWTTHGGGGGGVDD |
 |
 |
| Dataset #: 4 | Motif ID: 155 | Motif name: csCSCCdCCCcs |
| Original motif | Reverse complement motif |
| Consensus sequence: VBCCCCDCCCHV | Consensus sequence: VDGGGDGGGGBV |
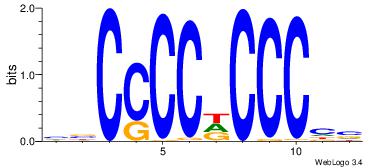 |
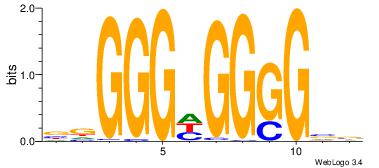 |
Best Matches for Motif ID 155 (Highest to Lowest)
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0 |
Alignment:
HBDRCCMCGCCCHHHD
--VBCCCCDCCCHV--
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.00350351 |
Alignment:
GKRGGGGGGGGGGBD
-VDGGGDGGGGBV--
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.00591405 |
Alignment:
BHHDYGGGGGGGGBVD
---VDGGGDGGGGBV-
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0128691 |
Alignment:
DHHBCCCCGCCAHHBHB
--VBCCCCDCCCHV---
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
 |
 |
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 0.0153192 |
Alignment:
HBBBCCGCCCCWHHHV
VBCCCCDCCCHV----
| Original motif | Reverse complement motif |
| Consensus sequence: HBBBCCGCCCCWHHHV | Consensus sequence: BHHHWGGGGCGGBBVH |
 |
 |
| Dataset #: 4 | Motif ID: 156 | Motif name: rgyGCCMyCTksTGGccd |
| Original motif | Reverse complement motif |
| Consensus sequence: RVYGCCCYCTKSTGGCHD | Consensus sequence: DDGCCASYAGMGGGCKVM |
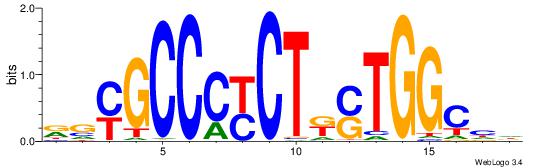 |
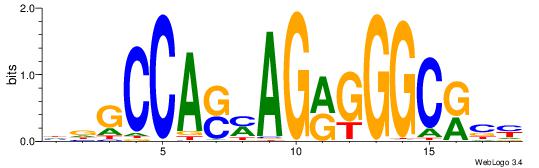 |
Best Matches for Motif ID 156 (Highest to Lowest)
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 18 |
| Similarity score: | 0.0420151 |
Alignment:
BADHBDHCGCCCMCGCAHHDBBV
-----RVYGCCCYCTKSTGGCHD
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0449867 |
Alignment:
DVMHHDHKGACCCTCCTSVCBH
----RVYGCCCYCTKSTGGCHD
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0484904 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
----RVYGCCCYCTKSTGGCHD
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0486343 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
RVYGCCCYCTKSTGGCHD-----
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.0531558 |
Alignment:
DDHBHBSTCACGTGAHBBDHHM
----DDGCCASYAGMGGGCKVM
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Dataset #: 4 | Motif ID: 157 | Motif name: wtATTTTTAww |
| Original motif | Reverse complement motif |
| Consensus sequence: DTATTTTTAWW | Consensus sequence: WWTAAAAATAD |
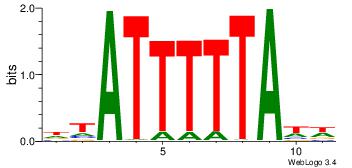 |
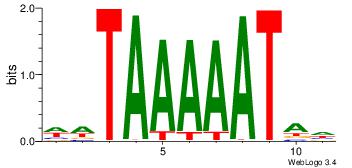 |
Best Matches for Motif ID 157 (Highest to Lowest)
| Motif ID: | UP00051 |
| Motif name: | Sox8_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0 |
Alignment:
DDDHDAACAATWBATDD
--WWTAAAAATAD----
| Original motif | Reverse complement motif |
| Consensus sequence: DDATBWATTGTTHHDDD | Consensus sequence: DDDHDAACAATWBATDD |
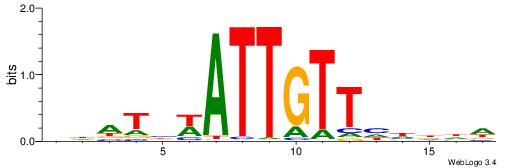 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 11 |
| Similarity score: | 1.71331e-05 |
Alignment:
WBHHHHHMDATTGTTHDHDVHH
-------DTATTTTTAWW----
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00096 |
| Motif name: | Sox13_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.00013394 |
Alignment:
DHDRGAACAATWWDHW
--WWTAAAAATAD---
| Original motif | Reverse complement motif |
| Consensus sequence: DHDRGAACAATWWDHW | Consensus sequence: WHDWWATTGTTCKDHD |
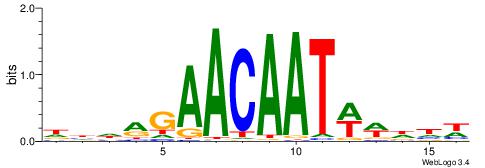 |
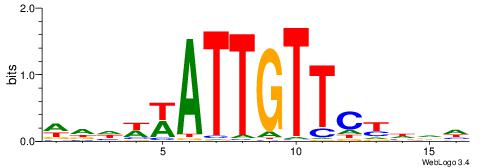 |
| Motif ID: | UP00091 |
| Motif name: | Sox5_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.00179252 |
Alignment:
DHDDWATTGTTCDDHD
---DTATTTTTAWW--
| Original motif | Reverse complement motif |
| Consensus sequence: DHDDGAACAATWDDHD | Consensus sequence: DHDDWATTGTTCDDHD |
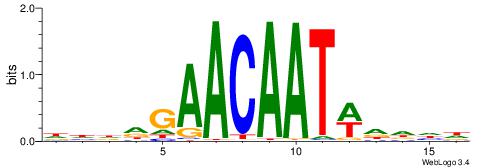 |
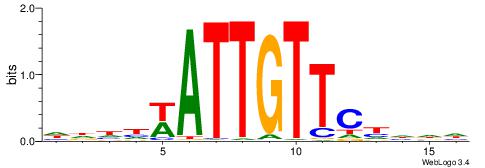 |
| Motif ID: | UP00213 |
| Motif name: | Hoxa9 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.00180638 |
Alignment:
WDTAWTTTWATGKCCGD
-DTATTTTTAWW-----
| Original motif | Reverse complement motif |
| Consensus sequence: DCGGYCATWAAAWTADW | Consensus sequence: WDTAWTTTWATGKCCGD |
 |
 |
| Dataset #: 4 | Motif ID: 158 | Motif name: grCCACwAGrk |
| Original motif | Reverse complement motif |
| Consensus sequence: DDCCACWAGRK | Consensus sequence: YMCTWGTGGHH |
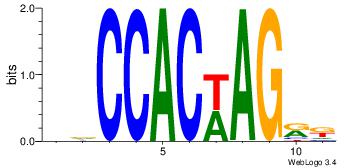 |
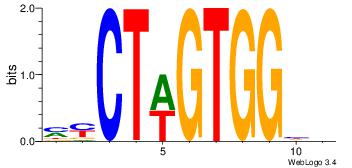 |
Best Matches for Motif ID 158 (Highest to Lowest)
| Motif ID: | UP00231 |
| Motif name: | Nkx2-2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0 |
Alignment:
HDVRCCACTTGARADBD
--DDCCACWAGRK----
| Original motif | Reverse complement motif |
| Consensus sequence: HDVRCCACTTGARADBD | Consensus sequence: DVDTKTCAAGTGGKBDH |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.00190925 |
Alignment:
DDASCACGTGBTBVDD
-DDCCACWAGRK----
| Original motif | Reverse complement motif |
| Consensus sequence: DDASCACGTGBTBVDD | Consensus sequence: HHVBABCACGTGSTHD |
 |
 |
| Motif ID: | UP00070 |
| Motif name: | Gcm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.00460463 |
Alignment:
BYMBKCCCCTATDVBHD
---DDCCACWAGRK---
| Original motif | Reverse complement motif |
| Consensus sequence: DDBVHATAGGGGRBRMB | Consensus sequence: BYMBKCCCCTATDVBHD |
 |
 |
| Motif ID: | UP00165 |
| Motif name: | Titf1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.00482349 |
Alignment:
AAHYTCAAGTGGBTBV
---YMCTWGTGGHH--
| Original motif | Reverse complement motif |
| Consensus sequence: BVABCCACTTGAMHTT | Consensus sequence: AAHYTCAAGTGGBTBV |
 |
 |
| Motif ID: | UP00249 |
| Motif name: | Nkx2-5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.00533429 |
Alignment:
HDADCCACTTRAAWTT
--DDCCACWAGRK---
| Original motif | Reverse complement motif |
| Consensus sequence: HDADCCACTTRAAWTT | Consensus sequence: AAWTTMAAGTGGHTDH |
 |
 |
| Dataset #: 4 | Motif ID: 159 | Motif name: kkAAGAGCAsy |
| Original motif | Reverse complement motif |
| Consensus sequence: DBAAGAGCAVH | Consensus sequence: HVTGCTCTTBH |
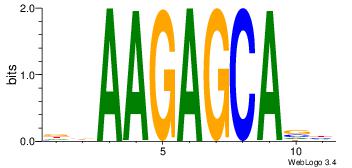 |
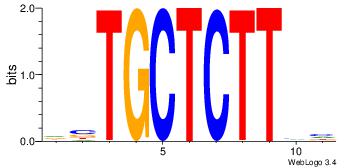 |
Best Matches for Motif ID 159 (Highest to Lowest)
| Motif ID: | UP00095 |
| Motif name: | Zfp691_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0 |
Alignment:
DDHBYGAGCACTGBHHB
-DBAAGAGCAVH-----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHVCAGTGCTCMBHDD | Consensus sequence: DDHBYGAGCACTGBHHB |
 |
 |
| Motif ID: | UP00048 |
| Motif name: | Rara |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.00374756 |
Alignment:
HHVGTGACCTTTDVDD
--HVTGCTCTTBH---
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCACVHD | Consensus sequence: HHVGTGACCTTTDVDD |
 |
 |
| Motif ID: | UP00035 |
| Motif name: | Hic1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.00417461 |
Alignment:
VHHDWTGGGCAMDHBH
--DBAAGAGCAVH---
| Original motif | Reverse complement motif |
| Consensus sequence: DBDDRTGCCCAWDHDV | Consensus sequence: VHHDWTGGGCAMDHBH |
 |
 |
| Motif ID: | UP00066 |
| Motif name: | Hnf4a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.00430492 |
Alignment:
HHAYTGGACTTTHDBV
--HVTGCTCTTBH---
| Original motif | Reverse complement motif |
| Consensus sequence: BBHHAAAGTCCAMTHH | Consensus sequence: HHAYTGGACTTTHDBV |
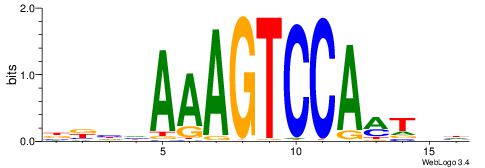 |
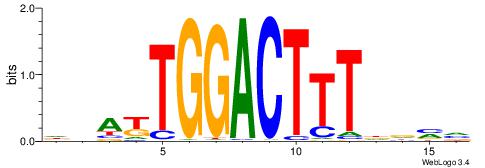 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.00467336 |
Alignment:
HHBVHTGACCTTGVDHD
---HVTGCTCTTBH---
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBCAAGGTCAHVBDH | Consensus sequence: HHBVHTGACCTTGVDHD |
 |
 |
| Dataset #: 4 | Motif ID: 160 | Motif name: brCAGGGCCrs |
| Original motif | Reverse complement motif |
| Consensus sequence: BVCAGGGCCVB | Consensus sequence: BBGGCCCTGBB |
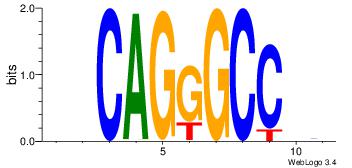 |
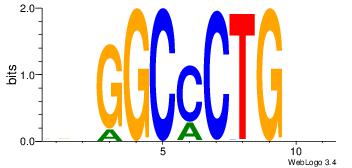 |
Best Matches for Motif ID 160 (Highest to Lowest)
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.00628499 |
Alignment:
VBHHVCAGGTGCDVDHD
---BVCAGGGCCVB---
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Motif ID: | UP00095 |
| Motif name: | Zfp691_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.00878794 |
Alignment:
BDHVCAGTGCTCMBHDD
--BVCAGGGCCVB----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHVCAGTGCTCMBHDD | Consensus sequence: DDHBYGAGCACTGBHHB |
 |
 |
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.00959728 |
Alignment:
BBBAVTGCAGTGBBVDD
-----BVCAGGGCCVB-
| Original motif | Reverse complement motif |
| Consensus sequence: DDBBBCACTGCABTBBB | Consensus sequence: BBBAVTGCAGTGBBVDD |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0163402 |
Alignment:
DHDBCAAGGTCAHVBDH
--BVCAGGGCCVB----
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBCAAGGTCAHVBDH | Consensus sequence: HHBVHTGACCTTGVDHD |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.0177751 |
Alignment:
DVMHHDHKGACCCTCCTSVCBH
------BBGGCCCTGBB-----
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Dataset #: 4 | Motif ID: 161 | Motif name: ssCGCwGCGss |
| Original motif | Reverse complement motif |
| Consensus sequence: VSCGCDGCGSB | Consensus sequence: BSCGCDGCGSV |
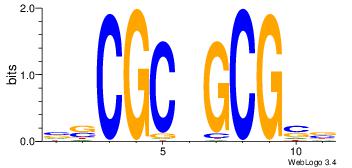 |
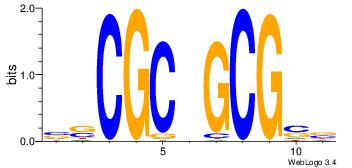 |
Best Matches for Motif ID 161 (Highest to Lowest)
| Motif ID: | UP00065 |
| Motif name: | Zfp161_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0 |
Alignment:
VVGCVMTGCGCGKC
-VSCGCDGCGSB--
| Original motif | Reverse complement motif |
| Consensus sequence: GYCGCGCARBGCVB | Consensus sequence: VVGCVMTGCGCGKC |
 |
 |
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.00206218 |
Alignment:
DDBBBCACTGCABTBBB
---VSCGCDGCGSB---
| Original motif | Reverse complement motif |
| Consensus sequence: DDBBBCACTGCABTBBB | Consensus sequence: BBBAVTGCAGTGBBVDD |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.010781 |
Alignment:
HDWHMHGACGCGCGTRBVVBHW
---VSCGCDGCGSB--------
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
 |
 |
| Motif ID: | UP00003 |
| Motif name: | E2F3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.0140886 |
Alignment:
HBHWYGGCGCCAMDVBB
VSCGCDGCGSB------
| Original motif | Reverse complement motif |
| Consensus sequence: HBHWYGGCGCCAMDVBB | Consensus sequence: BBBDYTGGCGCCKWDBD |
 |
 |
| Motif ID: | UP00001 |
| Motif name: | E2F2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0150699 |
Alignment:
HVBWYGGCGCCAADVBB
VSCGCDGCGSB------
| Original motif | Reverse complement motif |
| Consensus sequence: HVBWYGGCGCCAADVBB | Consensus sequence: BBBDTTGGCGCCKWVVD |
 |
 |
| Dataset #: 4 | Motif ID: 162 | Motif name: ccAsCCCCAcc |
| Original motif | Reverse complement motif |
| Consensus sequence: HVASCCCCABH | Consensus sequence: DBTGGGGSTVD |
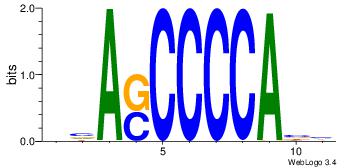 |
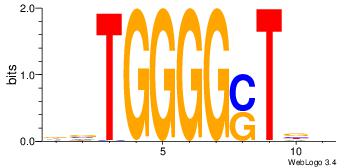 |
Best Matches for Motif ID 162 (Highest to Lowest)
| Motif ID: | UP00021 |
| Motif name: | Zfp281_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.00452097 |
Alignment:
VDDAHTGGGGGTHWYYD
---DBTGGGGSTVD---
| Original motif | Reverse complement motif |
| Consensus sequence: DKKWDACCCCCAHTDDV | Consensus sequence: VDDAHTGGGGGTHWYYD |
 |
 |
| Motif ID: | UP00024 |
| Motif name: | Glis2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.00896029 |
Alignment:
HKBBGGGGGGTCVHHH
--DBTGGGGSTVD---
| Original motif | Reverse complement motif |
| Consensus sequence: HHHVGACCCCCCVBRD | Consensus sequence: HKBBGGGGGGTCVHHH |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.010641 |
Alignment:
HVBCCCCCCCCMHHHB
---HVASCCCCABH--
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00070 |
| Motif name: | Gcm1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0169652 |
Alignment:
HVVHACCCGCATVDHD
--HVASCCCCABH---
| Original motif | Reverse complement motif |
| Consensus sequence: HVVHACCCGCATVDHD | Consensus sequence: DHDVATGCGGGTHVVH |
 |
 |
| Motif ID: | UP00047 |
| Motif name: | Zbtb7b_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0172076 |
Alignment:
VHWTTHGGGGGGVDD
----DBTGGGGSTVD
| Original motif | Reverse complement motif |
| Consensus sequence: DDVCCCCCCHAAWHB | Consensus sequence: VHWTTHGGGGGGVDD |
 |
 |
| Dataset #: 4 | Motif ID: 163 | Motif name: gwGGCCAGmAGAGGGCrby |
| Original motif | Reverse complement motif |
| Consensus sequence: VHGGCCAGMAGAGGGCRBY | Consensus sequence: KBKGCCCTCTYCTGGCCHV |
 |
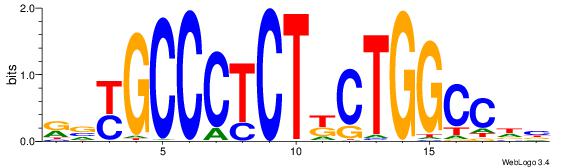 |
Best Matches for Motif ID 163 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 19 |
| Similarity score: | 0.0596242 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
VHGGCCAGMAGAGGGCRBY----
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 19 |
| Similarity score: | 0.0616662 |
Alignment:
HGDHVHKGACCACCCGGBGBGDB
----KBKGCCCTCTYCTGGCCHV
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 19 |
| Similarity score: | 0.0653251 |
Alignment:
VMYDHDGMCCHCCKBGVVAAVH
---KBKGCCCTCTYCTGGCCHV
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 19 |
| Similarity score: | 0.0654989 |
Alignment:
BBBDVVRGACCACCCAVGABBAB
----KBKGCCCTCTYCTGGCCHV
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 19 |
| Similarity score: | 0.066254 |
Alignment:
DDHBHBSTCACGTGAHBBDHHM
KBKGCCCTCTYCTGGCCHV---
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Dataset #: 4 | Motif ID: 164 | Motif name: asyAGrkGGCRGCAga |
| Original motif | Reverse complement motif |
| Consensus sequence: ASYAGRKGGCAGCABH | Consensus sequence: HBTGCTGCCYMCTKST |
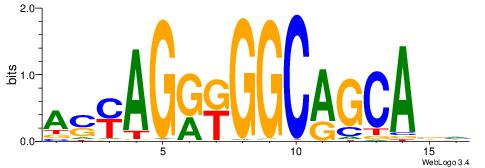 |
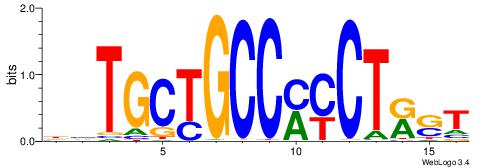 |
Best Matches for Motif ID 164 (Highest to Lowest)
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0281944 |
Alignment:
BVVHAGGGGGCGRDHHB
-ASYAGRKGGCAGCABH
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0303833 |
Alignment:
DBBHHTGACCCCTBBYV
HBTGCTGCCYMCTKST-
| Original motif | Reverse complement motif |
| Consensus sequence: VKBBAGGGGTCAHDBBH | Consensus sequence: DBBHHTGACCCCTBBYV |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0375379 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
------ASYAGRKGGCAGCABH-
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00088 |
| Motif name: | Plagl1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0402795 |
Alignment:
DBHGGGGGGTACCHHHD
ASYAGRKGGCAGCABH-
| Original motif | Reverse complement motif |
| Consensus sequence: DBHGGGGGGTACCHHHD | Consensus sequence: DHDDGGTACCCCCCHBH |
 |
 |
| Motif ID: | UP00066 |
| Motif name: | Hnf4a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0412697 |
Alignment:
BDBVMGGGGTCAABHDV
-ASYAGRKGGCAGCABH
| Original motif | Reverse complement motif |
| Consensus sequence: BDBVMGGGGTCAABHDV | Consensus sequence: BHHVTTGACCCCYVVDB |
 |
 |
| Dataset #: 4 | Motif ID: 165 | Motif name: wgGCCAshAGrGGGCrsy |
| Original motif | Reverse complement motif |
| Consensus sequence: HDGCCACHAGRGGGCRBY | Consensus sequence: KBKGCCCKCTHGTGGCHH |
 |
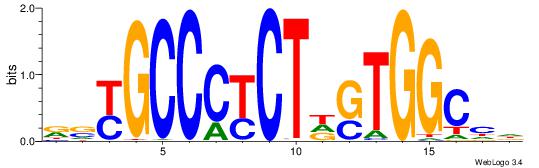 |
Best Matches for Motif ID 165 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.042684 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
-HDGCCACHAGRGGGCRBY----
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0461274 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
-----KBKGCCCKCTHGTGGCHH
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.0469346 |
Alignment:
DDHBHBSTCACGTGAHBBDHHM
KBKGCCCKCTHGTGGCHH----
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.0477902 |
Alignment:
HVGVSAGGAGGGTCYHHHHYVH
HDGCCACHAGRGGGCRBY----
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.0484261 |
Alignment:
HGDHVHKGACCACCCGGBGBGDB
----KBKGCCCKCTHGTGGCHH-
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Dataset #: 4 | Motif ID: 166 | Motif name: CasCAGrGGGCrsy |
| Original motif | Reverse complement motif |
| Consensus sequence: CACCAGRGGGCRSB | Consensus sequence: BSKGCCCKCTGGTG |
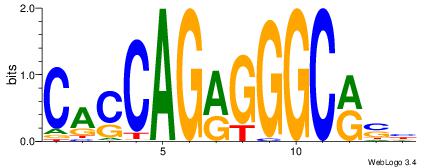 |
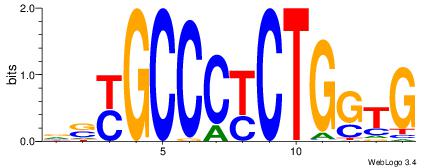 |
Best Matches for Motif ID 166 (Highest to Lowest)
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0233864 |
Alignment:
BVVHAGGGGGCGRDHHB
CACCAGRGGGCRSB---
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0278012 |
Alignment:
HVDDVCAGATGKYHBBV
--CACCAGRGGGCRSB-
| Original motif | Reverse complement motif |
| Consensus sequence: HVDDVCAGATGKYHBBV | Consensus sequence: VBBDMYCATCTGVHHBH |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0324879 |
Alignment:
VKBBAGGGGTCAHDBBH
CACCAGRGGGCRSB---
| Original motif | Reverse complement motif |
| Consensus sequence: VKBBAGGGGTCAHDBBH | Consensus sequence: DBBHHTGACCCCTBBYV |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0369901 |
Alignment:
VHTGCGGGGGCGGH
CACCAGRGGGCRSB
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0370867 |
Alignment:
BBYVVCAGCTGCBVHHD
--CACCAGRGGGCRSB-
| Original motif | Reverse complement motif |
| Consensus sequence: BBYVVCAGCTGCBVHHD | Consensus sequence: HHDVVGCAGCTGVBKVB |
 |
 |
| Dataset #: 4 | Motif ID: 167 | Motif name: rsyAGrkGGCGCCmyCTrsy |
| Original motif | Reverse complement motif |
| Consensus sequence: DSYAGRKGGCGCCMYCTRSH | Consensus sequence: HSKAGKYGGCGCCRMCTMSD |
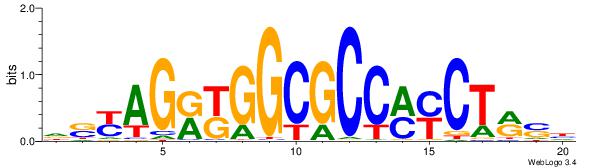 |
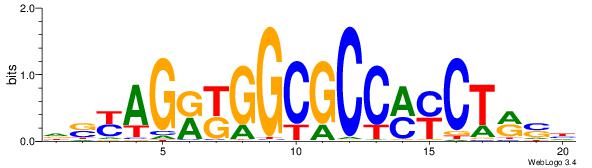 |
Best Matches for Motif ID 167 (Highest to Lowest)
| Motif ID: | UP00527 |
| Motif name: | Foxn4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 20 |
| Similarity score: | 0.0437543 |
Alignment:
TCDAMVBGACGCMAKDDDVBBD
--HSKAGKYGGCGCCRMCTMSD
| Original motif | Reverse complement motif |
| Consensus sequence: DBBVDDDRTRGCGTCBBYTHGA | Consensus sequence: TCDAMVBGACGCMAKDDDVBBD |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 20 |
| Similarity score: | 0.0440401 |
Alignment:
HVGVSAGGAGGGTCYHHHHYVH
--DSYAGRKGGCGCCMYCTRSH
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 20 |
| Similarity score: | 0.0496059 |
Alignment:
DVVTTVGTGGGHGGYAMHWHHHY
--DSYAGRKGGCGCCMYCTRSH-
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 20 |
| Similarity score: | 0.0498167 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
--HSKAGKYGGCGCCRMCTMSD
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 20 |
| Similarity score: | 0.05124 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
DSYAGRKGGCGCCMYCTRSH---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Dataset #: 4 | Motif ID: 168 | Motif name: yrcrGYGCCMyCTGGtG |
| Original motif | Reverse complement motif |
| Consensus sequence: HVCAGCGCCCYCTGGTG | Consensus sequence: CACCAGMGGGCGCTGBD |
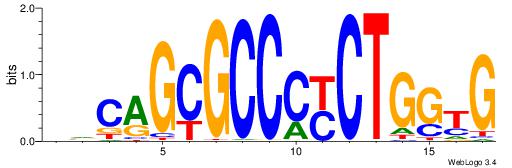 |
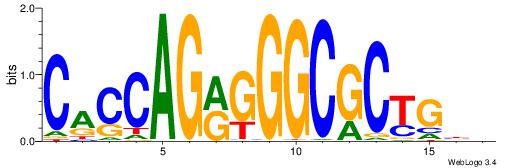 |
Best Matches for Motif ID 168 (Highest to Lowest)
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0436291 |
Alignment:
BDHHMCGCCCCCTHVBB
HVCAGCGCCCYCTGGTG
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 0.0524589 |
Alignment:
HGDHVHKGACCACCCGGBGBGDB
--HVCAGCGCCCYCTGGTG----
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 0.0526806 |
Alignment:
DDRMHBACGCGYGCGTDGBBDB
---CACCAGMGGGCGCTGBD--
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 17 |
| Similarity score: | 0.0534316 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
----CACCAGMGGGCGCTGBD--
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0549193 |
Alignment:
VKBBAGGGGTCAHDBBH
HVCAGCGCCCYCTGGTG
| Original motif | Reverse complement motif |
| Consensus sequence: VKBBAGGGGTCAHDBBH | Consensus sequence: DBBHHTGACCCCTBBYV |
 |
 |
| Dataset #: 4 | Motif ID: 169 | Motif name: yvTGCyGCCmCCwGgtG |
| Original motif | Reverse complement motif |
| Consensus sequence: BVTGCTGCCACCWGGDG | Consensus sequence: CDCCWGGTGGCAGCAVV |
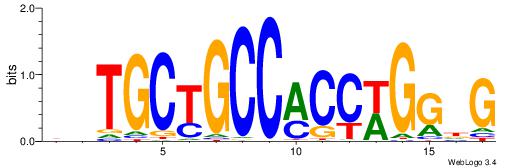 |
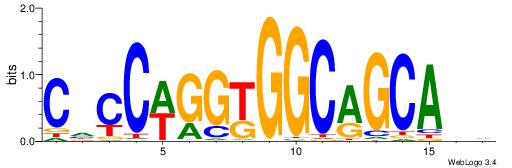 |
Best Matches for Motif ID 169 (Highest to Lowest)
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0458967 |
Alignment:
BDHHMCGCCCCCTHVBB
BVTGCTGCCACCWGGDG
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 17 |
| Similarity score: | 0.0491494 |
Alignment:
DVTTVVCVYGGHGGYCHHDMYB
----CDCCWGGTGGCAGCAVV-
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00047 |
| Motif name: | Zbtb7b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0516119 |
Alignment:
BDDRVGACCACCHBDVB
BVTGCTGCCACCWGGDG
| Original motif | Reverse complement motif |
| Consensus sequence: BDDRVGACCACCHBDVB | Consensus sequence: BBDVHGGTGGTCBKDDB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 0.0546478 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
-----CDCCWGGTGGCAGCAVV-
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 0.0549421 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
-BVTGCTGCCACCWGGDG----
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Dataset #: 4 | Motif ID: 170 | Motif name: ssGGCrsTGCrs |
| Original motif | Reverse complement motif |
| Consensus sequence: VVGGCRSTGCVB | Consensus sequence: BVGCASMGCCVV |
 |
 |
Best Matches for Motif ID 170 (Highest to Lowest)
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.00823722 |
Alignment:
DDBBBCACTGCABTBBB
-VVGGCRSTGCVB----
| Original motif | Reverse complement motif |
| Consensus sequence: DDBBBCACTGCABTBBB | Consensus sequence: BBBAVTGCAGTGBBVDD |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0121624 |
Alignment:
DHHBCCCCGCCAHHBHB
-BVGCASMGCCVV----
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
 |
 |
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0151043 |
Alignment:
HBDRCCMCGCCCHHHD
-BVGCASMGCCVV---
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00001 |
| Motif name: | E2F2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0208726 |
Alignment:
HVBWYGGCGCCAADVBB
-BVGCASMGCCVV----
| Original motif | Reverse complement motif |
| Consensus sequence: HVBWYGGCGCCAADVBB | Consensus sequence: BBBDTTGGCGCCKWVVD |
 |
 |
| Motif ID: | UP00003 |
| Motif name: | E2F3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 0.0216248 |
Alignment:
HBHWYGGCGCCAMDVBB
-BVGCASMGCCVV----
| Original motif | Reverse complement motif |
| Consensus sequence: HBHWYGGCGCCAMDVBB | Consensus sequence: BBBDYTGGCGCCKWDBD |
 |
 |
| Dataset #: 4 | Motif ID: 171 | Motif name: ysGTGGCCACsr |
| Original motif | Reverse complement motif |
| Consensus sequence: BVGTGGCCACBV | Consensus sequence: VBGTGGCCACVB |
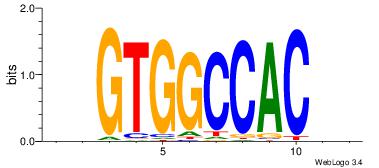 |
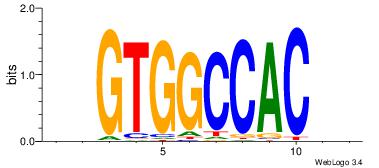 |
Best Matches for Motif ID 171 (Highest to Lowest)
| Motif ID: | UP00053 |
| Motif name: | Rxra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0 |
Alignment:
VHBHDGGGGTCAHBHBD
---VBGTGGCCACVB--
| Original motif | Reverse complement motif |
| Consensus sequence: DBHBDTGACCCCDHVHB | Consensus sequence: VHBHDGGGGTCAHBHBD |
 |
 |
| Motif ID: | UP00035 |
| Motif name: | Hic1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.00337191 |
Alignment:
VHHDWTGGGCAMDHBH
--BVGTGGCCACBV--
| Original motif | Reverse complement motif |
| Consensus sequence: DBDDRTGCCCAWDHDV | Consensus sequence: VHHDWTGGGCAMDHBH |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0057729 |
Alignment:
DBBHHTGACCCCTBBYV
--VBGTGGCCACVB---
| Original motif | Reverse complement motif |
| Consensus sequence: VKBBAGGGGTCAHDBBH | Consensus sequence: DBBHHTGACCCCTBBYV |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.00780464 |
Alignment:
BTBVTCVTGGGTGGTCMVVDVBB
--------BVGTGGCCACBV---
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00066 |
| Motif name: | Hnf4a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.00915211 |
Alignment:
BHHVTTGACCCCYVVDB
--BVGTGGCCACBV---
| Original motif | Reverse complement motif |
| Consensus sequence: BDBVMGGGGTCAABHDV | Consensus sequence: BHHVTTGACCCCYVVDB |
 |
 |