Best Matches in Database for Each Motif (Highest to Lowest)
| Dataset #: 1 | Motif ID: 1 | Motif name: Motif 1 |
| Original motif | Reverse complement motif |
| Consensus sequence: AGRKGGCR | Consensus sequence: KGCCYKCT |
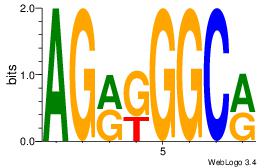 |
 |
Best Matches for Motif ID 1 (Highest to Lowest)
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
BVVHAGGGGGCGRDHHB
----AGRKGGCR-----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0152094 |
Alignment:
VKBBAGGGGTCAHDBBH
----AGRKGGCR-----
| Original motif | Reverse complement motif |
| Consensus sequence: VKBBAGGGGTCAHDBBH | Consensus sequence: DBBHHTGACCCCTBBYV |
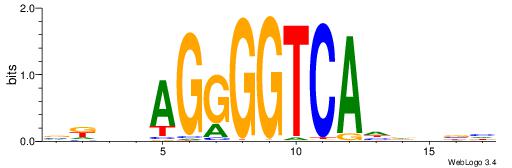 |
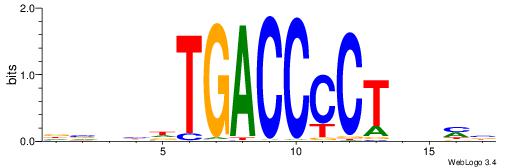 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0160089 |
Alignment:
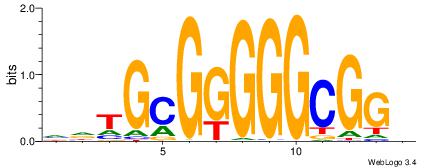
VHTGCGGGGGCGGH
----AGRKGGCR--
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
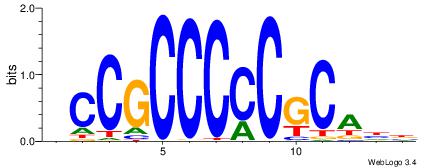 |
 |
| Motif ID: | UP00010 |
| Motif name: | Tcfap2b_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0244919 |
Alignment:
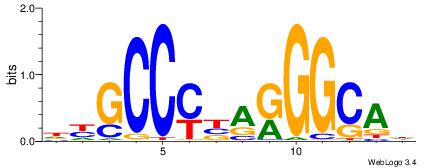
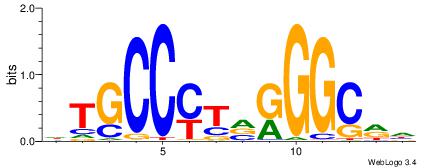
HTGCCMTVKGGCMD
-KGCCYKCT-----
| Original motif | Reverse complement motif |
| Consensus sequence: DYGCCYBARGGCAH | Consensus sequence: HTGCCMTVKGGCMD |
 |
 |
| Motif ID: | UP00070 |
| Motif name: | Gcm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0267457 |
Alignment:
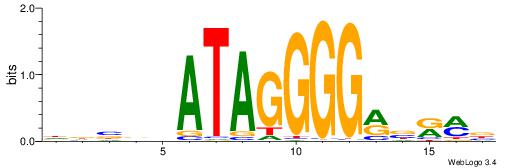
DDBVHATAGGGGRBRMB
-------AGRKGGCR--
| Original motif | Reverse complement motif |
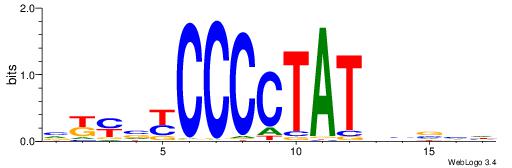
| Consensus sequence: DDBVHATAGGGGRBRMB | Consensus sequence: BYMBKCCCCTATDVBHD |
 |
 |
| Dataset #: 1 | Motif ID: 5 | Motif name: Motif 5 |
| Original motif | Reverse complement motif |
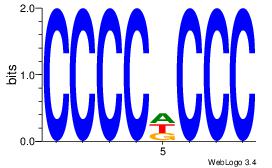
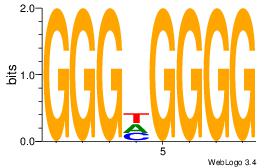
| Consensus sequence: CCCCDCCC | Consensus sequence: GGGDGGGG |
 |
 |
Best Matches for Motif ID 5 (Highest to Lowest)
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HBDRCCMCGCCCHHHD
----CCCCDCCC----
| Original motif | Reverse complement motif |
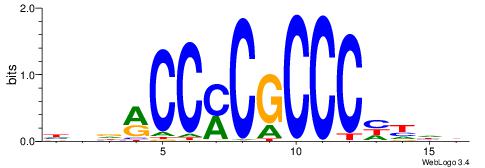
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
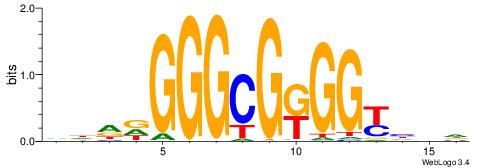 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0165797 |
Alignment:
GKRGGGGGGGGGGBD
-----GGGDGGGG--
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
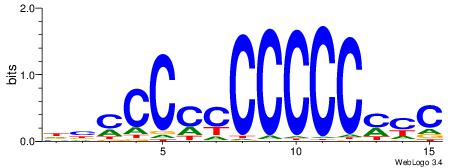 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.026633 |
Alignment:
BHHDYGGGGGGGGBVD
-----GGGDGGGG---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
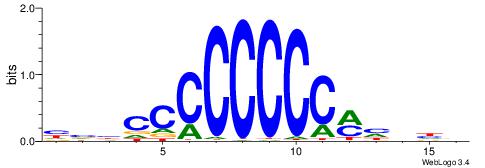 |
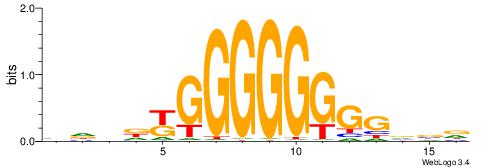 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0273225 |
Alignment:
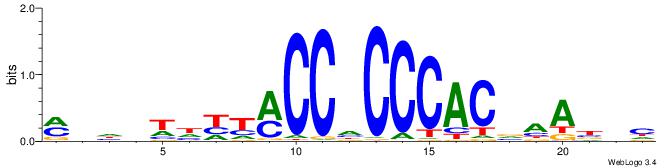
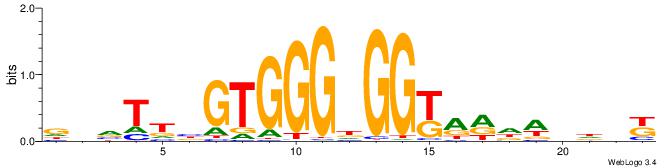
MHHHWHYTMCCHCCCACVAABVH
-------CCCCDCCC--------
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0364166 |
Alignment:
VHTGCGGGGGCGGH
-----GGGDGGGG-
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
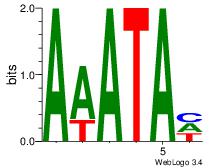
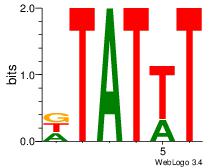
| Dataset #: 1 | Motif ID: 8 | Motif name: Motif 8 |
| Original motif | Reverse complement motif |
| Consensus sequence: AAATAH | Consensus sequence: DTATTT |
 |
 |
Best Matches for Motif ID 8 (Highest to Lowest)
| Motif ID: | UP00073 |
| Motif name: | Foxa2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
HHWAWGTAAAYAAAVDB
-------AAATAH----
| Original motif | Reverse complement motif |
| Consensus sequence: HHWAWGTAAAYAAAVDB | Consensus sequence: BDVTTTKTTTACWTWHH |
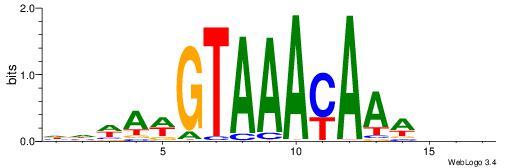 |
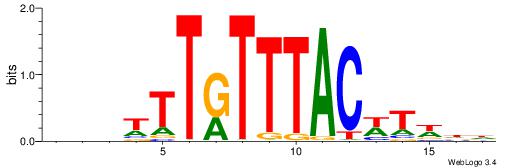 |
| Motif ID: | UP00166 |
| Motif name: | Barhl1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.00310877 |
Alignment:
DDVKTAATTGGTTDTH
---DTATTT-------
| Original motif | Reverse complement motif |
| Consensus sequence: HAHAACCAATTARBDH | Consensus sequence: DDVKTAATTGGTTDTH |
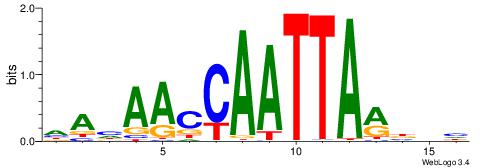 |
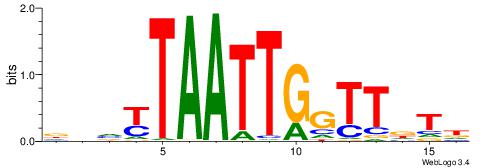 |
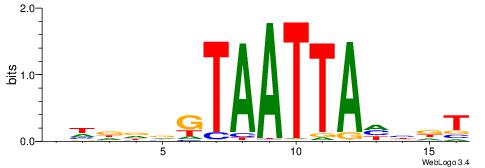
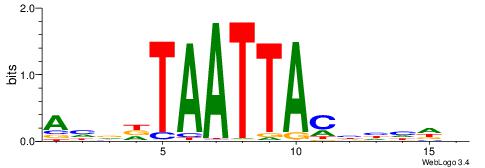
| Motif ID: | UP00242 |
| Motif name: | Hoxc8 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.00420475 |
Alignment:
BHDDDGTAATTAHHDT
-------AAATAH---
| Original motif | Reverse complement motif |
| Consensus sequence: BHDDDGTAATTAHHDT | Consensus sequence: AHDHTAATTACHHHHV |
 |
 |
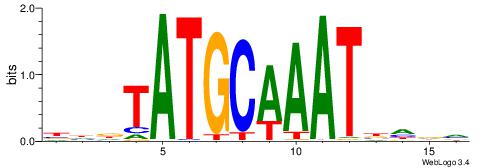
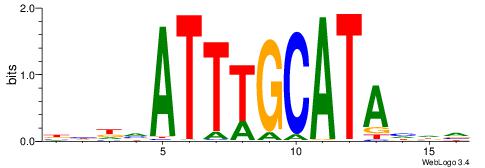
| Motif ID: | UP00179 |
| Motif name: | Pou2f3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.00508605 |
Alignment:
BHDTATGCAAATBHDV
--------AAATAH--
| Original motif | Reverse complement motif |
| Consensus sequence: BHDTATGCAAATBHDV | Consensus sequence: BHHVATTTGCATAHHV |
 |
 |
| Motif ID: | UP00202 |
| Motif name: | Dlx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.00517242 |
Alignment:
BHGVRDTAATTADY
-----DTATTT---
| Original motif | Reverse complement motif |
| Consensus sequence: BHGVRDTAATTADY | Consensus sequence: MDTAATTAHMBCHB |
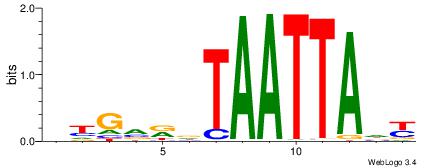 |
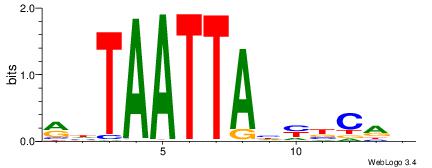 |
| Dataset #: 1 | Motif ID: 20 | Motif name: Motif 20 |
| Original motif | Reverse complement motif |
| Consensus sequence: CACGTR | Consensus sequence: MACGTG |
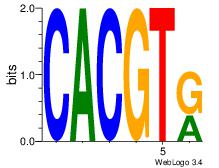 |
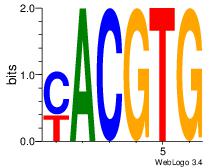 |
Best Matches for Motif ID 20 (Highest to Lowest)
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
DDHBHBSTCACGTGAHBBDHHM
--------CACGTR--------
| Original motif | Reverse complement motif |
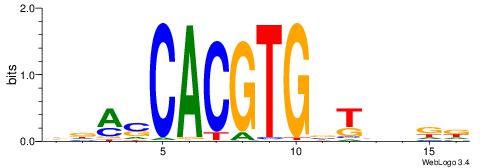
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
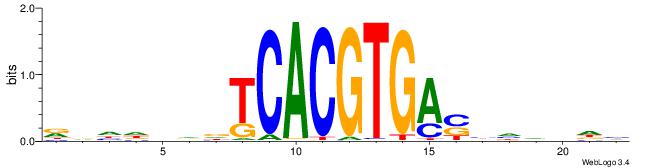 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.00504882 |
Alignment:
DDASCACGTGBTBVDD
----MACGTG------
| Original motif | Reverse complement motif |
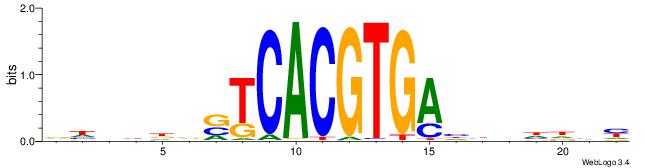
| Consensus sequence: DDASCACGTGBTBVDD | Consensus sequence: HHVBABCACGTGSTHD |
 |
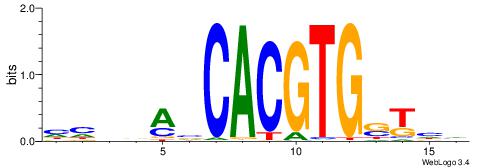 |
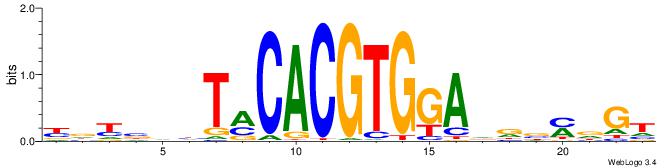
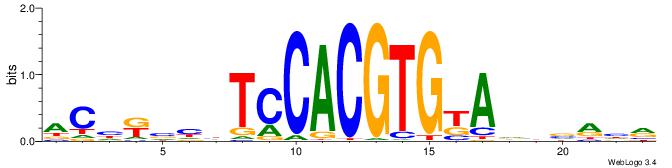
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.0050835 |
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
---------CACGTR--------
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
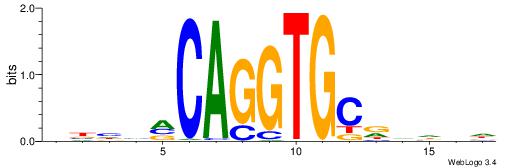
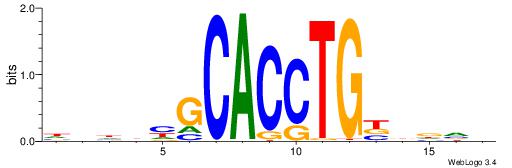
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.0336216 |
Alignment:
VBHHVCAGGTGCDVDHD
-----MACGTG------
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Motif ID: | UP00249 |
| Motif name: | Nkx2-5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.0339468 |
Alignment:
HDADCCACTTRAAWTT
-----CACGTR-----
| Original motif | Reverse complement motif |
| Consensus sequence: HDADCCACTTRAAWTT | Consensus sequence: AAWTTMAAGTGGHTDH |
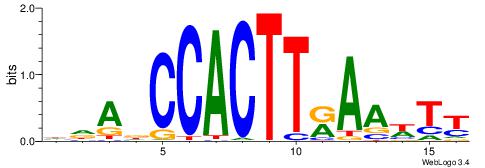 |
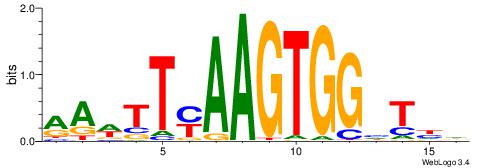 |
| Dataset #: 2 | Motif ID: 46 | Motif name: Motif 46 |
| Original motif | Reverse complement motif |
| Consensus sequence: AGRKGGCR | Consensus sequence: KGCCYKCT |
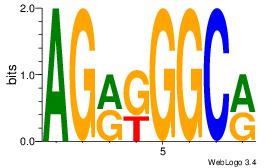 |
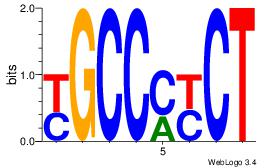 |
Best Matches for Motif ID 46 (Highest to Lowest)
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
BVVHAGGGGGCGRDHHB
----AGRKGGCR-----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0154192 |
Alignment:
VKBBAGGGGTCAHDBBH
----AGRKGGCR-----
| Original motif | Reverse complement motif |
| Consensus sequence: VKBBAGGGGTCAHDBBH | Consensus sequence: DBBHHTGACCCCTBBYV |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0160089 |
Alignment:
VHTGCGGGGGCGGH
----AGRKGGCR--
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
| Motif ID: | UP00010 |
| Motif name: | Tcfap2b_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0241611 |
Alignment:
HTGCCMTVKGGCMD
-KGCCYKCT-----
| Original motif | Reverse complement motif |
| Consensus sequence: DYGCCYBARGGCAH | Consensus sequence: HTGCCMTVKGGCMD |
 |
 |
| Motif ID: | UP00070 |
| Motif name: | Gcm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0267457 |
Alignment:
DDBVHATAGGGGRBRMB
-------AGRKGGCR--
| Original motif | Reverse complement motif |
| Consensus sequence: DDBVHATAGGGGRBRMB | Consensus sequence: BYMBKCCCCTATDVBHD |
 |
 |
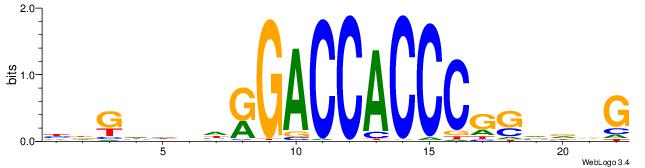
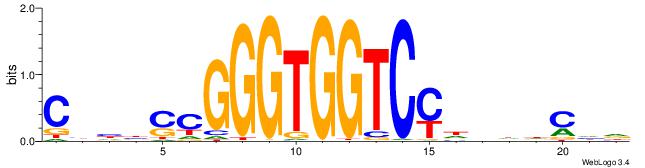
| Dataset #: 2 | Motif ID: 48 | Motif name: Motif 48 |
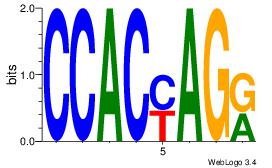
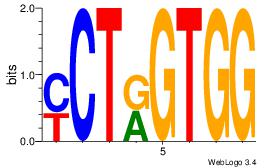
| Original motif | Reverse complement motif |
| Consensus sequence: CCACYAGR | Consensus sequence: MCTKGTGG |
 |
 |
Best Matches for Motif ID 48 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 10 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HGDHVHKGACCACCCGGBGBGDB
---------CCACYAGR------
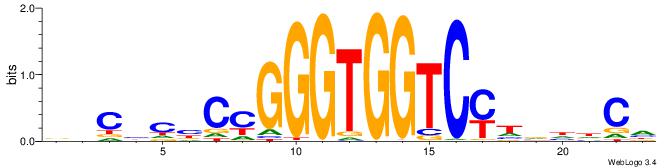
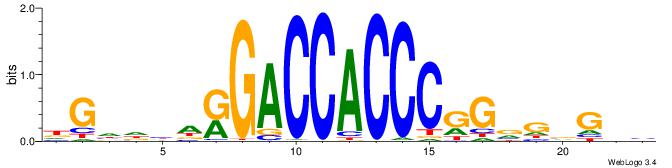
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
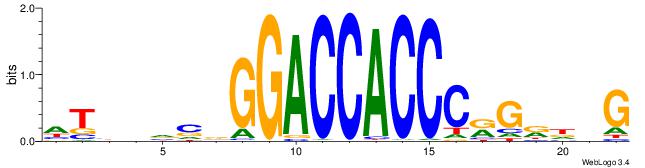
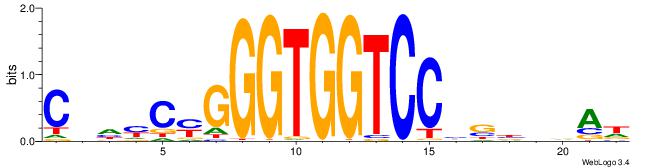
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 11 |
| Number of overlap: | 8 |
| Similarity score: | 0.000875703 |
Alignment:
CBDMCMGGGTGGTCCHVBVBAH
----MCTKGTGG----------
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
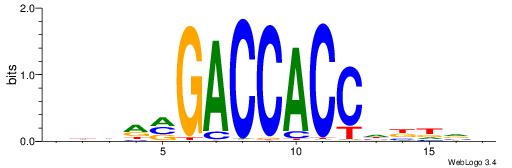
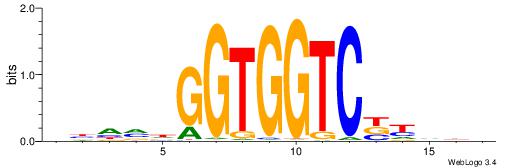
| Motif ID: | UP00047 |
| Motif name: | Zbtb7b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.00261558 |
Alignment:
BDDRVGACCACCHBDVB
-------CCACYAGR--
| Original motif | Reverse complement motif |
| Consensus sequence: BDDRVGACCACCHBDVB | Consensus sequence: BBDVHGGTGGTCBKDDB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.00296024 |
Alignment:
CVHBSCGGGTGGTCMHVHHCDH
----MCTKGTGG----------
| Original motif | Reverse complement motif |
| Consensus sequence: HDGHHBHRGACCACCCGSVDVG | Consensus sequence: CVHBSCGGGTGGTCMHVHHCDH |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0161251 |
Alignment:
VBHHVCAGGTGCDVDHD
----MCTKGTGG-----
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
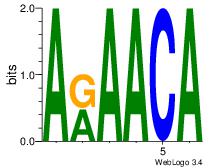
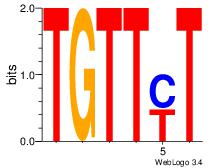
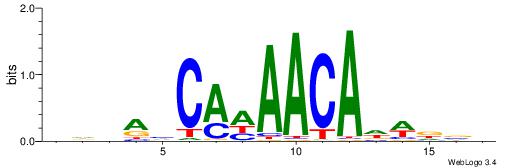
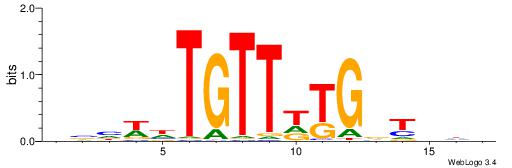
| Dataset #: 2 | Motif ID: 65 | Motif name: Motif 65 |
| Original motif | Reverse complement motif |
| Consensus sequence: ARAACA | Consensus sequence: TGTTMT |
 |
 |
Best Matches for Motif ID 65 (Highest to Lowest)
| Motif ID: | UP00101 |
| Motif name: | Sox12_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
DWTHAVAACAATHA
----ARAACA----
| Original motif | Reverse complement motif |
| Consensus sequence: THATTGTTVTHAWH | Consensus sequence: DWTHAVAACAATHA |
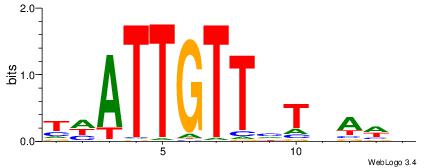 |
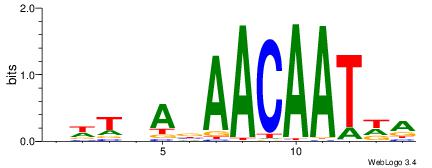 |
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.000395587 |
Alignment:
HBDDRAACAAAGRABHH
---ARAACA--------
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
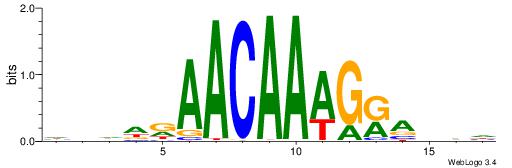 |
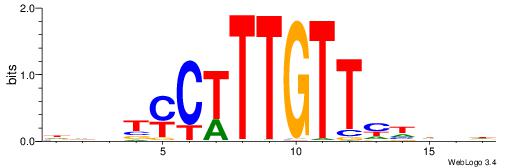 |
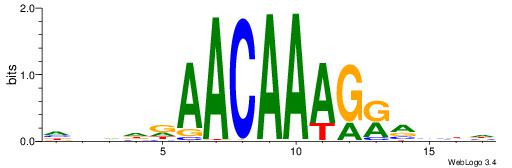
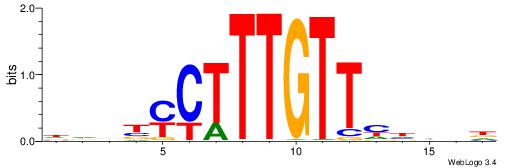
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.00357565 |
Alignment:
HHBBMCTTTGTTHDDBH
--------TGTTMT---
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
 |
 |
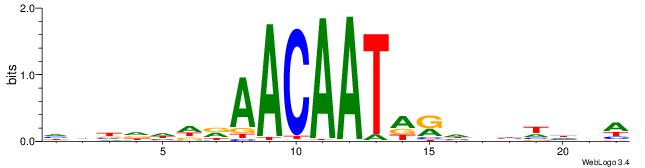
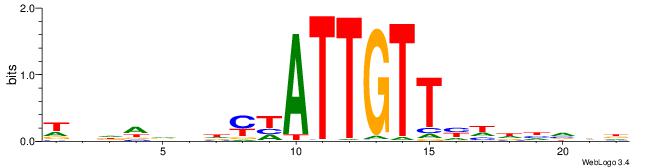
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.00753019 |
Alignment:
HHBDHDDAACAATDRHHHHHBW
-----ARAACA-----------
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00039 |
| Motif name: | Foxj3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0083324 |
Alignment:
DVHWDTGTTWTGDTBDH
-----TGTTMT------
| Original motif | Reverse complement motif |
| Consensus sequence: HDBAHCAWAACADWDVD | Consensus sequence: DVHWDTGTTWTGDTBDH |
 |
 |
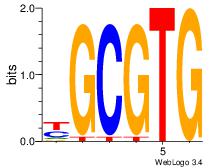
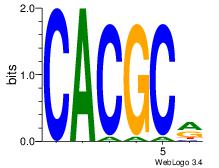
| Dataset #: 3 | Motif ID: 72 | Motif name: ArntAhr |
| Original motif | Reverse complement motif |
| Consensus sequence: YGCGTG | Consensus sequence: CACGCM |
 |
 |
Best Matches for Motif ID 72 (Highest to Lowest)
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
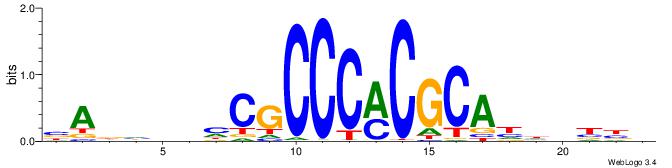
VVVDHHTGCGYGGGCGDHVHHTB
------YGCGTG-----------
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.00183587 |
Alignment:
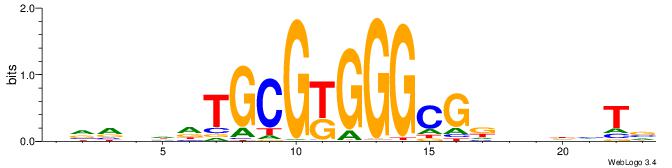
HHDGBCACGCCTWDB
-----CACGCM----
| Original motif | Reverse complement motif |
| Consensus sequence: BDWAGGCGTGBCHHD | Consensus sequence: HHDGBCACGCCTWDB |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.00348048 |
Alignment:
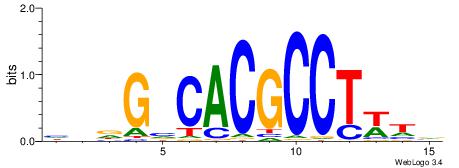
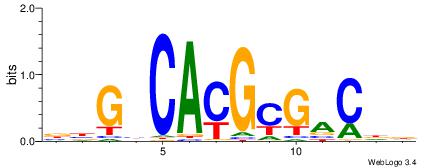
HDGTCGCGTGDCVB
----YGCGTG----
| Original motif | Reverse complement motif |
| Consensus sequence: BBGHCACGCGACDD | Consensus sequence: HDGTCGCGTGDCVB |
 |
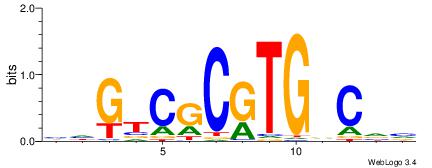 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.00526158 |
Alignment:
TCDAMVBGACGCMAKDDDVBBD
-------CACGCM---------
| Original motif | Reverse complement motif |
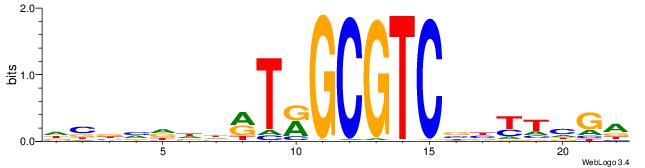
| Consensus sequence: DBBVDDDRTRGCGTCBBYTHGA | Consensus sequence: TCDAMVBGACGCMAKDDDVBBD |
 |
 |
| Motif ID: | UP00042 |
| Motif name: | Gm397_second |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.00558171 |
Alignment:
HBGYGTGTGTGCVRVD
---YGCGTG-------
| Original motif | Reverse complement motif |
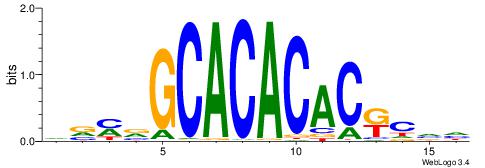
| Consensus sequence: DVMVGCACACACKCVH | Consensus sequence: HBGYGTGTGTGCVRVD |
 |
 |
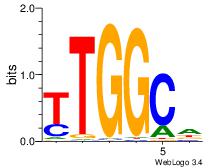
| Dataset #: 3 | Motif ID: 101 | Motif name: NFIC |
| Original motif | Reverse complement motif |
| Consensus sequence: TTGGCD | Consensus sequence: DGCCAA |
 |
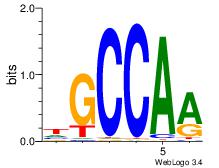 |
Best Matches for Motif ID 101 (Highest to Lowest)
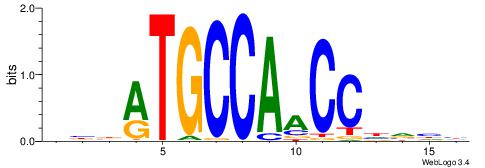
| Motif ID: | UP00035 |
| Motif name: | Hic1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
DBBATGCCAACCHVHH
----DGCCAA------
| Original motif | Reverse complement motif |
| Consensus sequence: DBBATGCCAACCHVHH | Consensus sequence: DDBHGGTTGGCATVBD |
 |
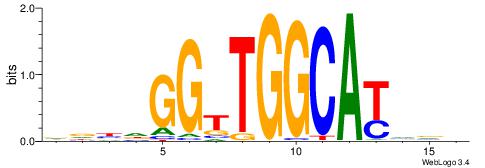 |
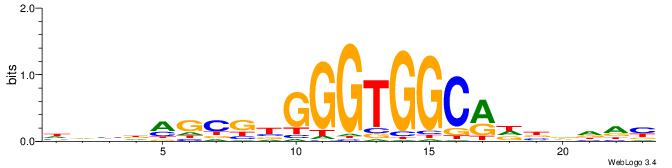
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 12 |
| Number of overlap: | 6 |
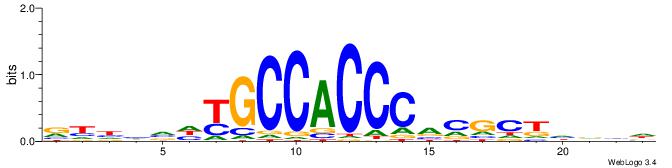
| Similarity score: | 0.0174701 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
------DGCCAA-----------
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00003 |
| Motif name: | E2F3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 6 |
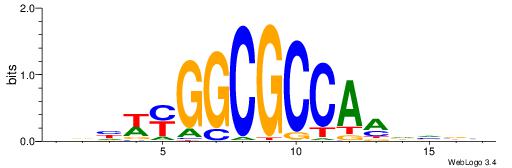
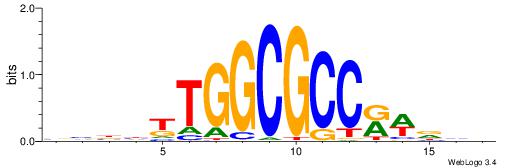
| Similarity score: | 0.0231918 |
Alignment:
BBBDYTGGCGCCKWDBD
----TTGGCD-------
| Original motif | Reverse complement motif |
| Consensus sequence: HBHWYGGCGCCAMDVBB | Consensus sequence: BBBDYTGGCGCCKWDBD |
 |
 |
| Motif ID: | UP00001 |
| Motif name: | E2F2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 6 |
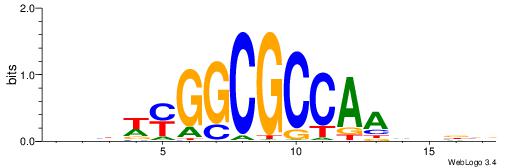
| Similarity score: | 0.0258343 |
Alignment:
BBBDTTGGCGCCKWVVD
----TTGGCD-------
| Original motif | Reverse complement motif |
| Consensus sequence: HVBWYGGCGCCAADVBB | Consensus sequence: BBBDTTGGCGCCKWVVD |
 |
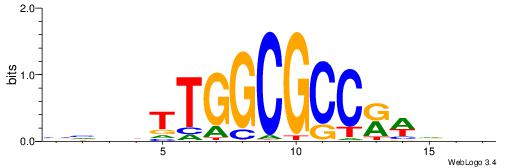 |
| Motif ID: | UP00060 |
| Motif name: | Max_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.0266372 |
Alignment:
HDGTCGCGTGDCVB
-------TTGGCD-
| Original motif | Reverse complement motif |
| Consensus sequence: BBGHCACGCGACDD | Consensus sequence: HDGTCGCGTGDCVB |
 |
 |
| Dataset #: 3 | Motif ID: 127 | Motif name: YY1 |
| Original motif | Reverse complement motif |
| Consensus sequence: RCCATB | Consensus sequence: BATGGM |
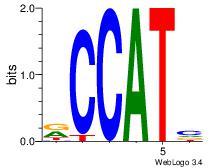 |
 |
Best Matches for Motif ID 127 (Highest to Lowest)
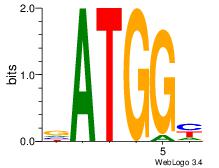
| Motif ID: | UP00033 |
| Motif name: | Zfp410_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
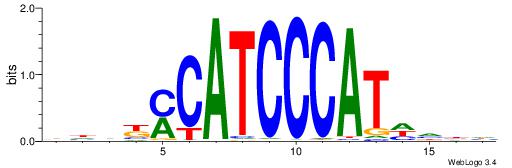
DVDDATGGGATGKMDDV
--------BATGGM---
| Original motif | Reverse complement motif |
| Consensus sequence: DVDDATGGGATGKMDDV | Consensus sequence: BDDYYCATCCCATDDBD |
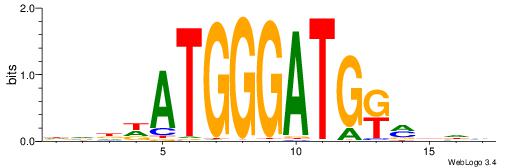 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.00378844 |
Alignment:
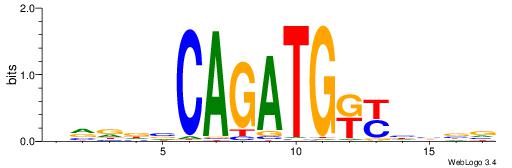
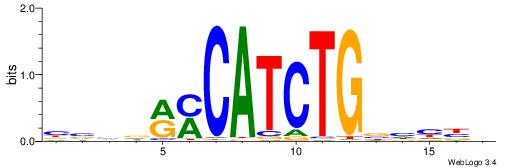
HVDDVCAGATGKYHBBV
-------BATGGM----
| Original motif | Reverse complement motif |
| Consensus sequence: HVDDVCAGATGKYHBBV | Consensus sequence: VBBDMYCATCTGVHHBH |
 |
 |
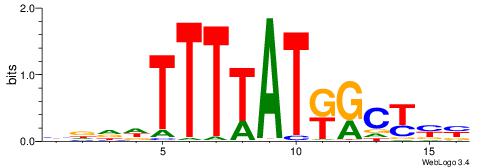
| Motif ID: | UP00207 |
| Motif name: | Hoxb9 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.0208704 |
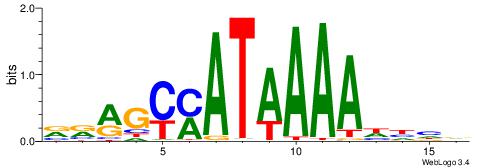
Alignment:
VRRGCMATAAAAHHHD
---RCCATB-------
| Original motif | Reverse complement motif |
| Consensus sequence: VRRGCMATAAAAHHHD | Consensus sequence: HDHHTTTTATRGCKMV |
 |
 |
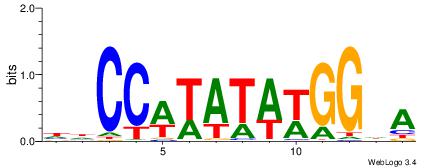
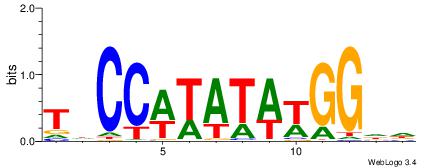
| Motif ID: | UP00077 |
| Motif name: | Srf_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.023319 |
Alignment:
HHCCWTATAWGGHA
-------BATGGM-
| Original motif | Reverse complement motif |
| Consensus sequence: HHCCWTATAWGGHA | Consensus sequence: THCCWTATAWGGHH |
 |
 |
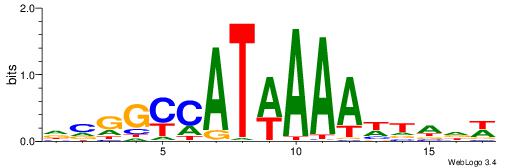
| Motif ID: | UP00213 |
| Motif name: | Hoxa9 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.0242984 |
Alignment:
DCGGYCATWAAAWTADW
---RCCATB--------
| Original motif | Reverse complement motif |
| Consensus sequence: DCGGYCATWAAAWTADW | Consensus sequence: WDTAWTTTWATGKCCGD |
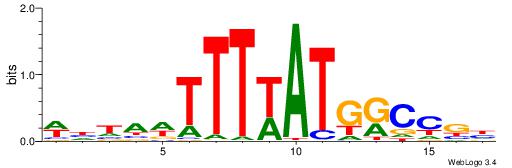
 |
 |