Top 10 Significant Motifs - Global Matching (Highest to Lowest)
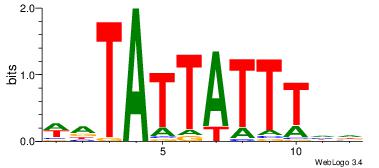
| Dataset #: 4 | Motif ID: 45 | Motif name: wbgTAAATAww |
| Original motif Consensus sequence: DBGTAAATAHD | Reverse complement motif Consensus sequence: DHTATTTACBD |
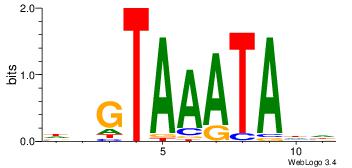 |
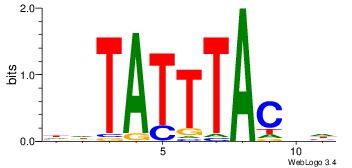 |
Best Matches for Top Significant Motif ID 45 (Highest to Lowest)
| Dataset #: | 5 |
| Motif ID: | 56 |
| Motif name: | TFM11 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 15 |
| Number of overlap: | 11 |
| Similarity score: | 0.0192768 |
Alignment:
TWVHWWWYTTTYTTTTTHTTTVWBH
--------------DHTATTTACBD
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA | Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 9 |
| Motif name: | Motif 9 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0250541 |
Alignment:
AATHATATWTHAAA
DHTATTTACBD---
| Original motif Consensus sequence: AATHATATWTHAAA | Reverse complement motif Consensus sequence: TTTDAWATATHATT |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 6 |
| Motif name: | Motif 6 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.027895 |
Alignment:
AATTYDGAARTAWW
---DBGTAAATAHD
| Original motif Consensus sequence: AATTYDGAARTAWW | Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 52 |
| Motif name: | TFM1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0283217 |
Alignment:
ABAAAAAAWHAAAAARAW
DBGTAAATAHD-------
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT | Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 53 |
| Motif name: | TFM3 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0308097 |
Alignment:
TWAAWTTVTGAAAAAHWW
------DBGTAAATAHD-
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA | Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 55 |
| Motif name: | TFM13 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0333874 |
Alignment:
ATKAAWTTTTRMAABAHHTW
-------DBGTAAATAHD--
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW | Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 4 |
| Motif name: | Motif 4 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0361187 |
Alignment:
AAAAWTTRCWT
DHTATTTACBD
| Original motif Consensus sequence: AAAAWTTRCWT | Reverse complement motif Consensus sequence: AWGKAAWTTTT |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 3 |
| Motif name: | Motif 3 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0410381 |
Alignment:
AWAAAWTWAAASWA
--DHTATTTACBD-
| Original motif Consensus sequence: AWAAAWTWAAASWA | Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 5 |
| Motif name: | Motif 5 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0477814 |
Alignment:
AWTAAATAYAATTT
---DHTATTTACBD
| Original motif Consensus sequence: AWTAAATAYAATTT | Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 31 |
| Motif name: | Pax5 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 10 |
| Number of overlap: | 11 |
| Similarity score: | 0.0512987 |
Alignment:
DGVBCABTGDWGCGKRRCSR
DBGTAAATAHD---------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR | Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
 |
 |
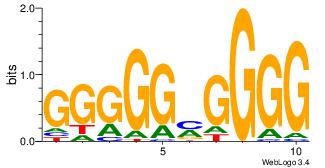
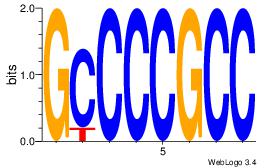
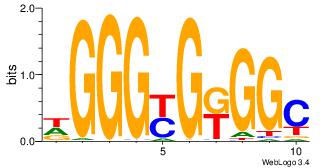
| Dataset #: 3 | Motif ID: 24 | Motif name: SP1 |
| Original motif Consensus sequence: CCCCKCCCCC | Reverse complement motif Consensus sequence: GGGGGYGGGG |
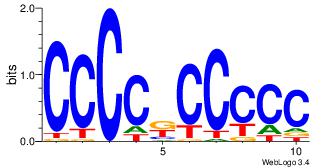 |
 |
Best Matches for Top Significant Motif ID 24 (Highest to Lowest)
| Dataset #: | 2 |
| Motif ID: | 7 |
| Motif name: | Motif 7 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.00451594 |
Alignment:
CSKCCCCGCCCCSY
---CCCCKCCCCC-
| Original motif Consensus sequence: CSKCCCCGCCCCSY | Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 36 |
| Motif name: | csGCCCCGCCCCsc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.00965796 |
Alignment:
HVGCCCCGCCCCBB
---CCCCKCCCCC-
| Original motif Consensus sequence: HVGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 38 |
| Motif name: | cccGCCCCGCCCCsb |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0127292 |
Alignment:
BCCGCCCCGCCCCBB
----CCCCKCCCCC-
| Original motif Consensus sequence: BCCGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 50 |
| Motif name: | TFF11 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0174405 |
Alignment:
GGMGGRGGCGGVGC
---GGGGGYGGGG-
| Original motif Consensus sequence: GGMGGRGGCGGVGC | Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 54 |
| Motif name: | TFM12 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0301548 |
Alignment:
CYYCBBCYYYTCCHCCTYYY
------CCCCKCCCCC----
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY | Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 51 |
| Motif name: | TFM2 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0329167 |
Alignment:
RGRGGAGRRGGHGGDG
------GGGGGYGGGG
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG | Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 49 |
| Motif name: | TFF1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0557738 |
Alignment:
GCVGCGGCBCCG
-CCCCKCCCCC-
| Original motif Consensus sequence: CGGVGCCGCVGC | Reverse complement motif Consensus sequence: GCVGCGGCBCCG |
 |
 |
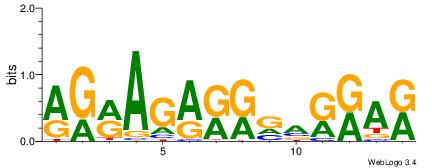
| Dataset #: | 2 |
| Motif ID: | 2 |
| Motif name: | Motif 2 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0648214 |
Alignment:
MTMMTCMMTCTKCK
----CCCCKCCCCC
| Original motif Consensus sequence: RGRAGARRGARRAR | Reverse complement motif Consensus sequence: MTMMTCMMTCTKCK |
 |
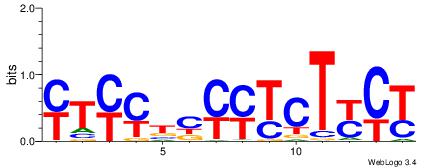 |
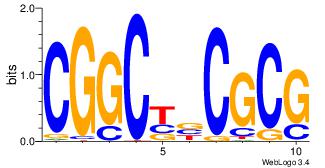
| Dataset #: | 5 |
| Motif ID: | 48 |
| Motif name: | TFW3 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0762976 |
Alignment:
CGGCYBCGCG
CCCCKCCCCC
| Original motif Consensus sequence: CGGCYBCGCG | Reverse complement motif Consensus sequence: CGCGBMGCCG |
 |
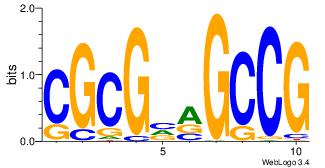 |
| Dataset #: | 4 |
| Motif ID: | 40 |
| Motif name: | kcACCTGCAgc |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0841987 |
Alignment:
GBTGCAGGTGB
GGGGGYGGGG-
| Original motif Consensus sequence: BCACCTGCABC | Reverse complement motif Consensus sequence: GBTGCAGGTGB |
 |
 |
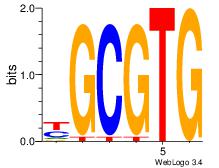
| Dataset #: 3 | Motif ID: 32 | Motif name: ArntAhr |
| Original motif Consensus sequence: YGCGTG | Reverse complement motif Consensus sequence: CACGCM |
 |
 |
Best Matches for Top Significant Motif ID 32 (Highest to Lowest)
| Dataset #: | 4 |
| Motif ID: | 36 |
| Motif name: | csGCCCCGCCCCsc |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0396403 |
Alignment:
HVGCCCCGCCCCBB
----CACGCM----
| Original motif Consensus sequence: HVGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
 |
 |
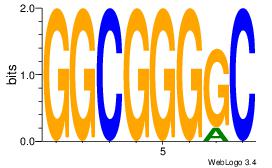
| Dataset #: | 1 |
| Motif ID: | 1 |
| Motif name: | Motif 1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.040625 |
Alignment:
GCCCCGCC
--CACGCM
| Original motif Consensus sequence: GGCGGGGC | Reverse complement motif Consensus sequence: GCCCCGCC |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 42 |
| Motif name: | sSGTCACGTGACSs |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0412037 |
Alignment:
SGGTCACGTGACCS
----YGCGTG----
| Original motif Consensus sequence: SGGTCACGTGACCS | Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 38 |
| Motif name: | cccGCCCCGCCCCsb |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0425089 |
Alignment:
BCCGCCCCGCCCCBB
-----CACGCM----
| Original motif Consensus sequence: BCCGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 7 |
| Motif name: | Motif 7 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0465867 |
Alignment:
MSGGGGCGGGGYSG
----YGCGTG----
| Original motif Consensus sequence: CSKCCCCGCCCCSY | Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 47 |
| Motif name: | TFW2 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.0469722 |
Alignment:
SCGCGCGG
-YGCGTG-
| Original motif Consensus sequence: SCGCGCGG | Reverse complement motif Consensus sequence: CCGCGCGS |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 48 |
| Motif name: | TFW3 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0586806 |
Alignment:
CGGCYBCGCG
----YGCGTG
| Original motif Consensus sequence: CGGCYBCGCG | Reverse complement motif Consensus sequence: CGCGBMGCCG |
 |
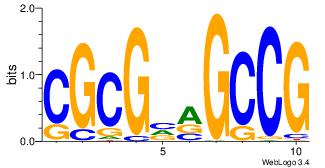 |
| Dataset #: | 4 |
| Motif ID: | 40 |
| Motif name: | kcACCTGCAgc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0637286 |
Alignment:
BCACCTGCABC
-YGCGTG----
| Original motif Consensus sequence: BCACCTGCABC | Reverse complement motif Consensus sequence: GBTGCAGGTGB |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 50 |
| Motif name: | TFF11 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.066121 |
Alignment:
GCVCCGCCMCCYCC
--CACGCM------
| Original motif Consensus sequence: GGMGGRGGCGGVGC | Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 46 |
| Motif name: | TFW1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0756826 |
Alignment:
CGCGAC
CACGCM
| Original motif Consensus sequence: GTCGCG | Reverse complement motif Consensus sequence: CGCGAC |
 |
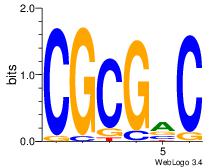 |
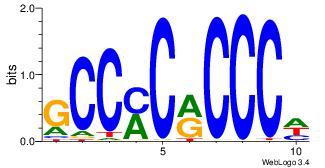
| Dataset #: 1 | Motif ID: 1 | Motif name: Motif 1 |
| Original motif Consensus sequence: GGCGGGGC | Reverse complement motif Consensus sequence: GCCCCGCC |
 |
 |
Best Matches for Top Significant Motif ID 1 (Highest to Lowest)
| Dataset #: | 4 |
| Motif ID: | 36 |
| Motif name: | csGCCCCGCCCCsc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
BBGGGGCGGGGCVD
----GGCGGGGC--
| Original motif Consensus sequence: HVGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 38 |
| Motif name: | cccGCCCCGCCCCsb |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.00250079 |
Alignment:
BCCGCCCCGCCCCBB
---GCCCCGCC----
| Original motif Consensus sequence: BCCGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 7 |
| Motif name: | Motif 7 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0149958 |
Alignment:
MSGGGGCGGGGYSG
----GGCGGGGC--
| Original motif Consensus sequence: CSKCCCCGCCCCSY | Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 50 |
| Motif name: | TFF11 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.0348969 |
Alignment:
GGMGGRGGCGGVGC
------GGCGGGGC
| Original motif Consensus sequence: GGMGGRGGCGGVGC | Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 27 |
| Motif name: | Klf4 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0349983 |
Alignment:
DGGGYGKGGC
--GGCGGGGC
| Original motif Consensus sequence: DGGGYGKGGC | Reverse complement motif Consensus sequence: GCCYCMCCCD |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 22 |
| Motif name: | Zfx |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0708224 |
Alignment:
VAGGCCBBGGCVBB
---GCCCCGCC---
| Original motif Consensus sequence: BBVGCCBVGGCCTV | Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 48 |
| Motif name: | TFW3 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.071681 |
Alignment:
CGCGBMGCCG
-GCCCCGCC-
| Original motif Consensus sequence: CGGCYBCGCG | Reverse complement motif Consensus sequence: CGCGBMGCCG |
 |
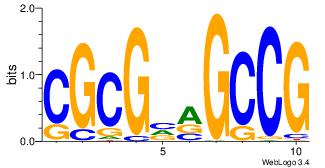 |
| Dataset #: | 5 |
| Motif ID: | 49 |
| Motif name: | TFF1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0803996 |
Alignment:
CGGVGCCGCVGC
--GCCCCGCC--
| Original motif Consensus sequence: CGGVGCCGCVGC | Reverse complement motif Consensus sequence: GCVGCGGCBCCG |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 31 |
| Motif name: | Pax5 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0853847 |
Alignment:
DGVBCABTGDWGCGKRRCSR
----------GGCGGGGC--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR | Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 23 |
| Motif name: | Egr1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.0878731 |
Alignment:
YCGCCCACGCH
---GCCCCGCC
| Original motif Consensus sequence: HGCGTGGGCGK | Reverse complement motif Consensus sequence: YCGCCCACGCH |
 |
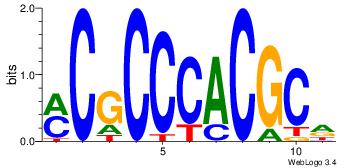 |
| Dataset #: | 5 |
| Motif ID: | 51 |
| Motif name: | TFM2 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0928252 |
Alignment:
CHCCBCCKMCTCCKCM
--GCCCCGCC------
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG | Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
 |
 |
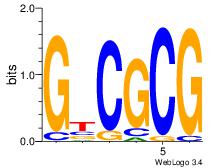
| Dataset #: 5 | Motif ID: 46 | Motif name: TFW1 |
| Original motif Consensus sequence: GTCGCG | Reverse complement motif Consensus sequence: CGCGAC |
 |
 |
Best Matches for Top Significant Motif ID 46 (Highest to Lowest)
| Dataset #: | 4 |
| Motif ID: | 42 |
| Motif name: | sSGTCACGTGACSs |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0375102 |
Alignment:
SGGTCACGTGACCS
--GTCGCG------
| Original motif Consensus sequence: SGGTCACGTGACCS | Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 33 |
| Motif name: | Mycn |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0537993 |
Alignment:
GCCACGTGSD
----GTCGCG
| Original motif Consensus sequence: HSCACGTGGC | Reverse complement motif Consensus sequence: GCCACGTGSD |
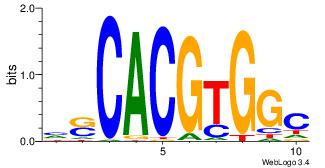 |
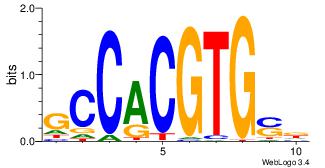 |
| Dataset #: | 4 |
| Motif ID: | 36 |
| Motif name: | csGCCCCGCCCCsc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0539 |
Alignment:
HVGCCCCGCCCCBB
--GTCGCG------
| Original motif Consensus sequence: HVGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 1 |
| Motif name: | Motif 1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0554263 |
Alignment:
GCCCCGCC
--GTCGCG
| Original motif Consensus sequence: GGCGGGGC | Reverse complement motif Consensus sequence: GCCCCGCC |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 38 |
| Motif name: | cccGCCCCGCCCCsb |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.0558733 |
Alignment:
BBGGGGCGGGGCGGB
-------GTCGCG--
| Original motif Consensus sequence: BCCGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 7 |
| Motif name: | Motif 7 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0644512 |
Alignment:
CSKCCCCGCCCCSY
--CGCGAC------
| Original motif Consensus sequence: CSKCCCCGCCCCSY | Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 34 |
| Motif name: | Myc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
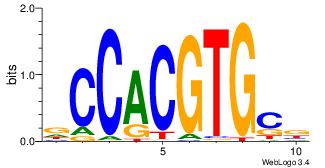
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0664905 |
Alignment:
DCCACGTGCV
----GTCGCG
| Original motif Consensus sequence: VGCACGTGGH | Reverse complement motif Consensus sequence: DCCACGTGCV |
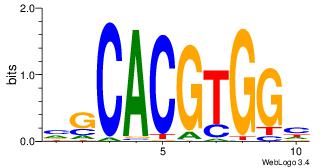 |
 |
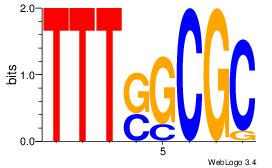
| Dataset #: | 3 |
| Motif ID: | 28 |
| Motif name: | E2F1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.0698654 |
Alignment:
TTTSGCGC
-GTCGCG-
| Original motif Consensus sequence: TTTSGCGC | Reverse complement motif Consensus sequence: GCGCSAAA |
 |
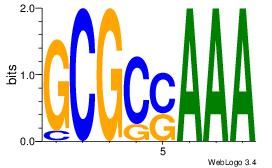 |
| Dataset #: | 3 |
| Motif ID: | 22 |
| Motif name: | Zfx |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.069909 |
Alignment:
BBVGCCBVGGCCTV
---GTCGCG-----
| Original motif Consensus sequence: BBVGCCBVGGCCTV | Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 31 |
| Motif name: | Pax5 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.072456 |
Alignment:
MSGKKRCGCWDCABTGBBCD
------CGCGAC--------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR | Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
 |
 |
| Dataset #: 5 | Motif ID: 47 | Motif name: TFW2 |
| Original motif Consensus sequence: SCGCGCGG | Reverse complement motif Consensus sequence: CCGCGCGS |
 |
 |
Best Matches for Top Significant Motif ID 47 (Highest to Lowest)
| Dataset #: | 4 |
| Motif ID: | 36 |
| Motif name: | csGCCCCGCCCCsc |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0456233 |
Alignment:
HVGCCCCGCCCCBB
-CCGCGCGS-----
| Original motif Consensus sequence: HVGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 38 |
| Motif name: | cccGCCCCGCCCCsb |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.045686 |
Alignment:
BBGGGGCGGGGCGGB
------SCGCGCGG-
| Original motif Consensus sequence: BCCGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 7 |
| Motif name: | Motif 7 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0504058 |
Alignment:
MSGGGGCGGGGYSG
-----SCGCGCGG-
| Original motif Consensus sequence: CSKCCCCGCCCCSY | Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 33 |
| Motif name: | Mycn |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0527532 |
Alignment:
HSCACGTGGC
-SCGCGCGG-
| Original motif Consensus sequence: HSCACGTGGC | Reverse complement motif Consensus sequence: GCCACGTGSD |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 34 |
| Motif name: | Myc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0574543 |
Alignment:
VGCACGTGGH
-SCGCGCGG-
| Original motif Consensus sequence: VGCACGTGGH | Reverse complement motif Consensus sequence: DCCACGTGCV |
 |
 |
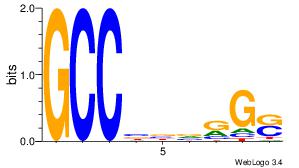
| Dataset #: | 3 |
| Motif ID: | 25 |
| Motif name: | TFAP2A |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.0678925 |
Alignment:
SCMVBBGGC
-SCGCGCGG
| Original motif Consensus sequence: GCCBBVRGS | Reverse complement motif Consensus sequence: SCMVBBGGC |
 |
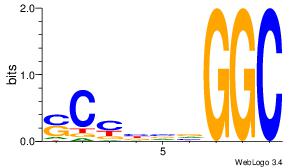 |
| Dataset #: | 3 |
| Motif ID: | 24 |
| Motif name: | SP1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.07018 |
Alignment:
CCCCKCCCCC
--CCGCGCGS
| Original motif Consensus sequence: CCCCKCCCCC | Reverse complement motif Consensus sequence: GGGGGYGGGG |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 22 |
| Motif name: | Zfx |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0710499 |
Alignment:
BBVGCCBVGGCCTV
--SCGCGCGG----
| Original motif Consensus sequence: BBVGCCBVGGCCTV | Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 27 |
| Motif name: | Klf4 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0768965 |
Alignment:
DGGGYGKGGC
-SCGCGCGG-
| Original motif Consensus sequence: DGGGYGKGGC | Reverse complement motif Consensus sequence: GCCYCMCCCD |
 |
 |
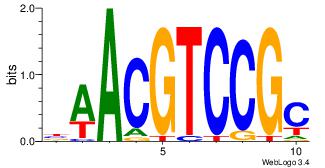
| Dataset #: | 3 |
| Motif ID: | 26 |
| Motif name: | MIZF |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0837414 |
Alignment:
GCGGACGTTV
--SCGCGCGG
| Original motif Consensus sequence: BAACGTCCGC | Reverse complement motif Consensus sequence: GCGGACGTTV |
 |
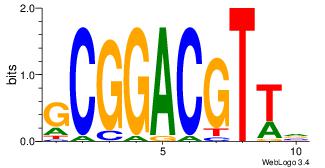 |
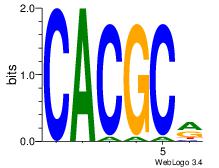
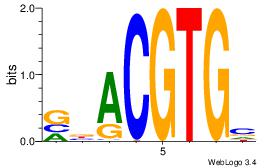
| Dataset #: 3 | Motif ID: 29 | Motif name: HIF1AARNT |
| Original motif Consensus sequence: VBACGTGV | Reverse complement motif Consensus sequence: VCACGTBV |
 |
 |
Best Matches for Top Significant Motif ID 29 (Highest to Lowest)
| Dataset #: | 4 |
| Motif ID: | 40 |
| Motif name: | kcACCTGCAgc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0295473 |
Alignment:
BCACCTGCABC
VBACGTGV---
| Original motif Consensus sequence: BCACCTGCABC | Reverse complement motif Consensus sequence: GBTGCAGGTGB |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 42 |
| Motif name: | sSGTCACGTGACSs |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0314837 |
Alignment:
SGGTCACGTGACCS
---VCACGTBV---
| Original motif Consensus sequence: SGGTCACGTGACCS | Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
 |
 |
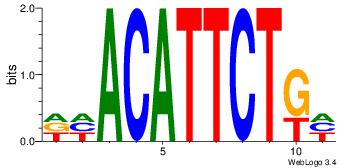
| Dataset #: | 4 |
| Motif ID: | 44 |
| Motif name: | dhACATTCTkh |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0640024 |
Alignment:
DHACATTCTGH
VCACGTBV---
| Original motif Consensus sequence: DHACATTCTGH | Reverse complement motif Consensus sequence: HCAGAATGTHD |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 47 |
| Motif name: | TFW2 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.0660185 |
Alignment:
CCGCGCGS
VBACGTGV
| Original motif Consensus sequence: SCGCGCGG | Reverse complement motif Consensus sequence: CCGCGCGS |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 50 |
| Motif name: | TFF11 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.067219 |
Alignment:
GCVCCGCCMCCYCC
------VCACGTBV
| Original motif Consensus sequence: GGMGGRGGCGGVGC | Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 51 |
| Motif name: | TFM2 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0698117 |
Alignment:
RGRGGAGRRGGHGGDG
---VBACGTGV-----
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG | Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 36 |
| Motif name: | csGCCCCGCCCCsc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0711168 |
Alignment:
BBGGGGCGGGGCVD
---VBACGTGV---
| Original motif Consensus sequence: HVGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 38 |
| Motif name: | cccGCCCCGCCCCsb |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0735453 |
Alignment:
BBGGGGCGGGGCGGB
---VBACGTGV----
| Original motif Consensus sequence: BCCGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 54 |
| Motif name: | TFM12 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 12 |
| Number of overlap: | 8 |
| Similarity score: | 0.0757612 |
Alignment:
KKKAGGDGGAKKMGBBGKMG
-VBACGTGV-----------
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY | Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 7 |
| Motif name: | Motif 7 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0775335 |
Alignment:
CSKCCCCGCCCCSY
---VCACGTBV---
| Original motif Consensus sequence: CSKCCCCGCCCCSY | Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
 |
 |
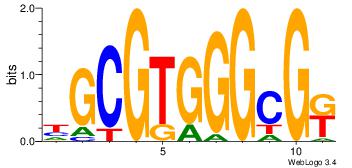
| Dataset #: 3 | Motif ID: 25 | Motif name: TFAP2A |
| Original motif Consensus sequence: GCCBBVRGS | Reverse complement motif Consensus sequence: SCMVBBGGC |
 |
 |
Best Matches for Top Significant Motif ID 25 (Highest to Lowest)
| Dataset #: | 5 |
| Motif ID: | 50 |
| Motif name: | TFF11 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 9 |
| Similarity score: | 0.0415732 |
Alignment:
GGMGGRGGCGGVGC
SCMVBBGGC-----
| Original motif Consensus sequence: GGMGGRGGCGGVGC | Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 48 |
| Motif name: | TFW3 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 9 |
| Similarity score: | 0.0552877 |
Alignment:
CGGCYBCGCG
-GCCBBVRGS
| Original motif Consensus sequence: CGGCYBCGCG | Reverse complement motif Consensus sequence: CGCGBMGCCG |
 |
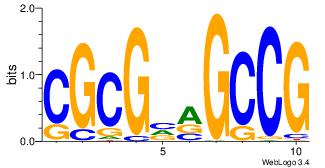 |
| Dataset #: | 5 |
| Motif ID: | 54 |
| Motif name: | TFM12 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 11 |
| Number of overlap: | 9 |
| Similarity score: | 0.0630429 |
Alignment:
KKKAGGDGGAKKMGBBGKMG
-SCMVBBGGC----------
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY | Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 49 |
| Motif name: | TFF1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 9 |
| Similarity score: | 0.0669772 |
Alignment:
GCVGCGGCBCCG
SCMVBBGGC---
| Original motif Consensus sequence: CGGVGCCGCVGC | Reverse complement motif Consensus sequence: GCVGCGGCBCCG |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 51 |
| Motif name: | TFM2 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 9 |
| Similarity score: | 0.069755 |
Alignment:
CHCCBCCKMCTCCKCM
SCMVBBGGC-------
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG | Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 2 |
| Motif name: | Motif 2 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 9 |
| Similarity score: | 0.0715961 |
Alignment:
MTMMTCMMTCTKCK
----SCMVBBGGC-
| Original motif Consensus sequence: RGRAGARRGARRAR | Reverse complement motif Consensus sequence: MTMMTCMMTCTKCK |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 38 |
| Motif name: | cccGCCCCGCCCCsb |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 9 |
| Similarity score: | 0.0719489 |
Alignment:
BCCGCCCCGCCCCBB
-SCMVBBGGC-----
| Original motif Consensus sequence: BCCGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 7 |
| Motif name: | Motif 7 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 9 |
| Similarity score: | 0.0781606 |
Alignment:
MSGGGGCGGGGYSG
GCCBBVRGS-----
| Original motif Consensus sequence: CSKCCCCGCCCCSY | Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 36 |
| Motif name: | csGCCCCGCCCCsc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 9 |
| Similarity score: | 0.0800026 |
Alignment:
BBGGGGCGGGGCVD
-----GCCBBVRGS
| Original motif Consensus sequence: HVGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 40 |
| Motif name: | kcACCTGCAgc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 9 |
| Similarity score: | 0.0857416 |
Alignment:
BCACCTGCABC
--GCCBBVRGS
| Original motif Consensus sequence: BCACCTGCABC | Reverse complement motif Consensus sequence: GBTGCAGGTGB |
 |
 |
| Dataset #: 4 | Motif ID: 41 | Motif name: wwAAATAATAtw |
| Original motif Consensus sequence: HDAAATAATADD | Reverse complement motif Consensus sequence: DDTATTATTTDH |
 |
 |
Best Matches for Top Significant Motif ID 41 (Highest to Lowest)
| Dataset #: | 2 |
| Motif ID: | 3 |
| Motif name: | Motif 3 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0536028 |
Alignment:
AWAAAWTWAAASWA
-HDAAATAATADD-
| Original motif Consensus sequence: AWAAAWTWAAASWA | Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 52 |
| Motif name: | TFM1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 12 |
| Similarity score: | 0.0565705 |
Alignment:
WTKTTTTTHWTTTTTTBT
------DDTATTATTTDH
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT | Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 56 |
| Motif name: | TFM11 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0575935 |
Alignment:
HDWVAAAHAAAAAMAAAMWWWHBWA
--HDAAATAATADD-----------
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA | Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 9 |
| Motif name: | Motif 9 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0591821 |
Alignment:
AATHATATWTHAAA
--DDTATTATTTDH
| Original motif Consensus sequence: AATHATATWTHAAA | Reverse complement motif Consensus sequence: TTTDAWATATHATT |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 55 |
| Motif name: | TFM13 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0712963 |
Alignment:
WAHHTVTTYKAAAAWTTRAT
--------HDAAATAATADD
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW | Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 5 |
| Motif name: | Motif 5 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0715278 |
Alignment:
AAATTKTATTTAWT
-DDTATTATTTDH-
| Original motif Consensus sequence: AWTAAATAYAATTT | Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 53 |
| Motif name: | TFM3 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0733516 |
Alignment:
WWHTTTTTCABAAWTTWA
------DDTATTATTTDH
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA | Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 16 |
| Motif name: | Motif 16 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0912037 |
Alignment:
TCAAAATKAWTTGT
--DDTATTATTTDH
| Original motif Consensus sequence: ACAAWTRATTTTGA | Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 6 |
| Motif name: | Motif 6 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0924576 |
Alignment:
WWTAKTTCDKAATT
DDTATTATTTDH--
| Original motif Consensus sequence: AATTYDGAARTAWW | Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 8 |
| Motif name: | Motif 8 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0989198 |
Alignment:
TGATTTTAWKATTT
-DDTATTATTTDH-
| Original motif Consensus sequence: AAATRWTAAAATCA | Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
 |
 |
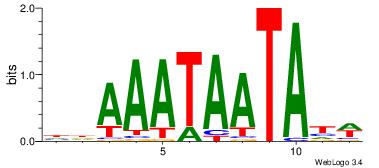
| Dataset #: 4 | Motif ID: 37 | Motif name: tkAAATAATAtw |
| Original motif Consensus sequence: HDAAATAATAHW | Reverse complement motif Consensus sequence: WHTATTATTTDH |
 |
 |
Best Matches for Top Significant Motif ID 37 (Highest to Lowest)
| Dataset #: | 2 |
| Motif ID: | 3 |
| Motif name: | Motif 3 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0522317 |
Alignment:
TWSTTTWAWTTTWT
-WHTATTATTTDH-
| Original motif Consensus sequence: AWAAAWTWAAASWA | Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 56 |
| Motif name: | TFM11 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0590505 |
Alignment:
TWVHWWWYTTTYTTTTTHTTTVWBH
-----------WHTATTATTTDH--
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA | Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 52 |
| Motif name: | TFM1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0637702 |
Alignment:
ABAAAAAAWHAAAAARAW
HDAAATAATAHW------
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT | Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 9 |
| Motif name: | Motif 9 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0641975 |
Alignment:
AATHATATWTHAAA
--WHTATTATTTDH
| Original motif Consensus sequence: AATHATATWTHAAA | Reverse complement motif Consensus sequence: TTTDAWATATHATT |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 5 |
| Motif name: | Motif 5 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0704475 |
Alignment:
AAATTKTATTTAWT
-WHTATTATTTDH-
| Original motif Consensus sequence: AWTAAATAYAATTT | Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 55 |
| Motif name: | TFM13 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0704475 |
Alignment:
ATKAAWTTTTRMAABAHHTW
WHTATTATTTDH--------
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW | Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 53 |
| Motif name: | TFM3 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0763819 |
Alignment:
WWHTTTTTCABAAWTTWA
------WHTATTATTTDH
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA | Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 16 |
| Motif name: | Motif 16 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0864198 |
Alignment:
TCAAAATKAWTTGT
--WHTATTATTTDH
| Original motif Consensus sequence: ACAAWTRATTTTGA | Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 6 |
| Motif name: | Motif 6 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.09076 |
Alignment:
WWTAKTTCDKAATT
WHTATTATTTDH--
| Original motif Consensus sequence: AATTYDGAARTAWW | Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
 |
 |
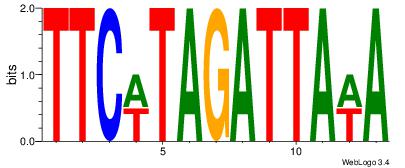
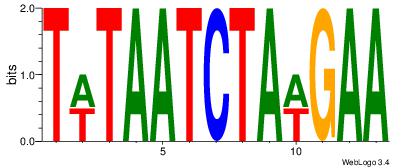
| Dataset #: | 2 |
| Motif ID: | 19 |
| Motif name: | Motif 19 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.096142 |
Alignment:
TWTAATCTAWGAA
WHTATTATTTDH-
| Original motif Consensus sequence: TTCWTAGATTAWA | Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
 |
 |