Top 10 Significant Motifs - Global Matching (Highest to Lowest)
| Dataset #: 4 |
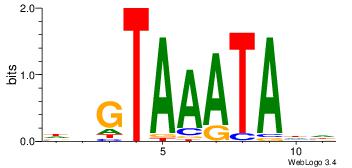
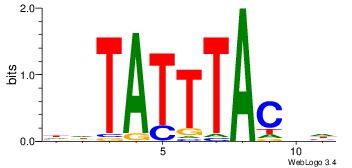
Motif ID: 45 |
Motif name: wbgTAAATAww |
| Original motif Consensus sequence: DBGTAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTACBD |
|
|
 |
 |
Best Matches for Top Significant Motif ID 45 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
15 |
| Number of overlap: |
11 |
| Similarity score: |
0.0192768 |
Alignment:
TWVHWWWYTTTYTTTTTHTTTVWBH
--------------DHTATTTACBD
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA |
Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0250541 |
Alignment:
AATHATATWTHAAA
DHTATTTACBD---
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.027895 |
Alignment:
AATTYDGAARTAWW
---DBGTAAATAHD
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
52 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0283217 |
Alignment:
ABAAAAAAWHAAAAARAW
DBGTAAATAHD-------
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT |
Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0308097 |
Alignment:
TWAAWTTVTGAAAAAHWW
------DBGTAAATAHD-
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA |
Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0333874 |
Alignment:
ATKAAWTTTTRMAABAHHTW
-------DBGTAAATAHD--
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0361187 |
Alignment:
AAAAWTTRCWT
DHTATTTACBD
| Original motif Consensus sequence: AAAAWTTRCWT |
Reverse complement motif Consensus sequence: AWGKAAWTTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0410381 |
Alignment:
AWAAAWTWAAASWA
--DHTATTTACBD-
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0477814 |
Alignment:
AWTAAATAYAATTT
---DHTATTTACBD
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
10 |
| Number of overlap: |
11 |
| Similarity score: |
0.0512987 |
Alignment:
DGVBCABTGDWGCGKRRCSR
DBGTAAATAHD---------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
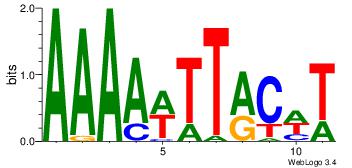
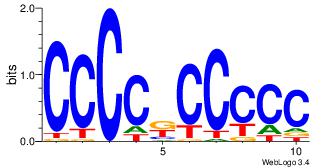
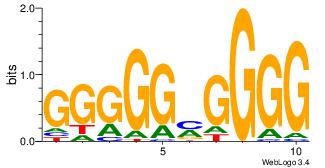
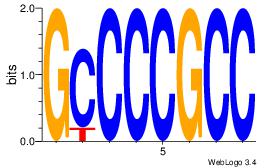
| Dataset #: 3 |
Motif ID: 24 |
Motif name: SP1 |
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
Best Matches for Top Significant Motif ID 24 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.00451594 |
Alignment:
CSKCCCCGCCCCSY
---CCCCKCCCCC-
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.00965796 |
Alignment:
HVGCCCCGCCCCBB
---CCCCKCCCCC-
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0127292 |
Alignment:
BCCGCCCCGCCCCBB
----CCCCKCCCCC-
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0174405 |
Alignment:
GGMGGRGGCGGVGC
---GGGGGYGGGG-
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
10 |
| Similarity score: |
0.0301548 |
Alignment:
CYYCBBCYYYTCCHCCTYYY
------CCCCKCCCCC----
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY |
Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0329167 |
Alignment:
RGRGGAGRRGGHGGDG
------GGGGGYGGGG
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG |
Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
49 |
| Motif name: |
TFF1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0557738 |
Alignment:
GCVGCGGCBCCG
-CCCCKCCCCC-
| Original motif Consensus sequence: CGGVGCCGCVGC |
Reverse complement motif Consensus sequence: GCVGCGGCBCCG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0648214 |
Alignment:
MTMMTCMMTCTKCK
----CCCCKCCCCC
| Original motif Consensus sequence: RGRAGARRGARRAR |
Reverse complement motif Consensus sequence: MTMMTCMMTCTKCK |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
48 |
| Motif name: |
TFW3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0762976 |
Alignment:
CGGCYBCGCG
CCCCKCCCCC
| Original motif Consensus sequence: CGGCYBCGCG |
Reverse complement motif Consensus sequence: CGCGBMGCCG |
|
|
 |
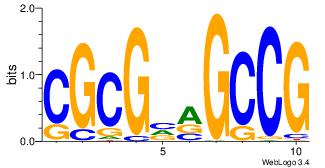 |
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0841987 |
Alignment:
GBTGCAGGTGB
GGGGGYGGGG-
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
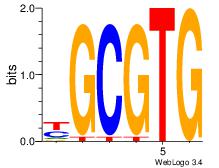
| Dataset #: 3 |
Motif ID: 32 |
Motif name: ArntAhr |
| Original motif Consensus sequence: YGCGTG |
Reverse complement motif Consensus sequence: CACGCM |
|
|
 |
 |
Best Matches for Top Significant Motif ID 32 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0396403 |
Alignment:
HVGCCCCGCCCCBB
----CACGCM----
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
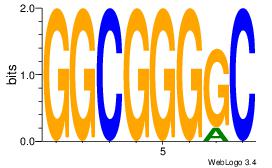
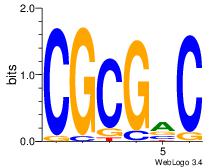
| Dataset #: |
1 |
| Motif ID: |
1 |
| Motif name: |
Motif 1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.040625 |
Alignment:
GCCCCGCC
--CACGCM
| Original motif Consensus sequence: GGCGGGGC |
Reverse complement motif Consensus sequence: GCCCCGCC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0412037 |
Alignment:
SGGTCACGTGACCS
----YGCGTG----
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0425089 |
Alignment:
BCCGCCCCGCCCCBB
-----CACGCM----
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0465867 |
Alignment:
MSGGGGCGGGGYSG
----YGCGTG----
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
47 |
| Motif name: |
TFW2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0469722 |
Alignment:
SCGCGCGG
-YGCGTG-
| Original motif Consensus sequence: SCGCGCGG |
Reverse complement motif Consensus sequence: CCGCGCGS |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
48 |
| Motif name: |
TFW3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0586806 |
Alignment:
CGGCYBCGCG
----YGCGTG
| Original motif Consensus sequence: CGGCYBCGCG |
Reverse complement motif Consensus sequence: CGCGBMGCCG |
|
|
 |
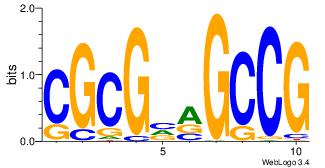 |
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0637286 |
Alignment:
BCACCTGCABC
-YGCGTG----
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.066121 |
Alignment:
GCVCCGCCMCCYCC
--CACGCM------
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
TFW1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0756826 |
Alignment:
CGCGAC
CACGCM
| Original motif Consensus sequence: GTCGCG |
Reverse complement motif Consensus sequence: CGCGAC |
|
|
 |
 |
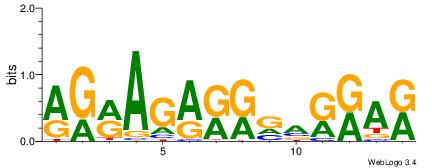
| Dataset #: 1 |
Motif ID: 1 |
Motif name: Motif 1 |
| Original motif Consensus sequence: GGCGGGGC |
Reverse complement motif Consensus sequence: GCCCCGCC |
|
|
 |
 |
Best Matches for Top Significant Motif ID 1 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0 |
Alignment:
BBGGGGCGGGGCVD
----GGCGGGGC--
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.00250079 |
Alignment:
BCCGCCCCGCCCCBB
---GCCCCGCC----
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0149958 |
Alignment:
MSGGGGCGGGGYSG
----GGCGGGGC--
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0348969 |
Alignment:
GGMGGRGGCGGVGC
------GGCGGGGC
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
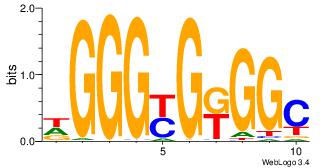
27 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0349983 |
Alignment:
DGGGYGKGGC
--GGCGGGGC
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0708224 |
Alignment:
VAGGCCBBGGCVBB
---GCCCCGCC---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
48 |
| Motif name: |
TFW3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.071681 |
Alignment:
CGCGBMGCCG
-GCCCCGCC-
| Original motif Consensus sequence: CGGCYBCGCG |
Reverse complement motif Consensus sequence: CGCGBMGCCG |
|
|
 |
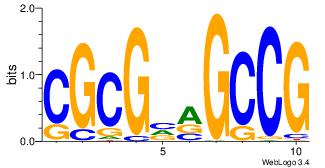 |
| Dataset #: |
5 |
| Motif ID: |
49 |
| Motif name: |
TFF1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0803996 |
Alignment:
CGGVGCCGCVGC
--GCCCCGCC--
| Original motif Consensus sequence: CGGVGCCGCVGC |
Reverse complement motif Consensus sequence: GCVGCGGCBCCG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0853847 |
Alignment:
DGVBCABTGDWGCGKRRCSR
----------GGCGGGGC--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
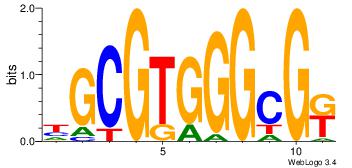
| Dataset #: |
3 |
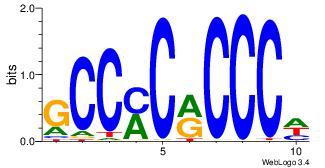
| Motif ID: |
23 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0878731 |
Alignment:
YCGCCCACGCH
---GCCCCGCC
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0928252 |
Alignment:
CHCCBCCKMCTCCKCM
--GCCCCGCC------
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG |
Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
|
|
 |
 |
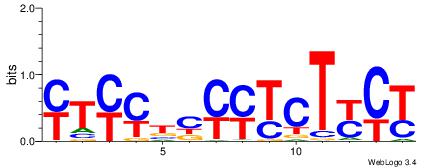
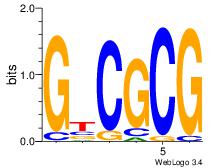
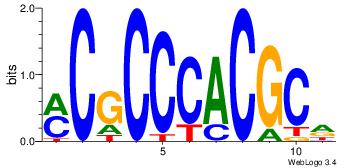
| Dataset #: 5 |
Motif ID: 46 |
Motif name: TFW1 |
| Original motif Consensus sequence: GTCGCG |
Reverse complement motif Consensus sequence: CGCGAC |
|
|
 |
 |
Best Matches for Top Significant Motif ID 46 (Highest to Lowest)
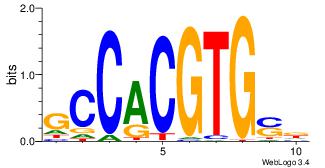
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
6 |
| Similarity score: |
0.0375102 |
Alignment:
SGGTCACGTGACCS
--GTCGCG------
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
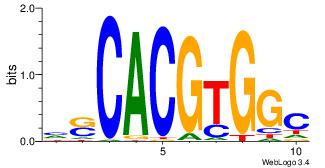
| Dataset #: |
3 |
| Motif ID: |
33 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0537993 |
Alignment:
GCCACGTGSD
----GTCGCG
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
6 |
| Similarity score: |
0.0539 |
Alignment:
HVGCCCCGCCCCBB
--GTCGCG------
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
1 |
| Motif name: |
Motif 1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0554263 |
Alignment:
GCCCCGCC
--GTCGCG
| Original motif Consensus sequence: GGCGGGGC |
Reverse complement motif Consensus sequence: GCCCCGCC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
6 |
| Similarity score: |
0.0558733 |
Alignment:
BBGGGGCGGGGCGGB
-------GTCGCG--
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
6 |
| Similarity score: |
0.0644512 |
Alignment:
CSKCCCCGCCCCSY
--CGCGAC------
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
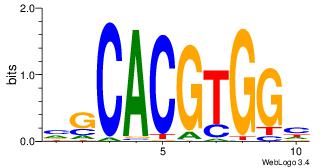
| Dataset #: |
3 |
| Motif ID: |
34 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0664905 |
Alignment:
DCCACGTGCV
----GTCGCG
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
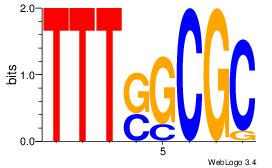
 |
| Dataset #: |
3 |
| Motif ID: |
28 |
| Motif name: |
E2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0698654 |
Alignment:
TTTSGCGC
-GTCGCG-
| Original motif Consensus sequence: TTTSGCGC |
Reverse complement motif Consensus sequence: GCGCSAAA |
|
|
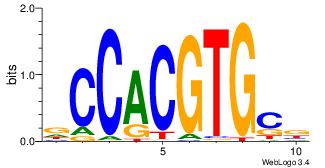
 |
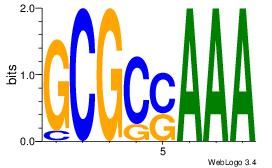 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
6 |
| Similarity score: |
0.069909 |
Alignment:
BBVGCCBVGGCCTV
---GTCGCG-----
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
6 |
| Similarity score: |
0.072456 |
Alignment:
MSGKKRCGCWDCABTGBBCD
------CGCGAC--------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
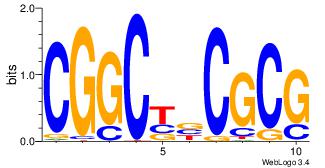
| Dataset #: 5 |
Motif ID: 47 |
Motif name: TFW2 |
| Original motif Consensus sequence: SCGCGCGG |
Reverse complement motif Consensus sequence: CCGCGCGS |
|
|
 |
 |
Best Matches for Top Significant Motif ID 47 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0456233 |
Alignment:
HVGCCCCGCCCCBB
-CCGCGCGS-----
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.045686 |
Alignment:
BBGGGGCGGGGCGGB
------SCGCGCGG-
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
8 |
| Similarity score: |
0.0504058 |
Alignment:
MSGGGGCGGGGYSG
-----SCGCGCGG-
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
33 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0527532 |
Alignment:
HSCACGTGGC
-SCGCGCGG-
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
34 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0574543 |
Alignment:
VGCACGTGGH
-SCGCGCGG-
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
25 |
| Motif name: |
TFAP2A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0678925 |
Alignment:
SCMVBBGGC
-SCGCGCGG
| Original motif Consensus sequence: GCCBBVRGS |
Reverse complement motif Consensus sequence: SCMVBBGGC |
|
|
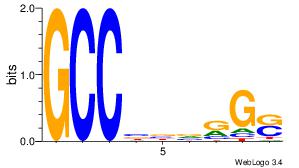 |
 |
| Dataset #: |
3 |
| Motif ID: |
24 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.07018 |
Alignment:
CCCCKCCCCC
--CCGCGCGS
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0710499 |
Alignment:
BBVGCCBVGGCCTV
--SCGCGCGG----
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
27 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0768965 |
Alignment:
DGGGYGKGGC
-SCGCGCGG-
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
26 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0837414 |
Alignment:
GCGGACGTTV
--SCGCGCGG
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
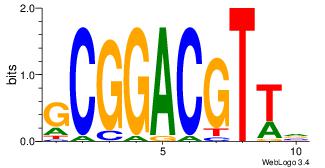 |
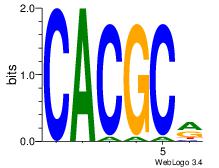
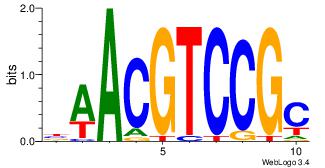
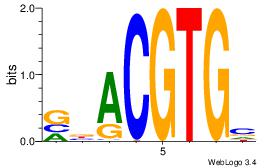
| Dataset #: 3 |
Motif ID: 29 |
Motif name: HIF1AARNT |
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
Best Matches for Top Significant Motif ID 29 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0295473 |
Alignment:
BCACCTGCABC
VBACGTGV---
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0314837 |
Alignment:
SGGTCACGTGACCS
---VCACGTBV---
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
4 |
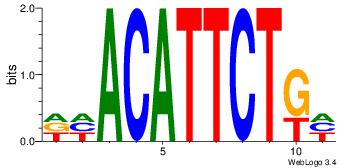
| Motif ID: |
44 |
| Motif name: |
dhACATTCTkh |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0640024 |
Alignment:
DHACATTCTGH
VCACGTBV---
| Original motif Consensus sequence: DHACATTCTGH |
Reverse complement motif Consensus sequence: HCAGAATGTHD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
47 |
| Motif name: |
TFW2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0660185 |
Alignment:
CCGCGCGS
VBACGTGV
| Original motif Consensus sequence: SCGCGCGG |
Reverse complement motif Consensus sequence: CCGCGCGS |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.067219 |
Alignment:
GCVCCGCCMCCYCC
------VCACGTBV
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
8 |
| Similarity score: |
0.0698117 |
Alignment:
RGRGGAGRRGGHGGDG
---VBACGTGV-----
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG |
Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0711168 |
Alignment:
BBGGGGCGGGGCVD
---VBACGTGV---
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0735453 |
Alignment:
BBGGGGCGGGGCGGB
---VBACGTGV----
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
12 |
| Number of overlap: |
8 |
| Similarity score: |
0.0757612 |
Alignment:
KKKAGGDGGAKKMGBBGKMG
-VBACGTGV-----------
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY |
Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0775335 |
Alignment:
CSKCCCCGCCCCSY
---VCACGTBV---
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
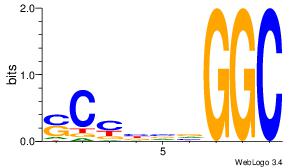
| Dataset #: 3 |
Motif ID: 25 |
Motif name: TFAP2A |
| Original motif Consensus sequence: GCCBBVRGS |
Reverse complement motif Consensus sequence: SCMVBBGGC |
|
|
 |
 |
Best Matches for Top Significant Motif ID 25 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0.0415732 |
Alignment:
GGMGGRGGCGGVGC
SCMVBBGGC-----
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
48 |
| Motif name: |
TFW3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0.0552877 |
Alignment:
CGGCYBCGCG
-GCCBBVRGS
| Original motif Consensus sequence: CGGCYBCGCG |
Reverse complement motif Consensus sequence: CGCGBMGCCG |
|
|
 |
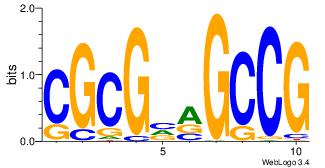 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
11 |
| Number of overlap: |
9 |
| Similarity score: |
0.0630429 |
Alignment:
KKKAGGDGGAKKMGBBGKMG
-SCMVBBGGC----------
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY |
Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
49 |
| Motif name: |
TFF1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0.0669772 |
Alignment:
GCVGCGGCBCCG
SCMVBBGGC---
| Original motif Consensus sequence: CGGVGCCGCVGC |
Reverse complement motif Consensus sequence: GCVGCGGCBCCG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0.069755 |
Alignment:
CHCCBCCKMCTCCKCM
SCMVBBGGC-------
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG |
Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.0715961 |
Alignment:
MTMMTCMMTCTKCK
----SCMVBBGGC-
| Original motif Consensus sequence: RGRAGARRGARRAR |
Reverse complement motif Consensus sequence: MTMMTCMMTCTKCK |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
9 |
| Similarity score: |
0.0719489 |
Alignment:
BCCGCCCCGCCCCBB
-SCMVBBGGC-----
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
9 |
| Similarity score: |
0.0781606 |
Alignment:
MSGGGGCGGGGYSG
GCCBBVRGS-----
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0.0800026 |
Alignment:
BBGGGGCGGGGCVD
-----GCCBBVRGS
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0.0857416 |
Alignment:
BCACCTGCABC
--GCCBBVRGS
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 41 |
Motif name: wwAAATAATAtw |
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
Best Matches for Top Significant Motif ID 41 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0536028 |
Alignment:
AWAAAWTWAAASWA
-HDAAATAATADD-
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
52 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
12 |
| Similarity score: |
0.0565705 |
Alignment:
WTKTTTTTHWTTTTTTBT
------DDTATTATTTDH
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT |
Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0575935 |
Alignment:
HDWVAAAHAAAAAMAAAMWWWHBWA
--HDAAATAATADD-----------
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA |
Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0591821 |
Alignment:
AATHATATWTHAAA
--DDTATTATTTDH
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0712963 |
Alignment:
WAHHTVTTYKAAAAWTTRAT
--------HDAAATAATADD
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0715278 |
Alignment:
AAATTKTATTTAWT
-DDTATTATTTDH-
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0733516 |
Alignment:
WWHTTTTTCABAAWTTWA
------DDTATTATTTDH
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA |
Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0912037 |
Alignment:
TCAAAATKAWTTGT
--DDTATTATTTDH
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0924576 |
Alignment:
WWTAKTTCDKAATT
DDTATTATTTDH--
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0989198 |
Alignment:
TGATTTTAWKATTT
-DDTATTATTTDH-
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
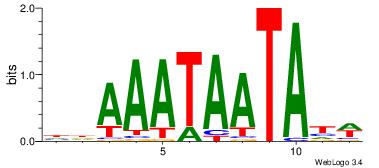
| Dataset #: 4 |
Motif ID: 37 |
Motif name: tkAAATAATAtw |
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
Best Matches for Top Significant Motif ID 37 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0522317 |
Alignment:
TWSTTTWAWTTTWT
-WHTATTATTTDH-
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0590505 |
Alignment:
TWVHWWWYTTTYTTTTTHTTTVWBH
-----------WHTATTATTTDH--
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA |
Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
52 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0637702 |
Alignment:
ABAAAAAAWHAAAAARAW
HDAAATAATAHW------
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT |
Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0641975 |
Alignment:
AATHATATWTHAAA
--WHTATTATTTDH
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0704475 |
Alignment:
AAATTKTATTTAWT
-WHTATTATTTDH-
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0704475 |
Alignment:
ATKAAWTTTTRMAABAHHTW
WHTATTATTTDH--------
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0763819 |
Alignment:
WWHTTTTTCABAAWTTWA
------WHTATTATTTDH
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA |
Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0864198 |
Alignment:
TCAAAATKAWTTGT
--WHTATTATTTDH
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.09076 |
Alignment:
WWTAKTTCDKAATT
WHTATTATTTDH--
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.096142 |
Alignment:
TWTAATCTAWGAA
WHTATTATTTDH-
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
Significant Motifs - Global and Local Matching (Highest to Lowest)
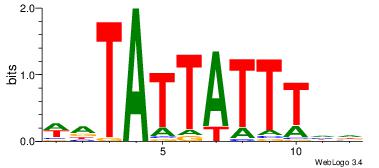
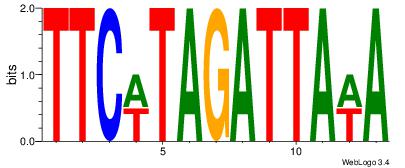
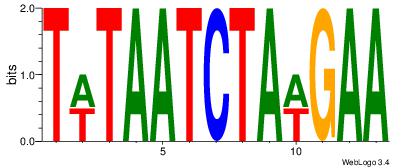
| Dataset #: 4 |
Motif ID: 45 |
Motif name: wbgTAAATAww |
| Original motif Consensus sequence: DBGTAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTACBD |
|
|
 |
 |
Best Matches for Significant Motif ID 45 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0162338 |
Alignment:
HDAAATAATAHW
-DBGTAAATAHD
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0164863 |
Alignment:
DDTATTATTTDH
DHTATTTACBD-
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
15 |
| Number of overlap: |
11 |
| Similarity score: |
0.0192768 |
Alignment:
TWVHWWWYTTTYTTTTTHTTTVWBH
--------------DHTATTTACBD
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA |
Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0250541 |
Alignment:
AATHATATWTHAAA
DHTATTTACBD---
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.027895 |
Alignment:
AATTYDGAARTAWW
---DBGTAAATAHD
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
52 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0283217 |
Alignment:
ABAAAAAAWHAAAAARAW
DBGTAAATAHD-------
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT |
Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0308097 |
Alignment:
TWAAWTTVTGAAAAAHWW
------DBGTAAATAHD-
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA |
Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
11 |
| Similarity score: |
0.0333874 |
Alignment:
ATKAAWTTTTRMAABAHHTW
-------DBGTAAATAHD--
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0361187 |
Alignment:
AAAAWTTRCWT
DHTATTTACBD
| Original motif Consensus sequence: AAAAWTTRCWT |
Reverse complement motif Consensus sequence: AWGKAAWTTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0410381 |
Alignment:
AWAAAWTWAAASWA
--DHTATTTACBD-
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 44 |
Motif name: dhACATTCTkh |
| Original motif Consensus sequence: DHACATTCTGH |
Reverse complement motif Consensus sequence: HCAGAATGTHD |
|
|
 |
 |
Best Matches for Significant Motif ID 44 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
11 |
| Similarity score: |
0.0217516 |
Alignment:
TWAAWTTVTGAAAAAHWW
DHACATTCTGH-------
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA |
Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0238442 |
Alignment:
BCACCTGCABC
DHACATTCTGH
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0258838 |
Alignment:
TCAAAATKAWTTGT
HCAGAATGTHD---
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0292508 |
Alignment:
DDTATTATTTDH
-HCAGAATGTHD
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
11 |
| Number of overlap: |
11 |
| Similarity score: |
0.0307858 |
Alignment:
TWVHWWWYTTTYTTTTTHTTTVWBH
----DHACATTCTGH----------
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA |
Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0357323 |
Alignment:
ATKAAWTTTTRMAABAHHTW
-DHACATTCTGH--------
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0364899 |
Alignment:
WHTATTATTTDH
-HCAGAATGTHD
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0400884 |
Alignment:
AATTYDGAARTAWW
HCAGAATGTHD---
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
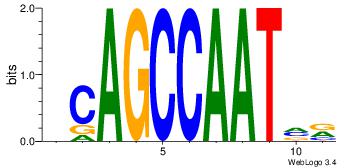
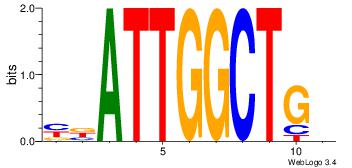
4 |
| Motif ID: |
39 |
| Motif name: |
kCAGCCAATmr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0412582 |
Alignment:
MBATTGGCTGH
DHACATTCTGH
| Original motif Consensus sequence: DCAGCCAATVR |
Reverse complement motif Consensus sequence: MBATTGGCTGH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
35 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0418896 |
Alignment:
CCGGAAGTGVV
HCAGAATGTHD
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
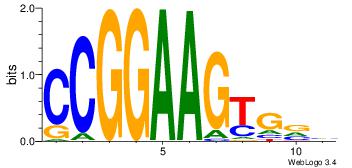 |
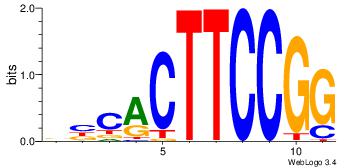 |
| Dataset #: 3 |
Motif ID: 24 |
Motif name: SP1 |
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
Best Matches for Significant Motif ID 24 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.00451594 |
Alignment:
CSKCCCCGCCCCSY
---CCCCKCCCCC-
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.00965796 |
Alignment:
BBGGGGCGGGGCVD
-GGGGGYGGGG---
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0127292 |
Alignment:
BCCGCCCCGCCCCBB
----CCCCKCCCCC-
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0174405 |
Alignment:
GGMGGRGGCGGVGC
---GGGGGYGGGG-
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0301548 |
Alignment:
CYYCBBCYYYTCCHCCTYYY
------CCCCKCCCCC----
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY |
Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0329167 |
Alignment:
RGRGGAGRRGGHGGDG
------GGGGGYGGGG
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG |
Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0540825 |
Alignment:
BBVGCCBVGGCCTV
----GGGGGYGGGG
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
49 |
| Motif name: |
TFF1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0557738 |
Alignment:
GCVGCGGCBCCG
-CCCCKCCCCC-
| Original motif Consensus sequence: CGGVGCCGCVGC |
Reverse complement motif Consensus sequence: GCVGCGGCBCCG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0600992 |
Alignment:
CCCCCTTGGGCCCC
-CCCCKCCCCC---
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
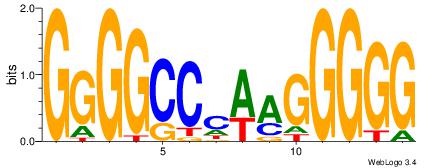 |
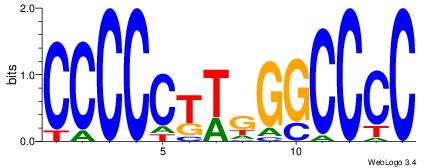 |
| Dataset #: |
3 |
| Motif ID: |
27 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0637183 |
Alignment:
GCCYCMCCCD
CCCCKCCCCC
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 32 |
Motif name: ArntAhr |
| Original motif Consensus sequence: YGCGTG |
Reverse complement motif Consensus sequence: CACGCM |
|
|
 |
 |
Best Matches for Significant Motif ID 32 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
23 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0 |
Alignment:
YCGCCCACGCH
-----CACGCM
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
29 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0112447 |
Alignment:
VBACGTGV
-YGCGTG-
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
33 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0377949 |
Alignment:
GCCACGTGSD
--YGCGTG--
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0396403 |
Alignment:
HVGCCCCGCCCCBB
----CACGCM----
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
34 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0399214 |
Alignment:
DCCACGTGCV
--YGCGTG--
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
1 |
| Motif name: |
Motif 1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.040625 |
Alignment:
GCCCCGCC
--CACGCM
| Original motif Consensus sequence: GGCGGGGC |
Reverse complement motif Consensus sequence: GCCCCGCC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0412037 |
Alignment:
SGGTCACGTGACCS
----YGCGTG----
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
6 |
| Similarity score: |
0.0425089 |
Alignment:
BCCGCCCCGCCCCBB
-----CACGCM----
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0465867 |
Alignment:
MSGGGGCGGGGYSG
----YGCGTG----
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
47 |
| Motif name: |
TFW2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0469722 |
Alignment:
SCGCGCGG
-YGCGTG-
| Original motif Consensus sequence: SCGCGCGG |
Reverse complement motif Consensus sequence: CCGCGCGS |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 40 |
Motif name: kcACCTGCAgc |
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
Best Matches for Significant Motif ID 40 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
43 |
| Motif name: |
wsTACwGTAsw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0215284 |
Alignment:
DBTACWGTAVH
BCACCTGCABC
| Original motif Consensus sequence: HVTACWGTABD |
Reverse complement motif Consensus sequence: DBTACWGTAVH |
|
|
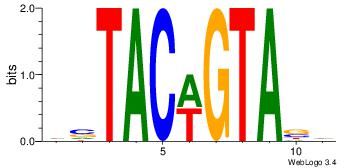 |
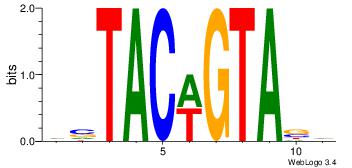 |
| Dataset #: |
4 |
| Motif ID: |
44 |
| Motif name: |
dhACATTCTkh |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0254134 |
Alignment:
DHACATTCTGH
BCACCTGCABC
| Original motif Consensus sequence: DHACATTCTGH |
Reverse complement motif Consensus sequence: HCAGAATGTHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.031338 |
Alignment:
SGGTCACGTGACCS
---BCACCTGCABC
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.034471 |
Alignment:
GCVCCGCCMCCYCC
BCACCTGCABC---
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0402518 |
Alignment:
HVGCCCCGCCCCBB
-BCACCTGCABC--
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0411257 |
Alignment:
CSKCCCCGCCCCSY
-BCACCTGCABC--
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
0.0420218 |
Alignment:
DGVBCABTGDWGCGKRRCSR
----GBTGCAGGTGB-----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
35 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0423275 |
Alignment:
CCGGAAGTGVV
GBTGCAGGTGB
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
39 |
| Motif name: |
kCAGCCAATmr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0439586 |
Alignment:
DCAGCCAATVR
GBTGCAGGTGB
| Original motif Consensus sequence: DCAGCCAATVR |
Reverse complement motif Consensus sequence: MBATTGGCTGH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0461444 |
Alignment:
BCCGCCCCGCCCCBB
--BCACCTGCABC--
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 43 |
Motif name: wsTACwGTAsw |
| Original motif Consensus sequence: HVTACWGTABD |
Reverse complement motif Consensus sequence: DBTACWGTAVH |
|
|
 |
 |
Best Matches for Significant Motif ID 43 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0199592 |
Alignment:
GBTGCAGGTGB
HVTACWGTABD
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0309343 |
Alignment:
DDTATTATTTDH
-HVTACWGTABD
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0337121 |
Alignment:
HDAAATAATAHW
DBTACWGTAVH-
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0353535 |
Alignment:
AATTYDGAARTAWW
HVTACWGTABD---
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
45 |
| Motif name: |
wbgTAAATAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.037518 |
Alignment:
DBGTAAATAHD
DBTACWGTAVH
| Original motif Consensus sequence: DBGTAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTACBD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0397727 |
Alignment:
TTTDAWATATHATT
DBTACWGTAVH---
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
0.0413797 |
Alignment:
TWAAWTTVTGAAAAAHWW
---HVTACWGTABD----
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA |
Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
11 |
| Similarity score: |
0.042298 |
Alignment:
DGVBCABTGDWGCGKRRCSR
-----HVTACWGTABD----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0445707 |
Alignment:
ATKAAWTTTTRMAABAHHTW
-------DBTACWGTAVH--
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0460859 |
Alignment:
TTCWTAGATTAWA
DBTACWGTAVH--
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 21 |
Motif name: Motif 21 |
| Original motif Consensus sequence: ATAAAA |
Reverse complement motif Consensus sequence: TTTTAT |
|
|
 |
 |
Best Matches for Significant Motif ID 21 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
6 |
| Similarity score: |
0 |
Alignment:
TTTTATTGTYAT
TTTTAT------
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
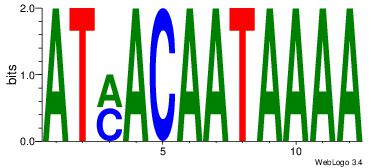 |
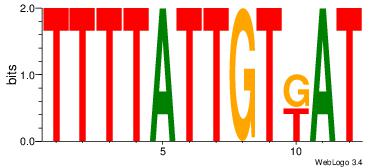 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0208333 |
Alignment:
TGATTTTAWKATTT
---TTTTAT-----
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0208333 |
Alignment:
AAATTKTATTTAWT
---TTTTAT-----
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0208333 |
Alignment:
TTTCATWAAAT
-ATAAAA----
| Original motif Consensus sequence: ATTTWATGAAA |
Reverse complement motif Consensus sequence: TTTCATWAAAT |
|
|
 |
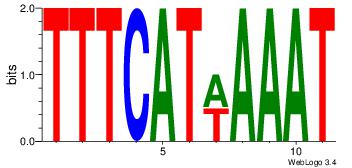 |
| Dataset #: |
2 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0416667 |
Alignment:
TTTCATAAWT
----TTTTAT
| Original motif Consensus sequence: TTTCATAAWT |
Reverse complement motif Consensus sequence: AWTTATGAAA |
|
|
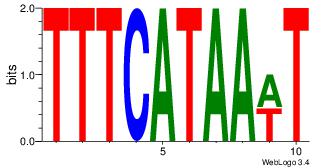 |
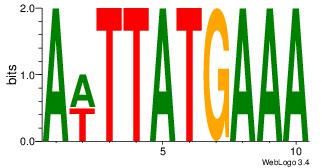 |
| Dataset #: |
2 |
| Motif ID: |
14 |
| Motif name: |
Motif 14 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0625 |
Alignment:
TTATTTWT
--TTTTAT
| Original motif Consensus sequence: AWAAATAA |
Reverse complement motif Consensus sequence: TTATTTWT |
|
|
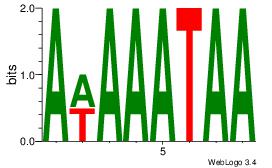 |
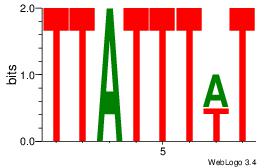 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
Motif 20 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0625 |
Alignment:
AAATCTTTYAA
-----TTTTAT
| Original motif Consensus sequence: TTMAAAGATTT |
Reverse complement motif Consensus sequence: AAATCTTTYAA |
|
|
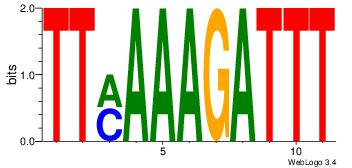 |
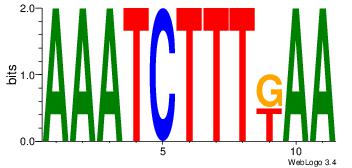 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0625 |
Alignment:
TTCWTAGATTAWA
-------ATAAAA
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0632716 |
Alignment:
DDTATTATTTDH
--TTTTAT----
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0689103 |
Alignment:
TWSTTTWAWTTTWT
---TTTTAT-----
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 29 |
Motif name: HIF1AARNT |
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
Best Matches for Significant Motif ID 29 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
33 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0123101 |
Alignment:
GCCACGTGSD
-VBACGTGV-
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
34 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0144491 |
Alignment:
DCCACGTGCV
-VBACGTGV-
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0295473 |
Alignment:
BCACCTGCABC
VBACGTGV---
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0314837 |
Alignment:
SGGTCACGTGACCS
---VCACGTBV---
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
26 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0377204 |
Alignment:
GCGGACGTTV
--VCACGTBV
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
8 |
| Similarity score: |
0.0556891 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-----VCACGTBV-------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
44 |
| Motif name: |
dhACATTCTkh |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0640024 |
Alignment:
DHACATTCTGH
VCACGTBV---
| Original motif Consensus sequence: DHACATTCTGH |
Reverse complement motif Consensus sequence: HCAGAATGTHD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
35 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0648153 |
Alignment:
CCGGAAGTGVV
--VCACGTBV-
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
47 |
| Motif name: |
TFW2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0660185 |
Alignment:
CCGCGCGS
VBACGTGV
| Original motif Consensus sequence: SCGCGCGG |
Reverse complement motif Consensus sequence: CCGCGCGS |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.067219 |
Alignment:
GCVCCGCCMCCYCC
------VCACGTBV
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 27 |
Motif name: Klf4 |
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
Best Matches for Significant Motif ID 27 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0152457 |
Alignment:
CSKCCCCGCCCCSY
--GCCYCMCCCD--
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0160185 |
Alignment:
BBGGGGCGGGGCVD
--DGGGYGKGGC--
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0194893 |
Alignment:
BBGGGGCGGGGCGGB
--DGGGYGKGGC---
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0402906 |
Alignment:
GGMGGRGGCGGVGC
----DGGGYGKGGC
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
10 |
| Similarity score: |
0.0556427 |
Alignment:
CYYCBBCYYYTCCHCCTYYY
----GCCYCMCCCD------
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY |
Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0606033 |
Alignment:
CHCCBCCKMCTCCKCM
----GCCYCMCCCD--
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG |
Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0623591 |
Alignment:
BBVGCCBVGGCCTV
-GCCYCMCCCD---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
24 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0663136 |
Alignment:
GGGGGYGGGG
DGGGYGKGGC
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
34 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0686848 |
Alignment:
VGCACGTGGH
DGGGYGKGGC
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
10 |
| Similarity score: |
0.06956 |
Alignment:
DGVBCABTGDWGCGKRRCSR
--------DGGGYGKGGC--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 35 |
Motif name: GABPA |
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
Best Matches for Significant Motif ID 35 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
0.0360984 |
Alignment:
CYYCBBCYYYTCCHCCTYYY
----VVCACTTCCGG-----
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY |
Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
0.0445566 |
Alignment:
RGRGGAGRRGGHGGDG
-CCGGAAGTGVV----
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG |
Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0446257 |
Alignment:
DGVBCABTGDWGCGKRRCSR
-------VVCACTTCCGG--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0457327 |
Alignment:
BBVGCCBVGGCCTV
VVCACTTCCGG---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0483841 |
Alignment:
BCACCTGCABC
VVCACTTCCGG
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
45 |
| Motif name: |
wbgTAAATAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0492127 |
Alignment:
DBGTAAATAHD
CCGGAAGTGVV
| Original motif Consensus sequence: DBGTAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTACBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
44 |
| Motif name: |
dhACATTCTkh |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0495154 |
Alignment:
DHACATTCTGH
VVCACTTCCGG
| Original motif Consensus sequence: DHACATTCTGH |
Reverse complement motif Consensus sequence: HCAGAATGTHD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0504656 |
Alignment:
GGMGGRGGCGGVGC
-CCGGAAGTGVV--
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0518418 |
Alignment:
AATTYDGAARTAWW
---CCGGAAGTGVV
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0554249 |
Alignment:
BCCGCCCCGCCCCBB
---VVCACTTCCGG-
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
Best Matches for Each Motif (Highest to Lowest)
| Dataset #: 1 |
Motif ID: 1 |
Motif name: Motif 1 |
| Original motif Consensus sequence: GGCGGGGC |
Reverse complement motif Consensus sequence: GCCCCGCC |
|
|
 |
 |
Best Matches for Motif ID 1 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0 |
Alignment:
BBGGGGCGGGGCVD
----GGCGGGGC--
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.00250079 |
Alignment:
BCCGCCCCGCCCCBB
---GCCCCGCC----
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0149958 |
Alignment:
MSGGGGCGGGGYSG
----GGCGGGGC--
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0348969 |
Alignment:
GGMGGRGGCGGVGC
------GGCGGGGC
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
27 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0349983 |
Alignment:
DGGGYGKGGC
--GGCGGGGC
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0708224 |
Alignment:
VAGGCCBBGGCVBB
---GCCCCGCC---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
48 |
| Motif name: |
TFW3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.071681 |
Alignment:
CGCGBMGCCG
-GCCCCGCC-
| Original motif Consensus sequence: CGGCYBCGCG |
Reverse complement motif Consensus sequence: CGCGBMGCCG |
|
|
 |
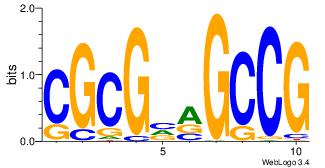 |
| Dataset #: |
5 |
| Motif ID: |
49 |
| Motif name: |
TFF1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0803996 |
Alignment:
CGGVGCCGCVGC
--GCCCCGCC--
| Original motif Consensus sequence: CGGVGCCGCVGC |
Reverse complement motif Consensus sequence: GCVGCGGCBCCG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0853847 |
Alignment:
DGVBCABTGDWGCGKRRCSR
----------GGCGGGGC--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
23 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0878731 |
Alignment:
YCGCCCACGCH
---GCCCCGCC
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0928252 |
Alignment:
CHCCBCCKMCTCCKCM
--GCCCCGCC------
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG |
Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 2 |
Motif name: Motif 2 |
| Original motif Consensus sequence: RGRAGARRGARRAR |
Reverse complement motif Consensus sequence: MTMMTCMMTCTKCK |
|
|
 |
 |
Best Matches for Motif ID 2 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.00948131 |
Alignment:
RGRGGAGRRGGHGGDG
--RGRAGARRGARRAR
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG |
Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
14 |
| Similarity score: |
0.047364 |
Alignment:
MSGKKRCGCWDCABTGBBCD
------MTMMTCMMTCTKCK
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
14 |
| Similarity score: |
0.0541717 |
Alignment:
HDWVAAAHAAAAAMAAAMWWWHBWA
---------RGRAGARRGARRAR--
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA |
Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0545069 |
Alignment:
CYYCBBCYYYTCCHCCTYYY
------MTMMTCMMTCTKCK
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY |
Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
52 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.0558935 |
Alignment:
ABAAAAAAWHAAAAARAW
-RGRAGARRGARRAR---
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT |
Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0689626 |
Alignment:
GGMGGRGGCGGVGC
RGRAGARRGARRAR
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.0775356 |
Alignment:
TWAAWTTVTGAAAAAHWW
-RGRAGARRGARRAR---
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA |
Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.57507 |
Alignment:
AWAAAWTWAAASWA-
-RGRAGARRGARRAR
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
12 |
| Similarity score: |
1.0763 |
Alignment:
ATKAAWTTTTRMAABAHHTW--
--------MTMMTCMMTCTKCK
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
1.57133 |
Alignment:
---MSGGGGCGGGGYSG
RGRAGARRGARRAR---
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 3 |
Motif name: Motif 3 |
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
Best Matches for Motif ID 3 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
14 |
| Similarity score: |
0.0137914 |
Alignment:
HDWVAAAHAAAAAMAAAMWWWHBWA
------AWAAAWTWAAASWA-----
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA |
Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
52 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0162962 |
Alignment:
ABAAAAAAWHAAAAARAW
----AWAAAWTWAAASWA
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT |
Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
14 |
| Similarity score: |
0.037679 |
Alignment:
ATKAAWTTTTRMAABAHHTW
--AWAAAWTWAAASWA----
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0423952 |
Alignment:
TGATTTTAWKATTT
TWSTTTWAWTTTWT
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
14 |
| Similarity score: |
0.0456419 |
Alignment:
TWAAWTTVTGAAAAAHWW
----AWAAAWTWAAASWA
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA |
Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0478896 |
Alignment:
AAATTKTATTTAWT
TWSTTTWAWTTTWT
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0624272 |
Alignment:
TTTDAWATATHATT
AWAAAWTWAAASWA
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0702111 |
Alignment:
WWTAKTTCDKAATT
TWSTTTWAWTTTWT
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.567825 |
Alignment:
-RGRAGARRGARRAR
AWAAAWTWAAASWA-
| Original motif Consensus sequence: RGRAGARRGARRAR |
Reverse complement motif Consensus sequence: MTMMTCMMTCTKCK |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.0432 |
Alignment:
--DDTATTATTTDH
TWSTTTWAWTTTWT
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 4 |
Motif name: Motif 4 |
| Original motif Consensus sequence: AAAAWTTRCWT |
Reverse complement motif Consensus sequence: AWGKAAWTTTT |
|
|
 |
 |
Best Matches for Motif ID 4 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
14 |
| Number of overlap: |
11 |
| Similarity score: |
0.0718583 |
Alignment:
HDWVAAAHAAAAAMAAAMWWWHBWA
-------------AAAAWTTRCWT-
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA |
Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0727073 |
Alignment:
ATKAAWTTTTRMAABAHHTW
AWGKAAWTTTT---------
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
52 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
0.0748297 |
Alignment:
WTKTTTTTHWTTTTTTBT
---AWGKAAWTTTT----
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT |
Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0786484 |
Alignment:
AAATTKTATTTAWT
AAAAWTTRCWT---
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0820375 |
Alignment:
AATHATATWTHAAA
AWGKAAWTTTT---
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.083433 |
Alignment:
TGATTTTAWKATTT
---AWGKAAWTTTT
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0852273 |
Alignment:
ACAAWTRATTTTGA
AAAAWTTRCWT---
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0858797 |
Alignment:
WWHTTTTTCABAAWTTWA
-------AWGKAAWTTTT
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA |
Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
45 |
| Motif name: |
wbgTAAATAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.086338 |
Alignment:
DHTATTTACBD
AAAAWTTRCWT
| Original motif Consensus sequence: DBGTAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTACBD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0867914 |
Alignment:
TWSTTTWAWTTTWT
--AWGKAAWTTTT-
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 5 |
Motif name: Motif 5 |
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
Best Matches for Motif ID 5 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.101648 |
Alignment:
TWSTTTWAWTTTWT
AAATTKTATTTAWT
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.107143 |
Alignment:
TGATTTTAWKATTT
AAATTKTATTTAWT
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
14 |
| Similarity score: |
0.109769 |
Alignment:
TWVHWWWYTTTYTTTTTHTTTVWBH
------AAATTKTATTTAWT-----
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA |
Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
52 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.110348 |
Alignment:
WTKTTTTTHWTTTTTTBT
-AAATTKTATTTAWT---
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT |
Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.111607 |
Alignment:
AATTYDGAARTAWW
AAATTKTATTTAWT
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
14 |
| Similarity score: |
0.124188 |
Alignment:
WWHTTTTTCABAAWTTWA
--AWTAAATAYAATTT--
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA |
Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.125 |
Alignment:
ACAAWTRATTTTGA
AAATTKTATTTAWT
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.596154 |
Alignment:
TTTDAWATATHATT-
-AWTAAATAYAATTT
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
12 |
| Similarity score: |
1.11736 |
Alignment:
WAHHTVTTYKAAAAWTTRAT--
--------AWTAAATAYAATTT
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
1.125 |
Alignment:
TWTAATCTAWGAA--
-AAATTKTATTTAWT
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 6 |
Motif name: Motif 6 |
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
Best Matches for Motif ID 6 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.0325081 |
Alignment:
WWHTTTTTCABAAWTTWA
-WWTAKTTCDKAATT---
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA |
Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
14 |
| Similarity score: |
0.0349702 |
Alignment:
ATKAAWTTTTRMAABAHHTW
----AATTYDGAARTAWW--
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
14 |
| Similarity score: |
0.0626838 |
Alignment:
TWVHWWWYTTTYTTTTTHTTTVWBH
---WWTAKTTCDKAATT--------
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA |
Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
52 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0667239 |
Alignment:
WTKTTTTTHWTTTTTTBT
-WWTAKTTCDKAATT---
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT |
Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0741071 |
Alignment:
AWTAAATAYAATTT
WWTAKTTCDKAATT
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0763393 |
Alignment:
TGATTTTAWKATTT
WWTAKTTCDKAATT
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0852679 |
Alignment:
TTTDAWATATHATT
WWTAKTTCDKAATT
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0864698 |
Alignment:
TWSTTTWAWTTTWT
WWTAKTTCDKAATT
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
1.06667 |
Alignment:
--TTCWTAGATTAWA
AATTYDGAARTAWW-
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.07587 |
Alignment:
WHTATTATTTDH--
WWTAKTTCDKAATT
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 7 |
Motif name: Motif 7 |
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
Best Matches for Motif ID 7 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0 |
Alignment:
HVGCCCCGCCCCBB
CSKCCCCGCCCCSY
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0012818 |
Alignment:
BCCGCCCCGCCCCBB
-CSKCCCCGCCCCSY
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.0537802 |
Alignment:
CYYCBBCYYYTCCHCCTYYY
---CSKCCCCGCCCCSY---
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY |
Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
14 |
| Similarity score: |
0.0662825 |
Alignment:
DGVBCABTGDWGCGKRRCSR
------MSGGGGCGGGGYSG
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0758609 |
Alignment:
CHCCBCCKMCTCCKCM
CSKCCCCGCCCCSY--
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG |
Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0829042 |
Alignment:
GCVCCGCCMCCYCC
CSKCCCCGCCCCSY
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0912118 |
Alignment:
VAGGCCBBGGCVBB
CSKCCCCGCCCCSY
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0948838 |
Alignment:
SGGTCACGTGACCS
MSGGGGCGGGGYSG
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.590412 |
Alignment:
CCCCCTTGGGCCCC-
-CSKCCCCGCCCCSY
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
49 |
| Motif name: |
TFF1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.07659 |
Alignment:
CGGVGCCGCVGC--
MSGGGGCGGGGYSG
| Original motif Consensus sequence: CGGVGCCGCVGC |
Reverse complement motif Consensus sequence: GCVGCGGCBCCG |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 8 |
Motif name: Motif 8 |
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
Best Matches for Motif ID 8 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0961539 |
Alignment:
TWSTTTWAWTTTWT
TGATTTTAWKATTT
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
52 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.10348 |
Alignment:
WTKTTTTTHWTTTTTTBT
-TGATTTTAWKATTT---
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT |
Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.107143 |
Alignment:
AAATTKTATTTAWT
TGATTTTAWKATTT
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
14 |
| Similarity score: |
0.111345 |
Alignment:
HDWVAAAHAAAAAMAAAMWWWHBWA
-----AAATRWTAAAATCA------
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA |
Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.11369 |
Alignment:
WAHHTVTTYKAAAAWTTRAT
---TGATTTTAWKATTT---
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.113839 |
Alignment:
WWTAKTTCDKAATT
TGATTTTAWKATTT
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.625 |
Alignment:
-AATHATATWTHAAA
AAATRWTAAAATCA-
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.10417 |
Alignment:
TTTTATTGTYAT--
TGATTTTAWKATTT
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
1.10417 |
Alignment:
TWTAATCTAWGAA--
-TGATTTTAWKATTT
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
1.125 |
Alignment:
ACAAWTRATTTTGA--
--AAATRWTAAAATCA
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 9 |
Motif name: Motif 9 |
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
Best Matches for Motif ID 9 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0332341 |
Alignment:
WAHHTVTTYKAAAAWTTRAT
------TTTDAWATATHATT
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0486562 |
Alignment:
WWHTTTTTCABAAWTTWA
---TTTDAWATATHATT-
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA |
Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
52 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.0528083 |
Alignment:
WTKTTTTTHWTTTTTTBT
---TTTDAWATATHATT-
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT |
Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.054155 |
Alignment:
HDWVAAAHAAAAAMAAAMWWWHBWA
TTTDAWATATHATT-----------
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA |
Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0675748 |
Alignment:
TWSTTTWAWTTTWT
AATHATATWTHAAA
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0741568 |
Alignment:
WWTAKTTCDKAATT
TTTDAWATATHATT
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.547543 |
Alignment:
-AAATTKTATTTAWT
AATHATATWTHAAA-
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.557158 |
Alignment:
-TTCWTAGATTAWA
AATHATATWTHAAA
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.576389 |
Alignment:
AAATRWTAAAATCA-
-AATHATATWTHAAA
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.03318 |
Alignment:
--HDAAATAATADD
TTTDAWATATHATT
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 10 |
Motif name: Motif 10 |
| Original motif Consensus sequence: TTTCATAAWT |
Reverse complement motif Consensus sequence: AWTTATGAAA |
|
|
 |
 |
Best Matches for Motif ID 10 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.01875 |
Alignment:
ATTTWATGAAA
-AWTTATGAAA
| Original motif Consensus sequence: ATTTWATGAAA |
Reverse complement motif Consensus sequence: TTTCATWAAAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0375 |
Alignment:
AATTYDGAARTAWW
----AWTTATGAAA
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.04375 |
Alignment:
TGATTTTAWKATTT
-TTTCATAAWT---
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
10 |
| Similarity score: |
0.0448864 |
Alignment:
TWAAWTTVTGAAAAAHWW
---AWTTATGAAA-----
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA |
Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0545833 |
Alignment:
ATKAAWTTTTRMAABAHHTW
----AWTTATGAAA------
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
17 |
| Motif name: |
Motif 17 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.05625 |
Alignment:
ATTCATTTTT
TTTCATAAWT
| Original motif Consensus sequence: AAAAATGAAT |
Reverse complement motif Consensus sequence: ATTCATTTTT |
|
|
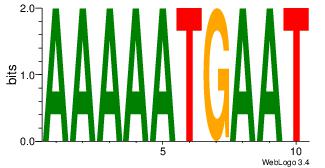 |
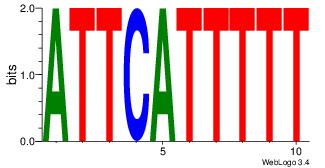 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.05625 |
Alignment:
AWTAAATAYAATTT
----AWTTATGAAA
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0604166 |
Alignment:
TTTDAWATATHATT
----TTTCATAAWT
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.06875 |
Alignment:
ATMACAATAAAA
--AWTTATGAAA
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0706731 |
Alignment:
AWAAAWTWAAASWA
----AWTTATGAAA
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 11 |
Motif name: Motif 11 |
| Original motif Consensus sequence: ATTTWATGAAA |
Reverse complement motif Consensus sequence: TTTCATWAAAT |
|
|
 |
 |
Best Matches for Motif ID 11 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0795455 |
Alignment:
AWTAAATAYAATTT
--TTTCATWAAAT-
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0795455 |
Alignment:
AAATRWTAAAATCA
--TTTCATWAAAT-
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
11 |
| Similarity score: |
0.0991736 |
Alignment:
TWAAWTTVTGAAAAAHWW
--ATTTWATGAAA-----
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA |
Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
Motif 18 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
TTATWAWCTAA
ATTTWATGAAA
| Original motif Consensus sequence: TTAGWTWATAA |
Reverse complement motif Consensus sequence: TTATWAWCTAA |
|
|
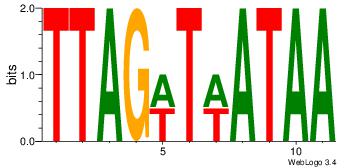 |
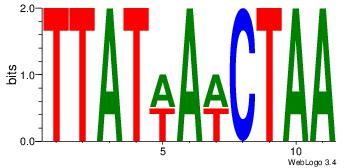 |
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
ATMACAATAAAA
-TTTCATWAAAT
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.109848 |
Alignment:
AATHATATWTHAAA
---ATTTWATGAAA
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
AATTYDGAARTAWW
-ATTTWATGAAA--
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
TWTAATCTAWGAA
TTTCATWAAAT--
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
AWAAAWTWAAASWA
ATTTWATGAAA---
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.114394 |
Alignment:
ATKAAWTTTTRMAABAHHTW
---ATTTWATGAAA------
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 12 |
Motif name: Motif 12 |
| Original motif Consensus sequence: AAAACAAA |
Reverse complement motif Consensus sequence: TTTGTTTT |
|
|
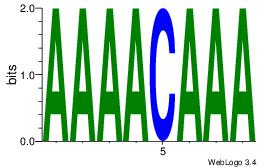 |
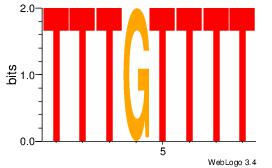 |
Best Matches for Motif ID 12 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0625 |
Alignment:
TTTTATTGTYAT
-TTTGTTTT---
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
17 |
| Motif name: |
Motif 17 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0625 |
Alignment:
AAAAATGAAT
-AAAACAAA-
| Original motif Consensus sequence: AAAAATGAAT |
Reverse complement motif Consensus sequence: ATTCATTTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
AWTAAATAYAATTT
----AAAACAAA--
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
TCAAAATKAWTTGT
--AAAACAAA----
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
14 |
| Motif name: |
Motif 14 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
AWAAATAA
AAAACAAA
| Original motif Consensus sequence: AWAAATAA |
Reverse complement motif Consensus sequence: TTATTTWT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
10 |
| Number of overlap: |
8 |
| Similarity score: |
0.0790441 |
Alignment:
HDWVAAAHAAAAAMAAAMWWWHBWA
---------AAAACAAA--------
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA |
Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0805288 |
Alignment:
AWAAAWTWAAASWA
--AAAACAAA----
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
52 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
8 |
| Similarity score: |
0.0873397 |
Alignment:
ABAAAAAAWHAAAAARAW
-----AAAACAAA-----
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT |
Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.09375 |
Alignment:
TGATTTTAWKATTT
----TTTGTTTT--
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
Motif 18 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.09375 |
Alignment:
TTAGWTWATAA
TTTGTTTT---
| Original motif Consensus sequence: TTAGWTWATAA |
Reverse complement motif Consensus sequence: TTATWAWCTAA |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 13 |
Motif name: Motif 13 |
| Original motif Consensus sequence: AAAGATTT |
Reverse complement motif Consensus sequence: AAATCTTT |
|
|
 |
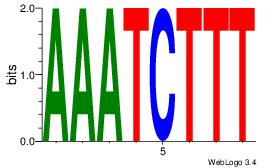 |
Best Matches for Motif ID 13 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
Motif 20 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0 |
Alignment:
AAATCTTTYAA
AAATCTTT---
| Original motif Consensus sequence: TTMAAAGATTT |
Reverse complement motif Consensus sequence: AAATCTTTYAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0625 |
Alignment:
ACAAWTRATTTTGA
---AAAGATTT---
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0625 |
Alignment:
AAATRWTAAAATCA
------AAATCTTT
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
AAATTKTATTTAWT
AAATCTTT------
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
TTCWTAGATTAWA
---AAAGATTT--
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0822369 |
Alignment:
AWGKAAWTTTT
---AAATCTTT
| Original motif Consensus sequence: AAAAWTTRCWT |
Reverse complement motif Consensus sequence: AWGKAAWTTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
17 |
| Motif name: |
Motif 17 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.09375 |
Alignment:
AAAAATGAAT
--AAATCTTT
| Original motif Consensus sequence: AAAAATGAAT |
Reverse complement motif Consensus sequence: ATTCATTTTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
44 |
| Motif name: |
dhACATTCTkh |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.104167 |
Alignment:
DHACATTCTGH
AAAGATTT---
| Original motif Consensus sequence: DHACATTCTGH |
Reverse complement motif Consensus sequence: HCAGAATGTHD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
14 |
| Motif name: |
Motif 14 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.109375 |
Alignment:
AWAAATAA
AAAGATTT
| Original motif Consensus sequence: AWAAATAA |
Reverse complement motif Consensus sequence: TTATTTWT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.110417 |
Alignment:
HDAAATAATAHW
--AAATCTTT--
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 14 |
Motif name: Motif 14 |
| Original motif Consensus sequence: AWAAATAA |
Reverse complement motif Consensus sequence: TTATTTWT |
|
|
 |
 |
Best Matches for Motif ID 14 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
17 |
| Motif name: |
Motif 17 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.046875 |
Alignment:
ATTCATTTTT
--TTATTTWT
| Original motif Consensus sequence: AAAAATGAAT |
Reverse complement motif Consensus sequence: ATTCATTTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
8 |
| Similarity score: |
0.046875 |
Alignment:
AAATTKTATTTAWT
-----TTATTTWT-
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0625 |
Alignment:
ACAAWTRATTTTGA
AWAAATAA------
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0625 |
Alignment:
ATMACAATAAAA
--AWAAATAA--
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0636574 |
Alignment:
HDAAATAATADD
AWAAATAA----
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0697917 |
Alignment:
WHTATTATTTDH
----TTATTTWT
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
52 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
WTKTTTTTHWTTTTTTBT
------TTATTTWT----
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT |
Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
TGATTTTAWKATTT
-----TTATTTWT-
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
Motif 18 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
TTAGWTWATAA
TTATTTWT---
| Original motif Consensus sequence: TTAGWTWATAA |
Reverse complement motif Consensus sequence: TTATWAWCTAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Motif 12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
AAAACAAA
AWAAATAA
| Original motif Consensus sequence: AAAACAAA |
Reverse complement motif Consensus sequence: TTTGTTTT |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 15 |
Motif name: Motif 15 |
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
Best Matches for Motif ID 15 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.104167 |
Alignment:
TGATTTTAWKATTT
TTTTATTGTYAT--
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
12 |
| Similarity score: |
0.109681 |
Alignment:
TWVHWWWYTTTYTTTTTHTTTVWBH
--------TTTTATTGTYAT-----
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA |
Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.114583 |
Alignment:
AWTAAATAYAATTT
-ATMACAATAAAA-
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.114583 |
Alignment:
AWAAAWTWAAASWA
ATMACAATAAAA--
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
52 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.115652 |
Alignment:
ABAAAAAAWHAAAAARAW
--ATMACAATAAAA----
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT |
Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.119792 |
Alignment:
AATTYDGAARTAWW
--ATMACAATAAAA
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.121528 |
Alignment:
TTTDAWATATHATT
-TTTTATTGTYAT-
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
12 |
| Similarity score: |
0.125 |
Alignment:
WWHTTTTTCABAAWTTWA
------ATMACAATAAAA
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA |
Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.590909 |
Alignment:
-TCAAAATKAWTTGT
ATMACAATAAAA---
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.602273 |
Alignment:
TTTCATWAAAT-
ATMACAATAAAA
| Original motif Consensus sequence: ATTTWATGAAA |
Reverse complement motif Consensus sequence: TTTCATWAAAT |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 16 |
Motif name: Motif 16 |
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
Best Matches for Motif ID 16 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.1 |
Alignment:
AWTAAATAYAATTT
TCAAAATKAWTTGT
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
13 |
| Similarity score: |
0.599359 |
Alignment:
ATKAAWTTTTRMAABAHHTW-
-------TCAAAATKAWTTGT
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
1.08264 |
Alignment:
TTTDAWATATHATT--
--TCAAAATKAWTTGT
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.08403 |
Alignment:
--WHTATTATTTDH
TCAAAATKAWTTGT
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.08881 |
Alignment:
--DDTATTATTTDH
TCAAAATKAWTTGT
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
1.1 |
Alignment:
TGATTTTAWKATTT--
--TCAAAATKAWTTGT
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
1.56591 |
Alignment:
TTTTATTGTYAT---
-ACAAWTRATTTTGA
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
1.56875 |
Alignment:
---AATTYDGAARTAWW
TCAAAATKAWTTGT---
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.57667 |
Alignment:
AAAAWTTRCWT---
ACAAWTRATTTTGA
| Original motif Consensus sequence: AAAAWTTRCWT |
Reverse complement motif Consensus sequence: AWGKAAWTTTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
44 |
| Motif name: |
dhACATTCTkh |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.57727 |
Alignment:
HCAGAATGTHD---
TCAAAATKAWTTGT
| Original motif Consensus sequence: DHACATTCTGH |
Reverse complement motif Consensus sequence: HCAGAATGTHD |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 17 |
Motif name: Motif 17 |
| Original motif Consensus sequence: AAAAATGAAT |
Reverse complement motif Consensus sequence: ATTCATTTTT |
|
|
 |
 |
Best Matches for Motif ID 17 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.05 |
Alignment:
ACAAWTRATTTTGA
-ATTCATTTTT---
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0875 |
Alignment:
TTTTATTGTYAT
ATTCATTTTT--
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0875 |
Alignment:
AWTAAATAYAATTT
---AAAAATGAAT-
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0875 |
Alignment:
AWTTATGAAA
AAAAATGAAT
| Original motif Consensus sequence: TTTCATAAWT |
Reverse complement motif Consensus sequence: AWTTATGAAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0947369 |
Alignment:
AWGKAAWTTTT
-ATTCATTTTT
| Original motif Consensus sequence: AAAAWTTRCWT |
Reverse complement motif Consensus sequence: AWGKAAWTTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.1 |
Alignment:
TGATTTTAWKATTT
---ATTCATTTTT-
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
44 |
| Motif name: |
dhACATTCTkh |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.1 |
Alignment:
DHACATTCTGH
ATTCATTTTT-
| Original motif Consensus sequence: DHACATTCTGH |
Reverse complement motif Consensus sequence: HCAGAATGTHD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
11 |
| Number of overlap: |
10 |
| Similarity score: |
0.103333 |
Alignment:
ATKAAWTTTTRMAABAHHTW
ATTCATTTTT----------
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.104808 |
Alignment:
TWSTTTWAWTTTWT
----ATTCATTTTT
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
52 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.105128 |
Alignment:
ABAAAAAAWHAAAAARAW
----AAAAATGAAT----
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT |
Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 18 |
Motif name: Motif 18 |
| Original motif Consensus sequence: TTAGWTWATAA |
Reverse complement motif Consensus sequence: TTATWAWCTAA |
|
|
 |
 |
Best Matches for Motif ID 18 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.09375 |
Alignment:
AATTYDGAARTAWW
-TTATWAWCTAA--
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
AAATRWTAAAATCA
TTATWAWCTAA---
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
TTTCATWAAAT
TTAGWTWATAA
| Original motif Consensus sequence: ATTTWATGAAA |
Reverse complement motif Consensus sequence: TTTCATWAAAT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
HDAAATAATAHW
TTAGWTWATAA-
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.104798 |
Alignment:
DDTATTATTTDH
-TTATWAWCTAA
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.108471 |
Alignment:
WWHTTTTTCABAAWTTWA
-------TTATWAWCTAA
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA |
Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.109848 |
Alignment:
TTTDAWATATHATT
-TTAGWTWATAA--
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.109848 |
Alignment:
ATKAAWTTTTRMAABAHHTW
------TTATWAWCTAA---
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
ATMACAATAAAA
TTATWAWCTAA-
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
TTCWTAGATTAWA
TTATWAWCTAA--
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 19 |
Motif name: Motif 19 |
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
Best Matches for Motif ID 19 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.0641025 |
Alignment:
AATHATATWTHAAA
-TTCWTAGATTAWA
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
13 |
| Similarity score: |
0.0762821 |
Alignment:
WAHHTVTTYKAAAAWTTRAT
------TTCWTAGATTAWA-
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
12 |
| Number of overlap: |
13 |
| Similarity score: |
0.0810709 |
Alignment:
HDWVAAAHAAAAAMAAAMWWWHBWA
-TWTAATCTAWGAA-----------
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA |
Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
13 |
| Similarity score: |
0.0815851 |
Alignment:
TWAAWTTVTGAAAAAHWW
-----TTCWTAGATTAWA
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA |
Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.5625 |
Alignment:
AATTYDGAARTAWW-
--TTCWTAGATTAWA
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.5625 |
Alignment:
-TGATTTTAWKATTT
TWTAATCTAWGAA--
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.577083 |
Alignment:
WHTATTATTTDH-
TWTAATCTAWGAA
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.58179 |
Alignment:
DDTATTATTTDH-
TWTAATCTAWGAA
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.583333 |
Alignment:
AWTAAATAYAATTT-
--TTCWTAGATTAWA
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
Motif 20 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.02652 |
Alignment:
TTMAAAGATTT--
TTCWTAGATTAWA
| Original motif Consensus sequence: TTMAAAGATTT |
Reverse complement motif Consensus sequence: AAATCTTTYAA |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 20 |
Motif name: Motif 20 |
| Original motif Consensus sequence: TTMAAAGATTT |
Reverse complement motif Consensus sequence: AAATCTTTYAA |
|
|
 |
 |
Best Matches for Motif ID 20 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0681818 |
Alignment:
AAATRWTAAAATCA
---AAATCTTTYAA
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0681818 |
Alignment:
TTCWTAGATTAWA
TTMAAAGATTT--
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
AWTAAATAYAATTT
--TTMAAAGATTT-
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
ACAAWTRATTTTGA
TTMAAAGATTT---
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.106061 |
Alignment:
TTTDAWATATHATT
--TTMAAAGATTT-
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
11 |
| Similarity score: |
0.110606 |
Alignment:
WAHHTVTTYKAAAAWTTRAT
-AAATCTTTYAA--------
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.110646 |
Alignment:
AWGKAAWTTTT
TTMAAAGATTT
| Original motif Consensus sequence: AAAAWTTRCWT |
Reverse complement motif Consensus sequence: AWGKAAWTTTT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
WWHTTTTTCABAAWTTWA
------TTMAAAGATTT-
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA |
Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.124242 |
Alignment:
WHTATTATTTDH
-AAATCTTTYAA
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.125 |
Alignment:
WWTAKTTCDKAATT
-AAATCTTTYAA--
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 21 |
Motif name: Motif 21 |
| Original motif Consensus sequence: ATAAAA |
Reverse complement motif Consensus sequence: TTTTAT |
|
|
 |
 |
Best Matches for Motif ID 21 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
6 |
| Similarity score: |
0 |
Alignment:
TTTTATTGTYAT
TTTTAT------
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0208333 |
Alignment:
TGATTTTAWKATTT
---TTTTAT-----
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0208333 |
Alignment:
AAATTKTATTTAWT
---TTTTAT-----
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0208333 |
Alignment:
TTTCATWAAAT
-ATAAAA----
| Original motif Consensus sequence: ATTTWATGAAA |
Reverse complement motif Consensus sequence: TTTCATWAAAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0416667 |
Alignment:
TTTCATAAWT
----TTTTAT
| Original motif Consensus sequence: TTTCATAAWT |
Reverse complement motif Consensus sequence: AWTTATGAAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
14 |
| Motif name: |
Motif 14 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0625 |
Alignment:
TTATTTWT
--TTTTAT
| Original motif Consensus sequence: AWAAATAA |
Reverse complement motif Consensus sequence: TTATTTWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
Motif 20 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0625 |
Alignment:
AAATCTTTYAA
-----TTTTAT
| Original motif Consensus sequence: TTMAAAGATTT |
Reverse complement motif Consensus sequence: AAATCTTTYAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0625 |
Alignment:
TTCWTAGATTAWA
-------ATAAAA
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0632716 |
Alignment:
DDTATTATTTDH
--TTTTAT----
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0689103 |
Alignment:
TWSTTTWAWTTTWT
---TTTTAT-----
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 22 |
Motif name: Zfx |
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
Best Matches for Motif ID 22 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0514008 |
Alignment:
BCCGCCCCGCCCCBB
BBVGCCBVGGCCTV-
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0590031 |
Alignment:
CHCCBCCKMCTCCKCM
-BBVGCCBVGGCCTV-
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG |
Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
14 |
| Similarity score: |
0.0611182 |
Alignment:
DGVBCABTGDWGCGKRRCSR
--BBVGCCBVGGCCTV----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
14 |
| Similarity score: |
0.0629383 |
Alignment:
KKKAGGDGGAKKMGBBGKMG
-----VAGGCCBBGGCVBB-
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY |
Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0654514 |
Alignment:
HVGCCCCGCCCCBB
VAGGCCBBGGCVBB
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0796879 |
Alignment:
CCCCCTTGGGCCCC
BBVGCCBVGGCCTV
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0799116 |
Alignment:
MSGGGGCGGGGYSG
BBVGCCBVGGCCTV
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.583338 |
Alignment:
-GCVCCGCCMCCYCC
BBVGCCBVGGCCTV-
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
49 |
| Motif name: |
TFF1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
1.55068 |
Alignment:
CGGVGCCGCVGC---
-BBVGCCBVGGCCTV
| Original motif Consensus sequence: CGGVGCCGCVGC |
Reverse complement motif Consensus sequence: GCVGCGGCBCCG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
35 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.57592 |
Alignment:
CCGGAAGTGVV---
VAGGCCBBGGCVBB
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 23 |
Motif name: Egr1 |
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
Best Matches for Motif ID 23 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0514701 |
Alignment:
GCVCCGCCMCCYCC
---YCGCCCACGCH
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
11 |
| Similarity score: |
0.0698683 |
Alignment:
CHCCBCCKMCTCCKCM
-----YCGCCCACGCH
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG |
Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0718056 |
Alignment:
KKKAGGDGGAKKMGBBGKMG
-HGCGTGGGCGK--------
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY |
Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
0.0730967 |
Alignment:
BBGGGGCGGGGCGGB
----HGCGTGGGCGK
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0764878 |
Alignment:
BBVGCCBVGGCCTV
--HGCGTGGGCGK-
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
11 |
| Similarity score: |
0.0765783 |
Alignment:
DGVBCABTGDWGCGKRRCSR
----HGCGTGGGCGK-----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0771215 |
Alignment:
CSKCCCCGCCCCSY
-YCGCCCACGCH--
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0825032 |
Alignment:
HVGCCCCGCCCCBB
-YCGCCCACGCH--
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
48 |
| Motif name: |
TFW3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.550222 |
Alignment:
CGCGBMGCCG-
HGCGTGGGCGK
| Original motif Consensus sequence: CGGCYBCGCG |
Reverse complement motif Consensus sequence: CGCGBMGCCG |
|
|
 |
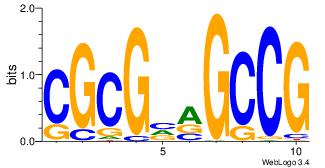 |
| Dataset #: |
3 |
| Motif ID: |
24 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.572401 |
Alignment:
GGGGGYGGGG-
HGCGTGGGCGK
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 24 |
Motif name: SP1 |
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
Best Matches for Motif ID 24 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.00451594 |
Alignment:
CSKCCCCGCCCCSY
---CCCCKCCCCC-
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.00965796 |
Alignment:
BBGGGGCGGGGCVD
-GGGGGYGGGG---
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0127292 |
Alignment:
BCCGCCCCGCCCCBB
----CCCCKCCCCC-
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0174405 |
Alignment:
GGMGGRGGCGGVGC
---GGGGGYGGGG-
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0301548 |
Alignment:
CYYCBBCYYYTCCHCCTYYY
------CCCCKCCCCC----
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY |
Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0329167 |
Alignment:
RGRGGAGRRGGHGGDG
------GGGGGYGGGG
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG |
Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0540825 |
Alignment:
BBVGCCBVGGCCTV
----GGGGGYGGGG
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
49 |
| Motif name: |
TFF1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0557738 |
Alignment:
GCVGCGGCBCCG
-CCCCKCCCCC-
| Original motif Consensus sequence: CGGVGCCGCVGC |
Reverse complement motif Consensus sequence: GCVGCGGCBCCG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0600992 |
Alignment:
CCCCCTTGGGCCCC
-CCCCKCCCCC---
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
27 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0637183 |
Alignment:
GCCYCMCCCD
CCCCKCCCCC
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 25 |
Motif name: TFAP2A |
| Original motif Consensus sequence: GCCBBVRGS |
Reverse complement motif Consensus sequence: SCMVBBGGC |
|
|
 |
 |
Best Matches for Motif ID 25 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
9 |
| Similarity score: |
0.0131247 |
Alignment:
BBVGCCBVGGCCTV
---SCMVBBGGC--
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
9 |
| Similarity score: |
0.0414588 |
Alignment:
GGGGCCCAAGGGGG
---GCCBBVRGS--
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0.0415732 |
Alignment:
GGMGGRGGCGGVGC
SCMVBBGGC-----
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
33 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.0550803 |
Alignment:
HSCACGTGGC
-SCMVBBGGC
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
48 |
| Motif name: |
TFW3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.0552877 |
Alignment:
CGGCYBCGCG
-GCCBBVRGS
| Original motif Consensus sequence: CGGCYBCGCG |
Reverse complement motif Consensus sequence: CGCGBMGCCG |
|
|
 |
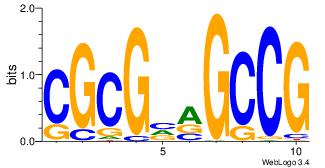 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
9 |
| Similarity score: |
0.0581255 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-------GCCBBVRGS----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
34 |
| Motif name: |
Myc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.0613455 |
Alignment:
VGCACGTGGH
-SCMVBBGGC
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
11 |
| Number of overlap: |
9 |
| Similarity score: |
0.0630429 |
Alignment:
KKKAGGDGGAKKMGBBGKMG
-SCMVBBGGC----------
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY |
Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
49 |
| Motif name: |
TFF1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0.0669772 |
Alignment:
GCVGCGGCBCCG
SCMVBBGGC---
| Original motif Consensus sequence: CGGVGCCGCVGC |
Reverse complement motif Consensus sequence: GCVGCGGCBCCG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
9 |
| Similarity score: |
0.069755 |
Alignment:
CHCCBCCKMCTCCKCM
SCMVBBGGC-------
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG |
Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 26 |
Motif name: MIZF |
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
Best Matches for Motif ID 26 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
35 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0459305 |
Alignment:
CCGGAAGTGVV
GCGGACGTTV-
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0778314 |
Alignment:
BCACCTGCABC
BAACGTCCGC-
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
44 |
| Motif name: |
dhACATTCTkh |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0794339 |
Alignment:
DHACATTCTGH
BAACGTCCGC-
| Original motif Consensus sequence: DHACATTCTGH |
Reverse complement motif Consensus sequence: HCAGAATGTHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0804061 |
Alignment:
SGGTCACGTGACCS
-GCGGACGTTV---
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0861006 |
Alignment:
GCVCCGCCMCCYCC
-BAACGTCCGC---
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
10 |
| Similarity score: |
0.0871006 |
Alignment:
CYYCBBCYYYTCCHCCTYYY
-----BAACGTCCGC-----
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY |
Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0877229 |
Alignment:
BCCGCCCCGCCCCBB
BAACGTCCGC-----
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
10 |
| Number of overlap: |
10 |
| Similarity score: |
0.0877672 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-GCGGACGTTV---------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0884513 |
Alignment:
HVGCCCCGCCCCBB
---BAACGTCCGC-
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
45 |
| Motif name: |
wbgTAAATAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0893149 |
Alignment:
DBGTAAATAHD
GCGGACGTTV-
| Original motif Consensus sequence: DBGTAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTACBD |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 27 |
Motif name: Klf4 |
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
Best Matches for Motif ID 27 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0152457 |
Alignment:
CSKCCCCGCCCCSY
--GCCYCMCCCD--
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0160185 |
Alignment:
BBGGGGCGGGGCVD
--DGGGYGKGGC--
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0194893 |
Alignment:
BBGGGGCGGGGCGGB
--DGGGYGKGGC---
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0402906 |
Alignment:
GGMGGRGGCGGVGC
----DGGGYGKGGC
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
10 |
| Similarity score: |
0.0556427 |
Alignment:
CYYCBBCYYYTCCHCCTYYY
----GCCYCMCCCD------
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY |
Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0606033 |
Alignment:
CHCCBCCKMCTCCKCM
----GCCYCMCCCD--
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG |
Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0623591 |
Alignment:
BBVGCCBVGGCCTV
-GCCYCMCCCD---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
24 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0663136 |
Alignment:
GGGGGYGGGG
DGGGYGKGGC
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
34 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0686848 |
Alignment:
VGCACGTGGH
DGGGYGKGGC
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
10 |
| Similarity score: |
0.06956 |
Alignment:
DGVBCABTGDWGCGKRRCSR
--------DGGGYGKGGC--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 28 |
Motif name: E2F1 |
| Original motif Consensus sequence: TTTSGCGC |
Reverse complement motif Consensus sequence: GCGCSAAA |
|
|
 |
 |
Best Matches for Motif ID 28 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0568827 |
Alignment:
GGGGCCCAAGGGGG
-GCGCSAAA-----
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
39 |
| Motif name: |
kCAGCCAATmr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0624791 |
Alignment:
DCAGCCAATVR
-GCGCSAAA--
| Original motif Consensus sequence: DCAGCCAATVR |
Reverse complement motif Consensus sequence: MBATTGGCTGH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0781129 |
Alignment:
GCVCCGCCMCCYCC
---GCGCSAAA---
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
23 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0784105 |
Alignment:
YCGCCCACGCH
GCGCSAAA---
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
49 |
| Motif name: |
TFF1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0798986 |
Alignment:
CGGVGCCGCVGC
--GCGCSAAA--
| Original motif Consensus sequence: CGGVGCCGCVGC |
Reverse complement motif Consensus sequence: GCVGCGGCBCCG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0836188 |
Alignment:
TTTTATTGTYAT
---TTTSGCGC-
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0836188 |
Alignment:
ATTTWATGAAA
-TTTSGCGC--
| Original motif Consensus sequence: ATTTWATGAAA |
Reverse complement motif Consensus sequence: TTTCATWAAAT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
19 |
| Number of overlap: |
7 |
| Similarity score: |
0.569963 |
Alignment:
-HDWVAAAHAAAAAMAAAMWWWHBWA
GCGCSAAA------------------
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA |
Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
7 |
| Similarity score: |
0.571005 |
Alignment:
-BBGGGGCGGGGCGGB
TTTSGCGC--------
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
48 |
| Motif name: |
TFW3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.571119 |
Alignment:
CGGCYBCGCG-
---TTTSGCGC
| Original motif Consensus sequence: CGGCYBCGCG |
Reverse complement motif Consensus sequence: CGCGBMGCCG |
|
|
 |
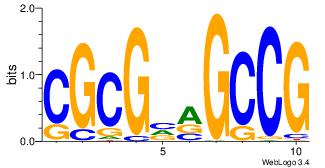 |
| Dataset #: 3 |
Motif ID: 29 |
Motif name: HIF1AARNT |
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
Best Matches for Motif ID 29 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
33 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0123101 |
Alignment:
GCCACGTGSD
-VBACGTGV-
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
34 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0144491 |
Alignment:
DCCACGTGCV
-VBACGTGV-
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0295473 |
Alignment:
BCACCTGCABC
VBACGTGV---
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0314837 |
Alignment:
SGGTCACGTGACCS
---VCACGTBV---
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
26 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0377204 |
Alignment:
GCGGACGTTV
--VCACGTBV
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
8 |
| Similarity score: |
0.0556891 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-----VCACGTBV-------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
44 |
| Motif name: |
dhACATTCTkh |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0640024 |
Alignment:
DHACATTCTGH
VCACGTBV---
| Original motif Consensus sequence: DHACATTCTGH |
Reverse complement motif Consensus sequence: HCAGAATGTHD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
35 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0648153 |
Alignment:
CCGGAAGTGVV
--VCACGTBV-
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
47 |
| Motif name: |
TFW2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0660185 |
Alignment:
CCGCGCGS
VBACGTGV
| Original motif Consensus sequence: SCGCGCGG |
Reverse complement motif Consensus sequence: CCGCGCGS |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.067219 |
Alignment:
GCVCCGCCMCCYCC
------VCACGTBV
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 30 |
Motif name: PLAG1 |
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
Best Matches for Motif ID 30 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
14 |
| Similarity score: |
0.0876918 |
Alignment:
CYYCBBCYYYTCCHCCTYYY
--CCCCCTTGGGCCCC----
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY |
Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0880716 |
Alignment:
CHCCBCCKMCTCCKCM
--CCCCCTTGGGCCCC
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG |
Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0886295 |
Alignment:
VAGGCCBBGGCVBB
GGGGCCCAAGGGGG
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0904945 |
Alignment:
BCCGCCCCGCCCCBB
CCCCCTTGGGCCCC-
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0943783 |
Alignment:
DGVBCABTGDWGCGKRRCSR
GGGGCCCAAGGGGG------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.588053 |
Alignment:
-MSGGGGCGGGGYSG
GGGGCCCAAGGGGG-
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.589357 |
Alignment:
GCVCCGCCMCCYCC-
-CCCCCTTGGGCCCC
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.595227 |
Alignment:
-HVGCCCCGCCCCBB
CCCCCTTGGGCCCC-
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
49 |
| Motif name: |
TFF1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.09276 |
Alignment:
CGGVGCCGCVGC--
GGGGCCCAAGGGGG
| Original motif Consensus sequence: CGGVGCCGCVGC |
Reverse complement motif Consensus sequence: GCVGCGGCBCCG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
24 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
2.07807 |
Alignment:
----GGGGGYGGGG
GGGGCCCAAGGGGG
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 31 |
Motif name: Pax5 |
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
Best Matches for Motif ID 31 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
20 |
| Similarity score: |
0.0373929 |
Alignment:
KKKAGGDGGAKKMGBBGKMG
DGVBCABTGDWGCGKRRCSR
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY |
Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
20 |
| Similarity score: |
0.0546306 |
Alignment:
HDWVAAAHAAAAAMAAAMWWWHBWA
DGVBCABTGDWGCGKRRCSR-----
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA |
Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
17 |
| Similarity score: |
1.5457 |
Alignment:
TWAAWTTVTGAAAAAHWW---
-DGVBCABTGDWGCGKRRCSR
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA |
Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
16 |
| Similarity score: |
2.0519 |
Alignment:
RGRGGAGRRGGHGGDG----
MSGKKRCGCWDCABTGBBCD
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG |
Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
52 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
16 |
| Similarity score: |
2.05456 |
Alignment:
ABAAAAAAWHAAAAARAW----
--DGVBCABTGDWGCGKRRCSR
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT |
Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
2.54483 |
Alignment:
BBGGGGCGGGGCGGB-----
MSGKKRCGCWDCABTGBBCD
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
15 |
| Similarity score: |
2.55478 |
Alignment:
WAHHTVTTYKAAAAWTTRAT-----
-----MSGKKRCGCWDCABTGBBCD
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.0203 |
Alignment:
------BBGGGGCGGGGCVD
DGVBCABTGDWGCGKRRCSR
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.02361 |
Alignment:
------CSKCCCCGCCCCSY
MSGKKRCGCWDCABTGBBCD
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.02394 |
Alignment:
------MTMMTCMMTCTKCK
MSGKKRCGCWDCABTGBBCD
| Original motif Consensus sequence: RGRAGARRGARRAR |
Reverse complement motif Consensus sequence: MTMMTCMMTCTKCK |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 32 |
Motif name: ArntAhr |
| Original motif Consensus sequence: YGCGTG |
Reverse complement motif Consensus sequence: CACGCM |
|
|
 |
 |
Best Matches for Motif ID 32 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
23 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0 |
Alignment:
YCGCCCACGCH
-----CACGCM
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
29 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0112447 |
Alignment:
VBACGTGV
-YGCGTG-
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
33 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0377949 |
Alignment:
GCCACGTGSD
--YGCGTG--
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0396403 |
Alignment:
HVGCCCCGCCCCBB
----CACGCM----
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
34 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0399214 |
Alignment:
DCCACGTGCV
--YGCGTG--
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
1 |
| Motif name: |
Motif 1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.040625 |
Alignment:
GCCCCGCC
--CACGCM
| Original motif Consensus sequence: GGCGGGGC |
Reverse complement motif Consensus sequence: GCCCCGCC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0412037 |
Alignment:
SGGTCACGTGACCS
----YGCGTG----
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
6 |
| Similarity score: |
0.0425089 |
Alignment:
BCCGCCCCGCCCCBB
-----CACGCM----
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0465867 |
Alignment:
MSGGGGCGGGGYSG
----YGCGTG----
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
47 |
| Motif name: |
TFW2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0469722 |
Alignment:
SCGCGCGG
-YGCGTG-
| Original motif Consensus sequence: SCGCGCGG |
Reverse complement motif Consensus sequence: CCGCGCGS |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 33 |
Motif name: Mycn |
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
Best Matches for Motif ID 33 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
34 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0 |
Alignment:
VGCACGTGGH
HSCACGTGGC
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0628441 |
Alignment:
SGGTCACGTGACCS
--HSCACGTGGC--
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
35 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0884396 |
Alignment:
VVCACTTCCGG
GCCACGTGSD-
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
27 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0893272 |
Alignment:
DGGGYGKGGC
HSCACGTGGC
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0895444 |
Alignment:
GGMGGRGGCGGVGC
--HSCACGTGGC--
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0899944 |
Alignment:
VAGGCCBBGGCVBB
---GCCACGTGSD-
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0921439 |
Alignment:
CCCCCTTGGGCCCC
----HSCACGTGGC
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.093472 |
Alignment:
HVGCCCCGCCCCBB
--GCCACGTGSD--
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0964516 |
Alignment:
MSGGGGCGGGGYSG
--HSCACGTGGC--
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0979367 |
Alignment:
BCCGCCCCGCCCCBB
---GCCACGTGSD--
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 34 |
Motif name: Myc |
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
Best Matches for Motif ID 34 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
33 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0 |
Alignment:
GCCACGTGSD
DCCACGTGCV
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0646601 |
Alignment:
SGGTCACGTGACCS
--DCCACGTGCV--
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
35 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0840341 |
Alignment:
VVCACTTCCGG
DCCACGTGCV-
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0851555 |
Alignment:
GGMGGRGGCGGVGC
--VGCACGTGGH--
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
27 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0883745 |
Alignment:
DGGGYGKGGC
VGCACGTGGH
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0925451 |
Alignment:
VAGGCCBBGGCVBB
---DCCACGTGCV-
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0928504 |
Alignment:
HVGCCCCGCCCCBB
--DCCACGTGCV--
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0930375 |
Alignment:
CCCCCTTGGGCCCC
----VGCACGTGGH
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0941376 |
Alignment:
MSGGGGCGGGGYSG
--VGCACGTGGH--
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0974172 |
Alignment:
BCCGCCCCGCCCCBB
---DCCACGTGCV--
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 35 |
Motif name: GABPA |
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
Best Matches for Motif ID 35 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
0.0360984 |
Alignment:
CYYCBBCYYYTCCHCCTYYY
----VVCACTTCCGG-----
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY |
Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
0.0445566 |
Alignment:
RGRGGAGRRGGHGGDG
-CCGGAAGTGVV----
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG |
Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0446257 |
Alignment:
DGVBCABTGDWGCGKRRCSR
-------VVCACTTCCGG--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0457327 |
Alignment:
BBVGCCBVGGCCTV
VVCACTTCCGG---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0483841 |
Alignment:
BCACCTGCABC
VVCACTTCCGG
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
45 |
| Motif name: |
wbgTAAATAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0492127 |
Alignment:
DBGTAAATAHD
CCGGAAGTGVV
| Original motif Consensus sequence: DBGTAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTACBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
44 |
| Motif name: |
dhACATTCTkh |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0495154 |
Alignment:
DHACATTCTGH
VVCACTTCCGG
| Original motif Consensus sequence: DHACATTCTGH |
Reverse complement motif Consensus sequence: HCAGAATGTHD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0504656 |
Alignment:
GGMGGRGGCGGVGC
-CCGGAAGTGVV--
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0518418 |
Alignment:
AATTYDGAARTAWW
---CCGGAAGTGVV
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0554249 |
Alignment:
BCCGCCCCGCCCCBB
---VVCACTTCCGG-
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 36 |
Motif name: csGCCCCGCCCCsc |
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
Best Matches for Motif ID 36 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0 |
Alignment:
BCCGCCCCGCCCCBB
-HVGCCCCGCCCCBB
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0117332 |
Alignment:
CSKCCCCGCCCCSY
HVGCCCCGCCCCBB
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0747029 |
Alignment:
MSGKKRCGCWDCABTGBBCD
------HVGCCCCGCCCCBB
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.0777991 |
Alignment:
CYYCBBCYYYTCCHCCTYYY
---HVGCCCCGCCCCBB---
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY |
Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
14 |
| Similarity score: |
0.0880609 |
Alignment:
RGRGGAGRRGGHGGDG
BBGGGGCGGGGCVD--
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG |
Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0884848 |
Alignment:
BBVGCCBVGGCCTV
BBGGGGCGGGGCVD
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0937271 |
Alignment:
GCVCCGCCMCCYCC
HVGCCCCGCCCCBB
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0971132 |
Alignment:
SGGTCACGTGACCS
HVGCCCCGCCCCBB
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.609319 |
Alignment:
GGGGCCCAAGGGGG-
-BBGGGGCGGGGCVD
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
49 |
| Motif name: |
TFF1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.08016 |
Alignment:
CGGVGCCGCVGC--
HVGCCCCGCCCCBB
| Original motif Consensus sequence: CGGVGCCGCVGC |
Reverse complement motif Consensus sequence: GCVGCGGCBCCG |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 37 |
Motif name: tkAAATAATAtw |
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
Best Matches for Motif ID 37 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0 |
Alignment:
HDAAATAATADD
HDAAATAATAHW
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0522317 |
Alignment:
TWSTTTWAWTTTWT
-WHTATTATTTDH-
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
12 |
| Number of overlap: |
12 |
| Similarity score: |
0.0590505 |
Alignment:
TWVHWWWYTTTYTTTTTHTTTVWBH
-----------WHTATTATTTDH--
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA |
Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
52 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0637702 |
Alignment:
ABAAAAAAWHAAAAARAW
HDAAATAATAHW------
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT |
Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0641975 |
Alignment:
AATHATATWTHAAA
--WHTATTATTTDH
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0704475 |
Alignment:
AAATTKTATTTAWT
-WHTATTATTTDH-
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
12 |
| Similarity score: |
0.0704475 |
Alignment:
WAHHTVTTYKAAAAWTTRAT
--------HDAAATAATAHW
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0763819 |
Alignment:
WWHTTTTTCABAAWTTWA
------WHTATTATTTDH
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA |
Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0864198 |
Alignment:
TCAAAATKAWTTGT
--WHTATTATTTDH
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.09076 |
Alignment:
WWTAKTTCDKAATT
WHTATTATTTDH--
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 38 |
Motif name: cccGCCCCGCCCCsb |
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
Best Matches for Motif ID 38 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
15 |
| Similarity score: |
0.0773345 |
Alignment:
CYYCBBCYYYTCCHCCTYYY
--BCCGCCCCGCCCCBB---
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY |
Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
15 |
| Similarity score: |
0.0869149 |
Alignment:
RGRGGAGRRGGHGGDG
BBGGGGCGGGGCGGB-
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG |
Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
0.0992376 |
Alignment:
MSGKKRCGCWDCABTGBBCD
BBGGGGCGGGGCGGB-----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.5 |
Alignment:
-HVGCCCCGCCCCBB
BCCGCCCCGCCCCBB
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.513015 |
Alignment:
-CSKCCCCGCCCCSY
BCCGCCCCGCCCCBB
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.574434 |
Alignment:
BBVGCCBVGGCCTV-
BCCGCCCCGCCCCBB
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.58333 |
Alignment:
GCVCCGCCMCCYCC-
BCCGCCCCGCCCCBB
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.601399 |
Alignment:
SGGTCACGTGACCS-
BBGGGGCGGGGCGGB
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.604586 |
Alignment:
GGGGCCCAAGGGGG-
BBGGGGCGGGGCGGB
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
49 |
| Motif name: |
TFF1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.58073 |
Alignment:
---GCVGCGGCBCCG
BCCGCCCCGCCCCBB
| Original motif Consensus sequence: CGGVGCCGCVGC |
Reverse complement motif Consensus sequence: GCVGCGGCBCCG |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 39 |
Motif name: kCAGCCAATmr |
| Original motif Consensus sequence: DCAGCCAATVR |
Reverse complement motif Consensus sequence: MBATTGGCTGH |
|
|
 |
 |
Best Matches for Motif ID 39 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0559269 |
Alignment:
WHTATTATTTDH
-MBATTGGCTGH
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
44 |
| Motif name: |
dhACATTCTkh |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0588235 |
Alignment:
HCAGAATGTHD
DCAGCCAATVR
| Original motif Consensus sequence: DHACATTCTGH |
Reverse complement motif Consensus sequence: HCAGAATGTHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0597643 |
Alignment:
DDTATTATTTDH
-MBATTGGCTGH
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0599548 |
Alignment:
BCACCTGCABC
MBATTGGCTGH
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0614719 |
Alignment:
GCVCCGCCMCCYCC
--DCAGCCAATVR-
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
11 |
| Similarity score: |
0.0635027 |
Alignment:
HDWVAAAHAAAAAMAAAMWWWHBWA
--------DCAGCCAATVR------
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA |
Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
Motif 18 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0661765 |
Alignment:
TTAGWTWATAA
DCAGCCAATVR
| Original motif Consensus sequence: TTAGWTWATAA |
Reverse complement motif Consensus sequence: TTATWAWCTAA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
35 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.56089 |
Alignment:
-CCGGAAGTGVV
DCAGCCAATVR-
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
45 |
| Motif name: |
wbgTAAATAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.561975 |
Alignment:
DHTATTTACBD-
-MBATTGGCTGH
| Original motif Consensus sequence: DBGTAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTACBD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.563709 |
Alignment:
VAGGCCBBGGCVBB-
----DCAGCCAATVR
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 40 |
Motif name: kcACCTGCAgc |
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
Best Matches for Motif ID 40 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
43 |
| Motif name: |
wsTACwGTAsw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0215284 |
Alignment:
DBTACWGTAVH
BCACCTGCABC
| Original motif Consensus sequence: HVTACWGTABD |
Reverse complement motif Consensus sequence: DBTACWGTAVH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
44 |
| Motif name: |
dhACATTCTkh |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0254134 |
Alignment:
DHACATTCTGH
BCACCTGCABC
| Original motif Consensus sequence: DHACATTCTGH |
Reverse complement motif Consensus sequence: HCAGAATGTHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.031338 |
Alignment:
SGGTCACGTGACCS
---BCACCTGCABC
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.034471 |
Alignment:
GCVCCGCCMCCYCC
BCACCTGCABC---
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0402518 |
Alignment:
HVGCCCCGCCCCBB
-BCACCTGCABC--
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0411257 |
Alignment:
CSKCCCCGCCCCSY
-BCACCTGCABC--
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
0.0420218 |
Alignment:
DGVBCABTGDWGCGKRRCSR
----GBTGCAGGTGB-----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
35 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0423275 |
Alignment:
CCGGAAGTGVV
GBTGCAGGTGB
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
39 |
| Motif name: |
kCAGCCAATmr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0439586 |
Alignment:
DCAGCCAATVR
GBTGCAGGTGB
| Original motif Consensus sequence: DCAGCCAATVR |
Reverse complement motif Consensus sequence: MBATTGGCTGH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0461444 |
Alignment:
BCCGCCCCGCCCCBB
--BCACCTGCABC--
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 41 |
Motif name: wwAAATAATAtw |
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
Best Matches for Motif ID 41 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0 |
Alignment:
HDAAATAATAHW
HDAAATAATADD
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0536028 |
Alignment:
AWAAAWTWAAASWA
-HDAAATAATADD-
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
52 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
12 |
| Similarity score: |
0.0565705 |
Alignment:
WTKTTTTTHWTTTTTTBT
------DDTATTATTTDH
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT |
Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0575935 |
Alignment:
HDWVAAAHAAAAAMAAAMWWWHBWA
--HDAAATAATADD-----------
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA |
Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0591821 |
Alignment:
AATHATATWTHAAA
--DDTATTATTTDH
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0712963 |
Alignment:
WAHHTVTTYKAAAAWTTRAT
--------HDAAATAATADD
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0715278 |
Alignment:
AAATTKTATTTAWT
-DDTATTATTTDH-
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0733516 |
Alignment:
WWHTTTTTCABAAWTTWA
------DDTATTATTTDH
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA |
Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0912037 |
Alignment:
TCAAAATKAWTTGT
--DDTATTATTTDH
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0924576 |
Alignment:
WWTAKTTCDKAATT
DDTATTATTTDH--
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 42 |
Motif name: sSGTCACGTGACSs |
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
Best Matches for Motif ID 42 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
14 |
| Similarity score: |
0.0518737 |
Alignment:
DGVBCABTGDWGCGKRRCSR
------SGGTCACGTGACCS
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0539097 |
Alignment:
BBGGGGCGGGGCVD
SGGTCACGTGACCS
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0581955 |
Alignment:
BCCGCCCCGCCCCBB
SGGTCACGTGACCS-
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0634135 |
Alignment:
MSGGGGCGGGGYSG
SGGTCACGTGACCS
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
1.05855 |
Alignment:
--GCVCCGCCMCCYCC
SGGTCACGTGACCS--
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
49 |
| Motif name: |
TFF1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.06047 |
Alignment:
--GCVGCGGCBCCG
SGGTCACGTGACCS
| Original motif Consensus sequence: CGGVGCCGCVGC |
Reverse complement motif Consensus sequence: GCVGCGGCBCCG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.54742 |
Alignment:
---BCACCTGCABC
SGGTCACGTGACCS
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
48 |
| Motif name: |
TFW3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
2.05201 |
Alignment:
----CGCGBMGCCG
SGGTCACGTGACCS
| Original motif Consensus sequence: CGGCYBCGCG |
Reverse complement motif Consensus sequence: CGCGBMGCCG |
|
|
 |
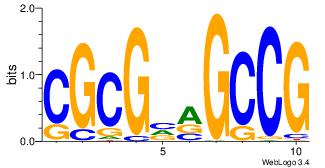 |
| Dataset #: |
3 |
| Motif ID: |
23 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
2.06209 |
Alignment:
----YCGCCCACGCH
SGGTCACGTGACCS-
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
7 |
| Similarity score: |
3.53253 |
Alignment:
CCCCCTTGGGCCCC-------
-------SGGTCACGTGACCS
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 43 |
Motif name: wsTACwGTAsw |
| Original motif Consensus sequence: HVTACWGTABD |
Reverse complement motif Consensus sequence: DBTACWGTAVH |
|
|
 |
 |
Best Matches for Motif ID 43 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0199592 |
Alignment:
GBTGCAGGTGB
HVTACWGTABD
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0309343 |
Alignment:
DDTATTATTTDH
-HVTACWGTABD
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0337121 |
Alignment:
HDAAATAATAHW
DBTACWGTAVH-
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0353535 |
Alignment:
AATTYDGAARTAWW
HVTACWGTABD---
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
45 |
| Motif name: |
wbgTAAATAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.037518 |
Alignment:
DBGTAAATAHD
DBTACWGTAVH
| Original motif Consensus sequence: DBGTAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTACBD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0397727 |
Alignment:
TTTDAWATATHATT
DBTACWGTAVH---
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
0.0413797 |
Alignment:
TWAAWTTVTGAAAAAHWW
---HVTACWGTABD----
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA |
Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
11 |
| Similarity score: |
0.042298 |
Alignment:
DGVBCABTGDWGCGKRRCSR
-----HVTACWGTABD----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0445707 |
Alignment:
ATKAAWTTTTRMAABAHHTW
-------DBTACWGTAVH--
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0460859 |
Alignment:
TTCWTAGATTAWA
DBTACWGTAVH--
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 44 |
Motif name: dhACATTCTkh |
| Original motif Consensus sequence: DHACATTCTGH |
Reverse complement motif Consensus sequence: HCAGAATGTHD |
|
|
 |
 |
Best Matches for Motif ID 44 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
11 |
| Similarity score: |
0.0217516 |
Alignment:
TWAAWTTVTGAAAAAHWW
DHACATTCTGH-------
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA |
Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0238442 |
Alignment:
BCACCTGCABC
DHACATTCTGH
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0258838 |
Alignment:
TCAAAATKAWTTGT
HCAGAATGTHD---
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0292508 |
Alignment:
DDTATTATTTDH
-HCAGAATGTHD
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
11 |
| Number of overlap: |
11 |
| Similarity score: |
0.0307858 |
Alignment:
TWVHWWWYTTTYTTTTTHTTTVWBH
----DHACATTCTGH----------
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA |
Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0357323 |
Alignment:
ATKAAWTTTTRMAABAHHTW
-DHACATTCTGH--------
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0364899 |
Alignment:
WHTATTATTTDH
-HCAGAATGTHD
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0400884 |
Alignment:
AATTYDGAARTAWW
HCAGAATGTHD---
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
39 |
| Motif name: |
kCAGCCAATmr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0412582 |
Alignment:
MBATTGGCTGH
DHACATTCTGH
| Original motif Consensus sequence: DCAGCCAATVR |
Reverse complement motif Consensus sequence: MBATTGGCTGH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
35 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0418896 |
Alignment:
CCGGAAGTGVV
HCAGAATGTHD
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 45 |
Motif name: wbgTAAATAww |
| Original motif Consensus sequence: DBGTAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTACBD |
|
|
 |
 |
Best Matches for Motif ID 45 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0162338 |
Alignment:
HDAAATAATAHW
-DBGTAAATAHD
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0164863 |
Alignment:
DDTATTATTTDH
DHTATTTACBD-
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
15 |
| Number of overlap: |
11 |
| Similarity score: |
0.0192768 |
Alignment:
TWVHWWWYTTTYTTTTTHTTTVWBH
--------------DHTATTTACBD
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA |
Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0250541 |
Alignment:
AATHATATWTHAAA
DHTATTTACBD---
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.027895 |
Alignment:
AATTYDGAARTAWW
---DBGTAAATAHD
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
52 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0283217 |
Alignment:
ABAAAAAAWHAAAAARAW
DBGTAAATAHD-------
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT |
Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0308097 |
Alignment:
TWAAWTTVTGAAAAAHWW
------DBGTAAATAHD-
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA |
Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
11 |
| Similarity score: |
0.0333874 |
Alignment:
ATKAAWTTTTRMAABAHHTW
-------DBGTAAATAHD--
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0361187 |
Alignment:
AAAAWTTRCWT
DHTATTTACBD
| Original motif Consensus sequence: AAAAWTTRCWT |
Reverse complement motif Consensus sequence: AWGKAAWTTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0410381 |
Alignment:
AWAAAWTWAAASWA
--DHTATTTACBD-
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 46 |
Motif name: TFW1 |
| Original motif Consensus sequence: GTCGCG |
Reverse complement motif Consensus sequence: CGCGAC |
|
|
 |
 |
Best Matches for Motif ID 46 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
48 |
| Motif name: |
TFW3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0350356 |
Alignment:
CGGCYBCGCG
----GTCGCG
| Original motif Consensus sequence: CGGCYBCGCG |
Reverse complement motif Consensus sequence: CGCGBMGCCG |
|
|
 |
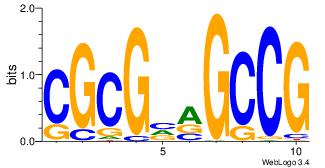 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0375102 |
Alignment:
SGGTCACGTGACCS
--GTCGCG------
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0381641 |
Alignment:
GCVCCGCCMCCYCC
----CGCGAC----
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
49 |
| Motif name: |
TFF1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
6 |
| Similarity score: |
0.0431161 |
Alignment:
CGGVGCCGCVGC
------CGCGAC
| Original motif Consensus sequence: CGGVGCCGCVGC |
Reverse complement motif Consensus sequence: GCVGCGGCBCCG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
47 |
| Motif name: |
TFW2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.052047 |
Alignment:
CCGCGCGS
-CGCGAC-
| Original motif Consensus sequence: SCGCGCGG |
Reverse complement motif Consensus sequence: CCGCGCGS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
33 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0537993 |
Alignment:
GCCACGTGSD
----GTCGCG
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0539 |
Alignment:
HVGCCCCGCCCCBB
--GTCGCG------
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
1 |
| Motif name: |
Motif 1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0554263 |
Alignment:
GCCCCGCC
--GTCGCG
| Original motif Consensus sequence: GGCGGGGC |
Reverse complement motif Consensus sequence: GCCCCGCC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
6 |
| Similarity score: |
0.0558733 |
Alignment:
BBGGGGCGGGGCGGB
-------GTCGCG--
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
6 |
| Similarity score: |
0.0644512 |
Alignment:
CSKCCCCGCCCCSY
--CGCGAC------
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 47 |
Motif name: TFW2 |
| Original motif Consensus sequence: SCGCGCGG |
Reverse complement motif Consensus sequence: CCGCGCGS |
|
|
 |
 |
Best Matches for Motif ID 47 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0456233 |
Alignment:
HVGCCCCGCCCCBB
-CCGCGCGS-----
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.045686 |
Alignment:
BBGGGGCGGGGCGGB
------SCGCGCGG-
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
8 |
| Similarity score: |
0.0504058 |
Alignment:
MSGGGGCGGGGYSG
-----SCGCGCGG-
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
33 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0527532 |
Alignment:
HSCACGTGGC
-SCGCGCGG-
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
34 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0574543 |
Alignment:
VGCACGTGGH
-SCGCGCGG-
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0583967 |
Alignment:
GCVCCGCCMCCYCC
---CCGCGCGS---
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
49 |
| Motif name: |
TFF1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.060966 |
Alignment:
CGGVGCCGCVGC
SCGCGCGG----
| Original motif Consensus sequence: CGGVGCCGCVGC |
Reverse complement motif Consensus sequence: GCVGCGGCBCCG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
25 |
| Motif name: |
TFAP2A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0678925 |
Alignment:
SCMVBBGGC
-SCGCGCGG
| Original motif Consensus sequence: GCCBBVRGS |
Reverse complement motif Consensus sequence: SCMVBBGGC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
24 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.07018 |
Alignment:
GGGGGYGGGG
SCGCGCGG--
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0710499 |
Alignment:
BBVGCCBVGGCCTV
--SCGCGCGG----
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 48 |
Motif name: TFW3 |
| Original motif Consensus sequence: CGGCYBCGCG |
Reverse complement motif Consensus sequence: CGCGBMGCCG |
|
|
 |
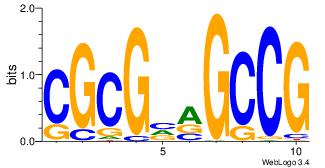 |
Best Matches for Motif ID 48 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
49 |
| Motif name: |
TFF1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0623879 |
Alignment:
CGGVGCCGCVGC
CGGCYBCGCG--
| Original motif Consensus sequence: CGGVGCCGCVGC |
Reverse complement motif Consensus sequence: GCVGCGGCBCCG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
23 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0630189 |
Alignment:
YCGCCCACGCH
CGGCYBCGCG-
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0645308 |
Alignment:
GGMGGRGGCGGVGC
CGCGBMGCCG----
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
11 |
| Number of overlap: |
10 |
| Similarity score: |
0.0714355 |
Alignment:
MSGKKRCGCWDCABTGBBCD
----------CGGCYBCGCG
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0717594 |
Alignment:
CSKCCCCGCCCCSY
CGGCYBCGCG----
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0718771 |
Alignment:
HVGCCCCGCCCCBB
CGGCYBCGCG----
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0719604 |
Alignment:
BCCGCCCCGCCCCBB
-CGGCYBCGCG----
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0766755 |
Alignment:
VAGGCCBBGGCVBB
--CGGCYBCGCG--
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
11 |
| Number of overlap: |
10 |
| Similarity score: |
0.0767689 |
Alignment:
KKKAGGDGGAKKMGBBGKMG
----------CGCGBMGCCG
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY |
Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
10 |
| Similarity score: |
0.0814831 |
Alignment:
CHCCBCCKMCTCCKCM
CGGCYBCGCG------
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG |
Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 49 |
Motif name: TFF1 |
| Original motif Consensus sequence: CGGVGCCGCVGC |
Reverse complement motif Consensus sequence: GCVGCGGCBCCG |
|
|
 |
 |
Best Matches for Motif ID 49 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0292081 |
Alignment:
GGMGGRGGCGGVGC
--CGGVGCCGCVGC
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0542674 |
Alignment:
HVGCCCCGCCCCBB
CGGVGCCGCVGC--
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
12 |
| Similarity score: |
0.0543272 |
Alignment:
KKKAGGDGGAKKMGBBGKMG
------CGGVGCCGCVGC--
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY |
Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
12 |
| Similarity score: |
0.0548371 |
Alignment:
BBGGGGCGGGGCGGB
---CGGVGCCGCVGC
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
12 |
| Similarity score: |
0.0556962 |
Alignment:
DGVBCABTGDWGCGKRRCSR
GCVGCGGCBCCG--------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0594165 |
Alignment:
CHCCBCCKMCTCCKCM
--GCVGCGGCBCCG--
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG |
Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0624366 |
Alignment:
CSKCCCCGCCCCSY
GCVGCGGCBCCG--
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0777862 |
Alignment:
SGGTCACGTGACCS
--CGGVGCCGCVGC
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0779228 |
Alignment:
BBVGCCBVGGCCTV
-GCVGCGGCBCCG-
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0809608 |
Alignment:
GGGGCCCAAGGGGG
CGGVGCCGCVGC--
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 50 |
Motif name: TFF11 |
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
Best Matches for Motif ID 50 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
14 |
| Similarity score: |
0.033716 |
Alignment:
RGRGGAGRRGGHGGDG
GGMGGRGGCGGVGC--
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG |
Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
14 |
| Similarity score: |
0.0386905 |
Alignment:
CYYCBBCYYYTCCHCCTYYY
-----GCVCCGCCMCCYCC-
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY |
Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0548198 |
Alignment:
BBGGGGCGGGGCGGB
GGMGGRGGCGGVGC-
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
14 |
| Similarity score: |
0.0573129 |
Alignment:
DGVBCABTGDWGCGKRRCSR
----GGMGGRGGCGGVGC--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0652173 |
Alignment:
BBGGGGCGGGGCVD
GGMGGRGGCGGVGC
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0661275 |
Alignment:
MSGGGGCGGGGYSG
GGMGGRGGCGGVGC
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0714286 |
Alignment:
RGRAGARRGARRAR
GGMGGRGGCGGVGC
| Original motif Consensus sequence: RGRAGARRGARRAR |
Reverse complement motif Consensus sequence: MTMMTCMMTCTKCK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.574939 |
Alignment:
-CCCCCTTGGGCCCC
GCVCCGCCMCCYCC-
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.577861 |
Alignment:
VAGGCCBBGGCVBB-
-GGMGGRGGCGGVGC
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
49 |
| Motif name: |
TFF1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.02659 |
Alignment:
--CGGVGCCGCVGC
GGMGGRGGCGGVGC
| Original motif Consensus sequence: CGGVGCCGCVGC |
Reverse complement motif Consensus sequence: GCVGCGGCBCCG |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 51 |
Motif name: TFM2 |
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG |
Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
|
|
 |
 |
Best Matches for Motif ID 51 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
16 |
| Similarity score: |
0.0331473 |
Alignment:
CYYCBBCYYYTCCHCCTYYY
CHCCBCCKMCTCCKCM----
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY |
Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
16 |
| Similarity score: |
0.0668155 |
Alignment:
DGVBCABTGDWGCGKRRCSR
-RGRGGAGRRGGHGGDG---
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
16 |
| Similarity score: |
0.0794752 |
Alignment:
HDWVAAAHAAAAAMAAAMWWWHBWA
-------RGRGGAGRRGGHGGDG--
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA |
Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
0.558405 |
Alignment:
BCCGCCCCGCCCCBB-
CHCCBCCKMCTCCKCM
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
1.01195 |
Alignment:
--RGRAGARRGARRAR
RGRGGAGRRGGHGGDG
| Original motif Consensus sequence: RGRAGARRGARRAR |
Reverse complement motif Consensus sequence: MTMMTCMMTCTKCK |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
1.03372 |
Alignment:
GGMGGRGGCGGVGC--
RGRGGAGRRGGHGGDG
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
1.05908 |
Alignment:
MSGGGGCGGGGYSG--
RGRGGAGRRGGHGGDG
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
1.05955 |
Alignment:
HVGCCCCGCCCCBB--
CHCCBCCKMCTCCKCM
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
1.07365 |
Alignment:
--CCCCCTTGGGCCCC
CHCCBCCKMCTCCKCM
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
2.06908 |
Alignment:
VAGGCCBBGGCVBB----
--RGRGGAGRRGGHGGDG
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 52 |
Motif name: TFM1 |
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT |
Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
|
|
 |
 |
Best Matches for Motif ID 52 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
18 |
| Similarity score: |
0 |
Alignment:
TWVHWWWYTTTYTTTTTHTTTVWBH
-----WTKTTTTTHWTTTTTTBT--
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA |
Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
18 |
| Similarity score: |
0.0364672 |
Alignment:
ATKAAWTTTTRMAABAHHTW
ABAAAAAAWHAAAAARAW--
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
0.0456974 |
Alignment:
WWHTTTTTCABAAWTTWA
WTKTTTTTHWTTTTTTBT
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA |
Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
17 |
| Similarity score: |
0.572783 |
Alignment:
CYYCBBCYYYTCCHCCTYYY-
---WTKTTTTTHWTTTTTTBT
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY |
Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
16 |
| Similarity score: |
1.07322 |
Alignment:
--MSGKKRCGCWDCABTGBBCD
WTKTTTTTHWTTTTTTBT----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.01877 |
Alignment:
----TWSTTTWAWTTTWT
WTKTTTTTHWTTTTTTBT
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.05025 |
Alignment:
----AATHATATWTHAAA
ABAAAAAAWHAAAAARAW
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.05187 |
Alignment:
MTMMTCMMTCTKCK----
WTKTTTTTHWTTTTTTBT
| Original motif Consensus sequence: RGRAGARRGARRAR |
Reverse complement motif Consensus sequence: MTMMTCMMTCTKCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.0566 |
Alignment:
AATTYDGAARTAWW----
ABAAAAAAWHAAAAARAW
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.07166 |
Alignment:
AAATRWTAAAATCA----
ABAAAAAAWHAAAAARAW
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 53 |
Motif name: TFM3 |
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA |
Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
|
|
 |
 |
Best Matches for Motif ID 53 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
18 |
| Similarity score: |
0.013876 |
Alignment:
ATKAAWTTTTRMAABAHHTW
-TWAAWTTVTGAAAAAHWW-
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
18 |
| Similarity score: |
0.0402327 |
Alignment:
TWVHWWWYTTTYTTTTTHTTTVWBH
----WWHTTTTTCABAAWTTWA---
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA |
Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
52 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
0.0432207 |
Alignment:
WTKTTTTTHWTTTTTTBT
WWHTTTTTCABAAWTTWA
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT |
Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
17 |
| Similarity score: |
0.561883 |
Alignment:
-MSGKKRCGCWDCABTGBBCD
WWHTTTTTCABAAWTTWA---
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.04564 |
Alignment:
----TWSTTTWAWTTTWT
WWHTTTTTCABAAWTTWA
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.04628 |
Alignment:
WWTAKTTCDKAATT----
TWAAWTTVTGAAAAAHWW
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.04784 |
Alignment:
AATHATATWTHAAA----
WWHTTTTTCABAAWTTWA
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
2.56928 |
Alignment:
-----RGRAGARRGARRAR
TWAAWTTVTGAAAAAHWW-
| Original motif Consensus sequence: RGRAGARRGARRAR |
Reverse complement motif Consensus sequence: MTMMTCMMTCTKCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
2.56949 |
Alignment:
-----TWTAATCTAWGAA
WWHTTTTTCABAAWTTWA
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
3.0422 |
Alignment:
------HDAAATAATADD
TWAAWTTVTGAAAAAHWW
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 54 |
Motif name: TFM12 |
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY |
Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
|
|
 |
 |
Best Matches for Motif ID 54 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
20 |
| Similarity score: |
0.0458155 |
Alignment:
MSGKKRCGCWDCABTGBBCD
CYYCBBCYYYTCCHCCTYYY
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
20 |
| Similarity score: |
0.0573204 |
Alignment:
TWVHWWWYTTTYTTTTTHTTTVWBH
CYYCBBCYYYTCCHCCTYYY-----
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA |
Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
52 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
17 |
| Similarity score: |
1.56255 |
Alignment:
---WTKTTTTTHWTTTTTTBT
CYYCBBCYYYTCCHCCTYYY-
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT |
Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
16 |
| Similarity score: |
2.01568 |
Alignment:
RGRGGAGRRGGHGGDG----
KKKAGGDGGAKKMGBBGKMG
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG |
Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
2.55221 |
Alignment:
-----BCCGCCCCGCCCCBB
CYYCBBCYYYTCCHCCTYYY
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.0395 |
Alignment:
------RGRAGARRGARRAR
KKKAGGDGGAKKMGBBGKMG
| Original motif Consensus sequence: RGRAGARRGARRAR |
Reverse complement motif Consensus sequence: MTMMTCMMTCTKCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.04463 |
Alignment:
------CSKCCCCGCCCCSY
CYYCBBCYYYTCCHCCTYYY
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.05024 |
Alignment:
------BBGGGGCGGGGCVD
KKKAGGDGGAKKMGBBGKMG
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.0542 |
Alignment:
GGMGGRGGCGGVGC------
KKKAGGDGGAKKMGBBGKMG
| Original motif Consensus sequence: GGMGGRGGCGGVGC |
Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.06074 |
Alignment:
GGGGCCCAAGGGGG------
KKKAGGDGGAKKMGBBGKMG
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 55 |
Motif name: TFM13 |
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
Best Matches for Motif ID 55 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
20 |
| Similarity score: |
0.0352451 |
Alignment:
TWVHWWWYTTTYTTTTTHTTTVWBH
----WAHHTVTTYKAAAAWTTRAT-
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA |
Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
1.02254 |
Alignment:
TWAAWTTVTGAAAAAHWW--
ATKAAWTTTTRMAABAHHTW
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA |
Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
52 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
1.02868 |
Alignment:
WTKTTTTTHWTTTTTTBT--
WAHHTVTTYKAAAAWTTRAT
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT |
Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
15 |
| Similarity score: |
2.56565 |
Alignment:
-----MSGKKRCGCWDCABTGBBCD
WAHHTVTTYKAAAAWTTRAT-----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.02278 |
Alignment:
------TTTDAWATATHATT
WAHHTVTTYKAAAAWTTRAT
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.04317 |
Alignment:
AWAAAWTWAAASWA------
ATKAAWTTTTRMAABAHHTW
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.05135 |
Alignment:
WWTAKTTCDKAATT------
WAHHTVTTYKAAAAWTTRAT
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
3.56273 |
Alignment:
TTCWTAGATTAWA-------
WAHHTVTTYKAAAAWTTRAT
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
3.56529 |
Alignment:
-------ACAAWTRATTTTGA
WAHHTVTTYKAAAAWTTRAT-
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
4.03399 |
Alignment:
--------WHTATTATTTDH
ATKAAWTTTTRMAABAHHTW
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 56 |
Motif name: TFM11 |
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA |
Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
|
|
 |
 |
Best Matches for Motif ID 56 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
20 |
| Similarity score: |
0.0486765 |
Alignment:
WAHHTVTTYKAAAAWTTRAT-----
TWVHWWWYTTTYTTTTTHTTTVWBH
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW |
Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
20 |
| Similarity score: |
0.0693284 |
Alignment:
KKKAGGDGGAKKMGBBGKMG-----
HDWVAAAHAAAAAMAAAMWWWHBWA
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY |
Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
20 |
| Similarity score: |
0.0750613 |
Alignment:
DGVBCABTGDWGCGKRRCSR-----
HDWVAAAHAAAAAMAAAMWWWHBWA
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
52 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
1.0232 |
Alignment:
-------ABAAAAAAWHAAAAARAW
HDWVAAAHAAAAAMAAAMWWWHBWA
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT |
Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
1.05388 |
Alignment:
WWHTTTTTCABAAWTTWA-------
TWVHWWWYTTTYTTTTTHTTTVWBH
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA |
Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.04236 |
Alignment:
TWSTTTWAWTTTWT-----------
TWVHWWWYTTTYTTTTTHTTTVWBH
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.05212 |
Alignment:
-----------RGRAGARRGARRAR
HDWVAAAHAAAAAMAAAMWWWHBWA
| Original motif Consensus sequence: RGRAGARRGARRAR |
Reverse complement motif Consensus sequence: MTMMTCMMTCTKCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.05326 |
Alignment:
AATHATATWTHAAA-----------
TWVHWWWYTTTYTTTTTHTTTVWBH
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.06919 |
Alignment:
WWTAKTTCDKAATT-----------
TWVHWWWYTTTYTTTTTHTTTVWBH
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
4.03464 |
Alignment:
HDAAATAATADD-------------
HDWVAAAHAAAAAMAAAMWWWHBWA
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |