Top 10 Significant Motifs - Global Matching (Highest to Lowest)
| Dataset #: 1 | Motif ID: 1 | Motif name: TFW1 |
| Original motif Consensus sequence: GTCGCG | Reverse complement motif Consensus sequence: CGCGAC |
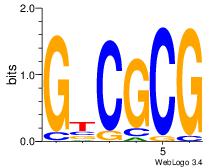 |
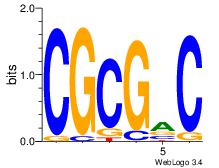 |
Best Matches for Top Significant Motif ID 1 (Highest to Lowest)
| Dataset #: | 2 |
| Motif ID: | 18 |
| Motif name: | sSGTCACGTGACSs |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0375102 |
Alignment:
SGGTCACGTGACCS
--GTCGCG------
| Original motif Consensus sequence: SGGTCACGTGACCS | Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
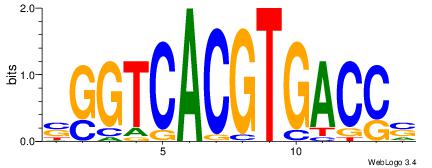 |
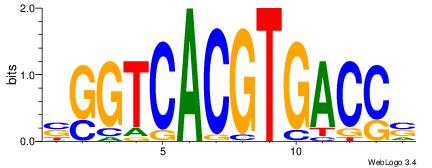 |
| Dataset #: | 2 |
| Motif ID: | 12 |
| Motif name: | csGCCCCGCCCCsc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0539 |
Alignment:
HVGCCCCGCCCCBB
--GTCGCG------
| Original motif Consensus sequence: HVGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
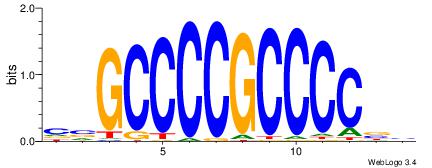 |
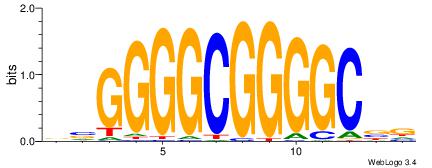 |
| Dataset #: | 2 |
| Motif ID: | 14 |
| Motif name: | cccGCCCCGCCCCsb |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.0558733 |
Alignment:
BCCGCCCCGCCCCBB
-------CGCGAC--
| Original motif Consensus sequence: BCCGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
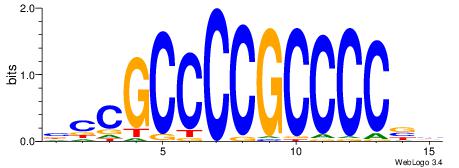 |
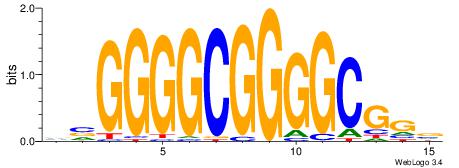 |
| Dataset #: | 2 |
| Motif ID: | 16 |
| Motif name: | kcACCTGCAgc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.0980422 |
Alignment:
GBTGCAGGTGB
GTCGCG-----
| Original motif Consensus sequence: BCACCTGCABC | Reverse complement motif Consensus sequence: GBTGCAGGTGB |
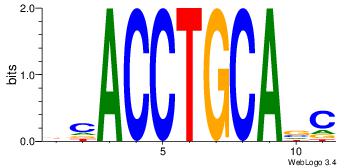 |
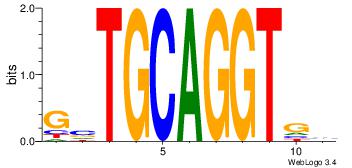 |
| Dataset #: | 2 |
| Motif ID: | 20 |
| Motif name: | dhACATTCTkh |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 4 |
| Similarity score: | 1.09467 |
Alignment:
HCAGAATGTHD--
-------GTCGCG
| Original motif Consensus sequence: DHACATTCTGH | Reverse complement motif Consensus sequence: HCAGAATGTHD |
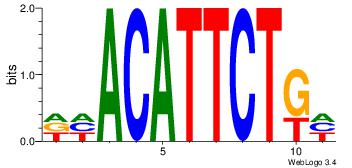 |
 |
| Dataset #: 1 | Motif ID: 3 | Motif name: TFW3 |
| Original motif Consensus sequence: CGGCYBCGCG | Reverse complement motif Consensus sequence: CGCGBMGCCG |
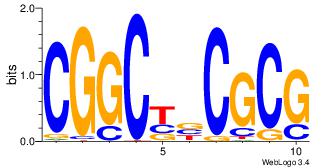 |
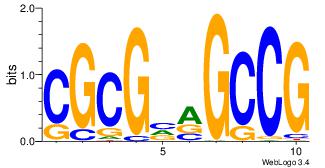 |
Best Matches for Top Significant Motif ID 3 (Highest to Lowest)
| Dataset #: | 2 |
| Motif ID: | 12 |
| Motif name: | csGCCCCGCCCCsc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0718771 |
Alignment:
HVGCCCCGCCCCBB
CGGCYBCGCG----
| Original motif Consensus sequence: HVGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 14 |
| Motif name: | cccGCCCCGCCCCsb |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0719604 |
Alignment:
BCCGCCCCGCCCCBB
-CGGCYBCGCG----
| Original motif Consensus sequence: BCCGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 18 |
| Motif name: | sSGTCACGTGACSs |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0895189 |
Alignment:
SGGTCACGTGACCS
----CGCGBMGCCG
| Original motif Consensus sequence: SGGTCACGTGACCS | Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 16 |
| Motif name: | kcACCTGCAgc |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 1.10343 |
Alignment:
BCACCTGCABC--
---CGCGBMGCCG
| Original motif Consensus sequence: BCACCTGCABC | Reverse complement motif Consensus sequence: GBTGCAGGTGB |
 |
 |
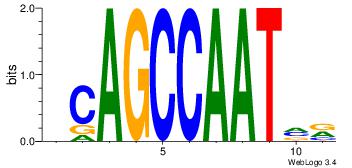
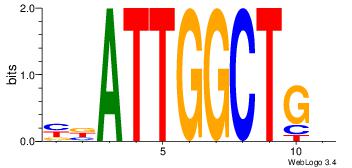
| Dataset #: | 2 |
| Motif ID: | 15 |
| Motif name: | kCAGCCAATmr |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 1.57732 |
Alignment:
---DCAGCCAATVR
CGCGBMGCCG----
| Original motif Consensus sequence: DCAGCCAATVR | Reverse complement motif Consensus sequence: MBATTGGCTGH |
 |
 |
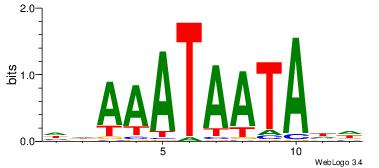
| Dataset #: 2 | Motif ID: 17 | Motif name: wwAAATAATAtw |
| Original motif Consensus sequence: HDAAATAATADD | Reverse complement motif Consensus sequence: DDTATTATTTDH |
 |
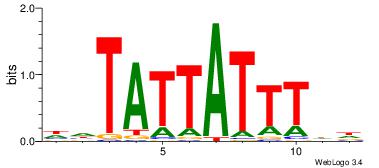 |
Best Matches for Top Significant Motif ID 17 (Highest to Lowest)
| Dataset #: | 1 |
| Motif ID: | 7 |
| Motif name: | TFM1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 12 |
| Similarity score: | 0.0565705 |
Alignment:
ABAAAAAAWHAAAAARAW
------HDAAATAATADD
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT | Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
 |
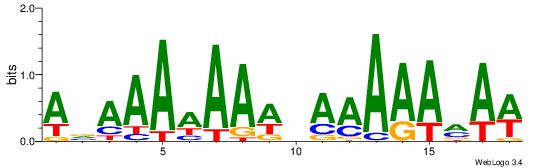 |
| Dataset #: | 1 |
| Motif ID: | 11 |
| Motif name: | TFM11 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0575935 |
Alignment:
HDWVAAAHAAAAAMAAAMWWWHBWA
--HDAAATAATADD-----------
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA | Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
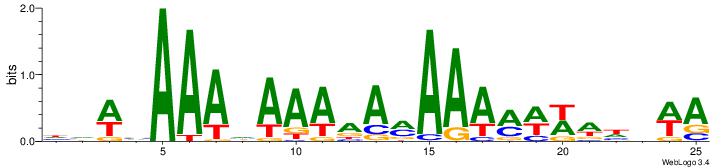 |
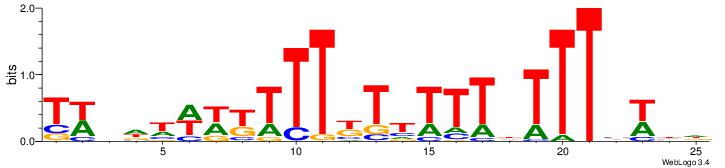 |
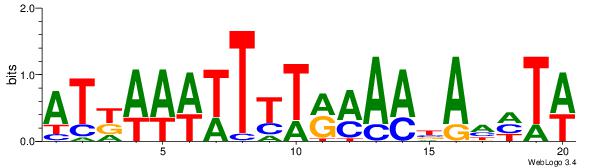
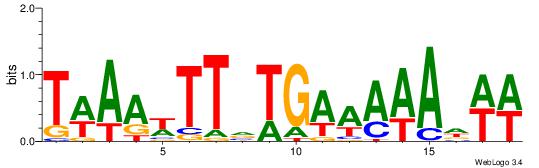
| Dataset #: | 1 |
| Motif ID: | 10 |
| Motif name: | TFM13 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0712963 |
Alignment:
ATKAAWTTTTRMAABAHHTW
--------DDTATTATTTDH
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW | Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
 |
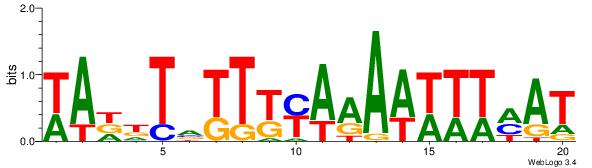 |
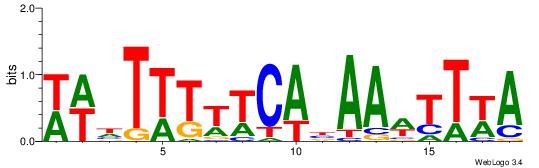
| Dataset #: | 1 |
| Motif ID: | 8 |
| Motif name: | TFM3 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0733516 |
Alignment:
TWAAWTTVTGAAAAAHWW
------HDAAATAATADD
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA | Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
 |
 |
| Dataset #: | 1 |
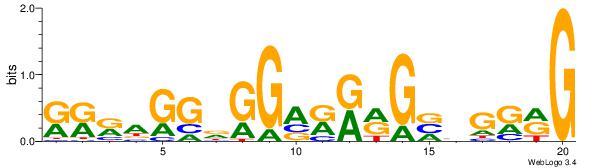
| Motif ID: | 9 |
| Motif name: | TFM12 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 16 |
| Number of overlap: | 5 |
| Similarity score: | 3.59569 |
Alignment:
-------KKKAGGDGGAKKMGBBGKMG
DDTATTATTTDH---------------
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY | Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
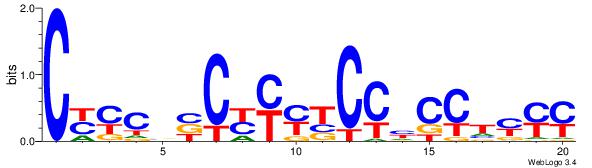 |
 |
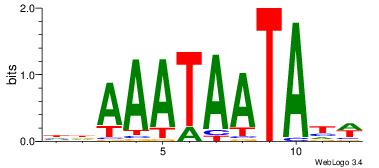
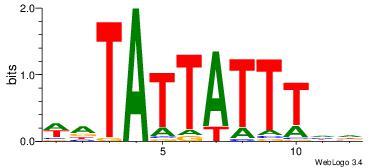
| Dataset #: 2 | Motif ID: 13 | Motif name: tkAAATAATAtw |
| Original motif Consensus sequence: HDAAATAATAHW | Reverse complement motif Consensus sequence: WHTATTATTTDH |
 |
 |
Best Matches for Top Significant Motif ID 13 (Highest to Lowest)
| Dataset #: | 1 |
| Motif ID: | 11 |
| Motif name: | TFM11 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 12 |
| Number of overlap: | 12 |
| Similarity score: | 0.0590505 |
Alignment:
TWVHWWWYTTTYTTTTTHTTTVWBH
-----------WHTATTATTTDH--
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA | Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 7 |
| Motif name: | TFM1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0637702 |
Alignment:
WTKTTTTTHWTTTTTTBT
WHTATTATTTDH------
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT | Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 10 |
| Motif name: | TFM13 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0704475 |
Alignment:
WAHHTVTTYKAAAAWTTRAT
HDAAATAATAHW--------
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW | Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 8 |
| Motif name: | TFM3 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0763819 |
Alignment:
TWAAWTTVTGAAAAAHWW
------HDAAATAATAHW
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA | Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 9 |
| Motif name: | TFM12 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 16 |
| Number of overlap: | 5 |
| Similarity score: | 3.59273 |
Alignment:
-------KKKAGGDGGAKKMGBBGKMG
WHTATTATTTDH---------------
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY | Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
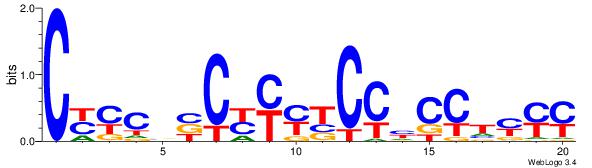 |
 |
| Dataset #: 1 | Motif ID: 2 | Motif name: TFW2 |
| Original motif Consensus sequence: SCGCGCGG | Reverse complement motif Consensus sequence: CCGCGCGS |
 |
 |
Best Matches for Top Significant Motif ID 2 (Highest to Lowest)
| Dataset #: | 2 |
| Motif ID: | 12 |
| Motif name: | csGCCCCGCCCCsc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0456233 |
Alignment:
BBGGGGCGGGGCVD
-SCGCGCGG-----
| Original motif Consensus sequence: HVGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 14 |
| Motif name: | cccGCCCCGCCCCsb |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.045686 |
Alignment:
BCCGCCCCGCCCCBB
------CCGCGCGS-
| Original motif Consensus sequence: BCCGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 18 |
| Motif name: | sSGTCACGTGACSs |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0988223 |
Alignment:
SGGTCACGTGACCS
---SCGCGCGG---
| Original motif Consensus sequence: SGGTCACGTGACCS | Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 16 |
| Motif name: | kcACCTGCAgc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 4 |
| Similarity score: | 2.08456 |
Alignment:
----GBTGCAGGTGB
SCGCGCGG-------
| Original motif Consensus sequence: BCACCTGCABC | Reverse complement motif Consensus sequence: GBTGCAGGTGB |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 15 |
| Motif name: | kCAGCCAATmr |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 4 |
| Similarity score: | 2.0901 |
Alignment:
MBATTGGCTGH----
-------CCGCGCGS
| Original motif Consensus sequence: DCAGCCAATVR | Reverse complement motif Consensus sequence: MBATTGGCTGH |
 |
 |
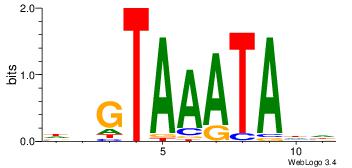
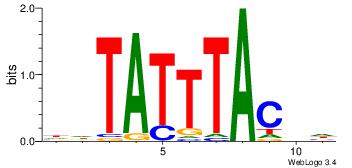
| Dataset #: 2 | Motif ID: 21 | Motif name: wbgTAAATAww |
| Original motif Consensus sequence: DBGTAAATAHD | Reverse complement motif Consensus sequence: DHTATTTACBD |
 |
 |
Best Matches for Top Significant Motif ID 21 (Highest to Lowest)
| Dataset #: | 1 |
| Motif ID: | 11 |
| Motif name: | TFM11 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 15 |
| Number of overlap: | 11 |
| Similarity score: | 0.0192768 |
Alignment:
TWVHWWWYTTTYTTTTTHTTTVWBH
--------------DHTATTTACBD
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA | Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 7 |
| Motif name: | TFM1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0283217 |
Alignment:
WTKTTTTTHWTTTTTTBT
DHTATTTACBD-------
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT | Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 8 |
| Motif name: | TFM3 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0308097 |
Alignment:
WWHTTTTTCABAAWTTWA
------DHTATTTACBD-
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA | Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 10 |
| Motif name: | TFM13 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0333874 |
Alignment:
ATKAAWTTTTRMAABAHHTW
-------DBGTAAATAHD--
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW | Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 9 |
| Motif name: | TFM12 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 14 |
| Number of overlap: | 7 |
| Similarity score: | 2.0544 |
Alignment:
----KKKAGGDGGAKKMGBBGKMG
DHTATTTACBD-------------
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY | Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
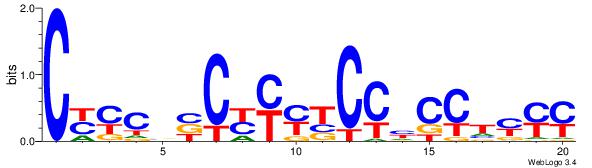 |
 |
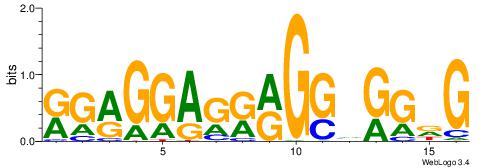
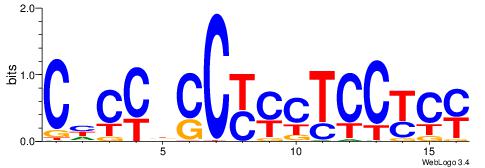
| Dataset #: | 1 |
| Motif ID: | 6 |
| Motif name: | TFM2 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 12 |
| Number of overlap: | 5 |
| Similarity score: | 3.05714 |
Alignment:
------CHCCBCCKMCTCCKCM
DHTATTTACBD-----------
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG | Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
 |
 |
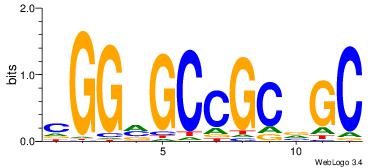
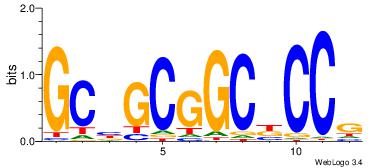
| Dataset #: | 1 |
| Motif ID: | 4 |
| Motif name: | TFF1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 4 |
| Similarity score: | 3.54896 |
Alignment:
-------GCVGCGGCBCCG
DHTATTTACBD--------
| Original motif Consensus sequence: CGGVGCCGCVGC | Reverse complement motif Consensus sequence: GCVGCGGCBCCG |
 |
 |
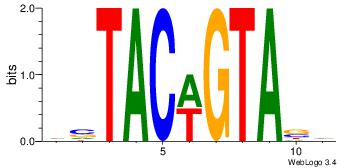
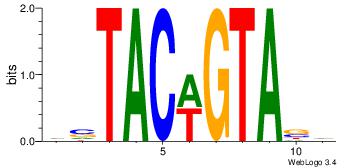
| Dataset #: 2 | Motif ID: 19 | Motif name: wsTACwGTAsw |
| Original motif Consensus sequence: HVTACWGTABD | Reverse complement motif Consensus sequence: DBTACWGTAVH |
 |
 |
Best Matches for Top Significant Motif ID 19 (Highest to Lowest)
| Dataset #: | 1 |
| Motif ID: | 8 |
| Motif name: | TFM3 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0413797 |
Alignment:
WWHTTTTTCABAAWTTWA
---DBTACWGTAVH----
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA | Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 10 |
| Motif name: | TFM13 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0445707 |
Alignment:
WAHHTVTTYKAAAAWTTRAT
-------HVTACWGTABD--
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW | Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 11 |
| Motif name: | TFM11 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0472742 |
Alignment:
HDWVAAAHAAAAAMAAAMWWWHBWA
---DBTACWGTAVH-----------
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA | Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 7 |
| Motif name: | TFM1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 10 |
| Number of overlap: | 9 |
| Similarity score: | 1.04742 |
Alignment:
WTKTTTTTHWTTTTTTBT--
---------DBTACWGTAVH
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT | Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 9 |
| Motif name: | TFM12 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 15 |
| Number of overlap: | 6 |
| Similarity score: | 2.54736 |
Alignment:
-----CYYCBBCYYYTCCHCCTYYY
DBTACWGTAVH--------------
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY | Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
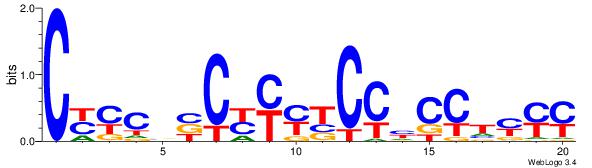 |
 |
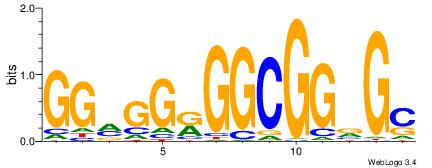
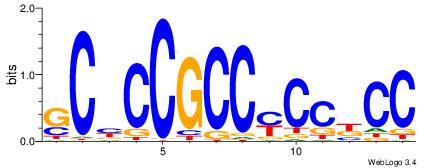
| Dataset #: | 1 |
| Motif ID: | 5 |
| Motif name: | TFF11 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 10 |
| Number of overlap: | 5 |
| Similarity score: | 3.04599 |
Alignment:
GGMGGRGGCGGVGC------
---------DBTACWGTAVH
| Original motif Consensus sequence: GGMGGRGGCGGVGC | Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 6 |
| Motif name: | TFM2 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 12 |
| Number of overlap: | 5 |
| Similarity score: | 3.04861 |
Alignment:
RGRGGAGRRGGHGGDG------
-----------DBTACWGTAVH
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG | Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
 |
 |
| Dataset #: 2 | Motif ID: 16 | Motif name: kcACCTGCAgc |
| Original motif Consensus sequence: BCACCTGCABC | Reverse complement motif Consensus sequence: GBTGCAGGTGB |
 |
 |
Best Matches for Top Significant Motif ID 16 (Highest to Lowest)
| Dataset #: | 1 |
| Motif ID: | 5 |
| Motif name: | TFF11 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0312321 |
Alignment:
GGMGGRGGCGGVGC
GBTGCAGGTGB---
| Original motif Consensus sequence: GGMGGRGGCGGVGC | Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 9 |
| Motif name: | TFM12 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0444938 |
Alignment:
KKKAGGDGGAKKMGBBGKMG
----GBTGCAGGTGB-----
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY | Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
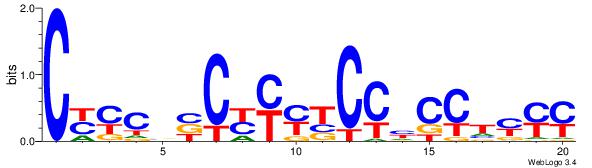 |
 |
| Dataset #: | 1 |
| Motif ID: | 6 |
| Motif name: | TFM2 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.538471 |
Alignment:
-RGRGGAGRRGGHGGDG
GBTGCAGGTGB------
| Original motif Consensus sequence: RGRGGAGRRGGHGGDG | Reverse complement motif Consensus sequence: CHCCBCCKMCTCCKCM |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 4 |
| Motif name: | TFF1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 1.54364 |
Alignment:
GCVGCGGCBCCG---
----BCACCTGCABC
| Original motif Consensus sequence: CGGVGCCGCVGC | Reverse complement motif Consensus sequence: GCVGCGGCBCCG |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 3 |
| Motif name: | TFW3 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 1.5466 |
Alignment:
---CGGCYBCGCG
GBTGCAGGTGB--
| Original motif Consensus sequence: CGGCYBCGCG | Reverse complement motif Consensus sequence: CGCGBMGCCG |
 |
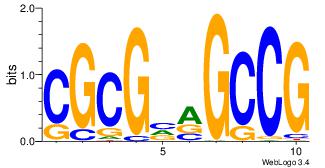 |
| Dataset #: | 1 |
| Motif ID: | 1 |
| Motif name: | TFW1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 2.54121 |
Alignment:
CGCGAC-----
BCACCTGCABC
| Original motif Consensus sequence: GTCGCG | Reverse complement motif Consensus sequence: CGCGAC |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 2 |
| Motif name: | TFW2 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 4 |
| Similarity score: | 3.53211 |
Alignment:
CCGCGCGS-------
----BCACCTGCABC
| Original motif Consensus sequence: SCGCGCGG | Reverse complement motif Consensus sequence: CCGCGCGS |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 11 |
| Motif name: | TFM11 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 22 |
| Number of overlap: | 4 |
| Similarity score: | 3.53789 |
Alignment:
-------TWVHWWWYTTTYTTTTTHTTTVWBH
GBTGCAGGTGB---------------------
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA | Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
 |
 |
| Dataset #: 1 | Motif ID: 4 | Motif name: TFF1 |
| Original motif Consensus sequence: CGGVGCCGCVGC | Reverse complement motif Consensus sequence: GCVGCGGCBCCG |
 |
 |
Best Matches for Top Significant Motif ID 4 (Highest to Lowest)
| Dataset #: | 2 |
| Motif ID: | 12 |
| Motif name: | csGCCCCGCCCCsc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0542674 |
Alignment:
HVGCCCCGCCCCBB
CGGVGCCGCVGC--
| Original motif Consensus sequence: HVGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 14 |
| Motif name: | cccGCCCCGCCCCsb |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0548371 |
Alignment:
BCCGCCCCGCCCCBB
---GCVGCGGCBCCG
| Original motif Consensus sequence: BCCGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 18 |
| Motif name: | sSGTCACGTGACSs |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0777862 |
Alignment:
SGGTCACGTGACCS
--GCVGCGGCBCCG
| Original motif Consensus sequence: SGGTCACGTGACCS | Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 16 |
| Motif name: | kcACCTGCAgc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 2.08028 |
Alignment:
----GBTGCAGGTGB
CGGVGCCGCVGC---
| Original motif Consensus sequence: BCACCTGCABC | Reverse complement motif Consensus sequence: GBTGCAGGTGB |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 15 |
| Motif name: | kCAGCCAATmr |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 5 |
| Similarity score: | 3.54126 |
Alignment:
-------DCAGCCAATVR
CGGVGCCGCVGC------
| Original motif Consensus sequence: DCAGCCAATVR | Reverse complement motif Consensus sequence: MBATTGGCTGH |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 20 |
| Motif name: | dhACATTCTkh |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 4 |
| Similarity score: | 4.07107 |
Alignment:
HCAGAATGTHD--------
-------GCVGCGGCBCCG
| Original motif Consensus sequence: DHACATTCTGH | Reverse complement motif Consensus sequence: HCAGAATGTHD |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 21 |
| Motif name: | wbgTAAATAww |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 4 |
| Similarity score: | 4.0742 |
Alignment:
DHTATTTACBD--------
-------GCVGCGGCBCCG
| Original motif Consensus sequence: DBGTAAATAHD | Reverse complement motif Consensus sequence: DHTATTTACBD |
 |
 |
| Dataset #: 2 | Motif ID: 20 | Motif name: dhACATTCTkh |
| Original motif Consensus sequence: DHACATTCTGH | Reverse complement motif Consensus sequence: HCAGAATGTHD |
 |
 |
Best Matches for Top Significant Motif ID 20 (Highest to Lowest)
| Dataset #: | 1 |
| Motif ID: | 8 |
| Motif name: | TFM3 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 11 |
| Similarity score: | 0.00522383 |
Alignment:
WWHTTTTTCABAAWTTWA
HCAGAATGTHD-------
| Original motif Consensus sequence: WWHTTTTTCABAAWTTWA | Reverse complement motif Consensus sequence: TWAAWTTVTGAAAAAHWW |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 11 |
| Motif name: | TFM11 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.014258 |
Alignment:
HDWVAAAHAAAAAMAAAMWWWHBWA
----HCAGAATGTHD----------
| Original motif Consensus sequence: HDWVAAAHAAAAAMAAAMWWWHBWA | Reverse complement motif Consensus sequence: TWVHWWWYTTTYTTTTTHTTTVWBH |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 10 |
| Motif name: | TFM13 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 11 |
| Similarity score: | 0.0192045 |
Alignment:
ATKAAWTTTTRMAABAHHTW
-DHACATTCTGH--------
| Original motif Consensus sequence: ATKAAWTTTTRMAABAHHTW | Reverse complement motif Consensus sequence: WAHHTVTTYKAAAAWTTRAT |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 7 |
| Motif name: | TFM1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 10 |
| Similarity score: | 0.519904 |
Alignment:
ABAAAAAAWHAAAAARAW-
--------HCAGAATGTHD
| Original motif Consensus sequence: WTKTTTTTHWTTTTTTBT | Reverse complement motif Consensus sequence: ABAAAAAAWHAAAAARAW |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 9 |
| Motif name: | TFM12 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 17 |
| Number of overlap: | 4 |
| Similarity score: | 3.5 |
Alignment:
-------CYYCBBCYYYTCCHCCTYYY
DHACATTCTGH----------------
| Original motif Consensus sequence: CYYCBBCYYYTCCHCCTYYY | Reverse complement motif Consensus sequence: KKKAGGDGGAKKMGBBGKMG |
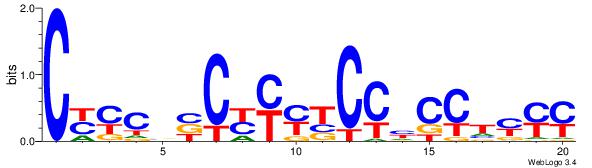 |
 |
| Dataset #: | 1 |
| Motif ID: | 4 |
| Motif name: | TFF1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 4 |
| Similarity score: | 3.51958 |
Alignment:
-------GCVGCGGCBCCG
HCAGAATGTHD--------
| Original motif Consensus sequence: CGGVGCCGCVGC | Reverse complement motif Consensus sequence: GCVGCGGCBCCG |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 1 |
| Motif name: | TFW1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 4 |
| Similarity score: | 3.52299 |
Alignment:
-------CGCGAC
DHACATTCTGH--
| Original motif Consensus sequence: GTCGCG | Reverse complement motif Consensus sequence: CGCGAC |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 5 |
| Motif name: | TFF11 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 11 |
| Number of overlap: | 4 |
| Similarity score: | 3.52554 |
Alignment:
-------GCVCCGCCMCCYCC
DHACATTCTGH----------
| Original motif Consensus sequence: GGMGGRGGCGGVGC | Reverse complement motif Consensus sequence: GCVCCGCCMCCYCC |
 |
 |