Top 10 Significant Motifs - Global Matching (Highest to Lowest)
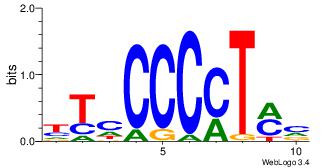
| Dataset #: 1 | Motif ID: 4 | Motif name: Motif 4 |
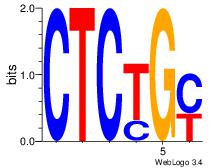
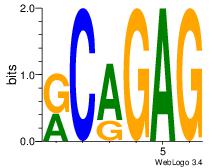
| Original motif Consensus sequence: CTCTGY | Reverse complement motif Consensus sequence: KCAGAG |
 |
 |
Best Matches for Top Significant Motif ID 4 (Highest to Lowest)
| Dataset #: | 2 |
| Motif ID: | 43 |
| Motif name: | NR1H2RXRA |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.012269 |
Alignment:
AAAGGTCAAAGGTCAAC
-----KCAGAG------
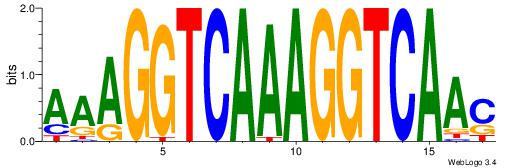
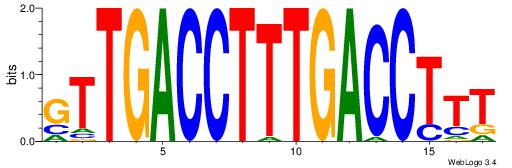
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC | Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 20 |
| Motif name: | PPARGRXRA |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.0140102 |
Alignment:
BTRGGDCARAGGKCA
-----KCAGAG----
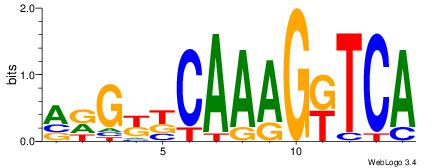
| Original motif Consensus sequence: BTRGGDCARAGGKCA | Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
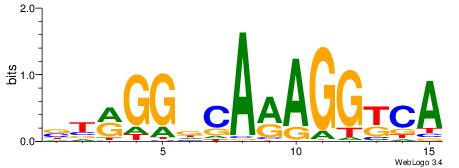 |
 |
| Dataset #: | 2 |
| Motif ID: | 22 |
| Motif name: | NR2F1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0186793 |
Alignment:
AKGYYCAAAGRTCA
----KCAGAG----
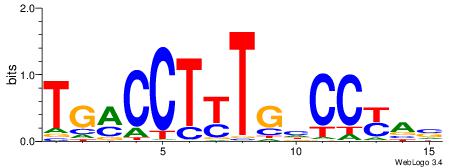
| Original motif Consensus sequence: TGAMCTTTGMMCYT | Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
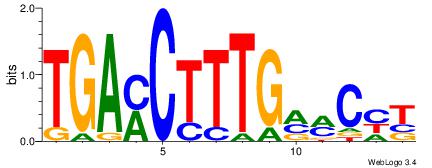 |
 |
| Dataset #: | 2 |
| Motif ID: | 34 |
| Motif name: | HNF4A |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.020602 |
Alignment:
RGGBCAAAGKYCA
---KCAGAG----
| Original motif Consensus sequence: RGGBCAAAGKYCA | Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
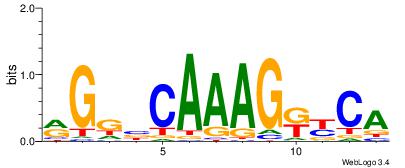 |
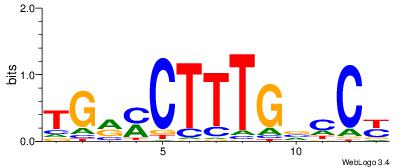 |
| Dataset #: | 2 |
| Motif ID: | 37 |
| Motif name: | MIZF |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0221665 |
Alignment:
BAACGTCCGC
----CTCTGY
| Original motif Consensus sequence: BAACGTCCGC | Reverse complement motif Consensus sequence: GCGGACGTTV |
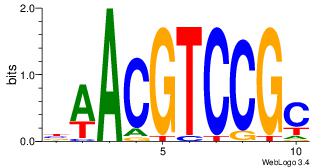 |
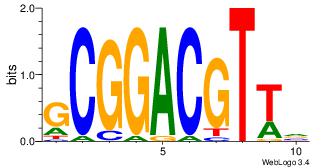 |
| Dataset #: | 2 |
| Motif ID: | 35 |
| Motif name: | INSM1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0255256 |
Alignment:
TGYCAGGGGGCR
--KCAGAG----
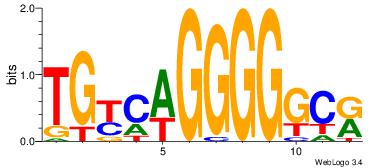
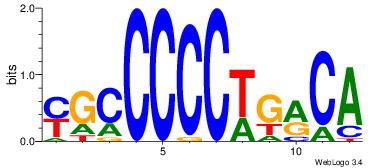
| Original motif Consensus sequence: TGYCAGGGGGCR | Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 16 |
| Motif name: | SP1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0349642 |
Alignment:
CCCCKCCCCC
CTCTGY----
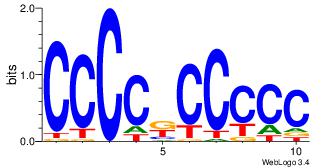
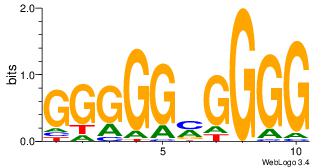
| Original motif Consensus sequence: CCCCKCCCCC | Reverse complement motif Consensus sequence: GGGGGYGGGG |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 19 |
| Motif name: | REST |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0376108 |
Alignment:
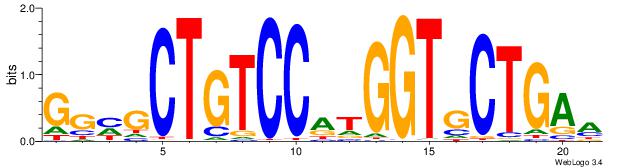
GGYGCTGTCCATGGTGCTGAA
--CTCTGY-------------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC | Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 12 |
| Motif name: | Zfx |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.0448449 |
Alignment:
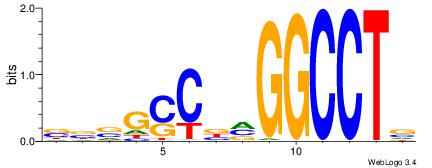
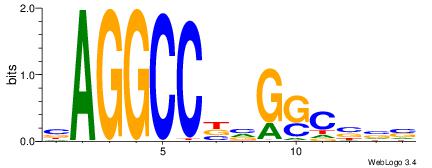
BBVGCCBVGGCCTV
---KCAGAG-----
| Original motif Consensus sequence: BBVGCCBVGGCCTV | Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 29 |
| Motif name: | ESR1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.0454269 |
Alignment:
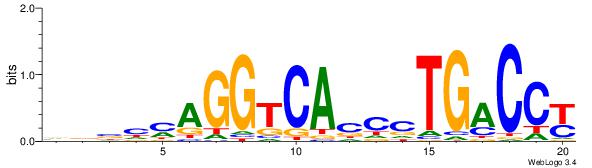
MGGTCAGGGTGACCTRDBHV
---KCAGAG-----------
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY | Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
 |
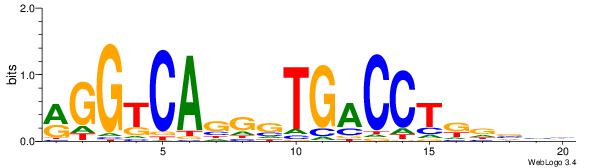 |
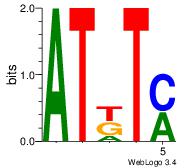
| Dataset #: 1 | Motif ID: 5 | Motif name: Motif 5 |
| Original motif Consensus sequence: ATKTM | Reverse complement motif Consensus sequence: RARAT |
 |
 |
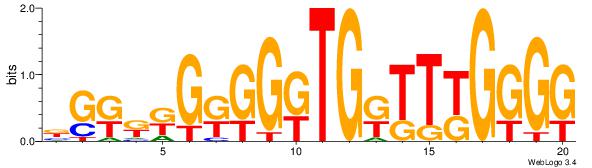
Best Matches for Top Significant Motif ID 5 (Highest to Lowest)
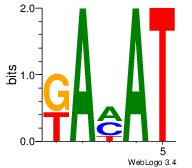
| Dataset #: | 2 |
| Motif ID: | 18 |
| Motif name: | RREB1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 10 |
| Number of overlap: | 5 |
| Similarity score: | 0.0256083 |
Alignment:
BGRRRGRGGRTGRTTYGGGG
------RARAT---------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV | Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
 |
 |
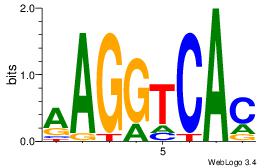
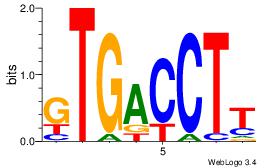
| Dataset #: | 2 |
| Motif ID: | 44 |
| Motif name: | NR4A2 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 5 |
| Similarity score: | 0.0276382 |
Alignment:
AAGGTCAC
RARAT---
| Original motif Consensus sequence: AAGGTCAC | Reverse complement motif Consensus sequence: GTGACCTT |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 22 |
| Motif name: | NR2F1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 5 |
| Similarity score: | 0.0279129 |
Alignment:
AKGYYCAAAGRTCA
-------RARAT--
| Original motif Consensus sequence: TGAMCTTTGMMCYT | Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 43 |
| Motif name: | NR1H2RXRA |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 10 |
| Number of overlap: | 5 |
| Similarity score: | 0.0292478 |
Alignment:
AAAGGTCAAAGGTCAAC
---------ATKTM---
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC | Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 37 |
| Motif name: | MIZF |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 5 |
| Similarity score: | 0.0292478 |
Alignment:
BAACGTCCGC
--ATKTM---
| Original motif Consensus sequence: BAACGTCCGC | Reverse complement motif Consensus sequence: GCGGACGTTV |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 20 |
| Motif name: | PPARGRXRA |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 5 |
| Similarity score: | 0.0305369 |
Alignment:
TGRCCTKTGHCCKAB
----ATKTM------
| Original motif Consensus sequence: BTRGGDCARAGGKCA | Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 19 |
| Motif name: | REST |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 13 |
| Number of overlap: | 5 |
| Similarity score: | 0.0315454 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
----ATKTM------------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC | Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
 |
 |
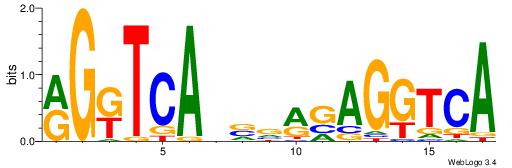
| Dataset #: | 2 |
| Motif ID: | 47 |
| Motif name: | RXRRAR_DR5 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 5 |
| Similarity score: | 0.0335956 |
Alignment:
RGKTCABVVRGAGGTCA
ATKTM------------
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA | Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
 |
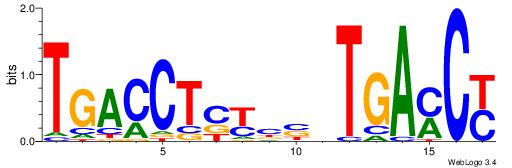 |
| Dataset #: | 2 |
| Motif ID: | 46 |
| Motif name: | RORA_1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 5 |
| Similarity score: | 0.0336052 |
Alignment:
AWVDAGGTCA
---RARAT--
| Original motif Consensus sequence: AWVDAGGTCA | Reverse complement motif Consensus sequence: TGACCTDVWT |
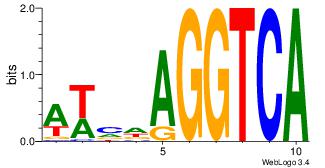 |
 |
| Dataset #: | 2 |
| Motif ID: | 34 |
| Motif name: | HNF4A |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 5 |
| Similarity score: | 0.0367105 |
Alignment:
RGGBCAAAGKYCA
-------ATKTM-
| Original motif Consensus sequence: RGGBCAAAGKYCA | Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
 |
 |
| Dataset #: 1 | Motif ID: 6 | Motif name: Motif 6 |
| Original motif Consensus sequence: CMGCRGC | Reverse complement motif Consensus sequence: GCKGCYG |
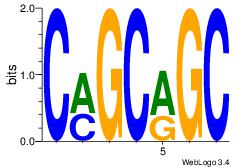 |
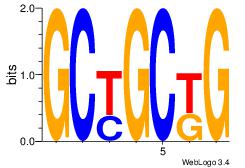 |
Best Matches for Top Significant Motif ID 6 (Highest to Lowest)
| Dataset #: | 2 |
| Motif ID: | 19 |
| Motif name: | REST |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 13 |
| Number of overlap: | 7 |
| Similarity score: | 0.0205607 |
Alignment:
TTCAGCACCATGGACAGCKCC
--CMGCRGC------------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC | Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 37 |
| Motif name: | MIZF |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 7 |
| Similarity score: | 0.0322623 |
Alignment:
GCGGACGTTV
GCKGCYG---
| Original motif Consensus sequence: BAACGTCCGC | Reverse complement motif Consensus sequence: GCGGACGTTV |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 12 |
| Motif name: | Zfx |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0.038091 |
Alignment:
BBVGCCBVGGCCTV
----CMGCRGC---
| Original motif Consensus sequence: BBVGCCBVGGCCTV | Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 15 |
| Motif name: | TFAP2A |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 7 |
| Similarity score: | 0.0393168 |
Alignment:
SCMVBBGGC
--CMGCRGC
| Original motif Consensus sequence: GCCBBVRGS | Reverse complement motif Consensus sequence: SCMVBBGGC |
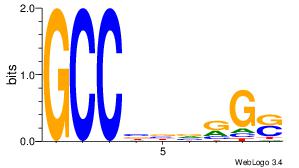 |
 |
| Dataset #: | 2 |
| Motif ID: | 45 |
| Motif name: | Pax5 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 7 |
| Similarity score: | 0.0448462 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-------GCKGCYG------
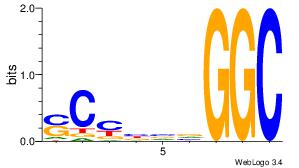
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR | Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 28 |
| Motif name: | Egr1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 7 |
| Similarity score: | 0.0453576 |
Alignment:
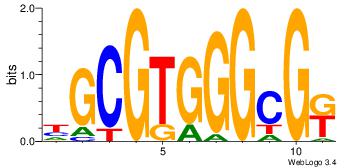
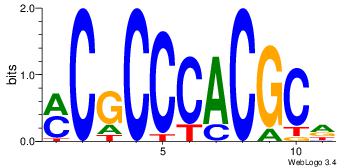
HGCGTGGGCGK
---GCKGCYG-
| Original motif Consensus sequence: HGCGTGGGCGK | Reverse complement motif Consensus sequence: YCGCCCACGCH |
 |
 |
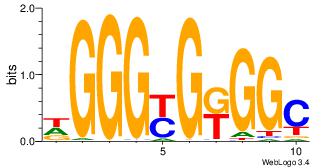
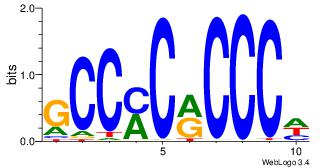
| Dataset #: | 2 |
| Motif ID: | 36 |
| Motif name: | Klf4 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 7 |
| Similarity score: | 0.0476958 |
Alignment:
GCCYCMCCCD
-CMGCRGC--
| Original motif Consensus sequence: DGGGYGKGGC | Reverse complement motif Consensus sequence: GCCYCMCCCD |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 16 |
| Motif name: | SP1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 7 |
| Similarity score: | 0.0490991 |
Alignment:
GGGGGYGGGG
GCKGCYG---
| Original motif Consensus sequence: CCCCKCCCCC | Reverse complement motif Consensus sequence: GGGGGYGGGG |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 18 |
| Motif name: | RREB1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.0498726 |
Alignment:
BGRRRGRGGRTGRTTYGGGG
-----GCKGCYG--------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV | Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 35 |
| Motif name: | INSM1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 7 |
| Similarity score: | 0.0511926 |
Alignment:
MGCCCCCTGMCA
---CMGCRGC--
| Original motif Consensus sequence: TGYCAGGGGGCR | Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
 |
 |
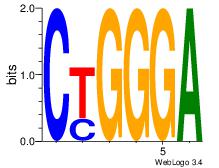
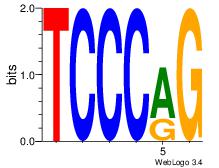
| Dataset #: 1 | Motif ID: 8 | Motif name: Motif 8 |
| Original motif Consensus sequence: CTGGGA | Reverse complement motif Consensus sequence: TCCCAG |
 |
 |
Best Matches for Top Significant Motif ID 8 (Highest to Lowest)
| Dataset #: | 2 |
| Motif ID: | 19 |
| Motif name: | REST |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.0392549 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
-------TCCCAG--------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC | Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
 |
 |
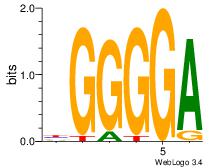
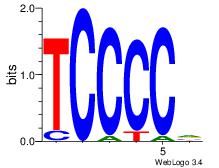
| Dataset #: | 2 |
| Motif ID: | 23 |
| Motif name: | MZF1_1-4 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0416319 |
Alignment:
TCCCCV
TCCCAG
| Original motif Consensus sequence: BGGGGA | Reverse complement motif Consensus sequence: TCCCCV |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 18 |
| Motif name: | RREB1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0509122 |
Alignment:
BGRRRGRGGRTGRTTYGGGG
--------------CTGGGA
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV | Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 35 |
| Motif name: | INSM1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.0557857 |
Alignment:
TGYCAGGGGGCR
---CTGGGA---
| Original motif Consensus sequence: TGYCAGGGGGCR | Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 13 |
| Motif name: | Zfp423 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.0624145 |
Alignment:
GYRYCCMTKGGYRSC
---TCCCAG------
| Original motif Consensus sequence: GSMMCCYARGGKKKC | Reverse complement motif Consensus sequence: GYRYCCMTKGGYRSC |
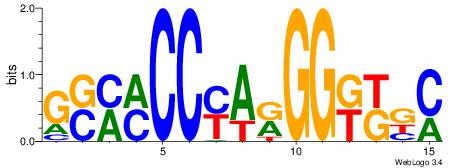 |
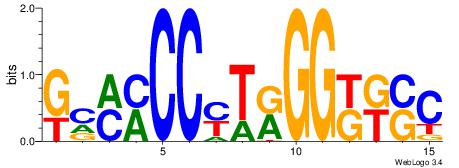 |
| Dataset #: | 2 |
| Motif ID: | 43 |
| Motif name: | NR1H2RXRA |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.0650784 |
Alignment:
GTTGACCTTTGACCTTT
------CTGGGA-----
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC | Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 48 |
| Motif name: | Tcfcp2l1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.0663804 |
Alignment:
CKGGDTBDMMCTGG
CTGGGA--------
| Original motif Consensus sequence: CCAGYYHVADCCRG | Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
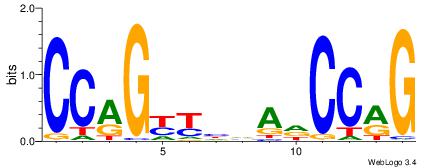 |
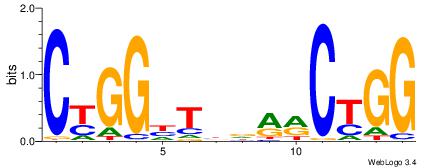 |
| Dataset #: | 2 |
| Motif ID: | 28 |
| Motif name: | Egr1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0678562 |
Alignment:
YCGCCCACGCH
--TCCCAG---
| Original motif Consensus sequence: HGCGTGGGCGK | Reverse complement motif Consensus sequence: YCGCCCACGCH |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 24 |
| Motif name: | Ar |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0714107 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
--------------TCCCAG--
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV | Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
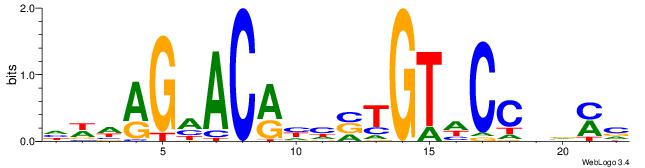 |
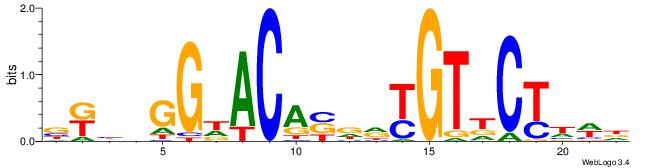 |
| Dataset #: | 2 |
| Motif ID: | 12 |
| Motif name: | Zfx |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.0718041 |
Alignment:
BBVGCCBVGGCCTV
-----CTGGGA---
| Original motif Consensus sequence: BBVGCCBVGGCCTV | Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
 |
 |
| Dataset #: 1 | Motif ID: 2 | Motif name: Motif 2 |
| Original motif Consensus sequence: AMACA | Reverse complement motif Consensus sequence: TGTRT |
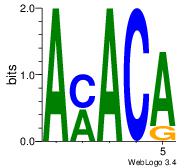 |
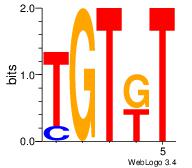 |
Best Matches for Top Significant Motif ID 2 (Highest to Lowest)
| Dataset #: | 2 |
| Motif ID: | 18 |
| Motif name: | RREB1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 11 |
| Number of overlap: | 5 |
| Similarity score: | 0.0243918 |
Alignment:
CCCCMAAMCAMCCMCMMMCV
-----AMACA----------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV | Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 24 |
| Motif name: | Ar |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 5 |
| Similarity score: | 0.0429089 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
----AMACA-------------
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV | Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 20 |
| Motif name: | PPARGRXRA |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.0595073 |
Alignment:
TGRCCTKTGHCCKAB
-----TGTRT-----
| Original motif Consensus sequence: BTRGGDCARAGGKCA | Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 26 |
| Motif name: | ArntAhr |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 5 |
| Similarity score: | 0.0600161 |
Alignment:
YGCGTG
TGTRT-
| Original motif Consensus sequence: YGCGTG | Reverse complement motif Consensus sequence: CACGCM |
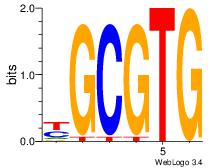 |
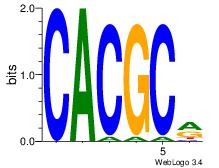 |
| Dataset #: | 2 |
| Motif ID: | 45 |
| Motif name: | Pax5 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 11 |
| Number of overlap: | 5 |
| Similarity score: | 0.0620995 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-----AMACA----------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR | Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 22 |
| Motif name: | NR2F1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 5 |
| Similarity score: | 0.0650242 |
Alignment:
AKGYYCAAAGRTCA
------AMACA---
| Original motif Consensus sequence: TGAMCTTTGMMCYT | Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 35 |
| Motif name: | INSM1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 5 |
| Similarity score: | 0.0670984 |
Alignment:
MGCCCCCTGMCA
-------AMACA
| Original motif Consensus sequence: TGYCAGGGGGCR | Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 33 |
| Motif name: | HIF1AARNT |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 5 |
| Similarity score: | 0.0700905 |
Alignment:
VCACGTBV
AMACA---
| Original motif Consensus sequence: VBACGTGV | Reverse complement motif Consensus sequence: VCACGTBV |
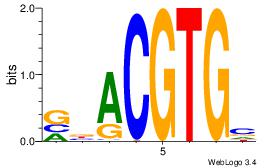 |
 |
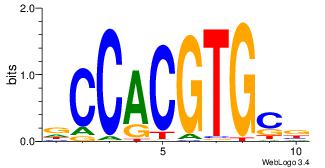
| Dataset #: | 2 |
| Motif ID: | 38 |
| Motif name: | Myc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 5 |
| Similarity score: | 0.0708148 |
Alignment:
VGCACGTGGH
-AMACA----
| Original motif Consensus sequence: VGCACGTGGH | Reverse complement motif Consensus sequence: DCCACGTGCV |
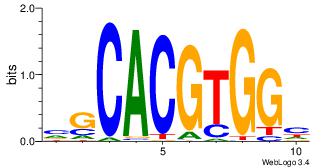 |
 |
| Dataset #: | 2 |
| Motif ID: | 34 |
| Motif name: | HNF4A |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 5 |
| Similarity score: | 0.071574 |
Alignment:
TGMYCTTTGBCCK
-----TGTRT---
| Original motif Consensus sequence: RGGBCAAAGKYCA | Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
 |
 |
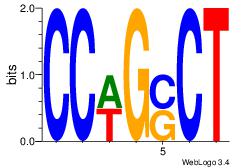
| Dataset #: 1 | Motif ID: 3 | Motif name: Motif 3 |
| Original motif Consensus sequence: AGSCWGG | Reverse complement motif Consensus sequence: CCWGSCT |
 |
 |
Best Matches for Top Significant Motif ID 3 (Highest to Lowest)
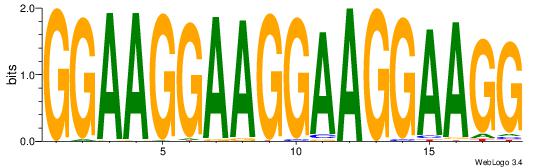
| Dataset #: | 2 |
| Motif ID: | 32 |
| Motif name: | EWSR1-FLI1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0.0395671 |
Alignment:
GGAAGGAAGGAAGGAAGG
-------AGSCWGG----
| Original motif Consensus sequence: GGAAGGAAGGAAGGAAGG | Reverse complement motif Consensus sequence: CCTTCCTTCCTTCCTTCC |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 35 |
| Motif name: | INSM1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 7 |
| Similarity score: | 0.0494688 |
Alignment:
MGCCCCCTGMCA
-----CCWGSCT
| Original motif Consensus sequence: TGYCAGGGGGCR | Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 48 |
| Motif name: | Tcfcp2l1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 7 |
| Similarity score: | 0.0669354 |
Alignment:
CCAGYYHVADCCRG
CCWGSCT-------
| Original motif Consensus sequence: CCAGYYHVADCCRG | Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 19 |
| Motif name: | REST |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 7 |
| Similarity score: | 0.0693747 |
Alignment:
TTCAGCACCATGGACAGCKCC
------AGSCWGG--------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC | Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 16 |
| Motif name: | SP1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 7 |
| Similarity score: | 0.0695377 |
Alignment:
CCCCKCCCCC
-CCWGSCT--
| Original motif Consensus sequence: CCCCKCCCCC | Reverse complement motif Consensus sequence: GGGGGYGGGG |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 12 |
| Motif name: | Zfx |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 7 |
| Similarity score: | 0.0720685 |
Alignment:
BBVGCCBVGGCCTV
------CCWGSCT-
| Original motif Consensus sequence: BBVGCCBVGGCCTV | Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 29 |
| Motif name: | ESR1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 13 |
| Number of overlap: | 7 |
| Similarity score: | 0.0771405 |
Alignment:
MGGTCAGGGTGACCTRDBHV
------------CCWGSCT-
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY | Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 46 |
| Motif name: | RORA_1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 7 |
| Similarity score: | 0.0772276 |
Alignment:
TGACCTDVWT
---CCWGSCT
| Original motif Consensus sequence: AWVDAGGTCA | Reverse complement motif Consensus sequence: TGACCTDVWT |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 36 |
| Motif name: | Klf4 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 7 |
| Similarity score: | 0.0798994 |
Alignment:
GCCYCMCCCD
---CCWGSCT
| Original motif Consensus sequence: DGGGYGKGGC | Reverse complement motif Consensus sequence: GCCYCMCCCD |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 39 |
| Motif name: | MYCMAX |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0.0839075 |
Alignment:
RASCACGTGGT
AGSCWGG----
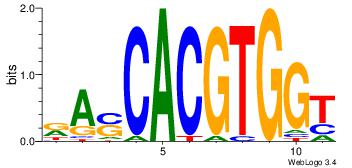
| Original motif Consensus sequence: RASCACGTGGT | Reverse complement motif Consensus sequence: ACCACGTGSTM |
 |
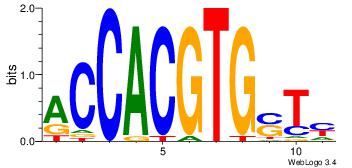 |
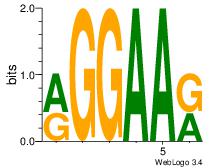
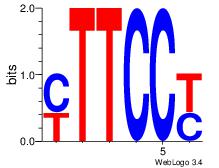
| Dataset #: 1 | Motif ID: 1 | Motif name: Motif 1 |
| Original motif Consensus sequence: RGGAAR | Reverse complement motif Consensus sequence: MTTCCK |
 |
 |
Best Matches for Top Significant Motif ID 1 (Highest to Lowest)
| Dataset #: | 2 |
| Motif ID: | 32 |
| Motif name: | EWSR1-FLI1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 12 |
| Number of overlap: | 6 |
| Similarity score: | 0.0146511 |
Alignment:
GGAAGGAAGGAAGGAAGG
-----------RGGAAR-
| Original motif Consensus sequence: GGAAGGAAGGAAGGAAGG | Reverse complement motif Consensus sequence: CCTTCCTTCCTTCCTTCC |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 37 |
| Motif name: | MIZF |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.0608243 |
Alignment:
BAACGTCCGC
---MTTCCK-
| Original motif Consensus sequence: BAACGTCCGC | Reverse complement motif Consensus sequence: GCGGACGTTV |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 41 |
| Motif name: | MZF1_5-13 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0753789 |
Alignment:
BKAGGGGDAD
----RGGAAR
| Original motif Consensus sequence: BKAGGGGDAD | Reverse complement motif Consensus sequence: BTHCCCCTYB |
 |
 |
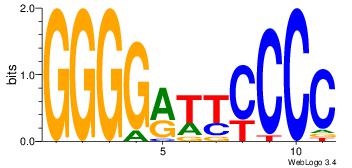
| Dataset #: | 2 |
| Motif ID: | 42 |
| Motif name: | NFKB1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.0757695 |
Alignment:
GGGGAAKCCCC
-RGGAAR----
| Original motif Consensus sequence: GGGGRTTCCCC | Reverse complement motif Consensus sequence: GGGGAAKCCCC |
 |
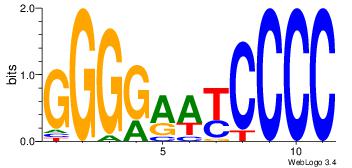 |
| Dataset #: | 2 |
| Motif ID: | 20 |
| Motif name: | PPARGRXRA |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0800484 |
Alignment:
BTRGGDCARAGGKCA
---RGGAAR------
| Original motif Consensus sequence: BTRGGDCARAGGKCA | Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 19 |
| Motif name: | REST |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.0829223 |
Alignment:
TTCAGCACCATGGACAGCKCC
----------RGGAAR-----
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC | Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 35 |
| Motif name: | INSM1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0832926 |
Alignment:
TGYCAGGGGGCR
------RGGAAR
| Original motif Consensus sequence: TGYCAGGGGGCR | Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 18 |
| Motif name: | RREB1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0835516 |
Alignment:
BGRRRGRGGRTGRTTYGGGG
----RGGAAR----------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV | Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 24 |
| Motif name: | Ar |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.0836451 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
---RGGAAR-------------
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV | Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
 |
 |
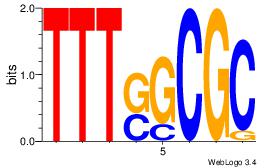
| Dataset #: | 2 |
| Motif ID: | 27 |
| Motif name: | E2F1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0842331 |
Alignment:
GCGCSAAA
--RGGAAR
| Original motif Consensus sequence: TTTSGCGC | Reverse complement motif Consensus sequence: GCGCSAAA |
 |
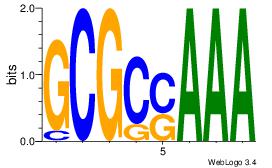 |
| Dataset #: | 2 |
| Motif ID: | 13 |
| Motif name: | Zfp423 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.0850287 |
Alignment:
GYRYCCMTKGGYRSC
--------RGGAAR-
| Original motif Consensus sequence: GSMMCCYARGGKKKC | Reverse complement motif Consensus sequence: GYRYCCMTKGGYRSC |
 |
 |
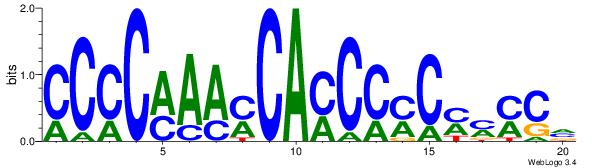
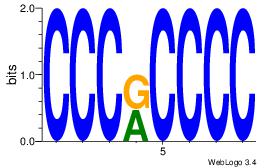
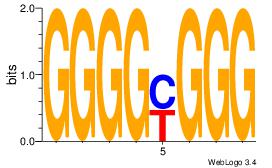
| Dataset #: 1 | Motif ID: 7 | Motif name: Motif 7 |
| Original motif Consensus sequence: CCCRCCCC | Reverse complement motif Consensus sequence: GGGGMGGG |
 |
 |
Best Matches for Top Significant Motif ID 7 (Highest to Lowest)
| Dataset #: | 2 |
| Motif ID: | 16 |
| Motif name: | SP1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0423611 |
Alignment:
CCCCKCCCCC
-CCCRCCCC-
| Original motif Consensus sequence: CCCCKCCCCC | Reverse complement motif Consensus sequence: GGGGGYGGGG |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 36 |
| Motif name: | Klf4 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.0461644 |
Alignment:
DGGGYGKGGC
GGGGMGGG--
| Original motif Consensus sequence: DGGGYGKGGC | Reverse complement motif Consensus sequence: GCCYCMCCCD |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 28 |
| Motif name: | Egr1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.0650374 |
Alignment:
YCGCCCACGCH
---CCCRCCCC
| Original motif Consensus sequence: HGCGTGGGCGK | Reverse complement motif Consensus sequence: YCGCCCACGCH |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 18 |
| Motif name: | RREB1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.076401 |
Alignment:
CCCCMAAMCAMCCMCMMMCV
------CCCRCCCC------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV | Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 42 |
| Motif name: | NFKB1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.0893429 |
Alignment:
GGGGAAKCCCC
GGGGMGGG---
| Original motif Consensus sequence: GGGGRTTCCCC | Reverse complement motif Consensus sequence: GGGGAAKCCCC |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 35 |
| Motif name: | INSM1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.0936832 |
Alignment:
TGYCAGGGGGCR
GGGGMGGG----
| Original motif Consensus sequence: TGYCAGGGGGCR | Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 32 |
| Motif name: | EWSR1-FLI1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0950969 |
Alignment:
GGAAGGAAGGAAGGAAGG
------GGGGMGGG----
| Original motif Consensus sequence: GGAAGGAAGGAAGGAAGG | Reverse complement motif Consensus sequence: CCTTCCTTCCTTCCTTCC |
 |
 |
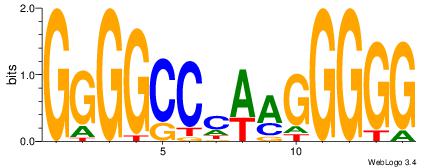
| Dataset #: | 2 |
| Motif ID: | 21 |
| Motif name: | PLAG1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.0998932 |
Alignment:
GGGGCCCAAGGGGG
GGGGMGGG------
| Original motif Consensus sequence: GGGGCCCAAGGGGG | Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
 |
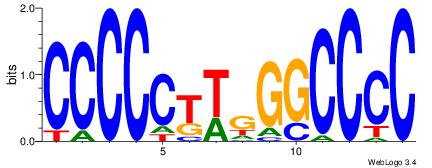 |
| Dataset #: | 2 |
| Motif ID: | 12 |
| Motif name: | Zfx |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.10221 |
Alignment:
BBVGCCBVGGCCTV
----CCCRCCCC--
| Original motif Consensus sequence: BBVGCCBVGGCCTV | Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 45 |
| Motif name: | Pax5 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.102497 |
Alignment:
DGVBCABTGDWGCGKRRCSR
--------GGGGMGGG----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR | Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
 |
 |
| Dataset #: 2 | Motif ID: 26 | Motif name: ArntAhr |
| Original motif Consensus sequence: YGCGTG | Reverse complement motif Consensus sequence: CACGCM |
 |
 |
Best Matches for Top Significant Motif ID 26 (Highest to Lowest)
| Dataset #: | 1 |
| Motif ID: | 11 |
| Motif name: | Motif 11 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0308396 |
Alignment:
TGTGTGTGTGTKTG
------YGCGTG--
| Original motif Consensus sequence: CAYACACACACACA | Reverse complement motif Consensus sequence: TGTGTGTGTGTKTG |
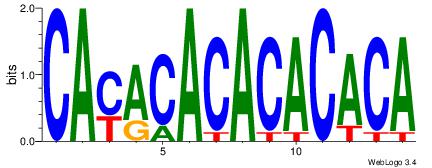 |
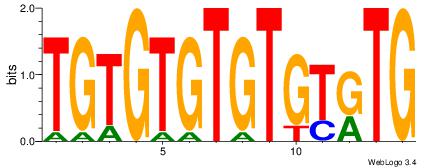 |
| Dataset #: | 1 |
| Motif ID: | 9 |
| Motif name: | Motif 9 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.0350575 |
Alignment:
CACACAVRCACACA
--------CACGCM
| Original motif Consensus sequence: CACACAVRCACACA | Reverse complement motif Consensus sequence: TGTGTGKVTGTGTG |
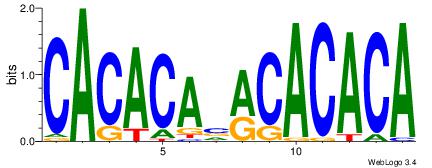 |
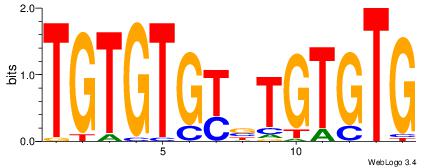 |
| Dataset #: | 1 |
| Motif ID: | 7 |
| Motif name: | Motif 7 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0586538 |
Alignment:
GGGGMGGG
YGCGTG--
| Original motif Consensus sequence: CCCRCCCC | Reverse complement motif Consensus sequence: GGGGMGGG |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 3 |
| Motif name: | Motif 3 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.0884818 |
Alignment:
CCWGSCT
-CACGCM
| Original motif Consensus sequence: AGSCWGG | Reverse complement motif Consensus sequence: CCWGSCT |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 2 |
| Motif name: | Motif 2 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 5 |
| Similarity score: | 0.563057 |
Alignment:
AMACA-
CACGCM
| Original motif Consensus sequence: AMACA | Reverse complement motif Consensus sequence: TGTRT |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 6 |
| Motif name: | Motif 6 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 5 |
| Similarity score: | 0.572774 |
Alignment:
-GCKGCYG
YGCGTG--
| Original motif Consensus sequence: CMGCRGC | Reverse complement motif Consensus sequence: GCKGCYG |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 8 |
| Motif name: | Motif 8 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 5 |
| Similarity score: | 0.581904 |
Alignment:
CTGGGA-
-YGCGTG
| Original motif Consensus sequence: CTGGGA | Reverse complement motif Consensus sequence: TCCCAG |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 4 |
| Motif name: | Motif 4 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 4 |
| Similarity score: | 1.06919 |
Alignment:
--KCAGAG
YGCGTG--
| Original motif Consensus sequence: CTCTGY | Reverse complement motif Consensus sequence: KCAGAG |
 |
 |
| Dataset #: 2 | Motif ID: 33 | Motif name: HIF1AARNT |
| Original motif Consensus sequence: VBACGTGV | Reverse complement motif Consensus sequence: VCACGTBV |
 |
 |
Best Matches for Top Significant Motif ID 33 (Highest to Lowest)
| Dataset #: | 1 |
| Motif ID: | 9 |
| Motif name: | Motif 9 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0745994 |
Alignment:
CACACAVRCACACA
-----VCACGTBV-
| Original motif Consensus sequence: CACACAVRCACACA | Reverse complement motif Consensus sequence: TGTGTGKVTGTGTG |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 11 |
| Motif name: | Motif 11 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0850889 |
Alignment:
CAYACACACACACA
---VCACGTBV---
| Original motif Consensus sequence: CAYACACACACACA | Reverse complement motif Consensus sequence: TGTGTGTGTGTKTG |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 7 |
| Motif name: | Motif 7 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 7 |
| Similarity score: | 0.588622 |
Alignment:
CCCRCCCC-
-VCACGTBV
| Original motif Consensus sequence: CCCRCCCC | Reverse complement motif Consensus sequence: GGGGMGGG |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 3 |
| Motif name: | Motif 3 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 5 |
| Similarity score: | 1.55675 |
Alignment:
CCWGSCT---
--VBACGTGV
| Original motif Consensus sequence: AGSCWGG | Reverse complement motif Consensus sequence: CCWGSCT |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 2 |
| Motif name: | Motif 2 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 5 |
| Similarity score: | 1.57238 |
Alignment:
TGTRT---
VBACGTGV
| Original motif Consensus sequence: AMACA | Reverse complement motif Consensus sequence: TGTRT |
 |
 |