Top 10 Significant Motifs - Global Matching (Highest to Lowest)
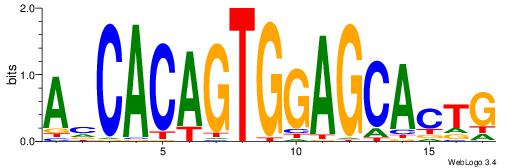
| Dataset #: 1 | Motif ID: 5 | Motif name: ZEB1 |
| Original motif Consensus sequence: CACCTD | Reverse complement motif Consensus sequence: HAGGTG |
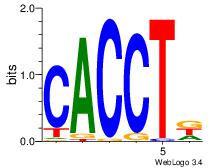 |
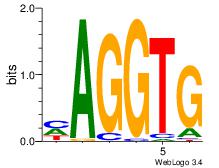 |
Best Matches for Top Significant Motif ID 5 (Highest to Lowest)
| Dataset #: | 2 |
| Motif ID: | 21 |
| Motif name: | AmTGTGACACCACAGT |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.0349782 |
Alignment:
AMTGTGACACCACAGT
-------CACCTD---
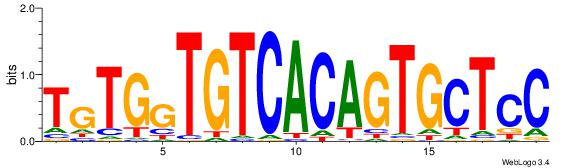
| Original motif Consensus sequence: AMTGTGACACCACAGT | Reverse complement motif Consensus sequence: ACTGTGGTGTCACART |
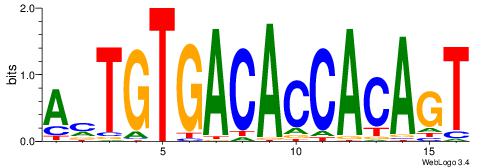 |
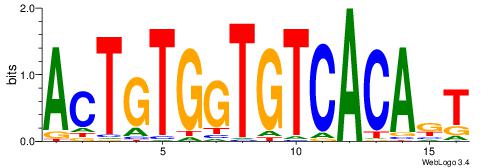 |
| Dataset #: | 2 |
| Motif ID: | 17 |
| Motif name: | TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.0362768 |
Alignment:
TGTGGTGTCACAGTGCTCC
-HAGGTG------------
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC | Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 27 |
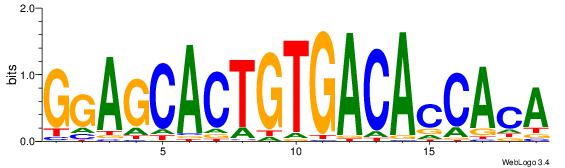
| Motif name: | AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 12 |
| Number of overlap: | 6 |
| Similarity score: | 0.0409613 |
Alignment:
AGTGCTCCACTGTGGTGTCACAGT
-----------HAGGTG-------
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT | Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 33 |
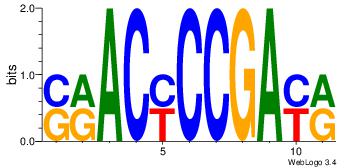
| Motif name: | srACyCCGAyr |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0426002 |
Alignment:
KKTCGGKGTKS
--HAGGTG---
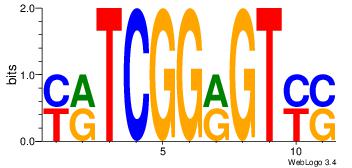
| Original motif Consensus sequence: SRACYCCGAYR | Reverse complement motif Consensus sequence: KKTCGGKGTKS |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 23 |
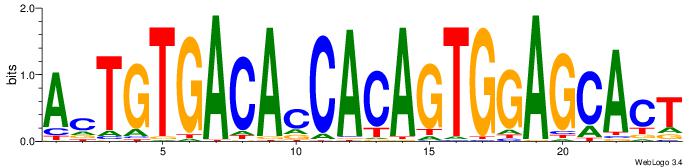
| Motif name: | TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.0496179 |
Alignment:
TGGAGCACTGTGACAVCACAGTGG
-------------CACCTD-----
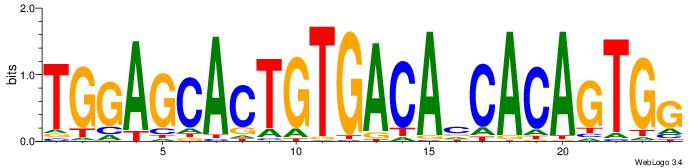
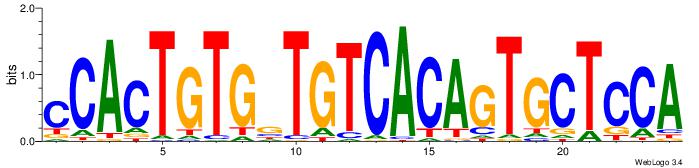
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG | Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 26 |
| Motif name: | CAcCACAGTGGAGCAct |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0538743 |
Alignment:
AGTGCTCCACTGTGDTG
-----------HAGGTG
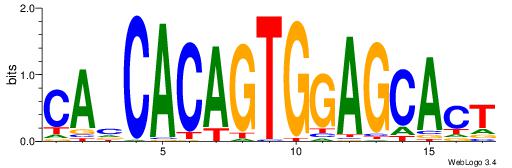
| Original motif Consensus sequence: CAHCACAGTGGAGCACT | Reverse complement motif Consensus sequence: AGTGCTCCACTGTGDTG |
 |
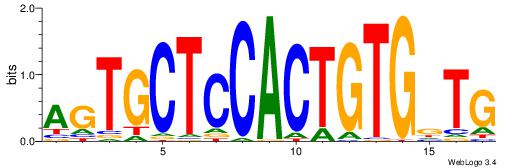 |
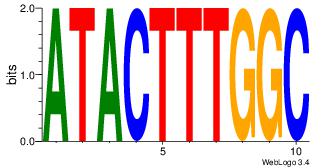
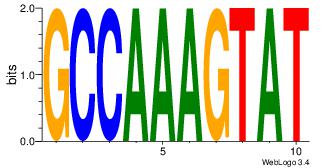
| Dataset #: | 2 |
| Motif ID: | 19 |
| Motif name: | atactttggc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.0553034 |
Alignment:
ATACTTTGGC
-CACCTD---
| Original motif Consensus sequence: ATACTTTGGC | Reverse complement motif Consensus sequence: GCCAAAGTAT |
 |
 |
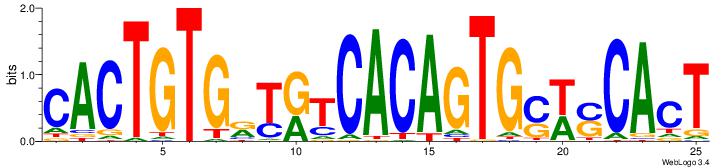
| Dataset #: | 2 |
| Motif ID: | 22 |
| Motif name: | CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 19 |
| Number of overlap: | 6 |
| Similarity score: | 0.0564649 |
Alignment:
AGTGSWSCACTGTGAMAMCACAGTG
------------------CACCTD-
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT | Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
 |
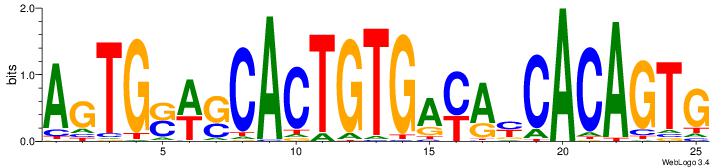 |
| Dataset #: | 2 |
| Motif ID: | 24 |
| Motif name: | ssCCCCGCSssk |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.0571854 |
Alignment:
SBCCCCGCCSBB
-----CACCTD-
| Original motif Consensus sequence: SBCCCCGCCSBB | Reverse complement motif Consensus sequence: BBSGGCGGGGBS |
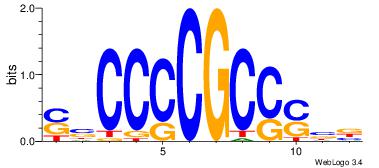 |
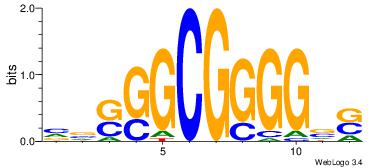 |
| Dataset #: | 2 |
| Motif ID: | 20 |
| Motif name: | CagTGCTCCACTGTGgT |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.0594701 |
Alignment:
ABCACAGTGGAGCACTG
----HAGGTG-------
| Original motif Consensus sequence: CAGTGCTCCACTGTGBT | Reverse complement motif Consensus sequence: ABCACAGTGGAGCACTG |
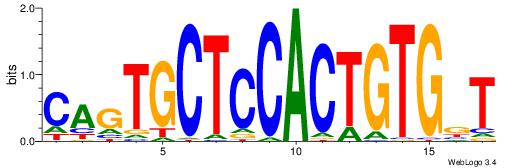 |
 |
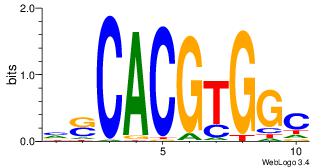
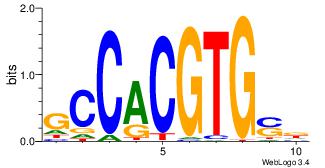
| Dataset #: 1 | Motif ID: 6 | Motif name: Mycn |
| Original motif Consensus sequence: HSCACGTGGC | Reverse complement motif Consensus sequence: GCCACGTGSD |
 |
 |
Best Matches for Top Significant Motif ID 6 (Highest to Lowest)
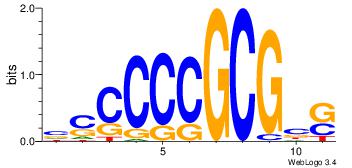
| Dataset #: | 2 |
| Motif ID: | 18 |
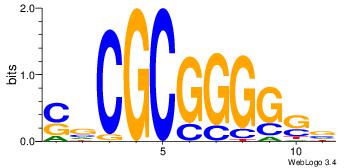
| Motif name: | sscCCCGCGcs |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0654819 |
Alignment:
BSCCCCGCGBS
-GCCACGTGSD
| Original motif Consensus sequence: BSCCCCGCGBS | Reverse complement motif Consensus sequence: SBCGCGGGGSB |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 24 |
| Motif name: | ssCCCCGCSssk |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0704988 |
Alignment:
BBSGGCGGGGBS
-HSCACGTGGC-
| Original motif Consensus sequence: SBCCCCGCCSBB | Reverse complement motif Consensus sequence: BBSGGCGGGGBS |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 22 |
| Motif name: | CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 10 |
| Number of overlap: | 10 |
| Similarity score: | 0.0883224 |
Alignment:
AGTGSWSCACTGTGAMAMCACAGTG
------GCCACGTGSD---------
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT | Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 20 |
| Motif name: | CagTGCTCCACTGTGgT |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0943014 |
Alignment:
ABCACAGTGGAGCACTG
-GCCACGTGSD------
| Original motif Consensus sequence: CAGTGCTCCACTGTGBT | Reverse complement motif Consensus sequence: ABCACAGTGGAGCACTG |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 27 |
| Motif name: | AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0949359 |
Alignment:
ACTGTGACACCACAGTGGAGCACT
---------GCCACGTGSD-----
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT | Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 23 |
| Motif name: | TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0955115 |
Alignment:
TGGAGCACTGTGACAVCACAGTGG
--------------HSCACGTGGC
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG | Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 26 |
| Motif name: | CAcCACAGTGGAGCAct |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0959628 |
Alignment:
CAHCACAGTGGAGCACT
--GCCACGTGSD-----
| Original motif Consensus sequence: CAHCACAGTGGAGCACT | Reverse complement motif Consensus sequence: AGTGCTCCACTGTGDTG |
 |
 |
| Dataset #: | 2 |
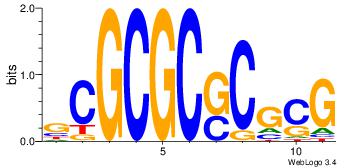
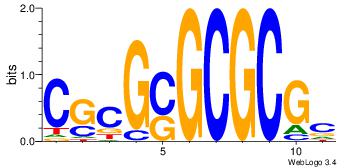
| Motif ID: | 32 |
| Motif name: | gCGCGCsCgsG |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0989932 |
Alignment:
GCGCGCSCGCG
-HSCACGTGGC
| Original motif Consensus sequence: GCGCGCSCGCG | Reverse complement motif Consensus sequence: CGCGSGCGCGC |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 17 |
| Motif name: | TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0991692 |
Alignment:
GGAGCACTGTGACACCACA
---GCCACGTGSD------
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC | Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
 |
 |
| Dataset #: | 2 |
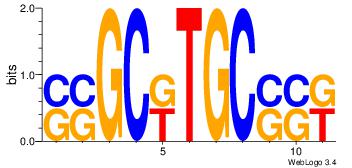
| Motif ID: | 28 |
| Motif name: | ssGCkTGCssk |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.101333 |
Alignment:
YSSGCAYGCSS
HSCACGTGGC-
| Original motif Consensus sequence: SSGCKTGCSSK | Reverse complement motif Consensus sequence: YSSGCAYGCSS |
 |
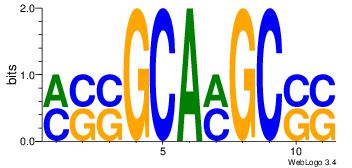 |
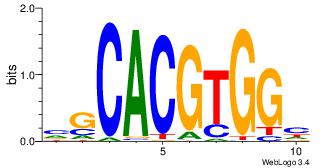
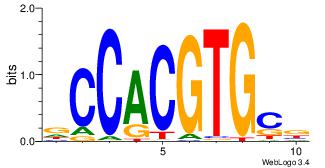
| Dataset #: 1 | Motif ID: 8 | Motif name: Myc |
| Original motif Consensus sequence: VGCACGTGGH | Reverse complement motif Consensus sequence: DCCACGTGCV |
 |
 |
Best Matches for Top Significant Motif ID 8 (Highest to Lowest)
| Dataset #: | 2 |
| Motif ID: | 18 |
| Motif name: | sscCCCGCGcs |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.064853 |
Alignment:
BSCCCCGCGBS
-DCCACGTGCV
| Original motif Consensus sequence: BSCCCCGCGBS | Reverse complement motif Consensus sequence: SBCGCGGGGSB |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 24 |
| Motif name: | ssCCCCGCSssk |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0728588 |
Alignment:
SBCCCCGCCSBB
-DCCACGTGCV-
| Original motif Consensus sequence: SBCCCCGCCSBB | Reverse complement motif Consensus sequence: BBSGGCGGGGBS |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 22 |
| Motif name: | CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0888909 |
Alignment:
CACTGTGRTRTCACAGTGSWSCACT
----------DCCACGTGCV-----
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT | Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 20 |
| Motif name: | CagTGCTCCACTGTGgT |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0955234 |
Alignment:
ABCACAGTGGAGCACTG
-DCCACGTGCV------
| Original motif Consensus sequence: CAGTGCTCCACTGTGBT | Reverse complement motif Consensus sequence: ABCACAGTGGAGCACTG |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 23 |
| Motif name: | TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 11 |
| Number of overlap: | 10 |
| Similarity score: | 0.0971125 |
Alignment:
TGGAGCACTGTGACAVCACAGTGG
----DCCACGTGCV----------
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG | Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
 |
 |
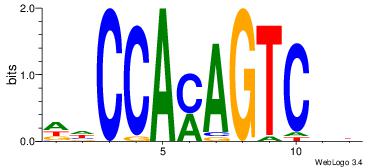
| Dataset #: | 2 |
| Motif ID: | 25 |
| Motif name: | wwCCAmAGTCmt |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0974038 |
Alignment:
VDGACTRTGGDD
-VGCACGTGGH-
| Original motif Consensus sequence: DDCCAMAGTCHB | Reverse complement motif Consensus sequence: VDGACTRTGGDD |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 26 |
| Motif name: | CAcCACAGTGGAGCAct |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0979703 |
Alignment:
CAHCACAGTGGAGCACT
--DCCACGTGCV-----
| Original motif Consensus sequence: CAHCACAGTGGAGCACT | Reverse complement motif Consensus sequence: AGTGCTCCACTGTGDTG |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 27 |
| Motif name: | AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0980018 |
Alignment:
ACTGTGACACCACAGTGGAGCACT
---------DCCACGTGCV-----
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT | Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 32 |
| Motif name: | gCGCGCsCgsG |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0989125 |
Alignment:
CGCGSGCGCGC
VGCACGTGGH-
| Original motif Consensus sequence: GCGCGCSCGCG | Reverse complement motif Consensus sequence: CGCGSGCGCGC |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 17 |
| Motif name: | TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0993196 |
Alignment:
GGAGCACTGTGACACCACA
---DCCACGTGCV------
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC | Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
 |
 |
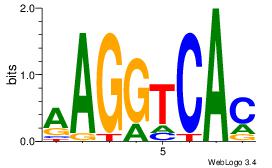
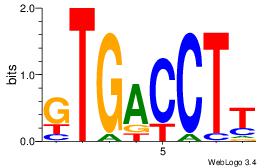
| Dataset #: 1 | Motif ID: 11 | Motif name: NR4A2 |
| Original motif Consensus sequence: AAGGTCAC | Reverse complement motif Consensus sequence: GTGACCTT |
 |
 |
Best Matches for Top Significant Motif ID 11 (Highest to Lowest)
| Dataset #: | 2 |
| Motif ID: | 22 |
| Motif name: | CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.0557243 |
Alignment:
CACTGTGRTRTCACAGTGSWSCACT
------AAGGTCAC-----------
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT | Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 23 |
| Motif name: | TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.060729 |
Alignment:
CCACTGTGVTGTCACAGTGCTCCA
-------AAGGTCAC---------
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG | Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 21 |
| Motif name: | AmTGTGACACCACAGT |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0645146 |
Alignment:
ACTGTGGTGTCACART
-----AAGGTCAC---
| Original motif Consensus sequence: AMTGTGACACCACAGT | Reverse complement motif Consensus sequence: ACTGTGGTGTCACART |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 17 |
| Motif name: | TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0695512 |
Alignment:
TGTGGTGTCACAGTGCTCC
---AAGGTCAC--------
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC | Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 27 |
| Motif name: | AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0709831 |
Alignment:
AGTGCTCCACTGTGGTGTCACAGT
-------------AAGGTCAC---
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT | Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
 |
 |
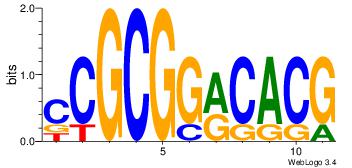
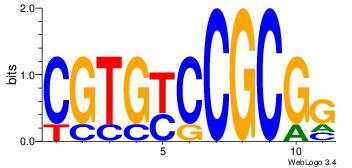
| Dataset #: | 2 |
| Motif ID: | 29 |
| Motif name: | cCGCGGrCACG |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0772292 |
Alignment:
CCGCGGRCACG
--AAGGTCAC-
| Original motif Consensus sequence: CCGCGGRCACG | Reverse complement motif Consensus sequence: CGTGKCCGCGG |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 25 |
| Motif name: | wwCCAmAGTCmt |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0788401 |
Alignment:
DDCCAMAGTCHB
----AAGGTCAC
| Original motif Consensus sequence: DDCCAMAGTCHB | Reverse complement motif Consensus sequence: VDGACTRTGGDD |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 20 |
| Motif name: | CagTGCTCCACTGTGgT |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 10 |
| Number of overlap: | 8 |
| Similarity score: | 0.0864039 |
Alignment:
ABCACAGTGGAGCACTG
---------GTGACCTT
| Original motif Consensus sequence: CAGTGCTCCACTGTGBT | Reverse complement motif Consensus sequence: ABCACAGTGGAGCACTG |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 28 |
| Motif name: | ssGCkTGCssk |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0.568556 |
Alignment:
SSGCKTGCSSK-
----GTGACCTT
| Original motif Consensus sequence: SSGCKTGCSSK | Reverse complement motif Consensus sequence: YSSGCAYGCSS |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 26 |
| Motif name: | CAcCACAGTGGAGCAct |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 11 |
| Number of overlap: | 7 |
| Similarity score: | 0.581814 |
Alignment:
CAHCACAGTGGAGCACT-
----------GTGACCTT
| Original motif Consensus sequence: CAHCACAGTGGAGCACT | Reverse complement motif Consensus sequence: AGTGCTCCACTGTGDTG |
 |
 |
| Dataset #: 2 | Motif ID: 25 | Motif name: wwCCAmAGTCmt |
| Original motif Consensus sequence: DDCCAMAGTCHB | Reverse complement motif Consensus sequence: VDGACTRTGGDD |
 |
 |
Best Matches for Top Significant Motif ID 25 (Highest to Lowest)
| Dataset #: | 1 |
| Motif ID: | 1 |
| Motif name: | HNF4A |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.032442 |
Alignment:
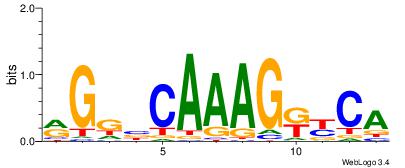
RGGBCAAAGKYCA
-DDCCAMAGTCHB
| Original motif Consensus sequence: RGGBCAAAGKYCA | Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 7 |
| Motif name: | REST |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0391228 |
Alignment:
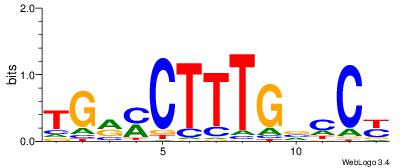
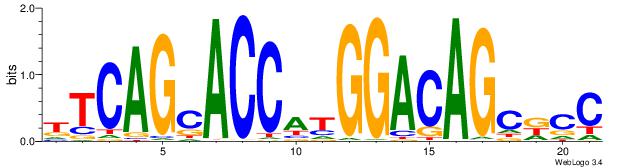
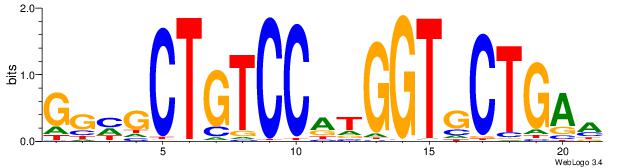
GGYGCTGTCCATGGTGCTGAA
---DDCCAMAGTCHB------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC | Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
 |
 |
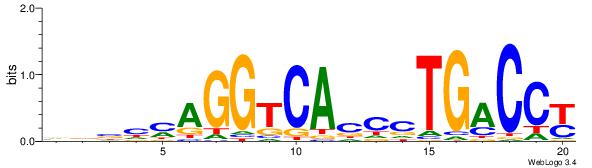
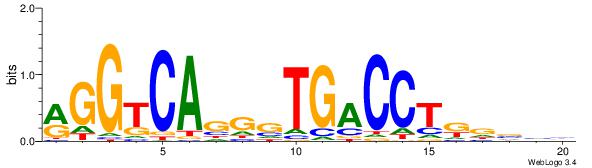
| Dataset #: | 1 |
| Motif ID: | 3 |
| Motif name: | ESR1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 12 |
| Similarity score: | 0.0407304 |
Alignment:
VDBHMAGGTCACCCTGACCY
DDCCAMAGTCHB--------
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY | Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
 |
 |
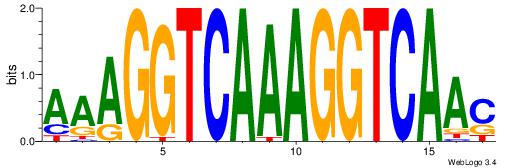
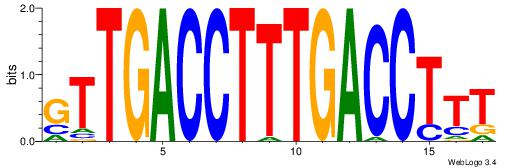
| Dataset #: | 1 |
| Motif ID: | 10 |
| Motif name: | NR1H2RXRA |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0463384 |
Alignment:
AAAGGTCAAAGGTCAAC
----DDCCAMAGTCHB-
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC | Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 14 |
| Motif name: | PPARGRXRA |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0480406 |
Alignment:
TGRCCTKTGHCCKAB
--DDCCAMAGTCHB-
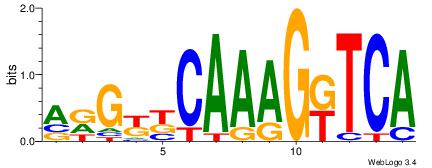
| Original motif Consensus sequence: BTRGGDCARAGGKCA | Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
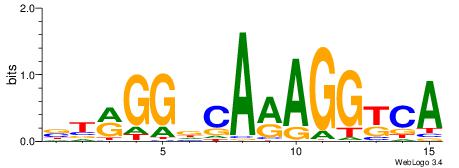 |
 |
| Dataset #: | 1 |
| Motif ID: | 2 |
| Motif name: | NR2F1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0506507 |
Alignment:
TGAMCTTTGMMCYT
--VDGACTRTGGDD
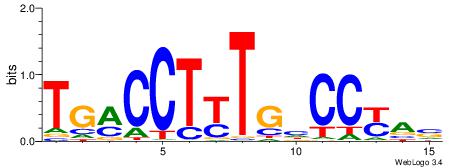
| Original motif Consensus sequence: TGAMCTTTGMMCYT | Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
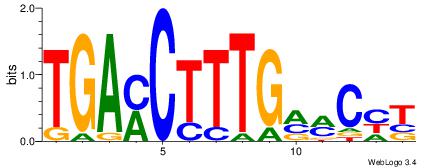 |
 |
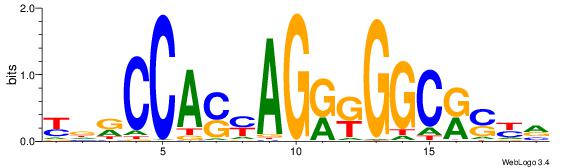
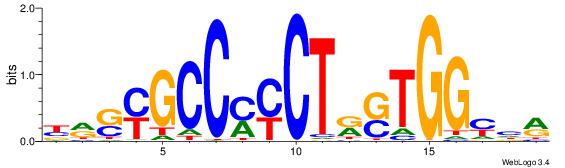
| Dataset #: | 1 |
| Motif ID: | 16 |
| Motif name: | CTCF |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 12 |
| Similarity score: | 0.0523578 |
Alignment:
BMSMGCCYMCTKSTGGMHM
DDCCAMAGTCHB-------
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV | Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
 |
 |
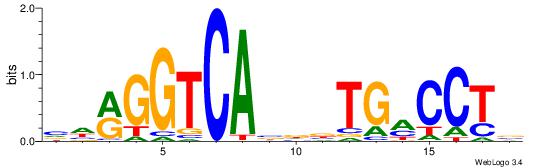
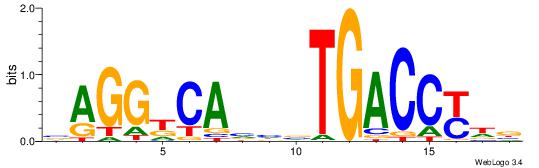
| Dataset #: | 1 |
| Motif ID: | 9 |
| Motif name: | ESR2 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0576723 |
Alignment:
VHRGGTCABDBTGMCCTB
---DDCCAMAGTCHB---
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB | Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 4 |
| Motif name: | Esrrb |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.525631 |
Alignment:
VBBYCAAGGTCA-
-DDCCAMAGTCHB
| Original motif Consensus sequence: VBBYCAAGGTCA | Reverse complement motif Consensus sequence: TGACCTTGMBBB |
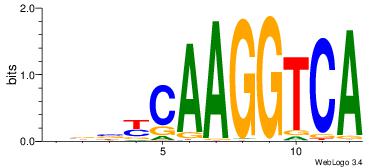 |
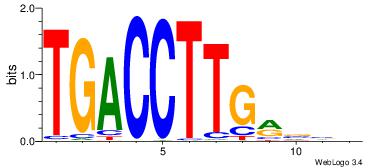 |
| Dataset #: | 1 |
| Motif ID: | 8 |
| Motif name: | Myc |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 1.0578 |
Alignment:
--VGCACGTGGH
VDGACTRTGGDD
| Original motif Consensus sequence: VGCACGTGGH | Reverse complement motif Consensus sequence: DCCACGTGCV |
 |
 |
| Dataset #: 2 | Motif ID: 28 | Motif name: ssGCkTGCssk |
| Original motif Consensus sequence: SSGCKTGCSSK | Reverse complement motif Consensus sequence: YSSGCAYGCSS |
 |
 |
Best Matches for Top Significant Motif ID 28 (Highest to Lowest)
| Dataset #: | 1 |
| Motif ID: | 16 |
| Motif name: | CTCF |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 11 |
| Similarity score: | 0.0349439 |
Alignment:
YDRCCASYAGRKGGCRSYV
-------SSGCKTGCSSK-
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV | Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 4 |
| Motif name: | Esrrb |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0358701 |
Alignment:
VBBYCAAGGTCA
-YSSGCAYGCSS
| Original motif Consensus sequence: VBBYCAAGGTCA | Reverse complement motif Consensus sequence: TGACCTTGMBBB |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 9 |
| Motif name: | ESR2 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0379074 |
Alignment:
VHRGGTCABDBTGMCCTB
-----YSSGCAYGCSS--
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB | Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 7 |
| Motif name: | REST |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 10 |
| Number of overlap: | 11 |
| Similarity score: | 0.0409322 |
Alignment:
TTCAGCACCATGGACAGCKCC
-YSSGCAYGCSS---------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC | Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 14 |
| Motif name: | PPARGRXRA |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.041419 |
Alignment:
BTRGGDCARAGGKCA
--YSSGCAYGCSS--
| Original motif Consensus sequence: BTRGGDCARAGGKCA | Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 1 |
| Motif name: | HNF4A |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0437981 |
Alignment:
RGGBCAAAGKYCA
--YSSGCAYGCSS
| Original motif Consensus sequence: RGGBCAAAGKYCA | Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 6 |
| Motif name: | Mycn |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.53887 |
Alignment:
GCCACGTGSD-
SSGCKTGCSSK
| Original motif Consensus sequence: HSCACGTGGC | Reverse complement motif Consensus sequence: GCCACGTGSD |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 8 |
| Motif name: | Myc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.542915 |
Alignment:
DCCACGTGCV-
SSGCKTGCSSK
| Original motif Consensus sequence: VGCACGTGGH | Reverse complement motif Consensus sequence: DCCACGTGCV |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 10 |
| Motif name: | NR1H2RXRA |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 10 |
| Number of overlap: | 8 |
| Similarity score: | 1.53583 |
Alignment:
---AAAGGTCAAAGGTCAAC
YSSGCAYGCSS---------
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC | Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 15 |
| Motif name: | Tcfcp2l1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 1.53704 |
Alignment:
---CCAGYYHVADCCRG
YSSGCAYGCSS------
| Original motif Consensus sequence: CCAGYYHVADCCRG | Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
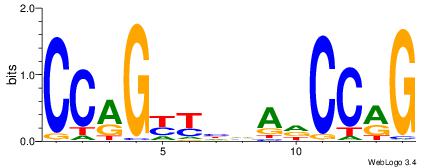 |
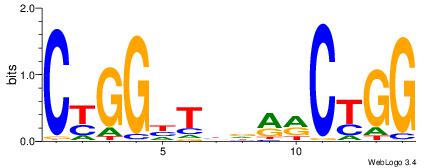 |
| Dataset #: 2 | Motif ID: 24 | Motif name: ssCCCCGCSssk |
| Original motif Consensus sequence: SBCCCCGCCSBB | Reverse complement motif Consensus sequence: BBSGGCGGGGBS |
 |
 |
Best Matches for Top Significant Motif ID 24 (Highest to Lowest)
| Dataset #: | 1 |
| Motif ID: | 3 |
| Motif name: | ESR1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.063076 |
Alignment:
MGGTCAGGGTGACCTRDBHV
-------BBSGGCGGGGBS-
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY | Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 16 |
| Motif name: | CTCF |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0642087 |
Alignment:
YDRCCASYAGRKGGCRSYV
-BBSGGCGGGGBS------
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV | Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 14 |
| Motif name: | PPARGRXRA |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0671548 |
Alignment:
TGRCCTKTGHCCKAB
-SBCCCCGCCSBB--
| Original motif Consensus sequence: BTRGGDCARAGGKCA | Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 9 |
| Motif name: | ESR2 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0734481 |
Alignment:
BAGGYCABHBTGACCKHV
-SBCCCCGCCSBB-----
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB | Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 7 |
| Motif name: | REST |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 10 |
| Number of overlap: | 12 |
| Similarity score: | 0.0751084 |
Alignment:
TTCAGCACCATGGACAGCKCC
BBSGGCGGGGBS---------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC | Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 4 |
| Motif name: | Esrrb |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.571364 |
Alignment:
VBBYCAAGGTCA-
-BBSGGCGGGGBS
| Original motif Consensus sequence: VBBYCAAGGTCA | Reverse complement motif Consensus sequence: TGACCTTGMBBB |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 1 |
| Motif name: | HNF4A |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.573202 |
Alignment:
-TGMYCTTTGBCCK
SBCCCCGCCSBB--
| Original motif Consensus sequence: RGGBCAAAGKYCA | Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 15 |
| Motif name: | Tcfcp2l1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 9 |
| Similarity score: | 1.54913 |
Alignment:
CKGGDTBDMMCTGG---
-----BBSGGCGGGGBS
| Original motif Consensus sequence: CCAGYYHVADCCRG | Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 6 |
| Motif name: | Mycn |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 9 |
| Similarity score: | 1.57507 |
Alignment:
---HSCACGTGGC
SBCCCCGCCSBB-
| Original motif Consensus sequence: HSCACGTGGC | Reverse complement motif Consensus sequence: GCCACGTGSD |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 8 |
| Motif name: | Myc |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 9 |
| Similarity score: | 1.57737 |
Alignment:
VGCACGTGGH---
-BBSGGCGGGGBS
| Original motif Consensus sequence: VGCACGTGGH | Reverse complement motif Consensus sequence: DCCACGTGCV |
 |
 |
| Dataset #: 2 | Motif ID: 29 | Motif name: cCGCGGrCACG |
| Original motif Consensus sequence: CCGCGGRCACG | Reverse complement motif Consensus sequence: CGTGKCCGCGG |
 |
 |
Best Matches for Top Significant Motif ID 29 (Highest to Lowest)
| Dataset #: | 1 |
| Motif ID: | 7 |
| Motif name: | REST |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0484441 |
Alignment:
TTCAGCACCATGGACAGCKCC
-------CCGCGGRCACG---
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC | Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 3 |
| Motif name: | ESR1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0486581 |
Alignment:
MGGTCAGGGTGACCTRDBHV
-------CGTGKCCGCGG--
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY | Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 16 |
| Motif name: | CTCF |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 11 |
| Similarity score: | 0.0588663 |
Alignment:
YDRCCASYAGRKGGCRSYV
-------CCGCGGRCACG-
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV | Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 14 |
| Motif name: | PPARGRXRA |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0629476 |
Alignment:
TGRCCTKTGHCCKAB
---CGTGKCCGCGG-
| Original motif Consensus sequence: BTRGGDCARAGGKCA | Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 9 |
| Motif name: | ESR2 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 10 |
| Similarity score: | 0.551597 |
Alignment:
BAGGYCABHBTGACCKHV-
--------CGTGKCCGCGG
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB | Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 8 |
| Motif name: | Myc |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 9 |
| Similarity score: | 1.0488 |
Alignment:
VGCACGTGGH--
-CGTGKCCGCGG
| Original motif Consensus sequence: VGCACGTGGH | Reverse complement motif Consensus sequence: DCCACGTGCV |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 6 |
| Motif name: | Mycn |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 9 |
| Similarity score: | 1.04892 |
Alignment:
HSCACGTGGC--
-CGTGKCCGCGG
| Original motif Consensus sequence: HSCACGTGGC | Reverse complement motif Consensus sequence: GCCACGTGSD |
 |
 |
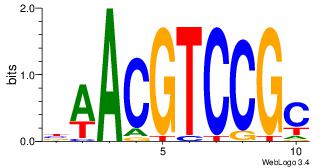
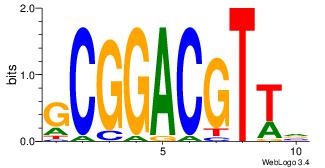
| Dataset #: | 1 |
| Motif ID: | 13 |
| Motif name: | MIZF |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 9 |
| Similarity score: | 1.04904 |
Alignment:
--BAACGTCCGC
CGTGKCCGCGG-
| Original motif Consensus sequence: BAACGTCCGC | Reverse complement motif Consensus sequence: GCGGACGTTV |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 4 |
| Motif name: | Esrrb |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 9 |
| Similarity score: | 1.05291 |
Alignment:
VBBYCAAGGTCA--
---CCGCGGRCACG
| Original motif Consensus sequence: VBBYCAAGGTCA | Reverse complement motif Consensus sequence: TGACCTTGMBBB |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 1 |
| Motif name: | HNF4A |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 1.55795 |
Alignment:
TGMYCTTTGBCCK---
-----CGTGKCCGCGG
| Original motif Consensus sequence: RGGBCAAAGKYCA | Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
 |
 |
| Dataset #: 1 | Motif ID: 4 | Motif name: Esrrb |
| Original motif Consensus sequence: VBBYCAAGGTCA | Reverse complement motif Consensus sequence: TGACCTTGMBBB |
 |
 |
Best Matches for Top Significant Motif ID 4 (Highest to Lowest)
| Dataset #: | 2 |
| Motif ID: | 17 |
| Motif name: | TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0772556 |
Alignment:
TGTGGTGTCACAGTGCTCC
-------TGACCTTGMBBB
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC | Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 23 |
| Motif name: | TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 12 |
| Number of overlap: | 12 |
| Similarity score: | 0.0785385 |
Alignment:
CCACTGTGVTGTCACAGTGCTCCA
-----------TGACCTTGMBBB-
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG | Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 22 |
| Motif name: | CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 11 |
| Number of overlap: | 12 |
| Similarity score: | 0.0832722 |
Alignment:
AGTGSWSCACTGTGAMAMCACAGTG
---VBBYCAAGGTCA----------
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT | Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 25 |
| Motif name: | wwCCAmAGTCmt |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.558919 |
Alignment:
-DDCCAMAGTCHB
VBBYCAAGGTCA-
| Original motif Consensus sequence: DDCCAMAGTCHB | Reverse complement motif Consensus sequence: VDGACTRTGGDD |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 28 |
| Motif name: | ssGCkTGCssk |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.581406 |
Alignment:
-SSGCKTGCSSK
TGACCTTGMBBB
| Original motif Consensus sequence: SSGCKTGCSSK | Reverse complement motif Consensus sequence: YSSGCAYGCSS |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 24 |
| Motif name: | ssCCCCGCSssk |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.583683 |
Alignment:
-SBCCCCGCCSBB
TGACCTTGMBBB-
| Original motif Consensus sequence: SBCCCCGCCSBB | Reverse complement motif Consensus sequence: BBSGGCGGGGBS |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 27 |
| Motif name: | AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 15 |
| Number of overlap: | 10 |
| Similarity score: | 1.08683 |
Alignment:
AGTGCTCCACTGTGGTGTCACAGT--
--------------VBBYCAAGGTCA
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT | Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 19 |
| Motif name: | atactttggc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 1.08726 |
Alignment:
--GCCAAAGTAT
VBBYCAAGGTCA
| Original motif Consensus sequence: ATACTTTGGC | Reverse complement motif Consensus sequence: GCCAAAGTAT |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 29 |
| Motif name: | cCGCGGrCACG |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 9 |
| Similarity score: | 1.57579 |
Alignment:
---CCGCGGRCACG
VBBYCAAGGTCA--
| Original motif Consensus sequence: CCGCGGRCACG | Reverse complement motif Consensus sequence: CGTGKCCGCGG |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 21 |
| Motif name: | AmTGTGACACCACAGT |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 10 |
| Number of overlap: | 7 |
| Similarity score: | 2.57595 |
Alignment:
-----AMTGTGACACCACAGT
VBBYCAAGGTCA---------
| Original motif Consensus sequence: AMTGTGACACCACAGT | Reverse complement motif Consensus sequence: ACTGTGGTGTCACART |
 |
 |
| Dataset #: 1 | Motif ID: 13 | Motif name: MIZF |
| Original motif Consensus sequence: BAACGTCCGC | Reverse complement motif Consensus sequence: GCGGACGTTV |
 |
 |
Best Matches for Top Significant Motif ID 13 (Highest to Lowest)
| Dataset #: | 2 |
| Motif ID: | 32 |
| Motif name: | gCGCGCsCgsG |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0651598 |
Alignment:
GCGCGCSCGCG
BAACGTCCGC-
| Original motif Consensus sequence: GCGCGCSCGCG | Reverse complement motif Consensus sequence: CGCGSGCGCGC |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 19 |
| Motif name: | atactttggc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0803571 |
Alignment:
ATACTTTGGC
BAACGTCCGC
| Original motif Consensus sequence: ATACTTTGGC | Reverse complement motif Consensus sequence: GCCAAAGTAT |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 29 |
| Motif name: | cCGCGGrCACG |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 9 |
| Similarity score: | 0.560913 |
Alignment:
CCGCGGRCACG-
--GCGGACGTTV
| Original motif Consensus sequence: CCGCGGRCACG | Reverse complement motif Consensus sequence: CGTGKCCGCGG |
 |
 |
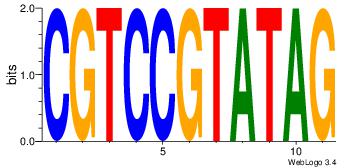
| Dataset #: | 2 |
| Motif ID: | 31 |
| Motif name: | ctatacggacg |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 9 |
| Similarity score: | 0.562302 |
Alignment:
-CGTCCGTATAG
GCGGACGTTV--
| Original motif Consensus sequence: CTATACGGACG | Reverse complement motif Consensus sequence: CGTCCGTATAG |
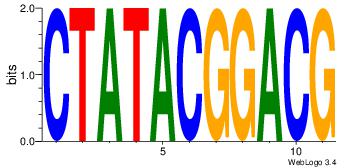 |
 |
| Dataset #: | 2 |
| Motif ID: | 25 |
| Motif name: | wwCCAmAGTCmt |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 9 |
| Similarity score: | 0.562428 |
Alignment:
DDCCAMAGTCHB-
---BAACGTCCGC
| Original motif Consensus sequence: DDCCAMAGTCHB | Reverse complement motif Consensus sequence: VDGACTRTGGDD |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 18 |
| Motif name: | sscCCCGCGcs |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 1.06446 |
Alignment:
SBCGCGGGGSB--
---GCGGACGTTV
| Original motif Consensus sequence: BSCCCCGCGBS | Reverse complement motif Consensus sequence: SBCGCGGGGSB |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 24 |
| Motif name: | ssCCCCGCSssk |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 1.07197 |
Alignment:
BBSGGCGGGGBS--
----GCGGACGTTV
| Original motif Consensus sequence: SBCCCCGCCSBB | Reverse complement motif Consensus sequence: BBSGGCGGGGBS |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 33 |
| Motif name: | srACyCCGAyr |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 1.55 |
Alignment:
SRACYCCGAYR---
----GCGGACGTTV
| Original motif Consensus sequence: SRACYCCGAYR | Reverse complement motif Consensus sequence: KKTCGGKGTKS |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 27 |
| Motif name: | AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 18 |
| Number of overlap: | 7 |
| Similarity score: | 1.5706 |
Alignment:
---AGTGCTCCACTGTGGTGTCACAGT
BAACGTCCGC-----------------
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT | Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 26 |
| Motif name: | CAcCACAGTGGAGCAct |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 11 |
| Number of overlap: | 7 |
| Similarity score: | 1.57176 |
Alignment:
CAHCACAGTGGAGCACT---
----------GCGGACGTTV
| Original motif Consensus sequence: CAHCACAGTGGAGCACT | Reverse complement motif Consensus sequence: AGTGCTCCACTGTGDTG |
 |
 |