Top 10 Significant Motifs - Global Matching (Highest to Lowest)
| Dataset #: 2 | Motif ID: 18 | Motif name: Motif 18 |
| Original motif Consensus sequence: AMACA | Reverse complement motif Consensus sequence: TGTRT |
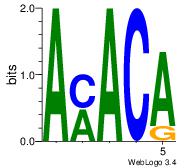 |
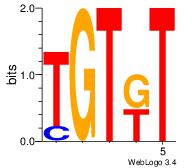 |
Best Matches for Top Significant Motif ID 18 (Highest to Lowest)
| Dataset #: | 1 |
| Motif ID: | 2 |
| Motif name: | Motif 2 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 5 |
| Similarity score: | 0 |
Alignment:
CACACACA
---AMACA
| Original motif Consensus sequence: CACACACA | Reverse complement motif Consensus sequence: TGTGTGTG |
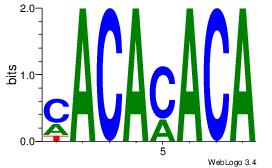 |
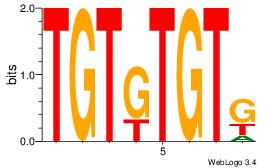 |
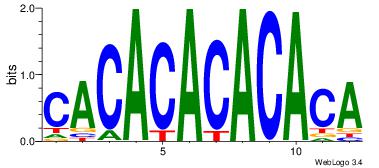
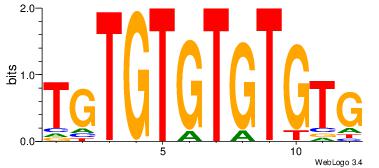
| Dataset #: | 4 |
| Motif ID: | 79 |
| Motif name: | CACACACACACA |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 5 |
| Similarity score: | 0.0148279 |
Alignment:
TGTGTGTGTGTG
--TGTRT-----
| Original motif Consensus sequence: CACACACACACA | Reverse complement motif Consensus sequence: TGTGTGTGTGTG |
 |
 |
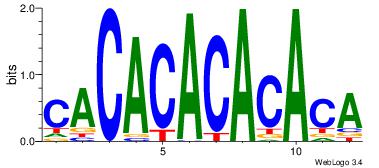
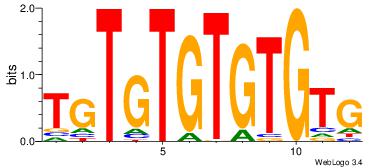
| Dataset #: | 4 |
| Motif ID: | 95 |
| Motif name: | CACACACACACA |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 5 |
| Similarity score: | 0.0228414 |
Alignment:
TGTGTGTGTGTG
----TGTRT---
| Original motif Consensus sequence: CACACACACACA | Reverse complement motif Consensus sequence: TGTGTGTGTGTG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 69 |
| Motif name: | aaACAAAACaa |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.0274014 |
Alignment:
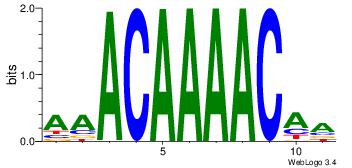
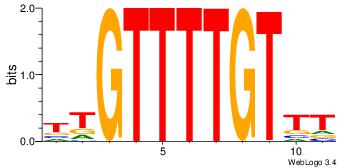
AAACAAAACAA
-----AMACA-
| Original motif Consensus sequence: AAACAAAACAA | Reverse complement motif Consensus sequence: TTGTTTTGTTT |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 86 |
| Motif name: | aaCAAAAACaa |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 5 |
| Similarity score: | 0.0291354 |
Alignment:
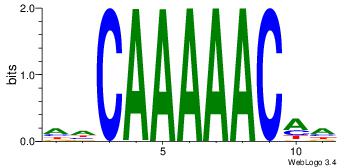
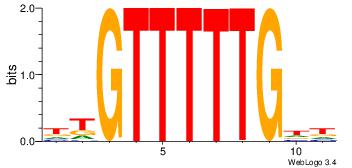
HHCAAAAACAH
-----AMACA-
| Original motif Consensus sequence: HHCAAAAACAH | Reverse complement motif Consensus sequence: HTGTTTTTGHH |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 97 |
| Motif name: | ryayAyAyrtGTRTrTry |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 5 |
| Similarity score: | 0.0293401 |
Alignment:
DHAYAYAYRTGTGTRTDH
---------TGTRT----
| Original motif Consensus sequence: DHAYAYAYRTGTGTRTDH | Reverse complement motif Consensus sequence: HDAMACACAKKTKTKTHD |
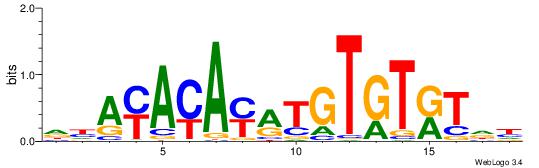 |
 |
| Dataset #: | 4 |
| Motif ID: | 100 |
| Motif name: | wrTGTATACAyw |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.0302831 |
Alignment:
HDTGTATACAHD
--TGTRT-----
| Original motif Consensus sequence: HDTGTATACAHD | Reverse complement motif Consensus sequence: DHTGTATACADH |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 5 |
| Motif name: | Motif 5 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 5 |
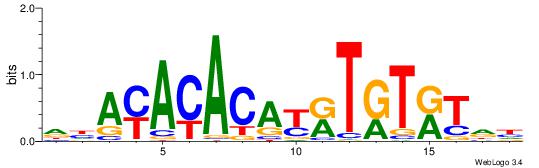
| Similarity score: | 0.0378142 |
Alignment:
AAAASAAA
-AMACA--
| Original motif Consensus sequence: AAAASAAA | Reverse complement motif Consensus sequence: TTTSTTTT |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 92 |
| Motif name: | tyTCTkTkTCyy |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 5 |
| Similarity score: | 0.0390126 |
Alignment:
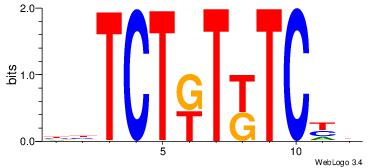
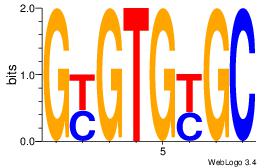
VHGARAYAGAVV
---AMACA----
| Original motif Consensus sequence: BBTCTKTKTCHB | Reverse complement motif Consensus sequence: VHGARAYAGAVV |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 14 |
| Motif name: | Motif 14 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 5 |
| Similarity score: | 0.042464 |
Alignment:
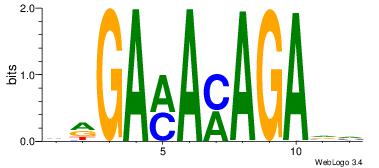
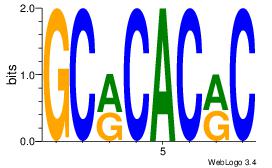
GKGTGKGC
-TGTRT--
| Original motif Consensus sequence: GCRCACRC | Reverse complement motif Consensus sequence: GKGTGKGC |
 |
 |
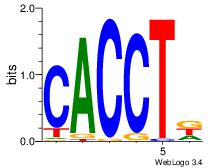
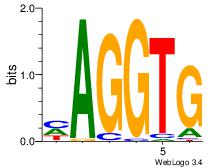
| Dataset #: 3 | Motif ID: 30 | Motif name: ZEB1 |
| Original motif Consensus sequence: CACCTD | Reverse complement motif Consensus sequence: HAGGTG |
 |
 |
Best Matches for Top Significant Motif ID 30 (Highest to Lowest)
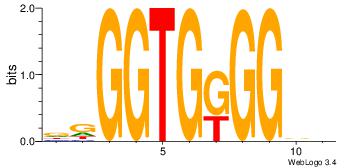
| Dataset #: | 4 |
| Motif ID: | 89 |
| Motif name: | ccCCmCACCcc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0148347 |
Alignment:
HBCCMCACCHH
-----CACCTD
| Original motif Consensus sequence: HBCCMCACCHH | Reverse complement motif Consensus sequence: DDGGTGRGGBD |
 |
 |
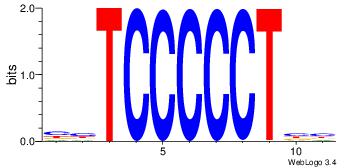
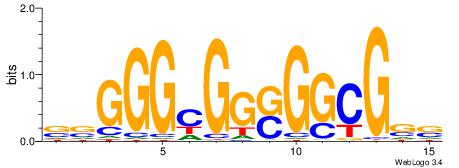
| Dataset #: | 4 |
| Motif ID: | 93 |
| Motif name: | rrAGGGGGArr |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0194512 |
Alignment:
VVAGGGGGAVV
-HAGGTG----
| Original motif Consensus sequence: VVAGGGGGAVV | Reverse complement motif Consensus sequence: VVTCCCCCTVV |
 |
 |
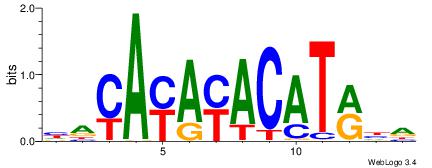
| Dataset #: | 4 |
| Motif ID: | 75 |
| Motif name: | yayAyAyACATrya |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0270731 |
Alignment:
HACAYAYACATRHD
------CACCTD--
| Original motif Consensus sequence: HACAYAYACATRHD | Reverse complement motif Consensus sequence: DHKATGTKTKTGTD |
 |
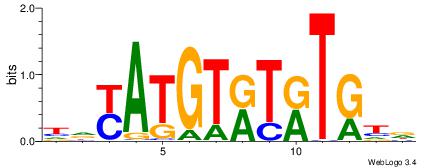 |
| Dataset #: | 4 |
| Motif ID: | 97 |
| Motif name: | ryayAyAyrtGTRTrTry |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.0321302 |
Alignment:
HDAMACACAKKTKTKTHD
-----CACCTD-------
| Original motif Consensus sequence: DHAYAYAYRTGTGTRTDH | Reverse complement motif Consensus sequence: HDAMACACAKKTKTKTHD |
 |
 |
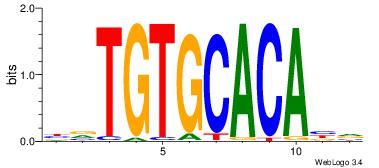
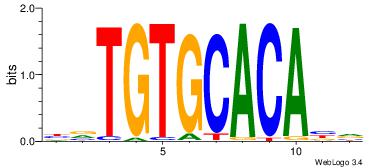
| Dataset #: | 4 |
| Motif ID: | 96 |
| Motif name: | yrTGTGCACAyr |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0342918 |
Alignment:
BVTGTGCACABV
HAGGTG------
| Original motif Consensus sequence: BVTGTGCACABV | Reverse complement motif Consensus sequence: BBTGTGCACAVV |
 |
 |
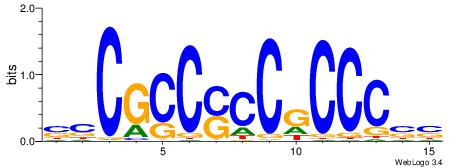
| Dataset #: | 4 |
| Motif ID: | 91 |
| Motif name: | ssCGCCsCCrCCCss |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.0456166 |
Alignment:
SSCGCCSCCGCCCSS
--------CACCTD-
| Original motif Consensus sequence: SSCGCCSCCGCCCSS | Reverse complement motif Consensus sequence: SSGGGCGGSGGCGSS |
 |
 |
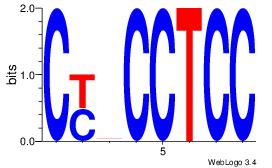
| Dataset #: | 1 |
| Motif ID: | 4 |
| Motif name: | Motif 4 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.0458702 |
Alignment:
CYBCCTCC
-CACCTD-
| Original motif Consensus sequence: CYBCCTCC | Reverse complement motif Consensus sequence: GGAGGVMG |
 |
 |
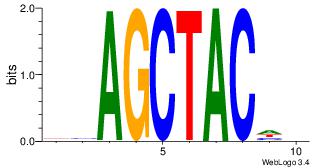
| Dataset #: | 4 |
| Motif ID: | 84 |
| Motif name: | wsAGCTACwy |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.0461351 |
Alignment:
VHGTAGCTVD
---CACCTD-
| Original motif Consensus sequence: DVAGCTACHB | Reverse complement motif Consensus sequence: VHGTAGCTVD |
 |
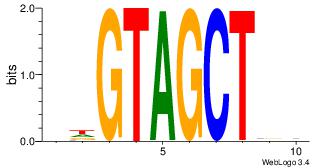 |
| Dataset #: | 4 |
| Motif ID: | 88 |
| Motif name: | ssCrCYyYCGss |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0554152 |
Alignment:
BBCRCCYCCGBB
--CACCTD----
| Original motif Consensus sequence: BBCRCCYCCGBB | Reverse complement motif Consensus sequence: BBCGGKGGMGBB |
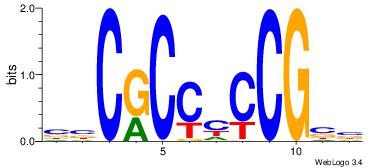 |
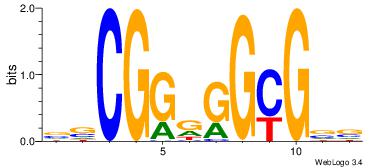 |
| Dataset #: | 4 |
| Motif ID: | 90 |
| Motif name: | ssCGsCwsCss |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0564656 |
Alignment:
BBCGSCWSCBB
-----CACCTD
| Original motif Consensus sequence: BBCGSCWSCBB | Reverse complement motif Consensus sequence: BBGSWGSCGBB |
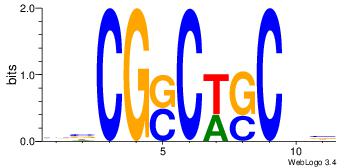 |
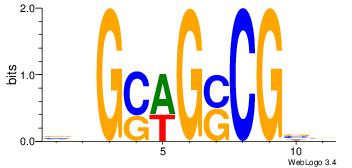 |
| Dataset #: 1 | Motif ID: 7 | Motif name: Motif 7 |
| Original motif Consensus sequence: ASASAGAA | Reverse complement motif Consensus sequence: TTCTSTST |
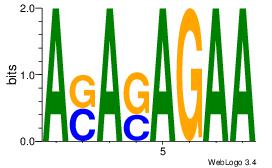 |
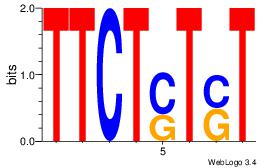 |
Best Matches for Top Significant Motif ID 7 (Highest to Lowest)
| Dataset #: | 4 |
| Motif ID: | 92 |
| Motif name: | tyTCTkTkTCyy |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.00355249 |
Alignment:
BBTCTKTKTCHB
-TTCTSTST---
| Original motif Consensus sequence: BBTCTKTKTCHB | Reverse complement motif Consensus sequence: VHGARAYAGAVV |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 69 |
| Motif name: | aaACAAAACaa |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.0313478 |
Alignment:
AAACAAAACAA
ASASAGAA---
| Original motif Consensus sequence: AAACAAAACAA | Reverse complement motif Consensus sequence: TTGTTTTGTTT |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 82 |
| Motif name: | rmACAGCAGsr |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.0319191 |
Alignment:
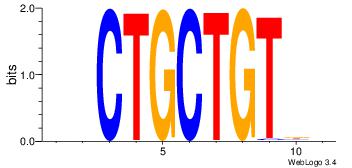
DVCTGCTGTVB
---TTCTSTST
| Original motif Consensus sequence: VVACAGCAGVD | Reverse complement motif Consensus sequence: DVCTGCTGTVB |
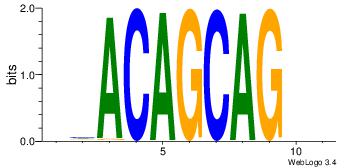 |
 |
| Dataset #: | 4 |
| Motif ID: | 95 |
| Motif name: | CACACACACACA |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0398795 |
Alignment:
TGTGTGTGTGTG
-TTCTSTST---
| Original motif Consensus sequence: CACACACACACA | Reverse complement motif Consensus sequence: TGTGTGTGTGTG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 79 |
| Motif name: | CACACACACACA |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0407034 |
Alignment:
TGTGTGTGTGTG
-TTCTSTST---
| Original motif Consensus sequence: CACACACACACA | Reverse complement motif Consensus sequence: TGTGTGTGTGTG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 102 |
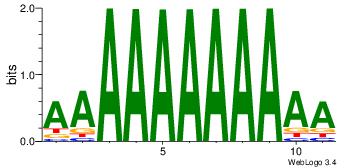
| Motif name: | AAAAAAAAAAA |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.0441956 |
Alignment:
AAAAAAAAAAA
ASASAGAA---
| Original motif Consensus sequence: AAAAAAAAAAA | Reverse complement motif Consensus sequence: TTTTTTTTTTT |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 26 |
| Motif name: | Motif 26 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0451851 |
Alignment:
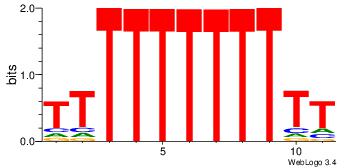
TKKTTTGTTTTTTT
----TTCTSTST--
| Original motif Consensus sequence: TKKTTTGTTTTTTT | Reverse complement motif Consensus sequence: AAAAAAACAAARRA |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 27 |
| Motif name: | Motif 27 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0493389 |
Alignment:
TGTGTGTGTGTKTG
-----TTCTSTST-
| Original motif Consensus sequence: CAYACACACACACA | Reverse complement motif Consensus sequence: TGTGTGTGTGTKTG |
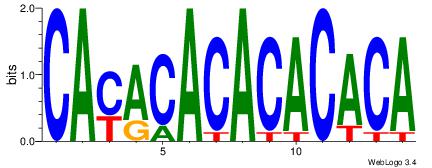 |
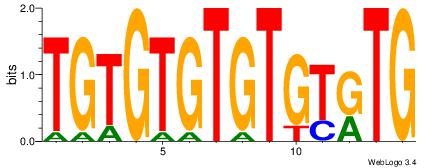 |
| Dataset #: | 2 |
| Motif ID: | 25 |
| Motif name: | Motif 25 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0494079 |
Alignment:
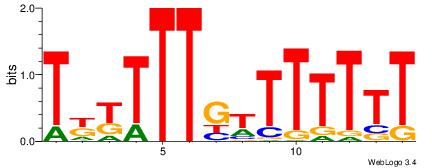
CACACAVRCACACA
-----ASASAGAA-
| Original motif Consensus sequence: CACACAVRCACACA | Reverse complement motif Consensus sequence: TGTGTGKVTGTGTG |
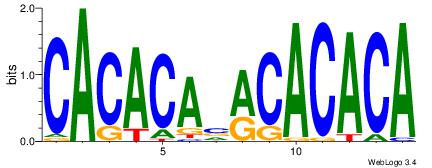 |
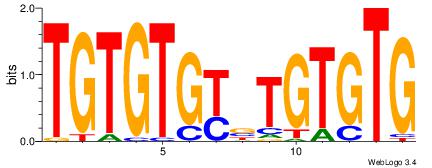 |
| Dataset #: | 3 |
| Motif ID: | 48 |
| Motif name: | EWSR1-FLI1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0559328 |
Alignment:
CCTTCCTTCCTTCCTTCC
---TTCTSTST-------
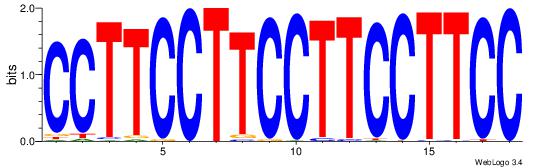
| Original motif Consensus sequence: GGAAGGAAGGAAGGAAGG | Reverse complement motif Consensus sequence: CCTTCCTTCCTTCCTTCC |
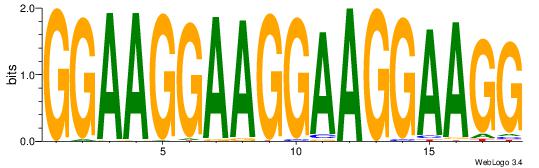 |
 |
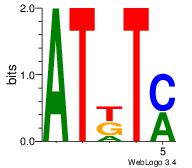
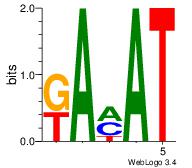
| Dataset #: 2 | Motif ID: 21 | Motif name: Motif 21 |
| Original motif Consensus sequence: ATKTM | Reverse complement motif Consensus sequence: RARAT |
 |
 |
Best Matches for Top Significant Motif ID 21 (Highest to Lowest)
| Dataset #: | 1 |
| Motif ID: | 15 |
| Motif name: | Motif 15 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 5 |
| Similarity score: | 0.0065927 |
Alignment:
CATTTKC
-ATKTM-
| Original motif Consensus sequence: GRAAATG | Reverse complement motif Consensus sequence: CATTTKC |
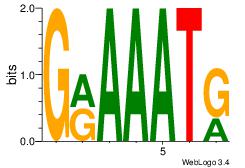 |
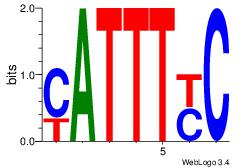 |
| Dataset #: | 1 |
| Motif ID: | 10 |
| Motif name: | Motif 10 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 5 |
| Similarity score: | 0.0217628 |
Alignment:
TAMATA
-RARAT
| Original motif Consensus sequence: TAMATA | Reverse complement motif Consensus sequence: TATRTA |
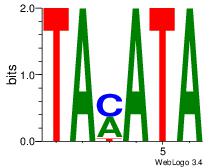 |
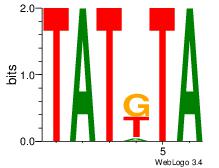 |
| Dataset #: | 4 |
| Motif ID: | 85 |
| Motif name: | wtATTTTTAww |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 5 |
| Similarity score: | 0.0314602 |
Alignment:
DDTAAAAATHD
----RARAT--
| Original motif Consensus sequence: DHATTTTTADD | Reverse complement motif Consensus sequence: DDTAAAAATHD |
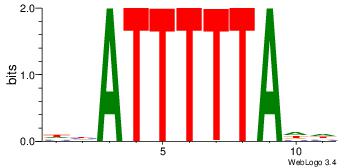 |
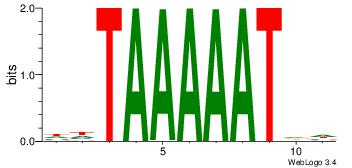 |
| Dataset #: | 4 |
| Motif ID: | 71 |
| Motif name: | wyATTTTAww |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 5 |
| Similarity score: | 0.0314958 |
Alignment:
DDTAAAATHD
---RARAT--
| Original motif Consensus sequence: DHATTTTADD | Reverse complement motif Consensus sequence: DDTAAAATHD |
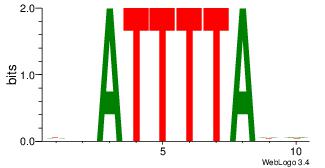 |
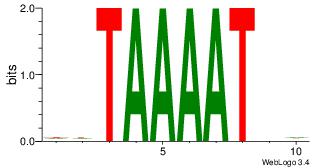 |
| Dataset #: | 4 |
| Motif ID: | 92 |
| Motif name: | tyTCTkTkTCyy |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.0326182 |
Alignment:
VHGARAYAGAVV
--RARAT-----
| Original motif Consensus sequence: BBTCTKTKTCHB | Reverse complement motif Consensus sequence: VHGARAYAGAVV |
 |
 |
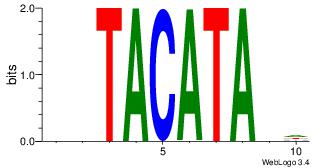
| Dataset #: | 4 |
| Motif ID: | 74 |
| Motif name: | twTACATAvw |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 5 |
| Similarity score: | 0.0335447 |
Alignment:
DBTATGTAHH
---ATKTM--
| Original motif Consensus sequence: HHTACATAVD | Reverse complement motif Consensus sequence: DBTATGTAHH |
 |
 |
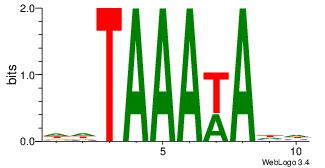
| Dataset #: | 4 |
| Motif ID: | 83 |
| Motif name: | wwTAAAwAww |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 5 |
| Similarity score: | 0.0514182 |
Alignment:
HHTWTTTADD
---ATKTM--
| Original motif Consensus sequence: DDTAAAWAHH | Reverse complement motif Consensus sequence: HHTWTTTADD |
 |
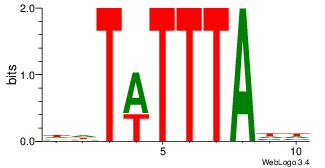 |
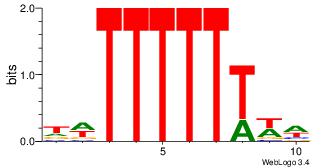
| Dataset #: | 4 |
| Motif ID: | 76 |
| Motif name: | wwWAAAAAwa |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 5 |
| Similarity score: | 0.0597589 |
Alignment:
WWAAAAAAWH
----RARAT-
| Original motif Consensus sequence: WWAAAAAAWH | Reverse complement motif Consensus sequence: HWTTTTTTWW |
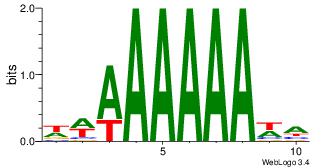 |
 |
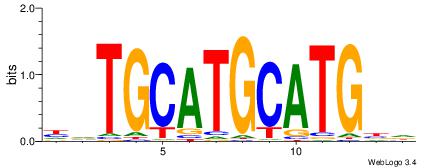
| Dataset #: | 4 |
| Motif ID: | 101 |
| Motif name: | AAAAAAAAAAA |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 10 |
| Number of overlap: | 5 |
| Similarity score: | 0.0609742 |
Alignment:
DVTGCATGCATGBH
---------ATKTM
| Original motif Consensus sequence: HVCATGCATGCABD | Reverse complement motif Consensus sequence: DVTGCATGCATGBH |
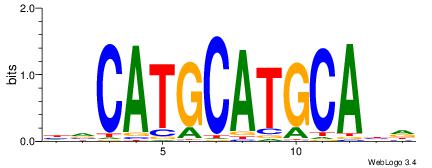 |
 |
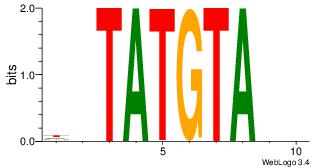
| Dataset #: | 4 |
| Motif ID: | 100 |
| Motif name: | wrTGTATACAyw |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 5 |
| Similarity score: | 0.0620607 |
Alignment:
DHTGTATACADH
-ATKTM------
| Original motif Consensus sequence: HDTGTATACAHD | Reverse complement motif Consensus sequence: DHTGTATACADH |
 |
 |
| Dataset #: 1 | Motif ID: 4 | Motif name: Motif 4 |
| Original motif Consensus sequence: CYBCCTCC | Reverse complement motif Consensus sequence: GGAGGVMG |
 |
 |
Best Matches for Top Significant Motif ID 4 (Highest to Lowest)
| Dataset #: | 3 |
| Motif ID: | 48 |
| Motif name: | EWSR1-FLI1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.022644 |
Alignment:
GGAAGGAAGGAAGGAAGG
-----GGAGGVMG-----
| Original motif Consensus sequence: GGAAGGAAGGAAGGAAGG | Reverse complement motif Consensus sequence: CCTTCCTTCCTTCCTTCC |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 32 |
| Motif name: | SP1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0247246 |
Alignment:
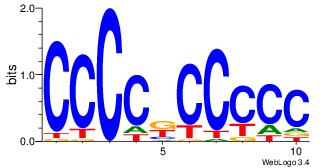
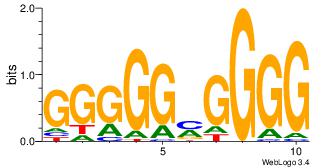
CCCCKCCCCC
--CYBCCTCC
| Original motif Consensus sequence: CCCCKCCCCC | Reverse complement motif Consensus sequence: GGGGGYGGGG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 93 |
| Motif name: | rrAGGGGGArr |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.0300296 |
Alignment:
VVTCCCCCTVV
---CYBCCTCC
| Original motif Consensus sequence: VVAGGGGGAVV | Reverse complement motif Consensus sequence: VVTCCCCCTVV |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 91 |
| Motif name: | ssCGCCsCCrCCCss |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0335629 |
Alignment:
SSCGCCSCCGCCCSS
----CYBCCTCC---
| Original motif Consensus sequence: SSCGCCSCCGCCCSS | Reverse complement motif Consensus sequence: SSGGGCGGSGGCGSS |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 52 |
| Motif name: | Klf4 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0354458 |
Alignment:
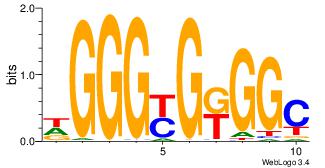
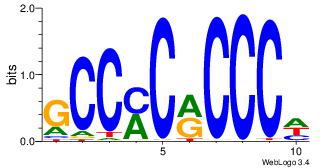
GCCYCMCCCD
-CYBCCTCC-
| Original motif Consensus sequence: DGGGYGKGGC | Reverse complement motif Consensus sequence: GCCYCMCCCD |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 88 |
| Motif name: | ssCrCYyYCGss |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.045829 |
Alignment:
BBCRCCYCCGBB
----CYBCCTCC
| Original motif Consensus sequence: BBCRCCYCCGBB | Reverse complement motif Consensus sequence: BBCGGKGGMGBB |
 |
 |
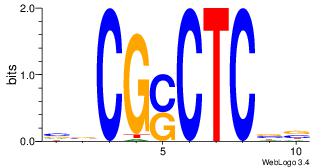
| Dataset #: | 4 |
| Motif ID: | 73 |
| Motif name: | ssCGsCTCss |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0472644 |
Alignment:
VVCGSCTCBB
-CYBCCTCC-
| Original motif Consensus sequence: VVCGSCTCBB | Reverse complement motif Consensus sequence: BBGAGSCGVV |
 |
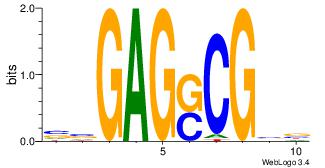 |
| Dataset #: | 3 |
| Motif ID: | 57 |
| Motif name: | MZF1_5-13 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.049303 |
Alignment:
BTHCCCCTYB
--CYBCCTCC
| Original motif Consensus sequence: BKAGGGGDAD | Reverse complement motif Consensus sequence: BTHCCCCTYB |
 |
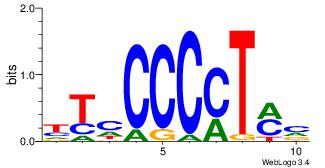 |
| Dataset #: | 3 |
| Motif ID: | 34 |
| Motif name: | RREB1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0528666 |
Alignment:
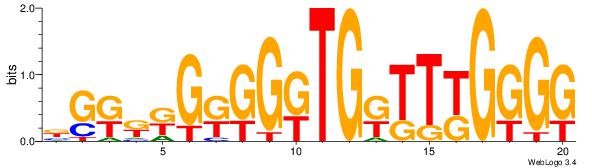
BGRRRGRGGRTGRTTYGGGG
----GGAGGVMG--------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV | Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 89 |
| Motif name: | ccCCmCACCcc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0549805 |
Alignment:
HBCCMCACCHH
-CYBCCTCC--
| Original motif Consensus sequence: HBCCMCACCHH | Reverse complement motif Consensus sequence: DDGGTGRGGBD |
 |
 |
| Dataset #: 1 | Motif ID: 2 | Motif name: Motif 2 |
| Original motif Consensus sequence: CACACACA | Reverse complement motif Consensus sequence: TGTGTGTG |
 |
 |
Best Matches for Top Significant Motif ID 2 (Highest to Lowest)
| Dataset #: | 4 |
| Motif ID: | 79 |
| Motif name: | CACACACACACA |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.00486551 |
Alignment:
CACACACACACA
--CACACACA--
| Original motif Consensus sequence: CACACACACACA | Reverse complement motif Consensus sequence: TGTGTGTGTGTG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 95 |
| Motif name: | CACACACACACA |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0100991 |
Alignment:
TGTGTGTGTGTG
----TGTGTGTG
| Original motif Consensus sequence: CACACACACACA | Reverse complement motif Consensus sequence: TGTGTGTGTGTG |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 27 |
| Motif name: | Motif 27 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0101254 |
Alignment:
TGTGTGTGTGTKTG
--TGTGTGTG----
| Original motif Consensus sequence: CAYACACACACACA | Reverse complement motif Consensus sequence: TGTGTGTGTGTKTG |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 25 |
| Motif name: | Motif 25 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.0226631 |
Alignment:
TGTGTGKVTGTGTG
TGTGTGTG------
| Original motif Consensus sequence: CACACAVRCACACA | Reverse complement motif Consensus sequence: TGTGTGKVTGTGTG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 97 |
| Motif name: | ryayAyAyrtGTRTrTry |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0393219 |
Alignment:
DHAYAYAYRTGTGTRTDH
---------TGTGTGTG-
| Original motif Consensus sequence: DHAYAYAYRTGTGTRTDH | Reverse complement motif Consensus sequence: HDAMACACAKKTKTKTHD |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 75 |
| Motif name: | yayAyAyACATrya |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0468047 |
Alignment:
HACAYAYACATRHD
--CACACACA----
| Original motif Consensus sequence: HACAYAYACATRHD | Reverse complement motif Consensus sequence: DHKATGTKTKTGTD |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 86 |
| Motif name: | aaCAAAAACaa |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0633924 |
Alignment:
HHCAAAAACAH
--CACACACA-
| Original motif Consensus sequence: HHCAAAAACAH | Reverse complement motif Consensus sequence: HTGTTTTTGHH |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 74 |
| Motif name: | twTACATAvw |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.0691437 |
Alignment:
DBTATGTAHH
TGTGTGTG--
| Original motif Consensus sequence: HHTACATAVD | Reverse complement motif Consensus sequence: DBTATGTAHH |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 34 |
| Motif name: | RREB1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.0752394 |
Alignment:
CCCCMAAMCAMCCMCMMMCV
------CACACACA------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV | Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 101 |
| Motif name: | AAAAAAAAAAA |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.0764586 |
Alignment:
DVTGCATGCATGBH
------TGTGTGTG
| Original motif Consensus sequence: HVCATGCATGCABD | Reverse complement motif Consensus sequence: DVTGCATGCATGBH |
 |
 |
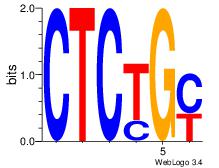
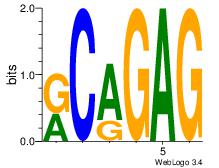
| Dataset #: 2 | Motif ID: 20 | Motif name: Motif 20 |
| Original motif Consensus sequence: CTCTGY | Reverse complement motif Consensus sequence: KCAGAG |
 |
 |
Best Matches for Top Significant Motif ID 20 (Highest to Lowest)
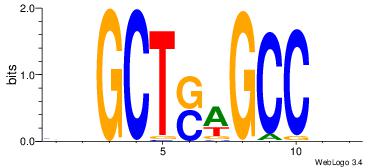
| Dataset #: | 4 |
| Motif ID: | 77 |
| Motif name: | ssGCTswGCCms |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.0323407 |
Alignment:
VBGCTSAGCCHB
---CTCTGY---
| Original motif Consensus sequence: VBGCTSAGCCHB | Reverse complement motif Consensus sequence: BDGGCTSAGCBV |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 7 |
| Motif name: | Motif 7 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0352938 |
Alignment:
ASASAGAA
--KCAGAG
| Original motif Consensus sequence: ASASAGAA | Reverse complement motif Consensus sequence: TTCTSTST |
 |
 |
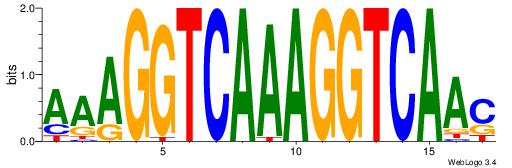
| Dataset #: | 3 |
| Motif ID: | 59 |
| Motif name: | NR1H2RXRA |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0398495 |
Alignment:
AAAGGTCAAAGGTCAAC
-----KCAGAG------
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC | Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
 |
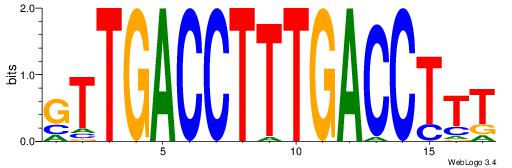 |
| Dataset #: | 3 |
| Motif ID: | 36 |
| Motif name: | PPARGRXRA |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.0415907 |
Alignment:
BTRGGDCARAGGKCA
-----KCAGAG----
| Original motif Consensus sequence: BTRGGDCARAGGKCA | Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 11 |
| Motif name: | Motif 11 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.0430865 |
Alignment:
CCGCCGCC
-CTCTGY-
| Original motif Consensus sequence: CCGCCGCC | Reverse complement motif Consensus sequence: GGCGGCGG |
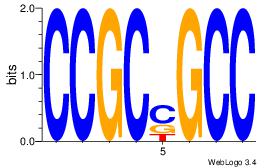 |
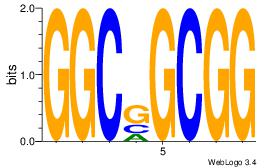 |
| Dataset #: | 4 |
| Motif ID: | 72 |
| Motif name: | ssCKCwsCGss |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.0439226 |
Alignment:
BVCGCWSCGBV
----CTCTGY-
| Original motif Consensus sequence: BVCGCWSCGBV | Reverse complement motif Consensus sequence: VBCGSWGCGVB |
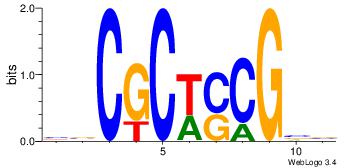 |
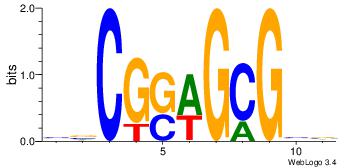 |
| Dataset #: | 3 |
| Motif ID: | 38 |
| Motif name: | NR2F1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0462597 |
Alignment:
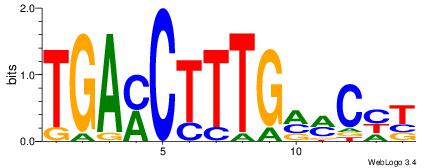
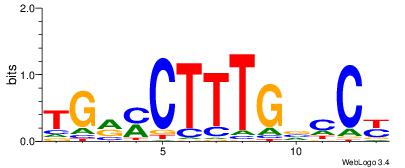
AKGYYCAAAGRTCA
----KCAGAG----
| Original motif Consensus sequence: TGAMCTTTGMMCYT | Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 92 |
| Motif name: | tyTCTkTkTCyy |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.0467436 |
Alignment:
BBTCTKTKTCHB
-CTCTGY-----
| Original motif Consensus sequence: BBTCTKTKTCHB | Reverse complement motif Consensus sequence: VHGARAYAGAVV |
 |
 |
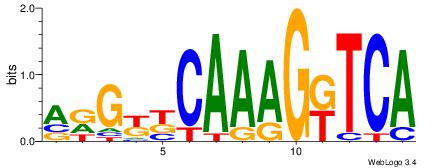
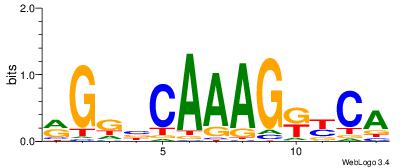
| Dataset #: | 3 |
| Motif ID: | 50 |
| Motif name: | HNF4A |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.0481824 |
Alignment:
RGGBCAAAGKYCA
---KCAGAG----
| Original motif Consensus sequence: RGGBCAAAGKYCA | Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 53 |
| Motif name: | MIZF |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.049747 |
Alignment:
BAACGTCCGC
----CTCTGY
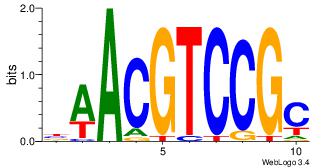
| Original motif Consensus sequence: BAACGTCCGC | Reverse complement motif Consensus sequence: GCGGACGTTV |
 |
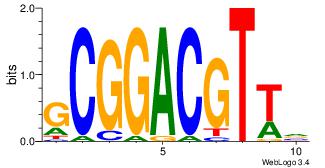 |
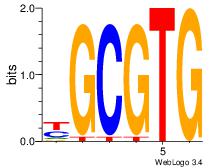
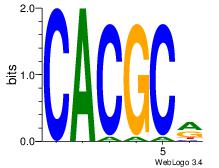
| Dataset #: 3 | Motif ID: 42 | Motif name: ArntAhr |
| Original motif Consensus sequence: YGCGTG | Reverse complement motif Consensus sequence: CACGCM |
 |
 |
Best Matches for Top Significant Motif ID 42 (Highest to Lowest)
| Dataset #: | 2 |
| Motif ID: | 27 |
| Motif name: | Motif 27 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0388571 |
Alignment:
TGTGTGTGTGTKTG
------YGCGTG--
| Original motif Consensus sequence: CAYACACACACACA | Reverse complement motif Consensus sequence: TGTGTGTGTGTKTG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 79 |
| Motif name: | CACACACACACA |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0420782 |
Alignment:
TGTGTGTGTGTG
YGCGTG------
| Original motif Consensus sequence: CACACACACACA | Reverse complement motif Consensus sequence: TGTGTGTGTGTG |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 25 |
| Motif name: | Motif 25 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0430749 |
Alignment:
CACACAVRCACACA
--------CACGCM
| Original motif Consensus sequence: CACACAVRCACACA | Reverse complement motif Consensus sequence: TGTGTGKVTGTGTG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 101 |
| Motif name: | AAAAAAAAAAA |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.043218 |
Alignment:
HVCATGCATGCABD
--CACGCM------
| Original motif Consensus sequence: HVCATGCATGCABD | Reverse complement motif Consensus sequence: DVTGCATGCATGBH |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 95 |
| Motif name: | CACACACACACA |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0440329 |
Alignment:
TGTGTGTGTGTG
----YGCGTG--
| Original motif Consensus sequence: CACACACACACA | Reverse complement motif Consensus sequence: TGTGTGTGTGTG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 96 |
| Motif name: | yrTGTGCACAyr |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0477579 |
Alignment:
BVTGTGCACABV
YGCGTG------
| Original motif Consensus sequence: BVTGTGCACABV | Reverse complement motif Consensus sequence: BBTGTGCACAVV |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 89 |
| Motif name: | ccCCmCACCcc |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0479115 |
Alignment:
HBCCMCACCHH
-----CACGCM
| Original motif Consensus sequence: HBCCMCACCHH | Reverse complement motif Consensus sequence: DDGGTGRGGBD |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 65 |
| Motif name: | ssCGrGCGss |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0487655 |
Alignment:
VBCGCMCGSV
----CACGCM
| Original motif Consensus sequence: VSCGRGCGBV | Reverse complement motif Consensus sequence: VBCGCMCGSV |
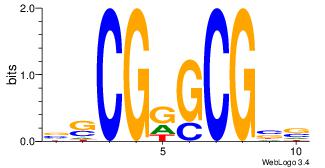 |
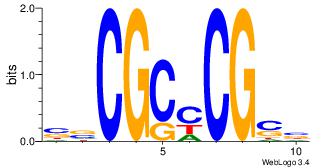 |
| Dataset #: | 1 |
| Motif ID: | 2 |
| Motif name: | Motif 2 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0505006 |
Alignment:
CACACACA
CACGCM--
| Original motif Consensus sequence: CACACACA | Reverse complement motif Consensus sequence: TGTGTGTG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 67 |
| Motif name: | ssGMsGRGCGss |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0538134 |
Alignment:
BBCGCCCSGCVV
CACGCM------
| Original motif Consensus sequence: VVGCSGGGCGBB | Reverse complement motif Consensus sequence: BBCGCCCSGCVV |
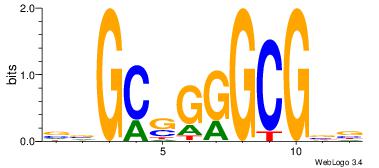 |
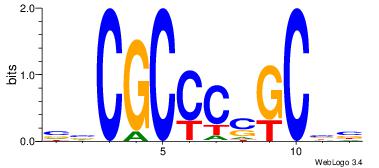 |
| Dataset #: 1 | Motif ID: 13 | Motif name: Motif 13 |
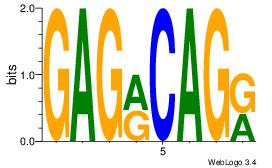
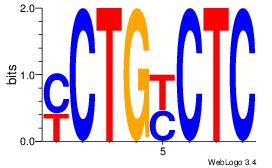
| Original motif Consensus sequence: GAGRCAGR | Reverse complement motif Consensus sequence: MCTGKCTC |
 |
 |
Best Matches for Top Significant Motif ID 13 (Highest to Lowest)
| Dataset #: | 4 |
| Motif ID: | 92 |
| Motif name: | tyTCTkTkTCyy |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0131318 |
Alignment:
VHGARAYAGAVV
--GAGRCAGR--
| Original motif Consensus sequence: BBTCTKTKTCHB | Reverse complement motif Consensus sequence: VHGARAYAGAVV |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 73 |
| Motif name: | ssCGsCTCss |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0334555 |
Alignment:
BBGAGSCGVV
--GAGRCAGR
| Original motif Consensus sequence: VVCGSCTCBB | Reverse complement motif Consensus sequence: BBGAGSCGVV |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 48 |
| Motif name: | EWSR1-FLI1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.0397014 |
Alignment:
GGAAGGAAGGAAGGAAGG
------GAGRCAGR----
| Original motif Consensus sequence: GGAAGGAAGGAAGGAAGG | Reverse complement motif Consensus sequence: CCTTCCTTCCTTCCTTCC |
 |
 |
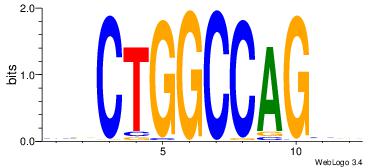
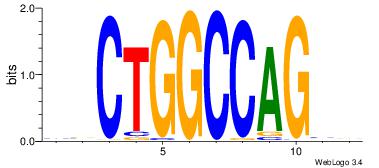
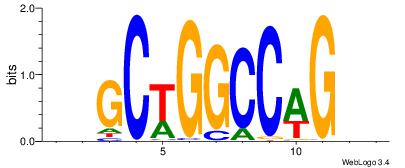
| Dataset #: | 4 |
| Motif ID: | 99 |
| Motif name: | sgCTGGCCAGcs |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0475948 |
Alignment:
VVCTGGCCAGBB
-MCTGKCTC---
| Original motif Consensus sequence: BBCTGGCCAGVV | Reverse complement motif Consensus sequence: VVCTGGCCAGBB |
 |
 |
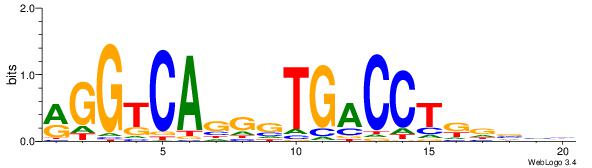
| Dataset #: | 3 |
| Motif ID: | 35 |
| Motif name: | REST |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.049302 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
---MCTGKCTC----------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC | Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
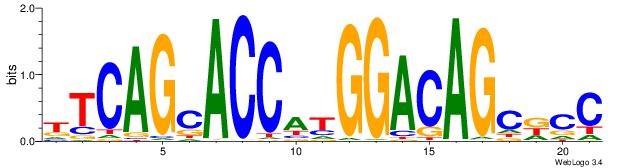 |
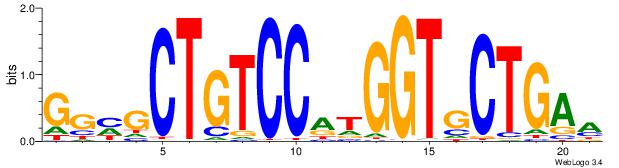 |
| Dataset #: | 2 |
| Motif ID: | 23 |
| Motif name: | Motif 23 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.0551203 |
Alignment:
GGGGMGGG
GAGRCAGR
| Original motif Consensus sequence: CCCRCCCC | Reverse complement motif Consensus sequence: GGGGMGGG |
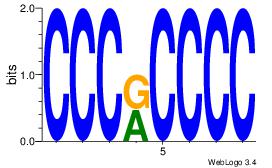 |
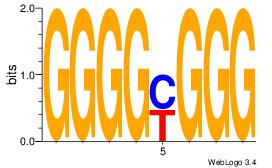 |
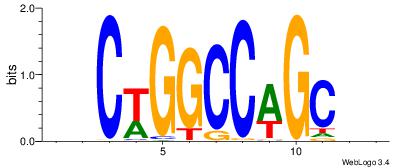
| Dataset #: | 4 |
| Motif ID: | 80 |
| Motif name: | ssCwGGCCwGCss |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.055328 |
Alignment:
BBCWGGCCAGCBB
-MCTGKCTC----
| Original motif Consensus sequence: BBCWGGCCAGCBB | Reverse complement motif Consensus sequence: BBGCTGGCCWGBB |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 93 |
| Motif name: | rrAGGGGGArr |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.0555608 |
Alignment:
VVAGGGGGAVV
---GAGRCAGR
| Original motif Consensus sequence: VVAGGGGGAVV | Reverse complement motif Consensus sequence: VVTCCCCCTVV |
 |
 |
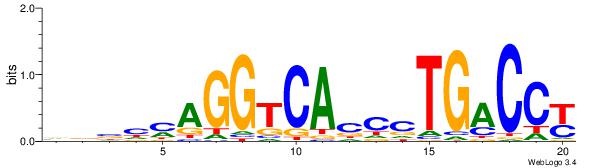
| Dataset #: | 3 |
| Motif ID: | 45 |
| Motif name: | ESR1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 13 |
| Number of overlap: | 8 |
| Similarity score: | 0.055859 |
Alignment:
VDBHMAGGTCACCCTGACCY
------------MCTGKCTC
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY | Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 36 |
| Motif name: | PPARGRXRA |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0596402 |
Alignment:
TGRCCTKTGHCCKAB
---MCTGKCTC----
| Original motif Consensus sequence: BTRGGDCARAGGKCA | Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
 |
 |
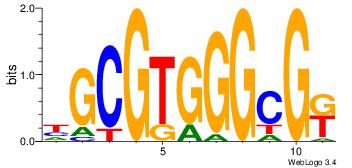
| Dataset #: 1 | Motif ID: 14 | Motif name: Motif 14 |
| Original motif Consensus sequence: GCRCACRC | Reverse complement motif Consensus sequence: GKGTGKGC |
 |
 |
Best Matches for Top Significant Motif ID 14 (Highest to Lowest)
| Dataset #: | 2 |
| Motif ID: | 27 |
| Motif name: | Motif 27 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0307099 |
Alignment:
CAYACACACACACA
---GCRCACRC---
| Original motif Consensus sequence: CAYACACACACACA | Reverse complement motif Consensus sequence: TGTGTGTGTGTKTG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 79 |
| Motif name: | CACACACACACA |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0357914 |
Alignment:
CACACACACACA
-GCRCACRC---
| Original motif Consensus sequence: CACACACACACA | Reverse complement motif Consensus sequence: TGTGTGTGTGTG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 95 |
| Motif name: | CACACACACACA |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0366948 |
Alignment:
CACACACACACA
-GCRCACRC---
| Original motif Consensus sequence: CACACACACACA | Reverse complement motif Consensus sequence: TGTGTGTGTGTG |
 |
 |
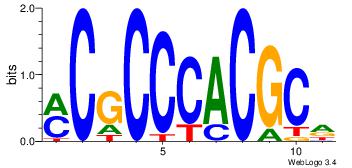
| Dataset #: | 3 |
| Motif ID: | 52 |
| Motif name: | Klf4 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.03983 |
Alignment:
DGGGYGKGGC
-GKGTGKGC-
| Original motif Consensus sequence: DGGGYGKGGC | Reverse complement motif Consensus sequence: GCCYCMCCCD |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 25 |
| Motif name: | Motif 25 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0418776 |
Alignment:
CACACAVRCACACA
-----GCRCACRC-
| Original motif Consensus sequence: CACACAVRCACACA | Reverse complement motif Consensus sequence: TGTGTGKVTGTGTG |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 44 |
| Motif name: | Egr1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0429433 |
Alignment:
HGCGTGGGCGK
-GKGTGKGC--
| Original motif Consensus sequence: HGCGTGGGCGK | Reverse complement motif Consensus sequence: YCGCCCACGCH |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 89 |
| Motif name: | ccCCmCACCcc |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0505252 |
Alignment:
DDGGTGRGGBD
-GKGTGKGC--
| Original motif Consensus sequence: HBCCMCACCHH | Reverse complement motif Consensus sequence: DDGGTGRGGBD |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 96 |
| Motif name: | yrTGTGCACAyr |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0537384 |
Alignment:
BVTGTGCACABV
---GCRCACRC-
| Original motif Consensus sequence: BVTGTGCACABV | Reverse complement motif Consensus sequence: BBTGTGCACAVV |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 65 |
| Motif name: | ssCGrGCGss |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.067702 |
Alignment:
VBCGCMCGSV
-GCRCACRC-
| Original motif Consensus sequence: VSCGRGCGBV | Reverse complement motif Consensus sequence: VBCGCMCGSV |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 91 |
| Motif name: | ssCGCCsCCrCCCss |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0685867 |
Alignment:
SSCGCCSCCGCCCSS
---GCRCACRC----
| Original motif Consensus sequence: SSCGCCSCCGCCCSS | Reverse complement motif Consensus sequence: SSGGGCGGSGGCGSS |
 |
 |