Top 10 Significant Motifs - Global Matching (Highest to Lowest)
| Dataset #: 5 | Motif ID: 84 | Motif name: TFW3 |
| Original motif Consensus sequence: GCACTG | Reverse complement motif Consensus sequence: CAGTGC |
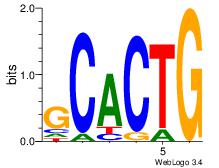 |
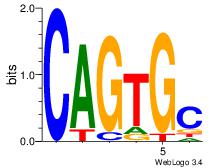 |
Best Matches for Top Significant Motif ID 84 (Highest to Lowest)
| Dataset #: | 4 |
| Motif ID: | 67 |
| Motif name: | TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
GGAGCACTGTGACACCACA
----------GCACTG---
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC | Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
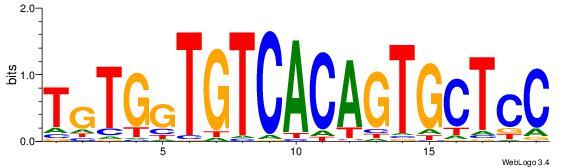 |
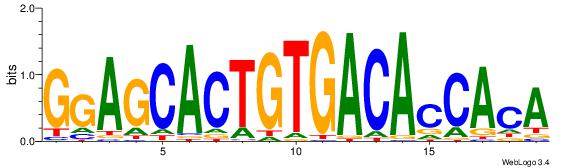 |
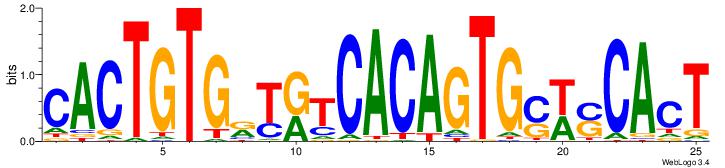
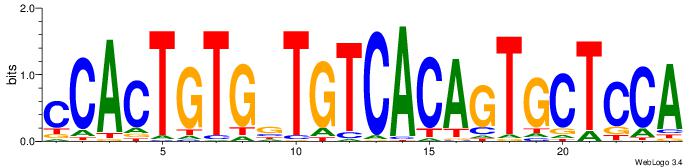
| Dataset #: | 4 |
| Motif ID: | 72 |
| Motif name: | CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.000794289 |
Alignment:
CACTGTGRTRTCACAGTGSWSCACT
------CAGTGC-------------
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT | Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
 |
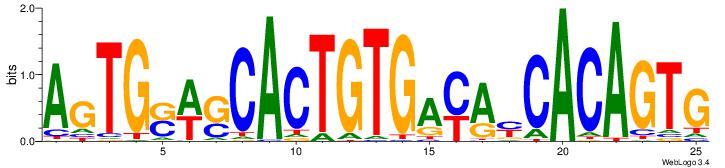 |
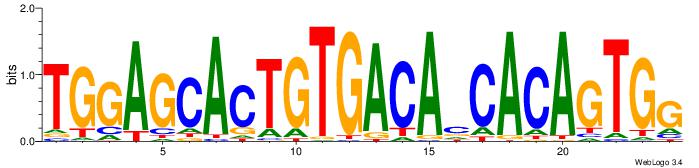
| Dataset #: | 4 |
| Motif ID: | 73 |
| Motif name: | TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.00482436 |
Alignment:
TGGAGCACTGTGACAVCACAGTGG
----GCACTG--------------
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG | Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 76 |
| Motif name: | CAcCACAGTGGAGCAct |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0201914 |
Alignment:
AGTGCTCCACTGTGDTG
-----GCACTG------
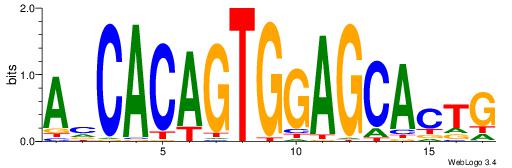
| Original motif Consensus sequence: CAHCACAGTGGAGCACT | Reverse complement motif Consensus sequence: AGTGCTCCACTGTGDTG |
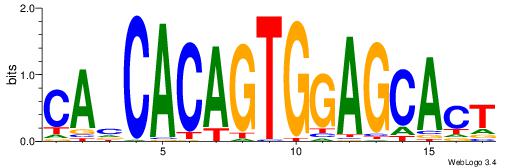 |
 |
| Dataset #: | 4 |
| Motif ID: | 70 |
| Motif name: | CagTGCTCCACTGTGgT |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.0205497 |
Alignment:
ABCACAGTGGAGCACTG
----CAGTGC-------
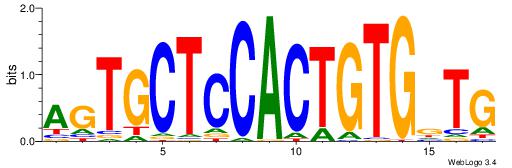
| Original motif Consensus sequence: CAGTGCTCCACTGTGBT | Reverse complement motif Consensus sequence: ABCACAGTGGAGCACTG |
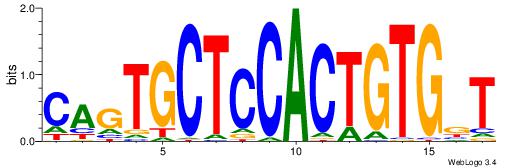 |
 |
| Dataset #: | 4 |
| Motif ID: | 77 |
| Motif name: | AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 13 |
| Number of overlap: | 6 |
| Similarity score: | 0.0290883 |
Alignment:
ACTGTGACACCACAGTGGAGCACT
------------CAGTGC------
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT | Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
 |
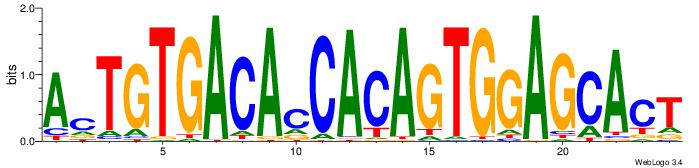 |
| Dataset #: | 3 |
| Motif ID: | 57 |
| Motif name: | REST |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 15 |
| Number of overlap: | 6 |
| Similarity score: | 0.039883 |
Alignment:
TTCAGCACCATGGACAGCKCC
-CAGTGC--------------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC | Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
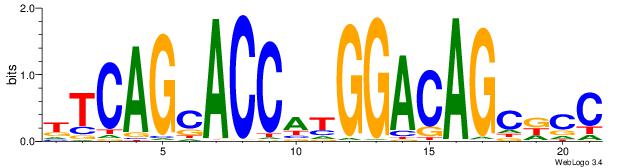 |
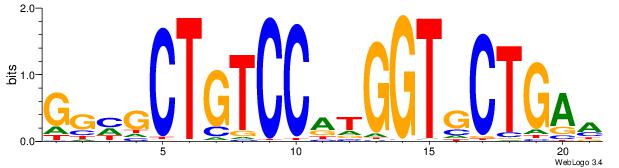 |
| Dataset #: | 3 |
| Motif ID: | 65 |
| Motif name: | Tcfcp2l1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.0447224 |
Alignment:
CCAGYYHVADCCRG
-------CAGTGC-
| Original motif Consensus sequence: CCAGYYHVADCCRG | Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
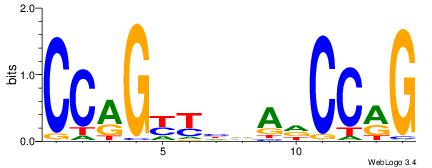 |
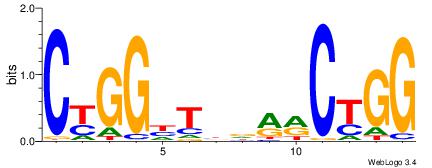 |
| Dataset #: | 1 |
| Motif ID: | 5 |
| Motif name: | Motif 5 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.0459818 |
Alignment:
GAKAGKTGGCTCWG
GCACTG--------
| Original motif Consensus sequence: CWGAGCCAYCTYTC | Reverse complement motif Consensus sequence: GAKAGKTGGCTCWG |
 |
 |
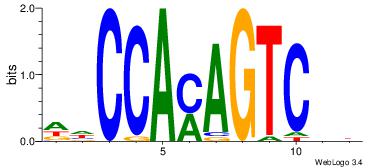
| Dataset #: | 4 |
| Motif ID: | 75 |
| Motif name: | wwCCAmAGTCmt |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.0587933 |
Alignment:
VDGACTRTGGDD
-----GCACTG-
| Original motif Consensus sequence: DDCCAMAGTCHB | Reverse complement motif Consensus sequence: VDGACTRTGGDD |
 |
 |
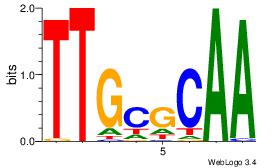
| Dataset #: 5 | Motif ID: 85 | Motif name: TFW1 |
| Original motif Consensus sequence: TTGCGCAA | Reverse complement motif Consensus sequence: TTGCGCAA |
 |
 |
Best Matches for Top Significant Motif ID 85 (Highest to Lowest)
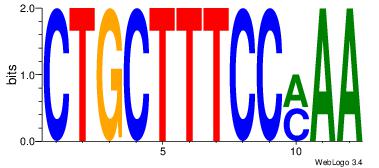
| Dataset #: | 1 |
| Motif ID: | 21 |
| Motif name: | Motif 21 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.0195312 |
Alignment:
TTYGGAAAGCAG
----TTGCGCAA
| Original motif Consensus sequence: CTGCTTTCCMAA | Reverse complement motif Consensus sequence: TTYGGAAAGCAG |
 |
 |
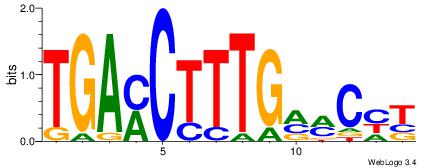
| Dataset #: | 3 |
| Motif ID: | 52 |
| Motif name: | NR2F1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.0276442 |
Alignment:
AKGYYCAAAGRTCA
TTGCGCAA------
| Original motif Consensus sequence: TGAMCTTTGMMCYT | Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
 |
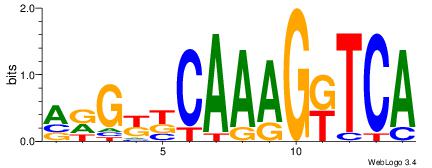 |
| Dataset #: | 3 |
| Motif ID: | 64 |
| Motif name: | PPARGRXRA |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.0277567 |
Alignment:
BTRGGDCARAGGKCA
------TTGCGCAA-
| Original motif Consensus sequence: BTRGGDCARAGGKCA | Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 28 |
| Motif name: | Motif 28 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0328125 |
Alignment:
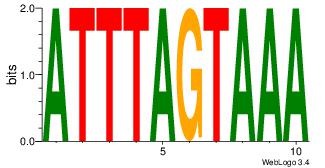
TTTACTAAAT
-TTGCGCAA-
| Original motif Consensus sequence: ATTTAGTAAA | Reverse complement motif Consensus sequence: TTTACTAAAT |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 76 |
| Motif name: | CAcCACAGTGGAGCAct |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0336914 |
Alignment:
CAHCACAGTGGAGCACT
-TTGCGCAA--------
| Original motif Consensus sequence: CAHCACAGTGGAGCACT | Reverse complement motif Consensus sequence: AGTGCTCCACTGTGDTG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 73 |
| Motif name: | TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 17 |
| Number of overlap: | 8 |
| Similarity score: | 0.0338471 |
Alignment:
TGGAGCACTGTGACAVCACAGTGG
TTGCGCAA----------------
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG | Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 77 |
| Motif name: | AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.034375 |
Alignment:
AGTGCTCCACTGTGGTGTCACAGT
---------------TTGCGCAA-
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT | Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 70 |
| Motif name: | CagTGCTCCACTGTGgT |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0354167 |
Alignment:
CAGTGCTCCACTGTGBT
-------TTGCGCAA--
| Original motif Consensus sequence: CAGTGCTCCACTGTGBT | Reverse complement motif Consensus sequence: ABCACAGTGGAGCACTG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 72 |
| Motif name: | CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0407242 |
Alignment:
CACTGTGRTRTCACAGTGSWSCACT
---------------TTGCGCAA--
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT | Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 41 |
| Motif name: | Motif 41 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0421875 |
Alignment:
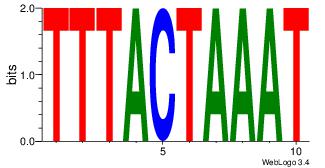
TTTCCTCCA
-TTGCGCAA
| Original motif Consensus sequence: TGGAGGAAA | Reverse complement motif Consensus sequence: TTTCCTCCA |
 |
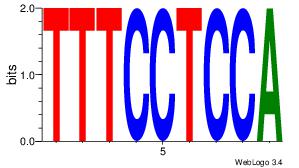 |
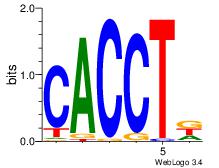
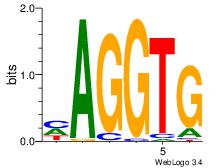
| Dataset #: 3 | Motif ID: 55 | Motif name: ZEB1 |
| Original motif Consensus sequence: CACCTD | Reverse complement motif Consensus sequence: HAGGTG |
 |
 |
Best Matches for Top Significant Motif ID 55 (Highest to Lowest)
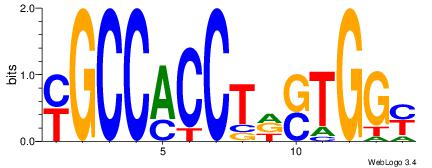
| Dataset #: | 2 |
| Motif ID: | 47 |
| Motif name: | Motif 47 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
YGCCACCTDSTGGY
---CACCTD-----
| Original motif Consensus sequence: YGCCACCTDSTGGY | Reverse complement motif Consensus sequence: KCCASDAGGTGGCK |
 |
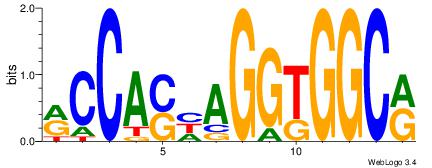 |
| Dataset #: | 1 |
| Motif ID: | 5 |
| Motif name: | Motif 5 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.00902841 |
Alignment:
CWGAGCCAYCTYTC
------CACCTD--
| Original motif Consensus sequence: CWGAGCCAYCTYTC | Reverse complement motif Consensus sequence: GAKAGKTGGCTCWG |
 |
 |
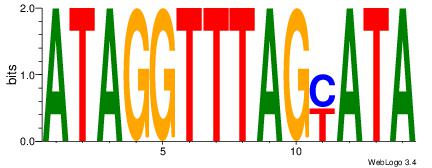
| Dataset #: | 1 |
| Motif ID: | 46 |
| Motif name: | Motif 46 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.0285279 |
Alignment:
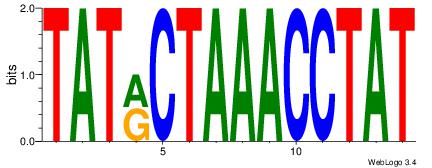
TATKCTAAACCTAT
-CACCTD-------
| Original motif Consensus sequence: ATAGGTTTAGYATA | Reverse complement motif Consensus sequence: TATKCTAAACCTAT |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 20 |
| Motif name: | Motif 20 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0346255 |
Alignment:
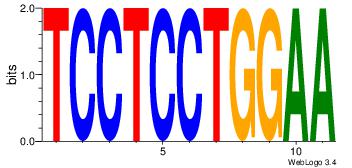
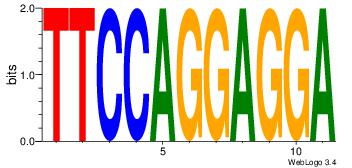
TCCTCCTGGAA
--CACCTD---
| Original motif Consensus sequence: TCCTCCTGGAA | Reverse complement motif Consensus sequence: TTCCAGGAGGA |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 12 |
| Motif name: | Motif 12 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.0376742 |
Alignment:
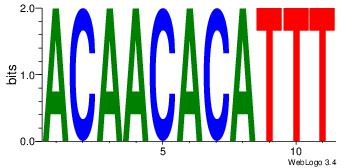
ACAACACATTT
----CACCTD-
| Original motif Consensus sequence: ACAACACATTT | Reverse complement motif Consensus sequence: AAATGTGTTGT |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 34 |
| Motif name: | Motif 34 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.0397068 |
Alignment:
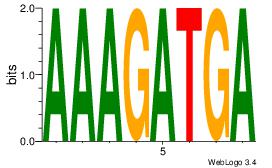
AAAGATGA
-HAGGTG-
| Original motif Consensus sequence: AAAGATGA | Reverse complement motif Consensus sequence: TCATCTTT |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 22 |
| Motif name: | Motif 22 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.0397068 |
Alignment:
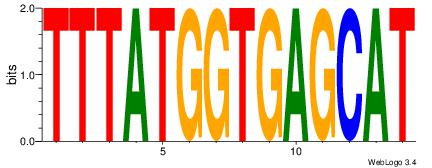
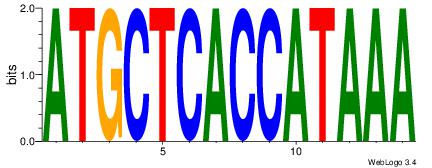
TTTATGGTGAGCAT
-----HAGGTG---
| Original motif Consensus sequence: TTTATGGTGAGCAT | Reverse complement motif Consensus sequence: ATGCTCACCATAAA |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 17 |
| Motif name: | Motif 17 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0458043 |
Alignment:
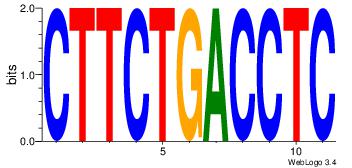
CTTCTGACCTC
-----CACCTD
| Original motif Consensus sequence: CTTCTGACCTC | Reverse complement motif Consensus sequence: GAGGTCAGAAG |
 |
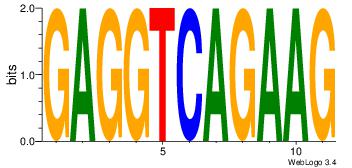 |
| Dataset #: | 5 |
| Motif ID: | 92 |
| Motif name: | TFM11 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0461561 |
Alignment:
CAMACACACACAYDCACACA
--------CACCTD------
| Original motif Consensus sequence: CAMACACACACAYDCACACA | Reverse complement motif Consensus sequence: TGTGTGDKTGTGTGTGTRTG |
 |
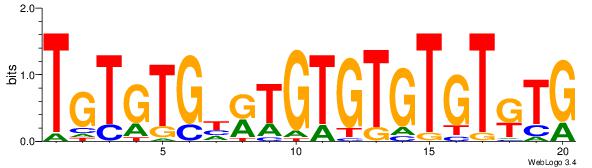 |
| Dataset #: | 5 |
| Motif ID: | 89 |
| Motif name: | TFM1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0467843 |
Alignment:
CACACACACACWCACACA
------CACCTD------
| Original motif Consensus sequence: CACACACACACWCACACA | Reverse complement motif Consensus sequence: TGTGTGWGTGTGTGTGTG |
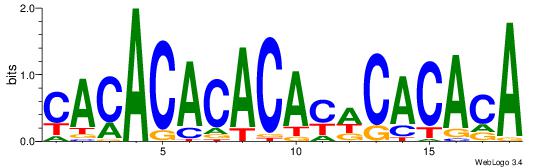 |
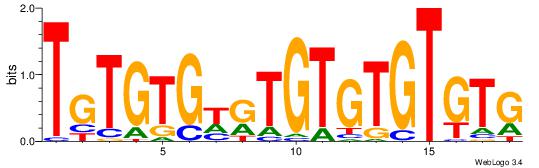 |
| Dataset #: 1 | Motif ID: 3 | Motif name: Motif 3 |
| Original motif Consensus sequence: HCAGRRVACASAG | Reverse complement motif Consensus sequence: CTSTGTBKMCTGH |
 |
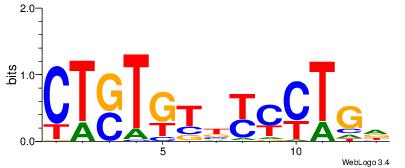 |
Best Matches for Top Significant Motif ID 3 (Highest to Lowest)
| Dataset #: | 5 |
| Motif ID: | 93 |
| Motif name: | TFM12 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 13 |
| Similarity score: | 0.00926418 |
Alignment:
RAGKGMAGVRRGSRCASAGV
------HCAGRRVACASAG-
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV | Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
 |
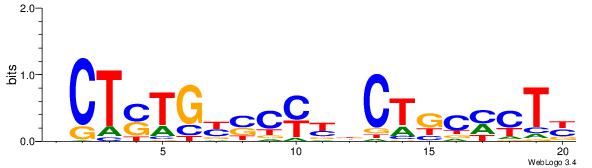 |
| Dataset #: | 3 |
| Motif ID: | 53 |
| Motif name: | ESR1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 13 |
| Similarity score: | 0.0328703 |
Alignment:
VDBHMAGGTCACCCTGACCY
---HCAGRRVACASAG----
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY | Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
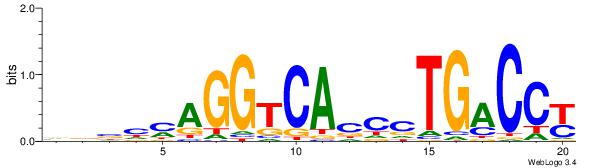 |
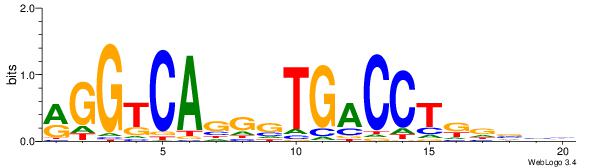 |
| Dataset #: | 4 |
| Motif ID: | 72 |
| Motif name: | CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 13 |
| Similarity score: | 0.0400966 |
Alignment:
AGTGSWSCACTGTGAMAMCACAGTG
----------HCAGRRVACASAG--
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT | Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
 |
 |
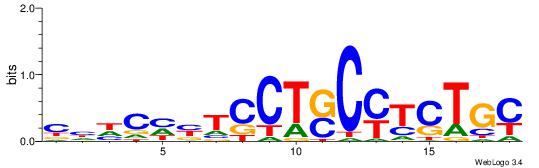
| Dataset #: | 5 |
| Motif ID: | 90 |
| Motif name: | TFM2 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 13 |
| Similarity score: | 0.0435274 |
Alignment:
GCAGAKGSAGGABDGHDG
--HCAGRRVACASAG---
| Original motif Consensus sequence: CHHCHBTCCTSCYTCTGC | Reverse complement motif Consensus sequence: GCAGAKGSAGGABDGHDG |
 |
 |
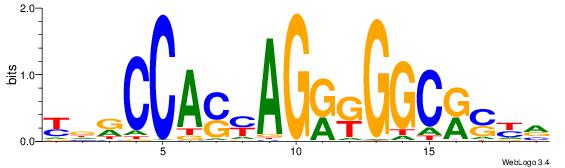
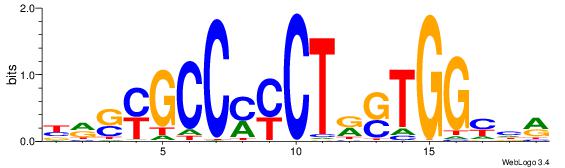
| Dataset #: | 3 |
| Motif ID: | 66 |
| Motif name: | CTCF |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 13 |
| Similarity score: | 0.0448341 |
Alignment:
YDRCCASYAGRKGGCRSYV
------HCAGRRVACASAG
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV | Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 73 |
| Motif name: | TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 13 |
| Similarity score: | 0.045698 |
Alignment:
CCACTGTGVTGTCACAGTGCTCCA
---CTSTGTBKMCTGH--------
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG | Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 64 |
| Motif name: | PPARGRXRA |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0457253 |
Alignment:
BTRGGDCARAGGKCA
HCAGRRVACASAG--
| Original motif Consensus sequence: BTRGGDCARAGGKCA | Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 65 |
| Motif name: | Tcfcp2l1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0477557 |
Alignment:
CKGGDTBDMMCTGG
CTSTGTBKMCTGH-
| Original motif Consensus sequence: CCAGYYHVADCCRG | Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
 |
 |
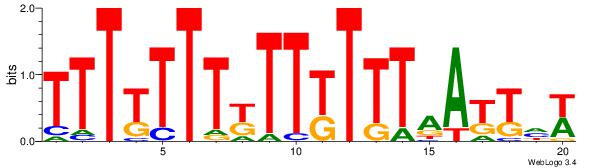
| Dataset #: | 5 |
| Motif ID: | 94 |
| Motif name: | TFM13 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 13 |
| Similarity score: | 0.0478998 |
Alignment:
WHAATTAAARAARAAAAAAA
----HCAGRRVACASAG---
| Original motif Consensus sequence: TTTTTTTKTTKTTTAATTHW | Reverse complement motif Consensus sequence: WHAATTAAARAARAAAAAAA |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 88 |
| Motif name: | TFF11 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.048132 |
Alignment:
HVCDBBDBCAGCAG
-HCAGRRVACASAG
| Original motif Consensus sequence: CTGCTGBDBBDGBD | Reverse complement motif Consensus sequence: HVCDBBDBCAGCAG |
 |
 |
| Dataset #: 2 | Motif ID: 48 | Motif name: Motif 48 |
| Original motif Consensus sequence: GCARRGGSAGS | Reverse complement motif Consensus sequence: SCTSCCKMTGC |
 |
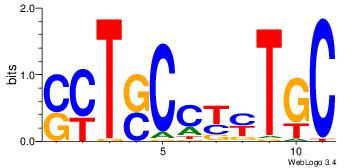 |
Best Matches for Top Significant Motif ID 48 (Highest to Lowest)
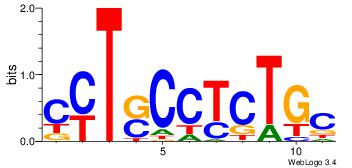
| Dataset #: | 1 |
| Motif ID: | 1 |
| Motif name: | Motif 1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0 |
Alignment:
GCAGAGGSAKG
GCARRGGSAGS
| Original motif Consensus sequence: CYTSCCTCTGC | Reverse complement motif Consensus sequence: GCAGAGGSAKG |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 90 |
| Motif name: | TFM2 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.000952693 |
Alignment:
GCAGAKGSAGGABDGHDG
GCARRGGSAGS-------
| Original motif Consensus sequence: CHHCHBTCCTSCYTCTGC | Reverse complement motif Consensus sequence: GCAGAKGSAGGABDGHDG |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 66 |
| Motif name: | CTCF |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.033658 |
Alignment:
YDRCCASYAGRKGGCRSYV
------GCARRGGSAGS--
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV | Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 88 |
| Motif name: | TFF11 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0410341 |
Alignment:
CTGCTGBDBBDGBD
--SCTSCCKMTGC-
| Original motif Consensus sequence: CTGCTGBDBBDGBD | Reverse complement motif Consensus sequence: HVCDBBDBCAGCAG |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 93 |
| Motif name: | TFM12 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0422459 |
Alignment:
RAGKGMAGVRRGSRCASAGV
----GCARRGGSAGS-----
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV | Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 65 |
| Motif name: | Tcfcp2l1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0533531 |
Alignment:
CCAGYYHVADCCRG
SCTSCCKMTGC---
| Original motif Consensus sequence: CCAGYYHVADCCRG | Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 57 |
| Motif name: | REST |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0578944 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
---SCTSCCKMTGC-------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC | Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 53 |
| Motif name: | ESR1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0620615 |
Alignment:
VDBHMAGGTCACCCTGACCY
--GCARRGGSAGS-------
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY | Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
 |
 |
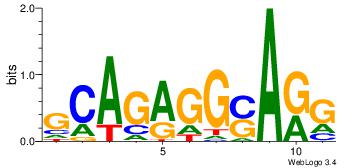
| Dataset #: | 1 |
| Motif ID: | 3 |
| Motif name: | Motif 3 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0627554 |
Alignment:
HCAGRRVACASAG
GCARRGGSAGS--
| Original motif Consensus sequence: HCAGRRVACASAG | Reverse complement motif Consensus sequence: CTSTGTBKMCTGH |
 |
 |
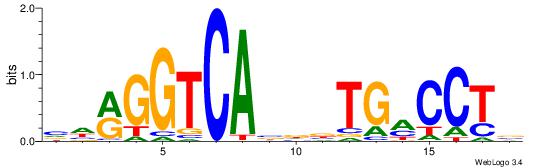
| Dataset #: | 3 |
| Motif ID: | 59 |
| Motif name: | ESR2 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0646979 |
Alignment:
VHRGGTCABDBTGMCCTB
---SCTSCCKMTGC----
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB | Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
 |
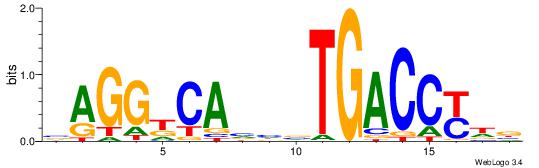 |
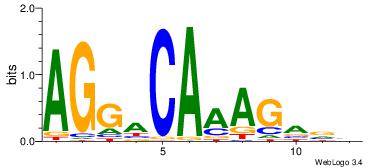
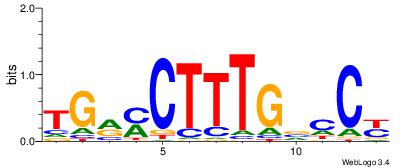
| Dataset #: 5 | Motif ID: 87 | Motif name: TFF1 |
| Original motif Consensus sequence: AGGACAAAGAVV | Reverse complement motif Consensus sequence: VVTCTTTGTCCT |
 |
 |
Best Matches for Top Significant Motif ID 87 (Highest to Lowest)
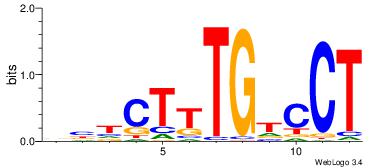
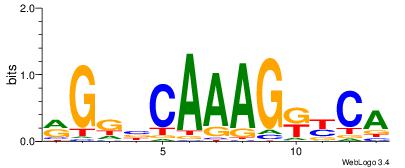
| Dataset #: | 3 |
| Motif ID: | 51 |
| Motif name: | HNF4A |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0111472 |
Alignment:
TGMYCTTTGBCCK
-VVTCTTTGTCCT
| Original motif Consensus sequence: RGGBCAAAGKYCA | Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 64 |
| Motif name: | PPARGRXRA |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0210867 |
Alignment:
BTRGGDCARAGGKCA
--AGGACAAAGAVV-
| Original motif Consensus sequence: BTRGGDCARAGGKCA | Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 59 |
| Motif name: | ESR2 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 0.0351633 |
Alignment:
BAGGYCABHBTGACCKHV
----VVTCTTTGTCCT--
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB | Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 53 |
| Motif name: | ESR1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 12 |
| Similarity score: | 0.039773 |
Alignment:
VDBHMAGGTCACCCTGACCY
VVTCTTTGTCCT--------
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY | Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 52 |
| Motif name: | NR2F1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0472374 |
Alignment:
AKGYYCAAAGRTCA
-AGGACAAAGAVV-
| Original motif Consensus sequence: TGAMCTTTGMMCYT | Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
 |
 |
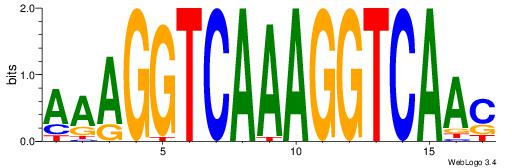
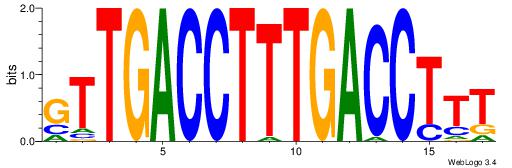
| Dataset #: | 3 |
| Motif ID: | 60 |
| Motif name: | NR1H2RXRA |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0472884 |
Alignment:
GTTGACCTTTGACCTTT
---VVTCTTTGTCCT--
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC | Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 72 |
| Motif name: | CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 12 |
| Similarity score: | 0.0530556 |
Alignment:
CACTGTGRTRTCACAGTGSWSCACT
-------AGGACAAAGAVV------
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT | Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 73 |
| Motif name: | TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 13 |
| Number of overlap: | 12 |
| Similarity score: | 0.0539204 |
Alignment:
TGGAGCACTGTGACAVCACAGTGG
------------AGGACAAAGAVV
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG | Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 57 |
| Motif name: | REST |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 12 |
| Similarity score: | 0.0558379 |
Alignment:
TTCAGCACCATGGACAGCKCC
---AGGACAAAGAVV------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC | Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 77 |
| Motif name: | AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 12 |
| Similarity score: | 0.0609269 |
Alignment:
AGTGCTCCACTGTGGTGTCACAGT
------VVTCTTTGTCCT------
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT | Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
 |
 |
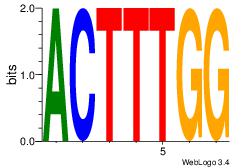
| Dataset #: 1 | Motif ID: 37 | Motif name: Motif 37 |
| Original motif Consensus sequence: ACTTTGG | Reverse complement motif Consensus sequence: CCAAAGT |
 |
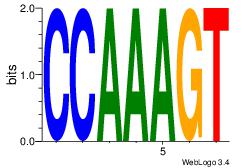 |
Best Matches for Top Significant Motif ID 37 (Highest to Lowest)
| Dataset #: | 4 |
| Motif ID: | 69 |
| Motif name: | atactttggc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 7 |
| Similarity score: | 0 |
Alignment:
ATACTTTGGC
--ACTTTGG-
| Original motif Consensus sequence: ATACTTTGGC | Reverse complement motif Consensus sequence: GCCAAAGTAT |
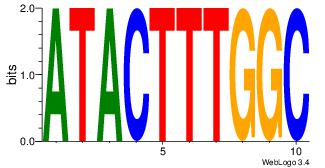 |
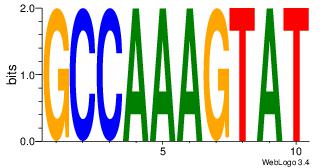 |
| Dataset #: | 4 |
| Motif ID: | 75 |
| Motif name: | wwCCAmAGTCmt |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 7 |
| Similarity score: | 0.0275974 |
Alignment:
DDCCAMAGTCHB
--CCAAAGT---
| Original motif Consensus sequence: DDCCAMAGTCHB | Reverse complement motif Consensus sequence: VDGACTRTGGDD |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 77 |
| Motif name: | AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 10 |
| Number of overlap: | 7 |
| Similarity score: | 0.056841 |
Alignment:
ACTGTGACACCACAGTGGAGCACT
---------CCAAAGT--------
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT | Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
 |
 |
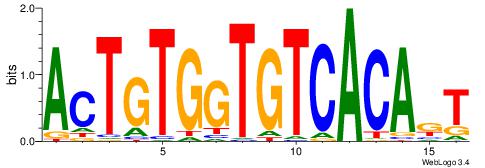
| Dataset #: | 4 |
| Motif ID: | 71 |
| Motif name: | AmTGTGACACCACAGT |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 7 |
| Similarity score: | 0.0597098 |
Alignment:
ACTGTGGTGTCACART
ACTTTGG---------
| Original motif Consensus sequence: AMTGTGACACCACAGT | Reverse complement motif Consensus sequence: ACTGTGGTGTCACART |
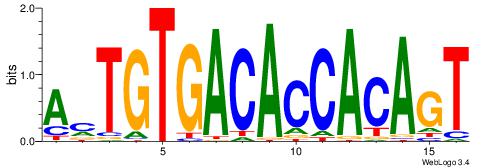 |
 |
| Dataset #: | 4 |
| Motif ID: | 76 |
| Motif name: | CAcCACAGTGGAGCAct |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 7 |
| Similarity score: | 0.061942 |
Alignment:
AGTGCTCCACTGTGDTG
--------ACTTTGG--
| Original motif Consensus sequence: CAHCACAGTGGAGCACT | Reverse complement motif Consensus sequence: AGTGCTCCACTGTGDTG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 70 |
| Motif name: | CagTGCTCCACTGTGgT |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 7 |
| Similarity score: | 0.0625 |
Alignment:
ABCACAGTGGAGCACTG
-CCAAAGT---------
| Original motif Consensus sequence: CAGTGCTCCACTGTGBT | Reverse complement motif Consensus sequence: ABCACAGTGGAGCACTG |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 52 |
| Motif name: | NR2F1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 7 |
| Similarity score: | 0.0686813 |
Alignment:
TGAMCTTTGMMCYT
---ACTTTGG----
| Original motif Consensus sequence: TGAMCTTTGMMCYT | Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 72 |
| Motif name: | CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 7 |
| Similarity score: | 0.0714286 |
Alignment:
CACTGTGRTRTCACAGTGSWSCACT
-ACTTTGG-----------------
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT | Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 73 |
| Motif name: | TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 7 |
| Similarity score: | 0.0714286 |
Alignment:
TGGAGCACTGTGACAVCACAGTGG
---------------CCAAAGT--
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG | Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 60 |
| Motif name: | NR1H2RXRA |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.0728571 |
Alignment:
GTTGACCTTTGACCTTT
-----ACTTTGG-----
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC | Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
 |
 |
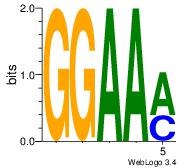
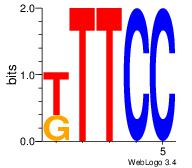
| Dataset #: 2 | Motif ID: 50 | Motif name: Motif 50 |
| Original motif Consensus sequence: GGAAM | Reverse complement motif Consensus sequence: YTTCC |
 |
 |
Best Matches for Top Significant Motif ID 50 (Highest to Lowest)
| Dataset #: | 1 |
| Motif ID: | 41 |
| Motif name: | Motif 41 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 5 |
| Similarity score: | 0.01875 |
Alignment:
TTTCCTCCA
YTTCC----
| Original motif Consensus sequence: TGGAGGAAA | Reverse complement motif Consensus sequence: TTTCCTCCA |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 21 |
| Motif name: | Motif 21 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 5 |
| Similarity score: | 0.01875 |
Alignment:
CTGCTTTCCMAA
---YTTCC----
| Original motif Consensus sequence: CTGCTTTCCMAA | Reverse complement motif Consensus sequence: TTYGGAAAGCAG |
 |
 |
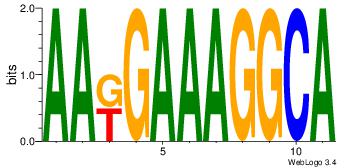
| Dataset #: | 1 |
| Motif ID: | 43 |
| Motif name: | Motif 43 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 5 |
| Similarity score: | 0.04375 |
Alignment:
AAKGAAAGGCA
--GGAAM----
| Original motif Consensus sequence: AAKGAAAGGCA | Reverse complement motif Consensus sequence: TGCCTTTCYTT |
 |
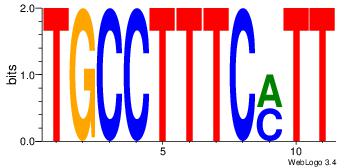 |
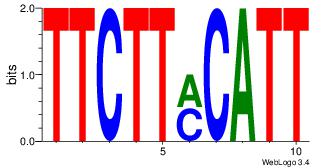
| Dataset #: | 1 |
| Motif ID: | 44 |
| Motif name: | Motif 44 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 5 |
| Similarity score: | 0.05625 |
Alignment:
TTCTTYCATT
--YTTCC---
| Original motif Consensus sequence: AATGKAAGAA | Reverse complement motif Consensus sequence: TTCTTYCATT |
 |
 |
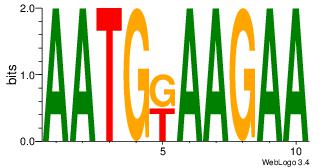
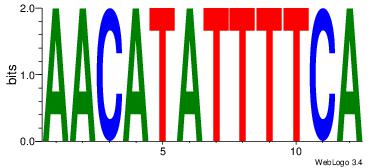
| Dataset #: | 1 |
| Motif ID: | 9 |
| Motif name: | Motif 9 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 5 |
| Similarity score: | 0.06875 |
Alignment:
AACATATTTTCA
YTTCC-------
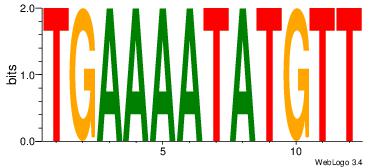
| Original motif Consensus sequence: AACATATTTTCA | Reverse complement motif Consensus sequence: TGAAAATATGTT |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 8 |
| Motif name: | Motif 8 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.06875 |
Alignment:
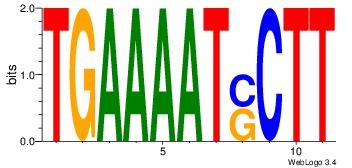
TGAAAATSCTT
-GGAAM-----
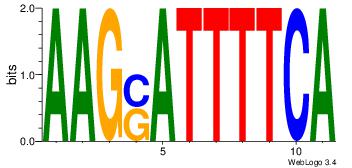
| Original motif Consensus sequence: TGAAAATSCTT | Reverse complement motif Consensus sequence: AAGSATTTTCA |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 28 |
| Motif name: | Motif 28 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.06875 |
Alignment:
TTTACTAAAT
YTTCC-----
| Original motif Consensus sequence: ATTTAGTAAA | Reverse complement motif Consensus sequence: TTTACTAAAT |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 19 |
| Motif name: | Motif 19 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 5 |
| Similarity score: | 0.06875 |
Alignment:
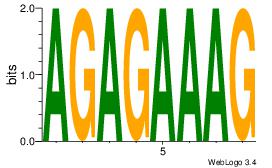
AGAGAAAG
--GGAAM-
| Original motif Consensus sequence: AGAGAAAG | Reverse complement motif Consensus sequence: CTTTCTCT |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 57 |
| Motif name: | REST |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.0747947 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
-----YTTCC-----------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC | Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
 |
 |
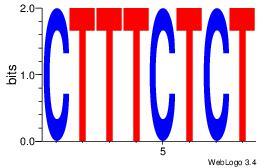
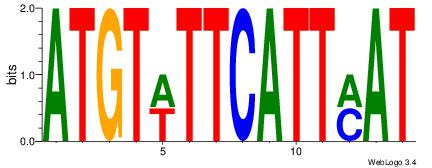
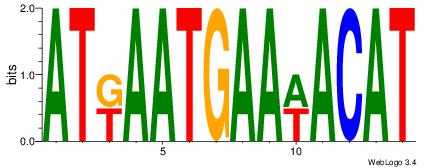
| Dataset #: | 1 |
| Motif ID: | 23 |
| Motif name: | Motif 23 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.075 |
Alignment:
ATGTWTTCATTMAT
-----YTTCC----
| Original motif Consensus sequence: ATGTWTTCATTMAT | Reverse complement motif Consensus sequence: ATYAATGAAWACAT |
 |
 |
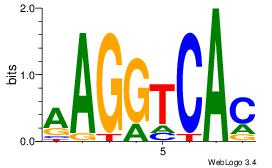
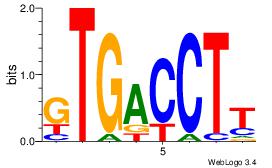
| Dataset #: 3 | Motif ID: 61 | Motif name: NR4A2 |
| Original motif Consensus sequence: AAGGTCAC | Reverse complement motif Consensus sequence: GTGACCTT |
 |
 |
Best Matches for Top Significant Motif ID 61 (Highest to Lowest)
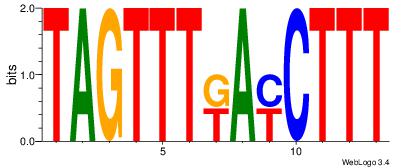
| Dataset #: | 1 |
| Motif ID: | 17 |
| Motif name: | Motif 17 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.0393171 |
Alignment:
CTTCTGACCTC
GTGACCTT---
| Original motif Consensus sequence: CTTCTGACCTC | Reverse complement motif Consensus sequence: GAGGTCAGAAG |
 |
 |
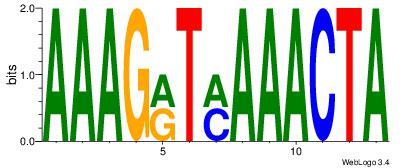
| Dataset #: | 1 |
| Motif ID: | 15 |
| Motif name: | Motif 15 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.040519 |
Alignment:
AAAGRTMAAACTA
-AAGGTCAC----
| Original motif Consensus sequence: AAAGRTMAAACTA | Reverse complement motif Consensus sequence: TAGTTTYAKCTTT |
 |
 |
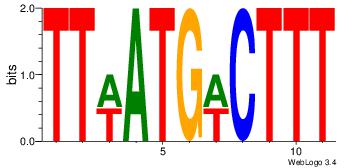
| Dataset #: | 1 |
| Motif ID: | 24 |
| Motif name: | Motif 24 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0539119 |
Alignment:
TTWATGWCTTT
---GTGACCTT
| Original motif Consensus sequence: AAAGWCATWAA | Reverse complement motif Consensus sequence: TTWATGWCTTT |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 72 |
| Motif name: | CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.0557243 |
Alignment:
CACTGTGRTRTCACAGTGSWSCACT
------AAGGTCAC-----------
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT | Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
 |
 |
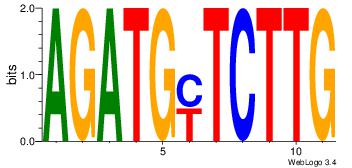
| Dataset #: | 1 |
| Motif ID: | 35 |
| Motif name: | Motif 35 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0566591 |
Alignment:
AGATGYTCTTG
-GTGACCTT--
| Original motif Consensus sequence: AGATGYTCTTG | Reverse complement motif Consensus sequence: CAAGAKCATCT |
 |
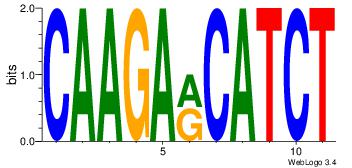 |
| Dataset #: | 5 |
| Motif ID: | 93 |
| Motif name: | TFM12 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 10 |
| Number of overlap: | 8 |
| Similarity score: | 0.0584971 |
Alignment:
RAGKGMAGVRRGSRCASAGV
---------AAGGTCAC---
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV | Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 73 |
| Motif name: | TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.060729 |
Alignment:
CCACTGTGVTGTCACAGTGCTCCA
-------AAGGTCAC---------
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG | Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 71 |
| Motif name: | AmTGTGACACCACAGT |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0645146 |
Alignment:
ACTGTGGTGTCACART
-----AAGGTCAC---
| Original motif Consensus sequence: AMTGTGACACCACAGT | Reverse complement motif Consensus sequence: ACTGTGGTGTCACART |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 22 |
| Motif name: | Motif 22 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.0664462 |
Alignment:
ATGCTCACCATAAA
------AAGGTCAC
| Original motif Consensus sequence: TTTATGGTGAGCAT | Reverse complement motif Consensus sequence: ATGCTCACCATAAA |
 |
 |
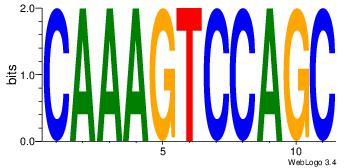
| Dataset #: | 1 |
| Motif ID: | 18 |
| Motif name: | Motif 18 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0674765 |
Alignment:
GCTGGACTTTG
-GTGACCTT--
| Original motif Consensus sequence: CAAAGTCCAGC | Reverse complement motif Consensus sequence: GCTGGACTTTG |
 |
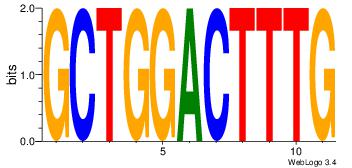 |
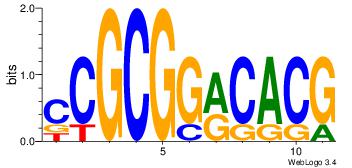
| Dataset #: 4 | Motif ID: 79 | Motif name: cCGCGGrCACG |
| Original motif Consensus sequence: CCGCGGRCACG | Reverse complement motif Consensus sequence: CGTGKCCGCGG |
 |
 |
Best Matches for Top Significant Motif ID 79 (Highest to Lowest)
| Dataset #: | 5 |
| Motif ID: | 88 |
| Motif name: | TFF11 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0478408 |
Alignment:
HVCDBBDBCAGCAG
---CGTGKCCGCGG
| Original motif Consensus sequence: CTGCTGBDBBDGBD | Reverse complement motif Consensus sequence: HVCDBBDBCAGCAG |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 57 |
| Motif name: | REST |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0484441 |
Alignment:
TTCAGCACCATGGACAGCKCC
-------CCGCGGRCACG---
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC | Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 53 |
| Motif name: | ESR1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0486581 |
Alignment:
MGGTCAGGGTGACCTRDBHV
-------CGTGKCCGCGG--
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY | Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 66 |
| Motif name: | CTCF |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 11 |
| Similarity score: | 0.0588663 |
Alignment:
YDRCCASYAGRKGGCRSYV
-------CCGCGGRCACG-
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV | Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 3 |
| Motif name: | Motif 3 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0597806 |
Alignment:
HCAGRRVACASAG
-CCGCGGRCACG-
| Original motif Consensus sequence: HCAGRRVACASAG | Reverse complement motif Consensus sequence: CTSTGTBKMCTGH |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 2 |
| Motif name: | Motif 2 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0624268 |
Alignment:
GTGTGTGTGTGTGT
-CGTGKCCGCGG--
| Original motif Consensus sequence: ACACACACACACAC | Reverse complement motif Consensus sequence: GTGTGTGTGTGTGT |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 93 |
| Motif name: | TFM12 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0625015 |
Alignment:
RAGKGMAGVRRGSRCASAGV
-----CCGCGGRCACG----
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV | Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 89 |
| Motif name: | TFM1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0628056 |
Alignment:
CACACACACACWCACACA
-----CCGCGGRCACG--
| Original motif Consensus sequence: CACACACACACWCACACA | Reverse complement motif Consensus sequence: TGTGTGWGTGTGTGTGTG |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 64 |
| Motif name: | PPARGRXRA |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0629476 |
Alignment:
TGRCCTKTGHCCKAB
---CGTGKCCGCGG-
| Original motif Consensus sequence: BTRGGDCARAGGKCA | Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 92 |
| Motif name: | TFM11 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0634175 |
Alignment:
CAMACACACACAYDCACACA
-------CCGCGGRCACG--
| Original motif Consensus sequence: CAMACACACACAYDCACACA | Reverse complement motif Consensus sequence: TGTGTGDKTGTGTGTGTRTG |
 |
 |