Top 10 Significant Motifs - Global Matching (Highest to Lowest)
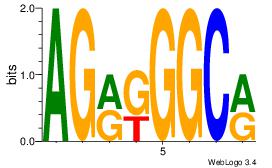
| Dataset #: 2 | Motif ID: 46 | Motif name: Motif 46 |
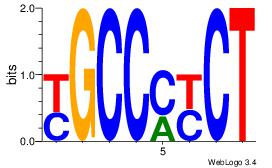
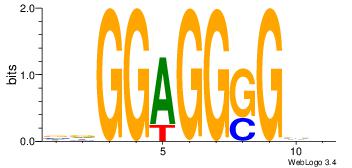
| Original motif Consensus sequence: AGRKGGCR | Reverse complement motif Consensus sequence: KGCCYKCT |
 |
 |
Best Matches for Top Significant Motif ID 46 (Highest to Lowest)
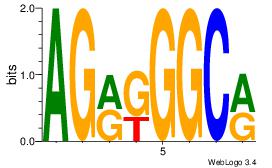
| Dataset #: | 1 |
| Motif ID: | 1 |
| Motif name: | Motif 1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
AGRKGGCR
AGRKGGCR
| Original motif Consensus sequence: AGRKGGCR | Reverse complement motif Consensus sequence: KGCCYKCT |
 |
 |
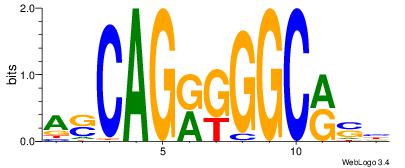
| Dataset #: | 4 |
| Motif ID: | 147 |
| Motif name: | asCAGrkGGCrsy |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.00651548 |
Alignment:
ASCAGRGGGCRSB
---AGRKGGCR--
| Original motif Consensus sequence: ASCAGRGGGCRSB | Reverse complement motif Consensus sequence: BSKGCCCMCTGST |
 |
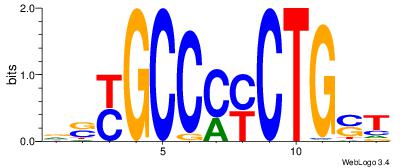 |
| Dataset #: | 4 |
| Motif ID: | 165 |
| Motif name: | wgGCCAshAGrGGGCrsy |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.0102443 |
Alignment:
KBKGCCCKCTHGTGGCHH
--KGCCYKCT--------
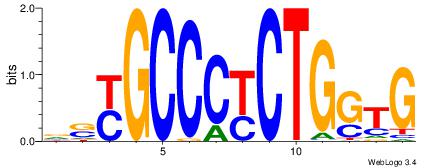
| Original motif Consensus sequence: HDGCCACHAGRGGGCRBY | Reverse complement motif Consensus sequence: KBKGCCCKCTHGTGGCHH |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 166 |
| Motif name: | CasCAGrGGGCrsy |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0104954 |
Alignment:
CACCAGRGGGCRSB
----AGRKGGCR--
| Original motif Consensus sequence: CACCAGRGGGCRSB | Reverse complement motif Consensus sequence: BSKGCCCKCTGGTG |
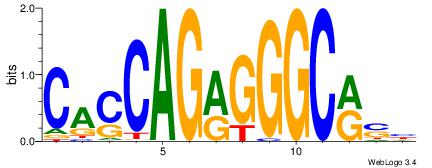 |
 |
| Dataset #: | 4 |
| Motif ID: | 163 |
| Motif name: | gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0165822 |
Alignment:
KBKGCCCTCTYCTGGCCHV
--KGCCYKCT---------
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY | Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
 |
 |
| Dataset #: 1 | Motif ID: 8 | Motif name: Motif 8 |
| Original motif Consensus sequence: AAATAH | Reverse complement motif Consensus sequence: DTATTT |
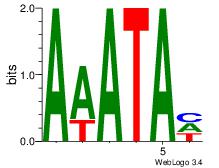 |
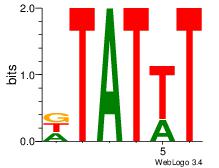 |
Best Matches for Top Significant Motif ID 8 (Highest to Lowest)
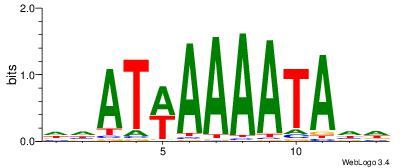
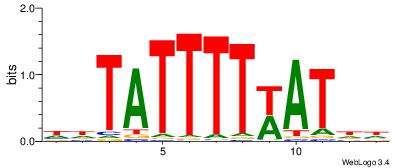
| Dataset #: | 4 |
| Motif ID: | 150 |
| Motif name: | waATwAAAATAww |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.00224204 |
Alignment:
DHATWAAAATAHD
------AAATAH-
| Original motif Consensus sequence: DHATWAAAATAHD | Reverse complement motif Consensus sequence: DHTATTTTWATHD |
 |
 |
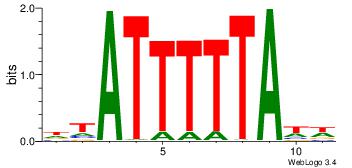
| Dataset #: | 4 |
| Motif ID: | 157 |
| Motif name: | wtATTTTTAww |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.00556446 |
Alignment:
WWTAAAAATAD
-----AAATAH
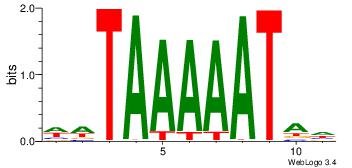
| Original motif Consensus sequence: DTATTTTTAWW | Reverse complement motif Consensus sequence: WWTAAAAATAD |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 68 |
| Motif name: | Motif 68 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0128826 |
Alignment:
WAAWAAAWAWWWAA
----AAATAH----
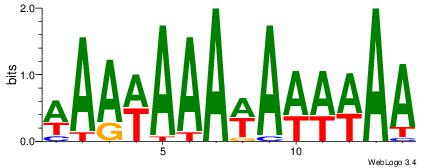
| Original motif Consensus sequence: WAAWAAAWAWWWAA | Reverse complement motif Consensus sequence: TTWWWTWTTTWTTW |
 |
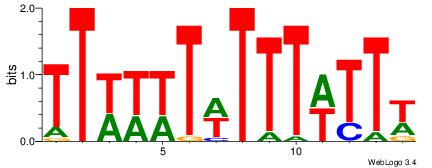 |
| Dataset #: | 2 |
| Motif ID: | 52 |
| Motif name: | Motif 52 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0167304 |
Alignment:
AAATAAAW
AAATAH--
| Original motif Consensus sequence: AAATAAAW | Reverse complement motif Consensus sequence: WTTTATTT |
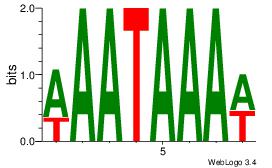 |
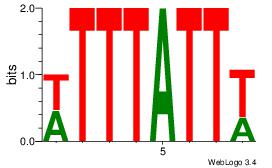 |
| Dataset #: | 3 |
| Motif ID: | 118 |
| Motif name: | SRF |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0210491 |
Alignment:
GCCCATATATGG
----AAATAH--
| Original motif Consensus sequence: GCCCATATATGG | Reverse complement motif Consensus sequence: CCATATATGGGC |
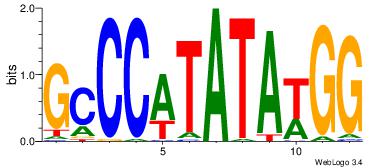 |
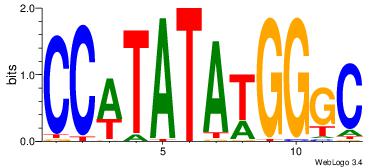 |
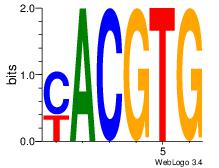
| Dataset #: 1 | Motif ID: 20 | Motif name: Motif 20 |
| Original motif Consensus sequence: CACGTR | Reverse complement motif Consensus sequence: MACGTG |
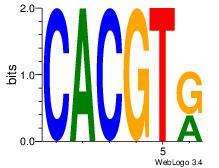 |
 |
Best Matches for Top Significant Motif ID 20 (Highest to Lowest)
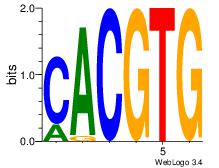
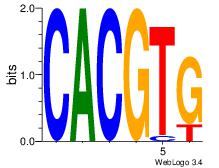
| Dataset #: | 3 |
| Motif ID: | 71 |
| Motif name: | Arnt |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
CACGTG
CACGTR
| Original motif Consensus sequence: CACGTG | Reverse complement motif Consensus sequence: CACGTG |
 |
 |
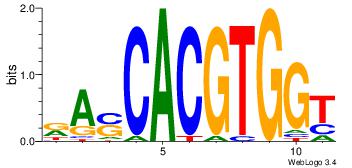
| Dataset #: | 3 |
| Motif ID: | 95 |
| Motif name: | MYCMAX |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.0102183 |
Alignment:
RASCACGTGGT
---MACGTG--
| Original motif Consensus sequence: RASCACGTGGT | Reverse complement motif Consensus sequence: ACCACGTGSTM |
 |
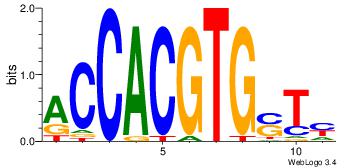 |
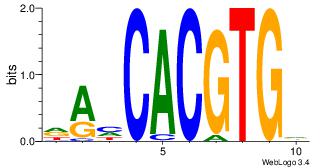
| Dataset #: | 3 |
| Motif ID: | 91 |
| Motif name: | MAX |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.011152 |
Alignment:
DAHCACGTGD
---CACGTR-
| Original motif Consensus sequence: DAHCACGTGD | Reverse complement motif Consensus sequence: BCACGTGDTD |
 |
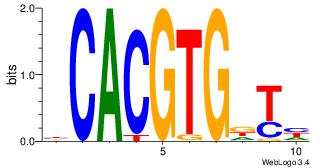 |
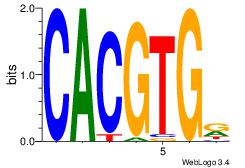
| Dataset #: | 3 |
| Motif ID: | 126 |
| Motif name: | USF1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.0131944 |
Alignment:
MCACGTG
-CACGTR
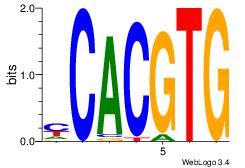
| Original motif Consensus sequence: CACGTGR | Reverse complement motif Consensus sequence: MCACGTG |
 |
 |
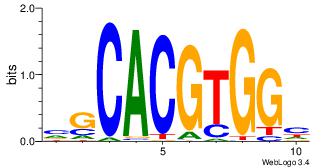
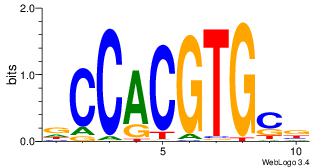
| Dataset #: | 3 |
| Motif ID: | 94 |
| Motif name: | Myc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0253396 |
Alignment:
DCCACGTGCV
--CACGTR--
| Original motif Consensus sequence: VGCACGTGGH | Reverse complement motif Consensus sequence: DCCACGTGCV |
 |
 |
| Dataset #: 1 | Motif ID: 1 | Motif name: Motif 1 |
| Original motif Consensus sequence: AGRKGGCR | Reverse complement motif Consensus sequence: KGCCYKCT |
 |
 |
Best Matches for Top Significant Motif ID 1 (Highest to Lowest)
| Dataset #: | 2 |
| Motif ID: | 46 |
| Motif name: | Motif 46 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
AGRKGGCR
AGRKGGCR
| Original motif Consensus sequence: AGRKGGCR | Reverse complement motif Consensus sequence: KGCCYKCT |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 147 |
| Motif name: | asCAGrkGGCrsy |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.00628523 |
Alignment:
ASCAGRGGGCRSB
---AGRKGGCR--
| Original motif Consensus sequence: ASCAGRGGGCRSB | Reverse complement motif Consensus sequence: BSKGCCCMCTGST |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 165 |
| Motif name: | wgGCCAshAGrGGGCrsy |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0102152 |
Alignment:
HDGCCACHAGRGGGCRBY
--------AGRKGGCR--
| Original motif Consensus sequence: HDGCCACHAGRGGGCRBY | Reverse complement motif Consensus sequence: KBKGCCCKCTHGTGGCHH |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 166 |
| Motif name: | CasCAGrGGGCrsy |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0106761 |
Alignment:
BSKGCCCKCTGGTG
--KGCCYKCT----
| Original motif Consensus sequence: CACCAGRGGGCRSB | Reverse complement motif Consensus sequence: BSKGCCCKCTGGTG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 163 |
| Motif name: | gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.016658 |
Alignment:
KBKGCCCTCTYCTGGCCHV
--KGCCYKCT---------
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY | Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
 |
 |
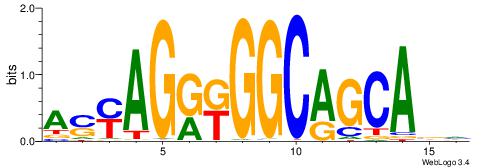
| Dataset #: | 4 |
| Motif ID: | 164 |
| Motif name: | asyAGrkGGCRGCAga |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0201849 |
Alignment:
ASYAGRKGGCAGCABH
---AGRKGGCR-----
| Original motif Consensus sequence: ASYAGRKGGCAGCABH | Reverse complement motif Consensus sequence: HBTGCTGCCYMCTKST |
 |
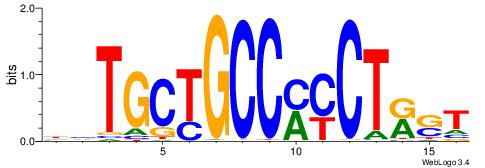 |
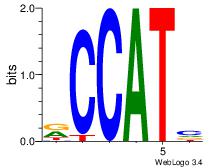
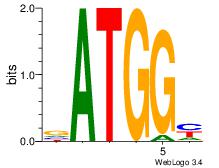
| Dataset #: 3 | Motif ID: 127 | Motif name: YY1 |
| Original motif Consensus sequence: RCCATB | Reverse complement motif Consensus sequence: BATGGM |
 |
 |
Best Matches for Top Significant Motif ID 127 (Highest to Lowest)
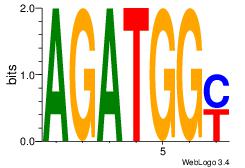
| Dataset #: | 2 |
| Motif ID: | 61 |
| Motif name: | Motif 61 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.00122549 |
Alignment:
AGATGGY
BATGGM-
| Original motif Consensus sequence: AGATGGY | Reverse complement motif Consensus sequence: KCCATCT |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 12 |
| Motif name: | Motif 12 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.00857843 |
Alignment:
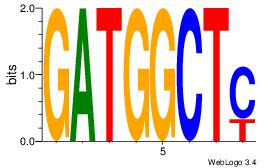
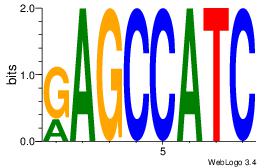
GATGGCTC
--BATGGM
| Original motif Consensus sequence: GATGGCTC | Reverse complement motif Consensus sequence: GAGCCATC |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 145 |
| Motif name: | grCCACyAGAkG |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.022036 |
Alignment:
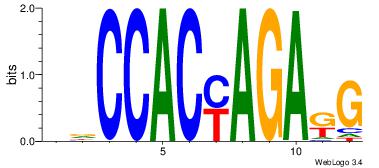
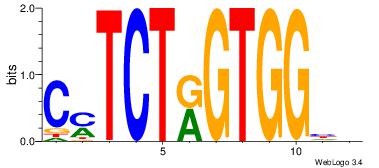
CYTCTKGTGGHH
-----BATGGM-
| Original motif Consensus sequence: DDCCACYAGAKG | Reverse complement motif Consensus sequence: CYTCTKGTGGHH |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 138 |
| Motif name: | grCCACyAGAkG |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.0223679 |
Alignment:
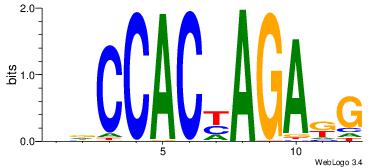
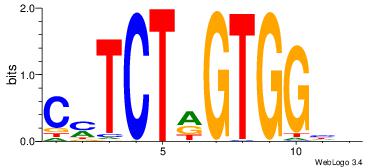
DDCCACYAGAKG
-RCCATB-----
| Original motif Consensus sequence: DDCCACYAGAKG | Reverse complement motif Consensus sequence: CYTCTMGTGGHH |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 156 |
| Motif name: | rgyGCCMyCTksTGGccd |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0275501 |
Alignment:
DDGCCASYAGMGGGCKVM
--RCCATB----------
| Original motif Consensus sequence: RVYGCCCYCTKSTGGCHD | Reverse complement motif Consensus sequence: DDGCCASYAGMGGGCKVM |
 |
 |
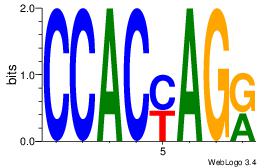
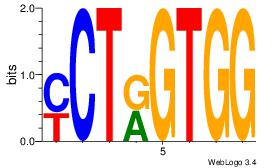
| Dataset #: 2 | Motif ID: 48 | Motif name: Motif 48 |
| Original motif Consensus sequence: CCACYAGR | Reverse complement motif Consensus sequence: MCTKGTGG |
 |
 |
Best Matches for Top Significant Motif ID 48 (Highest to Lowest)
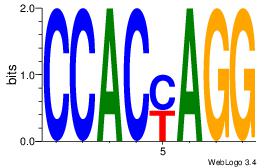
| Dataset #: | 1 |
| Motif ID: | 21 |
| Motif name: | Motif 21 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.00903109 |
Alignment:
CCACYAGG
CCACYAGR
| Original motif Consensus sequence: CCACYAGG | Reverse complement motif Consensus sequence: CCTKGTGG |
 |
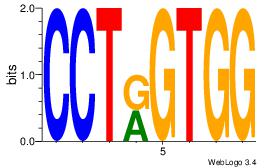 |
| Dataset #: | 4 |
| Motif ID: | 145 |
| Motif name: | grCCACyAGAkG |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0155354 |
Alignment:
DDCCACYAGAKG
--CCACYAGR--
| Original motif Consensus sequence: DDCCACYAGAKG | Reverse complement motif Consensus sequence: CYTCTKGTGGHH |
 |
 |
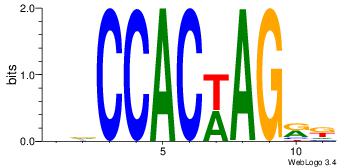
| Dataset #: | 4 |
| Motif ID: | 158 |
| Motif name: | grCCACwAGrk |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0192652 |
Alignment:
DDCCACWAGRK
--CCACYAGR-
| Original motif Consensus sequence: DDCCACWAGRK | Reverse complement motif Consensus sequence: YMCTWGTGGHH |
 |
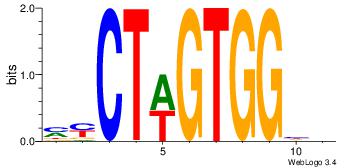 |
| Dataset #: | 4 |
| Motif ID: | 138 |
| Motif name: | grCCACyAGAkG |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0244381 |
Alignment:
DDCCACYAGAKG
--CCACYAGR--
| Original motif Consensus sequence: DDCCACYAGAKG | Reverse complement motif Consensus sequence: CYTCTMGTGGHH |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 74 |
| Motif name: | CTCF |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.0284404 |
Alignment:
BMSMGCCYMCTKSTGGMHM
--------MCTKGTGG---
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV | Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
 |
 |
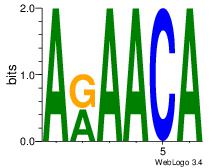
| Dataset #: 2 | Motif ID: 65 | Motif name: Motif 65 |
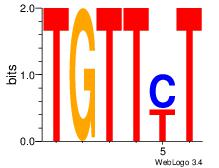
| Original motif Consensus sequence: ARAACA | Reverse complement motif Consensus sequence: TGTTMT |
 |
 |
Best Matches for Top Significant Motif ID 65 (Highest to Lowest)
| Dataset #: | 4 |
| Motif ID: | 141 |
| Motif name: | raCAAAACam |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.00122368 |
Alignment:
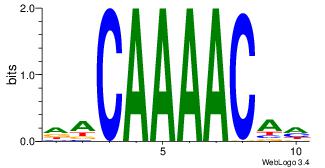
DACAAAACAH
---ARAACA-
| Original motif Consensus sequence: DACAAAACAH | Reverse complement motif Consensus sequence: HTGTTTTGTD |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 44 |
| Motif name: | Motif 44 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0101983 |
Alignment:
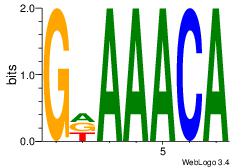
GDAAACA
-ARAACA
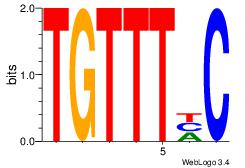
| Original motif Consensus sequence: GDAAACA | Reverse complement motif Consensus sequence: TGTTTDC |
 |
 |
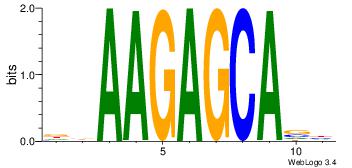
| Dataset #: | 4 |
| Motif ID: | 159 |
| Motif name: | kkAAGAGCAsy |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0232212 |
Alignment:
HVTGCTCTTBH
--TGTTMT---
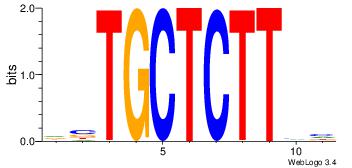
| Original motif Consensus sequence: DBAAGAGCAVH | Reverse complement motif Consensus sequence: HVTGCTCTTBH |
 |
 |
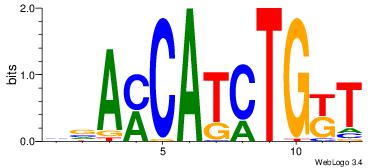
| Dataset #: | 3 |
| Motif ID: | 121 |
| Motif name: | TAL1TCF3 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0330056 |
Alignment:
HVAMCATCTGKT
ARAACA------
| Original motif Consensus sequence: HVAMCATCTGKT | Reverse complement motif Consensus sequence: ARCAGATGRTVD |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 2 |
| Motif name: | Motif 2 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0333069 |
Alignment:
AAAAHAAA
ARAACA--
| Original motif Consensus sequence: AAAAHAAA | Reverse complement motif Consensus sequence: TTTDTTTT |
 |
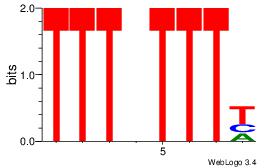 |
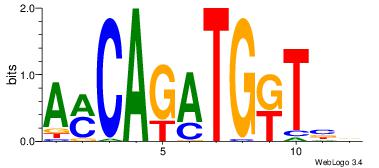
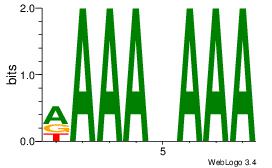
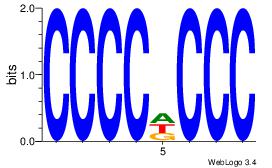
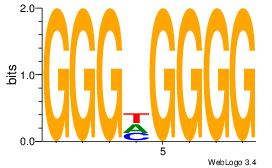
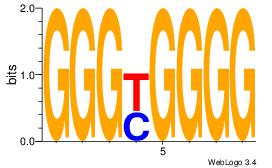
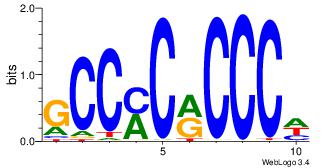
| Dataset #: 1 | Motif ID: 5 | Motif name: Motif 5 |
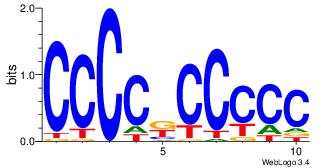
| Original motif Consensus sequence: CCCCDCCC | Reverse complement motif Consensus sequence: GGGDGGGG |
 |
 |
Best Matches for Top Significant Motif ID 5 (Highest to Lowest)
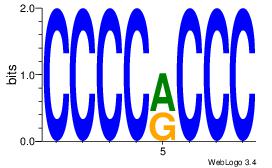
| Dataset #: | 2 |
| Motif ID: | 59 |
| Motif name: | Motif 59 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.00363191 |
Alignment:
CCCCRCCC
CCCCDCCC
| Original motif Consensus sequence: CCCCRCCC | Reverse complement motif Consensus sequence: GGGKGGGG |
 |
 |
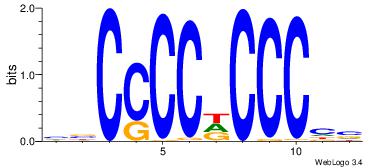
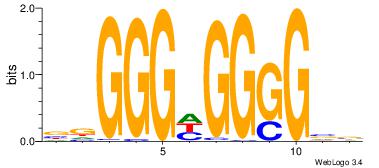
| Dataset #: | 4 |
| Motif ID: | 155 |
| Motif name: | csCSCCdCCCcs |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.00464015 |
Alignment:
VDGGGDGGGGBV
--GGGDGGGG--
| Original motif Consensus sequence: VBCCCCDCCCHV | Reverse complement motif Consensus sequence: VDGGGDGGGGBV |
 |
 |
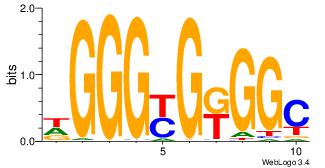
| Dataset #: | 3 |
| Motif ID: | 89 |
| Motif name: | Klf4 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0352587 |
Alignment:
GCCYCMCCCD
-CCCCDCCC-
| Original motif Consensus sequence: DGGGYGKGGC | Reverse complement motif Consensus sequence: GCCYCMCCCD |
 |
 |
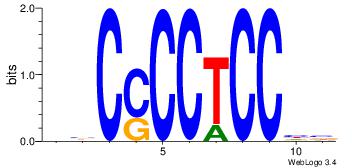
| Dataset #: | 4 |
| Motif ID: | 154 |
| Motif name: | csCsCCTCCcc |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0361349 |
Alignment:
BDGGAGGGGBV
-GGGDGGGG--
| Original motif Consensus sequence: VBCCCCTCCHB | Reverse complement motif Consensus sequence: BDGGAGGGGBV |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 116 |
| Motif name: | SP1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0368066 |
Alignment:
CCCCKCCCCC
CCCCDCCC--
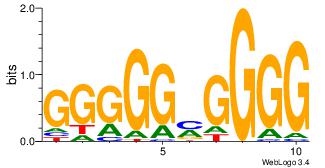
| Original motif Consensus sequence: CCCCKCCCCC | Reverse complement motif Consensus sequence: GGGGGYGGGG |
 |
 |
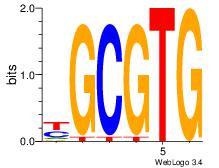
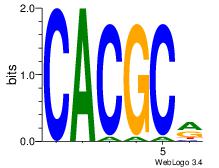
| Dataset #: 3 | Motif ID: 72 | Motif name: ArntAhr |
| Original motif Consensus sequence: YGCGTG | Reverse complement motif Consensus sequence: CACGCM |
 |
 |
Best Matches for Top Significant Motif ID 72 (Highest to Lowest)
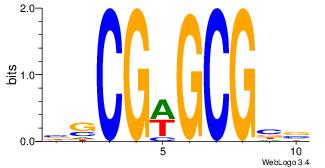
| Dataset #: | 4 |
| Motif ID: | 134 |
| Motif name: | ssCGwGCGss |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0128947 |
Alignment:
BSCGWGCGBV
YGCGTG----
| Original motif Consensus sequence: BSCGWGCGBV | Reverse complement motif Consensus sequence: VBCGCWCGSB |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 20 |
| Motif name: | Motif 20 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0206428 |
Alignment:
MACGTG
YGCGTG
| Original motif Consensus sequence: CACGTR | Reverse complement motif Consensus sequence: MACGTG |
 |
 |
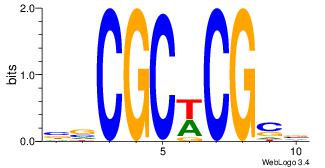
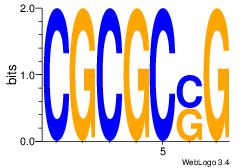
| Dataset #: | 1 |
| Motif ID: | 42 |
| Motif name: | Motif 42 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.0259081 |
Alignment:
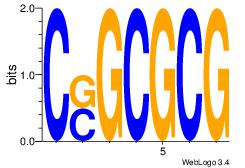
CSGCGCG
YGCGTG-
| Original motif Consensus sequence: CGCGCSG | Reverse complement motif Consensus sequence: CSGCGCG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 152 |
| Motif name: | yrCATGCAyr |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0279287 |
Alignment:
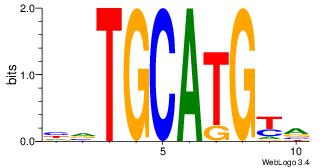
BRCATGCABD
--CACGCM--
| Original motif Consensus sequence: BRCATGCABD | Reverse complement motif Consensus sequence: HVTGCATGKV |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 57 |
| Motif name: | Motif 57 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.0315171 |
Alignment:
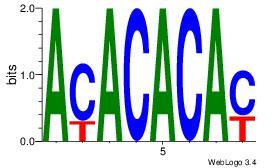
ACACACAY
-CACGCM-
| Original motif Consensus sequence: ACACACAY | Reverse complement motif Consensus sequence: KTGTGTGT |
 |
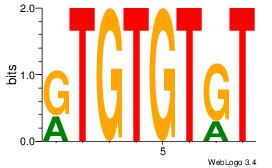 |
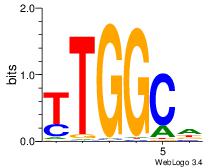
| Dataset #: 3 | Motif ID: 101 | Motif name: NFIC |
| Original motif Consensus sequence: TTGGCD | Reverse complement motif Consensus sequence: DGCCAA |
 |
 |
Best Matches for Top Significant Motif ID 101 (Highest to Lowest)
| Dataset #: | 4 |
| Motif ID: | 165 |
| Motif name: | wgGCCAshAGrGGGCrsy |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 12 |
| Number of overlap: | 6 |
| Similarity score: | 0.0215379 |
Alignment:
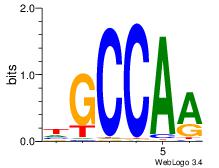
HDGCCACHAGRGGGCRBY
-DGCCAA-----------
| Original motif Consensus sequence: HDGCCACHAGRGGGCRBY | Reverse complement motif Consensus sequence: KBKGCCCKCTHGTGGCHH |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 156 |
| Motif name: | rgyGCCMyCTksTGGccd |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 12 |
| Number of overlap: | 6 |
| Similarity score: | 0.026631 |
Alignment:
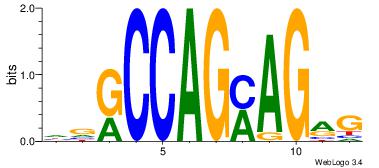
DDGCCASYAGMGGGCKVM
-DGCCAA-----------
| Original motif Consensus sequence: RVYGCCCYCTKSTGGCHD | Reverse complement motif Consensus sequence: DDGCCASYAGMGGGCKVM |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 151 |
| Motif name: | agrCCAGmAGrg |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.0273632 |
Alignment:
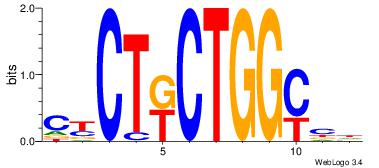
CKCTRCTGGCVH
-----TTGGCD-
| Original motif Consensus sequence: HVGCCAGMAGRG | Reverse complement motif Consensus sequence: CKCTRCTGGCVH |
 |
 |
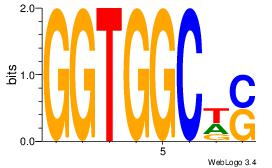
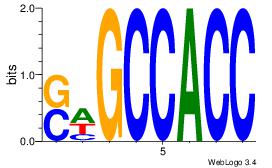
| Dataset #: | 1 |
| Motif ID: | 9 |
| Motif name: | Motif 9 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.0280507 |
Alignment:
SWGCCACC
-DGCCAA-
| Original motif Consensus sequence: GGTGGCWS | Reverse complement motif Consensus sequence: SWGCCACC |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 163 |
| Motif name: | gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 12 |
| Number of overlap: | 6 |
| Similarity score: | 0.0280858 |
Alignment:
KBKGCCCTCTYCTGGCCHV
-----------TTGGCD--
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY | Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
 |
 |