Best Matches for Each Motif (Highest to Lowest)
| Dataset #: 1 |
Motif ID: 1 |
Motif name: Motif 1 |
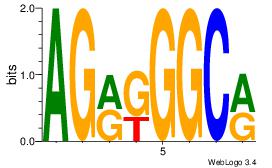
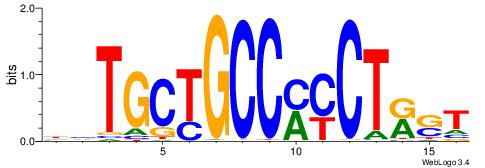
| Original motif Consensus sequence: AGRKGGCR |
Reverse complement motif Consensus sequence: KGCCYKCT |
|
|
 |
 |
Best Matches for Motif ID 1 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
46 |
| Motif name: |
Motif 46 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0 |
Alignment:
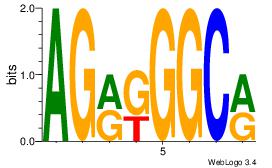
AGRKGGCR
AGRKGGCR
| Original motif Consensus sequence: AGRKGGCR |
Reverse complement motif Consensus sequence: KGCCYKCT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
147 |
| Motif name: |
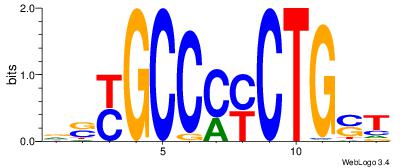
asCAGrkGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.00628523 |
Alignment:
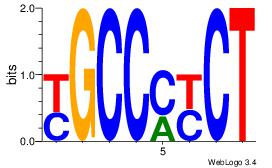
ASCAGRGGGCRSB
---AGRKGGCR--
| Original motif Consensus sequence: ASCAGRGGGCRSB |
Reverse complement motif Consensus sequence: BSKGCCCMCTGST |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
165 |
| Motif name: |
wgGCCAshAGrGGGCrsy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0102152 |
Alignment:
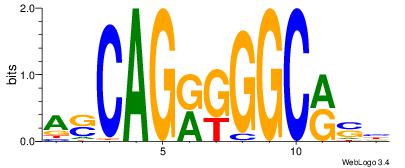
KBKGCCCKCTHGTGGCHH
--KGCCYKCT--------
| Original motif Consensus sequence: HDGCCACHAGRGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCKCTHGTGGCHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
166 |
| Motif name: |
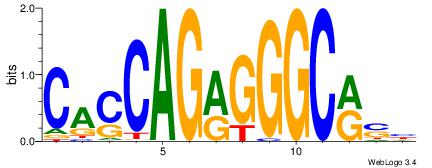
CasCAGrGGGCrsy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0106761 |
Alignment:
BSKGCCCKCTGGTG
--KGCCYKCT----
| Original motif Consensus sequence: CACCAGRGGGCRSB |
Reverse complement motif Consensus sequence: BSKGCCCKCTGGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
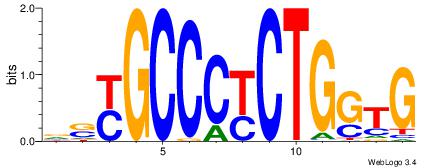
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.016658 |
Alignment:
KBKGCCCTCTYCTGGCCHV
--KGCCYKCT---------
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
164 |
| Motif name: |
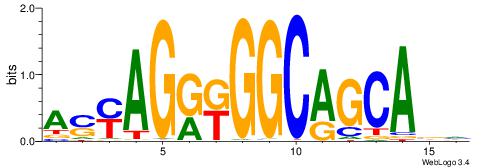
asyAGrkGGCRGCAga |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0201849 |
Alignment:
ASYAGRKGGCAGCABH
---AGRKGGCR-----
| Original motif Consensus sequence: ASYAGRKGGCAGCABH |
Reverse complement motif Consensus sequence: HBTGCTGCCYMCTKST |
|
|
 |
 |
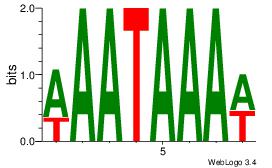
| Dataset #: 1 |
Motif ID: 2 |
Motif name: Motif 2 |
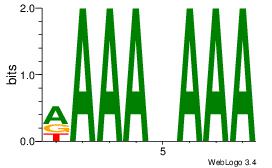
| Original motif Consensus sequence: AAAAHAAA |
Reverse complement motif Consensus sequence: TTTDTTTT |
|
|
 |
 |
Best Matches for Motif ID 2 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
68 |
| Motif name: |
Motif 68 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.034263 |
Alignment:
WAAWAAAWAWWWAA
---AAAAHAAA---
| Original motif Consensus sequence: WAAWAAAWAWWWAA |
Reverse complement motif Consensus sequence: TTWWWTWTTTWTTW |
|
|
 |
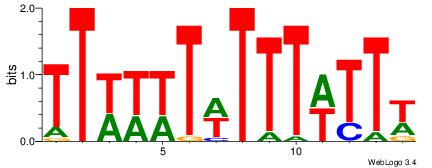 |
| Dataset #: |
4 |
| Motif ID: |
150 |
| Motif name: |
waATwAAAATAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
8 |
| Similarity score: |
0.061025 |
Alignment:
DHTATTTTWATHD
TTTDTTTT-----
| Original motif Consensus sequence: DHATWAAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTTWATHD |
|
|
 |
 |
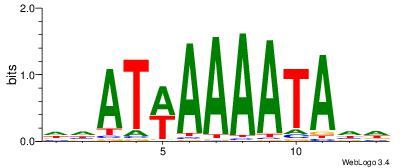
| Dataset #: |
2 |
| Motif ID: |
52 |
| Motif name: |
Motif 52 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0613251 |
Alignment:
AAATAAAW
AAAAHAAA
| Original motif Consensus sequence: AAATAAAW |
Reverse complement motif Consensus sequence: WTTTATTT |
|
|
 |
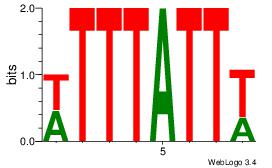 |
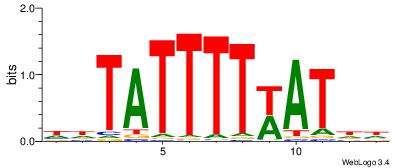
| Dataset #: |
4 |
| Motif ID: |
157 |
| Motif name: |
wtATTTTTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0669479 |
Alignment:
WWTAAAAATAD
AAAAHAAA---
| Original motif Consensus sequence: DTATTTTTAWW |
Reverse complement motif Consensus sequence: WWTAAAAATAD |
|
|
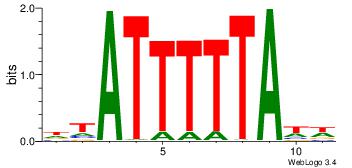 |
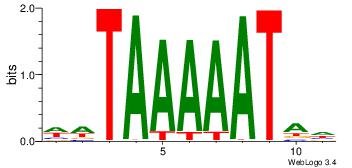 |
| Dataset #: |
4 |
| Motif ID: |
148 |
| Motif name: |
wwTwAAAAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0808845 |
Alignment:
DDTWAAAAHH
AAAAHAAA--
| Original motif Consensus sequence: DDTWAAAAHH |
Reverse complement motif Consensus sequence: HHTTTTWADD |
|
|
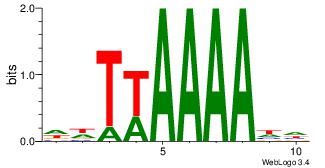 |
 |
| Dataset #: 1 |
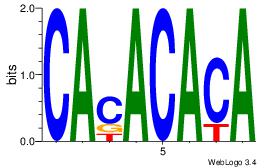
Motif ID: 3 |
Motif name: Motif 3 |
| Original motif Consensus sequence: CACACACA |
Reverse complement motif Consensus sequence: TGTGTGTG |
|
|
 |
 |
Best Matches for Motif ID 3 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
24 |
| Motif name: |
Motif 24 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0443933 |
Alignment:
TGTGGGTG
TGTGTGTG
| Original motif Consensus sequence: TGTGGGTG |
Reverse complement motif Consensus sequence: CACCCACA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
31 |
| Motif name: |
Motif 31 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0599548 |
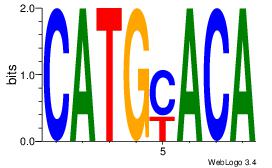
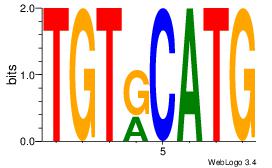
Alignment:
CATGYACA
CACACACA
| Original motif Consensus sequence: CATGYACA |
Reverse complement motif Consensus sequence: TGTKCATG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
120 |
| Motif name: |
T |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.065511 |
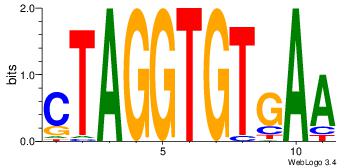
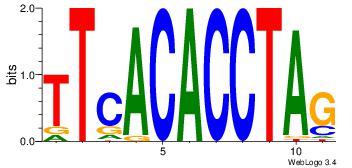
Alignment:
TTCACACCTAG
--CACACACA-
| Original motif Consensus sequence: CTAGGTGTGAA |
Reverse complement motif Consensus sequence: TTCACACCTAG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
64 |
| Motif name: |
Motif 64 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0681512 |
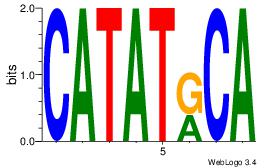
Alignment:
CATATRCA
CACACACA
| Original motif Consensus sequence: CATATRCA |
Reverse complement motif Consensus sequence: TGMATATG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
114 |
| Motif name: |
RUNX1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0795385 |
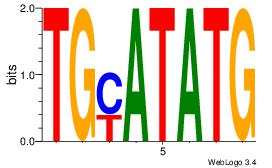
Alignment:
BBYTGTGGTTT
---TGTGTGTG
| Original motif Consensus sequence: BBYTGTGGTTT |
Reverse complement motif Consensus sequence: AAACCACAKVB |
|
|
 |
 |
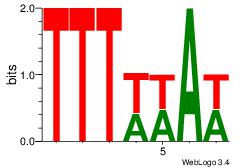
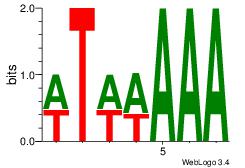
| Dataset #: 1 |
Motif ID: 4 |
Motif name: Motif 4 |
| Original motif Consensus sequence: TTTWWAW |
Reverse complement motif Consensus sequence: WTWWAAA |
|
|
 |
 |
Best Matches for Motif ID 4 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
148 |
| Motif name: |
wwTwAAAAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0.0204013 |
Alignment:
DDTWAAAAHH
-WTWWAAA--
| Original motif Consensus sequence: DDTWAAAAHH |
Reverse complement motif Consensus sequence: HHTTTTWADD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
150 |
| Motif name: |
waATwAAAATAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0.0245609 |
Alignment:
DHTATTTTWATHD
----TTTWWAW--
| Original motif Consensus sequence: DHATWAAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTTWATHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
157 |
| Motif name: |
wtATTTTTAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.028753 |
Alignment:
WWTAAAAATAD
-WTWWAAA---
| Original motif Consensus sequence: DTATTTTTAWW |
Reverse complement motif Consensus sequence: WWTAAAAATAD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
68 |
| Motif name: |
Motif 68 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.048084 |
Alignment:
WAAWAAAWAWWWAA
WTWWAAA-------
| Original motif Consensus sequence: WAAWAAAWAWWWAA |
Reverse complement motif Consensus sequence: TTWWWTWTTTWTTW |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
52 |
| Motif name: |
Motif 52 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0637767 |
Alignment:
AAATAAAW
WTWWAAA-
| Original motif Consensus sequence: AAATAAAW |
Reverse complement motif Consensus sequence: WTTTATTT |
|
|
 |
 |
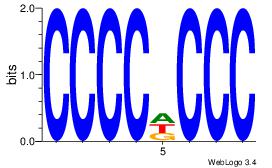
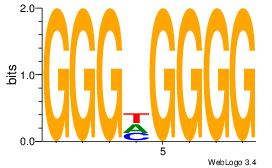
| Dataset #: 1 |
Motif ID: 5 |
Motif name: Motif 5 |
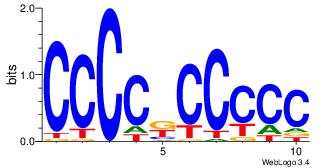
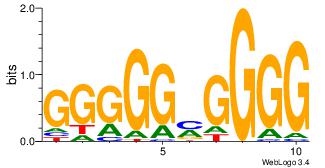
| Original motif Consensus sequence: CCCCDCCC |
Reverse complement motif Consensus sequence: GGGDGGGG |
|
|
 |
 |
Best Matches for Motif ID 5 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
59 |
| Motif name: |
Motif 59 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.00363191 |
Alignment:
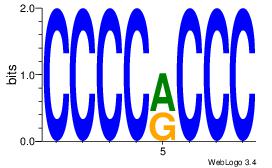
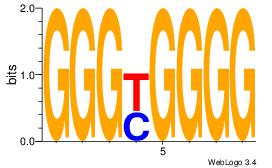
CCCCRCCC
CCCCDCCC
| Original motif Consensus sequence: CCCCRCCC |
Reverse complement motif Consensus sequence: GGGKGGGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
155 |
| Motif name: |
csCSCCdCCCcs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.00464015 |
Alignment:
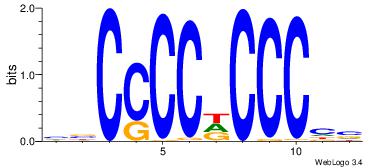
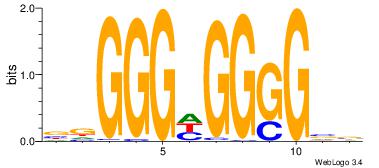
VBCCCCDCCCHV
--CCCCDCCC--
| Original motif Consensus sequence: VBCCCCDCCCHV |
Reverse complement motif Consensus sequence: VDGGGDGGGGBV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
89 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0352587 |
Alignment:
GCCYCMCCCD
-CCCCDCCC-
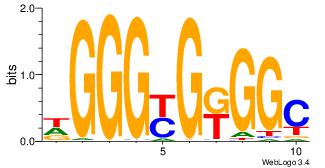
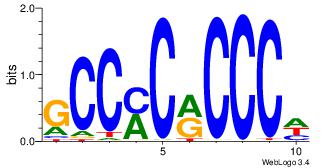
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
154 |
| Motif name: |
csCsCCTCCcc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0361349 |
Alignment:
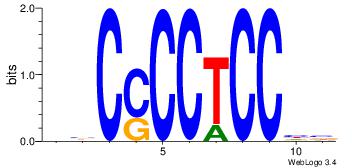
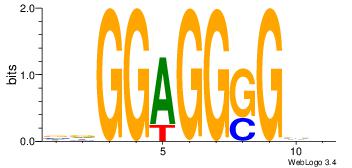
BDGGAGGGGBV
-GGGDGGGG--
| Original motif Consensus sequence: VBCCCCTCCHB |
Reverse complement motif Consensus sequence: BDGGAGGGGBV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
116 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0368066 |
Alignment:
CCCCKCCCCC
CCCCDCCC--
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 6 |
Motif name: Motif 6 |
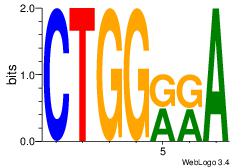
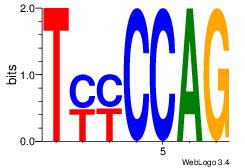
| Original motif Consensus sequence: CTGGRRA |
Reverse complement motif Consensus sequence: TMMCCAG |
|
|
 |
 |
Best Matches for Motif ID 6 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
40 |
| Motif name: |
Motif 40 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0326828 |
Alignment:
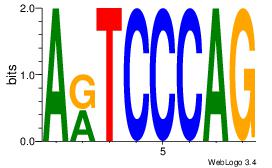
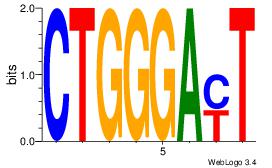
CTGGGAMT
CTGGRRA-
| Original motif Consensus sequence: ARTCCCAG |
Reverse complement motif Consensus sequence: CTGGGAMT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
69 |
| Motif name: |
Motif 69 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
7 |
| Similarity score: |
0.0472643 |
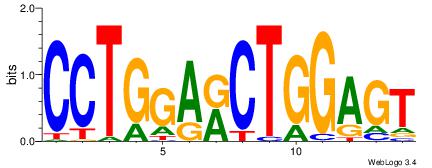
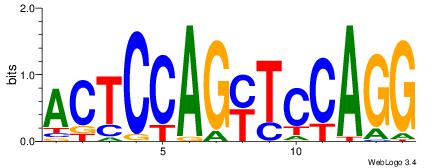
Alignment:
CCTGGARCTGGAGT
-CTGGRRA------
| Original motif Consensus sequence: CCTGGARCTGGAGT |
Reverse complement motif Consensus sequence: ACTCCAGMTCCAGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
85 |
| Motif name: |
Hand1Tcfe2a |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.0513687 |
Alignment:
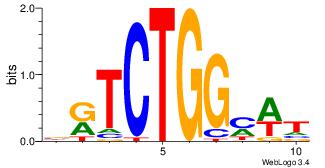
BRTCTGGMWT
---CTGGRRA
| Original motif Consensus sequence: BRTCTGGMWT |
Reverse complement motif Consensus sequence: AWRCCAGAMB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
151 |
| Motif name: |
agrCCAGmAGrg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
7 |
| Similarity score: |
0.0514331 |
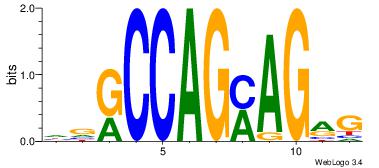
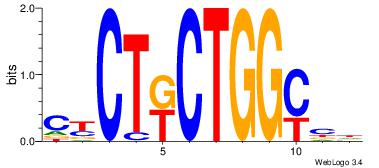
Alignment:
HVGCCAGMAGRG
TMMCCAG-----
| Original motif Consensus sequence: HVGCCAGMAGRG |
Reverse complement motif Consensus sequence: CKCTRCTGGCVH |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0553656 |
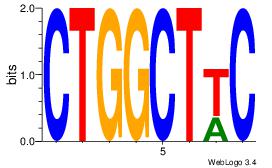
Alignment:
GWAGCCAG
-TMMCCAG
| Original motif Consensus sequence: CTGGCTWC |
Reverse complement motif Consensus sequence: GWAGCCAG |
|
|
 |
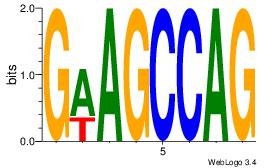 |
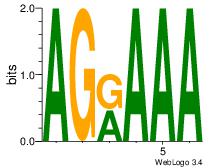
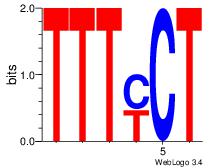
| Dataset #: 1 |
Motif ID: 7 |
Motif name: Motif 7 |
| Original motif Consensus sequence: AGRAAA |
Reverse complement motif Consensus sequence: TTTMCT |
|
|
 |
 |
Best Matches for Motif ID 7 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0617932 |
Alignment:
TTTDTTTT
TTTMCT--
| Original motif Consensus sequence: AAAAHAAA |
Reverse complement motif Consensus sequence: TTTDTTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
52 |
| Motif name: |
Motif 52 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0627423 |
Alignment:
WTTTATTT
-TTTMCT-
| Original motif Consensus sequence: AAATAAAW |
Reverse complement motif Consensus sequence: WTTTATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
68 |
| Motif name: |
Motif 68 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0627423 |
Alignment:
WAAWAAAWAWWWAA
-AGRAAA-------
| Original motif Consensus sequence: WAAWAAAWAWWWAA |
Reverse complement motif Consensus sequence: TTWWWTWTTTWTTW |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
32 |
| Motif name: |
Motif 32 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0627423 |
Alignment:
TTTATTY
TTTMCT-
| Original motif Consensus sequence: KAATAAA |
Reverse complement motif Consensus sequence: TTTATTY |
|
|
 |
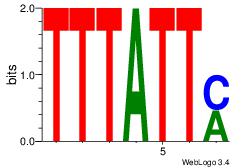 |
| Dataset #: |
3 |
| Motif ID: |
119 |
| Motif name: |
Stat3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.069443 |
Alignment:
TTCCAGGAAG
----AGRAAA
| Original motif Consensus sequence: TTCCAGGAAG |
Reverse complement motif Consensus sequence: CTTCCTGGAA |
|
|
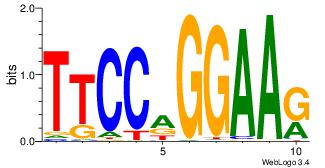 |
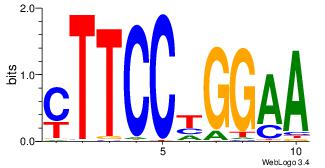 |
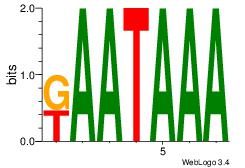
| Dataset #: 1 |
Motif ID: 8 |
Motif name: Motif 8 |
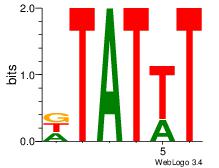
| Original motif Consensus sequence: AAATAH |
Reverse complement motif Consensus sequence: DTATTT |
|
|
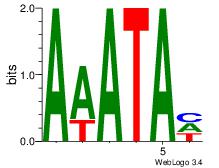 |
 |
Best Matches for Motif ID 8 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
150 |
| Motif name: |
waATwAAAATAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.00224204 |
Alignment:
DHATWAAAATAHD
------AAATAH-
| Original motif Consensus sequence: DHATWAAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTTWATHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
157 |
| Motif name: |
wtATTTTTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
6 |
| Similarity score: |
0.00556446 |
Alignment:
WWTAAAAATAD
-----AAATAH
| Original motif Consensus sequence: DTATTTTTAWW |
Reverse complement motif Consensus sequence: WWTAAAAATAD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
68 |
| Motif name: |
Motif 68 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0128826 |
Alignment:
WAAWAAAWAWWWAA
----AAATAH----
| Original motif Consensus sequence: WAAWAAAWAWWWAA |
Reverse complement motif Consensus sequence: TTWWWTWTTTWTTW |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
52 |
| Motif name: |
Motif 52 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0167304 |
Alignment:
AAATAAAW
AAATAH--
| Original motif Consensus sequence: AAATAAAW |
Reverse complement motif Consensus sequence: WTTTATTT |
|
|
 |
 |
| Dataset #: |
3 |
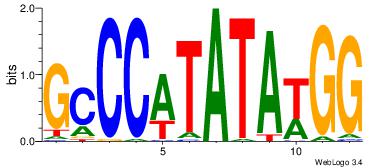
| Motif ID: |
118 |
| Motif name: |
SRF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0210491 |
Alignment:
GCCCATATATGG
----AAATAH--
| Original motif Consensus sequence: GCCCATATATGG |
Reverse complement motif Consensus sequence: CCATATATGGGC |
|
|
 |
 |
| Dataset #: 1 |
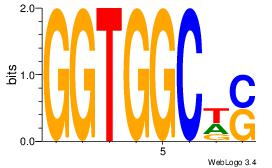
Motif ID: 9 |
Motif name: Motif 9 |
| Original motif Consensus sequence: GGTGGCWS |
Reverse complement motif Consensus sequence: SWGCCACC |
|
|
 |
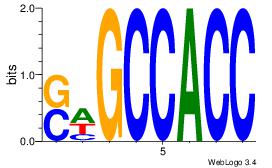 |
Best Matches for Motif ID 9 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
144 |
| Motif name: |
ctCTrsyGCCmCCTast |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0240374 |
Alignment:
ASTAGGYGGCMSCAGDD
----GGTGGCWS-----
| Original motif Consensus sequence: HDCTGSYGCCMCCTAST |
Reverse complement motif Consensus sequence: ASTAGGYGGCMSCAGDD |
|
|
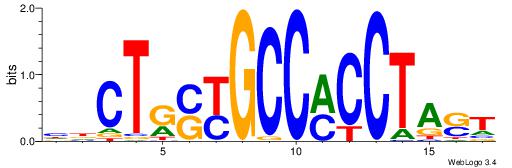 |
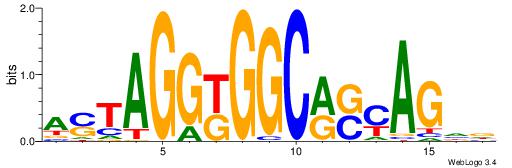 |
| Dataset #: |
4 |
| Motif ID: |
146 |
| Motif name: |
myrGYGCCmCCTast |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0280157 |
Alignment:
ASTAGGYGGCGCTBB
----GGTGGCWS---
| Original motif Consensus sequence: VBAGCGCCMCCTAST |
Reverse complement motif Consensus sequence: ASTAGGYGGCGCTBB |
|
|
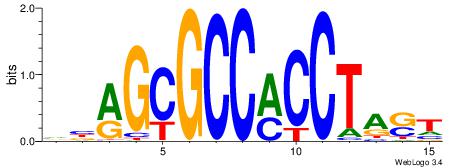 |
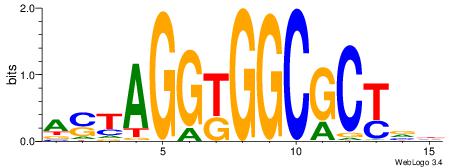 |
| Dataset #: |
4 |
| Motif ID: |
164 |
| Motif name: |
asyAGrkGGCRGCAga |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0360935 |
Alignment:
ASYAGRKGGCAGCABH
----GGTGGCWS----
| Original motif Consensus sequence: ASYAGRKGGCAGCABH |
Reverse complement motif Consensus sequence: HBTGCTGCCYMCTKST |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
147 |
| Motif name: |
asCAGrkGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0392839 |
Alignment:
ASCAGRGGGCRSB
----GGTGGCWS-
| Original motif Consensus sequence: ASCAGRGGGCRSB |
Reverse complement motif Consensus sequence: BSKGCCCMCTGST |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
10 |
| Number of overlap: |
8 |
| Similarity score: |
0.0429881 |
Alignment:
YDRCCASYAGRKGGCRSYV
---------GGTGGCWS--
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
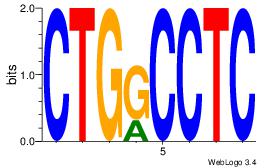
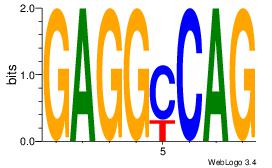
| Dataset #: 1 |
Motif ID: 10 |
Motif name: Motif 10 |
| Original motif Consensus sequence: CTGGCCTC |
Reverse complement motif Consensus sequence: GAGGCCAG |
|
|
 |
 |
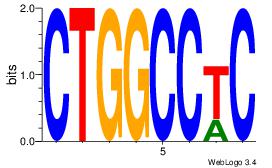
Best Matches for Motif ID 10 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0 |
Alignment:
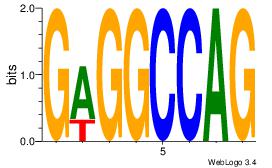
GAGGCCAG
GAGGCCAG
| Original motif Consensus sequence: CTGGCCTC |
Reverse complement motif Consensus sequence: GAGGCCAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0326888 |
Alignment:
CTGGCTWC
CTGGCCTC
| Original motif Consensus sequence: CTGGCTWC |
Reverse complement motif Consensus sequence: GWAGCCAG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
143 |
| Motif name: |
AgmAGAGGGCrscAGak |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.047812 |
Alignment:
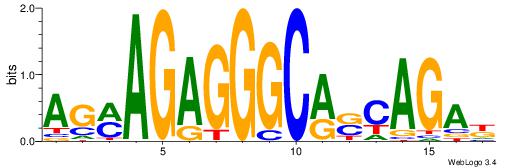
AGMAGAGGGCASCAGAK
----GAGGCCAG-----
| Original motif Consensus sequence: AGMAGAGGGCASCAGAK |
Reverse complement motif Consensus sequence: RTCTGSTGCCCTCTYCT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0506629 |
Alignment:
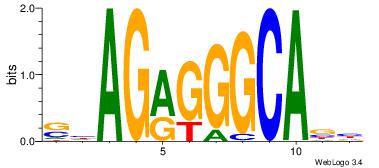
VHGGCCAGMAGAGGGCRBY
GAGGCCAG-----------
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
133 |
| Motif name: |
shAGrGGGCAgy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0509007 |
Alignment:
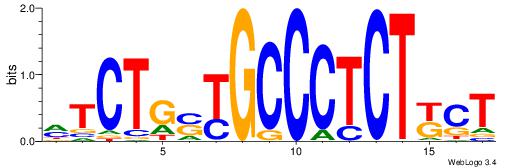
DBTGCCCKCTDS
-CTGGCCTC---
| Original motif Consensus sequence: SHAGRGGGCABH |
Reverse complement motif Consensus sequence: DBTGCCCKCTDS |
|
|
 |
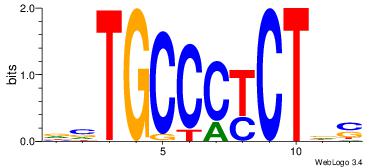 |
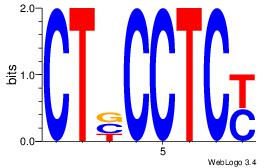
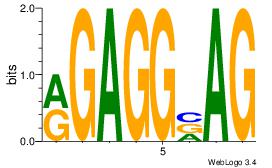
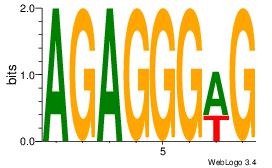
| Dataset #: 1 |
Motif ID: 11 |
Motif name: Motif 11 |
| Original motif Consensus sequence: CTBCCTCY |
Reverse complement motif Consensus sequence: MGAGGBAG |
|
|
 |
 |
Best Matches for Motif ID 11 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
63 |
| Motif name: |
Motif 63 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0297241 |
Alignment:
CWCCCTCT
CTBCCTCY
| Original motif Consensus sequence: AGAGGGWG |
Reverse complement motif Consensus sequence: CWCCCTCT |
|
|
 |
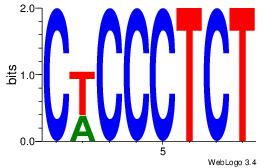 |
| Dataset #: |
1 |
| Motif ID: |
17 |
| Motif name: |
Motif 17 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0448422 |
Alignment:
CCDCCTCC
CTBCCTCY
| Original motif Consensus sequence: CCDCCTCC |
Reverse complement motif Consensus sequence: GGAGGDGG |
|
|
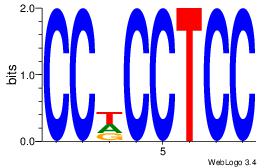 |
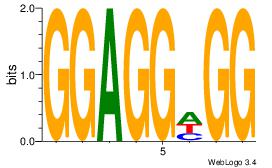 |
| Dataset #: |
4 |
| Motif ID: |
169 |
| Motif name: |
yvTGCyGCCmCCwGgtG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0620322 |
Alignment:
BVTGCTGCCACCWGGDG
----CTBCCTCY-----
| Original motif Consensus sequence: BVTGCTGCCACCWGGDG |
Reverse complement motif Consensus sequence: CDCCWGGTGGCAGCAVV |
|
|
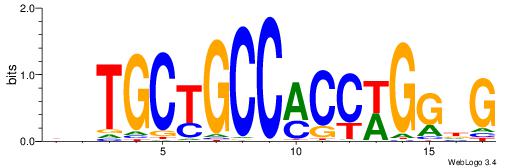 |
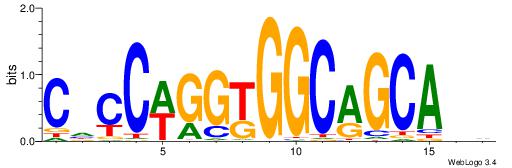 |
| Dataset #: |
4 |
| Motif ID: |
164 |
| Motif name: |
asyAGrkGGCRGCAga |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0623054 |
Alignment:
HBTGCTGCCYMCTKST
----CTBCCTCY----
| Original motif Consensus sequence: ASYAGRKGGCAGCABH |
Reverse complement motif Consensus sequence: HBTGCTGCCYMCTKST |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
154 |
| Motif name: |
csCsCCTCCcc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.067516 |
Alignment:
VBCCCCTCCHB
-CTBCCTCY--
| Original motif Consensus sequence: VBCCCCTCCHB |
Reverse complement motif Consensus sequence: BDGGAGGGGBV |
|
|
 |
 |
| Dataset #: 1 |
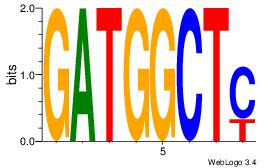
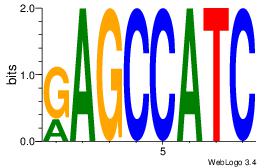
Motif ID: 12 |
Motif name: Motif 12 |
| Original motif Consensus sequence: GATGGCTC |
Reverse complement motif Consensus sequence: GAGCCATC |
|
|
 |
 |
Best Matches for Motif ID 12 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0446318 |
Alignment:
GGTGGCWS
GATGGCTC
| Original motif Consensus sequence: GGTGGCWS |
Reverse complement motif Consensus sequence: SWGCCACC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
149 |
| Motif name: |
asmAGRGGGCrCTGsmkc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.053856 |
Alignment:
DBTSCAGMGCCCTCTRST
------GAGCCATC----
| Original motif Consensus sequence: ASMAGAGGGCRCTGSABH |
Reverse complement motif Consensus sequence: DBTSCAGMGCCCTCTRST |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
146 |
| Motif name: |
myrGYGCCmCCTast |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0558657 |
Alignment:
ASTAGGYGGCGCTBB
----GATGGCTC---
| Original motif Consensus sequence: VBAGCGCCMCCTAST |
Reverse complement motif Consensus sequence: ASTAGGYGGCGCTBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
11 |
| Number of overlap: |
8 |
| Similarity score: |
0.0592323 |
Alignment:
VHGGCCAGMAGAGGGCRBY
----------GATGGCTC-
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
165 |
| Motif name: |
wgGCCAshAGrGGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0607886 |
Alignment:
HDGCCACHAGRGGGCRBY
---------GATGGCTC-
| Original motif Consensus sequence: HDGCCACHAGRGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCKCTHGTGGCHH |
|
|
 |
 |
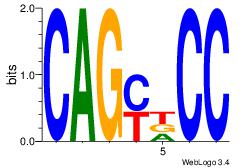
| Dataset #: 1 |
Motif ID: 13 |
Motif name: Motif 13 |
| Original motif Consensus sequence: CAGYDCC |
Reverse complement motif Consensus sequence: GGDKCTG |
|
|
 |
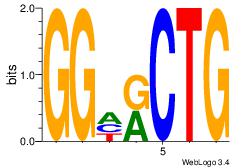
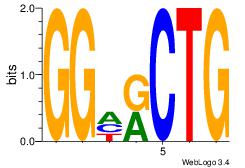 |
Best Matches for Motif ID 13 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
Motif 47 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0 |
Alignment:
CAGYDCC
CAGYDCC
| Original motif Consensus sequence: CAGYDCC |
Reverse complement motif Consensus sequence: GGDKCTG |
|
|
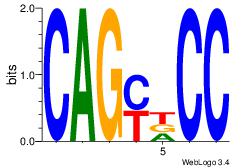 |
 |
| Dataset #: |
2 |
| Motif ID: |
53 |
| Motif name: |
Motif 53 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0294104 |
Alignment:
GGGATCTG
-GGDKCTG
| Original motif Consensus sequence: CAGATCCC |
Reverse complement motif Consensus sequence: GGGATCTG |
|
|
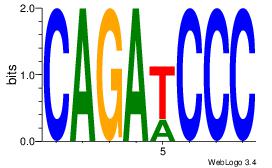 |
 |
| Dataset #: |
3 |
| Motif ID: |
113 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
13 |
| Number of overlap: |
7 |
| Similarity score: |
0.0500473 |
Alignment:
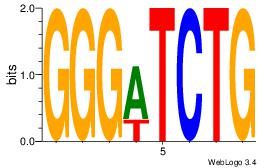
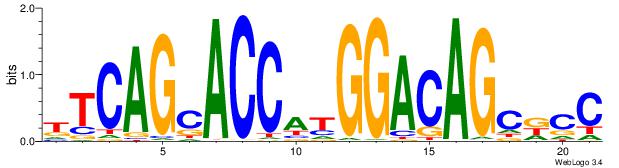
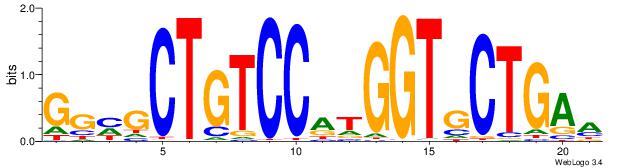
TTCAGCACCATGGACAGCKCC
--CAGYDCC------------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
22 |
| Motif name: |
Motif 22 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0533516 |
Alignment:
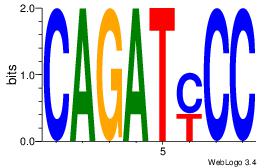
GGKATCTG
-GGDKCTG
| Original motif Consensus sequence: CAGATYCC |
Reverse complement motif Consensus sequence: GGKATCTG |
|
|
 |
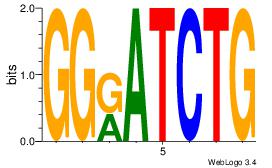 |
| Dataset #: |
4 |
| Motif ID: |
149 |
| Motif name: |
asmAGRGGGCrCTGsmkc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
7 |
| Similarity score: |
0.0558573 |
Alignment:
DBTSCAGMGCCCTCTRST
----CAGYDCC-------
| Original motif Consensus sequence: ASMAGAGGGCRCTGSABH |
Reverse complement motif Consensus sequence: DBTSCAGMGCCCTCTRST |
|
|
 |
 |
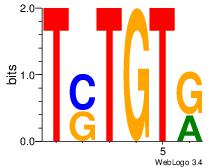
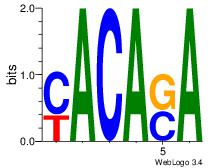
| Dataset #: 1 |
Motif ID: 14 |
Motif name: Motif 14 |
| Original motif Consensus sequence: TSTGTR |
Reverse complement motif Consensus sequence: MACASA |
|
|
 |
 |
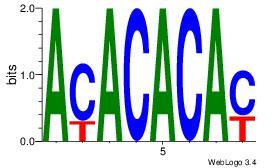
Best Matches for Motif ID 14 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
57 |
| Motif name: |
Motif 57 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0165773 |
Alignment:
KTGTGTGT
-TSTGTR-
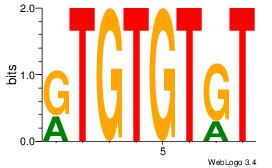
| Original motif Consensus sequence: ACACACAY |
Reverse complement motif Consensus sequence: KTGTGTGT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0165773 |
Alignment:
CACACACA
MACASA--
| Original motif Consensus sequence: CACACACA |
Reverse complement motif Consensus sequence: TGTGTGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
136 |
| Motif name: |
dwCAGAAGwh |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0401476 |
Alignment:
HDCTTCTGHD
----TSTGTR
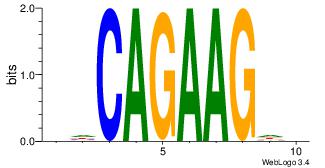
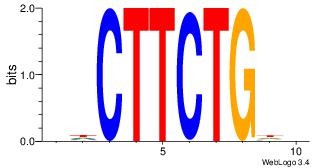
| Original motif Consensus sequence: DHCAGAAGDH |
Reverse complement motif Consensus sequence: HDCTTCTGHD |
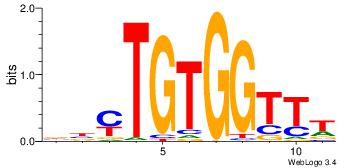
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
114 |
| Motif name: |
RUNX1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0445836 |
Alignment:
BBYTGTGGTTT
-TSTGTR----
| Original motif Consensus sequence: BBYTGTGGTTT |
Reverse complement motif Consensus sequence: AAACCACAKVB |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
24 |
| Motif name: |
Motif 24 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0456841 |
Alignment:
TGTGGGTG
--TSTGTR
| Original motif Consensus sequence: TGTGGGTG |
Reverse complement motif Consensus sequence: CACCCACA |
|
|
 |
 |
| Dataset #: 1 |
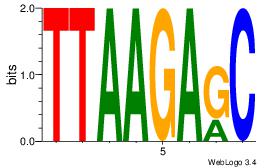
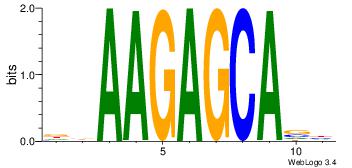
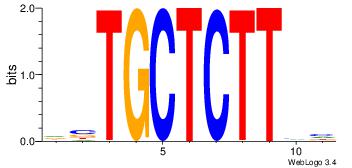
Motif ID: 15 |
Motif name: Motif 15 |
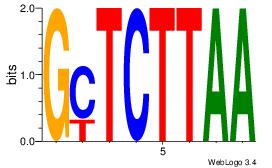
| Original motif Consensus sequence: GCTCTTAA |
Reverse complement motif Consensus sequence: TTAAGAGC |
|
|
 |
 |
Best Matches for Motif ID 15 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
159 |
| Motif name: |
kkAAGAGCAsy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0216416 |
Alignment:
DBAAGAGCAVH
TTAAGAGC---
| Original motif Consensus sequence: DBAAGAGCAVH |
Reverse complement motif Consensus sequence: HVTGCTCTTBH |
|
|
 |
 |
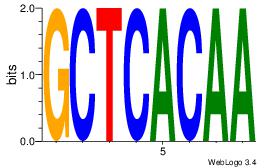
| Dataset #: |
2 |
| Motif ID: |
55 |
| Motif name: |
Motif 55 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0390835 |
Alignment:
GCTCACAA
GCTCTTAA
| Original motif Consensus sequence: GCTCACAA |
Reverse complement motif Consensus sequence: TTGTGAGC |
|
|
 |
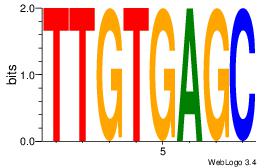 |
| Dataset #: |
4 |
| Motif ID: |
148 |
| Motif name: |
wwTwAAAAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0584188 |
Alignment:
HHTTTTWADD
GCTCTTAA--
| Original motif Consensus sequence: DDTWAAAAHH |
Reverse complement motif Consensus sequence: HHTTTTWADD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
120 |
| Motif name: |
T |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0666592 |
Alignment:
CTAGGTGTGAA
---GCTCTTAA
| Original motif Consensus sequence: CTAGGTGTGAA |
Reverse complement motif Consensus sequence: TTCACACCTAG |
|
|
 |
 |
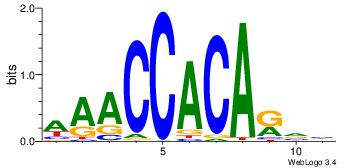
| Dataset #: |
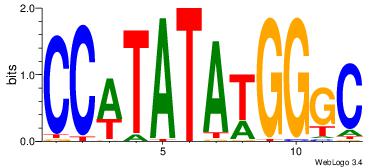
3 |
| Motif ID: |
118 |
| Motif name: |
SRF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0676161 |
Alignment:
CCATATATGGGC
----TTAAGAGC
| Original motif Consensus sequence: GCCCATATATGG |
Reverse complement motif Consensus sequence: CCATATATGGGC |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 16 |
Motif name: Motif 16 |
| Original motif Consensus sequence: GGAAGRR |
Reverse complement motif Consensus sequence: MMCTTCC |
|
|
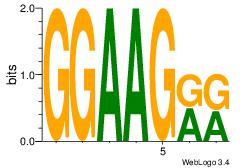 |
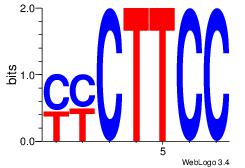 |
Best Matches for Motif ID 16 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
84 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0.0238137 |
Alignment:
VVCACTTCCGG
--MMCTTCC--
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
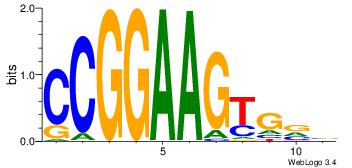 |
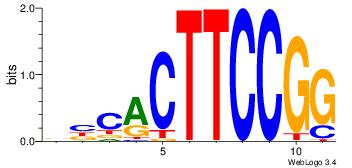 |
| Dataset #: |
1 |
| Motif ID: |
17 |
| Motif name: |
Motif 17 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0632856 |
Alignment:
GGAGGDGG
GGAAGRR-
| Original motif Consensus sequence: CCDCCTCC |
Reverse complement motif Consensus sequence: GGAGGDGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
136 |
| Motif name: |
dwCAGAAGwh |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0705782 |
Alignment:
HDCTTCTGHD
MMCTTCC---
| Original motif Consensus sequence: DHCAGAAGDH |
Reverse complement motif Consensus sequence: HDCTTCTGHD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
112 |
| Motif name: |
RELA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0712379 |
Alignment:
GGGRATTTCC
---MMCTTCC
| Original motif Consensus sequence: GGGRATTTCC |
Reverse complement motif Consensus sequence: GGAAATKCCC |
|
|
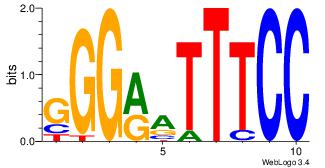 |
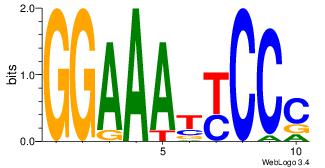 |
| Dataset #: |
4 |
| Motif ID: |
154 |
| Motif name: |
csCsCCTCCcc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0.0768131 |
Alignment:
BDGGAGGGGBV
--GGAAGRR--
| Original motif Consensus sequence: VBCCCCTCCHB |
Reverse complement motif Consensus sequence: BDGGAGGGGBV |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 17 |
Motif name: Motif 17 |
| Original motif Consensus sequence: CCDCCTCC |
Reverse complement motif Consensus sequence: GGAGGDGG |
|
|
 |
 |
Best Matches for Motif ID 17 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
154 |
| Motif name: |
csCsCCTCCcc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0325807 |
Alignment:
BDGGAGGGGBV
--GGAGGDGG-
| Original motif Consensus sequence: VBCCCCTCCHB |
Reverse complement motif Consensus sequence: BDGGAGGGGBV |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0441606 |
Alignment:
CTBCCTCY
CCDCCTCC
| Original motif Consensus sequence: CTBCCTCY |
Reverse complement motif Consensus sequence: MGAGGBAG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
155 |
| Motif name: |
csCSCCdCCCcs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0485525 |
Alignment:
VDGGGDGGGGBV
---GGAGGDGG-
| Original motif Consensus sequence: VBCCCCDCCCHV |
Reverse complement motif Consensus sequence: VDGGGDGGGGBV |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
27 |
| Motif name: |
Motif 27 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0499486 |
Alignment:
CSGCCGCC
CCDCCTCC
| Original motif Consensus sequence: CSGCCGCC |
Reverse complement motif Consensus sequence: GGCGGCSG |
|
|
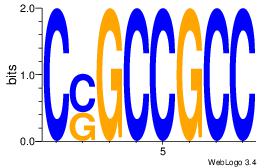 |
 |
| Dataset #: |
3 |
| Motif ID: |
116 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0500496 |
Alignment:
CCCCKCCCCC
--CCDCCTCC
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
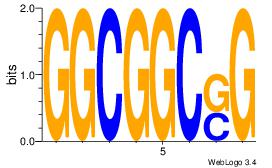
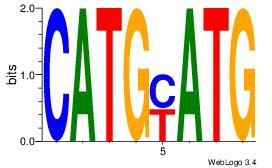
| Dataset #: 1 |
Motif ID: 18 |
Motif name: Motif 18 |
| Original motif Consensus sequence: CATGYATG |
Reverse complement motif Consensus sequence: CATKCATG |
|
|
 |
 |
Best Matches for Motif ID 18 (Highest to Lowest)
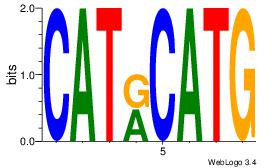
| Dataset #: |
4 |
| Motif ID: |
152 |
| Motif name: |
yrCATGCAyr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.058327 |
Alignment:
BRCATGCABD
--CATKCATG
| Original motif Consensus sequence: BRCATGCABD |
Reverse complement motif Consensus sequence: HVTGCATGKV |
|
|
 |
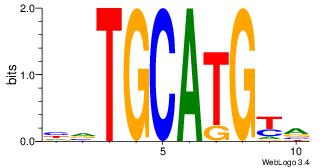 |
| Dataset #: |
1 |
| Motif ID: |
31 |
| Motif name: |
Motif 31 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0666244 |
Alignment:
CATGYACA
CATGYATG
| Original motif Consensus sequence: CATGYACA |
Reverse complement motif Consensus sequence: TGTKCATG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
125 |
| Motif name: |
TP53 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
8 |
| Similarity score: |
0.068413 |
Alignment:
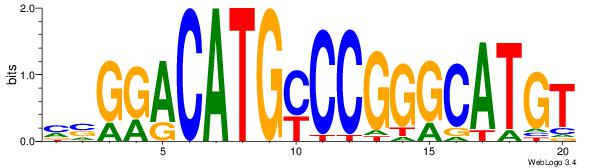
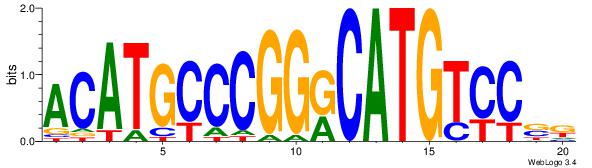
ACATGCCCGGKCATGTCCSR
-------CATKCATG-----
| Original motif Consensus sequence: MSGGACATGYCCGGGCATGT |
Reverse complement motif Consensus sequence: ACATGCCCGGKCATGTCCSR |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
118 |
| Motif name: |
SRF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0745698 |
Alignment:
CCATATATGGGC
-CATGYATG---
| Original motif Consensus sequence: GCCCATATATGG |
Reverse complement motif Consensus sequence: CCATATATGGGC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
131 |
| Motif name: |
znf143 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.105729 |
Alignment:
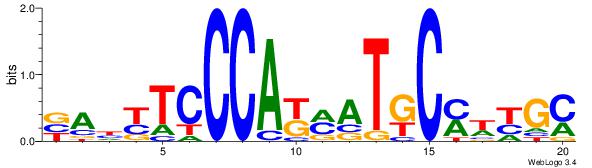
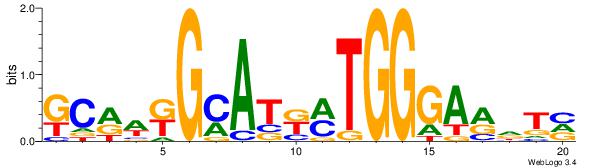
BAHYTCCCAKMATGCMWYGC
----------CATKCATG--
| Original motif Consensus sequence: BAHYTCCCAKMATGCMWYGC |
Reverse complement motif Consensus sequence: GCMWRGCATYRTGGGAMHTB |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 19 |
Motif name: Motif 19 |
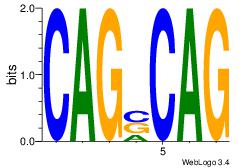
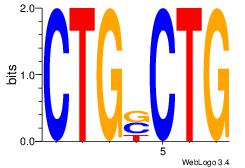
| Original motif Consensus sequence: CAGSCAG |
Reverse complement motif Consensus sequence: CTGSCTG |
|
|
 |
 |
Best Matches for Motif ID 19 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0196488 |
Alignment:
CTGGCTWC
CTGSCTG-
| Original motif Consensus sequence: CTGGCTWC |
Reverse complement motif Consensus sequence: GWAGCCAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
28 |
| Motif name: |
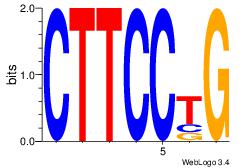
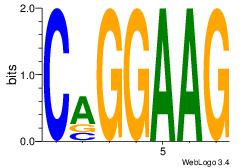
Motif 28 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0338273 |
Alignment:
CTTCCTG
CTGSCTG
| Original motif Consensus sequence: CTTCCTG |
Reverse complement motif Consensus sequence: CAGGAAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0430055 |
Alignment:
MGAGGBAG
CAGSCAG-
| Original motif Consensus sequence: CTBCCTCY |
Reverse complement motif Consensus sequence: MGAGGBAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0473229 |
Alignment:
CTGGCCTC
-CTGSCTG
| Original motif Consensus sequence: CTGGCCTC |
Reverse complement motif Consensus sequence: GAGGCCAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
25 |
| Motif name: |
Motif 25 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0479304 |
Alignment:
CTGRGTTC
CTGSCTG-
| Original motif Consensus sequence: CTGRGTTC |
Reverse complement motif Consensus sequence: GAACKCAG |
|
|
 |
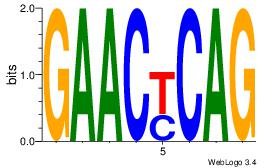 |
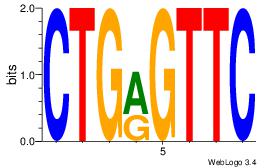
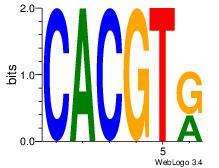
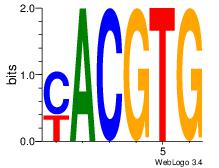
| Dataset #: 1 |
Motif ID: 20 |
Motif name: Motif 20 |
| Original motif Consensus sequence: CACGTR |
Reverse complement motif Consensus sequence: MACGTG |
|
|
 |
 |
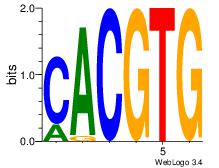
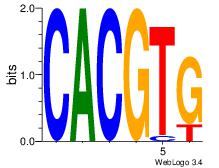
Best Matches for Motif ID 20 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
71 |
| Motif name: |
Arnt |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0 |
Alignment:
CACGTG
CACGTR
| Original motif Consensus sequence: CACGTG |
Reverse complement motif Consensus sequence: CACGTG |
|
|
 |
 |
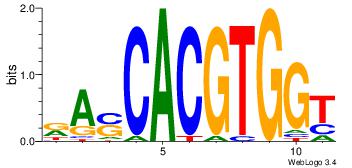
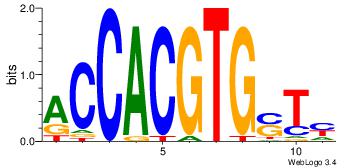
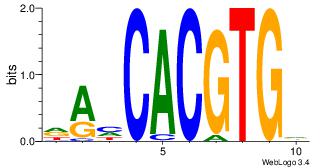
| Dataset #: |
3 |
| Motif ID: |
95 |
| Motif name: |
MYCMAX |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0102183 |
Alignment:
RASCACGTGGT
---MACGTG--
| Original motif Consensus sequence: RASCACGTGGT |
Reverse complement motif Consensus sequence: ACCACGTGSTM |
|
|
 |
 |
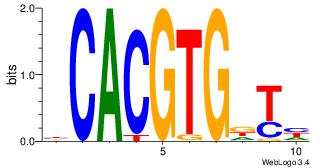
| Dataset #: |
3 |
| Motif ID: |
91 |
| Motif name: |
MAX |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.011152 |
Alignment:
DAHCACGTGD
---MACGTG-
| Original motif Consensus sequence: DAHCACGTGD |
Reverse complement motif Consensus sequence: BCACGTGDTD |
|
|
 |
 |
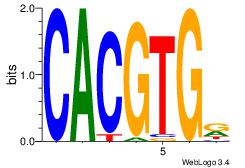
| Dataset #: |
3 |
| Motif ID: |
126 |
| Motif name: |
USF1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0131944 |
Alignment:
CACGTGR
MACGTG-
| Original motif Consensus sequence: CACGTGR |
Reverse complement motif Consensus sequence: MCACGTG |
|
|
 |
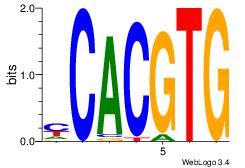 |
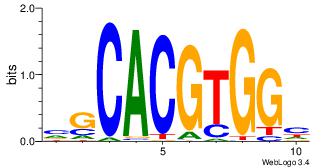
| Dataset #: |
3 |
| Motif ID: |
94 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0253396 |
Alignment:
DCCACGTGCV
--CACGTR--
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
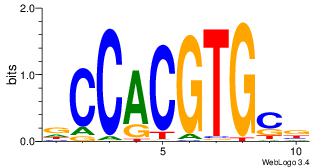 |
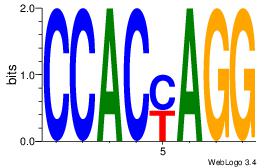
| Dataset #: 1 |
Motif ID: 21 |
Motif name: Motif 21 |
| Original motif Consensus sequence: CCACYAGG |
Reverse complement motif Consensus sequence: CCTKGTGG |
|
|
 |
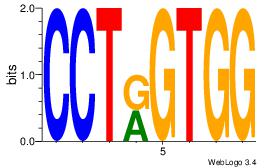 |
Best Matches for Motif ID 21 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
48 |
| Motif name: |
Motif 48 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.00903109 |
Alignment:
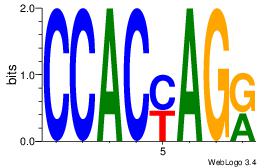
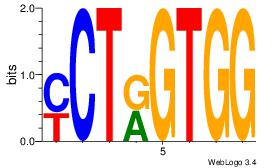
CCACYAGR
CCACYAGG
| Original motif Consensus sequence: CCACYAGR |
Reverse complement motif Consensus sequence: MCTKGTGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
145 |
| Motif name: |
grCCACyAGAkG |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.028408 |
Alignment:
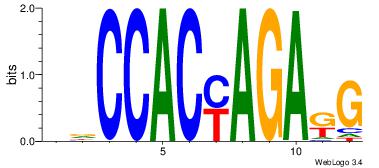
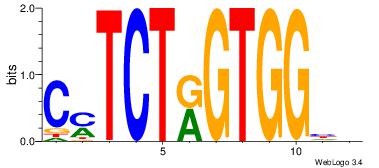
CYTCTKGTGGHH
--CCTKGTGG--
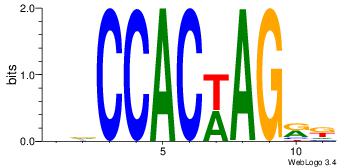
| Original motif Consensus sequence: DDCCACYAGAKG |
Reverse complement motif Consensus sequence: CYTCTKGTGGHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
158 |
| Motif name: |
grCCACwAGrk |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0314791 |
Alignment:
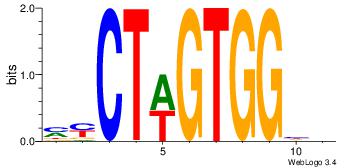
DDCCACWAGRK
--CCACYAGG-
| Original motif Consensus sequence: DDCCACWAGRK |
Reverse complement motif Consensus sequence: YMCTWGTGGHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
138 |
| Motif name: |
grCCACyAGAkG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0373754 |
Alignment:
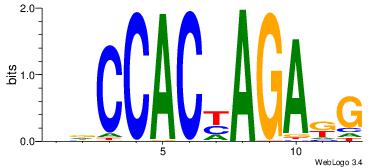
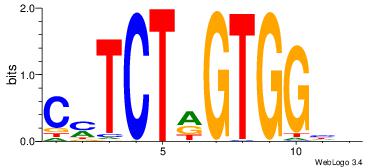
DDCCACYAGAKG
--CCACYAGG--
| Original motif Consensus sequence: DDCCACYAGAKG |
Reverse complement motif Consensus sequence: CYTCTMGTGGHH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
8 |
| Similarity score: |
0.0387762 |
Alignment:
BMSMGCCYMCTKSTGGMHM
--------CCTKGTGG---
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 22 |
Motif name: Motif 22 |
| Original motif Consensus sequence: CAGATYCC |
Reverse complement motif Consensus sequence: GGKATCTG |
|
|
 |
 |
Best Matches for Motif ID 22 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
53 |
| Motif name: |
Motif 53 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0 |
Alignment:
GGGATCTG
GGKATCTG
| Original motif Consensus sequence: CAGATCCC |
Reverse complement motif Consensus sequence: GGGATCTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
69 |
| Motif name: |
Motif 69 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.060058 |
Alignment:
ACTCCAGMTCCAGG
----CAGATYCC--
| Original motif Consensus sequence: CCTGGARCTGGAGT |
Reverse complement motif Consensus sequence: ACTCCAGMTCCAGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
149 |
| Motif name: |
asmAGRGGGCrCTGsmkc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.0603056 |
Alignment:
DBTSCAGMGCCCTCTRST
----CAGATYCC------
| Original motif Consensus sequence: ASMAGAGGGCRCTGSABH |
Reverse complement motif Consensus sequence: DBTSCAGMGCCCTCTRST |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
64 |
| Motif name: |
Motif 64 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0721296 |
Alignment:
CATATRCA
CAGATYCC
| Original motif Consensus sequence: CATATRCA |
Reverse complement motif Consensus sequence: TGMATATG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
160 |
| Motif name: |
brCAGGGCCrs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0730966 |
Alignment:
BBGGCCCTGBB
-GGKATCTG--
| Original motif Consensus sequence: BVCAGGGCCVB |
Reverse complement motif Consensus sequence: BBGGCCCTGBB |
|
|
 |
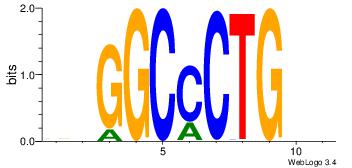 |
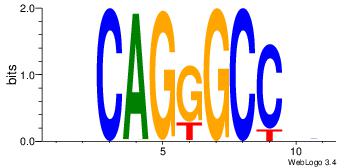
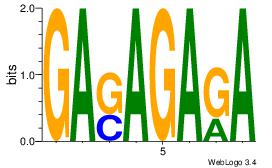
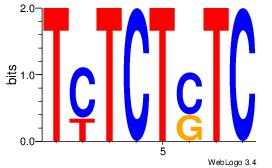
| Dataset #: 1 |
Motif ID: 23 |
Motif name: Motif 23 |
| Original motif Consensus sequence: GASAGAGA |
Reverse complement motif Consensus sequence: TCTCTSTC |
|
|
 |
 |
Best Matches for Motif ID 23 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
113 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
13 |
| Number of overlap: |
8 |
| Similarity score: |
0.0614501 |
Alignment:
TTCAGCACCATGGACAGCKCC
------------GASAGAGA-
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0761109 |
Alignment:
AAAAHAAA
GASAGAGA
| Original motif Consensus sequence: AAAAHAAA |
Reverse complement motif Consensus sequence: TTTDTTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
38 |
| Motif name: |
Motif 38 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.081673 |
Alignment:
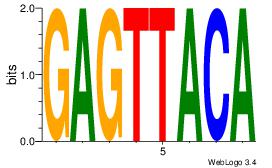
GAGTTACA
GASAGAGA
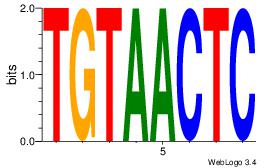
| Original motif Consensus sequence: GAGTTACA |
Reverse complement motif Consensus sequence: TGTAACTC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
51 |
| Motif name: |
Motif 51 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.081673 |
Alignment:
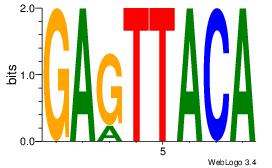
GAGTTACA
GASAGAGA
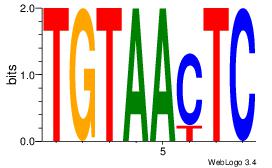
| Original motif Consensus sequence: GAGTTACA |
Reverse complement motif Consensus sequence: TGTAACTC |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0826138 |
Alignment:
CACACACA
GASAGAGA
| Original motif Consensus sequence: CACACACA |
Reverse complement motif Consensus sequence: TGTGTGTG |
|
|
 |
 |
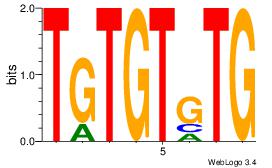
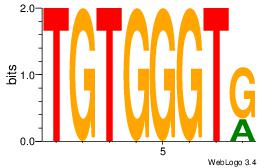
| Dataset #: 1 |
Motif ID: 24 |
Motif name: Motif 24 |
| Original motif Consensus sequence: TGTGGGTG |
Reverse complement motif Consensus sequence: CACCCACA |
|
|
 |
 |
Best Matches for Motif ID 24 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0589779 |
Alignment:
TGTGTGTG
TGTGGGTG
| Original motif Consensus sequence: CACACACA |
Reverse complement motif Consensus sequence: TGTGTGTG |
|
|
 |
 |
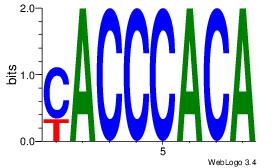
| Dataset #: |
4 |
| Motif ID: |
162 |
| Motif name: |
ccAsCCCCAcc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.060611 |
Alignment:
DBTGGGGSTVD
--TGTGGGTG-
| Original motif Consensus sequence: HVASCCCCABH |
Reverse complement motif Consensus sequence: DBTGGGGSTVD |
|
|
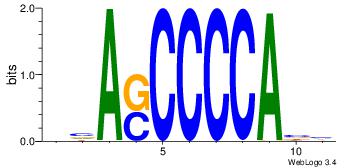 |
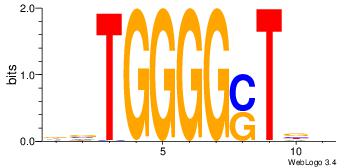 |
| Dataset #: |
3 |
| Motif ID: |
114 |
| Motif name: |
RUNX1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0649239 |
Alignment:
AAACCACAKVB
CACCCACA---
| Original motif Consensus sequence: BBYTGTGGTTT |
Reverse complement motif Consensus sequence: AAACCACAKVB |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
31 |
| Motif name: |
Motif 31 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0828996 |
Alignment:
CATGYACA
CACCCACA
| Original motif Consensus sequence: CATGYACA |
Reverse complement motif Consensus sequence: TGTKCATG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
34 |
| Motif name: |
Motif 34 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0843302 |
Alignment:
TGTGGYCA
TGTGGGTG
| Original motif Consensus sequence: TGKCCACA |
Reverse complement motif Consensus sequence: TGTGGYCA |
|
|
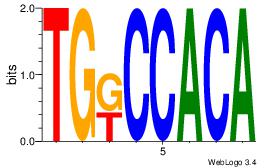 |
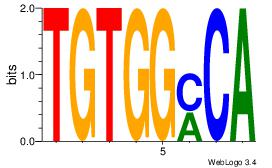 |
| Dataset #: 1 |
Motif ID: 25 |
Motif name: Motif 25 |
| Original motif Consensus sequence: CTGRGTTC |
Reverse complement motif Consensus sequence: GAACKCAG |
|
|
 |
 |
Best Matches for Motif ID 25 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0224867 |
Alignment:
CTGGCTWC
CTGRGTTC
| Original motif Consensus sequence: CTGGCTWC |
Reverse complement motif Consensus sequence: GWAGCCAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.034485 |
Alignment:
CTGGCCTC
CTGRGTTC
| Original motif Consensus sequence: CTGGCCTC |
Reverse complement motif Consensus sequence: GAGGCCAG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.052298 |
Alignment:
CTGGCCTC
CTGRGTTC
| Original motif Consensus sequence: CTGGCCTC |
Reverse complement motif Consensus sequence: GAGGCCAG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
107 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
8 |
| Similarity score: |
0.0551569 |
Alignment:
TGAMCTTTGMMCYT
-GAACKCAG-----
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
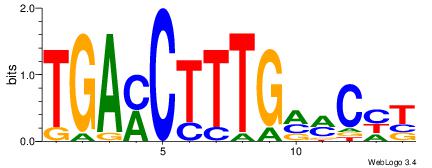 |
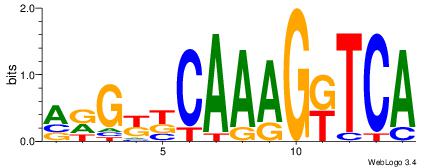 |
| Dataset #: |
1 |
| Motif ID: |
37 |
| Motif name: |
Motif 37 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0560348 |
Alignment:
CTGAGCYA
CTGRGTTC
| Original motif Consensus sequence: CTGAGCYA |
Reverse complement motif Consensus sequence: TKGCTCAG |
|
|
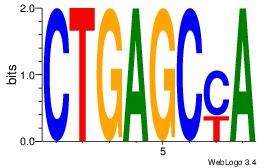 |
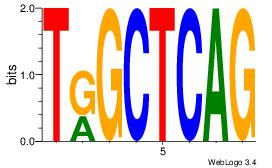 |
| Dataset #: 1 |
Motif ID: 26 |
Motif name: Motif 26 |
| Original motif Consensus sequence: CAGAGGAY |
Reverse complement motif Consensus sequence: KTCCTCTG |
|
|
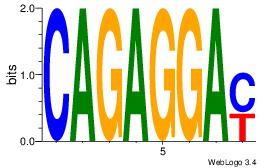 |
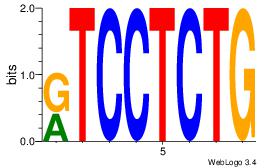 |
Best Matches for Motif ID 26 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
136 |
| Motif name: |
dwCAGAAGwh |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0359628 |
Alignment:
DHCAGAAGDH
--CAGAGGAY
| Original motif Consensus sequence: DHCAGAAGDH |
Reverse complement motif Consensus sequence: HDCTTCTGHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
166 |
| Motif name: |
CasCAGrGGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.036576 |
Alignment:
CACCAGRGGGCRSB
---CAGAGGAY---
| Original motif Consensus sequence: CACCAGRGGGCRSB |
Reverse complement motif Consensus sequence: BSKGCCCKCTGGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
143 |
| Motif name: |
AgmAGAGGGCrscAGak |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
8 |
| Similarity score: |
0.0438271 |
Alignment:
AGMAGAGGGCASCAGAK
--CAGAGGAY-------
| Original motif Consensus sequence: AGMAGAGGGCASCAGAK |
Reverse complement motif Consensus sequence: RTCTGSTGCCCTCTYCT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
168 |
| Motif name: |
yrcrGYGCCMyCTGGtG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.0450811 |
Alignment:
CACCAGMGGGCGCTGBD
---CAGAGGAY------
| Original motif Consensus sequence: HVCAGCGCCCYCTGGTG |
Reverse complement motif Consensus sequence: CACCAGMGGGCGCTGBD |
|
|
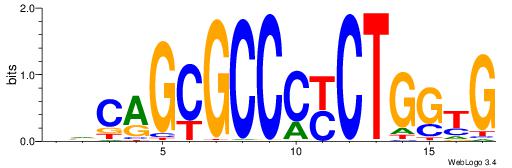 |
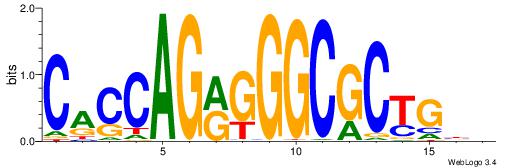 |
| Dataset #: |
4 |
| Motif ID: |
149 |
| Motif name: |
asmAGRGGGCrCTGsmkc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
8 |
| Similarity score: |
0.0460269 |
Alignment:
DBTSCAGMGCCCTCTRST
--------KTCCTCTG--
| Original motif Consensus sequence: ASMAGAGGGCRCTGSABH |
Reverse complement motif Consensus sequence: DBTSCAGMGCCCTCTRST |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 27 |
Motif name: Motif 27 |
| Original motif Consensus sequence: CSGCCGCC |
Reverse complement motif Consensus sequence: GGCGGCSG |
|
|
 |
 |
Best Matches for Motif ID 27 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
144 |
| Motif name: |
ctCTrsyGCCmCCTast |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0594103 |
Alignment:
ASTAGGYGGCMSCAGDD
----GGCGGCSG-----
| Original motif Consensus sequence: HDCTGSYGCCMCCTAST |
Reverse complement motif Consensus sequence: ASTAGGYGGCMSCAGDD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
169 |
| Motif name: |
yvTGCyGCCmCCwGgtG |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0661709 |
Alignment:
CDCCWGGTGGCAGCAVV
-----GGCGGCSG----
| Original motif Consensus sequence: BVTGCTGCCACCWGGDG |
Reverse complement motif Consensus sequence: CDCCWGGTGGCAGCAVV |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
17 |
| Motif name: |
Motif 17 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0678225 |
Alignment:
CCDCCTCC
CSGCCGCC
| Original motif Consensus sequence: CCDCCTCC |
Reverse complement motif Consensus sequence: GGAGGDGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
146 |
| Motif name: |
myrGYGCCmCCTast |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0682918 |
Alignment:
ASTAGGYGGCGCTBB
----GGCGGCSG---
| Original motif Consensus sequence: VBAGCGCCMCCTAST |
Reverse complement motif Consensus sequence: ASTAGGYGGCGCTBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
135 |
| Motif name: |
ssCGGCCGss |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0688339 |
Alignment:
BSCGGCCGSV
GGCGGCSG--
| Original motif Consensus sequence: BSCGGCCGSV |
Reverse complement motif Consensus sequence: VSCGGCCGSB |
|
|
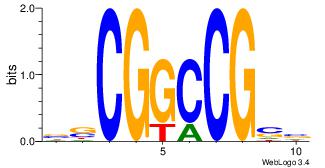 |
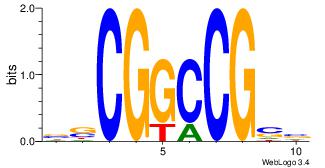 |
| Dataset #: 1 |
Motif ID: 28 |
Motif name: Motif 28 |
| Original motif Consensus sequence: CTTCCTG |
Reverse complement motif Consensus sequence: CAGGAAG |
|
|
 |
 |
Best Matches for Motif ID 28 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
119 |
| Motif name: |
Stat3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.0301014 |
Alignment:
CTTCCTGGAA
CTTCCTG---
| Original motif Consensus sequence: TTCCAGGAAG |
Reverse complement motif Consensus sequence: CTTCCTGGAA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
84 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0389741 |
Alignment:
CCGGAAGTGVV
CAGGAAG----
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
80 |
| Motif name: |
ELK4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.044949 |
Alignment:
ACCGGAAGT
-CAGGAAG-
| Original motif Consensus sequence: ACCGGAAGT |
Reverse complement motif Consensus sequence: ACTTCCGGT |
|
|
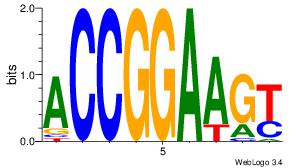 |
 |
| Dataset #: |
3 |
| Motif ID: |
79 |
| Motif name: |
ELK1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0709694 |
Alignment:
VDDCCGGAAR
---CAGGAAG
| Original motif Consensus sequence: VDDCCGGAAR |
Reverse complement motif Consensus sequence: MTTCCGGHBV |
|
|
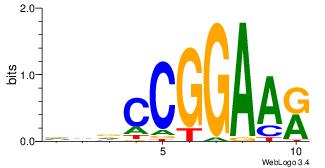 |
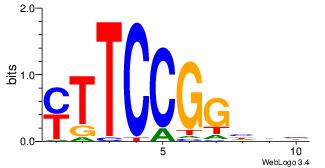 |
| Dataset #: |
1 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0713134 |
Alignment:
CAGSCAG
CAGGAAG
| Original motif Consensus sequence: CAGSCAG |
Reverse complement motif Consensus sequence: CTGSCTG |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 29 |
Motif name: Motif 29 |
| Original motif Consensus sequence: CATTTCY |
Reverse complement motif Consensus sequence: MGAAATG |
|
|
 |
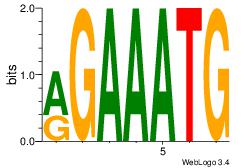 |
Best Matches for Motif ID 29 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
139 |
| Motif name: |
mkCTyTTCsg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0.0479916 |
Alignment:
HBGAAAAGBD
-MGAAATG--
| Original motif Consensus sequence: HBCTTTTCBD |
Reverse complement motif Consensus sequence: HBGAAAAGBD |
|
|
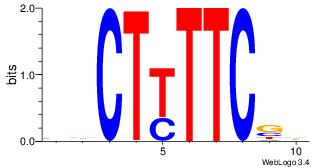 |
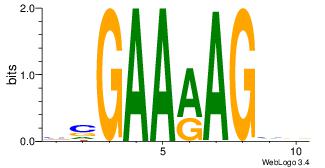 |
| Dataset #: |
2 |
| Motif ID: |
66 |
| Motif name: |
Motif 66 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.050303 |
Alignment:
TATAAATR
-MGAAATG
| Original motif Consensus sequence: TATAAATR |
Reverse complement motif Consensus sequence: KATTTATA |
|
|
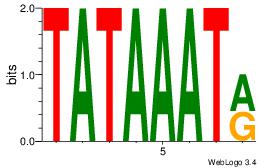 |
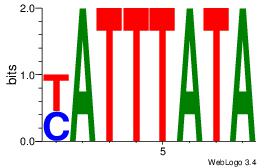 |
| Dataset #: |
3 |
| Motif ID: |
125 |
| Motif name: |
TP53 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
12 |
| Number of overlap: |
7 |
| Similarity score: |
0.058821 |
Alignment:
ACATGCCCGGKCATGTCCSR
-----------CATTTCY--
| Original motif Consensus sequence: MSGGACATGYCCGGGCATGT |
Reverse complement motif Consensus sequence: ACATGCCCGGKCATGTCCSR |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
112 |
| Motif name: |
RELA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0629654 |
Alignment:
GGAAATKCCC
MGAAATG---
| Original motif Consensus sequence: GGGRATTTCC |
Reverse complement motif Consensus sequence: GGAAATKCCC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
84 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0.0668119 |
Alignment:
VVCACTTCCGG
--CATTTCY--
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 30 |
Motif name: Motif 30 |
| Original motif Consensus sequence: CWGCAGC |
Reverse complement motif Consensus sequence: GCTGCWG |
|
|
 |
 |
Best Matches for Motif ID 30 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
97 |
| Motif name: |
Myf |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0232337 |
Alignment:
MRGCARCWGSWG
-----GCTGCWG
| Original motif Consensus sequence: MRGCARCWGSWG |
Reverse complement motif Consensus sequence: CWSCWGMTGCKR |
|
|
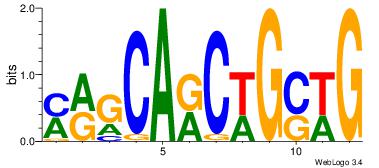 |
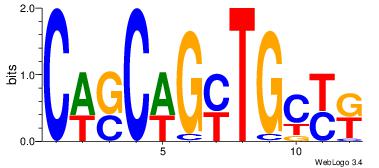 |
| Dataset #: |
1 |
| Motif ID: |
41 |
| Motif name: |
Motif 41 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.027698 |
Alignment:
YCAGCAGG
-CWGCAGC
| Original motif Consensus sequence: CCTGCTGK |
Reverse complement motif Consensus sequence: YCAGCAGG |
|
|
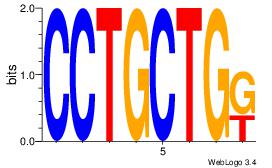 |
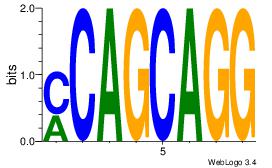 |
| Dataset #: |
3 |
| Motif ID: |
105 |
| Motif name: |
NHLH1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.032311 |
Alignment:
VCGCAGCTGCGV
CWGCAGC-----
| Original motif Consensus sequence: VCGCAGCTGCGB |
Reverse complement motif Consensus sequence: VCGCAGCTGCGV |
|
|
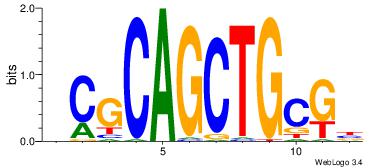 |
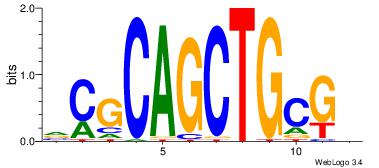 |
| Dataset #: |
1 |
| Motif ID: |
27 |
| Motif name: |
Motif 27 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.037602 |
Alignment:
GGCGGCSG
-GCTGCWG
| Original motif Consensus sequence: CSGCCGCC |
Reverse complement motif Consensus sequence: GGCGGCSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
151 |
| Motif name: |
agrCCAGmAGrg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
7 |
| Similarity score: |
0.0377961 |
Alignment:
CKCTRCTGGCVH
-GCTGCWG----
| Original motif Consensus sequence: HVGCCAGMAGRG |
Reverse complement motif Consensus sequence: CKCTRCTGGCVH |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 31 |
Motif name: Motif 31 |
| Original motif Consensus sequence: CATGYACA |
Reverse complement motif Consensus sequence: TGTKCATG |
|
|
 |
 |
Best Matches for Motif ID 31 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
125 |
| Motif name: |
TP53 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
8 |
| Similarity score: |
0.0607115 |
Alignment:
ACATGCCCGGKCATGTCCSR
-------TGTKCATG-----
| Original motif Consensus sequence: MSGGACATGYCCGGGCATGT |
Reverse complement motif Consensus sequence: ACATGCCCGGKCATGTCCSR |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
152 |
| Motif name: |
yrCATGCAyr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0609044 |
Alignment:
BRCATGCABD
--CATGYACA
| Original motif Consensus sequence: BRCATGCABD |
Reverse complement motif Consensus sequence: HVTGCATGKV |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
18 |
| Motif name: |
Motif 18 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0666244 |
Alignment:
CATGYATG
CATGYACA
| Original motif Consensus sequence: CATGYATG |
Reverse complement motif Consensus sequence: CATKCATG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
64 |
| Motif name: |
Motif 64 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0706964 |
Alignment:
CATATRCA
CATGYACA
| Original motif Consensus sequence: CATATRCA |
Reverse complement motif Consensus sequence: TGMATATG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
113 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
8 |
| Similarity score: |
0.0726387 |
Alignment:
TTCAGCACCATGGACAGCKCC
--------CATGYACA-----
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 32 |
Motif name: Motif 32 |
| Original motif Consensus sequence: KAATAAA |
Reverse complement motif Consensus sequence: TTTATTY |
|
|
 |
 |
Best Matches for Motif ID 32 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
52 |
| Motif name: |
Motif 52 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.017829 |
Alignment:
WTTTATTT
-TTTATTY
| Original motif Consensus sequence: AAATAAAW |
Reverse complement motif Consensus sequence: WTTTATTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0445453 |
Alignment:
TTTDTTTT
TTTATTY-
| Original motif Consensus sequence: AAAAHAAA |
Reverse complement motif Consensus sequence: TTTDTTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
68 |
| Motif name: |
Motif 68 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
7 |
| Similarity score: |
0.0470498 |
Alignment:
TTWWWTWTTTWTTW
-------TTTATTY
| Original motif Consensus sequence: WAAWAAAWAWWWAA |
Reverse complement motif Consensus sequence: TTWWWTWTTTWTTW |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
150 |
| Motif name: |
waATwAAAATAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0674341 |
Alignment:
DHTATTTTWATHD
------TTTATTY
| Original motif Consensus sequence: DHATWAAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTTWATHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
148 |
| Motif name: |
wwTwAAAAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.0811175 |
Alignment:
HHTTTTWADD
---TTTATTY
| Original motif Consensus sequence: DDTWAAAAHH |
Reverse complement motif Consensus sequence: HHTTTTWADD |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 33 |
Motif name: Motif 33 |
| Original motif Consensus sequence: CTGGCTWC |
Reverse complement motif Consensus sequence: GWAGCCAG |
|
|
 |
 |
Best Matches for Motif ID 33 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.011666 |
Alignment:
CTGGCCTC
CTGGCTWC
| Original motif Consensus sequence: CTGGCCTC |
Reverse complement motif Consensus sequence: GAGGCCAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.029479 |
Alignment:
CTGGCCTC
CTGGCTWC
| Original motif Consensus sequence: CTGGCCTC |
Reverse complement motif Consensus sequence: GAGGCCAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
25 |
| Motif name: |
Motif 25 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0400133 |
Alignment:
CTGRGTTC
CTGGCTWC
| Original motif Consensus sequence: CTGRGTTC |
Reverse complement motif Consensus sequence: GAACKCAG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.050611 |
Alignment:
KBKGCCCTCTYCTGGCCHV
-----------CTGGCTWC
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
133 |
| Motif name: |
shAGrGGGCAgy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0636793 |
Alignment:
DBTGCCCKCTDS
-CTGGCTWC---
| Original motif Consensus sequence: SHAGRGGGCABH |
Reverse complement motif Consensus sequence: DBTGCCCKCTDS |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 34 |
Motif name: Motif 34 |
| Original motif Consensus sequence: TGKCCACA |
Reverse complement motif Consensus sequence: TGTGGYCA |
|
|
 |
 |
Best Matches for Motif ID 34 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
171 |
| Motif name: |
ysGTGGCCACsr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0504663 |
Alignment:
VBGTGGCCACVB
---TGKCCACA-
| Original motif Consensus sequence: BVGTGGCCACBV |
Reverse complement motif Consensus sequence: VBGTGGCCACVB |
|
|
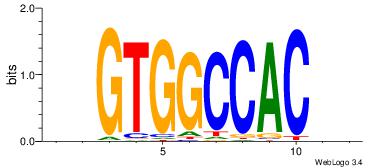 |
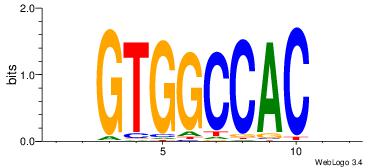 |
| Dataset #: |
3 |
| Motif ID: |
114 |
| Motif name: |
RUNX1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0690177 |
Alignment:
BBYTGTGGTTT
---TGTGGYCA
| Original motif Consensus sequence: BBYTGTGGTTT |
Reverse complement motif Consensus sequence: AAACCACAKVB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
165 |
| Motif name: |
wgGCCAshAGrGGGCrsy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
11 |
| Number of overlap: |
8 |
| Similarity score: |
0.083395 |
Alignment:
KBKGCCCKCTHGTGGCHH
----------TGTGGYCA
| Original motif Consensus sequence: HDGCCACHAGRGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCKCTHGTGGCHH |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
24 |
| Motif name: |
Motif 24 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0843302 |
Alignment:
CACCCACA
TGKCCACA
| Original motif Consensus sequence: TGTGGGTG |
Reverse complement motif Consensus sequence: CACCCACA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
12 |
| Number of overlap: |
8 |
| Similarity score: |
0.0872346 |
Alignment:
BMSMGCCYMCTKSTGGMHM
-----------TGTGGYCA
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 35 |
Motif name: Motif 35 |
| Original motif Consensus sequence: ACAACCAY |
Reverse complement motif Consensus sequence: KTGGTTGT |
|
|
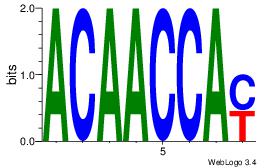 |
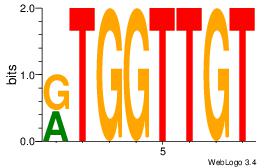 |
Best Matches for Motif ID 35 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
57 |
| Motif name: |
Motif 57 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0737654 |
Alignment:
ACACACAY
ACAACCAY
| Original motif Consensus sequence: ACACACAY |
Reverse complement motif Consensus sequence: KTGTGTGT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
113 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0887654 |
Alignment:
TTCAGCACCATGGACAGCKCC
---ACAACCAY----------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
120 |
| Motif name: |
T |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0890625 |
Alignment:
TTCACACCTAG
---ACAACCAY
| Original motif Consensus sequence: CTAGGTGTGAA |
Reverse complement motif Consensus sequence: TTCACACCTAG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
162 |
| Motif name: |
ccAsCCCCAcc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0921848 |
Alignment:
HVASCCCCABH
--ACAACCAY-
| Original motif Consensus sequence: HVASCCCCABH |
Reverse complement motif Consensus sequence: DBTGGGGSTVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
152 |
| Motif name: |
yrCATGCAyr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0932982 |
Alignment:
BRCATGCABD
-ACAACCAY-
| Original motif Consensus sequence: BRCATGCABD |
Reverse complement motif Consensus sequence: HVTGCATGKV |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 36 |
Motif name: Motif 36 |
| Original motif Consensus sequence: TCCTGGRA |
Reverse complement motif Consensus sequence: TMCCAGGA |
|
|
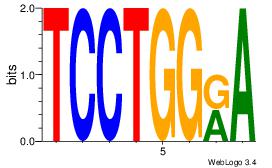 |
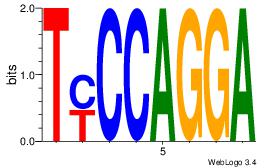 |
Best Matches for Motif ID 36 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
119 |
| Motif name: |
Stat3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0190833 |
Alignment:
CTTCCTGGAA
--TCCTGGRA
| Original motif Consensus sequence: TTCCAGGAAG |
Reverse complement motif Consensus sequence: CTTCCTGGAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
151 |
| Motif name: |
agrCCAGmAGrg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0609014 |
Alignment:
CKCTRCTGGCVH
---TCCTGGRA-
| Original motif Consensus sequence: HVGCCAGMAGRG |
Reverse complement motif Consensus sequence: CKCTRCTGGCVH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
131 |
| Motif name: |
znf143 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.061907 |
Alignment:
BAHYTCCCAKMATGCMWYGC
----TMCCAGGA--------
| Original motif Consensus sequence: BAHYTCCCAKMATGCMWYGC |
Reverse complement motif Consensus sequence: GCMWRGCATYRTGGGAMHTB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0667495 |
Alignment:
KBKGCCCTCTYCTGGCCHV
---------TCCTGGRA--
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
85 |
| Motif name: |
Hand1Tcfe2a |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0719458 |
Alignment:
AWRCCAGAMB
-TMCCAGGA-
| Original motif Consensus sequence: BRTCTGGMWT |
Reverse complement motif Consensus sequence: AWRCCAGAMB |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 37 |
Motif name: Motif 37 |
| Original motif Consensus sequence: CTGAGCYA |
Reverse complement motif Consensus sequence: TKGCTCAG |
|
|
 |
 |
Best Matches for Motif ID 37 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
100 |
| Motif name: |
NFE2L2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0542114 |
Alignment:
TGCTDWGTCAK
--CTGAGCYA-
| Original motif Consensus sequence: RTGACWHAGCA |
Reverse complement motif Consensus sequence: TGCTDWGTCAK |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
160 |
| Motif name: |
brCAGGGCCrs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0573364 |
Alignment:
BBGGCCCTGBB
-TKGCTCAG--
| Original motif Consensus sequence: BVCAGGGCCVB |
Reverse complement motif Consensus sequence: BBGGCCCTGBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
142 |
| Motif name: |
ctCTTAACyw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0660909 |
Alignment:
DDGTTAAGHD
TKGCTCAG--
| Original motif Consensus sequence: HHCTTAACHD |
Reverse complement motif Consensus sequence: DDGTTAAGHD |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
25 |
| Motif name: |
Motif 25 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0673416 |
Alignment:
CTGRGTTC
CTGAGCYA
| Original motif Consensus sequence: CTGRGTTC |
Reverse complement motif Consensus sequence: GAACKCAG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
111 |
| Motif name: |
PPARG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0687591 |
Alignment:
STAGGTCACBGTGACCYABT
----------CTGAGCYA--
| Original motif Consensus sequence: STAGGTCACBGTGACCYABT |
Reverse complement motif Consensus sequence: ABTMGGTCACBGTGACCTAS |
|
|
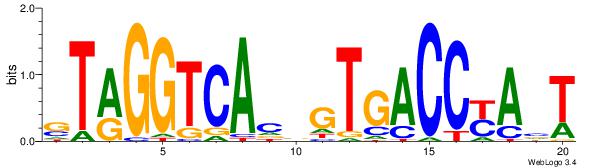 |
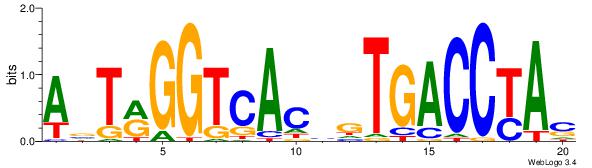 |
| Dataset #: 1 |
Motif ID: 38 |
Motif name: Motif 38 |
| Original motif Consensus sequence: GAGTTACA |
Reverse complement motif Consensus sequence: TGTAACTC |
|
|
 |
 |
Best Matches for Motif ID 38 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
51 |
| Motif name: |
Motif 51 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0 |
Alignment:
GAGTTACA
GAGTTACA
| Original motif Consensus sequence: GAGTTACA |
Reverse complement motif Consensus sequence: TGTAACTC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
66 |
| Motif name: |
Motif 66 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0742432 |
Alignment:
KATTTATA
GAGTTACA
| Original motif Consensus sequence: TATAAATR |
Reverse complement motif Consensus sequence: KATTTATA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
117 |
| Motif name: |
Spz1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0855279 |
Alignment:
AGGGTAWCAGC
-GAGTTACA--
| Original motif Consensus sequence: AGGGTAWCAGC |
Reverse complement motif Consensus sequence: GCTGWTACCCT |
|
|
 |
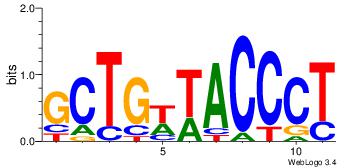 |
| Dataset #: |
2 |
| Motif ID: |
69 |
| Motif name: |
Motif 69 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0901267 |
Alignment:
CCTGGARCTGGAGT
--TGTAACTC----
| Original motif Consensus sequence: CCTGGARCTGGAGT |
Reverse complement motif Consensus sequence: ACTCCAGMTCCAGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
75 |
| Motif name: |
Ddit3Cebpa |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0913372 |
Alignment:
GGRATTGCAKHB
-GAGTTACA---
| Original motif Consensus sequence: VDRTGCAATMCC |
Reverse complement motif Consensus sequence: GGRATTGCAKHB |
|
|
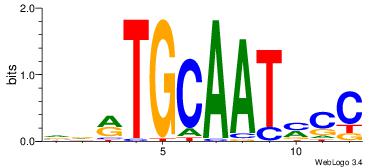 |
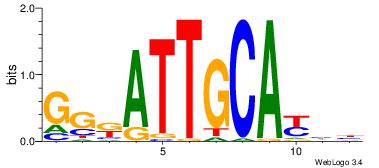 |
| Dataset #: 1 |
Motif ID: 39 |
Motif name: Motif 39 |
| Original motif Consensus sequence: BAGAAA |
Reverse complement motif Consensus sequence: TTTCTB |
|
|
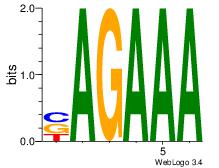 |
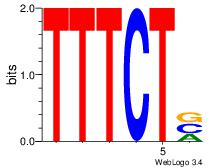 |
Best Matches for Motif ID 39 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
23 |
| Motif name: |
Motif 23 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0215381 |
Alignment:
GASAGAGA
--BAGAAA
| Original motif Consensus sequence: GASAGAGA |
Reverse complement motif Consensus sequence: TCTCTSTC |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
45 |
| Motif name: |
Motif 45 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0344566 |
Alignment:
GSAGAGA
-BAGAAA
| Original motif Consensus sequence: GSAGAGA |
Reverse complement motif Consensus sequence: TCTCTSC |
|
|
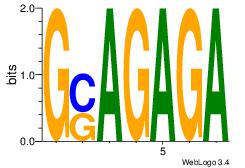 |
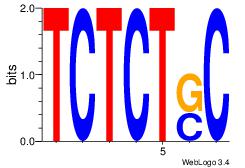 |
| Dataset #: |
4 |
| Motif ID: |
139 |
| Motif name: |
mkCTyTTCsg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0384118 |
Alignment:
HBCTTTTCBD
----TTTCTB
| Original motif Consensus sequence: HBCTTTTCBD |
Reverse complement motif Consensus sequence: HBGAAAAGBD |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0438306 |
Alignment:
TTTDTTTT
TTTCTB--
| Original motif Consensus sequence: AAAAHAAA |
Reverse complement motif Consensus sequence: TTTDTTTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
136 |
| Motif name: |
dwCAGAAGwh |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0496185 |
Alignment:
DHCAGAAGDH
--BAGAAA--
| Original motif Consensus sequence: DHCAGAAGDH |
Reverse complement motif Consensus sequence: HDCTTCTGHD |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 40 |
Motif name: Motif 40 |
| Original motif Consensus sequence: ARTCCCAG |
Reverse complement motif Consensus sequence: CTGGGAMT |
|
|
 |
 |
Best Matches for Motif ID 40 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
162 |
| Motif name: |
ccAsCCCCAcc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0511488 |
Alignment:
HVASCCCCABH
--ARTCCCAG-
| Original motif Consensus sequence: HVASCCCCABH |
Reverse complement motif Consensus sequence: DBTGGGGSTVD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
69 |
| Motif name: |
Motif 69 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0574096 |
Alignment:
CCTGGARCTGGAGT
-CTGGGAMT-----
| Original motif Consensus sequence: CCTGGARCTGGAGT |
Reverse complement motif Consensus sequence: ACTCCAGMTCCAGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
131 |
| Motif name: |
znf143 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
11 |
| Number of overlap: |
8 |
| Similarity score: |
0.0623563 |
Alignment:
GCMWRGCATYRTGGGAMHTB
----------CTGGGAMT--
| Original motif Consensus sequence: BAHYTCCCAKMATGCMWYGC |
Reverse complement motif Consensus sequence: GCMWRGCATYRTGGGAMHTB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
125 |
| Motif name: |
TP53 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
11 |
| Number of overlap: |
8 |
| Similarity score: |
0.0776137 |
Alignment:
MSGGACATGYCCGGGCATGT
----------CTGGGAMT--
| Original motif Consensus sequence: MSGGACATGYCCGGGCATGT |
Reverse complement motif Consensus sequence: ACATGCCCGGKCATGTCCSR |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
114 |
| Motif name: |
RUNX1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.080122 |
Alignment:
AAACCACAKVB
-ARTCCCAG--
| Original motif Consensus sequence: BBYTGTGGTTT |
Reverse complement motif Consensus sequence: AAACCACAKVB |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 41 |
Motif name: Motif 41 |
| Original motif Consensus sequence: CCTGCTGK |
Reverse complement motif Consensus sequence: YCAGCAGG |
|
|
 |
 |
Best Matches for Motif ID 41 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
151 |
| Motif name: |
agrCCAGmAGrg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.028602 |
Alignment:
HVGCCAGMAGRG
---YCAGCAGG-
| Original motif Consensus sequence: HVGCCAGMAGRG |
Reverse complement motif Consensus sequence: CKCTRCTGGCVH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
168 |
| Motif name: |
yrcrGYGCCMyCTGGtG |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0340048 |
Alignment:
CACCAGMGGGCGCTGBD
--YCAGCAGG-------
| Original motif Consensus sequence: HVCAGCGCCCYCTGGTG |
Reverse complement motif Consensus sequence: CACCAGMGGGCGCTGBD |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
21 |
| Motif name: |
Motif 21 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0342298 |
Alignment:
CCTKGTGG
CCTGCTGK
| Original motif Consensus sequence: CCACYAGG |
Reverse complement motif Consensus sequence: CCTKGTGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
166 |
| Motif name: |
CasCAGrGGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0366705 |
Alignment:
BSKGCCCKCTGGTG
----CCTGCTGK--
| Original motif Consensus sequence: CACCAGRGGGCRSB |
Reverse complement motif Consensus sequence: BSKGCCCKCTGGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0407982 |
Alignment:
VHGGCCAGMAGAGGGCRBY
----YCAGCAGG-------
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 42 |
Motif name: Motif 42 |
| Original motif Consensus sequence: CGCGCSG |
Reverse complement motif Consensus sequence: CSGCGCG |
|
|
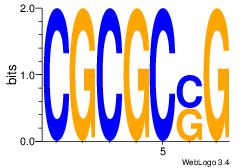 |
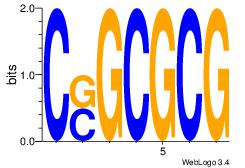 |
Best Matches for Motif ID 42 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
161 |
| Motif name: |
ssCGCwGCGss |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
7 |
| Similarity score: |
0.0226692 |
Alignment:
VSCGCDGCGSB
----CSGCGCG
| Original motif Consensus sequence: VSCGCDGCGSB |
Reverse complement motif Consensus sequence: BSCGCDGCGSV |
|
|
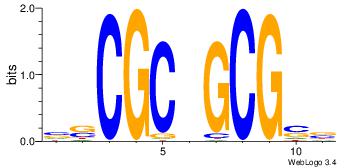 |
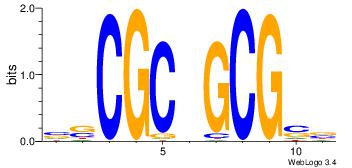 |
| Dataset #: |
4 |
| Motif ID: |
134 |
| Motif name: |
ssCGwGCGss |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0315768 |
Alignment:
BSCGWGCGBV
-CSGCGCG--
| Original motif Consensus sequence: BSCGWGCGBV |
Reverse complement motif Consensus sequence: VBCGCWCGSB |
|
|
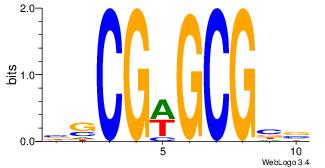 |
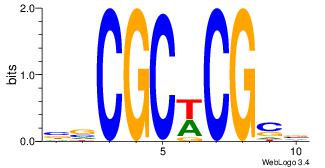 |
| Dataset #: |
4 |
| Motif ID: |
146 |
| Motif name: |
myrGYGCCmCCTast |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0.0331622 |
Alignment:
ASTAGGYGGCGCTBB
------CSGCGCG--
| Original motif Consensus sequence: VBAGCGCCMCCTAST |
Reverse complement motif Consensus sequence: ASTAGGYGGCGCTBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
168 |
| Motif name: |
yrcrGYGCCMyCTGGtG |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0.0399606 |
Alignment:
HVCAGCGCCCYCTGGTG
--CSGCGCG--------
| Original motif Consensus sequence: HVCAGCGCCCYCTGGTG |
Reverse complement motif Consensus sequence: CACCAGMGGGCGCTGBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
170 |
| Motif name: |
ssGGCrsTGCrs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.0456255 |
Alignment:
BVGCASMGCCVV
---CSGCGCG--
| Original motif Consensus sequence: VVGGCRSTGCVB |
Reverse complement motif Consensus sequence: BVGCASMGCCVV |
|
|
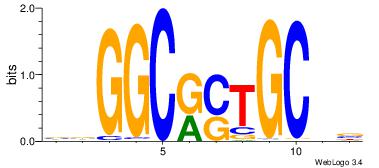 |
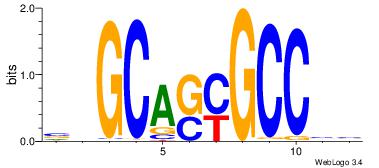 |
| Dataset #: 1 |
Motif ID: 43 |
Motif name: Motif 43 |
| Original motif Consensus sequence: ACATYTA |
Reverse complement motif Consensus sequence: TAMATGT |
|
|
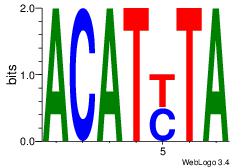 |
 |
Best Matches for Motif ID 43 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
120 |
| Motif name: |
T |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.0260628 |
Alignment:
TTCACACCTAG
---ACATYTA-
| Original motif Consensus sequence: CTAGGTGTGAA |
Reverse complement motif Consensus sequence: TTCACACCTAG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
58 |
| Motif name: |
Motif 58 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0342454 |
Alignment:
TACATGCA
TAMATGT-
| Original motif Consensus sequence: TACATGCA |
Reverse complement motif Consensus sequence: TGCATGTA |
|
|
 |
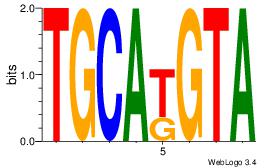 |
| Dataset #: |
2 |
| Motif ID: |
64 |
| Motif name: |
Motif 64 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0444069 |
Alignment:
TGMATATG
TAMATGT-
| Original motif Consensus sequence: CATATRCA |
Reverse complement motif Consensus sequence: TGMATATG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
152 |
| Motif name: |
yrCATGCAyr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0.0458993 |
Alignment:
HVTGCATGKV
--TAMATGT-
| Original motif Consensus sequence: BRCATGCABD |
Reverse complement motif Consensus sequence: HVTGCATGKV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
121 |
| Motif name: |
TAL1TCF3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.0525238 |
Alignment:
HVAMCATCTGKT
---ACATYTA--
| Original motif Consensus sequence: HVAMCATCTGKT |
Reverse complement motif Consensus sequence: ARCAGATGRTVD |
|
|
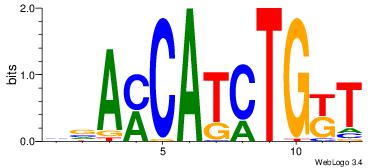 |
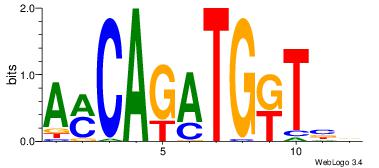 |
| Dataset #: 1 |
Motif ID: 44 |
Motif name: Motif 44 |
| Original motif Consensus sequence: GDAAACA |
Reverse complement motif Consensus sequence: TGTTTDC |
|
|
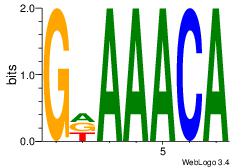 |
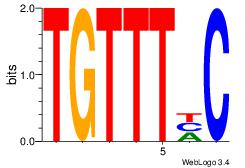 |
Best Matches for Motif ID 44 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
141 |
| Motif name: |
raCAAAACam |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0588603 |
Alignment:
DACAAAACAH
--GDAAACA-
| Original motif Consensus sequence: DACAAAACAH |
Reverse complement motif Consensus sequence: HTGTTTTGTD |
|
|
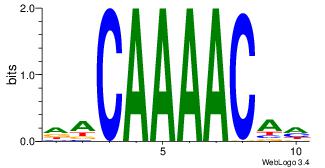 |
 |
| Dataset #: |
2 |
| Motif ID: |
68 |
| Motif name: |
Motif 68 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
7 |
| Similarity score: |
0.0651624 |
Alignment:
WAAWAAAWAWWWAA
--GDAAACA-----
| Original motif Consensus sequence: WAAWAAAWAWWWAA |
Reverse complement motif Consensus sequence: TTWWWTWTTTWTTW |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0667857 |
Alignment:
CTGGCTWC
TGTTTDC-
| Original motif Consensus sequence: CTGGCTWC |
Reverse complement motif Consensus sequence: GWAGCCAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
34 |
| Motif name: |
Motif 34 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.066941 |
Alignment:
TGKCCACA
-GDAAACA
| Original motif Consensus sequence: TGKCCACA |
Reverse complement motif Consensus sequence: TGTGGYCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
57 |
| Motif name: |
Motif 57 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0694703 |
Alignment:
KTGTGTGT
-TGTTTDC
| Original motif Consensus sequence: ACACACAY |
Reverse complement motif Consensus sequence: KTGTGTGT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 45 |
Motif name: Motif 45 |
| Original motif Consensus sequence: GSAGAGA |
Reverse complement motif Consensus sequence: TCTCTSC |
|
|
 |
 |
Best Matches for Motif ID 45 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
140 |
| Motif name: |
vkCKCTkCGk |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0315852 |
Alignment:
BCGYAGAGDV
--GSAGAGA-
| Original motif Consensus sequence: VDCTCTKCGB |
Reverse complement motif Consensus sequence: BCGYAGAGDV |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
23 |
| Motif name: |
Motif 23 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0364931 |
Alignment:
TCTCTSTC
TCTCTSC-
| Original motif Consensus sequence: GASAGAGA |
Reverse complement motif Consensus sequence: TCTCTSTC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
7 |
| Similarity score: |
0.0625376 |
Alignment:
VHGGCCAGMAGAGGGCRBY
-------GSAGAGA-----
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
143 |
| Motif name: |
AgmAGAGGGCrscAGak |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
10 |
| Number of overlap: |
7 |
| Similarity score: |
0.0633827 |
Alignment:
RTCTGSTGCCCTCTYCT
---------TCTCTSC-
| Original motif Consensus sequence: AGMAGAGGGCASCAGAK |
Reverse complement motif Consensus sequence: RTCTGSTGCCCTCTYCT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
139 |
| Motif name: |
mkCTyTTCsg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0693299 |
Alignment:
HBCTTTTCBD
-TCTCTSC--
| Original motif Consensus sequence: HBCTTTTCBD |
Reverse complement motif Consensus sequence: HBGAAAAGBD |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 46 |
Motif name: Motif 46 |
| Original motif Consensus sequence: AGRKGGCR |
Reverse complement motif Consensus sequence: KGCCYKCT |
|
|
 |
 |
Best Matches for Motif ID 46 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
1 |
| Motif name: |
Motif 1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0 |
Alignment:
AGRKGGCR
AGRKGGCR
| Original motif Consensus sequence: AGRKGGCR |
Reverse complement motif Consensus sequence: KGCCYKCT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
147 |
| Motif name: |
asCAGrkGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.00651548 |
Alignment:
ASCAGRGGGCRSB
---AGRKGGCR--
| Original motif Consensus sequence: ASCAGRGGGCRSB |
Reverse complement motif Consensus sequence: BSKGCCCMCTGST |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
165 |
| Motif name: |
wgGCCAshAGrGGGCrsy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
8 |
| Similarity score: |
0.0102443 |
Alignment:
KBKGCCCKCTHGTGGCHH
--KGCCYKCT--------
| Original motif Consensus sequence: HDGCCACHAGRGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCKCTHGTGGCHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
166 |
| Motif name: |
CasCAGrGGGCrsy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0104954 |
Alignment:
BSKGCCCKCTGGTG
--KGCCYKCT----
| Original motif Consensus sequence: CACCAGRGGGCRSB |
Reverse complement motif Consensus sequence: BSKGCCCKCTGGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0165822 |
Alignment:
KBKGCCCTCTYCTGGCCHV
--KGCCYKCT---------
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 47 |
Motif name: Motif 47 |
| Original motif Consensus sequence: CAGYDCC |
Reverse complement motif Consensus sequence: GGDKCTG |
|
|
 |
 |
Best Matches for Motif ID 47 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
13 |
| Motif name: |
Motif 13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0 |
Alignment:
GGDKCTG
GGDKCTG
| Original motif Consensus sequence: CAGYDCC |
Reverse complement motif Consensus sequence: GGDKCTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
53 |
| Motif name: |
Motif 53 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0288002 |
Alignment:
GGGATCTG
-GGDKCTG
| Original motif Consensus sequence: CAGATCCC |
Reverse complement motif Consensus sequence: GGGATCTG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
113 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
13 |
| Number of overlap: |
7 |
| Similarity score: |
0.0502666 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
------------GGDKCTG--
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
22 |
| Motif name: |
Motif 22 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0531399 |
Alignment:
GGKATCTG
-GGDKCTG
| Original motif Consensus sequence: CAGATYCC |
Reverse complement motif Consensus sequence: GGKATCTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
160 |
| Motif name: |
brCAGGGCCrs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0.0559697 |
Alignment:
BVCAGGGCCVB
--CAGYDCC--
| Original motif Consensus sequence: BVCAGGGCCVB |
Reverse complement motif Consensus sequence: BBGGCCCTGBB |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 48 |
Motif name: Motif 48 |
| Original motif Consensus sequence: CCACYAGR |
Reverse complement motif Consensus sequence: MCTKGTGG |
|
|
 |
 |
Best Matches for Motif ID 48 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
21 |
| Motif name: |
Motif 21 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.00903109 |
Alignment:
CCACYAGG
CCACYAGR
| Original motif Consensus sequence: CCACYAGG |
Reverse complement motif Consensus sequence: CCTKGTGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
145 |
| Motif name: |
grCCACyAGAkG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0155354 |
Alignment:
DDCCACYAGAKG
--CCACYAGR--
| Original motif Consensus sequence: DDCCACYAGAKG |
Reverse complement motif Consensus sequence: CYTCTKGTGGHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
158 |
| Motif name: |
grCCACwAGrk |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0192652 |
Alignment:
DDCCACWAGRK
--CCACYAGR-
| Original motif Consensus sequence: DDCCACWAGRK |
Reverse complement motif Consensus sequence: YMCTWGTGGHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
138 |
| Motif name: |
grCCACyAGAkG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0244381 |
Alignment:
DDCCACYAGAKG
--CCACYAGR--
| Original motif Consensus sequence: DDCCACYAGAKG |
Reverse complement motif Consensus sequence: CYTCTMGTGGHH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
8 |
| Similarity score: |
0.0284404 |
Alignment:
BMSMGCCYMCTKSTGGMHM
--------MCTKGTGG---
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 49 |
Motif name: Motif 49 |
| Original motif Consensus sequence: CTGGCCTC |
Reverse complement motif Consensus sequence: GAGGCCAG |
|
|
 |
 |
Best Matches for Motif ID 49 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0 |
Alignment:
CTGGCCTC
CTGGCCTC
| Original motif Consensus sequence: CTGGCCTC |
Reverse complement motif Consensus sequence: GAGGCCAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0148758 |
Alignment:
CTGGCTWC
CTGGCCTC
| Original motif Consensus sequence: CTGGCTWC |
Reverse complement motif Consensus sequence: GWAGCCAG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
12 |
| Number of overlap: |
8 |
| Similarity score: |
0.0362649 |
Alignment:
VHGGCCAGMAGAGGGCRBY
GAGGCCAG-----------
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
171 |
| Motif name: |
ysGTGGCCACsr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0504205 |
Alignment:
VBGTGGCCACVB
--CTGGCCTC--
| Original motif Consensus sequence: BVGTGGCCACBV |
Reverse complement motif Consensus sequence: VBGTGGCCACVB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
133 |
| Motif name: |
shAGrGGGCAgy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0509007 |
Alignment:
SHAGRGGGCABH
---GAGGCCAG-
| Original motif Consensus sequence: SHAGRGGGCABH |
Reverse complement motif Consensus sequence: DBTGCCCKCTDS |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 50 |
Motif name: Motif 50 |
| Original motif Consensus sequence: AGRAA |
Reverse complement motif Consensus sequence: TTMCT |
|
|
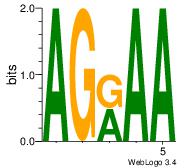 |
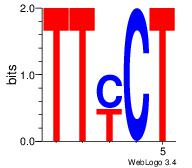 |
Best Matches for Motif ID 50 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
5 |
| Similarity score: |
0 |
Alignment:
AGRAAA
AGRAA-
| Original motif Consensus sequence: AGRAAA |
Reverse complement motif Consensus sequence: TTTMCT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
39 |
| Motif name: |
Motif 39 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
5 |
| Similarity score: |
0.0244128 |
Alignment:
BAGAAA
-AGRAA
| Original motif Consensus sequence: BAGAAA |
Reverse complement motif Consensus sequence: TTTCTB |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
28 |
| Motif name: |
Motif 28 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
5 |
| Similarity score: |
0.0419638 |
Alignment:
CAGGAAG
-AGRAA-
| Original motif Consensus sequence: CTTCCTG |
Reverse complement motif Consensus sequence: CAGGAAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
29 |
| Motif name: |
Motif 29 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
5 |
| Similarity score: |
0.0427966 |
Alignment:
CATTTCY
TTMCT--
| Original motif Consensus sequence: CATTTCY |
Reverse complement motif Consensus sequence: MGAAATG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
119 |
| Motif name: |
Stat3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
5 |
| Similarity score: |
0.0502276 |
Alignment:
TTCCAGGAAG
----AGRAA-
| Original motif Consensus sequence: TTCCAGGAAG |
Reverse complement motif Consensus sequence: CTTCCTGGAA |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 51 |
Motif name: Motif 51 |
| Original motif Consensus sequence: GAGTTACA |
Reverse complement motif Consensus sequence: TGTAACTC |
|
|
 |
 |
Best Matches for Motif ID 51 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
38 |
| Motif name: |
Motif 38 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0 |
Alignment:
GAGTTACA
GAGTTACA
| Original motif Consensus sequence: GAGTTACA |
Reverse complement motif Consensus sequence: TGTAACTC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
66 |
| Motif name: |
Motif 66 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0742432 |
Alignment:
KATTTATA
GAGTTACA
| Original motif Consensus sequence: TATAAATR |
Reverse complement motif Consensus sequence: KATTTATA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
75 |
| Motif name: |
Ddit3Cebpa |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0857192 |
Alignment:
GGRATTGCAKHB
-GAGTTACA---
| Original motif Consensus sequence: VDRTGCAATMCC |
Reverse complement motif Consensus sequence: GGRATTGCAKHB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
117 |
| Motif name: |
Spz1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0885417 |
Alignment:
AGGGTAWCAGC
-GAGTTACA--
| Original motif Consensus sequence: AGGGTAWCAGC |
Reverse complement motif Consensus sequence: GCTGWTACCCT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
112 |
| Motif name: |
RELA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0911946 |
Alignment:
GGGRATTTCC
--GAGTTACA
| Original motif Consensus sequence: GGGRATTTCC |
Reverse complement motif Consensus sequence: GGAAATKCCC |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 52 |
Motif name: Motif 52 |
| Original motif Consensus sequence: AAATAAAW |
Reverse complement motif Consensus sequence: WTTTATTT |
|
|
 |
 |
Best Matches for Motif ID 52 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
68 |
| Motif name: |
Motif 68 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0178205 |
Alignment:
TTWWWTWTTTWTTW
------WTTTATTT
| Original motif Consensus sequence: WAAWAAAWAWWWAA |
Reverse complement motif Consensus sequence: TTWWWTWTTTWTTW |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
150 |
| Motif name: |
waATwAAAATAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0502803 |
Alignment:
DHTATTTTWATHD
-----WTTTATTT
| Original motif Consensus sequence: DHATWAAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTTWATHD |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0573886 |
Alignment:
AAAAHAAA
AAATAAAW
| Original motif Consensus sequence: AAAAHAAA |
Reverse complement motif Consensus sequence: TTTDTTTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
148 |
| Motif name: |
wwTwAAAAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0617903 |
Alignment:
HHTTTTWADD
--WTTTATTT
| Original motif Consensus sequence: DDTWAAAAHH |
Reverse complement motif Consensus sequence: HHTTTTWADD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
157 |
| Motif name: |
wtATTTTTAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0718936 |
Alignment:
DTATTTTTAWW
--WTTTATTT-
| Original motif Consensus sequence: DTATTTTTAWW |
Reverse complement motif Consensus sequence: WWTAAAAATAD |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 53 |
Motif name: Motif 53 |
| Original motif Consensus sequence: CAGATCCC |
Reverse complement motif Consensus sequence: GGGATCTG |
|
|
 |
 |
Best Matches for Motif ID 53 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
22 |
| Motif name: |
Motif 22 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0 |
Alignment:
CAGATYCC
CAGATCCC
| Original motif Consensus sequence: CAGATYCC |
Reverse complement motif Consensus sequence: GGKATCTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
149 |
| Motif name: |
asmAGRGGGCrCTGsmkc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.0479917 |
Alignment:
DBTSCAGMGCCCTCTRST
----CAGATCCC------
| Original motif Consensus sequence: ASMAGAGGGCRCTGSABH |
Reverse complement motif Consensus sequence: DBTSCAGMGCCCTCTRST |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
160 |
| Motif name: |
brCAGGGCCrs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0605671 |
Alignment:
BBGGCCCTGBB
-GGGATCTG--
| Original motif Consensus sequence: BVCAGGGCCVB |
Reverse complement motif Consensus sequence: BBGGCCCTGBB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
69 |
| Motif name: |
Motif 69 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0606088 |
Alignment:
ACTCCAGMTCCAGG
----CAGATCCC--
| Original motif Consensus sequence: CCTGGARCTGGAGT |
Reverse complement motif Consensus sequence: ACTCCAGMTCCAGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
168 |
| Motif name: |
yrcrGYGCCMyCTGGtG |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.065385 |
Alignment:
CACCAGMGGGCGCTGBD
-------GGGATCTG--
| Original motif Consensus sequence: HVCAGCGCCCYCTGGTG |
Reverse complement motif Consensus sequence: CACCAGMGGGCGCTGBD |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 54 |
Motif name: Motif 54 |
| Original motif Consensus sequence: ATTYY |
Reverse complement motif Consensus sequence: MKAAT |
|
|
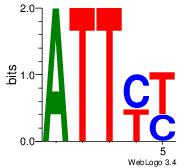 |
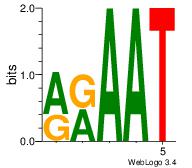 |
Best Matches for Motif ID 54 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
157 |
| Motif name: |
wtATTTTTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
5 |
| Similarity score: |
0.0491344 |
Alignment:
DTATTTTTAWW
--ATTYY----
| Original motif Consensus sequence: DTATTTTTAWW |
Reverse complement motif Consensus sequence: WWTAAAAATAD |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
29 |
| Motif name: |
Motif 29 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
5 |
| Similarity score: |
0.0551561 |
Alignment:
CATTTCY
-ATTYY-
| Original motif Consensus sequence: CATTTCY |
Reverse complement motif Consensus sequence: MGAAATG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
150 |
| Motif name: |
waATwAAAATAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
5 |
| Similarity score: |
0.0567288 |
Alignment:
DHTATTTTWATHD
---ATTYY-----
| Original motif Consensus sequence: DHATWAAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTTWATHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
148 |
| Motif name: |
wwTwAAAAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
5 |
| Similarity score: |
0.0727457 |
Alignment:
HHTTTTWADD
-ATTYY----
| Original motif Consensus sequence: DDTWAAAAHH |
Reverse complement motif Consensus sequence: HHTTTTWADD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
131 |
| Motif name: |
znf143 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
5 |
| Similarity score: |
0.0737392 |
Alignment:
BAHYTCCCAKMATGCMWYGC
--------MKAAT-------
| Original motif Consensus sequence: BAHYTCCCAKMATGCMWYGC |
Reverse complement motif Consensus sequence: GCMWRGCATYRTGGGAMHTB |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 55 |
Motif name: Motif 55 |
| Original motif Consensus sequence: GCTCACAA |
Reverse complement motif Consensus sequence: TTGTGAGC |
|
|
 |
 |
Best Matches for Motif ID 55 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.071643 |
Alignment:
GCTCTTAA
GCTCACAA
| Original motif Consensus sequence: GCTCTTAA |
Reverse complement motif Consensus sequence: TTAAGAGC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
57 |
| Motif name: |
Motif 57 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0907452 |
Alignment:
KTGTGTGT
TTGTGAGC
| Original motif Consensus sequence: ACACACAY |
Reverse complement motif Consensus sequence: KTGTGTGT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
145 |
| Motif name: |
grCCACyAGAkG |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0949146 |
Alignment:
CYTCTKGTGGHH
----TTGTGAGC
| Original motif Consensus sequence: DDCCACYAGAKG |
Reverse complement motif Consensus sequence: CYTCTKGTGGHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
158 |
| Motif name: |
grCCACwAGrk |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0951691 |
Alignment:
DDCCACWAGRK
GCTCACAA---
| Original motif Consensus sequence: DDCCACWAGRK |
Reverse complement motif Consensus sequence: YMCTWGTGGHH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
76 |
| Motif name: |
E2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.103125 |
Alignment:
GCGCSAAA
GCTCACAA
| Original motif Consensus sequence: TTTSGCGC |
Reverse complement motif Consensus sequence: GCGCSAAA |
|
|
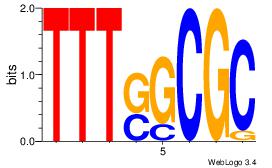 |
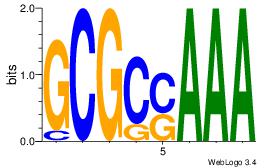 |
| Dataset #: 2 |
Motif ID: 56 |
Motif name: Motif 56 |
| Original motif Consensus sequence: CCACATGG |
Reverse complement motif Consensus sequence: CCATGTGG |
|
|
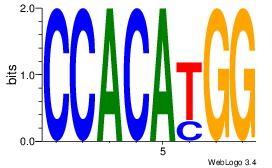 |
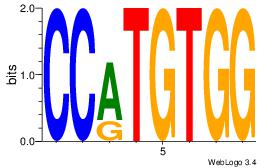 |
Best Matches for Motif ID 56 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
21 |
| Motif name: |
Motif 21 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0480363 |
Alignment:
CCTKGTGG
CCATGTGG
| Original motif Consensus sequence: CCACYAGG |
Reverse complement motif Consensus sequence: CCTKGTGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
48 |
| Motif name: |
Motif 48 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0609421 |
Alignment:
MCTKGTGG
CCATGTGG
| Original motif Consensus sequence: CCACYAGR |
Reverse complement motif Consensus sequence: MCTKGTGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
95 |
| Motif name: |
MYCMAX |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0616071 |
Alignment:
RASCACGTGGT
--CCACATGG-
| Original motif Consensus sequence: RASCACGTGGT |
Reverse complement motif Consensus sequence: ACCACGTGSTM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
96 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0634989 |
Alignment:
HSCACGTGGC
-CCACATGG-
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
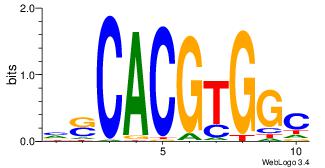 |
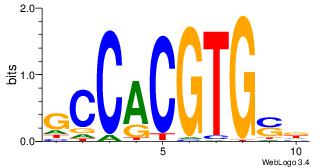 |
| Dataset #: |
4 |
| Motif ID: |
158 |
| Motif name: |
grCCACwAGrk |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0666667 |
Alignment:
DDCCACWAGRK
--CCACATGG-
| Original motif Consensus sequence: DDCCACWAGRK |
Reverse complement motif Consensus sequence: YMCTWGTGGHH |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 57 |
Motif name: Motif 57 |
| Original motif Consensus sequence: ACACACAY |
Reverse complement motif Consensus sequence: KTGTGTGT |
|
|
 |
 |
Best Matches for Motif ID 57 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
35 |
| Motif name: |
Motif 35 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0591808 |
Alignment:
ACAACCAY
ACACACAY
| Original motif Consensus sequence: ACAACCAY |
Reverse complement motif Consensus sequence: KTGGTTGT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
114 |
| Motif name: |
RUNX1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0712868 |
Alignment:
BBYTGTGGTTT
--KTGTGTGT-
| Original motif Consensus sequence: BBYTGTGGTTT |
Reverse complement motif Consensus sequence: AAACCACAKVB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
120 |
| Motif name: |
T |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0758601 |
Alignment:
TTCACACCTAG
-ACACACAY--
| Original motif Consensus sequence: CTAGGTGTGAA |
Reverse complement motif Consensus sequence: TTCACACCTAG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
55 |
| Motif name: |
Motif 55 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0761605 |
Alignment:
TTGTGAGC
KTGTGTGT
| Original motif Consensus sequence: GCTCACAA |
Reverse complement motif Consensus sequence: TTGTGAGC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
162 |
| Motif name: |
ccAsCCCCAcc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0776002 |
Alignment:
HVASCCCCABH
--ACACACAY-
| Original motif Consensus sequence: HVASCCCCABH |
Reverse complement motif Consensus sequence: DBTGGGGSTVD |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 58 |
Motif name: Motif 58 |
| Original motif Consensus sequence: TACATGCA |
Reverse complement motif Consensus sequence: TGCATGTA |
|
|
 |
 |
Best Matches for Motif ID 58 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
152 |
| Motif name: |
yrCATGCAyr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0160706 |
Alignment:
HVTGCATGKV
--TGCATGTA
| Original motif Consensus sequence: BRCATGCABD |
Reverse complement motif Consensus sequence: HVTGCATGKV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
125 |
| Motif name: |
TP53 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0595681 |
Alignment:
ACATGCCCGGKCATGTCCSR
---------TGCATGTA---
| Original motif Consensus sequence: MSGGACATGYCCGGGCATGT |
Reverse complement motif Consensus sequence: ACATGCCCGGKCATGTCCSR |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
64 |
| Motif name: |
Motif 64 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0642891 |
Alignment:
CATATRCA
TACATGCA
| Original motif Consensus sequence: CATATRCA |
Reverse complement motif Consensus sequence: TGMATATG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
36 |
| Motif name: |
Motif 36 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0792922 |
Alignment:
TCCTGGRA
TGCATGTA
| Original motif Consensus sequence: TCCTGGRA |
Reverse complement motif Consensus sequence: TMCCAGGA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
95 |
| Motif name: |
MYCMAX |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0822408 |
Alignment:
RASCACGTGGT
-TGCATGTA--
| Original motif Consensus sequence: RASCACGTGGT |
Reverse complement motif Consensus sequence: ACCACGTGSTM |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 59 |
Motif name: Motif 59 |
| Original motif Consensus sequence: CCCCRCCC |
Reverse complement motif Consensus sequence: GGGKGGGG |
|
|
 |
 |
Best Matches for Motif ID 59 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.00363191 |
Alignment:
CCCCDCCC
CCCCRCCC
| Original motif Consensus sequence: CCCCDCCC |
Reverse complement motif Consensus sequence: GGGDGGGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
155 |
| Motif name: |
csCSCCdCCCcs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0143924 |
Alignment:
VDGGGDGGGGBV
--GGGKGGGG--
| Original motif Consensus sequence: VBCCCCDCCCHV |
Reverse complement motif Consensus sequence: VDGGGDGGGGBV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
89 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0334874 |
Alignment:
GCCYCMCCCD
-CCCCRCCC-
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
116 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0421627 |
Alignment:
GGGGGYGGGG
--GGGKGGGG
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
154 |
| Motif name: |
csCsCCTCCcc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0467112 |
Alignment:
VBCCCCTCCHB
--CCCCRCCC-
| Original motif Consensus sequence: VBCCCCTCCHB |
Reverse complement motif Consensus sequence: BDGGAGGGGBV |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 60 |
Motif name: Motif 60 |
| Original motif Consensus sequence: CTGGAR |
Reverse complement motif Consensus sequence: MTCCAG |
|
|
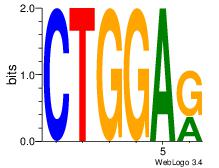 |
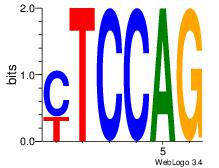 |
Best Matches for Motif ID 60 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.00500479 |
Alignment:
CTGGRRA
CTGGAR-
| Original motif Consensus sequence: CTGGRRA |
Reverse complement motif Consensus sequence: TMMCCAG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
69 |
| Motif name: |
Motif 69 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
6 |
| Similarity score: |
0.0149252 |
Alignment:
CCTGGARCTGGAGT
-------CTGGAR-
| Original motif Consensus sequence: CCTGGARCTGGAGT |
Reverse complement motif Consensus sequence: ACTCCAGMTCCAGG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
36 |
| Motif name: |
Motif 36 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0273394 |
Alignment:
TCCTGGRA
--CTGGAR
| Original motif Consensus sequence: TCCTGGRA |
Reverse complement motif Consensus sequence: TMCCAGGA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
151 |
| Motif name: |
agrCCAGmAGrg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
6 |
| Similarity score: |
0.0324331 |
Alignment:
CKCTRCTGGCVH
-----CTGGAR-
| Original motif Consensus sequence: HVGCCAGMAGRG |
Reverse complement motif Consensus sequence: CKCTRCTGGCVH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
85 |
| Motif name: |
Hand1Tcfe2a |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0370865 |
Alignment:
BRTCTGGMWT
---CTGGAR-
| Original motif Consensus sequence: BRTCTGGMWT |
Reverse complement motif Consensus sequence: AWRCCAGAMB |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 61 |
Motif name: Motif 61 |
| Original motif Consensus sequence: AGATGGY |
Reverse complement motif Consensus sequence: KCCATCT |
|
|
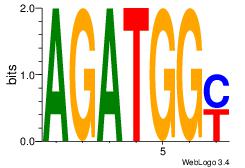 |
 |
Best Matches for Motif ID 61 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
63 |
| Motif name: |
Motif 63 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0339703 |
Alignment:
CWCCCTCT
-KCCATCT
| Original motif Consensus sequence: AGAGGGWG |
Reverse complement motif Consensus sequence: CWCCCTCT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
121 |
| Motif name: |
TAL1TCF3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.034782 |
Alignment:
ARCAGATGRTVD
---AGATGGY--
| Original motif Consensus sequence: HVAMCATCTGKT |
Reverse complement motif Consensus sequence: ARCAGATGRTVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.0365189 |
Alignment:
VHGGCCAGMAGAGGGCRBY
---------AGATGGY---
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
46 |
| Motif name: |
Motif 46 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0371728 |
Alignment:
AGRKGGCR
AGATGGY-
| Original motif Consensus sequence: AGRKGGCR |
Reverse complement motif Consensus sequence: KGCCYKCT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
1 |
| Motif name: |
Motif 1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0375558 |
Alignment:
AGRKGGCR
AGATGGY-
| Original motif Consensus sequence: AGRKGGCR |
Reverse complement motif Consensus sequence: KGCCYKCT |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 62 |
Motif name: Motif 62 |
| Original motif Consensus sequence: CGCCVCC |
Reverse complement motif Consensus sequence: GGVGGCG |
|
|
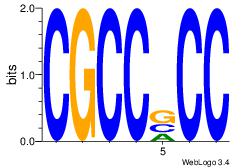 |
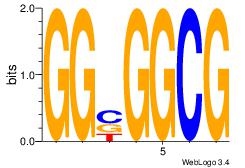 |
Best Matches for Motif ID 62 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
146 |
| Motif name: |
myrGYGCCmCCTast |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
7 |
| Similarity score: |
0.0315476 |
Alignment:
VBAGCGCCMCCTAST
----CGCCVCC----
| Original motif Consensus sequence: VBAGCGCCMCCTAST |
Reverse complement motif Consensus sequence: ASTAGGYGGCGCTBB |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
27 |
| Motif name: |
Motif 27 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0348335 |
Alignment:
CSGCCGCC
-CGCCVCC
| Original motif Consensus sequence: CSGCCGCC |
Reverse complement motif Consensus sequence: GGCGGCSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
137 |
| Motif name: |
rgCGCCmyCTgs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.0357527 |
Alignment:
SHAGKGGGCGCB
---GGVGGCG--
| Original motif Consensus sequence: VGCGCCCYCTDS |
Reverse complement motif Consensus sequence: SHAGKGGGCGCB |
|
|
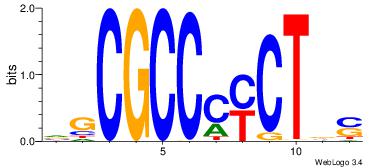 |
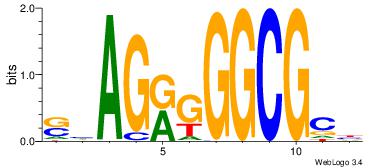 |
| Dataset #: |
4 |
| Motif ID: |
153 |
| Motif name: |
scAGrkGGCGcy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0.0403279 |
Alignment:
VGCGCCCMCTDS
--CGCCVCC---
| Original motif Consensus sequence: SHAGRGGGCGCB |
Reverse complement motif Consensus sequence: VGCGCCCMCTDS |
|
|
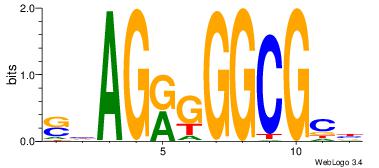 |
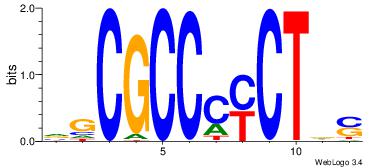 |
| Dataset #: |
4 |
| Motif ID: |
155 |
| Motif name: |
csCSCCdCCCcs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0.0439081 |
Alignment:
VBCCCCDCCCHV
--CGCCVCC---
| Original motif Consensus sequence: VBCCCCDCCCHV |
Reverse complement motif Consensus sequence: VDGGGDGGGGBV |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 63 |
Motif name: Motif 63 |
| Original motif Consensus sequence: AGAGGGWG |
Reverse complement motif Consensus sequence: CWCCCTCT |
|
|
 |
 |
Best Matches for Motif ID 63 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0323316 |
Alignment:
MGAGGBAG
AGAGGGWG
| Original motif Consensus sequence: CTBCCTCY |
Reverse complement motif Consensus sequence: MGAGGBAG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
149 |
| Motif name: |
asmAGRGGGCrCTGsmkc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
8 |
| Similarity score: |
0.040911 |
Alignment:
DBTSCAGMGCCCTCTRST
-------CWCCCTCT---
| Original motif Consensus sequence: ASMAGAGGGCRCTGSABH |
Reverse complement motif Consensus sequence: DBTSCAGMGCCCTCTRST |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
153 |
| Motif name: |
scAGrkGGCGcy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0435349 |
Alignment:
SHAGRGGGCGCB
--AGAGGGWG--
| Original motif Consensus sequence: SHAGRGGGCGCB |
Reverse complement motif Consensus sequence: VGCGCCCMCTDS |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0462891 |
Alignment:
VHGGCCAGMAGAGGGCRBY
---------AGAGGGWG--
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
137 |
| Motif name: |
rgCGCCmyCTgs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0481673 |
Alignment:
VGCGCCCYCTDS
--CWCCCTCT--
| Original motif Consensus sequence: VGCGCCCYCTDS |
Reverse complement motif Consensus sequence: SHAGKGGGCGCB |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 64 |
Motif name: Motif 64 |
| Original motif Consensus sequence: CATATRCA |
Reverse complement motif Consensus sequence: TGMATATG |
|
|
 |
 |
Best Matches for Motif ID 64 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
31 |
| Motif name: |
Motif 31 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0465487 |
Alignment:
CATGYACA
CATATRCA
| Original motif Consensus sequence: CATGYACA |
Reverse complement motif Consensus sequence: TGTKCATG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0585881 |
Alignment:
CACACACA
CATATRCA
| Original motif Consensus sequence: CACACACA |
Reverse complement motif Consensus sequence: TGTGTGTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
58 |
| Motif name: |
Motif 58 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0607109 |
Alignment:
TACATGCA
CATATRCA
| Original motif Consensus sequence: TACATGCA |
Reverse complement motif Consensus sequence: TGCATGTA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
152 |
| Motif name: |
yrCATGCAyr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0626249 |
Alignment:
BRCATGCABD
CATATRCA--
| Original motif Consensus sequence: BRCATGCABD |
Reverse complement motif Consensus sequence: HVTGCATGKV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
69 |
| Motif name: |
Motif 69 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0676075 |
Alignment:
ACTCCAGMTCCAGG
----CATATRCA--
| Original motif Consensus sequence: CCTGGARCTGGAGT |
Reverse complement motif Consensus sequence: ACTCCAGMTCCAGG |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 65 |
Motif name: Motif 65 |
| Original motif Consensus sequence: ARAACA |
Reverse complement motif Consensus sequence: TGTTMT |
|
|
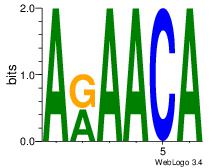 |
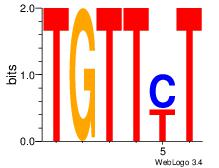 |
Best Matches for Motif ID 65 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
141 |
| Motif name: |
raCAAAACam |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.00122368 |
Alignment:
DACAAAACAH
---ARAACA-
| Original motif Consensus sequence: DACAAAACAH |
Reverse complement motif Consensus sequence: HTGTTTTGTD |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
44 |
| Motif name: |
Motif 44 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0101983 |
Alignment:
GDAAACA
-ARAACA
| Original motif Consensus sequence: GDAAACA |
Reverse complement motif Consensus sequence: TGTTTDC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
159 |
| Motif name: |
kkAAGAGCAsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0232212 |
Alignment:
DBAAGAGCAVH
---ARAACA--
| Original motif Consensus sequence: DBAAGAGCAVH |
Reverse complement motif Consensus sequence: HVTGCTCTTBH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
121 |
| Motif name: |
TAL1TCF3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
6 |
| Similarity score: |
0.0330056 |
Alignment:
HVAMCATCTGKT
ARAACA------
| Original motif Consensus sequence: HVAMCATCTGKT |
Reverse complement motif Consensus sequence: ARCAGATGRTVD |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0333069 |
Alignment:
AAAAHAAA
ARAACA--
| Original motif Consensus sequence: AAAAHAAA |
Reverse complement motif Consensus sequence: TTTDTTTT |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 66 |
Motif name: Motif 66 |
| Original motif Consensus sequence: TATAAATR |
Reverse complement motif Consensus sequence: KATTTATA |
|
|
 |
 |
Best Matches for Motif ID 66 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
157 |
| Motif name: |
wtATTTTTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.00950221 |
Alignment:
WWTAAAAATAD
--TATAAATR-
| Original motif Consensus sequence: DTATTTTTAWW |
Reverse complement motif Consensus sequence: WWTAAAAATAD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
150 |
| Motif name: |
waATwAAAATAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0358566 |
Alignment:
DHATWAAAATAHD
---TATAAATR--
| Original motif Consensus sequence: DHATWAAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTTWATHD |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
38 |
| Motif name: |
Motif 38 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0432007 |
Alignment:
TGTAACTC
TATAAATR
| Original motif Consensus sequence: GAGTTACA |
Reverse complement motif Consensus sequence: TGTAACTC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
51 |
| Motif name: |
Motif 51 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0432007 |
Alignment:
TGTAACTC
TATAAATR
| Original motif Consensus sequence: GAGTTACA |
Reverse complement motif Consensus sequence: TGTAACTC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
68 |
| Motif name: |
Motif 68 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
8 |
| Similarity score: |
0.0496717 |
Alignment:
WAAWAAAWAWWWAA
-TATAAATR-----
| Original motif Consensus sequence: WAAWAAAWAWWWAA |
Reverse complement motif Consensus sequence: TTWWWTWTTTWTTW |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 67 |
Motif name: Motif 67 |
| Original motif Consensus sequence: SYGCCCYCTDSTGG |
Reverse complement motif Consensus sequence: CCASHAGKGGGCKS |
|
|
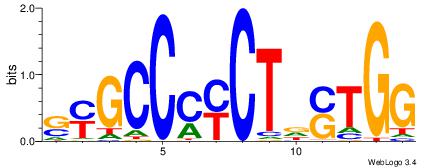 |
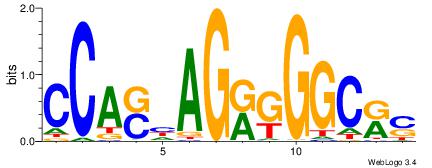 |
Best Matches for Motif ID 67 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
156 |
| Motif name: |
rgyGCCMyCTksTGGccd |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0 |
Alignment:
RVYGCCCYCTKSTGGCHD
-SYGCCCYCTDSTGG---
| Original motif Consensus sequence: RVYGCCCYCTKSTGGCHD |
Reverse complement motif Consensus sequence: DDGCCASYAGMGGGCKVM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
14 |
| Similarity score: |
0.0137353 |
Alignment:
BMSMGCCYMCTKSTGGMHM
--SYGCCCYCTDSTGG---
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
165 |
| Motif name: |
wgGCCAshAGrGGGCrsy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.0259772 |
Alignment:
HDGCCACHAGRGGGCRBY
---CCASHAGKGGGCKS-
| Original motif Consensus sequence: HDGCCACHAGRGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCKCTHGTGGCHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0316372 |
Alignment:
VHGGCCAGMAGAGGGCRBY
----CCASHAGKGGGCKS-
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
169 |
| Motif name: |
yvTGCyGCCmCCwGgtG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
14 |
| Similarity score: |
0.0786902 |
Alignment:
BVTGCTGCCACCWGGDG
-SYGCCCYCTDSTGG--
| Original motif Consensus sequence: BVTGCTGCCACCWGGDG |
Reverse complement motif Consensus sequence: CDCCWGGTGGCAGCAVV |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 68 |
Motif name: Motif 68 |
| Original motif Consensus sequence: WAAWAAAWAWWWAA |
Reverse complement motif Consensus sequence: TTWWWTWTTTWTTW |
|
|
 |
 |
Best Matches for Motif ID 68 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
150 |
| Motif name: |
waATwAAAATAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.0423083 |
Alignment:
-DHTATTTTWATHD
TTWWWTWTTTWTTW
| Original motif Consensus sequence: DHATWAAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTTWATHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
157 |
| Motif name: |
wtATTTTTAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.06057 |
Alignment:
DTATTTTTAWW---
TTWWWTWTTTWTTW
| Original motif Consensus sequence: DTATTTTTAWW |
Reverse complement motif Consensus sequence: WWTAAAAATAD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
148 |
| Motif name: |
wwTwAAAAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
1.54726 |
Alignment:
DDTWAAAAHH----
WAAWAAAWAWWWAA
| Original motif Consensus sequence: DDTWAAAAHH |
Reverse complement motif Consensus sequence: HHTTTTWADD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
141 |
| Motif name: |
raCAAAACam |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
1.55678 |
Alignment:
----HTGTTTTGTD
TTWWWTWTTTWTTW
| Original motif Consensus sequence: DACAAAACAH |
Reverse complement motif Consensus sequence: HTGTTTTGTD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
52 |
| Motif name: |
Motif 52 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
2.5 |
Alignment:
------WTTTATTT
TTWWWTWTTTWTTW
| Original motif Consensus sequence: AAATAAAW |
Reverse complement motif Consensus sequence: WTTTATTT |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 69 |
Motif name: Motif 69 |
| Original motif Consensus sequence: CCTGGARCTGGAGT |
Reverse complement motif Consensus sequence: ACTCCAGMTCCAGG |
|
|
 |
 |
Best Matches for Motif ID 69 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
156 |
| Motif name: |
rgyGCCMyCTksTGGccd |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.070946 |
Alignment:
DDGCCASYAGMGGGCKVM
ACTCCAGMTCCAGG----
| Original motif Consensus sequence: RVYGCCCYCTKSTGGCHD |
Reverse complement motif Consensus sequence: DDGCCASYAGMGGGCKVM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
149 |
| Motif name: |
asmAGRGGGCrCTGsmkc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
14 |
| Similarity score: |
0.073647 |
Alignment:
DBTSCAGMGCCCTCTRST
ACTCCAGMTCCAGG----
| Original motif Consensus sequence: ASMAGAGGGCRCTGSABH |
Reverse complement motif Consensus sequence: DBTSCAGMGCCCTCTRST |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0761843 |
Alignment:
VHGGCCAGMAGAGGGCRBY
-ACTCCAGMTCCAGG----
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
115 |
| Motif name: |
RXRRAR_DR5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0775577 |
Alignment:
RGKTCABVVRGAGGTCA
ACTCCAGMTCCAGG---
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
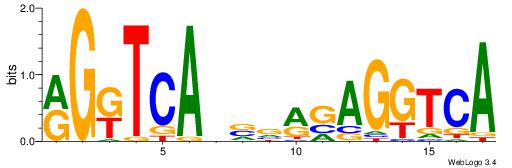 |
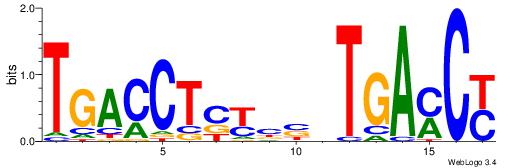 |
| Dataset #: |
2 |
| Motif ID: |
67 |
| Motif name: |
Motif 67 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0806144 |
Alignment:
CCASHAGKGGGCKS
CCTGGARCTGGAGT
| Original motif Consensus sequence: SYGCCCYCTDSTGG |
Reverse complement motif Consensus sequence: CCASHAGKGGGCKS |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 70 |
Motif name: Ar |
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
Best Matches for Motif ID 70 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
113 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
21 |
| Similarity score: |
0.0696331 |
Alignment:
-GGYGCTGTCCATGGTGCTGAA
VRVDGGHACAVDDKGTHCTDWH
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
111 |
| Motif name: |
PPARG |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
20 |
| Similarity score: |
0.55978 |
Alignment:
--STAGGTCACBGTGACCYABT
VRVDGGHACAVDDKGTHCTDWH
| Original motif Consensus sequence: STAGGTCACBGTGACCYABT |
Reverse complement motif Consensus sequence: ABTMGGTCACBGTGACCTAS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
109 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
20 |
| Similarity score: |
0.564914 |
Alignment:
--MSGKKRCGCWDCABTGBBCD
VRVDGGHACAVDDKGTHCTDWH
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
131 |
| Motif name: |
znf143 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
20 |
| Similarity score: |
0.566373 |
Alignment:
GCMWRGCATYRTGGGAMHTB--
VRVDGGHACAVDDKGTHCTDWH
| Original motif Consensus sequence: BAHYTCCCAKMATGCMWYGC |
Reverse complement motif Consensus sequence: GCMWRGCATYRTGGGAMHTB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
19 |
| Similarity score: |
1.05195 |
Alignment:
VDBHMAGGTCACCCTGACCY---
-VRVDGGHACAVDDKGTHCTDWH
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 71 |
Motif name: Arnt |
| Original motif Consensus sequence: CACGTG |
Reverse complement motif Consensus sequence: CACGTG |
|
|
 |
 |
Best Matches for Motif ID 71 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
20 |
| Motif name: |
Motif 20 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0 |
Alignment:
MACGTG
CACGTG
| Original motif Consensus sequence: CACGTR |
Reverse complement motif Consensus sequence: MACGTG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
91 |
| Motif name: |
MAX |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.00380329 |
Alignment:
BCACGTGDTD
-CACGTG---
| Original motif Consensus sequence: DAHCACGTGD |
Reverse complement motif Consensus sequence: BCACGTGDTD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
126 |
| Motif name: |
USF1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.00445689 |
Alignment:
MCACGTG
-CACGTG
| Original motif Consensus sequence: CACGTGR |
Reverse complement motif Consensus sequence: MCACGTG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
95 |
| Motif name: |
MYCMAX |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.00495292 |
Alignment:
RASCACGTGGT
---CACGTG--
| Original motif Consensus sequence: RASCACGTGGT |
Reverse complement motif Consensus sequence: ACCACGTGSTM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
94 |
| Motif name: |
Myc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0173209 |
Alignment:
DCCACGTGCV
--CACGTG--
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 72 |
Motif name: ArntAhr |
| Original motif Consensus sequence: YGCGTG |
Reverse complement motif Consensus sequence: CACGCM |
|
|
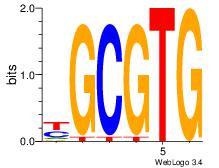 |
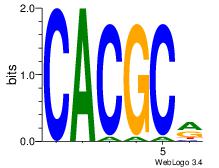 |
Best Matches for Motif ID 72 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
86 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0 |
Alignment:
VBACGTGV
-YGCGTG-
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
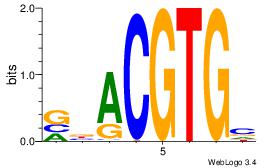 |
 |
| Dataset #: |
4 |
| Motif ID: |
134 |
| Motif name: |
ssCGwGCGss |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0128947 |
Alignment:
BSCGWGCGBV
YGCGTG----
| Original motif Consensus sequence: BSCGWGCGBV |
Reverse complement motif Consensus sequence: VBCGCWCGSB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
71 |
| Motif name: |
Arnt |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0203526 |
Alignment:
CACGTG
YGCGTG
| Original motif Consensus sequence: CACGTG |
Reverse complement motif Consensus sequence: CACGTG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
20 |
| Motif name: |
Motif 20 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0206428 |
Alignment:
MACGTG
YGCGTG
| Original motif Consensus sequence: CACGTR |
Reverse complement motif Consensus sequence: MACGTG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
126 |
| Motif name: |
USF1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0255609 |
Alignment:
MCACGTG
-YGCGTG
| Original motif Consensus sequence: CACGTGR |
Reverse complement motif Consensus sequence: MCACGTG |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 73 |
Motif name: CREB1 |
| Original motif Consensus sequence: TGACGTCA |
Reverse complement motif Consensus sequence: TGACGTCA |
|
|
 |
 |
Best Matches for Motif ID 73 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
92 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.00956439 |
Alignment:
GCGGACGTTV
--TGACGTCA
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
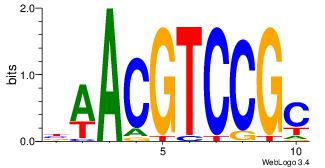 |
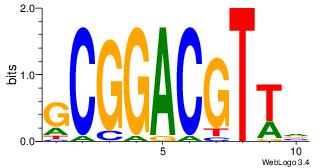 |
| Dataset #: |
3 |
| Motif ID: |
115 |
| Motif name: |
RXRRAR_DR5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
10 |
| Number of overlap: |
8 |
| Similarity score: |
0.0133811 |
Alignment:
TGACCTCKVVBTGAYCK
TGACGTCA---------
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0197902 |
Alignment:
VDBHMAGGTCACCCTGACCY
---TGACGTCA---------
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
82 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0208174 |
Alignment:
BAGGYCABHBTGACCKHV
----------TGACGTCA
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
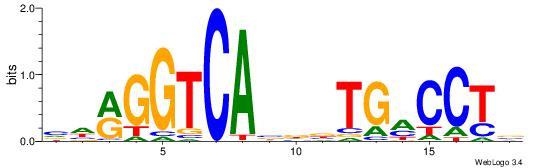 |
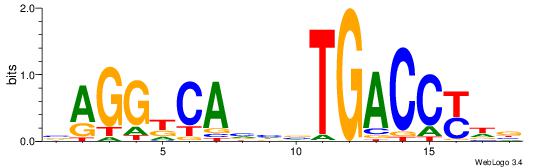 |
| Dataset #: |
1 |
| Motif ID: |
34 |
| Motif name: |
Motif 34 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0253747 |
Alignment:
TGKCCACA
TGACGTCA
| Original motif Consensus sequence: TGKCCACA |
Reverse complement motif Consensus sequence: TGTGGYCA |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 74 |
Motif name: CTCF |
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
Best Matches for Motif ID 74 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
70 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
19 |
| Similarity score: |
0.0903626 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
--BMSMGCCYMCTKSTGGMHM-
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
109 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
19 |
| Similarity score: |
0.0909586 |
Alignment:
DGVBCABTGDWGCGKRRCSR
YDRCCASYAGRKGGCRSYV-
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
19 |
| Similarity score: |
0.0943322 |
Alignment:
VDBHMAGGTCACCCTGACCY
-BMSMGCCYMCTKSTGGMHM
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
165 |
| Motif name: |
wgGCCAshAGrGGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
0.504548 |
Alignment:
HDGCCACHAGRGGGCRBY-
YDRCCASYAGRKGGCRSYV
| Original motif Consensus sequence: HDGCCACHAGRGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCKCTHGTGGCHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
156 |
| Motif name: |
rgyGCCMyCTksTGGccd |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
0.515938 |
Alignment:
-RVYGCCCYCTKSTGGCHD
BMSMGCCYMCTKSTGGMHM
| Original motif Consensus sequence: RVYGCCCYCTKSTGGCHD |
Reverse complement motif Consensus sequence: DDGCCASYAGMGGGCKVM |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 75 |
Motif name: Ddit3Cebpa |
| Original motif Consensus sequence: VDRTGCAATMCC |
Reverse complement motif Consensus sequence: GGRATTGCAKHB |
|
|
 |
 |
Best Matches for Motif ID 75 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
104 |
| Motif name: |
NFYA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.018401 |
Alignment:
BHVCKSATTGGMKBVV
---GGRATTGCAKHB-
| Original motif Consensus sequence: VBBRRCCAATSRGVDB |
Reverse complement motif Consensus sequence: BHVCKSATTGGMKBVV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
131 |
| Motif name: |
znf143 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
12 |
| Similarity score: |
0.037862 |
Alignment:
BAHYTCCCAKMATGCMWYGC
GGRATTGCAKHB--------
| Original motif Consensus sequence: BAHYTCCCAKMATGCMWYGC |
Reverse complement motif Consensus sequence: GCMWRGCATYRTGGGAMHTB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0416577 |
Alignment:
MGGTCAGGGTGACCTRDBHV
------VDRTGCAATMCC--
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
70 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0468764 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
----------GGRATTGCAKHB
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
109 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
12 |
| Similarity score: |
0.0480116 |
Alignment:
DGVBCABTGDWGCGKRRCSR
-------VDRTGCAATMCC-
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 76 |
Motif name: E2F1 |
| Original motif Consensus sequence: TTTSGCGC |
Reverse complement motif Consensus sequence: GCGCSAAA |
|
|
 |
 |
Best Matches for Motif ID 76 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
78 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0 |
Alignment:
TTTSGCGC
TTTSGCGC
| Original motif Consensus sequence: TTTSGCGC |
Reverse complement motif Consensus sequence: GCGCSAAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
143 |
| Motif name: |
AgmAGAGGGCrscAGak |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0915396 |
Alignment:
AGMAGAGGGCASCAGAK
--------GCGCSAAA-
| Original motif Consensus sequence: AGMAGAGGGCASCAGAK |
Reverse complement motif Consensus sequence: RTCTGSTGCCCTCTYCT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
144 |
| Motif name: |
ctCTrsyGCCmCCTast |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0943326 |
Alignment:
HDCTGSYGCCMCCTAST
-TTTSGCGC--------
| Original motif Consensus sequence: HDCTGSYGCCMCCTAST |
Reverse complement motif Consensus sequence: ASTAGGYGGCMSCAGDD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
167 |
| Motif name: |
rsyAGrkGGCGCCmyCTrsy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
8 |
| Similarity score: |
0.0955357 |
Alignment:
HSKAGKYGGCGCCRMCTMSD
--------GCGCSAAA----
| Original motif Consensus sequence: DSYAGRKGGCGCCMYCTRSH |
Reverse complement motif Consensus sequence: HSKAGKYGGCGCCRMCTMSD |
|
|
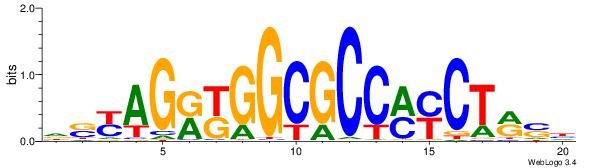 |
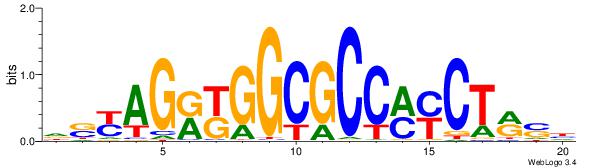 |
| Dataset #: |
4 |
| Motif ID: |
146 |
| Motif name: |
myrGYGCCmCCTast |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0975521 |
Alignment:
ASTAGGYGGCGCTBB
----TTTSGCGC---
| Original motif Consensus sequence: VBAGCGCCMCCTAST |
Reverse complement motif Consensus sequence: ASTAGGYGGCGCTBB |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 77 |
Motif name: EBF1 |
| Original motif Consensus sequence: MCCCMAGGGA |
Reverse complement motif Consensus sequence: TCCCTYGGGY |
|
|
 |
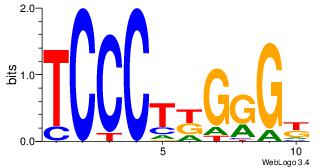 |
Best Matches for Motif ID 77 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
110 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0581959 |
Alignment:
CCCCCTTGGGCCCC
-TCCCTYGGGY---
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
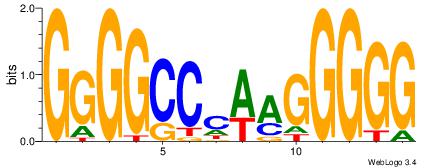 |
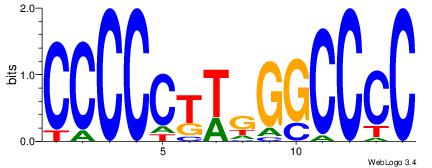 |
| Dataset #: |
3 |
| Motif ID: |
129 |
| Motif name: |
Zfp423 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.05861 |
Alignment:
GSMMCCYARGGKKKC
---MCCCMAGGGA--
| Original motif Consensus sequence: GSMMCCYARGGKKKC |
Reverse complement motif Consensus sequence: GYRYCCMTKGGYRSC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
166 |
| Motif name: |
CasCAGrGGGCrsy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0747791 |
Alignment:
BSKGCCCKCTGGTG
---TCCCTYGGGY-
| Original motif Consensus sequence: CACCAGRGGGCRSB |
Reverse complement motif Consensus sequence: BSKGCCCKCTGGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
168 |
| Motif name: |
yrcrGYGCCMyCTGGtG |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
10 |
| Similarity score: |
0.076024 |
Alignment:
HVCAGCGCCCYCTGGTG
------TCCCTYGGGY-
| Original motif Consensus sequence: HVCAGCGCCCYCTGGTG |
Reverse complement motif Consensus sequence: CACCAGMGGGCGCTGBD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
67 |
| Motif name: |
Motif 67 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.076343 |
Alignment:
SYGCCCYCTDSTGG
TCCCTYGGGY----
| Original motif Consensus sequence: SYGCCCYCTDSTGG |
Reverse complement motif Consensus sequence: CCASHAGKGGGCKS |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 78 |
Motif name: Egr1 |
| Original motif Consensus sequence: TTTSGCGC |
Reverse complement motif Consensus sequence: GCGCSAAA |
|
|
 |
 |
Best Matches for Motif ID 78 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
76 |
| Motif name: |
E2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0 |
Alignment:
TTTSGCGC
TTTSGCGC
| Original motif Consensus sequence: TTTSGCGC |
Reverse complement motif Consensus sequence: GCGCSAAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
143 |
| Motif name: |
AgmAGAGGGCrscAGak |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0915396 |
Alignment:
AGMAGAGGGCASCAGAK
--------GCGCSAAA-
| Original motif Consensus sequence: AGMAGAGGGCASCAGAK |
Reverse complement motif Consensus sequence: RTCTGSTGCCCTCTYCT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
144 |
| Motif name: |
ctCTrsyGCCmCCTast |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0943326 |
Alignment:
HDCTGSYGCCMCCTAST
-TTTSGCGC--------
| Original motif Consensus sequence: HDCTGSYGCCMCCTAST |
Reverse complement motif Consensus sequence: ASTAGGYGGCMSCAGDD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
167 |
| Motif name: |
rsyAGrkGGCGCCmyCTrsy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
8 |
| Similarity score: |
0.0955357 |
Alignment:
HSKAGKYGGCGCCRMCTMSD
--------GCGCSAAA----
| Original motif Consensus sequence: DSYAGRKGGCGCCMYCTRSH |
Reverse complement motif Consensus sequence: HSKAGKYGGCGCCRMCTMSD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
146 |
| Motif name: |
myrGYGCCmCCTast |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0975521 |
Alignment:
ASTAGGYGGCGCTBB
----TTTSGCGC---
| Original motif Consensus sequence: VBAGCGCCMCCTAST |
Reverse complement motif Consensus sequence: ASTAGGYGGCGCTBB |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 79 |
Motif name: ELK1 |
| Original motif Consensus sequence: VDDCCGGAAR |
Reverse complement motif Consensus sequence: MTTCCGGHBV |
|
|
 |
 |
Best Matches for Motif ID 79 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
85 |
| Motif name: |
Hand1Tcfe2a |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0406986 |
Alignment:
AWRCCAGAMB
VDDCCGGAAR
| Original motif Consensus sequence: BRTCTGGMWT |
Reverse complement motif Consensus sequence: AWRCCAGAMB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
119 |
| Motif name: |
Stat3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0421921 |
Alignment:
CTTCCTGGAA
MTTCCGGHBV
| Original motif Consensus sequence: TTCCAGGAAG |
Reverse complement motif Consensus sequence: CTTCCTGGAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
151 |
| Motif name: |
agrCCAGmAGrg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0478168 |
Alignment:
HVGCCAGMAGRG
VDDCCGGAAR--
| Original motif Consensus sequence: HVGCCAGMAGRG |
Reverse complement motif Consensus sequence: CKCTRCTGGCVH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
156 |
| Motif name: |
rgyGCCMyCTksTGGccd |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0493569 |
Alignment:
RVYGCCCYCTKSTGGCHD
--------MTTCCGGHBV
| Original motif Consensus sequence: RVYGCCCYCTKSTGGCHD |
Reverse complement motif Consensus sequence: DDGCCASYAGMGGGCKVM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
10 |
| Number of overlap: |
10 |
| Similarity score: |
0.0506531 |
Alignment:
BMSMGCCYMCTKSTGGMHM
---------MTTCCGGHBV
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 80 |
Motif name: ELK4 |
| Original motif Consensus sequence: ACCGGAAGT |
Reverse complement motif Consensus sequence: ACTTCCGGT |
|
|
 |
 |
Best Matches for Motif ID 80 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
151 |
| Motif name: |
agrCCAGmAGrg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
9 |
| Similarity score: |
0.0699758 |
Alignment:
HVGCCAGMAGRG
--ACCGGAAGT-
| Original motif Consensus sequence: HVGCCAGMAGRG |
Reverse complement motif Consensus sequence: CKCTRCTGGCVH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
136 |
| Motif name: |
dwCAGAAGwh |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0.0710473 |
Alignment:
DHCAGAAGDH
ACCGGAAGT-
| Original motif Consensus sequence: DHCAGAAGDH |
Reverse complement motif Consensus sequence: HDCTTCTGHD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
121 |
| Motif name: |
TAL1TCF3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
9 |
| Similarity score: |
0.0754906 |
Alignment:
HVAMCATCTGKT
---ACTTCCGGT
| Original motif Consensus sequence: HVAMCATCTGKT |
Reverse complement motif Consensus sequence: ARCAGATGRTVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
166 |
| Motif name: |
CasCAGrGGGCrsy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.0818647 |
Alignment:
BSKGCCCKCTGGTG
----ACTTCCGGT-
| Original motif Consensus sequence: CACCAGRGGGCRSB |
Reverse complement motif Consensus sequence: BSKGCCCKCTGGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
156 |
| Motif name: |
rgyGCCMyCTksTGGccd |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
9 |
| Similarity score: |
0.0852431 |
Alignment:
RVYGCCCYCTKSTGGCHD
-------ACTTCCGGT--
| Original motif Consensus sequence: RVYGCCCYCTKSTGGCHD |
Reverse complement motif Consensus sequence: DDGCCASYAGMGGGCKVM |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 81 |
Motif name: ESR1 |
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
Best Matches for Motif ID 81 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
109 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
20 |
| Similarity score: |
0.0624908 |
Alignment:
MSGKKRCGCWDCABTGBBCD
MGGTCAGGGTGACCTRDBHV
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
167 |
| Motif name: |
rsyAGrkGGCGCCmyCTrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
20 |
| Similarity score: |
0.0752066 |
Alignment:
DSYAGRKGGCGCCMYCTRSH
VDBHMAGGTCACCCTGACCY
| Original motif Consensus sequence: DSYAGRKGGCGCCMYCTRSH |
Reverse complement motif Consensus sequence: HSKAGKYGGCGCCRMCTMSD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
70 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
19 |
| Similarity score: |
0.556363 |
Alignment:
-HWDAGHACRHHVTGTHCCHVMV
MGGTCAGGGTGACCTRDBHV---
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
19 |
| Similarity score: |
0.579928 |
Alignment:
-YDRCCASYAGRKGGCRSYV
MGGTCAGGGTGACCTRDBHV
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
111 |
| Motif name: |
PPARG |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
18 |
| Similarity score: |
1.01936 |
Alignment:
STAGGTCACBGTGACCYABT--
--MGGTCAGGGTGACCTRDBHV
| Original motif Consensus sequence: STAGGTCACBGTGACCYABT |
Reverse complement motif Consensus sequence: ABTMGGTCACBGTGACCTAS |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 82 |
Motif name: ESR2 |
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
Best Matches for Motif ID 82 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
111 |
| Motif name: |
PPARG |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
0.0125377 |
Alignment:
ABTMGGTCACBGTGACCTAS
--BAGGYCABHBTGACCKHV
| Original motif Consensus sequence: STAGGTCACBGTGACCYABT |
Reverse complement motif Consensus sequence: ABTMGGTCACBGTGACCTAS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
70 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
18 |
| Similarity score: |
0.0399216 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
--BAGGYCABHBTGACCKHV--
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
131 |
| Motif name: |
znf143 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
18 |
| Similarity score: |
0.0692215 |
Alignment:
GCMWRGCATYRTGGGAMHTB
-BAGGYCABHBTGACCKHV-
| Original motif Consensus sequence: BAHYTCCCAKMATGCMWYGC |
Reverse complement motif Consensus sequence: GCMWRGCATYRTGGGAMHTB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
109 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
18 |
| Similarity score: |
0.0761757 |
Alignment:
MSGKKRCGCWDCABTGBBCD
BAGGYCABHBTGACCKHV--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
167 |
| Motif name: |
rsyAGrkGGCGCCmyCTrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
0.0808384 |
Alignment:
HSKAGKYGGCGCCRMCTMSD
VHRGGTCABDBTGMCCTB--
| Original motif Consensus sequence: DSYAGRKGGCGCCMYCTRSH |
Reverse complement motif Consensus sequence: HSKAGKYGGCGCCRMCTMSD |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 83 |
Motif name: Esrrb |
| Original motif Consensus sequence: VBBYCAAGGTCA |
Reverse complement motif Consensus sequence: TGACCTTGMBBB |
|
|
 |
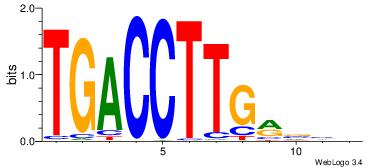 |
Best Matches for Motif ID 83 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
107 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0442611 |
Alignment:
AKGYYCAAAGRTCA
--VBBYCAAGGTCA
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
115 |
| Motif name: |
RXRRAR_DR5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0442659 |
Alignment:
TGACCTCKVVBTGAYCK
TGACCTTGMBBB-----
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
87 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0470808 |
Alignment:
RGGBCAAAGKYCA
-VBBYCAAGGTCA
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
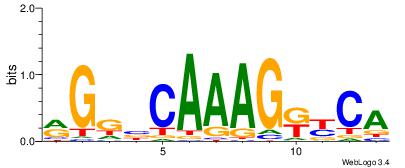 |
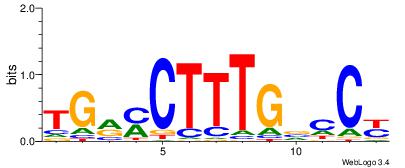 |
| Dataset #: |
3 |
| Motif ID: |
130 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0625961 |
Alignment:
VAGGCCBBGGCVBB
-TGACCTTGMBBB-
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
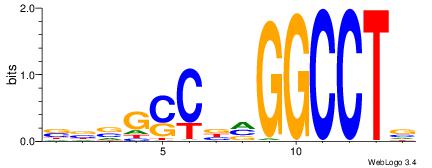 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
12 |
| Similarity score: |
0.0628484 |
Alignment:
VDBHMAGGTCACCCTGACCY
TGACCTTGMBBB--------
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 84 |
Motif name: GABPA |
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
Best Matches for Motif ID 84 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
154 |
| Motif name: |
csCsCCTCCcc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0708637 |
Alignment:
VBCCCCTCCHB
VVCACTTCCGG
| Original motif Consensus sequence: VBCCCCTCCHB |
Reverse complement motif Consensus sequence: BDGGAGGGGBV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
169 |
| Motif name: |
yvTGCyGCCmCCwGgtG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
11 |
| Similarity score: |
0.0756694 |
Alignment:
CDCCWGGTGGCAGCAVV
------CCGGAAGTGVV
| Original motif Consensus sequence: BVTGCTGCCACCWGGDG |
Reverse complement motif Consensus sequence: CDCCWGGTGGCAGCAVV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
67 |
| Motif name: |
Motif 67 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0810917 |
Alignment:
SYGCCCYCTDSTGG
VVCACTTCCGG---
| Original motif Consensus sequence: SYGCCCYCTDSTGG |
Reverse complement motif Consensus sequence: CCASHAGKGGGCKS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0813947 |
Alignment:
BMSMGCCYMCTKSTGGMHM
-----VVCACTTCCGG---
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
156 |
| Motif name: |
rgyGCCMyCTksTGGccd |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
0.0837237 |
Alignment:
RVYGCCCYCTKSTGGCHD
----VVCACTTCCGG---
| Original motif Consensus sequence: RVYGCCCYCTKSTGGCHD |
Reverse complement motif Consensus sequence: DDGCCASYAGMGGGCKVM |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 85 |
Motif name: Hand1Tcfe2a |
| Original motif Consensus sequence: BRTCTGGMWT |
Reverse complement motif Consensus sequence: AWRCCAGAMB |
|
|
 |
 |
Best Matches for Motif ID 85 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
151 |
| Motif name: |
agrCCAGmAGrg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0194367 |
Alignment:
HVGCCAGMAGRG
AWRCCAGAMB--
| Original motif Consensus sequence: HVGCCAGMAGRG |
Reverse complement motif Consensus sequence: CKCTRCTGGCVH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.027647 |
Alignment:
VHGGCCAGMAGAGGGCRBY
-AWRCCAGAMB--------
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
79 |
| Motif name: |
ELK1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0297824 |
Alignment:
MTTCCGGHBV
BRTCTGGMWT
| Original motif Consensus sequence: VDDCCGGAAR |
Reverse complement motif Consensus sequence: MTTCCGGHBV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
114 |
| Motif name: |
RUNX1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0323511 |
Alignment:
BBYTGTGGTTT
-BRTCTGGMWT
| Original motif Consensus sequence: BBYTGTGGTTT |
Reverse complement motif Consensus sequence: AAACCACAKVB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
156 |
| Motif name: |
rgyGCCMyCTksTGGccd |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
10 |
| Similarity score: |
0.0371584 |
Alignment:
RVYGCCCYCTKSTGGCHD
--------BRTCTGGMWT
| Original motif Consensus sequence: RVYGCCCYCTKSTGGCHD |
Reverse complement motif Consensus sequence: DDGCCASYAGMGGGCKVM |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 86 |
Motif name: HIF1AARNT |
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
Best Matches for Motif ID 86 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
91 |
| Motif name: |
MAX |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0166242 |
Alignment:
BCACGTGDTD
VCACGTBV--
| Original motif Consensus sequence: DAHCACGTGD |
Reverse complement motif Consensus sequence: BCACGTGDTD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
96 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0228069 |
Alignment:
GCCACGTGSD
-VBACGTGV-
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
94 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0249459 |
Alignment:
DCCACGTGCV
-VBACGTGV-
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
95 |
| Motif name: |
MYCMAX |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0254922 |
Alignment:
ACCACGTGSTM
-VCACGTBV--
| Original motif Consensus sequence: RASCACGTGGT |
Reverse complement motif Consensus sequence: ACCACGTGSTM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
125 |
| Motif name: |
TP53 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
11 |
| Number of overlap: |
8 |
| Similarity score: |
0.0362969 |
Alignment:
MSGGACATGYCCGGGCATGT
--VBACGTGV----------
| Original motif Consensus sequence: MSGGACATGYCCGGGCATGT |
Reverse complement motif Consensus sequence: ACATGCCCGGKCATGTCCSR |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 87 |
Motif name: HNF4A |
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
Best Matches for Motif ID 87 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
107 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.0127044 |
Alignment:
AKGYYCAAAGRTCA
-RGGBCAAAGKYCA
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
115 |
| Motif name: |
RXRRAR_DR5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.0388151 |
Alignment:
TGACCTCKVVBTGAYCK
TGMYCTTTGBCCK----
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
109 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
13 |
| Similarity score: |
0.0538871 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-------RGGBCAAAGKYCA
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
82 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
13 |
| Similarity score: |
0.0571246 |
Alignment:
VHRGGTCABDBTGMCCTB
--RGGBCAAAGKYCA---
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
70 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
13 |
| Similarity score: |
0.0577381 |
Alignment:
VRVDGGHACAVDDKGTHCTDWH
------TGMYCTTTGBCCK---
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 88 |
Motif name: INSM1 |
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
Best Matches for Motif ID 88 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
12 |
| Similarity score: |
0.0323447 |
Alignment:
BMSMGCCYMCTKSTGGMHM
---MGCCCCCTGMCA----
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
67 |
| Motif name: |
Motif 67 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0361443 |
Alignment:
CCASHAGKGGGCKS
-TGYCAGGGGGCR-
| Original motif Consensus sequence: SYGCCCYCTDSTGG |
Reverse complement motif Consensus sequence: CCASHAGKGGGCKS |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
166 |
| Motif name: |
CasCAGrGGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0386733 |
Alignment:
CACCAGRGGGCRSB
TGYCAGGGGGCR--
| Original motif Consensus sequence: CACCAGRGGGCRSB |
Reverse complement motif Consensus sequence: BSKGCCCKCTGGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
168 |
| Motif name: |
yrcrGYGCCMyCTGGtG |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0393748 |
Alignment:
HVCAGCGCCCYCTGGTG
-----MGCCCCCTGMCA
| Original motif Consensus sequence: HVCAGCGCCCYCTGGTG |
Reverse complement motif Consensus sequence: CACCAGMGGGCGCTGBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
156 |
| Motif name: |
rgyGCCMyCTksTGGccd |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0427424 |
Alignment:
RVYGCCCYCTKSTGGCHD
--MGCCCCCTGMCA----
| Original motif Consensus sequence: RVYGCCCYCTKSTGGCHD |
Reverse complement motif Consensus sequence: DDGCCASYAGMGGGCKVM |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 89 |
Motif name: Klf4 |
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
Best Matches for Motif ID 89 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
155 |
| Motif name: |
csCSCCdCCCcs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0234217 |
Alignment:
VDGGGDGGGGBV
-DGGGYGKGGC-
| Original motif Consensus sequence: VBCCCCDCCCHV |
Reverse complement motif Consensus sequence: VDGGGDGGGGBV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
154 |
| Motif name: |
csCsCCTCCcc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0429499 |
Alignment:
VBCCCCTCCHB
-GCCYCMCCCD
| Original motif Consensus sequence: VBCCCCTCCHB |
Reverse complement motif Consensus sequence: BDGGAGGGGBV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
149 |
| Motif name: |
asmAGRGGGCrCTGsmkc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0512984 |
Alignment:
ASMAGAGGGCRCTGSABH
-----DGGGYGKGGC---
| Original motif Consensus sequence: ASMAGAGGGCRCTGSABH |
Reverse complement motif Consensus sequence: DBTSCAGMGCCCTCTRST |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
146 |
| Motif name: |
myrGYGCCmCCTast |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0522764 |
Alignment:
VBAGCGCCMCCTAST
---GCCYCMCCCD--
| Original motif Consensus sequence: VBAGCGCCMCCTAST |
Reverse complement motif Consensus sequence: ASTAGGYGGCGCTBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
169 |
| Motif name: |
yvTGCyGCCmCCwGgtG |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0619219 |
Alignment:
BVTGCTGCCACCWGGDG
---GCCYCMCCCD----
| Original motif Consensus sequence: BVTGCTGCCACCWGGDG |
Reverse complement motif Consensus sequence: CDCCWGGTGGCAGCAVV |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 90 |
Motif name: Mafb |
| Original motif Consensus sequence: GCTGACDB |
Reverse complement motif Consensus sequence: BHGTCAGC |
|
|
 |
 |
Best Matches for Motif ID 90 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
149 |
| Motif name: |
asmAGRGGGCrCTGsmkc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0232655 |
Alignment:
ASMAGAGGGCRCTGSABH
----------GCTGACDB
| Original motif Consensus sequence: ASMAGAGGGCRCTGSABH |
Reverse complement motif Consensus sequence: DBTSCAGMGCCCTCTRST |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
111 |
| Motif name: |
PPARG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
10 |
| Number of overlap: |
8 |
| Similarity score: |
0.0322069 |
Alignment:
STAGGTCACBGTGACCYABT
---GCTGACDB---------
| Original motif Consensus sequence: STAGGTCACBGTGACCYABT |
Reverse complement motif Consensus sequence: ABTMGGTCACBGTGACCTAS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
82 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.035275 |
Alignment:
BAGGYCABHBTGACCKHV
--------GCTGACDB--
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
169 |
| Motif name: |
yvTGCyGCCmCCwGgtG |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.0358393 |
Alignment:
CDCCWGGTGGCAGCAVV
------BHGTCAGC---
| Original motif Consensus sequence: BVTGCTGCCACCWGGDG |
Reverse complement motif Consensus sequence: CDCCWGGTGGCAGCAVV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
115 |
| Motif name: |
RXRRAR_DR5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0388936 |
Alignment:
RGKTCABVVRGAGGTCA
-GCTGACDB--------
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 91 |
Motif name: MAX |
| Original motif Consensus sequence: DAHCACGTGD |
Reverse complement motif Consensus sequence: BCACGTGDTD |
|
|
 |
 |
Best Matches for Motif ID 91 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
95 |
| Motif name: |
MYCMAX |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0266106 |
Alignment:
RASCACGTGGT
DAHCACGTGD-
| Original motif Consensus sequence: RASCACGTGGT |
Reverse complement motif Consensus sequence: ACCACGTGSTM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
121 |
| Motif name: |
TAL1TCF3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0735692 |
Alignment:
ARCAGATGRTVD
-BCACGTGDTD-
| Original motif Consensus sequence: HVAMCATCTGKT |
Reverse complement motif Consensus sequence: ARCAGATGRTVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
158 |
| Motif name: |
grCCACwAGrk |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0740659 |
Alignment:
DDCCACWAGRK
DAHCACGTGD-
| Original motif Consensus sequence: DDCCACWAGRK |
Reverse complement motif Consensus sequence: YMCTWGTGGHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
138 |
| Motif name: |
grCCACyAGAkG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0824309 |
Alignment:
DDCCACYAGAKG
DAHCACGTGD--
| Original motif Consensus sequence: DDCCACYAGAKG |
Reverse complement motif Consensus sequence: CYTCTMGTGGHH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
105 |
| Motif name: |
NHLH1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0853797 |
Alignment:
VCGCAGCTGCGB
--BCACGTGDTD
| Original motif Consensus sequence: VCGCAGCTGCGB |
Reverse complement motif Consensus sequence: VCGCAGCTGCGV |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 92 |
Motif name: MIZF |
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
Best Matches for Motif ID 92 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
84 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0326212 |
Alignment:
VVCACTTCCGG
BAACGTCCGC-
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
168 |
| Motif name: |
yrcrGYGCCMyCTGGtG |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
10 |
| Similarity score: |
0.0553693 |
Alignment:
CACCAGMGGGCGCTGBD
-----GCGGACGTTV--
| Original motif Consensus sequence: HVCAGCGCCCYCTGGTG |
Reverse complement motif Consensus sequence: CACCAGMGGGCGCTGBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
149 |
| Motif name: |
asmAGRGGGCrCTGsmkc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0554206 |
Alignment:
DBTSCAGMGCCCTCTRST
----BAACGTCCGC----
| Original motif Consensus sequence: ASMAGAGGGCRCTGSABH |
Reverse complement motif Consensus sequence: DBTSCAGMGCCCTCTRST |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
95 |
| Motif name: |
MYCMAX |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0558866 |
Alignment:
ACCACGTGSTM
-BAACGTCCGC
| Original motif Consensus sequence: RASCACGTGGT |
Reverse complement motif Consensus sequence: ACCACGTGSTM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
82 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0567619 |
Alignment:
BAGGYCABHBTGACCKHV
--------GCGGACGTTV
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 93 |
Motif name: Myb |
| Original motif Consensus sequence: GRCVGTTG |
Reverse complement motif Consensus sequence: CAACVGMC |
|
|
 |
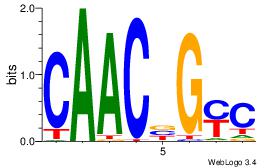 |
Best Matches for Motif ID 93 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
97 |
| Motif name: |
Myf |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0229626 |
Alignment:
MRGCARCWGSWG
---CAACVGMC-
| Original motif Consensus sequence: MRGCARCWGSWG |
Reverse complement motif Consensus sequence: CWSCWGMTGCKR |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
87 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0239903 |
Alignment:
RGGBCAAAGKYCA
-CAACVGMC----
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
121 |
| Motif name: |
TAL1TCF3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0259637 |
Alignment:
ARCAGATGRTVD
GRCVGTTG----
| Original motif Consensus sequence: HVAMCATCTGKT |
Reverse complement motif Consensus sequence: ARCAGATGRTVD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
114 |
| Motif name: |
RUNX1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0297264 |
Alignment:
AAACCACAKVB
---CAACVGMC
| Original motif Consensus sequence: BBYTGTGGTTT |
Reverse complement motif Consensus sequence: AAACCACAKVB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
104 |
| Motif name: |
NFYA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0310894 |
Alignment:
VBBRRCCAATSRGVDB
------CAACVGMC--
| Original motif Consensus sequence: VBBRRCCAATSRGVDB |
Reverse complement motif Consensus sequence: BHVCKSATTGGMKBVV |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 94 |
Motif name: Myc |
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
Best Matches for Motif ID 94 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
96 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0 |
Alignment:
HSCACGTGGC
VGCACGTGGH
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
95 |
| Motif name: |
MYCMAX |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0209359 |
Alignment:
RASCACGTGGT
-VGCACGTGGH
| Original motif Consensus sequence: RASCACGTGGT |
Reverse complement motif Consensus sequence: ACCACGTGSTM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
169 |
| Motif name: |
yvTGCyGCCmCCwGgtG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
10 |
| Similarity score: |
0.0628757 |
Alignment:
CDCCWGGTGGCAGCAVV
-VGCACGTGGH------
| Original motif Consensus sequence: BVTGCTGCCACCWGGDG |
Reverse complement motif Consensus sequence: CDCCWGGTGGCAGCAVV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
147 |
| Motif name: |
asCAGrkGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0675017 |
Alignment:
ASCAGRGGGCRSB
VGCACGTGGH---
| Original motif Consensus sequence: ASCAGRGGGCRSB |
Reverse complement motif Consensus sequence: BSKGCCCMCTGST |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
158 |
| Motif name: |
grCCACwAGrk |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0682475 |
Alignment:
DDCCACWAGRK
-DCCACGTGCV
| Original motif Consensus sequence: DDCCACWAGRK |
Reverse complement motif Consensus sequence: YMCTWGTGGHH |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 95 |
Motif name: MYCMAX |
| Original motif Consensus sequence: RASCACGTGGT |
Reverse complement motif Consensus sequence: ACCACGTGSTM |
|
|
 |
 |
Best Matches for Motif ID 95 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
121 |
| Motif name: |
TAL1TCF3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0652343 |
Alignment:
HVAMCATCTGKT
-RASCACGTGGT
| Original motif Consensus sequence: HVAMCATCTGKT |
Reverse complement motif Consensus sequence: ARCAGATGRTVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
158 |
| Motif name: |
grCCACwAGrk |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0786918 |
Alignment:
DDCCACWAGRK
RASCACGTGGT
| Original motif Consensus sequence: DDCCACWAGRK |
Reverse complement motif Consensus sequence: YMCTWGTGGHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
169 |
| Motif name: |
yvTGCyGCCmCCwGgtG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
11 |
| Similarity score: |
0.0804522 |
Alignment:
CDCCWGGTGGCAGCAVV
RASCACGTGGT------
| Original motif Consensus sequence: BVTGCTGCCACCWGGDG |
Reverse complement motif Consensus sequence: CDCCWGGTGGCAGCAVV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
97 |
| Motif name: |
Myf |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0843705 |
Alignment:
CWSCWGMTGCKR
-ACCACGTGSTM
| Original motif Consensus sequence: MRGCARCWGSWG |
Reverse complement motif Consensus sequence: CWSCWGMTGCKR |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
0.084904 |
Alignment:
YDRCCASYAGRKGGCRSYV
----RASCACGTGGT----
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 96 |
Motif name: Mycn |
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
Best Matches for Motif ID 96 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
94 |
| Motif name: |
Myc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0 |
Alignment:
DCCACGTGCV
GCCACGTGSD
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
95 |
| Motif name: |
MYCMAX |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0200467 |
Alignment:
RASCACGTGGT
-HSCACGTGGC
| Original motif Consensus sequence: RASCACGTGGT |
Reverse complement motif Consensus sequence: ACCACGTGSTM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
169 |
| Motif name: |
yvTGCyGCCmCCwGgtG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0581322 |
Alignment:
CDCCWGGTGGCAGCAVV
-HSCACGTGGC------
| Original motif Consensus sequence: BVTGCTGCCACCWGGDG |
Reverse complement motif Consensus sequence: CDCCWGGTGGCAGCAVV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
147 |
| Motif name: |
asCAGrkGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0621075 |
Alignment:
ASCAGRGGGCRSB
HSCACGTGGC---
| Original motif Consensus sequence: ASCAGRGGGCRSB |
Reverse complement motif Consensus sequence: BSKGCCCMCTGST |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
10 |
| Similarity score: |
0.0679017 |
Alignment:
YDRCCASYAGRKGGCRSYV
-----HSCACGTGGC----
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 97 |
Motif name: Myf |
| Original motif Consensus sequence: MRGCARCWGSWG |
Reverse complement motif Consensus sequence: CWSCWGMTGCKR |
|
|
 |
 |
Best Matches for Motif ID 97 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
105 |
| Motif name: |
NHLH1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0355504 |
Alignment:
VCGCAGCTGCGV
CWSCWGMTGCKR
| Original motif Consensus sequence: VCGCAGCTGCGB |
Reverse complement motif Consensus sequence: VCGCAGCTGCGV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
12 |
| Similarity score: |
0.0508517 |
Alignment:
YDRCCASYAGRKGGCRSYV
---CWSCWGMTGCKR----
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
166 |
| Motif name: |
CasCAGrGGGCrsy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.055783 |
Alignment:
CACCAGRGGGCRSB
CWSCWGMTGCKR--
| Original motif Consensus sequence: CACCAGRGGGCRSB |
Reverse complement motif Consensus sequence: BSKGCCCKCTGGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
156 |
| Motif name: |
rgyGCCMyCTksTGGccd |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.058463 |
Alignment:
DDGCCASYAGMGGGCKVM
----CWSCWGMTGCKR--
| Original motif Consensus sequence: RVYGCCCYCTKSTGGCHD |
Reverse complement motif Consensus sequence: DDGCCASYAGMGGGCKVM |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
69 |
| Motif name: |
Motif 69 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0591644 |
Alignment:
CCTGGARCTGGAGT
-MRGCARCWGSWG-
| Original motif Consensus sequence: CCTGGARCTGGAGT |
Reverse complement motif Consensus sequence: ACTCCAGMTCCAGG |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 98 |
Motif name: MZF1_1-4 |
| Original motif Consensus sequence: BGGGGA |
Reverse complement motif Consensus sequence: TCCCCV |
|
|
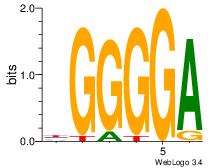 |
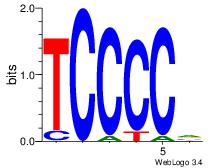 |
Best Matches for Motif ID 98 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
88 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.033712 |
Alignment:
MGCCCCCTGMCA
--TCCCCV----
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
77 |
| Motif name: |
EBF1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0339898 |
Alignment:
MCCCMAGGGA
----BGGGGA
| Original motif Consensus sequence: MCCCMAGGGA |
Reverse complement motif Consensus sequence: TCCCTYGGGY |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
155 |
| Motif name: |
csCSCCdCCCcs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0344568 |
Alignment:
VBCCCCDCCCHV
------TCCCCV
| Original motif Consensus sequence: VBCCCCDCCCHV |
Reverse complement motif Consensus sequence: VDGGGDGGGGBV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
99 |
| Motif name: |
MZF1_5-13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0375315 |
Alignment:
BKAGGGGDAD
---BGGGGA-
| Original motif Consensus sequence: BKAGGGGDAD |
Reverse complement motif Consensus sequence: BTHCCCCTYB |
|
|
 |
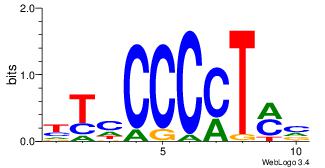 |
| Dataset #: |
4 |
| Motif ID: |
162 |
| Motif name: |
ccAsCCCCAcc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0465466 |
Alignment:
HVASCCCCABH
---TCCCCV--
| Original motif Consensus sequence: HVASCCCCABH |
Reverse complement motif Consensus sequence: DBTGGGGSTVD |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 99 |
Motif name: MZF1_5-13 |
| Original motif Consensus sequence: BKAGGGGDAD |
Reverse complement motif Consensus sequence: BTHCCCCTYB |
|
|
 |
 |
Best Matches for Motif ID 99 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
88 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0360532 |
Alignment:
TGYCAGGGGGCR
--BKAGGGGDAD
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
143 |
| Motif name: |
AgmAGAGGGCrscAGak |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0361803 |
Alignment:
RTCTGSTGCCCTCTYCT
------BTHCCCCTYB-
| Original motif Consensus sequence: AGMAGAGGGCASCAGAK |
Reverse complement motif Consensus sequence: RTCTGSTGCCCTCTYCT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
131 |
| Motif name: |
znf143 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0377199 |
Alignment:
GCMWRGCATYRTGGGAMHTB
--------BKAGGGGDAD--
| Original motif Consensus sequence: BAHYTCCCAKMATGCMWYGC |
Reverse complement motif Consensus sequence: GCMWRGCATYRTGGGAMHTB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
67 |
| Motif name: |
Motif 67 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0386182 |
Alignment:
CCASHAGKGGGCKS
-BKAGGGGDAD---
| Original motif Consensus sequence: SYGCCCYCTDSTGG |
Reverse complement motif Consensus sequence: CCASHAGKGGGCKS |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0397537 |
Alignment:
KBKGCCCTCTYCTGGCCHV
--BTHCCCCTYB-------
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 100 |
Motif name: NFE2L2 |
| Original motif Consensus sequence: RTGACWHAGCA |
Reverse complement motif Consensus sequence: TGCTDWGTCAK |
|
|
 |
 |
Best Matches for Motif ID 100 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
125 |
| Motif name: |
TP53 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0588881 |
Alignment:
ACATGCCCGGKCATGTCCSR
---TGCTDWGTCAK------
| Original motif Consensus sequence: MSGGACATGYCCGGGCATGT |
Reverse complement motif Consensus sequence: ACATGCCCGGKCATGTCCSR |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0627853 |
Alignment:
VDBHMAGGTCACCCTGACCY
--------TGCTDWGTCAK-
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
170 |
| Motif name: |
ssGGCrsTGCrs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0672435 |
Alignment:
VVGGCRSTGCVB
RTGACWHAGCA-
| Original motif Consensus sequence: VVGGCRSTGCVB |
Reverse complement motif Consensus sequence: BVGCASMGCCVV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
149 |
| Motif name: |
asmAGRGGGCrCTGsmkc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
11 |
| Similarity score: |
0.070397 |
Alignment:
ASMAGAGGGCRCTGSABH
-----RTGACWHAGCA--
| Original motif Consensus sequence: ASMAGAGGGCRCTGSABH |
Reverse complement motif Consensus sequence: DBTSCAGMGCCCTCTRST |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
109 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0725913 |
Alignment:
MSGKKRCGCWDCABTGBBCD
--RTGACWHAGCA-------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 101 |
Motif name: NFIC |
| Original motif Consensus sequence: TTGGCD |
Reverse complement motif Consensus sequence: DGCCAA |
|
|
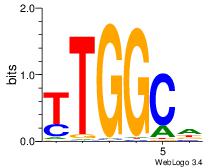 |
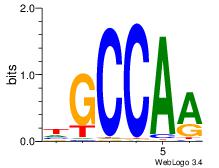 |
Best Matches for Motif ID 101 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
124 |
| Motif name: |
TLX1NFIC |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0 |
Alignment:
TTGGCHDBSTGCCA
TTGGCD--------
| Original motif Consensus sequence: TGGCASBDHGCCAA |
Reverse complement motif Consensus sequence: TTGGCHDBSTGCCA |
|
|
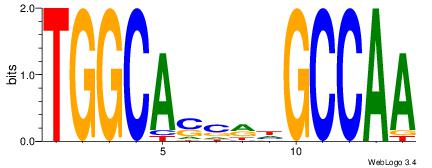 |
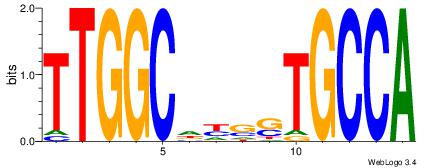 |
| Dataset #: |
3 |
| Motif ID: |
104 |
| Motif name: |
NFYA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.020508 |
Alignment:
VBBRRCCAATSRGVDB
---DGCCAA-------
| Original motif Consensus sequence: VBBRRCCAATSRGVDB |
Reverse complement motif Consensus sequence: BHVCKSATTGGMKBVV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
165 |
| Motif name: |
wgGCCAshAGrGGGCrsy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
12 |
| Number of overlap: |
6 |
| Similarity score: |
0.0215379 |
Alignment:
HDGCCACHAGRGGGCRBY
-DGCCAA-----------
| Original motif Consensus sequence: HDGCCACHAGRGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCKCTHGTGGCHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
156 |
| Motif name: |
rgyGCCMyCTksTGGccd |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.026631 |
Alignment:
DDGCCASYAGMGGGCKVM
-DGCCAA-----------
| Original motif Consensus sequence: RVYGCCCYCTKSTGGCHD |
Reverse complement motif Consensus sequence: DDGCCASYAGMGGGCKVM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
151 |
| Motif name: |
agrCCAGmAGrg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0273632 |
Alignment:
CKCTRCTGGCVH
-----TTGGCD-
| Original motif Consensus sequence: HVGCCAGMAGRG |
Reverse complement motif Consensus sequence: CKCTRCTGGCVH |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 102 |
Motif name: NF-kappaB |
| Original motif Consensus sequence: GGGRMTTYCC |
Reverse complement motif Consensus sequence: GGMAAYKCCC |
|
|
 |
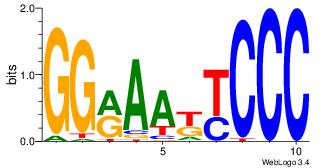 |
Best Matches for Motif ID 102 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
112 |
| Motif name: |
RELA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0219591 |
Alignment:
GGGRATTTCC
GGGRMTTYCC
| Original motif Consensus sequence: GGGRATTTCC |
Reverse complement motif Consensus sequence: GGAAATKCCC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
103 |
| Motif name: |
NFKB1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0264912 |
Alignment:
GGGGRTTCCCC
GGGRMTTYCC-
| Original motif Consensus sequence: GGGGRTTCCCC |
Reverse complement motif Consensus sequence: GGGGAAKCCCC |
|
|
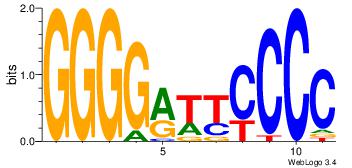 |
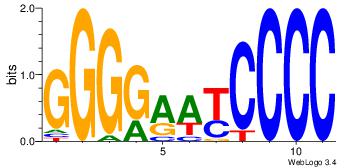 |
| Dataset #: |
3 |
| Motif ID: |
125 |
| Motif name: |
TP53 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
10 |
| Similarity score: |
0.0780263 |
Alignment:
MSGGACATGYCCGGGCATGT
--GGMAAYKCCC--------
| Original motif Consensus sequence: MSGGACATGYCCGGGCATGT |
Reverse complement motif Consensus sequence: ACATGCCCGGKCATGTCCSR |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
149 |
| Motif name: |
asmAGRGGGCrCTGsmkc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
10 |
| Similarity score: |
0.078539 |
Alignment:
DBTSCAGMGCCCTCTRST
--GGMAAYKCCC------
| Original motif Consensus sequence: ASMAGAGGGCRCTGSABH |
Reverse complement motif Consensus sequence: DBTSCAGMGCCCTCTRST |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
124 |
| Motif name: |
TLX1NFIC |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0847697 |
Alignment:
TGGCASBDHGCCAA
-GGMAAYKCCC---
| Original motif Consensus sequence: TGGCASBDHGCCAA |
Reverse complement motif Consensus sequence: TTGGCHDBSTGCCA |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 103 |
Motif name: NFKB1 |
| Original motif Consensus sequence: GGGGRTTCCCC |
Reverse complement motif Consensus sequence: GGGGAAKCCCC |
|
|
 |
 |
Best Matches for Motif ID 103 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
125 |
| Motif name: |
TP53 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.100899 |
Alignment:
MSGGACATGYCCGGGCATGT
-GGGGAAKCCCC--------
| Original motif Consensus sequence: MSGGACATGYCCGGGCATGT |
Reverse complement motif Consensus sequence: ACATGCCCGGKCATGTCCSR |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
122 |
| Motif name: |
Tcfcp2l1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.103405 |
Alignment:
CKGGDTBDMMCTGG
GGGGRTTCCCC---
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
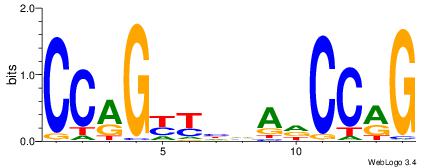 |
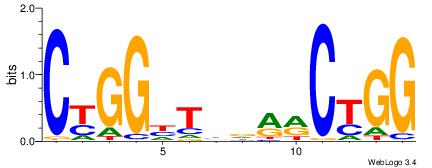 |
| Dataset #: |
4 |
| Motif ID: |
167 |
| Motif name: |
rsyAGrkGGCGCCmyCTrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
0.104275 |
Alignment:
DSYAGRKGGCGCCMYCTRSH
-----GGGGRTTCCCC----
| Original motif Consensus sequence: DSYAGRKGGCGCCMYCTRSH |
Reverse complement motif Consensus sequence: HSKAGKYGGCGCCRMCTMSD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
149 |
| Motif name: |
asmAGRGGGCrCTGsmkc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.109809 |
Alignment:
DBTSCAGMGCCCTCTRST
GGGGAAKCCCC-------
| Original motif Consensus sequence: ASMAGAGGGCRCTGSABH |
Reverse complement motif Consensus sequence: DBTSCAGMGCCCTCTRST |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
117 |
| Motif name: |
Spz1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.11048 |
Alignment:
AGGGTAWCAGC
GGGGAAKCCCC
| Original motif Consensus sequence: AGGGTAWCAGC |
Reverse complement motif Consensus sequence: GCTGWTACCCT |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 104 |
Motif name: NFYA |
| Original motif Consensus sequence: VBBRRCCAATSRGVDB |
Reverse complement motif Consensus sequence: BHVCKSATTGGMKBVV |
|
|
 |
 |
Best Matches for Motif ID 104 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
109 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
16 |
| Similarity score: |
0.0555608 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-VBBRRCCAATSRGVDB---
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
131 |
| Motif name: |
znf143 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
16 |
| Similarity score: |
0.0645227 |
Alignment:
BAHYTCCCAKMATGCMWYGC
-VBBRRCCAATSRGVDB---
| Original motif Consensus sequence: BAHYTCCCAKMATGCMWYGC |
Reverse complement motif Consensus sequence: GCMWRGCATYRTGGGAMHTB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
82 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
16 |
| Similarity score: |
0.0658537 |
Alignment:
BAGGYCABHBTGACCKHV
--BHVCKSATTGGMKBVV
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
16 |
| Similarity score: |
0.0705004 |
Alignment:
BMSMGCCYMCTKSTGGMHM
--BHVCKSATTGGMKBVV-
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
70 |
| Motif name: |
Ar |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
16 |
| Similarity score: |
0.0709164 |
Alignment:
VRVDGGHACAVDDKGTHCTDWH
----VBBRRCCAATSRGVDB--
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 105 |
Motif name: NHLH1 |
| Original motif Consensus sequence: VCGCAGCTGCGB |
Reverse complement motif Consensus sequence: VCGCAGCTGCGV |
|
|
 |
 |
Best Matches for Motif ID 105 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
97 |
| Motif name: |
Myf |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0173692 |
Alignment:
CWSCWGMTGCKR
VCGCAGCTGCGV
| Original motif Consensus sequence: MRGCARCWGSWG |
Reverse complement motif Consensus sequence: CWSCWGMTGCKR |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
109 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
12 |
| Similarity score: |
0.0355984 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-----VCGCAGCTGCGB---
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
170 |
| Motif name: |
ssGGCrsTGCrs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0376107 |
Alignment:
VVGGCRSTGCVB
VCGCAGCTGCGB
| Original motif Consensus sequence: VVGGCRSTGCVB |
Reverse complement motif Consensus sequence: BVGCASMGCCVV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
156 |
| Motif name: |
rgyGCCMyCTksTGGccd |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0465866 |
Alignment:
DDGCCASYAGMGGGCKVM
-VCGCAGCTGCGV-----
| Original motif Consensus sequence: RVYGCCCYCTKSTGGCHD |
Reverse complement motif Consensus sequence: DDGCCASYAGMGGGCKVM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
169 |
| Motif name: |
yvTGCyGCCmCCwGgtG |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0468076 |
Alignment:
CDCCWGGTGGCAGCAVV
----VCGCAGCTGCGV-
| Original motif Consensus sequence: BVTGCTGCCACCWGGDG |
Reverse complement motif Consensus sequence: CDCCWGGTGGCAGCAVV |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 106 |
Motif name: Nkx3-2 |
| Original motif Consensus sequence: BTRAGTGVV |
Reverse complement motif Consensus sequence: BVCACTKAH |
|
|
 |
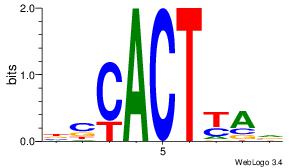 |
Best Matches for Motif ID 106 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
109 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
9 |
| Similarity score: |
0.0239583 |
Alignment:
MSGKKRCGCWDCABTGBBCD
----BVCACTKAH-------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
104 |
| Motif name: |
NFYA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
9 |
| Similarity score: |
0.0250758 |
Alignment:
VBBRRCCAATSRGVDB
---BVCACTKAH----
| Original motif Consensus sequence: VBBRRCCAATSRGVDB |
Reverse complement motif Consensus sequence: BHVCKSATTGGMKBVV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
84 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
9 |
| Similarity score: |
0.0255754 |
Alignment:
CCGGAAGTGVV
--BTRAGTGVV
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
114 |
| Motif name: |
RUNX1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0.0262083 |
Alignment:
AAACCACAKVB
--BVCACTKAH
| Original motif Consensus sequence: BBYTGTGGTTT |
Reverse complement motif Consensus sequence: AAACCACAKVB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
96 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0.0296502 |
Alignment:
HSCACGTGGC
-BVCACTKAH
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 107 |
Motif name: NR2F1 |
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
Best Matches for Motif ID 107 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
115 |
| Motif name: |
RXRRAR_DR5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.0610709 |
Alignment:
RGKTCABVVRGAGGTCA
---AKGYYCAAAGRTCA
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
82 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0695855 |
Alignment:
VHRGGTCABDBTGMCCTB
---AKGYYCAAAGRTCA-
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
113 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
14 |
| Similarity score: |
0.0703341 |
Alignment:
TTCAGCACCATGGACAGCKCC
--AKGYYCAAAGRTCA-----
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
104 |
| Motif name: |
NFYA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
14 |
| Similarity score: |
0.0715605 |
Alignment:
VBBRRCCAATSRGVDB
--TGAMCTTTGMMCYT
| Original motif Consensus sequence: VBBRRCCAATSRGVDB |
Reverse complement motif Consensus sequence: BHVCKSATTGGMKBVV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
109 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
14 |
| Similarity score: |
0.0727368 |
Alignment:
MSGKKRCGCWDCABTGBBCD
----TGAMCTTTGMMCYT--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 108 |
Motif name: NR4A2 |
| Original motif Consensus sequence: AAGGTCAC |
Reverse complement motif Consensus sequence: GTGACCTT |
|
|
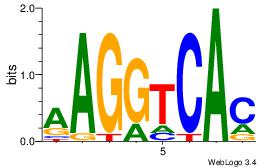 |
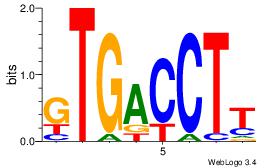 |
Best Matches for Motif ID 108 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0193781 |
Alignment:
VDBHMAGGTCACCCTGACCY
----AAGGTCAC--------
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
82 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
10 |
| Number of overlap: |
8 |
| Similarity score: |
0.0196918 |
Alignment:
BAGGYCABHBTGACCKHV
---------GTGACCTT-
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
111 |
| Motif name: |
PPARG |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
12 |
| Number of overlap: |
8 |
| Similarity score: |
0.0275817 |
Alignment:
ABTMGGTCACBGTGACCTAS
-----------GTGACCTT-
| Original motif Consensus sequence: STAGGTCACBGTGACCYABT |
Reverse complement motif Consensus sequence: ABTMGGTCACBGTGACCTAS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
70 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
13 |
| Number of overlap: |
8 |
| Similarity score: |
0.0486154 |
Alignment:
VRVDGGHACAVDDKGTHCTDWH
------------GTGACCTT--
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0525645 |
Alignment:
CTGGCCTC
GTGACCTT
| Original motif Consensus sequence: CTGGCCTC |
Reverse complement motif Consensus sequence: GAGGCCAG |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 109 |
Motif name: Pax5 |
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
Best Matches for Motif ID 109 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
20 |
| Similarity score: |
0.0458242 |
Alignment:
MGGTCAGGGTGACCTRDBHV
MSGKKRCGCWDCABTGBBCD
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
70 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
20 |
| Similarity score: |
0.052658 |
Alignment:
VRVDGGHACAVDDKGTHCTDWH
--MSGKKRCGCWDCABTGBBCD
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
167 |
| Motif name: |
rsyAGrkGGCGCCmyCTrsy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
20 |
| Similarity score: |
0.0660509 |
Alignment:
HSKAGKYGGCGCCRMCTMSD
MSGKKRCGCWDCABTGBBCD
| Original motif Consensus sequence: DSYAGRKGGCGCCMYCTRSH |
Reverse complement motif Consensus sequence: HSKAGKYGGCGCCRMCTMSD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
131 |
| Motif name: |
znf143 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
19 |
| Similarity score: |
0.552768 |
Alignment:
-BAHYTCCCAKMATGCMWYGC
MSGKKRCGCWDCABTGBBCD-
| Original motif Consensus sequence: BAHYTCCCAKMATGCMWYGC |
Reverse complement motif Consensus sequence: GCMWRGCATYRTGGGAMHTB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
19 |
| Similarity score: |
0.559888 |
Alignment:
YDRCCASYAGRKGGCRSYV-
DGVBCABTGDWGCGKRRCSR
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 110 |
Motif name: PLAG1 |
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
Best Matches for Motif ID 110 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
129 |
| Motif name: |
Zfp423 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0944264 |
Alignment:
GSMMCCYARGGKKKC
GGGGCCCAAGGGGG-
| Original motif Consensus sequence: GSMMCCYARGGKKKC |
Reverse complement motif Consensus sequence: GYRYCCMTKGGYRSC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
166 |
| Motif name: |
CasCAGrGGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0955683 |
Alignment:
BSKGCCCKCTGGTG
GGGGCCCAAGGGGG
| Original motif Consensus sequence: CACCAGRGGGCRSB |
Reverse complement motif Consensus sequence: BSKGCCCKCTGGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
14 |
| Similarity score: |
0.0963962 |
Alignment:
VHGGCCAGMAGAGGGCRBY
GGGGCCCAAGGGGG-----
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
165 |
| Motif name: |
wgGCCAshAGrGGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0984905 |
Alignment:
HDGCCACHAGRGGGCRBY
GGGGCCCAAGGGGG----
| Original motif Consensus sequence: HDGCCACHAGRGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCKCTHGTGGCHH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0985427 |
Alignment:
BMSMGCCYMCTKSTGGMHM
-----CCCCCTTGGGCCCC
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 111 |
Motif name: PPARG |
| Original motif Consensus sequence: STAGGTCACBGTGACCYABT |
Reverse complement motif Consensus sequence: ABTMGGTCACBGTGACCTAS |
|
|
 |
 |
Best Matches for Motif ID 111 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
70 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
20 |
| Similarity score: |
0.0567628 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
--ABTMGGTCACBGTGACCTAS
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
167 |
| Motif name: |
rsyAGrkGGCGCCmyCTrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
19 |
| Similarity score: |
0.574193 |
Alignment:
HSKAGKYGGCGCCRMCTMSD-
-STAGGTCACBGTGACCYABT
| Original motif Consensus sequence: DSYAGRKGGCGCCMYCTRSH |
Reverse complement motif Consensus sequence: HSKAGKYGGCGCCRMCTMSD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
82 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
1.00511 |
Alignment:
--BAGGYCABHBTGACCKHV
ABTMGGTCACBGTGACCTAS
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
18 |
| Similarity score: |
1.01193 |
Alignment:
--VDBHMAGGTCACCCTGACCY
ABTMGGTCACBGTGACCTAS--
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
109 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
18 |
| Similarity score: |
1.07268 |
Alignment:
MSGKKRCGCWDCABTGBBCD--
--STAGGTCACBGTGACCYABT
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 112 |
Motif name: RELA |
| Original motif Consensus sequence: GGGRATTTCC |
Reverse complement motif Consensus sequence: GGAAATKCCC |
|
|
 |
 |
Best Matches for Motif ID 112 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
102 |
| Motif name: |
NF-kappaB |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.00719219 |
Alignment:
GGMAAYKCCC
GGAAATKCCC
| Original motif Consensus sequence: GGGRMTTYCC |
Reverse complement motif Consensus sequence: GGMAAYKCCC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
103 |
| Motif name: |
NFKB1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0283179 |
Alignment:
GGGGRTTCCCC
GGGRATTTCC-
| Original motif Consensus sequence: GGGGRTTCCCC |
Reverse complement motif Consensus sequence: GGGGAAKCCCC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
125 |
| Motif name: |
TP53 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
10 |
| Similarity score: |
0.057648 |
Alignment:
MSGGACATGYCCGGGCATGT
--GGAAATKCCC--------
| Original motif Consensus sequence: MSGGACATGYCCGGGCATGT |
Reverse complement motif Consensus sequence: ACATGCCCGGKCATGTCCSR |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
113 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0666417 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
GGGRATTTCC-----------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
170 |
| Motif name: |
ssGGCrsTGCrs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0784221 |
Alignment:
VVGGCRSTGCVB
--GGAAATKCCC
| Original motif Consensus sequence: VVGGCRSTGCVB |
Reverse complement motif Consensus sequence: BVGCASMGCCVV |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 113 |
Motif name: REST |
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
Best Matches for Motif ID 113 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
70 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
21 |
| Similarity score: |
0.0685835 |
Alignment:
VRVDGGHACAVDDKGTHCTDWH
-GGYGCTGTCCATGGTGCTGAA
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
131 |
| Motif name: |
znf143 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
19 |
| Similarity score: |
1.0683 |
Alignment:
GCMWRGCATYRTGGGAMHTB--
-TTCAGCACCATGGACAGCKCC
| Original motif Consensus sequence: BAHYTCCCAKMATGCMWYGC |
Reverse complement motif Consensus sequence: GCMWRGCATYRTGGGAMHTB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
19 |
| Similarity score: |
1.07477 |
Alignment:
--VHGGCCAGMAGAGGGCRBY
TTCAGCACCATGGACAGCKCC
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
156 |
| Motif name: |
rgyGCCMyCTksTGGccd |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
1.57039 |
Alignment:
---DDGCCASYAGMGGGCKVM
TTCAGCACCATGGACAGCKCC
| Original motif Consensus sequence: RVYGCCCYCTKSTGGCHD |
Reverse complement motif Consensus sequence: DDGCCASYAGMGGGCKVM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
149 |
| Motif name: |
asmAGRGGGCrCTGsmkc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
1.57097 |
Alignment:
DBTSCAGMGCCCTCTRST---
GGYGCTGTCCATGGTGCTGAA
| Original motif Consensus sequence: ASMAGAGGGCRCTGSABH |
Reverse complement motif Consensus sequence: DBTSCAGMGCCCTCTRST |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 114 |
Motif name: RUNX1 |
| Original motif Consensus sequence: BBYTGTGGTTT |
Reverse complement motif Consensus sequence: AAACCACAKVB |
|
|
 |
 |
Best Matches for Motif ID 114 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
70 |
| Motif name: |
Ar |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0360379 |
Alignment:
VRVDGGHACAVDDKGTHCTDWH
-BBYTGTGGTTT----------
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
87 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0433682 |
Alignment:
RGGBCAAAGKYCA
--AAACCACAKVB
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
165 |
| Motif name: |
wgGCCAshAGrGGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
11 |
| Similarity score: |
0.0447401 |
Alignment:
KBKGCCCKCTHGTGGCHH
-------BBYTGTGGTTT
| Original motif Consensus sequence: HDGCCACHAGRGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCKCTHGTGGCHH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
88 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0456894 |
Alignment:
TGYCAGGGGGCR
-BBYTGTGGTTT
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
156 |
| Motif name: |
rgyGCCMyCTksTGGccd |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
11 |
| Similarity score: |
0.0459745 |
Alignment:
RVYGCCCYCTKSTGGCHD
-------BBYTGTGGTTT
| Original motif Consensus sequence: RVYGCCCYCTKSTGGCHD |
Reverse complement motif Consensus sequence: DDGCCASYAGMGGGCKVM |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 115 |
Motif name: RXRRAR_DR5 |
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
 |
 |
Best Matches for Motif ID 115 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
111 |
| Motif name: |
PPARG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
17 |
| Similarity score: |
0.0471437 |
Alignment:
STAGGTCACBGTGACCYABT
--RGKTCABVVRGAGGTCA-
| Original motif Consensus sequence: STAGGTCACBGTGACCYABT |
Reverse complement motif Consensus sequence: ABTMGGTCACBGTGACCTAS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
17 |
| Similarity score: |
0.0543729 |
Alignment:
MGGTCAGGGTGACCTRDBHV
---RGKTCABVVRGAGGTCA
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
82 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
17 |
| Similarity score: |
0.058015 |
Alignment:
VHRGGTCABDBTGMCCTB
-TGACCTCKVVBTGAYCK
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
109 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
17 |
| Similarity score: |
0.0584167 |
Alignment:
MSGKKRCGCWDCABTGBBCD
---TGACCTCKVVBTGAYCK
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
17 |
| Similarity score: |
0.0732845 |
Alignment:
KBKGCCCTCTYCTGGCCHV
--TGACCTCKVVBTGAYCK
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 116 |
Motif name: SP1 |
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
Best Matches for Motif ID 116 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
155 |
| Motif name: |
csCSCCdCCCcs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0226218 |
Alignment:
VBCCCCDCCCHV
--CCCCKCCCCC
| Original motif Consensus sequence: VBCCCCDCCCHV |
Reverse complement motif Consensus sequence: VDGGGDGGGGBV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
167 |
| Motif name: |
rsyAGrkGGCGCCmyCTrsy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
10 |
| Number of overlap: |
10 |
| Similarity score: |
0.0535317 |
Alignment:
HSKAGKYGGCGCCRMCTMSD
-GGGGGYGGGG---------
| Original motif Consensus sequence: DSYAGRKGGCGCCMYCTRSH |
Reverse complement motif Consensus sequence: HSKAGKYGGCGCCRMCTMSD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
10 |
| Number of overlap: |
10 |
| Similarity score: |
0.0604118 |
Alignment:
YDRCCASYAGRKGGCRSYV
GGGGGYGGGG---------
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
146 |
| Motif name: |
myrGYGCCmCCTast |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0650794 |
Alignment:
ASTAGGYGGCGCTBB
-GGGGGYGGGG----
| Original motif Consensus sequence: VBAGCGCCMCCTAST |
Reverse complement motif Consensus sequence: ASTAGGYGGCGCTBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
130 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0664832 |
Alignment:
VAGGCCBBGGCVBB
----CCCCKCCCCC
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 117 |
Motif name: Spz1 |
| Original motif Consensus sequence: AGGGTAWCAGC |
Reverse complement motif Consensus sequence: GCTGWTACCCT |
|
|
 |
 |
Best Matches for Motif ID 117 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
143 |
| Motif name: |
AgmAGAGGGCrscAGak |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
11 |
| Similarity score: |
0.0235542 |
Alignment:
RTCTGSTGCCCTCTYCT
-GCTGWTACCCT-----
| Original motif Consensus sequence: AGMAGAGGGCASCAGAK |
Reverse complement motif Consensus sequence: RTCTGSTGCCCTCTYCT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
164 |
| Motif name: |
asyAGrkGGCRGCAga |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
11 |
| Similarity score: |
0.0381204 |
Alignment:
ASYAGRKGGCAGCABH
-----AGGGTAWCAGC
| Original motif Consensus sequence: ASYAGRKGGCAGCABH |
Reverse complement motif Consensus sequence: HBTGCTGCCYMCTKST |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
70 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0401654 |
Alignment:
VRVDGGHACAVDDKGTHCTDWH
-GCTGWTACCCT----------
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
0.0458949 |
Alignment:
VDBHMAGGTCACCCTGACCY
-----GCTGWTACCCT----
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
122 |
| Motif name: |
Tcfcp2l1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0503934 |
Alignment:
CCAGYYHVADCCRG
---GCTGWTACCCT
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 118 |
Motif name: SRF |
| Original motif Consensus sequence: GCCCATATATGG |
Reverse complement motif Consensus sequence: CCATATATGGGC |
|
|
 |
 |
Best Matches for Motif ID 118 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
165 |
| Motif name: |
wgGCCAshAGrGGGCrsy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
12 |
| Similarity score: |
0.0548555 |
Alignment:
KBKGCCCKCTHGTGGCHH
----CCATATATGGGC--
| Original motif Consensus sequence: HDGCCACHAGRGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCKCTHGTGGCHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
12 |
| Similarity score: |
0.0556649 |
Alignment:
KBKGCCCTCTYCTGGCCHV
---GCCCATATATGG----
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
156 |
| Motif name: |
rgyGCCMyCTksTGGccd |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0558448 |
Alignment:
DDGCCASYAGMGGGCKVM
--GCCCATATATGG----
| Original motif Consensus sequence: RVYGCCCYCTKSTGGCHD |
Reverse complement motif Consensus sequence: DDGCCASYAGMGGGCKVM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
12 |
| Similarity score: |
0.0566321 |
Alignment:
YDRCCASYAGRKGGCRSYV
----CCATATATGGGC---
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
67 |
| Motif name: |
Motif 67 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0566949 |
Alignment:
CCASHAGKGGGCKS
CCATATATGGGC--
| Original motif Consensus sequence: SYGCCCYCTDSTGG |
Reverse complement motif Consensus sequence: CCASHAGKGGGCKS |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 119 |
Motif name: Stat3 |
| Original motif Consensus sequence: TTCCAGGAAG |
Reverse complement motif Consensus sequence: CTTCCTGGAA |
|
|
 |
 |
Best Matches for Motif ID 119 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
79 |
| Motif name: |
ELK1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0612721 |
Alignment:
MTTCCGGHBV
CTTCCTGGAA
| Original motif Consensus sequence: VDDCCGGAAR |
Reverse complement motif Consensus sequence: MTTCCGGHBV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
77 |
| Motif name: |
EBF1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0803002 |
Alignment:
TCCCTYGGGY
TTCCAGGAAG
| Original motif Consensus sequence: MCCCMAGGGA |
Reverse complement motif Consensus sequence: TCCCTYGGGY |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
88 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0834676 |
Alignment:
TGYCAGGGGGCR
TTCCAGGAAG--
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0852106 |
Alignment:
YDRCCASYAGRKGGCRSYV
-TTCCAGGAAG--------
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
125 |
| Motif name: |
TP53 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
10 |
| Similarity score: |
0.087969 |
Alignment:
MSGGACATGYCCGGGCATGT
-------CTTCCTGGAA---
| Original motif Consensus sequence: MSGGACATGYCCGGGCATGT |
Reverse complement motif Consensus sequence: ACATGCCCGGKCATGTCCSR |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 120 |
Motif name: T |
| Original motif Consensus sequence: CTAGGTGTGAA |
Reverse complement motif Consensus sequence: TTCACACCTAG |
|
|
 |
 |
Best Matches for Motif ID 120 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
144 |
| Motif name: |
ctCTrsyGCCmCCTast |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0559721 |
Alignment:
HDCTGSYGCCMCCTAST
-----TTCACACCTAG-
| Original motif Consensus sequence: HDCTGSYGCCMCCTAST |
Reverse complement motif Consensus sequence: ASTAGGYGGCMSCAGDD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
164 |
| Motif name: |
asyAGrkGGCRGCAga |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0641201 |
Alignment:
ASYAGRKGGCAGCABH
-CTAGGTGTGAA----
| Original motif Consensus sequence: ASYAGRKGGCAGCABH |
Reverse complement motif Consensus sequence: HBTGCTGCCYMCTKST |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
146 |
| Motif name: |
myrGYGCCmCCTast |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0654989 |
Alignment:
VBAGCGCCMCCTAST
---TTCACACCTAG-
| Original motif Consensus sequence: VBAGCGCCMCCTAST |
Reverse complement motif Consensus sequence: ASTAGGYGGCGCTBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
11 |
| Similarity score: |
0.0692022 |
Alignment:
YDRCCASYAGRKGGCRSYV
------CTAGGTGTGAA--
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
111 |
| Motif name: |
PPARG |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
10 |
| Number of overlap: |
11 |
| Similarity score: |
0.070764 |
Alignment:
ABTMGGTCACBGTGACCTAS
---------TTCACACCTAG
| Original motif Consensus sequence: STAGGTCACBGTGACCYABT |
Reverse complement motif Consensus sequence: ABTMGGTCACBGTGACCTAS |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 121 |
Motif name: TAL1TCF3 |
| Original motif Consensus sequence: HVAMCATCTGKT |
Reverse complement motif Consensus sequence: ARCAGATGRTVD |
|
|
 |
 |
Best Matches for Motif ID 121 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
147 |
| Motif name: |
asCAGrkGGCrsy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0388867 |
Alignment:
ASCAGRGGGCRSB
ARCAGATGRTVD-
| Original motif Consensus sequence: ASCAGRGGGCRSB |
Reverse complement motif Consensus sequence: BSKGCCCMCTGST |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
166 |
| Motif name: |
CasCAGrGGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0389554 |
Alignment:
BSKGCCCKCTGGTG
-HVAMCATCTGKT-
| Original motif Consensus sequence: CACCAGRGGGCRSB |
Reverse complement motif Consensus sequence: BSKGCCCKCTGGTG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
104 |
| Motif name: |
NFYA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
12 |
| Similarity score: |
0.0418463 |
Alignment:
BHVCKSATTGGMKBVV
---HVAMCATCTGKT-
| Original motif Consensus sequence: VBBRRCCAATSRGVDB |
Reverse complement motif Consensus sequence: BHVCKSATTGGMKBVV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
151 |
| Motif name: |
agrCCAGmAGrg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0439721 |
Alignment:
HVGCCAGMAGRG
HVAMCATCTGKT
| Original motif Consensus sequence: HVGCCAGMAGRG |
Reverse complement motif Consensus sequence: CKCTRCTGGCVH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
143 |
| Motif name: |
AgmAGAGGGCrscAGak |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
12 |
| Similarity score: |
0.0474478 |
Alignment:
AGMAGAGGGCASCAGAK
ARCAGATGRTVD-----
| Original motif Consensus sequence: AGMAGAGGGCASCAGAK |
Reverse complement motif Consensus sequence: RTCTGSTGCCCTCTYCT |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 122 |
Motif name: Tcfcp2l1 |
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
Best Matches for Motif ID 122 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
167 |
| Motif name: |
rsyAGrkGGCGCCmyCTrsy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
14 |
| Similarity score: |
0.0620246 |
Alignment:
DSYAGRKGGCGCCMYCTRSH
-----CKGGDTBDMMCTGG-
| Original motif Consensus sequence: DSYAGRKGGCGCCMYCTRSH |
Reverse complement motif Consensus sequence: HSKAGKYGGCGCCRMCTMSD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
70 |
| Motif name: |
Ar |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
14 |
| Similarity score: |
0.0642855 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
------CCAGYYHVADCCRG--
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
144 |
| Motif name: |
ctCTrsyGCCmCCTast |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0675705 |
Alignment:
HDCTGSYGCCMCCTAST
--CKGGDTBDMMCTGG-
| Original motif Consensus sequence: HDCTGSYGCCMCCTAST |
Reverse complement motif Consensus sequence: ASTAGGYGGCMSCAGDD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.0676841 |
Alignment:
MGGTCAGGGTGACCTRDBHV
---CCAGYYHVADCCRG---
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
130 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0677766 |
Alignment:
BBVGCCBVGGCCTV
CCAGYYHVADCCRG
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 123 |
Motif name: TFAP2A |
| Original motif Consensus sequence: GCCBBVRGS |
Reverse complement motif Consensus sequence: SCMVBBGGC |
|
|
 |
 |
Best Matches for Motif ID 123 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
130 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
9 |
| Similarity score: |
0.0365038 |
Alignment:
VAGGCCBBGGCVBB
---GCCBBVRGS--
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
137 |
| Motif name: |
rgCGCCmyCTgs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
9 |
| Similarity score: |
0.0578475 |
Alignment:
SHAGKGGGCGCB
SCMVBBGGC---
| Original motif Consensus sequence: VGCGCCCYCTDS |
Reverse complement motif Consensus sequence: SHAGKGGGCGCB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
124 |
| Motif name: |
TLX1NFIC |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
9 |
| Similarity score: |
0.0626983 |
Alignment:
TGGCASBDHGCCAA
--GCCBBVRGS---
| Original motif Consensus sequence: TGGCASBDHGCCAA |
Reverse complement motif Consensus sequence: TTGGCHDBSTGCCA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
147 |
| Motif name: |
asCAGrkGGCrsy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.0631625 |
Alignment:
ASCAGRGGGCRSB
-SCMVBBGGC---
| Original motif Consensus sequence: ASCAGRGGGCRSB |
Reverse complement motif Consensus sequence: BSKGCCCMCTGST |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
153 |
| Motif name: |
scAGrkGGCGcy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
9 |
| Similarity score: |
0.0640748 |
Alignment:
SHAGRGGGCGCB
SCMVBBGGC---
| Original motif Consensus sequence: SHAGRGGGCGCB |
Reverse complement motif Consensus sequence: VGCGCCCMCTDS |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 124 |
Motif name: TLX1NFIC |
| Original motif Consensus sequence: TGGCASBDHGCCAA |
Reverse complement motif Consensus sequence: TTGGCHDBSTGCCA |
|
|
 |
 |
Best Matches for Motif ID 124 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
14 |
| Similarity score: |
0.0758334 |
Alignment:
VDBHMAGGTCACCCTGACCY
------TGGCASBDHGCCAA
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
131 |
| Motif name: |
znf143 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.0763138 |
Alignment:
GCMWRGCATYRTGGGAMHTB
---TGGCASBDHGCCAA---
| Original motif Consensus sequence: BAHYTCCCAKMATGCMWYGC |
Reverse complement motif Consensus sequence: GCMWRGCATYRTGGGAMHTB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
82 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0777688 |
Alignment:
VHRGGTCABDBTGMCCTB
---TGGCASBDHGCCAA-
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
111 |
| Motif name: |
PPARG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.0838712 |
Alignment:
STAGGTCACBGTGACCYABT
---TGGCASBDHGCCAA---
| Original motif Consensus sequence: STAGGTCACBGTGACCYABT |
Reverse complement motif Consensus sequence: ABTMGGTCACBGTGACCTAS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
113 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
14 |
| Similarity score: |
0.0916353 |
Alignment:
TTCAGCACCATGGACAGCKCC
--TGGCASBDHGCCAA-----
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 125 |
Motif name: TP53 |
| Original motif Consensus sequence: MSGGACATGYCCGGGCATGT |
Reverse complement motif Consensus sequence: ACATGCCCGGKCATGTCCSR |
|
|
 |
 |
Best Matches for Motif ID 125 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
19 |
| Similarity score: |
0.0641029 |
Alignment:
-KBKGCCCTCTYCTGGCCHV
ACATGCCCGGKCATGTCCSR
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
131 |
| Motif name: |
znf143 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
19 |
| Similarity score: |
0.0730057 |
Alignment:
-BAHYTCCCAKMATGCMWYGC
ACATGCCCGGKCATGTCCSR-
| Original motif Consensus sequence: BAHYTCCCAKMATGCMWYGC |
Reverse complement motif Consensus sequence: GCMWRGCATYRTGGGAMHTB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
82 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
15 |
| Similarity score: |
2.07342 |
Alignment:
-----BAGGYCABHBTGACCKHV
MSGGACATGYCCGGGCATGT---
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
130 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.55908 |
Alignment:
------VAGGCCBBGGCVBB
ACATGCCCGGKCATGTCCSR
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
122 |
| Motif name: |
Tcfcp2l1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.56997 |
Alignment:
------CKGGDTBDMMCTGG
MSGGACATGYCCGGGCATGT
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 126 |
Motif name: USF1 |
| Original motif Consensus sequence: CACGTGR |
Reverse complement motif Consensus sequence: MCACGTG |
|
|
 |
 |
Best Matches for Motif ID 126 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
91 |
| Motif name: |
MAX |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0 |
Alignment:
DAHCACGTGD
-MCACGTG--
| Original motif Consensus sequence: DAHCACGTGD |
Reverse complement motif Consensus sequence: BCACGTGDTD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
95 |
| Motif name: |
MYCMAX |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0108143 |
Alignment:
ACCACGTGSTM
-MCACGTG---
| Original motif Consensus sequence: RASCACGTGGT |
Reverse complement motif Consensus sequence: ACCACGTGSTM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
94 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0198377 |
Alignment:
VGCACGTGGH
--CACGTGR-
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
96 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0207062 |
Alignment:
HSCACGTGGC
--CACGTGR-
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
86 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0436113 |
Alignment:
VBACGTGV
-MCACGTG
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 127 |
Motif name: YY1 |
| Original motif Consensus sequence: RCCATB |
Reverse complement motif Consensus sequence: BATGGM |
|
|
 |
 |
Best Matches for Motif ID 127 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
61 |
| Motif name: |
Motif 61 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.00122549 |
Alignment:
KCCATCT
-RCCATB
| Original motif Consensus sequence: AGATGGY |
Reverse complement motif Consensus sequence: KCCATCT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
12 |
| Motif name: |
Motif 12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.00857843 |
Alignment:
GAGCCATC
RCCATB--
| Original motif Consensus sequence: GATGGCTC |
Reverse complement motif Consensus sequence: GAGCCATC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
145 |
| Motif name: |
grCCACyAGAkG |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.022036 |
Alignment:
CYTCTKGTGGHH
-----BATGGM-
| Original motif Consensus sequence: DDCCACYAGAKG |
Reverse complement motif Consensus sequence: CYTCTKGTGGHH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0220417 |
Alignment:
BMSMGCCYMCTKSTGGMHM
-----------BATGGM--
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
138 |
| Motif name: |
grCCACyAGAkG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0223679 |
Alignment:
DDCCACYAGAKG
-RCCATB-----
| Original motif Consensus sequence: DDCCACYAGAKG |
Reverse complement motif Consensus sequence: CYTCTMGTGGHH |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 128 |
Motif name: ZEB1 |
| Original motif Consensus sequence: CACCTD |
Reverse complement motif Consensus sequence: HAGGTG |
|
|
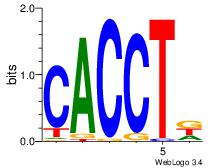 |
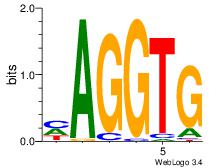 |
Best Matches for Motif ID 128 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
120 |
| Motif name: |
T |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0192581 |
Alignment:
TTCACACCTAG
-CACCTD----
| Original motif Consensus sequence: CTAGGTGTGAA |
Reverse complement motif Consensus sequence: TTCACACCTAG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
146 |
| Motif name: |
myrGYGCCmCCTast |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0325288 |
Alignment:
VBAGCGCCMCCTAST
-------CACCTD--
| Original motif Consensus sequence: VBAGCGCCMCCTAST |
Reverse complement motif Consensus sequence: ASTAGGYGGCGCTBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
6 |
| Similarity score: |
0.0341274 |
Alignment:
BMSMGCCYMCTKSTGGMHM
-------CACCTD------
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
164 |
| Motif name: |
asyAGrkGGCRGCAga |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
6 |
| Similarity score: |
0.0351031 |
Alignment:
ASYAGRKGGCAGCABH
--HAGGTG--------
| Original motif Consensus sequence: ASYAGRKGGCAGCABH |
Reverse complement motif Consensus sequence: HBTGCTGCCYMCTKST |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
144 |
| Motif name: |
ctCTrsyGCCmCCTast |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0356696 |
Alignment:
HDCTGSYGCCMCCTAST
---------CACCTD--
| Original motif Consensus sequence: HDCTGSYGCCMCCTAST |
Reverse complement motif Consensus sequence: ASTAGGYGGCMSCAGDD |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 129 |
Motif name: Zfp423 |
| Original motif Consensus sequence: GSMMCCYARGGKKKC |
Reverse complement motif Consensus sequence: GYRYCCMTKGGYRSC |
|
|
 |
 |
Best Matches for Motif ID 129 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
104 |
| Motif name: |
NFYA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
0.0358106 |
Alignment:
BHVCKSATTGGMKBVV
GYRYCCMTKGGYRSC-
| Original motif Consensus sequence: VBBRRCCAATSRGVDB |
Reverse complement motif Consensus sequence: BHVCKSATTGGMKBVV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
15 |
| Similarity score: |
0.0359705 |
Alignment:
BMSMGCCYMCTKSTGGMHM
--GYRYCCMTKGGYRSC--
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
156 |
| Motif name: |
rgyGCCMyCTksTGGccd |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
15 |
| Similarity score: |
0.0382225 |
Alignment:
RVYGCCCYCTKSTGGCHD
-GYRYCCMTKGGYRSC--
| Original motif Consensus sequence: RVYGCCCYCTKSTGGCHD |
Reverse complement motif Consensus sequence: DDGCCASYAGMGGGCKVM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
70 |
| Motif name: |
Ar |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
15 |
| Similarity score: |
0.0429662 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
---GSMMCCYARGGKKKC----
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
109 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
15 |
| Similarity score: |
0.0430294 |
Alignment:
DGVBCABTGDWGCGKRRCSR
--GSMMCCYARGGKKKC---
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 130 |
Motif name: Zfx |
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
Best Matches for Motif ID 130 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
149 |
| Motif name: |
asmAGRGGGCrCTGsmkc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0651255 |
Alignment:
DBTSCAGMGCCCTCTRST
BBVGCCBVGGCCTV----
| Original motif Consensus sequence: ASMAGAGGGCRCTGSABH |
Reverse complement motif Consensus sequence: DBTSCAGMGCCCTCTRST |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
109 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
14 |
| Similarity score: |
0.0724025 |
Alignment:
DGVBCABTGDWGCGKRRCSR
--BBVGCCBVGGCCTV----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
82 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0748557 |
Alignment:
VHRGGTCABDBTGMCCTB
---VAGGCCBBGGCVBB-
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
125 |
| Motif name: |
TP53 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
14 |
| Similarity score: |
0.0763693 |
Alignment:
ACATGCCCGGKCATGTCCSR
-----BBVGCCBVGGCCTV-
| Original motif Consensus sequence: MSGGACATGYCCGGGCATGT |
Reverse complement motif Consensus sequence: ACATGCCCGGKCATGTCCSR |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
168 |
| Motif name: |
yrcrGYGCCMyCTGGtG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0830122 |
Alignment:
HVCAGCGCCCYCTGGTG
BBVGCCBVGGCCTV---
| Original motif Consensus sequence: HVCAGCGCCCYCTGGTG |
Reverse complement motif Consensus sequence: CACCAGMGGGCGCTGBD |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 131 |
Motif name: znf143 |
| Original motif Consensus sequence: BAHYTCCCAKMATGCMWYGC |
Reverse complement motif Consensus sequence: GCMWRGCATYRTGGGAMHTB |
|
|
 |
 |
Best Matches for Motif ID 131 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
70 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
20 |
| Similarity score: |
0.0555912 |
Alignment:
VRVDGGHACAVDDKGTHCTDWH
GCMWRGCATYRTGGGAMHTB--
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
109 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
19 |
| Similarity score: |
0.554242 |
Alignment:
DGVBCABTGDWGCGKRRCSR-
-GCMWRGCATYRTGGGAMHTB
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
113 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
19 |
| Similarity score: |
0.558571 |
Alignment:
-GGYGCTGTCCATGGTGCTGAA
BAHYTCCCAKMATGCMWYGC--
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
125 |
| Motif name: |
TP53 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
19 |
| Similarity score: |
0.566085 |
Alignment:
MSGGACATGYCCGGGCATGT-
-GCMWRGCATYRTGGGAMHTB
| Original motif Consensus sequence: MSGGACATGYCCGGGCATGT |
Reverse complement motif Consensus sequence: ACATGCCCGGKCATGTCCSR |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
18 |
| Similarity score: |
1.05907 |
Alignment:
VDBHMAGGTCACCCTGACCY--
--BAHYTCCCAKMATGCMWYGC
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 132 |
Motif name: ZNF354C |
| Original motif Consensus sequence: MTCCAC |
Reverse complement motif Consensus sequence: GTGGAY |
|
|
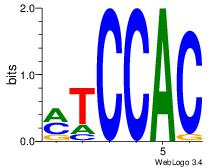 |
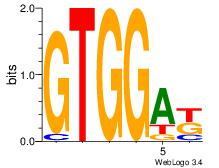 |
Best Matches for Motif ID 132 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
145 |
| Motif name: |
grCCACyAGAkG |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
6 |
| Similarity score: |
0.0253111 |
Alignment:
CYTCTKGTGGHH
------GTGGAY
| Original motif Consensus sequence: DDCCACYAGAKG |
Reverse complement motif Consensus sequence: CYTCTKGTGGHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
158 |
| Motif name: |
grCCACwAGrk |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0266594 |
Alignment:
YMCTWGTGGHH
-----GTGGAY
| Original motif Consensus sequence: DDCCACWAGRK |
Reverse complement motif Consensus sequence: YMCTWGTGGHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
138 |
| Motif name: |
grCCACyAGAkG |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
6 |
| Similarity score: |
0.0292621 |
Alignment:
CYTCTMGTGGHH
------GTGGAY
| Original motif Consensus sequence: DDCCACYAGAKG |
Reverse complement motif Consensus sequence: CYTCTMGTGGHH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
114 |
| Motif name: |
RUNX1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0470937 |
Alignment:
BBYTGTGGTTT
-GTGGAY----
| Original motif Consensus sequence: BBYTGTGGTTT |
Reverse complement motif Consensus sequence: AAACCACAKVB |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0507812 |
Alignment:
SWGCCACC
-MTCCAC-
| Original motif Consensus sequence: GGTGGCWS |
Reverse complement motif Consensus sequence: SWGCCACC |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 133 |
Motif name: shAGrGGGCAgy |
| Original motif Consensus sequence: SHAGRGGGCABH |
Reverse complement motif Consensus sequence: DBTGCCCKCTDS |
|
|
 |
 |
Best Matches for Motif ID 133 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
165 |
| Motif name: |
wgGCCAshAGrGGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
12 |
| Similarity score: |
0.00159897 |
Alignment:
HDGCCACHAGRGGGCRBY
------SHAGRGGGCABH
| Original motif Consensus sequence: HDGCCACHAGRGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCKCTHGTGGCHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
143 |
| Motif name: |
AgmAGAGGGCrscAGak |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
12 |
| Similarity score: |
0.00815796 |
Alignment:
AGMAGAGGGCASCAGAK
-SHAGRGGGCABH----
| Original motif Consensus sequence: AGMAGAGGGCASCAGAK |
Reverse complement motif Consensus sequence: RTCTGSTGCCCTCTYCT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
12 |
| Similarity score: |
0.0109424 |
Alignment:
VHGGCCAGMAGAGGGCRBY
-------SHAGRGGGCABH
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
166 |
| Motif name: |
CasCAGrGGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0124424 |
Alignment:
CACCAGRGGGCRSB
--SHAGRGGGCABH
| Original motif Consensus sequence: CACCAGRGGGCRSB |
Reverse complement motif Consensus sequence: BSKGCCCKCTGGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
147 |
| Motif name: |
asCAGrkGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0128393 |
Alignment:
ASCAGRGGGCRSB
-SHAGRGGGCABH
| Original motif Consensus sequence: ASCAGRGGGCRSB |
Reverse complement motif Consensus sequence: BSKGCCCMCTGST |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 134 |
Motif name: ssCGwGCGss |
| Original motif Consensus sequence: BSCGWGCGBV |
Reverse complement motif Consensus sequence: VBCGCWCGSB |
|
|
 |
 |
Best Matches for Motif ID 134 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
135 |
| Motif name: |
ssCGGCCGss |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0113669 |
Alignment:
BSCGGCCGSV
VBCGCWCGSB
| Original motif Consensus sequence: BSCGGCCGSV |
Reverse complement motif Consensus sequence: VSCGGCCGSB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
161 |
| Motif name: |
ssCGCwGCGss |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0473417 |
Alignment:
BSCGCDGCGSV
-BSCGWGCGBV
| Original motif Consensus sequence: VSCGCDGCGSB |
Reverse complement motif Consensus sequence: BSCGCDGCGSV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
152 |
| Motif name: |
yrCATGCAyr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0541916 |
Alignment:
BRCATGCABD
BSCGWGCGBV
| Original motif Consensus sequence: BRCATGCABD |
Reverse complement motif Consensus sequence: HVTGCATGKV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
158 |
| Motif name: |
grCCACwAGrk |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0546634 |
Alignment:
DDCCACWAGRK
-VBCGCWCGSB
| Original motif Consensus sequence: DDCCACWAGRK |
Reverse complement motif Consensus sequence: YMCTWGTGGHH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
109 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
10 |
| Similarity score: |
0.0551638 |
Alignment:
DGVBCABTGDWGCGKRRCSR
------BSCGWGCGBV----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 135 |
Motif name: ssCGGCCGss |
| Original motif Consensus sequence: BSCGGCCGSV |
Reverse complement motif Consensus sequence: VSCGGCCGSB |
|
|
 |
 |
Best Matches for Motif ID 135 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
134 |
| Motif name: |
ssCGwGCGss |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0 |
Alignment:
BSCGWGCGBV
VSCGGCCGSB
| Original motif Consensus sequence: BSCGWGCGBV |
Reverse complement motif Consensus sequence: VBCGCWCGSB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
171 |
| Motif name: |
ysGTGGCCACsr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0413265 |
Alignment:
BVGTGGCCACBV
-VSCGGCCGSB-
| Original motif Consensus sequence: BVGTGGCCACBV |
Reverse complement motif Consensus sequence: VBGTGGCCACVB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
82 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0424008 |
Alignment:
BAGGYCABHBTGACCKHV
--------VSCGGCCGSB
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
111 |
| Motif name: |
PPARG |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.046338 |
Alignment:
ABTMGGTCACBGTGACCTAS
-VSCGGCCGSB---------
| Original motif Consensus sequence: STAGGTCACBGTGACCYABT |
Reverse complement motif Consensus sequence: ABTMGGTCACBGTGACCTAS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
10 |
| Similarity score: |
0.0468951 |
Alignment:
MGGTCAGGGTGACCTRDBHV
-------VSCGGCCGSB---
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 136 |
Motif name: dwCAGAAGwh |
| Original motif Consensus sequence: DHCAGAAGDH |
Reverse complement motif Consensus sequence: HDCTTCTGHD |
|
|
 |
 |
Best Matches for Motif ID 136 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
151 |
| Motif name: |
agrCCAGmAGrg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.00562474 |
Alignment:
HVGCCAGMAGRG
--DHCAGAAGDH
| Original motif Consensus sequence: HVGCCAGMAGRG |
Reverse complement motif Consensus sequence: CKCTRCTGGCVH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
141 |
| Motif name: |
raCAAAACam |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0107169 |
Alignment:
DACAAAACAH
DHCAGAAGDH
| Original motif Consensus sequence: DACAAAACAH |
Reverse complement motif Consensus sequence: HTGTTTTGTD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
158 |
| Motif name: |
grCCACwAGrk |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0167426 |
Alignment:
DDCCACWAGRK
-DHCAGAAGDH
| Original motif Consensus sequence: DDCCACWAGRK |
Reverse complement motif Consensus sequence: YMCTWGTGGHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
139 |
| Motif name: |
mkCTyTTCsg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0207259 |
Alignment:
HBCTTTTCBD
HDCTTCTGHD
| Original motif Consensus sequence: HBCTTTTCBD |
Reverse complement motif Consensus sequence: HBGAAAAGBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0212719 |
Alignment:
VHGGCCAGMAGAGGGCRBY
---DHCAGAAGDH------
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 137 |
Motif name: rgCGCCmyCTgs |
| Original motif Consensus sequence: VGCGCCCYCTDS |
Reverse complement motif Consensus sequence: SHAGKGGGCGCB |
|
|
 |
 |
Best Matches for Motif ID 137 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
153 |
| Motif name: |
scAGrkGGCGcy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0 |
Alignment:
VGCGCCCMCTDS
VGCGCCCYCTDS
| Original motif Consensus sequence: SHAGRGGGCGCB |
Reverse complement motif Consensus sequence: VGCGCCCMCTDS |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
156 |
| Motif name: |
rgyGCCMyCTksTGGccd |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
12 |
| Similarity score: |
0.00894839 |
Alignment:
RVYGCCCYCTKSTGGCHD
VGCGCCCYCTDS------
| Original motif Consensus sequence: RVYGCCCYCTKSTGGCHD |
Reverse complement motif Consensus sequence: DDGCCASYAGMGGGCKVM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
168 |
| Motif name: |
yrcrGYGCCMyCTGGtG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0183868 |
Alignment:
HVCAGCGCCCYCTGGTG
---VGCGCCCYCTDS--
| Original motif Consensus sequence: HVCAGCGCCCYCTGGTG |
Reverse complement motif Consensus sequence: CACCAGMGGGCGCTGBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
146 |
| Motif name: |
myrGYGCCmCCTast |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0261893 |
Alignment:
VBAGCGCCMCCTAST
--VGCGCCCYCTDS-
| Original motif Consensus sequence: VBAGCGCCMCCTAST |
Reverse complement motif Consensus sequence: ASTAGGYGGCGCTBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
147 |
| Motif name: |
asCAGrkGGCrsy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0284224 |
Alignment:
ASCAGRGGGCRSB
-SHAGKGGGCGCB
| Original motif Consensus sequence: ASCAGRGGGCRSB |
Reverse complement motif Consensus sequence: BSKGCCCMCTGST |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 138 |
Motif name: grCCACyAGAkG |
| Original motif Consensus sequence: DDCCACYAGAKG |
Reverse complement motif Consensus sequence: CYTCTMGTGGHH |
|
|
 |
 |
Best Matches for Motif ID 138 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
145 |
| Motif name: |
grCCACyAGAkG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0 |
Alignment:
DDCCACYAGAKG
DDCCACYAGAKG
| Original motif Consensus sequence: DDCCACYAGAKG |
Reverse complement motif Consensus sequence: CYTCTKGTGGHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
165 |
| Motif name: |
wgGCCAshAGrGGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0317718 |
Alignment:
HDGCCACHAGRGGGCRBY
-DDCCACYAGAKG-----
| Original motif Consensus sequence: HDGCCACHAGRGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCKCTHGTGGCHH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
12 |
| Similarity score: |
0.0332329 |
Alignment:
YDRCCASYAGRKGGCRSYV
-DDCCACYAGAKG------
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
156 |
| Motif name: |
rgyGCCMyCTksTGGccd |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
12 |
| Similarity score: |
0.0390921 |
Alignment:
DDGCCASYAGMGGGCKVM
-DDCCACYAGAKG-----
| Original motif Consensus sequence: RVYGCCCYCTKSTGGCHD |
Reverse complement motif Consensus sequence: DDGCCASYAGMGGGCKVM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0478776 |
Alignment:
KBKGCCCTCTYCTGGCCHV
-----CYTCTMGTGGHH--
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 139 |
Motif name: mkCTyTTCsg |
| Original motif Consensus sequence: HBCTTTTCBD |
Reverse complement motif Consensus sequence: HBGAAAAGBD |
|
|
 |
 |
Best Matches for Motif ID 139 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
140 |
| Motif name: |
vkCKCTkCGk |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.000582108 |
Alignment:
VDCTCTKCGB
HBCTTTTCBD
| Original motif Consensus sequence: VDCTCTKCGB |
Reverse complement motif Consensus sequence: BCGYAGAGDV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
113 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
10 |
| Number of overlap: |
10 |
| Similarity score: |
0.0260893 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
---------HBCTTTTCBD--
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
136 |
| Motif name: |
dwCAGAAGwh |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0271307 |
Alignment:
DHCAGAAGDH
HBGAAAAGBD
| Original motif Consensus sequence: DHCAGAAGDH |
Reverse complement motif Consensus sequence: HDCTTCTGHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
142 |
| Motif name: |
ctCTTAACyw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0279851 |
Alignment:
HHCTTAACHD
HBCTTTTCBD
| Original motif Consensus sequence: HHCTTAACHD |
Reverse complement motif Consensus sequence: DDGTTAAGHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
141 |
| Motif name: |
raCAAAACam |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0368067 |
Alignment:
HTGTTTTGTD
HBCTTTTCBD
| Original motif Consensus sequence: DACAAAACAH |
Reverse complement motif Consensus sequence: HTGTTTTGTD |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 140 |
Motif name: vkCKCTkCGk |
| Original motif Consensus sequence: VDCTCTKCGB |
Reverse complement motif Consensus sequence: BCGYAGAGDV |
|
|
 |
 |
Best Matches for Motif ID 140 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
139 |
| Motif name: |
mkCTyTTCsg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.00351595 |
Alignment:
HBCTTTTCBD
VDCTCTKCGB
| Original motif Consensus sequence: HBCTTTTCBD |
Reverse complement motif Consensus sequence: HBGAAAAGBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
161 |
| Motif name: |
ssCGCwGCGss |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0202536 |
Alignment:
VSCGCDGCGSB
VDCTCTKCGB-
| Original motif Consensus sequence: VSCGCDGCGSB |
Reverse complement motif Consensus sequence: BSCGCDGCGSV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
170 |
| Motif name: |
ssGGCrsTGCrs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0403601 |
Alignment:
VVGGCRSTGCVB
--VDCTCTKCGB
| Original motif Consensus sequence: VVGGCRSTGCVB |
Reverse complement motif Consensus sequence: BVGCASMGCCVV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
154 |
| Motif name: |
csCsCCTCCcc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0407996 |
Alignment:
VBCCCCTCCHB
VDCTCTKCGB-
| Original motif Consensus sequence: VBCCCCTCCHB |
Reverse complement motif Consensus sequence: BDGGAGGGGBV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
109 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
10 |
| Similarity score: |
0.0447624 |
Alignment:
DGVBCABTGDWGCGKRRCSR
----BCGYAGAGDV------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 141 |
Motif name: raCAAAACam |
| Original motif Consensus sequence: DACAAAACAH |
Reverse complement motif Consensus sequence: HTGTTTTGTD |
|
|
 |
 |
Best Matches for Motif ID 141 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
136 |
| Motif name: |
dwCAGAAGwh |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0286451 |
Alignment:
DHCAGAAGDH
DACAAAACAH
| Original motif Consensus sequence: DHCAGAAGDH |
Reverse complement motif Consensus sequence: HDCTTCTGHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
142 |
| Motif name: |
ctCTTAACyw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0383253 |
Alignment:
DDGTTAAGHD
HTGTTTTGTD
| Original motif Consensus sequence: HHCTTAACHD |
Reverse complement motif Consensus sequence: DDGTTAAGHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
148 |
| Motif name: |
wwTwAAAAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0458158 |
Alignment:
DDTWAAAAHH
DACAAAACAH
| Original motif Consensus sequence: DDTWAAAAHH |
Reverse complement motif Consensus sequence: HHTTTTWADD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
157 |
| Motif name: |
wtATTTTTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0464408 |
Alignment:
WWTAAAAATAD
-DACAAAACAH
| Original motif Consensus sequence: DTATTTTTAWW |
Reverse complement motif Consensus sequence: WWTAAAAATAD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
139 |
| Motif name: |
mkCTyTTCsg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0483302 |
Alignment:
HBGAAAAGBD
DACAAAACAH
| Original motif Consensus sequence: HBCTTTTCBD |
Reverse complement motif Consensus sequence: HBGAAAAGBD |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 142 |
Motif name: ctCTTAACyw |
| Original motif Consensus sequence: HHCTTAACHD |
Reverse complement motif Consensus sequence: DDGTTAAGHD |
|
|
 |
 |
Best Matches for Motif ID 142 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
141 |
| Motif name: |
raCAAAACam |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0459536 |
Alignment:
HTGTTTTGTD
DDGTTAAGHD
| Original motif Consensus sequence: DACAAAACAH |
Reverse complement motif Consensus sequence: HTGTTTTGTD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
139 |
| Motif name: |
mkCTyTTCsg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0471369 |
Alignment:
HBCTTTTCBD
HHCTTAACHD
| Original motif Consensus sequence: HBCTTTTCBD |
Reverse complement motif Consensus sequence: HBGAAAAGBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
136 |
| Motif name: |
dwCAGAAGwh |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0550541 |
Alignment:
DHCAGAAGDH
DDGTTAAGHD
| Original motif Consensus sequence: DHCAGAAGDH |
Reverse complement motif Consensus sequence: HDCTTCTGHD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
83 |
| Motif name: |
Esrrb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0663739 |
Alignment:
VBBYCAAGGTCA
--DDGTTAAGHD
| Original motif Consensus sequence: VBBYCAAGGTCA |
Reverse complement motif Consensus sequence: TGACCTTGMBBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
82 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0691225 |
Alignment:
BAGGYCABHBTGACCKHV
-HHCTTAACHD-------
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 143 |
Motif name: AgmAGAGGGCrscAGak |
| Original motif Consensus sequence: AGMAGAGGGCASCAGAK |
Reverse complement motif Consensus sequence: RTCTGSTGCCCTCTYCT |
|
|
 |
 |
Best Matches for Motif ID 143 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
149 |
| Motif name: |
asmAGRGGGCrCTGsmkc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
17 |
| Similarity score: |
0.0311468 |
Alignment:
ASMAGAGGGCRCTGSABH
AGMAGAGGGCASCAGAK-
| Original motif Consensus sequence: ASMAGAGGGCRCTGSABH |
Reverse complement motif Consensus sequence: DBTSCAGMGCCCTCTRST |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
144 |
| Motif name: |
ctCTrsyGCCmCCTast |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
17 |
| Similarity score: |
0.0415124 |
Alignment:
HDCTGSYGCCMCCTAST
RTCTGSTGCCCTCTYCT
| Original motif Consensus sequence: HDCTGSYGCCMCCTAST |
Reverse complement motif Consensus sequence: ASTAGGYGGCMSCAGDD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
167 |
| Motif name: |
rsyAGrkGGCGCCmyCTrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
17 |
| Similarity score: |
0.0702327 |
Alignment:
DSYAGRKGGCGCCMYCTRSH
AGMAGAGGGCASCAGAK---
| Original motif Consensus sequence: DSYAGRKGGCGCCMYCTRSH |
Reverse complement motif Consensus sequence: HSKAGKYGGCGCCRMCTMSD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
17 |
| Similarity score: |
0.0915393 |
Alignment:
VDBHMAGGTCACCCTGACCY
AGMAGAGGGCASCAGAK---
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
164 |
| Motif name: |
asyAGrkGGCRGCAga |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
16 |
| Similarity score: |
0.531077 |
Alignment:
ASYAGRKGGCAGCABH-
AGMAGAGGGCASCAGAK
| Original motif Consensus sequence: ASYAGRKGGCAGCABH |
Reverse complement motif Consensus sequence: HBTGCTGCCYMCTKST |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 144 |
Motif name: ctCTrsyGCCmCCTast |
| Original motif Consensus sequence: HDCTGSYGCCMCCTAST |
Reverse complement motif Consensus sequence: ASTAGGYGGCMSCAGDD |
|
|
 |
 |
Best Matches for Motif ID 144 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
143 |
| Motif name: |
AgmAGAGGGCrscAGak |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
17 |
| Similarity score: |
0.0216483 |
Alignment:
AGMAGAGGGCASCAGAK
ASTAGGYGGCMSCAGDD
| Original motif Consensus sequence: AGMAGAGGGCASCAGAK |
Reverse complement motif Consensus sequence: RTCTGSTGCCCTCTYCT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
167 |
| Motif name: |
rsyAGrkGGCGCCmyCTrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
17 |
| Similarity score: |
0.0316423 |
Alignment:
DSYAGRKGGCGCCMYCTRSH
---HDCTGSYGCCMCCTAST
| Original motif Consensus sequence: DSYAGRKGGCGCCMYCTRSH |
Reverse complement motif Consensus sequence: HSKAGKYGGCGCCRMCTMSD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
149 |
| Motif name: |
asmAGRGGGCrCTGsmkc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
17 |
| Similarity score: |
0.040379 |
Alignment:
DBTSCAGMGCCCTCTRST
-HDCTGSYGCCMCCTAST
| Original motif Consensus sequence: ASMAGAGGGCRCTGSABH |
Reverse complement motif Consensus sequence: DBTSCAGMGCCCTCTRST |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
17 |
| Similarity score: |
0.0748345 |
Alignment:
MGGTCAGGGTGACCTRDBHV
---ASTAGGYGGCMSCAGDD
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
164 |
| Motif name: |
asyAGrkGGCRGCAga |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
16 |
| Similarity score: |
0.503247 |
Alignment:
ASYAGRKGGCAGCABH-
ASTAGGYGGCMSCAGDD
| Original motif Consensus sequence: ASYAGRKGGCAGCABH |
Reverse complement motif Consensus sequence: HBTGCTGCCYMCTKST |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 145 |
Motif name: grCCACyAGAkG |
| Original motif Consensus sequence: DDCCACYAGAKG |
Reverse complement motif Consensus sequence: CYTCTKGTGGHH |
|
|
 |
 |
Best Matches for Motif ID 145 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
138 |
| Motif name: |
grCCACyAGAkG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0 |
Alignment:
DDCCACYAGAKG
DDCCACYAGAKG
| Original motif Consensus sequence: DDCCACYAGAKG |
Reverse complement motif Consensus sequence: CYTCTMGTGGHH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0323771 |
Alignment:
YDRCCASYAGRKGGCRSYV
-DDCCACYAGAKG------
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
165 |
| Motif name: |
wgGCCAshAGrGGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
12 |
| Similarity score: |
0.0347955 |
Alignment:
HDGCCACHAGRGGGCRBY
-DDCCACYAGAKG-----
| Original motif Consensus sequence: HDGCCACHAGRGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCKCTHGTGGCHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
156 |
| Motif name: |
rgyGCCMyCTksTGGccd |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
12 |
| Similarity score: |
0.0388427 |
Alignment:
DDGCCASYAGMGGGCKVM
-DDCCACYAGAKG-----
| Original motif Consensus sequence: RVYGCCCYCTKSTGGCHD |
Reverse complement motif Consensus sequence: DDGCCASYAGMGGGCKVM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0502736 |
Alignment:
VHGGCCAGMAGAGGGCRBY
--DDCCACYAGAKG-----
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 146 |
Motif name: myrGYGCCmCCTast |
| Original motif Consensus sequence: VBAGCGCCMCCTAST |
Reverse complement motif Consensus sequence: ASTAGGYGGCGCTBB |
|
|
 |
 |
Best Matches for Motif ID 146 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
168 |
| Motif name: |
yrcrGYGCCMyCTGGtG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
15 |
| Similarity score: |
0.00670638 |
Alignment:
HVCAGCGCCCYCTGGTG
-VBAGCGCCMCCTAST-
| Original motif Consensus sequence: HVCAGCGCCCYCTGGTG |
Reverse complement motif Consensus sequence: CACCAGMGGGCGCTGBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
144 |
| Motif name: |
ctCTrsyGCCmCCTast |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
15 |
| Similarity score: |
0.0118351 |
Alignment:
HDCTGSYGCCMCCTAST
--VBAGCGCCMCCTAST
| Original motif Consensus sequence: HDCTGSYGCCMCCTAST |
Reverse complement motif Consensus sequence: ASTAGGYGGCMSCAGDD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
167 |
| Motif name: |
rsyAGrkGGCGCCmyCTrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
15 |
| Similarity score: |
0.0178333 |
Alignment:
DSYAGRKGGCGCCMYCTRSH
-----VBAGCGCCMCCTAST
| Original motif Consensus sequence: DSYAGRKGGCGCCMYCTRSH |
Reverse complement motif Consensus sequence: HSKAGKYGGCGCCRMCTMSD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
149 |
| Motif name: |
asmAGRGGGCrCTGsmkc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
0.0324399 |
Alignment:
DBTSCAGMGCCCTCTRST
---VBAGCGCCMCCTAST
| Original motif Consensus sequence: ASMAGAGGGCRCTGSABH |
Reverse complement motif Consensus sequence: DBTSCAGMGCCCTCTRST |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
169 |
| Motif name: |
yvTGCyGCCmCCwGgtG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
15 |
| Similarity score: |
0.0457506 |
Alignment:
BVTGCTGCCACCWGGDG
-VBAGCGCCMCCTAST-
| Original motif Consensus sequence: BVTGCTGCCACCWGGDG |
Reverse complement motif Consensus sequence: CDCCWGGTGGCAGCAVV |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 147 |
Motif name: asCAGrkGGCrsy |
| Original motif Consensus sequence: ASCAGRGGGCRSB |
Reverse complement motif Consensus sequence: BSKGCCCMCTGST |
|
|
 |
 |
Best Matches for Motif ID 147 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
166 |
| Motif name: |
CasCAGrGGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.00871755 |
Alignment:
CACCAGRGGGCRSB
-ASCAGRGGGCRSB
| Original motif Consensus sequence: CACCAGRGGGCRSB |
Reverse complement motif Consensus sequence: BSKGCCCKCTGGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
165 |
| Motif name: |
wgGCCAshAGrGGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.0301924 |
Alignment:
HDGCCACHAGRGGGCRBY
-----ASCAGRGGGCRSB
| Original motif Consensus sequence: HDGCCACHAGRGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCKCTHGTGGCHH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
13 |
| Similarity score: |
0.0305318 |
Alignment:
YDRCCASYAGRKGGCRSYV
-----ASCAGRGGGCRSB-
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
143 |
| Motif name: |
AgmAGAGGGCrscAGak |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
13 |
| Similarity score: |
0.0348963 |
Alignment:
AGMAGAGGGCASCAGAK
ASCAGRGGGCRSB----
| Original motif Consensus sequence: AGMAGAGGGCASCAGAK |
Reverse complement motif Consensus sequence: RTCTGSTGCCCTCTYCT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
164 |
| Motif name: |
asyAGrkGGCRGCAga |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.0366623 |
Alignment:
HBTGCTGCCYMCTKST
---BSKGCCCMCTGST
| Original motif Consensus sequence: ASYAGRKGGCAGCABH |
Reverse complement motif Consensus sequence: HBTGCTGCCYMCTKST |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 148 |
Motif name: wwTwAAAAww |
| Original motif Consensus sequence: DDTWAAAAHH |
Reverse complement motif Consensus sequence: HHTTTTWADD |
|
|
 |
 |
Best Matches for Motif ID 148 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
157 |
| Motif name: |
wtATTTTTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0143677 |
Alignment:
WWTAAAAATAD
DDTWAAAAHH-
| Original motif Consensus sequence: DTATTTTTAWW |
Reverse complement motif Consensus sequence: WWTAAAAATAD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
150 |
| Motif name: |
waATwAAAATAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0146704 |
Alignment:
DHATWAAAATAHD
-DDTWAAAAHH--
| Original motif Consensus sequence: DHATWAAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTTWATHD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
68 |
| Motif name: |
Motif 68 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0466767 |
Alignment:
WAAWAAAWAWWWAA
-DDTWAAAAHH---
| Original motif Consensus sequence: WAAWAAAWAWWWAA |
Reverse complement motif Consensus sequence: TTWWWTWTTTWTTW |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
141 |
| Motif name: |
raCAAAACam |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0504274 |
Alignment:
DACAAAACAH
DDTWAAAAHH
| Original motif Consensus sequence: DACAAAACAH |
Reverse complement motif Consensus sequence: HTGTTTTGTD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
142 |
| Motif name: |
ctCTTAACyw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0669752 |
Alignment:
HHCTTAACHD
DDTWAAAAHH
| Original motif Consensus sequence: HHCTTAACHD |
Reverse complement motif Consensus sequence: DDGTTAAGHD |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 149 |
Motif name: asmAGRGGGCrCTGsmkc |
| Original motif Consensus sequence: ASMAGAGGGCRCTGSABH |
Reverse complement motif Consensus sequence: DBTSCAGMGCCCTCTRST |
|
|
 |
 |
Best Matches for Motif ID 149 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
167 |
| Motif name: |
rsyAGrkGGCGCCmyCTrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
18 |
| Similarity score: |
0.0599593 |
Alignment:
DSYAGRKGGCGCCMYCTRSH
ASMAGAGGGCRCTGSABH--
| Original motif Consensus sequence: DSYAGRKGGCGCCMYCTRSH |
Reverse complement motif Consensus sequence: HSKAGKYGGCGCCRMCTMSD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
0.0884279 |
Alignment:
VDBHMAGGTCACCCTGACCY
--DBTSCAGMGCCCTCTRST
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
113 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
18 |
| Similarity score: |
0.0932033 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
DBTSCAGMGCCCTCTRST---
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
70 |
| Motif name: |
Ar |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
18 |
| Similarity score: |
0.0976041 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
ASMAGAGGGCRCTGSABH----
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
143 |
| Motif name: |
AgmAGAGGGCrscAGak |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
17 |
| Similarity score: |
0.529694 |
Alignment:
AGMAGAGGGCASCAGAK-
ASMAGAGGGCRCTGSABH
| Original motif Consensus sequence: AGMAGAGGGCASCAGAK |
Reverse complement motif Consensus sequence: RTCTGSTGCCCTCTYCT |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 150 |
Motif name: waATwAAAATAww |
| Original motif Consensus sequence: DHATWAAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTTWATHD |
|
|
 |
 |
Best Matches for Motif ID 150 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
68 |
| Motif name: |
Motif 68 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.0403377 |
Alignment:
TTWWWTWTTTWTTW
-DHTATTTTWATHD
| Original motif Consensus sequence: WAAWAAAWAWWWAA |
Reverse complement motif Consensus sequence: TTWWWTWTTTWTTW |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
70 |
| Motif name: |
Ar |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
10 |
| Number of overlap: |
13 |
| Similarity score: |
0.0805462 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
DHATWAAAATAHD---------
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
87 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.081608 |
Alignment:
RGGBCAAAGKYCA
DHATWAAAATAHD
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
104 |
| Motif name: |
NFYA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.0831397 |
Alignment:
BHVCKSATTGGMKBVV
--DHTATTTTWATHD-
| Original motif Consensus sequence: VBBRRCCAATSRGVDB |
Reverse complement motif Consensus sequence: BHVCKSATTGGMKBVV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
107 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.0856949 |
Alignment:
TGAMCTTTGMMCYT
DHTATTTTWATHD-
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 151 |
Motif name: agrCCAGmAGrg |
| Original motif Consensus sequence: HVGCCAGMAGRG |
Reverse complement motif Consensus sequence: CKCTRCTGGCVH |
|
|
 |
 |
Best Matches for Motif ID 151 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.00116274 |
Alignment:
VHGGCCAGMAGAGGGCRBY
-HVGCCAGMAGRG------
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
165 |
| Motif name: |
wgGCCAshAGrGGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0104581 |
Alignment:
HDGCCACHAGRGGGCRBY
HVGCCAGMAGRG------
| Original motif Consensus sequence: HDGCCACHAGRGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCKCTHGTGGCHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
156 |
| Motif name: |
rgyGCCMyCTksTGGccd |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0140707 |
Alignment:
RVYGCCCYCTKSTGGCHD
------CKCTRCTGGCVH
| Original motif Consensus sequence: RVYGCCCYCTKSTGGCHD |
Reverse complement motif Consensus sequence: DDGCCASYAGMGGGCKVM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0214576 |
Alignment:
YDRCCASYAGRKGGCRSYV
HVGCCAGMAGRG-------
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
143 |
| Motif name: |
AgmAGAGGGCrscAGak |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
12 |
| Similarity score: |
0.0481806 |
Alignment:
AGMAGAGGGCASCAGAK
-----HVGCCAGMAGRG
| Original motif Consensus sequence: AGMAGAGGGCASCAGAK |
Reverse complement motif Consensus sequence: RTCTGSTGCCCTCTYCT |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 152 |
Motif name: yrCATGCAyr |
| Original motif Consensus sequence: BRCATGCABD |
Reverse complement motif Consensus sequence: HVTGCATGKV |
|
|
 |
 |
Best Matches for Motif ID 152 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
75 |
| Motif name: |
Ddit3Cebpa |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0420406 |
Alignment:
VDRTGCAATMCC
-HVTGCATGKV-
| Original motif Consensus sequence: VDRTGCAATMCC |
Reverse complement motif Consensus sequence: GGRATTGCAKHB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
125 |
| Motif name: |
TP53 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0472223 |
Alignment:
ACATGCCCGGKCATGTCCSR
-------HVTGCATGKV---
| Original motif Consensus sequence: MSGGACATGYCCGGGCATGT |
Reverse complement motif Consensus sequence: ACATGCCCGGKCATGTCCSR |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
134 |
| Motif name: |
ssCGwGCGss |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.055087 |
Alignment:
BSCGWGCGBV
BRCATGCABD
| Original motif Consensus sequence: BSCGWGCGBV |
Reverse complement motif Consensus sequence: VBCGCWCGSB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
131 |
| Motif name: |
znf143 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0551971 |
Alignment:
GCMWRGCATYRTGGGAMHTB
----------BRCATGCABD
| Original motif Consensus sequence: BAHYTCCCAKMATGCMWYGC |
Reverse complement motif Consensus sequence: GCMWRGCATYRTGGGAMHTB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
159 |
| Motif name: |
kkAAGAGCAsy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0606344 |
Alignment:
HVTGCTCTTBH
HVTGCATGKV-
| Original motif Consensus sequence: DBAAGAGCAVH |
Reverse complement motif Consensus sequence: HVTGCTCTTBH |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 153 |
Motif name: scAGrkGGCGcy |
| Original motif Consensus sequence: SHAGRGGGCGCB |
Reverse complement motif Consensus sequence: VGCGCCCMCTDS |
|
|
 |
 |
Best Matches for Motif ID 153 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
137 |
| Motif name: |
rgCGCCmyCTgs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0 |
Alignment:
VGCGCCCYCTDS
VGCGCCCMCTDS
| Original motif Consensus sequence: VGCGCCCYCTDS |
Reverse complement motif Consensus sequence: SHAGKGGGCGCB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
147 |
| Motif name: |
asCAGrkGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0112071 |
Alignment:
ASCAGRGGGCRSB
-SHAGRGGGCGCB
| Original motif Consensus sequence: ASCAGRGGGCRSB |
Reverse complement motif Consensus sequence: BSKGCCCMCTGST |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
12 |
| Similarity score: |
0.0133899 |
Alignment:
BMSMGCCYMCTKSTGGMHM
-VGCGCCCMCTDS------
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
165 |
| Motif name: |
wgGCCAshAGrGGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0146442 |
Alignment:
HDGCCACHAGRGGGCRBY
------SHAGRGGGCGCB
| Original motif Consensus sequence: HDGCCACHAGRGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCKCTHGTGGCHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
166 |
| Motif name: |
CasCAGrGGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0200811 |
Alignment:
CACCAGRGGGCRSB
--SHAGRGGGCGCB
| Original motif Consensus sequence: CACCAGRGGGCRSB |
Reverse complement motif Consensus sequence: BSKGCCCKCTGGTG |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 154 |
Motif name: csCsCCTCCcc |
| Original motif Consensus sequence: VBCCCCTCCHB |
Reverse complement motif Consensus sequence: BDGGAGGGGBV |
|
|
 |
 |
Best Matches for Motif ID 154 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
155 |
| Motif name: |
csCSCCdCCCcs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0 |
Alignment:
VBCCCCDCCCHV
VBCCCCTCCHB-
| Original motif Consensus sequence: VBCCCCDCCCHV |
Reverse complement motif Consensus sequence: VDGGGDGGGGBV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
153 |
| Motif name: |
scAGrkGGCGcy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0415573 |
Alignment:
SHAGRGGGCGCB
BDGGAGGGGBV-
| Original motif Consensus sequence: SHAGRGGGCGCB |
Reverse complement motif Consensus sequence: VGCGCCCMCTDS |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
137 |
| Motif name: |
rgCGCCmyCTgs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0446252 |
Alignment:
SHAGKGGGCGCB
-BDGGAGGGGBV
| Original motif Consensus sequence: VGCGCCCYCTDS |
Reverse complement motif Consensus sequence: SHAGKGGGCGCB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
162 |
| Motif name: |
ccAsCCCCAcc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.049712 |
Alignment:
DBTGGGGSTVD
BDGGAGGGGBV
| Original motif Consensus sequence: HVASCCCCABH |
Reverse complement motif Consensus sequence: DBTGGGGSTVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
143 |
| Motif name: |
AgmAGAGGGCrscAGak |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0533559 |
Alignment:
AGMAGAGGGCASCAGAK
-----BDGGAGGGGBV-
| Original motif Consensus sequence: AGMAGAGGGCASCAGAK |
Reverse complement motif Consensus sequence: RTCTGSTGCCCTCTYCT |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 155 |
Motif name: csCSCCdCCCcs |
| Original motif Consensus sequence: VBCCCCDCCCHV |
Reverse complement motif Consensus sequence: VDGGGDGGGGBV |
|
|
 |
 |
Best Matches for Motif ID 155 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
137 |
| Motif name: |
rgCGCCmyCTgs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0604731 |
Alignment:
SHAGKGGGCGCB
VDGGGDGGGGBV
| Original motif Consensus sequence: VGCGCCCYCTDS |
Reverse complement motif Consensus sequence: SHAGKGGGCGCB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
153 |
| Motif name: |
scAGrkGGCGcy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0624586 |
Alignment:
VGCGCCCMCTDS
VBCCCCDCCCHV
| Original motif Consensus sequence: SHAGRGGGCGCB |
Reverse complement motif Consensus sequence: VGCGCCCMCTDS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0800509 |
Alignment:
BMSMGCCYMCTKSTGGMHM
------VBCCCCDCCCHV-
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
168 |
| Motif name: |
yrcrGYGCCMyCTGGtG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
12 |
| Similarity score: |
0.0810683 |
Alignment:
HVCAGCGCCCYCTGGTG
VBCCCCDCCCHV-----
| Original motif Consensus sequence: HVCAGCGCCCYCTGGTG |
Reverse complement motif Consensus sequence: CACCAGMGGGCGCTGBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
147 |
| Motif name: |
asCAGrkGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0823049 |
Alignment:
BSKGCCCMCTGST
-VBCCCCDCCCHV
| Original motif Consensus sequence: ASCAGRGGGCRSB |
Reverse complement motif Consensus sequence: BSKGCCCMCTGST |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 156 |
Motif name: rgyGCCMyCTksTGGccd |
| Original motif Consensus sequence: RVYGCCCYCTKSTGGCHD |
Reverse complement motif Consensus sequence: DDGCCASYAGMGGGCKVM |
|
|
 |
 |
Best Matches for Motif ID 156 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
165 |
| Motif name: |
wgGCCAshAGrGGGCrsy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
0.0156448 |
Alignment:
HDGCCACHAGRGGGCRBY
DDGCCASYAGMGGGCKVM
| Original motif Consensus sequence: HDGCCACHAGRGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCKCTHGTGGCHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
18 |
| Similarity score: |
0.0201276 |
Alignment:
VHGGCCAGMAGAGGGCRBY
-DDGCCASYAGMGGGCKVM
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
0.0207204 |
Alignment:
YDRCCASYAGRKGGCRSYV
-DDGCCASYAGMGGGCKVM
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
113 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
0.0950319 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
---RVYGCCCYCTKSTGGCHD
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
109 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
0.09559 |
Alignment:
MSGKKRCGCWDCABTGBBCD
--RVYGCCCYCTKSTGGCHD
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 157 |
Motif name: wtATTTTTAww |
| Original motif Consensus sequence: DTATTTTTAWW |
Reverse complement motif Consensus sequence: WWTAAAAATAD |
|
|
 |
 |
Best Matches for Motif ID 157 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
150 |
| Motif name: |
waATwAAAATAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0 |
Alignment:
DHTATTTTWATHD
-DTATTTTTAWW-
| Original motif Consensus sequence: DHATWAAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTTWATHD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
68 |
| Motif name: |
Motif 68 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0323073 |
Alignment:
WAAWAAAWAWWWAA
--WWTAAAAATAD-
| Original motif Consensus sequence: WAAWAAAWAWWWAA |
Reverse complement motif Consensus sequence: TTWWWTWTTTWTTW |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
87 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0727314 |
Alignment:
RGGBCAAAGKYCA
WWTAAAAATAD--
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
107 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0739182 |
Alignment:
AKGYYCAAAGRTCA
---WWTAAAAATAD
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
104 |
| Motif name: |
NFYA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
0.0755742 |
Alignment:
BHVCKSATTGGMKBVV
-DTATTTTTAWW----
| Original motif Consensus sequence: VBBRRCCAATSRGVDB |
Reverse complement motif Consensus sequence: BHVCKSATTGGMKBVV |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 158 |
Motif name: grCCACwAGrk |
| Original motif Consensus sequence: DDCCACWAGRK |
Reverse complement motif Consensus sequence: YMCTWGTGGHH |
|
|
 |
 |
Best Matches for Motif ID 158 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
138 |
| Motif name: |
grCCACyAGAkG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0013909 |
Alignment:
DDCCACYAGAKG
DDCCACWAGRK-
| Original motif Consensus sequence: DDCCACYAGAKG |
Reverse complement motif Consensus sequence: CYTCTMGTGGHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
145 |
| Motif name: |
grCCACyAGAkG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.00619044 |
Alignment:
DDCCACYAGAKG
DDCCACWAGRK-
| Original motif Consensus sequence: DDCCACYAGAKG |
Reverse complement motif Consensus sequence: CYTCTKGTGGHH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
11 |
| Similarity score: |
0.0223616 |
Alignment:
BMSMGCCYMCTKSTGGMHM
-------YMCTWGTGGHH-
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
165 |
| Motif name: |
wgGCCAshAGrGGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
11 |
| Similarity score: |
0.0228739 |
Alignment:
HDGCCACHAGRGGGCRBY
-DDCCACWAGRK------
| Original motif Consensus sequence: HDGCCACHAGRGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCKCTHGTGGCHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
151 |
| Motif name: |
agrCCAGmAGrg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0330864 |
Alignment:
HVGCCAGMAGRG
-DDCCACWAGRK
| Original motif Consensus sequence: HVGCCAGMAGRG |
Reverse complement motif Consensus sequence: CKCTRCTGGCVH |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 159 |
Motif name: kkAAGAGCAsy |
| Original motif Consensus sequence: DBAAGAGCAVH |
Reverse complement motif Consensus sequence: HVTGCTCTTBH |
|
|
 |
 |
Best Matches for Motif ID 159 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
160 |
| Motif name: |
brCAGGGCCrs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0212428 |
Alignment:
BBGGCCCTGBB
HVTGCTCTTBH
| Original motif Consensus sequence: BVCAGGGCCVB |
Reverse complement motif Consensus sequence: BBGGCCCTGBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
133 |
| Motif name: |
shAGrGGGCAgy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0322726 |
Alignment:
SHAGRGGGCABH
-DBAAGAGCAVH
| Original motif Consensus sequence: SHAGRGGGCABH |
Reverse complement motif Consensus sequence: DBTGCCCKCTDS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
70 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
12 |
| Number of overlap: |
11 |
| Similarity score: |
0.0353029 |
Alignment:
VRVDGGHACAVDDKGTHCTDWH
-----------HVTGCTCTTBH
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
75 |
| Motif name: |
Ddit3Cebpa |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0380063 |
Alignment:
VDRTGCAATMCC
HVTGCTCTTBH-
| Original motif Consensus sequence: VDRTGCAATMCC |
Reverse complement motif Consensus sequence: GGRATTGCAKHB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
168 |
| Motif name: |
yrcrGYGCCMyCTGGtG |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0432631 |
Alignment:
CACCAGMGGGCGCTGBD
------HVTGCTCTTBH
| Original motif Consensus sequence: HVCAGCGCCCYCTGGTG |
Reverse complement motif Consensus sequence: CACCAGMGGGCGCTGBD |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 160 |
Motif name: brCAGGGCCrs |
| Original motif Consensus sequence: BVCAGGGCCVB |
Reverse complement motif Consensus sequence: BBGGCCCTGBB |
|
|
 |
 |
Best Matches for Motif ID 160 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
168 |
| Motif name: |
yrcrGYGCCMyCTGGtG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0430085 |
Alignment:
HVCAGCGCCCYCTGGTG
BVCAGGGCCVB------
| Original motif Consensus sequence: HVCAGCGCCCYCTGGTG |
Reverse complement motif Consensus sequence: CACCAGMGGGCGCTGBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
170 |
| Motif name: |
ssGGCrsTGCrs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0438378 |
Alignment:
BVGCASMGCCVV
-BVCAGGGCCVB
| Original motif Consensus sequence: VVGGCRSTGCVB |
Reverse complement motif Consensus sequence: BVGCASMGCCVV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
159 |
| Motif name: |
kkAAGAGCAsy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0567379 |
Alignment:
HVTGCTCTTBH
BBGGCCCTGBB
| Original motif Consensus sequence: DBAAGAGCAVH |
Reverse complement motif Consensus sequence: HVTGCTCTTBH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
149 |
| Motif name: |
asmAGRGGGCrCTGsmkc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
11 |
| Similarity score: |
0.0583002 |
Alignment:
ASMAGAGGGCRCTGSABH
--BBGGCCCTGBB-----
| Original motif Consensus sequence: ASMAGAGGGCRCTGSABH |
Reverse complement motif Consensus sequence: DBTSCAGMGCCCTCTRST |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
113 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
11 |
| Number of overlap: |
11 |
| Similarity score: |
0.0603368 |
Alignment:
TTCAGCACCATGGACAGCKCC
BVCAGGGCCVB----------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 161 |
Motif name: ssCGCwGCGss |
| Original motif Consensus sequence: VSCGCDGCGSB |
Reverse complement motif Consensus sequence: BSCGCDGCGSV |
|
|
 |
 |
Best Matches for Motif ID 161 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
170 |
| Motif name: |
ssGGCrsTGCrs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0546471 |
Alignment:
VVGGCRSTGCVB
BSCGCDGCGSV-
| Original motif Consensus sequence: VVGGCRSTGCVB |
Reverse complement motif Consensus sequence: BVGCASMGCCVV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
169 |
| Motif name: |
yvTGCyGCCmCCwGgtG |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0565318 |
Alignment:
CDCCWGGTGGCAGCAVV
------BSCGCDGCGSV
| Original motif Consensus sequence: BVTGCTGCCACCWGGDG |
Reverse complement motif Consensus sequence: CDCCWGGTGGCAGCAVV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
149 |
| Motif name: |
asmAGRGGGCrCTGsmkc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0580603 |
Alignment:
DBTSCAGMGCCCTCTRST
BSCGCDGCGSV-------
| Original motif Consensus sequence: ASMAGAGGGCRCTGSABH |
Reverse complement motif Consensus sequence: DBTSCAGMGCCCTCTRST |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
154 |
| Motif name: |
csCsCCTCCcc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0592711 |
Alignment:
VBCCCCTCCHB
BSCGCDGCGSV
| Original motif Consensus sequence: VBCCCCTCCHB |
Reverse complement motif Consensus sequence: BDGGAGGGGBV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
155 |
| Motif name: |
csCSCCdCCCcs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0599572 |
Alignment:
VDGGGDGGGGBV
VSCGCDGCGSB-
| Original motif Consensus sequence: VBCCCCDCCCHV |
Reverse complement motif Consensus sequence: VDGGGDGGGGBV |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 162 |
Motif name: ccAsCCCCAcc |
| Original motif Consensus sequence: HVASCCCCABH |
Reverse complement motif Consensus sequence: DBTGGGGSTVD |
|
|
 |
 |
Best Matches for Motif ID 162 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
154 |
| Motif name: |
csCsCCTCCcc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0318935 |
Alignment:
BDGGAGGGGBV
DBTGGGGSTVD
| Original motif Consensus sequence: VBCCCCTCCHB |
Reverse complement motif Consensus sequence: BDGGAGGGGBV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
146 |
| Motif name: |
myrGYGCCmCCTast |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
0.0360618 |
Alignment:
VBAGCGCCMCCTAST
HVASCCCCABH----
| Original motif Consensus sequence: VBAGCGCCMCCTAST |
Reverse complement motif Consensus sequence: ASTAGGYGGCGCTBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
131 |
| Motif name: |
znf143 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
10 |
| Number of overlap: |
11 |
| Similarity score: |
0.0409371 |
Alignment:
BAHYTCCCAKMATGCMWYGC
HVASCCCCABH---------
| Original motif Consensus sequence: BAHYTCCCAKMATGCMWYGC |
Reverse complement motif Consensus sequence: GCMWRGCATYRTGGGAMHTB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
155 |
| Motif name: |
csCSCCdCCCcs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0412348 |
Alignment:
VDGGGDGGGGBV
-DBTGGGGSTVD
| Original motif Consensus sequence: VBCCCCDCCCHV |
Reverse complement motif Consensus sequence: VDGGGDGGGGBV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
111 |
| Motif name: |
PPARG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0446625 |
Alignment:
STAGGTCACBGTGACCYABT
---------HVASCCCCABH
| Original motif Consensus sequence: STAGGTCACBGTGACCYABT |
Reverse complement motif Consensus sequence: ABTMGGTCACBGTGACCTAS |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 163 |
Motif name: gwGGCCAGmAGAGGGCrby |
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
Best Matches for Motif ID 163 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
125 |
| Motif name: |
TP53 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
19 |
| Similarity score: |
0.0964434 |
Alignment:
ACATGCCCGGKCATGTCCSR
-KBKGCCCTCTYCTGGCCHV
| Original motif Consensus sequence: MSGGACATGYCCGGGCATGT |
Reverse complement motif Consensus sequence: ACATGCCCGGKCATGTCCSR |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
113 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
19 |
| Similarity score: |
0.104301 |
Alignment:
TTCAGCACCATGGACAGCKCC
--VHGGCCAGMAGAGGGCRBY
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
165 |
| Motif name: |
wgGCCAshAGrGGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
0.502859 |
Alignment:
-HDGCCACHAGRGGGCRBY
VHGGCCAGMAGAGGGCRBY
| Original motif Consensus sequence: HDGCCACHAGRGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCKCTHGTGGCHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
156 |
| Motif name: |
rgyGCCMyCTksTGGccd |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
0.525011 |
Alignment:
-RVYGCCCYCTKSTGGCHD
KBKGCCCTCTYCTGGCCHV
| Original motif Consensus sequence: RVYGCCCYCTKSTGGCHD |
Reverse complement motif Consensus sequence: DDGCCASYAGMGGGCKVM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
18 |
| Similarity score: |
0.529309 |
Alignment:
-YDRCCASYAGRKGGCRSYV
VHGGCCAGMAGAGGGCRBY-
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 164 |
Motif name: asyAGrkGGCRGCAga |
| Original motif Consensus sequence: ASYAGRKGGCAGCABH |
Reverse complement motif Consensus sequence: HBTGCTGCCYMCTKST |
|
|
 |
 |
Best Matches for Motif ID 164 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
144 |
| Motif name: |
ctCTrsyGCCmCCTast |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
16 |
| Similarity score: |
0.0253571 |
Alignment:
HDCTGSYGCCMCCTAST
HBTGCTGCCYMCTKST-
| Original motif Consensus sequence: HDCTGSYGCCMCCTAST |
Reverse complement motif Consensus sequence: ASTAGGYGGCMSCAGDD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
169 |
| Motif name: |
yvTGCyGCCmCCwGgtG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
16 |
| Similarity score: |
0.0299559 |
Alignment:
CDCCWGGTGGCAGCAVV
-ASYAGRKGGCAGCABH
| Original motif Consensus sequence: BVTGCTGCCACCWGGDG |
Reverse complement motif Consensus sequence: CDCCWGGTGGCAGCAVV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
143 |
| Motif name: |
AgmAGAGGGCrscAGak |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
16 |
| Similarity score: |
0.0333221 |
Alignment:
AGMAGAGGGCASCAGAK
ASYAGRKGGCAGCABH-
| Original motif Consensus sequence: AGMAGAGGGCASCAGAK |
Reverse complement motif Consensus sequence: RTCTGSTGCCCTCTYCT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
167 |
| Motif name: |
rsyAGrkGGCGCCmyCTrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
16 |
| Similarity score: |
0.0458621 |
Alignment:
DSYAGRKGGCGCCMYCTRSH
ASYAGRKGGCAGCABH----
| Original motif Consensus sequence: DSYAGRKGGCGCCMYCTRSH |
Reverse complement motif Consensus sequence: HSKAGKYGGCGCCRMCTMSD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
149 |
| Motif name: |
asmAGRGGGCrCTGsmkc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
16 |
| Similarity score: |
0.0529808 |
Alignment:
ASMAGAGGGCRCTGSABH
ASYAGRKGGCAGCABH--
| Original motif Consensus sequence: ASMAGAGGGCRCTGSABH |
Reverse complement motif Consensus sequence: DBTSCAGMGCCCTCTRST |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 165 |
Motif name: wgGCCAshAGrGGGCrsy |
| Original motif Consensus sequence: HDGCCACHAGRGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCKCTHGTGGCHH |
|
|
 |
 |
Best Matches for Motif ID 165 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
0.00922625 |
Alignment:
KBKGCCCTCTYCTGGCCHV
KBKGCCCKCTHGTGGCHH-
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
18 |
| Similarity score: |
0.0205802 |
Alignment:
YDRCCASYAGRKGGCRSYV
HDGCCACHAGRGGGCRBY-
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
156 |
| Motif name: |
rgyGCCMyCTksTGGccd |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
0.0268949 |
Alignment:
RVYGCCCYCTKSTGGCHD
KBKGCCCKCTHGTGGCHH
| Original motif Consensus sequence: RVYGCCCYCTKSTGGCHD |
Reverse complement motif Consensus sequence: DDGCCASYAGMGGGCKVM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
109 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
18 |
| Similarity score: |
0.103512 |
Alignment:
MSGKKRCGCWDCABTGBBCD
--KBKGCCCKCTHGTGGCHH
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
125 |
| Motif name: |
TP53 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
18 |
| Similarity score: |
0.105007 |
Alignment:
ACATGCCCGGKCATGTCCSR
-KBKGCCCKCTHGTGGCHH-
| Original motif Consensus sequence: MSGGACATGYCCGGGCATGT |
Reverse complement motif Consensus sequence: ACATGCCCGGKCATGTCCSR |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 166 |
Motif name: CasCAGrGGGCrsy |
| Original motif Consensus sequence: CACCAGRGGGCRSB |
Reverse complement motif Consensus sequence: BSKGCCCKCTGGTG |
|
|
 |
 |
Best Matches for Motif ID 166 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
165 |
| Motif name: |
wgGCCAshAGrGGGCrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
14 |
| Similarity score: |
0.015353 |
Alignment:
HDGCCACHAGRGGGCRBY
----CACCAGRGGGCRSB
| Original motif Consensus sequence: HDGCCACHAGRGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCKCTHGTGGCHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
14 |
| Similarity score: |
0.0273616 |
Alignment:
VHGGCCAGMAGAGGGCRBY
-----CACCAGRGGGCRSB
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
14 |
| Similarity score: |
0.0281629 |
Alignment:
YDRCCASYAGRKGGCRSYV
----CACCAGRGGGCRSB-
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
168 |
| Motif name: |
yrcrGYGCCMyCTGGtG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.03743 |
Alignment:
CACCAGMGGGCGCTGBD
CACCAGRGGGCRSB---
| Original motif Consensus sequence: HVCAGCGCCCYCTGGTG |
Reverse complement motif Consensus sequence: CACCAGMGGGCGCTGBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
156 |
| Motif name: |
rgyGCCMyCTksTGGccd |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
14 |
| Similarity score: |
0.043415 |
Alignment:
RVYGCCCYCTKSTGGCHD
----BSKGCCCKCTGGTG
| Original motif Consensus sequence: RVYGCCCYCTKSTGGCHD |
Reverse complement motif Consensus sequence: DDGCCASYAGMGGGCKVM |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 167 |
Motif name: rsyAGrkGGCGCCmyCTrsy |
| Original motif Consensus sequence: DSYAGRKGGCGCCMYCTRSH |
Reverse complement motif Consensus sequence: HSKAGKYGGCGCCRMCTMSD |
|
|
 |
 |
Best Matches for Motif ID 167 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
20 |
| Similarity score: |
0.0672057 |
Alignment:
VDBHMAGGTCACCCTGACCY
DSYAGRKGGCGCCMYCTRSH
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
109 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
20 |
| Similarity score: |
0.0747165 |
Alignment:
MSGKKRCGCWDCABTGBBCD
HSKAGKYGGCGCCRMCTMSD
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
70 |
| Motif name: |
Ar |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
19 |
| Similarity score: |
0.57268 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV-
---DSYAGRKGGCGCCMYCTRSH
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
111 |
| Motif name: |
PPARG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
19 |
| Similarity score: |
0.57362 |
Alignment:
-ABTMGGTCACBGTGACCTAS
DSYAGRKGGCGCCMYCTRSH-
| Original motif Consensus sequence: STAGGTCACBGTGACCYABT |
Reverse complement motif Consensus sequence: ABTMGGTCACBGTGACCTAS |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
149 |
| Motif name: |
asmAGRGGGCrCTGsmkc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
1.03519 |
Alignment:
ASMAGAGGGCRCTGSABH--
DSYAGRKGGCGCCMYCTRSH
| Original motif Consensus sequence: ASMAGAGGGCRCTGSABH |
Reverse complement motif Consensus sequence: DBTSCAGMGCCCTCTRST |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 168 |
Motif name: yrcrGYGCCMyCTGGtG |
| Original motif Consensus sequence: HVCAGCGCCCYCTGGTG |
Reverse complement motif Consensus sequence: CACCAGMGGGCGCTGBD |
|
|
 |
 |
Best Matches for Motif ID 168 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
169 |
| Motif name: |
yvTGCyGCCmCCwGgtG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
17 |
| Similarity score: |
0.0386724 |
Alignment:
BVTGCTGCCACCWGGDG
HVCAGCGCCCYCTGGTG
| Original motif Consensus sequence: BVTGCTGCCACCWGGDG |
Reverse complement motif Consensus sequence: CDCCWGGTGGCAGCAVV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
149 |
| Motif name: |
asmAGRGGGCrCTGsmkc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
17 |
| Similarity score: |
0.0542719 |
Alignment:
DBTSCAGMGCCCTCTRST
-CACCAGMGGGCGCTGBD
| Original motif Consensus sequence: ASMAGAGGGCRCTGSABH |
Reverse complement motif Consensus sequence: DBTSCAGMGCCCTCTRST |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
17 |
| Similarity score: |
0.0695994 |
Alignment:
VDBHMAGGTCACCCTGACCY
--HVCAGCGCCCYCTGGTG-
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
70 |
| Motif name: |
Ar |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
17 |
| Similarity score: |
0.074879 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
HVCAGCGCCCYCTGGTG-----
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
156 |
| Motif name: |
rgyGCCMyCTksTGGccd |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
17 |
| Similarity score: |
0.076934 |
Alignment:
RVYGCCCYCTKSTGGCHD
HVCAGCGCCCYCTGGTG-
| Original motif Consensus sequence: RVYGCCCYCTKSTGGCHD |
Reverse complement motif Consensus sequence: DDGCCASYAGMGGGCKVM |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 169 |
Motif name: yvTGCyGCCmCCwGgtG |
| Original motif Consensus sequence: BVTGCTGCCACCWGGDG |
Reverse complement motif Consensus sequence: CDCCWGGTGGCAGCAVV |
|
|
 |
 |
Best Matches for Motif ID 169 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
168 |
| Motif name: |
yrcrGYGCCMyCTGGtG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
17 |
| Similarity score: |
0.0263455 |
Alignment:
HVCAGCGCCCYCTGGTG
BVTGCTGCCACCWGGDG
| Original motif Consensus sequence: HVCAGCGCCCYCTGGTG |
Reverse complement motif Consensus sequence: CACCAGMGGGCGCTGBD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
17 |
| Similarity score: |
0.0578732 |
Alignment:
BMSMGCCYMCTKSTGGMHM
-BVTGCTGCCACCWGGDG-
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
167 |
| Motif name: |
rsyAGrkGGCGCCmyCTrsy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
17 |
| Similarity score: |
0.0596753 |
Alignment:
HSKAGKYGGCGCCRMCTMSD
--BVTGCTGCCACCWGGDG-
| Original motif Consensus sequence: DSYAGRKGGCGCCMYCTRSH |
Reverse complement motif Consensus sequence: HSKAGKYGGCGCCRMCTMSD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
156 |
| Motif name: |
rgyGCCMyCTksTGGccd |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
17 |
| Similarity score: |
0.0597938 |
Alignment:
RVYGCCCYCTKSTGGCHD
BVTGCTGCCACCWGGDG-
| Original motif Consensus sequence: RVYGCCCYCTKSTGGCHD |
Reverse complement motif Consensus sequence: DDGCCASYAGMGGGCKVM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
163 |
| Motif name: |
gwGGCCAGmAGAGGGCrby |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
17 |
| Similarity score: |
0.0605716 |
Alignment:
VHGGCCAGMAGAGGGCRBY
CDCCWGGTGGCAGCAVV--
| Original motif Consensus sequence: VHGGCCAGMAGAGGGCRBY |
Reverse complement motif Consensus sequence: KBKGCCCTCTYCTGGCCHV |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 170 |
Motif name: ssGGCrsTGCrs |
| Original motif Consensus sequence: VVGGCRSTGCVB |
Reverse complement motif Consensus sequence: BVGCASMGCCVV |
|
|
 |
 |
Best Matches for Motif ID 170 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
149 |
| Motif name: |
asmAGRGGGCrCTGsmkc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.038906 |
Alignment:
ASMAGAGGGCRCTGSABH
-----VVGGCRSTGCVB-
| Original motif Consensus sequence: ASMAGAGGGCRCTGSABH |
Reverse complement motif Consensus sequence: DBTSCAGMGCCCTCTRST |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
130 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0768177 |
Alignment:
BBVGCCBVGGCCTV
BVGCASMGCCVV--
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
155 |
| Motif name: |
csCSCCdCCCcs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0798188 |
Alignment:
VBCCCCDCCCHV
BVGCASMGCCVV
| Original motif Consensus sequence: VBCCCCDCCCHV |
Reverse complement motif Consensus sequence: VDGGGDGGGGBV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
144 |
| Motif name: |
ctCTrsyGCCmCCTast |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
12 |
| Similarity score: |
0.0800293 |
Alignment:
ASTAGGYGGCMSCAGDD
VVGGCRSTGCVB-----
| Original motif Consensus sequence: HDCTGSYGCCMCCTAST |
Reverse complement motif Consensus sequence: ASTAGGYGGCMSCAGDD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
146 |
| Motif name: |
myrGYGCCmCCTast |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0817044 |
Alignment:
VBAGCGCCMCCTAST
--BVGCASMGCCVV-
| Original motif Consensus sequence: VBAGCGCCMCCTAST |
Reverse complement motif Consensus sequence: ASTAGGYGGCGCTBB |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 171 |
Motif name: ysGTGGCCACsr |
| Original motif Consensus sequence: BVGTGGCCACBV |
Reverse complement motif Consensus sequence: VBGTGGCCACVB |
|
|
 |
 |
Best Matches for Motif ID 171 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
169 |
| Motif name: |
yvTGCyGCCmCCwGgtG |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0325062 |
Alignment:
BVTGCTGCCACCWGGDG
----VBGTGGCCACVB-
| Original motif Consensus sequence: BVTGCTGCCACCWGGDG |
Reverse complement motif Consensus sequence: CDCCWGGTGGCAGCAVV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0344223 |
Alignment:
MGGTCAGGGTGACCTRDBHV
------VBGTGGCCACVB--
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
111 |
| Motif name: |
PPARG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0381861 |
Alignment:
ABTMGGTCACBGTGACCTAS
--------BVGTGGCCACBV
| Original motif Consensus sequence: STAGGTCACBGTGACCYABT |
Reverse complement motif Consensus sequence: ABTMGGTCACBGTGACCTAS |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
146 |
| Motif name: |
myrGYGCCmCCTast |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0484805 |
Alignment:
ASTAGGYGGCGCTBB
BVGTGGCCACBV---
| Original motif Consensus sequence: VBAGCGCCMCCTAST |
Reverse complement motif Consensus sequence: ASTAGGYGGCGCTBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
170 |
| Motif name: |
ssGGCrsTGCrs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0498451 |
Alignment:
BVGCASMGCCVV
BVGTGGCCACBV
| Original motif Consensus sequence: VVGGCRSTGCVB |
Reverse complement motif Consensus sequence: BVGCASMGCCVV |
|
|
 |
 |













































































































































































































































































































































































