Top 10 Significant Motifs - Global Matching (Highest to Lowest)
| Dataset #: 4 | Motif ID: 45 | Motif name: wbgTAAATAww |
| Original motif Consensus sequence: DBGTAAATAHD | Reverse complement motif Consensus sequence: DHTATTTACBD |
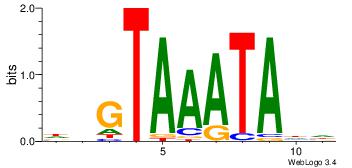 |
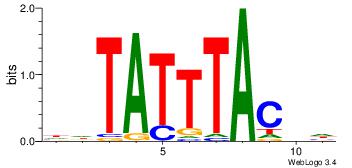 |
Best Matches for Top Significant Motif ID 45 (Highest to Lowest)
| Dataset #: | 2 |
| Motif ID: | 9 |
| Motif name: | Motif 9 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0177412 |
Alignment:
AATHATATWTHAAA
DHTATTTACBD---
| Original motif Consensus sequence: AATHATATWTHAAA | Reverse complement motif Consensus sequence: TTTDAWATATHATT |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 6 |
| Motif name: | Motif 6 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0205821 |
Alignment:
AATTYDGAARTAWW
---DBGTAAATAHD
| Original motif Consensus sequence: AATTYDGAARTAWW | Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 4 |
| Motif name: | Motif 4 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0288058 |
Alignment:
AWGKAAWTTTT
DBGTAAATAHD
| Original motif Consensus sequence: AAAAWTTRCWT | Reverse complement motif Consensus sequence: AWGKAAWTTTT |
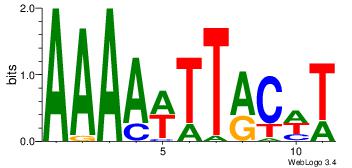 |
 |
| Dataset #: | 2 |
| Motif ID: | 3 |
| Motif name: | Motif 3 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0337252 |
Alignment:
AWAAAWTWAAASWA
--DHTATTTACBD-
| Original motif Consensus sequence: AWAAAWTWAAASWA | Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 5 |
| Motif name: | Motif 5 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0404685 |
Alignment:
AWTAAATAYAATTT
---DHTATTTACBD
| Original motif Consensus sequence: AWTAAATAYAATTT | Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 55 |
| Motif name: | Pax5 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 10 |
| Number of overlap: | 11 |
| Similarity score: | 0.0439858 |
Alignment:
DGVBCABTGDWGCGKRRCSR
DBGTAAATAHD---------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR | Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 31 |
| Motif name: | Pax5 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0439858 |
Alignment:
DGVBCABTGDWGCGKRRCSR
DBGTAAATAHD---------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR | Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 59 |
| Motif name: | GABPA |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0439962 |
Alignment:
CCGGAAGTGVV
DBGTAAATAHD
| Original motif Consensus sequence: CCGGAAGTGVV | Reverse complement motif Consensus sequence: VVCACTTCCGG |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 35 |
| Motif name: | GABPA |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0439962 |
Alignment:
CCGGAAGTGVV
DBGTAAATAHD
| Original motif Consensus sequence: CCGGAAGTGVV | Reverse complement motif Consensus sequence: VVCACTTCCGG |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 19 |
| Motif name: | Motif 19 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0477737 |
Alignment:
TWTAATCTAWGAA
-DHTATTTACBD-
| Original motif Consensus sequence: TTCWTAGATTAWA | Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
 |
 |
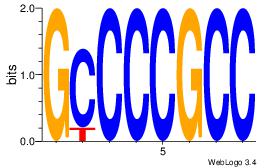
| Dataset #: 1 | Motif ID: 1 | Motif name: Motif 1 |
| Original motif Consensus sequence: GGCGGGGC | Reverse complement motif Consensus sequence: GCCCCGCC |
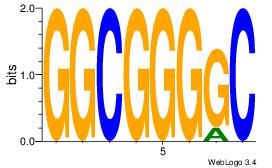 |
 |
Best Matches for Top Significant Motif ID 1 (Highest to Lowest)
| Dataset #: | 4 |
| Motif ID: | 36 |
| Motif name: | csGCCCCGCCCCsc |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HVGCCCCGCCCCBB
--GCCCCGCC----
| Original motif Consensus sequence: HVGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 38 |
| Motif name: | cccGCCCCGCCCCsb |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.00250079 |
Alignment:
BCCGCCCCGCCCCBB
---GCCCCGCC----
| Original motif Consensus sequence: BCCGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 7 |
| Motif name: | Motif 7 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0149958 |
Alignment:
MSGGGGCGGGGYSG
----GGCGGGGC--
| Original motif Consensus sequence: CSKCCCCGCCCCSY | Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 51 |
| Motif name: | Klf4 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0349983 |
Alignment:
DGGGYGKGGC
--GGCGGGGC
| Original motif Consensus sequence: DGGGYGKGGC | Reverse complement motif Consensus sequence: GCCYCMCCCD |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 27 |
| Motif name: | Klf4 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0349983 |
Alignment:
DGGGYGKGGC
--GGCGGGGC
| Original motif Consensus sequence: DGGGYGKGGC | Reverse complement motif Consensus sequence: GCCYCMCCCD |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 22 |
| Motif name: | Zfx |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0708224 |
Alignment:
VAGGCCBBGGCVBB
---GCCCCGCC---
| Original motif Consensus sequence: BBVGCCBVGGCCTV | Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 46 |
| Motif name: | Zfx |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0708224 |
Alignment:
VAGGCCBBGGCVBB
---GCCCCGCC---
| Original motif Consensus sequence: BBVGCCBVGGCCTV | Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 55 |
| Motif name: | Pax5 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0853847 |
Alignment:
DGVBCABTGDWGCGKRRCSR
----------GGCGGGGC--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR | Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 31 |
| Motif name: | Pax5 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0853847 |
Alignment:
DGVBCABTGDWGCGKRRCSR
----------GGCGGGGC--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR | Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 23 |
| Motif name: | Egr1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0878731 |
Alignment:
HGCGTGGGCGK
GGCGGGGC---
| Original motif Consensus sequence: HGCGTGGGCGK | Reverse complement motif Consensus sequence: YCGCCCACGCH |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 47 |
| Motif name: | Egr1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0878731 |
Alignment:
HGCGTGGGCGK
GGCGGGGC---
| Original motif Consensus sequence: HGCGTGGGCGK | Reverse complement motif Consensus sequence: YCGCCCACGCH |
 |
 |
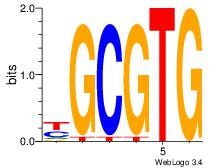
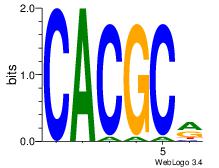
| Dataset #: 3 | Motif ID: 32 | Motif name: ArntAhr |
| Original motif Consensus sequence: YGCGTG | Reverse complement motif Consensus sequence: CACGCM |
 |
 |
Best Matches for Top Significant Motif ID 32 (Highest to Lowest)
| Dataset #: | 5 |
| Motif ID: | 56 |
| Motif name: | ArntAhr |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
YGCGTG
YGCGTG
| Original motif Consensus sequence: YGCGTG | Reverse complement motif Consensus sequence: CACGCM |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 47 |
| Motif name: | Egr1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0340278 |
Alignment:
HGCGTGGGCGK
YGCGTG-----
| Original motif Consensus sequence: HGCGTGGGCGK | Reverse complement motif Consensus sequence: YCGCCCACGCH |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 53 |
| Motif name: | HIF1AARNT |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.0452724 |
Alignment:
VBACGTGV
-YGCGTG-
| Original motif Consensus sequence: VBACGTGV | Reverse complement motif Consensus sequence: VCACGTBV |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 57 |
| Motif name: | Mycn |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0718227 |
Alignment:
GCCACGTGSD
--YGCGTG--
| Original motif Consensus sequence: HSCACGTGGC | Reverse complement motif Consensus sequence: GCCACGTGSD |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 36 |
| Motif name: | csGCCCCGCCCCsc |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0736681 |
Alignment:
HVGCCCCGCCCCBB
----CACGCM----
| Original motif Consensus sequence: HVGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 58 |
| Motif name: | Myc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0739492 |
Alignment:
DCCACGTGCV
--YGCGTG--
| Original motif Consensus sequence: VGCACGTGGH | Reverse complement motif Consensus sequence: DCCACGTGCV |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 1 |
| Motif name: | Motif 1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0746528 |
Alignment:
GCCCCGCC
--CACGCM
| Original motif Consensus sequence: GGCGGGGC | Reverse complement motif Consensus sequence: GCCCCGCC |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 42 |
| Motif name: | sSGTCACGTGACSs |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0752315 |
Alignment:
SGGTCACGTGACCS
----YGCGTG----
| Original motif Consensus sequence: SGGTCACGTGACCS | Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 38 |
| Motif name: | cccGCCCCGCCCCsb |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.0765366 |
Alignment:
BCCGCCCCGCCCCBB
-----CACGCM----
| Original motif Consensus sequence: BCCGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 7 |
| Motif name: | Motif 7 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0806145 |
Alignment:
MSGGGGCGGGGYSG
----YGCGTG----
| Original motif Consensus sequence: CSKCCCCGCCCCSY | Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
 |
 |
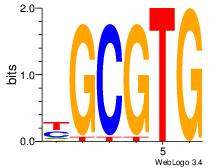
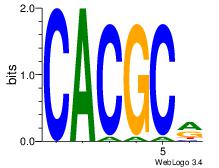
| Dataset #: 5 | Motif ID: 56 | Motif name: ArntAhr |
| Original motif Consensus sequence: YGCGTG | Reverse complement motif Consensus sequence: CACGCM |
 |
 |
Best Matches for Top Significant Motif ID 56 (Highest to Lowest)
| Dataset #: | 3 |
| Motif ID: | 32 |
| Motif name: | ArntAhr |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
YGCGTG
YGCGTG
| Original motif Consensus sequence: YGCGTG | Reverse complement motif Consensus sequence: CACGCM |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 23 |
| Motif name: | Egr1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 6 |
| Similarity score: | 0.0340278 |
Alignment:
YCGCCCACGCH
-----CACGCM
| Original motif Consensus sequence: HGCGTGGGCGK | Reverse complement motif Consensus sequence: YCGCCCACGCH |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 29 |
| Motif name: | HIF1AARNT |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.0452724 |
Alignment:
VBACGTGV
-YGCGTG-
| Original motif Consensus sequence: VBACGTGV | Reverse complement motif Consensus sequence: VCACGTBV |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 33 |
| Motif name: | Mycn |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0718227 |
Alignment:
HSCACGTGGC
--CACGCM--
| Original motif Consensus sequence: HSCACGTGGC | Reverse complement motif Consensus sequence: GCCACGTGSD |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 36 |
| Motif name: | csGCCCCGCCCCsc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0736681 |
Alignment:
BBGGGGCGGGGCVD
----YGCGTG----
| Original motif Consensus sequence: HVGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 34 |
| Motif name: | Myc |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0739492 |
Alignment:
VGCACGTGGH
--CACGCM--
| Original motif Consensus sequence: VGCACGTGGH | Reverse complement motif Consensus sequence: DCCACGTGCV |
 |
 |
| Dataset #: | 1 |
| Motif ID: | 1 |
| Motif name: | Motif 1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0746528 |
Alignment:
GCCCCGCC
--CACGCM
| Original motif Consensus sequence: GGCGGGGC | Reverse complement motif Consensus sequence: GCCCCGCC |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 42 |
| Motif name: | sSGTCACGTGACSs |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0752315 |
Alignment:
SGGTCACGTGACCS
----YGCGTG----
| Original motif Consensus sequence: SGGTCACGTGACCS | Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 38 |
| Motif name: | cccGCCCCGCCCCsb |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.0765366 |
Alignment:
BBGGGGCGGGGCGGB
-----YGCGTG----
| Original motif Consensus sequence: BCCGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 7 |
| Motif name: | Motif 7 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0806145 |
Alignment:
MSGGGGCGGGGYSG
----YGCGTG----
| Original motif Consensus sequence: CSKCCCCGCCCCSY | Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
 |
 |
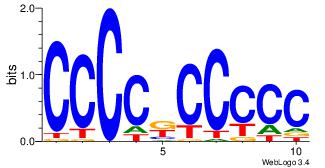
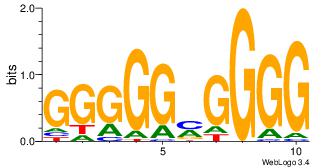
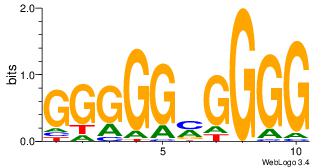
| Dataset #: 3 | Motif ID: 24 | Motif name: SP1 |
| Original motif Consensus sequence: CCCCKCCCCC | Reverse complement motif Consensus sequence: GGGGGYGGGG |
 |
 |
Best Matches for Top Significant Motif ID 24 (Highest to Lowest)
| Dataset #: | 5 |
| Motif ID: | 48 |
| Motif name: | SP1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
CCCCKCCCCC
CCCCKCCCCC
| Original motif Consensus sequence: CCCCKCCCCC | Reverse complement motif Consensus sequence: GGGGGYGGGG |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 7 |
| Motif name: | Motif 7 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.043504 |
Alignment:
CSKCCCCGCCCCSY
---CCCCKCCCCC-
| Original motif Consensus sequence: CSKCCCCGCCCCSY | Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 36 |
| Motif name: | csGCCCCGCCCCsc |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0486461 |
Alignment:
BBGGGGCGGGGCVD
-GGGGGYGGGG---
| Original motif Consensus sequence: HVGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 38 |
| Motif name: | cccGCCCCGCCCCsb |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0517173 |
Alignment:
BCCGCCCCGCCCCBB
----CCCCKCCCCC-
| Original motif Consensus sequence: BCCGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 46 |
| Motif name: | Zfx |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0930706 |
Alignment:
VAGGCCBBGGCVBB
----CCCCKCCCCC
| Original motif Consensus sequence: BBVGCCBVGGCCTV | Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 54 |
| Motif name: | PLAG1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0990873 |
Alignment:
CCCCCTTGGGCCCC
-CCCCKCCCCC---
| Original motif Consensus sequence: GGGGCCCAAGGGGG | Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 51 |
| Motif name: | Klf4 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.102706 |
Alignment:
GCCYCMCCCD
CCCCKCCCCC
| Original motif Consensus sequence: DGGGYGKGGC | Reverse complement motif Consensus sequence: GCCYCMCCCD |
 |
 |
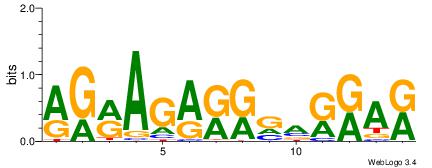
| Dataset #: | 2 |
| Motif ID: | 2 |
| Motif name: | Motif 2 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.103809 |
Alignment:
MTMMTCMMTCTKCK
----CCCCKCCCCC
| Original motif Consensus sequence: RGRAGARRGARRAR | Reverse complement motif Consensus sequence: MTMMTCMMTCTKCK |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 47 |
| Motif name: | Egr1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.106429 |
Alignment:
HGCGTGGGCGK
GGGGGYGGGG-
| Original motif Consensus sequence: HGCGTGGGCGK | Reverse complement motif Consensus sequence: YCGCCCACGCH |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 55 |
| Motif name: | Pax5 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 10 |
| Similarity score: | 0.11125 |
Alignment:
DGVBCABTGDWGCGKRRCSR
-------GGGGGYGGGG---
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR | Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
 |
 |
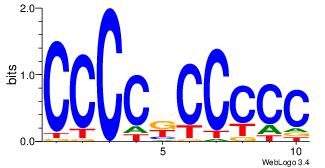
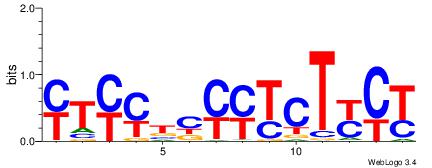
| Dataset #: 5 | Motif ID: 48 | Motif name: SP1 |
| Original motif Consensus sequence: CCCCKCCCCC | Reverse complement motif Consensus sequence: GGGGGYGGGG |
 |
 |
Best Matches for Top Significant Motif ID 48 (Highest to Lowest)
| Dataset #: | 3 |
| Motif ID: | 24 |
| Motif name: | SP1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
CCCCKCCCCC
CCCCKCCCCC
| Original motif Consensus sequence: CCCCKCCCCC | Reverse complement motif Consensus sequence: GGGGGYGGGG |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 7 |
| Motif name: | Motif 7 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.043504 |
Alignment:
CSKCCCCGCCCCSY
---CCCCKCCCCC-
| Original motif Consensus sequence: CSKCCCCGCCCCSY | Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 36 |
| Motif name: | csGCCCCGCCCCsc |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0486461 |
Alignment:
BBGGGGCGGGGCVD
-GGGGGYGGGG---
| Original motif Consensus sequence: HVGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 38 |
| Motif name: | cccGCCCCGCCCCsb |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0517173 |
Alignment:
BCCGCCCCGCCCCBB
----CCCCKCCCCC-
| Original motif Consensus sequence: BCCGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 22 |
| Motif name: | Zfx |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0930706 |
Alignment:
BBVGCCBVGGCCTV
----GGGGGYGGGG
| Original motif Consensus sequence: BBVGCCBVGGCCTV | Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 30 |
| Motif name: | PLAG1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0990873 |
Alignment:
GGGGCCCAAGGGGG
-GGGGGYGGGG---
| Original motif Consensus sequence: GGGGCCCAAGGGGG | Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 27 |
| Motif name: | Klf4 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.102706 |
Alignment:
DGGGYGKGGC
GGGGGYGGGG
| Original motif Consensus sequence: DGGGYGKGGC | Reverse complement motif Consensus sequence: GCCYCMCCCD |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 2 |
| Motif name: | Motif 2 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.103809 |
Alignment:
MTMMTCMMTCTKCK
----CCCCKCCCCC
| Original motif Consensus sequence: RGRAGARRGARRAR | Reverse complement motif Consensus sequence: MTMMTCMMTCTKCK |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 23 |
| Motif name: | Egr1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.106429 |
Alignment:
YCGCCCACGCH
CCCCKCCCCC-
| Original motif Consensus sequence: HGCGTGGGCGK | Reverse complement motif Consensus sequence: YCGCCCACGCH |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 31 |
| Motif name: | Pax5 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 10 |
| Similarity score: | 0.11125 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-------CCCCKCCCCC---
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR | Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
 |
 |
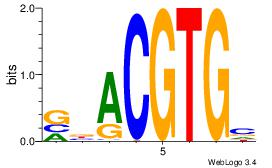
| Dataset #: 5 | Motif ID: 53 | Motif name: HIF1AARNT |
| Original motif Consensus sequence: VBACGTGV | Reverse complement motif Consensus sequence: VCACGTBV |
 |
 |
Best Matches for Top Significant Motif ID 53 (Highest to Lowest)
| Dataset #: | 3 |
| Motif ID: | 29 |
| Motif name: | HIF1AARNT |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
VBACGTGV
VBACGTGV
| Original motif Consensus sequence: VBACGTGV | Reverse complement motif Consensus sequence: VCACGTBV |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 33 |
| Motif name: | Mycn |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0575825 |
Alignment:
GCCACGTGSD
-VBACGTGV-
| Original motif Consensus sequence: HSCACGTGGC | Reverse complement motif Consensus sequence: GCCACGTGSD |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 34 |
| Motif name: | Myc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0597215 |
Alignment:
DCCACGTGCV
-VBACGTGV-
| Original motif Consensus sequence: VGCACGTGGH | Reverse complement motif Consensus sequence: DCCACGTGCV |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 40 |
| Motif name: | kcACCTGCAgc |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.0748197 |
Alignment:
GBTGCAGGTGB
---VCACGTBV
| Original motif Consensus sequence: BCACCTGCABC | Reverse complement motif Consensus sequence: GBTGCAGGTGB |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 42 |
| Motif name: | sSGTCACGTGACSs |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0767561 |
Alignment:
SGGTCACGTGACCS
---VBACGTGV---
| Original motif Consensus sequence: SGGTCACGTGACCS | Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
 |
 |
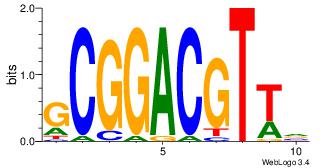
| Dataset #: | 3 |
| Motif ID: | 26 |
| Motif name: | MIZF |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0829928 |
Alignment:
GCGGACGTTV
--VCACGTBV
| Original motif Consensus sequence: BAACGTCCGC | Reverse complement motif Consensus sequence: GCGGACGTTV |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 31 |
| Motif name: | Pax5 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.100962 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-----VCACGTBV-------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR | Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
 |
 |
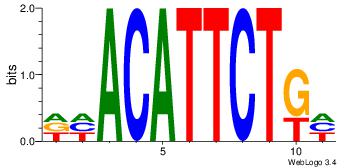
| Dataset #: | 4 |
| Motif ID: | 44 |
| Motif name: | dhACATTCTkh |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.109275 |
Alignment:
HCAGAATGTHD
VBACGTGV---
| Original motif Consensus sequence: DHACATTCTGH | Reverse complement motif Consensus sequence: HCAGAATGTHD |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 35 |
| Motif name: | GABPA |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.110088 |
Alignment:
VVCACTTCCGG
--VBACGTGV-
| Original motif Consensus sequence: CCGGAAGTGVV | Reverse complement motif Consensus sequence: VVCACTTCCGG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 36 |
| Motif name: | csGCCCCGCCCCsc |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.116389 |
Alignment:
HVGCCCCGCCCCBB
---VCACGTBV---
| Original motif Consensus sequence: HVGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
 |
 |
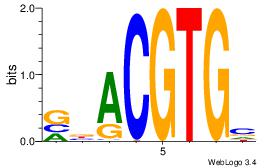
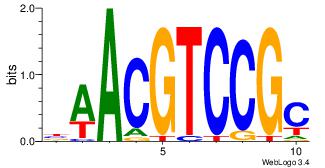
| Dataset #: 3 | Motif ID: 29 | Motif name: HIF1AARNT |
| Original motif Consensus sequence: VBACGTGV | Reverse complement motif Consensus sequence: VCACGTBV |
 |
 |
Best Matches for Top Significant Motif ID 29 (Highest to Lowest)
| Dataset #: | 5 |
| Motif ID: | 53 |
| Motif name: | HIF1AARNT |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
VBACGTGV
VBACGTGV
| Original motif Consensus sequence: VBACGTGV | Reverse complement motif Consensus sequence: VCACGTBV |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 57 |
| Motif name: | Mycn |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0575825 |
Alignment:
GCCACGTGSD
-VBACGTGV-
| Original motif Consensus sequence: HSCACGTGGC | Reverse complement motif Consensus sequence: GCCACGTGSD |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 58 |
| Motif name: | Myc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0597215 |
Alignment:
DCCACGTGCV
-VBACGTGV-
| Original motif Consensus sequence: VGCACGTGGH | Reverse complement motif Consensus sequence: DCCACGTGCV |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 40 |
| Motif name: | kcACCTGCAgc |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.0748197 |
Alignment:
GBTGCAGGTGB
---VCACGTBV
| Original motif Consensus sequence: BCACCTGCABC | Reverse complement motif Consensus sequence: GBTGCAGGTGB |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 42 |
| Motif name: | sSGTCACGTGACSs |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0767561 |
Alignment:
SGGTCACGTGACCS
---VCACGTBV---
| Original motif Consensus sequence: SGGTCACGTGACCS | Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 50 |
| Motif name: | MIZF |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0829928 |
Alignment:
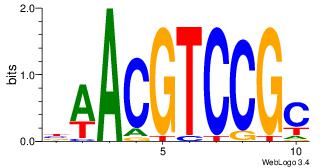
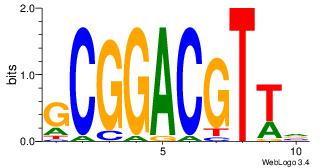
GCGGACGTTV
--VCACGTBV
| Original motif Consensus sequence: BAACGTCCGC | Reverse complement motif Consensus sequence: GCGGACGTTV |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 55 |
| Motif name: | Pax5 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.100962 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-----VCACGTBV-------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR | Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 44 |
| Motif name: | dhACATTCTkh |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.109275 |
Alignment:
DHACATTCTGH
VCACGTBV---
| Original motif Consensus sequence: DHACATTCTGH | Reverse complement motif Consensus sequence: HCAGAATGTHD |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 59 |
| Motif name: | GABPA |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.110088 |
Alignment:
CCGGAAGTGVV
--VCACGTBV-
| Original motif Consensus sequence: CCGGAAGTGVV | Reverse complement motif Consensus sequence: VVCACTTCCGG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 36 |
| Motif name: | csGCCCCGCCCCsc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.116389 |
Alignment:
BBGGGGCGGGGCVD
---VBACGTGV---
| Original motif Consensus sequence: HVGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
 |
 |
| Dataset #: 5 | Motif ID: 51 | Motif name: Klf4 |
| Original motif Consensus sequence: DGGGYGKGGC | Reverse complement motif Consensus sequence: GCCYCMCCCD |
 |
 |
Best Matches for Top Significant Motif ID 51 (Highest to Lowest)
| Dataset #: | 3 |
| Motif ID: | 27 |
| Motif name: | Klf4 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
DGGGYGKGGC
DGGGYGKGGC
| Original motif Consensus sequence: DGGGYGKGGC | Reverse complement motif Consensus sequence: GCCYCMCCCD |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 7 |
| Motif name: | Motif 7 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0516385 |
Alignment:
CSKCCCCGCCCCSY
--GCCYCMCCCD--
| Original motif Consensus sequence: CSKCCCCGCCCCSY | Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 36 |
| Motif name: | csGCCCCGCCCCsc |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0524114 |
Alignment:
HVGCCCCGCCCCBB
--GCCYCMCCCD--
| Original motif Consensus sequence: HVGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 38 |
| Motif name: | cccGCCCCGCCCCsb |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0558821 |
Alignment:
BCCGCCCCGCCCCBB
--GCCYCMCCCD---
| Original motif Consensus sequence: BCCGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 22 |
| Motif name: | Zfx |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0987519 |
Alignment:
BBVGCCBVGGCCTV
-GCCYCMCCCD---
| Original motif Consensus sequence: BBVGCCBVGGCCTV | Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 24 |
| Motif name: | SP1 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.102706 |
Alignment:
GGGGGYGGGG
DGGGYGKGGC
| Original motif Consensus sequence: CCCCKCCCCC | Reverse complement motif Consensus sequence: GGGGGYGGGG |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 34 |
| Motif name: | Myc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.105078 |
Alignment:
VGCACGTGGH
DGGGYGKGGC
| Original motif Consensus sequence: VGCACGTGGH | Reverse complement motif Consensus sequence: DCCACGTGCV |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 31 |
| Motif name: | Pax5 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 10 |
| Similarity score: | 0.105953 |
Alignment:
DGVBCABTGDWGCGKRRCSR
--------DGGGYGKGGC--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR | Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
 |
 |
| Dataset #: | 3 |
| Motif ID: | 33 |
| Motif name: | Mycn |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.10603 |
Alignment:
HSCACGTGGC
DGGGYGKGGC
| Original motif Consensus sequence: HSCACGTGGC | Reverse complement motif Consensus sequence: GCCACGTGSD |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 2 |
| Motif name: | Motif 2 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.110488 |
Alignment:
MTMMTCMMTCTKCK
-GCCYCMCCCD---
| Original motif Consensus sequence: RGRAGARRGARRAR | Reverse complement motif Consensus sequence: MTMMTCMMTCTKCK |
 |
 |
| Dataset #: 3 | Motif ID: 27 | Motif name: Klf4 |
| Original motif Consensus sequence: DGGGYGKGGC | Reverse complement motif Consensus sequence: GCCYCMCCCD |
 |
 |
Best Matches for Top Significant Motif ID 27 (Highest to Lowest)
| Dataset #: | 5 |
| Motif ID: | 51 |
| Motif name: | Klf4 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
DGGGYGKGGC
DGGGYGKGGC
| Original motif Consensus sequence: DGGGYGKGGC | Reverse complement motif Consensus sequence: GCCYCMCCCD |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 7 |
| Motif name: | Motif 7 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0516385 |
Alignment:
CSKCCCCGCCCCSY
--GCCYCMCCCD--
| Original motif Consensus sequence: CSKCCCCGCCCCSY | Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 36 |
| Motif name: | csGCCCCGCCCCsc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0524114 |
Alignment:
BBGGGGCGGGGCVD
--DGGGYGKGGC--
| Original motif Consensus sequence: HVGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
 |
 |
| Dataset #: | 4 |
| Motif ID: | 38 |
| Motif name: | cccGCCCCGCCCCsb |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0558821 |
Alignment:
BBGGGGCGGGGCGGB
--DGGGYGKGGC---
| Original motif Consensus sequence: BCCGCCCCGCCCCBB | Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 46 |
| Motif name: | Zfx |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0987519 |
Alignment:
VAGGCCBBGGCVBB
-DGGGYGKGGC---
| Original motif Consensus sequence: BBVGCCBVGGCCTV | Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 48 |
| Motif name: | SP1 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.102706 |
Alignment:
CCCCKCCCCC
GCCYCMCCCD
| Original motif Consensus sequence: CCCCKCCCCC | Reverse complement motif Consensus sequence: GGGGGYGGGG |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 58 |
| Motif name: | Myc |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.105078 |
Alignment:
VGCACGTGGH
DGGGYGKGGC
| Original motif Consensus sequence: VGCACGTGGH | Reverse complement motif Consensus sequence: DCCACGTGCV |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 55 |
| Motif name: | Pax5 |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 10 |
| Similarity score: | 0.105953 |
Alignment:
DGVBCABTGDWGCGKRRCSR
--------DGGGYGKGGC--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR | Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
 |
 |
| Dataset #: | 5 |
| Motif ID: | 57 |
| Motif name: | Mycn |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.10603 |
Alignment:
HSCACGTGGC
DGGGYGKGGC
| Original motif Consensus sequence: HSCACGTGGC | Reverse complement motif Consensus sequence: GCCACGTGSD |
 |
 |
| Dataset #: | 2 |
| Motif ID: | 2 |
| Motif name: | Motif 2 |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.110488 |
Alignment:
MTMMTCMMTCTKCK
-GCCYCMCCCD---
| Original motif Consensus sequence: RGRAGARRGARRAR | Reverse complement motif Consensus sequence: MTMMTCMMTCTKCK |
 |
 |