Top 10 Significant Motifs - Global Matching (Highest to Lowest)
| Dataset #: 4 |
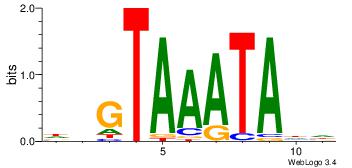
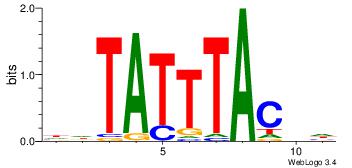
Motif ID: 45 |
Motif name: wbgTAAATAww |
| Original motif Consensus sequence: DBGTAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTACBD |
|
|
 |
 |
Best Matches for Top Significant Motif ID 45 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0177412 |
Alignment:
AATHATATWTHAAA
DHTATTTACBD---
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0205821 |
Alignment:
AATTYDGAARTAWW
---DBGTAAATAHD
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0288058 |
Alignment:
AWGKAAWTTTT
DBGTAAATAHD
| Original motif Consensus sequence: AAAAWTTRCWT |
Reverse complement motif Consensus sequence: AWGKAAWTTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0337252 |
Alignment:
AWAAAWTWAAASWA
--DHTATTTACBD-
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0404685 |
Alignment:
AWTAAATAYAATTT
---DHTATTTACBD
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
10 |
| Number of overlap: |
11 |
| Similarity score: |
0.0439858 |
Alignment:
DGVBCABTGDWGCGKRRCSR
DBGTAAATAHD---------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0439858 |
Alignment:
DGVBCABTGDWGCGKRRCSR
DBGTAAATAHD---------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
59 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0439962 |
Alignment:
CCGGAAGTGVV
DBGTAAATAHD
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
35 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0439962 |
Alignment:
CCGGAAGTGVV
DBGTAAATAHD
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0477737 |
Alignment:
TWTAATCTAWGAA
-DHTATTTACBD-
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
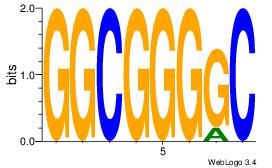
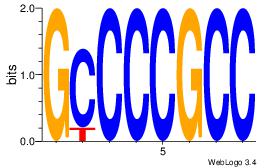
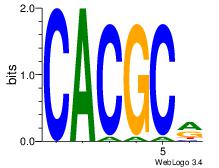
| Dataset #: 1 |
Motif ID: 1 |
Motif name: Motif 1 |
| Original motif Consensus sequence: GGCGGGGC |
Reverse complement motif Consensus sequence: GCCCCGCC |
|
|
 |
 |
Best Matches for Top Significant Motif ID 1 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0 |
Alignment:
HVGCCCCGCCCCBB
--GCCCCGCC----
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.00250079 |
Alignment:
BCCGCCCCGCCCCBB
---GCCCCGCC----
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0149958 |
Alignment:
MSGGGGCGGGGYSG
----GGCGGGGC--
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0349983 |
Alignment:
DGGGYGKGGC
--GGCGGGGC
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
27 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0349983 |
Alignment:
DGGGYGKGGC
--GGCGGGGC
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0708224 |
Alignment:
VAGGCCBBGGCVBB
---GCCCCGCC---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0708224 |
Alignment:
VAGGCCBBGGCVBB
---GCCCCGCC---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0853847 |
Alignment:
DGVBCABTGDWGCGKRRCSR
----------GGCGGGGC--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0853847 |
Alignment:
DGVBCABTGDWGCGKRRCSR
----------GGCGGGGC--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
23 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0878731 |
Alignment:
HGCGTGGGCGK
GGCGGGGC---
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
47 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0878731 |
Alignment:
HGCGTGGGCGK
GGCGGGGC---
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
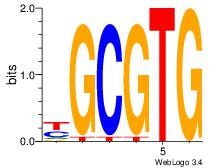
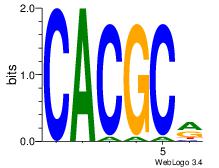
| Dataset #: 3 |
Motif ID: 32 |
Motif name: ArntAhr |
| Original motif Consensus sequence: YGCGTG |
Reverse complement motif Consensus sequence: CACGCM |
|
|
 |
 |
Best Matches for Top Significant Motif ID 32 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
ArntAhr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0 |
Alignment:
YGCGTG
YGCGTG
| Original motif Consensus sequence: YGCGTG |
Reverse complement motif Consensus sequence: CACGCM |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
47 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0340278 |
Alignment:
HGCGTGGGCGK
YGCGTG-----
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0452724 |
Alignment:
VBACGTGV
-YGCGTG-
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
57 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0718227 |
Alignment:
GCCACGTGSD
--YGCGTG--
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0736681 |
Alignment:
HVGCCCCGCCCCBB
----CACGCM----
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
58 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0739492 |
Alignment:
DCCACGTGCV
--YGCGTG--
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
1 |
| Motif name: |
Motif 1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0746528 |
Alignment:
GCCCCGCC
--CACGCM
| Original motif Consensus sequence: GGCGGGGC |
Reverse complement motif Consensus sequence: GCCCCGCC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0752315 |
Alignment:
SGGTCACGTGACCS
----YGCGTG----
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
6 |
| Similarity score: |
0.0765366 |
Alignment:
BCCGCCCCGCCCCBB
-----CACGCM----
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0806145 |
Alignment:
MSGGGGCGGGGYSG
----YGCGTG----
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
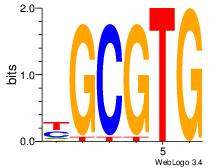
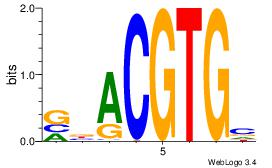
| Dataset #: 5 |
Motif ID: 56 |
Motif name: ArntAhr |
| Original motif Consensus sequence: YGCGTG |
Reverse complement motif Consensus sequence: CACGCM |
|
|
 |
 |
Best Matches for Top Significant Motif ID 56 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
32 |
| Motif name: |
ArntAhr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0 |
Alignment:
YGCGTG
YGCGTG
| Original motif Consensus sequence: YGCGTG |
Reverse complement motif Consensus sequence: CACGCM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
23 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0340278 |
Alignment:
YCGCCCACGCH
-----CACGCM
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
29 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0452724 |
Alignment:
VBACGTGV
-YGCGTG-
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
33 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0718227 |
Alignment:
HSCACGTGGC
--CACGCM--
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0736681 |
Alignment:
BBGGGGCGGGGCVD
----YGCGTG----
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
34 |
| Motif name: |
Myc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0739492 |
Alignment:
VGCACGTGGH
--CACGCM--
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
1 |
| Motif name: |
Motif 1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0746528 |
Alignment:
GCCCCGCC
--CACGCM
| Original motif Consensus sequence: GGCGGGGC |
Reverse complement motif Consensus sequence: GCCCCGCC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0752315 |
Alignment:
SGGTCACGTGACCS
----YGCGTG----
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
6 |
| Similarity score: |
0.0765366 |
Alignment:
BBGGGGCGGGGCGGB
-----YGCGTG----
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0806145 |
Alignment:
MSGGGGCGGGGYSG
----YGCGTG----
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
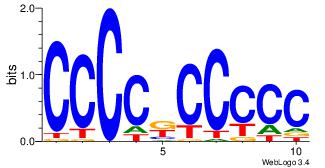
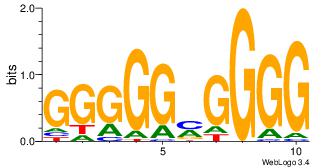
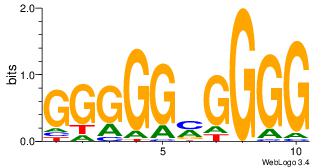
| Dataset #: 3 |
Motif ID: 24 |
Motif name: SP1 |
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
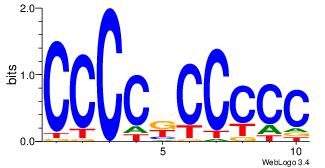
Best Matches for Top Significant Motif ID 24 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
48 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0 |
Alignment:
CCCCKCCCCC
CCCCKCCCCC
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.043504 |
Alignment:
CSKCCCCGCCCCSY
---CCCCKCCCCC-
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0486461 |
Alignment:
BBGGGGCGGGGCVD
-GGGGGYGGGG---
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0517173 |
Alignment:
BCCGCCCCGCCCCBB
----CCCCKCCCCC-
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0930706 |
Alignment:
VAGGCCBBGGCVBB
----CCCCKCCCCC
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0990873 |
Alignment:
CCCCCTTGGGCCCC
-CCCCKCCCCC---
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.102706 |
Alignment:
GCCYCMCCCD
CCCCKCCCCC
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
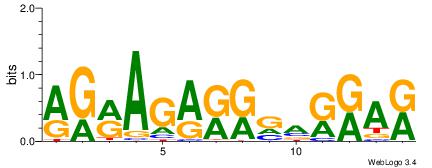
| Dataset #: |
2 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.103809 |
Alignment:
MTMMTCMMTCTKCK
----CCCCKCCCCC
| Original motif Consensus sequence: RGRAGARRGARRAR |
Reverse complement motif Consensus sequence: MTMMTCMMTCTKCK |
|
|
 |
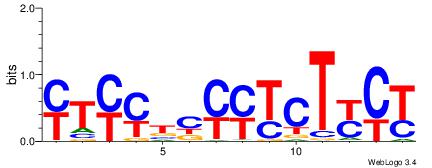 |
| Dataset #: |
5 |
| Motif ID: |
47 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.106429 |
Alignment:
HGCGTGGGCGK
GGGGGYGGGG-
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
10 |
| Similarity score: |
0.11125 |
Alignment:
DGVBCABTGDWGCGKRRCSR
-------GGGGGYGGGG---
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 48 |
Motif name: SP1 |
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
Best Matches for Top Significant Motif ID 48 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
24 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0 |
Alignment:
CCCCKCCCCC
CCCCKCCCCC
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.043504 |
Alignment:
CSKCCCCGCCCCSY
---CCCCKCCCCC-
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0486461 |
Alignment:
BBGGGGCGGGGCVD
-GGGGGYGGGG---
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0517173 |
Alignment:
BCCGCCCCGCCCCBB
----CCCCKCCCCC-
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0930706 |
Alignment:
BBVGCCBVGGCCTV
----GGGGGYGGGG
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0990873 |
Alignment:
GGGGCCCAAGGGGG
-GGGGGYGGGG---
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
27 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.102706 |
Alignment:
DGGGYGKGGC
GGGGGYGGGG
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.103809 |
Alignment:
MTMMTCMMTCTKCK
----CCCCKCCCCC
| Original motif Consensus sequence: RGRAGARRGARRAR |
Reverse complement motif Consensus sequence: MTMMTCMMTCTKCK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
23 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.106429 |
Alignment:
YCGCCCACGCH
CCCCKCCCCC-
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
10 |
| Similarity score: |
0.11125 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-------CCCCKCCCCC---
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 53 |
Motif name: HIF1AARNT |
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
Best Matches for Top Significant Motif ID 53 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
29 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0 |
Alignment:
VBACGTGV
VBACGTGV
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
33 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0575825 |
Alignment:
GCCACGTGSD
-VBACGTGV-
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
34 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0597215 |
Alignment:
DCCACGTGCV
-VBACGTGV-
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0748197 |
Alignment:
GBTGCAGGTGB
---VCACGTBV
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0767561 |
Alignment:
SGGTCACGTGACCS
---VBACGTGV---
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
26 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0829928 |
Alignment:
GCGGACGTTV
--VCACGTBV
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
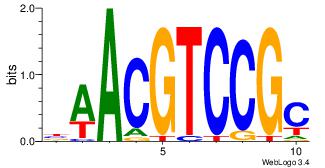 |
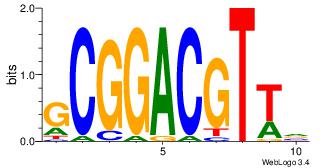 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
8 |
| Similarity score: |
0.100962 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-----VCACGTBV-------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
44 |
| Motif name: |
dhACATTCTkh |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.109275 |
Alignment:
HCAGAATGTHD
VBACGTGV---
| Original motif Consensus sequence: DHACATTCTGH |
Reverse complement motif Consensus sequence: HCAGAATGTHD |
|
|
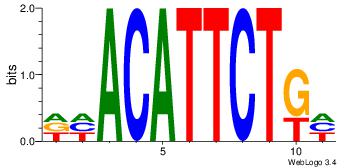 |
 |
| Dataset #: |
3 |
| Motif ID: |
35 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.110088 |
Alignment:
VVCACTTCCGG
--VBACGTGV-
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.116389 |
Alignment:
HVGCCCCGCCCCBB
---VCACGTBV---
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
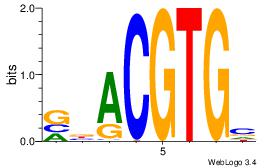
| Dataset #: 3 |
Motif ID: 29 |
Motif name: HIF1AARNT |
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
Best Matches for Top Significant Motif ID 29 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0 |
Alignment:
VBACGTGV
VBACGTGV
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
57 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0575825 |
Alignment:
GCCACGTGSD
-VBACGTGV-
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
58 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0597215 |
Alignment:
DCCACGTGCV
-VBACGTGV-
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0748197 |
Alignment:
GBTGCAGGTGB
---VCACGTBV
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0767561 |
Alignment:
SGGTCACGTGACCS
---VCACGTBV---
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0829928 |
Alignment:
GCGGACGTTV
--VCACGTBV
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
8 |
| Similarity score: |
0.100962 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-----VCACGTBV-------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
44 |
| Motif name: |
dhACATTCTkh |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.109275 |
Alignment:
DHACATTCTGH
VCACGTBV---
| Original motif Consensus sequence: DHACATTCTGH |
Reverse complement motif Consensus sequence: HCAGAATGTHD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
59 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.110088 |
Alignment:
CCGGAAGTGVV
--VCACGTBV-
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.116389 |
Alignment:
BBGGGGCGGGGCVD
---VBACGTGV---
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 51 |
Motif name: Klf4 |
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
Best Matches for Top Significant Motif ID 51 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
27 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0 |
Alignment:
DGGGYGKGGC
DGGGYGKGGC
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0516385 |
Alignment:
CSKCCCCGCCCCSY
--GCCYCMCCCD--
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0524114 |
Alignment:
HVGCCCCGCCCCBB
--GCCYCMCCCD--
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0558821 |
Alignment:
BCCGCCCCGCCCCBB
--GCCYCMCCCD---
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0987519 |
Alignment:
BBVGCCBVGGCCTV
-GCCYCMCCCD---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
24 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.102706 |
Alignment:
GGGGGYGGGG
DGGGYGKGGC
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
34 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.105078 |
Alignment:
VGCACGTGGH
DGGGYGKGGC
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
10 |
| Similarity score: |
0.105953 |
Alignment:
DGVBCABTGDWGCGKRRCSR
--------DGGGYGKGGC--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
33 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.10603 |
Alignment:
HSCACGTGGC
DGGGYGKGGC
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.110488 |
Alignment:
MTMMTCMMTCTKCK
-GCCYCMCCCD---
| Original motif Consensus sequence: RGRAGARRGARRAR |
Reverse complement motif Consensus sequence: MTMMTCMMTCTKCK |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 27 |
Motif name: Klf4 |
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
Best Matches for Top Significant Motif ID 27 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0 |
Alignment:
DGGGYGKGGC
DGGGYGKGGC
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0516385 |
Alignment:
CSKCCCCGCCCCSY
--GCCYCMCCCD--
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0524114 |
Alignment:
BBGGGGCGGGGCVD
--DGGGYGKGGC--
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0558821 |
Alignment:
BBGGGGCGGGGCGGB
--DGGGYGKGGC---
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0987519 |
Alignment:
VAGGCCBBGGCVBB
-DGGGYGKGGC---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
48 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.102706 |
Alignment:
CCCCKCCCCC
GCCYCMCCCD
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
58 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.105078 |
Alignment:
VGCACGTGGH
DGGGYGKGGC
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
10 |
| Similarity score: |
0.105953 |
Alignment:
DGVBCABTGDWGCGKRRCSR
--------DGGGYGKGGC--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
57 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.10603 |
Alignment:
HSCACGTGGC
DGGGYGKGGC
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.110488 |
Alignment:
MTMMTCMMTCTKCK
-GCCYCMCCCD---
| Original motif Consensus sequence: RGRAGARRGARRAR |
Reverse complement motif Consensus sequence: MTMMTCMMTCTKCK |
|
|
 |
 |
Significant Motifs - Global and Local Matching (Highest to Lowest)
| Dataset #: 4 |
Motif ID: 45 |
Motif name: wbgTAAATAww |
| Original motif Consensus sequence: DBGTAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTACBD |
|
|
 |
 |
Best Matches for Significant Motif ID 45 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.00892084 |
Alignment:
HDAAATAATAHW
-DBGTAAATAHD
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.00917337 |
Alignment:
DDTATTATTTDH
DHTATTTACBD-
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0177412 |
Alignment:
AATHATATWTHAAA
DHTATTTACBD---
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0205821 |
Alignment:
AATTYDGAARTAWW
---DBGTAAATAHD
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0288058 |
Alignment:
AAAAWTTRCWT
DHTATTTACBD
| Original motif Consensus sequence: AAAAWTTRCWT |
Reverse complement motif Consensus sequence: AWGKAAWTTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0337252 |
Alignment:
AWAAAWTWAAASWA
--DHTATTTACBD-
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
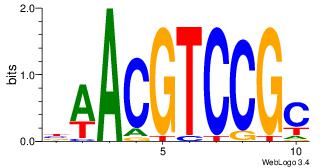
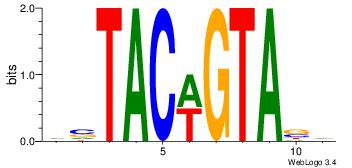
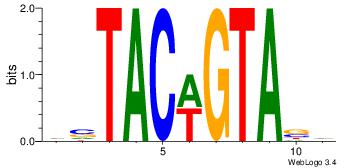
| Dataset #: |
4 |
| Motif ID: |
43 |
| Motif name: |
wsTACwGTAsw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0399273 |
Alignment:
HVTACWGTABD
DHTATTTACBD
| Original motif Consensus sequence: HVTACWGTABD |
Reverse complement motif Consensus sequence: DBTACWGTAVH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0404685 |
Alignment:
AWTAAATAYAATTT
---DHTATTTACBD
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
10 |
| Number of overlap: |
11 |
| Similarity score: |
0.0439858 |
Alignment:
DGVBCABTGDWGCGKRRCSR
DBGTAAATAHD---------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0439858 |
Alignment:
DGVBCABTGDWGCGKRRCSR
DBGTAAATAHD---------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 44 |
Motif name: dhACATTCTkh |
| Original motif Consensus sequence: DHACATTCTGH |
Reverse complement motif Consensus sequence: HCAGAATGTHD |
|
|
 |
 |
Best Matches for Significant Motif ID 44 (Highest to Lowest)
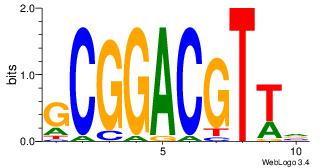
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0238442 |
Alignment:
BCACCTGCABC
DHACATTCTGH
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
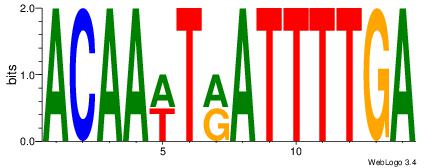
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0258838 |
Alignment:
TCAAAATKAWTTGT
HCAGAATGTHD---
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
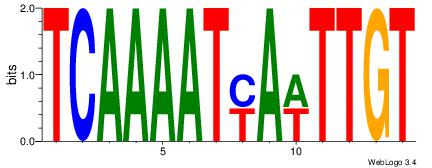 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0292508 |
Alignment:
DDTATTATTTDH
-HCAGAATGTHD
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0364899 |
Alignment:
WHTATTATTTDH
-HCAGAATGTHD
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0400884 |
Alignment:
AATTYDGAARTAWW
HCAGAATGTHD---
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
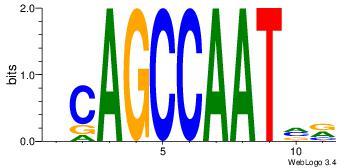
| Dataset #: |
4 |
| Motif ID: |
39 |
| Motif name: |
kCAGCCAATmr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0412582 |
Alignment:
MBATTGGCTGH
DHACATTCTGH
| Original motif Consensus sequence: DCAGCCAATVR |
Reverse complement motif Consensus sequence: MBATTGGCTGH |
|
|
 |
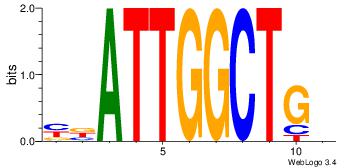 |
| Dataset #: |
5 |
| Motif ID: |
59 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0418896 |
Alignment:
VVCACTTCCGG
DHACATTCTGH
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
35 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0418896 |
Alignment:
CCGGAAGTGVV
HCAGAATGTHD
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0448232 |
Alignment:
TTTTATTGTYAT
HCAGAATGTHD-
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.045689 |
Alignment:
RGRAGARRGARRAR
--HCAGAATGTHD-
| Original motif Consensus sequence: RGRAGARRGARRAR |
Reverse complement motif Consensus sequence: MTMMTCMMTCTKCK |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 40 |
Motif name: kcACCTGCAgc |
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
Best Matches for Significant Motif ID 40 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
43 |
| Motif name: |
wsTACwGTAsw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0215284 |
Alignment:
DBTACWGTAVH
BCACCTGCABC
| Original motif Consensus sequence: HVTACWGTABD |
Reverse complement motif Consensus sequence: DBTACWGTAVH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
44 |
| Motif name: |
dhACATTCTkh |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0254134 |
Alignment:
DHACATTCTGH
BCACCTGCABC
| Original motif Consensus sequence: DHACATTCTGH |
Reverse complement motif Consensus sequence: HCAGAATGTHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.031338 |
Alignment:
SGGTCACGTGACCS
---BCACCTGCABC
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0402518 |
Alignment:
HVGCCCCGCCCCBB
-BCACCTGCABC--
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0411257 |
Alignment:
CSKCCCCGCCCCSY
-BCACCTGCABC--
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
11 |
| Similarity score: |
0.0420218 |
Alignment:
MSGKKRCGCWDCABTGBBCD
----BCACCTGCABC-----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
0.0420218 |
Alignment:
DGVBCABTGDWGCGKRRCSR
----GBTGCAGGTGB-----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
35 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0423275 |
Alignment:
CCGGAAGTGVV
GBTGCAGGTGB
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
59 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0423275 |
Alignment:
VVCACTTCCGG
BCACCTGCABC
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
39 |
| Motif name: |
kCAGCCAATmr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0439586 |
Alignment:
DCAGCCAATVR
GBTGCAGGTGB
| Original motif Consensus sequence: DCAGCCAATVR |
Reverse complement motif Consensus sequence: MBATTGGCTGH |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 21 |
Motif name: Motif 21 |
| Original motif Consensus sequence: ATAAAA |
Reverse complement motif Consensus sequence: TTTTAT |
|
|
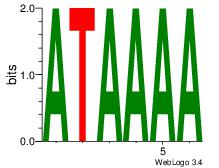 |
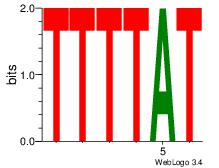 |
Best Matches for Significant Motif ID 21 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0 |
Alignment:
TTTTATTGTYAT
TTTTAT------
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0208333 |
Alignment:
AAATTKTATTTAWT
---TTTTAT-----
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0208333 |
Alignment:
TTTCATWAAAT
-ATAAAA----
| Original motif Consensus sequence: ATTTWATGAAA |
Reverse complement motif Consensus sequence: TTTCATWAAAT |
|
|
 |
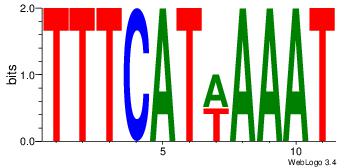 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0208333 |
Alignment:
TGATTTTAWKATTT
---TTTTAT-----
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0416667 |
Alignment:
TTTCATAAWT
----TTTTAT
| Original motif Consensus sequence: TTTCATAAWT |
Reverse complement motif Consensus sequence: AWTTATGAAA |
|
|
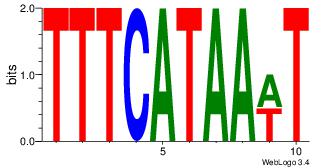 |
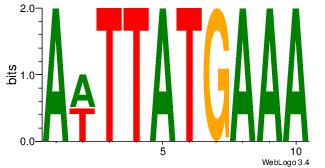 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0625 |
Alignment:
TWTAATCTAWGAA
TTTTAT-------
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
14 |
| Motif name: |
Motif 14 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0625 |
Alignment:
TTATTTWT
--TTTTAT
| Original motif Consensus sequence: AWAAATAA |
Reverse complement motif Consensus sequence: TTATTTWT |
|
|
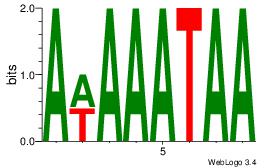 |
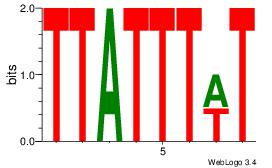 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
Motif 20 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0625 |
Alignment:
AAATCTTTYAA
-----TTTTAT
| Original motif Consensus sequence: TTMAAAGATTT |
Reverse complement motif Consensus sequence: AAATCTTTYAA |
|
|
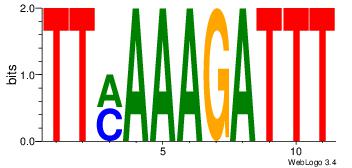 |
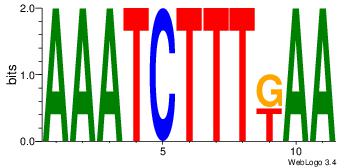 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0632716 |
Alignment:
DDTATTATTTDH
--TTTTAT----
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0689103 |
Alignment:
TWSTTTWAWTTTWT
---TTTTAT-----
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 32 |
Motif name: ArntAhr |
| Original motif Consensus sequence: YGCGTG |
Reverse complement motif Consensus sequence: CACGCM |
|
|
 |
 |
Best Matches for Significant Motif ID 32 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
ArntAhr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0 |
Alignment:
YGCGTG
YGCGTG
| Original motif Consensus sequence: YGCGTG |
Reverse complement motif Consensus sequence: CACGCM |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
47 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0340278 |
Alignment:
YCGCCCACGCH
-----CACGCM
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
23 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0340278 |
Alignment:
HGCGTGGGCGK
YGCGTG-----
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
29 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0452724 |
Alignment:
VBACGTGV
-YGCGTG-
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0452724 |
Alignment:
VBACGTGV
-YGCGTG-
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
57 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0718227 |
Alignment:
GCCACGTGSD
--YGCGTG--
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
33 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0718227 |
Alignment:
GCCACGTGSD
--YGCGTG--
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0736681 |
Alignment:
HVGCCCCGCCCCBB
----CACGCM----
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
34 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0739492 |
Alignment:
DCCACGTGCV
--YGCGTG--
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
58 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0739492 |
Alignment:
DCCACGTGCV
--YGCGTG--
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 56 |
Motif name: ArntAhr |
| Original motif Consensus sequence: YGCGTG |
Reverse complement motif Consensus sequence: CACGCM |
|
|
 |
 |
Best Matches for Significant Motif ID 56 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
32 |
| Motif name: |
ArntAhr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0 |
Alignment:
YGCGTG
YGCGTG
| Original motif Consensus sequence: YGCGTG |
Reverse complement motif Consensus sequence: CACGCM |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
47 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0340278 |
Alignment:
YCGCCCACGCH
-----CACGCM
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
23 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0340278 |
Alignment:
HGCGTGGGCGK
YGCGTG-----
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0452724 |
Alignment:
VBACGTGV
-YGCGTG-
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
29 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0452724 |
Alignment:
VBACGTGV
-YGCGTG-
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
33 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0718227 |
Alignment:
HSCACGTGGC
--CACGCM--
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
57 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0718227 |
Alignment:
GCCACGTGSD
--YGCGTG--
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0736681 |
Alignment:
BBGGGGCGGGGCVD
----YGCGTG----
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
58 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0739492 |
Alignment:
DCCACGTGCV
--YGCGTG--
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
34 |
| Motif name: |
Myc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0739492 |
Alignment:
VGCACGTGGH
--CACGCM--
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 10 |
Motif name: Motif 10 |
| Original motif Consensus sequence: TTTCATAAWT |
Reverse complement motif Consensus sequence: AWTTATGAAA |
|
|
 |
 |
Best Matches for Significant Motif ID 10 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.01875 |
Alignment:
ATTTWATGAAA
-AWTTATGAAA
| Original motif Consensus sequence: ATTTWATGAAA |
Reverse complement motif Consensus sequence: TTTCATWAAAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0375 |
Alignment:
AATTYDGAARTAWW
----AWTTATGAAA
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.04375 |
Alignment:
TGATTTTAWKATTT
-TTTCATAAWT---
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.05625 |
Alignment:
AWTAAATAYAATTT
----AWTTATGAAA
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
17 |
| Motif name: |
Motif 17 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.05625 |
Alignment:
ATTCATTTTT
TTTCATAAWT
| Original motif Consensus sequence: AAAAATGAAT |
Reverse complement motif Consensus sequence: ATTCATTTTT |
|
|
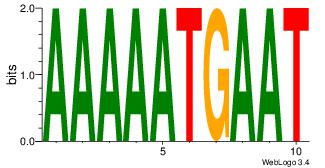 |
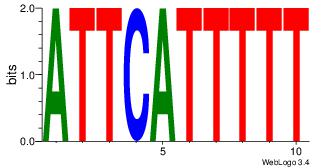 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0604166 |
Alignment:
TTTDAWATATHATT
----TTTCATAAWT
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.06875 |
Alignment:
TTTTATTGTYAT
TTTCATAAWT--
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0706731 |
Alignment:
AWAAAWTWAAASWA
----AWTTATGAAA
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
Motif 18 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.08125 |
Alignment:
TTAGWTWATAA
TTTCATAAWT-
| Original motif Consensus sequence: TTAGWTWATAA |
Reverse complement motif Consensus sequence: TTATWAWCTAA |
|
|
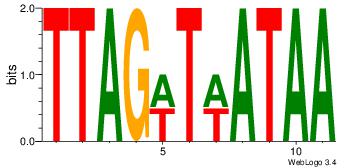 |
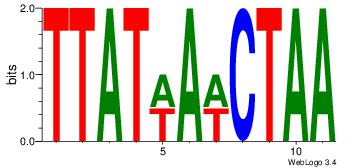 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.08125 |
Alignment:
TWTAATCTAWGAA
TTTCATAAWT---
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 1 |
Motif name: Motif 1 |
| Original motif Consensus sequence: GGCGGGGC |
Reverse complement motif Consensus sequence: GCCCCGCC |
|
|
 |
 |
Best Matches for Significant Motif ID 1 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0 |
Alignment:
HVGCCCCGCCCCBB
--GCCCCGCC----
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.00250079 |
Alignment:
BCCGCCCCGCCCCBB
---GCCCCGCC----
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0149958 |
Alignment:
MSGGGGCGGGGYSG
----GGCGGGGC--
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0349983 |
Alignment:
DGGGYGKGGC
--GGCGGGGC
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
27 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0349983 |
Alignment:
DGGGYGKGGC
--GGCGGGGC
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0708224 |
Alignment:
VAGGCCBBGGCVBB
---GCCCCGCC---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0708224 |
Alignment:
VAGGCCBBGGCVBB
---GCCCCGCC---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0853847 |
Alignment:
DGVBCABTGDWGCGKRRCSR
----------GGCGGGGC--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0853847 |
Alignment:
DGVBCABTGDWGCGKRRCSR
----------GGCGGGGC--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
23 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0878731 |
Alignment:
HGCGTGGGCGK
GGCGGGGC---
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
47 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0878731 |
Alignment:
HGCGTGGGCGK
GGCGGGGC---
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 29 |
Motif name: HIF1AARNT |
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
Best Matches for Significant Motif ID 29 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0 |
Alignment:
VBACGTGV
VBACGTGV
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
57 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0575825 |
Alignment:
GCCACGTGSD
-VBACGTGV-
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
33 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0575825 |
Alignment:
GCCACGTGSD
-VBACGTGV-
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
58 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0597215 |
Alignment:
DCCACGTGCV
-VBACGTGV-
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
34 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0597215 |
Alignment:
DCCACGTGCV
-VBACGTGV-
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0748197 |
Alignment:
BCACCTGCABC
VBACGTGV---
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0767561 |
Alignment:
SGGTCACGTGACCS
---VCACGTBV---
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0829928 |
Alignment:
GCGGACGTTV
--VCACGTBV
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
26 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0829928 |
Alignment:
GCGGACGTTV
--VCACGTBV
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
8 |
| Similarity score: |
0.100962 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-----VCACGTBV-------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 53 |
Motif name: HIF1AARNT |
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
Best Matches for Significant Motif ID 53 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
29 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0 |
Alignment:
VBACGTGV
VBACGTGV
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
57 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0575825 |
Alignment:
GCCACGTGSD
-VBACGTGV-
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
33 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0575825 |
Alignment:
GCCACGTGSD
-VBACGTGV-
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
58 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0597215 |
Alignment:
DCCACGTGCV
-VBACGTGV-
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
34 |
| Motif name: |
Myc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0597215 |
Alignment:
VGCACGTGGH
-VCACGTBV-
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0748197 |
Alignment:
GBTGCAGGTGB
---VCACGTBV
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0767561 |
Alignment:
SGGTCACGTGACCS
---VBACGTGV---
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
26 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0829928 |
Alignment:
GCGGACGTTV
--VCACGTBV
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0829928 |
Alignment:
GCGGACGTTV
--VCACGTBV
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
8 |
| Similarity score: |
0.100962 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-----VCACGTBV-------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
Best Matches for Each Motif (Highest to Lowest)
| Dataset #: 1 |
Motif ID: 1 |
Motif name: Motif 1 |
| Original motif Consensus sequence: GGCGGGGC |
Reverse complement motif Consensus sequence: GCCCCGCC |
|
|
 |
 |
Best Matches for Motif ID 1 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0 |
Alignment:
HVGCCCCGCCCCBB
--GCCCCGCC----
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.00250079 |
Alignment:
BCCGCCCCGCCCCBB
---GCCCCGCC----
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0149958 |
Alignment:
MSGGGGCGGGGYSG
----GGCGGGGC--
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0349983 |
Alignment:
DGGGYGKGGC
--GGCGGGGC
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
27 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0349983 |
Alignment:
DGGGYGKGGC
--GGCGGGGC
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0708224 |
Alignment:
VAGGCCBBGGCVBB
---GCCCCGCC---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0708224 |
Alignment:
VAGGCCBBGGCVBB
---GCCCCGCC---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0853847 |
Alignment:
DGVBCABTGDWGCGKRRCSR
----------GGCGGGGC--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0853847 |
Alignment:
DGVBCABTGDWGCGKRRCSR
----------GGCGGGGC--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
23 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0878731 |
Alignment:
HGCGTGGGCGK
GGCGGGGC---
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
47 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0878731 |
Alignment:
HGCGTGGGCGK
GGCGGGGC---
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 2 |
Motif name: Motif 2 |
| Original motif Consensus sequence: RGRAGARRGARRAR |
Reverse complement motif Consensus sequence: MTMMTCMMTCTKCK |
|
|
 |
 |
Best Matches for Motif ID 2 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
14 |
| Similarity score: |
0.0197704 |
Alignment:
MSGKKRCGCWDCABTGBBCD
------MTMMTCMMTCTKCK
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
14 |
| Similarity score: |
0.0197704 |
Alignment:
MSGKKRCGCWDCABTGBBCD
------MTMMTCMMTCTKCK
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.547476 |
Alignment:
AWAAAWTWAAASWA-
-RGRAGARRGARRAR
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
1.54373 |
Alignment:
---MSGGGGCGGGGYSG
RGRAGARRGARRAR---
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
1.54718 |
Alignment:
BBGGGGCGGGGCVD---
---RGRAGARRGARRAR
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
24 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
2.0297 |
Alignment:
----GGGGGYGGGG
RGRAGARRGARRAR
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
48 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
2.0297 |
Alignment:
----GGGGGYGGGG
RGRAGARRGARRAR
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
45 |
| Motif name: |
wbgTAAATAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
2.03935 |
Alignment:
----DHTATTTACBD
MTMMTCMMTCTKCK-
| Original motif Consensus sequence: DBGTAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTACBD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
2.04042 |
Alignment:
----GGGGCCCAAGGGGG
RGRAGARRGARRAR----
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
2.04042 |
Alignment:
----GGGGCCCAAGGGGG
RGRAGARRGARRAR----
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 3 |
Motif name: Motif 3 |
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
Best Matches for Motif ID 3 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0394231 |
Alignment:
TGATTTTAWKATTT
TWSTTTWAWTTTWT
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0449176 |
Alignment:
AAATTKTATTTAWT
TWSTTTWAWTTTWT
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0594551 |
Alignment:
TTTDAWATATHATT
AWAAAWTWAAASWA
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.067239 |
Alignment:
WWTAKTTCDKAATT
TWSTTTWAWTTTWT
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.564853 |
Alignment:
-RGRAGARRGARRAR
AWAAAWTWAAASWA-
| Original motif Consensus sequence: RGRAGARRGARRAR |
Reverse complement motif Consensus sequence: MTMMTCMMTCTKCK |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.04022 |
Alignment:
HDAAATAATADD--
AWAAAWTWAAASWA
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.04989 |
Alignment:
--HDAAATAATAHW
AWAAAWTWAAASWA
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.05785 |
Alignment:
ATMACAATAAAA--
AWAAAWTWAAASWA
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
45 |
| Motif name: |
wbgTAAATAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.55185 |
Alignment:
---DBGTAAATAHD
AWAAAWTWAAASWA
| Original motif Consensus sequence: DBGTAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTACBD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.55323 |
Alignment:
---AWGKAAWTTTT
TWSTTTWAWTTTWT
| Original motif Consensus sequence: AAAAWTTRCWT |
Reverse complement motif Consensus sequence: AWGKAAWTTTT |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 4 |
Motif name: Motif 4 |
| Original motif Consensus sequence: AAAAWTTRCWT |
Reverse complement motif Consensus sequence: AWGKAAWTTTT |
|
|
 |
 |
Best Matches for Motif ID 4 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0786484 |
Alignment:
AAATTKTATTTAWT
AAAAWTTRCWT---
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0820375 |
Alignment:
AATHATATWTHAAA
AWGKAAWTTTT---
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.083433 |
Alignment:
TGATTTTAWKATTT
---AWGKAAWTTTT
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0852273 |
Alignment:
ACAAWTRATTTTGA
AAAAWTTRCWT---
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
45 |
| Motif name: |
wbgTAAATAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.086338 |
Alignment:
DHTATTTACBD
AAAAWTTRCWT
| Original motif Consensus sequence: DBGTAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTACBD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0867914 |
Alignment:
TWSTTTWAWTTTWT
--AWGKAAWTTTT-
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
Motif 20 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0941986 |
Alignment:
AAATCTTTYAA
AAAAWTTRCWT
| Original motif Consensus sequence: TTMAAAGATTT |
Reverse complement motif Consensus sequence: AAATCTTTYAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.097632 |
Alignment:
HDAAATAATADD
AAAAWTTRCWT-
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.102771 |
Alignment:
HDAAATAATAHW
AAAAWTTRCWT-
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.103618 |
Alignment:
AATTYDGAARTAWW
---AWGKAAWTTTT
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 5 |
Motif name: Motif 5 |
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
Best Matches for Motif ID 5 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.101648 |
Alignment:
TWSTTTWAWTTTWT
AAATTKTATTTAWT
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.107143 |
Alignment:
TGATTTTAWKATTT
AAATTKTATTTAWT
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.111607 |
Alignment:
AATTYDGAARTAWW
AAATTKTATTTAWT
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.125 |
Alignment:
ACAAWTRATTTTGA
AAATTKTATTTAWT
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.596154 |
Alignment:
TTTDAWATATHATT-
-AWTAAATAYAATTT
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
1.125 |
Alignment:
--TTCWTAGATTAWA
AWTAAATAYAATTT-
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.125 |
Alignment:
--TTTTATTGTYAT
AAATTKTATTTAWT
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.57955 |
Alignment:
---TTTCATWAAAT
AAATTKTATTTAWT
| Original motif Consensus sequence: ATTTWATGAAA |
Reverse complement motif Consensus sequence: TTTCATWAAAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.5951 |
Alignment:
AWGKAAWTTTT---
AWTAAATAYAATTT
| Original motif Consensus sequence: AAAAWTTRCWT |
Reverse complement motif Consensus sequence: AWGKAAWTTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
Motif 20 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.61364 |
Alignment:
AAATCTTTYAA---
AAATTKTATTTAWT
| Original motif Consensus sequence: TTMAAAGATTT |
Reverse complement motif Consensus sequence: AAATCTTTYAA |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 6 |
Motif name: Motif 6 |
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
Best Matches for Motif ID 6 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0741071 |
Alignment:
AWTAAATAYAATTT
WWTAKTTCDKAATT
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0763393 |
Alignment:
TGATTTTAWKATTT
WWTAKTTCDKAATT
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0852679 |
Alignment:
TTTDAWATATHATT
WWTAKTTCDKAATT
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0864698 |
Alignment:
TWSTTTWAWTTTWT
WWTAKTTCDKAATT
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
1.06667 |
Alignment:
--TTCWTAGATTAWA
AATTYDGAARTAWW-
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.07587 |
Alignment:
WHTATTATTTDH--
WWTAKTTCDKAATT
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.07757 |
Alignment:
DDTATTATTTDH--
WWTAKTTCDKAATT
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.08229 |
Alignment:
TTTTATTGTYAT--
WWTAKTTCDKAATT
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
1.55625 |
Alignment:
ACAAWTRATTTTGA---
---WWTAKTTCDKAATT
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
45 |
| Motif name: |
wbgTAAATAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.55706 |
Alignment:
---DBGTAAATAHD
AATTYDGAARTAWW
| Original motif Consensus sequence: DBGTAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTACBD |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 7 |
Motif name: Motif 7 |
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
Best Matches for Motif ID 7 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0 |
Alignment:
HVGCCCCGCCCCBB
CSKCCCCGCCCCSY
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0012818 |
Alignment:
BCCGCCCCGCCCCBB
-CSKCCCCGCCCCSY
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
14 |
| Similarity score: |
0.0662825 |
Alignment:
DGVBCABTGDWGCGKRRCSR
------MSGGGGCGGGGYSG
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
14 |
| Similarity score: |
0.0662825 |
Alignment:
DGVBCABTGDWGCGKRRCSR
------MSGGGGCGGGGYSG
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0912118 |
Alignment:
VAGGCCBBGGCVBB
CSKCCCCGCCCCSY
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0912118 |
Alignment:
VAGGCCBBGGCVBB
CSKCCCCGCCCCSY
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0948838 |
Alignment:
SGGTCACGTGACCS
MSGGGGCGGGGYSG
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.590412 |
Alignment:
CCCCCTTGGGCCCC-
-CSKCCCCGCCCCSY
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.590412 |
Alignment:
CCCCCTTGGGCCCC-
-CSKCCCCGCCCCSY
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
23 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.58474 |
Alignment:
YCGCCCACGCH---
CSKCCCCGCCCCSY
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 8 |
Motif name: Motif 8 |
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
Best Matches for Motif ID 8 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0961539 |
Alignment:
TWSTTTWAWTTTWT
TGATTTTAWKATTT
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.107143 |
Alignment:
AAATTKTATTTAWT
TGATTTTAWKATTT
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.113839 |
Alignment:
WWTAKTTCDKAATT
TGATTTTAWKATTT
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.625 |
Alignment:
-AATHATATWTHAAA
AAATRWTAAAATCA-
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.10417 |
Alignment:
--ATMACAATAAAA
AAATRWTAAAATCA
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
1.10417 |
Alignment:
TWTAATCTAWGAA--
-TGATTTTAWKATTT
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
1.125 |
Alignment:
--TCAAAATKAWTTGT
TGATTTTAWKATTT--
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
Motif 20 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.56818 |
Alignment:
---TTMAAAGATTT
TGATTTTAWKATTT
| Original motif Consensus sequence: TTMAAAGATTT |
Reverse complement motif Consensus sequence: AAATCTTTYAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.59091 |
Alignment:
---TTTCATWAAAT
TGATTTTAWKATTT
| Original motif Consensus sequence: ATTTWATGAAA |
Reverse complement motif Consensus sequence: TTTCATWAAAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.59988 |
Alignment:
---AWGKAAWTTTT
TGATTTTAWKATTT
| Original motif Consensus sequence: AAAAWTTRCWT |
Reverse complement motif Consensus sequence: AWGKAAWTTTT |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 9 |
Motif name: Motif 9 |
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
Best Matches for Motif ID 9 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0675748 |
Alignment:
TWSTTTWAWTTTWT
AATHATATWTHAAA
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0741568 |
Alignment:
WWTAKTTCDKAATT
TTTDAWATATHATT
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.547543 |
Alignment:
-AAATTKTATTTAWT
AATHATATWTHAAA-
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.557158 |
Alignment:
-TTCWTAGATTAWA
AATHATATWTHAAA
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.576389 |
Alignment:
AAATRWTAAAATCA-
-AATHATATWTHAAA
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.03318 |
Alignment:
--HDAAATAATADD
TTTDAWATATHATT
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.03819 |
Alignment:
--HDAAATAATAHW
TTTDAWATATHATT
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
1.05903 |
Alignment:
--TCAAAATKAWTTGT
TTTDAWATATHATT--
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
45 |
| Motif name: |
wbgTAAATAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.54311 |
Alignment:
---DHTATTTACBD
AATHATATWTHAAA
| Original motif Consensus sequence: DBGTAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTACBD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.54987 |
Alignment:
AAAAWTTRCWT---
TTTDAWATATHATT
| Original motif Consensus sequence: AAAAWTTRCWT |
Reverse complement motif Consensus sequence: AWGKAAWTTTT |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 10 |
Motif name: Motif 10 |
| Original motif Consensus sequence: TTTCATAAWT |
Reverse complement motif Consensus sequence: AWTTATGAAA |
|
|
 |
 |
Best Matches for Motif ID 10 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.01875 |
Alignment:
ATTTWATGAAA
-AWTTATGAAA
| Original motif Consensus sequence: ATTTWATGAAA |
Reverse complement motif Consensus sequence: TTTCATWAAAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0375 |
Alignment:
AATTYDGAARTAWW
----AWTTATGAAA
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.04375 |
Alignment:
TGATTTTAWKATTT
-TTTCATAAWT---
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.05625 |
Alignment:
AWTAAATAYAATTT
----AWTTATGAAA
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
17 |
| Motif name: |
Motif 17 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.05625 |
Alignment:
ATTCATTTTT
TTTCATAAWT
| Original motif Consensus sequence: AAAAATGAAT |
Reverse complement motif Consensus sequence: ATTCATTTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0604166 |
Alignment:
TTTDAWATATHATT
----TTTCATAAWT
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.06875 |
Alignment:
TTTTATTGTYAT
TTTCATAAWT--
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0706731 |
Alignment:
AWAAAWTWAAASWA
----AWTTATGAAA
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
Motif 18 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.08125 |
Alignment:
TTAGWTWATAA
TTTCATAAWT-
| Original motif Consensus sequence: TTAGWTWATAA |
Reverse complement motif Consensus sequence: TTATWAWCTAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.08125 |
Alignment:
TWTAATCTAWGAA
TTTCATAAWT---
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 11 |
Motif name: Motif 11 |
| Original motif Consensus sequence: ATTTWATGAAA |
Reverse complement motif Consensus sequence: TTTCATWAAAT |
|
|
 |
 |
Best Matches for Motif ID 11 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0795455 |
Alignment:
AWTAAATAYAATTT
--TTTCATWAAAT-
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0795455 |
Alignment:
AAATRWTAAAATCA
--TTTCATWAAAT-
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
Motif 18 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
TTATWAWCTAA
ATTTWATGAAA
| Original motif Consensus sequence: TTAGWTWATAA |
Reverse complement motif Consensus sequence: TTATWAWCTAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
TTTTATTGTYAT
ATTTWATGAAA-
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.109848 |
Alignment:
AATHATATWTHAAA
---ATTTWATGAAA
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
TTCWTAGATTAWA
--ATTTWATGAAA
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
AATTYDGAARTAWW
-ATTTWATGAAA--
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
TWSTTTWAWTTTWT
---TTTCATWAAAT
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.123737 |
Alignment:
HDAAATAATADD
TTTCATWAAAT-
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.55 |
Alignment:
-TTTCATAAWT
TTTCATWAAAT
| Original motif Consensus sequence: TTTCATAAWT |
Reverse complement motif Consensus sequence: AWTTATGAAA |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 12 |
Motif name: Motif 12 |
| Original motif Consensus sequence: AAAACAAA |
Reverse complement motif Consensus sequence: TTTGTTTT |
|
|
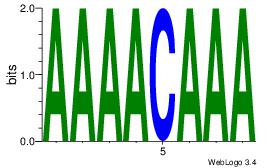 |
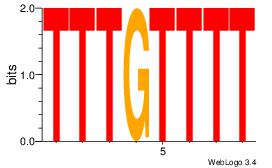 |
Best Matches for Motif ID 12 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0625 |
Alignment:
ATMACAATAAAA
---AAAACAAA-
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
17 |
| Motif name: |
Motif 17 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0625 |
Alignment:
AAAAATGAAT
-AAAACAAA-
| Original motif Consensus sequence: AAAAATGAAT |
Reverse complement motif Consensus sequence: ATTCATTTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
AAATTKTATTTAWT
--TTTGTTTT----
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
ACAAWTRATTTTGA
----TTTGTTTT--
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
14 |
| Motif name: |
Motif 14 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
AWAAATAA
AAAACAAA
| Original motif Consensus sequence: AWAAATAA |
Reverse complement motif Consensus sequence: TTATTTWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0805288 |
Alignment:
AWAAAWTWAAASWA
--AAAACAAA----
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
Motif 18 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.09375 |
Alignment:
TTAGWTWATAA
TTTGTTTT---
| Original motif Consensus sequence: TTAGWTWATAA |
Reverse complement motif Consensus sequence: TTATWAWCTAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.09375 |
Alignment:
AAATRWTAAAATCA
--AAAACAAA----
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.101562 |
Alignment:
WWTAKTTCDKAATT
-TTTGTTTT-----
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.103618 |
Alignment:
AAAAWTTRCWT
AAAACAAA---
| Original motif Consensus sequence: AAAAWTTRCWT |
Reverse complement motif Consensus sequence: AWGKAAWTTTT |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 13 |
Motif name: Motif 13 |
| Original motif Consensus sequence: AAAGATTT |
Reverse complement motif Consensus sequence: AAATCTTT |
|
|
 |
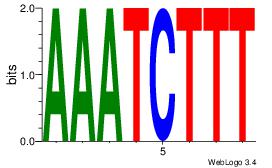 |
Best Matches for Motif ID 13 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
Motif 20 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0 |
Alignment:
TTMAAAGATTT
---AAAGATTT
| Original motif Consensus sequence: TTMAAAGATTT |
Reverse complement motif Consensus sequence: AAATCTTTYAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0625 |
Alignment:
ACAAWTRATTTTGA
---AAAGATTT---
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.0625 |
Alignment:
AAATRWTAAAATCA
------AAATCTTT
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
TTCWTAGATTAWA
---AAAGATTT--
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
AAATTKTATTTAWT
AAATCTTT------
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0822369 |
Alignment:
AWGKAAWTTTT
---AAATCTTT
| Original motif Consensus sequence: AAAAWTTRCWT |
Reverse complement motif Consensus sequence: AWGKAAWTTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
17 |
| Motif name: |
Motif 17 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.09375 |
Alignment:
AAAAATGAAT
AAAGATTT--
| Original motif Consensus sequence: AAAAATGAAT |
Reverse complement motif Consensus sequence: ATTCATTTTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
44 |
| Motif name: |
dhACATTCTkh |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.104167 |
Alignment:
HCAGAATGTHD
---AAATCTTT
| Original motif Consensus sequence: DHACATTCTGH |
Reverse complement motif Consensus sequence: HCAGAATGTHD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
14 |
| Motif name: |
Motif 14 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.109375 |
Alignment:
AWAAATAA
AAAGATTT
| Original motif Consensus sequence: AWAAATAA |
Reverse complement motif Consensus sequence: TTATTTWT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.110417 |
Alignment:
HDAAATAATAHW
--AAATCTTT--
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 14 |
Motif name: Motif 14 |
| Original motif Consensus sequence: AWAAATAA |
Reverse complement motif Consensus sequence: TTATTTWT |
|
|
 |
 |
Best Matches for Motif ID 14 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
17 |
| Motif name: |
Motif 17 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.046875 |
Alignment:
ATTCATTTTT
--TTATTTWT
| Original motif Consensus sequence: AAAAATGAAT |
Reverse complement motif Consensus sequence: ATTCATTTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.046875 |
Alignment:
AAATTKTATTTAWT
-----TTATTTWT-
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0625 |
Alignment:
ATMACAATAAAA
--AWAAATAA--
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0625 |
Alignment:
TCAAAATKAWTTGT
------TTATTTWT
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0636574 |
Alignment:
DDTATTATTTDH
----TTATTTWT
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0697917 |
Alignment:
WHTATTATTTDH
----TTATTTWT
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
TGATTTTAWKATTT
TTATTTWT------
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
Motif 18 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
TTAGWTWATAA
TTATTTWT---
| Original motif Consensus sequence: TTAGWTWATAA |
Reverse complement motif Consensus sequence: TTATWAWCTAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Motif 12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
AAAACAAA
AWAAATAA
| Original motif Consensus sequence: AAAACAAA |
Reverse complement motif Consensus sequence: TTTGTTTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
45 |
| Motif name: |
wbgTAAATAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0825893 |
Alignment:
DBGTAAATAHD
--AWAAATAA-
| Original motif Consensus sequence: DBGTAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTACBD |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 15 |
Motif name: Motif 15 |
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
Best Matches for Motif ID 15 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.104167 |
Alignment:
TGATTTTAWKATTT
TTTTATTGTYAT--
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.114583 |
Alignment:
AWTAAATAYAATTT
-ATMACAATAAAA-
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.114583 |
Alignment:
AWAAAWTWAAASWA
ATMACAATAAAA--
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.119792 |
Alignment:
AATTYDGAARTAWW
--ATMACAATAAAA
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.121528 |
Alignment:
AATHATATWTHAAA
-ATMACAATAAAA-
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.590909 |
Alignment:
-TCAAAATKAWTTGT
ATMACAATAAAA---
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.602273 |
Alignment:
-ATTTWATGAAA
TTTTATTGTYAT
| Original motif Consensus sequence: ATTTWATGAAA |
Reverse complement motif Consensus sequence: TTTCATWAAAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.613636 |
Alignment:
-TWTAATCTAWGAA
TTTTATTGTYAT--
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
Motif 18 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.613636 |
Alignment:
TTAGWTWATAA-
TTTTATTGTYAT
| Original motif Consensus sequence: TTAGWTWATAA |
Reverse complement motif Consensus sequence: TTATWAWCTAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.613636 |
Alignment:
-WHTATTATTTDH
TTTTATTGTYAT-
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 16 |
Motif name: Motif 16 |
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
Best Matches for Motif ID 16 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.1 |
Alignment:
AWTAAATAYAATTT
TCAAAATKAWTTGT
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
1.08264 |
Alignment:
--AATHATATWTHAAA
ACAAWTRATTTTGA--
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.08403 |
Alignment:
--WHTATTATTTDH
TCAAAATKAWTTGT
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.08881 |
Alignment:
--DDTATTATTTDH
TCAAAATKAWTTGT
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
1.1 |
Alignment:
--AAATRWTAAAATCA
ACAAWTRATTTTGA--
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
1.56591 |
Alignment:
TTTTATTGTYAT---
-ACAAWTRATTTTGA
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
1.56875 |
Alignment:
---AATTYDGAARTAWW
TCAAAATKAWTTGT---
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.57667 |
Alignment:
AAAAWTTRCWT---
ACAAWTRATTTTGA
| Original motif Consensus sequence: AAAAWTTRCWT |
Reverse complement motif Consensus sequence: AWGKAAWTTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
Motif 20 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.57727 |
Alignment:
TTMAAAGATTT---
ACAAWTRATTTTGA
| Original motif Consensus sequence: TTMAAAGATTT |
Reverse complement motif Consensus sequence: AAATCTTTYAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
44 |
| Motif name: |
dhACATTCTkh |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.57727 |
Alignment:
HCAGAATGTHD---
TCAAAATKAWTTGT
| Original motif Consensus sequence: DHACATTCTGH |
Reverse complement motif Consensus sequence: HCAGAATGTHD |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 17 |
Motif name: Motif 17 |
| Original motif Consensus sequence: AAAAATGAAT |
Reverse complement motif Consensus sequence: ATTCATTTTT |
|
|
 |
 |
Best Matches for Motif ID 17 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.05 |
Alignment:
ACAAWTRATTTTGA
-ATTCATTTTT---
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0875 |
Alignment:
ATMACAATAAAA
--AAAAATGAAT
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0875 |
Alignment:
AWTTATGAAA
AAAAATGAAT
| Original motif Consensus sequence: TTTCATAAWT |
Reverse complement motif Consensus sequence: AWTTATGAAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0875 |
Alignment:
AWTAAATAYAATTT
---AAAAATGAAT-
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0947369 |
Alignment:
AWGKAAWTTTT
-ATTCATTTTT
| Original motif Consensus sequence: AAAAWTTRCWT |
Reverse complement motif Consensus sequence: AWGKAAWTTTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
44 |
| Motif name: |
dhACATTCTkh |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.1 |
Alignment:
DHACATTCTGH
ATTCATTTTT-
| Original motif Consensus sequence: DHACATTCTGH |
Reverse complement motif Consensus sequence: HCAGAATGTHD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.1 |
Alignment:
TGATTTTAWKATTT
---ATTCATTTTT-
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.104808 |
Alignment:
TWSTTTWAWTTTWT
----ATTCATTTTT
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.10625 |
Alignment:
TTTDAWATATHATT
AAAAATGAAT----
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
Motif 20 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.1125 |
Alignment:
AAATCTTTYAA
-ATTCATTTTT
| Original motif Consensus sequence: TTMAAAGATTT |
Reverse complement motif Consensus sequence: AAATCTTTYAA |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 18 |
Motif name: Motif 18 |
| Original motif Consensus sequence: TTAGWTWATAA |
Reverse complement motif Consensus sequence: TTATWAWCTAA |
|
|
 |
 |
Best Matches for Motif ID 18 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.09375 |
Alignment:
AATTYDGAARTAWW
-TTATWAWCTAA--
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
AAATRWTAAAATCA
TTATWAWCTAA---
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
TTTCATWAAAT
TTAGWTWATAA
| Original motif Consensus sequence: ATTTWATGAAA |
Reverse complement motif Consensus sequence: TTTCATWAAAT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
WHTATTATTTDH
-TTATWAWCTAA
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.104798 |
Alignment:
DDTATTATTTDH
-TTATWAWCTAA
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.109848 |
Alignment:
TTTDAWATATHATT
-TTAGWTWATAA--
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
TWTAATCTAWGAA
--TTAGWTWATAA
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
ATMACAATAAAA
-TTAGWTWATAA
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
AAATTKTATTTAWT
--TTATWAWCTAA-
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.118881 |
Alignment:
AWAAAWTWAAASWA
-TTAGWTWATAA--
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 19 |
Motif name: Motif 19 |
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
Best Matches for Motif ID 19 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.0641025 |
Alignment:
AATHATATWTHAAA
-TTCWTAGATTAWA
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.5625 |
Alignment:
AATTYDGAARTAWW-
--TTCWTAGATTAWA
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.5625 |
Alignment:
-TGATTTTAWKATTT
TWTAATCTAWGAA--
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.577083 |
Alignment:
WHTATTATTTDH-
TWTAATCTAWGAA
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.58179 |
Alignment:
DDTATTATTTDH-
TWTAATCTAWGAA
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.583333 |
Alignment:
AWTAAATAYAATTT-
--TTCWTAGATTAWA
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
Motif 20 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.02652 |
Alignment:
TTMAAAGATTT--
TTCWTAGATTAWA
| Original motif Consensus sequence: TTMAAAGATTT |
Reverse complement motif Consensus sequence: AAATCTTTYAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.07197 |
Alignment:
TTTCATWAAAT--
TWTAATCTAWGAA
| Original motif Consensus sequence: ATTTWATGAAA |
Reverse complement motif Consensus sequence: TTTCATWAAAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
Motif 18 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.07197 |
Alignment:
--TTATWAWCTAA
TTCWTAGATTAWA
| Original motif Consensus sequence: TTAGWTWATAA |
Reverse complement motif Consensus sequence: TTATWAWCTAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
1.07197 |
Alignment:
--ATMACAATAAAA
TTCWTAGATTAWA-
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 20 |
Motif name: Motif 20 |
| Original motif Consensus sequence: TTMAAAGATTT |
Reverse complement motif Consensus sequence: AAATCTTTYAA |
|
|
 |
 |
Best Matches for Motif ID 20 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0681818 |
Alignment:
TTCWTAGATTAWA
TTMAAAGATTT--
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0681818 |
Alignment:
AAATRWTAAAATCA
---AAATCTTTYAA
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
ACAAWTRATTTTGA
-AAATCTTTYAA--
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
AAATTKTATTTAWT
-AAATCTTTYAA--
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.106061 |
Alignment:
TTTDAWATATHATT
--TTMAAAGATTT-
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.110646 |
Alignment:
AWGKAAWTTTT
TTMAAAGATTT
| Original motif Consensus sequence: AAAAWTTRCWT |
Reverse complement motif Consensus sequence: AWGKAAWTTTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.124242 |
Alignment:
WHTATTATTTDH
-AAATCTTTYAA
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.125 |
Alignment:
ATMACAATAAAA
-AAATCTTTYAA
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.125 |
Alignment:
WWTAKTTCDKAATT
-AAATCTTTYAA--
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.6 |
Alignment:
ATTTWATGAAA-
-AAATCTTTYAA
| Original motif Consensus sequence: ATTTWATGAAA |
Reverse complement motif Consensus sequence: TTTCATWAAAT |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 21 |
Motif name: Motif 21 |
| Original motif Consensus sequence: ATAAAA |
Reverse complement motif Consensus sequence: TTTTAT |
|
|
 |
 |
Best Matches for Motif ID 21 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0 |
Alignment:
TTTTATTGTYAT
TTTTAT------
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0208333 |
Alignment:
AAATTKTATTTAWT
---TTTTAT-----
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0208333 |
Alignment:
TTTCATWAAAT
-ATAAAA----
| Original motif Consensus sequence: ATTTWATGAAA |
Reverse complement motif Consensus sequence: TTTCATWAAAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0208333 |
Alignment:
TGATTTTAWKATTT
---TTTTAT-----
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0416667 |
Alignment:
TTTCATAAWT
----TTTTAT
| Original motif Consensus sequence: TTTCATAAWT |
Reverse complement motif Consensus sequence: AWTTATGAAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0625 |
Alignment:
TWTAATCTAWGAA
TTTTAT-------
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
14 |
| Motif name: |
Motif 14 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0625 |
Alignment:
TTATTTWT
--TTTTAT
| Original motif Consensus sequence: AWAAATAA |
Reverse complement motif Consensus sequence: TTATTTWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
Motif 20 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0625 |
Alignment:
AAATCTTTYAA
-----TTTTAT
| Original motif Consensus sequence: TTMAAAGATTT |
Reverse complement motif Consensus sequence: AAATCTTTYAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0632716 |
Alignment:
DDTATTATTTDH
--TTTTAT----
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0689103 |
Alignment:
TWSTTTWAWTTTWT
---TTTTAT-----
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 22 |
Motif name: Zfx |
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
Best Matches for Motif ID 22 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0 |
Alignment:
BBVGCCBVGGCCTV
BBVGCCBVGGCCTV
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.089972 |
Alignment:
BCCGCCCCGCCCCBB
BBVGCCBVGGCCTV-
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
14 |
| Similarity score: |
0.0996894 |
Alignment:
DGVBCABTGDWGCGKRRCSR
--BBVGCCBVGGCCTV----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
14 |
| Similarity score: |
0.0996894 |
Alignment:
DGVBCABTGDWGCGKRRCSR
--BBVGCCBVGGCCTV----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.104023 |
Alignment:
HVGCCCCGCCCCBB
VAGGCCBBGGCVBB
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.118259 |
Alignment:
CCCCCTTGGGCCCC
BBVGCCBVGGCCTV
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.118259 |
Alignment:
CCCCCTTGGGCCCC
BBVGCCBVGGCCTV
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.118483 |
Alignment:
MSGGGGCGGGGYSG
BBVGCCBVGGCCTV
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
59 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.6145 |
Alignment:
CCGGAAGTGVV---
VAGGCCBBGGCVBB
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
35 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.6145 |
Alignment:
CCGGAAGTGVV---
VAGGCCBBGGCVBB
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 23 |
Motif name: Egr1 |
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
Best Matches for Motif ID 23 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
47 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0 |
Alignment:
HGCGTGGGCGK
HGCGTGGGCGK
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
0.107124 |
Alignment:
BCCGCCCCGCCCCBB
YCGCCCACGCH----
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.110516 |
Alignment:
BBVGCCBVGGCCTV
--HGCGTGGGCGK-
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.110516 |
Alignment:
BBVGCCBVGGCCTV
--HGCGTGGGCGK-
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
11 |
| Similarity score: |
0.110606 |
Alignment:
DGVBCABTGDWGCGKRRCSR
----HGCGTGGGCGK-----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
0.110606 |
Alignment:
DGVBCABTGDWGCGKRRCSR
----HGCGTGGGCGK-----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.111149 |
Alignment:
CSKCCCCGCCCCSY
-YCGCCCACGCH--
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.116531 |
Alignment:
HVGCCCCGCCCCBB
-YCGCCCACGCH--
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
24 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.606429 |
Alignment:
GGGGGYGGGG-
HGCGTGGGCGK
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
48 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.606429 |
Alignment:
GGGGGYGGGG-
HGCGTGGGCGK
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 24 |
Motif name: SP1 |
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
Best Matches for Motif ID 24 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
48 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0 |
Alignment:
CCCCKCCCCC
CCCCKCCCCC
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.043504 |
Alignment:
CSKCCCCGCCCCSY
---CCCCKCCCCC-
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0486461 |
Alignment:
HVGCCCCGCCCCBB
---CCCCKCCCCC-
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0517173 |
Alignment:
BCCGCCCCGCCCCBB
----CCCCKCCCCC-
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0930706 |
Alignment:
BBVGCCBVGGCCTV
----GGGGGYGGGG
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0930706 |
Alignment:
VAGGCCBBGGCVBB
----CCCCKCCCCC
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0990873 |
Alignment:
CCCCCTTGGGCCCC
-CCCCKCCCCC---
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0990873 |
Alignment:
CCCCCTTGGGCCCC
-CCCCKCCCCC---
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.102706 |
Alignment:
GCCYCMCCCD
CCCCKCCCCC
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
27 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.102706 |
Alignment:
GCCYCMCCCD
CCCCKCCCCC
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 25 |
Motif name: TFAP2A |
| Original motif Consensus sequence: GCCBBVRGS |
Reverse complement motif Consensus sequence: SCMVBBGGC |
|
|
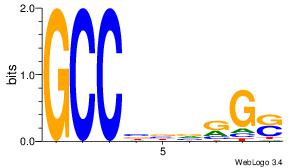 |
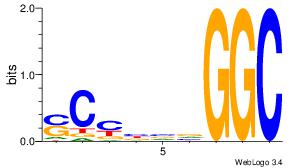 |
Best Matches for Motif ID 25 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
49 |
| Motif name: |
TFAP2A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0 |
Alignment:
GCCBBVRGS
GCCBBVRGS
| Original motif Consensus sequence: GCCBBVRGS |
Reverse complement motif Consensus sequence: SCMVBBGGC |
|
|
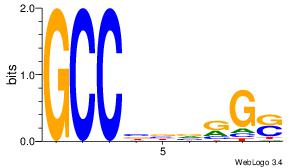 |
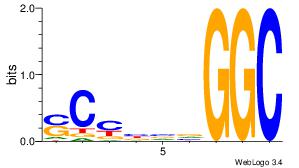 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
9 |
| Similarity score: |
0.0516959 |
Alignment:
BBVGCCBVGGCCTV
---SCMVBBGGC--
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
9 |
| Similarity score: |
0.0516959 |
Alignment:
VAGGCCBBGGCVBB
---GCCBBVRGS--
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
9 |
| Similarity score: |
0.08003 |
Alignment:
GGGGCCCAAGGGGG
---GCCBBVRGS--
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
9 |
| Similarity score: |
0.08003 |
Alignment:
GGGGCCCAAGGGGG
---GCCBBVRGS--
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
57 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.0936515 |
Alignment:
HSCACGTGGC
-SCMVBBGGC
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
33 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.0936515 |
Alignment:
HSCACGTGGC
-SCMVBBGGC
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
9 |
| Similarity score: |
0.0966967 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-------GCCBBVRGS----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
9 |
| Similarity score: |
0.0966967 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-------GCCBBVRGS----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
58 |
| Motif name: |
Myc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.0999167 |
Alignment:
VGCACGTGGH
-SCMVBBGGC
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 26 |
Motif name: MIZF |
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
Best Matches for Motif ID 26 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0 |
Alignment:
BAACGTCCGC
BAACGTCCGC
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
59 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0810799 |
Alignment:
CCGGAAGTGVV
GCGGACGTTV-
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
35 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0810799 |
Alignment:
CCGGAAGTGVV
GCGGACGTTV-
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.112981 |
Alignment:
BCACCTGCABC
BAACGTCCGC-
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
44 |
| Motif name: |
dhACATTCTkh |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.114583 |
Alignment:
DHACATTCTGH
BAACGTCCGC-
| Original motif Consensus sequence: DHACATTCTGH |
Reverse complement motif Consensus sequence: HCAGAATGTHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.115556 |
Alignment:
SGGTCACGTGACCS
-GCGGACGTTV---
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.122872 |
Alignment:
BCCGCCCCGCCCCBB
BAACGTCCGC-----
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
10 |
| Number of overlap: |
10 |
| Similarity score: |
0.122917 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-GCGGACGTTV---------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.122917 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-GCGGACGTTV---------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.123601 |
Alignment:
HVGCCCCGCCCCBB
---BAACGTCCGC-
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 27 |
Motif name: Klf4 |
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
Best Matches for Motif ID 27 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0 |
Alignment:
DGGGYGKGGC
DGGGYGKGGC
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0516385 |
Alignment:
CSKCCCCGCCCCSY
--GCCYCMCCCD--
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0524114 |
Alignment:
BBGGGGCGGGGCVD
--DGGGYGKGGC--
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0558821 |
Alignment:
BBGGGGCGGGGCGGB
--DGGGYGKGGC---
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0987519 |
Alignment:
VAGGCCBBGGCVBB
-DGGGYGKGGC---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0987519 |
Alignment:
BBVGCCBVGGCCTV
-GCCYCMCCCD---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
24 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.102706 |
Alignment:
GGGGGYGGGG
DGGGYGKGGC
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
48 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.102706 |
Alignment:
CCCCKCCCCC
GCCYCMCCCD
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
34 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.105078 |
Alignment:
VGCACGTGGH
DGGGYGKGGC
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
58 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.105078 |
Alignment:
VGCACGTGGH
DGGGYGKGGC
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 28 |
Motif name: E2F1 |
| Original motif Consensus sequence: TTTSGCGC |
Reverse complement motif Consensus sequence: GCGCSAAA |
|
|
 |
 |
Best Matches for Motif ID 28 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
52 |
| Motif name: |
E2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0 |
Alignment:
TTTSGCGC
TTTSGCGC
| Original motif Consensus sequence: TTTSGCGC |
Reverse complement motif Consensus sequence: GCGCSAAA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0982639 |
Alignment:
GGGGCCCAAGGGGG
-GCGCSAAA-----
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0982639 |
Alignment:
GGGGCCCAAGGGGG
-GCGCSAAA-----
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
39 |
| Motif name: |
kCAGCCAATmr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.10386 |
Alignment:
DCAGCCAATVR
-GCGCSAAA--
| Original motif Consensus sequence: DCAGCCAATVR |
Reverse complement motif Consensus sequence: MBATTGGCTGH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
47 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.119792 |
Alignment:
YCGCCCACGCH
GCGCSAAA---
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
23 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.119792 |
Alignment:
YCGCCCACGCH
GCGCSAAA---
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.125 |
Alignment:
ATMACAATAAAA
-GCGCSAAA---
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.125 |
Alignment:
ATTTWATGAAA
-TTTSGCGC--
| Original motif Consensus sequence: ATTTWATGAAA |
Reverse complement motif Consensus sequence: TTTCATWAAAT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
7 |
| Similarity score: |
0.612386 |
Alignment:
-BBGGGGCGGGGCGGB
TTTSGCGC--------
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
7 |
| Similarity score: |
0.616684 |
Alignment:
-BBGGGGCGGGGCVD
TTTSGCGC-------
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 29 |
Motif name: HIF1AARNT |
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
Best Matches for Motif ID 29 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0 |
Alignment:
VBACGTGV
VBACGTGV
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
57 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0575825 |
Alignment:
GCCACGTGSD
-VBACGTGV-
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
33 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0575825 |
Alignment:
GCCACGTGSD
-VBACGTGV-
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
58 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0597215 |
Alignment:
DCCACGTGCV
-VBACGTGV-
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
34 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0597215 |
Alignment:
DCCACGTGCV
-VBACGTGV-
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0748197 |
Alignment:
BCACCTGCABC
VBACGTGV---
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0767561 |
Alignment:
SGGTCACGTGACCS
---VCACGTBV---
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0829928 |
Alignment:
GCGGACGTTV
--VCACGTBV
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
26 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0829928 |
Alignment:
GCGGACGTTV
--VCACGTBV
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
8 |
| Similarity score: |
0.100962 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-----VCACGTBV-------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 30 |
Motif name: PLAG1 |
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
Best Matches for Motif ID 30 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0 |
Alignment:
GGGGCCCAAGGGGG
GGGGCCCAAGGGGG
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.118259 |
Alignment:
VAGGCCBBGGCVBB
GGGGCCCAAGGGGG
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.118259 |
Alignment:
BBVGCCBVGGCCTV
CCCCCTTGGGCCCC
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.120124 |
Alignment:
BCCGCCCCGCCCCBB
CCCCCTTGGGCCCC-
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.124008 |
Alignment:
DGVBCABTGDWGCGKRRCSR
GGGGCCCAAGGGGG------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.124008 |
Alignment:
DGVBCABTGDWGCGKRRCSR
GGGGCCCAAGGGGG------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.617683 |
Alignment:
-MSGGGGCGGGGYSG
GGGGCCCAAGGGGG-
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.624856 |
Alignment:
-HVGCCCCGCCCCBB
CCCCCTTGGGCCCC-
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
48 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
2.1077 |
Alignment:
----CCCCKCCCCC
CCCCCTTGGGCCCC
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
24 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
2.1077 |
Alignment:
----GGGGGYGGGG
GGGGCCCAAGGGGG
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 31 |
Motif name: Pax5 |
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
Best Matches for Motif ID 31 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
20 |
| Similarity score: |
0 |
Alignment:
DGVBCABTGDWGCGKRRCSR
DGVBCABTGDWGCGKRRCSR
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
2.61478 |
Alignment:
BBGGGGCGGGGCGGB-----
MSGKKRCGCWDCABTGBBCD
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.09024 |
Alignment:
------BBGGGGCGGGGCVD
DGVBCABTGDWGCGKRRCSR
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.09355 |
Alignment:
------CSKCCCCGCCCCSY
MSGKKRCGCWDCABTGBBCD
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.09388 |
Alignment:
------MTMMTCMMTCTKCK
MSGKKRCGCWDCABTGBBCD
| Original motif Consensus sequence: RGRAGARRGARRAR |
Reverse complement motif Consensus sequence: MTMMTCMMTCTKCK |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.11062 |
Alignment:
------SGGTCACGTGACCS
DGVBCABTGDWGCGKRRCSR
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.12401 |
Alignment:
GGGGCCCAAGGGGG------
DGVBCABTGDWGCGKRRCSR
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.12401 |
Alignment:
GGGGCCCAAGGGGG------
DGVBCABTGDWGCGKRRCSR
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
3.6245 |
Alignment:
-------VAGGCCBBGGCVBB
DGVBCABTGDWGCGKRRCSR-
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
3.6245 |
Alignment:
-------BBVGCCBVGGCCTV
MSGKKRCGCWDCABTGBBCD-
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 32 |
Motif name: ArntAhr |
| Original motif Consensus sequence: YGCGTG |
Reverse complement motif Consensus sequence: CACGCM |
|
|
 |
 |
Best Matches for Motif ID 32 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
56 |
| Motif name: |
ArntAhr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0 |
Alignment:
YGCGTG
YGCGTG
| Original motif Consensus sequence: YGCGTG |
Reverse complement motif Consensus sequence: CACGCM |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
47 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0340278 |
Alignment:
YCGCCCACGCH
-----CACGCM
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
23 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0340278 |
Alignment:
HGCGTGGGCGK
YGCGTG-----
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
29 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0452724 |
Alignment:
VBACGTGV
-YGCGTG-
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0452724 |
Alignment:
VBACGTGV
-YGCGTG-
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
57 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0718227 |
Alignment:
GCCACGTGSD
--YGCGTG--
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
33 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0718227 |
Alignment:
GCCACGTGSD
--YGCGTG--
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0736681 |
Alignment:
HVGCCCCGCCCCBB
----CACGCM----
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
34 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0739492 |
Alignment:
DCCACGTGCV
--YGCGTG--
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
58 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0739492 |
Alignment:
DCCACGTGCV
--YGCGTG--
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 33 |
Motif name: Mycn |
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
Best Matches for Motif ID 33 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
57 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0 |
Alignment:
HSCACGTGGC
HSCACGTGGC
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
58 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0167031 |
Alignment:
VGCACGTGGH
HSCACGTGGC
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
34 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0167031 |
Alignment:
VGCACGTGGH
HSCACGTGGC
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0795472 |
Alignment:
SGGTCACGTGACCS
--HSCACGTGGC--
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
59 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.105143 |
Alignment:
VVCACTTCCGG
GCCACGTGSD-
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
35 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.105143 |
Alignment:
VVCACTTCCGG
GCCACGTGSD-
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
27 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.10603 |
Alignment:
DGGGYGKGGC
HSCACGTGGC
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.10603 |
Alignment:
DGGGYGKGGC
HSCACGTGGC
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.106697 |
Alignment:
VAGGCCBBGGCVBB
---GCCACGTGSD-
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.106697 |
Alignment:
VAGGCCBBGGCVBB
---GCCACGTGSD-
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 34 |
Motif name: Myc |
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
Best Matches for Motif ID 34 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
58 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0 |
Alignment:
VGCACGTGGH
VGCACGTGGH
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
57 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0167031 |
Alignment:
GCCACGTGSD
DCCACGTGCV
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
33 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0167031 |
Alignment:
GCCACGTGSD
DCCACGTGCV
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0813632 |
Alignment:
SGGTCACGTGACCS
--DCCACGTGCV--
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
59 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.100737 |
Alignment:
VVCACTTCCGG
DCCACGTGCV-
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
35 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.100737 |
Alignment:
VVCACTTCCGG
DCCACGTGCV-
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.105078 |
Alignment:
DGGGYGKGGC
VGCACGTGGH
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
27 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.105078 |
Alignment:
DGGGYGKGGC
VGCACGTGGH
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.109248 |
Alignment:
VAGGCCBBGGCVBB
---DCCACGTGCV-
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.109248 |
Alignment:
VAGGCCBBGGCVBB
---DCCACGTGCV-
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 35 |
Motif name: GABPA |
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
Best Matches for Motif ID 35 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
59 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0 |
Alignment:
CCGGAAGTGVV
CCGGAAGTGVV
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.113389 |
Alignment:
DGVBCABTGDWGCGKRRCSR
-------VVCACTTCCGG--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.113389 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-------CCGGAAGTGVV--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.114496 |
Alignment:
BBVGCCBVGGCCTV
VVCACTTCCGG---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.114496 |
Alignment:
VAGGCCBBGGCVBB
CCGGAAGTGVV---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.117147 |
Alignment:
BCACCTGCABC
VVCACTTCCGG
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
45 |
| Motif name: |
wbgTAAATAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.117976 |
Alignment:
DBGTAAATAHD
CCGGAAGTGVV
| Original motif Consensus sequence: DBGTAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTACBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
44 |
| Motif name: |
dhACATTCTkh |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.118278 |
Alignment:
DHACATTCTGH
VVCACTTCCGG
| Original motif Consensus sequence: DHACATTCTGH |
Reverse complement motif Consensus sequence: HCAGAATGTHD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.120605 |
Alignment:
AATTYDGAARTAWW
---CCGGAAGTGVV
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.124188 |
Alignment:
BCCGCCCCGCCCCBB
---VVCACTTCCGG-
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 36 |
Motif name: csGCCCCGCCCCsc |
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
Best Matches for Motif ID 36 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0 |
Alignment:
BBGGGGCGGGGCGGB
BBGGGGCGGGGCVD-
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0117332 |
Alignment:
CSKCCCCGCCCCSY
HVGCCCCGCCCCBB
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0747029 |
Alignment:
DGVBCABTGDWGCGKRRCSR
------BBGGGGCGGGGCVD
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0747029 |
Alignment:
MSGKKRCGCWDCABTGBBCD
------HVGCCCCGCCCCBB
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0884848 |
Alignment:
BBVGCCBVGGCCTV
BBGGGGCGGGGCVD
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0884848 |
Alignment:
VAGGCCBBGGCVBB
HVGCCCCGCCCCBB
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0971132 |
Alignment:
SGGTCACGTGACCS
HVGCCCCGCCCCBB
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.609319 |
Alignment:
GGGGCCCAAGGGGG-
-BBGGGGCGGGGCVD
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.609319 |
Alignment:
CCCCCTTGGGCCCC-
-HVGCCCCGCCCCBB
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
47 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.60337 |
Alignment:
YCGCCCACGCH---
HVGCCCCGCCCCBB
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 37 |
Motif name: tkAAATAATAtw |
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
Best Matches for Motif ID 37 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0 |
Alignment:
HDAAATAATADD
HDAAATAATAHW
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0522317 |
Alignment:
TWSTTTWAWTTTWT
-WHTATTATTTDH-
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0641975 |
Alignment:
AATHATATWTHAAA
--WHTATTATTTDH
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0704475 |
Alignment:
AAATTKTATTTAWT
-WHTATTATTTDH-
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0864198 |
Alignment:
TCAAAATKAWTTGT
--WHTATTATTTDH
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.09076 |
Alignment:
WWTAKTTCDKAATT
WHTATTATTTDH--
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.096142 |
Alignment:
TWTAATCTAWGAA
WHTATTATTTDH-
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
45 |
| Motif name: |
wbgTAAATAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.560292 |
Alignment:
DHTATTTACBD-
WHTATTATTTDH
| Original motif Consensus sequence: DBGTAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTACBD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
Motif 18 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.579665 |
Alignment:
-TTATWAWCTAA
WHTATTATTTDH
| Original motif Consensus sequence: TTAGWTWATAA |
Reverse complement motif Consensus sequence: TTATWAWCTAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
43 |
| Motif name: |
wsTACwGTAsw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.587493 |
Alignment:
-DBTACWGTAVH
HDAAATAATAHW
| Original motif Consensus sequence: HVTACWGTABD |
Reverse complement motif Consensus sequence: DBTACWGTAVH |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 38 |
Motif name: cccGCCCCGCCCCsb |
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
Best Matches for Motif ID 38 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
0.0992376 |
Alignment:
MSGKKRCGCWDCABTGBBCD
BBGGGGCGGGGCGGB-----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
0.0992376 |
Alignment:
MSGKKRCGCWDCABTGBBCD
BBGGGGCGGGGCGGB-----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.5 |
Alignment:
BBGGGGCGGGGCVD-
BBGGGGCGGGGCGGB
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.513015 |
Alignment:
-CSKCCCCGCCCCSY
BCCGCCCCGCCCCBB
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.574434 |
Alignment:
BBVGCCBVGGCCTV-
BCCGCCCCGCCCCBB
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.574434 |
Alignment:
BBVGCCBVGGCCTV-
BCCGCCCCGCCCCBB
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.601399 |
Alignment:
SGGTCACGTGACCS-
BBGGGGCGGGGCGGB
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.604586 |
Alignment:
GGGGCCCAAGGGGG-
BBGGGGCGGGGCGGB
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.604586 |
Alignment:
CCCCCTTGGGCCCC-
BCCGCCCCGCCCCBB
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
47 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
2.09159 |
Alignment:
YCGCCCACGCH----
BCCGCCCCGCCCCBB
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 39 |
Motif name: kCAGCCAATmr |
| Original motif Consensus sequence: DCAGCCAATVR |
Reverse complement motif Consensus sequence: MBATTGGCTGH |
|
|
 |
 |
Best Matches for Motif ID 39 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0559269 |
Alignment:
WHTATTATTTDH
-MBATTGGCTGH
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
44 |
| Motif name: |
dhACATTCTkh |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0588235 |
Alignment:
HCAGAATGTHD
DCAGCCAATVR
| Original motif Consensus sequence: DHACATTCTGH |
Reverse complement motif Consensus sequence: HCAGAATGTHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0597643 |
Alignment:
DDTATTATTTDH
-MBATTGGCTGH
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0599548 |
Alignment:
BCACCTGCABC
MBATTGGCTGH
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
Motif 18 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0661765 |
Alignment:
TTAGWTWATAA
DCAGCCAATVR
| Original motif Consensus sequence: TTAGWTWATAA |
Reverse complement motif Consensus sequence: TTATWAWCTAA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
59 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.56089 |
Alignment:
-CCGGAAGTGVV
DCAGCCAATVR-
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
35 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.56089 |
Alignment:
-CCGGAAGTGVV
DCAGCCAATVR-
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
45 |
| Motif name: |
wbgTAAATAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.561975 |
Alignment:
DHTATTTACBD-
-MBATTGGCTGH
| Original motif Consensus sequence: DBGTAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTACBD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.563709 |
Alignment:
BBVGCCBVGGCCTV-
----MBATTGGCTGH
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.563709 |
Alignment:
VAGGCCBBGGCVBB-
----DCAGCCAATVR
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 40 |
Motif name: kcACCTGCAgc |
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
Best Matches for Motif ID 40 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
43 |
| Motif name: |
wsTACwGTAsw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0215284 |
Alignment:
DBTACWGTAVH
BCACCTGCABC
| Original motif Consensus sequence: HVTACWGTABD |
Reverse complement motif Consensus sequence: DBTACWGTAVH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
44 |
| Motif name: |
dhACATTCTkh |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0254134 |
Alignment:
DHACATTCTGH
BCACCTGCABC
| Original motif Consensus sequence: DHACATTCTGH |
Reverse complement motif Consensus sequence: HCAGAATGTHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.031338 |
Alignment:
SGGTCACGTGACCS
---BCACCTGCABC
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0402518 |
Alignment:
HVGCCCCGCCCCBB
-BCACCTGCABC--
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0411257 |
Alignment:
CSKCCCCGCCCCSY
-BCACCTGCABC--
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
11 |
| Similarity score: |
0.0420218 |
Alignment:
MSGKKRCGCWDCABTGBBCD
----BCACCTGCABC-----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
0.0420218 |
Alignment:
DGVBCABTGDWGCGKRRCSR
----GBTGCAGGTGB-----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
35 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0423275 |
Alignment:
CCGGAAGTGVV
GBTGCAGGTGB
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
59 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0423275 |
Alignment:
VVCACTTCCGG
BCACCTGCABC
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
39 |
| Motif name: |
kCAGCCAATmr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0439586 |
Alignment:
DCAGCCAATVR
GBTGCAGGTGB
| Original motif Consensus sequence: DCAGCCAATVR |
Reverse complement motif Consensus sequence: MBATTGGCTGH |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 41 |
Motif name: wwAAATAATAtw |
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
Best Matches for Motif ID 41 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0 |
Alignment:
HDAAATAATAHW
HDAAATAATADD
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0536028 |
Alignment:
AWAAAWTWAAASWA
-HDAAATAATADD-
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0591821 |
Alignment:
AATHATATWTHAAA
--DDTATTATTTDH
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0715278 |
Alignment:
AAATTKTATTTAWT
-DDTATTATTTDH-
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0912037 |
Alignment:
TCAAAATKAWTTGT
--DDTATTATTTDH
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0924576 |
Alignment:
WWTAKTTCDKAATT
DDTATTATTTDH--
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0989198 |
Alignment:
TGATTTTAWKATTT
-DDTATTATTTDH-
| Original motif Consensus sequence: AAATRWTAAAATCA |
Reverse complement motif Consensus sequence: TGATTTTAWKATTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.100849 |
Alignment:
TWTAATCTAWGAA
DDTATTATTTDH-
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
45 |
| Motif name: |
wbgTAAATAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.560545 |
Alignment:
DHTATTTACBD-
DDTATTATTTDH
| Original motif Consensus sequence: DBGTAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTACBD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
Motif 18 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.58219 |
Alignment:
-TTATWAWCTAA
DDTATTATTTDH
| Original motif Consensus sequence: TTAGWTWATAA |
Reverse complement motif Consensus sequence: TTATWAWCTAA |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 42 |
Motif name: sSGTCACGTGACSs |
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
Best Matches for Motif ID 42 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
14 |
| Similarity score: |
0.0353836 |
Alignment:
DGVBCABTGDWGCGKRRCSR
------SGGTCACGTGACCS
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
14 |
| Similarity score: |
0.0353836 |
Alignment:
DGVBCABTGDWGCGKRRCSR
------SGGTCACGTGACCS
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0374195 |
Alignment:
BBGGGGCGGGGCVD
SGGTCACGTGACCS
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0417054 |
Alignment:
BCCGCCCCGCCCCBB
SGGTCACGTGACCS-
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0469233 |
Alignment:
MSGGGGCGGGGYSG
SGGTCACGTGACCS
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.53093 |
Alignment:
---BCACCTGCABC
SGGTCACGTGACCS
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
23 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
2.0456 |
Alignment:
----YCGCCCACGCH
SGGTCACGTGACCS-
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
47 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
2.0456 |
Alignment:
----HGCGTGGGCGK
SGGTCACGTGACCS-
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
7 |
| Similarity score: |
3.51604 |
Alignment:
CCCCCTTGGGCCCC-------
-------SGGTCACGTGACCS
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
7 |
| Similarity score: |
3.51604 |
Alignment:
CCCCCTTGGGCCCC-------
-------SGGTCACGTGACCS
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 43 |
Motif name: wsTACwGTAsw |
| Original motif Consensus sequence: HVTACWGTABD |
Reverse complement motif Consensus sequence: DBTACWGTAVH |
|
|
 |
 |
Best Matches for Motif ID 43 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.00144069 |
Alignment:
GBTGCAGGTGB
HVTACWGTABD
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0124158 |
Alignment:
DDTATTATTTDH
-HVTACWGTABD
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0151936 |
Alignment:
WHTATTATTTDH
-HVTACWGTABD
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.016835 |
Alignment:
AATTYDGAARTAWW
HVTACWGTABD---
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
45 |
| Motif name: |
wbgTAAATAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0189995 |
Alignment:
DBGTAAATAHD
DBTACWGTAVH
| Original motif Consensus sequence: DBGTAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTACBD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0212542 |
Alignment:
TTTDAWATATHATT
DBTACWGTAVH---
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
11 |
| Similarity score: |
0.0237795 |
Alignment:
DGVBCABTGDWGCGKRRCSR
-----HVTACWGTABD----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
11 |
| Similarity score: |
0.0237795 |
Alignment:
DGVBCABTGDWGCGKRRCSR
-----HVTACWGTABD----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0275673 |
Alignment:
TTCWTAGATTAWA
DBTACWGTAVH--
| Original motif Consensus sequence: TTCWTAGATTAWA |
Reverse complement motif Consensus sequence: TWTAATCTAWGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.525634 |
Alignment:
-AAAAWTTRCWT
HVTACWGTABD-
| Original motif Consensus sequence: AAAAWTTRCWT |
Reverse complement motif Consensus sequence: AWGKAAWTTTT |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 44 |
Motif name: dhACATTCTkh |
| Original motif Consensus sequence: DHACATTCTGH |
Reverse complement motif Consensus sequence: HCAGAATGTHD |
|
|
 |
 |
Best Matches for Motif ID 44 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0238442 |
Alignment:
BCACCTGCABC
DHACATTCTGH
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0258838 |
Alignment:
TCAAAATKAWTTGT
HCAGAATGTHD---
| Original motif Consensus sequence: ACAAWTRATTTTGA |
Reverse complement motif Consensus sequence: TCAAAATKAWTTGT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0292508 |
Alignment:
DDTATTATTTDH
-HCAGAATGTHD
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0364899 |
Alignment:
WHTATTATTTDH
-HCAGAATGTHD
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0400884 |
Alignment:
AATTYDGAARTAWW
HCAGAATGTHD---
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
39 |
| Motif name: |
kCAGCCAATmr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0412582 |
Alignment:
MBATTGGCTGH
DHACATTCTGH
| Original motif Consensus sequence: DCAGCCAATVR |
Reverse complement motif Consensus sequence: MBATTGGCTGH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
59 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0418896 |
Alignment:
VVCACTTCCGG
DHACATTCTGH
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
35 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0418896 |
Alignment:
CCGGAAGTGVV
HCAGAATGTHD
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0448232 |
Alignment:
TTTTATTGTYAT
HCAGAATGTHD-
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.045689 |
Alignment:
RGRAGARRGARRAR
--HCAGAATGTHD-
| Original motif Consensus sequence: RGRAGARRGARRAR |
Reverse complement motif Consensus sequence: MTMMTCMMTCTKCK |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 45 |
Motif name: wbgTAAATAww |
| Original motif Consensus sequence: DBGTAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTACBD |
|
|
 |
 |
Best Matches for Motif ID 45 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
37 |
| Motif name: |
tkAAATAATAtw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.00892084 |
Alignment:
HDAAATAATAHW
-DBGTAAATAHD
| Original motif Consensus sequence: HDAAATAATAHW |
Reverse complement motif Consensus sequence: WHTATTATTTDH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
41 |
| Motif name: |
wwAAATAATAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.00917337 |
Alignment:
DDTATTATTTDH
DHTATTTACBD-
| Original motif Consensus sequence: HDAAATAATADD |
Reverse complement motif Consensus sequence: DDTATTATTTDH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0177412 |
Alignment:
AATHATATWTHAAA
DHTATTTACBD---
| Original motif Consensus sequence: AATHATATWTHAAA |
Reverse complement motif Consensus sequence: TTTDAWATATHATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0205821 |
Alignment:
AATTYDGAARTAWW
---DBGTAAATAHD
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0288058 |
Alignment:
AAAAWTTRCWT
DHTATTTACBD
| Original motif Consensus sequence: AAAAWTTRCWT |
Reverse complement motif Consensus sequence: AWGKAAWTTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0337252 |
Alignment:
AWAAAWTWAAASWA
--DHTATTTACBD-
| Original motif Consensus sequence: AWAAAWTWAAASWA |
Reverse complement motif Consensus sequence: TWSTTTWAWTTTWT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
43 |
| Motif name: |
wsTACwGTAsw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0399273 |
Alignment:
HVTACWGTABD
DHTATTTACBD
| Original motif Consensus sequence: HVTACWGTABD |
Reverse complement motif Consensus sequence: DBTACWGTAVH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0404685 |
Alignment:
AWTAAATAYAATTT
---DHTATTTACBD
| Original motif Consensus sequence: AWTAAATAYAATTT |
Reverse complement motif Consensus sequence: AAATTKTATTTAWT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
10 |
| Number of overlap: |
11 |
| Similarity score: |
0.0439858 |
Alignment:
DGVBCABTGDWGCGKRRCSR
DBGTAAATAHD---------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0439858 |
Alignment:
DGVBCABTGDWGCGKRRCSR
DBGTAAATAHD---------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 46 |
Motif name: Zfx |
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
Best Matches for Motif ID 46 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0 |
Alignment:
BBVGCCBVGGCCTV
BBVGCCBVGGCCTV
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.089972 |
Alignment:
BCCGCCCCGCCCCBB
BBVGCCBVGGCCTV-
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
14 |
| Similarity score: |
0.0996894 |
Alignment:
DGVBCABTGDWGCGKRRCSR
--BBVGCCBVGGCCTV----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
14 |
| Similarity score: |
0.0996894 |
Alignment:
DGVBCABTGDWGCGKRRCSR
--BBVGCCBVGGCCTV----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.104023 |
Alignment:
BBGGGGCGGGGCVD
BBVGCCBVGGCCTV
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.118259 |
Alignment:
GGGGCCCAAGGGGG
VAGGCCBBGGCVBB
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.118259 |
Alignment:
CCCCCTTGGGCCCC
BBVGCCBVGGCCTV
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.118483 |
Alignment:
MSGGGGCGGGGYSG
BBVGCCBVGGCCTV
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
35 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.6145 |
Alignment:
VVCACTTCCGG---
BBVGCCBVGGCCTV
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
59 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.6145 |
Alignment:
CCGGAAGTGVV---
VAGGCCBBGGCVBB
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 47 |
Motif name: Egr1 |
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
Best Matches for Motif ID 47 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
23 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0 |
Alignment:
HGCGTGGGCGK
HGCGTGGGCGK
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
0.107124 |
Alignment:
BBGGGGCGGGGCGGB
HGCGTGGGCGK----
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.110516 |
Alignment:
BBVGCCBVGGCCTV
--HGCGTGGGCGK-
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.110516 |
Alignment:
BBVGCCBVGGCCTV
--HGCGTGGGCGK-
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
11 |
| Similarity score: |
0.110606 |
Alignment:
DGVBCABTGDWGCGKRRCSR
----HGCGTGGGCGK-----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
0.110606 |
Alignment:
DGVBCABTGDWGCGKRRCSR
----HGCGTGGGCGK-----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.111149 |
Alignment:
CSKCCCCGCCCCSY
-YCGCCCACGCH--
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.116531 |
Alignment:
BBGGGGCGGGGCVD
-HGCGTGGGCGK--
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
24 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.606429 |
Alignment:
CCCCKCCCCC-
YCGCCCACGCH
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
48 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.606429 |
Alignment:
GGGGGYGGGG-
HGCGTGGGCGK
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 48 |
Motif name: SP1 |
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
Best Matches for Motif ID 48 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
24 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0 |
Alignment:
CCCCKCCCCC
CCCCKCCCCC
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.043504 |
Alignment:
MSGGGGCGGGGYSG
-GGGGGYGGGG---
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0486461 |
Alignment:
HVGCCCCGCCCCBB
---CCCCKCCCCC-
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0517173 |
Alignment:
BBGGGGCGGGGCGGB
-GGGGGYGGGG----
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0930706 |
Alignment:
BBVGCCBVGGCCTV
----GGGGGYGGGG
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0930706 |
Alignment:
BBVGCCBVGGCCTV
----GGGGGYGGGG
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0990873 |
Alignment:
GGGGCCCAAGGGGG
-GGGGGYGGGG---
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0990873 |
Alignment:
CCCCCTTGGGCCCC
-CCCCKCCCCC---
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.102706 |
Alignment:
GCCYCMCCCD
CCCCKCCCCC
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
27 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.102706 |
Alignment:
DGGGYGKGGC
GGGGGYGGGG
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 49 |
Motif name: TFAP2A |
| Original motif Consensus sequence: GCCBBVRGS |
Reverse complement motif Consensus sequence: SCMVBBGGC |
|
|
 |
 |
Best Matches for Motif ID 49 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
25 |
| Motif name: |
TFAP2A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0 |
Alignment:
GCCBBVRGS
GCCBBVRGS
| Original motif Consensus sequence: GCCBBVRGS |
Reverse complement motif Consensus sequence: SCMVBBGGC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
9 |
| Similarity score: |
0.0516959 |
Alignment:
BBVGCCBVGGCCTV
---SCMVBBGGC--
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
9 |
| Similarity score: |
0.0516959 |
Alignment:
BBVGCCBVGGCCTV
---SCMVBBGGC--
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
9 |
| Similarity score: |
0.08003 |
Alignment:
GGGGCCCAAGGGGG
---GCCBBVRGS--
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
9 |
| Similarity score: |
0.08003 |
Alignment:
GGGGCCCAAGGGGG
---GCCBBVRGS--
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
57 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.0936515 |
Alignment:
HSCACGTGGC
-SCMVBBGGC
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
33 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.0936515 |
Alignment:
GCCACGTGSD
-GCCBBVRGS
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
9 |
| Similarity score: |
0.0966967 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-------GCCBBVRGS----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
9 |
| Similarity score: |
0.0966967 |
Alignment:
DGVBCABTGDWGCGKRRCSR
-------SCMVBBGGC----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
34 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.0999167 |
Alignment:
DCCACGTGCV
-GCCBBVRGS
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 50 |
Motif name: MIZF |
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
Best Matches for Motif ID 50 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
26 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0 |
Alignment:
BAACGTCCGC
BAACGTCCGC
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
59 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0810799 |
Alignment:
CCGGAAGTGVV
GCGGACGTTV-
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
35 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0810799 |
Alignment:
VVCACTTCCGG
BAACGTCCGC-
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.112981 |
Alignment:
BCACCTGCABC
BAACGTCCGC-
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
44 |
| Motif name: |
dhACATTCTkh |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.114583 |
Alignment:
DHACATTCTGH
BAACGTCCGC-
| Original motif Consensus sequence: DHACATTCTGH |
Reverse complement motif Consensus sequence: HCAGAATGTHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.115556 |
Alignment:
SGGTCACGTGACCS
-GCGGACGTTV---
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.122872 |
Alignment:
BCCGCCCCGCCCCBB
BAACGTCCGC-----
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.122917 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-GCGGACGTTV---------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.122917 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-GCGGACGTTV---------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.123601 |
Alignment:
HVGCCCCGCCCCBB
---BAACGTCCGC-
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 51 |
Motif name: Klf4 |
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
Best Matches for Motif ID 51 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
27 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0 |
Alignment:
DGGGYGKGGC
DGGGYGKGGC
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0516385 |
Alignment:
CSKCCCCGCCCCSY
--GCCYCMCCCD--
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0524114 |
Alignment:
HVGCCCCGCCCCBB
--GCCYCMCCCD--
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0558821 |
Alignment:
BCCGCCCCGCCCCBB
--GCCYCMCCCD---
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0987519 |
Alignment:
BBVGCCBVGGCCTV
-GCCYCMCCCD---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0987519 |
Alignment:
BBVGCCBVGGCCTV
-GCCYCMCCCD---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
48 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.102706 |
Alignment:
GGGGGYGGGG
DGGGYGKGGC
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
24 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.102706 |
Alignment:
GGGGGYGGGG
DGGGYGKGGC
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
58 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.105078 |
Alignment:
VGCACGTGGH
DGGGYGKGGC
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
34 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.105078 |
Alignment:
VGCACGTGGH
DGGGYGKGGC
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 52 |
Motif name: E2F1 |
| Original motif Consensus sequence: TTTSGCGC |
Reverse complement motif Consensus sequence: GCGCSAAA |
|
|
 |
 |
Best Matches for Motif ID 52 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
28 |
| Motif name: |
E2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0 |
Alignment:
TTTSGCGC
TTTSGCGC
| Original motif Consensus sequence: TTTSGCGC |
Reverse complement motif Consensus sequence: GCGCSAAA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0982639 |
Alignment:
CCCCCTTGGGCCCC
-TTTSGCGC-----
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0982639 |
Alignment:
GGGGCCCAAGGGGG
-GCGCSAAA-----
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
39 |
| Motif name: |
kCAGCCAATmr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.10386 |
Alignment:
DCAGCCAATVR
--GCGCSAAA-
| Original motif Consensus sequence: DCAGCCAATVR |
Reverse complement motif Consensus sequence: MBATTGGCTGH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
23 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.119792 |
Alignment:
YCGCCCACGCH
GCGCSAAA---
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
47 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.119792 |
Alignment:
YCGCCCACGCH
GCGCSAAA---
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.125 |
Alignment:
ATTTWATGAAA
-TTTSGCGC--
| Original motif Consensus sequence: ATTTWATGAAA |
Reverse complement motif Consensus sequence: TTTCATWAAAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.125 |
Alignment:
ATMACAATAAAA
-GCGCSAAA---
| Original motif Consensus sequence: ATMACAATAAAA |
Reverse complement motif Consensus sequence: TTTTATTGTYAT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
7 |
| Similarity score: |
0.612386 |
Alignment:
-BCCGCCCCGCCCCBB
GCGCSAAA--------
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
7 |
| Similarity score: |
0.616684 |
Alignment:
-HVGCCCCGCCCCBB
GCGCSAAA-------
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 53 |
Motif name: HIF1AARNT |
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
Best Matches for Motif ID 53 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
29 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0 |
Alignment:
VBACGTGV
VBACGTGV
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
57 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0575825 |
Alignment:
GCCACGTGSD
-VBACGTGV-
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
33 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0575825 |
Alignment:
GCCACGTGSD
-VBACGTGV-
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
58 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0597215 |
Alignment:
DCCACGTGCV
-VBACGTGV-
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
34 |
| Motif name: |
Myc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0597215 |
Alignment:
VGCACGTGGH
-VCACGTBV-
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0748197 |
Alignment:
GBTGCAGGTGB
---VCACGTBV
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0767561 |
Alignment:
SGGTCACGTGACCS
---VBACGTGV---
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
26 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0829928 |
Alignment:
GCGGACGTTV
--VCACGTBV
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
50 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0829928 |
Alignment:
GCGGACGTTV
--VCACGTBV
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
8 |
| Similarity score: |
0.100962 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-----VCACGTBV-------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 54 |
Motif name: PLAG1 |
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
Best Matches for Motif ID 54 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0 |
Alignment:
GGGGCCCAAGGGGG
GGGGCCCAAGGGGG
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.118259 |
Alignment:
VAGGCCBBGGCVBB
GGGGCCCAAGGGGG
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.118259 |
Alignment:
VAGGCCBBGGCVBB
GGGGCCCAAGGGGG
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.120124 |
Alignment:
BBGGGGCGGGGCGGB
GGGGCCCAAGGGGG-
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.124008 |
Alignment:
DGVBCABTGDWGCGKRRCSR
GGGGCCCAAGGGGG------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.124008 |
Alignment:
DGVBCABTGDWGCGKRRCSR
GGGGCCCAAGGGGG------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.617683 |
Alignment:
-MSGGGGCGGGGYSG
GGGGCCCAAGGGGG-
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.624856 |
Alignment:
-BBGGGGCGGGGCVD
GGGGCCCAAGGGGG-
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
48 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
2.1077 |
Alignment:
----GGGGGYGGGG
GGGGCCCAAGGGGG
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
24 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
2.1077 |
Alignment:
----GGGGGYGGGG
GGGGCCCAAGGGGG
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 55 |
Motif name: Pax5 |
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
Best Matches for Motif ID 55 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
20 |
| Similarity score: |
0 |
Alignment:
DGVBCABTGDWGCGKRRCSR
DGVBCABTGDWGCGKRRCSR
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
2.61478 |
Alignment:
BBGGGGCGGGGCGGB-----
MSGKKRCGCWDCABTGBBCD
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.09024 |
Alignment:
------HVGCCCCGCCCCBB
MSGKKRCGCWDCABTGBBCD
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.09355 |
Alignment:
------CSKCCCCGCCCCSY
MSGKKRCGCWDCABTGBBCD
| Original motif Consensus sequence: CSKCCCCGCCCCSY |
Reverse complement motif Consensus sequence: MSGGGGCGGGGYSG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.09388 |
Alignment:
------MTMMTCMMTCTKCK
MSGKKRCGCWDCABTGBBCD
| Original motif Consensus sequence: RGRAGARRGARRAR |
Reverse complement motif Consensus sequence: MTMMTCMMTCTKCK |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.11062 |
Alignment:
------SGGTCACGTGACCS
DGVBCABTGDWGCGKRRCSR
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
54 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.12401 |
Alignment:
GGGGCCCAAGGGGG------
DGVBCABTGDWGCGKRRCSR
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
30 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.12401 |
Alignment:
GGGGCCCAAGGGGG------
DGVBCABTGDWGCGKRRCSR
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
3.6245 |
Alignment:
-------VAGGCCBBGGCVBB
DGVBCABTGDWGCGKRRCSR-
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
3.6245 |
Alignment:
-------VAGGCCBBGGCVBB
DGVBCABTGDWGCGKRRCSR-
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 56 |
Motif name: ArntAhr |
| Original motif Consensus sequence: YGCGTG |
Reverse complement motif Consensus sequence: CACGCM |
|
|
 |
 |
Best Matches for Motif ID 56 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
32 |
| Motif name: |
ArntAhr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0 |
Alignment:
YGCGTG
YGCGTG
| Original motif Consensus sequence: YGCGTG |
Reverse complement motif Consensus sequence: CACGCM |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
47 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0340278 |
Alignment:
YCGCCCACGCH
-----CACGCM
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
23 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0340278 |
Alignment:
HGCGTGGGCGK
YGCGTG-----
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
53 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0452724 |
Alignment:
VBACGTGV
-YGCGTG-
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
29 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0452724 |
Alignment:
VBACGTGV
-YGCGTG-
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
33 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0718227 |
Alignment:
HSCACGTGGC
--CACGCM--
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
57 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0718227 |
Alignment:
GCCACGTGSD
--YGCGTG--
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
36 |
| Motif name: |
csGCCCCGCCCCsc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0736681 |
Alignment:
BBGGGGCGGGGCVD
----YGCGTG----
| Original motif Consensus sequence: HVGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCVD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
58 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0739492 |
Alignment:
DCCACGTGCV
--YGCGTG--
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
34 |
| Motif name: |
Myc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0739492 |
Alignment:
VGCACGTGGH
--CACGCM--
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 57 |
Motif name: Mycn |
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
Best Matches for Motif ID 57 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
33 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0 |
Alignment:
HSCACGTGGC
HSCACGTGGC
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
58 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0167031 |
Alignment:
VGCACGTGGH
HSCACGTGGC
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
34 |
| Motif name: |
Myc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0167031 |
Alignment:
DCCACGTGCV
GCCACGTGSD
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0795472 |
Alignment:
SGGTCACGTGACCS
--HSCACGTGGC--
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
35 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.105143 |
Alignment:
VVCACTTCCGG
GCCACGTGSD-
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
59 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.105143 |
Alignment:
VVCACTTCCGG
GCCACGTGSD-
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.10603 |
Alignment:
DGGGYGKGGC
HSCACGTGGC
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
27 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.10603 |
Alignment:
DGGGYGKGGC
HSCACGTGGC
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.106697 |
Alignment:
VAGGCCBBGGCVBB
---GCCACGTGSD-
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.106697 |
Alignment:
VAGGCCBBGGCVBB
---GCCACGTGSD-
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 58 |
Motif name: Myc |
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
Best Matches for Motif ID 58 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
34 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0 |
Alignment:
VGCACGTGGH
VGCACGTGGH
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
57 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0167031 |
Alignment:
HSCACGTGGC
VGCACGTGGH
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
33 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0167031 |
Alignment:
GCCACGTGSD
DCCACGTGCV
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
42 |
| Motif name: |
sSGTCACGTGACSs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0813632 |
Alignment:
SGGTCACGTGACCS
--VGCACGTGGH--
| Original motif Consensus sequence: SGGTCACGTGACCS |
Reverse complement motif Consensus sequence: SGGTCACGTGACCS |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
59 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.100737 |
Alignment:
VVCACTTCCGG
DCCACGTGCV-
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
35 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.100737 |
Alignment:
VVCACTTCCGG
DCCACGTGCV-
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
51 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.105078 |
Alignment:
DGGGYGKGGC
VGCACGTGGH
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
27 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.105078 |
Alignment:
DGGGYGKGGC
VGCACGTGGH
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.109248 |
Alignment:
VAGGCCBBGGCVBB
---DCCACGTGCV-
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.109248 |
Alignment:
VAGGCCBBGGCVBB
---DCCACGTGCV-
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 59 |
Motif name: GABPA |
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
Best Matches for Motif ID 59 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
35 |
| Motif name: |
GABPA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0 |
Alignment:
CCGGAAGTGVV
CCGGAAGTGVV
| Original motif Consensus sequence: CCGGAAGTGVV |
Reverse complement motif Consensus sequence: VVCACTTCCGG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
55 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
11 |
| Similarity score: |
0.113389 |
Alignment:
DGVBCABTGDWGCGKRRCSR
-------VVCACTTCCGG--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
31 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.113389 |
Alignment:
DGVBCABTGDWGCGKRRCSR
-------VVCACTTCCGG--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
22 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.114496 |
Alignment:
BBVGCCBVGGCCTV
VVCACTTCCGG---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
46 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.114496 |
Alignment:
BBVGCCBVGGCCTV
VVCACTTCCGG---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
40 |
| Motif name: |
kcACCTGCAgc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.117147 |
Alignment:
GBTGCAGGTGB
CCGGAAGTGVV
| Original motif Consensus sequence: BCACCTGCABC |
Reverse complement motif Consensus sequence: GBTGCAGGTGB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
45 |
| Motif name: |
wbgTAAATAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.117976 |
Alignment:
DBGTAAATAHD
CCGGAAGTGVV
| Original motif Consensus sequence: DBGTAAATAHD |
Reverse complement motif Consensus sequence: DHTATTTACBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
44 |
| Motif name: |
dhACATTCTkh |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.118278 |
Alignment:
HCAGAATGTHD
CCGGAAGTGVV
| Original motif Consensus sequence: DHACATTCTGH |
Reverse complement motif Consensus sequence: HCAGAATGTHD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.120605 |
Alignment:
AATTYDGAARTAWW
---CCGGAAGTGVV
| Original motif Consensus sequence: AATTYDGAARTAWW |
Reverse complement motif Consensus sequence: WWTAKTTCDKAATT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
38 |
| Motif name: |
cccGCCCCGCCCCsb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.124188 |
Alignment:
BBGGGGCGGGGCGGB
---CCGGAAGTGVV-
| Original motif Consensus sequence: BCCGCCCCGCCCCBB |
Reverse complement motif Consensus sequence: BBGGGGCGGGGCGGB |
|
|
 |
 |





































































































































































































































































