Best Matches in Database for Each Motif (Highest to Lowest)
| Dataset #: 1 | Motif ID: 1 | Motif name: Motif 1 |
| Original motif | Reverse complement motif |
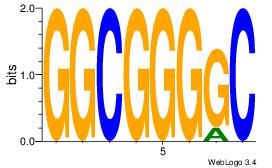
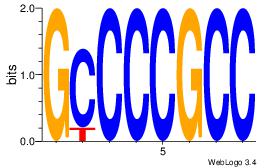
| Consensus sequence: GGCGGGGC | Consensus sequence: GCCCCGCC |
 |
 |
Best Matches for Motif ID 1 (Highest to Lowest)
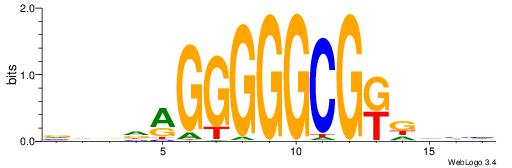
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HBDRCCMCGCCCHHHD
---GCCCCGCC-----
| Original motif | Reverse complement motif |
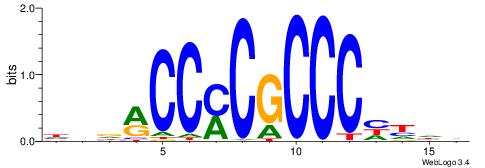
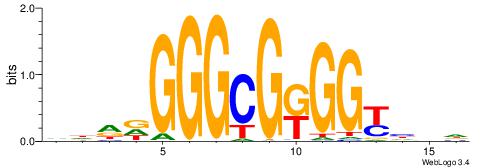
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.00617043 |
Alignment:
BHBHDTGGCGGGGBDHD
------GGCGGGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
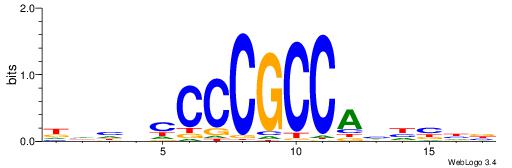 |
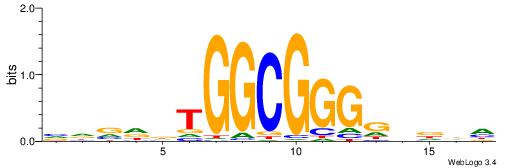 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0145996 |
Alignment:
BDWAGGCGTGBCHHD
----GGCGGGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: BDWAGGCGTGBCHHD | Consensus sequence: HHDGBCACGCCTWDB |
 |
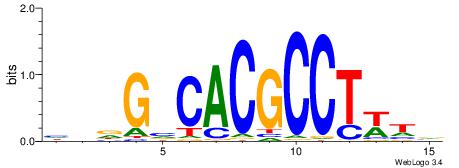 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.0233506 |
Alignment:
BDHHMCGCCCCCTHVBB
-GCCCCGCC--------
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
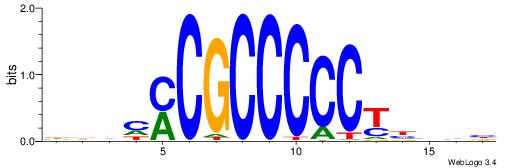 |
 |
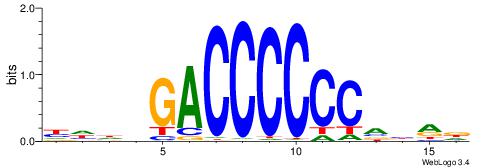
| Motif ID: | UP00024 |
| Motif name: | Glis2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0291232 |
Alignment:
HHHVGACCCCCCVBRD
----GCCCCGCC----
| Original motif | Reverse complement motif |
| Consensus sequence: HHHVGACCCCCCVBRD | Consensus sequence: HKBBGGGGGGTCVHHH |
 |
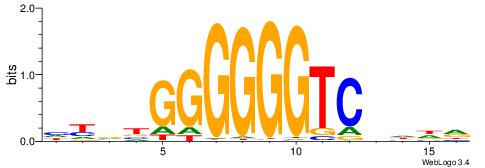 |
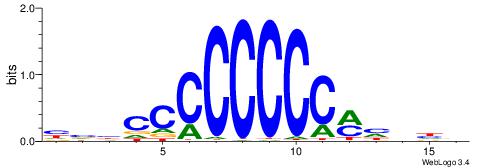
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0338469 |
Alignment:
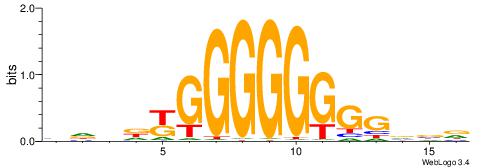
BHHDYGGGGGGGGBVD
-----GGCGGGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0361147 |
Alignment:
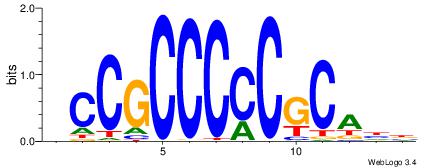
VHTGCGGGGGCGGH
---GGCGGGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0374146 |
Alignment:
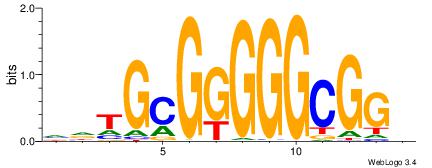
DTVBDDRGACCACCCAGDWDGBH
-------GCCCCGCC--------
| Original motif | Reverse complement motif |
| Consensus sequence: DTVBDDRGACCACCCAGDWDGBH | Consensus sequence: DBCDWHCTGGGTGGTCMDHBBAH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.0376381 |
Alignment:
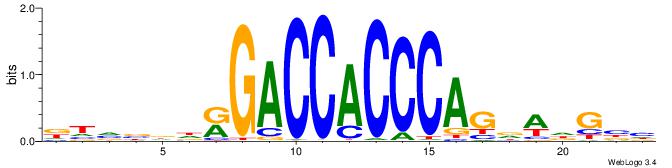
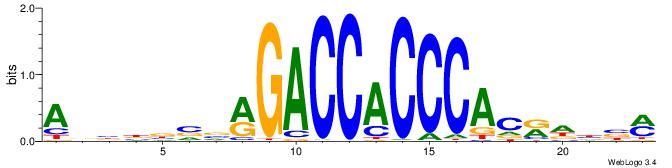
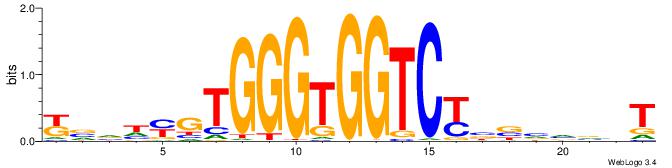
ABBBBVVRGACCACCCACRDBBM
--------GCCCCGCC-------
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0392043 |
Alignment:
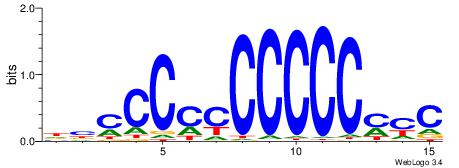
GKRGGGGGGGGGGBD
---GGCGGGGC----
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
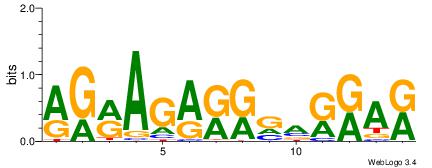
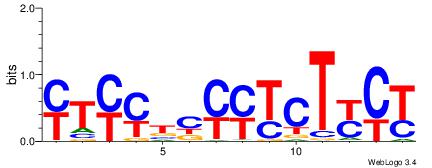
| Dataset #: 2 | Motif ID: 2 | Motif name: Motif 2 |
| Original motif | Reverse complement motif |
| Consensus sequence: RGRAGARRGARRAR | Consensus sequence: MTMMTCMMTCTKCK |
 |
 |
Best Matches for Motif ID 2 (Highest to Lowest)
| Motif ID: | UP00013 |
| Motif name: | Gabpa_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0378622 |
Alignment:
DBVVDRSGGAWKVHKG
RGRAGARRGARRAR--
| Original motif | Reverse complement motif |
| Consensus sequence: CYDBYWTCCSMHBVVH | Consensus sequence: DBVVDRSGGAWKVHKG |
 |
 |
| Motif ID: | UP00101 |
| Motif name: | Sox12_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.038148 |
Alignment:
VVDBASACAAAGRADH
-RGRAGARRGARRAR-
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBASACAAAGRADH | Consensus sequence: HDTMCTTTGTSTVDBB |
 |
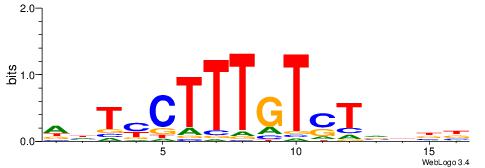 |
| Motif ID: | UP00058 |
| Motif name: | Tcf3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.039618 |
Alignment:
DVBSBDAHAHAHAHH
RGRAGARRGARRAR-
| Original motif | Reverse complement motif |
| Consensus sequence: DVBSBDAHAHAHAHH | Consensus sequence: HHTHTHTHTDBSBVD |
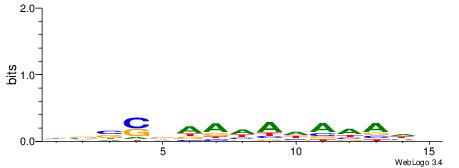 |
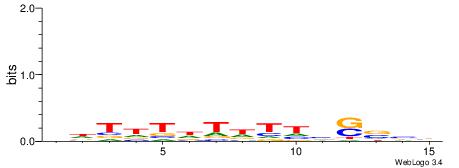 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 14 |
| Similarity score: | 0.0407385 |
Alignment:
MWSMBAARRATGCDCABHATGD
MTMMTCMMTCTKCK--------
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
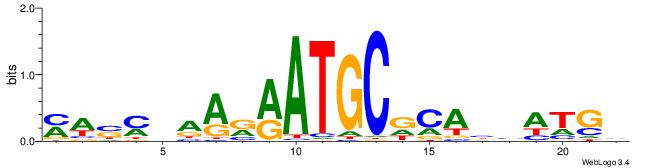 |
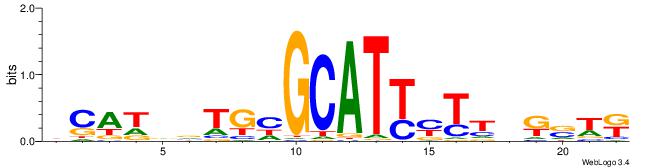 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0409403 |
Alignment:
BVVHAGGGGGCGRDHHB
-RGRAGARRGARRAR--
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0411988 |
Alignment:
BHVDCCCCDCCCBDBH
RGRAGARRGARRAR--
| Original motif | Reverse complement motif |
| Consensus sequence: BHVDCCCCDCCCBDBH | Consensus sequence: HVDVGGGHGGGGDBHB |
 |
 |
| Motif ID: | UP00059 |
| Motif name: | Arid5a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0412606 |
Alignment:
BDYDVAATACGADDBVV
---RGRAGARRGARRAR
| Original motif | Reverse complement motif |
| Consensus sequence: BDYDVAATACGADDBVV | Consensus sequence: BBVDDTCGTATTVDMHB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0419643 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
--RGRAGARRGARRAR------
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00077 |
| Motif name: | Srf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0420085 |
Alignment:
VBDDAAAAAAAAMVBHH
---RGRAGARRGARRAR
| Original motif | Reverse complement motif |
| Consensus sequence: VBDDAAAAAAAAMVBHH | Consensus sequence: HHVBYTTTTTTTTDDVV |
 |
 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0430662 |
Alignment:
DBCCCCCCCCCCMYC
RGRAGARRGARRAR-
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
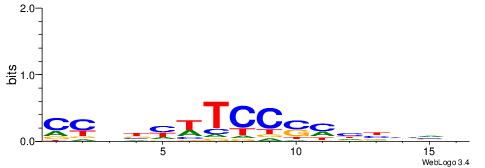
| Dataset #: 2 | Motif ID: 3 | Motif name: Motif 3 |
| Original motif | Reverse complement motif |
| Consensus sequence: AWAAAWTWAAASWA | Consensus sequence: TWSTTTWAWTTTWT |
 |
 |
Best Matches for Motif ID 3 (Highest to Lowest)
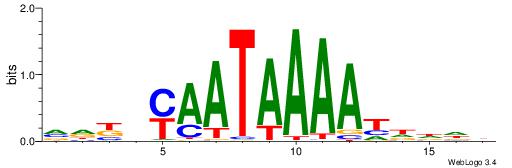
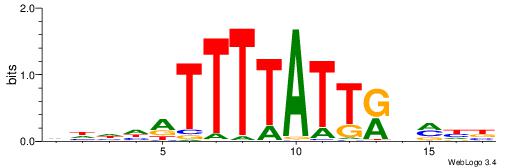
| Motif ID: | UP00121 |
| Motif name: | Hoxd10 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0232094 |
Alignment:
HDDDATTTTATTKVRDB
--TWSTTTWAWTTTWT-
| Original motif | Reverse complement motif |
| Consensus sequence: VDKVYAATAAAATDDDH | Consensus sequence: HDDDATTTTATTKVRDB |
 |
 |
| Motif ID: | UP00077 |
| Motif name: | Srf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.024404 |
Alignment:
VBDDAAAAAAAAMVBHH
--AWAAAWTWAAASWA-
| Original motif | Reverse complement motif |
| Consensus sequence: VBDDAAAAAAAAMVBHH | Consensus sequence: HHVBYTTTTTTTTDDVV |
 |
 |
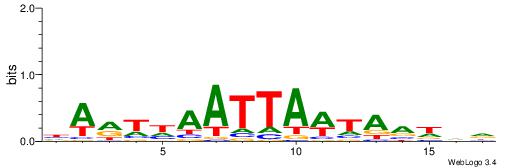
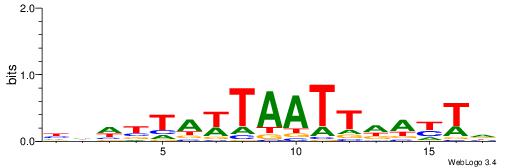
| Motif ID: | UP00255 |
| Motif name: | Dbx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0273798 |
Alignment:
HARTHAATTAWTAVDHD
--AWAAAWTWAAASWA-
| Original motif | Reverse complement motif |
| Consensus sequence: HARTHAATTAWTAVDHD | Consensus sequence: DHDBTAWTAATTHAKTH |
 |
 |
| Motif ID: | UP00090 |
| Motif name: | Elf3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0293574 |
Alignment:
DBTHHAAAAAAAAVHDH
--AWAAAWTWAAASWA-
| Original motif | Reverse complement motif |
| Consensus sequence: DBTHHAAAAAAAAVHDH | Consensus sequence: DDHBTTTTTTTTHDAVH |
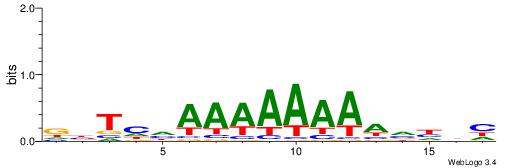 |
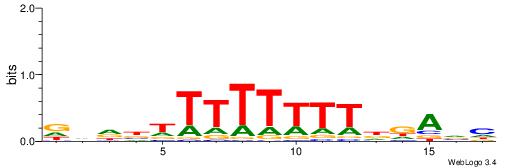 |
| Motif ID: | UP00037 |
| Motif name: | Zfp105_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0316804 |
Alignment:
HDYAAAHAAMAADHD
AWAAAWTWAAASWA-
| Original motif | Reverse complement motif |
| Consensus sequence: HDYAAAHAAMAADHD | Consensus sequence: HHHTTRTTDTTTKDH |
 |
 |
| Motif ID: | UP00134 |
| Motif name: | Hoxb13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.031986 |
Alignment:
VDHWTTTTATTGGDKB
-TWSTTTWAWTTTWT-
| Original motif | Reverse complement motif |
| Consensus sequence: VRHCCAATAAAAWHHV | Consensus sequence: VDHWTTTTATTGGDKB |
 |
 |
| Motif ID: | UP00078 |
| Motif name: | Arid3a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0323368 |
Alignment:
DHHYTTTAATTAAHBBV
TWSTTTWAWTTTWT---
| Original motif | Reverse complement motif |
| Consensus sequence: VBBHTTAATTAAAMHHH | Consensus sequence: DHHYTTTAATTAAHBBV |
 |
 |
| Motif ID: | UP00004 |
| Motif name: | Sox14_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0327495 |
Alignment:
DDDWATTATAATTHDH
--TWSTTTWAWTTTWT
| Original motif | Reverse complement motif |
| Consensus sequence: DHHAATTATAATWDDH | Consensus sequence: DDDWATTATAATTHDH |
 |
 |
| Motif ID: | UP00180 |
| Motif name: | Hoxd13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0332268 |
Alignment:
HDHHTTTTATTKKHDD
-TWSTTTWAWTTTWT-
| Original motif | Reverse complement motif |
| Consensus sequence: HDHYYAATAAAAHHHH | Consensus sequence: HDHHTTTTATTKKHDD |
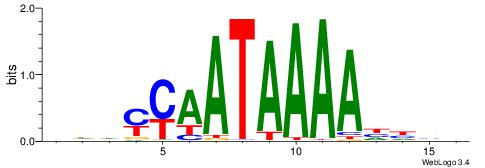 |
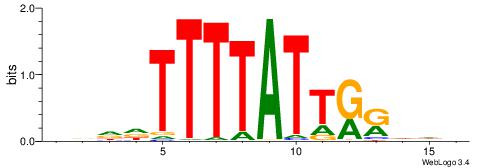 |
| Motif ID: | UP00097 |
| Motif name: | Mtf1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.034592 |
Alignment:
HDATAHDAAAHADV
AWAAAWTWAAASWA
| Original motif | Reverse complement motif |
| Consensus sequence: HDATAHDAAAHADV | Consensus sequence: VDTHTTTHHTATDH |
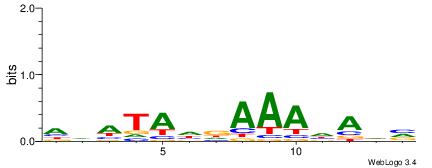 |
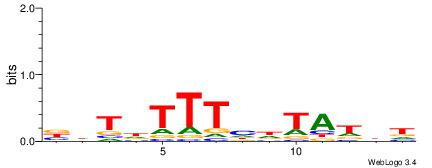 |
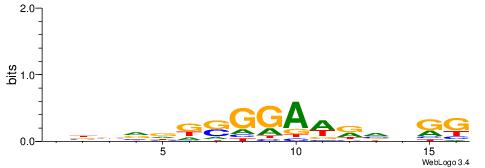
| Dataset #: 2 | Motif ID: 4 | Motif name: Motif 4 |
| Original motif | Reverse complement motif |
| Consensus sequence: AAAAWTTRCWT | Consensus sequence: AWGKAAWTTTT |
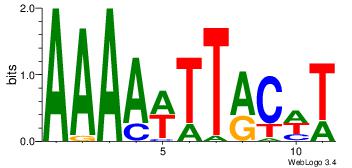 |
 |
Best Matches for Motif ID 4 (Highest to Lowest)
| Motif ID: | UP00044 |
| Motif name: | Mafk_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.022187 |
Alignment:
VHAAAAADTGCAHBR
--AAAAWTTRCWT--
| Original motif | Reverse complement motif |
| Consensus sequence: VHAAAAADTGCAHBR | Consensus sequence: MBHTGCADTTTTTHV |
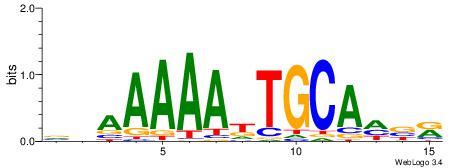 |
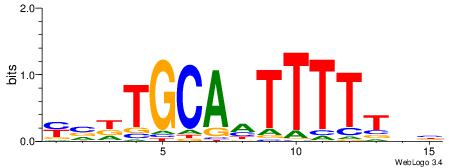 |
| Motif ID: | UP00075 |
| Motif name: | Sox15_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0318877 |
Alignment:
DHVKMWATTGTTHDHDH
AWGKAAWTTTT------
| Original motif | Reverse complement motif |
| Consensus sequence: HDDDDAACAATWRRBHD | Consensus sequence: DHVKMWATTGTTHDHDH |
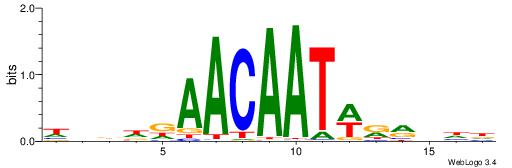 |
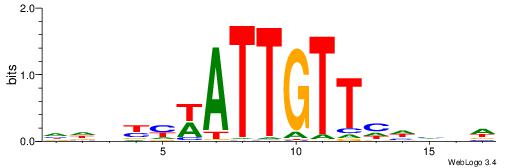 |
| Motif ID: | UP00091 |
| Motif name: | Sox5_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0333897 |
Alignment:
DHDDWATTGTTCDDHD
AWGKAAWTTTT-----
| Original motif | Reverse complement motif |
| Consensus sequence: DHDDGAACAATWDDHD | Consensus sequence: DHDDWATTGTTCDDHD |
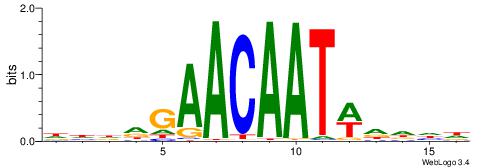 |
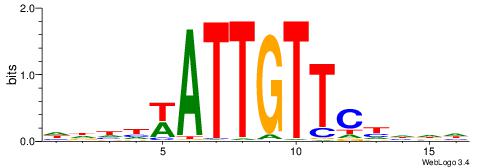 |
| Motif ID: | UP00014 |
| Motif name: | Sox17_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0352391 |
Alignment:
DDWTBAATTGTTDDB
-AWGKAAWTTTT---
| Original motif | Reverse complement motif |
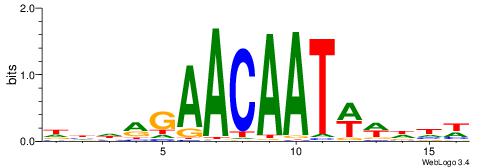
| Consensus sequence: VDDAACAATTVAWHD | Consensus sequence: DDWTBAATTGTTDDB |
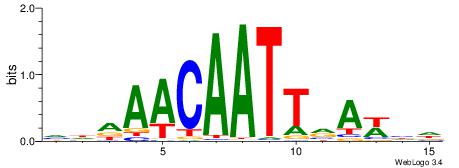 |
 |
| Motif ID: | UP00069 |
| Motif name: | Sox1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0357052 |
Alignment:
HBDTAATTGTTABBV
AWGKAAWTTTT----
| Original motif | Reverse complement motif |
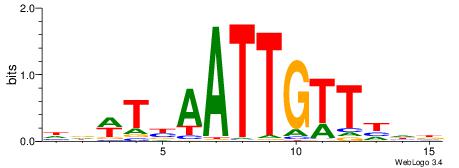
| Consensus sequence: HBDTAATTGTTABBV | Consensus sequence: VBVTAACAATTADVD |
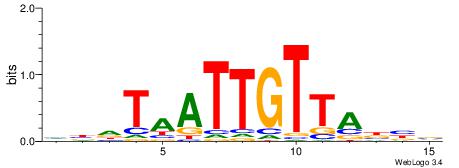 |
 |
| Motif ID: | UP00096 |
| Motif name: | Sox13_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0361647 |
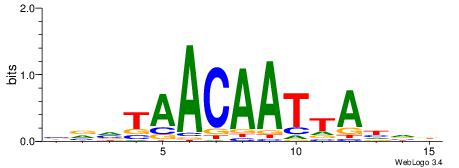
Alignment:
WHDWWATTGTTCKDHD
AWGKAAWTTTT-----
| Original motif | Reverse complement motif |
| Consensus sequence: DHDRGAACAATWWDHW | Consensus sequence: WHDWWATTGTTCKDHD |
 |
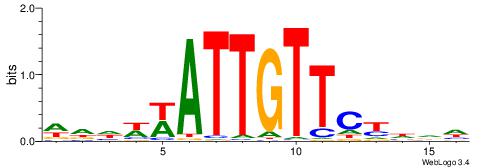 |
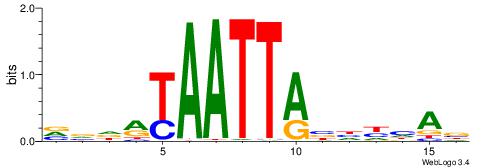
| Motif ID: | UP00126 |
| Motif name: | Dlx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0370352 |
Alignment:
VVDRTAATTABHHHAB
--AAAAWTTRCWT---
| Original motif | Reverse complement motif |
| Consensus sequence: VVDRTAATTABHHHAB | Consensus sequence: BTDHHBTAATTAKDVV |
 |
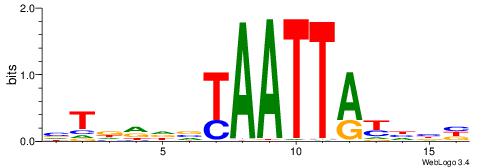 |
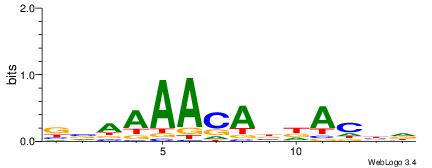
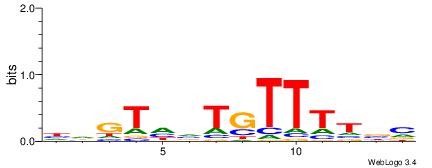
| Motif ID: | UP00074 |
| Motif name: | Isgf3g_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0371842 |
Alignment:
BVAAAACABDACHD
---AAAAWTTRCWT
| Original motif | Reverse complement motif |
| Consensus sequence: BVAAAACABDACHD | Consensus sequence: DHGTDVTGTTTTVB |
 |
 |
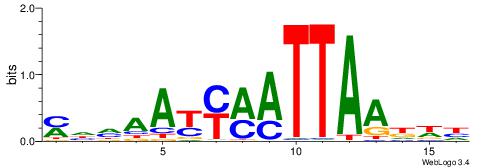
| Motif ID: | UP00170 |
| Motif name: | Isl2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0377846 |
Alignment:
HWDTTAATYKMTTHHR
-AWGKAAWTTTT----
| Original motif | Reverse complement motif |
| Consensus sequence: MHHAAYYMATTAADWH | Consensus sequence: HWDTTAATYKMTTHHR |
 |
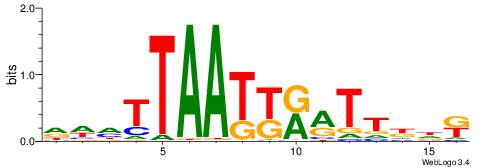 |
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0386497 |
Alignment:
HHBBMCTTTGTTHDDBH
AWGKAAWTTTT------
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
 |
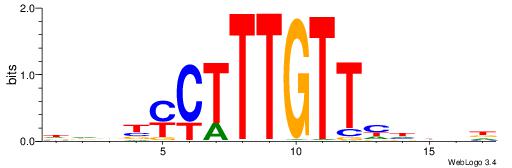 |
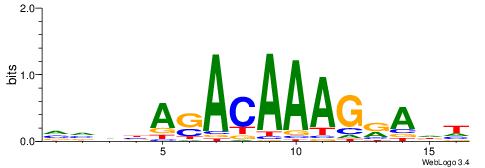
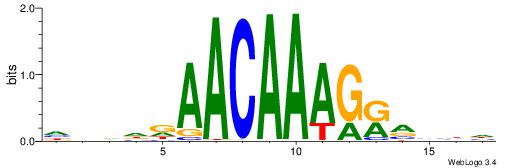
| Dataset #: 2 | Motif ID: 5 | Motif name: Motif 5 |
| Original motif | Reverse complement motif |
| Consensus sequence: AWTAAATAYAATTT | Consensus sequence: AAATTKTATTTAWT |
 |
 |
Best Matches for Motif ID 5 (Highest to Lowest)
| Motif ID: | UP00121 |
| Motif name: | Hoxd10 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0258732 |
Alignment:
HDDDATTTTATTKVRDB
--AAATTKTATTTAWT-
| Original motif | Reverse complement motif |
| Consensus sequence: VDKVYAATAAAATDDDH | Consensus sequence: HDDDATTTTATTKVRDB |
 |
 |
| Motif ID: | UP00169 |
| Motif name: | Lmx1b |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0275967 |
Alignment:
VDWWWTTAATTAATDHB
AAATTKTATTTAWT---
| Original motif | Reverse complement motif |
| Consensus sequence: VDWWWTTAATTAATDHB | Consensus sequence: BHDATTAATTAAWWWHB |
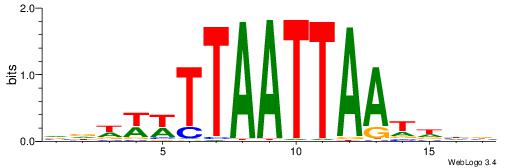 |
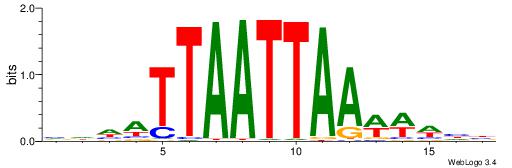 |
| Motif ID: | UP00212 |
| Motif name: | Lhx5 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0276125 |
Alignment:
VDHWTTTAATTAATHHV
AAATTKTATTTAWT---
| Original motif | Reverse complement motif |
| Consensus sequence: VDHATTAATTAAAWHHB | Consensus sequence: VDHWTTTAATTAATHHV |
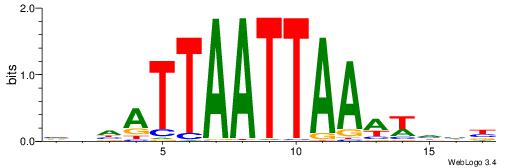 |
 |
| Motif ID: | UP00016 |
| Motif name: | Sry_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.027741 |
Alignment:
BHDHATTATAATDDDV
AWTAAATAYAATTT--
| Original motif | Reverse complement motif |
| Consensus sequence: BHDHATTATAATDDDV | Consensus sequence: VDDDATTATAATHDHV |
 |
 |
| Motif ID: | UP00180 |
| Motif name: | Hoxd13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0290926 |
Alignment:
HDHHTTTTATTKKHDD
-AAATTKTATTTAWT-
| Original motif | Reverse complement motif |
| Consensus sequence: HDHYYAATAAAAHHHH | Consensus sequence: HDHHTTTTATTKKHDD |
 |
 |
| Motif ID: | UP00004 |
| Motif name: | Sox14_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.029483 |
Alignment:
DHHAATTATAATWDDH
AWTAAATAYAATTT--
| Original motif | Reverse complement motif |
| Consensus sequence: DHHAATTATAATWDDH | Consensus sequence: DDDWATTATAATTHDH |
 |
 |
| Motif ID: | UP00130 |
| Motif name: | Lhx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.029893 |
Alignment:
VHHWWTTAATTAATTDV
AAATTKTATTTAWT---
| Original motif | Reverse complement motif |
| Consensus sequence: VDAATTAATTAAWWHHB | Consensus sequence: VHHWWTTAATTAATTDV |
 |
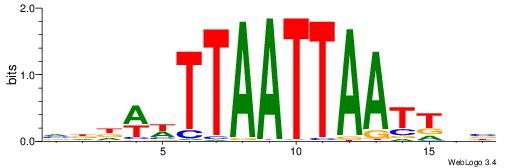 |
| Motif ID: | UP00134 |
| Motif name: | Hoxb13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.030701 |
Alignment:
VDHWTTTTATTGGDKB
-AAATTKTATTTAWT-
| Original motif | Reverse complement motif |
| Consensus sequence: VRHCCAATAAAAWHHV | Consensus sequence: VDHWTTTTATTGGDKB |
 |
 |
| Motif ID: | UP00266 |
| Motif name: | Prrx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0314917 |
Alignment:
AVDAKKTAATTAGTTVV
AAATTKTATTTAWT---
| Original motif | Reverse complement motif |
| Consensus sequence: VBAACTAATTARYTDVT | Consensus sequence: AVDAKKTAATTAGTTVV |
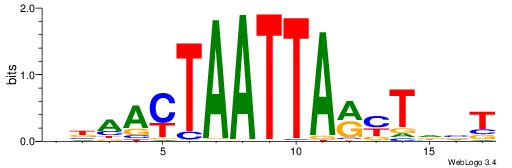 |
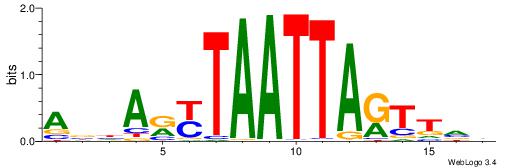 |
| Motif ID: | UP00262 |
| Motif name: | Lhx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0319088 |
Alignment:
BHTWWTTAATTAATDVV
AAATTKTATTTAWT---
| Original motif | Reverse complement motif |
| Consensus sequence: VVDATTAATTAAWWAHB | Consensus sequence: BHTWWTTAATTAATDVV |
 |
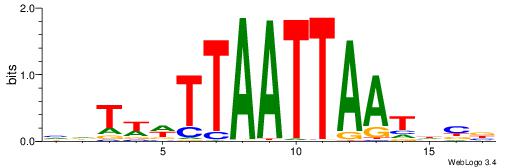 |
| Dataset #: 2 | Motif ID: 6 | Motif name: Motif 6 |
| Original motif | Reverse complement motif |
| Consensus sequence: AATTYDGAARTAWW | Consensus sequence: WWTAKTTCDKAATT |
 |
 |
Best Matches for Motif ID 6 (Highest to Lowest)
| Motif ID: | UP00015 |
| Motif name: | Ehf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0435331 |
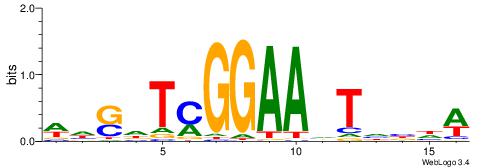
Alignment:
WDSHTYGGAAVTDVDW
-AATTYDGAARTAWW-
| Original motif | Reverse complement motif |
| Consensus sequence: WDVDABTTCCKAHSDW | Consensus sequence: WDSHTYGGAAVTDVDW |
 |
 |
| Motif ID: | UP00044 |
| Motif name: | Mafk_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0443465 |
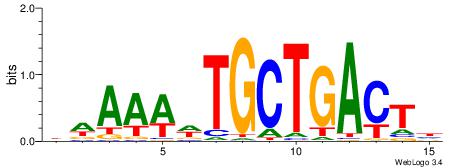
Alignment:
HAGTCAGCAWTTTWD
AATTYDGAARTAWW-
| Original motif | Reverse complement motif |
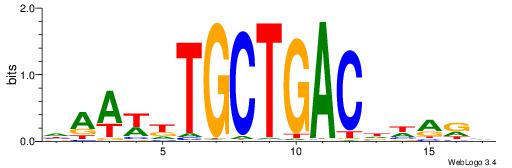
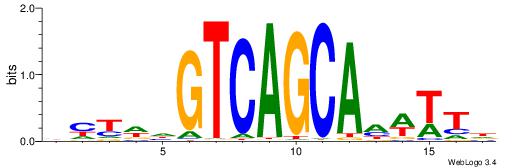
| Consensus sequence: DWAAAWTGCTGACTH | Consensus sequence: HAGTCAGCAWTTTWD |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0477209 |
Alignment:
HDTYTTGTTWTKDDDT
--WWTAKTTCDKAATT
| Original motif | Reverse complement motif |
| Consensus sequence: ADDDYAWAACAAMAHH | Consensus sequence: HDTYTTGTTWTKDDDT |
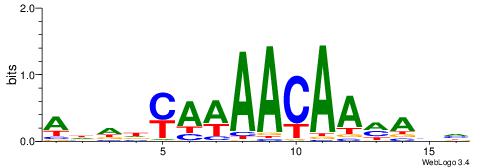 |
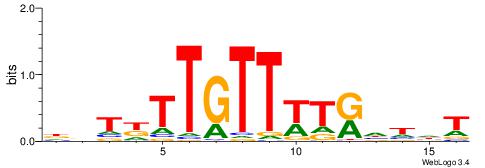 |
| Motif ID: | UP00255 |
| Motif name: | Dbx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0481558 |
Alignment:
DHDBTAWTAATTHAKTH
--WWTAKTTCDKAATT-
| Original motif | Reverse complement motif |
| Consensus sequence: HARTHAATTAWTAVDHD | Consensus sequence: DHDBTAWTAATTHAKTH |
 |
 |
| Motif ID: | UP00097 |
| Motif name: | Mtf1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0495241 |
Alignment:
VDTHTTTHHTATDH
WWTAKTTCDKAATT
| Original motif | Reverse complement motif |
| Consensus sequence: HDATAHDAAAHADV | Consensus sequence: VDTHTTTHHTATDH |
 |
 |
| Motif ID: | UP00262 |
| Motif name: | Lhx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0497972 |
Alignment:
BHTWWTTAATTAATDVV
-WWTAKTTCDKAATT--
| Original motif | Reverse complement motif |
| Consensus sequence: VVDATTAATTAAWWAHB | Consensus sequence: BHTWWTTAATTAATDVV |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 14 |
| Similarity score: | 0.0511395 |
Alignment:
WBHHHHHMDATTGTTHDHDVHH
--------WWTAKTTCDKAATT
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
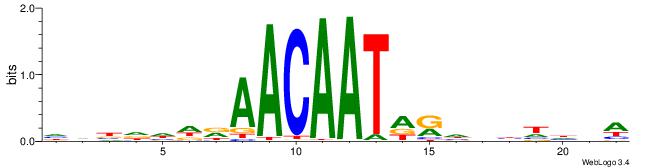 |
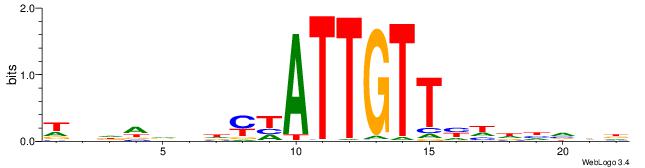 |
| Motif ID: | UP00045 |
| Motif name: | Mafb_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0517233 |
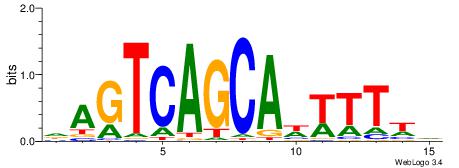
Alignment:
DMTWDGTCAGCADWTTH
---AATTYDGAARTAWW
| Original motif | Reverse complement motif |
| Consensus sequence: HAAWDTGCTGACDWARH | Consensus sequence: DMTWDGTCAGCADWTTH |
 |
 |
| Motif ID: | UP00130 |
| Motif name: | Lhx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0519872 |
Alignment:
VHHWWTTAATTAATTDV
-WWTAKTTCDKAATT--
| Original motif | Reverse complement motif |
| Consensus sequence: VDAATTAATTAAWWHHB | Consensus sequence: VHHWWTTAATTAATTDV |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0521736 |
Alignment:
DWAATRTAAACAAWVVB
AATTYDGAARTAWW---
| Original motif | Reverse complement motif |
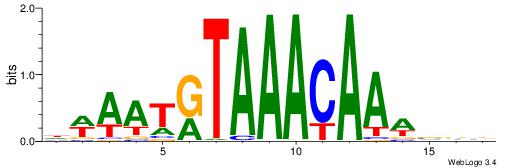
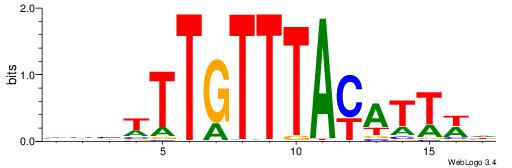
| Consensus sequence: DWAATRTAAACAAWVVB | Consensus sequence: VVVWTTGTTTAMATTWD |
 |
 |
| Dataset #: 2 | Motif ID: 7 | Motif name: Motif 7 |
| Original motif | Reverse complement motif |
| Consensus sequence: CSKCCCCGCCCCSY | Consensus sequence: MSGGGGCGGGGYSG |
 |
 |
Best Matches for Motif ID 7 (Highest to Lowest)
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.000548354 |
Alignment:
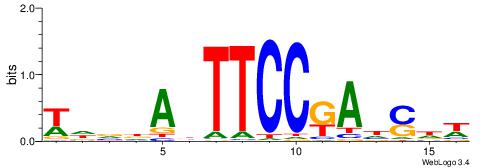
DHHDGGGCGRGGKHBH
-MSGGGGCGGGGYSG-
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.00741351 |
Alignment:
DHHBCCCCGCCAHHBHB
-CSKCCCCGCCCCSY--
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
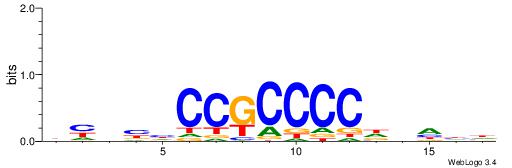
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0114155 |
Alignment:
HVBCCCCCCCCMHHHB
CSKCCCCGCCCCSY--
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0133301 |
Alignment:
DBCCCCCCCCCCMYC
-CSKCCCCGCCCCSY
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
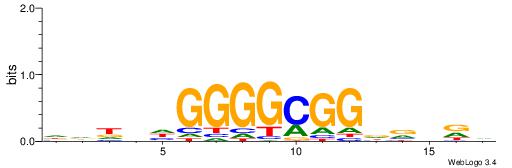
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0158143 |
Alignment:
HHHHDGGGGCGGDBHDV
---MSGGGGCGGGGYSG
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
 |
 |
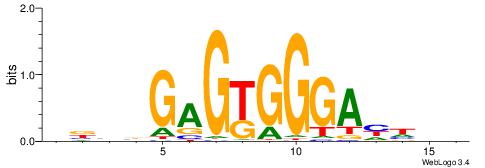
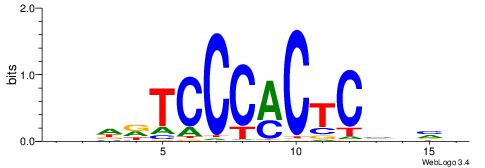
| Motif ID: | UP00007 |
| Motif name: | Egr1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0242521 |
Alignment:
BHHDTCCCACTCBDBV
-CSKCCCCGCCCCSY-
| Original motif | Reverse complement motif |
| Consensus sequence: BBHBGAGTGGGAHHDB | Consensus sequence: BHHDTCCCACTCBDBV |
 |
 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0244083 |
Alignment:
BHVDCCCCDCCCBDBH
-CSKCCCCGCCCCSY-
| Original motif | Reverse complement motif |
| Consensus sequence: BHVDCCCCDCCCBDBH | Consensus sequence: HVDVGGGHGGGGDBHB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 14 |
| Similarity score: | 0.0264047 |
Alignment:
YBVDMGTGGGTGGTCKVVBVBBT
----MSGGGGCGGGGYSG-----
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 14 |
| Similarity score: | 0.0276053 |
Alignment:
HHBSHTCGTGRGTGGKGDBVWMV
------MSGGGGCGGGGYSG---
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
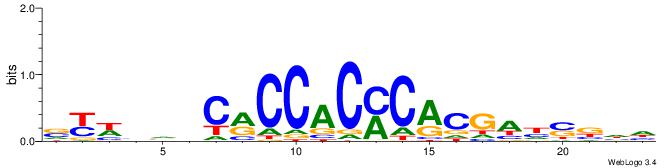 |
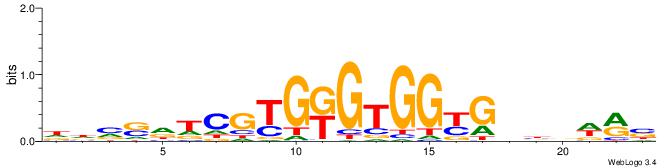 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 14 |
| Similarity score: | 0.0290826 |
Alignment:
BTBVTCVTGGGTGGTCMVVDVBB
-----MSGGGGCGGGGYSG----
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
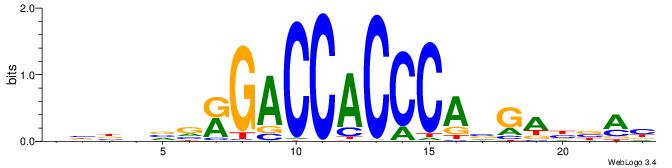 |
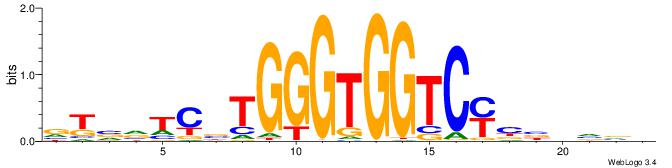 |
| Dataset #: 2 | Motif ID: 8 | Motif name: Motif 8 |
| Original motif | Reverse complement motif |
| Consensus sequence: AAATRWTAAAATCA | Consensus sequence: TGATTTTAWKATTT |
 |
 |
Best Matches for Motif ID 8 (Highest to Lowest)
| Motif ID: | UP00173 |
| Motif name: | Hoxc13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0325737 |
Alignment:
DHWTTTTAMGAGBDHH
TGATTTTAWKATTT--
| Original motif | Reverse complement motif |
| Consensus sequence: HHDBCTCRTAAAAWHD | Consensus sequence: DHWTTTTAMGAGBDHH |
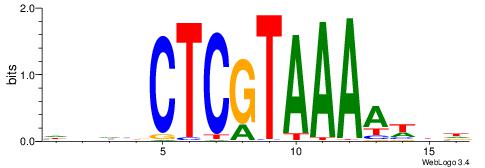
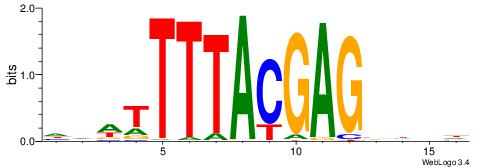
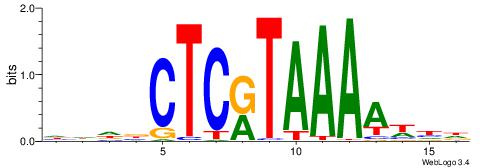 |
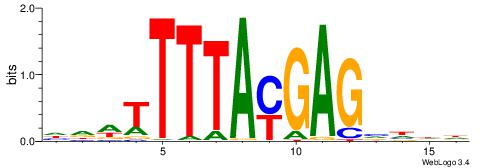 |
| Motif ID: | UP00235 |
| Motif name: | Hoxc11 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.036026 |
Alignment:
HDHTTTTACGACDTTH
TGATTTTAWKATTT--
| Original motif | Reverse complement motif |
| Consensus sequence: HAADGTCGTAAAAHDD | Consensus sequence: HDHTTTTACGACDTTH |
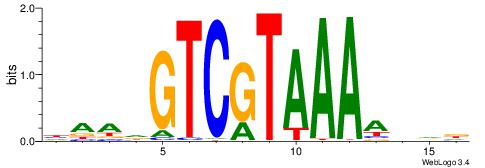 |
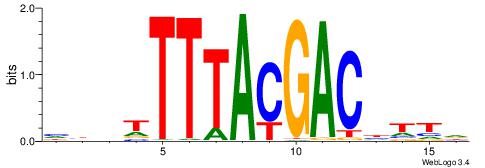 |
| Motif ID: | UP00183 |
| Motif name: | Hoxa13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0380573 |
Alignment:
DVWTTTTACGAGBHDH
TGATTTTAWKATTT--
| Original motif | Reverse complement motif |
| Consensus sequence: HDHBCTCGTAAAAWBD | Consensus sequence: DVWTTTTACGAGBHDH |
 |
 |
| Motif ID: | UP00246 |
| Motif name: | Hoxa11 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.039136 |
Alignment:
DDDTTTTACGACDTDH
TGATTTTAWKATTT--
| Original motif | Reverse complement motif |
| Consensus sequence: HDADGTCGTAAAAHDD | Consensus sequence: DDDTTTTACGACDTDH |
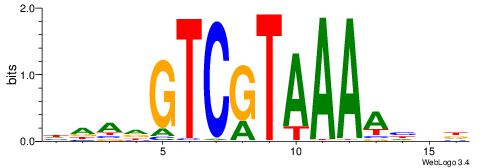 |
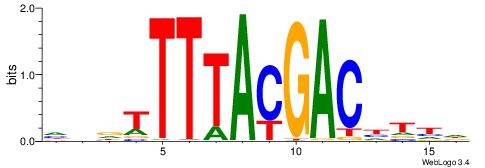 |
| Motif ID: | UP00180 |
| Motif name: | Hoxd13 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0391372 |
Alignment:
HDHYYAATAAAAHHHH
-AAATRWTAAAATCA-
| Original motif | Reverse complement motif |
| Consensus sequence: HDHYYAATAAAAHHHH | Consensus sequence: HDHHTTTTATTKKHDD |
 |
 |
| Motif ID: | UP00117 |
| Motif name: | Hoxd11 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.039319 |
Alignment:
DDDHTTTTACGACVDHH
-TGATTTTAWKATTT--
| Original motif | Reverse complement motif |
| Consensus sequence: HHDVGTCGTAAAAHHHD | Consensus sequence: DDDHTTTTACGACVDHH |
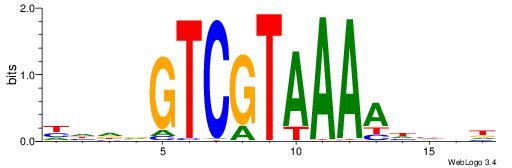 |
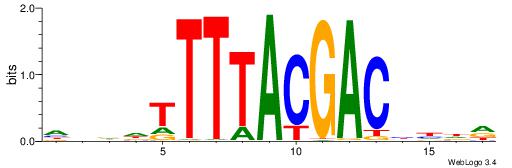 |
| Motif ID: | UP00207 |
| Motif name: | Hoxb9 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0397774 |
Alignment:
VRRGCMATAAAAHHHD
-AAATRWTAAAATCA-
| Original motif | Reverse complement motif |
| Consensus sequence: VRRGCMATAAAAHHHD | Consensus sequence: HDHHTTTTATRGCKMV |
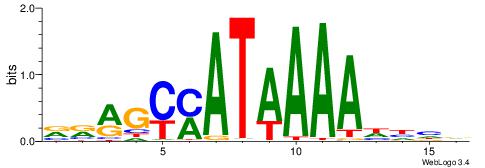 |
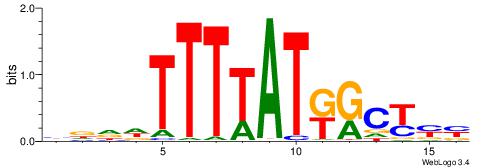 |
| Motif ID: | UP00121 |
| Motif name: | Hoxd10 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0397808 |
Alignment:
VDKVYAATAAAATDDDH
-AAATRWTAAAATCA--
| Original motif | Reverse complement motif |
| Consensus sequence: VDKVYAATAAAATDDDH | Consensus sequence: HDDDATTTTATTKVRDB |
 |
 |
| Motif ID: | UP00177 |
| Motif name: | Hoxd12 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0400538 |
Alignment:
DHDHTTTTACGACVDHD
-TGATTTTAWKATTT--
| Original motif | Reverse complement motif |
| Consensus sequence: HHDVGTCGTAAAAHHHD | Consensus sequence: DHDHTTTTACGACVDHD |
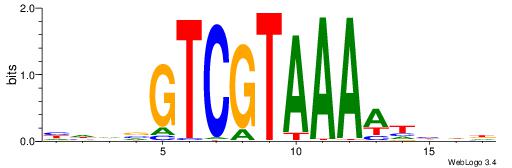 |
 |
| Motif ID: | UP00016 |
| Motif name: | Sry_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0409601 |
Alignment:
VDDDATTATAATHDHV
AAATRWTAAAATCA--
| Original motif | Reverse complement motif |
| Consensus sequence: BHDHATTATAATDDDV | Consensus sequence: VDDDATTATAATHDHV |
 |
 |
| Dataset #: 2 | Motif ID: 9 | Motif name: Motif 9 |
| Original motif | Reverse complement motif |
| Consensus sequence: AATHATATWTHAAA | Consensus sequence: TTTDAWATATHATT |
 |
 |
Best Matches for Motif ID 9 (Highest to Lowest)
| Motif ID: | UP00090 |
| Motif name: | Elf3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0521335 |
Alignment:
DBTHHAAAAAAAAVHDH
--TTTDAWATATHATT-
| Original motif | Reverse complement motif |
| Consensus sequence: DBTHHAAAAAAAAVHDH | Consensus sequence: DDHBTTTTTTTTHDAVH |
 |
 |
| Motif ID: | UP00255 |
| Motif name: | Dbx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0522843 |
Alignment:
DHDBTAWTAATTHAKTH
--TTTDAWATATHATT-
| Original motif | Reverse complement motif |
| Consensus sequence: HARTHAATTAWTAVDHD | Consensus sequence: DHDBTAWTAATTHAKTH |
 |
 |
| Motif ID: | UP00091 |
| Motif name: | Sox5_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0548459 |
Alignment:
DHDCATMATTDASDD
AATHATATWTHAAA-
| Original motif | Reverse complement motif |
| Consensus sequence: DHDCATMATTDASDD | Consensus sequence: DHSTDAATYATGDHD |
 |
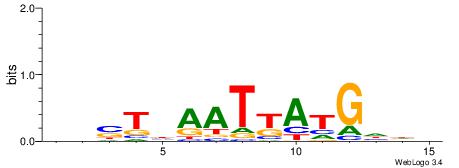 |
| Motif ID: | UP00121 |
| Motif name: | Hoxd10 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0549685 |
Alignment:
VDKVYAATAAAATDDDH
---AATHATATWTHAAA
| Original motif | Reverse complement motif |
| Consensus sequence: VDKVYAATAAAATDDDH | Consensus sequence: HDDDATTTTATTKVRDB |
 |
 |
| Motif ID: | UP00078 |
| Motif name: | Arid3a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0553164 |
Alignment:
VBBHTTAATTAAAMHHH
AATHATATWTHAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: VBBHTTAATTAAAMHHH | Consensus sequence: DHHYTTTAATTAAHBBV |
 |
 |
| Motif ID: | UP00169 |
| Motif name: | Lmx1b |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0554285 |
Alignment:
BHDATTAATTAAWWWHB
---TTTDAWATATHATT
| Original motif | Reverse complement motif |
| Consensus sequence: VDWWWTTAATTAATDHB | Consensus sequence: BHDATTAATTAAWWWHB |
 |
 |
| Motif ID: | UP00141 |
| Motif name: | Vsx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0554615 |
Alignment:
VHTDHTTAATTAAMHVG
AATHATATWTHAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: CVHRTTAATTAAHDAHB | Consensus sequence: VHTDHTTAATTAAMHVG |
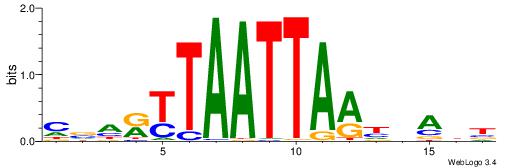 |
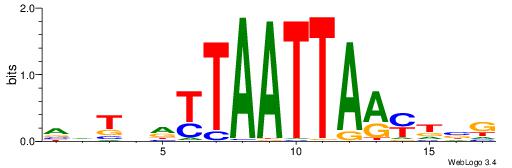 |
| Motif ID: | UP00024 |
| Motif name: | Glis2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0558193 |
Alignment:
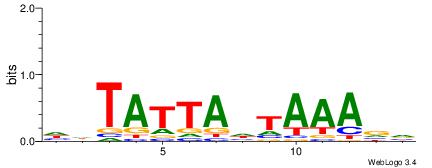
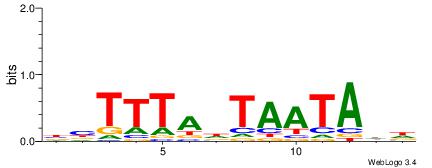
BVTTTAHTAATADH
TTTDAWATATHATT
| Original motif | Reverse complement motif |
| Consensus sequence: HDTATTAHTAAAVV | Consensus sequence: BVTTTAHTAATADH |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0563339 |
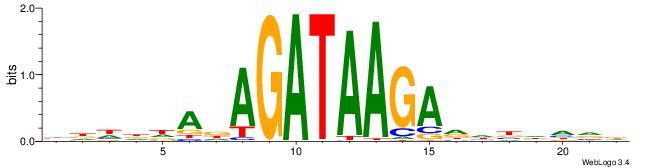
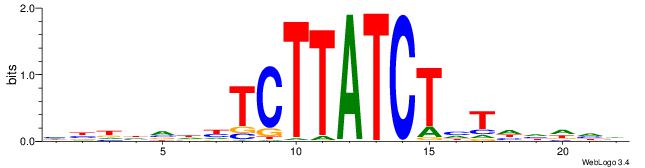
Alignment:
HBHDVHDTCTTATCTHTDDHDV
-------TTTDAWATATHATT-
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Motif ID: | UP00194 |
| Motif name: | Irx4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0570128 |
Alignment:
DBTWTWACATGTWDHDW
--TTTDAWATATHATT-
| Original motif | Reverse complement motif |
| Consensus sequence: WDHDWACATGTWAWABD | Consensus sequence: DBTWTWACATGTWDHDW |
 |
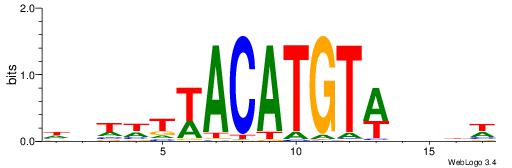 |
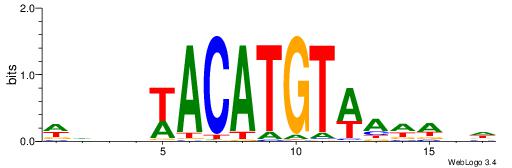
| Dataset #: 2 | Motif ID: 10 | Motif name: Motif 10 |
| Original motif | Reverse complement motif |
| Consensus sequence: TTTCATAAWT | Consensus sequence: AWTTATGAAA |
 |
 |
Best Matches for Motif ID 10 (Highest to Lowest)
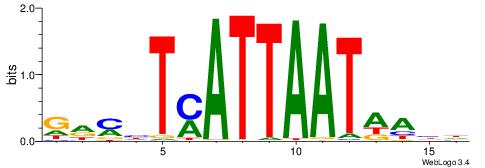
| Motif ID: | UP00118 |
| Motif name: | Pou4f3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.017789 |
Alignment:
GHYHTYATTAATAAVH
--TTTCATAAWT----
| Original motif | Reverse complement motif |
| Consensus sequence: HVTTATTAATKADKHC | Consensus sequence: GHYHTYATTAATAAVH |
 |
 |
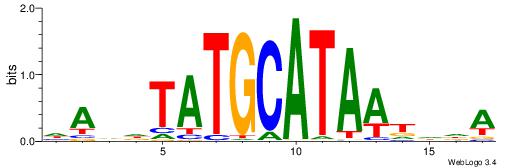
| Motif ID: | UP00211 |
| Motif name: | Pou3f3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0192304 |
Alignment:
HADDTATGCATAATDDA
-----TTTCATAAWT--
| Original motif | Reverse complement motif |
| Consensus sequence: HADDTATGCATAATDDA | Consensus sequence: TDDATTATGCATADDTH |
 |
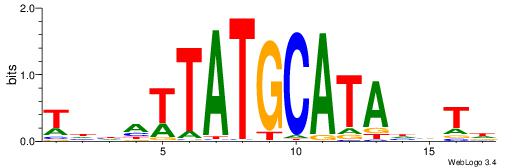 |
| Motif ID: | UP00179 |
| Motif name: | Pou2f3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
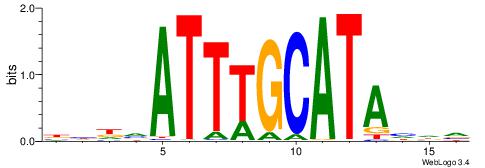
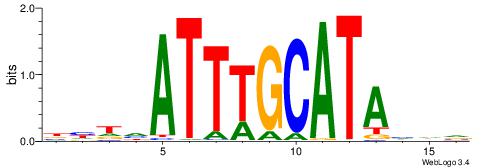
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0193076 |
Alignment:
BHHVATTTGCATAHHV
AWTTATGAAA------
| Original motif | Reverse complement motif |
| Consensus sequence: BHDTATGCAAATBHDV | Consensus sequence: BHHVATTTGCATAHHV |
 |
 |
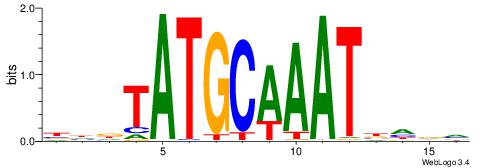
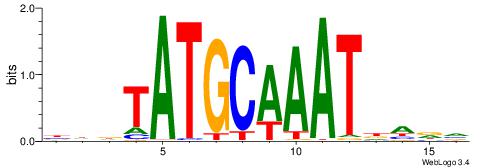
| Motif ID: | UP00191 |
| Motif name: | Pou2f2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0200501 |
Alignment:
BHDTATGCAAATDHDV
------TTTCATAAWT
| Original motif | Reverse complement motif |
| Consensus sequence: BHDTATGCAAATDHDV | Consensus sequence: BHHDATTTGCATAHHV |
 |
 |
| Motif ID: | UP00130 |
| Motif name: | Lhx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.020314 |
Alignment:
VHHWWTTAATTAATTDV
----TTTCATAAWT---
| Original motif | Reverse complement motif |
| Consensus sequence: VDAATTAATTAAWWHHB | Consensus sequence: VHHWWTTAATTAATTDV |
 |
 |
| Motif ID: | UP00212 |
| Motif name: | Lhx5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0203295 |
Alignment:
VDHWTTTAATTAATHHV
----TTTCATAAWT---
| Original motif | Reverse complement motif |
| Consensus sequence: VDHATTAATTAAAWHHB | Consensus sequence: VDHWTTTAATTAATHHV |
 |
 |
| Motif ID: | UP00169 |
| Motif name: | Lmx1b |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0232405 |
Alignment:
VDWWWTTAATTAATDHB
----TTTCATAAWT---
| Original motif | Reverse complement motif |
| Consensus sequence: VDWWWTTAATTAATDHB | Consensus sequence: BHDATTAATTAAWWWHB |
 |
 |
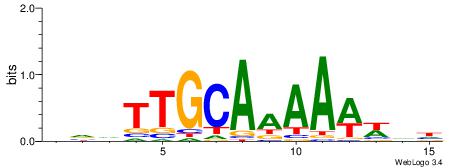
| Motif ID: | UP00045 |
| Motif name: | Mafb_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0250697 |
Alignment:
HVDTTGCAAAAWWDH
---TTTCATAAWT--
| Original motif | Reverse complement motif |
| Consensus sequence: HVDTTGCAAAAWWDH | Consensus sequence: HDWWTTTTGCAADBD |
 |
 |
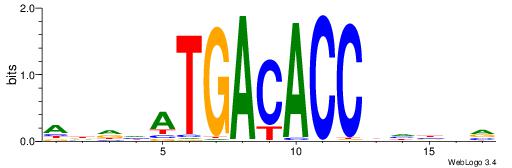
| Motif ID: | UP00199 |
| Motif name: | Six4 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0256033 |
Alignment:
ADDHATGACACCBHBBV
-------TTTCATAAWT
| Original motif | Reverse complement motif |
| Consensus sequence: ADDHATGACACCBHBBV | Consensus sequence: BBVHVGGTGTCATHDDT |
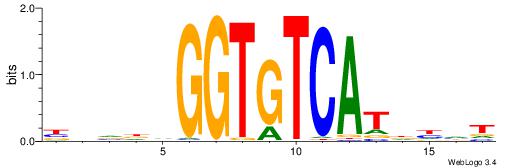
 |
 |
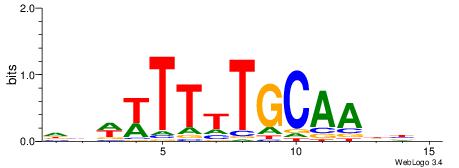
| Motif ID: | UP00173 |
| Motif name: | Hoxc13 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0262559 |
Alignment:
HHDBCTCRTAAAAWHD
---TTTCATAAWT---
| Original motif | Reverse complement motif |
| Consensus sequence: HHDBCTCRTAAAAWHD | Consensus sequence: DHWTTTTAMGAGBDHH |
 |
 |
| Dataset #: 2 | Motif ID: 11 | Motif name: Motif 11 |
| Original motif | Reverse complement motif |
| Consensus sequence: ATTTWATGAAA | Consensus sequence: TTTCATWAAAT |
 |
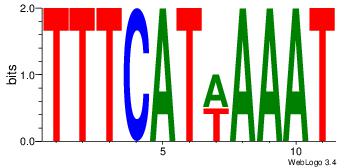 |
Best Matches for Motif ID 11 (Highest to Lowest)
| Motif ID: | UP00130 |
| Motif name: | Lhx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0235917 |
Alignment:
VHHWWTTAATTAATTDV
----TTTCATWAAAT--
| Original motif | Reverse complement motif |
| Consensus sequence: VDAATTAATTAAWWHHB | Consensus sequence: VHHWWTTAATTAATTDV |
 |
 |
| Motif ID: | UP00212 |
| Motif name: | Lhx5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0238582 |
Alignment:
VDHWTTTAATTAATHHV
---ATTTWATGAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: VDHATTAATTAAAWHHB | Consensus sequence: VDHWTTTAATTAATHHV |
 |
 |
| Motif ID: | UP00173 |
| Motif name: | Hoxc13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.024458 |
Alignment:
HHDBCTCRTAAAAWHD
---TTTCATWAAAT--
| Original motif | Reverse complement motif |
| Consensus sequence: HHDBCTCRTAAAAWHD | Consensus sequence: DHWTTTTAMGAGBDHH |
 |
 |
| Motif ID: | UP00169 |
| Motif name: | Lmx1b |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0247146 |
Alignment:
VDWWWTTAATTAATDHB
----TTTCATWAAAT--
| Original motif | Reverse complement motif |
| Consensus sequence: VDWWWTTAATTAATDHB | Consensus sequence: BHDATTAATTAAWWWHB |
 |
 |
| Motif ID: | UP00188 |
| Motif name: | Lmx1a |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0250542 |
Alignment:
CVHHTTAATTAAWWWHB
--ATTTWATGAAA----
| Original motif | Reverse complement motif |
| Consensus sequence: CVHHTTAATTAAWWWHB | Consensus sequence: BDWWWTTAATTAAHHVG |
 |
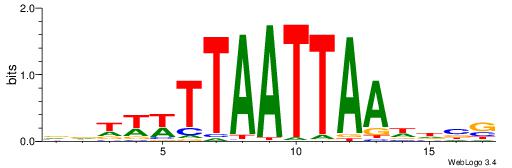 |
| Motif ID: | UP00197 |
| Motif name: | Hoxc9 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0275369 |
Alignment:
VDHHTTAATGACCBHV
--ATTTWATGAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: VDVGGTCATTAAHHDB | Consensus sequence: VDHHTTAATGACCBHV |
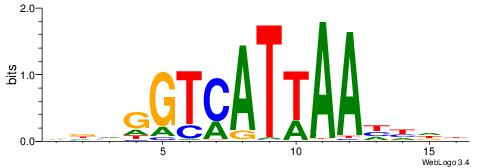 |
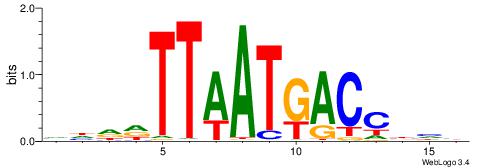 |
| Motif ID: | UP00117 |
| Motif name: | Hoxd11 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.028378 |
Alignment:
DDDHTTTTACGACVDHH
---ATTTWATGAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: HHDVGTCGTAAAAHHHD | Consensus sequence: DDDHTTTTACGACVDHH |
 |
 |
| Motif ID: | UP00213 |
| Motif name: | Hoxa9 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0286621 |
Alignment:
DCGGYCATWAAAWTADW
--TTTCATWAAAT----
| Original motif | Reverse complement motif |
| Consensus sequence: DCGGYCATWAAAWTADW | Consensus sequence: WDTAWTTTWATGKCCGD |
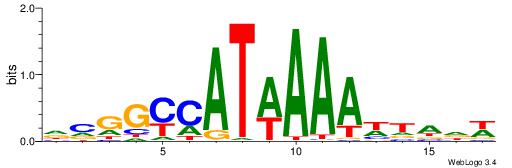 |
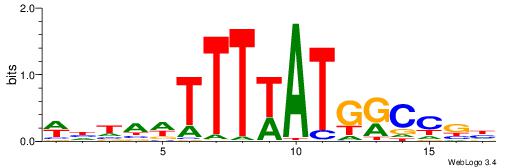 |
| Motif ID: | UP00121 |
| Motif name: | Hoxd10 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0288394 |
Alignment:
VDKVYAATAAAATDDDH
--TTTCATWAAAT----
| Original motif | Reverse complement motif |
| Consensus sequence: VDKVYAATAAAATDDDH | Consensus sequence: HDDDATTTTATTKVRDB |
 |
 |
| Motif ID: | UP00183 |
| Motif name: | Hoxa13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.028941 |
Alignment:
HDHBCTCGTAAAAWBD
---TTTCATWAAAT--
| Original motif | Reverse complement motif |
| Consensus sequence: HDHBCTCGTAAAAWBD | Consensus sequence: DVWTTTTACGAGBHDH |
 |
 |
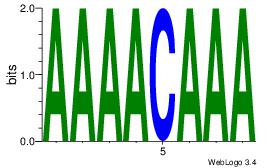
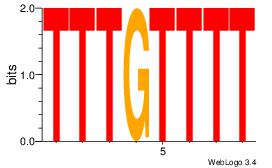
| Dataset #: 2 | Motif ID: 12 | Motif name: Motif 12 |
| Original motif | Reverse complement motif |
| Consensus sequence: AAAACAAA | Consensus sequence: TTTGTTTT |
 |
 |
Best Matches for Motif ID 12 (Highest to Lowest)
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HBDDDAACAAAGRVBHH
---AAAACAAA------
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
 |
 |
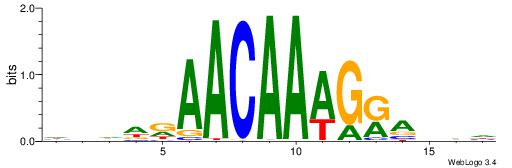
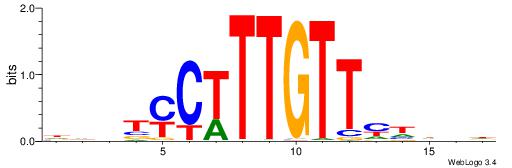
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.000282957 |
Alignment:
HBDDRAACAAAGRABHH
---AAAACAAA------
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
 |
 |
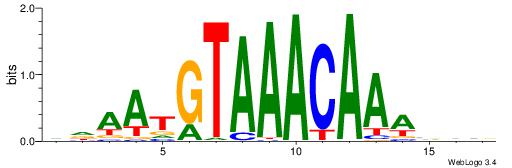
| Motif ID: | UP00025 |
| Motif name: | Foxk1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.00309128 |
Alignment:
BBVTTTGTTTACATTDD
---TTTGTTTT------
| Original motif | Reverse complement motif |
| Consensus sequence: DDAATGTAAACAAAVVB | Consensus sequence: BBVTTTGTTTACATTDD |
 |
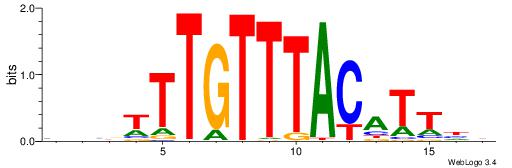 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.00620509 |
Alignment:
VVVWTTGTTTAMATTWD
---TTTGTTTT------
| Original motif | Reverse complement motif |
| Consensus sequence: DWAATRTAAACAAWVVB | Consensus sequence: VVVWTTGTTTAMATTWD |
 |
 |
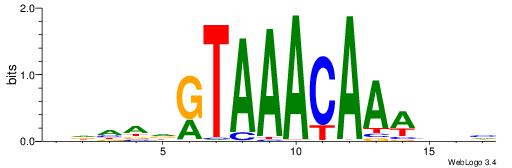
| Motif ID: | UP00039 |
| Motif name: | Foxj3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.00680733 |
Alignment:
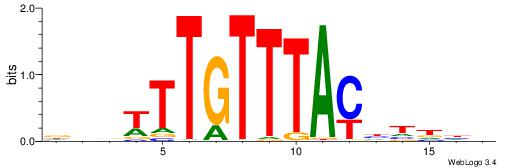
HDHADGTAAACAAAVVH
------AAAACAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: HDHADGTAAACAAAVVH | Consensus sequence: DVVTTTGTTTACDTHDH |
 |
 |
| Motif ID: | UP00041 |
| Motif name: | Foxj1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0130585 |
Alignment:
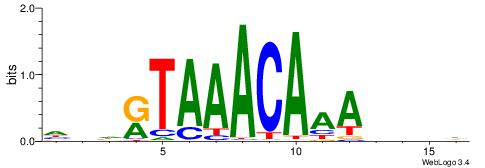
DDDHTTTGTTTAMHDH
----TTTGTTTT----
| Original motif | Reverse complement motif |
| Consensus sequence: HDHRTAAACAAAHDDD | Consensus sequence: DDDHTTTGTTTAMHDH |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0137227 |
Alignment:
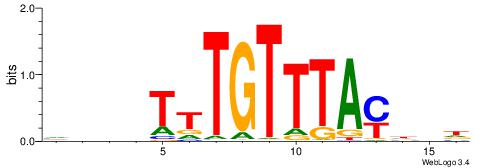
ADDDYAWAACAAMAHH
-----AAAACAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: ADDDYAWAACAAMAHH | Consensus sequence: HDTYTTGTTWTKDDDT |
 |
 |
| Motif ID: | UP00073 |
| Motif name: | Foxa2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0160723 |
Alignment:
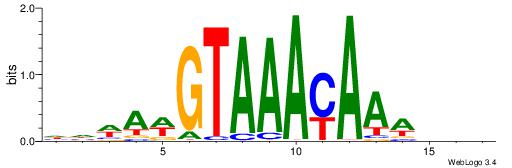
BDVTTTKTTTACWTWHH
---TTTGTTTT------
| Original motif | Reverse complement motif |
| Consensus sequence: HHWAWGTAAAYAAAVDB | Consensus sequence: BDVTTTKTTTACWTWHH |
 |
 |
| Motif ID: | UP00101 |
| Motif name: | Sox12_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0161675 |
Alignment:
DWTHAVAACAATHA
----AAAACAAA--
| Original motif | Reverse complement motif |
| Consensus sequence: THATTGTTVTHAWH | Consensus sequence: DWTHAVAACAATHA |
 |
 |
| Motif ID: | UP00039 |
| Motif name: | Foxj3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.0164735 |
Alignment:
DVHWDTGTTWTGDTBDH
---TTTGTTTT------
| Original motif | Reverse complement motif |
| Consensus sequence: HDBAHCAWAACADWDVD | Consensus sequence: DVHWDTGTTWTGDTBDH |
 |
 |
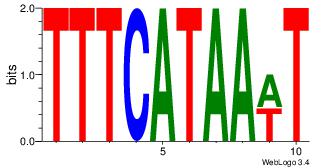
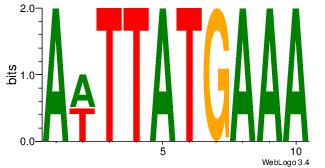
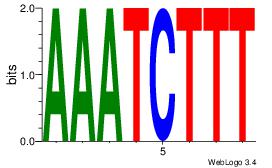
| Dataset #: 2 | Motif ID: 13 | Motif name: Motif 13 |
| Original motif | Reverse complement motif |
| Consensus sequence: AAAGATTT | Consensus sequence: AAATCTTT |
 |
 |
Best Matches for Motif ID 13 (Highest to Lowest)
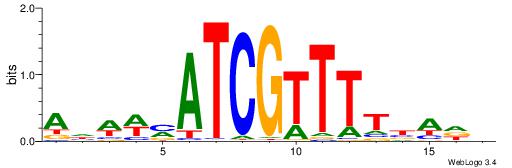
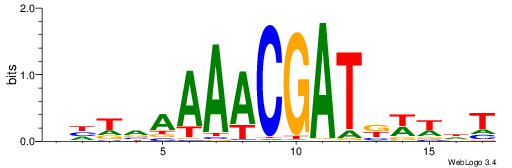
| Motif ID: | UP00114 |
| Motif name: | Homez |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.00327586 |
Alignment:
AHWWMATCGTTTTBADD
---AAATCTTT------
| Original motif | Reverse complement motif |
| Consensus sequence: AHWWMATCGTTTTBADD | Consensus sequence: HDTVAAAACGATRWWHT |
 |
 |
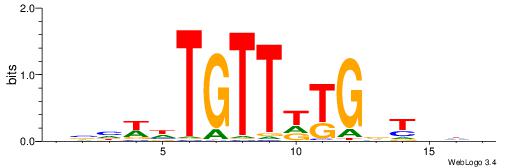
| Motif ID: | UP00148 |
| Motif name: | Hdx |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.0107572 |
Alignment:
DDKDHGAAATCAHHBHV
------AAATCTTT---
| Original motif | Reverse complement motif |
| Consensus sequence: DDKDHGAAATCAHHBHV | Consensus sequence: BDBDHTGATTTCDHYDD |
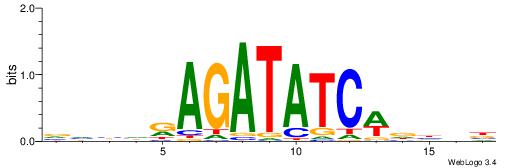
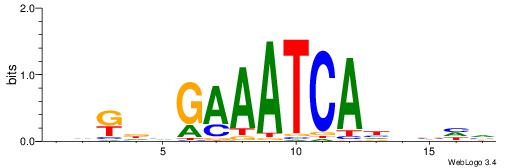 |
 |
| Motif ID: | UP00080 |
| Motif name: | Gata5_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.0119733 |
Alignment:
VVBHRAGATATCWDHBB
---AAAGATTT------
| Original motif | Reverse complement motif |
| Consensus sequence: VVBHRAGATATCWDHBB | Consensus sequence: VVHHWGATATCTMHBBV |
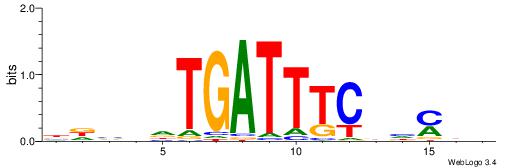
 |
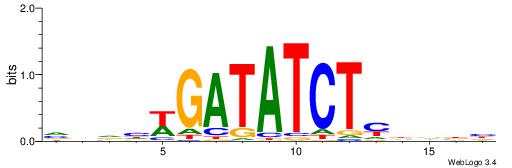 |
| Motif ID: | UP00059 |
| Motif name: | Arid5a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0140725 |
Alignment:
HDTDVCAATATTMV
-----AAAGATTT-
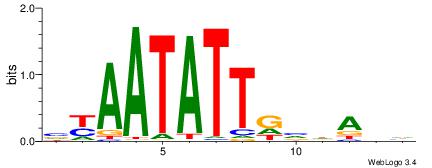
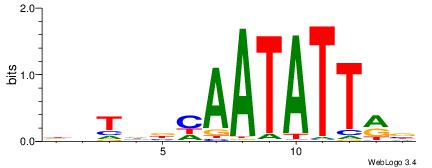
| Original motif | Reverse complement motif |
| Consensus sequence: VYAATATTGVDADH | Consensus sequence: HDTDVCAATATTMV |
 |
 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.0151544 |
Alignment:
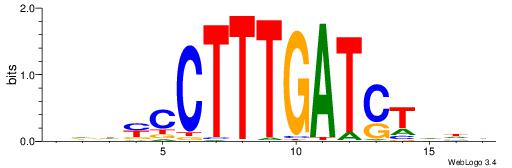
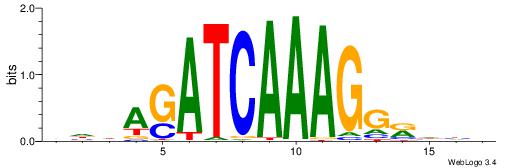
VDDAGATCAAAGGKDDD
--------AAAGATTT-
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
 |
 |
| Motif ID: | UP00208 |
| Motif name: | Obox5_3963.2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.015824 |
Alignment:
VRVAGGGATTAAHTVWV
---AAAGATTT------
| Original motif | Reverse complement motif |
| Consensus sequence: VRVAGGGATTAAHTVWV | Consensus sequence: VWBAHTTAATCCCTVKB |
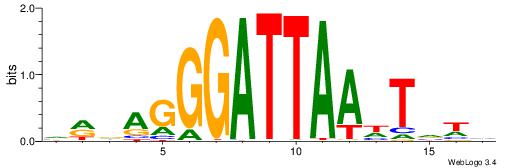 |
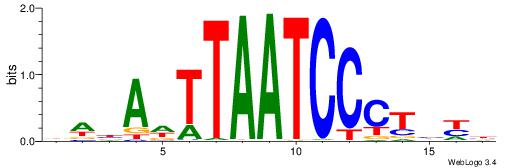 |
| Motif ID: | UP00167 |
| Motif name: | En1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0159173 |
Alignment:
VHRKTAATTAGTTVVV
-----AAATCTTT---
| Original motif | Reverse complement motif |
| Consensus sequence: VVVAACTAATTARKDV | Consensus sequence: VHRKTAATTAGTTVVV |
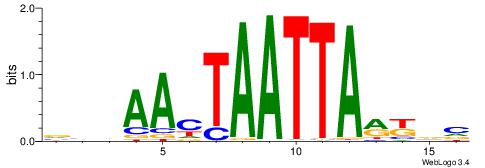 |
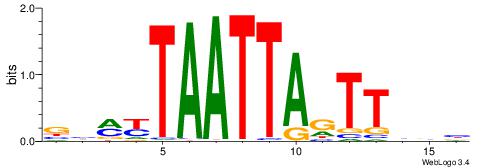 |
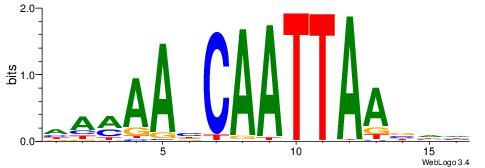
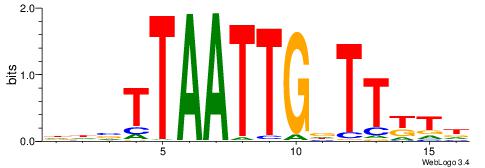
| Motif ID: | UP00145 |
| Motif name: | Barhl2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0164085 |
Alignment:
HHBTTAATTGDTTYTH
-----AAATCTTT---
| Original motif | Reverse complement motif |
| Consensus sequence: HAMAAHCAATTAABHH | Consensus sequence: HHBTTAATTGDTTYTH |
 |
 |
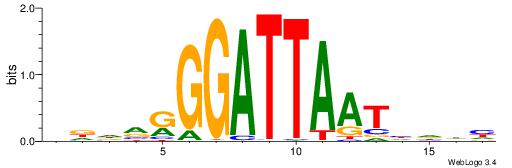
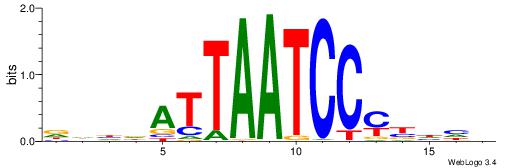
| Motif ID: | UP00125 |
| Motif name: | Pitx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0166506 |
Alignment:
DDDVRGGATTAATHDDH
---AAAGATTT------
| Original motif | Reverse complement motif |
| Consensus sequence: DDDVRGGATTAATHDDH | Consensus sequence: DDDDATTAATCCMBDHD |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0204535 |
Alignment:
HBHDVHDTCTTATCTHTDDHDV
---------AAATCTTT-----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Dataset #: 2 | Motif ID: 14 | Motif name: Motif 14 |
| Original motif | Reverse complement motif |
| Consensus sequence: AWAAATAA | Consensus sequence: TTATTTWT |
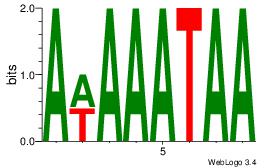 |
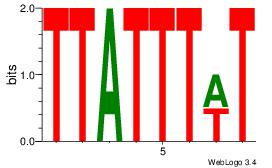 |
Best Matches for Motif ID 14 (Highest to Lowest)
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.00506466 |
Alignment:
HBHDVHDTCTTATCTHTDDHDV
---------TTATTTWT-----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.00546278 |
Alignment:
DWAATRTAAACAAWVVB
-----AWAAATAA----
| Original motif | Reverse complement motif |
| Consensus sequence: DWAATRTAAACAAWVVB | Consensus sequence: VVVWTTGTTTAMATTWD |
 |
 |
| Motif ID: | UP00118 |
| Motif name: | Pou4f3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.00832205 |
Alignment:
GHYHTYATTAATAAVH
----TTATTTWT----
| Original motif | Reverse complement motif |
| Consensus sequence: HVTTATTAATKADKHC | Consensus sequence: GHYHTYATTAATAAVH |
 |
 |
| Motif ID: | UP00037 |
| Motif name: | Zfp105_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.00912866 |
Alignment:
HHHTTRTTDTTTKDH
---TTATTTWT----
| Original motif | Reverse complement motif |
| Consensus sequence: HDYAAAHAAMAADHD | Consensus sequence: HHHTTRTTDTTTKDH |
 |
 |
| Motif ID: | UP00039 |
| Motif name: | Foxj3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0105685 |
Alignment:
HDHADGTAAACAAAVVH
-----AWAAATAA----
| Original motif | Reverse complement motif |
| Consensus sequence: HDHADGTAAACAAAVVH | Consensus sequence: DVVTTTGTTTACDTHDH |
 |
 |
| Motif ID: | UP00025 |
| Motif name: | Foxk1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0141517 |
Alignment:
DDAATGTAAACAAAVVB
-----AWAAATAA----
| Original motif | Reverse complement motif |
| Consensus sequence: DDAATGTAAACAAAVVB | Consensus sequence: BBVTTTGTTTACATTDD |
 |
 |
| Motif ID: | UP00180 |
| Motif name: | Hoxd13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0142824 |
Alignment:
HDHHTTTTATTKKHDD
------TTATTTWT--
| Original motif | Reverse complement motif |
| Consensus sequence: HDHYYAATAAAAHHHH | Consensus sequence: HDHHTTTTATTKKHDD |
 |
 |
| Motif ID: | UP00073 |
| Motif name: | Foxa2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0155566 |
Alignment:
HHWAWGTAAAYAAAVDB
-----AWAAATAA----
| Original motif | Reverse complement motif |
| Consensus sequence: HHWAWGTAAAYAAAVDB | Consensus sequence: BDVTTTKTTTACWTWHH |
 |
 |
| Motif ID: | UP00200 |
| Motif name: | Nkx6-1_2825.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0158439 |
Alignment:
DVHAATTAATTAMYBBG
-AWAAATAA--------
| Original motif | Reverse complement motif |
| Consensus sequence: DVHAATTAATTAMYBBG | Consensus sequence: CBVMRTAATTAATTHBH |
 |
 |
| Motif ID: | UP00105 |
| Motif name: | Pou3f4 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0159057 |
Alignment:
DVHTAATTAATTADDDB
--AWAAATAA-------
| Original motif | Reverse complement motif |
| Consensus sequence: DVHTAATTAATTADDDB | Consensus sequence: BDDDTAATTAATTAHBD |
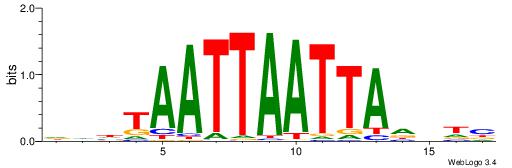 |
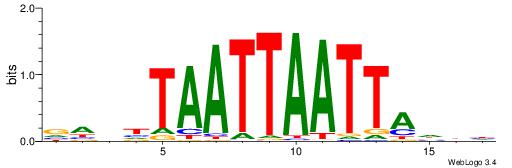 |
| Dataset #: 2 | Motif ID: 15 | Motif name: Motif 15 |
| Original motif | Reverse complement motif |
| Consensus sequence: ATMACAATAAAA | Consensus sequence: TTTTATTGTYAT |
 |
 |
Best Matches for Motif ID 15 (Highest to Lowest)
| Motif ID: | UP00180 |
| Motif name: | Hoxd13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.00878905 |
Alignment:
HDHHTTTTATTKKHDD
----TTTTATTGTYAT
| Original motif | Reverse complement motif |
| Consensus sequence: HDHYYAATAAAAHHHH | Consensus sequence: HDHHTTTTATTKKHDD |
 |
 |
| Motif ID: | UP00134 |
| Motif name: | Hoxb13 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0162131 |
Alignment:
VRHCCAATAAAAWHHV
ATMACAATAAAA----
| Original motif | Reverse complement motif |
| Consensus sequence: VRHCCAATAAAAWHHV | Consensus sequence: VDHWTTTTATTGGDKB |
 |
 |
| Motif ID: | UP00121 |
| Motif name: | Hoxd10 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0246749 |
Alignment:
VDKVYAATAAAATDDDH
ATMACAATAAAA-----
| Original motif | Reverse complement motif |
| Consensus sequence: VDKVYAATAAAATDDDH | Consensus sequence: HDDDATTTTATTKVRDB |
 |
 |
| Motif ID: | UP00091 |
| Motif name: | Sox5_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0247927 |
Alignment:
DHDDWATTGTTCDDHD
-TTTTATTGTYAT---
| Original motif | Reverse complement motif |
| Consensus sequence: DHDDGAACAATWDDHD | Consensus sequence: DHDDWATTGTTCDDHD |
 |
 |
| Motif ID: | UP00207 |
| Motif name: | Hoxb9 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 0.0248315 |
Alignment:
VRRGCMATAAAAHHHD
ATMACAATAAAA----
| Original motif | Reverse complement motif |
| Consensus sequence: VRRGCMATAAAAHHHD | Consensus sequence: HDHHTTTTATRGCKMV |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 12 |
| Similarity score: | 0.0268436 |
Alignment:
HHBDHDDAACAATDRHHHHHBW
-----ATMACAATAAAA-----
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00051 |
| Motif name: | Sox8_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.02954 |
Alignment:
DDATBWATTGTTHHDDD
--TTTTATTGTYAT---
| Original motif | Reverse complement motif |
| Consensus sequence: DDATBWATTGTTHHDDD | Consensus sequence: DDDHDAACAATWBATDD |
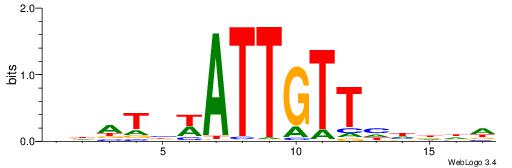 |
 |
| Motif ID: | UP00064 |
| Motif name: | Sox18_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0314917 |
Alignment:
HHDHABAACAATHBWV
TTTTATTGTYAT----
| Original motif | Reverse complement motif |
| Consensus sequence: BWBHATTGTTBTHDHH | Consensus sequence: HHDHABAACAATHBWV |
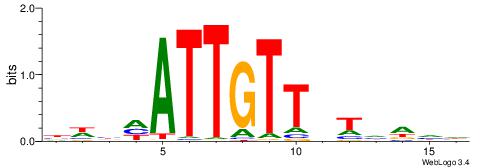 |
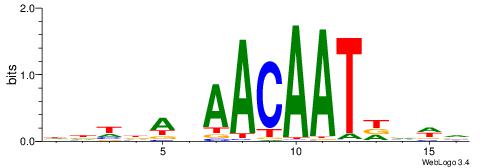 |
| Motif ID: | UP00096 |
| Motif name: | Sox13_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0318712 |
Alignment:
DHDRGAACAATWWDHW
---ATMACAATAAAA-
| Original motif | Reverse complement motif |
| Consensus sequence: DHDRGAACAATWWDHW | Consensus sequence: WHDWWATTGTTCKDHD |
 |
 |
| Motif ID: | UP00014 |
| Motif name: | Sox17_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0332895 |
Alignment:
DDWTBAATTGTTDDB
--TTTTATTGTYAT-
| Original motif | Reverse complement motif |
| Consensus sequence: VDDAACAATTVAWHD | Consensus sequence: DDWTBAATTGTTDDB |
 |
 |
| Dataset #: 2 | Motif ID: 16 | Motif name: Motif 16 |
| Original motif | Reverse complement motif |
| Consensus sequence: ACAAWTRATTTTGA | Consensus sequence: TCAAAATKAWTTGT |
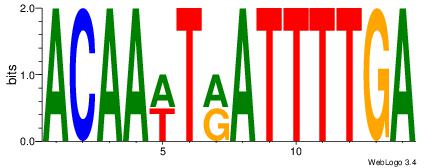 |
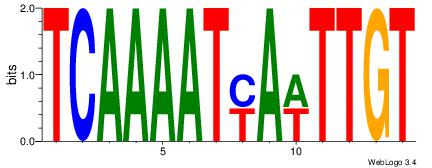 |
Best Matches for Motif ID 16 (Highest to Lowest)
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0530035 |
Alignment:
HHBDHDDAACAATDRHHHHHBW
--------ACAAWTRATTTTGA
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00158 |
| Motif name: | Pou1f1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.059328 |
Alignment:
BDHDTAATTAATTADBH
-TCAAAATKAWTTGT--
| Original motif | Reverse complement motif |
| Consensus sequence: DVDTAATTAATTADDDB | Consensus sequence: BDHDTAATTAATTADBH |
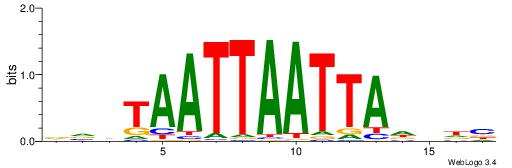 |
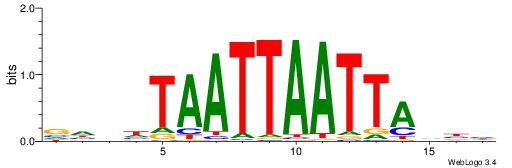 |
| Motif ID: | UP00128 |
| Motif name: | Pou3f2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0618019 |
Alignment:
HHDVMTAATTAATTDDV
--TCAAAATKAWTTGT-
| Original motif | Reverse complement motif |
| Consensus sequence: VDDAATTAATTARBDHD | Consensus sequence: HHDVMTAATTAATTDDV |
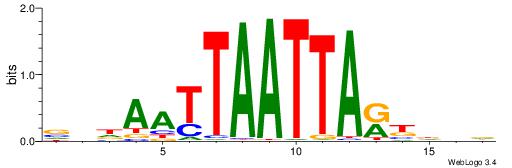 |
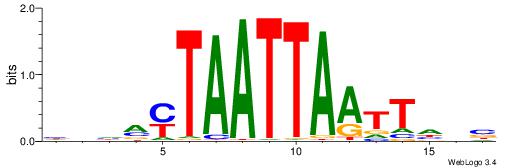 |
| Motif ID: | UP00105 |
| Motif name: | Pou3f4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0620147 |
Alignment:
BDDDTAATTAATTAHBD
-TCAAAATKAWTTGT--
| Original motif | Reverse complement motif |
| Consensus sequence: DVHTAATTAATTADDDB | Consensus sequence: BDDDTAATTAATTAHBD |
 |
 |
| Motif ID: | UP00200 |
| Motif name: | Nkx6-1_2825.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0623004 |
Alignment:
CBVMRTAATTAATTHBH
--TCAAAATKAWTTGT-
| Original motif | Reverse complement motif |
| Consensus sequence: DVHAATTAATTAMYBBG | Consensus sequence: CBVMRTAATTAATTHBH |
 |
 |
| Motif ID: | UP00167 |
| Motif name: | En1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0634894 |
Alignment:
VVVAACTAATTARKDV
-ACAAWTRATTTTGA-
| Original motif | Reverse complement motif |
| Consensus sequence: VVVAACTAATTARKDV | Consensus sequence: VHRKTAATTAGTTVVV |
 |
 |
| Motif ID: | UP00146 |
| Motif name: | Pou6f1_3733.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0650456 |
Alignment:
VHHVATAATGAGSTDDH
--TCAAAATKAWTTGT-
| Original motif | Reverse complement motif |
| Consensus sequence: VHHVATAATGAGSTDDH | Consensus sequence: DHDASCTCATTATVHHB |
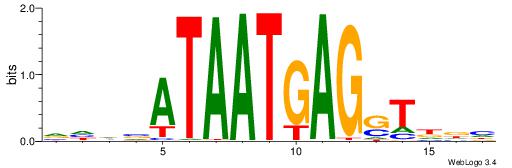 |
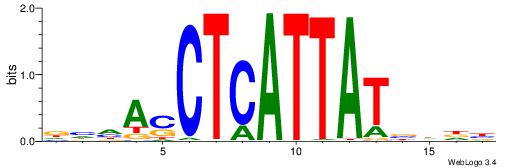 |
| Motif ID: | UP00145 |
| Motif name: | Barhl2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0651148 |
Alignment:
HAMAAHCAATTAABHH
-ACAAWTRATTTTGA-
| Original motif | Reverse complement motif |
| Consensus sequence: HAMAAHCAATTAABHH | Consensus sequence: HHBTTAATTGDTTYTH |
 |
 |
| Motif ID: | UP00168 |
| Motif name: | Hoxd8 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0654698 |
Alignment:
HADCMATTAATTADTTA
--ACAAWTRATTTTGA-
| Original motif | Reverse complement motif |
| Consensus sequence: TAADTAATTAATRGHTH | Consensus sequence: HADCMATTAATTADTTA |
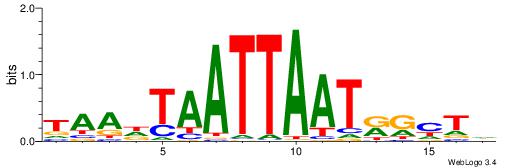 |
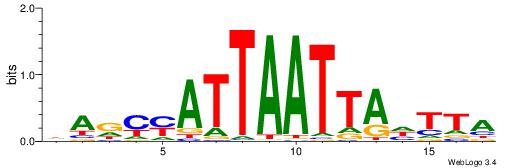 |
| Motif ID: | UP00129 |
| Motif name: | Pou3f1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0654983 |
Alignment:
DVBTAATTAATTAABTH
--ACAAWTRATTTTGA-
| Original motif | Reverse complement motif |
| Consensus sequence: DVBTAATTAATTAABTH | Consensus sequence: DAVTTAATTAATTAVBD |
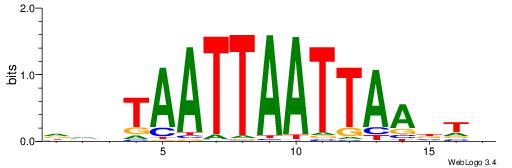 |
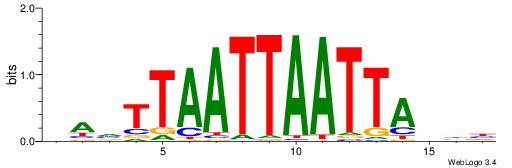 |
| Dataset #: 2 | Motif ID: 17 | Motif name: Motif 17 |
| Original motif | Reverse complement motif |
| Consensus sequence: AAAAATGAAT | Consensus sequence: ATTCATTTTT |
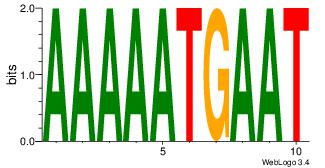 |
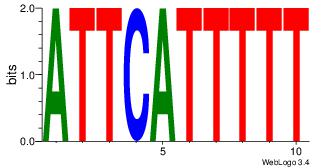 |
Best Matches for Motif ID 17 (Highest to Lowest)
| Motif ID: | UP00055 |
| Motif name: | Hbp1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.00216896 |
Alignment:
HHDWTSAATGAATDDB
---AAAAATGAAT---
| Original motif | Reverse complement motif |
| Consensus sequence: HHDWTSAATGAATDDB | Consensus sequence: VDHATTCATTSAWDDH |
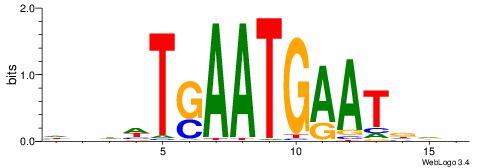 |
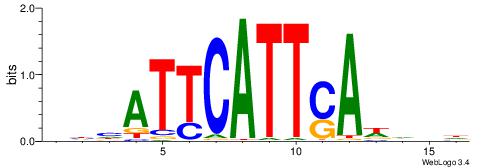 |
| Motif ID: | UP00145 |
| Motif name: | Barhl2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.010273 |
Alignment:
HAMAAHCAATTAABHH
AAAAATGAAT------
| Original motif | Reverse complement motif |
| Consensus sequence: HAMAAHCAATTAABHH | Consensus sequence: HHBTTAATTGDTTYTH |
 |
 |
| Motif ID: | UP00171 |
| Motif name: | Msx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0106868 |
Alignment:
HHVTTAATTKGTTHTV
--ATTCATTTTT----
| Original motif | Reverse complement motif |
| Consensus sequence: VAHAACYAATTAABHH | Consensus sequence: HHVTTAATTKGTTHTV |
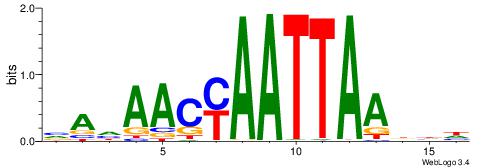 |
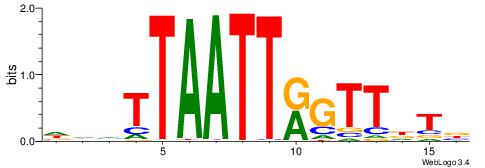 |
| Motif ID: | UP00044 |
| Motif name: | Mafk_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0131738 |
Alignment:
MBHTGCADTTTTTHV
--ATTCATTTTT---
| Original motif | Reverse complement motif |
| Consensus sequence: VHAAAAADTGCAHBR | Consensus sequence: MBHTGCADTTTTTHV |
 |
 |
| Motif ID: | UP00105 |
| Motif name: | Pou3f4 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0157045 |
Alignment:
DVHTAATTAATTADDDB
-AAAAATGAAT------
| Original motif | Reverse complement motif |
| Consensus sequence: DVHTAATTAATTADDDB | Consensus sequence: BDDDTAATTAATTAHBD |
 |
 |
| Motif ID: | UP00168 |
| Motif name: | Hoxd8 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0158473 |
Alignment:
HADCMATTAATTADTTA
-----ATTCATTTTT--
| Original motif | Reverse complement motif |
| Consensus sequence: TAADTAATTAATRGHTH | Consensus sequence: HADCMATTAATTADTTA |
 |
 |
| Motif ID: | UP00158 |
| Motif name: | Pou1f1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0161731 |
Alignment:
DVDTAATTAATTADDDB
-AAAAATGAAT------
| Original motif | Reverse complement motif |
| Consensus sequence: DVDTAATTAATTADDDB | Consensus sequence: BDHDTAATTAATTADBH |
 |
 |
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0163433 |
Alignment:
HBDDRAACAAAGRABHH
-----AAAAATGAAT--
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
 |
 |
| Motif ID: | UP00129 |
| Motif name: | Pou3f1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0163434 |
Alignment:
DVBTAATTAATTAABTH
-AAAAATGAAT------
| Original motif | Reverse complement motif |
| Consensus sequence: DVBTAATTAATTAABTH | Consensus sequence: DAVTTAATTAATTAVBD |
 |
 |
| Motif ID: | UP00200 |
| Motif name: | Nkx6-1_2825.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0167348 |
Alignment:
CBVMRTAATTAATTHBH
---AAAAATGAAT----
| Original motif | Reverse complement motif |
| Consensus sequence: DVHAATTAATTAMYBBG | Consensus sequence: CBVMRTAATTAATTHBH |
 |
 |
| Dataset #: 2 | Motif ID: 18 | Motif name: Motif 18 |
| Original motif | Reverse complement motif |
| Consensus sequence: TTAGWTWATAA | Consensus sequence: TTATWAWCTAA |
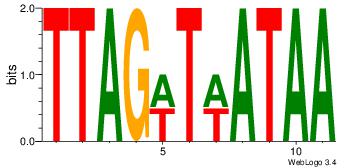 |
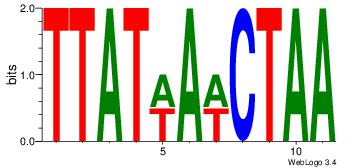 |
Best Matches for Motif ID 18 (Highest to Lowest)
| Motif ID: | UP00262 |
| Motif name: | Lhx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0358953 |
Alignment:
VVDATTAATTAAWWAHB
----TTAGWTWATAA--
| Original motif | Reverse complement motif |
| Consensus sequence: VVDATTAATTAAWWAHB | Consensus sequence: BHTWWTTAATTAATDVV |
 |
 |
| Motif ID: | UP00129 |
| Motif name: | Pou3f1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.038042 |
Alignment:
DAVTTAATTAATTAVBD
---TTAGWTWATAA---
| Original motif | Reverse complement motif |
| Consensus sequence: DVBTAATTAATTAABTH | Consensus sequence: DAVTTAATTAATTAVBD |
 |
 |
| Motif ID: | UP00082 |
| Motif name: | Zfp187_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0384021 |
Alignment:
BTATGTACTAATWH
---TTAGWTWATAA
| Original motif | Reverse complement motif |
| Consensus sequence: BTATGTACTAATWH | Consensus sequence: HWATTAGTACATAV |
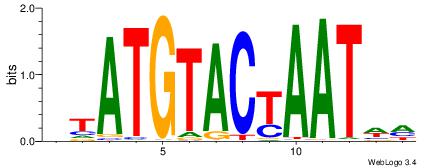 |
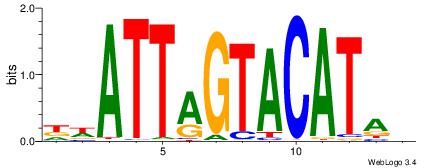 |
| Motif ID: | UP00146 |
| Motif name: | Pou6f1_1731.2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.0387939 |
Alignment:
DHDASCTCATTAWVVHV
------TTATWAWCTAA
| Original motif | Reverse complement motif |
| Consensus sequence: VHVVWTAATGAGSTDDH | Consensus sequence: DHDASCTCATTAWVVHV |
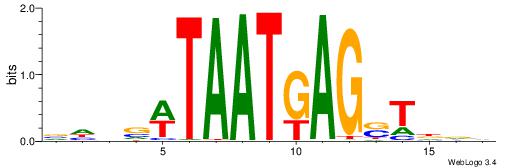 |
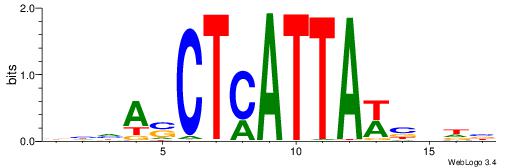 |
| Motif ID: | UP00169 |
| Motif name: | Lmx1b |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0395035 |
Alignment:
VDWWWTTAATTAATDHB
--TTATWAWCTAA----
| Original motif | Reverse complement motif |
| Consensus sequence: VDWWWTTAATTAATDHB | Consensus sequence: BHDATTAATTAAWWWHB |
 |
 |
| Motif ID: | UP00196 |
| Motif name: | Hoxa4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0396152 |
Alignment:
DDTHATTAATTAACKBG
--TTATWAWCTAA----
| Original motif | Reverse complement motif |
| Consensus sequence: DDTHATTAATTAACKBG | Consensus sequence: CVRGTTAATTAATHADH |
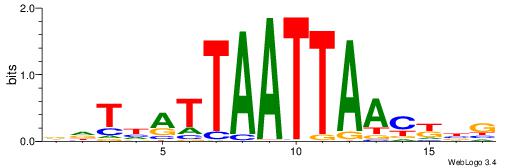 |
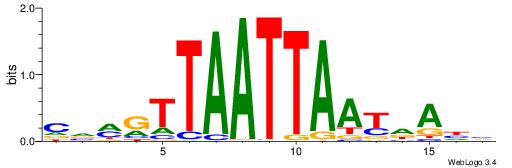 |
| Motif ID: | UP00142 |
| Motif name: | Uncx4.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.03971 |
Alignment:
VDHDATTAATTAABVVG
--TTATWAWCTAA----
| Original motif | Reverse complement motif |
| Consensus sequence: VDHDATTAATTAABVVG | Consensus sequence: CVVBTTAATTAATDHDV |
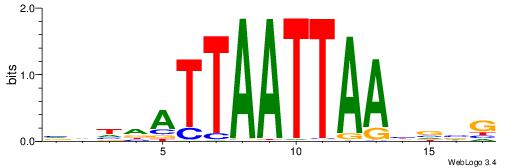 |
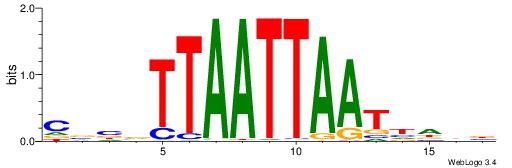 |
| Motif ID: | UP00264 |
| Motif name: | Hoxa1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0401139 |
Alignment:
MBDRTAATTAGCTHAD
----TTATWAWCTAA-
| Original motif | Reverse complement motif |
| Consensus sequence: HTDAGCTAATTAMHBY | Consensus sequence: MBDRTAATTAGCTHAD |
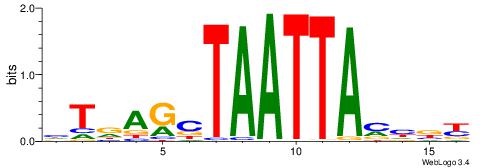 |
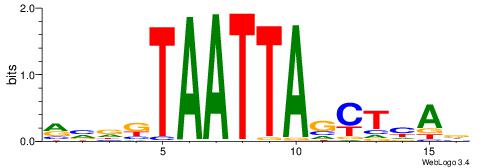 |
| Motif ID: | UP00023 |
| Motif name: | Sox30_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0405076 |
Alignment:
VVVYATTATAATBDHA
-----TTATWAWCTAA
| Original motif | Reverse complement motif |
| Consensus sequence: THDBATTATAATMVVV | Consensus sequence: VVVYATTATAATBDHA |
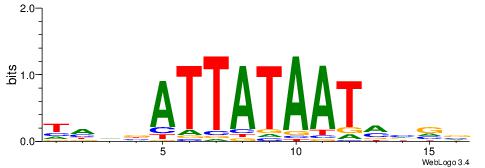 |
 |
| Motif ID: | UP00130 |
| Motif name: | Lhx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0405397 |
Alignment:
VDAATTAATTAAWWHHB
----TTAGWTWATAA--
| Original motif | Reverse complement motif |
| Consensus sequence: VDAATTAATTAAWWHHB | Consensus sequence: VHHWWTTAATTAATTDV |
 |
 |
| Dataset #: 2 | Motif ID: 19 | Motif name: Motif 19 |
| Original motif | Reverse complement motif |
| Consensus sequence: TTCWTAGATTAWA | Consensus sequence: TWTAATCTAWGAA |
 |
 |
Best Matches for Motif ID 19 (Highest to Lowest)
| Motif ID: | UP00208 |
| Motif name: | Obox5_2284.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 13 |
| Similarity score: | 0.0233931 |
Alignment:
DHHAHTTAATCCCKVDH
----TWTAATCTAWGAA
| Original motif | Reverse complement motif |
| Consensus sequence: HDVRGGGATTAAHTHHH | Consensus sequence: DHHAHTTAATCCCKVDH |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 13 |
| Similarity score: | 0.0251817 |
Alignment:
HBHDVHDTCTTATCTHTDDHDV
-------TWTAATCTAWGAA--
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Motif ID: | UP00208 |
| Motif name: | Obox5_3963.2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 13 |
| Similarity score: | 0.026815 |
Alignment:
VWBAHTTAATCCCTVKB
----TWTAATCTAWGAA
| Original motif | Reverse complement motif |
| Consensus sequence: VRVAGGGATTAAHTVWV | Consensus sequence: VWBAHTTAATCCCTVKB |
 |
 |
| Motif ID: | UP00188 |
| Motif name: | Lmx1a |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.02879 |
Alignment:
BDWWWTTAATTAAHHVG
-TTCWTAGATTAWA---
| Original motif | Reverse complement motif |
| Consensus sequence: CVHHTTAATTAAWWWHB | Consensus sequence: BDWWWTTAATTAAHHVG |
 |
 |
| Motif ID: | UP00109 |
| Motif name: | Obox6 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.028848 |
Alignment:
AHAADCGGATTAWHB
-TTCWTAGATTAWA-
| Original motif | Reverse complement motif |
| Consensus sequence: AHAADCGGATTAWHB | Consensus sequence: BHWTAATCCGDTTHT |
 |
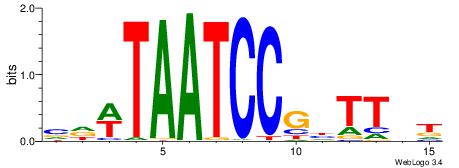 |
| Motif ID: | UP00262 |
| Motif name: | Lhx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0301327 |
Alignment:
BHTWWTTAATTAATDVV
-TTCWTAGATTAWA---
| Original motif | Reverse complement motif |
| Consensus sequence: VVDATTAATTAAWWAHB | Consensus sequence: BHTWWTTAATTAATDVV |
 |
 |
| Motif ID: | UP00112 |
| Motif name: | Gsc |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 13 |
| Similarity score: | 0.0301882 |
Alignment:
BHDMSGGATTAMMDWBH
TTCWTAGATTAWA----
| Original motif | Reverse complement motif |
| Consensus sequence: HVWHRYTAATCCSYDHV | Consensus sequence: BHDMSGGATTAMMDWBH |
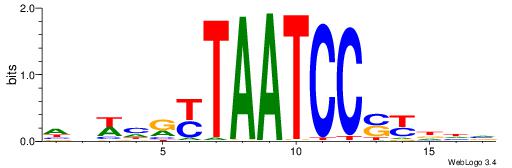 |
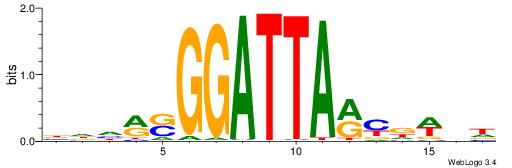 |
| Motif ID: | UP00169 |
| Motif name: | Lmx1b |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 13 |
| Similarity score: | 0.0307426 |
Alignment:
BHDATTAATTAAWWWHB
---TWTAATCTAWGAA-
| Original motif | Reverse complement motif |
| Consensus sequence: VDWWWTTAATTAATDHB | Consensus sequence: BHDATTAATTAAWWWHB |
 |
 |
| Motif ID: | UP00129 |
| Motif name: | Pou3f1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0308362 |
Alignment:
DVBTAATTAATTAABTH
-TWTAATCTAWGAA---
| Original motif | Reverse complement motif |
| Consensus sequence: DVBTAATTAATTAABTH | Consensus sequence: DAVTTAATTAATTAVBD |
 |
 |
| Motif ID: | UP00267 |
| Motif name: | Otx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.031113 |
Alignment:
DDHHATTAATCCCKDHD
----TWTAATCTAWGAA
| Original motif | Reverse complement motif |
| Consensus sequence: DDDRGGGATTAATHDDH | Consensus sequence: DDHHATTAATCCCKDHD |
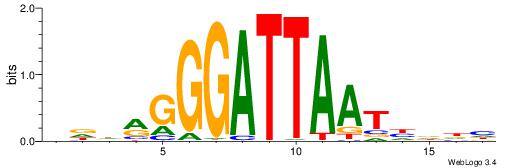 |
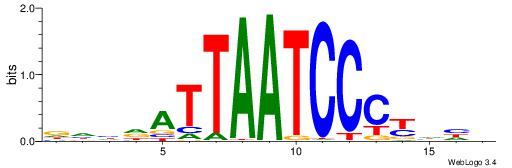 |
| Dataset #: 2 | Motif ID: 20 | Motif name: Motif 20 |
| Original motif | Reverse complement motif |
| Consensus sequence: TTMAAAGATTT | Consensus sequence: AAATCTTTYAA |
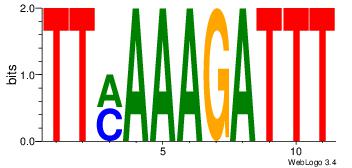 |
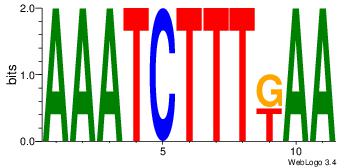 |
Best Matches for Motif ID 20 (Highest to Lowest)
| Motif ID: | UP00145 |
| Motif name: | Barhl2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0251004 |
Alignment:
HAMAAHCAATTAABHH
--AAATCTTTYAA---
| Original motif | Reverse complement motif |
| Consensus sequence: HAMAAHCAATTAABHH | Consensus sequence: HHBTTAATTGDTTYTH |
 |
 |
| Motif ID: | UP00258 |
| Motif name: | Tgif2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0274736 |
Alignment:
AABBAGCTGTCAAWVV
--AAATCTTTYAA---
| Original motif | Reverse complement motif |
| Consensus sequence: AABBAGCTGTCAAWVV | Consensus sequence: VBWTTGACAGCTVBTT |
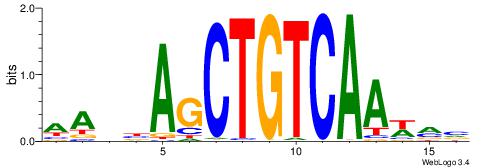 |
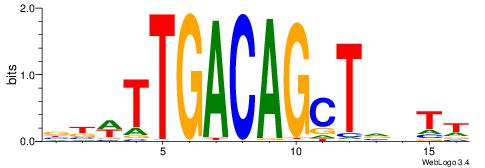 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0280825 |
Alignment:
VDDAGATCAAAGGKDDD
-----TTMAAAGATTT-
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
 |
 |
| Motif ID: | UP00203 |
| Motif name: | Pknox1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0285392 |
Alignment:
DRVSASCTGTCAAWVV
--AAATCTTTYAA---
| Original motif | Reverse complement motif |
| Consensus sequence: DRVSASCTGTCAAWVV | Consensus sequence: VVWTTGACAGSTSBKD |
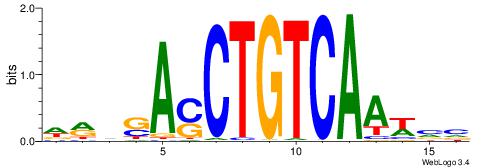 |
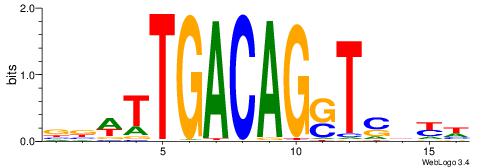 |
| Motif ID: | UP00186 |
| Motif name: | Meis1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0294621 |
Alignment:
WAVBASCTGTCAAWVH
--AAATCTTTYAA---
| Original motif | Reverse complement motif |
| Consensus sequence: WAVBASCTGTCAAWVH | Consensus sequence: DBWTTGACAGSTBVTW |
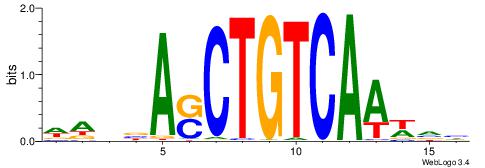 |
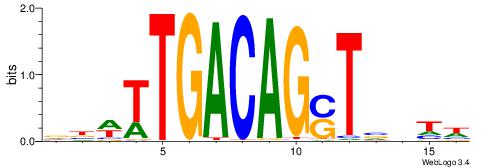 |
| Motif ID: | UP00226 |
| Motif name: | Mrg1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0296127 |
Alignment:
WAVBASCTGTCAAWHB
--AAATCTTTYAA---
| Original motif | Reverse complement motif |
| Consensus sequence: WAVBASCTGTCAAWHB | Consensus sequence: BHWTTGACAGSTBBTW |
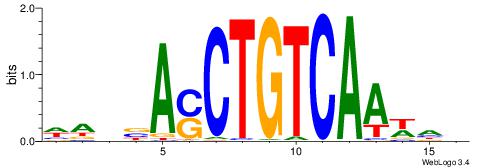 |
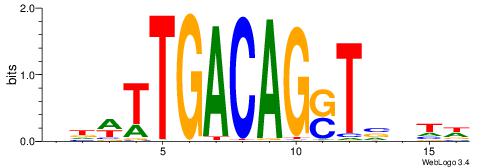 |
| Motif ID: | UP00055 |
| Motif name: | Hbp1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.030034 |
Alignment:
VDHATTCATTSAWDDH
--AAATCTTTYAA---
| Original motif | Reverse complement motif |
| Consensus sequence: HHDWTSAATGAATDDB | Consensus sequence: VDHATTCATTSAWDDH |
 |
 |
| Motif ID: | UP00148 |
| Motif name: | Hdx |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0308277 |
Alignment:
BDBDHTGATTTCDHYDD
------AAATCTTTYAA
| Original motif | Reverse complement motif |
| Consensus sequence: DDKDHGAAATCAHHBHV | Consensus sequence: BDBDHTGATTTCDHYDD |
 |
 |
| Motif ID: | UP00059 |
| Motif name: | Arid5a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0312649 |
Alignment:
HDTDVCAATATTMV
--TTMAAAGATTT-
| Original motif | Reverse complement motif |
| Consensus sequence: VYAATATTGVDADH | Consensus sequence: HDTDVCAATATTMV |
 |
 |
| Motif ID: | UP00012 |
| Motif name: | Bbx_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0314419 |
Alignment:
HWBTTCAWTGAADTH
---TTMAAAGATTT-
| Original motif | Reverse complement motif |
| Consensus sequence: HADTTCAWTGAABWD | Consensus sequence: HWBTTCAWTGAADTH |
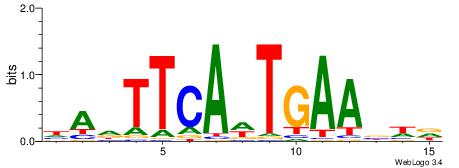 |
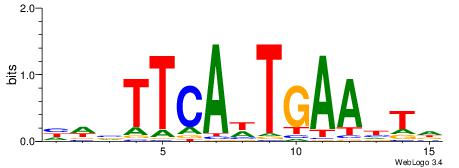 |
| Dataset #: 2 | Motif ID: 21 | Motif name: Motif 21 |
| Original motif | Reverse complement motif |
| Consensus sequence: ATAAAA | Consensus sequence: TTTTAT |
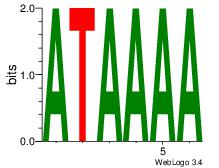 |
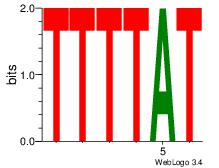 |
Best Matches for Motif ID 21 (Highest to Lowest)
| Motif ID: | UP00180 |
| Motif name: | Hoxd13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
HDHHTTTTATTKKHDD
----TTTTAT------
| Original motif | Reverse complement motif |
| Consensus sequence: HDHYYAATAAAAHHHH | Consensus sequence: HDHHTTTTATTKKHDD |
 |
 |
| Motif ID: | UP00134 |
| Motif name: | Hoxb13 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.00902808 |
Alignment:
VRHCCAATAAAAWHHV
------ATAAAA----
| Original motif | Reverse complement motif |
| Consensus sequence: VRHCCAATAAAAWHHV | Consensus sequence: VDHWTTTTATTGGDKB |
 |
 |
| Motif ID: | UP00207 |
| Motif name: | Hoxb9 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.00908997 |
Alignment:
VRRGCMATAAAAHHHD
------ATAAAA----
| Original motif | Reverse complement motif |
| Consensus sequence: VRRGCMATAAAAHHHD | Consensus sequence: HDHHTTTTATRGCKMV |
 |
 |
| Motif ID: | UP00121 |
| Motif name: | Hoxd10 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0141617 |
Alignment:
VDKVYAATAAAATDDDH
------ATAAAA-----
| Original motif | Reverse complement motif |
| Consensus sequence: VDKVYAATAAAATDDDH | Consensus sequence: HDDDATTTTATTKVRDB |
 |
 |
| Motif ID: | UP00133 |
| Motif name: | Cdx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.0156625 |
Alignment:
DVVGGYMATAAAAHKH
-------ATAAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: DVVGGYMATAAAAHKH | Consensus sequence: HRHTTTTATYMCCVBD |
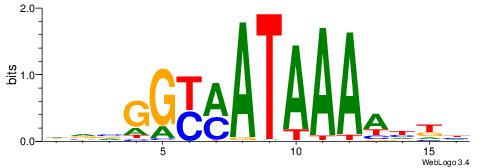 |
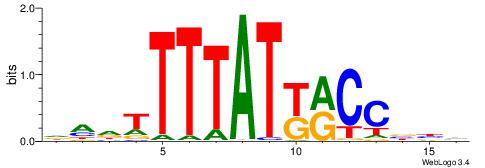 |
| Motif ID: | UP00213 |
| Motif name: | Hoxa9 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0225404 |
Alignment:
WDTAWTTTWATGKCCGD
-----TTTTAT------
| Original motif | Reverse complement motif |
| Consensus sequence: DCGGYCATWAAAWTADW | Consensus sequence: WDTAWTTTWATGKCCGD |
 |
 |
| Motif ID: | UP00217 |
| Motif name: | Hoxa10 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0259142 |
Alignment:
WDDATTTTATTMCBHD
----TTTTAT------
| Original motif | Reverse complement motif |
| Consensus sequence: DHBGYAATAAAATDHW | Consensus sequence: WDDATTTTATTMCBHD |
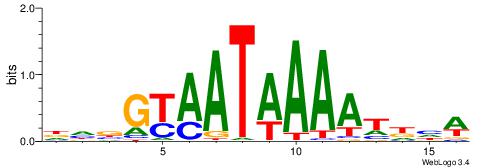 |
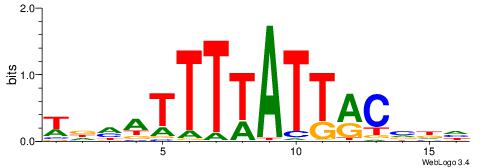 |
| Motif ID: | UP00240 |
| Motif name: | Cdx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.026022 |
Alignment:
DDVGGTMATAAAABKH
-------ATAAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: DDVGGTMATAAAABKH | Consensus sequence: HRVTTTTATYACCBDD |
 |
 |
| Motif ID: | UP00173 |
| Motif name: | Hoxc13 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.033825 |
Alignment:
HHDBCTCRTAAAAWHD
-------ATAAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: HHDBCTCRTAAAAWHD | Consensus sequence: DHWTTTTAMGAGBDHH |
 |
 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.0353549 |
Alignment:
DDDYCCTTTGATCTDDV
------TTTTAT-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
 |
 |
| Dataset #: 3 | Motif ID: 22 | Motif name: Zfx |
| Original motif | Reverse complement motif |
| Consensus sequence: BBVGCCBVGGCCTV | Consensus sequence: VAGGCCBBGGCVBB |
 |
 |
Best Matches for Motif ID 22 (Highest to Lowest)
| Motif ID: | UP00005 |
| Motif name: | Tcfap2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0596136 |
Alignment:
HHRCCBBWGGDCDB
BBVGCCBVGGCCTV
| Original motif | Reverse complement motif |
| Consensus sequence: HHRCCBBWGGDCDB | Consensus sequence: BDGHCCWBVGGKDH |
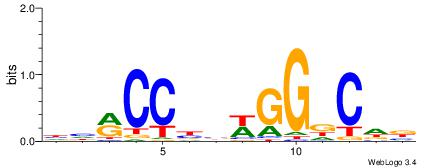 |
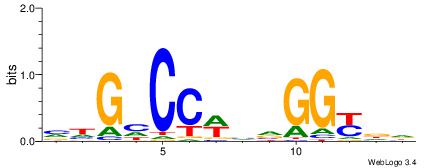 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0603516 |
Alignment:
ABBBBVVRGACCACCCACRDBBM
BBVGCCBVGGCCTV---------
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0604694 |
Alignment:
BDHHMCGCCCCCTHVBB
---VAGGCCBBGGCVBB
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0606067 |
Alignment:
HBDRCCMCGCCCHHHD
BBVGCCBVGGCCTV--
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 14 |
| Similarity score: | 0.0606878 |
Alignment:
HVGVSAGGAGGGTCYHHHHYVH
--------VAGGCCBBGGCVBB
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
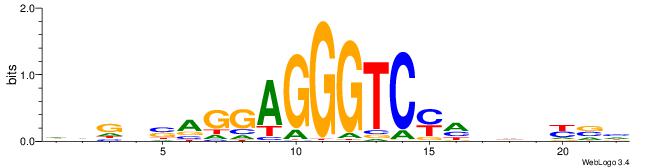 |
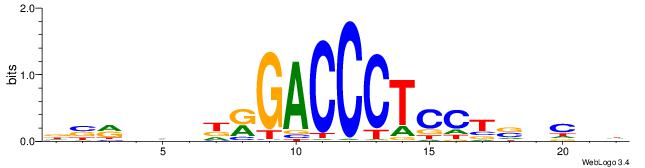 |
| Motif ID: | UP00087 |
| Motif name: | Tcfap2c_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0608063 |
Alignment:
HWGCCCTDGGGCDB
VAGGCCBBGGCVBB
| Original motif | Reverse complement motif |
| Consensus sequence: BHGCCCDAGGGCWD | Consensus sequence: HWGCCCTDGGGCDB |
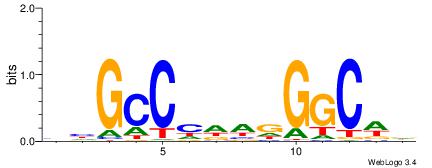 |
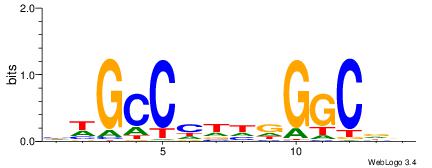 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 14 |
| Similarity score: | 0.0615001 |
Alignment:
DVTTVVCVYGGHGGYCHHDMYB
----BBVGCCBVGGCCTV----
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00010 |
| Motif name: | Tcfap2b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0624288 |
Alignment:
HHKGCCYSRGGCWAD
-BBVGCCBVGGCCTV
| Original motif | Reverse complement motif |
| Consensus sequence: DTWGCCKSMGGCRHH | Consensus sequence: HHKGCCYSRGGCWAD |
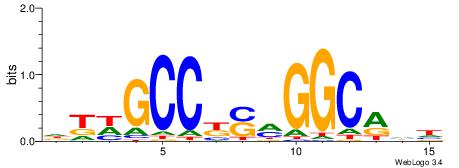 |
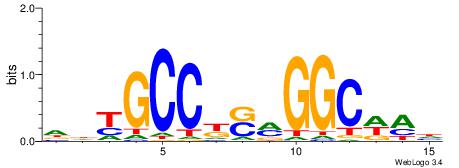 |
| Motif ID: | UP00082 |
| Motif name: | Zfp187_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0627664 |
Alignment:
VHMDGGACVAGGRCDH
VAGGCCBBGGCVBB--
| Original motif | Reverse complement motif |
| Consensus sequence: DDGMCCTBGTCCHYHV | Consensus sequence: VHMDGGACVAGGRCDH |
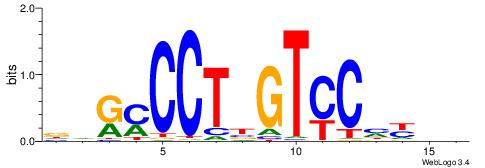 |
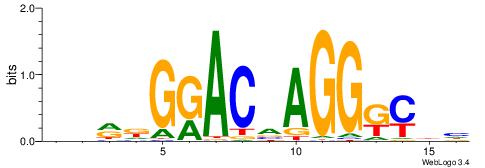 |
| Motif ID: | UP00009 |
| Motif name: | Nr2f2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0636529 |
Alignment:
HVBVTGACCCSGBVBV
--VAGGCCBBGGCVBB
| Original motif | Reverse complement motif |
| Consensus sequence: VBVBCSGGGTCAVBBH | Consensus sequence: HVBVTGACCCSGBVBV |
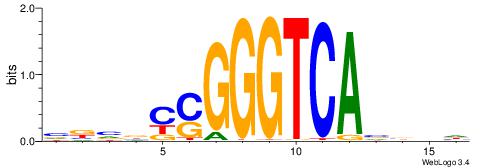 |
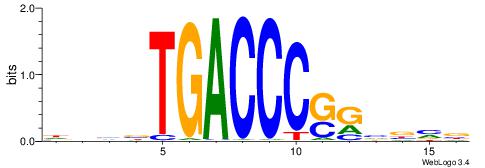 |
| Dataset #: 3 | Motif ID: 23 | Motif name: Egr1 |
| Original motif | Reverse complement motif |
| Consensus sequence: HGCGTGGGCGK | Consensus sequence: YCGCCCACGCH |
 |
 |
Best Matches for Motif ID 23 (Highest to Lowest)
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
------HGCGTGGGCGK------
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
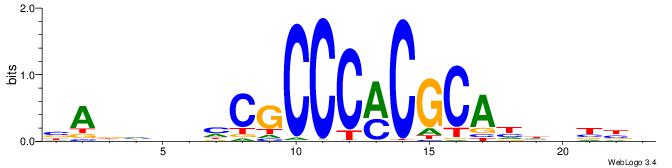 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.00727533 |
Alignment:
HCCGCCCCCGCAHB
-YCGCCCACGCH--
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 10 |
| Number of overlap: | 11 |
| Similarity score: | 0.0262579 |
Alignment:
MHHHWHYTMCCHCCCACVAABVH
---------YCGCCCACGCH---
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
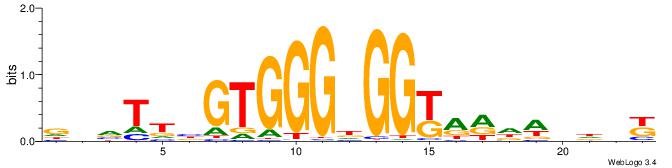 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0297053 |
Alignment:
YBVDMGTGGGTGGTCKVVBVBBT
--HGCGTGGGCGK----------
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0318144 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
----HGCGTGGGCGK--------
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
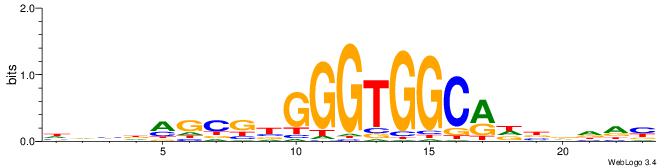 |
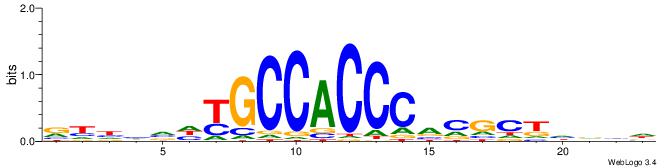 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0345009 |
Alignment:
BVVHAGGGGGCGRDHHB
--HGCGTGGGCGK----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0353502 |
Alignment:
BTBVTCVTGGGTGGTCMVVDVBB
---HGCGTGGGCGK---------
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0360229 |
Alignment:
BHHDTCCCACTCBDBV
--YCGCCCACGCH---
| Original motif | Reverse complement motif |
| Consensus sequence: BBHBGAGTGGGAHHDB | Consensus sequence: BHHDTCCCACTCBDBV |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 11 |
| Similarity score: | 0.0362743 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
--------YCGCCCACGCH----
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00097 |
| Motif name: | Mtf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0386944 |
Alignment:
HDYTTGCACACGDHBH
---YCGCCCACGCH--
| Original motif | Reverse complement motif |
| Consensus sequence: DBDHCGTGTGCAAMDH | Consensus sequence: HDYTTGCACACGDHBH |
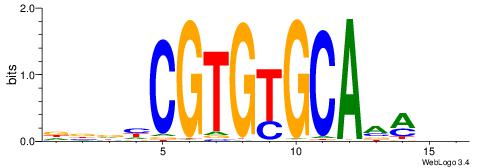 |
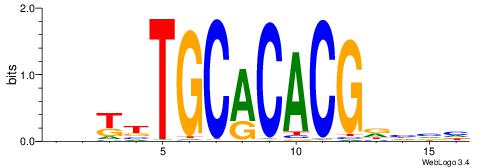 |
| Dataset #: 3 | Motif ID: 24 | Motif name: SP1 |
| Original motif | Reverse complement motif |
| Consensus sequence: CCCCKCCCCC | Consensus sequence: GGGGGYGGGG |
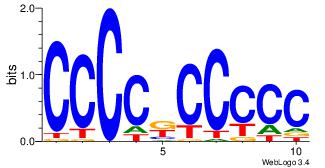 |
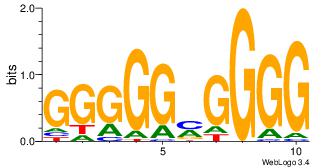 |
Best Matches for Motif ID 24 (Highest to Lowest)
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0231018 |
Alignment:
DBCCCCCCCCCCMYC
--CCCCKCCCCC---
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0303207 |
Alignment:
HBDRCCMCGCCCHHHD
----CCCCKCCCCC--
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0323007 |
Alignment:
HHHHDGGGGCGGDBHDV
----GGGGGYGGGG---
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
 |
 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0340973 |
Alignment:
HVDVGGGHGGGGDBHB
--GGGGGYGGGG----
| Original motif | Reverse complement motif |
| Consensus sequence: BHVDCCCCDCCCBDBH | Consensus sequence: HVDVGGGHGGGGDBHB |
 |
 |
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0351051 |
Alignment:
BHHHWGGGGCGGBBVH
----GGGGGYGGGG--
| Original motif | Reverse complement motif |
| Consensus sequence: HBBBCCGCCCCWHHHV | Consensus sequence: BHHHWGGGGCGGBBVH |
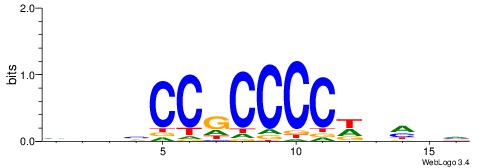 |
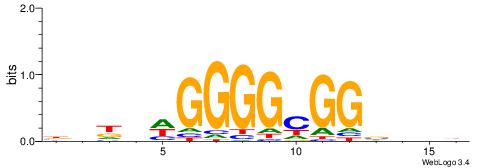 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0390437 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
------GGGGGYGGGG------
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0393358 |
Alignment:
HHBSHTCGTGRGTGGKGDBVWMV
-------GGGGGYGGGG------
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0413863 |
Alignment:
DHHBCCCCGCCAHHBHB
----CCCCKCCCCC---
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0418947 |
Alignment:
HVBCCCCCCCCMHHHB
---CCCCKCCCCC---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0420708 |
Alignment:
BDHHMCGCCCCCTHVBB
--CCCCKCCCCC-----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Dataset #: 3 | Motif ID: 25 | Motif name: TFAP2A |
| Original motif | Reverse complement motif |
| Consensus sequence: GCCBBVRGS | Consensus sequence: SCMVBBGGC |
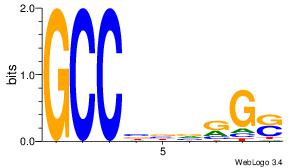 |
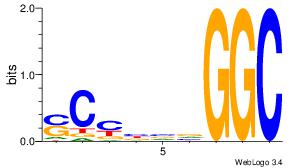 |
Best Matches for Motif ID 25 (Highest to Lowest)
| Motif ID: | UP00010 |
| Motif name: | Tcfap2b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 9 |
| Similarity score: | 0.0115546 |
Alignment:
HHKGCCYSRGGCWAD
---GCCBBVRGS---
| Original motif | Reverse complement motif |
| Consensus sequence: DTWGCCKSMGGCRHH | Consensus sequence: HHKGCCYSRGGCWAD |
 |
 |
| Motif ID: | UP00010 |
| Motif name: | Tcfap2b_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 9 |
| Similarity score: | 0.0150447 |
Alignment:
DYGCCYBARGGCAH
--GCCBBVRGS---
| Original motif | Reverse complement motif |
| Consensus sequence: DYGCCYBARGGCAH | Consensus sequence: HTGCCMTVKGGCMD |
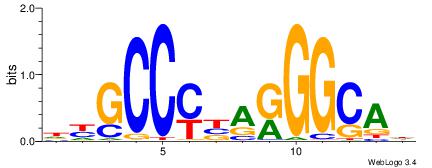 |
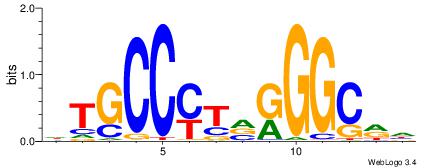 |
| Motif ID: | UP00005 |
| Motif name: | Tcfap2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 9 |
| Similarity score: | 0.0151527 |
Alignment:
WDHSCCYSRGGSRAD
---GCCBBVRGS---
| Original motif | Reverse complement motif |
| Consensus sequence: WDHSCCYSRGGSRAD | Consensus sequence: DTMSCCKSMGGSHDW |
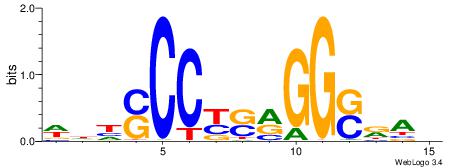 |
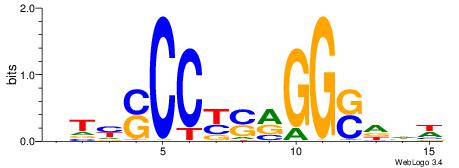 |
| Motif ID: | UP00087 |
| Motif name: | Tcfap2c_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 9 |
| Similarity score: | 0.016909 |
Alignment:
WHHSCCYSRGGSDAD
---GCCBBVRGS---
| Original motif | Reverse complement motif |
| Consensus sequence: WHHSCCYSRGGSDAD | Consensus sequence: DTHSCCKSMGGSHHW |
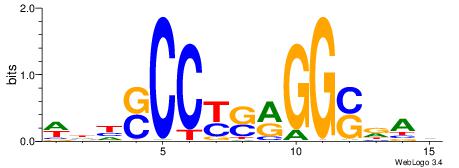 |
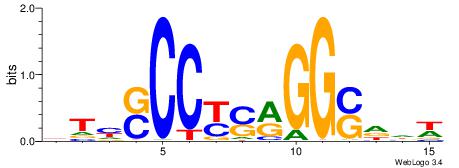 |
| Motif ID: | UP00087 |
| Motif name: | Tcfap2c_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 9 |
| Similarity score: | 0.0170259 |
Alignment:
BHGCCCDAGGGCWD
--GCCBBVRGS---
| Original motif | Reverse complement motif |
| Consensus sequence: BHGCCCDAGGGCWD | Consensus sequence: HWGCCCTDGGGCDB |
 |
 |
| Motif ID: | UP00028 |
| Motif name: | Tcfap2e_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 9 |
| Similarity score: | 0.0198854 |
Alignment:
HHHGCCTSAGGCDAD
---GCCBBVRGS---
| Original motif | Reverse complement motif |
| Consensus sequence: HHHGCCTSAGGCDAD | Consensus sequence: DTHGCCTSAGGCHHH |
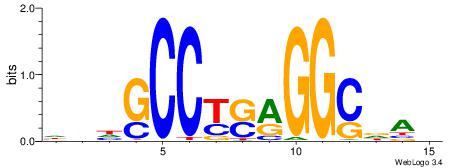 |
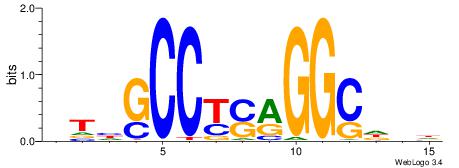 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 9 |
| Similarity score: | 0.0203681 |
Alignment:
BHBHDTGGCGGGGBDHD
SCMVBBGGC--------
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
 |
 |
| Motif ID: | UP00003 |
| Motif name: | E2F3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 9 |
| Similarity score: | 0.0217064 |
Alignment:
BBBDYTGGCGCCKWDBD
SCMVBBGGC--------
| Original motif | Reverse complement motif |
| Consensus sequence: HBHWYGGCGCCAMDVBB | Consensus sequence: BBBDYTGGCGCCKWDBD |
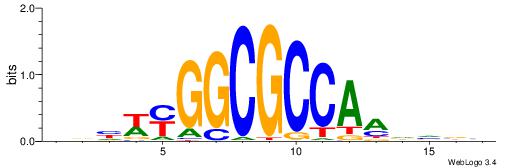 |
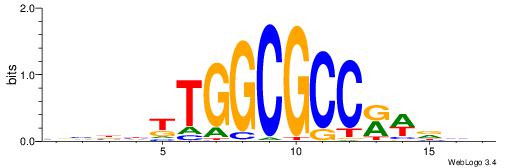 |
| Motif ID: | UP00005 |
| Motif name: | Tcfap2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 9 |
| Similarity score: | 0.0234775 |
Alignment:
HHRCCBBWGGDCDB
--GCCBBVRGS---
| Original motif | Reverse complement motif |
| Consensus sequence: HHRCCBBWGGDCDB | Consensus sequence: BDGHCCWBVGGKDH |
 |
 |
| Motif ID: | UP00093 |
| Motif name: | Klf7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 9 |
| Similarity score: | 0.026642 |
Alignment:
VVDBATACGCCCHAHHD
--------GCCBBVRGS
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBATACGCCCHAHHD | Consensus sequence: DHDTHGGGCGTATBHBB |
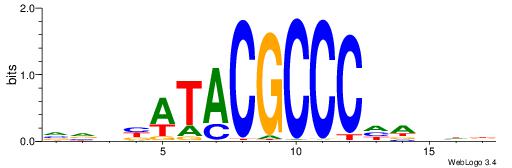 |
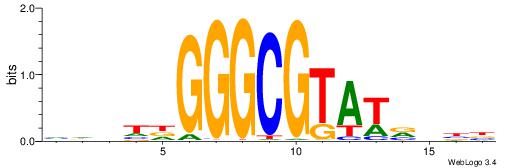 |
| Dataset #: 3 | Motif ID: 26 | Motif name: MIZF |
| Original motif | Reverse complement motif |
| Consensus sequence: BAACGTCCGC | Consensus sequence: GCGGACGTTV |
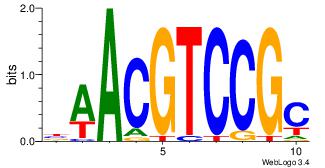 |
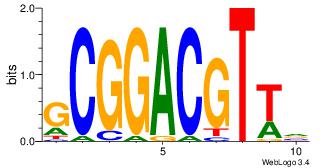 |
Best Matches for Motif ID 26 (Highest to Lowest)
| Motif ID: | UP00419 |
| Motif name: | Spic |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
DWACTTCCRCTTTB
BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: VAAAGMGGAAGTWD | Consensus sequence: DWACTTCCRCTTTB |
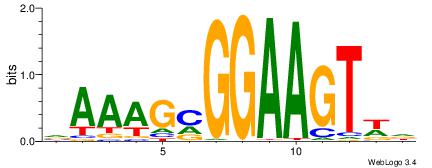 |
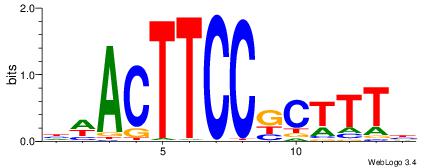 |
| Motif ID: | UP00409 |
| Motif name: | Elf5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.00379999 |
Alignment:
BWAHSMGGAAGTWD
BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: BWAHSMGGAAGTWD | Consensus sequence: DWACTTCCRSDTWV |
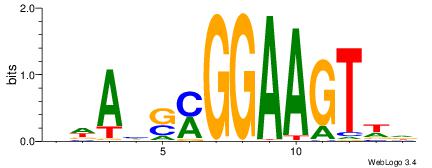 |
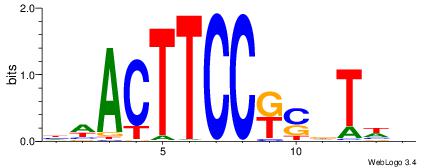 |
| Motif ID: | UP00413 |
| Motif name: | Elf4 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0124101 |
Alignment:
DBBHACTTCCGGKWTH
--BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGKWTH | Consensus sequence: HAWYCCGGAAGTHBBD |
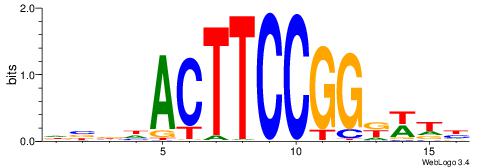 |
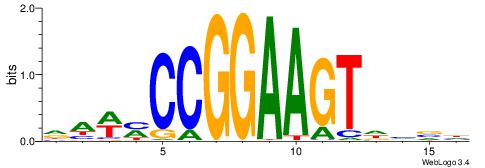 |
| Motif ID: | UP00404 |
| Motif name: | Elf2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0128348 |
Alignment:
DBBHACTTCCGGDWDB
--BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGDWDB | Consensus sequence: BDWDCCGGAAGTHBBD |
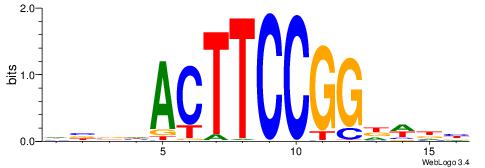 |
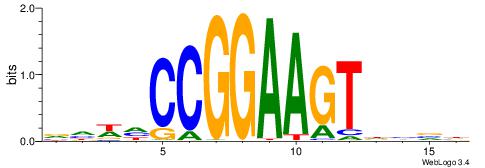 |
| Motif ID: | UP00085 |
| Motif name: | Sfpi1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0141516 |
Alignment:
HDACTTCCBCWTDV
BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: BDAWGVGGAAGTDH | Consensus sequence: HDACTTCCBCWTDV |
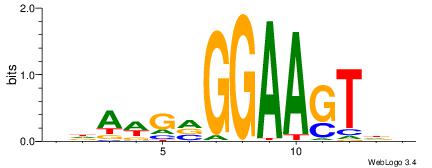 |
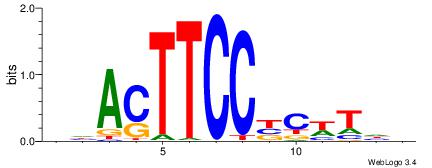 |
| Motif ID: | UP00423 |
| Motif name: | Gm5454 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0144665 |
Alignment:
DVHACCGGAAGTHDDV
----GCGGACGTTV--
| Original motif | Reverse complement motif |
| Consensus sequence: VDHHACTTCCGGTHVH | Consensus sequence: DVHACCGGAAGTHDDV |
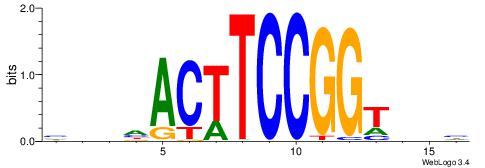 |
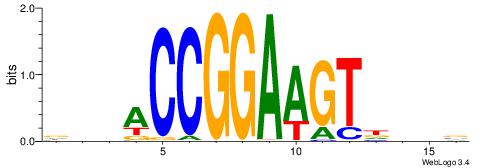 |
| Motif ID: | UP00412 |
| Motif name: | Etv5 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0155825 |
Alignment:
DHHACCGGAWGTTDDV
----GCGGACGTTV--
| Original motif | Reverse complement motif |
| Consensus sequence: VDHAACWTCCGGTHHH | Consensus sequence: DHHACCGGAWGTTDDV |
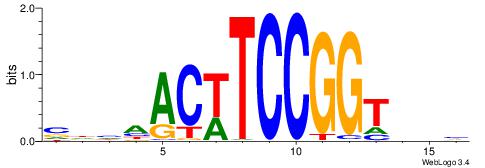 |
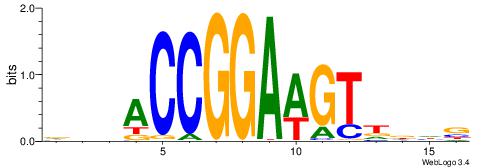 |
| Motif ID: | UP00015 |
| Motif name: | Ehf_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0162018 |
Alignment:
VBDABCCGGAAGTDD
-----GCGGACGTTV
| Original motif | Reverse complement motif |
| Consensus sequence: VBDABCCGGAAGTDD | Consensus sequence: DDACTTCCGGBTHBB |
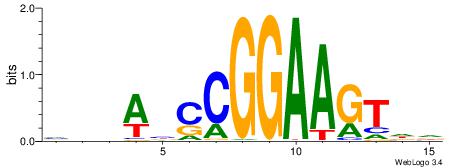 |
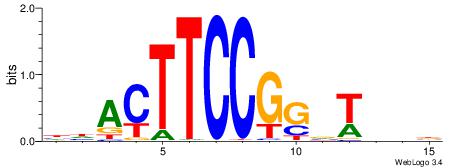 |
| Motif ID: | UP00418 |
| Motif name: | Etv6 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0171234 |
Alignment:
DVHBTACTTCCGGHTHB
---BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: DVHBTACTTCCGGHTHB | Consensus sequence: BHAHCCGGAAGTABDVD |
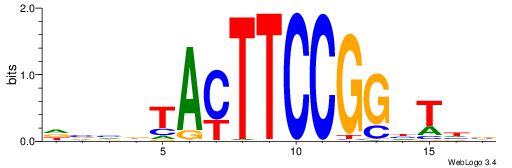 |
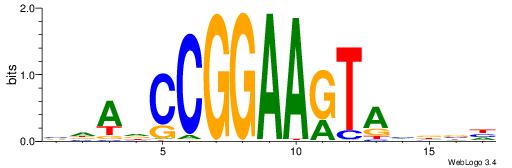 |
| Motif ID: | UP00421 |
| Motif name: | Etv1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0184147 |
Alignment:
MVHBACCGGAAGTVDVH
-----GCGGACGTTV--
| Original motif | Reverse complement motif |
| Consensus sequence: MVHBACCGGAAGTVDVH | Consensus sequence: HBDBACTTCCGGTBHVY |
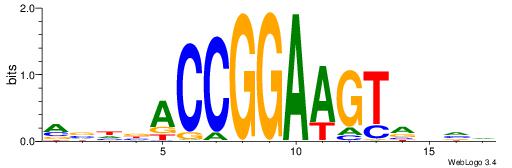 |
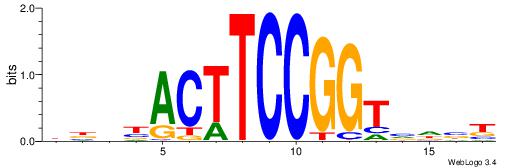 |
| Dataset #: 3 | Motif ID: 27 | Motif name: Klf4 |
| Original motif | Reverse complement motif |
| Consensus sequence: DGGGYGKGGC | Consensus sequence: GCCYCMCCCD |
 |
 |
Best Matches for Motif ID 27 (Highest to Lowest)
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
DHHDGGGCGRGGKHBH
---DGGGYGKGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0153308 |
Alignment:
DBCCCCCCCCCCMYC
---GCCYCMCCCD--
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0186791 |
Alignment:
BDWAGGCGTGBCHHD
--DGGGYGKGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: BDWAGGCGTGBCHHD | Consensus sequence: HHDGBCACGCCTWDB |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0218399 |
Alignment:
HVBCCCCCCCCMHHHB
--GCCYCMCCCD----
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0249586 |
Alignment:
HCCGCCCCCGCAHB
GCCYCMCCCD----
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0299159 |
Alignment:
BVVHAGGGGGCGRDHHB
------DGGGYGKGGC-
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0308595 |
Alignment:
BHBHDTGGCGGGGBDHD
----DGGGYGKGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
 |
 |
| Motif ID: | UP00047 |
| Motif name: | Zbtb7b_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0345891 |
Alignment:
VHWTTHGGGGGGVDD
GCCYCMCCCD-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDVCCCCCCHAAWHB | Consensus sequence: VHWTTHGGGGGGVDD |
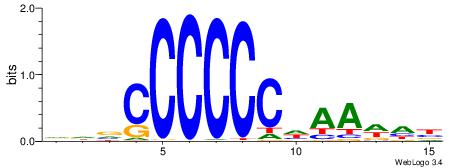 |
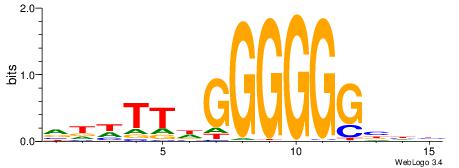 |
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0346735 |
Alignment:
HBBBCCGCCCCWHHHV
--GCCYCMCCCD----
| Original motif | Reverse complement motif |
| Consensus sequence: HBBBCCGCCCCWHHHV | Consensus sequence: BHHHWGGGGCGGBBVH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 10 |
| Similarity score: | 0.0351496 |
Alignment:
MHHHWHYTMCCHCCCACVAABVH
------GCCYCMCCCD-------
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Dataset #: 3 | Motif ID: 28 | Motif name: E2F1 |
| Original motif | Reverse complement motif |
| Consensus sequence: TTTSGCGC | Consensus sequence: GCGCSAAA |
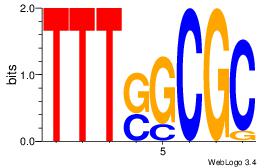 |
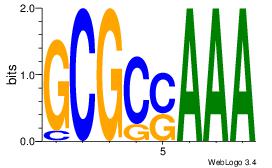 |
Best Matches for Motif ID 28 (Highest to Lowest)
| Motif ID: | UP00001 |
| Motif name: | E2F2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
BBBDTTGGCGCCKWVVD
---TTTSGCGC------
| Original motif | Reverse complement motif |
| Consensus sequence: HVBWYGGCGCCAADVBB | Consensus sequence: BBBDTTGGCGCCKWVVD |
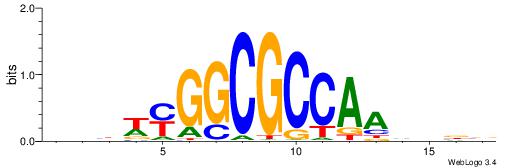 |
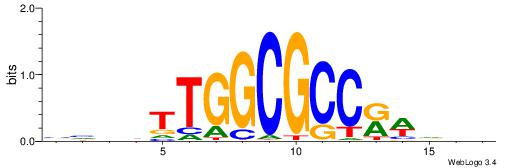 |
| Motif ID: | UP00003 |
| Motif name: | E2F3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.00182028 |
Alignment:
HBHWYGGCGCCAMDVBB
------GCGCSAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: HBHWYGGCGCCAMDVBB | Consensus sequence: BBBDYTGGCGCCKWDBD |
 |
 |
| Motif ID: | UP00072 |
| Motif name: | IRC900814_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0237639 |
Alignment:
DDHTACGACAAAWDDB
----GCGCSAAA----
| Original motif | Reverse complement motif |
| Consensus sequence: DDHTACGACAAAWDDB | Consensus sequence: BHDWTTTGTCGTAHDD |
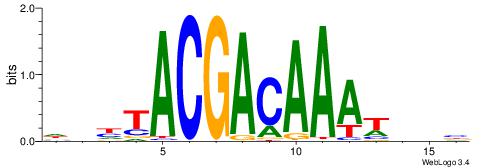 |
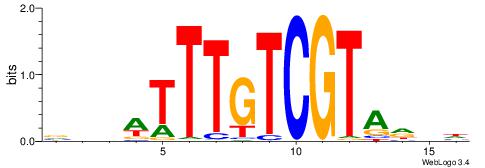 |
| Motif ID: | UP00003 |
| Motif name: | E2F3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0275636 |
Alignment:
WHSGCGCGCCHTDHB
-----GCGCSAAA--
| Original motif | Reverse complement motif |
| Consensus sequence: VHDADGGCGCGCSHW | Consensus sequence: WHSGCGCGCCHTDHB |
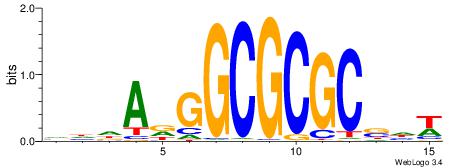 |
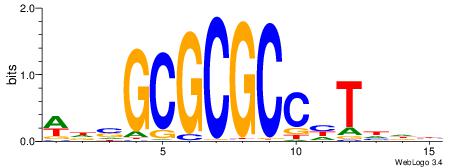 |
| Motif ID: | UP00065 |
| Motif name: | Zfp161_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.028109 |
Alignment:
DMMDGMGCGCGCGCHD
--------GCGCSAAA
| Original motif | Reverse complement motif |
| Consensus sequence: DDGCGCGCGCRCHYRD | Consensus sequence: DMMDGMGCGCGCGCHD |
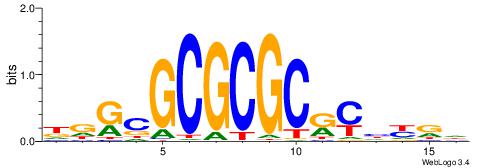 |
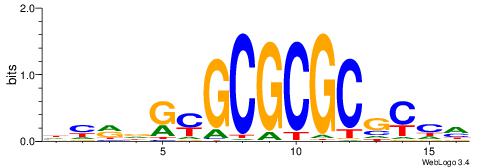 |
| Motif ID: | UP00001 |
| Motif name: | E2F2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0296302 |
Alignment:
HHVGCGCGCCKTDHB
-----GCGCSAAA--
| Original motif | Reverse complement motif |
| Consensus sequence: VHDARGGCGCGCVHH | Consensus sequence: HHVGCGCGCCKTDHB |
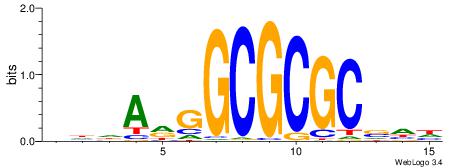 |
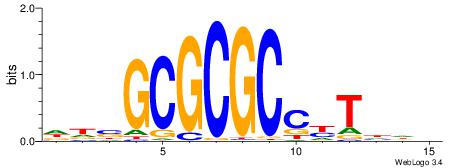 |
| Motif ID: | UP00068 |
| Motif name: | Eomes_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.0348969 |
Alignment:
BVHDAGGTGTGAAAHHH
------GCGCSAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: BVHDAGGTGTGAAAHHH | Consensus sequence: HHHTTTCACACCTDHBV |
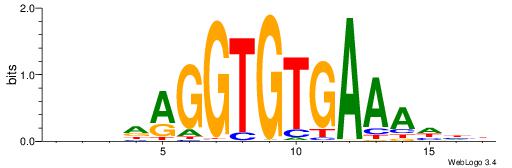 |
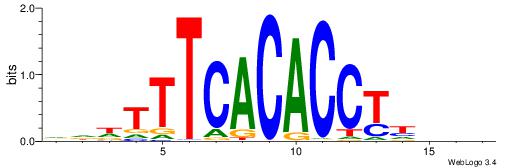 |
| Motif ID: | UP00011 |
| Motif name: | Irf6_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0368278 |
Alignment:
DHDBTKGTTTCGHTHDD
-------TTTSGCGC--
| Original motif | Reverse complement motif |
| Consensus sequence: HDDAHCGAAACYAVDDD | Consensus sequence: DHDBTKGTTTCGHTHDD |
 |
 |
| Motif ID: | UP00049 |
| Motif name: | Sp100_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0383813 |
Alignment:
DWTTTCCGHHWAWD
-----GCGCSAAA-
| Original motif | Reverse complement motif |
| Consensus sequence: DWTWHHCGGAAAWD | Consensus sequence: DWTTTCCGHHWAWD |
 |
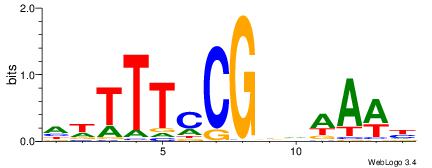 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.0406002 |
Alignment:
TCDAMVBGACGCMAKDDDVBBD
--------GCGCSAAA------
| Original motif | Reverse complement motif |
| Consensus sequence: DBBVDDDRTRGCGTCBBYTHGA | Consensus sequence: TCDAMVBGACGCMAKDDDVBBD |
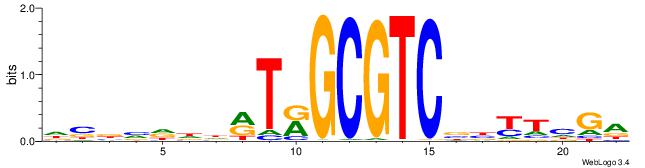 |
 |
| Dataset #: 3 | Motif ID: 29 | Motif name: HIF1AARNT |
| Original motif | Reverse complement motif |
| Consensus sequence: VBACGTGV | Consensus sequence: VCACGTBV |
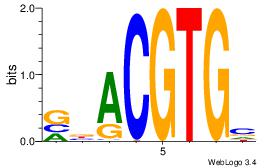 |
 |
Best Matches for Motif ID 29 (Highest to Lowest)
| Motif ID: | UP00097 |
| Motif name: | Mtf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HDYTTGCACACGDHBH
-------VCACGTBV-
| Original motif | Reverse complement motif |
| Consensus sequence: DBDHCGTGTGCAAMDH | Consensus sequence: HDYTTGCACACGDHBH |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.000574345 |
Alignment:
HHVBABCACGTGSTHD
-----VCACGTBV---
| Original motif | Reverse complement motif |
| Consensus sequence: DDASCACGTGBTBVDD | Consensus sequence: HHVBABCACGTGSTHD |
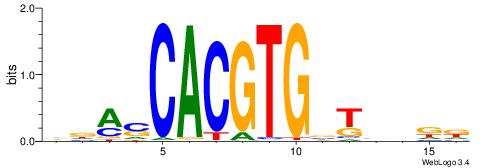 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.00642455 |
Alignment:
YDYBDHTMCACGTGGADDBMDGT
-------VBACGTGV--------
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00084 |
| Motif name: | Gmeb1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0131171 |
Alignment:
BVVDVRYACGTRYVBHB
-----VBACGTGV----
| Original motif | Reverse complement motif |
| Consensus sequence: BHBBKKACGTMMVDBVB | Consensus sequence: BVVDVRYACGTRYVBHB |
 |
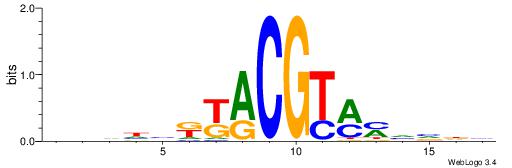 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.014006 |
Alignment:
RDHDBVDTCACGTGASBHVHDH
-------VCACGTBV-------
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
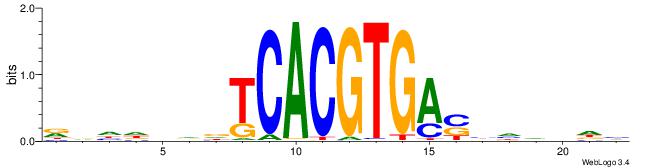 |
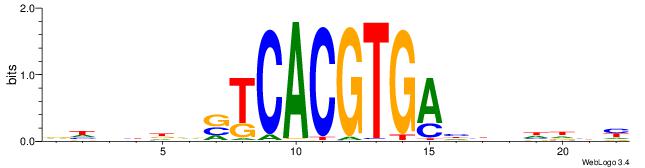 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0184296 |
Alignment:
VBHHVCAGGTGCDVDHD
----VBACGTGV-----
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
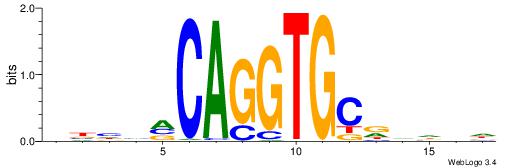 |
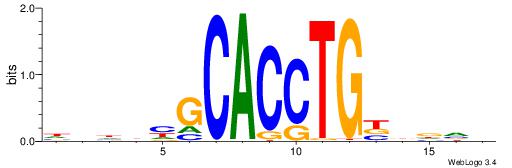 |
| Motif ID: | UP00068 |
| Motif name: | Eomes_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0188154 |
Alignment:
VVBVCKACACCTHVBV
------VCACGTBV--
| Original motif | Reverse complement motif |
| Consensus sequence: VBVDAGGTGTYGVBBV | Consensus sequence: VVBVCKACACCTHVBV |
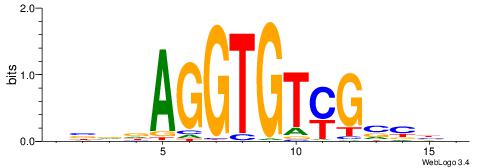 |
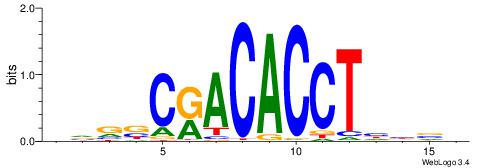 |
| Motif ID: | UP00026 |
| Motif name: | Zscan4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.0189568 |
Alignment:
DHVDTGTGCACAYAHDH
-VBACGTGV--------
| Original motif | Reverse complement motif |
| Consensus sequence: DHVDTGTGCACAYAHDH | Consensus sequence: HDHTMTGTGCACADVHD |
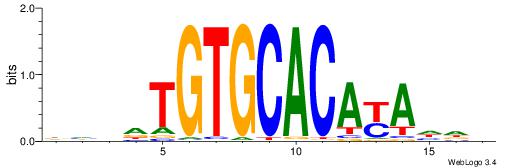 |
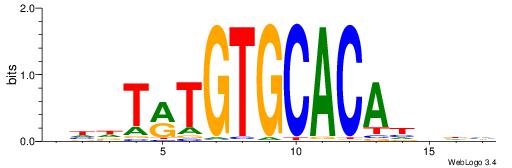 |
| Motif ID: | UP00042 |
| Motif name: | Gm397_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0198987 |
Alignment:
HHRTATGTGCACATHHD
--VBACGTGV-------
| Original motif | Reverse complement motif |
| Consensus sequence: HHDATGTGCACATAMDH | Consensus sequence: HHRTATGTGCACATHHD |
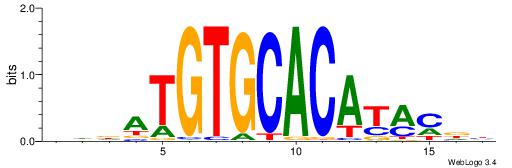 |
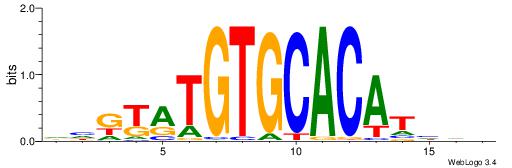 |
| Motif ID: | UP00060 |
| Motif name: | Max_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.021967 |
Alignment:
BBGHCACGCGACDD
---VCACGTBV---
| Original motif | Reverse complement motif |
| Consensus sequence: BBGHCACGCGACDD | Consensus sequence: HDGTCGCGTGDCVB |
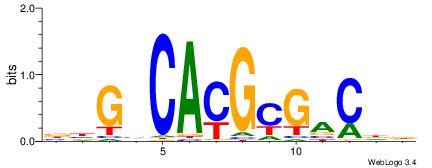 |
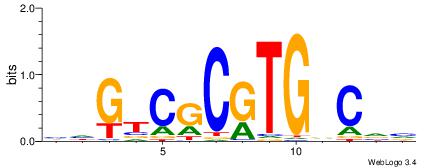 |
| Dataset #: 3 | Motif ID: 30 | Motif name: PLAG1 |
| Original motif | Reverse complement motif |
| Consensus sequence: GGGGCCCAAGGGGG | Consensus sequence: CCCCCTTGGGCCCC |
 |
 |
Best Matches for Motif ID 30 (Highest to Lowest)
| Motif ID: | UP00102 |
| Motif name: | Zic1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0499525 |
Alignment:
CCCCCCCGGGGGHB
CCCCCTTGGGCCCC
| Original motif | Reverse complement motif |
| Consensus sequence: BHCCCCCGGGGGGG | Consensus sequence: CCCCCCCGGGGGHB |
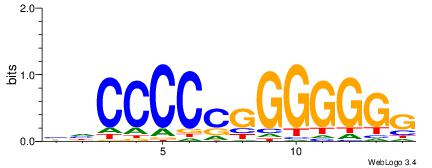 |
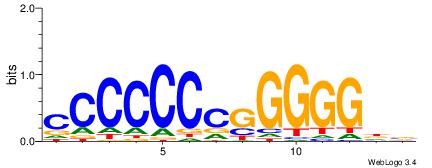 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0540613 |
Alignment:
BBBDVVRGACCACCCAVGABBAB
---------CCCCCTTGGGCCCC
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00057 |
| Motif name: | Zic2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0548411 |
Alignment:
VCCCCCCCGGGGGRB
-CCCCCTTGGGCCCC
| Original motif | Reverse complement motif |
| Consensus sequence: BMCCCCCGGGGGGGB | Consensus sequence: VCCCCCCCGGGGGRB |
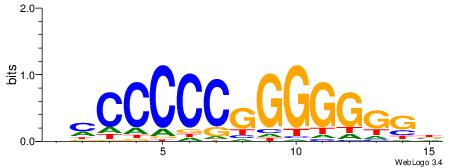 |
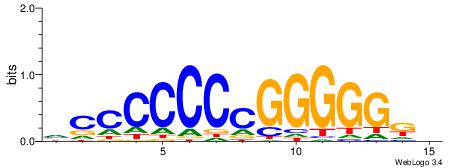 |
| Motif ID: | UP00005 |
| Motif name: | Tcfap2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0556785 |
Alignment:
HHRCCBBWGGDCDB
GGGGCCCAAGGGGG
| Original motif | Reverse complement motif |
| Consensus sequence: HHRCCBBWGGDCDB | Consensus sequence: BDGHCCWBVGGKDH |
 |
 |
| Motif ID: | UP00006 |
| Motif name: | Zic3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.056976 |
Alignment:
VCCCCCCCGGGGGRB
-CCCCCTTGGGCCCC
| Original motif | Reverse complement motif |
| Consensus sequence: BMCCCCCGGGGGGGB | Consensus sequence: VCCCCCCCGGGGGRB |
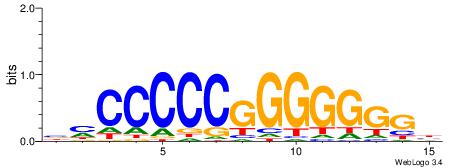 |
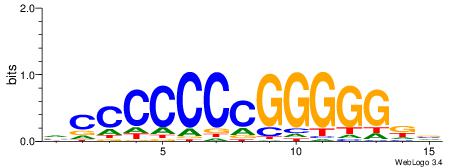 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 14 |
| Similarity score: | 0.0573495 |
Alignment:
HDGHHBHRGACCACCCGSVDVG
CCCCCTTGGGCCCC--------
| Original motif | Reverse complement motif |
| Consensus sequence: HDGHHBHRGACCACCCGSVDVG | Consensus sequence: CVHBSCGGGTGGTCMHVHHCDH |
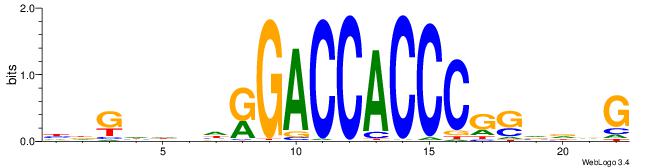 |
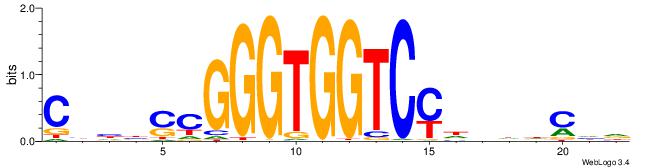 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 14 |
| Similarity score: | 0.0579022 |
Alignment:
VMYDHDGMCCHCCKBGVVAAVH
--------CCCCCTTGGGCCCC
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 14 |
| Similarity score: | 0.0580662 |
Alignment:
DVMHHDHKGACCCTCCTSVCBH
CCCCCTTGGGCCCC--------
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0597453 |
Alignment:
HGDHVHKGACCACCCGGBGBGDB
---------CCCCCTTGGGCCCC
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
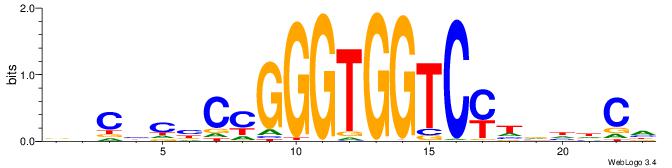 |
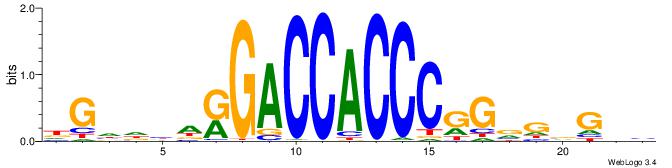 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0598088 |
Alignment:
DBCCCCCCCCCCMYC
-CCCCCTTGGGCCCC
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Dataset #: 3 | Motif ID: 31 | Motif name: Pax5 |
| Original motif | Reverse complement motif |
| Consensus sequence: DGVBCABTGDWGCGKRRCSR | Consensus sequence: MSGKKRCGCWDCABTGBBCD |
 |
 |
Best Matches for Motif ID 31 (Highest to Lowest)
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 20 |
| Similarity score: | 0.0499537 |
Alignment:
DDRMHBACGCGYGCGTDGBBDB
-DGVBCABTGDWGCGKRRCSR-
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
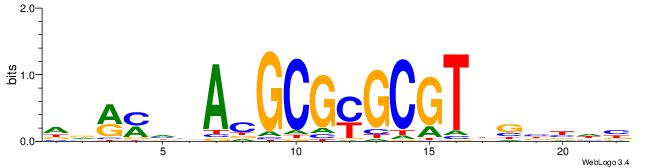 |
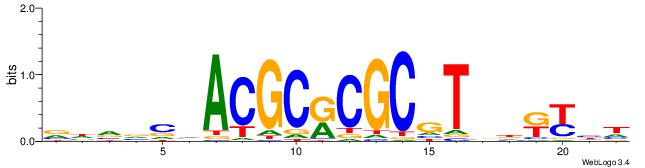 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 20 |
| Similarity score: | 0.0520196 |
Alignment:
WHVBVVMACGCGCGTCDYHWDH
DGVBCABTGDWGCGKRRCSR--
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
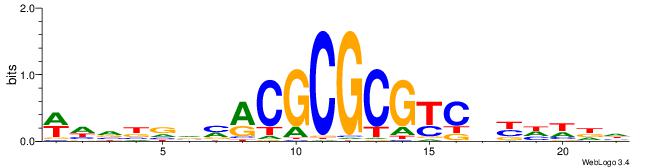 |
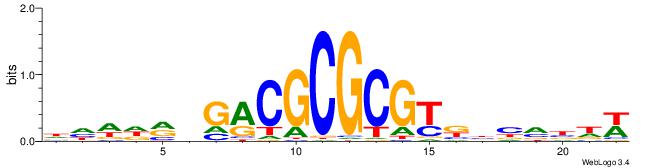 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 20 |
| Similarity score: | 0.0542764 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
-DGVBCABTGDWGCGKRRCSR-
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 20 |
| Similarity score: | 0.0560636 |
Alignment:
DCATDBTGHGCATKMTTBRSWR
--MSGKKRCGCWDCABTGBBCD
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 20 |
| Similarity score: | 0.0595671 |
Alignment:
HHBSHTCGTGRGTGGKGDBVWMV
DGVBCABTGDWGCGKRRCSR---
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 20 |
| Similarity score: | 0.0602559 |
Alignment:
HADTBVADGATGCATCMTGMVHB
DGVBCABTGDWGCGKRRCSR---
| Original motif | Reverse complement motif |
| Consensus sequence: HADTBVADGATGCATCMTGMVHB | Consensus sequence: VHVRCAYGATGCATCDTVVADTH |
 |
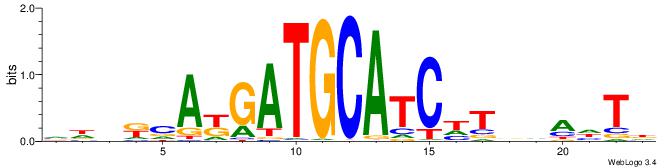 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 20 |
| Similarity score: | 0.0607506 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
DGVBCABTGDWGCGKRRCSR---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 20 |
| Similarity score: | 0.0611227 |
Alignment:
BDBBVBVGTAWCCAAGVBHBAGB
MSGKKRCGCWDCABTGBBCD---
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
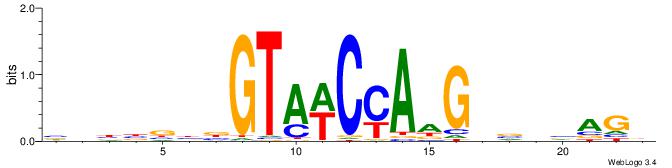 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 20 |
| Similarity score: | 0.0611361 |
Alignment:
RDHDBVDTCACGTGASBHVHDH
--MSGKKRCGCWDCABTGBBCD
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 20 |
| Similarity score: | 0.061403 |
Alignment:
GTVBDAATTGTVTGHGDDHKVB
DGVBCABTGDWGCGKRRCSR--
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
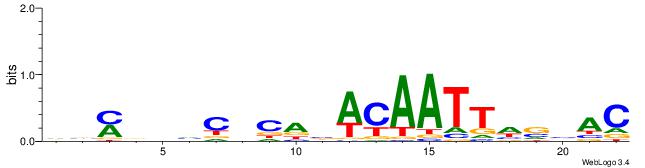 |
| Dataset #: 3 | Motif ID: 32 | Motif name: ArntAhr |
| Original motif | Reverse complement motif |
| Consensus sequence: YGCGTG | Consensus sequence: CACGCM |
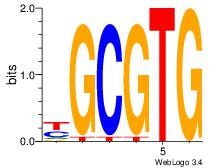 |
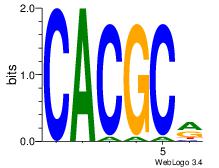 |
Best Matches for Motif ID 32 (Highest to Lowest)
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
------YGCGTG-----------
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.00183587 |
Alignment:
HHDGBCACGCCTWDB
-----CACGCM----
| Original motif | Reverse complement motif |
| Consensus sequence: BDWAGGCGTGBCHHD | Consensus sequence: HHDGBCACGCCTWDB |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.00348048 |
Alignment:
HDGTCGCGTGDCVB
----YGCGTG----
| Original motif | Reverse complement motif |
| Consensus sequence: BBGHCACGCGACDD | Consensus sequence: HDGTCGCGTGDCVB |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.00526158 |
Alignment:
TCDAMVBGACGCMAKDDDVBBD
-------CACGCM---------
| Original motif | Reverse complement motif |
| Consensus sequence: DBBVDDDRTRGCGTCBBYTHGA | Consensus sequence: TCDAMVBGACGCMAKDDDVBBD |
 |
 |
| Motif ID: | UP00042 |
| Motif name: | Gm397_second |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.00558171 |
Alignment:
HBGYGTGTGTGCVRVD
---YGCGTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: DVMVGCACACACKCVH | Consensus sequence: HBGYGTGTGTGCVRVD |
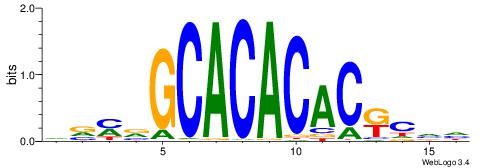 |
 |
| Motif ID: | UP00097 |
| Motif name: | Mtf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.00702416 |
Alignment:
HDYTTGCACACGDHBH
--------CACGCM--
| Original motif | Reverse complement motif |
| Consensus sequence: DBDHCGTGTGCAAMDH | Consensus sequence: HDYTTGCACACGDHBH |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.0123233 |
Alignment:
DDRMHBACGCGYGCGTDGBBDB
-------YGCGTG---------
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0141579 |
Alignment:
HCCGCCCCCGCAHB
------CACGCM--
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 11 |
| Number of overlap: | 6 |
| Similarity score: | 0.0144485 |
Alignment:
BHCBCBCCGGGTGGTCYHVHDCH
-------YGCGTG----------
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 10 |
| Number of overlap: | 6 |
| Similarity score: | 0.0172109 |
Alignment:
VHYTCCDWCCGCGTCMMTHWHV
---------YGCGTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
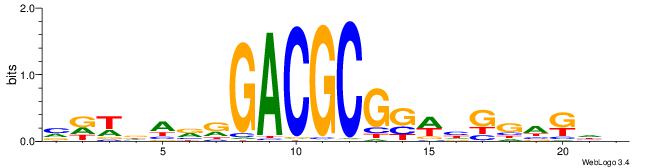 |
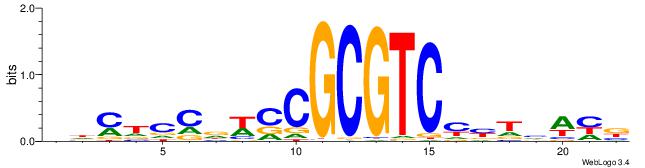 |
| Dataset #: 3 | Motif ID: 33 | Motif name: Mycn |
| Original motif | Reverse complement motif |
| Consensus sequence: HSCACGTGGC | Consensus sequence: GCCACGTGSD |
 |
 |
Best Matches for Motif ID 33 (Highest to Lowest)
| Motif ID: | UP00060 |
| Motif name: | Max_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
DDASCACGTGBTBVDD
--HSCACGTGGC----
| Original motif | Reverse complement motif |
| Consensus sequence: DDASCACGTGBTBVDD | Consensus sequence: HHVBABCACGTGSTHD |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.00783793 |
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
-------GCCACGTGSD------
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0132858 |
Alignment:
DDHBHBSTCACGTGAHBBDHHM
------GCCACGTGSD------
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0227933 |
Alignment:
VBHHVCAGGTGCDVDHD
---GCCACGTGSD----
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0257011 |
Alignment:
BBGHCACGCGACDD
--HSCACGTGGC--
| Original motif | Reverse complement motif |
| Consensus sequence: BBGHCACGCGACDD | Consensus sequence: HDGTCGCGTGDCVB |
 |
 |
| Motif ID: | UP00165 |
| Motif name: | Titf1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0344833 |
Alignment:
BVABCCACTTGAMHTT
---GCCACGTGSD---
| Original motif | Reverse complement motif |
| Consensus sequence: BVABCCACTTGAMHTT | Consensus sequence: AAHYTCAAGTGGBTBV |
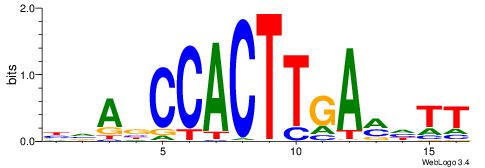 |
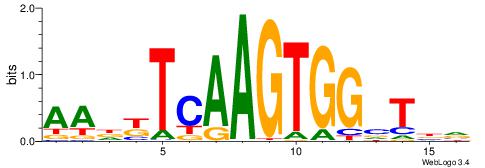 |
| Motif ID: | UP00231 |
| Motif name: | Nkx2-2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.035228 |
Alignment:
HDVRCCACTTGARADBD
---GCCACGTGSD----
| Original motif | Reverse complement motif |
| Consensus sequence: HDVRCCACTTGARADBD | Consensus sequence: DVDTKTCAAGTGGKBDH |
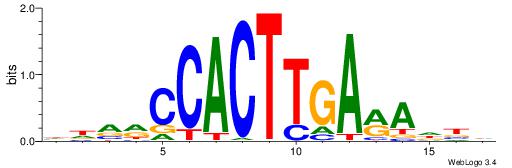 |
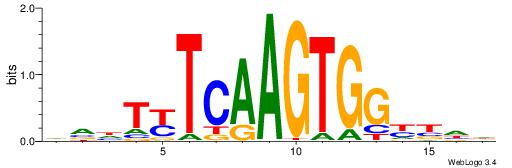 |
| Motif ID: | UP00084 |
| Motif name: | Gmeb1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0358581 |
Alignment:
BVVDVRYACGTRYVBHB
----GCCACGTGSD---
| Original motif | Reverse complement motif |
| Consensus sequence: BHBBKKACGTMMVDBVB | Consensus sequence: BVVDVRYACGTRYVBHB |
 |
 |
| Motif ID: | UP00084 |
| Motif name: | Gmeb1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.03725 |
Alignment:
HHMACKACGTMGBVCD
---HSCACGTGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: DGVBCRACGTYGTYHH | Consensus sequence: HHMACKACGTMGBVCD |
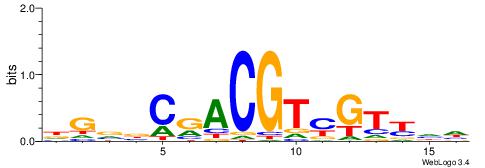 |
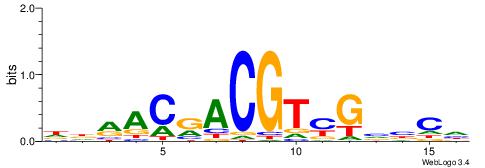 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0383187 |
Alignment:
WHVBVVMACGCGCGTCDYHWDH
------HSCACGTGGC------
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
 |
 |
| Dataset #: 3 | Motif ID: 34 | Motif name: Myc |
| Original motif | Reverse complement motif |
| Consensus sequence: VGCACGTGGH | Consensus sequence: DCCACGTGCV |
 |
 |
Best Matches for Motif ID 34 (Highest to Lowest)
| Motif ID: | UP00060 |
| Motif name: | Max_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
DDASCACGTGBTBVDD
--VGCACGTGGH----
| Original motif | Reverse complement motif |
| Consensus sequence: DDASCACGTGBTBVDD | Consensus sequence: HHVBABCACGTGSTHD |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 10 |
| Similarity score: | 0.00631885 |
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
-------DCCACGTGCV------
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.00821668 |
Alignment:
DDHBHBSTCACGTGAHBBDHHM
------DCCACGTGCV------
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0140005 |
Alignment:
DHDBHGCACCTGBDDVB
----VGCACGTGGH---
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0308912 |
Alignment:
BBGHCACGCGACDD
--VGCACGTGGH--
| Original motif | Reverse complement motif |
| Consensus sequence: BBGHCACGCGACDD | Consensus sequence: HDGTCGCGTGDCVB |
 |
 |
| Motif ID: | UP00165 |
| Motif name: | Titf1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0311723 |
Alignment:
BVABCCACTTGAMHTT
---DCCACGTGCV---
| Original motif | Reverse complement motif |
| Consensus sequence: BVABCCACTTGAMHTT | Consensus sequence: AAHYTCAAGTGGBTBV |
 |
 |
| Motif ID: | UP00249 |
| Motif name: | Nkx2-5 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.031308 |
Alignment:
HDADCCACTTRAAWTT
---DCCACGTGCV---
| Original motif | Reverse complement motif |
| Consensus sequence: HDADCCACTTRAAWTT | Consensus sequence: AAWTTMAAGTGGHTDH |
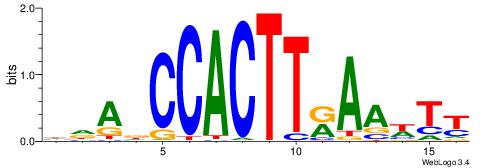 |
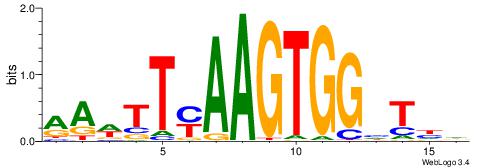 |
| Motif ID: | UP00231 |
| Motif name: | Nkx2-2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0334845 |
Alignment:
HDVRCCACTTGARADBD
---DCCACGTGCV----
| Original motif | Reverse complement motif |
| Consensus sequence: HDVRCCACTTGARADBD | Consensus sequence: DVDTKTCAAGTGGKBDH |
 |
 |
| Motif ID: | UP00147 |
| Motif name: | Nkx2-6 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0345548 |
Alignment:
DDADCCACTTAAVHTT
---DCCACGTGCV---
| Original motif | Reverse complement motif |
| Consensus sequence: DDADCCACTTAAVHTT | Consensus sequence: AAHVTTAAGTGGHTDD |
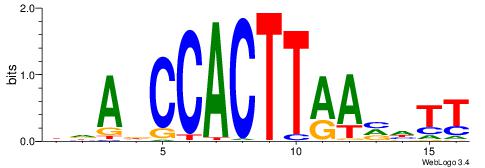 |
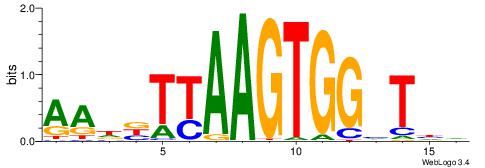 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0363818 |
Alignment:
VDWDARRGACGCGGWHGGAKHV
-----VGCACGTGGH-------
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Dataset #: 3 | Motif ID: 35 | Motif name: GABPA |
| Original motif | Reverse complement motif |
| Consensus sequence: CCGGAAGTGVV | Consensus sequence: VVCACTTCCGG |
 |
 |
Best Matches for Motif ID 35 (Highest to Lowest)
| Motif ID: | UP00013 |
| Motif name: | Gabpa_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0 |
Alignment:
BHHHACTTCCGGTHHBD
-VVCACTTCCGG-----
| Original motif | Reverse complement motif |
| Consensus sequence: HVHHACCGGAAGTDHHV | Consensus sequence: BHHHACTTCCGGTHHBD |
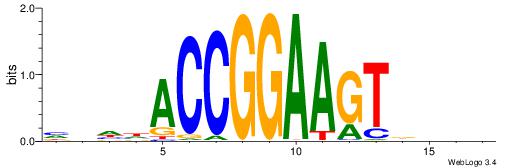 |
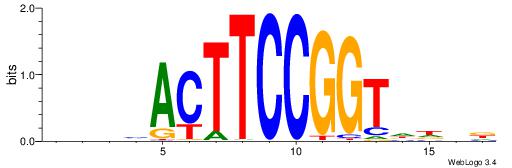 |
| Motif ID: | UP00410 |
| Motif name: | Elk1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.000136484 |
Alignment:
VVHBACCGGAAGTDBHD
-----CCGGAAGTGVV-
| Original motif | Reverse complement motif |
| Consensus sequence: VVHBACCGGAAGTDBHD | Consensus sequence: DHBDACTTCCGGTVHVB |
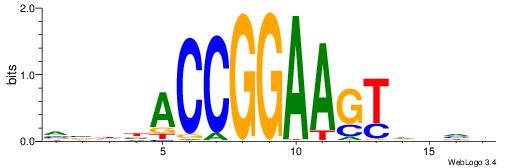 |
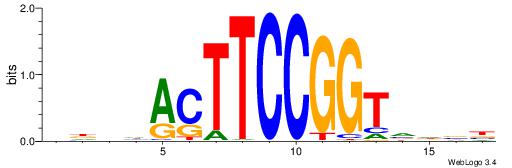 |
| Motif ID: | UP00420 |
| Motif name: | Elk3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.000219706 |
Alignment:
DVBBACCGGAAGYDVVV
-----CCGGAAGTGVV-
| Original motif | Reverse complement motif |
| Consensus sequence: DVBBACCGGAAGYDVVV | Consensus sequence: VBVDMCTTCCGGTVBVD |
 |
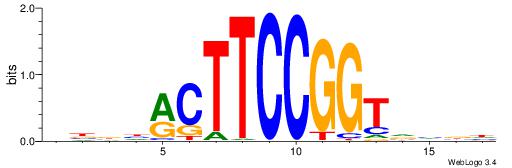 |
| Motif ID: | UP00421 |
| Motif name: | Etv1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.000932023 |
Alignment:
MVHBACCGGAAGTVDVH
-----CCGGAAGTGVV-
| Original motif | Reverse complement motif |
| Consensus sequence: MVHBACCGGAAGTVDVH | Consensus sequence: HBDBACTTCCGGTBHVY |
 |
 |
| Motif ID: | UP00404 |
| Motif name: | Elf2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.00527483 |
Alignment:
DBBHACTTCCGGDWDB
-VVCACTTCCGG----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGDWDB | Consensus sequence: BDWDCCGGAAGTHBBD |
 |
 |
| Motif ID: | UP00414 |
| Motif name: | Ets1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.00805613 |
Alignment:
BDDDHACCGGAARTVDD
------CCGGAAGTGVV
| Original motif | Reverse complement motif |
| Consensus sequence: BDDDHACCGGAARTVDD | Consensus sequence: DDBAMTTCCGGTDHDHB |
 |
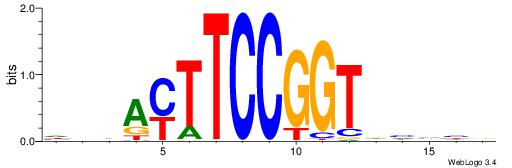 |
| Motif ID: | UP00423 |
| Motif name: | Gm5454 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.00933976 |
Alignment:
VDHHACTTCCGGTHVH
-VVCACTTCCGG----
| Original motif | Reverse complement motif |
| Consensus sequence: VDHHACTTCCGGTHVH | Consensus sequence: DVHACCGGAAGTHDDV |
 |
 |
| Motif ID: | UP00413 |
| Motif name: | Elf4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.00992792 |
Alignment:
DBBHACTTCCGGKWTH
-VVCACTTCCGG----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGKWTH | Consensus sequence: HAWYCCGGAAGTHBBD |
 |
 |
| Motif ID: | UP00418 |
| Motif name: | Etv6 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0114183 |
Alignment:
DVHBTACTTCCGGHTHB
--VVCACTTCCGG----
| Original motif | Reverse complement motif |
| Consensus sequence: DVHBTACTTCCGGHTHB | Consensus sequence: BHAHCCGGAAGTABDVD |
 |
 |
| Motif ID: | UP00411 |
| Motif name: | Erg |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0122484 |
Alignment:
BDDACCGGAAKTVBVB
----CCGGAAGTGVV-
| Original motif | Reverse complement motif |
| Consensus sequence: VBBVAYTTCCGGTDDB | Consensus sequence: BDDACCGGAAKTVBVB |
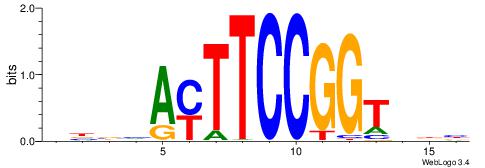 |
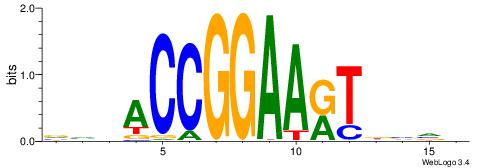 |
| Dataset #: 4 | Motif ID: 36 | Motif name: csGCCCCGCCCCsc |
| Original motif | Reverse complement motif |
| Consensus sequence: HVGCCCCGCCCCBB | Consensus sequence: BBGGGGCGGGGCVD |
 |
 |
Best Matches for Motif ID 36 (Highest to Lowest)
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0 |
Alignment:
HBDRCCMCGCCCHHHD
-HVGCCCCGCCCCBB-
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0106621 |
Alignment:
DHHBCCCCGCCAHHBHB
-HVGCCCCGCCCCBB--
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
 |
 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.017761 |
Alignment:
GKRGGGGGGGGGGBD
BBGGGGCGGGGCVD-
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0181905 |
Alignment:
HVBCCCCCCCCMHHHB
HVGCCCCGCCCCBB--
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0225788 |
Alignment:
BHHBHCCGCCCCDHHHH
HVGCCCCGCCCCBB---
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0277585 |
Alignment:
BDWAGGCGTGBCHHD
BBGGGGCGGGGCVD-
| Original motif | Reverse complement motif |
| Consensus sequence: BDWAGGCGTGBCHHD | Consensus sequence: HHDGBCACGCCTWDB |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0318885 |
Alignment:
BHHDTCCCACTCBDBV
-HVGCCCCGCCCCBB-
| Original motif | Reverse complement motif |
| Consensus sequence: BBHBGAGTGGGAHHDB | Consensus sequence: BHHDTCCCACTCBDBV |
 |
 |
| Motif ID: | UP00024 |
| Motif name: | Glis2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0330753 |
Alignment:
HKBBGGGGGGTCVHHH
BBGGGGCGGGGCVD--
| Original motif | Reverse complement motif |
| Consensus sequence: HHHVGACCCCCCVBRD | Consensus sequence: HKBBGGGGGGTCVHHH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 14 |
| Similarity score: | 0.0334003 |
Alignment:
CVHBSCGGGTGGTCMHVHHCDH
---BBGGGGCGGGGCVD-----
| Original motif | Reverse complement motif |
| Consensus sequence: HDGHHBHRGACCACCCGSVDVG | Consensus sequence: CVHBSCGGGTGGTCMHVHHCDH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0351614 |
Alignment:
CBDMCMGGGTGGTCCHVBVBAH
---BBGGGGCGGGGCVD-----
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
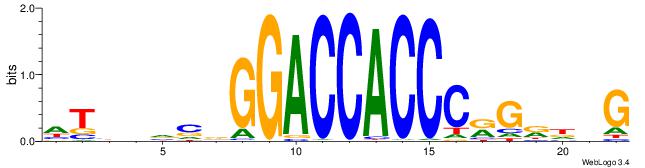 |
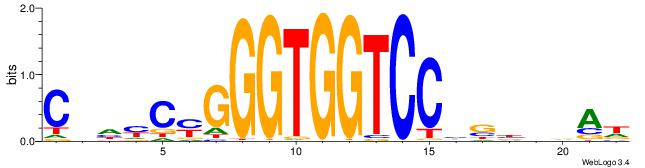 |
| Dataset #: 4 | Motif ID: 37 | Motif name: tkAAATAATAtw |
| Original motif | Reverse complement motif |
| Consensus sequence: HDAAATAATAHW | Consensus sequence: WHTATTATTTDH |
 |
 |
Best Matches for Motif ID 37 (Highest to Lowest)
| Motif ID: | UP00121 |
| Motif name: | Hoxd10 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.00353196 |
Alignment:
HDDDATTTTATTKVRDB
---WHTATTATTTDH--
| Original motif | Reverse complement motif |
| Consensus sequence: VDKVYAATAAAATDDDH | Consensus sequence: HDDDATTTTATTKVRDB |
 |
 |
| Motif ID: | UP00244 |
| Motif name: | Tlx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.00646023 |
Alignment:
WWDTTATTAATTHATTV
--WHTATTATTTDH---
| Original motif | Reverse complement motif |
| Consensus sequence: BAATHAATTAATAAHWW | Consensus sequence: WWDTTATTAATTHATTV |
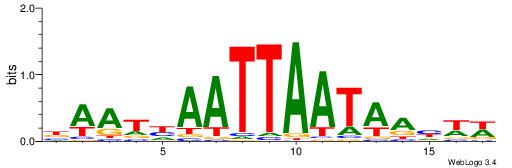 |
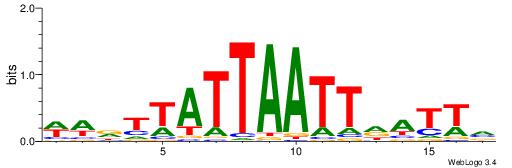 |
| Motif ID: | UP00051 |
| Motif name: | Sox8_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0120674 |
Alignment:
DDDHDAACAATWBATDD
--HDAAATAATAHW---
| Original motif | Reverse complement motif |
| Consensus sequence: DDATBWATTGTTHHDDD | Consensus sequence: DDDHDAACAATWBATDD |
 |
 |
| Motif ID: | UP00180 |
| Motif name: | Hoxd13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0123772 |
Alignment:
HDHHTTTTATTKKHDD
--WHTATTATTTDH--
| Original motif | Reverse complement motif |
| Consensus sequence: HDHYYAATAAAAHHHH | Consensus sequence: HDHHTTTTATTKKHDD |
 |
 |
| Motif ID: | UP00037 |
| Motif name: | Zfp105_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0129559 |
Alignment:
HHHTTRTTDTTTKDH
--WHTATTATTTDH-
| Original motif | Reverse complement motif |
| Consensus sequence: HDYAAAHAAMAADHD | Consensus sequence: HHHTTRTTDTTTKDH |
 |
 |
| Motif ID: | UP00064 |
| Motif name: | Sox18_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0130138 |
Alignment:
HHDHABAACAATHBWV
---HDAAATAATAHW-
| Original motif | Reverse complement motif |
| Consensus sequence: BWBHATTGTTBTHDHH | Consensus sequence: HHDHABAACAATHBWV |
 |
 |
| Motif ID: | UP00255 |
| Motif name: | Dbx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0131034 |
Alignment:
HARTHAATTAWTAVDHD
---HDAAATAATAHW--
| Original motif | Reverse complement motif |
| Consensus sequence: HARTHAATTAWTAVDHD | Consensus sequence: DHDBTAWTAATTHAKTH |
 |
 |
| Motif ID: | UP00024 |
| Motif name: | Glis2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0140433 |
Alignment:
BVTTTAHTAATADH
WHTATTATTTDH--
| Original motif | Reverse complement motif |
| Consensus sequence: HDTATTAHTAAAVV | Consensus sequence: BVTTTAHTAATADH |
 |
 |
| Motif ID: | UP00075 |
| Motif name: | Sox15_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0143532 |
Alignment:
DHVKMWATTGTTHDHDH
---WHTATTATTTDH--
| Original motif | Reverse complement motif |
| Consensus sequence: HDDDDAACAATWRRBHD | Consensus sequence: DHVKMWATTGTTHDHDH |
 |
 |
| Motif ID: | UP00096 |
| Motif name: | Sox13_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.014607 |
Alignment:
DHDRGAACAATWWDHW
--HDAAATAATAHW--
| Original motif | Reverse complement motif |
| Consensus sequence: DHDRGAACAATWWDHW | Consensus sequence: WHDWWATTGTTCKDHD |
 |
 |
| Dataset #: 4 | Motif ID: 38 | Motif name: cccGCCCCGCCCCsb |
| Original motif | Reverse complement motif |
| Consensus sequence: BCCGCCCCGCCCCBB | Consensus sequence: BBGGGGCGGGGCGGB |
 |
 |
Best Matches for Motif ID 38 (Highest to Lowest)
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.00481788 |
Alignment:
HBDRCCMCGCCCHHHD
BCCGCCCCGCCCCBB-
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0108117 |
Alignment:
DHHBCCCCGCCAHHBHB
BCCGCCCCGCCCCBB--
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
 |
 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0154152 |
Alignment:
GKRGGGGGGGGGGBD
BBGGGGCGGGGCGGB
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.018287 |
Alignment:
HVBCCCCCCCCMHHHB
BCCGCCCCGCCCCBB-
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0247447 |
Alignment:
BDWAGGCGTGBCHHD
BBGGGGCGGGGCGGB
| Original motif | Reverse complement motif |
| Consensus sequence: BDWAGGCGTGBCHHD | Consensus sequence: HHDGBCACGCCTWDB |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0279869 |
Alignment:
BHHDTCCCACTCBDBV
BCCGCCCCGCCCCBB-
| Original motif | Reverse complement motif |
| Consensus sequence: BBHBGAGTGGGAHHDB | Consensus sequence: BHHDTCCCACTCBDBV |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 15 |
| Similarity score: | 0.0312526 |
Alignment:
CVHBSCGGGTGGTCMHVHHCDH
---BBGGGGCGGGGCGGB----
| Original motif | Reverse complement motif |
| Consensus sequence: HDGHHBHRGACCACCCGSVDVG | Consensus sequence: CVHBSCGGGTGGTCMHVHHCDH |
 |
 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0318321 |
Alignment:
BHVDCCCCDCCCBDBH
BCCGCCCCGCCCCBB-
| Original motif | Reverse complement motif |
| Consensus sequence: BHVDCCCCDCCCBDBH | Consensus sequence: HVDVGGGHGGGGDBHB |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 15 |
| Similarity score: | 0.0340857 |
Alignment:
VHYTCCDWCCGCGTCMMTHWHV
----BCCGCCCCGCCCCBB---
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 15 |
| Similarity score: | 0.0345185 |
Alignment:
CBDMCMGGGTGGTCCHVBVBAH
---BBGGGGCGGGGCGGB----
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Dataset #: 4 | Motif ID: 39 | Motif name: kCAGCCAATmr |
| Original motif | Reverse complement motif |
| Consensus sequence: DCAGCCAATVR | Consensus sequence: MBATTGGCTGH |
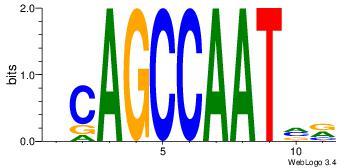 |
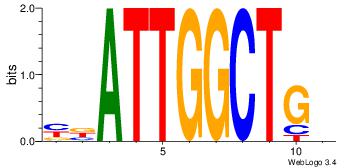 |
Best Matches for Motif ID 39 (Highest to Lowest)
| Motif ID: | UP00033 |
| Motif name: | Zfp410_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.000708728 |
Alignment:
BDDYYCATCCCATDDBD
----DCAGCCAATVR--
| Original motif | Reverse complement motif |
| Consensus sequence: DVDDATGGGATGKMDDV | Consensus sequence: BDDYYCATCCCATDDBD |
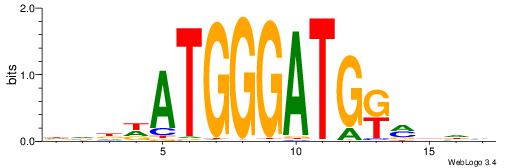 |
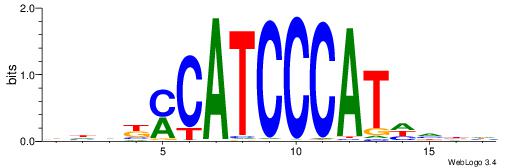 |
| Motif ID: | UP00004 |
| Motif name: | Sox14_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.00658734 |
Alignment:
BKBASACAATRGHBB
-DCAGCCAATVR---
| Original motif | Reverse complement motif |
| Consensus sequence: BKBASACAATRGHBB | Consensus sequence: BBDCMATTGTSTBRB |
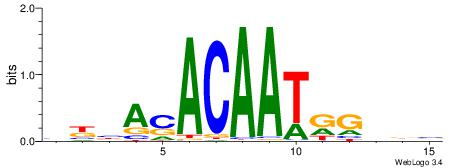 |
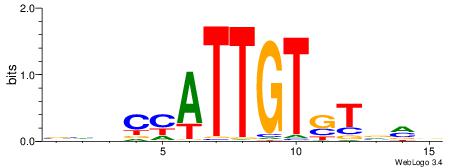 |
| Motif ID: | UP00249 |
| Motif name: | Nkx2-5 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.00805715 |
Alignment:
AAWTTMAAGTGGHTDH
-----MBATTGGCTGH
| Original motif | Reverse complement motif |
| Consensus sequence: HDADCCACTTRAAWTT | Consensus sequence: AAWTTMAAGTGGHTDH |
 |
 |
| Motif ID: | UP00147 |
| Motif name: | Nkx2-6 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0128272 |
Alignment:
DDADCCACTTAAVHTT
DCAGCCAATVR-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDADCCACTTAAVHTT | Consensus sequence: AAHVTTAAGTGGHTDD |
 |
 |
| Motif ID: | UP00165 |
| Motif name: | Titf1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0131729 |
Alignment:
AAHYTCAAGTGGBTBV
-----MBATTGGCTGH
| Original motif | Reverse complement motif |
| Consensus sequence: BVABCCACTTGAMHTT | Consensus sequence: AAHYTCAAGTGGBTBV |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 11 |
| Similarity score: | 0.0131873 |
Alignment:
HHBDHDDAACAATDRHHHHHBW
----DCAGCCAATVR-------
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00258 |
| Motif name: | Tgif2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0132199 |
Alignment:
AABBAGCTGTCAAWVV
-----DCAGCCAATVR
| Original motif | Reverse complement motif |
| Consensus sequence: AABBAGCTGTCAAWVV | Consensus sequence: VBWTTGACAGCTVBTT |
 |
 |
| Motif ID: | UP00205 |
| Motif name: | Pknox2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0138143 |
Alignment:
VHATTGACAGSKSVTH
MBATTGGCTGH-----
| Original motif | Reverse complement motif |
| Consensus sequence: HAVSRSCTGTCAATHB | Consensus sequence: VHATTGACAGSKSVTH |
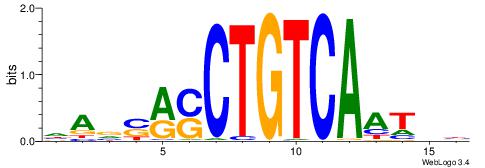 |
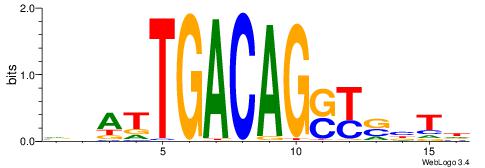 |
| Motif ID: | UP00023 |
| Motif name: | Sox30_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0141895 |
Alignment:
HHHHMHATTGTTBDHD
----MBATTGGCTGH-
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBAACAATDRHHHH | Consensus sequence: HHHHMHATTGTTBDHD |
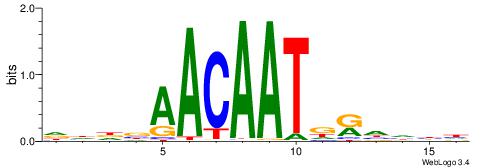 |
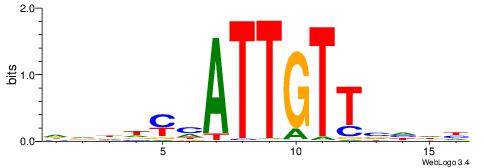 |
| Motif ID: | UP00227 |
| Motif name: | Duxl |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0154208 |
Alignment:
YRACCCAATCAACVVHV
DCAGCCAATVR------
| Original motif | Reverse complement motif |
| Consensus sequence: YRACCCAATCAACVVHV | Consensus sequence: VHVVGTTGATTGGGTMK |
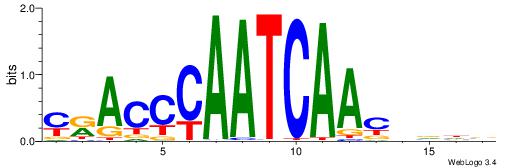 |
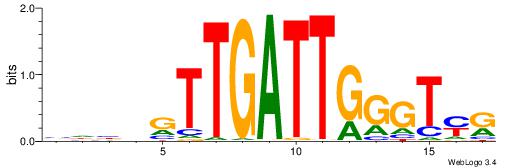 |
| Dataset #: 4 | Motif ID: 40 | Motif name: kcACCTGCAgc |
| Original motif | Reverse complement motif |
| Consensus sequence: BCACCTGCABC | Consensus sequence: GBTGCAGGTGB |
 |
 |
Best Matches for Motif ID 40 (Highest to Lowest)
| Motif ID: | UP00070 |
| Motif name: | Gcm1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0 |
Alignment:
DHDVATGCGGGTHVVH
---GBTGCAGGTGB--
| Original motif | Reverse complement motif |
| Consensus sequence: HVVHACCCGCATVDHD | Consensus sequence: DHDVATGCGGGTHVVH |
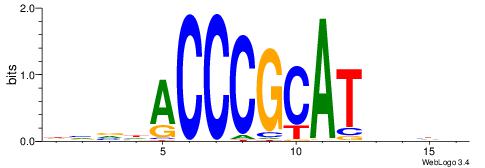 |
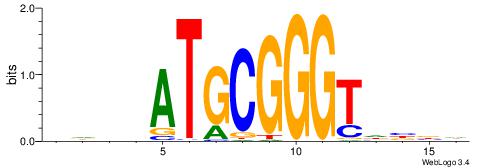 |
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.000503863 |
Alignment:
BBBAVTGCAGTGBBVDD
---GBTGCAGGTGB---
| Original motif | Reverse complement motif |
| Consensus sequence: DDBBBCACTGCABTBBB | Consensus sequence: BBBAVTGCAGTGBBVDD |
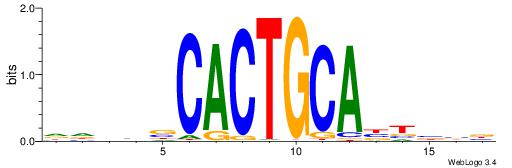 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.000557448 |
Alignment:
DHDBHGCACCTGBDDVB
-----BCACCTGCABC-
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.00842747 |
Alignment:
BBYVVCAGCTGCBVHHD
----BCACCTGCABC--
| Original motif | Reverse complement motif |
| Consensus sequence: BBYVVCAGCTGCBVHHD | Consensus sequence: HHDVVGCAGCTGVBKVB |
 |
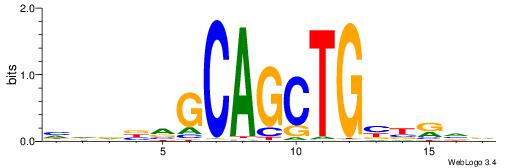 |
| Motif ID: | UP00420 |
| Motif name: | Elk3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0111732 |
Alignment:
DVBBACCGGAAGYDVVV
--BCACCTGCABC----
| Original motif | Reverse complement motif |
| Consensus sequence: DVBBACCGGAAGYDVVV | Consensus sequence: VBVDMCTTCCGGTVBVD |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 11 |
| Similarity score: | 0.0122671 |
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
--------BCACCTGCABC----
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00092 |
| Motif name: | Myb_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.015733 |
Alignment:
BDACCAACTGCCRBBH
---BCACCTGCABC--
| Original motif | Reverse complement motif |
| Consensus sequence: BDACCAACTGCCRBBH | Consensus sequence: DBVKGGCAGTTGGTHB |
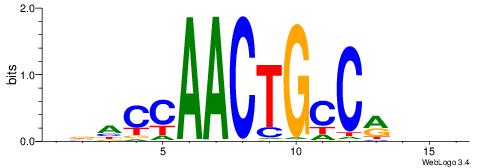 |
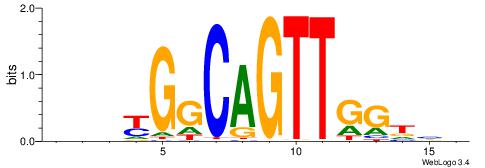 |
| Motif ID: | UP00236 |
| Motif name: | Irx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0163141 |
Alignment:
WVWDDTACATGTADDDW
---GBTGCAGGTGB---
| Original motif | Reverse complement motif |
| Consensus sequence: WDDDTACATGTADDWBW | Consensus sequence: WVWDDTACATGTADDDW |
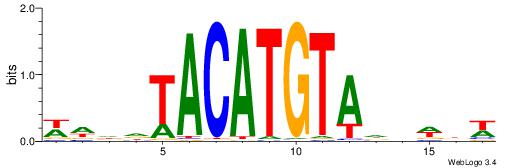 |
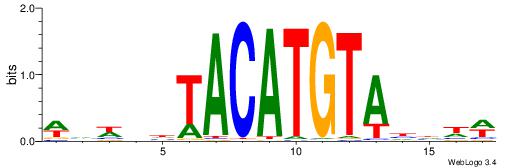 |
| Motif ID: | UP00223 |
| Motif name: | Irx3_0920.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0165255 |
Alignment:
DDWWBTACATGTADDDD
----BCACCTGCABC--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDDTACATGTAVWWHD | Consensus sequence: DDWWBTACATGTADDDD |
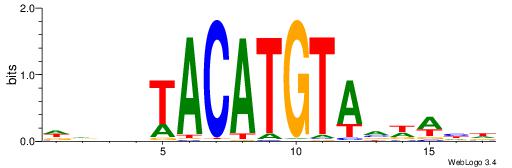 |
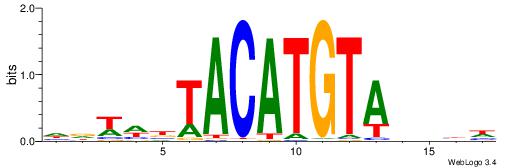 |
| Motif ID: | UP00057 |
| Motif name: | Zic2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0169178 |
Alignment:
DVBCYTGCTGBGDDD
BCACCTGCABC----
| Original motif | Reverse complement motif |
| Consensus sequence: HHDCVCAGCAKGVVD | Consensus sequence: DVBCYTGCTGBGDDD |
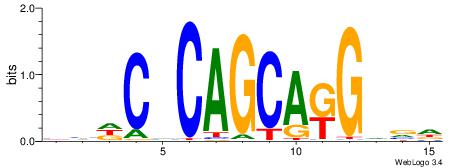 |
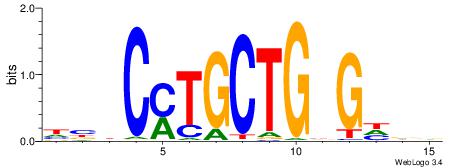 |
| Dataset #: 4 | Motif ID: 41 | Motif name: wwAAATAATAtw |
| Original motif | Reverse complement motif |
| Consensus sequence: HDAAATAATADD | Consensus sequence: DDTATTATTTDH |
 |
 |
Best Matches for Motif ID 41 (Highest to Lowest)
| Motif ID: | UP00244 |
| Motif name: | Tlx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.00576686 |
Alignment:
WWDTTATTAATTHATTV
--DDTATTATTTDH---
| Original motif | Reverse complement motif |
| Consensus sequence: BAATHAATTAATAAHWW | Consensus sequence: WWDTTATTAATTHATTV |
 |
 |
| Motif ID: | UP00121 |
| Motif name: | Hoxd10 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.00673636 |
Alignment:
HDDDATTTTATTKVRDB
---DDTATTATTTDH--
| Original motif | Reverse complement motif |
| Consensus sequence: VDKVYAATAAAATDDDH | Consensus sequence: HDDDATTTTATTKVRDB |
 |
 |
| Motif ID: | UP00180 |
| Motif name: | Hoxd13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.00685179 |
Alignment:
HDHHTTTTATTKKHDD
--DDTATTATTTDH--
| Original motif | Reverse complement motif |
| Consensus sequence: HDHYYAATAAAAHHHH | Consensus sequence: HDHHTTTTATTKKHDD |
 |
 |
| Motif ID: | UP00073 |
| Motif name: | Foxa2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0108327 |
Alignment:
VVDAATAACAAAVVB
HDAAATAATADD---
| Original motif | Reverse complement motif |
| Consensus sequence: VVDAATAACAAAVVB | Consensus sequence: BVVTTTGTTATTDBB |
 |
 |
| Motif ID: | UP00134 |
| Motif name: | Hoxb13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0121723 |
Alignment:
VDHWTTTTATTGGDKB
--DDTATTATTTDH--
| Original motif | Reverse complement motif |
| Consensus sequence: VRHCCAATAAAAWHHV | Consensus sequence: VDHWTTTTATTGGDKB |
 |
 |
| Motif ID: | UP00255 |
| Motif name: | Dbx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0133808 |
Alignment:
HARTHAATTAWTAVDHD
---HDAAATAATADD--
| Original motif | Reverse complement motif |
| Consensus sequence: HARTHAATTAWTAVDHD | Consensus sequence: DHDBTAWTAATTHAKTH |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 12 |
| Similarity score: | 0.0144702 |
Alignment:
HHBDHDDAACAATDRHHHHHBW
----HDAAATAATADD------
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00051 |
| Motif name: | Sox8_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0145614 |
Alignment:
DDATBWATTGTTHHDDD
---DDTATTATTTDH--
| Original motif | Reverse complement motif |
| Consensus sequence: DDATBWATTGTTHHDDD | Consensus sequence: DDDHDAACAATWBATDD |
 |
 |
| Motif ID: | UP00024 |
| Motif name: | Glis2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0148858 |
Alignment:
HDTATTAHTAAAVV
--HDAAATAATADD
| Original motif | Reverse complement motif |
| Consensus sequence: HDTATTAHTAAAVV | Consensus sequence: BVTTTAHTAATADH |
 |
 |
| Motif ID: | UP00096 |
| Motif name: | Sox13_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.014968 |
Alignment:
DHDRGAACAATWWDHW
--HDAAATAATADD--
| Original motif | Reverse complement motif |
| Consensus sequence: DHDRGAACAATWWDHW | Consensus sequence: WHDWWATTGTTCKDHD |
 |
 |
| Dataset #: 4 | Motif ID: 42 | Motif name: sSGTCACGTGACSs |
| Original motif | Reverse complement motif |
| Consensus sequence: SGGTCACGTGACCS | Consensus sequence: SGGTCACGTGACCS |
 |
 |
Best Matches for Motif ID 42 (Highest to Lowest)
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 14 |
| Similarity score: | 0 |
Alignment:
RDHDBVDTCACGTGASBHVHDH
----SGGTCACGTGACCS----
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0226905 |
Alignment:
DDASCACGTGBTBVDD
--SGGTCACGTGACCS
| Original motif | Reverse complement motif |
| Consensus sequence: DDASCACGTGBTBVDD | Consensus sequence: HHVBABCACGTGSTHD |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0240371 |
Alignment:
BBGHCACGCGACDD
SGGTCACGTGACCS
| Original motif | Reverse complement motif |
| Consensus sequence: BBGHCACGCGACDD | Consensus sequence: HDGTCGCGTGDCVB |
 |
 |
| Motif ID: | UP00103 |
| Motif name: | Jundm2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0253986 |
Alignment:
BBGATGACGTCAYCVH
-SGGTCACGTGACCS-
| Original motif | Reverse complement motif |
| Consensus sequence: BBGATGACGTCAYCVH | Consensus sequence: HVGMTGACGTCATCBB |
 |
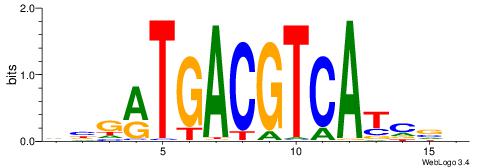 |
| Motif ID: | UP00020 |
| Motif name: | Atf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0259163 |
Alignment:
DBDRTGACGTCAYHRD
-SGGTCACGTGACCS-
| Original motif | Reverse complement motif |
| Consensus sequence: DBDRTGACGTCAYHRD | Consensus sequence: DMDMTGACGTCAKHBD |
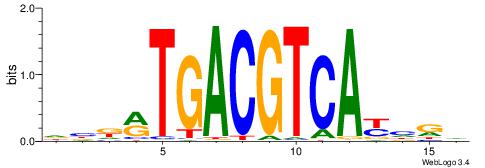 |
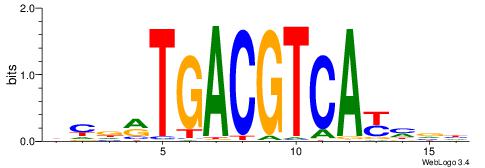 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 14 |
| Similarity score: | 0.0403911 |
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
-----SGGTCACGTGACCS----
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00036 |
| Motif name: | Myf6_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0440464 |
Alignment:
VVDBAACAGBTGBYHB
--SGGTCACGTGACCS
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBAACAGBTGBYHB | Consensus sequence: BDKVCABCTGTTBDBV |
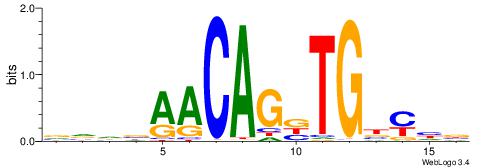 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0471124 |
Alignment:
VBBDMYCATCTGVHHBH
--SGGTCACGTGACCS-
| Original motif | Reverse complement motif |
| Consensus sequence: HVDDVCAGATGKYHBBV | Consensus sequence: VBBDMYCATCTGVHHBH |
 |
 |
| Motif ID: | UP00165 |
| Motif name: | Titf1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0471926 |
Alignment:
BVABCCACTTGAMHTT
-SGGTCACGTGACCS-
| Original motif | Reverse complement motif |
| Consensus sequence: BVABCCACTTGAMHTT | Consensus sequence: AAHYTCAAGTGGBTBV |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0506329 |
Alignment:
DHDBHGCACCTGBDDVB
--SGGTCACGTGACCS-
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Dataset #: 4 | Motif ID: 43 | Motif name: wsTACwGTAsw |
| Original motif | Reverse complement motif |
| Consensus sequence: HVTACWGTABD | Consensus sequence: DBTACWGTAVH |
 |
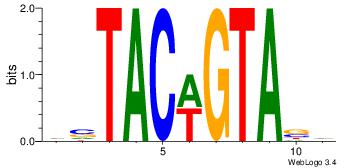 |
Best Matches for Motif ID 43 (Highest to Lowest)
| Motif ID: | UP00052 |
| Motif name: | Osr2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0 |
Alignment:
HHHHGCTACKGTHVHH
----DBTACWGTAVH-
| Original motif | Reverse complement motif |
| Consensus sequence: HHVHACRGTAGCHHHD | Consensus sequence: HHHHGCTACKGTHVHH |
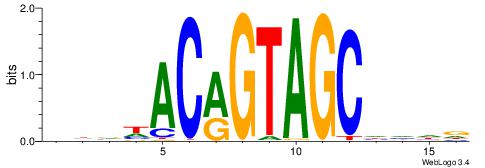 |
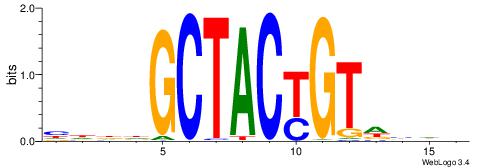 |
| Motif ID: | UP00027 |
| Motif name: | Osr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.00480574 |
Alignment:
DHBHGCTACKGTHDHH
----DBTACWGTAVH-
| Original motif | Reverse complement motif |
| Consensus sequence: HHDHACRGTAGCHVHD | Consensus sequence: DHBHGCTACKGTHDHH |
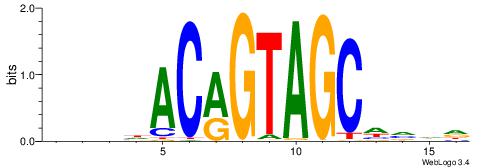 |
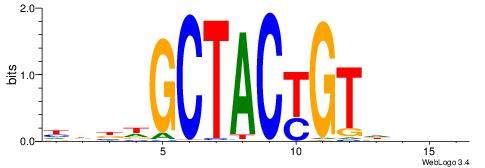 |
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0165381 |
Alignment:
BBBAVTGCAGTGBBVDD
---HVTACWGTABD---
| Original motif | Reverse complement motif |
| Consensus sequence: DDBBBCACTGCABTBBB | Consensus sequence: BBBAVTGCAGTGBBVDD |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.0248017 |
Alignment:
HHBDHDDAACAATDRHHHHHBW
-----DBTACWGTAVH------
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00023 |
| Motif name: | Sox30_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0294174 |
Alignment:
HHHHMHATTGTTBDHD
---HVTACWGTABD--
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBAACAATDRHHHH | Consensus sequence: HHHHMHATTGTTBDHD |
 |
 |
| Motif ID: | UP00075 |
| Motif name: | Sox15_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0301544 |
Alignment:
DHVKMWATTGTTHDHDH
---HVTACWGTABD---
| Original motif | Reverse complement motif |
| Consensus sequence: HDDDDAACAATWRRBHD | Consensus sequence: DHVKMWATTGTTHDHDH |
 |
 |
| Motif ID: | UP00016 |
| Motif name: | Sry_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0310991 |
Alignment:
VHVHDATTGTTMHDDDH
--DBTACWGTAVH----
| Original motif | Reverse complement motif |
| Consensus sequence: HHDHDRAACAATDDVHV | Consensus sequence: VHVHDATTGTTMHDDDH |
 |
 |
| Motif ID: | UP00064 |
| Motif name: | Sox18_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.031364 |
Alignment:
BWBHATTGTTBTHDHH
-DBTACWGTAVH----
| Original motif | Reverse complement motif |
| Consensus sequence: BWBHATTGTTBTHDHH | Consensus sequence: HHDHABAACAATHBWV |
 |
 |
| Motif ID: | UP00092 |
| Motif name: | Myb_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0314236 |
Alignment:
HDDDHAACCGTTAHHHD
---DBTACWGTAVH---
| Original motif | Reverse complement motif |
| Consensus sequence: HDDDHAACCGTTAHHHD | Consensus sequence: DHHHTAACGGTTHHHDH |
 |
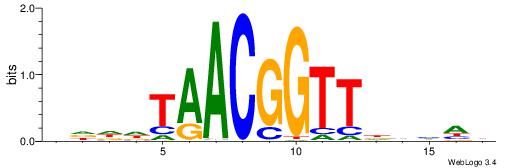 |
| Motif ID: | UP00071 |
| Motif name: | Sox21_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0319794 |
Alignment:
VVYBWATTGTTCVBHDV
--HVTACWGTABD----
| Original motif | Reverse complement motif |
| Consensus sequence: VVYBWATTGTTCVBHDV | Consensus sequence: BDDBVGAACAATWBMBV |
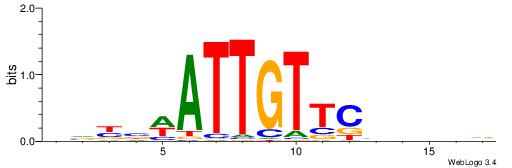 |
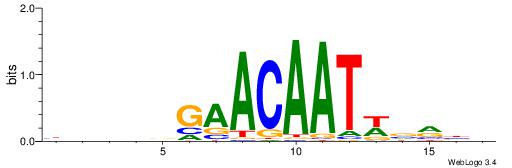 |
| Dataset #: 4 | Motif ID: 44 | Motif name: dhACATTCTkh |
| Original motif | Reverse complement motif |
| Consensus sequence: DHACATTCTGH | Consensus sequence: HCAGAATGTHD |
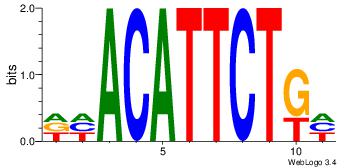 |
 |
Best Matches for Motif ID 44 (Highest to Lowest)
| Motif ID: | UP00038 |
| Motif name: | Spdef_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.00581553 |
Alignment:
VDWYTAGGATGTHVDH
---HCAGAATGTHD--
| Original motif | Reverse complement motif |
| Consensus sequence: DDBHACATCCTAKWDV | Consensus sequence: VDWYTAGGATGTHVDH |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 11 |
| Similarity score: | 0.00582895 |
Alignment:
DCATDBTGHGCATKMTTBRSWR
-------DHACATTCTGH----
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
 |
 |
| Motif ID: | UP00023 |
| Motif name: | Sox30_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.00613057 |
Alignment:
HHHHMHATTGTTBDHD
--DHACATTCTGH---
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBAACAATDRHHHH | Consensus sequence: HHHHMHATTGTTBDHD |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0119275 |
Alignment:
WBHHHHHMDATTGTTHDHDVHH
-----DHACATTCTGH------
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0122803 |
Alignment:
HHBTMCTTTGTTMDDVH
--DHACATTCTGH----
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
 |
 |
| Motif ID: | UP00250 |
| Motif name: | Irx5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0132772 |
Alignment:
WVTDHWACATGTWHDDD
----DHACATTCTGH--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDHWACATGTWHDABW | Consensus sequence: WVTDHWACATGTWHDDD |
 |
 |
| Motif ID: | UP00057 |
| Motif name: | Zic2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0141401 |
Alignment:
DVBCYTGCTGBGDDD
----HCAGAATGTHD
| Original motif | Reverse complement motif |
| Consensus sequence: HHDCVCAGCAKGVVD | Consensus sequence: DVBCYTGCTGBGDDD |
 |
 |
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0143902 |
Alignment:
HHBBMCTTTGTTHDDBH
--DHACATTCTGH----
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
 |
 |
| Motif ID: | UP00004 |
| Motif name: | Sox14_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0151959 |
Alignment:
BKBASACAATRGHBB
---HCAGAATGTHD-
| Original motif | Reverse complement motif |
| Consensus sequence: BKBASACAATRGHBB | Consensus sequence: BBDCMATTGTSTBRB |
 |
 |
| Motif ID: | UP00064 |
| Motif name: | Sox18_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.015273 |
Alignment:
BWBHATTGTTBTHDHH
DHACATTCTGH-----
| Original motif | Reverse complement motif |
| Consensus sequence: BWBHATTGTTBTHDHH | Consensus sequence: HHDHABAACAATHBWV |
 |
 |
| Dataset #: 4 | Motif ID: 45 | Motif name: wbgTAAATAww |
| Original motif | Reverse complement motif |
| Consensus sequence: DBGTAAATAHD | Consensus sequence: DHTATTTACBD |
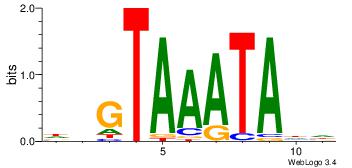 |
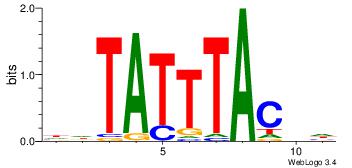 |
Best Matches for Motif ID 45 (Highest to Lowest)
| Motif ID: | UP00242 |
| Motif name: | Hoxc8 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0 |
Alignment:
AHDHTAATTACHHHHV
--DHTATTTACBD---
| Original motif | Reverse complement motif |
| Consensus sequence: BHDDDGTAATTAHHDT | Consensus sequence: AHDHTAATTACHHHHV |
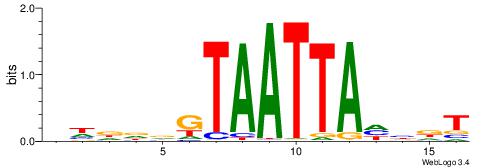 |
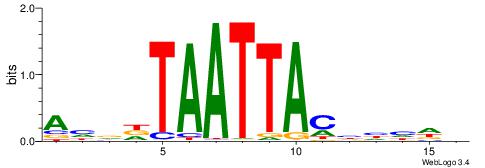 |
| Motif ID: | UP00073 |
| Motif name: | Foxa2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.00453517 |
Alignment:
HHWAWGTAAAYAAAVDB
---DBGTAAATAHD---
| Original motif | Reverse complement motif |
| Consensus sequence: HHWAWGTAAAYAAAVDB | Consensus sequence: BDVTTTKTTTACWTWHH |
 |
 |
| Motif ID: | UP00039 |
| Motif name: | Foxj3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.00490457 |
Alignment:
HDHADGTAAACAAAVVH
---DBGTAAATAHD---
| Original motif | Reverse complement motif |
| Consensus sequence: HDHADGTAAACAAAVVH | Consensus sequence: DVVTTTGTTTACDTHDH |
 |
 |
| Motif ID: | UP00041 |
| Motif name: | Foxj1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.00656547 |
Alignment:
HDHRTAAACAAAHDDD
-DBGTAAATAHD----
| Original motif | Reverse complement motif |
| Consensus sequence: HDHRTAAACAAAHDDD | Consensus sequence: DDDHTTTGTTTAMHDH |
 |
 |
| Motif ID: | UP00182 |
| Motif name: | Hoxa6 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0086354 |
Alignment:
DTHHGGTAATTAYCYT
---DBGTAAATAHD--
| Original motif | Reverse complement motif |
| Consensus sequence: AMGKTAATTACCHHAD | Consensus sequence: DTHHGGTAATTAYCYT |
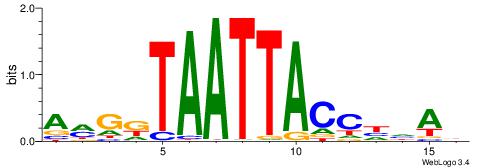 |
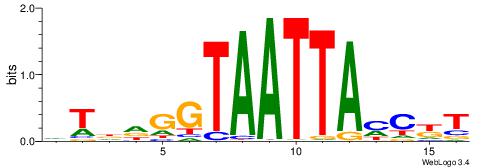 |
| Motif ID: | UP00260 |
| Motif name: | Hoxc6 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0092156 |
Alignment:
BRHVKTAATTAMBDVVD
---DHTATTTACBD---
| Original motif | Reverse complement motif |
| Consensus sequence: BRHVKTAATTAMBDVVD | Consensus sequence: DBBDVYTAATTARBHKB |
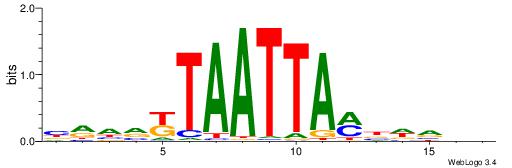 |
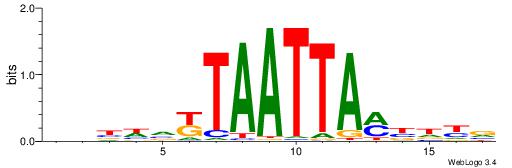 |
| Motif ID: | UP00164 |
| Motif name: | Hoxa7_2668.2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.00942246 |
Alignment:
VVHDDTAATTAHTDDBC
---DHTATTTACBD---
| Original motif | Reverse complement motif |
| Consensus sequence: VVHDDTAATTAHTDDBC | Consensus sequence: GBDDAHTAATTADHHVV |
 |
 |
| Motif ID: | UP00259 |
| Motif name: | Hoxb6 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.00949005 |
Alignment:
VDVGTAATTACBDAWW
-DBGTAAATAHD----
| Original motif | Reverse complement motif |
| Consensus sequence: WWTDBGTAATTACVDB | Consensus sequence: VDVGTAATTACBDAWW |
 |
 |
| Motif ID: | UP00254 |
| Motif name: | Pou2f1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.00998499 |
Alignment:
WABKTAATTAHKHHHD
-DBGTAAATAHD----
| Original motif | Reverse complement motif |
| Consensus sequence: DHDHRHTAATTARBTW | Consensus sequence: WABKTAATTAHKHHHD |
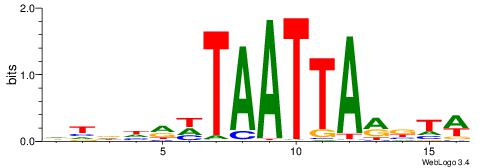 |
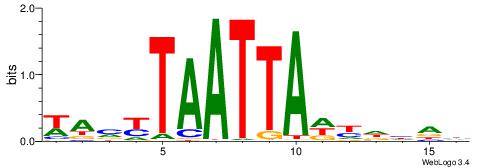 |
| Motif ID: | UP00110 |
| Motif name: | Dlx4 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0107458 |
Alignment:
GHVRGTAATTATDVVGV
--DBGTAAATAHD----
| Original motif | Reverse complement motif |
| Consensus sequence: BCVVDATAATTACMVHC | Consensus sequence: GHVRGTAATTATDVVGV |
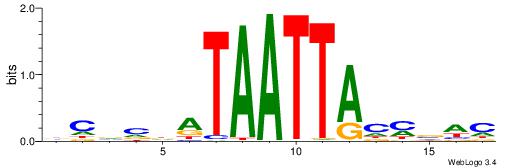 |
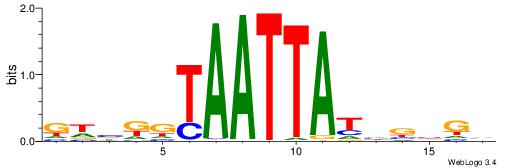 |
| Dataset #: 5 | Motif ID: 46 | Motif name: Zfx |
| Original motif | Reverse complement motif |
| Consensus sequence: BBVGCCBVGGCCTV | Consensus sequence: VAGGCCBBGGCVBB |
 |
 |
Best Matches for Motif ID 46 (Highest to Lowest)
| Motif ID: | UP00005 |
| Motif name: | Tcfap2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0596136 |
Alignment:
HHRCCBBWGGDCDB
BBVGCCBVGGCCTV
| Original motif | Reverse complement motif |
| Consensus sequence: HHRCCBBWGGDCDB | Consensus sequence: BDGHCCWBVGGKDH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0603516 |
Alignment:
ABBBBVVRGACCACCCACRDBBM
BBVGCCBVGGCCTV---------
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0604694 |
Alignment:
BDHHMCGCCCCCTHVBB
---VAGGCCBBGGCVBB
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0606067 |
Alignment:
HBDRCCMCGCCCHHHD
BBVGCCBVGGCCTV--
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 14 |
| Similarity score: | 0.0606878 |
Alignment:
HVGVSAGGAGGGTCYHHHHYVH
--------VAGGCCBBGGCVBB
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Motif ID: | UP00087 |
| Motif name: | Tcfap2c_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0608063 |
Alignment:
HWGCCCTDGGGCDB
VAGGCCBBGGCVBB
| Original motif | Reverse complement motif |
| Consensus sequence: BHGCCCDAGGGCWD | Consensus sequence: HWGCCCTDGGGCDB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 14 |
| Similarity score: | 0.0615001 |
Alignment:
DVTTVVCVYGGHGGYCHHDMYB
----BBVGCCBVGGCCTV----
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00010 |
| Motif name: | Tcfap2b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0624288 |
Alignment:
HHKGCCYSRGGCWAD
-BBVGCCBVGGCCTV
| Original motif | Reverse complement motif |
| Consensus sequence: DTWGCCKSMGGCRHH | Consensus sequence: HHKGCCYSRGGCWAD |
 |
 |
| Motif ID: | UP00082 |
| Motif name: | Zfp187_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0627664 |
Alignment:
DDGMCCTBGTCCHYHV
VAGGCCBBGGCVBB--
| Original motif | Reverse complement motif |
| Consensus sequence: DDGMCCTBGTCCHYHV | Consensus sequence: VHMDGGACVAGGRCDH |
 |
 |
| Motif ID: | UP00009 |
| Motif name: | Nr2f2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0636529 |
Alignment:
HVBVTGACCCSGBVBV
--VAGGCCBBGGCVBB
| Original motif | Reverse complement motif |
| Consensus sequence: VBVBCSGGGTCAVBBH | Consensus sequence: HVBVTGACCCSGBVBV |
 |
 |
| Dataset #: 5 | Motif ID: 47 | Motif name: Egr1 |
| Original motif | Reverse complement motif |
| Consensus sequence: HGCGTGGGCGK | Consensus sequence: YCGCCCACGCH |
 |
 |
Best Matches for Motif ID 47 (Highest to Lowest)
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
------HGCGTGGGCGK------
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.00727533 |
Alignment:
HCCGCCCCCGCAHB
-YCGCCCACGCH--
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0262579 |
Alignment:
MHHHWHYTMCCHCCCACVAABVH
---------YCGCCCACGCH---
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 11 |
| Number of overlap: | 11 |
| Similarity score: | 0.0297053 |
Alignment:
YBVDMGTGGGTGGTCKVVBVBBT
--HGCGTGGGCGK----------
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0318144 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
----HGCGTGGGCGK--------
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0345009 |
Alignment:
BVVHAGGGGGCGRDHHB
--HGCGTGGGCGK----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0353502 |
Alignment:
BTBVTCVTGGGTGGTCMVVDVBB
---HGCGTGGGCGK---------
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0360229 |
Alignment:
BHHDTCCCACTCBDBV
--YCGCCCACGCH---
| Original motif | Reverse complement motif |
| Consensus sequence: BBHBGAGTGGGAHHDB | Consensus sequence: BHHDTCCCACTCBDBV |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 11 |
| Similarity score: | 0.0362743 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
--------YCGCCCACGCH----
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00097 |
| Motif name: | Mtf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0386944 |
Alignment:
HDYTTGCACACGDHBH
---YCGCCCACGCH--
| Original motif | Reverse complement motif |
| Consensus sequence: DBDHCGTGTGCAAMDH | Consensus sequence: HDYTTGCACACGDHBH |
 |
 |
| Dataset #: 5 | Motif ID: 48 | Motif name: SP1 |
| Original motif | Reverse complement motif |
| Consensus sequence: CCCCKCCCCC | Consensus sequence: GGGGGYGGGG |
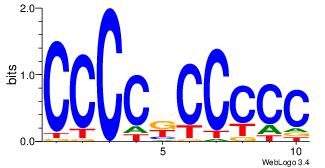 |
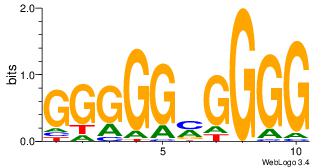 |
Best Matches for Motif ID 48 (Highest to Lowest)
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0231018 |
Alignment:
DBCCCCCCCCCCMYC
--CCCCKCCCCC---
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0303207 |
Alignment:
HBDRCCMCGCCCHHHD
----CCCCKCCCCC--
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0323007 |
Alignment:
HHHHDGGGGCGGDBHDV
----GGGGGYGGGG---
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
 |
 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0340973 |
Alignment:
HVDVGGGHGGGGDBHB
--GGGGGYGGGG----
| Original motif | Reverse complement motif |
| Consensus sequence: BHVDCCCCDCCCBDBH | Consensus sequence: HVDVGGGHGGGGDBHB |
 |
 |
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0351051 |
Alignment:
BHHHWGGGGCGGBBVH
----GGGGGYGGGG--
| Original motif | Reverse complement motif |
| Consensus sequence: HBBBCCGCCCCWHHHV | Consensus sequence: BHHHWGGGGCGGBBVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0390437 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
------GGGGGYGGGG------
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0393358 |
Alignment:
HHBSHTCGTGRGTGGKGDBVWMV
-------GGGGGYGGGG------
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0413863 |
Alignment:
DHHBCCCCGCCAHHBHB
----CCCCKCCCCC---
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0418947 |
Alignment:
HVBCCCCCCCCMHHHB
---CCCCKCCCCC---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0420708 |
Alignment:
BDHHMCGCCCCCTHVBB
--CCCCKCCCCC-----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Dataset #: 5 | Motif ID: 49 | Motif name: TFAP2A |
| Original motif | Reverse complement motif |
| Consensus sequence: GCCBBVRGS | Consensus sequence: SCMVBBGGC |
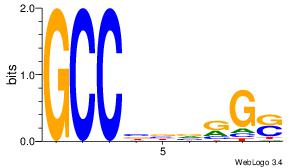 |
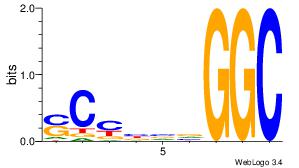 |
Best Matches for Motif ID 49 (Highest to Lowest)
| Motif ID: | UP00010 |
| Motif name: | Tcfap2b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 9 |
| Similarity score: | 0.0115546 |
Alignment:
HHKGCCYSRGGCWAD
---GCCBBVRGS---
| Original motif | Reverse complement motif |
| Consensus sequence: DTWGCCKSMGGCRHH | Consensus sequence: HHKGCCYSRGGCWAD |
 |
 |
| Motif ID: | UP00010 |
| Motif name: | Tcfap2b_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 9 |
| Similarity score: | 0.0150447 |
Alignment:
DYGCCYBARGGCAH
--GCCBBVRGS---
| Original motif | Reverse complement motif |
| Consensus sequence: DYGCCYBARGGCAH | Consensus sequence: HTGCCMTVKGGCMD |
 |
 |
| Motif ID: | UP00005 |
| Motif name: | Tcfap2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 9 |
| Similarity score: | 0.0151527 |
Alignment:
WDHSCCYSRGGSRAD
---GCCBBVRGS---
| Original motif | Reverse complement motif |
| Consensus sequence: WDHSCCYSRGGSRAD | Consensus sequence: DTMSCCKSMGGSHDW |
 |
 |
| Motif ID: | UP00087 |
| Motif name: | Tcfap2c_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 9 |
| Similarity score: | 0.016909 |
Alignment:
WHHSCCYSRGGSDAD
---GCCBBVRGS---
| Original motif | Reverse complement motif |
| Consensus sequence: WHHSCCYSRGGSDAD | Consensus sequence: DTHSCCKSMGGSHHW |
 |
 |
| Motif ID: | UP00087 |
| Motif name: | Tcfap2c_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 9 |
| Similarity score: | 0.0170259 |
Alignment:
BHGCCCDAGGGCWD
--GCCBBVRGS---
| Original motif | Reverse complement motif |
| Consensus sequence: BHGCCCDAGGGCWD | Consensus sequence: HWGCCCTDGGGCDB |
 |
 |
| Motif ID: | UP00028 |
| Motif name: | Tcfap2e_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 9 |
| Similarity score: | 0.0198854 |
Alignment:
HHHGCCTSAGGCDAD
---GCCBBVRGS---
| Original motif | Reverse complement motif |
| Consensus sequence: HHHGCCTSAGGCDAD | Consensus sequence: DTHGCCTSAGGCHHH |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 9 |
| Similarity score: | 0.0203681 |
Alignment:
BHBHDTGGCGGGGBDHD
SCMVBBGGC--------
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
 |
 |
| Motif ID: | UP00003 |
| Motif name: | E2F3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 9 |
| Similarity score: | 0.0217064 |
Alignment:
BBBDYTGGCGCCKWDBD
SCMVBBGGC--------
| Original motif | Reverse complement motif |
| Consensus sequence: HBHWYGGCGCCAMDVBB | Consensus sequence: BBBDYTGGCGCCKWDBD |
 |
 |
| Motif ID: | UP00005 |
| Motif name: | Tcfap2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 9 |
| Similarity score: | 0.0234775 |
Alignment:
HHRCCBBWGGDCDB
--GCCBBVRGS---
| Original motif | Reverse complement motif |
| Consensus sequence: HHRCCBBWGGDCDB | Consensus sequence: BDGHCCWBVGGKDH |
 |
 |
| Motif ID: | UP00093 |
| Motif name: | Klf7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 9 |
| Similarity score: | 0.026642 |
Alignment:
VVDBATACGCCCHAHHD
--------GCCBBVRGS
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBATACGCCCHAHHD | Consensus sequence: DHDTHGGGCGTATBHBB |
 |
 |
| Dataset #: 5 | Motif ID: 50 | Motif name: MIZF |
| Original motif | Reverse complement motif |
| Consensus sequence: BAACGTCCGC | Consensus sequence: GCGGACGTTV |
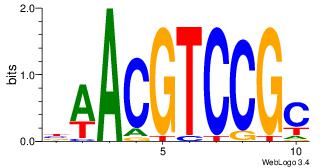 |
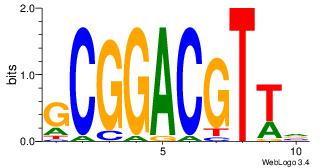 |
Best Matches for Motif ID 50 (Highest to Lowest)
| Motif ID: | UP00419 |
| Motif name: | Spic |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
DWACTTCCRCTTTB
BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: VAAAGMGGAAGTWD | Consensus sequence: DWACTTCCRCTTTB |
 |
 |
| Motif ID: | UP00409 |
| Motif name: | Elf5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.00379999 |
Alignment:
BWAHSMGGAAGTWD
BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: BWAHSMGGAAGTWD | Consensus sequence: DWACTTCCRSDTWV |
 |
 |
| Motif ID: | UP00413 |
| Motif name: | Elf4 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0124101 |
Alignment:
DBBHACTTCCGGKWTH
--BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGKWTH | Consensus sequence: HAWYCCGGAAGTHBBD |
 |
 |
| Motif ID: | UP00404 |
| Motif name: | Elf2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0128348 |
Alignment:
DBBHACTTCCGGDWDB
--BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGDWDB | Consensus sequence: BDWDCCGGAAGTHBBD |
 |
 |
| Motif ID: | UP00085 |
| Motif name: | Sfpi1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0141516 |
Alignment:
HDACTTCCBCWTDV
BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: BDAWGVGGAAGTDH | Consensus sequence: HDACTTCCBCWTDV |
 |
 |
| Motif ID: | UP00423 |
| Motif name: | Gm5454 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0144665 |
Alignment:
DVHACCGGAAGTHDDV
----GCGGACGTTV--
| Original motif | Reverse complement motif |
| Consensus sequence: VDHHACTTCCGGTHVH | Consensus sequence: DVHACCGGAAGTHDDV |
 |
 |
| Motif ID: | UP00412 |
| Motif name: | Etv5 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0155825 |
Alignment:
DHHACCGGAWGTTDDV
----GCGGACGTTV--
| Original motif | Reverse complement motif |
| Consensus sequence: VDHAACWTCCGGTHHH | Consensus sequence: DHHACCGGAWGTTDDV |
 |
 |
| Motif ID: | UP00015 |
| Motif name: | Ehf_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0162018 |
Alignment:
VBDABCCGGAAGTDD
-----GCGGACGTTV
| Original motif | Reverse complement motif |
| Consensus sequence: VBDABCCGGAAGTDD | Consensus sequence: DDACTTCCGGBTHBB |
 |
 |
| Motif ID: | UP00418 |
| Motif name: | Etv6 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0171234 |
Alignment:
DVHBTACTTCCGGHTHB
---BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: DVHBTACTTCCGGHTHB | Consensus sequence: BHAHCCGGAAGTABDVD |
 |
 |
| Motif ID: | UP00421 |
| Motif name: | Etv1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0184147 |
Alignment:
MVHBACCGGAAGTVDVH
-----GCGGACGTTV--
| Original motif | Reverse complement motif |
| Consensus sequence: MVHBACCGGAAGTVDVH | Consensus sequence: HBDBACTTCCGGTBHVY |
 |
 |
| Dataset #: 5 | Motif ID: 51 | Motif name: Klf4 |
| Original motif | Reverse complement motif |
| Consensus sequence: DGGGYGKGGC | Consensus sequence: GCCYCMCCCD |
 |
 |
Best Matches for Motif ID 51 (Highest to Lowest)
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
DHHDGGGCGRGGKHBH
---DGGGYGKGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0153308 |
Alignment:
DBCCCCCCCCCCMYC
---GCCYCMCCCD--
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0186791 |
Alignment:
BDWAGGCGTGBCHHD
--DGGGYGKGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: BDWAGGCGTGBCHHD | Consensus sequence: HHDGBCACGCCTWDB |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0218399 |
Alignment:
HVBCCCCCCCCMHHHB
--GCCYCMCCCD----
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0249586 |
Alignment:
VHTGCGGGGGCGGH
GCCYCMCCCD----
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0299159 |
Alignment:
BVVHAGGGGGCGRDHHB
------DGGGYGKGGC-
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0308595 |
Alignment:
BHBHDTGGCGGGGBDHD
----DGGGYGKGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
 |
 |
| Motif ID: | UP00047 |
| Motif name: | Zbtb7b_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0345891 |
Alignment:
VHWTTHGGGGGGVDD
GCCYCMCCCD-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDVCCCCCCHAAWHB | Consensus sequence: VHWTTHGGGGGGVDD |
 |
 |
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0346735 |
Alignment:
HBBBCCGCCCCWHHHV
--GCCYCMCCCD----
| Original motif | Reverse complement motif |
| Consensus sequence: HBBBCCGCCCCWHHHV | Consensus sequence: BHHHWGGGGCGGBBVH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 10 |
| Similarity score: | 0.0351496 |
Alignment:
MHHHWHYTMCCHCCCACVAABVH
------GCCYCMCCCD-------
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Dataset #: 5 | Motif ID: 52 | Motif name: E2F1 |
| Original motif | Reverse complement motif |
| Consensus sequence: TTTSGCGC | Consensus sequence: GCGCSAAA |
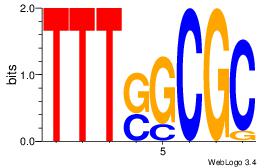 |
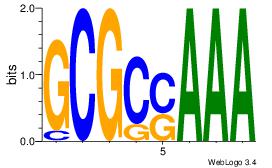 |
Best Matches for Motif ID 52 (Highest to Lowest)
| Motif ID: | UP00001 |
| Motif name: | E2F2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HVBWYGGCGCCAADVBB
------GCGCSAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBWYGGCGCCAADVBB | Consensus sequence: BBBDTTGGCGCCKWVVD |
 |
 |
| Motif ID: | UP00003 |
| Motif name: | E2F3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.00182028 |
Alignment:
HBHWYGGCGCCAMDVBB
------GCGCSAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: HBHWYGGCGCCAMDVBB | Consensus sequence: BBBDYTGGCGCCKWDBD |
 |
 |
| Motif ID: | UP00072 |
| Motif name: | IRC900814_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0237639 |
Alignment:
BHDWTTTGTCGTAHDD
----TTTSGCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: DDHTACGACAAAWDDB | Consensus sequence: BHDWTTTGTCGTAHDD |
 |
 |
| Motif ID: | UP00003 |
| Motif name: | E2F3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0275636 |
Alignment:
WHSGCGCGCCHTDHB
-----GCGCSAAA--
| Original motif | Reverse complement motif |
| Consensus sequence: VHDADGGCGCGCSHW | Consensus sequence: WHSGCGCGCCHTDHB |
 |
 |
| Motif ID: | UP00065 |
| Motif name: | Zfp161_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.028109 |
Alignment:
DMMDGMGCGCGCGCHD
--------GCGCSAAA
| Original motif | Reverse complement motif |
| Consensus sequence: DDGCGCGCGCRCHYRD | Consensus sequence: DMMDGMGCGCGCGCHD |
 |
 |
| Motif ID: | UP00001 |
| Motif name: | E2F2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0296302 |
Alignment:
HHVGCGCGCCKTDHB
-----GCGCSAAA--
| Original motif | Reverse complement motif |
| Consensus sequence: VHDARGGCGCGCVHH | Consensus sequence: HHVGCGCGCCKTDHB |
 |
 |
| Motif ID: | UP00068 |
| Motif name: | Eomes_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.0348969 |
Alignment:
BVHDAGGTGTGAAAHHH
------GCGCSAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: BVHDAGGTGTGAAAHHH | Consensus sequence: HHHTTTCACACCTDHBV |
 |
 |
| Motif ID: | UP00011 |
| Motif name: | Irf6_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0368278 |
Alignment:
DHDBTKGTTTCGHTHDD
-------TTTSGCGC--
| Original motif | Reverse complement motif |
| Consensus sequence: HDDAHCGAAACYAVDDD | Consensus sequence: DHDBTKGTTTCGHTHDD |
 |
 |
| Motif ID: | UP00049 |
| Motif name: | Sp100_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0383813 |
Alignment:
DWTTTCCGHHWAWD
-----GCGCSAAA-
| Original motif | Reverse complement motif |
| Consensus sequence: DWTWHHCGGAAAWD | Consensus sequence: DWTTTCCGHHWAWD |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.0406002 |
Alignment:
TCDAMVBGACGCMAKDDDVBBD
--------GCGCSAAA------
| Original motif | Reverse complement motif |
| Consensus sequence: DBBVDDDRTRGCGTCBBYTHGA | Consensus sequence: TCDAMVBGACGCMAKDDDVBBD |
 |
 |
| Dataset #: 5 | Motif ID: 53 | Motif name: HIF1AARNT |
| Original motif | Reverse complement motif |
| Consensus sequence: VBACGTGV | Consensus sequence: VCACGTBV |
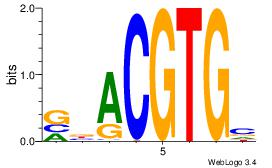 |
 |
Best Matches for Motif ID 53 (Highest to Lowest)
| Motif ID: | UP00097 |
| Motif name: | Mtf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HDYTTGCACACGDHBH
-------VCACGTBV-
| Original motif | Reverse complement motif |
| Consensus sequence: DBDHCGTGTGCAAMDH | Consensus sequence: HDYTTGCACACGDHBH |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.000574345 |
Alignment:
DDASCACGTGBTBVDD
---VBACGTGV-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDASCACGTGBTBVDD | Consensus sequence: HHVBABCACGTGSTHD |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.00642455 |
Alignment:
YDYBDHTMCACGTGGADDBMDGT
-------VBACGTGV--------
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00084 |
| Motif name: | Gmeb1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0131171 |
Alignment:
BVVDVRYACGTRYVBHB
-----VBACGTGV----
| Original motif | Reverse complement motif |
| Consensus sequence: BHBBKKACGTMMVDBVB | Consensus sequence: BVVDVRYACGTRYVBHB |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.014006 |
Alignment:
RDHDBVDTCACGTGASBHVHDH
-------VCACGTBV-------
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0184296 |
Alignment:
VBHHVCAGGTGCDVDHD
----VBACGTGV-----
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Motif ID: | UP00068 |
| Motif name: | Eomes_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0188154 |
Alignment:
VVBVCKACACCTHVBV
------VCACGTBV--
| Original motif | Reverse complement motif |
| Consensus sequence: VBVDAGGTGTYGVBBV | Consensus sequence: VVBVCKACACCTHVBV |
 |
 |
| Motif ID: | UP00026 |
| Motif name: | Zscan4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.0189568 |
Alignment:
DHVDTGTGCACAYAHDH
-VBACGTGV--------
| Original motif | Reverse complement motif |
| Consensus sequence: DHVDTGTGCACAYAHDH | Consensus sequence: HDHTMTGTGCACADVHD |
 |
 |
| Motif ID: | UP00042 |
| Motif name: | Gm397_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0198987 |
Alignment:
HHRTATGTGCACATHHD
--VBACGTGV-------
| Original motif | Reverse complement motif |
| Consensus sequence: HHDATGTGCACATAMDH | Consensus sequence: HHRTATGTGCACATHHD |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.021967 |
Alignment:
BBGHCACGCGACDD
---VCACGTBV---
| Original motif | Reverse complement motif |
| Consensus sequence: BBGHCACGCGACDD | Consensus sequence: HDGTCGCGTGDCVB |
 |
 |
| Dataset #: 5 | Motif ID: 54 | Motif name: PLAG1 |
| Original motif | Reverse complement motif |
| Consensus sequence: GGGGCCCAAGGGGG | Consensus sequence: CCCCCTTGGGCCCC |
 |
 |
Best Matches for Motif ID 54 (Highest to Lowest)
| Motif ID: | UP00102 |
| Motif name: | Zic1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0499525 |
Alignment:
CCCCCCCGGGGGHB
CCCCCTTGGGCCCC
| Original motif | Reverse complement motif |
| Consensus sequence: BHCCCCCGGGGGGG | Consensus sequence: CCCCCCCGGGGGHB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0540613 |
Alignment:
BBBDVVRGACCACCCAVGABBAB
---------CCCCCTTGGGCCCC
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00057 |
| Motif name: | Zic2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0548411 |
Alignment:
BMCCCCCGGGGGGGB
-CCCCCTTGGGCCCC
| Original motif | Reverse complement motif |
| Consensus sequence: BMCCCCCGGGGGGGB | Consensus sequence: VCCCCCCCGGGGGRB |
 |
 |
| Motif ID: | UP00005 |
| Motif name: | Tcfap2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0556785 |
Alignment:
HHRCCBBWGGDCDB
GGGGCCCAAGGGGG
| Original motif | Reverse complement motif |
| Consensus sequence: HHRCCBBWGGDCDB | Consensus sequence: BDGHCCWBVGGKDH |
 |
 |
| Motif ID: | UP00006 |
| Motif name: | Zic3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.056976 |
Alignment:
BMCCCCCGGGGGGGB
-CCCCCTTGGGCCCC
| Original motif | Reverse complement motif |
| Consensus sequence: BMCCCCCGGGGGGGB | Consensus sequence: VCCCCCCCGGGGGRB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 14 |
| Similarity score: | 0.0573495 |
Alignment:
HDGHHBHRGACCACCCGSVDVG
CCCCCTTGGGCCCC--------
| Original motif | Reverse complement motif |
| Consensus sequence: HDGHHBHRGACCACCCGSVDVG | Consensus sequence: CVHBSCGGGTGGTCMHVHHCDH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 14 |
| Similarity score: | 0.0579022 |
Alignment:
VMYDHDGMCCHCCKBGVVAAVH
--------CCCCCTTGGGCCCC
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 14 |
| Similarity score: | 0.0580662 |
Alignment:
DVMHHDHKGACCCTCCTSVCBH
CCCCCTTGGGCCCC--------
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0597453 |
Alignment:
HGDHVHKGACCACCCGGBGBGDB
---------CCCCCTTGGGCCCC
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0598088 |
Alignment:
GKRGGGGGGGGGGBD
-CCCCCTTGGGCCCC
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Dataset #: 5 | Motif ID: 55 | Motif name: Pax5 |
| Original motif | Reverse complement motif |
| Consensus sequence: DGVBCABTGDWGCGKRRCSR | Consensus sequence: MSGKKRCGCWDCABTGBBCD |
 |
 |
Best Matches for Motif ID 55 (Highest to Lowest)
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 20 |
| Similarity score: | 0.0499537 |
Alignment:
DDRMHBACGCGYGCGTDGBBDB
-DGVBCABTGDWGCGKRRCSR-
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 20 |
| Similarity score: | 0.0520196 |
Alignment:
WHVBVVMACGCGCGTCDYHWDH
DGVBCABTGDWGCGKRRCSR--
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 20 |
| Similarity score: | 0.0542764 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
-DGVBCABTGDWGCGKRRCSR-
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 20 |
| Similarity score: | 0.0560636 |
Alignment:
DCATDBTGHGCATKMTTBRSWR
--MSGKKRCGCWDCABTGBBCD
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 20 |
| Similarity score: | 0.0595671 |
Alignment:
HHBSHTCGTGRGTGGKGDBVWMV
DGVBCABTGDWGCGKRRCSR---
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 20 |
| Similarity score: | 0.0602559 |
Alignment:
HADTBVADGATGCATCMTGMVHB
DGVBCABTGDWGCGKRRCSR---
| Original motif | Reverse complement motif |
| Consensus sequence: HADTBVADGATGCATCMTGMVHB | Consensus sequence: VHVRCAYGATGCATCDTVVADTH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 20 |
| Similarity score: | 0.0607506 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
DGVBCABTGDWGCGKRRCSR---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 20 |
| Similarity score: | 0.0611227 |
Alignment:
BDBBVBVGTAWCCAAGVBHBAGB
MSGKKRCGCWDCABTGBBCD---
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 20 |
| Similarity score: | 0.0611361 |
Alignment:
RDHDBVDTCACGTGASBHVHDH
--MSGKKRCGCWDCABTGBBCD
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 20 |
| Similarity score: | 0.061403 |
Alignment:
GTVBDAATTGTVTGHGDDHKVB
DGVBCABTGDWGCGKRRCSR--
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Dataset #: 5 | Motif ID: 56 | Motif name: ArntAhr |
| Original motif | Reverse complement motif |
| Consensus sequence: YGCGTG | Consensus sequence: CACGCM |
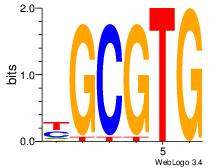 |
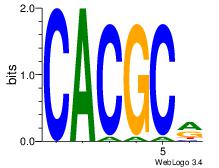 |
Best Matches for Motif ID 56 (Highest to Lowest)
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
------YGCGTG-----------
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.00183587 |
Alignment:
HHDGBCACGCCTWDB
-----CACGCM----
| Original motif | Reverse complement motif |
| Consensus sequence: BDWAGGCGTGBCHHD | Consensus sequence: HHDGBCACGCCTWDB |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.00348048 |
Alignment:
HDGTCGCGTGDCVB
----YGCGTG----
| Original motif | Reverse complement motif |
| Consensus sequence: BBGHCACGCGACDD | Consensus sequence: HDGTCGCGTGDCVB |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.00526158 |
Alignment:
TCDAMVBGACGCMAKDDDVBBD
-------CACGCM---------
| Original motif | Reverse complement motif |
| Consensus sequence: DBBVDDDRTRGCGTCBBYTHGA | Consensus sequence: TCDAMVBGACGCMAKDDDVBBD |
 |
 |
| Motif ID: | UP00042 |
| Motif name: | Gm397_second |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.00558171 |
Alignment:
HBGYGTGTGTGCVRVD
---YGCGTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: DVMVGCACACACKCVH | Consensus sequence: HBGYGTGTGTGCVRVD |
 |
 |
| Motif ID: | UP00097 |
| Motif name: | Mtf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.00702416 |
Alignment:
HDYTTGCACACGDHBH
--------CACGCM--
| Original motif | Reverse complement motif |
| Consensus sequence: DBDHCGTGTGCAAMDH | Consensus sequence: HDYTTGCACACGDHBH |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.0123233 |
Alignment:
DDRMHBACGCGYGCGTDGBBDB
-------YGCGTG---------
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0141579 |
Alignment:
HCCGCCCCCGCAHB
------CACGCM--
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 11 |
| Number of overlap: | 6 |
| Similarity score: | 0.0144485 |
Alignment:
BHCBCBCCGGGTGGTCYHVHDCH
-------YGCGTG----------
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 10 |
| Number of overlap: | 6 |
| Similarity score: | 0.0172109 |
Alignment:
VHYTCCDWCCGCGTCMMTHWHV
---------YGCGTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Dataset #: 5 | Motif ID: 57 | Motif name: Mycn |
| Original motif | Reverse complement motif |
| Consensus sequence: HSCACGTGGC | Consensus sequence: GCCACGTGSD |
 |
 |
Best Matches for Motif ID 57 (Highest to Lowest)
| Motif ID: | UP00060 |
| Motif name: | Max_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
DDASCACGTGBTBVDD
--HSCACGTGGC----
| Original motif | Reverse complement motif |
| Consensus sequence: DDASCACGTGBTBVDD | Consensus sequence: HHVBABCACGTGSTHD |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.00783793 |
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
-------GCCACGTGSD------
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0132858 |
Alignment:
DDHBHBSTCACGTGAHBBDHHM
------GCCACGTGSD------
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0227933 |
Alignment:
VBHHVCAGGTGCDVDHD
---GCCACGTGSD----
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0257011 |
Alignment:
BBGHCACGCGACDD
--HSCACGTGGC--
| Original motif | Reverse complement motif |
| Consensus sequence: BBGHCACGCGACDD | Consensus sequence: HDGTCGCGTGDCVB |
 |
 |
| Motif ID: | UP00165 |
| Motif name: | Titf1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0344833 |
Alignment:
BVABCCACTTGAMHTT
---GCCACGTGSD---
| Original motif | Reverse complement motif |
| Consensus sequence: BVABCCACTTGAMHTT | Consensus sequence: AAHYTCAAGTGGBTBV |
 |
 |
| Motif ID: | UP00231 |
| Motif name: | Nkx2-2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.035228 |
Alignment:
HDVRCCACTTGARADBD
---GCCACGTGSD----
| Original motif | Reverse complement motif |
| Consensus sequence: HDVRCCACTTGARADBD | Consensus sequence: DVDTKTCAAGTGGKBDH |
 |
 |
| Motif ID: | UP00084 |
| Motif name: | Gmeb1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0358581 |
Alignment:
BVVDVRYACGTRYVBHB
----GCCACGTGSD---
| Original motif | Reverse complement motif |
| Consensus sequence: BHBBKKACGTMMVDBVB | Consensus sequence: BVVDVRYACGTRYVBHB |
 |
 |
| Motif ID: | UP00084 |
| Motif name: | Gmeb1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.03725 |
Alignment:
HHMACKACGTMGBVCD
---HSCACGTGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: DGVBCRACGTYGTYHH | Consensus sequence: HHMACKACGTMGBVCD |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0383187 |
Alignment:
WHVBVVMACGCGCGTCDYHWDH
------HSCACGTGGC------
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
 |
 |
| Dataset #: 5 | Motif ID: 58 | Motif name: Myc |
| Original motif | Reverse complement motif |
| Consensus sequence: VGCACGTGGH | Consensus sequence: DCCACGTGCV |
 |
 |
Best Matches for Motif ID 58 (Highest to Lowest)
| Motif ID: | UP00060 |
| Motif name: | Max_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
DDASCACGTGBTBVDD
--VGCACGTGGH----
| Original motif | Reverse complement motif |
| Consensus sequence: DDASCACGTGBTBVDD | Consensus sequence: HHVBABCACGTGSTHD |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 10 |
| Similarity score: | 0.00631885 |
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
-------DCCACGTGCV------
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.00821668 |
Alignment:
DDHBHBSTCACGTGAHBBDHHM
------DCCACGTGCV------
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0140005 |
Alignment:
DHDBHGCACCTGBDDVB
----VGCACGTGGH---
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0308912 |
Alignment:
BBGHCACGCGACDD
--VGCACGTGGH--
| Original motif | Reverse complement motif |
| Consensus sequence: BBGHCACGCGACDD | Consensus sequence: HDGTCGCGTGDCVB |
 |
 |
| Motif ID: | UP00165 |
| Motif name: | Titf1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0311723 |
Alignment:
BVABCCACTTGAMHTT
---DCCACGTGCV---
| Original motif | Reverse complement motif |
| Consensus sequence: BVABCCACTTGAMHTT | Consensus sequence: AAHYTCAAGTGGBTBV |
 |
 |
| Motif ID: | UP00249 |
| Motif name: | Nkx2-5 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.031308 |
Alignment:
HDADCCACTTRAAWTT
---DCCACGTGCV---
| Original motif | Reverse complement motif |
| Consensus sequence: HDADCCACTTRAAWTT | Consensus sequence: AAWTTMAAGTGGHTDH |
 |
 |
| Motif ID: | UP00231 |
| Motif name: | Nkx2-2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0334845 |
Alignment:
HDVRCCACTTGARADBD
---DCCACGTGCV----
| Original motif | Reverse complement motif |
| Consensus sequence: HDVRCCACTTGARADBD | Consensus sequence: DVDTKTCAAGTGGKBDH |
 |
 |
| Motif ID: | UP00147 |
| Motif name: | Nkx2-6 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0345548 |
Alignment:
DDADCCACTTAAVHTT
---DCCACGTGCV---
| Original motif | Reverse complement motif |
| Consensus sequence: DDADCCACTTAAVHTT | Consensus sequence: AAHVTTAAGTGGHTDD |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 10 |
| Similarity score: | 0.0363818 |
Alignment:
VDWDARRGACGCGGWHGGAKHV
-----VGCACGTGGH-------
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Dataset #: 5 | Motif ID: 59 | Motif name: GABPA |
| Original motif | Reverse complement motif |
| Consensus sequence: CCGGAAGTGVV | Consensus sequence: VVCACTTCCGG |
 |
 |
Best Matches for Motif ID 59 (Highest to Lowest)
| Motif ID: | UP00013 |
| Motif name: | Gabpa_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0 |
Alignment:
BHHHACTTCCGGTHHBD
-VVCACTTCCGG-----
| Original motif | Reverse complement motif |
| Consensus sequence: HVHHACCGGAAGTDHHV | Consensus sequence: BHHHACTTCCGGTHHBD |
 |
 |
| Motif ID: | UP00410 |
| Motif name: | Elk1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.000136484 |
Alignment:
VVHBACCGGAAGTDBHD
-----CCGGAAGTGVV-
| Original motif | Reverse complement motif |
| Consensus sequence: VVHBACCGGAAGTDBHD | Consensus sequence: DHBDACTTCCGGTVHVB |
 |
 |
| Motif ID: | UP00420 |
| Motif name: | Elk3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.000219706 |
Alignment:
DVBBACCGGAAGYDVVV
-----CCGGAAGTGVV-
| Original motif | Reverse complement motif |
| Consensus sequence: DVBBACCGGAAGYDVVV | Consensus sequence: VBVDMCTTCCGGTVBVD |
 |
 |
| Motif ID: | UP00421 |
| Motif name: | Etv1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.000932023 |
Alignment:
MVHBACCGGAAGTVDVH
-----CCGGAAGTGVV-
| Original motif | Reverse complement motif |
| Consensus sequence: MVHBACCGGAAGTVDVH | Consensus sequence: HBDBACTTCCGGTBHVY |
 |
 |
| Motif ID: | UP00404 |
| Motif name: | Elf2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.00527483 |
Alignment:
DBBHACTTCCGGDWDB
-VVCACTTCCGG----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGDWDB | Consensus sequence: BDWDCCGGAAGTHBBD |
 |
 |
| Motif ID: | UP00414 |
| Motif name: | Ets1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.00805613 |
Alignment:
BDDDHACCGGAARTVDD
------CCGGAAGTGVV
| Original motif | Reverse complement motif |
| Consensus sequence: BDDDHACCGGAARTVDD | Consensus sequence: DDBAMTTCCGGTDHDHB |
 |
 |
| Motif ID: | UP00423 |
| Motif name: | Gm5454 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.00933976 |
Alignment:
VDHHACTTCCGGTHVH
-VVCACTTCCGG----
| Original motif | Reverse complement motif |
| Consensus sequence: VDHHACTTCCGGTHVH | Consensus sequence: DVHACCGGAAGTHDDV |
 |
 |
| Motif ID: | UP00413 |
| Motif name: | Elf4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.00992792 |
Alignment:
DBBHACTTCCGGKWTH
-VVCACTTCCGG----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGKWTH | Consensus sequence: HAWYCCGGAAGTHBBD |
 |
 |
| Motif ID: | UP00418 |
| Motif name: | Etv6 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0114183 |
Alignment:
DVHBTACTTCCGGHTHB
--VVCACTTCCGG----
| Original motif | Reverse complement motif |
| Consensus sequence: DVHBTACTTCCGGHTHB | Consensus sequence: BHAHCCGGAAGTABDVD |
 |
 |
| Motif ID: | UP00411 |
| Motif name: | Erg |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0122484 |
Alignment:
BDDACCGGAAKTVBVB
----CCGGAAGTGVV-
| Original motif | Reverse complement motif |
| Consensus sequence: VBBVAYTTCCGGTDDB | Consensus sequence: BDDACCGGAAKTVBVB |
 |
 |