Best Matches in Database for Each Motif (Highest to Lowest)
| Dataset #: 1 | Motif ID: 1 | Motif name: Motif 1 |
| Original motif | Reverse complement motif |
| Consensus sequence: GGCGGGGC | Consensus sequence: GCCCCGCC |
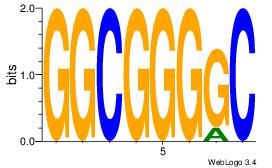 |
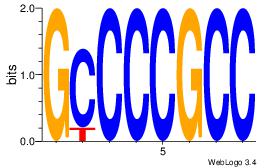 |
Best Matches for Motif ID 1 (Highest to Lowest)
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HBDRCCMCGCCCHHHD
---GCCCCGCC-----
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
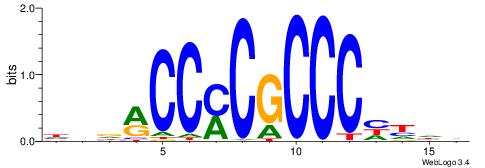 |
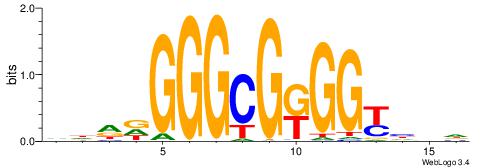 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.00617043 |
Alignment:
BHBHDTGGCGGGGBDHD
------GGCGGGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
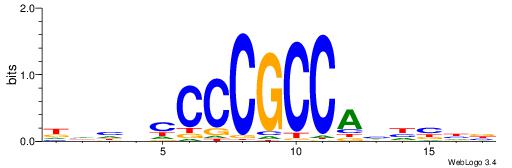 |
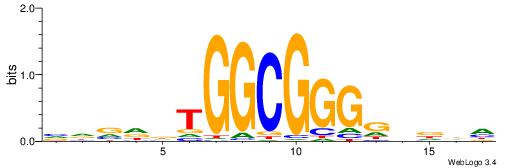 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0145996 |
Alignment:
BDWAGGCGTGBCHHD
----GGCGGGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: BDWAGGCGTGBCHHD | Consensus sequence: HHDGBCACGCCTWDB |
 |
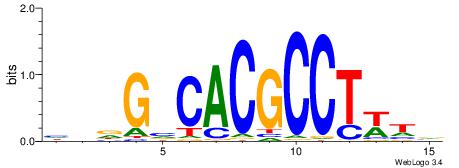 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0233506 |
Alignment:
BDHHMCGCCCCCTHVBB
-GCCCCGCC--------
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
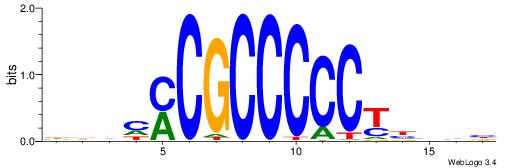 |
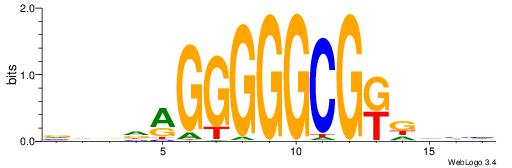 |
| Motif ID: | UP00024 |
| Motif name: | Glis2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0291232 |
Alignment:
HHHVGACCCCCCVBRD
----GCCCCGCC----
| Original motif | Reverse complement motif |
| Consensus sequence: HHHVGACCCCCCVBRD | Consensus sequence: HKBBGGGGGGTCVHHH |
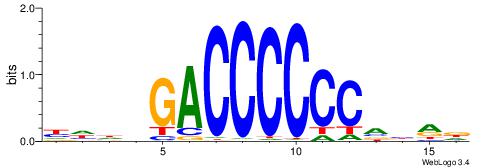 |
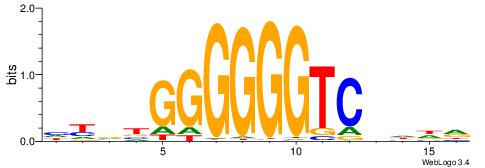 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0338469 |
Alignment:
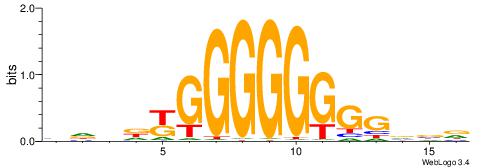
BHHDYGGGGGGGGBVD
-----GGCGGGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
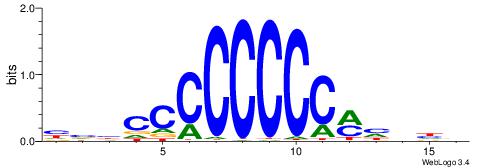 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0361147 |
Alignment:
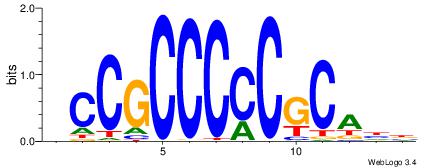
VHTGCGGGGGCGGH
---GGCGGGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0374146 |
Alignment:
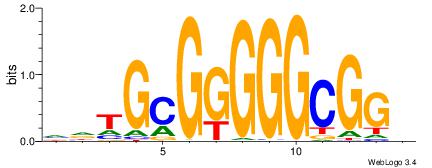
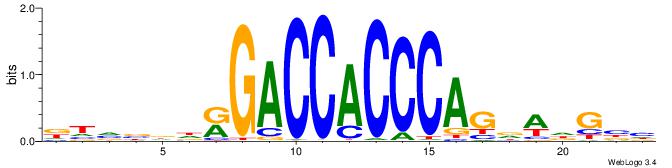
DTVBDDRGACCACCCAGDWDGBH
-------GCCCCGCC--------
| Original motif | Reverse complement motif |
| Consensus sequence: DTVBDDRGACCACCCAGDWDGBH | Consensus sequence: DBCDWHCTGGGTGGTCMDHBBAH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.0376381 |
Alignment:
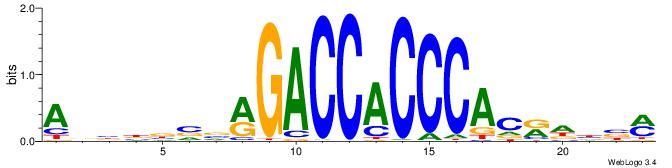
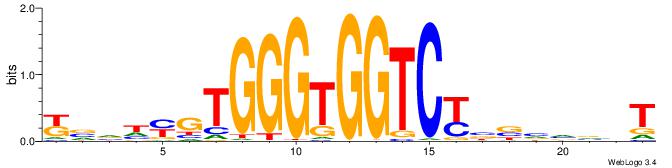
ABBBBVVRGACCACCCACRDBBM
--------GCCCCGCC-------
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0392043 |
Alignment:
GKRGGGGGGGGGGBD
---GGCGGGGC----
| Original motif | Reverse complement motif |
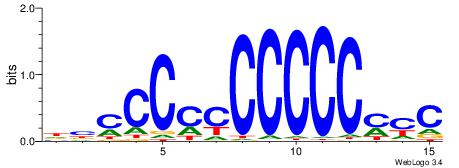
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
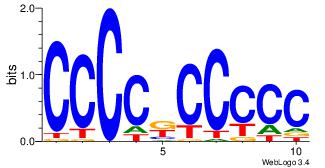
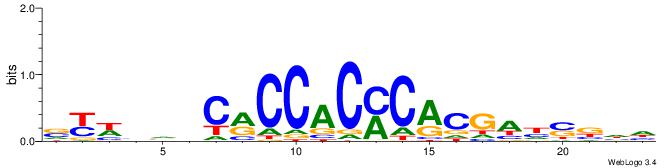
| Dataset #: 3 | Motif ID: 24 | Motif name: SP1 |
| Original motif | Reverse complement motif |
| Consensus sequence: CCCCKCCCCC | Consensus sequence: GGGGGYGGGG |
 |
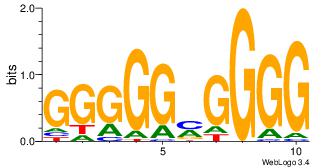 |
Best Matches for Motif ID 24 (Highest to Lowest)
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0231018 |
Alignment:
DBCCCCCCCCCCMYC
--CCCCKCCCCC---
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0303207 |
Alignment:
HBDRCCMCGCCCHHHD
----CCCCKCCCCC--
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0323007 |
Alignment:
HHHHDGGGGCGGDBHDV
----GGGGGYGGGG---
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
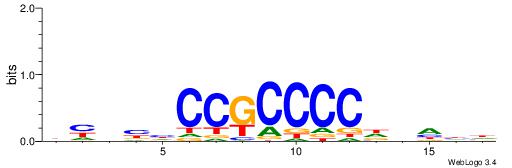 |
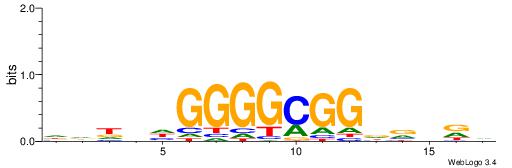 |
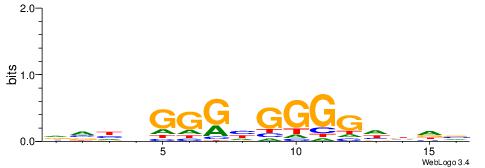
| Motif ID: | UP00099 |
| Motif name: | Ascl2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0340973 |
Alignment:
HVDVGGGHGGGGDBHB
--GGGGGYGGGG----
| Original motif | Reverse complement motif |
| Consensus sequence: BHVDCCCCDCCCBDBH | Consensus sequence: HVDVGGGHGGGGDBHB |
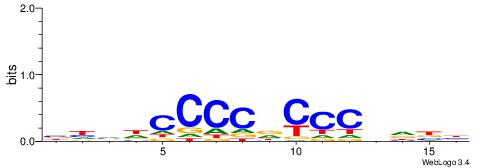 |
 |
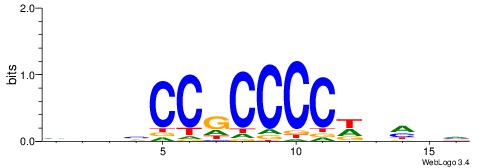
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0351051 |
Alignment:
BHHHWGGGGCGGBBVH
----GGGGGYGGGG--
| Original motif | Reverse complement motif |
| Consensus sequence: HBBBCCGCCCCWHHHV | Consensus sequence: BHHHWGGGGCGGBBVH |
 |
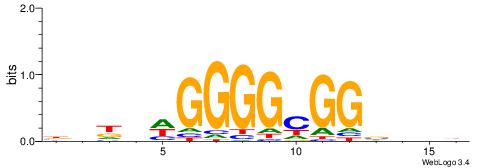 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0390437 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
------GGGGGYGGGG------
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
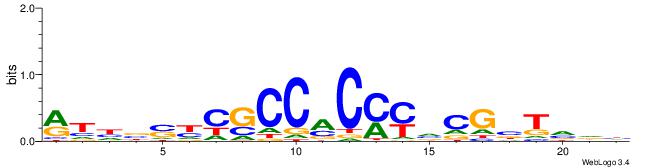
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0393358 |
Alignment:
HHBSHTCGTGRGTGGKGDBVWMV
-------GGGGGYGGGG------
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0413863 |
Alignment:
DHHBCCCCGCCAHHBHB
----CCCCKCCCCC---
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0418947 |
Alignment:
HVBCCCCCCCCMHHHB
---CCCCKCCCCC---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0420708 |
Alignment:
BDHHMCGCCCCCTHVBB
--CCCCKCCCCC-----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
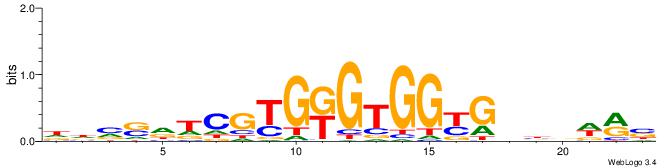
| Dataset #: 3 | Motif ID: 27 | Motif name: Klf4 |
| Original motif | Reverse complement motif |
| Consensus sequence: DGGGYGKGGC | Consensus sequence: GCCYCMCCCD |
 |
 |
Best Matches for Motif ID 27 (Highest to Lowest)
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
DHHDGGGCGRGGKHBH
---DGGGYGKGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0153308 |
Alignment:
DBCCCCCCCCCCMYC
---GCCYCMCCCD--
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0186791 |
Alignment:
BDWAGGCGTGBCHHD
--DGGGYGKGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: BDWAGGCGTGBCHHD | Consensus sequence: HHDGBCACGCCTWDB |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0218399 |
Alignment:
HVBCCCCCCCCMHHHB
--GCCYCMCCCD----
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0249586 |
Alignment:
VHTGCGGGGGCGGH
GCCYCMCCCD----
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0299159 |
Alignment:
BVVHAGGGGGCGRDHHB
------DGGGYGKGGC-
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0308595 |
Alignment:
BHBHDTGGCGGGGBDHD
----DGGGYGKGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
 |
 |
| Motif ID: | UP00047 |
| Motif name: | Zbtb7b_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0345891 |
Alignment:
DDVCCCCCCHAAWHB
GCCYCMCCCD-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDVCCCCCCHAAWHB | Consensus sequence: VHWTTHGGGGGGVDD |
 |
 |
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0346735 |
Alignment:
HBBBCCGCCCCWHHHV
--GCCYCMCCCD----
| Original motif | Reverse complement motif |
| Consensus sequence: HBBBCCGCCCCWHHHV | Consensus sequence: BHHHWGGGGCGGBBVH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 10 |
| Similarity score: | 0.0351496 |
Alignment:
MHHHWHYTMCCHCCCACVAABVH
------GCCYCMCCCD-------
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
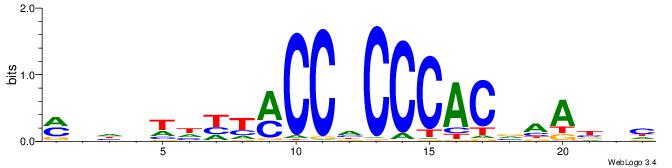 |
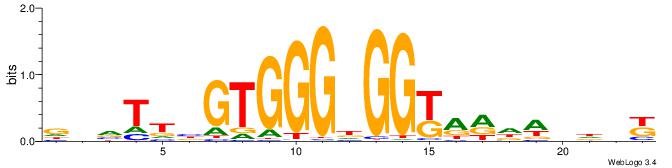 |
| Dataset #: 3 | Motif ID: 29 | Motif name: HIF1AARNT |
| Original motif | Reverse complement motif |
| Consensus sequence: VBACGTGV | Consensus sequence: VCACGTBV |
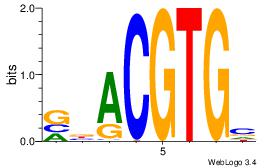 |
 |
Best Matches for Motif ID 29 (Highest to Lowest)
| Motif ID: | UP00097 |
| Motif name: | Mtf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
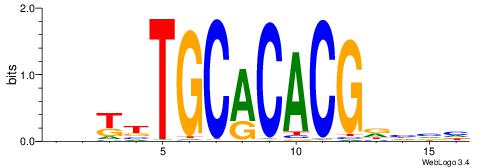
HDYTTGCACACGDHBH
-------VCACGTBV-
| Original motif | Reverse complement motif |
| Consensus sequence: DBDHCGTGTGCAAMDH | Consensus sequence: HDYTTGCACACGDHBH |
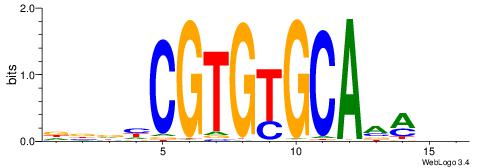 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.000574345 |
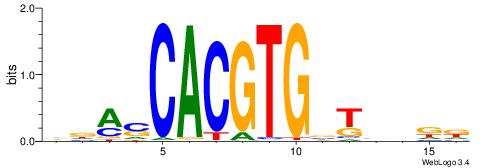
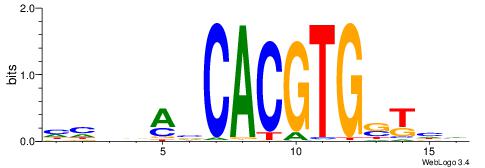
Alignment:
DDASCACGTGBTBVDD
---VBACGTGV-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDASCACGTGBTBVDD | Consensus sequence: HHVBABCACGTGSTHD |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.00642455 |
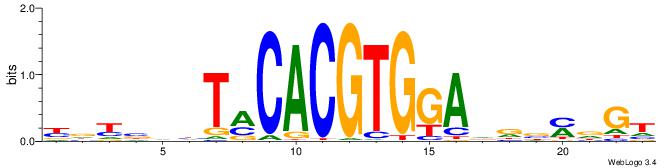
Alignment:
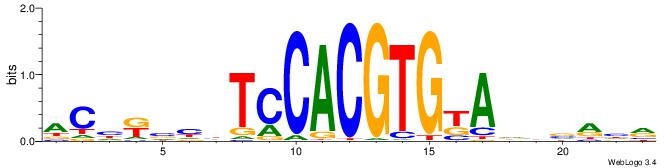
YDYBDHTMCACGTGGADDBMDGT
-------VBACGTGV--------
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00084 |
| Motif name: | Gmeb1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0131171 |
Alignment:
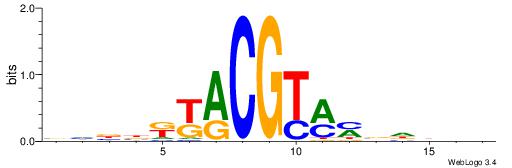
BVVDVRYACGTRYVBHB
-----VBACGTGV----
| Original motif | Reverse complement motif |
| Consensus sequence: BHBBKKACGTMMVDBVB | Consensus sequence: BVVDVRYACGTRYVBHB |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.014006 |
Alignment:
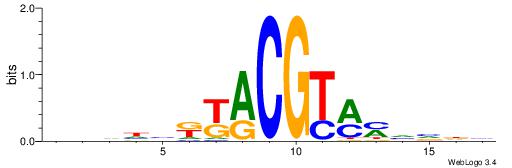
RDHDBVDTCACGTGASBHVHDH
-------VCACGTBV-------
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
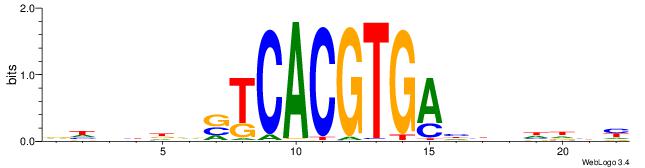 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0184296 |
Alignment:
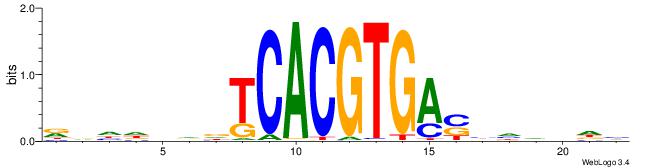
VBHHVCAGGTGCDVDHD
----VBACGTGV-----
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
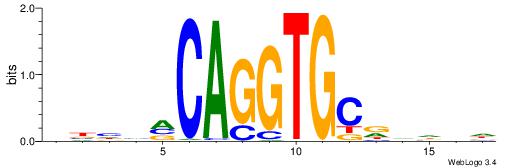 |
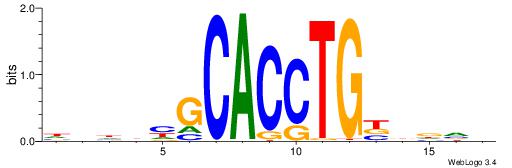 |
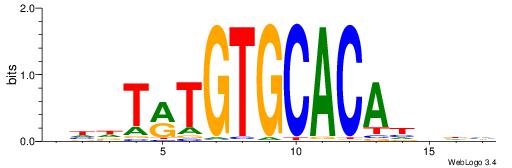
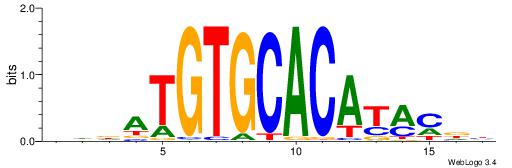
| Motif ID: | UP00068 |
| Motif name: | Eomes_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0188154 |
Alignment:
VVBVCKACACCTHVBV
------VCACGTBV--
| Original motif | Reverse complement motif |
| Consensus sequence: VBVDAGGTGTYGVBBV | Consensus sequence: VVBVCKACACCTHVBV |
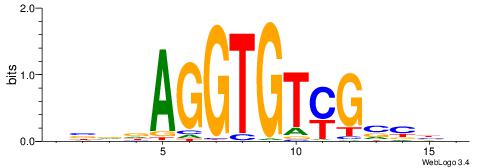 |
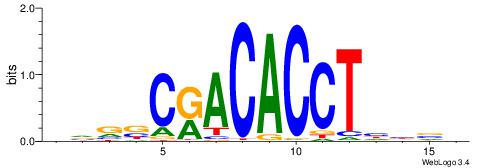 |
| Motif ID: | UP00026 |
| Motif name: | Zscan4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.0189568 |
Alignment:
DHVDTGTGCACAYAHDH
-VBACGTGV--------
| Original motif | Reverse complement motif |
| Consensus sequence: DHVDTGTGCACAYAHDH | Consensus sequence: HDHTMTGTGCACADVHD |
 |
 |
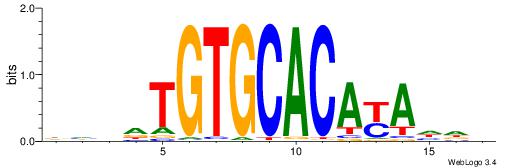
| Motif ID: | UP00042 |
| Motif name: | Gm397_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0198987 |
Alignment:
HHRTATGTGCACATHHD
--VBACGTGV-------
| Original motif | Reverse complement motif |
| Consensus sequence: HHDATGTGCACATAMDH | Consensus sequence: HHRTATGTGCACATHHD |
 |
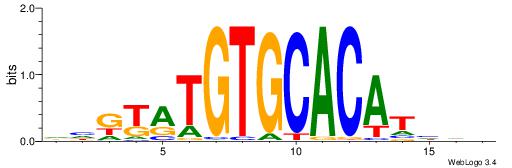 |
| Motif ID: | UP00060 |
| Motif name: | Max_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.021967 |
Alignment:
BBGHCACGCGACDD
---VCACGTBV---
| Original motif | Reverse complement motif |
| Consensus sequence: BBGHCACGCGACDD | Consensus sequence: HDGTCGCGTGDCVB |
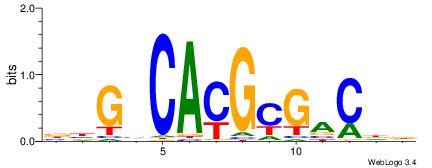 |
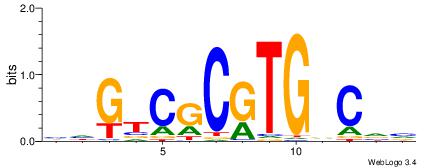 |
| Dataset #: 3 | Motif ID: 32 | Motif name: ArntAhr |
| Original motif | Reverse complement motif |
| Consensus sequence: YGCGTG | Consensus sequence: CACGCM |
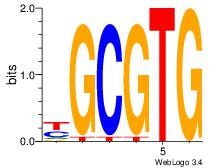 |
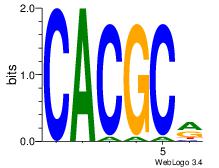 |
Best Matches for Motif ID 32 (Highest to Lowest)
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
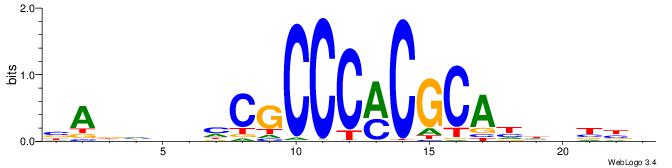
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
------YGCGTG-----------
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.00183587 |
Alignment:
HHDGBCACGCCTWDB
-----CACGCM----
| Original motif | Reverse complement motif |
| Consensus sequence: BDWAGGCGTGBCHHD | Consensus sequence: HHDGBCACGCCTWDB |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.00348048 |
Alignment:
HDGTCGCGTGDCVB
----YGCGTG----
| Original motif | Reverse complement motif |
| Consensus sequence: BBGHCACGCGACDD | Consensus sequence: HDGTCGCGTGDCVB |
 |
 |
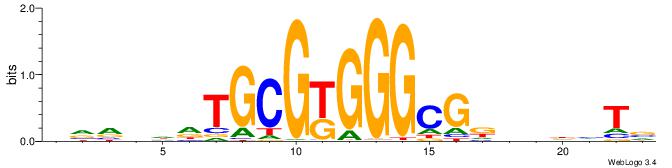
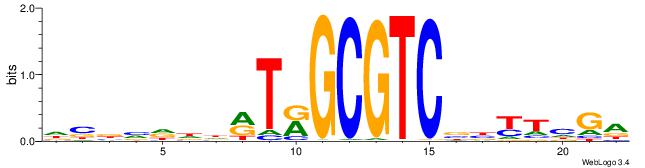
| Motif ID: | UP00527 |
| Motif name: | Foxn4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.00526158 |
Alignment:
TCDAMVBGACGCMAKDDDVBBD
-------CACGCM---------
| Original motif | Reverse complement motif |
| Consensus sequence: DBBVDDDRTRGCGTCBBYTHGA | Consensus sequence: TCDAMVBGACGCMAKDDDVBBD |
 |
 |
| Motif ID: | UP00042 |
| Motif name: | Gm397_second |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.00558171 |
Alignment:
HBGYGTGTGTGCVRVD
---YGCGTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: DVMVGCACACACKCVH | Consensus sequence: HBGYGTGTGTGCVRVD |
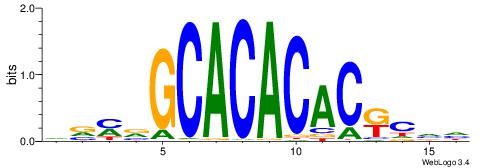 |
 |
| Motif ID: | UP00097 |
| Motif name: | Mtf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.00702416 |
Alignment:
HDYTTGCACACGDHBH
--------CACGCM--
| Original motif | Reverse complement motif |
| Consensus sequence: DBDHCGTGTGCAAMDH | Consensus sequence: HDYTTGCACACGDHBH |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.0123233 |
Alignment:
DDRMHBACGCGYGCGTDGBBDB
-------YGCGTG---------
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
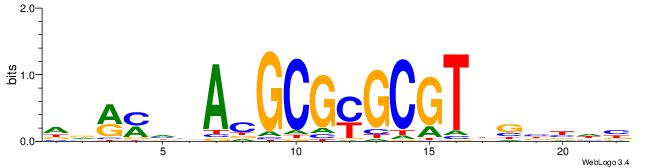 |
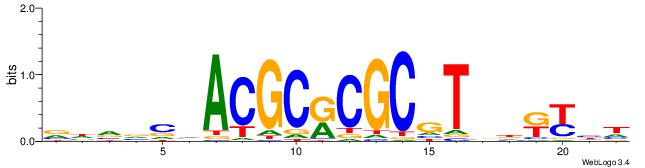 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0141579 |
Alignment:
HCCGCCCCCGCAHB
------CACGCM--
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 11 |
| Number of overlap: | 6 |
| Similarity score: | 0.0144485 |
Alignment:
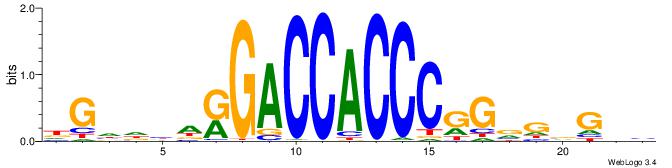
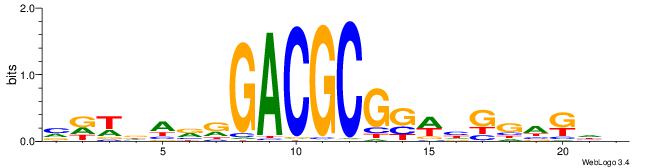
BHCBCBCCGGGTGGTCYHVHDCH
-------YGCGTG----------
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
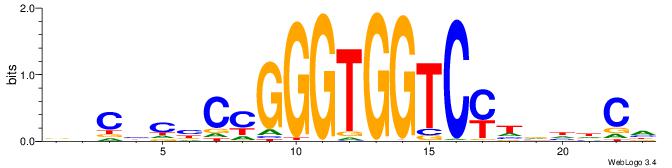 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.0172109 |
Alignment:
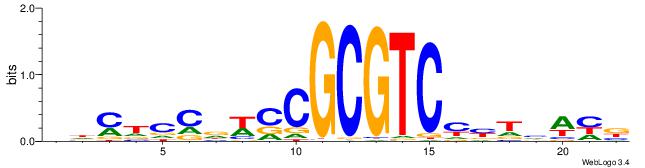
VHYTCCDWCCGCGTCMMTHWHV
---------YGCGTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
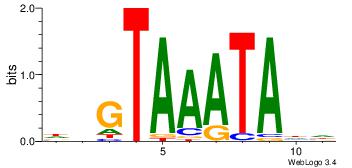
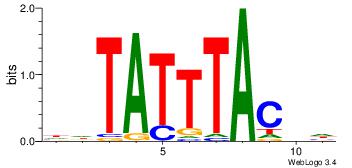
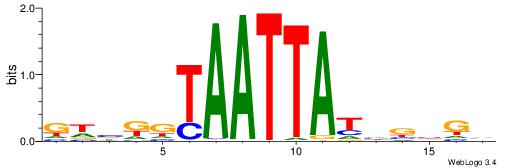
| Dataset #: 4 | Motif ID: 45 | Motif name: wbgTAAATAww |
| Original motif | Reverse complement motif |
| Consensus sequence: DBGTAAATAHD | Consensus sequence: DHTATTTACBD |
 |
 |
Best Matches for Motif ID 45 (Highest to Lowest)
| Motif ID: | UP00242 |
| Motif name: | Hoxc8 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0 |
Alignment:
AHDHTAATTACHHHHV
--DHTATTTACBD---
| Original motif | Reverse complement motif |
| Consensus sequence: BHDDDGTAATTAHHDT | Consensus sequence: AHDHTAATTACHHHHV |
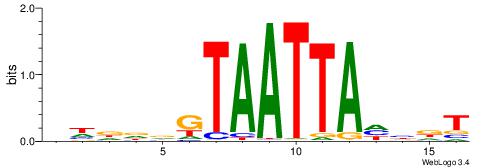 |
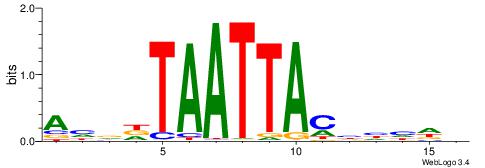 |
| Motif ID: | UP00073 |
| Motif name: | Foxa2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.00453517 |
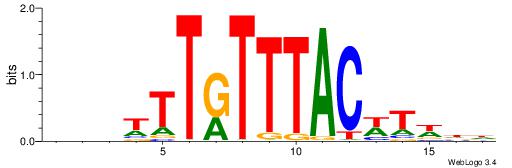
Alignment:
HHWAWGTAAAYAAAVDB
---DBGTAAATAHD---
| Original motif | Reverse complement motif |
| Consensus sequence: HHWAWGTAAAYAAAVDB | Consensus sequence: BDVTTTKTTTACWTWHH |
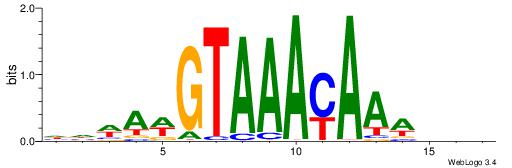 |
 |
| Motif ID: | UP00039 |
| Motif name: | Foxj3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.00490457 |
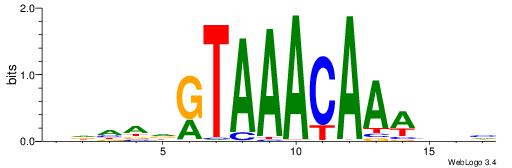
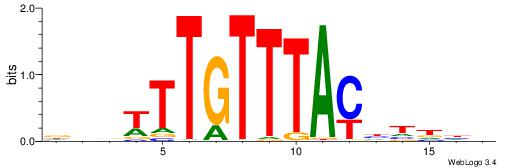
Alignment:
HDHADGTAAACAAAVVH
---DBGTAAATAHD---
| Original motif | Reverse complement motif |
| Consensus sequence: HDHADGTAAACAAAVVH | Consensus sequence: DVVTTTGTTTACDTHDH |
 |
 |
| Motif ID: | UP00041 |
| Motif name: | Foxj1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.00656547 |
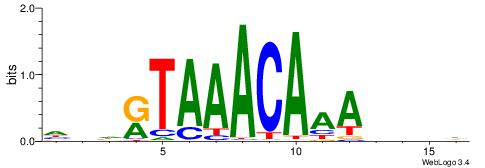
Alignment:
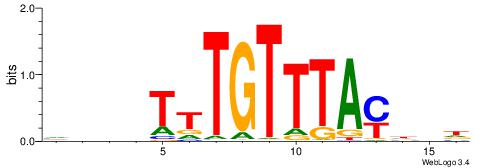
HDHRTAAACAAAHDDD
-DBGTAAATAHD----
| Original motif | Reverse complement motif |
| Consensus sequence: HDHRTAAACAAAHDDD | Consensus sequence: DDDHTTTGTTTAMHDH |
 |
 |
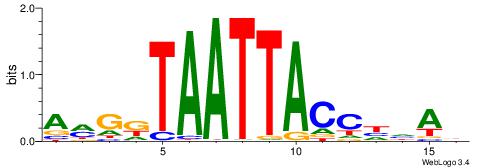
| Motif ID: | UP00182 |
| Motif name: | Hoxa6 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0086354 |
Alignment:
DTHHGGTAATTAYCYT
---DBGTAAATAHD--
| Original motif | Reverse complement motif |
| Consensus sequence: AMGKTAATTACCHHAD | Consensus sequence: DTHHGGTAATTAYCYT |
 |
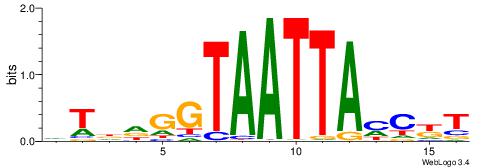 |
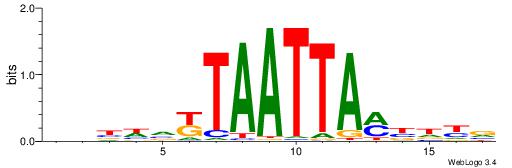
| Motif ID: | UP00260 |
| Motif name: | Hoxc6 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0092156 |
Alignment:
BRHVKTAATTAMBDVVD
---DHTATTTACBD---
| Original motif | Reverse complement motif |
| Consensus sequence: BRHVKTAATTAMBDVVD | Consensus sequence: DBBDVYTAATTARBHKB |
 |
 |
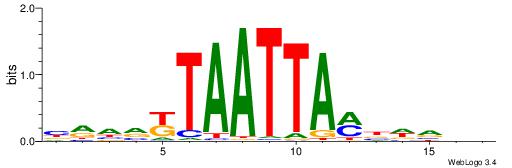
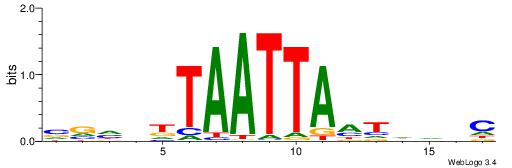
| Motif ID: | UP00164 |
| Motif name: | Hoxa7_2668.2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.00942246 |
Alignment:
VVHDDTAATTAHTDDBC
---DHTATTTACBD---
| Original motif | Reverse complement motif |
| Consensus sequence: VVHDDTAATTAHTDDBC | Consensus sequence: GBDDAHTAATTADHHVV |
 |
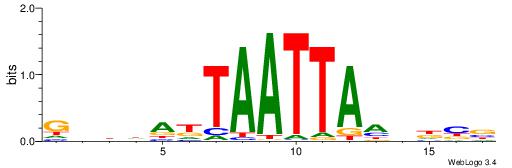 |
| Motif ID: | UP00259 |
| Motif name: | Hoxb6 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.00949005 |
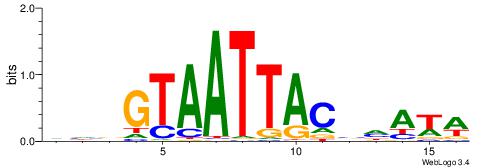
Alignment:
VDVGTAATTACBDAWW
-DBGTAAATAHD----
| Original motif | Reverse complement motif |
| Consensus sequence: WWTDBGTAATTACVDB | Consensus sequence: VDVGTAATTACBDAWW |
 |
 |
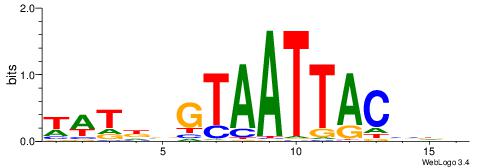
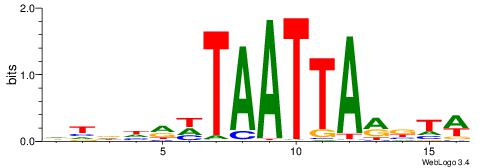
| Motif ID: | UP00254 |
| Motif name: | Pou2f1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.00998499 |
Alignment:
WABKTAATTAHKHHHD
-DBGTAAATAHD----
| Original motif | Reverse complement motif |
| Consensus sequence: DHDHRHTAATTARBTW | Consensus sequence: WABKTAATTAHKHHHD |
 |
 |
| Motif ID: | UP00110 |
| Motif name: | Dlx4 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0107458 |
Alignment:
GHVRGTAATTATDVVGV
--DBGTAAATAHD----
| Original motif | Reverse complement motif |
| Consensus sequence: BCVVDATAATTACMVHC | Consensus sequence: GHVRGTAATTATDVVGV |
 |
 |
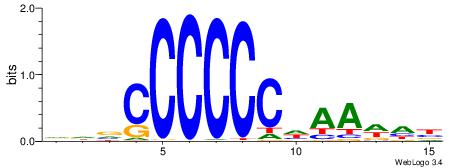
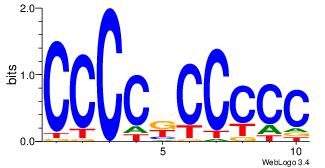
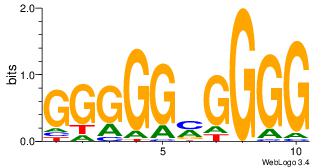
| Dataset #: 5 | Motif ID: 48 | Motif name: SP1 |
| Original motif | Reverse complement motif |
| Consensus sequence: CCCCKCCCCC | Consensus sequence: GGGGGYGGGG |
 |
 |
Best Matches for Motif ID 48 (Highest to Lowest)
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0231018 |
Alignment:
DBCCCCCCCCCCMYC
--CCCCKCCCCC---
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0303207 |
Alignment:
HBDRCCMCGCCCHHHD
----CCCCKCCCCC--
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0323007 |
Alignment:
HHHHDGGGGCGGDBHDV
----GGGGGYGGGG---
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
 |
 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0340973 |
Alignment:
HVDVGGGHGGGGDBHB
--GGGGGYGGGG----
| Original motif | Reverse complement motif |
| Consensus sequence: BHVDCCCCDCCCBDBH | Consensus sequence: HVDVGGGHGGGGDBHB |
 |
 |
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0351051 |
Alignment:
BHHHWGGGGCGGBBVH
----GGGGGYGGGG--
| Original motif | Reverse complement motif |
| Consensus sequence: HBBBCCGCCCCWHHHV | Consensus sequence: BHHHWGGGGCGGBBVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0390437 |
Alignment:
HDBABCGBKRGYGGCGMSBHAK
------GGGGGYGGGG------
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0393358 |
Alignment:
HHBSHTCGTGRGTGGKGDBVWMV
-------GGGGGYGGGG------
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0413863 |
Alignment:
DHHBCCCCGCCAHHBHB
----CCCCKCCCCC---
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0418947 |
Alignment:
HVBCCCCCCCCMHHHB
---CCCCKCCCCC---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0420708 |
Alignment:
BDHHMCGCCCCCTHVBB
--CCCCKCCCCC-----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
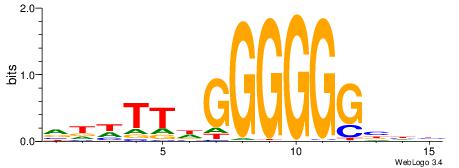
| Dataset #: 5 | Motif ID: 51 | Motif name: Klf4 |
| Original motif | Reverse complement motif |
| Consensus sequence: DGGGYGKGGC | Consensus sequence: GCCYCMCCCD |
 |
 |
Best Matches for Motif ID 51 (Highest to Lowest)
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
DHHDGGGCGRGGKHBH
---DGGGYGKGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0153308 |
Alignment:
DBCCCCCCCCCCMYC
---GCCYCMCCCD--
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0186791 |
Alignment:
BDWAGGCGTGBCHHD
--DGGGYGKGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: BDWAGGCGTGBCHHD | Consensus sequence: HHDGBCACGCCTWDB |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0218399 |
Alignment:
HVBCCCCCCCCMHHHB
--GCCYCMCCCD----
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0249586 |
Alignment:
VHTGCGGGGGCGGH
GCCYCMCCCD----
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0299159 |
Alignment:
BVVHAGGGGGCGRDHHB
------DGGGYGKGGC-
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0308595 |
Alignment:
BHBHDTGGCGGGGBDHD
----DGGGYGKGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
 |
 |
| Motif ID: | UP00047 |
| Motif name: | Zbtb7b_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0345891 |
Alignment:
VHWTTHGGGGGGVDD
GCCYCMCCCD-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDVCCCCCCHAAWHB | Consensus sequence: VHWTTHGGGGGGVDD |
 |
 |
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0346735 |
Alignment:
HBBBCCGCCCCWHHHV
--GCCYCMCCCD----
| Original motif | Reverse complement motif |
| Consensus sequence: HBBBCCGCCCCWHHHV | Consensus sequence: BHHHWGGGGCGGBBVH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 10 |
| Similarity score: | 0.0351496 |
Alignment:
MHHHWHYTMCCHCCCACVAABVH
------GCCYCMCCCD-------
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
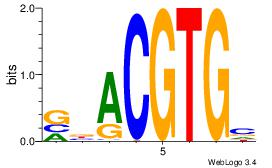
| Dataset #: 5 | Motif ID: 53 | Motif name: HIF1AARNT |
| Original motif | Reverse complement motif |
| Consensus sequence: VBACGTGV | Consensus sequence: VCACGTBV |
 |
 |
Best Matches for Motif ID 53 (Highest to Lowest)
| Motif ID: | UP00097 |
| Motif name: | Mtf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HDYTTGCACACGDHBH
-------VCACGTBV-
| Original motif | Reverse complement motif |
| Consensus sequence: DBDHCGTGTGCAAMDH | Consensus sequence: HDYTTGCACACGDHBH |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.000574345 |
Alignment:
DDASCACGTGBTBVDD
---VBACGTGV-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDASCACGTGBTBVDD | Consensus sequence: HHVBABCACGTGSTHD |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.00642455 |
Alignment:
YDYBDHTMCACGTGGADDBMDGT
-------VBACGTGV--------
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00084 |
| Motif name: | Gmeb1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0131171 |
Alignment:
BVVDVRYACGTRYVBHB
-----VBACGTGV----
| Original motif | Reverse complement motif |
| Consensus sequence: BHBBKKACGTMMVDBVB | Consensus sequence: BVVDVRYACGTRYVBHB |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.014006 |
Alignment:
RDHDBVDTCACGTGASBHVHDH
-------VCACGTBV-------
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0184296 |
Alignment:
VBHHVCAGGTGCDVDHD
----VBACGTGV-----
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Motif ID: | UP00068 |
| Motif name: | Eomes_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0188154 |
Alignment:
VVBVCKACACCTHVBV
------VCACGTBV--
| Original motif | Reverse complement motif |
| Consensus sequence: VBVDAGGTGTYGVBBV | Consensus sequence: VVBVCKACACCTHVBV |
 |
 |
| Motif ID: | UP00026 |
| Motif name: | Zscan4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.0189568 |
Alignment:
DHVDTGTGCACAYAHDH
-VBACGTGV--------
| Original motif | Reverse complement motif |
| Consensus sequence: DHVDTGTGCACAYAHDH | Consensus sequence: HDHTMTGTGCACADVHD |
 |
 |
| Motif ID: | UP00042 |
| Motif name: | Gm397_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0198987 |
Alignment:
HHRTATGTGCACATHHD
--VBACGTGV-------
| Original motif | Reverse complement motif |
| Consensus sequence: HHDATGTGCACATAMDH | Consensus sequence: HHRTATGTGCACATHHD |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.021967 |
Alignment:
BBGHCACGCGACDD
---VCACGTBV---
| Original motif | Reverse complement motif |
| Consensus sequence: BBGHCACGCGACDD | Consensus sequence: HDGTCGCGTGDCVB |
 |
 |
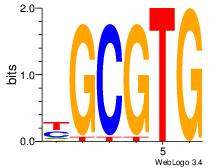
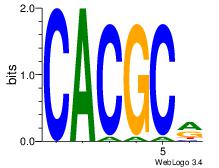
| Dataset #: 5 | Motif ID: 56 | Motif name: ArntAhr |
| Original motif | Reverse complement motif |
| Consensus sequence: YGCGTG | Consensus sequence: CACGCM |
 |
 |
Best Matches for Motif ID 56 (Highest to Lowest)
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
------YGCGTG-----------
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.00183587 |
Alignment:
HHDGBCACGCCTWDB
-----CACGCM----
| Original motif | Reverse complement motif |
| Consensus sequence: BDWAGGCGTGBCHHD | Consensus sequence: HHDGBCACGCCTWDB |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.00348048 |
Alignment:
HDGTCGCGTGDCVB
----YGCGTG----
| Original motif | Reverse complement motif |
| Consensus sequence: BBGHCACGCGACDD | Consensus sequence: HDGTCGCGTGDCVB |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.00526158 |
Alignment:
TCDAMVBGACGCMAKDDDVBBD
-------CACGCM---------
| Original motif | Reverse complement motif |
| Consensus sequence: DBBVDDDRTRGCGTCBBYTHGA | Consensus sequence: TCDAMVBGACGCMAKDDDVBBD |
 |
 |
| Motif ID: | UP00042 |
| Motif name: | Gm397_second |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.00558171 |
Alignment:
HBGYGTGTGTGCVRVD
---YGCGTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: DVMVGCACACACKCVH | Consensus sequence: HBGYGTGTGTGCVRVD |
 |
 |
| Motif ID: | UP00097 |
| Motif name: | Mtf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.00702416 |
Alignment:
HDYTTGCACACGDHBH
--------CACGCM--
| Original motif | Reverse complement motif |
| Consensus sequence: DBDHCGTGTGCAAMDH | Consensus sequence: HDYTTGCACACGDHBH |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.0123233 |
Alignment:
DDRMHBACGCGYGCGTDGBBDB
-------YGCGTG---------
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0141579 |
Alignment:
HCCGCCCCCGCAHB
------CACGCM--
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 11 |
| Number of overlap: | 6 |
| Similarity score: | 0.0144485 |
Alignment:
BHCBCBCCGGGTGGTCYHVHDCH
-------YGCGTG----------
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.0172109 |
Alignment:
VHYTCCDWCCGCGTCMMTHWHV
---------YGCGTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |