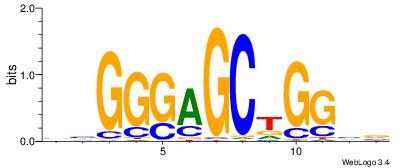Best Matches for Each Motif (Highest to Lowest)
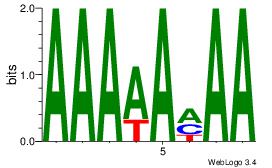
| Dataset #: 1 |
Motif ID: 1 |
Motif name: Motif 1 |
| Original motif Consensus sequence: AAAAAMAA |
Reverse complement motif Consensus sequence: TTYTTTTT |
|
|
 |
 |
Best Matches for Motif ID 1 (Highest to Lowest)
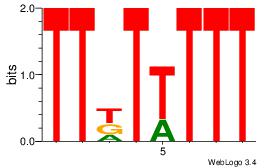
| Dataset #: |
2 |
| Motif ID: |
60 |
| Motif name: |
Motif 60 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0315879 |
Alignment:
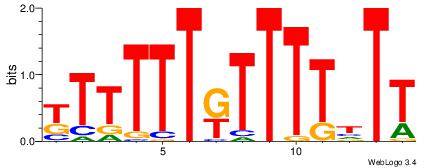
AADAAAAMAAAAAM
--AAAAAMAA----
| Original motif Consensus sequence: AADAAAAMAAAAAM |
Reverse complement motif Consensus sequence: YTTTTTRTTTTDTT |
|
|
 |
 |
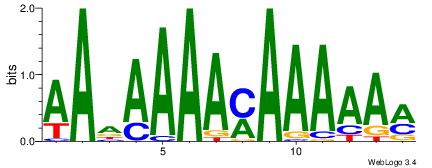
| Dataset #: |
1 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0375279 |
Alignment:
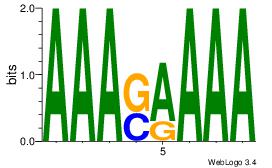
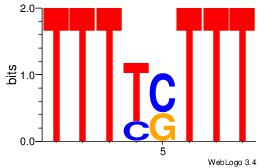
AAASAAAA
AAAAAMAA
| Original motif Consensus sequence: AAASAAAA |
Reverse complement motif Consensus sequence: TTTTSTTT |
|
|
 |
 |
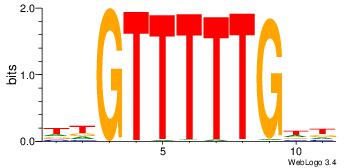
| Dataset #: |
4 |
| Motif ID: |
109 |
| Motif name: |
aaCAAAAACaa |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0446212 |
Alignment:
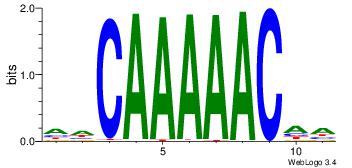
HHCAAAAACHH
---AAAAAMAA
| Original motif Consensus sequence: HHCAAAAACHH |
Reverse complement motif Consensus sequence: HHGTTTTTGHH |
|
|
 |
 |
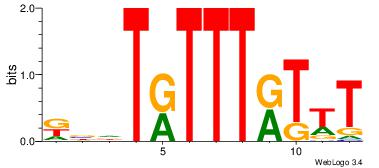
| Dataset #: |
3 |
| Motif ID: |
67 |
| Motif name: |
FOXI1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0453228 |
Alignment:
AAAMAAAMADBY
--AAAAAMAA--
| Original motif Consensus sequence: KBDTRTTTRTTT |
Reverse complement motif Consensus sequence: AAAMAAAMADBY |
|
|
 |
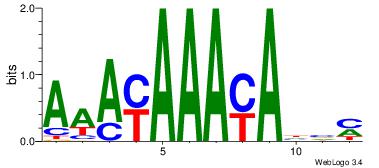 |
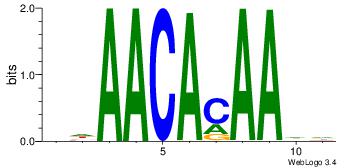
| Dataset #: |
4 |
| Motif ID: |
114 |
| Motif name: |
awAACAcAAwa |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0467797 |
Alignment:
DDAACACAADV
-AAAAAMAA--
| Original motif Consensus sequence: DDAACACAADV |
Reverse complement motif Consensus sequence: BDTTGTGTTDD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
108 |
| Motif name: |
wwATkTTTAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0570668 |
Alignment:
DHATKTTTAHH
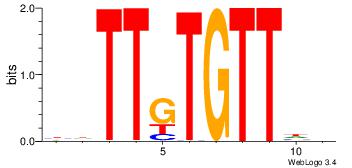
TTYTTTTT---
| Original motif Consensus sequence: DHATKTTTAHH |
Reverse complement motif Consensus sequence: HHTAAARATHD |
|
|
 |
 |
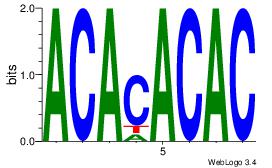
| Dataset #: 1 |
Motif ID: 2 |
Motif name: Motif 2 |
| Original motif Consensus sequence: ACACACAC |
Reverse complement motif Consensus sequence: GTGTGTGT |
|
|
 |
 |
Best Matches for Motif ID 2 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
58 |
| Motif name: |
Motif 58 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
8 |
| Similarity score: |
0.0337866 |
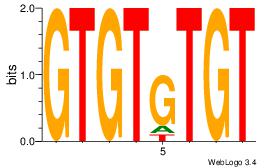
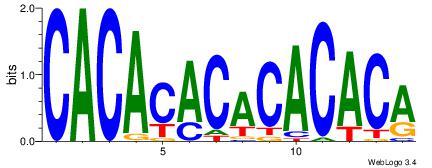
Alignment:
TGTGTGTGTGTGTG
-----GTGTGTGT-
| Original motif Consensus sequence: CACACACACACACA |
Reverse complement motif Consensus sequence: TGTGTGTGTGTGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
117 |
| Motif name: |
yaCATAYACay |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0556866 |
Alignment:
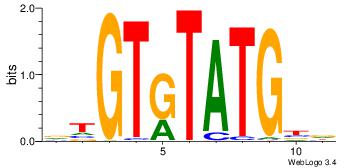
DTGTGTATGBD
--GTGTGTGT-
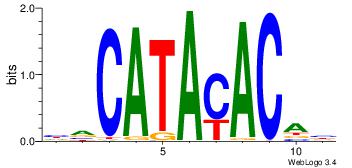
| Original motif Consensus sequence: HVCATACACAH |
Reverse complement motif Consensus sequence: DTGTGTATGBD |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
12 |
| Motif name: |
Motif 12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0626247 |
Alignment:
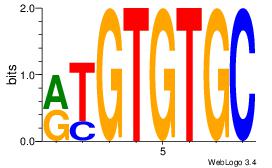
GCACACAY
ACACACAC
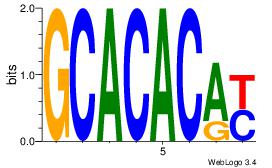
| Original motif Consensus sequence: GCACACAY |
Reverse complement motif Consensus sequence: MTGTGTGC |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
21 |
| Motif name: |
Motif 21 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0696577 |
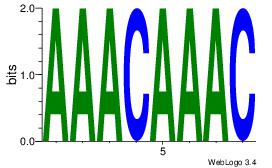
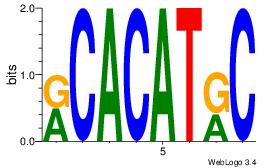
Alignment:
AAACAAAC
ACACACAC
| Original motif Consensus sequence: AAACAAAC |
Reverse complement motif Consensus sequence: GTTTGTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
37 |
| Motif name: |
Motif 37 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.07313 |
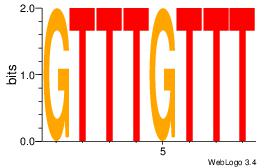
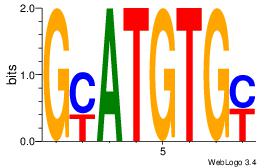
Alignment:
GYATGTGY
GTGTGTGT
| Original motif Consensus sequence: GYATGTGY |
Reverse complement motif Consensus sequence: KCACATKC |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 3 |
Motif name: Motif 3 |
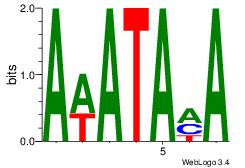
| Original motif Consensus sequence: AWATAMA |
Reverse complement motif Consensus sequence: TYTATWT |
|
|
 |
 |
Best Matches for Motif ID 3 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
1 |
| Motif name: |
Motif 1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.00716822 |
Alignment:
AAAAAMAA
AWATAMA-
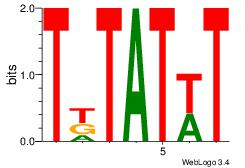
| Original motif Consensus sequence: AAAAAMAA |
Reverse complement motif Consensus sequence: TTYTTTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
49 |
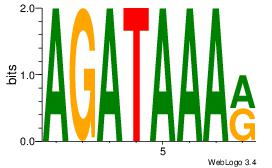
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.021456 |
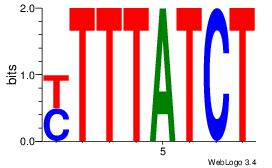
Alignment:
KTTTATCT
-TYTATWT
| Original motif Consensus sequence: AGATAAAR |
Reverse complement motif Consensus sequence: KTTTATCT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
67 |
| Motif name: |
FOXI1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
7 |
| Similarity score: |
0.0260643 |
Alignment:
KBDTRTTTRTTT
-----TYTATWT
| Original motif Consensus sequence: KBDTRTTTRTTT |
Reverse complement motif Consensus sequence: AAAMAAAMADBY |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
60 |
| Motif name: |
Motif 60 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
7 |
| Similarity score: |
0.031739 |
Alignment:
YTTTTTRTTTTDTT
---TYTATWT----
| Original motif Consensus sequence: AADAAAAMAAAAAM |
Reverse complement motif Consensus sequence: YTTTTTRTTTTDTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
90 |
| Motif name: |
ymTACATAyw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.034879 |
Alignment:
DBTATGTAHH
TYTATWT---
| Original motif Consensus sequence: HHTACATABD |
Reverse complement motif Consensus sequence: DBTATGTAHH |
|
|
 |
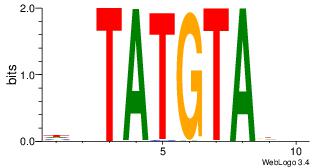 |
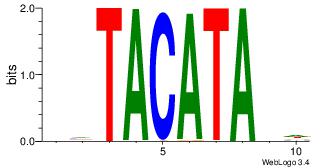
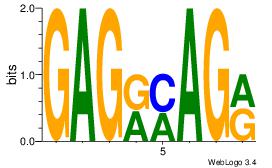
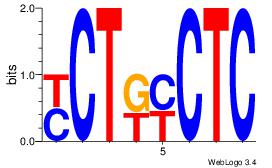
| Dataset #: 1 |
Motif ID: 4 |
Motif name: Motif 4 |
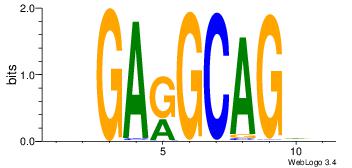
| Original motif Consensus sequence: GAGRMAGR |
Reverse complement motif Consensus sequence: KCTRMCTC |
|
|
 |
 |
Best Matches for Motif ID 4 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
116 |
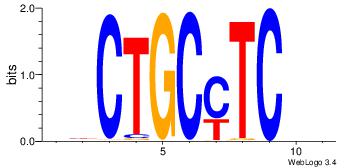
| Motif name: |
dyCTGCyTCyc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0229288 |
Alignment:
BHGAKGCAGHH
--GAGRMAGR-
| Original motif Consensus sequence: DHCTGCYTCHB |
Reverse complement motif Consensus sequence: BHGAKGCAGHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
113 |
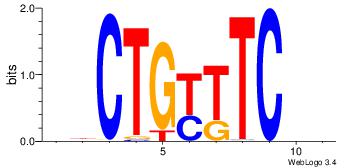
| Motif name: |
htCTGyKTCbt |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0339962 |
Alignment:
HVGAAMCAGVH
--GAGRMAGR-
| Original motif Consensus sequence: HBCTGYTTCBH |
Reverse complement motif Consensus sequence: HVGAAMCAGVH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
112 |
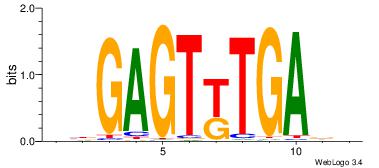
| Motif name: |
ctGAGTkTGAgg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0422491 |
Alignment:
HHTCARACTCVV
--KCTRMCTC--
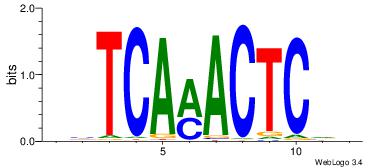
| Original motif Consensus sequence: VBGAGTKTGADD |
Reverse complement motif Consensus sequence: HHTCARACTCVV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
121 |
| Motif name: |
ctGAGTCATCkb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0508307 |
Alignment:
BHGATGACTCDD
--KCTRMCTC--
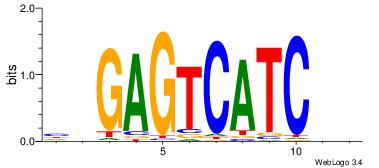
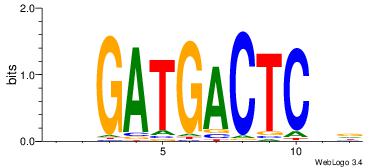
| Original motif Consensus sequence: HDGAGTCATCDB |
Reverse complement motif Consensus sequence: BHGATGACTCDD |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
18 |
| Motif name: |
Motif 18 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0519824 |
Alignment:
GRRAGAGA
GAGRMAGR
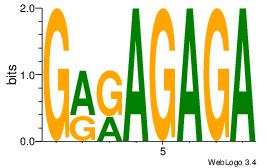
| Original motif Consensus sequence: GRRAGAGA |
Reverse complement motif Consensus sequence: TCTCTMKC |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 5 |
Motif name: Motif 5 |
| Original motif Consensus sequence: AAASAAAA |
Reverse complement motif Consensus sequence: TTTTSTTT |
|
|
 |
 |
Best Matches for Motif ID 5 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
60 |
| Motif name: |
Motif 60 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0229557 |
Alignment:
AADAAAAMAAAAAM
----AAASAAAA--
| Original motif Consensus sequence: AADAAAAMAAAAAM |
Reverse complement motif Consensus sequence: YTTTTTRTTTTDTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
1 |
| Motif name: |
Motif 1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0330694 |
Alignment:
TTYTTTTT
TTTTSTTT
| Original motif Consensus sequence: AAAAAMAA |
Reverse complement motif Consensus sequence: TTYTTTTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
89 |
| Motif name: |
aaACAAAACaa |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0345209 |
Alignment:
BHGTTTTGTTT
---TTTTSTTT
| Original motif Consensus sequence: AAACAAAACHV |
Reverse complement motif Consensus sequence: BHGTTTTGTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
21 |
| Motif name: |
Motif 21 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0346745 |
Alignment:
AAACAAAC
AAASAAAA
| Original motif Consensus sequence: AAACAAAC |
Reverse complement motif Consensus sequence: GTTTGTTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
65 |
| Motif name: |
Foxd3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0440489 |
Alignment:
AAAHAAACAWTC
AAASAAAA----
| Original motif Consensus sequence: GAWTGTTTDTTT |
Reverse complement motif Consensus sequence: AAAHAAACAWTC |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 6 |
Motif name: Motif 6 |
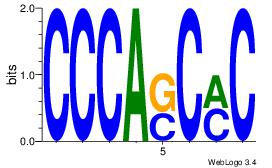
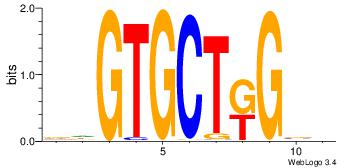
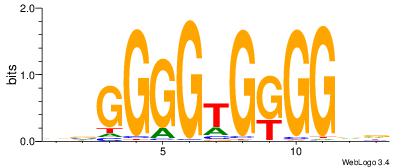
| Original motif Consensus sequence: CCCASCMC |
Reverse complement motif Consensus sequence: GYGSTGGG |
|
|
 |
 |
Best Matches for Motif ID 6 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
22 |
| Motif name: |
Motif 22 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.00727797 |
Alignment:
MCCACCAC
CCCASCMC
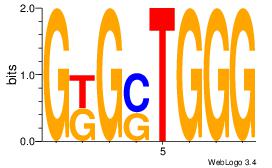
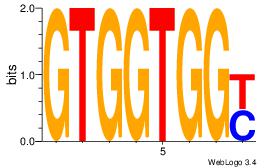
| Original motif Consensus sequence: GTGGTGGY |
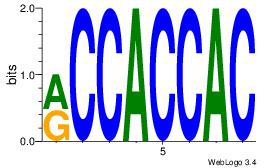
Reverse complement motif Consensus sequence: MCCACCAC |
|
|
 |
 |
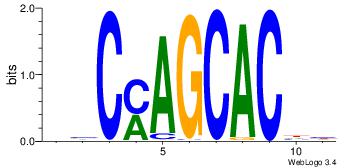
| Dataset #: |
4 |
| Motif ID: |
115 |
| Motif name: |
ysCmAGCACwy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0212781 |
Alignment:
DHGTGCTRGVH
--GYGSTGGG-
| Original motif Consensus sequence: HVCMAGCACHH |
Reverse complement motif Consensus sequence: DHGTGCTRGVH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
111 |
| Motif name: |
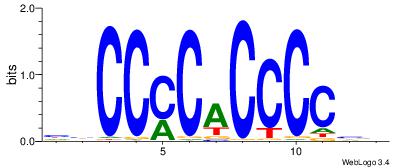
ccCCmCaCCCCcc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0266863 |
Alignment:
HBCCCCACCCCBH
---CCCASCMC--
| Original motif Consensus sequence: HBCCCCACCCCBH |
Reverse complement motif Consensus sequence: DBGGGGTGGGGBD |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
42 |
| Motif name: |
Motif 42 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.030487 |
Alignment:
GTTCTKGG
GYGSTGGG
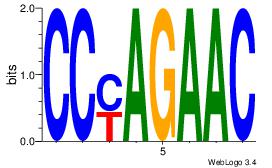
| Original motif Consensus sequence: CCYAGAAC |
Reverse complement motif Consensus sequence: GTTCTKGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
93 |
| Motif name: |
cmCARCACTwr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.030511 |
Alignment:
BHCAGCACTHV
CCCASCMC---
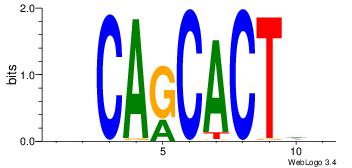
| Original motif Consensus sequence: BHCAGCACTHV |
Reverse complement motif Consensus sequence: VHAGTGCTGHB |
|
|
 |
 |
| Dataset #: 1 |
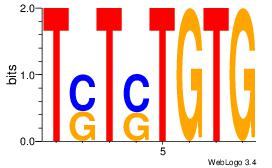
Motif ID: 7 |
Motif name: Motif 7 |
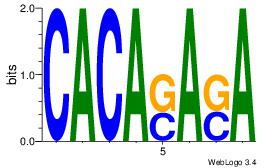
| Original motif Consensus sequence: CACASASA |
Reverse complement motif Consensus sequence: TSTSTGTG |
|
|
 |
 |
Best Matches for Motif ID 7 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
58 |
| Motif name: |
Motif 58 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.0271648 |
Alignment:
TGTGTGTGTGTGTG
------TSTSTGTG
| Original motif Consensus sequence: CACACACACACACA |
Reverse complement motif Consensus sequence: TGTGTGTGTGTGTG |
|
|
 |
 |
| Dataset #: |
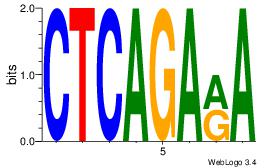
1 |
| Motif ID: |
41 |
| Motif name: |
Motif 41 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0361772 |
Alignment:
CTCAGARA
CACASASA
| Original motif Consensus sequence: CTCAGARA |
Reverse complement motif Consensus sequence: TKTCTGAG |
|
|
 |
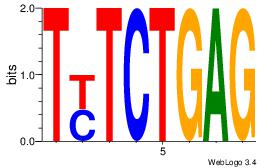 |
| Dataset #: |
4 |
| Motif ID: |
117 |
| Motif name: |
yaCATAYACay |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0572872 |
Alignment:
DTGTGTATGBD
-TSTSTGTG--
| Original motif Consensus sequence: HVCATACACAH |
Reverse complement motif Consensus sequence: DTGTGTATGBD |
|
|
 |
 |
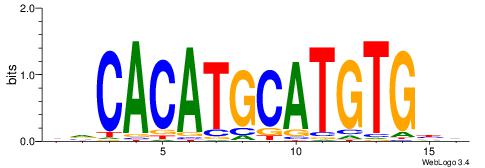
| Dataset #: |
4 |
| Motif ID: |
118 |
| Motif name: |
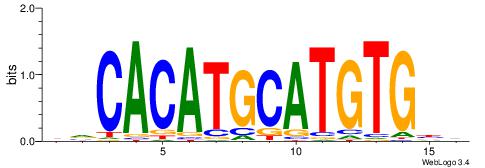
yrCACATGCATGTGyr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.0627214 |
Alignment:
HVCACATGCATGTGBD
--CACASASA------
| Original motif Consensus sequence: HVCACATGCATGTGBD |
Reverse complement motif Consensus sequence: DVCACATGCATGTGBH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
114 |
| Motif name: |
awAACAcAAwa |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0715956 |
Alignment:
DDAACACAADV
--CACASASA-
| Original motif Consensus sequence: DDAACACAADV |
Reverse complement motif Consensus sequence: BDTTGTGTTDD |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 8 |
Motif name: Motif 8 |
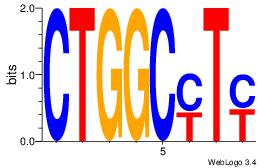
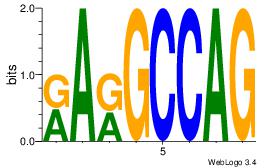
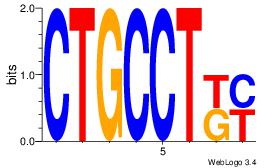
| Original motif Consensus sequence: CTGGCYTY |
Reverse complement motif Consensus sequence: KAKGCCAG |
|
|
 |
 |
Best Matches for Motif ID 8 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
97 |
| Motif name: |
sgArGCCACg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0352649 |
Alignment:
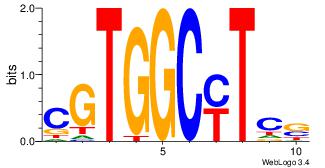
SGARGCCACG
-KAKGCCAG-
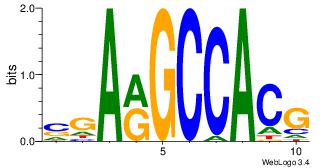
| Original motif Consensus sequence: SGARGCCACG |
Reverse complement motif Consensus sequence: CGTGGCKTCS |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
92 |
| Motif name: |
cgAKGyCAcg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0406315 |
Alignment:
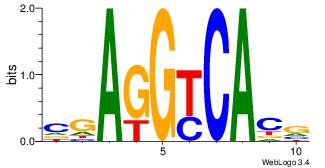
CGAGGYCACV
-KAKGCCAG-
| Original motif Consensus sequence: CGAGGYCACV |
Reverse complement motif Consensus sequence: VGTGMCCTCG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
27 |
| Motif name: |
Motif 27 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0419581 |
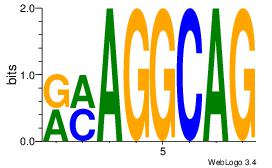
Alignment:
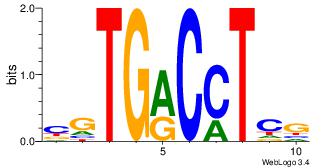
CTGCCTKY
CTGGCYTY
| Original motif Consensus sequence: CTGCCTKY |
Reverse complement motif Consensus sequence: KRAGGCAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
31 |
| Motif name: |
Motif 31 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0519508 |
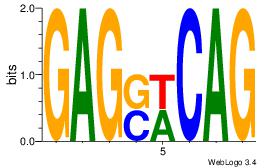
Alignment:
CTGWSCTC
CTGGCYTY
| Original motif Consensus sequence: CTGWSCTC |
Reverse complement motif Consensus sequence: GAGSWCAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0648749 |
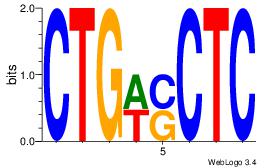
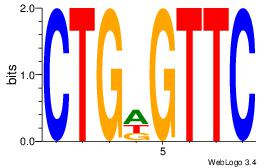
Alignment:
CTGDGTTC
CTGGCYTY
| Original motif Consensus sequence: CTGDGTTC |
Reverse complement motif Consensus sequence: GAACDCAG |
|
|
 |
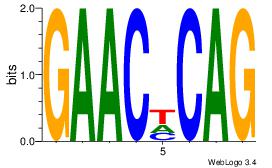 |
| Dataset #: 1 |
Motif ID: 9 |
Motif name: Motif 9 |
| Original motif Consensus sequence: TTTWWAAA |
Reverse complement motif Consensus sequence: TTTWWAAA |
|
|
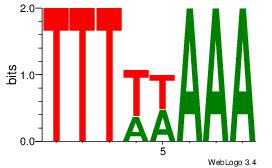 |
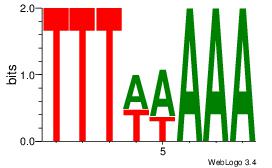 |
Best Matches for Motif ID 9 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
120 |
| Motif name: |
wtTTTwAAAaw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0 |
Alignment:
HHTTTWAAADD
-TTTWWAAA--
| Original motif Consensus sequence: HHTTTWAAADD |
Reverse complement motif Consensus sequence: DDTTTWAAAHH |
|
|
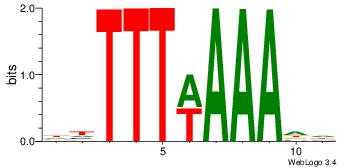 |
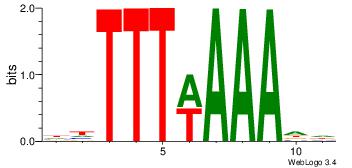 |
| Dataset #: |
4 |
| Motif ID: |
108 |
| Motif name: |
wwATkTTTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0472911 |
Alignment:
DHATKTTTAHH
---TTTWWAAA
| Original motif Consensus sequence: DHATKTTTAHH |
Reverse complement motif Consensus sequence: HHTAAARATHD |
|
|
 |
 |
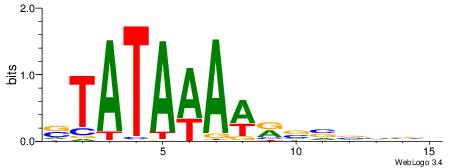
| Dataset #: |
3 |
| Motif ID: |
62 |
| Motif name: |
TBP |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0552987 |
Alignment:
STATAAAWRVVVVBV
-TTTWWAAA------
| Original motif Consensus sequence: STATAAAWRVVVVBV |
Reverse complement motif Consensus sequence: VBVVVVMWTTTATAS |
|
|
 |
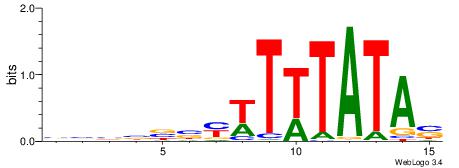 |
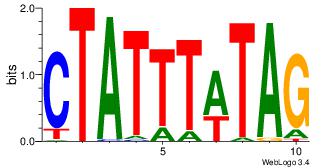
| Dataset #: |
3 |
| Motif ID: |
61 |
| Motif name: |
MEF2A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0641646 |
Alignment:
CTATTTWTAG
-TTTWWAAA-
| Original motif Consensus sequence: CTATTTWTAG |
Reverse complement motif Consensus sequence: CTAWAAATAG |
|
|
 |
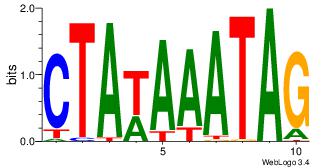 |
| Dataset #: |
4 |
| Motif ID: |
91 |
| Motif name: |
wtATTTTAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0724871 |
Alignment:
HWTAAAATHD
TTTWWAAA--
| Original motif Consensus sequence: DHATTTTAWH |
Reverse complement motif Consensus sequence: HWTAAAATHD |
|
|
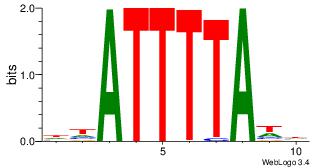 |
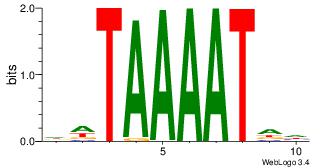 |
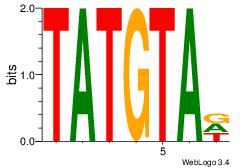
| Dataset #: 1 |
Motif ID: 10 |
Motif name: Motif 10 |
| Original motif Consensus sequence: HTACATA |
Reverse complement motif Consensus sequence: TATGTAD |
|
|
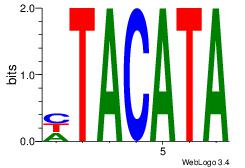 |
 |
Best Matches for Motif ID 10 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
90 |
| Motif name: |
ymTACATAyw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0 |
Alignment:
HHTACATABD
-HTACATA--
| Original motif Consensus sequence: HHTACATABD |
Reverse complement motif Consensus sequence: DBTATGTAHH |
|
|
 |
 |
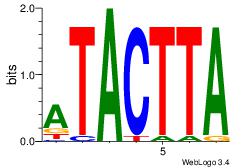
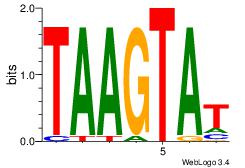
| Dataset #: |
3 |
| Motif ID: |
69 |
| Motif name: |
NKX3-1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0460238 |
Alignment:
TAAGTAT
TATGTAD
| Original motif Consensus sequence: ATACTTA |
Reverse complement motif Consensus sequence: TAAGTAT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
114 |
| Motif name: |
awAACAcAAwa |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.0619118 |
Alignment:
BDTTGTGTTDD
---TATGTAD-
| Original motif Consensus sequence: DDAACACAADV |
Reverse complement motif Consensus sequence: BDTTGTGTTDD |
|
|
 |
 |
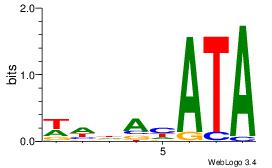
| Dataset #: |
3 |
| Motif ID: |
63 |
| Motif name: |
FOXL1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0663597 |
Alignment:
WHDAHATA
-HTACATA
| Original motif Consensus sequence: WHDAHATA |
Reverse complement motif Consensus sequence: TATDTDHW |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
108 |
| Motif name: |
wwATkTTTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.0688346 |
Alignment:
HHTAAARATHD
-HTACATA---
| Original motif Consensus sequence: DHATKTTTAHH |
Reverse complement motif Consensus sequence: HHTAAARATHD |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 11 |
Motif name: Motif 11 |
| Original motif Consensus sequence: CTGDGTTC |
Reverse complement motif Consensus sequence: GAACDCAG |
|
|
 |
 |
Best Matches for Motif ID 11 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
31 |
| Motif name: |
Motif 31 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0558383 |
Alignment:
CTGWSCTC
CTGDGTTC
| Original motif Consensus sequence: CTGWSCTC |
Reverse complement motif Consensus sequence: GAGSWCAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
12 |
| Motif name: |
Motif 12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0780567 |
Alignment:
MTGTGTGC
CTGDGTTC
| Original motif Consensus sequence: GCACACAY |
Reverse complement motif Consensus sequence: MTGTGTGC |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
27 |
| Motif name: |
Motif 27 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0849877 |
Alignment:
KRAGGCAG
GAACDCAG
| Original motif Consensus sequence: CTGCCTKY |
Reverse complement motif Consensus sequence: KRAGGCAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0874939 |
Alignment:
CTGGCYTY
CTGDGTTC
| Original motif Consensus sequence: CTGGCYTY |
Reverse complement motif Consensus sequence: KAKGCCAG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
114 |
| Motif name: |
awAACAcAAwa |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0916924 |
Alignment:
DDAACACAADV
-GAACDCAG--
| Original motif Consensus sequence: DDAACACAADV |
Reverse complement motif Consensus sequence: BDTTGTGTTDD |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 12 |
Motif name: Motif 12 |
| Original motif Consensus sequence: GCACACAY |
Reverse complement motif Consensus sequence: MTGTGTGC |
|
|
 |
 |
Best Matches for Motif ID 12 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0388776 |
Alignment:
ACACACAC
GCACACAY
| Original motif Consensus sequence: ACACACAC |
Reverse complement motif Consensus sequence: GTGTGTGT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
58 |
| Motif name: |
Motif 58 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
8 |
| Similarity score: |
0.0509001 |
Alignment:
CACACACACACACA
-GCACACAY-----
| Original motif Consensus sequence: CACACACACACACA |
Reverse complement motif Consensus sequence: TGTGTGTGTGTGTG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0543096 |
Alignment:
GAACDCAG
GCACACAY
| Original motif Consensus sequence: CTGDGTTC |
Reverse complement motif Consensus sequence: GAACDCAG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
126 |
| Motif name: |
yvTGCAGsCAcg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0573852 |
Alignment:
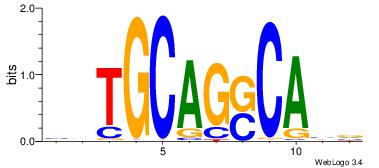
BVTGCAGSCABV
---GCACACAY-
| Original motif Consensus sequence: BVTGCAGSCABV |
Reverse complement motif Consensus sequence: VBTGSCTGCAVB |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
37 |
| Motif name: |
Motif 37 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0626478 |
Alignment:
KCACATKC
GCACACAY
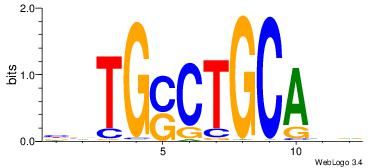
| Original motif Consensus sequence: GYATGTGY |
Reverse complement motif Consensus sequence: KCACATKC |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 13 |
Motif name: Motif 13 |
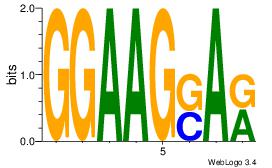
| Original motif Consensus sequence: GGAAGSAR |
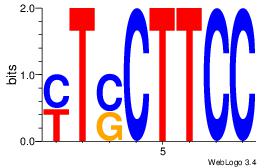
Reverse complement motif Consensus sequence: MTSCTTCC |
|
|
 |
 |
Best Matches for Motif ID 13 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
17 |
| Motif name: |
Motif 17 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0653924 |
Alignment:
GGGAGGRR
GGAAGSAR
| Original motif Consensus sequence: GGGAGGRR |
Reverse complement motif Consensus sequence: MMCCTCCC |
|
|
 |
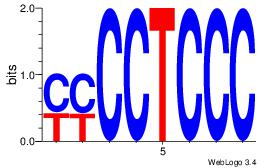 |
| Dataset #: |
4 |
| Motif ID: |
116 |
| Motif name: |
dyCTGCyTCyc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0796555 |
Alignment:
DHCTGCYTCHB
--MTSCTTCC-
| Original motif Consensus sequence: DHCTGCYTCHB |
Reverse complement motif Consensus sequence: BHGAKGCAGHH |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
26 |
| Motif name: |
Motif 26 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0803708 |
Alignment:
RGARGAAA
GGAAGSAR
| Original motif Consensus sequence: RGARGAAA |
Reverse complement motif Consensus sequence: TTTCKTCK |
|
|
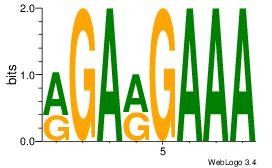 |
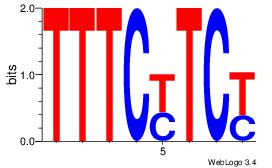 |
| Dataset #: |
4 |
| Motif ID: |
113 |
| Motif name: |
htCTGyKTCbt |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0857132 |
Alignment:
HBCTGYTTCBH
--MTSCTTCC-
| Original motif Consensus sequence: HBCTGYTTCBH |
Reverse complement motif Consensus sequence: HVGAAMCAGVH |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
27 |
| Motif name: |
Motif 27 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0948401 |
Alignment:
KRAGGCAG
GGAAGSAR
| Original motif Consensus sequence: CTGCCTKY |
Reverse complement motif Consensus sequence: KRAGGCAG |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 14 |
Motif name: Motif 14 |
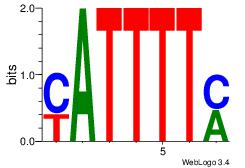
| Original motif Consensus sequence: KAAAATR |
Reverse complement motif Consensus sequence: MATTTTY |
|
|
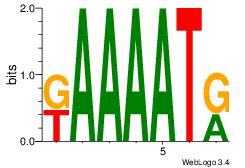 |
 |
Best Matches for Motif ID 14 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
91 |
| Motif name: |
wtATTTTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0107308 |
Alignment:
HWTAAAATHD
--KAAAATR-
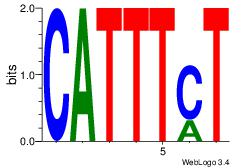
| Original motif Consensus sequence: DHATTTTAWH |
Reverse complement motif Consensus sequence: HWTAAAATHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
108 |
| Motif name: |
wwATkTTTAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0228401 |
Alignment:
DHATKTTTAHH
-MATTTTY---
| Original motif Consensus sequence: DHATKTTTAHH |
Reverse complement motif Consensus sequence: HHTAAARATHD |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
44 |
| Motif name: |
Motif 44 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0324742 |
Alignment:
CATTTCT
MATTTTY
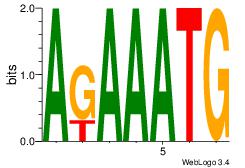
| Original motif Consensus sequence: AGAAATG |
Reverse complement motif Consensus sequence: CATTTCT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
87 |
| Motif name: |
SOX9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0488777 |
Alignment:
CMATTGTTH
-MATTTTY-
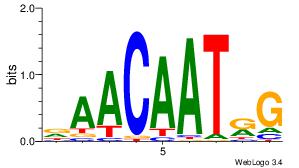
| Original motif Consensus sequence: DAACAATRG |
Reverse complement motif Consensus sequence: CMATTGTTH |
|
|
 |
 |
| Dataset #: |
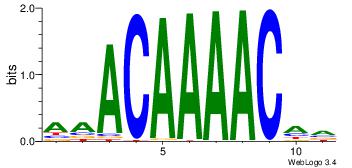
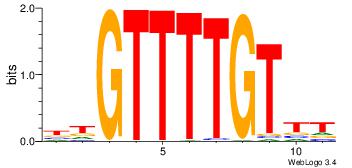
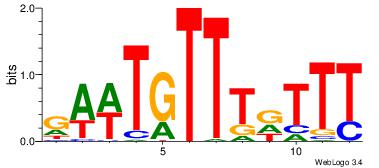
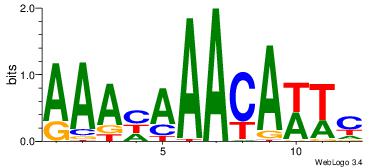
4 |
| Motif ID: |
109 |
| Motif name: |
aaCAAAAACaa |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0510773 |
Alignment:
HHGTTTTTGHH
-MATTTTY---
| Original motif Consensus sequence: HHCAAAAACHH |
Reverse complement motif Consensus sequence: HHGTTTTTGHH |
|
|
 |
 |
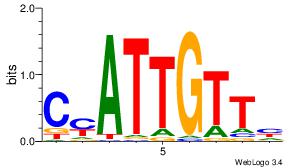
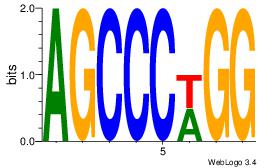
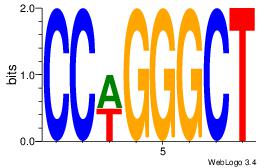
| Dataset #: 1 |
Motif ID: 15 |
Motif name: Motif 15 |
| Original motif Consensus sequence: AGCCCWGG |
Reverse complement motif Consensus sequence: CCWGGGCT |
|
|
 |
 |
Best Matches for Motif ID 15 (Highest to Lowest)
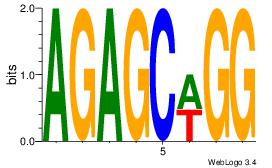
| Dataset #: |
1 |
| Motif ID: |
51 |
| Motif name: |
Motif 51 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0387232 |
Alignment:
AGAGCWGG
AGCCCWGG
| Original motif Consensus sequence: AGAGCWGG |
Reverse complement motif Consensus sequence: CCWGCTCT |
|
|
 |
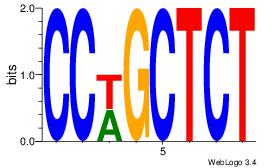 |
| Dataset #: |
4 |
| Motif ID: |
104 |
| Motif name: |
cgCCCwGGScsg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0422017 |
Alignment:
VDCCCWGGGCBB
AGCCCWGG----
| Original motif Consensus sequence: VDCCCWGGGCBB |
Reverse complement motif Consensus sequence: BBGCCCWGGGHV |
|
|
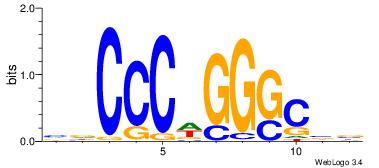 |
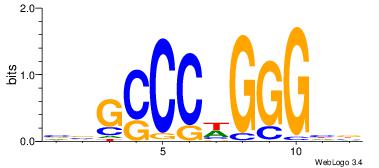 |
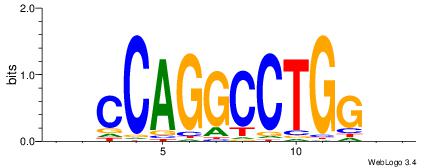
| Dataset #: |
4 |
| Motif ID: |
122 |
| Motif name: |
hsCCAGGCCTGGsr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0482193 |
Alignment:
HVCCAGGCCTGGBD
--CCWGGGCT----
| Original motif Consensus sequence: HVCCAGGCCTGGBD |
Reverse complement motif Consensus sequence: HBCCAGGCCTGGVD |
|
|
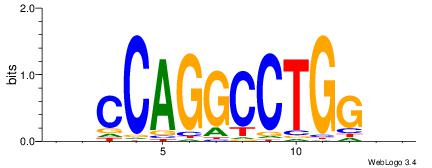 |
 |
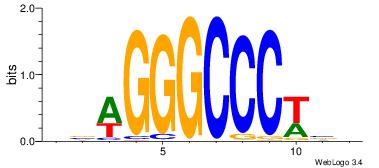
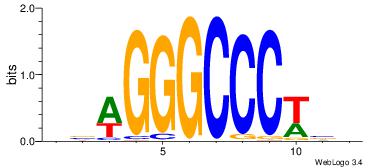
| Dataset #: |
4 |
| Motif ID: |
124 |
| Motif name: |
cswGGGCCCwsg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0580747 |
Alignment:
BBWGGGCCCWVV
----AGCCCWGG
| Original motif Consensus sequence: VVWGGGCCCWBB |
Reverse complement motif Consensus sequence: BBWGGGCCCWVV |
|
|
 |
 |
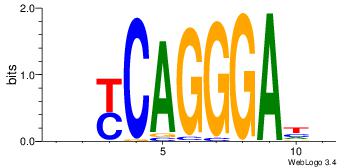
| Dataset #: |
4 |
| Motif ID: |
127 |
| Motif name: |
rryCAGGGAyw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0593634 |
Alignment:
DDYCAGGGAHD
--CCWGGGCT-
| Original motif Consensus sequence: DDYCAGGGAHD |
Reverse complement motif Consensus sequence: DHTCCCTGMDD |
|
|
 |
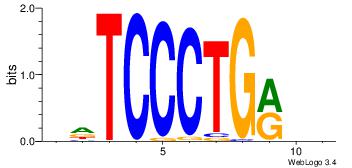 |
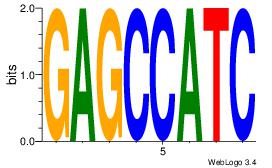
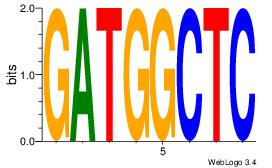
| Dataset #: 1 |
Motif ID: 16 |
Motif name: Motif 16 |
| Original motif Consensus sequence: GAGCCATC |
Reverse complement motif Consensus sequence: GATGGCTC |
|
|
 |
 |
Best Matches for Motif ID 16 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
121 |
| Motif name: |
ctGAGTCATCkb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0217603 |
Alignment:
HDGAGTCATCDB
--GAGCCATC--
| Original motif Consensus sequence: HDGAGTCATCDB |
Reverse complement motif Consensus sequence: BHGATGACTCDD |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0643454 |
Alignment:
KCTRMCTC
GATGGCTC
| Original motif Consensus sequence: GAGRMAGR |
Reverse complement motif Consensus sequence: KCTRMCTC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
116 |
| Motif name: |
dyCTGCyTCyc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0716659 |
Alignment:
BHGAKGCAGHH
--GAGCCATC-
| Original motif Consensus sequence: DHCTGCYTCHB |
Reverse complement motif Consensus sequence: BHGAKGCAGHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
124 |
| Motif name: |
cswGGGCCCwsg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0726629 |
Alignment:
BBWGGGCCCWVV
---GAGCCATC-
| Original motif Consensus sequence: VVWGGGCCCWBB |
Reverse complement motif Consensus sequence: BBWGGGCCCWVV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
97 |
| Motif name: |
sgArGCCACg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0739514 |
Alignment:
CGTGGCKTCS
GATGGCTC--
| Original motif Consensus sequence: SGARGCCACG |
Reverse complement motif Consensus sequence: CGTGGCKTCS |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 17 |
Motif name: Motif 17 |
| Original motif Consensus sequence: GGGAGGRR |
Reverse complement motif Consensus sequence: MMCCTCCC |
|
|
 |
 |
Best Matches for Motif ID 17 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
13 |
| Motif name: |
Motif 13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0153924 |
Alignment:
GGAAGSAR
GGGAGGRR
| Original motif Consensus sequence: GGAAGSAR |
Reverse complement motif Consensus sequence: MTSCTTCC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
111 |
| Motif name: |
ccCCmCaCCCCcc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0180858 |
Alignment:
HBCCCCACCCCBH
--MMCCTCCC---
| Original motif Consensus sequence: HBCCCCACCCCBH |
Reverse complement motif Consensus sequence: DBGGGGTGGGGBD |
|
|
 |
 |
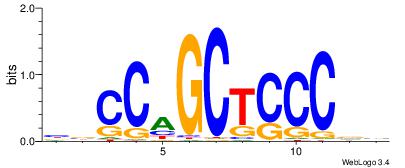
| Dataset #: |
4 |
| Motif ID: |
103 |
| Motif name: |
csCCaGCTCCCgs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0339203 |
Alignment:
BBCCAGCTCCCVB
---MMCCTCCC--
| Original motif Consensus sequence: BBCCAGCTCCCVB |
Reverse complement motif Consensus sequence: BVGGGAGCTGGBB |
|
|
 |
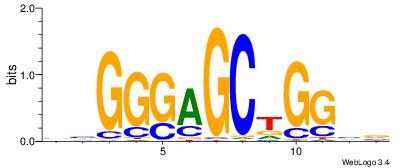 |
| Dataset #: |
4 |
| Motif ID: |
110 |
| Motif name: |
cmCCACACCcm |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0373897 |
Alignment:
HVCCACACCBV
MMCCTCCC---
| Original motif Consensus sequence: HVCCACACCBV |
Reverse complement motif Consensus sequence: VBGGTGTGGBD |
|
|
 |
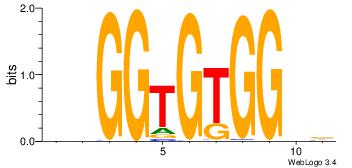 |
| Dataset #: |
4 |
| Motif ID: |
99 |
| Motif name: |
graGGGGGArr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0403428 |
Alignment:
DVAGGGGGADD
---GGGAGGRR
| Original motif Consensus sequence: DVAGGGGGADD |
Reverse complement motif Consensus sequence: HDTCCCCCTVH |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 18 |
Motif name: Motif 18 |
| Original motif Consensus sequence: GRRAGAGA |
Reverse complement motif Consensus sequence: TCTCTMKC |
|
|
 |
 |
Best Matches for Motif ID 18 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0524813 |
Alignment:
GAGRMAGR
GRRAGAGA
| Original motif Consensus sequence: GAGRMAGR |
Reverse complement motif Consensus sequence: KCTRMCTC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
113 |
| Motif name: |
htCTGyKTCbt |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0543958 |
Alignment:
HVGAAMCAGVH
--GRRAGAGA-
| Original motif Consensus sequence: HBCTGYTTCBH |
Reverse complement motif Consensus sequence: HVGAAMCAGVH |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
17 |
| Motif name: |
Motif 17 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0594801 |
Alignment:
GGGAGGRR
GRRAGAGA
| Original motif Consensus sequence: GGGAGGRR |
Reverse complement motif Consensus sequence: MMCCTCCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
59 |
| Motif name: |
Motif 59 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0602419 |
Alignment:
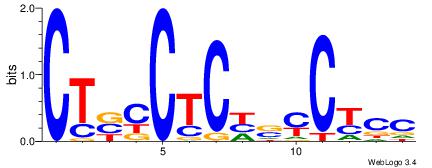
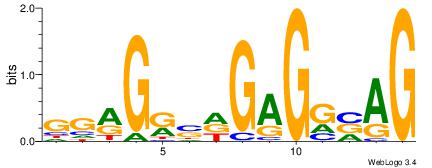
CTBCCTCHSYCYCC
---TCTCTMKC---
| Original motif Consensus sequence: CTBCCTCHSYCYCC |
Reverse complement motif Consensus sequence: GGMGKSHGAGGBAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0675926 |
Alignment:
CACASASA
GRRAGAGA
| Original motif Consensus sequence: CACASASA |
Reverse complement motif Consensus sequence: TSTSTGTG |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 19 |
Motif name: Motif 19 |
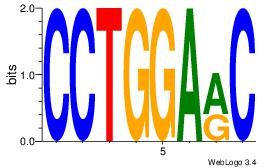
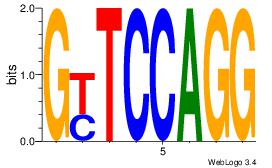
| Original motif Consensus sequence: CCTGGARC |
Reverse complement motif Consensus sequence: GKTCCAGG |
|
|
 |
 |
Best Matches for Motif ID 19 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
42 |
| Motif name: |
Motif 42 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0610815 |
Alignment:
CCYAGAAC
CCTGGARC
| Original motif Consensus sequence: CCYAGAAC |
Reverse complement motif Consensus sequence: GTTCTKGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
84 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0814394 |
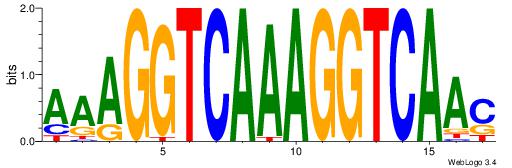
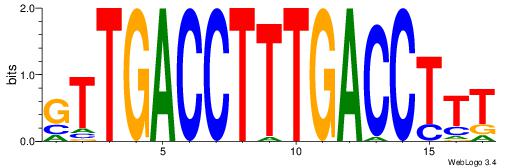
Alignment:
AAAGGTCAAAGGTCAAC
---GKTCCAGG------
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
20 |
| Motif name: |
Motif 20 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0952394 |
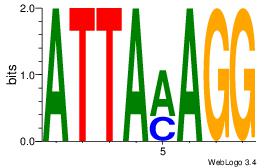
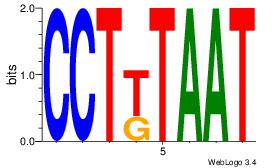
Alignment:
ATTAMAGG
GKTCCAGG
| Original motif Consensus sequence: ATTAMAGG |
Reverse complement motif Consensus sequence: CCTYTAAT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
102 |
| Motif name: |
ssCkGGYCCCsg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0966701 |
Alignment:
VVGGGGCCRGBB
---GKTCCAGG-
| Original motif Consensus sequence: BBCKGGCCCCVV |
Reverse complement motif Consensus sequence: VVGGGGCCRGBB |
|
|
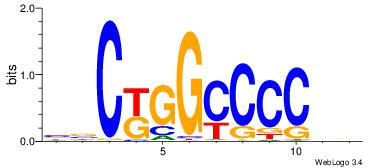 |
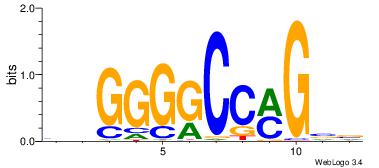 |
| Dataset #: |
1 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0972534 |
Alignment:
CCWGGGCT
CCTGGARC
| Original motif Consensus sequence: AGCCCWGG |
Reverse complement motif Consensus sequence: CCWGGGCT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 20 |
Motif name: Motif 20 |
| Original motif Consensus sequence: ATTAMAGG |
Reverse complement motif Consensus sequence: CCTYTAAT |
|
|
 |
 |
Best Matches for Motif ID 20 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
84 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
8 |
| Similarity score: |
0.0699525 |
Alignment:
GTTGACCTTTGACCTTT
-----CCTYTAAT----
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0931208 |
Alignment:
CCTGGARC
CCTYTAAT
| Original motif Consensus sequence: CCTGGARC |
Reverse complement motif Consensus sequence: GKTCCAGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
Lhx3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0993275 |
Alignment:
BMWTAATTAATTW
---CCTYTAAT--
| Original motif Consensus sequence: WAATTAATTAWYB |
Reverse complement motif Consensus sequence: BMWTAATTAATTW |
|
|
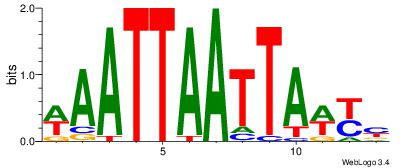 |
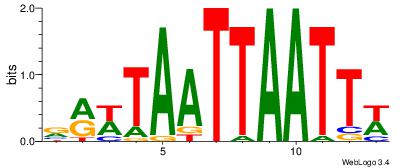 |
| Dataset #: |
3 |
| Motif ID: |
83 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.100049 |
Alignment:
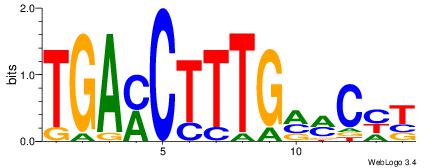
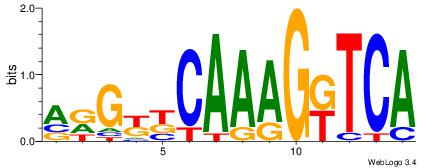
TGAMCTTTGMMCYT
---CCTYTAAT---
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
108 |
| Motif name: |
wwATkTTTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.101957 |
Alignment:
HHTAAARATHD
ATTAMAGG---
| Original motif Consensus sequence: DHATKTTTAHH |
Reverse complement motif Consensus sequence: HHTAAARATHD |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 21 |
Motif name: Motif 21 |
| Original motif Consensus sequence: AAACAAAC |
Reverse complement motif Consensus sequence: GTTTGTTT |
|
|
 |
 |
Best Matches for Motif ID 21 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
60 |
| Motif name: |
Motif 60 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0392428 |
Alignment:
AADAAAAMAAAAAM
----AAACAAAC--
| Original motif Consensus sequence: AADAAAAMAAAAAM |
Reverse complement motif Consensus sequence: YTTTTTRTTTTDTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
67 |
| Motif name: |
FOXI1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0439051 |
Alignment:
AAAMAAAMADBY
AAACAAAC----
| Original motif Consensus sequence: KBDTRTTTRTTT |
Reverse complement motif Consensus sequence: AAAMAAAMADBY |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0458289 |
Alignment:
AAASAAAA
AAACAAAC
| Original motif Consensus sequence: AAASAAAA |
Reverse complement motif Consensus sequence: TTTTSTTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
65 |
| Motif name: |
Foxd3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0463073 |
Alignment:
AAAHAAACAWTC
AAACAAAC----
| Original motif Consensus sequence: GAWTGTTTDTTT |
Reverse complement motif Consensus sequence: AAAHAAACAWTC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
89 |
| Motif name: |
aaACAAAACaa |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0522284 |
Alignment:
BHGTTTTGTTT
---GTTTGTTT
| Original motif Consensus sequence: AAACAAAACHV |
Reverse complement motif Consensus sequence: BHGTTTTGTTT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 22 |
Motif name: Motif 22 |
| Original motif Consensus sequence: GTGGTGGY |
Reverse complement motif Consensus sequence: MCCACCAC |
|
|
 |
 |
Best Matches for Motif ID 22 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0503283 |
Alignment:
GYGSTGGG
GTGGTGGY
| Original motif Consensus sequence: CCCASCMC |
Reverse complement motif Consensus sequence: GYGSTGGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
115 |
| Motif name: |
ysCmAGCACwy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0623778 |
Alignment:
HVCMAGCACHH
-MCCACCAC--
| Original motif Consensus sequence: HVCMAGCACHH |
Reverse complement motif Consensus sequence: DHGTGCTRGVH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
93 |
| Motif name: |
cmCARCACTwr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0725856 |
Alignment:
BHCAGCACTHV
MCCACCAC---
| Original motif Consensus sequence: BHCAGCACTHV |
Reverse complement motif Consensus sequence: VHAGTGCTGHB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
101 |
| Motif name: |
csCACCCCgg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0756613 |
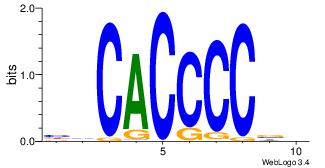
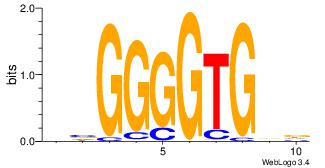
Alignment:
HVCACCCCBB
MCCACCAC--
| Original motif Consensus sequence: HVCACCCCBB |
Reverse complement motif Consensus sequence: BBGGGGTGVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
110 |
| Motif name: |
cmCCACACCcm |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0823897 |
Alignment:
HVCCACACCBV
-MCCACCAC--
| Original motif Consensus sequence: HVCCACACCBV |
Reverse complement motif Consensus sequence: VBGGTGTGGBD |
|
|
 |
 |
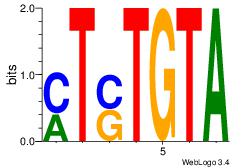
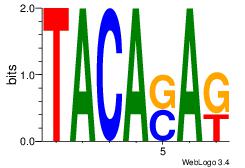
| Dataset #: 1 |
Motif ID: 23 |
Motif name: Motif 23 |
| Original motif Consensus sequence: MTSTGTA |
Reverse complement motif Consensus sequence: TACASAR |
|
|
 |
 |
Best Matches for Motif ID 23 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0172404 |
Alignment:
TSTSTGTG
MTSTGTA-
| Original motif Consensus sequence: CACASASA |
Reverse complement motif Consensus sequence: TSTSTGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
90 |
| Motif name: |
ymTACATAyw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0191193 |
Alignment:
HHTACATABD
--TACASAR-
| Original motif Consensus sequence: HHTACATABD |
Reverse complement motif Consensus sequence: DBTATGTAHH |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
12 |
| Motif name: |
Motif 12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.03188 |
Alignment:
GCACACAY
TACASAR-
| Original motif Consensus sequence: GCACACAY |
Reverse complement motif Consensus sequence: MTGTGTGC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
114 |
| Motif name: |
awAACAcAAwa |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0.0346685 |
Alignment:
DDAACACAADV
--TACASAR--
| Original motif Consensus sequence: DDAACACAADV |
Reverse complement motif Consensus sequence: BDTTGTGTTDD |
|
|
 |
 |
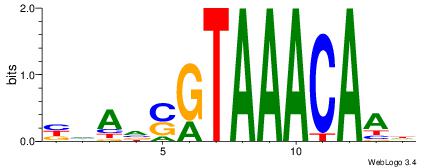
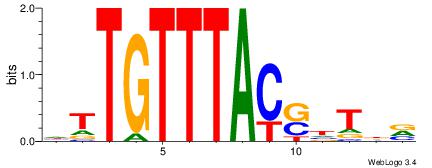
| Dataset #: |
3 |
| Motif ID: |
75 |
| Motif name: |
FOXF2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0372723 |
Alignment:
BHADSGTAAACAAD
------TACASAR-
| Original motif Consensus sequence: BHADSGTAAACAAD |
Reverse complement motif Consensus sequence: DTTGTTTACSBTBB |
|
|
 |
 |
| Dataset #: 1 |
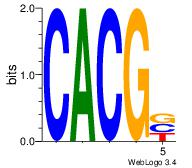
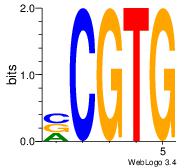
Motif ID: 24 |
Motif name: Motif 24 |
| Original motif Consensus sequence: CACGB |
Reverse complement motif Consensus sequence: BCGTG |
|
|
 |
 |
Best Matches for Motif ID 24 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
56 |
| Motif name: |
Motif 56 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
5 |
| Similarity score: |
0 |
Alignment:
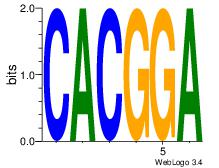
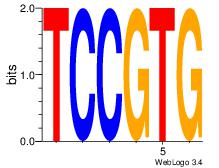
TCCGTG
-BCGTG
| Original motif Consensus sequence: CACGGA |
Reverse complement motif Consensus sequence: TCCGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
115 |
| Motif name: |
ysCmAGCACwy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
5 |
| Similarity score: |
0.0186397 |
Alignment:
DHGTGCTRGVH
BCGTG------
| Original motif Consensus sequence: HVCMAGCACHH |
Reverse complement motif Consensus sequence: DHGTGCTRGVH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
107 |
| Motif name: |
crCGyGCRcg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
5 |
| Similarity score: |
0.019278 |
Alignment:
CGCGTGCACG
-BCGTG----
| Original motif Consensus sequence: CGCGTGCACG |
Reverse complement motif Consensus sequence: CGTGCACGCG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
12 |
| Motif name: |
Motif 12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
5 |
| Similarity score: |
0.0246242 |
Alignment:
GCACACAY
---CACGB
| Original motif Consensus sequence: GCACACAY |
Reverse complement motif Consensus sequence: MTGTGTGC |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
5 |
| Similarity score: |
0.0336642 |
Alignment:
CACASASA
CACGB---
| Original motif Consensus sequence: CACASASA |
Reverse complement motif Consensus sequence: TSTSTGTG |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 25 |
Motif name: Motif 25 |
| Original motif Consensus sequence: TTTAWW |
Reverse complement motif Consensus sequence: WWTAAA |
|
|
 |
 |
Best Matches for Motif ID 25 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
91 |
| Motif name: |
wtATTTTAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0116421 |
Alignment:
HWTAAAATHD
WWTAAA----
| Original motif Consensus sequence: DHATTTTAWH |
Reverse complement motif Consensus sequence: HWTAAAATHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
108 |
| Motif name: |
wwATkTTTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0118254 |
Alignment:
DHATKTTTAHH
-----TTTAWW
| Original motif Consensus sequence: DHATKTTTAHH |
Reverse complement motif Consensus sequence: HHTAAARATHD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
70 |
| Motif name: |
Foxq1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0158082 |
Alignment:
HATTGTTTATW
-----TTTAWW
| Original motif Consensus sequence: HATTGTTTATW |
Reverse complement motif Consensus sequence: WATAAACAATH |
|
|
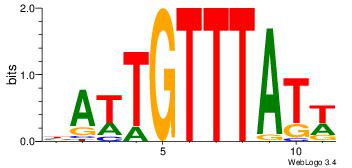 |
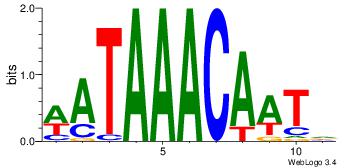 |
| Dataset #: |
1 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0300892 |
Alignment:
AWATAMA
WWTAAA-
| Original motif Consensus sequence: AWATAMA |
Reverse complement motif Consensus sequence: TYTATWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
55 |
| Motif name: |
Motif 55 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0307675 |
Alignment:
TTTAAAA
TTTAWW-
| Original motif Consensus sequence: TTTAAAA |
Reverse complement motif Consensus sequence: TTTTAAA |
|
|
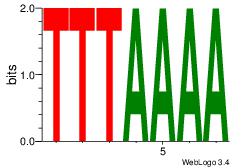 |
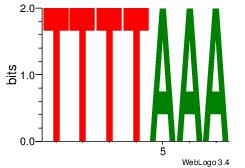 |
| Dataset #: 1 |
Motif ID: 26 |
Motif name: Motif 26 |
| Original motif Consensus sequence: RGARGAAA |
Reverse complement motif Consensus sequence: TTTCKTCK |
|
|
 |
 |
Best Matches for Motif ID 26 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
71 |
| Motif name: |
Evi1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0776207 |
Alignment:
TBTTATCTTMTCTT
---TTTCKTCK---
| Original motif Consensus sequence: AAGAYAAGATAABA |
Reverse complement motif Consensus sequence: TBTTATCTTMTCTT |
|
|
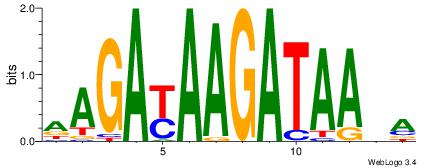 |
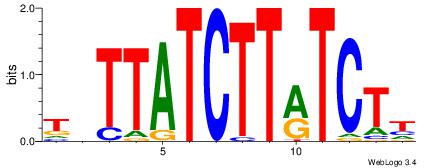 |
| Dataset #: |
1 |
| Motif ID: |
13 |
| Motif name: |
Motif 13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0795772 |
Alignment:
GGAAGSAR
RGARGAAA
| Original motif Consensus sequence: GGAAGSAR |
Reverse complement motif Consensus sequence: MTSCTTCC |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0833075 |
Alignment:
AAASAAAA
RGARGAAA
| Original motif Consensus sequence: AAASAAAA |
Reverse complement motif Consensus sequence: TTTTSTTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
89 |
| Motif name: |
aaACAAAACaa |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.087026 |
Alignment:
BHGTTTTGTTT
---TTTCKTCK
| Original motif Consensus sequence: AAACAAAACHV |
Reverse complement motif Consensus sequence: BHGTTTTGTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
60 |
| Motif name: |
Motif 60 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0876612 |
Alignment:
YTTTTTRTTTTDTT
---TTTCKTCK---
| Original motif Consensus sequence: AADAAAAMAAAAAM |
Reverse complement motif Consensus sequence: YTTTTTRTTTTDTT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 27 |
Motif name: Motif 27 |
| Original motif Consensus sequence: CTGCCTKY |
Reverse complement motif Consensus sequence: KRAGGCAG |
|
|
 |
 |
Best Matches for Motif ID 27 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
39 |
| Motif name: |
Motif 39 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0247901 |
Alignment:
CTGYCTGT
CTGCCTKY
| Original motif Consensus sequence: ACAGMCAG |
Reverse complement motif Consensus sequence: CTGYCTGT |
|
|
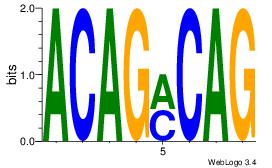 |
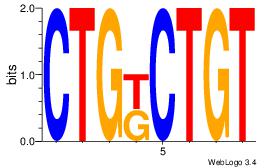 |
| Dataset #: |
4 |
| Motif ID: |
116 |
| Motif name: |
dyCTGCyTCyc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0249073 |
Alignment:
BHGAKGCAGHH
-KRAGGCAG--
| Original motif Consensus sequence: DHCTGCYTCHB |
Reverse complement motif Consensus sequence: BHGAKGCAGHH |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0407877 |
Alignment:
CTGGCYTY
CTGCCTKY
| Original motif Consensus sequence: CTGGCYTY |
Reverse complement motif Consensus sequence: KAKGCCAG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
126 |
| Motif name: |
yvTGCAGsCAcg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0601303 |
Alignment:
VBTGSCTGCAVB
-CTGCCTKY---
| Original motif Consensus sequence: BVTGCAGSCABV |
Reverse complement motif Consensus sequence: VBTGSCTGCAVB |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0611983 |
Alignment:
GAACDCAG
KRAGGCAG
| Original motif Consensus sequence: CTGDGTTC |
Reverse complement motif Consensus sequence: GAACDCAG |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 28 |
Motif name: Motif 28 |
| Original motif Consensus sequence: AVAGAAA |
Reverse complement motif Consensus sequence: TTTCTVT |
|
|
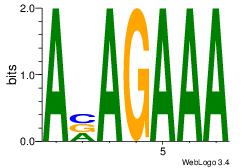 |
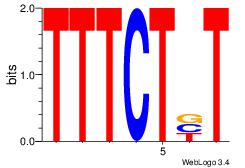 |
Best Matches for Motif ID 28 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.034392 |
Alignment:
AAASAAAA
AVAGAAA-
| Original motif Consensus sequence: AAASAAAA |
Reverse complement motif Consensus sequence: TTTTSTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0458576 |
Alignment:
AGATAAAR
AVAGAAA-
| Original motif Consensus sequence: AGATAAAR |
Reverse complement motif Consensus sequence: KTTTATCT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
21 |
| Motif name: |
Motif 21 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0461542 |
Alignment:
AAACAAAC
AVAGAAA-
| Original motif Consensus sequence: AAACAAAC |
Reverse complement motif Consensus sequence: GTTTGTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
18 |
| Motif name: |
Motif 18 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.047492 |
Alignment:
TCTCTMKC
TTTCTVT-
| Original motif Consensus sequence: GRRAGAGA |
Reverse complement motif Consensus sequence: TCTCTMKC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
89 |
| Motif name: |
aaACAAAACaa |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
7 |
| Similarity score: |
0.055017 |
Alignment:
AAACAAAACHV
AVAGAAA----
| Original motif Consensus sequence: AAACAAAACHV |
Reverse complement motif Consensus sequence: BHGTTTTGTTT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 29 |
Motif name: Motif 29 |
| Original motif Consensus sequence: CCMGCCCC |
Reverse complement motif Consensus sequence: GGGGCYGG |
|
|
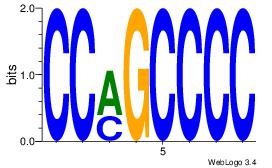 |
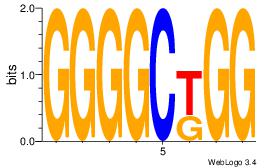 |
Best Matches for Motif ID 29 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
103 |
| Motif name: |
csCCaGCTCCCgs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0420068 |
Alignment:
BVGGGAGCTGGBB
---GGGGCYGG--
| Original motif Consensus sequence: BBCCAGCTCCCVB |
Reverse complement motif Consensus sequence: BVGGGAGCTGGBB |
|
|
 |
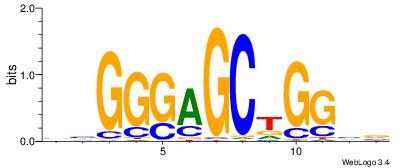 |
| Dataset #: |
4 |
| Motif ID: |
119 |
| Motif name: |
cbCCAGCTCmyk |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0430311 |
Alignment:
BBCCAGCTCMBD
--CCMGCCCC--
| Original motif Consensus sequence: BBCCAGCTCMBD |
Reverse complement motif Consensus sequence: HBYGAGCTGGBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
102 |
| Motif name: |
ssCkGGYCCCsg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0462922 |
Alignment:
VVGGGGCCRGBB
--GGGGCYGG--
| Original motif Consensus sequence: BBCKGGCCCCVV |
Reverse complement motif Consensus sequence: VVGGGGCCRGBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
111 |
| Motif name: |
ccCCmCaCCCCcc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.051245 |
Alignment:
HBCCCCACCCCBH
---CCMGCCCC--
| Original motif Consensus sequence: HBCCCCACCCCBH |
Reverse complement motif Consensus sequence: DBGGGGTGGGGBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
115 |
| Motif name: |
ysCmAGCACwy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0556862 |
Alignment:
HVCMAGCACHH
--CCMGCCCC-
| Original motif Consensus sequence: HVCMAGCACHH |
Reverse complement motif Consensus sequence: DHGTGCTRGVH |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 30 |
Motif name: Motif 30 |
| Original motif Consensus sequence: CAGGARGM |
Reverse complement motif Consensus sequence: YCMTCCTG |
|
|
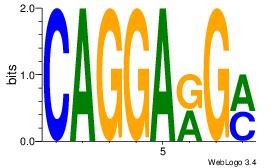 |
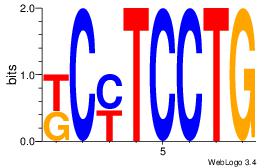 |
Best Matches for Motif ID 30 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
47 |
| Motif name: |
Motif 47 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0890875 |
Alignment:
KGCTGCTG
YCMTCCTG
| Original motif Consensus sequence: KGCTGCTG |
Reverse complement motif Consensus sequence: CAGCAGCR |
|
|
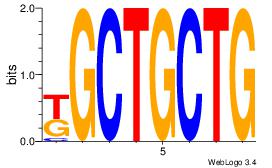 |
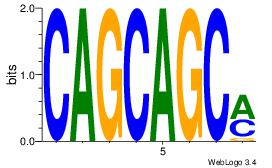 |
| Dataset #: |
1 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0957719 |
Alignment:
GAGRMAGR
CAGGARGM
| Original motif Consensus sequence: GAGRMAGR |
Reverse complement motif Consensus sequence: KCTRMCTC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
98 |
| Motif name: |
myyCCCACmTGCmyr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0969866 |
Alignment:
HBCCCCACMTGCMHV
---YCMTCCTG----
| Original motif Consensus sequence: HBCCCCACMTGCMHV |
Reverse complement motif Consensus sequence: VDRGCAYGTGGGGVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
127 |
| Motif name: |
rryCAGGGAyw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0975352 |
Alignment:
DDYCAGGGAHD
---CAGGARGM
| Original motif Consensus sequence: DDYCAGGGAHD |
Reverse complement motif Consensus sequence: DHTCCCTGMDD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
99 |
| Motif name: |
graGGGGGArr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0989909 |
Alignment:
DVAGGGGGADD
-CAGGARGM--
| Original motif Consensus sequence: DVAGGGGGADD |
Reverse complement motif Consensus sequence: HDTCCCCCTVH |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 31 |
Motif name: Motif 31 |
| Original motif Consensus sequence: CTGWSCTC |
Reverse complement motif Consensus sequence: GAGSWCAG |
|
|
 |
 |
Best Matches for Motif ID 31 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.023129 |
Alignment:
GAACDCAG
GAGSWCAG
| Original motif Consensus sequence: CTGDGTTC |
Reverse complement motif Consensus sequence: GAACDCAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0418605 |
Alignment:
KAKGCCAG
GAGSWCAG
| Original motif Consensus sequence: CTGGCYTY |
Reverse complement motif Consensus sequence: KAKGCCAG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
92 |
| Motif name: |
cgAKGyCAcg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0464751 |
Alignment:
CGAGGYCACV
-GAGSWCAG-
| Original motif Consensus sequence: CGAGGYCACV |
Reverse complement motif Consensus sequence: VGTGMCCTCG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
84 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0568298 |
Alignment:
AAAGGTCAAAGGTCAAC
--------GAGSWCAG-
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
119 |
| Motif name: |
cbCCAGCTCmyk |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0705619 |
Alignment:
HBYGAGCTGGBB
-CTGWSCTC---
| Original motif Consensus sequence: BBCCAGCTCMBD |
Reverse complement motif Consensus sequence: HBYGAGCTGGBB |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 32 |
Motif name: Motif 32 |
| Original motif Consensus sequence: AVATGGC |
Reverse complement motif Consensus sequence: GCCATVT |
|
|
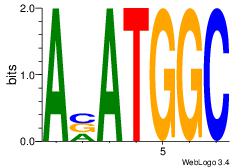 |
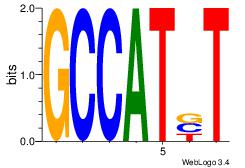 |
Best Matches for Motif ID 32 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
124 |
| Motif name: |
cswGGGCCCwsg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
7 |
| Similarity score: |
0.0595251 |
Alignment:
VVWGGGCCCWBB
AVATGGC-----
| Original motif Consensus sequence: VVWGGGCCCWBB |
Reverse complement motif Consensus sequence: BBWGGGCCCWVV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
121 |
| Motif name: |
ctGAGTCATCkb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0676952 |
Alignment:
HDGAGTCATCDB
----GCCATVT-
| Original motif Consensus sequence: HDGAGTCATCDB |
Reverse complement motif Consensus sequence: BHGATGACTCDD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
84 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
7 |
| Similarity score: |
0.0701366 |
Alignment:
AAAGGTCAAAGGTCAAC
-------AVATGGC---
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
119 |
| Motif name: |
cbCCAGCTCmyk |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
7 |
| Similarity score: |
0.0704204 |
Alignment:
BBCCAGCTCMBD
-GCCATVT----
| Original motif Consensus sequence: BBCCAGCTCMBD |
Reverse complement motif Consensus sequence: HBYGAGCTGGBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
98 |
| Motif name: |
myyCCCACmTGCmyr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0.0736077 |
Alignment:
HBCCCCACMTGCMHV
------AVATGGC--
| Original motif Consensus sequence: HBCCCCACMTGCMHV |
Reverse complement motif Consensus sequence: VDRGCAYGTGGGGVD |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 33 |
Motif name: Motif 33 |
| Original motif Consensus sequence: CMTTTCC |
Reverse complement motif Consensus sequence: GGAAARG |
|
|
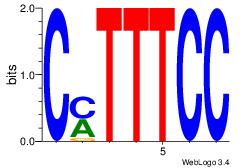 |
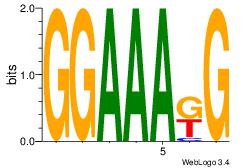 |
Best Matches for Motif ID 33 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
46 |
| Motif name: |
Motif 46 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0542683 |
Alignment:
TSTTTCC
CMTTTCC
| Original motif Consensus sequence: GGAAASA |
Reverse complement motif Consensus sequence: TSTTTCC |
|
|
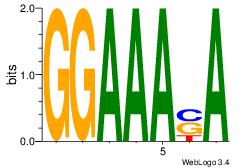 |
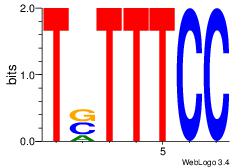 |
| Dataset #: |
1 |
| Motif ID: |
44 |
| Motif name: |
Motif 44 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0667064 |
Alignment:
CATTTCT
CMTTTCC
| Original motif Consensus sequence: AGAAATG |
Reverse complement motif Consensus sequence: CATTTCT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
88 |
| Motif name: |
ygCTyTTCsg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0.0693505 |
Alignment:
BCGAAMAGHH
-GGAAARG--
| Original motif Consensus sequence: HDCTYTTCGB |
Reverse complement motif Consensus sequence: BCGAAMAGHH |
|
|
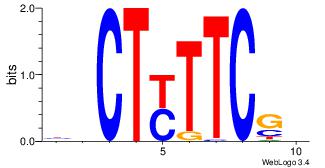 |
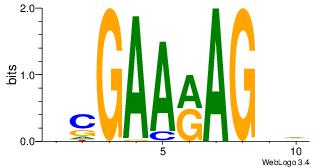 |
| Dataset #: |
1 |
| Motif ID: |
38 |
| Motif name: |
Motif 38 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0734352 |
Alignment:
GGAGMAGG
GGAAARG-
| Original motif Consensus sequence: CCTYCTCC |
Reverse complement motif Consensus sequence: GGAGMAGG |
|
|
 |
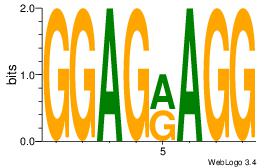 |
| Dataset #: |
1 |
| Motif ID: |
13 |
| Motif name: |
Motif 13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0861393 |
Alignment:
GGAAGSAR
-GGAAARG
| Original motif Consensus sequence: GGAAGSAR |
Reverse complement motif Consensus sequence: MTSCTTCC |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 34 |
Motif name: Motif 34 |
| Original motif Consensus sequence: ATATTTA |
Reverse complement motif Consensus sequence: TAAATAT |
|
|
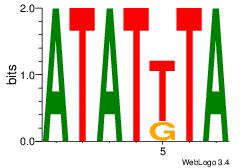 |
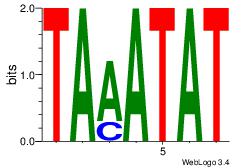 |
Best Matches for Motif ID 34 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
108 |
| Motif name: |
wwATkTTTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0.0294375 |
Alignment:
DHATKTTTAHH
--ATATTTA--
| Original motif Consensus sequence: DHATKTTTAHH |
Reverse complement motif Consensus sequence: HHTAAARATHD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
82 |
| Motif name: |
HNF1B |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0.033217 |
Alignment:
GTTAAWKATTAA
--TAAATAT---
| Original motif Consensus sequence: TTAATRWTTAAC |
Reverse complement motif Consensus sequence: GTTAAWKATTAA |
|
|
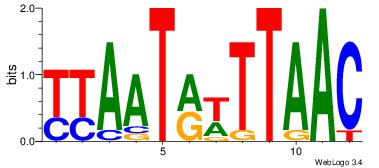 |
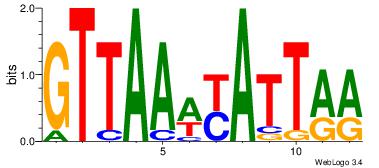 |
| Dataset #: |
4 |
| Motif ID: |
90 |
| Motif name: |
ymTACATAyw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0399459 |
Alignment:
DBTATGTAHH
-ATATTTA--
| Original motif Consensus sequence: HHTACATABD |
Reverse complement motif Consensus sequence: DBTATGTAHH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
61 |
| Motif name: |
MEF2A |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0440271 |
Alignment:
CTAWAAATAG
---TAAATAT
| Original motif Consensus sequence: CTATTTWTAG |
Reverse complement motif Consensus sequence: CTAWAAATAG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
69 |
| Motif name: |
NKX3-1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0453202 |
Alignment:
ATACTTA
ATATTTA
| Original motif Consensus sequence: ATACTTA |
Reverse complement motif Consensus sequence: TAAGTAT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 35 |
Motif name: Motif 35 |
| Original motif Consensus sequence: KGCTGGS |
Reverse complement motif Consensus sequence: SCCAGCR |
|
|
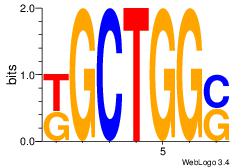 |
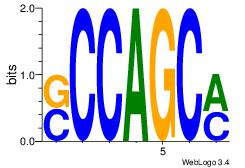 |
Best Matches for Motif ID 35 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
115 |
| Motif name: |
ysCmAGCACwy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.0239923 |
Alignment:
DHGTGCTRGVH
---KGCTGGS-
| Original motif Consensus sequence: HVCMAGCACHH |
Reverse complement motif Consensus sequence: DHGTGCTRGVH |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0288353 |
Alignment:
CCCASCMC
-SCCAGCR
| Original motif Consensus sequence: CCCASCMC |
Reverse complement motif Consensus sequence: GYGSTGGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
119 |
| Motif name: |
cbCCAGCTCmyk |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0386119 |
Alignment:
HBYGAGCTGGBB
----KGCTGGS-
| Original motif Consensus sequence: BBCCAGCTCMBD |
Reverse complement motif Consensus sequence: HBYGAGCTGGBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
93 |
| Motif name: |
cmCARCACTwr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
7 |
| Similarity score: |
0.0467269 |
Alignment:
BHCAGCACTHV
SCCAGCR----
| Original motif Consensus sequence: BHCAGCACTHV |
Reverse complement motif Consensus sequence: VHAGTGCTGHB |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
22 |
| Motif name: |
Motif 22 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.04839 |
Alignment:
GTGGTGGY
-KGCTGGS
| Original motif Consensus sequence: GTGGTGGY |
Reverse complement motif Consensus sequence: MCCACCAC |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 36 |
Motif name: Motif 36 |
| Original motif Consensus sequence: CATGGTGG |
Reverse complement motif Consensus sequence: CCACCATG |
|
|
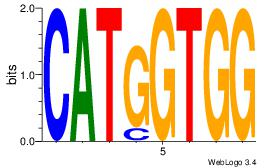 |
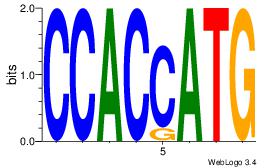 |
Best Matches for Motif ID 36 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
98 |
| Motif name: |
myyCCCACmTGCmyr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0987507 |
Alignment:
VDRGCAYGTGGGGVD
----CATGGTGG---
| Original motif Consensus sequence: HBCCCCACMTGCMHV |
Reverse complement motif Consensus sequence: VDRGCAYGTGGGGVD |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
30 |
| Motif name: |
Motif 30 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.100683 |
Alignment:
YCMTCCTG
CCACCATG
| Original motif Consensus sequence: CAGGARGM |
Reverse complement motif Consensus sequence: YCMTCCTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
115 |
| Motif name: |
ysCmAGCACwy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.106759 |
Alignment:
DHGTGCTRGVH
-CATGGTGG--
| Original motif Consensus sequence: HVCMAGCACHH |
Reverse complement motif Consensus sequence: DHGTGCTRGVH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
118 |
| Motif name: |
yrCACATGCATGTGyr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.106836 |
Alignment:
DVCACATGCATGTGBH
----CATGGTGG----
| Original motif Consensus sequence: HVCACATGCATGTGBD |
Reverse complement motif Consensus sequence: DVCACATGCATGTGBH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
101 |
| Motif name: |
csCACCCCgg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.10804 |
Alignment:
BBGGGGTGVD
-CATGGTGG-
| Original motif Consensus sequence: HVCACCCCBB |
Reverse complement motif Consensus sequence: BBGGGGTGVD |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 37 |
Motif name: Motif 37 |
| Original motif Consensus sequence: GYATGTGY |
Reverse complement motif Consensus sequence: KCACATKC |
|
|
 |
 |
Best Matches for Motif ID 37 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
118 |
| Motif name: |
yrCACATGCATGTGyr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
8 |
| Similarity score: |
0.0125979 |
Alignment:
HVCACATGCATGTGBD
-------GYATGTGY-
| Original motif Consensus sequence: HVCACATGCATGTGBD |
Reverse complement motif Consensus sequence: DVCACATGCATGTGBH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
98 |
| Motif name: |
myyCCCACmTGCmyr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.035887 |
Alignment:
VDRGCAYGTGGGGVD
---GYATGTGY----
| Original motif Consensus sequence: HBCCCCACMTGCMHV |
Reverse complement motif Consensus sequence: VDRGCAYGTGGGGVD |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0415984 |
Alignment:
ACACACAC
KCACATKC
| Original motif Consensus sequence: ACACACAC |
Reverse complement motif Consensus sequence: GTGTGTGT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
58 |
| Motif name: |
Motif 58 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0521491 |
Alignment:
TGTGTGTGTGTGTG
-GYATGTGY-----
| Original motif Consensus sequence: CACACACACACACA |
Reverse complement motif Consensus sequence: TGTGTGTGTGTGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
90 |
| Motif name: |
ymTACATAyw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0544369 |
Alignment:
DBTATGTAHH
-GYATGTGY-
| Original motif Consensus sequence: HHTACATABD |
Reverse complement motif Consensus sequence: DBTATGTAHH |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 38 |
Motif name: Motif 38 |
| Original motif Consensus sequence: CCTYCTCC |
Reverse complement motif Consensus sequence: GGAGMAGG |
|
|
 |
 |
Best Matches for Motif ID 38 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
51 |
| Motif name: |
Motif 51 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0329729 |
Alignment:
AGAGCWGG
GGAGMAGG
| Original motif Consensus sequence: AGAGCWGG |
Reverse complement motif Consensus sequence: CCWGCTCT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
88 |
| Motif name: |
ygCTyTTCsg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0476921 |
Alignment:
BCGAAMAGHH
-GGAGMAGG-
| Original motif Consensus sequence: HDCTYTTCGB |
Reverse complement motif Consensus sequence: BCGAAMAGHH |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
29 |
| Motif name: |
Motif 29 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0493297 |
Alignment:
GGGGCYGG
GGAGMAGG
| Original motif Consensus sequence: CCMGCCCC |
Reverse complement motif Consensus sequence: GGGGCYGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
103 |
| Motif name: |
csCCaGCTCCCgs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0526885 |
Alignment:
BVGGGAGCTGGBB
---GGAGMAGG--
| Original motif Consensus sequence: BBCCAGCTCCCVB |
Reverse complement motif Consensus sequence: BVGGGAGCTGGBB |
|
|
 |
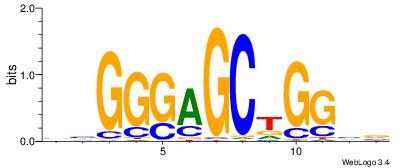 |
| Dataset #: |
4 |
| Motif ID: |
119 |
| Motif name: |
cbCCAGCTCmyk |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0576001 |
Alignment:
HBYGAGCTGGBB
--GGAGMAGG--
| Original motif Consensus sequence: BBCCAGCTCMBD |
Reverse complement motif Consensus sequence: HBYGAGCTGGBB |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 39 |
Motif name: Motif 39 |
| Original motif Consensus sequence: ACAGMCAG |
Reverse complement motif Consensus sequence: CTGYCTGT |
|
|
 |
 |
Best Matches for Motif ID 39 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
27 |
| Motif name: |
Motif 27 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0307023 |
Alignment:
KRAGGCAG
ACAGMCAG
| Original motif Consensus sequence: CTGCCTKY |
Reverse complement motif Consensus sequence: KRAGGCAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0593235 |
Alignment:
ACACACAC
ACAGMCAG
| Original motif Consensus sequence: ACACACAC |
Reverse complement motif Consensus sequence: GTGTGTGT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
58 |
| Motif name: |
Motif 58 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
8 |
| Similarity score: |
0.0634902 |
Alignment:
TGTGTGTGTGTGTG
-----CTGYCTGT-
| Original motif Consensus sequence: CACACACACACACA |
Reverse complement motif Consensus sequence: TGTGTGTGTGTGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
116 |
| Motif name: |
dyCTGCyTCyc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0653685 |
Alignment:
DHCTGCYTCHB
--CTGYCTGT-
| Original motif Consensus sequence: DHCTGCYTCHB |
Reverse complement motif Consensus sequence: BHGAKGCAGHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
113 |
| Motif name: |
htCTGyKTCbt |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0674965 |
Alignment:
HVGAAMCAGVH
-ACAGMCAG--
| Original motif Consensus sequence: HBCTGYTTCBH |
Reverse complement motif Consensus sequence: HVGAAMCAGVH |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 40 |
Motif name: Motif 40 |
| Original motif Consensus sequence: CCACASC |
Reverse complement motif Consensus sequence: GSTGTGG |
|
|
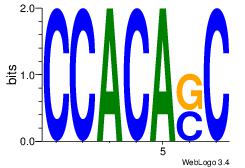 |
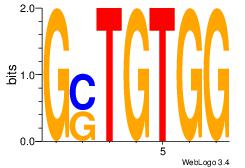 |
Best Matches for Motif ID 40 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
110 |
| Motif name: |
cmCCACACCcm |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0.0381642 |
Alignment:
VBGGTGTGGBD
--GSTGTGG--
| Original motif Consensus sequence: HVCCACACCBV |
Reverse complement motif Consensus sequence: VBGGTGTGGBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
111 |
| Motif name: |
ccCCmCaCCCCcc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
7 |
| Similarity score: |
0.0665887 |
Alignment:
HBCCCCACCCCBH
--CCACASC----
| Original motif Consensus sequence: HBCCCCACCCCBH |
Reverse complement motif Consensus sequence: DBGGGGTGGGGBD |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
22 |
| Motif name: |
Motif 22 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0714286 |
Alignment:
GTGGTGGY
-GSTGTGG
| Original motif Consensus sequence: GTGGTGGY |
Reverse complement motif Consensus sequence: MCCACCAC |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0745048 |
Alignment:
GYGSTGGG
GSTGTGG-
| Original motif Consensus sequence: CCCASCMC |
Reverse complement motif Consensus sequence: GYGSTGGG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
12 |
| Motif name: |
Motif 12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0820312 |
Alignment:
MTGTGTGC
-GSTGTGG
| Original motif Consensus sequence: GCACACAY |
Reverse complement motif Consensus sequence: MTGTGTGC |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 41 |
Motif name: Motif 41 |
| Original motif Consensus sequence: CTCAGARA |
Reverse complement motif Consensus sequence: TKTCTGAG |
|
|
 |
 |
Best Matches for Motif ID 41 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0342598 |
Alignment:
CACASASA
CTCAGARA
| Original motif Consensus sequence: CACASASA |
Reverse complement motif Consensus sequence: TSTSTGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
117 |
| Motif name: |
yaCATAYACay |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0601983 |
Alignment:
DTGTGTATGBD
---TKTCTGAG
| Original motif Consensus sequence: HVCATACACAH |
Reverse complement motif Consensus sequence: DTGTGTATGBD |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
42 |
| Motif name: |
Motif 42 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0645699 |
Alignment:
GTTCTKGG
TKTCTGAG
| Original motif Consensus sequence: CCYAGAAC |
Reverse complement motif Consensus sequence: GTTCTKGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
127 |
| Motif name: |
rryCAGGGAyw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0655288 |
Alignment:
DDYCAGGGAHD
-CTCAGARA--
| Original motif Consensus sequence: DDYCAGGGAHD |
Reverse complement motif Consensus sequence: DHTCCCTGMDD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
58 |
| Motif name: |
Motif 58 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.0667371 |
Alignment:
TGTGTGTGTGTGTG
------TKTCTGAG
| Original motif Consensus sequence: CACACACACACACA |
Reverse complement motif Consensus sequence: TGTGTGTGTGTGTG |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 42 |
Motif name: Motif 42 |
| Original motif Consensus sequence: CCYAGAAC |
Reverse complement motif Consensus sequence: GTTCTKGG |
|
|
 |
 |
Best Matches for Motif ID 42 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0610815 |
Alignment:
CCTGGARC
CCYAGAAC
| Original motif Consensus sequence: CCTGGARC |
Reverse complement motif Consensus sequence: GKTCCAGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
115 |
| Motif name: |
ysCmAGCACwy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0690213 |
Alignment:
HVCMAGCACHH
-CCYAGAAC--
| Original motif Consensus sequence: HVCMAGCACHH |
Reverse complement motif Consensus sequence: DHGTGCTRGVH |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0735374 |
Alignment:
GYGSTGGG
GTTCTKGG
| Original motif Consensus sequence: CCCASCMC |
Reverse complement motif Consensus sequence: GYGSTGGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
109 |
| Motif name: |
aaCAAAAACaa |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0769138 |
Alignment:
HHGTTTTTGHH
--GTTCTKGG-
| Original motif Consensus sequence: HHCAAAAACHH |
Reverse complement motif Consensus sequence: HHGTTTTTGHH |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
41 |
| Motif name: |
Motif 41 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0911976 |
Alignment:
CTCAGARA
CCYAGAAC
| Original motif Consensus sequence: CTCAGARA |
Reverse complement motif Consensus sequence: TKTCTGAG |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 43 |
Motif name: Motif 43 |
| Original motif Consensus sequence: CTCTGCCY |
Reverse complement motif Consensus sequence: KGGCAGAG |
|
|
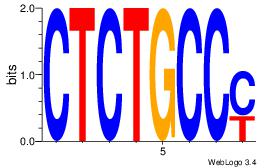 |
 |
Best Matches for Motif ID 43 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
84 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.0750223 |
Alignment:
GTTGACCTTTGACCTTT
------CTCTGCCY---
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
59 |
| Motif name: |
Motif 59 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0776916 |
Alignment:
CTBCCTCHSYCYCC
----CTCTGCCY--
| Original motif Consensus sequence: CTBCCTCHSYCYCC |
Reverse complement motif Consensus sequence: GGMGKSHGAGGBAG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
116 |
| Motif name: |
dyCTGCyTCyc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0799777 |
Alignment:
DHCTGCYTCHB
CTCTGCCY---
| Original motif Consensus sequence: DHCTGCYTCHB |
Reverse complement motif Consensus sequence: BHGAKGCAGHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
88 |
| Motif name: |
ygCTyTTCsg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0904749 |
Alignment:
HDCTYTTCGB
--CTCTGCCY
| Original motif Consensus sequence: HDCTYTTCGB |
Reverse complement motif Consensus sequence: BCGAAMAGHH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
83 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0905992 |
Alignment:
TGAMCTTTGMMCYT
----CTCTGCCY--
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 44 |
Motif name: Motif 44 |
| Original motif Consensus sequence: AGAAATG |
Reverse complement motif Consensus sequence: CATTTCT |
|
|
 |
 |
Best Matches for Motif ID 44 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
14 |
| Motif name: |
Motif 14 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0447866 |
Alignment:
MATTTTY
CATTTCT
| Original motif Consensus sequence: KAAAATR |
Reverse complement motif Consensus sequence: MATTTTY |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
108 |
| Motif name: |
wwATkTTTAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.0513825 |
Alignment:
DHATKTTTAHH
-CATTTCT---
| Original motif Consensus sequence: DHATKTTTAHH |
Reverse complement motif Consensus sequence: HHTAAARATHD |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0527225 |
Alignment:
GGAAARG
AGAAATG
| Original motif Consensus sequence: CMTTTCC |
Reverse complement motif Consensus sequence: GGAAARG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
61 |
| Motif name: |
MEF2A |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0603872 |
Alignment:
CTATTTWTAG
-CATTTCT--
| Original motif Consensus sequence: CTATTTWTAG |
Reverse complement motif Consensus sequence: CTAWAAATAG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
71 |
| Motif name: |
Evi1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0740469 |
Alignment:
TBTTATCTTMTCTT
------CATTTCT-
| Original motif Consensus sequence: AAGAYAAGATAABA |
Reverse complement motif Consensus sequence: TBTTATCTTMTCTT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 45 |
Motif name: Motif 45 |
| Original motif Consensus sequence: ATTATTAW |
Reverse complement motif Consensus sequence: WTAATAAT |
|
|
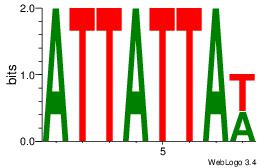 |
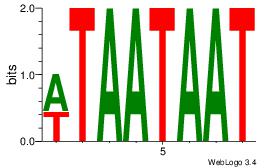 |
Best Matches for Motif ID 45 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
100 |
| Motif name: |
wwTAATAAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0312128 |
Alignment:
DDTTATTAHD
-ATTATTAW-
| Original motif Consensus sequence: DHTAATAADD |
Reverse complement motif Consensus sequence: DDTTATTAHD |
|
|
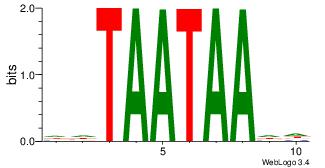 |
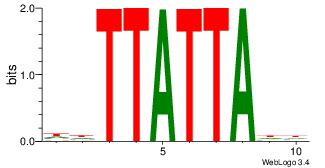 |
| Dataset #: |
4 |
| Motif ID: |
108 |
| Motif name: |
wwATkTTTAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0503721 |
Alignment:
HHTAAARATHD
-WTAATAAT--
| Original motif Consensus sequence: DHATKTTTAHH |
Reverse complement motif Consensus sequence: HHTAAARATHD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
68 |
| Motif name: |
HNF1A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0585066 |
Alignment:
GYTAATDATTAACC
----ATTATTAW--
| Original motif Consensus sequence: GGTTAATDATTAMC |
Reverse complement motif Consensus sequence: GYTAATDATTAACC |
|
|
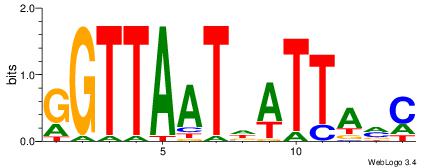 |
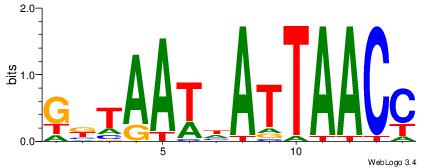 |
| Dataset #: |
3 |
| Motif ID: |
82 |
| Motif name: |
HNF1B |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0743796 |
Alignment:
TTAATRWTTAAC
WTAATAAT----
| Original motif Consensus sequence: TTAATRWTTAAC |
Reverse complement motif Consensus sequence: GTTAAWKATTAA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
Lhx3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0744439 |
Alignment:
WAATTAATTAWYB
WTAATAAT-----
| Original motif Consensus sequence: WAATTAATTAWYB |
Reverse complement motif Consensus sequence: BMWTAATTAATTW |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 46 |
Motif name: Motif 46 |
| Original motif Consensus sequence: GGAAASA |
Reverse complement motif Consensus sequence: TSTTTCC |
|
|
 |
 |
Best Matches for Motif ID 46 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
13 |
| Motif name: |
Motif 13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0428804 |
Alignment:
GGAAGSAR
GGAAASA-
| Original motif Consensus sequence: GGAAGSAR |
Reverse complement motif Consensus sequence: MTSCTTCC |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0542683 |
Alignment:
GGAAARG
GGAAASA
| Original motif Consensus sequence: CMTTTCC |
Reverse complement motif Consensus sequence: GGAAARG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
75 |
| Motif name: |
FOXF2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
7 |
| Similarity score: |
0.0636714 |
Alignment:
BHADSGTAAACAAD
-----GGAAASA--
| Original motif Consensus sequence: BHADSGTAAACAAD |
Reverse complement motif Consensus sequence: DTTGTTTACSBTBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
86 |
| Motif name: |
FOXO3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0652994 |
Alignment:
KGTAAACA
-GGAAASA
| Original motif Consensus sequence: KGTAAACA |
Reverse complement motif Consensus sequence: TGTTTACR |
|
|
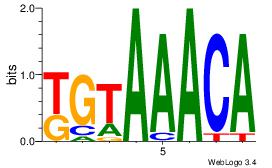 |
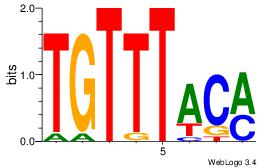 |
| Dataset #: |
4 |
| Motif ID: |
113 |
| Motif name: |
htCTGyKTCbt |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.0693025 |
Alignment:
HBCTGYTTCBH
---TSTTTCC-
| Original motif Consensus sequence: HBCTGYTTCBH |
Reverse complement motif Consensus sequence: HVGAAMCAGVH |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 47 |
Motif name: Motif 47 |
| Original motif Consensus sequence: KGCTGCTG |
Reverse complement motif Consensus sequence: CAGCAGCR |
|
|
 |
 |
Best Matches for Motif ID 47 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
105 |
| Motif name: |
ccGCAGCCss |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0572744 |
Alignment:
BBGGCTGCGV
--KGCTGCTG
| Original motif Consensus sequence: VCGCAGCCBB |
Reverse complement motif Consensus sequence: BBGGCTGCGV |
|
|
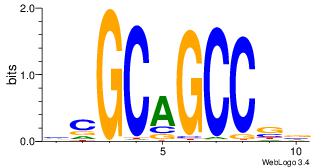 |
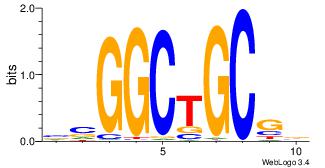 |
| Dataset #: |
4 |
| Motif ID: |
115 |
| Motif name: |
ysCmAGCACwy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0621107 |
Alignment:
DHGTGCTRGVH
KGCTGCTG---
| Original motif Consensus sequence: HVCMAGCACHH |
Reverse complement motif Consensus sequence: DHGTGCTRGVH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
93 |
| Motif name: |
cmCARCACTwr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0664426 |
Alignment:
VHAGTGCTGHB
-KGCTGCTG--
| Original motif Consensus sequence: BHCAGCACTHV |
Reverse complement motif Consensus sequence: VHAGTGCTGHB |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
30 |
| Motif name: |
Motif 30 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0671183 |
Alignment:
YCMTCCTG
KGCTGCTG
| Original motif Consensus sequence: CAGGARGM |
Reverse complement motif Consensus sequence: YCMTCCTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
116 |
| Motif name: |
dyCTGCyTCyc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0696714 |
Alignment:
DHCTGCYTCHB
KGCTGCTG---
| Original motif Consensus sequence: DHCTGCYTCHB |
Reverse complement motif Consensus sequence: BHGAKGCAGHH |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 48 |
Motif name: Motif 48 |
| Original motif Consensus sequence: ACACWG |
Reverse complement motif Consensus sequence: CWGTGT |
|
|
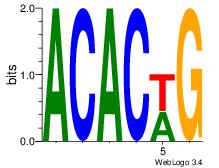 |
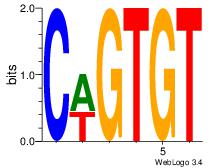 |
Best Matches for Motif ID 48 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
12 |
| Motif name: |
Motif 12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0450356 |
Alignment:
MTGTGTGC
CWGTGT--
| Original motif Consensus sequence: GCACACAY |
Reverse complement motif Consensus sequence: MTGTGTGC |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0534035 |
Alignment:
CTGDGTTC
--CWGTGT
| Original motif Consensus sequence: CTGDGTTC |
Reverse complement motif Consensus sequence: GAACDCAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
23 |
| Motif name: |
Motif 23 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0615752 |
Alignment:
MTSTGTA
-CWGTGT
| Original motif Consensus sequence: MTSTGTA |
Reverse complement motif Consensus sequence: TACASAR |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.065723 |
Alignment:
CACASASA
-ACACWG-
| Original motif Consensus sequence: CACASASA |
Reverse complement motif Consensus sequence: TSTSTGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
117 |
| Motif name: |
yaCATAYACay |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
6 |
| Similarity score: |
0.0663715 |
Alignment:
DTGTGTATGBD
CWGTGT-----
| Original motif Consensus sequence: HVCATACACAH |
Reverse complement motif Consensus sequence: DTGTGTATGBD |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 49 |
Motif name: Motif 49 |
| Original motif Consensus sequence: AGATAAAR |
Reverse complement motif Consensus sequence: KTTTATCT |
|
|
 |
 |
Best Matches for Motif ID 49 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
71 |
| Motif name: |
Evi1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0168651 |
Alignment:
TBTTATCTTMTCTT
KTTTATCT------
| Original motif Consensus sequence: AAGAYAAGATAABA |
Reverse complement motif Consensus sequence: TBTTATCTTMTCTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
67 |
| Motif name: |
FOXI1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0451069 |
Alignment:
KBDTRTTTRTTT
----KTTTATCT
| Original motif Consensus sequence: KBDTRTTTRTTT |
Reverse complement motif Consensus sequence: AAAMAAAMADBY |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
60 |
| Motif name: |
Motif 60 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0469577 |
Alignment:
YTTTTTRTTTTDTT
--KTTTATCT----
| Original motif Consensus sequence: AADAAAAMAAAAAM |
Reverse complement motif Consensus sequence: YTTTTTRTTTTDTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
21 |
| Motif name: |
Motif 21 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0471231 |
Alignment:
GTTTGTTT
KTTTATCT
| Original motif Consensus sequence: AAACAAAC |
Reverse complement motif Consensus sequence: GTTTGTTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
84 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
10 |
| Number of overlap: |
8 |
| Similarity score: |
0.0483731 |
Alignment:
GTTGACCTTTGACCTTT
KTTTATCT---------
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 50 |
Motif name: Motif 50 |
| Original motif Consensus sequence: GAAGAGCA |
Reverse complement motif Consensus sequence: TGCTCTTC |
|
|
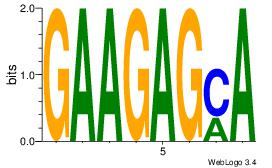 |
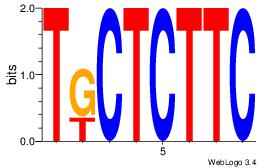 |
Best Matches for Motif ID 50 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
88 |
| Motif name: |
ygCTyTTCsg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0206791 |
Alignment:
HDCTYTTCGB
TGCTCTTC--
| Original motif Consensus sequence: HDCTYTTCGB |
Reverse complement motif Consensus sequence: BCGAAMAGHH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
71 |
| Motif name: |
Evi1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0485428 |
Alignment:
TBTTATCTTMTCTT
--TGCTCTTC----
| Original motif Consensus sequence: AAGAYAAGATAABA |
Reverse complement motif Consensus sequence: TBTTATCTTMTCTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
83 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0680446 |
Alignment:
AKGYYCAAAGRTCA
------GAAGAGCA
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0707433 |
Alignment:
AAASAAAA
GAAGAGCA
| Original motif Consensus sequence: AAASAAAA |
Reverse complement motif Consensus sequence: TTTTSTTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
114 |
| Motif name: |
awAACAcAAwa |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0709876 |
Alignment:
DDAACACAADV
-GAAGAGCA--
| Original motif Consensus sequence: DDAACACAADV |
Reverse complement motif Consensus sequence: BDTTGTGTTDD |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 51 |
Motif name: Motif 51 |
| Original motif Consensus sequence: AGAGCWGG |
Reverse complement motif Consensus sequence: CCWGCTCT |
|
|
 |
 |
Best Matches for Motif ID 51 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
119 |
| Motif name: |
cbCCAGCTCmyk |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0568763 |
Alignment:
BBCCAGCTCMBD
--CCWGCTCT--
| Original motif Consensus sequence: BBCCAGCTCMBD |
Reverse complement motif Consensus sequence: HBYGAGCTGGBB |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
38 |
| Motif name: |
Motif 38 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0627656 |
Alignment:
CCTYCTCC
CCWGCTCT
| Original motif Consensus sequence: CCTYCTCC |
Reverse complement motif Consensus sequence: GGAGMAGG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0634208 |
Alignment:
AGCCCWGG
AGAGCWGG
| Original motif Consensus sequence: AGCCCWGG |
Reverse complement motif Consensus sequence: CCWGGGCT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
103 |
| Motif name: |
csCCaGCTCCCgs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0769435 |
Alignment:
BVGGGAGCTGGBB
---AGAGCWGG--
| Original motif Consensus sequence: BBCCAGCTCCCVB |
Reverse complement motif Consensus sequence: BVGGGAGCTGGBB |
|
|
 |
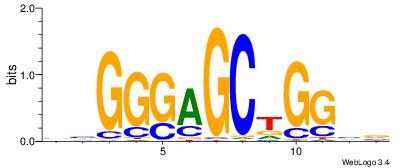 |
| Dataset #: |
4 |
| Motif ID: |
93 |
| Motif name: |
cmCARCACTwr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0785511 |
Alignment:
BHCAGCACTHV
-CCWGCTCT--
| Original motif Consensus sequence: BHCAGCACTHV |
Reverse complement motif Consensus sequence: VHAGTGCTGHB |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 52 |
Motif name: Motif 52 |
| Original motif Consensus sequence: AGVCAGG |
Reverse complement motif Consensus sequence: CCTGVCT |
|
|
 |
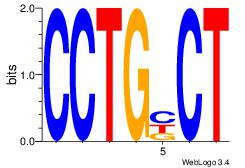 |
Best Matches for Motif ID 52 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0426638 |
Alignment:
GAGRMAGR
-AGVCAGG
| Original motif Consensus sequence: GAGRMAGR |
Reverse complement motif Consensus sequence: KCTRMCTC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
116 |
| Motif name: |
dyCTGCyTCyc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.0621766 |
Alignment:
DHCTGCYTCHB
-CCTGVCT---
| Original motif Consensus sequence: DHCTGCYTCHB |
Reverse complement motif Consensus sequence: BHGAKGCAGHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
127 |
| Motif name: |
rryCAGGGAyw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
7 |
| Similarity score: |
0.0705096 |
Alignment:
DHTCCCTGMDD
----CCTGVCT
| Original motif Consensus sequence: DDYCAGGGAHD |
Reverse complement motif Consensus sequence: DHTCCCTGMDD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
113 |
| Motif name: |
htCTGyKTCbt |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.0753705 |
Alignment:
HVGAAMCAGVH
---AGVCAGG-
| Original motif Consensus sequence: HBCTGYTTCBH |
Reverse complement motif Consensus sequence: HVGAAMCAGVH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
126 |
| Motif name: |
yvTGCAGsCAcg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.077286 |
Alignment:
BVTGCAGSCABV
-----AGVCAGG
| Original motif Consensus sequence: BVTGCAGSCABV |
Reverse complement motif Consensus sequence: VBTGSCTGCAVB |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 53 |
Motif name: Motif 53 |
| Original motif Consensus sequence: TSTGTR |
Reverse complement motif Consensus sequence: MACASA |
|
|
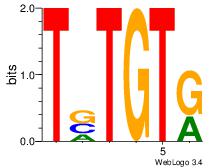 |
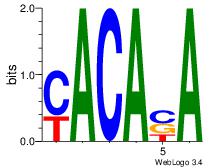 |
Best Matches for Motif ID 53 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.00228262 |
Alignment:
TSTSTGTG
TSTGTR--
| Original motif Consensus sequence: CACASASA |
Reverse complement motif Consensus sequence: TSTSTGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
114 |
| Motif name: |
awAACAcAAwa |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0122629 |
Alignment:
DDAACACAADV
--MACASA---
| Original motif Consensus sequence: DDAACACAADV |
Reverse complement motif Consensus sequence: BDTTGTGTTDD |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
23 |
| Motif name: |
Motif 23 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0137023 |
Alignment:
MTSTGTA
-TSTGTR
| Original motif Consensus sequence: MTSTGTA |
Reverse complement motif Consensus sequence: TACASAR |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0252334 |
Alignment:
GTGTGTGT
-TSTGTR-
| Original motif Consensus sequence: ACACACAC |
Reverse complement motif Consensus sequence: GTGTGTGT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
58 |
| Motif name: |
Motif 58 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.025412 |
Alignment:
CACACACACACACA
MACASA--------
| Original motif Consensus sequence: CACACACACACACA |
Reverse complement motif Consensus sequence: TGTGTGTGTGTGTG |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 54 |
Motif name: Motif 54 |
| Original motif Consensus sequence: RGAAA |
Reverse complement motif Consensus sequence: TTTCK |
|
|
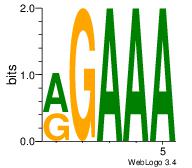 |
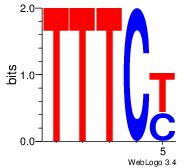 |
Best Matches for Motif ID 54 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
26 |
| Motif name: |
Motif 26 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
5 |
| Similarity score: |
0 |
Alignment:
TTTCKTCK
TTTCK---
| Original motif Consensus sequence: RGARGAAA |
Reverse complement motif Consensus sequence: TTTCKTCK |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
28 |
| Motif name: |
Motif 28 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
5 |
| Similarity score: |
0.0199405 |
Alignment:
AVAGAAA
--RGAAA
| Original motif Consensus sequence: AVAGAAA |
Reverse complement motif Consensus sequence: TTTCTVT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
5 |
| Similarity score: |
0.0284722 |
Alignment:
CMTTTCC
TTTCK--
| Original motif Consensus sequence: CMTTTCC |
Reverse complement motif Consensus sequence: GGAAARG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
46 |
| Motif name: |
Motif 46 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
5 |
| Similarity score: |
0.0284722 |
Alignment:
GGAAASA
RGAAA--
| Original motif Consensus sequence: GGAAASA |
Reverse complement motif Consensus sequence: TSTTTCC |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
44 |
| Motif name: |
Motif 44 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
5 |
| Similarity score: |
0.0339244 |
Alignment:
CATTTCT
--TTTCK
| Original motif Consensus sequence: AGAAATG |
Reverse complement motif Consensus sequence: CATTTCT |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 55 |
Motif name: Motif 55 |
| Original motif Consensus sequence: TTTAAAA |
Reverse complement motif Consensus sequence: TTTTAAA |
|
|
 |
 |
Best Matches for Motif ID 55 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
120 |
| Motif name: |
wtTTTwAAAaw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0.00286145 |
Alignment:
HHTTTWAAADD
--TTTAAAA--
| Original motif Consensus sequence: HHTTTWAAADD |
Reverse complement motif Consensus sequence: DDTTTWAAAHH |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0142562 |
Alignment:
TTTWWAAA
-TTTAAAA
| Original motif Consensus sequence: TTTWWAAA |
Reverse complement motif Consensus sequence: TTTWWAAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
91 |
| Motif name: |
wtATTTTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.036821 |
Alignment:
HWTAAAATHD
TTTAAAA---
| Original motif Consensus sequence: DHATTTTAWH |
Reverse complement motif Consensus sequence: HWTAAAATHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
108 |
| Motif name: |
wwATkTTTAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0428268 |
Alignment:
DHATKTTTAHH
----TTTTAAA
| Original motif Consensus sequence: DHATKTTTAHH |
Reverse complement motif Consensus sequence: HHTAAARATHD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
62 |
| Motif name: |
TBP |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
7 |
| Similarity score: |
0.058454 |
Alignment:
VBVVVVMWTTTATAS
-------TTTTAAA-
| Original motif Consensus sequence: STATAAAWRVVVVBV |
Reverse complement motif Consensus sequence: VBVVVVMWTTTATAS |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 56 |
Motif name: Motif 56 |
| Original motif Consensus sequence: CACGGA |
Reverse complement motif Consensus sequence: TCCGTG |
|
|
 |
 |
Best Matches for Motif ID 56 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
127 |
| Motif name: |
rryCAGGGAyw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0151346 |
Alignment:
DDYCAGGGAHD
---CACGGA--
| Original motif Consensus sequence: DDYCAGGGAHD |
Reverse complement motif Consensus sequence: DHTCCCTGMDD |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0281094 |
Alignment:
CACASASA
CACGGA--
| Original motif Consensus sequence: CACASASA |
Reverse complement motif Consensus sequence: TSTSTGTG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
41 |
| Motif name: |
Motif 41 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.051968 |
Alignment:
TKTCTGAG
--TCCGTG
| Original motif Consensus sequence: CTCAGARA |
Reverse complement motif Consensus sequence: TKTCTGAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.051968 |
Alignment:
CCTGGARC
CACGGA--
| Original motif Consensus sequence: CCTGGARC |
Reverse complement motif Consensus sequence: GKTCCAGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
53 |
| Motif name: |
Motif 53 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0528198 |
Alignment:
TSTGTR
TCCGTG
| Original motif Consensus sequence: TSTGTR |
Reverse complement motif Consensus sequence: MACASA |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 57 |
Motif name: Motif 57 |
| Original motif Consensus sequence: SAGGAA |
Reverse complement motif Consensus sequence: TTCCTS |
|
|
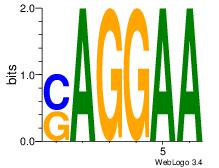 |
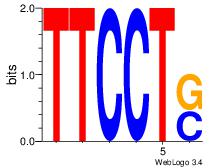 |
Best Matches for Motif ID 57 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
30 |
| Motif name: |
Motif 30 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0409967 |
Alignment:
CAGGARGM
SAGGAA--
| Original motif Consensus sequence: CAGGARGM |
Reverse complement motif Consensus sequence: YCMTCCTG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
26 |
| Motif name: |
Motif 26 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0466104 |
Alignment:
RGARGAAA
-SAGGAA-
| Original motif Consensus sequence: RGARGAAA |
Reverse complement motif Consensus sequence: TTTCKTCK |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
28 |
| Motif name: |
Motif 28 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0548389 |
Alignment:
TTTCTVT
TTCCTS-
| Original motif Consensus sequence: AVAGAAA |
Reverse complement motif Consensus sequence: TTTCTVT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
27 |
| Motif name: |
Motif 27 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0627861 |
Alignment:
KRAGGCAG
-SAGGAA-
| Original motif Consensus sequence: CTGCCTKY |
Reverse complement motif Consensus sequence: KRAGGCAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0659124 |
Alignment:
GAGRMAGR
SAGGAA--
| Original motif Consensus sequence: GAGRMAGR |
Reverse complement motif Consensus sequence: KCTRMCTC |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 58 |
Motif name: Motif 58 |
| Original motif Consensus sequence: CACACACACACACA |
Reverse complement motif Consensus sequence: TGTGTGTGTGTGTG |
|
|
 |
 |
Best Matches for Motif ID 58 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
118 |
| Motif name: |
yrCACATGCATGTGyr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
14 |
| Similarity score: |
0.0942589 |
Alignment:
HVCACATGCATGTGBD
--CACACACACACACA
| Original motif Consensus sequence: HVCACATGCATGTGBD |
Reverse complement motif Consensus sequence: DVCACATGCATGTGBH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
59 |
| Motif name: |
Motif 59 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0950431 |
Alignment:
GGMGKSHGAGGBAG
TGTGTGTGTGTGTG
| Original motif Consensus sequence: CTBCCTCHSYCYCC |
Reverse complement motif Consensus sequence: GGMGKSHGAGGBAG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
98 |
| Motif name: |
myyCCCACmTGCmyr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0967648 |
Alignment:
HBCCCCACMTGCMHV
-CACACACACACACA
| Original motif Consensus sequence: HBCCCCACMTGCMHV |
Reverse complement motif Consensus sequence: VDRGCAYGTGGGGVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
111 |
| Motif name: |
ccCCmCaCCCCcc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.595051 |
Alignment:
-HBCCCCACCCCBH
CACACACACACACA
| Original motif Consensus sequence: HBCCCCACCCCBH |
Reverse complement motif Consensus sequence: DBGGGGTGGGGBD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
60 |
| Motif name: |
Motif 60 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.597407 |
Alignment:
-AADAAAAMAAAAAM
CACACACACACACA-
| Original motif Consensus sequence: AADAAAAMAAAAAM |
Reverse complement motif Consensus sequence: YTTTTTRTTTTDTT |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 59 |
Motif name: Motif 59 |
| Original motif Consensus sequence: CTBCCTCHSYCYCC |
Reverse complement motif Consensus sequence: GGMGKSHGAGGBAG |
|
|
 |
 |
Best Matches for Motif ID 59 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
Pax4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
13 |
| Number of overlap: |
14 |
| Similarity score: |
0.0440476 |
Alignment:
RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC
------------CTBCCTCHSYCYCC----
| Original motif Consensus sequence: RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC |
Reverse complement motif Consensus sequence: GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
98 |
| Motif name: |
myyCCCACmTGCmyr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.064469 |
Alignment:
HBCCCCACMTGCMHV
-CTBCCTCHSYCYCC
| Original motif Consensus sequence: HBCCCCACMTGCMHV |
Reverse complement motif Consensus sequence: VDRGCAYGTGGGGVD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
58 |
| Motif name: |
Motif 58 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0727936 |
Alignment:
TGTGTGTGTGTGTG
GGMGKSHGAGGBAG
| Original motif Consensus sequence: CACACACACACACA |
Reverse complement motif Consensus sequence: TGTGTGTGTGTGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
111 |
| Motif name: |
ccCCmCaCCCCcc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.53768 |
Alignment:
-HBCCCCACCCCBH
CTBCCTCHSYCYCC
| Original motif Consensus sequence: HBCCCCACCCCBH |
Reverse complement motif Consensus sequence: DBGGGGTGGGGBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
103 |
| Motif name: |
csCCaGCTCCCgs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.558794 |
Alignment:
-BVGGGAGCTGGBB
GGMGKSHGAGGBAG
| Original motif Consensus sequence: BBCCAGCTCCCVB |
Reverse complement motif Consensus sequence: BVGGGAGCTGGBB |
|
|
 |
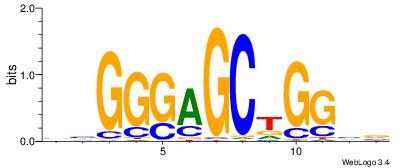 |
| Dataset #: 2 |
Motif ID: 60 |
Motif name: Motif 60 |
| Original motif Consensus sequence: AADAAAAMAAAAAM |
Reverse complement motif Consensus sequence: YTTTTTRTTTTDTT |
|
|
 |
 |
Best Matches for Motif ID 60 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
71 |
| Motif name: |
Evi1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0695286 |
Alignment:
AAGAYAAGATAABA
AADAAAAMAAAAAM
| Original motif Consensus sequence: AAGAYAAGATAABA |
Reverse complement motif Consensus sequence: TBTTATCTTMTCTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
Pax4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
15 |
| Number of overlap: |
14 |
| Similarity score: |
0.0795461 |
Alignment:
RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC
--AADAAAAMAAAAAM--------------
| Original motif Consensus sequence: RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC |
Reverse complement motif Consensus sequence: GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
62 |
| Motif name: |
TBP |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0901534 |
Alignment:
STATAAAWRVVVVBV
-AADAAAAMAAAAAM
| Original motif Consensus sequence: STATAAAWRVVVVBV |
Reverse complement motif Consensus sequence: VBVVVVMWTTTATAS |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
125 |
| Motif name: |
wwTAATwwATTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0954049 |
Alignment:
DDTAATWWATTAHH
AADAAAAMAAAAAM
| Original motif Consensus sequence: DDTAATWWATTAHH |
Reverse complement motif Consensus sequence: HHTAATWWATTADD |
|
|
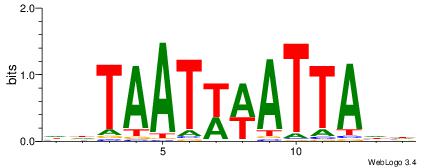 |
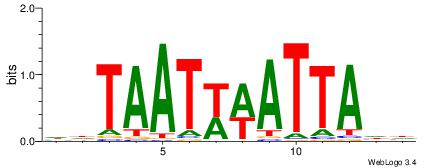 |
| Dataset #: |
3 |
| Motif ID: |
75 |
| Motif name: |
FOXF2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0959909 |
Alignment:
BHADSGTAAACAAD
AADAAAAMAAAAAM
| Original motif Consensus sequence: BHADSGTAAACAAD |
Reverse complement motif Consensus sequence: DTTGTTTACSBTBB |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 61 |
Motif name: MEF2A |
| Original motif Consensus sequence: CTATTTWTAG |
Reverse complement motif Consensus sequence: CTAWAAATAG |
|
|
 |
 |
Best Matches for Motif ID 61 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
62 |
| Motif name: |
TBP |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
10 |
| Similarity score: |
0.0368263 |
Alignment:
STATAAAWRVVVVBV
CTAWAAATAG-----
| Original motif Consensus sequence: STATAAAWRVVVVBV |
Reverse complement motif Consensus sequence: VBVVVVMWTTTATAS |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
108 |
| Motif name: |
wwATkTTTAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0401238 |
Alignment:
HHTAAARATHD
-CTAWAAATAG
| Original motif Consensus sequence: DHATKTTTAHH |
Reverse complement motif Consensus sequence: HHTAAARATHD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
67 |
| Motif name: |
FOXI1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.056188 |
Alignment:
KBDTRTTTRTTT
--CTATTTWTAG
| Original motif Consensus sequence: KBDTRTTTRTTT |
Reverse complement motif Consensus sequence: AAAMAAAMADBY |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
68 |
| Motif name: |
HNF1A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0684894 |
Alignment:
GGTTAATDATTAMC
--CTATTTWTAG--
| Original motif Consensus sequence: GGTTAATDATTAMC |
Reverse complement motif Consensus sequence: GYTAATDATTAACC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
91 |
| Motif name: |
wtATTTTAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0714984 |
Alignment:
HWTAAAATHD
CTAWAAATAG
| Original motif Consensus sequence: DHATTTTAWH |
Reverse complement motif Consensus sequence: HWTAAAATHD |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 62 |
Motif name: TBP |
| Original motif Consensus sequence: STATAAAWRVVVVBV |
Reverse complement motif Consensus sequence: VBVVVVMWTTTATAS |
|
|
 |
 |
Best Matches for Motif ID 62 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
Pax4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
0.050528 |
Alignment:
RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC
STATAAAWRVVVVBV---------------
| Original motif Consensus sequence: RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC |
Reverse complement motif Consensus sequence: GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
125 |
| Motif name: |
wwTAATwwATTAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.581919 |
Alignment:
DDTAATWWATTAHH-
VBVVVVMWTTTATAS
| Original motif Consensus sequence: DDTAATWWATTAHH |
Reverse complement motif Consensus sequence: HHTAATWWATTADD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
68 |
| Motif name: |
HNF1A |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.583461 |
Alignment:
-GYTAATDATTAACC
VBVVVVMWTTTATAS
| Original motif Consensus sequence: GGTTAATDATTAMC |
Reverse complement motif Consensus sequence: GYTAATDATTAACC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
60 |
| Motif name: |
Motif 60 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.58354 |
Alignment:
-AADAAAAMAAAAAM
STATAAAWRVVVVBV
| Original motif Consensus sequence: AADAAAAMAAAAAM |
Reverse complement motif Consensus sequence: YTTTTTRTTTTDTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
98 |
| Motif name: |
myyCCCACmTGCmyr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.587864 |
Alignment:
-HBCCCCACMTGCMHV
VBVVVVMWTTTATAS-
| Original motif Consensus sequence: HBCCCCACMTGCMHV |
Reverse complement motif Consensus sequence: VDRGCAYGTGGGGVD |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 63 |
Motif name: FOXL1 |
| Original motif Consensus sequence: WHDAHATA |
Reverse complement motif Consensus sequence: TATDTDHW |
|
|
 |
 |
Best Matches for Motif ID 63 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
75 |
| Motif name: |
FOXF2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.0287322 |
Alignment:
BHADSGTAAACAAD
WHDAHATA------
| Original motif Consensus sequence: BHADSGTAAACAAD |
Reverse complement motif Consensus sequence: DTTGTTTACSBTBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
90 |
| Motif name: |
ymTACATAyw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0294054 |
Alignment:
DBTATGTAHH
--TATDTDHW
| Original motif Consensus sequence: HHTACATABD |
Reverse complement motif Consensus sequence: DBTATGTAHH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
74 |
| Motif name: |
FOXC1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.036287 |
Alignment:
VBHVHGTA
WHDAHATA
| Original motif Consensus sequence: VBHVHGTA |
Reverse complement motif Consensus sequence: TACHBHBV |
|
|
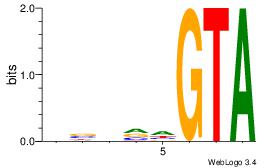 |
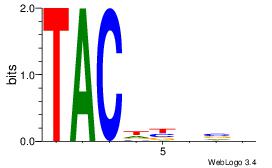 |
| Dataset #: |
3 |
| Motif ID: |
61 |
| Motif name: |
MEF2A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0377453 |
Alignment:
CTAWAAATAG
-WHDAHATA-
| Original motif Consensus sequence: CTATTTWTAG |
Reverse complement motif Consensus sequence: CTAWAAATAG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
76 |
| Motif name: |
Sox2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0399271 |
Alignment:
CCWTTGTYATGCAAA
-------TATDTDHW
| Original motif Consensus sequence: CCWTTGTYATGCAAA |
Reverse complement motif Consensus sequence: TTTGCATMACAAWGG |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 64 |
Motif name: ARID3A |
| Original motif Consensus sequence: ATYAAA |
Reverse complement motif Consensus sequence: TTTMAT |
|
|
 |
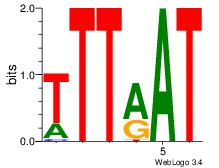 |
Best Matches for Motif ID 64 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
106 |
| Motif name: |
wtAATTAAtw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0114914 |
Alignment:
DTAATTAATH
---ATYAAA-
| Original motif Consensus sequence: DTAATTAATH |
Reverse complement motif Consensus sequence: HATTAATTAD |
|
|
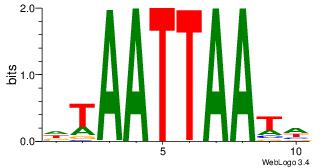 |
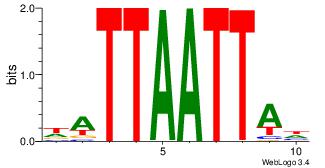 |
| Dataset #: |
1 |
| Motif ID: |
20 |
| Motif name: |
Motif 20 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.015396 |
Alignment:
ATTAMAGG
ATYAAA--
| Original motif Consensus sequence: ATTAMAGG |
Reverse complement motif Consensus sequence: CCTYTAAT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
Lhx3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
6 |
| Similarity score: |
0.0225727 |
Alignment:
BMWTAATTAATTW
-----TTTMAT--
| Original motif Consensus sequence: WAATTAATTAWYB |
Reverse complement motif Consensus sequence: BMWTAATTAATTW |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0340365 |
Alignment:
AGATAAAR
--ATYAAA
| Original motif Consensus sequence: AGATAAAR |
Reverse complement motif Consensus sequence: KTTTATCT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
25 |
| Motif name: |
Motif 25 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0368439 |
Alignment:
WWTAAA
ATYAAA
| Original motif Consensus sequence: TTTAWW |
Reverse complement motif Consensus sequence: WWTAAA |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 65 |
Motif name: Foxd3 |
| Original motif Consensus sequence: GAWTGTTTDTTT |
Reverse complement motif Consensus sequence: AAAHAAACAWTC |
|
|
 |
 |
Best Matches for Motif ID 65 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
67 |
| Motif name: |
FOXI1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.00335618 |
Alignment:
AAAMAAAMADBY
AAAHAAACAWTC
| Original motif Consensus sequence: KBDTRTTTRTTT |
Reverse complement motif Consensus sequence: AAAMAAAMADBY |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
60 |
| Motif name: |
Motif 60 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0177558 |
Alignment:
YTTTTTRTTTTDTT
GAWTGTTTDTTT--
| Original motif Consensus sequence: AADAAAAMAAAAAM |
Reverse complement motif Consensus sequence: YTTTTTRTTTTDTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
Lhx3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0424677 |
Alignment:
BMWTAATTAATTW
GAWTGTTTDTTT-
| Original motif Consensus sequence: WAATTAATTAWYB |
Reverse complement motif Consensus sequence: BMWTAATTAATTW |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
Pax4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
16 |
| Number of overlap: |
12 |
| Similarity score: |
0.0493847 |
Alignment:
RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC
---AAAHAAACAWTC---------------
| Original motif Consensus sequence: RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC |
Reverse complement motif Consensus sequence: GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
68 |
| Motif name: |
HNF1A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0526352 |
Alignment:
GGTTAATDATTAMC
GAWTGTTTDTTT--
| Original motif Consensus sequence: GGTTAATDATTAMC |
Reverse complement motif Consensus sequence: GYTAATDATTAACC |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 66 |
Motif name: Lhx3 |
| Original motif Consensus sequence: WAATTAATTAWYB |
Reverse complement motif Consensus sequence: BMWTAATTAATTW |
|
|
 |
 |
Best Matches for Motif ID 66 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
68 |
| Motif name: |
HNF1A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.0814534 |
Alignment:
GYTAATDATTAACC
-WAATTAATTAWYB
| Original motif Consensus sequence: GGTTAATDATTAMC |
Reverse complement motif Consensus sequence: GYTAATDATTAACC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
62 |
| Motif name: |
TBP |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
13 |
| Similarity score: |
0.100645 |
Alignment:
VBVVVVMWTTTATAS
--BMWTAATTAATTW
| Original motif Consensus sequence: STATAAAWRVVVVBV |
Reverse complement motif Consensus sequence: VBVVVVMWTTTATAS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
71 |
| Motif name: |
Evi1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.102129 |
Alignment:
TBTTATCTTMTCTT
BMWTAATTAATTW-
| Original motif Consensus sequence: AAGAYAAGATAABA |
Reverse complement motif Consensus sequence: TBTTATCTTMTCTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
125 |
| Motif name: |
wwTAATwwATTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.102843 |
Alignment:
DDTAATWWATTAHH
-WAATTAATTAWYB
| Original motif Consensus sequence: DDTAATWWATTAHH |
Reverse complement motif Consensus sequence: HHTAATWWATTADD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
60 |
| Motif name: |
Motif 60 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.106545 |
Alignment:
AADAAAAMAAAAAM
-BMWTAATTAATTW
| Original motif Consensus sequence: AADAAAAMAAAAAM |
Reverse complement motif Consensus sequence: YTTTTTRTTTTDTT |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 67 |
Motif name: FOXI1 |
| Original motif Consensus sequence: KBDTRTTTRTTT |
Reverse complement motif Consensus sequence: AAAMAAAMADBY |
|
|
 |
 |
Best Matches for Motif ID 67 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
65 |
| Motif name: |
Foxd3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0238603 |
Alignment:
AAAHAAACAWTC
AAAMAAAMADBY
| Original motif Consensus sequence: GAWTGTTTDTTT |
Reverse complement motif Consensus sequence: AAAHAAACAWTC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
60 |
| Motif name: |
Motif 60 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.035724 |
Alignment:
YTTTTTRTTTTDTT
KBDTRTTTRTTT--
| Original motif Consensus sequence: AADAAAAMAAAAAM |
Reverse complement motif Consensus sequence: YTTTTTRTTTTDTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
Lhx3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0611272 |
Alignment:
WAATTAATTAWYB
AAAMAAAMADBY-
| Original motif Consensus sequence: WAATTAATTAWYB |
Reverse complement motif Consensus sequence: BMWTAATTAATTW |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
Pax4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0613545 |
Alignment:
RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC
--AAAMAAAMADBY----------------
| Original motif Consensus sequence: RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC |
Reverse complement motif Consensus sequence: GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
62 |
| Motif name: |
TBP |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0644285 |
Alignment:
VBVVVVMWTTTATAS
--KBDTRTTTRTTT-
| Original motif Consensus sequence: STATAAAWRVVVVBV |
Reverse complement motif Consensus sequence: VBVVVVMWTTTATAS |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 68 |
Motif name: HNF1A |
| Original motif Consensus sequence: GGTTAATDATTAMC |
Reverse complement motif Consensus sequence: GYTAATDATTAACC |
|
|
 |
 |
Best Matches for Motif ID 68 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
125 |
| Motif name: |
wwTAATwwATTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0430148 |
Alignment:
DDTAATWWATTAHH
GGTTAATDATTAMC
| Original motif Consensus sequence: DDTAATWWATTAHH |
Reverse complement motif Consensus sequence: HHTAATWWATTADD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
84 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0678001 |
Alignment:
AAAGGTCAAAGGTCAAC
---GGTTAATDATTAMC
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
Pax4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
17 |
| Number of overlap: |
14 |
| Similarity score: |
0.0691946 |
Alignment:
GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM
----------------GYTAATDATTAACC
| Original motif Consensus sequence: RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC |
Reverse complement motif Consensus sequence: GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
62 |
| Motif name: |
TBP |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0722834 |
Alignment:
VBVVVVMWTTTATAS
-GYTAATDATTAACC
| Original motif Consensus sequence: STATAAAWRVVVVBV |
Reverse complement motif Consensus sequence: VBVVVVMWTTTATAS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
76 |
| Motif name: |
Sox2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.077247 |
Alignment:
CCWTTGTYATGCAAA
GGTTAATDATTAMC-
| Original motif Consensus sequence: CCWTTGTYATGCAAA |
Reverse complement motif Consensus sequence: TTTGCATMACAAWGG |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 69 |
Motif name: NKX3-1 |
| Original motif Consensus sequence: ATACTTA |
Reverse complement motif Consensus sequence: TAAGTAT |
|
|
 |
 |
Best Matches for Motif ID 69 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
90 |
| Motif name: |
ymTACATAyw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0 |
Alignment:
HHTACATABD
-ATACTTA--
| Original motif Consensus sequence: HHTACATABD |
Reverse complement motif Consensus sequence: DBTATGTAHH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
Lhx3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
7 |
| Similarity score: |
0.000214177 |
Alignment:
WAATTAATTAWYB
--TAAGTAT----
| Original motif Consensus sequence: WAATTAATTAWYB |
Reverse complement motif Consensus sequence: BMWTAATTAATTW |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.00274846 |
Alignment:
TATGTAD
TAAGTAT
| Original motif Consensus sequence: HTACATA |
Reverse complement motif Consensus sequence: TATGTAD |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
34 |
| Motif name: |
Motif 34 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.00883486 |
Alignment:
ATATTTA
ATACTTA
| Original motif Consensus sequence: ATATTTA |
Reverse complement motif Consensus sequence: TAAATAT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
106 |
| Motif name: |
wtAATTAAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.0100679 |
Alignment:
HATTAATTAD
---TAAGTAT
| Original motif Consensus sequence: DTAATTAATH |
Reverse complement motif Consensus sequence: HATTAATTAD |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 70 |
Motif name: Foxq1 |
| Original motif Consensus sequence: HATTGTTTATW |
Reverse complement motif Consensus sequence: WATAAACAATH |
|
|
 |
 |
Best Matches for Motif ID 70 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
67 |
| Motif name: |
FOXI1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0247019 |
Alignment:
KBDTRTTTRTTT
HATTGTTTATW-
| Original motif Consensus sequence: KBDTRTTTRTTT |
Reverse complement motif Consensus sequence: AAAMAAAMADBY |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
108 |
| Motif name: |
wwATkTTTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0341803 |
Alignment:
DHATKTTTAHH
HATTGTTTATW
| Original motif Consensus sequence: DHATKTTTAHH |
Reverse complement motif Consensus sequence: HHTAAARATHD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
65 |
| Motif name: |
Foxd3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0354607 |
Alignment:
GAWTGTTTDTTT
HATTGTTTATW-
| Original motif Consensus sequence: GAWTGTTTDTTT |
Reverse complement motif Consensus sequence: AAAHAAACAWTC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
60 |
| Motif name: |
Motif 60 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0611511 |
Alignment:
YTTTTTRTTTTDTT
-HATTGTTTATW--
| Original motif Consensus sequence: AADAAAAMAAAAAM |
Reverse complement motif Consensus sequence: YTTTTTRTTTTDTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
114 |
| Motif name: |
awAACAcAAwa |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0621125 |
Alignment:
BDTTGTGTTDD
HATTGTTTATW
| Original motif Consensus sequence: DDAACACAADV |
Reverse complement motif Consensus sequence: BDTTGTGTTDD |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 71 |
Motif name: Evi1 |
| Original motif Consensus sequence: AAGAYAAGATAABA |
Reverse complement motif Consensus sequence: TBTTATCTTMTCTT |
|
|
 |
 |
Best Matches for Motif ID 71 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
60 |
| Motif name: |
Motif 60 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0493882 |
Alignment:
AADAAAAMAAAAAM
AAGAYAAGATAABA
| Original motif Consensus sequence: AADAAAAMAAAAAM |
Reverse complement motif Consensus sequence: YTTTTTRTTTTDTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
125 |
| Motif name: |
wwTAATwwATTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.072295 |
Alignment:
DDTAATWWATTAHH
AAGAYAAGATAABA
| Original motif Consensus sequence: DDTAATWWATTAHH |
Reverse complement motif Consensus sequence: HHTAATWWATTADD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
Pax4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
14 |
| Similarity score: |
0.0763889 |
Alignment:
RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC
----AAGAYAAGATAABA------------
| Original motif Consensus sequence: RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC |
Reverse complement motif Consensus sequence: GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
Lhx3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.564942 |
Alignment:
BMWTAATTAATTW-
TBTTATCTTMTCTT
| Original motif Consensus sequence: WAATTAATTAWYB |
Reverse complement motif Consensus sequence: BMWTAATTAATTW |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
75 |
| Motif name: |
FOXF2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.568274 |
Alignment:
-DTTGTTTACSBTBB
TBTTATCTTMTCTT-
| Original motif Consensus sequence: BHADSGTAAACAAD |
Reverse complement motif Consensus sequence: DTTGTTTACSBTBB |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 72 |
Motif name: Nkx2-5 |
| Original motif Consensus sequence: WTAAKTG |
Reverse complement motif Consensus sequence: CARTTAW |
|
|
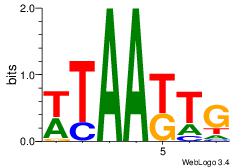 |
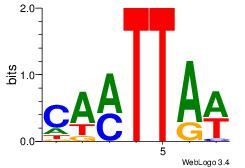 |
Best Matches for Motif ID 72 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
Lhx3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
7 |
| Similarity score: |
0.0204974 |
Alignment:
WAATTAATTAWYB
------CARTTAW
| Original motif Consensus sequence: WAATTAATTAWYB |
Reverse complement motif Consensus sequence: BMWTAATTAATTW |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
125 |
| Motif name: |
wwTAATwwATTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.021519 |
Alignment:
HHTAATWWATTADD
-WTAAKTG------
| Original motif Consensus sequence: DDTAATWWATTAHH |
Reverse complement motif Consensus sequence: HHTAATWWATTADD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
106 |
| Motif name: |
wtAATTAAtw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0.0245852 |
Alignment:
HATTAATTAD
--WTAAKTG-
| Original motif Consensus sequence: DTAATTAATH |
Reverse complement motif Consensus sequence: HATTAATTAD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
68 |
| Motif name: |
HNF1A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
7 |
| Similarity score: |
0.0356285 |
Alignment:
GGTTAATDATTAMC
--WTAAKTG-----
| Original motif Consensus sequence: GGTTAATDATTAMC |
Reverse complement motif Consensus sequence: GYTAATDATTAACC |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
45 |
| Motif name: |
Motif 45 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0367872 |
Alignment:
WTAATAAT
-WTAAKTG
| Original motif Consensus sequence: ATTATTAW |
Reverse complement motif Consensus sequence: WTAATAAT |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 73 |
Motif name: SRY |
| Original motif Consensus sequence: BDWAACAAT |
Reverse complement motif Consensus sequence: ATTGTTWDB |
|
|
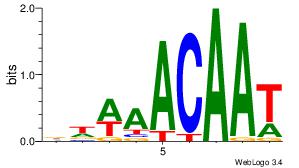 |
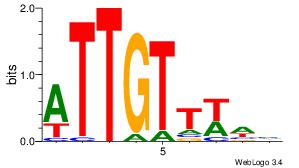 |
Best Matches for Motif ID 73 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
76 |
| Motif name: |
Sox2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
9 |
| Similarity score: |
0.0274533 |
Alignment:
TTTGCATMACAAWGG
----BDWAACAAT--
| Original motif Consensus sequence: CCWTTGTYATGCAAA |
Reverse complement motif Consensus sequence: TTTGCATMACAAWGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
65 |
| Motif name: |
Foxd3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.0332484 |
Alignment:
AAAHAAACAWTC
-BDWAACAAT--
| Original motif Consensus sequence: GAWTGTTTDTTT |
Reverse complement motif Consensus sequence: AAAHAAACAWTC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
75 |
| Motif name: |
FOXF2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0.0379812 |
Alignment:
DTTGTTTACSBTBB
ATTGTTWDB-----
| Original motif Consensus sequence: BHADSGTAAACAAD |
Reverse complement motif Consensus sequence: DTTGTTTACSBTBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
70 |
| Motif name: |
Foxq1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.0407369 |
Alignment:
WATAAACAATH
-BDWAACAAT-
| Original motif Consensus sequence: HATTGTTTATW |
Reverse complement motif Consensus sequence: WATAAACAATH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
96 |
| Motif name: |
mdAAcCAACG |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0.044154 |
Alignment:
CGTTGGTTDB
-ATTGTTWDB
| Original motif Consensus sequence: VDAACCAACG |
Reverse complement motif Consensus sequence: CGTTGGTTDB |
|
|
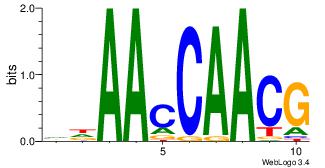 |
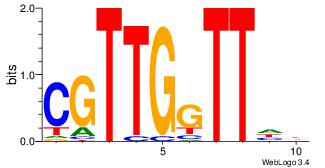 |
| Dataset #: 3 |
Motif ID: 74 |
Motif name: FOXC1 |
| Original motif Consensus sequence: VBHVHGTA |
Reverse complement motif Consensus sequence: TACHBHBV |
|
|
 |
 |
Best Matches for Motif ID 74 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
75 |
| Motif name: |
FOXF2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0250969 |
Alignment:
DTTGTTTACSBTBB
------TACHBHBV
| Original motif Consensus sequence: BHADSGTAAACAAD |
Reverse complement motif Consensus sequence: DTTGTTTACSBTBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
63 |
| Motif name: |
FOXL1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0421733 |
Alignment:
WHDAHATA
VBHVHGTA
| Original motif Consensus sequence: WHDAHATA |
Reverse complement motif Consensus sequence: TATDTDHW |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
90 |
| Motif name: |
ymTACATAyw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0422828 |
Alignment:
HHTACATABD
--TACHBHBV
| Original motif Consensus sequence: HHTACATABD |
Reverse complement motif Consensus sequence: DBTATGTAHH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
62 |
| Motif name: |
TBP |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0452131 |
Alignment:
STATAAAWRVVVVBV
--TACHBHBV-----
| Original motif Consensus sequence: STATAAAWRVVVVBV |
Reverse complement motif Consensus sequence: VBVVVVMWTTTATAS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
Pax4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
14 |
| Number of overlap: |
8 |
| Similarity score: |
0.046771 |
Alignment:
GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM
---------VBHVHGTA-------------
| Original motif Consensus sequence: RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC |
Reverse complement motif Consensus sequence: GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 75 |
Motif name: FOXF2 |
| Original motif Consensus sequence: BHADSGTAAACAAD |
Reverse complement motif Consensus sequence: DTTGTTTACSBTBB |
|
|
 |
 |
Best Matches for Motif ID 75 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
125 |
| Motif name: |
wwTAATwwATTAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0789966 |
Alignment:
DDTAATWWATTAHH
DTTGTTTACSBTBB
| Original motif Consensus sequence: DDTAATWWATTAHH |
Reverse complement motif Consensus sequence: HHTAATWWATTADD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
Pax4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
14 |
| Similarity score: |
0.0801361 |
Alignment:
GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM
--------------BHADSGTAAACAAD--
| Original motif Consensus sequence: RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC |
Reverse complement motif Consensus sequence: GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
76 |
| Motif name: |
Sox2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0829475 |
Alignment:
TTTGCATMACAAWGG
BHADSGTAAACAAD-
| Original motif Consensus sequence: CCWTTGTYATGCAAA |
Reverse complement motif Consensus sequence: TTTGCATMACAAWGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
60 |
| Motif name: |
Motif 60 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0886208 |
Alignment:
AADAAAAMAAAAAM
BHADSGTAAACAAD
| Original motif Consensus sequence: AADAAAAMAAAAAM |
Reverse complement motif Consensus sequence: YTTTTTRTTTTDTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
80 |
| Motif name: |
Pou5f1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0891042 |
Alignment:
ATTTGCATHACAAWG
-DTTGTTTACSBTBB
| Original motif Consensus sequence: CWTTGTHATGCAAAT |
Reverse complement motif Consensus sequence: ATTTGCATHACAAWG |
|
|
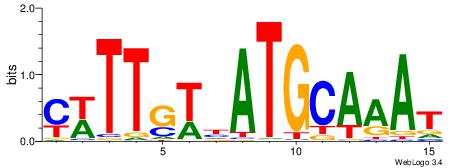 |
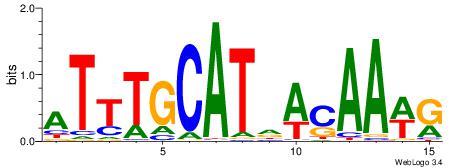 |
| Dataset #: 3 |
Motif ID: 76 |
Motif name: Sox2 |
| Original motif Consensus sequence: CCWTTGTYATGCAAA |
Reverse complement motif Consensus sequence: TTTGCATMACAAWGG |
|
|
 |
 |
Best Matches for Motif ID 76 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
Pax4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
15 |
| Similarity score: |
0.0816521 |
Alignment:
RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC
-------TTTGCATMACAAWGG--------
| Original motif Consensus sequence: RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC |
Reverse complement motif Consensus sequence: GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
80 |
| Motif name: |
Pou5f1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.5 |
Alignment:
ATTTGCATHACAAWG-
-TTTGCATMACAAWGG
| Original motif Consensus sequence: CWTTGTHATGCAAAT |
Reverse complement motif Consensus sequence: ATTTGCATHACAAWG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
125 |
| Motif name: |
wwTAATwwATTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.584157 |
Alignment:
-HHTAATWWATTADD
CCWTTGTYATGCAAA
| Original motif Consensus sequence: DDTAATWWATTAHH |
Reverse complement motif Consensus sequence: HHTAATWWATTADD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
75 |
| Motif name: |
FOXF2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.585609 |
Alignment:
DTTGTTTACSBTBB-
CCWTTGTYATGCAAA
| Original motif Consensus sequence: BHADSGTAAACAAD |
Reverse complement motif Consensus sequence: DTTGTTTACSBTBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
68 |
| Motif name: |
HNF1A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.59033 |
Alignment:
GGTTAATDATTAMC-
CCWTTGTYATGCAAA
| Original motif Consensus sequence: GGTTAATDATTAMC |
Reverse complement motif Consensus sequence: GYTAATDATTAACC |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 77 |
Motif name: Sox5 |
| Original motif Consensus sequence: HAACAAT |
Reverse complement motif Consensus sequence: ATTGTTH |
|
|
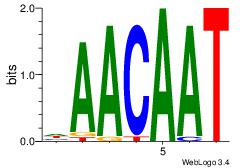 |
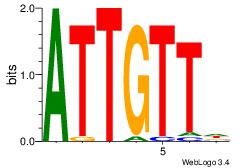 |
Best Matches for Motif ID 77 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
87 |
| Motif name: |
SOX9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0 |
Alignment:
CMATTGTTH
--ATTGTTH
| Original motif Consensus sequence: DAACAATRG |
Reverse complement motif Consensus sequence: CMATTGTTH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
73 |
| Motif name: |
SRY |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0011033 |
Alignment:
ATTGTTWDB
ATTGTTH--
| Original motif Consensus sequence: BDWAACAAT |
Reverse complement motif Consensus sequence: ATTGTTWDB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
76 |
| Motif name: |
Sox2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
7 |
| Similarity score: |
0.022869 |
Alignment:
CCWTTGTYATGCAAA
------ATTGTTH--
| Original motif Consensus sequence: CCWTTGTYATGCAAA |
Reverse complement motif Consensus sequence: TTTGCATMACAAWGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
70 |
| Motif name: |
Foxq1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.0242658 |
Alignment:
WATAAACAATH
-HAACAAT---
| Original motif Consensus sequence: HATTGTTTATW |
Reverse complement motif Consensus sequence: WATAAACAATH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
89 |
| Motif name: |
aaACAAAACaa |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0278776 |
Alignment:
AAACAAAACHV
HAACAAT----
| Original motif Consensus sequence: AAACAAAACHV |
Reverse complement motif Consensus sequence: BHGTTTTGTTT |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 78 |
Motif name: RORA_1 |
| Original motif Consensus sequence: AWVDAGGTCA |
Reverse complement motif Consensus sequence: TGACCTDVWT |
|
|
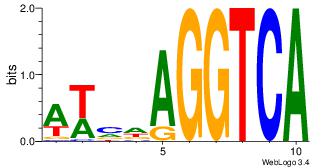 |
 |
Best Matches for Motif ID 78 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
84 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.05425 |
Alignment:
AAAGGTCAAAGGTCAAC
-----AWVDAGGTCA--
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
83 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0654808 |
Alignment:
AKGYYCAAAGRTCA
----AWVDAGGTCA
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
119 |
| Motif name: |
cbCCAGCTCmyk |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0688859 |
Alignment:
HBYGAGCTGGBB
--TGACCTDVWT
| Original motif Consensus sequence: BBCCAGCTCMBD |
Reverse complement motif Consensus sequence: HBYGAGCTGGBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
122 |
| Motif name: |
hsCCAGGCCTGGsr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0826087 |
Alignment:
HVCCAGGCCTGGBD
----TGACCTDVWT
| Original motif Consensus sequence: HVCCAGGCCTGGBD |
Reverse complement motif Consensus sequence: HBCCAGGCCTGGVD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
76 |
| Motif name: |
Sox2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0865719 |
Alignment:
TTTGCATMACAAWGG
----AWVDAGGTCA-
| Original motif Consensus sequence: CCWTTGTYATGCAAA |
Reverse complement motif Consensus sequence: TTTGCATMACAAWGG |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 79 |
Motif name: Prrx2 |
| Original motif Consensus sequence: AATTA |
Reverse complement motif Consensus sequence: TAATT |
|
|
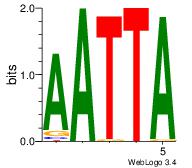 |
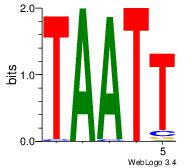 |
Best Matches for Motif ID 79 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
106 |
| Motif name: |
wtAATTAAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
5 |
| Similarity score: |
0.00550847 |
Alignment:
HATTAATTAD
---TAATT--
| Original motif Consensus sequence: DTAATTAATH |
Reverse complement motif Consensus sequence: HATTAATTAD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
Lhx3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
5 |
| Similarity score: |
0.00864407 |
Alignment:
BMWTAATTAATTW
-------TAATT-
| Original motif Consensus sequence: WAATTAATTAWYB |
Reverse complement motif Consensus sequence: BMWTAATTAATTW |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
85 |
| Motif name: |
Nobox |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
5 |
| Similarity score: |
0.0115968 |
Alignment:
TAATTRSY
TAATT---
| Original motif Consensus sequence: TAATTRSY |
Reverse complement motif Consensus sequence: MSMAATTA |
|
|
 |
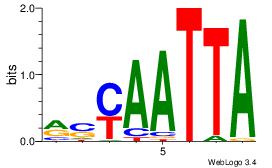 |
| Dataset #: |
4 |
| Motif ID: |
125 |
| Motif name: |
wwTAATwwATTAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
5 |
| Similarity score: |
0.0417489 |
Alignment:
HHTAATWWATTADD
--TAATT-------
| Original motif Consensus sequence: DDTAATWWATTAHH |
Reverse complement motif Consensus sequence: HHTAATWWATTADD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
68 |
| Motif name: |
HNF1A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
5 |
| Similarity score: |
0.0434826 |
Alignment:
GYTAATDATTAACC
---AATTA------
| Original motif Consensus sequence: GGTTAATDATTAMC |
Reverse complement motif Consensus sequence: GYTAATDATTAACC |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 80 |
Motif name: Pou5f1 |
| Original motif Consensus sequence: CWTTGTHATGCAAAT |
Reverse complement motif Consensus sequence: ATTTGCATHACAAWG |
|
|
 |
 |
Best Matches for Motif ID 80 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
Pax4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
15 |
| Similarity score: |
0.0828317 |
Alignment:
RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC
----ATTTGCATHACAAWG-----------
| Original motif Consensus sequence: RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC |
Reverse complement motif Consensus sequence: GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
76 |
| Motif name: |
Sox2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.5 |
Alignment:
CCWTTGTYATGCAAA-
-CWTTGTHATGCAAAT
| Original motif Consensus sequence: CCWTTGTYATGCAAA |
Reverse complement motif Consensus sequence: TTTGCATMACAAWGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
75 |
| Motif name: |
FOXF2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.591766 |
Alignment:
-DTTGTTTACSBTBB
ATTTGCATHACAAWG
| Original motif Consensus sequence: BHADSGTAAACAAD |
Reverse complement motif Consensus sequence: DTTGTTTACSBTBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
98 |
| Motif name: |
myyCCCACmTGCmyr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.592463 |
Alignment:
HBCCCCACMTGCMHV-
-CWTTGTHATGCAAAT
| Original motif Consensus sequence: HBCCCCACMTGCMHV |
Reverse complement motif Consensus sequence: VDRGCAYGTGGGGVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
118 |
| Motif name: |
yrCACATGCATGTGyr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
13 |
| Similarity score: |
1.07966 |
Alignment:
--DVCACATGCATGTGBH
CWTTGTHATGCAAAT---
| Original motif Consensus sequence: HVCACATGCATGTGBD |
Reverse complement motif Consensus sequence: DVCACATGCATGTGBH |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 81 |
Motif name: Pax4 |
| Original motif Consensus sequence: RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC |
Reverse complement motif Consensus sequence: GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM |
|
|
 |
 |
Best Matches for Motif ID 81 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
62 |
| Motif name: |
TBP |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
0.0350174 |
Alignment:
STATAAAWRVVVVBV---------------
RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC
| Original motif Consensus sequence: STATAAAWRVVVVBV |
Reverse complement motif Consensus sequence: VBVVVVMWTTTATAS |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
98 |
| Motif name: |
myyCCCACmTGCmyr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
0.0514106 |
Alignment:
---------------HBCCCCACMTGCMHV
RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC
| Original motif Consensus sequence: HBCCCCACMTGCMHV |
Reverse complement motif Consensus sequence: VDRGCAYGTGGGGVD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
59 |
| Motif name: |
Motif 59 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.542018 |
Alignment:
----------------CTBCCTCHSYCYCC
RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC
| Original motif Consensus sequence: CTBCCTCHSYCYCC |
Reverse complement motif Consensus sequence: GGMGKSHGAGGBAG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
125 |
| Motif name: |
wwTAATwwATTAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.557646 |
Alignment:
----------------HHTAATWWATTADD
GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM
| Original motif Consensus sequence: DDTAATWWATTAHH |
Reverse complement motif Consensus sequence: HHTAATWWATTADD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
68 |
| Motif name: |
HNF1A |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.564862 |
Alignment:
----------------GYTAATDATTAACC
GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM
| Original motif Consensus sequence: GGTTAATDATTAMC |
Reverse complement motif Consensus sequence: GYTAATDATTAACC |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 82 |
Motif name: HNF1B |
| Original motif Consensus sequence: TTAATRWTTAAC |
Reverse complement motif Consensus sequence: GTTAAWKATTAA |
|
|
 |
 |
Best Matches for Motif ID 82 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
68 |
| Motif name: |
HNF1A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0255432 |
Alignment:
GYTAATDATTAACC
-TTAATRWTTAAC-
| Original motif Consensus sequence: GGTTAATDATTAMC |
Reverse complement motif Consensus sequence: GYTAATDATTAACC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
125 |
| Motif name: |
wwTAATwwATTAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0657822 |
Alignment:
DDTAATWWATTAHH
-GTTAAWKATTAA-
| Original motif Consensus sequence: DDTAATWWATTAHH |
Reverse complement motif Consensus sequence: HHTAATWWATTADD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
84 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0685194 |
Alignment:
AAAGGTCAAAGGTCAAC
----GTTAAWKATTAA-
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
Lhx3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0745842 |
Alignment:
WAATTAATTAWYB
TTAATRWTTAAC-
| Original motif Consensus sequence: WAATTAATTAWYB |
Reverse complement motif Consensus sequence: BMWTAATTAATTW |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
60 |
| Motif name: |
Motif 60 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0845958 |
Alignment:
AADAAAAMAAAAAM
-GTTAAWKATTAA-
| Original motif Consensus sequence: AADAAAAMAAAAAM |
Reverse complement motif Consensus sequence: YTTTTTRTTTTDTT |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 83 |
Motif name: NR2F1 |
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
Best Matches for Motif ID 83 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
84 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0174588 |
Alignment:
GTTGACCTTTGACCTTT
--TGAMCTTTGMMCYT-
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
Pax4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
14 |
| Similarity score: |
0.0713206 |
Alignment:
RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC
-------TGAMCTTTGMMCYT---------
| Original motif Consensus sequence: RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC |
Reverse complement motif Consensus sequence: GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
98 |
| Motif name: |
myyCCCACmTGCmyr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0745302 |
Alignment:
HBCCCCACMTGCMHV
AKGYYCAAAGRTCA-
| Original motif Consensus sequence: HBCCCCACMTGCMHV |
Reverse complement motif Consensus sequence: VDRGCAYGTGGGGVD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
71 |
| Motif name: |
Evi1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
1.06838 |
Alignment:
--TBTTATCTTMTCTT
TGAMCTTTGMMCYT--
| Original motif Consensus sequence: AAGAYAAGATAABA |
Reverse complement motif Consensus sequence: TBTTATCTTMTCTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
68 |
| Motif name: |
HNF1A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
1.06886 |
Alignment:
GGTTAATDATTAMC--
--TGAMCTTTGMMCYT
| Original motif Consensus sequence: GGTTAATDATTAMC |
Reverse complement motif Consensus sequence: GYTAATDATTAACC |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 84 |
Motif name: NR1H2RXRA |
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
Best Matches for Motif ID 84 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
68 |
| Motif name: |
HNF1A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0896514 |
Alignment:
---GGTTAATDATTAMC
AAAGGTCAAAGGTCAAC
| Original motif Consensus sequence: GGTTAATDATTAMC |
Reverse complement motif Consensus sequence: GYTAATDATTAACC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
75 |
| Motif name: |
FOXF2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.595213 |
Alignment:
----DTTGTTTACSBTBB
GTTGACCTTTGACCTTT-
| Original motif Consensus sequence: BHADSGTAAACAAD |
Reverse complement motif Consensus sequence: DTTGTTTACSBTBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
76 |
| Motif name: |
Sox2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
12 |
| Similarity score: |
1.0817 |
Alignment:
-----TTTGCATMACAAWGG
AAAGGTCAAAGGTCAAC---
| Original motif Consensus sequence: CCWTTGTYATGCAAA |
Reverse complement motif Consensus sequence: TTTGCATMACAAWGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
82 |
| Motif name: |
HNF1B |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.0906 |
Alignment:
-----TTAATRWTTAAC
AAAGGTCAAAGGTCAAC
| Original motif Consensus sequence: TTAATRWTTAAC |
Reverse complement motif Consensus sequence: GTTAAWKATTAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
119 |
| Motif name: |
cbCCAGCTCmyk |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.09978 |
Alignment:
HBYGAGCTGGBB-----
GTTGACCTTTGACCTTT
| Original motif Consensus sequence: BBCCAGCTCMBD |
Reverse complement motif Consensus sequence: HBYGAGCTGGBB |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 85 |
Motif name: Nobox |
| Original motif Consensus sequence: TAATTRSY |
Reverse complement motif Consensus sequence: MSMAATTA |
|
|
 |
 |
Best Matches for Motif ID 85 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
Lhx3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0419408 |
Alignment:
WAATTAATTAWYB
----TAATTRSY-
| Original motif Consensus sequence: WAATTAATTAWYB |
Reverse complement motif Consensus sequence: BMWTAATTAATTW |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
125 |
| Motif name: |
wwTAATwwATTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.0591612 |
Alignment:
HHTAATWWATTADD
------TAATTRSY
| Original motif Consensus sequence: DDTAATWWATTAHH |
Reverse complement motif Consensus sequence: HHTAATWWATTADD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
62 |
| Motif name: |
TBP |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0604219 |
Alignment:
VBVVVVMWTTTATAS
----MSMAATTA---
| Original motif Consensus sequence: STATAAAWRVVVVBV |
Reverse complement motif Consensus sequence: VBVVVVMWTTTATAS |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
106 |
| Motif name: |
wtAATTAAtw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0649275 |
Alignment:
DTAATTAATH
-TAATTRSY-
| Original motif Consensus sequence: DTAATTAATH |
Reverse complement motif Consensus sequence: HATTAATTAD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
68 |
| Motif name: |
HNF1A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0652412 |
Alignment:
GGTTAATDATTAMC
------TAATTRSY
| Original motif Consensus sequence: GGTTAATDATTAMC |
Reverse complement motif Consensus sequence: GYTAATDATTAACC |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 86 |
Motif name: FOXO3 |
| Original motif Consensus sequence: KGTAAACA |
Reverse complement motif Consensus sequence: TGTTTACR |
|
|
 |
 |
Best Matches for Motif ID 86 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
75 |
| Motif name: |
FOXF2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0236411 |
Alignment:
DTTGTTTACSBTBB
--TGTTTACR----
| Original motif Consensus sequence: BHADSGTAAACAAD |
Reverse complement motif Consensus sequence: DTTGTTTACSBTBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
70 |
| Motif name: |
Foxq1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0397436 |
Alignment:
HATTGTTTATW
TGTTTACR---
| Original motif Consensus sequence: HATTGTTTATW |
Reverse complement motif Consensus sequence: WATAAACAATH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
108 |
| Motif name: |
wwATkTTTAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.054763 |
Alignment:
DHATKTTTAHH
---TGTTTACR
| Original motif Consensus sequence: DHATKTTTAHH |
Reverse complement motif Consensus sequence: HHTAAARATHD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
67 |
| Motif name: |
FOXI1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0656948 |
Alignment:
AAAMAAAMADBY
---KGTAAACA-
| Original motif Consensus sequence: KBDTRTTTRTTT |
Reverse complement motif Consensus sequence: AAAMAAAMADBY |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
61 |
| Motif name: |
MEF2A |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0727619 |
Alignment:
CTATTTWTAG
-TGTTTACR-
| Original motif Consensus sequence: CTATTTWTAG |
Reverse complement motif Consensus sequence: CTAWAAATAG |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 87 |
Motif name: SOX9 |
| Original motif Consensus sequence: DAACAATRG |
Reverse complement motif Consensus sequence: CMATTGTTH |
|
|
 |
 |
Best Matches for Motif ID 87 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
76 |
| Motif name: |
Sox2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
9 |
| Similarity score: |
0.018186 |
Alignment:
TTTGCATMACAAWGG
DAACAATRG------
| Original motif Consensus sequence: CCWTTGTYATGCAAA |
Reverse complement motif Consensus sequence: TTTGCATMACAAWGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
Pax4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0.0583456 |
Alignment:
RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC
DAACAATRG---------------------
| Original motif Consensus sequence: RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC |
Reverse complement motif Consensus sequence: GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
89 |
| Motif name: |
aaACAAAACaa |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0.0599655 |
Alignment:
AAACAAAACHV
DAACAATRG--
| Original motif Consensus sequence: AAACAAAACHV |
Reverse complement motif Consensus sequence: BHGTTTTGTTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
108 |
| Motif name: |
wwATkTTTAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
9 |
| Similarity score: |
0.0642936 |
Alignment:
DHATKTTTAHH
CMATTGTTH--
| Original motif Consensus sequence: DHATKTTTAHH |
Reverse complement motif Consensus sequence: HHTAAARATHD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
61 |
| Motif name: |
MEF2A |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0.0646581 |
Alignment:
CTATTTWTAG
-CMATTGTTH
| Original motif Consensus sequence: CTATTTWTAG |
Reverse complement motif Consensus sequence: CTAWAAATAG |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 88 |
Motif name: ygCTyTTCsg |
| Original motif Consensus sequence: HDCTYTTCGB |
Reverse complement motif Consensus sequence: BCGAAMAGHH |
|
|
 |
 |
Best Matches for Motif ID 88 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
113 |
| Motif name: |
htCTGyKTCbt |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0278763 |
Alignment:
HVGAAMCAGVH
-BCGAAMAGHH
| Original motif Consensus sequence: HBCTGYTTCBH |
Reverse complement motif Consensus sequence: HVGAAMCAGVH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
67 |
| Motif name: |
FOXI1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0294361 |
Alignment:
AAAMAAAMADBY
--BCGAAMAGHH
| Original motif Consensus sequence: KBDTRTTTRTTT |
Reverse complement motif Consensus sequence: AAAMAAAMADBY |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
109 |
| Motif name: |
aaCAAAAACaa |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0382901 |
Alignment:
HHCAAAAACHH
-BCGAAMAGHH
| Original motif Consensus sequence: HHCAAAAACHH |
Reverse complement motif Consensus sequence: HHGTTTTTGHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
91 |
| Motif name: |
wtATTTTAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0393404 |
Alignment:
HWTAAAATHD
BCGAAMAGHH
| Original motif Consensus sequence: DHATTTTAWH |
Reverse complement motif Consensus sequence: HWTAAAATHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
119 |
| Motif name: |
cbCCAGCTCmyk |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0400249 |
Alignment:
HBYGAGCTGGBB
-BCGAAMAGHH-
| Original motif Consensus sequence: BBCCAGCTCMBD |
Reverse complement motif Consensus sequence: HBYGAGCTGGBB |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 89 |
Motif name: aaACAAAACaa |
| Original motif Consensus sequence: AAACAAAACHV |
Reverse complement motif Consensus sequence: BHGTTTTGTTT |
|
|
 |
 |
Best Matches for Motif ID 89 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
109 |
| Motif name: |
aaCAAAAACaa |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.00582159 |
Alignment:
HHGTTTTTGHH
BHGTTTTGTTT
| Original motif Consensus sequence: HHCAAAAACHH |
Reverse complement motif Consensus sequence: HHGTTTTTGHH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
67 |
| Motif name: |
FOXI1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0320461 |
Alignment:
KBDTRTTTRTTT
BHGTTTTGTTT-
| Original motif Consensus sequence: KBDTRTTTRTTT |
Reverse complement motif Consensus sequence: AAAMAAAMADBY |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
60 |
| Motif name: |
Motif 60 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.039575 |
Alignment:
AADAAAAMAAAAAM
--AAACAAAACHV-
| Original motif Consensus sequence: AADAAAAMAAAAAM |
Reverse complement motif Consensus sequence: YTTTTTRTTTTDTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
75 |
| Motif name: |
FOXF2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0401124 |
Alignment:
BHADSGTAAACAAD
--AAACAAAACHV-
| Original motif Consensus sequence: BHADSGTAAACAAD |
Reverse complement motif Consensus sequence: DTTGTTTACSBTBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
108 |
| Motif name: |
wwATkTTTAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0486334 |
Alignment:
DHATKTTTAHH
BHGTTTTGTTT
| Original motif Consensus sequence: DHATKTTTAHH |
Reverse complement motif Consensus sequence: HHTAAARATHD |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 90 |
Motif name: ymTACATAyw |
| Original motif Consensus sequence: HHTACATABD |
Reverse complement motif Consensus sequence: DBTATGTAHH |
|
|
 |
 |
Best Matches for Motif ID 90 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
114 |
| Motif name: |
awAACAcAAwa |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0626629 |
Alignment:
BDTTGTGTTDD
-DBTATGTAHH
| Original motif Consensus sequence: DDAACACAADV |
Reverse complement motif Consensus sequence: BDTTGTGTTDD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
108 |
| Motif name: |
wwATkTTTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0690336 |
Alignment:
HHTAAARATHD
HHTACATABD-
| Original motif Consensus sequence: DHATKTTTAHH |
Reverse complement motif Consensus sequence: HHTAAARATHD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
67 |
| Motif name: |
FOXI1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0763781 |
Alignment:
AAAMAAAMADBY
-HHTACATABD-
| Original motif Consensus sequence: KBDTRTTTRTTT |
Reverse complement motif Consensus sequence: AAAMAAAMADBY |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
75 |
| Motif name: |
FOXF2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0772253 |
Alignment:
DTTGTTTACSBTBB
DBTATGTAHH----
| Original motif Consensus sequence: BHADSGTAAACAAD |
Reverse complement motif Consensus sequence: DTTGTTTACSBTBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
100 |
| Motif name: |
wwTAATAAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0773471 |
Alignment:
DDTTATTAHD
DBTATGTAHH
| Original motif Consensus sequence: DHTAATAADD |
Reverse complement motif Consensus sequence: DDTTATTAHD |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 91 |
Motif name: wtATTTTAww |
| Original motif Consensus sequence: DHATTTTAWH |
Reverse complement motif Consensus sequence: HWTAAAATHD |
|
|
 |
 |
Best Matches for Motif ID 91 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
100 |
| Motif name: |
wwTAATAAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0307554 |
Alignment:
DHTAATAADD
HWTAAAATHD
| Original motif Consensus sequence: DHTAATAADD |
Reverse complement motif Consensus sequence: DDTTATTAHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
108 |
| Motif name: |
wwATkTTTAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.031047 |
Alignment:
HHTAAARATHD
-HWTAAAATHD
| Original motif Consensus sequence: DHATKTTTAHH |
Reverse complement motif Consensus sequence: HHTAAARATHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
106 |
| Motif name: |
wtAATTAAtw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0371798 |
Alignment:
DTAATTAATH
DHATTTTAWH
| Original motif Consensus sequence: DTAATTAATH |
Reverse complement motif Consensus sequence: HATTAATTAD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
95 |
| Motif name: |
rmATGCTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0404266 |
Alignment:
DHATGCTADH
DHATTTTAWH
| Original motif Consensus sequence: DHATGCTADH |
Reverse complement motif Consensus sequence: HDTAGCATHD |
|
|
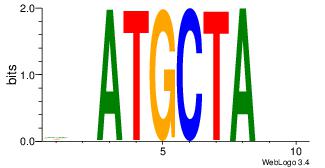 |
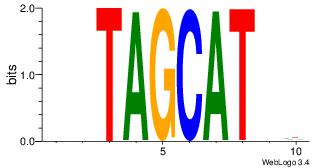 |
| Dataset #: |
4 |
| Motif ID: |
89 |
| Motif name: |
aaACAAAACaa |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0448101 |
Alignment:
BHGTTTTGTTT
-DHATTTTAWH
| Original motif Consensus sequence: AAACAAAACHV |
Reverse complement motif Consensus sequence: BHGTTTTGTTT |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 92 |
Motif name: cgAKGyCAcg |
| Original motif Consensus sequence: CGAGGYCACV |
Reverse complement motif Consensus sequence: VGTGMCCTCG |
|
|
 |
 |
Best Matches for Motif ID 92 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
97 |
| Motif name: |
sgArGCCACg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.00323214 |
Alignment:
SGARGCCACG
CGAGGYCACV
| Original motif Consensus sequence: SGARGCCACG |
Reverse complement motif Consensus sequence: CGTGGCKTCS |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
107 |
| Motif name: |
crCGyGCRcg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.039949 |
Alignment:
CGTGCACGCG
VGTGMCCTCG
| Original motif Consensus sequence: CGCGTGCACG |
Reverse complement motif Consensus sequence: CGTGCACGCG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
84 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
10 |
| Similarity score: |
0.0444539 |
Alignment:
GTTGACCTTTGACCTTT
VGTGMCCTCG-------
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
95 |
| Motif name: |
rmATGCTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0540906 |
Alignment:
DHATGCTADH
CGAGGYCACV
| Original motif Consensus sequence: DHATGCTADH |
Reverse complement motif Consensus sequence: HDTAGCATHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
119 |
| Motif name: |
cbCCAGCTCmyk |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0546695 |
Alignment:
HBYGAGCTGGBB
VGTGMCCTCG--
| Original motif Consensus sequence: BBCCAGCTCMBD |
Reverse complement motif Consensus sequence: HBYGAGCTGGBB |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 93 |
Motif name: cmCARCACTwr |
| Original motif Consensus sequence: BHCAGCACTHV |
Reverse complement motif Consensus sequence: VHAGTGCTGHB |
|
|
 |
 |
Best Matches for Motif ID 93 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
119 |
| Motif name: |
cbCCAGCTCmyk |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.00936148 |
Alignment:
BBCCAGCTCMBD
-BHCAGCACTHV
| Original motif Consensus sequence: BBCCAGCTCMBD |
Reverse complement motif Consensus sequence: HBYGAGCTGGBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
110 |
| Motif name: |
cmCCACACCcm |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0121475 |
Alignment:
HVCCACACCBV
BHCAGCACTHV
| Original motif Consensus sequence: HVCCACACCBV |
Reverse complement motif Consensus sequence: VBGGTGTGGBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
121 |
| Motif name: |
ctGAGTCATCkb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0295417 |
Alignment:
HDGAGTCATCDB
-VHAGTGCTGHB
| Original motif Consensus sequence: HDGAGTCATCDB |
Reverse complement motif Consensus sequence: BHGATGACTCDD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
122 |
| Motif name: |
hsCCAGGCCTGGsr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0336897 |
Alignment:
HBCCAGGCCTGGVD
--VHAGTGCTGHB-
| Original motif Consensus sequence: HVCCAGGCCTGGBD |
Reverse complement motif Consensus sequence: HBCCAGGCCTGGVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
103 |
| Motif name: |
csCCaGCTCCCgs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0373286 |
Alignment:
BBCCAGCTCCCVB
-BHCAGCACTHV-
| Original motif Consensus sequence: BBCCAGCTCCCVB |
Reverse complement motif Consensus sequence: BVGGGAGCTGGBB |
|
|
 |
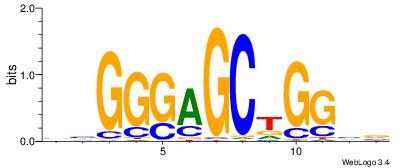 |
| Dataset #: 4 |
Motif ID: 94 |
Motif name: yrTCWCAAyr |
| Original motif Consensus sequence: HVTCACAAYD |
Reverse complement motif Consensus sequence: HKTTGTGABH |
|
|
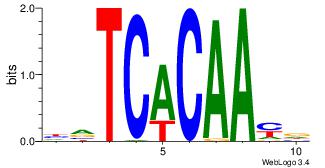 |
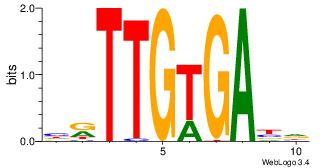 |
Best Matches for Motif ID 94 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
114 |
| Motif name: |
awAACAcAAwa |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.00495535 |
Alignment:
BDTTGTGTTDD
HKTTGTGABH-
| Original motif Consensus sequence: DDAACACAADV |
Reverse complement motif Consensus sequence: BDTTGTGTTDD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
76 |
| Motif name: |
Sox2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.00762497 |
Alignment:
CCWTTGTYATGCAAA
----HKTTGTGABH-
| Original motif Consensus sequence: CCWTTGTYATGCAAA |
Reverse complement motif Consensus sequence: TTTGCATMACAAWGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
100 |
| Motif name: |
wwTAATAAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0114769 |
Alignment:
DDTTATTAHD
HKTTGTGABH
| Original motif Consensus sequence: DHTAATAADD |
Reverse complement motif Consensus sequence: DDTTATTAHD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
80 |
| Motif name: |
Pou5f1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
10 |
| Similarity score: |
0.0127574 |
Alignment:
CWTTGTHATGCAAAT
-----HKTTGTGABH
| Original motif Consensus sequence: CWTTGTHATGCAAAT |
Reverse complement motif Consensus sequence: ATTTGCATHACAAWG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
89 |
| Motif name: |
aaACAAAACaa |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0208363 |
Alignment:
AAACAAAACHV
HVTCACAAYD-
| Original motif Consensus sequence: AAACAAAACHV |
Reverse complement motif Consensus sequence: BHGTTTTGTTT |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 95 |
Motif name: rmATGCTAww |
| Original motif Consensus sequence: DHATGCTADH |
Reverse complement motif Consensus sequence: HDTAGCATHD |
|
|
 |
 |
Best Matches for Motif ID 95 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
91 |
| Motif name: |
wtATTTTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.00863735 |
Alignment:
DHATTTTAWH
DHATGCTADH
| Original motif Consensus sequence: DHATTTTAWH |
Reverse complement motif Consensus sequence: HWTAAAATHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
115 |
| Motif name: |
ysCmAGCACwy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0142021 |
Alignment:
DHGTGCTRGVH
DHATGCTADH-
| Original motif Consensus sequence: HVCMAGCACHH |
Reverse complement motif Consensus sequence: DHGTGCTRGVH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
80 |
| Motif name: |
Pou5f1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0215325 |
Alignment:
CWTTGTHATGCAAAT
-----DHATGCTADH
| Original motif Consensus sequence: CWTTGTHATGCAAAT |
Reverse complement motif Consensus sequence: ATTTGCATHACAAWG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
123 |
| Motif name: |
asMAAGCTTKst |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0234838 |
Alignment:
HBCAAGCTTGVD
-DHATGCTADH-
| Original motif Consensus sequence: HBCAAGCTTGVD |
Reverse complement motif Consensus sequence: DVCAAGCTTGBH |
|
|
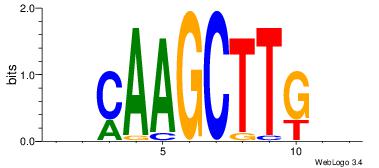 |
 |
| Dataset #: |
4 |
| Motif ID: |
93 |
| Motif name: |
cmCARCACTwr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0266703 |
Alignment:
VHAGTGCTGHB
DHATGCTADH-
| Original motif Consensus sequence: BHCAGCACTHV |
Reverse complement motif Consensus sequence: VHAGTGCTGHB |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 96 |
Motif name: mdAAcCAACG |
| Original motif Consensus sequence: VDAACCAACG |
Reverse complement motif Consensus sequence: CGTTGGTTDB |
|
|
 |
 |
Best Matches for Motif ID 96 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
97 |
| Motif name: |
sgArGCCACg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0161902 |
Alignment:
SGARGCCACG
VDAACCAACG
| Original motif Consensus sequence: SGARGCCACG |
Reverse complement motif Consensus sequence: CGTGGCKTCS |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
60 |
| Motif name: |
Motif 60 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0244161 |
Alignment:
YTTTTTRTTTTDTT
--CGTTGGTTDB--
| Original motif Consensus sequence: AADAAAAMAAAAAM |
Reverse complement motif Consensus sequence: YTTTTTRTTTTDTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
106 |
| Motif name: |
wtAATTAAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0244247 |
Alignment:
HATTAATTAD
CGTTGGTTDB
| Original motif Consensus sequence: DTAATTAATH |
Reverse complement motif Consensus sequence: HATTAATTAD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
70 |
| Motif name: |
Foxq1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0266383 |
Alignment:
HATTGTTTATW
-CGTTGGTTDB
| Original motif Consensus sequence: HATTGTTTATW |
Reverse complement motif Consensus sequence: WATAAACAATH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
113 |
| Motif name: |
htCTGyKTCbt |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0296542 |
Alignment:
HVGAAMCAGVH
-VDAACCAACG
| Original motif Consensus sequence: HBCTGYTTCBH |
Reverse complement motif Consensus sequence: HVGAAMCAGVH |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 97 |
Motif name: sgArGCCACg |
| Original motif Consensus sequence: SGARGCCACG |
Reverse complement motif Consensus sequence: CGTGGCKTCS |
|
|
 |
 |
Best Matches for Motif ID 97 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
92 |
| Motif name: |
cgAKGyCAcg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0020736 |
Alignment:
CGAGGYCACV
SGARGCCACG
| Original motif Consensus sequence: CGAGGYCACV |
Reverse complement motif Consensus sequence: VGTGMCCTCG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
96 |
| Motif name: |
mdAAcCAACG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0408537 |
Alignment:
VDAACCAACG
SGARGCCACG
| Original motif Consensus sequence: VDAACCAACG |
Reverse complement motif Consensus sequence: CGTTGGTTDB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
95 |
| Motif name: |
rmATGCTAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0472561 |
Alignment:
HDTAGCATHD
CGTGGCKTCS
| Original motif Consensus sequence: DHATGCTADH |
Reverse complement motif Consensus sequence: HDTAGCATHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
126 |
| Motif name: |
yvTGCAGsCAcg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.057998 |
Alignment:
VBTGSCTGCAVB
CGTGGCKTCS--
| Original motif Consensus sequence: BVTGCAGSCABV |
Reverse complement motif Consensus sequence: VBTGSCTGCAVB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
116 |
| Motif name: |
dyCTGCyTCyc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0617012 |
Alignment:
BHGAKGCAGHH
-SGARGCCACG
| Original motif Consensus sequence: DHCTGCYTCHB |
Reverse complement motif Consensus sequence: BHGAKGCAGHH |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 98 |
Motif name: myyCCCACmTGCmyr |
| Original motif Consensus sequence: HBCCCCACMTGCMHV |
Reverse complement motif Consensus sequence: VDRGCAYGTGGGGVD |
|
|
 |
 |
Best Matches for Motif ID 98 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
Pax4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
15 |
| Similarity score: |
0.0499349 |
Alignment:
RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC
-------HBCCCCACMTGCMHV--------
| Original motif Consensus sequence: RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC |
Reverse complement motif Consensus sequence: GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
118 |
| Motif name: |
yrCACATGCATGTGyr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
0.0613869 |
Alignment:
HVCACATGCATGTGBD
-HBCCCCACMTGCMHV
| Original motif Consensus sequence: HVCACATGCATGTGBD |
Reverse complement motif Consensus sequence: DVCACATGCATGTGBH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
122 |
| Motif name: |
hsCCAGGCCTGGsr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.538726 |
Alignment:
HVCCAGGCCTGGBD-
HBCCCCACMTGCMHV
| Original motif Consensus sequence: HVCCAGGCCTGGBD |
Reverse complement motif Consensus sequence: HBCCAGGCCTGGVD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
59 |
| Motif name: |
Motif 59 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.560094 |
Alignment:
-CTBCCTCHSYCYCC
HBCCCCACMTGCMHV
| Original motif Consensus sequence: CTBCCTCHSYCYCC |
Reverse complement motif Consensus sequence: GGMGKSHGAGGBAG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
58 |
| Motif name: |
Motif 58 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.57014 |
Alignment:
-CACACACACACACA
HBCCCCACMTGCMHV
| Original motif Consensus sequence: CACACACACACACA |
Reverse complement motif Consensus sequence: TGTGTGTGTGTGTG |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 99 |
Motif name: graGGGGGArr |
| Original motif Consensus sequence: DVAGGGGGADD |
Reverse complement motif Consensus sequence: HDTCCCCCTVH |
|
|
 |
 |
Best Matches for Motif ID 99 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
127 |
| Motif name: |
rryCAGGGAyw |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0288287 |
Alignment:
DDYCAGGGAHD
DVAGGGGGADD
| Original motif Consensus sequence: DDYCAGGGAHD |
Reverse complement motif Consensus sequence: DHTCCCTGMDD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
59 |
| Motif name: |
Motif 59 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0416239 |
Alignment:
GGMGKSHGAGGBAG
---DVAGGGGGADD
| Original motif Consensus sequence: CTBCCTCHSYCYCC |
Reverse complement motif Consensus sequence: GGMGKSHGAGGBAG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
110 |
| Motif name: |
cmCCACACCcm |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0422985 |
Alignment:
HVCCACACCBV
HDTCCCCCTVH
| Original motif Consensus sequence: HVCCACACCBV |
Reverse complement motif Consensus sequence: VBGGTGTGGBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
112 |
| Motif name: |
ctGAGTkTGAgg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.042367 |
Alignment:
VBGAGTKTGADD
-DVAGGGGGADD
| Original motif Consensus sequence: VBGAGTKTGADD |
Reverse complement motif Consensus sequence: HHTCARACTCVV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
111 |
| Motif name: |
ccCCmCaCCCCcc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0428468 |
Alignment:
DBGGGGTGGGGBD
DVAGGGGGADD--
| Original motif Consensus sequence: HBCCCCACCCCBH |
Reverse complement motif Consensus sequence: DBGGGGTGGGGBD |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 100 |
Motif name: wwTAATAAww |
| Original motif Consensus sequence: DHTAATAADD |
Reverse complement motif Consensus sequence: DDTTATTAHD |
|
|
 |
 |
Best Matches for Motif ID 100 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
91 |
| Motif name: |
wtATTTTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0241522 |
Alignment:
HWTAAAATHD
DHTAATAADD
| Original motif Consensus sequence: DHATTTTAWH |
Reverse complement motif Consensus sequence: HWTAAAATHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
108 |
| Motif name: |
wwATkTTTAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0255442 |
Alignment:
DHATKTTTAHH
-DDTTATTAHD
| Original motif Consensus sequence: DHATKTTTAHH |
Reverse complement motif Consensus sequence: HHTAAARATHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
125 |
| Motif name: |
wwTAATwwATTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0362107 |
Alignment:
HHTAATWWATTADD
DHTAATAADD----
| Original motif Consensus sequence: DDTAATWWATTAHH |
Reverse complement motif Consensus sequence: HHTAATWWATTADD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
106 |
| Motif name: |
wtAATTAAtw |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0364833 |
Alignment:
HATTAATTAD
DDTTATTAHD
| Original motif Consensus sequence: DTAATTAATH |
Reverse complement motif Consensus sequence: HATTAATTAD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
109 |
| Motif name: |
aaCAAAAACaa |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0471554 |
Alignment:
HHCAAAAACHH
DHTAATAADD-
| Original motif Consensus sequence: HHCAAAAACHH |
Reverse complement motif Consensus sequence: HHGTTTTTGHH |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 101 |
Motif name: csCACCCCgg |
| Original motif Consensus sequence: HVCACCCCBB |
Reverse complement motif Consensus sequence: BBGGGGTGVD |
|
|
 |
 |
Best Matches for Motif ID 101 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
111 |
| Motif name: |
ccCCmCaCCCCcc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0 |
Alignment:
HBCCCCACCCCBH
---HVCACCCCBB
| Original motif Consensus sequence: HBCCCCACCCCBH |
Reverse complement motif Consensus sequence: DBGGGGTGGGGBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
110 |
| Motif name: |
cmCCACACCcm |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.00930301 |
Alignment:
HVCCACACCBV
-HVCACCCCBB
| Original motif Consensus sequence: HVCCACACCBV |
Reverse complement motif Consensus sequence: VBGGTGTGGBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
99 |
| Motif name: |
graGGGGGArr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0256956 |
Alignment:
HDTCCCCCTVH
-HVCACCCCBB
| Original motif Consensus sequence: DVAGGGGGADD |
Reverse complement motif Consensus sequence: HDTCCCCCTVH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
119 |
| Motif name: |
cbCCAGCTCmyk |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.033038 |
Alignment:
HBYGAGCTGGBB
-BBGGGGTGVD-
| Original motif Consensus sequence: BBCCAGCTCMBD |
Reverse complement motif Consensus sequence: HBYGAGCTGGBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
93 |
| Motif name: |
cmCARCACTwr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0377738 |
Alignment:
BHCAGCACTHV
HVCACCCCBB-
| Original motif Consensus sequence: BHCAGCACTHV |
Reverse complement motif Consensus sequence: VHAGTGCTGHB |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 102 |
Motif name: ssCkGGYCCCsg |
| Original motif Consensus sequence: BBCKGGCCCCVV |
Reverse complement motif Consensus sequence: VVGGGGCCRGBB |
|
|
 |
 |
Best Matches for Motif ID 102 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
103 |
| Motif name: |
csCCaGCTCCCgs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.00699729 |
Alignment:
BVGGGAGCTGGBB
-VVGGGGCCRGBB
| Original motif Consensus sequence: BBCCAGCTCCCVB |
Reverse complement motif Consensus sequence: BVGGGAGCTGGBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
124 |
| Motif name: |
cswGGGCCCwsg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.00901563 |
Alignment:
VVWGGGCCCWBB
BBCKGGCCCCVV
| Original motif Consensus sequence: VVWGGGCCCWBB |
Reverse complement motif Consensus sequence: BBWGGGCCCWVV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
111 |
| Motif name: |
ccCCmCaCCCCcc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.015463 |
Alignment:
DBGGGGTGGGGBD
VVGGGGCCRGBB-
| Original motif Consensus sequence: HBCCCCACCCCBH |
Reverse complement motif Consensus sequence: DBGGGGTGGGGBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
122 |
| Motif name: |
hsCCAGGCCTGGsr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0222107 |
Alignment:
HVCCAGGCCTGGBD
-BBCKGGCCCCVV-
| Original motif Consensus sequence: HVCCAGGCCTGGBD |
Reverse complement motif Consensus sequence: HBCCAGGCCTGGVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
119 |
| Motif name: |
cbCCAGCTCmyk |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0277516 |
Alignment:
BBCCAGCTCMBD
BBCKGGCCCCVV
| Original motif Consensus sequence: BBCCAGCTCMBD |
Reverse complement motif Consensus sequence: HBYGAGCTGGBB |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 103 |
Motif name: csCCaGCTCCCgs |
| Original motif Consensus sequence: BBCCAGCTCCCVB |
Reverse complement motif Consensus sequence: BVGGGAGCTGGBB |
|
|
 |
 |
Best Matches for Motif ID 103 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
98 |
| Motif name: |
myyCCCACmTGCmyr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
13 |
| Similarity score: |
0.0331906 |
Alignment:
HBCCCCACMTGCMHV
--BBCCAGCTCCCVB
| Original motif Consensus sequence: HBCCCCACMTGCMHV |
Reverse complement motif Consensus sequence: VDRGCAYGTGGGGVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
122 |
| Motif name: |
hsCCAGGCCTGGsr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.033684 |
Alignment:
HVCCAGGCCTGGBD
-BVGGGAGCTGGBB
| Original motif Consensus sequence: HVCCAGGCCTGGBD |
Reverse complement motif Consensus sequence: HBCCAGGCCTGGVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
111 |
| Motif name: |
ccCCmCaCCCCcc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.0344113 |
Alignment:
HBCCCCACCCCBH
BBCCAGCTCCCVB
| Original motif Consensus sequence: HBCCCCACCCCBH |
Reverse complement motif Consensus sequence: DBGGGGTGGGGBD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
Pax4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
13 |
| Similarity score: |
0.0424871 |
Alignment:
GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM
--BVGGGAGCTGGBB---------------
| Original motif Consensus sequence: RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC |
Reverse complement motif Consensus sequence: GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
59 |
| Motif name: |
Motif 59 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.0502084 |
Alignment:
GGMGKSHGAGGBAG
-BVGGGAGCTGGBB
| Original motif Consensus sequence: CTBCCTCHSYCYCC |
Reverse complement motif Consensus sequence: GGMGKSHGAGGBAG |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 104 |
Motif name: cgCCCwGGScsg |
| Original motif Consensus sequence: VDCCCWGGGCBB |
Reverse complement motif Consensus sequence: BBGCCCWGGGHV |
|
|
 |
 |
Best Matches for Motif ID 104 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
111 |
| Motif name: |
ccCCmCaCCCCcc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0295528 |
Alignment:
HBCCCCACCCCBH
-VDCCCWGGGCBB
| Original motif Consensus sequence: HBCCCCACCCCBH |
Reverse complement motif Consensus sequence: DBGGGGTGGGGBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
126 |
| Motif name: |
yvTGCAGsCAcg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0306486 |
Alignment:
BVTGCAGSCABV
VDCCCWGGGCBB
| Original motif Consensus sequence: BVTGCAGSCABV |
Reverse complement motif Consensus sequence: VBTGSCTGCAVB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
103 |
| Motif name: |
csCCaGCTCCCgs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0384576 |
Alignment:
BBCCAGCTCCCVB
VDCCCWGGGCBB-
| Original motif Consensus sequence: BBCCAGCTCCCVB |
Reverse complement motif Consensus sequence: BVGGGAGCTGGBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
98 |
| Motif name: |
myyCCCACmTGCmyr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
12 |
| Similarity score: |
0.0393753 |
Alignment:
HBCCCCACMTGCMHV
---VDCCCWGGGCBB
| Original motif Consensus sequence: HBCCCCACMTGCMHV |
Reverse complement motif Consensus sequence: VDRGCAYGTGGGGVD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
59 |
| Motif name: |
Motif 59 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0399857 |
Alignment:
CTBCCTCHSYCYCC
--VDCCCWGGGCBB
| Original motif Consensus sequence: CTBCCTCHSYCYCC |
Reverse complement motif Consensus sequence: GGMGKSHGAGGBAG |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 105 |
Motif name: ccGCAGCCss |
| Original motif Consensus sequence: VCGCAGCCBB |
Reverse complement motif Consensus sequence: BBGGCTGCGV |
|
|
 |
 |
Best Matches for Motif ID 105 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
126 |
| Motif name: |
yvTGCAGsCAcg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0 |
Alignment:
VBTGSCTGCAVB
-BBGGCTGCGV-
| Original motif Consensus sequence: BVTGCAGSCABV |
Reverse complement motif Consensus sequence: VBTGSCTGCAVB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
103 |
| Motif name: |
csCCaGCTCCCgs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0101194 |
Alignment:
BVGGGAGCTGGBB
BBGGCTGCGV---
| Original motif Consensus sequence: BBCCAGCTCCCVB |
Reverse complement motif Consensus sequence: BVGGGAGCTGGBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
102 |
| Motif name: |
ssCkGGYCCCsg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0260007 |
Alignment:
BBCKGGCCCCVV
BBGGCTGCGV--
| Original motif Consensus sequence: BBCKGGCCCCVV |
Reverse complement motif Consensus sequence: VVGGGGCCRGBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
119 |
| Motif name: |
cbCCAGCTCmyk |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0296738 |
Alignment:
HBYGAGCTGGBB
--BBGGCTGCGV
| Original motif Consensus sequence: BBCCAGCTCMBD |
Reverse complement motif Consensus sequence: HBYGAGCTGGBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
116 |
| Motif name: |
dyCTGCyTCyc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0303133 |
Alignment:
DHCTGCYTCHB
-BBGGCTGCGV
| Original motif Consensus sequence: DHCTGCYTCHB |
Reverse complement motif Consensus sequence: BHGAKGCAGHH |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 106 |
Motif name: wtAATTAAtw |
| Original motif Consensus sequence: DTAATTAATH |
Reverse complement motif Consensus sequence: HATTAATTAD |
|
|
 |
 |
Best Matches for Motif ID 106 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
Lhx3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0302494 |
Alignment:
BMWTAATTAATTW
-DTAATTAATH--
| Original motif Consensus sequence: WAATTAATTAWYB |
Reverse complement motif Consensus sequence: BMWTAATTAATTW |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
125 |
| Motif name: |
wwTAATwwATTAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0551105 |
Alignment:
HHTAATWWATTADD
---HATTAATTAD-
| Original motif Consensus sequence: DDTAATWWATTAHH |
Reverse complement motif Consensus sequence: HHTAATWWATTADD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
91 |
| Motif name: |
wtATTTTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0562809 |
Alignment:
DHATTTTAWH
DTAATTAATH
| Original motif Consensus sequence: DHATTTTAWH |
Reverse complement motif Consensus sequence: HWTAAAATHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
100 |
| Motif name: |
wwTAATAAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0621876 |
Alignment:
DDTTATTAHD
HATTAATTAD
| Original motif Consensus sequence: DHTAATAADD |
Reverse complement motif Consensus sequence: DDTTATTAHD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
68 |
| Motif name: |
HNF1A |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0690833 |
Alignment:
GGTTAATDATTAMC
----HATTAATTAD
| Original motif Consensus sequence: GGTTAATDATTAMC |
Reverse complement motif Consensus sequence: GYTAATDATTAACC |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 107 |
Motif name: crCGyGCRcg |
| Original motif Consensus sequence: CGCGTGCACG |
Reverse complement motif Consensus sequence: CGTGCACGCG |
|
|
 |
 |
Best Matches for Motif ID 107 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
92 |
| Motif name: |
cgAKGyCAcg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0293056 |
Alignment:
VGTGMCCTCG
CGTGCACGCG
| Original motif Consensus sequence: CGAGGYCACV |
Reverse complement motif Consensus sequence: VGTGMCCTCG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
115 |
| Motif name: |
ysCmAGCACwy |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0326988 |
Alignment:
DHGTGCTRGVH
-CGTGCACGCG
| Original motif Consensus sequence: HVCMAGCACHH |
Reverse complement motif Consensus sequence: DHGTGCTRGVH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
98 |
| Motif name: |
myyCCCACmTGCmyr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.033442 |
Alignment:
HBCCCCACMTGCMHV
-----CGCGTGCACG
| Original motif Consensus sequence: HBCCCCACMTGCMHV |
Reverse complement motif Consensus sequence: VDRGCAYGTGGGGVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
126 |
| Motif name: |
yvTGCAGsCAcg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0362108 |
Alignment:
VBTGSCTGCAVB
--CGCGTGCACG
| Original motif Consensus sequence: BVTGCAGSCABV |
Reverse complement motif Consensus sequence: VBTGSCTGCAVB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
102 |
| Motif name: |
ssCkGGYCCCsg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0421604 |
Alignment:
VVGGGGCCRGBB
--CGTGCACGCG
| Original motif Consensus sequence: BBCKGGCCCCVV |
Reverse complement motif Consensus sequence: VVGGGGCCRGBB |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 108 |
Motif name: wwATkTTTAww |
| Original motif Consensus sequence: DHATKTTTAHH |
Reverse complement motif Consensus sequence: HHTAAARATHD |
|
|
 |
 |
Best Matches for Motif ID 108 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
70 |
| Motif name: |
Foxq1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0381631 |
Alignment:
HATTGTTTATW
DHATKTTTAHH
| Original motif Consensus sequence: HATTGTTTATW |
Reverse complement motif Consensus sequence: WATAAACAATH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
109 |
| Motif name: |
aaCAAAAACaa |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0416262 |
Alignment:
HHCAAAAACHH
HHTAAARATHD
| Original motif Consensus sequence: HHCAAAAACHH |
Reverse complement motif Consensus sequence: HHGTTTTTGHH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
67 |
| Motif name: |
FOXI1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0453405 |
Alignment:
KBDTRTTTRTTT
DHATKTTTAHH-
| Original motif Consensus sequence: KBDTRTTTRTTT |
Reverse complement motif Consensus sequence: AAAMAAAMADBY |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
113 |
| Motif name: |
htCTGyKTCbt |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0594403 |
Alignment:
HBCTGYTTCBH
DHATKTTTAHH
| Original motif Consensus sequence: HBCTGYTTCBH |
Reverse complement motif Consensus sequence: HVGAAMCAGVH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
62 |
| Motif name: |
TBP |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0630171 |
Alignment:
VBVVVVMWTTTATAS
-DHATKTTTAHH---
| Original motif Consensus sequence: STATAAAWRVVVVBV |
Reverse complement motif Consensus sequence: VBVVVVMWTTTATAS |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 109 |
Motif name: aaCAAAAACaa |
| Original motif Consensus sequence: HHCAAAAACHH |
Reverse complement motif Consensus sequence: HHGTTTTTGHH |
|
|
 |
 |
Best Matches for Motif ID 109 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
89 |
| Motif name: |
aaACAAAACaa |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.00652588 |
Alignment:
BHGTTTTGTTT
HHGTTTTTGHH
| Original motif Consensus sequence: AAACAAAACHV |
Reverse complement motif Consensus sequence: BHGTTTTGTTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
117 |
| Motif name: |
yaCATAYACay |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0113253 |
Alignment:
DTGTGTATGBD
HHGTTTTTGHH
| Original motif Consensus sequence: HVCATACACAH |
Reverse complement motif Consensus sequence: DTGTGTATGBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
108 |
| Motif name: |
wwATkTTTAww |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0251184 |
Alignment:
DHATKTTTAHH
HHGTTTTTGHH
| Original motif Consensus sequence: DHATKTTTAHH |
Reverse complement motif Consensus sequence: HHTAAARATHD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
75 |
| Motif name: |
FOXF2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0278186 |
Alignment:
BHADSGTAAACAAD
--HHCAAAAACHH-
| Original motif Consensus sequence: BHADSGTAAACAAD |
Reverse complement motif Consensus sequence: DTTGTTTACSBTBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
67 |
| Motif name: |
FOXI1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0320335 |
Alignment:
KBDTRTTTRTTT
-HHGTTTTTGHH
| Original motif Consensus sequence: KBDTRTTTRTTT |
Reverse complement motif Consensus sequence: AAAMAAAMADBY |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 110 |
Motif name: cmCCACACCcm |
| Original motif Consensus sequence: HVCCACACCBV |
Reverse complement motif Consensus sequence: VBGGTGTGGBD |
|
|
 |
 |
Best Matches for Motif ID 110 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
111 |
| Motif name: |
ccCCmCaCCCCcc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.00944953 |
Alignment:
HBCCCCACCCCBH
HVCCACACCBV--
| Original motif Consensus sequence: HBCCCCACCCCBH |
Reverse complement motif Consensus sequence: DBGGGGTGGGGBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
93 |
| Motif name: |
cmCARCACTwr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0342522 |
Alignment:
BHCAGCACTHV
HVCCACACCBV
| Original motif Consensus sequence: BHCAGCACTHV |
Reverse complement motif Consensus sequence: VHAGTGCTGHB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
98 |
| Motif name: |
myyCCCACmTGCmyr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0444 |
Alignment:
HBCCCCACMTGCMHV
HVCCACACCBV----
| Original motif Consensus sequence: HBCCCCACMTGCMHV |
Reverse complement motif Consensus sequence: VDRGCAYGTGGGGVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
99 |
| Motif name: |
graGGGGGArr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0501309 |
Alignment:
DVAGGGGGADD
VBGGTGTGGBD
| Original motif Consensus sequence: DVAGGGGGADD |
Reverse complement motif Consensus sequence: HDTCCCCCTVH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
116 |
| Motif name: |
dyCTGCyTCyc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0510757 |
Alignment:
BHGAKGCAGHH
VBGGTGTGGBD
| Original motif Consensus sequence: DHCTGCYTCHB |
Reverse complement motif Consensus sequence: BHGAKGCAGHH |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 111 |
Motif name: ccCCmCaCCCCcc |
| Original motif Consensus sequence: HBCCCCACCCCBH |
Reverse complement motif Consensus sequence: DBGGGGTGGGGBD |
|
|
 |
 |
Best Matches for Motif ID 111 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
98 |
| Motif name: |
myyCCCACmTGCmyr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.0348277 |
Alignment:
HBCCCCACMTGCMHV
--HBCCCCACCCCBH
| Original motif Consensus sequence: HBCCCCACMTGCMHV |
Reverse complement motif Consensus sequence: VDRGCAYGTGGGGVD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
59 |
| Motif name: |
Motif 59 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.0360633 |
Alignment:
CTBCCTCHSYCYCC
-HBCCCCACCCCBH
| Original motif Consensus sequence: CTBCCTCHSYCYCC |
Reverse complement motif Consensus sequence: GGMGKSHGAGGBAG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
103 |
| Motif name: |
csCCaGCTCCCgs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.0413801 |
Alignment:
BBCCAGCTCCCVB
HBCCCCACCCCBH
| Original motif Consensus sequence: BBCCAGCTCCCVB |
Reverse complement motif Consensus sequence: BVGGGAGCTGGBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
Pax4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
13 |
| Similarity score: |
0.0432691 |
Alignment:
RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC
---------------HBCCCCACCCCBH--
| Original motif Consensus sequence: RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC |
Reverse complement motif Consensus sequence: GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
122 |
| Motif name: |
hsCCAGGCCTGGsr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.0554645 |
Alignment:
HBCCAGGCCTGGVD
HBCCCCACCCCBH-
| Original motif Consensus sequence: HVCCAGGCCTGGBD |
Reverse complement motif Consensus sequence: HBCCAGGCCTGGVD |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 112 |
Motif name: ctGAGTkTGAgg |
| Original motif Consensus sequence: VBGAGTKTGADD |
Reverse complement motif Consensus sequence: HHTCARACTCVV |
|
|
 |
 |
Best Matches for Motif ID 112 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
121 |
| Motif name: |
ctGAGTCATCkb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.00842365 |
Alignment:
BHGATGACTCDD
HHTCARACTCVV
| Original motif Consensus sequence: HDGAGTCATCDB |
Reverse complement motif Consensus sequence: BHGATGACTCDD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
111 |
| Motif name: |
ccCCmCaCCCCcc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0219353 |
Alignment:
DBGGGGTGGGGBD
VBGAGTKTGADD-
| Original motif Consensus sequence: HBCCCCACCCCBH |
Reverse complement motif Consensus sequence: DBGGGGTGGGGBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
98 |
| Motif name: |
myyCCCACmTGCmyr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0363368 |
Alignment:
HBCCCCACMTGCMHV
-HHTCARACTCVV--
| Original motif Consensus sequence: HBCCCCACMTGCMHV |
Reverse complement motif Consensus sequence: VDRGCAYGTGGGGVD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
Pax4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
16 |
| Number of overlap: |
12 |
| Similarity score: |
0.0370187 |
Alignment:
RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC
---HHTCARACTCVV---------------
| Original motif Consensus sequence: RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC |
Reverse complement motif Consensus sequence: GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
119 |
| Motif name: |
cbCCAGCTCmyk |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0389051 |
Alignment:
BBCCAGCTCMBD
HHTCARACTCVV
| Original motif Consensus sequence: BBCCAGCTCMBD |
Reverse complement motif Consensus sequence: HBYGAGCTGGBB |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 113 |
Motif name: htCTGyKTCbt |
| Original motif Consensus sequence: HBCTGYTTCBH |
Reverse complement motif Consensus sequence: HVGAAMCAGVH |
|
|
 |
 |
Best Matches for Motif ID 113 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
116 |
| Motif name: |
dyCTGCyTCyc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0 |
Alignment:
DHCTGCYTCHB
HBCTGYTTCBH
| Original motif Consensus sequence: DHCTGCYTCHB |
Reverse complement motif Consensus sequence: BHGAKGCAGHH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
108 |
| Motif name: |
wwATkTTTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0453878 |
Alignment:
DHATKTTTAHH
HBCTGYTTCBH
| Original motif Consensus sequence: DHATKTTTAHH |
Reverse complement motif Consensus sequence: HHTAAARATHD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
114 |
| Motif name: |
awAACAcAAwa |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0496817 |
Alignment:
BDTTGTGTTDD
HBCTGYTTCBH
| Original motif Consensus sequence: DDAACACAADV |
Reverse complement motif Consensus sequence: BDTTGTGTTDD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
121 |
| Motif name: |
ctGAGTCATCkb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0512287 |
Alignment:
HDGAGTCATCDB
HVGAAMCAGVH-
| Original motif Consensus sequence: HDGAGTCATCDB |
Reverse complement motif Consensus sequence: BHGATGACTCDD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
67 |
| Motif name: |
FOXI1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0525328 |
Alignment:
AAAMAAAMADBY
-HVGAAMCAGVH
| Original motif Consensus sequence: KBDTRTTTRTTT |
Reverse complement motif Consensus sequence: AAAMAAAMADBY |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 114 |
Motif name: awAACAcAAwa |
| Original motif Consensus sequence: DDAACACAADV |
Reverse complement motif Consensus sequence: BDTTGTGTTDD |
|
|
 |
 |
Best Matches for Motif ID 114 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
60 |
| Motif name: |
Motif 60 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0409581 |
Alignment:
AADAAAAMAAAAAM
-DDAACACAADV--
| Original motif Consensus sequence: AADAAAAMAAAAAM |
Reverse complement motif Consensus sequence: YTTTTTRTTTTDTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
71 |
| Motif name: |
Evi1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0441291 |
Alignment:
TBTTATCTTMTCTT
BDTTGTGTTDD---
| Original motif Consensus sequence: AAGAYAAGATAABA |
Reverse complement motif Consensus sequence: TBTTATCTTMTCTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
67 |
| Motif name: |
FOXI1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0515608 |
Alignment:
KBDTRTTTRTTT
-BDTTGTGTTDD
| Original motif Consensus sequence: KBDTRTTTRTTT |
Reverse complement motif Consensus sequence: AAAMAAAMADBY |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
76 |
| Motif name: |
Sox2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0542339 |
Alignment:
CCWTTGTYATGCAAA
---BDTTGTGTTDD-
| Original motif Consensus sequence: CCWTTGTYATGCAAA |
Reverse complement motif Consensus sequence: TTTGCATMACAAWGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
117 |
| Motif name: |
yaCATAYACay |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0554944 |
Alignment:
DTGTGTATGBD
BDTTGTGTTDD
| Original motif Consensus sequence: HVCATACACAH |
Reverse complement motif Consensus sequence: DTGTGTATGBD |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 115 |
Motif name: ysCmAGCACwy |
| Original motif Consensus sequence: HVCMAGCACHH |
Reverse complement motif Consensus sequence: DHGTGCTRGVH |
|
|
 |
 |
Best Matches for Motif ID 115 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
119 |
| Motif name: |
cbCCAGCTCmyk |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.00720731 |
Alignment:
BBCCAGCTCMBD
HVCMAGCACHH-
| Original motif Consensus sequence: BBCCAGCTCMBD |
Reverse complement motif Consensus sequence: HBYGAGCTGGBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
117 |
| Motif name: |
yaCATAYACay |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0367273 |
Alignment:
HVCATACACAH
HVCMAGCACHH
| Original motif Consensus sequence: HVCATACACAH |
Reverse complement motif Consensus sequence: DTGTGTATGBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
103 |
| Motif name: |
csCCaGCTCCCgs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0374067 |
Alignment:
BBCCAGCTCCCVB
HVCMAGCACHH--
| Original motif Consensus sequence: BBCCAGCTCCCVB |
Reverse complement motif Consensus sequence: BVGGGAGCTGGBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
123 |
| Motif name: |
asMAAGCTTKst |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0385747 |
Alignment:
HBCAAGCTTGVD
HVCMAGCACHH-
| Original motif Consensus sequence: HBCAAGCTTGVD |
Reverse complement motif Consensus sequence: DVCAAGCTTGBH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
102 |
| Motif name: |
ssCkGGYCCCsg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0395809 |
Alignment:
VVGGGGCCRGBB
-DHGTGCTRGVH
| Original motif Consensus sequence: BBCKGGCCCCVV |
Reverse complement motif Consensus sequence: VVGGGGCCRGBB |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 116 |
Motif name: dyCTGCyTCyc |
| Original motif Consensus sequence: DHCTGCYTCHB |
Reverse complement motif Consensus sequence: BHGAKGCAGHH |
|
|
 |
 |
Best Matches for Motif ID 116 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
113 |
| Motif name: |
htCTGyKTCbt |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0 |
Alignment:
HBCTGYTTCBH
DHCTGCYTCHB
| Original motif Consensus sequence: HBCTGYTTCBH |
Reverse complement motif Consensus sequence: HVGAAMCAGVH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
121 |
| Motif name: |
ctGAGTCATCkb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0409177 |
Alignment:
HDGAGTCATCDB
BHGAKGCAGHH-
| Original motif Consensus sequence: HDGAGTCATCDB |
Reverse complement motif Consensus sequence: BHGATGACTCDD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
110 |
| Motif name: |
cmCCACACCcm |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0482758 |
Alignment:
VBGGTGTGGBD
BHGAKGCAGHH
| Original motif Consensus sequence: HVCCACACCBV |
Reverse complement motif Consensus sequence: VBGGTGTGGBD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
102 |
| Motif name: |
ssCkGGYCCCsg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0492303 |
Alignment:
BBCKGGCCCCVV
DHCTGCYTCHB-
| Original motif Consensus sequence: BBCKGGCCCCVV |
Reverse complement motif Consensus sequence: VVGGGGCCRGBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
119 |
| Motif name: |
cbCCAGCTCmyk |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0496606 |
Alignment:
BBCCAGCTCMBD
-DHCTGCYTCHB
| Original motif Consensus sequence: BBCCAGCTCMBD |
Reverse complement motif Consensus sequence: HBYGAGCTGGBB |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 117 |
Motif name: yaCATAYACay |
| Original motif Consensus sequence: HVCATACACAH |
Reverse complement motif Consensus sequence: DTGTGTATGBD |
|
|
 |
 |
Best Matches for Motif ID 117 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
109 |
| Motif name: |
aaCAAAAACaa |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0144352 |
Alignment:
HHGTTTTTGHH
DTGTGTATGBD
| Original motif Consensus sequence: HHCAAAAACHH |
Reverse complement motif Consensus sequence: HHGTTTTTGHH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
75 |
| Motif name: |
FOXF2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0390604 |
Alignment:
DTTGTTTACSBTBB
-DTGTGTATGBD--
| Original motif Consensus sequence: BHADSGTAAACAAD |
Reverse complement motif Consensus sequence: DTTGTTTACSBTBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
112 |
| Motif name: |
ctGAGTkTGAgg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0405823 |
Alignment:
HHTCARACTCVV
HVCATACACAH-
| Original motif Consensus sequence: VBGAGTKTGADD |
Reverse complement motif Consensus sequence: HHTCARACTCVV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
115 |
| Motif name: |
ysCmAGCACwy |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0423793 |
Alignment:
HVCMAGCACHH
HVCATACACAH
| Original motif Consensus sequence: HVCMAGCACHH |
Reverse complement motif Consensus sequence: DHGTGCTRGVH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
58 |
| Motif name: |
Motif 58 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0462842 |
Alignment:
CACACACACACACA
HVCATACACAH---
| Original motif Consensus sequence: CACACACACACACA |
Reverse complement motif Consensus sequence: TGTGTGTGTGTGTG |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 118 |
Motif name: yrCACATGCATGTGyr |
| Original motif Consensus sequence: HVCACATGCATGTGBD |
Reverse complement motif Consensus sequence: DVCACATGCATGTGBH |
|
|
 |
 |
Best Matches for Motif ID 118 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
98 |
| Motif name: |
myyCCCACmTGCmyr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
0.0676289 |
Alignment:
-HBCCCCACMTGCMHV
HVCACATGCATGTGBD
| Original motif Consensus sequence: HBCCCCACMTGCMHV |
Reverse complement motif Consensus sequence: VDRGCAYGTGGGGVD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
58 |
| Motif name: |
Motif 58 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.573876 |
Alignment:
--CACACACACACACA
HVCACATGCATGTGBD
| Original motif Consensus sequence: CACACACACACACA |
Reverse complement motif Consensus sequence: TGTGTGTGTGTGTG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
80 |
| Motif name: |
Pou5f1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
13 |
| Similarity score: |
1.06805 |
Alignment:
ATTTGCATHACAAWG---
--HVCACATGCATGTGBD
| Original motif Consensus sequence: CWTTGTHATGCAAAT |
Reverse complement motif Consensus sequence: ATTTGCATHACAAWG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
76 |
| Motif name: |
Sox2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
13 |
| Similarity score: |
1.07558 |
Alignment:
CCWTTGTYATGCAAA---
--DVCACATGCATGTGBH
| Original motif Consensus sequence: CCWTTGTYATGCAAA |
Reverse complement motif Consensus sequence: TTTGCATMACAAWGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
126 |
| Motif name: |
yvTGCAGsCAcg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.55231 |
Alignment:
BVTGCAGSCABV----
HVCACATGCATGTGBD
| Original motif Consensus sequence: BVTGCAGSCABV |
Reverse complement motif Consensus sequence: VBTGSCTGCAVB |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 119 |
Motif name: cbCCAGCTCmyk |
| Original motif Consensus sequence: BBCCAGCTCMBD |
Reverse complement motif Consensus sequence: HBYGAGCTGGBB |
|
|
 |
 |
Best Matches for Motif ID 119 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
103 |
| Motif name: |
csCCaGCTCCCgs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.00141375 |
Alignment:
BBCCAGCTCCCVB
BBCCAGCTCMBD-
| Original motif Consensus sequence: BBCCAGCTCCCVB |
Reverse complement motif Consensus sequence: BVGGGAGCTGGBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
123 |
| Motif name: |
asMAAGCTTKst |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0040263 |
Alignment:
DVCAAGCTTGBH
HBYGAGCTGGBB
| Original motif Consensus sequence: HBCAAGCTTGVD |
Reverse complement motif Consensus sequence: DVCAAGCTTGBH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
122 |
| Motif name: |
hsCCAGGCCTGGsr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0363393 |
Alignment:
HVCCAGGCCTGGBD
BBCCAGCTCMBD--
| Original motif Consensus sequence: HVCCAGGCCTGGBD |
Reverse complement motif Consensus sequence: HBCCAGGCCTGGVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
102 |
| Motif name: |
ssCkGGYCCCsg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0378472 |
Alignment:
BBCKGGCCCCVV
BBCCAGCTCMBD
| Original motif Consensus sequence: BBCKGGCCCCVV |
Reverse complement motif Consensus sequence: VVGGGGCCRGBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
111 |
| Motif name: |
ccCCmCaCCCCcc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0419313 |
Alignment:
HBCCCCACCCCBH
-BBCCAGCTCMBD
| Original motif Consensus sequence: HBCCCCACCCCBH |
Reverse complement motif Consensus sequence: DBGGGGTGGGGBD |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 120 |
Motif name: wtTTTwAAAaw |
| Original motif Consensus sequence: HHTTTWAAADD |
Reverse complement motif Consensus sequence: DDTTTWAAAHH |
|
|
 |
 |
Best Matches for Motif ID 120 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
108 |
| Motif name: |
wwATkTTTAww |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0769117 |
Alignment:
HHTAAARATHD
HHTTTWAAADD
| Original motif Consensus sequence: DHATKTTTAHH |
Reverse complement motif Consensus sequence: HHTAAARATHD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
75 |
| Motif name: |
FOXF2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0772648 |
Alignment:
DTTGTTTACSBTBB
DDTTTWAAAHH---
| Original motif Consensus sequence: BHADSGTAAACAAD |
Reverse complement motif Consensus sequence: DTTGTTTACSBTBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
Lhx3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0876632 |
Alignment:
BMWTAATTAATTW
--DDTTTWAAAHH
| Original motif Consensus sequence: WAATTAATTAWYB |
Reverse complement motif Consensus sequence: BMWTAATTAATTW |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
62 |
| Motif name: |
TBP |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0881502 |
Alignment:
STATAAAWRVVVVBV
HHTTTWAAADD----
| Original motif Consensus sequence: STATAAAWRVVVVBV |
Reverse complement motif Consensus sequence: VBVVVVMWTTTATAS |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
114 |
| Motif name: |
awAACAcAAwa |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.090069 |
Alignment:
BDTTGTGTTDD
HHTTTWAAADD
| Original motif Consensus sequence: DDAACACAADV |
Reverse complement motif Consensus sequence: BDTTGTGTTDD |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 121 |
Motif name: ctGAGTCATCkb |
| Original motif Consensus sequence: HDGAGTCATCDB |
Reverse complement motif Consensus sequence: BHGATGACTCDD |
|
|
 |
 |
Best Matches for Motif ID 121 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
112 |
| Motif name: |
ctGAGTkTGAgg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0322402 |
Alignment:
HHTCARACTCVV
BHGATGACTCDD
| Original motif Consensus sequence: VBGAGTKTGADD |
Reverse complement motif Consensus sequence: HHTCARACTCVV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
122 |
| Motif name: |
hsCCAGGCCTGGsr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0510995 |
Alignment:
HBCCAGGCCTGGVD
-HDGAGTCATCDB-
| Original motif Consensus sequence: HVCCAGGCCTGGBD |
Reverse complement motif Consensus sequence: HBCCAGGCCTGGVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
102 |
| Motif name: |
ssCkGGYCCCsg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0569923 |
Alignment:
BBCKGGCCCCVV
BHGATGACTCDD
| Original motif Consensus sequence: BBCKGGCCCCVV |
Reverse complement motif Consensus sequence: VVGGGGCCRGBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
123 |
| Motif name: |
asMAAGCTTKst |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0601978 |
Alignment:
DVCAAGCTTGBH
HDGAGTCATCDB
| Original motif Consensus sequence: HBCAAGCTTGVD |
Reverse complement motif Consensus sequence: DVCAAGCTTGBH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
Pax4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
11 |
| Number of overlap: |
12 |
| Similarity score: |
0.0641337 |
Alignment:
RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC
----------BHGATGACTCDD--------
| Original motif Consensus sequence: RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC |
Reverse complement motif Consensus sequence: GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 122 |
Motif name: hsCCAGGCCTGGsr |
| Original motif Consensus sequence: HVCCAGGCCTGGBD |
Reverse complement motif Consensus sequence: HBCCAGGCCTGGVD |
|
|
 |
 |
Best Matches for Motif ID 122 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
98 |
| Motif name: |
myyCCCACmTGCmyr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0170832 |
Alignment:
HBCCCCACMTGCMHV
HVCCAGGCCTGGBD-
| Original motif Consensus sequence: HBCCCCACMTGCMHV |
Reverse complement motif Consensus sequence: VDRGCAYGTGGGGVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
118 |
| Motif name: |
yrCACATGCATGTGyr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0217283 |
Alignment:
DVCACATGCATGTGBH
-HBCCAGGCCTGGVD-
| Original motif Consensus sequence: HVCACATGCATGTGBD |
Reverse complement motif Consensus sequence: DVCACATGCATGTGBH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
Pax4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
14 |
| Similarity score: |
0.0375854 |
Alignment:
RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC
--------HBCCAGGCCTGGVD--------
| Original motif Consensus sequence: RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC |
Reverse complement motif Consensus sequence: GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
103 |
| Motif name: |
csCCaGCTCCCgs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.516252 |
Alignment:
-BBCCAGCTCCCVB
HBCCAGGCCTGGVD
| Original motif Consensus sequence: BBCCAGCTCCCVB |
Reverse complement motif Consensus sequence: BVGGGAGCTGGBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
111 |
| Motif name: |
ccCCmCaCCCCcc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.531063 |
Alignment:
DBGGGGTGGGGBD-
HVCCAGGCCTGGBD
| Original motif Consensus sequence: HBCCCCACCCCBH |
Reverse complement motif Consensus sequence: DBGGGGTGGGGBD |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 123 |
Motif name: asMAAGCTTKst |
| Original motif Consensus sequence: HBCAAGCTTGVD |
Reverse complement motif Consensus sequence: DVCAAGCTTGBH |
|
|
 |
 |
Best Matches for Motif ID 123 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
119 |
| Motif name: |
cbCCAGCTCmyk |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0 |
Alignment:
HBYGAGCTGGBB
DVCAAGCTTGBH
| Original motif Consensus sequence: BBCCAGCTCMBD |
Reverse complement motif Consensus sequence: HBYGAGCTGGBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
122 |
| Motif name: |
hsCCAGGCCTGGsr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.025923 |
Alignment:
HBCCAGGCCTGGVD
-DVCAAGCTTGBH-
| Original motif Consensus sequence: HVCCAGGCCTGGBD |
Reverse complement motif Consensus sequence: HBCCAGGCCTGGVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
103 |
| Motif name: |
csCCaGCTCCCgs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0387894 |
Alignment:
BBCCAGCTCCCVB
HBCAAGCTTGVD-
| Original motif Consensus sequence: BBCCAGCTCCCVB |
Reverse complement motif Consensus sequence: BVGGGAGCTGGBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
71 |
| Motif name: |
Evi1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0417163 |
Alignment:
TBTTATCTTMTCTT
--HBCAAGCTTGVD
| Original motif Consensus sequence: AAGAYAAGATAABA |
Reverse complement motif Consensus sequence: TBTTATCTTMTCTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
121 |
| Motif name: |
ctGAGTCATCkb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0518025 |
Alignment:
BHGATGACTCDD
HBCAAGCTTGVD
| Original motif Consensus sequence: HDGAGTCATCDB |
Reverse complement motif Consensus sequence: BHGATGACTCDD |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 124 |
Motif name: cswGGGCCCwsg |
| Original motif Consensus sequence: VVWGGGCCCWBB |
Reverse complement motif Consensus sequence: BBWGGGCCCWVV |
|
|
 |
 |
Best Matches for Motif ID 124 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
102 |
| Motif name: |
ssCkGGYCCCsg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0170635 |
Alignment:
BBCKGGCCCCVV
VVWGGGCCCWBB
| Original motif Consensus sequence: BBCKGGCCCCVV |
Reverse complement motif Consensus sequence: VVGGGGCCRGBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
119 |
| Motif name: |
cbCCAGCTCmyk |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0400014 |
Alignment:
HBYGAGCTGGBB
VVWGGGCCCWBB
| Original motif Consensus sequence: BBCCAGCTCMBD |
Reverse complement motif Consensus sequence: HBYGAGCTGGBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
126 |
| Motif name: |
yvTGCAGsCAcg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0429334 |
Alignment:
BVTGCAGSCABV
VVWGGGCCCWBB
| Original motif Consensus sequence: BVTGCAGSCABV |
Reverse complement motif Consensus sequence: VBTGSCTGCAVB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
122 |
| Motif name: |
hsCCAGGCCTGGsr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0467887 |
Alignment:
HVCCAGGCCTGGBD
-VVWGGGCCCWBB-
| Original motif Consensus sequence: HVCCAGGCCTGGBD |
Reverse complement motif Consensus sequence: HBCCAGGCCTGGVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
103 |
| Motif name: |
csCCaGCTCCCgs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0482771 |
Alignment:
BBCCAGCTCCCVB
-BBWGGGCCCWVV
| Original motif Consensus sequence: BBCCAGCTCCCVB |
Reverse complement motif Consensus sequence: BVGGGAGCTGGBB |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 125 |
Motif name: wwTAATwwATTAww |
| Original motif Consensus sequence: DDTAATWWATTAHH |
Reverse complement motif Consensus sequence: HHTAATWWATTADD |
|
|
 |
 |
Best Matches for Motif ID 125 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
68 |
| Motif name: |
HNF1A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0447485 |
Alignment:
GGTTAATDATTAMC
DDTAATWWATTAHH
| Original motif Consensus sequence: GGTTAATDATTAMC |
Reverse complement motif Consensus sequence: GYTAATDATTAACC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
81 |
| Motif name: |
Pax4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0620115 |
Alignment:
GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM
---------------HHTAATWWATTADD-
| Original motif Consensus sequence: RAAWAWTWDCMVHHVBBHHHVHMHCMCYMC |
Reverse complement motif Consensus sequence: GRKGRGDRHVHDDVBBHHBRGDWAWTWTTM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
75 |
| Motif name: |
FOXF2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0703085 |
Alignment:
BHADSGTAAACAAD
HHTAATWWATTADD
| Original motif Consensus sequence: BHADSGTAAACAAD |
Reverse complement motif Consensus sequence: DTTGTTTACSBTBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
62 |
| Motif name: |
TBP |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0724742 |
Alignment:
STATAAAWRVVVVBV
HHTAATWWATTADD-
| Original motif Consensus sequence: STATAAAWRVVVVBV |
Reverse complement motif Consensus sequence: VBVVVVMWTTTATAS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
76 |
| Motif name: |
Sox2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0728076 |
Alignment:
TTTGCATMACAAWGG
-DDTAATWWATTAHH
| Original motif Consensus sequence: CCWTTGTYATGCAAA |
Reverse complement motif Consensus sequence: TTTGCATMACAAWGG |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 126 |
Motif name: yvTGCAGsCAcg |
| Original motif Consensus sequence: BVTGCAGSCABV |
Reverse complement motif Consensus sequence: VBTGSCTGCAVB |
|
|
 |
 |
Best Matches for Motif ID 126 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
104 |
| Motif name: |
cgCCCwGGScsg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0284753 |
Alignment:
VDCCCWGGGCBB
BVTGCAGSCABV
| Original motif Consensus sequence: VDCCCWGGGCBB |
Reverse complement motif Consensus sequence: BBGCCCWGGGHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
118 |
| Motif name: |
yrCACATGCATGTGyr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0340354 |
Alignment:
HVCACATGCATGTGBD
BVTGCAGSCABV----
| Original motif Consensus sequence: HVCACATGCATGTGBD |
Reverse complement motif Consensus sequence: DVCACATGCATGTGBH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
98 |
| Motif name: |
myyCCCACmTGCmyr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0351986 |
Alignment:
VDRGCAYGTGGGGVD
---BVTGCAGSCABV
| Original motif Consensus sequence: HBCCCCACMTGCMHV |
Reverse complement motif Consensus sequence: VDRGCAYGTGGGGVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
124 |
| Motif name: |
cswGGGCCCwsg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0357442 |
Alignment:
VVWGGGCCCWBB
BVTGCAGSCABV
| Original motif Consensus sequence: VVWGGGCCCWBB |
Reverse complement motif Consensus sequence: BBWGGGCCCWVV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
103 |
| Motif name: |
csCCaGCTCCCgs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0445722 |
Alignment:
BBCCAGCTCCCVB
VBTGSCTGCAVB-
| Original motif Consensus sequence: BBCCAGCTCCCVB |
Reverse complement motif Consensus sequence: BVGGGAGCTGGBB |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 127 |
Motif name: rryCAGGGAyw |
| Original motif Consensus sequence: DDYCAGGGAHD |
Reverse complement motif Consensus sequence: DHTCCCTGMDD |
|
|
 |
 |
Best Matches for Motif ID 127 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
99 |
| Motif name: |
graGGGGGArr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0283253 |
Alignment:
DVAGGGGGADD
DDYCAGGGAHD
| Original motif Consensus sequence: DVAGGGGGADD |
Reverse complement motif Consensus sequence: HDTCCCCCTVH |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
126 |
| Motif name: |
yvTGCAGsCAcg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0326364 |
Alignment:
VBTGSCTGCAVB
DHTCCCTGMDD-
| Original motif Consensus sequence: BVTGCAGSCABV |
Reverse complement motif Consensus sequence: VBTGSCTGCAVB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
104 |
| Motif name: |
cgCCCwGGScsg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0424475 |
Alignment:
VDCCCWGGGCBB
-DDYCAGGGAHD
| Original motif Consensus sequence: VDCCCWGGGCBB |
Reverse complement motif Consensus sequence: BBGCCCWGGGHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
122 |
| Motif name: |
hsCCAGGCCTGGsr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0451737 |
Alignment:
HVCCAGGCCTGGBD
DDYCAGGGAHD---
| Original motif Consensus sequence: HVCCAGGCCTGGBD |
Reverse complement motif Consensus sequence: HBCCAGGCCTGGVD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
119 |
| Motif name: |
cbCCAGCTCmyk |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0514394 |
Alignment:
BBCCAGCTCMBD
DDYCAGGGAHD-
| Original motif Consensus sequence: BBCCAGCTCMBD |
Reverse complement motif Consensus sequence: HBYGAGCTGGBB |
|
|
 |
 |