Best Matches in Database for Each Motif (Highest to Lowest)
| Dataset #: 1 | Motif ID: 6 | Motif name: Motif 6 |
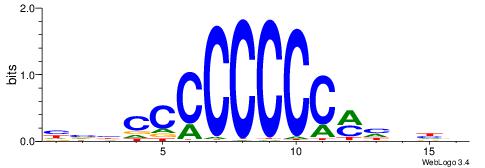
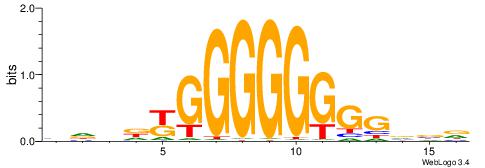
| Original motif | Reverse complement motif |
| Consensus sequence: CCCASCMC | Consensus sequence: GYGSTGGG |
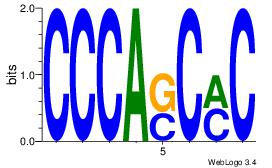 |
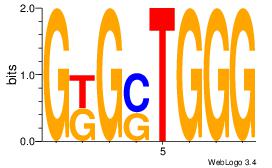 |
Best Matches for Motif ID 6 (Highest to Lowest)
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.00921145 |
Alignment:
HBDRCCMCGCCCHHHD
----CCCASCMC----
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
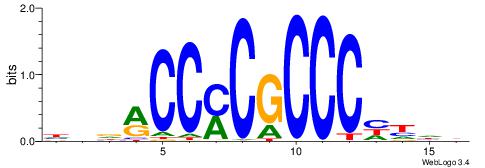 |
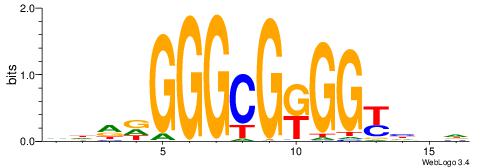 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 13 |
| Number of overlap: | 8 |
| Similarity score: | 0.00924564 |
Alignment:
DBCDWHCTGGGTGGTCMDHBBAH
---GYGSTGGG------------
| Original motif | Reverse complement motif |
| Consensus sequence: DTVBDDRGACCACCCAGDWDGBH | Consensus sequence: DBCDWHCTGGGTGGTCMDHBBAH |
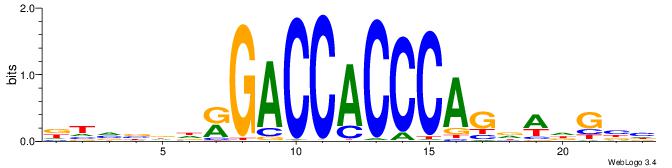 |
 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0121799 |
Alignment:
DBCCCCCCCCCCMYC
--CCCASCMC-----
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
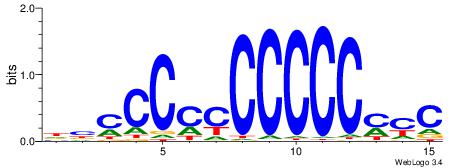 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.014495 |
Alignment:
BDDYYCATCCCATDDBD
---CCCASCMC------
| Original motif | Reverse complement motif |
| Consensus sequence: DVDDATGGGATGKMDDV | Consensus sequence: BDDYYCATCCCATDDBD |
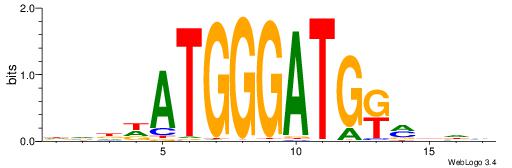 |
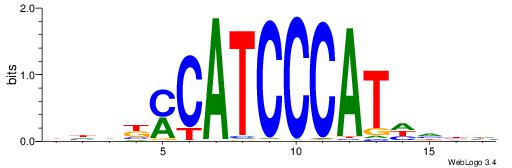 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0175119 |
Alignment:
BHHDYGGGGGGGGBVD
---GYGSTGGG-----
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
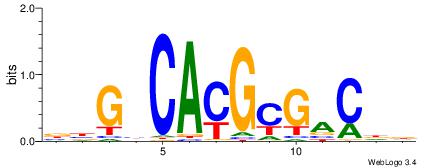
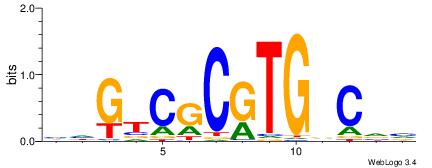
| Dataset #: 1 | Motif ID: 24 | Motif name: Motif 24 |
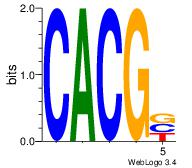
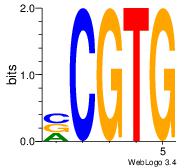
| Original motif | Reverse complement motif |
| Consensus sequence: CACGB | Consensus sequence: BCGTG |
 |
 |
Best Matches for Motif ID 24 (Highest to Lowest)
| Motif ID: | UP00097 |
| Motif name: | Mtf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 5 |
| Similarity score: | 0 |
Alignment:
HDYTTGCACACGDHBH
--------CACGB---
| Original motif | Reverse complement motif |
| Consensus sequence: DBDHCGTGTGCAAMDH | Consensus sequence: HDYTTGCACACGDHBH |
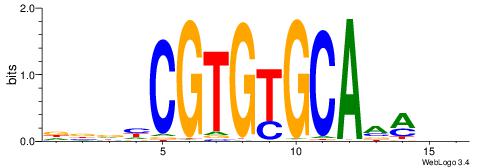 |
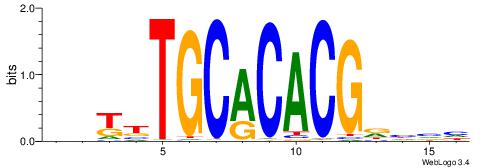 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 11 |
| Number of overlap: | 5 |
| Similarity score: | 0.0184689 |
Alignment:
YDYBDHTMCACGTGGADDBMDGT
--------CACGB----------
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
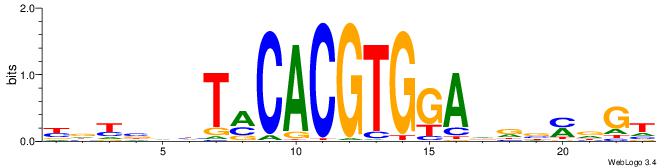 |
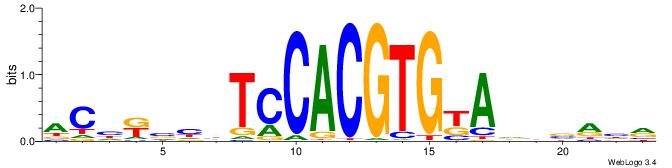 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 5 |
| Similarity score: | 0.0193222 |
Alignment:
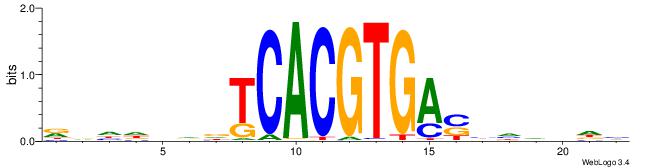
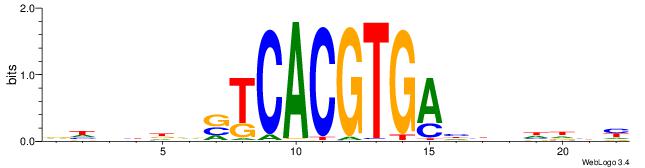
RDHDBVDTCACGTGASBHVHDH
--------CACGB---------
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 5 |
| Similarity score: | 0.0231779 |
Alignment:
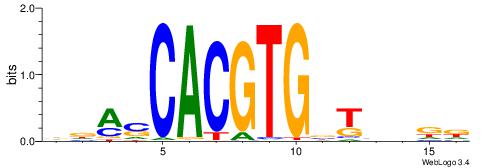
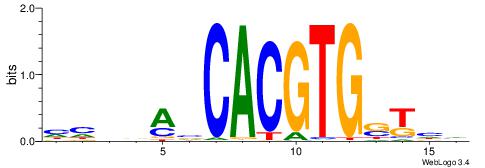
HHVBABCACGTGSTHD
------CACGB-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDASCACGTGBTBVDD | Consensus sequence: HHVBABCACGTGSTHD |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.02459 |
Alignment:
HDGTCGCGTGDCVB
-----BCGTG----
| Original motif | Reverse complement motif |
| Consensus sequence: BBGHCACGCGACDD | Consensus sequence: HDGTCGCGTGDCVB |
 |
 |
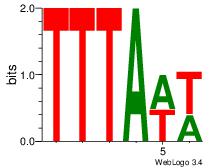
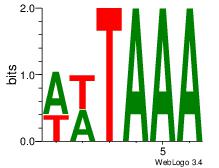
| Dataset #: 1 | Motif ID: 25 | Motif name: Motif 25 |
| Original motif | Reverse complement motif |
| Consensus sequence: TTTAWW | Consensus sequence: WWTAAA |
 |
 |
Best Matches for Motif ID 25 (Highest to Lowest)
| Motif ID: | UP00180 |
| Motif name: | Hoxd13 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
HDHHTTTTATTKKHDD
-----TTTAWW-----
| Original motif | Reverse complement motif |
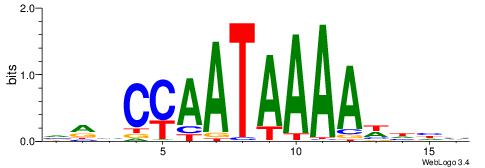
| Consensus sequence: HDHYYAATAAAAHHHH | Consensus sequence: HDHHTTTTATTKKHDD |
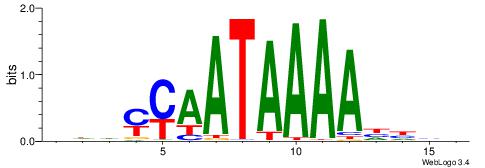 |
 |
| Motif ID: | UP00134 |
| Motif name: | Hoxb13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.00930561 |
Alignment:
VRHCCAATAAAAWHHV
-----WWTAAA-----
| Original motif | Reverse complement motif |
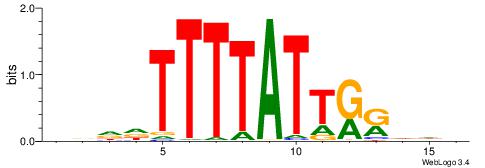
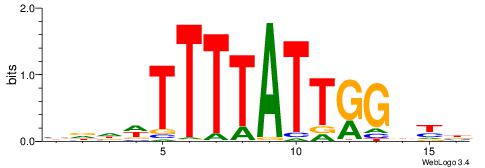
| Consensus sequence: VRHCCAATAAAAWHHV | Consensus sequence: VDHWTTTTATTGGDKB |
 |
 |
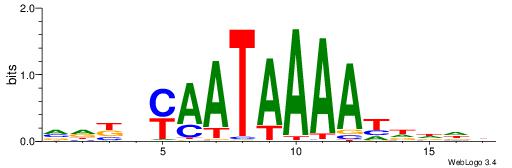
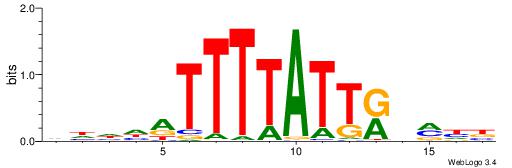
| Motif ID: | UP00121 |
| Motif name: | Hoxd10 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.0106116 |
Alignment:
HDDDATTTTATTKVRDB
------TTTAWW-----
| Original motif | Reverse complement motif |
| Consensus sequence: VDKVYAATAAAATDDDH | Consensus sequence: HDDDATTTTATTKVRDB |
 |
 |
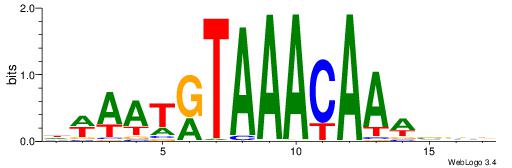
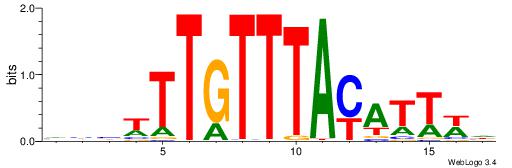
| Motif ID: | UP00061 |
| Motif name: | Foxl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.0135319 |
Alignment:
DWAATRTAAACAAWVVB
----WWTAAA-------
| Original motif | Reverse complement motif |
| Consensus sequence: DWAATRTAAACAAWVVB | Consensus sequence: VVVWTTGTTTAMATTWD |
 |
 |
| Motif ID: | UP00133 |
| Motif name: | Cdx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0138323 |
Alignment:
HRHTTTTATYMCCVBD
----TTTAWW------
| Original motif | Reverse complement motif |
| Consensus sequence: DVVGGYMATAAAAHKH | Consensus sequence: HRHTTTTATYMCCVBD |
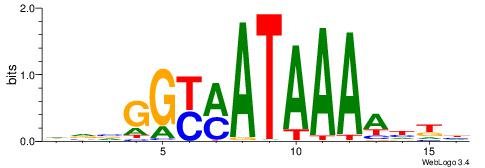 |
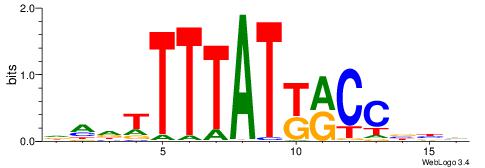 |
| Dataset #: 2 | Motif ID: 53 | Motif name: Motif 53 |
| Original motif | Reverse complement motif |
| Consensus sequence: TSTGTR | Consensus sequence: MACASA |
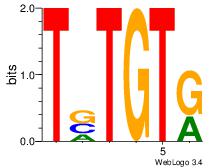 |
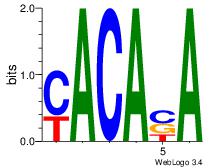 |
Best Matches for Motif ID 53 (Highest to Lowest)
| Motif ID: | UP00026 |
| Motif name: | Zscan4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
BHHHTTGTGTGCKHVD
-----TSTGTR-----
| Original motif | Reverse complement motif |
| Consensus sequence: HVHRGCACACAAHHHV | Consensus sequence: BHHHTTGTGTGCKHVD |
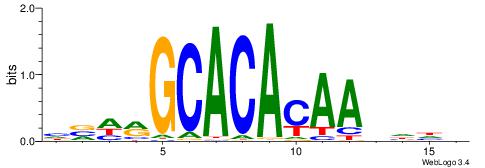 |
 |
| Motif ID: | UP00042 |
| Motif name: | Gm397_second |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.00423357 |
Alignment:
HBGYGTGTGTGCVRVD
-----TSTGTR-----
| Original motif | Reverse complement motif |
| Consensus sequence: DVMVGCACACACKCVH | Consensus sequence: HBGYGTGTGTGCVRVD |
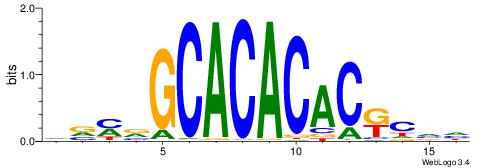 |
 |
| Motif ID: | UP00026 |
| Motif name: | Zscan4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.00980483 |
Alignment:
HDHTMTGTGCACADVHD
---TSTGTR--------
| Original motif | Reverse complement motif |
| Consensus sequence: DHVDTGTGCACAYAHDH | Consensus sequence: HDHTMTGTGCACADVHD |
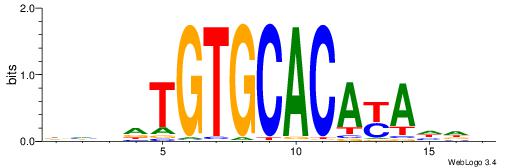 |
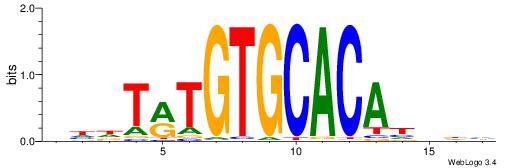 |
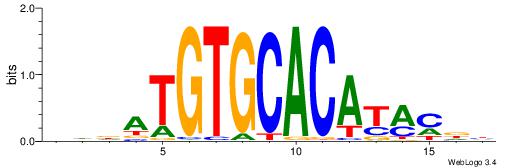
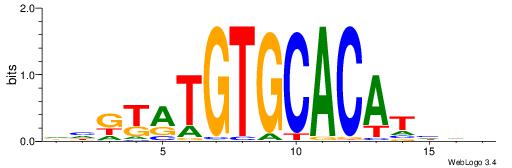
| Motif ID: | UP00042 |
| Motif name: | Gm397_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.0166332 |
Alignment:
HHRTATGTGCACATHHD
---TSTGTR--------
| Original motif | Reverse complement motif |
| Consensus sequence: HHDATGTGCACATAMDH | Consensus sequence: HHRTATGTGCACATHHD |
 |
 |
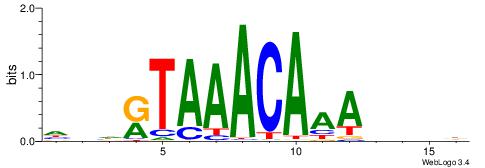
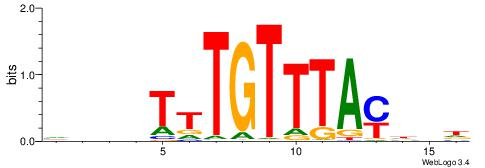
| Motif ID: | UP00041 |
| Motif name: | Foxj1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0213483 |
Alignment:
HDHRTAAACAAAHDDD
------MACASA----
| Original motif | Reverse complement motif |
| Consensus sequence: HDHRTAAACAAAHDDD | Consensus sequence: DDDHTTTGTTTAMHDH |
 |
 |
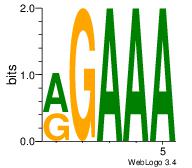
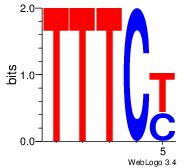
| Dataset #: 2 | Motif ID: 54 | Motif name: Motif 54 |
| Original motif | Reverse complement motif |
| Consensus sequence: RGAAA | Consensus sequence: TTTCK |
 |
 |
Best Matches for Motif ID 54 (Highest to Lowest)
| Motif ID: | UP00018 |
| Motif name: | Irf4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0 |
Alignment:
TBDKGTTTCGMTDHV
-----TTTCK-----
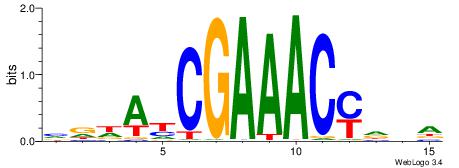
| Original motif | Reverse complement motif |
| Consensus sequence: VDDAYCGAAACYDVA | Consensus sequence: TBDKGTTTCGMTDHV |
 |
 |
| Motif ID: | UP00074 |
| Motif name: | Isgf3g_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 5 |
| Similarity score: | 0.00225908 |
Alignment:
TDAGTTTCGWTTKDV
----TTTCK------
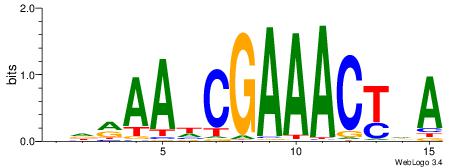
| Original motif | Reverse complement motif |
| Consensus sequence: VDRAAWCGAAACTDA | Consensus sequence: TDAGTTTCGWTTKDV |
 |
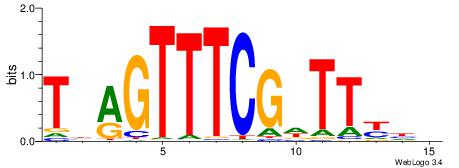 |
| Motif ID: | UP00040 |
| Motif name: | Irf5_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 5 |
| Similarity score: | 0.00434877 |
Alignment:
BDKGTTTCGKTWDDD
----TTTCK------
| Original motif | Reverse complement motif |
| Consensus sequence: DDDWAYCGAAACYDV | Consensus sequence: BDKGTTTCGKTWDDD |
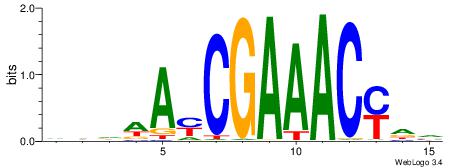 |
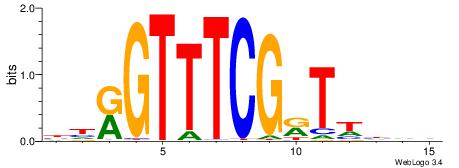 |
| Motif ID: | UP00011 |
| Motif name: | Irf6_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.0136873 |
Alignment:
DHDBTKGTTTCGHTHDD
-------TTTCK-----
| Original motif | Reverse complement motif |
| Consensus sequence: HDDAHCGAAACYAVDDD | Consensus sequence: DHDBTKGTTTCGHTHDD |
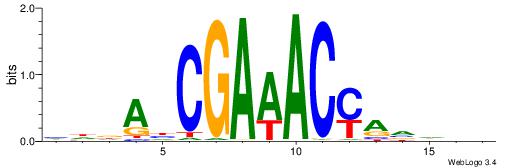 |
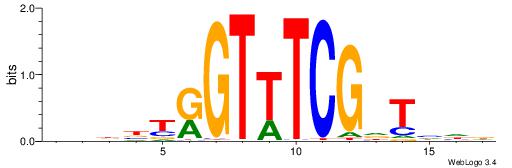 |
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 10 |
| Number of overlap: | 5 |
| Similarity score: | 0.0141224 |
Alignment:
DCTTTCKAGGAATHHV
--TTTCK---------
| Original motif | Reverse complement motif |
| Consensus sequence: DCTTTCKAGGAATHHV | Consensus sequence: VHHATTCCTYGAAAGD |
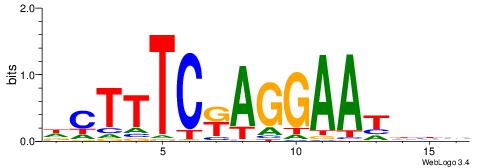 |
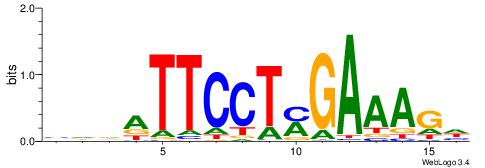 |
| Dataset #: 2 | Motif ID: 55 | Motif name: Motif 55 |
| Original motif | Reverse complement motif |
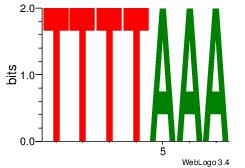
| Consensus sequence: TTTAAAA | Consensus sequence: TTTTAAA |
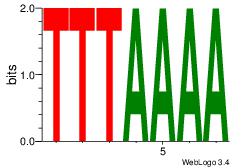 |
 |
Best Matches for Motif ID 55 (Highest to Lowest)
| Motif ID: | UP00173 |
| Motif name: | Hoxc13 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0 |
Alignment:
DHWTTTTAMGAGBDHH
----TTTAAAA-----
| Original motif | Reverse complement motif |
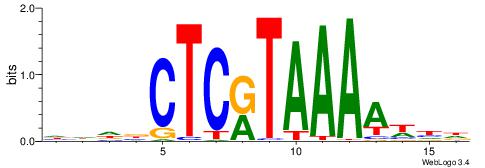
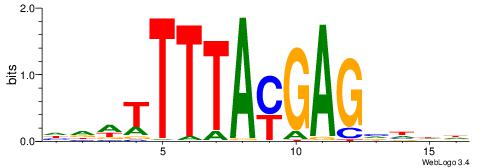
| Consensus sequence: HHDBCTCRTAAAAWHD | Consensus sequence: DHWTTTTAMGAGBDHH |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 7 |
| Similarity score: | 0.00269372 |
Alignment:
VVVWTTGTTTAMATTWD
-------TTTAAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: DWAATRTAAACAAWVVB | Consensus sequence: VVVWTTGTTTAMATTWD |
 |
 |
| Motif ID: | UP00245 |
| Motif name: | Hoxc10 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0.00395798 |
Alignment:
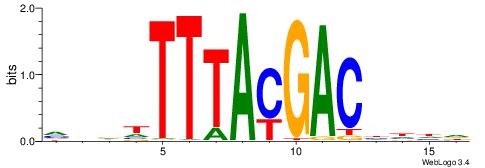
VBDDTTTACGACDDDH
----TTTAAAA-----
| Original motif | Reverse complement motif |
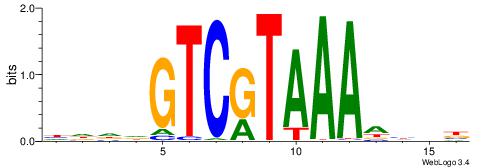
| Consensus sequence: HDDDGTCGTAAADHBB | Consensus sequence: VBDDTTTACGACDDDH |
 |
 |
| Motif ID: | UP00246 |
| Motif name: | Hoxa11 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.00543955 |
Alignment:
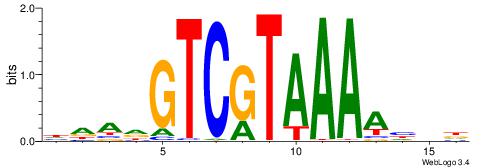
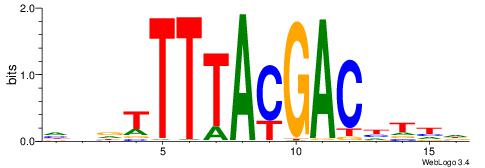
DDDTTTTACGACDTDH
----TTTAAAA-----
| Original motif | Reverse complement motif |
| Consensus sequence: HDADGTCGTAAAAHDD | Consensus sequence: DDDTTTTACGACDTDH |
 |
 |
| Motif ID: | UP00183 |
| Motif name: | Hoxa13 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0.00548253 |
Alignment:
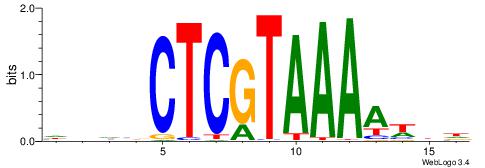
DVWTTTTACGAGBHDH
----TTTAAAA-----
| Original motif | Reverse complement motif |
| Consensus sequence: HDHBCTCGTAAAAWBD | Consensus sequence: DVWTTTTACGAGBHDH |
 |
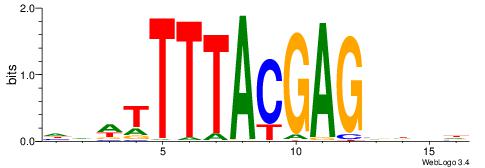 |
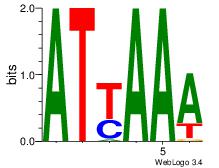
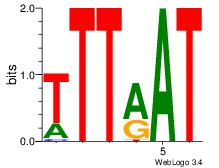
| Dataset #: 3 | Motif ID: 64 | Motif name: ARID3A |
| Original motif | Reverse complement motif |
| Consensus sequence: ATYAAA | Consensus sequence: TTTMAT |
 |
 |
Best Matches for Motif ID 64 (Highest to Lowest)
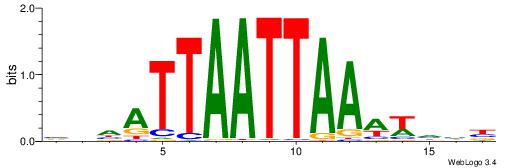
| Motif ID: | UP00212 |
| Motif name: | Lhx5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
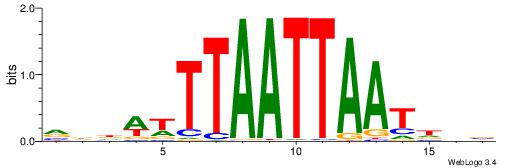
VDHATTAATTAAAWHHB
-------ATYAAA----
| Original motif | Reverse complement motif |
| Consensus sequence: VDHATTAATTAAAWHHB | Consensus sequence: VDHWTTTAATTAATHHV |
 |
 |
| Motif ID: | UP00130 |
| Motif name: | Lhx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.00146194 |
Alignment:
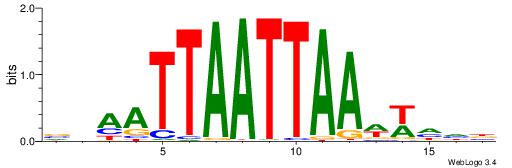
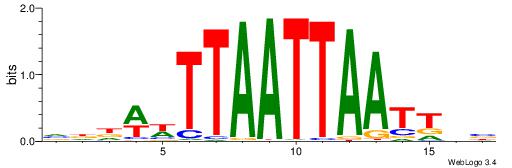
VHHWWTTAATTAATTDV
----TTTMAT-------
| Original motif | Reverse complement motif |
| Consensus sequence: VDAATTAATTAAWWHHB | Consensus sequence: VHHWWTTAATTAATTDV |
 |
 |
| Motif ID: | UP00169 |
| Motif name: | Lmx1b |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.00211796 |
Alignment:
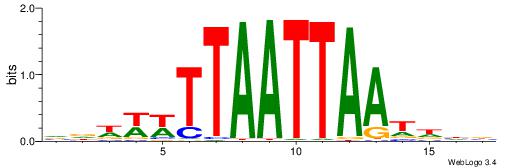
BHDATTAATTAAWWWHB
-------ATYAAA----
| Original motif | Reverse complement motif |
| Consensus sequence: VDWWWTTAATTAATDHB | Consensus sequence: BHDATTAATTAAWWWHB |
 |
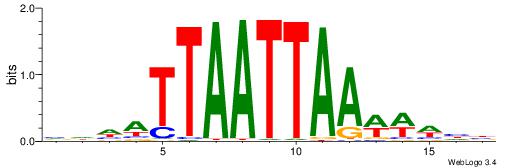 |
| Motif ID: | UP00213 |
| Motif name: | Hoxa9 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.00270371 |
Alignment:
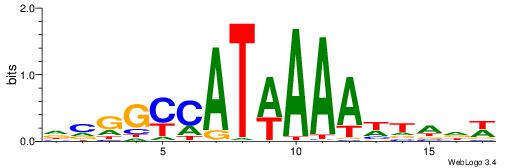
DCGGYCATWAAAWTADW
------ATYAAA-----
| Original motif | Reverse complement motif |
| Consensus sequence: DCGGYCATWAAAWTADW | Consensus sequence: WDTAWTTTWATGKCCGD |
 |
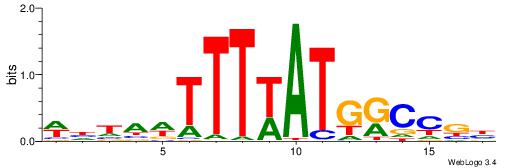 |
| Motif ID: | UP00188 |
| Motif name: | Lmx1a |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.00429494 |
Alignment:
BDWWWTTAATTAAHHVG
----TTTMAT-------
| Original motif | Reverse complement motif |
| Consensus sequence: CVHHTTAATTAAWWWHB | Consensus sequence: BDWWWTTAATTAAHHVG |
 |
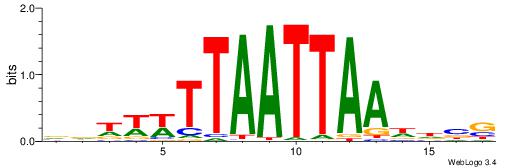 |
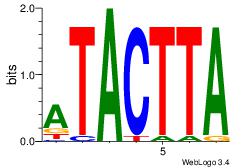
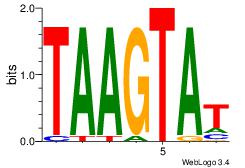
| Dataset #: 3 | Motif ID: 69 | Motif name: NKX3-1 |
| Original motif | Reverse complement motif |
| Consensus sequence: ATACTTA | Consensus sequence: TAAGTAT |
 |
 |
Best Matches for Motif ID 69 (Highest to Lowest)
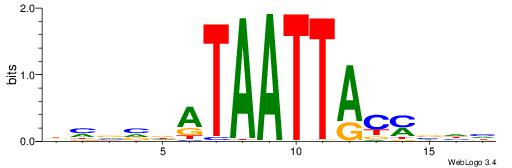
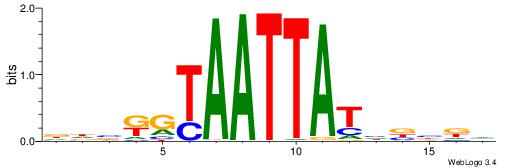
| Motif ID: | UP00154 |
| Motif name: | Dlx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0 |
Alignment:
DHVRGTAATTATHVVDV
-----TAAGTAT-----
| Original motif | Reverse complement motif |
| Consensus sequence: BHVVDATAATTACMVHH | Consensus sequence: DHVRGTAATTATHVVDV |
 |
 |
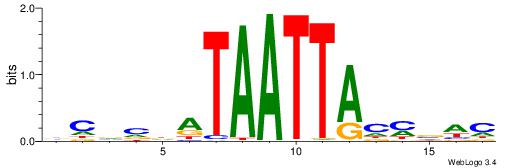
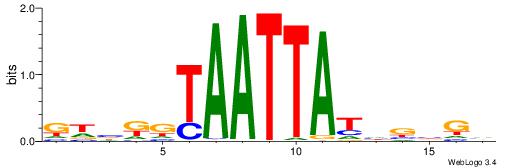
| Motif ID: | UP00110 |
| Motif name: | Dlx4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.000606776 |
Alignment:
GHVRGTAATTATDVVGV
-----TAAGTAT-----
| Original motif | Reverse complement motif |
| Consensus sequence: BCVVDATAATTACMVHC | Consensus sequence: GHVRGTAATTATDVVGV |
 |
 |
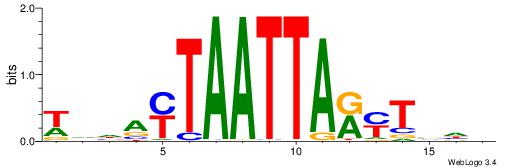
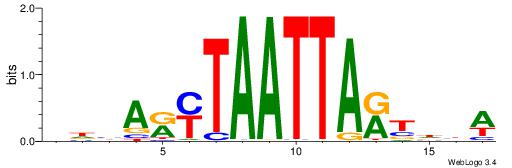
| Motif ID: | UP00108 |
| Motif name: | Alx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 7 |
| Similarity score: | 0.00319583 |
Alignment:
VHVAKMTAATTAKTHBW
------TAAGTAT----
| Original motif | Reverse complement motif |
| Consensus sequence: WVHAYTAATTARYTVHV | Consensus sequence: VHVAKMTAATTAKTHBW |
 |
 |
| Motif ID: | UP00202 |
| Motif name: | Dlx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 7 |
| Similarity score: | 0.00693044 |
Alignment:
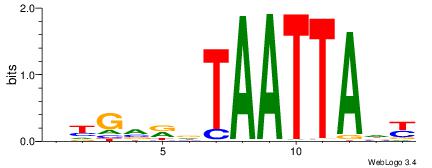
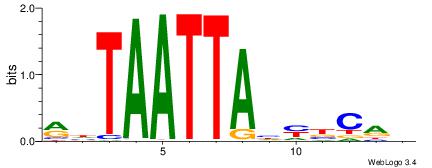
BHGVRDTAATTADY
-----ATACTTA--
| Original motif | Reverse complement motif |
| Consensus sequence: BHGVRDTAATTADY | Consensus sequence: MDTAATTAHMBCHB |
 |
 |
| Motif ID: | UP00214 |
| Motif name: | Hoxb5 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 7 |
| Similarity score: | 0.00733424 |
Alignment:
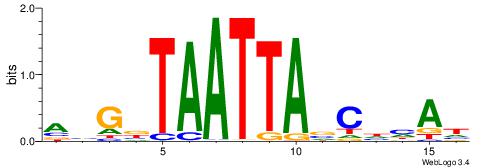
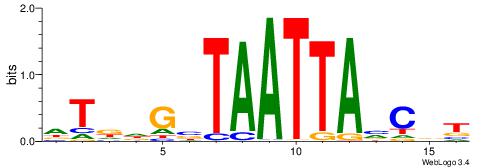
HTDHGVTAATTABCVT
------TAAGTAT---
| Original motif | Reverse complement motif |
| Consensus sequence: AVGBTAATTAVCHHAH | Consensus sequence: HTDHGVTAATTABCVT |
 |
 |
| Dataset #: 4 | Motif ID: 94 | Motif name: yrTCWCAAyr |
| Original motif | Reverse complement motif |
| Consensus sequence: HVTCACAAYD | Consensus sequence: HKTTGTGABH |
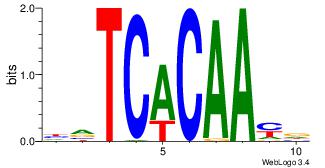 |
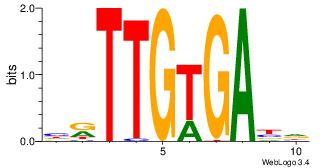 |
Best Matches for Motif ID 94 (Highest to Lowest)
| Motif ID: | UP00004 |
| Motif name: | Sox14_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.00180568 |
Alignment:
BBDCMATTGTSTBRB
----HKTTGTGABH-
| Original motif | Reverse complement motif |
| Consensus sequence: BKBASACAATRGHBB | Consensus sequence: BBDCMATTGTSTBRB |
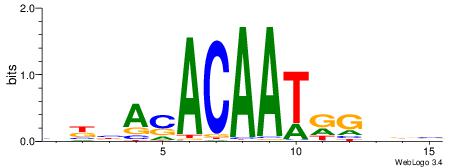 |
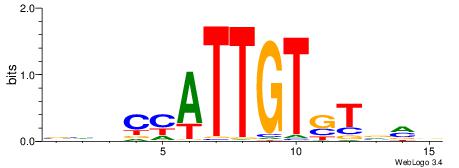 |
| Motif ID: | UP00041 |
| Motif name: | Foxj1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.00349348 |
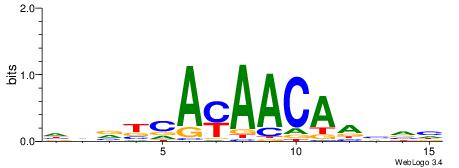
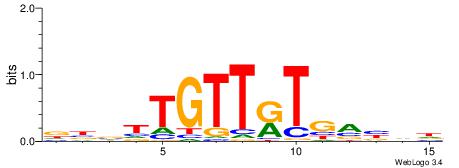
Alignment:
HBVBCAYAACAABVH
-HVTCACAAYD----
| Original motif | Reverse complement motif |
| Consensus sequence: HBVBCAYAACAABVH | Consensus sequence: DBBTTGTTKTGVVVH |
 |
 |
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.00703247 |
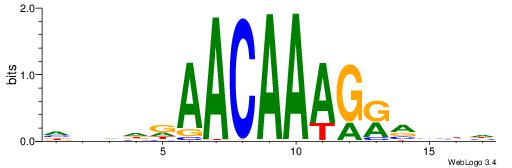
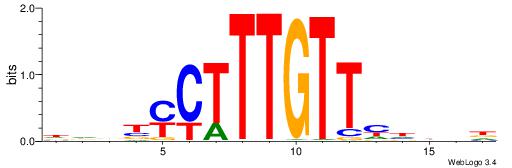
Alignment:
HHBBMCTTTGTTHDDBH
-----HKTTGTGABH--
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
 |
 |
| Motif ID: | UP00069 |
| Motif name: | Sox1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.00766951 |
Alignment:
HBDTAATTGTTABBV
----HKTTGTGABH-
| Original motif | Reverse complement motif |
| Consensus sequence: HBDTAATTGTTABBV | Consensus sequence: VBVTAACAATTADVD |
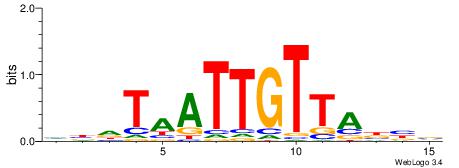 |
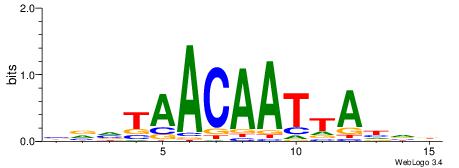 |
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.00853696 |
Alignment:
HHBTMCTTTGTTMDDVH
-----HKTTGTGABH--
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
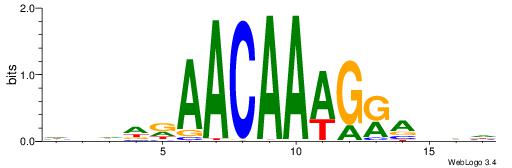 |
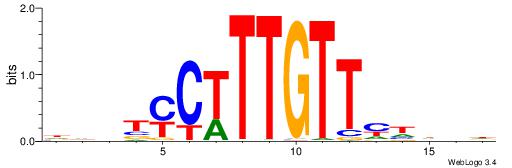 |
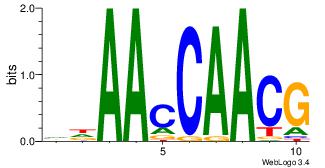
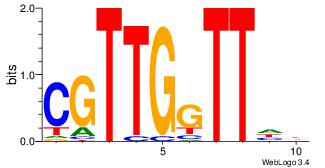
| Dataset #: 4 | Motif ID: 96 | Motif name: mdAAcCAACG |
| Original motif | Reverse complement motif |
| Consensus sequence: VDAACCAACG | Consensus sequence: CGTTGGTTDB |
 |
 |
Best Matches for Motif ID 96 (Highest to Lowest)
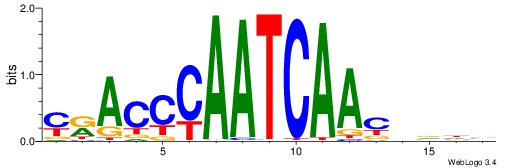
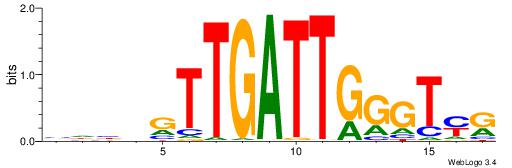
| Motif ID: | UP00227 |
| Motif name: | Duxl |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.00355394 |
Alignment:
VHVVGTTGATTGGGTMK
---CGTTGGTTDB----
| Original motif | Reverse complement motif |
| Consensus sequence: YRACCCAATCAACVVHV | Consensus sequence: VHVVGTTGATTGGGTMK |
 |
 |
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.00855132 |
Alignment:
HBDDRAACAAAGRABHH
--VDAACCAACG-----
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
 |
 |
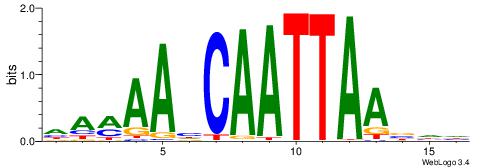
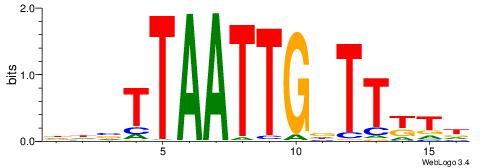
| Motif ID: | UP00145 |
| Motif name: | Barhl2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0107325 |
Alignment:
HAMAAHCAATTAABHH
-VDAACCAACG-----
| Original motif | Reverse complement motif |
| Consensus sequence: HAMAAHCAATTAABHH | Consensus sequence: HHBTTAATTGDTTYTH |
 |
 |
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0110249 |
Alignment:
HBDDDAACAAAGRVBHH
--VDAACCAACG-----
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
 |
 |
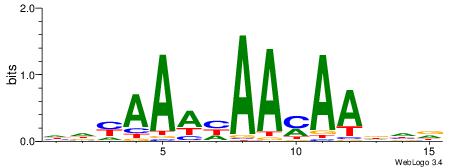
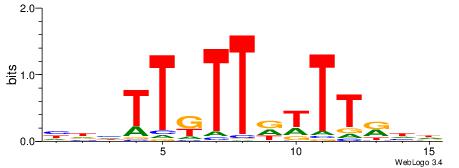
| Motif ID: | UP00037 |
| Motif name: | Zfp105_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0135248 |
Alignment:
HHHTTRTTDTTTKDH
----CGTTGGTTDB-
| Original motif | Reverse complement motif |
| Consensus sequence: HDYAAAHAAMAADHD | Consensus sequence: HHHTTRTTDTTTKDH |
 |
 |