Best Matches in Database for Each Motif (Highest to Lowest)
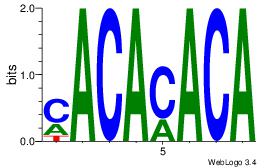
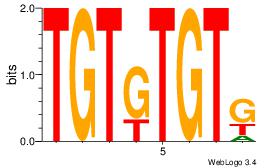
| Dataset #: 1 | Motif ID: 2 | Motif name: Motif 2 |
| Original motif | Reverse complement motif |
| Consensus sequence: CACACACA | Consensus sequence: TGTGTGTG |
 |
 |
Best Matches for Motif ID 2 (Highest to Lowest)
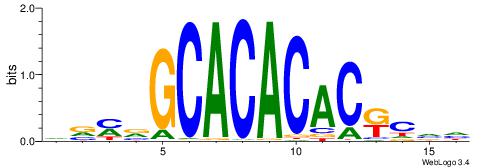
| Motif ID: | UP00042 |
| Motif name: | Gm397_second |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HBGYGTGTGTGCVRVD
---TGTGTGTG-----
| Original motif | Reverse complement motif |
| Consensus sequence: DVMVGCACACACKCVH | Consensus sequence: HBGYGTGTGTGCVRVD |
 |
 |
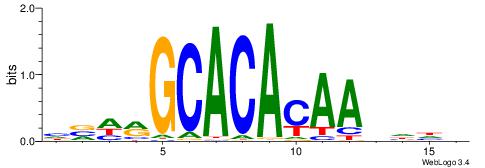
| Motif ID: | UP00026 |
| Motif name: | Zscan4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0178637 |
Alignment:
BHHHTTGTGTGCKHVD
-----TGTGTGTG---
| Original motif | Reverse complement motif |
| Consensus sequence: HVHRGCACACAAHHHV | Consensus sequence: BHHHTTGTGTGCKHVD |
 |
 |
| Motif ID: | UP00004 |
| Motif name: | Sox14_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0291592 |
Alignment:
BBDCMATTGTSTBRB
-------TGTGTGTG
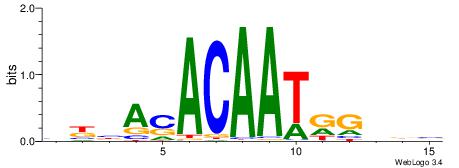
| Original motif | Reverse complement motif |
| Consensus sequence: BKBASACAATRGHBB | Consensus sequence: BBDCMATTGTSTBRB |
 |
 |
| Motif ID: | UP00042 |
| Motif name: | Gm397_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.0316665 |
Alignment:
HHRTATGTGCACATHHD
-TGTGTGTG--------
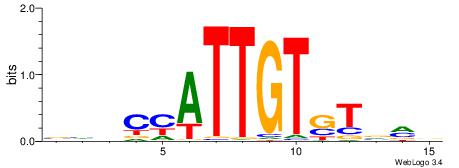
| Original motif | Reverse complement motif |
| Consensus sequence: HHDATGTGCACATAMDH | Consensus sequence: HHRTATGTGCACATHHD |
 |
 |
| Motif ID: | UP00026 |
| Motif name: | Zscan4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0327662 |
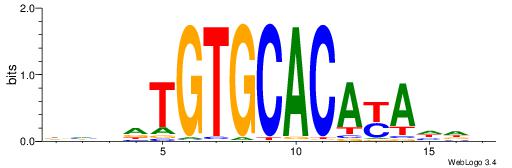
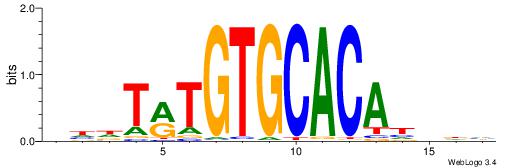
Alignment:
DHVDTGTGCACAYAHDH
--------CACACACA-
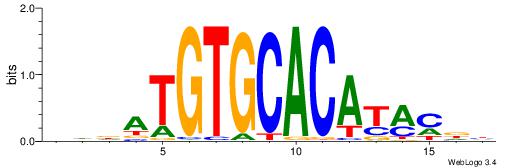
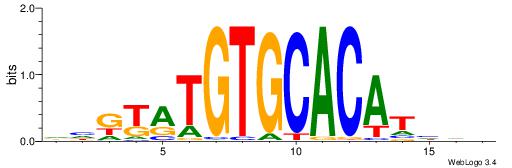
| Original motif | Reverse complement motif |
| Consensus sequence: DHVDTGTGCACAYAHDH | Consensus sequence: HDHTMTGTGCACADVHD |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0329902 |
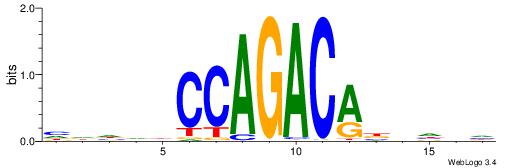
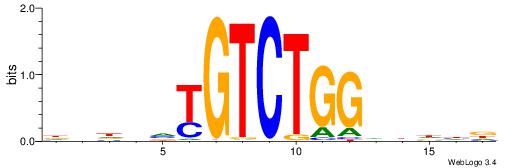
Alignment:
HVHBVTGTCTGGDDHDD
-----TGTGTGTG----
| Original motif | Reverse complement motif |
| Consensus sequence: HDHDDCCAGACABBHVH | Consensus sequence: HVHBVTGTCTGGDDHDD |
 |
 |
| Motif ID: | UP00041 |
| Motif name: | Foxj1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.0361089 |
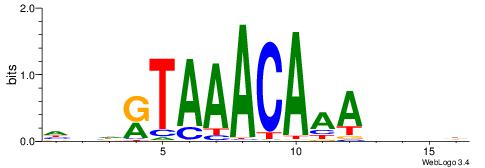
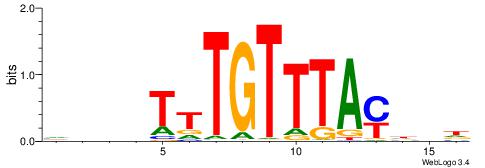
Alignment:
HDHRTAAACAAAHDDD
--CACACACA------
| Original motif | Reverse complement motif |
| Consensus sequence: HDHRTAAACAAAHDDD | Consensus sequence: DDDHTTTGTTTAMHDH |
 |
 |
| Motif ID: | UP00101 |
| Motif name: | Sox12_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.039766 |
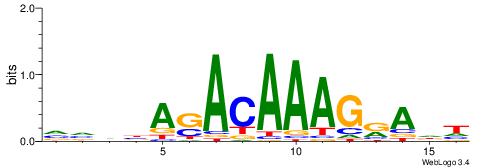
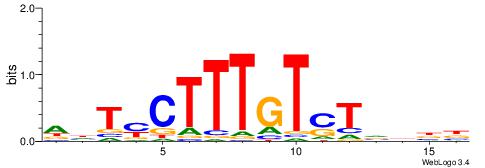
Alignment:
HDTMCTTTGTSTVDBB
-----TGTGTGTG---
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBASACAAAGRADH | Consensus sequence: HDTMCTTTGTSTVDBB |
 |
 |
| Motif ID: | UP00039 |
| Motif name: | Foxj3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0414454 |
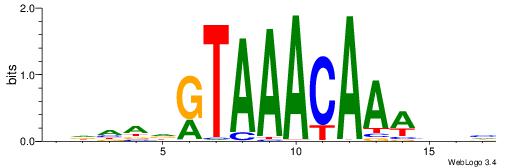
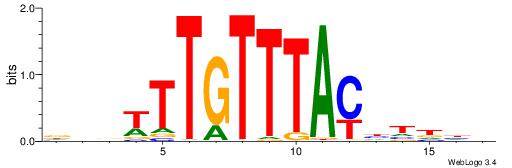
Alignment:
HDHADGTAAACAAAVVH
----CACACACA-----
| Original motif | Reverse complement motif |
| Consensus sequence: HDHADGTAAACAAAVVH | Consensus sequence: DVVTTTGTTTACDTHDH |
 |
 |
| Motif ID: | UP00070 |
| Motif name: | Gcm1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0420768 |
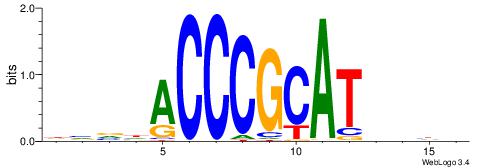
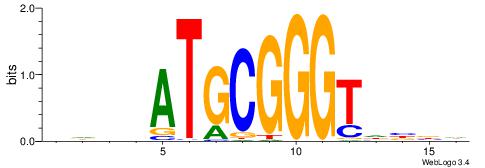
Alignment:
HVVHACCCGCATVDHD
---CACACACA-----
| Original motif | Reverse complement motif |
| Consensus sequence: HVVHACCCGCATVDHD | Consensus sequence: DHDVATGCGGGTHVVH |
 |
 |
| Dataset #: 1 | Motif ID: 4 | Motif name: Motif 4 |
| Original motif | Reverse complement motif |
| Consensus sequence: CYBCCTCC | Consensus sequence: GGAGGVMG |
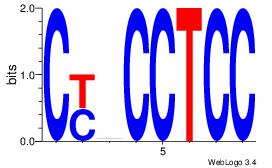 |
 |
Best Matches for Motif ID 4 (Highest to Lowest)
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0047591 |
Alignment:
DBCCCCCCCCCCMYC
----CYBCCTCC---
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
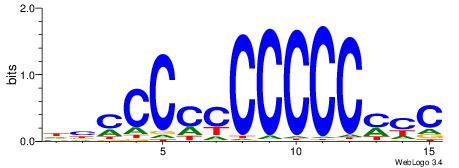 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.0211234 |
Alignment:
MHHHWHYTMCCHCCCACVAABVH
------CYBCCTCC---------
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
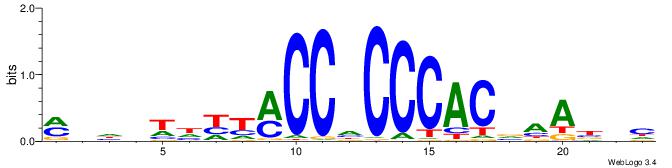 |
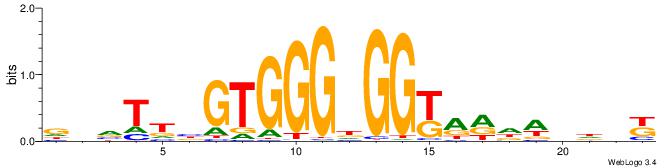 |
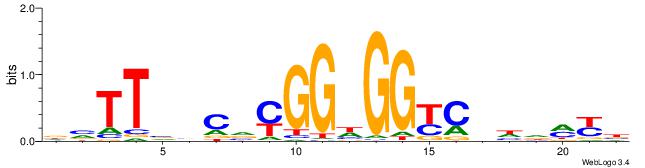
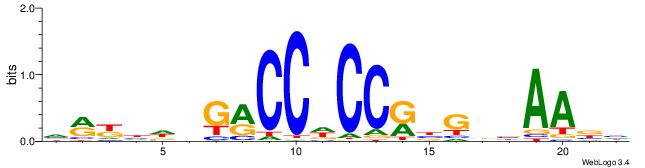
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0214303 |
Alignment:
DVTTVVCVYGGHGGYCHHDMYB
---------GGAGGVMG-----
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
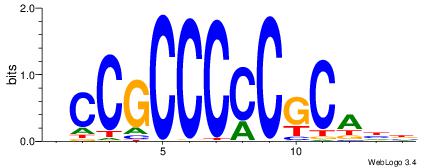
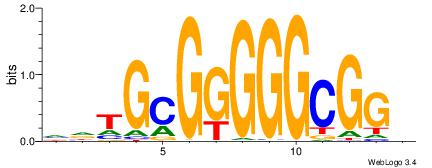
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0215351 |
Alignment:
HCCGCCCCCGCAHB
-CYBCCTCC-----
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0262858 |
Alignment:
BDDYYCATCCCATDDBD
--CYBCCTCC-------
| Original motif | Reverse complement motif |
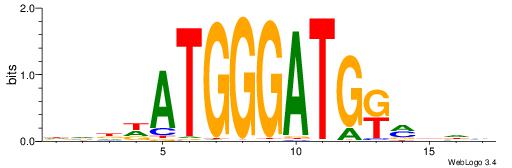
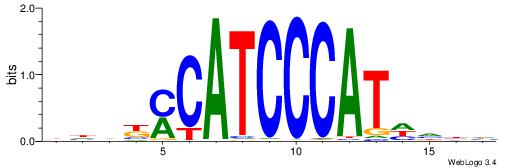
| Consensus sequence: DVDDATGGGATGKMDDV | Consensus sequence: BDDYYCATCCCATDDBD |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0273549 |
Alignment:
HVBCCCCCCCCMHHHB
---CYBCCTCC-----
| Original motif | Reverse complement motif |
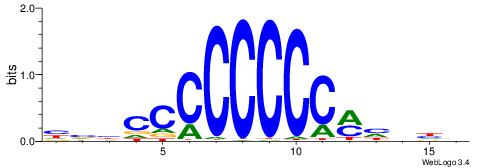
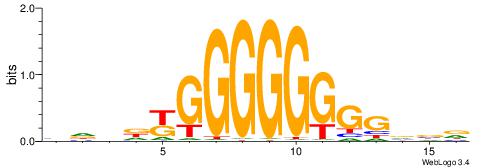
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00047 |
| Motif name: | Zbtb7b_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.0294721 |
Alignment:
VHWTTHGGGGGGVDD
------GGAGGVMG-
| Original motif | Reverse complement motif |
| Consensus sequence: DDVCCCCCCHAAWHB | Consensus sequence: VHWTTHGGGGGGVDD |
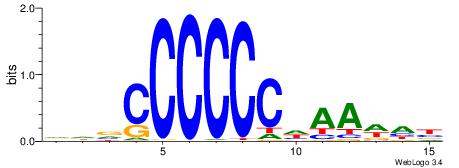 |
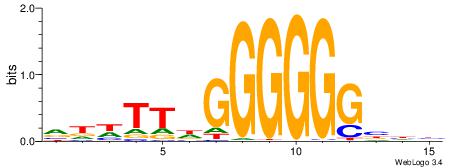 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0296935 |
Alignment:
YDYBDHTMCACGTGGADDBMDGT
-------------GGAGGVMG--
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
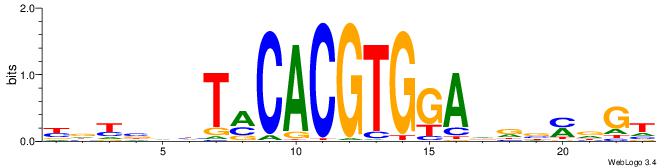 |
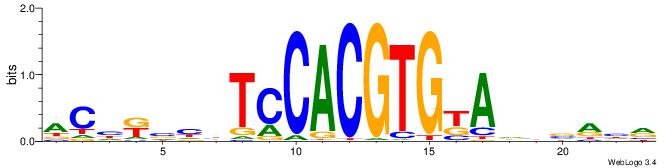 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0314233 |
Alignment:
HVKDCCGGGGGGWADDD
-------GGAGGVMG--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDTWCCCCCCGGDRVH | Consensus sequence: HVKDCCGGGGGGWADDD |
 |
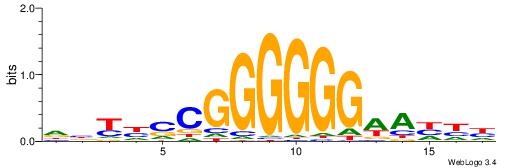 |
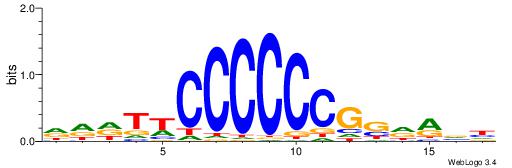
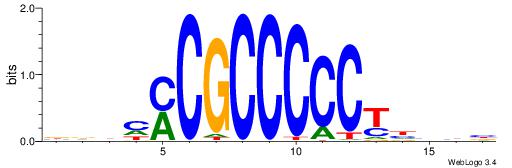
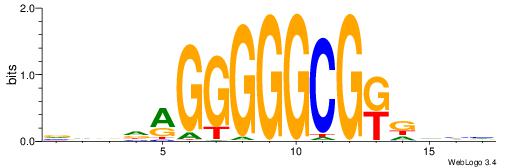
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0324346 |
Alignment:
BDHHMCGCCCCCTHVBB
----CYBCCTCC-----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Dataset #: 1 | Motif ID: 7 | Motif name: Motif 7 |
| Original motif | Reverse complement motif |
| Consensus sequence: ASASAGAA | Consensus sequence: TTCTSTST |
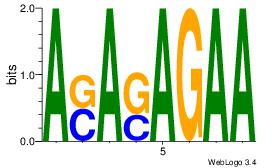 |
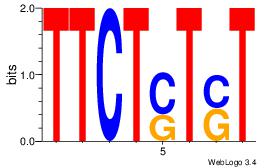 |
Best Matches for Motif ID 7 (Highest to Lowest)
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
BDHDDADAGATAAGADHBDHVD
-----ASASAGAA---------
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
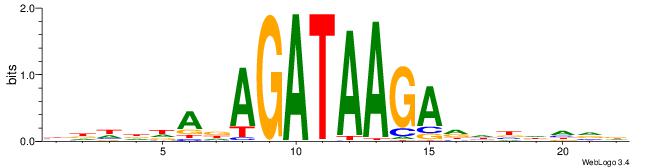 |
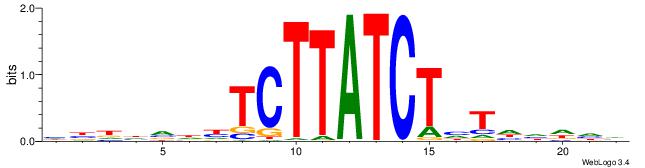 |
| Motif ID: | UP00040 |
| Motif name: | Irf5_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.00562375 |
Alignment:
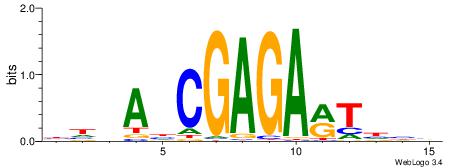
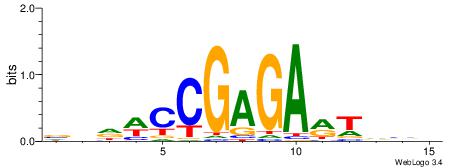
BHDAHCGAGARTHBV
---ASASAGAA----
| Original motif | Reverse complement motif |
| Consensus sequence: BHDAHCGAGARTHBV | Consensus sequence: VBHAKTCTCGHTHHV |
 |
 |
| Motif ID: | UP00101 |
| Motif name: | Sox12_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.0120605 |
Alignment:
VVDBASACAAAGRADH
--ASASAGAA------
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBASACAAAGRADH | Consensus sequence: HDTMCTTTGTSTVDBB |
 |
 |
| Motif ID: | UP00018 |
| Motif name: | Irf4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.017471 |
Alignment:
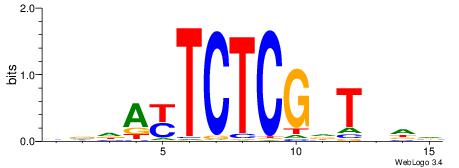
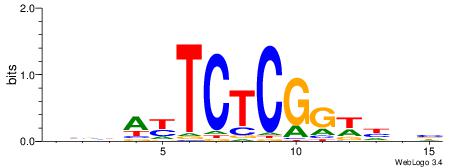
BHHWCCGAGAMTVVB
---ASASAGAA----
| Original motif | Reverse complement motif |
| Consensus sequence: VVBAYTCTCGGWHDB | Consensus sequence: BHHWCCGAGAMTVVB |
 |
 |
| Motif ID: | UP00090 |
| Motif name: | Elf3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.018291 |
Alignment:
TDAMTTCCYYDTD
----TTCTSTST-
| Original motif | Reverse complement motif |
| Consensus sequence: DAHMMGGAARTDA | Consensus sequence: TDAMTTCCYYDTD |
 |
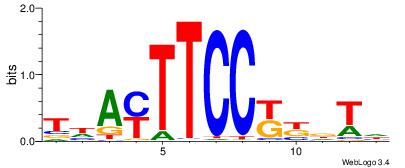 |
| Motif ID: | UP00004 |
| Motif name: | Sox14_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0185351 |
Alignment:
BKBASACAATRGHBB
-ASASAGAA------
| Original motif | Reverse complement motif |
| Consensus sequence: BKBASACAATRGHBB | Consensus sequence: BBDCMATTGTSTBRB |
 |
 |
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0187871 |
Alignment:
HBDDRAACAAAGRABHH
------ASASAGAA---
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
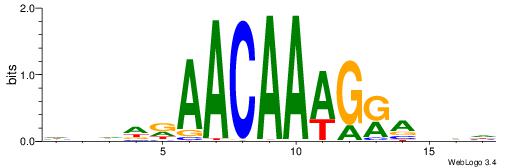 |
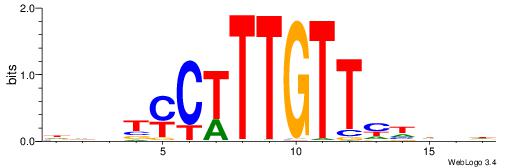 |
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.0188246 |
Alignment:
HBDDDAACAAAGRVBHH
------ASASAGAA---
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
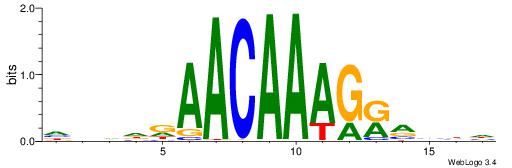 |
 |
| Motif ID: | UP00409 |
| Motif name: | Elf5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0192614 |
Alignment:
BWAHSMGGAAGTWD
--ASASAGAA----
| Original motif | Reverse complement motif |
| Consensus sequence: BWAHSMGGAAGTWD | Consensus sequence: DWACTTCCRSDTWV |
 |
 |
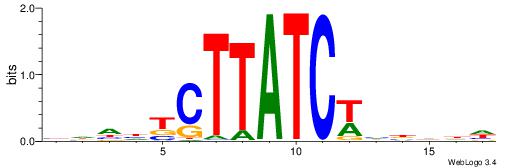
| Motif ID: | UP00100 |
| Motif name: | Gata6_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0192685 |
Alignment:
VHDDTCTTATCWHBDHD
------TTCTSTST---
| Original motif | Reverse complement motif |
| Consensus sequence: DHDVDWGATAAGADDHV | Consensus sequence: VHDDTCTTATCWHBDHD |
 |
 |
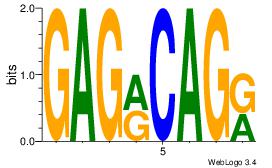
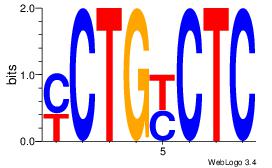
| Dataset #: 1 | Motif ID: 13 | Motif name: Motif 13 |
| Original motif | Reverse complement motif |
| Consensus sequence: GAGRCAGR | Consensus sequence: MCTGKCTC |
 |
 |
Best Matches for Motif ID 13 (Highest to Lowest)
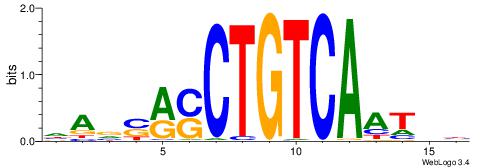
| Motif ID: | UP00205 |
| Motif name: | Pknox2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HAVSRSCTGTCAATHB
-----MCTGKCTC---
| Original motif | Reverse complement motif |
| Consensus sequence: HAVSRSCTGTCAATHB | Consensus sequence: VHATTGACAGSKSVTH |
 |
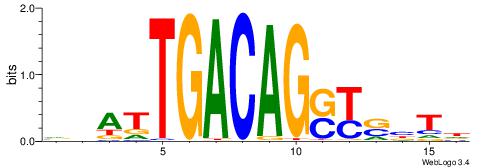 |
| Motif ID: | UP00203 |
| Motif name: | Pknox1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0024357 |
Alignment:
DRVSASCTGTCAAWVV
-----MCTGKCTC---
| Original motif | Reverse complement motif |
| Consensus sequence: DRVSASCTGTCAAWVV | Consensus sequence: VVWTTGACAGSTSBKD |
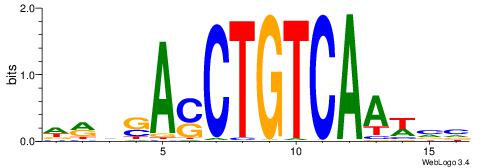 |
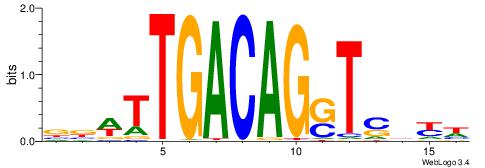 |
| Motif ID: | UP00226 |
| Motif name: | Mrg1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.00532831 |
Alignment:
WAVBASCTGTCAAWHB
-----MCTGKCTC---
| Original motif | Reverse complement motif |
| Consensus sequence: WAVBASCTGTCAAWHB | Consensus sequence: BHWTTGACAGSTBBTW |
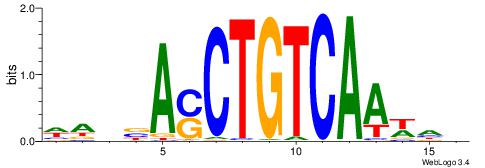 |
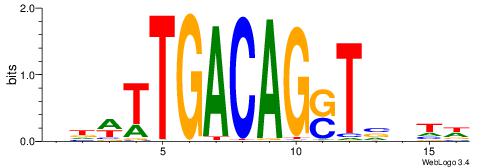 |
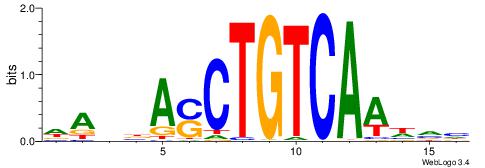
| Motif ID: | UP00210 |
| Motif name: | Mrg2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.00847279 |
Alignment:
DADBASCTGTCAAHVH
-----MCTGKCTC---
| Original motif | Reverse complement motif |
| Consensus sequence: DADBASCTGTCAAHVH | Consensus sequence: DBHTTGACAGSTVDTD |
 |
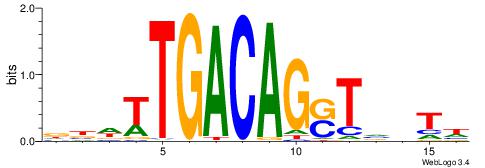 |
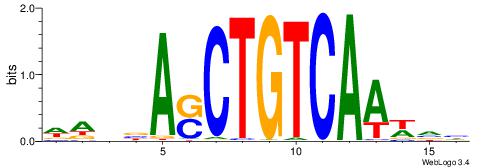
| Motif ID: | UP00186 |
| Motif name: | Meis1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.00869293 |
Alignment:
WAVBASCTGTCAAWVH
-----MCTGKCTC---
| Original motif | Reverse complement motif |
| Consensus sequence: WAVBASCTGTCAAWVH | Consensus sequence: DBWTTGACAGSTBVTW |
 |
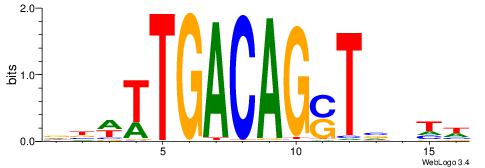 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.00919674 |
Alignment:
HDHDDCCAGACABBHVH
------GAGRCAGR---
| Original motif | Reverse complement motif |
| Consensus sequence: HDHDDCCAGACABBHVH | Consensus sequence: HVHBVTGTCTGGDDHDD |
 |
 |
| Motif ID: | UP00103 |
| Motif name: | Jundm2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.00968214 |
Alignment:
HDDKTGASTCAKHRDB
-----GAGRCAGR---
| Original motif | Reverse complement motif |
| Consensus sequence: VDKDRTGASTCAYHDH | Consensus sequence: HDDKTGASTCAKHRDB |
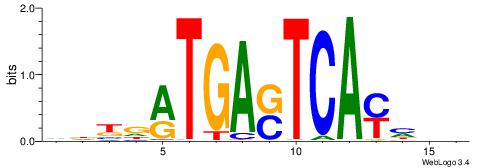 |
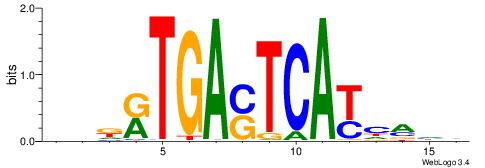 |
| Motif ID: | UP00095 |
| Motif name: | Zfp691_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0110909 |
Alignment:
DABBWGACTCCTHDVHV
--MCTGKCTC-------
| Original motif | Reverse complement motif |
| Consensus sequence: DABBWGACTCCTHDVHV | Consensus sequence: VHBDDAGGAGTCWBBTD |
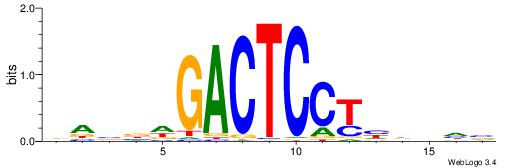 |
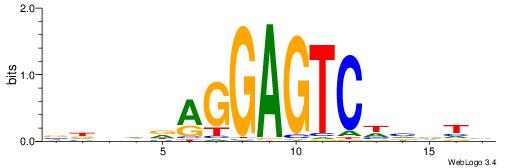 |
| Motif ID: | UP00122 |
| Motif name: | Tgif1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0125857 |
Alignment:
DABWWTGACAGCTVVBD
----GAGRCAGR-----
| Original motif | Reverse complement motif |
| Consensus sequence: DABWWTGACAGCTVVBD | Consensus sequence: DBVVAGCTGTCAWWVTH |
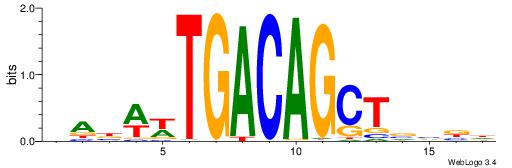 |
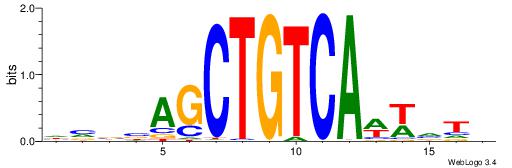 |
| Motif ID: | UP00258 |
| Motif name: | Tgif2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0145433 |
Alignment:
AABBAGCTGTCAAWVV
-----MCTGKCTC---
| Original motif | Reverse complement motif |
| Consensus sequence: AABBAGCTGTCAAWVV | Consensus sequence: VBWTTGACAGCTVBTT |
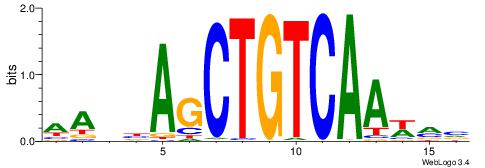 |
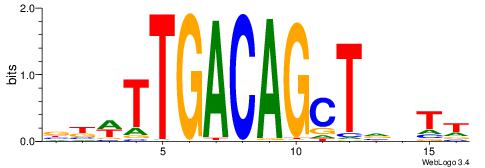 |
| Dataset #: 1 | Motif ID: 14 | Motif name: Motif 14 |
| Original motif | Reverse complement motif |
| Consensus sequence: GCRCACRC | Consensus sequence: GKGTGKGC |
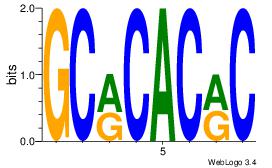 |
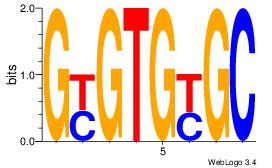 |
Best Matches for Motif ID 14 (Highest to Lowest)
| Motif ID: | UP00042 |
| Motif name: | Gm397_second |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
DVMVGCACACACKCVH
----GCRCACRC----
| Original motif | Reverse complement motif |
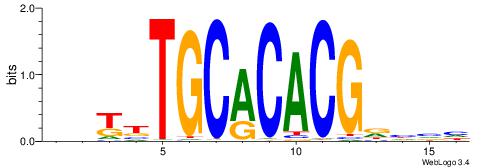
| Consensus sequence: DVMVGCACACACKCVH | Consensus sequence: HBGYGTGTGTGCVRVD |
 |
 |
| Motif ID: | UP00097 |
| Motif name: | Mtf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0261244 |
Alignment:
HDYTTGCACACGDHBH
-----GCRCACRC---
| Original motif | Reverse complement motif |
| Consensus sequence: DBDHCGTGTGCAAMDH | Consensus sequence: HDYTTGCACACGDHBH |
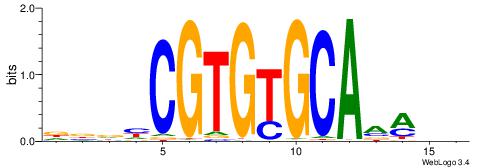 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.0329776 |
Alignment:
BHCBCBCCGGGTGGTCYHVHDCH
--------GKGTGKGC-------
| Original motif | Reverse complement motif |
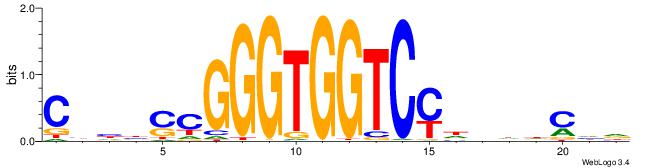
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
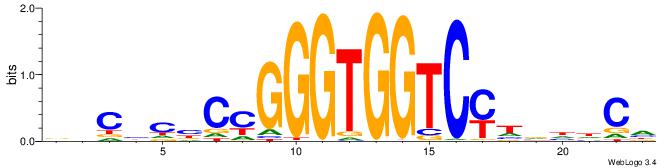 |
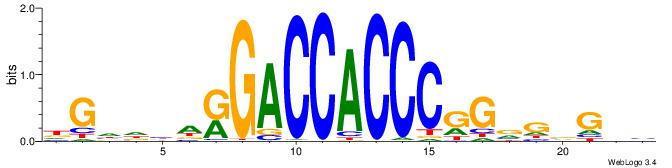 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.0341116 |
Alignment:
CVHBSCGGGTGGTCMHVHHCDH
------GKGTGKGC--------
| Original motif | Reverse complement motif |
| Consensus sequence: HDGHHBHRGACCACCCGSVDVG | Consensus sequence: CVHBSCGGGTGGTCMHVHHCDH |
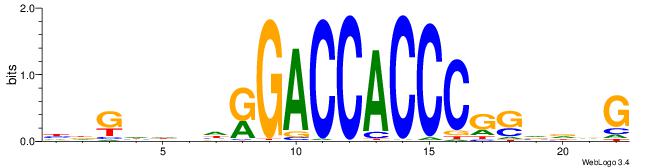 |
 |
| Motif ID: | UP00026 |
| Motif name: | Zscan4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0351584 |
Alignment:
HVHRGCACACAAHHHV
----GCRCACRC----
| Original motif | Reverse complement motif |
| Consensus sequence: HVHRGCACACAAHHHV | Consensus sequence: BHHHTTGTGTGCKHVD |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.0353979 |
Alignment:
DBCDWHCTGGGTGGTCMDHBBAH
--------GKGTGKGC-------
| Original motif | Reverse complement motif |
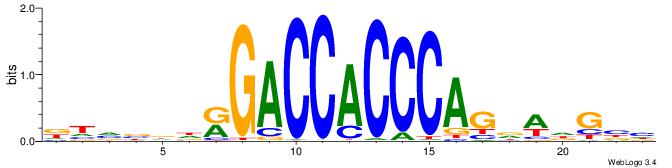
| Consensus sequence: DTVBDDRGACCACCCAGDWDGBH | Consensus sequence: DBCDWHCTGGGTGGTCMDHBBAH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0359411 |
Alignment:
BTBVTCVTGGGTGGTCMVVDVBB
--------GKGTGKGC-------
| Original motif | Reverse complement motif |
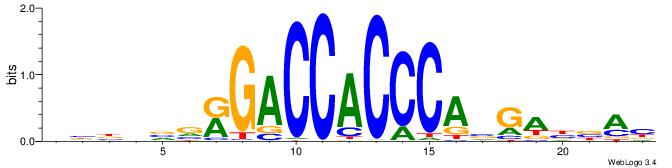
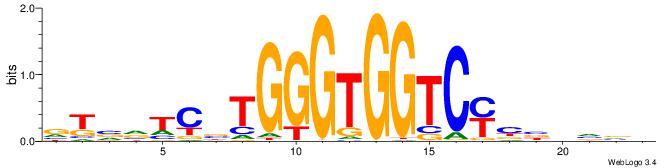
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.0360788 |
Alignment:
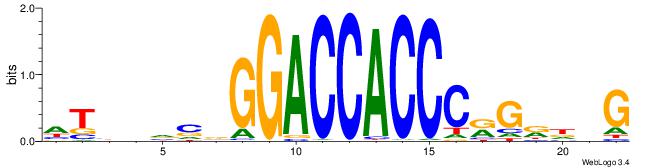
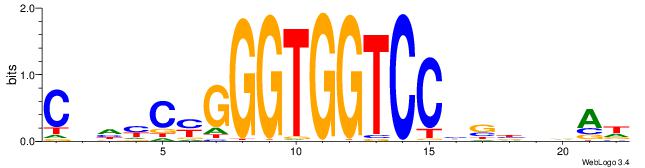
CBDMCMGGGTGGTCCHVBVBAH
------GKGTGKGC--------
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Motif ID: | UP00042 |
| Motif name: | Gm397_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 8 |
| Similarity score: | 0.0376684 |
Alignment:
HHDATGTGCACATAMDH
-------GCRCACRC--
| Original motif | Reverse complement motif |
| Consensus sequence: HHDATGTGCACATAMDH | Consensus sequence: HHRTATGTGCACATHHD |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.0381663 |
Alignment:
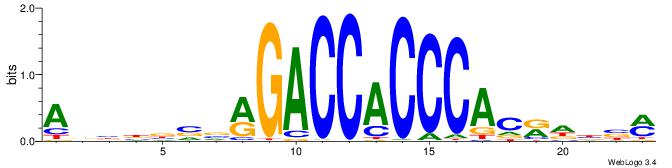
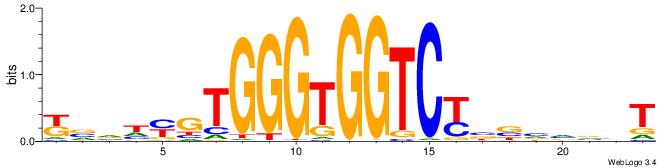
YBVDMGTGGGTGGTCKVVBVBBT
-------GKGTGKGC--------
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Dataset #: 2 | Motif ID: 18 | Motif name: Motif 18 |
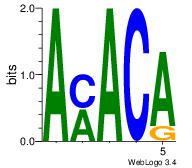
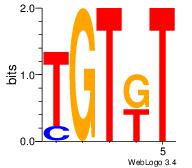
| Original motif | Reverse complement motif |
| Consensus sequence: AMACA | Consensus sequence: TGTRT |
 |
 |
Best Matches for Motif ID 18 (Highest to Lowest)
| Motif ID: | UP00042 |
| Motif name: | Gm397_second |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 5 |
| Similarity score: | 0 |
Alignment:
DVMVGCACACACKCVH
------AMACA-----
| Original motif | Reverse complement motif |
| Consensus sequence: DVMVGCACACACKCVH | Consensus sequence: HBGYGTGTGTGCVRVD |
 |
 |
| Motif ID: | UP00025 |
| Motif name: | Foxk1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 5 |
| Similarity score: | 0.00605402 |
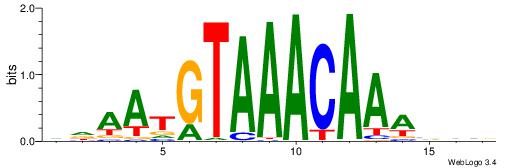
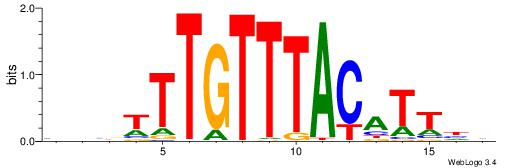
Alignment:
DDAATGTAAACAAAVVB
-------AMACA-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDAATGTAAACAAAVVB | Consensus sequence: BBVTTTGTTTACATTDD |
 |
 |
| Motif ID: | UP00041 |
| Motif name: | Foxj1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.00753267 |
Alignment:
HDHRTAAACAAAHDDD
-----AMACA------
| Original motif | Reverse complement motif |
| Consensus sequence: HDHRTAAACAAAHDDD | Consensus sequence: DDDHTTTGTTTAMHDH |
 |
 |
| Motif ID: | UP00039 |
| Motif name: | Foxj3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 5 |
| Similarity score: | 0.00965634 |
Alignment:
HDHADGTAAACAAAVVH
-------AMACA-----
| Original motif | Reverse complement motif |
| Consensus sequence: HDHADGTAAACAAAVVH | Consensus sequence: DVVTTTGTTTACDTHDH |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.00989492 |
Alignment:
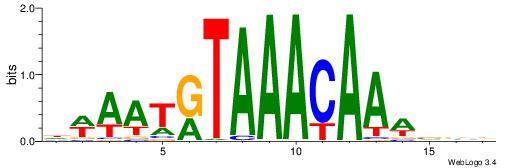
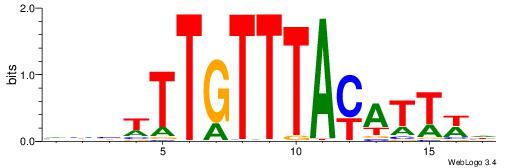
DWAATRTAAACAAWVVB
-------AMACA-----
| Original motif | Reverse complement motif |
| Consensus sequence: DWAATRTAAACAAWVVB | Consensus sequence: VVVWTTGTTTAMATTWD |
 |
 |
| Motif ID: | UP00004 |
| Motif name: | Sox14_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 5 |
| Similarity score: | 0.0116453 |
Alignment:
BBDCMATTGTSTBRB
-------TGTRT---
| Original motif | Reverse complement motif |
| Consensus sequence: BKBASACAATRGHBB | Consensus sequence: BBDCMATTGTSTBRB |
 |
 |
| Motif ID: | UP00073 |
| Motif name: | Foxa2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.0143122 |
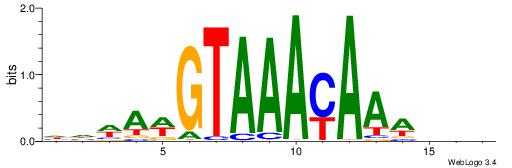
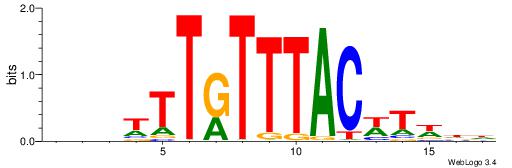
Alignment:
HHWAWGTAAAYAAAVDB
-------AMACA-----
| Original motif | Reverse complement motif |
| Consensus sequence: HHWAWGTAAAYAAAVDB | Consensus sequence: BDVTTTKTTTACWTWHH |
 |
 |
| Motif ID: | UP00026 |
| Motif name: | Zscan4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.0153228 |
Alignment:
BHHHTTGTGTGCKHVD
-----TGTRT------
| Original motif | Reverse complement motif |
| Consensus sequence: HVHRGCACACAAHHHV | Consensus sequence: BHHHTTGTGTGCKHVD |
 |
 |
| Motif ID: | UP00011 |
| Motif name: | Irf6_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.024195 |
Alignment:
DHDBTKGTTTCGHTHDD
-----TGTRT-------
| Original motif | Reverse complement motif |
| Consensus sequence: HDDAHCGAAACYAVDDD | Consensus sequence: DHDBTKGTTTCGHTHDD |
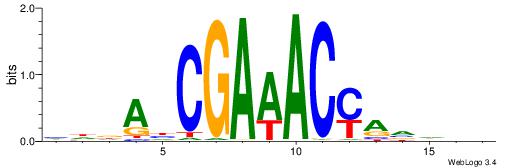 |
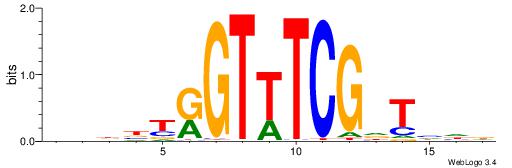 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.0255974 |
Alignment:
HDHDDCCAGACABBHVH
-------AMACA-----
| Original motif | Reverse complement motif |
| Consensus sequence: HDHDDCCAGACABBHVH | Consensus sequence: HVHBVTGTCTGGDDHDD |
 |
 |
| Dataset #: 2 | Motif ID: 20 | Motif name: Motif 20 |
| Original motif | Reverse complement motif |
| Consensus sequence: CTCTGY | Consensus sequence: KCAGAG |
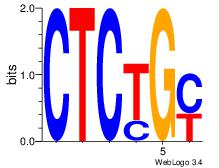 |
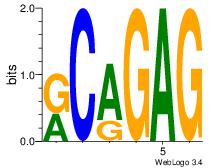 |
Best Matches for Motif ID 20 (Highest to Lowest)
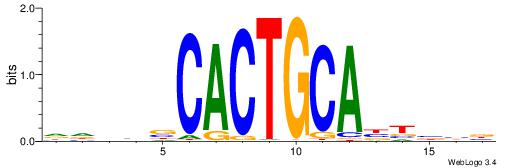
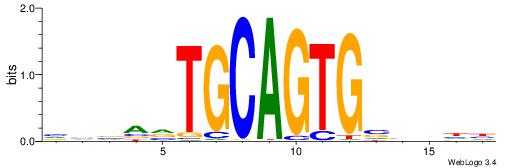
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
BBBAVTGCAGTGBBVDD
------KCAGAG-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDBBBCACTGCABTBBB | Consensus sequence: BBBAVTGCAGTGBBVDD |
 |
 |
| Motif ID: | UP00018 |
| Motif name: | Irf4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.00610915 |
Alignment:
BHHWCCGAGAMTVVB
---KCAGAG------
| Original motif | Reverse complement motif |
| Consensus sequence: VVBAYTCTCGGWHDB | Consensus sequence: BHHWCCGAGAMTVVB |
 |
 |
| Motif ID: | UP00095 |
| Motif name: | Zfp691_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.00648219 |
Alignment:
VHBDDAGGAGTCWBBTD
----KCAGAG-------
| Original motif | Reverse complement motif |
| Consensus sequence: DABBWGACTCCTHDVHV | Consensus sequence: VHBDDAGGAGTCWBBTD |
 |
 |
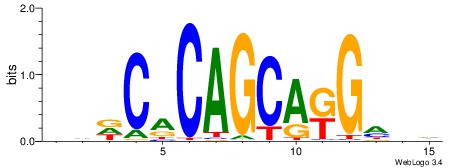
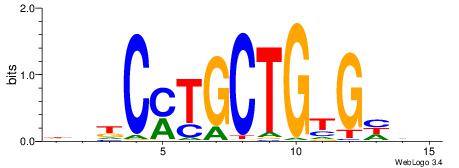
| Motif ID: | UP00006 |
| Motif name: | Zic3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.00853125 |
Alignment:
DDDCACAGCAKGHVD
--KCAGAG-------
| Original motif | Reverse complement motif |
| Consensus sequence: DDDCACAGCAKGHVD | Consensus sequence: DVHCYTGCTGTGHDH |
 |
 |
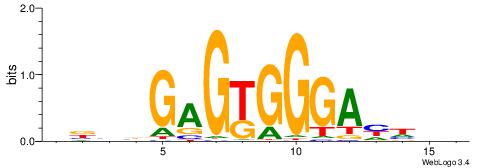
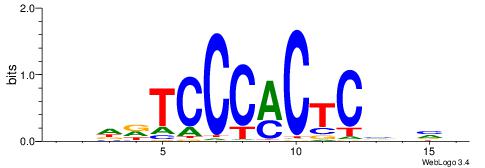
| Motif ID: | UP00007 |
| Motif name: | Egr1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 6 |
| Similarity score: | 0.00895004 |
Alignment:
BHHDTCCCACTCBDBV
---------CTCTGY-
| Original motif | Reverse complement motif |
| Consensus sequence: BBHBGAGTGGGAHHDB | Consensus sequence: BHHDTCCCACTCBDBV |
 |
 |
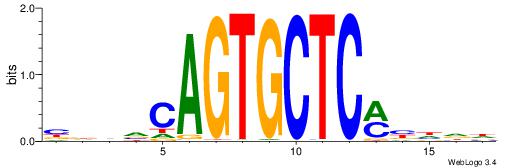
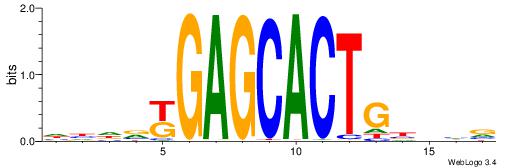
| Motif ID: | UP00095 |
| Motif name: | Zfp691_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.00955889 |
Alignment:
DDHBYGAGCACTGBHHB
--------CTCTGY---
| Original motif | Reverse complement motif |
| Consensus sequence: BDHVCAGTGCTCMBHDD | Consensus sequence: DDHBYGAGCACTGBHHB |
 |
 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.0106184 |
Alignment:
VDDAGATCAAAGGKDDD
------KCAGAG-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
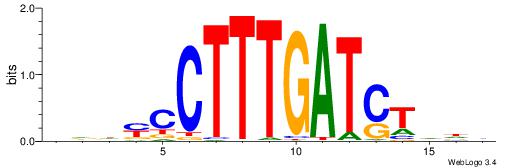 |
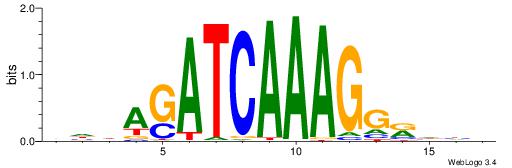 |
| Motif ID: | UP00102 |
| Motif name: | Zic1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.0122635 |
Alignment:
BHDCVCAGCAGGHVD
--KCAGAG-------
| Original motif | Reverse complement motif |
| Consensus sequence: BHDCVCAGCAGGHVD | Consensus sequence: DVHCCTGCTGBGDDB |
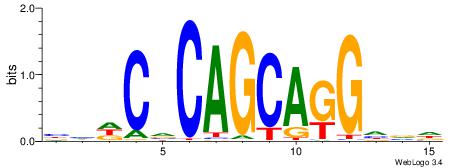 |
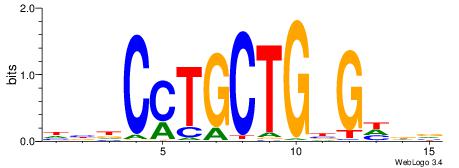 |
| Motif ID: | UP00083 |
| Motif name: | Tcf7l2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.0131237 |
Alignment:
BDDASATCAAAGGMDDD
------KCAGAG-----
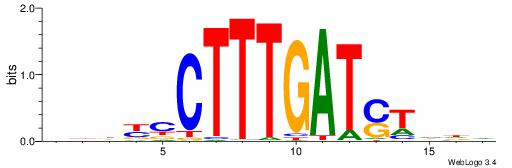
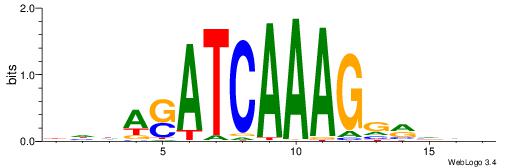
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATSTDDV | Consensus sequence: BDDASATCAAAGGMDDD |
 |
 |
| Motif ID: | UP00057 |
| Motif name: | Zic2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0136526 |
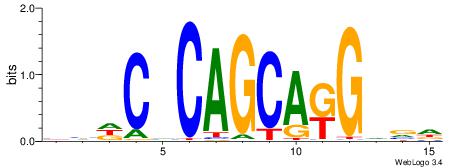
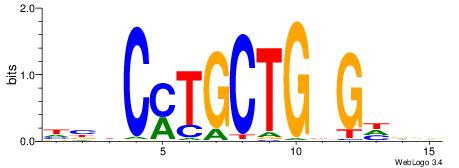
Alignment:
HHDCVCAGCAKGVVD
--KCAGAG-------
| Original motif | Reverse complement motif |
| Consensus sequence: HHDCVCAGCAKGVVD | Consensus sequence: DVBCYTGCTGBGDDD |
 |
 |
| Dataset #: 2 | Motif ID: 21 | Motif name: Motif 21 |
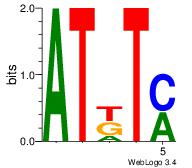
| Original motif | Reverse complement motif |
| Consensus sequence: ATKTM | Consensus sequence: RARAT |
 |
 |
Best Matches for Motif ID 21 (Highest to Lowest)
| Motif ID: | UP00080 |
| Motif name: | Gata5_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 5 |
| Similarity score: | 0 |
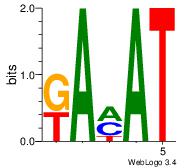
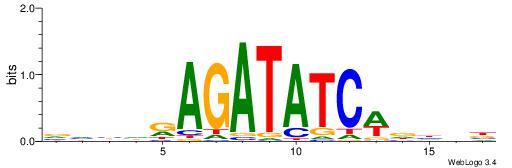
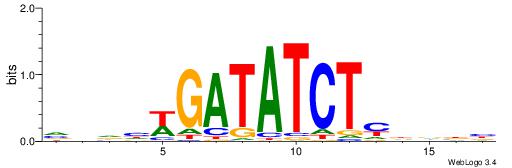
Alignment:
VVBHRAGATATCWDHBB
----RARAT--------
| Original motif | Reverse complement motif |
| Consensus sequence: VVBHRAGATATCWDHBB | Consensus sequence: VVHHWGATATCTMHBBV |
 |
 |
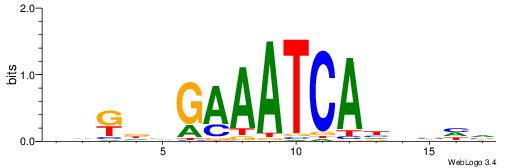
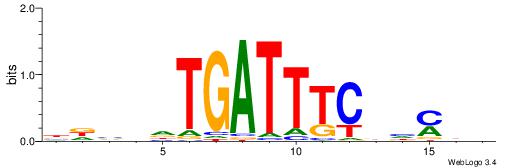
| Motif ID: | UP00148 |
| Motif name: | Hdx |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 5 |
| Similarity score: | 0.00279496 |
Alignment:
DDKDHGAAATCAHHBHV
-----RARAT-------
| Original motif | Reverse complement motif |
| Consensus sequence: DDKDHGAAATCAHHBHV | Consensus sequence: BDBDHTGATTTCDHYDD |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 12 |
| Number of overlap: | 5 |
| Similarity score: | 0.00967655 |
Alignment:
BDHDDADAGATAAGADHBDHVD
------RARAT-----------
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
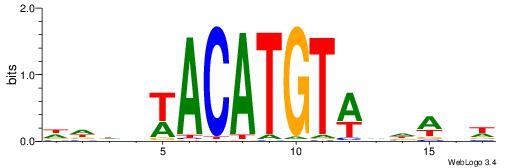
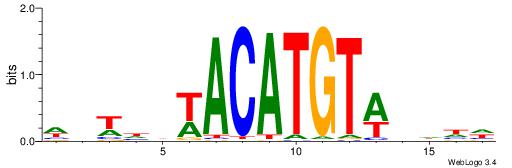
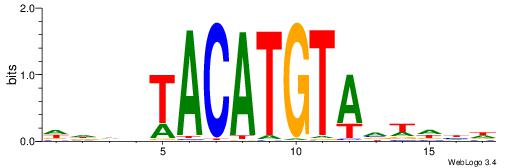
| Motif ID: | UP00250 |
| Motif name: | Irx5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.0116566 |
Alignment:
DDDHWACATGTWHDABW
-------ATKTM-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDHWACATGTWHDABW | Consensus sequence: WVTDHWACATGTWHDDD |
 |
 |
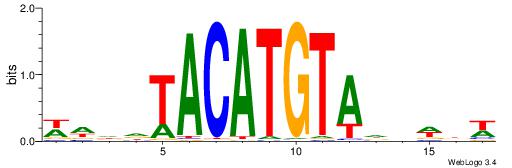
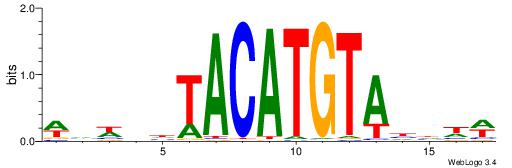
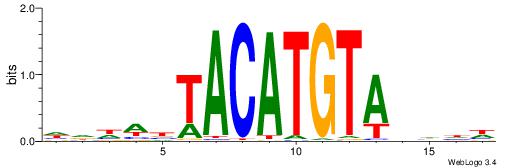
| Motif ID: | UP00236 |
| Motif name: | Irx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.0120196 |
Alignment:
WDDDTACATGTADDWBW
-------ATKTM-----
| Original motif | Reverse complement motif |
| Consensus sequence: WDDDTACATGTADDWBW | Consensus sequence: WVWDDTACATGTADDDW |
 |
 |
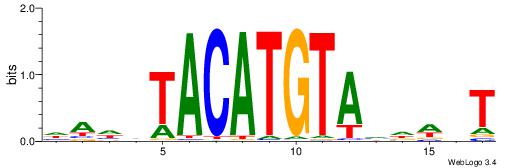
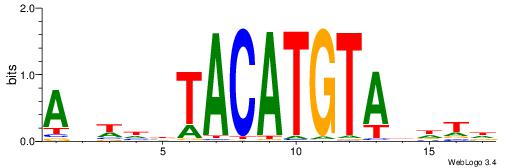
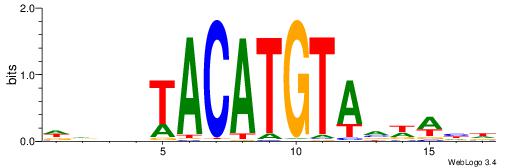
| Motif ID: | UP00150 |
| Motif name: | Irx6 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 5 |
| Similarity score: | 0.0120601 |
Alignment:
HAHDTACATGTAVDWHT
-------ATKTM-----
| Original motif | Reverse complement motif |
| Consensus sequence: HAHDTACATGTAVDWHT | Consensus sequence: AHWDBTACATGTADHTH |
 |
 |
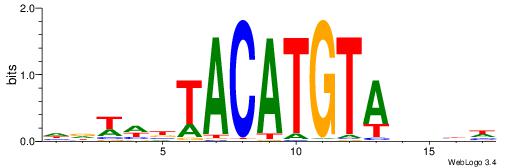
| Motif ID: | UP00223 |
| Motif name: | Irx3_2226.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 5 |
| Similarity score: | 0.0124884 |
Alignment:
DDDHTACATGTAVWDHD
-------ATKTM-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDHTACATGTAVWDHD | Consensus sequence: DHDWBTACATGTAHDDD |
 |
 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 10 |
| Number of overlap: | 5 |
| Similarity score: | 0.0150044 |
Alignment:
DVDDATGGGATGKMDDV
---------ATKTM---
| Original motif | Reverse complement motif |
| Consensus sequence: DVDDATGGGATGKMDDV | Consensus sequence: BDDYYCATCCCATDDBD |
 |
 |
| Motif ID: | UP00223 |
| Motif name: | Irx3_0920.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.0150192 |
Alignment:
DDDDTACATGTAVWWHD
-------ATKTM-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDDTACATGTAVWWHD | Consensus sequence: DDWWBTACATGTADDDD |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 12 |
| Number of overlap: | 5 |
| Similarity score: | 0.0166075 |
Alignment:
MWSMBAARRATGCDCABHATGD
------RARAT-----------
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
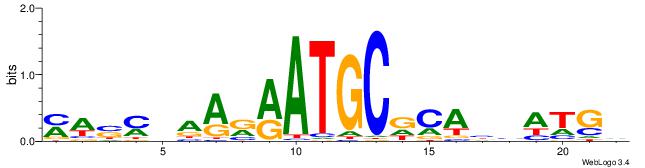 |
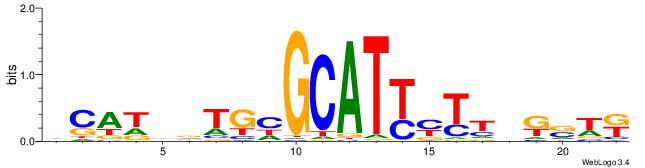 |
| Dataset #: 3 | Motif ID: 30 | Motif name: ZEB1 |
| Original motif | Reverse complement motif |
| Consensus sequence: CACCTD | Consensus sequence: HAGGTG |
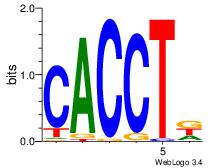 |
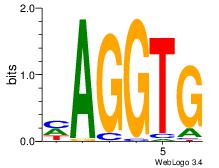 |
Best Matches for Motif ID 30 (Highest to Lowest)
| Motif ID: | UP00068 |
| Motif name: | Eomes_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
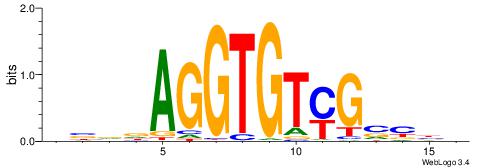
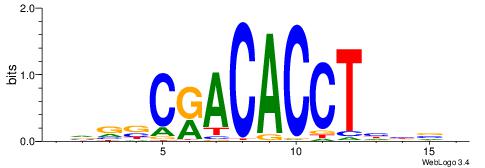
VVBVCKACACCTHVBV
-------CACCTD---
| Original motif | Reverse complement motif |
| Consensus sequence: VBVDAGGTGTYGVBBV | Consensus sequence: VVBVCKACACCTHVBV |
 |
 |
| Motif ID: | UP00199 |
| Motif name: | Six4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.00433068 |
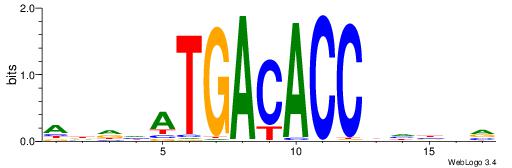
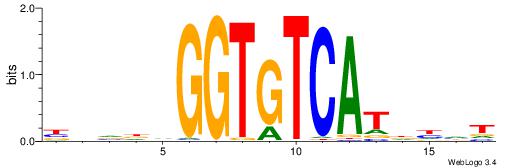
Alignment:
BBVHVGGTGTCATHDDT
---HAGGTG--------
| Original motif | Reverse complement motif |
| Consensus sequence: ADDHATGACACCBHBBV | Consensus sequence: BBVHVGGTGTCATHDDT |
 |
 |
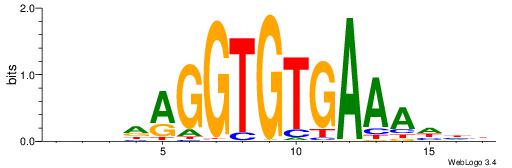
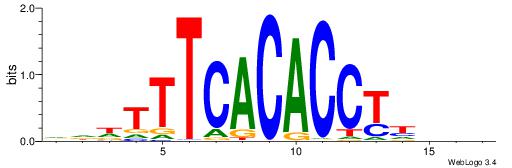
| Motif ID: | UP00068 |
| Motif name: | Eomes_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.00524081 |
Alignment:
HHHTTTCACACCTDHBV
--------CACCTD---
| Original motif | Reverse complement motif |
| Consensus sequence: BVHDAGGTGTGAAAHHH | Consensus sequence: HHHTTTCACACCTDHBV |
 |
 |
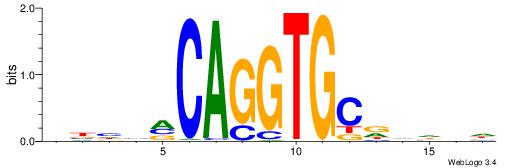
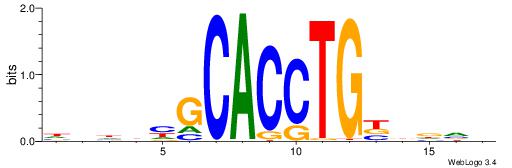
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.00911301 |
Alignment:
DHDBHGCACCTGBDDVB
------CACCTD-----
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Motif ID: | UP00047 |
| Motif name: | Zbtb7b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.0233556 |
Alignment:
BDDRVGACCACCHBDVB
--------CACCTD---
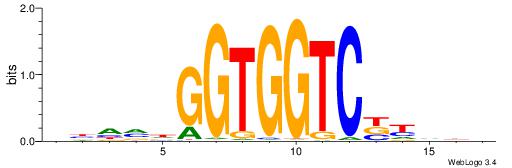
| Original motif | Reverse complement motif |
| Consensus sequence: BDDRVGACCACCHBDVB | Consensus sequence: BBDVHGGTGGTCBKDDB |
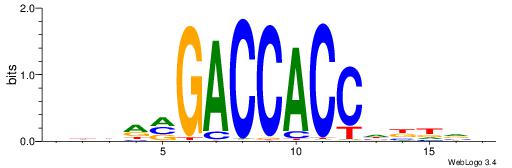 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 11 |
| Number of overlap: | 6 |
| Similarity score: | 0.0233928 |
Alignment:
BHCBCBCCGGGTGGTCYHVHDCH
-------HAGGTG----------
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.0254338 |
Alignment:
HTBVVVDGGACCACCCRGRDBG
-----------CACCTD-----
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.0290143 |
Alignment:
HDGHHBHRGACCACCCGSVDVG
-----------CACCTD-----
| Original motif | Reverse complement motif |
| Consensus sequence: HDGHHBHRGACCACCCGSVDVG | Consensus sequence: CVHBSCGGGTGGTCMHVHHCDH |
 |
 |
| Motif ID: | UP00036 |
| Motif name: | Myf6_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0309673 |
Alignment:
VVDBAACAGBTGBYHB
------HAGGTG----
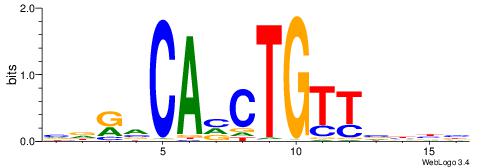
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBAACAGBTGBYHB | Consensus sequence: BDKVCABCTGTTBDBV |
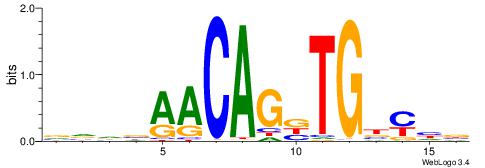 |
 |
| Motif ID: | UP00249 |
| Motif name: | Nkx2-5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.031502 |
Alignment:
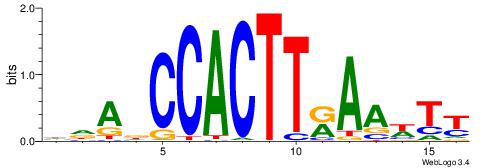
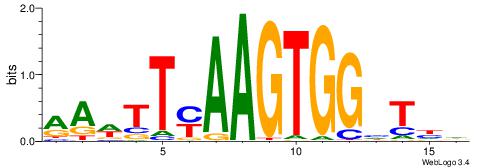
HDADCCACTTRAAWTT
-----CACCTD-----
| Original motif | Reverse complement motif |
| Consensus sequence: HDADCCACTTRAAWTT | Consensus sequence: AAWTTMAAGTGGHTDH |
 |
 |
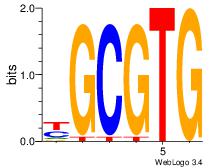
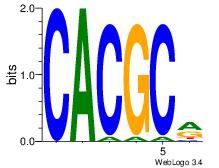
| Dataset #: 3 | Motif ID: 42 | Motif name: ArntAhr |
| Original motif | Reverse complement motif |
| Consensus sequence: YGCGTG | Consensus sequence: CACGCM |
 |
 |
Best Matches for Motif ID 42 (Highest to Lowest)
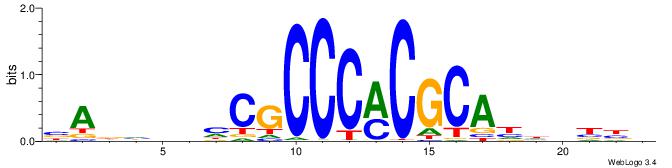
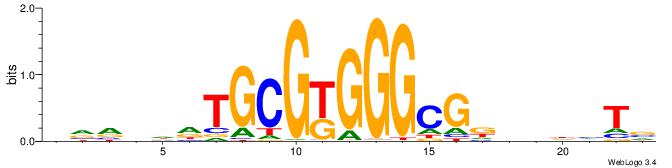
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
------YGCGTG-----------
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
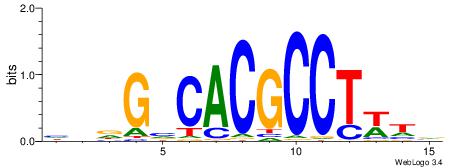
| Motif ID: | UP00002 |
| Motif name: | Sp4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.00183587 |
Alignment:
HHDGBCACGCCTWDB
-----CACGCM----
| Original motif | Reverse complement motif |
| Consensus sequence: BDWAGGCGTGBCHHD | Consensus sequence: HHDGBCACGCCTWDB |
 |
 |
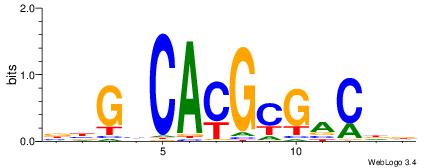
| Motif ID: | UP00060 |
| Motif name: | Max_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.00348048 |
Alignment:
HDGTCGCGTGDCVB
----YGCGTG----
| Original motif | Reverse complement motif |
| Consensus sequence: BBGHCACGCGACDD | Consensus sequence: HDGTCGCGTGDCVB |
 |
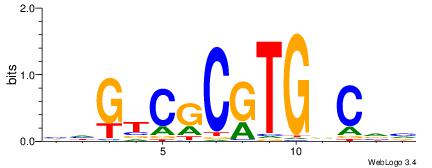 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.00526158 |
Alignment:
TCDAMVBGACGCMAKDDDVBBD
-------CACGCM---------
| Original motif | Reverse complement motif |
| Consensus sequence: DBBVDDDRTRGCGTCBBYTHGA | Consensus sequence: TCDAMVBGACGCMAKDDDVBBD |
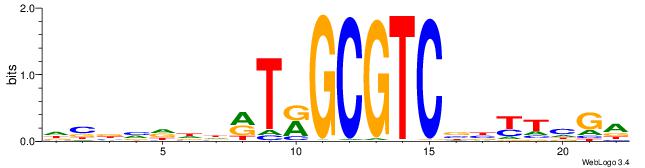 |
 |
| Motif ID: | UP00042 |
| Motif name: | Gm397_second |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.00558171 |
Alignment:
HBGYGTGTGTGCVRVD
---YGCGTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: DVMVGCACACACKCVH | Consensus sequence: HBGYGTGTGTGCVRVD |
 |
 |
| Motif ID: | UP00097 |
| Motif name: | Mtf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.00702416 |
Alignment:
HDYTTGCACACGDHBH
--------CACGCM--
| Original motif | Reverse complement motif |
| Consensus sequence: DBDHCGTGTGCAAMDH | Consensus sequence: HDYTTGCACACGDHBH |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.0123233 |
Alignment:
DDRMHBACGCGYGCGTDGBBDB
-------YGCGTG---------
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
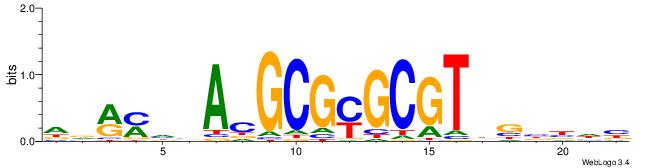 |
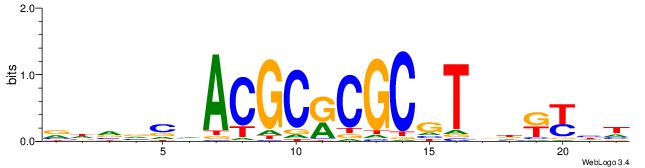 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0141579 |
Alignment:
HCCGCCCCCGCAHB
------CACGCM--
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 11 |
| Number of overlap: | 6 |
| Similarity score: | 0.0144485 |
Alignment:
BHCBCBCCGGGTGGTCYHVHDCH
-------YGCGTG----------
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.0172109 |
Alignment:
VHYTCCDWCCGCGTCMMTHWHV
---------YGCGTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
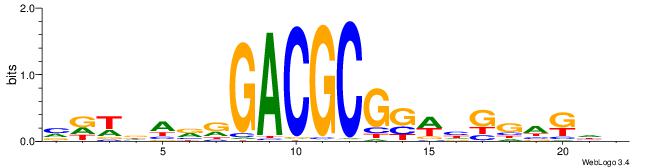 |
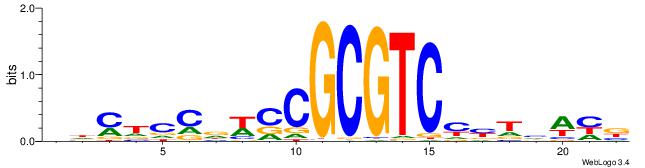 |