Best Matches for Each Motif (Highest to Lowest)
| Dataset #: 1 |
Motif ID: 1 |
Motif name: Motif 1 |
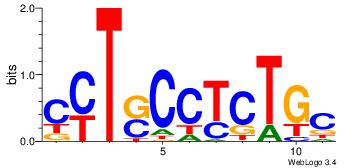
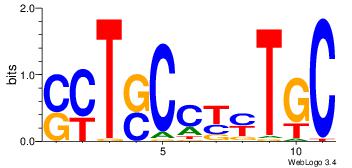
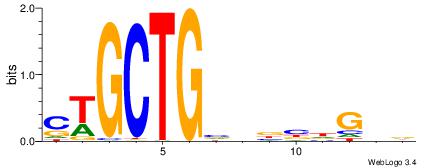
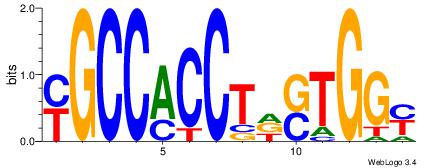
| Original motif Consensus sequence: CYTSCCTCTGC |
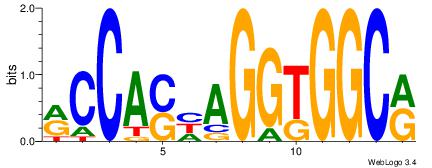
Reverse complement motif Consensus sequence: GCAGAGGSAKG |
|
|
 |
 |
Best Matches for Motif ID 1 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
90 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
11 |
| Similarity score: |
0 |
Alignment:
CHHCHBTCCTSCYTCTGC
-------CYTSCCTCTGC
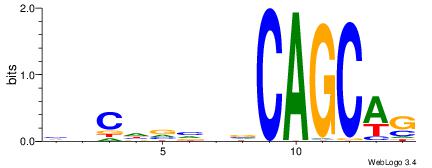
| Original motif Consensus sequence: CHHCHBTCCTSCYTCTGC |
Reverse complement motif Consensus sequence: GCAGAKGSAGGABDGHDG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
48 |
| Motif name: |
Motif 48 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0204179 |
Alignment:
SCTSCCKMTGC
CYTSCCTCTGC
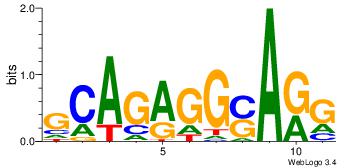
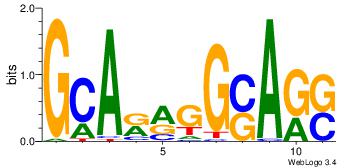
| Original motif Consensus sequence: GCARRGGSAGS |
Reverse complement motif Consensus sequence: SCTSCCKMTGC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
88 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0606594 |
Alignment:
CTGCTGBDBBDGBD
--CYTSCCTCTGC-
| Original motif Consensus sequence: CTGCTGBDBBDGBD |
Reverse complement motif Consensus sequence: HVCDBBDBCAGCAG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
11 |
| Similarity score: |
0.0635403 |
Alignment:
RAGKGMAGVRRGSRCASAGV
----GCAGAGGSAKG-----
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0656459 |
Alignment:
YDRCCASYAGRKGGCRSYV
------GCAGAGGSAKG--
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0782887 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
---CYTSCCTCTGC-------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0788385 |
Alignment:
MGGTCAGGGTGACCTRDBHV
--------CYTSCCTCTGC-
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
64 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0823596 |
Alignment:
TGRCCTKTGHCCKAB
---CYTSCCTCTGC-
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
65 |
| Motif name: |
Tcfcp2l1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0826758 |
Alignment:
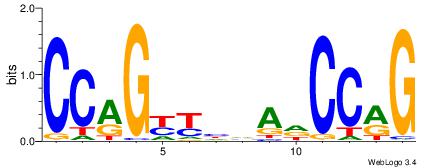
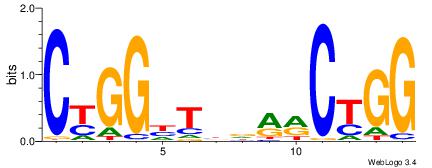
CCAGYYHVADCCRG
CYTSCCTCTGC---
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
Motif 47 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0840726 |
Alignment:
YGCCACCTDSTGGY
--CYTSCCTCTGC-
| Original motif Consensus sequence: YGCCACCTDSTGGY |
Reverse complement motif Consensus sequence: KCCASDAGGTGGCK |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
74 |
| Motif name: |
ssCCCCGCSssk |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0843055 |
Alignment:
BBSGGCGGGGBS
-GCAGAGGSAKG
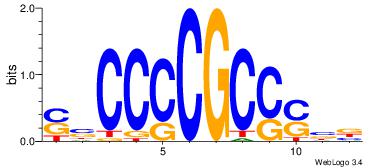
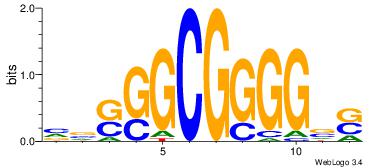
| Original motif Consensus sequence: SBCCCCGCCSBB |
Reverse complement motif Consensus sequence: BBSGGCGGGGBS |
|
|
 |
 |
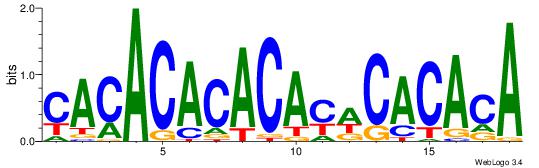
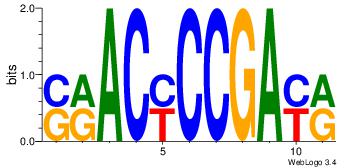
| Dataset #: 1 |
Motif ID: 2 |
Motif name: Motif 2 |
| Original motif Consensus sequence: ACACACACACACAC |
Reverse complement motif Consensus sequence: GTGTGTGTGTGTGT |
|
|
 |
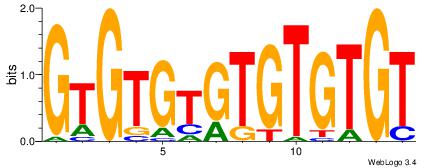 |
Best Matches for Motif ID 2 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
89 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0 |
Alignment:
TGTGTGWGTGTGTGTGTG
-GTGTGTGTGTGTGT---
| Original motif Consensus sequence: CACACACACACWCACACA |
Reverse complement motif Consensus sequence: TGTGTGWGTGTGTGTGTG |
|
|
 |
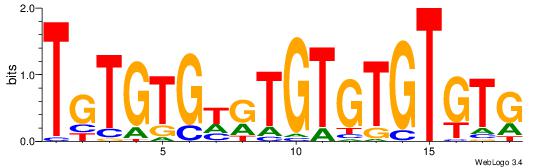 |
| Dataset #: |
5 |
| Motif ID: |
92 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.00332777 |
Alignment:
CAMACACACACAYDCACACA
-----ACACACACACACAC-
| Original motif Consensus sequence: CAMACACACACAYDCACACA |
Reverse complement motif Consensus sequence: TGTGTGDKTGTGTGTGTRTG |
|
|
 |
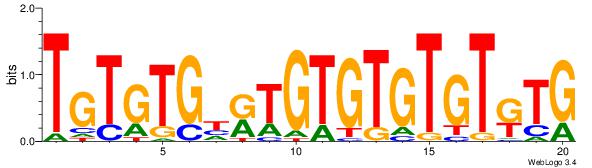 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0819161 |
Alignment:
AGTGSWSCACTGTGAMAMCACAGTG
-GTGTGTGTGTGTGT----------
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
91 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.082111 |
Alignment:
KTTTTTTKTTTTTTDAAB
-GTGTGTGTGTGTGT---
| Original motif Consensus sequence: KTTTTTTKTTTTTTDAAB |
Reverse complement motif Consensus sequence: BTTHAAAAAAYAAAAAAR |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.531086 |
Alignment:
TGTGTSWGTBTSTG-
-GTGTGTGTGTGTGT
| Original motif Consensus sequence: CASABACWSACACA |
Reverse complement motif Consensus sequence: TGTGTSWGTBTSTG |
|
|
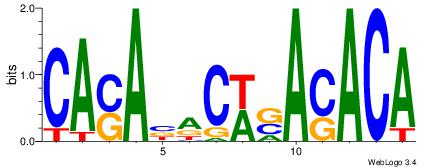 |
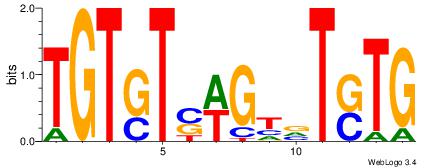 |
| Dataset #: |
5 |
| Motif ID: |
94 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
13 |
| Similarity score: |
0.576988 |
Alignment:
-TTTTTTTKTTKTTTAATTHW
GTGTGTGTGTGTGT-------
| Original motif Consensus sequence: TTTTTTTKTTKTTTAATTHW |
Reverse complement motif Consensus sequence: WHAATTAAARAARAAAAAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.579965 |
Alignment:
-TTTRKTTTHTTTTH
GTGTGTGTGTGTGT-
| Original motif Consensus sequence: HAAAAHAAARMAAA |
Reverse complement motif Consensus sequence: TTTRKTTTHTTTTH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
88 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
1.5772 |
Alignment:
CTGCTGBDBBDGBD---
---GTGTGTGTGTGTGT
| Original motif Consensus sequence: CTGCTGBDBBDGBD |
Reverse complement motif Consensus sequence: HVCDBBDBCAGCAG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
87 |
| Motif name: |
TFF1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
1.57782 |
Alignment:
AGGACAAAGAVV---
-ACACACACACACAC
| Original motif Consensus sequence: AGGACAAAGAVV |
Reverse complement motif Consensus sequence: VVTCTTTGTCCT |
|
|
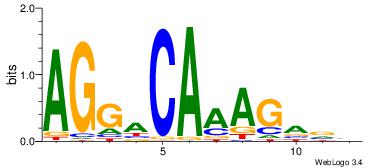 |
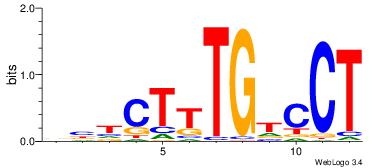 |
| Dataset #: |
4 |
| Motif ID: |
83 |
| Motif name: |
srACyCCGAyr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.58018 |
Alignment:
---KKTCGGKGTKS
GTGTGTGTGTGTGT
| Original motif Consensus sequence: SRACYCCGAYR |
Reverse complement motif Consensus sequence: KKTCGGKGTKS |
|
|
 |
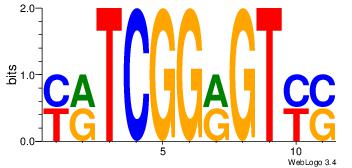 |
| Dataset #: 1 |
Motif ID: 3 |
Motif name: Motif 3 |
| Original motif Consensus sequence: HCAGRRVACASAG |
Reverse complement motif Consensus sequence: CTSTGTBKMCTGH |
|
|
 |
 |
Best Matches for Motif ID 3 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
13 |
| Similarity score: |
0.00926418 |
Alignment:
RAGKGMAGVRRGSRCASAGV
------HCAGRRVACASAG-
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
13 |
| Similarity score: |
0.0328703 |
Alignment:
VDBHMAGGTCACCCTGACCY
---HCAGRRVACASAG----
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
13 |
| Similarity score: |
0.0400966 |
Alignment:
AGTGSWSCACTGTGAMAMCACAGTG
----------HCAGRRVACASAG--
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
90 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
13 |
| Similarity score: |
0.0435274 |
Alignment:
GCAGAKGSAGGABDGHDG
--HCAGRRVACASAG---
| Original motif Consensus sequence: CHHCHBTCCTSCYTCTGC |
Reverse complement motif Consensus sequence: GCAGAKGSAGGABDGHDG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
13 |
| Similarity score: |
0.0448341 |
Alignment:
YDRCCASYAGRKGGCRSYV
------HCAGRRVACASAG
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
13 |
| Similarity score: |
0.045698 |
Alignment:
CCACTGTGVTGTCACAGTGCTCCA
---CTSTGTBKMCTGH--------
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
64 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.0457253 |
Alignment:
BTRGGDCARAGGKCA
HCAGRRVACASAG--
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
65 |
| Motif name: |
Tcfcp2l1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.0477557 |
Alignment:
CKGGDTBDMMCTGG
CTSTGTBKMCTGH-
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
94 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
13 |
| Similarity score: |
0.0478998 |
Alignment:
WHAATTAAARAARAAAAAAA
----HCAGRRVACASAG---
| Original motif Consensus sequence: TTTTTTTKTTKTTTAATTHW |
Reverse complement motif Consensus sequence: WHAATTAAARAARAAAAAAA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
88 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.048132 |
Alignment:
HVCDBBDBCAGCAG
-HCAGRRVACASAG
| Original motif Consensus sequence: CTGCTGBDBBDGBD |
Reverse complement motif Consensus sequence: HVCDBBDBCAGCAG |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 4 |
Motif name: Motif 4 |
| Original motif Consensus sequence: HAAAAHAAARMAAA |
Reverse complement motif Consensus sequence: TTTRKTTTHTTTTH |
|
|
 |
 |
Best Matches for Motif ID 4 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
94 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
14 |
| Similarity score: |
0.0333546 |
Alignment:
WHAATTAAARAARAAAAAAA
-----HAAAAHAAARMAAA-
| Original motif Consensus sequence: TTTTTTTKTTKTTTAATTHW |
Reverse complement motif Consensus sequence: WHAATTAAARAARAAAAAAA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
91 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.0408482 |
Alignment:
BTTHAAAAAAYAAAAAAR
---HAAAAHAAARMAAA-
| Original motif Consensus sequence: KTTTTTTKTTTTTTDAAB |
Reverse complement motif Consensus sequence: BTTHAAAAAAYAAAAAAR |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0486607 |
Alignment:
AAAAAAAAAAAAAT
HAAAAHAAARMAAA
| Original motif Consensus sequence: AAAAAAAAAAAAAT |
Reverse complement motif Consensus sequence: ATTTTTTTTTTTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0540179 |
Alignment:
TTTAATGTYTTTWA
TTTRKTTTHTTTTH
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
92 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
14 |
| Similarity score: |
0.0882898 |
Alignment:
TGTGTGDKTGTGTGTGTRTG
--TTTRKTTTHTTTTH----
| Original motif Consensus sequence: CAMACACACACAYDCACACA |
Reverse complement motif Consensus sequence: TGTGTGDKTGTGTGTGTRTG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
89 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0933036 |
Alignment:
CACACACACACWCACACA
HAAAAHAAARMAAA----
| Original motif Consensus sequence: CACACACACACWCACACA |
Reverse complement motif Consensus sequence: TGTGTGWGTGTGTGTGTG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0942815 |
Alignment:
CASABACWSACACA
HAAAAHAAARMAAA
| Original motif Consensus sequence: CASABACWSACACA |
Reverse complement motif Consensus sequence: TGTGTSWGTBTSTG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
14 |
| Similarity score: |
0.096284 |
Alignment:
RAGKGMAGVRRGSRCASAGV
----HAAAAHAAARMAAA--
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
45 |
| Motif name: |
Motif 45 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.563221 |
Alignment:
-TAASTCTWTTTTAA
TTTRKTTTHTTTTH-
| Original motif Consensus sequence: TAASTCTWTTTTAA |
Reverse complement motif Consensus sequence: TTAAAAWAGASTTA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.572837 |
Alignment:
AAAGRTMAAACTA-
HAAAAHAAARMAAA
| Original motif Consensus sequence: AAAGRTMAAACTA |
Reverse complement motif Consensus sequence: TAGTTTYAKCTTT |
|
|
 |
 |
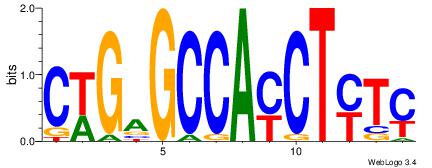
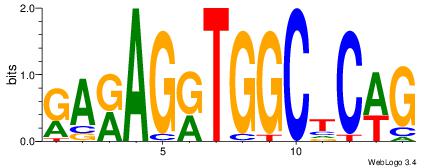
| Dataset #: 1 |
Motif ID: 5 |
Motif name: Motif 5 |
| Original motif Consensus sequence: CWGAGCCAYCTYTC |
Reverse complement motif Consensus sequence: GAKAGKTGGCTCWG |
|
|
 |
 |
Best Matches for Motif ID 5 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0474988 |
Alignment:
YDRCCASYAGRKGGCRSYV
-----GAKAGKTGGCTCWG
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
14 |
| Similarity score: |
0.0554531 |
Alignment:
CCACTGTGVTGTCACAGTGCTCCA
---GAKAGKTGGCTCWG-------
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.0576317 |
Alignment:
VCTSTGKSCMKBCTRCYCTK
---CWGAGCCAYCTYTC---
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0576632 |
Alignment:
ACTGTGACACCACAGTGGAGCACT
-CWGAGCCAYCTYTC---------
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
71 |
| Motif name: |
AmTGTGACACCACAGT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0578361 |
Alignment:
AMTGTGACACCACAGT
-CWGAGCCAYCTYTC-
| Original motif Consensus sequence: AMTGTGACACCACAGT |
Reverse complement motif Consensus sequence: ACTGTGGTGTCACART |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
14 |
| Similarity score: |
0.065312 |
Alignment:
AGTGSWSCACTGTGAMAMCACAGTG
---------CWGAGCCAYCTYTC--
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
59 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0658518 |
Alignment:
VHRGGTCABDBTGMCCTB
CWGAGCCAYCTYTC----
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
14 |
| Similarity score: |
0.0671359 |
Alignment:
MGGTCAGGGTGACCTRDBHV
----CWGAGCCAYCTYTC--
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
13 |
| Similarity score: |
0.538291 |
Alignment:
GGAGCACTGTGACACCACA-
------CWGAGCCAYCTYTC
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
65 |
| Motif name: |
Tcfcp2l1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.560622 |
Alignment:
-CKGGDTBDMMCTGG
GAKAGKTGGCTCWG-
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 6 |
Motif name: Motif 6 |
| Original motif Consensus sequence: CASABACWSACACA |
Reverse complement motif Consensus sequence: TGTGTSWGTBTSTG |
|
|
 |
 |
Best Matches for Motif ID 6 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
92 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0383879 |
Alignment:
TGTGTGDKTGTGTGTGTRTG
TGTGTSWGTBTSTG------
| Original motif Consensus sequence: CAMACACACACAYDCACACA |
Reverse complement motif Consensus sequence: TGTGTGDKTGTGTGTGTRTG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
89 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0411352 |
Alignment:
TGTGTGWGTGTGTGTGTG
TGTGTSWGTBTSTG----
| Original motif Consensus sequence: CACACACACACWCACACA |
Reverse complement motif Consensus sequence: TGTGTGWGTGTGTGTGTG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
14 |
| Similarity score: |
0.0803592 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
-----TGTGTSWGTBTSTG--
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
59 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.0865284 |
Alignment:
VHRGGTCABDBTGMCCTB
-TGTGTSWGTBTSTG---
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.0874787 |
Alignment:
TGTGGTGTCACAGTGCTCC
---TGTGTSWGTBTSTG--
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
94 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
14 |
| Similarity score: |
0.0913053 |
Alignment:
TTTTTTTKTTKTTTAATTHW
-TGTGTSWGTBTSTG-----
| Original motif Consensus sequence: TTTTTTTKTTKTTTAATTHW |
Reverse complement motif Consensus sequence: WHAATTAAARAARAAAAAAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.0930634 |
Alignment:
CCACTGTGVTGTCACAGTGCTCCA
-------TGTGTSWGTBTSTG---
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0933886 |
Alignment:
HAAAAHAAARMAAA
CASABACWSACACA
| Original motif Consensus sequence: HAAAAHAAARMAAA |
Reverse complement motif Consensus sequence: TTTRKTTTHTTTTH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
91 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0935906 |
Alignment:
KTTTTTTKTTTTTTDAAB
TGTGTSWGTBTSTG----
| Original motif Consensus sequence: KTTTTTTKTTTTTTDAAB |
Reverse complement motif Consensus sequence: BTTHAAAAAAYAAAAAAR |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
11 |
| Number of overlap: |
14 |
| Similarity score: |
0.0939701 |
Alignment:
ACTGTGACACCACAGTGGAGCACT
----------CASABACWSACACA
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
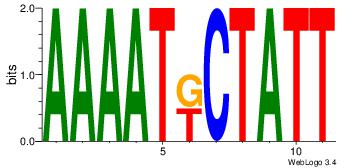
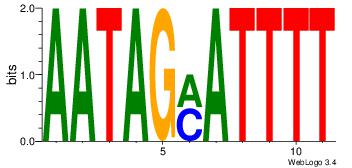
| Dataset #: 1 |
Motif ID: 7 |
Motif name: Motif 7 |
| Original motif Consensus sequence: AAAATKCTATT |
Reverse complement motif Consensus sequence: AATAGYATTTT |
|
|
 |
 |
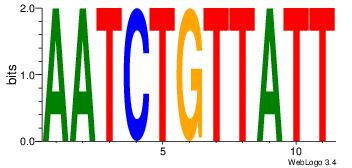
Best Matches for Motif ID 7 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
25 |
| Motif name: |
Motif 25 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0795455 |
Alignment:
AATCTGTTATT
AAAATKCTATT
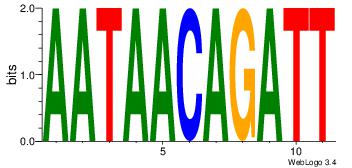
| Original motif Consensus sequence: AATAACAGATT |
Reverse complement motif Consensus sequence: AATCTGTTATT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
13 |
| Motif name: |
Motif 13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0795455 |
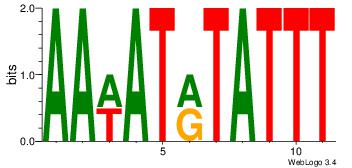
Alignment:
AAATAKATWTT
AAAATKCTATT
| Original motif Consensus sequence: AAWATRTATTT |
Reverse complement motif Consensus sequence: AAATAKATWTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
45 |
| Motif name: |
Motif 45 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0909091 |
Alignment:
TAASTCTWTTTTAA
-AATAGYATTTT--
| Original motif Consensus sequence: TAASTCTWTTTTAA |
Reverse complement motif Consensus sequence: TTAAAAWAGASTTA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
12 |
| Motif name: |
Motif 12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
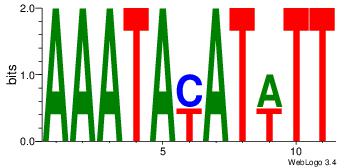
Alignment:
AAATGTGTTGT
AATAGYATTTT
| Original motif Consensus sequence: ACAACACATTT |
Reverse complement motif Consensus sequence: AAATGTGTTGT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
TWAAAKACATTAAA
AAAATKCTATT---
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
TAGTTTYAKCTTT
-AATAGYATTTT-
| Original motif Consensus sequence: AAAGRTMAAACTA |
Reverse complement motif Consensus sequence: TAGTTTYAKCTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
ATTTTTTTTTTTTT
AATAGYATTTT---
| Original motif Consensus sequence: AAAAAAAAAAAAAT |
Reverse complement motif Consensus sequence: ATTTTTTTTTTTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
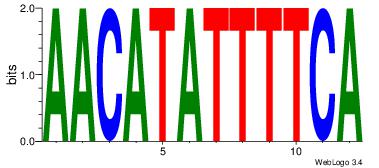
Alignment:
TGAAAATATGTT
-AAAATKCTATT
| Original motif Consensus sequence: AACATATTTTCA |
Reverse complement motif Consensus sequence: TGAAAATATGTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
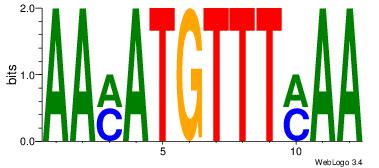
Alignment:
TTYAAACATYTT
-AATAGYATTTT
| Original motif Consensus sequence: AAMATGTTTMAA |
Reverse complement motif Consensus sequence: TTYAAACATYTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
AAAMATTGTTT
AAAATKCTATT
| Original motif Consensus sequence: AAAMATTGTTT |
Reverse complement motif Consensus sequence: AAACAATYTTT |
|
|
 |
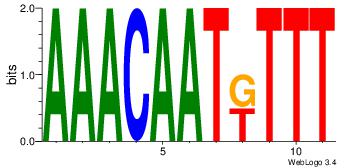 |
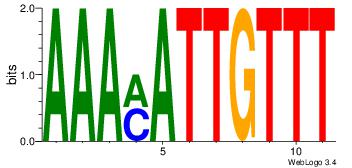
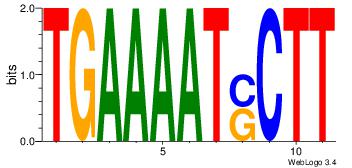
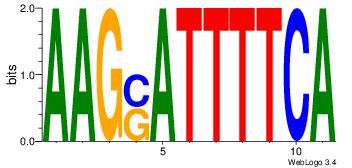
| Dataset #: 1 |
Motif ID: 8 |
Motif name: Motif 8 |
| Original motif Consensus sequence: TGAAAATSCTT |
Reverse complement motif Consensus sequence: AAGSATTTTCA |
|
|
 |
 |
Best Matches for Motif ID 8 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0681818 |
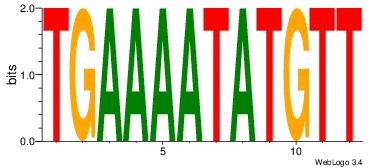
Alignment:
TGAAAATATGTT
TGAAAATSCTT-
| Original motif Consensus sequence: AACATATTTTCA |
Reverse complement motif Consensus sequence: TGAAAATATGTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0909091 |
Alignment:
TTTAATGTYTTTWA
---AAGSATTTTCA
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
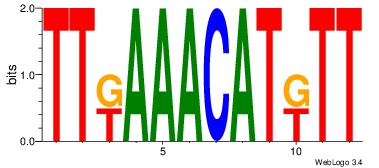
Alignment:
AAMATGTTTMAA
AAGSATTTTCA-
| Original motif Consensus sequence: AAMATGTTTMAA |
Reverse complement motif Consensus sequence: TTYAAACATYTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
AAACAATYTTT
TGAAAATSCTT
| Original motif Consensus sequence: AAAMATTGTTT |
Reverse complement motif Consensus sequence: AAACAATYTTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.111179 |
Alignment:
TGGAGCACTGTGACAVCACAGTGG
--AAGSATTTTCA-----------
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
11 |
| Similarity score: |
0.113005 |
Alignment:
TGTGGTGTCACAGTGCTCC
-------TGAAAATSCTT-
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
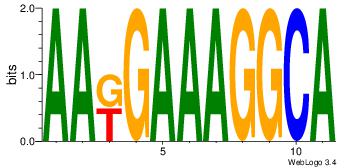
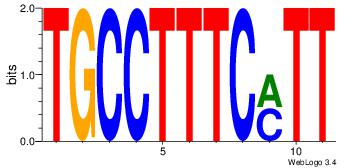
43 |
| Motif name: |
Motif 43 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
TGCCTTTCYTT
TGAAAATSCTT
| Original motif Consensus sequence: AAKGAAAGGCA |
Reverse complement motif Consensus sequence: TGCCTTTCYTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
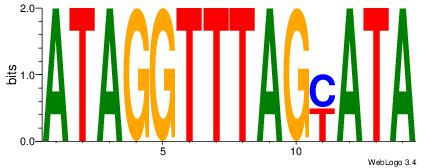
46 |
| Motif name: |
Motif 46 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
TATKCTAAACCTAT
--TGAAAATSCTT-
| Original motif Consensus sequence: ATAGGTTTAGYATA |
Reverse complement motif Consensus sequence: TATKCTAAACCTAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
45 |
| Motif name: |
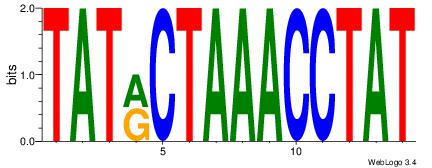
Motif 45 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
TTAAAAWAGASTTA
--TGAAAATSCTT-
| Original motif Consensus sequence: TAASTCTWTTTTAA |
Reverse complement motif Consensus sequence: TTAAAAWAGASTTA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
24 |
| Motif name: |
Motif 24 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.125 |
Alignment:
TTWATGWCTTT
TGAAAATSCTT
| Original motif Consensus sequence: AAAGWCATWAA |
Reverse complement motif Consensus sequence: TTWATGWCTTT |
|
|
 |
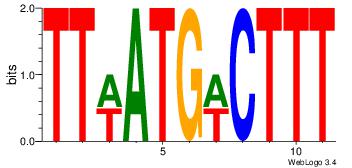 |
| Dataset #: 1 |
Motif ID: 9 |
Motif name: Motif 9 |
| Original motif Consensus sequence: AACATATTTTCA |
Reverse complement motif Consensus sequence: TGAAAATATGTT |
|
|
 |
 |
Best Matches for Motif ID 9 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0729167 |
Alignment:
AAMATGTTTMAA
AACATATTTTCA
| Original motif Consensus sequence: AAMATGTTTMAA |
Reverse complement motif Consensus sequence: TTYAAACATYTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
45 |
| Motif name: |
Motif 45 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.09375 |
Alignment:
TAASTCTWTTTTAA
--AACATATTTTCA
| Original motif Consensus sequence: TAASTCTWTTTTAA |
Reverse complement motif Consensus sequence: TTAAAAWAGASTTA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.104167 |
Alignment:
TWAAAKACATTAAA
-AACATATTTTCA-
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
12 |
| Similarity score: |
0.120305 |
Alignment:
AGTGCTCCACTGTGGTGTCACAGT
--------AACATATTTTCA----
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
89 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
12 |
| Similarity score: |
0.12128 |
Alignment:
CACACACACACWCACACA
--AACATATTTTCA----
| Original motif Consensus sequence: CACACACACACWCACACA |
Reverse complement motif Consensus sequence: TGTGTGWGTGTGTGTGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
71 |
| Motif name: |
AmTGTGACACCACAGT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
12 |
| Similarity score: |
0.12207 |
Alignment:
AMTGTGACACCACAGT
----TGAAAATATGTT
| Original motif Consensus sequence: AMTGTGACACCACAGT |
Reverse complement motif Consensus sequence: ACTGTGGTGTCACART |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
11 |
| Number of overlap: |
12 |
| Similarity score: |
0.122466 |
Alignment:
CCACTGTGVTGTCACAGTGCTCCA
--AACATATTTTCA----------
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
92 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
12 |
| Similarity score: |
0.123397 |
Alignment:
CAMACACACACAYDCACACA
----AACATATTTTCA----
| Original motif Consensus sequence: CAMACACACACAYDCACACA |
Reverse complement motif Consensus sequence: TGTGTGDKTGTGTGTGTRTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.124008 |
Alignment:
AGTGSWSCACTGTGAMAMCACAGTG
------------TGAAAATATGTT-
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.125 |
Alignment:
ATTTTTTTTTTTTT
AACATATTTTCA--
| Original motif Consensus sequence: AAAAAAAAAAAAAT |
Reverse complement motif Consensus sequence: ATTTTTTTTTTTTT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 10 |
Motif name: Motif 10 |
| Original motif Consensus sequence: AAAMATTGTTT |
Reverse complement motif Consensus sequence: AAACAATYTTT |
|
|
 |
 |
Best Matches for Motif ID 10 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
45 |
| Motif name: |
Motif 45 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0700758 |
Alignment:
TTAAAAWAGASTTA
---AAAMATTGTTT
| Original motif Consensus sequence: TAASTCTWTTTTAA |
Reverse complement motif Consensus sequence: TTAAAAWAGASTTA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
13 |
| Motif name: |
Motif 13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0700758 |
Alignment:
AAWATRTATTT
AAAMATTGTTT
| Original motif Consensus sequence: AAWATRTATTT |
Reverse complement motif Consensus sequence: AAATAKATWTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0814394 |
Alignment:
TGAAAATATGTT
AAAMATTGTTT-
| Original motif Consensus sequence: AACATATTTTCA |
Reverse complement motif Consensus sequence: TGAAAATATGTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0814394 |
Alignment:
AAGSATTTTCA
AAAMATTGTTT
| Original motif Consensus sequence: TGAAAATSCTT |
Reverse complement motif Consensus sequence: AAGSATTTTCA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
25 |
| Motif name: |
Motif 25 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0814394 |
Alignment:
AATAACAGATT
AAAMATTGTTT
| Original motif Consensus sequence: AATAACAGATT |
Reverse complement motif Consensus sequence: AATCTGTTATT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.092803 |
Alignment:
AAAATKCTATT
AAAMATTGTTT
| Original motif Consensus sequence: AAAATKCTATT |
Reverse complement motif Consensus sequence: AATAGYATTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.092803 |
Alignment:
TTTAATGTYTTTWA
--AAAMATTGTTT-
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.092803 |
Alignment:
AAAAAAAAAAAAAT
---AAACAATYTTT
| Original motif Consensus sequence: AAAAAAAAAAAAAT |
Reverse complement motif Consensus sequence: ATTTTTTTTTTTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
29 |
| Motif name: |
Motif 29 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.092803 |
Alignment:
ATTTATTGCTA
AAAMATTGTTT
| Original motif Consensus sequence: ATTTATTGCTA |
Reverse complement motif Consensus sequence: TAGCAATAAAT |
|
|
 |
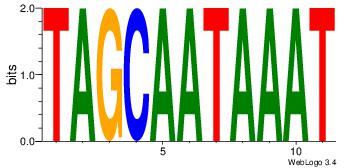 |
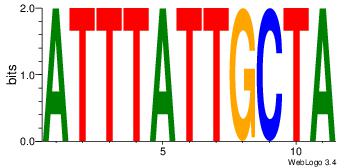
| Dataset #: |
1 |
| Motif ID: |
12 |
| Motif name: |
Motif 12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.092803 |
Alignment:
AAATGTGTTGT
AAAMATTGTTT
| Original motif Consensus sequence: ACAACACATTT |
Reverse complement motif Consensus sequence: AAATGTGTTGT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 11 |
Motif name: Motif 11 |
| Original motif Consensus sequence: AAMATGTTTMAA |
Reverse complement motif Consensus sequence: TTYAAACATYTT |
|
|
 |
 |
Best Matches for Motif ID 11 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0479167 |
Alignment:
AACATATTTTCA
AAMATGTTTMAA
| Original motif Consensus sequence: AACATATTTTCA |
Reverse complement motif Consensus sequence: TGAAAATATGTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.06875 |
Alignment:
TTTAATGTYTTTWA
TTYAAACATYTT--
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
45 |
| Motif name: |
Motif 45 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0791667 |
Alignment:
TTAAAAWAGASTTA
TTYAAACATYTT--
| Original motif Consensus sequence: TAASTCTWTTTTAA |
Reverse complement motif Consensus sequence: TTAAAAWAGASTTA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
91 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
12 |
| Similarity score: |
0.0830729 |
Alignment:
BTTHAAAAAAYAAAAAAR
-TTYAAACATYTT-----
| Original motif Consensus sequence: KTTTTTTKTTTTTTDAAB |
Reverse complement motif Consensus sequence: BTTHAAAAAAYAAAAAAR |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
94 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
12 |
| Similarity score: |
0.0970238 |
Alignment:
WHAATTAAARAARAAAAAAA
-------AAMATGTTTMAA-
| Original motif Consensus sequence: TTTTTTTKTTKTTTAATTHW |
Reverse complement motif Consensus sequence: WHAATTAAARAARAAAAAAA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
60 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
12 |
| Similarity score: |
0.0983333 |
Alignment:
AAAGGTCAAAGGTCAAC
----TTYAAACATYTT-
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.1 |
Alignment:
AAAAAAAAAAAAAT
-AAMATGTTTMAA-
| Original motif Consensus sequence: AAAAAAAAAAAAAT |
Reverse complement motif Consensus sequence: ATTTTTTTTTTTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
46 |
| Motif name: |
Motif 46 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.565909 |
Alignment:
TATKCTAAACCTAT-
---TTYAAACATYTT
| Original motif Consensus sequence: ATAGGTTTAGYATA |
Reverse complement motif Consensus sequence: TATKCTAAACCTAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
13 |
| Motif name: |
Motif 13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.565909 |
Alignment:
AAWATRTATTT-
AAMATGTTTMAA
| Original motif Consensus sequence: AAWATRTATTT |
Reverse complement motif Consensus sequence: AAATAKATWTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
12 |
| Motif name: |
Motif 12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.577273 |
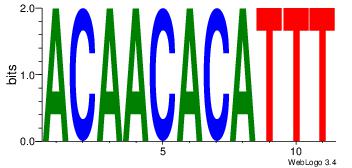
Alignment:
-AAATGTGTTGT
AAMATGTTTMAA
| Original motif Consensus sequence: ACAACACATTT |
Reverse complement motif Consensus sequence: AAATGTGTTGT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 12 |
Motif name: Motif 12 |
| Original motif Consensus sequence: ACAACACATTT |
Reverse complement motif Consensus sequence: AAATGTGTTGT |
|
|
 |
 |
Best Matches for Motif ID 12 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
11 |
| Similarity score: |
0.074904 |
Alignment:
AGTGCTCCACTGTGGTGTCACAGT
-------AAATGTGTTGT------
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0795455 |
Alignment:
TGGAGCACTGTGACAVCACAGTGG
------------ACAACACATTT-
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0795455 |
Alignment:
TWAAAKACATTAAA
-ACAACACATTT--
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0891053 |
Alignment:
AGTGSWSCACTGTGAMAMCACAGTG
--------------ACAACACATTT
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
13 |
| Motif name: |
Motif 13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0909091 |
Alignment:
AAWATRTATTT
ACAACACATTT
| Original motif Consensus sequence: AAWATRTATTT |
Reverse complement motif Consensus sequence: AAATAKATWTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
45 |
| Motif name: |
Motif 45 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0909091 |
Alignment:
TTAAAAWAGASTTA
--ACAACACATTT-
| Original motif Consensus sequence: TAASTCTWTTTTAA |
Reverse complement motif Consensus sequence: TTAAAAWAGASTTA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
AATAGYATTTT
AAATGTGTTGT
| Original motif Consensus sequence: AAAATKCTATT |
Reverse complement motif Consensus sequence: AATAGYATTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
46 |
| Motif name: |
Motif 46 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
ATAGGTTTAGYATA
AAATGTGTTGT---
| Original motif Consensus sequence: ATAGGTTTAGYATA |
Reverse complement motif Consensus sequence: TATKCTAAACCTAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
AAMATGTTTMAA
AAATGTGTTGT-
| Original motif Consensus sequence: AAMATGTTTMAA |
Reverse complement motif Consensus sequence: TTYAAACATYTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
AACATATTTTCA
AAATGTGTTGT-
| Original motif Consensus sequence: AACATATTTTCA |
Reverse complement motif Consensus sequence: TGAAAATATGTT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 13 |
Motif name: Motif 13 |
| Original motif Consensus sequence: AAWATRTATTT |
Reverse complement motif Consensus sequence: AAATAKATWTT |
|
|
 |
 |
Best Matches for Motif ID 13 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
45 |
| Motif name: |
Motif 45 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0795455 |
Alignment:
TTAAAAWAGASTTA
--AAWATRTATTT-
| Original motif Consensus sequence: TAASTCTWTTTTAA |
Reverse complement motif Consensus sequence: TTAAAAWAGASTTA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0795455 |
Alignment:
TGAAAATATGTT
-AAATAKATWTT
| Original motif Consensus sequence: AACATATTTTCA |
Reverse complement motif Consensus sequence: TGAAAATATGTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0795455 |
Alignment:
AATAGYATTTT
AAWATRTATTT
| Original motif Consensus sequence: AAAATKCTATT |
Reverse complement motif Consensus sequence: AATAGYATTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0909091 |
Alignment:
TWAAAKACATTAAA
--AAATAKATWTT-
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
12 |
| Motif name: |
Motif 12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0909091 |
Alignment:
ACAACACATTT
AAWATRTATTT
| Original motif Consensus sequence: ACAACACATTT |
Reverse complement motif Consensus sequence: AAATGTGTTGT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
24 |
| Motif name: |
Motif 24 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0909091 |
Alignment:
TTWATGWCTTT
AAWATRTATTT
| Original motif Consensus sequence: AAAGWCATWAA |
Reverse complement motif Consensus sequence: TTWATGWCTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0909091 |
Alignment:
AAMATGTTTMAA
AAWATRTATTT-
| Original motif Consensus sequence: AAMATGTTTMAA |
Reverse complement motif Consensus sequence: TTYAAACATYTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0909091 |
Alignment:
AAAMATTGTTT
AAWATRTATTT
| Original motif Consensus sequence: AAAMATTGTTT |
Reverse complement motif Consensus sequence: AAACAATYTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
25 |
| Motif name: |
Motif 25 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0909091 |
Alignment:
AATCTGTTATT
AAWATRTATTT
| Original motif Consensus sequence: AATAACAGATT |
Reverse complement motif Consensus sequence: AATCTGTTATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
AAAAAAAAAAAAAT
---AAATAKATWTT
| Original motif Consensus sequence: AAAAAAAAAAAAAT |
Reverse complement motif Consensus sequence: ATTTTTTTTTTTTT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 14 |
Motif name: Motif 14 |
| Original motif Consensus sequence: TAATKATATAA |
Reverse complement motif Consensus sequence: TTATATYATTA |
|
|
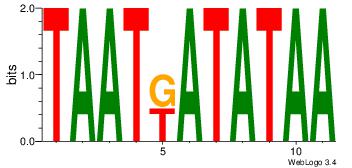 |
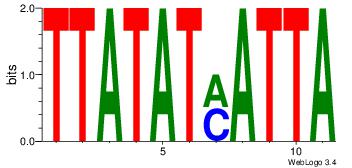 |
Best Matches for Motif ID 14 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0482955 |
Alignment:
TTTAATGTYTTTWA
--TTATATYATTA-
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
94 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
11 |
| Similarity score: |
0.0629058 |
Alignment:
WHAATTAAARAARAAAAAAA
-TAATKATATAA--------
| Original motif Consensus sequence: TTTTTTTKTTKTTTAATTHW |
Reverse complement motif Consensus sequence: WHAATTAAARAARAAAAAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
23 |
| Motif name: |
Motif 23 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0710227 |
Alignment:
ATGTWTTCATTMAT
-TTATATYATTA--
| Original motif Consensus sequence: ATGTWTTCATTMAT |
Reverse complement motif Consensus sequence: ATYAATGAAWACAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
29 |
| Motif name: |
Motif 29 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0710227 |
Alignment:
ATTTATTGCTA
TTATATYATTA
| Original motif Consensus sequence: ATTTATTGCTA |
Reverse complement motif Consensus sequence: TAGCAATAAAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0710227 |
Alignment:
TWTTGACAYAGA
TAATKATATAA-
| Original motif Consensus sequence: TCTKTGTCAAWA |
Reverse complement motif Consensus sequence: TWTTGACAYAGA |
|
|
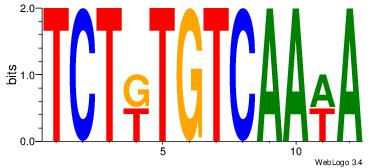 |
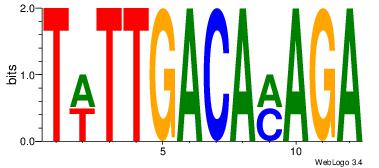 |
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0710227 |
Alignment:
TGAAAATATGTT
-TAATKATATAA
| Original motif Consensus sequence: AACATATTTTCA |
Reverse complement motif Consensus sequence: TGAAAATATGTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0710227 |
Alignment:
ATTTTTTTTTTTTT
---TTATATYATTA
| Original motif Consensus sequence: AAAAAAAAAAAAAT |
Reverse complement motif Consensus sequence: ATTTTTTTTTTTTT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
91 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
11 |
| Similarity score: |
0.0752841 |
Alignment:
KTTTTTTKTTTTTTDAAB
-----TTATATYATTA--
| Original motif Consensus sequence: KTTTTTTKTTTTTTDAAB |
Reverse complement motif Consensus sequence: BTTHAAAAAAYAAAAAAR |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0778409 |
Alignment:
TTTRKTTTHTTTTH
-TTATATYATTA--
| Original motif Consensus sequence: HAAAAHAAARMAAA |
Reverse complement motif Consensus sequence: TTTRKTTTHTTTTH |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
45 |
| Motif name: |
Motif 45 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0823864 |
Alignment:
TAASTCTWTTTTAA
TAATKATATAA---
| Original motif Consensus sequence: TAASTCTWTTTTAA |
Reverse complement motif Consensus sequence: TTAAAAWAGASTTA |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 15 |
Motif name: Motif 15 |
| Original motif Consensus sequence: AAAGRTMAAACTA |
Reverse complement motif Consensus sequence: TAGTTTYAKCTTT |
|
|
 |
 |
Best Matches for Motif ID 15 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
46 |
| Motif name: |
Motif 46 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.0865385 |
Alignment:
ATAGGTTTAGYATA
-TAGTTTYAKCTTT
| Original motif Consensus sequence: ATAGGTTTAGYATA |
Reverse complement motif Consensus sequence: TATKCTAAACCTAT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
60 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.0938462 |
Alignment:
AAAGGTCAAAGGTCAAC
AAAGRTMAAACTA----
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.0961538 |
Alignment:
ATTTTTTTTTTTTT
-TAGTTTYAKCTTT
| Original motif Consensus sequence: AAAAAAAAAAAAAT |
Reverse complement motif Consensus sequence: ATTTTTTTTTTTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.100962 |
Alignment:
HAAAAHAAARMAAA
AAAGRTMAAACTA-
| Original motif Consensus sequence: HAAAAHAAARMAAA |
Reverse complement motif Consensus sequence: TTTRKTTTHTTTTH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
94 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
13 |
| Similarity score: |
0.101648 |
Alignment:
WHAATTAAARAARAAAAAAA
------AAAGRTMAAACTA-
| Original motif Consensus sequence: TTTTTTTKTTKTTTAATTHW |
Reverse complement motif Consensus sequence: WHAATTAAARAARAAAAAAA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
91 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.110577 |
Alignment:
KTTTTTTKTTTTTTDAAB
TAGTTTYAKCTTT-----
| Original motif Consensus sequence: KTTTTTTKTTTTTTDAAB |
Reverse complement motif Consensus sequence: BTTHAAAAAAYAAAAAAR |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
23 |
| Motif name: |
Motif 23 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.125 |
Alignment:
ATGTWTTCATTMAT
-TAGTTTYAKCTTT
| Original motif Consensus sequence: ATGTWTTCATTMAT |
Reverse complement motif Consensus sequence: ATYAATGAAWACAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
45 |
| Motif name: |
Motif 45 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.125 |
Alignment:
TTAAAAWAGASTTA
-AAAGRTMAAACTA
| Original motif Consensus sequence: TAASTCTWTTTTAA |
Reverse complement motif Consensus sequence: TTAAAAWAGASTTA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.125 |
Alignment:
GAKAGKTGGCTCWG
AAAGRTMAAACTA-
| Original motif Consensus sequence: CWGAGCCAYCTYTC |
Reverse complement motif Consensus sequence: GAKAGKTGGCTCWG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
22 |
| Motif name: |
Motif 22 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.614583 |
Alignment:
-ATGCTCACCATAAA
AAAGRTMAAACTA--
| Original motif Consensus sequence: TTTATGGTGAGCAT |
Reverse complement motif Consensus sequence: ATGCTCACCATAAA |
|
|
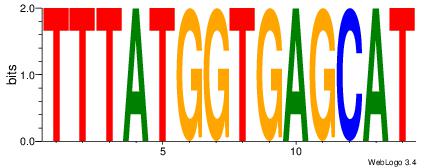 |
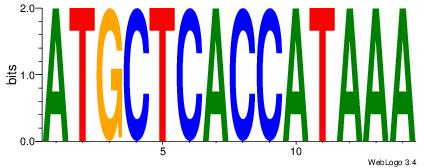 |
| Dataset #: 1 |
Motif ID: 16 |
Motif name: Motif 16 |
| Original motif Consensus sequence: TCTKTGTCAAWA |
Reverse complement motif Consensus sequence: TWTTGACAYAGA |
|
|
 |
 |
Best Matches for Motif ID 16 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
12 |
| Similarity score: |
0.100225 |
Alignment:
TGGAGCACTGTGACAVCACAGTGG
------TCTKTGTCAAWA------
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
12 |
| Similarity score: |
0.100694 |
Alignment:
AGTGSWSCACTGTGAMAMCACAGTG
--------TCTKTGTCAAWA-----
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.102141 |
Alignment:
TGTGGTGTCACAGTGCTCC
--TWTTGACAYAGA-----
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
13 |
| Number of overlap: |
12 |
| Similarity score: |
0.103433 |
Alignment:
AGTGCTCCACTGTGGTGTCACAGT
------------TWTTGACAYAGA
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
71 |
| Motif name: |
AmTGTGACACCACAGT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.104492 |
Alignment:
AMTGTGACACCACAGT
TCTKTGTCAAWA----
| Original motif Consensus sequence: AMTGTGACACCACAGT |
Reverse complement motif Consensus sequence: ACTGTGGTGTCACART |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
91 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
12 |
| Similarity score: |
0.11849 |
Alignment:
KTTTTTTKTTTTTTDAAB
----TCTKTGTCAAWA--
| Original motif Consensus sequence: KTTTTTTKTTTTTTDAAB |
Reverse complement motif Consensus sequence: BTTHAAAAAAYAAAAAAR |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
94 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
12 |
| Similarity score: |
0.119048 |
Alignment:
TTTTTTTKTTKTTTAATTHW
-------TCTKTGTCAAWA-
| Original motif Consensus sequence: TTTTTTTKTTKTTTAATTHW |
Reverse complement motif Consensus sequence: WHAATTAAARAARAAAAAAA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
64 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
12 |
| Similarity score: |
0.121043 |
Alignment:
TGRCCTKTGHCCKAB
---TCTKTGTCAAWA
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
60 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.121667 |
Alignment:
GTTGACCTTTGACCTTT
-----TCTKTGTCAAWA
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.125 |
Alignment:
TWAAAKACATTAAA
-TWTTGACAYAGA-
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 17 |
Motif name: Motif 17 |
| Original motif Consensus sequence: CTTCTGACCTC |
Reverse complement motif Consensus sequence: GAGGTCAGAAG |
|
|
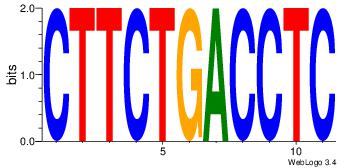 |
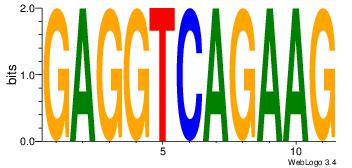 |
Best Matches for Motif ID 17 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
60 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
11 |
| Similarity score: |
0.0348052 |
Alignment:
AAAGGTCAAAGGTCAAC
-GAGGTCAGAAG-----
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
64 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0792232 |
Alignment:
BTRGGDCARAGGKCA
-GAGGTCAGAAG---
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
24 |
| Motif name: |
Motif 24 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0793506 |
Alignment:
TTWATGWCTTT
CTTCTGACCTC
| Original motif Consensus sequence: AAAGWCATWAA |
Reverse complement motif Consensus sequence: TTWATGWCTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
21 |
| Motif name: |
Motif 21 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0793506 |
Alignment:
CTGCTTTCCMAA
CTTCTGACCTC-
| Original motif Consensus sequence: CTGCTTTCCMAA |
Reverse complement motif Consensus sequence: TTYGGAAAGCAG |
|
|
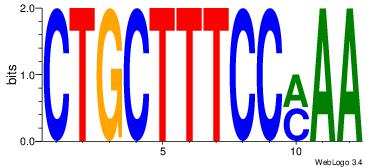 |
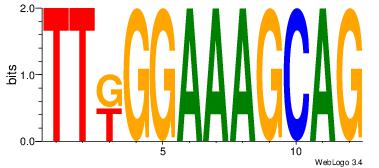 |
| Dataset #: |
1 |
| Motif ID: |
22 |
| Motif name: |
Motif 22 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0793506 |
Alignment:
ATGCTCACCATAAA
CTTCTGACCTC---
| Original motif Consensus sequence: TTTATGGTGAGCAT |
Reverse complement motif Consensus sequence: ATGCTCACCATAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0793506 |
Alignment:
TAGTTTYAKCTTT
-CTTCTGACCTC-
| Original motif Consensus sequence: AAAGRTMAAACTA |
Reverse complement motif Consensus sequence: TAGTTTYAKCTTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
59 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0794143 |
Alignment:
BAGGYCABHBTGACCKHV
------CTTCTGACCTC-
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0799909 |
Alignment:
ACTGTGACACCACAGTGGAGCACT
CTTCTGACCTC-------------
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
71 |
| Motif name: |
AmTGTGACACCACAGT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
11 |
| Similarity score: |
0.0800609 |
Alignment:
ACTGTGGTGTCACART
-----GAGGTCAGAAG
| Original motif Consensus sequence: AMTGTGACACCACAGT |
Reverse complement motif Consensus sequence: ACTGTGGTGTCACART |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0877439 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
-CTTCTGACCTC---------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 18 |
Motif name: Motif 18 |
| Original motif Consensus sequence: CAAAGTCCAGC |
Reverse complement motif Consensus sequence: GCTGGACTTTG |
|
|
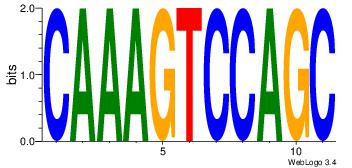 |
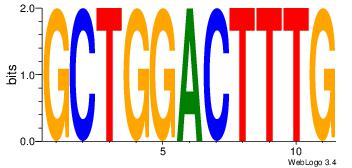 |
Best Matches for Motif ID 18 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
60 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
11 |
| Similarity score: |
0.0763636 |
Alignment:
AAAGGTCAAAGGTCAAC
------CAAAGTCCAGC
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
70 |
| Motif name: |
CagTGCTCCACTGTGgT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0780303 |
Alignment:
ABCACAGTGGAGCACTG
--CAAAGTCCAGC----
| Original motif Consensus sequence: CAGTGCTCCACTGTGBT |
Reverse complement motif Consensus sequence: ABCACAGTGGAGCACTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
76 |
| Motif name: |
CAcCACAGTGGAGCAct |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0795455 |
Alignment:
AGTGCTCCACTGTGDTG
---GCTGGACTTTG---
| Original motif Consensus sequence: CAHCACAGTGGAGCACT |
Reverse complement motif Consensus sequence: AGTGCTCCACTGTGDTG |
|
|
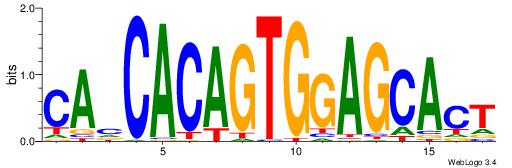 |
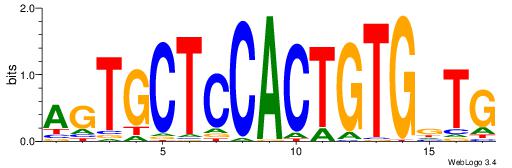 |
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
11 |
| Number of overlap: |
11 |
| Similarity score: |
0.0797055 |
Alignment:
ACTGTGACACCACAGTGGAGCACT
----------CAAAGTCCAGC---
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
12 |
| Number of overlap: |
11 |
| Similarity score: |
0.0894661 |
Alignment:
AGTGSWSCACTGTGAMAMCACAGTG
---GCTGGACTTTG-----------
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.10258 |
Alignment:
CCACTGTGVTGTCACAGTGCTCCA
------------CAAAGTCCAGC-
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
11 |
| Similarity score: |
0.107008 |
Alignment:
TGTGGTGTCACAGTGCTCC
--------CAAAGTCCAGC
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.110715 |
Alignment:
TTCAGCACCATGGACAGCKCC
-------CAAAGTCCAGC---
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
AAAGRTMAAACTA
CAAAGTCCAGC--
| Original motif Consensus sequence: AAAGRTMAAACTA |
Reverse complement motif Consensus sequence: TAGTTTYAKCTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.120455 |
Alignment:
HAAAAHAAARMAAA
CAAAGTCCAGC---
| Original motif Consensus sequence: HAAAAHAAARMAAA |
Reverse complement motif Consensus sequence: TTTRKTTTHTTTTH |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 19 |
Motif name: Motif 19 |
| Original motif Consensus sequence: AGAGAAAG |
Reverse complement motif Consensus sequence: CTTTCTCT |
|
|
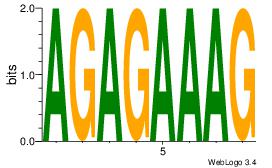 |
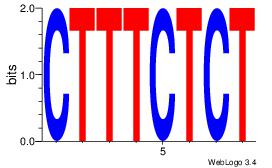 |
Best Matches for Motif ID 19 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
43 |
| Motif name: |
Motif 43 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.046875 |
Alignment:
TGCCTTTCYTT
---CTTTCTCT
| Original motif Consensus sequence: AAKGAAAGGCA |
Reverse complement motif Consensus sequence: TGCCTTTCYTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
21 |
| Motif name: |
Motif 21 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
CTGCTTTCCMAA
---CTTTCTCT-
| Original motif Consensus sequence: CTGCTTTCCMAA |
Reverse complement motif Consensus sequence: TTYGGAAAGCAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
44 |
| Motif name: |
Motif 44 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
TTCTTYCATT
--CTTTCTCT
| Original motif Consensus sequence: AATGKAAGAA |
Reverse complement motif Consensus sequence: TTCTTYCATT |
|
|
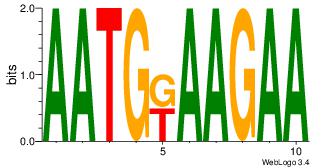 |
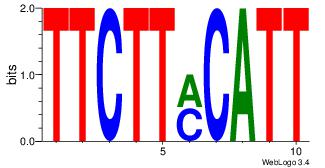 |
| Dataset #: |
1 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
TWTTGACAYAGA
-AGAGAAAG---
| Original motif Consensus sequence: TCTKTGTCAAWA |
Reverse complement motif Consensus sequence: TWTTGACAYAGA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.09375 |
Alignment:
AAAGRTMAAACTA
--AGAGAAAG---
| Original motif Consensus sequence: AAAGRTMAAACTA |
Reverse complement motif Consensus sequence: TAGTTTYAKCTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.09375 |
Alignment:
AAAAAAAAAAAAAT
AGAGAAAG------
| Original motif Consensus sequence: AAAAAAAAAAAAAT |
Reverse complement motif Consensus sequence: ATTTTTTTTTTTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
41 |
| Motif name: |
Motif 41 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.09375 |
Alignment:
TTTCCTCCA
CTTTCTCT-
| Original motif Consensus sequence: TGGAGGAAA |
Reverse complement motif Consensus sequence: TTTCCTCCA |
|
|
 |
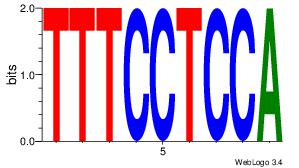 |
| Dataset #: |
5 |
| Motif ID: |
94 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
10 |
| Number of overlap: |
8 |
| Similarity score: |
0.0959821 |
Alignment:
WHAATTAAARAARAAAAAAA
---------AGAGAAAG---
| Original motif Consensus sequence: TTTTTTTKTTKTTTAATTHW |
Reverse complement motif Consensus sequence: WHAATTAAARAARAAAAAAA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
60 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.09625 |
Alignment:
AAAGGTCAAAGGTCAAC
---AGAGAAAG------
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.101562 |
Alignment:
HAAAAHAAARMAAA
--AGAGAAAG----
| Original motif Consensus sequence: HAAAAHAAARMAAA |
Reverse complement motif Consensus sequence: TTTRKTTTHTTTTH |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 20 |
Motif name: Motif 20 |
| Original motif Consensus sequence: TCCTCCTGGAA |
Reverse complement motif Consensus sequence: TTCCAGGAGGA |
|
|
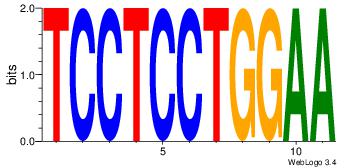 |
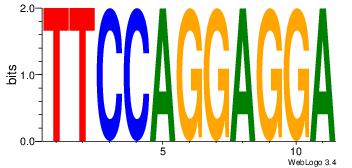 |
Best Matches for Motif ID 20 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
11 |
| Similarity score: |
0.102062 |
Alignment:
BMSMGCCYMCTKSTGGMHM
-------TCCTCCTGGAA-
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
Motif 47 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.121753 |
Alignment:
YGCCACCTDSTGGY
-TCCTCCTGGAA--
| Original motif Consensus sequence: YGCCACCTDSTGGY |
Reverse complement motif Consensus sequence: KCCASDAGGTGGCK |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
21 |
| Motif name: |
Motif 21 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.125 |
Alignment:
TTYGGAAAGCAG
TTCCAGGAGGA-
| Original motif Consensus sequence: CTGCTTTCCMAA |
Reverse complement motif Consensus sequence: TTYGGAAAGCAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
23 |
| Motif name: |
Motif 23 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.125 |
Alignment:
ATYAATGAAWACAT
TTCCAGGAGGA---
| Original motif Consensus sequence: ATGTWTTCATTMAT |
Reverse complement motif Consensus sequence: ATYAATGAAWACAT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
56 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.612329 |
Alignment:
GCCACGTGSD-
TCCTCCTGGAA
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
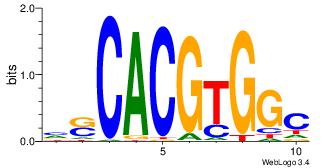 |
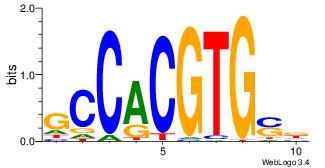 |
| Dataset #: |
1 |
| Motif ID: |
27 |
| Motif name: |
Motif 27 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.6125 |
Alignment:
ATTCTGTRAAG-
-TTCCAGGAGGA
| Original motif Consensus sequence: ATTCTGTRAAG |
Reverse complement motif Consensus sequence: CTTKACAGAAT |
|
|
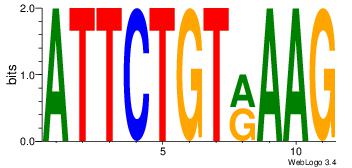 |
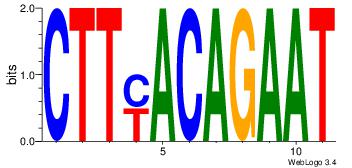 |
| Dataset #: |
3 |
| Motif ID: |
58 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.615419 |
Alignment:
DCCACGTGCV-
TCCTCCTGGAA
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
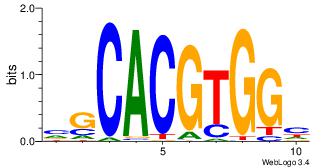 |
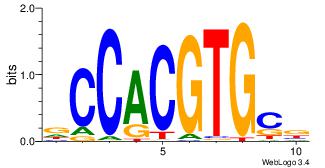 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
12 |
| Number of overlap: |
10 |
| Similarity score: |
0.62065 |
Alignment:
-TTCAGCACCATGGACAGCKCC
TTCCAGGAGGA-----------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
30 |
| Motif name: |
Motif 30 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.625 |
Alignment:
TTGCTGCTTTT-
-TCCTCCTGGAA
| Original motif Consensus sequence: TTGCTGCTTTT |
Reverse complement motif Consensus sequence: AAAAGCAGCAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
22 |
| Motif name: |
Motif 22 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.625 |
Alignment:
ATGCTCACCATAAA-
----TCCTCCTGGAA
| Original motif Consensus sequence: TTTATGGTGAGCAT |
Reverse complement motif Consensus sequence: ATGCTCACCATAAA |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 21 |
Motif name: Motif 21 |
| Original motif Consensus sequence: CTGCTTTCCMAA |
Reverse complement motif Consensus sequence: TTYGGAAAGCAG |
|
|
 |
 |
Best Matches for Motif ID 21 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
22 |
| Motif name: |
Motif 22 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.075 |
Alignment:
ATGCTCACCATAAA
CTGCTTTCCMAA--
| Original motif Consensus sequence: TTTATGGTGAGCAT |
Reverse complement motif Consensus sequence: ATGCTCACCATAAA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
91 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
12 |
| Similarity score: |
0.0854167 |
Alignment:
BTTHAAAAAAYAAAAAAR
-TTYGGAAAGCAG-----
| Original motif Consensus sequence: KTTTTTTKTTTTTTDAAB |
Reverse complement motif Consensus sequence: BTTHAAAAAAYAAAAAAR |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
12 |
| Similarity score: |
0.0868982 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
-CTGCTTTCCMAA--------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
71 |
| Motif name: |
AmTGTGACACCACAGT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
12 |
| Similarity score: |
0.0938802 |
Alignment:
ACTGTGGTGTCACART
---CTGCTTTCCMAA-
| Original motif Consensus sequence: AMTGTGACACCACAGT |
Reverse complement motif Consensus sequence: ACTGTGGTGTCACART |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
12 |
| Number of overlap: |
12 |
| Similarity score: |
0.0952465 |
Alignment:
AGTGCTCCACTGTGGTGTCACAGT
-----------CTGCTTTCCMAA-
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
12 |
| Similarity score: |
0.0978041 |
Alignment:
CCACTGTGVTGTCACAGTGCTCCA
-----CTGCTTTCCMAA-------
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.100752 |
Alignment:
TGTGGTGTCACAGTGCTCC
-CTGCTTTCCMAA------
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
23 |
| Motif name: |
Motif 23 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.10625 |
Alignment:
ATGTWTTCATTMAT
CTGCTTTCCMAA--
| Original motif Consensus sequence: ATGTWTTCATTMAT |
Reverse complement motif Consensus sequence: ATYAATGAAWACAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
46 |
| Motif name: |
Motif 46 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.10625 |
Alignment:
TATKCTAAACCTAT
-CTGCTTTCCMAA-
| Original motif Consensus sequence: ATAGGTTTAGYATA |
Reverse complement motif Consensus sequence: TATKCTAAACCTAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
43 |
| Motif name: |
Motif 43 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.583523 |
Alignment:
AAKGAAAGGCA-
TTYGGAAAGCAG
| Original motif Consensus sequence: AAKGAAAGGCA |
Reverse complement motif Consensus sequence: TGCCTTTCYTT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 22 |
Motif name: Motif 22 |
| Original motif Consensus sequence: TTTATGGTGAGCAT |
Reverse complement motif Consensus sequence: ATGCTCACCATAAA |
|
|
 |
 |
Best Matches for Motif ID 22 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
23 |
| Motif name: |
Motif 23 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.125 |
Alignment:
ATYAATGAAWACAT
TTTATGGTGAGCAT
| Original motif Consensus sequence: ATGTWTTCATTMAT |
Reverse complement motif Consensus sequence: ATYAATGAAWACAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
46 |
| Motif name: |
Motif 46 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.586538 |
Alignment:
TATKCTAAACCTAT-
-ATGCTCACCATAAA
| Original motif Consensus sequence: ATAGGTTTAGYATA |
Reverse complement motif Consensus sequence: TATKCTAAACCTAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.625 |
Alignment:
TTTAATGTYTTTWA-
-TTTATGGTGAGCAT
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
21 |
| Motif name: |
Motif 21 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.09375 |
Alignment:
--CTGCTTTCCMAA
ATGCTCACCATAAA
| Original motif Consensus sequence: CTGCTTTCCMAA |
Reverse complement motif Consensus sequence: TTYGGAAAGCAG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
60 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
12 |
| Similarity score: |
1.11167 |
Alignment:
AAAGGTCAAAGGTCAAC--
-----TTTATGGTGAGCAT
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
1.11458 |
Alignment:
TAGTTTYAKCTTT--
-TTTATGGTGAGCAT
| Original motif Consensus sequence: AAAGRTMAAACTA |
Reverse complement motif Consensus sequence: TAGTTTYAKCTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
31 |
| Motif name: |
Motif 31 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.61364 |
Alignment:
---TTGTTGATTTT
TTTATGGTGAGCAT
| Original motif Consensus sequence: TTGTTGATTTT |
Reverse complement motif Consensus sequence: AAAATCAACAA |
|
|
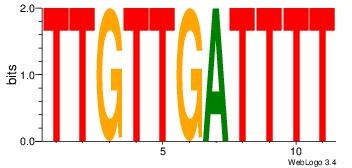 |
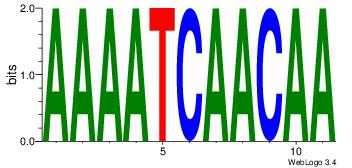 |
| Dataset #: |
1 |
| Motif ID: |
27 |
| Motif name: |
Motif 27 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.61364 |
Alignment:
---CTTKACAGAAT
ATGCTCACCATAAA
| Original motif Consensus sequence: ATTCTGTRAAG |
Reverse complement motif Consensus sequence: CTTKACAGAAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
17 |
| Motif name: |
Motif 17 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.61364 |
Alignment:
---CTTCTGACCTC
ATGCTCACCATAAA
| Original motif Consensus sequence: CTTCTGACCTC |
Reverse complement motif Consensus sequence: GAGGTCAGAAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
24 |
| Motif name: |
Motif 24 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.625 |
Alignment:
TTWATGWCTTT---
TTTATGGTGAGCAT
| Original motif Consensus sequence: AAAGWCATWAA |
Reverse complement motif Consensus sequence: TTWATGWCTTT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 23 |
Motif name: Motif 23 |
| Original motif Consensus sequence: ATGTWTTCATTMAT |
Reverse complement motif Consensus sequence: ATYAATGAAWACAT |
|
|
 |
 |
Best Matches for Motif ID 23 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0825893 |
Alignment:
ATTTTTTTTTTTTT
ATGTWTTCATTMAT
| Original motif Consensus sequence: AAAAAAAAAAAAAT |
Reverse complement motif Consensus sequence: ATTTTTTTTTTTTT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
94 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.0876913 |
Alignment:
WHAATTAAARAARAAAAAAA
---ATYAATGAAWACAT---
| Original motif Consensus sequence: TTTTTTTKTTKTTTAATTHW |
Reverse complement motif Consensus sequence: WHAATTAAARAARAAAAAAA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
91 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.0959821 |
Alignment:
BTTHAAAAAAYAAAAAAR
-ATYAATGAAWACAT---
| Original motif Consensus sequence: KTTTTTTKTTTTTTDAAB |
Reverse complement motif Consensus sequence: BTTHAAAAAAYAAAAAAR |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.109375 |
Alignment:
TWAAAKACATTAAA
ATGTWTTCATTMAT
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
22 |
| Motif name: |
Motif 22 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.109375 |
Alignment:
ATGCTCACCATAAA
ATGTWTTCATTMAT
| Original motif Consensus sequence: TTTATGGTGAGCAT |
Reverse complement motif Consensus sequence: ATGCTCACCATAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
45 |
| Motif name: |
Motif 45 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.590144 |
Alignment:
TAASTCTWTTTTAA-
-ATGTWTTCATTMAT
| Original motif Consensus sequence: TAASTCTWTTTTAA |
Reverse complement motif Consensus sequence: TTAAAAWAGASTTA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.601683 |
Alignment:
HAAAAHAAARMAAA-
-ATYAATGAAWACAT
| Original motif Consensus sequence: HAAAAHAAARMAAA |
Reverse complement motif Consensus sequence: TTTRKTTTHTTTTH |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.609375 |
Alignment:
-AAAGRTMAAACTA
ATYAATGAAWACAT
| Original motif Consensus sequence: AAAGRTMAAACTA |
Reverse complement motif Consensus sequence: TAGTTTYAKCTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
46 |
| Motif name: |
Motif 46 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
1.09896 |
Alignment:
--TATKCTAAACCTAT
ATYAATGAAWACAT--
| Original motif Consensus sequence: ATAGGTTTAGYATA |
Reverse complement motif Consensus sequence: TATKCTAAACCTAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
21 |
| Motif name: |
Motif 21 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.10938 |
Alignment:
CTGCTTTCCMAA--
ATGTWTTCATTMAT
| Original motif Consensus sequence: CTGCTTTCCMAA |
Reverse complement motif Consensus sequence: TTYGGAAAGCAG |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 24 |
Motif name: Motif 24 |
| Original motif Consensus sequence: AAAGWCATWAA |
Reverse complement motif Consensus sequence: TTWATGWCTTT |
|
|
 |
 |
Best Matches for Motif ID 24 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0340909 |
Alignment:
TWAAAKACATTAAA
--AAAGWCATWAA-
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
31 |
| Motif name: |
Motif 31 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0795455 |
Alignment:
AAAATCAACAA
AAAGWCATWAA
| Original motif Consensus sequence: TTGTTGATTTT |
Reverse complement motif Consensus sequence: AAAATCAACAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0909091 |
Alignment:
AAAAAAAAAAAAAT
AAAGWCATWAA---
| Original motif Consensus sequence: AAAAAAAAAAAAAT |
Reverse complement motif Consensus sequence: ATTTTTTTTTTTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
30 |
| Motif name: |
Motif 30 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0909091 |
Alignment:
AAAAGCAGCAA
AAAGWCATWAA
| Original motif Consensus sequence: TTGCTGCTTTT |
Reverse complement motif Consensus sequence: AAAAGCAGCAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
13 |
| Motif name: |
Motif 13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0909091 |
Alignment:
AAATAKATWTT
AAAGWCATWAA
| Original motif Consensus sequence: AAWATRTATTT |
Reverse complement motif Consensus sequence: AAATAKATWTT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
91 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
11 |
| Similarity score: |
0.0951705 |
Alignment:
KTTTTTTKTTTTTTDAAB
--TTWATGWCTTT-----
| Original motif Consensus sequence: KTTTTTTKTTTTTTDAAB |
Reverse complement motif Consensus sequence: BTTHAAAAAAYAAAAAAR |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0977273 |
Alignment:
TTTRKTTTHTTTTH
-TTWATGWCTTT--
| Original motif Consensus sequence: HAAAAHAAARMAAA |
Reverse complement motif Consensus sequence: TTTRKTTTHTTTTH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
94 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
11 |
| Similarity score: |
0.099026 |
Alignment:
TTTTTTTKTTKTTTAATTHW
-TTWATGWCTTT--------
| Original motif Consensus sequence: TTTTTTTKTTKTTTAATTHW |
Reverse complement motif Consensus sequence: WHAATTAAARAARAAAAAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
TAGTTTYAKCTTT
--TTWATGWCTTT
| Original motif Consensus sequence: AAAGRTMAAACTA |
Reverse complement motif Consensus sequence: TAGTTTYAKCTTT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
89 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
0.112013 |
Alignment:
CACACACACACWCACACA
---AAAGWCATWAA----
| Original motif Consensus sequence: CACACACACACWCACACA |
Reverse complement motif Consensus sequence: TGTGTGWGTGTGTGTGTG |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 25 |
Motif name: Motif 25 |
| Original motif Consensus sequence: AATAACAGATT |
Reverse complement motif Consensus sequence: AATCTGTTATT |
|
|
 |
 |
Best Matches for Motif ID 25 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0482955 |
Alignment:
AATAGYATTTT
AATAACAGATT
| Original motif Consensus sequence: AAAATKCTATT |
Reverse complement motif Consensus sequence: AATAGYATTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0596591 |
Alignment:
ATTTTTTTTTTTTT
AATCTGTTATT---
| Original motif Consensus sequence: AAAAAAAAAAAAAT |
Reverse complement motif Consensus sequence: ATTTTTTTTTTTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
13 |
| Motif name: |
Motif 13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0596591 |
Alignment:
AAATAKATWTT
AATAACAGATT
| Original motif Consensus sequence: AAWATRTATTT |
Reverse complement motif Consensus sequence: AAATAKATWTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
45 |
| Motif name: |
Motif 45 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0596591 |
Alignment:
TAASTCTWTTTTAA
--AATCTGTTATT-
| Original motif Consensus sequence: TAASTCTWTTTTAA |
Reverse complement motif Consensus sequence: TTAAAAWAGASTTA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
27 |
| Motif name: |
Motif 27 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0596591 |
Alignment:
CTTKACAGAAT
AATAACAGATT
| Original motif Consensus sequence: ATTCTGTRAAG |
Reverse complement motif Consensus sequence: CTTKACAGAAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0710227 |
Alignment:
TWAAAKACATTAAA
AATAACAGATT---
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0710227 |
Alignment:
AAAMATTGTTT
AATAACAGATT
| Original motif Consensus sequence: AAAMATTGTTT |
Reverse complement motif Consensus sequence: AAACAATYTTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
11 |
| Similarity score: |
0.0798255 |
Alignment:
AGTGCTCCACTGTGGTGTCACAGT
--------AATCTGTTATT-----
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0823864 |
Alignment:
AAAGRTMAAACTA
-AATAACAGATT-
| Original motif Consensus sequence: AAAGRTMAAACTA |
Reverse complement motif Consensus sequence: TAGTTTYAKCTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0823864 |
Alignment:
TTYAAACATYTT
AATAACAGATT-
| Original motif Consensus sequence: AAMATGTTTMAA |
Reverse complement motif Consensus sequence: TTYAAACATYTT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 26 |
Motif name: Motif 26 |
| Original motif Consensus sequence: CTGTTTTWAT |
Reverse complement motif Consensus sequence: ATWAAAACAG |
|
|
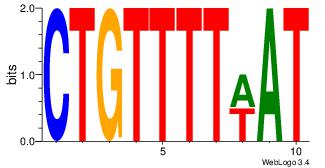 |
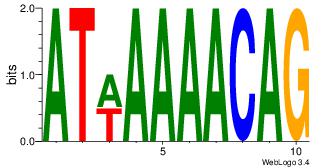 |
Best Matches for Motif ID 26 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
23 |
| Motif name: |
Motif 23 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0625 |
Alignment:
ATGTWTTCATTMAT
CTGTTTTWAT----
| Original motif Consensus sequence: ATGTWTTCATTMAT |
Reverse complement motif Consensus sequence: ATYAATGAAWACAT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
94 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
10 |
| Similarity score: |
0.0821429 |
Alignment:
WHAATTAAARAARAAAAAAA
---ATWAAAACAG-------
| Original motif Consensus sequence: TTTTTTTKTTKTTTAATTHW |
Reverse complement motif Consensus sequence: WHAATTAAARAARAAAAAAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0875 |
Alignment:
ATTTTTTTTTTTTT
---CTGTTTTWAT-
| Original motif Consensus sequence: AAAAAAAAAAAAAT |
Reverse complement motif Consensus sequence: ATTTTTTTTTTTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0875 |
Alignment:
TTTAATGTYTTTWA
----CTGTTTTWAT
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0875 |
Alignment:
TCTKTGTCAAWA
-CTGTTTTWAT-
| Original motif Consensus sequence: TCTKTGTCAAWA |
Reverse complement motif Consensus sequence: TWTTGACAYAGA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.09 |
Alignment:
TTTRKTTTHTTTTH
---CTGTTTTWAT-
| Original motif Consensus sequence: HAAAAHAAARMAAA |
Reverse complement motif Consensus sequence: TTTRKTTTHTTTTH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
91 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.096875 |
Alignment:
KTTTTTTKTTTTTTDAAB
-----CTGTTTTWAT---
| Original motif Consensus sequence: KTTTTTTKTTTTTTDAAB |
Reverse complement motif Consensus sequence: BTTHAAAAAAYAAAAAAR |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
21 |
| Motif name: |
Motif 21 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.1 |
Alignment:
CTGCTTTCCMAA
CTGTTTTWAT--
| Original motif Consensus sequence: CTGCTTTCCMAA |
Reverse complement motif Consensus sequence: TTYGGAAAGCAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
45 |
| Motif name: |
Motif 45 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.1125 |
Alignment:
TTAAAAWAGASTTA
-ATWAAAACAG---
| Original motif Consensus sequence: TAASTCTWTTTTAA |
Reverse complement motif Consensus sequence: TTAAAAWAGASTTA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
25 |
| Motif name: |
Motif 25 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.1125 |
Alignment:
AATCTGTTATT
CTGTTTTWAT-
| Original motif Consensus sequence: AATAACAGATT |
Reverse complement motif Consensus sequence: AATCTGTTATT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 27 |
Motif name: Motif 27 |
| Original motif Consensus sequence: ATTCTGTRAAG |
Reverse complement motif Consensus sequence: CTTKACAGAAT |
|
|
 |
 |
Best Matches for Motif ID 27 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
25 |
| Motif name: |
Motif 25 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0596591 |
Alignment:
AATCTGTTATT
ATTCTGTRAAG
| Original motif Consensus sequence: AATAACAGATT |
Reverse complement motif Consensus sequence: AATCTGTTATT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
91 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0696023 |
Alignment:
KTTTTTTKTTTTTTDAAB
-------ATTCTGTRAAG
| Original motif Consensus sequence: KTTTTTTKTTTTTTDAAB |
Reverse complement motif Consensus sequence: BTTHAAAAAAYAAAAAAR |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0710227 |
Alignment:
TWTTGACAYAGA
-CTTKACAGAAT
| Original motif Consensus sequence: TCTKTGTCAAWA |
Reverse complement motif Consensus sequence: TWTTGACAYAGA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
81 |
| Motif name: |
ctatacggacg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0710227 |
Alignment:
CGTCCGTATAG
ATTCTGTRAAG
| Original motif Consensus sequence: CTATACGGACG |
Reverse complement motif Consensus sequence: CGTCCGTATAG |
|
|
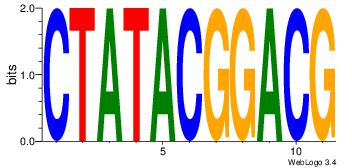 |
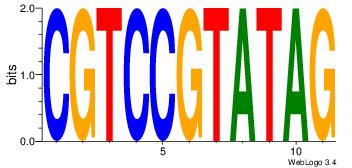 |
| Dataset #: |
5 |
| Motif ID: |
94 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.080763 |
Alignment:
TTTTTTTKTTKTTTAATTHW
-------ATTCTGTRAAG--
| Original motif Consensus sequence: TTTTTTTKTTKTTTAATTHW |
Reverse complement motif Consensus sequence: WHAATTAAARAARAAAAAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
22 |
| Motif name: |
Motif 22 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0823864 |
Alignment:
TTTATGGTGAGCAT
ATTCTGTRAAG---
| Original motif Consensus sequence: TTTATGGTGAGCAT |
Reverse complement motif Consensus sequence: ATGCTCACCATAAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
71 |
| Motif name: |
AmTGTGACACCACAGT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0901989 |
Alignment:
AMTGTGACACCACAGT
--CTTKACAGAAT---
| Original motif Consensus sequence: AMTGTGACACCACAGT |
Reverse complement motif Consensus sequence: ACTGTGGTGTCACART |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
70 |
| Motif name: |
CagTGCTCCACTGTGgT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
11 |
| Similarity score: |
0.0910985 |
Alignment:
ABCACAGTGGAGCACTG
-ATTCTGTRAAG-----
| Original motif Consensus sequence: CAGTGCTCCACTGTGBT |
Reverse complement motif Consensus sequence: ABCACAGTGGAGCACTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
12 |
| Number of overlap: |
11 |
| Similarity score: |
0.0921495 |
Alignment:
ACTGTGACACCACAGTGGAGCACT
--CTTKACAGAAT-----------
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
11 |
| Similarity score: |
0.0925215 |
Alignment:
TGGAGCACTGTGACAVCACAGTGG
--------CTTKACAGAAT-----
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 28 |
Motif name: Motif 28 |
| Original motif Consensus sequence: ATTTAGTAAA |
Reverse complement motif Consensus sequence: TTTACTAAAT |
|
|
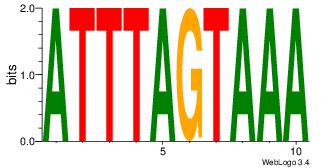 |
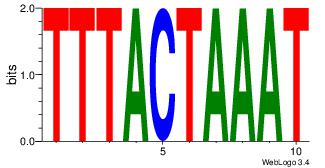 |
Best Matches for Motif ID 28 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
27 |
| Motif name: |
Motif 27 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0625 |
Alignment:
CTTKACAGAAT
-TTTACTAAAT
| Original motif Consensus sequence: ATTCTGTRAAG |
Reverse complement motif Consensus sequence: CTTKACAGAAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
46 |
| Motif name: |
Motif 46 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0625 |
Alignment:
ATAGGTTTAGYATA
----ATTTAGTAAA
| Original motif Consensus sequence: ATAGGTTTAGYATA |
Reverse complement motif Consensus sequence: TATKCTAAACCTAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0875 |
Alignment:
TWAAAKACATTAAA
----ATTTAGTAAA
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
91 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
10 |
| Similarity score: |
0.09375 |
Alignment:
BTTHAAAAAAYAAAAAAR
-TTTACTAAAT-------
| Original motif Consensus sequence: KTTTTTTKTTTTTTDAAB |
Reverse complement motif Consensus sequence: BTTHAAAAAAYAAAAAAR |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
23 |
| Motif name: |
Motif 23 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.1 |
Alignment:
ATGTWTTCATTMAT
TTTACTAAAT----
| Original motif Consensus sequence: ATGTWTTCATTMAT |
Reverse complement motif Consensus sequence: ATYAATGAAWACAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
29 |
| Motif name: |
Motif 29 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.1 |
Alignment:
ATTTATTGCTA
ATTTAGTAAA-
| Original motif Consensus sequence: ATTTATTGCTA |
Reverse complement motif Consensus sequence: TAGCAATAAAT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
94 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.105357 |
Alignment:
WHAATTAAARAARAAAAAAA
--ATTTAGTAAA--------
| Original motif Consensus sequence: TTTTTTTKTTKTTTAATTHW |
Reverse complement motif Consensus sequence: WHAATTAAARAARAAAAAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.1125 |
Alignment:
TCTKTGTCAAWA
ATTTAGTAAA--
| Original motif Consensus sequence: TCTKTGTCAAWA |
Reverse complement motif Consensus sequence: TWTTGACAYAGA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
14 |
| Motif name: |
Motif 14 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.1125 |
Alignment:
TAATKATATAA
-ATTTAGTAAA
| Original motif Consensus sequence: TAATKATATAA |
Reverse complement motif Consensus sequence: TTATATYATTA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.115 |
Alignment:
TTTRKTTTHTTTTH
TTTACTAAAT----
| Original motif Consensus sequence: HAAAAHAAARMAAA |
Reverse complement motif Consensus sequence: TTTRKTTTHTTTTH |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 29 |
Motif name: Motif 29 |
| Original motif Consensus sequence: ATTTATTGCTA |
Reverse complement motif Consensus sequence: TAGCAATAAAT |
|
|
 |
 |
Best Matches for Motif ID 29 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0909091 |
Alignment:
AAAAAAAAAAAAAT
---TAGCAATAAAT
| Original motif Consensus sequence: AAAAAAAAAAAAAT |
Reverse complement motif Consensus sequence: ATTTTTTTTTTTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
23 |
| Motif name: |
Motif 23 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
ATGTWTTCATTMAT
ATTTATTGCTA---
| Original motif Consensus sequence: ATGTWTTCATTMAT |
Reverse complement motif Consensus sequence: ATYAATGAAWACAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
14 |
| Motif name: |
Motif 14 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
TAATKATATAA
TAGCAATAAAT
| Original motif Consensus sequence: TAATKATATAA |
Reverse complement motif Consensus sequence: TTATATYATTA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
AAAGRTMAAACTA
-TAGCAATAAAT-
| Original motif Consensus sequence: AAAGRTMAAACTA |
Reverse complement motif Consensus sequence: TAGTTTYAKCTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
TGAAAATATGTT
TAGCAATAAAT-
| Original motif Consensus sequence: AACATATTTTCA |
Reverse complement motif Consensus sequence: TGAAAATATGTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
AAAMATTGTTT
ATTTATTGCTA
| Original motif Consensus sequence: AAAMATTGTTT |
Reverse complement motif Consensus sequence: AAACAATYTTT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
94 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
TTTTTTTKTTKTTTAATTHW
---ATTTATTGCTA------
| Original motif Consensus sequence: TTTTTTTKTTKTTTAATTHW |
Reverse complement motif Consensus sequence: WHAATTAAARAARAAAAAAA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
91 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.120739 |
Alignment:
BTTHAAAAAAYAAAAAAR
-------TAGCAATAAAT
| Original motif Consensus sequence: KTTTTTTKTTTTTTDAAB |
Reverse complement motif Consensus sequence: BTTHAAAAAAYAAAAAAR |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.125 |
Alignment:
TTTAATGTYTTTWA
ATTTATTGCTA---
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
45 |
| Motif name: |
Motif 45 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.125 |
Alignment:
TTAAAAWAGASTTA
TAGCAATAAAT---
| Original motif Consensus sequence: TAASTCTWTTTTAA |
Reverse complement motif Consensus sequence: TTAAAAWAGASTTA |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 30 |
Motif name: Motif 30 |
| Original motif Consensus sequence: TTGCTGCTTTT |
Reverse complement motif Consensus sequence: AAAAGCAGCAA |
|
|
 |
 |
Best Matches for Motif ID 30 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
31 |
| Motif name: |
Motif 31 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0142045 |
Alignment:
TTGTTGATTTT
TTGCTGCTTTT
| Original motif Consensus sequence: TTGTTGATTTT |
Reverse complement motif Consensus sequence: AAAATCAACAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0482955 |
Alignment:
TTTAATGTYTTTWA
-TTGCTGCTTTT--
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0573864 |
Alignment:
TTTRKTTTHTTTTH
--TTGCTGCTTTT-
| Original motif Consensus sequence: HAAAAHAAARMAAA |
Reverse complement motif Consensus sequence: TTTRKTTTHTTTTH |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
24 |
| Motif name: |
Motif 24 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0596591 |
Alignment:
TTWATGWCTTT
TTGCTGCTTTT
| Original motif Consensus sequence: AAAGWCATWAA |
Reverse complement motif Consensus sequence: TTWATGWCTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0596591 |
Alignment:
AAAAAAAAAAAAAT
AAAAGCAGCAA---
| Original motif Consensus sequence: AAAAAAAAAAAAAT |
Reverse complement motif Consensus sequence: ATTTTTTTTTTTTT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
91 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
11 |
| Similarity score: |
0.0610795 |
Alignment:
KTTTTTTKTTTTTTDAAB
--TTGCTGCTTTT-----
| Original motif Consensus sequence: KTTTTTTKTTTTTTDAAB |
Reverse complement motif Consensus sequence: BTTHAAAAAAYAAAAAAR |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
94 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0629058 |
Alignment:
TTTTTTTKTTKTTTAATTHW
-TTGCTGCTTTT--------
| Original motif Consensus sequence: TTTTTTTKTTKTTTAATTHW |
Reverse complement motif Consensus sequence: WHAATTAAARAARAAAAAAA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
88 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0756642 |
Alignment:
CTGCTGBDBBDGBD
TTGCTGCTTTT---
| Original motif Consensus sequence: CTGCTGBDBBDGBD |
Reverse complement motif Consensus sequence: HVCDBBDBCAGCAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
25 |
| Motif name: |
Motif 25 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0823864 |
Alignment:
AATCTGTTATT
TTGCTGCTTTT
| Original motif Consensus sequence: AATAACAGATT |
Reverse complement motif Consensus sequence: AATCTGTTATT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
45 |
| Motif name: |
Motif 45 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0823864 |
Alignment:
TTAAAAWAGASTTA
---AAAAGCAGCAA
| Original motif Consensus sequence: TAASTCTWTTTTAA |
Reverse complement motif Consensus sequence: TTAAAAWAGASTTA |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 31 |
Motif name: Motif 31 |
| Original motif Consensus sequence: TTGTTGATTTT |
Reverse complement motif Consensus sequence: AAAATCAACAA |
|
|
 |
 |
Best Matches for Motif ID 31 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
30 |
| Motif name: |
Motif 30 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0454545 |
Alignment:
TTGCTGCTTTT
TTGTTGATTTT
| Original motif Consensus sequence: TTGCTGCTTTT |
Reverse complement motif Consensus sequence: AAAAGCAGCAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0681818 |
Alignment:
ATTTTTTTTTTTTT
--TTGTTGATTTT-
| Original motif Consensus sequence: AAAAAAAAAAAAAT |
Reverse complement motif Consensus sequence: ATTTTTTTTTTTTT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
94 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0746753 |
Alignment:
TTTTTTTKTTKTTTAATTHW
-TTGTTGATTTT--------
| Original motif Consensus sequence: TTTTTTTKTTKTTTAATTHW |
Reverse complement motif Consensus sequence: WHAATTAAARAARAAAAAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0795455 |
Alignment:
TTTAATGTYTTTWA
-TTGTTGATTTT--
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
24 |
| Motif name: |
Motif 24 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0795455 |
Alignment:
TTWATGWCTTT
TTGTTGATTTT
| Original motif Consensus sequence: AAAGWCATWAA |
Reverse complement motif Consensus sequence: TTWATGWCTTT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
91 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0809659 |
Alignment:
KTTTTTTKTTTTTTDAAB
--TTGTTGATTTT-----
| Original motif Consensus sequence: KTTTTTTKTTTTTTDAAB |
Reverse complement motif Consensus sequence: BTTHAAAAAAYAAAAAAR |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0852273 |
Alignment:
TTTRKTTTHTTTTH
--TTGTTGATTTT-
| Original motif Consensus sequence: HAAAAHAAARMAAA |
Reverse complement motif Consensus sequence: TTTRKTTTHTTTTH |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
13 |
| Motif name: |
Motif 13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
AAWATRTATTT
AAAATCAACAA
| Original motif Consensus sequence: AAWATRTATTT |
Reverse complement motif Consensus sequence: AAATAKATWTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
22 |
| Motif name: |
Motif 22 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
ATGCTCACCATAAA
AAAATCAACAA---
| Original motif Consensus sequence: TTTATGGTGAGCAT |
Reverse complement motif Consensus sequence: ATGCTCACCATAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
45 |
| Motif name: |
Motif 45 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
TAASTCTWTTTTAA
-TTGTTGATTTT--
| Original motif Consensus sequence: TAASTCTWTTTTAA |
Reverse complement motif Consensus sequence: TTAAAAWAGASTTA |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 32 |
Motif name: Motif 32 |
| Original motif Consensus sequence: TTACWGTTT |
Reverse complement motif Consensus sequence: AAACWGTAA |
|
|
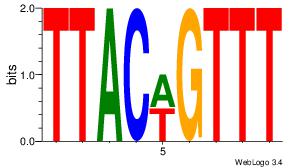 |
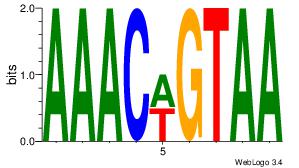 |
Best Matches for Motif ID 32 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
9 |
| Similarity score: |
0.0347222 |
Alignment:
TTTAATGTYTTTWA
-TTACWGTTT----
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
25 |
| Motif name: |
Motif 25 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0.0486111 |
Alignment:
AATAACAGATT
--TTACWGTTT
| Original motif Consensus sequence: AATAACAGATT |
Reverse complement motif Consensus sequence: AATCTGTTATT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
30 |
| Motif name: |
Motif 30 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
9 |
| Similarity score: |
0.0486111 |
Alignment:
TTGCTGCTTTT
TTACWGTTT--
| Original motif Consensus sequence: TTGCTGCTTTT |
Reverse complement motif Consensus sequence: AAAAGCAGCAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
27 |
| Motif name: |
Motif 27 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0.0625 |
Alignment:
CTTKACAGAAT
TTACWGTTT--
| Original motif Consensus sequence: ATTCTGTRAAG |
Reverse complement motif Consensus sequence: CTTKACAGAAT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
91 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
9 |
| Similarity score: |
0.0729167 |
Alignment:
KTTTTTTKTTTTTTDAAB
--TTACWGTTT-------
| Original motif Consensus sequence: KTTTTTTKTTTTTTDAAB |
Reverse complement motif Consensus sequence: BTTHAAAAAAYAAAAAAR |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.0763889 |
Alignment:
AATAGYATTTT
-TTACWGTTT-
| Original motif Consensus sequence: AAAATKCTATT |
Reverse complement motif Consensus sequence: AATAGYATTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
13 |
| Motif name: |
Motif 13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0.0763889 |
Alignment:
AAATAKATWTT
TTACWGTTT--
| Original motif Consensus sequence: AAWATRTATTT |
Reverse complement motif Consensus sequence: AAATAKATWTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0.0763889 |
Alignment:
AAAAAAAAAAAAAT
AAACWGTAA-----
| Original motif Consensus sequence: AAAAAAAAAAAAAT |
Reverse complement motif Consensus sequence: ATTTTTTTTTTTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
45 |
| Motif name: |
Motif 45 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0.0763889 |
Alignment:
TTAAAAWAGASTTA
-----AAACWGTAA
| Original motif Consensus sequence: TAASTCTWTTTTAA |
Reverse complement motif Consensus sequence: TTAAAAWAGASTTA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
24 |
| Motif name: |
Motif 24 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
9 |
| Similarity score: |
0.0763889 |
Alignment:
TTWATGWCTTT
--TTACWGTTT
| Original motif Consensus sequence: AAAGWCATWAA |
Reverse complement motif Consensus sequence: TTWATGWCTTT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 33 |
Motif name: Motif 33 |
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
Best Matches for Motif ID 33 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0821429 |
Alignment:
HAAAAHAAARMAAA
TWAAAKACATTAAA
| Original motif Consensus sequence: HAAAAHAAARMAAA |
Reverse complement motif Consensus sequence: TTTRKTTTHTTTTH |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
94 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0918367 |
Alignment:
TTTTTTTKTTKTTTAATTHW
-TTTAATGTYTTTWA-----
| Original motif Consensus sequence: TTTTTTTKTTKTTTAATTHW |
Reverse complement motif Consensus sequence: WHAATTAAARAARAAAAAAA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
91 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.09375 |
Alignment:
KTTTTTTKTTTTTTDAAB
-TTTAATGTYTTTWA---
| Original motif Consensus sequence: KTTTTTTKTTTTTTDAAB |
Reverse complement motif Consensus sequence: BTTHAAAAAAYAAAAAAR |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.107143 |
Alignment:
ATTTTTTTTTTTTT
TTTAATGTYTTTWA
| Original motif Consensus sequence: AAAAAAAAAAAAAT |
Reverse complement motif Consensus sequence: ATTTTTTTTTTTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
23 |
| Motif name: |
Motif 23 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.125 |
Alignment:
ATYAATGAAWACAT
TTTAATGTYTTTWA
| Original motif Consensus sequence: ATGTWTTCATTMAT |
Reverse complement motif Consensus sequence: ATYAATGAAWACAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
45 |
| Motif name: |
Motif 45 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.576923 |
Alignment:
-TAASTCTWTTTTAA
TTTAATGTYTTTWA-
| Original motif Consensus sequence: TAASTCTWTTTTAA |
Reverse complement motif Consensus sequence: TTAAAAWAGASTTA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
22 |
| Motif name: |
Motif 22 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.625 |
Alignment:
-TTTATGGTGAGCAT
TTTAATGTYTTTWA-
| Original motif Consensus sequence: TTTATGGTGAGCAT |
Reverse complement motif Consensus sequence: ATGCTCACCATAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.09375 |
Alignment:
AAMATGTTTMAA--
TWAAAKACATTAAA
| Original motif Consensus sequence: AAMATGTTTMAA |
Reverse complement motif Consensus sequence: TTYAAACATYTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
14 |
| Number of overlap: |
12 |
| Similarity score: |
1.12483 |
Alignment:
AGTGSWSCACTGTGAMAMCACAGTG--
-------------TWAAAKACATTAAA
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
1.125 |
Alignment:
--AAAGRTMAAACTA
TWAAAKACATTAAA-
| Original motif Consensus sequence: AAAGRTMAAACTA |
Reverse complement motif Consensus sequence: TAGTTTYAKCTTT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 34 |
Motif name: Motif 34 |
| Original motif Consensus sequence: AAAGATGA |
Reverse complement motif Consensus sequence: TCATCTTT |
|
|
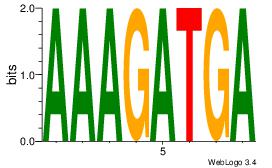 |
 |
Best Matches for Motif ID 34 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
8 |
| Similarity score: |
0.03125 |
Alignment:
TAGTTTYAKCTTT
-----TCATCTTT
| Original motif Consensus sequence: AAAGRTMAAACTA |
Reverse complement motif Consensus sequence: TAGTTTYAKCTTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
80 |
| Motif name: |
taaacgatgcc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0625 |
Alignment:
TAAACGATGCC
--AAAGATGA-
| Original motif Consensus sequence: TAAACGATGCC |
Reverse complement motif Consensus sequence: GGCATCGTTTA |
|
|
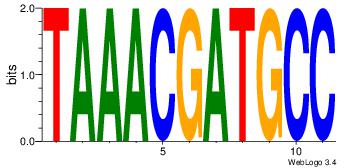 |
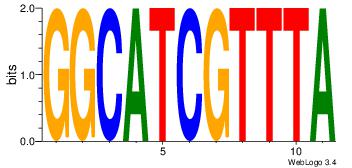 |
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0625 |
Alignment:
TGAAAATATGTT
-AAAGATGA---
| Original motif Consensus sequence: AACATATTTTCA |
Reverse complement motif Consensus sequence: TGAAAATATGTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
60 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.06375 |
Alignment:
AAAGGTCAAAGGTCAAC
-------AAAGATGA--
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
52 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.0697115 |
Alignment:
AKGYYCAAAGRTCA
------AAAGATGA
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
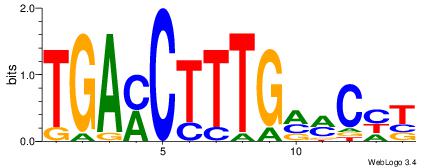 |
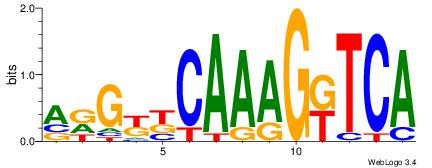 |
| Dataset #: |
1 |
| Motif ID: |
45 |
| Motif name: |
Motif 45 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
TAASTCTWTTTTAA
----TCATCTTT--
| Original motif Consensus sequence: TAASTCTWTTTTAA |
Reverse complement motif Consensus sequence: TTAAAAWAGASTTA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
35 |
| Motif name: |
Motif 35 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
CAAGAKCATCT
AAAGATGA---
| Original motif Consensus sequence: AGATGYTCTTG |
Reverse complement motif Consensus sequence: CAAGAKCATCT |
|
|
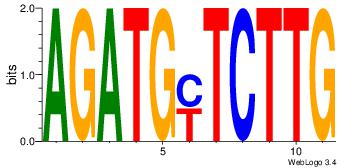 |
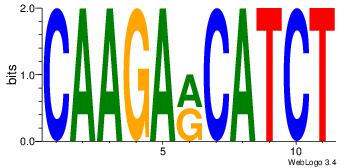 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
TWAAAKACATTAAA
--AAAGATGA----
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
TTYAAACATYTT
-AAAGATGA---
| Original motif Consensus sequence: AAMATGTTTMAA |
Reverse complement motif Consensus sequence: TTYAAACATYTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
AAACAATYTTT
---TCATCTTT
| Original motif Consensus sequence: AAAMATTGTTT |
Reverse complement motif Consensus sequence: AAACAATYTTT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 35 |
Motif name: Motif 35 |
| Original motif Consensus sequence: AGATGYTCTTG |
Reverse complement motif Consensus sequence: CAAGAKCATCT |
|
|
 |
 |
Best Matches for Motif ID 35 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.092803 |
Alignment:
TTTAATGTYTTTWA
--AGATGYTCTTG-
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
11 |
| Similarity score: |
0.0964903 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
--AGATGYTCTTG--------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0999053 |
Alignment:
GAKAGKTGGCTCWG
AGATGYTCTTG---
| Original motif Consensus sequence: CWGAGCCAYCTYTC |
Reverse complement motif Consensus sequence: GAKAGKTGGCTCWG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
88 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.101926 |
Alignment:
CTGCTGBDBBDGBD
-AGATGYTCTTG--
| Original motif Consensus sequence: CTGCTGBDBBDGBD |
Reverse complement motif Consensus sequence: HVCDBBDBCAGCAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.104167 |
Alignment:
TAGTTTYAKCTTT
AGATGYTCTTG--
| Original motif Consensus sequence: AAAGRTMAAACTA |
Reverse complement motif Consensus sequence: TAGTTTYAKCTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.104167 |
Alignment:
AACATATTTTCA
-AGATGYTCTTG
| Original motif Consensus sequence: AACATATTTTCA |
Reverse complement motif Consensus sequence: TGAAAATATGTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.104167 |
Alignment:
ATTTTTTTTTTTTT
AGATGYTCTTG---
| Original motif Consensus sequence: AAAAAAAAAAAAAT |
Reverse complement motif Consensus sequence: ATTTTTTTTTTTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
13 |
| Motif name: |
Motif 13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.104167 |
Alignment:
AAATAKATWTT
CAAGAKCATCT
| Original motif Consensus sequence: AAWATRTATTT |
Reverse complement motif Consensus sequence: AAATAKATWTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
23 |
| Motif name: |
Motif 23 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.104167 |
Alignment:
ATGTWTTCATTMAT
AGATGYTCTTG---
| Original motif Consensus sequence: ATGTWTTCATTMAT |
Reverse complement motif Consensus sequence: ATYAATGAAWACAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
12 |
| Motif name: |
Motif 12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.104167 |
Alignment:
ACAACACATTT
CAAGAKCATCT
| Original motif Consensus sequence: ACAACACATTT |
Reverse complement motif Consensus sequence: AAATGTGTTGT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 36 |
Motif name: Motif 36 |
| Original motif Consensus sequence: TGGTGTCA |
Reverse complement motif Consensus sequence: TGACACCA |
|
|
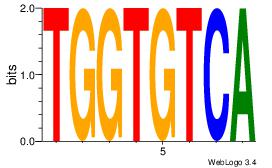 |
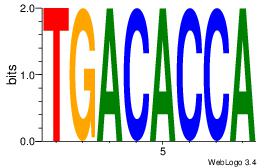 |
Best Matches for Motif ID 36 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
71 |
| Motif name: |
AmTGTGACACCACAGT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0205078 |
Alignment:
ACTGTGGTGTCACART
----TGGTGTCA----
| Original motif Consensus sequence: AMTGTGACACCACAGT |
Reverse complement motif Consensus sequence: ACTGTGGTGTCACART |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
13 |
| Number of overlap: |
8 |
| Similarity score: |
0.0246479 |
Alignment:
ACTGTGACACCACAGTGGAGCACT
----TGACACCA------------
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0338542 |
Alignment:
TGTGGTGTCACAGTGCTCC
--TGGTGTCA---------
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.035473 |
Alignment:
CCACTGTGVTGTCACAGTGCTCCA
------TGGTGTCA----------
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
13 |
| Number of overlap: |
8 |
| Similarity score: |
0.0575397 |
Alignment:
AGTGSWSCACTGTGAMAMCACAGTG
------------TGACACCA-----
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
23 |
| Motif name: |
Motif 23 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
ATGTWTTCATTMAT
-TGGTGTCA-----
| Original motif Consensus sequence: ATGTWTTCATTMAT |
Reverse complement motif Consensus sequence: ATYAATGAAWACAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
AAAGRTMAAACTA
TGACACCA-----
| Original motif Consensus sequence: AAAGRTMAAACTA |
Reverse complement motif Consensus sequence: TAGTTTYAKCTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
TCTKTGTCAAWA
-TGGTGTCA---
| Original motif Consensus sequence: TCTKTGTCAAWA |
Reverse complement motif Consensus sequence: TWTTGACAYAGA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
27 |
| Motif name: |
Motif 27 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
CTTKACAGAAT
--TGACACCA-
| Original motif Consensus sequence: ATTCTGTRAAG |
Reverse complement motif Consensus sequence: CTTKACAGAAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
Motif 47 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0915178 |
Alignment:
YGCCACCTDSTGGY
TGACACCA------
| Original motif Consensus sequence: YGCCACCTDSTGGY |
Reverse complement motif Consensus sequence: KCCASDAGGTGGCK |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 37 |
Motif name: Motif 37 |
| Original motif Consensus sequence: ACTTTGG |
Reverse complement motif Consensus sequence: CCAAAGT |
|
|
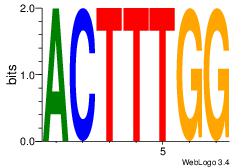 |
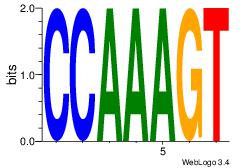 |
Best Matches for Motif ID 37 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
69 |
| Motif name: |
atactttggc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0 |
Alignment:
GCCAAAGTAT
-CCAAAGT--
| Original motif Consensus sequence: ATACTTTGGC |
Reverse complement motif Consensus sequence: GCCAAAGTAT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
75 |
| Motif name: |
wwCCAmAGTCmt |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.0275974 |
Alignment:
DDCCAMAGTCHB
--CCAAAGT---
| Original motif Consensus sequence: DDCCAMAGTCHB |
Reverse complement motif Consensus sequence: VDGACTRTGGDD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
10 |
| Number of overlap: |
7 |
| Similarity score: |
0.056841 |
Alignment:
ACTGTGACACCACAGTGGAGCACT
---------CCAAAGT--------
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
71 |
| Motif name: |
AmTGTGACACCACAGT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
10 |
| Number of overlap: |
7 |
| Similarity score: |
0.0597098 |
Alignment:
ACTGTGGTGTCACART
ACTTTGG---------
| Original motif Consensus sequence: AMTGTGACACCACAGT |
Reverse complement motif Consensus sequence: ACTGTGGTGTCACART |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
76 |
| Motif name: |
CAcCACAGTGGAGCAct |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
7 |
| Similarity score: |
0.061942 |
Alignment:
AGTGCTCCACTGTGDTG
--------ACTTTGG--
| Original motif Consensus sequence: CAHCACAGTGGAGCACT |
Reverse complement motif Consensus sequence: AGTGCTCCACTGTGDTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
70 |
| Motif name: |
CagTGCTCCACTGTGgT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0625 |
Alignment:
ABCACAGTGGAGCACTG
-CCAAAGT---------
| Original motif Consensus sequence: CAGTGCTCCACTGTGBT |
Reverse complement motif Consensus sequence: ABCACAGTGGAGCACTG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
52 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.0686813 |
Alignment:
TGAMCTTTGMMCYT
---ACTTTGG----
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0714286 |
Alignment:
CACTGTGRTRTCACAGTGSWSCACT
-ACTTTGG-----------------
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0.0714286 |
Alignment:
TGGAGCACTGTGACAVCACAGTGG
---------------CCAAAGT--
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
60 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
7 |
| Similarity score: |
0.0728571 |
Alignment:
GTTGACCTTTGACCTTT
-----ACTTTGG-----
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 38 |
Motif name: Motif 38 |
| Original motif Consensus sequence: ATKAYTTTG |
Reverse complement motif Consensus sequence: CAAAKTYAT |
|
|
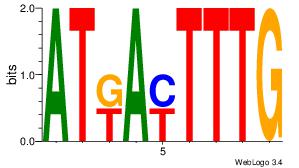 |
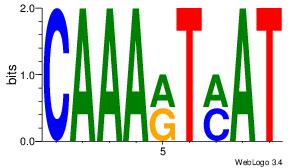 |
Best Matches for Motif ID 38 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
9 |
| Similarity score: |
0.0486111 |
Alignment:
TTTAATGTYTTTWA
----ATKAYTTTG-
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
35 |
| Motif name: |
Motif 35 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0.0625 |
Alignment:
CAAGAKCATCT
CAAAKTYAT--
| Original motif Consensus sequence: AGATGYTCTTG |
Reverse complement motif Consensus sequence: CAAGAKCATCT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
18 |
| Motif name: |
Motif 18 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
9 |
| Similarity score: |
0.0625 |
Alignment:
GCTGGACTTTG
ATKAYTTTG--
| Original motif Consensus sequence: CAAAGTCCAGC |
Reverse complement motif Consensus sequence: GCTGGACTTTG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.0625 |
Alignment:
TGAAAATSCTT
-CAAAKTYAT-
| Original motif Consensus sequence: TGAAAATSCTT |
Reverse complement motif Consensus sequence: AAGSATTTTCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
9 |
| Similarity score: |
0.0625 |
Alignment:
ATTTTTTTTTTTTT
ATKAYTTTG-----
| Original motif Consensus sequence: AAAAAAAAAAAAAT |
Reverse complement motif Consensus sequence: ATTTTTTTTTTTTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
75 |
| Motif name: |
wwCCAmAGTCmt |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0.0650253 |
Alignment:
DDCCAMAGTCHB
---CAAAKTYAT
| Original motif Consensus sequence: DDCCAMAGTCHB |
Reverse complement motif Consensus sequence: VDGACTRTGGDD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
69 |
| Motif name: |
atactttggc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0.0763889 |
Alignment:
GCCAAAGTAT
-CAAAKTYAT
| Original motif Consensus sequence: ATACTTTGGC |
Reverse complement motif Consensus sequence: GCCAAAGTAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.0763889 |
Alignment:
AATAGYATTTT
-ATKAYTTTG-
| Original motif Consensus sequence: AAAATKCTATT |
Reverse complement motif Consensus sequence: AATAGYATTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
14 |
| Motif name: |
Motif 14 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
9 |
| Similarity score: |
0.0763889 |
Alignment:
TAATKATATAA
--ATKAYTTTG
| Original motif Consensus sequence: TAATKATATAA |
Reverse complement motif Consensus sequence: TTATATYATTA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
46 |
| Motif name: |
Motif 46 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0.0763889 |
Alignment:
TATKCTAAACCTAT
-----CAAAKTYAT
| Original motif Consensus sequence: ATAGGTTTAGYATA |
Reverse complement motif Consensus sequence: TATKCTAAACCTAT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 39 |
Motif name: Motif 39 |
| Original motif Consensus sequence: GCTGTGAC |
Reverse complement motif Consensus sequence: GTCACAGC |
|
|
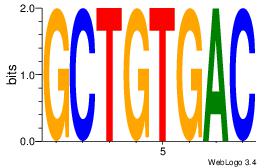 |
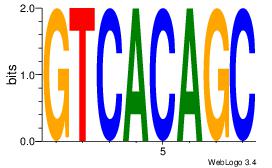 |
Best Matches for Motif ID 39 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
8 |
| Similarity score: |
0.0546875 |
Alignment:
GGAGCACTGTGACACCACA
-----GCTGTGAC------
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
71 |
| Motif name: |
AmTGTGACACCACAGT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
8 |
| Similarity score: |
0.0551758 |
Alignment:
ACTGTGGTGTCACART
--------GTCACAGC
| Original motif Consensus sequence: AMTGTGACACCACAGT |
Reverse complement motif Consensus sequence: ACTGTGGTGTCACART |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
17 |
| Number of overlap: |
8 |
| Similarity score: |
0.0580986 |
Alignment:
ACTGTGACACCACAGTGGAGCACT
GCTGTGAC----------------
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
11 |
| Number of overlap: |
8 |
| Similarity score: |
0.058277 |
Alignment:
CCACTGTGVTGTCACAGTGCTCCA
----------GTCACAGC------
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
60 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.06375 |
Alignment:
AAAGGTCAAAGGTCAAC
----GTCACAGC-----
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
8 |
| Similarity score: |
0.0659722 |
Alignment:
AGTGSWSCACTGTGAMAMCACAGTG
--------GCTGTGAC---------
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
TCTKTGTCAAWA
GCTGTGAC----
| Original motif Consensus sequence: TCTKTGTCAAWA |
Reverse complement motif Consensus sequence: TWTTGACAYAGA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
27 |
| Motif name: |
Motif 27 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
ATTCTGTRAAG
--GCTGTGAC-
| Original motif Consensus sequence: ATTCTGTRAAG |
Reverse complement motif Consensus sequence: CTTKACAGAAT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
88 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0926496 |
Alignment:
HVCDBBDBCAGCAG
----GTCACAGC--
| Original motif Consensus sequence: CTGCTGBDBBDGBD |
Reverse complement motif Consensus sequence: HVCDBBDBCAGCAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
25 |
| Motif name: |
Motif 25 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.09375 |
Alignment:
AATCTGTTATT
-GCTGTGAC--
| Original motif Consensus sequence: AATAACAGATT |
Reverse complement motif Consensus sequence: AATCTGTTATT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 40 |
Motif name: Motif 40 |
| Original motif Consensus sequence: AAGTTCA |
Reverse complement motif Consensus sequence: TGAACTT |
|
|
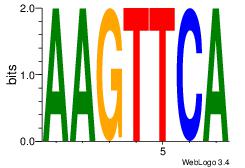 |
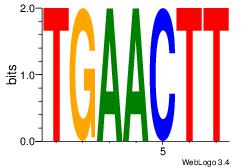 |
Best Matches for Motif ID 40 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
18 |
| Motif name: |
Motif 18 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0.0357143 |
Alignment:
CAAAGTCCAGC
--AAGTTCA--
| Original motif Consensus sequence: CAAAGTCCAGC |
Reverse complement motif Consensus sequence: GCTGGACTTTG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
60 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0.0371429 |
Alignment:
AAAGGTCAAAGGTCAAC
--------AAGTTCA--
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
52 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
7 |
| Similarity score: |
0.043956 |
Alignment:
TGAMCTTTGMMCYT
TGAACTT-------
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
54 |
| Motif name: |
Esrrb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
7 |
| Similarity score: |
0.0492088 |
Alignment:
TGACCTTGMBBB
TGAACTT-----
| Original motif Consensus sequence: VBBYCAAGGTCA |
Reverse complement motif Consensus sequence: TGACCTTGMBBB |
|
|
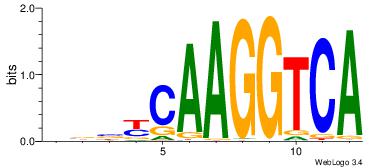 |
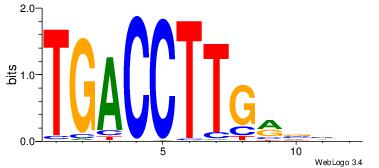 |
| Dataset #: |
1 |
| Motif ID: |
35 |
| Motif name: |
Motif 35 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0535714 |
Alignment:
AGATGYTCTTG
-TGAACTT---
| Original motif Consensus sequence: AGATGYTCTTG |
Reverse complement motif Consensus sequence: CAAGAKCATCT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
7 |
| Similarity score: |
0.0535714 |
Alignment:
AAAGRTMAAACTA
-AAGTTCA-----
| Original motif Consensus sequence: AAAGRTMAAACTA |
Reverse complement motif Consensus sequence: TAGTTTYAKCTTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
61 |
| Motif name: |
NR4A2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0596546 |
Alignment:
GTGACCTT
-TGAACTT
| Original motif Consensus sequence: AAGGTCAC |
Reverse complement motif Consensus sequence: GTGACCTT |
|
|
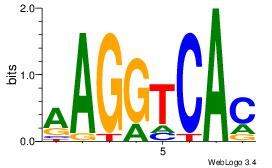 |
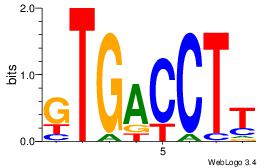 |
| Dataset #: |
3 |
| Motif ID: |
62 |
| Motif name: |
RORA_1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0614286 |
Alignment:
TGACCTDVWT
TGAACTT---
| Original motif Consensus sequence: AWVDAGGTCA |
Reverse complement motif Consensus sequence: TGACCTDVWT |
|
|
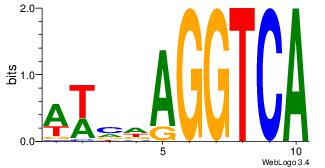 |
 |
| Dataset #: |
5 |
| Motif ID: |
86 |
| Motif name: |
TFW2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.0674603 |
Alignment:
YGGACTTACG
TGAACTT---
| Original motif Consensus sequence: YGGACTTACG |
Reverse complement motif Consensus sequence: CGTAAGTCCK |
|
|
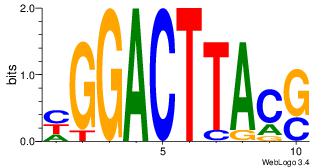 |
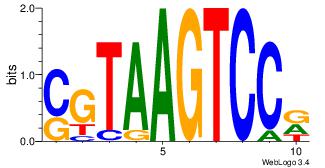 |
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
7 |
| Similarity score: |
0.0714286 |
Alignment:
TGAAAATATGTT
TGAACTT-----
| Original motif Consensus sequence: AACATATTTTCA |
Reverse complement motif Consensus sequence: TGAAAATATGTT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 41 |
Motif name: Motif 41 |
| Original motif Consensus sequence: TGGAGGAAA |
Reverse complement motif Consensus sequence: TTTCCTCCA |
|
|
 |
 |
Best Matches for Motif ID 41 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
28 |
| Motif name: |
Motif 28 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0.0645833 |
Alignment:
TTTACTAAAT
TTTCCTCCA-
| Original motif Consensus sequence: ATTTAGTAAA |
Reverse complement motif Consensus sequence: TTTACTAAAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
23 |
| Motif name: |
Motif 23 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.0645833 |
Alignment:
ATYAATGAAWACAT
----TGGAGGAAA-
| Original motif Consensus sequence: ATGTWTTCATTMAT |
Reverse complement motif Consensus sequence: ATYAATGAAWACAT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
70 |
| Motif name: |
CagTGCTCCACTGTGgT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
9 |
| Similarity score: |
0.068287 |
Alignment:
ABCACAGTGGAGCACTG
-------TGGAGGAAA-
| Original motif Consensus sequence: CAGTGCTCCACTGTGBT |
Reverse complement motif Consensus sequence: ABCACAGTGGAGCACTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0.0692782 |
Alignment:
ACTGTGACACCACAGTGGAGCACT
---------------TGGAGGAAA
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
76 |
| Motif name: |
CAcCACAGTGGAGCAct |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
9 |
| Similarity score: |
0.0697917 |
Alignment:
AGTGCTCCACTGTGDTG
TTTCCTCCA--------
| Original motif Consensus sequence: CAHCACAGTGGAGCACT |
Reverse complement motif Consensus sequence: AGTGCTCCACTGTGDTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
16 |
| Number of overlap: |
9 |
| Similarity score: |
0.0844782 |
Alignment:
CCACTGTGVTGTCACAGTGCTCCA
---------------TTTCCTCCA
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
20 |
| Motif name: |
Motif 20 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.0923611 |
Alignment:
TTCCAGGAGGA
-TGGAGGAAA-
| Original motif Consensus sequence: TCCTCCTGGAA |
Reverse complement motif Consensus sequence: TTCCAGGAGGA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
29 |
| Motif name: |
Motif 29 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.0923611 |
Alignment:
TAGCAATAAAT
-TGGAGGAAA-
| Original motif Consensus sequence: ATTTATTGCTA |
Reverse complement motif Consensus sequence: TAGCAATAAAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
46 |
| Motif name: |
Motif 46 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
9 |
| Similarity score: |
0.0923611 |
Alignment:
TATKCTAAACCTAT
TTTCCTCCA-----
| Original motif Consensus sequence: ATAGGTTTAGYATA |
Reverse complement motif Consensus sequence: TATKCTAAACCTAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
22 |
| Motif name: |
Motif 22 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.0923611 |
Alignment:
ATGCTCACCATAAA
-TTTCCTCCA----
| Original motif Consensus sequence: TTTATGGTGAGCAT |
Reverse complement motif Consensus sequence: ATGCTCACCATAAA |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 42 |
Motif name: Motif 42 |
| Original motif Consensus sequence: TGAATGAA |
Reverse complement motif Consensus sequence: TTCATTCA |
|
|
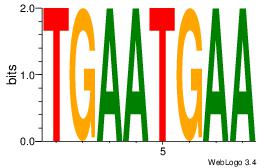 |
 |
Best Matches for Motif ID 42 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
23 |
| Motif name: |
Motif 23 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.015625 |
Alignment:
ATYAATGAAWACAT
-----TGAATGAA-
| Original motif Consensus sequence: ATGTWTTCATTMAT |
Reverse complement motif Consensus sequence: ATYAATGAAWACAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
44 |
| Motif name: |
Motif 44 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.046875 |
Alignment:
TTCTTYCATT
TTCATTCA--
| Original motif Consensus sequence: AATGKAAGAA |
Reverse complement motif Consensus sequence: TTCTTYCATT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
41 |
| Motif name: |
Motif 41 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0625 |
Alignment:
TGGAGGAAA
TGAATGAA-
| Original motif Consensus sequence: TGGAGGAAA |
Reverse complement motif Consensus sequence: TTTCCTCCA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
TWAAAKACATTAAA
-----TTCATTCA-
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
27 |
| Motif name: |
Motif 27 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
ATTCTGTRAAG
--TTCATTCA-
| Original motif Consensus sequence: ATTCTGTRAAG |
Reverse complement motif Consensus sequence: CTTKACAGAAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.078125 |
Alignment:
TTYAAACATYTT
TTCATTCA----
| Original motif Consensus sequence: AAMATGTTTMAA |
Reverse complement motif Consensus sequence: TTYAAACATYTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
21 |
| Motif name: |
Motif 21 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.09375 |
Alignment:
TTYGGAAAGCAG
-TGAATGAA---
| Original motif Consensus sequence: CTGCTTTCCMAA |
Reverse complement motif Consensus sequence: TTYGGAAAGCAG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
80 |
| Motif name: |
taaacgatgcc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.09375 |
Alignment:
TAAACGATGCC
TGAATGAA---
| Original motif Consensus sequence: TAAACGATGCC |
Reverse complement motif Consensus sequence: GGCATCGTTTA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.09375 |
Alignment:
AACATATTTTCA
TTCATTCA----
| Original motif Consensus sequence: AACATATTTTCA |
Reverse complement motif Consensus sequence: TGAAAATATGTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
24 |
| Motif name: |
Motif 24 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.09375 |
Alignment:
TTWATGWCTTT
---TGAATGAA
| Original motif Consensus sequence: AAAGWCATWAA |
Reverse complement motif Consensus sequence: TTWATGWCTTT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 43 |
Motif name: Motif 43 |
| Original motif Consensus sequence: AAKGAAAGGCA |
Reverse complement motif Consensus sequence: TGCCTTTCYTT |
|
|
 |
 |
Best Matches for Motif ID 43 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
TTTRKTTTHTTTTH
-TGCCTTTCYTT--
| Original motif Consensus sequence: HAAAAHAAARMAAA |
Reverse complement motif Consensus sequence: TTTRKTTTHTTTTH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
ATTTTTTTTTTTTT
---TGCCTTTCYTT
| Original motif Consensus sequence: AAAAAAAAAAAAAT |
Reverse complement motif Consensus sequence: ATTTTTTTTTTTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
21 |
| Motif name: |
Motif 21 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
CTGCTTTCCMAA
TGCCTTTCYTT-
| Original motif Consensus sequence: CTGCTTTCCMAA |
Reverse complement motif Consensus sequence: TTYGGAAAGCAG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
94 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.112013 |
Alignment:
TTTTTTTKTTKTTTAATTHW
--TGCCTTTCYTT-------
| Original motif Consensus sequence: TTTTTTTKTTKTTTAATTHW |
Reverse complement motif Consensus sequence: WHAATTAAARAARAAAAAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
23 |
| Motif name: |
Motif 23 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
ATGTWTTCATTMAT
---TGCCTTTCYTT
| Original motif Consensus sequence: ATGTWTTCATTMAT |
Reverse complement motif Consensus sequence: ATYAATGAAWACAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
AAGSATTTTCA
AAKGAAAGGCA
| Original motif Consensus sequence: TGAAAATSCTT |
Reverse complement motif Consensus sequence: AAGSATTTTCA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
45 |
| Motif name: |
Motif 45 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
TAASTCTWTTTTAA
TGCCTTTCYTT---
| Original motif Consensus sequence: TAASTCTWTTTTAA |
Reverse complement motif Consensus sequence: TTAAAAWAGASTTA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
24 |
| Motif name: |
Motif 24 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
TTWATGWCTTT
TGCCTTTCYTT
| Original motif Consensus sequence: AAAGWCATWAA |
Reverse complement motif Consensus sequence: TTWATGWCTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
TWAAAKACATTAAA
-AAKGAAAGGCA--
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
54 |
| Motif name: |
Esrrb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.116255 |
Alignment:
VBBYCAAGGTCA
-AAKGAAAGGCA
| Original motif Consensus sequence: VBBYCAAGGTCA |
Reverse complement motif Consensus sequence: TGACCTTGMBBB |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 44 |
Motif name: Motif 44 |
| Original motif Consensus sequence: AATGKAAGAA |
Reverse complement motif Consensus sequence: TTCTTYCATT |
|
|
 |
 |
Best Matches for Motif ID 44 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
23 |
| Motif name: |
Motif 23 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0541667 |
Alignment:
ATYAATGAAWACAT
-AATGKAAGAA---
| Original motif Consensus sequence: ATGTWTTCATTMAT |
Reverse complement motif Consensus sequence: ATYAATGAAWACAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
14 |
| Motif name: |
Motif 14 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0541667 |
Alignment:
TTATATYATTA
TTCTTYCATT-
| Original motif Consensus sequence: TAATKATATAA |
Reverse complement motif Consensus sequence: TTATATYATTA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
25 |
| Motif name: |
Motif 25 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0666667 |
Alignment:
AATCTGTTATT
-TTCTTYCATT
| Original motif Consensus sequence: AATAACAGATT |
Reverse complement motif Consensus sequence: AATCTGTTATT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0666667 |
Alignment:
ATTTTTTTTTTTTT
-TTCTTYCATT---
| Original motif Consensus sequence: AAAAAAAAAAAAAT |
Reverse complement motif Consensus sequence: ATTTTTTTTTTTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
17 |
| Motif name: |
Motif 17 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0666667 |
Alignment:
CTTCTGACCTC
TTCTTYCATT-
| Original motif Consensus sequence: CTTCTGACCTC |
Reverse complement motif Consensus sequence: GAGGTCAGAAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
43 |
| Motif name: |
Motif 43 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0666667 |
Alignment:
TGCCTTTCYTT
-TTCTTYCATT
| Original motif Consensus sequence: AAKGAAAGGCA |
Reverse complement motif Consensus sequence: TGCCTTTCYTT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
94 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
10 |
| Similarity score: |
0.0684524 |
Alignment:
WHAATTAAARAARAAAAAAA
--AATGKAAGAA--------
| Original motif Consensus sequence: TTTTTTTKTTKTTTAATTHW |
Reverse complement motif Consensus sequence: WHAATTAAARAARAAAAAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0754167 |
Alignment:
TTTRKTTTHTTTTH
-TTCTTYCATT---
| Original motif Consensus sequence: HAAAAHAAARMAAA |
Reverse complement motif Consensus sequence: TTTRKTTTHTTTTH |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
24 |
| Motif name: |
Motif 24 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0791667 |
Alignment:
AAAGWCATWAA
AATGKAAGAA-
| Original motif Consensus sequence: AAAGWCATWAA |
Reverse complement motif Consensus sequence: TTWATGWCTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
31 |
| Motif name: |
Motif 31 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0791667 |
Alignment:
AAAATCAACAA
AATGKAAGAA-
| Original motif Consensus sequence: TTGTTGATTTT |
Reverse complement motif Consensus sequence: AAAATCAACAA |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 45 |
Motif name: Motif 45 |
| Original motif Consensus sequence: TAASTCTWTTTTAA |
Reverse complement motif Consensus sequence: TTAAAAWAGASTTA |
|
|
 |
 |
Best Matches for Motif ID 45 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
94 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
14 |
| Similarity score: |
0.100765 |
Alignment:
WHAATTAAARAARAAAAAAA
----TTAAAAWAGASTTA--
| Original motif Consensus sequence: TTTTTTTKTTKTTTAATTHW |
Reverse complement motif Consensus sequence: WHAATTAAARAARAAAAAAA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
91 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
14 |
| Similarity score: |
0.104911 |
Alignment:
BTTHAAAAAAYAAAAAAR
--TTAAAAWAGASTTA--
| Original motif Consensus sequence: KTTTTTTKTTTTTTDAAB |
Reverse complement motif Consensus sequence: BTTHAAAAAAYAAAAAAR |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.576923 |
Alignment:
-TWAAAKACATTAAA
TTAAAAWAGASTTA-
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.591346 |
Alignment:
HAAAAHAAARMAAA-
-TTAAAAWAGASTTA
| Original motif Consensus sequence: HAAAAHAAARMAAA |
Reverse complement motif Consensus sequence: TTTRKTTTHTTTTH |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
23 |
| Motif name: |
Motif 23 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.605769 |
Alignment:
ATYAATGAAWACAT-
-TTAAAAWAGASTTA
| Original motif Consensus sequence: ATGTWTTCATTMAT |
Reverse complement motif Consensus sequence: ATYAATGAAWACAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
46 |
| Motif name: |
Motif 46 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.605769 |
Alignment:
ATAGGTTTAGYATA-
-TAASTCTWTTTTAA
| Original motif Consensus sequence: ATAGGTTTAGYATA |
Reverse complement motif Consensus sequence: TATKCTAAACCTAT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.605769 |
Alignment:
-ATTTTTTTTTTTTT
TAASTCTWTTTTAA-
| Original motif Consensus sequence: AAAAAAAAAAAAAT |
Reverse complement motif Consensus sequence: ATTTTTTTTTTTTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
13 |
| Number of overlap: |
13 |
| Similarity score: |
0.620726 |
Alignment:
AGTGSWSCACTGTGAMAMCACAGTG-
------------TTAAAAWAGASTTA
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.625 |
Alignment:
AAAGRTMAAACTA-
TTAAAAWAGASTTA
| Original motif Consensus sequence: AAAGRTMAAACTA |
Reverse complement motif Consensus sequence: TAGTTTYAKCTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.09375 |
Alignment:
TGAAAATATGTT--
TTAAAAWAGASTTA
| Original motif Consensus sequence: AACATATTTTCA |
Reverse complement motif Consensus sequence: TGAAAATATGTT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 46 |
Motif name: Motif 46 |
| Original motif Consensus sequence: ATAGGTTTAGYATA |
Reverse complement motif Consensus sequence: TATKCTAAACCTAT |
|
|
 |
 |
Best Matches for Motif ID 46 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.0865385 |
Alignment:
-AAAGRTMAAACTA
TATKCTAAACCTAT
| Original motif Consensus sequence: AAAGRTMAAACTA |
Reverse complement motif Consensus sequence: TAGTTTYAKCTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
22 |
| Motif name: |
Motif 22 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.0865385 |
Alignment:
TTTATGGTGAGCAT-
-ATAGGTTTAGYATA
| Original motif Consensus sequence: TTTATGGTGAGCAT |
Reverse complement motif Consensus sequence: ATGCTCACCATAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
45 |
| Motif name: |
Motif 45 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.105769 |
Alignment:
-TAASTCTWTTTTAA
ATAGGTTTAGYATA-
| Original motif Consensus sequence: TAASTCTWTTTTAA |
Reverse complement motif Consensus sequence: TTAAAAWAGASTTA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
23 |
| Motif name: |
Motif 23 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.614583 |
Alignment:
ATYAATGAAWACAT--
--TATKCTAAACCTAT
| Original motif Consensus sequence: ATGTWTTCATTMAT |
Reverse complement motif Consensus sequence: ATYAATGAAWACAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.625 |
Alignment:
TTTAATGTYTTTWA--
--ATAGGTTTAGYATA
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
1.09091 |
Alignment:
---TTYAAACATYTT
TATKCTAAACCTAT-
| Original motif Consensus sequence: AAMATGTTTMAA |
Reverse complement motif Consensus sequence: TTYAAACATYTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
80 |
| Motif name: |
taaacgatgcc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.10227 |
Alignment:
---GGCATCGTTTA
ATAGGTTTAGYATA
| Original motif Consensus sequence: TAAACGATGCC |
Reverse complement motif Consensus sequence: GGCATCGTTTA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
12 |
| Motif name: |
Motif 12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.10227 |
Alignment:
ACAACACATTT---
TATKCTAAACCTAT
| Original motif Consensus sequence: ACAACACATTT |
Reverse complement motif Consensus sequence: AAATGTGTTGT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
18 |
| Motif name: |
Motif 18 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.125 |
Alignment:
---GCTGGACTTTG
TATKCTAAACCTAT
| Original motif Consensus sequence: CAAAGTCCAGC |
Reverse complement motif Consensus sequence: GCTGGACTTTG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
1.125 |
Alignment:
---TGAAAATATGTT
TATKCTAAACCTAT-
| Original motif Consensus sequence: AACATATTTTCA |
Reverse complement motif Consensus sequence: TGAAAATATGTT |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 47 |
Motif name: Motif 47 |
| Original motif Consensus sequence: YGCCACCTDSTGGY |
Reverse complement motif Consensus sequence: KCCASDAGGTGGCK |
|
|
 |
 |
Best Matches for Motif ID 47 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
14 |
| Similarity score: |
0.0209936 |
Alignment:
BMSMGCCYMCTKSTGGMHM
---YGCCACCTDSTGGY--
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
70 |
| Motif name: |
CagTGCTCCACTGTGgT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.0790816 |
Alignment:
CAGTGCTCCACTGTGBT
---YGCCACCTDSTGGY
| Original motif Consensus sequence: CAGTGCTCCACTGTGBT |
Reverse complement motif Consensus sequence: ABCACAGTGGAGCACTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
76 |
| Motif name: |
CAcCACAGTGGAGCAct |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0804767 |
Alignment:
AGTGCTCCACTGTGDTG
--YGCCACCTDSTGGY-
| Original motif Consensus sequence: CAHCACAGTGGAGCACT |
Reverse complement motif Consensus sequence: AGTGCTCCACTGTGDTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
14 |
| Similarity score: |
0.0836896 |
Alignment:
AGTGCTCCACTGTGGTGTCACAGT
--YGCCACCTDSTGGY--------
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.0913786 |
Alignment:
RAGKGMAGVRRGSRCASAGV
---KCCASDAGGTGGCK---
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
90 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
13 |
| Similarity score: |
0.582451 |
Alignment:
CHHCHBTCCTSCYTCTGC-
-----YGCCACCTDSTGGY
| Original motif Consensus sequence: CHHCHBTCCTSCYTCTGC |
Reverse complement motif Consensus sequence: GCAGAKGSAGGABDGHDG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
13 |
| Similarity score: |
0.590483 |
Alignment:
MGGTCAGGGTGACCTRDBHV-
-------YGCCACCTDSTGGY
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
54 |
| Motif name: |
Esrrb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.08988 |
Alignment:
VBBYCAAGGTCA--
KCCASDAGGTGGCK
| Original motif Consensus sequence: VBBYCAAGGTCA |
Reverse complement motif Consensus sequence: TGACCTTGMBBB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
13 |
| Number of overlap: |
12 |
| Similarity score: |
1.09162 |
Alignment:
--CCACTGTGVTGTCACAGTGCTCCA
YGCCACCTDSTGGY------------
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
74 |
| Motif name: |
ssCCCCGCSssk |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.09358 |
Alignment:
SBCCCCGCCSBB--
YGCCACCTDSTGGY
| Original motif Consensus sequence: SBCCCCGCCSBB |
Reverse complement motif Consensus sequence: BBSGGCGGGGBS |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 48 |
Motif name: Motif 48 |
| Original motif Consensus sequence: GCARRGGSAGS |
Reverse complement motif Consensus sequence: SCTSCCKMTGC |
|
|
 |
 |
Best Matches for Motif ID 48 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
1 |
| Motif name: |
Motif 1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0 |
Alignment:
GCAGAGGSAKG
GCARRGGSAGS
| Original motif Consensus sequence: CYTSCCTCTGC |
Reverse complement motif Consensus sequence: GCAGAGGSAKG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
90 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.000952693 |
Alignment:
GCAGAKGSAGGABDGHDG
GCARRGGSAGS-------
| Original motif Consensus sequence: CHHCHBTCCTSCYTCTGC |
Reverse complement motif Consensus sequence: GCAGAKGSAGGABDGHDG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.033658 |
Alignment:
YDRCCASYAGRKGGCRSYV
------GCARRGGSAGS--
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
88 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0410341 |
Alignment:
CTGCTGBDBBDGBD
--SCTSCCKMTGC-
| Original motif Consensus sequence: CTGCTGBDBBDGBD |
Reverse complement motif Consensus sequence: HVCDBBDBCAGCAG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
11 |
| Similarity score: |
0.0422459 |
Alignment:
RAGKGMAGVRRGSRCASAGV
----GCARRGGSAGS-----
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
65 |
| Motif name: |
Tcfcp2l1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0533531 |
Alignment:
CCAGYYHVADCCRG
SCTSCCKMTGC---
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0578944 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
---SCTSCCKMTGC-------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0620615 |
Alignment:
VDBHMAGGTCACCCTGACCY
--GCARRGGSAGS-------
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0627554 |
Alignment:
HCAGRRVACASAG
GCARRGGSAGS--
| Original motif Consensus sequence: HCAGRRVACASAG |
Reverse complement motif Consensus sequence: CTSTGTBKMCTGH |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
59 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
0.0646979 |
Alignment:
VHRGGTCABDBTGMCCTB
---SCTSCCKMTGC----
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 49 |
Motif name: Motif 49 |
| Original motif Consensus sequence: AAAAAAAAAAAAAT |
Reverse complement motif Consensus sequence: ATTTTTTTTTTTTT |
|
|
 |
 |
Best Matches for Motif ID 49 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
94 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0548469 |
Alignment:
WHAATTAAARAARAAAAAAA
------AAAAAAAAAAAAAT
| Original motif Consensus sequence: TTTTTTTKTTKTTTAATTHW |
Reverse complement motif Consensus sequence: WHAATTAAARAARAAAAAAA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
91 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0602679 |
Alignment:
KTTTTTTKTTTTTTDAAB
ATTTTTTTTTTTTT----
| Original motif Consensus sequence: KTTTTTTKTTTTTTDAAB |
Reverse complement motif Consensus sequence: BTTHAAAAAAYAAAAAAR |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0767857 |
Alignment:
HAAAAHAAARMAAA
AAAAAAAAAAAAAT
| Original motif Consensus sequence: HAAAAHAAARMAAA |
Reverse complement motif Consensus sequence: TTTRKTTTHTTTTH |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
23 |
| Motif name: |
Motif 23 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0982143 |
Alignment:
ATYAATGAAWACAT
AAAAAAAAAAAAAT
| Original motif Consensus sequence: ATGTWTTCATTMAT |
Reverse complement motif Consensus sequence: ATYAATGAAWACAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.107143 |
Alignment:
TWAAAKACATTAAA
AAAAAAAAAAAAAT
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.596154 |
Alignment:
-TAGTTTYAKCTTT
ATTTTTTTTTTTTT
| Original motif Consensus sequence: AAAGRTMAAACTA |
Reverse complement motif Consensus sequence: TAGTTTYAKCTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
45 |
| Motif name: |
Motif 45 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.605769 |
Alignment:
TTAAAAWAGASTTA-
-AAAAAAAAAAAAAT
| Original motif Consensus sequence: TAASTCTWTTTTAA |
Reverse complement motif Consensus sequence: TTAAAAWAGASTTA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.125 |
Alignment:
TGAAAATATGTT--
AAAAAAAAAAAAAT
| Original motif Consensus sequence: AACATATTTTCA |
Reverse complement motif Consensus sequence: TGAAAATATGTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.125 |
Alignment:
--TTYAAACATYTT
ATTTTTTTTTTTTT
| Original motif Consensus sequence: AAMATGTTTMAA |
Reverse complement motif Consensus sequence: TTYAAACATYTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
31 |
| Motif name: |
Motif 31 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.56818 |
Alignment:
TTGTTGATTTT---
ATTTTTTTTTTTTT
| Original motif Consensus sequence: TTGTTGATTTT |
Reverse complement motif Consensus sequence: AAAATCAACAA |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 50 |
Motif name: Motif 50 |
| Original motif Consensus sequence: GGAAM |
Reverse complement motif Consensus sequence: YTTCC |
|
|
 |
 |
Best Matches for Motif ID 50 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
41 |
| Motif name: |
Motif 41 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
5 |
| Similarity score: |
0.01875 |
Alignment:
TTTCCTCCA
YTTCC----
| Original motif Consensus sequence: TGGAGGAAA |
Reverse complement motif Consensus sequence: TTTCCTCCA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
21 |
| Motif name: |
Motif 21 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
5 |
| Similarity score: |
0.01875 |
Alignment:
CTGCTTTCCMAA
---YTTCC----
| Original motif Consensus sequence: CTGCTTTCCMAA |
Reverse complement motif Consensus sequence: TTYGGAAAGCAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
43 |
| Motif name: |
Motif 43 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
5 |
| Similarity score: |
0.04375 |
Alignment:
TGCCTTTCYTT
----YTTCC--
| Original motif Consensus sequence: AAKGAAAGGCA |
Reverse complement motif Consensus sequence: TGCCTTTCYTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
44 |
| Motif name: |
Motif 44 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
5 |
| Similarity score: |
0.05625 |
Alignment:
TTCTTYCATT
--YTTCC---
| Original motif Consensus sequence: AATGKAAGAA |
Reverse complement motif Consensus sequence: TTCTTYCATT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
5 |
| Similarity score: |
0.06875 |
Alignment:
AAGSATTTTCA
------YTTCC
| Original motif Consensus sequence: TGAAAATSCTT |
Reverse complement motif Consensus sequence: AAGSATTTTCA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
5 |
| Similarity score: |
0.06875 |
Alignment:
TGAAAATATGTT
-------GGAAM
| Original motif Consensus sequence: AACATATTTTCA |
Reverse complement motif Consensus sequence: TGAAAATATGTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
19 |
| Motif name: |
Motif 19 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
5 |
| Similarity score: |
0.06875 |
Alignment:
CTTTCTCT
-YTTCC--
| Original motif Consensus sequence: AGAGAAAG |
Reverse complement motif Consensus sequence: CTTTCTCT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
28 |
| Motif name: |
Motif 28 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
5 |
| Similarity score: |
0.06875 |
Alignment:
TTTACTAAAT
YTTCC-----
| Original motif Consensus sequence: ATTTAGTAAA |
Reverse complement motif Consensus sequence: TTTACTAAAT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
12 |
| Number of overlap: |
5 |
| Similarity score: |
0.0747947 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
-----YTTCC-----------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
23 |
| Motif name: |
Motif 23 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
5 |
| Similarity score: |
0.075 |
Alignment:
ATYAATGAAWACAT
----GGAAM-----
| Original motif Consensus sequence: ATGTWTTCATTMAT |
Reverse complement motif Consensus sequence: ATYAATGAAWACAT |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 51 |
Motif name: HNF4A |
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
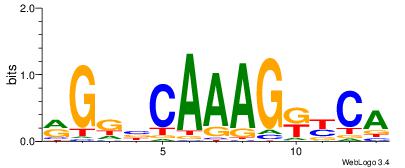 |
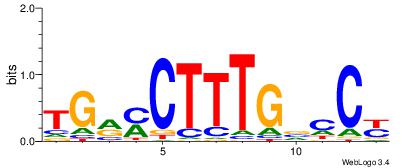 |
Best Matches for Motif ID 51 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
64 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
13 |
| Similarity score: |
0 |
Alignment:
BTRGGDCARAGGKCA
--RGGBCAAAGKYCA
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
52 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.0227413 |
Alignment:
AKGYYCAAAGRTCA
-RGGBCAAAGKYCA
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
60 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
13 |
| Similarity score: |
0.0423023 |
Alignment:
AAAGGTCAAAGGTCAAC
--RGGBCAAAGKYCA--
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
13 |
| Similarity score: |
0.0557654 |
Alignment:
CACTGTGRTRTCACAGTGSWSCACT
-------RGGBCAAAGKYCA-----
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.0609093 |
Alignment:
VCTSTGKSCMKBCTRCYCTK
------TGMYCTTTGBCCK-
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
13 |
| Similarity score: |
0.0656443 |
Alignment:
CCACTGTGVTGTCACAGTGCTCCA
--------RGGBCAAAGKYCA---
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
59 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
13 |
| Similarity score: |
0.0671615 |
Alignment:
VHRGGTCABDBTGMCCTB
--RGGBCAAAGKYCA---
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
13 |
| Similarity score: |
0.0681338 |
Alignment:
TGTGGTGTCACAGTGCTCC
----RGGBCAAAGKYCA--
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.0711137 |
Alignment:
MGGTCAGGGTGACCTRDBHV
RGGBCAAAGKYCA-------
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
13 |
| Similarity score: |
0.0741922 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
-----TGMYCTTTGBCCK---
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 52 |
Motif name: NR2F1 |
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
Best Matches for Motif ID 52 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
60 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0289973 |
Alignment:
GTTGACCTTTGACCTTT
--TGAMCTTTGMMCYT-
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
64 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0393667 |
Alignment:
TGRCCTKTGHCCKAB
TGAMCTTTGMMCYT-
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
14 |
| Similarity score: |
0.0713032 |
Alignment:
AGTGSWSCACTGTGAMAMCACAGTG
-----TGAMCTTTGMMCYT------
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
14 |
| Similarity score: |
0.0742967 |
Alignment:
RAGKGMAGVRRGSRCASAGV
--AKGYYCAAAGRTCA----
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
59 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.0825859 |
Alignment:
BAGGYCABHBTGACCKHV
---TGAMCTTTGMMCYT-
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
14 |
| Similarity score: |
0.0833345 |
Alignment:
TTCAGCACCATGGACAGCKCC
--AKGYYCAAAGRTCA-----
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
14 |
| Similarity score: |
0.0852669 |
Alignment:
TGGAGCACTGTGACAVCACAGTGG
---TGAMCTTTGMMCYT-------
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.0855082 |
Alignment:
GGAGCACTGTGACACCACA
--TGAMCTTTGMMCYT---
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
51 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.525587 |
Alignment:
-TGMYCTTTGBCCK
TGAMCTTTGMMCYT
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
13 |
| Similarity score: |
0.581407 |
Alignment:
VDBHMAGGTCACCCTGACCY-
-------TGAMCTTTGMMCYT
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 53 |
Motif name: ESR1 |
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
Best Matches for Motif ID 53 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
20 |
| Similarity score: |
0.0722586 |
Alignment:
CCACTGTGVTGTCACAGTGCTCCA
---VDBHMAGGTCACCCTGACCY-
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
20 |
| Similarity score: |
0.0818971 |
Alignment:
CACTGTGRTRTCACAGTGSWSCACT
--VDBHMAGGTCACCCTGACCY---
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
19 |
| Similarity score: |
0.553964 |
Alignment:
-VCTSTGKSCMKBCTRCYCTK
VDBHMAGGTCACCCTGACCY-
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
19 |
| Similarity score: |
0.566376 |
Alignment:
-TGTGGTGTCACAGTGCTCC
VDBHMAGGTCACCCTGACCY
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
19 |
| Similarity score: |
0.579928 |
Alignment:
-BMSMGCCYMCTKSTGGMHM
VDBHMAGGTCACCCTGACCY
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
59 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
17 |
| Similarity score: |
1.50287 |
Alignment:
---VHRGGTCABDBTGMCCTB
VDBHMAGGTCACCCTGACCY-
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
90 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
17 |
| Similarity score: |
1.55953 |
Alignment:
CHHCHBTCCTSCYTCTGC---
-VDBHMAGGTCACCCTGACCY
| Original motif Consensus sequence: CHHCHBTCCTSCYTCTGC |
Reverse complement motif Consensus sequence: GCAGAKGSAGGABDGHDG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
89 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
17 |
| Similarity score: |
1.5827 |
Alignment:
---CACACACACACWCACACA
VDBHMAGGTCACCCTGACCY-
| Original motif Consensus sequence: CACACACACACWCACACA |
Reverse complement motif Consensus sequence: TGTGTGWGTGTGTGTGTG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
16 |
| Similarity score: |
2.07409 |
Alignment:
TTCAGCACCATGGACAGCKCC----
-----VDBHMAGGTCACCCTGACCY
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
71 |
| Motif name: |
AmTGTGACACCACAGT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
15 |
| Similarity score: |
2.57517 |
Alignment:
ACTGTGGTGTCACART-----
-VDBHMAGGTCACCCTGACCY
| Original motif Consensus sequence: AMTGTGACACCACAGT |
Reverse complement motif Consensus sequence: ACTGTGGTGTCACART |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 54 |
Motif name: Esrrb |
| Original motif Consensus sequence: VBBYCAAGGTCA |
Reverse complement motif Consensus sequence: TGACCTTGMBBB |
|
|
 |
 |
Best Matches for Motif ID 54 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
52 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0613872 |
Alignment:
TGAMCTTTGMMCYT
--TGACCTTGMBBB
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
64 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
12 |
| Similarity score: |
0.0641161 |
Alignment:
BTRGGDCARAGGKCA
---VBBYCAAGGTCA
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
51 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0642069 |
Alignment:
RGGBCAAAGKYCA
-VBBYCAAGGTCA
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
60 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0678238 |
Alignment:
AAAGGTCAAAGGTCAAC
---VBBYCAAGGTCA--
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
12 |
| Similarity score: |
0.0799744 |
Alignment:
VDBHMAGGTCACCCTGACCY
TGACCTTGMBBB--------
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
59 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
12 |
| Similarity score: |
0.0865873 |
Alignment:
VHRGGTCABDBTGMCCTB
-----TGACCTTGMBBB-
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
12 |
| Similarity score: |
0.0884496 |
Alignment:
TTCAGCACCATGGACAGCKCC
----VBBYCAAGGTCA-----
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0943816 |
Alignment:
TGTGGTGTCACAGTGCTCC
-------TGACCTTGMBBB
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
12 |
| Number of overlap: |
12 |
| Similarity score: |
0.0956646 |
Alignment:
CCACTGTGVTGTCACAGTGCTCCA
-----------TGACCTTGMBBB-
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
11 |
| Number of overlap: |
12 |
| Similarity score: |
0.100398 |
Alignment:
AGTGSWSCACTGTGAMAMCACAGTG
---VBBYCAAGGTCA----------
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 55 |
Motif name: ZEB1 |
| Original motif Consensus sequence: CACCTD |
Reverse complement motif Consensus sequence: HAGGTG |
|
|
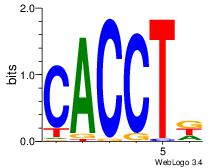 |
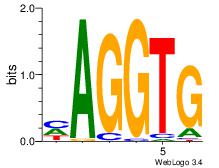 |
Best Matches for Motif ID 55 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
Motif 47 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
6 |
| Similarity score: |
0 |
Alignment:
YGCCACCTDSTGGY
---CACCTD-----
| Original motif Consensus sequence: YGCCACCTDSTGGY |
Reverse complement motif Consensus sequence: KCCASDAGGTGGCK |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.00902841 |
Alignment:
CWGAGCCAYCTYTC
------CACCTD--
| Original motif Consensus sequence: CWGAGCCAYCTYTC |
Reverse complement motif Consensus sequence: GAKAGKTGGCTCWG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
6 |
| Similarity score: |
0.0118423 |
Alignment:
YDRCCASYAGRKGGCRSYV
-------HAGGTG------
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
62 |
| Motif name: |
RORA_1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0187312 |
Alignment:
TGACCTDVWT
-CACCTD---
| Original motif Consensus sequence: AWVDAGGTCA |
Reverse complement motif Consensus sequence: TGACCTDVWT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
6 |
| Similarity score: |
0.0189156 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
----------HAGGTG-----
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
54 |
| Motif name: |
Esrrb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0248883 |
Alignment:
VBBYCAAGGTCA
-HAGGTG-----
| Original motif Consensus sequence: VBBYCAAGGTCA |
Reverse complement motif Consensus sequence: TGACCTTGMBBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
60 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0278775 |
Alignment:
GTTGACCTTTGACCTTT
----------CACCTD-
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
46 |
| Motif name: |
Motif 46 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
6 |
| Similarity score: |
0.0285279 |
Alignment:
TATKCTAAACCTAT
-CACCTD-------
| Original motif Consensus sequence: ATAGGTTTAGYATA |
Reverse complement motif Consensus sequence: TATKCTAAACCTAT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
64 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
6 |
| Similarity score: |
0.0310859 |
Alignment:
TGRCCTKTGHCCKAB
-CACCTD--------
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
61 |
| Motif name: |
NR4A2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0334919 |
Alignment:
GTGACCTT
--CACCTD
| Original motif Consensus sequence: AAGGTCAC |
Reverse complement motif Consensus sequence: GTGACCTT |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 56 |
Motif name: Mycn |
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
Best Matches for Motif ID 56 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
58 |
| Motif name: |
Myc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0 |
Alignment:
DCCACGTGCV
GCCACGTGSD
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
68 |
| Motif name: |
sscCCCGCGcs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0654819 |
Alignment:
BSCCCCGCGBS
-GCCACGTGSD
| Original motif Consensus sequence: BSCCCCGCGBS |
Reverse complement motif Consensus sequence: SBCGCGGGGSB |
|
|
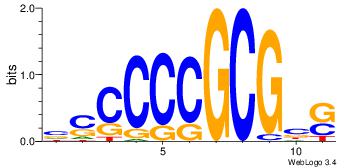 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
10 |
| Similarity score: |
0.0679017 |
Alignment:
YDRCCASYAGRKGGCRSYV
-----HSCACGTGGC----
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
74 |
| Motif name: |
ssCCCCGCSssk |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0704988 |
Alignment:
BBSGGCGGGGBS
-HSCACGTGGC-
| Original motif Consensus sequence: SBCCCCGCCSBB |
Reverse complement motif Consensus sequence: BBSGGCGGGGBS |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
Motif 47 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0733001 |
Alignment:
KCCASDAGGTGGCK
-HSCACGTGGC---
| Original motif Consensus sequence: YGCCACCTDSTGGY |
Reverse complement motif Consensus sequence: KCCASDAGGTGGCK |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
10 |
| Number of overlap: |
10 |
| Similarity score: |
0.0883224 |
Alignment:
AGTGSWSCACTGTGAMAMCACAGTG
------GCCACGTGSD---------
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.089863 |
Alignment:
MGGTCAGGGTGACCTRDBHV
--------GCCACGTGSD--
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
70 |
| Motif name: |
CagTGCTCCACTGTGgT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
10 |
| Similarity score: |
0.0943014 |
Alignment:
ABCACAGTGGAGCACTG
-GCCACGTGSD------
| Original motif Consensus sequence: CAGTGCTCCACTGTGBT |
Reverse complement motif Consensus sequence: ABCACAGTGGAGCACTG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
51 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0947124 |
Alignment:
TGMYCTTTGBCCK
-GCCACGTGSD--
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
10 |
| Similarity score: |
0.0949359 |
Alignment:
ACTGTGACACCACAGTGGAGCACT
---------GCCACGTGSD-----
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 57 |
Motif name: REST |
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
Best Matches for Motif ID 57 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
20 |
| Similarity score: |
0.0656473 |
Alignment:
-VCTSTGKSCMKBCTRCYCTK
GGYGCTGTCCATGGTGCTGAA
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
19 |
| Similarity score: |
0.560523 |
Alignment:
--AGTGCTCCACTGTGGTGTCACAGT
GGYGCTGTCCATGGTGCTGAA-----
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
16 |
| Similarity score: |
2.06264 |
Alignment:
-----VDBHMAGGTCACCCTGACCY
TTCAGCACCATGGACAGCKCC----
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
90 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
14 |
| Similarity score: |
3.06651 |
Alignment:
CHHCHBTCCTSCYTCTGC-------
----GGYGCTGTCCATGGTGCTGAA
| Original motif Consensus sequence: CHHCHBTCCTSCYTCTGC |
Reverse complement motif Consensus sequence: GCAGAKGSAGGABDGHDG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
70 |
| Motif name: |
CagTGCTCCACTGTGgT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
3.07032 |
Alignment:
-------ABCACAGTGGAGCACTG
GGYGCTGTCCATGGTGCTGAA---
| Original motif Consensus sequence: CAGTGCTCCACTGTGBT |
Reverse complement motif Consensus sequence: ABCACAGTGGAGCACTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
71 |
| Motif name: |
AmTGTGACACCACAGT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
14 |
| Similarity score: |
3.07054 |
Alignment:
ACTGTGGTGTCACART-------
--GGYGCTGTCCATGGTGCTGAA
| Original motif Consensus sequence: AMTGTGACACCACAGT |
Reverse complement motif Consensus sequence: ACTGTGGTGTCACART |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
12 |
| Number of overlap: |
13 |
| Similarity score: |
3.54841 |
Alignment:
TGGAGCACTGTGACAVCACAGTGG--------
-----------TTCAGCACCATGGACAGCKCC
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
59 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
13 |
| Similarity score: |
3.55156 |
Alignment:
BAGGYCABHBTGACCKHV--------
-----GGYGCTGTCCATGGTGCTGAA
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
51 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
3.56375 |
Alignment:
TGMYCTTTGBCCK--------
GGYGCTGTCCATGGTGCTGAA
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
64 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
13 |
| Similarity score: |
3.56574 |
Alignment:
--------BTRGGDCARAGGKCA
TTCAGCACCATGGACAGCKCC--
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 58 |
Motif name: Myc |
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
Best Matches for Motif ID 58 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
56 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0 |
Alignment:
HSCACGTGGC
VGCACGTGGH
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
68 |
| Motif name: |
sscCCCGCGcs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.064853 |
Alignment:
BSCCCCGCGBS
-DCCACGTGCV
| Original motif Consensus sequence: BSCCCCGCGBS |
Reverse complement motif Consensus sequence: SBCGCGGGGSB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
74 |
| Motif name: |
ssCCCCGCSssk |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0728588 |
Alignment:
SBCCCCGCCSBB
-DCCACGTGCV-
| Original motif Consensus sequence: SBCCCCGCCSBB |
Reverse complement motif Consensus sequence: BBSGGCGGGGBS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
10 |
| Similarity score: |
0.0754257 |
Alignment:
YDRCCASYAGRKGGCRSYV
--DCCACGTGCV-------
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
Motif 47 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0792377 |
Alignment:
KCCASDAGGTGGCK
-VGCACGTGGH---
| Original motif Consensus sequence: YGCCACCTDSTGGY |
Reverse complement motif Consensus sequence: KCCASDAGGTGGCK |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
10 |
| Similarity score: |
0.0888909 |
Alignment:
CACTGTGRTRTCACAGTGSWSCACT
----------DCCACGTGCV-----
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
51 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0934488 |
Alignment:
TGMYCTTTGBCCK
---VGCACGTGGH
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
10 |
| Similarity score: |
0.0940438 |
Alignment:
MGGTCAGGGTGACCTRDBHV
--------DCCACGTGCV--
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
70 |
| Motif name: |
CagTGCTCCACTGTGgT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
10 |
| Similarity score: |
0.0955234 |
Alignment:
ABCACAGTGGAGCACTG
-DCCACGTGCV------
| Original motif Consensus sequence: CAGTGCTCCACTGTGBT |
Reverse complement motif Consensus sequence: ABCACAGTGGAGCACTG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
59 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0968598 |
Alignment:
VHRGGTCABDBTGMCCTB
----VGCACGTGGH----
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 59 |
Motif name: ESR2 |
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
Best Matches for Motif ID 59 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
18 |
| Similarity score: |
0.0691727 |
Alignment:
VCTSTGKSCMKBCTRCYCTK
--VHRGGTCABDBTGMCCTB
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
18 |
| Similarity score: |
0.0717757 |
Alignment:
CCACTGTGVTGTCACAGTGCTCCA
------VHRGGTCABDBTGMCCTB
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
18 |
| Similarity score: |
0.0815903 |
Alignment:
AGTGSWSCACTGTGAMAMCACAGTG
-VHRGGTCABDBTGMCCTB------
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
18 |
| Similarity score: |
0.0916961 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
-VHRGGTCABDBTGMCCTB--
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
17 |
| Similarity score: |
0.511812 |
Alignment:
VDBHMAGGTCACCCTGACCY-
---VHRGGTCABDBTGMCCTB
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
17 |
| Similarity score: |
0.569595 |
Alignment:
TGTGGTGTCACAGTGCTCC-
--VHRGGTCABDBTGMCCTB
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
90 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
17 |
| Similarity score: |
0.583467 |
Alignment:
GCAGAKGSAGGABDGHDG-
-VHRGGTCABDBTGMCCTB
| Original motif Consensus sequence: CHHCHBTCCTSCYTCTGC |
Reverse complement motif Consensus sequence: GCAGAKGSAGGABDGHDG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
64 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
1.56699 |
Alignment:
BTRGGDCARAGGKCA---
VHRGGTCABDBTGMCCTB
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.08635 |
Alignment:
CWGAGCCAYCTYTC----
VHRGGTCABDBTGMCCTB
| Original motif Consensus sequence: CWGAGCCAYCTYTC |
Reverse complement motif Consensus sequence: GAKAGKTGGCTCWG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
52 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.08921 |
Alignment:
----TGAMCTTTGMMCYT
VHRGGTCABDBTGMCCTB
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 60 |
Motif name: NR1H2RXRA |
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
Best Matches for Motif ID 60 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
17 |
| Similarity score: |
0.0911991 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
---GTTGACCTTTGACCTTT-
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
64 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
1.05945 |
Alignment:
BTRGGDCARAGGKCA--
AAAGGTCAAAGGTCAAC
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
2.0701 |
Alignment:
AAAGRTMAAACTA----
AAAGGTCAAAGGTCAAC
| Original motif Consensus sequence: AAAGRTMAAACTA |
Reverse complement motif Consensus sequence: TAGTTTYAKCTTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
13 |
| Similarity score: |
2.08514 |
Alignment:
----VDBHMAGGTCACCCTGACCY
AAAGGTCAAAGGTCAAC-------
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
22 |
| Motif name: |
Motif 22 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
2.58792 |
Alignment:
-----TTTATGGTGAGCAT
AAAGGTCAAAGGTCAAC--
| Original motif Consensus sequence: TTTATGGTGAGCAT |
Reverse complement motif Consensus sequence: ATGCTCACCATAAA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
12 |
| Similarity score: |
2.58894 |
Alignment:
RAGKGMAGVRRGSRCASAGV-----
--------AAAGGTCAAAGGTCAAC
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
16 |
| Motif name: |
Motif 16 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
2.59792 |
Alignment:
-----TWTTGACAYAGA
AAAGGTCAAAGGTCAAC
| Original motif Consensus sequence: TCTKTGTCAAWA |
Reverse complement motif Consensus sequence: TWTTGACAYAGA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
13 |
| Number of overlap: |
12 |
| Similarity score: |
2.59961 |
Alignment:
AGTGCTCCACTGTGGTGTCACAGT-----
------------AAAGGTCAAAGGTCAAC
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
71 |
| Motif name: |
AmTGTGACACCACAGT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
12 |
| Similarity score: |
2.59967 |
Alignment:
ACTGTGGTGTCACART-----
----AAAGGTCAAAGGTCAAC
| Original motif Consensus sequence: AMTGTGACACCACAGT |
Reverse complement motif Consensus sequence: ACTGTGGTGTCACART |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
18 |
| Motif name: |
Motif 18 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
3.05261 |
Alignment:
------CAAAGTCCAGC
AAAGGTCAAAGGTCAAC
| Original motif Consensus sequence: CAAAGTCCAGC |
Reverse complement motif Consensus sequence: GCTGGACTTTG |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 61 |
Motif name: NR4A2 |
| Original motif Consensus sequence: AAGGTCAC |
Reverse complement motif Consensus sequence: GTGACCTT |
|
|
 |
 |
Best Matches for Motif ID 61 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
60 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
8 |
| Similarity score: |
0.0167759 |
Alignment:
AAAGGTCAAAGGTCAAC
-AAGGTCAC--------
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0195235 |
Alignment:
VDBHMAGGTCACCCTGACCY
----AAGGTCAC--------
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
59 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0198372 |
Alignment:
BAGGYCABHBTGACCKHV
---------GTGACCTT-
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
17 |
| Motif name: |
Motif 17 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0393171 |
Alignment:
CTTCTGACCTC
GTGACCTT---
| Original motif Consensus sequence: CTTCTGACCTC |
Reverse complement motif Consensus sequence: GAGGTCAGAAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.040519 |
Alignment:
AAAGRTMAAACTA
-AAGGTCAC----
| Original motif Consensus sequence: AAAGRTMAAACTA |
Reverse complement motif Consensus sequence: TAGTTTYAKCTTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
64 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.0495607 |
Alignment:
BTRGGDCARAGGKCA
-AAGGTCAC------
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
24 |
| Motif name: |
Motif 24 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0539119 |
Alignment:
TTWATGWCTTT
---GTGACCTT
| Original motif Consensus sequence: AAAGWCATWAA |
Reverse complement motif Consensus sequence: TTWATGWCTTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.0557243 |
Alignment:
CACTGTGRTRTCACAGTGSWSCACT
------AAGGTCAC-----------
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
52 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.0564874 |
Alignment:
AKGYYCAAAGRTCA
------AAGGTCAC
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
35 |
| Motif name: |
Motif 35 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0566591 |
Alignment:
AGATGYTCTTG
-GTGACCTT--
| Original motif Consensus sequence: AGATGYTCTTG |
Reverse complement motif Consensus sequence: CAAGAKCATCT |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 62 |
Motif name: RORA_1 |
| Original motif Consensus sequence: AWVDAGGTCA |
Reverse complement motif Consensus sequence: TGACCTDVWT |
|
|
 |
 |
Best Matches for Motif ID 62 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
54 |
| Motif name: |
Esrrb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.053151 |
Alignment:
VBBYCAAGGTCA
--AWVDAGGTCA
| Original motif Consensus sequence: VBBYCAAGGTCA |
Reverse complement motif Consensus sequence: TGACCTTGMBBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0655351 |
Alignment:
MGGTCAGGGTGACCTRDBHV
---------TGACCTDVWT-
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
60 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
10 |
| Similarity score: |
0.078 |
Alignment:
AAAGGTCAAAGGTCAAC
-----AWVDAGGTCA--
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
52 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0892308 |
Alignment:
TGAMCTTTGMMCYT
----TGACCTDVWT
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
64 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.09043 |
Alignment:
BTRGGDCARAGGKCA
-----AWVDAGGTCA
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
10 |
| Similarity score: |
0.0993108 |
Alignment:
TTCAGCACCATGGACAGCKCC
------AWVDAGGTCA-----
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
88 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.103528 |
Alignment:
CTGCTGBDBBDGBD
----TGACCTDVWT
| Original motif Consensus sequence: CTGCTGBDBBDGBD |
Reverse complement motif Consensus sequence: HVCDBBDBCAGCAG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
51 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.10409 |
Alignment:
RGGBCAAAGKYCA
---AWVDAGGTCA
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.106859 |
Alignment:
RAGKGMAGVRRGSRCASAGV
------AWVDAGGTCA----
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
22 |
| Motif name: |
Motif 22 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.111 |
Alignment:
ATGCTCACCATAAA
----TGACCTDVWT
| Original motif Consensus sequence: TTTATGGTGAGCAT |
Reverse complement motif Consensus sequence: ATGCTCACCATAAA |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 63 |
Motif name: MIZF |
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
Best Matches for Motif ID 63 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
18 |
| Motif name: |
Motif 18 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0591071 |
Alignment:
CAAAGTCCAGC
BAACGTCCGC-
| Original motif Consensus sequence: CAAAGTCCAGC |
Reverse complement motif Consensus sequence: GCTGGACTTTG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
59 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0605777 |
Alignment:
BAGGYCABHBTGACCKHV
--------GCGGACGTTV
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
10 |
| Similarity score: |
0.0614567 |
Alignment:
VDBHMAGGTCACCCTGACCY
---BAACGTCCGC-------
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
60 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0646071 |
Alignment:
AAAGGTCAAAGGTCAAC
-------BAACGTCCGC
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
82 |
| Motif name: |
gCGCGCsCgsG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0651598 |
Alignment:
GCGCGCSCGCG
BAACGTCCGC-
| Original motif Consensus sequence: GCGCGCSCGCG |
Reverse complement motif Consensus sequence: CGCGSGCGCGC |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
10 |
| Number of overlap: |
10 |
| Similarity score: |
0.0697767 |
Alignment:
BMSMGCCYMCTKSTGGMHM
BAACGTCCGC---------
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0708149 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
---------BAACGTCCGC--
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
88 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0753571 |
Alignment:
CTGCTGBDBBDGBD
--GCGGACGTTV--
| Original motif Consensus sequence: CTGCTGBDBBDGBD |
Reverse complement motif Consensus sequence: HVCDBBDBCAGCAG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
10 |
| Similarity score: |
0.0780684 |
Alignment:
VCTSTGKSCMKBCTRCYCTK
------BAACGTCCGC----
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
1 |
| Motif name: |
Motif 1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0797585 |
Alignment:
CYTSCCTCTGC
-BAACGTCCGC
| Original motif Consensus sequence: CYTSCCTCTGC |
Reverse complement motif Consensus sequence: GCAGAGGSAKG |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 64 |
Motif name: PPARGRXRA |
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
Best Matches for Motif ID 64 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
60 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
15 |
| Similarity score: |
0.0442974 |
Alignment:
AAAGGTCAAAGGTCAAC
BTRGGDCARAGGKCA--
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
59 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
15 |
| Similarity score: |
0.0602408 |
Alignment:
VHRGGTCABDBTGMCCTB
BTRGGDCARAGGKCA---
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
15 |
| Similarity score: |
0.0615418 |
Alignment:
RAGKGMAGVRRGSRCASAGV
-BTRGGDCARAGGKCA----
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
15 |
| Similarity score: |
0.0723092 |
Alignment:
VDBHMAGGTCACCCTGACCY
---BTRGGDCARAGGKCA--
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
15 |
| Similarity score: |
0.0741352 |
Alignment:
CACTGTGRTRTCACAGTGSWSCACT
-----BTRGGDCARAGGKCA-----
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
15 |
| Similarity score: |
0.0786357 |
Alignment:
YDRCCASYAGRKGGCRSYV
-BTRGGDCARAGGKCA---
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
90 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
15 |
| Similarity score: |
0.0790185 |
Alignment:
CHHCHBTCCTSCYTCTGC
TGRCCTKTGHCCKAB---
| Original motif Consensus sequence: CHHCHBTCCTSCYTCTGC |
Reverse complement motif Consensus sequence: GCAGAKGSAGGABDGHDG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
15 |
| Similarity score: |
0.0796159 |
Alignment:
TTCAGCACCATGGACAGCKCC
-BTRGGDCARAGGKCA-----
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
15 |
| Similarity score: |
0.0813414 |
Alignment:
CCACTGTGVTGTCACAGTGCTCCA
------BTRGGDCARAGGKCA---
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
15 |
| Similarity score: |
0.0818655 |
Alignment:
TGTGGTGTCACAGTGCTCC
--BTRGGDCARAGGKCA--
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 65 |
Motif name: Tcfcp2l1 |
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
Best Matches for Motif ID 65 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.0453398 |
Alignment:
MGGTCAGGGTGACCTRDBHV
---CCAGYYHVADCCRG---
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.0491111 |
Alignment:
YDRCCASYAGRKGGCRSYV
---CCAGYYHVADCCRG--
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0502168 |
Alignment:
VCTSTGKSCMKBCTRCYCTK
-CCAGYYHVADCCRG-----
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
90 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0514558 |
Alignment:
GCAGAKGSAGGABDGHDG
----CKGGDTBDMMCTGG
| Original motif Consensus sequence: CHHCHBTCCTSCYTCTGC |
Reverse complement motif Consensus sequence: GCAGAKGSAGGABDGHDG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
88 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0546308 |
Alignment:
HVCDBBDBCAGCAG
CCAGYYHVADCCRG
| Original motif Consensus sequence: CTGCTGBDBBDGBD |
Reverse complement motif Consensus sequence: HVCDBBDBCAGCAG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
59 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0568174 |
Alignment:
VHRGGTCABDBTGMCCTB
----CKGGDTBDMMCTGG
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
64 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0569399 |
Alignment:
TGRCCTKTGHCCKAB
-CKGGDTBDMMCTGG
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.542892 |
Alignment:
CTSTGTBKMCTGH-
CKGGDTBDMMCTGG
| Original motif Consensus sequence: HCAGRRVACASAG |
Reverse complement motif Consensus sequence: CTSTGTBKMCTGH |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.546848 |
Alignment:
GAKAGKTGGCTCWG-
-CKGGDTBDMMCTGG
| Original motif Consensus sequence: CWGAGCCAYCTYTC |
Reverse complement motif Consensus sequence: GAKAGKTGGCTCWG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
70 |
| Motif name: |
CagTGCTCCACTGTGgT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
13 |
| Similarity score: |
0.551995 |
Alignment:
ABCACAGTGGAGCACTG-
----CKGGDTBDMMCTGG
| Original motif Consensus sequence: CAGTGCTCCACTGTGBT |
Reverse complement motif Consensus sequence: ABCACAGTGGAGCACTG |
|
|
 |
 |
| Dataset #: 3 |
Motif ID: 66 |
Motif name: CTCF |
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
Best Matches for Motif ID 66 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
19 |
| Similarity score: |
0.066318 |
Alignment:
VCTSTGKSCMKBCTRCYCTK
BMSMGCCYMCTKSTGGMHM-
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
19 |
| Similarity score: |
0.0687764 |
Alignment:
MGGTCAGGGTGACCTRDBHV
-YDRCCASYAGRKGGCRSYV
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
18 |
| Similarity score: |
0.566688 |
Alignment:
ACTGTGACACCACAGTGGAGCACT-
------YDRCCASYAGRKGGCRSYV
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
18 |
| Similarity score: |
0.56949 |
Alignment:
-AGTGSWSCACTGTGAMAMCACAGTG
BMSMGCCYMCTKSTGGMHM-------
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
90 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
0.571172 |
Alignment:
-CHHCHBTCCTSCYTCTGC
BMSMGCCYMCTKSTGGMHM
| Original motif Consensus sequence: CHHCHBTCCTSCYTCTGC |
Reverse complement motif Consensus sequence: GCAGAKGSAGGABDGHDG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
70 |
| Motif name: |
CagTGCTCCACTGTGgT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
17 |
| Similarity score: |
1.05559 |
Alignment:
--ABCACAGTGGAGCACTG
YDRCCASYAGRKGGCRSYV
| Original motif Consensus sequence: CAGTGCTCCACTGTGBT |
Reverse complement motif Consensus sequence: ABCACAGTGGAGCACTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
76 |
| Motif name: |
CAcCACAGTGGAGCAct |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
15 |
| Similarity score: |
2.06507 |
Alignment:
----CAHCACAGTGGAGCACT
YDRCCASYAGRKGGCRSYV--
| Original motif Consensus sequence: CAHCACAGTGGAGCACT |
Reverse complement motif Consensus sequence: AGTGCTCCACTGTGDTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
10 |
| Number of overlap: |
15 |
| Similarity score: |
2.07021 |
Alignment:
TGGAGCACTGTGACAVCACAGTGG----
---------YDRCCASYAGRKGGCRSYV
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.54791 |
Alignment:
-----CWGAGCCAYCTYTC
BMSMGCCYMCTKSTGGMHM
| Original motif Consensus sequence: CWGAGCCAYCTYTC |
Reverse complement motif Consensus sequence: GAKAGKTGGCTCWG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
Motif 47 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.56483 |
Alignment:
KCCASDAGGTGGCK-----
YDRCCASYAGRKGGCRSYV
| Original motif Consensus sequence: YGCCACCTDSTGGY |
Reverse complement motif Consensus sequence: KCCASDAGGTGGCK |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 67 |
Motif name: TgTGgTGTCACAGTGCTCC |
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
Best Matches for Motif ID 67 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
19 |
| Similarity score: |
0 |
Alignment:
CCACTGTGVTGTCACAGTGCTCCA
----TGTGGTGTCACAGTGCTCC-
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
19 |
| Similarity score: |
0.0217259 |
Alignment:
AGTGSWSCACTGTGAMAMCACAGTG
---GGAGCACTGTGACACCACA---
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
19 |
| Similarity score: |
0.074816 |
Alignment:
ACTGTGACACCACAGTGGAGCACT
--TGTGGTGTCACAGTGCTCC---
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
19 |
| Similarity score: |
0.0857262 |
Alignment:
VDBHMAGGTCACCCTGACCY
-TGTGGTGTCACAGTGCTCC
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
18 |
| Similarity score: |
0.595281 |
Alignment:
-RAGKGMAGVRRGSRCASAGV
GGAGCACTGTGACACCACA--
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
59 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
17 |
| Similarity score: |
1.08001 |
Alignment:
--VHRGGTCABDBTGMCCTB
TGTGGTGTCACAGTGCTCC-
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
70 |
| Motif name: |
CagTGCTCCACTGTGgT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
17 |
| Similarity score: |
1.08847 |
Alignment:
CAGTGCTCCACTGTGBT--
TGTGGTGTCACAGTGCTCC
| Original motif Consensus sequence: CAGTGCTCCACTGTGBT |
Reverse complement motif Consensus sequence: ABCACAGTGGAGCACTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
71 |
| Motif name: |
AmTGTGACACCACAGT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
16 |
| Similarity score: |
1.59452 |
Alignment:
ACTGTGGTGTCACART---
GGAGCACTGTGACACCACA
| Original motif Consensus sequence: AMTGTGACACCACAGT |
Reverse complement motif Consensus sequence: ACTGTGGTGTCACART |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
92 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
16 |
| Similarity score: |
1.60014 |
Alignment:
---TGTGTGDKTGTGTGTGTRTG
TGTGGTGTCACAGTGCTCC----
| Original motif Consensus sequence: CAMACACACACAYDCACACA |
Reverse complement motif Consensus sequence: TGTGTGDKTGTGTGTGTRTG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
89 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
16 |
| Similarity score: |
1.60282 |
Alignment:
---TGTGTGWGTGTGTGTGTG
TGTGGTGTCACAGTGCTCC--
| Original motif Consensus sequence: CACACACACACWCACACA |
Reverse complement motif Consensus sequence: TGTGTGWGTGTGTGTGTG |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 68 |
Motif name: sscCCCGCGcs |
| Original motif Consensus sequence: BSCCCCGCGBS |
Reverse complement motif Consensus sequence: SBCGCGGGGSB |
|
|
 |
 |
Best Matches for Motif ID 68 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
74 |
| Motif name: |
ssCCCCGCSssk |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0 |
Alignment:
SBCCCCGCCSBB
BSCCCCGCGBS-
| Original motif Consensus sequence: SBCCCCGCCSBB |
Reverse complement motif Consensus sequence: BBSGGCGGGGBS |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
82 |
| Motif name: |
gCGCGCsCgsG |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0417227 |
Alignment:
CGCGSGCGCGC
SBCGCGGGGSB
| Original motif Consensus sequence: GCGCGCSCGCG |
Reverse complement motif Consensus sequence: CGCGSGCGCGC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
78 |
| Motif name: |
ssGCkTGCssk |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0523123 |
Alignment:
SSGCKTGCSSK
BSCCCCGCGBS
| Original motif Consensus sequence: SSGCKTGCSSK |
Reverse complement motif Consensus sequence: YSSGCAYGCSS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
11 |
| Similarity score: |
0.0564004 |
Alignment:
YDRCCASYAGRKGGCRSYV
--SBCGCGGGGSB------
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
90 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0666605 |
Alignment:
CHHCHBTCCTSCYTCTGC
----BSCCCCGCGBS---
| Original motif Consensus sequence: CHHCHBTCCTSCYTCTGC |
Reverse complement motif Consensus sequence: GCAGAKGSAGGABDGHDG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
11 |
| Similarity score: |
0.0677432 |
Alignment:
RAGKGMAGVRRGSRCASAGV
----SBCGCGGGGSB-----
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0734979 |
Alignment:
MGGTCAGGGTGACCTRDBHV
BSCCCCGCGBS---------
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
79 |
| Motif name: |
cCGCGGrCACG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0757081 |
Alignment:
CCGCGGRCACG
BSCCCCGCGBS
| Original motif Consensus sequence: CCGCGGRCACG |
Reverse complement motif Consensus sequence: CGTGKCCGCGG |
|
|
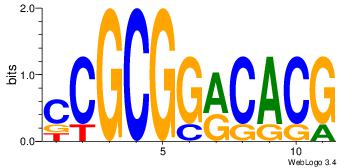 |
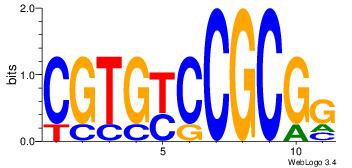 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0758097 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
SBCGCGGGGSB----------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
1 |
| Motif name: |
Motif 1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0788714 |
Alignment:
GCAGAGGSAKG
SBCGCGGGGSB
| Original motif Consensus sequence: CYTSCCTCTGC |
Reverse complement motif Consensus sequence: GCAGAGGSAKG |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 69 |
Motif name: atactttggc |
| Original motif Consensus sequence: ATACTTTGGC |
Reverse complement motif Consensus sequence: GCCAAAGTAT |
|
|
 |
 |
Best Matches for Motif ID 69 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
75 |
| Motif name: |
wwCCAmAGTCmt |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0840909 |
Alignment:
DDCCAMAGTCHB
-GCCAAAGTAT-
| Original motif Consensus sequence: DDCCAMAGTCHB |
Reverse complement motif Consensus sequence: VDGACTRTGGDD |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
80 |
| Motif name: |
taaacgatgcc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.1 |
Alignment:
GGCATCGTTTA
GCCAAAGTAT-
| Original motif Consensus sequence: TAAACGATGCC |
Reverse complement motif Consensus sequence: GGCATCGTTTA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
60 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.102 |
Alignment:
AAAGGTCAAAGGTCAAC
---GCCAAAGTAT----
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.109722 |
Alignment:
GGAGCACTGTGACACCACA
------ATACTTTGGC---
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
10 |
| Similarity score: |
0.109921 |
Alignment:
CACTGTGRTRTCACAGTGSWSCACT
---------GCCAAAGTAT------
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
51 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.110821 |
Alignment:
TGMYCTTTGBCCK
--ATACTTTGGC-
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
52 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.111538 |
Alignment:
TGAMCTTTGMMCYT
-ATACTTTGGC---
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.111824 |
Alignment:
CCACTGTGVTGTCACAGTGCTCCA
----------GCCAAAGTAT----
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
10 |
| Similarity score: |
0.112324 |
Alignment:
ACTGTGACACCACAGTGGAGCACT
--------GCCAAAGTAT------
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
46 |
| Motif name: |
Motif 46 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.1125 |
Alignment:
TATKCTAAACCTAT
---ATACTTTGGC-
| Original motif Consensus sequence: ATAGGTTTAGYATA |
Reverse complement motif Consensus sequence: TATKCTAAACCTAT |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 70 |
Motif name: CagTGCTCCACTGTGgT |
| Original motif Consensus sequence: CAGTGCTCCACTGTGBT |
Reverse complement motif Consensus sequence: ABCACAGTGGAGCACTG |
|
|
 |
 |
Best Matches for Motif ID 70 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
17 |
| Similarity score: |
0.0957051 |
Alignment:
BMSMGCCYMCTKSTGGMHM
--CAGTGCTCCACTGTGBT
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
17 |
| Similarity score: |
0.0975871 |
Alignment:
TTCAGCACCATGGACAGCKCC
-ABCACAGTGGAGCACTG---
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
17 |
| Similarity score: |
0.0980868 |
Alignment:
TGTGGTGTCACAGTGCTCC
CAGTGCTCCACTGTGBT--
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
17 |
| Similarity score: |
0.099784 |
Alignment:
AGTGCTCCACTGTGGTGTCACAGT
-----ABCACAGTGGAGCACTG--
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
17 |
| Similarity score: |
0.104226 |
Alignment:
AGTGSWSCACTGTGAMAMCACAGTG
-----ABCACAGTGGAGCACTG---
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
17 |
| Similarity score: |
0.107548 |
Alignment:
TGGAGCACTGTGACAVCACAGTGG
---ABCACAGTGGAGCACTG----
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
76 |
| Motif name: |
CAcCACAGTGGAGCAct |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
16 |
| Similarity score: |
0.5 |
Alignment:
-AGTGCTCCACTGTGDTG
CAGTGCTCCACTGTGBT-
| Original motif Consensus sequence: CAHCACAGTGGAGCACT |
Reverse complement motif Consensus sequence: AGTGCTCCACTGTGDTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
71 |
| Motif name: |
AmTGTGACACCACAGT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
16 |
| Similarity score: |
0.607943 |
Alignment:
-AMTGTGACACCACAGT
CAGTGCTCCACTGTGBT
| Original motif Consensus sequence: AMTGTGACACCACAGT |
Reverse complement motif Consensus sequence: ACTGTGGTGTCACART |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
Motif 47 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
1.59821 |
Alignment:
---YGCCACCTDSTGGY
CAGTGCTCCACTGTGBT
| Original motif Consensus sequence: YGCCACCTDSTGGY |
Reverse complement motif Consensus sequence: KCCASDAGGTGGCK |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
13 |
| Similarity score: |
2.09999 |
Alignment:
VCTSTGKSCMKBCTRCYCTK----
-------CAGTGCTCCACTGTGBT
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 71 |
Motif name: AmTGTGACACCACAGT |
| Original motif Consensus sequence: AMTGTGACACCACAGT |
Reverse complement motif Consensus sequence: ACTGTGGTGTCACART |
|
|
 |
 |
Best Matches for Motif ID 71 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
16 |
| Similarity score: |
0.00066365 |
Alignment:
ACTGTGACACCACAGTGGAGCACT
AMTGTGACACCACAGT--------
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
16 |
| Similarity score: |
0.00238202 |
Alignment:
CCACTGTGVTGTCACAGTGCTCCA
--ACTGTGGTGTCACART------
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
16 |
| Similarity score: |
0.0203916 |
Alignment:
CACTGTGRTRTCACAGTGSWSCACT
-ACTGTGGTGTCACART--------
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
16 |
| Similarity score: |
0.0957574 |
Alignment:
GGAGCACTGTGACACCACA
ACTGTGGTGTCACART---
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
70 |
| Motif name: |
CagTGCTCCACTGTGgT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
16 |
| Similarity score: |
0.0995605 |
Alignment:
CAGTGCTCCACTGTGBT
-AMTGTGACACCACAGT
| Original motif Consensus sequence: CAGTGCTCCACTGTGBT |
Reverse complement motif Consensus sequence: ABCACAGTGGAGCACTG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
16 |
| Similarity score: |
0.10061 |
Alignment:
RAGKGMAGVRRGSRCASAGV
----AMTGTGACACCACAGT
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
76 |
| Motif name: |
CAcCACAGTGGAGCAct |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
16 |
| Similarity score: |
0.102539 |
Alignment:
AGTGCTCCACTGTGDTG
AMTGTGACACCACAGT-
| Original motif Consensus sequence: CAHCACAGTGGAGCACT |
Reverse complement motif Consensus sequence: AGTGCTCCACTGTGDTG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
15 |
| Similarity score: |
0.595756 |
Alignment:
-MGGTCAGGGTGACCTRDBHV
AMTGTGACACCACAGT-----
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
14 |
| Similarity score: |
1.10257 |
Alignment:
--GGYGCTGTCCATGGTGCTGAA
ACTGTGGTGTCACART-------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
88 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
1.59198 |
Alignment:
HVCDBBDBCAGCAG---
-AMTGTGACACCACAGT
| Original motif Consensus sequence: CTGCTGBDBBDGBD |
Reverse complement motif Consensus sequence: HVCDBBDBCAGCAG |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 72 |
Motif name: CACTGTGrYrtCACAGTGswsCAcT |
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
Best Matches for Motif ID 72 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
24 |
| Similarity score: |
0.0299166 |
Alignment:
-ACTGTGACACCACAGTGGAGCACT
CACTGTGRTRTCACAGTGSWSCACT
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
23 |
| Similarity score: |
0.506166 |
Alignment:
--TGGAGCACTGTGACAVCACAGTGG
AGTGSWSCACTGTGAMAMCACAGTG-
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
19 |
| Similarity score: |
2.59185 |
Alignment:
RAGKGMAGVRRGSRCASAGV------
-AGTGSWSCACTGTGAMAMCACAGTG
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
18 |
| Similarity score: |
3.08766 |
Alignment:
MGGTCAGGGTGACCTRDBHV-------
--CACTGTGRTRTCACAGTGSWSCACT
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
18 |
| Similarity score: |
3.09362 |
Alignment:
BMSMGCCYMCTKSTGGMHM-------
-AGTGSWSCACTGTGAMAMCACAGTG
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
18 |
| Similarity score: |
3.09504 |
Alignment:
GGAGCACTGTGACACCACA-------
-CACTGTGRTRTCACAGTGSWSCACT
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
76 |
| Motif name: |
CAcCACAGTGGAGCAct |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
17 |
| Similarity score: |
3.52969 |
Alignment:
--------CAHCACAGTGGAGCACT
CACTGTGRTRTCACAGTGSWSCACT
| Original motif Consensus sequence: CAHCACAGTGGAGCACT |
Reverse complement motif Consensus sequence: AGTGCTCCACTGTGDTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
70 |
| Motif name: |
CagTGCTCCACTGTGgT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
16 |
| Similarity score: |
4.02785 |
Alignment:
---------CAGTGCTCCACTGTGBT
AGTGSWSCACTGTGAMAMCACAGTG-
| Original motif Consensus sequence: CAGTGCTCCACTGTGBT |
Reverse complement motif Consensus sequence: ABCACAGTGGAGCACTG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
90 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
15 |
| Similarity score: |
4.59058 |
Alignment:
----------CHHCHBTCCTSCYTCTGC
CACTGTGRTRTCACAGTGSWSCACT---
| Original motif Consensus sequence: CHHCHBTCCTSCYTCTGC |
Reverse complement motif Consensus sequence: GCAGAKGSAGGABDGHDG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
59 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
13 |
| Similarity score: |
5.58821 |
Alignment:
VHRGGTCABDBTGMCCTB------------
-----AGTGSWSCACTGTGAMAMCACAGTG
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 73 |
Motif name: TGGAGCACTGTGACAcCACAGTGg |
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
Best Matches for Motif ID 73 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
23 |
| Similarity score: |
0.0136536 |
Alignment:
CACTGTGRTRTCACAGTGSWSCACT-
--CCACTGTGVTGTCACAGTGCTCCA
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
22 |
| Similarity score: |
0.560082 |
Alignment:
--ACTGTGACACCACAGTGGAGCACT
CCACTGTGVTGTCACAGTGCTCCA--
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
59 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
2.5833 |
Alignment:
------BAGGYCABHBTGACCKHV
TGGAGCACTGTGACAVCACAGTGG
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
90 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
2.60363 |
Alignment:
CHHCHBTCCTSCYTCTGC------
CCACTGTGVTGTCACAGTGCTCCA
| Original motif Consensus sequence: CHHCHBTCCTSCYTCTGC |
Reverse complement motif Consensus sequence: GCAGAKGSAGGABDGHDG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
17 |
| Similarity score: |
3.10239 |
Alignment:
MGGTCAGGGTGACCTRDBHV-------
---CCACTGTGVTGTCACAGTGCTCCA
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
17 |
| Similarity score: |
3.10268 |
Alignment:
RAGKGMAGVRRGSRCASAGV-------
---TGGAGCACTGTGACAVCACAGTGG
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
16 |
| Similarity score: |
3.56015 |
Alignment:
GGAGCACTGTGACACCACA--------
---CCACTGTGVTGTCACAGTGCTCCA
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
70 |
| Motif name: |
CagTGCTCCACTGTGgT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
16 |
| Similarity score: |
3.58845 |
Alignment:
ABCACAGTGGAGCACTG--------
-CCACTGTGVTGTCACAGTGCTCCA
| Original motif Consensus sequence: CAGTGCTCCACTGTGBT |
Reverse complement motif Consensus sequence: ABCACAGTGGAGCACTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
76 |
| Motif name: |
CAcCACAGTGGAGCAct |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
15 |
| Similarity score: |
4.07113 |
Alignment:
AGTGCTCCACTGTGDTG---------
--TGGAGCACTGTGACAVCACAGTGG
| Original motif Consensus sequence: CAHCACAGTGGAGCACT |
Reverse complement motif Consensus sequence: AGTGCTCCACTGTGDTG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
15 |
| Similarity score: |
4.10183 |
Alignment:
---------YDRCCASYAGRKGGCRSYV
TGGAGCACTGTGACAVCACAGTGG----
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 74 |
Motif name: ssCCCCGCSssk |
| Original motif Consensus sequence: SBCCCCGCCSBB |
Reverse complement motif Consensus sequence: BBSGGCGGGGBS |
|
|
 |
 |
Best Matches for Motif ID 74 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
90 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0606622 |
Alignment:
CHHCHBTCCTSCYTCTGC
----SBCCCCGCCSBB--
| Original motif Consensus sequence: CHHCHBTCCTSCYTCTGC |
Reverse complement motif Consensus sequence: GCAGAKGSAGGABDGHDG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.063076 |
Alignment:
MGGTCAGGGTGACCTRDBHV
-------BBSGGCGGGGBS-
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0642087 |
Alignment:
YDRCCASYAGRKGGCRSYV
-BBSGGCGGGGBS------
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
64 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0671548 |
Alignment:
TGRCCTKTGHCCKAB
-SBCCCCGCCSBB--
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
12 |
| Similarity score: |
0.0674545 |
Alignment:
RAGKGMAGVRRGSRCASAGV
---BBSGGCGGGGBS-----
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
59 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0734481 |
Alignment:
BAGGYCABHBTGACCKHV
-SBCCCCGCCSBB-----
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
10 |
| Number of overlap: |
12 |
| Similarity score: |
0.0751084 |
Alignment:
TTCAGCACCATGGACAGCKCC
BBSGGCGGGGBS---------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
Motif 47 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.078885 |
Alignment:
YGCCACCTDSTGGY
SBCCCCGCCSBB--
| Original motif Consensus sequence: YGCCACCTDSTGGY |
Reverse complement motif Consensus sequence: KCCASDAGGTGGCK |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
68 |
| Motif name: |
sscCCCGCGcs |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.5 |
Alignment:
BSCCCCGCGBS-
SBCCCCGCCSBB
| Original motif Consensus sequence: BSCCCCGCGBS |
Reverse complement motif Consensus sequence: SBCGCGGGGSB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
82 |
| Motif name: |
gCGCGCsCgsG |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.548902 |
Alignment:
-CGCGSGCGCGC
BBSGGCGGGGBS
| Original motif Consensus sequence: GCGCGCSCGCG |
Reverse complement motif Consensus sequence: CGCGSGCGCGC |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 75 |
Motif name: wwCCAmAGTCmt |
| Original motif Consensus sequence: DDCCAMAGTCHB |
Reverse complement motif Consensus sequence: VDGACTRTGGDD |
|
|
 |
 |
Best Matches for Motif ID 75 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
12 |
| Similarity score: |
0.0656866 |
Alignment:
CACTGTGRTRTCACAGTGSWSCACT
--------DDCCAMAGTCHB-----
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
70 |
| Motif name: |
CagTGCTCCACTGTGgT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
12 |
| Similarity score: |
0.0676046 |
Alignment:
ABCACAGTGGAGCACTG
VDGACTRTGGDD-----
| Original motif Consensus sequence: CAGTGCTCCACTGTGBT |
Reverse complement motif Consensus sequence: ABCACAGTGGAGCACTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
76 |
| Motif name: |
CAcCACAGTGGAGCAct |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
12 |
| Similarity score: |
0.0684312 |
Alignment:
AGTGCTCCACTGTGDTG
-----VDGACTRTGGDD
| Original motif Consensus sequence: CAHCACAGTGGAGCACT |
Reverse complement motif Consensus sequence: AGTGCTCCACTGTGDTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
12 |
| Similarity score: |
0.0695137 |
Alignment:
AGTGCTCCACTGTGGTGTCACAGT
-----VDGACTRTGGDD-------
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
51 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0717637 |
Alignment:
RGGBCAAAGKYCA
-DDCCAMAGTCHB
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
92 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
12 |
| Similarity score: |
0.0751124 |
Alignment:
CAMACACACACAYDCACACA
-DDCCAMAGTCHB-------
| Original motif Consensus sequence: CAMACACACACAYDCACACA |
Reverse complement motif Consensus sequence: TGTGTGDKTGTGTGTGTRTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
12 |
| Similarity score: |
0.075392 |
Alignment:
CCACTGTGVTGTCACAGTGCTCCA
---DDCCAMAGTCHB---------
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
12 |
| Similarity score: |
0.0784446 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
---DDCCAMAGTCHB------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0786999 |
Alignment:
TGTGGTGTCACAGTGCTCC
-----DDCCAMAGTCHB--
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0800522 |
Alignment:
VDBHMAGGTCACCCTGACCY
DDCCAMAGTCHB--------
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 76 |
Motif name: CAcCACAGTGGAGCAct |
| Original motif Consensus sequence: CAHCACAGTGGAGCACT |
Reverse complement motif Consensus sequence: AGTGCTCCACTGTGDTG |
|
|
 |
 |
Best Matches for Motif ID 76 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
17 |
| Similarity score: |
0.00742502 |
Alignment:
ACTGTGACACCACAGTGGAGCACT
-------CAHCACAGTGGAGCACT
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
17 |
| Similarity score: |
0.0456803 |
Alignment:
CACTGTGRTRTCACAGTGSWSCACT
--------CAHCACAGTGGAGCACT
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
17 |
| Similarity score: |
0.0894884 |
Alignment:
TTCAGCACCATGGACAGCKCC
--CAHCACAGTGGAGCACT--
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
17 |
| Similarity score: |
0.0962536 |
Alignment:
GGAGCACTGTGACACCACA
-CAHCACAGTGGAGCACT-
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
17 |
| Similarity score: |
0.0978126 |
Alignment:
YDRCCASYAGRKGGCRSYV
-CAHCACAGTGGAGCACT-
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
17 |
| Similarity score: |
0.106708 |
Alignment:
CCACTGTGVTGTCACAGTGCTCCA
-----AGTGCTCCACTGTGDTG--
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
70 |
| Motif name: |
CagTGCTCCACTGTGgT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
16 |
| Similarity score: |
0.5 |
Alignment:
-CAGTGCTCCACTGTGBT
AGTGCTCCACTGTGDTG-
| Original motif Consensus sequence: CAGTGCTCCACTGTGBT |
Reverse complement motif Consensus sequence: ABCACAGTGGAGCACTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
71 |
| Motif name: |
AmTGTGACACCACAGT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
16 |
| Similarity score: |
0.610921 |
Alignment:
ACTGTGGTGTCACART-
CAHCACAGTGGAGCACT
| Original motif Consensus sequence: AMTGTGACACCACAGT |
Reverse complement motif Consensus sequence: ACTGTGGTGTCACART |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
75 |
| Motif name: |
wwCCAmAGTCmt |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
2.5839 |
Alignment:
-----VDGACTRTGGDD
AGTGCTCCACTGTGDTG
| Original motif Consensus sequence: DDCCAMAGTCHB |
Reverse complement motif Consensus sequence: VDGACTRTGGDD |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
12 |
| Similarity score: |
2.60274 |
Alignment:
VCTSTGKSCMKBCTRCYCTK-----
--------AGTGCTCCACTGTGDTG
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 77 |
Motif name: AgTGCTCCACTGTGgTGTCACAgT |
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
Best Matches for Motif ID 77 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
24 |
| Similarity score: |
0.0409586 |
Alignment:
AGTGSWSCACTGTGAMAMCACAGTG
-AGTGCTCCACTGTGGTGTCACAGT
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
22 |
| Similarity score: |
1.06364 |
Alignment:
CCACTGTGVTGTCACAGTGCTCCA--
--ACTGTGACACCACAGTGGAGCACT
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
19 |
| Similarity score: |
2.59599 |
Alignment:
TTCAGCACCATGGACAGCKCC-----
--ACTGTGACACCACAGTGGAGCACT
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
18 |
| Similarity score: |
3.10123 |
Alignment:
------GGAGCACTGTGACACCACA
ACTGTGACACCACAGTGGAGCACT-
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
18 |
| Similarity score: |
3.10186 |
Alignment:
------BMSMGCCYMCTKSTGGMHM
AGTGCTCCACTGTGGTGTCACAGT-
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
76 |
| Motif name: |
CAcCACAGTGGAGCAct |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
17 |
| Similarity score: |
3.50248 |
Alignment:
-------AGTGCTCCACTGTGDTG
AGTGCTCCACTGTGGTGTCACAGT
| Original motif Consensus sequence: CAHCACAGTGGAGCACT |
Reverse complement motif Consensus sequence: AGTGCTCCACTGTGDTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
70 |
| Motif name: |
CagTGCTCCACTGTGgT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
16 |
| Similarity score: |
4 |
Alignment:
CAGTGCTCCACTGTGBT--------
-AGTGCTCCACTGTGGTGTCACAGT
| Original motif Consensus sequence: CAGTGCTCCACTGTGBT |
Reverse complement motif Consensus sequence: ABCACAGTGGAGCACTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
71 |
| Motif name: |
AmTGTGACACCACAGT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
16 |
| Similarity score: |
4.0041 |
Alignment:
--------AMTGTGACACCACAGT
ACTGTGACACCACAGTGGAGCACT
| Original motif Consensus sequence: AMTGTGACACCACAGT |
Reverse complement motif Consensus sequence: ACTGTGGTGTCACART |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
16 |
| Similarity score: |
4.10485 |
Alignment:
RAGKGMAGVRRGSRCASAGV--------
----ACTGTGACACCACAGTGGAGCACT
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
15 |
| Similarity score: |
4.59996 |
Alignment:
MGGTCAGGGTGACCTRDBHV---------
-----ACTGTGACACCACAGTGGAGCACT
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 78 |
Motif name: ssGCkTGCssk |
| Original motif Consensus sequence: SSGCKTGCSSK |
Reverse complement motif Consensus sequence: YSSGCAYGCSS |
|
|
 |
 |
Best Matches for Motif ID 78 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
82 |
| Motif name: |
gCGCGCsCgsG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0230178 |
Alignment:
GCGCGCSCGCG
SSGCKTGCSSK
| Original motif Consensus sequence: GCGCGCSCGCG |
Reverse complement motif Consensus sequence: CGCGSGCGCGC |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
74 |
| Motif name: |
ssCCCCGCSssk |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.036087 |
Alignment:
BBSGGCGGGGBS
-YSSGCAYGCSS
| Original motif Consensus sequence: SBCCCCGCCSBB |
Reverse complement motif Consensus sequence: BBSGGCGGGGBS |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
68 |
| Motif name: |
sscCCCGCGcs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0375477 |
Alignment:
SBCGCGGGGSB
YSSGCAYGCSS
| Original motif Consensus sequence: BSCCCCGCGBS |
Reverse complement motif Consensus sequence: SBCGCGGGGSB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
79 |
| Motif name: |
cCGCGGrCACG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0461039 |
Alignment:
CCGCGGRCACG
SSGCKTGCSSK
| Original motif Consensus sequence: CCGCGGRCACG |
Reverse complement motif Consensus sequence: CGTGKCCGCGG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
21 |
| Motif name: |
Motif 21 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0529221 |
Alignment:
CTGCTTTCCMAA
SSGCKTGCSSK-
| Original motif Consensus sequence: CTGCTTTCCMAA |
Reverse complement motif Consensus sequence: TTYGGAAAGCAG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
11 |
| Similarity score: |
0.0533963 |
Alignment:
YDRCCASYAGRKGGCRSYV
-------SSGCKTGCSSK-
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
54 |
| Motif name: |
Esrrb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0543224 |
Alignment:
VBBYCAAGGTCA
-YSSGCAYGCSS
| Original motif Consensus sequence: VBBYCAAGGTCA |
Reverse complement motif Consensus sequence: TGACCTTGMBBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
59 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0563598 |
Alignment:
VHRGGTCABDBTGMCCTB
-----YSSGCAYGCSS--
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
10 |
| Number of overlap: |
11 |
| Similarity score: |
0.0593846 |
Alignment:
TTCAGCACCATGGACAGCKCC
-YSSGCAYGCSS---------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
64 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0598714 |
Alignment:
BTRGGDCARAGGKCA
--YSSGCAYGCSS--
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 79 |
Motif name: cCGCGGrCACG |
| Original motif Consensus sequence: CCGCGGRCACG |
Reverse complement motif Consensus sequence: CGTGKCCGCGG |
|
|
 |
 |
Best Matches for Motif ID 79 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
82 |
| Motif name: |
gCGCGCsCgsG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.010473 |
Alignment:
GCGCGCSCGCG
CCGCGGRCACG
| Original motif Consensus sequence: GCGCGCSCGCG |
Reverse complement motif Consensus sequence: CGCGSGCGCGC |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
88 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0478408 |
Alignment:
HVCDBBDBCAGCAG
---CGTGKCCGCGG
| Original motif Consensus sequence: CTGCTGBDBBDGBD |
Reverse complement motif Consensus sequence: HVCDBBDBCAGCAG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0484441 |
Alignment:
TTCAGCACCATGGACAGCKCC
-------CCGCGGRCACG---
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0486581 |
Alignment:
MGGTCAGGGTGACCTRDBHV
-------CGTGKCCGCGG--
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
78 |
| Motif name: |
ssGCkTGCssk |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0503056 |
Alignment:
SSGCKTGCSSK
CCGCGGRCACG
| Original motif Consensus sequence: SSGCKTGCSSK |
Reverse complement motif Consensus sequence: YSSGCAYGCSS |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
74 |
| Motif name: |
ssCCCCGCSssk |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.055777 |
Alignment:
BBSGGCGGGGBS
CGTGKCCGCGG-
| Original motif Consensus sequence: SBCCCCGCCSBB |
Reverse complement motif Consensus sequence: BBSGGCGGGGBS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
11 |
| Similarity score: |
0.0588663 |
Alignment:
YDRCCASYAGRKGGCRSYV
-------CCGCGGRCACG-
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0597806 |
Alignment:
HCAGRRVACASAG
-CCGCGGRCACG-
| Original motif Consensus sequence: HCAGRRVACASAG |
Reverse complement motif Consensus sequence: CTSTGTBKMCTGH |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0624268 |
Alignment:
GTGTGTGTGTGTGT
-CGTGKCCGCGG--
| Original motif Consensus sequence: ACACACACACACAC |
Reverse complement motif Consensus sequence: GTGTGTGTGTGTGT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
0.0625015 |
Alignment:
RAGKGMAGVRRGSRCASAGV
-----CCGCGGRCACG----
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 80 |
Motif name: taaacgatgcc |
| Original motif Consensus sequence: TAAACGATGCC |
Reverse complement motif Consensus sequence: GGCATCGTTTA |
|
|
 |
 |
Best Matches for Motif ID 80 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
46 |
| Motif name: |
Motif 46 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
TATKCTAAACCTAT
---TAAACGATGCC
| Original motif Consensus sequence: ATAGGTTTAGYATA |
Reverse complement motif Consensus sequence: TATKCTAAACCTAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
TGAAAATATGTT
TAAACGATGCC-
| Original motif Consensus sequence: AACATATTTTCA |
Reverse complement motif Consensus sequence: TGAAAATATGTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
23 |
| Motif name: |
Motif 23 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
ATGTWTTCATTMAT
-GGCATCGTTTA--
| Original motif Consensus sequence: ATGTWTTCATTMAT |
Reverse complement motif Consensus sequence: ATYAATGAAWACAT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
60 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.113636 |
Alignment:
GTTGACCTTTGACCTTT
----TAAACGATGCC--
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
52 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.118881 |
Alignment:
TGAMCTTTGMMCYT
TAAACGATGCC---
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.125 |
Alignment:
TTYAAACATYTT
TAAACGATGCC-
| Original motif Consensus sequence: AAMATGTTTMAA |
Reverse complement motif Consensus sequence: TTYAAACATYTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
45 |
| Motif name: |
Motif 45 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.125 |
Alignment:
TTAAAAWAGASTTA
--TAAACGATGCC-
| Original motif Consensus sequence: TAASTCTWTTTTAA |
Reverse complement motif Consensus sequence: TTAAAAWAGASTTA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.5875 |
Alignment:
AAACAATYTTT-
-TAAACGATGCC
| Original motif Consensus sequence: AAAMATTGTTT |
Reverse complement motif Consensus sequence: AAACAATYTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
30 |
| Motif name: |
Motif 30 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.6 |
Alignment:
AAAAGCAGCAA-
-TAAACGATGCC
| Original motif Consensus sequence: TTGCTGCTTTT |
Reverse complement motif Consensus sequence: AAAAGCAGCAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
69 |
| Motif name: |
atactttggc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.6 |
Alignment:
-ATACTTTGGC
TAAACGATGCC
| Original motif Consensus sequence: ATACTTTGGC |
Reverse complement motif Consensus sequence: GCCAAAGTAT |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 81 |
Motif name: ctatacggacg |
| Original motif Consensus sequence: CTATACGGACG |
Reverse complement motif Consensus sequence: CGTCCGTATAG |
|
|
 |
 |
Best Matches for Motif ID 81 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
27 |
| Motif name: |
Motif 27 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.102273 |
Alignment:
CTTKACAGAAT
CTATACGGACG
| Original motif Consensus sequence: ATTCTGTRAAG |
Reverse complement motif Consensus sequence: CTTKACAGAAT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
83 |
| Motif name: |
srACyCCGAyr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.125 |
Alignment:
SRACYCCGAYR
CTATACGGACG
| Original motif Consensus sequence: SRACYCCGAYR |
Reverse complement motif Consensus sequence: KKTCGGKGTKS |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
46 |
| Motif name: |
Motif 46 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.6 |
Alignment:
-TATKCTAAACCTAT
CTATACGGACG----
| Original motif Consensus sequence: ATAGGTTTAGYATA |
Reverse complement motif Consensus sequence: TATKCTAAACCTAT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
89 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
10 |
| Similarity score: |
0.624107 |
Alignment:
TGTGTGWGTGTGTGTGTG-
--------CGTCCGTATAG
| Original motif Consensus sequence: CACACACACACWCACACA |
Reverse complement motif Consensus sequence: TGTGTGWGTGTGTGTGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
80 |
| Motif name: |
taaacgatgcc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.625 |
Alignment:
GGCATCGTTTA-
-CGTCCGTATAG
| Original motif Consensus sequence: TAAACGATGCC |
Reverse complement motif Consensus sequence: GGCATCGTTTA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
20 |
| Motif name: |
Motif 20 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.625 |
Alignment:
TTCCAGGAGGA-
-CTATACGGACG
| Original motif Consensus sequence: TCCTCCTGGAA |
Reverse complement motif Consensus sequence: TTCCAGGAGGA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
22 |
| Motif name: |
Motif 22 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.625 |
Alignment:
ATGCTCACCATAAA-
----CGTCCGTATAG
| Original motif Consensus sequence: TTTATGGTGAGCAT |
Reverse complement motif Consensus sequence: ATGCTCACCATAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
28 |
| Motif name: |
Motif 28 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.625 |
Alignment:
-ATTTAGTAAA
CGTCCGTATAG
| Original motif Consensus sequence: ATTTAGTAAA |
Reverse complement motif Consensus sequence: TTTACTAAAT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
21 |
| Motif name: |
Motif 21 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
9 |
| Similarity score: |
1.09722 |
Alignment:
--TTYGGAAAGCAG
CGTCCGTATAG---
| Original motif Consensus sequence: CTGCTTTCCMAA |
Reverse complement motif Consensus sequence: TTYGGAAAGCAG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
63 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
1.10694 |
Alignment:
GCGGACGTTV--
-CGTCCGTATAG
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 82 |
Motif name: gCGCGCsCgsG |
| Original motif Consensus sequence: GCGCGCSCGCG |
Reverse complement motif Consensus sequence: CGCGSGCGCGC |
|
|
 |
 |
Best Matches for Motif ID 82 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
79 |
| Motif name: |
cCGCGGrCACG |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0131931 |
Alignment:
CCGCGGRCACG
GCGCGCSCGCG
| Original motif Consensus sequence: CCGCGGRCACG |
Reverse complement motif Consensus sequence: CGTGKCCGCGG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
78 |
| Motif name: |
ssGCkTGCssk |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0299395 |
Alignment:
SSGCKTGCSSK
GCGCGCSCGCG
| Original motif Consensus sequence: SSGCKTGCSSK |
Reverse complement motif Consensus sequence: YSSGCAYGCSS |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
68 |
| Motif name: |
sscCCCGCGcs |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0338798 |
Alignment:
SBCGCGGGGSB
CGCGSGCGCGC
| Original motif Consensus sequence: BSCCCCGCGBS |
Reverse complement motif Consensus sequence: SBCGCGGGGSB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
74 |
| Motif name: |
ssCCCCGCSssk |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0410595 |
Alignment:
BBSGGCGGGGBS
-CGCGSGCGCGC
| Original motif Consensus sequence: SBCCCCGCCSBB |
Reverse complement motif Consensus sequence: BBSGGCGGGGBS |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
11 |
| Similarity score: |
0.0537298 |
Alignment:
BMSMGCCYMCTKSTGGMHM
------GCGCGCSCGCG--
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
11 |
| Similarity score: |
0.0651602 |
Alignment:
MGGTCAGGGTGACCTRDBHV
---GCGCGCSCGCG------
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
88 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0669956 |
Alignment:
HVCDBBDBCAGCAG
---CGCGSGCGCGC
| Original motif Consensus sequence: CTGCTGBDBBDGBD |
Reverse complement motif Consensus sequence: HVCDBBDBCAGCAG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
63 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.55601 |
Alignment:
BAACGTCCGC-
GCGCGCSCGCG
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
Motif 47 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.557768 |
Alignment:
-YGCCACCTDSTGGY
GCGCGCSCGCG----
| Original motif Consensus sequence: YGCCACCTDSTGGY |
Reverse complement motif Consensus sequence: KCCASDAGGTGGCK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
58 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.561823 |
Alignment:
DCCACGTGCV-
GCGCGCSCGCG
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: 4 |
Motif ID: 83 |
Motif name: srACyCCGAyr |
| Original motif Consensus sequence: SRACYCCGAYR |
Reverse complement motif Consensus sequence: KKTCGGKGTKS |
|
|
 |
 |
Best Matches for Motif ID 83 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
92 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
11 |
| Similarity score: |
0.0337509 |
Alignment:
TGTGTGDKTGTGTGTGTRTG
--KKTCGGKGTKS-------
| Original motif Consensus sequence: CAMACACACACAYDCACACA |
Reverse complement motif Consensus sequence: TGTGTGDKTGTGTGTGTRTG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0342809 |
Alignment:
MGGTCAGGGTGACCTRDBHV
-KKTCGGKGTKS--------
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
89 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
11 |
| Similarity score: |
0.0348124 |
Alignment:
TGTGTGWGTGTGTGTGTG
------KKTCGGKGTKS-
| Original motif Consensus sequence: CACACACACACWCACACA |
Reverse complement motif Consensus sequence: TGTGTGWGTGTGTGTGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
0.0372475 |
Alignment:
CCACTGTGVTGTCACAGTGCTCCA
---------KKTCGGKGTKS----
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
59 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0379796 |
Alignment:
BAGGYCABHBTGACCKHV
---SRACYCCGAYR----
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
11 |
| Similarity score: |
0.0381944 |
Alignment:
GGAGCACTGTGACACCACA
-----SRACYCCGAYR---
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.039412 |
Alignment:
TGTGTSWGTBTSTG
--KKTCGGKGTKS-
| Original motif Consensus sequence: CASABACWSACACA |
Reverse complement motif Consensus sequence: TGTGTSWGTBTSTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
11 |
| Similarity score: |
0.0397727 |
Alignment:
AGTGSWSCACTGTGAMAMCACAGTG
--------SRACYCCGAYR------
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
0.0402801 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
------KKTCGGKGTKS----
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0438762 |
Alignment:
GTGTGTGTGTGTGT
-KKTCGGKGTKS--
| Original motif Consensus sequence: ACACACACACACAC |
Reverse complement motif Consensus sequence: GTGTGTGTGTGTGT |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 84 |
Motif name: TFW3 |
| Original motif Consensus sequence: GCACTG |
Reverse complement motif Consensus sequence: CAGTGC |
|
|
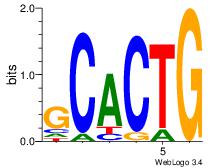 |
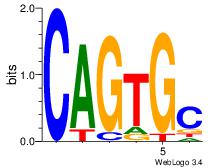 |
Best Matches for Motif ID 84 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
11 |
| Number of overlap: |
6 |
| Similarity score: |
0 |
Alignment:
GGAGCACTGTGACACCACA
----------GCACTG---
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
6 |
| Similarity score: |
0.000794289 |
Alignment:
AGTGSWSCACTGTGAMAMCACAGTG
-------------GCACTG------
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.00482436 |
Alignment:
CCACTGTGVTGTCACAGTGCTCCA
--------------CAGTGC----
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
76 |
| Motif name: |
CAcCACAGTGGAGCAct |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
6 |
| Similarity score: |
0.0201914 |
Alignment:
CAHCACAGTGGAGCACT
------CAGTGC-----
| Original motif Consensus sequence: CAHCACAGTGGAGCACT |
Reverse complement motif Consensus sequence: AGTGCTCCACTGTGDTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
70 |
| Motif name: |
CagTGCTCCACTGTGgT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
6 |
| Similarity score: |
0.0205497 |
Alignment:
ABCACAGTGGAGCACTG
----CAGTGC-------
| Original motif Consensus sequence: CAGTGCTCCACTGTGBT |
Reverse complement motif Consensus sequence: ABCACAGTGGAGCACTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
13 |
| Number of overlap: |
6 |
| Similarity score: |
0.0290883 |
Alignment:
AGTGCTCCACTGTGGTGTCACAGT
------GCACTG------------
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
15 |
| Number of overlap: |
6 |
| Similarity score: |
0.039883 |
Alignment:
TTCAGCACCATGGACAGCKCC
-CAGTGC--------------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
65 |
| Motif name: |
Tcfcp2l1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
6 |
| Similarity score: |
0.0447224 |
Alignment:
CCAGYYHVADCCRG
-------CAGTGC-
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
5 |
| Motif name: |
Motif 5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
6 |
| Similarity score: |
0.0459818 |
Alignment:
GAKAGKTGGCTCWG
GCACTG--------
| Original motif Consensus sequence: CWGAGCCAYCTYTC |
Reverse complement motif Consensus sequence: GAKAGKTGGCTCWG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
75 |
| Motif name: |
wwCCAmAGTCmt |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
6 |
| Similarity score: |
0.0587933 |
Alignment:
VDGACTRTGGDD
-----GCACTG-
| Original motif Consensus sequence: DDCCAMAGTCHB |
Reverse complement motif Consensus sequence: VDGACTRTGGDD |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 85 |
Motif name: TFW1 |
| Original motif Consensus sequence: TTGCGCAA |
Reverse complement motif Consensus sequence: TTGCGCAA |
|
|
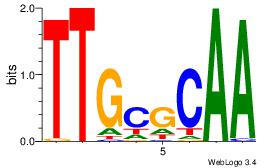 |
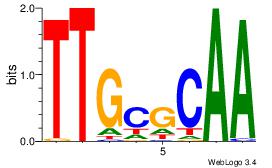 |
Best Matches for Motif ID 85 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
21 |
| Motif name: |
Motif 21 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0195312 |
Alignment:
TTYGGAAAGCAG
----TTGCGCAA
| Original motif Consensus sequence: CTGCTTTCCMAA |
Reverse complement motif Consensus sequence: TTYGGAAAGCAG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
52 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0276442 |
Alignment:
AKGYYCAAAGRTCA
TTGCGCAA------
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
64 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.0277567 |
Alignment:
BTRGGDCARAGGKCA
------TTGCGCAA-
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
28 |
| Motif name: |
Motif 28 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0328125 |
Alignment:
TTTACTAAAT
-TTGCGCAA-
| Original motif Consensus sequence: ATTTAGTAAA |
Reverse complement motif Consensus sequence: TTTACTAAAT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
76 |
| Motif name: |
CAcCACAGTGGAGCAct |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0336914 |
Alignment:
CAHCACAGTGGAGCACT
-TTGCGCAA--------
| Original motif Consensus sequence: CAHCACAGTGGAGCACT |
Reverse complement motif Consensus sequence: AGTGCTCCACTGTGDTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
17 |
| Number of overlap: |
8 |
| Similarity score: |
0.0338471 |
Alignment:
TGGAGCACTGTGACAVCACAGTGG
TTGCGCAA----------------
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
77 |
| Motif name: |
AgTGCTCCACTGTGgTGTCACAgT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
16 |
| Number of overlap: |
8 |
| Similarity score: |
0.034375 |
Alignment:
AGTGCTCCACTGTGGTGTCACAGT
---------------TTGCGCAA-
| Original motif Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT |
Reverse complement motif Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
70 |
| Motif name: |
CagTGCTCCACTGTGgT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0354167 |
Alignment:
CAGTGCTCCACTGTGBT
-------TTGCGCAA--
| Original motif Consensus sequence: CAGTGCTCCACTGTGBT |
Reverse complement motif Consensus sequence: ABCACAGTGGAGCACTG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
91 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0359375 |
Alignment:
KTTTTTTKTTTTTTDAAB
---------TTGCGCAA-
| Original motif Consensus sequence: KTTTTTTKTTTTTTDAAB |
Reverse complement motif Consensus sequence: BTTHAAAAAAYAAAAAAR |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0407242 |
Alignment:
CACTGTGRTRTCACAGTGSWSCACT
---------------TTGCGCAA--
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 86 |
Motif name: TFW2 |
| Original motif Consensus sequence: YGGACTTACG |
Reverse complement motif Consensus sequence: CGTAAGTCCK |
|
|
 |
 |
Best Matches for Motif ID 86 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
75 |
| Motif name: |
wwCCAmAGTCmt |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0441919 |
Alignment:
VDGACTRTGGDD
--YGGACTTACG
| Original motif Consensus sequence: DDCCAMAGTCHB |
Reverse complement motif Consensus sequence: VDGACTRTGGDD |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
54 |
| Motif name: |
Esrrb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0583503 |
Alignment:
VBBYCAAGGTCA
CGTAAGTCCK--
| Original motif Consensus sequence: VBBYCAAGGTCA |
Reverse complement motif Consensus sequence: TGACCTTGMBBB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
51 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.063039 |
Alignment:
TGMYCTTTGBCCK
---YGGACTTACG
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0662938 |
Alignment:
TTCAGCACCATGGACAGCKCC
----------YGGACTTACG-
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
52 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0672009 |
Alignment:
TGAMCTTTGMMCYT
YGGACTTACG----
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0684067 |
Alignment:
VDBHMAGGTCACCCTGACCY
--------CGTAAGTCCK--
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
64 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0692256 |
Alignment:
TGRCCTKTGHCCKAB
---CGTAAGTCCK--
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0694444 |
Alignment:
AAGSATTTTCA
YGGACTTACG-
| Original motif Consensus sequence: TGAAAATSCTT |
Reverse complement motif Consensus sequence: AAGSATTTTCA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
87 |
| Motif name: |
TFF1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0707746 |
Alignment:
VVTCTTTGTCCT
--CGTAAGTCCK
| Original motif Consensus sequence: AGGACAAAGAVV |
Reverse complement motif Consensus sequence: VVTCTTTGTCCT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
60 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
10 |
| Similarity score: |
0.0713889 |
Alignment:
GTTGACCTTTGACCTTT
------YGGACTTACG-
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 87 |
Motif name: TFF1 |
| Original motif Consensus sequence: AGGACAAAGAVV |
Reverse complement motif Consensus sequence: VVTCTTTGTCCT |
|
|
 |
 |
Best Matches for Motif ID 87 (Highest to Lowest)
| Dataset #: |
3 |
| Motif ID: |
51 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0111472 |
Alignment:
TGMYCTTTGBCCK
-VVTCTTTGTCCT
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
64 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0210867 |
Alignment:
BTRGGDCARAGGKCA
--AGGACAAAGAVV-
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
59 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
12 |
| Similarity score: |
0.0351633 |
Alignment:
BAGGYCABHBTGACCKHV
----VVTCTTTGTCCT--
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
12 |
| Similarity score: |
0.039773 |
Alignment:
VDBHMAGGTCACCCTGACCY
VVTCTTTGTCCT--------
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
12 |
| Similarity score: |
0.0408272 |
Alignment:
RAGKGMAGVRRGSRCASAGV
-AGGACAAAGAVV-------
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
52 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0472374 |
Alignment:
AKGYYCAAAGRTCA
-AGGACAAAGAVV-
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
60 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
12 |
| Similarity score: |
0.0472884 |
Alignment:
GTTGACCTTTGACCTTT
---VVTCTTTGTCCT--
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
12 |
| Similarity score: |
0.0530556 |
Alignment:
CACTGTGRTRTCACAGTGSWSCACT
-------AGGACAAAGAVV------
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
13 |
| Number of overlap: |
12 |
| Similarity score: |
0.0539204 |
Alignment:
TGGAGCACTGTGACAVCACAGTGG
------------AGGACAAAGAVV
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
12 |
| Similarity score: |
0.0558379 |
Alignment:
TTCAGCACCATGGACAGCKCC
---AGGACAAAGAVV------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 88 |
Motif name: TFF11 |
| Original motif Consensus sequence: CTGCTGBDBBDGBD |
Reverse complement motif Consensus sequence: HVCDBBDBCAGCAG |
|
|
 |
 |
Best Matches for Motif ID 88 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
90 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.03576 |
Alignment:
GCAGAKGSAGGABDGHDG
-CTGCTGBDBBDGBD---
| Original motif Consensus sequence: CHHCHBTCCTSCYTCTGC |
Reverse complement motif Consensus sequence: GCAGAKGSAGGABDGHDG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.0381494 |
Alignment:
RAGKGMAGVRRGSRCASAGV
---HVCDBBDBCAGCAG---
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
14 |
| Similarity score: |
0.0491354 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
------CTGCTGBDBBDGBD-
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0521061 |
Alignment:
VDBHMAGGTCACCCTGACCY
-----HVCDBBDBCAGCAG-
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
59 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0565525 |
Alignment:
VHRGGTCABDBTGMCCTB
-CTGCTGBDBBDGBD---
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
64 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0584136 |
Alignment:
BTRGGDCARAGGKCA
-HVCDBBDBCAGCAG
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
14 |
| Similarity score: |
0.0586124 |
Alignment:
GGAGCACTGTGACACCACA
----HVCDBBDBCAGCAG-
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
14 |
| Similarity score: |
0.060085 |
Alignment:
CCACTGTGVTGTCACAGTGCTCCA
-----CTGCTGBDBBDGBD-----
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
65 |
| Motif name: |
Tcfcp2l1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0669051 |
Alignment:
CKGGDTBDMMCTGG
CTGCTGBDBBDGBD
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0670544 |
Alignment:
BMSMGCCYMCTKSTGGMHM
-HVCDBBDBCAGCAG----
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 89 |
Motif name: TFM1 |
| Original motif Consensus sequence: CACACACACACWCACACA |
Reverse complement motif Consensus sequence: TGTGTGWGTGTGTGTGTG |
|
|
 |
 |
Best Matches for Motif ID 89 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
92 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
18 |
| Similarity score: |
0 |
Alignment:
CAMACACACACAYDCACACA
--CACACACACACWCACACA
| Original motif Consensus sequence: CAMACACACACAYDCACACA |
Reverse complement motif Consensus sequence: TGTGTGDKTGTGTGTGTRTG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
91 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
0.099254 |
Alignment:
KTTTTTTKTTTTTTDAAB
TGTGTGWGTGTGTGTGTG
| Original motif Consensus sequence: KTTTTTTKTTTTTTDAAB |
Reverse complement motif Consensus sequence: BTTHAAAAAAYAAAAAAR |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
17 |
| Similarity score: |
0.598911 |
Alignment:
VDBHMAGGTCACCCTGACCY-
---CACACACACACWCACACA
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
16 |
| Similarity score: |
1.09969 |
Alignment:
GGAGCACTGTGACACCACA--
---CACACACACACWCACACA
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
94 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
15 |
| Similarity score: |
1.59953 |
Alignment:
---TTTTTTTKTTKTTTAATTHW
TGTGTGWGTGTGTGTGTG-----
| Original motif Consensus sequence: TTTTTTTKTTKTTTAATTHW |
Reverse complement motif Consensus sequence: WHAATTAAARAARAAAAAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.04528 |
Alignment:
----CASABACWSACACA
CACACACACACWCACACA
| Original motif Consensus sequence: CASABACWSACACA |
Reverse complement motif Consensus sequence: TGTGTSWGTBTSTG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.09655 |
Alignment:
HAAAAHAAARMAAA----
CACACACACACWCACACA
| Original motif Consensus sequence: HAAAAHAAARMAAA |
Reverse complement motif Consensus sequence: TTTRKTTTHTTTTH |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
2.51954 |
Alignment:
ACACACACACACAC-----
-CACACACACACWCACACA
| Original motif Consensus sequence: ACACACACACACAC |
Reverse complement motif Consensus sequence: GTGTGTGTGTGTGT |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
59 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
13 |
| Similarity score: |
2.59375 |
Alignment:
-----BAGGYCABHBTGACCKHV
CACACACACACWCACACA-----
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
13 |
| Similarity score: |
2.59669 |
Alignment:
TTCAGCACCATGGACAGCKCC-----
--------CACACACACACWCACACA
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 90 |
Motif name: TFM2 |
| Original motif Consensus sequence: CHHCHBTCCTSCYTCTGC |
Reverse complement motif Consensus sequence: GCAGAKGSAGGABDGHDG |
|
|
 |
 |
Best Matches for Motif ID 90 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
18 |
| Similarity score: |
0.0675327 |
Alignment:
RAGKGMAGVRRGSRCASAGV
--GCAGAKGSAGGABDGHDG
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
18 |
| Similarity score: |
0.0741766 |
Alignment:
VDBHMAGGTCACCCTGACCY
-CHHCHBTCCTSCYTCTGC-
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
0.0893246 |
Alignment:
YDRCCASYAGRKGGCRSYV
-GCAGAKGSAGGABDGHDG
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
0.0901636 |
Alignment:
TGGAGCACTGTGACAVCACAGTGG
GCAGAKGSAGGABDGHDG------
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
59 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
17 |
| Similarity score: |
0.58153 |
Alignment:
-BAGGYCABHBTGACCKHV
CHHCHBTCCTSCYTCTGC-
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
64 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
1.58383 |
Alignment:
BTRGGDCARAGGKCA---
GCAGAKGSAGGABDGHDG
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
11 |
| Number of overlap: |
15 |
| Similarity score: |
1.5846 |
Alignment:
CACTGTGRTRTCACAGTGSWSCACT---
----------CHHCHBTCCTSCYTCTGC
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
88 |
| Motif name: |
TFF11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.05771 |
Alignment:
----HVCDBBDBCAGCAG
CHHCHBTCCTSCYTCTGC
| Original motif Consensus sequence: CTGCTGBDBBDGBD |
Reverse complement motif Consensus sequence: HVCDBBDBCAGCAG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
65 |
| Motif name: |
Tcfcp2l1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.08379 |
Alignment:
----CKGGDTBDMMCTGG
GCAGAKGSAGGABDGHDG
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
14 |
| Similarity score: |
2.08495 |
Alignment:
----TTCAGCACCATGGACAGCKCC
GCAGAKGSAGGABDGHDG-------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 91 |
Motif name: TFM3 |
| Original motif Consensus sequence: KTTTTTTKTTTTTTDAAB |
Reverse complement motif Consensus sequence: BTTHAAAAAAYAAAAAAR |
|
|
 |
 |
Best Matches for Motif ID 91 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
94 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
0.00694444 |
Alignment:
TTTTTTTKTTKTTTAATTHW
KTTTTTTKTTTTTTDAAB--
| Original motif Consensus sequence: TTTTTTTKTTKTTTAATTHW |
Reverse complement motif Consensus sequence: WHAATTAAARAARAAAAAAA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
89 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
0.0750248 |
Alignment:
CACACACACACWCACACA
BTTHAAAAAAYAAAAAAR
| Original motif Consensus sequence: CACACACACACWCACACA |
Reverse complement motif Consensus sequence: TGTGTGWGTGTGTGTGTG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
92 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
15 |
| Similarity score: |
1.57541 |
Alignment:
TGTGTGDKTGTGTGTGTRTG---
-----KTTTTTTKTTTTTTDAAB
| Original motif Consensus sequence: CAMACACACACAYDCACACA |
Reverse complement motif Consensus sequence: TGTGTGDKTGTGTGTGTRTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.01116 |
Alignment:
AAAAAAAAAAAAAT----
BTTHAAAAAAYAAAAAAR
| Original motif Consensus sequence: AAAAAAAAAAAAAT |
Reverse complement motif Consensus sequence: ATTTTTTTTTTTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.02946 |
Alignment:
----TTTRKTTTHTTTTH
KTTTTTTKTTTTTTDAAB
| Original motif Consensus sequence: HAAAAHAAARMAAA |
Reverse complement motif Consensus sequence: TTTRKTTTHTTTTH |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.0692 |
Alignment:
TTTAATGTYTTTWA----
KTTTTTTKTTTTTTDAAB
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.0735 |
Alignment:
CASABACWSACACA----
BTTHAAAAAAYAAAAAAR
| Original motif Consensus sequence: CASABACWSACACA |
Reverse complement motif Consensus sequence: TGTGTSWGTBTSTG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
45 |
| Motif name: |
Motif 45 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.07366 |
Alignment:
TAASTCTWTTTTAA----
KTTTTTTKTTTTTTDAAB
| Original motif Consensus sequence: TAASTCTWTTTTAA |
Reverse complement motif Consensus sequence: TTAAAAWAGASTTA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
15 |
| Motif name: |
Motif 15 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
2.56147 |
Alignment:
-----TAGTTTYAKCTTT
KTTTTTTKTTTTTTDAAB
| Original motif Consensus sequence: AAAGRTMAAACTA |
Reverse complement motif Consensus sequence: TAGTTTYAKCTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
23 |
| Motif name: |
Motif 23 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
2.56748 |
Alignment:
-----ATYAATGAAWACAT
BTTHAAAAAAYAAAAAAR-
| Original motif Consensus sequence: ATGTWTTCATTMAT |
Reverse complement motif Consensus sequence: ATYAATGAAWACAT |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 92 |
Motif name: TFM11 |
| Original motif Consensus sequence: CAMACACACACAYDCACACA |
Reverse complement motif Consensus sequence: TGTGTGDKTGTGTGTGTRTG |
|
|
 |
 |
Best Matches for Motif ID 92 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
94 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
19 |
| Similarity score: |
0.0921695 |
Alignment:
TTTTTTTKTTKTTTAATTHW-
-TGTGTGDKTGTGTGTGTRTG
| Original motif Consensus sequence: TTTTTTTKTTKTTTAATTHW |
Reverse complement motif Consensus sequence: WHAATTAAARAARAAAAAAA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
89 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
0.5 |
Alignment:
TGTGTGWGTGTGTGTGTG--
TGTGTGDKTGTGTGTGTRTG
| Original motif Consensus sequence: CACACACACACWCACACA |
Reverse complement motif Consensus sequence: TGTGTGWGTGTGTGTGTG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
16 |
| Similarity score: |
1.597 |
Alignment:
GGAGCACTGTGACACCACA----
---CAMACACACACAYDCACACA
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
91 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
15 |
| Similarity score: |
2.09964 |
Alignment:
-----BTTHAAAAAAYAAAAAAR
CAMACACACACAYDCACACA---
| Original motif Consensus sequence: KTTTTTTKTTTTTTDAAB |
Reverse complement motif Consensus sequence: BTTHAAAAAAYAAAAAAR |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.54253 |
Alignment:
TGTGTSWGTBTSTG------
TGTGTGDKTGTGTGTGTRTG
| Original motif Consensus sequence: CASABACWSACACA |
Reverse complement motif Consensus sequence: TGTGTSWGTBTSTG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
14 |
| Similarity score: |
2.59651 |
Alignment:
RAGKGMAGVRRGSRCASAGV------
------CAMACACACACAYDCACACA
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.59861 |
Alignment:
HAAAAHAAARMAAA------
CAMACACACACAYDCACACA
| Original motif Consensus sequence: HAAAAHAAARMAAA |
Reverse complement motif Consensus sequence: TTTRKTTTHTTTTH |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
3.02049 |
Alignment:
ACACACACACACAC-------
-CAMACACACACAYDCACACA
| Original motif Consensus sequence: ACACACACACACAC |
Reverse complement motif Consensus sequence: GTGTGTGTGTGTGT |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
90 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
13 |
| Similarity score: |
3.09283 |
Alignment:
GCAGAKGSAGGABDGHDG-------
-----TGTGTGDKTGTGTGTGTRTG
| Original motif Consensus sequence: CHHCHBTCCTSCYTCTGC |
Reverse complement motif Consensus sequence: GCAGAKGSAGGABDGHDG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
3.09411 |
Alignment:
HCAGRRVACASAG-------
CAMACACACACAYDCACACA
| Original motif Consensus sequence: HCAGRRVACASAG |
Reverse complement motif Consensus sequence: CTSTGTBKMCTGH |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 93 |
Motif name: TFM12 |
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
Best Matches for Motif ID 93 (Highest to Lowest)
| Dataset #: |
4 |
| Motif ID: |
73 |
| Motif name: |
TGGAGCACTGTGACAcCACAGTGg |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
20 |
| Similarity score: |
0.0627086 |
Alignment:
TGGAGCACTGTGACAVCACAGTGG
--RAGKGMAGVRRGSRCASAGV--
| Original motif Consensus sequence: TGGAGCACTGTGACAVCACAGTGG |
Reverse complement motif Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
72 |
| Motif name: |
CACTGTGrYrtCACAGTGswsCAcT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
20 |
| Similarity score: |
0.063541 |
Alignment:
CACTGTGRTRTCACAGTGSWSCACT
----VCTSTGKSCMKBCTRCYCTK-
| Original motif Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT |
Reverse complement motif Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
57 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
20 |
| Similarity score: |
0.0650732 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
-VCTSTGKSCMKBCTRCYCTK
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
53 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
19 |
| Similarity score: |
0.541945 |
Alignment:
MGGTCAGGGTGACCTRDBHV-
-RAGKGMAGVRRGSRCASAGV
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
66 |
| Motif name: |
CTCF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
19 |
| Similarity score: |
0.565451 |
Alignment:
BMSMGCCYMCTKSTGGMHM-
VCTSTGKSCMKBCTRCYCTK
| Original motif Consensus sequence: YDRCCASYAGRKGGCRSYV |
Reverse complement motif Consensus sequence: BMSMGCCYMCTKSTGGMHM |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
59 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
1.04822 |
Alignment:
--BAGGYCABHBTGACCKHV
RAGKGMAGVRRGSRCASAGV
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
90 |
| Motif name: |
TFM2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
1.04851 |
Alignment:
--CHHCHBTCCTSCYTCTGC
VCTSTGKSCMKBCTRCYCTK
| Original motif Consensus sequence: CHHCHBTCCTSCYTCTGC |
Reverse complement motif Consensus sequence: GCAGAKGSAGGABDGHDG |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
18 |
| Similarity score: |
1.06391 |
Alignment:
TGTGGTGTCACAGTGCTCC--
-VCTSTGKSCMKBCTRCYCTK
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
94 |
| Motif name: |
TFM13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
18 |
| Similarity score: |
1.06828 |
Alignment:
WHAATTAAARAARAAAAAAA--
--RAGKGMAGVRRGSRCASAGV
| Original motif Consensus sequence: TTTTTTTKTTKTTTAATTHW |
Reverse complement motif Consensus sequence: WHAATTAAARAARAAAAAAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
71 |
| Motif name: |
AmTGTGACACCACAGT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
16 |
| Similarity score: |
2.06801 |
Alignment:
----AMTGTGACACCACAGT
RAGKGMAGVRRGSRCASAGV
| Original motif Consensus sequence: AMTGTGACACCACAGT |
Reverse complement motif Consensus sequence: ACTGTGGTGTCACART |
|
|
 |
 |
| Dataset #: 5 |
Motif ID: 94 |
Motif name: TFM13 |
| Original motif Consensus sequence: TTTTTTTKTTKTTTAATTHW |
Reverse complement motif Consensus sequence: WHAATTAAARAARAAAAAAA |
|
|
 |
 |
Best Matches for Motif ID 94 (Highest to Lowest)
| Dataset #: |
5 |
| Motif ID: |
92 |
| Motif name: |
TFM11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
19 |
| Similarity score: |
0.0813331 |
Alignment:
-CAMACACACACAYDCACACA
WHAATTAAARAARAAAAAAA-
| Original motif Consensus sequence: CAMACACACACAYDCACACA |
Reverse complement motif Consensus sequence: TGTGTGDKTGTGTGTGTRTG |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
91 |
| Motif name: |
TFM3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
0.520337 |
Alignment:
KTTTTTTKTTTTTTDAAB--
TTTTTTTKTTKTTTAATTHW
| Original motif Consensus sequence: KTTTTTTKTTTTTTDAAB |
Reverse complement motif Consensus sequence: BTTHAAAAAAYAAAAAAR |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
93 |
| Motif name: |
TFM12 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
18 |
| Similarity score: |
0.585681 |
Alignment:
--VCTSTGKSCMKBCTRCYCTK
TTTTTTTKTTKTTTAATTHW--
| Original motif Consensus sequence: RAGKGMAGVRRGSRCASAGV |
Reverse complement motif Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
|
|
 |
 |
| Dataset #: |
5 |
| Motif ID: |
89 |
| Motif name: |
TFM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
15 |
| Similarity score: |
2.08869 |
Alignment:
CACACACACACWCACACA-----
---WHAATTAAARAARAAAAAAA
| Original motif Consensus sequence: CACACACACACWCACACA |
Reverse complement motif Consensus sequence: TGTGTGWGTGTGTGTGTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
49 |
| Motif name: |
Motif 49 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.51913 |
Alignment:
AAAAAAAAAAAAAT------
WHAATTAAARAARAAAAAAA
| Original motif Consensus sequence: AAAAAAAAAAAAAT |
Reverse complement motif Consensus sequence: ATTTTTTTTTTTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.53469 |
Alignment:
------TTTRKTTTHTTTTH
TTTTTTTKTTKTTTAATTHW
| Original motif Consensus sequence: HAAAAHAAARMAAA |
Reverse complement motif Consensus sequence: TTTRKTTTHTTTTH |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
33 |
| Motif name: |
Motif 33 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.5727 |
Alignment:
TTTAATGTYTTTWA------
TTTTTTTKTTKTTTAATTHW
| Original motif Consensus sequence: TTTAATGTYTTTWA |
Reverse complement motif Consensus sequence: TWAAAKACATTAAA |
|
|
 |
 |
| Dataset #: |
4 |
| Motif ID: |
67 |
| Motif name: |
TgTGgTGTCACAGTGCTCC |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
14 |
| Similarity score: |
2.5844 |
Alignment:
------TGTGGTGTCACAGTGCTCC
TTTTTTTKTTKTTTAATTHW-----
| Original motif Consensus sequence: TGTGGTGTCACAGTGCTCC |
Reverse complement motif Consensus sequence: GGAGCACTGTGACACCACA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
45 |
| Motif name: |
Motif 45 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.58546 |
Alignment:
------TTAAAAWAGASTTA
WHAATTAAARAARAAAAAAA
| Original motif Consensus sequence: TAASTCTWTTTTAA |
Reverse complement motif Consensus sequence: TTAAAAWAGASTTA |
|
|
 |
 |
| Dataset #: |
3 |
| Motif ID: |
64 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
2.58746 |
Alignment:
TGRCCTKTGHCCKAB------
-TTTTTTTKTTKTTTAATTHW
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |



































































































































































































































































































