Best Matches in Database for Each Motif (Highest to Lowest)
| Dataset #: 1 | Motif ID: 1 | Motif name: Motif 1 |
| Original motif | Reverse complement motif |
| Consensus sequence: CYTSCCTCTGC | Consensus sequence: GCAGAGGSAKG |
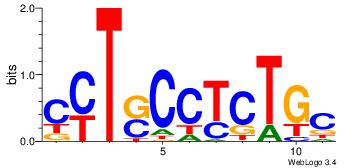 |
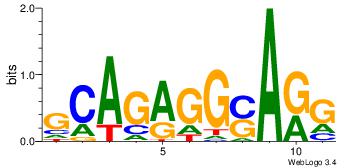 |
Best Matches for Motif ID 1 (Highest to Lowest)
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0313012 |
Alignment:
HBBBCCGCCCCWHHHV
---CYTSCCTCTGC--
| Original motif | Reverse complement motif |
| Consensus sequence: HBBBCCGCCCCWHHHV | Consensus sequence: BHHHWGGGGCGGBBVH |
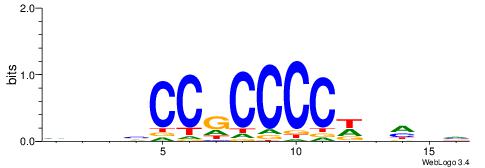 |
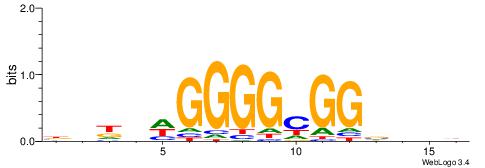 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0317134 |
Alignment:
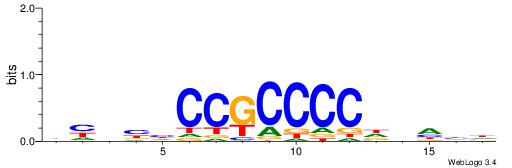
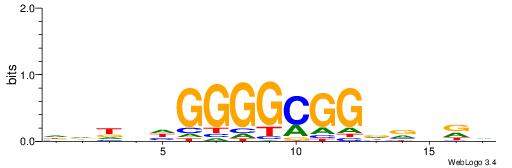
BHHBHCCGCCCCDHHHH
----CYTSCCTCTGC--
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
 |
 |
| Motif ID: | UP00013 |
| Motif name: | Gabpa_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0322746 |
Alignment:
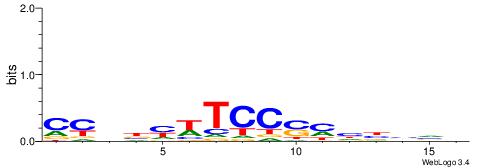
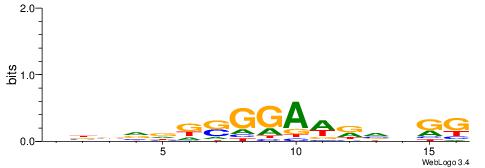
CYDBYWTCCSMHBVVH
----CYTSCCTCTGC-
| Original motif | Reverse complement motif |
| Consensus sequence: CYDBYWTCCSMHBVVH | Consensus sequence: DBVVDRSGGAWKVHKG |
 |
 |
| Motif ID: | UP00010 |
| Motif name: | Tcfap2b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0336419 |
Alignment:
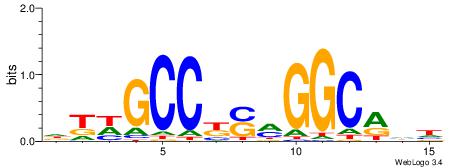
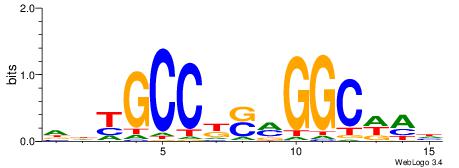
HHKGCCYSRGGCWAD
CYTSCCTCTGC----
| Original motif | Reverse complement motif |
| Consensus sequence: DTWGCCKSMGGCRHH | Consensus sequence: HHKGCCYSRGGCWAD |
 |
 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0343277 |
Alignment:
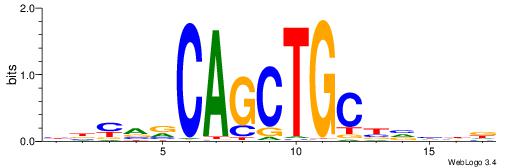
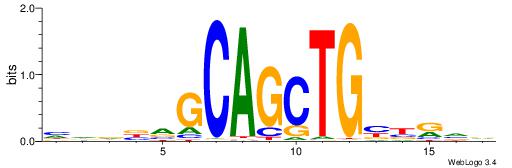
HHDVVGCAGCTGVBKVB
-----GCAGAGGSAKG-
| Original motif | Reverse complement motif |
| Consensus sequence: BBYVVCAGCTGCBVHHD | Consensus sequence: HHDVVGCAGCTGVBKVB |
 |
 |
| Motif ID: | UP00096 |
| Motif name: | Sox13_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0359879 |
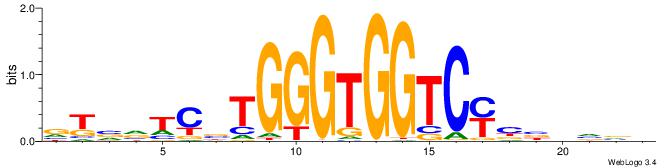
Alignment:
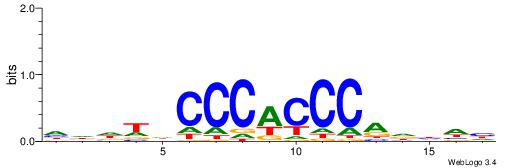
DDDHTGGGTGGGHAHHD
------CYTSCCTCTGC
| Original motif | Reverse complement motif |
| Consensus sequence: DDDHTGGGTGGGHAHHD | Consensus sequence: DHHTHCCCACCCAHDDH |
 |
 |
| Motif ID: | UP00085 |
| Motif name: | Sfpi1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.036078 |
Alignment:
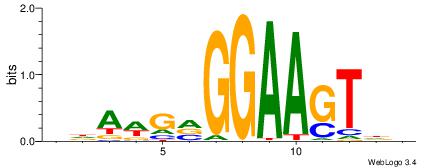
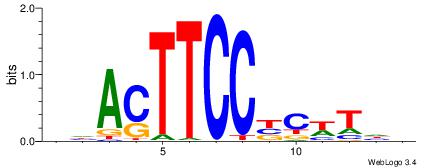
HDACTTCCBCWTDV
---CYTSCCTCTGC
| Original motif | Reverse complement motif |
| Consensus sequence: BDAWGVGGAAGTDH | Consensus sequence: HDACTTCCBCWTDV |
 |
 |
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0369028 |
Alignment:
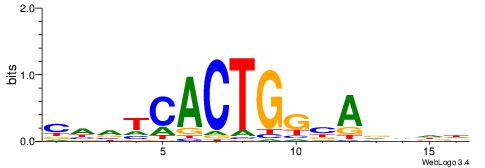
DDHHTBCCAGTGABHG
--CYTSCCTCTGC---
| Original motif | Reverse complement motif |
| Consensus sequence: CHVTCACTGGBADHDD | Consensus sequence: DDHHTBCCAGTGABHG |
 |
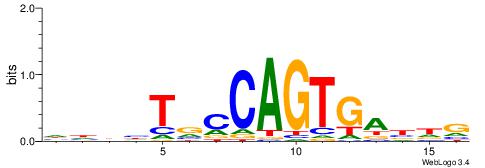 |
| Motif ID: | UP00052 |
| Motif name: | Osr2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0370849 |
Alignment:
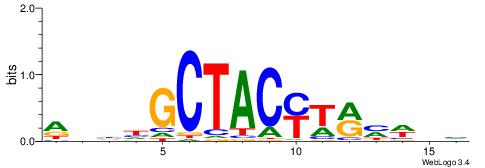
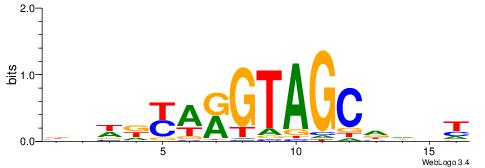
VDWRKAKGTAGCHHDT
-GCAGAGGSAKG----
| Original motif | Reverse complement motif |
| Consensus sequence: AHHHGCTACYTRMWHV | Consensus sequence: VDWRKAKGTAGCHHDT |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 11 |
| Similarity score: | 0.0381821 |
Alignment:
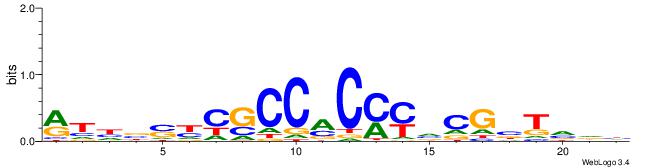
RTHBSYCGCCMCMYVCGBTVDH
-------CYTSCCTCTGC----
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
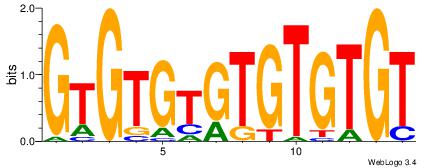
| Dataset #: 1 | Motif ID: 2 | Motif name: Motif 2 |
| Original motif | Reverse complement motif |
| Consensus sequence: ACACACACACACAC | Consensus sequence: GTGTGTGTGTGTGT |
 |
 |
Best Matches for Motif ID 2 (Highest to Lowest)
| Motif ID: | UP00042 |
| Motif name: | Gm397_second |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0391499 |
Alignment:
HBGYGTGTGTGCVRVD
GTGTGTGTGTGTGT--
| Original motif | Reverse complement motif |
| Consensus sequence: DVMVGCACACACKCVH | Consensus sequence: HBGYGTGTGTGCVRVD |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 10 |
| Number of overlap: | 14 |
| Similarity score: | 0.043857 |
Alignment:
MHHHWHYTMCCHCCCACVAABVH
---------ACACACACACACAC
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
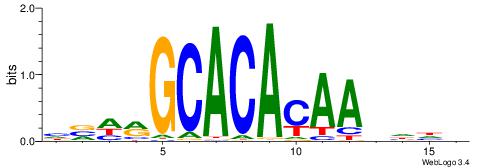
| Motif ID: | UP00026 |
| Motif name: | Zscan4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0468808 |
Alignment:
BHHHTTGTGTGCKHVD
GTGTGTGTGTGTGT--
| Original motif | Reverse complement motif |
| Consensus sequence: HVHRGCACACAAHHHV | Consensus sequence: BHHHTTGTGTGCKHVD |
 |
 |
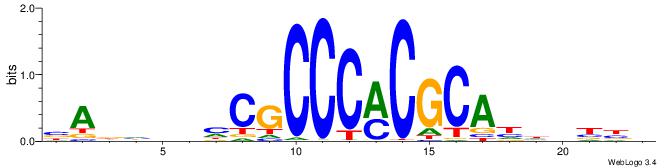
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 14 |
| Similarity score: | 0.0479337 |
Alignment:
BADHBDHCGCCCMCGCAHHDBBV
------ACACACACACACAC---
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
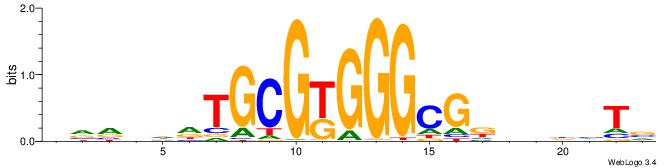 |
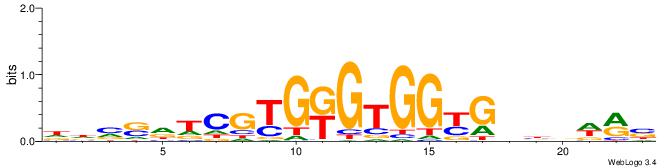
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 14 |
| Similarity score: | 0.0485959 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
-GTGTGTGTGTGTGT--------
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
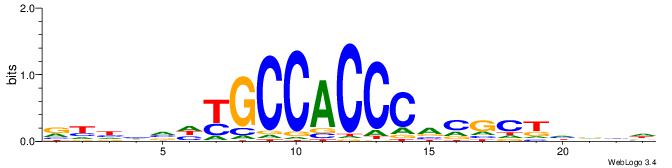 |
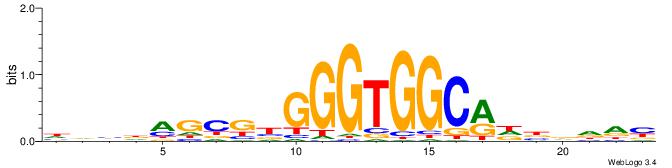
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 14 |
| Similarity score: | 0.0526961 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
--ACACACACACACAC-------
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
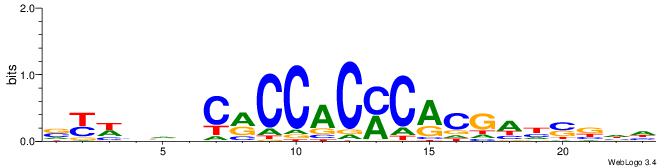
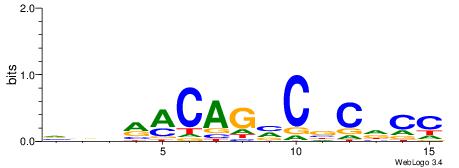
| Motif ID: | UP00036 |
| Motif name: | Myf6_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0540872 |
Alignment:
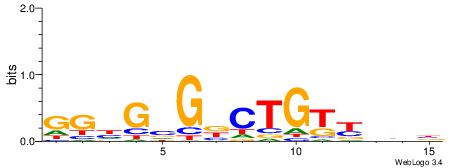
VVVRACAGVCBCDCC
ACACACACACACAC-
| Original motif | Reverse complement motif |
| Consensus sequence: VVVRACAGVCBCDCC | Consensus sequence: GGDGBGVCTGTKVVB |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 14 |
| Similarity score: | 0.0550075 |
Alignment:
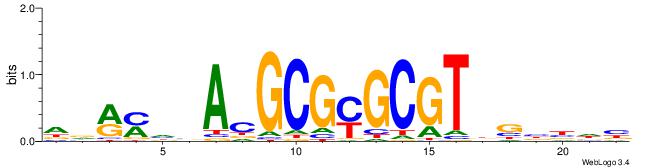
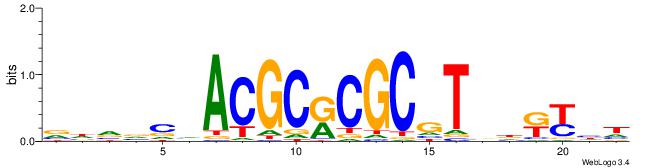
DDRMHBACGCGYGCGTDGBBDB
ACACACACACACAC--------
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.055413 |
Alignment:
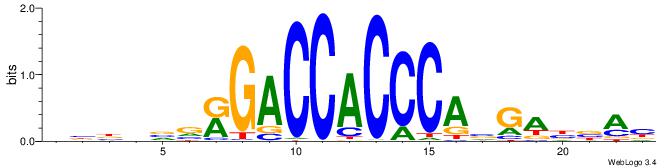
BBBDVVRGACCACCCAVGABBAB
---------ACACACACACACAC
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00096 |
| Motif name: | Sox13_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0582564 |
Alignment:
DDDHTGGGTGGGHAHHD
ACACACACACACAC---
| Original motif | Reverse complement motif |
| Consensus sequence: DDDHTGGGTGGGHAHHD | Consensus sequence: DHHTHCCCACCCAHDDH |
 |
 |
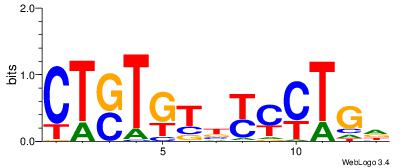
| Dataset #: 1 | Motif ID: 3 | Motif name: Motif 3 |
| Original motif | Reverse complement motif |
| Consensus sequence: HCAGRRVACASAG | Consensus sequence: CTSTGTBKMCTGH |
 |
 |
Best Matches for Motif ID 3 (Highest to Lowest)
| Motif ID: | UP00028 |
| Motif name: | Tcfap2e_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0445121 |
Alignment:
HDCDBRAAAAAADD
-HCAGRRVACASAG
| Original motif | Reverse complement motif |
| Consensus sequence: HDCDBRAAAAAADD | Consensus sequence: DDTTTTTTMBDGDH |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 10 |
| Number of overlap: | 13 |
| Similarity score: | 0.0448793 |
Alignment:
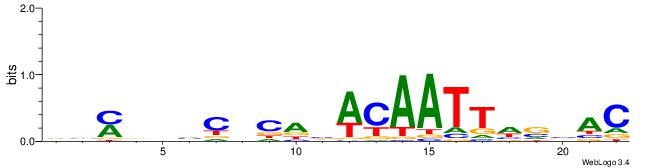
GTVBDAATTGTVTGHGDDHKVB
---------CTSTGTBKMCTGH
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00041 |
| Motif name: | Foxj1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0454552 |
Alignment:
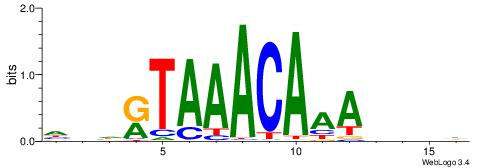
DDDHTTTGTTTAMHDH
---CTSTGTBKMCTGH
| Original motif | Reverse complement motif |
| Consensus sequence: HDHRTAAACAAAHDDD | Consensus sequence: DDDHTTTGTTTAMHDH |
 |
 |
| Motif ID: | UP00016 |
| Motif name: | Sry_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0481412 |
Alignment:
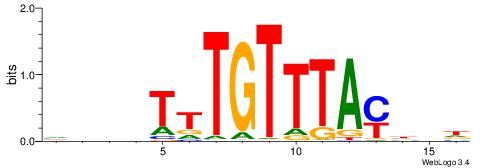
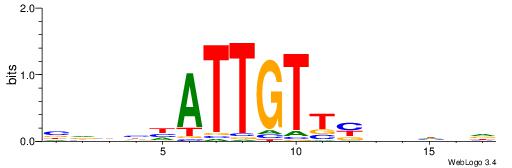
HHDHDRAACAATDDVHV
HCAGRRVACASAG----
| Original motif | Reverse complement motif |
| Consensus sequence: HHDHDRAACAATDDVHV | Consensus sequence: VHVHDATTGTTMHDDDH |
 |
 |
| Motif ID: | UP00077 |
| Motif name: | Srf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 13 |
| Similarity score: | 0.0545707 |
Alignment:
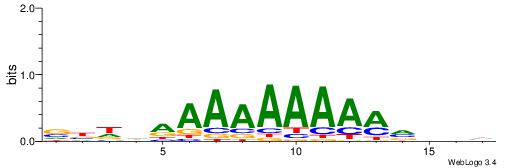
VBDDAAAAAAAAMVBHH
-HCAGRRVACASAG---
| Original motif | Reverse complement motif |
| Consensus sequence: VBDDAAAAAAAAMVBHH | Consensus sequence: HHVBYTTTTTTTTDDVV |
 |
 |
| Motif ID: | UP00058 |
| Motif name: | Tcf3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0553073 |
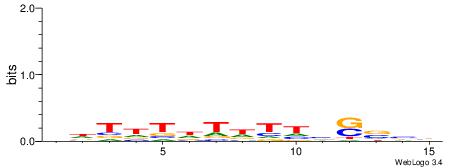
Alignment:
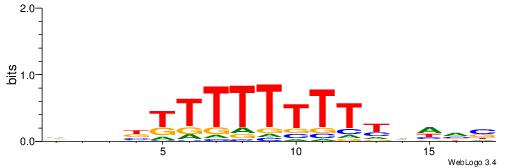
DVBSBDAHAHAHAHH
-HCAGRRVACASAG-
| Original motif | Reverse complement motif |
| Consensus sequence: DVBSBDAHAHAHAHH | Consensus sequence: HHTHTHTHTDBSBVD |
 |
 |
| Motif ID: | UP00091 |
| Motif name: | Sox5_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0562585 |
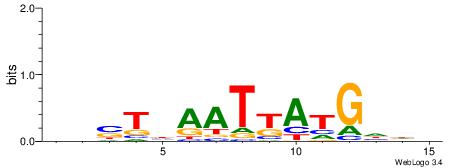
Alignment:
DHDCATMATTDASDD
--HCAGRRVACASAG
| Original motif | Reverse complement motif |
| Consensus sequence: DHDCATMATTDASDD | Consensus sequence: DHSTDAATYATGDHD |
 |
 |
| Motif ID: | UP00012 |
| Motif name: | Bbx_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 13 |
| Similarity score: | 0.0564984 |
Alignment:
VHVHBTGTTAACVHWVH
--CTSTGTBKMCTGH--
| Original motif | Reverse complement motif |
| Consensus sequence: HVWHBGTTAACABHBDV | Consensus sequence: VHVHBTGTTAACVHWVH |
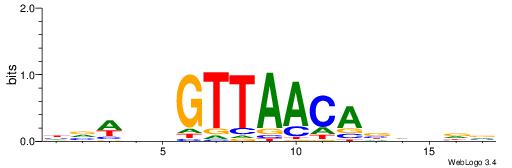 |
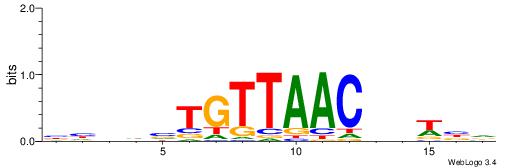 |
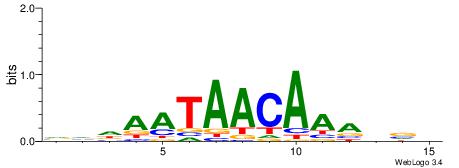
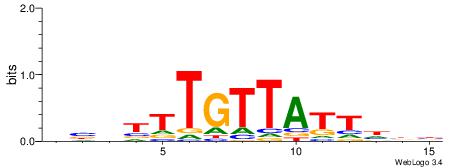
| Motif ID: | UP00073 |
| Motif name: | Foxa2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 13 |
| Similarity score: | 0.0571073 |
Alignment:
VVDAATAACAAAVVB
HCAGRRVACASAG--
| Original motif | Reverse complement motif |
| Consensus sequence: VVDAATAACAAAVVB | Consensus sequence: BVVTTTGTTATTDBB |
 |
 |
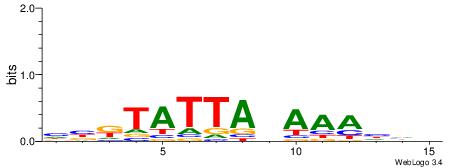
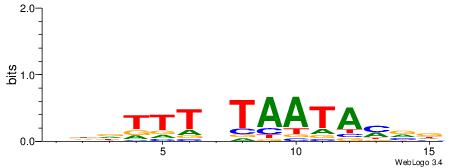
| Motif ID: | UP00054 |
| Motif name: | Tcf7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 13 |
| Similarity score: | 0.0574997 |
Alignment:
HHDTTTVTAATAYBD
CTSTGTBKMCTGH--
| Original motif | Reverse complement motif |
| Consensus sequence: HBKTATTABAAAHHH | Consensus sequence: HHDTTTVTAATAYBD |
 |
 |
| Dataset #: 1 | Motif ID: 4 | Motif name: Motif 4 |
| Original motif | Reverse complement motif |
| Consensus sequence: HAAAAHAAARMAAA | Consensus sequence: TTTRKTTTHTTTTH |
 |
 |
Best Matches for Motif ID 4 (Highest to Lowest)
| Motif ID: | UP00025 |
| Motif name: | Foxk1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0212263 |
Alignment:
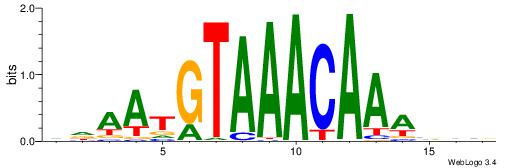
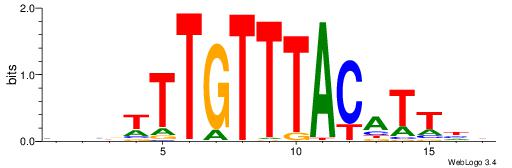
BBVTTTGTTTACATTDD
---TTTRKTTTHTTTTH
| Original motif | Reverse complement motif |
| Consensus sequence: DDAATGTAAACAAAVVB | Consensus sequence: BBVTTTGTTTACATTDD |
 |
 |
| Motif ID: | UP00077 |
| Motif name: | Srf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0216448 |
Alignment:
VBDDAAAAAAAAMVBHH
---HAAAAHAAARMAAA
| Original motif | Reverse complement motif |
| Consensus sequence: VBDDAAAAAAAAMVBHH | Consensus sequence: HHVBYTTTTTTTTDDVV |
 |
 |
| Motif ID: | UP00039 |
| Motif name: | Foxj3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0216826 |
Alignment:
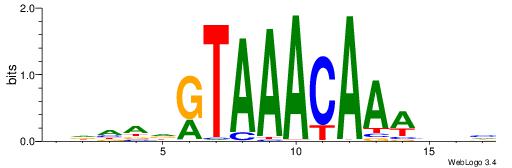
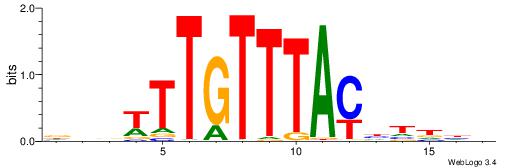
DVVTTTGTTTACDTHDH
---TTTRKTTTHTTTTH
| Original motif | Reverse complement motif |
| Consensus sequence: HDHADGTAAACAAAVVH | Consensus sequence: DVVTTTGTTTACDTHDH |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0232085 |
Alignment:
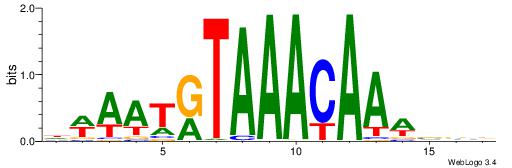
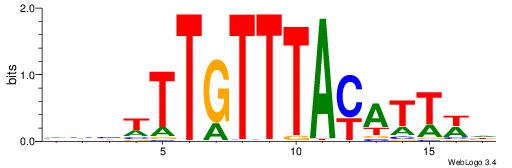
VVVWTTGTTTAMATTWD
---TTTRKTTTHTTTTH
| Original motif | Reverse complement motif |
| Consensus sequence: DWAATRTAAACAAWVVB | Consensus sequence: VVVWTTGTTTAMATTWD |
 |
 |
| Motif ID: | UP00073 |
| Motif name: | Foxa2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.025394 |
Alignment:
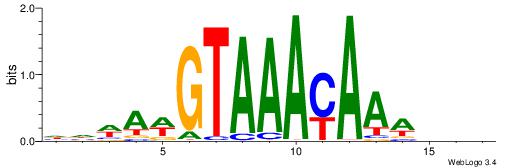
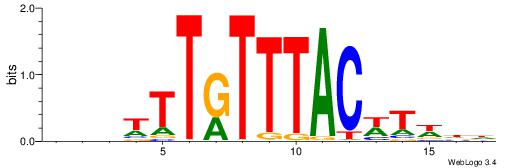
BDVTTTKTTTACWTWHH
---TTTRKTTTHTTTTH
| Original motif | Reverse complement motif |
| Consensus sequence: HHWAWGTAAAYAAAVDB | Consensus sequence: BDVTTTKTTTACWTWHH |
 |
 |
| Motif ID: | UP00025 |
| Motif name: | Foxk1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0257356 |
Alignment:
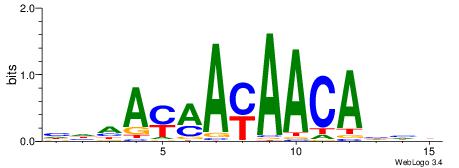
HDDTGTTKTTKTHHD
-TTTRKTTTHTTTTH
| Original motif | Reverse complement motif |
| Consensus sequence: HHHAYAAYAACAHHH | Consensus sequence: HDDTGTTKTTKTHHD |
 |
 |
| Motif ID: | UP00037 |
| Motif name: | Zfp105_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0280765 |
Alignment:
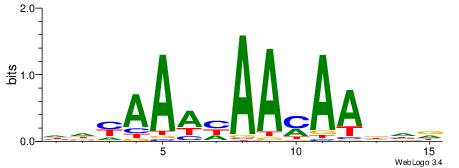
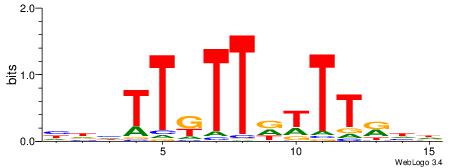
HHHTTRTTDTTTKDH
-TTTRKTTTHTTTTH
| Original motif | Reverse complement motif |
| Consensus sequence: HDYAAAHAAMAADHD | Consensus sequence: HHHTTRTTDTTTKDH |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0284931 |
Alignment:
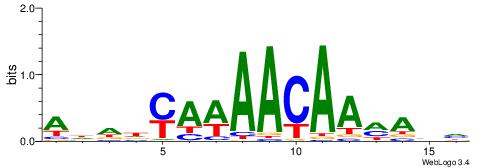
HDTYTTGTTWTKDDDT
--TTTRKTTTHTTTTH
| Original motif | Reverse complement motif |
| Consensus sequence: ADDDYAWAACAAMAHH | Consensus sequence: HDTYTTGTTWTKDDDT |
 |
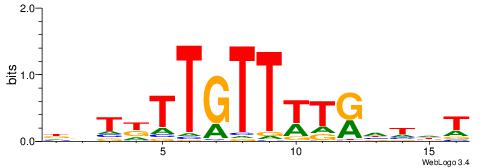 |
| Motif ID: | UP00145 |
| Motif name: | Barhl2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.031537 |
Alignment:
HHBTTAATTGDTTYTH
--TTTRKTTTHTTTTH
| Original motif | Reverse complement motif |
| Consensus sequence: HAMAAHCAATTAABHH | Consensus sequence: HHBTTAATTGDTTYTH |
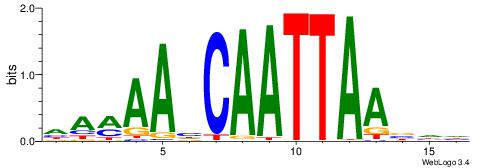 |
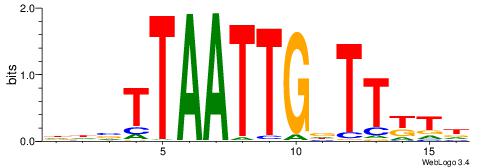 |
| Motif ID: | UP00090 |
| Motif name: | Elf3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0324424 |
Alignment:
DBTHHAAAAAAAAVHDH
---HAAAAHAAARMAAA
| Original motif | Reverse complement motif |
| Consensus sequence: DBTHHAAAAAAAAVHDH | Consensus sequence: DDHBTTTTTTTTHDAVH |
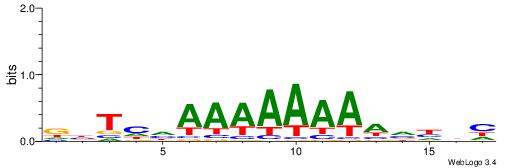 |
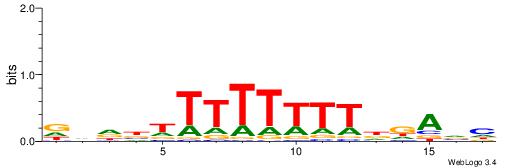 |
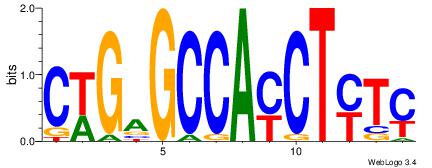
| Dataset #: 1 | Motif ID: 5 | Motif name: Motif 5 |
| Original motif | Reverse complement motif |
| Consensus sequence: CWGAGCCAYCTYTC | Consensus sequence: GAKAGKTGGCTCWG |
 |
 |
Best Matches for Motif ID 5 (Highest to Lowest)
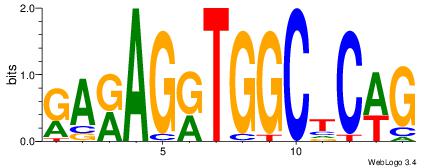
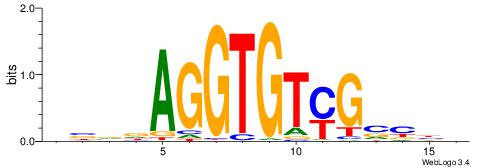
| Motif ID: | UP00068 |
| Motif name: | Eomes_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0120339 |
Alignment:
VBVDAGGTGTYGVBBV
-GAKAGKTGGCTCWG-
| Original motif | Reverse complement motif |
| Consensus sequence: VBVDAGGTGTYGVBBV | Consensus sequence: VVBVCKACACCTHVBV |
 |
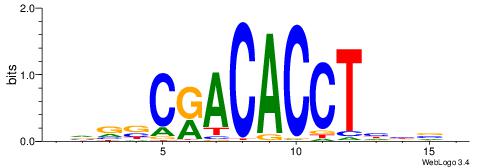 |
| Motif ID: | UP00035 |
| Motif name: | Hic1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0137859 |
Alignment:
DBBATGCCAACCHVHH
-CWGAGCCAYCTYTC-
| Original motif | Reverse complement motif |
| Consensus sequence: DBBATGCCAACCHVHH | Consensus sequence: DDBHGGTTGGCATVBD |
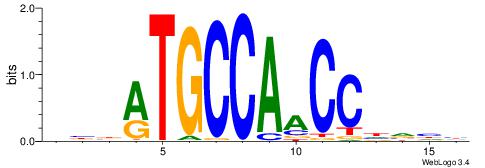 |
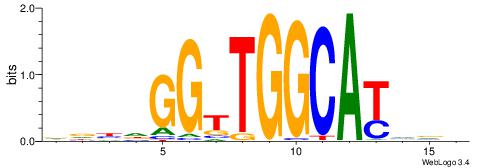 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.017234 |
Alignment:
HHHHDVVGTTGCTAHGGDHBAHH
--GAKAGKTGGCTCWG-------
| Original motif | Reverse complement motif |
| Consensus sequence: HDTBHHCCHTAGCAACVVDHHHH | Consensus sequence: HHHHDVVGTTGCTAHGGDHBAHH |
 |
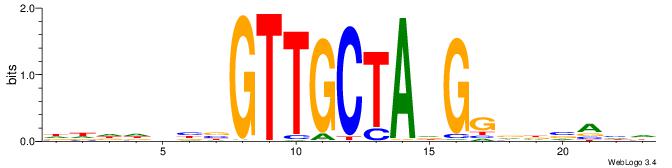 |
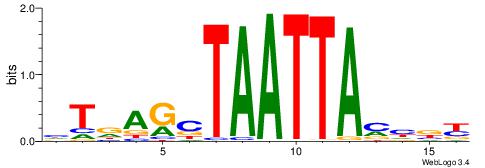
| Motif ID: | UP00264 |
| Motif name: | Hoxa1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0174544 |
Alignment:
MBDRTAATTAGCTHAD
--GAKAGKTGGCTCWG
| Original motif | Reverse complement motif |
| Consensus sequence: HTDAGCTAATTAMHBY | Consensus sequence: MBDRTAATTAGCTHAD |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 14 |
| Similarity score: | 0.0187141 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
---CWGAGCCAYCTYTC------
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
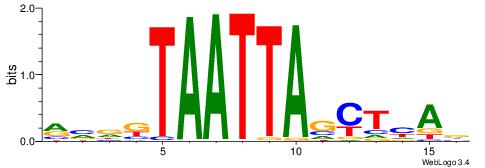
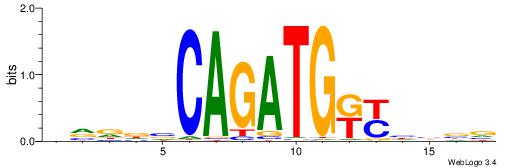
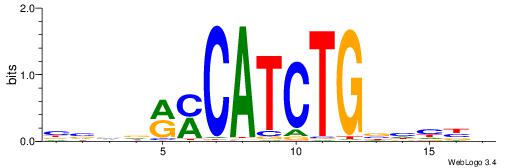
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0198398 |
Alignment:
HVDDVCAGATGKYHBBV
---GAKAGKTGGCTCWG
| Original motif | Reverse complement motif |
| Consensus sequence: HVDDVCAGATGKYHBBV | Consensus sequence: VBBDMYCATCTGVHHBH |
 |
 |
| Motif ID: | UP00052 |
| Motif name: | Osr2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0203442 |
Alignment:
VDWRKAKGTAGCHHDT
--GAKAGKTGGCTCWG
| Original motif | Reverse complement motif |
| Consensus sequence: AHHHGCTACYTRMWHV | Consensus sequence: VDWRKAKGTAGCHHDT |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 14 |
| Similarity score: | 0.0208381 |
Alignment:
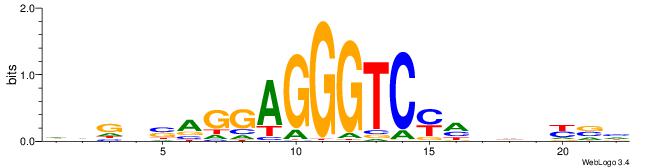
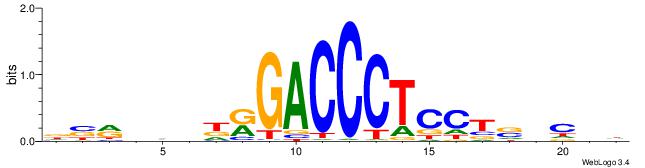
DVMHHDHKGACCCTCCTSVCBH
------CWGAGCCAYCTYTC--
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Motif ID: | UP00095 |
| Motif name: | Zfp691_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0209451 |
Alignment:
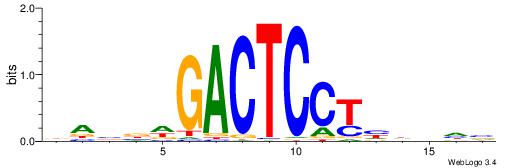
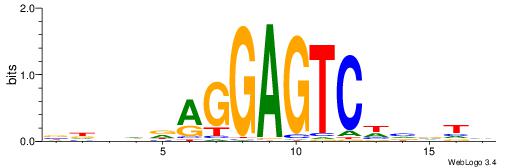
VHBDDAGGAGTCWBBTD
--GAKAGKTGGCTCWG-
| Original motif | Reverse complement motif |
| Consensus sequence: DABBWGACTCCTHDVHV | Consensus sequence: VHBDDAGGAGTCWBBTD |
 |
 |
| Motif ID: | UP00228 |
| Motif name: | Bapx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0212049 |
Alignment:
DTBVTTAAGTGGTTHHV
---GAKAGKTGGCTCWG
| Original motif | Reverse complement motif |
| Consensus sequence: VHHAACCACTTAAVVAH | Consensus sequence: DTBVTTAAGTGGTTHHV |
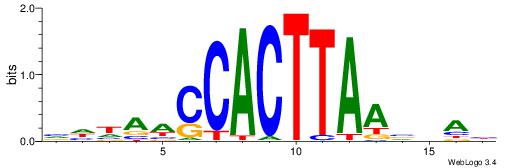 |
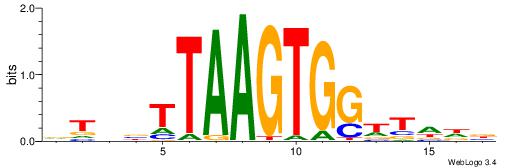 |
| Dataset #: 1 | Motif ID: 6 | Motif name: Motif 6 |
| Original motif | Reverse complement motif |
| Consensus sequence: CASABACWSACACA | Consensus sequence: TGTGTSWGTBTSTG |
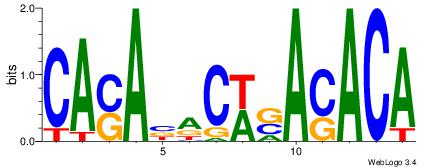 |
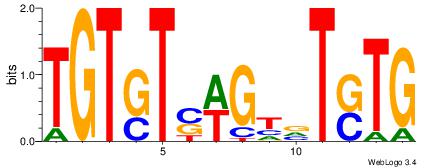 |
Best Matches for Motif ID 6 (Highest to Lowest)
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0357045 |
Alignment:
VVYDDDCHCAVACAATTDBVAC
CASABACWSACACA--------
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
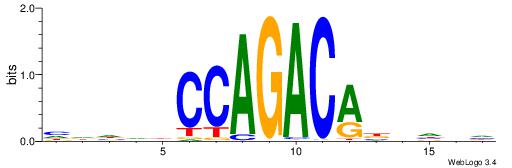
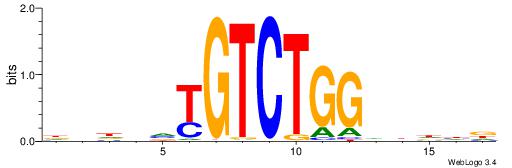
| Motif ID: | UP00000 |
| Motif name: | Smad3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0482294 |
Alignment:
HVHBVTGTCTGGDDHDD
---TGTGTSWGTBTSTG
| Original motif | Reverse complement motif |
| Consensus sequence: HDHDDCCAGACABBHVH | Consensus sequence: HVHBVTGTCTGGDDHDD |
 |
 |
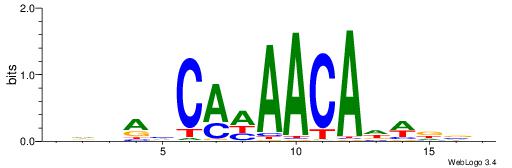
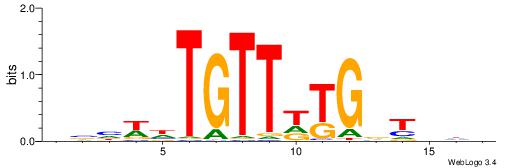
| Motif ID: | UP00039 |
| Motif name: | Foxj3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0493845 |
Alignment:
HDBAHCAWAACADWDVD
CASABACWSACACA---
| Original motif | Reverse complement motif |
| Consensus sequence: HDBAHCAWAACADWDVD | Consensus sequence: DVHWDTGTTWTGDTBDH |
 |
 |
| Motif ID: | UP00025 |
| Motif name: | Foxk1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0507431 |
Alignment:
HHHAYAAYAACAHHH
CASABACWSACACA-
| Original motif | Reverse complement motif |
| Consensus sequence: HHHAYAAYAACAHHH | Consensus sequence: HDDTGTTKTTKTHHD |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 14 |
| Similarity score: | 0.0524108 |
Alignment:
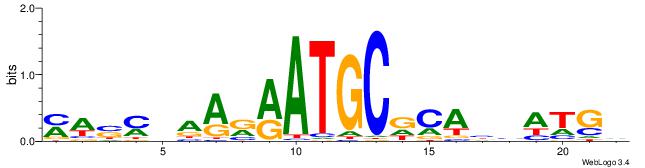
MWSMBAARRATGCDCABHATGD
CASABACWSACACA--------
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0538393 |
Alignment:
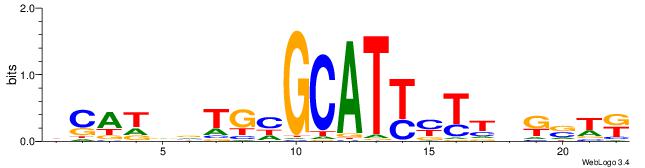
HADTBVADGATGCATCMTGMVHB
CASABACWSACACA---------
| Original motif | Reverse complement motif |
| Consensus sequence: HADTBVADGATGCATCMTGMVHB | Consensus sequence: VHVRCAYGATGCATCDTVVADTH |
 |
 |
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0538699 |
Alignment:
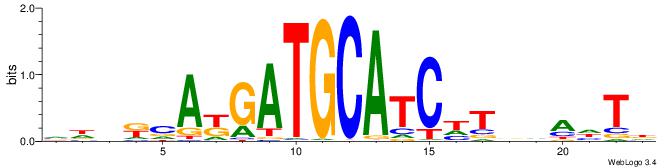
DDHHTBCCAGTGABHG
--TGTGTSWGTBTSTG
| Original motif | Reverse complement motif |
| Consensus sequence: CHVTCACTGGBADHDD | Consensus sequence: DDHHTBCCAGTGABHG |
 |
 |
| Motif ID: | UP00014 |
| Motif name: | Sox17_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0541456 |
Alignment:
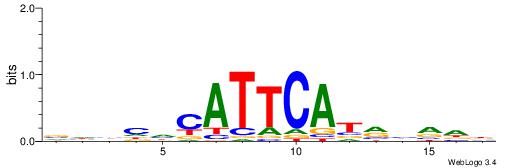
VHBHHCATTCATRHVDD
---CASABACWSACACA
| Original motif | Reverse complement motif |
| Consensus sequence: VHBHHCATTCATRHVDD | Consensus sequence: DDBDKATGAATGHDBHV |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0541653 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
---------CASABACWSACACA
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0545034 |
Alignment:
BDBBVBVGTAWCCAAGVBHBAGB
--------TGTGTSWGTBTSTG-
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
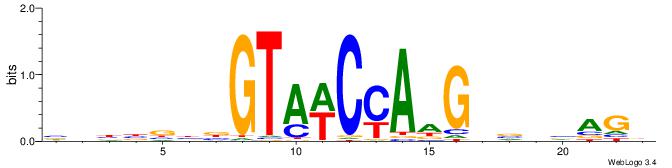 |
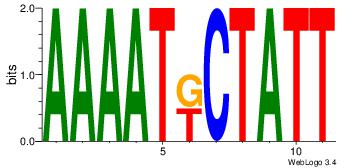
| Dataset #: 1 | Motif ID: 7 | Motif name: Motif 7 |
| Original motif | Reverse complement motif |
| Consensus sequence: AAAATKCTATT | Consensus sequence: AATAGYATTTT |
 |
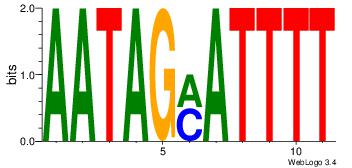 |
Best Matches for Motif ID 7 (Highest to Lowest)
| Motif ID: | UP00145 |
| Motif name: | Barhl2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0314776 |
Alignment:
HHBTTAATTGDTTYTH
-----AATAGYATTTT
| Original motif | Reverse complement motif |
| Consensus sequence: HAMAAHCAATTAABHH | Consensus sequence: HHBTTAATTGDTTYTH |
 |
 |
| Motif ID: | UP00180 |
| Motif name: | Hoxd13 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0388915 |
Alignment:
HDHHTTTTATTKKHDD
AAAATKCTATT-----
| Original motif | Reverse complement motif |
| Consensus sequence: HDHYYAATAAAAHHHH | Consensus sequence: HDHHTTTTATTKKHDD |
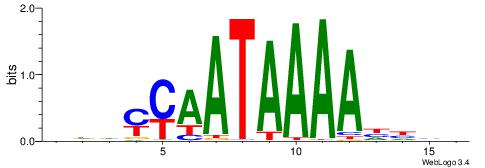 |
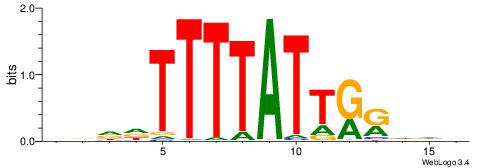 |
| Motif ID: | UP00016 |
| Motif name: | Sry_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.039378 |
Alignment:
VDDDATTATAATHDHV
-AAAATKCTATT----
| Original motif | Reverse complement motif |
| Consensus sequence: BHDHATTATAATDDDV | Consensus sequence: VDDDATTATAATHDHV |
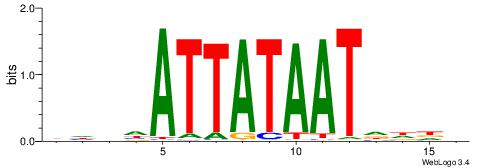 |
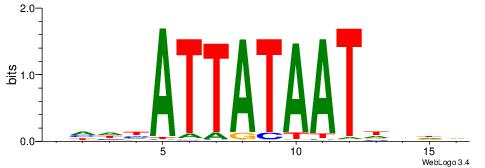 |
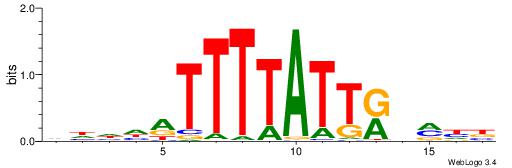
| Motif ID: | UP00121 |
| Motif name: | Hoxd10 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.039567 |
Alignment:
HDDDATTTTATTKVRDB
-AAAATKCTATT-----
| Original motif | Reverse complement motif |
| Consensus sequence: VDKVYAATAAAATDDDH | Consensus sequence: HDDDATTTTATTKVRDB |
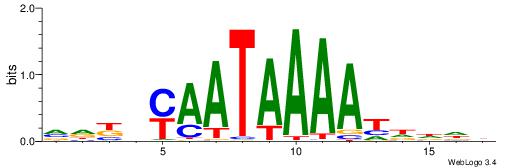 |
 |
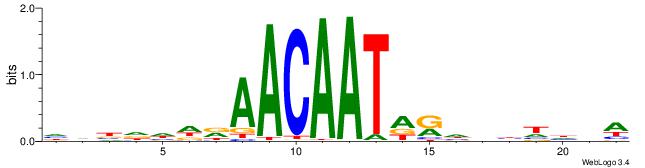
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 11 |
| Number of overlap: | 11 |
| Similarity score: | 0.0403247 |
Alignment:
WBHHHHHMDATTGTTHDHDVHH
-AAAATKCTATT----------
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
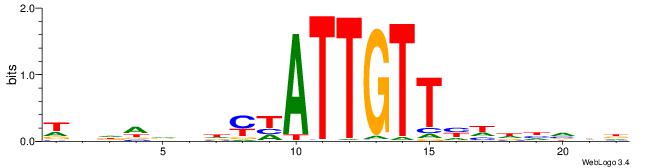 |
| Motif ID: | UP00038 |
| Motif name: | Spdef_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0405096 |
Alignment:
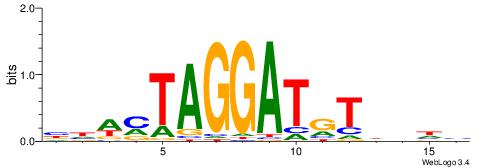
DDBHACATCCTAKWDV
---AAAATKCTATT--
| Original motif | Reverse complement motif |
| Consensus sequence: DDBHACATCCTAKWDV | Consensus sequence: VDWYTAGGATGTHVDH |
 |
 |
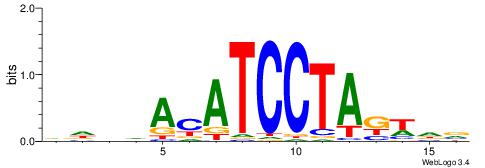
| Motif ID: | UP00191 |
| Motif name: | Pou2f2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0406764 |
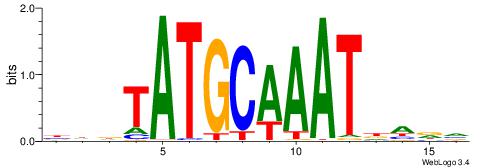
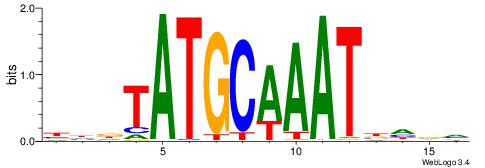
Alignment:
BHHDATTTGCATAHHV
----AATAGYATTTT-
| Original motif | Reverse complement motif |
| Consensus sequence: BHDTATGCAAATDHDV | Consensus sequence: BHHDATTTGCATAHHV |
 |
 |
| Motif ID: | UP00179 |
| Motif name: | Pou2f3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0407045 |
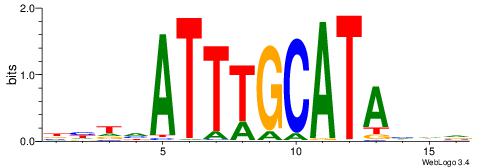
Alignment:
BHHVATTTGCATAHHV
----AATAGYATTTT-
| Original motif | Reverse complement motif |
| Consensus sequence: BHDTATGCAAATBHDV | Consensus sequence: BHHVATTTGCATAHHV |
 |
 |
| Motif ID: | UP00023 |
| Motif name: | Sox30_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0413872 |
Alignment:
HHHHMHATTGTTBDHD
AATAGYATTTT-----
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBAACAATDRHHHH | Consensus sequence: HHHHMHATTGTTBDHD |
 |
 |
| Motif ID: | UP00166 |
| Motif name: | Barhl1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0427435 |
Alignment:
HAHAACCAATTARBDH
AAAATKCTATT-----
| Original motif | Reverse complement motif |
| Consensus sequence: HAHAACCAATTARBDH | Consensus sequence: DDVKTAATTGGTTDTH |
 |
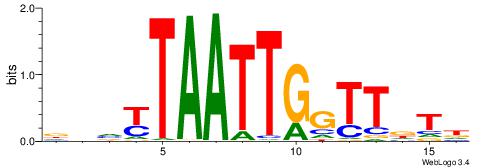 |
| Dataset #: 1 | Motif ID: 8 | Motif name: Motif 8 |
| Original motif | Reverse complement motif |
| Consensus sequence: TGAAAATSCTT | Consensus sequence: AAGSATTTTCA |
 |
 |
Best Matches for Motif ID 8 (Highest to Lowest)
| Motif ID: | UP00179 |
| Motif name: | Pou2f3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0170413 |
Alignment:
BHHVATTTGCATAHHV
AAGSATTTTCA-----
| Original motif | Reverse complement motif |
| Consensus sequence: BHDTATGCAAATBHDV | Consensus sequence: BHHVATTTGCATAHHV |
 |
 |
| Motif ID: | UP00191 |
| Motif name: | Pou2f2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0186797 |
Alignment:
BHHDATTTGCATAHHV
AAGSATTTTCA-----
| Original motif | Reverse complement motif |
| Consensus sequence: BHDTATGCAAATDHDV | Consensus sequence: BHHDATTTGCATAHHV |
 |
 |
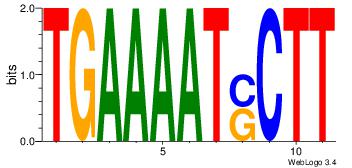
| Motif ID: | UP00044 |
| Motif name: | Mafk_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.025968 |
Alignment:
DWAAAWTGCTGACTH
TGAAAATSCTT----
| Original motif | Reverse complement motif |
| Consensus sequence: DWAAAWTGCTGACTH | Consensus sequence: HAGTCAGCAWTTTWD |
 |
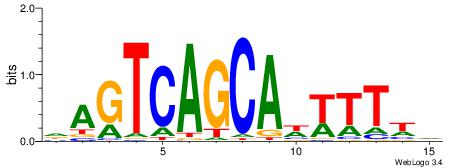 |
| Motif ID: | UP00389 |
| Motif name: | Nkx3-1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0281427 |
Alignment:
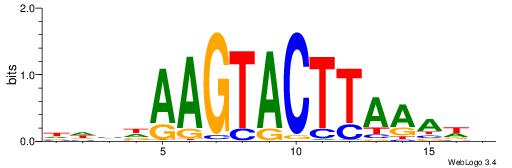
BDTTTAAGTACTTDDDD
--TGAAAATSCTT----
| Original motif | Reverse complement motif |
| Consensus sequence: DDHDAAGTACTTAAADB | Consensus sequence: BDTTTAAGTACTTDDDD |
 |
 |
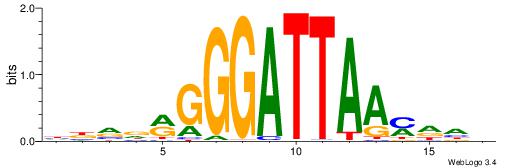
| Motif ID: | UP00153 |
| Motif name: | Pitx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.02834 |
Alignment:
HBVDRGGGATTAAMDHH
----AAGSATTTTCA--
| Original motif | Reverse complement motif |
| Consensus sequence: HBVDRGGGATTAAMDHH | Consensus sequence: HHDRTTAATCCCKHBVH |
 |
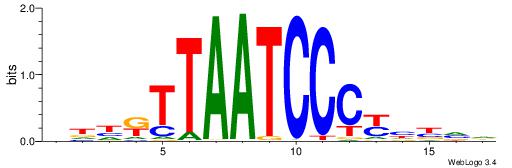 |
| Motif ID: | UP00219 |
| Motif name: | Cutl1_3494.2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0294239 |
Alignment:
DADTGATMATCABTD
---TGAAAATSCTT-
| Original motif | Reverse complement motif |
| Consensus sequence: DAVTGATRATCAHTD | Consensus sequence: DADTGATMATCABTD |
 |
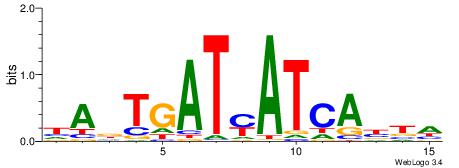 |
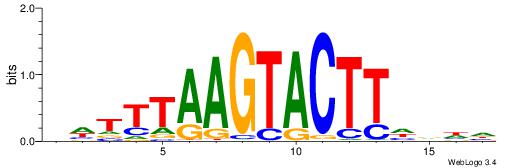
| Motif ID: | UP00119 |
| Motif name: | Nkx2-9 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0300831 |
Alignment:
HDTTBAAGTACTTDAAA
-----AAGSATTTTCA-
| Original motif | Reverse complement motif |
| Consensus sequence: TTTDAAGTACTTVAADH | Consensus sequence: HDTTBAAGTACTTDAAA |
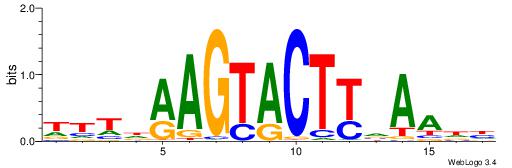 |
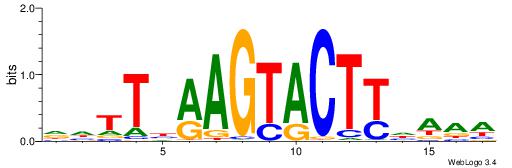 |
| Motif ID: | UP00190 |
| Motif name: | Nkx2-3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0302066 |
Alignment:
HHTTAAGTACTTAAMK
----AAGSATTTTCA-
| Original motif | Reverse complement motif |
| Consensus sequence: YYTTAAGTACTTAAHD | Consensus sequence: HHTTAAGTACTTAAMK |
 |
 |
| Motif ID: | UP00038 |
| Motif name: | Spdef_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0318364 |
Alignment:
VDWYTAGGATGTHVDH
----AAGSATTTTCA-
| Original motif | Reverse complement motif |
| Consensus sequence: DDBHACATCCTAKWDV | Consensus sequence: VDWYTAGGATGTHVDH |
 |
 |
| Motif ID: | UP00080 |
| Motif name: | Gata5_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0333422 |
Alignment:
DDHBKCTTATCAGDDDH
-AAGSATTTTCA-----
| Original motif | Reverse complement motif |
| Consensus sequence: HDDDCTGATAAGRVDDD | Consensus sequence: DDHBKCTTATCAGDDDH |
 |
 |
| Dataset #: 1 | Motif ID: 9 | Motif name: Motif 9 |
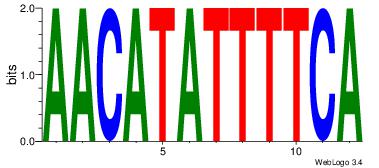
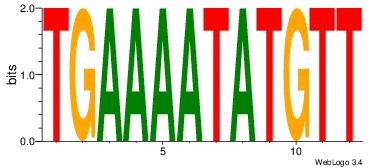
| Original motif | Reverse complement motif |
| Consensus sequence: AACATATTTTCA | Consensus sequence: TGAAAATATGTT |
 |
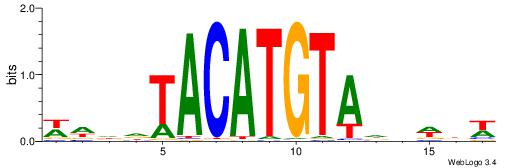
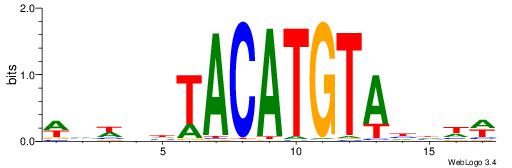
 |
Best Matches for Motif ID 9 (Highest to Lowest)
| Motif ID: | UP00250 |
| Motif name: | Irx5 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 12 |
| Similarity score: | 0.0253217 |
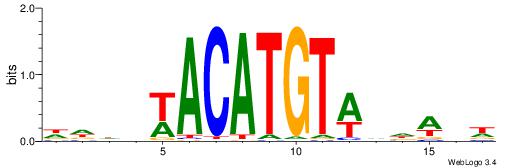
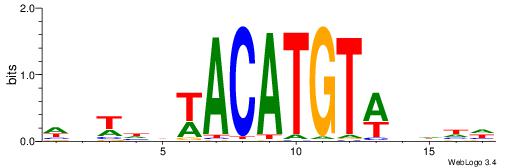
Alignment:
DDDHWACATGTWHDABW
TGAAAATATGTT-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDHWACATGTWHDABW | Consensus sequence: WVTDHWACATGTWHDDD |
 |
 |
| Motif ID: | UP00236 |
| Motif name: | Irx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0267342 |
Alignment:
WDDDTACATGTADDWBW
TGAAAATATGTT-----
| Original motif | Reverse complement motif |
| Consensus sequence: WDDDTACATGTADDWBW | Consensus sequence: WVWDDTACATGTADDDW |
 |
 |
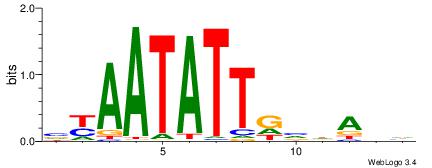
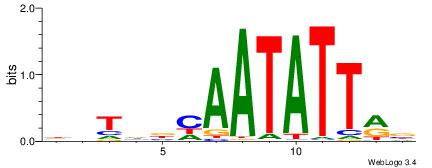
| Motif ID: | UP00059 |
| Motif name: | Arid5a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0285822 |
Alignment:
HDTDVCAATATTMV
--TGAAAATATGTT
| Original motif | Reverse complement motif |
| Consensus sequence: VYAATATTGVDADH | Consensus sequence: HDTDVCAATATTMV |
 |
 |
| Motif ID: | UP00194 |
| Motif name: | Irx4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0297419 |
Alignment:
WDHDWACATGTWAWABD
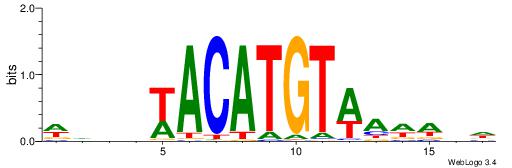
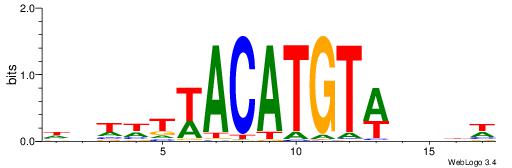
TGAAAATATGTT-----
| Original motif | Reverse complement motif |
| Consensus sequence: WDHDWACATGTWAWABD | Consensus sequence: DBTWTWACATGTWDHDW |
 |
 |
| Motif ID: | UP00223 |
| Motif name: | Irx3_0920.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
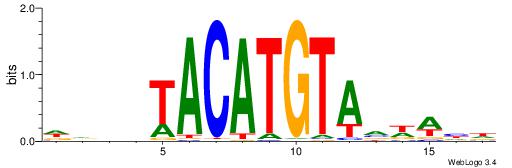
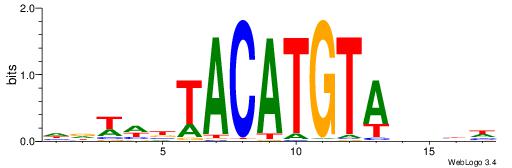
| Number of overlap: | 12 |
| Similarity score: | 0.029857 |
Alignment:
DDDDTACATGTAVWWHD
TGAAAATATGTT-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDDTACATGTAVWWHD | Consensus sequence: DDWWBTACATGTADDDD |
 |
 |
| Motif ID: | UP00223 |
| Motif name: | Irx3_2226.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 12 |
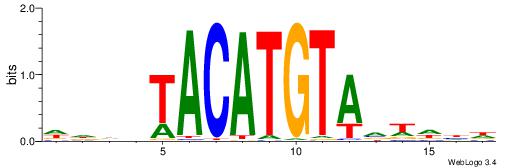
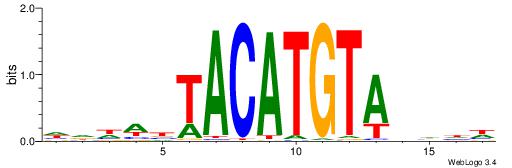
| Similarity score: | 0.0299906 |
Alignment:
DDDHTACATGTAVWDHD
TGAAAATATGTT-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDHTACATGTAVWDHD | Consensus sequence: DHDWBTACATGTAHDDD |
 |
 |
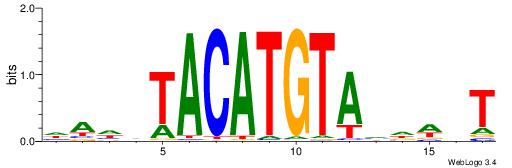
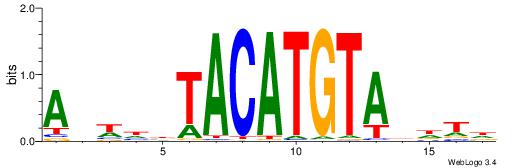
| Motif ID: | UP00150 |
| Motif name: | Irx6 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0303256 |
Alignment:
AHWDBTACATGTADHTH
TGAAAATATGTT-----
| Original motif | Reverse complement motif |
| Consensus sequence: HAHDTACATGTAVDWHT | Consensus sequence: AHWDBTACATGTADHTH |
 |
 |
| Motif ID: | UP00171 |
| Motif name: | Msx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0315999 |
Alignment:
HHVTTAATTKGTTHTV
-TGAAAATATGTT---
| Original motif | Reverse complement motif |
| Consensus sequence: VAHAACYAATTAABHH | Consensus sequence: HHVTTAATTKGTTHTV |
 |
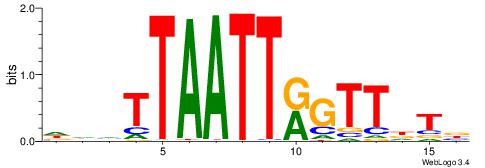 |
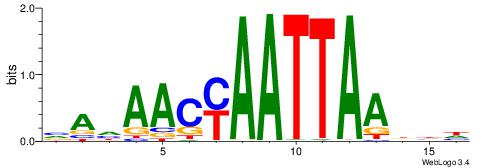
| Motif ID: | UP00146 |
| Motif name: | Pou6f1_3733.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0330849 |
Alignment:
DHDASCTCATTATVHHB
---AACATATTTTCA--
| Original motif | Reverse complement motif |
| Consensus sequence: VHHVATAATGAGSTDDH | Consensus sequence: DHDASCTCATTATVHHB |
 |
 |
| Motif ID: | UP00080 |
| Motif name: | Gata5_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0334048 |
Alignment:
HDDDCTGATAAGRVDDD
AACATATTTTCA-----
| Original motif | Reverse complement motif |
| Consensus sequence: HDDDCTGATAAGRVDDD | Consensus sequence: DDHBKCTTATCAGDDDH |
 |
 |
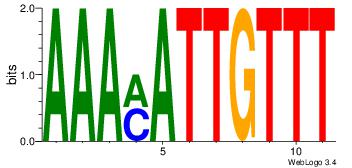
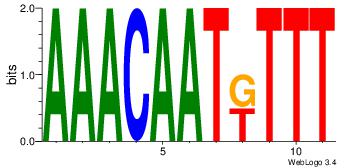
| Dataset #: 1 | Motif ID: 10 | Motif name: Motif 10 |
| Original motif | Reverse complement motif |
| Consensus sequence: AAAMATTGTTT | Consensus sequence: AAACAATYTTT |
 |
 |
Best Matches for Motif ID 10 (Highest to Lowest)
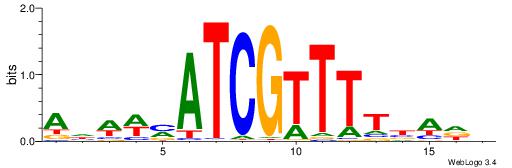
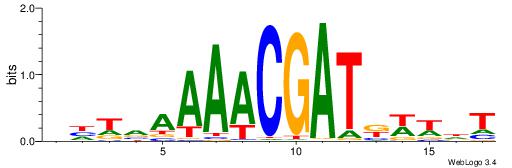
| Motif ID: | UP00114 |
| Motif name: | Homez |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0148981 |
Alignment:
AHWWMATCGTTTTBADD
-AAAMATTGTTT-----
| Original motif | Reverse complement motif |
| Consensus sequence: AHWWMATCGTTTTBADD | Consensus sequence: HDTVAAAACGATRWWHT |
 |
 |
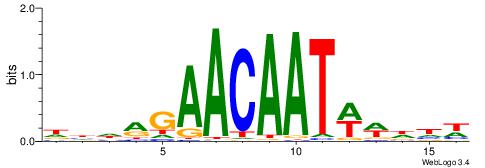
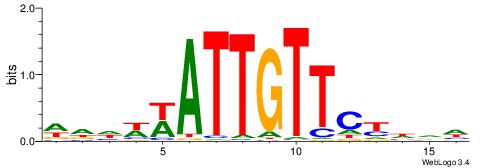
| Motif ID: | UP00096 |
| Motif name: | Sox13_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0154717 |
Alignment:
WHDWWATTGTTCKDHD
-AAAMATTGTTT----
| Original motif | Reverse complement motif |
| Consensus sequence: DHDRGAACAATWWDHW | Consensus sequence: WHDWWATTGTTCKDHD |
 |
 |
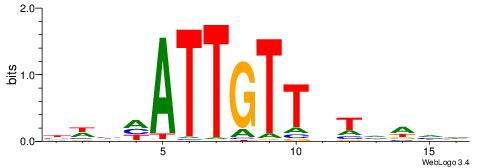
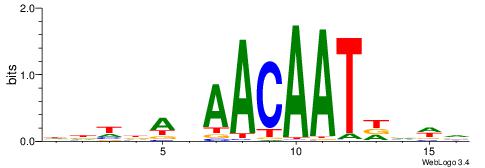
| Motif ID: | UP00064 |
| Motif name: | Sox18_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0200897 |
Alignment:
HHDHABAACAATHBWV
-----AAACAATYTTT
| Original motif | Reverse complement motif |
| Consensus sequence: BWBHATTGTTBTHDHH | Consensus sequence: HHDHABAACAATHBWV |
 |
 |
| Motif ID: | UP00062 |
| Motif name: | Sox4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.020377 |
Alignment:
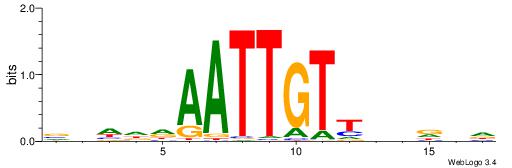
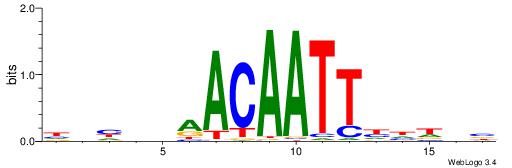
DDHVDAACAATTDDHVV
----AAACAATYTTT--
| Original motif | Reverse complement motif |
| Consensus sequence: VVHDDAATTGTTDVDDD | Consensus sequence: DDHVDAACAATTDDHVV |
 |
 |
| Motif ID: | UP00051 |
| Motif name: | Sox8_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0208116 |
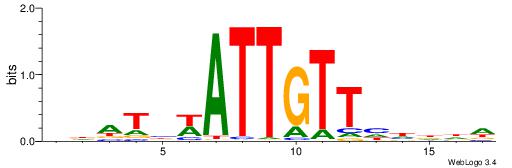
Alignment:
DDDHDAACAATWBATDD
----AAACAATYTTT--
| Original motif | Reverse complement motif |
| Consensus sequence: DDATBWATTGTTHHDDD | Consensus sequence: DDDHDAACAATWBATDD |
 |
 |
| Motif ID: | UP00023 |
| Motif name: | Sox30_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0235407 |
Alignment:
HHHHMHATTGTTBDHD
--AAAMATTGTTT---
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBAACAATDRHHHH | Consensus sequence: HHHHMHATTGTTBDHD |
 |
 |
| Motif ID: | UP00091 |
| Motif name: | Sox5_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0236834 |
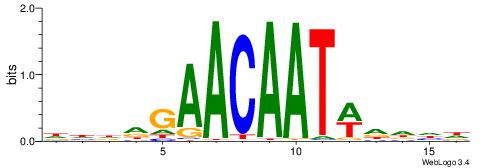
Alignment:
DHDDGAACAATWDDHD
----AAACAATYTTT-
| Original motif | Reverse complement motif |
| Consensus sequence: DHDDGAACAATWDDHD | Consensus sequence: DHDDWATTGTTCDDHD |
 |
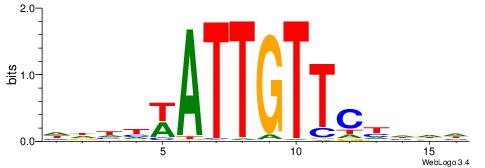 |
| Motif ID: | UP00014 |
| Motif name: | Sox17_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0256157 |
Alignment:
VDDAACAATTVAWHD
--AAACAATYTTT--
| Original motif | Reverse complement motif |
| Consensus sequence: VDDAACAATTVAWHD | Consensus sequence: DDWTBAATTGTTDDB |
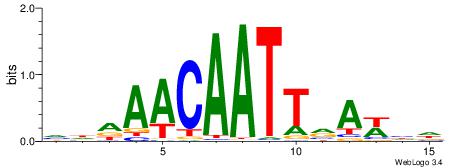 |
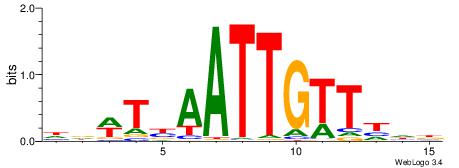 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0264034 |
Alignment:
WBHHHHHMDATTGTTHDHDVHH
-----AAAMATTGTTT------
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
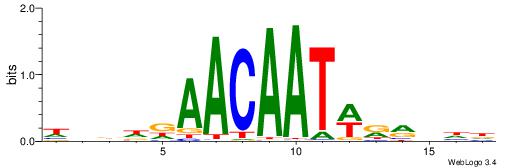
| Motif ID: | UP00075 |
| Motif name: | Sox15_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0265966 |
Alignment:
DHVKMWATTGTTHDHDH
--AAAMATTGTTT----
| Original motif | Reverse complement motif |
| Consensus sequence: HDDDDAACAATWRRBHD | Consensus sequence: DHVKMWATTGTTHDHDH |
 |
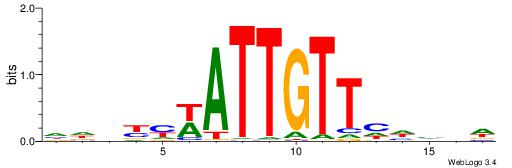 |
| Dataset #: 1 | Motif ID: 11 | Motif name: Motif 11 |
| Original motif | Reverse complement motif |
| Consensus sequence: AAMATGTTTMAA | Consensus sequence: TTYAAACATYTT |
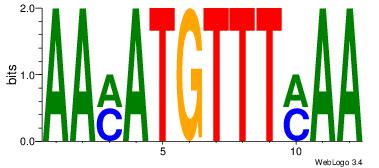 |
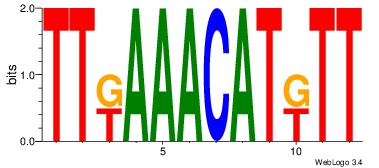 |
Best Matches for Motif ID 11 (Highest to Lowest)
| Motif ID: | UP00250 |
| Motif name: | Irx5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 12 |
| Similarity score: | 0.00569393 |
Alignment:
WVTDHWACATGTWHDDD
-----AAMATGTTTMAA
| Original motif | Reverse complement motif |
| Consensus sequence: DDDHWACATGTWHDABW | Consensus sequence: WVTDHWACATGTWHDDD |
 |
 |
| Motif ID: | UP00236 |
| Motif name: | Irx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.00753568 |
Alignment:
WVWDDTACATGTADDDW
-----AAMATGTTTMAA
| Original motif | Reverse complement motif |
| Consensus sequence: WDDDTACATGTADDWBW | Consensus sequence: WVWDDTACATGTADDDW |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 0.00927938 |
Alignment:
VVVWTTGTTTAMATTWD
-AAMATGTTTMAA----
| Original motif | Reverse complement motif |
| Consensus sequence: DWAATRTAAACAAWVVB | Consensus sequence: VVVWTTGTTTAMATTWD |
 |
 |
| Motif ID: | UP00150 |
| Motif name: | Irx6 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0104366 |
Alignment:
HAHDTACATGTAVDWHT
----AAMATGTTTMAA-
| Original motif | Reverse complement motif |
| Consensus sequence: HAHDTACATGTAVDWHT | Consensus sequence: AHWDBTACATGTADHTH |
 |
 |
| Motif ID: | UP00194 |
| Motif name: | Irx4 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0110074 |
Alignment:
WDHDWACATGTWAWABD
----AAMATGTTTMAA-
| Original motif | Reverse complement motif |
| Consensus sequence: WDHDWACATGTWAWABD | Consensus sequence: DBTWTWACATGTWDHDW |
 |
 |
| Motif ID: | UP00068 |
| Motif name: | Eomes_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0113539 |
Alignment:
HHHTTTCACACCTDHBV
----TTYAAACATYTT-
| Original motif | Reverse complement motif |
| Consensus sequence: BVHDAGGTGTGAAAHHH | Consensus sequence: HHHTTTCACACCTDHBV |
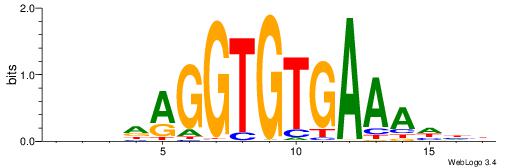 |
 |
| Motif ID: | UP00223 |
| Motif name: | Irx3_2226.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 12 |
| Similarity score: | 0.0118824 |
Alignment:
DHDWBTACATGTAHDDD
-----AAMATGTTTMAA
| Original motif | Reverse complement motif |
| Consensus sequence: DDDHTACATGTAVWDHD | Consensus sequence: DHDWBTACATGTAHDDD |
 |
 |
| Motif ID: | UP00223 |
| Motif name: | Irx3_0920.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0132149 |
Alignment:
DDDDTACATGTAVWWHD
-----AAMATGTTTMAA
| Original motif | Reverse complement motif |
| Consensus sequence: DDDDTACATGTAVWWHD | Consensus sequence: DDWWBTACATGTADDDD |
 |
 |
| Motif ID: | UP00025 |
| Motif name: | Foxk1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 0.0142458 |
Alignment:
BBVTTTGTTTACATTDD
-AAMATGTTTMAA----
| Original motif | Reverse complement motif |
| Consensus sequence: DDAATGTAAACAAAVVB | Consensus sequence: BBVTTTGTTTACATTDD |
 |
 |
| Motif ID: | UP00039 |
| Motif name: | Foxj3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 0.0210343 |
Alignment:
DVVTTTGTTTACDTHDH
-AAMATGTTTMAA----
| Original motif | Reverse complement motif |
| Consensus sequence: HDHADGTAAACAAAVVH | Consensus sequence: DVVTTTGTTTACDTHDH |
 |
 |
| Dataset #: 1 | Motif ID: 12 | Motif name: Motif 12 |
| Original motif | Reverse complement motif |
| Consensus sequence: ACAACACATTT | Consensus sequence: AAATGTGTTGT |
 |
 |
Best Matches for Motif ID 12 (Highest to Lowest)
| Motif ID: | UP00033 |
| Motif name: | Zfp410_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0181 |
Alignment:
DVDDATGGGATGKMDDV
--AAATGTGTTGT----
| Original motif | Reverse complement motif |
| Consensus sequence: DVDDATGGGATGKMDDV | Consensus sequence: BDDYYCATCCCATDDBD |
 |
 |
| Motif ID: | UP00068 |
| Motif name: | Eomes_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0202807 |
Alignment:
VBVDAGGTGTYGVBBV
--AAATGTGTTGT---
| Original motif | Reverse complement motif |
| Consensus sequence: VBVDAGGTGTYGVBBV | Consensus sequence: VVBVCKACACCTHVBV |
 |
 |
| Motif ID: | UP00171 |
| Motif name: | Msx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0211208 |
Alignment:
HHVTTAATTKGTTHTV
----AAATGTGTTGT-
| Original motif | Reverse complement motif |
| Consensus sequence: VAHAACYAATTAABHH | Consensus sequence: HHVTTAATTKGTTHTV |
 |
 |
| Motif ID: | UP00072 |
| Motif name: | IRC900814_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0216511 |
Alignment:
BHDWTTTGTCGTAHDD
-AAATGTGTTGT----
| Original motif | Reverse complement motif |
| Consensus sequence: DDHTACGACAAAWDDB | Consensus sequence: BHDWTTTGTCGTAHDD |
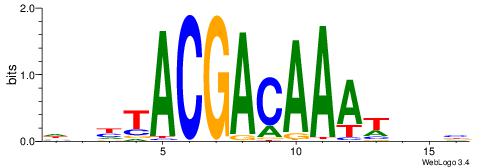 |
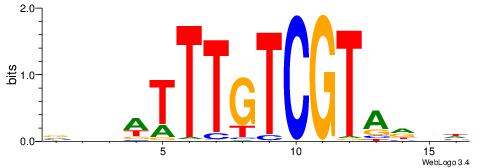 |
| Motif ID: | UP00166 |
| Motif name: | Barhl1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0230664 |
Alignment:
DDVKTAATTGGTTDTH
----AAATGTGTTGT-
| Original motif | Reverse complement motif |
| Consensus sequence: HAHAACCAATTARBDH | Consensus sequence: DDVKTAATTGGTTDTH |
 |
 |
| Motif ID: | UP00045 |
| Motif name: | Mafb_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.0231414 |
Alignment:
HAAWDTGCTGACDWARH
AAATGTGTTGT------
| Original motif | Reverse complement motif |
| Consensus sequence: HAAWDTGCTGACDWARH | Consensus sequence: DMTWDGTCAGCADWTTH |
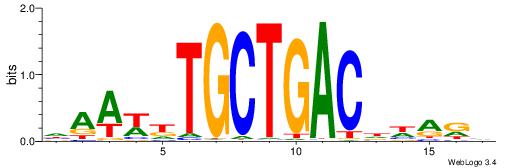 |
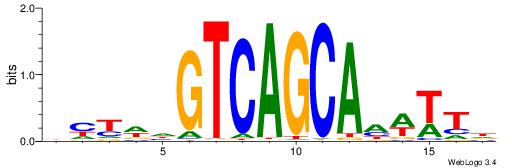 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0270722 |
Alignment:
YBVDMGTGGGTGGTCKVVBVBBT
---AAATGTGTTGT---------
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
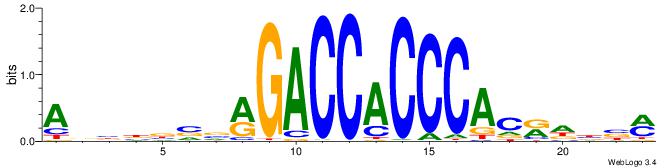 |
 |
| Motif ID: | UP00236 |
| Motif name: | Irx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0272203 |
Alignment:
WVWDDTACATGTADDDW
------AAATGTGTTGT
| Original motif | Reverse complement motif |
| Consensus sequence: WDDDTACATGTADDWBW | Consensus sequence: WVWDDTACATGTADDDW |
 |
 |
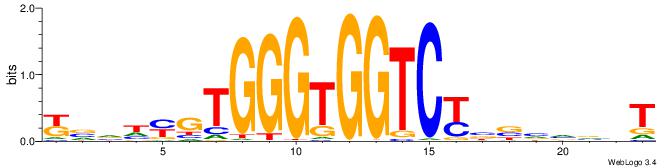
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 11 |
| Similarity score: | 0.0274347 |
Alignment:
DTVBDDRGACCACCCAGDWDGBH
--------ACAACACATTT----
| Original motif | Reverse complement motif |
| Consensus sequence: DTVBDDRGACCACCCAGDWDGBH | Consensus sequence: DBCDWHCTGGGTGGTCMDHBBAH |
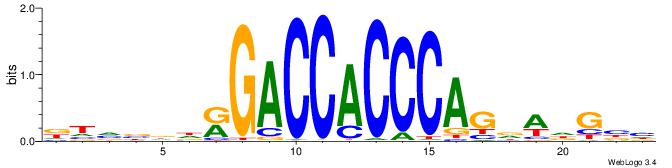 |
 |
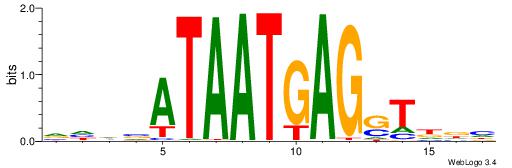
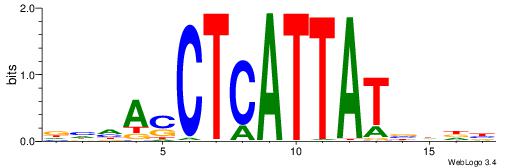
| Motif ID: | UP00146 |
| Motif name: | Pou6f1_3733.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0275808 |
Alignment:
VHHVATAATGAGSTDDH
-----AAATGTGTTGT-
| Original motif | Reverse complement motif |
| Consensus sequence: VHHVATAATGAGSTDDH | Consensus sequence: DHDASCTCATTATVHHB |
 |
 |
| Dataset #: 1 | Motif ID: 13 | Motif name: Motif 13 |
| Original motif | Reverse complement motif |
| Consensus sequence: AAWATRTATTT | Consensus sequence: AAATAKATWTT |
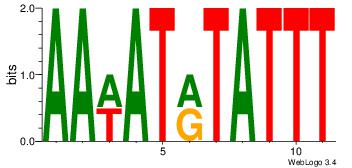 |
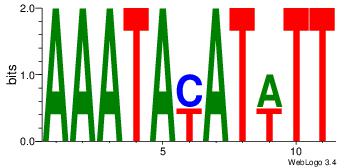 |
Best Matches for Motif ID 13 (Highest to Lowest)
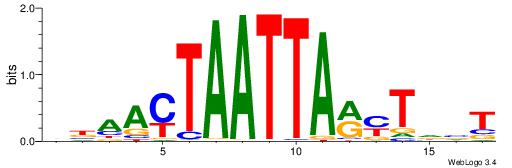
| Motif ID: | UP00266 |
| Motif name: | Prrx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.0142423 |
Alignment:
AVDAKKTAATTAGTTVV
AAWATRTATTT------
| Original motif | Reverse complement motif |
| Consensus sequence: VBAACTAATTARYTDVT | Consensus sequence: AVDAKKTAATTAGTTVV |
 |
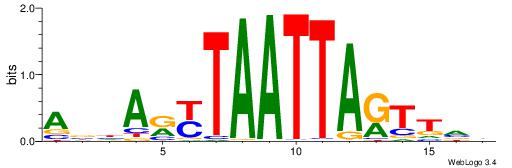 |
| Motif ID: | UP00150 |
| Motif name: | Irx6 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.020069 |
Alignment:
AHWDBTACATGTADHTH
-----AAWATRTATTT-
| Original motif | Reverse complement motif |
| Consensus sequence: HAHDTACATGTAVDWHT | Consensus sequence: AHWDBTACATGTADHTH |
 |
 |
| Motif ID: | UP00236 |
| Motif name: | Irx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0214423 |
Alignment:
WVWDDTACATGTADDDW
-----AAWATRTATTT-
| Original motif | Reverse complement motif |
| Consensus sequence: WDDDTACATGTADDWBW | Consensus sequence: WVWDDTACATGTADDDW |
 |
 |
| Motif ID: | UP00261 |
| Motif name: | Lhx4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0230284 |
Alignment:
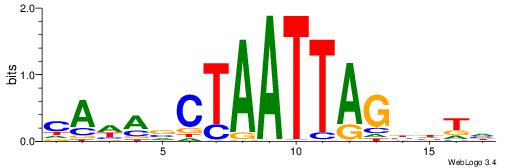
DADVCTAATTAGBTWTG
------AAATAKATWTT
| Original motif | Reverse complement motif |
| Consensus sequence: DADVCTAATTAGBTWTG | Consensus sequence: CAWABCTAATTAGBDTD |
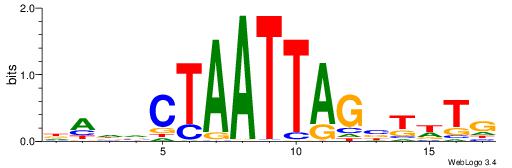 |
 |
| Motif ID: | UP00170 |
| Motif name: | Isl2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0233945 |
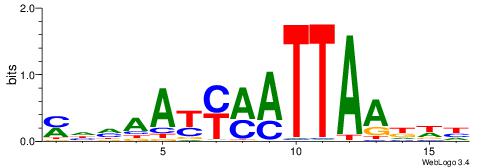
Alignment:
HWDTTAATYKMTTHHR
----AAATAKATWTT-
| Original motif | Reverse complement motif |
| Consensus sequence: MHHAAYYMATTAADWH | Consensus sequence: HWDTTAATYKMTTHHR |
 |
 |
| Motif ID: | UP00212 |
| Motif name: | Lhx5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.0236276 |
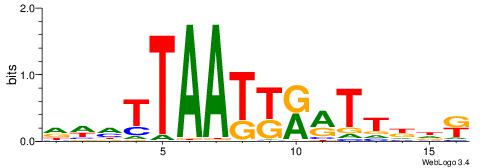
Alignment:
VDHWTTTAATTAATHHV
AAWATRTATTT------
| Original motif | Reverse complement motif |
| Consensus sequence: VDHATTAATTAAAWHHB | Consensus sequence: VDHWTTTAATTAATHHV |
 |
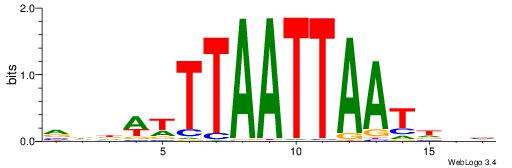 |
| Motif ID: | UP00108 |
| Motif name: | Alx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.0243258 |
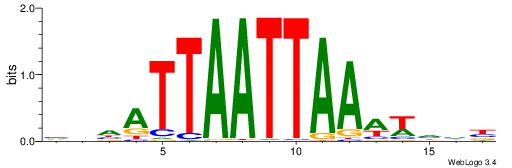
Alignment:
VHVAKMTAATTAKTHBW
AAWATRTATTT------
| Original motif | Reverse complement motif |
| Consensus sequence: WVHAYTAATTARYTVHV | Consensus sequence: VHVAKMTAATTAKTHBW |
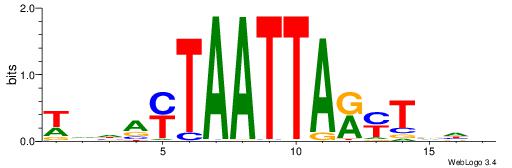 |
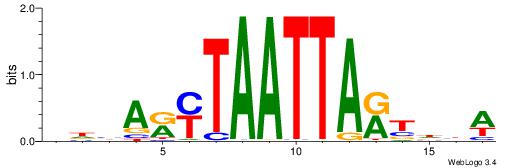 |
| Motif ID: | UP00171 |
| Motif name: | Msx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0254704 |
Alignment:
HHVTTAATTKGTTHTV
----AAATAKATWTT-
| Original motif | Reverse complement motif |
| Consensus sequence: VAHAACYAATTAABHH | Consensus sequence: HHVTTAATTKGTTHTV |
 |
 |
| Motif ID: | UP00250 |
| Motif name: | Irx5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0259267 |
Alignment:
WVTDHWACATGTWHDDD
-----AAWATRTATTT-
| Original motif | Reverse complement motif |
| Consensus sequence: DDDHWACATGTWHDABW | Consensus sequence: WVTDHWACATGTWHDDD |
 |
 |
| Motif ID: | UP00223 |
| Motif name: | Irx3_2226.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0263428 |
Alignment:
DDDHTACATGTAVWDHD
----AAWATRTATTT--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDHTACATGTAVWDHD | Consensus sequence: DHDWBTACATGTAHDDD |
 |
 |
| Dataset #: 1 | Motif ID: 14 | Motif name: Motif 14 |
| Original motif | Reverse complement motif |
| Consensus sequence: TAATKATATAA | Consensus sequence: TTATATYATTA |
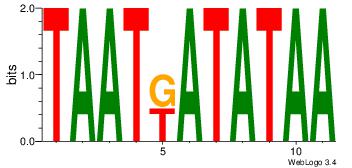 |
 |
Best Matches for Motif ID 14 (Highest to Lowest)
| Motif ID: | UP00264 |
| Motif name: | Hoxa1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0229661 |
Alignment:
MBDRTAATTAGCTHAD
----TAATKATATAA-
| Original motif | Reverse complement motif |
| Consensus sequence: HTDAGCTAATTAMHBY | Consensus sequence: MBDRTAATTAGCTHAD |
 |
 |
| Motif ID: | UP00129 |
| Motif name: | Pou3f1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.02767 |
Alignment:
DVBTAATTAATTAABTH
---TAATKATATAA---
| Original motif | Reverse complement motif |
| Consensus sequence: DVBTAATTAATTAABTH | Consensus sequence: DAVTTAATTAATTAVBD |
 |
 |
| Motif ID: | UP00212 |
| Motif name: | Lhx5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0295456 |
Alignment:
VDHATTAATTAAAWHHB
-----TAATKATATAA-
| Original motif | Reverse complement motif |
| Consensus sequence: VDHATTAATTAAAWHHB | Consensus sequence: VDHWTTTAATTAATHHV |
 |
 |
| Motif ID: | UP00130 |
| Motif name: | Lhx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0295647 |
Alignment:
VDAATTAATTAAWWHHB
-----TAATKATATAA-
| Original motif | Reverse complement motif |
| Consensus sequence: VDAATTAATTAAWWHHB | Consensus sequence: VHHWWTTAATTAATTDV |
 |
 |
| Motif ID: | UP00247 |
| Motif name: | Pax4 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0309932 |
Alignment:
DDSBKMTAATTAKDBVW
------TAATKATATAA
| Original motif | Reverse complement motif |
| Consensus sequence: WVVDYTAATTARYBSDH | Consensus sequence: DDSBKMTAATTAKDBVW |
 |
 |
| Motif ID: | UP00108 |
| Motif name: | Alx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.0319078 |
Alignment:
VHVAKMTAATTAKTHBW
------TAATKATATAA
| Original motif | Reverse complement motif |
| Consensus sequence: WVHAYTAATTARYTVHV | Consensus sequence: VHVAKMTAATTAKTHBW |
 |
 |
| Motif ID: | UP00105 |
| Motif name: | Pou3f4 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0323543 |
Alignment:
DVHTAATTAATTADDDB
---TAATKATATAA---
| Original motif | Reverse complement motif |
| Consensus sequence: DVHTAATTAATTADDDB | Consensus sequence: BDDDTAATTAATTAHBD |
 |
 |
| Motif ID: | UP00202 |
| Motif name: | Dlx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0327446 |
Alignment:
MDTAATTAHMBCHB
--TAATKATATAA-
| Original motif | Reverse complement motif |
| Consensus sequence: BHGVRDTAATTADY | Consensus sequence: MDTAATTAHMBCHB |
 |
 |
| Motif ID: | UP00128 |
| Motif name: | Pou3f2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0328898 |
Alignment:
HHDVMTAATTAATTDDV
-----TAATKATATAA-
| Original motif | Reverse complement motif |
| Consensus sequence: VDDAATTAATTARBDHD | Consensus sequence: HHDVMTAATTAATTDDV |
 |
 |
| Motif ID: | UP00266 |
| Motif name: | Prrx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.0331473 |
Alignment:
AVDAKKTAATTAGTTVV
------TAATKATATAA
| Original motif | Reverse complement motif |
| Consensus sequence: VBAACTAATTARYTDVT | Consensus sequence: AVDAKKTAATTAGTTVV |
 |
 |
| Dataset #: 1 | Motif ID: 15 | Motif name: Motif 15 |
| Original motif | Reverse complement motif |
| Consensus sequence: AAAGRTMAAACTA | Consensus sequence: TAGTTTYAKCTTT |
 |
 |
Best Matches for Motif ID 15 (Highest to Lowest)
| Motif ID: | UP00074 |
| Motif name: | Isgf3g_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0359564 |
Alignment:
TDAGTTTCGWTTKDV
-TAGTTTYAKCTTT-
| Original motif | Reverse complement motif |
| Consensus sequence: VDRAAWCGAAACTDA | Consensus sequence: TDAGTTTCGWTTKDV |
 |
 |
| Motif ID: | UP00083 |
| Motif name: | Tcf7l2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 13 |
| Similarity score: | 0.0371142 |
Alignment:
BDDASATCAAAGGMDDD
-AAAGRTMAAACTA---
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATSTDDV | Consensus sequence: BDDASATCAAAGGMDDD |
 |
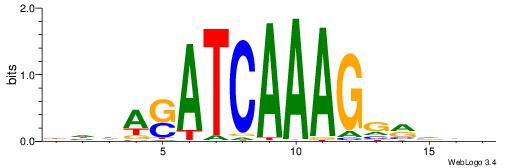 |
| Motif ID: | UP00054 |
| Motif name: | Tcf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 13 |
| Similarity score: | 0.0377174 |
Alignment:
BHHASATCAAAGGAHHH
-AAAGRTMAAACTA---
| Original motif | Reverse complement motif |
| Consensus sequence: BHHASATCAAAGGAHHH | Consensus sequence: HHHTCCTTTGATSTHHV |
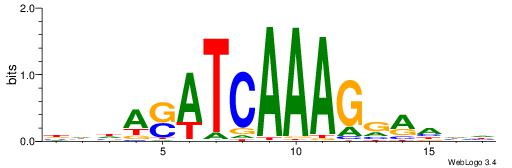 |
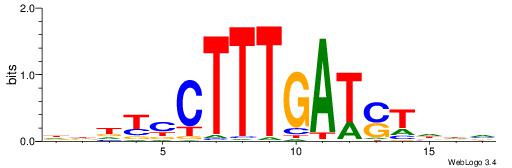 |
| Motif ID: | UP00058 |
| Motif name: | Tcf3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 13 |
| Similarity score: | 0.0394685 |
Alignment:
HHHASATCAAAGVRHHH
-AAAGRTMAAACTA---
| Original motif | Reverse complement motif |
| Consensus sequence: HHHASATCAAAGVRHHH | Consensus sequence: HHHKVCTTTGATSTHHH |
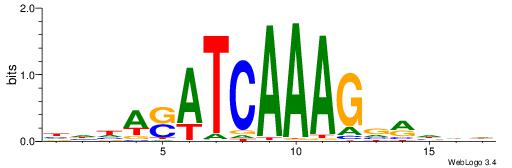 |
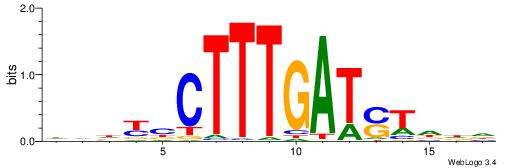 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 13 |
| Similarity score: | 0.0406011 |
Alignment:
HBHDVHDTCTTATCTHTDDHDV
----TAGTTTYAKCTTT-----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
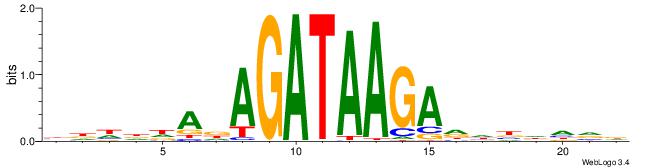 |
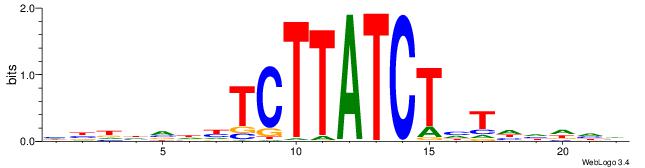 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 13 |
| Similarity score: | 0.0411941 |
Alignment:
VDDAGATCAAAGGKDDD
-AAAGRTMAAACTA---
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
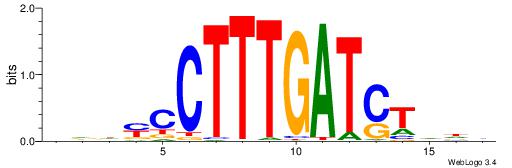 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.042799 |
Alignment:
DHDBCAAGGTCAHVBDH
----AAAGRTMAAACTA
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBCAAGGTCAHVBDH | Consensus sequence: HHBVHTGACCTTGVDHD |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0473603 |
Alignment:
DWAATRTAAACAAWVVB
-AAAGRTMAAACTA---
| Original motif | Reverse complement motif |
| Consensus sequence: DWAATRTAAACAAWVVB | Consensus sequence: VVVWTTGTTTAMATTWD |
 |
 |
| Motif ID: | UP00040 |
| Motif name: | Irf5_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0474515 |
Alignment:
BDKGTTTCGKTWDDD
-TAGTTTYAKCTTT-
| Original motif | Reverse complement motif |
| Consensus sequence: DDDWAYCGAAACYDV | Consensus sequence: BDKGTTTCGKTWDDD |
 |
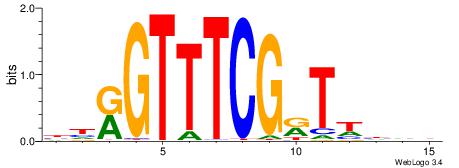 |
| Motif ID: | UP00199 |
| Motif name: | Six4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0477024 |
Alignment:
BBVHVGGTGTCATHDDT
----TAGTTTYAKCTTT
| Original motif | Reverse complement motif |
| Consensus sequence: ADDHATGACACCBHBBV | Consensus sequence: BBVHVGGTGTCATHDDT |
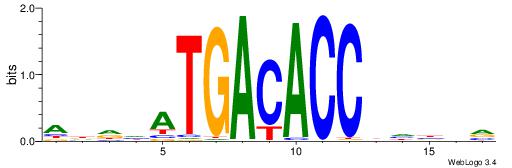 |
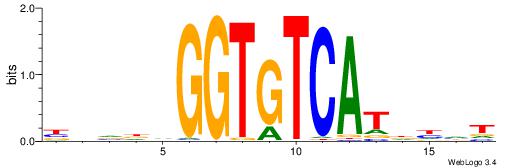 |
| Dataset #: 1 | Motif ID: 16 | Motif name: Motif 16 |
| Original motif | Reverse complement motif |
| Consensus sequence: TCTKTGTCAAWA | Consensus sequence: TWTTGACAYAGA |
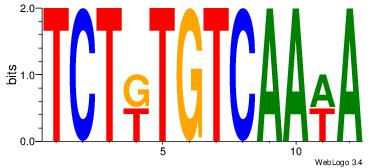 |
 |
Best Matches for Motif ID 16 (Highest to Lowest)
| Motif ID: | UP00258 |
| Motif name: | Tgif2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.00294541 |
Alignment:
VBWTTGACAGCTVBTT
-TWTTGACAYAGA---
| Original motif | Reverse complement motif |
| Consensus sequence: AABBAGCTGTCAAWVV | Consensus sequence: VBWTTGACAGCTVBTT |
 |
 |
| Motif ID: | UP00210 |
| Motif name: | Mrg2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.00709656 |
Alignment:
DBHTTGACAGSTVDTD
-TWTTGACAYAGA---
| Original motif | Reverse complement motif |
| Consensus sequence: DADBASCTGTCAAHVH | Consensus sequence: DBHTTGACAGSTVDTD |
 |
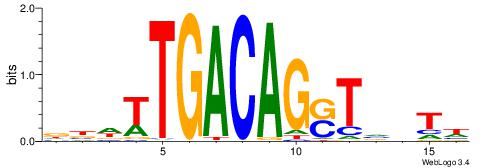 |
| Motif ID: | UP00186 |
| Motif name: | Meis1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.00883873 |
Alignment:
WAVBASCTGTCAAWVH
---TCTKTGTCAAWA-
| Original motif | Reverse complement motif |
| Consensus sequence: WAVBASCTGTCAAWVH | Consensus sequence: DBWTTGACAGSTBVTW |
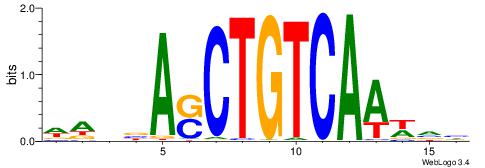 |
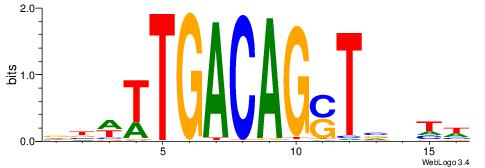 |
| Motif ID: | UP00226 |
| Motif name: | Mrg1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.00900015 |
Alignment:
WAVBASCTGTCAAWHB
---TCTKTGTCAAWA-
| Original motif | Reverse complement motif |
| Consensus sequence: WAVBASCTGTCAAWHB | Consensus sequence: BHWTTGACAGSTBBTW |
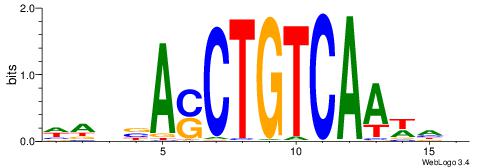 |
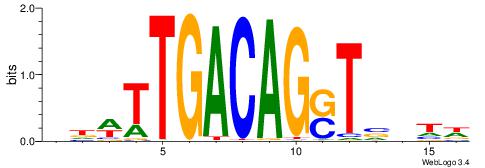 |
| Motif ID: | UP00122 |
| Motif name: | Tgif1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0128037 |
Alignment:
DABWWTGACAGCTVVBD
--TWTTGACAYAGA---
| Original motif | Reverse complement motif |
| Consensus sequence: DABWWTGACAGCTVVBD | Consensus sequence: DBVVAGCTGTCAWWVTH |
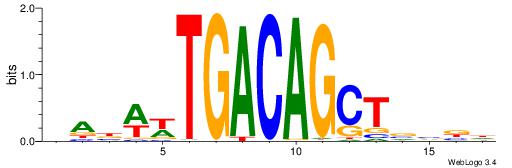 |
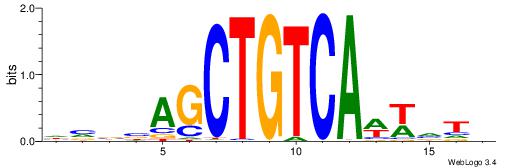 |
| Motif ID: | UP00073 |
| Motif name: | Foxa2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0145754 |
Alignment:
BDVTTTKTTTACWTWHH
-----TWTTGACAYAGA
| Original motif | Reverse complement motif |
| Consensus sequence: HHWAWGTAAAYAAAVDB | Consensus sequence: BDVTTTKTTTACWTWHH |
 |
 |
| Motif ID: | UP00205 |
| Motif name: | Pknox2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0147372 |
Alignment:
VHATTGACAGSKSVTH
-TWTTGACAYAGA---
| Original motif | Reverse complement motif |
| Consensus sequence: HAVSRSCTGTCAATHB | Consensus sequence: VHATTGACAGSKSVTH |
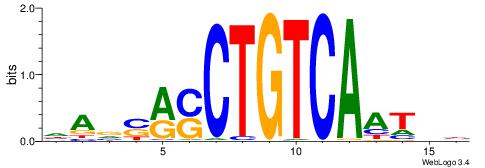 |
 |
| Motif ID: | UP00203 |
| Motif name: | Pknox1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0147793 |
Alignment:
DRVSASCTGTCAAWVV
---TCTKTGTCAAWA-
| Original motif | Reverse complement motif |
| Consensus sequence: DRVSASCTGTCAAWVV | Consensus sequence: VVWTTGACAGSTSBKD |
 |
 |
| Motif ID: | UP00045 |
| Motif name: | Mafb_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0150474 |
Alignment:
DMTWDGTCAGCADWTTH
-----TWTTGACAYAGA
| Original motif | Reverse complement motif |
| Consensus sequence: HAAWDTGCTGACDWARH | Consensus sequence: DMTWDGTCAGCADWTTH |
 |
 |
| Motif ID: | UP00103 |
| Motif name: | Jundm2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0167106 |
Alignment:
HDDKTGASTCAKHRDB
-TWTTGACAYAGA---
| Original motif | Reverse complement motif |
| Consensus sequence: VDKDRTGASTCAYHDH | Consensus sequence: HDDKTGASTCAKHRDB |
 |
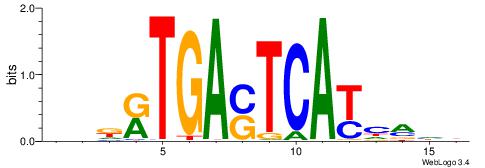 |
| Dataset #: 1 | Motif ID: 17 | Motif name: Motif 17 |
| Original motif | Reverse complement motif |
| Consensus sequence: CTTCTGACCTC | Consensus sequence: GAGGTCAGAAG |
 |
 |
Best Matches for Motif ID 17 (Highest to Lowest)
| Motif ID: | UP00079 |
| Motif name: | Esrra_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0 |
Alignment:
VKBBAGGGGTCAHDBBH
-----GAGGTCAGAAG-
| Original motif | Reverse complement motif |
| Consensus sequence: VKBBAGGGGTCAHDBBH | Consensus sequence: DBBHHTGACCCCTBBYV |
 |
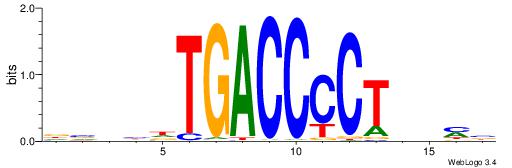 |
| Motif ID: | UP00045 |
| Motif name: | Mafb_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.00315419 |
Alignment:
DMTWDGTCAGCADWTTH
--GAGGTCAGAAG----
| Original motif | Reverse complement motif |
| Consensus sequence: HAAWDTGCTGACDWARH | Consensus sequence: DMTWDGTCAGCADWTTH |
 |
 |
| Motif ID: | UP00009 |
| Motif name: | Nr2f2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.00435047 |
Alignment:
BHVDTGACCTTTDVDD
-----GAGGTCAGAAG
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCAHVHB | Consensus sequence: BHVDTGACCTTTDVDD |
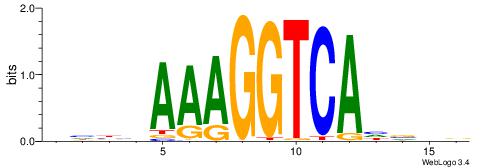 |
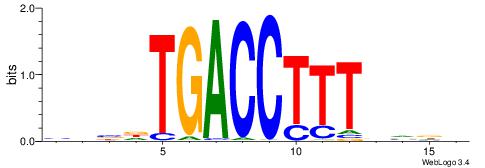 |
| Motif ID: | UP00048 |
| Motif name: | Rara |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.00499607 |
Alignment:
HHVGTGACCTTTDVDD
-----GAGGTCAGAAG
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCACVHD | Consensus sequence: HHVGTGACCTTTDVDD |
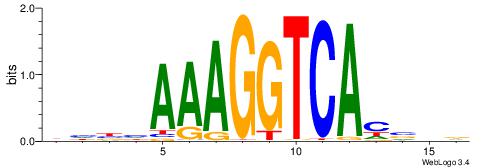 |
 |
| Motif ID: | UP00053 |
| Motif name: | Rxra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.00503678 |
Alignment:
DBHBDTGACCCCDHVHB
-CTTCTGACCTC-----
| Original motif | Reverse complement motif |
| Consensus sequence: DBHBDTGACCCCDHVHB | Consensus sequence: VHBHDGGGGTCAHBHBD |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.00855607 |
Alignment:
HHBVHTGACCTTGVDHD
-CTTCTGACCTC-----
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBCAAGGTCAHVBDH | Consensus sequence: HHBVHTGACCTTGVDHD |
 |
 |
| Motif ID: | UP00020 |
| Motif name: | Atf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0106472 |
Alignment:
DMDMTGACGTCAKHBD
-----GAGGTCAGAAG
| Original motif | Reverse complement motif |
| Consensus sequence: DBDRTGACGTCAYHRD | Consensus sequence: DMDMTGACGTCAKHBD |
 |
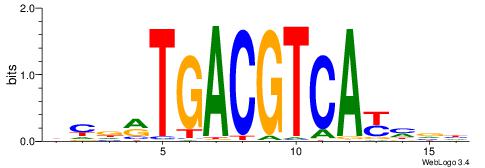 |
| Motif ID: | UP00066 |
| Motif name: | Hnf4a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0108194 |
Alignment:
BDBVMGGGGTCAABHDV
-----GAGGTCAGAAG-
| Original motif | Reverse complement motif |
| Consensus sequence: BDBVMGGGGTCAABHDV | Consensus sequence: BHHVTTGACCCCYVVDB |
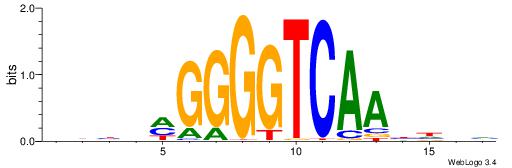 |
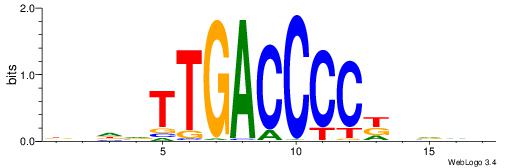 |
| Motif ID: | UP00068 |
| Motif name: | Eomes_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0131175 |
Alignment:
BVHDAGGTGTGAAAHHH
---GAGGTCAGAAG---
| Original motif | Reverse complement motif |
| Consensus sequence: BVHDAGGTGTGAAAHHH | Consensus sequence: HHHTTTCACACCTDHBV |
 |
 |
| Motif ID: | UP00103 |
| Motif name: | Jundm2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0132197 |
Alignment:
HVGMTGACGTCATCBB
CTTCTGACCTC-----
| Original motif | Reverse complement motif |
| Consensus sequence: BBGATGACGTCAYCVH | Consensus sequence: HVGMTGACGTCATCBB |
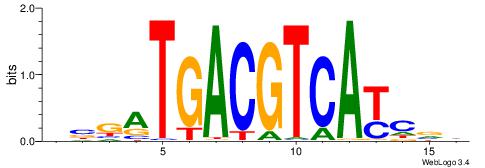 |
 |
| Dataset #: 1 | Motif ID: 18 | Motif name: Motif 18 |
| Original motif | Reverse complement motif |
| Consensus sequence: CAAAGTCCAGC | Consensus sequence: GCTGGACTTTG |
 |
 |
Best Matches for Motif ID 18 (Highest to Lowest)
| Motif ID: | UP00066 |
| Motif name: | Hnf4a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0 |
Alignment:
HHAYTGGACTTTHDBV
--GCTGGACTTTG---
| Original motif | Reverse complement motif |
| Consensus sequence: BBHHAAAGTCCAMTHH | Consensus sequence: HHAYTGGACTTTHDBV |
 |
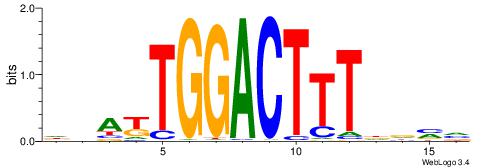 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0313848 |
Alignment:
HHBVHTGACCTTGVDHD
--GCTGGACTTTG----
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBCAAGGTCAHVBDH | Consensus sequence: HHBVHTGACCTTGVDHD |
 |
 |
| Motif ID: | UP00048 |
| Motif name: | Rara |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0347594 |
Alignment:
DHBHAAAGGTCACVHD
---CAAAGTCCAGC--
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCACVHD | Consensus sequence: HHVGTGACCTTTDVDD |
 |
 |
| Motif ID: | UP00009 |
| Motif name: | Nr2f2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0448871 |
Alignment:
DHBHAAAGGTCAHVHB
---CAAAGTCCAGC--
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCAHVHB | Consensus sequence: BHVDTGACCTTTDVDD |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 10 |
| Number of overlap: | 11 |
| Similarity score: | 0.0461676 |
Alignment:
BHCBCBCCGGGTGGTCYHVHDCH
---------GCTGGACTTTG---
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
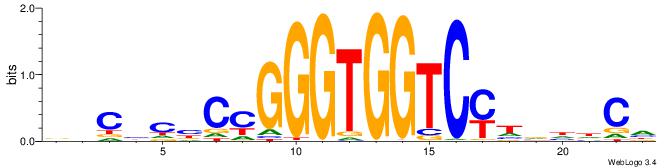 |
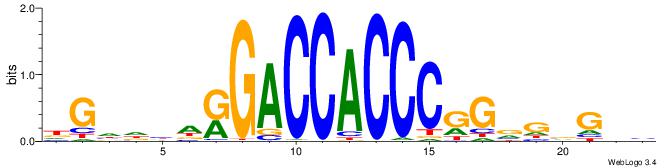 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0468432 |
Alignment:
DTVBDDRGACCACCCAGDWDGBH
---CAAAGTCCAGC---------
| Original motif | Reverse complement motif |
| Consensus sequence: DTVBDDRGACCACCCAGDWDGBH | Consensus sequence: DBCDWHCTGGGTGGTCMDHBBAH |
 |
 |
| Motif ID: | UP00047 |
| Motif name: | Zbtb7b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0474554 |
Alignment:
BBDVHGGTGGTCBKDDB
-----GCTGGACTTTG-
| Original motif | Reverse complement motif |
| Consensus sequence: BDDRVGACCACCHBDVB | Consensus sequence: BBDVHGGTGGTCBKDDB |
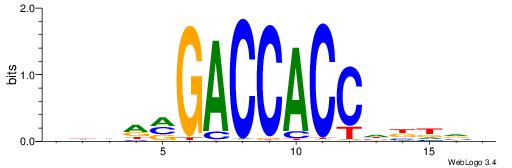 |
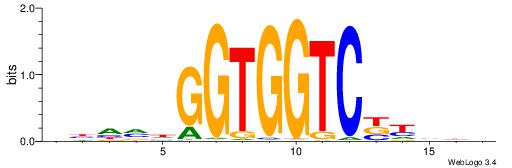 |
| Motif ID: | UP00416 |
| Motif name: | Fli1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0475257 |
Alignment:
DHDACCGGAAKTBBVV
---GCTGGACTTTG--
| Original motif | Reverse complement motif |
| Consensus sequence: VBBVAYTTCCGGTDDH | Consensus sequence: DHDACCGGAAKTBBVV |
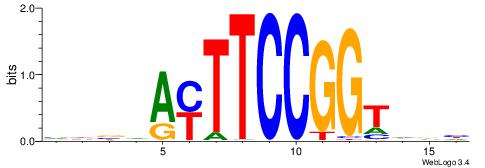 |
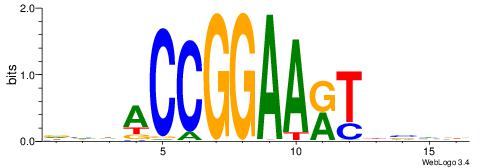 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0476315 |
Alignment:
HDGHHBHRGACCACCCGSVDVG
----CAAAGTCCAGC-------
| Original motif | Reverse complement motif |
| Consensus sequence: HDGHHBHRGACCACCCGSVDVG | Consensus sequence: CVHBSCGGGTGGTCMHVHHCDH |
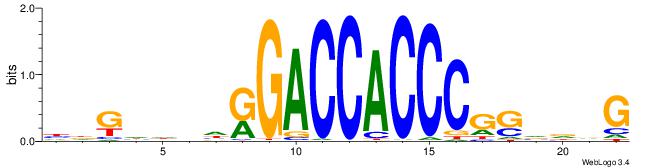 |
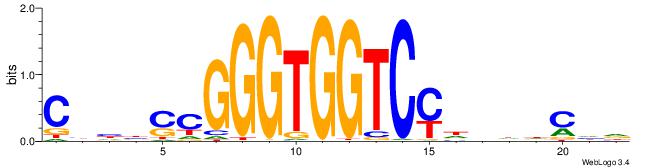 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 11 |
| Similarity score: | 0.0492402 |
Alignment:
ABBBBVVRGACCACCCACRDBBM
----CAAAGTCCAGC--------
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Dataset #: 1 | Motif ID: 19 | Motif name: Motif 19 |
| Original motif | Reverse complement motif |
| Consensus sequence: AGAGAAAG | Consensus sequence: CTTTCTCT |
 |
 |
Best Matches for Motif ID 19 (Highest to Lowest)
| Motif ID: | UP00101 |
| Motif name: | Sox12_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
VVDBASACAAAGRADH
----AGAGAAAG----
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBASACAAAGRADH | Consensus sequence: HDTMCTTTGTSTVDBB |
 |
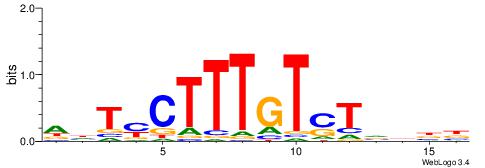 |
| Motif ID: | UP00419 |
| Motif name: | Spic |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.00218499 |
Alignment:
VAAAGMGGAAGTWD
---AGAGAAAG---
| Original motif | Reverse complement motif |
| Consensus sequence: VAAAGMGGAAGTWD | Consensus sequence: DWACTTCCRCTTTB |
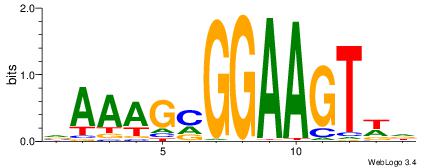 |
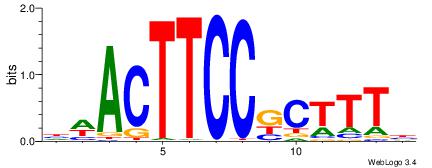 |
| Motif ID: | UP00074 |
| Motif name: | Isgf3g_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0130365 |
Alignment:
TDAGTTTCGWTTKDV
---CTTTCTCT----
| Original motif | Reverse complement motif |
| Consensus sequence: VDRAAWCGAAACTDA | Consensus sequence: TDAGTTTCGWTTKDV |
 |
 |
| Motif ID: | UP00085 |
| Motif name: | Sfpi1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.013531 |
Alignment:
BDAWGVGGAAGTDH
---AGAGAAAG---
| Original motif | Reverse complement motif |
| Consensus sequence: BDAWGVGGAAGTDH | Consensus sequence: HDACTTCCBCWTDV |
 |
 |
| Motif ID: | UP00018 |
| Motif name: | Irf4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0150506 |
Alignment:
TBDKGTTTCGMTDHV
----CTTTCTCT---
| Original motif | Reverse complement motif |
| Consensus sequence: VDDAYCGAAACYDVA | Consensus sequence: TBDKGTTTCGMTDHV |
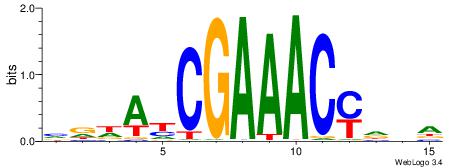 |
 |
| Motif ID: | UP00409 |
| Motif name: | Elf5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0153654 |
Alignment:
BWAHSMGGAAGTWD
---AGAGAAAG---
| Original motif | Reverse complement motif |
| Consensus sequence: BWAHSMGGAAGTWD | Consensus sequence: DWACTTCCRSDTWV |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0177379 |
Alignment:
BDHDDADAGATAAGADHBDHVD
-------AGAGAAAG-------
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.017774 |
Alignment:
HBDDDAACAAAGRVBHH
----AGAGAAAG-----
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
 |
 |
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.019138 |
Alignment:
HBDDRAACAAAGRABHH
----AGAGAAAG-----
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
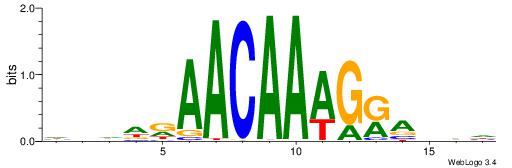 |
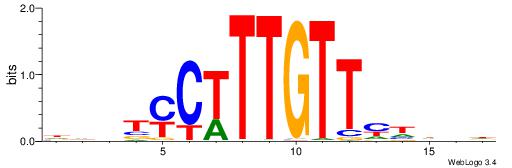 |
| Motif ID: | UP00040 |
| Motif name: | Irf5_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0202543 |
Alignment:
BDKGTTTCGKTWDDD
---CTTTCTCT----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDWAYCGAAACYDV | Consensus sequence: BDKGTTTCGKTWDDD |
 |
 |
| Dataset #: 1 | Motif ID: 20 | Motif name: Motif 20 |
| Original motif | Reverse complement motif |
| Consensus sequence: TCCTCCTGGAA | Consensus sequence: TTCCAGGAGGA |
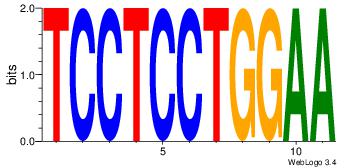 |
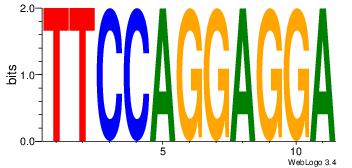 |
Best Matches for Motif ID 20 (Highest to Lowest)
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 11 |
| Similarity score: | 0.0142508 |
Alignment:
YDYBDHTMCACGTGGADDBMDGT
-----TTCCAGGAGGA-------
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
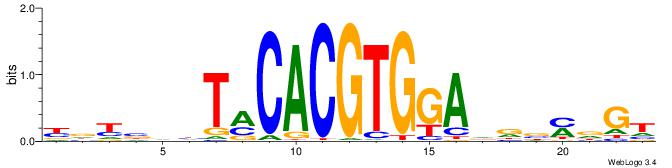 |
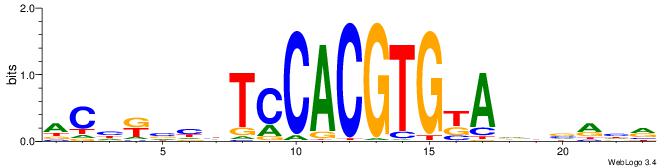 |
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0219794 |
Alignment:
VHHATTCCTYGAAAGD
--TCCTCCTGGAA---
| Original motif | Reverse complement motif |
| Consensus sequence: DCTTTCKAGGAATHHV | Consensus sequence: VHHATTCCTYGAAAGD |
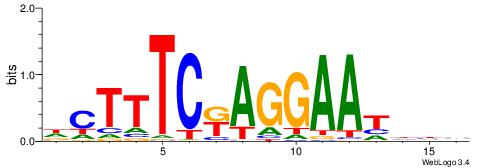 |
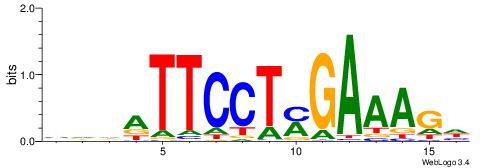 |
| Motif ID: | UP00102 |
| Motif name: | Zic1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0220915 |
Alignment:
DVHCCTGCTGBGDDB
--TCCTCCTGGAA--
| Original motif | Reverse complement motif |
| Consensus sequence: BHDCVCAGCAGGHVD | Consensus sequence: DVHCCTGCTGBGDDB |
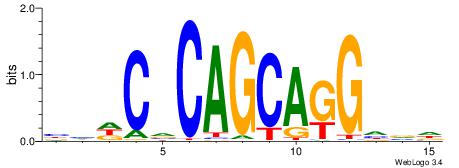 |
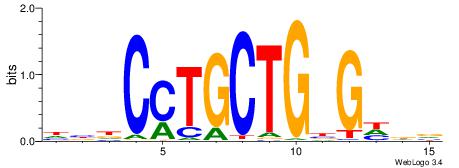 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0240277 |
Alignment:
DDDTWCCCCCCGGDRVH
----TCCTCCTGGAA--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDTWCCCCCCGGDRVH | Consensus sequence: HVKDCCGGGGGGWADDD |
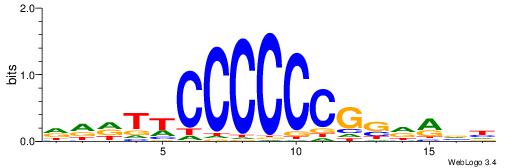 |
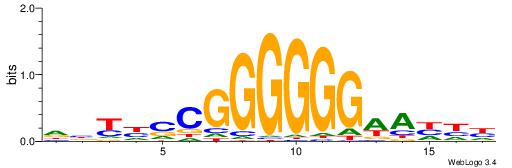 |
| Motif ID: | UP00057 |
| Motif name: | Zic2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0255161 |
Alignment:
DVBCYTGCTGBGDDD
--TCCTCCTGGAA--
| Original motif | Reverse complement motif |
| Consensus sequence: HHDCVCAGCAKGVVD | Consensus sequence: DVBCYTGCTGBGDDD |
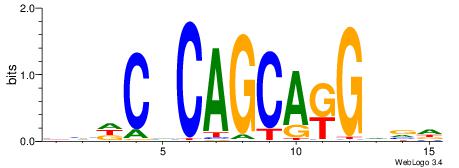 |
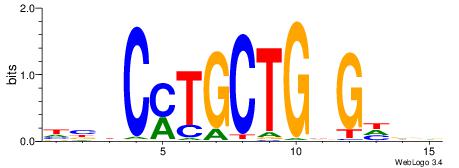 |
| Motif ID: | UP00420 |
| Motif name: | Elk3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.0258834 |
Alignment:
VBVDMCTTCCGGTVBVD
------TTCCAGGAGGA
| Original motif | Reverse complement motif |
| Consensus sequence: DVBBACCGGAAGYDVVV | Consensus sequence: VBVDMCTTCCGGTVBVD |
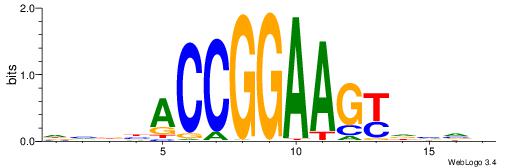 |
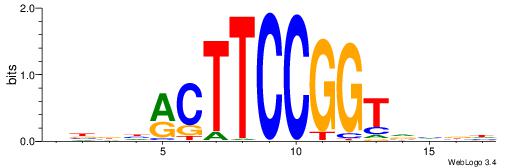 |
| Motif ID: | UP00006 |
| Motif name: | Zic3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0261265 |
Alignment:
DVHCYTGCTGTGHDH
--TCCTCCTGGAA--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDCACAGCAKGHVD | Consensus sequence: DVHCYTGCTGTGHDH |
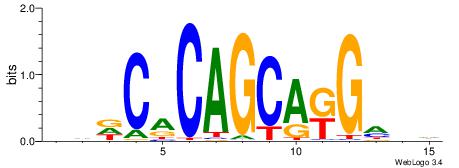 |
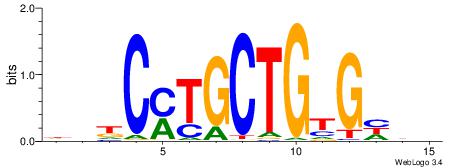 |
| Motif ID: | UP00013 |
| Motif name: | Gabpa_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0280194 |
Alignment:
BHHHACTTCCGGTHHBD
------TTCCAGGAGGA
| Original motif | Reverse complement motif |
| Consensus sequence: HVHHACCGGAAGTDHHV | Consensus sequence: BHHHACTTCCGGTHHBD |
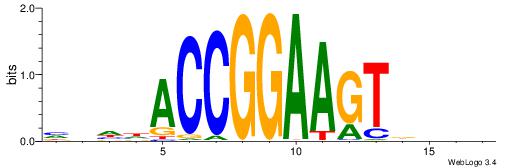 |
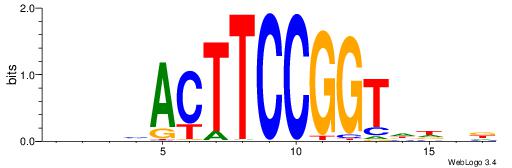 |
| Motif ID: | UP00410 |
| Motif name: | Elk1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0292751 |
Alignment:
DHBDACTTCCGGTVHVB
------TTCCAGGAGGA
| Original motif | Reverse complement motif |
| Consensus sequence: VVHBACCGGAAGTDBHD | Consensus sequence: DHBDACTTCCGGTVHVB |
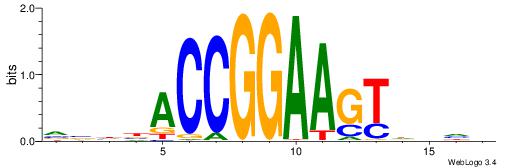 |
 |
| Motif ID: | UP00183 |
| Motif name: | Hoxa13 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0307181 |
Alignment:
HDHBCTCGTAAAAWBD
--TCCTCCTGGAA---
| Original motif | Reverse complement motif |
| Consensus sequence: HDHBCTCGTAAAAWBD | Consensus sequence: DVWTTTTACGAGBHDH |
 |
 |
| Dataset #: 1 | Motif ID: 21 | Motif name: Motif 21 |
| Original motif | Reverse complement motif |
| Consensus sequence: CTGCTTTCCMAA | Consensus sequence: TTYGGAAAGCAG |
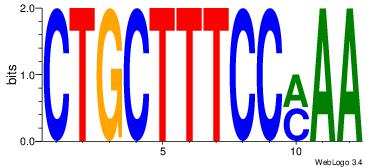 |
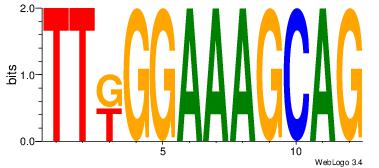 |
Best Matches for Motif ID 21 (Highest to Lowest)
| Motif ID: | UP00018 |
| Motif name: | Irf4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0328573 |
Alignment:
TBDKGTTTCGMTDHV
-CTGCTTTCCMAA--
| Original motif | Reverse complement motif |
| Consensus sequence: VDDAYCGAAACYDVA | Consensus sequence: TBDKGTTTCGMTDHV |
 |
 |
| Motif ID: | UP00015 |
| Motif name: | Ehf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0339295 |
Alignment:
WDSHTYGGAAVTDVDW
---TTYGGAAAGCAG-
| Original motif | Reverse complement motif |
| Consensus sequence: WDVDABTTCCKAHSDW | Consensus sequence: WDSHTYGGAAVTDVDW |
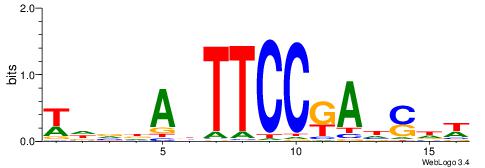 |
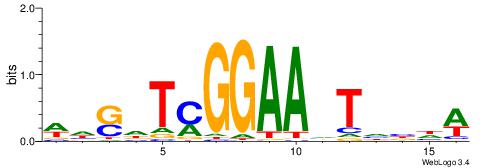 |
| Motif ID: | UP00045 |
| Motif name: | Mafb_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0362612 |
Alignment:
DMTWDGTCAGCADWTTH
-TTYGGAAAGCAG----
| Original motif | Reverse complement motif |
| Consensus sequence: HAAWDTGCTGACDWARH | Consensus sequence: DMTWDGTCAGCADWTTH |
 |
 |
| Motif ID: | UP00122 |
| Motif name: | Tgif1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0373175 |
Alignment:
DBVVAGCTGTCAWWVTH
---CTGCTTTCCMAA--
| Original motif | Reverse complement motif |
| Consensus sequence: DABWWTGACAGCTVVBD | Consensus sequence: DBVVAGCTGTCAWWVTH |
 |
 |
| Motif ID: | UP00258 |
| Motif name: | Tgif2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0385167 |
Alignment:
AABBAGCTGTCAAWVV
---CTGCTTTCCMAA-
| Original motif | Reverse complement motif |
| Consensus sequence: AABBAGCTGTCAAWVV | Consensus sequence: VBWTTGACAGCTVBTT |
 |
 |
| Motif ID: | UP00148 |
| Motif name: | Hdx |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0391678 |
Alignment:
DDKDHGAAATCAHHBHV
-TTYGGAAAGCAG----
| Original motif | Reverse complement motif |
| Consensus sequence: DDKDHGAAATCAHHBHV | Consensus sequence: BDBDHTGATTTCDHYDD |
 |
 |
| Motif ID: | UP00040 |
| Motif name: | Irf5_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.04057 |
Alignment:
DDDWAYCGAAACYDV
---TTYGGAAAGCAG
| Original motif | Reverse complement motif |
| Consensus sequence: DDDWAYCGAAACYDV | Consensus sequence: BDKGTTTCGKTWDDD |
 |
 |
| Motif ID: | UP00192 |
| Motif name: | Six1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 0.0414168 |
Alignment:
AVHDHTGATACCCMDHH
----CTGCTTTCCMAA-
| Original motif | Reverse complement motif |
| Consensus sequence: DHDRGGGTATCAHDHBT | Consensus sequence: AVHDHTGATACCCMDHH |
 |
 |
| Motif ID: | UP00159 |
| Motif name: | Six2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 0.0420524 |
Alignment:
AHWDGTGATACCCMRTD
----CTGCTTTCCMAA-
| Original motif | Reverse complement motif |
| Consensus sequence: DAKRGGGTATCACDWHT | Consensus sequence: AHWDGTGATACCCMRTD |
 |
 |
| Motif ID: | UP00235 |
| Motif name: | Hoxc11 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 0.0421437 |
Alignment:
HAADGTCGTAAAAHDD
----TTYGGAAAGCAG
| Original motif | Reverse complement motif |
| Consensus sequence: HAADGTCGTAAAAHDD | Consensus sequence: HDHTTTTACGACDTTH |
 |
 |
| Dataset #: 1 | Motif ID: 22 | Motif name: Motif 22 |
| Original motif | Reverse complement motif |
| Consensus sequence: TTTATGGTGAGCAT | Consensus sequence: ATGCTCACCATAAA |
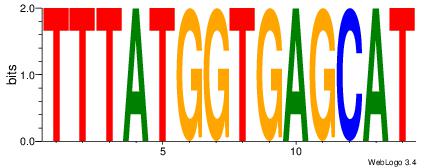 |
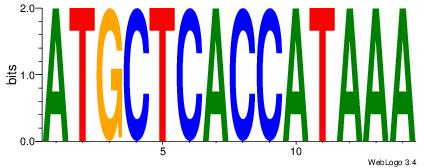 |
Best Matches for Motif ID 22 (Highest to Lowest)
| Motif ID: | UP00146 |
| Motif name: | Pou6f1_3733.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0269204 |
Alignment:
DHDASCTCATTATVHHB
--ATGCTCACCATAAA-
| Original motif | Reverse complement motif |
| Consensus sequence: VHHVATAATGAGSTDDH | Consensus sequence: DHDASCTCATTATVHHB |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 10 |
| Number of overlap: | 13 |
| Similarity score: | 0.511393 |
Alignment:
-DCATDBTGHGCATKMTTBRSWR
TTTATGGTGAGCAT---------
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
 |
 |
| Motif ID: | UP00045 |
| Motif name: | Mafb_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 13 |
| Similarity score: | 0.520342 |
Alignment:
HAAWDTGCTGACDWARH-
----ATGCTCACCATAAA
| Original motif | Reverse complement motif |
| Consensus sequence: HAAWDTGCTGACDWARH | Consensus sequence: DMTWDGTCAGCADWTTH |
 |
 |
| Motif ID: | UP00097 |
| Motif name: | Mtf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.520462 |
Alignment:
DBDHCGTGTGCAAMDH-
-TTTATGGTGAGCAT--
| Original motif | Reverse complement motif |
| Consensus sequence: DBDHCGTGTGCAAMDH | Consensus sequence: HDYTTGCACACGDHBH |
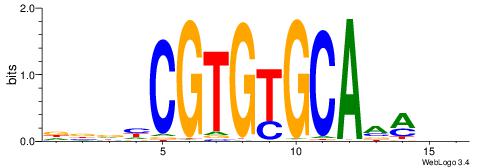 |
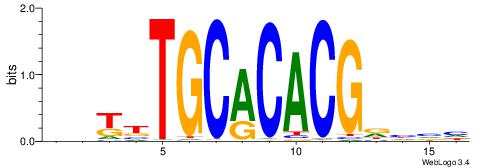 |
| Motif ID: | UP00035 |
| Motif name: | Hic1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 13 |
| Similarity score: | 0.524109 |
Alignment:
-DDBHGGTTGGCATVBD
TTTATGGTGAGCAT---
| Original motif | Reverse complement motif |
| Consensus sequence: DBBATGCCAACCHVHH | Consensus sequence: DDBHGGTTGGCATVBD |
 |
 |
| Motif ID: | UP00072 |
| Motif name: | IRC900814_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 1.01715 |
Alignment:
--DDHTACGACAAAWDDB
--TTTATGGTGAGCAT--
| Original motif | Reverse complement motif |
| Consensus sequence: DDHTACGACAAAWDDB | Consensus sequence: BHDWTTTGTCGTAHDD |
 |
 |
| Motif ID: | UP00191 |
| Motif name: | Pou2f2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 1.0174 |
Alignment:
BHDTATGCAAATDHDV--
--TTTATGGTGAGCAT--
| Original motif | Reverse complement motif |
| Consensus sequence: BHDTATGCAAATDHDV | Consensus sequence: BHHDATTTGCATAHHV |
 |
 |
| Motif ID: | UP00179 |
| Motif name: | Pou2f3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 1.01782 |
Alignment:
BHDTATGCAAATBHDV--
--TTTATGGTGAGCAT--
| Original motif | Reverse complement motif |
| Consensus sequence: BHDTATGCAAATBHDV | Consensus sequence: BHHVATTTGCATAHHV |
 |
 |
| Motif ID: | UP00133 |
| Motif name: | Cdx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 1.02211 |
Alignment:
--DVVGGYMATAAAAHKH
ATGCTCACCATAAA----
| Original motif | Reverse complement motif |
| Consensus sequence: DVVGGYMATAAAAHKH | Consensus sequence: HRHTTTTATYMCCVBD |
 |
 |
| Motif ID: | UP00173 |
| Motif name: | Hoxc13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 1.02921 |
Alignment:
DHWTTTTAMGAGBDHH--
--ATGCTCACCATAAA--
| Original motif | Reverse complement motif |
| Consensus sequence: HHDBCTCRTAAAAWHD | Consensus sequence: DHWTTTTAMGAGBDHH |
 |
 |
| Dataset #: 1 | Motif ID: 23 | Motif name: Motif 23 |
| Original motif | Reverse complement motif |
| Consensus sequence: ATGTWTTCATTMAT | Consensus sequence: ATYAATGAAWACAT |
 |
 |
Best Matches for Motif ID 23 (Highest to Lowest)
| Motif ID: | UP00130 |
| Motif name: | Lhx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0285638 |
Alignment:
VDAATTAATTAAWWHHB
---ATYAATGAAWACAT
| Original motif | Reverse complement motif |
| Consensus sequence: VDAATTAATTAAWWHHB | Consensus sequence: VHHWWTTAATTAATTDV |
 |
 |
| Motif ID: | UP00212 |
| Motif name: | Lhx5 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0306386 |
Alignment:
VDHATTAATTAAAWHHB
---ATYAATGAAWACAT
| Original motif | Reverse complement motif |
| Consensus sequence: VDHATTAATTAAAWHHB | Consensus sequence: VDHWTTTAATTAATHHV |
 |
 |
| Motif ID: | UP00169 |
| Motif name: | Lmx1b |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0343199 |
Alignment:
VDWWWTTAATTAATDHB
ATGTWTTCATTMAT---
| Original motif | Reverse complement motif |
| Consensus sequence: VDWWWTTAATTAATDHB | Consensus sequence: BHDATTAATTAAWWWHB |
 |
 |
| Motif ID: | UP00262 |
| Motif name: | Lhx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0349569 |
Alignment:
VVDATTAATTAAWWAHB
---ATYAATGAAWACAT
| Original motif | Reverse complement motif |
| Consensus sequence: VVDATTAATTAAWWAHB | Consensus sequence: BHTWWTTAATTAATDVV |
 |
 |
| Motif ID: | UP00244 |
| Motif name: | Tlx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0361912 |
Alignment:
WWDTTATTAATTHATTV
-ATGTWTTCATTMAT--
| Original motif | Reverse complement motif |
| Consensus sequence: BAATHAATTAATAAHWW | Consensus sequence: WWDTTATTAATTHATTV |
 |
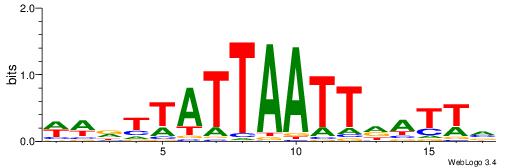 |
| Motif ID: | UP00188 |
| Motif name: | Lmx1a |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0375132 |
Alignment:
BDWWWTTAATTAAHHVG
---ATYAATGAAWACAT
| Original motif | Reverse complement motif |
| Consensus sequence: CVHHTTAATTAAWWWHB | Consensus sequence: BDWWWTTAATTAAHHVG |
 |
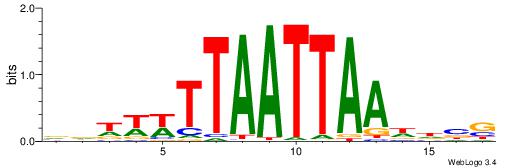 |
| Motif ID: | UP00155 |
| Motif name: | Hmx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0386004 |
Alignment:
VVAAGCAATTAAHVVDT
---ATYAATGAAWACAT
| Original motif | Reverse complement motif |
| Consensus sequence: VVAAGCAATTAAHVVDT | Consensus sequence: ADBVHTTAATTGCTTVB |
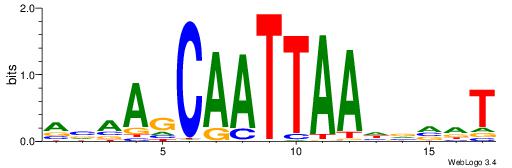 |
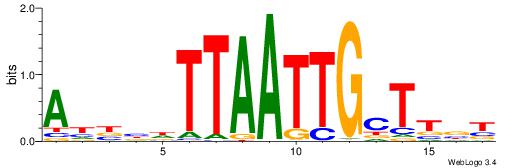 |
| Motif ID: | UP00157 |
| Motif name: | Hmx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0390217 |
Alignment:
VBVAGCAATTAAHVVAT
---ATYAATGAAWACAT
| Original motif | Reverse complement motif |
| Consensus sequence: VBVAGCAATTAAHVVAT | Consensus sequence: ATBVHTTAATTGCTBBB |
 |
 |
| Motif ID: | UP00172 |
| Motif name: | Prop1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0397917 |
Alignment:
VVVVTTAATTAABDDDC
---ATYAATGAAWACAT
| Original motif | Reverse complement motif |
| Consensus sequence: VVVVTTAATTAABDDDC | Consensus sequence: GDDDBTTAATTAABBVV |
 |
 |
| Motif ID: | UP00237 |
| Motif name: | Otp |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.041444 |
Alignment:
CHDMTTAATTAAKDHVV
---ATYAATGAAWACAT
| Original motif | Reverse complement motif |
| Consensus sequence: VVHDRTTAATTAAYDDG | Consensus sequence: CHDMTTAATTAAKDHVV |
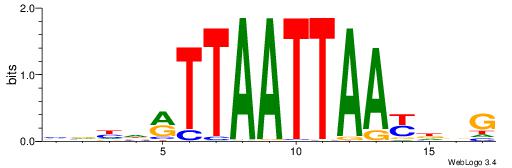 |
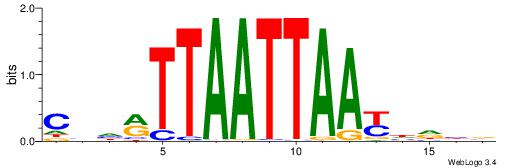 |
| Dataset #: 1 | Motif ID: 24 | Motif name: Motif 24 |
| Original motif | Reverse complement motif |
| Consensus sequence: AAAGWCATWAA | Consensus sequence: TTWATGWCTTT |
 |
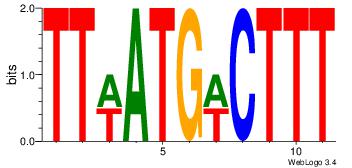 |
Best Matches for Motif ID 24 (Highest to Lowest)
| Motif ID: | UP00246 |
| Motif name: | Hoxa11 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.011591 |
Alignment:
HDADGTCGTAAAAHDD
-AAAGWCATWAA----
| Original motif | Reverse complement motif |
| Consensus sequence: HDADGTCGTAAAAHDD | Consensus sequence: DDDTTTTACGACDTDH |
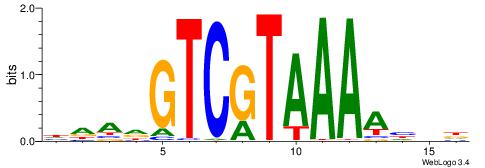 |
 |
| Motif ID: | UP00235 |
| Motif name: | Hoxc11 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0124232 |
Alignment:
HAADGTCGTAAAAHDD
-AAAGWCATWAA----
| Original motif | Reverse complement motif |
| Consensus sequence: HAADGTCGTAAAAHDD | Consensus sequence: HDHTTTTACGACDTTH |
 |
 |
| Motif ID: | UP00207 |
| Motif name: | Hoxb9 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0163525 |
Alignment:
VRRGCMATAAAAHHHD
AAAGWCATWAA-----
| Original motif | Reverse complement motif |
| Consensus sequence: VRRGCMATAAAAHHHD | Consensus sequence: HDHHTTTTATRGCKMV |
 |
 |
| Motif ID: | UP00197 |
| Motif name: | Hoxc9 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0176256 |
Alignment:
VDHHTTAATGACCBHV
----TTWATGWCTTT-
| Original motif | Reverse complement motif |
| Consensus sequence: VDVGGTCATTAAHHDB | Consensus sequence: VDHHTTAATGACCBHV |
 |
 |
| Motif ID: | UP00245 |
| Motif name: | Hoxc10 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.01781 |
Alignment:
HDDDGTCGTAAADHBB
-AAAGWCATWAA----
| Original motif | Reverse complement motif |
| Consensus sequence: HDDDGTCGTAAADHBB | Consensus sequence: VBDDTTTACGACDDDH |
 |
 |
| Motif ID: | UP00117 |
| Motif name: | Hoxd11 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0206572 |
Alignment:
HHDVGTCGTAAAAHHHD
-AAAGWCATWAA-----
| Original motif | Reverse complement motif |
| Consensus sequence: HHDVGTCGTAAAAHHHD | Consensus sequence: DDDHTTTTACGACVDHH |
 |
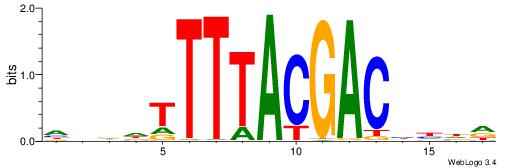 |
| Motif ID: | UP00240 |
| Motif name: | Cdx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0231269 |
Alignment:
DDVGGTMATAAAABKH
-AAAGWCATWAA----
| Original motif | Reverse complement motif |
| Consensus sequence: DDVGGTMATAAAABKH | Consensus sequence: HRVTTTTATYACCBDD |
 |
 |
| Motif ID: | UP00213 |
| Motif name: | Hoxa9 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0241179 |
Alignment:
DCGGYCATWAAAWTADW
AAAGWCATWAA------
| Original motif | Reverse complement motif |
| Consensus sequence: DCGGYCATWAAAWTADW | Consensus sequence: WDTAWTTTWATGKCCGD |
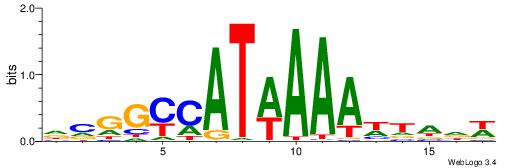 |
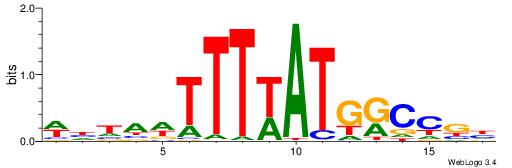 |
| Motif ID: | UP00133 |
| Motif name: | Cdx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0264981 |
Alignment:
DVVGGYMATAAAAHKH
-AAAGWCATWAA----
| Original motif | Reverse complement motif |
| Consensus sequence: DVVGGYMATAAAAHKH | Consensus sequence: HRHTTTTATYMCCVBD |
 |
 |
| Motif ID: | UP00177 |
| Motif name: | Hoxd12 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0271901 |
Alignment:
HHDVGTCGTAAAAHHHD
-AAAGWCATWAA-----
| Original motif | Reverse complement motif |
| Consensus sequence: HHDVGTCGTAAAAHHHD | Consensus sequence: DHDHTTTTACGACVDHD |
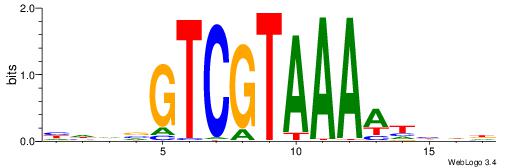 |
 |
| Dataset #: 1 | Motif ID: 25 | Motif name: Motif 25 |
| Original motif | Reverse complement motif |
| Consensus sequence: AATAACAGATT | Consensus sequence: AATCTGTTATT |
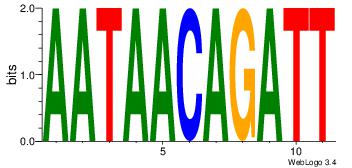 |
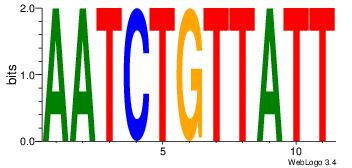 |
Best Matches for Motif ID 25 (Highest to Lowest)
| Motif ID: | UP00186 |
| Motif name: | Meis1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0336973 |
Alignment:
DBWTTGACAGSTBVTW
--AATAACAGATT---
| Original motif | Reverse complement motif |
| Consensus sequence: WAVBASCTGTCAAWVH | Consensus sequence: DBWTTGACAGSTBVTW |
 |
 |
| Motif ID: | UP00226 |
| Motif name: | Mrg1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0361504 |
Alignment:
BHWTTGACAGSTBBTW
--AATAACAGATT---
| Original motif | Reverse complement motif |
| Consensus sequence: WAVBASCTGTCAAWHB | Consensus sequence: BHWTTGACAGSTBBTW |
 |
 |
| Motif ID: | UP00122 |
| Motif name: | Tgif1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0367255 |
Alignment:
DBVVAGCTGTCAWWVTH
---AATCTGTTATT---
| Original motif | Reverse complement motif |
| Consensus sequence: DABWWTGACAGCTVVBD | Consensus sequence: DBVVAGCTGTCAWWVTH |
 |
 |
| Motif ID: | UP00203 |
| Motif name: | Pknox1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0373644 |
Alignment:
VVWTTGACAGSTSBKD
--AATAACAGATT---
| Original motif | Reverse complement motif |
| Consensus sequence: DRVSASCTGTCAAWVV | Consensus sequence: VVWTTGACAGSTSBKD |
 |
 |
| Motif ID: | UP00258 |
| Motif name: | Tgif2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0400385 |
Alignment:
AABBAGCTGTCAAWVV
---AATCTGTTATT--
| Original motif | Reverse complement motif |
| Consensus sequence: AABBAGCTGTCAAWVV | Consensus sequence: VBWTTGACAGCTVBTT |
 |
 |
| Motif ID: | UP00166 |
| Motif name: | Barhl1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0408157 |
Alignment:
DDVKTAATTGGTTDTH
-----AATCTGTTATT
| Original motif | Reverse complement motif |
| Consensus sequence: HAHAACCAATTARBDH | Consensus sequence: DDVKTAATTGGTTDTH |
 |
 |
| Motif ID: | UP00171 |
| Motif name: | Msx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.040907 |
Alignment:
HHVTTAATTKGTTHTV
-----AATCTGTTATT
| Original motif | Reverse complement motif |
| Consensus sequence: VAHAACYAATTAABHH | Consensus sequence: HHVTTAATTKGTTHTV |
 |
 |
| Motif ID: | UP00109 |
| Motif name: | Obox6 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0411003 |
Alignment:
AHAADCGGATTAWHB
AATAACAGATT----
| Original motif | Reverse complement motif |
| Consensus sequence: AHAADCGGATTAWHB | Consensus sequence: BHWTAATCCGDTTHT |
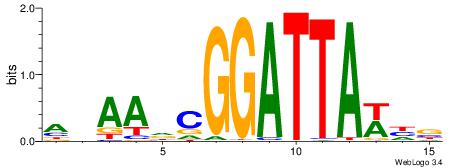 |
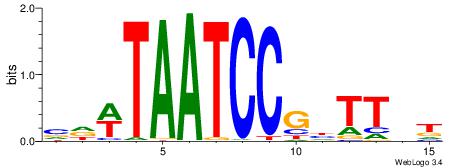 |
| Motif ID: | UP00205 |
| Motif name: | Pknox2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0430579 |
Alignment:
VHATTGACAGSKSVTH
--AATAACAGATT---
| Original motif | Reverse complement motif |
| Consensus sequence: HAVSRSCTGTCAATHB | Consensus sequence: VHATTGACAGSKSVTH |
 |
 |
| Motif ID: | UP00081 |
| Motif name: | Mybl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0431149 |
Alignment:
HDDDHAACCGTTAHHHD
----AATCTGTTATT--
| Original motif | Reverse complement motif |
| Consensus sequence: HDDDHAACCGTTAHHHD | Consensus sequence: DHHHTAACGGTTHDHDH |
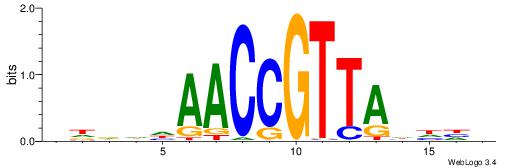 |
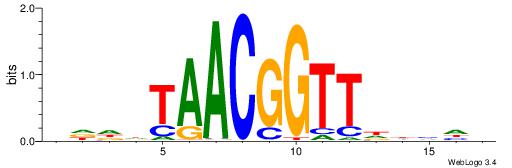 |
| Dataset #: 1 | Motif ID: 26 | Motif name: Motif 26 |
| Original motif | Reverse complement motif |
| Consensus sequence: CTGTTTTWAT | Consensus sequence: ATWAAAACAG |
 |
 |
Best Matches for Motif ID 26 (Highest to Lowest)
| Motif ID: | UP00207 |
| Motif name: | Hoxb9 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.030167 |
Alignment:
VRRGCMATAAAAHHHD
------ATWAAAACAG
| Original motif | Reverse complement motif |
| Consensus sequence: VRRGCMATAAAAHHHD | Consensus sequence: HDHHTTTTATRGCKMV |
 |
 |
| Motif ID: | UP00213 |
| Motif name: | Hoxa9 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0323471 |
Alignment:
DCGGYCATWAAAWTADW
------ATWAAAACAG-
| Original motif | Reverse complement motif |
| Consensus sequence: DCGGYCATWAAAWTADW | Consensus sequence: WDTAWTTTWATGKCCGD |
 |
 |
| Motif ID: | UP00134 |
| Motif name: | Hoxb13 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0362663 |
Alignment:
VDHWTTTTATTGGDKB
CTGTTTTWAT------
| Original motif | Reverse complement motif |
| Consensus sequence: VRHCCAATAAAAWHHV | Consensus sequence: VDHWTTTTATTGGDKB |
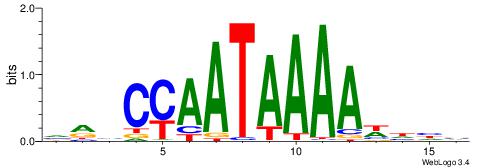 |
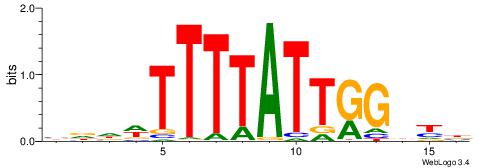 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0372816 |
Alignment:
VVVWTTGTTTAMATTWD
----CTGTTTTWAT---
| Original motif | Reverse complement motif |
| Consensus sequence: DWAATRTAAACAAWVVB | Consensus sequence: VVVWTTGTTTAMATTWD |
 |
 |
| Motif ID: | UP00025 |
| Motif name: | Foxk1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0386784 |
Alignment:
BBVTTTGTTTACATTDD
----CTGTTTTWAT---
| Original motif | Reverse complement motif |
| Consensus sequence: DDAATGTAAACAAAVVB | Consensus sequence: BBVTTTGTTTACATTDD |
 |
 |
| Motif ID: | UP00180 |
| Motif name: | Hoxd13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0390272 |
Alignment:
HDHYYAATAAAAHHHH
------ATWAAAACAG
| Original motif | Reverse complement motif |
| Consensus sequence: HDHYYAATAAAAHHHH | Consensus sequence: HDHHTTTTATTKKHDD |
 |
 |
| Motif ID: | UP00025 |
| Motif name: | Foxk1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0396761 |
Alignment:
HDDTGTTKTTKTHHD
--CTGTTTTWAT---
| Original motif | Reverse complement motif |
| Consensus sequence: HHHAYAAYAACAHHH | Consensus sequence: HDDTGTTKTTKTHHD |
 |
 |
| Motif ID: | UP00121 |
| Motif name: | Hoxd10 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0450256 |
Alignment:
HDDDATTTTATTKVRDB
-CTGTTTTWAT------
| Original motif | Reverse complement motif |
| Consensus sequence: VDKVYAATAAAATDDDH | Consensus sequence: HDDDATTTTATTKVRDB |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0451436 |
Alignment:
ADDDYAWAACAAMAHH
--ATWAAAACAG----
| Original motif | Reverse complement motif |
| Consensus sequence: ADDDYAWAACAAMAHH | Consensus sequence: HDTYTTGTTWTKDDDT |
 |
 |
| Motif ID: | UP00101 |
| Motif name: | Sox12_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0452358 |
Alignment:
DWTHAVAACAATHA
-ATWAAAACAG---
| Original motif | Reverse complement motif |
| Consensus sequence: THATTGTTVTHAWH | Consensus sequence: DWTHAVAACAATHA |
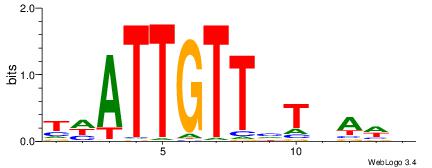 |
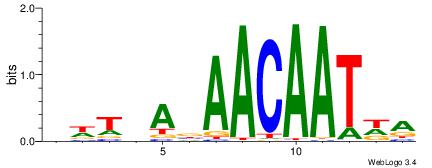 |
| Dataset #: 1 | Motif ID: 27 | Motif name: Motif 27 |
| Original motif | Reverse complement motif |
| Consensus sequence: ATTCTGTRAAG | Consensus sequence: CTTKACAGAAT |
 |
 |
Best Matches for Motif ID 27 (Highest to Lowest)
| Motif ID: | UP00193 |
| Motif name: | Rhox11_2205.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.022933 |
Alignment:
AVDHCGCTGTWAWDVVW
---ATTCTGTRAAG---
| Original motif | Reverse complement motif |
| Consensus sequence: AVDHCGCTGTWAWDVVW | Consensus sequence: WVVHWTWACAGCGHHVT |
 |
 |
| Motif ID: | UP00186 |
| Motif name: | Meis1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0231555 |
Alignment:
DBWTTGACAGSTBVTW
--CTTKACAGAAT---
| Original motif | Reverse complement motif |
| Consensus sequence: WAVBASCTGTCAAWVH | Consensus sequence: DBWTTGACAGSTBVTW |
 |
 |
| Motif ID: | UP00258 |
| Motif name: | Tgif2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0232158 |
Alignment:
VBWTTGACAGCTVBTT
--CTTKACAGAAT---
| Original motif | Reverse complement motif |
| Consensus sequence: AABBAGCTGTCAAWVV | Consensus sequence: VBWTTGACAGCTVBTT |
 |
 |
| Motif ID: | UP00012 |
| Motif name: | Bbx_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.023325 |
Alignment:
HADTTCAWTGAABWD
--ATTCTGTRAAG--
| Original motif | Reverse complement motif |
| Consensus sequence: HADTTCAWTGAABWD | Consensus sequence: HWBTTCAWTGAADTH |
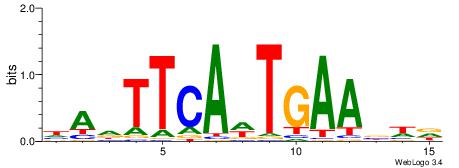 |
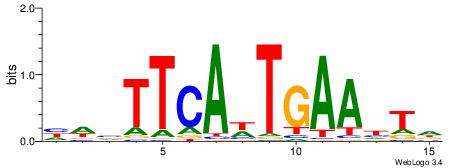 |
| Motif ID: | UP00226 |
| Motif name: | Mrg1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0239764 |
Alignment:
BHWTTGACAGSTBBTW
--CTTKACAGAAT---
| Original motif | Reverse complement motif |
| Consensus sequence: WAVBASCTGTCAAWHB | Consensus sequence: BHWTTGACAGSTBBTW |
 |
 |
| Motif ID: | UP00203 |
| Motif name: | Pknox1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.025386 |
Alignment:
VVWTTGACAGSTSBKD
--CTTKACAGAAT---
| Original motif | Reverse complement motif |
| Consensus sequence: DRVSASCTGTCAAWVV | Consensus sequence: VVWTTGACAGSTSBKD |
 |
 |
| Motif ID: | UP00193 |
| Motif name: | Rhox11_1765.2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0259448 |
Alignment:
AVDVCGCTGTWAWDVVW
---ATTCTGTRAAG---
| Original motif | Reverse complement motif |
| Consensus sequence: AVDVCGCTGTWAWDVVW | Consensus sequence: WVVHWTWACAGCGBHBT |
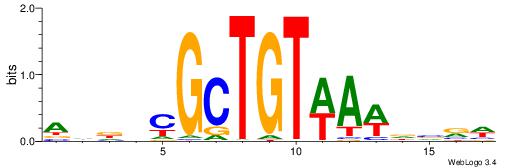 |
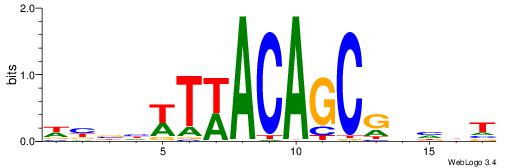 |
| Motif ID: | UP00205 |
| Motif name: | Pknox2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0264392 |
Alignment:
VHATTGACAGSKSVTH
--CTTKACAGAAT---
| Original motif | Reverse complement motif |
| Consensus sequence: HAVSRSCTGTCAATHB | Consensus sequence: VHATTGACAGSKSVTH |
 |
 |
| Motif ID: | UP00055 |
| Motif name: | Hbp1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0287967 |
Alignment:
HHDWTSAATGAATDDB
--CTTKACAGAAT---
| Original motif | Reverse complement motif |
| Consensus sequence: HHDWTSAATGAATDDB | Consensus sequence: VDHATTCATTSAWDDH |
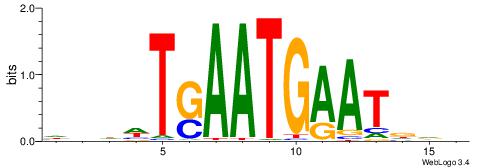 |
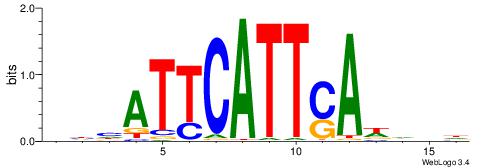 |
| Motif ID: | UP00210 |
| Motif name: | Mrg2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0289027 |
Alignment:
DBHTTGACAGSTVDTD
--CTTKACAGAAT---
| Original motif | Reverse complement motif |
| Consensus sequence: DADBASCTGTCAAHVH | Consensus sequence: DBHTTGACAGSTVDTD |
 |
 |
| Dataset #: 1 | Motif ID: 28 | Motif name: Motif 28 |
| Original motif | Reverse complement motif |
| Consensus sequence: ATTTAGTAAA | Consensus sequence: TTTACTAAAT |
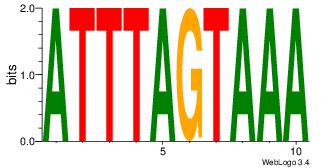 |
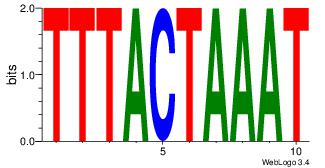 |
Best Matches for Motif ID 28 (Highest to Lowest)
| Motif ID: | UP00130 |
| Motif name: | Lhx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.00271264 |
Alignment:
VHHWWTTAATTAATTDV
----TTTACTAAAT---
| Original motif | Reverse complement motif |
| Consensus sequence: VDAATTAATTAAWWHHB | Consensus sequence: VHHWWTTAATTAATTDV |
 |
 |
| Motif ID: | UP00082 |
| Motif name: | Zfp187_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.00275365 |
Alignment:
HWATTAGTACATAV
-ATTTAGTAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: BTATGTACTAATWH | Consensus sequence: HWATTAGTACATAV |
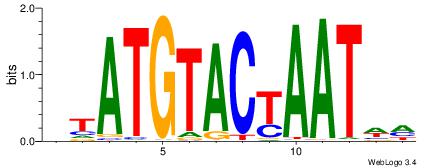 |
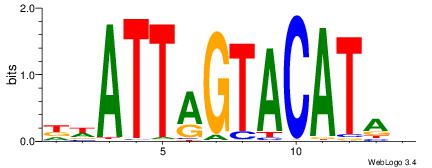 |
| Motif ID: | UP00212 |
| Motif name: | Lhx5 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.00285069 |
Alignment:
VDHWTTTAATTAATHHV
----TTTACTAAAT---
| Original motif | Reverse complement motif |
| Consensus sequence: VDHATTAATTAAAWHHB | Consensus sequence: VDHWTTTAATTAATHHV |
 |
 |
| Motif ID: | UP00169 |
| Motif name: | Lmx1b |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.00676865 |
Alignment:
BHDATTAATTAAWWWHB
---ATTTAGTAAA----
| Original motif | Reverse complement motif |
| Consensus sequence: VDWWWTTAATTAATDHB | Consensus sequence: BHDATTAATTAAWWWHB |
 |
 |
| Motif ID: | UP00177 |
| Motif name: | Hoxd12 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.00747245 |
Alignment:
DHDHTTTTACGACVDHD
-----TTTACTAAAT--
| Original motif | Reverse complement motif |
| Consensus sequence: HHDVGTCGTAAAAHHHD | Consensus sequence: DHDHTTTTACGACVDHD |
 |
 |
| Motif ID: | UP00135 |
| Motif name: | Hoxc12 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0076403 |
Alignment:
DDVHWTTTACGACCKHH
-----TTTACTAAAT--
| Original motif | Reverse complement motif |
| Consensus sequence: HHRGGTCGTAAAWHBDH | Consensus sequence: DDVHWTTTACGACCKHH |
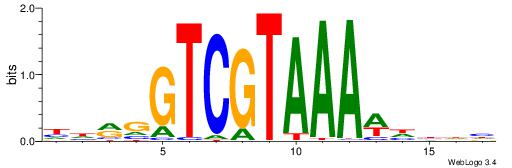 |
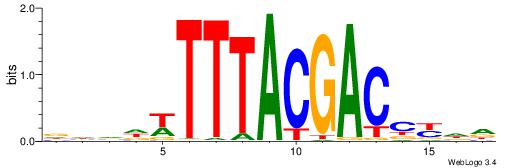 |
| Motif ID: | UP00117 |
| Motif name: | Hoxd11 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.00864633 |
Alignment:
DDDHTTTTACGACVDHH
-----TTTACTAAAT--
| Original motif | Reverse complement motif |
| Consensus sequence: HHDVGTCGTAAAAHHHD | Consensus sequence: DDDHTTTTACGACVDHH |
 |
 |
| Motif ID: | UP00142 |
| Motif name: | Uncx4.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0100967 |
Alignment:
CVVBTTAATTAATDHDV
---TTTACTAAAT----
| Original motif | Reverse complement motif |
| Consensus sequence: VDHDATTAATTAABVVG | Consensus sequence: CVVBTTAATTAATDHDV |
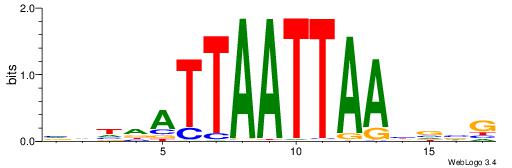 |
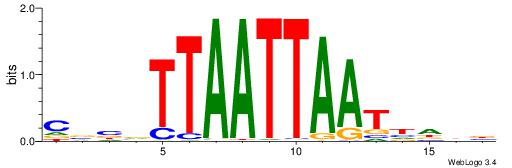 |
| Motif ID: | UP00235 |
| Motif name: | Hoxc11 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.010176 |
Alignment:
HDHTTTTACGACDTTH
----TTTACTAAAT--
| Original motif | Reverse complement motif |
| Consensus sequence: HAADGTCGTAAAAHDD | Consensus sequence: HDHTTTTACGACDTTH |
 |
 |
| Motif ID: | UP00246 |
| Motif name: | Hoxa11 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0105914 |
Alignment:
DDDTTTTACGACDTDH
----TTTACTAAAT--
| Original motif | Reverse complement motif |
| Consensus sequence: HDADGTCGTAAAAHDD | Consensus sequence: DDDTTTTACGACDTDH |
 |
 |
| Dataset #: 1 | Motif ID: 29 | Motif name: Motif 29 |
| Original motif | Reverse complement motif |
| Consensus sequence: ATTTATTGCTA | Consensus sequence: TAGCAATAAAT |
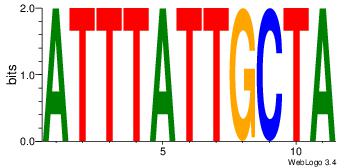 |
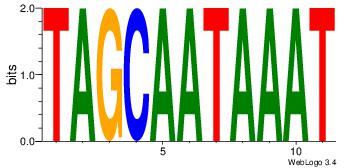 |
Best Matches for Motif ID 29 (Highest to Lowest)
| Motif ID: | UP00104 |
| Motif name: | Hmx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.00115362 |
Alignment:
VVVAGCAATTAADGVDB
--TAGCAATAAAT----
| Original motif | Reverse complement motif |
| Consensus sequence: VVVAGCAATTAADGVDB | Consensus sequence: VDBCDTTAATTGCTBVB |
 |
 |
| Motif ID: | UP00155 |
| Motif name: | Hmx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.00544084 |
Alignment:
ADBVHTTAATTGCTTVB
----ATTTATTGCTA--
| Original motif | Reverse complement motif |
| Consensus sequence: VVAAGCAATTAAHVVDT | Consensus sequence: ADBVHTTAATTGCTTVB |
 |
 |
| Motif ID: | UP00157 |
| Motif name: | Hmx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.00624569 |
Alignment:
VBVAGCAATTAAHVVAT
--TAGCAATAAAT----
| Original motif | Reverse complement motif |
| Consensus sequence: VBVAGCAATTAAHVVAT | Consensus sequence: ATBVHTTAATTGCTBBB |
 |
 |
| Motif ID: | UP00263 |
| Motif name: | Hoxb8 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.00742166 |
Alignment:
HVBDGYAATTAATADW
--TAGCAATAAAT---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDGYAATTAATADW | Consensus sequence: WDTATTAATTKCHBVH |
 |
 |
| Motif ID: | UP00133 |
| Motif name: | Cdx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0101377 |
Alignment:
DVVGGYMATAAAAHKH
--TAGCAATAAAT---
| Original motif | Reverse complement motif |
| Consensus sequence: DVVGGYMATAAAAHKH | Consensus sequence: HRHTTTTATYMCCVBD |
 |
 |
| Motif ID: | UP00207 |
| Motif name: | Hoxb9 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0107409 |
Alignment:
VRRGCMATAAAAHHHD
-TAGCAATAAAT----
| Original motif | Reverse complement motif |
| Consensus sequence: VRRGCMATAAAAHHHD | Consensus sequence: HDHHTTTTATRGCKMV |
 |
 |
| Motif ID: | UP00145 |
| Motif name: | Barhl2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0138179 |
Alignment:
HAMAAHCAATTAABHH
---TAGCAATAAAT--
| Original motif | Reverse complement motif |
| Consensus sequence: HAMAAHCAATTAABHH | Consensus sequence: HHBTTAATTGDTTYTH |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 10 |
| Number of overlap: | 11 |
| Similarity score: | 0.0142512 |
Alignment:
HDTBHHCCHTAGCAACVVDHHHH
---------TAGCAATAAAT---
| Original motif | Reverse complement motif |
| Consensus sequence: HDTBHHCCHTAGCAACVVDHHHH | Consensus sequence: HHHHDVVGTTGCTAHGGDHBAHH |
 |
 |
| Motif ID: | UP00180 |
| Motif name: | Hoxd13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.01435 |
Alignment:
HDHYYAATAAAAHHHH
-TAGCAATAAAT----
| Original motif | Reverse complement motif |
| Consensus sequence: HDHYYAATAAAAHHHH | Consensus sequence: HDHHTTTTATTKKHDD |
 |
 |
| Motif ID: | UP00168 |
| Motif name: | Hoxd8 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0169109 |
Alignment:
HADCMATTAATTADTTA
TAGCAATAAAT------
| Original motif | Reverse complement motif |
| Consensus sequence: TAADTAATTAATRGHTH | Consensus sequence: HADCMATTAATTADTTA |
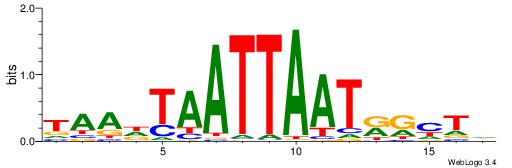 |
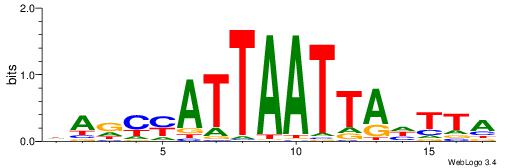 |
| Dataset #: 1 | Motif ID: 30 | Motif name: Motif 30 |
| Original motif | Reverse complement motif |
| Consensus sequence: TTGCTGCTTTT | Consensus sequence: AAAAGCAGCAA |
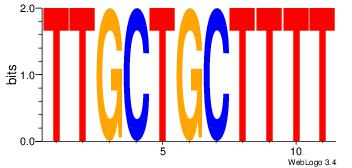 |
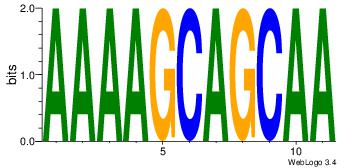 |
Best Matches for Motif ID 30 (Highest to Lowest)
| Motif ID: | UP00027 |
| Motif name: | Osr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.00770725 |
Alignment:
DHBHGCTACKGTHDHH
--TTGCTGCTTTT---
| Original motif | Reverse complement motif |
| Consensus sequence: HHDHACRGTAGCHVHD | Consensus sequence: DHBHGCTACKGTHDHH |
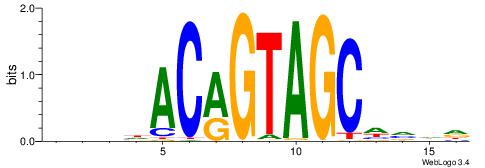 |
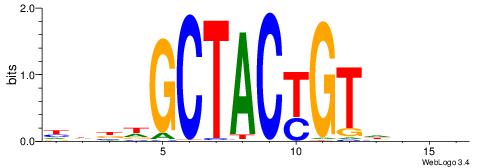 |
| Motif ID: | UP00044 |
| Motif name: | Mafk_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0113442 |
Alignment:
MBHTGCADTTTTTHV
--TTGCTGCTTTT--
| Original motif | Reverse complement motif |
| Consensus sequence: VHAAAAADTGCAHBR | Consensus sequence: MBHTGCADTTTTTHV |
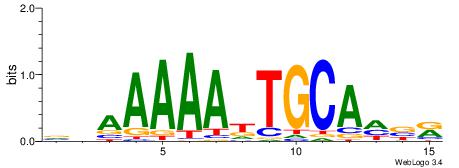 |
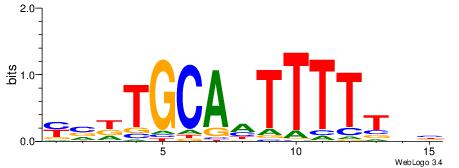 |
| Motif ID: | UP00052 |
| Motif name: | Osr2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0117169 |
Alignment:
HHHHGCTACKGTHVHH
--TTGCTGCTTTT---
| Original motif | Reverse complement motif |
| Consensus sequence: HHVHACRGTAGCHHHD | Consensus sequence: HHHHGCTACKGTHVHH |
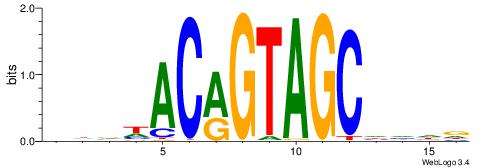 |
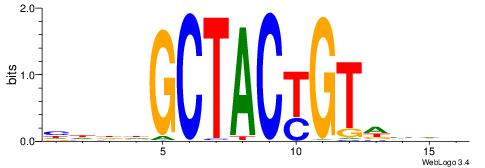 |
| Motif ID: | UP00045 |
| Motif name: | Mafb_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0149918 |
Alignment:
HAAWDTGCTGACDWARH
----TTGCTGCTTTT--
| Original motif | Reverse complement motif |
| Consensus sequence: HAAWDTGCTGACDWARH | Consensus sequence: DMTWDGTCAGCADWTTH |
 |
 |
| Motif ID: | UP00037 |
| Motif name: | Zfp105_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0204182 |
Alignment:
HHHTTRTTDTTTKDH
---TTGCTGCTTTT-
| Original motif | Reverse complement motif |
| Consensus sequence: HDYAAAHAAMAADHD | Consensus sequence: HHHTTRTTDTTTKDH |
 |
 |
| Motif ID: | UP00057 |
| Motif name: | Zic2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0211824 |
Alignment:
DVBCYTGCTGBGDDD
----TTGCTGCTTTT
| Original motif | Reverse complement motif |
| Consensus sequence: HHDCVCAGCAKGVVD | Consensus sequence: DVBCYTGCTGBGDDD |
 |
 |
| Motif ID: | UP00145 |
| Motif name: | Barhl2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0241392 |
Alignment:
HHBTTAATTGDTTYTH
----TTGCTGCTTTT-
| Original motif | Reverse complement motif |
| Consensus sequence: HAMAAHCAATTAABHH | Consensus sequence: HHBTTAATTGDTTYTH |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 11 |
| Similarity score: | 0.0241736 |
Alignment:
TCDAMVBGACGCMAKDDDVBBD
---AAAAGCAGCAA--------
| Original motif | Reverse complement motif |
| Consensus sequence: DBBVDDDRTRGCGTCBBYTHGA | Consensus sequence: TCDAMVBGACGCMAKDDDVBBD |
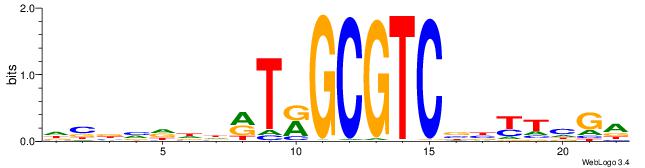 |
 |
| Motif ID: | UP00102 |
| Motif name: | Zic1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0245177 |
Alignment:
DVHCCTGCTGBGDDB
----TTGCTGCTTTT
| Original motif | Reverse complement motif |
| Consensus sequence: BHDCVCAGCAGGHVD | Consensus sequence: DVHCCTGCTGBGDDB |
 |
 |
| Motif ID: | UP00185 |
| Motif name: | Pbx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.0245464 |
Alignment:
BVDWHWTTGATGRKTKH
------TTGCTGCTTTT
| Original motif | Reverse complement motif |
| Consensus sequence: HYAYMCATCAAWHWDVV | Consensus sequence: BVDWHWTTGATGRKTKH |
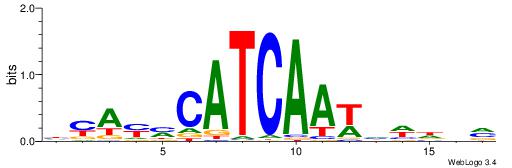 |
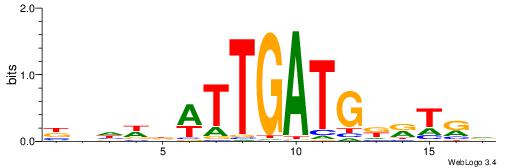 |
| Dataset #: 1 | Motif ID: 31 | Motif name: Motif 31 |
| Original motif | Reverse complement motif |
| Consensus sequence: TTGTTGATTTT | Consensus sequence: AAAATCAACAA |
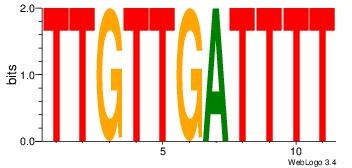 |
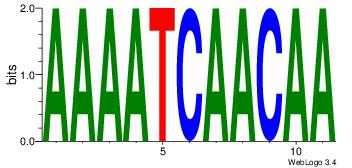 |
Best Matches for Motif ID 31 (Highest to Lowest)
| Motif ID: | UP00061 |
| Motif name: | Foxl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0 |
Alignment:
DWAATRTAAACAAWVVB
--AAAATCAACAA----
| Original motif | Reverse complement motif |
| Consensus sequence: DWAATRTAAACAAWVVB | Consensus sequence: VVVWTTGTTTAMATTWD |
 |
 |
| Motif ID: | UP00025 |
| Motif name: | Foxk1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.00256455 |
Alignment:
DDAATGTAAACAAAVVB
--AAAATCAACAA----
| Original motif | Reverse complement motif |
| Consensus sequence: DDAATGTAAACAAAVVB | Consensus sequence: BBVTTTGTTTACATTDD |
 |
 |
| Motif ID: | UP00039 |
| Motif name: | Foxj3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.00497188 |
Alignment:
HDHADGTAAACAAAVVH
--AAAATCAACAA----
| Original motif | Reverse complement motif |
| Consensus sequence: HDHADGTAAACAAAVVH | Consensus sequence: DVVTTTGTTTACDTHDH |
 |
 |
| Motif ID: | UP00227 |
| Motif name: | Duxl |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.00721745 |
Alignment:
VHVVGTTGATTGGGTMK
--TTGTTGATTTT----
| Original motif | Reverse complement motif |
| Consensus sequence: YRACCCAATCAACVVHV | Consensus sequence: VHVVGTTGATTGGGTMK |
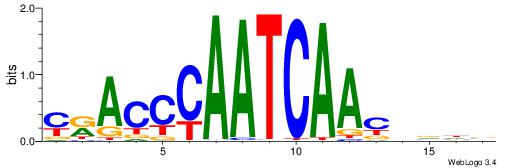 |
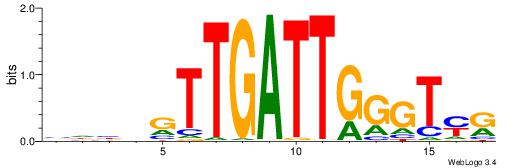 |
| Motif ID: | UP00037 |
| Motif name: | Zfp105_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0101946 |
Alignment:
HHHTTRTTDTTTKDH
---TTGTTGATTTT-
| Original motif | Reverse complement motif |
| Consensus sequence: HDYAAAHAAMAADHD | Consensus sequence: HHHTTRTTDTTTKDH |
 |
 |
| Motif ID: | UP00073 |
| Motif name: | Foxa2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0135831 |
Alignment:
HHWAWGTAAAYAAAVDB
--AAAATCAACAA----
| Original motif | Reverse complement motif |
| Consensus sequence: HHWAWGTAAAYAAAVDB | Consensus sequence: BDVTTTKTTTACWTWHH |
 |
 |
| Motif ID: | UP00145 |
| Motif name: | Barhl2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0139419 |
Alignment:
HAMAAHCAATTAABHH
-AAAATCAACAA----
| Original motif | Reverse complement motif |
| Consensus sequence: HAMAAHCAATTAABHH | Consensus sequence: HHBTTAATTGDTTYTH |
 |
 |
| Motif ID: | UP00041 |
| Motif name: | Foxj1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0164816 |
Alignment:
HDHRTAAACAAAHDDD
AAAATCAACAA-----
| Original motif | Reverse complement motif |
| Consensus sequence: HDHRTAAACAAAHDDD | Consensus sequence: DDDHTTTGTTTAMHDH |
 |
 |
| Motif ID: | UP00025 |
| Motif name: | Foxk1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0220198 |
Alignment:
HDDTGTTKTTKTHHD
--TTGTTGATTTT--
| Original motif | Reverse complement motif |
| Consensus sequence: HHHAYAAYAACAHHH | Consensus sequence: HDDTGTTKTTKTHHD |
 |
 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0242838 |
Alignment:
DHVDTGATTGATVBDV
----TTGTTGATTTT-
| Original motif | Reverse complement motif |
| Consensus sequence: VDVVATCAATCAHBHD | Consensus sequence: DHVDTGATTGATVBDV |
 |
 |
| Dataset #: 1 | Motif ID: 32 | Motif name: Motif 32 |
| Original motif | Reverse complement motif |
| Consensus sequence: TTACWGTTT | Consensus sequence: AAACWGTAA |
 |
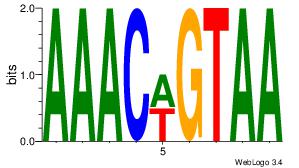 |
Best Matches for Motif ID 32 (Highest to Lowest)
| Motif ID: | UP00052 |
| Motif name: | Osr2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 9 |
| Similarity score: | 0.0202622 |
Alignment:
HHVHACRGTAGCHHHD
--AAACWGTAA-----
| Original motif | Reverse complement motif |
| Consensus sequence: HHVHACRGTAGCHHHD | Consensus sequence: HHHHGCTACKGTHVHH |
 |
 |
| Motif ID: | UP00027 |
| Motif name: | Osr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 9 |
| Similarity score: | 0.0205632 |
Alignment:
HHDHACRGTAGCHVHD
--AAACWGTAA-----
| Original motif | Reverse complement motif |
| Consensus sequence: HHDHACRGTAGCHVHD | Consensus sequence: DHBHGCTACKGTHDHH |
 |
 |
| Motif ID: | UP00073 |
| Motif name: | Foxa2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 9 |
| Similarity score: | 0.0212082 |
Alignment:
HHWAWGTAAAYAAAVDB
--------TTACWGTTT
| Original motif | Reverse complement motif |
| Consensus sequence: HHWAWGTAAAYAAAVDB | Consensus sequence: BDVTTTKTTTACWTWHH |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 9 |
| Similarity score: | 0.0213447 |
Alignment:
VVVWTTGTTTAMATTWD
--------TTACWGTTT
| Original motif | Reverse complement motif |
| Consensus sequence: DWAATRTAAACAAWVVB | Consensus sequence: VVVWTTGTTTAMATTWD |
 |
 |
| Motif ID: | UP00081 |
| Motif name: | Mybl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 9 |
| Similarity score: | 0.0237771 |
Alignment:
DHHHTAACGGTTHDHDH
----TTACWGTTT----
| Original motif | Reverse complement motif |
| Consensus sequence: HDDDHAACCGTTAHHHD | Consensus sequence: DHHHTAACGGTTHDHDH |
 |
 |
| Motif ID: | UP00025 |
| Motif name: | Foxk1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 9 |
| Similarity score: | 0.0240192 |
Alignment:
BBVTTTGTTTACATTDD
--------TTACWGTTT
| Original motif | Reverse complement motif |
| Consensus sequence: DDAATGTAAACAAAVVB | Consensus sequence: BBVTTTGTTTACATTDD |
 |
 |
| Motif ID: | UP00186 |
| Motif name: | Meis1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 9 |
| Similarity score: | 0.0245505 |
Alignment:
DBWTTGACAGSTBVTW
----TTACWGTTT---
| Original motif | Reverse complement motif |
| Consensus sequence: WAVBASCTGTCAAWVH | Consensus sequence: DBWTTGACAGSTBVTW |
 |
 |
| Motif ID: | UP00092 |
| Motif name: | Myb_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 9 |
| Similarity score: | 0.0258406 |
Alignment:
HDDDHAACCGTTAHHHD
----AAACWGTAA----
| Original motif | Reverse complement motif |
| Consensus sequence: HDDDHAACCGTTAHHHD | Consensus sequence: DHHHTAACGGTTHHHDH |
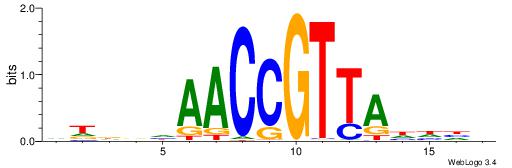 |
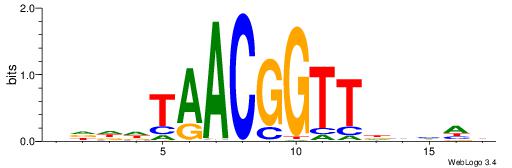 |
| Motif ID: | UP00226 |
| Motif name: | Mrg1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 9 |
| Similarity score: | 0.0267732 |
Alignment:
BHWTTGACAGSTBBTW
----TTACWGTTT---
| Original motif | Reverse complement motif |
| Consensus sequence: WAVBASCTGTCAAWHB | Consensus sequence: BHWTTGACAGSTBBTW |
 |
 |
| Motif ID: | UP00203 |
| Motif name: | Pknox1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 9 |
| Similarity score: | 0.0277187 |
Alignment:
VVWTTGACAGSTSBKD
----TTACWGTTT---
| Original motif | Reverse complement motif |
| Consensus sequence: DRVSASCTGTCAAWVV | Consensus sequence: VVWTTGACAGSTSBKD |
 |
 |
| Dataset #: 1 | Motif ID: 33 | Motif name: Motif 33 |
| Original motif | Reverse complement motif |
| Consensus sequence: TTTAATGTYTTTWA | Consensus sequence: TWAAAKACATTAAA |
 |
 |
Best Matches for Motif ID 33 (Highest to Lowest)
| Motif ID: | UP00171 |
| Motif name: | Msx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0143635 |
Alignment:
HHVTTAATTKGTTHTV
--TTTAATGTYTTTWA
| Original motif | Reverse complement motif |
| Consensus sequence: VAHAACYAATTAABHH | Consensus sequence: HHVTTAATTKGTTHTV |
 |
 |
| Motif ID: | UP00145 |
| Motif name: | Barhl2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0149596 |
Alignment:
HHBTTAATTGDTTYTH
--TTTAATGTYTTTWA
| Original motif | Reverse complement motif |
| Consensus sequence: HAMAAHCAATTAABHH | Consensus sequence: HHBTTAATTGDTTYTH |
 |
 |
| Motif ID: | UP00170 |
| Motif name: | Isl2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0226802 |
Alignment:
HWDTTAATYKMTTHHR
--TTTAATGTYTTTWA
| Original motif | Reverse complement motif |
| Consensus sequence: MHHAAYYMATTAADWH | Consensus sequence: HWDTTAATYKMTTHHR |
 |
 |
| Motif ID: | UP00119 |
| Motif name: | Nkx2-9 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0229232 |
Alignment:
HDTTBAAGTACTTDAAA
---TWAAAKACATTAAA
| Original motif | Reverse complement motif |
| Consensus sequence: TTTDAAGTACTTVAADH | Consensus sequence: HDTTBAAGTACTTDAAA |
 |
 |
| Motif ID: | UP00216 |
| Motif name: | Obox1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0255021 |
Alignment:
DHDGTTAATCCCCKTDD
TWAAAKACATTAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: DDARGGGGATTAACDHH | Consensus sequence: DHDGTTAATCCCCKTDD |
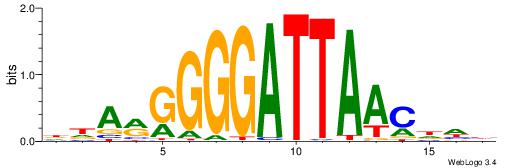 |
 |
| Motif ID: | UP00051 |
| Motif name: | Sox8_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0266643 |
Alignment:
DDATBWATTGTTHHDDD
---TTTAATGTYTTTWA
| Original motif | Reverse complement motif |
| Consensus sequence: DDATBWATTGTTHHDDD | Consensus sequence: DDDHDAACAATWBATDD |
 |
 |
| Motif ID: | UP00091 |
| Motif name: | Sox5_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0288082 |
Alignment:
DHDDWATTGTTCDDHD
--TTTAATGTYTTTWA
| Original motif | Reverse complement motif |
| Consensus sequence: DHDDGAACAATWDDHD | Consensus sequence: DHDDWATTGTTCDDHD |
 |
 |
| Motif ID: | UP00096 |
| Motif name: | Sox13_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0301291 |
Alignment:
WHDWWATTGTTCKDHD
--TTTAATGTYTTTWA
| Original motif | Reverse complement motif |
| Consensus sequence: DHDRGAACAATWWDHW | Consensus sequence: WHDWWATTGTTCKDHD |
 |
 |
| Motif ID: | UP00166 |
| Motif name: | Barhl1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0303352 |
Alignment:
DDVKTAATTGGTTDTH
--TTTAATGTYTTTWA
| Original motif | Reverse complement motif |
| Consensus sequence: HAHAACCAATTARBDH | Consensus sequence: DDVKTAATTGGTTDTH |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 14 |
| Similarity score: | 0.0304836 |
Alignment:
BDHDDADAGATAAGADHBDHVD
-TWAAAKACATTAAA-------
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Dataset #: 1 | Motif ID: 34 | Motif name: Motif 34 |
| Original motif | Reverse complement motif |
| Consensus sequence: AAAGATGA | Consensus sequence: TCATCTTT |
 |
 |
Best Matches for Motif ID 34 (Highest to Lowest)
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0235901 |
Alignment:
HBHDVHDTCTTATCTHTDDHDV
---------TCATCTTT-----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Motif ID: | UP00048 |
| Motif name: | Rara |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0248583 |
Alignment:
HHVGTGACCTTTDVDD
----TCATCTTT----
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCACVHD | Consensus sequence: HHVGTGACCTTTDVDD |
 |
 |
| Motif ID: | UP00009 |
| Motif name: | Nr2f2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0320787 |
Alignment:
BHVDTGACCTTTDVDD
----TCATCTTT----
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCAHVHB | Consensus sequence: BHVDTGACCTTTDVDD |
 |
 |
| Motif ID: | UP00114 |
| Motif name: | Homez |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0353395 |
Alignment:
HDTVAAAACGATRWWHT
------AAAGATGA---
| Original motif | Reverse complement motif |
| Consensus sequence: AHWWMATCGTTTTBADD | Consensus sequence: HDTVAAAACGATRWWHT |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0375145 |
Alignment:
VBBDMYCATCTGVHHBH
-----TCATCTTT----
| Original motif | Reverse complement motif |
| Consensus sequence: HVDDVCAGATGKYHBBV | Consensus sequence: VBBDMYCATCTGVHHBH |
 |
 |
| Motif ID: | UP00100 |
| Motif name: | Gata6_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0391681 |
Alignment:
VHDDTCTTATCWHBDHD
------TCATCTTT---
| Original motif | Reverse complement motif |
| Consensus sequence: DHDVDWGATAAGADDHV | Consensus sequence: VHDDTCTTATCWHBDHD |
 |
 |
| Motif ID: | UP00419 |
| Motif name: | Spic |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0437411 |
Alignment:
VAAAGMGGAAGTWD
-AAAGATGA-----
| Original motif | Reverse complement motif |
| Consensus sequence: VAAAGMGGAAGTWD | Consensus sequence: DWACTTCCRCTTTB |
 |
 |
| Motif ID: | UP00199 |
| Motif name: | Six4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.0467222 |
Alignment:
BBVHVGGTGTCATHDDT
---------TCATCTTT
| Original motif | Reverse complement motif |
| Consensus sequence: ADDHATGACACCBHBBV | Consensus sequence: BBVHVGGTGTCATHDDT |
 |
 |
| Motif ID: | UP00219 |
| Motif name: | Cutl1_3494.2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.0483168 |
Alignment:
DAVTGATRATCAHTD
-AAAGATGA------
| Original motif | Reverse complement motif |
| Consensus sequence: DAVTGATRATCAHTD | Consensus sequence: DADTGATMATCABTD |
 |
 |
| Motif ID: | UP00208 |
| Motif name: | Obox5_3963.2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0488248 |
Alignment:
VRVAGGGATTAAHTVWV
---AAAGATGA------
| Original motif | Reverse complement motif |
| Consensus sequence: VRVAGGGATTAAHTVWV | Consensus sequence: VWBAHTTAATCCCTVKB |
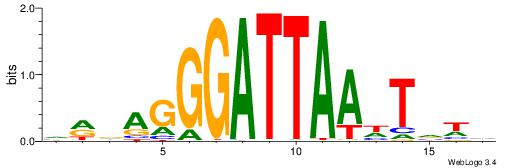 |
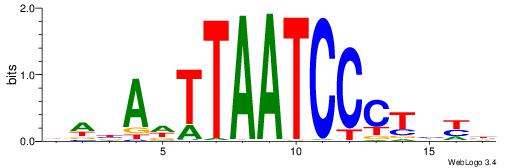 |
| Dataset #: 1 | Motif ID: 35 | Motif name: Motif 35 |
| Original motif | Reverse complement motif |
| Consensus sequence: AGATGYTCTTG | Consensus sequence: CAAGAKCATCT |
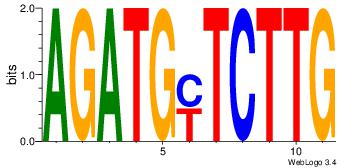 |
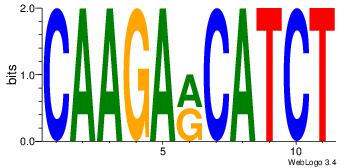 |
Best Matches for Motif ID 35 (Highest to Lowest)
| Motif ID: | UP00079 |
| Motif name: | Esrra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0140765 |
Alignment:
DHDBCAAGGTCAHVBDH
----CAAGAKCATCT--
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBCAAGGTCAHVBDH | Consensus sequence: HHBVHTGACCTTGVDHD |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.0175222 |
Alignment:
HVDDVCAGATGKYHBBV
------AGATGYTCTTG
| Original motif | Reverse complement motif |
| Consensus sequence: HVDDVCAGATGKYHBBV | Consensus sequence: VBBDMYCATCTGVHHBH |
 |
 |
| Motif ID: | UP00019 |
| Motif name: | Zbtb12_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0209646 |
Alignment:
BDHRBGTTCTAGATCVB
-AGATGYTCTTG-----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHRBGTTCTAGATCVB | Consensus sequence: BBGATCTAGAACBKHDB |
 |
 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.0302699 |
Alignment:
BBYVVCAGCTGCBVHHD
------AGATGYTCTTG
| Original motif | Reverse complement motif |
| Consensus sequence: BBYVVCAGCTGCBVHHD | Consensus sequence: HHDVVGCAGCTGVBKVB |
 |
 |
| Motif ID: | UP00048 |
| Motif name: | Rara |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0317546 |
Alignment:
DHBHAAAGGTCACVHD
----CAAGAKCATCT-
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCACVHD | Consensus sequence: HHVGTGACCTTTDVDD |
 |
 |
| Motif ID: | UP00025 |
| Motif name: | Foxk1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0329048 |
Alignment:
HDDTGTTKTTKTHHD
AGATGYTCTTG----
| Original motif | Reverse complement motif |
| Consensus sequence: HHHAYAAYAACAHHH | Consensus sequence: HDDTGTTKTTKTHHD |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 11 |
| Similarity score: | 0.0350601 |
Alignment:
BHCBCBCCGGGTGGTCYHVHDCH
--------AGATGYTCTTG----
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0366699 |
Alignment:
HBHDVBWGATHBTATCRVBHHH
------AGATGYTCTTG-----
| Original motif | Reverse complement motif |
| Consensus sequence: HBHDVBWGATHBTATCRVBHHH | Consensus sequence: HHHBBMGATAVHATCWVVDHVH |
 |
 |
| Motif ID: | UP00047 |
| Motif name: | Zbtb7b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0367981 |
Alignment:
BBDVHGGTGGTCBKDDB
----AGATGYTCTTG--
| Original motif | Reverse complement motif |
| Consensus sequence: BDDRVGACCACCHBDVB | Consensus sequence: BBDVHGGTGGTCBKDDB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.038145 |
Alignment:
HTBVVVDGGACCACCCRGRDBG
-----CAAGAKCATCT------
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
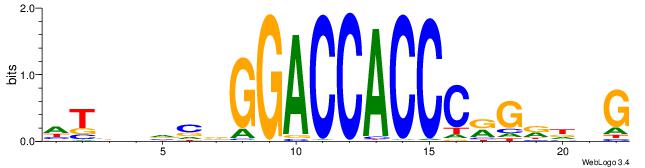 |
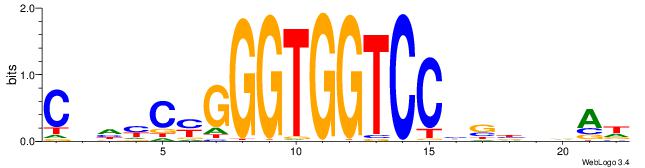 |
| Dataset #: 1 | Motif ID: 36 | Motif name: Motif 36 |
| Original motif | Reverse complement motif |
| Consensus sequence: TGGTGTCA | Consensus sequence: TGACACCA |
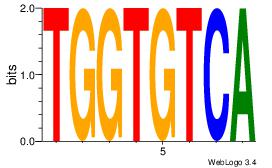 |
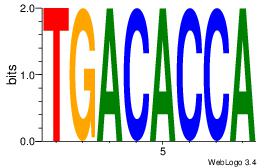 |
Best Matches for Motif ID 36 (Highest to Lowest)
| Motif ID: | UP00199 |
| Motif name: | Six4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
ADDHATGACACCBHBBV
-----TGACACCA----
| Original motif | Reverse complement motif |
| Consensus sequence: ADDHATGACACCBHBBV | Consensus sequence: BBVHVGGTGTCATHDDT |
 |
 |
| Motif ID: | UP00388 |
| Motif name: | Six6_2267.4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0335611 |
Alignment:
AHWDHTGATACCCKATD
-----TGACACCA----
| Original motif | Reverse complement motif |
| Consensus sequence: DATRGGGTATCAHDWHT | Consensus sequence: AHWDHTGATACCCKATD |
 |
 |
| Motif ID: | UP00159 |
| Motif name: | Six2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0342894 |
Alignment:
AHWDGTGATACCCMRTD
-----TGACACCA----
| Original motif | Reverse complement motif |
| Consensus sequence: DAKRGGGTATCACDWHT | Consensus sequence: AHWDGTGATACCCMRTD |
 |
 |
| Motif ID: | UP00053 |
| Motif name: | Rxra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.034602 |
Alignment:
DBHBDTGACCCCDHVHB
-----TGACACCA----
| Original motif | Reverse complement motif |
| Consensus sequence: DBHBDTGACCCCDHVHB | Consensus sequence: VHBHDGGGGTCAHBHBD |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0349175 |
Alignment:
VKBBAGGGGTCAHDBBH
----TGGTGTCA-----
| Original motif | Reverse complement motif |
| Consensus sequence: VKBBAGGGGTCAHDBBH | Consensus sequence: DBBHHTGACCCCTBBYV |
 |
 |
| Motif ID: | UP00122 |
| Motif name: | Tgif1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0354712 |
Alignment:
DABWWTGACAGCTVVBD
-----TGACACCA----
| Original motif | Reverse complement motif |
| Consensus sequence: DABWWTGACAGCTVVBD | Consensus sequence: DBVVAGCTGTCAWWVTH |
 |
 |
| Motif ID: | UP00192 |
| Motif name: | Six1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0355547 |
Alignment:
AVHDHTGATACCCMDHH
-----TGACACCA----
| Original motif | Reverse complement motif |
| Consensus sequence: DHDRGGGTATCAHDHBT | Consensus sequence: AVHDHTGATACCCMDHH |
 |
 |
| Motif ID: | UP00388 |
| Motif name: | Six6_2267.5 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.035579 |
Alignment:
AHWHWTGATACCCKDDD
-----TGACACCA----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDRGGGTATCAWHWHT | Consensus sequence: AHWHWTGATACCCKDDD |
 |
 |
| Motif ID: | UP00068 |
| Motif name: | Eomes_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0362156 |
Alignment:
BVHDAGGTGTGAAAHHH
----TGGTGTCA-----
| Original motif | Reverse complement motif |
| Consensus sequence: BVHDAGGTGTGAAAHHH | Consensus sequence: HHHTTTCACACCTDHBV |
 |
 |
| Motif ID: | UP00258 |
| Motif name: | Tgif2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0373826 |
Alignment:
VBWTTGACAGCTVBTT
----TGACACCA----
| Original motif | Reverse complement motif |
| Consensus sequence: AABBAGCTGTCAAWVV | Consensus sequence: VBWTTGACAGCTVBTT |
 |
 |
| Dataset #: 1 | Motif ID: 37 | Motif name: Motif 37 |
| Original motif | Reverse complement motif |
| Consensus sequence: ACTTTGG | Consensus sequence: CCAAAGT |
 |
 |
Best Matches for Motif ID 37 (Highest to Lowest)
| Motif ID: | UP00066 |
| Motif name: | Hnf4a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 7 |
| Similarity score: | 0 |
Alignment:
BBHHAAAGTCCAMTHH
--CCAAAGT-------
| Original motif | Reverse complement motif |
| Consensus sequence: BBHHAAAGTCCAMTHH | Consensus sequence: HHAYTGGACTTTHDBV |
 |
 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 7 |
| Similarity score: | 0.0128381 |
Alignment:
VDDAGATCAAAGGKDDD
------CCAAAGT----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 7 |
| Similarity score: | 0.0140846 |
Alignment:
DHDBCAAGGTCAHVBDH
---CCAAAGT-------
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBCAAGGTCAHVBDH | Consensus sequence: HHBVHTGACCTTGVDHD |
 |
 |
| Motif ID: | UP00083 |
| Motif name: | Tcf7l2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0.0141156 |
Alignment:
BDDASATCAAAGGMDDD
------CCAAAGT----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATSTDDV | Consensus sequence: BDDASATCAAAGGMDDD |
 |
 |
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0.0159798 |
Alignment:
HHBBMCTTTGTTHDDBH
----ACTTTGG------
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
 |
 |
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0.0181605 |
Alignment:
HHBTMCTTTGTTMDDVH
----ACTTTGG------
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
 |
 |
| Motif ID: | UP00058 |
| Motif name: | Tcf3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0.0200761 |
Alignment:
HHHKVCTTTGATSTHHH
----ACTTTGG------
| Original motif | Reverse complement motif |
| Consensus sequence: HHHASATCAAAGVRHHH | Consensus sequence: HHHKVCTTTGATSTHHH |
 |
 |
| Motif ID: | UP00054 |
| Motif name: | Tcf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0.0230245 |
Alignment:
HHHTCCTTTGATSTHHV
----ACTTTGG------
| Original motif | Reverse complement motif |
| Consensus sequence: BHHASATCAAAGGAHHH | Consensus sequence: HHHTCCTTTGATSTHHV |
 |
 |
| Motif ID: | UP00413 |
| Motif name: | Elf4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0.0283328 |
Alignment:
HAWYCCGGAAGTHBBD
-----CCAAAGT----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGKWTH | Consensus sequence: HAWYCCGGAAGTHBBD |
 |
 |
| Motif ID: | UP00418 |
| Motif name: | Etv6 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.0287289 |
Alignment:
BHAHCCGGAAGTABDVD
-----CCAAAGT-----
| Original motif | Reverse complement motif |
| Consensus sequence: DVHBTACTTCCGGHTHB | Consensus sequence: BHAHCCGGAAGTABDVD |
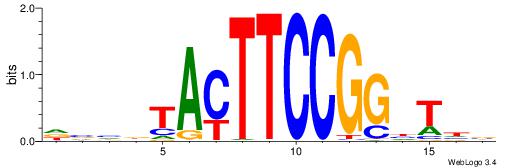 |
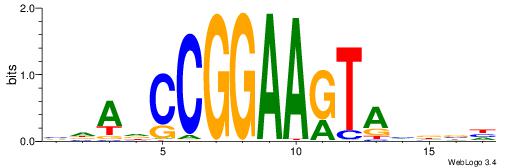 |
| Dataset #: 1 | Motif ID: 38 | Motif name: Motif 38 |
| Original motif | Reverse complement motif |
| Consensus sequence: ATKAYTTTG | Consensus sequence: CAAAKTYAT |
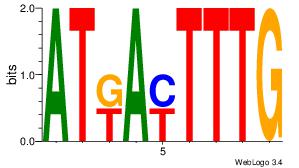 |
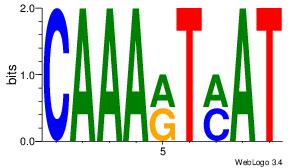 |
Best Matches for Motif ID 38 (Highest to Lowest)
| Motif ID: | UP00079 |
| Motif name: | Esrra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 9 |
| Similarity score: | 0.0159205 |
Alignment:
HHBVHTGACCTTGVDHD
----ATKAYTTTG----
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBCAAGGTCAHVBDH | Consensus sequence: HHBVHTGACCTTGVDHD |
 |
 |
| Motif ID: | UP00054 |
| Motif name: | Tcf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 9 |
| Similarity score: | 0.0182772 |
Alignment:
HHHTCCTTTGATSTHHV
-ATKAYTTTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: BHHASATCAAAGGAHHH | Consensus sequence: HHHTCCTTTGATSTHHV |
 |
 |
| Motif ID: | UP00058 |
| Motif name: | Tcf3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 9 |
| Similarity score: | 0.01865 |
Alignment:
HHHKVCTTTGATSTHHH
-ATKAYTTTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: HHHASATCAAAGVRHHH | Consensus sequence: HHHKVCTTTGATSTHHH |
 |
 |
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 9 |
| Similarity score: | 0.0192718 |
Alignment:
HHBBMCTTTGTTHDDBH
-ATKAYTTTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
 |
 |
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 9 |
| Similarity score: | 0.022524 |
Alignment:
HHBTMCTTTGTTMDDVH
-ATKAYTTTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
 |
 |
| Motif ID: | UP00200 |
| Motif name: | Nkx6-1_2825.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 9 |
| Similarity score: | 0.0239738 |
Alignment:
DVHAATTAATTAMYBBG
--------ATKAYTTTG
| Original motif | Reverse complement motif |
| Consensus sequence: DVHAATTAATTAMYBBG | Consensus sequence: CBVMRTAATTAATTHBH |
 |
 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 9 |
| Similarity score: | 0.0240474 |
Alignment:
VDDAGATCAAAGGKDDD
-------CAAAKTYAT-
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
 |
 |
| Motif ID: | UP00083 |
| Motif name: | Tcf7l2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 9 |
| Similarity score: | 0.0253434 |
Alignment:
DDDYCCTTTGATSTDDV
-ATKAYTTTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATSTDDV | Consensus sequence: BDDASATCAAAGGMDDD |
 |
 |
| Motif ID: | UP00167 |
| Motif name: | En1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 9 |
| Similarity score: | 0.0259902 |
Alignment:
VHRKTAATTAGTTVVV
------ATKAYTTTG-
| Original motif | Reverse complement motif |
| Consensus sequence: VVVAACTAATTARKDV | Consensus sequence: VHRKTAATTAGTTVVV |
 |
 |
| Motif ID: | UP00238 |
| Motif name: | Nkx6-3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 9 |
| Similarity score: | 0.0262036 |
Alignment:
DVHWATTAATTAVTBBB
--------ATKAYTTTG
| Original motif | Reverse complement motif |
| Consensus sequence: DVHWATTAATTAVTBBB | Consensus sequence: BVVAVTAATTAATWHBH |
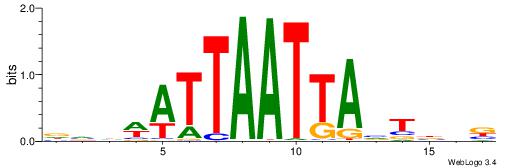 |
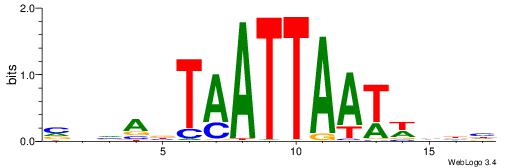 |
| Dataset #: 1 | Motif ID: 39 | Motif name: Motif 39 |
| Original motif | Reverse complement motif |
| Consensus sequence: GCTGTGAC | Consensus sequence: GTCACAGC |
 |
 |
Best Matches for Motif ID 39 (Highest to Lowest)
| Motif ID: | UP00122 |
| Motif name: | Tgif1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
DABWWTGACAGCTVVBD
----GTCACAGC-----
| Original motif | Reverse complement motif |
| Consensus sequence: DABWWTGACAGCTVVBD | Consensus sequence: DBVVAGCTGTCAWWVTH |
 |
 |
| Motif ID: | UP00258 |
| Motif name: | Tgif2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.00241817 |
Alignment:
AABBAGCTGTCAAWVV
-----GCTGTGAC---
| Original motif | Reverse complement motif |
| Consensus sequence: AABBAGCTGTCAAWVV | Consensus sequence: VBWTTGACAGCTVBTT |
 |
 |
| Motif ID: | UP00006 |
| Motif name: | Zic3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 8 |
| Similarity score: | 0.00553853 |
Alignment:
DDDCACAGCAKGHVD
-GTCACAGC------
| Original motif | Reverse complement motif |
| Consensus sequence: DDDCACAGCAKGHVD | Consensus sequence: DVHCYTGCTGTGHDH |
 |
 |
| Motif ID: | UP00068 |
| Motif name: | Eomes_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.00578189 |
Alignment:
BVHDAGGTGTGAAAHHH
-----GCTGTGAC----
| Original motif | Reverse complement motif |
| Consensus sequence: BVHDAGGTGTGAAAHHH | Consensus sequence: HHHTTTCACACCTDHBV |
 |
 |
| Motif ID: | UP00193 |
| Motif name: | Rhox11_2205.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.00629536 |
Alignment:
AVDHCGCTGTWAWDVVW
-----GCTGTGAC----
| Original motif | Reverse complement motif |
| Consensus sequence: AVDHCGCTGTWAWDVVW | Consensus sequence: WVVHWTWACAGCGHHVT |
 |
 |
| Motif ID: | UP00193 |
| Motif name: | Rhox11_1765.2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.00666342 |
Alignment:
AVDVCGCTGTWAWDVVW
-----GCTGTGAC----
| Original motif | Reverse complement motif |
| Consensus sequence: AVDVCGCTGTWAWDVVW | Consensus sequence: WVVHWTWACAGCGBHBT |
 |
 |
| Motif ID: | UP00205 |
| Motif name: | Pknox2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.00747692 |
Alignment:
HAVSRSCTGTCAATHB
-----GCTGTGAC---
| Original motif | Reverse complement motif |
| Consensus sequence: HAVSRSCTGTCAATHB | Consensus sequence: VHATTGACAGSKSVTH |
 |
 |
| Motif ID: | UP00102 |
| Motif name: | Zic1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.00877453 |
Alignment:
BHDCVCAGCAGGHVD
-GTCACAGC------
| Original motif | Reverse complement motif |
| Consensus sequence: BHDCVCAGCAGGHVD | Consensus sequence: DVHCCTGCTGBGDDB |
 |
 |
| Motif ID: | UP00186 |
| Motif name: | Meis1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.00956623 |
Alignment:
WAVBASCTGTCAAWVH
-----GCTGTGAC---
| Original motif | Reverse complement motif |
| Consensus sequence: WAVBASCTGTCAAWVH | Consensus sequence: DBWTTGACAGSTBVTW |
 |
 |
| Motif ID: | UP00226 |
| Motif name: | Mrg1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0124611 |
Alignment:
WAVBASCTGTCAAWHB
-----GCTGTGAC---
| Original motif | Reverse complement motif |
| Consensus sequence: WAVBASCTGTCAAWHB | Consensus sequence: BHWTTGACAGSTBBTW |
 |
 |
| Dataset #: 1 | Motif ID: 40 | Motif name: Motif 40 |
| Original motif | Reverse complement motif |
| Consensus sequence: AAGTTCA | Consensus sequence: TGAACTT |
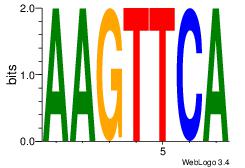 |
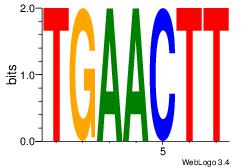 |
Best Matches for Motif ID 40 (Highest to Lowest)
| Motif ID: | UP00048 |
| Motif name: | Rara |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0 |
Alignment:
DHBHAAAGGTCACVHD
-----AAGTTCA----
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCACVHD | Consensus sequence: HHVGTGACCTTTDVDD |
 |
 |
| Motif ID: | UP00066 |
| Motif name: | Hnf4a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.00516503 |
Alignment:
HHAYTGGACTTTHDBV
----TGAACTT-----
| Original motif | Reverse complement motif |
| Consensus sequence: BBHHAAAGTCCAMTHH | Consensus sequence: HHAYTGGACTTTHDBV |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.00773155 |
Alignment:
DHDBCAAGGTCAHVBDH
-----AAGTTCA-----
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBCAAGGTCAHVBDH | Consensus sequence: HHBVHTGACCTTGVDHD |
 |
 |
| Motif ID: | UP00009 |
| Motif name: | Nr2f2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0.00909243 |
Alignment:
DHBHAAAGGTCAHVHB
-----AAGTTCA----
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCAHVHB | Consensus sequence: BHVDTGACCTTTDVDD |
 |
 |
| Motif ID: | UP00019 |
| Motif name: | Zbtb12_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 7 |
| Similarity score: | 0.035752 |
Alignment:
BBGATCTAGAACBKHDB
-------TGAACTT---
| Original motif | Reverse complement motif |
| Consensus sequence: BDHRBGTTCTAGATCVB | Consensus sequence: BBGATCTAGAACBKHDB |
 |
 |
| Motif ID: | UP00222 |
| Motif name: | Tcf2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 7 |
| Similarity score: | 0.0379796 |
Alignment:
ABDBCTAGTTAACVBBD
-----AAGTTCA-----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBBGTTAACTAGBHBT | Consensus sequence: ABDBCTAGTTAACVBBD |
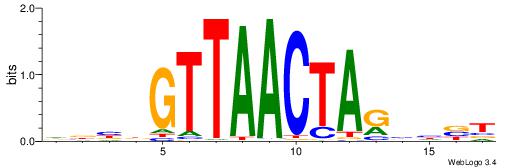 |
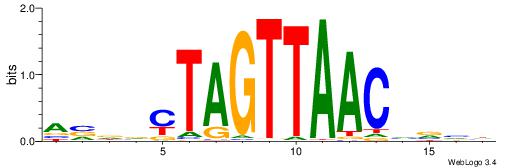 |
| Motif ID: | UP00042 |
| Motif name: | Gm397_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 7 |
| Similarity score: | 0.0388625 |
Alignment:
HHRTATGTGCACATHHD
----AAGTTCA------
| Original motif | Reverse complement motif |
| Consensus sequence: HHDATGTGCACATAMDH | Consensus sequence: HHRTATGTGCACATHHD |
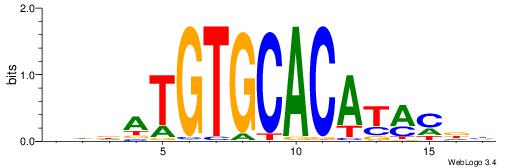 |
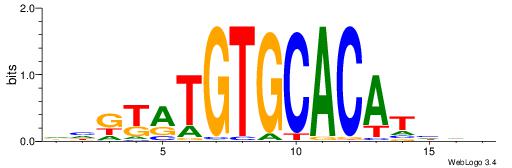 |
| Motif ID: | UP00413 |
| Motif name: | Elf4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 7 |
| Similarity score: | 0.0421261 |
Alignment:
DBBHACTTCCGGKWTH
-TGAACTT--------
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGKWTH | Consensus sequence: HAWYCCGGAAGTHBBD |
 |
 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 7 |
| Similarity score: | 0.0424658 |
Alignment:
DDDYCCTTTGATCTDDV
--------TGAACTT--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
 |
 |
| Motif ID: | UP00026 |
| Motif name: | Zscan4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 7 |
| Similarity score: | 0.0427674 |
Alignment:
HDHTMTGTGCACADVHD
----AAGTTCA------
| Original motif | Reverse complement motif |
| Consensus sequence: DHVDTGTGCACAYAHDH | Consensus sequence: HDHTMTGTGCACADVHD |
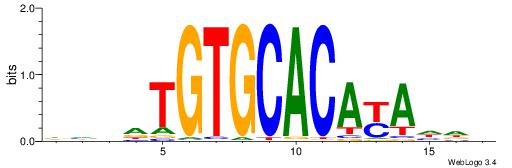 |
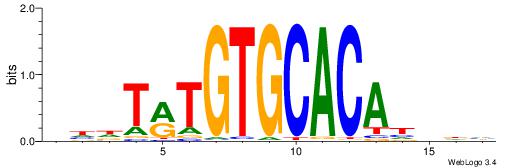 |
| Dataset #: 1 | Motif ID: 41 | Motif name: Motif 41 |
| Original motif | Reverse complement motif |
| Consensus sequence: TGGAGGAAA | Consensus sequence: TTTCCTCCA |
 |
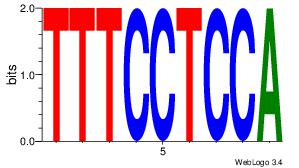 |
Best Matches for Motif ID 41 (Highest to Lowest)
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 9 |
| Similarity score: | 0 |
Alignment:
VHHATTCCTYGAAAGD
---TTTCCTCCA----
| Original motif | Reverse complement motif |
| Consensus sequence: DCTTTCKAGGAATHHV | Consensus sequence: VHHATTCCTYGAAAGD |
 |
 |
| Motif ID: | UP00409 |
| Motif name: | Elf5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 9 |
| Similarity score: | 0.0103598 |
Alignment:
BWAHSMGGAAGTWD
--TGGAGGAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: BWAHSMGGAAGTWD | Consensus sequence: DWACTTCCRSDTWV |
 |
 |
| Motif ID: | UP00090 |
| Motif name: | Elf3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 9 |
| Similarity score: | 0.0126625 |
Alignment:
DAHMMGGAARTDA
-TGGAGGAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: DAHMMGGAARTDA | Consensus sequence: TDAMTTCCYYDTD |
 |
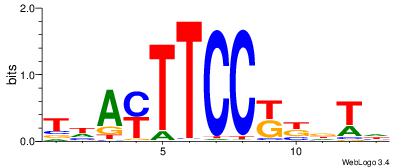 |
| Motif ID: | UP00085 |
| Motif name: | Sfpi1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 9 |
| Similarity score: | 0.0131092 |
Alignment:
BDAWGVGGAAGTDH
--TGGAGGAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: BDAWGVGGAAGTDH | Consensus sequence: HDACTTCCBCWTDV |
 |
 |
| Motif ID: | UP00055 |
| Motif name: | Hbp1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 9 |
| Similarity score: | 0.0140163 |
Alignment:
HHDWTSAATGAATDDB
----TGGAGGAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: HHDWTSAATGAATDDB | Consensus sequence: VDHATTCATTSAWDDH |
 |
 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 9 |
| Similarity score: | 0.0142598 |
Alignment:
DKKWDACCCCCAHTDDV
---TTTCCTCCA-----
| Original motif | Reverse complement motif |
| Consensus sequence: DKKWDACCCCCAHTDDV | Consensus sequence: VDDAHTGGGGGTHWYYD |
 |
 |
| Motif ID: | UP00208 |
| Motif name: | Obox5_2284.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 9 |
| Similarity score: | 0.0161902 |
Alignment:
DHHAHTTAATCCCKVDH
----TTTCCTCCA----
| Original motif | Reverse complement motif |
| Consensus sequence: HDVRGGGATTAAHTHHH | Consensus sequence: DHHAHTTAATCCCKVDH |
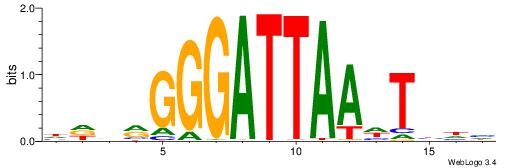 |
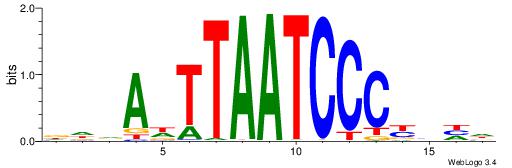 |
| Motif ID: | UP00419 |
| Motif name: | Spic |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 9 |
| Similarity score: | 0.0168157 |
Alignment:
VAAAGMGGAAGTWD
--TGGAGGAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: VAAAGMGGAAGTWD | Consensus sequence: DWACTTCCRCTTTB |
 |
 |
| Motif ID: | UP00208 |
| Motif name: | Obox5_3963.2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 9 |
| Similarity score: | 0.0175071 |
Alignment:
VWBAHTTAATCCCTVKB
----TTTCCTCCA----
| Original motif | Reverse complement motif |
| Consensus sequence: VRVAGGGATTAAHTVWV | Consensus sequence: VWBAHTTAATCCCTVKB |
 |
 |
| Motif ID: | UP00082 |
| Motif name: | Zfp187_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 9 |
| Similarity score: | 0.017867 |
Alignment:
HWATTAGTACATAV
--TTTCCTCCA---
| Original motif | Reverse complement motif |
| Consensus sequence: BTATGTACTAATWH | Consensus sequence: HWATTAGTACATAV |
 |
 |
| Dataset #: 1 | Motif ID: 42 | Motif name: Motif 42 |
| Original motif | Reverse complement motif |
| Consensus sequence: TGAATGAA | Consensus sequence: TTCATTCA |
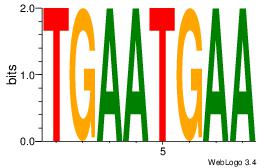 |
 |
Best Matches for Motif ID 42 (Highest to Lowest)
| Motif ID: | UP00055 |
| Motif name: | Hbp1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HHDWTSAATGAATDDB
----TGAATGAA----
| Original motif | Reverse complement motif |
| Consensus sequence: HHDWTSAATGAATDDB | Consensus sequence: VDHATTCATTSAWDDH |
 |
 |
| Motif ID: | UP00118 |
| Motif name: | Pou4f3 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0420896 |
Alignment:
HVTTATTAATKADKHC
-----TGAATGAA---
| Original motif | Reverse complement motif |
| Consensus sequence: HVTTATTAATKADKHC | Consensus sequence: GHYHTYATTAATAAVH |
 |
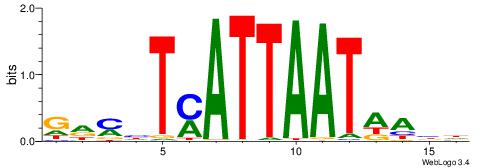 |
| Motif ID: | UP00237 |
| Motif name: | Otp |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0426251 |
Alignment:
CHDMTTAATTAAKDHVV
----TTCATTCA-----
| Original motif | Reverse complement motif |
| Consensus sequence: VVHDRTTAATTAAYDDG | Consensus sequence: CHDMTTAATTAAKDHVV |
 |
 |
| Motif ID: | UP00130 |
| Motif name: | Lhx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0427891 |
Alignment:
VHHWWTTAATTAATTDV
-----TTCATTCA----
| Original motif | Reverse complement motif |
| Consensus sequence: VDAATTAATTAAWWHHB | Consensus sequence: VHHWWTTAATTAATTDV |
 |
 |
| Motif ID: | UP00212 |
| Motif name: | Lhx5 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.044202 |
Alignment:
VDHWTTTAATTAATHHV
-----TTCATTCA----
| Original motif | Reverse complement motif |
| Consensus sequence: VDHATTAATTAAAWHHB | Consensus sequence: VDHWTTTAATTAATHHV |
 |
 |
| Motif ID: | UP00012 |
| Motif name: | Bbx_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0451439 |
Alignment:
HWBTTCAWTGAADTH
---TTCATTCA----
| Original motif | Reverse complement motif |
| Consensus sequence: HADTTCAWTGAABWD | Consensus sequence: HWBTTCAWTGAADTH |
 |
 |
| Motif ID: | UP00142 |
| Motif name: | Uncx4.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0455949 |
Alignment:
CVVBTTAATTAATDHDV
----TGAATGAA-----
| Original motif | Reverse complement motif |
| Consensus sequence: VDHDATTAATTAABVVG | Consensus sequence: CVVBTTAATTAATDHDV |
 |
 |
| Motif ID: | UP00172 |
| Motif name: | Prop1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0460946 |
Alignment:
GDDDBTTAATTAABBVV
-----TTCATTCA----
| Original motif | Reverse complement motif |
| Consensus sequence: VVVVTTAATTAABDDDC | Consensus sequence: GDDDBTTAATTAABBVV |
 |
 |
| Motif ID: | UP00169 |
| Motif name: | Lmx1b |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.048175 |
Alignment:
BHDATTAATTAAWWWHB
----TGAATGAA-----
| Original motif | Reverse complement motif |
| Consensus sequence: VDWWWTTAATTAATDHB | Consensus sequence: BHDATTAATTAAWWWHB |
 |
 |
| Motif ID: | UP00149 |
| Motif name: | Phox2b |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0491281 |
Alignment:
CBDVTTAATTAATTBVV
----TTCATTCA-----
| Original motif | Reverse complement motif |
| Consensus sequence: VVBAATTAATTAABDBG | Consensus sequence: CBDVTTAATTAATTBVV |
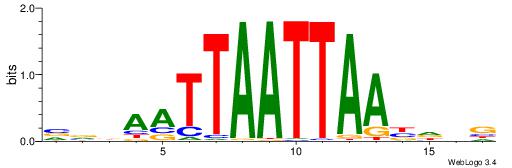 |
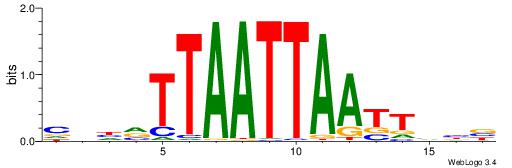 |
| Dataset #: 1 | Motif ID: 43 | Motif name: Motif 43 |
| Original motif | Reverse complement motif |
| Consensus sequence: AAKGAAAGGCA | Consensus sequence: TGCCTTTCYTT |
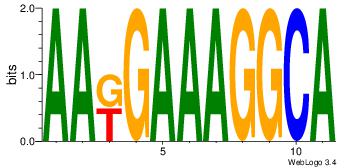 |
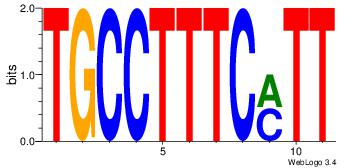 |
Best Matches for Motif ID 43 (Highest to Lowest)
| Motif ID: | UP00074 |
| Motif name: | Isgf3g_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.012358 |
Alignment:
TDAGTTTCGWTTKDV
TGCCTTTCYTT----
| Original motif | Reverse complement motif |
| Consensus sequence: VDRAAWCGAAACTDA | Consensus sequence: TDAGTTTCGWTTKDV |
 |
 |
| Motif ID: | UP00083 |
| Motif name: | Tcf7l2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.014351 |
Alignment:
BDDASATCAAAGGMDDD
----AAKGAAAGGCA--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATSTDDV | Consensus sequence: BDDASATCAAAGGMDDD |
 |
 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.015991 |
Alignment:
VDDAGATCAAAGGKDDD
----AAKGAAAGGCA--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
 |
 |
| Motif ID: | UP00054 |
| Motif name: | Tcf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0221257 |
Alignment:
BHHASATCAAAGGAHHH
----AAKGAAAGGCA--
| Original motif | Reverse complement motif |
| Consensus sequence: BHHASATCAAAGGAHHH | Consensus sequence: HHHTCCTTTGATSTHHV |
 |
 |
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0233703 |
Alignment:
HHBTMCTTTGTTMDDVH
--TGCCTTTCYTT----
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
 |
 |
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0234975 |
Alignment:
HHBBMCTTTGTTHDDBH
--TGCCTTTCYTT----
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
 |
 |
| Motif ID: | UP00058 |
| Motif name: | Tcf3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0239255 |
Alignment:
HHHASATCAAAGVRHHH
----AAKGAAAGGCA--
| Original motif | Reverse complement motif |
| Consensus sequence: HHHASATCAAAGVRHHH | Consensus sequence: HHHKVCTTTGATSTHHH |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0239781 |
Alignment:
DHDBCAAGGTCAHVBDH
-AAKGAAAGGCA-----
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBCAAGGTCAHVBDH | Consensus sequence: HHBVHTGACCTTGVDHD |
 |
 |
| Motif ID: | UP00044 |
| Motif name: | Mafk_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0264082 |
Alignment:
MBHTGCADTTTTTHV
---TGCCTTTCYTT-
| Original motif | Reverse complement motif |
| Consensus sequence: VHAAAAADTGCAHBR | Consensus sequence: MBHTGCADTTTTTHV |
 |
 |
| Motif ID: | UP00217 |
| Motif name: | Hoxa10 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0273937 |
Alignment:
DHBGYAATAAAATDHW
-----AAKGAAAGGCA
| Original motif | Reverse complement motif |
| Consensus sequence: DHBGYAATAAAATDHW | Consensus sequence: WDDATTTTATTMCBHD |
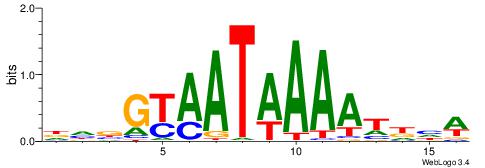 |
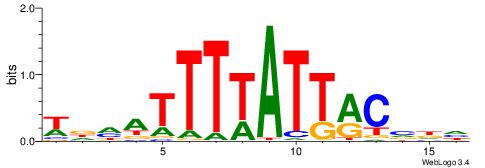 |
| Dataset #: 1 | Motif ID: 44 | Motif name: Motif 44 |
| Original motif | Reverse complement motif |
| Consensus sequence: AATGKAAGAA | Consensus sequence: TTCTTYCATT |
 |
 |
Best Matches for Motif ID 44 (Highest to Lowest)
| Motif ID: | UP00082 |
| Motif name: | Zfp187_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0293208 |
Alignment:
BTATGTACTAATWH
-AATGKAAGAA---
| Original motif | Reverse complement motif |
| Consensus sequence: BTATGTACTAATWH | Consensus sequence: HWATTAGTACATAV |
 |
 |
| Motif ID: | UP00055 |
| Motif name: | Hbp1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0314105 |
Alignment:
VDHATTCATTSAWDDH
TTCTTYCATT------
| Original motif | Reverse complement motif |
| Consensus sequence: HHDWTSAATGAATDDB | Consensus sequence: VDHATTCATTSAWDDH |
 |
 |
| Motif ID: | UP00025 |
| Motif name: | Foxk1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0385918 |
Alignment:
DDAATGTAAACAAAVVB
--AATGKAAGAA-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDAATGTAAACAAAVVB | Consensus sequence: BBVTTTGTTTACATTDD |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0392784 |
Alignment:
DWAATRTAAACAAWVVB
--AATGKAAGAA-----
| Original motif | Reverse complement motif |
| Consensus sequence: DWAATRTAAACAAWVVB | Consensus sequence: VVVWTTGTTTAMATTWD |
 |
 |
| Motif ID: | UP00157 |
| Motif name: | Hmx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0433812 |
Alignment:
ATBVHTTAATTGCTBBB
-TTCTTYCATT------
| Original motif | Reverse complement motif |
| Consensus sequence: VBVAGCAATTAAHVVAT | Consensus sequence: ATBVHTTAATTGCTBBB |
 |
 |
| Motif ID: | UP00083 |
| Motif name: | Tcf7l2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0436699 |
Alignment:
DDDYCCTTTGATSTDDV
---TTCTTYCATT----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATSTDDV | Consensus sequence: BDDASATCAAAGGMDDD |
 |
 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0444106 |
Alignment:
DDDYCCTTTGATCTDDV
---TTCTTYCATT----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
 |
 |
| Motif ID: | UP00155 |
| Motif name: | Hmx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0454888 |
Alignment:
ADBVHTTAATTGCTTVB
-TTCTTYCATT------
| Original motif | Reverse complement motif |
| Consensus sequence: VVAAGCAATTAAHVVDT | Consensus sequence: ADBVHTTAATTGCTTVB |
 |
 |
| Motif ID: | UP00134 |
| Motif name: | Hoxb13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0479046 |
Alignment:
VDHWTTTTATTGGDKB
-TTCTTYCATT-----
| Original motif | Reverse complement motif |
| Consensus sequence: VRHCCAATAAAAWHHV | Consensus sequence: VDHWTTTTATTGGDKB |
 |
 |
| Motif ID: | UP00180 |
| Motif name: | Hoxd13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0481894 |
Alignment:
HDHHTTTTATTKKHDD
-TTCTTYCATT-----
| Original motif | Reverse complement motif |
| Consensus sequence: HDHYYAATAAAAHHHH | Consensus sequence: HDHHTTTTATTKKHDD |
 |
 |
| Dataset #: 1 | Motif ID: 45 | Motif name: Motif 45 |
| Original motif | Reverse complement motif |
| Consensus sequence: TAASTCTWTTTTAA | Consensus sequence: TTAAAAWAGASTTA |
 |
 |
Best Matches for Motif ID 45 (Highest to Lowest)
| Motif ID: | UP00114 |
| Motif name: | Homez |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0426024 |
Alignment:
AHWWMATCGTTTTBADD
--TAASTCTWTTTTAA-
| Original motif | Reverse complement motif |
| Consensus sequence: AHWWMATCGTTTTBADD | Consensus sequence: HDTVAAAACGATRWWHT |
 |
 |
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.04935 |
Alignment:
HHBBMCTTTGTTHDDBH
---TTAAAAWAGASTTA
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.542468 |
Alignment:
DWAATRTAAACAAWVVB-
-TAASTCTWTTTTAA---
| Original motif | Reverse complement motif |
| Consensus sequence: DWAATRTAAACAAWVVB | Consensus sequence: VVVWTTGTTTAMATTWD |
 |
 |
| Motif ID: | UP00172 |
| Motif name: | Prop1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 13 |
| Similarity score: | 0.545182 |
Alignment:
VVVVTTAATTAABDDDC-
----TTAAAAWAGASTTA
| Original motif | Reverse complement motif |
| Consensus sequence: VVVVTTAATTAABDDDC | Consensus sequence: GDDDBTTAATTAABBVV |
 |
 |
| Motif ID: | UP00212 |
| Motif name: | Lhx5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 13 |
| Similarity score: | 0.547386 |
Alignment:
-VDHWTTTAATTAATHHV
TAASTCTWTTTTAA----
| Original motif | Reverse complement motif |
| Consensus sequence: VDHATTAATTAAAWHHB | Consensus sequence: VDHWTTTAATTAATHHV |
 |
 |
| Motif ID: | UP00237 |
| Motif name: | Otp |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 13 |
| Similarity score: | 0.548444 |
Alignment:
CHDMTTAATTAAKDHVV-
----TTAAAAWAGASTTA
| Original motif | Reverse complement motif |
| Consensus sequence: VVHDRTTAATTAAYDDG | Consensus sequence: CHDMTTAATTAAKDHVV |
 |
 |
| Motif ID: | UP00130 |
| Motif name: | Lhx3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.548839 |
Alignment:
-VHHWWTTAATTAATTDV
---TTAAAAWAGASTTA-
| Original motif | Reverse complement motif |
| Consensus sequence: VDAATTAATTAAWWHHB | Consensus sequence: VHHWWTTAATTAATTDV |
 |
 |
| Motif ID: | UP00145 |
| Motif name: | Barhl2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 1.03054 |
Alignment:
--HAMAAHCAATTAABHH
--TAASTCTWTTTTAA--
| Original motif | Reverse complement motif |
| Consensus sequence: HAMAAHCAATTAABHH | Consensus sequence: HHBTTAATTGDTTYTH |
 |
 |
| Motif ID: | UP00109 |
| Motif name: | Obox6 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 1.0344 |
Alignment:
--AHAADCGGATTAWHB
TTAAAAWAGASTTA---
| Original motif | Reverse complement motif |
| Consensus sequence: AHAADCGGATTAWHB | Consensus sequence: BHWTAATCCGDTTHT |
 |
 |
| Motif ID: | UP00166 |
| Motif name: | Barhl1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 1.03746 |
Alignment:
--HAHAACCAATTARBDH
TTAAAAWAGASTTA----
| Original motif | Reverse complement motif |
| Consensus sequence: HAHAACCAATTARBDH | Consensus sequence: DDVKTAATTGGTTDTH |
 |
 |
| Dataset #: 1 | Motif ID: 46 | Motif name: Motif 46 |
| Original motif | Reverse complement motif |
| Consensus sequence: ATAGGTTTAGYATA | Consensus sequence: TATKCTAAACCTAT |
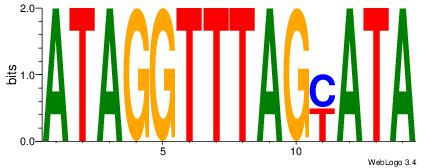 |
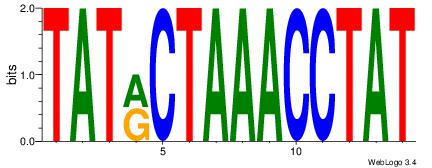 |
Best Matches for Motif ID 46 (Highest to Lowest)
| Motif ID: | UP00111 |
| Motif name: | Dmbx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0364989 |
Alignment:
WDRWHMGGATTADKBHD
---ATAGGTTTAGYATA
| Original motif | Reverse complement motif |
| Consensus sequence: WDRWHMGGATTADKBHD | Consensus sequence: DHBRDTAATCCRDWKHW |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0393329 |
Alignment:
DWAATRTAAACAAWVVB
-TATKCTAAACCTAT--
| Original motif | Reverse complement motif |
| Consensus sequence: DWAATRTAAACAAWVVB | Consensus sequence: VVVWTTGTTTAMATTWD |
 |
 |
| Motif ID: | UP00068 |
| Motif name: | Eomes_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0394759 |
Alignment:
VVBVCKACACCTHVBV
TATKCTAAACCTAT--
| Original motif | Reverse complement motif |
| Consensus sequence: VBVDAGGTGTYGVBBV | Consensus sequence: VVBVCKACACCTHVBV |
 |
 |
| Motif ID: | UP00112 |
| Motif name: | Gsc |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0411853 |
Alignment:
HVWHRYTAATCCSYDHV
-TATKCTAAACCTAT--
| Original motif | Reverse complement motif |
| Consensus sequence: HVWHRYTAATCCSYDHV | Consensus sequence: BHDMSGGATTAMMDWBH |
 |
 |
| Motif ID: | UP00018 |
| Motif name: | Irf4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.041683 |
Alignment:
VDDAYCGAAACYDVA
ATAGGTTTAGYATA-
| Original motif | Reverse complement motif |
| Consensus sequence: VDDAYCGAAACYDVA | Consensus sequence: TBDKGTTTCGMTDHV |
 |
 |
| Motif ID: | UP00025 |
| Motif name: | Foxk1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0422263 |
Alignment:
DDAATGTAAACAAAVVB
-TATKCTAAACCTAT--
| Original motif | Reverse complement motif |
| Consensus sequence: DDAATGTAAACAAAVVB | Consensus sequence: BBVTTTGTTTACATTDD |
 |
 |
| Motif ID: | UP00011 |
| Motif name: | Irf6_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.042495 |
Alignment:
DHDBTKGTTTCGHTHDD
--ATAGGTTTAGYATA-
| Original motif | Reverse complement motif |
| Consensus sequence: HDDAHCGAAACYAVDDD | Consensus sequence: DHDBTKGTTTCGHTHDD |
 |
 |
| Motif ID: | UP00125 |
| Motif name: | Pitx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0436835 |
Alignment:
DDDVRGGATTAATHDDH
--ATAGGTTTAGYATA-
| Original motif | Reverse complement motif |
| Consensus sequence: DDDVRGGATTAATHDDH | Consensus sequence: DDDDATTAATCCMBDHD |
 |
 |
| Motif ID: | UP00229 |
| Motif name: | Otx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0443397 |
Alignment:
DDDRGGGATTAATWHHB
--ATAGGTTTAGYATA-
| Original motif | Reverse complement motif |
| Consensus sequence: DDDRGGGATTAATWHHB | Consensus sequence: VHHWATTAATCCCMDHH |
 |
 |
| Motif ID: | UP00153 |
| Motif name: | Pitx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.044637 |
Alignment:
HBVDRGGGATTAAMDHH
---ATAGGTTTAGYATA
| Original motif | Reverse complement motif |
| Consensus sequence: HBVDRGGGATTAAMDHH | Consensus sequence: HHDRTTAATCCCKHBVH |
 |
 |
| Dataset #: 2 | Motif ID: 47 | Motif name: Motif 47 |
| Original motif | Reverse complement motif |
| Consensus sequence: YGCCACCTDSTGGY | Consensus sequence: KCCASDAGGTGGCK |
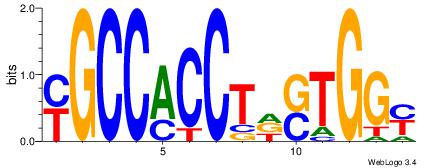 |
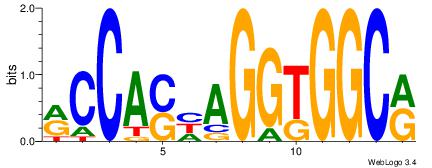 |
Best Matches for Motif ID 47 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 14 |
| Similarity score: | 0.0145956 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
---KCCASDAGGTGGCK------
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 14 |
| Similarity score: | 0.0188184 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
------YGCCACCTDSTGGY--
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 14 |
| Similarity score: | 0.0231104 |
Alignment:
HGDHVHKGACCACCCGGBGBGDB
-------YGCCACCTDSTGGY--
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.025419 |
Alignment:
DDDTWCCCCCCGGDRVH
---YGCCACCTDSTGGY
| Original motif | Reverse complement motif |
| Consensus sequence: DDDTWCCCCCCGGDRVH | Consensus sequence: HVKDCCGGGGGGWADDD |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0278794 |
Alignment:
HDGHHBHRGACCACCCGSVDVG
--------YGCCACCTDSTGGY
| Original motif | Reverse complement motif |
| Consensus sequence: HDGHHBHRGACCACCCGSVDVG | Consensus sequence: CVHBSCGGGTGGTCMHVHHCDH |
 |
 |
| Motif ID: | UP00006 |
| Motif name: | Zic3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0283962 |
Alignment:
BMCCCCCGGGGGGGB
YGCCACCTDSTGGY-
| Original motif | Reverse complement motif |
| Consensus sequence: BMCCCCCGGGGGGGB | Consensus sequence: VCCCCCCCGGGGGRB |
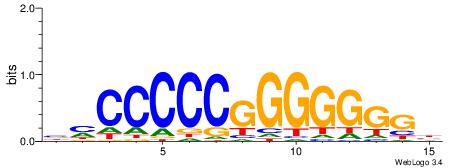 |
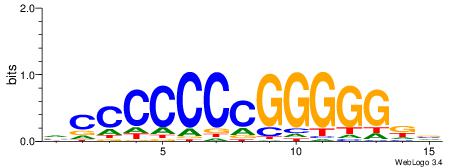 |
| Motif ID: | UP00102 |
| Motif name: | Zic1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0287857 |
Alignment:
BHCCCCCGGGGGGG
YGCCACCTDSTGGY
| Original motif | Reverse complement motif |
| Consensus sequence: BHCCCCCGGGGGGG | Consensus sequence: CCCCCCCGGGGGHB |
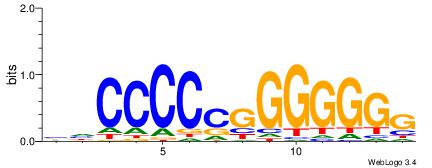 |
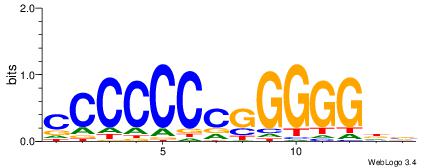 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 14 |
| Similarity score: | 0.0289988 |
Alignment:
MHHHWHYTMCCHCCCACVAABVH
-------YGCCACCTDSTGGY--
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 14 |
| Similarity score: | 0.0297857 |
Alignment:
HHBSHTCGTGRGTGGKGDBVWMV
---KCCASDAGGTGGCK------
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00057 |
| Motif name: | Zic2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0302183 |
Alignment:
BMCCCCCGGGGGGGB
YGCCACCTDSTGGY-
| Original motif | Reverse complement motif |
| Consensus sequence: BMCCCCCGGGGGGGB | Consensus sequence: VCCCCCCCGGGGGRB |
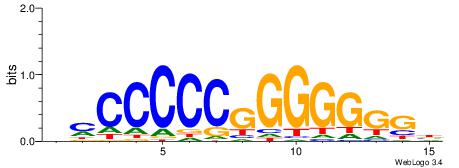 |
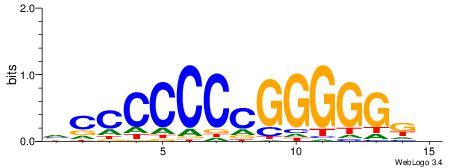 |
| Dataset #: 2 | Motif ID: 48 | Motif name: Motif 48 |
| Original motif | Reverse complement motif |
| Consensus sequence: GCARRGGSAGS | Consensus sequence: SCTSCCKMTGC |
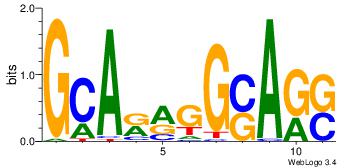 |
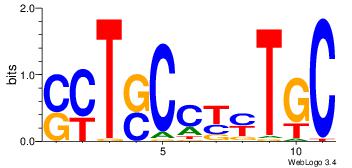 |
Best Matches for Motif ID 48 (Highest to Lowest)
| Motif ID: | UP00153 |
| Motif name: | Pitx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.0454693 |
Alignment:
HHDRTTAATCCCKHBVH
------SCTSCCKMTGC
| Original motif | Reverse complement motif |
| Consensus sequence: HBVDRGGGATTAAMDHH | Consensus sequence: HHDRTTAATCCCKHBVH |
 |
 |
| Motif ID: | UP00085 |
| Motif name: | Sfpi1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0458219 |
Alignment:
BDAWGVGGAAGTDH
GCARRGGSAGS---
| Original motif | Reverse complement motif |
| Consensus sequence: BDAWGVGGAAGTDH | Consensus sequence: HDACTTCCBCWTDV |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 11 |
| Similarity score: | 0.0463674 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
-------SCTSCCKMTGC----
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0465857 |
Alignment:
BADHBDHCGCCCMCGCAHHDBBV
-----SCTSCCKMTGC-------
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0473355 |
Alignment:
BBYVVCAGCTGCBVHHD
-SCTSCCKMTGC-----
| Original motif | Reverse complement motif |
| Consensus sequence: BBYVVCAGCTGCBVHHD | Consensus sequence: HHDVVGCAGCTGVBKVB |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0487126 |
Alignment:
VHYTCCDWCCGCGTCMMTHWHV
-SCTSCCKMTGC----------
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00010 |
| Motif name: | Tcfap2b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0497623 |
Alignment:
DTWGCCKSMGGCRHH
----GCARRGGSAGS
| Original motif | Reverse complement motif |
| Consensus sequence: DTWGCCKSMGGCRHH | Consensus sequence: HHKGCCYSRGGCWAD |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0507547 |
Alignment:
DVMHHDHKGACCCTCCTSVCBH
-----------SCTSCCKMTGC
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Motif ID: | UP00055 |
| Motif name: | Hbp1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0508648 |
Alignment:
HBTHVACAATGGGMDHD
-----GCARRGGSAGS-
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCCATTGTVHABH | Consensus sequence: HBTHVACAATGGGMDHD |
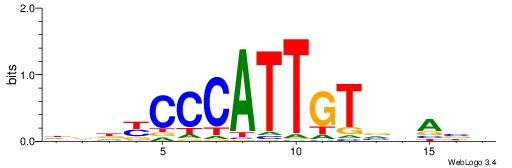 |
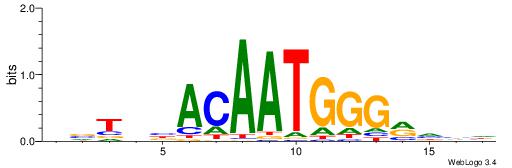 |
| Motif ID: | UP00005 |
| Motif name: | Tcfap2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0512938 |
Alignment:
WDHSCCYSRGGSRAD
----GCARRGGSAGS
| Original motif | Reverse complement motif |
| Consensus sequence: WDHSCCYSRGGSRAD | Consensus sequence: DTMSCCKSMGGSHDW |
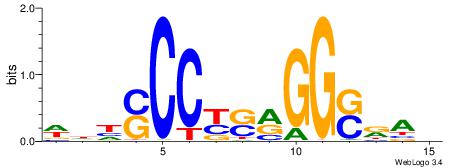 |
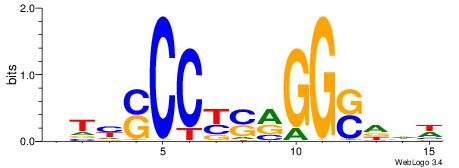 |
| Dataset #: 2 | Motif ID: 49 | Motif name: Motif 49 |
| Original motif | Reverse complement motif |
| Consensus sequence: AAAAAAAAAAAAAT | Consensus sequence: ATTTTTTTTTTTTT |
 |
 |
Best Matches for Motif ID 49 (Highest to Lowest)
| Motif ID: | UP00037 |
| Motif name: | Zfp105_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0361736 |
Alignment:
HHHTTRTTDTTTKDH
ATTTTTTTTTTTTT-
| Original motif | Reverse complement motif |
| Consensus sequence: HDYAAAHAAMAADHD | Consensus sequence: HHHTTRTTDTTTKDH |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0391439 |
Alignment:
DWAATRTAAACAAWVVB
-AAAAAAAAAAAAAT--
| Original motif | Reverse complement motif |
| Consensus sequence: DWAATRTAAACAAWVVB | Consensus sequence: VVVWTTGTTTAMATTWD |
 |
 |
| Motif ID: | UP00077 |
| Motif name: | Srf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0416036 |
Alignment:
VBDDAAAAAAAAMVBHH
--AAAAAAAAAAAAAT-
| Original motif | Reverse complement motif |
| Consensus sequence: VBDDAAAAAAAAMVBHH | Consensus sequence: HHVBYTTTTTTTTDDVV |
 |
 |
| Motif ID: | UP00073 |
| Motif name: | Foxa2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0420875 |
Alignment:
BDVTTTKTTTACWTWHH
--ATTTTTTTTTTTTT-
| Original motif | Reverse complement motif |
| Consensus sequence: HHWAWGTAAAYAAAVDB | Consensus sequence: BDVTTTKTTTACWTWHH |
 |
 |
| Motif ID: | UP00180 |
| Motif name: | Hoxd13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0424132 |
Alignment:
HDHHTTTTATTKKHDD
--ATTTTTTTTTTTTT
| Original motif | Reverse complement motif |
| Consensus sequence: HDHYYAATAAAAHHHH | Consensus sequence: HDHHTTTTATTKKHDD |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0424666 |
Alignment:
HDTYTTGTTWTKDDDT
-ATTTTTTTTTTTTT-
| Original motif | Reverse complement motif |
| Consensus sequence: ADDDYAWAACAAMAHH | Consensus sequence: HDTYTTGTTWTKDDDT |
 |
 |
| Motif ID: | UP00025 |
| Motif name: | Foxk1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0430608 |
Alignment:
BBVTTTGTTTACATTDD
--ATTTTTTTTTTTTT-
| Original motif | Reverse complement motif |
| Consensus sequence: DDAATGTAAACAAAVVB | Consensus sequence: BBVTTTGTTTACATTDD |
 |
 |
| Motif ID: | UP00134 |
| Motif name: | Hoxb13 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0435537 |
Alignment:
VDHWTTTTATTGGDKB
--ATTTTTTTTTTTTT
| Original motif | Reverse complement motif |
| Consensus sequence: VRHCCAATAAAAWHHV | Consensus sequence: VDHWTTTTATTGGDKB |
 |
 |
| Motif ID: | UP00025 |
| Motif name: | Foxk1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.043907 |
Alignment:
HDDTGTTKTTKTHHD
ATTTTTTTTTTTTT-
| Original motif | Reverse complement motif |
| Consensus sequence: HHHAYAAYAACAHHH | Consensus sequence: HDDTGTTKTTKTHHD |
 |
 |
| Motif ID: | UP00039 |
| Motif name: | Foxj3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.045165 |
Alignment:
HDHADGTAAACAAAVVH
-AAAAAAAAAAAAAT--
| Original motif | Reverse complement motif |
| Consensus sequence: HDHADGTAAACAAAVVH | Consensus sequence: DVVTTTGTTTACDTHDH |
 |
 |
| Dataset #: 2 | Motif ID: 50 | Motif name: Motif 50 |
| Original motif | Reverse complement motif |
| Consensus sequence: GGAAM | Consensus sequence: YTTCC |
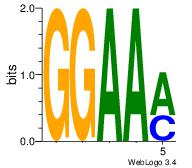 |
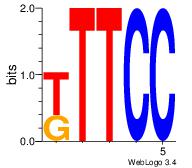 |
Best Matches for Motif ID 50 (Highest to Lowest)
| Motif ID: | UP00424 |
| Motif name: | Gm4881 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0 |
Alignment:
VBBHAYTTCCGGTDDD
-----YTTCC------
| Original motif | Reverse complement motif |
| Consensus sequence: VBBHAYTTCCGGTDDD | Consensus sequence: DHDACCGGAAMTHBVB |
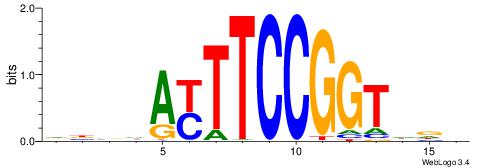 |
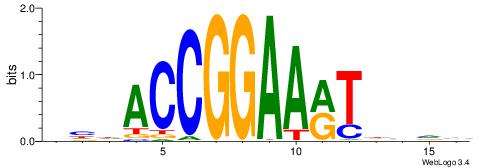 |
| Motif ID: | UP00416 |
| Motif name: | Fli1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.00163794 |
Alignment:
VBBVAYTTCCGGTDDH
-----YTTCC------
| Original motif | Reverse complement motif |
| Consensus sequence: VBBVAYTTCCGGTDDH | Consensus sequence: DHDACCGGAAKTBBVV |
 |
 |
| Motif ID: | UP00422 |
| Motif name: | Etv3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.00234412 |
Alignment:
VBBHAYTTCCGGTHDH
-----YTTCC------
| Original motif | Reverse complement motif |
| Consensus sequence: VBBHAYTTCCGGTHDH | Consensus sequence: DHHACCGGAAKTHBBB |
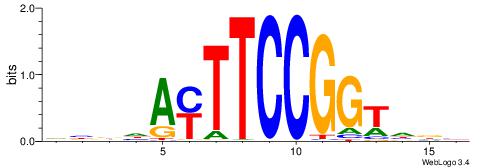 |
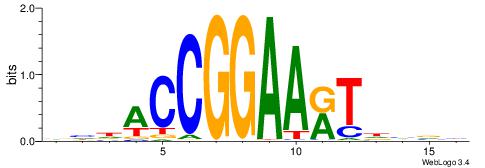 |
| Motif ID: | UP00411 |
| Motif name: | Erg |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.00589761 |
Alignment:
VBBVAYTTCCGGTDDB
-----YTTCC------
| Original motif | Reverse complement motif |
| Consensus sequence: VBBVAYTTCCGGTDDB | Consensus sequence: BDDACCGGAAKTVBVB |
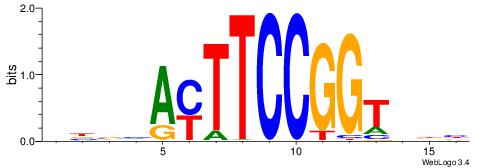 |
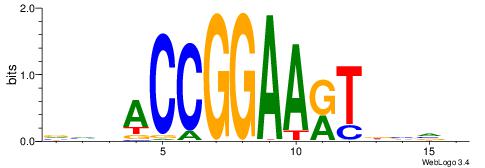 |
| Motif ID: | UP00418 |
| Motif name: | Etv6 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 5 |
| Similarity score: | 0.00691345 |
Alignment:
DVHBTACTTCCGGHTHB
------YTTCC------
| Original motif | Reverse complement motif |
| Consensus sequence: DVHBTACTTCCGGHTHB | Consensus sequence: BHAHCCGGAAGTABDVD |
 |
 |
| Motif ID: | UP00415 |
| Motif name: | Elk4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 5 |
| Similarity score: | 0.00722176 |
Alignment:
VBBHRYTTCCGGTDDH
-----YTTCC------
| Original motif | Reverse complement motif |
| Consensus sequence: VBBHRYTTCCGGTDDH | Consensus sequence: DHDACCGGAAKKHBVB |
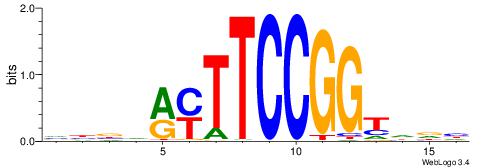 |
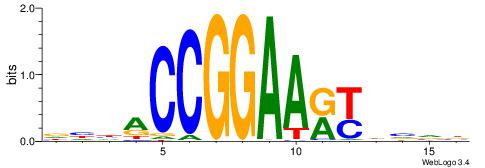 |
| Motif ID: | UP00413 |
| Motif name: | Elf4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 5 |
| Similarity score: | 0.0153114 |
Alignment:
DBBHACTTCCGGKWTH
-----YTTCC------
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGKWTH | Consensus sequence: HAWYCCGGAAGTHBBD |
 |
 |
| Motif ID: | UP00404 |
| Motif name: | Elf2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 5 |
| Similarity score: | 0.0153458 |
Alignment:
DBBHACTTCCGGDWDB
-----YTTCC------
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGDWDB | Consensus sequence: BDWDCCGGAAGTHBBD |
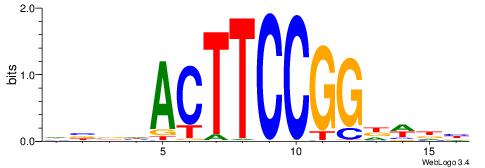 |
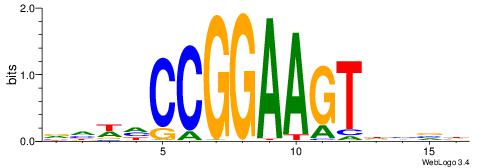 |
| Motif ID: | UP00417 |
| Motif name: | Etv4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 5 |
| Similarity score: | 0.0219684 |
Alignment:
VBBHAYWTCCGGTDDB
-----YTTCC------
| Original motif | Reverse complement motif |
| Consensus sequence: VBBHAYWTCCGGTDDB | Consensus sequence: BHHACCGGAWKTHBVV |
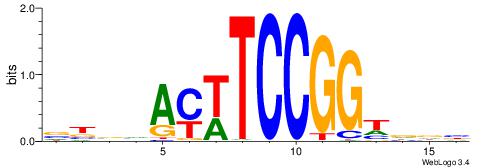 |
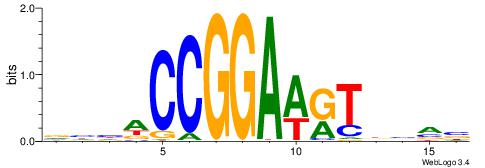 |
| Motif ID: | UP00419 |
| Motif name: | Spic |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 5 |
| Similarity score: | 0.0224017 |
Alignment:
DWACTTCCRCTTTB
---YTTCC------
| Original motif | Reverse complement motif |
| Consensus sequence: VAAAGMGGAAGTWD | Consensus sequence: DWACTTCCRCTTTB |
 |
 |
| Dataset #: 3 | Motif ID: 51 | Motif name: HNF4A |
| Original motif | Reverse complement motif |
| Consensus sequence: RGGBCAAAGKYCA | Consensus sequence: TGMYCTTTGBCCK |
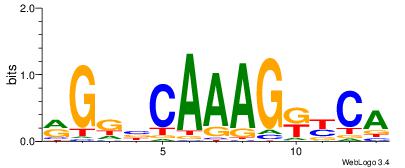 |
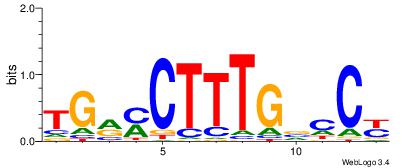 |
Best Matches for Motif ID 51 (Highest to Lowest)
| Motif ID: | UP00058 |
| Motif name: | Tcf3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0357776 |
Alignment:
HHHASATCAAAGVRHHH
---RGGBCAAAGKYCA-
| Original motif | Reverse complement motif |
| Consensus sequence: HHHASATCAAAGVRHHH | Consensus sequence: HHHKVCTTTGATSTHHH |
 |
 |
| Motif ID: | UP00054 |
| Motif name: | Tcf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0357792 |
Alignment:
BHHASATCAAAGGAHHH
---RGGBCAAAGKYCA-
| Original motif | Reverse complement motif |
| Consensus sequence: BHHASATCAAAGGAHHH | Consensus sequence: HHHTCCTTTGATSTHHV |
 |
 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 13 |
| Similarity score: | 0.0359011 |
Alignment:
VDDAGATCAAAGGKDDD
---RGGBCAAAGKYCA-
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
 |
 |
| Motif ID: | UP00083 |
| Motif name: | Tcf7l2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0360786 |
Alignment:
BDDASATCAAAGGMDDD
---RGGBCAAAGKYCA-
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATSTDDV | Consensus sequence: BDDASATCAAAGGMDDD |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 13 |
| Similarity score: | 0.0405764 |
Alignment:
HVDDVCAGATGKYHBBV
-RGGBCAAAGKYCA---
| Original motif | Reverse complement motif |
| Consensus sequence: HVDDVCAGATGKYHBBV | Consensus sequence: VBBDMYCATCTGVHHBH |
 |
 |
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0409915 |
Alignment:
HHBBMCTTTGTTHDDBH
-TGMYCTTTGBCCK---
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
 |
 |
| Motif ID: | UP00101 |
| Motif name: | Sox12_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0413218 |
Alignment:
HDTMCTTTGTSTVDBB
TGMYCTTTGBCCK---
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBASACAAAGRADH | Consensus sequence: HDTMCTTTGTSTVDBB |
 |
 |
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 13 |
| Similarity score: | 0.0421105 |
Alignment:
HHBTMCTTTGTTMDDVH
-TGMYCTTTGBCCK---
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
 |
 |
| Motif ID: | UP00086 |
| Motif name: | Irf3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0442004 |
Alignment:
DDAGAADGGDSCDV
RGGBCAAAGKYCA-
| Original motif | Reverse complement motif |
| Consensus sequence: DDAGAADGGDSCDV | Consensus sequence: BHGSDCCDTTCTHH |
 |
 |
| Motif ID: | UP00082 |
| Motif name: | Zfp187_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 13 |
| Similarity score: | 0.0446884 |
Alignment:
DDGMCCTBGTCCHYHV
---RGGBCAAAGKYCA
| Original motif | Reverse complement motif |
| Consensus sequence: DDGMCCTBGTCCHYHV | Consensus sequence: VHMDGGACVAGGRCDH |
 |
 |
| Dataset #: 3 | Motif ID: 52 | Motif name: NR2F1 |
| Original motif | Reverse complement motif |
| Consensus sequence: TGAMCTTTGMMCYT | Consensus sequence: AKGYYCAAAGRTCA |
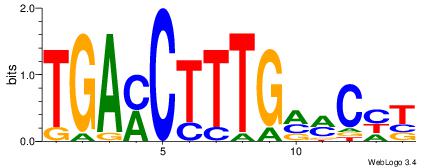 |
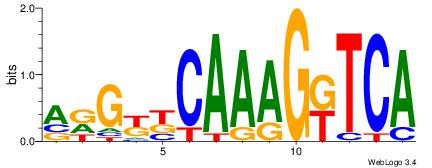 |
Best Matches for Motif ID 52 (Highest to Lowest)
| Motif ID: | UP00083 |
| Motif name: | Tcf7l2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0452364 |
Alignment:
DDDYCCTTTGATSTDDV
-TGAMCTTTGMMCYT--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATSTDDV | Consensus sequence: BDDASATCAAAGGMDDD |
 |
 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0453449 |
Alignment:
VDDAGATCAAAGGKDDD
--AKGYYCAAAGRTCA-
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 14 |
| Similarity score: | 0.0461358 |
Alignment:
DVMHHDHKGACCCTCCTSVCBH
-------TGAMCTTTGMMCYT-
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Motif ID: | UP00054 |
| Motif name: | Tcf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0461768 |
Alignment:
BHHASATCAAAGGAHHH
--AKGYYCAAAGRTCA-
| Original motif | Reverse complement motif |
| Consensus sequence: BHHASATCAAAGGAHHH | Consensus sequence: HHHTCCTTTGATSTHHV |
 |
 |
| Motif ID: | UP00058 |
| Motif name: | Tcf3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0473628 |
Alignment:
HHHASATCAAAGVRHHH
--AKGYYCAAAGRTCA-
| Original motif | Reverse complement motif |
| Consensus sequence: HHHASATCAAAGVRHHH | Consensus sequence: HHHKVCTTTGATSTHHH |
 |
 |
| Motif ID: | UP00082 |
| Motif name: | Zfp187_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0480188 |
Alignment:
DDGMCCTBGTCCHYHV
TGAMCTTTGMMCYT--
| Original motif | Reverse complement motif |
| Consensus sequence: DDGMCCTBGTCCHYHV | Consensus sequence: VHMDGGACVAGGRCDH |
 |
 |
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0495447 |
Alignment:
HHBBMCTTTGTTHDDBH
-TGAMCTTTGMMCYT--
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
 |
 |
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0499033 |
Alignment:
HHBTMCTTTGTTMDDVH
-TGAMCTTTGMMCYT--
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
 |
 |
| Motif ID: | UP00023 |
| Motif name: | Sox30_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0511534 |
Alignment:
DHDBAACAATDRHHHH
--TGAMCTTTGMMCYT
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBAACAATDRHHHH | Consensus sequence: HHHHMHATTGTTBDHD |
 |
 |
| Motif ID: | UP00005 |
| Motif name: | Tcfap2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0516973 |
Alignment:
BDGHCCWBVGGKDH
AKGYYCAAAGRTCA
| Original motif | Reverse complement motif |
| Consensus sequence: HHRCCBBWGGDCDB | Consensus sequence: BDGHCCWBVGGKDH |
 |
 |
| Dataset #: 3 | Motif ID: 53 | Motif name: ESR1 |
| Original motif | Reverse complement motif |
| Consensus sequence: VDBHMAGGTCACCCTGACCY | Consensus sequence: MGGTCAGGGTGACCTRDBHV |
 |
 |
Best Matches for Motif ID 53 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 20 |
| Similarity score: | 0.0658405 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
VDBHMAGGTCACCCTGACCY---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 20 |
| Similarity score: | 0.0689748 |
Alignment:
BDBBVBVGTAWCCAAGVBHBAGB
VDBHMAGGTCACCCTGACCY---
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 20 |
| Similarity score: | 0.0701247 |
Alignment:
CBDMCMGGGTGGTCCHVBVBAH
MGGTCAGGGTGACCTRDBHV--
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 20 |
| Similarity score: | 0.0711267 |
Alignment:
HVGVSAGGAGGGTCYHHHHYVH
MGGTCAGGGTGACCTRDBHV--
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 20 |
| Similarity score: | 0.0742427 |
Alignment:
CVHBSCGGGTGGTCMHVHHCDH
-VDBHMAGGTCACCCTGACCY-
| Original motif | Reverse complement motif |
| Consensus sequence: HDGHHBHRGACCACCCGSVDVG | Consensus sequence: CVHBSCGGGTGGTCMHVHHCDH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 20 |
| Similarity score: | 0.0747053 |
Alignment:
ABBBBVVRGACCACCCACRDBBM
--VDBHMAGGTCACCCTGACCY-
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 20 |
| Similarity score: | 0.0749165 |
Alignment:
BHCBCBCCGGGTGGTCYHVHDCH
---VDBHMAGGTCACCCTGACCY
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 20 |
| Similarity score: | 0.0759307 |
Alignment:
BBBDVVRGACCACCCAVGABBAB
-VDBHMAGGTCACCCTGACCY--
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 20 |
| Similarity score: | 0.0762689 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
-MGGTCAGGGTGACCTRDBHV--
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 20 |
| Similarity score: | 0.0780113 |
Alignment:
GTVBDAATTGTVTGHGDDHKVB
VDBHMAGGTCACCCTGACCY--
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Dataset #: 3 | Motif ID: 54 | Motif name: Esrrb |
| Original motif | Reverse complement motif |
| Consensus sequence: VBBYCAAGGTCA | Consensus sequence: TGACCTTGMBBB |
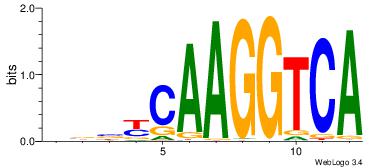 |
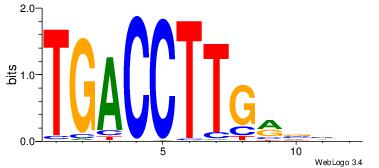 |
Best Matches for Motif ID 54 (Highest to Lowest)
| Motif ID: | UP00079 |
| Motif name: | Esrra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 12 |
| Similarity score: | 0 |
Alignment:
HHBVHTGACCTTGVDHD
-----TGACCTTGMBBB
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBCAAGGTCAHVBDH | Consensus sequence: HHBVHTGACCTTGVDHD |
 |
 |
| Motif ID: | UP00009 |
| Motif name: | Nr2f2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 0.0205471 |
Alignment:
DHBHAAAGGTCAHVHB
VBBYCAAGGTCA----
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCAHVHB | Consensus sequence: BHVDTGACCTTTDVDD |
 |
 |
| Motif ID: | UP00048 |
| Motif name: | Rara |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 0.0214372 |
Alignment:
DHBHAAAGGTCACVHD
VBBYCAAGGTCA----
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCACVHD | Consensus sequence: HHVGTGACCTTTDVDD |
 |
 |
| Motif ID: | UP00009 |
| Motif name: | Nr2f2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 0.0430759 |
Alignment:
HVBVTGACCCSGBVBV
----TGACCTTGMBBB
| Original motif | Reverse complement motif |
| Consensus sequence: VBVBCSGGGTCAVBBH | Consensus sequence: HVBVTGACCCSGBVBV |
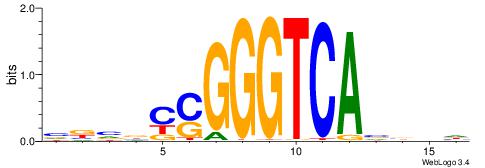 |
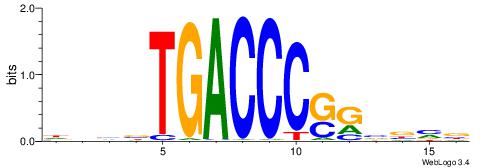 |
| Motif ID: | UP00066 |
| Motif name: | Hnf4a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0451776 |
Alignment:
BDBVMGGGGTCAABHDV
VBBYCAAGGTCA-----
| Original motif | Reverse complement motif |
| Consensus sequence: BDBVMGGGGTCAABHDV | Consensus sequence: BHHVTTGACCCCYVVDB |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 12 |
| Similarity score: | 0.0482496 |
Alignment:
VKBBAGGGGTCAHDBBH
VBBYCAAGGTCA-----
| Original motif | Reverse complement motif |
| Consensus sequence: VKBBAGGGGTCAHDBBH | Consensus sequence: DBBHHTGACCCCTBBYV |
 |
 |
| Motif ID: | UP00053 |
| Motif name: | Rxra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0499025 |
Alignment:
DBHBDTGACCCCDHVHB
-----TGACCTTGMBBB
| Original motif | Reverse complement motif |
| Consensus sequence: DBHBDTGACCCCDHVHB | Consensus sequence: VHBHDGGGGTCAHBHBD |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 12 |
| Similarity score: | 0.0525354 |
Alignment:
DVMHHDHKGACCCTCCTSVCBH
-------TGACCTTGMBBB---
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Motif ID: | UP00066 |
| Motif name: | Hnf4a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0580553 |
Alignment:
HHAYTGGACTTTHDBV
----TGACCTTGMBBB
| Original motif | Reverse complement motif |
| Consensus sequence: BBHHAAAGTCCAMTHH | Consensus sequence: HHAYTGGACTTTHDBV |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 12 |
| Similarity score: | 0.0609738 |
Alignment:
BDBBVBVGTAWCCAAGVBHBAGB
--------TGACCTTGMBBB---
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Dataset #: 3 | Motif ID: 55 | Motif name: ZEB1 |
| Original motif | Reverse complement motif |
| Consensus sequence: CACCTD | Consensus sequence: HAGGTG |
 |
 |
Best Matches for Motif ID 55 (Highest to Lowest)
| Motif ID: | UP00068 |
| Motif name: | Eomes_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
VVBVCKACACCTHVBV
-------CACCTD---
| Original motif | Reverse complement motif |
| Consensus sequence: VBVDAGGTGTYGVBBV | Consensus sequence: VVBVCKACACCTHVBV |
 |
 |
| Motif ID: | UP00199 |
| Motif name: | Six4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.00433068 |
Alignment:
BBVHVGGTGTCATHDDT
---HAGGTG--------
| Original motif | Reverse complement motif |
| Consensus sequence: ADDHATGACACCBHBBV | Consensus sequence: BBVHVGGTGTCATHDDT |
 |
 |
| Motif ID: | UP00068 |
| Motif name: | Eomes_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.00524081 |
Alignment:
HHHTTTCACACCTDHBV
--------CACCTD---
| Original motif | Reverse complement motif |
| Consensus sequence: BVHDAGGTGTGAAAHHH | Consensus sequence: HHHTTTCACACCTDHBV |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.00911301 |
Alignment:
DHDBHGCACCTGBDDVB
------CACCTD-----
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Motif ID: | UP00047 |
| Motif name: | Zbtb7b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 6 |
| Similarity score: | 0.0233556 |
Alignment:
BDDRVGACCACCHBDVB
--------CACCTD---
| Original motif | Reverse complement motif |
| Consensus sequence: BDDRVGACCACCHBDVB | Consensus sequence: BBDVHGGTGGTCBKDDB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 11 |
| Number of overlap: | 6 |
| Similarity score: | 0.0233928 |
Alignment:
BHCBCBCCGGGTGGTCYHVHDCH
-------HAGGTG----------
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.0254338 |
Alignment:
HTBVVVDGGACCACCCRGRDBG
-----------CACCTD-----
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.0290143 |
Alignment:
HDGHHBHRGACCACCCGSVDVG
-----------CACCTD-----
| Original motif | Reverse complement motif |
| Consensus sequence: HDGHHBHRGACCACCCGSVDVG | Consensus sequence: CVHBSCGGGTGGTCMHVHHCDH |
 |
 |
| Motif ID: | UP00036 |
| Motif name: | Myf6_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0309673 |
Alignment:
VVDBAACAGBTGBYHB
------HAGGTG----
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBAACAGBTGBYHB | Consensus sequence: BDKVCABCTGTTBDBV |
 |
 |
| Motif ID: | UP00249 |
| Motif name: | Nkx2-5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 6 |
| Similarity score: | 0.031502 |
Alignment:
HDADCCACTTRAAWTT
-----CACCTD-----
| Original motif | Reverse complement motif |
| Consensus sequence: HDADCCACTTRAAWTT | Consensus sequence: AAWTTMAAGTGGHTDH |
 |
 |
| Dataset #: 3 | Motif ID: 56 | Motif name: Mycn |
| Original motif | Reverse complement motif |
| Consensus sequence: HSCACGTGGC | Consensus sequence: GCCACGTGSD |
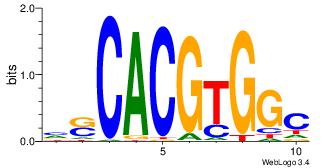 |
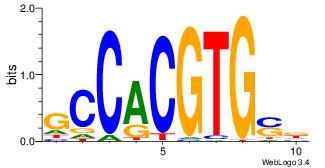 |
Best Matches for Motif ID 56 (Highest to Lowest)
| Motif ID: | UP00060 |
| Motif name: | Max_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
DDASCACGTGBTBVDD
--HSCACGTGGC----
| Original motif | Reverse complement motif |
| Consensus sequence: DDASCACGTGBTBVDD | Consensus sequence: HHVBABCACGTGSTHD |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.00783793 |
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
-------GCCACGTGSD------
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0132858 |
Alignment:
DDHBHBSTCACGTGAHBBDHHM
------GCCACGTGSD------
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
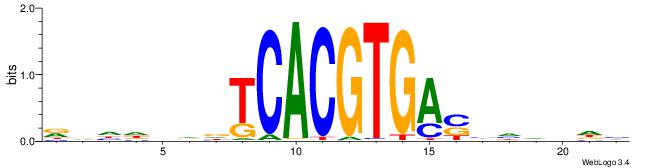 |
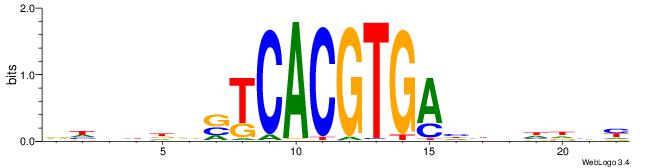 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0227933 |
Alignment:
VBHHVCAGGTGCDVDHD
---GCCACGTGSD----
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0257011 |
Alignment:
BBGHCACGCGACDD
--HSCACGTGGC--
| Original motif | Reverse complement motif |
| Consensus sequence: BBGHCACGCGACDD | Consensus sequence: HDGTCGCGTGDCVB |
 |
 |
| Motif ID: | UP00165 |
| Motif name: | Titf1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0344833 |
Alignment:
BVABCCACTTGAMHTT
---GCCACGTGSD---
| Original motif | Reverse complement motif |
| Consensus sequence: BVABCCACTTGAMHTT | Consensus sequence: AAHYTCAAGTGGBTBV |
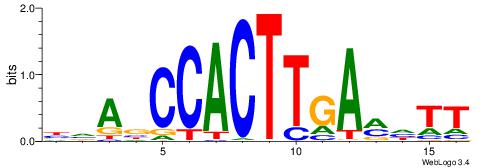 |
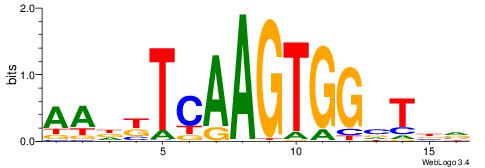 |
| Motif ID: | UP00231 |
| Motif name: | Nkx2-2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.035228 |
Alignment:
HDVRCCACTTGARADBD
---GCCACGTGSD----
| Original motif | Reverse complement motif |
| Consensus sequence: HDVRCCACTTGARADBD | Consensus sequence: DVDTKTCAAGTGGKBDH |
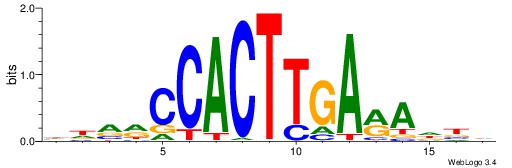 |
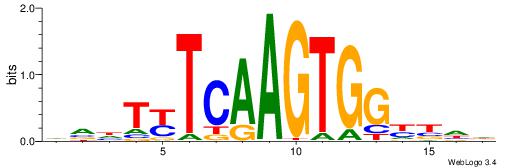 |
| Motif ID: | UP00084 |
| Motif name: | Gmeb1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0358581 |
Alignment:
BVVDVRYACGTRYVBHB
----GCCACGTGSD---
| Original motif | Reverse complement motif |
| Consensus sequence: BHBBKKACGTMMVDBVB | Consensus sequence: BVVDVRYACGTRYVBHB |
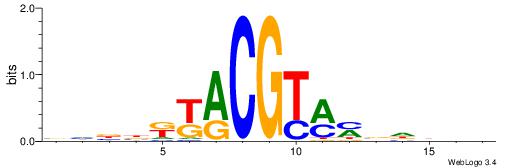 |
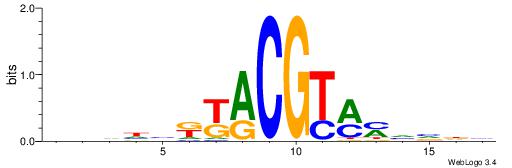 |
| Motif ID: | UP00084 |
| Motif name: | Gmeb1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.03725 |
Alignment:
HHMACKACGTMGBVCD
---HSCACGTGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: DGVBCRACGTYGTYHH | Consensus sequence: HHMACKACGTMGBVCD |
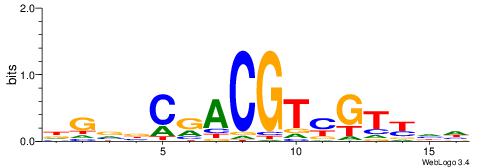 |
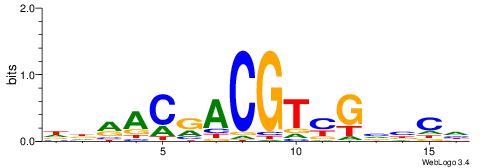 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.0383187 |
Alignment:
WHVBVVMACGCGCGTCDYHWDH
------HSCACGTGGC------
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
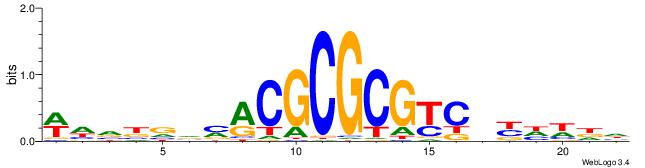 |
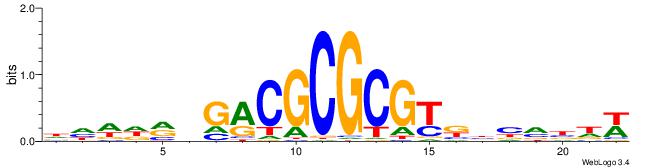 |
| Dataset #: 3 | Motif ID: 57 | Motif name: REST |
| Original motif | Reverse complement motif |
| Consensus sequence: TTCAGCACCATGGACAGCKCC | Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
 |
 |
Best Matches for Motif ID 57 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 21 |
| Similarity score: | 0.0699178 |
Alignment:
BBBDVVRGACCACCCAVGABBAB
--TTCAGCACCATGGACAGCKCC
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 21 |
| Similarity score: | 0.0734685 |
Alignment:
VMYDHDGMCCHCCKBGVVAAVH
GGYGCTGTCCATGGTGCTGAA-
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
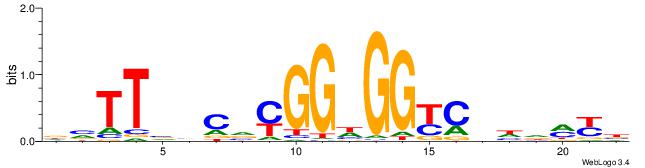 |
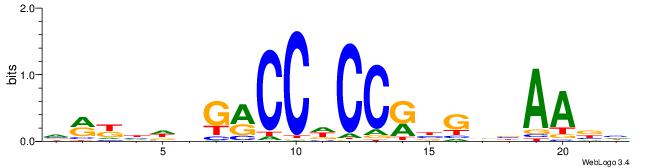 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 21 |
| Similarity score: | 0.0740652 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
-TTCAGCACCATGGACAGCKCC-
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 21 |
| Similarity score: | 0.0750152 |
Alignment:
MHHHWHYTMCCHCCCACVAABVH
--TTCAGCACCATGGACAGCKCC
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 21 |
| Similarity score: | 0.0751449 |
Alignment:
BHCBCBCCGGGTGGTCYHVHDCH
GGYGCTGTCCATGGTGCTGAA--
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 21 |
| Similarity score: | 0.075832 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
-TTCAGCACCATGGACAGCKCC-
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 21 |
| Similarity score: | 0.0773446 |
Alignment:
DTVBDDRGACCACCCAGDWDGBH
--TTCAGCACCATGGACAGCKCC
| Original motif | Reverse complement motif |
| Consensus sequence: DTVBDDRGACCACCCAGDWDGBH | Consensus sequence: DBCDWHCTGGGTGGTCMDHBBAH |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 21 |
| Similarity score: | 0.0775367 |
Alignment:
YDYBDHTMCACGTGGADDBMDGT
--TTCAGCACCATGGACAGCKCC
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 21 |
| Similarity score: | 0.077977 |
Alignment:
DCATDBTGHGCATKMTTBRSWR
-GGYGCTGTCCATGGTGCTGAA
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 21 |
| Similarity score: | 0.0785969 |
Alignment:
BADHBDHCGCCCMCGCAHHDBBV
--GGYGCTGTCCATGGTGCTGAA
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Dataset #: 3 | Motif ID: 58 | Motif name: Myc |
| Original motif | Reverse complement motif |
| Consensus sequence: VGCACGTGGH | Consensus sequence: DCCACGTGCV |
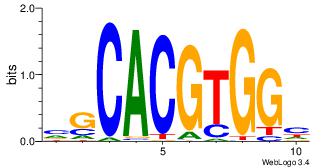 |
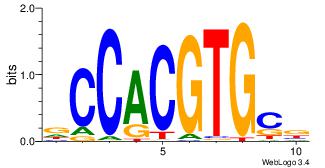 |
Best Matches for Motif ID 58 (Highest to Lowest)
| Motif ID: | UP00060 |
| Motif name: | Max_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
DDASCACGTGBTBVDD
--VGCACGTGGH----
| Original motif | Reverse complement motif |
| Consensus sequence: DDASCACGTGBTBVDD | Consensus sequence: HHVBABCACGTGSTHD |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 10 |
| Similarity score: | 0.00631885 |
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
-------DCCACGTGCV------
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 10 |
| Similarity score: | 0.00821668 |
Alignment:
DDHBHBSTCACGTGAHBBDHHM
------DCCACGTGCV------
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0140005 |
Alignment:
DHDBHGCACCTGBDDVB
----VGCACGTGGH---
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0308912 |
Alignment:
BBGHCACGCGACDD
--VGCACGTGGH--
| Original motif | Reverse complement motif |
| Consensus sequence: BBGHCACGCGACDD | Consensus sequence: HDGTCGCGTGDCVB |
 |
 |
| Motif ID: | UP00165 |
| Motif name: | Titf1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0311723 |
Alignment:
BVABCCACTTGAMHTT
---DCCACGTGCV---
| Original motif | Reverse complement motif |
| Consensus sequence: BVABCCACTTGAMHTT | Consensus sequence: AAHYTCAAGTGGBTBV |
 |
 |
| Motif ID: | UP00249 |
| Motif name: | Nkx2-5 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.031308 |
Alignment:
HDADCCACTTRAAWTT
---DCCACGTGCV---
| Original motif | Reverse complement motif |
| Consensus sequence: HDADCCACTTRAAWTT | Consensus sequence: AAWTTMAAGTGGHTDH |
 |
 |
| Motif ID: | UP00231 |
| Motif name: | Nkx2-2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0334845 |
Alignment:
HDVRCCACTTGARADBD
---DCCACGTGCV----
| Original motif | Reverse complement motif |
| Consensus sequence: HDVRCCACTTGARADBD | Consensus sequence: DVDTKTCAAGTGGKBDH |
 |
 |
| Motif ID: | UP00147 |
| Motif name: | Nkx2-6 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0345548 |
Alignment:
DDADCCACTTAAVHTT
---DCCACGTGCV---
| Original motif | Reverse complement motif |
| Consensus sequence: DDADCCACTTAAVHTT | Consensus sequence: AAHVTTAAGTGGHTDD |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0363818 |
Alignment:
VDWDARRGACGCGGWHGGAKHV
-----VGCACGTGGH-------
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Dataset #: 3 | Motif ID: 59 | Motif name: ESR2 |
| Original motif | Reverse complement motif |
| Consensus sequence: VHRGGTCABDBTGMCCTB | Consensus sequence: BAGGYCABHBTGACCKHV |
 |
 |
Best Matches for Motif ID 59 (Highest to Lowest)
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0443894 |
Alignment:
GTVBDAATTGTVTGHGDDHKVB
---VHRGGTCABDBTGMCCTB-
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.049219 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
---VHRGGTCABDBTGMCCTB--
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0509182 |
Alignment:
DDHBHBSTCACGTGAHBBDHHM
--VHRGGTCABDBTGMCCTB--
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.0527075 |
Alignment:
DVTTVVCVYGGHGGYCHHDMYB
-BAGGYCABHBTGACCKHV---
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.0541044 |
Alignment:
WBHHHHHMDATTGTTHDHDVHH
-BAGGYCABHBTGACCKHV---
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0541108 |
Alignment:
VCTBHBVCTTGGWTACVVVVVDB
--BAGGYCABHBTGACCKHV---
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.0543733 |
Alignment:
VDWDARRGACGCGGWHGGAKHV
---VHRGGTCABDBTGMCCTB-
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.0548087 |
Alignment:
BHCBCBCCGGGTGGTCYHVHDCH
-BAGGYCABHBTGACCKHV----
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.055453 |
Alignment:
DCATDBTGHGCATKMTTBRSWR
----VHRGGTCABDBTGMCCTB
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0566426 |
Alignment:
HBHDVBWGATHBTATCRVBHHH
-VHRGGTCABDBTGMCCTB---
| Original motif | Reverse complement motif |
| Consensus sequence: HBHDVBWGATHBTATCRVBHHH | Consensus sequence: HHHBBMGATAVHATCWVVDHVH |
 |
 |
| Dataset #: 3 | Motif ID: 60 | Motif name: NR1H2RXRA |
| Original motif | Reverse complement motif |
| Consensus sequence: AAAGGTCAAAGGTCAAC | Consensus sequence: GTTGACCTTTGACCTTT |
 |
 |
Best Matches for Motif ID 60 (Highest to Lowest)
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0538452 |
Alignment:
DVMHHDHKGACCCTCCTSVCBH
-----GTTGACCTTTGACCTTT
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0545098 |
Alignment:
BDHDDADAGATAAGADHBDHVD
-----AAAGGTCAAAGGTCAAC
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.521114 |
Alignment:
VDDAGATCAAAGGKDDD-
-AAAGGTCAAAGGTCAAC
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
 |
 |
| Motif ID: | UP00083 |
| Motif name: | Tcf7l2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.527825 |
Alignment:
-DDDYCCTTTGATSTDDV
GTTGACCTTTGACCTTT-
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATSTDDV | Consensus sequence: BDDASATCAAAGGMDDD |
 |
 |
| Motif ID: | UP00054 |
| Motif name: | Tcf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.531154 |
Alignment:
BHHASATCAAAGGAHHH-
-AAAGGTCAAAGGTCAAC
| Original motif | Reverse complement motif |
| Consensus sequence: BHHASATCAAAGGAHHH | Consensus sequence: HHHTCCTTTGATSTHHV |
 |
 |
| Motif ID: | UP00058 |
| Motif name: | Tcf3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.531876 |
Alignment:
-HHHASATCAAAGVRHHH
AAAGGTCAAAGGTCAAC-
| Original motif | Reverse complement motif |
| Consensus sequence: HHHASATCAAAGVRHHH | Consensus sequence: HHHKVCTTTGATSTHHH |
 |
 |
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.544403 |
Alignment:
HBDDRAACAAAGRABHH-
-AAAGGTCAAAGGTCAAC
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
 |
 |
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.544565 |
Alignment:
-HHBBMCTTTGTTHDDBH
GTTGACCTTTGACCTTT-
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
 |
 |
| Motif ID: | UP00019 |
| Motif name: | Zbtb12_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.550511 |
Alignment:
-BBGATCTAGAACBKHDB
GTTGACCTTTGACCTTT-
| Original motif | Reverse complement motif |
| Consensus sequence: BDHRBGTTCTAGATCVB | Consensus sequence: BBGATCTAGAACBKHDB |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.552064 |
Alignment:
-HVDDVCAGATGKYHBBV
AAAGGTCAAAGGTCAAC-
| Original motif | Reverse complement motif |
| Consensus sequence: HVDDVCAGATGKYHBBV | Consensus sequence: VBBDMYCATCTGVHHBH |
 |
 |
| Dataset #: 3 | Motif ID: 61 | Motif name: NR4A2 |
| Original motif | Reverse complement motif |
| Consensus sequence: AAGGTCAC | Consensus sequence: GTGACCTT |
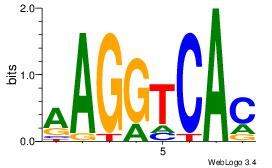 |
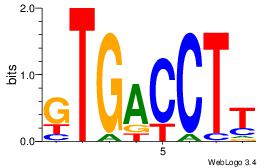 |
Best Matches for Motif ID 61 (Highest to Lowest)
| Motif ID: | UP00048 |
| Motif name: | Rara |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0 |
Alignment:
HHVGTGACCTTTDVDD
---GTGACCTT-----
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCACVHD | Consensus sequence: HHVGTGACCTTTDVDD |
 |
 |
| Motif ID: | UP00009 |
| Motif name: | Nr2f2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0074527 |
Alignment:
DHBHAAAGGTCAHVHB
-----AAGGTCAC---
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCAHVHB | Consensus sequence: BHVDTGACCTTTDVDD |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.01542 |
Alignment:
DHDBCAAGGTCAHVBDH
-----AAGGTCAC----
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBCAAGGTCAHVBDH | Consensus sequence: HHBVHTGACCTTGVDHD |
 |
 |
| Motif ID: | UP00009 |
| Motif name: | Nr2f2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0395644 |
Alignment:
VBVBCSGGGTCAVBBH
-----AAGGTCAC---
| Original motif | Reverse complement motif |
| Consensus sequence: VBVBCSGGGTCAVBBH | Consensus sequence: HVBVTGACCCSGBVBV |
 |
 |
| Motif ID: | UP00082 |
| Motif name: | Zfp187_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 8 |
| Similarity score: | 0.0407874 |
Alignment:
DDGMCCTBGTCCHYHV
GTGACCTT--------
| Original motif | Reverse complement motif |
| Consensus sequence: DDGMCCTBGTCCHYHV | Consensus sequence: VHMDGGACVAGGRCDH |
 |
 |
| Motif ID: | UP00053 |
| Motif name: | Rxra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0422933 |
Alignment:
DBHBDTGACCCCDHVHB
----GTGACCTT-----
| Original motif | Reverse complement motif |
| Consensus sequence: DBHBDTGACCCCDHVHB | Consensus sequence: VHBHDGGGGTCAHBHBD |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0447444 |
Alignment:
VKBBAGGGGTCAHDBBH
-----AAGGTCAC----
| Original motif | Reverse complement motif |
| Consensus sequence: VKBBAGGGGTCAHDBBH | Consensus sequence: DBBHHTGACCCCTBBYV |
 |
 |
| Motif ID: | UP00066 |
| Motif name: | Hnf4a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0481418 |
Alignment:
BBHHAAAGTCCAMTHH
-----AAGGTCAC---
| Original motif | Reverse complement motif |
| Consensus sequence: BBHHAAAGTCCAMTHH | Consensus sequence: HHAYTGGACTTTHDBV |
 |
 |
| Motif ID: | UP00197 |
| Motif name: | Hoxc9 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0497679 |
Alignment:
VDHHTTAATGACCBHV
-------GTGACCTT-
| Original motif | Reverse complement motif |
| Consensus sequence: VDVGGTCATTAAHHDB | Consensus sequence: VDHHTTAATGACCBHV |
 |
 |
| Motif ID: | UP00035 |
| Motif name: | Hic1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0500062 |
Alignment:
DBDDRTGCCCAWDHDV
----GTGACCTT----
| Original motif | Reverse complement motif |
| Consensus sequence: DBDDRTGCCCAWDHDV | Consensus sequence: VHHDWTGGGCAMDHBH |
 |
 |
| Dataset #: 3 | Motif ID: 62 | Motif name: RORA_1 |
| Original motif | Reverse complement motif |
| Consensus sequence: AWVDAGGTCA | Consensus sequence: TGACCTDVWT |
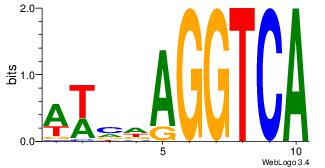 |
 |
Best Matches for Motif ID 62 (Highest to Lowest)
| Motif ID: | UP00009 |
| Motif name: | Nr2f2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0154749 |
Alignment:
DHBHAAAGGTCAHVHB
--AWVDAGGTCA----
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCAHVHB | Consensus sequence: BHVDTGACCTTTDVDD |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0176661 |
Alignment:
HHBVHTGACCTTGVDHD
-----TGACCTDVWT--
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBCAAGGTCAHVBDH | Consensus sequence: HHBVHTGACCTTGVDHD |
 |
 |
| Motif ID: | UP00048 |
| Motif name: | Rara |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0189599 |
Alignment:
DHBHAAAGGTCACVHD
--AWVDAGGTCA----
| Original motif | Reverse complement motif |
| Consensus sequence: DHBHAAAGGTCACVHD | Consensus sequence: HHVGTGACCTTTDVDD |
 |
 |
| Motif ID: | UP00009 |
| Motif name: | Nr2f2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0331999 |
Alignment:
VBVBCSGGGTCAVBBH
--AWVDAGGTCA----
| Original motif | Reverse complement motif |
| Consensus sequence: VBVBCSGGGTCAVBBH | Consensus sequence: HVBVTGACCCSGBVBV |
 |
 |
| Motif ID: | UP00066 |
| Motif name: | Hnf4a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0341771 |
Alignment:
BDBVMGGGGTCAABHDV
--AWVDAGGTCA-----
| Original motif | Reverse complement motif |
| Consensus sequence: BDBVMGGGGTCAABHDV | Consensus sequence: BHHVTTGACCCCYVVDB |
 |
 |
| Motif ID: | UP00053 |
| Motif name: | Rxra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0363566 |
Alignment:
DBHBDTGACCCCDHVHB
-----TGACCTDVWT--
| Original motif | Reverse complement motif |
| Consensus sequence: DBHBDTGACCCCDHVHB | Consensus sequence: VHBHDGGGGTCAHBHBD |
 |
 |
| Motif ID: | UP00079 |
| Motif name: | Esrra_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0391284 |
Alignment:
VKBBAGGGGTCAHDBBH
--AWVDAGGTCA-----
| Original motif | Reverse complement motif |
| Consensus sequence: VKBBAGGGGTCAHDBBH | Consensus sequence: DBBHHTGACCCCTBBYV |
 |
 |
| Motif ID: | UP00035 |
| Motif name: | Hic1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 10 |
| Similarity score: | 0.0444205 |
Alignment:
DBDDRTGCCCAWDHDV
-----TGACCTDVWT-
| Original motif | Reverse complement motif |
| Consensus sequence: DBDDRTGCCCAWDHDV | Consensus sequence: VHHDWTGGGCAMDHBH |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0461434 |
Alignment:
RDHDBVDTCACGTGASBHVHDH
AWVDAGGTCA------------
| Original motif | Reverse complement motif |
| Consensus sequence: RDHDBVDTCACGTGASBHVHDH | Consensus sequence: DDHBHBSTCACGTGAHBBDHHM |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0489571 |
Alignment:
BHCBCBCCGGGTGGTCYHVHDCH
-----------TGACCTDVWT--
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Dataset #: 3 | Motif ID: 63 | Motif name: MIZF |
| Original motif | Reverse complement motif |
| Consensus sequence: BAACGTCCGC | Consensus sequence: GCGGACGTTV |
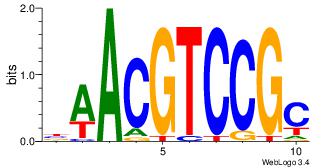 |
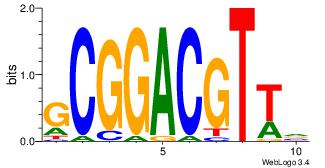 |
Best Matches for Motif ID 63 (Highest to Lowest)
| Motif ID: | UP00419 |
| Motif name: | Spic |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
DWACTTCCRCTTTB
BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: VAAAGMGGAAGTWD | Consensus sequence: DWACTTCCRCTTTB |
 |
 |
| Motif ID: | UP00409 |
| Motif name: | Elf5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.00379999 |
Alignment:
BWAHSMGGAAGTWD
BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: BWAHSMGGAAGTWD | Consensus sequence: DWACTTCCRSDTWV |
 |
 |
| Motif ID: | UP00413 |
| Motif name: | Elf4 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0124101 |
Alignment:
DBBHACTTCCGGKWTH
--BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGKWTH | Consensus sequence: HAWYCCGGAAGTHBBD |
 |
 |
| Motif ID: | UP00404 |
| Motif name: | Elf2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0128348 |
Alignment:
DBBHACTTCCGGDWDB
--BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGDWDB | Consensus sequence: BDWDCCGGAAGTHBBD |
 |
 |
| Motif ID: | UP00085 |
| Motif name: | Sfpi1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 10 |
| Similarity score: | 0.0141516 |
Alignment:
BDAWGVGGAAGTDH
BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: BDAWGVGGAAGTDH | Consensus sequence: HDACTTCCBCWTDV |
 |
 |
| Motif ID: | UP00423 |
| Motif name: | Gm5454 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0144665 |
Alignment:
DVHACCGGAAGTHDDV
----GCGGACGTTV--
| Original motif | Reverse complement motif |
| Consensus sequence: VDHHACTTCCGGTHVH | Consensus sequence: DVHACCGGAAGTHDDV |
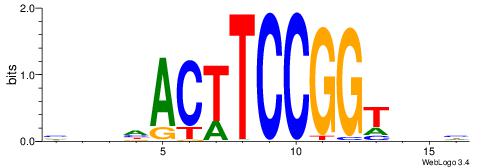 |
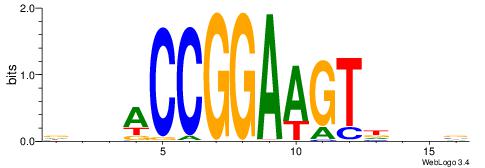 |
| Motif ID: | UP00412 |
| Motif name: | Etv5 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0155825 |
Alignment:
DHHACCGGAWGTTDDV
----GCGGACGTTV--
| Original motif | Reverse complement motif |
| Consensus sequence: VDHAACWTCCGGTHHH | Consensus sequence: DHHACCGGAWGTTDDV |
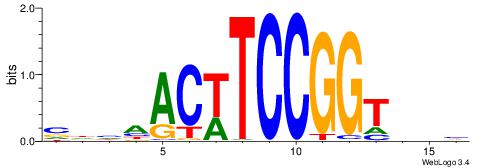 |
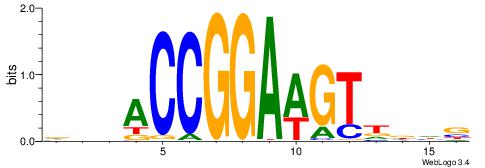 |
| Motif ID: | UP00015 |
| Motif name: | Ehf_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0162018 |
Alignment:
VBDABCCGGAAGTDD
-----GCGGACGTTV
| Original motif | Reverse complement motif |
| Consensus sequence: VBDABCCGGAAGTDD | Consensus sequence: DDACTTCCGGBTHBB |
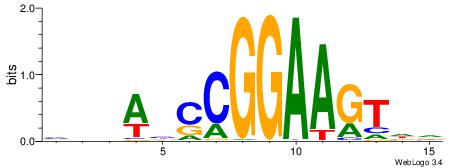 |
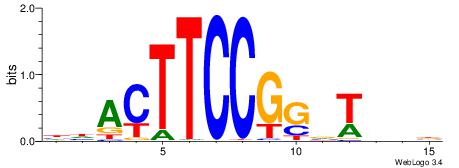 |
| Motif ID: | UP00418 |
| Motif name: | Etv6 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0171234 |
Alignment:
DVHBTACTTCCGGHTHB
---BAACGTCCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: DVHBTACTTCCGGHTHB | Consensus sequence: BHAHCCGGAAGTABDVD |
 |
 |
| Motif ID: | UP00421 |
| Motif name: | Etv1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0184147 |
Alignment:
MVHBACCGGAAGTVDVH
-----GCGGACGTTV--
| Original motif | Reverse complement motif |
| Consensus sequence: MVHBACCGGAAGTVDVH | Consensus sequence: HBDBACTTCCGGTBHVY |
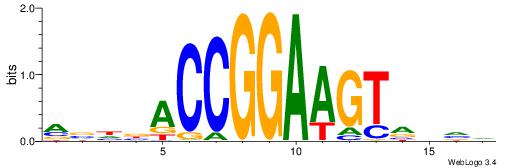 |
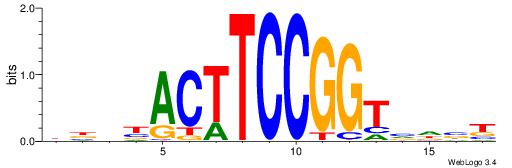 |
| Dataset #: 3 | Motif ID: 64 | Motif name: PPARGRXRA |
| Original motif | Reverse complement motif |
| Consensus sequence: BTRGGDCARAGGKCA | Consensus sequence: TGRCCTKTGHCCKAB |
 |
 |
Best Matches for Motif ID 64 (Highest to Lowest)
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0304954 |
Alignment:
VDDAGATCAAAGGKDDD
-BTRGGDCARAGGKCA-
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
 |
 |
| Motif ID: | UP00058 |
| Motif name: | Tcf3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0319475 |
Alignment:
HHHASATCAAAGVRHHH
-BTRGGDCARAGGKCA-
| Original motif | Reverse complement motif |
| Consensus sequence: HHHASATCAAAGVRHHH | Consensus sequence: HHHKVCTTTGATSTHHH |
 |
 |
| Motif ID: | UP00054 |
| Motif name: | Tcf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.032779 |
Alignment:
BHHASATCAAAGGAHHH
-BTRGGDCARAGGKCA-
| Original motif | Reverse complement motif |
| Consensus sequence: BHHASATCAAAGGAHHH | Consensus sequence: HHHTCCTTTGATSTHHV |
 |
 |
| Motif ID: | UP00082 |
| Motif name: | Zfp187_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0334185 |
Alignment:
VHMDGGACVAGGRCDH
BTRGGDCARAGGKCA-
| Original motif | Reverse complement motif |
| Consensus sequence: DDGMCCTBGTCCHYHV | Consensus sequence: VHMDGGACVAGGRCDH |
 |
 |
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0352539 |
Alignment:
HBDDDAACAAAGRVBHH
-BTRGGDCARAGGKCA-
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
 |
 |
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0358782 |
Alignment:
HBDDRAACAAAGRABHH
-BTRGGDCARAGGKCA-
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
 |
 |
| Motif ID: | UP00101 |
| Motif name: | Sox12_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0374729 |
Alignment:
HDTMCTTTGTSTVDBB
TGRCCTKTGHCCKAB-
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBASACAAAGRADH | Consensus sequence: HDTMCTTTGTSTVDBB |
 |
 |
| Motif ID: | UP00083 |
| Motif name: | Tcf7l2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0379757 |
Alignment:
BDDASATCAAAGGMDDD
-BTRGGDCARAGGKCA-
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATSTDDV | Consensus sequence: BDDASATCAAAGGMDDD |
 |
 |
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 15 |
| Similarity score: | 0.0382733 |
Alignment:
DHHDGGGCGRGGKHBH
BTRGGDCARAGGKCA-
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
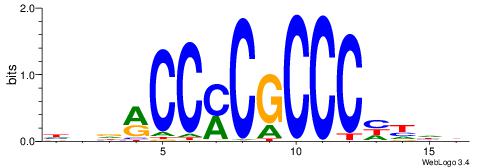 |
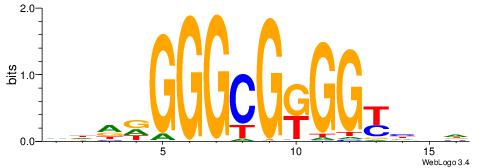 |
| Motif ID: | UP00055 |
| Motif name: | Hbp1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 15 |
| Similarity score: | 0.0397168 |
Alignment:
DDDYCCCATTGTVHABH
BTRGGDCARAGGKCA--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCCATTGTVHABH | Consensus sequence: HBTHVACAATGGGMDHD |
 |
 |
| Dataset #: 3 | Motif ID: 65 | Motif name: Tcfcp2l1 |
| Original motif | Reverse complement motif |
| Consensus sequence: CCAGYYHVADCCRG | Consensus sequence: CKGGDTBDMMCTGG |
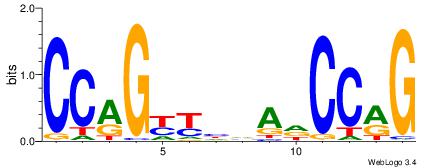 |
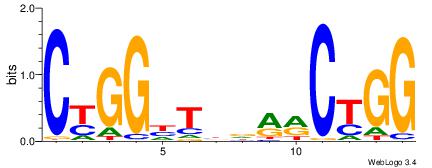 |
Best Matches for Motif ID 65 (Highest to Lowest)
| Motif ID: | UP00032 |
| Motif name: | Gata3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 14 |
| Similarity score: | 0.0527249 |
Alignment:
HBHDVBWGATHBTATCRVBHHH
----CCAGYYHVADCCRG----
| Original motif | Reverse complement motif |
| Consensus sequence: HBHDVBWGATHBTATCRVBHHH | Consensus sequence: HHHBBMGATAVHATCWVVDHVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0551691 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
--------CCAGYYHVADCCRG
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00005 |
| Motif name: | Tcfap2a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0560983 |
Alignment:
HHRCCBBWGGDCDB
CCAGYYHVADCCRG
| Original motif | Reverse complement motif |
| Consensus sequence: HHRCCBBWGGDCDB | Consensus sequence: BDGHCCWBVGGKDH |
 |
 |
| Motif ID: | UP00013 |
| Motif name: | Gabpa_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0566467 |
Alignment:
CYDBYWTCCSMHBVVH
CCAGYYHVADCCRG--
| Original motif | Reverse complement motif |
| Consensus sequence: CYDBYWTCCSMHBVVH | Consensus sequence: DBVVDRSGGAWKVHKG |
 |
 |
| Motif ID: | UP00075 |
| Motif name: | Sox15_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0577636 |
Alignment:
BBVAATGDMATTHVA
-CCAGYYHVADCCRG
| Original motif | Reverse complement motif |
| Consensus sequence: BBVAATGDMATTHVA | Consensus sequence: TVDAATYDCATTVVV |
 |
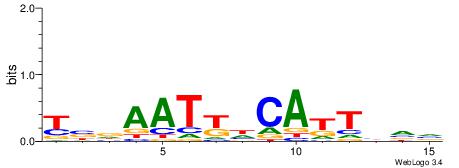 |
| Motif ID: | UP00096 |
| Motif name: | Sox13_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 14 |
| Similarity score: | 0.0587893 |
Alignment:
DDDHTGGGTGGGHAHHD
---CKGGDTBDMMCTGG
| Original motif | Reverse complement motif |
| Consensus sequence: DDDHTGGGTGGGHAHHD | Consensus sequence: DHHTHCCCACCCAHDDH |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0590238 |
Alignment:
VVYDDDCHCAVACAATTDBVAC
--CCAGYYHVADCCRG------
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 14 |
| Similarity score: | 0.0596904 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
--------CCAGYYHVADCCRG-
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0597695 |
Alignment:
BBBDVVRGACCACCCAVGABBAB
---------CCAGYYHVADCCRG
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 14 |
| Similarity score: | 0.0598357 |
Alignment:
HVGVSAGGAGGGTCYHHHHYVH
-------CKGGDTBDMMCTGG-
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Dataset #: 3 | Motif ID: 66 | Motif name: CTCF |
| Original motif | Reverse complement motif |
| Consensus sequence: YDRCCASYAGRKGGCRSYV | Consensus sequence: BMSMGCCYMCTKSTGGMHM |
 |
 |
Best Matches for Motif ID 66 (Highest to Lowest)
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 19 |
| Similarity score: | 0.0409369 |
Alignment:
BADHBDHCGCCCMCGCAHHDBBV
----BMSMGCCYMCTKSTGGMHM
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 19 |
| Similarity score: | 0.0409815 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
-YDRCCASYAGRKGGCRSYV---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 19 |
| Similarity score: | 0.0431951 |
Alignment:
VMYDHDGMCCHCCKBGVVAAVH
---BMSMGCCYMCTKSTGGMHM
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 19 |
| Similarity score: | 0.0469794 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
---BMSMGCCYMCTKSTGGMHM
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 19 |
| Similarity score: | 0.0482204 |
Alignment:
BHCBCBCCGGGTGGTCYHVHDCH
YDRCCASYAGRKGGCRSYV----
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 19 |
| Similarity score: | 0.0491604 |
Alignment:
HHBSHTCGTGRGTGGKGDBVWMV
-YDRCCASYAGRKGGCRSYV---
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 19 |
| Similarity score: | 0.0511535 |
Alignment:
BBBDVVRGACCACCCAVGABBAB
---BMSMGCCYMCTKSTGGMHM-
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 19 |
| Similarity score: | 0.0520741 |
Alignment:
HVGVSAGGAGGGTCYHHHHYVH
YDRCCASYAGRKGGCRSYV---
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 19 |
| Similarity score: | 0.0532334 |
Alignment:
DVVTTVGTGGGHGGYAMHWHHHY
YDRCCASYAGRKGGCRSYV----
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 19 |
| Similarity score: | 0.0535675 |
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
--BMSMGCCYMCTKSTGGMHM--
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Dataset #: 4 | Motif ID: 67 | Motif name: TgTGgTGTCACAGTGCTCC |
| Original motif | Reverse complement motif |
| Consensus sequence: TGTGGTGTCACAGTGCTCC | Consensus sequence: GGAGCACTGTGACACCACA |
 |
 |
Best Matches for Motif ID 67 (Highest to Lowest)
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 19 |
| Similarity score: | 0.043204 |
Alignment:
BDBBVBVGTAWCCAAGVBHBAGB
---TGTGGTGTCACAGTGCTCC-
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 18 |
| Similarity score: | 0.553914 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB-
-----TGTGGTGTCACAGTGCTCC
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00192 |
| Motif name: | Six1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 1.04528 |
Alignment:
AVHDHTGATACCCMDHH--
--GGAGCACTGTGACACCACA
| Original motif | Reverse complement motif |
| Consensus sequence: DHDRGGGTATCAHDHBT | Consensus sequence: AVHDHTGATACCCMDHH |
 |
 |
| Motif ID: | UP00193 |
| Motif name: | Rhox11_2205.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 1.0459 |
Alignment:
--AVDHCGCTGTWAWDVVW
TGTGGTGTCACAGTGCTCC
| Original motif | Reverse complement motif |
| Consensus sequence: AVDHCGCTGTWAWDVVW | Consensus sequence: WVVHWTWACAGCGHHVT |
 |
 |
| Motif ID: | UP00193 |
| Motif name: | Rhox11_1765.2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 1.0462 |
Alignment:
--WVVHWTWACAGCGBHBT
TGTGGTGTCACAGTGCTCC
| Original motif | Reverse complement motif |
| Consensus sequence: AVDVCGCTGTWAWDVVW | Consensus sequence: WVVHWTWACAGCGBHBT |
 |
 |
| Motif ID: | UP00068 |
| Motif name: | Eomes_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 1.04631 |
Alignment:
--HHHTTTCACACCTDHBV
GGAGCACTGTGACACCACA--
| Original motif | Reverse complement motif |
| Consensus sequence: BVHDAGGTGTGAAAHHH | Consensus sequence: HHHTTTCACACCTDHBV |
 |
 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 1.04807 |
Alignment:
VDDAGATCAAAGGKDDD--
-GGAGCACTGTGACACCACA
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 1.04831 |
Alignment:
YBVDMGTGGGTGGTCKVVBVBBT--
--GGAGCACTGTGACACCACA----
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Motif ID: | UP00053 |
| Motif name: | Rxra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 1.04867 |
Alignment:
--DBHBDTGACCCCDHVHB
TGTGGTGTCACAGTGCTCC--
| Original motif | Reverse complement motif |
| Consensus sequence: DBHBDTGACCCCDHVHB | Consensus sequence: VHBHDGGGGTCAHBHBD |
 |
 |
| Motif ID: | UP00388 |
| Motif name: | Six6_2267.4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 1.0491 |
Alignment:
AHWDHTGATACCCKATD--
--GGAGCACTGTGACACCACA
| Original motif | Reverse complement motif |
| Consensus sequence: DATRGGGTATCAHDWHT | Consensus sequence: AHWDHTGATACCCKATD |
 |
 |
| Dataset #: 4 | Motif ID: 68 | Motif name: sscCCCGCGcs |
| Original motif | Reverse complement motif |
| Consensus sequence: BSCCCCGCGBS | Consensus sequence: SBCGCGGGGSB |
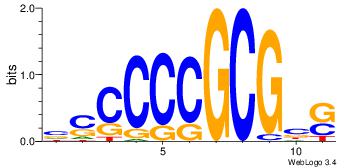 |
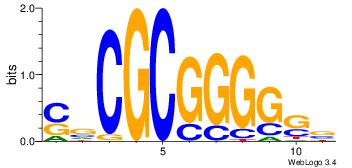 |
Best Matches for Motif ID 68 (Highest to Lowest)
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.00934222 |
Alignment:
BHBHDTGGCGGGGBDHD
----SBCGCGGGGSB--
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
 |
 |
| Motif ID: | UP00006 |
| Motif name: | Zic3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0105793 |
Alignment:
BMCCCCCGGGGGGGB
-BSCCCCGCGBS---
| Original motif | Reverse complement motif |
| Consensus sequence: BMCCCCCGGGGGGGB | Consensus sequence: VCCCCCCCGGGGGRB |
 |
 |
| Motif ID: | UP00057 |
| Motif name: | Zic2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0112489 |
Alignment:
VCCCCCCCGGGGGRB
---SBCGCGGGGSB-
| Original motif | Reverse complement motif |
| Consensus sequence: BMCCCCCGGGGGGGB | Consensus sequence: VCCCCCCCGGGGGRB |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.014467 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
------SBCGCGGGGSB------
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0150561 |
Alignment:
HDWHMHGACGCGCGTRBVVBHW
--------SBCGCGGGGSB---
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
 |
 |
| Motif ID: | UP00065 |
| Motif name: | Zfp161_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0169049 |
Alignment:
DMMDGMGCGCGCGCHD
--BSCCCCGCGBS---
| Original motif | Reverse complement motif |
| Consensus sequence: DDGCGCGCGCRCHYRD | Consensus sequence: DMMDGMGCGCGCGCHD |
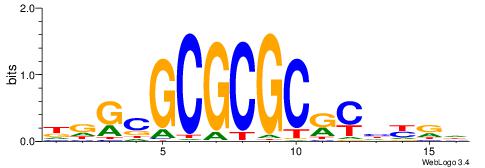 |
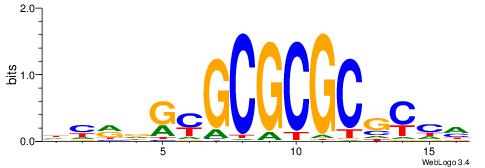 |
| Motif ID: | UP00102 |
| Motif name: | Zic1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0186112 |
Alignment:
BHCCCCCGGGGGGG
-BSCCCCGCGBS--
| Original motif | Reverse complement motif |
| Consensus sequence: BHCCCCCGGGGGGG | Consensus sequence: CCCCCCCGGGGGHB |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 11 |
| Similarity score: | 0.0192401 |
Alignment:
VHYTCCDWCCGCGTCMMTHWHV
----BSCCCCGCGBS-------
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0204596 |
Alignment:
BHHDYGGGGGGGGBVD
--SBCGCGGGGSB---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
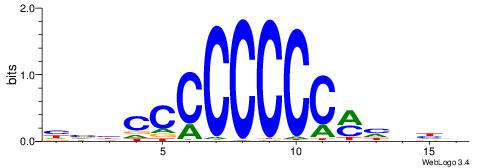 |
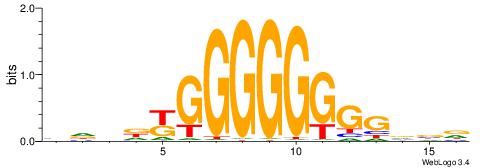 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0232114 |
Alignment:
DDRMHBACGCGYGCGTDGBBDB
-------SBCGCGGGGSB----
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
 |
 |
| Dataset #: 4 | Motif ID: 69 | Motif name: atactttggc |
| Original motif | Reverse complement motif |
| Consensus sequence: ATACTTTGGC | Consensus sequence: GCCAAAGTAT |
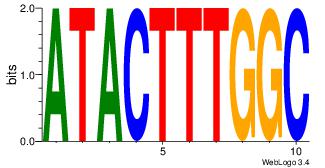 |
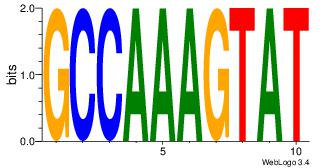 |
Best Matches for Motif ID 69 (Highest to Lowest)
| Motif ID: | UP00156 |
| Motif name: | Msx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0221581 |
Alignment:
ADHKCTAATTKGTCDBV
----ATACTTTGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: VVDGACYAATTAGYDHT | Consensus sequence: ADHKCTAATTKGTCDBV |
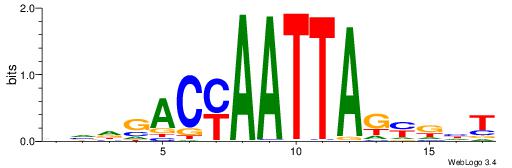 |
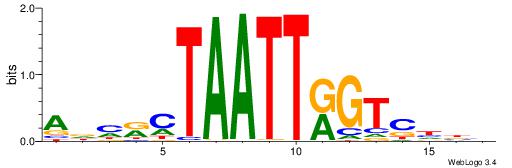 |
| Motif ID: | UP00101 |
| Motif name: | Sox12_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.027519 |
Alignment:
HDTMCTTTGTSTVDBB
-ATACTTTGGC-----
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBASACAAAGRADH | Consensus sequence: HDTMCTTTGTSTVDBB |
 |
 |
| Motif ID: | UP00066 |
| Motif name: | Hnf4a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0285786 |
Alignment:
BBHHAAAGTCCAMTHH
-GCCAAAGTAT-----
| Original motif | Reverse complement motif |
| Consensus sequence: BBHHAAAGTCCAMTHH | Consensus sequence: HHAYTGGACTTTHDBV |
 |
 |
| Motif ID: | UP00418 |
| Motif name: | Etv6 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.030488 |
Alignment:
BHAHCCGGAAGTABDVD
----GCCAAAGTAT---
| Original motif | Reverse complement motif |
| Consensus sequence: DVHBTACTTCCGGHTHB | Consensus sequence: BHAHCCGGAAGTABDVD |
 |
 |
| Motif ID: | UP00083 |
| Motif name: | Tcf7l2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0314685 |
Alignment:
BDDASATCAAAGGMDDD
-----GCCAAAGTAT--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATSTDDV | Consensus sequence: BDDASATCAAAGGMDDD |
 |
 |
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0317293 |
Alignment:
HHBTMCTTTGTTMDDVH
--ATACTTTGGC-----
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
 |
 |
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.031914 |
Alignment:
HHBBMCTTTGTTHDDBH
--ATACTTTGGC-----
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
 |
 |
| Motif ID: | UP00035 |
| Motif name: | Hic1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0323012 |
Alignment:
DBBATGCCAACCHVHH
-----GCCAAAGTAT-
| Original motif | Reverse complement motif |
| Consensus sequence: DBBATGCCAACCHVHH | Consensus sequence: DDBHGGTTGGCATVBD |
 |
 |
| Motif ID: | UP00146 |
| Motif name: | Pou6f1_3733.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.032571 |
Alignment:
VHHVATAATGAGSTDDH
----ATACTTTGGC---
| Original motif | Reverse complement motif |
| Consensus sequence: VHHVATAATGAGSTDDH | Consensus sequence: DHDASCTCATTATVHHB |
 |
 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0330221 |
Alignment:
DDDYCCTTTGATCTDDV
--ATACTTTGGC-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
 |
 |
| Dataset #: 4 | Motif ID: 70 | Motif name: CagTGCTCCACTGTGgT |
| Original motif | Reverse complement motif |
| Consensus sequence: CAGTGCTCCACTGTGBT | Consensus sequence: ABCACAGTGGAGCACTG |
 |
 |
Best Matches for Motif ID 70 (Highest to Lowest)
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 17 |
| Similarity score: | 0.0419306 |
Alignment:
CBDMCMGGGTGGTCCHVBVBAH
--ABCACAGTGGAGCACTG---
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0501904 |
Alignment:
HHBDHDDAACAATDRHHHHHBW
-----ABCACAGTGGAGCACTG
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 0.0504778 |
Alignment:
HGDHVHKGACCACCCGGBGBGDB
--CAGTGCTCCACTGTGBT----
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0513094 |
Alignment:
DDDTWCCCCCCGGDRVH
CAGTGCTCCACTGTGBT
| Original motif | Reverse complement motif |
| Consensus sequence: DDDTWCCCCCCGGDRVH | Consensus sequence: HVKDCCGGGGGGWADDD |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 0.0516506 |
Alignment:
VMYDHDGMCCHCCKBGVVAAVH
-CAGTGCTCCACTGTGBT----
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 0.0523515 |
Alignment:
VHVRCAYGATGCATCDTVVADTH
--ABCACAGTGGAGCACTG----
| Original motif | Reverse complement motif |
| Consensus sequence: HADTBVADGATGCATCMTGMVHB | Consensus sequence: VHVRCAYGATGCATCDTVVADTH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 0.053668 |
Alignment:
CVHBSCGGGTGGTCMHVHHCDH
--ABCACAGTGGAGCACTG---
| Original motif | Reverse complement motif |
| Consensus sequence: HDGHHBHRGACCACCCGSVDVG | Consensus sequence: CVHBSCGGGTGGTCMHVHHCDH |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 17 |
| Similarity score: | 0.0539016 |
Alignment:
YDYBDHTMCACGTGGADDBMDGT
-----ABCACAGTGGAGCACTG-
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00042 |
| Motif name: | Gm397_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0558205 |
Alignment:
HHRTATGTGCACATHHD
ABCACAGTGGAGCACTG
| Original motif | Reverse complement motif |
| Consensus sequence: HHDATGTGCACATAMDH | Consensus sequence: HHRTATGTGCACATHHD |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 0.0558674 |
Alignment:
BDVBCDACGCKCGCGTBHRKHD
----CAGTGCTCCACTGTGBT-
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
 |
 |
| Dataset #: 4 | Motif ID: 71 | Motif name: AmTGTGACACCACAGT |
| Original motif | Reverse complement motif |
| Consensus sequence: AMTGTGACACCACAGT | Consensus sequence: ACTGTGGTGTCACART |
 |
 |
Best Matches for Motif ID 71 (Highest to Lowest)
| Motif ID: | UP00020 |
| Motif name: | Atf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0273934 |
Alignment:
DBDRTGACGTCAYHRD
AMTGTGACACCACAGT
| Original motif | Reverse complement motif |
| Consensus sequence: DBDRTGACGTCAYHRD | Consensus sequence: DMDMTGACGTCAKHBD |
 |
 |
| Motif ID: | UP00159 |
| Motif name: | Six2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0303245 |
Alignment:
AHWDGTGATACCCMRTD
-AMTGTGACACCACAGT
| Original motif | Reverse complement motif |
| Consensus sequence: DAKRGGGTATCACDWHT | Consensus sequence: AHWDGTGATACCCMRTD |
 |
 |
| Motif ID: | UP00103 |
| Motif name: | Jundm2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0344928 |
Alignment:
BBGATGACGTCAYCVH
AMTGTGACACCACAGT
| Original motif | Reverse complement motif |
| Consensus sequence: BBGATGACGTCAYCVH | Consensus sequence: HVGMTGACGTCATCBB |
 |
 |
| Motif ID: | UP00122 |
| Motif name: | Tgif1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0348371 |
Alignment:
DBVVAGCTGTCAWWVTH
ACTGTGGTGTCACART-
| Original motif | Reverse complement motif |
| Consensus sequence: DABWWTGACAGCTVVBD | Consensus sequence: DBVVAGCTGTCAWWVTH |
 |
 |
| Motif ID: | UP00195 |
| Motif name: | Six3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0349083 |
Alignment:
DATRGGGTATCAHHWHT
ACTGTGGTGTCACART-
| Original motif | Reverse complement motif |
| Consensus sequence: DATRGGGTATCAHHWHT | Consensus sequence: AHWHDTGATACCCKATH |
 |
 |
| Motif ID: | UP00388 |
| Motif name: | Six6_2267.4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0354092 |
Alignment:
DATRGGGTATCAHDWHT
ACTGTGGTGTCACART-
| Original motif | Reverse complement motif |
| Consensus sequence: DATRGGGTATCAHDWHT | Consensus sequence: AHWDHTGATACCCKATD |
 |
 |
| Motif ID: | UP00192 |
| Motif name: | Six1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0354313 |
Alignment:
AVHDHTGATACCCMDHH
ACTGTGGTGTCACART-
| Original motif | Reverse complement motif |
| Consensus sequence: DHDRGGGTATCAHDHBT | Consensus sequence: AVHDHTGATACCCMDHH |
 |
 |
| Motif ID: | UP00199 |
| Motif name: | Six4 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0355151 |
Alignment:
ADDHATGACACCBHBBV
-AMTGTGACACCACAGT
| Original motif | Reverse complement motif |
| Consensus sequence: ADDHATGACACCBHBBV | Consensus sequence: BBVHVGGTGTCATHDDT |
 |
 |
| Motif ID: | UP00053 |
| Motif name: | Rxra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 16 |
| Similarity score: | 0.0362492 |
Alignment:
DBHBDTGACCCCDHVHB
-AMTGTGACACCACAGT
| Original motif | Reverse complement motif |
| Consensus sequence: DBHBDTGACCCCDHVHB | Consensus sequence: VHBHDGGGGTCAHBHBD |
 |
 |
| Motif ID: | UP00388 |
| Motif name: | Six6_2267.5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 16 |
| Similarity score: | 0.0374881 |
Alignment:
DDDRGGGTATCAWHWHT
-AMTGTGACACCACAGT
| Original motif | Reverse complement motif |
| Consensus sequence: DDDRGGGTATCAWHWHT | Consensus sequence: AHWHWTGATACCCKDDD |
 |
 |
| Dataset #: 4 | Motif ID: 72 | Motif name: CACTGTGrYrtCACAGTGswsCAcT |
| Original motif | Reverse complement motif |
| Consensus sequence: CACTGTGRTRTCACAGTGSWSCACT | Consensus sequence: AGTGSWSCACTGTGAMAMCACAGTG |
 |
 |
Best Matches for Motif ID 72 (Highest to Lowest)
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 21 |
| Similarity score: | 0.0558366 |
Alignment:
----HHBDHDDAACAATDRHHHHHBW
CACTGTGRTRTCACAGTGSWSCACT-
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 20 |
| Similarity score: | 0.55477 |
Alignment:
VMYDHDGMCCHCCKBGVVAAVH-----
--AGTGSWSCACTGTGAMAMCACAGTG
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 19 |
| Similarity score: | 1.05722 |
Alignment:
DBCDWHCTGGGTGGTCMDHBBAH------
------AGTGSWSCACTGTGAMAMCACAGTG
| Original motif | Reverse complement motif |
| Consensus sequence: DTVBDDRGACCACCCAGDWDGBH | Consensus sequence: DBCDWHCTGGGTGGTCMDHBBAH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 19 |
| Similarity score: | 1.05946 |
Alignment:
HGDHVHKGACCACCCGGBGBGDB------
------AGTGSWSCACTGTGAMAMCACAGTG
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 18 |
| Similarity score: | 1.5496 |
Alignment:
HTBVVVDGGACCACCCRGRDBG-------
-------CACTGTGRTRTCACAGTGSWSCACT
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 18 |
| Similarity score: | 1.55149 |
Alignment:
HADTBVADGATGCATCMTGMVHB-------
-------CACTGTGRTRTCACAGTGSWSCACT
| Original motif | Reverse complement motif |
| Consensus sequence: HADTBVADGATGCATCMTGMVHB | Consensus sequence: VHVRCAYGATGCATCDTVVADTH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 18 |
| Similarity score: | 1.55607 |
Alignment:
-------CVHBSCGGGTGGTCMHVHHCDH
AGTGSWSCACTGTGAMAMCACAGTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: HDGHHBHRGACCACCCGSVDVG | Consensus sequence: CVHBSCGGGTGGTCMHVHHCDH |
 |
 |
| Motif ID: | UP00228 |
| Motif name: | Bapx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 17 |
| Similarity score: | 2.04247 |
Alignment:
--------DTBVTTAAGTGGTTHHV
AGTGSWSCACTGTGAMAMCACAGTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: VHHAACCACTTAAVVAH | Consensus sequence: DTBVTTAAGTGGTTHHV |
 |
 |
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 17 |
| Similarity score: | 2.04316 |
Alignment:
DDBBBCACTGCABTBBB--------
------CACTGTGRTRTCACAGTGSWSCACT
| Original motif | Reverse complement motif |
| Consensus sequence: DDBBBCACTGCABTBBB | Consensus sequence: BBBAVTGCAGTGBBVDD |
 |
 |
| Motif ID: | UP00193 |
| Motif name: | Rhox11_1765.2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 17 |
| Similarity score: | 2.04346 |
Alignment:
WVVHWTWACAGCGBHBT--------
---AGTGSWSCACTGTGAMAMCACAGTG
| Original motif | Reverse complement motif |
| Consensus sequence: AVDVCGCTGTWAWDVVW | Consensus sequence: WVVHWTWACAGCGBHBT |
 |
 |
| Dataset #: 4 | Motif ID: 73 | Motif name: TGGAGCACTGTGACAcCACAGTGg |
| Original motif | Reverse complement motif |
| Consensus sequence: TGGAGCACTGTGACAVCACAGTGG | Consensus sequence: CCACTGTGVTGTCACAGTGCTCCA |
 |
 |
Best Matches for Motif ID 73 (Highest to Lowest)
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 21 |
| Similarity score: | 0.0391954 |
Alignment:
BDBBVBVGTAWCCAAGVBHBAGB---
---CCACTGTGVTGTCACAGTGCTCCA
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 21 |
| Similarity score: | 0.0400271 |
Alignment:
YBVDMGTGGGTGGTCKVVBVBBT---
--CCACTGTGVTGTCACAGTGCTCCA
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 20 |
| Similarity score: | 0.535486 |
Alignment:
----DBCDWHCTGGGTGGTCMDHBBAH
CCACTGTGVTGTCACAGTGCTCCA----
| Original motif | Reverse complement motif |
| Consensus sequence: DTVBDDRGACCACCCAGDWDGBH | Consensus sequence: DBCDWHCTGGGTGGTCMDHBBAH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 20 |
| Similarity score: | 0.536069 |
Alignment:
----BTBVTCVTGGGTGGTCMVVDVBB
CCACTGTGVTGTCACAGTGCTCCA----
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 20 |
| Similarity score: | 0.539704 |
Alignment:
----HGDHVHKGACCACCCGGBGBGDB
TGGAGCACTGTGACAVCACAGTGG---
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 18 |
| Similarity score: | 1.53819 |
Alignment:
------DVTTVVCVYGGHGGYCHHDMYB
TGGAGCACTGTGACAVCACAGTGG------
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00122 |
| Motif name: | Tgif1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 2.02534 |
Alignment:
-------DABWWTGACAGCTVVBD
TGGAGCACTGTGACAVCACAGTGG--
| Original motif | Reverse complement motif |
| Consensus sequence: DABWWTGACAGCTVVBD | Consensus sequence: DBVVAGCTGTCAWWVTH |
 |
 |
| Motif ID: | UP00159 |
| Motif name: | Six2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 2.02571 |
Alignment:
AHWDGTGATACCCMRTD-------
--CCACTGTGVTGTCACAGTGCTCCA
| Original motif | Reverse complement motif |
| Consensus sequence: DAKRGGGTATCACDWHT | Consensus sequence: AHWDGTGATACCCMRTD |
 |
 |
| Motif ID: | UP00192 |
| Motif name: | Six1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 17 |
| Similarity score: | 2.02892 |
Alignment:
AVHDHTGATACCCMDHH-------
--CCACTGTGVTGTCACAGTGCTCCA
| Original motif | Reverse complement motif |
| Consensus sequence: DHDRGGGTATCAHDHBT | Consensus sequence: AVHDHTGATACCCMDHH |
 |
 |
| Motif ID: | UP00076 |
| Motif name: | Rfxdc2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 17 |
| Similarity score: | 2.0292 |
Alignment:
HDDVSGTRTCMADGBVB-------
---CCACTGTGVTGTCACAGTGCTCCA
| Original motif | Reverse complement motif |
| Consensus sequence: BBVCDTRGAKACSVDDH | Consensus sequence: HDDVSGTRTCMADGBVB |
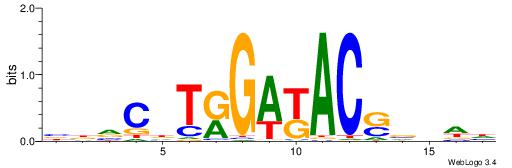 |
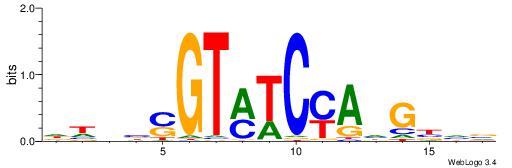 |
| Dataset #: 4 | Motif ID: 74 | Motif name: ssCCCCGCSssk |
| Original motif | Reverse complement motif |
| Consensus sequence: SBCCCCGCCSBB | Consensus sequence: BBSGGCGGGGBS |
 |
 |
Best Matches for Motif ID 74 (Highest to Lowest)
| Motif ID: | UP00000 |
| Motif name: | Smad3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0114847 |
Alignment:
DHHBCCCCGCCAHHBHB
--SBCCCCGCCSBB---
| Original motif | Reverse complement motif |
| Consensus sequence: DHHBCCCCGCCAHHBHB | Consensus sequence: BHBHDTGGCGGGGBDHD |
 |
 |
| Motif ID: | UP00093 |
| Motif name: | Klf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0165262 |
Alignment:
HBDRCCMCGCCCHHHD
--SBCCCCGCCSBB--
| Original motif | Reverse complement motif |
| Consensus sequence: HBDRCCMCGCCCHHHD | Consensus sequence: DHHDGGGCGRGGKHBH |
 |
 |
| Motif ID: | UP00022 |
| Motif name: | Zfp740_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0276137 |
Alignment:
HVBCCCCCCCCMHHHB
--SBCCCCGCCSBB--
| Original motif | Reverse complement motif |
| Consensus sequence: HVBCCCCCCCCMHHHB | Consensus sequence: BHHDYGGGGGGGGBVD |
 |
 |
| Motif ID: | UP00003 |
| Motif name: | E2F3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0328578 |
Alignment:
WHSGCGCGCCHTDHB
-SBCCCCGCCSBB--
| Original motif | Reverse complement motif |
| Consensus sequence: VHDADGGCGCGCSHW | Consensus sequence: WHSGCGCGCCHTDHB |
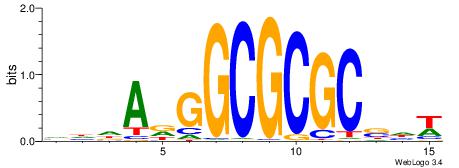 |
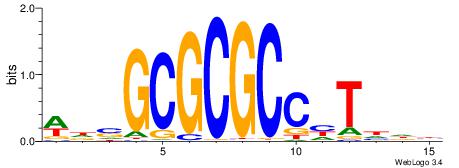 |
| Motif ID: | UP00033 |
| Motif name: | Zfp410_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0336734 |
Alignment:
BHHBHCCGCCCCDHHHH
-SBCCCCGCCSBB----
| Original motif | Reverse complement motif |
| Consensus sequence: BHHBHCCGCCCCDHHHH | Consensus sequence: HHHHDGGGGCGGDBHDV |
 |
 |
| Motif ID: | UP00001 |
| Motif name: | E2F2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0345737 |
Alignment:
HHVGCGCGCCKTDHB
-SBCCCCGCCSBB--
| Original motif | Reverse complement motif |
| Consensus sequence: VHDARGGCGCGCVHH | Consensus sequence: HHVGCGCGCCKTDHB |
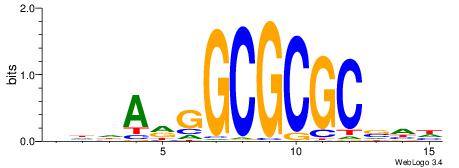 |
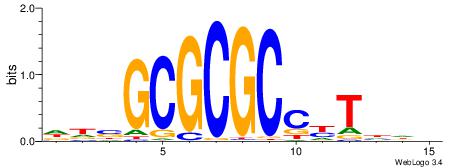 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0347468 |
Alignment:
HVDVGGGHGGGGDBHB
--BBSGGCGGGGBS--
| Original motif | Reverse complement motif |
| Consensus sequence: BHVDCCCCDCCCBDBH | Consensus sequence: HVDVGGGHGGGGDBHB |
 |
 |
| Motif ID: | UP00043 |
| Motif name: | Bcl6b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0358124 |
Alignment:
BHHHWGGGGCGGBBVH
--BBSGGCGGGGBS--
| Original motif | Reverse complement motif |
| Consensus sequence: HBBBCCGCCCCWHHHV | Consensus sequence: BHHHWGGGGCGGBBVH |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 12 |
| Similarity score: | 0.0365354 |
Alignment:
BADHBDHCGCCCMCGCAHHDBBV
--------SBCCCCGCCSBB---
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 12 |
| Similarity score: | 0.0389727 |
Alignment:
HDWHMHGACGCGCGTRBVVBHW
-----BBSGGCGGGGBS-----
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
 |
 |
| Dataset #: 4 | Motif ID: 75 | Motif name: wwCCAmAGTCmt |
| Original motif | Reverse complement motif |
| Consensus sequence: DDCCAMAGTCHB | Consensus sequence: VDGACTRTGGDD |
 |
 |
Best Matches for Motif ID 75 (Highest to Lowest)
| Motif ID: | UP00079 |
| Motif name: | Esrra_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 12 |
| Similarity score: | 0.00927341 |
Alignment:
DHDBCAAGGTCAHVBDH
-DDCCAMAGTCHB----
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBCAAGGTCAHVBDH | Consensus sequence: HHBVHTGACCTTGVDHD |
 |
 |
| Motif ID: | UP00066 |
| Motif name: | Hnf4a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0157554 |
Alignment:
BBHHAAAGTCCAMTHH
DDCCAMAGTCHB----
| Original motif | Reverse complement motif |
| Consensus sequence: BBHHAAAGTCCAMTHH | Consensus sequence: HHAYTGGACTTTHDBV |
 |
 |
| Motif ID: | UP00023 |
| Motif name: | Sox30_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0218941 |
Alignment:
DHDBAACAATDRHHHH
--VDGACTRTGGDD--
| Original motif | Reverse complement motif |
| Consensus sequence: DHDBAACAATDRHHHH | Consensus sequence: HHHHMHATTGTTBDHD |
 |
 |
| Motif ID: | UP00028 |
| Motif name: | Tcfap2e_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.023236 |
Alignment:
DTHGCCTSAGGCHHH
--DDCCAMAGTCHB-
| Original motif | Reverse complement motif |
| Consensus sequence: HHHGCCTSAGGCDAD | Consensus sequence: DTHGCCTSAGGCHHH |
 |
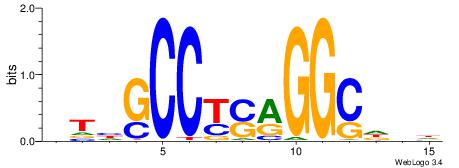 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 12 |
| Similarity score: | 0.0235871 |
Alignment:
HHBDHDDAACAATDRHHHHHBW
-----VDGACTRTGGDD-----
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00095 |
| Motif name: | Zfp691_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0240897 |
Alignment:
VHBDDAGGAGTCWBBTD
--DDCCAMAGTCHB---
| Original motif | Reverse complement motif |
| Consensus sequence: DABBWGACTCCTHDVHV | Consensus sequence: VHBDDAGGAGTCWBBTD |
 |
 |
| Motif ID: | UP00004 |
| Motif name: | Sox14_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.025783 |
Alignment:
BKBASACAATRGHBB
--VDGACTRTGGDD-
| Original motif | Reverse complement motif |
| Consensus sequence: BKBASACAATRGHBB | Consensus sequence: BBDCMATTGTSTBRB |
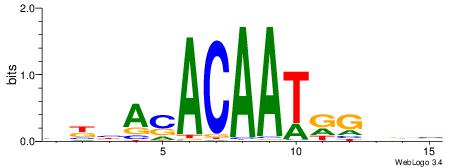 |
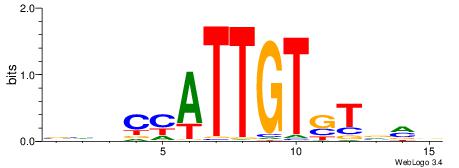 |
| Motif ID: | UP00081 |
| Motif name: | Mybl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.026472 |
Alignment:
HDDDHAACCGTTAHHHD
---VDGACTRTGGDD--
| Original motif | Reverse complement motif |
| Consensus sequence: HDDDHAACCGTTAHHHD | Consensus sequence: DHHHTAACGGTTHDHDH |
 |
 |
| Motif ID: | UP00052 |
| Motif name: | Osr2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.027027 |
Alignment:
HHHHGCTACKGTHVHH
---DDCCAMAGTCHB-
| Original motif | Reverse complement motif |
| Consensus sequence: HHVHACRGTAGCHHHD | Consensus sequence: HHHHGCTACKGTHVHH |
 |
 |
| Motif ID: | UP00000 |
| Motif name: | Smad3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0270662 |
Alignment:
HVHBVTGTCTGGDDHDD
--VDGACTRTGGDD---
| Original motif | Reverse complement motif |
| Consensus sequence: HDHDDCCAGACABBHVH | Consensus sequence: HVHBVTGTCTGGDDHDD |
 |
 |
| Dataset #: 4 | Motif ID: 76 | Motif name: CAcCACAGTGGAGCAct |
| Original motif | Reverse complement motif |
| Consensus sequence: CAHCACAGTGGAGCACT | Consensus sequence: AGTGCTCCACTGTGDTG |
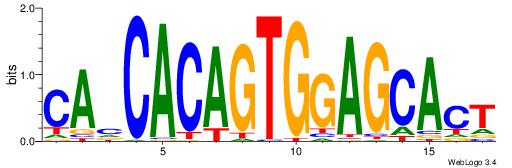 |
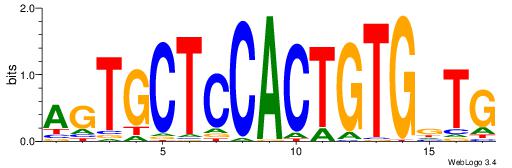 |
Best Matches for Motif ID 76 (Highest to Lowest)
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 0.0381337 |
Alignment:
CBDMCMGGGTGGTCCHVBVBAH
-CAHCACAGTGGAGCACT----
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0393208 |
Alignment:
BBBAVTGCAGTGBBVDD
AGTGCTCCACTGTGDTG
| Original motif | Reverse complement motif |
| Consensus sequence: DDBBBCACTGCABTBBB | Consensus sequence: BBBAVTGCAGTGBBVDD |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 0.0441166 |
Alignment:
HHBDHDDAACAATDRHHHHHBW
----CAHCACAGTGGAGCACT-
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 17 |
| Similarity score: | 0.0477617 |
Alignment:
HGDHVHKGACCACCCGGBGBGDB
---AGTGCTCCACTGTGDTG---
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00231 |
| Motif name: | Nkx2-2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0479423 |
Alignment:
HDVRCCACTTGARADBD
CAHCACAGTGGAGCACT
| Original motif | Reverse complement motif |
| Consensus sequence: HDVRCCACTTGARADBD | Consensus sequence: DVDTKTCAAGTGGKBDH |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 17 |
| Similarity score: | 0.0484681 |
Alignment:
VHVRCAYGATGCATCDTVVADTH
-CAHCACAGTGGAGCACT-----
| Original motif | Reverse complement motif |
| Consensus sequence: HADTBVADGATGCATCMTGMVHB | Consensus sequence: VHVRCAYGATGCATCDTVVADTH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.0487437 |
Alignment:
HHBSHTCGTGRGTGGKGDBVWMV
AGTGCTCCACTGTGDTG------
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00046 |
| Motif name: | Tcfe2a_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.048891 |
Alignment:
DHDBHGCACCTGBDDVB
AGTGCTCCACTGTGDTG
| Original motif | Reverse complement motif |
| Consensus sequence: VBHHVCAGGTGCDVDHD | Consensus sequence: DHDBHGCACCTGBDDVB |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 17 |
| Similarity score: | 0.0491352 |
Alignment:
CVHBSCGGGTGGTCMHVHHCDH
-CAHCACAGTGGAGCACT----
| Original motif | Reverse complement motif |
| Consensus sequence: HDGHHBHRGACCACCCGSVDVG | Consensus sequence: CVHBSCGGGTGGTCMHVHHCDH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 17 |
| Similarity score: | 0.0515016 |
Alignment:
DVTTVVCVYGGHGGYCHHDMYB
---CAHCACAGTGGAGCACT--
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Dataset #: 4 | Motif ID: 77 | Motif name: AgTGCTCCACTGTGgTGTCACAgT |
| Original motif | Reverse complement motif |
| Consensus sequence: AGTGCTCCACTGTGGTGTCACAGT | Consensus sequence: ACTGTGACACCACAGTGGAGCACT |
 |
 |
Best Matches for Motif ID 77 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 23 |
| Similarity score: | 0.0509731 |
Alignment:
BBBDVVRGACCACCCAVGABBAB-
ACTGTGACACCACAGTGGAGCACT
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 21 |
| Similarity score: | 1.0488 |
Alignment:
DTVBDDRGACCACCCAGDWDGBH---
---AGTGCTCCACTGTGGTGTCACAGT
| Original motif | Reverse complement motif |
| Consensus sequence: DTVBDDRGACCACCCAGDWDGBH | Consensus sequence: DBCDWHCTGGGTGGTCMDHBBAH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 21 |
| Similarity score: | 1.05087 |
Alignment:
HGDHVHKGACCACCCGGBGBGDB---
--ACTGTGACACCACAGTGGAGCACT
| Original motif | Reverse complement motif |
| Consensus sequence: BHCBCBCCGGGTGGTCYHVHDCH | Consensus sequence: HGDHVHKGACCACCCGGBGBGDB |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 18 |
| Similarity score: | 2.54748 |
Alignment:
CBDMCMGGGTGGTCCHVBVBAH------
------ACTGTGACACCACAGTGGAGCACT
| Original motif | Reverse complement motif |
| Consensus sequence: HTBVVVDGGACCACCCRGRDBG | Consensus sequence: CBDMCMGGGTGGTCCHVBVBAH |
 |
 |
| Motif ID: | UP00045 |
| Motif name: | Mafb_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 3.0367 |
Alignment:
-------HAAWDTGCTGACDWARH
AGTGCTCCACTGTGGTGTCACAGT
| Original motif | Reverse complement motif |
| Consensus sequence: HAAWDTGCTGACDWARH | Consensus sequence: DMTWDGTCAGCADWTTH |
 |
 |
| Motif ID: | UP00055 |
| Motif name: | Hbp1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 17 |
| Similarity score: | 3.04028 |
Alignment:
-------DDDYCCCATTGTVHABH
AGTGCTCCACTGTGGTGTCACAGT------
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCCATTGTVHABH | Consensus sequence: HBTHVACAATGGGMDHD |
 |
 |
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 17 |
| Similarity score: | 3.04347 |
Alignment:
-------DDBBBCACTGCABTBBB
AGTGCTCCACTGTGGTGTCACAGT-------
| Original motif | Reverse complement motif |
| Consensus sequence: DDBBBCACTGCABTBBB | Consensus sequence: BBBAVTGCAGTGBBVDD |
 |
 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 17 |
| Similarity score: | 3.04662 |
Alignment:
VDDAGATCAAAGGKDDD-------
---ACTGTGACACCACAGTGGAGCACT
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
 |
 |
| Motif ID: | UP00070 |
| Motif name: | Gcm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 3.04663 |
Alignment:
DDBVHATAGGGGRBRMB-------
-AGTGCTCCACTGTGGTGTCACAGT
| Original motif | Reverse complement motif |
| Consensus sequence: DDBVHATAGGGGRBRMB | Consensus sequence: BYMBKCCCCTATDVBHD |
 |
 |
| Motif ID: | UP00148 |
| Motif name: | Hdx |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 17 |
| Similarity score: | 3.04907 |
Alignment:
-------BDBDHTGATTTCDHYDD
ACTGTGACACCACAGTGGAGCACT-------
| Original motif | Reverse complement motif |
| Consensus sequence: DDKDHGAAATCAHHBHV | Consensus sequence: BDBDHTGATTTCDHYDD |
 |
 |
| Dataset #: 4 | Motif ID: 78 | Motif name: ssGCkTGCssk |
| Original motif | Reverse complement motif |
| Consensus sequence: SSGCKTGCSSK | Consensus sequence: YSSGCAYGCSS |
 |
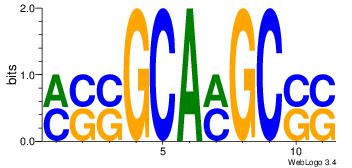 |
Best Matches for Motif ID 78 (Highest to Lowest)
| Motif ID: | UP00003 |
| Motif name: | E2F3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.00463328 |
Alignment:
VHDADGGCGCGCSHW
----SSGCKTGCSSK
| Original motif | Reverse complement motif |
| Consensus sequence: VHDADGGCGCGCSHW | Consensus sequence: WHSGCGCGCCHTDHB |
 |
 |
| Motif ID: | UP00094 |
| Motif name: | Zfp128_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.00482124 |
Alignment:
BHHDKGGCGTACCCBHD
----SSGCKTGCSSK--
| Original motif | Reverse complement motif |
| Consensus sequence: BHHDKGGCGTACCCBHD | Consensus sequence: DHVGGGTACGCCRDHDV |
 |
 |
| Motif ID: | UP00035 |
| Motif name: | Hic1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.00562779 |
Alignment:
VHHDWTGGGCAMDHBH
-----YSSGCAYGCSS
| Original motif | Reverse complement motif |
| Consensus sequence: DBDDRTGCCCAWDHDV | Consensus sequence: VHHDWTGGGCAMDHBH |
 |
 |
| Motif ID: | UP00065 |
| Motif name: | Zfp161_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.00595984 |
Alignment:
DDGCGCGCGCRCHYRD
--SSGCKTGCSSK---
| Original motif | Reverse complement motif |
| Consensus sequence: DDGCGCGCGCRCHYRD | Consensus sequence: DMMDGMGCGCGCGCHD |
 |
 |
| Motif ID: | UP00001 |
| Motif name: | E2F2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.00821725 |
Alignment:
HHVGCGCGCCKTDHB
-SSGCKTGCSSK---
| Original motif | Reverse complement motif |
| Consensus sequence: VHDARGGCGCGCVHH | Consensus sequence: HHVGCGCGCCKTDHB |
 |
 |
| Motif ID: | UP00060 |
| Motif name: | Max_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0109259 |
Alignment:
HDGTCGCGTGDCVB
---SSGCKTGCSSK
| Original motif | Reverse complement motif |
| Consensus sequence: BBGHCACGCGACDD | Consensus sequence: HDGTCGCGTGDCVB |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0150199 |
Alignment:
VDWDARRGACGCGGWHGGAKHV
---YSSGCAYGCSS--------
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0160778 |
Alignment:
HHDGBCACGCCTWDB
-YSSGCAYGCSS---
| Original motif | Reverse complement motif |
| Consensus sequence: BDWAGGCGTGBCHHD | Consensus sequence: HHDGBCACGCCTWDB |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0171602 |
Alignment:
HDWHMHGACGCGCGTRBVVBHW
---------SSGCKTGCSSK--
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0172713 |
Alignment:
DCATDBTGHGCATKMTTBRSWR
------YSSGCAYGCSS-----
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
 |
 |
| Dataset #: 4 | Motif ID: 79 | Motif name: cCGCGGrCACG |
| Original motif | Reverse complement motif |
| Consensus sequence: CCGCGGRCACG | Consensus sequence: CGTGKCCGCGG |
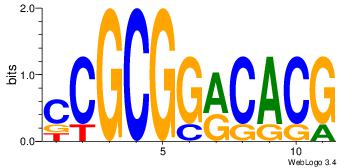 |
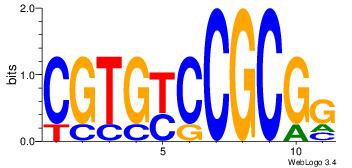 |
Best Matches for Motif ID 79 (Highest to Lowest)
| Motif ID: | UP00065 |
| Motif name: | Zfp161_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0220206 |
Alignment:
DDGCGCGCGCRCHYRD
----CCGCGGRCACG-
| Original motif | Reverse complement motif |
| Consensus sequence: DDGCGCGCGCRCHYRD | Consensus sequence: DMMDGMGCGCGCGCHD |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 11 |
| Similarity score: | 0.024575 |
Alignment:
VHYTCCDWCCGCGTCMMTHWHV
---CGTGKCCGCGG--------
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00042 |
| Motif name: | Gm397_second |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0279366 |
Alignment:
DVMVGCACACACKCVH
--CCGCGGRCACG---
| Original motif | Reverse complement motif |
| Consensus sequence: DVMVGCACACACKCVH | Consensus sequence: HBGYGTGTGTGCVRVD |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0285381 |
Alignment:
VHTGCGGGGGCGGH
-CCGCGGRCACG--
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
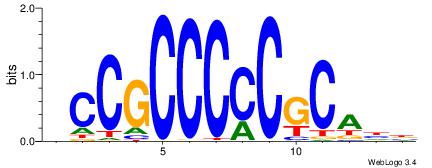 |
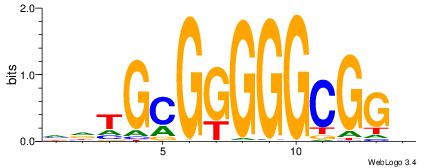 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 11 |
| Similarity score: | 0.0335378 |
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
--------CCGCGGRCACG----
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0340428 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
-------CCGCGGRCACG-----
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00065 |
| Motif name: | Zfp161_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0345855 |
Alignment:
GYCGCGCARBGCVB
-CCGCGGRCACG--
| Original motif | Reverse complement motif |
| Consensus sequence: GYCGCGCARBGCVB | Consensus sequence: VVGCVMTGCGCGKC |
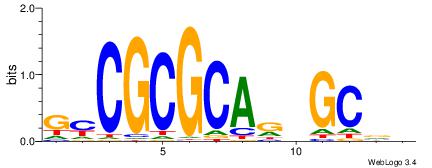 |
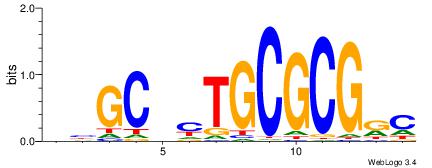 |
| Motif ID: | UP00097 |
| Motif name: | Mtf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0350434 |
Alignment:
DBDHCGTGTGCAAMDH
---CCGCGGRCACG--
| Original motif | Reverse complement motif |
| Consensus sequence: DBDHCGTGTGCAAMDH | Consensus sequence: HDYTTGCACACGDHBH |
 |
 |
| Motif ID: | UP00076 |
| Motif name: | Rfxdc2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0359928 |
Alignment:
HDDVSGTRTCMADGBVB
----CGTGKCCGCGG--
| Original motif | Reverse complement motif |
| Consensus sequence: BBVCDTRGAKACSVDDH | Consensus sequence: HDDVSGTRTCMADGBVB |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0361963 |
Alignment:
HDWHMHGACGCGCGTRBVVBHW
---------CCGCGGRCACG--
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
 |
 |
| Dataset #: 4 | Motif ID: 80 | Motif name: taaacgatgcc |
| Original motif | Reverse complement motif |
| Consensus sequence: TAAACGATGCC | Consensus sequence: GGCATCGTTTA |
 |
 |
Best Matches for Motif ID 80 (Highest to Lowest)
| Motif ID: | UP00114 |
| Motif name: | Homez |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0207645 |
Alignment:
HDTVAAAACGATRWWHT
----TAAACGATGCC--
| Original motif | Reverse complement motif |
| Consensus sequence: AHWWMATCGTTTTBADD | Consensus sequence: HDTVAAAACGATRWWHT |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0294316 |
Alignment:
VVVWTTGTTTAMATTWD
GGCATCGTTTA------
| Original motif | Reverse complement motif |
| Consensus sequence: DWAATRTAAACAAWVVB | Consensus sequence: VVVWTTGTTTAMATTWD |
 |
 |
| Motif ID: | UP00039 |
| Motif name: | Foxj3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.030091 |
Alignment:
HDHADGTAAACAAAVVH
------TAAACGATGCC
| Original motif | Reverse complement motif |
| Consensus sequence: HDHADGTAAACAAAVVH | Consensus sequence: DVVTTTGTTTACDTHDH |
 |
 |
| Motif ID: | UP00025 |
| Motif name: | Foxk1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.0306614 |
Alignment:
BBVTTTGTTTACATTDD
GGCATCGTTTA------
| Original motif | Reverse complement motif |
| Consensus sequence: DDAATGTAAACAAAVVB | Consensus sequence: BBVTTTGTTTACATTDD |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 10 |
| Number of overlap: | 11 |
| Similarity score: | 0.0342477 |
Alignment:
DBBVDDDRTRGCGTCBBYTHGA
---------GGCATCGTTTA--
| Original motif | Reverse complement motif |
| Consensus sequence: DBBVDDDRTRGCGTCBBYTHGA | Consensus sequence: TCDAMVBGACGCMAKDDDVBBD |
 |
 |
| Motif ID: | UP00040 |
| Motif name: | Irf5_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0345017 |
Alignment:
BDKGTTTCGKTWDDD
--GGCATCGTTTA--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDWAYCGAAACYDV | Consensus sequence: BDKGTTTCGKTWDDD |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0351616 |
Alignment:
VHVRCAYGATGCATCDTVVADTH
--TAAACGATGCC----------
| Original motif | Reverse complement motif |
| Consensus sequence: HADTBVADGATGCATCMTGMVHB | Consensus sequence: VHVRCAYGATGCATCDTVVADTH |
 |
 |
| Motif ID: | UP00232 |
| Motif name: | Dobox4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0371486 |
Alignment:
DAWKGGGTATCTAWHWH
-----GGCATCGTTTA-
| Original motif | Reverse complement motif |
| Consensus sequence: HWHWTAGATACCCYWTD | Consensus sequence: DAWKGGGTATCTAWHWH |
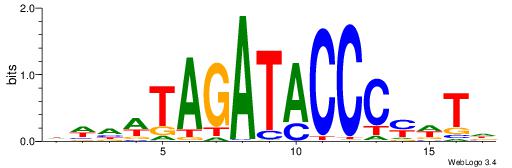 |
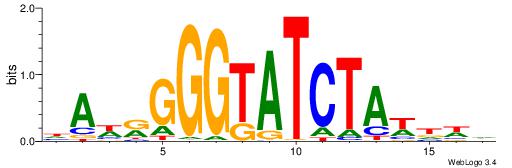 |
| Motif ID: | UP00073 |
| Motif name: | Foxa2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.0384798 |
Alignment:
BDVTTTKTTTACWTWHH
GGCATCGTTTA------
| Original motif | Reverse complement motif |
| Consensus sequence: HHWAWGTAAAYAAAVDB | Consensus sequence: BDVTTTKTTTACWTWHH |
 |
 |
| Motif ID: | UP00011 |
| Motif name: | Irf6_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.0388532 |
Alignment:
DHDBTKGTTTCGHTHDD
-----GGCATCGTTTA-
| Original motif | Reverse complement motif |
| Consensus sequence: HDDAHCGAAACYAVDDD | Consensus sequence: DHDBTKGTTTCGHTHDD |
 |
 |
| Dataset #: 4 | Motif ID: 81 | Motif name: ctatacggacg |
| Original motif | Reverse complement motif |
| Consensus sequence: CTATACGGACG | Consensus sequence: CGTCCGTATAG |
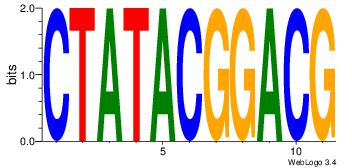 |
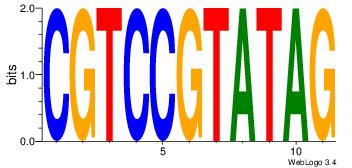 |
Best Matches for Motif ID 81 (Highest to Lowest)
| Motif ID: | UP00038 |
| Motif name: | Spdef_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.027939 |
Alignment:
VVHHATCCGGATGKHV
--CTATACGGACG---
| Original motif | Reverse complement motif |
| Consensus sequence: VHRCATCCGGATHHBB | Consensus sequence: VVHHATCCGGATGKHV |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0308121 |
Alignment:
VDWDARRGACGCGGWHGGAKHV
CTATACGGACG-----------
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00413 |
| Motif name: | Elf4 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0328673 |
Alignment:
HAWYCCGGAAGTHBBD
CTATACGGACG-----
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGKWTH | Consensus sequence: HAWYCCGGAAGTHBBD |
 |
 |
| Motif ID: | UP00418 |
| Motif name: | Etv6 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0334005 |
Alignment:
BHAHCCGGAAGTABDVD
------CGTCCGTATAG
| Original motif | Reverse complement motif |
| Consensus sequence: DVHBTACTTCCGGHTHB | Consensus sequence: BHAHCCGGAAGTABDVD |
 |
 |
| Motif ID: | UP00409 |
| Motif name: | Elf5 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0336391 |
Alignment:
BWAHSMGGAAGTWD
CTATACGGACG---
| Original motif | Reverse complement motif |
| Consensus sequence: BWAHSMGGAAGTWD | Consensus sequence: DWACTTCCRSDTWV |
 |
 |
| Motif ID: | UP00097 |
| Motif name: | Mtf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0346533 |
Alignment:
DBDHCGTGTGCAAMDH
----CGTCCGTATAG-
| Original motif | Reverse complement motif |
| Consensus sequence: DBDHCGTGTGCAAMDH | Consensus sequence: HDYTTGCACACGDHBH |
 |
 |
| Motif ID: | UP00135 |
| Motif name: | Hoxc12 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0360998 |
Alignment:
DDVHWTTTACGACCKHH
----CTATACGGACG--
| Original motif | Reverse complement motif |
| Consensus sequence: HHRGGTCGTAAAWHBDH | Consensus sequence: DDVHWTTTACGACCKHH |
 |
 |
| Motif ID: | UP00084 |
| Motif name: | Gmeb1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0365466 |
Alignment:
BVVDVRYACGTRYVBHB
----CGTCCGTATAG--
| Original motif | Reverse complement motif |
| Consensus sequence: BHBBKKACGTMMVDBVB | Consensus sequence: BVVDVRYACGTRYVBHB |
 |
 |
| Motif ID: | UP00015 |
| Motif name: | Ehf_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0367131 |
Alignment:
VBDABCCGGAAGTDD
-CTATACGGACG---
| Original motif | Reverse complement motif |
| Consensus sequence: VBDABCCGGAAGTDD | Consensus sequence: DDACTTCCGGBTHBB |
 |
 |
| Motif ID: | UP00421 |
| Motif name: | Etv1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0373 |
Alignment:
HBDBACTTCCGGTBHVY
-----CGTCCGTATAG-
| Original motif | Reverse complement motif |
| Consensus sequence: MVHBACCGGAAGTVDVH | Consensus sequence: HBDBACTTCCGGTBHVY |
 |
 |
| Dataset #: 4 | Motif ID: 82 | Motif name: gCGCGCsCgsG |
| Original motif | Reverse complement motif |
| Consensus sequence: GCGCGCSCGCG | Consensus sequence: CGCGSGCGCGC |
 |
 |
Best Matches for Motif ID 82 (Highest to Lowest)
| Motif ID: | UP00065 |
| Motif name: | Zfp161_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0 |
Alignment:
DDGCGCGCGCRCHYRD
-CGCGSGCGCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: DDGCGCGCGCRCHYRD | Consensus sequence: DMMDGMGCGCGCGCHD |
 |
 |
| Motif ID: | UP00003 |
| Motif name: | E2F3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0102009 |
Alignment:
WHSGCGCGCCHTDHB
CGCGSGCGCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: VHDADGGCGCGCSHW | Consensus sequence: WHSGCGCGCCHTDHB |
 |
 |
| Motif ID: | UP00001 |
| Motif name: | E2F2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 11 |
| Similarity score: | 0.0124455 |
Alignment:
HHVGCGCGCCKTDHB
CGCGSGCGCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: VHDARGGCGCGCVHH | Consensus sequence: HHVGCGCGCCKTDHB |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 11 |
| Similarity score: | 0.0192204 |
Alignment:
WHVBVVMACGCGCGTCDYHWDH
----CGCGSGCGCGC-------
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 11 |
| Similarity score: | 0.0231982 |
Alignment:
DDRMHBACGCGYGCGTDGBBDB
-------CGCGSGCGCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
 |
 |
| Motif ID: | UP00007 |
| Motif name: | Egr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0289369 |
Alignment:
HCCGCCCCCGCAHB
-GCGCGCSCGCG--
| Original motif | Reverse complement motif |
| Consensus sequence: HCCGCCCCCGCAHB | Consensus sequence: VHTGCGGGGGCGGH |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.0297254 |
Alignment:
BADHBDHCGCCCMCGCAHHDBBV
------GCGCGCSCGCG------
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00088 |
| Motif name: | Plagl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 11 |
| Similarity score: | 0.0316913 |
Alignment:
BHDGGGGSSCCCCBVV
-CGCGSGCGCGC----
| Original motif | Reverse complement motif |
| Consensus sequence: BHDGGGGSSCCCCBVV | Consensus sequence: VBVGGGGSSCCCCHHV |
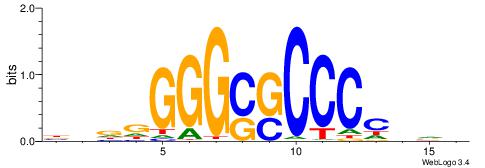 |
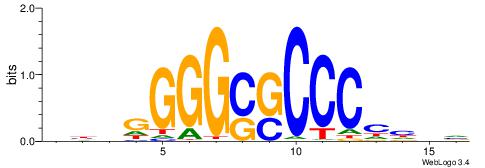 |
| Motif ID: | UP00002 |
| Motif name: | Sp4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0323627 |
Alignment:
BVVHAGGGGGCGRDHHB
----CGCGSGCGCGC--
| Original motif | Reverse complement motif |
| Consensus sequence: BDHHMCGCCCCCTHVBB | Consensus sequence: BVVHAGGGGGCGRDHHB |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 11 |
| Similarity score: | 0.039112 |
Alignment:
VHYTCCDWCCGCGTCMMTHWHV
-----CGCGSGCGCGC------
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Dataset #: 4 | Motif ID: 83 | Motif name: srACyCCGAyr |
| Original motif | Reverse complement motif |
| Consensus sequence: SRACYCCGAYR | Consensus sequence: KKTCGGKGTKS |
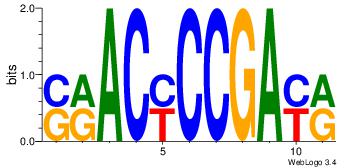 |
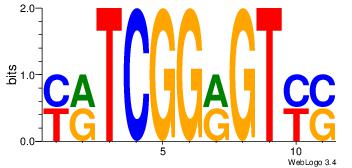 |
Best Matches for Motif ID 83 (Highest to Lowest)
| Motif ID: | UP00024 |
| Motif name: | Glis2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0129914 |
Alignment:
HKBBGGGGGGTCVHHH
--KKTCGGKGTKS---
| Original motif | Reverse complement motif |
| Consensus sequence: HHHVGACCCCCCVBRD | Consensus sequence: HKBBGGGGGGTCVHHH |
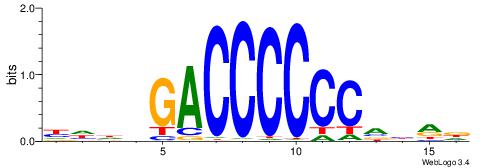 |
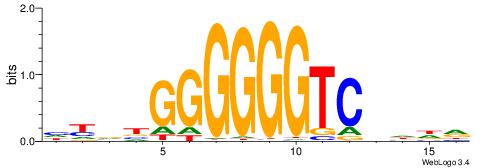 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 11 |
| Similarity score: | 0.013032 |
Alignment:
VDWDARRGACGCGGWHGGAKHV
------SRACYCCGAYR-----
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00070 |
| Motif name: | Gcm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0132459 |
Alignment:
BYMBKCCCCTATDVBHD
--SRACYCCGAYR----
| Original motif | Reverse complement motif |
| Consensus sequence: DDBVHATAGGGGRBRMB | Consensus sequence: BYMBKCCCCTATDVBHD |
 |
 |
| Motif ID: | UP00070 |
| Motif name: | Gcm1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0135374 |
Alignment:
HVVHACCCGCATVDHD
--SRACYCCGAYR---
| Original motif | Reverse complement motif |
| Consensus sequence: HVVHACCCGCATVDHD | Consensus sequence: DHDVATGCGGGTHVVH |
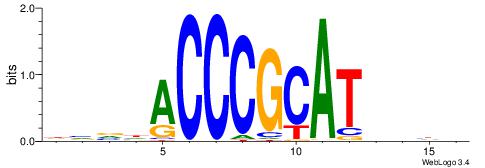 |
 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 11 |
| Similarity score: | 0.0136908 |
Alignment:
DKKWDACCCCCAHTDDV
---SRACYCCGAYR---
| Original motif | Reverse complement motif |
| Consensus sequence: DKKWDACCCCCAHTDDV | Consensus sequence: VDDAHTGGGGGTHWYYD |
 |
 |
| Motif ID: | UP00047 |
| Motif name: | Zbtb7b_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 11 |
| Similarity score: | 0.0146089 |
Alignment:
VHWTTHGGGGGGVDD
--KKTCGGKGTKS--
| Original motif | Reverse complement motif |
| Consensus sequence: DDVCCCCCCHAAWHB | Consensus sequence: VHWTTHGGGGGGVDD |
 |
 |
| Motif ID: | UP00040 |
| Motif name: | Irf5_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0148195 |
Alignment:
BDKGTTTCGKTWDDD
----KKTCGGKGTKS
| Original motif | Reverse complement motif |
| Consensus sequence: DDDWAYCGAAACYDV | Consensus sequence: BDKGTTTCGKTWDDD |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 11 |
| Similarity score: | 0.014859 |
Alignment:
ABBBBVVRGACCACCCACRDBBM
--------SRACYCCGAYR----
| Original motif | Reverse complement motif |
| Consensus sequence: ABBBBVVRGACCACCCACRDBBM | Consensus sequence: YBVDMGTGGGTGGTCKVVBVBBT |
 |
 |
| Motif ID: | UP00021 |
| Motif name: | Zfp281_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 11 |
| Similarity score: | 0.0153296 |
Alignment:
GKRGGGGGGGGGGBD
KKTCGGKGTKS----
| Original motif | Reverse complement motif |
| Consensus sequence: DBCCCCCCCCCCMYC | Consensus sequence: GKRGGGGGGGGGGBD |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 10 |
| Number of overlap: | 11 |
| Similarity score: | 0.0155548 |
Alignment:
DBCDWHCTGGGTGGTCMDHBBAH
---KKTCGGKGTKS---------
| Original motif | Reverse complement motif |
| Consensus sequence: DTVBDDRGACCACCCAGDWDGBH | Consensus sequence: DBCDWHCTGGGTGGTCMDHBBAH |
 |
 |
| Dataset #: 5 | Motif ID: 84 | Motif name: TFW3 |
| Original motif | Reverse complement motif |
| Consensus sequence: GCACTG | Consensus sequence: CAGTGC |
 |
 |
Best Matches for Motif ID 84 (Highest to Lowest)
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0 |
Alignment:
BBBAVTGCAGTGBBVDD
-------CAGTGC----
| Original motif | Reverse complement motif |
| Consensus sequence: DDBBBCACTGCABTBBB | Consensus sequence: BBBAVTGCAGTGBBVDD |
 |
 |
| Motif ID: | UP00095 |
| Motif name: | Zfp691_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.020963 |
Alignment:
DDHBYGAGCACTGBHHB
-------GCACTG----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHVCAGTGCTCMBHDD | Consensus sequence: DDHBYGAGCACTGBHHB |
 |
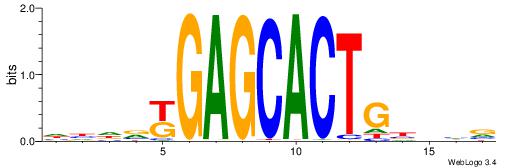 |
| Motif ID: | UP00031 |
| Motif name: | Zbtb3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.0248871 |
Alignment:
DDHHTBCCAGTGABHG
-------CAGTGC---
| Original motif | Reverse complement motif |
| Consensus sequence: CHVTCACTGGBADHDD | Consensus sequence: DDHHTBCCAGTGABHG |
 |
 |
| Motif ID: | UP00065 |
| Motif name: | Zfp161_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 6 |
| Similarity score: | 0.0303167 |
Alignment:
GYCGCGCARBGCVB
------CAGTGC--
| Original motif | Reverse complement motif |
| Consensus sequence: GYCGCGCARBGCVB | Consensus sequence: VVGCVMTGCGCGKC |
 |
 |
| Motif ID: | UP00006 |
| Motif name: | Zic3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0420592 |
Alignment:
DVHCYTGCTGTGHDH
-------CAGTGC--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDCACAGCAKGHVD | Consensus sequence: DVHCYTGCTGTGHDH |
 |
 |
| Motif ID: | UP00102 |
| Motif name: | Zic1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 6 |
| Similarity score: | 0.0499082 |
Alignment:
BHDCVCAGCAGGHVD
--GCACTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: BHDCVCAGCAGGHVD | Consensus sequence: DVHCCTGCTGBGDDB |
 |
 |
| Motif ID: | UP00231 |
| Motif name: | Nkx2-2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.0507672 |
Alignment:
DVDTKTCAAGTGGKBDH
-------CAGTGC----
| Original motif | Reverse complement motif |
| Consensus sequence: HDVRCCACTTGARADBD | Consensus sequence: DVDTKTCAAGTGGKBDH |
 |
 |
| Motif ID: | UP00107 |
| Motif name: | Nkx2-4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 6 |
| Similarity score: | 0.0519322 |
Alignment:
AAWYTMAAGTGGVTDH
------CAGTGC----
| Original motif | Reverse complement motif |
| Consensus sequence: HDAVCCACTTRAMWTT | Consensus sequence: AAWYTMAAGTGGVTDH |
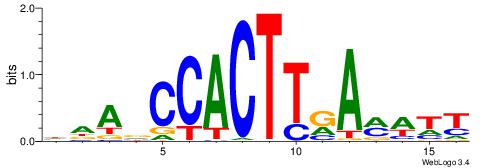 |
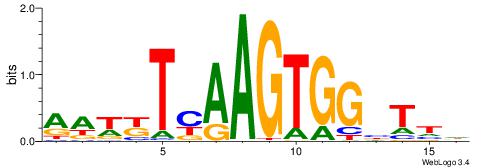 |
| Motif ID: | UP00193 |
| Motif name: | Rhox11_2205.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 6 |
| Similarity score: | 0.0530479 |
Alignment:
WVVHWTWACAGCGHHVT
--------CAGTGC---
| Original motif | Reverse complement motif |
| Consensus sequence: AVDHCGCTGTWAWDVVW | Consensus sequence: WVVHWTWACAGCGHHVT |
 |
 |
| Motif ID: | UP00057 |
| Motif name: | Zic2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 6 |
| Similarity score: | 0.0539075 |
Alignment:
HHDCVCAGCAKGVVD
--GCACTG-------
| Original motif | Reverse complement motif |
| Consensus sequence: HHDCVCAGCAKGVVD | Consensus sequence: DVBCYTGCTGBGDDD |
 |
 |
| Dataset #: 5 | Motif ID: 85 | Motif name: TFW1 |
| Original motif | Reverse complement motif |
| Consensus sequence: TTGCGCAA | Consensus sequence: TTGCGCAA |
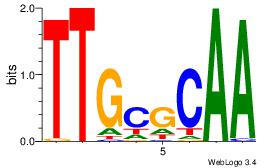 |
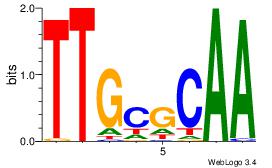 |
Best Matches for Motif ID 85 (Highest to Lowest)
| Motif ID: | UP00045 |
| Motif name: | Mafb_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.00957029 |
Alignment:
HDWWTTTTGCAADBD
----TTGCGCAA---
| Original motif | Reverse complement motif |
| Consensus sequence: HVDTTGCAAAAWWDH | Consensus sequence: HDWWTTTTGCAADBD |
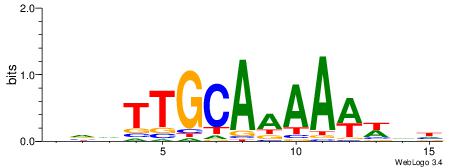 |
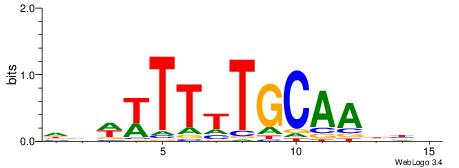 |
| Motif ID: | UP00097 |
| Motif name: | Mtf1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 8 |
| Similarity score: | 0.0184619 |
Alignment:
DBDHCGTGTGCAAMDH
-----TTGCGCAA---
| Original motif | Reverse complement motif |
| Consensus sequence: DBDHCGTGTGCAAMDH | Consensus sequence: HDYTTGCACACGDHBH |
 |
 |
| Motif ID: | UP00259 |
| Motif name: | Hoxb6 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0226004 |
Alignment:
WWTDBGTAATTACVDB
-TTGCGCAA-------
| Original motif | Reverse complement motif |
| Consensus sequence: WWTDBGTAATTACVDB | Consensus sequence: VDVGTAATTACBDAWW |
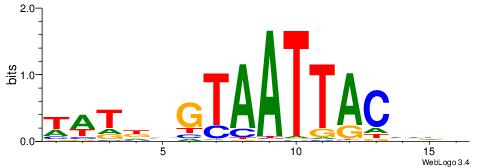 |
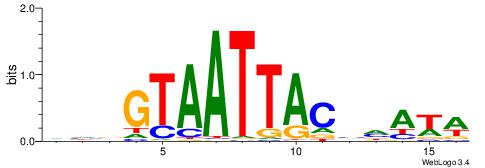 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 8 |
| Similarity score: | 0.0239956 |
Alignment:
DCATDBTGHGCATKMTTBRSWR
-----TTGCGCAA---------
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
 |
 |
| Motif ID: | UP00035 |
| Motif name: | Hic1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 8 |
| Similarity score: | 0.0261465 |
Alignment:
VHHDWTGGGCAMDHBH
----TTGCGCAA----
| Original motif | Reverse complement motif |
| Consensus sequence: DBDDRTGCCCAWDHDV | Consensus sequence: VHHDWTGGGCAMDHBH |
 |
 |
| Motif ID: | UP00004 |
| Motif name: | Sox14_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0272314 |
Alignment:
BKBASACAATRGHBB
-TTGCGCAA------
| Original motif | Reverse complement motif |
| Consensus sequence: BKBASACAATRGHBB | Consensus sequence: BBDCMATTGTSTBRB |
 |
 |
| Motif ID: | UP00055 |
| Motif name: | Hbp1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 9 |
| Number of overlap: | 8 |
| Similarity score: | 0.0278778 |
Alignment:
HBTHVACAATGGGMDHD
-TTGCGCAA--------
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCCATTGTVHABH | Consensus sequence: HBTHVACAATGGGMDHD |
 |
 |
| Motif ID: | UP00138 |
| Motif name: | Bsx |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.0281625 |
Alignment:
VVRDYAATTRBYBVAD
-------TTGCGCAA-
| Original motif | Reverse complement motif |
| Consensus sequence: VVRDYAATTRBYBVAD | Consensus sequence: HTVVKBKAATTMHMBV |
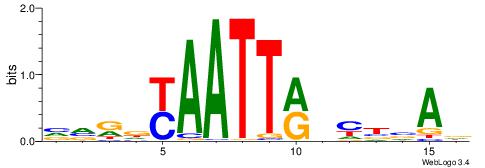 |
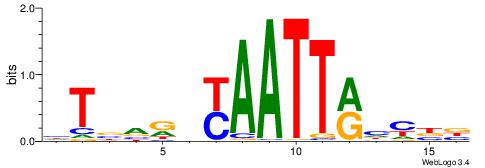 |
| Motif ID: | UP00126 |
| Motif name: | Dlx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 8 |
| Number of overlap: | 8 |
| Similarity score: | 0.030109 |
Alignment:
VVDRTAATTABHHHAB
-------TTGCGCAA-
| Original motif | Reverse complement motif |
| Consensus sequence: VVDRTAATTABHHHAB | Consensus sequence: BTDHHBTAATTAKDVV |
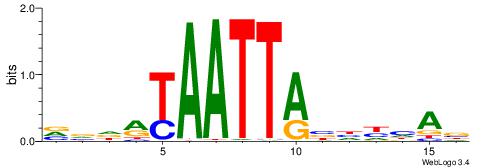 |
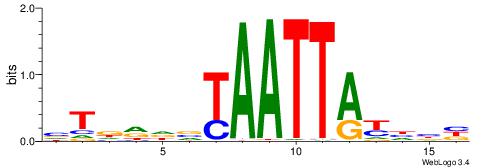 |
| Motif ID: | UP00182 |
| Motif name: | Hoxa6 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 8 |
| Similarity score: | 0.0308523 |
Alignment:
AMGKTAATTACCHHAD
-------TTGCGCAA-
| Original motif | Reverse complement motif |
| Consensus sequence: AMGKTAATTACCHHAD | Consensus sequence: DTHHGGTAATTAYCYT |
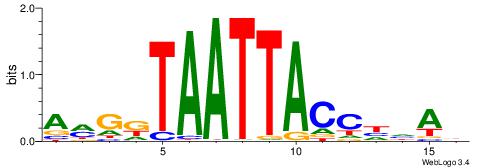 |
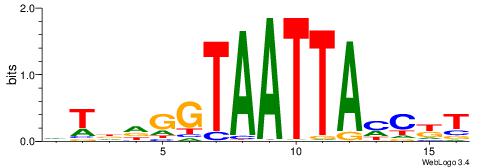 |
| Dataset #: 5 | Motif ID: 86 | Motif name: TFW2 |
| Original motif | Reverse complement motif |
| Consensus sequence: YGGACTTACG | Consensus sequence: CGTAAGTCCK |
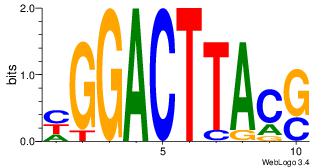 |
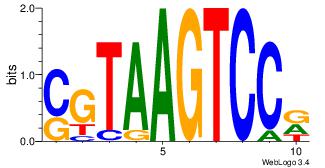 |
Best Matches for Motif ID 86 (Highest to Lowest)
| Motif ID: | UP00066 |
| Motif name: | Hnf4a_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0 |
Alignment:
HHAYTGGACTTTHDBV
----YGGACTTACG--
| Original motif | Reverse complement motif |
| Consensus sequence: BBHHAAAGTCCAMTHH | Consensus sequence: HHAYTGGACTTTHDBV |
 |
 |
| Motif ID: | UP00259 |
| Motif name: | Hoxb6 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0133265 |
Alignment:
VDVGTAATTACBDAWW
--YGGACTTACG----
| Original motif | Reverse complement motif |
| Consensus sequence: WWTDBGTAATTACVDB | Consensus sequence: VDVGTAATTACBDAWW |
 |
 |
| Motif ID: | UP00228 |
| Motif name: | Bapx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 10 |
| Similarity score: | 0.0145352 |
Alignment:
DTBVTTAAGTGGTTHHV
---CGTAAGTCCK----
| Original motif | Reverse complement motif |
| Consensus sequence: VHHAACCACTTAAVVAH | Consensus sequence: DTBVTTAAGTGGTTHHV |
 |
 |
| Motif ID: | UP00190 |
| Motif name: | Nkx2-3 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0167419 |
Alignment:
HHTTAAGTACTTAAMK
-CGTAAGTCCK-----
| Original motif | Reverse complement motif |
| Consensus sequence: YYTTAAGTACTTAAHD | Consensus sequence: HHTTAAGTACTTAAMK |
 |
 |
| Motif ID: | UP00389 |
| Motif name: | Nkx3-1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 10 |
| Similarity score: | 0.0201669 |
Alignment:
BDTTTAAGTACTTDDDD
--CGTAAGTCCK-----
| Original motif | Reverse complement motif |
| Consensus sequence: DDHDAAGTACTTAAADB | Consensus sequence: BDTTTAAGTACTTDDDD |
 |
 |
| Motif ID: | UP00418 |
| Motif name: | Etv6 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0206612 |
Alignment:
BHAHCCGGAAGTABDVD
-----YGGACTTACG--
| Original motif | Reverse complement motif |
| Consensus sequence: DVHBTACTTCCGGHTHB | Consensus sequence: BHAHCCGGAAGTABDVD |
 |
 |
| Motif ID: | UP00112 |
| Motif name: | Gsc |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0208429 |
Alignment:
BHDMSGGATTAMMDWBH
----YGGACTTACG---
| Original motif | Reverse complement motif |
| Consensus sequence: HVWHRYTAATCCSYDHV | Consensus sequence: BHDMSGGATTAMMDWBH |
 |
 |
| Motif ID: | UP00093 |
| Motif name: | Klf7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 10 |
| Similarity score: | 0.0217998 |
Alignment:
DHDTHGGGCGTATBHBB
----YGGACTTACG---
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBATACGCCCHAHHD | Consensus sequence: DHDTHGGGCGTATBHBB |
 |
 |
| Motif ID: | UP00404 |
| Motif name: | Elf2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0220676 |
Alignment:
BDWDCCGGAAGTHBBD
-----CGTAAGTCCK-
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGDWDB | Consensus sequence: BDWDCCGGAAGTHBBD |
 |
 |
| Motif ID: | UP00413 |
| Motif name: | Elf4 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 10 |
| Similarity score: | 0.0221416 |
Alignment:
HAWYCCGGAAGTHBBD
-----CGTAAGTCCK-
| Original motif | Reverse complement motif |
| Consensus sequence: DBBHACTTCCGGKWTH | Consensus sequence: HAWYCCGGAAGTHBBD |
 |
 |
| Dataset #: 5 | Motif ID: 87 | Motif name: TFF1 |
| Original motif | Reverse complement motif |
| Consensus sequence: AGGACAAAGAVV | Consensus sequence: VVTCTTTGTCCT |
 |
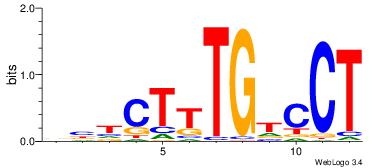 |
Best Matches for Motif ID 87 (Highest to Lowest)
| Motif ID: | UP00062 |
| Motif name: | Sox4_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0258536 |
Alignment:
HBDDDAACAAAGRVBHH
---AGGACAAAGAVV--
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDDAACAAAGRVBHH | Consensus sequence: HHBBMCTTTGTTHDDBH |
 |
 |
| Motif ID: | UP00072 |
| Motif name: | IRC900814_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0258585 |
Alignment:
DDHTACGACAAAWDDB
----AGGACAAAGAVV
| Original motif | Reverse complement motif |
| Consensus sequence: DDHTACGACAAAWDDB | Consensus sequence: BHDWTTTGTCGTAHDD |
 |
 |
| Motif ID: | UP00030 |
| Motif name: | Sox11_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0284165 |
Alignment:
HBDDRAACAAAGRABHH
---AGGACAAAGAVV--
| Original motif | Reverse complement motif |
| Consensus sequence: HBDDRAACAAAGRABHH | Consensus sequence: HHBTMCTTTGTTMDDVH |
 |
 |
| Motif ID: | UP00101 |
| Motif name: | Sox12_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 12 |
| Similarity score: | 0.0317868 |
Alignment:
VVDBASACAAAGRADH
---AGGACAAAGAVV-
| Original motif | Reverse complement motif |
| Consensus sequence: VVDBASACAAAGRADH | Consensus sequence: HDTMCTTTGTSTVDBB |
 |
 |
| Motif ID: | UP00067 |
| Motif name: | Lef1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0318041 |
Alignment:
VDDAGATCAAAGGKDDD
---AGGACAAAGAVV--
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATCTDDV | Consensus sequence: VDDAGATCAAAGGKDDD |
 |
 |
| Motif ID: | UP00058 |
| Motif name: | Tcf3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 12 |
| Similarity score: | 0.0329392 |
Alignment:
HHHASATCAAAGVRHHH
---AGGACAAAGAVV--
| Original motif | Reverse complement motif |
| Consensus sequence: HHHASATCAAAGVRHHH | Consensus sequence: HHHKVCTTTGATSTHHH |
 |
 |
| Motif ID: | UP00054 |
| Motif name: | Tcf7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0344802 |
Alignment:
BHHASATCAAAGGAHHH
---AGGACAAAGAVV--
| Original motif | Reverse complement motif |
| Consensus sequence: BHHASATCAAAGGAHHH | Consensus sequence: HHHTCCTTTGATSTHHV |
 |
 |
| Motif ID: | UP00026 |
| Motif name: | Zscan4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0344812 |
Alignment:
BHHHTTGTGTGCKHVD
-VVTCTTTGTCCT---
| Original motif | Reverse complement motif |
| Consensus sequence: HVHRGCACACAAHHHV | Consensus sequence: BHHHTTGTGTGCKHVD |
 |
 |
| Motif ID: | UP00083 |
| Motif name: | Tcf7l2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 12 |
| Similarity score: | 0.0344876 |
Alignment:
DDDYCCTTTGATSTDDV
--VVTCTTTGTCCT---
| Original motif | Reverse complement motif |
| Consensus sequence: DDDYCCTTTGATSTDDV | Consensus sequence: BDDASATCAAAGGMDDD |
 |
 |
| Motif ID: | UP00020 |
| Motif name: | Atf1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 12 |
| Similarity score: | 0.0352035 |
Alignment:
VDATGACGADYADH
--AGGACAAAGAVV
| Original motif | Reverse complement motif |
| Consensus sequence: VDATGACGADYADH | Consensus sequence: DDTMDTCGTCATDV |
 |
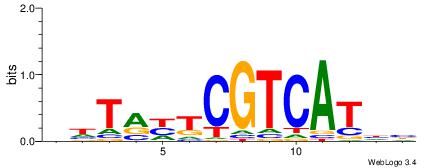 |
| Dataset #: 5 | Motif ID: 88 | Motif name: TFF11 |
| Original motif | Reverse complement motif |
| Consensus sequence: CTGCTGBDBBDGBD | Consensus sequence: HVCDBBDBCAGCAG |
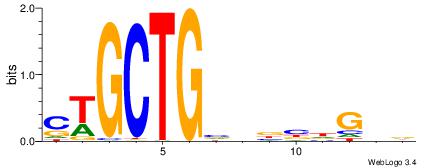 |
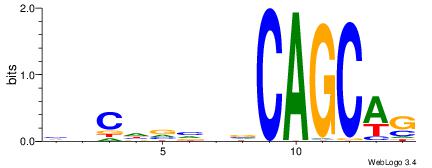 |
Best Matches for Motif ID 88 (Highest to Lowest)
| Motif ID: | UP00193 |
| Motif name: | Rhox11_1765.2 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.054015 |
Alignment:
WVVHWTWACAGCGBHBT
HVCDBBDBCAGCAG---
| Original motif | Reverse complement motif |
| Consensus sequence: AVDVCGCTGTWAWDVVW | Consensus sequence: WVVHWTWACAGCGBHBT |
 |
 |
| Motif ID: | UP00193 |
| Motif name: | Rhox11_2205.1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0543852 |
Alignment:
WVVHWTWACAGCGHHVT
HVCDBBDBCAGCAG---
| Original motif | Reverse complement motif |
| Consensus sequence: AVDHCGCTGTWAWDVVW | Consensus sequence: WVVHWTWACAGCGHHVT |
 |
 |
| Motif ID: | UP00027 |
| Motif name: | Osr1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0554469 |
Alignment:
HHDHACRGTAGCHVHD
--CTGCTGBDBBDGBD
| Original motif | Reverse complement motif |
| Consensus sequence: HHDHACRGTAGCHVHD | Consensus sequence: DHBHGCTACKGTHDHH |
 |
 |
| Motif ID: | UP00052 |
| Motif name: | Osr2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0576412 |
Alignment:
VDWRKAKGTAGCHHDT
HVCDBBDBCAGCAG--
| Original motif | Reverse complement motif |
| Consensus sequence: AHHHGCTACYTRMWHV | Consensus sequence: VDWRKAKGTAGCHHDT |
 |
 |
| Motif ID: | UP00099 |
| Motif name: | Ascl2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0581121 |
Alignment:
HHDVVGCAGCTGVBKVB
HVCDBBDBCAGCAG---
| Original motif | Reverse complement motif |
| Consensus sequence: BBYVVCAGCTGCBVHHD | Consensus sequence: HHDVVGCAGCTGVBKVB |
 |
 |
| Motif ID: | UP00027 |
| Motif name: | Osr1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0586391 |
Alignment:
VHDHKWKGTAGCHHDK
HVCDBBDBCAGCAG--
| Original motif | Reverse complement motif |
| Consensus sequence: RHHHGCTACYWRHDHV | Consensus sequence: VHDHKWKGTAGCHHDK |
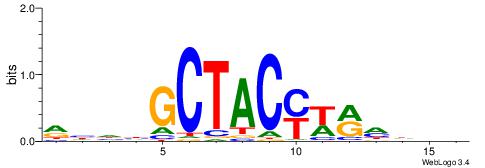 |
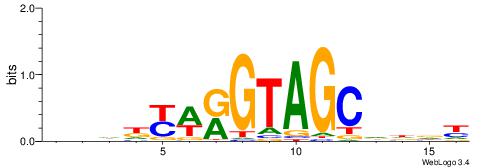 |
| Motif ID: | UP00122 |
| Motif name: | Tgif1 |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0592876 |
Alignment:
DBVVAGCTGTCAWWVTH
HVCDBBDBCAGCAG---
| Original motif | Reverse complement motif |
| Consensus sequence: DABWWTGACAGCTVVBD | Consensus sequence: DBVVAGCTGTCAWWVTH |
 |
 |
| Motif ID: | UP00052 |
| Motif name: | Osr2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 14 |
| Similarity score: | 0.0610328 |
Alignment:
HHHHGCTACKGTHVHH
--CTGCTGBDBBDGBD
| Original motif | Reverse complement motif |
| Consensus sequence: HHVHACRGTAGCHHHD | Consensus sequence: HHHHGCTACKGTHVHH |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 9 |
| Number of overlap: | 14 |
| Similarity score: | 0.0628447 |
Alignment:
HHHHDVVGTTGCTAHGGDHBAHH
--------CTGCTGBDBBDGBD-
| Original motif | Reverse complement motif |
| Consensus sequence: HDTBHHCCHTAGCAACVVDHHHH | Consensus sequence: HHHHDVVGTTGCTAHGGDHBAHH |
 |
 |
| Motif ID: | UP00047 |
| Motif name: | Zbtb7b_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 14 |
| Similarity score: | 0.0632938 |
Alignment:
BBDVHGGTGGTCBKDDB
---CTGCTGBDBBDGBD
| Original motif | Reverse complement motif |
| Consensus sequence: BDDRVGACCACCHBDVB | Consensus sequence: BBDVHGGTGGTCBKDDB |
 |
 |
| Dataset #: 5 | Motif ID: 89 | Motif name: TFM1 |
| Original motif | Reverse complement motif |
| Consensus sequence: CACACACACACWCACACA | Consensus sequence: TGTGTGWGTGTGTGTGTG |
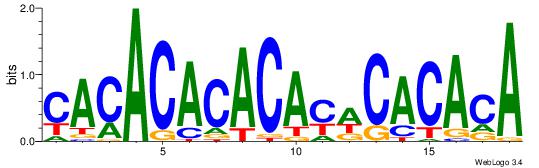 |
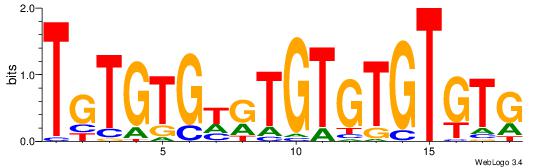 |
Best Matches for Motif ID 89 (Highest to Lowest)
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 18 |
| Similarity score: | 0.0442064 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
-----CACACACACACWCACACA
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0448118 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
TGTGTGWGTGTGTGTGTG-----
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0469936 |
Alignment:
VVYDDDCHCAVACAATTDBVAC
TGTGTGWGTGTGTGTGTG----
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0474757 |
Alignment:
MHHHWHYTMCCHCCCACVAABVH
--CACACACACACWCACACA---
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 18 |
| Similarity score: | 0.0483565 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
---CACACACACACWCACACA-
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 6 |
| Number of overlap: | 18 |
| Similarity score: | 0.0502008 |
Alignment:
BADHBDHCGCCCMCGCAHHDBBV
-----CACACACACACWCACACA
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0557575 |
Alignment:
BDVBCDACGCKCGCGTBHRKHD
---TGTGTGWGTGTGTGTGTG-
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
 |
 |
| Motif ID: | UP00096 |
| Motif name: | Sox13_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.549115 |
Alignment:
-DDDHTGGGTGGGHAHHD
CACACACACACWCACACA
| Original motif | Reverse complement motif |
| Consensus sequence: DDDHTGGGTGGGHAHHD | Consensus sequence: DHHTHCCCACCCAHDDH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 7 |
| Number of overlap: | 17 |
| Similarity score: | 0.550082 |
Alignment:
BBBDVVRGACCACCCAVGABBAB-
------CACACACACACWCACACA
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00228 |
| Motif name: | Bapx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 0.553591 |
Alignment:
-VHHAACCACTTAAVVAH
CACACACACACWCACACA-
| Original motif | Reverse complement motif |
| Consensus sequence: VHHAACCACTTAAVVAH | Consensus sequence: DTBVTTAAGTGGTTHHV |
 |
 |
| Dataset #: 5 | Motif ID: 90 | Motif name: TFM2 |
| Original motif | Reverse complement motif |
| Consensus sequence: CHHCHBTCCTSCYTCTGC | Consensus sequence: GCAGAKGSAGGABDGHDG |
 |
 |
Best Matches for Motif ID 90 (Highest to Lowest)
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0498793 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
CHHCHBTCCTSCYTCTGC----
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0564418 |
Alignment:
HADTBVADGATGCATCMTGMVHB
-----GCAGAKGSAGGABDGHDG
| Original motif | Reverse complement motif |
| Consensus sequence: HADTBVADGATGCATCMTGMVHB | Consensus sequence: VHVRCAYGATGCATCDTVVADTH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 5 |
| Number of overlap: | 18 |
| Similarity score: | 0.0565957 |
Alignment:
DVMHHDHKGACCCTCCTSVCBH
CHHCHBTCCTSCYTCTGC----
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 18 |
| Similarity score: | 0.0573354 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
CHHCHBTCCTSCYTCTGC-----
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 6 |
| Number of overlap: | 18 |
| Similarity score: | 0.0598613 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
CHHCHBTCCTSCYTCTGC-----
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0598775 |
Alignment:
MHHHWHYTMCCHCCCACVAABVH
--CHHCHBTCCTSCYTCTGC---
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0606451 |
Alignment:
GTVBDAATTGTVTGHGDDHKVB
--GCAGAKGSAGGABDGHDG--
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0626287 |
Alignment:
VMYDHDGMCCHCCKBGVVAAVH
-CHHCHBTCCTSCYTCTGC---
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.0626567 |
Alignment:
VHYTCCDWCCGCGTCMMTHWHV
-CHHCHBTCCTSCYTCTGC---
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 18 |
| Similarity score: | 0.064761 |
Alignment:
HBHDVHDTCTTATCTHTDDHDV
-CHHCHBTCCTSCYTCTGC---
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Dataset #: 5 | Motif ID: 91 | Motif name: TFM3 |
| Original motif | Reverse complement motif |
| Consensus sequence: KTTTTTTKTTTTTTDAAB | Consensus sequence: BTTHAAAAAAYAAAAAAR |
 |
 |
Best Matches for Motif ID 91 (Highest to Lowest)
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0426153 |
Alignment:
BDHDDADAGATAAGADHBDHVD
BTTHAAAAAAYAAAAAAR----
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 18 |
| Similarity score: | 0.0478771 |
Alignment:
HHBDHDDAACAATDRHHHHHBW
--BTTHAAAAAAYAAAAAAR--
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 18 |
| Similarity score: | 0.0564691 |
Alignment:
VVYDDDCHCAVACAATTDBVAC
BTTHAAAAAAYAAAAAAR----
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00077 |
| Motif name: | Srf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.52452 |
Alignment:
-VBDDAAAAAAAAMVBHH
KTTTTTTKTTTTTTDAAB
| Original motif | Reverse complement motif |
| Consensus sequence: VBDDAAAAAAAAMVBHH | Consensus sequence: HHVBYTTTTTTTTDDVV |
 |
 |
| Motif ID: | UP00090 |
| Motif name: | Elf3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.531868 |
Alignment:
-DDHBTTTTTTTTHDAVH
KTTTTTTKTTTTTTDAAB
| Original motif | Reverse complement motif |
| Consensus sequence: DBTHHAAAAAAAAVHDH | Consensus sequence: DDHBTTTTTTTTHDAVH |
 |
 |
| Motif ID: | UP00121 |
| Motif name: | Hoxd10 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 0.539211 |
Alignment:
-HDDDATTTTATTKVRDB
KTTTTTTKTTTTTTDAAB
| Original motif | Reverse complement motif |
| Consensus sequence: VDKVYAATAAAATDDDH | Consensus sequence: HDDDATTTTATTKVRDB |
 |
 |
| Motif ID: | UP00073 |
| Motif name: | Foxa2_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 0.539687 |
Alignment:
HHWAWGTAAAYAAAVDB-
-KTTTTTTKTTTTTTDAAB
| Original motif | Reverse complement motif |
| Consensus sequence: HHWAWGTAAAYAAAVDB | Consensus sequence: BDVTTTKTTTACWTWHH |
 |
 |
| Motif ID: | UP00039 |
| Motif name: | Foxj3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 0.542507 |
Alignment:
-DVVTTTGTTTACDTHDH
BTTHAAAAAAYAAAAAAR-
| Original motif | Reverse complement motif |
| Consensus sequence: HDHADGTAAACAAAVVH | Consensus sequence: DVVTTTGTTTACDTHDH |
 |
 |
| Motif ID: | UP00061 |
| Motif name: | Foxl1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 0.547029 |
Alignment:
-VVVWTTGTTTAMATTWD
BTTHAAAAAAYAAAAAAR-
| Original motif | Reverse complement motif |
| Consensus sequence: DWAATRTAAACAAWVVB | Consensus sequence: VVVWTTGTTTAMATTWD |
 |
 |
| Motif ID: | UP00051 |
| Motif name: | Sox8_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 0.547298 |
Alignment:
-DDDHDAACAATWBATDD
BTTHAAAAAAYAAAAAAR-
| Original motif | Reverse complement motif |
| Consensus sequence: DDATBWATTGTTHHDDD | Consensus sequence: DDDHDAACAATWBATDD |
 |
 |
| Dataset #: 5 | Motif ID: 92 | Motif name: TFM11 |
| Original motif | Reverse complement motif |
| Consensus sequence: CAMACACACACAYDCACACA | Consensus sequence: TGTGTGDKTGTGTGTGTRTG |
 |
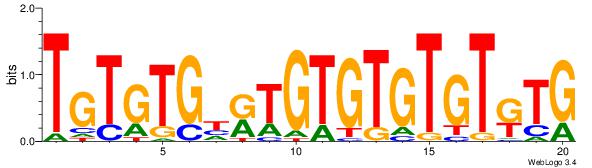 |
Best Matches for Motif ID 92 (Highest to Lowest)
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 20 |
| Similarity score: | 0.0426758 |
Alignment:
HVBDAGCGBGGGTGGCRDBDDDB
TGTGTGDKTGTGTGTGTRTG---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 20 |
| Similarity score: | 0.0439623 |
Alignment:
GTVBDAATTGTVTGHGDDHKVB
TGTGTGDKTGTGTGTGTRTG--
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 20 |
| Similarity score: | 0.0449001 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
---CAMACACACACAYDCACACA
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 20 |
| Similarity score: | 0.0492241 |
Alignment:
MHHHWHYTMCCHCCCACVAABVH
CAMACACACACAYDCACACA---
| Original motif | Reverse complement motif |
| Consensus sequence: MHHHWHYTMCCHCCCACVAABVH | Consensus sequence: DVVTTVGTGGGHGGYAMHWHHHY |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 20 |
| Similarity score: | 0.0524009 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
-CAMACACACACAYDCACACA-
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 20 |
| Similarity score: | 0.0527926 |
Alignment:
BADHBDHCGCCCMCGCAHHDBBV
---CAMACACACACAYDCACACA
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00050 |
| Motif name: | Bhlhb2_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 20 |
| Similarity score: | 0.0547661 |
Alignment:
ACHRBHDTCCACGTGYAHHBMHM
-CAMACACACACAYDCACACA--
| Original motif | Reverse complement motif |
| Consensus sequence: YDYBDHTMCACGTGGADDBMDGT | Consensus sequence: ACHRBHDTCCACGTGYAHHBMHM |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 20 |
| Similarity score: | 0.0575159 |
Alignment:
DDRMHBACGCGYGCGTDGBBDB
-CAMACACACACAYDCACACA-
| Original motif | Reverse complement motif |
| Consensus sequence: DDRMHBACGCGYGCGTDGBBDB | Consensus sequence: BDVBCDACGCKCGCGTBHRKHD |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v015681_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 5 |
| Number of overlap: | 19 |
| Similarity score: | 0.551038 |
Alignment:
BBBDVVRGACCACCCAVGABBAB-
----CAMACACACACAYDCACACA
| Original motif | Reverse complement motif |
| Consensus sequence: BBBDVVRGACCACCCAVGABBAB | Consensus sequence: BTBVTCVTGGGTGGTCMVVDVBB |
 |
 |
| Motif ID: | UP00527 |
| Motif name: | Foxn4_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 19 |
| Similarity score: | 0.551577 |
Alignment:
VDWDARRGACGCGGWHGGAKHV-
---CAMACACACACAYDCACACA
| Original motif | Reverse complement motif |
| Consensus sequence: VDWDARRGACGCGGWHGGAKHV | Consensus sequence: VHYTCCDWCCGCGTCMMTHWHV |
 |
 |
| Dataset #: 5 | Motif ID: 93 | Motif name: TFM12 |
| Original motif | Reverse complement motif |
| Consensus sequence: RAGKGMAGVRRGSRCASAGV | Consensus sequence: VCTSTGKSCMKBCTRCYCTK |
 |
 |
Best Matches for Motif ID 93 (Highest to Lowest)
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 20 |
| Similarity score: | 0.0465838 |
Alignment:
MWSMBAARRATGCDCABHATGD
RAGKGMAGVRRGSRCASAGV--
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 3 |
| Number of overlap: | 20 |
| Similarity score: | 0.0472219 |
Alignment:
DVMHHDHKGACCCTCCTSVCBH
--VCTSTGKSCMKBCTRCYCTK
| Original motif | Reverse complement motif |
| Consensus sequence: HVGVSAGGAGGGTCYHHHHYVH | Consensus sequence: DVMHHDHKGACCCTCCTSVCBH |
 |
 |
| Motif ID: | UP00539 |
| Motif name: | Gli2_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 20 |
| Similarity score: | 0.0477526 |
Alignment:
RTHBSYCGCCMCMYVCGBTVDH
VCTSTGKSCMKBCTRCYCTK--
| Original motif | Reverse complement motif |
| Consensus sequence: RTHBSYCGCCMCMYVCGBTVDH | Consensus sequence: HDBABCGBKRGYGGCGMSBHAK |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 20 |
| Similarity score: | 0.0479772 |
Alignment:
VVYDDDCHCAVACAATTDBVAC
VCTSTGKSCMKBCTRCYCTK--
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00538 |
| Motif name: | Gli1_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 20 |
| Similarity score: | 0.0487318 |
Alignment:
BDDHVDKGCCACCCVCGCTDBVH
VCTSTGKSCMKBCTRCYCTK---
| Original motif | Reverse complement motif |
| Consensus sequence: HVBDAGCGBGGGTGGCRDBDDDB | Consensus sequence: BDDHVDKGCCACCCVCGCTDBVH |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v016060_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 20 |
| Similarity score: | 0.0495675 |
Alignment:
VYWVVHCRCCACMCACGAHSBHH
VCTSTGKSCMKBCTRCYCTK---
| Original motif | Reverse complement motif |
| Consensus sequence: VYWVVHCRCCACMCACGAHSBHH | Consensus sequence: HHBSHTCGTGRGTGGKGDBVWMV |
 |
 |
| Motif ID: | UP00540 |
| Motif name: | Gli3_v015681_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 3 |
| Number of overlap: | 20 |
| Similarity score: | 0.0506701 |
Alignment:
VMYDHDGMCCHCCKBGVVAAVH
VCTSTGKSCMKBCTRCYCTK--
| Original motif | Reverse complement motif |
| Consensus sequence: DVTTVVCVYGGHGGYCHHDMYB | Consensus sequence: VMYDHDGMCCHCCKBGVVAAVH |
 |
 |
| Motif ID: | UP00400 |
| Motif name: | Zif268 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 20 |
| Similarity score: | 0.0532516 |
Alignment:
VVVDHHTGCGYGGGCGDHVHHTB
RAGKGMAGVRRGSRCASAGV---
| Original motif | Reverse complement motif |
| Consensus sequence: BADHBDHCGCCCMCGCAHHDBBV | Consensus sequence: VVVDHHTGCGYGGGCGDHVHHTB |
 |
 |
| Motif ID: | UP00526 |
| Motif name: | Foxn1_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 20 |
| Similarity score: | 0.0536082 |
Alignment:
HDWHMHGACGCGCGTRBVVBHW
-VCTSTGKSCMKBCTRCYCTK-
| Original motif | Reverse complement motif |
| Consensus sequence: WHVBVVMACGCGCGTCDYHWDH | Consensus sequence: HDWHMHGACGCGCGTRBVVBHW |
 |
 |
| Motif ID: | UP00098 |
| Motif name: | Rfx3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 20 |
| Similarity score: | 0.0554568 |
Alignment:
BDBBVBVGTAWCCAAGVBHBAGB
---RAGKGMAGVRRGSRCASAGV
| Original motif | Reverse complement motif |
| Consensus sequence: VCTBHBVCTTGGWTACVVVVVDB | Consensus sequence: BDBBVBVGTAWCCAAGVBHBAGB |
 |
 |
| Dataset #: 5 | Motif ID: 94 | Motif name: TFM13 |
| Original motif | Reverse complement motif |
| Consensus sequence: TTTTTTTKTTKTTTAATTHW | Consensus sequence: WHAATTAAARAARAAAAAAA |
 |
 |
Best Matches for Motif ID 94 (Highest to Lowest)
| Motif ID: | UP00032 |
| Motif name: | Gata3_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 2 |
| Number of overlap: | 20 |
| Similarity score: | 0.0558709 |
Alignment:
BDHDDADAGATAAGADHBDHVD
-WHAATTAAARAARAAAAAAA-
| Original motif | Reverse complement motif |
| Consensus sequence: BDHDDADAGATAAGADHBDHVD | Consensus sequence: HBHDVHDTCTTATCTHTDDHDV |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_primary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 20 |
| Similarity score: | 0.0604961 |
Alignment:
WBHHHHHMDATTGTTHDHDVHH
WHAATTAAARAARAAAAAAA--
| Original motif | Reverse complement motif |
| Consensus sequence: HHBDHDDAACAATDRHHHHHBW | Consensus sequence: WBHHHHHMDATTGTTHDHDVHH |
 |
 |
| Motif ID: | UP00032 |
| Motif name: | Gata3_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 20 |
| Similarity score: | 0.0784238 |
Alignment:
HBHDVBWGATHBTATCRVBHHH
-TTTTTTTKTTKTTTAATTHW-
| Original motif | Reverse complement motif |
| Consensus sequence: HBHDVBWGATHBTATCRVBHHH | Consensus sequence: HHHBBMGATAVHATCWVVDHVH |
 |
 |
| Motif ID: | UP00034 |
| Motif name: | Sox7_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 20 |
| Similarity score: | 0.0790353 |
Alignment:
GTVBDAATTGTVTGHGDDHKVB
--TTTTTTTKTTKTTTAATTHW
| Original motif | Reverse complement motif |
| Consensus sequence: GTVBDAATTGTVTGHGDDHKVB | Consensus sequence: VVYDDDCHCAVACAATTDBVAC |
 |
 |
| Motif ID: | UP00528 |
| Motif name: | Foxm1_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Backward |
| Position number: | 4 |
| Number of overlap: | 19 |
| Similarity score: | 0.574613 |
Alignment:
-MWSMBAARRATGCDCABHATGD
WHAATTAAARAARAAAAAAA---
| Original motif | Reverse complement motif |
| Consensus sequence: MWSMBAARRATGCDCABHATGD | Consensus sequence: DCATDBTGHGCATKMTTBRSWR |
 |
 |
| Motif ID: | UP00077 |
| Motif name: | Srf_secondary |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Original Motif |
| Direction: | Forward |
| Position number: | 4 |
| Number of overlap: | 17 |
| Similarity score: | 1.5444 |
Alignment:
HHVBYTTTTTTTTDDVV---
---WHAATTAAARAARAAAAAAA
| Original motif | Reverse complement motif |
| Consensus sequence: VBDDAAAAAAAAMVBHH | Consensus sequence: HHVBYTTTTTTTTDDVV |
 |
 |
| Motif ID: | UP00262 |
| Motif name: | Lhx1 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 1.54972 |
Alignment:
---VVDATTAATTAAWWAHB
TTTTTTTKTTKTTTAATTHW
| Original motif | Reverse complement motif |
| Consensus sequence: VVDATTAATTAAWWAHB | Consensus sequence: BHTWWTTAATTAATDVV |
 |
 |
| Motif ID: | UP00188 |
| Motif name: | Lmx1a |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 1.55162 |
Alignment:
---BDWWWTTAATTAAHHVG
TTTTTTTKTTKTTTAATTHW
| Original motif | Reverse complement motif |
| Consensus sequence: CVHHTTAATTAAWWWHB | Consensus sequence: BDWWWTTAATTAAHHVG |
 |
 |
| Motif ID: | UP00244 |
| Motif name: | Tlx2 |
| Species: | Mus musculus |
| Matching format of first motif: | Original Motif |
| Matching format of second motif: | Reverse Complement |
| Direction: | Backward |
| Position number: | 2 |
| Number of overlap: | 17 |
| Similarity score: | 1.55254 |
Alignment:
---BAATHAATTAATAAHWW
TTTTTTTKTTKTTTAATTHW-
| Original motif | Reverse complement motif |
| Consensus sequence: BAATHAATTAATAAHWW | Consensus sequence: WWDTTATTAATTHATTV |
 |
 |
| Motif ID: | UP00169 |
| Motif name: | Lmx1b |
| Species: | Mus musculus |
| Matching format of first motif: | Reverse Complement |
| Matching format of second motif: | Reverse Complement |
| Direction: | Forward |
| Position number: | 1 |
| Number of overlap: | 17 |
| Similarity score: | 1.55258 |
Alignment:
BHDATTAATTAAWWWHB---
WHAATTAAARAARAAAAAAA
| Original motif | Reverse complement motif |
| Consensus sequence: VDWWWTTAATTAATDHB | Consensus sequence: BHDATTAATTAAWWWHB |
 |
 |