Top 10 Significant Motifs - Global Matching (Highest to Lowest)
| Dataset #: 1 |
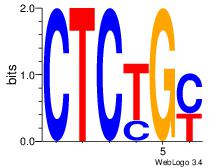
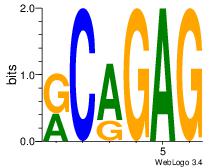
Motif ID: 4 |
Motif name: Motif 4 |
| Original motif Consensus sequence: CTCTGY |
Reverse complement motif Consensus sequence: KCAGAG |
|
|
 |
 |
Best Matches for Top Significant Motif ID 4 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
43 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
6 |
| Similarity score: |
0.012269 |
Alignment:
AAAGGTCAAAGGTCAAC
-----KCAGAG------
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
6 |
| Similarity score: |
0.0140102 |
Alignment:
BTRGGDCARAGGKCA
-----KCAGAG----
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
22 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0186793 |
Alignment:
AKGYYCAAAGRTCA
----KCAGAG----
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
34 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.020602 |
Alignment:
RGGBCAAAGKYCA
---KCAGAG----
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
37 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0221665 |
Alignment:
BAACGTCCGC
----CTCTGY
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0255256 |
Alignment:
TGYCAGGGGGCR
--KCAGAG----
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0349642 |
Alignment:
CCCCKCCCCC
CTCTGY----
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0376108 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
--CTCTGY-------------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
6 |
| Similarity score: |
0.0448449 |
Alignment:
BBVGCCBVGGCCTV
---KCAGAG-----
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0454269 |
Alignment:
MGGTCAGGGTGACCTRDBHV
---KCAGAG-----------
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: 1 |
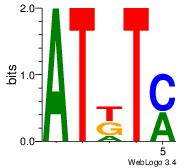
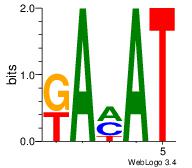
Motif ID: 5 |
Motif name: Motif 5 |
| Original motif Consensus sequence: ATKTM |
Reverse complement motif Consensus sequence: RARAT |
|
|
 |
 |
Best Matches for Top Significant Motif ID 5 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
10 |
| Number of overlap: |
5 |
| Similarity score: |
0.0256083 |
Alignment:
BGRRRGRGGRTGRTTYGGGG
------RARAT---------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
44 |
| Motif name: |
NR4A2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
5 |
| Similarity score: |
0.0276382 |
Alignment:
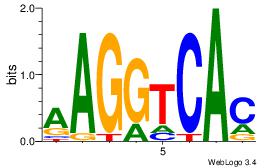
AAGGTCAC
RARAT---
| Original motif Consensus sequence: AAGGTCAC |
Reverse complement motif Consensus sequence: GTGACCTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
22 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
5 |
| Similarity score: |
0.0279129 |
Alignment:
AKGYYCAAAGRTCA
-------RARAT--
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
37 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
5 |
| Similarity score: |
0.0292478 |
Alignment:
BAACGTCCGC
--ATKTM---
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
43 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
10 |
| Number of overlap: |
5 |
| Similarity score: |
0.0292478 |
Alignment:
AAAGGTCAAAGGTCAAC
---------ATKTM---
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
5 |
| Similarity score: |
0.0305369 |
Alignment:
TGRCCTKTGHCCKAB
----ATKTM------
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
5 |
| Similarity score: |
0.0315454 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
----ATKTM------------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
RXRRAR_DR5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
5 |
| Similarity score: |
0.0335956 |
Alignment:
RGKTCABVVRGAGGTCA
ATKTM------------
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
46 |
| Motif name: |
RORA_1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
5 |
| Similarity score: |
0.0336052 |
Alignment:
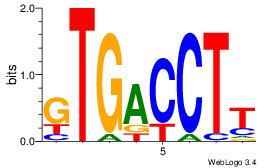
AWVDAGGTCA
---RARAT--
| Original motif Consensus sequence: AWVDAGGTCA |
Reverse complement motif Consensus sequence: TGACCTDVWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
34 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
5 |
| Similarity score: |
0.0367105 |
Alignment:
RGGBCAAAGKYCA
-------ATKTM-
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
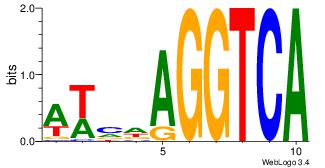
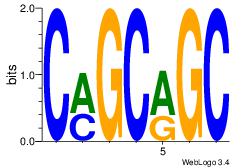
| Dataset #: 1 |
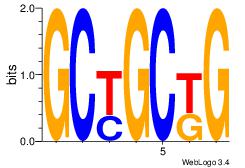
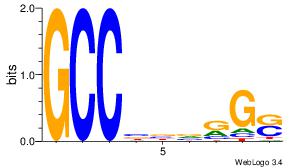
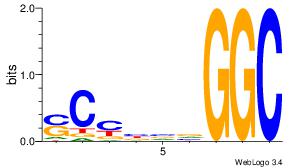
Motif ID: 6 |
Motif name: Motif 6 |
| Original motif Consensus sequence: CMGCRGC |
Reverse complement motif Consensus sequence: GCKGCYG |
|
|
 |
 |
Best Matches for Top Significant Motif ID 6 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
13 |
| Number of overlap: |
7 |
| Similarity score: |
0.0205607 |
Alignment:
TTCAGCACCATGGACAGCKCC
--CMGCRGC------------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
37 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0322623 |
Alignment:
GCGGACGTTV
GCKGCYG---
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
7 |
| Similarity score: |
0.038091 |
Alignment:
BBVGCCBVGGCCTV
----CMGCRGC---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
TFAP2A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0393168 |
Alignment:
SCMVBBGGC
--CMGCRGC
| Original motif Consensus sequence: GCCBBVRGS |
Reverse complement motif Consensus sequence: SCMVBBGGC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
7 |
| Similarity score: |
0.0448462 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-------GCKGCYG------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
28 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0453576 |
Alignment:
HGCGTGGGCGK
---GCKGCYG-
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
36 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0476958 |
Alignment:
GCCYCMCCCD
-CMGCRGC--
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0490991 |
Alignment:
GGGGGYGGGG
GCKGCYG---
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
7 |
| Similarity score: |
0.0498726 |
Alignment:
BGRRRGRGGRTGRTTYGGGG
-----GCKGCYG--------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.0511926 |
Alignment:
MGCCCCCTGMCA
---CMGCRGC--
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: 1 |
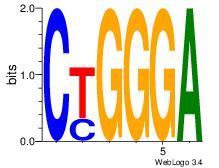
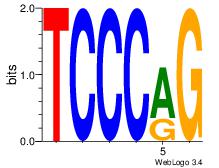
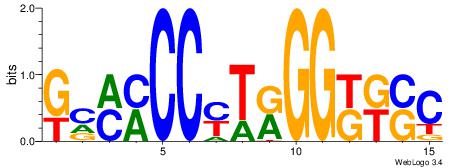
Motif ID: 8 |
Motif name: Motif 8 |
| Original motif Consensus sequence: CTGGGA |
Reverse complement motif Consensus sequence: TCCCAG |
|
|
 |
 |
Best Matches for Top Significant Motif ID 8 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
6 |
| Similarity score: |
0.0392549 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
-------TCCCAG--------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
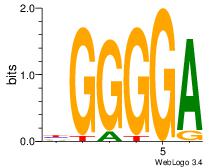
2 |
| Motif ID: |
23 |
| Motif name: |
MZF1_1-4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0416319 |
Alignment:
TCCCCV
TCCCAG
| Original motif Consensus sequence: BGGGGA |
Reverse complement motif Consensus sequence: TCCCCV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0509122 |
Alignment:
BGRRRGRGGRTGRTTYGGGG
--------------CTGGGA
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0557857 |
Alignment:
TGYCAGGGGGCR
---CTGGGA---
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
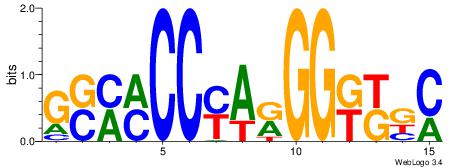
| Dataset #: |
2 |
| Motif ID: |
13 |
| Motif name: |
Zfp423 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
6 |
| Similarity score: |
0.0624145 |
Alignment:
GYRYCCMTKGGYRSC
---TCCCAG------
| Original motif Consensus sequence: GSMMCCYARGGKKKC |
Reverse complement motif Consensus sequence: GYRYCCMTKGGYRSC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
43 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
6 |
| Similarity score: |
0.0650784 |
Alignment:
AAAGGTCAAAGGTCAAC
-----TCCCAG------
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
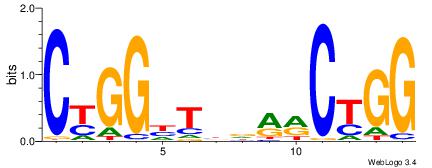
| Dataset #: |
2 |
| Motif ID: |
48 |
| Motif name: |
Tcfcp2l1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0663804 |
Alignment:
CKGGDTBDMMCTGG
CTGGGA--------
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
28 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0678562 |
Alignment:
YCGCCCACGCH
--TCCCAG---
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0714107 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
--------------TCCCAG--
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0718041 |
Alignment:
BBVGCCBVGGCCTV
-----CTGGGA---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
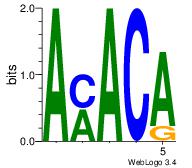
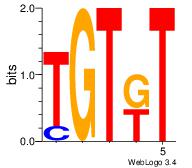
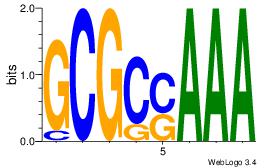
| Dataset #: 1 |
Motif ID: 2 |
Motif name: Motif 2 |
| Original motif Consensus sequence: AMACA |
Reverse complement motif Consensus sequence: TGTRT |
|
|
 |
 |
Best Matches for Top Significant Motif ID 2 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
11 |
| Number of overlap: |
5 |
| Similarity score: |
0.0243918 |
Alignment:
CCCCMAAMCAMCCMCMMMCV
-----AMACA----------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
5 |
| Similarity score: |
0.0429089 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
----AMACA-------------
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
5 |
| Similarity score: |
0.0595073 |
Alignment:
TGRCCTKTGHCCKAB
-----TGTRT-----
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
26 |
| Motif name: |
ArntAhr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
5 |
| Similarity score: |
0.0600161 |
Alignment:
YGCGTG
TGTRT-
| Original motif Consensus sequence: YGCGTG |
Reverse complement motif Consensus sequence: CACGCM |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
11 |
| Number of overlap: |
5 |
| Similarity score: |
0.0620995 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-----AMACA----------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
22 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
5 |
| Similarity score: |
0.0650242 |
Alignment:
AKGYYCAAAGRTCA
------AMACA---
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
5 |
| Similarity score: |
0.0670984 |
Alignment:
MGCCCCCTGMCA
-------AMACA
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
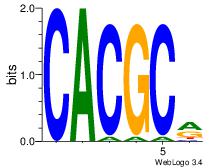
| Dataset #: |
2 |
| Motif ID: |
33 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
5 |
| Similarity score: |
0.0700905 |
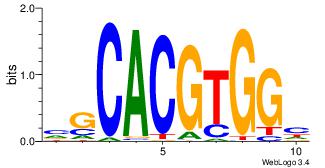
Alignment:
VCACGTBV
AMACA---
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
38 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
5 |
| Similarity score: |
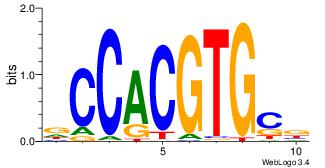
0.0708148 |
Alignment:
VGCACGTGGH
-AMACA----
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
34 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
5 |
| Similarity score: |
0.071574 |
Alignment:
TGMYCTTTGBCCK
-----TGTRT---
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
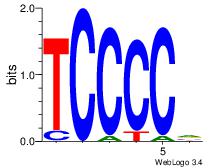
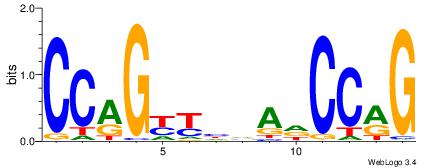
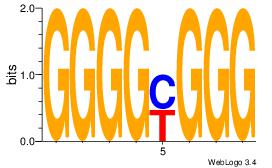
| Dataset #: 1 |
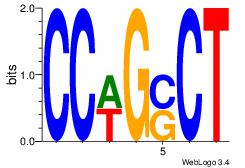
Motif ID: 3 |
Motif name: Motif 3 |
| Original motif Consensus sequence: AGSCWGG |
Reverse complement motif Consensus sequence: CCWGSCT |
|
|
 |
 |
Best Matches for Top Significant Motif ID 3 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
32 |
| Motif name: |
EWSR1-FLI1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
7 |
| Similarity score: |
0.0395671 |
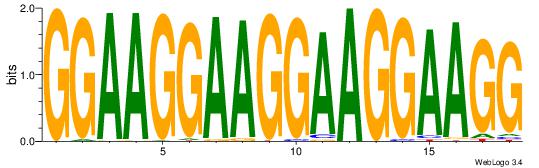
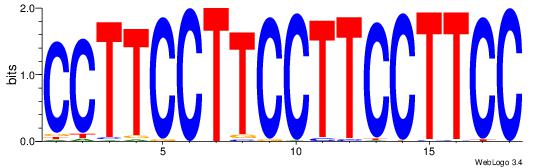
Alignment:
GGAAGGAAGGAAGGAAGG
-------AGSCWGG----
| Original motif Consensus sequence: GGAAGGAAGGAAGGAAGG |
Reverse complement motif Consensus sequence: CCTTCCTTCCTTCCTTCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0494688 |
Alignment:
MGCCCCCTGMCA
-----CCWGSCT
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
48 |
| Motif name: |
Tcfcp2l1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0669354 |
Alignment:
CCAGYYHVADCCRG
CCWGSCT-------
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
7 |
| Similarity score: |
0.0693747 |
Alignment:
TTCAGCACCATGGACAGCKCC
------AGSCWGG--------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0.0695377 |
Alignment:
CCCCKCCCCC
-CCWGSCT--
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
7 |
| Similarity score: |
0.0720685 |
Alignment:
BBVGCCBVGGCCTV
------CCWGSCT-
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
13 |
| Number of overlap: |
7 |
| Similarity score: |
0.0771405 |
Alignment:
MGGTCAGGGTGACCTRDBHV
------------CCWGSCT-
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
46 |
| Motif name: |
RORA_1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.0772276 |
Alignment:
TGACCTDVWT
---CCWGSCT
| Original motif Consensus sequence: AWVDAGGTCA |
Reverse complement motif Consensus sequence: TGACCTDVWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
36 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0798994 |
Alignment:
GCCYCMCCCD
---CCWGSCT
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
39 |
| Motif name: |
MYCMAX |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
7 |
| Similarity score: |
0.0839075 |
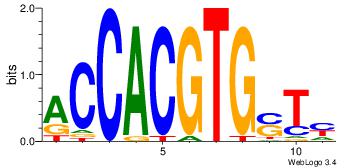
Alignment:
RASCACGTGGT
AGSCWGG----
| Original motif Consensus sequence: RASCACGTGGT |
Reverse complement motif Consensus sequence: ACCACGTGSTM |
|
|
 |
 |
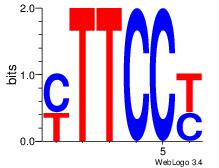
| Dataset #: 1 |
Motif ID: 1 |
Motif name: Motif 1 |
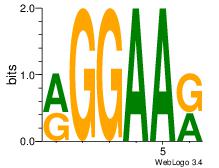
| Original motif Consensus sequence: RGGAAR |
Reverse complement motif Consensus sequence: MTTCCK |
|
|
 |
 |
Best Matches for Top Significant Motif ID 1 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
32 |
| Motif name: |
EWSR1-FLI1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
12 |
| Number of overlap: |
6 |
| Similarity score: |
0.0146511 |
Alignment:
GGAAGGAAGGAAGGAAGG
-----------RGGAAR-
| Original motif Consensus sequence: GGAAGGAAGGAAGGAAGG |
Reverse complement motif Consensus sequence: CCTTCCTTCCTTCCTTCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
37 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0608243 |
Alignment:
BAACGTCCGC
---MTTCCK-
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
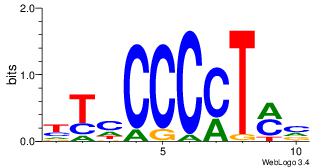
| Dataset #: |
2 |
| Motif ID: |
41 |
| Motif name: |
MZF1_5-13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0753789 |
Alignment:
BKAGGGGDAD
----RGGAAR
| Original motif Consensus sequence: BKAGGGGDAD |
Reverse complement motif Consensus sequence: BTHCCCCTYB |
|
|
 |
 |
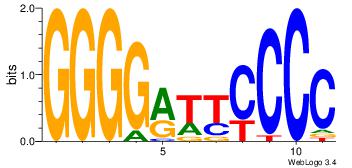
| Dataset #: |
2 |
| Motif ID: |
42 |
| Motif name: |
NFKB1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0757695 |
Alignment:
GGGGAAKCCCC
-RGGAAR----
| Original motif Consensus sequence: GGGGRTTCCCC |
Reverse complement motif Consensus sequence: GGGGAAKCCCC |
|
|
 |
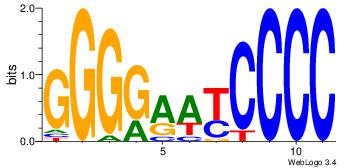 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
6 |
| Similarity score: |
0.0800484 |
Alignment:
BTRGGDCARAGGKCA
---RGGAAR------
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
6 |
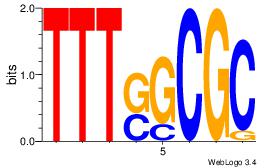
| Similarity score: |
0.0829223 |
Alignment:
TTCAGCACCATGGACAGCKCC
----------RGGAAR-----
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0832926 |
Alignment:
TGYCAGGGGGCR
------RGGAAR
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0835516 |
Alignment:
BGRRRGRGGRTGRTTYGGGG
----RGGAAR----------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0836451 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
---RGGAAR-------------
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
27 |
| Motif name: |
E2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0842331 |
Alignment:
GCGCSAAA
--RGGAAR
| Original motif Consensus sequence: TTTSGCGC |
Reverse complement motif Consensus sequence: GCGCSAAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
13 |
| Motif name: |
Zfp423 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0850287 |
Alignment:
GYRYCCMTKGGYRSC
--------RGGAAR-
| Original motif Consensus sequence: GSMMCCYARGGKKKC |
Reverse complement motif Consensus sequence: GYRYCCMTKGGYRSC |
|
|
 |
 |
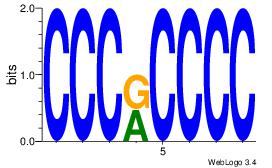
| Dataset #: 1 |
Motif ID: 7 |
Motif name: Motif 7 |
| Original motif Consensus sequence: CCCRCCCC |
Reverse complement motif Consensus sequence: GGGGMGGG |
|
|
 |
 |
Best Matches for Top Significant Motif ID 7 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0423611 |
Alignment:
CCCCKCCCCC
-CCCRCCCC-
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
36 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0461644 |
Alignment:
DGGGYGKGGC
GGGGMGGG--
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
28 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0650374 |
Alignment:
YCGCCCACGCH
---CCCRCCCC
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.076401 |
Alignment:
CCCCMAAMCAMCCMCMMMCV
------CCCRCCCC------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
42 |
| Motif name: |
NFKB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0893429 |
Alignment:
GGGGAAKCCCC
GGGGMGGG---
| Original motif Consensus sequence: GGGGRTTCCCC |
Reverse complement motif Consensus sequence: GGGGAAKCCCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0936832 |
Alignment:
TGYCAGGGGGCR
GGGGMGGG----
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
32 |
| Motif name: |
EWSR1-FLI1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.0950969 |
Alignment:
GGAAGGAAGGAAGGAAGG
------GGGGMGGG----
| Original motif Consensus sequence: GGAAGGAAGGAAGGAAGG |
Reverse complement motif Consensus sequence: CCTTCCTTCCTTCCTTCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
21 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0998932 |
Alignment:
GGGGCCCAAGGGGG
GGGGMGGG------
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.10221 |
Alignment:
BBVGCCBVGGCCTV
----CCCRCCCC--
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.102497 |
Alignment:
DGVBCABTGDWGCGKRRCSR
--------GGGGMGGG----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
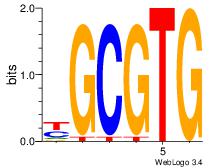
 |
| Dataset #: 2 |
Motif ID: 26 |
Motif name: ArntAhr |
| Original motif Consensus sequence: YGCGTG |
Reverse complement motif Consensus sequence: CACGCM |
|
|
 |
 |
Best Matches for Top Significant Motif ID 26 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
6 |
| Similarity score: |
0.0308396 |
Alignment:
TGTGTGTGTGTKTG
------YGCGTG--
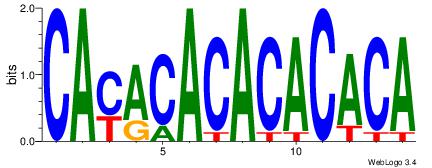
| Original motif Consensus sequence: CAYACACACACACA |
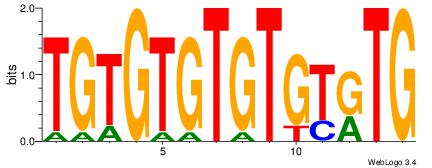
Reverse complement motif Consensus sequence: TGTGTGTGTGTKTG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0350575 |
Alignment:
CACACAVRCACACA
--------CACGCM
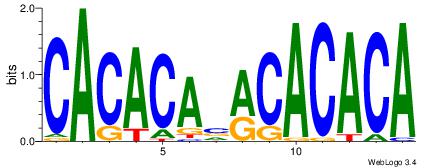
| Original motif Consensus sequence: CACACAVRCACACA |
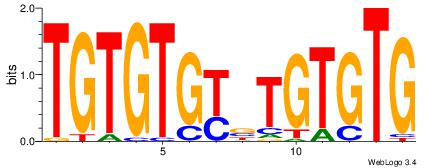
Reverse complement motif Consensus sequence: TGTGTGKVTGTGTG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0586538 |
Alignment:
GGGGMGGG
YGCGTG--
| Original motif Consensus sequence: CCCRCCCC |
Reverse complement motif Consensus sequence: GGGGMGGG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0884818 |
Alignment:
CCWGSCT
-CACGCM
| Original motif Consensus sequence: AGSCWGG |
Reverse complement motif Consensus sequence: CCWGSCT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
5 |
| Similarity score: |
0.563057 |
Alignment:
AMACA-
CACGCM
| Original motif Consensus sequence: AMACA |
Reverse complement motif Consensus sequence: TGTRT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
6 |
| Motif name: |
Motif 6 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
5 |
| Similarity score: |
0.572774 |
Alignment:
-GCKGCYG
YGCGTG--
| Original motif Consensus sequence: CMGCRGC |
Reverse complement motif Consensus sequence: GCKGCYG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
5 |
| Similarity score: |
0.581904 |
Alignment:
CTGGGA-
-YGCGTG
| Original motif Consensus sequence: CTGGGA |
Reverse complement motif Consensus sequence: TCCCAG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
4 |
| Motif name: |
Motif 4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
4 |
| Similarity score: |
1.06919 |
Alignment:
--KCAGAG
YGCGTG--
| Original motif Consensus sequence: CTCTGY |
Reverse complement motif Consensus sequence: KCAGAG |
|
|
 |
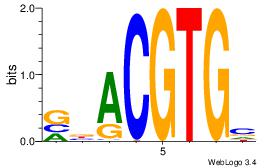
 |
| Dataset #: 2 |
Motif ID: 33 |
Motif name: HIF1AARNT |
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
Best Matches for Top Significant Motif ID 33 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0745994 |
Alignment:
CACACAVRCACACA
-----VCACGTBV-
| Original motif Consensus sequence: CACACAVRCACACA |
Reverse complement motif Consensus sequence: TGTGTGKVTGTGTG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0850889 |
Alignment:
CAYACACACACACA
---VCACGTBV---
| Original motif Consensus sequence: CAYACACACACACA |
Reverse complement motif Consensus sequence: TGTGTGTGTGTKTG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.588622 |
Alignment:
CCCRCCCC-
-VCACGTBV
| Original motif Consensus sequence: CCCRCCCC |
Reverse complement motif Consensus sequence: GGGGMGGG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
5 |
| Similarity score: |
1.55675 |
Alignment:
CCWGSCT---
--VBACGTGV
| Original motif Consensus sequence: AGSCWGG |
Reverse complement motif Consensus sequence: CCWGSCT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
2 |
| Motif name: |
Motif 2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
5 |
| Similarity score: |
1.57238 |
Alignment:
TGTRT---
VBACGTGV
| Original motif Consensus sequence: AMACA |
Reverse complement motif Consensus sequence: TGTRT |
|
|
 |
 |
Significant Motifs - Global and Local Matching (Highest to Lowest)
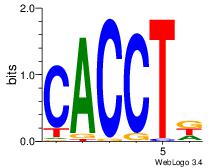
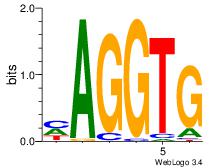
| Dataset #: 2 |
Motif ID: 14 |
Motif name: ZEB1 |
| Original motif Consensus sequence: CACCTD |
Reverse complement motif Consensus sequence: HAGGTG |
|
|
 |
 |
Best Matches for Significant Motif ID 14 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
46 |
| Motif name: |
RORA_1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0141289 |
Alignment:
TGACCTDVWT
-CACCTD---
| Original motif Consensus sequence: AWVDAGGTCA |
Reverse complement motif Consensus sequence: TGACCTDVWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
6 |
| Similarity score: |
0.0143133 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
----------HAGGTG-----
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
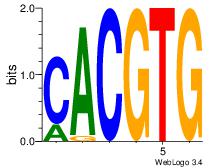
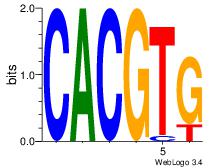
| Dataset #: |
2 |
| Motif ID: |
25 |
| Motif name: |
Arnt |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0207244 |
Alignment:
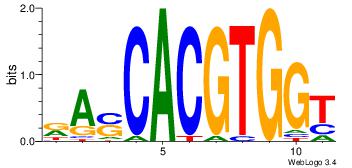
CACGTG
CACCTD
| Original motif Consensus sequence: CACGTG |
Reverse complement motif Consensus sequence: CACGTG |
|
|
 |
 |
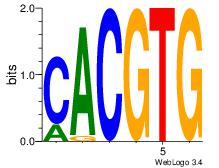
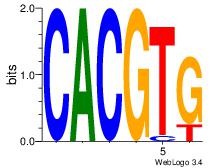
| Dataset #: |
2 |
| Motif ID: |
31 |
| Motif name: |
Esrrb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0207244 |
Alignment:
CACGTG
HAGGTG
| Original motif Consensus sequence: CACGTG |
Reverse complement motif Consensus sequence: CACGTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
43 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0232753 |
Alignment:
GTTGACCTTTGACCTTT
----------CACCTD-
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
33 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0237889 |
Alignment:
VCACGTBV
-CACCTD-
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0262123 |
Alignment:
TGYCAGGGGGCR
---HAGGTG---
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
6 |
| Similarity score: |
0.0264837 |
Alignment:
TGRCCTKTGHCCKAB
-CACCTD--------
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
39 |
| Motif name: |
MYCMAX |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0269744 |
Alignment:
ACCACGTGSTM
--CACCTD---
| Original motif Consensus sequence: RASCACGTGGT |
Reverse complement motif Consensus sequence: ACCACGTGSTM |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
44 |
| Motif name: |
NR4A2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0288897 |
Alignment:
GTGACCTT
--CACCTD
| Original motif Consensus sequence: AAGGTCAC |
Reverse complement motif Consensus sequence: GTGACCTT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 4 |
Motif name: Motif 4 |
| Original motif Consensus sequence: CTCTGY |
Reverse complement motif Consensus sequence: KCAGAG |
|
|
 |
 |
Best Matches for Significant Motif ID 4 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
43 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
6 |
| Similarity score: |
0.012269 |
Alignment:
AAAGGTCAAAGGTCAAC
-----KCAGAG------
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0140102 |
Alignment:
BTRGGDCARAGGKCA
-----KCAGAG----
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
22 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0186793 |
Alignment:
AKGYYCAAAGRTCA
----KCAGAG----
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
34 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.020602 |
Alignment:
RGGBCAAAGKYCA
---KCAGAG----
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
37 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0221665 |
Alignment:
BAACGTCCGC
----CTCTGY
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0255256 |
Alignment:
TGYCAGGGGGCR
--KCAGAG----
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0349642 |
Alignment:
CCCCKCCCCC
CTCTGY----
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0376108 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
--CTCTGY-------------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0446026 |
Alignment:
TCCCAG
KCAGAG
| Original motif Consensus sequence: CTGGGA |
Reverse complement motif Consensus sequence: TCCCAG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0448449 |
Alignment:
BBVGCCBVGGCCTV
---KCAGAG-----
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 5 |
Motif name: Motif 5 |
| Original motif Consensus sequence: ATKTM |
Reverse complement motif Consensus sequence: RARAT |
|
|
 |
 |
Best Matches for Significant Motif ID 5 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
10 |
| Number of overlap: |
5 |
| Similarity score: |
0.0256083 |
Alignment:
BGRRRGRGGRTGRTTYGGGG
------RARAT---------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
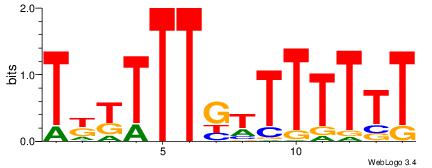
1 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
5 |
| Similarity score: |
0.0260117 |
Alignment:
AAAAAAACAAARRA
-RARAT--------
| Original motif Consensus sequence: TKKTTTGTTTTTTT |
Reverse complement motif Consensus sequence: AAAAAAACAAARRA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
44 |
| Motif name: |
NR4A2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
5 |
| Similarity score: |
0.0276382 |
Alignment:
AAGGTCAC
RARAT---
| Original motif Consensus sequence: AAGGTCAC |
Reverse complement motif Consensus sequence: GTGACCTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
22 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
5 |
| Similarity score: |
0.0279129 |
Alignment:
AKGYYCAAAGRTCA
-------RARAT--
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
37 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
5 |
| Similarity score: |
0.0292478 |
Alignment:
BAACGTCCGC
--ATKTM---
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
43 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
10 |
| Number of overlap: |
5 |
| Similarity score: |
0.0292478 |
Alignment:
AAAGGTCAAAGGTCAAC
---------ATKTM---
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
5 |
| Similarity score: |
0.0305369 |
Alignment:
TGRCCTKTGHCCKAB
----ATKTM------
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
5 |
| Similarity score: |
0.0315454 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
----ATKTM------------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
RXRRAR_DR5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
5 |
| Similarity score: |
0.0335956 |
Alignment:
RGKTCABVVRGAGGTCA
ATKTM------------
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
46 |
| Motif name: |
RORA_1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
5 |
| Similarity score: |
0.0336052 |
Alignment:
AWVDAGGTCA
---RARAT--
| Original motif Consensus sequence: AWVDAGGTCA |
Reverse complement motif Consensus sequence: TGACCTDVWT |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 26 |
Motif name: ArntAhr |
| Original motif Consensus sequence: YGCGTG |
Reverse complement motif Consensus sequence: CACGCM |
|
|
 |
 |
Best Matches for Significant Motif ID 26 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
28 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0 |
Alignment:
YCGCCCACGCH
-----CACGCM
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
33 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0112447 |
Alignment:
VBACGTGV
-YGCGTG-
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
6 |
| Similarity score: |
0.0308396 |
Alignment:
TGTGTGTGTGTKTG
------YGCGTG--
| Original motif Consensus sequence: CAYACACACACACA |
Reverse complement motif Consensus sequence: TGTGTGTGTGTKTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
31 |
| Motif name: |
Esrrb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0315972 |
Alignment:
CACGTG
YGCGTG
| Original motif Consensus sequence: CACGTG |
Reverse complement motif Consensus sequence: CACGTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
25 |
| Motif name: |
Arnt |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0315972 |
Alignment:
CACGTG
YGCGTG
| Original motif Consensus sequence: CACGTG |
Reverse complement motif Consensus sequence: CACGTG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
6 |
| Similarity score: |
0.0350575 |
Alignment:
TGTGTGKVTGTGTG
YGCGTG--------
| Original motif Consensus sequence: CACACAVRCACACA |
Reverse complement motif Consensus sequence: TGTGTGKVTGTGTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
39 |
| Motif name: |
MYCMAX |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0376488 |
Alignment:
RASCACGTGGT
---YGCGTG--
| Original motif Consensus sequence: RASCACGTGGT |
Reverse complement motif Consensus sequence: ACCACGTGSTM |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
40 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0377949 |
Alignment:
GCCACGTGSD
--YGCGTG--
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
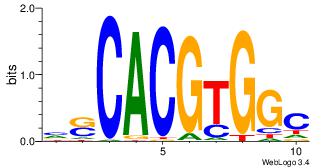 |
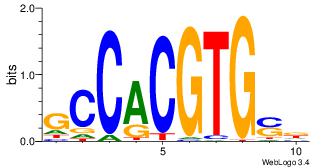 |
| Dataset #: |
2 |
| Motif ID: |
38 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0399214 |
Alignment:
DCCACGTGCV
--YGCGTG--
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
6 |
| Similarity score: |
0.0433081 |
Alignment:
BGRRRGRGGRTGRTTYGGGG
--------YGCGTG------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 6 |
Motif name: Motif 6 |
| Original motif Consensus sequence: CMGCRGC |
Reverse complement motif Consensus sequence: GCKGCYG |
|
|
 |
 |
Best Matches for Significant Motif ID 6 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
13 |
| Number of overlap: |
7 |
| Similarity score: |
0.0205607 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
------------GCKGCYG--
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
37 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0322623 |
Alignment:
GCGGACGTTV
GCKGCYG---
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.038091 |
Alignment:
BBVGCCBVGGCCTV
----CMGCRGC---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
TFAP2A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0.0393168 |
Alignment:
SCMVBBGGC
--CMGCRGC
| Original motif Consensus sequence: GCCBBVRGS |
Reverse complement motif Consensus sequence: SCMVBBGGC |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0443244 |
Alignment:
CCCRCCCC
-CMGCRGC
| Original motif Consensus sequence: CCCRCCCC |
Reverse complement motif Consensus sequence: GGGGMGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
7 |
| Similarity score: |
0.0448462 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-------GCKGCYG------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
28 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0453576 |
Alignment:
HGCGTGGGCGK
---GCKGCYG-
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
36 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0476958 |
Alignment:
GCCYCMCCCD
-CMGCRGC--
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0490991 |
Alignment:
GGGGGYGGGG
GCKGCYG---
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
7 |
| Similarity score: |
0.0498726 |
Alignment:
BGRRRGRGGRTGRTTYGGGG
-----GCKGCYG--------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 2 |
Motif name: Motif 2 |
| Original motif Consensus sequence: AMACA |
Reverse complement motif Consensus sequence: TGTRT |
|
|
 |
 |
Best Matches for Significant Motif ID 2 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
5 |
| Similarity score: |
0 |
Alignment:
TGTGTGTGTGTKTG
----TGTRT-----
| Original motif Consensus sequence: CAYACACACACACA |
Reverse complement motif Consensus sequence: TGTGTGTGTGTKTG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
10 |
| Number of overlap: |
5 |
| Similarity score: |
0.0040753 |
Alignment:
CACACAVRCACACA
---------AMACA
| Original motif Consensus sequence: CACACAVRCACACA |
Reverse complement motif Consensus sequence: TGTGTGKVTGTGTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
11 |
| Number of overlap: |
5 |
| Similarity score: |
0.0243918 |
Alignment:
CCCCMAAMCAMCCMCMMMCV
-----AMACA----------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
5 |
| Similarity score: |
0.0394919 |
Alignment:
AAAAAAACAAARRA
----AMACA-----
| Original motif Consensus sequence: TKKTTTGTTTTTTT |
Reverse complement motif Consensus sequence: AAAAAAACAAARRA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
5 |
| Similarity score: |
0.0429089 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
----AMACA-------------
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
5 |
| Similarity score: |
0.0595073 |
Alignment:
TGRCCTKTGHCCKAB
-----TGTRT-----
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
26 |
| Motif name: |
ArntAhr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
5 |
| Similarity score: |
0.0600161 |
Alignment:
YGCGTG
TGTRT-
| Original motif Consensus sequence: YGCGTG |
Reverse complement motif Consensus sequence: CACGCM |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
5 |
| Similarity score: |
0.0620995 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-----AMACA----------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
22 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
5 |
| Similarity score: |
0.0650242 |
Alignment:
AKGYYCAAAGRTCA
------AMACA---
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
5 |
| Similarity score: |
0.0670984 |
Alignment:
TGYCAGGGGGCR
TGTRT-------
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 23 |
Motif name: MZF1_1-4 |
| Original motif Consensus sequence: BGGGGA |
Reverse complement motif Consensus sequence: TCCCCV |
|
|
 |
 |
Best Matches for Significant Motif ID 23 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0276389 |
Alignment:
MGCCCCCTGMCA
--TCCCCV----
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
41 |
| Motif name: |
MZF1_5-13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0314583 |
Alignment:
BKAGGGGDAD
---BGGGGA-
| Original motif Consensus sequence: BKAGGGGDAD |
Reverse complement motif Consensus sequence: BTHCCCCTYB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
36 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0406514 |
Alignment:
GCCYCMCCCD
TCCCCV----
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
21 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
6 |
| Similarity score: |
0.0412963 |
Alignment:
GGGGCCCAAGGGGG
--------BGGGGA
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0426517 |
Alignment:
TCCCAG
TCCCCV
| Original motif Consensus sequence: CTGGGA |
Reverse complement motif Consensus sequence: TCCCAG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0464881 |
Alignment:
GGGGGYGGGG
----BGGGGA
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0493561 |
Alignment:
CCCCMAAMCAMCCMCMMMCV
----TCCCCV----------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
42 |
| Motif name: |
NFKB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0521759 |
Alignment:
GGGGRTTCCCC
-----TCCCCV
| Original motif Consensus sequence: GGGGRTTCCCC |
Reverse complement motif Consensus sequence: GGGGAAKCCCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
11 |
| Number of overlap: |
6 |
| Similarity score: |
0.0581528 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
-----TCCCCV----------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0585417 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
--------------TCCCCV--
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 44 |
Motif name: NR4A2 |
| Original motif Consensus sequence: AAGGTCAC |
Reverse complement motif Consensus sequence: GTGACCTT |
|
|
 |
 |
Best Matches for Significant Motif ID 44 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
43 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0167759 |
Alignment:
AAAGGTCAAAGGTCAAC
-AAGGTCAC--------
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0195235 |
Alignment:
VDBHMAGGTCACCCTGACCY
----AAGGTCAC--------
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
30 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0198372 |
Alignment:
VHRGGTCABDBTGMCCTB
-AAGGTCAC---------
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
13 |
| Number of overlap: |
8 |
| Similarity score: |
0.0487608 |
Alignment:
VRVDGGHACAVDDKGTHCTDWH
------------GTGACCTT--
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.0495607 |
Alignment:
BTRGGDCARAGGKCA
-AAGGTCAC------
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
22 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.0564874 |
Alignment:
AKGYYCAAAGRTCA
------AAGGTCAC
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0594649 |
Alignment:
TTCAGCACCATGGACAGCKCC
---------AAGGTCAC----
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0631514 |
Alignment:
BBVGCCBVGGCCTV
GTGACCTT------
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0720656 |
Alignment:
BGRRRGRGGRTGRTTYGGGG
---------GTGACCTT---
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
48 |
| Motif name: |
Tcfcp2l1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0740998 |
Alignment:
CKGGDTBDMMCTGG
-----GTGACCTT-
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 15 |
Motif name: TFAP2A |
| Original motif Consensus sequence: GCCBBVRGS |
Reverse complement motif Consensus sequence: SCMVBBGGC |
|
|
 |
 |
Best Matches for Significant Motif ID 15 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
9 |
| Similarity score: |
0.0131247 |
Alignment:
BBVGCCBVGGCCTV
---SCMVBBGGC--
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
21 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
9 |
| Similarity score: |
0.0414588 |
Alignment:
GGGGCCCAAGGGGG
---GCCBBVRGS--
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
13 |
| Motif name: |
Zfp423 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
9 |
| Similarity score: |
0.0438157 |
Alignment:
GYRYCCMTKGGYRSC
---SCMVBBGGC---
| Original motif Consensus sequence: GSMMCCYARGGKKKC |
Reverse complement motif Consensus sequence: GYRYCCMTKGGYRSC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.0450672 |
Alignment:
MGGTCAGGGTGACCTRDBHV
----------GCCBBVRGS-
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
30 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
9 |
| Similarity score: |
0.0531668 |
Alignment:
BAGGYCABHBTGACCKHV
-----SCMVBBGGC----
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
40 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.0550803 |
Alignment:
HSCACGTGGC
-SCMVBBGGC
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
9 |
| Similarity score: |
0.0581255 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-------GCCBBVRGS----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
RXRRAR_DR5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
9 |
| Similarity score: |
0.0606062 |
Alignment:
RGKTCABVVRGAGGTCA
--GCCBBVRGS------
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
38 |
| Motif name: |
Myc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.0613455 |
Alignment:
VGCACGTGGH
-SCMVBBGGC
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
48 |
| Motif name: |
Tcfcp2l1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
9 |
| Similarity score: |
0.0631412 |
Alignment:
CCAGYYHVADCCRG
---GCCBBVRGS--
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 25 |
Motif name: Arnt |
| Original motif Consensus sequence: CACGTG |
Reverse complement motif Consensus sequence: CACGTG |
|
|
 |
 |
Best Matches for Significant Motif ID 25 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
31 |
| Motif name: |
Esrrb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0 |
Alignment:
CACGTG
CACGTG
| Original motif Consensus sequence: CACGTG |
Reverse complement motif Consensus sequence: CACGTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
39 |
| Motif name: |
MYCMAX |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0143849 |
Alignment:
ACCACGTGSTM
--CACGTG---
| Original motif Consensus sequence: RASCACGTGGT |
Reverse complement motif Consensus sequence: ACCACGTGSTM |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
38 |
| Motif name: |
Myc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0267529 |
Alignment:
DCCACGTGCV
--CACGTG--
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
40 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0272641 |
Alignment:
HSCACGTGGC
--CACGTG--
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
33 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0347756 |
Alignment:
VBACGTGV
-CACGTG-
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
26 |
| Motif name: |
ArntAhr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.065625 |
Alignment:
YGCGTG
CACGTG
| Original motif Consensus sequence: YGCGTG |
Reverse complement motif Consensus sequence: CACGCM |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
14 |
| Motif name: |
ZEB1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0689533 |
Alignment:
HAGGTG
CACGTG
| Original motif Consensus sequence: CACCTD |
Reverse complement motif Consensus sequence: HAGGTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
28 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0784722 |
Alignment:
HGCGTGGGCGK
CACGTG-----
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
37 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0854167 |
Alignment:
BAACGTCCGC
-CACGTG---
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
21 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
6 |
| Similarity score: |
0.105093 |
Alignment:
GGGGCCCAAGGGGG
------CACGTG--
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
Best Matches for Each Motif (Highest to Lowest)
| Dataset #: 1 |
Motif ID: 1 |
Motif name: Motif 1 |
| Original motif Consensus sequence: RGGAAR |
Reverse complement motif Consensus sequence: MTTCCK |
|
|
 |
 |
Best Matches for Motif ID 1 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
32 |
| Motif name: |
EWSR1-FLI1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
12 |
| Number of overlap: |
6 |
| Similarity score: |
0.0146511 |
Alignment:
CCTTCCTTCCTTCCTTCC
-MTTCCK-----------
| Original motif Consensus sequence: GGAAGGAAGGAAGGAAGG |
Reverse complement motif Consensus sequence: CCTTCCTTCCTTCCTTCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
37 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0608243 |
Alignment:
BAACGTCCGC
---MTTCCK-
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
41 |
| Motif name: |
MZF1_5-13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0753789 |
Alignment:
BKAGGGGDAD
----RGGAAR
| Original motif Consensus sequence: BKAGGGGDAD |
Reverse complement motif Consensus sequence: BTHCCCCTYB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
42 |
| Motif name: |
NFKB1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0757695 |
Alignment:
GGGGAAKCCCC
-RGGAAR----
| Original motif Consensus sequence: GGGGRTTCCCC |
Reverse complement motif Consensus sequence: GGGGAAKCCCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
6 |
| Similarity score: |
0.0800484 |
Alignment:
BTRGGDCARAGGKCA
---RGGAAR------
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0827261 |
Alignment:
GGGGMGGG
RGGAAR--
| Original motif Consensus sequence: CCCRCCCC |
Reverse complement motif Consensus sequence: GGGGMGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
6 |
| Similarity score: |
0.0829223 |
Alignment:
TTCAGCACCATGGACAGCKCC
----------RGGAAR-----
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
6 |
| Similarity score: |
0.0832926 |
Alignment:
TGYCAGGGGGCR
------RGGAAR
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0835516 |
Alignment:
BGRRRGRGGRTGRTTYGGGG
----RGGAAR----------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0836451 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
---RGGAAR-------------
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
27 |
| Motif name: |
E2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0842331 |
Alignment:
GCGCSAAA
--RGGAAR
| Original motif Consensus sequence: TTTSGCGC |
Reverse complement motif Consensus sequence: GCGCSAAA |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 2 |
Motif name: Motif 2 |
| Original motif Consensus sequence: AMACA |
Reverse complement motif Consensus sequence: TGTRT |
|
|
 |
 |
Best Matches for Motif ID 2 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
5 |
| Similarity score: |
0 |
Alignment:
TGTGTGTGTGTKTG
----TGTRT-----
| Original motif Consensus sequence: CAYACACACACACA |
Reverse complement motif Consensus sequence: TGTGTGTGTGTKTG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
10 |
| Number of overlap: |
5 |
| Similarity score: |
0.0040753 |
Alignment:
CACACAVRCACACA
---------AMACA
| Original motif Consensus sequence: CACACAVRCACACA |
Reverse complement motif Consensus sequence: TGTGTGKVTGTGTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
11 |
| Number of overlap: |
5 |
| Similarity score: |
0.0243918 |
Alignment:
CCCCMAAMCAMCCMCMMMCV
-----AMACA----------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
5 |
| Similarity score: |
0.0394919 |
Alignment:
AAAAAAACAAARRA
----AMACA-----
| Original motif Consensus sequence: TKKTTTGTTTTTTT |
Reverse complement motif Consensus sequence: AAAAAAACAAARRA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
5 |
| Similarity score: |
0.0429089 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
----AMACA-------------
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
5 |
| Similarity score: |
0.0595073 |
Alignment:
TGRCCTKTGHCCKAB
-----TGTRT-----
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
26 |
| Motif name: |
ArntAhr |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
5 |
| Similarity score: |
0.0600161 |
Alignment:
YGCGTG
TGTRT-
| Original motif Consensus sequence: YGCGTG |
Reverse complement motif Consensus sequence: CACGCM |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
5 |
| Similarity score: |
0.0620995 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-----AMACA----------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
22 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
5 |
| Similarity score: |
0.0650242 |
Alignment:
AKGYYCAAAGRTCA
------AMACA---
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
5 |
| Similarity score: |
0.0670984 |
Alignment:
TGYCAGGGGGCR
TGTRT-------
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 3 |
Motif name: Motif 3 |
| Original motif Consensus sequence: AGSCWGG |
Reverse complement motif Consensus sequence: CCWGSCT |
|
|
 |
 |
Best Matches for Motif ID 3 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
32 |
| Motif name: |
EWSR1-FLI1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
7 |
| Similarity score: |
0.0395671 |
Alignment:
GGAAGGAAGGAAGGAAGG
-------AGSCWGG----
| Original motif Consensus sequence: GGAAGGAAGGAAGGAAGG |
Reverse complement motif Consensus sequence: CCTTCCTTCCTTCCTTCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0494688 |
Alignment:
MGCCCCCTGMCA
-----CCWGSCT
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
48 |
| Motif name: |
Tcfcp2l1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0669354 |
Alignment:
CCAGYYHVADCCRG
CCWGSCT-------
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
7 |
| Similarity score: |
0.0693747 |
Alignment:
TTCAGCACCATGGACAGCKCC
------AGSCWGG--------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0.0695377 |
Alignment:
CCCCKCCCCC
-CCWGSCT--
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
7 |
| Similarity score: |
0.0720685 |
Alignment:
BBVGCCBVGGCCTV
------CCWGSCT-
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0742342 |
Alignment:
CCCRCCCC
CCWGSCT-
| Original motif Consensus sequence: CCCRCCCC |
Reverse complement motif Consensus sequence: GGGGMGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0771405 |
Alignment:
MGGTCAGGGTGACCTRDBHV
------------CCWGSCT-
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
46 |
| Motif name: |
RORA_1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.0772276 |
Alignment:
TGACCTDVWT
---CCWGSCT
| Original motif Consensus sequence: AWVDAGGTCA |
Reverse complement motif Consensus sequence: TGACCTDVWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
36 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0798994 |
Alignment:
GCCYCMCCCD
---CCWGSCT
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 4 |
Motif name: Motif 4 |
| Original motif Consensus sequence: CTCTGY |
Reverse complement motif Consensus sequence: KCAGAG |
|
|
 |
 |
Best Matches for Motif ID 4 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
43 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
6 |
| Similarity score: |
0.012269 |
Alignment:
AAAGGTCAAAGGTCAAC
-----KCAGAG------
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0140102 |
Alignment:
BTRGGDCARAGGKCA
-----KCAGAG----
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
22 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0186793 |
Alignment:
AKGYYCAAAGRTCA
----KCAGAG----
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
34 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.020602 |
Alignment:
RGGBCAAAGKYCA
---KCAGAG----
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
37 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0221665 |
Alignment:
BAACGTCCGC
----CTCTGY
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0255256 |
Alignment:
TGYCAGGGGGCR
--KCAGAG----
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0349642 |
Alignment:
CCCCKCCCCC
CTCTGY----
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0376108 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
--CTCTGY-------------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0446026 |
Alignment:
TCCCAG
KCAGAG
| Original motif Consensus sequence: CTGGGA |
Reverse complement motif Consensus sequence: TCCCAG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0448449 |
Alignment:
BBVGCCBVGGCCTV
---KCAGAG-----
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 5 |
Motif name: Motif 5 |
| Original motif Consensus sequence: ATKTM |
Reverse complement motif Consensus sequence: RARAT |
|
|
 |
 |
Best Matches for Motif ID 5 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
10 |
| Number of overlap: |
5 |
| Similarity score: |
0.0256083 |
Alignment:
BGRRRGRGGRTGRTTYGGGG
------RARAT---------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
5 |
| Similarity score: |
0.0260117 |
Alignment:
AAAAAAACAAARRA
-RARAT--------
| Original motif Consensus sequence: TKKTTTGTTTTTTT |
Reverse complement motif Consensus sequence: AAAAAAACAAARRA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
44 |
| Motif name: |
NR4A2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
5 |
| Similarity score: |
0.0276382 |
Alignment:
AAGGTCAC
RARAT---
| Original motif Consensus sequence: AAGGTCAC |
Reverse complement motif Consensus sequence: GTGACCTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
22 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
5 |
| Similarity score: |
0.0279129 |
Alignment:
AKGYYCAAAGRTCA
-------RARAT--
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
37 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
5 |
| Similarity score: |
0.0292478 |
Alignment:
BAACGTCCGC
--ATKTM---
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
43 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
10 |
| Number of overlap: |
5 |
| Similarity score: |
0.0292478 |
Alignment:
AAAGGTCAAAGGTCAAC
---------ATKTM---
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
5 |
| Similarity score: |
0.0305369 |
Alignment:
TGRCCTKTGHCCKAB
----ATKTM------
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
5 |
| Similarity score: |
0.0315454 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
----ATKTM------------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
RXRRAR_DR5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
5 |
| Similarity score: |
0.0335956 |
Alignment:
RGKTCABVVRGAGGTCA
ATKTM------------
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
46 |
| Motif name: |
RORA_1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
5 |
| Similarity score: |
0.0336052 |
Alignment:
AWVDAGGTCA
---RARAT--
| Original motif Consensus sequence: AWVDAGGTCA |
Reverse complement motif Consensus sequence: TGACCTDVWT |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 6 |
Motif name: Motif 6 |
| Original motif Consensus sequence: CMGCRGC |
Reverse complement motif Consensus sequence: GCKGCYG |
|
|
 |
 |
Best Matches for Motif ID 6 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
13 |
| Number of overlap: |
7 |
| Similarity score: |
0.0205607 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
------------GCKGCYG--
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
37 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0322623 |
Alignment:
GCGGACGTTV
GCKGCYG---
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
0.038091 |
Alignment:
BBVGCCBVGGCCTV
----CMGCRGC---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
15 |
| Motif name: |
TFAP2A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
7 |
| Similarity score: |
0.0393168 |
Alignment:
SCMVBBGGC
--CMGCRGC
| Original motif Consensus sequence: GCCBBVRGS |
Reverse complement motif Consensus sequence: SCMVBBGGC |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0443244 |
Alignment:
CCCRCCCC
-CMGCRGC
| Original motif Consensus sequence: CCCRCCCC |
Reverse complement motif Consensus sequence: GGGGMGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
7 |
| Similarity score: |
0.0448462 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-------GCKGCYG------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
28 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0453576 |
Alignment:
HGCGTGGGCGK
---GCKGCYG-
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
36 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
0.0476958 |
Alignment:
GCCYCMCCCD
-CMGCRGC--
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
0.0490991 |
Alignment:
GGGGGYGGGG
GCKGCYG---
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
7 |
| Similarity score: |
0.0498726 |
Alignment:
BGRRRGRGGRTGRTTYGGGG
-----GCKGCYG--------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 7 |
Motif name: Motif 7 |
| Original motif Consensus sequence: CCCRCCCC |
Reverse complement motif Consensus sequence: GGGGMGGG |
|
|
 |
 |
Best Matches for Motif ID 7 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0423611 |
Alignment:
CCCCKCCCCC
-CCCRCCCC-
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
36 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0461644 |
Alignment:
DGGGYGKGGC
GGGGMGGG--
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
28 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0650374 |
Alignment:
YCGCCCACGCH
---CCCRCCCC
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.076401 |
Alignment:
CCCCMAAMCAMCCMCMMMCV
-CCCRCCCC-----------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
42 |
| Motif name: |
NFKB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0893429 |
Alignment:
GGGGAAKCCCC
GGGGMGGG---
| Original motif Consensus sequence: GGGGRTTCCCC |
Reverse complement motif Consensus sequence: GGGGAAKCCCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0936832 |
Alignment:
TGYCAGGGGGCR
GGGGMGGG----
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
32 |
| Motif name: |
EWSR1-FLI1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0950969 |
Alignment:
GGAAGGAAGGAAGGAAGG
------GGGGMGGG----
| Original motif Consensus sequence: GGAAGGAAGGAAGGAAGG |
Reverse complement motif Consensus sequence: CCTTCCTTCCTTCCTTCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
21 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0998932 |
Alignment:
GGGGCCCAAGGGGG
GGGGMGGG------
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.10221 |
Alignment:
BBVGCCBVGGCCTV
----CCCRCCCC--
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.102497 |
Alignment:
DGVBCABTGDWGCGKRRCSR
--------GGGGMGGG----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 8 |
Motif name: Motif 8 |
| Original motif Consensus sequence: CTGGGA |
Reverse complement motif Consensus sequence: TCCCAG |
|
|
 |
 |
Best Matches for Motif ID 8 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
6 |
| Similarity score: |
0.0392549 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
-------TCCCAG--------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
23 |
| Motif name: |
MZF1_1-4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0416319 |
Alignment:
TCCCCV
TCCCAG
| Original motif Consensus sequence: BGGGGA |
Reverse complement motif Consensus sequence: TCCCCV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0509122 |
Alignment:
BGRRRGRGGRTGRTTYGGGG
--------------CTGGGA
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0557857 |
Alignment:
TGYCAGGGGGCR
---CTGGGA---
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
13 |
| Motif name: |
Zfp423 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
6 |
| Similarity score: |
0.0624145 |
Alignment:
GYRYCCMTKGGYRSC
---TCCCAG------
| Original motif Consensus sequence: GSMMCCYARGGKKKC |
Reverse complement motif Consensus sequence: GYRYCCMTKGGYRSC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
43 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
6 |
| Similarity score: |
0.0650784 |
Alignment:
AAAGGTCAAAGGTCAAC
-----TCCCAG------
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
48 |
| Motif name: |
Tcfcp2l1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
6 |
| Similarity score: |
0.0663804 |
Alignment:
CKGGDTBDMMCTGG
CTGGGA--------
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
28 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0678562 |
Alignment:
YCGCCCACGCH
--TCCCAG---
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0714107 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
--------------TCCCAG--
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0718041 |
Alignment:
BBVGCCBVGGCCTV
-----CTGGGA---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 9 |
Motif name: Motif 9 |
| Original motif Consensus sequence: CACACAVRCACACA |
Reverse complement motif Consensus sequence: TGTGTGKVTGTGTG |
|
|
 |
 |
Best Matches for Motif ID 9 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0170175 |
Alignment:
CAYACACACACACA
CACACAVRCACACA
| Original motif Consensus sequence: CAYACACACACACA |
Reverse complement motif Consensus sequence: TGTGTGTGTGTKTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
14 |
| Similarity score: |
0.0868786 |
Alignment:
CCCCMAAMCAMCCMCMMMCV
----CACACAVRCACACA--
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.578949 |
Alignment:
TKKTTTGTTTTTTT-
-TGTGTGKVTGTGTG
| Original motif Consensus sequence: TKKTTTGTTTTTTT |
Reverse complement motif Consensus sequence: AAAAAAACAAARRA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
17 |
| Motif name: |
RXRAVDR |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
13 |
| Similarity score: |
0.593978 |
Alignment:
TGAACKCBWTGACCC-
--CACACAVRCACACA
| Original motif Consensus sequence: GGGTCAWBGRGTTCA |
Reverse complement motif Consensus sequence: TGAACKCBWTGACCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
11 |
| Number of overlap: |
12 |
| Similarity score: |
1.09291 |
Alignment:
--VRVDGGHACAVDDKGTHCTDWH
CACACAVRCACACA----------
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
28 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.57424 |
Alignment:
HGCGTGGGCGK---
TGTGTGKVTGTGTG
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
10 |
| Number of overlap: |
11 |
| Similarity score: |
1.58704 |
Alignment:
VDBHMAGGTCACCCTGACCY---
---------CACACAVRCACACA
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
2.08086 |
Alignment:
----TGYCAGGGGGCR
TGTGTGKVTGTGTG--
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
36 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
2.0884 |
Alignment:
DGGGYGKGGC----
TGTGTGKVTGTGTG
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
34 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
2.09226 |
Alignment:
TGMYCTTTGBCCK----
---TGTGTGKVTGTGTG
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 10 |
Motif name: Motif 10 |
| Original motif Consensus sequence: TKKTTTGTTTTTTT |
Reverse complement motif Consensus sequence: AAAAAAACAAARRA |
|
|
 |
 |
Best Matches for Motif ID 10 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
32 |
| Motif name: |
EWSR1-FLI1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
14 |
| Similarity score: |
0.0424269 |
Alignment:
GGAAGGAAGGAAGGAAGG
--AAAAAAACAAARRA--
| Original motif Consensus sequence: GGAAGGAAGGAAGGAAGG |
Reverse complement motif Consensus sequence: CCTTCCTTCCTTCCTTCC |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.529035 |
Alignment:
-CACACAVRCACACA
AAAAAAACAAARRA-
| Original motif Consensus sequence: CACACAVRCACACA |
Reverse complement motif Consensus sequence: TGTGTGKVTGTGTG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.531594 |
Alignment:
-TGTGTGTGTGTKTG
TKKTTTGTTTTTTT-
| Original motif Consensus sequence: CAYACACACACACA |
Reverse complement motif Consensus sequence: TGTGTGTGTGTKTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
43 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
13 |
| Similarity score: |
0.535643 |
Alignment:
-AAAGGTCAAAGGTCAAC
AAAAAAACAAARRA----
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
13 |
| Similarity score: |
0.543043 |
Alignment:
-BTRGGDCARAGGKCA
AAAAAAACAAARRA--
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
10 |
| Number of overlap: |
13 |
| Similarity score: |
0.544053 |
Alignment:
VRVDGGHACAVDDKGTHCTDWH-
---------TKKTTTGTTTTTTT
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
22 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
1.04059 |
Alignment:
--AKGYYCAAAGRTCA
AAAAAAACAAARRA--
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
34 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
1.53429 |
Alignment:
---RGGBCAAAGKYCA
AAAAAAACAAARRA--
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
10 |
| Number of overlap: |
11 |
| Similarity score: |
1.54022 |
Alignment:
---DGVBCABTGDWGCGKRRCSR
AAAAAAACAAARRA---------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
11 |
| Number of overlap: |
10 |
| Similarity score: |
2.0484 |
Alignment:
----CCCCMAAMCAMCCMCMMMCV
AAAAAAACAAARRA----------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: 1 |
Motif ID: 11 |
Motif name: Motif 11 |
| Original motif Consensus sequence: CAYACACACACACA |
Reverse complement motif Consensus sequence: TGTGTGTGTGTKTG |
|
|
 |
 |
Best Matches for Motif ID 11 (Highest to Lowest)
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0170175 |
Alignment:
CACACAVRCACACA
CAYACACACACACA
| Original motif Consensus sequence: CACACAVRCACACA |
Reverse complement motif Consensus sequence: TGTGTGKVTGTGTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
14 |
| Similarity score: |
0.0837439 |
Alignment:
CCCCMAAMCAMCCMCMMMCV
----CAYACACACACACA--
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.581509 |
Alignment:
AAAAAAACAAARRA-
-CAYACACACACACA
| Original motif Consensus sequence: TKKTTTGTTTTTTT |
Reverse complement motif Consensus sequence: AAAAAAACAAARRA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.09362 |
Alignment:
--TGYCAGGGGGCR
TGTGTGTGTGTKTG
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
28 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
1.56977 |
Alignment:
HGCGTGGGCGK---
TGTGTGTGTGTKTG
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
10 |
| Number of overlap: |
11 |
| Similarity score: |
1.59795 |
Alignment:
VDBHMAGGTCACCCTGACCY---
---------CAYACACACACACA
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
36 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
2.08683 |
Alignment:
DGGGYGKGGC----
TGTGTGTGTGTKTG
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
2.59204 |
Alignment:
-----GGGGGYGGGG
TGTGTGTGTGTKTG-
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
3.09205 |
Alignment:
------CCCRCCCC
CAYACACACACACA
| Original motif Consensus sequence: CCCRCCCC |
Reverse complement motif Consensus sequence: GGGGMGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
33 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
7 |
| Similarity score: |
3.58699 |
Alignment:
-------VBACGTGV
TGTGTGTGTGTKTG-
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 12 |
Motif name: Zfx |
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
Best Matches for Motif ID 12 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
14 |
| Similarity score: |
0.0611182 |
Alignment:
DGVBCABTGDWGCGKRRCSR
--BBVGCCBVGGCCTV----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
30 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0635714 |
Alignment:
BAGGYCABHBTGACCKHV
---BBVGCCBVGGCCTV-
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
14 |
| Similarity score: |
0.0722733 |
Alignment:
MGGTCAGGGTGACCTRDBHV
--BBVGCCBVGGCCTV----
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
48 |
| Motif name: |
Tcfcp2l1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0732898 |
Alignment:
CCAGYYHVADCCRG
BBVGCCBVGGCCTV
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
13 |
| Motif name: |
Zfp423 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0747812 |
Alignment:
GYRYCCMTKGGYRSC
-VAGGCCBBGGCVBB
| Original motif Consensus sequence: GSMMCCYARGGKKKC |
Reverse complement motif Consensus sequence: GYRYCCMTKGGYRSC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
21 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0796879 |
Alignment:
CCCCCTTGGGCCCC
BBVGCCBVGGCCTV
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
14 |
| Similarity score: |
0.0812253 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
----VAGGCCBBGGCVBB---
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.0832838 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
-----VAGGCCBBGGCVBB---
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0836605 |
Alignment:
TGRCCTKTGHCCKAB
BBVGCCBVGGCCTV-
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
RXRRAR_DR5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
13 |
| Similarity score: |
0.570818 |
Alignment:
TGACCTCKVVBTGAYCK-
----BBVGCCBVGGCCTV
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 13 |
Motif name: Zfp423 |
| Original motif Consensus sequence: GSMMCCYARGGKKKC |
Reverse complement motif Consensus sequence: GYRYCCMTKGGYRSC |
|
|
 |
 |
Best Matches for Motif ID 13 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
15 |
| Similarity score: |
0.0400042 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
---GSMMCCYARGGKKKC----
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
15 |
| Similarity score: |
0.0400673 |
Alignment:
DGVBCABTGDWGCGKRRCSR
--GSMMCCYARGGKKKC---
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
15 |
| Similarity score: |
0.0410001 |
Alignment:
TTCAGCACCATGGACAGCKCC
---GYRYCCMTKGGYRSC---
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
17 |
| Motif name: |
RXRAVDR |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
0.0465067 |
Alignment:
GGGTCAWBGRGTTCA
GYRYCCMTKGGYRSC
| Original motif Consensus sequence: GGGTCAWBGRGTTCA |
Reverse complement motif Consensus sequence: TGAACKCBWTGACCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
21 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.527928 |
Alignment:
GGGGCCCAAGGGGG-
GSMMCCYARGGKKKC
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.53991 |
Alignment:
-VAGGCCBBGGCVBB
GYRYCCMTKGGYRSC
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
13 |
| Similarity score: |
1.03254 |
Alignment:
--VDBHMAGGTCACCCTGACCY
GYRYCCMTKGGYRSC-------
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
13 |
| Similarity score: |
1.04952 |
Alignment:
BGRRRGRGGRTGRTTYGGGG--
-------GYRYCCMTKGGYRSC
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
1.54193 |
Alignment:
---TGYCAGGGGGCR
GSMMCCYARGGKKKC
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
48 |
| Motif name: |
Tcfcp2l1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
2.04218 |
Alignment:
CKGGDTBDMMCTGG----
---GYRYCCMTKGGYRSC
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 14 |
Motif name: ZEB1 |
| Original motif Consensus sequence: CACCTD |
Reverse complement motif Consensus sequence: HAGGTG |
|
|
 |
 |
Best Matches for Motif ID 14 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
46 |
| Motif name: |
RORA_1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0141289 |
Alignment:
TGACCTDVWT
-CACCTD---
| Original motif Consensus sequence: AWVDAGGTCA |
Reverse complement motif Consensus sequence: TGACCTDVWT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
6 |
| Similarity score: |
0.0143133 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
----------HAGGTG-----
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
25 |
| Motif name: |
Arnt |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0207244 |
Alignment:
CACGTG
CACCTD
| Original motif Consensus sequence: CACGTG |
Reverse complement motif Consensus sequence: CACGTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
31 |
| Motif name: |
Esrrb |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0207244 |
Alignment:
CACGTG
HAGGTG
| Original motif Consensus sequence: CACGTG |
Reverse complement motif Consensus sequence: CACGTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
43 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0232753 |
Alignment:
GTTGACCTTTGACCTTT
----------CACCTD-
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
33 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0237889 |
Alignment:
VCACGTBV
-CACCTD-
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0262123 |
Alignment:
TGYCAGGGGGCR
---HAGGTG---
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
6 |
| Similarity score: |
0.0264837 |
Alignment:
TGRCCTKTGHCCKAB
-CACCTD--------
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
39 |
| Motif name: |
MYCMAX |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0269744 |
Alignment:
ACCACGTGSTM
--CACCTD---
| Original motif Consensus sequence: RASCACGTGGT |
Reverse complement motif Consensus sequence: ACCACGTGSTM |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
44 |
| Motif name: |
NR4A2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0288897 |
Alignment:
GTGACCTT
--CACCTD
| Original motif Consensus sequence: AAGGTCAC |
Reverse complement motif Consensus sequence: GTGACCTT |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 15 |
Motif name: TFAP2A |
| Original motif Consensus sequence: GCCBBVRGS |
Reverse complement motif Consensus sequence: SCMVBBGGC |
|
|
 |
 |
Best Matches for Motif ID 15 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
9 |
| Similarity score: |
0.0131247 |
Alignment:
BBVGCCBVGGCCTV
---SCMVBBGGC--
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
21 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
9 |
| Similarity score: |
0.0414588 |
Alignment:
GGGGCCCAAGGGGG
---GCCBBVRGS--
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
13 |
| Motif name: |
Zfp423 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
9 |
| Similarity score: |
0.0438157 |
Alignment:
GYRYCCMTKGGYRSC
---SCMVBBGGC---
| Original motif Consensus sequence: GSMMCCYARGGKKKC |
Reverse complement motif Consensus sequence: GYRYCCMTKGGYRSC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.0450672 |
Alignment:
MGGTCAGGGTGACCTRDBHV
----------GCCBBVRGS-
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
30 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
9 |
| Similarity score: |
0.0531668 |
Alignment:
BAGGYCABHBTGACCKHV
-----SCMVBBGGC----
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
40 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.0550803 |
Alignment:
HSCACGTGGC
-SCMVBBGGC
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
9 |
| Similarity score: |
0.0581255 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-------GCCBBVRGS----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
RXRRAR_DR5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
9 |
| Similarity score: |
0.0606062 |
Alignment:
RGKTCABVVRGAGGTCA
--GCCBBVRGS------
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
38 |
| Motif name: |
Myc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.0613455 |
Alignment:
VGCACGTGGH
-SCMVBBGGC
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
48 |
| Motif name: |
Tcfcp2l1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
9 |
| Similarity score: |
0.0631412 |
Alignment:
CCAGYYHVADCCRG
---GCCBBVRGS--
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 16 |
Motif name: SP1 |
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
Best Matches for Motif ID 16 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0646465 |
Alignment:
BGRRRGRGGRTGRTTYGGGG
-GGGGGYGGGG---------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0664832 |
Alignment:
BBVGCCBVGGCCTV
----GGGGGYGGGG
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
21 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0725 |
Alignment:
CCCCCTTGGGCCCC
-CCCCKCCCCC---
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0731995 |
Alignment:
TGRCCTKTGHCCKAB
--CCCCKCCCCC---
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
36 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0761191 |
Alignment:
GCCYCMCCCD
CCCCKCCCCC
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
28 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0798413 |
Alignment:
HGCGTGGGCGK
GGGGGYGGGG-
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
10 |
| Similarity score: |
0.0846627 |
Alignment:
DGVBCABTGDWGCGKRRCSR
-------GGGGGYGGGG---
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
41 |
| Motif name: |
MZF1_5-13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0884127 |
Alignment:
BKAGGGGDAD
GGGGGYGGGG
| Original motif Consensus sequence: BKAGGGGDAD |
Reverse complement motif Consensus sequence: BTHCCCCTYB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.089127 |
Alignment:
TGYCAGGGGGCR
GGGGGYGGGG--
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
32 |
| Motif name: |
EWSR1-FLI1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
10 |
| Similarity score: |
0.0893651 |
Alignment:
CCTTCCTTCCTTCCTTCC
---CCCCKCCCCC-----
| Original motif Consensus sequence: GGAAGGAAGGAAGGAAGG |
Reverse complement motif Consensus sequence: CCTTCCTTCCTTCCTTCC |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 17 |
Motif name: RXRAVDR |
| Original motif Consensus sequence: GGGTCAWBGRGTTCA |
Reverse complement motif Consensus sequence: TGAACKCBWTGACCC |
|
|
 |
 |
Best Matches for Motif ID 17 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
15 |
| Similarity score: |
0.0566667 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
---TGAACKCBWTGACCC----
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
30 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
15 |
| Similarity score: |
0.0617507 |
Alignment:
VHRGGTCABDBTGMCCTB
--GGGTCAWBGRGTTCA-
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
RXRRAR_DR5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
15 |
| Similarity score: |
0.062029 |
Alignment:
RGKTCABVVRGAGGTCA
GGGTCAWBGRGTTCA--
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
0.0714525 |
Alignment:
VDBHMAGGTCACCCTGACCY
-----TGAACKCBWTGACCC
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
13 |
| Motif name: |
Zfp423 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
0.0849495 |
Alignment:
GSMMCCYARGGKKKC
TGAACKCBWTGACCC
| Original motif Consensus sequence: GSMMCCYARGGKKKC |
Reverse complement motif Consensus sequence: GYRYCCMTKGGYRSC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
43 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
15 |
| Similarity score: |
0.0866667 |
Alignment:
AAAGGTCAAAGGTCAAC
--GGGTCAWBGRGTTCA
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
21 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
1.06201 |
Alignment:
GGGGCCCAAGGGGG--
-GGGTCAWBGRGTTCA
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
13 |
| Similarity score: |
1.07837 |
Alignment:
BTRGGDCARAGGKCA--
--GGGTCAWBGRGTTCA
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
34 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
1.0801 |
Alignment:
RGGBCAAAGKYCA--
GGGTCAWBGRGTTCA
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
22 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
1.08482 |
Alignment:
AKGYYCAAAGRTCA--
-GGGTCAWBGRGTTCA
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 18 |
Motif name: RREB1 |
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
Best Matches for Motif ID 18 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
19 |
| Similarity score: |
0.0874679 |
Alignment:
-MGGTCAGGGTGACCTRDBHV
BGRRRGRGGRTGRTTYGGGG-
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
19 |
| Similarity score: |
0.0927532 |
Alignment:
-HWDAGHACRHHVTGTHCCHVMV
CCCCMAAMCAMCCMCMMMCV---
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
30 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
0.593122 |
Alignment:
BAGGYCABHBTGACCKHV--
BGRRRGRGGRTGRTTYGGGG
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
17 |
| Similarity score: |
1.09431 |
Alignment:
---DGVBCABTGDWGCGKRRCSR
BGRRRGRGGRTGRTTYGGGG---
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
21 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.5804 |
Alignment:
CCCCCTTGGGCCCC------
CCCCMAAMCAMCCMCMMMCV
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.58596 |
Alignment:
------CACACAVRCACACA
CCCCMAAMCAMCCMCMMMCV
| Original motif Consensus sequence: CACACAVRCACACA |
Reverse complement motif Consensus sequence: TGTGTGKVTGTGTG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.59077 |
Alignment:
------CAYACACACACACA
CCCCMAAMCAMCCMCMMMCV
| Original motif Consensus sequence: CAYACACACACACA |
Reverse complement motif Consensus sequence: TGTGTGTGTGTKTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
RXRRAR_DR5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
13 |
| Similarity score: |
3.07964 |
Alignment:
RGKTCABVVRGAGGTCA-------
----BGRRRGRGGRTGRTTYGGGG
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
13 |
| Similarity score: |
3.08033 |
Alignment:
BTRGGDCARAGGKCA-------
--BGRRRGRGGRTGRTTYGGGG
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
48 |
| Motif name: |
Tcfcp2l1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
3.08771 |
Alignment:
-------CCAGYYHVADCCRG
CCCCMAAMCAMCCMCMMMCV-
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 19 |
Motif name: REST |
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
Best Matches for Motif ID 19 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
21 |
| Similarity score: |
0.0685835 |
Alignment:
VRVDGGHACAVDDKGTHCTDWH
-GGYGCTGTCCATGGTGCTGAA
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
18 |
| Similarity score: |
1.577 |
Alignment:
MSGKKRCGCWDCABTGBBCD---
--GGYGCTGTCCATGGTGCTGAA
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
16 |
| Similarity score: |
2.56863 |
Alignment:
-----VDBHMAGGTCACCCTGACCY
TTCAGCACCATGGACAGCKCC----
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
30 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
13 |
| Similarity score: |
4.05754 |
Alignment:
BAGGYCABHBTGACCKHV--------
-----GGYGCTGTCCATGGTGCTGAA
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
34 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
4.06973 |
Alignment:
TGMYCTTTGBCCK--------
GGYGCTGTCCATGGTGCTGAA
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
13 |
| Similarity score: |
4.07172 |
Alignment:
--------BTRGGDCARAGGKCA
TTCAGCACCATGGACAGCKCC--
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
48 |
| Motif name: |
Tcfcp2l1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
4.57098 |
Alignment:
CCAGYYHVADCCRG---------
--GGYGCTGTCCATGGTGCTGAA
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
4.57482 |
Alignment:
---------BBVGCCBVGGCCTV
GGYGCTGTCCATGGTGCTGAA--
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
17 |
| Motif name: |
RXRAVDR |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
5.05758 |
Alignment:
TGAACKCBWTGACCC----------
----GGYGCTGTCCATGGTGCTGAA
| Original motif Consensus sequence: GGGTCAWBGRGTTCA |
Reverse complement motif Consensus sequence: TGAACKCBWTGACCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
RXRRAR_DR5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
11 |
| Similarity score: |
5.05947 |
Alignment:
TGACCTCKVVBTGAYCK----------
------GGYGCTGTCCATGGTGCTGAA
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 20 |
Motif name: PPARGRXRA |
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
Best Matches for Motif ID 20 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
43 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
15 |
| Similarity score: |
0.0442974 |
Alignment:
AAAGGTCAAAGGTCAAC
BTRGGDCARAGGKCA--
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
RXRRAR_DR5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
0.0500583 |
Alignment:
RGKTCABVVRGAGGTCA
--BTRGGDCARAGGKCA
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
30 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
15 |
| Similarity score: |
0.0602408 |
Alignment:
VHRGGTCABDBTGMCCTB
BTRGGDCARAGGKCA---
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
15 |
| Similarity score: |
0.0714981 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
-------TGRCCTKTGHCCKAB
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
15 |
| Similarity score: |
0.0720255 |
Alignment:
MSGKKRCGCWDCABTGBBCD
---TGRCCTKTGHCCKAB--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
15 |
| Similarity score: |
0.0723092 |
Alignment:
VDBHMAGGTCACCCTGACCY
---BTRGGDCARAGGKCA--
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
15 |
| Similarity score: |
0.0755648 |
Alignment:
CCCCMAAMCAMCCMCMMMCV
-TGRCCTKTGHCCKAB----
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
15 |
| Similarity score: |
0.0796159 |
Alignment:
TTCAGCACCATGGACAGCKCC
-BTRGGDCARAGGKCA-----
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
22 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.536521 |
Alignment:
TGAMCTTTGMMCYT-
TGRCCTKTGHCCKAB
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
21 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.575248 |
Alignment:
-GGGGCCCAAGGGGG
BTRGGDCARAGGKCA
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 21 |
Motif name: PLAG1 |
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
Best Matches for Motif ID 21 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
13 |
| Motif name: |
Zfp423 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0944264 |
Alignment:
GSMMCCYARGGKKKC
GGGGCCCAAGGGGG-
| Original motif Consensus sequence: GSMMCCYARGGKKKC |
Reverse complement motif Consensus sequence: GYRYCCMTKGGYRSC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
14 |
| Similarity score: |
0.102814 |
Alignment:
BGRRRGRGGRTGRTTYGGGG
GGGGCCCAAGGGGG------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.107206 |
Alignment:
BTRGGDCARAGGKCA
-GGGGCCCAAGGGGG
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.111315 |
Alignment:
VAGGCCBBGGCVBB
GGGGCCCAAGGGGG
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
48 |
| Motif name: |
Tcfcp2l1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.115151 |
Alignment:
CCAGYYHVADCCRG
CCCCCTTGGGCCCC
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
RXRRAR_DR5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.116287 |
Alignment:
RGKTCABVVRGAGGTCA
GGGGCCCAAGGGGG---
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.117063 |
Alignment:
DGVBCABTGDWGCGKRRCSR
GGGGCCCAAGGGGG------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
17 |
| Motif name: |
RXRAVDR |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
13 |
| Similarity score: |
0.590064 |
Alignment:
-GGGTCAWBGRGTTCA
GGGGCCCAAGGGGG--
| Original motif Consensus sequence: GGGTCAWBGRGTTCA |
Reverse complement motif Consensus sequence: TGAACKCBWTGACCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
34 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.603385 |
Alignment:
-RGGBCAAAGKYCA
GGGGCCCAAGGGGG
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
22 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.613207 |
Alignment:
-AKGYYCAAAGRTCA
GGGGCCCAAGGGGG-
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 22 |
Motif name: NR2F1 |
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
Best Matches for Motif ID 22 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
43 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0174588 |
Alignment:
GTTGACCTTTGACCTTT
--TGAMCTTTGMMCYT-
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0278283 |
Alignment:
BTRGGDCARAGGKCA
AKGYYCAAAGRTCA-
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
RXRRAR_DR5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.0625328 |
Alignment:
RGKTCABVVRGAGGTCA
---AKGYYCAAAGRTCA
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
30 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0710475 |
Alignment:
BAGGYCABHBTGACCKHV
---TGAMCTTTGMMCYT-
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
14 |
| Similarity score: |
0.071796 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
--TGAMCTTTGMMCYT-----
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
14 |
| Similarity score: |
0.0741987 |
Alignment:
MSGKKRCGCWDCABTGBBCD
----TGAMCTTTGMMCYT--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
34 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.514048 |
Alignment:
-RGGBCAAAGKYCA
AKGYYCAAAGRTCA
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.568042 |
Alignment:
BBVGCCBVGGCCTV-
-AKGYYCAAAGRTCA
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
13 |
| Similarity score: |
0.569869 |
Alignment:
VDBHMAGGTCACCCTGACCY-
-------TGAMCTTTGMMCYT
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
17 |
| Motif name: |
RXRAVDR |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
13 |
| Similarity score: |
0.572226 |
Alignment:
-TGAACKCBWTGACCC
TGAMCTTTGMMCYT--
| Original motif Consensus sequence: GGGTCAWBGRGTTCA |
Reverse complement motif Consensus sequence: TGAACKCBWTGACCC |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 23 |
Motif name: MZF1_1-4 |
| Original motif Consensus sequence: BGGGGA |
Reverse complement motif Consensus sequence: TCCCCV |
|
|
 |
 |
Best Matches for Motif ID 23 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0276389 |
Alignment:
MGCCCCCTGMCA
--TCCCCV----
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
41 |
| Motif name: |
MZF1_5-13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0314583 |
Alignment:
BKAGGGGDAD
---BGGGGA-
| Original motif Consensus sequence: BKAGGGGDAD |
Reverse complement motif Consensus sequence: BTHCCCCTYB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
36 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0406514 |
Alignment:
GCCYCMCCCD
TCCCCV----
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
21 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
6 |
| Similarity score: |
0.0412963 |
Alignment:
GGGGCCCAAGGGGG
--------BGGGGA
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
8 |
| Motif name: |
Motif 8 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0426517 |
Alignment:
TCCCAG
TCCCCV
| Original motif Consensus sequence: CTGGGA |
Reverse complement motif Consensus sequence: TCCCAG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0464881 |
Alignment:
GGGGGYGGGG
----BGGGGA
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
6 |
| Similarity score: |
0.0493561 |
Alignment:
CCCCMAAMCAMCCMCMMMCV
----TCCCCV----------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
42 |
| Motif name: |
NFKB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0521759 |
Alignment:
GGGGRTTCCCC
-----TCCCCV
| Original motif Consensus sequence: GGGGRTTCCCC |
Reverse complement motif Consensus sequence: GGGGAAKCCCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
11 |
| Number of overlap: |
6 |
| Similarity score: |
0.0581528 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
-----TCCCCV----------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0585417 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
--------------TCCCCV--
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 24 |
Motif name: Ar |
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
Best Matches for Motif ID 24 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
21 |
| Similarity score: |
0.0696331 |
Alignment:
-GGYGCTGTCCATGGTGCTGAA
VRVDGGHACAVDDKGTHCTDWH
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
20 |
| Similarity score: |
0.564914 |
Alignment:
--MSGKKRCGCWDCABTGBBCD
VRVDGGHACAVDDKGTHCTDWH
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
19 |
| Similarity score: |
1.05195 |
Alignment:
VDBHMAGGTCACCCTGACCY---
-VRVDGGHACAVDDKGTHCTDWH
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
19 |
| Similarity score: |
1.07661 |
Alignment:
CCCCMAAMCAMCCMCMMMCV---
-HWDAGHACRHHVTGTHCCHVMV
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
3.0649 |
Alignment:
-------BTRGGDCARAGGKCA
VRVDGGHACAVDDKGTHCTDWH
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
30 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
14 |
| Similarity score: |
3.57597 |
Alignment:
--------VHRGGTCABDBTGMCCTB
VRVDGGHACAVDDKGTHCTDWH----
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
34 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
4.07506 |
Alignment:
---------TGMYCTTTGBCCK
HWDAGHACRHHVTGTHCCHVMV
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
4.07511 |
Alignment:
---------AAAAAAACAAARRA
HWDAGHACRHHVTGTHCCHVMV-
| Original motif Consensus sequence: TKKTTTGTTTTTTT |
Reverse complement motif Consensus sequence: AAAAAAACAAARRA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
48 |
| Motif name: |
Tcfcp2l1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
4.0791 |
Alignment:
CKGGDTBDMMCTGG---------
-VRVDGGHACAVDDKGTHCTDWH
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
4.57405 |
Alignment:
TGTGTGKVTGTGTG----------
--HWDAGHACRHHVTGTHCCHVMV
| Original motif Consensus sequence: CACACAVRCACACA |
Reverse complement motif Consensus sequence: TGTGTGKVTGTGTG |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 25 |
Motif name: Arnt |
| Original motif Consensus sequence: CACGTG |
Reverse complement motif Consensus sequence: CACGTG |
|
|
 |
 |
Best Matches for Motif ID 25 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
31 |
| Motif name: |
Esrrb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0 |
Alignment:
CACGTG
CACGTG
| Original motif Consensus sequence: CACGTG |
Reverse complement motif Consensus sequence: CACGTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
39 |
| Motif name: |
MYCMAX |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0143849 |
Alignment:
ACCACGTGSTM
--CACGTG---
| Original motif Consensus sequence: RASCACGTGGT |
Reverse complement motif Consensus sequence: ACCACGTGSTM |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
38 |
| Motif name: |
Myc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0267529 |
Alignment:
DCCACGTGCV
--CACGTG--
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
40 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0272641 |
Alignment:
HSCACGTGGC
--CACGTG--
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
33 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0347756 |
Alignment:
VBACGTGV
-CACGTG-
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
26 |
| Motif name: |
ArntAhr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.065625 |
Alignment:
YGCGTG
CACGTG
| Original motif Consensus sequence: YGCGTG |
Reverse complement motif Consensus sequence: CACGCM |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
14 |
| Motif name: |
ZEB1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0689533 |
Alignment:
HAGGTG
CACGTG
| Original motif Consensus sequence: CACCTD |
Reverse complement motif Consensus sequence: HAGGTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
28 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0784722 |
Alignment:
HGCGTGGGCGK
CACGTG-----
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
37 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0854167 |
Alignment:
BAACGTCCGC
-CACGTG---
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
21 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
6 |
| Similarity score: |
0.105093 |
Alignment:
GGGGCCCAAGGGGG
------CACGTG--
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 26 |
Motif name: ArntAhr |
| Original motif Consensus sequence: YGCGTG |
Reverse complement motif Consensus sequence: CACGCM |
|
|
 |
 |
Best Matches for Motif ID 26 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
28 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0 |
Alignment:
YCGCCCACGCH
-----CACGCM
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
33 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0112447 |
Alignment:
VBACGTGV
-YGCGTG-
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
6 |
| Similarity score: |
0.0308396 |
Alignment:
TGTGTGTGTGTKTG
------YGCGTG--
| Original motif Consensus sequence: CAYACACACACACA |
Reverse complement motif Consensus sequence: TGTGTGTGTGTKTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
31 |
| Motif name: |
Esrrb |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0315972 |
Alignment:
CACGTG
YGCGTG
| Original motif Consensus sequence: CACGTG |
Reverse complement motif Consensus sequence: CACGTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
25 |
| Motif name: |
Arnt |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0315972 |
Alignment:
CACGTG
YGCGTG
| Original motif Consensus sequence: CACGTG |
Reverse complement motif Consensus sequence: CACGTG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
6 |
| Similarity score: |
0.0350575 |
Alignment:
TGTGTGKVTGTGTG
YGCGTG--------
| Original motif Consensus sequence: CACACAVRCACACA |
Reverse complement motif Consensus sequence: TGTGTGKVTGTGTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
39 |
| Motif name: |
MYCMAX |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0376488 |
Alignment:
RASCACGTGGT
---YGCGTG--
| Original motif Consensus sequence: RASCACGTGGT |
Reverse complement motif Consensus sequence: ACCACGTGSTM |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
40 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0377949 |
Alignment:
GCCACGTGSD
--YGCGTG--
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
38 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0399214 |
Alignment:
DCCACGTGCV
--YGCGTG--
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
6 |
| Similarity score: |
0.0433081 |
Alignment:
BGRRRGRGGRTGRTTYGGGG
--------YGCGTG------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 27 |
Motif name: E2F1 |
| Original motif Consensus sequence: TTTSGCGC |
Reverse complement motif Consensus sequence: GCGCSAAA |
|
|
 |
 |
Best Matches for Motif ID 27 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
21 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0255644 |
Alignment:
CCCCCTTGGGCCCC
-TTTSGCGC-----
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
8 |
| Similarity score: |
0.0280436 |
Alignment:
TGRCCTKTGHCCKAB
--TTTSGCGC-----
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
34 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0283733 |
Alignment:
RGGBCAAAGKYCA
GCGCSAAA-----
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
43 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0298005 |
Alignment:
AAAGGTCAAAGGTCAAC
--GCGCSAAA-------
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
22 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0369159 |
Alignment:
TGAMCTTTGMMCYT
-----TTTSGCGC-
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
13 |
| Motif name: |
Zfp423 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.0423573 |
Alignment:
GSMMCCYARGGKKKC
------GCGCSAAA-
| Original motif Consensus sequence: GSMMCCYARGGKKKC |
Reverse complement motif Consensus sequence: GYRYCCMTKGGYRSC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
28 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0470922 |
Alignment:
YCGCCCACGCH
GCGCSAAA---
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0486459 |
Alignment:
TKKTTTGTTTTTTT
---TTTSGCGC---
| Original motif Consensus sequence: TKKTTTGTTTTTTT |
Reverse complement motif Consensus sequence: AAAAAAACAAARRA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
13 |
| Number of overlap: |
8 |
| Similarity score: |
0.0497437 |
Alignment:
BGRRRGRGGRTGRTTYGGGG
TTTSGCGC------------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
1 |
| Motif name: |
Motif 1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
1.03392 |
Alignment:
--MTTCCK
TTTSGCGC
| Original motif Consensus sequence: RGGAAR |
Reverse complement motif Consensus sequence: MTTCCK |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 28 |
Motif name: Egr1 |
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
Best Matches for Motif ID 28 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.0550907 |
Alignment:
BGRRRGRGGRTGRTTYGGGG
-------HGCGTGGGCGK--
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.062391 |
Alignment:
CAYACACACACACA
YCGCCCACGCH---
| Original motif Consensus sequence: CAYACACACACACA |
Reverse complement motif Consensus sequence: TGTGTGTGTGTKTG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0668604 |
Alignment:
CACACAVRCACACA
YCGCCCACGCH---
| Original motif Consensus sequence: CACACAVRCACACA |
Reverse complement motif Consensus sequence: TGTGTGKVTGTGTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
13 |
| Motif name: |
Zfp423 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0673496 |
Alignment:
GSMMCCYARGGKKKC
---YCGCCCACGCH-
| Original motif Consensus sequence: GSMMCCYARGGKKKC |
Reverse complement motif Consensus sequence: GYRYCCMTKGGYRSC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.0764878 |
Alignment:
BBVGCCBVGGCCTV
--HGCGTGGGCGK-
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
11 |
| Similarity score: |
0.0765783 |
Alignment:
DGVBCABTGDWGCGKRRCSR
----HGCGTGGGCGK-----
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0830177 |
Alignment:
TGYCAGGGGGCR
HGCGTGGGCGK-
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
34 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.0857144 |
Alignment:
TGMYCTTTGBCCK
--HGCGTGGGCGK
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.572401 |
Alignment:
CCCCKCCCCC-
YCGCCCACGCH
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
36 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.580823 |
Alignment:
DGGGYGKGGC-
HGCGTGGGCGK
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 29 |
Motif name: ESR1 |
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
Best Matches for Motif ID 29 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
20 |
| Similarity score: |
0.0624908 |
Alignment:
MSGKKRCGCWDCABTGBBCD
MGGTCAGGGTGACCTRDBHV
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
19 |
| Similarity score: |
0.556363 |
Alignment:
-HWDAGHACRHHVTGTHCCHVMV
MGGTCAGGGTGACCTRDBHV---
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
19 |
| Similarity score: |
0.575732 |
Alignment:
BGRRRGRGGRTGRTTYGGGG-
-MGGTCAGGGTGACCTRDBHV
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
30 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
17 |
| Similarity score: |
1.50287 |
Alignment:
---VHRGGTCABDBTGMCCTB
VDBHMAGGTCACCCTGACCY-
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
RXRRAR_DR5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
17 |
| Similarity score: |
1.55839 |
Alignment:
---TGACCTCKVVBTGAYCK
VDBHMAGGTCACCCTGACCY
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
16 |
| Similarity score: |
2.07409 |
Alignment:
TTCAGCACCATGGACAGCKCC----
-----VDBHMAGGTCACCCTGACCY
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
17 |
| Motif name: |
RXRAVDR |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
2.56536 |
Alignment:
-----GGGTCAWBGRGTTCA
MGGTCAGGGTGACCTRDBHV
| Original motif Consensus sequence: GGGTCAWBGRGTTCA |
Reverse complement motif Consensus sequence: TGAACKCBWTGACCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
3.07055 |
Alignment:
------TGRCCTKTGHCCKAB
VDBHMAGGTCACCCTGACCY-
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
3.56429 |
Alignment:
VAGGCCBBGGCVBB-------
-MGGTCAGGGTGACCTRDBHV
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
13 |
| Motif name: |
Zfp423 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
13 |
| Similarity score: |
3.56489 |
Alignment:
GSMMCCYARGGKKKC-------
--MGGTCAGGGTGACCTRDBHV
| Original motif Consensus sequence: GSMMCCYARGGKKKC |
Reverse complement motif Consensus sequence: GYRYCCMTKGGYRSC |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 30 |
Motif name: ESR2 |
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
Best Matches for Motif ID 30 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
18 |
| Similarity score: |
0.0488603 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
--BAGGYCABHBTGACCKHV--
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
18 |
| Similarity score: |
0.0851144 |
Alignment:
MSGKKRCGCWDCABTGBBCD
BAGGYCABHBTGACCKHV--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
0.0903249 |
Alignment:
BGRRRGRGGRTGRTTYGGGG
BAGGYCABHBTGACCKHV--
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
18 |
| Similarity score: |
0.0916961 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
-VHRGGTCABDBTGMCCTB--
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
17 |
| Similarity score: |
0.511812 |
Alignment:
VDBHMAGGTCACCCTGACCY-
---VHRGGTCABDBTGMCCTB
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
RXRRAR_DR5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
17 |
| Similarity score: |
0.570972 |
Alignment:
-RGKTCABVVRGAGGTCA
BAGGYCABHBTGACCKHV
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
1.56699 |
Alignment:
BTRGGDCARAGGKCA---
VHRGGTCABDBTGMCCTB
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.07891 |
Alignment:
VAGGCCBBGGCVBB----
BAGGYCABHBTGACCKHV
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
22 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.08921 |
Alignment:
----TGAMCTTTGMMCYT
VHRGGTCABDBTGMCCTB
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
48 |
| Motif name: |
Tcfcp2l1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
2.09109 |
Alignment:
----CCAGYYHVADCCRG
BAGGYCABHBTGACCKHV
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 31 |
Motif name: Esrrb |
| Original motif Consensus sequence: CACGTG |
Reverse complement motif Consensus sequence: CACGTG |
|
|
 |
 |
Best Matches for Motif ID 31 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
25 |
| Motif name: |
Arnt |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0 |
Alignment:
CACGTG
CACGTG
| Original motif Consensus sequence: CACGTG |
Reverse complement motif Consensus sequence: CACGTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
39 |
| Motif name: |
MYCMAX |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0143849 |
Alignment:
ACCACGTGSTM
--CACGTG---
| Original motif Consensus sequence: RASCACGTGGT |
Reverse complement motif Consensus sequence: ACCACGTGSTM |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
38 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0267529 |
Alignment:
VGCACGTGGH
--CACGTG--
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
40 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.0272641 |
Alignment:
GCCACGTGSD
--CACGTG--
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
33 |
| Motif name: |
HIF1AARNT |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
6 |
| Similarity score: |
0.0347756 |
Alignment:
VCACGTBV
-CACGTG-
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
26 |
| Motif name: |
ArntAhr |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.065625 |
Alignment:
YGCGTG
CACGTG
| Original motif Consensus sequence: YGCGTG |
Reverse complement motif Consensus sequence: CACGCM |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
14 |
| Motif name: |
ZEB1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0689533 |
Alignment:
HAGGTG
CACGTG
| Original motif Consensus sequence: CACCTD |
Reverse complement motif Consensus sequence: HAGGTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
28 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
6 |
| Similarity score: |
0.0784722 |
Alignment:
HGCGTGGGCGK
CACGTG-----
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
37 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
6 |
| Similarity score: |
0.0854167 |
Alignment:
BAACGTCCGC
-CACGTG---
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
21 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
6 |
| Similarity score: |
0.105093 |
Alignment:
GGGGCCCAAGGGGG
------CACGTG--
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 32 |
Motif name: EWSR1-FLI1 |
| Original motif Consensus sequence: GGAAGGAAGGAAGGAAGG |
Reverse complement motif Consensus sequence: CCTTCCTTCCTTCCTTCC |
|
|
 |
 |
Best Matches for Motif ID 32 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
43 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
12 |
| Similarity score: |
1.09472 |
Alignment:
------AAAGGTCAAAGGTCAAC
GGAAGGAAGGAAGGAAGG-----
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
1.1007 |
Alignment:
------TKKTTTGTTTTTTT
CCTTCCTTCCTTCCTTCC--
| Original motif Consensus sequence: TKKTTTGTTTTTTT |
Reverse complement motif Consensus sequence: AAAAAAACAAARRA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
11 |
| Number of overlap: |
10 |
| Similarity score: |
2.09833 |
Alignment:
--------CCCCMAAMCAMCCMCMMMCV
CCTTCCTTCCTTCCTTCC----------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
2.58264 |
Alignment:
GGGGGYGGGG---------
-GGAAGGAAGGAAGGAAGG
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
37 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
3.0896 |
Alignment:
----------BAACGTCCGC
CCTTCCTTCCTTCCTTCC--
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
3.09107 |
Alignment:
----------CCCRCCCC
CCTTCCTTCCTTCCTTCC
| Original motif Consensus sequence: CCCRCCCC |
Reverse complement motif Consensus sequence: GGGGMGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
13 |
| Number of overlap: |
8 |
| Similarity score: |
3.09836 |
Alignment:
MGGTCAGGGTGACCTRDBHV----------
------------GGAAGGAAGGAAGGAAGG
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
36 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
3.10206 |
Alignment:
----------DGGGYGKGGC
GGAAGGAAGGAAGGAAGG--
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
3 |
| Motif name: |
Motif 3 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
7 |
| Similarity score: |
3.55161 |
Alignment:
-----------AGSCWGG
GGAAGGAAGGAAGGAAGG
| Original motif Consensus sequence: AGSCWGG |
Reverse complement motif Consensus sequence: CCWGSCT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
46 |
| Motif name: |
RORA_1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
7 |
| Similarity score: |
3.58428 |
Alignment:
TGACCTDVWT-----------
---CCTTCCTTCCTTCCTTCC
| Original motif Consensus sequence: AWVDAGGTCA |
Reverse complement motif Consensus sequence: TGACCTDVWT |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 33 |
Motif name: HIF1AARNT |
| Original motif Consensus sequence: VBACGTGV |
Reverse complement motif Consensus sequence: VCACGTBV |
|
|
 |
 |
Best Matches for Motif ID 33 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
40 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0228069 |
Alignment:
GCCACGTGSD
-VBACGTGV-
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
38 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0249459 |
Alignment:
DCCACGTGCV
-VBACGTGV-
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
39 |
| Motif name: |
MYCMAX |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0254922 |
Alignment:
ACCACGTGSTM
-VCACGTBV--
| Original motif Consensus sequence: RASCACGTGGT |
Reverse complement motif Consensus sequence: ACCACGTGSTM |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
37 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0482171 |
Alignment:
GCGGACGTTV
--VCACGTBV
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0533654 |
Alignment:
VRVDGGHACAVDDKGTHCTDWH
----VBACGTGV----------
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
8 |
| Similarity score: |
0.0661859 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-----VCACGTBV-------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
8 |
| Similarity score: |
0.0666776 |
Alignment:
CCCCMAAMCAMCCMCMMMCV
-----VCACGTBV-------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
41 |
| Motif name: |
MZF1_5-13 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
0.0703926 |
Alignment:
BKAGGGGDAD
VBACGTGV--
| Original motif Consensus sequence: BKAGGGGDAD |
Reverse complement motif Consensus sequence: BTHCCCCTYB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
30 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
8 |
| Similarity score: |
0.0716055 |
Alignment:
BAGGYCABHBTGACCKHV
-----VBACGTGV-----
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
RXRRAR_DR5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0726789 |
Alignment:
TGACCTCKVVBTGAYCK
------VBACGTGV---
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 34 |
Motif name: HNF4A |
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
Best Matches for Motif ID 34 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
13 |
| Similarity score: |
0 |
Alignment:
BTRGGDCARAGGKCA
--RGGBCAAAGKYCA
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
22 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
0.0227413 |
Alignment:
TGAMCTTTGMMCYT
-TGMYCTTTGBCCK
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
43 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
13 |
| Similarity score: |
0.0423023 |
Alignment:
AAAGGTCAAAGGTCAAC
--RGGBCAAAGKYCA--
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
RXRRAR_DR5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.048852 |
Alignment:
TGACCTCKVVBTGAYCK
TGMYCTTTGBCCK----
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
13 |
| Similarity score: |
0.063924 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-------RGGBCAAAGKYCA
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
30 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
13 |
| Similarity score: |
0.0671615 |
Alignment:
VHRGGTCABDBTGMCCTB
--RGGBCAAAGKYCA---
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
13 |
| Similarity score: |
0.067775 |
Alignment:
VRVDGGHACAVDDKGTHCTDWH
------TGMYCTTTGBCCK---
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.0711137 |
Alignment:
VDBHMAGGTCACCCTGACCY
TGMYCTTTGBCCK-------
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
21 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
0.0714266 |
Alignment:
GGGGCCCAAGGGGG
-RGGBCAAAGKYCA
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
13 |
| Similarity score: |
0.0741922 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
-----TGMYCTTTGBCCK---
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 35 |
Motif name: INSM1 |
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
Best Matches for Motif ID 35 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
12 |
| Similarity score: |
0.0472454 |
Alignment:
BTRGGDCARAGGKCA
---TGYCAGGGGGCR
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0499162 |
Alignment:
BGRRRGRGGRTGRTTYGGGG
--------TGYCAGGGGGCR
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
13 |
| Motif name: |
Zfp423 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0538619 |
Alignment:
GYRYCCMTKGGYRSC
-MGCCCCCTGMCA--
| Original motif Consensus sequence: GSMMCCYARGGKKKC |
Reverse complement motif Consensus sequence: GYRYCCMTKGGYRSC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
34 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0551858 |
Alignment:
RGGBCAAAGKYCA
-TGYCAGGGGGCR
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.0630846 |
Alignment:
VDBHMAGGTCACCCTGACCY
-TGYCAGGGGGCR-------
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
6 |
| Number of overlap: |
12 |
| Similarity score: |
0.0679087 |
Alignment:
DGVBCABTGDWGCGKRRCSR
-----TGYCAGGGGGCR---
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
12 |
| Similarity score: |
0.069037 |
Alignment:
VAGGCCBBGGCVBB
-MGCCCCCTGMCA-
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0692502 |
Alignment:
CAYACACACACACA
--MGCCCCCTGMCA
| Original motif Consensus sequence: CAYACACACACACA |
Reverse complement motif Consensus sequence: TGTGTGTGTGTKTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
43 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
12 |
| Similarity score: |
0.0696795 |
Alignment:
AAAGGTCAAAGGTCAAC
---TGYCAGGGGGCR--
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
22 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
12 |
| Similarity score: |
0.0705128 |
Alignment:
TGAMCTTTGMMCYT
--MGCCCCCTGMCA
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 36 |
Motif name: Klf4 |
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
Best Matches for Motif ID 36 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.065355 |
Alignment:
BGRRRGRGGRTGRTTYGGGG
----------DGGGYGKGGC
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0734166 |
Alignment:
BBVGCCBVGGCCTV
-GCCYCMCCCD---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0773711 |
Alignment:
GGGGGYGGGG
DGGGYGKGGC
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
11 |
| Motif name: |
Motif 11 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0781179 |
Alignment:
CAYACACACACACA
--GCCYCMCCCD--
| Original motif Consensus sequence: CAYACACACACACA |
Reverse complement motif Consensus sequence: TGTGTGTGTGTKTG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0785684 |
Alignment:
MGCCCCCTGMCA
--GCCYCMCCCD
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
38 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0797423 |
Alignment:
VGCACGTGGH
DGGGYGKGGC
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
10 |
| Similarity score: |
0.0806175 |
Alignment:
DGVBCABTGDWGCGKRRCSR
--------DGGGYGKGGC--
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
40 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.080695 |
Alignment:
HSCACGTGGC
DGGGYGKGGC
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
48 |
| Motif name: |
Tcfcp2l1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0807235 |
Alignment:
CCAGYYHVADCCRG
---GCCYCMCCCD-
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
28 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.089515 |
Alignment:
YCGCCCACGCH
-GCCYCMCCCD
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 37 |
Motif name: MIZF |
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
Best Matches for Motif ID 37 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
39 |
| Motif name: |
MYCMAX |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0390724 |
Alignment:
ACCACGTGSTM
-BAACGTCCGC
| Original motif Consensus sequence: RASCACGTGGT |
Reverse complement motif Consensus sequence: ACCACGTGSTM |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
30 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0399477 |
Alignment:
BAGGYCABHBTGACCKHV
--------GCGGACGTTV
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
10 |
| Similarity score: |
0.0408267 |
Alignment:
VDBHMAGGTCACCCTGACCY
---BAACGTCCGC-------
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0432688 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
---------BAACGTCCGC---
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
43 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0439771 |
Alignment:
AAAGGTCAAAGGTCAAC
-------BAACGTCCGC
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0501848 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
---------BAACGTCCGC--
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0576438 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-GCGGACGTTV---------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
38 |
| Motif name: |
Myc |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.535015 |
Alignment:
DCCACGTGCV-
-BAACGTCCGC
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
28 |
| Motif name: |
Egr1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
9 |
| Similarity score: |
0.537968 |
Alignment:
-HGCGTGGGCGK
BAACGTCCGC--
| Original motif Consensus sequence: HGCGTGGGCGK |
Reverse complement motif Consensus sequence: YCGCCCACGCH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
40 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
0.540549 |
Alignment:
GCCACGTGSD-
-BAACGTCCGC
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 38 |
Motif name: Myc |
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
Best Matches for Motif ID 38 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
40 |
| Motif name: |
Mycn |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0 |
Alignment:
HSCACGTGGC
VGCACGTGGH
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
39 |
| Motif name: |
MYCMAX |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0209359 |
Alignment:
RASCACGTGGT
-VGCACGTGGH
| Original motif Consensus sequence: RASCACGTGGT |
Reverse complement motif Consensus sequence: ACCACGTGSTM |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0844808 |
Alignment:
MGCCCCCTGMCA
-DCCACGTGCV-
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
36 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0883745 |
Alignment:
DGGGYGKGGC
VGCACGTGGH
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0925451 |
Alignment:
VAGGCCBBGGCVBB
---DCCACGTGCV-
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
21 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0930375 |
Alignment:
CCCCCTTGGGCCCC
----VGCACGTGGH
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
13 |
| Motif name: |
Zfp423 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0930519 |
Alignment:
GYRYCCMTKGGYRSC
---DCCACGTGCV--
| Original motif Consensus sequence: GSMMCCYARGGKKKC |
Reverse complement motif Consensus sequence: GYRYCCMTKGGYRSC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
34 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0934488 |
Alignment:
TGMYCTTTGBCCK
---VGCACGTGGH
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.0940438 |
Alignment:
MGGTCAGGGTGACCTRDBHV
--------DCCACGTGCV--
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
10 |
| Similarity score: |
0.0966137 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
------DCCACGTGCV------
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 39 |
Motif name: MYCMAX |
| Original motif Consensus sequence: RASCACGTGGT |
Reverse complement motif Consensus sequence: ACCACGTGSTM |
|
|
 |
 |
Best Matches for Motif ID 39 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
11 |
| Similarity score: |
0.0862644 |
Alignment:
CCCCMAAMCAMCCMCMMMCV
---ACCACGTGSTM------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
10 |
| Number of overlap: |
11 |
| Similarity score: |
0.0877525 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
--RASCACGTGGT---------
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
13 |
| Motif name: |
Zfp423 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.0933483 |
Alignment:
GSMMCCYARGGKKKC
-ACCACGTGSTM---
| Original motif Consensus sequence: GSMMCCYARGGKKKC |
Reverse complement motif Consensus sequence: GYRYCCMTKGGYRSC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
21 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.10358 |
Alignment:
CCCCCTTGGGCCCC
ACCACGTGSTM---
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.104527 |
Alignment:
TGYCAGGGGGCR
RASCACGTGGT-
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.105174 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
--------ACCACGTGSTM--
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
30 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.105681 |
Alignment:
VHRGGTCABDBTGMCCTB
---RASCACGTGGT----
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
11 |
| Similarity score: |
0.107782 |
Alignment:
MGGTCAGGGTGACCTRDBHV
--------ACCACGTGSTM-
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
RXRRAR_DR5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
3 |
| Number of overlap: |
11 |
| Similarity score: |
0.108615 |
Alignment:
TGACCTCKVVBTGAYCK
--ACCACGTGSTM----
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.110189 |
Alignment:
BBVGCCBVGGCCTV
RASCACGTGGT---
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 40 |
Motif name: Mycn |
| Original motif Consensus sequence: HSCACGTGGC |
Reverse complement motif Consensus sequence: GCCACGTGSD |
|
|
 |
 |
Best Matches for Motif ID 40 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
38 |
| Motif name: |
Myc |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0 |
Alignment:
DCCACGTGCV
GCCACGTGSD
| Original motif Consensus sequence: VGCACGTGGH |
Reverse complement motif Consensus sequence: DCCACGTGCV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
39 |
| Motif name: |
MYCMAX |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0200467 |
Alignment:
RASCACGTGGT
-HSCACGTGGC
| Original motif Consensus sequence: RASCACGTGGT |
Reverse complement motif Consensus sequence: ACCACGTGSTM |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0834824 |
Alignment:
MGCCCCCTGMCA
-GCCACGTGSD-
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
36 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0893272 |
Alignment:
DGGGYGKGGC
HSCACGTGGC
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.089863 |
Alignment:
MGGTCAGGGTGACCTRDBHV
--------GCCACGTGSD--
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0899944 |
Alignment:
VAGGCCBBGGCVBB
---GCCACGTGSD-
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
21 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0921439 |
Alignment:
CCCCCTTGGGCCCC
----HSCACGTGGC
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
34 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0947124 |
Alignment:
TGMYCTTTGBCCK
-GCCACGTGSD--
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0954444 |
Alignment:
TGRCCTKTGHCCKAB
-GCCACGTGSD----
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
30 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0964708 |
Alignment:
VHRGGTCABDBTGMCCTB
----HSCACGTGGC----
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 41 |
Motif name: MZF1_5-13 |
| Original motif Consensus sequence: BKAGGGGDAD |
Reverse complement motif Consensus sequence: BTHCCCCTYB |
|
|
 |
 |
Best Matches for Motif ID 41 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
35 |
| Motif name: |
INSM1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0360532 |
Alignment:
TGYCAGGGGGCR
--BKAGGGGDAD
| Original motif Consensus sequence: TGYCAGGGGGCR |
Reverse complement motif Consensus sequence: MGCCCCCTGMCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0428241 |
Alignment:
DGVBCABTGDWGCGKRRCSR
-------BKAGGGGDAD---
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0470779 |
Alignment:
BTRGGDCARAGGKCA
BKAGGGGDAD-----
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
13 |
| Motif name: |
Zfp423 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0524832 |
Alignment:
GSMMCCYARGGKKKC
-----BKAGGGGDAD
| Original motif Consensus sequence: GSMMCCYARGGKKKC |
Reverse complement motif Consensus sequence: GYRYCCMTKGGYRSC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0548506 |
Alignment:
CCCCMAAMCAMCCMCMMMCV
-BTHCCCCTYB---------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
34 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0568244 |
Alignment:
TGMYCTTTGBCCK
BTHCCCCTYB---
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
RXRRAR_DR5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0581547 |
Alignment:
TGACCTCKVVBTGAYCK
BTHCCCCTYB-------
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
13 |
| Number of overlap: |
10 |
| Similarity score: |
0.0610532 |
Alignment:
VRVDGGHACAVDDKGTHCTDWH
------------BKAGGGGDAD
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
10 |
| Similarity score: |
0.0642057 |
Alignment:
VDBHMAGGTCACCCTGACCY
--BKAGGGGDAD--------
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0692824 |
Alignment:
CCCCKCCCCC
BTHCCCCTYB
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 42 |
Motif name: NFKB1 |
| Original motif Consensus sequence: GGGGRTTCCCC |
Reverse complement motif Consensus sequence: GGGGAAKCCCC |
|
|
 |
 |
Best Matches for Motif ID 42 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
48 |
| Motif name: |
Tcfcp2l1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
11 |
| Similarity score: |
0.103405 |
Alignment:
CKGGDTBDMMCTGG
GGGGRTTCCCC---
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
11 |
| Similarity score: |
0.112926 |
Alignment:
TTCAGCACCATGGACAGCKCC
GGGGAAKCCCC----------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
9 |
| Number of overlap: |
11 |
| Similarity score: |
0.116162 |
Alignment:
MSGKKRCGCWDCABTGBBCD
-GGGGAAKCCCC--------
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
11 |
| Similarity score: |
0.117826 |
Alignment:
CCCCMAAMCAMCCMCMMMCV
-----GGGGAAKCCCC----
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
36 |
| Motif name: |
Klf4 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.616368 |
Alignment:
GCCYCMCCCD-
GGGGRTTCCCC
| Original motif Consensus sequence: DGGGYGKGGC |
Reverse complement motif Consensus sequence: GCCYCMCCCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
21 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
6 |
| Number of overlap: |
9 |
| Similarity score: |
1.11497 |
Alignment:
--GGGGCCCAAGGGGG
GGGGRTTCCCC-----
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
16 |
| Motif name: |
SP1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
9 |
| Similarity score: |
1.11585 |
Alignment:
--GGGGGYGGGG
GGGGRTTCCCC-
| Original motif Consensus sequence: CCCCKCCCCC |
Reverse complement motif Consensus sequence: GGGGGYGGGG |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
7 |
| Motif name: |
Motif 7 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
1.59629 |
Alignment:
GGGGMGGG---
GGGGAAKCCCC
| Original motif Consensus sequence: CCCRCCCC |
Reverse complement motif Consensus sequence: GGGGMGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
13 |
| Motif name: |
Zfp423 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
8 |
| Similarity score: |
1.59848 |
Alignment:
GSMMCCYARGGKKKC---
-------GGGGRTTCCCC
| Original motif Consensus sequence: GSMMCCYARGGKKKC |
Reverse complement motif Consensus sequence: GYRYCCMTKGGYRSC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
41 |
| Motif name: |
MZF1_5-13 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
8 |
| Similarity score: |
1.61306 |
Alignment:
---BKAGGGGDAD
GGGGAAKCCCC--
| Original motif Consensus sequence: BKAGGGGDAD |
Reverse complement motif Consensus sequence: BTHCCCCTYB |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 43 |
Motif name: NR1H2RXRA |
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
Best Matches for Motif ID 43 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
17 |
| Similarity score: |
0.0911991 |
Alignment:
GGYGCTGTCCATGGTGCTGAA
---GTTGACCTTTGACCTTT-
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
1.05945 |
Alignment:
BTRGGDCARAGGKCA--
AAAGGTCAAAGGTCAAC
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
17 |
| Motif name: |
RXRAVDR |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
1.09792 |
Alignment:
--GGGTCAWBGRGTTCA
AAAGGTCAAAGGTCAAC
| Original motif Consensus sequence: GGGTCAWBGRGTTCA |
Reverse complement motif Consensus sequence: TGAACKCBWTGACCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
13 |
| Similarity score: |
2.08514 |
Alignment:
----VDBHMAGGTCACCCTGACCY
AAAGGTCAAAGGTCAAC-------
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
10 |
| Motif name: |
Motif 10 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
13 |
| Similarity score: |
2.08845 |
Alignment:
TKKTTTGTTTTTTT----
-GTTGACCTTTGACCTTT
| Original motif Consensus sequence: TKKTTTGTTTTTTT |
Reverse complement motif Consensus sequence: AAAAAAACAAARRA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
32 |
| Motif name: |
EWSR1-FLI1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
12 |
| Similarity score: |
2.59335 |
Alignment:
GGAAGGAAGGAAGGAAGG-----
------AAAGGTCAAAGGTCAAC
| Original motif Consensus sequence: GGAAGGAAGGAAGGAAGG |
Reverse complement motif Consensus sequence: CCTTCCTTCCTTCCTTCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
12 |
| Number of overlap: |
11 |
| Similarity score: |
3.09595 |
Alignment:
VRVDGGHACAVDDKGTHCTDWH------
-----------GTTGACCTTTGACCTTT
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
30 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
9 |
| Number of overlap: |
10 |
| Similarity score: |
3.56221 |
Alignment:
BAGGYCABHBTGACCKHV-------
--------GTTGACCTTTGACCTTT
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
37 |
| Motif name: |
MIZF |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
3.5855 |
Alignment:
-------BAACGTCCGC
AAAGGTCAAAGGTCAAC
| Original motif Consensus sequence: BAACGTCCGC |
Reverse complement motif Consensus sequence: GCGGACGTTV |
|
|
 |
 |
| Dataset #: |
1 |
| Motif ID: |
9 |
| Motif name: |
Motif 9 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
3.59839 |
Alignment:
CACACAVRCACACA-------
----AAAGGTCAAAGGTCAAC
| Original motif Consensus sequence: CACACAVRCACACA |
Reverse complement motif Consensus sequence: TGTGTGKVTGTGTG |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 44 |
Motif name: NR4A2 |
| Original motif Consensus sequence: AAGGTCAC |
Reverse complement motif Consensus sequence: GTGACCTT |
|
|
 |
 |
Best Matches for Motif ID 44 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
43 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0167759 |
Alignment:
AAAGGTCAAAGGTCAAC
-AAGGTCAC--------
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0195235 |
Alignment:
VDBHMAGGTCACCCTGACCY
----AAGGTCAC--------
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
30 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0198372 |
Alignment:
VHRGGTCABDBTGMCCTB
-AAGGTCAC---------
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
13 |
| Number of overlap: |
8 |
| Similarity score: |
0.0487608 |
Alignment:
VRVDGGHACAVDDKGTHCTDWH
------------GTGACCTT--
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.0495607 |
Alignment:
BTRGGDCARAGGKCA
-AAGGTCAC------
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
22 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
8 |
| Similarity score: |
0.0564874 |
Alignment:
AKGYYCAAAGRTCA
------AAGGTCAC
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
5 |
| Number of overlap: |
8 |
| Similarity score: |
0.0594649 |
Alignment:
TTCAGCACCATGGACAGCKCC
---------AAGGTCAC----
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
8 |
| Similarity score: |
0.0631514 |
Alignment:
BBVGCCBVGGCCTV
GTGACCTT------
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
8 |
| Similarity score: |
0.0720656 |
Alignment:
BGRRRGRGGRTGRTTYGGGG
---------GTGACCTT---
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
48 |
| Motif name: |
Tcfcp2l1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
8 |
| Similarity score: |
0.0740998 |
Alignment:
CKGGDTBDMMCTGG
-----GTGACCTT-
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 45 |
Motif name: Pax5 |
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
Best Matches for Motif ID 45 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
20 |
| Similarity score: |
0.0382347 |
Alignment:
MGGTCAGGGTGACCTRDBHV
MSGKKRCGCWDCABTGBBCD
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
20 |
| Similarity score: |
0.0450685 |
Alignment:
VRVDGGHACAVDDKGTHCTDWH
--MSGKKRCGCWDCABTGBBCD
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
30 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
18 |
| Similarity score: |
1.05192 |
Alignment:
BAGGYCABHBTGACCKHV--
MSGKKRCGCWDCABTGBBCD
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
4 |
| Number of overlap: |
18 |
| Similarity score: |
1.0582 |
Alignment:
--GGYGCTGTCCATGGTGCTGAA
MSGKKRCGCWDCABTGBBCD---
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
RXRRAR_DR5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
17 |
| Similarity score: |
1.53818 |
Alignment:
---TGACCTCKVVBTGAYCK
MSGKKRCGCWDCABTGBBCD
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
17 |
| Similarity score: |
1.55831 |
Alignment:
CCCCMAAMCAMCCMCMMMCV---
---MSGKKRCGCWDCABTGBBCD
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
2.55126 |
Alignment:
-----TGRCCTKTGHCCKAB
DGVBCABTGDWGCGKRRCSR
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
21 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.05866 |
Alignment:
GGGGCCCAAGGGGG------
DGVBCABTGDWGCGKRRCSR
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
22 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
3.05897 |
Alignment:
------AKGYYCAAAGRTCA
MSGKKRCGCWDCABTGBBCD
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
34 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
3.53748 |
Alignment:
-------TGMYCTTTGBCCK
DGVBCABTGDWGCGKRRCSR
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 46 |
Motif name: RORA_1 |
| Original motif Consensus sequence: AWVDAGGTCA |
Reverse complement motif Consensus sequence: TGACCTDVWT |
|
|
 |
 |
Best Matches for Motif ID 46 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
10 |
| Similarity score: |
0.0417851 |
Alignment:
MGGTCAGGGTGACCTRDBHV
---------TGACCTDVWT-
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
17 |
| Motif name: |
RXRAVDR |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.05325 |
Alignment:
GGGTCAWBGRGTTCA
-----AWVDAGGTCA
| Original motif Consensus sequence: GGGTCAWBGRGTTCA |
Reverse complement motif Consensus sequence: TGAACKCBWTGACCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
43 |
| Motif name: |
NR1H2RXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
3 |
| Number of overlap: |
10 |
| Similarity score: |
0.05425 |
Alignment:
AAAGGTCAAAGGTCAAC
-----AWVDAGGTCA--
| Original motif Consensus sequence: AAAGGTCAAAGGTCAAC |
Reverse complement motif Consensus sequence: GTTGACCTTTGACCTTT |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
RXRRAR_DR5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.0600761 |
Alignment:
TGACCTCKVVBTGAYCK
TGACCTDVWT-------
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
22 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
10 |
| Similarity score: |
0.0654808 |
Alignment:
TGAMCTTTGMMCYT
----TGACCTDVWT
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
10 |
| Similarity score: |
0.06668 |
Alignment:
BTRGGDCARAGGKCA
-----AWVDAGGTCA
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
10 |
| Similarity score: |
0.0755608 |
Alignment:
TTCAGCACCATGGACAGCKCC
------AWVDAGGTCA-----
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
34 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
10 |
| Similarity score: |
0.0803396 |
Alignment:
RGGBCAAAGKYCA
---AWVDAGGTCA
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
13 |
| Number of overlap: |
10 |
| Similarity score: |
0.0814167 |
Alignment:
VRVDGGHACAVDDKGTHCTDWH
------------AWVDAGGTCA
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
10 |
| Similarity score: |
0.0901591 |
Alignment:
CCCCMAAMCAMCCMCMMMCV
---TGACCTDVWT-------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 47 |
Motif name: RXRRAR_DR5 |
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
 |
 |
Best Matches for Motif ID 47 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
17 |
| Similarity score: |
0.0554256 |
Alignment:
MGGTCAGGGTGACCTRDBHV
---RGKTCABVVRGAGGTCA
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
30 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
2 |
| Number of overlap: |
17 |
| Similarity score: |
0.0590677 |
Alignment:
VHRGGTCABDBTGMCCTB
-TGACCTCKVVBTGAYCK
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
17 |
| Similarity score: |
0.0594693 |
Alignment:
MSGKKRCGCWDCABTGBBCD
---TGACCTCKVVBTGAYCK
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
17 |
| Similarity score: |
0.0751876 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
----TGACCTCKVVBTGAYCK-
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
1.0449 |
Alignment:
--BTRGGDCARAGGKCA
RGKTCABVVRGAGGTCA
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
17 |
| Motif name: |
RXRAVDR |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
15 |
| Similarity score: |
1.05297 |
Alignment:
GGGTCAWBGRGTTCA--
RGKTCABVVRGAGGTCA
| Original motif Consensus sequence: GGGTCAWBGRGTTCA |
Reverse complement motif Consensus sequence: TGAACKCBWTGACCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
22 |
| Motif name: |
NR2F1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
1.56607 |
Alignment:
---TGAMCTTTGMMCYT
TGACCTCKVVBTGAYCK
| Original motif Consensus sequence: TGAMCTTTGMMCYT |
Reverse complement motif Consensus sequence: AKGYYCAAAGRTCA |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
21 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
1.57917 |
Alignment:
GGGGCCCAAGGGGG---
RGKTCABVVRGAGGTCA
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
34 |
| Motif name: |
HNF4A |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
13 |
| Similarity score: |
2.0437 |
Alignment:
TGMYCTTTGBCCK----
TGACCTCKVVBTGAYCK
| Original motif Consensus sequence: RGGBCAAAGKYCA |
Reverse complement motif Consensus sequence: TGMYCTTTGBCCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
8 |
| Number of overlap: |
13 |
| Similarity score: |
2.06494 |
Alignment:
----CCCCMAAMCAMCCMCMMMCV
TGACCTCKVVBTGAYCK-------
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: 2 |
Motif ID: 48 |
Motif name: Tcfcp2l1 |
| Original motif Consensus sequence: CCAGYYHVADCCRG |
Reverse complement motif Consensus sequence: CKGGDTBDMMCTGG |
|
|
 |
 |
Best Matches for Motif ID 48 (Highest to Lowest)
| Dataset #: |
2 |
| Motif ID: |
24 |
| Motif name: |
Ar |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
7 |
| Number of overlap: |
14 |
| Similarity score: |
0.0679942 |
Alignment:
HWDAGHACRHHVTGTHCCHVMV
------CCAGYYHVADCCRG--
| Original motif Consensus sequence: HWDAGHACRHHVTGTHCCHVMV |
Reverse complement motif Consensus sequence: VRVDGGHACAVDDKGTHCTDWH |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
29 |
| Motif name: |
ESR1 |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Forward |
| Position number: |
4 |
| Number of overlap: |
14 |
| Similarity score: |
0.0713928 |
Alignment:
MGGTCAGGGTGACCTRDBHV
---CCAGYYHVADCCRG---
| Original motif Consensus sequence: VDBHMAGGTCACCCTGACCY |
Reverse complement motif Consensus sequence: MGGTCAGGGTGACCTRDBHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
12 |
| Motif name: |
Zfx |
| Matching format of first motif: |
Original Motif |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0714852 |
Alignment:
BBVGCCBVGGCCTV
CCAGYYHVADCCRG
| Original motif Consensus sequence: BBVGCCBVGGCCTV |
Reverse complement motif Consensus sequence: VAGGCCBBGGCVBB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
18 |
| Motif name: |
RREB1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
2 |
| Number of overlap: |
14 |
| Similarity score: |
0.0777364 |
Alignment:
BGRRRGRGGRTGRTTYGGGG
-----CKGGDTBDMMCTGG-
| Original motif Consensus sequence: CCCCMAAMCAMCCMCMMMCV |
Reverse complement motif Consensus sequence: BGRRRGRGGRTGRTTYGGGG |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
21 |
| Motif name: |
PLAG1 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0817193 |
Alignment:
GGGGCCCAAGGGGG
CKGGDTBDMMCTGG
| Original motif Consensus sequence: GGGGCCCAAGGGGG |
Reverse complement motif Consensus sequence: CCCCCTTGGGCCCC |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
30 |
| Motif name: |
ESR2 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0828703 |
Alignment:
VHRGGTCABDBTGMCCTB
----CKGGDTBDMMCTGG
| Original motif Consensus sequence: VHRGGTCABDBTGMCCTB |
Reverse complement motif Consensus sequence: BAGGYCABHBTGACCKHV |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
20 |
| Motif name: |
PPARGRXRA |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Reverse Complement |
| Direction: |
Backward |
| Position number: |
1 |
| Number of overlap: |
14 |
| Similarity score: |
0.0829929 |
Alignment:
TGRCCTKTGHCCKAB
-CKGGDTBDMMCTGG
| Original motif Consensus sequence: BTRGGDCARAGGKCA |
Reverse complement motif Consensus sequence: TGRCCTKTGHCCKAB |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
45 |
| Motif name: |
Pax5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
8 |
| Number of overlap: |
13 |
| Similarity score: |
0.575635 |
Alignment:
DGVBCABTGDWGCGKRRCSR-
-------CKGGDTBDMMCTGG
| Original motif Consensus sequence: DGVBCABTGDWGCGKRRCSR |
Reverse complement motif Consensus sequence: MSGKKRCGCWDCABTGBBCD |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
47 |
| Motif name: |
RXRRAR_DR5 |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Forward |
| Position number: |
5 |
| Number of overlap: |
13 |
| Similarity score: |
0.578477 |
Alignment:
RGKTCABVVRGAGGTCA-
----CKGGDTBDMMCTGG
| Original motif Consensus sequence: RGKTCABVVRGAGGTCA |
Reverse complement motif Consensus sequence: TGACCTCKVVBTGAYCK |
|
|
 |
 |
| Dataset #: |
2 |
| Motif ID: |
19 |
| Motif name: |
REST |
| Matching format of first motif: |
Reverse Complement |
| Matching format of second motif: |
Original Motif |
| Direction: |
Backward |
| Position number: |
10 |
| Number of overlap: |
12 |
| Similarity score: |
1.07716 |
Alignment:
--TTCAGCACCATGGACAGCKCC
CKGGDTBDMMCTGG---------
| Original motif Consensus sequence: TTCAGCACCATGGACAGCKCC |
Reverse complement motif Consensus sequence: GGYGCTGTCCATGGTGCTGAA |
|
|
 |
 |





























































































































































































































